Abstract
The dried blood spot (DBS) is increasingly used for the hepatitis C virus (HCV) screening. Our objective was to perform a meta-analysis of the methodology for HCV screening in DBS samples, particularly in the type of diagnostic assay used. We performed a meta-analysis of all eligible studies published to date (March 2018). The literature search revealed 26 studies: 21 for detection of anti-HCV antibodies and 10 for detection of HCV-RNA. Statistical analyses were performed using Meta-DiSc and STATA (MIDAS module). For detection of HCV antibodies, pooled diagnostic accuracy measures were as follows: sensitivity 96.1%, specificity 99.2%, positive likelihood ratio (PLR) 105, negative likelihood ratio (NLR) 0.04, diagnostic odds ratio (DOR) 2692.9, and summary receiver operating characteristic (SROC) 0.997 ± 0.001. For detection of HCV-RNA, the pooled diagnostic accuracy measures were as follows: sensitivity 97.8%, specificity 99.2%, PLR 44.8, NLR 0.04, DOR 1966.9, and SROC 0.996 ± 0.013. Similar values of pooled diagnostic accuracy measures were found according to the type of anti-HCV antibody detection assay (enzyme-linked immunosorbent assay, rapid diagnostic test, and chemiluminescence assays) and HCV-RNA detection assay (real-time polymerase chain reaction and transcription-mediated amplification). The analysis of external validity showed a high negative predicted value (NPV) for both approaches, but a low positive predicted value (PPV) when prevalence was < 10%, particularly in HCV-RNA tests. Finally, this meta-analysis is subject to limitations, especially publication bias and significant heterogeneity between studies. In conclusion, HCV screening in DBS samples has an outstanding diagnostic performance, with no relevant differences between the techniques used. However, external validity may be limited when the HCV prevalence is low.
Subject terms: Hepatitis, Viral infection
Introduction
About 71 million people have chronic Hepatitis C virus (HCV) infection and around 80% are undiagnosed1, thus leading to the development of liver disease and/or transmission of HCV infection to others unknowingly2–4. Furthermore, HCV diagnosis remains problematic for persons from low- and middle -income countries (LMICs) and difficult-to-access populations in developed countries (people who inject drugs [PWID], homeless people, immigrants, and sex workers), where very few individuals have access to diagnosis2,5,6.
The standard HCV diagnosis requires an initial serological test, followed by a confirmatory nucleic acid test (NAT) for the detection of HCV ribonucleic acid (RNA) in serum/plasma samples obtained during routine venous blood collection4,6,7. The tests require high-cost facilities and equipment (not always available in resource-limited settings) and specialized personnel and adequate infrastructures for the collection, transport, and storage of venous blood samples6. In addition, some patients, such as PWIDs, may have limited venous access, which further hampers diagnosis2.
Several strategies have been proposed to overcome some of these limitations2,5,8. One is the use of dried blood spots (DBS), which are obtained by finger puncture and depositing the blood drops on a filter paper. Such an approach can be used for HCV diagnosis in serological tests (anti-HCV antibodies) and in virological tests (HCV-RNA)9. DBS facilitates the sampling process by avoiding venipuncture and removing the need to separate plasma samples. In addition, DBS samples are highly stable at room temperature, and it is not necessary to maintain the cold chain for the storage of the samples and transport to the processing laboratory10. These advantages have made DBS sampling a promising approach to HCV screening and epidemiological surveillance in LMICs and risk groups11–14.
DBS is increasingly used for HCV screening, although the sensitivity and specificity of this approach for hepatitis C remain uncertain. Additionally, the laboratory methodology used for the analysis of DBS samples is very diverse, covering a wide range of settings15–17. In recent years, several systematic reviews and meta-analyses on this topic have been published15–19. However, to our knowledge, none has performed a detailed analysis of the diagnostic accuracy of laboratory-based screening for HCV. Therefore, our aim was to carefully analyze the diagnostic performance of the methodology that enables the detection of HCV infection in DBS samples, particularly in the type of diagnostic assay used, by conducting a meta-analysis of all eligible studies published to date (March 2018).
Material and Methods
The meta-analysis was conducted following guidelines on systematic reviews and meta-analyses (PRISMA; see Supplemental File (SF) 1)20.
Search strategy
Relevant studies were identified by a literature search in PubMed, Scopus, Embase, Lilacs, Web of Science, and the Cochrane Library with the following terms: (“hepatitis C” OR HCV) AND (DBS OR Whatman OR “filter paper” OR “Dried blood spot” OR “Dried blood filter” OR “Dried blood” OR “Dried sample”) AND (sensitivity OR specificity OR “Positive Predictive Value” OR “Negative Predictive Value” OR AUROC OR AUCROC OR diagnostic OR screening) NOT (review). We also reviewed the reference lists of several previously published reviews on HCV screening in DBS. The information contained in this report is based on articles published before March 2018.
Study selection
The inclusion and exclusion criteria were established before proceeding to the search and review. The inclusion criteria were as follows: (1) studies had to have evaluated the detection of anti-HCV antibodies and/or HCV-RNA in DBS; (2) results for DBS had to have been compared with a reference method (gold standard) using serum, plasma, or whole blood samples; (3) there had to be enough available data to construct 2 × 2 tables and to calculate the number of true positives (TP), true negatives (TN), false positives (FP), and false negatives (FN). The articles excluded were as follows: (1) studies whose objective differed from that of the meta-analysis; (2) editorial comments, reviews, opinion letters, and conference proceedings; (3) studies with insufficient data to estimate the sensitivity and/or specificity of the techniques evaluated; and (4) studies in which all samples were not tested using at least a reference test or in which a reference test was performed only on a subset of samples (only positive, negative, or discordant results).
We based our selection of eligible articles on careful screening of the title and abstract; when an article fulfilled the inclusion criteria, the full text was examined, and data were extracted. Moreover, when a study included different subgroups with various diagnostic tests, only those subgroups that met the inclusion criteria were included in our meta-analysis. In the case of antibody detection assays, if a receiver operating characteristic (ROC) curve analysis was performed to determine the optimal cut-off point, we only included the data obtained with the cut-off point established by the manufacturer for the use of serum or plasma samples. However, some investigators also determined others cut-off points based on ROC curve analysis, which were excluded from the meta-analysis.
Data extraction
Data were extracted independently by 2 investigators (B.A. and S.V.M.) and then cross-checked. When data were unclear or required assumptions to be made, other investigators (S.R. and M.A.J.S.) were consulted to reach a consensus. The values of TP, TN, FP, and FN corresponding to the tests evaluated in each article were extracted in order to create 2×2 contingency tables and calculate sensitivity and specificity. When this information was not explicitly reported, we contacted the corresponding author to request the data. If we did not receive the necessary data, the study was excluded.
Quality assessment
Study quality and risk of bias were evaluated using QUADAS-2 (Quality Assessment of Diagnostic Accuracy studies 2)21, which is designed to evaluate the quality of primary diagnostic accuracy studies through 4 key domains (patient selection, index test, reference standard, and flow and timing). Each domain was assessed in terms of risk of bias (low, high, or unclear) and, in the first 3 domains, concerns about applicability (low, high, or unclear) were also considered. Two reviewers (B.A. and S.V.M.) independently assessed the study characteristics and methodological quality.
Statistical analysis
Data were analyzed using Meta-DiSc 1.422 and STATA (version 14, STATA Corp., Texas USA) with the MIDAS module. Analyses were performed according to the HCV detection method. First, we performed the analysis of HCV-antibody detection studies and HCV-RNA detection studies separately. Next, we performed a subgroup analysis within each of the above categories: anti-HCV antibody detection (enzyme-linked immunosorbent assay [ELISA], rapid diagnostic tests (RDTs), and chemiluminescence assays [CLA]) and HCV-RNA detection (real-time polymerase chain reaction [PCR] and transcription-mediated amplification [TMA]). Analyses were performed only when 3 or more articles were available.
We calculated the pooled estimates of sensitivity, specificity, positive predicted value (PPV), negative predicted value (NPV), positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio (DOR), and their corresponding 95% confidence intervals (95%CI) using the accuracy data (TP, FP, FN, and TN) extracted from each eligible study. We used a random effects model (DerSimonian-Laird method), since there was significant between-study variability23. In order to facilitate analysis, a value of 0.5 was added to those cells that contained the value zero.
The data were displayed graphically on forest-plots, summary ROC (SROC) curves, and likelihood ratio scatter plots. Heterogeneity was measured using the Cochran’s Q test and the inconsistency index (I2). The Cochran’s Q was calculated as the weighted sum of squared differences between individual study effects and the pooled effect across studies and P-values were calculated by comparing the statistic with a χ2 distribution with k-1 degrees of freedom (where k is the number of studies). The I² statistic describes the percentage of total variation across studies due to heterogeneity rather than sampling error (chance). A p-value ≤ 0.05 and an I2 value ≥ 50% indicated significant heterogeneity. Sensitivity analyses were performed by sequential omission of individual outlier studies to investigate the influence of each study on the pooled estimates. In addition, we performed a meta-regression analysis for anti-HCV antibody detection assays and HCV-RNA detection assays with the aim of defining the potential effect of different covariates on diagnostic accuracy measures. For this purpose, we used the restricted maximum likelihood method and weighted least-squares model, with weights obtained from the study sizes. The covariates analyzed were as follows: study performed after 2010, study performed in LMICs, HIV coinfection, type of HCV detection test, capillary or venous DBS samples, and anti-HCV or HCV-RNA prevalence. A statistically significant linear relationship was one with a p-value ≤ 0.05. Publication bias was assessed using Deek’s funnel plot, and a P value < 0.05 indicated the presence of publication bias.
Results
Search results
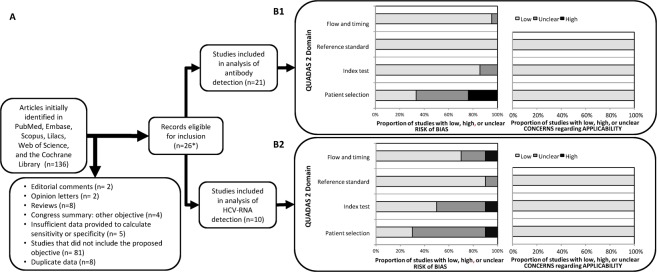
The literature search yielded 136 articles (Fig. 1A), of which 110 were excluded because they did not meet the inclusion criteria or presented an exclusion criterion. The remaining 26 eligible studies comprised 21 that were selected for the anti-HCV antibody detection meta-analysis9,24–43 and 10 for the HCV-RNA detection meta-analysis9,24,27,38,39,44–48. Five studies were included in both analyses9,24,27,38,39.
Figure 1.
Summary of study selection process (A) and quality assessment using the QUADAS-2 tool (B): (B1) Results of quality assessment of anti-HCV antibody detection studies; (B2) Results of quality assessment of HCV-RNA detection studies. Abbreviations: HCV, hepatitis C virus; RNA, ribonucleic acid; QUADAS, quality assessment of diagnostic accuracy studies.
Article characteristics
The main characteristics of the articles are summarized in Table 1 for the anti-HCV antibody detection meta-analysis and Table 2 for the HCV-RNA detection meta-analysis. The publication year of the studies ranged from 2002 to 2018. A total of 9679 DBS samples from 9679 individuals were included for anti-HCV antibody detection, and 2201 DBS samples from 2015 individuals were included for HCV-RNA detection. The median prevalence was 51.1% for anti-HCV antibodies (p25th = 24.3%; p75th = 64.3%) and 61.8% for HCV-RNA (p25th = 60.5%; p75th = 70.1%). Eleven studies provided information about HCV genotypes in infected individuals9,24,32,36,37,40,41,45–48. Only 7 studies included the HIV/HCV coinfection status24,28,29,31,37,38,47. Most of the studies (n = 15) were based on DBS samples obtained by spotting whole venous blood on filter paper, 12 were based on capillary DBS samples24,25,27,29,31,33,37,38,41–43,49, 2 on DBS samples obtained by either capillary or venous blood33,41, and 1 on mock DBS made by diluting the Second WHO International Standard for HCV-RNA in negative blood46.
Table 1.
Characteristics of the Studies Included in Meta-Analysis for Antibody Detection.
| Year | Author | HCV-GT | HIV status | Country | Age (years) | Males (%) | Assay in DBS samples | Gold standard | Sample (n) | HCV-infected (n) | Se (%) | Sp (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ELISA | ||||||||||||
| 2013 | Brandao et al.25 | — | No | Brazil | 40 | 43.8 | Monolisa HCV AgAb ULTRA Assay (Bio-Rad) | Monolisa HCV AgAb ULTRA Assay (Bio-Rad) | 386 | 40 | 95 | 100 |
| 2013 | Brandao et al.25 | — | No | Brazil | 40 | 43.8 | Murex HCV AgAb combination EIA (Diasorin) | Murex HCV AgAb combination EIA (DiaSorin) | 386 | 40 | 82.5 | 98 |
| 2006 | Croom et al.26 | — | No | Australia | — | — | Monolisa anti-HCV PLUS Version 2 EIA (Bio-Rad) | Monolisa anti-HCV PLUS Version 2 EIA (Bio-Rad) | 183 | 75 | 100 | 100 |
| 2014 | Dokubo el al27 | — | No | USA | < 30 | — | ELISA HCV Version 3.0 (Ortho-Clinical Diagnostics) | Third-generation ELISA or CLA (Unspecified) | 148 | 77 | 70.1 | 100 |
| 2017 | Flores et al.28 | — | Yes | Brazil | 50.9 | 52 | Murex anti-HCV (DiaSorin) | Murex anti-HCV (DiaSorin) | 524 | 278 | 92.1 | 99.2 |
| 2003 | Judd et al.29 | — | Yes | UK | — | — | HCV 3.0 SAVe ELISA (Ortho-Clinical Diagnostics) | HCV 3.0 SAVe ELISA (Ortho-Clinical Diagnostics) | 633 | 252 | 99.2 | 100 |
| 2013 | Kania et al.30 | — | No | Burkina Faso | 29.8 | 37.2 | Monolisa HCV Ab-Ag ULTRA Assay (Bio-Rad) | Monolisa HCV Ab-Ag ULTRA Assay (Bio-Rad) | 218 | 5 | 100 | 100 |
| 2012 | Larrat et al.31 | — | Yes | France | 46.5 | 61.9 | Monolisa HCV Ab-Ag ULTRA Assay (Bio-Rad) | Monolisa HCV Ab-Ag ULTRA Assay (Bio-Rad) | 201 | 113 | 98.2 | 100 |
| 2013 | Lima41 | — | No | Brazil | 38.5 | 39.8 | Murex anti-HCV v.4.0, (DiaSorin) | Murex anti-HCV v.4.0 (DiaSorin) | 1043 | 7 | 57.1 | 100 |
| 2013 | Lima41 | — | No | Brazil | 38.5 | 39.8 | Murex anti-HCV v.4.0, (DiaSorin) | Murex anti-HCV v.4.0 (DiaSorin) | 491 | 5 | 40.0 | 99.8 |
| 2013 | Lima41 | — | No | Brazil | 38.5 | 39.8 | Murex anti-HCV v.4.0, (DiaSorin) | Murex anti-HCV v.4.0 (DiaSorin) | 254 | 228 | 91.7 | 92.3 |
| 2016 | Marques et al.32 | 1b, 1a, 3 | No | Brazil | 47.3 | 43.5 | HCV Ab (RADIM diagnostic) | HCV Ab (RADIM diagnostic) | 99 | 59 | 94.9 | 100 |
| 2012 | Marques et al.33 | — | No | Brazil | 40 | 43.8 | HCV Ab (RADIM diagnostic) | HCV Ab (RADIM diagnostic) | 411 | 40 | 97.5 | 99.5 |
| 2012 | Marques et al.33 | No | Brazil | 40 | 43.8 | ETI-AB-HCVK-4 (DiaSorin) | ETI-AB-HCVK-4 (DiaSorin) | 411 | 45 | 88.9 | 96.1 | |
| 1999 | McCarron et al.34 | — | No | UK | Monolisa anti-HCV (Sanofi Pasteur) | AxSYM HCV version 3.0 (Abbott Diagnostics) | 220 | 108 | 100 | 87.5 | ||
| 2014 | Nandagopal et al.35 | — | No | India | — | — | Anti-HCV ELISA (Murex Biotech S.A) | Anti-HCV ELISA (Murex Biotech S.A) | 60 | 31 | 100 | 100 |
| 2001 | O’Brien42 | No | USA | 44.8 | 38 | Third-generation EIA in DBS | Third-generation EIA in blood | 1090 | 404 | 98.3 | 100 | |
| 2012 | Rice et al.43 | — | No | UK | — | — | Chiron V3.0 SaVE assay (Ortho-Clinical Diagnostics) | Unspecified | 93 | 59 | 98.3 | 100 |
| 2016 | Soulier et al.9 | 1, 2, 3, 4, 5a, 6 | No | France | 54 | 55.2 | aHCV VITROS ECi (Ortho-Clinical Diagnostics) | aHCV VITROS ECi (Ortho-Clinical Diagnostics) | 511 | 341 | 99.1 | 98.2 |
| 2015 | Tejada-Strop et al.36 | 1a, 1c, 2, 3 | No | USA | — | — | HCV 3.0 EIA (Ortho-Clinical Diagnostics) | HCV 3.0 EIA (Ortho-Clinical Diagnostics) | 103 | 52 | 90 | 100 |
| 2010 | Tuaillon et al.37 | 1, 2, 3, 4 | Yes | France | — | — | HCV 3.0 EIA (Ortho-Clinical Diagnostics) | HCV 3.0 EIA (Ortho-Clinical Diagnostics) | 200 | 100 | 99 | 98 |
| 2018 | Vázquez-Morón et al.24 | 1a, 1b, 2, 3, 4, 5 | Yes | Spain | 44.3 | 66.9 | Murex anti-HCV kit, v.4.0 (DiaSorin,) | ADVIA Centaur® HCV assay | 139 | 108 | 92.6 | 100 |
| Chemiluminescence immunoassays | ||||||||||||
| 2016 | Mössner et al.38 | — | Yes | Denmark | — | — | Architect anti-HCV assay (Abbott Diagnostics) | Architect anti-HCV assay (Abbott Diagnostics) | 404 | 116 | 96.6 | 100 |
| 2013 | Ross et al.39 | — | No | Germany | — | — | Architect anti-HCV assay (Abbott Diagnostics) | Architect anti-HCV assay (Abbott Diagnostics) | 339 | 179 | 97.8 | 100 |
| 2015 | Tejada-Strop et al.36 | 1a, 1c, 2, 3 | No | USA | — | — | VITROS anti-HCV IgG CLA (Ortho-Clinical Diagnostics) | VITROS anti-HCV IgG CLA (Ortho-Clinical Diagnostics) | 103 | 52 | 92 | 100 |
| Rapid diagnostic tests | ||||||||||||
| 2012 | Larrat et al.31 | — | Yes | France | 46.5 | 61.9 | OraQuick HCV Rapid Antibody Test (OraSure Technologies) | Monolisa HCV AbAg ULTRA Assay (Bio-Rad) | 201 | 113 | 97.4 | 100 |
| 2016 | Poiteau et al.40 | 1a, 1b, 2, 3a, 4, 6 | No | France | — | — | OraQuick HCV Rapid Antibody Test (OraSure Technologies) | aHCV VITROS ECi (Ortho-Clinical Diagnostics | 207 | 139 | 100 | 100 |
| 2016 | Poiteau et al.40 | 1a, 1b, 2, 3a, 4, 6 | No | France | — | — | First Response® HCV Card Test (Premier Medical Corporation Ltd) | aHCV VITROS ECi (Ortho-Clinical Diagnostics | 207 | 139 | 99.3 | 100 |
| 2016 | Poiteau et al.40 | 1a, 1b, 2, 3a, 4, 6 | No | France | — | — | Assure HCV Rapid Test (MP Diagnostics) | aHCV VITROS ECi (Ortho-Clinical Diagnostics | 207 | 139 | 98.6 | 100 |
| 2016 | Poiteau et al.40 | 1a, 1b, 2, 3a, 4, 6 | No | France | — | — | MultiSure HCV Antibody Assay (MP Diagnostics) | aHCV VITROS ECi (Ortho-Clinical Diagnostics) | 207 | 139 | 98.6 | 100 |
Abbreviations: Ab, antibody; Ag, antigen; Anti-HCV, HCV antibodies; CLA, chemiluminescence assay; DBD, dried blood spot; EIA, enzyme immunoassay; ELISA, enzyme-linked immunosorbent assay; GT, genotype; HCV, hepatitis C virus; HIV, human immunodeficiency virus; Se, sensitivity; Sp, specificity; TMA, transcription mediated amplification.
Table 2.
Characteristics of Studies Included in the Meta-Analysis With Respect to HCV-RNA Detection.
| Year | Author | HCV-GT | HIV | Country | Age (years) | Males (%) | Assay in DBS samples | Gold standard | Sample (n) | HCV-infected (n) | Se (%) | Sp (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2012 | Bennett et al.46 | 1a, 2b, 3a, 4a, 5, 6a | No | United Kingdom | — | — | TaqMan real-time PCR (In-house 2-step RT and PCR) | Real-time RT-PCR: m2000 rt (Abbott Molecular) | 80 | 57 | 100 | 95.8 |
| 2010 | De Crignis et al.47 | 1a, 1b, 2a, 3a | Yes | Italy | — | — | SYBR Green real-time RT-PCR for HCV and HIV detection | Real-time RT-PCR: VERSANT HCV RNA 3.0 b-DNA Assay (Siemens) | 25 | 16 | 93.8 | 100 |
| 2014 | Dokubo et al.27 | — | No | USA | < 30 | — | dHCV TMA (Novartis Vaccines and Diagnostics) | TMA: Procleix Ultrio Assay (Novartis Vaccines and Diagnostics) s | 132 | 48 | 89.6 | 100 |
| 2016 | Mössner et al.38 | — | Yes | Denmark | — | — | TMA: Procleix Ultrio Elite assay (Novartis Vaccines and Diagnostics) | TMA: Procleix Ultrio Elite Assay (Novartis Vaccines and Diagnostics) | 107 | 85 | 95.3 | 95.5 |
| 2013 | Ross et al.39 | — | No | Germany | — | — | TMA: VERSANT HCV RNA Qualitative Assay (Siemens) | TMA: VERSANT HCV RNA Qualitative Assay (Siemens) | 150 | 100 | 100 | 100 |
| 2018 | Saludes et al.45 | 1, 2, 3, 4 | No | Spain | — | — | QuantiFast Pathogen RT-PCR + IC Kit (QIAGEN) | Abbott RealTime HCV (Abbott Molecular) | 82 | 38 | 100 | 100 |
| 2012 | Santos et al.44 | No | Brazil | — | — | In-house qPCR | In-house qPCR | 168 | 101 | 98 | 94.3 | |
| 2002 | Solmone et al.48 | 1b, 2a, 2c, 3a, 4c, 4d | No | Italy | — | — | In-house 2-step RT and PCR | Amplicor HCV Monitor (Roche Molecular) TMA: VERSANT HCV RNA Qualitative Assay (Siemens) | 55 | 34 | 100 | 100 |
| 2002 | Solmone et al.48 | 1b, 2a, 2c, 3a, 4c, 4d | No | Italy | — | — | TMA Unspedified. | Amplicor HCV Monitor (Roche Molecular) TMA: VERSANT HCV RNA Qualitative Assay (Siemens) | 55 | 34 | 100 | 100 |
| 2016 | Soulier et al.9 | 1, 2, 3, 4, 5a, 6 | No | France | 54 | 55,2 | Real-time RT-PCR: Cobas Ampliprep/Cobas TaqMan HCV version 2 (Roche Molecular) | Real-time RT-PCR: Cobas Ampliprep/Cobas TaqMan HCV version 2 (Roche Molecular) | 511 | 315 | 97.1 | 100 |
| 2016 | Soulier et al.9 | 1, 2, 3, 4, 5a, 6 | No | France | 54 | 55,2 | Real-time RT-PCR: m2000rt (Abbott Molecular) | Real-time RT-PCR: m2000rt (Abbott Molecular) | 511 | 314 | 98.1 | 100 |
| 2018 | Vázquez-Morón et al.24 | 1a, 1b, 2, 3, 4, 5 | Yes | Spain | 44.3 | 66.9 | Quantitec SYBR Green RT-PCR One Step kit (Qiagen) | VERSANT HCV RNA 1.0 Assay | 139 | 108 | 99.1 | 100 |
Abbreviations: dHCV TMA, discriminatory HCV transcription-mediated amplification assay; GT, genotype; HCV, hepatitis C virus; HIV, human immunodeficiency virus; PCR, polymerase chain reaction; RT, reverse transcription; Se, sensitivity; Sp, specificity; TMA, transcription-mediated amplification.
Quality was assessed using QUADAS-2 (Fig. 1 and SF 2). Quality scores are summarized in Fig. 1B1 for the anti-HCV antibody detection assays and Fig. 1B2 for the HCV-RNA detection assays. The overall quality of the studies was moderate and most did not use a random or consecutive sampling method. Additionally, a significant percentage of studies did not hide the results of the reference test or did not report blinding of laboratory personnel to the results of the reference test. As for concerns about the applicability of the studies, the risk of bias was low in all cases.
Meta-analysis of anti-HCV antibody detection in DBS samples
A total of 30 anti-HCV antibodies assays from 21 different studies were analyzed: 22 ELISA9,24–37,41–43, 5 RDT31,40, and 3 CLA36,38,39. Publication bias was detected using Deek’s funnel plot asymmetry test (P = 0.001) (SF 3).
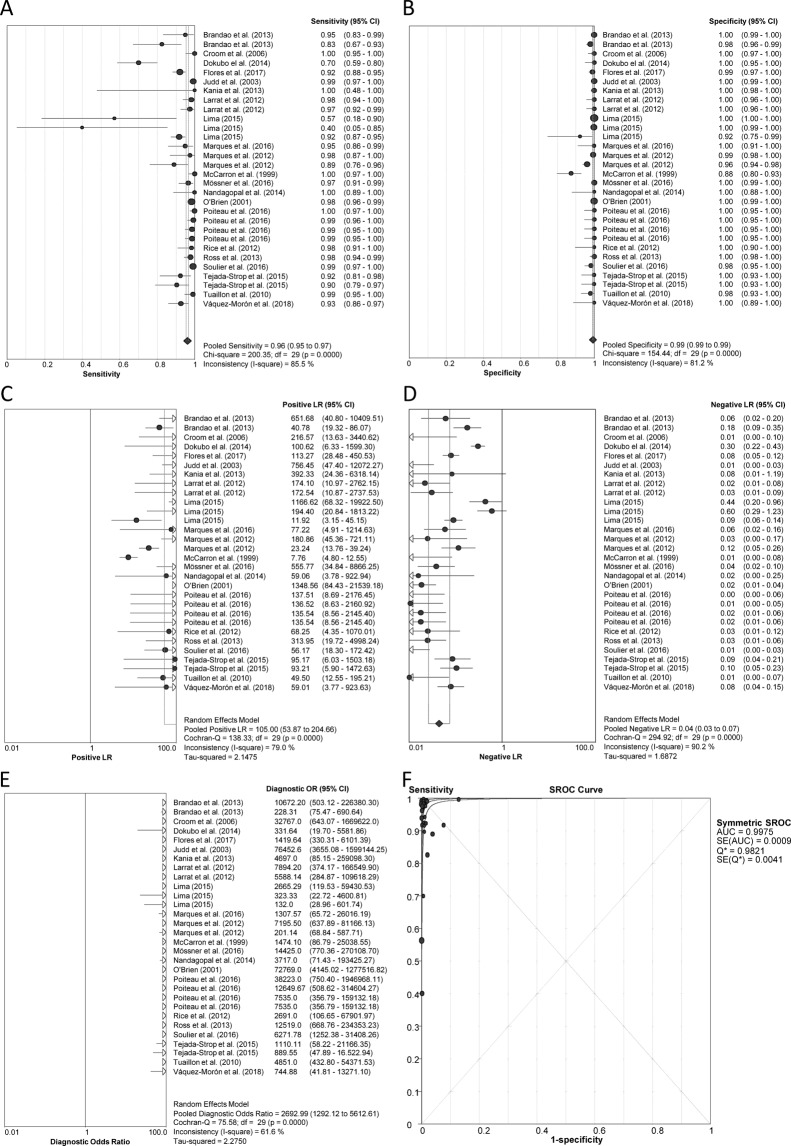
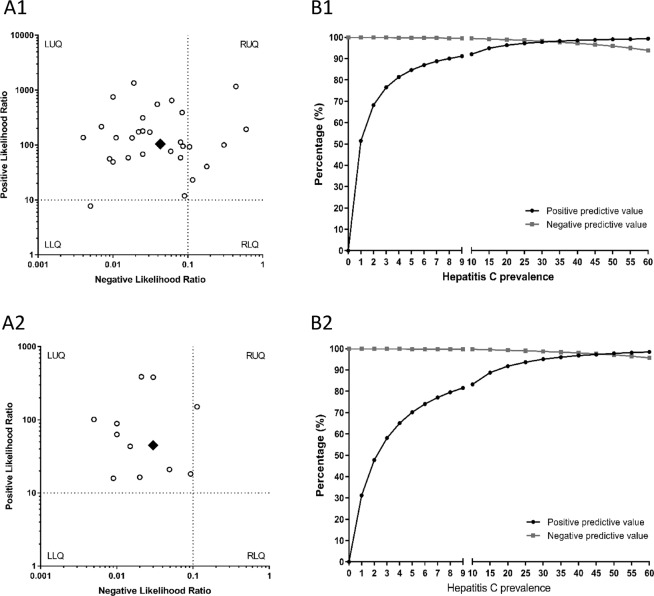
Pooled diagnostic accuracy measures are shown in Fig. 2. Sensitivity was 96.1% (95%CI = 95.4%; 96.7%), specificity 99.2% (95%CI = 99.0%; 99.4%), PLR 105.0 (95%CI = 53.8; 204.6), NLR 0.04 (95%CI = 0.03; 0.07), DOR 2692.9 (95%CI = 1292.1; 5612.6), and SROC 0.997 ± 0.001, indicating high diagnostic accuracy. All statistical measures showed values of I2 > 50% (P < 0.05), suggesting substantial heterogeneity. However, according to the sensitivity analysis, no individual study had a significant influence on pooled estimates. The likelihood scatter-plot (Fig. 3A1) shows a matrix of values of PLR and NLR. Most studies were in the left upper quadrant (LUQ), which means that anti-HCV antibody detection tests from DBS samples are useful for confirmation and exclusion of exposure to HCV. We also calculated the theoretical values of PPV and NPV using pooled values of PLR (105.0) and NLR (0.04) and plotted them against increasing anti-HCV antibody prevalence (Fig. 3B1, full description in SF 4). We found a PPV of 84.7% for a 5% prevalence and 92.1% for a 10% prevalence, whereas NPV continued with values above 93% when prevalence reached 60%.
Figure 2.
Anti-HCV antibody assays. Forest plot of sensitivity (A), specificity (B), positive LR (C), negative LR (D), diagnostic odds ratio (E), and SROC plot (F). Abbreviations: LR, likelihood ratio; SROC, summary of receiver operating characteristic; HCV, hepatitis C virus.
Figure 3.
The likelihood scatter-plot (A1 & A2) and theoretical values of positive predicted value (PPV) and negative predicted value (NPV) against increasing anti-HCV antibody prevalence values (B1) and HCV-RNA prevalence (B2). Abbreviations: HCV, hepatitis C virus; RNA, ribonucleic acid; LUQ, left upper quadrant; RUQ, right upper quadrant; LUQ, left lower quadrant; LUQ, right lower quadrant.
We also performed an analysis according to the type of anti-HCV antibody detection assay: ELISA (SF 5), RDT (SF 1), and CLA (SF 7). Diagnostic accuracy measures were similar for ELISA, RDT, and CLA, and in turn similar to the global estimates for anti-HCV antibody detection (Fig. 2). For ELISA, PPV was 82.5% for a 5% prevalence and 90.8% for a 10% prevalence; for RDT, PPV was 88.3% for a 5% prevalence and 94.1% for a 10% prevalence; and for CLA, PPV was 93.1% for a 5% prevalence and 96.6% for a 10% prevalence, whereas NPV was ≥ 91% for ELISA, RDT, and CLA when prevalence reached 60% (SF 4).
Meta-regression results are shown in SF 7. In the univariate analysis, we found significant values for studies performed after 2010 (p = 0.041) and in LMICs (p < 0.001). However, in the multivariate analysis, none of the factors had a significant potential effect on the diagnostic accuracy of the anti-HCV antibody detection assays.
Meta-analysis of HCV-RNA detection in DBS samples
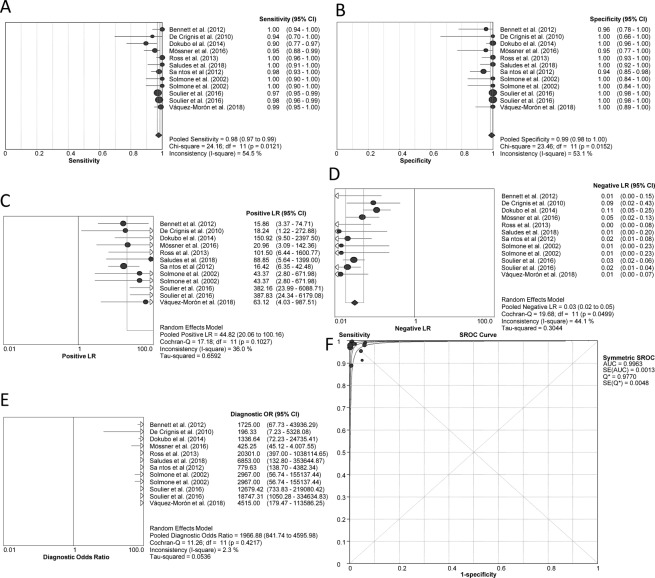
A total of 12 HCV-RNA detection assays from 10 different studies were analyzed: 9 PCR assays9,24,39,45–49 and 3 TMA assays27,38,48. Deek’s funnel plot asymmetry test revealed publication bias (P = 0.009) (SF 9).
Pooled diagnostic accuracy measures are shown in Fig. 4. Sensitivity was 97.8% (95%CI = 96.8%; 98.5%), specificity 99.2% (95%CI = 98.3%; 99.7%), PLR 44.8 (95%CI = 20.1; 100.2), NLR 0.04 (95%CI = 0.02; 0.06), DOR 1966.9 (95%CI = 841.7; 4596.0), and SROC 0.996 ± 0.013, indicating high diagnostic accuracy. Sensitivity and specificity showed I2 > 50% (P < 0.05), suggesting considerable heterogeneity, whereas the other statistical measures had I2 < 50%. No individual study showed a significant influence on pooled estimates in the sensitivity analyses. The likelihood scatter-plot (Fig. 3A2) also showed that most of the studies were in the LUQ. Thus, HCV-RNA detection tests in DBS samples are also useful for confirmation and exclusion of HCV infection. The theoretical values of PPV and NPV were calculated using the pooled PLR (44.82) and NLR (0.03) values and plotted against increasing HCV-RNA prevalence values (Fig. 3B2, full description in SF 4). We found a PPV of 70.2% for a 5% prevalence and 83.3% for a 10% prevalence, whereas NPV continued with values above 95% when prevalence reached 60%.
Figure 4.
HCV-RNA detection assays. Forest plot of sensitivity (A), specificity (B), positive LR (C), negative LR (D), diagnostic odds ratio (E), and SROC plot (F). Abbreviations: LR, likelihood ratio; SROC, summary of receiver operating characteristic; HCV, hepatitis C virus; RNA, ribonucleic acid.
We also performed an analysis according to the type of HCV-RNA detection assay: PCR (SF 10) and TMA (SF 11). Pooled diagnostic accuracy measures were similar between PCR and TMA, and in turn similar to global estimates for HCV-RNA detection (Fig. 4). For PCR, PPV was 72.3% for a 5% prevalence and 84.7% for a 10% prevalence; for TMA, PPV was 68.1% for a 5% prevalence and 81.9% for a 10% prevalence, whereas NPV was > 90% for PCR and TMA when prevalence reached 60% (SF 4).
Meta-regression results are shown in SF 12. We found no significant values in the univariate or the multivariate analysis. Therefore, no factors showed a significant potential effect on the diagnostic accuracy of the HCV-RNA detection assays.
Discussion
In this meta-analysis, we found that the internal validity of HCV screening tests in DBS samples was very high: pooled sensitivity values were > 96% and specificity values were > 99%. The diagnostic performance of anti-HCV antibody tests (ELISA, RDT, and CLA) was slightly lower than that of HCV-RNA tests (PCR and TMA), with the latter having higher sensitivity values. However, SROC values were > 0.985 in all of the tests analyzed.
Likelihood ratios summarize the ability of a test to predict the probability of disease. The positive value confirms the reliability of a positive test, whereas the negative value calls a normal result into question. Additionally, the characteristics of diagnostic tests (relating to the detection or exclusion of the condition of interest) may be described in terms of likelihood scatter-plots and the threshold values used to recommend a test for clinical use, namely, 0.1 for NLR and 10.0 for PLR50. In our study, most of studies were in the LUQ (NLR < 0.1 and PLR > 10.0), indicating that HCV screening tests in DBS samples may be useful for either confirmation or exclusion of exposure to HCV and active infection.
We also evaluated theoretical values of PPV and NPV, which were calculated to evaluate the external validity of HCV screening, against the increasing prevalence of HCV (see SF 3). We found similar NPV values owing to the high specificity of both approaches. It should be noted that NPV values were > 99% for an HCV prevalence < 10% and did not undergo a marked decline at higher prevalence values (NPV > 90% at 60% HCV prevalence), thus indicating that HCV screening could be used in DBS samples owing to the low number of false negatives it produces. However, PPV values were not uniform in all the techniques. The PPV for anti-HCV antibody tests was higher than for HCV-RNA tests, particularly when the prevalence of HCV was lower than 10%. Furthermore, RDT and CLA assays showed higher PPV values than ELISA assays, whereas in HCV-RNA assays, PCR showed higher PPV values than TMA assays when prevalence was < 10%. When we evaluated the possible use of these tests with an HCV prevalence < 5%, we found PPV values < 90% for anti-HCV antibody tests (except CLA) and < 85% for HCV-RNA, both of which are highly undesirable for diagnostic assays in the general population of most countries (HCV prevalence < 2.5%)51. In contrast, at an HCV prevalence > 10%, the PPV was for anti-HCV antibody tests > 90%, which is within acceptable limits for HCV screening in DBS samples and could be used in high-prevalence populations, such as PWID, prisoners or ex-prisoners, sex workers, and MSM3. In these key populations, screening for and treatment of hepatitis C and other interventions that reduce HCV transmission seem to be cost-effective52–55. In addition, use of DBS may increase uptake of HCV testing19, thus enabling scale-up of HCV treatment and other interventions to minimize HCV transmission54,55. However, further research on the cost-effectiveness of HCV screening in DBS samples in at-risk populations is needed before its use can be generalized. Additionally, it would be applicable to the general population in countries with a high prevalence of HCV, such as Egypt (14.7%), Cameroon (13.8%), Burundi (11.3%), and Uzbekistan (11.3%), where significant iatrogenic transmission has been reported51. Therefore, HCV screening in DBS samples with anti-HCV antibody tests may be an ideal diagnostic assay in endemic areas with a higher HCV prevalence, especially in populations where there is limited access to health services, laboratory facilities, or rapid diagnostic tests. However, HCV-RNA tests showed PPV values < 90%, particularly TMA assays (with 81.9% when prevalence was 10%), which would not be appropriate for HCV screening as a single laboratory test. However, it should improve when using the reference two-step HCV screening algorithm based on serological screening followed by molecular confirmation of serology positives. Thus, HCV-RNA tests should be used with some reservations, depending on the technique applied and the expected HCV prevalence.
The accuracy of a systematic review depends on the quality of the studies included. The quality of the articles included in the present meta-analysis was moderate, since many items regarding the risk of bias were lacking or unclear. Some of the main drawbacks were that most of the studies were cross-sectional studies, whose design resembled that of case-control studies, thus providing little information about the reference standard and index test and unclear data on flow and timing information. Moreover, a high percentage of DBS samples were not collected at the point of care under real-world conditions. Therefore, the number of high-quality articles was moderate, and an analysis based only on high-quality studies (ie, with a low risk of bias) could not be performed.
In this meta-analysis, we evaluated the heterogeneity by the tests of Cochran’s Q and I². However, Cochran’s Q test is known to be poor at detecting true heterogeneity among studies, particularly when meta-analyses include small numbers of studies; whereas this test may have excessive power when there are many studies, particularly when those studies are large56. Moreover, I² test is a preferable measurement to evaluate the heterogeneity because it is easy to calculate, has values between 0 and 100 (intuitive interpretation), focuses attention on the effect of any heterogeneity on the meta-analysis, is independent of the number of studies included in the meta-analysis and the type of outcome data56. In our study, we found significant heterogeneity among the studies, which is a common and expected characteristic in a meta-analysis of diagnostic tests. To our knowledge, there is no real consensus on how to interpret the heterogeneity measures in meta-analysis. In the opinion of many researchers, this depends on the type of meta-analysis. When performing a meta-analysis of clinical trials that evaluate the effects of a treatment and that included studies for estimating the same outcome, the heterogeneity should be low. However, the situation is different when pooling results from epidemiological studies. In our case, we performed a meta-analysis of epidemiological studies that had different designs and objectives, and which measured also different outcomes. In this type of meta-analysis, there is no way to control all possible confounding factors, so substantial heterogeneity between the studies can be expected. In this situation, a random effects analysis should be performed on the assumption that the different studies measures different things23. This heterogeneity may have been due to a large series of factors that could have influenced our results, such as sample size, reference tests, screening tests, HCV prevalence, HCV viral load, HCV genotype, HIV coinfection, DBS elution method, RNA extraction method, HCV-RNA amplification technique, type of PCR used, and the region of the amplified HCV genome. Some of these factors were analyzed, although others were not owing to the lack of data in the articles included. We analyzed whether heterogeneity was due to differences between the types of tests used in each study. All pooled measures for anti-HCV antibody assays showed high heterogeneity. A subgroup analysis was then performed according to the technique used (ELISA, TDR, or CLA) to investigate potential sources of heterogeneity. Thus, the HCV screening test used seemed to be responsible for part of the heterogeneity, since RDT and CLA became homogeneous once the analysis was stratified by the type of test. However, heterogeneity was maintained in the ELISA group, probably because of the high variability in the ELISA tests used by the authors. Furthermore, HCV-RNA tests were less heterogeneous, and when the HCV-RNA techniques (PCR or TMA) were analyzed separately, we found higher heterogeneity in TMA than in PCR. On the other hand, a meta-regression analysis found that several factors had no influence on the diagnostic accuracy of HCV screening. No individual study showed significant influence on pooled estimates when sensitivity analyses were performed, thus indicating the reliability and stability of our results.
Generalizability of HCV screening in DBS samples remains limited in specific circumstances. On the one hand, no uniform protocols were applied in most of studies included in our meta-analysis. For example, the HCV screening kits were generally from different companies, and when they were from the same company, they could have been subject to changes in the manufacturing process over the years. Manufacturers should formally validate their DBS assays for use with their commercial assays. Also important is the optimal pre-analytical treatment of DBS samples in relation to storage conditions (eg, temperature, humidity) and sample elution. Manufacturers should establish protocols for optimal pre-analytical treatment of DBS specimens. Further studies are needed to demonstrate the stability of DBS under different conditions. In our meta-analysis, a significant percentage of studies (35%) did not report information about storage conditions, with the result that sample storage conditions were not included in the meta-regression. In addition, the specifications of the storage conditions were very different, thus making it difficult to create a variable that can be entered into the meta-analysis. When the articles included this information, the storage conditions were very far from real-world conditions in LMICs, because most studies were from high-income countries. Future studies on diagnostic accuracy should assess the impact of environmental conditions common to low-resource field settings.
The intrinsic validity of a diagnostic test is obtained by calculating the values of sensitivity and specificity. Therefore, the diagnostic tests analyzed in this meta-analysis were quite valid, since the sensitivity was > 96% and specificity was > 99%. Only a small margin of improvement remains to raise the sensitivity to 99–100%, which could be achieved with assays and methodologies more appropriate for the use of DBS in the HCV screening. Moreover, predictive values provide relevant information when making a clinical decision about a specific test result, but it has the limitation that they depend on the prevalence of the disease to be diagnosed in the study population. Thus, the low prevalence of HCV infection ( < 5%) affects the external validity of the tests since affects the positive and negative predictive values.
In two recent meta-analyses, Lange et al.15,16 evaluated the diagnostic accuracy of HCV screening according to the type of laboratory methodology. However, in our opinion, our meta-analysis has a series of merits that provide a differential added value. In contrast to Lange et al., we showed pooled diagnostic accuracy measures (sensitivity, specificity, PPV, NPV, PLR, NLR, DOR, and SROC) according to the type of laboratory assay used for HCV screening (anti-HCV antibody assays [ELISA, TDR, and CLA] and HCV-RNA tests [PCR and TMA]) in the same meta-analysis. We showed both the internal validity and the external validity (PPV and NPV against the increasing prevalence of HCV) of the techniques used. We also analyzed publication bias and the impact of covariables on effect size using regression-based techniques.
Finally, in order to ensure that our results were interpreted appropriately, a series of points had to be taken into account. Firstly, this meta-analysis revealed a publication bias, indicating that there was a higher tendency to publish studies with favorable results than studies with unfavorable results. Secondly, the number of studies in some subgroup analyses was small, thus potentially leading to weak results. Thirdly, the fact that most of the studies included were conducted in Western countries and South America does not prevent DBS samples from being used in other parts of the world (eg, Asia), provided similar technology and protocols are applied. Fourthly, we have not done a study of cost-effectiveness, which is an important consideration in most public health interventions, especially in LMICs.
In the current era of direct-acting antivirals for HCV treatment, there is a growing need for rapid, sensitive, and specific identification of HCV-infected individuals to enable effective prompt HCV therapy, implement HCV-specific infection control measures, and decrease HCV prevalence. HCV screening tests in DBS samples may have the potential to meet some of these needs, although their current limitations in diagnostic performance should be considered. More sensitive assays must be developed to increase the potential of DBS as an HCV screening tool.
Conclusions
In conclusion, HCV screening from DBS samples has an outstanding diagnostic performance, with no relevant differences in diagnostic accuracy according to the type of technique used for detection of anti-HCV antibodies or HCV-RNA. However, external validity may be limited when the prevalence of HCV is low. This approach could be useful for the implementation of national programs for the control of HCV infections in LMICs and in difficult-to-access populations. Further studies to determine diagnostic accuracy in real-world settings are needed.
Supplementary information
Acknowledgements
We thank the authors of the studies included in this review for providing additional information upon request. The authors thank Thomas O’Boyle for writing assistance during the preparation of the manuscript. This study was supported by grants from Instituto de Salud Carlos III [grant numbers PI17CIII/00003 and RD16CIII/0002/0002], and a research grant from Merck Sharpe & Dohme (MISP IIS#54846). No funding bodies had any role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author Contributions
Sonia Vázquez-Morón: investigation, methodology, writing – original draft. Beatriz Ardizone: investigation, methodology, writing – original draft. María A. Jiménez-Sousa: investigation, methodology, writing – review and editing. José M. Bellón: methodology: statistical analysis. Pablo Ryan: writing – review and editing. Salvador Resino: conceptualization, formal analysis, writing – original draft, supervision
Data Availability
All relevant data are within the paper and its Supporting Information files.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Sonia Vázquez-Morón and Beatriz Ardizone Jiménez contributed equally.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-41139-8.
References
- 1.WHO. Global Hepatitis Report 2017. Geneva: World Health Organization. 83 (World Health Organization, Geneva, Switzerland 2017).
- 2.Easterbrook PJ, Group WGD. Who to test and how to test for chronic hepatitis C infection–2016 WHO testing guidance for low-and middle-income countries. J. Hepatol. 2016;65:S46–S66. doi: 10.1016/j.jhep.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 3.Midgard H, et al. HCV epidemiology in high-risk groups and the risk of reinfection. J. Hepatol. 2016;65:S33–45. doi: 10.1016/j.jhep.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 4.AASLD IDSA HCV. Guidance Panel. Hepatitis C guidance: AASLD-IDSA recommendations for testing, managing, and treating adults infected with hepatitis C virus. Hepatology. 2015;62:932–954. doi: 10.1002/hep.27950. [DOI] [PubMed] [Google Scholar]
- 5.Thursz M, Lacombe K. Breaking Down Barriers to Care in Hepatitis C Virus Infection. J. Infect. Dis. 2016;213:1055–1056. doi: 10.1093/infdis/jiv427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Easterbrook PJ, Roberts T, Sands A, Peeling R. Diagnosis of viral hepatitis. Curr Opin. HIV AIDS. 2017;12:302–314. doi: 10.1097/COH.0000000000000370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Testing CDC. Testing for HCV infection: an update of guidance for clinicians and laboratorians. MMWR. 2013;62:362. [PMC free article] [PubMed] [Google Scholar]
- 8.WHO. Guidelines for the screening, care and treatment of persons with chronic hepatitis C infection. (World Health Organization 2016). [PubMed]
- 9.Soulier A, et al. Dried Blood Spots: A Tool to Ensure Broad Access to Hepatitis C Screening, Diagnosis, and Treatment Monitoring. J. Infect. Dis. 2016;213:1087–1095. doi: 10.1093/infdis/jiv423. [DOI] [PubMed] [Google Scholar]
- 10.UNITAID. HEPATITIS C Diagnostics Technology Landscape. 86 (World Health Organization, Geneva 2015).
- 11.Parr JB, et al. An Efficient, Large-Scale Survey of Hepatitis C Viremia in the Democratic Republic of the Congo Using Dried Blood Spots. Clin. Infect. Dis. 2018;66:254–260. doi: 10.1093/cid/cix771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hope VD, et al. Injection into the jugular vein among people who inject drugs in the United Kingdom: Prevalence, associated factors and harms. Int J Drug Policy. 2017;46:28–33. doi: 10.1016/j.drugpo.2017.05.005. [DOI] [PubMed] [Google Scholar]
- 13.Iversen J, et al. Estimating the cascade of hepatitis C testing, care and treatment among people who inject drugs in Australia. Int J Drug Policy. 2017;47:77–85. doi: 10.1016/j.drugpo.2017.05.022. [DOI] [PubMed] [Google Scholar]
- 14.Clifford GM, et al. Hepatitis C virus seroprevalence in the general female population of 9 countries in Europe, Asia and Africa. Infect. Agent. Cancer. 2017;12:9. doi: 10.1186/s13027-017-0121-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lange B, et al. Diagnostic accuracy of serological diagnosis of hepatitis C and B using dried blood spot samples (DBS): two systematic reviews and meta-analyses. BMC Infect. Dis. 2017;17:700. doi: 10.1186/s12879-017-2777-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lange B, et al. Diagnostic accuracy of detection and quantification of HBV-DNA and HCV-RNA using dried blood spot (DBS) samples - a systematic review and meta-analysis. BMC Infect. Dis. 2017;17:693. doi: 10.1186/s12879-017-2776-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muzembo BA, Mbendi NC, Nakayama SF. Systematic review with meta-analysis: performance of dried blood spots for hepatitis C antibodies detection. Public Health. 2017;153:128–136. doi: 10.1016/j.puhe.2017.08.008. [DOI] [PubMed] [Google Scholar]
- 18.Greenman J, Roberts T, Cohn J, Messac L. Dried blood spot in the genotyping, quantification and storage of HCV RNA: a systematic literature review. J. Viral Hepat. 2015;22:353–361. doi: 10.1111/jvh.12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones L, et al. Effectiveness of interventions to increase hepatitis C testing uptake among high-risk groups: a systematic review. Eur. J. Public Health. 2014;24:781–788. doi: 10.1093/eurpub/ckt156. [DOI] [PubMed] [Google Scholar]
- 20.Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. J. Clin. Epidemiol. 2009;62:1006–1012. doi: 10.1016/j.jclinepi.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 21.Whiting PF, et al. Quadas-2: A revised tool for the quality assessment of diagnostic accuracy studies. Ann. Intern. Med. 2011;155:529–536. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 22.Zamora J, Abraira V, Muriel A, Khan K, Coomarasamy A. Meta-DiSc: a software for meta-analysis of test accuracy data. BMC Med. Res. Methodol. 2006;6:31. doi: 10.1186/1471-2288-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Macaskill, P., Gatsonis, C., Deeks, J., Harbord, R. & Takwoingi, Y. Cochrane handbook for systematic reviews of diagnostic test accuracy. Version 1. 0. London: The Cochrane Collaboration (2010).
- 24.Vazquez-Moron S, et al. Evaluation of dried blood spot samples for screening of hepatitis C and human immunodeficiency virus in a real-world setting. Sci. Rep. 2018;8:1858. doi: 10.1038/s41598-018-20312-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brandao CP, et al. Simultaneous detection of hepatitis C virus antigen and antibodies in dried blood spots. J. Clin. Virol. 2013;57:98–102. doi: 10.1016/j.jcv.2013.02.014. [DOI] [PubMed] [Google Scholar]
- 26.Croom HA, et al. Commercial enzyme immunoassay adapted for the detection of antibodies to hepatitis C virus in dried blood spots. J. Clin. Virol. 2006;36:68–71. doi: 10.1016/j.jcv.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 27.Dokubo EK, et al. Comparison of Hepatitis C Virus RNA and antibody detection in dried blood spots and plasma specimens. J. Clin. Virol. 2014;59:223–227. doi: 10.1016/j.jcv.2014.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flores GL, et al. Performance of ANTI-HCV testing in dried blood spots and saliva according to HIV status. J. Med. Virol. 2017;89:1435–1441. doi: 10.1002/jmv.24777. [DOI] [PubMed] [Google Scholar]
- 29.Judd A, et al. Evaluation of a modified commercial assay in detecting antibody to hepatitis C virus in oral fluids and dried blood spots. J. Med. Virol. 2003;71:49–55. doi: 10.1002/jmv.10463. [DOI] [PubMed] [Google Scholar]
- 30.Kania D, et al. Combining rapid diagnostic tests and dried blood spot assays for point-of-care testing of human immunodeficiency virus, hepatitis B and hepatitis C infections in Burkina Faso, West Africa. Clin. Microbiol. Infect. 2013;19:E533–541. doi: 10.1111/1469-0691.12292. [DOI] [PubMed] [Google Scholar]
- 31.Larrat S, et al. Performance of an antigen-antibody combined assay for hepatitis C virus testing without venipuncture. J. Clin. Virol. 2012;55:220–225. doi: 10.1016/j.jcv.2012.07.016. [DOI] [PubMed] [Google Scholar]
- 32.Marques BL, et al. Evaluation of dried blood spot samples for hepatitis C virus detection and quantification. J. Clin. Virol. 2016;82:139–144. doi: 10.1016/j.jcv.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 33.Marques BL, et al. Dried blood spot samples: optimization of commercial EIAs for hepatitis C antibody detection and stability under different storage conditions. J. Med. Virol. 2012;84:1600–1607. doi: 10.1002/jmv.23379. [DOI] [PubMed] [Google Scholar]
- 34.McCarron B, et al. Hepatitis C antibody detection in dried blood spots. J. Viral Hepat. 1999;6:453–456. doi: 10.1046/j.1365-2893.1999.00197.x. [DOI] [PubMed] [Google Scholar]
- 35.Nandagopal P, et al. Evaluation of dried blood spot as an alternative specimen for the diagnosis of anti-HCV in resource-limited settings. Indian J. Med. Microbiol. 2014;32:208–210. doi: 10.4103/0255-0857.129867. [DOI] [PubMed] [Google Scholar]
- 36.Tejada-Strop A, et al. Disparate detection outcomes for anti-HCV IgG and HCV RNA in dried blood spots. J. Virol. Methods. 2015;212:66–70. doi: 10.1016/j.jviromet.2014.10.018. [DOI] [PubMed] [Google Scholar]
- 37.Tuaillon E, et al. Dried blood spot for hepatitis C virus serology and molecular testing. Hepatology. 2010;51:752–758. doi: 10.1002/hep.23407. [DOI] [PubMed] [Google Scholar]
- 38.Mossner BK, et al. Dried blood spots, valid screening for viral hepatitis and human immunodeficiency virus in real-life. World J. Gastroenterol. 2016;22:7604–7612. doi: 10.3748/wjg.v22.i33.7604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ross RS, et al. Detection of infections with hepatitis B virus, hepatitis C virus, and human immunodeficiency virus by analyses of dried blood spots–performance characteristics of the ARCHITECT system and two commercial assays for nucleic acid amplification. Virol J. 2013;10:72. doi: 10.1186/1743-422x-10-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poiteau L, et al. Performance of rapid diagnostic tests for the detection of antibodies to hepatitis C virus in whole blood collected on dried blood spots. J. Viral Hepat. 2016;23:399–401. doi: 10.1111/jvh.12501. [DOI] [PubMed] [Google Scholar]
- 41.Lima, M. P. d. Aplicabilidade do sangue seco em papel de filtro para estudos de prevalência da infecção pelo vírus da hepatite C em grupos [master’s thesis on the internet], Instituto Oswaldo Cruz (2015).
- 42.O’Brien JM, et al. Detection of Hepatitis C Antibody With At-Home Collection Kits Using an Innovative Laboratory Algorithm. Infectious Diseases in Clinical Practice. 2001;10:474–480. doi: 10.1097/00019048-200112000-00004. [DOI] [Google Scholar]
- 43.Rice P, Abou-Saleh MT. Detecting Antibodies to Hepatitis C in Injecting Drug Users: A Comparative Study Between Saliva, Serum, and Dried Blood Spot Tests. Addictive Disorders & Their Treatment. 2012;11:76–83. doi: 10.1097/ADT.0b013e31822afd5c. [DOI] [Google Scholar]
- 44.Santos C, et al. The use of real-time PCR to detect hepatitis C virus RNA in dried blood spots from Brazilian patients infected chronically. J. Virol. Methods. 2012;179:17–20. doi: 10.1016/j.jviromet.2011.06.012. [DOI] [PubMed] [Google Scholar]
- 45.Saludes V, et al. Community-based screening of hepatitis C with a one-step RNA detection algorithm from dried-blood spots: Analysis of key populations in Barcelona, Spain. J. Viral Hepat. 2018;25:236–244. doi: 10.1111/jvh.12809. [DOI] [PubMed] [Google Scholar]
- 46.Bennett S, et al. Detection of hepatitis C virus RNA in dried blood spots. J. Clin. Virol. 2012;54:106–109. doi: 10.1016/j.jcv.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 47.De Crignis E, Re MC, Cimatti L, Zecchi L, Gibellini D. HIV-1 and HCV detection in dried blood spots by SYBR Green multiplex real-time RT-PCR. J. Virol. Methods. 2010;165:51–56. doi: 10.1016/j.jviromet.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 48.Solmone M, et al. Simple and reliable method for detection and genotyping of hepatitis C virus RNA in dried blood spots stored at room temperature. J. Clin. Microbiol. 2002;40:3512–3514. doi: 10.1128/JCM.40.9.3512-3514.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Santos C, et al. The use of real-time PCR to detect hepatitis C virus RNA in dried blood spots from Brazilian patients infected chronically. Journal of Virological Methods. 2012;179:17–20. doi: 10.1016/j.jviromet.2011.06.012. [DOI] [PubMed] [Google Scholar]
- 50.Stengel D, Bauwens K, Sehouli J, Ekkernkamp A, Porzsolt F. A likelihood ratio approach to meta-analysis of diagnostic studies. J. Med. Screen. 2003;10:47–51. doi: 10.1258/096914103321610806. [DOI] [PubMed] [Google Scholar]
- 51.Petruzziello A, Marigliano S, Loquercio G, Cozzolino A, Cacciapuoti C. Global epidemiology of hepatitis C virus infection: An up-date of the distribution and circulation of hepatitis C virus genotypes. World J. Gastroenterol. 2016;22:7824–7840. doi: 10.3748/wjg.v22.i34.7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fraser H, et al. Model projections on the impact of HCV treatment in the prevention of HCV transmission among people who inject drugs in Europe. J. Hepatol. 2018;68:402–411. doi: 10.1016/j.jhep.2017.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Platt L, et al. Needle syringe programmes and opioid substitution therapy for preventing hepatitis C transmission in people who inject drugs. Cochrane Database Syst Rev. 2017;9:CD012021. doi: 10.1002/14651858.CD012021.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martin NK, et al. Prioritization of HCV treatment in the direct-acting antiviral era: An economic evaluation. J. Hepatol. 2016;65:17–25. doi: 10.1016/j.jhep.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martin Natasha K, Hickman Matthew, Miners Alec, Hutchinson Sharon J, Taylor Avril, Vickerman Peter. Cost-effectiveness of HCV case-finding for people who inject drugs via dried blood spot testing in specialist addiction services and prisons. BMJ Open. 2013;3(8):e003153. doi: 10.1136/bmjopen-2013-003153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.