Abstract
Distinct combinations of transcription factors are necessary to elicit cell fate changes in embryonic development. Yet within each group of fate-changing transcription factors, a subset called ‘pioneer factors’ are dominant in their ability to engage silent, unmarked chromatin and initiate the recruitment of other factors, thereby imparting new function to regulatory DNA sequences. Recent studies have shown that pioneer factors are also crucial for cellular reprogramming and that they are implicated in the marked changes in gene regulatory networks that occur in various cancers. Here, we provide an overview of the contexts in which pioneer factors function, how they can target silent genes, and their limitations at regions of heterochromatin. Understanding how pioneer factors regulate gene expression greatly enhances our understanding of how specific developmental lineages are established as well as how cell fates can be manipulated.
KEY WORDS: Cell fate control, Pioneer factors, Transcription
Summary: This Development at a Glance article provides an overview of pioneer transcription factors including their role in lineage specification in vivo, and how they can be used to manipulate cell fate in vitro.
Introduction
A differentiated, multicellular organism is made up of diverse cell types that are induced and maintained by cell type-specific chromatin states and patterns of gene expression. When new cell fates are induced during embryonic development, tissue regeneration or cell reprogramming, regulatory transcription factors must suppress genes specific to the original cell fate and activate genes specific to the new cell fate. Rather than one regulatory protein being singularly responsible for determining each cell type, it is well established that particular combinations of transcription factors elicit cell fate changes and maintain cell identity (Yamamoto, 1985). But although groups of transcription factors elicit cell fate changes, each member of a group does not function identically in the mechanism of changing gene expression patterns. Data derived from chromatin-binding studies in embryos, biochemical reconstitution of chromatin in vitro, genomics, and genetics have revealed that certain transcription factors, termed ‘pioneer factors’, have the distinct ability to target nucleosomal DNA sites at silent genes that have not been marked for activity, and thus initiate the process whereby a collective of regulatory proteins can assemble at particular sequences and activate genes specific to a new fate. In this way, pioneer factors are fundamental to development and reprogramming, and have even been shown to play a role in cancer. Here and in the accompanying poster, we provide an overview of pioneer factors by summarizing their role in cell fate specification and conversion, as well as the molecular basis for their activity. Understanding how pioneer factors interact with chromatin and the limits of their activity will enhance our ability to manipulate cell fate control for diverse research and therapeutic purposes.
The molecular basis of pioneer factor activity
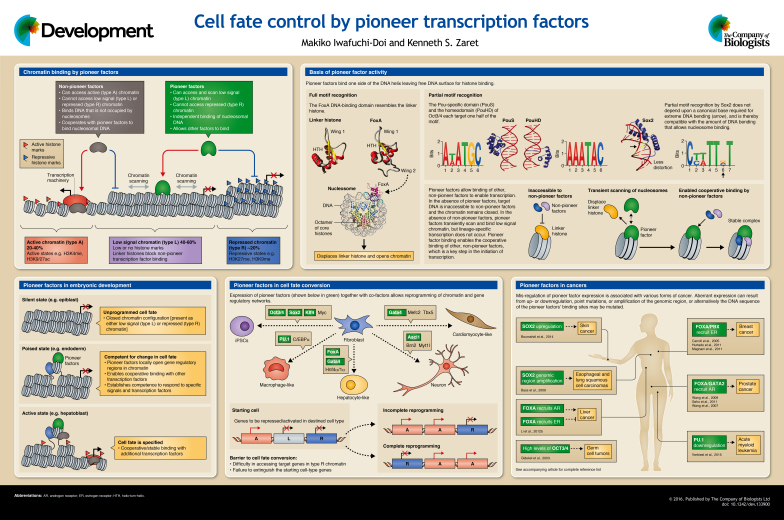
Gene regulation occurs in the context of chromatin, the complex of DNA and histone proteins that makes up the chromosomes. Within the chromatin, cellular DNA is wrapped nearly twice around an octamer of the four core histones to form arrays of nucleosomes. Linker histones bind to nucleosomes and stabilize condensed, repressive states. Chromatin can become ‘opened’ at gene regulatory regions, which allows RNA polymerase to function. This type of open chromatin is known as active chromatin, or chromatin type A. After a domain becomes open, it can typically accommodate the binding of any transcription factor and is accompanied by ‘active’ covalent modifications of the core histones, including histone H3 methylation on lysine 4 (H3K4me) and H3K9 and H3K27 acetylation (The ENCODE Project Consortium, 2011; Kharchenko et al., 2011). At promoters, these modifications can flank a nucleosome-free region immediately upstream of the transcription start site.
Large-scale chromatin-mapping studies have revealed that about half of the genome in a given cell possesses vast stretches of ‘low signal state’ chromatin that does not accumulate either active or repressive histone modifications (Kharchenko et al., 2011; Ho et al., 2014; Roadmap Epigenomics Consortium et al., 2015), and thus can be considered as unprogrammed. This type of chromatin is called low signal chromatin, or chromatin type L. Such unprogrammed chromatin is likely to be bound non-specifically by linker histones, which are nearly as abundant as core histones and are inherently repressive to transcription. Although many transcription factors cannot access target sites in the type L chromatin, pioneer factors are indeed capable of targeting this type of chromatin (Soufi et al., 2012; van Oevelen et al., 2015). In contrast to active and low signal chromatin, actively repressed chromatin domains – also known as repressed chromatin or chromatin type R – are inhibitory to the binding of most transcription factors, including pioneer factors, and thus strongly repress transcription. Such closed domains are typically accompanied by repressive covalent modifications of the core histones, including H3K9 or H3K27 methylation, although usually type R domains do not possess both marks at the same time, thus reflecting the different mechanisms of chromatin repression (Ho et al., 2014; Becker et al., 2016).
A central question regarding changes in cell fate is how genes in silent, low signal chromatin (type L) and repressed chromatin (type R) can be accessed and activated, despite the fact that the DNA is nucleosomal and thus inherently closed to most binding factors. The ability to target low signal chromatin is based on the inherent capacity of pioneer factors to recognize their target DNA sequences on the nucleosome (Cirillo et al., 1998), either by full or partial motif recognition. The FoxA DNA-binding domain (DBD) possesses a ‘winged helix’ structure that resembles the globular, nucleosomal binding domain of linker histone, binding to the full motif on one side of a DNA helix and leaving the other side of DNA free to bind core histones (Clark et al., 1993; Ramakrishnan et al., 1993; Cirillo et al., 1998). In vivo genetic and in vitro biochemical studies indicate that FoxA binding displaces linker histones from the local chromatin and keeps nucleosomes accessible (Cirillo et al., 1998; Iwafuchi-Doi et al., 2016). Furthermore, the C-terminal domain of FoxA can bind directly to core histone proteins and is required for opening chromatin, without ATP or ATP-dependent chromatin remodelers (Cirillo et al., 2002). In contrast to the full motif recognition binding of FoxA, some pioneer factors recognize only a partial motif. Oct3/4 (Pou5f1), Klf4 and basic helix-loop-helix type pioneer factors, such as Ascl1, target a partial DNA sequence of their canonical binding motifs on DNA, which is compatible with nucleosome binding (Soufi et al., 2015). It is now possible to predict a transcription factor's pioneer activity by extensive computational analysis of genomic DNA motifs and the factor's 3D structure (Soufi et al., 2015). In addition, pioneer factors can be identified empirically by their ability to target nucleosomal DNA in vivo (Soufi et al., 2015) and/or elicit an open region of chromatin (Sherwood et al., 2014).
Despite the importance of pioneer factor binding for initiating changes in cell fate, the process itself is insufficient to induce changes in gene expression directly. For this to take place, pioneer factor binding must occur cooperatively with other factors that, by themselves, cannot initiate such events in silent chromatin. Indeed, FoxA can recruit other activators or repressors to their target sites (Carroll et al., 2005; Zhang et al., 2005; Sekiya and Zaret, 2007; Li et al., 2012a; Iwafuchi-Doi et al., 2016). In Caenorhabditis elegans development, PHA-4, a homolog of FoxA, frequently binds promoters and recruits RNA polymerase II (Hsu et al., 2015). Thus, the initial chromatin-binding activity of pioneer factors is followed by the assembly of activating or repressing complexes, as dictated by the local DNA sequence and the presence of factors that are able to bind and further modify the chromatin.
Pioneer factors and developmental competence
The emergence of distinct cell types during embryonic development results from an intricate and dynamic cascade of regulatory changes in gene expression. Pioneer factors are likely to play a role in establishing a competence for many different cell fates, and indeed have already been shown to be fundamental in many developmental contexts, such as Pax7 for pituitary melanotrope development and PU.1 (Spi1) for myeloid and lymphoid development (summarized by Iwafuchi-Doi and Zaret, 2014). In Drosophila embryos, the maternal transcription factor Zelda (Vielfaltig – FlyBase) plays a primary role in zygotic genome activation following fertilization (Liang et al., 2008). Zelda is present in nuclei earlier than other master regulators, such as Bicoid and Dorsal, and locally opens inactive regulatory regions to allow other transcription factors to bind (Li et al., 2014; Schulz et al., 2015; Sun et al., 2015). Similarly, maternal Class V POU factors (e.g. Oct3/4 and Pou5f3) and SoxB1 factors (e.g. Sox2 and Sox19) are primary regulators of zygotic genome activation in mouse (Foygel et al., 2008; Pan and Schultz, 2011) and zebrafish (Lee et al., 2013; Leichsenring et al., 2013).
The TALE family of homeodomain transcription factors, including pre-B cell leukemia homeobox (Pbx) and Meis homebox, are important co-factors of Hox proteins. They engage the hoxb1a promoter during early zebrafish embryogenesis and recruit chromatin-modifying enzymes and RNA polymerase II to establish a poised state. Hoxb1b is then recruited and activates the hoxb1a promoter at a later stage (Choe et al., 2014). A recent study showed that Hoxa2 is recruited to a subset of sites that are pre-bound by Meis, which specifies the second branchial arches (Amin et al., 2015). Pbx also acts as a pioneer factor to enable the action of MyoD in specifying muscle fate (Berkes et al., 2004).
Pioneer factors FoxA and GATA
As a paradigm, the pioneer factors FoxA and GATA have been studied in great detail. Direct assessment of transcription factor occupancy on silent, liver-specific genes in early mouse embryos revealed that binding sites for FoxA and GATA transcription factors, but not sites for various other factors expressed in the liver lineage, were occupied in the undifferentiated foregut endoderm (Gualdi et al., 1996). Upon induction of the liver bud, other factors became engaged with the chromatin to initiate the liver-specific gene expression program. FoxA and GATA proteins are also expressed in areas outside the prospective liver region, for example in the medial-posterior endoderm. Here, their chromatin-binding activity still endows competence for the induction of liver genes, but this does not eventuate owing to the restrictive mesodermal interactions that normally inhibit the liver fate in this tissue (Bossard and Zaret, 2000; McLin et al., 2007).
Early evidence for the pioneer factor activity of FoxA and GATA came from attempts to model their binding with purified components in vitro (Cirillo et al., 1998; Cirillo and Zaret, 1999). Various liver-specific transcription factors were tested for their ability to bind to their sites on nucleosomal DNA and to engage nucleosome arrays harboring their binding sites, where the arrays were compacted by binding of linker histone. Remarkably, only purified FoxA protein could engage its target sequence on nucleosomes and enhance GATA factor binding, whereas the other factors tested could not. Furthermore, FoxA protein, and to a lesser extent Gata4, but not other liver-specific factors, could engage their target sites on the compacted nucleosome arrays and create a local open domain of chromatin, independent of nucleosome remodelers. Indeed, this is how the name ‘pioneer factors’ was coined (Cirillo et al., 2002).
The occupation by FoxA and GATA of a liver-specific enhancer in endoderm chromatin prior to the specification of liver cells, and the fact that at least FoxA could target nucleosomal DNA, suggested that these proteins functioned as competence factors (Zaret, 1999). Consistent with this, genetic inactivation of FoxA genes in the endoderm resulted in a failure of the endoderm to initiate hepatic differentiation (Lee et al., 2005). In C. elegans, when the FoxA homolog PHA-4 is first expressed it occupies target sequences of highest affinity, but as its expression is elevated during endoderm development, PHA-4 begins to occupy lower affinity targets, indicating that its action in binding to chromatin is concentration dependent (Gaudet and Mango, 2002). Recent genomics studies of pancreatic differentiation of human embryonic stem cells showed that FOXA binding occurs at genes for various endodermal fates in the endodermal intermediate stage when competence is acquired (Wang et al., 2015), consistent with the original hypothesis that pioneer factor binding imparts the competence to elicit fate changes. Emphasizing the utility of the mechanism, FoxA and GATA factors appear to be a crucial component of the network ‘kernel’ that specifies endoderm competence in all metazoans, over half a billion years of evolution (Davidson and Erwin, 2006).
Pioneer factors in cell fate conversions
To date, diverse cell fate conversions have been reported by various combinations of transcription factors, typically including pioneer factors. The most dramatic example is the reprogramming of fibroblasts into induced pluripotent stem cells (iPSCs) by only four transcription factors: Oct3/4, Sox2, Klf4 and c-Myc (Takahashi and Yamanaka, 2006). Of these factors, Oct3/4, Sox2 and Klf4 act as pioneer factors in that they can access silent, low signal state chromatin, regardless of whether they bind together or alone (Soufi et al., 2012, 2015). By contrast, c-Myc alone prefers to bind to active chromatin, but can also bind low signal chromatin sites when cooperating with the other factors (Soufi et al., 2012, 2015). There are more examples of a combination of cell type-specific pioneer factors and co-factors directing cell conversion: PU.1 and C/EBPa can reprogram macrophage-like cells from fibroblasts (Feng et al., 2008; van Oevelen et al., 2015); Gata4, Mef2c, Tbx5, Hand2 and Nkx2-5 can reprogram cardiomyocyte-like cells from fibroblasts (Ieda et al., 2010; Addis et al., 2013); Ascl1, Brn2 (Pou3f2) and Myt1l can reprogram functional glutaminergic neurons from fibroblasts (Vierbuchen et al., 2010); and FoxA, Gata4 and Hnf4α/1α can reprogram hepatocyte-like cells from fibroblasts (Huang et al., 2011; Sekiya and Suzuki, 2011). In all of these cases, at least one component of the factor combination plays a central role as a pioneer factor: PU.1 (macrophage-like cells), Gata4 (cardiomyocyte-like cells), Ascl1 (glutaminergic neurons) and FoxA (hepatocyte-like cells) (summarized by Iwafuchi-Doi and Zaret, 2014). For instance, Ascl1 binds silent, low signal chromatin and recruits Brn2 to target sites, and Brn2 is primarily required for the later stage of cell conversion by contributing to cell maturation (Wapinski et al., 2013). These findings demonstrate that even when the factors are expressed simultaneously, they function in a hierarchical manner, and that pioneer factors act first to establish a competence for a specific cell fate, which is followed by co-factor binding to instruct further differentiation.
Pioneer factors in cancers
As pioneer factors play a primary role in gene regulation, it is no surprise that their mis-regulation can compromise human health. Indeed, in many forms of cancer, pioneer factors are up- or downregulated, mutated or amplified in their genomic region, or alternatively the DNA sequence of the pioneer factors' binding sites is mutated. In esophageal and lung squamous cell carcinomas, chromosome segments containing SOX2 are often amplified (Bass et al., 2009). In the case of skin squamous-cell carcinoma, Sox2 is the most upregulated transcription factor, and conditional deletion of Sox2 markedly decreases skin tumor formation (Boumahdi et al., 2014). FoxA and GATA factors are involved in a variety of hormone-dependent cancers, such as estrogen receptor (ER)-positive breast cancer, androgen receptor (AR)-positive prostate cancer, and ER-dependent resistance and AR-mediated facilitation of liver cancer, and FOXA levels correlate well with clinical outcomes (reviewed by Jozwik and Carroll, 2012). Furthermore, single nucleotide polymorphisms at FOXA binding sites reduce binding of FOXA and ER in liver and correlate with hepatocellular carcinoma development in female patients (Li et al., 2012b).
Thus far, non-nuclear receptor transcription factors, such as pioneer factors, have been considered ‘undruggable’ targets for cancer treatment, but they could be attractive targets that avoid the issues of drug resistance that usually occur when targeting intracellular signaling pathways (Johnston and Carroll, 2015). The FoxM1-DNA interaction, which is upregulated in a wide range of cancers, is inhibited by direct interaction with a natural product called thiostepton (Hegde et al., 2011). Although findings such as these may hold promise for future cancer treatments, a better understanding of how pioneer factors function will be required in order to target them reliably for therapeutic breakthroughs.
Conclusions
Pioneer factors are among the master regulators of cell fate. They function by initiating chromatin targeting events on nucleosomal DNA, typically in low signal chromatin regions where the presence of linker histones represses transcription. The local exposure of chromatin brought about by pioneer factor binding allows other, non-pioneer transcription factors to access nucleosomal DNA, which in turn drives lineage-specific gene expression and selection of cell fate. The ability of pioneer factors to target silent genes and allow other factors to bind provides a mechanistic explanation for the long-standing phenomenon of developmental competence, in which a tissue gains the potential to execute a cell fate decision. However, pioneer factors do not occupy all in silico target sites in the genome; they are actively excluded from heterochromatic domains spanned by H3K9me2/3 (type R chromatin) (Lupien et al., 2008; Soufi et al., 2012), among others. By presenting a barrier to factor binding, heterochromatic, repressive domains provide a means for cells to stably retain their fate (Becker et al., 2016). We suggest that a possible reason for cell conversions being typically of a low efficiency and failing to shut off their initial genetic program (Cahan et al., 2014) may relate to the inefficiency of reprogramming factors in engaging with heterochromatic domains that span genes for which expression is required for the desired cell type. It is possible to alter chromatin state broadly by applying small molecules to target chromatin-modifying enzymes, but such changes will occur globally throughout the genome. We speculate that understanding how cell type-specific heterochromatic domains are established and how pioneer factors can overcome such barriers during development will provide more targeted ways to manipulate cell fate in health and disease. More broadly, further work in the field should be aimed at understanding how different pioneer factors target silent, low signal-state chromatin and how heterochromatic features at highly repressed chromatin might block pioneer factor binding. These detailed mechanistic insights will pave the way for the future ability to program and reprogram cell fates at will.
Acknowledgements
We thank Eileen Hulme for help in preparing the manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
Work on pioneer factors was supported by the National Institutes of Health [GM36477 to K.S.Z.] M.I.-D. was supported by postdoctoral fellowships from the Japan Society for the Promotion of Science and the Naito, Astellas and Uehara foundations.
Development at a Glance
A high-resolution version of the poster is available for downloading in the online version of this article at http://dev.biologists.org/content/143/11/1833/F1.poster.jpg
References
- Addis R. C., Ifkovits J. L., Pinto F., Kellam L. D., Esteso P., Rentschler S., Christoforou N., Epstein J. A. and Gearhart J. D. (2013). Optimization of direct fibroblast reprogramming to cardiomyocytes using calcium activity as a functional measure of success. J. Mol. Cell. Cardiol. 60, 97-106. 10.1016/j.yjmcc.2013.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin S., Donaldson I. J., Zannino D. A., Hensman J., Rattray M., Losa M., Spitz F., Ladam F., Sagerström C. and Bobola N. (2015). Hoxa2 selectively enhances Meis binding to change a branchial arch ground state. Dev. Cell 32, 265-277. 10.1016/j.devcel.2014.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass A. J., Watanabe H., Mermel C. H., Yu S., Perner S., Verhaak R. G., Kim S. Y., Wardwell L., Tamayo P., Gat-Viks I. et al. (2009). SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat. Genet. 41, 1238-1242. 10.1038/ng.465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J. S., Nicetto D. and Zaret K. S. (2016). H3K9me3-dependent heterochromatin: barrier to cell fate changes. Trends Genet. 32, 29-41. 10.1016/j.tig.2015.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkes C. A., Bergstrom D. A., Penn B. H., Seaver K. J., Knoepfler P. S. and Tapscott S. J. (2004). Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol. Cell 14, 465-477. 10.1016/S1097-2765(04)00260-6 [DOI] [PubMed] [Google Scholar]
- Bossard P. and Zaret K. S. (2000). Repressive and restrictive mesodermal interactions with gut endoderm: possible relation to Meckel's Diverticulum. Development 127, 4915-4923. [DOI] [PubMed] [Google Scholar]
- Boumahdi S., Driessens G., Lapouge G., Rorive S., Nassar D., Le Mercier M., Delatte B., Caauwe A., Lenglez S., Nkusi E. et al. (2014). SOX2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma. Nature 511, 246-250. 10.1038/nature13305 [DOI] [PubMed] [Google Scholar]
- Cahan P., Li H., Morris S. A., Lummertz da Rocha E., Daley G. Q. and Collins J. J. (2014). CellNet: network biology applied to stem cell engineering. Cell 158, 903-915. 10.1016/j.cell.2014.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll J. S., Liu X. S., Brodsky A. S., Li W., Meyer C. A., Szary A. J., Eeckhoute J., Shao W., Hestermann E. V., Geistlinger T. R. et al. (2005). Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122, 33-43. 10.1016/j.cell.2005.05.008 [DOI] [PubMed] [Google Scholar]
- Choe S.-K., Ladam F. and Sagerström C. G. (2014). TALE factors poise promoters for activation by Hox proteins. Dev. Cell 28, 203-211. 10.1016/j.devcel.2013.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo L. A. and Zaret K. S. (1999). An early developmental transcription factor complex that is more stable on nucleosome core particles than on free DNA. Mol. Cell 4, 961-969. 10.1016/S1097-2765(00)80225-7 [DOI] [PubMed] [Google Scholar]
- Cirillo L. A., McPherson C. E., Bossard P., Stevens K., Cherian S., Shim E.-Y., Clark K. L., Burley S. K. and Zaret K. S. (1998). Binding of the winged-helix transcription factor HNF3 to a linker histone site on the nucleosome. EMBO J. 17, 244-254. 10.1093/emboj/17.1.244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo L. A., Lin F. R., Cuesta I., Friedman D., Jarnik M. and Zaret K. S. (2002). Opening of compacted chromatin by early developmental transcription factors HNF3 (FOXA) and GATA-4. Mol. Cell 9, 279-289. 10.1016/S1097-2765(02)00459-8 [DOI] [PubMed] [Google Scholar]
- Clark K. L., Halay E. D., Lai E. and Burley S. K. (1993). Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature 364, 412-420. 10.1038/364412a0 [DOI] [PubMed] [Google Scholar]
- Davidson E. H. and Erwin D. H. (2006). Gene regulatory networks and the evolution of animal body plans. Science 311, 796-800. 10.1126/science.1113832 [DOI] [PubMed] [Google Scholar]
- Feng R., Desbordes S. C., Xie H., Tillo E. S., Pixley F., Stanley E. R. and Graf T. (2008). PU.1 and C/EBPalpha/beta convert fibroblasts into macrophage-like cells. Proc. Natl. Acad. Sci. USA 105, 6057-6062. 10.1073/pnas.0711961105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foygel K., Choi B., Jun S., Leong D. E., Lee A., Wong C. C., Zuo E., Eckart M., Reijo Pera R. A., Wong W. H. et al. (2008). A novel and critical role for Oct4 as a regulator of the maternal-embryonic transition. PLoS ONE 3, e4109 10.1371/journal.pone.0004109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet J. and Mango S. E. (2002). Regulation of Organogenesis by the Caenorhabditis elegans FoxA protein PHA-4. Science 295, 821-825. 10.1126/science.1065175 [DOI] [PubMed] [Google Scholar]
- Gidekel S., Pizov G., Bergman Y., and Pikarsky E. (2003). Oct-3/4 is a dose-dependent oncogenic fate determinant. Cancer Cell 4, 361-370. 10.1016/S1535-6108(03)00270-8 [DOI] [PubMed] [Google Scholar]
- Gualdi R., Bossard P., Zheng M., Hamada Y., Coleman J. R. and Zaret K. S. (1996). Hepatic specification of the gut endoderm in vitro: cell signaling and transcriptional control. Genes Dev. 10, 1670-1682. 10.1101/gad.10.13.1670 [DOI] [PubMed] [Google Scholar]
- Hegde N. S., Sanders D. A., Rodriguez R. and Balasubramanian S. (2011). The transcription factor FOXM1 is a cellular target of the natural product thiostrepton. Nat. Chem. 3, 725-731. 10.1038/nchem.1114 [DOI] [PubMed] [Google Scholar]
- Ho J. W. K., Jung Y. L., Liu T., Alver B. H., Lee S., Ikegami K., Sohn K.-A., Minoda A., Tolstorukov M. Y., Appert A. et al. (2014). Comparative analysis of metazoan chromatin organization. Nature 512, 449-452. 10.1038/nature13415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu H.-T., Chen H.-M., Yang Z., Wang J., Lee N. K., Burger A., Zaret K., Liu T., Levine E. and Mango S. E. (2015). Recruitment of RNA polymerase II by the pioneer transcription factor PHA-4. Science 348, 1372-1376. 10.1126/science.aab1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P., He Z., Ji S., Sun H., Xiang D., Liu C., Hu Y., Wang X. and Hui L. (2011). Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature 475, 386-389. 10.1038/nature10116 [DOI] [PubMed] [Google Scholar]
- Hurtado A., Holmes K. A., Ross-Innes C. S., Schmidt D. and Carroll J. S. (2011). FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat. Genet. 43, 27-33. 10.1038/ng.730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieda M., Fu J.-D., Delgado-Olguin P., Vedantham V., Hayashi Y., Bruneau B. G. and Srivastava D. (2010). Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 142, 375-386. 10.1016/j.cell.2010.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwafuchi-Doi M. and Zaret K. S. (2014). Pioneer transcription factors in cell reprogramming. Genes Dev. 28, 2679-2692. 10.1101/gad.253443.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwafuchi-Doi M., Donahue G., Kakumanu A., Watts J. A., Mahony S., Pugh B. F., Lee D., Kaestner K. H. and Zaret K. S. (2016). The pioneer transcription factor FoxA maintains an accessible nucleosome configuration at enhancers for tissue-specific gene activation. Mol. Cell 62, 79-91. 10.1016/j.molcel.2016.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston S. J. and Carroll J. S. (2015). Transcription factors and chromatin proteins as therapeutic targets in cancer. Biochim. Biophys. Acta 1855, 183-192. 10.1016/j.bbcan.2015.02.002 [DOI] [PubMed] [Google Scholar]
- Jozwik K. M. and Carroll J. S. (2012). Pioneer factors in hormone-dependent cancers. Nat. Rev. Cancer 12, 381-385. 10.1038/nrc3263 [DOI] [PubMed] [Google Scholar]
- Kharchenko P. V., Alekseyenko A. A., Schwartz Y. B., Minoda A., Riddle N. C., Ernst J., Sabo P. J., Larschan E., Gorchakov A. A., Gu T. et al. (2011). Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature 471, 480-485. 10.1038/nature09725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. S., Friedman J. R., Fulmer J. T. and Kaestner K. H. (2005). The initiation of liver development is dependent on Foxa transcription factors. Nature 435, 944-947. 10.1038/nature03649 [DOI] [PubMed] [Google Scholar]
- Lee M. T., Bonneau A. R., Takacs C. M., Bazzini A. A., DiVito K. R., Fleming E. S. and Giraldez A. J. (2013). Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature 503, 360-364. 10.1038/nature12632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leichsenring M., Maes J., Mossner R., Driever W. and Onichtchouk D. (2013). Pou5f1 transcription factor controls zygotic gene activation in vertebrates. Science 341, 1005-1009. 10.1126/science.1242527 [DOI] [PubMed] [Google Scholar]
- Li G., Ruan X., Auerbach R. K., Sandhu K. S., Zheng M., Wang P., Poh H. M., Goh Y., Lim J., Zhang J. et al. (2012a). Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell 148, 84-98. 10.1016/j.cell.2011.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Tuteja G., Schug J. and Kaestner K. H. (2012b). Foxa1 and Foxa2 are essential for sexual dimorphism in liver cancer. Cell 148, 72-83. 10.1016/j.cell.2011.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.-Y., Harrison M. M., Villalta J. E., Kaplan T. and Eisen M. B. (2014). Establishment of regions of genomic activity during the Drosophila maternal to zygotic transition. Elife 3, e03737 10.7554/eLife.03737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H.-L., Nien C.-Y., Liu H.-Y., Metzstein M. M., Kirov N. and Rushlow C. (2008). The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature 456, 400-403. 10.1038/nature07388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupien M., Eeckhoute J., Meyer C. A., Wang Q., Zhang Y., Li W., Carroll J. S., Liu X. S. and Brown M. (2008). FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 132, 958-970. 10.1016/j.cell.2008.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnani L., Ballantyne E. B., Zhang X. and Lupien M. (2011). PBX1 genomic pioneer function drives ERalpha signaling underlying progression in breast cancer. PLoS Genet. 7, e1002368 10.1371/journal.pgen.1002368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLin V. A., Rankin S. A. and Zorn A. M. (2007). Repression of Wnt/beta-catenin signaling in the anterior endoderm is essential for liver and pancreas development. Development 134, 2207-2217. 10.1242/dev.001230 [DOI] [PubMed] [Google Scholar]
- Pan H. and Schultz R. M. (2011). Sox2 modulates reprogramming of gene expression in two-cell mouse embryos. Biol. Reprod. 85, 409-416. 10.1095/biolreprod.111.090886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan V., Finch J. T., Graziano V., Lee P. L. and Sweet R. M. (1993). Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature 362, 219-223. 10.1038/362219a0 [DOI] [PubMed] [Google Scholar]
- Roadmap Epigenomics Consortium, Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J. et al. (2015). Integrative analysis of 111 reference human epigenomes. Nature 518, 317-330. 10.1038/nature14248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu B., Laakso M., Ovaska K., Mirtti T., Lundin J., Rannikko A., Sankila A., Turunen J. P., Lundin M. et al. (2011). Dual role of FoxA1 in androgen receptor binding to chromatin, androgen signalling and prostate cancer. EMBO J. 30, 3962-3976. 10.1038/emboj.2011.328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz K. N., Bondra E. R., Moshe A., Villalta J. E., Lieb J. D., Kaplan T., McKay D. J. and Harrison M. M. (2015). Zelda is differentially required for chromatin accessibility, transcription factor binding, and gene expression in the early Drosophila embryo. Genome Res. 25, 1715-1726. 10.1101/gr.192682.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya S. and Suzuki A. (2011). Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature 475, 390-393. 10.1038/nature10263 [DOI] [PubMed] [Google Scholar]
- Sekiya T. and Zaret K. S. (2007). Repression by Groucho/TLE/Grg proteins: genomic site recruitment generates compacted chromatin in vitro and impairs activator binding in vivo. Mol. Cell 28, 291-303. 10.1016/j.molcel.2007.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood R. I., Hashimoto T., O'Donnell C. W., Lewis S., Barkal A. A., van Hoff J. P., Karun V., Jaakkola T. and Gifford D. K. (2014). Discovery of directional and nondirectional pioneer transcription factors by modeling DNase profile magnitude and shape. Nat. Biotechnol. 32, 171-178. 10.1038/nbt.2798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A., Donahue G. and Zaret K. S. (2012). Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell 151, 994-1004. 10.1016/j.cell.2012.09.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A., Garcia M. F., Jaroszewicz A., Osman N., Pellegrini M. and Zaret K. S. (2015). Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell 161, 555-568. 10.1016/j.cell.2015.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Nien C.-Y., Chen K., Liu H.-Y., Johnston J., Zeitlinger J. and Rushlow C. (2015). Zelda overcomes the high intrinsic nucleosome barrier at enhancers during Drosophila zygotic genome activation. Genome Res. 25, 1703-1714. 10.1101/gr.192542.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K. and Yamanaka S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663-676. 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. (2011). A user's guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol. 9, e1001046 10.1371/journal.pbio.1001046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oevelen C., Collombet S., Vicent G., Hoogenkamp M., Lepoivre C., Badeaux A., Bussmann L., Sardina J. L., Thieffry D., Beato M. et al. (2015). C/EBPalpha activates pre-existing and de novo macrophage enhancers during induced pre-B cell transdifferentiation and myelopoiesis. Stem Cell Rep. 5, 232-247. 10.1016/j.stemcr.2015.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbiest T., Bouffler S., Nutt S. L. and Badie C. (2015). PU.1 downregulation in murine radiation-induced acute myeloid leukaemia (AML): from molecular mechanism to human AML. Carcinogenesis 36, 413-419. 10.1093/carcin/bgv016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T., Ostermeier A., Pang Z. P., Kokubu Y., Südhof T. C. and Wernig M. (2010). Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463, 1035-1041. 10.1038/nature08797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Li W., Liu X. S., Carroll J. S., Jänne O. A., Keeton E. K., Chinnaiyan A. M., Pienta K. J. and Brown M. (2007). A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol. Cell 27, 380-392. 10.1016/j.molcel.2007.05.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Li W., Zhang Y., Yuan X., Xu K., Yu J., Chen Z., Beroukhim R., Wang H., Lupien M. et al. (2009). Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell 138, 245-256. 10.1016/j.cell.2009.04.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A., Yue F., Li Y., Xie R., Harper T., Patel N. A., Muth K., Palmer J., Qiu Y., Wang J. et al. (2015). Epigenetic priming of enhancers predicts developmental competence of hESC-derived endodermal lineage intermediates. Cell Stem Cell 16, 386-399. 10.1016/j.stem.2015.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski O. L., Vierbuchen T., Qu K., Lee Q. Y., Chanda S., Fuentes D. R., Giresi P. G., Ng Y. H., Marro S., Neff N. F. et al. (2013). Hierarchical mechanisms for direct reprogramming of fibroblasts to neurons. Cell 155, 621-635. 10.1016/j.cell.2013.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K. R. (1985). Steroid receptor regulated transcription of specific genes and gene networks. Annu. Rev. Genet. 19, 209-252. 10.1146/annurev.ge.19.120185.001233 [DOI] [PubMed] [Google Scholar]
- Zaret K. (1999). Developmental competence of the gut endoderm: genetic potentiation by GATA and HNF3/fork head proteins. Dev. Biol. 209, 1-10. 10.1006/dbio.1999.9228 [DOI] [PubMed] [Google Scholar]
- Zhang H., Roberts D. N. and Cairns B. R. (2005). Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell 123, 219-231. 10.1016/j.cell.2005.08.036 [DOI] [PMC free article] [PubMed] [Google Scholar]