Abstract
Epigenetic regulation by the type II protein arginine methyltransferase, PRMT5, plays an essential role in the control of cancer cell proliferation and tumorigenesis. In this report, we investigate the relationship between PRMT5 and WNT/β-CATENIN as well as AKT/GSK3β proliferative signaling in three different types of non-Hodgkin's lymphoma cell lines, clinical samples, and mouse primary lymphoma cells. We show that PRMT5 stimulates WNT/β-CATENIN signaling through direct epigenetic silencing of pathway antagonists, AXIN2 and WIF1, and indirect activation of AKT/GSK3β signaling. PRMT5 inhibition with either shRNA-mediated knockdown or a specific small molecule PRMT5 inhibitor, CMP-5, not only leads to derepression of WNT antagonists and decreased levels of active phospho-AKT (Thr-450 and Ser-473) and inactive phospho-GSK3β (Ser-9) but also results in decreased transcription of WNT/β-CATENIN target genes, CYCLIN D1, c-MYC, and SURVIVIN, and enhanced lymphoma cell death. Furthermore, PRMT5 inhibition leads to reduced recruitment of co-activators CBP, p300, and MLL1, as well as enhanced recruitment of co-repressors HDAC2 and LSD1 to the WNT/β-CATENIN target gene promoters. These results indicate that PRMT5 governs expression of prosurvival genes by promoting WNT/β-CATENIN and AKT/GSK3β proliferative signaling and that its inhibition induces lymphoma cell death, which warrants further clinical evaluation.
Keywords: protein arginine N-methyltransferase 5 (PRMT5), Wnt signaling, Akt PKB, lymphoma, histone, non-Hodgkin's lymphoma, protein arginine methyltransferase 5 (PRMT5), protein kinase B (AKT)/glycogen synthase kinase 3β, WNT antagonists, WNT/β-catenin signaling
Introduction
B-cell transformation can take place during different stages of B-cell development, and molecular profiling has shown that B-cell malignancies are extremely heterogeneous. Mantle cell lymphoma (MCL)2 is a subtype of pre-germinal center non-Hodgkin's lymphoma (NHL) characterized by translocation t(11;14)(q13;q32) detected in virtually 100% of cases by fluorescence in situ hybridization, which juxtaposes the CYCLIN D1 (CCND-1) gene at 11q13 to the immunoglobulin heavy chain locus at 14q32 (1–3). MCL is associated with poor prognosis and is considered incurable (4). Similarly, diffuse large B-cell lymphoma (DLBCL) is the most common NHL worldwide that has been divided by global gene expression profiling into three different types: germinal center B-cell (GCB)-DLBCL, activated B-cell–like (ABC)-DLBCL, and primary mediastinal B-cell lymphoma (5, 6). Different chromosomal translocations have been identified in DLBCLs, including BCL2 and BCL6 rearrangements, which are mutually exclusive (7–9). Despite these genetic variations, there are common traits that unify lymphoma cell types. For example, our previous work has shown that protein arginine methyltransferase 5 (PRMT5) is highly expressed in B-cell lymphomas of different types, including MCL, GCB-DLBCL, and ABC-DLBCL (10–12), and that it is involved in B-cell immortalization and maintenance of the malignant phenotype (12, 13). These findings support the notion that PRMT5 controls proliferation and survival of lymphoma cells through regulation of conserved growth regulatory pathways.
Histone methylation is an important epigenetic modification that controls chromatin structure and gene expression and has been implicated in cancer development (14–16). However, limited knowledge exists regarding the role of epigenetic modifications and their potential impact on NHL pathogenesis. Although the role of histone lysine methylation has been extensively studied, histone arginine methylation remains largely unexplored. PRMT5 is a type II arginine methyltransferase that can target histones H3 and H4 and alter gene expression programs in a significant way to contribute to cellular transformation (10, 12, 13, 17). PRMT5 is also capable of methylating nonhistone proteins such as p53 and E2F1, and the direct outcome of these modifications is aberrant expression of specific target genes regulated by these transcription factors, accompanied by enhanced cancer cell growth (18, 19). We have previously shown that PRMT5 is selectively overexpressed in NHL cells and that it is a major prosurvival factor that prompts growth of lymphoma cells (10). We have also shown mechanistically that PRMT5 promotes lymphoma cell growth and survival through negative regulation of the RB/E2F tumor suppressor pathway (11). In a follow-up study, we demonstrated that PRMT5 promotes polycomb repressor complex 2 (PRC2) expression through inactivation of the RB/E2F pathway using a two-pronged approach, which consists of RB1 hyperphosphorylation by CYCLIN D1-CDK4/CDK6 and direct PRMT5-mediated epigenetic silencing of RBL2 expression (12). Although the relationship between PRMT5 and the RB-E2F–signaling pathway has been established, we still do not know the mechanism by which PRMT5 controls expression of prosurvival factors CYCLIN D1, c-MYC, and SURVIVIN. Because all three genes are bona fide β-CATENIN targets, we focused our investigation on the molecular mechanisms by which PRMT5 promotes WNT/β-CATENIN–proliferative signaling.
The WNT (Wingless related integration site)-signaling pathway is evolutionarily conserved and can be triggered by a large number of WNT proteins, which play important roles during proliferation, differentiation, and development (20–26). WNTs are secreted glycoproteins rich in cysteine that activate receptor-mediated signaling pathways. Four different pathways are activated by WNT signaling: the canonical WNT/β-CATENIN cascade; the noncanonical planar cell polarity; WNT/Ca2+; and protein kinase A pathways (25, 27). Binding of WNT molecules to a cognate receptor composed of a seven-pass transmembrane Frizzled receptor and associated single-pass, low-density lipoprotein receptor-related (LRP5/6) proteins located on the cell surface can, in the canonical (WNT/β-CATENIN) pathway, activate intracellular Disheveled protein and recruit AXIN proteins to the cellular membrane. Consequently, the AXIN–APC–GSK3β–CK1α destruction complex becomes inactive, and cytosolic levels of β-CATENIN increase, which in turn leads to translocation of β-CATENIN into the nucleus. Upon entering the nucleus, β-CATENIN interacts with members of the T-cell factor (TCF)/lymphoid enhancer factor (LEF) transcription factor family as well as B-cell CLL/lymphoma 9 (BCL9) and Pygopus, displaces GROUCHO/transducin-like enhancer of split/HDAC1 repressor proteins, and stimulates target gene expression through recruitment of chromatin remodelers such as CBP/p300, MLL1/MLL2, and BRG1 (20, 21, 26).
Aberrations due to genetic deletions, point mutations, and epigenetic silencing of WNT inhibitor genes result in uncontrolled activation and continuous WNT/β-CATENIN proliferative signaling (20, 22, 25). Dysregulation of the WNT/β-CATENIN–signaling pathway is a hallmark of several types of tumors, including lymphoma, leukemia, colorectal, and hepatocellular carcinoma (28). Although it is known that DNA methylation in the promoter region of WNT/β-CATENIN antagonists results in their transcriptional silencing, and is associated with activation of β-CATENIN/TCF/LEF target genes and tumor development, little is known about the role played by PRMT5 in this process. Using patient-derived NHL cell lines, we show that PRMT5 promotes WNT/β-CATENIN signaling through direct transcriptional repression of two major antagonists, AXIN2 and WIF1, and indirect activation of AKT/GSK3β signaling. PRMT5 inhibition with either short hairpin RNA (shRNA)-mediated knockdown or a PRMT5-specific inhibitor, compound 5 (CMP-5), leads to AXIN2 and WIF1 re-expression as well as inactivation of phospho-AKT (Thr-450 and Ser-473) and reactivation of GSK3β. The net outcome of these molecular changes is decreased expression of the WNT/β-CATENIN target genes CYCLIN D1, c-MYC, and SURVIVIN and induced lymphoma cell death. Recruitment studies show that PRMT5 inhibition results in loss of H3(Me2)R8 and H4(Me2)R3 methylation marks in the promoter region of AXIN2 and WIF1. In addition, remarkable changes occur at the CYCLIN D1, c-MYC, and SURVIVIN promoters, which consist of a decrease in recruitment of co-activators CBP, p300, and MLL1, and loss of their induced epigenetic marks H3K9Ac, H3K14Ac, and H3(Me3)K4, which is accompanied by enhanced binding of co-repressors HDAC2 and LSD1. These results have also been confirmed in NHL patient samples and mouse primary lymphoma cells. Therefore, our work identifies PRMT5 as a novel regulator of the AKT/GSK3β and WNT/β-CATENIN–signaling pathways in cells derived from multiple lymphoma histologic subtypes, and it further validates the importance of targeting PRMT5 in aggressive lymphomas.
Results
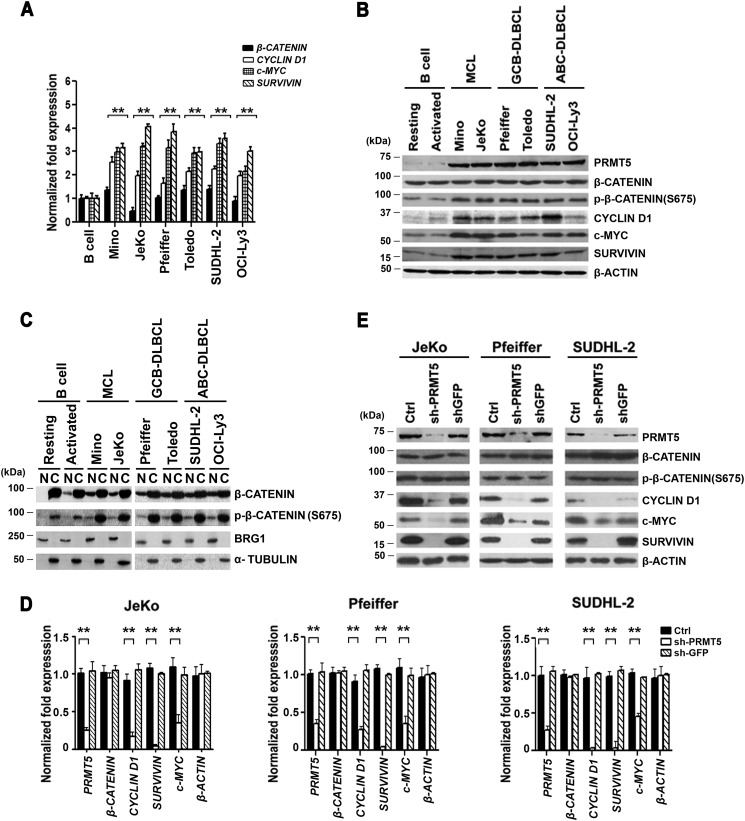
PRMT5 regulates expression of WNT/β-CATENIN target genes
We have previously shown that PRMT5 protein levels are low in normal B cells and that its levels are enhanced as a result of decreased expression of specific miRNAs (10, 11). We have also reported previously that PRMT5 promotes lymphoma cell growth and survival through inactivation of the retinoblastoma tumor suppressor proteins, RB1 and RBL2, and up-regulation of the PRC2 (12). Our previous work also showed that although RB1 inhibition is due to hyperphosphorylation by CYCLIN D1–CDK4/CDK6, RBL2 inactivation is a direct result of PRMT5-mediated epigenetic silencing. In addition, evaluation of protein levels revealed a strong correlation between increased PRMT5 expression and elevated levels of CYCLIN D1, a known WNT/β-CATENIN target gene (12). In light of these results and findings, which show that PRMT5 is essential for c-MYC–driven oncogenesis (29), we hypothesized that PRMT5 might affect lymphoma cell growth and proliferation by promoting WNT/β-CATENIN signaling.
To verify whether PRMT5 is involved in regulating WNT/β-CATENIN signaling, we measured the mRNA and protein levels of WNT/β-CATENIN downstream target genes, CYCLIN D1, c-MYC, and SURVIVIN (Fig. 1). Two distinct patient-derived cell lines were used to assess the relationship between PRMT5 and WNT/β-CATENIN target genes in each of the three different lymphoma cell types selected, including MCL (Mino and JeKo), GCB-DLBCL (Pfeiffer and Toledo), and ABC-DLBCL (SUDHL-2 and OCI-Ly3). We found that although there was no significant change in β-CATENIN mRNA and protein expression in both normal and transformed B cells, there was an increase in CYCLIN D1 (1.7–2.5-fold, p < 10−3), c-MYC (2–3.3-fold, p < 10−3), and SURVIVIN (3–4-fold, p < 10−3) mRNA levels as well as protein expression in transformed B cells compared with control normal resting or activated CD19+ B cells (Fig. 1, A and B). Consistent with our previous findings, PRMT5 protein levels were also elevated in all three lymphoma cell types. Interestingly, the positive correlation between PRMT5 protein expression and enhanced levels of WNT/Β-CATENIN target genes was paralleled by an increase in the amount of nuclear and transcriptionally active phospho-β-CATENIN (Ser-675) (Fig. 1C), suggesting that PRMT5 promotes lymphoma cell growth and survival via activation of the WNT/β-CATENIN–signaling pathway.
Figure 1.
PRMT5 regulates WNT/β-CATENIN signaling in patient-derived lymphoma cell lines. A, levels of PRMT5, β-CATENIN, CYCLIN D1, c-MYC, and SURVIVIN mRNAs were measured in normal B (resting or activated), pre-GCB MCL (Mino and JeKo), GCB-DLBCL (Pfeiffer and Toledo), and post-GCB ABC-DLBCL (SUDHL-2 and OCI-Ly3) cells by real-time RT-PCR using gene-specific primers and probe sets. The values represent the average from three biological replicates with three technical replicates each and are reported as mean ± S.D. 18S rRNA was used as an internal control. B, RIPA extracts (20 μg) from either normal or transformed B cells were analyzed by Western blotting using the indicated antibodies. β-ACTIN serves as a loading control and is used in both B and Fig. 2D because these data resulted from the same experiment. This experiment was repeated two times, and representative blots are shown. C, approximately 25 μg of either nuclear (N) or cytosolic (C) extracts were prepared from either normal resting or activated B cells, as well as the indicated lymphoma cell lines, and proteins were detected using the indicated antibodies. Both BRG1 and α-TUBULIN served as control for nuclear and cytosolic fractionation. D, levels of PRMT5, β-CATENIN, CYCLIN D1, c-MYC, and SURVIVIN mRNAs were measured by real-time RT-PCR using total RNA isolated from either uninfected (Ctrl) JeKo, Pfeiffer, and SUDHL-2 cells, or lymphoma cells infected with shGFP or shPRMT5 lentivirus. Specific primers and probe sets were used to detect each of the indicated genes. This experiment was conducted using three biological replicates with three technical replicates as described in A. ** indicates p values < 10−3. E, RIPA extracts (20 μg) were prepared from control-uninfected and -infected (shGFP or shPRMT5) lymphoma cell lines and analyzed by immunoblotting using the indicated antibodies. β-ACTIN served as a loading control.
To investigate the extent to which PRMT5 influences WNT/β-CATENIN target gene expression, we inhibited its expression by shRNA-mediated knockdown in JeKo, Pfeiffer, and SUDHL-2 cells and monitored mRNA and protein expression of CYCLIN D1, c-MYC, and SURVIVIN. We had previously shown that PRMT5 knockdown using two distinct shRNAs (shPRMT5-1 and shPRMT5-2) achieves similar reduction in PRMT5 mRNA and protein levels (12). Therefore, we opted to use only shPRMT5-1 in this study. Both mRNA and protein analyses showed that reduced PRMT5 expression results in repression of CYCLIN D1 (3–5-fold, p < 10−3), c-MYC (2.3–3-fold, p < 10−3), and SURVIVIN (26–50-fold, p < 10−4) mRNA levels as well as protein expression in all three lymphoma cell types compared with control uninfected lymphoma cells or lymphoma cells infected with lentivirus that expresses control shGFP (Fig. 1, D and E). To further confirm these results, we used the PRMT5-specific inhibitor, compound 5 (CMP-5), which has recently been characterized (13), to treat JeKo, Pfeiffer, and SUDHL-2 cell lines (Fig. S1, A and B). In agreement with the PRMT5 knockdown experiments, there was a clear decrease in CYCLIN D1, c-MYC, and SURVIVIN mRNA and protein expression in all three lymphoma cell types, but not in control lymphoma cells treated with either DMSO or control CMP-6. Furthermore, when we analyzed proliferation and survival of JeKo, Pfeiffer, and SUDHL-2 cells treated with CMP-5, there was a clear inhibition of cell growth by day 2, which became maximal by day 6, and culminated in enhanced cell death in all three lymphoma cell types as evidenced by the increase in the number of annexin V-positive cells (Fig. S1, C and D). Collectively, these results show that PRMT5 controls expression of prosurvival factors, CYCLIN D1, c-MYC, and SURVIVIN, and that its inhibition results in their suppression and induced lymphoma cell death.
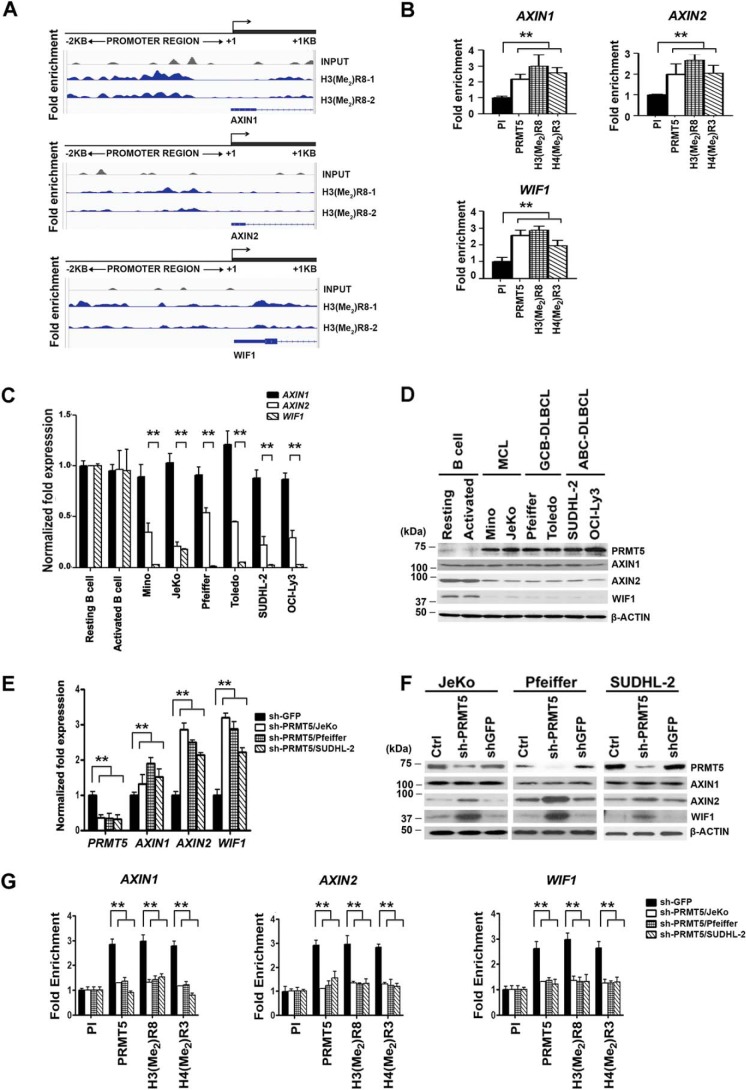
PRMT5 epigenetically represses WNT/β-CATENIN antagonists, AXIN2 and WIF1
Given the importance of WNT/β-CATENIN signaling in the regulation of cellular growth and proliferation, we wanted to determine whether PRMT5 was directly involved in controlling expression of pathway antagonists. To evaluate the genome-wide distribution of PRMT5, ChIP-Seq assay was carried out using immune anti-H3(Me2)R8 antibody to immunoprecipitate cross-linked chromatin from Pfeiffer cells. Among the common target genes identified (Table S1), we found increased H3R8 methylation in the promoter region (−2 to +1 kb) of AXIN1, AXIN2, and WIF1 compared with control input DNA (Fig. 2A). To confirm the ChIP-Seq results, ChIP assays followed by real-time PCR analyses were conducted on cross-linked chromatin immunoprecipitated from the three lymphoma cell lines using either pre-immune or immune anti-PRMT5, anti-H3(Me2)R8, and anti-H4(Me2)R3 (Fig. 2B and Fig. S2, A and B). Consistent with the ChIP-Seq results, there was 2–3-fold (p < 10−4) increased PRMT5 recruitment and enrichment of its epigenetic marks in the promoter region of AXIN1, AXIN2, and WIF1. Moreover, inspection of the amplified region in the promoter region of all three WNT/β-CATENIN antagonists revealed that there are conserved DNA-binding sites for p53, PAX5, TFII-I, and estrogen receptor-α (ER-α) transcription factor (Table S2).
Figure 2.
PRMT5 epigenetically regulates AXIN2 and WIF1, and its inhibition reactivates both target genes. A, cross-linked chromatin from Pfeiffer cells was immunoprecipitated using anti-H3(Me2)R8 antibody and analyzed by ChIP-Seq as described under “Experimental procedures.” Enrichment of H3(Me2)R8 on AXIN1, AXIN2, and WIF1 promoter sequences spanning −2 kb to +1 kb was determined using two biological replicates (H3(Me2)R8-1, and H3(Me2)R8-2). Peak detection for H3(Me2)R8 enrichment is reported in blue color compared with control input, which is reported in gray color. B, ChIP assays were performed on cross-linked chromatin from Pfeiffer cells using pre-immune (PI), anti-PRMT5, anti-H3(Me2)R8, or anti-H4(Me2)R3 antibody. ChIP experiments were carried out using two biological replicates with three technical replicates, and AXIN1, AXIN2, and WIF1 promoter sequences were detected by real-time PCR. Fold enrichment with each antibody was calculated relative to the PI sample. Data in each graph represent the mean ± S.D. C, levels of AXIN1, AXIN2, and WIF1 mRNAs were measured in normal B cells (resting and activated) and the indicated transformed B cells by real-time RT-PCR using gene-specific primers and probe sets. This experiment was conducted using three biological replicates with three technical replicates, and 18S rRNA was used as control. The data shown represent the mean ± S.D. D, RIPA extracts (20 μg) from normal (resting and activated) and transformed B cells were analyzed by Western blotting using the indicated antibodies. Anti-β-ACTIN was used to show equal loading. β-ACTIN loading control in both B and D are the same because these data resulted from the same experiment. E, levels of PRMT5, AXIN1, AXIN2, and WIF1 mRNAs were measured using total RNA from either control (Ctrl) uninfected JeKo, Pfeiffer, and SUDHL-2 cells or lymphoma cells infected with shGFP or shPRMT5 lentivirus as described in C. ** indicates p values < 10−3. F, AXIN1, AXIN2, and WIF1 protein levels were analyzed by Western blotting using RIPA extracts (20 μg) prepared from control-uninfected and -infected (shGFP or shPRMT5) JeKo, Pfeiffer, and SUDHL-2 lymphoma cell lines 72 h post-infection. Anti-β-ACTIN was used to show equal loading. G, ChIP assays were carried out using cross-linked chromatin from either control shGFP- or shPRMT5-infected lymphoma cell lines. Enrichment of PRMT5 and its induced epigenetic marks was determined using the indicated antibodies, and PI antibody was used as a control. This experiment was repeated using three biological replicates with three technical replicates each, and the data were plotted as in B.
In light of these results, we proceeded to evaluate mRNA and protein expression of WNT/β-CATENIN antagonists in both control resting or activated B cells and transformed B-cell lines (Fig. 2, C and D). Both AXIN2 and WIF1 mRNAs and proteins exhibited 2–20-fold (p < 10−3) reduced expression profiles in lymphoma cells compared with control B cells, whereas AXIN1 mRNA and protein levels were unaffected in normal and transformed B cells. To verify whether PRMT5 inhibition alters expression of AXIN2 and WIF1, we knocked down its expression in JeKo, Pfeiffer, and SUDHL-2 cells and monitored their mRNA and protein levels (Fig. 2, E and F). Both AXIN2 and WIF1 showed 2–3-fold (p < 10−3) derepression at the mRNA level, which was accompanied by an increase in their protein expression compared with control uninfected or shGFP-infected lymphoma cells. Like control β-ACTIN, levels of AXIN1 mRNA and protein did not show any significant change. Similarly, when we used CMP-5 to inhibit PRMT5, there was 1.8–3-fold (p < 10−3) derepression of both AXIN2 and WIF1 mRNAs, and a moderate increase in their protein expression (Fig. S3, A and B).
Having found that PRMT5 is enriched in the promoter region of all three WNT/β-CATENIN antagonist genes, we sought to monitor its recruitment in both control and PRMT5 knockdown as well as CMP-5–treated lymphoma cell lines (Fig. 2G and Fig. S3C). PRMT5 recruitment and enrichment of its epigenetic marks were abolished in both PRMT5 knockdown cells as well as CMP-5–treated lymphoma cell lines compared with control shGFP-infected and DMSO-treated lymphoma cells. Taken together, these results indicate that PRMT5 epigenetically suppresses expression of AXIN2 and WIF1 to promote WNT/β-CATENIN signaling and target gene expression.
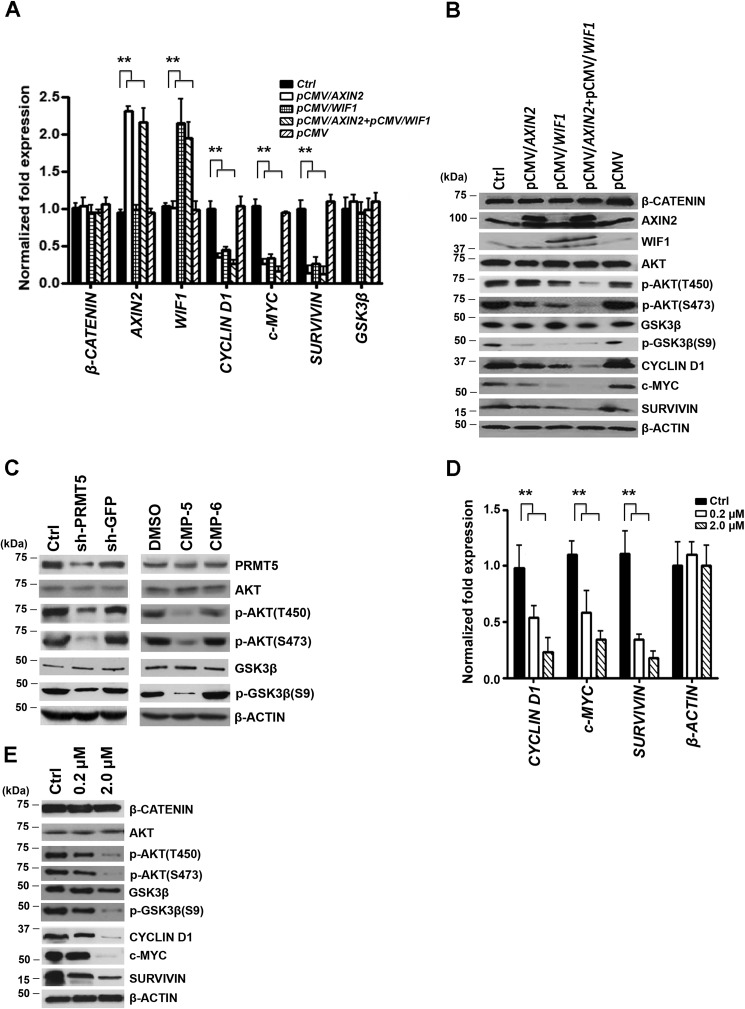
AXIN2 and/or WIF1 re-expression lead to reduced AKT/GSK3β phosphorylation and CYCLIN D1, c-MYC, and SURVIVIN expression
To assess the role played by AXIN2 and WIF1 in WNT/β-CATENIN signaling, we ectopically restored their expression either individually or in combination in Pfeiffer cells and monitored expression of CYCLIN D1, c-MYC, and SURVIVIN (Fig. 3). Total RNA as well as whole-cell extracts were collected and examined for mRNA and protein expression. Re-expression of either AXIN2 or WIF1 triggered a reduction in CYCLIN D1 (2.2–2.9-fold, p < 10−4), c-MYC (3–3.8-fold, p < 10−4), and SURVIVIN (3–7.7-fold, p < 10−3) mRNA levels (Fig. 3, A and B), as well as a decrease in their protein expression (Fig. 3C). Remarkably, co-expression of both WNT/β-CATENIN antagonists resulted in a more pronounced decrease (3.8–8.3-fold, p < 10−4) in WNT/β-CATENIN target gene mRNA levels, which was accompanied by a more pronounced decrease in their protein expression.
Figure 3.
Re-expression of AXIN2 and/or WIF1 inhibits WNT/β-CATENIN target gene expression through inactivation of AKT. A, Pfeiffer cells were transfected with either control (Ctrl) vector (pCMV-entry) or pCMV-entry/AXIN2 and/or pCMV-entry/WIF1 for 72 h, and mRNA levels of AXIN2, WIF1, CYCLIN D1, c-MYC, and SURVIVIN were analyzed by real-time RT-PCR. This experiment was conducted using three biological replicates with three technical replicates each, and β-ACTIN was used as control. The data shown represent the mean ± S.D., and ** indicates p values < 10−3. B, RIPA extracts (20 μg) prepared from either control-untransfected or -transfected Pfeiffer cells were analyzed by immunoblotting using the indicated antibodies. β-ACTIN was detected to show equal loading. C, levels of both active phospho-AKT (Thr-450 or Ser-473) and inactive AKT as well as active GSK3β or inactive phospho-GSK3β (Ser-9) were determined by immunoblotting using RIPA extracts (20 μg) prepared from Pfeiffer cells infected with lentivirus that expresses either control shGFP or shPRMT5 and Pfeiffer cells treated with either control DMSO or CMP-5. Both uninfected and CMP-6–treated Pfeiffer cells were used as internal controls. RIPA cell extracts were prepared 72 h post-infection or 48 h post-treatment. D, total RNA was prepared from Pfeiffer cells treated for 48 h with either control DMSO or increasing concentrations of AKT inhibitor IV (0.2 and 2 μm), and the mRNA levels of CYCLIN D1, c-MYC, and SURVIVIN were evaluated by real-time RT-PCR using gene-specific primers and probe sets as described in A. E, Pfeiffer cells were treated as in D, and RIPA extracts (20 μg) were analyzed by Western blotting using the indicated antibodies. β-ACTIN was detected to show equal loading.
It is well established that canonical WNT signaling regulates the amount of transcriptionally active β-CATENIN (30). It is also known that AKT (protein kinase B), which is activated in many tumor cell types, cross-talks with WNT/β-CATENIN signaling through phosphorylation and inactivation of glycogen synthase kinase 3β (GSK3β), thereby promoting nuclear translocation of transcriptionally competent phospho-β-CATENIN (Ser-675) (31, 32). In light of these studies, we evaluated expression and phosphorylation of AKT/GSK3β in control and transfected Pfeiffer cells (Fig. 3B). Re-expression of either AXIN2 or WIF1 brought about a slight decrease in the level of phosphorylated AKT (Thr-450 and/or Ser-473); however, in the presence of both WNT antagonists there was a significant drop in active AKT as measured by dephosphorylation of Thr-450 and Ser-473 (33). Similarly, re-expression of either AXIN2 and or WIF1 led to decreased levels of phospho-GSK3β (Ser-9), which directly reflects inactivation of AKT. Because our results showed that PRMT5 inhibition derepresses AXIN2 and WIF1, we wanted to determine whether PRMT5 knockdown or PRMT5 inhibition with CMP-5 has the same effect on AKT/GSK3β phosphorylation (Fig. 3C). In fact, both treatments decreased phosphorylation of AKT at Thr-450 and Ser-473, as well as phosphorylation of GSK3β at Ser-9, confirming the AXIN2 and WIF1 re-expression results.
To further evaluate the contribution of AKT/GSK3β signaling to the regulation of WNT/β-CATENIN target gene expression, we used increasing concentrations of AKT inhibitor IV (0.2 and 2 μm) to treat Pfeiffer cells, and we monitored CYCLIN D1, c-MYC, and SURVIVIN mRNA and protein expression (Fig. 3, D and E). Although a low concentration of AKT inhibitor IV (0.2 μm) caused a 2–3-fold decrease (p < 10−3) in mRNA expression of all three WNT/β-CATENIN target genes, there was no significant drop in their protein expression. However, when 2 μm of AKT inhibitor IV was used, there was a 4-fold decrease (p < 10−4) in mRNA levels and a remarkable drop in CYCLIN D1, c-MYC, and SURVIVIN protein expression. Similarly, phosphorylation of both AKT (Thr-450/Ser-473) and GSK3β was also significantly decreased in the presence of 2 μm of AKT inhibitor IV. These findings suggest that both AXIN2 and WIF1 work in concert to down-regulate expression of CYCLIN D1, c-MYC, and SURVIVIN through inactivation of AKT/GSK3β and WNT/β-CATENIN signaling.
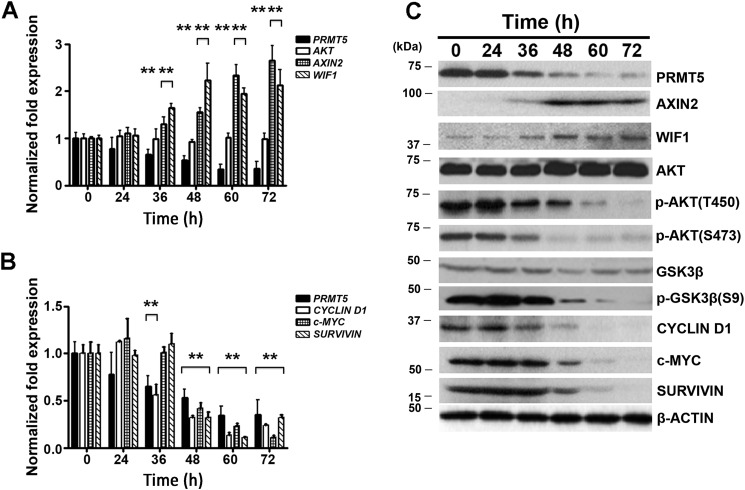
Kinetics of AXIN2 and WIF1 derepression coincide with AKT/GSK3β dephosphorylation and silencing of WNT/β-CATENIN target gene expression
Having found that PRMT5 inhibition restores expression of both AXIN2 and WIF1 and results in dephosphorylation of AKT/GSK3β, we sought to determine the chronology in which these events occur. A time-course analysis was carried out after either knocking down PRMT5 expression or inhibiting its activity (Fig. 4 and Fig. S4). When PRMT5 levels were reduced by short hairpin-mediated knockdown, AXIN2 and WIF1 mRNA levels increased 36 h post-infection (1.5-fold, p < 10−3) and reached 2–2.2-fold (p < 10−3) derepression by 72 h post-infection (Fig. 4A). This change in mRNA levels correlated with an increase in AXIN2 and WIF1 protein levels (Fig. 4C). In accord with these results, there was a drop in CYCLIN D1, c-MYC, and SURVIVIN mRNA levels, which started 36–48 h post-infection and became maximal 72 h post-infection (Fig. 4B). However, the decrease in WNT/β-CATENIN target gene protein expression did not become apparent until 48–60 h post-infection. Moreover, analysis of AKT/GSK3β phosphorylation revealed that there was a time concordance between AXIN2 and WIF1 derepression and AKT/GSK3β dephosphorylation (Fig. 4C).
Figure 4.
PRMT5 knockdown triggers WNT/β-CATENIN target gene suppression through re-expression of AXIN2 and WIF1 and inactivation of AKT signaling. Pfeiffer cells were infected with lentivirus that expresses shPRMT5, and mRNA levels of PRMT5, AKT, AXIN2, and WIF1 (A), and PRMT5, CYCLIN D1, c-MYC, and SURVIVIN (B) were measured by real-time RT-PCR at different time points using 18S rRNA as internal control. For comparison, mRNA levels were also determined before infection at time 0 h. Values represent the average of three biological replicates used with three technical replicates each and are reported as mean ± S.D. C, RIPA extracts (20 μg) were prepared from control uninfected (0 h) and infected (shPRMT5) Pfeiffer cell lines and analyzed by immunoblotting using the indicated antibodies. β-ACTIN was detected to show equal loading.
Similar results were observed when PRMT5 was inhibited with CMP-5; however, the kinetics of re-expression for AXIN2 (1.4–2.5-fold, p < 10−4) and WIF1 (1.5–2.9-fold, p < 10−4) mRNAs were faster (1–3 h post-infection) compared with cells where PRMT5 expression was knocked down (Fig. S4A). In addition, there was a concomitant increase in AXIN2 and WIF1 protein levels, which was more pronounced for WIF1 1-h post-treatment (Fig. S4C). When we examined CYCLIN D1, c-MYC, and SURVIVIN mRNA and protein expression, we found that their decrease started 6–12 h post-treatment, further confirming that AXIN2 and WIF1 re-expression precedes inhibition of WNT/β-CATENIN target gene expression (Fig. S4, B and C). Our results also showed that dephosphorylation of AKT/GSK3β occurred immediately (3 h post-treatment) after restored expression of AXIN2 and WIF1, suggesting that both antagonists work in concert to inactivate AKT/GSK3β and WNT/β-CATENIN signaling.
Promoters of WNT/β-CATENIN target genes undergo specific lysine deacetylation and demethylation upon PRMT5 inhibition
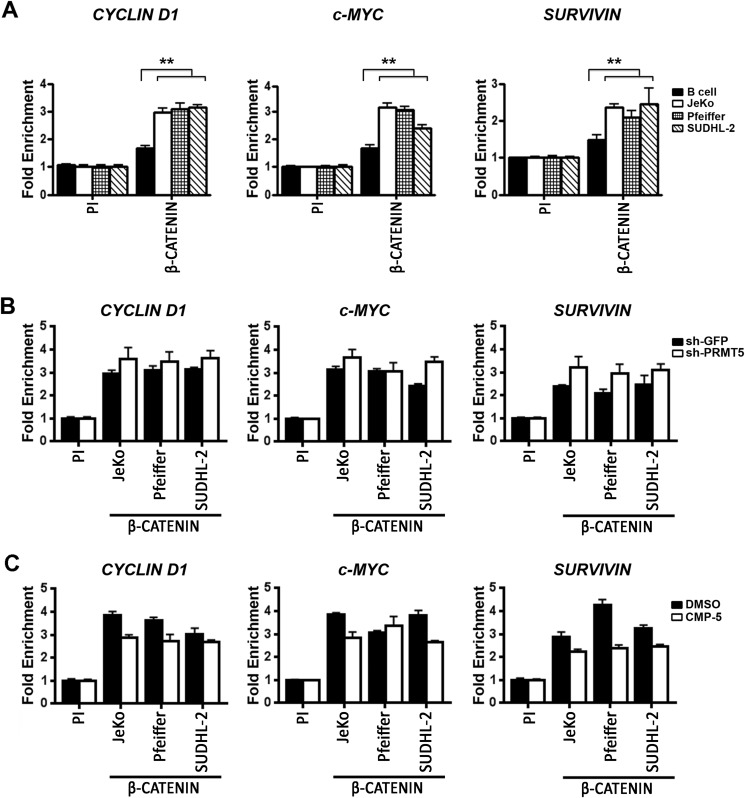
Transcriptional regulation of CYCLIN D1, c-MYC, and SURVIVIN is known to be up-regulated through binding of β-CATENIN to their promoters (34). The fact that we have found that either PRMT5 knockdown or inhibition of its methyltransferase activity led to a significant decrease in WNT/β-CATENIN target gene expression, we set out to examine the dynamic changes that occurred in their promoter region (Fig. 5 and Fig. 6). First, we monitored recruitment of β-CATENIN to the promoter region of CYCLIN D1, c-MYC, and SURVIVIN in both normal and transformed B cells (Fig. 5). Binding of β-CATENIN to the promoter proximal region of all three WNT/β-CATENIN target genes was enriched 3-fold (p < 10−4) in lymphoma cells compared with control pre-immune antibody and 1.9-fold (p < 10−3) compared with control normal B cells (Fig. 5A). However, when PRMT5 was either knocked down (Fig. 5B) or inhibited (Fig. 5C), there was no significant change in β-CATENIN recruitment to the promoter regions of CYCLIN D1, c-MYC, and SURVIVIN.
Figure 5.
β-CATENIN recruitment to the promoter region of CYCLIN D1, c-MYC, and SURVIVIN is increased in lymphoma cells and is unaffected by PRMT5 knockdown. A, ChIP assays were conducted using cross-linked chromatin from either normal or the indicated transformed B cells using PI or immune anti-β-CATENIN antibody. This experiment was conducted using three biological replicates with three technical replicates each, and CYCLIN D1, c-MYC, and SURVIVIN promoter sequences were detected by real-time PCR. Fold enrichment was calculated relative to the PI sample, and the data in each graph are represented as mean ± S.D. B, cross-linked chromatin from the indicated lymphoma cells infected with lentivirus that expresses either control shGFP or shPRMT5 was immunoprecipitated using PI or immune anti-β-CATENIN antibody. The values in each graph were generated using two biological replicates with three technical replicates each and are represented as described in A. C, JeKo, Pfeiffer, and SUDHL-2 cells were treated with either DMSO or CMP-5, and cross-linked chromatin was immunoprecipitated with either PI or immune anti-β-CATENIN antibody. CYCLIN D1, c-MYC, and SURVIVIN promoter sequences were detected as described in B.
Figure 6.
PRMT5 knockdown alters recruitment of co-activators and co-repressors to the WNT/β-CATENIN target genes. A and B, cross-linked chromatin from Pfeiffer cells infected with lentivirus that expresses either control shGFP or shPRMT5 was immunoprecipitated using the indicated antibodies, and PI antibody was used as a control. ChIP assays were carried out using two biological replicates with three technical replicates each. Fold enrichment with each antibody was calculated relative to the PI sample, and the data in each graph are represented as mean ± S.D.
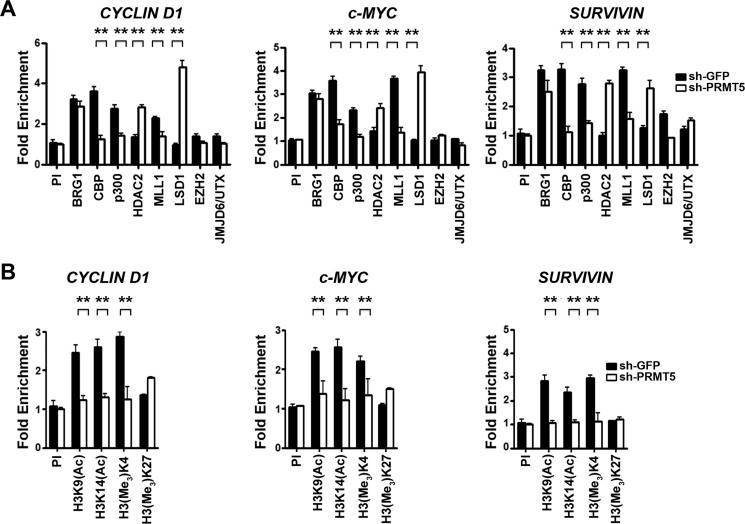
Because binding of β-CATENIN induces transcription of target genes through specific association with a wide variety of transcriptional co-activators (30, 34), we measured recruitment of hallmark interacting proteins in the promoter region of CYCLIN D1, c-MYC, and SURVIVIN in both control and PRMT5 knockdown lymphoma cells (Fig. 6 and Fig. S5). Among the proteins examined were BRG1, the catalytic subunit of hSWI/SNF chromatin remodeling complex, CBP, and p300 histone acetyltransferases, which acetylate histone H3 lysine 9 (H3K9) and H3K14, and MLL1, which specializes in methylating H3K4 and is associated with transcriptional activation. ChIP assays were also used to examine histones H3 and H4 post-translational modifications, as well as recruitment of co-repressor proteins, including HDAC2, which antagonizes CBP and p300, and LSD1, which demethylates H3K4 and is associated with transcriptional repression. Recruitment of EZH2, the catalytic subunit of the PRC2, which can induce transcriptional silencing via H3K27 methylation, and its antagonistic demethylase, JMJD6/UTX, was also examined.
Cross-linked chromatin from Pfeiffer cells infected with lentivirus that expresses either control shGFP or shPRMT5 was immunoprecipitated using either pre-immune or specific immune antibodies, and protein binding as well as histone modification in the promoter region of WNT/β-CATENIN target genes were evaluated by real-time PCR (Fig. 6). Although there was no significant change in recruitment of BRG1, EZH2, and JMJD6/UTX, binding of CBP to the promoter region of CYCLIND1, c-MYC, and SURVIVIN decreased by 2–3-fold (p < 10−3) in PRMT5 knockdown cells (Fig. 6A). Similarly, recruitment of p300 and MLL1 was also reduced by 2-fold (p < 10−4) and 1.7–2.7-fold (p < 10−3), respectively, in PRMT5 knockdown cells. However, association of both HDAC2 and LSD1 co-repressor proteins with the promoter region of WNT/β-CATENIN target genes was enhanced by 1.7–2.8-fold (p < 10−3) and 2–5-fold (p < 10−3), respectively. When we examined histones H3 and H4 acetylation and methylation marks, we found a positive correlation between loss of co-activator recruitment and decreased amounts of epigenetic marks associated with gene activation (Fig. 6B). All three histone marks associated with transcriptional activation were reduced by 1.8–2.8-fold (p < 10−3) for H3K9(Ac), 2–2.2-fold (p < 10−3) for H3K14(Ac), and 1.6–2.6-fold (p < 10−4) for H3(Me3)K4 in the promoter region of WNT/β-CATENIN target genes. This decrease in H3K4 methylation was not observed in the case of H3K27, highlighting the specificity of the dynamic changes that occur at the CYCLIN D1, c-MYC, and SURVIVIN promoters. Similar results were observed when PRMT5 was knocked down in JeKo and SUDHL-2 cells (Fig. S5, A and D), suggesting that a common mechanism is used to inhibit transcription of CYCLIN D1, c-MYC, and SURVIVIN in all three lymphoma cell lines.
PRMT5-mediated epigenetic silencing of AXIN2 and WIF1 correlates with constitutive activation of AKT/GSK3β and WNT/β-CATENIN signaling in NHL clinical samples
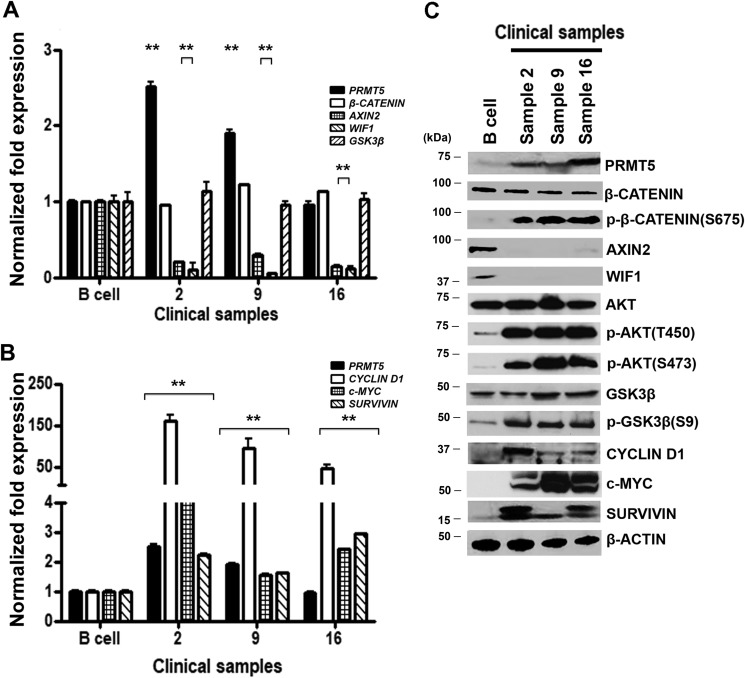
Our findings using three different types of patient-derived lymphoma cell lines clearly showed that elevated levels of PRMT5 trigger suppression of WNT/β-CATENIN antagonists, which in turn contribute to constitutive activation of AKT/GSK3β and WNT/β-CATENIN proliferative signaling. Based on these results, we wanted to determine whether these molecular changes also occur in primary NHL tumor cells (Fig. 7). Both total mRNA and protein extracts were prepared from either control normal CD19+ B lymphocytes or three NHL clinical samples, and expression of PRMT5 as well as AXIN2 and WIF1 was evaluated. Although PRMT5 mRNA levels were either unchanged or slightly increased by 1.9–2.5-fold (p < 10−2) in two of the three primary tumors compared with normal control B cells, PRMT5 protein expression was noticeably higher in all three primary tumors (Fig. 7, A and C). In contrast, expression of AXIN2 and WIF1 mRNAs was suppressed 3.3–6.7-fold (p < 10−2 to 10−3) and 8.3–20-fold (p < 10−2) in primary tumor cells, respectively. Consistent with these results, AXIN2 and WIF1 protein expressions were also highly suppressed in primary tumor cells.
Figure 7.
PRMT5 overexpression correlates with enhanced WNT/β-CATENIN and AKT/GSK3β signaling in NHL clinical samples. Levels of PRMT5, β-CATENIN, AXIN2, WIF1, GSK3β (A) and CYCLIN D1, c-MYC, and SURVIVIN (B) mRNAs were measured in normal B cells and NHL patient samples (2, 9, and 16) by real-time RT-PCR using gene-specific primers and probe sets. This experiment was conducted using three biological replicates with three technical replicates each, and the values represent the mean ± S.D. 18S rRNA was used as internal control. C, RIPA extracts (40 μg) prepared from normal B cells and NHL patient samples were analyzed by immunoblotting using the indicated antibodies, and β-ACTIN was detected to show equal loading.
Because both AKT/GSK3β and WNT/β-CATENIN signalings were active in patient-derived lymphoma cell lines, we investigated the phosphorylation status of AKT, GSK3β, and β-CATENIN (Fig. 7C). AKT activation as measured by phosphorylation of Thr-450 and Ser-473 was significantly increased in primary tumor cells compared with normal B cells and correlated with enhanced phosphorylation and inactivation of GSK3β (Ser-9). Next, we checked levels of transcriptionally active phospho-β-CATENIN (Ser-675) as well as WNT/β-CATENIN target genes. We found that the levels of phospho-β-CATENIN (Ser-675) were elevated in all three clinical samples, indicating that WNT/β-CATENIN proliferative signaling is activated. We also discovered that except for CYCLIN D1, which showed extremely high mRNA levels that ranged from a 47- to 162-fold (p < 10−3) increase compared with normal B cells, c-MYC and SURVIVIN mRNAs levels were 1.6–4.6-fold (p < 10−3) and 1.6–2.9-fold higher in primary tumors cells, respectively (Fig. 7B). In line with this increase in mRNA levels, CYCLIN D1, c-MYC, and SURVIVIN protein expression was also elevated in all three clinical samples compared with normal B cells. These results support our findings with patient-derived lymphoma cell lines and further confirm the positive correlation between enhanced PRMT5 expression, constitutive AKT/GSK3β and WNT/β-CATENIN signaling, and elevated expression of CYCLIN D1, c-MYC, and SURVIVIN prosurvival factors.
PRMT5 inhibition induces mouse primary lymphoma cell death through inactivation of AKT/GSK3β and WNT/β-CATENIN proliferative signaling
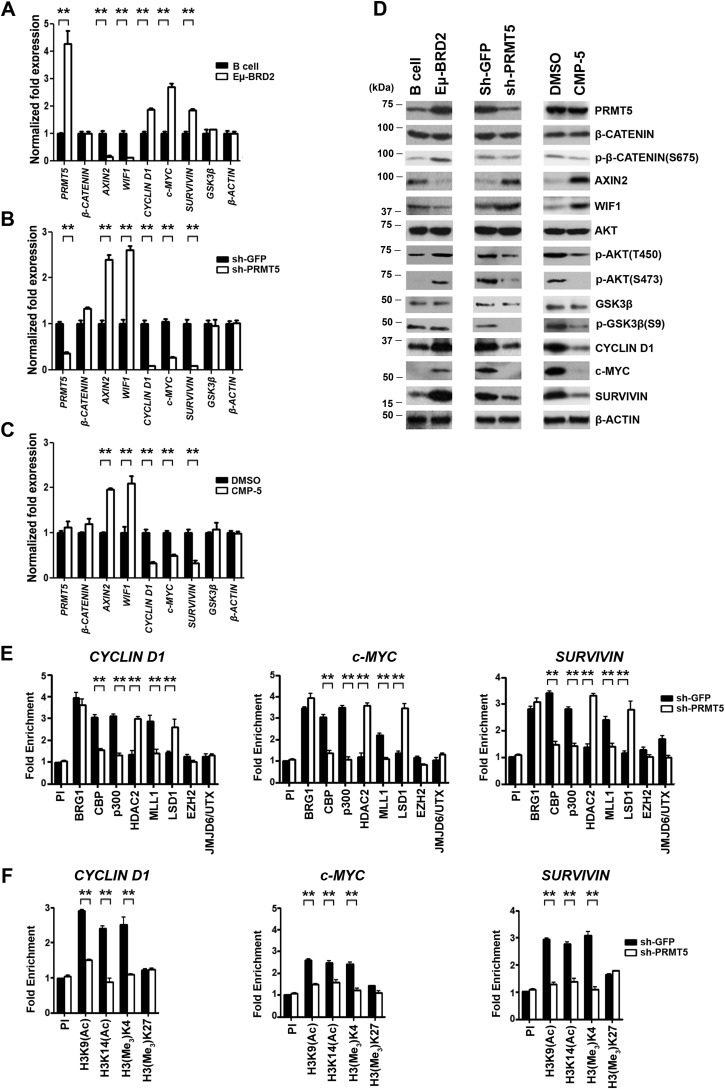
We have shown that PRMT5 promotes lymphoma cell growth and survival by suppressing expression of WNT antagonists and enhancing expression of WNT/β-CATENIN target genes in three different types of patient-derived lymphoma cell lines as well as clinical samples. In light of these results, we sought to determine whether the mechanism by which PRMT5 enables human lymphoma cells to proliferate was also used in mouse primary lymphoma cells derived from the Eμ-BRD2 transgenic mouse (35). Transcriptome analysis of Eμ-BRD2 primary tumor cells revealed that the gene expression program induced in this type of murine lymphoma is similar to human DLBCL. Therefore, we checked expression of Prmt5, Axin2, Wif1, and WNT/β-CATENIN target genes, and we also evaluated the phosphorylation status of AKT, GSK3β, and β-CATENIN (Fig. 8). Consistent with our findings in patient-derived cell lines and clinical samples, Prmt5 mRNA was increased 4.3-fold (p < 10−3), and PRMT5 protein levels were also elevated in Eμ-BRD2 primary tumor cells compared with normal B cells (Fig. 8, A and D, left panel). Similarly, Cyclin D1 (1.9-fold, p < 10−3), c-MYC (2.7-fold, p < 10−3), and Survivin (1.8-fold, p < 10−3) mRNA levels were increased in mouse primary lymphoma cells compared normal B cells, and the same was true for their protein levels. When we examined expression of WNT/β-CATENIN antagonists, we found that both Axin2 and Wif1 mRNAs were suppressed 7.7- and 8.3-fold, respectively. However, at the protein level, AXIN2 was suppressed more significantly in primary lymphoma cells compared with normal B cells, whereas WIF1 did not show only any decrease in Eμ-BRD2 lymphoma cells. When we evaluated phosphorylation of AKT, GSK3β, and β-CATENIN, we found that whereas AKT (Thr-450) phosphorylation was only moderately enhanced, both AKT (Ser-473) and β-CATENIN (Ser-675) phosphorylations were significantly induced in lymphoma cells. In contrast, phosphorylation of GSK3β (Ser-9) was unchanged in normal and mouse primary Eμ-BRD2 tumor cells.
Figure 8.
PRMT5 knockdown or inhibition results in Axin2 and Wif1 derepression and inactivation of WNT/β-CATENIN and AKT/GSK3β signaling in mouse primary lymphoma cells. A, real-time RT-PCR was performed on total RNA from normal mouse B cells and Eμ-BRD2 cells, and steady-state mRNA levels of the indicated target genes were measured using gene-specific primer sets and probes. B, Eμ-BRD2 cells were activated by adding recombinant human interleukin-4 (15 ng/ml) and goat anti-human IgG + IgM (15 μg/ml) before infection with lentivirus that expresses either control shGFP–GFP or shPRMT5–GFP. Total RNA was isolated 72 h post-infection, and the mRNA levels of the indicated genes were measured by real-time RT-PCR using gene-specific primers and probe sets. C, total RNA was isolated from Eμ-BRD2 cells treated with either control DMSO or CMP-5, and mRNA levels of the indicated target genes were measured 48 h post-treatment as described in B. D, RIPA extracts (40 μg) were prepared from the indicated cells and analyzed by immunoblotting using the specified antibodies. β-ACTIN serves as a loading control. E and F, ChIP assays were carried out using cross-linked chromatin from activated Eμ-BRD2 cells infected with lentivirus that expresses either shGFP or shPRMT5. The values in each graph were generated from two biological replicates with three technical replicates and are plotted as mean ± S.D.
To verify whether PRMT5 controls expression of Cyclin D1, c-Myc, and Survivin through modulation of AKT/GSK3β and WNT/β-CATENIN signaling, we proceeded to inhibit PRMT5 using either shPRMT5-mediated knockdown (Fig. 8, B and D, middle panel) or CMP-5–mediated inhibition (Fig. 8, C and D, right panel). Prmt5 knockdown resulted in a 2.4- and 2.6-fold (p < 10−4) derepression of Axin2 and Wif1 mRNA levels, respectively, which was consistent with the observed increase in their protein expression compared with control shGFP (Fig. 8D). In addition, transcription of WNT/β-CATENIN target genes was reduced 11.6-fold (p < 10−3) for both Cyclin D1 and Survivin and 3.8-fold (p < 10−3) for c-Myc. At the protein level, there was a decrease in WNT/β-CATENIN target gene expression, which was accompanied by dephosphorylation of AKT (Ser-473) and GSK3β (Ser-9). Similar changes in mRNA and protein expression of WNT/β-CATENIN antagonists and target genes were observed when primary lymphoma cells were treated with the PRMT5 inhibitor (Fig. 8, C and D, right panel). Both Axin2 and Wif1 mRNAs were derepressed 2-fold (p < 10−3), which was clearly reflected by the increase in their protein expression. Furthermore, Cyclin D1 and Survivin mRNA levels were inhibited by 3.1-fold, whereas c-Myc mRNA was reduced by 2-fold in treated primary lymphoma cells compared with nontreated cells. These negative changes in WNT/β-CATENIN target gene mRNA levels were detected at the protein level as well. When phosphorylation of AKT and GSK3β was assessed, there was a substantial drop in phospho-AKT (Thr-450 and Ser-473) and phospho-GSK3β (Ser-9).
To determine whether the changes in WNT/β-CATENIN target gene expression were governed by the same set of chromatin remodeling enzymes, we carried out ChIP experiments to monitor binding of co-activator and co-repressor enzymes in the promoter region of Cyclin D1, c-Myc, and Survivin (Fig. 8, E and F). Recruitment of CBP, p300, and MLL1 to the Cyclin D1 promoter region was reduced by 2–2.4-fold (p < 10−3) in Prmt5 knockdown cells, and association of HDAC2 and LSD1 was enhanced by 2.1- and 1.9-fold (p < 10−3), respectively (Fig. 8E). At the c-Myc promoter, co-activator binding was reduced by 2.1-fold (p < 10−2) for CBP, 3.2-fold (p < 10−3) for p300, and 2.2-fold (p < 10−3) for MLL1. In addition, co-repressor recruitment was stimulated by 3- and 2.5-fold (p < 10−3) for HDAC2 and LSD1, respectively. Similar recruitment changes were observed at the Survivin promoter, where association of co-activators decreased by 1.7–2.3-fold (p < 10−3), and recruitment of co-repressors was enhanced by 2.3-fold (p < 10−3). Consistent with these molecular changes, we found that there was a decrease in histone H3 acetylation (H3K9Ac and H3K14Ac) and methylation (H3Me3K4) in the promoter region of all three WNT/β-CATENIN target genes (Fig. 8F). At the Cyclin D1 promoter, H3K9/K14 acetylation and H3K4 methylation marks were reduced by 2–2.8-fold (p < 10−3), whereas at the c-Myc and Survivin promoters, a decrease in both acetylation and methylation marks varied between 1.6- and 2-fold (p < 10−3) and 2- and 3-fold (p < 10−3), respectively. These fluctuations in epigenetic marks were specific, because neither recruitment of EZH2 nor enrichment of its induced H3(Me3)K27 mark showed any changes in control shGFP and shPRMT5 knockdown cells.
Discussion
The impact of PRMT5 overexpression on cancer cell growth and survival has been documented in several cancer cell types, and its mechanism of action appears to be diverse enough to modulate a wide range of growth regulatory pathways (16). We have previously explored the role played by PRMT5 in controlling cancer cell growth using different patient-derived lymphoma cell lines, and we were able to determine that PRMT5 acts upstream of the CYCLIN D1/CDK4/CDK6 and RB/E2F pathway (12). In light of these findings, which showed that PRMT5 knockdown or inhibition results in CYCLIN D1 down-regulation and induced cell death, and the fact that our ChIP-Seq and real-time ChIP results indicated that PRMT5 is enriched in the promoter region of WNT/β-CATENIN antagonist genes, we investigated the effect of PRMT5 inhibition on WNT/β-CATENIN signaling and downstream target gene expression.
Our results show that there is a positive correlation between PRMT5 overexpression and increased levels of CYCLIN D1, c-MYC, and SURVIVIN protein levels in three different lymphoma cell types (Fig. 1). This correlation was confirmed using NHL clinical samples and mouse primary lymphoma cells (Figs. 7 and 8). When PRMT5 was either knocked down or inhibited, levels of all three WNT/β-CATENIN target genes decreased significantly both at the mRNA and protein levels (Fig. 1 and Fig. S1). In addition, our ChIP-Seq results showed that PRMT5 binds to the promoter region of three WNT/β-CATENIN antagonist genes, AXIN1, AXIN2, and WIF1 (Fig. 2 and Fig. S2). Validation by real-time ChIP assays confirmed our findings and prompted us to examine expression of all three PRMT5 target genes in patient-derived lymphoma cell lines, NHL clinical samples, and mouse primary lymphoma cells. We discovered that although all three WNT/β-CATENIN antagonists were repressed at the mRNA level, only AXIN2 and WIF1 were suppressed at the protein level, suggesting that there are other mechanisms involved in regulating AXIN1 expression. To evaluate the role of PRMT5 in transcriptional repression of AXIN1, AXIN2, and WIF1, we inhibited its methyltransferase activity by either shRNA-mediated knockdown (Fig. 2, E–G) or CMP-5 treatment (Fig. S3). We found that PRMT5 recruitment to the promoter region of WNT/β-CATENIN antagonist genes was abolished under both treatment regimens. Consequently, AXIN2 and WIF1 were derepressed at the mRNA and protein levels; however, AXIN1 showed only a modest increase at the mRNA level (<2-fold) and no change at the protein level, providing more evidence for the presence of other mechanisms involved in its regulation.
In fact, previous work showed that AXIN1 is down-regulated in lung and colorectal cancer cells through promoter DNA hypermethylation, raising the possibility that DNA methylation is involved in regulating AXIN1 gene expression (36, 37). Another interesting point worth mentioning is that a recent study in colorectal cancer argued that there are differential functional roles of AXIN1 and AXIN2 in the β-CATENIN destruction complex (38). The results from this investigation revealed that AXIN1 does not affect destruction complex-induced β-CATENIN degradation, whereas AXIN2 depletion impairs the function of the β-CATENIN destruction complex, implying that AXIN2 plays a more important role in regulating WNT/β-CATENIN signaling in colorectal cancer. Our findings show that AXIN2 becomes derepressed upon PRMT5 inhibition, indicating that this might enhance the function of the β-CATENIN destruction complex and lead to decreased levels of β-CATENIN. However, our results did not show any changes at the level of steady-state levels of β-CATENIN or phospho-β-CATENIN (Ser-675) when PRMT5 was either knocked down or inhibited in patient-derived cell lines (Fig. 1E) or mouse primary lymphoma cells (Fig. 8D). There was also no change in the amount of total β-CATENIN when AXIN2 was re-expressed in lymphoma cells (Fig. 3B). Similarly, there was no decrease in the level of chromatin-bound β-CATENIN as measured by real-time ChIP assays (Fig. 5), suggesting that down-regulation of WNT/β-CATENIN signaling in lymphoma cells is effected through other means.
Prior work has shown that AKT is hyperactivated in DLBCL patients (39) and that AKT inactivates GSK3β through serine 9 phosphorylation to promote mTOR signaling and translation of WNT/β-CATENIN target genes (31, 32, 34). In light of the cross-talk that exists between WNT/β-CATENIN and AKT/GSK3β signaling, we assessed the contribution of AXIN2 and WIF1 to their regulation and expression of CYCLIN D1, c-MYC, and SURVIVIN. Ectopic expression of AXIN2 and/or WIF1 showed that although each antagonist alone had a moderate effect on AKT/GSK3β signaling and expression of WNT/β-CATENIN target genes, their co-expression had a more dramatic effect (Fig. 3, A and B). These results indicate that both AXIN2 and WIF1 work in concert to inactivate AKT, which in turn leads to GSK3β activation and down-regulation of WNT/β-CATENIN target gene expression. Coincidentally, when we tested the effect of derepressed and excreted WIF1 from the cell culture media of lymphoma cells, which were either knocked down for PRMT5 expression or inhibited with CMP-5, we discovered that WIF1 was capable of inactivating AKT/GSK3β signaling and shutting down CYCLIN D1, c-MYC, and SURVIVIN expression (Fig. S6 and supporting Experimental procedures). However, when WIF1 was neutralized using a specific WIF1 antibody, its inhibitory effect was abolished, highlighting the specificity of its biological function (Fig. S6, B and D). Furthermore, when we evaluated the effect of AKT inactivation on WNT/β-CATENIN target gene expression, we found that AKT inhibition was sufficient to bring about down-regulation of CYCLIN D1, c-MYC, and SURVIVIN (Fig. 3, D and E). Similar results were also observed when PRMT5 was either knocked down or inhibited, although the kinetics for GSK3β dephosphorylation/reactivation and suppression of WNT/β-CATENIN target gene expression were much more rapid using CMP-5 compared with shPRMT5 (Fig. 4 and Fig. S4).
The protein expression profile of AXIN2 and WIF1 was analyzed in different patient-derived lymphoma cell lines in the presence and absence of PRMT5 activity, and it indicates that the mechanism by which PRMT5 promotes lymphoma cell growth and survival is conserved among different lymphoma cell types. In addition, when we examined the mRNA and protein levels of both WNT/β-CATENIN antagonists and downstream target genes in NHL clinical samples (Fig. 7) and mouse primary lymphoma cells (Fig. 8), we found the same inverse correlation in their expression as determined for the patient-derived cell lines. In addition, there was hyperphosphorylation of β-CATENIN (Ser-675), AKT (Thr-450/Ser-473), and GSK3β (Ser-9), which was consistent with our findings with the three different lymphoma cell types. The positive correlation between PRMT5 overexpression and hyperactivation of AKT/GSK3β and WNT/β-CATENIN signaling in lymphoma cell lines, clinical samples, and mouse primary lymphoma cells and the loss of AKT/GSK3β phosphorylation and CYCLIN D1, c-MYC, and SURVIVIN expression upon PRMT5 knockdown or inhibition argue that PRMT5 controls a complex network of interacting growth regulatory pathways. Through suppression of AXIN2 and WIF1, PRMT5 is able to promote AKT/GSK3β signaling, which advertently leads to enhanced transcription and translation of CYCLIN D1, c-MYC, and SURVIVIN.
Regulation of chromatin structure through histone modification is one of the major mechanisms by which nucleosomal DNA is either made more or less accessible during gene transcription. Upon translocation to the nucleus, β-CATENIN complexes with one of the members of the TCF of transcription factors and recruits many transcriptional co-activators to the promoter region of WNT/β-CATENIN target genes. We have monitored binding of both co-activators as well as co-repressors to the promoter region of all three WNT/β-CATENIN target genes in three different lymphoma cell lines (Fig. 6 and Fig. S5) as well as mouse primary lymphoma cells (Fig. 8, E and F). We have shown that in the presence of PRMT5, there is preponderant recruitment of histone acetyltransferases, CBP and p300, and histone H3K4-specific methyltransferase MLL1. This is consistent with the enriched levels of both histone H3K9/K14 acetylation and H3K4 methylation, which are known to be associated with activated transcription. However, when PRMT5 was knocked down, recruitment of co-activators was lost and replaced with enhanced recruitment of co-repressors, HDAC2 and LSD1, which can deacetylate and demethylate histone H3 marks, respectively.
To gain further insight into how differential recruitment of co-activators and co-repressors was effected, we measured β-CATENIN recruitment to the promoter region of CYCLIN D1, c-MYC, and SURVIVIN in both normal B cells and lymphoma cell lines (Fig. 5A). Unexpectedly, we found that despite increased β-CATENIN recruitment to the promoter region of WNT/β-CATENIN target genes in lymphoma cell lines, its binding was unchanged when PRMT5 was either knocked down or inhibited (Fig. 5, B and C). It is conceivable that the dynamic changes that occur in the promoter region of all three WNT/β-CATENIN target genes are due to direct AKT-mediated phosphorylation of MLL1 and or CBP/p300. Alternatively, AKT might lead to phosphorylation of subunits within co-activator complexes that include MLL1 and CBP/p300. There is supporting evidence for both scenarios; for instance, previous studies have shown that phosphorylation of retinoblastoma-binding protein 5 (RbBP5), a component of the MLL1-containing COMPASS complex, might be the way by which PI3K/AKT stimulates MLL1 activity (40). In another investigation, using a rat model of hormone-induced prostate cancer, it was demonstrated that PI3K/AKT signaling activates MLL1, which in turn leads to H3K4 hypermethylation and up-regulation of 32 genes (41). Analysis of the mechanism by which MLL1 is switched on in this case revealed that upon PI3K/AKT activation, MLL1 undergoes Taspase-1–dependent cleavage into N- and C-terminal fragments, which associate to form the catalytically active form of MLL1N320/C180. More interestingly, evaluation of H3K4 trimethylation revealed that this mark was enriched near the start of transcription for several target genes involved in signal transduction, cell cycle regulation, and oncogenesis. This is consistent with our results, which show that AKT is activated in lymphoma cell lines, clinical samples, and mouse primary lymphoma cells that express elevated levels of PRMT5 and that MLL1 recruitment and enrichment of its induced H3(Me3)K4 mark are increased in the promoter region WNT/β-CATENIN target genes (Fig. 6 and Fig. S5). However, when PRMT5 is either inhibited or knocked down, AKT becomes inactive resulting in reduced MLL1 recruitment and H3(Me3)K4 methylation in the promoter region of CYCLIN D1, c-MYC, and SURVIVIN. A more detailed investigation, which is beyond the scope of this study, is required to determine whether AKT activates MLL1 either through Taspase-1–mediated cleavage or through phosphorylation of a subunit within the MLL complex in lymphoma cells.
Interaction between AKT and CBP/p300 has been documented, and it is clear that the outcome is not always positive as illustrated by CBP inactivation due to its AKT-mediated phosphorylation on threonine 1871, which results in a decrease in H3K18 acetylation and inactivation of target genes (42). However, PI3K/AKT signaling has also been shown to promote association between Smad3 and CBP and to enhance its CBP-mediated acetylation, which leads to target gene activation (43). Furthermore, phosphorylation of p300 on serine 1384 by AKT has been shown to enhance its acetyltransferase activity and results in increased acetylation of its substrates (44). Because p300 can act on both histone and nonhistone proteins, it is clear that all downstream target proteins will be impacted as evidenced by the p300-mediated ADA3 (Alteration/Deficiency in Activation 3) acetylation, which results in stimulation of cell cycle progression and inhibition of apoptosis (44). Another mechanism by which p300 co-activates gene expression is through its direct acetylation of promoter histones. Although we cannot rule out the possibility that CBP and p300 acetylate other components within the transcriptional complex assembled through recruitment of β-CATENIN, our findings show positive correlation between increased binding of CBP/p300 and enhanced histones H3K9 and K14 acetylation in the presence of PRMT5 (Fig. 6). We have also observed that upon PRMT5 inhibition or knockdown, both AKT inactivation and loss of CBP/p300 recruitment were noted in the promoter region of CYCLIN D1, c-MYC, and SURVIVIN. These changes in phosphorylation and acetylation of probably both histone and nonhistone proteins could provide an opportunity for antagonistic enzymes such as HDAC2 and LSD1 to be recruited to the promoter region of WNT/β-CATENIN target genes and to change the epigenetic landscape from a transcriptionally active to an inactive form. Therefore, the ability of PRMT5 to activate major kinases involved in regulating cell growth and proliferation, so that they can modulate the activity of other epigenetic programmers, is another way by which this versatile arginine methyltransferase reprograms the epigenome of cancer cells.
Experimental procedures
Plasmid construction
PRMT5 knockdown was achieved as described previously (12) using lentiviral constructs for expression of either control GFP (pLenti X2 DEST/shGFP), shPRMT5-1 (pLenti X2 DEST/shPRMT5-1), which was used to infect patient-derived lymphoma cell lines, or GFP-shPRMT5-1 (pLenti-CMV-GFP-DEST/shPRMT5-1), which was used to infect mouse primary Eμ-BRD2 lymphoma cells. Plasmids pLenti X2 DEST/shPRMT5-1 and pLenti-CMV-GFP-DEST/shPRMT5-1 were constructed using sense (5′-GATCCCGCCCAGTTTGAGATGCCTTATGTGTGCTGTCCATAAGGCATCTCAAACTGGGCTTTTTGGAAA-3′) and antisense (5′-AGCTTTTCCAAAAAGCCCAGTTTGAGATGCCTTATGGACAGCACACATAAGGCATCTCAAACTGGGCGG-3′) oligonucleotides, which were designed to include sticky ends compatible with BglII and HindIII, and were cloned into the BglII–HindIII-linearized pENTER/pTER+ vector. Positive clones were identified by EcoRI or EcoRV digestion and confirmed by DNA sequencing. Next, plasmid pENTER/pTER+/shPRMT5-1 was individually recombined into either the pLenti X2 DEST or pLenti-CMV-GFP-DEST vector, as described previously (12, 45), and positive clones were identified by EcoRV digestion in the case of pLenti X2 DEST/shPRMT5-1 or by EcoRI digestion in the case of pLenti-CMV-GFP-DEST/shPRMT5, followed by DNA sequencing. Plasmids pCMV6-entry, pCMV6-entry-AXIN2, and pCMV-entry-WIF1 were purchased from Origene Technologies, Inc., Rockville, MD.
Cell culture, B cell isolation, infection, and transfection assays
Patient-derived lymphoma cell lines and mouse primary Eμ-BRD2 lymphoma cells were grown in RPMI 1640 medium supplemented with 10% FBS and 1 mm sodium pyruvate. Normal human B cells were isolated from tonsilar tissues obtained from Nationwide Children's Hospital through the Cooperative Human Tissue Network, as described previously (10, 11). To activate resting B cells, purified B lymphocytes were resuspended at a density of 5 × 106 cells/ml in RPMI 1640 medium containing 10% FBS, and then cells were induced to enter the cell cycle by adding 15 ng/ml recombinant human interleukin-4 (Thermo Fisher Scientific, Waltham, MA) and 15 μg/ml of goat anti-human IgG + IgM (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA). To generate lentiviral particles that express either control shGFP or shPRMT5 RNA, 15 μg of either control plasmid pLenti X2 DEST/shGFP, plasmid pLenti X2 DEST/shPRMT5-1, or pLenti-CMV-GFP-DEST/shPRMT5 was transfected along with 15 μg of pLP1, 6 μg of pLP2, and 3 μg of pVSG into 5 × 106 293T cells, which were seeded 1 day before transfection. Approximately 72 h post-transfection, control shGFP or shPRMT5 lentiviral supernatant was harvested, clarified by centrifugation at 2,000 rpm for 4 min, and used to infect patient-derived lymphoma cell lines or mouse primary Eμ-BRD2 lymphoma cells as described previously (12). Briefly, 1 × 105 lymphoma cells were resuspended in RPMI 1640 medium containing 10% FBS and were infected with 400 μl of either shGFP or shPRMT5 lentiviral supernatant, which was pre-incubated with 10 μg of Polybrene (Sigma). Total RNA, whole-cell extract, and cross-linked chromatin were prepared 72 h post-infection.
To determine the effect of PRMT5 inhibition on WNT/β-CATENIN antagonist and target gene expression, an equal number (8 × 105) of lymphoma cells was treated with either control DMSO or CMP-5 (50 μm), and total RNA, whole-cell extract, and cross-linked chromatin were prepared 48 h post-treatment. To evaluate the effect of AXIN2 and WIF1 re-expression on AKT/GSK3β and WNT/β-CATENIN signaling, ∼2 μg of either plasmid pCMV6-entry, pCMV6-entry-AXIN2, pCMV-entry-WIF1, or pCMV6-entry-AXIN2 and pCMV-entry-WIF1 was mixed with 100 μl of Amaxa Cell Line Nucleofector Kit V (catalog no. VCA-1003) before electroporation into 1 × 106 lymphoma cells as specified by the manufacturer (Lonza Co., Allendale, NJ). Total RNA and whole-cell extract were prepared 72 h post-transfection.
Cell proliferation and fluorescence activated cell sorting (FACS)
To evaluate the impact of PRMT5 inhibition on lymphoma cell growth and proliferation, an equal number of lymphoma cells (8 × 105) was treated with either control DMSO or CMP-5 (50 μm), and viable cells were counted every 2 days for 6 days using trypan blue dye exclusion. To determine the percent of lymphoma cells undergoing apoptosis, control untreated lymphoma cells or lymphoma cells treated with either DMSO or CMP-5 were harvested 72 h post-treatment and were stained with anti-annexin V antibody and propidium iodide as specified by the manufacturer (BD Biosciences). Next, samples were analyzed on a Beckman Coulter FC500 flow cytometer.
Lymphoma clinical samples and animal studies
All studies using patient lymphoma samples, which had no patient identifiers, abide by the declaration of Helsinki principles and were approved by the Ohio State University Comprehensive Cancer Center Institutional Review Board (IRB protocol no. 1997CO194) and conducted in agreement with the approved guidelines (IBC protocol no. 2006R0017-R1-AM6). Similarly, all animal studies were performed in compliance with guidelines approved by the Federal and the Ohio State University Institutional Animal Care and Use Committee (IACUC protocol no. 2009A0094-R3).
RT-PCR and ChIP assays
Total RNA was isolated using TRIzol reagent (Thermo Fisher Scientific, Waltham, MA), and real-time RT-PCR was performed as described previously (12). To measure mRNA levels of target genes, real-time PCR was carried out using the Applied Biosystems TaqMan assay in a 10-μl reaction, as described previously (10). To normalize mRNA levels, levels of 18S rRNA were measured in both control and test cell lines using 1× pre-mixed 18S primer/probe set (Applied Biosystems, Inc., Foster City, CA). To monitor recruitment to target genes, ChIP assays were performed using cross-linked chromatin from either normal or transformed B cells, and when PRMT5 inhibition was performed, cross-linked chromatin was prepared from either untreated cells or treated lymphoma cells, as described previously (12). Chromatin was analyzed by agarose gel electrophoresis to ensure that DNA fragment size did not exceed 500 bp, and chromatin samples were pre-cleared with 30 μl of protein A magnetic beads, which were treated overnight with blocking buffer (0.2 mg/ml sheared salmon sperm DNA, 0.5 mg/ml BSA) before incubation with either pre-immune or immune antibodies. The primer sets and probes used in real-time RT-PCR assays are listed in Table 1, and the primer sets and probes used in real-time ChIP assays are listed in Table 2.
Table 1.
Real time RT-PCR primer and probe sets
| Primer | Forward sequence | Reverse sequence | Probe |
|---|---|---|---|
| PRMT5 | 5′-TATGTGGTACGGCTGCACA3-′ | 5′-TGGCTGAAGGTGAAACAGG3-′ | 31 |
| AXIN1 | 5′-AGCTCTCCGAGACAGAGACAA-3′ | 5′-ACAACGATGCTGTCACACG-3′ | 6 |
| AXIN2 | 5′-GCTAGGAGTGCGTTCATGGT-3′ | 5′-AGGGACGTAGTGCAAAGCAT-3′ | 8 |
| WIF1 | 5′-CCAGGGGAGCCTCTGTTCAA-3′ | 5′-TTGGGTTCATGGCAGGTT-3′ | 76 |
| β-CATENIN | 5′-TTAAAAAGCCAGTTTGGGTAAAA-3′ | 5′-CCCACCCTACCAACCAAGT-3′ | 13 |
| β-ACTIN | 5′-GGTAGACGCGATCTGTTGG-3′ | 5′-GGCATGGAATCAACCTCAAC-3′ | 6 |
| CYCLIN D1 | 5′-GAAGATCGTCGCCACCTG-3′ | 5′-GACCTCCTCCTCGCACTTCT-3′ | 67 |
| c-MYC | 5′-CACCAGCAGCGACTCTGA-3′ | 5′-GATCCAGACTCTGACCTTTTG-3′ | 34 |
| SURVIVIN | 5′-GCCCAGTGTTTCTTCTGCTT-3′ | 5′-CCGGACGAATGCTTTTTATG-3′ | 11 |
| AKT | 5′-GGCTATTGTGAAGGAGGGTTG-3′ | 5′-TCCTTGTAGCCAATGAAGGTG-3′ | 69 |
| GSK3β | 5′-GACATTTCACCTCAGGAGTGC-3′ | 5′-GTTAGTCGGCAGTTGGTGT-3′ | 67 |
| Prmt5 | 5′-GCTGTCACCTGAGTGTCTGG-3′ | 5′-GATGCTCACGCCATCATCT-3′ | 99 |
| Axin1 | 5′-GGGCCCCCTCAAGTAGAC-3′ | 5′-CCCTCCAAGATCCATACCTG-3′ | 13 |
| Axin2 | 5′-CTGGTGTGGGTCGCTACAG-3′ | 5′-TGACACTGCTGATGGTGGTAG-3′ | 77 |
| Wif1 | 5′-TTAAGTGAAGGCGTGTGTCG-3′ | 5′-GCAGACACTGCAATAAGAGG-3′ | 40 |
| β-Catenin | 5′-TTCCTATGGGAACAGTCGAAG-3′ | 5′-TTGTATTGTTACTCCTCGACCAAA-3′ | 93 |
| β-Actin | 5′-GCAAACATCCCCAAATT-3′ | 5′-TTTCATGGATACTTGGAATGACTA-3′ | 5 |
| Cyclin D1 | 5′-CTCCTCTTCGCACTTCTGCT-3′ | 5′-GAGATTGTGCCATCCATGC-3′ | 67 |
| c-Myc | 5′-TCTTCCTCATCTTCTTGCTCTTC-3′ | 5′-CCTAGTGCTGCATGAGGAGA-3′ | 77 |
| Survivin | 5′-CCTGGCCCACAAGTATCACTA-3′ | 5′-GGACTTAGGGAACAAAGGAACC-3′ | 56 |
| Gsk3β | 5′-CAAGAAGAGCCATCATGTCG-3′ | 5′-TGGTTACCTGCTGCCATCT-3′ | 10 |
Table 2.
Real-time ChIP primer and probe sets
| Primer | Forward sequence | Reverse sequence | Probe |
|---|---|---|---|
| AXIN1 | 5′-ACTCCTGGGAGCCTGCTT-3′ | 5′-CAGTACATCGGAGGGCAGTC-3′ | 81 |
| AXIN2 | 5′-TCTTGCCTTCCTCTCACCTT-3′ | 5′-CACACTTTTCGGGGTTGG-3′ | 24 |
| WIF1 | 5′-CGGAGGGTCAGGTACAGCTA-3′ | 5′-CCCTGATAACCCGGCAGT-3′ | 81 |
| CYCLIN D1 | 5′-CGGGCTTTGATCTTTGCTTA-3′ | 5′-TCTGCTGCTCGCTGCTACT-3′ | 1 |
| c-MYC | 5′-GAAATTGGGAACTCCGTGTG-3′ | 5′-CTAGGGCGAGAGGGAGGTT-3′ | 53 |
| SURVIVIN | 5′-AAGGAGGAGTTTGCCCTGAG-3′ | 5′-GGGCCACTACCGTGATAAGA-3′ | 24 |
| Cyclin D1 | 5′-CTAGCTGTCCTCCTGTCCAGA-3′ | 5′-GGGCTTCTTTCCCTAAGAGG-3′ | 10 |
| c-Myc | 5′-TCATGCTGCGCTATTACTGTTT-3′ | 5′-CCTCTGCTTTGGGAACTCG-3′ | 63 |
| Survivin | 5′-GGAGGACCCTCTTAGGGAAA-3′ | 5′-AATCGCTCGGACTGCTTCT-3′ | 48 |
ChIP-Seq library construction, data processing, and analysis
For ChIP-Seq assays, cross-linked chromatin prepared from 5 × 106 normal B, Pfeiffer, or SUDHL-2 cells was immunoprecipitated with immune anti-H3(Me2)R8, which has been characterized previously (10, 17). For all cell types, ChIP assays were carried out as described previously (10, 11). Duplicate ChIP-Seq experiments were performed using cross-linked chromatin from two different batches of cells cultured on separate days. Before initiating library construction, the immunoprecipitated DNA was checked for enrichment of H3R8 symmetric methylation on two established PRMT5 target promoters, ST7 and RBL2, in ChIP versus input samples using real-time PCR. Next, libraries were created using Illumina Hi-Seq 2000 Prep Kit (Illumina, Inc., San Diego). DNA fragments that ranged in size from 300 to 400 bp were selected by agarose gel electrophoresis after the adapter ligation step, followed by 15 amplification cycles. Once again, real-time PCR was performed to confirm enrichment of control ST7 and RBL2 promoter target sequences. The ChIP-Seq DNA libraries were then sequenced using a high-throughput Illumina GAIIx sequencer, and the DNA sequence reads were aligned to the UCSC human genome assembly HG18 using the Eland pipeline (Illumina, Inc., San Diego). For analysis, the anti-H3(Me2)R8 ChIP-Seq data from normal and transformed B cells was used to determine the genome-wide binding sites of PRMT5. First, the DNA sequence reads were aligned to the human reference genome (GRCh37) with Rsubread version 1.22.2 (46), and duplicates were marked with Picard tools version 1.94 (http://broadinstitute.github.io/picard/),3 and peak calling was performed with MACS2 version 2.1.1 (47). Consensus peaks and read counts around transcriptional start sites were generated using a series of in-house shell and R scripts utilizing NGSplot tool (48), GenomicRanges (49), and ChIPpeakAnno (50) packages.
Antibodies and Western blotting assay
Protein extraction, immunoblotting, and immunoprecipitation were performed as described previously (10, 12, 51). Briefly, whole-cell extracts were prepared in RIPA buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.5 mm DTT, 1 mm phenylmethylsulfonyl fluoride) supplemented with protease and phosphatase inhibitors (10 μg/ml benzamidine hydrochloride, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 2.25 μg/ml pepstatin A, 10 mm β-glycerophosphate, 1 mm sodium orthovanadate, 50 mm sodium fluoride). Next, proteins were separated on an 8–12% SDS-PAGE, transferred onto polyvinylidene difluoride membrane, and detected by enhanced chemiluminescence using Amersham Biosciences ECL Western blotting detection reagent (GE Healthcare). For protein detection, antibodies against BRG1, PRMT5, and its epigenetic marks were used as described previously (10). Polyclonal antibodies against CBP (sc-7300), p300 (sc-48343), CYCLIN D1 (sc-718), c-MYC (sc-40), and WIF1 (sc-25520) were purchased from Santa Cruz Biotechnology, Inc., Dallas, TX. Anti-SURVIVIN (catalog no. 2808), anti-β-CATENIN (catalog no. 8480), anti-p-β-CATENIN (Ser-675) (catalog no. 4176), anti-AKT (catalog no. 4691), anti-p-AKT (Thr-450) (catalog no. 9267), and anti-p-AKT (Ser-473) (catalog no. 4060) antibodies were purchased from Cell Signaling Technology, Danvers, MA; and anti-AXIN1 (ab131372), anti-AXIN2 (ab32197), anti-H3(Me3)K4 (ab8580), anti-H3(Me3)K9 (ab8898), anti-H3(Me3)K27 (ab6002), anti-LSD1 (ab17721), anti-JMJD6/UTX (ab50720), and anti-HDAC2 (ab32117) antibodies were purchased from Abcam, Cambridge, UK. Both anti-H3K9(Ac) (07-352) and anti-H3K14(Ac) (07-360) antibodies were purchased from EMD Millipore, Bedford, MA. The anti-MLL1 (39829) antibody was purchased from Active Motif, Carlsbad, CA, and the anti-β-ACTIN (SAB5500001) antibody was purchased from Sigma.
Statistical analysis
All real-time RT-PCR, ChIP, and immunoblotting experiments were conducted at least three times using different samples, and the results were expressed as the mean ± S.D. Paired t tests and analysis of variance followed by Sidak multiple-comparison adjusted t tests were used to generate p values for comparisons between two samples.
Author contributions
J. C. and V. K. formal analysis; J. C., V. K., and S. S. validation; J. C. and V. K. investigation; J. C., V. K., and S. S. visualization; R. A. B. and S. S. resources; R. A. B. and S. S. supervision; R. A. B. and S. S. funding acquisition; S. S. conceptualization; S. S. writing-original draft; S. S. project administration; S. S. writing-review and editing; S. S. prepared the first draft of the manuscript, which was reviewed by all co-authors, and prepared the final draft of both manuscript and figures.
Supplementary Material
Acknowledgments
We thank A. N. Imbalzano for providing lentiviral expression vectors, G. Denis for kindly providing mouse primary Eμ-BRD2 lymphoma cells and advice on their use, and O. H. Gulcin for help with ChIP-Seq data analysis.
This work was supported by Leukemia and Lymphoma Society Award MCL7001-18 (to R. A. B.) and National Priorities Research Program Grant NPRP8-617-3-131 from the National Research Fund (a member of Qatar Foundation) (to S. S). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S6, Tables S1 and S2, and supporting Experimental procedures.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- MCL
- mantle cell lymphoma
- NHL
- non-Hodgkin's lymphoma
- DLBCL
- diffuse large B-cell lymphoma
- GCB
- germinal center B cell
- ABC
- activated B-cell–like
- TCF
- T cell factor
- LEF
- lymphoid enhancer factor
- FBS
- fetal bovine serum
- PI
- pre-immune
- sh
- short hairpin
- PI3K
- phosphatidylinositol 3-kinase.
References
- 1. Kuppers R., Klein U., Hansmann M. L., and Rajewsky K. (1999) Cellular origin of human B-cell lymphoma. N. Engl. J. Med. 341, 1520–1529 10.1056/NEJM199911113412007 [DOI] [PubMed] [Google Scholar]
- 2. Stevenson F. K., Sahota S. S., Ottensmeier C. H., Zhu D., Forconi F., and Hamblin T. J. (2001) The occurrence and significance of V gene mutations in B cell-derived human malignancy. Adv. Cancer Res. 83, 81–116 10.1016/S0065-230X(01)83004-9 [DOI] [PubMed] [Google Scholar]
- 3. Nogai H., Dörken B., and Lenz G. (2011) Pathogenesis of non-Hodgkin's lymphoma. J. Clin. Oncol. 29, 1803–1811 10.1200/JCO.2010.33.3252 [DOI] [PubMed] [Google Scholar]
- 4. Swerdlow S. H., Campo E., Seto M., and Müller-Hemrelink H. K. (2008) in WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues (Swerdlow S. H., Campo E., Harris N. L., Jaffe E. S., Pileri S. A., Stein H., Thiele J., and Vardiman J. W., eds) pp. 229–232. IARC Publishing Group, Lyon, France [Google Scholar]
- 5. Alizadeh A. A., Eisen M. B., Davis R. E., Ma C., Lossos I. S., Rosenwald A., Boldrick J. C., Sabet H., Tran T., Yu X., Powell J. I., Yang L., Marti G. E., Moore T., Hudson J. Jr., et al. (2000) Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403, 503–511 10.1038/35000501 [DOI] [PubMed] [Google Scholar]
- 6. Rosenwald A., Wright G., Chan W. C., Connors J. M., Campo E., Fisher R. I., Gascoyne R. D., Muller-Hermelink H. K., Smeland E. B., Giltnane J. M., Hurt E. M., Zhao H., Averett L., Yang L., et al. (2002) The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N. Engl. J. Med. 346, 1937–1947 10.1056/NEJMoa012914 [DOI] [PubMed] [Google Scholar]
- 7. Pezzella F., Gatter K. C., and Mason D. Y. (1989) Detection of 14;18 chromosomal translocation in paraffin-embedded lymphoma tissue. Lancet 1, 779–780 [DOI] [PubMed] [Google Scholar]
- 8. Motlló C., Grau J., Juncà J., Ruiz N., Mate J. L., Orna E., Navarro J. T., Vives S., Sancho J. M., Esteban D., Granada I., Feliu E., Ribera J. M., and Millà F. (2010) Translocation (3;8)(q27;q24) in two cases of triple hit lymphoma. Cancer Genet. Cytogenet. 203, 328–332 10.1016/j.cancergencyto.2010.08.018 [DOI] [PubMed] [Google Scholar]
- 9. Torres C. H., and Tirado C. A. (2018) Two double-hit lymphomas cases: a molecular cytogenetic approach. J. Assoc. Genet. Technol. 44, 141–145 [PubMed] [Google Scholar]
- 10. Pal S., Baiocchi R. A., Byrd J. C., Grever M. R., Jacob S. T., and Sif S. (2007) Low levels of miR-92b/96 induce PRMT5 translation and H3R8/H4R3 methylation in mantle cell lymphoma. EMBO J. 26, 3558–3569 10.1038/sj.emboj.7601794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang L., Pal S., and Sif S. (2008) Protein arginine methyltransferase 5 suppresses the transcription of the RB family of tumor suppressors in leukemia and lymphoma cells. Mol. Cell. Biol. 28, 6262–6277 10.1128/MCB.00923-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chung J., Karkhanis V., Tae S., Yan F., Smith P., Ayers L. W., Agostinelli C., Pileri S., Denis G. V., Baiocchi R. A., and Sif S. (2013) Protein arginine methyltransferase 5 (PRMT5) inhibition induces lymphoma cell death through reactivation of the retinoblastoma tumor suppressor pathway and polycomb repressor complex 2 (PRC2) silencing. J. Biol. Chem. 288, 35534–35547 10.1074/jbc.M113.510669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Alinari L., Mahasenan K. V., Yan F., Karkhanis V., Chung J. H., Smith E. M., Quinion C., Smith P. L., Kim L., Patton J. T., Lapalombella R., Yu B., Wu Y., Roy S., De Leo A., et al. (2015) Selective inhibition of protein arginine methyltransferase 5 blocks initiation and maintenance of B-cell transformation. Blood 125, 2530–2543 10.1182/blood-2014-12-619783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Karkhanis V., Hu Y. J., Baiocchi R. A., Imbalzano A. N., and Sif S. (2011) Versatility of PRMT5-induced methylation in growth control and development. Trends Biochem. Sci. 36, 633–641 10.1016/j.tibs.2011.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang Y., and Bedford M. T. (2013) Protein arginine methyltransferases and cancer. Nat. Rev. Cancer 13, 37–50 10.1038/nrc3409 [DOI] [PubMed] [Google Scholar]
- 16. Shailesh H., Zakaria Z. Z., Baiocchi R., and Sif S. (2018) Protein arginine methyltransferase 5 (PRMT5) dysregulation in cancer. Oncotarget 9, 36705–36718 10.18632/oncotarget.26404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pal S., Vishwanath S. N., Erdjument-Bromage H., Tempst P., and Sif S. (2004) Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol. Cell. Biol. 24, 9630–9645 10.1128/MCB.24.21.9630-9645.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jansson M., Durant S. T., Cho E. C., Sheahan S., Edelmann M., Kessler B., and La Thangue N. B. (2008) Arginine methylation regulates the p53 response. Nat. Cell Biol. 10, 1431–1439 10.1038/ncb1802 [DOI] [PubMed] [Google Scholar]
- 19. Cho E. C., Zheng S., Munro S., Liu G., Carr S. M., Moehlenbrink J., Lu Y. C., Stimson L., Khan O., Konietzny R., McGouran J., Coutts A. S., Kessler B., Kerr D. J., and Thangue N. B. (2012) Arginine methylation controls growth regulation by E2F-1. EMBO J. 31, 1785–1797 10.1038/emboj.2012.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Logan C. Y., and Nusse R. (2004) The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 20, 781–810 10.1146/annurev.cellbio.20.010403.113126 [DOI] [PubMed] [Google Scholar]
- 21. Willert K., and Jones K. A. (2006) Wnt signaling: is the party in the nucleus? Genes Dev. 20, 1394–1404 10.1101/gad.1424006 [DOI] [PubMed] [Google Scholar]
- 22. Klaus A., and Birchmeier W. (2008) Wnt signaling and its impact on development and cancer. Nat. Rev. Cancer 8, 387–398 10.1038/nrc2389 [DOI] [PubMed] [Google Scholar]
- 23. Angers S., and Moon R. T. (2009) Proximal events in Wnt signal transduction. Nat. Rev. Mol. Cell Biol. 10, 468–477 10.1038/nrm2717 [DOI] [PubMed] [Google Scholar]
- 24. Lai S. L., Chien A. J., and Moon R. T. (2009) Wnt/Fz signaling and the cytoskeleton: potential roles in tumorigenesis. Cell Res. 19, 532–545 10.1038/cr.2009.41 [DOI] [PubMed] [Google Scholar]
- 25. Mosimann C., Hausmann G., and Basler K. (2009) β-Catenin hits chromatin: regulation of Wnt target gene activation. Nat. Rev. Mol. Cell Biol. 10, 276–286 10.1038/nrm2654 [DOI] [PubMed] [Google Scholar]
- 26. Niehrs C. (2012) The complex world of WNT receptor signaling. Nat. Rev. Mol. Cell Biol. 13, 767–779 10.1038/nrm3470 [DOI] [PubMed] [Google Scholar]
- 27. Henderson B. R. (2000) Nuclear-cytoplasmic shuttling of APC regulates β-catenin subcellular localization and turnover. Nat. Cell Biol. 2, 653–660 10.1038/35023605 [DOI] [PubMed] [Google Scholar]
- 28. Valenta T., Hausmann G., and Basler K. (2012) The many faces and functions of β-catenin. EMBO J. 31, 2714–2736 10.1038/emboj.2012.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li Y., Chitnis N., Nakagawa H., Kita Y., Natsugoe S., Yang Y., Li Z., Wasik M., Klein-Szanto A. J., Rustgi A. K., and Diehl J. A. (2015) PRMT5 is required for lymphomagenesis triggered by multiple oncogenic drivers. Cancer Discov. 5, 288–303 10.1158/2159-8290.CD-14-0625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. MacDonald B. T., Tamai K., and He X. (2009) Wnt/β-catenin signaling: components, mechanisms, and diseases. Dev. Cell 17, 9–26 10.1016/j.devcel.2009.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cross D. A., Alessi D. R., Cohen P., Andjelkovich M., and Hemmings B. A. (1995) Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature 378, 785–789 10.1038/378785a0 [DOI] [PubMed] [Google Scholar]
- 32. Fukumoto S., Hsieh C. M., Maemura K., Layne M. D., Yet S. F., Lee K. H., Matsui T., Rosenzweig A., Taylor W. G., Rubin J. S., Perrella M. A., and Lee M. E. (2001) Akt participation in the Wnt signaling pathway through Dishevelled. J. Biol. Chem. 276, 17479–17483 10.1074/jbc.C000880200 [DOI] [PubMed] [Google Scholar]
- 33. Hart J. R., and Vogt P. K. (2011) Phosphorylation of AKT: a mutational analysis. Oncotarget 2, 467–476 10.18632/oncotarget.293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McCubrey J. A., Steelman L. S., Bertrand F. E., Davis N. M., Abrams S. L., Montalto G., D'Assoro A. B., Libra M., Nicoletti F., Maestro R., Basecke J., Cocco L., Cervello M., and Martelli A. M. (2014) Multifaceted roles of GSK-3 and Wnt/β-catenin in hematopoiesis and leukemogenesis: opportunities for therapeutic intervention. Leukemia 28, 15–33 10.1038/leu.2013.184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lenburg M. E., Sinha A., Faller D. V., and Denis G. V. (2007) Tumor-specific and proliferation specific gene expression typifies murine transgenic B cell lymphomagenesis. J. Biol. Chem. 282, 4803–4811 10.1074/jbc.M605870200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lin S. Y., Yeh K. T., Chen W. T., Chen H. C., Chen S. T., Chiou H. Y., and Chang J. G. (2004) Promoter CpG methylation of tumor suppressor genes in colorectal cancer and its relationship to clinical features. Oncol. Rep. 11, 341–348 [PubMed] [Google Scholar]
- 37. Yang L. H., Xu H. T., Li Q. C., Jiang G. Y., Zhang X. P., Zhao H. Y., Xu K., and Wang E. H. (2013) Abnormal hypermethylation and clinicopathological significance of Axin gene in lung cancer. Tumor Biol. 34, 749–757 10.1007/s13277-012-0604-z [DOI] [PubMed] [Google Scholar]
- 38. Thorvaldsen T. E., Pedersen N. M., Wenzel E. M., and Stenmark H. (2017) Differential roles of AXIN1 and AXIN2 in tankyrase inhibitor-induced formation of degradasomes and β-catenin degradation. PLoS ONE 12, e0170508 10.1371/journal.pone.0170508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang J., Xu-Monette Z. Y., Jabbar K. J., Shen Q., Manyam G. C., Tzankov A., Visco C., Wang J., Montes-Moreno S., Dybkaer K., Tam W., Bhagat G., His E. D., van Krieken J. H., Ponzoni M., et al. (2017) AKT hyperactivation and the potential of AKT-targeted therapy in diffuse large B-cell lymphoma. Am. J. Pathol. 187, 1700–1716 10.1016/j.ajpath.2017.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang P., Chaturvedi C. P., Tremblay V., Cramet M., Brunzelle J. S., Skiniotis G., Brand M., Shilatifard A., and Couture J. F. (2015) A phosphorylation switch on RbBP5 regulates histone H3 Lys4 methylation. Genes Dev. 29, 123–128 10.1101/gad.254870.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang Q., Trevino L. S., Wong R. L., Medvedovic M., Chen J., Ho S. M., Shen J., Foulds C. E., Coarfa C., O'Malley B. W., Shilatifard A., and Walker C. L. (2016) Reprogramming of the epigenome by MLL1 links early-life environmental exposures to prostate cancer risk. Mol. Endocrinol. 30, 856–871 10.1210/me.2015-1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu Y., Xing Z. B., Zhang J. H., and Fang Y. (2013) Akt kinase targets the association of CBP with histone H3 to regulate the acetylation of lysine 18. FEBS Lett. 587, 847–853 10.1016/j.febslet.2013.02.023 [DOI] [PubMed] [Google Scholar]
- 43. Das F., Ghosh-Choudhury N., Venkatesan B., Li X., Mahimainathan L., and Choudhury G. G. (2008) Akt kinase targets association of CBP with SMAD3 to regulate TGFβ-induced expression of plasminogen activator inhibitor-1. J. Cell. Physiol. 214, 513–527 10.1002/jcp.21236 [DOI] [PubMed] [Google Scholar]
- 44. Srivastava S., Mohibi S., Mirza S., Band H., and Band V. (2017) Epidermal growth factor receptor activation promotes ADA3 acetylation through the AKT-p300 pathway. Cell Cycle 16, 1515–1525 10.1080/15384101.2017.1339846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Karkhanis V., Wang L., Tae S., Hu Y. J., Imbalzano A. N., and Sif S. (2012) Protein arginine methyltransferase 7 regulates cellular response to DNA damage by methylating promoter histones H2A and H4 of the polymerase δ catalytic subunit gene, POLD1. J. Biol. Chem. 287, 29801–29814 10.1074/jbc.M112.378281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liao Y., Smyth G. K., and Shi W. (2013) The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 41, e108 10.1093/nar/gkt214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nusbaum C., Myers R. M., Brown M., Li W., and Liu X. S. (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shen L., Shao N. Y., Liu X., Maze I., Feng J., and Nestler E. J. (2013) diffReps: detecting differential chromatin modification sites from ChIP-seq data with biological replicates. PLoS ONE 8, e65598 10.1371/journal.pone.0065598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lawrence M., Huber W., Pagès H., Aboyoun P., Carlson M., Gentleman R., Morgan M. T., and Carey V. J. (2013) Software for computing and annotating genomic ranges. PLoS Comput. Biol. 9, e1003118 10.1371/journal.pcbi.1003118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhu L. J., Gazin C., Lawson N. D., Pagès H., Lin S. M., Lapointe D. S., and Green M. R. (2010) ChIPpeakAnno: a bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics 11, 237 10.1186/1471-2105-11-237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tae S., Karkhanis V., Velasco K., Yaneva M., Erdjument-Bromage H., Tempst P., and Sif S. (2011) Bromodomain protein 7 interacts with PRMT5 and PRC2, and is involved in transcriptional repression of their target genes. Nucleic Acids Res. 39, 5424–5438 10.1093/nar/gkr170 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.