Summary
The skin is colonized by a diverse microbiome intricately involved in various molecular and cellular processes within the skin and beyond. UV radiation is known to induce profound changes in the skin and modulate the immune response. However, the role of the microbiome in UV-induced immune suppression has been overlooked. By employing the standard model of contact hypersensitivity (using germ-free mice) we found diminished UV-induced systemic immune suppression in the presence of microbiome. Upon UV exposure, we found enhanced epidermal hyperplasia and neutrophilic infiltration in the presence and enhanced numbers of mast cells and monocyte or macrophages in the absence of microbiome. Transcriptome analysis revealed a predominant expression of cytokine genes related to pro-inflammatory milieu in the presence versus immunosuppressive milieu (with increased interleukin-10) in the absence of microbiome. Collectively, microbiome abrogates the immunosuppressive response to UV by modulating gene expression and cellular microenvironment of the skin.
Subject Areas: Dermatology, Radiation Biology, Immune Response, Microbiome
Graphical Abstract
Highlights
-
•
Epidermal and immune response to UV is dependent on skin microbiome
-
•
Increased neutrophilic infiltration and expression of IL-1β in SPF mice after UV-R
-
•
Elevated macrophage infiltration and expression of IL-10 in GF mice after UV-R
-
•
Skin microbiome diminishes UV-induced immune suppression to contact allergen DNFB
Dermatology; Radiation Biology; Immune Response; Microbiome
Introduction
The human skin is colonized by a diverse collection of microbes including bacteria, fungi, archaea, mites, and viruses. The majority of these microbes are commensals or transients that live in a mutualistic relationship with the skin's immune system, and recent studies indicate that the skin microbes influence gene expression in the skin (Meisel et al., 2018) and are involved in educating and modulating its immune response (Belkaid and Segre, 2014). Dysbiosis in the skin microbiome is linked to many skin pathologies such as acne, atopic dermatitis, and psoriasis (Zeeuwen et al., 2013).
Ultraviolet radiation (UV-R), on the other hand, has long been known to affect immune response, playing a role in skin carcinogenesis and in certain inflammatory skin diseases including photodermatoses and photoaggravated conditions (Wolf et al., 2016b). UV-R is known to induce innate immunity and suppress adaptive immune responses in healthy individuals (Schwarz, 2010). UV-R-induced immune suppression is mediated through T cells (Schwarz, 2010), and the immunomodulating properties of UV-R are most often investigated by using the contact hypersensitivity (CHS) model in mice and in humans (Fourtanier et al., 2005, Kelly et al., 2000). Furthermore, UV exposure is known to induce changes within the transcriptome of the skin and affect the gene expression of various biological processes that contribute to immune response (Sesto et al., 2002, Shen et al., 2016). Interestingly, a recent study reports the contribution of the skin microbiome to differential regulation of gene expression in the skin, most notably for the genes encoding Toll-like receptors (TLRs) and antimicrobial peptides (AMPs) and genes related to the interleukin (IL)-1 family (Meisel et al., 2018). The skin microbiome is also involved in regulating genes responsible for epidermal differentiation and development and in influencing wound healing (Canesso et al., 2014, Meisel et al., 2018).
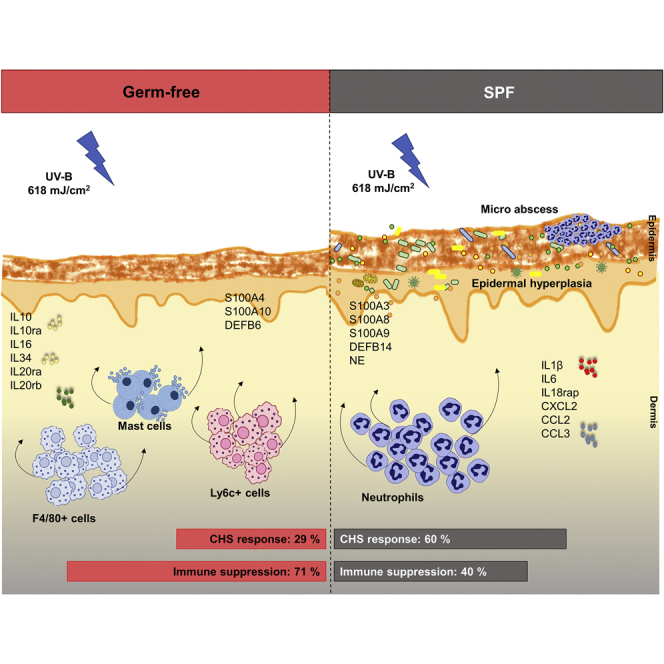
Although previous studies have shown the broad influence of UV-B on immune function, they have overlooked the potential contribution of the skin microbiome to this phenomenon (Patra et al., 2016, Patra et al., 2018). Therefore we employed the model of UV-induced suppression of induction of CHS in sterile, germ-free (GF) mice (totally devoid of microbiome) and found that the skin microbiome inhibited UV-induced immune suppression. Furthermore, in our transcriptome and histological analyses, we observed that a single UV dose induced several pro-inflammatory genes such as IL-1β, IL-6, and IL-18rap; increased microabscesses and neutrophilic infiltration in specific pathogen free (SPF) mouse skin versus induction of anti-inflammatory genes such as IL-10, IL-10ra, IL-20rb, and IL-7r; and increased numbers of mast cells, macrophages, and IL-10+ cells in GF skin. Collectively, our results show that the skin microbiome affords immune protection by orchestrating local cellular and innate immune responses to UV.
Results
Skin Microbiome Limits UV-Induced Suppression of Systemic Adaptive Immune Response
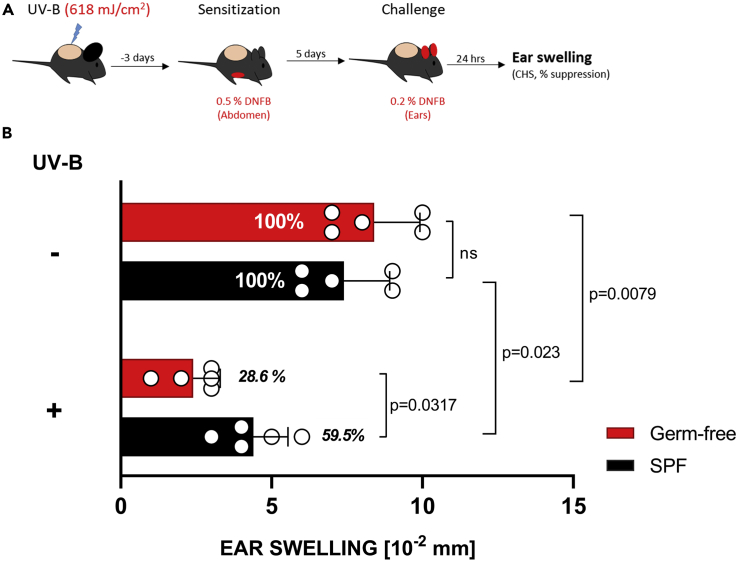
UV-B is known to inhibit CHS responses to haptens such as dinitrofluorobenzene (DNFB). However, the role of skin microbiome in this induction of immune response has not yet been studied. We irradiated mice on the shaved dorsal skin (ears covered with black electrical tape) with a dose of 618 mJ/cm2 (equaling two minimal inflammatory doses; Figure S2). Three days later, the mice were sensitized on the abdomen with 0.5% DNFB and challenged on the ears 5 days later with 0.2% DNFB (Figure 1A). The skin thickness was measured before and after challenge, and CHS response (as determined by skin swelling) was calculated for the different experimental groups. The presence of skin microbiome inhibited UV-induced suppression of induction of CHS in SPF mice exposed to UV-B before sensitization with DNFB. GF mice showed a significantly reduced CHS response compared with SPF mice (28.6% versus 59.5%) (Figure 1B). These results show that the absence of skin microbiome at the time of UV-B exposure enhanced immune suppression.
Figure 1.
Skin Microbiome Inhibits UV-B-Induced Suppression of Induction of CHS
(A) GF or control SPF mice were subjected to CHS assay as described in detail in Transparent Methods.
(B) The ear swelling differences between UV-B-irradiated and non-irradiated mice, with relatively reduced CHS response in GF compared with SPF (28.6% versus 59.5%), indicated immune protection by microbiome. Data shown represent mean ± SEM. N = 5 mice per experimental group; p value determined by Mann-Whitney test.
See also Figure S2.
Local Cellular and Innate Immune Responses Differ between GF and SPF Skin after UV Exposure
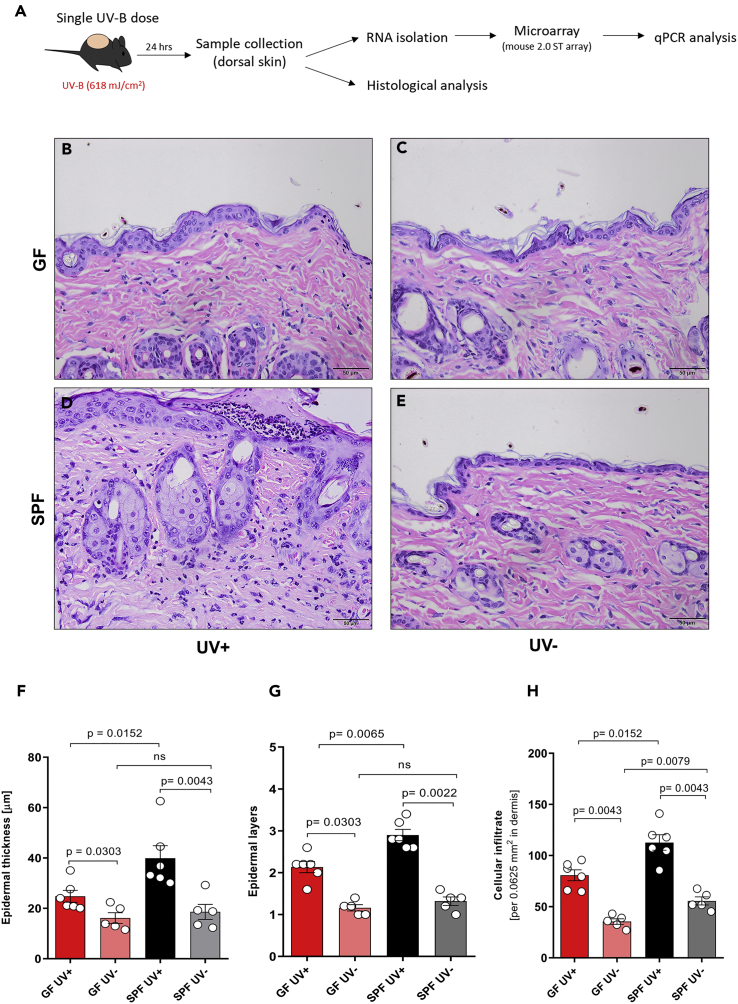
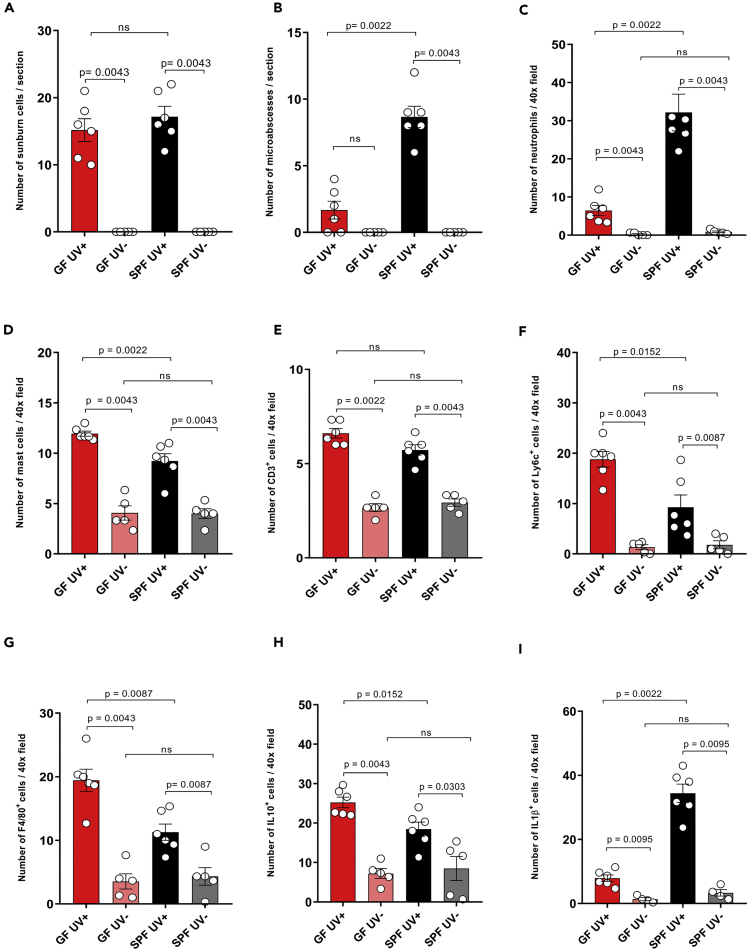
UV exposure leads to a series of cellular changes within the skin and systemic immune suppression, which is mediated by release of various immunosuppressive mediators (such as IL-10) and infiltration of the skin by immune cells such as monocytes and macrophages. To understand the cellular events occurring early after UV exposure, we took tissue samples from the UV-irradiated dorsal skin of GF and SPF mice 24 h after exposure (Figure 2A). Histological analysis of dorsal skin sections (Figures 2B–2E) revealed that UV-irradiated skin of both GF and SPF mice showed an increase in epidermal thickness, epidermal layers, and cellular infiltrate compared with the unexposed skin (Figures 2F–2H). No significant difference in epidermal thickness and layers was seen between unexposed GF and SPF skin, well in line with a recently published report (Meisel et al., 2018), but baseline density in cellular infiltrate was slightly, although significantly, increased in the skin of unexposed SPF mice compared with that of GF mice. Strikingly, UV-B exposure significantly increased the epidermal thickness (Figure 2F), epidermal layers (Figure 2G), and cellular infiltrate (Figure 2H) to a greater degree in SPF mice than in GF mice. This differential epidermal response to UV could be due to a UV-induced cellular response in the sterile environment of GF mice, resulting in only host-derived damage-associated molecular patterns, whereas in SPF mice host-derived and microbial-associated molecular patterns might amplify the epidermal response to UV exposure (Canesso et al., 2014). This is consistent with our observations that UV exposure boosted neutrophilic infiltration in SPF mice to a greater degree than it did in GF mice, as evidenced by the increased numbers of microabscesses (built up mainly by neutrophils) in the epidermis (Figure 3B) and densely scattered neutrophils in the dermis (Figure 3C). No significant differences in neutrophil numbers were observed in the unexposed skin of GF versus SPF skin. In parallel, UV-exposed GF skin showed increased numbers of mast cells, Ly6c+ cells (activated macrophages in inflammatory tissues), and F4/80+ cells (matured tissue macrophages) in the dermis, compared with SPF skin (Figures 3D, 3F, and 3G). Moreover, the anti-inflammatory cytokine IL-10 (Figure 3H) was expressed at significantly higher levels in UV-exposed GF skin than in SPF skin (mostly in the cellular infiltrate in the dermis), despite a slightly higher baseline expression of this cytokine in SPF mice. Indeed, it has been recently reported that the microbiome can itself trigger the expression of IL-10 in SPF mice (Meisel et al., 2018) (Figure 3H). No significant difference was observed in the numbers of sunburn cells and CD3+ cells (most likely dendritic epidermal T cells) in UV-exposed and unexposed GF skin compared with SPF skin (Figures 3A and 3E). Notably, the initial cellular response to UV-B differed between GF and SPF mice. SPF mice showed increased epidermal hyperplasia and dermal cellular infiltration, despite no overall difference in DNA damage as measured in terms of numbers of sunburn cells. Furthermore, dendritic epidermal γδ T cells are known to be activated after exposure to UV and to be involved in limiting DNA damage (MacLeod et al., 2014); however, we found no significant differences in epidermal CD3+ cells between UV-exposed GF and SPF skin. Collectively, exposure to UV leads to a pro-inflammatory environment with increased epidermal response and neutrophilic infiltration and IL-1β expression (Figures 3I and 6C) in the presence of microbiome. On the other hand, UV exposure to the skin in the absence of microbiome leads to an anti-inflammatory environment with increased mast cells, monocytes or macrophages, and IL-10 expression that is involved in immune suppression. This indicates that the presence of microbiome orchestrates the response of the skin to UV-B exposure.
Figure 2.
Microbiome Contributes to Increased Cellular Response to UV-B
(A) A single dose of UV-B (618 mJ/cm2) was administered to the shaved dorsal skin of GF (n = 6) and SPF (n = 6) mice 24 h before tissue collection.
(B–H) (B–E) Representative hematoxylin and eosin stainings of formalin-fixed, paraffin-embedded skin sections are shown and indicate a UV-induced increase in (F) epidermal thickness, (G) epidermal layers, and (H) cellular infiltrate to a greater degree in SPF mice than in GF mice. Data pooled from two separate experiments. Data shown represent mean ± SEM. p value determined by Mann-Whitney test.
Figure 3.
UV Exposure Boosts the Infiltration of Skin by Neutrophils and IL-1β in SPF Mice but by Macrophages and IL-10+ Cells in GF Mice
(A–I) Quantitative analysis showed highly increased numbers of (B) epidermal microabscesses and (C) increased neutrophil infiltration and (I) IL-1β+ cells in UV-exposed SPF versus GF skin. In contrast, UV-exposed GF skin showed higher numbers of (D) mast cells (i.e., cells with enlarged cytoplasm and multiple granules), (F) Ly6c, (G) F4/80, and (H) IL10 + cells. Besides a slight increase of IL10 + cells in the skin of SPF mice, no differences were seen in baseline levels of (A) sunburn cells and (E) CD3+ cells between SPF and GF skin. Data pooled from two separate experiments. Data shown represent mean ± SEM; N = 6 mice for UV-irradiated group and N = 5 for unirradiated group; p value determined by Mann-Whitney test. See also Figure S1.
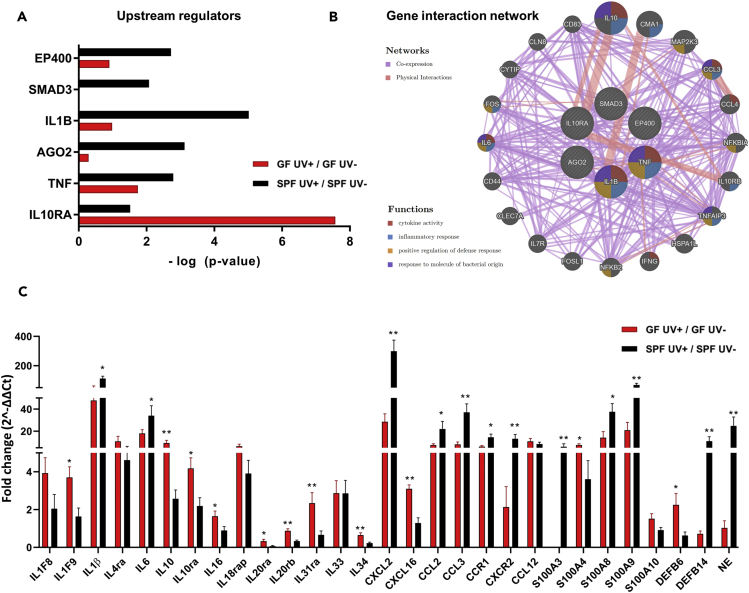
Figure 6.
Effect of UV-B on Various Upstream Regulators and Gene Interaction
(A) Predicted upstream regulators enriched in UV-B-treated GF and SPF skin identified by microarray analysis. The upstream regulators (filtered by prediction activation state and p < 0.05) are plotted based on their -log (p value).
(B) Gene interaction network of upstream regulators was visualized using geneMANIA. The genes involved in various functions are represented by colors. Co-expression and physical interactions are also shown in the network.
(C) qPCR analysis was done for selected genes from the dataset and gene network of upstream regulators. qPCR was performed in triplicates. Data were pooled from two separate experiments.
Data shown represent mean ± SEM; N = 6 mice for UV-irradiated group and N = 5 for unirradiated group; p value determined by Mann-Whitney test. *p = 0.0332; **p = 0.0021. See also Tables S1 and S4.
UV-B Modulates Cutaneous Gene Expression in GF and SPF Skin
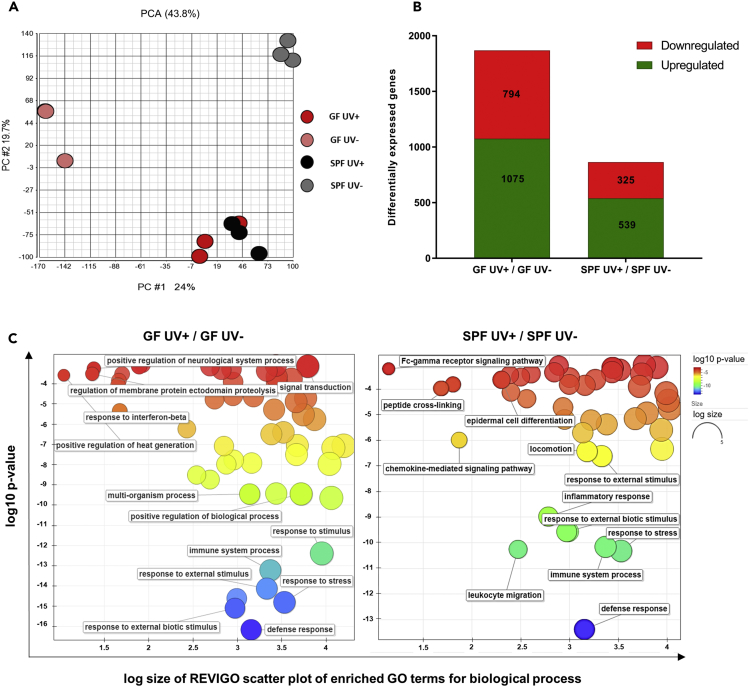
To further understand the effect of UV on immune response in the presence or absence of the microbiome, we examined gene expression in the UV-exposed and unexposed skin of GF and SPF mice by means of microarray analysis. Biological replicates of UV-exposed GF and SPF skin clustered together as demonstrated by principal-component analysis (Figure 4A) compared with the unexposed groups. To analyze up- and downregulated gene expression, the distribution was set to false discovery rate 5%, p < 0.05, and fold change of ±2. In total, 1,075 genes were upregulated and 794 genes were downregulated in response to UV-B treatment in GF mice. In comparison, 539 genes were upregulated and 325 genes were downregulated in SPF mice (Figure 4B). In this regard, a recent study has revealed differential gene expression (2,820 genes) using RNA sequencing in GF and SPF skin (Meisel et al., 2018), indicating that the microbiome can regulate gene expression within the skin. In our microarray analysis, we observed a similar number of differentially expressed genes (2,414 genes) in unexposed GF skin compared with SPF skin. Furthermore, differences in biological processes (plotted as treemap in Figure S3 and Data S4) resulting from differentially expressed genes in GF versus SPF skin (determined by microarray), such as regulation of epidermal development, cell division, and cytokinesis were consistent with previously published data (produced by RNA sequencing) by Meisel et al. (Meisel et al., 2018). Gene Ontology (GO) terms of the filtered genes were obtained using gene ontology enrichment analysis and visualization tool (Eden et al., 2009) (see Data S1), and the resulting GO terms and p values were used as input for REVIGO (Supek et al., 2011) for visualizing the scatterplots for biological processes (Figure 4C) in GF and SPF mice. The biological process GO terms enriched in the gene set contained terms related to “defense response” (GO: 0006952), “immune system process” (GO: 0002376), “response to external stimulus” (GO: 0009650), “leukocyte migration” (GO: 0050900), and “chemokine-mediated signaling pathway” (GO: 0070098). The full list of GO terms and associated genes is given in Data S2 and S3. In addition, UV-induced differentially expressed genes contributing to disease and functions by ingenuity pathway analysis are given in Data S5 and S6 for GF and SPF skin, respectively. These results indicate that UV-B acts differentially in regulating the gene expression depending on the skin microbiome.
Figure 4.
UV-B Modulates Gene Expression in GF and SPF Skin
(A) Two-dimensional principal-component analysis was done using Partek. Each point corresponds to a specific sample hybridized on one microarray containing 53,617 probesets. Points that are closer together (GF UV+ and SPF UV+) represent similar transcriptomic profile; points that are farther apart (GF UV- and SPF UV-) represent dissimilar transcriptomic profile.
(B) The numbers of UV-B-induced differentially expressed upregulated or downregulated genes with p < 0.05 and FC ±2 are plotted for GF and SPF skin.
(C) GO enrichment analysis summarized and plotted using REVIGO scatterplots for biological processes involving UV-induced differentially expressed genes in GF and SPF skin. The scatterplot shows the clusters in two-dimensional space by applying multidimensional scaling for the input GO terms. Log10 p values of GO terms are shown on the y axis (indicated as colors) and log size (frequency of the GO term in the database, bubbles of more general terms being larger) on the x axis.
Differential Expression of Interleukins, Chemokines, AMPs, and Other Innate Immune Response Genes in GF and SPF Skin after UV-B Irradiation
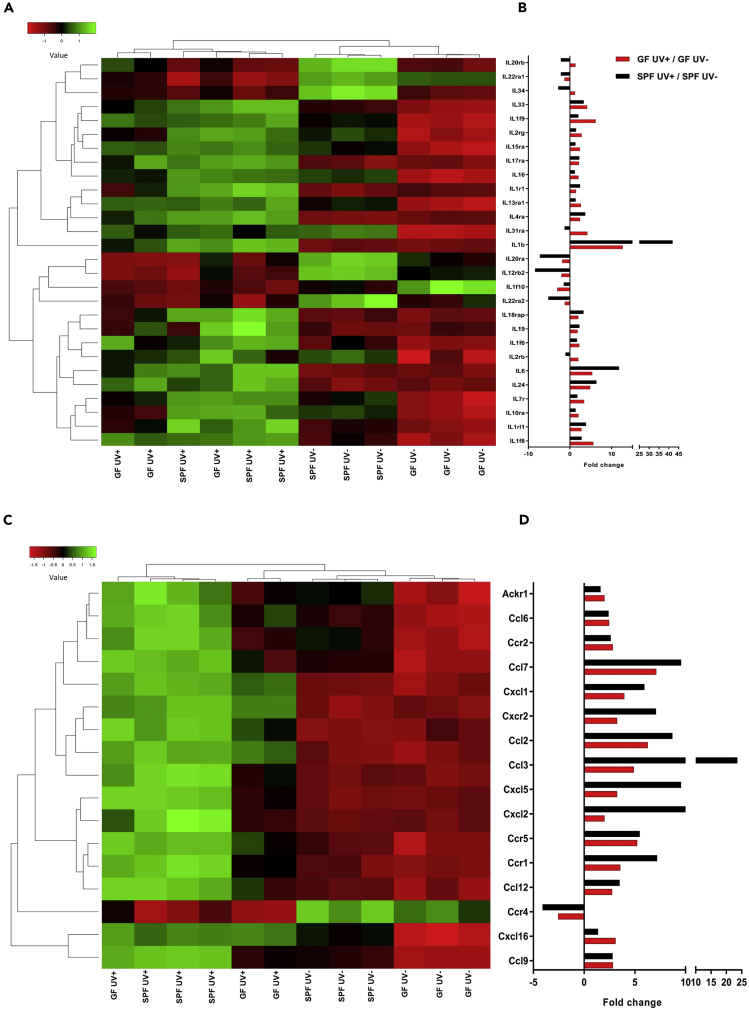
Innate immune genes related to cytokines (ILs and chemokines) and AMPs among others are crucial for inducing UV's immunological effects. These cytokines are involved in signaling pathways that lead to the activation of various immune cells infiltrating the skin. Furthermore, AMPs are key molecules in maintaining healthy homeostasis between the microbiome and immune system of the skin. Of note, UV-R is known to induce production of various AMPs in the skin (Glaser et al., 2009). We analyzed the expression of various cytokines, AMPs, TLRs, and serotonin-related genes within our dataset. Gene intensity values of ILs with p < 0.05 and fold change ±2 were used to plot the expression heatmap of each mouse group. Genes are clustered by average linkage, and distance was measured using the Euclidean method. Fold change values are plotted for GF UV+/GF UV− and SPF UV+/SPF UV− groups. The most notable changes in gene expression were seen for IL-20rb, IL-22ra1, IL-34, IL-20ra, IL-12rb2, IL-1f10, and IL-22ra2, which were differentially expressed without any UV treatment (Figure 5A). However, after a single UV-B dose, we observed significant flipping of the expression of these genes in both GF and SPF skin (Figure 5B). Genes for IL-1β, IL-6, and IL-24 were highly expressed in SPF compared with GF skin after UV-B treatment. On the other hand, IL-20ra, IL-12rb2, IL-22ra1, IL-22ra2, IL-34, and IL-20rb were downregulated by UV-B treatment to a greater degree in SPF skin than in GF skin. However, the gene expression of IL-20rb, IL-34, IL-1f9, IL-2rg, IL-15ra, IL-16, IL-13ra1, IL-31ra, IL-1f6, IL-2rb, IL-7r, IL-10ra, and IL-1f8 was increased in GF skin compared with SPF skin (Figure 5B). Furthermore, the genes for most chemokines were upregulated after UV-B exposure in both GF and SPF skin. However, we observed a complete flipping of Ccr4 expression in both GF and SPF skin (Figure 5C). The expression of almost all the genes for chemokines was higher in SPF skin; the few exceptions included CXCL16, CCL6, CCR2, CCR5, and CCL9, which were expressed similarly in both GF and SPF skin. The expression of other genes such as CCL7, CXCL1, CXCR2, CCL2, CCL3, CXCL5, CXCL2, CCR1, and CCL12 was significantly higher in SPF skin than in GF skin after UV-B treatment (Figures 5D and 6C). Innate immune genes such as AMPs, TLRs, and serotonin-related genes were differentially regulated by UV-B in GF and SPF skin (see Table S1). We found a total of 11 genes for AMPs (S100 family, DEFB, and RNase) to be differentially expressed in GF and SPF skin after UV-B exposure. In addition, TLR13 and TLR1 were significantly upregulated in SPF skin compared with GF skin. Furthermore, serotonin signaling receptor genes HTR2A and SLC6A4 were upregulated in GF skin compared with SPF skin. These data suggested that UV-B exposure resulted in differential expression of various cytokines and innate immune response genes depending upon the presence or absence of skin microbiome, thereby creating in UV-exposed skin an anti-inflammatory (IL-10, IL-10ra, IL-20rb, and IL-7r) environment in GF or pro-inflammatory (IL-1β, IL-6, and IL-18rap) environment in SPF mice.
Figure 5.
UV Induces Differential Expression of Interleukins and Chemokines in GF Versus SPF Skin
(A–D) (A and C) Robust multi-array average (RMA) intensities from microarray analyses were used to construct heatmaps. The genes are clustered by average linkage, and distance was measured using the Euclidean method. Fold change in gene expression for various (B) interleukins and (D) chemokines due to UV-B treatment in GF and SPF skin is shown in the bar graph. N = 3 mice per experimental group.
Potential Upstream Transcription Regulator Genes in UV-B-Exposed GF and SPF Skin and Their Gene Interaction Network
Upstream regulators within Ingenuity Pathway Analysis analyzes linkage to differentially expressed genes through coordinated expression and identifies potential upstream regulators that have been experimentally observed to participate in gene expression (Kramer et al., 2014). These upstream regulator genes are capable of affecting the expression of other genes or molecules that can be helpful to illuminate the biological activities occurring after UV exposure in GF and SPF skin. Our analyses of significant upstream regulators in GF and SPF skin response to UV-B exposure identified six genes: EP400, SMAD3, IL-1β, AGO2, TNF, and IL10RA (Figure 6A). We found that EP400, SMAD3, IL-1β, AGO2, and TNF were highly upregulated in SPF skin compared with GF. Intriguingly, after UV-B exposure, IL-10RA was highly activated in GF compared with SPF skin. In contrast, the expression of pro-inflammatory molecule IL-1B was more pronounced in UV-B-irradiated SPF than in GF skin. We further used the GeneMANIA (Warde-Farley et al., 2010) to predict the interactions and functional association of the upstream regulator genes. We found several target genes including IL-10, CMA1, MAP2K3, CCL3, CCL4, NFKBIA, IL10RB, TNFAIP3, HSPA1L, IFNG, NFKB2, FOSL1, IL7R, CLEC7A, CD44, IL6, FOS, CYTIP, CLN8, and CD83 that were interacting physically as identified by linking of two gene products in protein-protein interaction studies or were co-expressed with similar expression levels across conditions. Notably, these genes are involved in cytokine activity, inflammatory response, positive regulation of defense response, and response to molecules of bacterial origin (Figure 6B). To confirm microarray data we performed qPCR analysis of the genes within our dataset and of target genes observed in the gene network of upstream regulators and confirmed significantly higher expression of IL-1F9, IL-10, IL-10ra, IL-16, IL-20ra, IL-20rb, IL-31ra, IL-34, CXCL16, S100A4, and DEFB6 in GF skin than in SPF skin after UV-B exposure. Consistently, gene expression of IL-1β, IL-6, CXCL2, CCL2, CCL3, CCR1, CXCR2, S100A3, S100A8, S100A9, DEFB14, and NE was downregulated by UV-B in GF skin compared with SPF skin (Figure 6C). Comparing GF and SPF mice, these data suggest that UV exposure leads to differential expression of various upstream regulator genes such as IL-1β and IL-10RA, which can affect the transcription of other genes of the networks shown in Figure 6B.
Discussion
The impact of UV-R on the immune system was first described in landmark studies conducted by Margaret Kripke and colleagues during the 1970s (Fisher and Kripke, 1977). These studies showed that mice lost the ability to reject subcutaneously implanted immunogenic tumors in the immunosuppressive environment created after UV exposure. To illuminate the immune suppression effects of UV-R, CHS and delayed type hypersensitivity (DTH) models were established in which immune responses were found to be suppressed via the mediation of specific regulatory T cells (Schwarz, 2010) and B cells (Liu et al., 2018). We now show that the microbiome of the skin profoundly modulates the effect of UV-B on the immune system.
As demonstrated by our studies in GF and control SPF mice, the presence of microbiome protected against UV-induced immune suppression as measured by the suppression of induction of CHS (Figure 1). Indeed, our transcriptome analysis revealed the differential regulation of genes after UV exposure in the presence or absence of microbiome, resulting in a predominance of pro-inflammatory cytokines such as IL-1β, IL-6, and IL-18rap versus immunosuppressive cytokines such as IL-10, IL-10ra, IL-20rb, and IL-7r (Figures 3, 5 and 6C). This agrees with recent work showing that microbes or microbial products can induce immunoregulatory effects (Belkaid and Segre, 2014, Harrison et al., 2019, Lai et al., 2009), protect against UV-induced skin neoplasia (Nakatsuji et al., 2018), and modulate gene expression in the skin and can influence the epidermal development and differentiation (Meisel et al., 2018) and wound healing (Canesso et al., 2014). Moreover, it is known that certain skin-resident microbes and microbial products can regulate the expression of AMPs (Gallo and Hooper, 2012). Furthermore, UV-R can also directly induce the expression of certain AMPs within the skin (Glaser et al., 2009), possibly indirectly by the production of pro-inflammatory cytokines. We found significant modulation of AMP expression in UV-exposed GF compared with SPF skin (Table S1). This modulation of AMPs in GF skin may greatly contribute to cellular and humoral cytokine production and immune response in the skin (Lande et al., 2007). UV-exposed GF skin showed an increased expression of TLR13 (Table S1), which is an important site-specific mouse macrophage receptor (Kolter et al., 2016). In addition, we observed increased macrophage (Ly6c+ and F4/80 + cells) infiltration and IL10+ cells in UV-exposed GF skin compared with SPF skin (Figures 3F–3H and S1). Intriguingly, macrophages can produce large amounts of the anti-inflammatory cytokine IL-10, which we observed to be upregulated in UV-exposed GF skin. Moreover, serotonin signaling genes HTR2A and SLC6A4 were significantly upregulated in GF skin compared with SPF skin (Table S1). As serotonin signaling is crucial in UV-induced systemic immune suppression, but not local inflammation (Wolf et al., 2016a), this supports our observation of increased UV-induced immune suppression, but weaker epidermal response and cellular infiltration, in GF versus SPF mice. A previous study has shown that sensitization through UV-exposed skin induces regulatory T cells (Tregs) and that UV-induced Tregs suppress immune response via production of IL-10 (Maeda et al., 2008). GF mice are known to have increased number of cutaneous Foxp3+ Tregs compared with SPF mice (Naik et al., 2012).
After exposure of skin to UV-R, IL-10 is secreted to some extent by keratinocytes, but a large amount of IL-10 is produced by bone marrow-derived macrophages infiltrating the skin (Enk et al., 1995, Kang et al., 1994). In our gene expression, gene interaction, and immunohistochemical analyses (Figures 3 and S1), the skin of UV-exposed GF mice showed high expression of IL-10 and its receptors (IL-10RA, IL-10RB) compared with SPF mouse skin (Figures 5A and 6C). IL-10 inhibits the production of IL-1α, IL-1β, and IL-6 by monocytes (de Waal Malefyt et al., 1991); this is consistent with our data showing increased expression of IL-10 and its receptor IL-10ra and reduced expression of IL-1β and IL-6 in UV-exposed GF skin and an opposite effect in SPF skin, namely, increased expression of IL-1β (linked to CHS; Watanabe et al., 2007) and IL-6 and reduced expression of IL-10 (Figures 3, 5 and, 6C). Interestingly, IL-20rb was seen to be upregulated in UV-exposed GF skin but downregulated in SPF skin. IL-20rb is known to play a major role in signaling related to cutaneous inflammatory response and also to reduce production of IL-1β (Myles et al., 2013). Previous research has shown that commensal microbes can augment IL-1 signaling and subsequently promote effector T cell functions (Naik et al., 2012). Furthermore, UV exposure is known to increase IL-1β expression in the skin (Feldmeyer et al., 2007); our data now indicate that this increase is augmented in the presence of microbes (Figures 3I, 6C, and S1). Moreover, IL-34, which is known to stimulate monocyte and macrophage development and proliferation, was highly expressed in UV-exposed GF skin and downregulated in SPF skin (Figures 5 and 6C). IL-7r, which is known to block innate and adaptive inflammatory responses (Willis et al., 2012), was highly expressed in the skin of UV-exposed GF mice compared with SPF mice.
Alongside these findings, we also observed that UV exposure increased the expression of various chemokines in SPF skin compared with GF skin. We found increased expression of CCL7, CXCL1, CXCR2, CCL2, CCL3, CXCL5, CXCL2, CCR1, and CCL12 (Figures 5 and 6C), which goes well in line with previously published research (Dawes et al., 2011) and suggests that UV exposure induces chemokine production, which in turn mediates the recruitment and activation of immune cells. We also observed high expression of pro-inflammatory chemokines CCL2 and CCL3 induced by UV-B in SPF skin. Furthermore, we observed in SPF skin increased levels of CXCL5, which is known to be involved in UV-B-induced hypersensitivity in humans and rats and causes local inflammation by helping neutrophils and macrophages infiltrate the skin (Dawes et al., 2011). This is consistent with our observation of increased numbers of microabscesses (which could be aggravated by UV, Nakaguma et al., 1995, in the presence of microbes, Horton et al., 2015), neutrophils, gene expression of neutrophil elastase (NE) (Doring, 1994), and overall cellular infiltrate in SPF compared with GF skin (Figures 3C and 6C). CCR4, which is expressed on Foxp3+ Tregs, is critical for suppressing cutaneous inflammatory response, and Ccr4−/− mice exhibit a stronger CHS response than wild-type mice (Sells and Hwang, 2010). We observed that UV-B suppressed the expression of CCR4 slightly more in SPF skin than in GF skin (Figure 5). A decrease of CCR4 expression could impair DC-T-cell interaction and ultimately lead to enhanced inflammation in the skin (Lehtimaki et al., 2010, Sells and Hwang, 2010). Our data are consistent with the observation that topical application of Staphylococcus epidermidis on the skin results in modulation of cytokines such as CCL3, CCR2, CXCL2, IL-18rap, IL-1β, IL-6 (Naik et al., 2015). We observed these cytokines to be upregulated in UV-exposed SPF versus GF skin (Figures 5 and 6C).
Taken together, our findings suggest that UV-B induces a local pro-inflammatory environment in the skin in the presence of microbes, but promotes an anti-inflammatory environment with an immunosuppressive state in the absence of microbes. These findings are crucial for understanding UV-induced skin carcinogenesis and are in line with a recent report (Nakatsuji et al., 2018) showing a protective effect against skin cancer by the common skin commensal Staphylococcus epidermidis.
Limitations of the Study
One limitation of the GF model is that it cannot separate the effects of distal gut microbes from those of skin-resident microbes. The gut microbiome could influence the response of the skin through circulating microbial products or metabolites and thus further modulate the immune response (O'Neill et al., 2016). Indeed, GF mice showed altered lymphoid organ development in the gut (Cebra, 1999), but no morphological flaws in the skin (Chen et al., 2018, Naik et al., 2012). However, the exact degree to which gut microbiome could influence skin immunity is unknown. The fact that we observed increased immune suppression not only in GF mice but also in topical disinfected SPF mice by using chlorhexidine (data not shown) strongly indicates that the skin microbiome can directly modulate the immune response to UV-B, independent of the gut microbiome. That said, the skin microbiome may have potential physiological implications for other body sites such as gut, which has not been investigated here. Interestingly, a recent study showed that UV irradiation of skin resulted in alteration of intestinal (Jung et al., 2017) and fecal microbiome (Ghaly et al., 2018). Moreover, diet alone can also influence the functions of skin microbiome in triggering inflammation (Ridaura et al., 2018).
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We thank Johanna Aspsäter and Josefine Rosén, CGFR, KI, for their support for the GF animal experiments and Gerlinde Mayer and Isabella Perchthaler for technical support. We thank Dr. Marc Vocanson (CIRI, INSERM, Lyon) for critical reading and Jude Richard, Austin, TX, for editing the manuscript. This work was supported by the Austrian Science Fund FWF (W1241) and the Medical University of Graz through the Ph.D. Program Molecular Fundamentals of Inflammation (DK-MOLIN).
Author Contributions
V.P. and P.W. designed the study. V.P. performed all the experiments. V.P. and P.W. analyzed the data of the manuscript. K.W. performed microarray and data acquisition. V.A. was involved in administrative, technical, and material support for experiments involving GF animals. V.P. drafted the manuscript, and all authors critically revised the manuscript for important intellectual content. P.W. obtained the funding and supervised the study.
Declaration of Interests
The authors declare no competing interests.
Published: May 31, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2019.04.026.
Data and Software Availability
All data needed to evaluate the conclusions in the article are present in the article or the Supplemental Information. The microarray data have been deposited in the Gene Expression Omnibus (GEO) of the National Center for Biotechnology Information and are accessible through GEO Series accession number GSE117359.
Supplemental Information
References
- Belkaid Y., Segre J.A. Dialogue between skin microbiota and immunity. Science. 2014;346:954–959. doi: 10.1126/science.1260144. [DOI] [PubMed] [Google Scholar]
- Canesso M.C., Vieira A.T., Castro T.B., Schirmer B.G., Cisalpino D., Martins F.S., Rachid M.A., Nicoli J.R., Teixeira M.M., Barcelos L.S. Skin wound healing is accelerated and scarless in the absence of commensal microbiota. J. Immunol. 2014;193:5171–5180. doi: 10.4049/jimmunol.1400625. [DOI] [PubMed] [Google Scholar]
- Cebra J.J. Influences of microbiota on intestinal immune system development. Am. J. Clin. Nutr. 1999;69:1046S–1051S. doi: 10.1093/ajcn/69.5.1046s. [DOI] [PubMed] [Google Scholar]
- Chen Y.E., Fischbach M.A., Belkaid Y. Skin microbiota-host interactions. Nature. 2018;553:427–436. doi: 10.1038/nature25177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawes J.M., Calvo M., Perkins J.R., Paterson K.J., Kiesewetter H., Hobbs C., Kaan T.K., Orengo C., Bennett D.L., McMahon S.B. CXCL5 mediates UVB irradiation-induced pain. Sci. Transl. Med. 2011;3:90ra60. doi: 10.1126/scitranslmed.3002193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Waal Malefyt R., Abrams J., Bennett B., Figdor C.G., de Vries J.E. Interleukin 10(IL-10) inhibits cytokine synthesis by human monocytes: an autoregulatory role of IL-10 produced by monocytes. J. Exp. Med. 1991;174:1209–1220. doi: 10.1084/jem.174.5.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doring G. The role of neutrophil elastase in chronic inflammation. Am. J. Respir. Crit. Care Med. 1994;150:S114–S117. doi: 10.1164/ajrccm/150.6_Pt_2.S114. [DOI] [PubMed] [Google Scholar]
- Eden E., Navon R., Steinfeld I., Lipson D., Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enk C.D., Sredni D., Blauvelt A., Katz S.I. Induction of IL-10 gene expression in human keratinocytes by UVB exposure in vivo and in vitro. J. Immunol. 1995;154:4851–4856. [PubMed] [Google Scholar]
- Feldmeyer L., Keller M., Niklaus G., Hohl D., Werner S., Beer H.D. The inflammasome mediates UVB-induced activation and secretion of interleukin-1beta by keratinocytes. Curr. Biol. 2007;17:1140–1145. doi: 10.1016/j.cub.2007.05.074. [DOI] [PubMed] [Google Scholar]
- Fisher M.S., Kripke M.L. Systemic alteration induced in mice by ultraviolet light irradiation and its relationship to ultraviolet carcinogenesis. Proc. Natl. Acad. Sci. U S A. 1977;74:1688–1692. doi: 10.1073/pnas.74.4.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourtanier A., Moyal D., Maccario J., Compan D., Wolf P., Quehenberger F., Cooper K., Baron E., Halliday G., Poon T. Measurement of sunscreen immune protection factors in humans: a consensus paper. J. Invest. Dermatol. 2005;125:403–409. doi: 10.1111/j.0022-202X.2005.23857.x. [DOI] [PubMed] [Google Scholar]
- Gallo R.L., Hooper L.V. Epithelial antimicrobial defence of the skin and intestine. Nat. Rev. Immunol. 2012;12:503–516. doi: 10.1038/nri3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaly S., Kaakoush N.O., Lloyd F., Gordon L., Forest C., Lawrance I.C., Hart P.H. Ultraviolet Irradiation of skin alters the faecal microbiome independently of vitamin D in mice. Nutrients. 2018;10:1069. doi: 10.3390/nu10081069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser R., Navid F., Schuller W., Jantschitsch C., Harder J., Schroder J.M., Schwarz A., Schwarz T. UV-B radiation induces the expression of antimicrobial peptides in human keratinocytes in vitro and in vivo. J. Allergy Clin. Immunol. 2009;123:1117–1123. doi: 10.1016/j.jaci.2009.01.043. [DOI] [PubMed] [Google Scholar]
- Harrison O.J., Linehan J.L., Shih H.Y., Bouladoux N., Han S.J., Smelkinson M., Sen S.K., Byrd A.L., Enamorado M., Yao C. Commensal-specific T cell plasticity promotes rapid tissue adaptation to injury. Science. 2019;363:eaat6280. doi: 10.1126/science.aat6280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton J.M., Gao Z., Sullivan D.M., Shopsin B., Perez-Perez G.I., Blaser M.J. The cutaneous microbiome in outpatients presenting with acute skin abscesses. J. Infect. Dis. 2015;211:1895–1904. doi: 10.1093/infdis/jiv003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung E.S., Park H.M., Hyun S.M., Shon J.C., Singh D., Liu K.H., Whon T.W., Bae J.W., Hwang J.S., Lee C.H. The green tea modulates large intestinal microbiome and exo/endogenous metabolome altered through chronic UVB-exposure. PLoS One. 2017;12:e0187154. doi: 10.1371/journal.pone.0187154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang K., Hammerberg C., Meunier L., Cooper K.D. CD11b+ macrophages that infiltrate human epidermis after in vivo ultraviolet exposure potently produce IL-10 and represent the major secretory source of epidermal IL-10 protein. J. Immunol. 1994;153:5256–5264. [PubMed] [Google Scholar]
- Kelly D.A., Young A.R., McGregor J.M., Seed P.T., Potten C.S., Walker S.L. Sensitivity to sunburn is associated with susceptibility to ultraviolet radiation–induced suppression of cutaneous cell–mediated immunity. J. Exp. Med. 2000;191:561–566. doi: 10.1084/jem.191.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolter J., Feuerstein R., Spoeri E., Gharun K., Elling R., Trieu-Cuot P., Goldmann T., Waskow C., Chen Z.J., Kirschning C.J. Streptococci engage TLR13 on myeloid cells in a site-specific fashion. J. Immunol. 2016;196:2733–2741. doi: 10.4049/jimmunol.1501014. [DOI] [PubMed] [Google Scholar]
- Kramer A., Green J., Pollard J., Jr., Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y., Di Nardo A., Nakatsuji T., Leichtle A., Yang Y., Cogen A.L., Wu Z.R., Hooper L.V., Schmidt R.R., von Aulock S. Commensal bacteria regulate Toll-like receptor 3-dependent inflammation after skin injury. Nat. Med. 2009;15:1377–1382. doi: 10.1038/nm.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R., Gregorio J., Facchinetti V., Chatterjee B., Wang Y.H., Homey B., Cao W., Wang Y.H., Su B., Nestle F.O. Plasmacytoid dendritic cells sense self-DNA coupled with antimicrobial peptide. Nature. 2007;449:564–569. doi: 10.1038/nature06116. [DOI] [PubMed] [Google Scholar]
- Lehtimaki S., Tillander S., Puustinen A., Matikainen S., Nyman T., Fyhrquist N., Savinko T., Majuri M.L., Wolff H., Alenius H. Absence of CCR4 exacerbates skin inflammation in an oxazolone-induced contact hypersensitivity model. J. Invest. Dermatol. 2010;130:2743–2751. doi: 10.1038/jid.2010.208. [DOI] [PubMed] [Google Scholar]
- Liu X., Huang H., Gao H., Wu X., Zhang W., Yu B., Dou X. Regulatory B cells induced by ultraviolet B through toll-like receptor 4 signalling contribute to the suppression of contact hypersensitivity responses in mice. Contact Dermatitis. 2018;78:117–130. doi: 10.1111/cod.12913. [DOI] [PubMed] [Google Scholar]
- MacLeod A.S., Rudolph R., Corriden R., Ye I., Garijo O., Havran W.L. Skin-resident T cells sense ultraviolet radiation-induced injury and contribute to DNA repair. J. Immunol. 2014;192:5695–5702. doi: 10.4049/jimmunol.1303297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda A., Beissert S., Schwarz T., Schwarz A. Phenotypic and functional characterization of ultraviolet radiation-induced regulatory T cells. J. Immunol. 2008;180:3065–3071. doi: 10.4049/jimmunol.180.5.3065. [DOI] [PubMed] [Google Scholar]
- Meisel J.S., Sfyroera G., Bartow-McKenney C., Gimblet C., Bugayev J., Horwinski J., Kim B., Brestoff J.R., Tyldsley A.S., Zheng Q. Commensal microbiota modulate gene expression in the skin. Microbiome. 2018;6:20. doi: 10.1186/s40168-018-0404-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myles I.A., Fontecilla N.M., Valdez P.A., Vithayathil P.J., Naik S., Belkaid Y., Ouyang W., Datta S.K. Signaling via the IL-20 receptor inhibits cutaneous production of IL-1beta and IL-17A to promote infection with methicillin-resistant Staphylococcus aureus. Nat. Immunol. 2013;14:804–811. doi: 10.1038/ni.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik S., Bouladoux N., Linehan J.L., Han S.J., Harrison O.J., Wilhelm C., Conlan S., Himmelfarb S., Byrd A.L., Deming C. Commensal-dendritic-cell interaction specifies a unique protective skin immune signature. Nature. 2015;520:104–108. doi: 10.1038/nature14052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik S., Bouladoux N., Wilhelm C., Molloy M.J., Salcedo R., Kastenmuller W., Deming C., Quinones M., Koo L., Conlan S. Compartmentalized control of skin immunity by resident commensals. Science. 2012;337:1115–1119. doi: 10.1126/science.1225152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaguma H., Kambara T., Yamamoto T. Rat ultraviolet ray B photodermatitis: an experimental model of psoriasis vulgaris. Int. J. Exp. Pathol. 1995;76:65–73. [PMC free article] [PubMed] [Google Scholar]
- Nakatsuji T., Chen T.H., Butcher A.M., Trzoss L.L., Nam S.J., Shirakawa K.T., Zhou W., Oh J., Otto M., Fenical W. A commensal strain of Staphylococcus epidermidis protects against skin neoplasia. Sci. Adv. 2018;4:eaao4502. doi: 10.1126/sciadv.aao4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill C.A., Monteleone G., McLaughlin J.T., Paus R. The gut-skin axis in health and disease: a paradigm with therapeutic implications. Bioessays. 2016;38:1167–1176. doi: 10.1002/bies.201600008. [DOI] [PubMed] [Google Scholar]
- Patra V., Byrne S.N., Wolf P. The skin microbiome: is it affected by UV-induced immune suppression? Front. Microbiol. 2016;7:1235. doi: 10.3389/fmicb.2016.01235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra V., Laoubi L., Nicolas J.F., Vocanson M., Wolf P. A perspective on the interplay of ultraviolet-radiation, skin microbiome and skin resident memory TCRalphabeta+ cells. Front. Med. (Lausanne) 2018;5:166. doi: 10.3389/fmed.2018.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridaura V.K., Bouladoux N., Claesen J., Chen Y.E., Byrd A.L., Constantinides M.G., Merrill E.D., Tamoutounour S., Fischbach M.A., Belkaid Y. Contextual control of skin immunity and inflammation by Corynebacterium. J. Exp. Med. 2018;215:785–799. doi: 10.1084/jem.20171079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz T. The dark and the sunny sides of UVR-induced immunosuppression: photoimmunology revisited. J. Invest. Dermatol. 2010;130:49–54. doi: 10.1038/jid.2009.217. [DOI] [PubMed] [Google Scholar]
- Sells R.E., Hwang S.T. Paradoxical increase in skin inflammation in the absence of CCR4. J. Invest. Dermatol. 2010;130:2697–2699. doi: 10.1038/jid.2010.292. [DOI] [PubMed] [Google Scholar]
- Sesto A., Navarro M., Burslem F., Jorcano J.L. Analysis of the ultraviolet B response in primary human keratinocytes using oligonucleotide microarrays. Proc. Natl. Acad. Sci. U S A. 2002;99:2965–2970. doi: 10.1073/pnas.052678999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Kim A.L., Du R., Liu L. Transcriptome analysis identifies the dysregulation of ultraviolet target genes in human skin cancers. PLoS One. 2016;11:e0163054. doi: 10.1371/journal.pone.0163054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F., Bosnjak M., Skunca N., Smuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warde-Farley D., Donaldson S.L., Comes O., Zuberi K., Badrawi R., Chao P., Franz M., Grouios C., Kazi F., Lopes C.T. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe H., Gaide O., Petrilli V., Martinon F., Contassot E., Roques S., Kummer J.A., Tschopp J., French L.E. Activation of the IL-1beta-processing inflammasome is involved in contact hypersensitivity. J. Invest. Dermatol. 2007;127:1956–1963. doi: 10.1038/sj.jid.5700819. [DOI] [PubMed] [Google Scholar]
- Willis C.R., Seamons A., Maxwell J., Treuting P.M., Nelson L., Chen G., Phelps S., Smith C.L., Brabb T., Iritani B.M. Interleukin-7 receptor blockade suppresses adaptive and innate inflammatory responses in experimental colitis. J. Inflamm. (Lond.) 2012;9:39. doi: 10.1186/1476-9255-9-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf P., Byrne S.N., Limon-Flores A.Y., Hoefler G., Ullrich S.E. Serotonin signalling is crucial in the induction of PUVA-induced systemic suppression of delayed-type hypersensitivity but not local apoptosis or inflammation of the skin. Exp. Dermatol. 2016;25:537–543. doi: 10.1111/exd.12990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf P., Weger W., Patra V., Gruber-Wackernagel A., Byrne S.N. Desired response to phototherapy vs photoaggravation in psoriasis: what makes the difference? Exp. Dermatol. 2016;25:937–944. doi: 10.1111/exd.13137. [DOI] [PubMed] [Google Scholar]
- Zeeuwen P.L., Kleerebezem M., Timmerman H.M., Schalkwijk J. Microbiome and skin diseases. Curr. Opin. Allergy Clin. Immunol. 2013;13:514–520. doi: 10.1097/ACI.0b013e328364ebeb. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.