Abstract
Despite the central importance of germ cells for transmission of genetic material, our understanding of the molecular programs that control primordial germ cell (PGC) specification and differentiation are limited. Here, we present findings that X chromosome NonDisjunction factor-1 (XND-1), known for its role in regulating meiotic crossover formation, is an early determinant of germ cell fates in Caenorhabditis elegans. xnd-1 mutant embryos display a novel ‘one PGC’ phenotype as a result of G2 cell cycle arrest of the P4 blastomere. Larvae and adults display smaller germ lines and reduced brood size consistent with a role for XND-1 in germ cell proliferation. Maternal XND-1 proteins are found in the P4 lineage and are exclusively localized to the nucleus in PGCs, Z2 and Z3. Zygotic XND-1 turns on shortly thereafter, at the ∼300-cell stage, making XND-1 the earliest zygotically expressed gene in worm PGCs. Strikingly, a subset of xnd-1 mutants lack germ cells, a phenotype shared with nos-2, a member of the conserved Nanos family of germline determinants. We generated a nos-2 null allele and show that nos-2; xnd-1 double mutants display synthetic sterility. Further removal of nos-1 leads to almost complete sterility, with the vast majority of animals without germ cells. Sterility in xnd-1 mutants is correlated with an increase in transcriptional activation-associated histone modification and aberrant expression of somatic transgenes. Together, these data strongly suggest that xnd-1 defines a new branch for PGC development that functions redundantly with nos-2 and nos-1 to promote germline fates by maintaining transcriptional quiescence and regulating germ cell proliferation.
KEY WORDS: C. elegans, Nanos, XND-1, Germ line, Primordial germ cells, Proliferation
Summary: The chromatin-associated protein XND-1 is identified as a key regulator of a novel pathway for primordial germ cell differentiation in C. elegans.
INTRODUCTION
For sexually reproducing metazoans, the ability to transfer genetic information to the next generation resides in the germ cells. During early embryogenesis, the germ lineage inherits germ plasm, the specialized cytoplasm containing maternally encoded mRNAs and proteins, and gives rise to the primordial germ cell (PGC) lineage. A common theme emerging from germ cell studies in C. elegans, Drosophila and mouse is that transcriptional repression is crucial for PGC specification (Nakamura and Seydoux, 2008; Pirrotta, 2002; Seydoux and Schedl, 2001). In C. elegans and Drosophila, transcriptional repression is achieved by two means. Firstly, by direct inhibition of RNA polymerase II transcriptional initiation and elongation functions (Seydoux and Dunn, 1997; Van Doren et al., 1998). And secondly, by regulation of chromatin states, specifically inhibition of histone H3 lysine 4 methylation (H3K4me2), a mark of active chromatin, and retention of histone H3 lysine 9 methylation (H3K9me) for repressed chromatin (reviewed by Nakamura and Seydoux, 2008). This repression is thought to be essential in preventing the activation of somatic genes within the germ line. Indeed, aberrant accumulation of H3K4me2 and loss of H3K9me in the germ line has been correlated with aberrant transcriptional activation and progressive loss of fertility (Katz et al., 2009; Kerr et al., 2014).
In C. elegans, germ plasm is segregated through four asymmetric cell divisions into germline blastomere, known as P4. This then divides symmetrically to give rise to PGCs, Z2 and Z3 (Sulston et al., 1983), which replicate their DNA and arrest in the G2 stage of the cell cycle for the remainder of embryogenesis (Fukuyama et al., 2006). In this fashion, PGCs are primed to proliferate when the larva hatches and senses food. Germ cells enter meiosis midway through the third larval stage (L3) and, in the hermaphrodite, the first germ cells differentiate into sperm before switching to oogenesis in L4 (Kimble and White, 1981; Sulston et al., 1983). Although the chromatin of the P lineage has high levels of H3K4me2 (Schaner et al., 2003), it is transcriptionally quiescent because PIE-1 blocks phosphorylation of the RNA polymerase II CTD (Seydoux and Dunn, 1997). Z2 and Z3 maintain transcriptional quiescence by losing marks of active chromatin (Schaner et al., 2003) and attaining a unique condensed chromatin appearance (Nakamura and Seydoux, 2008). Despite high levels of repressive chromatin marks, zygotic expression of germline genes can be observed during embryogenesis, with the earliest zygotic transcripts seen at the ∼550 cell stage. Early transcripts include those for P granule components (pgl-1) and the Nanos ortholog, nos-1 (Kawasaki et al., 2004; Subramaniam and Seydoux, 1999).
Members of the Nanos gene family have emerged as conserved determinants of germline development (Tsuda et al., 2003). The founding member of this family, the Drosophila maternal effect gene nanos, was identified for its role in embryonic patterning (Wang and Lehmann, 1991) and later shown to have roles in PGC specification and migration during embryogenesis (Forbes and Lehmann, 1998). Zygotically expressed Nanos is required for the differentiation of germline stem cells in the adult gonad (Forbes and Lehmann, 1998; Kobayashi et al., 1996). Nanos is an RNA-binding protein which functions together with Pumilio to inhibit translation initiation (Sonoda and Wharton, 1999), at least in part by recruiting the CCR4-NOT deadenylation complex to target genes (Kadyrova et al., 2007). Mouse NANOS2 also interacts with CCR4-NOT, where it has been proposed to trigger the degradation of RNAs involved in meiosis (Suzuki et al., 2010, 2012). C. elegans possesses four Nanos homologs, nos-1, nos-2, nos-3 and T02G5.11. nos-2 is maternally deposited and functions embryonically to ensure incorporation of PGCs into the somatic gonad. It also functions redundantly with nos-1 to promote PGC proliferation and survival during larval development (Subramaniam and Seydoux, 1999). The functional targets of C. elegans NOS-1 and NOS-2 that are required for germ cell differentiation remain elusive and the partial sterile phenotypes seen in double mutants intimate the existence of additional germ cell determinants.
Previously, we identified xnd-1 as a chromatin-associated protein that regulates X chromosome crossover formation (Wagner et al., 2010). Other phenotypes associated with xnd-1, including sterility and smaller germline size, were suggestive of a broader role in germline development. Here, we report that xnd-1 is a key regulator of germ cell development in C. elegans. xnd-1 mutant embryos have defects in P4 division, P granule segregation, and PGC migration. In addition, we show that XND-1 is one of the first proteins turned on in the PGCs, at the ∼300 cell stage. This zygotic protein is required for PGC proliferation in addition to its later meiotic role. Double mutants of xnd-1 and nos-2 exhibit a synthetic sterile phenotype with a large proportion of animals containing no or severely reduced numbers of germ cells. The sterility in xnd-1 single and double mutants is accompanied by an increase in H3K4me2 in PGCs, suggesting that aberrant transcriptional activation might underlie the increased sterility in these animals. These studies therefore identify XND-1 as one of the earliest markers of PGCs and show that it functions in parallel to previously characterized PGCs determinants, thus defining a novel pathway for germ cell differentiation.
RESULTS
XND-1 is among the earliest proteins to be expressed in newly born PGCs
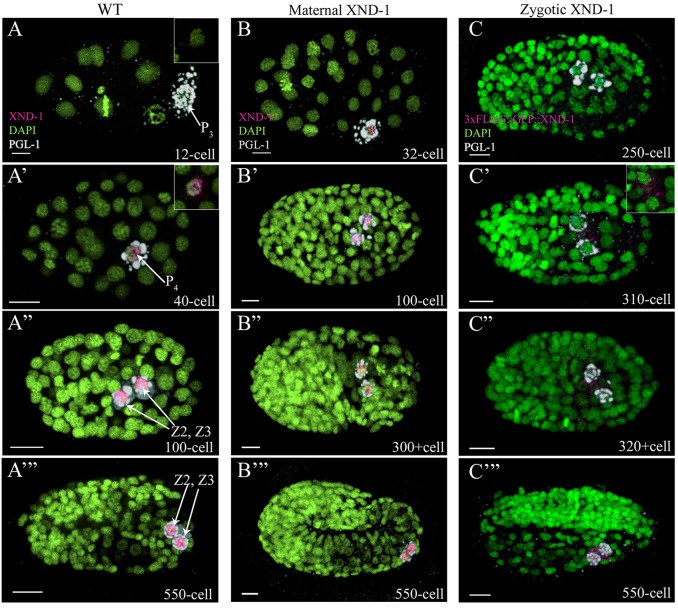
The xnd-1 gene of C. elegans regulates meiotic crossover formation consistent with its expression from the mitotic tip of the germ line through the late pachytene region (Wagner et al., 2010). However, the sterility and reduced brood sizes associated with xnd-1 suggest a more pleiotropic role in germline development. Therefore, we set out to examine XND-1 expression throughout development using our previously described anti-XND-1 antibodies (Wagner et al., 2010) with anti-PGL-1 antibodies to mark a core component of the germ plasm (Kawasaki et al., 1998, 2004). The antibodies were specific to XND-1 in embryos, as well as in the adult germ line, as shown by lack of staining in xnd-1 mutants (Fig. S1A-A″). No XND-1 was visible in <28-cell stage embryos (Fig. 1A); it was first detected in the cytoplasm and nucleus of P4 (Fig. 1A′). Chromatin association of XND-1 is readily apparent at this stage. This early embryo expression is consistent with mRNA expression data that indicated xnd-1 mRNA is maternally deposited and preferentially enriched in P4 (NEXTDB; http://nematode.lab.nig.ac.jp). We note that the in situ data suggests that xnd-1 mRNAs are found in all embryonic blastomeres, raising the possibility that post-transcriptional mechanisms repress xnd-1 both in the early embryo and in later, somatic blastomeres. Consistent with this hypothesis, a targeted genetic screen in our lab has identified mutants that derepress XND-1 in somatic blastomeres (M.R. and J.L.Y., unpublished data). Just prior to the division of P4, XND-1 localization became predominantly nuclear and remained chromatin-associated for the remainder of development and into the adult. (Fig. 1A″,A‴; Fig. S1B).
Fig. 1.
XND-1 is expressed in primordial germ cells. (A-C) C. elegans embryos stained with anti-XND-1 (magenta), anti-PGL-1 (white) to mark the germ lineage, and DAPI (green) to show embryonic nuclei. All embryos are oriented with anterior to the left. (A-A‴) Wild-type embryos showed XND-1 expression in P4 and its daughters, Z2 and Z3. (A) Inset shows that XND-1 was absent in the germline blastomere of 12-cell stage embryos, (A′-A‴) whereas its accumulation and expression was seen in the cytoplasm and the nucleus of the P4 cell at the 40-cell stage. (B) xnd-1 M+Z− embryos showing maternal XND-1 expression similar to that in wild type. (C) Zygotic expression of XND-1 was assessed by staining embryos resulting from a cross of 3×FLAG::GFP::xnd-1 transgenic males with wild-type hermaphrodites. XND-1 was not detected until the 310-cell stage (C′), at which time its accumulation was seen in the cytoplasm of PGCs. Shortly thereafter, at the ∼320+cell stage, XND-1 localization became nuclear (C″,C‴). Scale bars: 5 µm.
We also generated transgenic animals containing stably integrated transgenes expressing an N-terminus fusion of 3×FLAG::GFP to XND-1. The transgenic lines rescued the low brood and high embryonic lethality defects of xnd-1(ok709), confirming they are fully functional (Table S1). Transgenic lines showed identical expression patterns to that of immunostaining; GFP was discernible only in the PGCs during embryonic development (Fig. S2A,B), expression persisted exclusively in the germline nuclei throughout larval development, and adult expression was observed in the mitotic zone through the late pachytene region with an autosomal bias (Fig. S2C) (Wagner et al., 2010). XND-1 protein abruptly disappears from the late pachytene region and remains absent in mature oocytes and sperm (Fig. S1B, Fig. S2C). No somatic expression was observed with the transgenes or by immunostaining, confirming our prior conclusion from genetic studies that xnd-1 function appears limited to the germ line (Wagner et al., 2010).
xnd-1 regulates germ cell proliferation
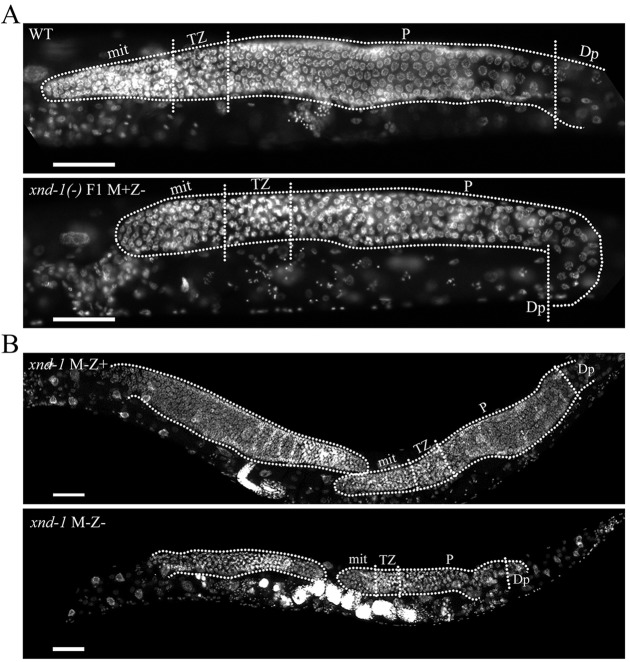
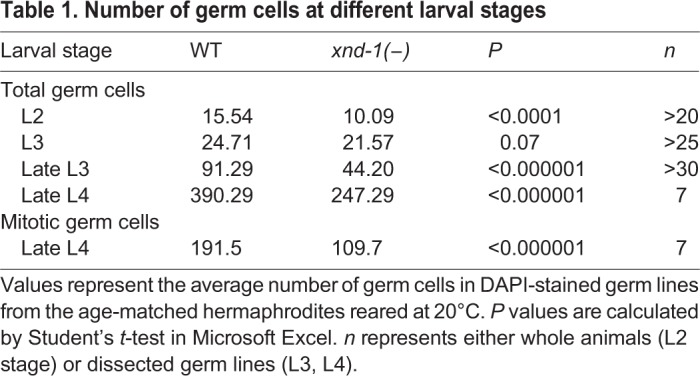
We previously reported that xnd-1 mutant animals had a decreased brood size (Wagner et al., 2010). Here, we confirm and extend these results; our wild-type strain laid an average of ∼292 embryos (n=20), whereas xnd-1 mutants laid an average of only ∼89 embryos (n=34) (Student's t-test, P<0.0001). DAPI staining of whole-mounted animals revealed that the adult germ line was significantly smaller than that of wild type (Fig. 2A), with approximately one-third fewer germ cells compared with wild type at the late L4 stage (Table 1). The small germline size was apparent across the entire xnd-1 population and was so severe in ∼1% of animals that mutant animals were devoid of germ cells. Therefore, we hypothesized that xnd-1 mutants might have a general defect in germline proliferation. A difference in germ cell number was apparent as early as L2 and the extent of this difference increased as animals progressed through larval development (Table 1). By late L4, the pool of proliferating germ cells in xnd-1 mutants was 43% less than in wild type (see Mitotic germ cells in Table 1). This analysis supports the hypothesis that xnd-1 promotes germ cell proliferation during larval stages.
Fig. 2.
xnd-1 is essential for germ cell proliferation. Photomicrograph of age-matched adult animals stained with DAPI to show the germ lines (dotted lines, distal to the left). (A) Zygotic loss of xnd-1 led to reduced germline size. Compare wild type (top) with the smaller xnd-1 M+Z− (bottom). (B) Comparison of germline size in xnd-1 M−Z+ (top) versus xnd-1 M−Z− (bottom) revealed XND-1 zygotic rescue of germ cell proliferation defects. Anterior is to the left. Scale bars: 20 µm. mit, mitotic zone; TZ, transition zone; P, pachytene; Dp, diplotene.
Table 1.
Number of germ cells at different larval stages
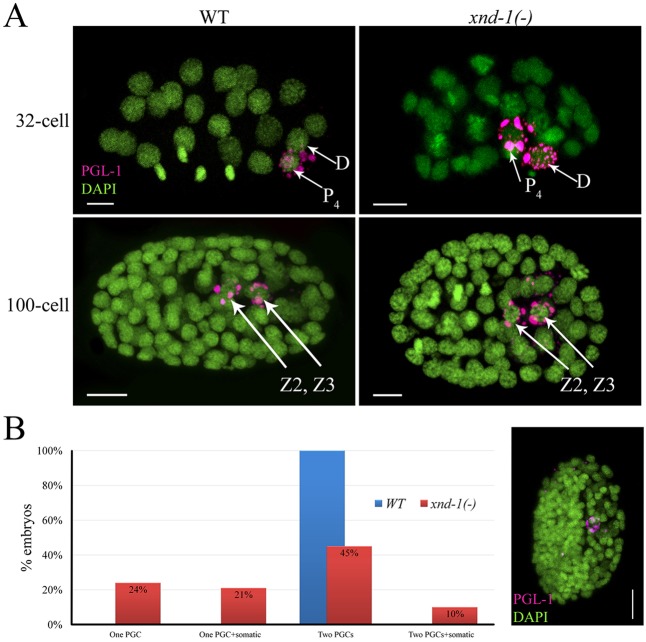
The expression pattern of XND-1 in the early germ cell precursors led us to hypothesize that xnd-1 is required for proper specification and/or differentiation of these cells. We therefore examined PGCs of xnd-1 M−Z− (maternal and zygotic null) mutant embryos. In wild type, PGL-1 is almost exclusively found in PGCs, although faint staining can be seen in the D cell of 28-cell to 60-cell stage embryos (Fig. 3A). In contrast to wild type, xnd-1 mutants showed robust PGL-1 staining in the D cell in nearly 45% (31/69) of embryos (Fig. 3A). The missegregated P granules persisted in somatic blastomeres until the 550-cell stage (compare Fig. S3A,D with S3B,E) and sometimes into L1 larvae. Despite the missegregation of P granules, these cells do not appear to adopt a PGC fate (Strome et al., 1995), as we cannot detect missegregation of PIE-1 or NOS-2 into these cells (data not shown), nor do they express the meiotic marker HTP-3 (Petrella et al., 2011) (Fig. S3B,B′). Furthermore, consistent with published results (Gallo et al., 2010), we found that the missegregation of P granules did not alter specification of the PGC, and they appropriately expressed germ cell markers (data not shown). Nonetheless, we cannot rule out the possibility that the missegregation of P granule components contributes to the reduced fecundity and sterility phenotypes of xnd-1 mutations.
Fig. 3.
xnd-1 mutant embryos missegregate P granules and exhibit PGCs defects. (A) C. elegans embryos stained with anti-PGL-1 (magenta) and DAPI (green). Anterior is to the left. xnd-1 mutant embryos missegregate P granules into the D cell and its daughters. (B) Classification of 100+ cell stage xnd-1(−) embryos based upon the number of blastomeres having PGL-1 staining, shown in magenta in photomicrograph to the right of the graph. (1) One PGC: Embryos in this class have only one PGL-1-positive cell. (2) One PGC+somatic: Embryos have one blastomere with strong PGL-1 staining (PGC) and one or more additional blastomeres with weak PGL-1 staining (presumptive somatic cell). (3) Two PGCs: Embryos with two PGL-1-positive cells. (4) Two PGCs+somatic: Embryos in this class have two blastomeres with strong PGL-1 staining and one or more blastomeres with weak PGL-1 staining. Scale bars: 5 µm.
In wild type, P4 divides at the ∼88-cell stage, after which Z2 and Z3 stay closely apposed, without dividing, throughout the remainder of embryogenesis. To examine if PGC differentiation is normal in xnd-1 mutant embryos, we analyzed the PGCs in >100-cell stage embryos. In contrast to wild type, in which 100% of PGCs divided, xnd-1 M−Z− mutant embryos exhibited multiple defects in PGC development (Fig. 3B). We classified xnd-1 mutant embryos based upon the number of blastomeres expressing PGL-1, and distinguished PGCs from other PGL-1-positive blastomeres by co-staining with both anti-HTP-3 to visualize the meiotic axis (Petrella et al., 2011) (Fig. S3A-C), and anti-MYO-3 mAb 5-6, to mark the muscle cells that arise from the D blastomere (Miller et al., 1983) (Fig. S3D-F). The most penetrant phenotype was a ‘one PGC’ defect, in which nearly half of post-gastrulation xnd-1 mutant embryos only contain a single PGC (Fig. 3B; Fig. S3B,E). This is the first description of a one PGC mutant phenotype in C. elegans. We also frequently observed late-stage embryos with mislocalized PGCs (7%) (Fig. S3F), enlarged nuclear morphology (Fig. S3G, inset) and aberrant muscle morphology that appear to be inviable. A minor subset of late stage xnd-1 embryos (<1%) showed premature proliferation of the PGCs based on anti-HTP-3 staining (Fig. S3C). Both of these latter phenotypes were not detected in L1 larvae (data not shown).
Mutation of xnd-1 conferred high embryonic lethality (∼50%), at least in part as a result of aneuploidy caused by defects in crossover formation (Wagner et al., 2010). We reasoned that if the one PGC phenotype is a secondary consequence of the meiotic defects, then we would observe a similar PGC phenotype in embryos of him-5(ok1896) animals, which share meiotic crossover defects with xnd-1 (Meneely et al., 2012). Similar to wild type, however, him-5 mutant embryos always contained two PGCs (Table 2). In addition, him-5 mutant embryos neither exhibited P granule segregation defects nor other overt PGC defects. Thus, we conclude that the PGC phenotypes of xnd-1 mutants are not a consequence of upstream meiotic deficits.
Table 2.
Frequency of the one PGC phenotype in >100-cell stage embryos
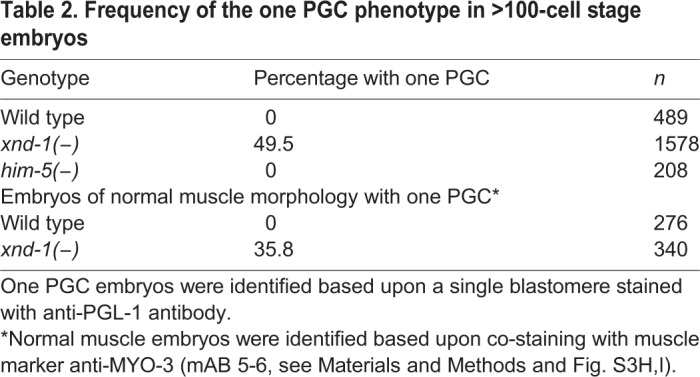
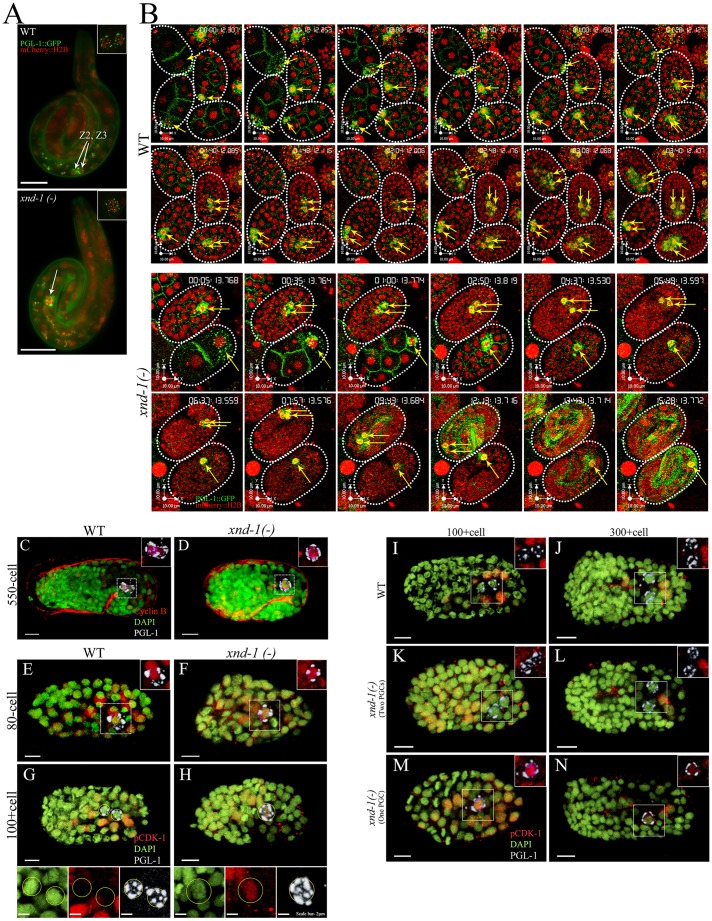
We further confirmed that the one PGC phenotype was visible in L1 larvae that ultimately develop into viable adults. Using PGL-1::GFP to mark PGCs and HIS-58::mCherry to mark all embryonic nuclei, we observed that wild-type larvae always contained two PGCs, whereas 29% of xnd-1 mutant larvae had a single PGC (Fig. 4A; Table 3). We note that this frequency is lower than in embryos. This could be explained if a subset of the single PGCs divide during late embryogenesis, after the onset of morphogenetic movements. However, time-lapse movies (Fig. 4B; Movies 1,2; discussed below) show no evidence of delayed division. Instead, we favor an alternative explanation that one PGC embryos are more prone to embryonic lethality. When we removed the embryos that displayed defective muscle morphology (Fig. S3H,I) from our analysis, the frequency of one PGC embryos dropped to ∼35%, similar to the frequency reported here for L1 larvae (Table 2). These observations support the conclusion that xnd-1 plays a crucial role in normal PGC development.
Fig. 4.
xnd-1 regulates the PGC cell cycle. (A) L1 larvae expressing PGL-1::GFP (green) and mCherry::H2B (red). L1 larvae with two (top) or one PGCs (bottom) can easily be identified. Insets show maximum projection z-stacks of the PGCs. Arrows point to the PGCs. (B) Snapshots from time-lapse movies of wild-type and xnd-1(–) embryos expressing PGL-1::GFP (green) to mark germline blastomeres (yellow arrows) and mCherry::H2B (red) to mark nuclei. Time stamp is in h:m:s:ms format. (C,D) Wild-type and xnd-1(−) embryos of 550-cell stage stained with anti-PGL-1 to mark the PGCs (white), anti-CYB-1 to detect Cyclin B (red) and DAPI to mark nuclei (green). (E-N) Wild-type and xnd-1(−) embryos of indicated cell stages stained with anti-PGL-1 to mark the PGC (white), anti-pCDK-1 to mark cells arrested in G2 (red) and DAPI to mark nuclei (green). Wild-type or xnd-1(−) embryos (>100-cell stage) with two PGCs never showed accumulation of pCDK-1 in the PGC (G,I-L), whereas xnd-1(−) embryos (>100-cell stage) with one PGC showed strong PGC accumulation of pCDK-1 (H,M,N). Scale bars: 5 µm.
Table 3.
Frequency of L1 larvae with one PGC

xnd-1 regulates the PGC cell cycle
Larvae with one PGC might arise if either the P4 blastomere fails to divide or if one of the PGCs died or was lost during embryogenesis. To distinguish between these possibilities, we captured three-dimensional time-lapse (4D) movies of embryogenesis in xnd-1 mutant animals that were expressing PGL-1::GFP and HIS-58::mCherry. Our movies showed that P4 divided into Z2 and Z3 at the ∼88 cell stage in wild-type embryos (n>10) (Fig. 4B; Movie 1). In xnd-1 mutants, however, P4 divided normally in only about half of the embryos, whereas P4 failed to divide in the remainder (n>15) (Fig. 4B; Movie 2). This did not appear to reflect a transient delay in division as we did not observe cell divisions through the ‘pretzel’ stage (∼550 cells) when morphogenetic movements commence. Furthermore, many L1 larvae with a single PGC are observed (Fig. 4A) suggesting these embryos hatch without further division of the germ cell precursor.
To analyze further the nature of cell cycle arrest, we examined several possible reasons for the single PGC arrest in xnd-1. In metazoans, entry into mitosis is regulated by the combination of nuclear accumulation of Cyclin B and removal of inhibitory phosphorylation (on Thr14 and Tyr15) from CDK-1 by CDC25 phosphatase (O'Farrell, 2001; Takizawa and Morgan, 2000). Using anti-CYB-1 antibodies (Rahman et al., 2014; Shakes et al., 2009), we observed no differences in the accumulation of CYB-1 in one PGC xnd-1 embryos compared with wild type (Fig. 4C,D). Thus, the one PGC arrest appears to be independent of CYB-1 accumulation. We next monitored CDK-1 activity using an antibody against mammalian Cdk1 pTyr15 (pCDK-1) that cross-reacts with the corresponding phospho-epitope of C. elegans NCC-1 (CDK-1) (Hachet et al., 2007). In wild type, P4 cells stain positive for pCDK-1 (Fig. 4E,F), as expected for G2 cells just prior to division (Norbury et al., 1991). However, after division, no staining for pCDK-1 was detected in Z2 and Z3 (Fig. 4G,I-L). Because prior studies based on quantification of DAPI signal have shown that Z2 and Z3 arrest in G2 (Fukuyama et al., 2006), the lack of pCDK-1 staining suggests that the PGCs are arrested in a distinct sub-stage of G2. In xnd-1 mutant embryos, pCDK-1 staining is normal in P4 and in two PGCs embryos. By contrast, xnd-1 mutant embryos with a single PGC retained high levels of pCDK-1 late into embryogenesis (Fig. 4H,M-N), consistent with the 4D imaging data that a defect in P4 division leads to the one PGC phenotype.
Although the reduction in fertility was characteristic of all xnd-1 mutant animals, we wanted to determine whether there was a phenotypic consequence on fertility of being born with a single PGC. Prior studies showed that upon laser ablation of either Z2 or Z3 during early larval growth, the remaining PGC still gave rise to two germline arms, but the overall germline size was reduced (Kimble and White, 1981). Therefore, we assayed brood sizes from xnd-1 mutant animals that had been sorted as L1 larvae based on the presence of one or two PGCs. Although the entire population of xnd-1 animals exhibited reduced fecundity compared with wild type (see above), xnd-1 adults developing from L1 larvae with two PGCs exhibited higher average broods (average brood size ∼109) than those developing from L1 larvae with only one PGC (average brood size ∼39, Student's t-test, P<0.0001). These data suggest that animals born lacking a PGC exhibit reduced fecundity, irrespective of baseline fecundity. We further demonstrated that the one PGC defect of xnd-1 mutants is heritable; whereas only 28% of hatched larvae from two PGC xnd-1 mutant animals showed one PGC, 67% of hatched larvae (a significant increase, Fisher's exact test, P<0.0001) from one PGC animals also had one PGC.
Maternal xnd-1 is crucial for embryonic PGC development
Because zygotic transcription is not active in P4 (Seydoux and Dunn, 1997) and maternally deposited mRNAs serve as the sole source of newly translated proteins, we wanted to define the individual roles of maternally and zygotically expressed XND-1 proteins. Because it is only after Z2 and Z3 are born that PGCs become capable of initiating new transcription, we expected that the early expression of XND-1 would reflect translation from maternally contributed mRNA and that zygotic XND-1 proteins would not be observed until later. When xnd-1 heterozygous mothers were crossed to xnd-1 mutant males (Fig. S4A), both the M−Z− (maternal and zygotic null) and M+Z− (zygotic null) offspring showed XND-1 expression in PGCs as early as the 28-cell stage and throughout all of embryogenesis (Fig. 1B). Thus, maternally contributed xnd-1 mRNAs account for the P4 and early Z2 and Z3 expression of xnd-1. To analyze the function of maternal XND-1 proteins, we examined xnd-1 M+Z− mutant embryos arising from the balanced xnd-1/hT2 mothers (for crosses see Fig. S4A). PGC development in these embryos was normal, however, the adult germ line in these individuals was markedly reduced in size (Fig. 2A). Thus, we infer that maternally deposited XND-1 products are sufficient to support embryonic PGC divisions, but insufficient to promote normal germline growth (Fig. 2A). These results support a direct role for xnd-1 in controlling PGC proliferation.
Zygotic expression of xnd-1 promotes germline proliferation
We then performed reciprocal crosses, crossing xnd-1 mutant mothers with wild-type males, to monitor zygotic expression (Fig. S4C-F). Antibody staining of the resultant M−Z+ (maternal null) revealed expression of XND-1 in 250-cell stage embryos, where it appeared first in the cytoplasm of Z2 and Z3 and then became predominantly nuclear (Fig. S4E,F). To rule out that early expression of XND-1 was not a result of loss of maternal xnd-1 function, we used a similar crossing strategy to introduce 3×FLAG::GFP::XND-1 transgenes into wild-type embryos (Fig. S4B; Fig. 1C). Immunolocalization of the XND-1 fusion proteins revealed expression in the ∼300-cell stage PGCs. These results indicate that XND-1 is one of the earliest genes to be expressed in PGCs and suggests that transcriptional activation in the PGC begins earlier than had previously been anticipated.
To determine the functional contributions of zygotically expressed XND-1, we asked whether zygotic xnd-1 could rescue the one PGC embryonic phenotype. PGC defects in xnd-1 M−Z+ embryos were indistinguishable from xnd-1 M−Z− embryos (compare Fig. S4D-F with Fig. S3B,C,E,G), suggesting that maternal loss of xnd-1 is solely responsible for the one PGC phenotype. Loss of xnd-1 function also causes significant embryonic lethality, in part as a result of meiotic defects (Wagner et al., 2010) that could not be rescued by zygotic xnd-1 expression (58% embryonic lethality in M−Z+ versus 67% in M−Z−, Student's t-test, P=0.3). We next hatched F1 embryos expressing zygotic xnd-1 and grew them to adulthood to examine fertility and germline size (Fig. 2B; Table 4). In contrast to the partial sterility and reduced germline sizes seen in xnd-1 M-Z- mutant animals (Fig. 5, Fig. 2B; Table S2), all F1 xnd-1 M−Z+ adults were fertile (n>50) with germlines sizes within the normal range (Fig. 2B; Table 4). Brood sizes were also substantially increased in the xnd-1 M−Z+ animals (244 progeny in xnd-1 M−Z+ versus 89 in xnd-1 M−Z−, Student's t-test, P<5.0×10−12) and these animals manifested no meiotic defects (normal pachytene nuclei progression, six DAPI-stained bodies at diakinesis, and absence of males in M−Z+ broods). These results strongly suggest a role for maternal XND-1 in embryonic PGC development and for zygotic XND-1 in germ cell proliferation during larval development and adult fertility.
Table 4.
Comparison of adult gonad size
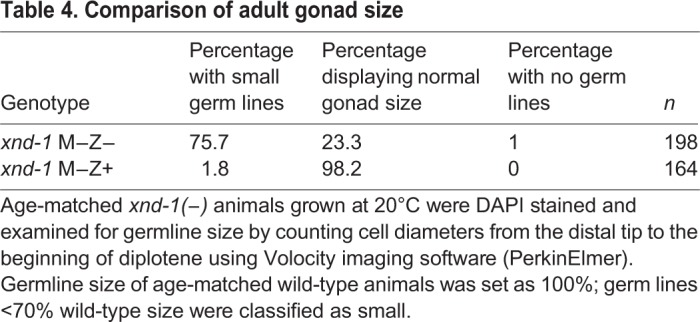
Fig. 5.
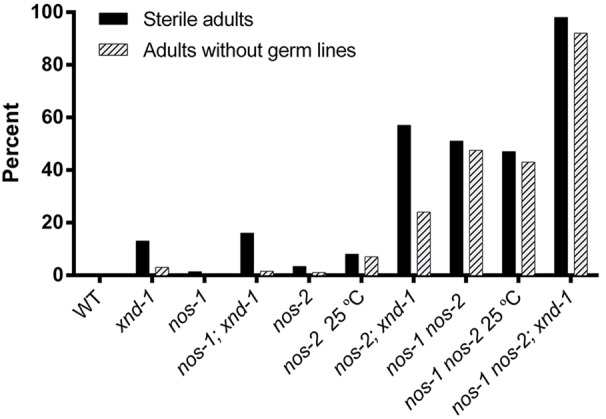
xnd-1(−) and nos-2(−) animals are sterile and lack germ lines. Graph of sterility and percentage of no germ cell animals of the indicated genotypes (See Table S2 for more details).
nos-1 nos-2 double null mutants exhibit partial sterility
Nanos homologs are conserved determinants of germline fates (Tsuda et al., 2003). In C. elegans, RNA interference revealed functional redundancy between the Nanos homologs nos-1 and nos-2 for germline development; the majority of nos-1(RNAi) nos-2(RNAi) had defects in germ cell proliferation leading to adults with empty gonads (Subramaniam and Seydoux, 1999). The striking similarities in expression and mutant phenotypes between nos-1 nos-2 and xnd-1 led us to pursue a detailed analysis of their interaction. As a first step, however, we needed to create a functional nos-2 null allele; nos-2 resides within an intron of the essential meiotic gene him-14/msh-4 and the nos-2(ok230) allele removes all of the nos-2 coding region as well as part of the flanking exons in him-14 (Fig. S5A). Because mutation of him-14 leads to a high rate of aneuploidy and resultant embryonic lethality, this allele cannot be analyzed for nos-2 phenotypes. To create a nos-2 allele, we chose a fosmid rescue approach of him-14&nos-2(ok230), creating a transgenic line with a fosmid clone containing the entire him-14 locus with the nos-2 coding sequence replaced with yfp [hereafter referred to as nos-2(−)]. Although him-14&nos-2(ok230) exhibits ∼98% embryonic lethality, nos-2(−) animals manifest only ∼2% embryonic lethality (Table S3), revealing near complete rescue of the him-14 nondisjunction phenotype. This strain provides the first characterization of a nos-2 null mutant in C. elegans.
We next sought to confirm the role of nos-1 nos-2 in germline differentiation as revealed by maternal-effect sterility and a germ cell-less phenotype. The average brood size of nos-2(−) was lower than that of wild type: 204.5 vs 295.9, respectively (Table S3). nos-2(−) exhibited mislocalization and premature proliferation of PGCs, as reported in nos-2 RNAi (Subramaniam and Seydoux, 1999). In addition, we noticed that the premature proliferation of PGCs associated with nos-2 loss of function begins during mid-embryogenesis (Fig. S5B). nos-2(−) mutant animals exhibited only 3% sterility, with ∼1% animals lacking any germ line at 20°C (Fig. 5; Table S2). These phenotypes became more severe upon growth at 25°C, with 8% sterility and ∼7% animals with no germ line (Fig. 5; Table S2). nos-1(gv5) null mutants are fertile and do not exhibit overt germline defects (Subramaniam and Seydoux, 1999 and confirmed in this study).
When we examined nos-1(gv5) nos-2(−) double mutants, we observed a substantial increase in sterility with nearly half of the animals lacking germ cells (Fig. 5; Table S2) in addition to a further decrease in brood size (Table S3). Sterility in nos-1(gv5) nos-2(−) did not increase at the higher growth temperature of 25°C (Fig. 5; Table S2). We note that sterility in both the nos-2(−) and nos-1 nos-2 double mutant is substantially lower than had been reported with RNAi (Katz et al., 2009; Subramaniam and Seydoux, 1999; Zanin et al., 2010), supporting the claim that nos-2(RNAi) might have off-target effects or that activation of RNA interference in nos-2 mutants exacerbates its phenotype.
nos-1, nos-2 and xnd-1 in a parallel pathway to control germline differentiation
To determine if xnd-1 shared functions with nos-2, we first investigated whether either gene affects the other's expression. Using antibody staining, we saw no abrogation of NOS-2 or XND-1 expression in the respective mutants (Fig. S5C,D). We could not examine NOS-1 expression as a result of unavailability of functional antibodies. We then explored the epistatic relationship between xnd-1 and the Nanos homologs by constructing nos-1; xnd-1 and nos-2; xnd-1 double mutants and nos-1 nos-2; xnd-1 triple mutants and characterizing their sterility and germline phenotypes. nos-1; xnd-1 double mutants revealed no enhancement of sterility compared with xnd-1 alone (Fig. 5; Table S2). By contrast, nos-2; xnd-1 mutants revealed a synergistic interaction with close to 57% sterile progeny (Fig. 5; Table S2). This analysis suggests that nos-2 and xnd-1 function in redundant pathways controlling germline differentiation.
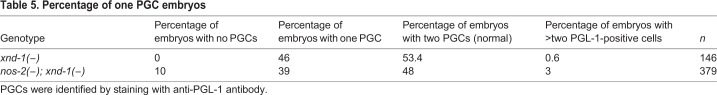
More detailed analysis of the mutant embryos revealed no change in the percentage of two PGC animals between xnd-1 and nos-2; xnd-1 mutants and a plurality of remaining embryos contained only one PGC. Unlike in either single mutant, however, in nearly 10% of double mutant embryos, no PGC was discernible by PGL-1 staining (Table 5) or nos-2p::YFP expression (not shown). These data suggest that nos-2 and xnd-1 function in distinct pathways controlling germline differentiation and might also redundantly influence PGC fate specification.
Table 5.
Percentage of one PGC embryos
The lack of interaction between nos-1 and xnd-1, but their mutual enhancement of nos-2, suggested that nos-1 and xnd-1 might function in the same genetic pathway. We reasoned that if nos-1 and xnd-1 function together, then the sterility of nos-1 nos-2; xnd-1 triple mutants would be indistinguishable from nos-2; xnd-1 double mutants, whereas if they regulate distinct pathways, then the triple mutant should produce more sterile progeny than either double mutant alone. As shown in Fig. 5, the nos-1 nos-2; xnd-1 triple mutants produced 98% sterile progeny, 92% of which contained no germ cells in the adult gonad, versus 57% in nos-2; xnd-1 double mutants. This result reveals distinct roles for each of these genes in germline differentiation, with xnd-1 and nos-1 each having overlapping functions with nos-2.
xnd-1 late embryos show premature activation of transcription in the PGCs
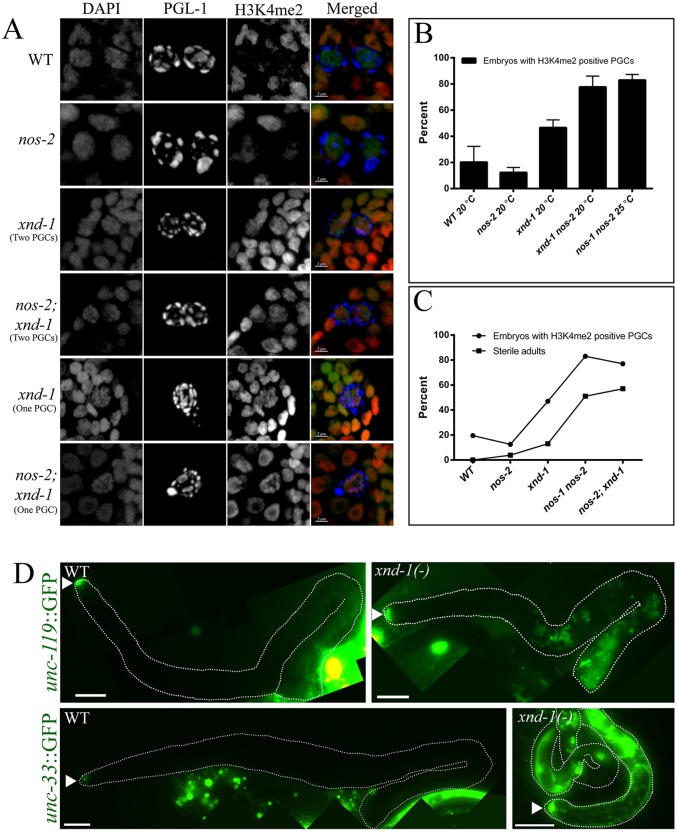
Transcriptional quiescence in PGCs is important for their specification and to maintain germ cell fate (reviewed by Strome and Lehmann, 2007). Accordingly, mutants that fail to maintain germ cell identity frequently reveal an increase in H3K4me2 staining in PGCs. Retention of H3K4me2 in Z2 and Z3 has been associated with lower brood sizes and increased sterility in mutants, affecting normal accumulation and removal of H3K4me2 (Katz et al., 2009; Schaner et al., 2003; Strome and Lehmann, 2007). Therefore, we wished to address whether the germline defects in xnd-1 mutants are associated with altered H3K4me2 accumulation in PGCs. Because xnd-1 mutants laid embryos that produced either one or two PGCs, we characterized these classes independently. H3K4me2 was assessed by antibody staining of 2-fold stage embryos (Fig. 6A). In wild type, H3K4me2 was detected in only one-fifth of PGCs, (Fig. 6B; Table S4). In xnd-1 mutants, over one-third of the two PGC embryos and over one half of the one PGC embryos showed accumulation of H3K4me2 (Fig. 6A,B; Table S4). In addition, we found that nos-2 mutants did not differ significantly from wild type (both <20%), consistent with the mild fertility defects in this mutant. By contrast, nos-2; xnd-1 double mutants showed an increased percentage of embryos with inappropriate PGC accumulation of H3K4me2 (82% of one PGC and ∼72% of two PGCs embryos) consistent with their increased sterility (∼57%) (Table S4). nos-1 nos-2 double mutants also showed >80% embryos with H3K4me2-positive PGCs, again correlated with higher maternal effect sterility (∼51%). Thus, we found a correlation between the inappropriate accumulation of H3K4me2 in PGCs and the extent of sterility (Fig. 6C).
Fig. 6.
xnd-1(−) PGCs show hallmarks of aberrant transcriptional activation. (A-C) H3K4me2 persists and accumulates in Z2/Z3 of xnd-1 and nos-2 late embryos. (A) Photomicrographs show close-ups of PGCs in >100-cell embryos of the indicated genotypes. Embryos were stained with anti-PGL-1 to mark PGCs (blue), anti-H3K4me2 (red) and DAPI to mark nuclei (green). Scale bars: 2 µm. (B) The percentage of embryos with H3K4me2-positive PGCs for the indicated genotypes. Data are represented as mean±s.d. (C) The correlation between the percentage embryos with H3K4me2-positive PGCs with the percentage sterility for the indicated genotypes. (D) unc-119::GFP and unc-33::GFP expression in the germ lines of wild-type and xnd-1 M−Z− adults. Germ lines are outlined and arrowheads point to the distal tip cells. Both transgenes are aberrantly expressed in xnd-1 mutant animals from mid-pachytene through diplotene. Adult gonad images were obtained as multiple images and then merged to create a final image of an entire gonad. Scale bars: 20 µm.
The aberrant accumulation of H3K4me2 in xnd-1 mutant PGCs raised the possibility that transcription is inappropriately activated in these germ lines. We utilized two pan-neuronal reporter transgenes, unc-119::GFP and unc-33::GFP (Ciosk et al., 2006; Patel et al., 2012; Tursun et al., 2011; Updike et al., 2014) to assay somatic gene expression in the germ line. Both reporters, as expected, were undetectable in wild-type or xnd-1/+ germ lines. By contrast, ∼15% (n=52) of xnd-1 M−Z− adults showed misexpression of unc-119::GFP in the germ line, usually in groups of cells in the mid-pachytene region (Fig. 6D). Similar aberrant expression of unc-33::GFP was observed in xnd-1 M−Z− germ lines. Taken together, these results suggest that xnd-1 sterility might result, at least in part, from an inability of the germ cells to maintain the germline fate as revealed by misexpression of neuronal markers in the adult germ line.
DISCUSSION
Proper specification and differentiation of the germ line is essential for fecundity, yet the genes required for elaboration of germ line fates are still poorly understood. We have identified XND-1 as a key determinant of germ cell fate, functioning redundantly with NOS-2 and NOS-1 to promote germ cell growth and maintenance.
XND-1 is a key regulator of germ cell development
We have identified C. elegans XND-1 protein as one of the first markers of PGC. Both antibody staining and GFP transgenes revealed exclusive localization of XND-1 proteins to germline progenitors. In this regard, XND-1 becomes a member of a very exclusive group of proteins that are translated in the P4 cell at the time of its birth (as opposed to restricted there by asymmetric divisions) and which continue to be expressed in Z2 and Z3. Indeed, the only other known member of this exclusive group is the Nanos homolog nos-2 (Subramaniam and Seydoux, 1999). The first XND-1 proteins observed in the embryo are translated from maternally deposited mRNAs in the P4 cell and are maintained in PGCs beyond the three-fold stage. Zygotic XND-1 expression begins at around the 300-cell stage, making XND-1 one of the earliest zygotically expressed PGC genes.
Consistent with its localization and timing of expression, xnd-1 contributes to multiple aspects of PGC differentiation. In addition to influencing the P4 division (discussed below), xnd-1 is also required for the accurate segregation of P granule components and for promoting normal germ cell number. The missegregation of PGL-1 into somatic blastomeres is reminiscent of mes-1 mutant embryos (Strome et al., 1995), raising the possibility that mes-1 functions in the same pathway as xnd-1. We could not, however, confirm whether MES-1 expression is affected in xnd-1 embryos as a result of technical issues with the available antibody. Therefore, future studies aimed at understanding the epistatic relationship between xnd-1 and mes-1 will be informative.
Post-embryogenesis, xnd-1 affects germ cell number and the ultimate size of the adult germ line, thereby promoting fecundity. The reduction in larval germ cell numbers in the xnd-1 mutant is consistent with a continued role for xnd-1 in PGC proliferation. One possibility is that xnd-1 directs the expression of key cell cycle regulators necessary for PGC development. XND-1 protein is chromatin-associated, but its molecular function still remains unclear. Recent studies have revealed that xnd-1 influences the expression of the meiotic proteins HIM-5 (Meneely et al., 2012) and CRA-1 (Gao et al., 2015). Thus, xnd-1 might function directly as a transcription factor. Alternatively, xnd-1 might indirectly control gene expression by modulating histone post-translational modifications, e.g. H3K4me2 in the embryonic germ line and H2AK5ac in the adult germ line. Future studies to determine the genome-wide binding sites of XND-1 and correlate these sites with changes in gene expression throughout development promise to shed light on the mechanism(s) by which XND-1 controls various aspects of germline growth and differentiation.
Germ cell pools can be determined during embryogenesis
Our results provide new insights into the regulation of the PGC cell cycle and its importance in controlling fertility. xnd-1 is the first C. elegans mutant to confer a ‘one PGC’ phenotype, which correlates with the reduced overall fecundity of these animals. In addition, the offspring of animals harboring a single PGC are themselves more likely to have only one PGC. In related studies, we have observed that xnd-1 mutants express a mortal germline phenotype, going sterile after tens of generations (J.L.Y., unpublished). This phenotype could be driven in part by a death spiral induced by the accumulation of the one PGC animals in the population. xnd-1 encodes a chromatin-associated protein and xnd-1 mutant animals are defective in accumulation of H3K4me2 in embryonic PGCs and H2AK5ac in the adult germ line (Fig. 6A) (Wagner et al., 2010). Together, these observations raise the possibility that these epigenetic marks transduce information about the reproductive health of the mother to alter the fecundity of the offspring.
Time-lapse movies of PGCs demonstrate that the one PGC phenotype arises from a defect in P4 cell division (Fig. 4H,M,N; Movie 2). The normal accumulation of Cyclin B and persistence of p-CDK-1 in these embryos supports our observation that the one PGC is arrested in G2. Quantification of H2B::GFP signal in one PGCs of L1 larvae indicates ∼4C DNA content (M.R. and J.L.Y., unpublished). The sustained cell cycle arrest in the single PGC might result from misregulation of either Wee kinase (wee-1.3) or CDC25 phosphatase. Alternatively, the recent observation that transcriptional activation in Z2 and Z3 activates the damage checkpoint (Butuči et al., 2015), together with our data that PGCs in xnd-1 animals prematurely activates transcription, raise the possibility that the G2 arrest results from activation of the DNA damage-dependent checkpoint.
Although the single PGCs seem to arise from arrested P4 cells, they do not completely retain P4 identity. P4 is transcriptionally quiescent, but the single PGCs, like Z2 and Z3, activated zygotic expression of XND-1 proteins (Fig. S4D,E). Furthermore, the presence of replete – albeit smaller – germ lines in L2 adult xnd-1 mutants reveals that the P4 arrest is transient and that the larval proliferative program is initiated in one PGC animals. Surprisingly, the addition of zygotic xnd-1 (M−Z+ animals) supported wild-type germline development, suggesting that the deficit in embryonic PGC number can be overcome during the larval divisions. This is in striking contrast to the laser ablation studies in which loss of Z2 or Z3 in L1 larvae was sufficient to reduce germ line size.
Multiple pathways regulate germ cell differentiation
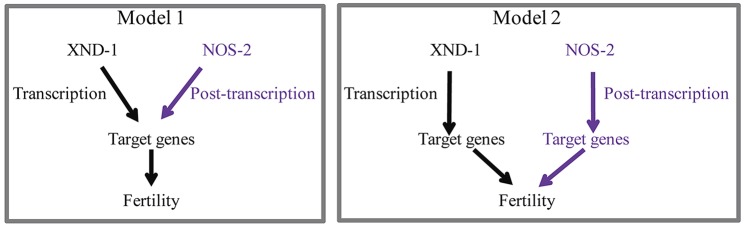
These studies demonstrate that xnd-1 is required for proper germline development and becomes essential for germ cell development in the absence of nos-1 and nos-2. Our characterization of the first genetic null mutant of nos-2 reveals additional layers of redundancy for germ cell differentiation than had previously been appreciated. These studies point to at least three major pathways controlling germ cell differentiation, and possibly a fourth, given the existence of – albeit a small fraction – fertile nos-1 nos-2; xnd-1 mutant animals. It will be particularly interesting to investigate whether nos-3 or T02G5.11 contributes to fertility in these backgrounds. Each of these branches appears to impinge on gene expression, as changes in H3K4me2 are observed in each of the double mutant combinations, but whether the effect on transcriptional competence is direct or indirect is currently unknown. Because nanos interacts with translation factors in Drosophila, it is tempting to speculate that xnd-1 and nos-2 might share target loci, with xnd-1 reducing expression through direct transcriptional regulation or changes in chromatin architecture and nos-2 then further reducing expression of these target genes post-transcriptionally. Alternatively, xnd-1 and nos-2 might each regulate a unique set of genes, both of which are required to retain germ cell fate (see model in Fig. 7). Because direct targets of nos-2 and nos-1 are currently unknown, future studies to identify and determine their targets may reveal the underlying cause of sterility as a result of loss of nos-1 nos-2 double and loss of xnd-1 over nos-2 and nos-1 nos-2.
Fig. 7.
Models for the control of germline fates by XND-1 and NOS-2. In Model 1, XND-1 and NOS-2 regulate common targets via different modes of regulation, transcriptional regulation by XND-1 and post-transcriptional regulation by NOS-2. In Model 2, XND-1 and NOS-2 regulate distinct sets of targets genes which are both required to confer fertility.
MATERIALS AND METHODS
C. elegans strains
Protocols for nematode caring and feeding have been described (Brenner, 1974). Unless indicated, growth was at 20°C. The strains utilized in this study are listed in Table S5. Standard genetic crosses were used to make double and triple mutants.
Constructing nos-2(−) strain
Fosmid clone WRM069bE04 was recombineered using galK selection to replace the nos-2 coding sequence and introns with yfp, as described (Tursun et al., 2009). yfp containing FRT-flanked galK within an intron was amplified from pBALU2 and recombineered into WRM069bE04 to replace nos-2. Specific primers used are provided in supplementary materials and methods.
Subcloned fosmid DNA (fJN059) was introduced into unc-119(ed3) worms using microparticle bombardment (Praitis et al., 2001) to produce transgene insertion xnIs273. Subsequently, xnIs273 was crossed into a him-14&nos-2(ok230) background to produce nos-2(−) mutants.
Immunostaining and microscopy
Fixation and immunolocalization protocols are described elsewhere (Chan et al., 2003), but can also be found together with details of the antibodies used in supplementary materials and methods.
Images were acquired with a Nikon A1R confocal microscope (Nikon Instruments) with 0.2-µm sections. Embryos were staged either by counting the total numbers of blastomeres or by assessing the embryonic morphology during embryonic elongation stage. Adult gonad images were obtained as multiple images and then merged to create a final image of an entire gonad using Photoshop CS5 Extended.
RNAi
RNAi clones from the Ahringer bacterial feeding library were verified and used as described previously (Kamath and Ahringer, 2003). Unavailable coding regions were independently PCR amplified and inserted into the RNAi feeding vector pSV2 (Mainpal et al., 2011), sequenced, and introduced into Escherichia coli HT115. These E. coli clones were spread on NGM plates containing 1 mM IPTG and 50 µg/ml kanamycin, and used for inducing RNAi by the feeding method (Timmons et al., 2001).
Time-lapse imaging
Four-dimensional movies were acquired using a Nikon TiE wide-field fluorescent microscope and 63×1.4 NA objective. z-stacks of 1.5 µm covering from top to bottom of developing embryos were captured every 5 min, starting from a two-cell stage embryo until they reached ∼300-cell stage or hatched, and division of PGC was observed.
Sterility assay
All genotypes were age matched and synchronized by allowing gravid adults to lay on freshly seeded OP50 plates for 3 h and growing worms at 20°C until day two of adulthood (∼115 h post-laying). Adults were collected and washed with M9 in microcentrifuge tubes, then fixed and stained in 95% ethanol with DNA-binding dye 4,6-diamidino-2-phenylindole (DAPI) at 4°C overnight. Fixed animals were washed twice with M9 and mounted in ProLong Gold antifading agent (Molecular Probes, Cat# P36935) on glass slides. Slides were examined with a Nikon wide-field TiE fluorescent microscope using a 10× or 20× non-oil lens. Gravid adults were counted as fertile, animals without embryos were classified as sterile. Sterile animals were further subdivided by whether they had two gonad arms, one gonad arm, or no germ line with very few germ cells in stunted gonads.
Zygotic and maternal requirement
Crosses to examine maternal and zygotic expression and function are shown in Fig. S4. Briefly, to examine the maternal expression of XND-1, xnd-1/hT2 was crossed into a fog-2 mutant to obtain an obligate male/female stock. xnd-1/hT2; fog-2 (M+Z+ for xnd-1) females were crossed with either xnd-1 or xnd-1; fog-2 males (M−Z−). Embryos from the resulting cross were either M+Z+ (GFP positive) or M+Z− (GFP negative). Expression and perdurance of XND-1 was examined by immunostaining in GFP negative embryos, which were only expressing the maternal copy of xnd-1. For zygotic expression, xnd-1; fog-2 females (M−Z−) were crossed with either wild-type or fog-2 males (M+Z+) to give M−Z+ embryos. All embryos were immunostained and examined for XND-1 expression.
Isolation of one PGC larvae
xnd-1 mutant gravid hermaphrodites expressing PGL-1::GFP and mCherry::HIS-58 to mark germ cells were bleached to collect embryos. Harvested embryos were hatched on a low speed shaker in M9 media (no food) at room temperature for ∼18 h. Hatched L1 larvae were mounted on a glass slide in M9 and larvae with one PGC and two PGCs were manually sorted with a pulled glass pipette using GFP and mCherry fluorescent markers under a Nikon wide-field fluorescent microscope using a 40× dry objective.
Brood size analysis
For brood size analysis of one- and two-PGC animals, sorted L1 larvae were grown until they reached L4, then individually plated and grown for another 4 days. All hatched progeny from individually plated animals were counted as their viable brood.
To determine the total number of eggs laid, L4 animals were individually plated on NGM plates seeded with E. coli OP50 and transferred to newly seeded plates every 24 h until laying stopped. Total number of eggs and hatched worms were counted from all plates to determine the total laid progeny.
Acknowledgements
The authors wish to extend thanks to the Caenorhabditis Genetics Center, funded by NIH Office of Research Infrastructure Programs [P40 OD010440], for the distribution of C. elegans strains used in this study. They also want to thank Tim Schedl (Washington University School of Medicine, MO, USA) for sharing the nos-1 nos-2 double mutant strain, David Greenstein (University of Minnesota, MN, USA) and Al Fisher (UTHSCSA, TX, USA) for sharing recombineering reagents, Susan Strome (University of California Santa Cruz, CA, USA) for sharing anti-PGL-1 antibodies, K. Subramaniam (Indian Institute of Technology - Madras, Chennai, India) for sharing anti-NOS-2 antibodies, and Karen Bennett (Johns Hopkins University School of Medicine, MD, USA) for sharing anti-GLH-1 antibodies. The authors extend their gratitude to Arjumand Ghazi and members of the Yanowitz lab for discussion and careful reading of the paper.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
R.M. and J.L.Y. designed the experiments. J.N. designed and constructed the nos-2 fosmids and transgenic line. R.M. performed the experiments and R.M. and J.L.Y. analyzed the data. R.M. and J.L.Y. wrote the manuscript and J.N. edited it.
Funding
This work is supported by funding from the National Institutes of Health [R01 GM104007 to J.L.Y. and R21HD058953 to J.N.]; Pennsylvania Department of Health, Research Formula Funds (J.L.Y.); a MARS fellowship at Magee-Womens Research Institute (J.L.Y.); and a Magee-Womens Research Institute post-doctoral fellowship (R.M.). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.125732/-/DC1
References
- Brenner S. (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butuči M., Williams A. B., Wong M. M., Kramer B. and Michael W. M. (2015). Zygotic genome activation triggers chromosome damage and checkpoint signaling in C. elegans primordial germ cells. Dev. Cell 34, 85-95. 10.1016/j.devcel.2015.04.019 [DOI] [PubMed] [Google Scholar]
- Chan R. C., Chan A., Jeon M., Wu T. F., Pasqualone D., Rougvie A. E. and Meyer B. J. (2003). Chromosome cohesion is regulated by a clock gene paralogue TIM-1. Nature 423, 1002-1009. 10.1038/nature01697 [DOI] [PubMed] [Google Scholar]
- Ciosk R., DePalma M. and Priess J. R. (2006). Translational regulators maintain totipotency in the Caenorhabditis elegans germline. Science 311, 851-853. 10.1126/science.1122491 [DOI] [PubMed] [Google Scholar]
- Forbes A. and Lehmann R. (1998). Nanos and Pumilio have critical roles in the development and function of Drosophila germline stem cells. Development 125, 679-690. [DOI] [PubMed] [Google Scholar]
- Fukuyama M., Rougvie A. E. and Rothman J. H. (2006). C. elegans DAF-18/PTEN mediates nutrient-dependent arrest of cell cycle and growth in the germline. Curr. Biol. 16, 773-779. 10.1016/j.cub.2006.02.073 [DOI] [PubMed] [Google Scholar]
- Gallo C. M., Wang J. T., Motegi F. and Seydoux G. (2010). Cytoplasmic partitioning of P granule components is not required to specify the germline in C. elegans. Science 330, 1685-1689. 10.1126/science.1193697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Kim H.-M., Elia A. E., Elledge S. J. and Colaiácovo M. P. (2015). NatB Domain-containing CRA-1 antagonizes hydrolase ACER-1 linking Acetyl-CoA metabolism to the initiation of recombination during C. elegans meiosis. PLoS Genet. 11, e1005029 10.1371/journal.pgen.1005029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachet V., Canard C. and Gönczy P. (2007). Centrosomes promote timely mitotic entry in C. elegans embryos. Dev. Cell 12, 531-541. 10.1016/j.devcel.2007.02.015 [DOI] [PubMed] [Google Scholar]
- Kadyrova L. Y., Habara Y., Lee T. H. and Wharton R. P. (2007). Translational control of maternal Cyclin B mRNA by Nanos in the Drosophila germline. Development 134, 1519-1527. 10.1242/dev.002212 [DOI] [PubMed] [Google Scholar]
- Kamath R. S. and Ahringer J. (2003). Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30, 313-321. 10.1016/S1046-2023(03)00050-1 [DOI] [PubMed] [Google Scholar]
- Katz D. J., Edwards T. M., Reinke V. and Kelly W. G. (2009). A C. elegans LSD1 demethylase contributes to germline immortality by reprogramming epigenetic memory. Cell 137, 308-320. 10.1016/j.cell.2009.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki I., Shim Y.-H., Kirchner J., Kaminker J., Wood W. B. and Strome S. (1998). PGL-1, a predicted RNA-binding component of germ granules, is essential for fertility in C. elegans. Cell 94, 635-645. 10.1016/S0092-8674(00)81605-0 [DOI] [PubMed] [Google Scholar]
- Kawasaki I., Amiri A., Fan Y., Meyer N., Dunkelbarger S., Motohashi T., Karashima T., Bossinger O. and Strome S. (2004). The PGL family proteins associate with germ granules and function redundantly in Caenorhabditis elegans germline development. Genetics 167, 645-661. 10.1534/genetics.103.023093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr S. C., Ruppersburg C. C., Francis J. W. and Katz D. J. (2014). SPR-5 and MET-2 function cooperatively to reestablish an epigenetic ground state during passage through the germ line. Proc. Natl. Acad. Sci. USA 111, 9509-9514. 10.1073/pnas.1321843111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J. E. and White J. G. (1981). On the control of germ cell development in Caenorhabditis elegans. Dev. Biol. 81, 208-219. 10.1016/0012-1606(81)90284-0 [DOI] [PubMed] [Google Scholar]
- Kobayashi S., Yamada M., Asaoka M. and Kitamura T. (1996). Essential role of the posterior morphogen nanos for germline development in Drosophila. Nature 380, 708-711. 10.1038/380708a0 [DOI] [PubMed] [Google Scholar]
- Mainpal R., Priti A. and Subramaniam K. (2011). PUF-8 suppresses the somatic transcription factor PAL-1 expression in C. elegans germline stem cells. Dev. Biol. 360, 195-207. 10.1016/j.ydbio.2011.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meneely P. M., McGovern O. L., Heinis F. I. and Yanowitz J. L. (2012). Crossover distribution and frequency are regulated by him-5 in Caenorhabditis elegans. Genetics 190, 1251-1266. 10.1534/genetics.111.137463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D. M., Ortiz I., Berliner G. C. and Epstein H. F. (1983). Differential localization of two myosins within nematode thick filaments. Cell 34, 477-490. 10.1016/0092-8674(83)90381-1 [DOI] [PubMed] [Google Scholar]
- Nakamura A. and Seydoux G. (2008). Less is more: specification of the germline by transcriptional repression. Development 135, 3817-3827. 10.1242/dev.022434 [DOI] [PubMed] [Google Scholar]
- Norbury C., Blow J. and Nurse P. (1991). Regulatory phosphorylation of the p34cdc2 protein kinase in vertebrates. EMBO J. 10, 3321-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Farrell P. H. (2001). Triggering the all-or-nothing switch into mitosis. Trends Cell Biol. 11, 512-519. 10.1016/S0962-8924(01)02142-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel T., Tursun B., Rahe D. P. and Hobert O. (2012). Removal of Polycomb repressive complex 2 makes C. elegans germ cells susceptible to direct conversion into specific somatic cell types. Cell Rep. 2, 1178-1186. 10.1016/j.celrep.2012.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrella L. N., Wang W., Spike C. A., Rechtsteiner A., Reinke V. and Strome S. (2011). synMuv B proteins antagonize germline fate in the intestine and ensure C. elegans survival. Development 138, 1069-1079. 10.1242/dev.059501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrotta V. (2002). Silence in the germ. Cell 110, 661-664. 10.1016/S0092-8674(02)00967-4 [DOI] [PubMed] [Google Scholar]
- Praitis V., Casey E., Collar D. and Austin J. (2001). Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics 157, 1217-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M. M., Rosu S., Joseph-Strauss D. and Cohen-Fix O. (2014). Down-regulation of tricarboxylic acid (TCA) cycle genes blocks progression through the first mitotic division in Caenorhabditis elegans embryos. Proc. Natl. Acad. Sci. USA 111, 2602-2607. 10.1073/pnas.1311635111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaner C. E., Deshpande G., Schedl P. D. and Kelly W. G. (2003). A conserved chromatin architecture marks and maintains the restricted germ cell lineage in worms and flies. Dev. Cell 5, 747-757. 10.1016/S1534-5807(03)00327-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux G. and Dunn M. A. (1997). Transcriptionally repressed germ cells lack a subpopulation of phosphorylated RNA polymerase II in early embryos of Caenorhabditis elegans and Drosophila melanogaster. Development 124, 2191-2201. [DOI] [PubMed] [Google Scholar]
- Seydoux G. and Schedl T. (2001). The germline in C. elegans: origins, proliferation, and silencing. Int. Rev. Cytol. 203, 139-185. 10.1016/S0074-7696(01)03006-6 [DOI] [PubMed] [Google Scholar]
- Shakes D. C., Wu J.-c., Sadler P. L., LaPrade K., Moore L. L., Noritake A. and Chu D. S. (2009). Spermatogenesis-specific features of the meiotic program in Caenorhabditis elegans. PLoS Genet. 5, e1000611 10.1371/journal.pgen.1000611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda J. and Wharton R. P. (1999). Recruitment of Nanos to hunchback mRNA by Pumilio. Genes Dev. 13, 2704-2712. 10.1101/gad.13.20.2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome S. and Lehmann R. (2007). Germ versus soma decisions: lessons from flies and worms. Science 316, 392-393. 10.1126/science.1140846 [DOI] [PubMed] [Google Scholar]
- Strome S., Martin P., Schierenberg E. and Paulsen J. (1995). Transformation of the germ line into muscle in mes-1 mutant embryos of C. elegans. Development 121, 2961-2972. [DOI] [PubMed] [Google Scholar]
- Subramaniam K. and Seydoux G. (1999). nos-1 and nos-2, two genes related to Drosophila nanos, regulate primordial germ cell development and survival in Caenorhabditis elegans. Development 126, 4861-4871. [DOI] [PubMed] [Google Scholar]
- Sulston J. E., Schierenberg E., White J. G. and Thomson J. N. (1983). The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100, 64-119. 10.1016/0012-1606(83)90201-4 [DOI] [PubMed] [Google Scholar]
- Suzuki A., Igarashi K., Aisaki K.-i., Kanno J. and Saga Y. (2010). NANOS2 interacts with the CCR4-NOT deadenylation complex and leads to suppression of specific RNAs. Proc. Natl. Acad. Sci. USA 107, 3594-3599. 10.1073/pnas.0908664107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A., Saba R., Miyoshi K., Morita Y. and Saga Y. (2012). Interaction between NANOS2 and the CCR4-NOT deadenylation complex is essential for male germ cell development in mouse. PLoS ONE 7, e33558 10.1371/journal.pone.0033558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa C. G. and Morgan D. O. (2000). Control of mitosis by changes in the subcellular location of cyclin-B1-Cdk1 and Cdc25C. Curr. Opin. Cell Biol. 12, 658-665. 10.1016/S0955-0674(00)00149-6 [DOI] [PubMed] [Google Scholar]
- Timmons L., Court D. L. and Fire A. (2001). Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263, 103-112. 10.1016/S0378-1119(00)00579-5 [DOI] [PubMed] [Google Scholar]
- Tsuda M., Sasaoka Y., Kiso M., Abe K., Haraguchi S., Kobayashi S. and Saga Y. (2003). Conserved role of nanos proteins in germ cell development. Science 301, 1239-1241. 10.1126/science.1085222 [DOI] [PubMed] [Google Scholar]
- Tursun B., Cochella L., Carrera I. and Hobert O. (2009). A toolkit and robust pipeline for the generation of fosmid-based reporter genes in C. elegans. PLoS ONE 4, e4625 10.1371/journal.pone.0004625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tursun B., Patel T., Kratsios P. and Hobert O. (2011). Direct conversion of C. elegans germ cells into specific neuron types. Science 331, 304-308. 10.1126/science.1199082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updike D. L., Knutson A. K., Egelhofer T. A., Campbell A. C. and Strome S. (2014). Germ-granule components prevent somatic development in the C. elegans germline. Curr. Biol. 24, 970-975. 10.1016/j.cub.2014.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Doren M., Williamson A. L. and Lehmann R. (1998). Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr. Biol. 8, 243-246. 10.1016/S0960-9822(98)70091-0 [DOI] [PubMed] [Google Scholar]
- Wagner C. R., Kuervers L., Baillie D. L. and Yanowitz J. L. (2010). xnd-1 regulates the global recombination landscape in Caenorhabditis elegans. Nature 467, 839-843. 10.1038/nature09429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. and Lehmann R. (1991). Nanos is the localized posterior determinant in Drosophila. Cell 66, 637-647. 10.1016/0092-8674(91)90110-K [DOI] [PubMed] [Google Scholar]
- Zanin E., Pacquelet A., Scheckel C., Ciosk R. and Gotta M. (2010). LARP-1 promotes oogenesis by repressing fem-3 in the C. elegans germline. J. Cell Sci. 123, 2717-2724. 10.1242/jcs.066761 [DOI] [PubMed] [Google Scholar]