Abstract
An efficient immunosurveillance of CD8+ T cells in the periphery depends on positive/negative selection of thymocytes and thus on the dynamics of antigen degradation and epitope production by thymoproteasome and immunoproteasome in the thymus. Although studies in mouse systems have shown how thymoproteasome activity differs from that of immunoproteasome and strongly impacts the T cell repertoire, the proteolytic dynamics and the regulation of human thymoproteasome are unknown. By combining biochemical and computational modeling approaches, we show here that human 20S thymoproteasome and immunoproteasome differ not only in the proteolytic activity of the catalytic sites but also in the peptide transport. These differences impinge upon the quantity of peptide products rather than where the substrates are cleaved. The comparison of the two human 20S proteasome isoforms depicts different processing of antigens that are associated to tumors and autoimmune diseases.
Keywords: proteasome, protein degradation, antigen processing, computational biology, bioinformatics, thymoproteasome, thymus, proteolysis
Introduction
The immune system constantly patrols the human body to detect pathological situations. An important role in this is played by CD8+ T cells, which recognize and kill infected and aberrant cells. Because of their high specificity and cytotoxic activity, CD8+ T cells are intensely investigated as tools and/or targets of immunotherapies against infection, autoimmunity, and cancer (1–3).
CD8+ T cells recognize, via their T cell receptor coupled to a CD8 molecule, a specific epitope presented in the cleft of major histocompatibility complex class I (MHC-I)4 molecules (4). The cytotoxic CD8+ T cells are primed in lymph nodes. The naïve CD8+ T cells arrive in the lymph nodes from the thymus. In this latter organ, thymocytes, prior to becoming mature naïve CD8+ T cells, undergo a series of maturation/selection processes called central tolerance. According to one of the most accepted models, central tolerance can be summarized in two selection steps. Initially, thymocytes undergo a positive selection step, which takes place in the thymic cortex, leading to the survival and maturation of double-positive thymocytes that express T cell receptors with intermediate affinity and/or avidity for MHC-I–peptide complexes. Afterward, in the thymic medulla, thymocytes undergo the negative selection step, which leads to the elimination of thymocytes recognizing self-peptide–MHC complexes with a high affinity (5).
The large majority of peptides bound to MHC-I molecules and recognized by CD8+ T lymphocytes are generated by proteasome, which is the final effector of the ubiquitin–proteasome system (6). This barrel-shaped protease can break proteins and release the peptide fragments or religate them, thereby forming new (spliced) peptides with sequences that do not recapitulate the parental protein (4). The proteasome is a multisubunit enzyme, which has a 20S proteasome as the core and various proteins bound at both sides of its gate, where they play a regulatory role (4). The 26S proteasome, comprising a 20S proteasome core coupled to a 19S regulatory complex, is often the most active form of proteasome, with an increasing amount of evidence suggesting that the 20S proteasome is independently functional and both degrades and activates proteins in cells (4, 7, 8). The 20S proteasome is constituted of four rings, two α rings at the apexes and two β rings forming the central chamber. Each ring has seven distinct subunits. Each β ring carries three catalytic (i.e. β1, β2, and β5) subunits, which have distinct preferences for peptide sequence motifs (9). Human cells can express different isoforms of catalytic subunits, which are incorporated in distinct proteasome isoforms. Standard proteasome (s-proteasome) contains β1, β2, and β5 subunits. Immunoproteasome (i-proteasome) contains β1i, β2i, and β5i subunits and is present in immune cells (constitutively) as well as in cells exposed to inflammatory milieu. The majority of cells express a mixed-type proteasome population where both s- and i-proteasome subunits are present in various amount (4). A decade ago, Murata et al. (10) identified the so-called thymoproteasome (t-proteasome), which carries the β1i, β2i, and β5t subunits and has so far only been detected in the thymus.
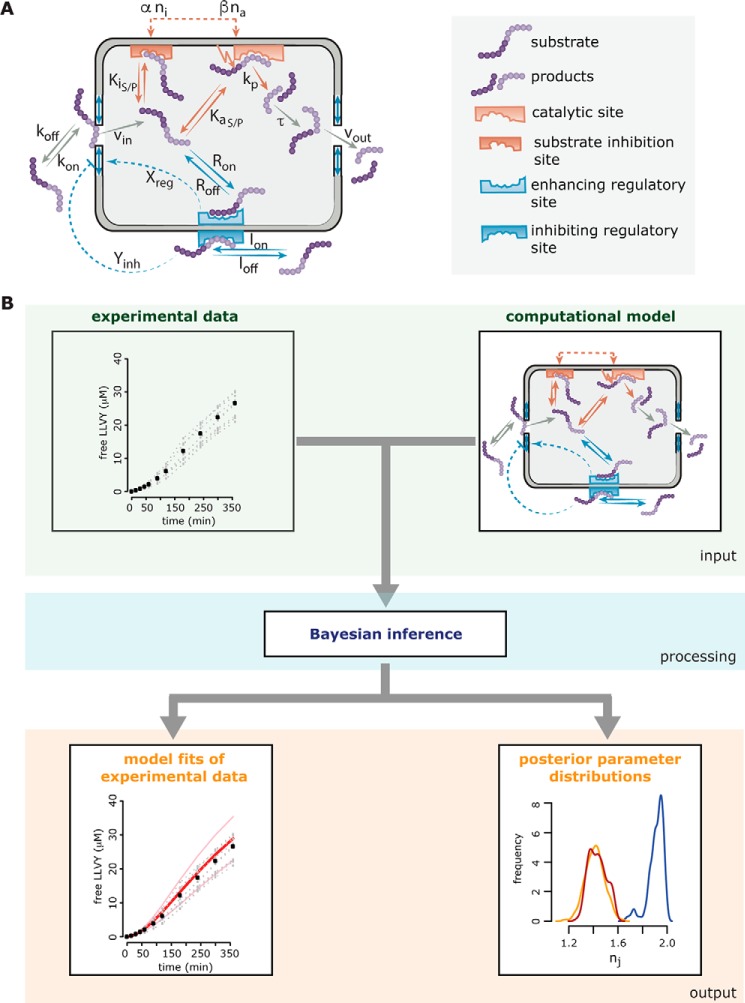
To investigate the dynamics and its regulation of peptide hydrolysis by the 20S proteasome and to elucidate differences between s- and i-proteasomes, we previously developed a computational mechanistic model of proteasome peptide degradation (11) (Fig. 1A). The model was constructed and compared with many competing models in a Bayesian model selection framework (12, 13) and finally challenged with further experimentation.
Figure 1.
Overview of the computational modeling approach describing the proteasome proteolytic dynamics. In A, the schematic of the compartmentalized proteasome model proposed by Liepe et al. (11) is shown. The model describes all relevant steps involved in substrate degradation. These include (i) peptide transport steps (peptide binding close to the outer site of the gate, peptide influx into the chamber, peptide translocation inside the chamber, and peptide efflux out of the chamber), (ii) substrate hydrolysis steps (peptide binding to the active site and subsequent hydrolysis and peptide binding to the noncatalytic inhibitor site), and (iii) transport regulation (peptide binding to the noncatalytic enhancer site, peptide binding to the noncatalytic inhibitor site, and resulting effects on the conformation of the proteasome gate). The gray chamber represents a simplification of the 20S proteasome catalytic chamber with openings to the outside. The substrate and product peptides (purple) can enter the 20S proteasome chamber upon binding to the outer face of the gate, interact with the regulatory and catalytic sites inside the chamber, and leave the proteasome chamber upon translocation to the proximity of the inner face of the gate. Gray arrows indicate the transport of substrate and product peptides. The orange arrow denotes the hydrolysis reaction where a substrate peptide is transformed into product peptide, thereby releasing the fluorophore. Enhancing regulatory sites inside the chamber are shown in blue with the dashed arrows indicating their effect (transport-enhancing gate conformation). The inhibiting regulatory site outside the chamber is shown in light blue with the dashed arrow indicating its effect (transport-inhibiting gate conformation). The catalytic site consists of an active site (light orange) and an inactive modifier site (dark orange). For details of the model equations, model setup, and model parameters, please refer to Liepe et al. (11). In B, the schematic of Bayesian inference is sketched. Computational models describing biological systems are often parameterized. These parameters can be abstract, or as is the case here, they can be kinetic parameters with a direct physical translation. To learn anything from the model, it is necessary to calibrate the model against experimental data. This can be done in many fashions; however, in the last decade Bayesian inference techniques proved to be powerful model calibration tools. One of the advantages of Bayesian inference is that it estimates not only the model parameters but also their uncertainty. In general, experimental data are collected and a computational model is formulated. Both are then used as input for the Bayesian inference algorithm (here approximate Bayesian computation). The basic concept of the algorithm is to test all sorts of combinations of parameters (through a defined sampling scheme) and simulate the model with those parameter combinations. If the model simulations correspond well to the experimental data, the corresponding parameter combination is accepted; otherwise the parameter combination is discarded. This is done repeatedly until a certain number of accepted parameter combinations is reached, which then construct the so-called posterior parameter distribution. This posterior distribution contains all information about the separate model parameters as well as their dependences among each other. The outputs of Bayesian inference are therefore the model fits of the experimental data and the posterior parameter distributions. Calibrating the same model to experimental data generated under different conditions (here different proteasome isoforms) allows us to compare the obtained posterior parameter distributions and detect which model parameters differ for the different conditions and which parameters are not influenced.
Specifically, a series of models was constructed with increasing complexity, starting from the simplest Michaelis–Menten model and ignoring the structural properties of the proteasome. Kinetic time-course data tracking the degradation of short fluorogenic peptides by 20S proteasomes were used to test whether the constructed models were able to produce the experimentally observed kinetics. The latter data and further experimentation were used to yield insight into possible dynamics of the proteasome and guide the development of competing mechanistic models with increasing complexity. These models can be seen as representing competing hypotheses, allowing selection of the hypothesis that best justifies our experimental data. In contrast to many hypothesis-testing techniques, Bayesian model selection allows us to not only reject a likely wrong hypothesis but also rank competing models given experimental data (14, 15).
Model development and model selection have two aspects to consider: the model structure and the model parameters. Although the model structure can be seen as a map, the model parameters essentially identify where in that map the dynamics of the model can occur. It is therefore of interest to define both model structure and model parameters, or in Bayesian terms the posterior model distribution and the posterior parameter distribution (16, 17). Our Bayesian model selection framework allows us to determine both the best model structure and the model parameters, each with prior knowledge, i.e. the original model distribution (the construction of a set of competing models and our initial confidence in them) and the original parameter distribution (the allowed values of a kinetic parameter). Both the suggested model structure and the suggested model parameters will contain uncertainty, which can be assessed from the posterior distribution (18, 19). Roughly speaking, the broader a posterior parameter distribution is, the less information the experimental data contains about this parameter and thus the higher the uncertainty. This parameter uncertainty will be carried into model predictions. Note that in some cases not all model parameters must be inferred with low uncertainty to make precise model predictions (20). This is because the system's dynamics may not be susceptible to alterations in those parameters.
The applied Bayesian model selection framework allowed us to determine the best model of 10 constructed competing models; however, this does not guarantee the accuracy of the winning model. A crucial step during model development is model validation, whereby independent experimental data, which were not used for model development or model calibration, are used to challenge the chosen model. At this point, it is important to note that any model is a simplification of the true biochemical system and that in many cases a model is developed to explain certain aspects of the system but not all (21). Assumptions and simplifications often dictate under which conditions the system can be described by the developed model. For example, in our 20S proteasome model, we do not include proteasome activators such as PA28αβ and 19S complexes, which may significantly alter the observed dynamics and its regulation. Model validation can help to elucidate the limits and predictive potential of the model.
In most cases, the motivation behind model development is to generate a model to predict system behavior that cannot be observed experimentally. However, here we have created a model that tests different mechanistic hypotheses and derives kinetic parameters to in turn characterize different proteasome isoforms. As we showed that proteasome isoforms differ quantitatively but not qualitatively in the peptides they generate (22) (at least with the sensitivity allowed by the assays applied), we can therefore assume that the overall model structure is the same for different isoforms and that only the kinetic parameters differ. These differences can be acquired by comparing the marginal posterior parameter distributions in a practical rather than statistical manner. That is, differences are detected if the distributions to be compared overlap only slightly and their values with the highest densities clearly vary. On the contrary, if the parameter distributions to be compared cover the same range of possible parameter values, then either the data do not contain sufficient information to detect differences, or the parameters indeed do not differ.
In our previous study, parameter inference and subsequent comparison of posterior distributions of the kinetic model parameters (Fig. 1B) showed that 20S s- and i-proteasomes differ in the activity of their catalytic sites, in peptide transport along their inner channels, and in the transport regulation dynamics. Because the peptide transport often seems to be the rate-limiting step of the overall peptide-bond hydrolysis (11), such differences can impinge upon the degradation rate of specific proteins and the generation efficiency of specific antigenic peptides. Quantitative differences in antigenic peptide production, after the downstream antigen presentation steps, can result in an impaired or enhanced CD8+ T cell response in vivo (22–25).
The efficiency of the antigenic peptide generation by i-proteasome plays a key role in the negative selection of thymocytes because medullary professional antigen-presenting cells mainly express this proteasome isoform (5). The cortical thymic epithelial cells, on the contrary, mainly express the t-proteasome, although s-proteasome catalytic subunits have also been detected (10, 26). t-proteasome influences the CD8+ T cell repertoire and the response to infection in mice (10, 27–29). Some evidence hints toward a unique t-proteasome proteolytic activity, which, in mice, would lead to the generation of t-proteasome–specific antigenic peptides with peculiar features promoting the positive selection of thymocytes (5, 28, 30). As a consequence, the difference between the proteolytic dynamics of human i- and t-proteasomes is supposed to have a large impact on the central tolerance and, thus, on the T cell repertoire and the efficacy of CD8+ T cells to recognize infected or aberrant cells and eliminate them in the human body.
To study in which aspects of proteolytic dynamics the two human proteasome isoforms diverge, we have coupled biochemical experiments to bioinformatics analyses. We have made use of the previously developed computational model to infer the kinetic parameters of human 20S t-proteasomes compared with human 20S s- and i-proteasomes. We have taken advantage of Bayesian inference to obtain posterior parameter distributions that capture not only the most plausible parameter values but also the information and uncertainty carried by the experimental data. The latter is of particular interest when aiming to detect differences between the proteasome isoforms. Based on the inferred kinetic parameters, we have performed model simulations to identify the rate-limiting steps and peptide transport dynamics of human 20S t-proteasome.
In a second step of the study, we have investigated the quantitative differences between the three 20S proteasome isoforms in substrate cleavage-site preferences and epitope production. Because of the immunological implications that such differences can have, we have used synthetic polypeptides substrates derived from tumors and multiple sclerosis, which are two examples of diseases where the MHC-I–presented epitopes are therapeutically relevant.
Results
Human 20S t-proteasome differs from s- and i-proteasomes in its proteolytic dynamics
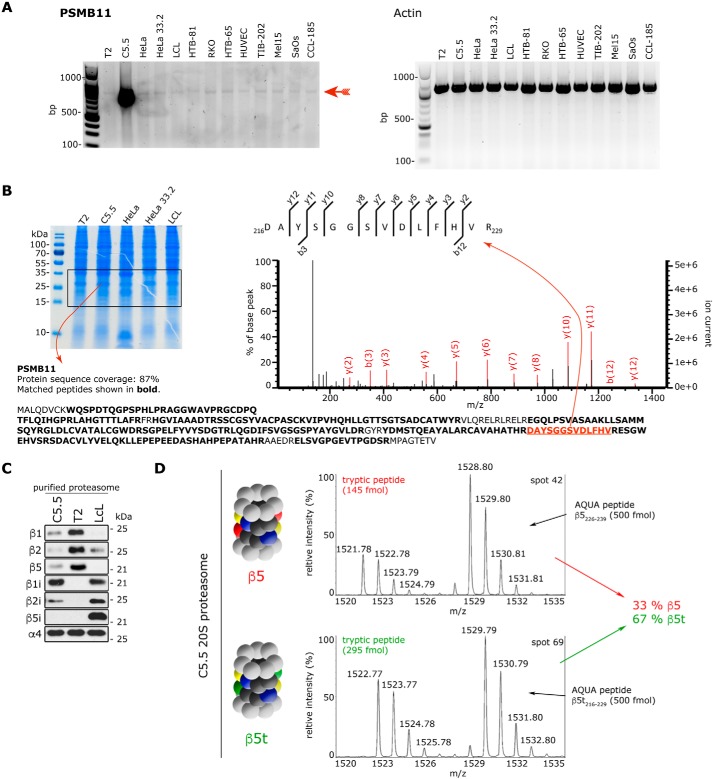
Human β5t subunit has been previously detected in different forms of human thymoma (31, 32), which are tumors originating from the epithelial cells of the thymus. We have tested whether the t-proteasome subunit was detectable in other cancer-derived cell lines. The mRNA of PSMB11, which is the gene encoding the human β5t proteasome subunit, is also detectable in several tumor-derived or immortalized cell lines by RT-PCR (Fig. 2A); however, its expression does not lead to a detectable quantity of β5t subunit through the use of a standard proteomics strategy (Fig. 2B). This result confirms that in humans the expression of the β5t proteasome subunit also seems to be limited to the thymus.
Figure 2.
Catalytic β subunit expression in different human cell lines. A, RT-PCR products specific for PSMB11 (β5t subunit) or actin (as control) obtained from different immortalized or tumor-derived human cell lines. The PSMB11-specific band is marked with a red arrow. Specificity has been confirmed by cloning and sequencing the cDNA extracted from the band. B, MS identification of the β5t subunit in the cell lysate of the same cell lines shown in A. Cell lysates have been separated on an SDS gel and stained with Coomassie (left panel; only some representative cell samples are shown), the proteins framed in the picture have been cut and digested by trypsin, and the β5t subunit–specific peptides have been detected by MS. Only for the C5.5 cell line have we identified the β5t subunit products (marked with bold letters; 19 specific peptides depicted below the gel) with significant MS/MS spectra (the spectrum of one of them is depicted in the right panel). C, Western blot assays for proteasome catalytic subunits, which have been carried out after separation of 0.5 μg of purified 20S proteasomes by SDS-PAGE, are shown. D, relative quantification of the subunits β5 (PSB5_human) and β5t (PSB11_human) of purified 20S proteasome after absolute quantification with AQUA peptides. Shown are representative MALDI mass spectra of the tryptic peptide 226DAYSGGAVNLYHVR239 with [M + H]+exp = 1521.78 and its heavy analogue with [M + H]+exp = 1528.80 of β5 subunit at spot 42 and 216DAYSGGSVDLFHVR229 with [M + H]+exp = 1522.77 and its heavy analogue with [M + H]+exp = 1529.79 of β5t subunit at spot 69. The calculation of the absolute amount of the tryptic peptides is performed by comparison of the MS peak areas with those of the corresponding AQUA peptides.
Therefore, not having access to enough human cortical thymic epithelial cells, we generated a cell line, C5.5. This cell line is derived from the human lymphoblastoid cell line T2. The C5.5 cell line expresses mainly the β1i, β2i, and β5t subunits, with a β5:β5t subunit ratio of 1:2.5 according to our quantitative proteomics analysis carried out with AQUA peptides (Fig. 2, C and D). This mixed-type 20S proteasome, here referred to as t-proteasome, purified from the C5.5 cell line has been compared in our study with either the 20S s-proteasome derived from parental T2 or the intermediate-type 20S proteasome purified from Epstein–Barr virus—immortalized lymphoblastoid cell lines (LCLs) that has a β5:β5i subunit ratio of 1:2.5 or larger (22) and has often been used as an example of i-proteasome.
To investigate the proteolytic dynamics of human 20S t-proteasome as compared with 20S s- and i-proteasomes, we have adopted an approach that integrates an extensive set of in vitro degradation kinetics with computational modeling (Fig. 1, A and B). We have first purified 20S proteasomes from T2, LCL, and C5.5 cells and used these proteasomes to perform in vitro degradation kinetics of the short fluorogenic peptides Suc-LLVY-MCA, and Z-LLE-MCA, two substrates specific for the chymotrypsin-like and caspase-like activity of proteasomes. The MCA group of these substrates is released upon endopeptidase cleavage by proteasomes, and its fluorescence can be measured quantitatively. Murata et al. (10) showed that the degradation rate of Suc-LLVY-MCA, which is mainly carried out by the β5/β5i/β5t subunits, diverges between mouse i- and t-proteasomes. In contrast, the cleavage rate of Z-LLE-MCA, which is mainly carried out by the β1/β1i subunits, should not be different between human i- and t-proteasomes because they carry similar amounts of the β1i subunit (Fig. 2C). Furthermore, although these short fluorogenic substrates do not recapitulate the full substrate specificity of proteasomes (22), they have been successfully used to discriminate between 20S s- and i-proteasome dynamics by the development of a computational model and its calibration with time-course data (11) (Fig. 1). We have here applied the same computational modeling and model calibration approach on Z-LLE-MCA and Suc-LLVY-MCA digestion kinetics of 20S s-, i-, and t-proteasomes. Using Bayesian inference techniques, we have obtained model fits to the experimental data by estimating the model parameters, resulting in a posterior parameter distribution for each 20S proteasome isoform (Figs. S1 and S2). The posterior parameter distribution contains information on possible kinetic parameter values able to explain the experimental data sets and their relationship with each other. The kinetic differences between 20S s- and i-proteasomes confirm our previous results (11) and therefore the correct setup of our study (Fig. 3, A and B).
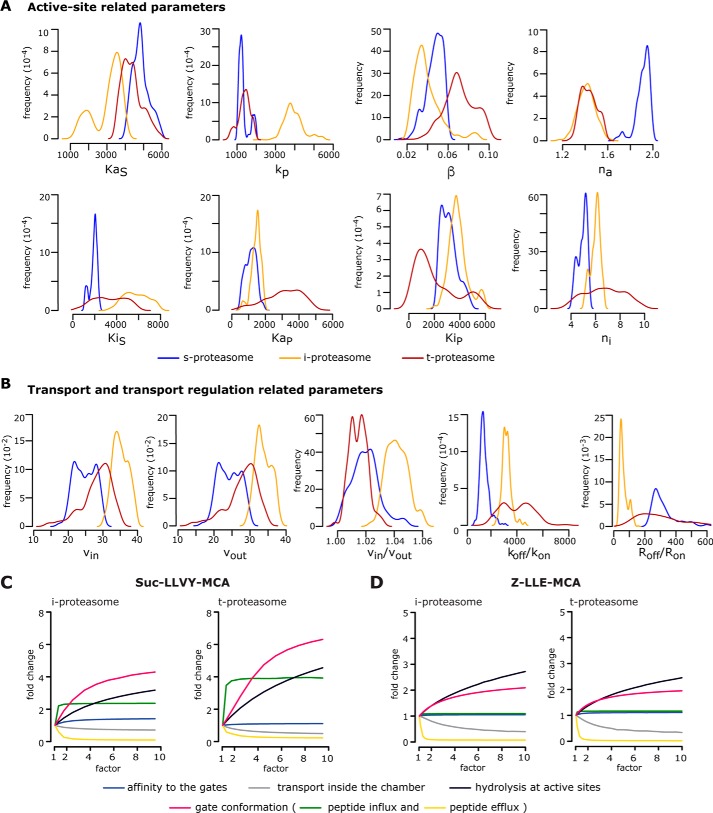
Figure 3.
Computational model parameters and rate-limiting steps of proteasome isoform dynamics. A and B, marginal posterior parameter distributions obtained by calibrating the proteasome kinetic model against experimental data (n = 3–5) derived from the degradation of the substrate Suc-LLVY-MCA by 20S s-, i-, and t-proteasomes. Parameters are grouped into active site–related parameters (A) and transport- and transport regulation–related parameters (B). Briefly, KaS and KaP are the dissociation constant of substrate (S) and product (P) to active site(s); kp is the peptide-bond hydrolysis rate at active site(s); β is the factor by which kp is multiplied upon binding to inhibitory site(s); na and ni are the Hill coefficients for binding to the active site(s) and the inhibitor site(s); KiS and KiP are the dissociation constants of substrate (S) and product (P) to inhibitor site(s); vin and vout are the peptide influx and efflux rates, and vin/vout is their ratio; koff/kon is the ratio between the dissociation and the association rates to the gate; and Roff/Ron is the ratio between the unbinding and binding rates to the enhancing regulator site(s). The meaning of all parameters is depicted in Fig. 1 and Table 1. C and D, analysis of rate-limiting steps in 20S i- and t-proteasomes. Depicted is the -fold change of product formation (y axis) upon increase (by a factor of; x axis) of a specific reaction step for the degradation of Suc-LLVY-MCA (C) and Z-LLE-MCA (D) as simulated by our computational model of the 20S proteasome dynamics. The initial substrate concentration for this analysis is 160 μm, and the -fold change is determined after 60-min reaction relative to the experimentally measured proteasome kinetics (factor = 1). The mean of 1000 in silico predictions (colored lines) is plotted over time for the degradation of the substrates with the same initial substrate concentrations as in the experiments. The rate-limiting steps are those for which the increase leads to the largest -fold change.
Regarding the comparison of the two proteasome isoforms mainly involved in the positive/negative selection, no difference emerges between 20S t- and i-proteasomes when the estimated parameter distributions obtained from Z-LLE-MCA digestions are compared (Fig. S3). On the contrary, in the Suc-LLVY-MCA degradation kinetics, we have found differences related to the active-site parameters between these two 20S proteasome isoforms (Fig. 3A). Most apparent is the hydrolysis strength, kp (for explanations, see Table 1), which appears to be ∼4-fold smaller in 20S t-proteasome compared with 20S i-proteasome and therefore recapitulates the kp observed in s-proteasome. Also, the dissociation constant of the peptide to the substrate-binding site (KaS), which is slightly increased in 20S t-proteasome compared with 20S i-proteasome, results again in a similar KaS as observed in 20S s-proteasome (Fig. 3A). In addition, the peptide transport and transport regulation parameters differ, indicating that the subunit exchange has not only local but also global effects on 20S proteasome dynamics (Fig. 3B).
Table 1.
List of mathematical model parameters
The table and the meaning of the parameters have been previously published by Liepe et al. (11), and they correspond to the parameters indicated in Fig. 1A.
| Peptide-bond hydrolysis | |
| kp | Peptide-bond hydrolysis rate at active site(s) |
| KaS, KaP | Dissociation constant of substrate (S) and product (P) to active site(s) |
| na | Hill coefficient for binding to active site(s) |
| KiS and KiP | Dissociation constant of substrate (S) and product (P) to inhibitor site(s) |
| ni | Hill coefficient for binding to inhibitor site(s) |
| α | Factor, by which KaS, KaP, KiS, and KiP are multiplied |
| β | Factor, by which kp is multiplied upon binding to inhibitory site(s) |
| Transport | |
| kon | Association rate to the gate |
| koff | Dissociation rate to from gate |
| vin | Peptide influx rate |
| τ | Peptide translocation rate inside the chamber |
| vout | Peptide efflux rate |
| C | Capacity (maximum number of molecules inside the chamber) |
| Transport regulation | |
| Ron | Binding rate to the enhancing regulator site(s) |
| Roff | Unbinding rate to the enhancing regulator site(s) |
| Xenh | Strength of enhancing regulator site(s) |
| Ion | Binding rate to the inhibiting regulator site(s) outside the chamber |
| Ioff | Unbinding rate to the inhibiting regulator site(s) outside the chamber |
| H | Coefficient for binding to inhibiting regulator site(s) outside the chamber |
| Yin | Strength of inhibiting regulator site(s) |
Peptide transport dominates the substrate degradation in human t-proteasome
By studying inferred posterior parameter distribution, we can determine the rate-limiting steps of the reaction. Previously, we showed that for both 20S s- and i-proteasomes, the gate conformation, which determines the peptide transport (influx and efflux), is often the rate-limiting step (11). By in silico simulations, we have now found that although several kinetic parameters significantly differ between 20S i- and t-proteasome, for the Suc-LLVY-MCA substrate the rate-limiting step of 20S t- and i-proteasomes is the peptide transport (Fig. 3C), whereas for the Z-LLE-MCA substrate the rate-limiting steps are primarily the peptide-bond hydrolysis at the active site and secondarily the peptide transport (Fig. 3D).
Peptide transport also regulates how much substrate and product are located inside the proteasome chamber over time (11). The local substrate concentration around the active site Thr1 inside the proteasome chamber then strongly influences peptide hydrolysis. For a fast substrate turnover, the peptide flux through the chamber should be large enough to allow sufficient supply of new substrate molecules and sufficient efflux of product molecules. Furthermore, the peptide flux should be such that the substrate concentration around the active site is high enough to obtain reaction velocities at a level of vmax but low enough to avoid substrate inhibition (11).
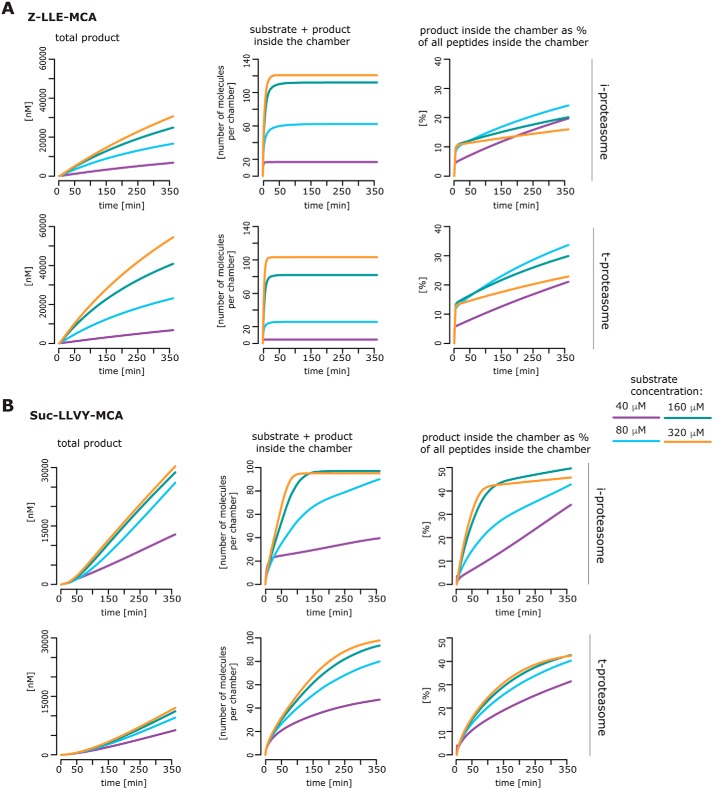
Taking this into account, differences in peptide transport between proteasome isoforms and thus in the filling dynamics of the proteasome should strongly influence the observed substrate degradation rates. To investigate this aspect, we have used the estimated posterior parameter distributions for 20S i- and t-proteasomes to calculate in silico the amount of substrate and products inside the proteasome chamber over time. In the case of Z-LLE-MCA, our simulations show only minor differences in the filling kinetics between 20S i- and t-proteasomes (Fig. 4A). In contrast, in the case of Suc-LLVY-MCA, our simulations suggest that the chamber of 20S t-proteasome is filled more slowly with substrate and product molecules than the 20S i-proteasome chamber (Fig. 4B). For both 20S proteasome isoforms, our simulations suggest that, in our experimental conditions, after 6 h of reaction an equal proportion of substrate and product molecules is present inside the chamber. However, this equal proportion is reached faster by 20S i-proteasome than 20S t-proteasome, indicating a stronger peptide-bond hydrolysis activity by 20S i-proteasome, which could be reflected by its higher kp value (Fig. 3A). On the contrary, the peptide-bond hydrolysis of the Z-LLE-MCA substrate does not differ between these two isoforms, and it is much lower compared with that of the Suc-LLVY-MCA substrate. In summary, these analyses highlight that the overall substrate degradation differences between human 20S i- and t-proteasome cannot be explained only by differences in the active-site subunits, but they can result from dynamical differences that regulate the peptide transport efficiencies.
Figure 4.
Simulation of the substrate and product dynamics inside the chambers of the human s-, i-, and t-proteasomes. The mean of 1000 in silico predictions (colored lines) is plotted over time for the degradation of the substrate Z-LLE-MCA (A) or Suc-LLVY-MCA (B) with the same initial substrate concentrations as used in the independent experiments (n = 3–5; see “Material and methods”). The simulation has been performed for 20S i- and t-proteasomes. The number of peptide molecules (product and substrate) and the relative amount of products versus total amount of peptides inside the chambers over time have been computed by the computational model and are based on the estimated posterior parameter distributions for each substrate and proteasome isoform.
20S proteasome isoforms quantitatively differ in substrate cleavage-site strength and the generation of self-epitopes
To introduce a further degree of complexity in our experimental approach, we have analyzed, by mass spectrometry (MS), digests obtained after incubation of 20S proteasomes with eight synthetic polypeptides, three derived from the melanoma-associated antigen gp100PMEL17 and five from myelin sheath proteins, which are the main autoantigens attacked by CD8+ T cells in multiple sclerosis (Table S1). We have selected these substrates because of their immunogenicity toward CD8+ T cells associated with either tumor recognition or autoimmune response against oligodendrocytes in multiple sclerosis. In other words, epitopes derived from these antigenic sequences are known to be detected at the cell surface by autoreactive CD8+ T cells. These lymphocytes specifically detect tumor-associated antigens, e.g. gp100, and multiple sclerosis–associated antigens, e.g. myelin oligodendrocyte glycoprotein (MOG) and myelin basic protein (MBP), which can be expressed in medullary thymic antigen-presenting cells. Nonetheless, they survived negative selection in the thymus and are present in the periphery as shown in various studies (33–37).
By MS analysis of the peptide products generated by 20S s-, i-, and t-proteasomes, we have identified 510 nonspliced and 67 spliced peptide products. All spliced and nonspliced peptide products are quantifiable in the digestions. All 20S proteasome isoforms cleave the substrates between the same residues.
The fact that any of the three 20S proteasome isoforms do not use even one substrate cleavage site that is not used by the other two proteasome isoforms does not exclude that substrate cleavage sites can be preferentially used by one of the 20S proteasome isoforms. We have verified this hypothesis through investigation of proteasome-mediated digestion kinetics of four substrates (i.e. gp100201–230, gp10035–57, MOG172–202, and MBP102–129). We have adopted a quantitative strategy in our MS analysis, using the quantification with minimal effort (QME) methodology. QME estimates the absolute content of spliced and nonspliced peptide products based on their MS peak area, measured in the digestion probe (38). QME can also estimate the frequency of use of each substrate cleavage site by proteasomes, i.e. in the substrate cleavage-site strength (SCS).
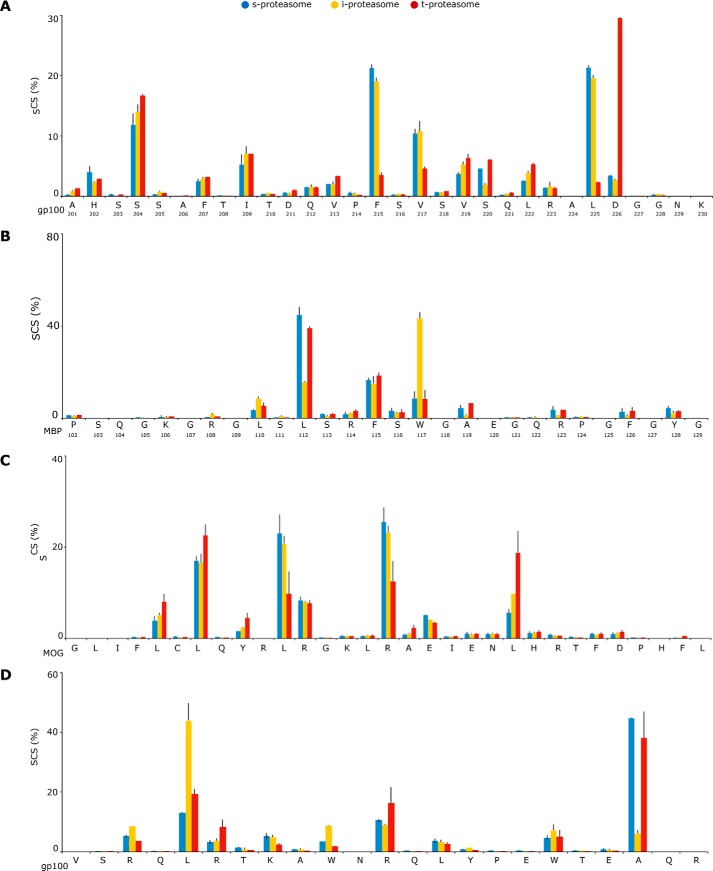
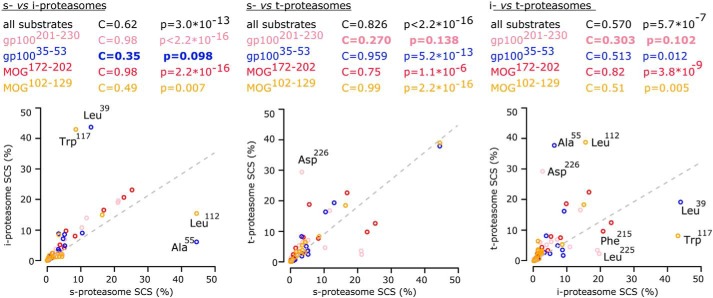
According to our hypothesis, we have observed quantitative differences in the substrate cleavage site predominantly used between the three 20S proteasome isoforms (Fig. 5). There are no amino acids that are clearly preferred by one of the 20S proteasome isoforms rather than the others for peptide-bond hydrolysis, likely because the peptide sequence motifs (8–10 residues) surrounding the cleavage site influence the frequency of usage of that cleavage site. For instance, the gp100 Leu225 residue is seldom used by 20S t-proteasome (Fig. 5A), although the MBP Leu112 (Fig. 5B) and the MOG Leu193 residues are scarcely used by 20S i-proteasome (Fig. 5C), which conversely prefers the gp100 Leu39 residue (Fig. 5D). For a more systematic comparison of the SCSs of the 20S proteasome isoforms, we performed pairwise correlations between their SCSs. We have found significant pairwise correlations between SCSs of all three 20S proteasome isoforms, thereby indicating that their overall catalytic activity is comparable (Fig. 6). However, when we analyzed the SCS of specific substrates, differences emerge. These differences are due to specific substrate cleavage sites, which are used by all three 20S proteasome isoforms although with divergent frequencies (e.g. gp100 Phe215, Leu225, Asp226, Leu39, and Ala55).
Figure 5.
Different proteasome isoforms preferentially use different substrate cleavage sites. A–D, the relative frequency of the substrate cleavage-site usage (i.e. the SCS of the synthetic substrates gp100201–230 (A), MBP102–129 (B), MOG172–202 (C), and gp10035–53 (D) by 20S s-, i-, and t-proteasome are shown as the mean of two to four independent experiments (error bars are the S.D.) measured two to three times. Quantitative analyses are done by applying QME to the MS measurements.
Figure 6.
Proteasome isoforms overall share a common substrate cleavage-site usage with specific differences. The correlation between the SCSs of the synthetic substrates gp100201–230 (pink circles), gp10035–53 (blue circles), MBP102–129 (green circles), and MOG172–202 (black circles) generated in in vitro digestion kinetics by human 20S s-, i-, or t-proteasomes is shown. The SCSs are compared in proteasomes pairwise. The computed Pearson's correlation coefficients and the tests for significance of these correlations are reported. Nonsignificant correlations are marked in bold and depict SCSs that differ between the pair of compared 20S proteasome types. The substrate cleavage-sites which the use of is more remarkably varied between 20S proteasome isoforms are labeled.
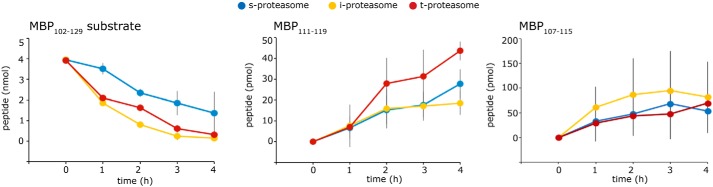
These quantitative differences in SCSs are also reflected in the generation of specific peptide products, including some that have already been shown to be epitopes recognized by CD8+ T cells. For instance, the epitope MBP111–119, which is an HLA-A*02:01–binding epitope recognized by multiple sclerosis and healthy donor patients (39–43), is better generated by 20S t-proteasome than 20S i-proteasome. However, this phenomenon is epitope-specific because the generation of the epitope MBP107–115 (41) is not favored when carried out by human 20S t-proteasome as compared with 20S i-proteasome (Fig. 7).
Figure 7.
Proteasome isoforms generate MBP epitopes with different kinetics. The degradation kinetics of the synthetic substrates MBP102–129 by 20S s-, i-, and t-proteasomes and the generation kinetics of the epitopes MBP111–119 and MBP107–115 are shown. The mean of two to four independent experiments (error bars are the S.D.) measured in duplicate is shown. Quantitative analyses are done by applying QME to the MS measurements.
Discussion
Diverging from what was shown for mouse 20S t-proteasome (30), human 20S t-proteasome does not seem to possess a unique proteolytic activity in processing the self-antigen substrates included in this study and in our experimental conditions. We show here, however, that human 20S t- and i-proteasomes differ in their catalytic activity, peptide transport, and transport regulation. The differences in the peptide transport are particularly relevant because the latter is often the rate-limiting step, at least in the degradation of short peptides, as also shown here for human 20S t-proteasome. Differences in the peptide transport imply alterations of the overall 20S proteasome dynamics and long-range effects over the entire proteasome chamber due to the incorporation of the β5t subunit. As a consequence, the quantity of peptides produced by the β1i and β2i subunits (present in both t- and i-proteasomes) can differ in the case that they are incorporated into either 20S t- or i-proteasome because the peptide transport, and therefore the concentration and dynamics of peptides in these proteolytic pockets, could be altered too.
These dissimilarities in catalytic activity, peptide transport, and transport regulation can explain the variable preferences of human proteasome isoforms for specific substrate-cleavage sites. The fact that we have not observed cleavage sites only used by either 20S t- or i-proteasome could not be explained by the presence of standard catalytic subunits in our 20S t- and i-proteasome preparations. Indeed, if a substrate cleavage site is used only by 20S i-proteasome, for instance, we shall not detect it in the 20S t-proteasome digestions and vice versa. Therefore, although in our experimental setup we cannot exclude the existence of substrate cleavage sites exclusively used by human 20S s-proteasome, we can exclude substrate cleavage sites exclusively used by either 20S i- or t-proteasome in the sequences analyzed in this study, as previously demonstrated already for human and mouse 20S i-proteasomes (22). This does not exclude the possibility that improving MS sensitivity could show a qualitative difference in the spliced and nonspliced products between 20S proteasome isoforms, and this also does not provide direct information about differences between 20S proteasome isoforms coupled to 19S and other regulatory complexes.
We have observed, however, strong variation from proteasome isoform to proteasome isoform in the quantity of peptides produced, including MHC-I–binding epitopes, as shown here for the epitope MBP111–119. These quantitative differences in peptide generation can lead to a negligible presentation of epitopes at the cell surface as demonstrated in previous studies where mouse 20S s- and i-proteasomes have been compared (22, 24, 25). Our observations, therefore, are compatible with the hypothesis that cells expressing human thymoproteasome (both 20S proteasomes and 20S proteasomes coupled to regulatory complexes) could present on the cell surface some specific “private” epitopes, which could be directly involved in the positive selection of thymocytes (5). These private epitopes were not presented by cells expressing i-proteasome, although they were produced in the intracellular space, because of the progressive reduction of the peptide amounts during the steps of the antigen presentation pathway (44). Different proteolytic dynamics between human i- and t-proteasomes could also result in significant variation in the turnover of specific antigens and therefore in the antigenic landscape of cells expressing either t- or i-proteasome, which would further favor the “private epitopes” hypothesis.
Our study also shows that 20S t-proteasomes can generate spliced peptides, which are thought to represent a large portion of the MHC-I immunopeptidome, the peptides bound to MHC-I molecules (45–47). The generation of spliced epitopes by t- and i-proteasomes in the thymus could strongly impinge upon our models of central tolerance and discrimination between self and nonself by our immune system (48).
Materials and methods
Cell lines
To generate a cell line stably overexpressing the proteasome β5t subunit (protein, A5LHX3; gene, PSMB11), the thymic cDNA was transcribed from human thymic total RNA (Clontech) using a Transcriptor First Strand cDNA Synthesis kit (Roche Applied Science) according to the manufacturer's instructions. For amplification of the human β5t sequence (PSMB11) by PCR, the following primers were used: Fw, 5′-gggatggctctgcaggatgtgtgc, and Rev, 5′-ctcacaccgtctcagtccctgc. The PCR product was first inserted into pcDNA3.1(+) (Thermo Fisher Scientific) and then recloned into a pSG5 vector (Stratagene) via EcoRI/BamHI. T2 cells were cultured in RPMI 1640 medium supplemented with fetal bovine serum to a final concentration of 10% and 2 mm l-glutamine in a 5% CO2 atmosphere. Cells were transfected with pSG5/PSMB11, pSG5/PSMB9 (lab stock), and pSVneo in equal amounts using Amaxa Cell line Nucleofector® Kit C (Lonza). Stable transfected cells were selected with 1 mg/ml G418, and the expression was controlled by PCR and Western blotting. The positive cells were isolated and cultured again. The clone C5.5, which was transfected with β5t (PSMB11) and β1i (PSMB9) subunits, was selected for the study.
To verify the endogenous expression of the β5t subunit, the following cell lines were grown in basal Iscove's medium supplemented with 10% fetal calf serum, 2 mm glutamine, and 100 units/ml penicillin, 0.1 mg/ml streptomycin in a 5% CO2 atmosphere: (i) T2 and C5.5 cell lines; (ii) LCLs; (iii) HeLa cells and clones 33.2 and 21.2, the latter of which are stably transfected with HLA-A*02:01 and β1i+β2i+β5i subunits or β1i+β2i subunits, respectively (49); (iv) DU145 prostate carcinoma purchased from ATCC (ATCC® HTB-81TM); (v) RKO human colon carcinoma (ATCC CRL-2577); (vi) MeWo malignant melanoma (ATCC HTB-65TM); (vii) human umbilical vein endothelial cells; (viii) THP1 acute monocytic leukemia (ATCC TIB-202TM); (ix) Mel15 malignant melanoma (50); (x) SaOs human bone osteosarcoma (ATCC HTB-85); and (xi) A549 adenocarcinomic human alveolar basal epithelial cells (ATCC CCL-185TM).
Western blotting and β5t subunit identification and quantification
Proteasome subunits have been revealed by Western blot assays as described previously (22) using anti-α4 (1:10,000; in-house), anti-β1 (1:1000; Enzo Life Science), anti-β2 (1:1000; Enzo Life Science), anti-β5 (1:1000; Abcam), anti-β1i (1:20,000; in-house), anti-β2i (1:3000; Thermo Fisher), anti-β5i (1:1000; Santa Cruz Biotechnology), and anti-FLAG (1:1000; Sigma-Aldrich). For the identification of β5t subunit in different cell lines (Fig. 2B), bands have been excised from the SDS-PAGE gel, washed with 200 mm ammonium bicarbonate in acetonitrile/water (50:50, v/v) and 50 mm ammonium bicarbonate in acetonitrile/water (5:95, v/v) (digestion buffer), shrunk by dehydration in acetonitrile, and dried. For reduction of cysteine residues, 50 μl of 10 mm DTT in 100 mm ammonium bicarbonate have been added, and the samples have been incubated for 45 min at 56 °C. After three washes with water, 50 μl of 55 mm iodoacetamide in 50 mm ammonium bicarbonate have been added. Incubation has been performed at room temperature in the dark for 20 min. The gel bands have been washed with water and digestion buffer and dried in acetonitrile after shrinking. 25 μl of 0.1 g/liter trypsin (Promega) in digestion buffer has been added and incubated at 37 °C overnight. Trypsin-digested peptides have been extracted by one change of 25 μl of 0.5% TFA in acetonitrile/water (70:30, v/v) and 25 μl of acetonitrile (10 min for each) at room temperature, combined, and dried down.
LC-MS/MS analyses of peptides have been performed as follows. The sample has been concentrated for 4 min on a trap column (PepMap C18, 5 mm × 300 μm × 5 μm, 100 Å, Thermo Fisher Scientific) with 2:98 (v/v) acetonitrile/water containing 0.1% (v/v) TFA at a flow rate of 30 μl/min and then analyzed by nanoscale LC-MS/MS measurements using a Q Exactive Plus mass spectrometer coupled with an Ultimate 3000 RSLCnano (Thermo Fisher Scientific). The system comprised a 75-μm inner diameter × 250 mm nano-LC column (Acclaim PepMap C18, 2 μm, 100 Å; Thermo Fisher Scientific). Mobile phase A was 0.1% (v/v) formic acid in water, and mobile phase B was 80:20 (v/v) acetonitrile/water containing 0.1% (v/v) formic acid. The elution has been carried out using a gradient of 3–43% mobile phase B in 80 min with a flow rate of 300 nl/min. Full MS spectra (m/z 350–1,600) have been acquired at a resolution of 70,000 (full width at half-maximum) followed by a data-dependent MS/MS fragmentation of the top10 precursor ions (resolution, 17,500; 1+ charge state excluded; isolation window of 1.6 m/z; normalized collision energy of 27%; dissociation method, higher-energy collisional dissociation). The maximum ion injection time for MS scans has been set to 50 ms, and that for MS/MS scans was set to 120 ms. Background ions at m/z 391.2843 and 445.1200 act as lock mass. Protein identifications have been performed with Mascot software version 2.6.1. (Matrix Science Ltd., London, UK). Data have been searched against Swiss-Prot (July 2017; taxonomy, human; 20,215 sequences) and a contaminant database (247 sequences). The following parameters have been set: enzyme, trypsin/P with one missed cleavage; static modification, carbamidomethylation (Cys); variable modifications, protein N-acetylation and oxidation (Met); mass tolerances for MS and MS/MS, 5 ppm and 0.02 Da. Proteins have been accepted as identified if at least two unique peptides provided a Mascot MS/MS score for identity (p < 0.01).
For relative protein quantification of β5 and β5t subunits, two heavy AQUA peptides (Table S2) have been synthesized based on Fmoc solid-phase chemistry, i.e. the heavy analogue peptide 226DAYSGGAVNLYHVR239 (with [M + H]+exp = 1528.80) of human β5 subunit and the heavy analogue peptide 216DAYSGGSVDLFHVR229 (with [M + H]+exp = 1529.79) of the human β5t subunit. During peptide synthesis, the heavy isotope–labeled amino acid [U-13C6,15N]leucine-N-Fmoc from Euriso-Top GmbH (Saarbrücken, Germany) has been incorporated. Estimation of exact peptide amount has been performed by amino acid analysis (Gennaxon Bioscience, Ulm Germany).
β5/β5t subunit quantification of purified C5.5 20S proteasome reported in Fig. 2D has been performed after LC separation of tryptic peptides on a MALDI-TOF/TOF mass spectrometer. In the assay (n = 2), 5 μg of proteasome has been reduced, alkylated, and digested with trypsin as described elsewhere (51). Aliquots of the sample spiked with 500 fmol of each AQUA peptide have been analyzed by LC-MS/MS on a 4700 Proteomics Analyzer (AB SCIEX, Framingham, MS) off-line coupled with a Dionex UltiMate 3000 RSLC system and Probot fractionation device (Thermo Scientific, Idstein, Germany) as described previously (52). The calculation of the absolute amount of the tryptic peptides has been performed by comparison of the MS peak areas with those of the corresponding AQUA peptides. Based on the absolute amounts, the relative ratio β5:β5t has been determined as follows: β5t/(β5 + β5t).
RT-PCR and validation
Total RNA has been isolated from human cell lines using a High Pure RNA Isolation kit (Roche Applied Science) in the presence of DNase according to the manufacturer's protocol. The cDNA has been obtained from 1 μg of total RNA using a Primer Script RT Reagent kit (TaKaRa Bio Inc.). The RT-PCR has been performed with 1 μl of cDNAs for 25 cycles at an annealing temperature of 56 °C. For the RT-PCR specific for the huPSMB11 gene, the primers huPSMB11 Fw (5′-gggatggctctgcaggatgtgtgc) and huPSMB11 Rev (5′-ctcacaccgtctcagtccctgc) have been used, thereby generating a transcript of 907 bp. To control the efficiency of RT-PCR, actin has been amplified from the cDNA by RT-PCR using the following actin-specific primers: actin Fw, 5′-ctcaccatggatgatatcg; and actin Rev, 5′-tcgtcatactcctgcttgctg. For the verification of the β5t-subunit's RT-PCR products, the PCR products have been eluted from the agarose gel and inserted into pCR2.1 Topo® (Thermo Fisher) and sequenced by using the T7 primer by LCG Genomics GmbH (LGC Group). The latter step has confirmed the specificity of the amplified sequence marked in Fig. 2A.
Computational analysis of the proteasome dynamics
20S proteasome degradation dynamics have been assessed using an integrative modeling approach with Bayesian model calibration to the in vitro degradation of the short fluorogenic peptides Suc-LLVY-MCA and Z-LLE-MCA. In vitro degradation kinetics (n = 3–5) have been performed with different substrate concentration (0–480 μm) in 100 μl of TEAD buffer (20 mm Tris, 1 mm EDTA, 1 mm NaN3, 1 mm DTT, pH 7.2) at 37 °C as described previously (22). For the analysis, we have used the computational model of 20S proteasome activity published previously (11). In essence, by applying an approximate Bayesian computation to fit the model to the experimental data, we have obtained posterior parameter distributions for each proteasome isoform. The comparison of the marginal posterior parameter distributions has allowed us to detect differences in the kinetic parameters between the different isoforms. For the Bayesian inference, we have used the package ABC-SysBio (12) implemented in Python with GPU support (53). The prior distributions for all parameters were uniform as described previously (11). Furthermore, all other algorithm parameters have been kept as in Liepe et al. (11). The posterior analysis has been carried out in R (54). Computation of rate-limiting steps has been performed as described in Liepe et al. (11).
20S proteasome purification
20S proteasomes have been purified from T2, C5.5, and LCLs as described previously (55). Proteasome concentration has been measured by Bradford staining and verified by Coomassie staining in an SDS-PAGE gel as shown elsewhere (56). The purity of the standardized proteasome preparations has been shown previously (22). LCL and C5.5 cell lines mainly express human i-proteasome (22, 57) or t-proteasomes, respectively (Fig. 2). LCL proteasome has often been used as an example of i-proteasome in several previous studies (22, 38, 39, 50, 57–61). T2 cell line expresses only s-proteasome (Fig. 2). The three proteasomes have been purified in parallel to minimize artifacts due to the purification or storage conditions.
Peptides and in vitro digestion of synthetic polypeptides
The sequence enumeration for the polypeptide substrates is reported in Table S1. All peptides have been synthesized using Fmoc solid-phase chemistry. Synthetic polypeptides (20–40 μm) have been digested by 1–3 μg of 20S proteasomes in 100 μl of TEAD buffer over time at 37 °C as described previously (22).
Quantitative analysis of peptide products by QME and MS
Liquid chromatography–MS analyses of polypeptide digestion products have been performed as described previously (52) with the ESI-ion trap instrument DECA XP MAX (Thermo Fisher Scientific). Database searching has been performed using SpliceMet ProteaJ (52). Quantification of peptides produced in the experiments has been carried out by applying the QME method to the LC-MS analyses. QME estimates the absolute content of spliced and nonspliced peptide products based on their MS peak area measured in the digestion probe. QME is an optimization tool that makes use of the law of mass conservation and MS instrument features. The QME algorithm parameters were empirically computed in our previous study (38) and have been applied here. By applying QME, we have also calculated the SCS, which describes the relative frequencies of proteasome cleavage after any given residue of the synthetic polypeptide substrate. SCS values shown in this study are the average of SCS measured in kinetic assays.
Statistical analysis
The SCS correlation has been done using Pearson's correlation coefficient. Significance tests have been performed, testing for association between paired samples based on Pearson's correlation coefficient. All data analysis has been implemented in R (54). Sample size and number of replicates are disclosed in the figure and table captions.
Data and software availability
Data sets are available at the online archive Mendeley (http://dx.doi.org/10.17632/zjhhc2bm9p.1).5 Data sets linked to the computational model of proteasome dynamics and published by Liepe et al. (11) are available at http://datadryad.org/resource/doi:10.5061/dryad.nk453.5 The statistical background behind the model inference presented in this study is described in Liepe et al. (12, 13), and its implementation can be found at ABC-SysBio (https://sourceforge.net/projects/abc-sysbio/).5
Author contributions
U. K., M. M., and J. L. conceptualization; U. K., M. M., and J. L. data curation; U. K., K. J., M. P. S., M. M., and J. L. supervision; U. K., S. S., K. T.-T., A. N., M. M., and J. L. investigation; U. K., C. K., K. J., M. P. S., M. M., and J. L. methodology; U. K., M. P. S., M. M., and J. L. writing-review and editing; P. H. and J. L. resources; M. P. S., M. M., and J. L. funding acquisition; M. M. and J. L. formal analysis; M. M. and J. L. validation; M. M. and J. L. visualization; M. M. and J. L. writing-original draft; M. M. project administration; J. L. software.
Supplementary Material
Acknowledgments
We thank C. Keller, P. Kunert, and B. Brecht-Jachan for technical assistance (Charité Berlin) and W. Scott and A. Abbas (King's College London) for proofreading the text. We thank the Shared Facility Mass Spectrometry of the Charité for support in acquisition of the data and A. Goede for the use of QME software.
Note added in proof
Correlations and p values were missing from Fig. 6 in the version of this article that was published as a Paper in Press on March 26, 2019. This has now been corrected and does not affect the results or conclusions of this work.
This work was supported in part by Cancer Research UK King's Health Partners Centre at King's College London (Development Fund 2018) (to M. M.), Berlin Institute of Health Grant CRG1-TP1 (to P. M. Kloetzel). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S3 and Tables S1 and S2.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party-hosted site.
- MHC-I
- major histocompatibility complex class I
- s-proteasome
- standard proteasome
- i-proteasome
- immunoproteasome
- t-proteasome
- thymoproteasome
- AQUA
- absolute quantification
- LCL
- lymphoblastoid cell line
- Suc
- succinyl
- MCA
- 4-methylcoumaryl-7-amide
- Z
- benzyloxycarbonyl
- MOG
- myelin oligodendrocyte glycoprotein
- MBP
- myelin basic protein
- QME
- quantification with minimal effort
- SCS
- substrate cleavage-site strength
- Fw
- forward
- Rev
- reverse
- Fmoc
- N-(9-fluorenyl)methoxycarbonyl.
References
- 1. Pearson H., Daouda T., Granados D. P., Durette C., Bonneil E., Courcelles M., Rodenbrock A., Laverdure J. P., Côté C., Mader S., Lemieux S., Thibault P., and Perreault C. (2016) MHC class I-associated peptides derive from selective regions of the human genome. J. Clin. Investig. 126, 4690–4701 10.1172/JCI88590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gubin M. M., Zhang X., Schuster H., Caron E., Ward J. P., Noguchi T., Ivanova Y., Hundal J., Arthur C. D., Krebber W. J., Mulder G. E., Toebes M., Vesely M. D., Lam S. S., Korman A. J., et al. (2014) Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature 515, 577–581 10.1038/nature13988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jaravine V., Raffegerst S., Schendel D. J., and Frishman D. (2017) Assessment of cancer and virus antigens for cross-reactivity in human tissues. Bioinformatics 33, 104–111 10.1093/bioinformatics/btw567 [DOI] [PubMed] [Google Scholar]
- 4. Mishto M., and Liepe J. (2017) Post-translational peptide splicing and T cell responses. Trends Immunol. 38, 904–915 10.1016/j.it.2017.07.011 [DOI] [PubMed] [Google Scholar]
- 5. Klein L., Kyewski B., Allen P. M., and Hogquist K. A. (2014) Positive and negative selection of the T cell repertoire: what thymocytes see (and don't see). Nat. Rev. Immunol. 14, 377–391 10.1038/nri3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Weissman A. M., Shabek N., and Ciechanover A. (2011) The predator becomes the prey: regulating the ubiquitin system by ubiquitylation and degradation. Nat. Rev. Mol. Cell Biol. 12, 605–620 10.1038/nrm3173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Solomon H., Bräuning B., Fainer I., Ben-Nissan G., Rabani S., Goldfinger N., Moscovitz O., Shakked Z., Rotter V., and Sharon M. (2017) Post-translational regulation of p53 function through 20S proteasome-mediated cleavage. Cell Death Differ. 24, 2187–2198 10.1038/cdd.2017.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Olshina M. A., Ben-Nissan G., and Sharon M. (2018) Functional regulation of proteins by 20S proteasome proteolytic processing. Cell Cycle 17, 393–394 10.1080/15384101.2017.1414682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schrader J., Henneberg F., Mata R. A., Tittmann K., Schneider T. R., Stark H., Bourenkov G., and Chari A. (2016) The inhibition mechanism of human 20S proteasomes enables next-generation inhibitor design. Science 353, 594–598 10.1126/science.aaf8993 [DOI] [PubMed] [Google Scholar]
- 10. Murata S., Sasaki K., Kishimoto T., Niwa S., Hayashi H., Takahama Y., and Tanaka K. (2007) Regulation of CD8+ T cell development by thymus-specific proteasomes. Science 316, 1349–1353 10.1126/science.1141915 [DOI] [PubMed] [Google Scholar]
- 11. Liepe J., Holzhütter H. G., Bellavista E., Kloetzel P. M., Stumpf M. P., and Mishto M. (2015) Quantitative time-resolved analysis reveals intricate, differential regulation of standard- and immuno-proteasomes. Elife 4, e07545 10.7554/eLife.07545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liepe J., Kirk P., Filippi S., Toni T., Barnes C. P., and Stumpf M. P. (2014) A framework for parameter estimation and model selection from experimental data in systems biology using approximate Bayesian computation. Nat. Protoc. 9, 439–456 10.1038/nprot.2014.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liepe J., Barnes C., Cule E., Erguler K., Kirk P., Toni T., and Stumpf M. P. (2010) ABC-SysBio—approximate Bayesian computation in Python with GPU support. Bioinformatics 26, 1797–1799 10.1093/bioinformatics/btq278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vyshemirsky V., and Girolami M. A. (2008) Bayesian ranking of biochemical system models. Bioinformatics 24, 833–839 10.1093/bioinformatics/btm607 [DOI] [PubMed] [Google Scholar]
- 15. Toni T., and Stumpf M. P. (2010) Simulation-based model selection for dynamical systems in systems and population biology. Bioinformatics 26, 104–110 10.1093/bioinformatics/btp619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Toni T., Welch D., Strelkowa N., Ipsen A., and Stumpf M. P. (2009) Approximate Bayesian computation scheme for parameter inference and model selection in dynamical systems. J. R. Soc. Interface 6, 187–202 10.1098/rsif.2008.0172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Putter H., Heisterkamp S. H., Lange J. M., and de Wolf F. (2002) A Bayesian approach to parameter estimation in HIV dynamical models. Stat. Med. 21, 2199–2214 10.1002/sim.1211 [DOI] [PubMed] [Google Scholar]
- 18. Gutenkunst R. N., Waterfall J. J., Casey F. P., Brown K. S., Myers C. R., and Sethna J. P. (2007) Universally sloppy parameter sensitivities in systems biology models. PLoS Comput. Biol. 3, 1871–1878 10.1371/journal.pcbi.0030189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Erguler K., and Stumpf M. P. (2011) Practical limits for reverse engineering of dynamical systems: a statistical analysis of sensitivity and parameter inferability in systems biology models. Mol. Biosyst. 7, 1593–1602 10.1039/c0mb00107d [DOI] [PubMed] [Google Scholar]
- 20. Liepe J., Filippi S., Komorowski M., and Stumpf M. P. (2013) Maximizing the information content of experiments in systems biology. PLoS Comput. Biol. 9, e1002888 10.1371/journal.pcbi.1002888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nurse P., and Hayles J. (2011) The cell in an era of systems biology. Cell 144, 850–854 10.1016/j.cell.2011.02.045 [DOI] [PubMed] [Google Scholar]
- 22. Mishto M., Liepe J., Textoris-Taube K., Keller C., Henklein P., Weberruß M., Dahlmann B., Enenkel C., Voigt A., Kuckelkorn U., Stumpf M. P., and Kloetzel P. M. (2014) Proteasome isoforms exhibit only quantitative differences in cleavage and epitope generation. Eur. J. Immunol. 44, 3508–3521 10.1002/eji.201444902 [DOI] [PubMed] [Google Scholar]
- 23. Zanker D., Waithman J., Yewdell J. W., and Chen W. (2013) Mixed proteasomes function to increase viral peptide diversity and broaden antiviral CD8+ T cell responses. J. Immunol. 191, 52–59 10.4049/jimmunol.1300802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kincaid E. Z., Che J. W., York I., Escobar H., Reyes-Vargas E., Delgado J. C., Welsh R. M., Karow M. L., Murphy A. J., Valenzuela D. M., Yancopoulos G. D., and Rock K. L. (2011) Mice completely lacking immunoproteasomes show major changes in antigen presentation. Nat. Immunol. 13, 129–135 10.1038/ni.2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Deol P., Zaiss D. M., Monaco J. J., and Sijts A. J. (2007) Rates of processing determine the immunogenicity of immunoproteasome-generated epitopes. J. Immunol. 178, 7557–7562 10.4049/jimmunol.178.12.7557 [DOI] [PubMed] [Google Scholar]
- 26. Nil A., Firat E., Sobek V., Eichmann K., and Niedermann G. (2004) Expression of housekeeping and immunoproteasome subunit genes is differentially regulated in positively and negatively selecting thymic stroma subsets. Eur. J. Immunol. 34, 2681–2689 10.1002/eji.200425032 [DOI] [PubMed] [Google Scholar]
- 27. Takada K., Van Laethem F., Xing Y., Akane K., Suzuki H., Murata S., Tanaka K., Jameson S. C., Singer A., and Takahama Y. (2015) TCR affinity for thymoproteasome-dependent positively selecting peptides conditions antigen responsiveness in CD8+ T cells. Nat. Immunol. 16, 1069–1076 10.1038/ni.3237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nitta T., Murata S., Sasaki K., Fujii H., Ripen A. M., Ishimaru N., Koyasu S., Tanaka K., and Takahama Y. (2010) Thymoproteasome shapes immunocompetent repertoire of CD8+ T cells. Immunity 32, 29–40 10.1016/j.immuni.2009.10.009 [DOI] [PubMed] [Google Scholar]
- 29. Kincaid E. Z., Murata S., Tanaka K., and Rock K. L. (2016) Specialized proteasome subunits have an essential role in the thymic selection of CD8+ T cells. Nat. Immunol. 17, 938–945 10.1038/ni.3480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sasaki K., Takada K., Ohte Y., Kondo H., Sorimachi H., Tanaka K., Takahama Y., and Murata S. (2015) Thymoproteasomes produce unique peptide motifs for positive selection of CD8+ T cells. Nat. Commun. 6, 7484 10.1038/ncomms8484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yamada Y., Tomaru U., Ishizu A., Kiuchi T., Marukawa K., Matsuno Y., and Kasahara M. (2011) Expression of proteasome subunit beta5t in thymic epithelial tumors. Am. J. Surg. Pathol. 35, 1296–1304 10.1097/PAS.0b013e3182237f5d [DOI] [PubMed] [Google Scholar]
- 32. Yamada Y., Tomaru U., Ishizu A., Kiuchi T., Kasahara M., and Matsuno Y. (2014) Expression of thymoproteasome subunit β5t in type AB thymoma. J. Clin. Pathol. 67, 276–278 10.1136/jclinpath-2013-201930 [DOI] [PubMed] [Google Scholar]
- 33. Takase H., Yu C. R., Mahdi R. M., Douek D. C., Dirusso G. B., Midgley F. M., Dogra R., Allende G., Rosenkranz E., Pugliese A., Egwuagu C. E., and Gery I. (2005) Thymic expression of peripheral tissue antigens in humans: a remarkable variability among individuals. Int. Immunol. 17, 1131–1140 10.1093/intimm/dxh275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Adamopoulou E., Tenzer S., Hillen N., Klug P., Rota I. A., Tietz S., Gebhardt M., Stevanovic S., Schild H., Tolosa E., Melms A., and Stoeckle C. (2013) Exploring the MHC-peptide matrix of central tolerance in the human thymus. Nat. Commun. 4, 2039 10.1038/ncomms3039 [DOI] [PubMed] [Google Scholar]
- 35. Perchellet A., Stromnes I., Pang J. M., and Goverman J. (2004) CD8+ T cells maintain tolerance to myelin basic protein by 'epitope theft'. Nat. Immunol. 5, 606–614 10.1038/ni1073 [DOI] [PubMed] [Google Scholar]
- 36. Perchellet A., Brabb T., and Goverman J. M. (2008) Crosspresentation by nonhematopoietic and direct presentation by hematopoietic cells induce central tolerance to myelin basic protein. Proc. Natl. Acad. Sci. U.S.A. 105, 14040–14045 10.1073/pnas.0804970105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Friese M. A., Jakobsen K. B., Friis L., Etzensperger R., Craner M. J., McMahon R. M., Jensen L. T., Huygelen V., Jones E. Y., Bell J. I., and Fugger L. (2008) Opposing effects of HLA class I molecules in tuning autoreactive CD8+ T cells in multiple sclerosis. Nat. Med. 14, 1227–1235 10.1038/nm.1881 [DOI] [PubMed] [Google Scholar]
- 38. Mishto M., Goede A., Taube K. T., Keller C., Janek K., Henklein P., Niewienda A., Kloss A., Gohlke S., Dahlmann B., Enenkel C., and Kloetzel P. M. (2012) Driving forces of proteasome-catalyzed peptide splicing in yeast and humans. Mol. Cell. Proteomics 11, 1008–1023 10.1074/mcp.M112.020164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mishto M., Bellavista E., Ligorio C., Textoris-Taube K., Santoro A., Giordano M., D'Alfonso S., Listì F., Nacmias B., Cellini E., Leone M., Grimaldi L. M., Fenoglio C., Esposito F., Martinelli-Boneschi F., et al. (2010) Immunoproteasome LMP2 60HH variant alters MBP epitope generation and reduces the risk to develop multiple sclerosis in Italian female population. PLoS One 5, e9287 10.1371/journal.pone.0009287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jurewicz A., Biddison W. E., and Antel J. P. (1998) MHC class I-restricted lysis of human oligodendrocytes by myelin basic protein peptide-specific CD8 T lymphocytes. J. Immunol. 160, 3056–3059 [PubMed] [Google Scholar]
- 41. Berthelot L., Laplaud D. A., Pettré S., Ballet C., Michel L., Hillion S., Braudeau C., Connan F., Lefrère F., Wiertlewski S., Guillet J. G., Brouard S., Choppin J., and Soulillou J. P. (2008) Blood CD8+ T cell responses against myelin determinants in multiple sclerosis and healthy individuals. Eur. J. Immunol. 38, 1889–1899 10.1002/eji.200838023 [DOI] [PubMed] [Google Scholar]
- 42. Zang Y. C., Li S., Rivera V. M., Hong J., Robinson R. R., Breitbach W. T., Killian J., and Zhang J. Z. (2004) Increased CD8+ cytotoxic T cell responses to myelin basic protein in multiple sclerosis. J. Immunol. 172, 5120–5127 10.4049/jimmunol.172.8.5120 [DOI] [PubMed] [Google Scholar]
- 43. Tsuchida T., Parker K. C., Turner R. V., McFarland H. F., Coligan J. E., and Biddison W. E. (1994) Autoreactive CD8+ T-cell responses to human myelin protein-derived peptides. Proc. Natl. Acad. Sci. U.S.A. 91, 10859–10863 10.1073/pnas.91.23.10859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Princiotta M. F., Finzi D., Qian S. B., Gibbs J., Schuchmann S., Buttgereit F., Bennink J. R., and Yewdell J. W. (2003) Quantitating protein synthesis, degradation, and endogenous antigen processing. Immunity 18, 343–354 10.1016/S1074-7613(03)00051-7 [DOI] [PubMed] [Google Scholar]
- 45. Liepe J., Marino F., Sidney J., Jeko A., Bunting D. E., Sette A., Kloetzel P. M., Stumpf M. P., Heck A. J., and Mishto M. (2016) A large fraction of HLA class I ligands are proteasome-generated spliced peptides. Science 354, 354–358 10.1126/science.aaf4384 [DOI] [PubMed] [Google Scholar]
- 46. Faridi P., Li C., Ramarathinam S. H., Vivian J. P., Illing P. T., Mifsud N. A., Ayala R., Song J., Gearing L. J., Hertzog P. J., Ternette N., Rossjohn J., Croft N. P., and Purcell A. W. (2018) A subset of HLA-I peptides are not genomically templated: Evidence for cis- and trans-spliced peptide ligands. Sci. Immunol. 3, eaar3947 10.1126/sciimmunol.aar3947 [DOI] [PubMed] [Google Scholar]
- 47. Liepe J., Sidney J., Lorenz F. K. M., Sette A., and Mishto M. (2019) Mapping the MHC class I-spliced immunopeptidome of cancer cells. Cancer Immunol. Res. 7, 62–76 10.1158/2326-6066.CIR-18-0424 [DOI] [PubMed] [Google Scholar]
- 48. Liepe J., Ovaa H., and Mishto M. (2018) Why do proteases mess up with antigen presentation by re-shuffling antigen sequences? Curr. Opin. Immunol. 52, 81–86 10.1016/j.coi.2018.04.016 [DOI] [PubMed] [Google Scholar]
- 49. Keller M., Ebstein F., Bürger E., Textoris-Taube K., Gorny X., Urban S., Zhao F., Dannenberg T., Sucker A., Keller C., Saveanu L., Krüger E., Rothkötter H. J., Dahlmann B., Henklein P., et al. (2015) The proteasome immunosubunits, PA28 and ER-aminopeptidase 1 protect melanoma cells from efficient MART-126–35-specific T-cell recognition. Eur. J. Immunol. 45, 3257–3268 10.1002/eji.201445243 [DOI] [PubMed] [Google Scholar]
- 50. Ebstein F., Textoris-Taube K., Keller C., Golnik R., Vigneron N., Van den Eynde B. J., Schuler-Thurner B., Schadendorf D., Lorenz F. K., Uckert W., Urban S., Lehmann A., Albrecht-Koepke N., Janek K., Henklein P., et al. (2016) Proteasomes generate spliced epitopes by two different mechanisms and as efficiently as non-spliced epitopes. Sci. Rep. 6, 24032 10.1038/srep24032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ebner F., Hepworth M. R., Rausch S., Janek K., Niewienda A., Kühl A., Henklein P., Lucius R., Hamelmann E., and Hartmann S. (2014) Therapeutic potential of larval excretory/secretory proteins of the pig whipworm Trichuris suis in allergic disease. Allergy 69, 1489–1497 10.1111/all.12496 [DOI] [PubMed] [Google Scholar]
- 52. Liepe J., Mishto M., Textoris-Taube K., Janek K., Keller C., Henklein P., Kloetzel P. M., and Zaikin A. (2010) The 20S proteasome splicing activity discovered by SpliceMet. PLoS Comput. Biol. 6, e1000830 10.1371/journal.pcbi.1000830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhou Y., Liepe J., Sheng X., Stumpf M. P., and Barnes C. (2011) GPU accelerated biochemical network simulation. Bioinformatics 27, 874–876 10.1093/bioinformatics/btr015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. R Core Team (2014) R: a language and environment for statistical computing, R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 55. Peters B., Janek K., Kuckelkorn U., and Holzhütter H. G. (2002) Assessment of proteasomal cleavage probabilities from kinetic analysis of time-dependent product formation. J. Mol. Biol. 318, 847–862 10.1016/S0022-2836(02)00167-5 [DOI] [PubMed] [Google Scholar]
- 56. Mishto M., Raza M. L., de Biase D., Ravizza T., Vasuri F., Martucci M., Keller C., Bellavista E., Buchholz T. J., Kloetzel P. M., Pession A., Vezzani A., and Heinemann U. (2015) The immunoproteasome β5i subunit is a key contributor to ictogenesis in a rat model of chronic epilepsy. Brain Behav. Immun. 49, 188–196 10.1016/j.bbi.2015.05.007 [DOI] [PubMed] [Google Scholar]
- 57. Mishto M., Santoro A., Bellavista E., Sessions R., Textoris-Taube K., Dal Piaz F., Carrard G., Forti K., Salvioli S., Friguet B., Kloetzel P. M., Rivett A. J., and Franceschi C. (2006) A structural model of 20S immunoproteasomes: effect of LMP2 codon 60 polymorphism on expression, activity, intracellular localisation and insight into the regulatory mechanisms. Biol. Chem. 387, 417–429 10.1515/BC.2006.056 [DOI] [PubMed] [Google Scholar]
- 58. Textoris-Taube K., Keller C., Liepe J., Henklein P., Sidney J., Sette A., Kloetzel P. M., and Mishto M. (2015) The T210M substitution in the HLA-a*02:01 gp100 epitope strongly affects overall proteasomal cleavage site usage and antigen processing. J. Biol. Chem. 290, 30417–30428 10.1074/jbc.M115.695189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gaczynska M., Rock K. L., and Goldberg A. L. (1993) γ-Interferon and expression of MHC genes regulate peptide hydrolysis by proteasomes. Nature 365, 264–267 10.1038/365264a0 [DOI] [PubMed] [Google Scholar]
- 60. Toes R. E., Nussbaum A. K., Degermann S., Schirle M., Emmerich N. P., Kraft M., Laplace C., Zwinderman A., Dick T. P., Müller J., Schönfisch B., Schmid C., Fehling H. J., Stevanovic S., Rammensee H. G., et al. (2001) Discrete cleavage motifs of constitutive and immunoproteasomes revealed by quantitative analysis of cleavage products. J. Exp. Med. 194, 1–12 10.1084/jem.194.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gaczynska M., Rock K. L., Spies T., and Goldberg A. L. (1994) Peptidase activities of proteasomes are differentially regulated by the major histocompatibility complex-encoded genes for LMP2 and LMP7. Proc. Natl. Acad. Sci. U.S.A. 91, 9213–9217 10.1073/pnas.91.20.9213 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data sets are available at the online archive Mendeley (http://dx.doi.org/10.17632/zjhhc2bm9p.1).5 Data sets linked to the computational model of proteasome dynamics and published by Liepe et al. (11) are available at http://datadryad.org/resource/doi:10.5061/dryad.nk453.5 The statistical background behind the model inference presented in this study is described in Liepe et al. (12, 13), and its implementation can be found at ABC-SysBio (https://sourceforge.net/projects/abc-sysbio/).5