Abstract
Background and Purpose
Protein inhibitor of activated STAT1 (PIAS1) is phosphorylated by IKKα at Ser90 in a PIAS1 E3 ligase activity‐dependent manner. Whether PIAS1 is also phosphorylated at other residues and the functional significance of these additional phosphorylation events are not known. The transcription factor Elk‐1 remains SUMOylated under basal conditions, but the role of Elk‐1 SUMOylation in brain is unknown. Here, we examined the functional significance of PIAS1‐mediated Elk‐1 SUMOylation in Alzheimer's disease (AD) using the APP/PS1 mouse model of AD and amyloid β (Aβ) microinjections in vivo.
Experimental Approach
Novel phosphorylation site(s) on PIAS1 were identified by LC–MS/MS, and MAPK/ERK‐mediated phosphorylation of Elk‐1 demonstrated using in vitro kinase assays. Elk‐1 SUMOylation by PIAS1 in brain was determined using in vitro SUMOylation assays. Apoptosis in hippocampus was assessed by measuring GADD45α expression by western blotting, and apoptosis of hippocampal neurons in APP/PS1 mice was assessed by TUNEL assay.
Key Results
Using LC–MS/MS, we identified a novel MAPK/ERK‐mediated phosphorylation site on PIAS1 at Ser503 and showed this phosphorylation determines PIAS1 E3 ligase activity. In rat brain, Elk‐1 was SUMOylated by PIAS1, which decreased Elk‐1 phosphorylation and down‐regulated GADD45α expression. Moreover, lentiviral‐mediated transduction of Elk‐1‐SUMO1 reduced the number of hippocampal apoptotic neurons in APP/PS1 mice.
Conclusions and Implications
MAPK/ERK‐mediated phosphorylation of PIAS1 at Ser503 determines PIAS1 E3 ligase activity. Moreover, PIAS1 mediates SUMOylation of Elk‐1, which functions as an endogenous defence mechanism against Aβ toxicity in vivo. Targeting Elk‐1 SUMOylation could be considered a novel therapeutic strategy against AD.
What is already known
Elk‐1 is SUMO‐modified in the cell under basal conditions.
PIAS1 is phosphorylated by IKKalpha at Ser‐90.
What this study adds
PIAS1 Ser‐503 phosphorylation by MAPK/ERK determines PIAS1 E3 ligase activity.
Elk‐1 SUMOylation by PIAS1 functions as an endogenous defense mechanism against amyloid‐beta toxicity in APP/PS1 mice.
What is the clinical significance
Targeting Elk‐1 SUMOylation could be a novel therapeutic strategy against AD.
Abbreviations
- AD
Alzheimer's disease
- APP
amyloid precursor protein
- Aβ
amyloid β
- GADD45α
growth arrest and DNA damage‐inducible 45α
- PIAS1
protein inhibitor of activated STAT1
- SUMO
small ubiquitin‐like modifier
1. INTRODUCTION
One of the two pathological hallmarks of Alzheimer's disease (AD) in the brains of AD patients is the accumulation of senile plaques, composed largely of amyloid β (Aβ) peptides (Aβ1–40 and Aβ1–42). Aβ, generated by sequential proteolytic cleavage of amyloid precursor protein (APP) by β‐secretase and γ‐secretase (De Strooper & Annaert, 2000), initiates a detrimental cascade that increases lipid peroxidation, free radical production, caspase activation, and DNA damage, eventually leading to neuronal death (Butterfield, Drake, Pocernich, & Castegna, 2001; Dickson, 2004; Hardy & Selkoe, 2002). However, neurons have the capability of developing endogenous defence mechanisms to cope with Aβ toxicity. For example, soluble APPα has been shown to promote cell survival through activation of neuroprotectin D1 (Lukiw & Bazan, 2006). In addition, we have previously shown that acute Aβ exposure increases the expression of Mcl‐1, which provides neuroprotection through activation of the MAPK/ERK‐SGK (serum and glucocorticoid‐inducible kinase)‐STAT1/STAT2 signalling pathway (Hsu, Chiu, Tai, Ma, & Lee, 2009). More recently, we found that acute Aβ exposure induces the expression of protein inhibitor of activated STAT1 (PIAS1), which enhances the SUMOylation of histone deacetylase 1 and increases the expression of neprilysin to provide endogenous neuroprotection against Aβ toxicity (Tao, Hsu, Ma, Cheng, & Lee, 2017). However, there may be other protective mechanisms that remain to be explored.
PIAS1, identified as an inhibitor of STAT1, has been shown to block the DNA‐binding activity of STAT1 and inhibit its transcriptional activity in response to cytokine stimulation (Liao, Fu, & Shuai, 2000; Liu et al., 1998). PIAS1 also inhibits IFN‐inducible gene expression and plays an important role in the innate immune response through negative regulation of STAT1 (Liu et al., 2004). PIAS1 is also a small ubiquitin‐like modifier (SUMO) E3 ligase that facilitates the transfer of the SUMO molecule from UBC9 (E2) to substrate proteins and increases SUMOylation specificity (Gareau & Lima, 2010; Schmidt & Muller, 2003). PIAS1 was found to enhance the SUMOylation of Sp3 and suppress its transcriptional activity (Sapetschnig et al., 2002). PIAS1 and PIASxβ can also SUMOylate c‐Jun and down‐regulate AP1 transcriptional activity (Bossis et al., 2005). In the brain, PIAS1 was found to facilitate learning and memory performance in rats by enhancing the SUMOylation of several transcription factors, including STAT1, CREB, and Smad4 (Chen, Hsu, Ma, Tai, & Lee, 2014; Hsu, Ma, Liu, & Lee, 2017; Tai, Hsu, Liu, Ma, & Lee, 2011). In the context of the present study, PIAS1 was shown to enhance the SUMOylation of Hes‐1 to inhibit apoptosis (Chiou, Liu, Lin, & Lee, 2014). But whether PIAS1 also promotes the SUMOylation of other transcription factors that play a neuroprotective role against Aβ toxicity remains to be investigated.
Ternary complex factors represent a subfamily of ETS domain transcription factors. Elk‐1, one of three ternary complex factors in mammals, is found in both cytoplasmic and nuclear compartments as well as processes of adult rat brain neurons (Sgambato et al., 1998) and plays a role in the regulation of cell growth, differentiation, migration, and survival (for review, see Kasza, 2013). Elk‐1 is phosphorylated directly by MAPKs at Ser383 and Ser389 (Janknecht, Ernst, Pingoud, & Nordheim, 1993). In the nervous system, Elk‐1 phosphorylation by MAPK/ERK (extracellular signal‐regulated kinase) was found to be involved in contextual fear memory formation in mice (Sananbenesi, Fischer, Schrick, Spiess, & Radulovic, 2002). Avoidance learning was found to activate Elk‐1 and stimulate c‐Fos production in the rat hippocampus (Cammarota et al., 2000). Moreover, high‐frequency tetanic stimulation of the perforant path induces LTP concomitant with Elk‐1 hyperphosphorylation and zif268 expression (Davis, Vanhoutte, Pages, Caboche, & Laroche, 2000). Induction of long‐term depression also results in Elk‐1 phosphorylation (Thiels, Kanterewicz, Norman, Trzaskos, & Klann, 2002). In addition to being phosphorylated, Elk‐1 is also post‐translationally modified by conjugation of the SUMO molecule to the R motif of Elk‐1, which represses the transcriptional activity of Elk‐1 (S. H. Yang, Bumpass, Perkins, & Sharrocks, 2002; S. H. Yang, Jaffray, Hay, & Sharrocks, 2003). SUMOylation of Elk‐1 also regulates Elk‐1 nuclear retention and reduces its nucleo‐cytoplasmic shuttling (Salinas et al., 2004). Upon activation of MAPK signalling pathways, Elk‐1 is phosphorylated and de‐SUMOylated, which allows Elk‐1 to exert its transcriptional activity (S. H. Yang et al., 2003). Thus, the regulation of Elk‐1 transcriptional activity reflects the interplay between Elk‐1 phosphorylation and Elk‐1 SUMOylation. However, although the role of Elk‐1 phosphorylation in neuronal plasticity has been examined, little is known about the role and function of Elk‐1 SUMOylation in the brain, apart from the fact that Elk‐1 remains SUMOylated under resting conditions. The aim of the present study was to examine the role and mechanism of Elk‐1 SUMOylation. We found that PIAS1 phosphorylation mediates Elk‐1 SUMOylation and that this modification functions as an endogenous defence mechanism in the protection against Aβ toxicity in vivo.
2. METHODS
2.1. Animals
Adult male Sprague–Dawley rats (2–3 months old, 250–350 g, [RRID:MGI:5651135]), adult male and female wild‐type mice (strain name: C57BL/6J, stock number: 000664; 9–10 months old, 38–43 g, [RRID:IMSR_JAX:000664]), and adult male and female APP/PS1 transgenic mice (strain name: B6.Cg‐Tg (APPswe,PSEN1dE9)85Dbo/Mmjax, stock number: 005864; 9–10 months old, 38–43 g, [RRID:MGI:5701399]) were used in this study. The APP/PS1 mice and WT mice were purchased from Jackson Laboratory (Bar Harbor, ME, USA). The reason for using rats is because repeated drug injections are applied in one experiment that requires cannulation for precise localization of the injection area, but it is not feasible to do this cannulation in mice. Moreover, the amount of tissue lysate obtained from the mouse CA1 area is not enough for the SUMOylation assay. But we have tested our hypothesis in APP/PS1 mice for the last part of this study. Animals were bred at the Animal Facility of the Institute of Biomedical Sciences (IBMS), Academia Sinica, Taiwan. They were housed and maintained on a 12/12 hr light/dark cycle (lights on at 8:00 am) with food and water continuously available. Animal studies are reported in compliance with the ARRIVE guidelines (Kilkenny, Browne, Cuthill, Emerson, & Altman, 2010) and with the recommendations made by the British Journal of Pharmacology. Experimental procedures follow the Guidelines of Animal Use and Care of the National Institute of Health (NIH) and were approved by the Animal Committee of IBMS, Academia Sinica.
2.2. Randomization and blinding
Blinding and random assignment of animals to different groups in this study were similar to that of another study (Anton et al., 2018) and in accordance of the guidelines of BJP. The wild‐type animals were randomly divided to control and experimental groups. For the APP/PS1 mice, they were randomly divided to different subgroups and received different lentiviral injections. Blinding was adopted as much as possible. Drugs, plasmid DNA, and lentiviral vector injections were made by one experimenter. Tissue collection was performed by two different experimenters. Biochemical assays and TUNEL staining were conducted by different experimenters, excluding the experimenter doing the drug and DNA injections. Quantification of blots and statistical analysis were performed by two different investigators.
2.3. Intra‐hippocampal drug infusion, plasmid DNA transfection, and siRNA injection
Rats were anaesthetized with pentobarbital (40 mg·kg−1, i.p.) and subjected to stereotaxic surgery. Two 23‐gauge, stainless steel, thin‐wall cannulae were implanted bilaterally to the CA1 area of rat brain at the following coordinates: 3.5 mm posterior to the bregma, ±2.5 mm lateral to the midline, and 3.4 mm ventral to the skull surface. After recovery from the surgery, Aβ (20 μg·μl−1) and U0126 (2.8 or 1.4 μg·μl−1) were directly injected to the CA1 area at a rate of 0.1 μl·min−1. A total of 0.7 μl was injected to each side. For experiments that only require transient plasmid DNA expression in the CA1 area, the non‐viral vector transfection reagent polyethyleneimine (PEI) was used, because we have previously demonstrated that PEI is efficient at plasmid transfection, and it is not toxic to hippocampal neurons (Chao, Ma, & Lee, 2011). Before injection, plasmid DNA was diluted in 5% glucose to a stock concentration of 2.77 μg·μl−1. Branched PEI of 25 kDa (Sigma‐Aldrich) was diluted to 0.1 M concentration in 5% glucose. Immediately before injection, 0.1 M PEI was added to the DNA solution (0.45 μl PEI and 0.55 μl plasmid DNA) to reach a ratio of PEI nitrogen per DNA phosphate equal to 10. The final concentration of the plasmid DNA was 1.5 μg·μl−1. For PIAS1 siRNA preparation, it was diluted in distilled water and stored at a stock concentration of 20 pmol·μl−1. Before injection, 0.1 M PEI was added to PIAS1 siRNA (1:1, v/v), and the final concentration of PIAS1 siRNA was 10 pmol·μl−1. The sequences for PIAS1 siRNA were 5′‐UCCGGAUCAUUCUAGAGCUtt‐3′ (sense) and 5′‐AGCUCUAGAAUGAUCCGGAtt‐3′ (antisense). The Silencer Negative Control number 1 siRNA (control siRNA) was used as a control. These were the siRNAs with sequences that do not target any gene product (Ambion, Austin, TX, USA). PIAS1 siRNA and control siRNA were synthesized by Ambion. The mixture was subjected to vortex for 30 s and allowed to equilibrate for 15 min before injection. The injection volume was 0.7 μl each side. The injection rate was 0.1 μl·min−1. Animals were killed 48 hr after plasmid DNA transfection or siRNA injection, and their CA1 tissue was extracted for biochemical assays. For drug infusion, Aβ (20 μg·μl−1) and U0126 (2.8 or 1.4 μg μl−1) were directly injected to the CA1 area at a rate of 0.1 μl·min−1. A total of 0.7 μl was injected to each side. Animals were killed at different time intervals, and their CA1 tissue was extracted for biochemical assays.
2.4. Intra‐hippocampal lentiviral vector transduction
The WT and APP/PS1 mice were anaesthetized with pentobarbital (40 mg·kg−1, i.p.) and subjected to stereotaxic surgery with direct injection of lentivirus particle (pLenti‐Flag‐tagged Elk‐1, pLenti‐Flag‐tagged Elk‐1‐sumo1, or pLenti‐Tri‐cistronic vector) to their CA1 area (without cannulation) at the following coordinates: −1.8 mm posterior to the bregma, ±1.3 mm lateral to the midline, and −2.1 mm ventral to the skull surface. Before injection, the lentivirus particles were diluted in PBS (pH 7.4) to a titre of 3.5 × 107 μl−1 at least but with comparable concentrations. The volume for each lentiviral vector injection was 0.25 μl each side. The injection rate was 0.1 μl min−1.
2.5. Elk‐1 SUMOylation assay in hippocampal tissue
Hippocampal CA1 tissue lysates were prepared in the same way as that prepared for Western blots. For IP Elk‐1, the clarified lysate (0.5 mg) was immunoprecipitated with 3 μl of anti‐Elk‐1 antibody at 4°C overnight. The protein A agarose beads (30 μl, 50% slurry, GE Healthcare) were added to the IP reaction product to catch the immune complex at 4°C for 3 hr. The immune complex on beads was washed three times with washing buffer containing 50 mM Tris–HCl (pH 7.4), 150 mM NaCl, 2 mM EDTA, and 0.5% IGEPAL CA‐630 and subjected to the SUMOylation reaction with the addition of recombinant PIAS1 protein (3 μl, Catalogue No. BML‐UW9960, Enzo Life Sciences, Farmingdale, NY, USA), E1 (1 μl), E2 (1 μl), and the SUMO1 (0.5 μl) proteins provided in the kit. The SUMO protease inhibitor N‐ethylmaleimide was also added to the reaction. SUMOylation assay was performed using the SUMO linkTM kit according to the manufacturer's instructions (Active Motif) and boiled in sample buffer at 95°C for 10 min. The SUMOylation product was then subjected to 8% SDS‐PAGE and then transferred onto the PVDF membrane. The membrane was immunoblotted with anti‐Elk‐1 antibody or anti‐SUMO1 antibody. For determination of endogenous Elk‐1 SUMOylation by PIAS1, no recombinant PIAS1 protein was added to the reaction (Figure 5c). The immuno‐related procedures used comply with the recommendations made by the British Journal of Pharmacology.
Figure 5.
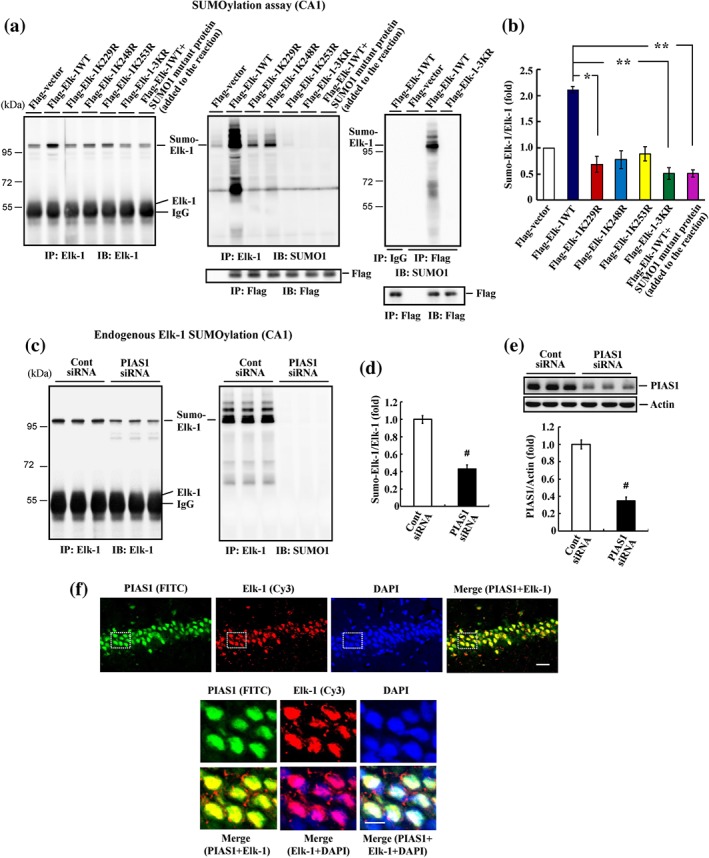
Elk‐1 is SUMO‐modified by PIAS1 endogenously in the hippocampus. (a) Flag‐vector, Flag‐Elk‐1WT (with or without SUMO1 mutant protein added to the reaction), individual Flag‐Elk‐1 mutant plasmid, or Flag‐Elk‐1 3KR mutant plasmid was transfected to rat hippocampal CA1 area, and Elk‐1 SUMOylation was determined by SUMOylation assay 48 hr later. Anti‐Elk‐1 antibody (left) and anti‐SUMO1 antibody (middle) were used for immunoblotting after immunoprecipitation with anti‐Elk‐1 antibody. Immunoprecipitation and western blot using anti‐Flag antibody confirm the transfection and expression of Flag‐tagged plasmids (middle‐lower panel). Flag‐vector, Flag‐Elk‐1WT, or Flag‐Elk‐1 3KR plasmid was transfected to rat CA1 area, and tissue lysate was immunoprecipitated with anti‐Flag antibody and immunoblotted with anti‐SUMO1 antibody. An additional group of rats received Flag‐Elk‐1WT transfection but was immunoprecipitated with IgG to serve as the control. Tissue lysate was similarly immunoprecipitated and immunoblotted with anti‐Flag antibody to confirm the transfection and expression and Flag‐tagged plasmids (right‐lower panel). (b) The quantified result of Elk‐1 SUMOylation (n = 5 different animals for each group). * P < 0.05 for Elk‐1K229R versus Elk‐1WT group and ** P < 0.05 for Elk‐1 3KR versus Elk‐1WT group and for Elk‐1WT + SUMO1 mutant protein versus Elk‐1WT group. (c) Control siRNA or PIAS1 siRNA was transfected to the rat CA1 area, and endogenous Elk‐1 SUMOylation was determined 48 hr later as described above but without the addition of the recombinant PIAS1 protein. (d) The quantified result of Elk‐1 SUMOylation (n = 5 different animals for each group). # P < 0.05. (e) Western blot of PIAS1 expression and the quantified result (n = 5 different animals for each group). # P < 0.05. (f) Immunohistochemistry showing PIAS1 (green) and Elk‐1 (red) are both present in the nucleus of the same neurons in the CA1 area of the rat brain (n = 3 different animals for each group). The lower panel is the magnification of the selected area from the upper panel. Scale bar is 25 μm for the upper panel and 10 μm for the lower panel. Data are mean ± SEM
2.6. TUNEL staining
Wild‐type or APP/PS1 mice received pLenti‐Flag‐tagged Elk‐1, pLenti‐Flag‐tagged Elk‐1‐sumo1, or pLenti‐Tri‐cistronic vector transduction as described above, and their CA1 tissue was subjected to TUNEL staining for detection of apoptotic cells according to the manufacturer's protocols (Millipore). This was achieved by using the Apoptag plus peroxidase in situ apoptosis detection kit. Briefly, brain sections (30‐μm thickness) were fixed in 4% paraformaldehyde for 10 min and were permeabilized with pre‐chilled EtOH/CH3COOH (2:1) for 10 min at −20°C followed by reacting with 3% H2O2 for 5 min to remove the endogenous peroxidase activity. The sections were then incubated with the TdT enzyme for 1 hr at 37°C followed by incubation with anti‐digoxigenin peroxidase conjugate for 30 min. After the specimen had been washed with PBS, 3,3‐diaminobenzidine peroxidase substrate was applied to the specimen for 3 to 6 min for colour development. Apoptotic nuclei developed a brown colour with 3,3‐diaminobenzidine staining. The slides were then counterstained with methyl blue dye for visualization of total cells. Cells were examined under a Leica DM IL LED light microscope.
2.7. Western blot
Cell lysates were resolved by 8~12% SDS‐PAGE and transferred onto the PVDF membrane (Millipore). Blots were blocked for 1hr in 2%BSA and then washed. Western blot was conducted using the following antibodies: anti‐PIAS1 (1:10000, Cat#2474‐1, rabbit monoclonal IgG, Epitomics, Burlingame, CA), anti‐Elk‐1 (1:2000, Cat#sc‐355, rabbit polyclonal IgG, Santa Cruz Biotechnology, Santa Cruz, CA, RRID:AB_631429), anti‐p383‐Elk‐1 (1:2000, Cat#PAB25225, rabbit polyclonal IgG, Abnova, Taipei, Taiwan), anti‐GADD45α (1:2000, Cat#GTX54090, rabbit polyclonal IgG, GeneTex, San Antonio, TX), anti‐SUMO1 (1:10000, part#101897 from SUMOlink kit, Cat#40120, Active Motif, Carlsbad, CA), anti‐p53 (1:10000, part#100853 from SUMOlink kit, Cat#40120, Active Motif), anti‐Flag M2 (1:5000, Cat#F1804, mouse monoclonal IgG, Sigma‐Aldrich, RRID:AB_2637089), anti‐Myc (1:5000, Cat#05‐419, mouse monoclonal IgG1, Millipore, RRID:AB_309725), anti‐GFP (1:5000, Cat#2955, mouse monoclonal IgG2a, Cell Signaling, Danvers, MA, RRID:AB_1196614), anti‐ERK1/2 (1:5000, Cat#4695, rabbit monoclonal IgG, Cell Signaling, RRID:AB_390779), anti‐phospho‐ERK1/2 (1:5000, Cat#4376, rabbit monoclonal IgG, Cell Signaling, RRID:AB_331772), anti‐Akt (1:5000, Cat#9272, rabbit polyclonal IgG, Cell Signaling, RRID:AB_329827), anti‐His (1:5000, Cat#05‐531, mouse monoclonal IgG2a, Millipore, RRID:AB_309786), anti‐GST (1:5000, Cat#05‐311, mouse monoclonal IgG, Millipore, RRID:AB_309675) and anti‐actin (1:100000, Cat#MAB1501, mouse monoclonal IgG2b, Millipore, RRID:AB_2223041). The secondary antibody used was HRP‐conjugated goat‐anti rabbit IgG antibody (1:8000, Cat#111‐035‐003, Jackson ImmunoResearch, West Grove, PA) or goat‐anti mouse IgG antibody (1:8000, Cat#115‐035‐003, Jackson ImmunoResearch). Membrane was developed by reacting with chemiluminescence HRP substrate (Millipore) and exposed to the LAS‐3000 image system (Fujifilm, Tokyo, Japan) for visualization of protein bands. The protein bands were quantified by using Image J Software (verison 1.50i, National Institutes of Health, Bethesda, MD). The band densities values of protein were normalized to the internal control β‐actin or Elk‐1. The data obtained from control groups and expressed as fold change relative to the control group. Western blots follow the guidelines of Alexander et al. (2018).
For generation of the phospho(p)‐Ser503 PIAS1 antibody, the peptide containing the HQASPVSRTPSLPA sequence was synthesized by the LTK BioLaboratories Company (Taoyuan, Taiwan) and injected into the rabbit to obtain the custom‐made polyclonal antibody. Our tests showec this antibody is suitable for Western blot experiments.
2.8. Data and statistical analysis
For each animal experiment carried out in vivo (except the TUNEL staining), at least five different animals were included in each group. This is adopted based on our previous results that an n number of 5 clearly demonstrates the effect of Aβ and other treatment‐mediated signalling changes and endogenous protein SUMOylation alterations when comparisons were made between groups (Tai et al., 2016; Tao et al., 2017). This is also for the purpose of carrying out statistical analysis according to the guidelines of BJP (Curtis et al., 2018). For the TUNEL assay carried out in vivo, only three different animals were included in each group. This is adopted based on our previous experience that an n number of 3 clearly distinguishes cell apoptosis between groups (Chiou et al., 2014). Further, we have quantified the apoptotic cells in three different tissue sections for each animal. That means we have obtained data from nine tissue sections for each group. For the experiments carried out in vitro, the repeat numbers are two, three, or four for each group. This is because some of these experiments are technical repeats, such as Figure 1a, or the control group has no signals, such as Figure 3a,b. This is also based on our previous experience that these n numbers reliably identify the candidate SUMOylation residues by using LC–MS/MS chromatography and effectively distinguish the different levels of phosphorylation, SUMOylation, and gene expression between groups (Lin, Liu, & Lee, 2016; Tai et al., 2016). For the Western blot and SUMOylation experiments, the results are expressed as “fold difference” compared to the corresponding control value, and the control value was normalized to 1.0. The reason for this normalization is because these results (protein bands) were resolved from the gel, whereas the intensity of protein bands for the same treatment is different on different gels. Direct comparison cannot be made from different gels.
Figure 1.
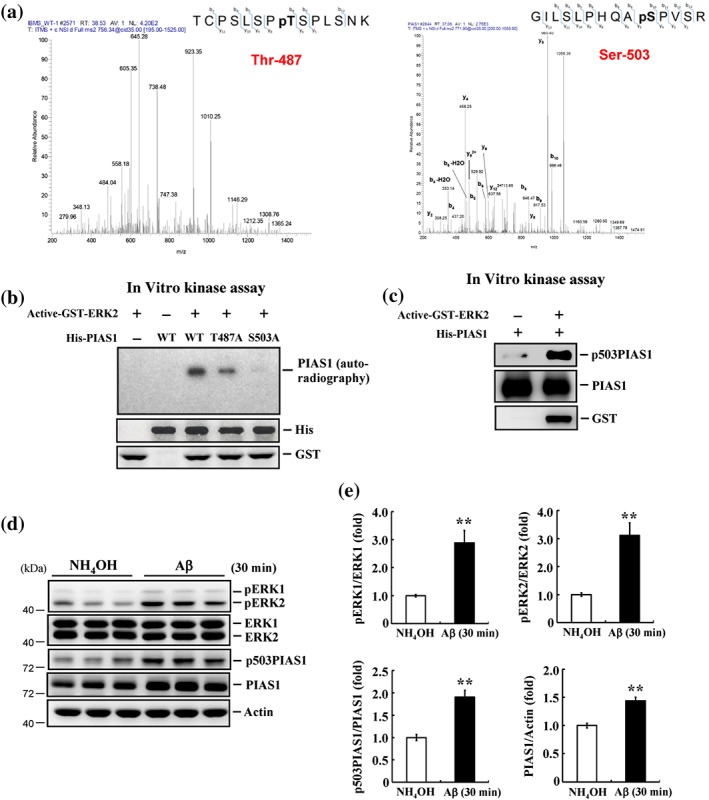
PIAS1 is phosphorylated at Thr487 and Ser503 by MAPK/ERK and acute Aβ induces MAPK/ERK activation and PIAS1 Ser503 phosphorylation. (a) LC–MS/MS chromatography prediction of potential phosphorylation sites on PIAS1 at Thr487 (left) and Ser503 (right). (b) His‐tagged PIAS1WT, PIAS1T487A mutant, and PIAS1S503A mutant recombinant proteins (1 μg each) were incubated with activated GST‐ERK2 protein (30 ng) and 6 μCi of [γ‐32P] ATP (100 μM) for 30 min for kinase reaction and Western blot using anti‐His and anti‐GST antibody. (c) His‐tagged PIAS1 fusion protein (1 μg) was incubated with or without activated GST‐ERK2 (30 ng) and ATP (100 μM) for 30 min for kinase reaction and Western blot. Antibodies for p503PIAS1, PIAS1, and GST were used for Western blot. The p503PIAS1 antibody recognizes phosphorylated PIAS1 only. Results are from two independent experiments for (a), (b), and (c). (d) NH4OH (1%) or Aβ (14 μg) was injected to rat hippocampal CA1 area, and the expression level of pERK1/2, ERK1/2, pSer503 PIAS1, and PIAS1 was determined by Western blot 30 min later. (e) The quantified results of the above measures (n = 6 different animals for each group). ** P < 0.05 for Aβ versus NH4OH group. Data are expressed as mean ± SEM
Figure 3.
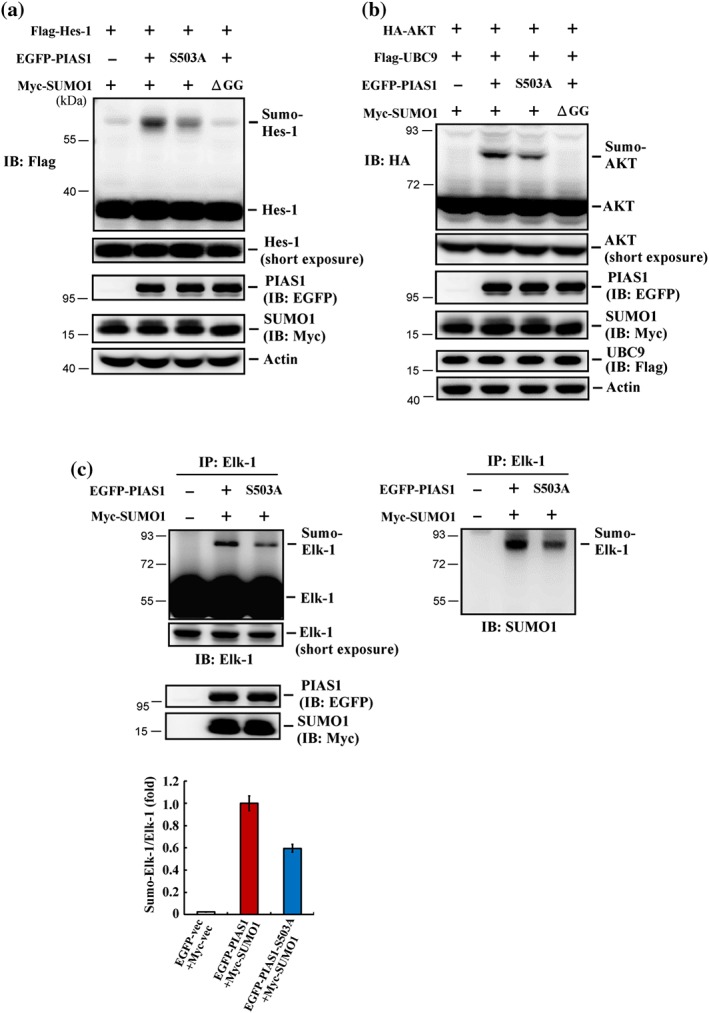
Blockade of PIAS1 Ser503 phosphorylation decreases the SUMO E3 ligase activity of PIAS1 in HEK293T cells. (a) Flag‐Hes‐1 plasmid, EGFP‐PIAS1 (or EGFP‐PIAS1S503A) plasmid, and Myc‐SUMO1 (or Myc‐SUMO1ΔGG) plasmid were co‐transfected to HEK293T cells, and Hes‐1 SUMOylation was determined by Western blot 48 hr later using anti‐Flag antibody. Experiments are in two repeats. (b) HA‐Akt plasmid, Flag‐UBC9 plasmid, together with EGFP‐PIAS1 (or EGFP‐PIAS1S503A) plasmid and Myc‐SUMO1 (or Myc‐SUMO1ΔGG) plasmid were co‐transfected to HEK293T cells, and Akt SUMOylation was determined by Western blot 48 hr later using anti‐HA antibody. Results are from two independent repeats. (c) EGFP‐PIAS1 (or EGFP‐PIAS1S503A) plasmid was co‐transfected with Myc‐SUMO1 plasmid to HEK293T cells. Elk‐1 SUMOylation was determined 48 hr later by immunoprecipitation with anti‐Elk‐1 antibody and immunoblotting with anti‐Elk‐1 antibody (upper‐left panel). Four independent experiments were carried out, and the quantified result is shown in the left‐lower panel. Data are mean ± SEM. The membrane was stripped and re‐blotting with anti‐SUMO1 antibody (right panel)
The data and statistical analysis comply with the recommendations on experimental design and analysis in pharmacology (Curtis et al., 2018). Data from experiments of n values ≧5 were analysed by Student's t test (two groups) or one‐way ANOVA (three or more groups) when there was no significant variance inhomogeneity. Post hoc Newman–Keuls comparisons were further conducted only if the ANOVA value reached a statistically significant level. For those experiments of n values ≧ 5 but with very small or no SEM values in the control group with mean value normalized to 1.0, non‐parametric Mann–Whitney U test (two groups) or Kruskal–Wallis one‐way ANOVA (three or more groups) were used. Further comparisons after the Kruskal–Wallis test were made by the post hoc Dunn's method. For experiments of n value <5, no statistical analysis was performed. Values of P < 0.05 were considered statistically significant.
2.9. Materials
Recombinant human Aβ (1‐42) protein was purchased from GenScript (Piscataway, NJ). It was dissolved in 1% NH4OH before injection. The MEK inhibitor U0126 was purchased from Millipore (Bedford, MA). It was dissolved in 15% DMSO and diluted with PBS before injection. Nerve growth factor (NGF) was purchased from Sigma‐Aldrich (St. Louis, MO). It was dissolved in PBS before injection.
2.10. Nomenclature of targets and ligands
Key protein targets and ligands in this article are hyperlinked to corresponding entries in http://www.guidetopharmacology.org, the common portal for data from the IUPHAR/BPS Guide to PHARMACOLOGY (Harding et al., 2018), and are permanently archived in the Concise Guide to PHARMACOLOGY 2017/18 (Alexander, Fabbro, et al., 2017; Alexander, Kelly, et al., 2017).
3. RESULTS
3.1. PIAS1 is phosphorylated at Thr487 and Ser503 by MAPK/ERK, and acute Aβ exposure in vivo induces MAPK/ERK activation and PIAS1 Ser503 phosphorylation
A previous study showed that PIAS1 is phosphorylated by IKKα at Ser90 and that this reduces inflammation induced by pro‐inflammatory stimuli (Liu et al., 2007). But whether PIAS1 is phosphorylated at other residues and what physiological functions these other phosphorylation events might mediate are not known. Here, we used LC–MS/MS analysis to identify candidate phosphorylation residues on PIAS1. This analysis revealed two phosphorylation sites on PIAS1: Thr487 and Ser503 (Figure 1a). Because MAPK/ERK signalling is known to mediate neuroprotection against glutamate toxicity and Aβ toxicity (Almeida et al., 2005; Hsu et al., 2009), we next performed in vitro kinase assays to determine whether PIAS1 could be phosphorylated by MAPK/ERK at these two residues. To this end, we incubated active GST‐tagged ERK2 protein with His‐tagged WT (PIAS1WT) or mutant (PIAS1T487A and PIAS1S503A) recombinant PIAS1 protein. This analysis revealed that PIAS1WT was phosphorylated by active ERK2, but phosphorylation of the T487A and S503A mutant proteins, especially the latter, was clearly reduced (Figure 1b). Because the phosphorylation signal was diminished to a greater degree in the S503A mutant protein, we targeted Ser503, generating a polyclonal antibody that specifically recognizes this phosphorylated residue. In vitro kinase assays followed by western blotting using the phospho‐Ser503 PIAS1 antibody revealed that ERK2 directly phosphorylated PIAS1 at Ser503 (Figure 1c). It has been previously shown that acute administration of Aβ activates MPAK/ERK and induces an increase in PIAS1 expression in the hippocampus that mediates neuroprotection (Tao et al., 2017). Because ERK2 phosphorylates PIAS1 at Ser503, we examined whether Aβ induces PIAS1 phosphorylation at Ser503. Rats were acutely exposed to NH4OH or Aβ (14 μg) in vivo by directly injecting these agents into the hippocampal CA1 area. Western blotting performed 30 min later revealed that acute Aβ exposure induced activation of ERK1 and ERK2. It also increased the level of Ser503‐phosphorylated PIAS1 and the level of PIAS1 expression (Figure 1d,e).
3.2. PIAS1 phosphorylation at Ser503 induced by Aβ injection is mediated by MAPK/ERK activation
The above results show that acute Aβ exposure induces MAPK/ERK activation and PIAS1 Ser503 phosphorylation but do not establish whether this phosphorylation of PIAS1 is mediated by MAPK/ERK. To address this, we first investigated whether inhibition of MAPK/ERK activity with U0126 decreased the level of Ser503‐phosphorylated PIAS1. Rats were divided into two groups and administered an intra‐hippocampal injection of DMSO or U0126 (2 μg). After 1 hr, hippocampal CA1 tissue was analysed by Western blotting. These analyses revealed that U0126 (2 μg) decreased the levels of phosphorylated ERK1 and ERK2. It also decreased levels of Ser503‐phosphorylated PIAS1 and the expression level of PIAS1 (Figure 2a,b). Using a sub‐threshold dose of U0126 (1 μg; Y. C. Yang, Ma, Liu, & Lee, 2011), we then examined block of PIAS1 Ser503 phosphorylation following acute administration of Aβ. Rats received DMSO (15%) + NH4OH (1%), DMSO + Aβ (14 μg), U0126 (1 μg) + NH4OH, or U0126 + Aβ injections into their CA1 area. Subsequent Western blot analyses revealed that acute Aβ exposure consistently increased the phosphorylation level of ERK1, ERK2, and Ser503 PIAS1, as well as the expression level of PIAS1. Notably, these effects of Aβ were blocked by 1 μg U0126, a dose that did not affect these measures by itself (Figure 2c,d).
Figure 2.
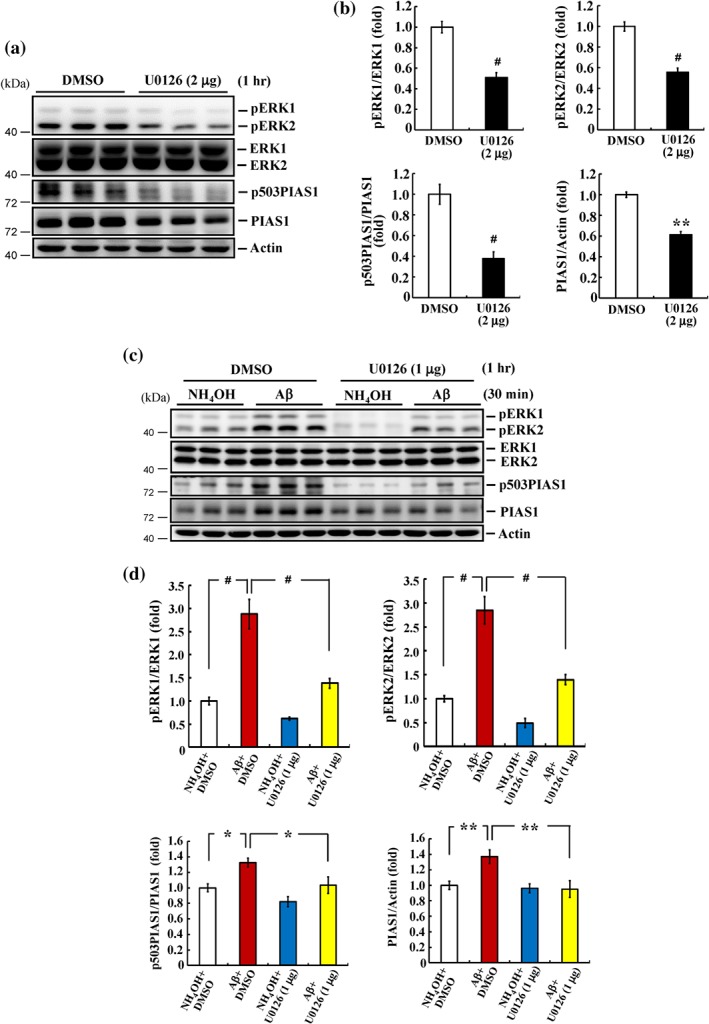
Aβ induction of PIAS1 phosphorylation at Ser503 is mediated through MAPK/ERK activation. (a) DMSO (15%) or U0126 (2 μg) was injected to rat hippocampal CA1 area, and the expression of pERK1/2, ERK1/2, pSer503 PIAS1, and PIAS1 was determined by Western blot 1 hr later. (b) The quantified results of these measures (n = 6 different animals for each group). ** P < 0.05 and # P < 0.05 for U0126 versus DMSO group. (c) DMSO (15%) or U0126 (1 μg) together with NH4OH (1%) or Aβ (14 μg) was injected to rat hippocampal CA1 area 30 min apart, and the expression of pERK1/2, ERK1/2, pSer503 PIAS1, and PIAS1 was determined by Western blot 30 min after NH4OH or Aβ injection. (d) The quantified results of these measures (n = 6 different animals for each group). * P < 0.05, ** P < 0.05, and # P < 0.05 comparing Aβ + DMSO versus NH4OH + DMSO group and * P < 0.05, ** P < 0.05, and # P < 0.05 comparing Aβ + U0126 versus Aβ + DMSO group. Data are expressed as mean ± SEM
3.3. Blockade of PIAS1 Ser503 phosphorylation decreases the SUMO E3 ligase activity of PIAS1 in HEK293T cells
Because PIAS1 is a SUMO E3 ligase, we next examined whether phosphorylation of PIAS1 at Ser503 affects its SUMO E3 ligase activity. The known PIAS1 substrates Hes‐1 (Chiou et al., 2014) and Akt (Lin et al., 2016) were used as positive controls. HEK293T cells were co‐transfected with a Flag‐Hes‐1 plasmid and EGFP‐PIAS1 (or EGFP‐PIAS1S503A) and Myc‐SUMO1 (or Myc‐SUMO1ΔGG) plasmids, after which Hes‐1 SUMOylation was examined by western blotting using an anti‐Flag antibody. This analysis revealed that Hes‐1 was SUMOylated upon co‐transfection of Flag‐Hes‐1, Myc‐SUMO1, and EGFP‐PIAS1WT but was decreased by co‐transfection of EGFP‐PIAS1S503A and completely blocked by co‐transfection of Myc‐SUMO1ΔGG (Figure 3a). Using an anti‐HA antibody in Western blot analyses, we further found that Akt was SUMOylated in HEK293T cells co‐transfected with HA‐Akt, Flag‐UBC9, EGFP‐PIAS1, and Myc‐SUMO1 plasmids. However, Akt SUMOylation was diminished in cells co‐transfected with EGFP‐PIAS1S503A instead of EGFP‐PIAS1WT and completely blocked in cells co‐transfected with Myc‐SUMO1ΔGG instead of Myc‐SUMO1 (Figure 3b). Next, we determined whether PIAS1 Ser503 phosphorylation affects Elk‐1 SUMOylation. Accordingly, we co‐transfected HEK293T cells with Myc‐SUMO1 plasmid together with EGFP‐PIAS1WT or EGFP‐PIAS1S503A plasmid and assessed Elk‐1 SUMOylation by immunoprecipitating Elk‐1 followed by immunoblotting for Elk‐1. This analysis showed that Elk‐1 was SUMOylated in cells co‐transfected with EGFP‐PIAS1WT and Myc‐SUMO1 plasmids but was significantly diminished in cells co‐transfected with EGFP‐PIAS1S503A instead of EGFP‐PIAS1WT (Figure 3c, left panel). To confirm that the up‐shifted band in Figure 3c is indeed SUMOylated Elk‐1, we stripped the membrane and re‐blotted with an anti‐SUMO1 antibody. This analysis revealed a SUMOylated Elk‐1 band at the same position on the gel and showed that this band was similarly diminished in cells transfected with EGFP‐PIAS1S503A instead of EGFP‐PIAS1WT (Figure 3c, right panel).
3.4. Elk‐1 is SUMOylated by PIAS1 at three lysine residues in HEK293T cells
The above results indicate that Elk‐1 could be SUMOylated by PIAS1 in HEK293T cells. Next, we screened for additional candidate SUMO sites on rat Elk‐1 using SUMO2.0 software. This analysis predicted four candidate lysine residues: Lys‐229, Lys‐248, Lys‐253, and Lys‐270 (corresponding to Lys‐230, Lys‐249, Lys‐254, and Lys‐271, respectively, in humans), three of which conformed to the consensus SUMO‐substrate motif (Figure 4a). We next generated individual mutants for each of these residues and co‐transfected HEK293T cells with each mutant or WT Elk‐1 (Flag‐tagged), together with EGFP‐PIAS1 and Myc‐SUMO1 plasmids. A subsequent western blot analysis revealed a SUMOylated Elk‐1 band in cells transfected with Flag‐Elk‐1WT plasmid and further showed that the intensity of this band was decreased in cells transfected with Flag‐Elk‐1K229R, Flag‐Elk‐1K248R, or Flag‐Elk‐1K253R mutant. Elk‐1 SUMOylation was not affected by transfection of Flag‐Elk‐1K270R (Figure 4b). Quantification of these results is shown in Figure 4c. We then generated a triple‐mutant Elk‐1 construct containing K229R, K248R, and K253R (Elk‐1 3KR) and transfected HEK293T cells with this plasmid or Elk‐1WT plasmid. We found that transfection of cells with Elk‐1 3KR completely blocked Elk‐1 SUMOylation (Figure 4d); quantification of these results is shown in Figure 4e. Because ubiquitination also takes place at lysine residues, we examined whether Elk‐1 ubiquitination also occurs at these three residues. Accordingly, HEK293T cells were co‐transfected with Flag‐tagged Elk‐1WT or Elk‐1 3KR plasmid together with a His‐ubiquitin plasmid, after which Elk‐1 was immunoprecipitated, and immunoprecipitates were probed by immunoblotting with an anti‐His antibody. This analysis revealed that the intensities of bands corresponding to ubiquitinated Elk‐1 were similar between cells transfected with Flag‐Elk‐1WT plasmid and those transfected with Flag‐Elk‐1 3KR plasmid (Figure S1).
Figure 4.
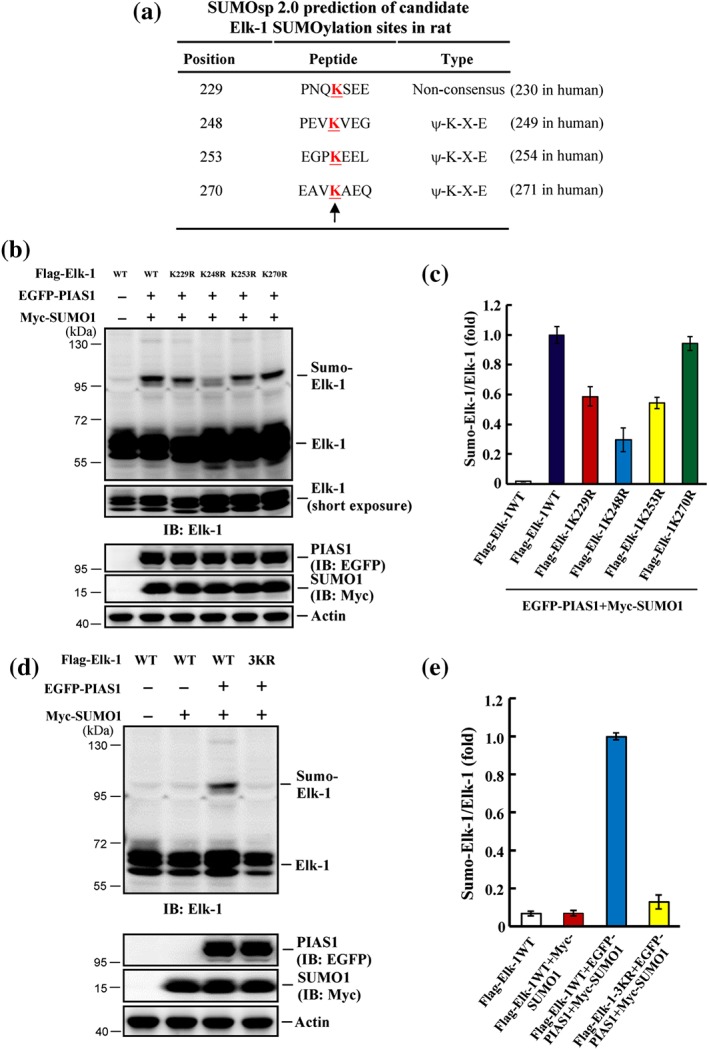
Elk‐1 is SUMO‐modified by PIAS1 at three lysine residues in HEK293T cells. (a) SUMO2.0 software prediction of candidate SUMO acceptors on Elk‐1. The “K” letters indicated by the arrow represent the candidate SUMO sites. (b) Flag‐Elk‐1WT plasmid and individual Elk‐1 mutant plasmid at K229, K248, K253, and K270 together with EGFP‐PIAS1 plasmid and Myc‐SUMO1 plasmid were transfected to HEK293T cells, and Elk‐1 SUMOylation was determined by Western blot 48 hr later using anti‐Elk‐1 antibody. (c) The quantified result of Elk‐1 SUMOylation was from three independent experiments. (d) Flag‐Elk‐1WT or Flag‐Elk‐1 3KR plasmid was co‐transfected with (or without) EGFP‐PIAS1 plasmid, and Myc‐SUMO1 plasmid to HEK293T cells and Elk‐1 SUMOylation was determined by Western blot 48 hr later using anti‐Elk‐1 antibody. (e) The quantified result of Elk‐1 SUMOylation from three independent experiments. Data are mean ± SEM
3.5. Elk‐1 is SUMOylated by endogenous PIAS1 in the hippocampus
The above results showed that Elk‐1 could be SUMOylated by PIAS1 at Lys‐229, Lys‐248, and Lys‐253 in a cellular expression system, but whether Elk‐1 is SUMOylated by PIAS1 at these residues in the brain is not known. To address this issue, we transfected the rat CA1 area with Flag‐vector control, Flag‐Elk‐1WT, individual Flag‐tagged Elk‐1 point mutants, or Flag‐Elk‐1 3KR mutant and then performed SUMOylation assays. All groups contained recombinant SUMO1 protein except for the last group, in which recombinant SUMO1 mutant protein was included to block the SUMOylation reaction. This experimental paradigm is similar to that in experiments presented in Figure 3a, in which cells were transfected with Myc‐SUMO1ΔGG plasmid to block SUMO conjugation. This analysis revealed that Elk‐1 was consistently SUMOylated in the CA1 area following transfection of the Elk‐1WT plasmid. However, it was diminished following transfection with individual Elk‐1 mutant plasmids and was completely blocked by transfection of the Elk‐1 3KR plasmid. SUMOylation was similarly blocked by co‐transfection of the Elk‐1WT plasmid together with the SUMO1 mutant protein (Figure 5a, left panel). Quantification of these result is shown in Figure 5b. Similar results were observed when the membrane was stripped and re‐blotted with an anti‐SUMO1 antibody (Figure 5a, middle panel). Western blotting using an anti‐Flag antibody confirmed the transfection and expression of Flag‐tagged plasmids (Figure 5a, middle‐lower panel). To further confirm that Elk‐1 SUMOylation is blocked by transfection of Elk‐1 3KR, we performed an additional experiment in which animals were transfected with Flag‐vector, Flag‐Elk‐1WT, or Flag‐Elk‐1 3KR. CA1 tissue lysates from rats in each group were immunoprecipitated with an anti‐Flag antibody and immunoblotted with an anti‐SUMO1 antibody. Another group of animals was also transfected with Flag‐Elk‐1WT but was immunoprecipitated with IgG as a negative control. This experiment revealed that Elk‐1 was SUMOylated following transfection of Flag‐Elk‐1WT plasmid, but Elk‐1 SUMOylation was completely blocked in animals transfected with the Flag‐Elk‐1 3KR plasmid. As expected, Elk‐1 SUMOylation was not observed in tissue lysates from the Flag‐Elk‐1WT‐transfected group immunoprecipitated with IgG (Figure 5a, right panel). Western blotting using an anti‐Flag antibody confirmed the transfection and expression of Flag‐tagged plasmids (Figure 5a, right‐lower panel).
We next sought to determine whether Elk‐1 is SUMOylated by endogenous PIAS1. To this end, we transfected the CA1 area of rats with control siRNA or PIAS1 siRNA (8 pmol) and determined SUMOylation of endogenous Elk‐1 by immunoprecipitating the tissue lysates with an anti‐Elk‐1 antibody and immunoblotting with an anti‐Elk‐1 antibody or anti‐SUMO1 antibody. The results indicated that siRNA‐mediated PIAS1 knockdown significantly decreased the level of endogenous Elk‐1 SUMOylation (Figure 5c). Quantified of Elk‐1 SUMOylation results are shown in Figure 5d. The effectiveness of PIAS1 siRNA transfection was confirmed by the marked decrease in PIAS1 expression (Figure 5e).
Next, we examined whether PIAS1 and Elk‐1 are present in the same neurons in the hippocampus. PIAS1 and Elk‐1 expressions in tissue sections containing the CA1 region from the rat brain were assessed by immunohistochemistry; nuclei were counterstained with DAPI. These experiments revealed PIAS1 (green), Elk‐1 (red), and DAPI (blue) immunofluorescence in the same neurons in the CA1 area (Figure 5f, upper panel). When CA1 neurons were visualized at a higher magnification, Elk‐1 was found to be present in both cytoplasmic and nuclear compartments, but PIAS1 was co‐localized with Elk‐1 only in the nuclear compartment of the same neurons (Figure 5f, lower panel).
3.6. Acute Aβ increases Elk‐1 SUMOylation in the hippocampus in a manner that depends on PIAS1 Ser503 phosphorylation
We demonstrated above that acute Aβ exposure increases the level of Ser503‐phosphorylated PIAS1 in the hippocampus. Because Ser503‐phosphorylated PIAS1 mediates Elk‐1 SUMOylation, it is conceivable that acute Aβ exposure would also induce Elk‐1 SUMOylation. To test this hypothesis, we acutely injected the CA1 region of rats with NH4OH or Aβ and determined Elk‐1 SUMOylation 1 hr later by immunoprecipitating the membrane fraction of tissue lysates with anti‐Elk‐1 antibody and immunoblotting with anti‐Elk‐1 antibody or anti‐SUMO1 antibody. The results showed that acute Aβ injection markedly increased Elk‐1 SUMOylation (Figure 6a); quantification of these results is shown in Figure 6b. Next, we examined whether blockade of PIAS1 Ser503 phosphorylation prevented Elk‐1 SUMOylation. Accordingly, the rat CA1 area was transfected with Flag‐vector (control), Flag‐PIAS1WT, or Flag‐PIAS1S503A plasmid, and Elk‐1 SUMOylation was determined by immunoblotting with an anti‐Elk‐1 antibody (Figure 6c, left panel) or anti‐SUMO1 antibody (Figure 6c, right panel). These experiments revealed that transfection of PIAS1WT plasmid significantly increased the level of Elk‐1 SUMOylation, whereas transfection of the PIAS1S503A plasmid completely blocked Elk‐1 SUMOylation (Figure 6c). Quantification of results is shown in Figure 6d. Immunoprecipitation and immunoblotting using an anti‐Flag antibody confirmed plasmid transfection and equal expression of the Flag‐tagged PIAS1 plasmids (Figure 6c, left‐lower panel).
Figure 6.
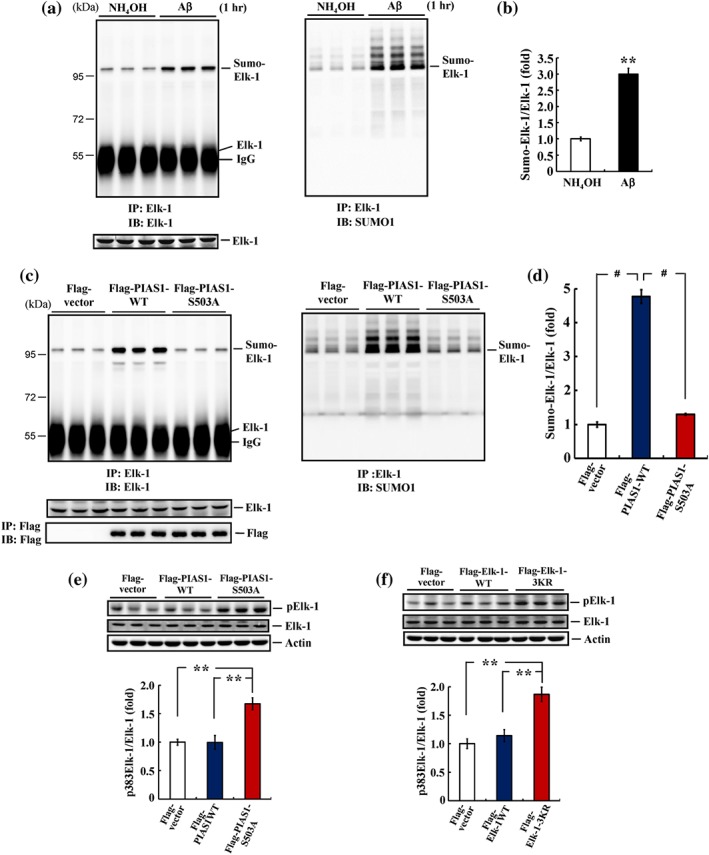
Acute Aβ increases Elk‐1 SUMOylation and Elk‐1 SUMOylation is PIAS1 Ser503 phosphorylation‐dependent in the hippocampus. (a) NH4OH (1%) or Aβ (14 μg) was injected to rat hippocampal CA1 area, and Elk‐1 SUMOylation was determined 1 hr later as described above. Anti‐Elk‐1 antibody (left) and anti‐SUMO1 antibody (right) were used for immunoblotting after immunoprecipitation with anti‐Elk‐1 antibody. (b) The quantified result of Elk‐1 SUMOylation (n = 6 different animals for each group). ** P < 0.05. (c) Flag‐vector, Flag‐PIAS1WT, or Flag‐PIAS1S503A plasmid was transfected to rat CA1 area, and Elk‐1 SUMOylation was determined 48 hr later. (d) The quantified result of Elk‐1 SUMOylation (n = 6 different animals for each group). # P < 0.05 for Flag‐PIAS1WT versus Flag‐vector group and # P < 0.05 for Flag‐PIAS1S503A versus Flag‐PIAS1WT group. (e) A separate group of animals also received the same plasmid transfections as described in (c) and Elk‐1 Ser382 (Ser383 in human) phosphorylation was determined 48 hr later (n = 5 different animals for each group). ** P < 0.05 for Flag‐PIAS1S503A versus Flag‐vector group and ** P < 0.05 for Flag‐PIAS1S503A versus Flag‐PIAS1WT group. (f) Flag‐vector, Flag‐Elk‐1WT, or Flag‐Elk‐1 3KR plasmid was transfected to rat CA1 area, and Elk‐1 Ser382 (Ser383 in human) phosphorylation was determined 48 hr later (n = 5 different animals for each group). ** P < 0.05 for Flag‐Elk‐1 3KR versus Flag‐vector group and ** P < 0.05 for Flag‐Elk‐1 3KR versus Flag‐Elk‐1WT group. Data are mean ± SEM
To further examine the interplay between Elk‐1 SUMOylation and Elk‐1 phosphorylation, we examined whether blockade of PIAS1 phosphorylation at Ser503 alters the phosphorylation level of Elk‐1. Accordingly, the rat CA1 area was transfected with Flag‐vector, Flag‐PIAS1WT, or Flag‐PIAS1S503A plasmid, and Elk‐1 phosphorylation at Ser‐382 (Ser‐383 in human) was examined by western blotting. We found that transfection of PIAS1WT had no apparent effect on Elk‐1 phosphorylation, whereas transfection of PIAS1S503A markedly increased the phosphorylation level of Elk‐1 (Figure 6e). Elk‐1 is SUMOylated under resting conditions and remains inactive (S. H. Yang et al., 2003). Thus, we also examined whether blockade of Elk‐1 SUMOylation altered the phosphorylation level of Elk‐1. To this end, the rat CA1 area was transfected with Flag‐vector, Flag‐Elk‐1WT, or Flag‐Elk‐1 3KR plasmid, and Elk‐1 phosphorylation at Ser‐382 was examined by Western blotting. These experiments showed no apparent effect of Elk‐1WT transfection on Elk‐1 phosphorylation but demonstrated that transfection of Elk‐1 3KR significantly increased the phosphorylation level of Elk‐1 (Figure 6f).
3.7. Both PIAS1 Ser503 phosphorylation and Elk‐1 SUMOylation down‐regulate GADD45α expression
Results from the above experiments illustrate the relationship between acute Aβ exposure and PIAS1 Ser503 phosphorylation and Elk‐1 SUMOylation in the hippocampus. Next, we examined the functional significance of this signalling pathway. Growth arrest and DNA damage‐inducible 45α (GADD45α) is involved in various cellular functions and is an important stress sensor in the cell (Moskalev et al., 2012; Salvador, Brown‐Clay, & Fornace, 2013). Of greater relevance to the present study, GADD45α has been suggested to play a role in apoptosis; consistent with this, down‐regulation of GADD45α expression increases cell survival (Chiou et al., 2014; Sheikh, Hollander, & Fornance, 2000). Moreover, GADD45α expression was found to be regulated by Elk‐1 signalling induced by arsenic stimulation (Shi, Sutariya, Bishayee, & Bhatia, 2014). On the basis of these observations, we first examined whether GADD45α expression is regulated by acute Aβ exposure. Animals were injected in the CA1 area with NH4OH (1%) or Aβ (14 μg), and GADD45α expression was examined by western blotting 1 hr later. The results revealed that acute administration of Aβ decreased GADD45α expression levels by approximately 50%; it also consistently increased PIAS1 expression levels (Figure 7a). Because GADD45α has been suggested to mediate apoptosis, we also examined the effect of Aβ on GADD45α expression at a later stage when the toxicity of Aβ has been established. To this end, we similarly injected the CA1 area of a separate group of animals with NH4OH (1%) or Aβ (14 μg), but this time, we examined GADD45α expression 14 days later. We found that Aβ significantly increased the expression level of GADD45α at this time point (Figure S2). Next, we examined whether GADD45α expression is regulated by PIAS1 Ser503 phosphorylation and Elk‐1 SUMOylation. In these experiments, animals were transfected in their CA1 area with Flag‐vector, Flag‐PIAS1WT, or Flag‐PIAS1S503A plasmid, and GADD45α expression was examined by Western blotting. The results showed that transfection of Flag‐PIAS1WT decreased GADD45α expression, whereas transfection of Flag‐PIAS1S503A increased it (Figure 7b). In the next experiment, the CA1 area of animals was transfected with Flag‐vector, Flag‐Elk‐1WT, or Flag‐Elk‐1 3KR, and GADD45α expression was examined by western blotting. These experiments showed that transfection of Flag‐Elk‐1WT decreased GADD45α expression, but transfection of Flag‐Elk‐1 3KR increased it (Figure 7c).
Figure 7.
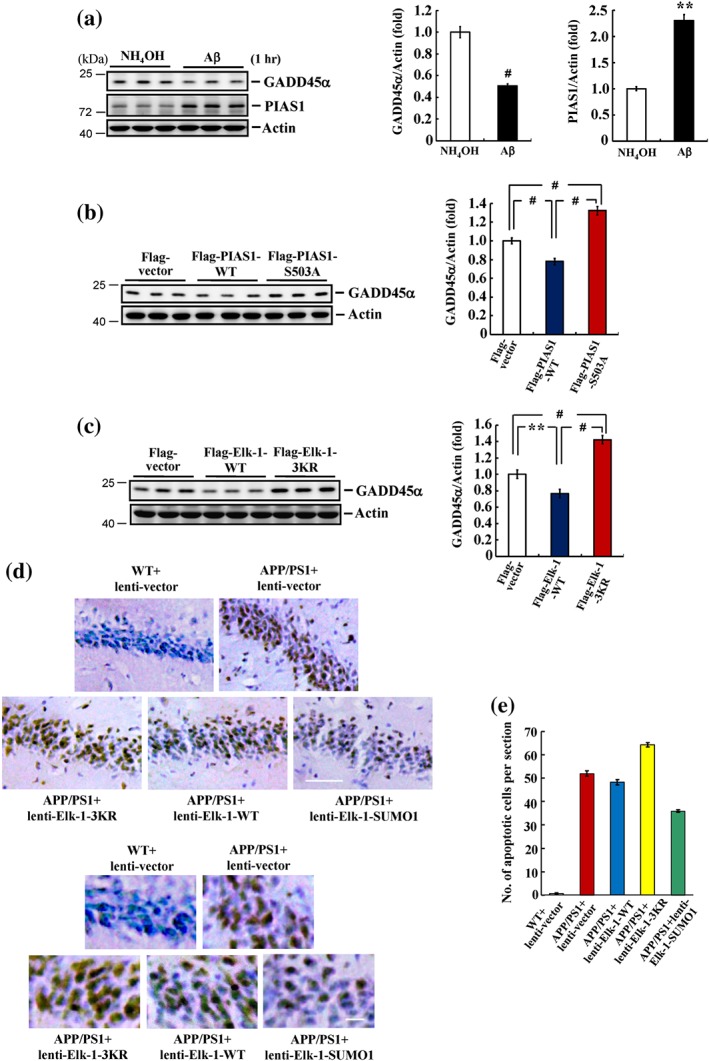
PIAS1 Ser503 phosphorylation and Elk‐1 SUMOylation both down‐regulate GADD45α expression. (a) NH4OH (1%) or Aβ (14 μg) was injected to rat hippocampal CA1 area, and GADD45α and PIAS1 expressions were determined by Western blot 1 hr later (n = 6 different animals for each group). ** P < 0.05 for Aβ versus NH4OH group. (b) Flag‐vector, Flag‐PIAS1WT, or Flag‐PIAS1S503A plasmid was transfected to CA1 area, and GADD45α expression was determined by Western blot 48 hr later (n = 6 different animals for each group). # P < 0.05 for Flag‐PIAS1WT group versus Flag‐vector group, # P < 0.05 for Flag‐PIAS1S503A group versus Flag‐PIAS1WT group, and # P < 0.05 for Flag‐PIAS1S503A group versus Flag‐vector group. (c) Flag‐vector, Flag‐Elk‐1WT, or Flag‐Elk‐1 3KR plasmid was transfected to CA1 area, and GADD45α expression was determined by Western blot 48 hr later (n = 6 different animals for each group). ** P < 0.05 for Flag‐Elk‐1WT group versus Flag‐vector group, # P < 0.05 for Flag‐Elk‐1 3KR group versus Flag‐Elk‐1WT group, and # P < 0.05 for Flag‐Elk‐1 3KR group versus Flag‐vector group. (d) WT and APP/PS1 mice received lenti‐vector, lenti‐Elk‐1WT, lenti‐Elk‐1 3KR, or lenti‐Elk‐1‐SUMO1 fusion vector transduction to their CA1 area, and TUNEL assay was performed 14 days later. Scale bar equals 50 μm for the upper panel and 12.5 μm for the lower panel. (e) The quantified result of TUNEL staining (n = 3 different animals for each group). Data are mean ± SEM
3.8. Elk‐1 SUMOylation rescues hippocampal neurons from apoptosis in APP/PS1 mice
The above result showed that blockade of Elk‐1 SUMOylation increases GADD45α expression. Here, we further examined the role of Elk‐1 SUMOylation in the apoptosis of hippocampal neurons in the APP/PS1 mouse model of AD using TUNEL staining. For these experiments, mice (10 months old) were divided into the following five groups: (a) WT mice transduced with control lenti‐vector, (b) APP/PS1 mice transduced with control lenti‐vector, (c) APP/PS1 mice transduced with lenti‐Elk‐1WT, (d) APP/PS1 mice transduced with lenti‐Elk‐1 3KR, and (e) APP/PS1 mice transduced with lenti‐Elk‐1‐SUMO1. TUNEL assays were performed 14 days after transduction. Almost no apoptotic cells were detectable in WT animals transduced with lenti‐vector; however, the number of apoptotic cells was dramatically increased in control APP/PS1 mice transduced with lenti‐vector and was further increased in APP/PS1 mice transduced with lenti‐Elk‐1 3KR. The extent of cell apoptosis in APP/PS1 mice transduced with lenti‐Elk‐1WT was similar to that in APP/PS1 mice transduced with lenti‐vector; however, transduction of APP/PS1 mice with lenti‐Elk‐1‐SUMO1 markedly decreased the number of apoptotic cells compared with APP/PS1 mice transduced with lenti‐vector (Figure 7d). Quantification of these results is shown in Figure 7e.
4. DISCUSSION
In this study, we examined the role of Elk‐1 SUMOylation in the hippocampus in the context of AD and found that Elk‐1 SUMOylation functions as an endogenous defence mechanism against acute Aβ‐induced toxicity and apoptosis in APP/PS1 mice. Our results showed that Elk‐1 could be SUMOylated by PIAS1 at Lys229, Lys248, and Lys253 in the rat brain, consistent with previous reports that Elk‐1 is SUMOylated at Lys230, Lys249, and Lys254 in human cells (Salinas et al., 2004; S. H. Yang et al., 2003). In addition, we identified a novel phosphorylation site on PIAS1 (Ser503) that is phosphorylated by MAPK/ERK and found that PIAS1 E3 ligase activity in the brain is dependent on Ser503 phosphorylation. An earlier study found that PIAS1 is phosphorylated by IKKα at Ser90 and showed that PIAS1 phosphorylation at this residue is PIAS1 E3 ligase‐activity dependent (Liu et al., 2007). Taken together, these observations suggest an important role for PIAS1 phosphorylation at Ser503, reflecting its subsequent regulation of PIAS1 E3 ligase activity and PIAS1 phosphorylation by other kinases, such as IKKα. Unlike the case in the brain, we found that transfection of HEK293T cells with a PIAS1S503A mutant did not completely block Elk‐1 SUMOylation. One possible explanation for this apparent discrepancy is that Elk‐1 SUMOylation in HEK293T cells may also be regulated, at least in part, by PIAS1 phosphorylation at Thr487, consistent with our demonstration that Thr487 is another phosphorylation site in PIAS1. Alternatively, Elk‐1 may be SUMOylated by other PIAS family proteins in addition to PIAS1 in HEK293T cells, whereas PIAS1 may play a predominant role in the brain.
Our immunohistochemical results revealed that, under resting conditions, Elk‐1 is mostly localized to the nucleus of hippocampal neurons, where it is co‐localized with PIAS1, although some cytoplasmic distribution is also observed. This result is congruent with previous findings that Elk‐1 is SUMOylated under basal conditions (S. H. Yang et al., 2003) and that SUMOylation is responsible for the nuclear retention of Elk‐1 (Salinas et al., 2004). We also found that blockade of Elk‐1 SUMOylation increased the basal phosphorylation level of Elk‐1 and GADD45α expressions in hippocampal neurons. These results are consistent with a previous report that Elk‐1 SUMOylation inactivates Elk‐1 and suppresses its transcriptional activity (S. H. Yang et al., 2003). They are also congruent with the observation that Elk‐1 signalling regulates GADD45α expression (Shi et al., 2014). On the other hand, although Elk‐1 is SUMOylated under basal conditions, the endogenous levels of SUMOylated Elk‐1 are very low, raising possible questions about its relevance to the physiological functions examined here. However, protein SUMOylation at low levels has been found to produce significant biological effects. For example, low‐level SUMOylation of the DNA‐modifying enzyme, thymine‐DNA glycosylase, plays a critical role in its enzymatic function (Hardeland, Steinacher, Jiricny, & Schar, 2002). Moreover, SUMOylation often occurs in a small percentage of a protein population, but such low‐level SUMOylation of a few proteins was shown to be very important in the regulation of DNA damage responses, including DNA repair (Johnson, 2004; Sarangi & Zhao, 2015). In addition, the basal level of Elk‐1 SUMOylation could be different in different cell types. In this context, we found here that endogenous Elk‐1 SUMOylation was detectable in the brain but not in HEK293T cells. In another study, endogenous Elk‐1 SUMOylation was observed in nuclear extracts of HeLa cells but not COS‐7 cells (S. H. Yang et al., 2003).
In this study, we found that Elk‐1 SUMOylation was enhanced (and Elk‐1 phosphorylation was suppressed) 1 hr after acute Aβ injection, whereas GADD45α expression was decreased, effects that were presumably mediated by Aβ‐induced activation of MAPK/ERK and subsequent phosphorylation of PIAS1 at Ser503. These results appear to be at odds with earlier reports showing that Elk‐1 is a direct target of MAPK/ERK and that MAPK/ERK phosphorylation of Elk‐1 increases the transcriptional activity of Elk‐1 (Janknecht et al., 1993; Besnard, Galan‐Rodriguez, Vanhoutte, & Caboche, 2011 for review). One possible explanation is that MAPK/ERK phosphorylation of Elk‐1 may be transient, but MAPK/ERK‐mediated Elk‐1 SUMOylation is sustained for a longer period, resulting in a net decrease in Elk‐1 transcriptional activity. Another possibility is that acute Aβ treatment may also increase the activity of other kinases besides MAPK/ERK that may also phosphorylate PIAS1 at Ser503 and produce a synergistic Elk‐1 SUMOylation‐enhancing effect. The decrease in Elk‐1 transcription activity produced by these events together may exceed the increase in Elk‐1 transcription activity that results from activation of MAPK/ERK. For example, we previously reported that acute Aβ also increases the phosphorylation level of Akt (Tao et al., 2017), and it has been previously suggested that PI3K/Akt signalling mediates neuroprotection against glutamate‐induced apoptosis (Almeida et al., 2005). Whether PI3K also phosphorylates PIAS1 at Ser503 remains to be examined.
In this study, we found that acute Aβ decreased GADD45α expression 1 hr later. This decrease, which is presumably caused by a decrease in Elk‐1 transcription, serves as an endogenous neuroprotective mechanism against acute Aβ toxicity. This result is generally compatible with a previous report that Gadd45 is an Aβ‐responsive gene (Santiard‐Baron et al., 1999). Although this latter study found that Gadd45 mRNA levels were dramatically increased 6 hr after Aβ exposure, Gadd45 mRNA expression tended to be decreased 1 hr after Aβ exposure. The reason we chose the 1 hr time point is because we previously found that Aβ induction of PIAS1 expression peaks at 1 hr (Tao et al., 2017). In addition, endogenous defence mechanisms are evoked within 1 hr after Aβ insult (Hsu et al., 2009; Tao et al., 2017). On the other hand, we also found that Aβ increased GADD45α expression after 14 days, a time point at which Aβ toxicity has been established by different mechanisms (Figure S2).
In this study, we found that PIAS1 suppressed GADD45α expression through SUMOylation of Elk‐1. But this result does not exclude the possibility that PIAS1 might down‐regulate the expression of GADD45α through other transcription factors or mechanisms. For example, we have previously demonstrated that PIAS1 increases cell survival through enhanced SUMOylation of Hes‐1 and enhanced suppressive effects of Hes‐1 on GADD45α expression (Chiou et al., 2014). Further, it has been found that NF‐κB signalling, which increases GADD45α expression and thereby mediates cell death, is negatively regulated by PIAS1 in response to UV B irradiation (Liu et al., 2005; Thyss et al., 2005). But the regulatory mechanism could be more complicated. JNK has been suggested to mediate oxidative stress responses, including the transcription of GADD45α and that of other ROS‐sensitive genes. PIAS1 was found to bi‐directionally regulate JNK‐dependent oxidative stress responses, at least in part through inhibition of JNK‐mediated gene expression, as evidenced by the fact that knockdown of PIAS1 enhances H2O2‐induced GADD45α expression (Leitao, Jones, & Brosens, 2011). On the other hand, we also found that transduction of lenti‐Elk‐1 3KR into APP/PS1 mice increased the number of apoptotic cells compared with control lenti‐vector‐transduced APP/PS1 mice, whereas transduction of lenti‐Elk‐1‐SUMO1 into these same mice reduced the number of apoptotic cells. Although transfection of Elk‐1 3KR increased GADD45α expression, it is not possible to determine from these results whether the protective effect of Elk‐1‐SUMO1 transfection against cell death is definitely mediated by a decrease in GADD45α expression. The involvement of other genes is also possible. For example, p21 has been implicated in cell apoptosis in response to sodium arsenite (NaASO2) exposure, and Elk‐1 directly and transcriptionally activates p21 expression (Shin, Kim, Lim, & Lee, 2011). Because Elk‐1 SUMOylation decreases Elk‐1 transcriptional activity, it would presumably also decrease p21 expression and apoptosis. The protective effect of Elk‐1 SUMOylation could also be indirect. Elk‐1 has been found to induce expression of Bax, another apoptotic gene, following exposure to NaASO2, an effect that is mediated by Egr‐1 (Shin et al., 2011). Similarly, Elk‐1 SUMOylation is expected to decrease Bax expression and cell apoptosis through decreased Elk‐1 activity.
Elk‐1 is SUMOylated under basal conditions, but the role and function of Elk‐1 SUMOylation in the brain had been previously unknown. Our results provide the first demonstration that acute Aβ exposure enhances Elk‐1 SUMOylation through MAPK/ERK‐mediated phosphorylation of PIAS1 Ser503 within a short time after Aβ injection. This leads to decreased Elk‐1 transcription and GADD45α expression. This signalling pathway serves as an endogenous defence mechanism against acute Aβ toxicity (Figure S3). We further found that elevated Elk‐1 SUMOylation rescues hippocampal neurons from apoptosis in APP/PS1 mice. Taken together, these results not only demonstrate the functional significance of Elk‐1 SUMOylation under resting conditions but also suggest that targeting Elk‐1 SUMOylation could be a novel strategy for providing neuroprotection against AD.
AUTHOR CONTRIBUTIONS
S.Y.L. and E.H.Y.L. designed the experiments and wrote the manuscript. S.Y.L. constructed the plasmids; S.Y.L. and Y.L.M. conducted most of the biochemical and animal experiments. W.L.H. performed the SUMOylation assay. H.Y.C. and S.Y.L. performed the LC–MS/MS analyses. S.Y.L., Y.L.M., and W.L.H. performed the statistical analyses.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
DECLARATION OF TRANSPARENCY AND SCIENTIFIC RIGOUR
This Declaration acknowledges that this paper adheres to the principles for transparent reporting and scientific rigour of preclinical research as stated in the BJP guidelines for Design & Analysis, Immunoblotting and Immunochemistry, and Animal Experimentation, and as recommended by funding agencies, publishers and other organisations engaged with supporting research.
Supporting information
Figure S1. Lys‐229, Lys‐248 and Lys‐253 are not candidate ubiquitination sites on Elk‐1.
Flag‐Elk‐1 or Flag‐Elk‐1 3KR plasmid was co‐transfected with His‐ubiquitin plasmid to HEK293T cells and cell lysates were immunoprecipitated with anti‐Flag antibody and immunoblotted with anti‐Flag (left) or anti‐His (right) antibody for examination of Elk‐1 ubiquitination. Experiments are in two independent repeats.
Figure S2. Acute Aβ increases GADD45α expression 14 days later.
NH4OH (1%) or Aβ (14 μg) was injected to rat hippocampal CA1 area and GADD45α expression was determined by western blot 14 days later (n=6 different animals for each group). # P < 0.05 for Aβ vs. NH4OH group. Data are mean ± SEM.
Figure S3. A schematic diagram showing the signaling pathway induced by acute Aβ in hippocampal neurons that serves as an endogenous defense mechanism
Table S1. All the statistics for non‐parametric tests (Mann‐Whitney U test and Kruskal‐Wallis one‐way ANOVA)
Data S1. Supplementary Methods
ACKNOWLEDGEMENTS
This work was supported by a grant from Ministry of Science and Technology in Taiwan (MOST 107‐2320‐B‐001‐020) and research fund from the Institute of Biomedical Sciences (IBMS), Academia Sinica, Taiwan. Thanks are given to the Animal Core, Confocal Core, and Proteomic Core Facilities of IBMS.
Liu S‐Y, Ma Y‐L, Hsu W‐L, Chiou H‐Y, Lee EHY. Protein inhibitor of activated STAT1 Ser503 phosphorylation‐mediated Elk‐1 SUMOylation promotes neuronal survival in APP/PS1 mice. Br J Pharmacol. 2019;176:1793–1810. 10.1111/bph.14656
REFERENCES
- Alexander, S. P. H. , Fabbro, D. , Kelly, E. , Marrion, N. , Peters, J. A. , Benson, H. E. , … CGTP Collaborators . (2017). The Concise Guide to PHARMACOLOGY 2015/16: Enzymes. British Journal of Pharmacology, 172, 6024–6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, S. P. H. , Kelly, E. , Marrion, N. , Peters, J. A. , Benson, H. E. , Faccenda, E. , … CGTP Collaborators . (2017). The Concise Guide to pharmacology 2015/16: Overview. British Journal of Pharmacology, 172, 5734–5143. [Google Scholar]
- Alexander, S. P. H. , Roberts, R. E. , Broughton, B. R. S. , Sobey, C. G. , George, C. H. , Stanford, S. C. , … Ahluwalia, A. (2018). Goal and practicalities of immunoblotting and immunohistochemistry: A guide for submission to the British Journal of Pharmacology . British Journal of Pharmacology, 175, 407–411. 10.1111/bph.14112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida, R. D. , Manadas, B. J. , Melo, C. V. , Gomes, J. R. , Mendes, C. S. , Graos, M. M. , … Duarte, C. B. (2005). Neuroprotection by BDNF against glutamate‐induced apoptotic cell death is mediated by ERK and PI3‐kinase pathways. Cell Death and Differentiation, 12, 1329–1343. 10.1038/sj.cdd.4401662 [DOI] [PubMed] [Google Scholar]
- Anton, M. , Rodriguez‐Gonzalez, A. , Ballesta, A. , Gonzalez, N. , del Pozo, A. , de Fonseca, F. R. , … Orio, L. (2018). Alcohol binge disrupts the rat intestinal barrier: The partial protective role of oleoylethanolamide. British Journal of Pharmacology, 175, 4464–4479. 10.1111/bph.14501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard, A. , Galan‐Rodriguez, B. , Vanhoutte, P. , & Caboche, J. (2011). Elk‐1 a transcription factor with multiple facets in the brain. Frontiers in Neuroscience, 5, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossis, G. , Malnou, C. E. , Farras, R. , Andermarcher, E. , Hipskind, R. , Rodriguez, M. , … Piechaczyk, M. (2005). Down‐regulation of c‐Fos/c‐Jun AP‐1 dimer activity by SUMOylation. Molecular and Cellular Biology, 25, 6964–6979. 10.1128/MCB.25.16.6964-6979.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterfield, D. A. , Drake, J. , Pocernich, C. , & Castegna, A. (2001). Evidence of oxidative damage in Alzheimer's disease brain: Central role for amyloid β‐peptide. Trends in Molecular Medicine, 7, 548–554. 10.1016/S1471-4914(01)02173-6 [DOI] [PubMed] [Google Scholar]
- Cammarota, M. , Bevilaqua, L. R. , Ardenghi, P. , Paratcha, G. , Levi de Stein, M. , Izquierdo, I. , & Medina, J. H. (2000). Learning‐associated activation of nuclear MAPK, CREB and Elk‐1, along with Fos production, in the rat hippocampus after a one‐trial avoidance learning: Abolition by NMDA receptor blockade. Brain Research. Molecular Brain Research, 76, 36–46. 10.1016/S0169-328X(99)00329-0 [DOI] [PubMed] [Google Scholar]
- Chao, C. C. , Ma, Y. L. , & Lee, E. H. Y. (2011). Brain‐derived neurotrophic factor enhances Bcl‐xL expression through protein kinase casein kinase 2‐activated and nuclear factor κB‐mediated pathway in rat hippocampus. Brain Pathology, 21, 150–162. 10.1111/j.1750-3639.2010.00431.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. C. , Hsu, W. L. , Ma, Y. L. , Tai, D. J. , & Lee, E. H. Y. (2014). CREB SUMOylation by the E3 ligase PIAS1 enhances spatial memory. The Journal of Neuroscience, 34, 9574–9589. 10.1523/JNEUROSCI.4302-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou, H. Y. , Liu, S. Y. , Lin, C. H. , & Lee, E. H. Y. (2014). Hes‐1 SUMOylation by protein inhibitor of activated STAT1 enhances the suppressing effect of Hes‐1 on GADD45α expression to increase cell survival. Journal of Biomedical Science, 21, 53 10.1186/1423-0127-21-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis, M. J. , Alexander, S. , Cirino, G. , Docherty, J. R. , George, C. H. , Giembycz, M. A. , … Ahluwalia, A. (2018). Experimental design and analysis and their reporting II: Updated and simplified guidance for authors and peer reviewers. British Journal of Pharmacology, 175, 987–993. 10.1111/bph.14153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, S. , Vanhoutte, P. , Pages, C. , Caboche, J. , & Laroche, S. (2000). The MAPK/ERK cascade targets both Elk‐1 and cAMP response element‐binding protein to control long‐term potentiation‐dependent gene expression in the dentate gyrus in vivo. The Journal of Neuroscience, 20, 4563–4572. 10.1523/JNEUROSCI.20-12-04563.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Strooper, B. , & Annaert, W. (2000). Proteolytic processing and cell biological functions of the amyloid precursor protein. Journal of Cell Science, 113 (Pt 11), 1857–1870. [DOI] [PubMed] [Google Scholar]
- Dickson, D. W. (2004). Apoptotic mechanisms in Alzheimer neurofibrillary degeneration: Cause or effect? The Journal of Clinical Investigation, 114, 23–27. 10.1172/JCI22317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gareau, J. R. , & Lima, C. D. (2010). The SUMO pathway: Emerging mechanisms that shape specificity, conjugation and recognition. Nature Reviews. Molecular Cell Biology, 11, 861–871. 10.1038/nrm3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardeland, U. , Steinacher, R. , Jiricny, J. , & Schar, P. (2002). Modification of the human thymine‐DNA glycosylase by ubiquitin‐like proteins facilitates enzymatic turnover. The EMBO Journal, 21, 1456–1464. 10.1093/emboj/21.6.1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding, S. D. , Sharman, J. L. , Faccenda, E. , Southan, C. , Pawson, A. J. , Ireland, S. , … NC‐IUPHAR (2018). The IUPHAR/BPS guide to pharmacology in 2018: Updates and expansion to encompass the new guide to immunopharmacology. Nucleic Acids Research, 46, D1091–D1106. 10.1093/nar/gkx1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy, J. , & Selkoe, D. J. (2002). The amyloid hypothesis of Alzheimer's disease: Progress and problems on the road to therapeutics. Science, 297, 353–356. 10.1126/science.1072994 [DOI] [PubMed] [Google Scholar]
- Hsu, W. L. , Chiu, T. H. , Tai, D. J. , Ma, Y. L. , & Lee, E. H. Y. (2009). A novel defense mechanism that is activated on amyloid‐β insult to mediate cell survival: Role of SGK1‐STAT1/STAT2 signaling. Cell Death and Differentiation, 16, 1515–1529. 10.1038/cdd.2009.91 [DOI] [PubMed] [Google Scholar]
- Hsu, W. L. , Ma, Y. L. , Liu, Y. C. , & Lee, E. H. Y. (2017). Smad4 SUMOylation is essential for memory formation through upregulation of the skeletal myopathy gene TPM2. BMC Biology, 15, 112 10.1186/s12915-017-0452-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janknecht, R. , Ernst, W. H. , Pingoud, V. , & Nordheim, A. (1993). Activation of ternary complex factor Elk‐1 by MAP kinases. The EMBO Journal, 12, 5097–5104. 10.1002/j.1460-2075.1993.tb06204.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, E. S. (2004). Protein modification by SUMO. Annual Review of Biochemistry, 73, 355–382. 10.1146/annurev.biochem.73.011303.074118 [DOI] [PubMed] [Google Scholar]
- Kasza, A. (2013). Signal‐dependent Elk‐1 target genes involved in transcript processing and cell migration. Biochimica et Biophysica Acta, 1829, 1026–1033. 10.1016/j.bbagrm.2013.05.004 [DOI] [PubMed] [Google Scholar]
- Kilkenny, C. , Browne, W. , Cuthill, I. C. , Emerson, M. , & Altman, D. G. (2010). Animal research: reporting in vivo experiments: The ARRIVE guidelines. British Journal of Pharmacology, 160, 1577–1579. 10.1111/j.1476-5381.2010.00872.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitao, B. B. , Jones, M. C. , & Brosens, J. J. (2011). The SUMOE3‐ligase PIAS1 couples reactive oxygen species‐dependent JNK activation to oxidative cell death. FASEB J, 25, 3416–3425. 10.1096/fj.11-186346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, J. , Fu, Y. , & Shuai, K. (2000). Distinct roles of the NH2‐ and COOH‐terminal domains of the protein inhibitor of activated signal transducer and activator of transcription (STAT) 1 (PIAS1) in cytokine‐induced PIAS1‐Stat1 interaction. Proceedings of the National Academy of Sciences of the United States of America, 97, 5267–5272. 10.1073/pnas.97.10.5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C. H. , Liu, S. Y. , & Lee, E. H. Y. (2016). SUMO modification of Akt regulates global SUMOylation and substrate SUMOylation specificity through Akt phosphorylation of Ubc9 and SUMO1. Oncogene, 35, 595–607. 10.1038/onc.2015.115 [DOI] [PubMed] [Google Scholar]
- Liu, B. , Liao, J. , Rao, X. , Kushner, S. A. , Chung, C. D. , Chang, D. D. , & Shuai, K. (1998). Inhibition of Stat1‐mediated gene activation by PIAS1. Proceedings of the National Academy of Sciences of the United States of America, 95, 10626–10631. 10.1073/pnas.95.18.10626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, B. , Mink, S. , Wong, K. A. , Stein, N. , Getman, C. , Dempsey, P. W. , … Shuai, K. (2004). PIAS1 selectively inhibits interferon‐inducible genes and is important in innate immunity. Nature Immunology, 5, 891–898. [DOI] [PubMed] [Google Scholar]
- Liu, B. , Yang, R. , Wong, K. A. , Getman, C. , Stein, N. , Teitell, M. A. , … Shuai, K. (2005). Negative regulation of NF‐κB signaling by PIAS1. Molecular and Cellular Biology, 25, 1113–1123. 10.1128/MCB.25.3.1113-1123.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, B. , Yang, Y. , Chernishof, V. , Loo, R. R. , Jang, H. , Tahk, S. , … Shuai, K. (2007). Proinflammatory stimuli induce IKKα‐mediated phosphorylation of PIAS1 to restrict inflammation and immunity. Cell, 129, 903–914. 10.1016/j.cell.2007.03.056 [DOI] [PubMed] [Google Scholar]
- Lukiw, W. J. , & Bazan, N. G. (2006). Survival signalling in Alzheimer's disease. Biochemical Society Transactions, 34, 1277–1282. 10.1042/BST0341277 [DOI] [PubMed] [Google Scholar]
- Moskalev, A. A. , Smit‐McBride, Z. , Shaposhnikov, M. V. , Plyusnina, E. N. , Zhavoronkov, A. , Budovsky, A. , … Fraifeld, V. E. (2012). Gadd45 proteins: Relevance to aging, longevity and age‐related pathologies. Ageing Research Reviews, 11, 51–66. 10.1016/j.arr.2011.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas, S. , Briancon‐Marjollet, A. , Bossis, G. , Lopez, M. A. , Piechaczyk, M. , Jariel‐Encontre, I. , … Hipskind, R. A. (2004). SUMOylation regulates nucleo‐cytoplasmic shuttling of Elk‐1. The Journal of Cell Biology, 165, 767–773. 10.1083/jcb.200310136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvador, J. M. , Brown‐Clay, J. D. , & Fornace, A. J. Jr. (2013). Gadd45 in stress signaling, cell cycle control, and apoptosis. Advances in Experimental Medicine and Biology, 793, 1–19. 10.1007/978-1-4614-8289-5_1 [DOI] [PubMed] [Google Scholar]
- Sananbenesi, F. , Fischer, A. , Schrick, C. , Spiess, J. , & Radulovic, J. (2002). Phosphorylation of hippocampal Erk‐1/2, Elk‐1, and p90‐Rsk‐1 during contextual fear conditioning: Interactions between Erk‐1/2 and Elk‐1. Molecular and Cellular Neurosciences, 21, 463–476. 10.1006/mcne.2002.1188 [DOI] [PubMed] [Google Scholar]
- Santiard‐Baron, D. , Gosset, P. , Nicole, A. , Sinet, P. M. , Christen, Y. , & Ceballos‐Picot, I. (1999). Identification of β‐amyloid‐responsive genes by RNA differential display: Early induction of a DNA damage‐inducible gene, gadd45. Experimental Neurology, 158, 206–213. 10.1006/exnr.1999.7076 [DOI] [PubMed] [Google Scholar]
- Sapetschnig, A. , Rischitor, G. , Braun, H. , Doll, A. , Schergaut, M. , Melchior, F. , & Suske, G. (2002). Transcription factor Sp3 is silenced through SUMO modification by PIAS1. The EMBO Journal, 21, 5206–5215. 10.1093/emboj/cdf510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarangi, P. , & Zhao, X. (2015). SUMO‐mediated regulation of DNA damage repair and responses. Trends in Biochemical Sciences, 40, 233–242. 10.1016/j.tibs.2015.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, D. , & Muller, S. (2003). PIAS/SUMO: New partners in transcriptional regulation. Cellular and Molecular Life Sciences, 60, 2561–2574. 10.1007/s00018-003-3129-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgambato, V. , Vanhoutte, P. , Pages, C. , Rogard, M. , Hipskind, R. , Besson, M. J. , & Caboche, J. (1998). In vivo expression and regulation of Elk‐1, a target of the extracellular‐regulated kinase signaling pathway, in the adult rat brain. The Journal of Neuroscience, 18, 214–226. 10.1523/JNEUROSCI.18-01-00214.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh, M. S. , Hollander, M. C. , & Fornance, A. J. Jr. (2000). Role of Gadd45 in apoptosis. Biochemical Pharmacology, 59, 43–45. 10.1016/S0006-2952(99)00291-9 [DOI] [PubMed] [Google Scholar]
- Shi, Q. , Sutariya, V. , Bishayee, A. , & Bhatia, D. (2014). Sequential activation of Elk‐1/Egr‐1/GADD45α by arsenic. Oncotarget, 5, 3862–3870. 10.18632/oncotarget.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, S. Y. , Kim, C. G. , Lim, Y. , & Lee, Y. H. (2011). The ETS family transcription factor Elk‐1 regulates induction of the cell cycle‐regulatory gene p21 and the Bax gene in sodium arsenite‐exposed human keratinocyte HaCa T cells. The Journal of Biological Chemistry, 286, 26860–26872. 10.1074/jbc.M110.216721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai, D. J. C. , Hsu, W. L. , Liu, Y. C. , Ma, Y. L. , & Lee, E. H. Y. (2011). Novel role and mechanism of protein inhibitor of activated STAT1 in spatial learning. The EMBO Journal, 30, 205–220. 10.1038/emboj.2010.290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai, D. J. C. , Liu, Y. C. , Hsu, W. L. , Ma, Y. L. , Cheng, S. J. , Liu, S. Y. , & Lee, E. H. Y. (2016). MeCP2 SUMOylation rescues Mecp2 mutant‐induced behavioral deficits in a mouse model of Rett syndrome. Nature Communications, 7, 10552 10.1038/ncomms10552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, C. C. , Hsu, W. L. , Ma, Y. L. , Cheng, S. J. , & Lee, E. H. Y. (2017). Epigenetic regulation of HDAC1 SUMOylation as an endogenous neuroprotection against Aβ toxicity in a mouse model of Alzheimer's disease. Cell Death and Differentiation, 24, 597–614. 10.1038/cdd.2016.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiels, E. , Kanterewicz, B. I. , Norman, E. D. , Trzaskos, J. M. , & Klann, E. (2002). Long‐term depression in the adult hippocampus in vivo involves activation of extracellular signal‐regulated kinase and phosphorylation of Elk‐1. The Journal of Neuroscience, 22, 2054–2062. 10.1523/JNEUROSCI.22-06-02054.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyss, R. , Virolle, V. , Imbert, V. , Peyron, J. F. , Aberdam, D. , & Virolle, T. (2005). NF‐κB/Egr‐1/Gadd45 are sequentially activated upon UVB irradiation to mediate epidermal cell death. The EMBO Journal, 24, 128–137. 10.1038/sj.emboj.7600501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S. H. , Bumpass, D. C. , Perkins, N. D. , & Sharrocks, A. D. (2002). The ETS domain transcription factor Elk‐1 contains a novel class of repression domain. Molecular and Cellular Biology, 22, 5036–5046. 10.1128/MCB.22.14.5036-5046.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S. H. , Jaffray, E. , Hay, R. T. , & Sharrocks, A. D. (2003). Dynamic interplay of the SUMO and ERK pathways in regulating Elk‐1 transcriptional activity. Molecular Cell, 12, 63–74. 10.1016/S1097-2765(03)00265-X [DOI] [PubMed] [Google Scholar]
- Yang, Y. C. , Ma, Y. L. , Liu, W. T. , & Lee, E. H. Y. (2011). Laminin‐β1 impairs spatial learning through inhibition of ERK/MAPK and SGK1 signaling. Neuropsychopharmacology, 36, 2571–2586. 10.1038/npp.2011.148 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Lys‐229, Lys‐248 and Lys‐253 are not candidate ubiquitination sites on Elk‐1.
Flag‐Elk‐1 or Flag‐Elk‐1 3KR plasmid was co‐transfected with His‐ubiquitin plasmid to HEK293T cells and cell lysates were immunoprecipitated with anti‐Flag antibody and immunoblotted with anti‐Flag (left) or anti‐His (right) antibody for examination of Elk‐1 ubiquitination. Experiments are in two independent repeats.
Figure S2. Acute Aβ increases GADD45α expression 14 days later.
NH4OH (1%) or Aβ (14 μg) was injected to rat hippocampal CA1 area and GADD45α expression was determined by western blot 14 days later (n=6 different animals for each group). # P < 0.05 for Aβ vs. NH4OH group. Data are mean ± SEM.
Figure S3. A schematic diagram showing the signaling pathway induced by acute Aβ in hippocampal neurons that serves as an endogenous defense mechanism
Table S1. All the statistics for non‐parametric tests (Mann‐Whitney U test and Kruskal‐Wallis one‐way ANOVA)
Data S1. Supplementary Methods


