Abstract
How divergent genetic systems regulate a common pathway during the development of two serial structures, forelimbs and hindlimbs, is not well understood. Specifically, HAND2 has been shown to regulate Shh directly to initiate its expression in the posterior margin of the limb mesenchyme. Although the Hand2-Shh morphoregulatory system operates in both the forelimb and hindlimb bud, a recent analysis suggested that its upstream regulation is different in the forelimb and hindlimb bud. A combination of all four Hox9 genes is required for Hand2 expression in the forelimb-forming region; however, it remains elusive what genetic system regulates the Hand2-Shh pathway in the hindlimb-forming region. By conditional inactivation of Islet1 in the hindlimb-forming region using the Hoxb6Cre transgene, we show that Islet1 is required for establishing the posterior hindlimb field, but not the forelimb field, upstream of the Hand2-Shh pathway. Inactivation of Islet1 caused the loss of posterior structures in the distal and proximal regions, specifically in the hindlimb. We found that Hand2 expression was downregulated in the hindlimb field and that Shh expression was severely impaired in the hindlimb bud. In the Hoxb6Cre; Islet1 mutant pelvis, the proximal element that is formed in a Shh-independent manner, displayed complementary defects in comparison with Pitx1–/– hindlimbs. This suggests that Islet1 and Pitx1 function in parallel during girdle development in hindlimbs, which is in contrast with the known requirement for Tbx5 in girdle development in forelimbs. Our studies have identified a role for Islet1 in hindlimb-specific development and have revealed Islet1 functions in two distinct processes: regulation upstream of the Hand2-Shh pathway and contributions to girdle development.
Keywords: Islet1, Hand2, Shh, Limb, Forelimb-hindlimb, Pitx1-Tbx4, Mouse
INTRODUCTION
Vertebrate limb buds emerge as paired protrusions in the lateral plate mesoderm (LPM), forelimb buds located anteriorly and hindlimb buds located posteriorly. The molecular and genetic systems for correct patterning and growth of the limb bud have been studied extensively, mainly in mouse and chick model systems, and have demonstrated that the forelimb and hindlimb buds share most of their developmental programs (Zeller et al., 2009). Studies of various animal models, such as chondrichthyes (cartilaginous fishes) and teleost fish, have illustrated that limb/fin developmental systems are evolutionarily conserved, and suggested lateral fin folds as the origin of paired appendages (Tanaka et al., 2002; Yonei-Tamura et al., 2008). Throughout the development of the limb and fin in animals examined so far, SHH is a central factor (Dahn et al., 2007; Krauss et al., 1993; Riddle et al., 1993). Shh encodes a secreted cell-cell signaling molecule expressed at the posterior margin, which defines the mesenchymal zone of polarizing activity (ZPA) in both forelimb and hindlimb buds (Riddle et al., 1993). SHH regulates anterior-posterior patterning of digits and distal outgrowth of the limb bud (Chiang et al., 1996; Riddle et al., 1993; Yang et al., 1997). Several models have been proposed to explain the functions of Shh during limb development, such as the growth-morphogen model, the temporal expansion model and the biphasic model (Harfe et al., 2004; Towers et al., 2008; Zhu et al., 2008). In all these models, a central requirement is the localized expression of Shh in the posterior mesenchyme, which is tightly regulated. Recent analyses have uncovered the upstream regulatory system controlling Shh expression. Of particular importance is the basic helix-loop-helix transcription factor HAND2, which is expressed broadly in the LPM before limb outgrowth, but its subsequent expression becomes confined to the posterior region of both forelimb- and hindlimb-forming regions (Charite et al., 2000). HAND2 activates Shh transcription in the ZPA by directly binding to the far upstream limb bud-specific cis-regulatory element in the Shh landscape (Galli et al., 2010). In the absence of Hand2, Shh activation is disrupted specifically in the limb bud, and the limb skeleton of Hand2 conditional knockout embryos phenocopies that of Shh–/– embryos (Galli et al., 2010). Conversely, ectopic expression of Hand2 induces a small ectopic anterior Shh expression domain (Fernandez-Teran et al., 2000; McFadden et al., 2002). Given that crucial functions of Shh in limb development are linked to its expression in the posterior limb bud, the elucidation of the genetic mechanisms upstream of the Hand2-Shh interactions is important.
A recent report has demonstrated a striking difference in the upstream regulation of limb-specific Hand2-Shh pathways. Inactivating all four Hox9 paralogs (Hoxa9, Hoxb9, Hoxc9, Hoxd9) resulted in the loss of both Hand2 and Shh expression in mouse forelimb buds (Xu and Wellik, 2011). However, expression of the four Hox9 genes overlaps in the forelimb-but not hindlimb-forming area. Consistent with this expression pattern, Hox9 genes are required only in the forelimb-forming area, whereas hindlimb development proceeds normally and Hand2 and Shh are expressed normally in hindlimb buds of mouse embryos lacking all four Hox9 paralogs. This observation indicates that these two homologous tissues, the fore- and hindlimb bud; use different mechanisms to enable Hand2-dependent activation of Shh expression at the posterior margin. Furthermore, the hindlimb bud-specific mechanism acting upstream of this Hand2-Shh pathway remained thus far elusive.
Given that the overlapping expression of four Hox9 genes is detected in the forelimb-forming area, it is conceivable that gene(s) selectively expressed in the hindlimb-forming area function upstream of the Hand2-Shh pathway. Knockout studies of genes expressed specifically in the hindlimb-forming region, such as Tbx4, Pitx1 and Hoxc10, failed to demonstrate an involvement in regulation of the Hand2-Shh pathway. In Tbx4–/– embryos, Hand2 remains expressed in the hindlimb-forming region (Naiche and Papaioannou, 2003). Pitx1–/– embryos develop small hindlimbs with five digits, except for the loss of the anterior-most digit in an inbred 129sv genetic background, which pointed to the absence of significant defects in the Hand2-Shh pathway. Pitx1–/– embryos also exhibited a specific loss of the ilium, the anterior segment of the pelvic girdle, indicating its contribution to development of the anterior-proximal element (Lanctot et al., 1999; Marcil et al., 2003; Szeto et al., 1999). Inactivating Hoxc10, which marks the hindlimb forming region along with other Hox10 genes (Hoxa10 and Hoxd10) resulted in axial transformations, but only stylopod development was affected (McIntyre et al., 2007; Wellik, 2007; Wellik and Capecchi, 2003). These studies indicate that Tbx4, Pitx1 and Hoxc10, in combination with other Hox10 genes, are not required to regulate the Hand2-Shh pathway in the hindlimb.
In this study, we identify Islet1 (Isl1 – Mouse Genome Informatics), a LIM-homeodomain protein, as a key upstream regulator that controls the Hand2-Shh pathway specifically in hindlimb buds. Islet1 expression is restricted to hindlimb progenitors, but not to forelimb progenitors (Kawakami et al., 2011; Yang et al., 2006). Our recent analysis demonstrated that Tcre-mediated early inactivation of Islet1 resulted in a complete failure to initiate hindlimb bud development (Kawakami et al., 2011). This phenotype was caused by the lack of proliferation of hindlimb progenitors in the LPM and failure to activate the Fgf10-Fgf8 feedback loop formation. In this article, we conditionally inactivated Islet1 by using the Hoxb6Cre line, which causes Cre-dependent recombination in LPM later than Tcre (Lowe et al., 2000). This strategy appeared to bypass the early requirement of Islet1, and a large fraction of mutant embryos developed hindlimb buds. The mutant hindlimb buds exhibited downregulation of Shh expression, which is preceded by downregulation of Hand2 expression in the hindlimb-forming region. Our data reveal a novel Islet1-dependent genetic mechanism that controls specifically hindlimb bud development. This mechanism differs from the Hox9-dependent control of early forelimb bud development but converges at the level of Hand2 expression. Moreover, our results point to independent functions of Islet1 and Pitx1 in the patterning of distinct segments of the pelvic girdle in hindlimbs, in contrast to the contribution of Tbx5 to the development of the shoulder girdle as a whole in forelimbs (Rallis et al., 2003).
MATERIALS AND METHODS
Mouse lines
The Islet1flox/flox (Song et al., 2009; Sun et al., 2008), Hoxb6Cre (Lowe et al., 2000), Prx1Cre (Logan et al., 2002) and Hand2+/– (Galli et al., 2010) mouse lines were published previously. Islet1+/– mice were generated by germline recombination of Islet1flox allele by the CMV-Cre line. Islet1flox/flox, Hoxb6Cre; Islet1+/– and Hand2+/– were maintained on a mixed genetic background. Skeletal preparation was carried out as previously published (Kawakami et al., 2009). All of the animal breeding and procedures in the Kawakami laboratory were performed according to the approval by the Institutional Animal Care and Use Committee of University of Minnesota. Studies involving mice in the Zeller laboratory were performed in accordance with Swiss law after being approved by the Joint Commission on Experiments involving Animals of the Cantons of Argovia and both Basel.
Gene expression analysis
In situ hybridization was performed according to a standard procedure (Bluske et al., 2009; Kawakami et al., 2009; Wilkinson, 1993). For quantitative RT-PCR (qRT-PCR) analysis, we collected RNA from the hindlimb bud of each embryo at embryonic day (E) 10.5 using Trizol (Invitrogen), reverse-transcribed 1 μg total RNA with oligo dT primer and Superscript III (Invitrogen) and analyzed with an Eppendorf Mastercycler by the TaqMan method following manufacturer’s instructions (Applied Biosystems). Probes used were Shh (Mm_00436528_m1), Gli1 (Mm_00494645_m1), Ptch1 (Mm_00436026_m1), Pitx1 (Mm_00440824_m1) and Tbx4 (Mm_00550372_m1), and TATA-binding protein (Tbp) transcripts were used as an internal control (Mm_00446973_m1). Relative transcript levels were normalized using Tbp as internal standard and the average of specific transcript levels of control embryos was set to 100% (n=6 embryos). In the case of analysis of Shh transcript, owing to undetectable levels of Shh at E10.5 in mutant hindlimb buds, we pooled hindlimb buds from three E11.5 embryos with identical genotype as a group. Four groups of control and mutant embryos were analyzed. Statistical significance was examined by the independent t-test.
Immunofluorescence and histochemistry
Anti-ISLET1 immunofluorescence was carried out according to a standard method (Kawakami et al., 2011) with mouse anti-ISLET1 (39.4D5, Developmental Studies Hybridoma Bank). Detection was carried out using an Alexa488-anti-mouse IgG and a Zeiss LSM710 laser scanning confocal microscope or an HRP-anti-mouse IgG and a Zeiss Axioskop2 compound microscope. For double detection of Hand2 mRNA and ISLET1, section in situ hybridization was performed first with FITC-labeled Hand2 probe, then, sections were incubated with anti-ISLET1, and signals were detected by Alexa488-anti-FITC and Alexa594-anti-mouse IgG. The fluorescent signals were detected using a Zeiss LSM710 laser scanning confocal microscope.
RESULTS
Conditional inactivation of Islet1 causes hindlimb defects
Islet1 is expressed in the posterior embryo, including the hindlimb-forming region (Yang et al., 2006). Development of Islet1-deficient embryos arrests at E9.5 with severe cardiac defects (Cai et al., 2003; Pfaff et al., 1996), which precedes development of hindlimb buds. Thus, we conditionally inactivated Islet1 in the LPM, where hindlimb bud precursors arise. Cre-mediated recombination by Hoxb6Cre has been shown to take place in the LPM before outgrowth of the hindlimb bud (Li et al., 2005; Lowe et al., 2000). To establish that this approach eliminates ISLET1 from the hindlimb-forming region prior to the onset of hindlimb budding, we examined ISLET1 immunoreactivity in the LPM of the hindlimb-forming region in mutant embryos (Hoxb6CreTg/+; Islet1flox/–, hereafter referred to as Hoxb6Cre; Isl1 cKO) at E9.25-9.5 (20-24 somite stage). Compared with wild-type embryos (n=3), ISLET1 was undetectable in LPM of the hindlimb-forming region in four out of nine Hoxb6Cre; Isl1 cKO embryos (supplementary material Fig. S1D). Three out of nine embryos showed a faint signal in the LPM (supplementary material Fig. S1C), and two out of nine embryos showed clearly detectable ISLET1 signal (supplementary material Fig. S1B), although it was reduced in comparison with wild-type controls. These results indicate that Hoxb6Cre inactivates Islet1 with variable efficiency resulting in variable hypomorphic phenotypes. In particular, the early hindlimb initiation defects observed in Tcre; Isl1 mutants (Kawakami et al., 2011) are bypassed in a significant fraction of embryos, thereby allowing analysis of altered early hindlimb bud development.
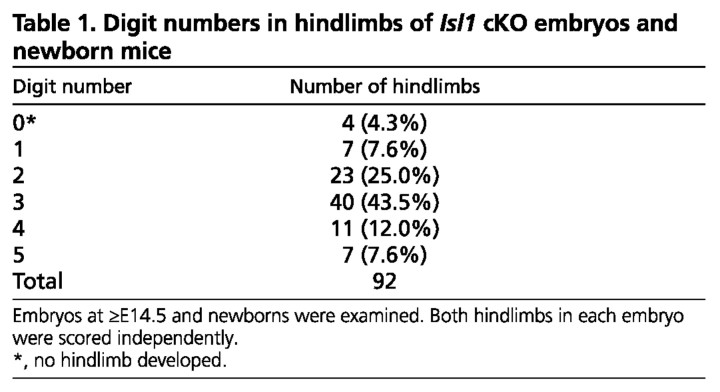
Hoxb6Cre; Isl1 cKO embryos exhibited defects in hindlimb development with varying severity in digit loss (Table 1, Fig. 1). In the most severe cases, no hindlimbs formed (4.3%; Fig. 1G) and only the pelvic girdle, which attaches the leg to vertebrae, was apparent (Fig. 1I). It is most likely that this phenotype reflected the morphological consequences of disrupting the Islet1-dependent activation of the Fgf10-Fgf8 feedback loop that is normally required for initiation of limb outgrowth (Kawakami et al., 2011). In the pelvic girdle, the ilium was formed, but the pubis and ischium were not patterned (Fig. 1W). However, in a fraction of Hoxb6Cre; Isl1 mutant embryos, this complete disruption of hindlimb development was bypassed by hypomorphic ISLET1 levels (supplementary material Fig. S1; n=5/9). Almost half of mutant embryos (43.5%) displayed hindlimbs with three digits and one zeugopodal bone in combination with a malformed pelvic girdle (Fig. 1D,O-S). Less severely affected hindlimbs with four digits were also observed (for details, see Table 1 and Fig. 1T-1V). In these mutants, the most posterior digit 5 was always missing, whereas the most anterior digit 1 formed. The morphology of the other digits probably corresponded to digit 2-4, providing further evidence for the hypomorphic nature of the conditional inactivation (supplementary material Fig. S1). A similar variation of digit loss was observed in the hindlimb of Hand2 cKO embryos (Galli et al., 2010), suggesting that the heterogeneity of inactivation could be the cause of the variation in phenotype. The single zeugopodal element observed in Hoxb6Cre; Isl1 cKO hindlimbs articulates with the femur (Fig. 1M,R) and, thus, is likely to correspond to the tibia, which indicates that the posterior zeugopod element (fibula) was lost. All mutants with one or two digits also exhibited similar phenotypes in the zeugopod and pelvic girdle, although mutants with four digits showed variable phenotypes in these elements. Thus, the majority of mutants exhibited similar skeletal defects with variable number of digits. No skeletal defects were observed in mutant forelimbs (Fig. 1B,E,H) as Hoxb6Cre-mediated recombination takes place only in the posterior half of the forelimb field (Lowe et al., 2000). This is consistent with our recent analysis of Tcre; Isl1 cKO embryos, in which Islet1 is inactivated in the forelimb-field, but with no alteration of forelimb bud development by E10.0 (Kawakami et al., 2011). Moreover, inactivating Islet1 using the Prx1Cre transgene, a limb mesenchyme-specific deleter, did not alter development of the forelimb skeleton (n=10 at E15.5 and n=8 in neonates; supplementary material Fig. S2). In summary, this analysis reveals the specific but variable loss of posterior skeletal elements in hindlimbs of a large fraction of Hoxb6Cre; Isl1 cKO embryos. These phenotypes are reminiscent of the limb skeletal defects caused by variable reduction of Shh transcript levels (Galli et al., 2010).
Table 1.
Digit numbers in hindlimbs of Isl1 cKO embryos and newborn mice
Fig. 1.
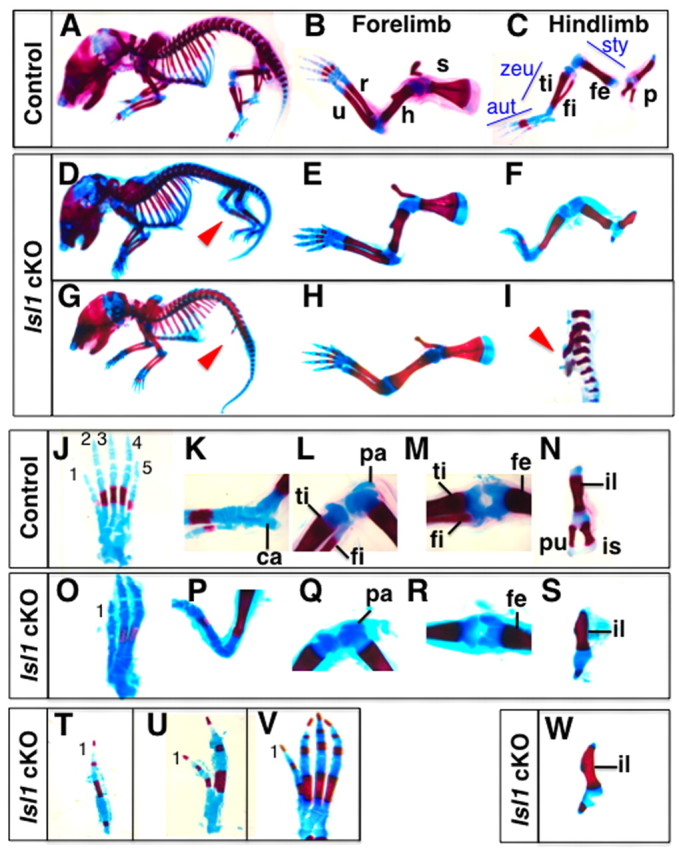
Hindlimb-specific defects in Hoxb6Cre; Islet1 conditional knockout mice. Skeletal preparations of newborn control (Hoxb6CreTg/+; Islet1flox/+; A-C,J-N) and mutant (Hoxb6CreTg/+; Islet1flox/-; D-I,O-W) mice. (A-C) Lateral views of the control mouse (A). In the forelimb (B), the scapula (s), humerus (h), radius (r) and ulna (u) are indicated, and in the hindlimb (C), pelvic girdle (p), femur (fe), tibia (ti) and fibula (fi) are indicated. Aut, autopod; sty, stylopod; zeu, zeugopod. (D-I) Lateral views of Isl1 cKO newborn with three digits in a leg (D-F) and a mutant lacking hindlimbs (G-I). In both cases, defects specific to the hindlimb (red arrowheads) were observed (D,G). Forelimbs formed normally (E,H) but only one zeugopodal bone formed in a mutant (F). In the mutant lacking hindlimbs, only the pelvic girdle is present (arrowhead, I) (J-N) Dorsal view of the hindlimb autopod (J), lateral views of the ankle (K) and knee (L) and dorsal views of the knee (M) and pelvic girdle (N) of a control mouse. Digits are numbered 1-5. The calcaneus (ca) in the ankle, and the patella (pa) in the knee are structures characteristic for hindlimbs. (O-S) Dorsal view of the hindlimb autopod with three digits (O). The calcaneus is missing in the ankle (P), but the patella is present in the knee (Q). The knee articulation in Isl1 cKO hindlimbs is similar to that in controls (R). The mutant pelvic girdle consists of an ilium (il) located anteriorly, whereas ischium (is) and pubis (pu) failed to develop. (T-W) Mutants with one digit (T), two digits (U) or four digits (V) were also obtained. In all cases the most anterior digit 1 was present and the most posterior digit (digit 5) was lost (O,T-V). In case of hindlimb aplasia, a pelvic girdle with a morphology similar to the other phenotypic groups formed (W).
Significant reduction of Shh expression in the hindlimb bud of Hoxb6Cre; Isl1 cKO embryos
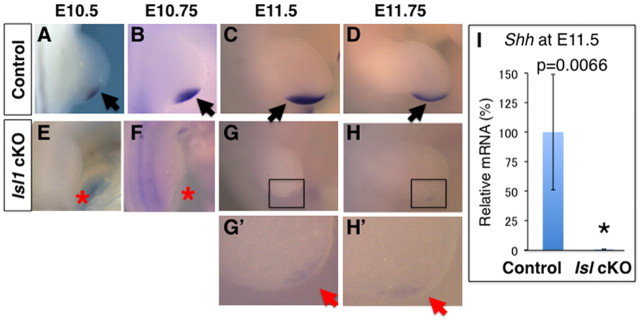
Shh is a crucial factor for regulating anterior-posterior patterning and progenitor expansion in the developing limb bud (Towers and Tickle, 2009; Zeller et al., 2009). Thus, we examined Shh expression by in situ hybridization. Because the morphological phenotype shows variation, we examined three to four mutant embryos at each stage, and analyzed mutant embryos with smaller hindlimb buds and comparable forelimb bud size in comparison with wild-type controls. At E10.5 (n=4) and E10.75 (n=3), when Shh expression normally expands within the posterior mesenchyme of wild-type limb buds (Fig. 2A,B), no Shh expression was detected in Hoxb6Cre; Isl1 cKO hindlimb buds (Fig. 2E,F). At E11.5-11.75, when Shh is expressed strongly in the distal-posterior mesenchyme of wild-type limb buds (Fig. 2C,D), only very low (n=3) or no (n=1) Shh expression was seen in mutant hindlimb buds (Fig. 2G-H′,I). Collectively, these results demonstrated that in Hoxb6Cre; Isl1 cKO hindlimb buds, Shh expression is very much lowered or even lost from early hindlimb bud stages onwards.
Fig. 2.
Shh expression is downregulated in Hoxb6Cre; Isl1 cKO hindlimb buds. (A-D) Shh expression in the control hindlimb bud at the stages indicated. Shh expression initiates in a small posterior mesenchymal domain (A) and expands distally (B,C) and is downregulated in advanced stages (D). Black arrowheads indicate normal expression. (E-H′) In mutant hindlimb buds, no Shh expression was detected at E10.5 (E) and E10.75 (F). At E11.5 (G,G′) and E11.75 (H,H′), very low levels of Shh transcripts were detected. G′ and H′ are magnified views of the boxed areas in G and H. Red arrowheads and asterisks indicate reduced and no expression, respectively. (I) qRT-PCR analysis of Shh transcripts in hindlimbs at E11.5. The relative Shh levels in mutant hindlimbs (n=4, 0.6%) in comparison with control littermate hindlimb buds (n=4) is shown as average ±s.d.
Reduced levels of SHH signaling in hindlimb buds of Hoxb6Cre; Isl1 cKO embryos
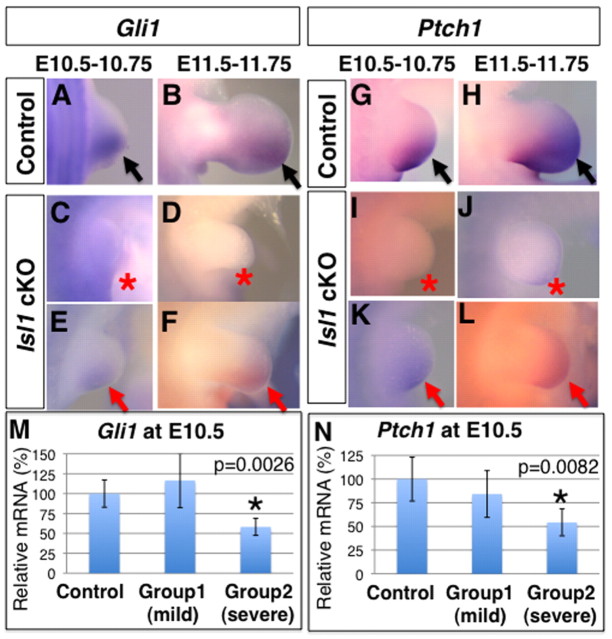
Loss of Shh results in the development of an autopod with a rudimentary anterior digit (Chiang et al., 1996), which was also observed in 7.6% of all Hoxb6Cre; Isl1 cKO hindlimbs. However, the hypomorphic nature of the digit loss suggests that SHH signal transduction might occur even at low levels of Shh expression. Therefore, expression of Gli1 and patched homolog 1 (Ptch1), two transcriptional targets of SHH signaling, was examined. Consistent with absent or reduced levels of Shh expression in Hoxb6Cre; Isl1 cKO hindlimb buds, Gli1 and Ptch1 expression was variable. In particular at E10.5-10.75, Gli1 (n=2/4) and Ptch1 (n=2/4) were not expressed in half of all hindlimb buds (Fig. 3A,C,G,I), and low levels were detected in the others (Fig. 3E,K). This was similar at E11.5-11.75 for both Gli1 and Ptch1 expression (Fig. 3B,D,F,H,J,L). qRT-PCR analysis at E10.5 revealed that there were two groups of embryos. In a subset of mutant hindlimb buds (n=6), we detected Gli1 and Ptch1 transcripts levels similar to those in control littermates (n=6). However, in a distinct subset (n=4), Gli1 (58.3%, P=0.0026) and Ptch1 (54.3%, P=0.0082) expression was significantly reduced in comparison with control littermates (Fig. 3M,N). The significant degree of variation of Gli1 and Patch1 transcript levels observed by qRT-PCR analysis agrees with the variable expression detected by RNA in situ hybridization. Data-based mechanistic models of how SHH controls limb bud patterning, such as the biphasic and temporal expansion models (Harfe et al., 2004; Zhu et al., 2008), depend crucially on high levels of SHH signaling for specification of posterior digits. The lack of the posterior-most digit 5 in most Hoxb6Cre; Isl1 cKO hindlimb buds (n=83/88) is consistent with the observed low levels of SHH signal transduction. Taken together, our results suggest that, owing to the variability in inactivating the conditional Islet1 allele, the levels of Shh transcripts and SHH signal transduction are also variable. This provides a molecular explanation for the variable penetrance of the skeletal phenotypes in hindlimb buds.
Fig. 3.
Reduction of SHH signaling in Hoxb6Cre; Isl1 cKO hindlimb buds. (A-L) Lateral views of Gli1 (A-F) and Ptch1 (G-L) expression in control (A,B,G,H) and Hoxb6Cre; Isl1 cKO (C-F,I-L) hindlimb buds. Black arrowheads indicate normal expression, red arrowheads and asterisks indicate reduced and no expression, respectively. (A-F) Gli1 expression in the posterior mesenchyme at E10.5-10.75 (A) and 11.5-11.75 (B) was either not detected (C,D) or significantly downregulated (E,F) in Isl1 cKO hindlimbs. (G-L). Ptch1 expression in the posterior mesenchyme at E10.5-10.75 (G) and 11.5-11.75 (H) was either not detected (I,J) or significantly downregulated (K,L) in Isl1 cKO hindlimbs. (M,N) qRT-PCR analysis of Gli1 (M) and Ptch1 (N) transcript levels in hindlimb buds at E10.5. The relative transcript levels in two groups of mutants (n=6 in group1, n=4 in group2) in comparison with controls (n=6) are shown as average ±s.d. Asterisks indicate statistically significant changes.
Additional transcriptional targets of SHH signal transduction include Hoxd13 in the distal mesenchyme (Riddle et al., 1993) and Fgf4 in the apical ectodermal ridge (AER; as part of the SHH/GREM1/FGF feedback loop) (Zuniga et al., 1999). In particular, Hoxd13 is expressed by the autopod primordia, with the exception of the anterior-most domain in wild-type limb buds (supplementary material Fig. S3A). In Hoxb6Cre; Isl1 cKO hindlimb buds, the posterior domain of Hoxd13 was reduced (supplementary material Fig. S3G). Fgf4 expression, which is restricted to the posterior half of the AER, was also more restricted in Hoxb6Cre; Isl1 cKO hindlimb buds. (supplementary material Fig. S3B,H). Likewise, the posterior expression domains of Tbx2 and Tbx3 were also reduced in Hoxb6Cre; Isl1 cKO hindlimb buds, whereas anterior expression was upregulated (supplementary material Fig. S3C,D,I,J), similar to Shh–/– forelimb buds (Galli et al., 2010). Furthermore, the normally anteriorly restricted expression of Alx4 and Irx3 expanded posteriorly in Hoxb6Cre; Isl1 cKO hindlimb buds (supplementary material Fig. S3E,F,K,L). These expression patterns pointed to a reduction of posterior gene expression and an expansion of anterior gene expression owing to the drastic lowering of SHH signal transduction in Hoxb6Cre; Isl1 cKO hindlimb buds.
Reduced expression and activity of the Fgf10-Fgf8 feedback loop and cell cycle-related genes in hindlimb buds of Hoxb6Cre; Isl1 cKO embryos
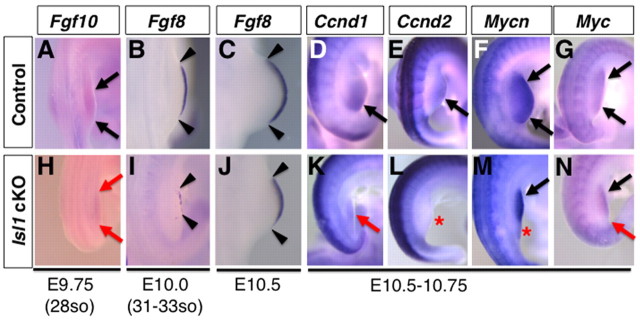
Distal outgrowth of limb buds requires maintenance of mesenchymal proliferation by the mesenchymal Fgf10 to ectodermal Fgf8 feedback loop (Capdevila and Izpisua Belmonte, 2001). Thus, we examined expression of these genes during early stages of hindlimb bud outgrowth. Fgf10 expression was reduced in Hoxb6Cre; Isl1 cKO hindlimb buds at E9.75 in comparison with controls (n=4/5; Fig. 4A,H). Similarly, Fgf8 was detected in a narrower AER domain in mutant than control hindlimb buds at E10.0 (n=3/3; Fig. 4B,I). The AER-Fgf8 expression domain expanded in E10.5 mutant hindlimb buds, but remained smaller than in control hindlimb buds (n=2/3; Fig. 4C,J). The reduced expression of both mesenchymal Fgf10 and AER Fgf8 pointed to reduced Fgf10-Fgf8 feedback loop activity, which, in combination with reduced SHH signal transduction, is the likely cause underlying the smaller hindlimb buds in Hoxb6Cre; Isl1 cKO embryos.
Fig. 4.
Reduced expression of Fgf10, Fgf8, cyclin type D and Myc genes in Hoxb6Cre; Isl1 cKO hindlimb buds. (A-N) Expression of Fgf10 (A,H), Fgf8 (B,C,I,J), Ccnd1 (D,K), Ccnd2 (E,L), Mycn (F,M) and Myc (G,N) in control (A-G) and Hoxb6Cre; Isl1 cKO (H-N) hindlimb buds at the indicated stages. Mesenchymal Fgf10 expression in mutant hindlimb buds (H, red arrows) was detected at lower levels in comparison with controls at E9.75 (A, black arrows). AER-Fgf8 expression in mutant hindlimb buds (I,J) was detected in a narrower than normal expression domain (B,C). Arrowheads in B, C, I and J indicate the anterior and posterior edges of the AER-Fgf8 expression domain. (D-G,K-N) In comparison with controls (black arrows in D-G), Ccnd1 expression was reduced in posterior mesenchyme of hindlimb buds (K, red arrow). Also the expression of Ccnd2 was significantly downregulated (L, red asterisk). In the posterior mesenchyme, Mycn expression was downregulated (M, red asterisk) and Myc expression was weaker (N, red arrow), whereas their expression in the anterior was not significantly altered (black arrows).
Consistent with this idea, we found that expression of D type cyclin genes and Myc genes was lowered preferentially in the posterior mesenchyme of Hoxb6Cre; Isl1 cKO hindlimb buds (Fig. 4D-G,K-N). These genes are known targets of SHH and FGF signaling, and are involved in cell cycle progression (Mill et al., 2005; Roy and Ingham, 2002; ten Berge et al., 2008). Alterations in expression of these genes are also consistent with the loss of the tibia and digit 5 as evidenced by the skeletal preparations.
Proximal patterning defects in Hoxb6Cre; Isl1 cKO hindlimb buds
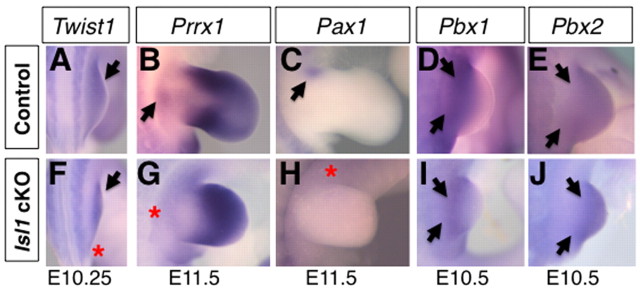
Hoxb6Cre; Isl1 cKO hindlimb skeletons also exhibited proximal defects (Fig. 1N,S,W) as in particular the pubis and ischium, two posterior segments of the pelvic girdle, were missing. As Shh–/– limbs develop normal proximal structures (Chiang et al., 2001; Kraus et al., 2001), the proximal defects are likely to be due to alterations other than the downregulation of Shh. Therefore, we examined expression of some of genes involved in the development of the pelvic girdle (Capellini et al., 2011). Indeed, the expression of Twist1 and Prrx1 in the proximal hindlimb bud was reduced and Pax1 expression was lost in Hoxb6Cre; Isl1 cKO hindlimb buds (Fig. 5A-C,F-H). These observations are consistent with the requirement of these three genes for development of the pubis, pubic symphysis and ischium (Krawchuk et al., 2010; Kuijper et al., 2005; ten Berge et al., 1998). By contrast, expression of Pbx1 and Pbx2 in the proximal region of mutant hindlimbs was similar to that of control embryos (Fig. 5D,E,I,J). Pbx genes are important for development of the ilium (Capellini et al., 2011), which was not affected in Hoxb6Cre; Isl1 cKO hindlimbs. These results indicate that Islet1 regulates the genetic programs that regulate the development of the proximal-posterior structures independently from its requirement for correct activation of the SHH pathway.
Fig. 5.
Altered pelvis marker gene expression in Hoxb6Cre; Isl1 cKO hindlimb buds. (A-J) Analysis of Twist1 (A,F), Prrx1 (B,G), Pax1 (C,H), Pbx1 (D,I) and Pbx2 (E,J) expression in control (A-E) and mutant (F-J) hindlimb buds at E10.25-11.5. Twist1 expression is detected both in control (A) and mutant (F) hindlimb buds (arrow), but its posterior-proximal expression is downregulated in mutant hindlimb buds (asterisk, F). Prrx1 expression in the proximal region in control hindlimb buds (B, arrow) is lacking from mutant hindlimb buds (G, asterisk). Pax1 is normally expressed in the proximal-anterior region (C, arrow) and is lost from mutant hindlimb buds (H, asterisk). Pbx1 (D,I) and Pbx2 (E,J) is expressed in both control and mutant hindlimb buds (arrows).
Islet1 appears to not be required for establishing hindlimb bud-specific characters
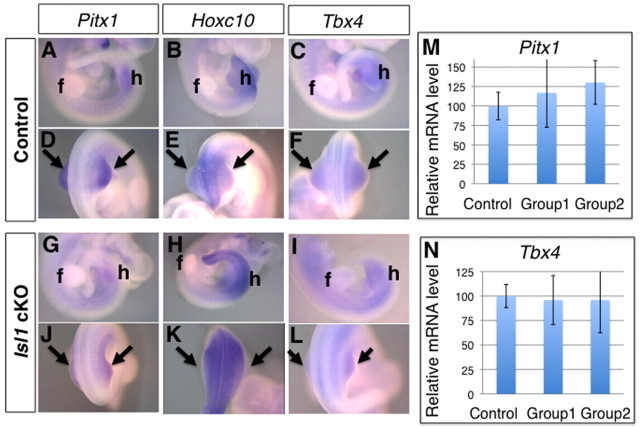
Pitx1, a homeodomain protein, is required and sufficient for determining hindlimb-specific characters (DeLaurier et al., 2006; Lanctot et al., 1999; Logan and Tabin, 1999; Szeto et al., 1999), whereas the role of Tbx4, a T-box transcription factor, in hindlimb specification remains controversial (Minguillon et al., 2005; Naiche and Papaioannou, 2007; Ouimette et al., 2010). Localized expression of Islet1 in hindlimb progenitors (Yang et al., 2006) suggests that it might be involved in establishing hindlimb-specific characters. Examination of the skeletal elements in Hoxb6Cre; Isl1 cKO hindlimbs revealed the normal knee articulation and presence of the patella, an element specific to hindlimbs (Fig. 1L,Q). However, the calcaneus, a hindlimb-specific structure in the autopod was missing (Fig. 1K,P). The latter aplasia might be secondary to the autopod hypoplasia rather than indicative of losing hindlimb-specific characters. Thus, we addressed further the possible contribution of Islet1 to establishing hindlimb-specific characters by analyzing the expression of hindlimb-specific genes, such as Pitx1, Tbx4 and Hoxc10, in Hoxb6Cre; Isl1 cKO embryos at E10.5 (Fig. 6). In situ hybridization failed to reveal any significant differences between wild-type and mutant hindlimb buds. In addition, we examined expression of Pitx1 and Tbx4 at E10.5 by qRT-PCR. In both group 1 mutants (Gli1 and Ptch1 levels not significantly altered, Fig. 3M,N) and group 2 mutants (Gli1 and Ptch1 levels downregulated, Fig. 3M,N), levels of Pitx1 and Tbx4 were comparable to controls (Fig. 6M,N). These results showed that the expression of hindlimb bud-specific genes was not altered in Hoxb6Cre; Isl1 cKO embryos and indicated that Islet1 is not required to establish and/or maintain hindlimb-specific character after activation of the Fgf10-Fgf8 feedback loop (Kawakami et al., 2011).
Fig. 6.
Hindlimb bud-specific gene expression is maintained in Hoxb6Cre; Isl1 cKO embryos. (A-L) Expression of Pitx1 (A,D,G,J), Hoxc10 (B,E,H,K) and Tbx4 (C,F,I,L) in control (A-F) and Hoxb6Cre; Isl1 cKO (G-L) embryos at E10.5. Lateral views (A-C,G-I) and dorsal views (D-F,J-L) are shown. Pitx1 was detected both in control (A,D) and Isl1 cKO (G,J) hindlimb buds. Hoxc10 was detected in control (B,E) and Isl1 cKO (H,K) hindlimb buds. Tbx4 was detected in control (C,F) and Isl1 cKO hindlimb buds (I,L). Arrows point to the expression in the hindlimb bud. f, forelimb buds; h, hindlimb buds. (M,N) qRT-PCR analysis of Pitx1 (M) and Tbx4 (N) transcripts in hindlimbs at E10.5. The relative transcript levels in two groups of mutants (n=6 in group1, n=4 in group2) in comparison with controls (n=6) are shown as average ±s.d. The same samples as for the analysis shown in Fig. 3 are used.
Islet1 is a hindlimb-specific upstream regulator of the Hand2-Shh morphoregulatory network
Islet1 expression is downregulated during initiation of hindlimb bud outgrowth (Yang et al., 2006) (Fig. 7) and does not overlap Shh expression, which suggests that Islet1 probably controls a regulator(s) of Shh expression. Molecular analysis of Hand2-deficient limb buds has shown that the HAND2 transcriptional regulator is required for activation Shh expression in the ZPA during the early phase of limb development (Charite et al., 2000; Galli et al., 2010). In particular, conditional Hand2 inactivation disrupts Shh activation and the resulting skeletal phenotypes are strikingly similar to Shh–/– limbs (Galli et al., 2010). Thus, we investigated whether Islet1 regulates Hand2 upstream of Shh expression. We first examined the ISLET1 protein distribution in hindlimb buds, and discovered that ISLET1 proteins persisted after its transcription had been downregulated (Fig. 7). ISLET1-positive cells were detected in a broad region of posterior hindlimb buds at E9.75 (27-28 somite stage), and remained in the ventral part of posterior hindlimb buds at E10.0 (30-31 somite stage). We found that the cytosolic Hand2 transcripts and nuclear ISLET1 signal colocalize in a single confocal plane within the mesenchymal cells of posterior hindlimbs at E9.75 (Fig. 8A-D). Therefore, we comparatively analyzed Hand2 expression at E9.75 (26-28 somites) in the hindlimb-forming region.
Fig. 7.
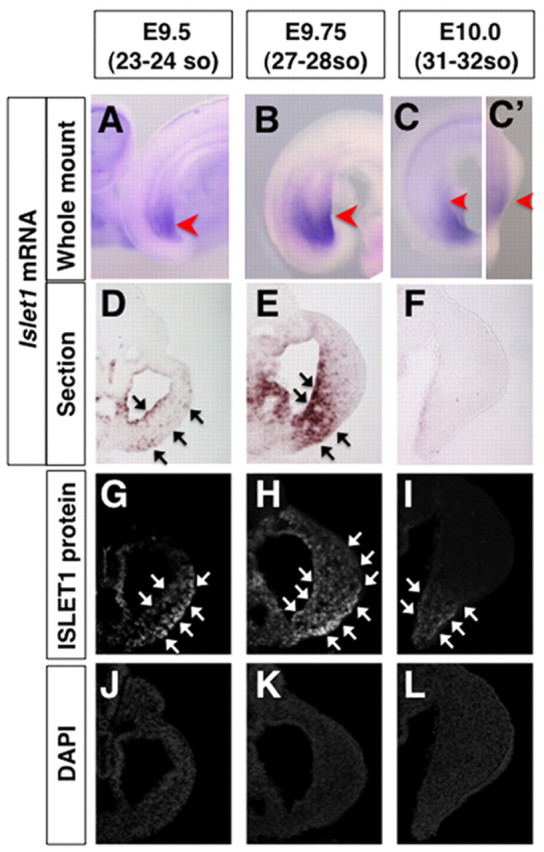
Spatial distribution of Islet1 mRNA and ISLET1 protein in hindlimb buds. (A-C′) RNA in situ hybridization showing the expression of Islet1 in the hindlimb field and bud. Red arrowheads indicate the approximate positions of sections shown in panels D-L. Dorsal-lateral views of the E9.5 (A) and E9.75 (B) embryos show the expression of Islet1 in the posterior hindlimb field. Lateral (C) and dorsal (C′) views of the E10.0 embryo show absence of Islet1 expression in hindlimb buds. (D-L) Islet1 section RNA in situ hybridization (D-F) and ISLET1 protein immunostaining (G-I) using adjacent sections, and DAPI analysis (J-L) of the sections shown in panels G-I. Immunoreactive ISLET1 proteins (G,H) were detected in more cells than were Islet1 transcripts (D,E) at E9.5 and E9.75. Islet1 mRNA was barely detectable in hindlimb buds at E10.0 (F), whereas ISLET1 proteins remained in the ventral part of the posterior hindlimb bud (I). Black and white arrows indicate Islet1 and ISLET1 positive areas, respectively.
Fig. 8.
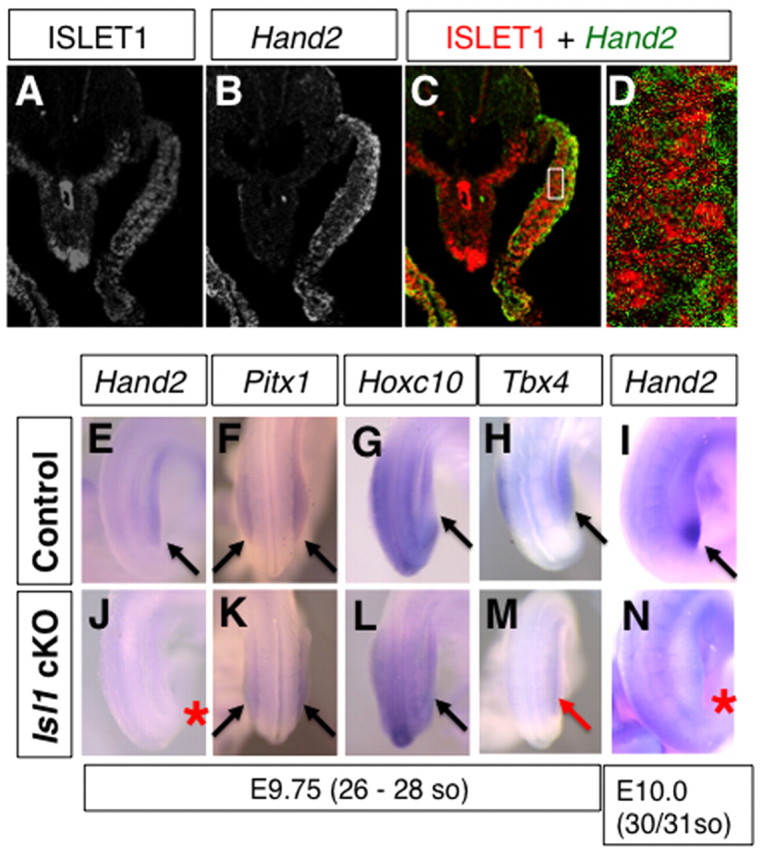
Islet1 acts upstream of Hand2 in the hindlimb-field and prior to Shh activation. (A-D) Colocalization of Hand2 mRNA and ISLET1. ISLET1 immunoreactivity (A), Hand2 mRNA by in situ hybridization (B) and a merged image (C) using the same section of a hindlimb field at E9.75. (D) Higher magnification view of the LPM (boxed in C) shows nuclear signal of ISLET1 (red) and cytoplasmic Hand2 mRNA signal (green) in the same cells. (E-N) Control (E-I) and mutant (J-N) embryos at E9.75 (26-28 somite stage) and E10.0 (30/31 somite stage) were examined for gene expression. Black arrows point to normal expression, and red arrow and asterisks point to reduced and lack of expression, respectively. Hand2 expression in the mutant hindlimb field (J) is severely downregulated in comparison with controls (E). Pitx1 (F,K) and Hoxc10 (G,L) were detected in both the control (F,G) and mutant (K,L) hindlimb field. Tbx4 expression in mutants (M) was lower than that in controls (H). The characteristic expression of Hand2 in the posterior mesenchyme (I) was absent in mutant hindlimb buds at E10.0 (N).
In contrast to wild-type controls, Hand2 expression was not detected in hindlimb buds of Hoxb6Cre; Isl1 cKO embryos at E9.75 (26-28 somite stage, n=2/3; Fig. 8E,J). At E10.0 (30/31 somite stage), when Hand2 expression was restricted to posterior mesenchyme in wild-type hindlimb buds (Fig. 8I), its expression could still not be detected in hindlimb buds of Hoxb6Cre; Isl1 cKO embryos (n=3/3; Fig. 8N). By contrast, the expression of other hindlimb field markers, such as Tbx4, Pitx1 and Hoxc10, was readily detected at E9.75, although Tbx4 levels appeared to be reduced (Fig. 8F-H,K-M). The expression of these hindlimb-field markers suggested that the hindlimb field is correctly specified whereas the expression of Hand2 was specifically lost in Hoxb6Cre; Isl1 cKO embryos. These results demonstrate that Islet1 acts upstream of the Hand2-Shh morphoregulatory network in hindlimb buds.
Hand2 expression is activated in a broad region of the LPM as early as E8.5 (Charite et al., 2000), raising the possibility that Hand2 and Islet1 interact in hindlimb progenitors prior to Shh activation. In the hindlimb-forming region of Hand2–/– embryos Islet1 was expressed normally (supplementary material Fig. S4A-C,F-H), which indicated that Hand2 did not regulate Islet1 expression. As the expression domain of Hand2 in the LPM by E9.5 is broader than that of Islet1, any interactions of Islet1 and Hand2 would be likely to take place in the hindlimb field at the time of bud initiation. Taken together, our study points to a hierarchical mechanism in which Islet1 induces Hand2 specifically in the hindlimb bud mesenchyme and HAND2 in turn participates in activation of Shh expression and, thereby, the posterior organizer region.
DISCUSSION
Two phases of Islet1 functions: initiation of hindlimb bud development and establishment of the posterior hindlimb-field upstream of Hand2
In this article, we identified a novel role of Islet1 during posterior hindlimb field development in the mouse embryo. Islet1 expression is initiated in a discrete posterior region of the embryo as early as E8.5, and continues to be expressed in the hindlimb-forming region (Yang et al., 2006). Islet1 transcripts are downregulated in hindlimb buds by E10.0 (Fig. 7). Our recent study (Kawakami et al., 2011) and this study reveal that Islet1 has two roles during this time window. Early inactivation of Islet1 by the Tcre line resulted in complete disruption of initiating the Fgf10-Fgf8 feedback loop and hindlimb bud outgrowth (Kawakami et al., 2011). Use of Hoxb6Cre, which results in later and more variable recombination than using Tcre (Lowe et al., 2000) (supplementary material Fig. S1), together with stability of ISLET1 proteins (Fig. 7), did not interfere with the earlier requirement of Islet1 in most cases and allowed us to study a second, later role for Islet1 in establishing the posterior hindlimb field. Weaker but detectable expression of Fgf10 and Fgf8 in Hoxb6Cre; Isl1 cKO hindlimb buds than in control (Fig. 4) illustrates bypassing of the early requirement of Islet1. However, a small fraction of Hoxb6Cre; Isl1 cKO embryos lacked hindlimbs (Fig. 1G-I), probably owing to variability in the timing of recombination.
Our Hoxb6Cre-mediated conditional knockout analysis demonstrated that Islet1 functions upstream of the Hand2-Shh morphoregulatory system (Fig. 9A). Previous fate-mapping analysis has shown that Islet1-expressing progenitors in the hindlimb-forming region contribute to mesenchymal tissue of the hindlimb bud in a posterior to anterior gradient (Yang et al., 2006). This observation is consistent with our finding that Hand2 expression in the posterior hindlimb-field was lost in Hoxb6Cre; Isl1 cKO embryos. The importance of regulating Hand2 specifically during hindlimb development has also been illustrated for dolphins (Thewissen et al., 2006), which are modern mammalian cetaceans that normally lack hindlimbs. Interestingly, hindlimb bud development is initiated in dolphin embryos, but regresses owing to lack of Shh expression by the ZPA, which results in failure to maintain the AER, and thereby limb bud outgrowth (Capdevila and Izpisua Belmonte, 2001; Johnson and Tabin, 1997). Dolphin embryos lack Hand2 expression specifically in the hindlimb region, whereas it is maintained in the forelimb region. These alterations bear similarities to the effects on hindlimb bud development of Hoxb6Cre; Isl1 cKO embryos (this study), which suggests that disruption of Islet1-mediated regulation of Hand2 in the hindlimb field might underlie the disruption of hindlimb bud development in dolphin embryos. Therefore, differential regulation of Hand2 in forelimb- and hindlimb-forming regions is likely to be essential for species-specific variations in early limb bud development in different species.
Fig. 9.
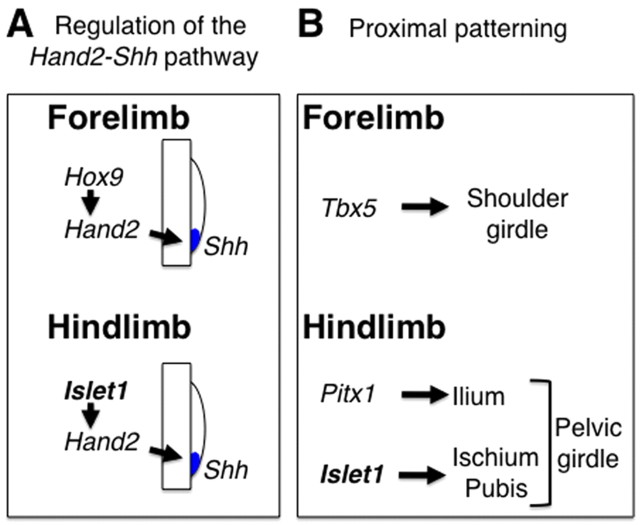
Scheme of the divergent genetic systems that control the Hand2-Shh morphoregulatory system and the proximal skeleton in fore- and hindlimb buds. (A) Axial Hox9 genes regulate the Hand2-Shh pathway during initiation of forelimb bud development. The present study reveals the hindlimb-specific regulation of the common Hand2-Shh pathway by Islet1. (B) A forelimb-field specific gene, Tbx5, is required for development of the shoulder girdle. The present study reveals that development of different segments of the pelvic girdle is controlled by either Islet1 or Pitx1.
The nascent limb field is pre-patterned by antagonistic interaction between Hand2 in the posterior mesenchyme and Gli3 in anterior mesenchyme (te Welscher et al., 2002). Our current study shows that in the early hindlimb bud, Islet1 participates in this process upstream of Hand2. In contrast to the widespread Hand2 expression in the LPM (Charite et al., 2000), Islet1 expression is restricted to the posterior part of the embryo proper and the hindlimb-forming territory (Yang et al., 2006). As Islet1 is more restricted than Hand2, the Hand2 expression must also be regulated by factors other than ISLET1. Thus, spatial restriction of Islet1 expression in the hindlimb-field might induce polarization of the posterior hindlimb field or early bud mesenchyme by activating the Hand2-Shh pathway specifically in the posterior domain.
Interestingly, Islet1 and Hand2 are also important for development of the second heart field. Mouse embryos lacking Islet1 fail to develop heart structures derived from the second heart field (Cai et al., 2003), survival and expansion of which requires Hand2 function (Tsuchihashi et al., 2011). Although the mechanisms by which Hand2 is regulated in the second heart field remains unknown, it is tempting to speculate that Islet1-mediated regulation of Hand2 expression is a shared feature of hindlimb and heart development.
LIM-homeodomain proteins during limb development
Studies to date have identified four LIM-homeodomain proteins that function in limb development. These include Islet1 (this study) (Kawakami et al., 2011), Lmx1b (Chen et al., 1998; Dreyer et al., 1998), Lhx2 and Lhx9 (Tzchori et al., 2009). Among these, Islet1 functions specifically during early hindlimb bud development, whereas the others participate in both forelimb and hindlimb bud development. Detailed genetic analysis of Lhx2, Lhx9, Lmx1b and their co-factor Ldb1 established that these genes cooperate in an overlapping manner to maintain the FGF10-FGF8 and the SHH/GREM1/AER-FGF signaling feedback loops (Tzchori et al., 2009). Inactivating several of these LIM-homeodomain factors together results in a failure to maintain Shh and localized Grem1 expression owing to the disrupted feedback regulation. By contrast, our data suggest that Islet1 acts upstream of these feedback loops, which reveals its distinct functions in comparison to the other LIM-homeodomain proteins.
Relationship between Islet1 and the Pitx1-Tbx4 pathway
Our recent study showed that early inactivation of Islet1 by Tcre caused reduction, but not abolishment, of Tbx4 expression without significantly altering Pitx1 expression in the LPM (Kawakami et al., 2011). Contrary to this, the expression of Tbx4 was not changed in hindlimb buds of Hoxb6Cre; Isl1 cKO embryos. Thus, the regulation of Tbx4 by ISLET1 appears to be limited to the period preceding hindlimb bud outgrowth, whereas Pitx1 appears to be a major regulator of Tbx4 in developing hindlimb buds (Lanctot et al., 1999). This is consistent with the observed expression of Islet1 in the hindlimb field and early bud as its expression is terminated by E10.0.
The defects in pelvic girdle development in Hoxb6Cre; Isl1 cKO embryos and Pitx1–/– embryos are distinct. In Hoxb6Cre; Isl1 cKO embryos, the ilium developed, whereas it was missing in Pitx1–/– embryos. By contrast, Hoxb6Cre; Isl1 cKO embryos lacked the pubis and ischium, which developed in Pitx1–/– embryos (Lanctot et al., 1999; Szeto et al., 1999). Thus, Islet1 and the Pitx1-Tbx4 pathway seem to regulate the development of different segments of the pelvic girdle in a parallel manner. This is in contrast to development of the shoulder girdle. Tbx5 is specifically expressed in the forelimb field (Gibson-Brown et al., 1996) and its conditional inactivation causes loss of the entire shoulder girdle (Rallis et al., 2003). Thus, the genetic program for girdle development in hindlimbs appears to be regulated in a distinct manner by Islet1 and the Pitx1-Tbx4 pathway, whereas Tbx5 regulates the entire genetic program for girdle development in forelimbs (Fig. 9B).
Evolutionary aspects of the regulation of the Hand2-Shh morphoregulatory system during paired appendage development
Growing evidence shows that early fore- and hindlimb development are controlled by different genetic systems (Abu-Daya et al., 2011; Agarwal et al., 2003; Itou et al., 2011; Rallis et al., 2003; Robertson et al., 2007). Our data, together with the recent analysis of the functions of Hox9 genes (Xu and Wellik, 2011), provide insight into the genetic disparities that underlie the establishment of the posterior mesenchymal organizer in the fore- and hindlimb buds. In the Hox9 quadruple knockout, Hand2 and Shh expression were not activated in forelimb buds, but hindlimb development progressed normally. It was envisaged that a similar scenario involving more posterior Hox paralogs would control Hand2 and Shh in early hindlimb buds. However, no Hox genes that would regulate the early expression of Hand2 in hindlimb buds have been identified (Wellik and Capecchi, 2003). Our data show that Islet1 fulfills a role similar to Hox9 paralogs and functions as a regulator of the Hand2-Shh morphoregulatory network in early hindlimb buds (Fig. 9A). Current evolutionary models of the origin of vertebrate appendages suggest that ancestral fin folds acquired Shh expression in posterior mesenchyme, which enabled development of the fin bud (Dahn et al., 2007; Tanaka et al., 2002; Yonei-Tamura et al., 2008). Given that the Hand2-Shh morphoregulatory network controls both teleost pectoral fin and tetrapod limb bud development (Galli et al., 2010; Gibert et al., 2006), and as both genes are expressed in fin buds of cartilaginous fish (Dahn et al., 2007; Tanaka et al., 2002; Yonei-Tamura et al., 2008), the direct interaction of HAND2 with Shh cis-regulatory regions would define an evolutionarily conserved module for initiation of appendage development (Charite et al., 2000; Galli et al., 2010; Yelon et al., 2000). The apparent differences in the genetic systems (Xu and Wellik, 2011) (this study) that control fore- and hindlimb bud induction indicate that these two types of paired appendages might have arisen by differential control of the Hand2-Shh module.
Acknowledgments
We thank Dr Michael Kuehn for providing the Hoxb6Cre line; Dr David Zarkower for the use of the Mastercycler machine; Dr Michael B. O’Connor for the use of the Zeiss LSM710; Dr Thomas Neufeld for the use of Zeiss Axioskop2; and Austin Johnson and Jenna Richter for excellent technical help. We also thank Dr Licia Selleri for sharing probes; Dr Naoyuki Wada for critical reading; and Developmental Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biology.
Footnotes
Funding
J.I. was partly supported by a fellowship from the Cell Science Research Foundation. This work is supported by the Minnesota Medical Foundation [3962-9211-09 to Y.K.] and American Cancer Society Institutional Research Grant [IRG-58-001-52-IRG04 to Y.K.] and the Swiss National Science Foundation [310003A_130803 to R.Z.].
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.073056/-/DC1
References
- Abu-Daya A., Nishimoto S., Fairclough L., Mohun T. J., Logan M. P., Zimmerman L. B. (2011). The secreted integrin ligand nephronectin is necessary for forelimb formation in Xenopus tropicalis. Dev. Biol. 349, 204–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal P., Wylie J. N., Galceran J., Arkhitko O., Li C., Deng C., Grosschedl R., Bruneau B. G. (2003). Tbx5 is essential for forelimb bud initiation following patterning of the limb field in the mouse embryo. Development 130, 623–633. [DOI] [PubMed] [Google Scholar]
- Bluske K. K., Kawakami Y., Koyano-Nakagawa N., Nakagawa Y. (2009). Differential activity of Wnt/beta-catenin signaling in the embryonic mouse thalamus. Dev. Dyn. 238, 3297–3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai C. L., Liang X., Shi Y., Chu P. H., Pfaff S. L., Chen J., Evans S. (2003). Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev. Cell 5, 877–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capdevila J., Izpisua Belmonte J. C. (2001). Patterning mechanisms controlling vertebrate limb development. Annu. Rev. Cell Dev. Biol. 17, 87–132. [DOI] [PubMed] [Google Scholar]
- Capellini T. D., Handschuh K., Quintana L., Ferretti E., Di Giacomo G., Fantini S., Vaccari G., Clarke S. L., Wenger A. M., Bejerano G., et al. (2011). Control of pelvic girdle development by genes of the Pbx family and Emx2. Dev. Dyn. 240, 1173–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charite J., McFadden D. G., Olson E. N. (2000). The bHLH transcription factor dHAND controls Sonic hedgehog expression and establishment of the zone of polarizing activity during limb development. Development 127, 2461–2470. [DOI] [PubMed] [Google Scholar]
- Chen H., Lun Y., Ovchinnikov D., Kokubo H., Oberg K. C., Pepicelli C. V., Gan L., Lee B., Johnson R. L. (1998). Limb and kidney defects in Lmx1b mutant mice suggest an involvement of LMX1B in human nail patella syndrome. Nat. Genet. 19, 51–55. [DOI] [PubMed] [Google Scholar]
- Chiang C., Litingtung Y., Lee E., Young K. E., Corden J. L., Westphal H., Beachy P. A. (1996). Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature 383, 407–413. [DOI] [PubMed] [Google Scholar]
- Chiang C., Litingtung Y., Harris M. P., Simandl B. K., Li Y., Beachy P. A., Fallon J. F. (2001). Manifestation of the limb prepattern: limb development in the absence of sonic hedgehog function. Dev. Biol. 236, 421–435. [DOI] [PubMed] [Google Scholar]
- Dahn R. D., Davis M. C., Pappano W. N., Shubin N. H. (2007). Sonic hedgehog function in chondrichthyan fins and the evolution of appendage patterning. Nature 445, 311–314. [DOI] [PubMed] [Google Scholar]
- DeLaurier A., Schweitzer R., Logan M. (2006). Pitx1 determines the morphology of muscle, tendon, and bones of the hindlimb. Dev. Biol. 299, 22–34. [DOI] [PubMed] [Google Scholar]
- Dreyer S. D., Zhou G., Baldini A., Winterpacht A., Zabel B., Cole W., Johnson R. L., Lee B. (1998). Mutations in LMX1B cause abnormal skeletal patterning and renal dysplasia in nail patella syndrome. Nat. Genet. 19, 47–50. [DOI] [PubMed] [Google Scholar]
- Fernandez-Teran M., Piedra M. E., Kathiriya I. S., Srivastava D., Rodriguez-Rey J. C., Ros M. A. (2000). Role of dHAND in the anterior-posterior polarization of the limb bud: implications for the Sonic hedgehog pathway. Development 127, 2133–2142. [DOI] [PubMed] [Google Scholar]
- Galli A., Robay D., Osterwalder M., Bao X., Benazet J. D., Tariq M., Paro R., Mackem S., Zeller R. (2010). Distinct roles of Hand2 in initiating polarity and posterior Shh expression during the onset of mouse limb bud development. PLoS Genet. 6, e1000901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibert Y., Gajewski A., Meyer A., Begemann G. (2006). Induction and prepatterning of the zebrafish pectoral fin bud requires axial retinoic acid signaling. Development 133, 2649–2659. [DOI] [PubMed] [Google Scholar]
- Gibson-Brown J. J., Agulnik S. I., Chapman D. L., Alexiou M., Garvey N., Silver L. M., Papaioannou V. E. (1996). Evidence of a role for T-box genes in the evolution of limb morphogenesis and the specification of forelimb/hindlimb identity. Mech. Dev. 56, 93–101. [DOI] [PubMed] [Google Scholar]
- Harfe B. D., Scherz P. J., Nissim S., Tian H., McMahon A. P., Tabin C. J. (2004). Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell 118, 517–528. [DOI] [PubMed] [Google Scholar]
- Itou J., Taniguchi N., Oishi I., Kawakami H., Lotz M., Kawakami Y. (2011). HMGB factors are required for posterior digit development through integrating signaling pathway activities. Dev. Dyn. 240, 1151–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. L., Tabin C. J. (1997). Molecular models for vertebrate limb development. Cell 90, 979–990. [DOI] [PubMed] [Google Scholar]
- Kawakami Y., Uchiyama Y., Rodriguez Esteban C., Inenaga T., Koyano-Nakagawa N., Kawakami H., Marti M., Kmita M., Monaghan-Nichols P., Nishinakamura R., et al. (2009). Sall genes regulate region-specific morphogenesis in the mouse limb by modulating Hox activities. Development 136, 585–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami Y., Marti M., Kawakami H., Itou J., Quach T., Johnson A., Sahara S., O’Leary D. D., Nakagawa Y., Lewandoski M., et al. (2011). Islet1-mediated activation of the beta-catenin pathway is necessary for hindlimb initiation in mice. Development 138, 4465–4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus P., Fraidenraich D., Loomis C. A. (2001). Some distal limb structures develop in mice lacking Sonic hedgehog signaling. Mech. Dev. 100, 45–58. [DOI] [PubMed] [Google Scholar]
- Krauss S., Concordet J. P., Ingham P. W. (1993). A functionally conserved homolog of the Drosophila segment polarity gene hh is expressed in tissues with polarizing activity in zebrafish embryos. Cell 75, 1431–1444. [DOI] [PubMed] [Google Scholar]
- Krawchuk D., Weiner S. J., Chen Y. T., Lu B. C., Costantini F., Behringer R. R., Laufer E. (2010). Twist1 activity thresholds define multiple functions in limb development. Dev. Biol. 347, 133–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijper S., Beverdam A., Kroon C., Brouwer A., Candille S., Barsh G., Meijlink F. (2005). Genetics of shoulder girdle formation: roles of Tbx15 and aristaless-like genes. Development 132, 1601–1610. [DOI] [PubMed] [Google Scholar]
- Lanctot C., Moreau A., Chamberland M., Tremblay M. L., Drouin J. (1999). Hindlimb patterning and mandible development require the Ptx1 gene. Development 126, 1805–1810. [DOI] [PubMed] [Google Scholar]
- Li C., Xu X., Nelson D. K., Williams T., Kuehn M. R., Deng C. X. (2005). FGFR1 function at the earliest stages of mouse limb development plays an indispensable role in subsequent autopod morphogenesis. Development 132, 4755–4764. [DOI] [PubMed] [Google Scholar]
- Logan M., Tabin C. J. (1999). Role of Pitx1 upstream of Tbx4 in specification of hindlimb identity. Science 283, 1736–1739. [DOI] [PubMed] [Google Scholar]
- Logan M., Martin J. F., Nagy A., Lobe C., Olson E. N., Tabin C. J. (2002). Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis 33, 77–80. [DOI] [PubMed] [Google Scholar]
- Lowe L. A., Yamada S., Kuehn M. R. (2000). HoxB6-Cre transgenic mice express Cre recombinase in extra-embryonic mesoderm, in lateral plate and limb mesoderm and at the midbrain/hindbrain junction. Genesis 26, 118–120. [DOI] [PubMed] [Google Scholar]
- Marcil A., Dumontier E., Chamberland M., Camper S. A., Drouin J. (2003). Pitx1 and Pitx2 are required for development of hindlimb buds. Development 130, 45–55. [DOI] [PubMed] [Google Scholar]
- McFadden D. G., McAnally J., Richardson J. A., Charite J., Olson E. N. (2002). Misexpression of dHAND induces ectopic digits in the developing limb bud in the absence of direct DNA binding. Development 129, 3077–3088. [DOI] [PubMed] [Google Scholar]
- McIntyre D. C., Rakshit S., Yallowitz A. R., Loken L., Jeannotte L., Capecchi M. R., Wellik D. M. (2007). Hox patterning of the vertebrate rib cage. Development 134, 2981–2989. [DOI] [PubMed] [Google Scholar]
- Mill P., Mo R., Hu M. C., Dagnino L., Rosenblum N. D., Hui C. C. (2005). Shh controls epithelial proliferation via independent pathways that converge on N-Myc. Dev. Cell 9, 293–303. [DOI] [PubMed] [Google Scholar]
- Minguillon C., Del Buono J., Logan M. P. (2005). Tbx5 and Tbx4 are not sufficient to determine limb-specific morphologies but have common roles in initiating limb outgrowth. Dev. Cell 8, 75–84. [DOI] [PubMed] [Google Scholar]
- Naiche L. A., Papaioannou V. E. (2003). Loss of Tbx4 blocks hindlimb development and affects vascularization and fusion of the allantois. Development 130, 2681–2693. [DOI] [PubMed] [Google Scholar]
- Naiche L. A., Papaioannou V. E. (2007). Tbx4 is not required for hindlimb identity or post-bud hindlimb outgrowth. Development 134, 93–103. [DOI] [PubMed] [Google Scholar]
- Ouimette J.-F., Jolin M. L., L’honore A., Gifuni A., Drouin J. (2010). Divergent transcriptional activities determine limb identity. Nat. Commun. 1, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaff S. L., Mendelsohn M., Stewart C. L., Edlund T., Jessell T. M. (1996). Requirement for LIM homeobox gene Isl1 in motor neuron generation reveals a motor neuron-dependent step in interneuron differentiation. Cell 84, 309–320. [DOI] [PubMed] [Google Scholar]
- Rallis C., Bruneau B. G., Del Buono J., Seidman C. E., Seidman J. G., Nissim S., Tabin C. J., Logan M. P. (2003). Tbx5 is required for forelimb bud formation and continued outgrowth. Development 130, 2741–2751. [DOI] [PubMed] [Google Scholar]
- Riddle R. D., Johnson R. L., Laufer E., Tabin C. (1993). Sonic hedgehog mediates the polarizing activity of the ZPA. Cell 75, 1401–1416. [DOI] [PubMed] [Google Scholar]
- Robertson E. J., Charatsi I., Joyner C. J., Koonce C. H., Morgan M., Islam A., Paterson C., Lejsek E., Arnold S. J., Kallies A., et al. (2007). Blimp1 regulates development of the posterior forelimb, caudal pharyngeal arches, heart and sensory vibrissae in mice. Development 134, 4335–4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S., Ingham P. W. (2002). Hedgehogs tryst with the cell cycle. J. Cell Sci. 115, 4393–4397. [DOI] [PubMed] [Google Scholar]
- Song M. R., Sun Y., Bryson A., Gill G. N., Evans S. M., Pfaff S. L. (2009). Islet-to-LMO stoichiometries control the function of transcription complexes that specify motor neuron and V2a interneuron identity. Development 136, 2923–2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Dykes I. M., Liang X., Eng S. R., Evans S. M., Turner E. E. (2008). A central role for Islet1 in sensory neuron development linking sensory and spinal gene regulatory programs. Nat. Neurosci. 11, 1283–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szeto D. P., Rodriguez-Esteban C., Ryan A. K., O’Connell S. M., Liu F., Kioussi C., Gleiberman A. S., Izpisua-Belmonte J. C., Rosenfeld M. G. (1999). Role of the Bicoid-related homeodomain factor Pitx1 in specifying hindlimb morphogenesis and pituitary development. Genes Dev. 13, 484–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Munsterberg A., Anderson W. G., Prescott A. R., Hazon N., Tickle C. (2002). Fin development in a cartilaginous fish and the origin of vertebrate limbs. Nature 416, 527–531. [DOI] [PubMed] [Google Scholar]
- te Welscher P., Fernandez-Teran M., Ros M. A., Zeller R. (2002). Mutual genetic antagonism involving GLI3 and dHAND prepatterns the vertebrate limb bud mesenchyme prior to SHH signaling. Genes Dev. 16, 421–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Berge D., Brouwer A., Korving J., Martin J. F., Meijlink F. (1998). Prx1 and Prx2 in skeletogenesis: roles in the craniofacial region, inner ear and limbs. Development 125, 3831–3842. [DOI] [PubMed] [Google Scholar]
- ten Berge D., Brugmann S. A., Helms J. A., Nusse R. (2008). Wnt and FGF signals interact to coordinate growth with cell fate specification during limb development. Development 135, 3247–3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thewissen J. G., Cohn M. J., Stevens L. S., Bajpai S., Heyning J., Horton W. E., Jr (2006). Developmental basis for hind-limb loss in dolphins and origin of the cetacean bodyplan. Proc. Natl. Acad. Sci. USA 103, 8414–8418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towers M., Tickle C. (2009). Growing models of vertebrate limb development. Development 136, 179–190. [DOI] [PubMed] [Google Scholar]
- Towers M., Mahood R., Yin Y., Tickle C. (2008). Integration of growth and specification in chick wing digit-patterning. Nature 452, 882–886. [DOI] [PubMed] [Google Scholar]
- Tsuchihashi T., Maeda J., Shin C. H., Ivey K. N., Black B. L., Olson E. N., Yamagishi H., Srivastava D. (2011). Hand2 function in second heart field progenitors is essential for cardiogenesis. Dev. Biol. 351, 62–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzchori I., Day T. F., Carolan P. J., Zhao Y., Wassif C. A., Li L., Lewandoski M., Gorivodsky M., Love P. E., Porter F. D., et al. (2009). LIM homeobox transcription factors integrate signaling events that control three-dimensional limb patterning and growth. Development 136, 1375–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellik D. M. (2007). Hox patterning of the vertebrate axial skeleton. Dev. Dyn. 236, 2454–2463. [DOI] [PubMed] [Google Scholar]
- Wellik D. M., Capecchi M. R. (2003). Hox10 and Hox11 genes are required to globally pattern the mammalian skeleton. Science 301, 363–367. [DOI] [PubMed] [Google Scholar]
- Wilkinson D. G. (1993). Whole mount in situ hybridization of vertebrate embryos. In In Situ Hybridization. Oxford: Oxford University Press. [Google Scholar]
- Xu B., Wellik D. M. (2011). Axial Hox9 activity establishes the posterior field in the developing forelimb. Proc. Natl. Acad. Sci. USA 108, 4888–4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Cai C. L., Lin L., Qyang Y., Chung C., Monteiro R. M., Mummery C. L., Fishman G. I., Cogen A., Evans S. (2006). Isl1Cre reveals a common Bmp pathway in heart and limb development. Development 133, 1575–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Drossopoulou G., Chuang P. T., Duprez D., Marti E., Bumcrot D., Vargesson N., Clarke J., Niswander L., McMahon A., et al. (1997). Relationship between dose, distance and time in Sonic Hedgehog-mediated regulation of anteroposterior polarity in the chick limb. Development 124, 4393–4404. [DOI] [PubMed] [Google Scholar]
- Yelon D., Ticho B., Halpern M. E., Ruvinsky I., Ho R. K., Silver L. M., Stainier D. Y. (2000). The bHLH transcription factor hand2 plays parallel roles in zebrafish heart and pectoral fin development. Development 127, 2573–2582. [DOI] [PubMed] [Google Scholar]
- Yonei-Tamura S., Abe G., Tanaka Y., Anno H., Noro M., Ide H., Aono H., Kuraishi R., Osumi N., Kuratani S., et al. (2008). Competent stripes for diverse positions of limbs/fins in gnathostome embryos. Evol. Dev. 10, 737–745. [DOI] [PubMed] [Google Scholar]
- Zeller R., Lopez-Rios J., Zuniga A. (2009). Vertebrate limb bud development: moving towards integrative analysis of organogenesis. Nat. Rev. Genet. 10, 845–858. [DOI] [PubMed] [Google Scholar]
- Zhu J., Nakamura E., Nguyen M. T., Bao X., Akiyama H., Mackem S. (2008). Uncoupling Sonic hedgehog control of pattern and expansion of the developing limb bud. Dev. Cell 14, 624–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga A., Haramis A. P., McMahon A. P., Zeller R. (1999). Signal relay by BMP antagonism controls the SHH/FGF4 feedback loop in vertebrate limb buds. Nature 401, 598–602. [DOI] [PubMed] [Google Scholar]