Abstract
Background:
Familial hypercholesterolemia (FH) is characterized by inherited high levels of low-density lipoprotein cholesterol (LDL-C) and premature coronary heart disease (CHD). Over a thousand low-frequency variants in LDLR, APOB and PCSK9 have been implicated in FH but few have been examined at the population level. We aim to estimate the phenotypic effects of a subset of FH variants on LDL-C and clinical outcomes among 331,107 multi-ethnic participants.
Methods:
We examined the individual and collective association between putatively pathogenic FH variants included on the MVP biobank array and the maximum LDL-C level over an interval of 15 years (maxLDL). We assessed the collective effect on clinical outcomes by leveraging data from 61.7 million clinical encounters.
Results:
We found 8 out of 16 putatively pathogenic FH variants with ≥30 observed carriers to be significantly associated with elevated maxLDL (9.4–80.2 mg/dL). Phenotypic effects were similar for European and African Americans despite substantial differences in carrier frequencies. Based on observed effects on maxLDL, we identified a total of 748 carriers (1:443) who had elevated maxLDL (36.5±1.4 mg/dL, p=1.2×10−152), and higher prevalence of clinical diagnoses related to hypercholesterolemia and CHD in a phenome-wide scan. Adjusted for maxLDL, FH variants collectively associated with higher prevalence of CHD (odds ratio, 1.59 [95% CI 1.36–1.86], p=1.1×10−8) but not peripheral artery disease.
Conclusions:
The distribution and phenotypic effects of putatively pathogenic FH variants were heterogeneous within and across variants. More robust evidence of genotype-phenotype associations of FH variants in multi-ethnic populations is needed to accurately infer at-risk individuals from genetic screening.
Keywords: familial hypercholesterolemia, low-density lipoprotein cholesterol, coronary heart disease, race and ethnicity, Genetic, Association Studies, Cardiovascular Disease, Lipids and Cholesterol
Introduction
Familial hypercholesterolemia (FH) is a common genetic disorder affecting approximately 1 in 250 adults.1 The condition causes lifelong elevated levels of low-density lipoprotein cholesterol (LDL-C), which substantially increases the risk for developing atherosclerotic cardiovascular disease, including early-onset disease. A recent study suggests that carriers of FH variants have an elevated risk for incident coronary heart disease (CHD) that is independent of a single measure of LDL-C.2 Treating FH with statins at younger ages can greatly reduce the risk of atherosclerotic cardiovascular disease related morbidity.3 Based on the National Institute for Health and Clinical Excellence guideline,4 FH screening is classified as a tier 1 genomic application.5
Many ‘causal’ mutations for FH have been reported and >1,000 variants have been documented in the LDL receptor (LDLR) gene alone. The functional effect of these variants appears to be highly variable, as estimated by LDLR activities and LDL-C in affected families.6, 7 The advancement of genotyping technologies enables accurate and cost-effective genetic screening of FH variants in large population and assessment of their individual effects. Previously, rare FH variants based on functional annotation were typically grouped together to assess their joint phenotypic effects on LDL-C.2, 8
The phenotypic effects of most FH variants have been assessed either in pedigrees without proper population controls or in population samples without sufficient sample size to assess phenotypic effects of individual variants. The majority of published genetic studies of FH were conducted in samples of European ancestry. While the prevalence of FH is higher in African Americans,1 the knowledge of FH variants in minority populations is very limited. To determine LDL raising effects, and to better understand the distribution of rare FH variants, we examined “pathogenic” and “likely pathogenic” (P/LP) FH variants, and their associations with LDL cholesterol levels and clinical outcomes among >300,000 multi-ethnic veterans in the Million Veteran Program (MVP).9
Methods
The individual level data of veteran participants will not be made available to other researchers without approval from the VA institutional review board.
Participants of multiple ethnicities were recruited from approximately 50 VA healthcare facilities across the United States.9 Individuals consented to a blood draw and to have their DNA extracted for genomic profiling and linked to their full electronic health record within the VA. Both MVP biobank and this analysis were approved by the VA institutional review boards.
The phenotypic and genotypic measures of the MVP participants were previously described.10 The detailed methods are available as supplemental material.
Results
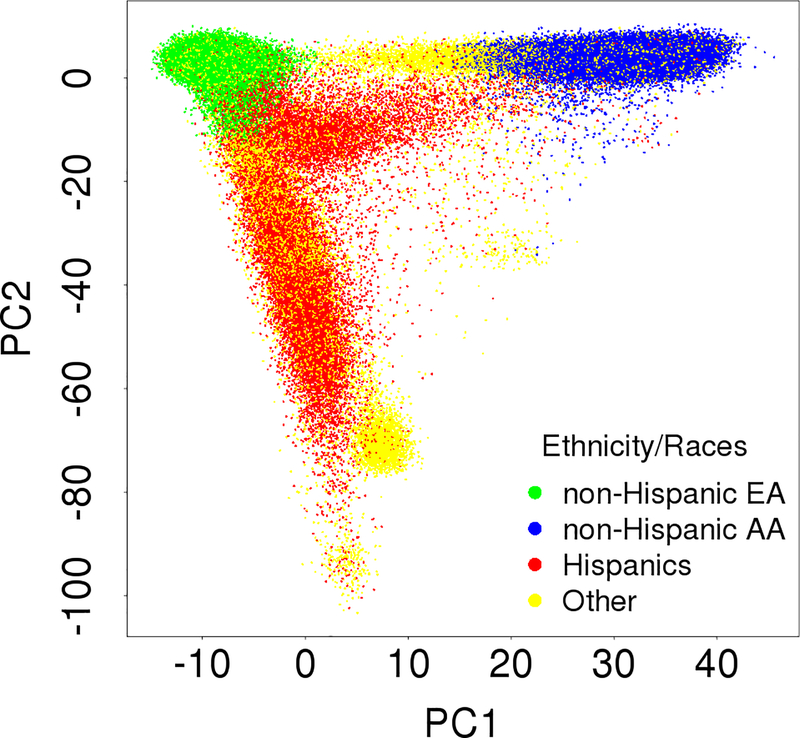
Among the multi-ethnic MVP cohort (summarized in Table 1), 331,107 veterans (mean age of 58.8 years old, 91.6% male) have both cleaned genotype and phenotype data. The MVP participants represented major continental ancestries of African, European, Asian and Native American (Figure 1). The self-reported Hispanics had heterogeneous genetic background from all major ancestries indicated by the first two principal components (Figure 1), and because of this and the low number of pathogenic variants in this group, Hispanics were not included in the current analyses.
Table 1.
Characteristics of the MVP samples stratified by non-Hispanic European Americans and African Americans.
| Non-Hispanic European Americans (N=231,481) | Non-Hispanic African Americans (N=64,929) | |
|---|---|---|
| Age at Enrollment (years) mean ± SD | 64.5 ±13.1 | 58.3 ± 11.9 |
| Gender n (%) | 215,240 (93.0%) | 56,752 (87.4%) |
| BMI (kg/m2) mean ± SD | 30.11 ± 5.88 | 30.25 ± 6.15 |
| Obesity, n (%) | 95,963 (41.5%) | 28,604 (44.1%) |
| Never smoker, n (%) | 62,716 (27.1%) | 19,265 (29.7%) |
| Former smoker, n (%) | 126,183 (54.5%) | 28,272 (43.5%) |
| maxLDL-C (mg/dL) mean ± SD | 139.2 ± 38.1 | 142.0 ± 40.3 |
| minHDL-C (mg/dL) mean ± SD | 36.1 ± 11.5 | 38.9 ± 12.8 |
| maxTG (mg/dL) median ± IQR | 214 ± 181 | 180 ± 154 |
| maxTC (mg/dL) mean ± SD | 219.2 ± 47.1 | 220.9 ± 46.8 |
| Statin use prior to maxLDL-C, n (%) | 9,864 (4.3%) | 2,650 (4.1%) |
| CHD*, n (%) | 57,782 (25.0%) | 9,764 (15.0%) |
| PAD*, n (%) | 17,305 (7.5%) | 3,956 (6.1%) |
| Hypertension*, n (%) | 149,634 (64.6%) | 45,642 (70.3%) |
defined using inpatient and outpatient ICD-9 and Current Procedural Terminology (CPT) codes available in EHR data by August 25th, 2017.
SD: standard deviation; IQR: Interquartile Range; LDL-C, Low-Density Lipoprotein Cholesterol; HDL-C, High-Density Lipoprotein Cholesterol; TG, Triglycerides; TC, Total Cholesterol; CHD: coronary heart disease; PAD: peripheral artery disease.
Figure 1.
Principal component (PC) plot of the 331,107 MVP multi-ethnic participants included in the analysis of FH variants.
We identified 58 FH variants classified as pathogenic or likely pathogenic (P/LP) in ClinVar on the MVP biobank array, of which 57 FH variants were not monomorphic (supplementary table 1). Although all of these FH variants had at least one submission annotated as P/LP, they were classified into four clinical significance categories in ClinVar, 4 as “Pathogenic”, 16 as “Pathogenic/Likely pathogenic”, 10 as “Likely pathogenic”, and 27 as “Conflicting interpretations of pathogenicity”. We observed at least 30 carriers for 16 variants (12 variants in LDLR, 2 in APOB, and 2 in PCSK9) which were individually assessed for association with LDL-C (Table 2). The number of carriers observed for the remaining 41 FH variants ranged from 1 to 27.
Table 2:
Ixndividual FH variants (≥30 carriers) associated with elevated maxLDL-C among 331,107 multi-ethnic veterans.
| SNP ID | Chr: basepair position | Gene | Effective Allele | Amino Acid Substitution | Number of Carriers | Beta±SE | p-value | maxLDL≥190 (%) |
|---|---|---|---|---|---|---|---|---|
| rs141502002 | 1:55524222 | PCSK9 | T | Arg469Trp | 1,147 | 9.4±1.1 | 3.71×10−17 | 17.9% |
| rs12713559 | 2:21229068 | APOB | A | Arg3558Cys | 366 | 9.9±2.0 | 6.32×10−7 | 15.3% |
| rs5742904 | 2:21229160 | APOB | T | Arg3527Gln | 256 | 43.5±2.4 | 3.40×10−75 | 42.2% |
| rs768563000 | 19:11217264 | LDLR | A | Glu240Ter | 34 | 20.3±6.5 | 1.8×10−3 | 26.5% |
| rs151207122 | 19:11218157 | LDLR | T | Arg303Trp | 49 | 36.2±5.4 | 2.16×10−11 | 36.7% |
| rs121908030 | 19:11218160 | LDLR | A | Asp304Tyr | 30 | 80.2±6.9 | 4.67×10−31 | 66.7% |
| rs201573863 | 19:11231154 | LDLR | T | Pro699Leu | 51 | 44.0±5.3 | 1.03×10−16 | 51.0% |
| rs137853964 | 19:11240278 | LDLR | A | Val827Ile | 328 | 12.6±2.1 | 1.67×10−9 | 18.0% |
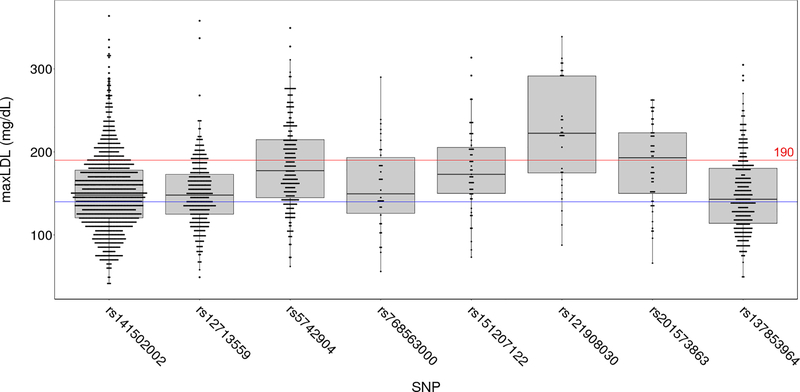
A total of 8 of these 16 P/LP variants, including 5 in LDLR, 2 in APOB, and 1 in PCSK9 were significantly associated with elevated level of maxLDL compared to non-carriers after correction for multiple testing (p<0.05/16) (Table 2). Among these 8 variants, we observed a wide range of effects on the mean maxLDL (+9.4 to +80.2 mg/dL) and a wide range of proportion of individuals with a maxLDL >190 mg/dL (15% to 67%) (Table 2). These effects of individual FH variants were highly heterogeneous (I2=97.8% [95% CI 97.1%−98.3%], p<0.001). The distribution of maxLDL among carriers of each FH variant was also highly variable (Figure 2), with some carriers having maxLDL levels lower than 100 mg/dL. We also observed that variants with the largest effect sizes (e.g., rs121908030, rs5742904, rs201573863 and rs151207122) were relatively less common (Table 2). Carriers of the remaining less common variants (<30 carriers for each) collectively demonstrated a mean maxLDL that was increased by 27.3±2.1 mg/dL compared to non-carriers (p=2.8×10−39).
Figure 2.
Distribution of maxLDL levels of LDL-raising variants among the MVP participants. Red horizontal line (190 mg/dL) indicates the threshold for severe hypercholesterolemia; blue horizontal line (140 mg/dL) represents the mean maxLDL of 331,107 multi-ethnic participants.
We examined the phenotypic effects and distribution of individual P/LP variants among 214,455 MVP participants of European Ancestry and 57,850 MVP participants of African Ancestry (Table 3). Because no single pathogenic variant had sufficient number of carriers (allele count of 30 or greater) in the MVP Hispanic Americans to assess the genetic effects, we did not include Hispanic Americans in ethnicity-specific analysis. All except one (rs12713559) of the P/LP variants associated with maxLDL had carrier frequencies that were more than threefold difference in prevalence between European and African ancestries (Table 3). One variant in PCSK9 (rs141502002) and two variants in LDLR (rs121908030 and rs151207122) were predominantly observed among African American (AA) participants, with a frequency >70 times greater than that in European American (EA) participants. For example, rs121908030 (LDLR), which had the largest effect on maxLDL among all tested P/LP variants, was observed only once among EAs, and was 85 times more frequent among AA participants than for EA participants.
Table 3.
Ethnicity-specific effects and frequencies of FH variants associated with elevated maxLDL.
| SNP ID | Gene | A1 | Amino Acid Substitution | European Ancestry N=214,455 | African Ancestry N=57,850 | Carrier Freq Ratio (AA/EA) | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Carriers | Beta±SE | p-value | Carriers | Beta±SE | p-value | |||||
| rs141502002 | PCSK9 | T | Arg469Trp | 23 | 3.1±7.8 | 0.69 | 932 | 10.3±1.3 | 3.79×10−15 | 150 |
| rs12713559 | APOB | A | Arg3558Cys | 248 | 11.0±2.4 | 3.20×10−6 | 52 | 10.2±5.6 | 6.61×10−2 | 0.78 |
| rs5742904 | APOB | T | Arg3527Gln | 212 | 44.5±2.6 | 1.07×10−67 | 15 | 50.7±10.3 | 9.66×10−7 | 0.26 |
| rs768563000 | LDLR | A | Glu240Ter | 30 | 26.6±6.8 | 9.01×10−5 | 2 | NA | NA | 0.25 |
| rs151207122 | LDLR | T | Arg303Trp | 2 | NA | NA | 42 | 37.3±6.2 | 1.62×10−9 | 78 |
| rs121908030 | LDLR | A | Asp304Tyr | 1 | NA | NA | 23 | 87.7±8.3 | 8.44×10−26 | 85 |
| rs201573863 | LDLR | T | Pro699Leu | 11 | NA | NA | 29 | 38.8±7.4 | 1.86×10−7 | 9.8 |
| rs137853964 | LDLR | A | Val827Ile | 243 | 13.9±2.4 | 5.75×10−9 | 5 | NA | NA | 0.08 |
Including all variants associated with elevated maxLDL (9.4–80.2 mg/dL), we identified 2,594 carriers (1 in 128). However, the phenotypic effects of three variants, rs141502002 (PCSK9), rs12713559 (APOB) and rs137853964 (LDLR), were moderate (increased maxLDL 9.4–12.6 mg/dL) and not considered as “pathogenic” for FH in follow-up analyses. Of the remaining variants that increase maxLDL by >20 mg/dL, there were 748 veterans carrying these more deleterious FH variants among 331,107 participants (1 in 443). The ten most frequent variants accounted for 71.0% of genotyped pathogenic FH variant carriers. The carrier rates of these pathogenic FH variants were 1:462 among 214,455 participants of European ancestry, and 1:362 among 57,850 participants of African ancestry. Among 32,174 individuals with maxLDL above 190 mg/dL, 1 in 117 carried one of pathogenic FH variants.
We further examined the combined effect of pathogenic FH variants that substantially increase maxLDL (mean increase > 20 mg/dL) within each of the 3 FH genes. Carriers of FH variants within LDLR, APOB, and PCSK9, were associated with an increase (>30 mg/dL) of maxLDL (Table 4). We found the highest mean effect on maxLDL in carriers of APOB FH variants. Carriers of any of these FH variants had a maxLDL that was 36.5±1.4 mg/dL greater than non-carriers (p=1.2×10−152). Ethnicity-specific association analysis revealed that the gene specific and join effects of P/LP variants were similar for individuals of European and African ancestries (Table 4).
Table 4.
Collective associations of pathogenic FH variants with maxLDL levels among the multi-ethnic, European ancestry (EA) and African ancestry (AA) participants.
| All N=331,107 | EA N=214,455 | AA N=57,850 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Carriers | Beta±SE | p-value | Carriers | Beta±SE | p-value | Carriers | Beta±SE | p-value | |
| APOB | 275 | 42.3±2.3 | 2.10×10−76 | 225 | 43.8±2.5 | 9.79×10−70 | 16 | 46.5±10.0 | 3.37×10−6 |
| LDLR | 451 | 32.0±1.8 | 2.44×10−76 | 226 | 28.3±2.5 | 3.95×10−30 | 142 | 41.7±3.4 | 2.28×10−35 |
| PCSK9 | 22 | 33.9±8.1 | 2.66×10−5 | 13 | 35.2±10.3 | 6.64×10−4 | 2 | 105.2±28.3 | 2.04×10−4 |
| ALL | 748 | 36.5±1.4 | 1.22×10−152 | 464 | 36.1±1.7 | 1.97×10−96 | 160 | 43.0±3.2 | 5.00×10−42 |
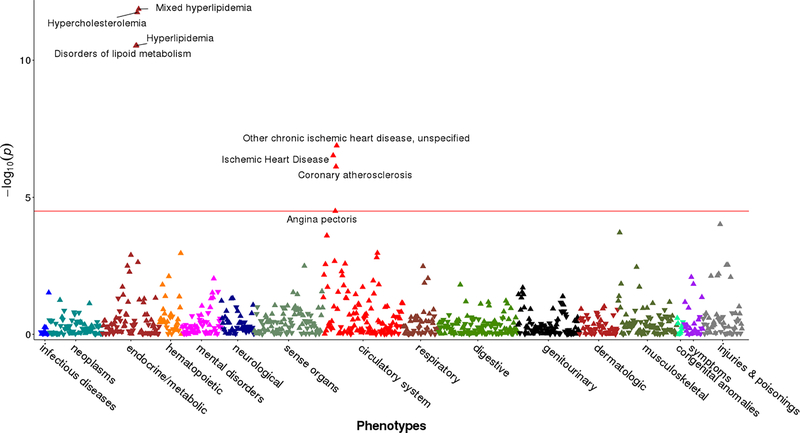
Overall there were 748 carriers of deleterious FH variants. These individuals had a mean age of 58.5 years old and 91.7% were male. We determined the collective impact of these deleterious FH variants on a wide range of clinical outcomes through a phenome-wide association study (PheWAS) by leveraging data from 61.7 million clinical encounters across 3 million patient-years of health data. Among >200,000 participants with European ancestry, we examined 1,171 diagnoses within 17 categories of disease based on ICD-9 codes. Compared to non-carriers, carriers of FH variants had a statistically significantly (p < 0.05/1,171 or 4.3 × 10−5) higher prevalence of clinical diagnoses related to hypercholesterolemia and CHD-related diagnoses including ischemic heart disease, coronary atherosclerosis, and angina pectoris (Figure 3). We further examined the associations between collective FH variants and the prevalence of CHD and peripheral arterial disease (PAD) as determined by a more refined definition of these outcomes leveraging elements of the EHR. Adjusted for age, sex and the top 10 principal components for population structure, FH variants remained collectively associated with a greater prevalence of CHD (odds ratio of 1.68, 95% CI 1.43–1.97, p=1.65×10−10), but not PAD (odds ratio of 1.09, 95% CI 0.86–1.39, p=0.45). With additional adjustment for maxLDL, the odds ratio for CHD decreased slightly but remained significant (odds ratio of 1.59, CI 1.36–1.86, p=1.12×10−8).
Figure 3.
The genetic associations between pathogenic FH variants and clinical outcomes in a phenome-wide association (PheWAS) analysis. Red horizontal line represents Bonferroni corrected p-value of 0.05. Positive association denoted by triangle pointing up (▲), and negative association by triangle pointing down (▼).
A majority (603 or 80.6%) of the 748 carriers of pathogenic FH variants had been prescribed a statin at least once within the VA healthcare system at any time prior to or on the day of their enrollment into MVP. Overall the participants who had ever been prescribed statin had mean maxLDL of 186.2 (SD of 56.3) mg/dL, 49.9 mg/dL higher than those without any record of statin prescription. Only 51 (6.8%) had a record of statin use within 90 days prior to the date of maxLDL measures. A total of 468 FH variant carriers (62.6%) on current therapy (prescription within 3 months of enrollment), had an average treated LDL-C level of 124.2 (SD of 46.7) mg/dL, 64.2 mg/dL lower than their maxLDL. The remaining 145 (19.4%) carriers, who had no record of statin use, had lower level of maxLDL (136.3 (SD of 39.4) mg/dL, p<2.2×10−16).
To explore the phenotypic effects of other rare variants potentially related to FH, we examined 102 additional variants in APOB, LDLR and PCSK9 with MAF < 0.01 and MAC > 30 in the MVP. After correction for multiple testing, we identified six variants (three in APOB and three in LDLR) significantly associated with elevated maxLDL (p-value < 0.05/102=4.9×10−4). Carrying each rare allele on average increased 2.5 to 24.6 mg/dL maxLDL among the MVP participants (Table 5). Four variants (three in APOB and one in LDLR) coded for missense mutations. Notably, the synonymous variant in LDLR (rs368889457) had the largest effect size (24.6±4.9 mg/dL, p-value=6.60×10−7) and lowest MAF (9.44×10−5).
Table 5.
Non-ClinVar P/LP rare variants in APOB and PCSK9 associated with elevated maxLDL among multi-ethnic veterans.
| SNP ID | rs ID | Chr:basepair | Gene | Function | Effective allele | MAF | Beta±SE | p-value |
|---|---|---|---|---|---|---|---|---|
| AX-83185920 | rs151333262 | 2:21225111 | APOB | missense | T | 1.04×10−4 | 24.1±4.5 | 9.58×10−8 |
| AX-40824279 | rs12720854 | 2:21229905 | APOB | missense | C | 0.005859 | 2.5±0.6 | 7.27×10−5 |
| AX-83134365 | rs72653060 | 2:21257697 | APOB | missense | C | 0.000278 | 22.7±2.7 | 1.42×10−16 |
| AX-86688464 | rs368889457 | 19:11224355 | LDLR | synonymous | A | 9.44×10−5 | 24.6±4.9 | 6.60×10−7 |
| AX-56828551 | rs17248882 | 19:11227525 | LDLR | intron | A | 0.001998 | 4.5±1.0 | 1.89×10−5 |
| AX-32617011 | rs45508991 | 19:11233886 | LDLR | missense | T | 0.005736 | 4.4±0.6 | 2.39×10−12 |
Discussion
The phenotypic expression of FH on LDL-C and the age of onset and severity of disease varies substantially by the causal gene and individual variants.7, 8, 11, 12 Previous studies have focused on reporting either effects of selected FH variants, pedigrees or case groups without consideration of proper population controls, or compared results to population samples without sufficient sample size to assess individual variants. In this study, we overcame these limitations by analyzing the effect of 57 FH variants annotated as pathogenic among 331,107 subjects receiving care within the VA health care system. We confirmed a number of low-frequency pathogenic variants raising LDL-C of MVP participants. Among these pathogenic variants, APOB gene on average carried the most severe mutations among three tested genes. However, such observation was restricted to the variants measured on the MVP genotyping array. The generalization of the observation to the entirety APOB variants, should be cautious, and requires more complete coverage of genetic mutations in future population studies. We observed notable heterogeneity in the effects of these variants on LDL-C, suggesting the annotation of their pathogenicity is imperfect. Since most P/LP variants for FH have not been validated at a population level, concluding pathogenicity on the basis of a variant’s putative disruption of the gene’s function, or its protein product may be insufficient.
We investigated the distribution of FH variants among participants of European (n=214,455) and African ancestry (n=57,850) separately to address the limited knowledge of FH variant distribution among AAs in comparison to EAs.13 We found that all except one P/LP variants associated with maxLDL had carrier frequencies that were more than threefold different between the European and African ancestries. Several variants, including the one with the largest effect on maxLDL among all tested P/LP variants (rs121908030 of LDLR), were predominantly observed among AAs. Additionally, the joint carrier frequency of deleterious FH variants in African ancestry (1:362) is also higher than that in European ancestry (1:462). Such differences in FH variant distribution between races, demand in-depth genotyping efforts to discover and validate FH variants and other rare pathogenic variants among very large multi-ethnic populations.
The current estimated prevalence of FH is approximately ~1:250 in the US 1 and other Western countries.14–17 Recent exome-sequencing based screenings have reported frequencies for FH carriers of 1:222 8 and 1:211,2 in an EHR-based cohort and a CAD case-control population, respectively. However, the latter study included only 7% African Americans. In this multi-ethnic study, the calculated FH carrier frequency was dependent on the inclusion and exclusion criteria of individual variants and race/ethnicity. The overall results were substantially influenced by a few common variants. When three variants with moderate LDL-raising effects were included, the FH carrier frequencies were 1:128, 1:234 and 1:50 among all, European alone, and African American alone participants, respectively. Notably, the FH carrier frequency among Europeans is consistent with previous studies (1:211) that included up to 7% African Americans.2 A single variant rs141502002 (PSCK9), which is associated with moderately increased maxLDL (9.4±1.1 mg/dL, p=3.7×10−17), was predominantly observed among African ancestry, and accounted for 81% of LDL-raising FH variant carriers in this ethnic group. After excluding the three variants with moderate effects on maxLDL, we estimated a lower FH carrier frequency (1:443) using a more stringent inclusion criteria of LDL-raising FH variants (mean increase >20 mg/dL) in this microarray-based genotyping study.
Using a PheWAS approach, we explored the phenotypic effects of FH variants for >1,000 clinical diagnoses in the EHR. We identified that FH variant carriers with elevated risk for diagnoses related to CHD or hypercholesterolemia, but no obvious pleiotropic effects on other clinical diagnosis. Although we assembled one of the largest samples of FH carriers, the number of some less frequent clinical diagnoses may partially explain why we did not observe strong pleiotropic effect of FH variants on other disease outcomes. Using refined phenotyping algorithms of CHD and PAD, we confirmed the genetic association with CHD with and without adjustment of maxLDL, but not with PAD. Since FH carriers in the MVP achieved comparable LDL-C to non-carriers after statin treatment, the residual risk for CHD adjusted for maxLDL suggests that earlier and more aggressive treatments maybe needed to further reduce the CHD risk among FH carriers.
Our study has several important limitations. First, our genotyping platform limited the assessment of the phenotypic effects of FH variants genotyped on the MVP biobank array, but did not include all documented FH variants. However, the number of identified carriers with moderate to more severe form of FH variants should account for a large proportion of all deleterious FH variants, most having extremely low frequency in the population. Large scale sequencing studies may ultimately be necessary to provide more complete identification of FH variants in multi-ethnic populations. Second, our study included predominantly male veterans (<10% women) over the age of 50 years, which is not representative of the general population. Future larger-scale genomic studies of FH need to cover broader demographics, especially younger participants,18 since severe FH often leads to greater CVD risk in young and middle-aged adults. Lastly, the older age of MVP participants may introduce ascertainment bias by not including individuals with more severe clinical outcomes (e.g., death) at a younger age. Although we were not able to quantify such bias using available data, such a loss of severe FH patients may have resulted in an observed phenotypic effect of one or more FH variants that is lower than the true overall effect of that variant on LDL-C.
Both genetic and environmental factors can potentially modify the effects of FH variants on LDL-C. Since there were no previous reports of such gene-gene or gene-environment interactions for FH pathogenicity, we assumed the independence between pathogenic variants and other genetic (e.g., protective variants) or environmental factors in present study. Other molecular factors such as lipoprotein (a) may also influence the genetic effects of FH variants, particularly in regards to race differences. Because lipoprotein (a) was not systematically measured, we did not examine its influence on FH variants in present study.
FH patients have elevated LDL-C throughout life but are underdiagnosed and undertreated in the general population. Effective treatment to reduce the risk for developing atherosclerotic diseases requires timely diagnosis at the youngest age possible. Universal screening through gene sequencing has the potential to identify all individuals with familial hypercholesterolemia, but is not yet feasible or cost effective. Since FH is an autosomal dominant disorder, targeted screening (e.g., cascade screening 19 and child-parent screening20, 21) among the family members of index cases may be more efficient. Positive genotyping of pathogenic FH variants is often used as a critical component of the diagnosis.18, 22, 23
The precision of a genetic diagnosis depends on accurate annotation of pathogenicity. Our findings of heterogeneous phenotypic effects within and across individual FH variants highlight the limitations in the ability to correctly classify the pathogenicity of many of these variants hindering a clinician’s optimal interpretation and application of the results of genetic tests for FH. Given the wide range of individual FH variants’ effects on LDL-C, we suggest including additional components of the evidence to better annotate the pathogenicity in current databases such as ClinVar. Such annotation components should add the summary statistics from family-based or population-based studies of individual disease-causing variants, to assist the researchers to accurately assess and rank the evidence of pathogenicity beyond putatively functional changes. Broadly, we recommend extra caution when interpreting the potential effect on LDL-C of a purported FH variant that has not been examined in large-scale population studies. This extra caution includes the search for additional evidence of pathogenicity through the careful documentation of a family history of hyperlipidemia and CHD. Importantly, both cholesterol-based and genotype-based testing for the diagnosis of FH should continue to be used.24 Among individuals with no family history or other pathognomonic clinical signs of FH, cholesterol level may serve as a primary testing, with gene testing adding to CVD risk prediction in certain individuals carrying validated pathogenic variants.
Supplementary Material
Acknowledgements:
This research is based on data from the Million Veteran Program, Office of Research and Development, Veterans Health Administration. This publication does not represent the views of the Department of Veterans Affairs or the United States Government. We would like to thank all Veteran participants in the MVP for donating their samples, information, and time to this project. In addition, we would like to thank all the MVP staff working in the various operational domains including the biorepository, recruitment sites, VA Central Office, and clinicians for all their efforts to ensure the success of the MVP.
Sources of Funding: This research was supported by award I01-BX003340 and I01-BX002641 from the Department of Veterans Affairs. YVS is partially supported by NIH grant R01NR013520.
Appendix: Million Veteran Program: Consortium
MVP Executive Committee
Co-Chair: J. Michael Gaziano, M.D., M.P.H.
Co-Chair: Rachel Ramoni, D.M.D., Sc.D.
Jim Breeling, M.D. (ex-officio)
Kyong-Mi Chang, M.D.
Grant Huang, Ph.D.
Sumitra Muralidhar, Ph.D.
Christopher J. O’Donnell, M.D., M.P.H.
Philip S. Tsao, Ph.D.
MVP Program Office
Sumitra Muralidhar, Ph.D.
Jennifer Moser, Ph.D.
MVP Recruitment/Enrollment
Recruitment/Enrollment Director/Deputy Director, Boston – Stacey B. Whitbourne, Ph.D.; Jessica V. Brewer, M.P.H.
- MVP Coordinating Centers
- Clinical Epidemiology Research Center (CERC), West Haven – John Concato,
- M.D., M.P.H.
- Cooperative Studies Program Clinical Research Pharmacy Coordinating Center,
- Albuquerque - Stuart Warren, J.D., Pharm D.; Dean P. Argyres, M.S. o Genomics Coordinating Center, Palo Alto – Philip S. Tsao, Ph.D.
- Massachusetts Veterans Epidemiology Research Information Center
- (MAVERIC), Boston - J. Michael Gaziano, M.D., M.P.H.
- MVP Information Center, Canandaigua – Brady Stephens, M.S.
Core Biorepository, Boston – Mary T. Brophy M.D., M.P.H.; Donald E. Humphries, Ph.D.
MVP Informatics, Boston – Nhan Do, M.D.; Shahpoor Shayan
Data Operations/Analytics, Boston – Xuan-Mai T. Nguyen, Ph.D.
MVP Science
Genomics - Christopher J. O’Donnell, M.D., M.P.H.; Saiju Pyarajan Ph.D.; Philip S. Tsao, Ph.D.
Phenomics - Kelly Cho, M.P.H, Ph.D.
Data and Computational Sciences – Saiju Pyarajan, Ph.D.
Statistical Genetics – Elizabeth Hauser, Ph.D.; Yan Sun, Ph.D.; Hongyu Zhao, Ph.D.
MVP Local Site Investigators
Atlanta VA Medical Center (Peter Wilson)
Bay Pines VA Healthcare System (Rachel McArdle)
Birmingham V A Medical Center (Louis Dellitalia)
Cincinnati VA Medical Center (John Harley)
Clement J. Zablocki VA Medical Center (Jeffrey Whittle)
Durham V A Medical Center (Jean Beckham)
Edith Nourse Rogers Memorial Veterans Hospital (John Wells)
Edward Hines, Jr. VA Medical Center (Salvador Gutierrez)
Fayetteville V A Medical Center (Gretchen Gibson)
VA Health Care Upstate New York (Laurence Kaminsky)
New Mexico VA Health Care System (Gerardo Villareal)
VA Boston Healthcare System (Scott Kinlay)
VA Western New York Healthcare System (Junzhe Xu)
Ralph H. Johnson VA Medical Center (Mark Hamner)
Wm. Jennings Bryan Dorn VA Medical Center (Kathlyn Sue Haddock)
VA North Texas Health Care System (Sujata Bhushan)
Hampton VA Medical Center (Pran Iruvanti)
Hunter Holmes McGuire VA Medical Center (Michael Godschalk)
Iowa City VA Health Care System (Zuhair Ballas)
Jack C. Montgomery VA Medical Center (Malcolm Buford)
James A. Haley Veterans’ Hospital (Stephen Mastorides)
Louisville V A Medical Center (Jon Klein)
Manchester V A Medical Center (Nora Ratcliffe)
Miami VA Health Care System (Hermes Florez)
Michael E. DeBakey VA Medical Center (Alan Swann)
Minneapolis VA Health Care System (Maureen Murdoch)
N. FL/S. GA Veterans Health System (Peruvemba Sriram)
Northport VA Medical Center (Shing Shing Yeh)
Overton Brooks VA Medical Center (Ronald Washburn)
Philadelphia V A Medical Center (Darshana Jhala)
Phoenix VA Health Care System (Samuel Aguayo)
Portland VA Medical Center (David Cohen)
Providence V A Medical Center (Satish Sharma)
Richard Roudebush VA Medical Center (John Callaghan)
Salem VA Medical Center (Kris Ann Oursler)
San Francisco VA Health Care System (Mary Whooley)
South Texas Veterans Health Care System (Sunil Ahuja)
Southeast Louisiana Veterans Health Care System (Amparo Gutierrez)
Southern Arizona VA Health Care System (Ronald Schifman)
Sioux Falls VA Health Care System (Jennifer Greco)
St. Louis VA Health Care System (Michael Rauchman)
Syracuse VA Medical Center (Richard Servatius)
VA Eastern Kansas Health Care System (Mary Oehlert)
VA Greater Los Angeles Health Care System (Agnes Wallbom)
VA Loma Linda Healthcare System (Ronald Fernando)
VA Long Beach Healthcare System (Timothy Morgan)
VA Maine Healthcare System (Todd Stapley)
VA New York Harbor Healthcare System (Scott Sherman)
VA Pacific Islands Health Care System (Gwenevere Anderson)
VA Palo Alto Health Care System (Philip Tsao)
VA Pittsburgh Health Care System (Elif Sonel)
VA Puget Sound Health Care System (Edward Boyko)
VA Salt Lake City Health Care System (Laurence Meyer)
VA San Diego Healthcare System (Samir Gupta)
VA Southern Nevada Healthcare System (Joseph Fayad)
VA Tennessee Valley Healthcare System (Adriana Hung)
Washington DC VA Medical Center (Jack Lichy)
W.G. (Bill) Hefner VA Medical Center (Robin Hurley)
White River Junction VA Medical Center (Brooks Robey)
William S. Middleton Memorial Veterans Hospital (Robert Striker)
Footnotes
Disclosures: None
References:
- 1.de Ferranti SD, et al. Prevalence of Familial Hypercholesterolemia in the 1999 to 2012 United States National Health and Nutrition Examination Surveys (NHANES). Circulation. 2016;133:1067–72. [DOI] [PubMed] [Google Scholar]
- 2.Khera AV, et al. Diagnostic Yield and Clinical Utility of Sequencing Familial Hypercholesterolemia Genes in Patients With Severe Hypercholesterolemia. J Am Coll Cardiol. 2016;67:2578–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Versmissen J, et al. Efficacy of statins in familial hypercholesterolaemia: a long term cohort study. BMJ. 2008;337:a2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wierzbicki AS, et al. Familial hypercholesterolaemia: summary of NICE guidance. BMJ. 2008;337:a1095. [DOI] [PubMed] [Google Scholar]
- 5.Dotson WD, et al. Prioritizing genomic applications for action by level of evidence: a horizon-scanning method. Clin Pharmacol Ther. 2014;95:394–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertolini S, et al. Clinical expression of familial hypercholesterolemia in clusters of mutations of the LDL receptor gene that cause a receptor-defective or receptor-negative phenotype. Arterioscler Thromb Vasc Biol. 2000;20:E41–52. [DOI] [PubMed] [Google Scholar]
- 7.Jansen AC, et al. Phenotypic variability in familial hypercholesterolaemia: an update. Curr Opin Lipidol. 2002;13:165–71. [DOI] [PubMed] [Google Scholar]
- 8.Abul-Husn NS, et al. Genetic identification of familial hypercholesterolemia within a single U.S. health care system. Science. 2016;354. [DOI] [PubMed] [Google Scholar]
- 9.Gaziano JM, et al. Million Veteran Program: A mega-biobank to study genetic influences on health and disease. J Clin Epidemiol 2016;70:214–23. [DOI] [PubMed] [Google Scholar]
- 10.Klarin D, et al. Genetics of blood lipids among ~300,000 multi-ethnic participants of the Million Veteran Program. Nat Genet. 2018;50:1514–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Humphries SE, et al. Genetic causes of familial hypercholesterolaemia in patients in the UK: relation to plasma lipid levels and coronary heart disease risk. J Med Genet. 2006;43:943–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huijgen R, et al. Cardiovascular risk in relation to functionality of sequence variants in the gene coding for the low-density lipoprotein receptor: a study among 29,365 individuals tested for 64 specific low-density lipoprotein-receptor sequence variants. Eur Heart J. 2012;33:2325–30. [DOI] [PubMed] [Google Scholar]
- 13.Wright ML, et al. A perspective for sequencing familial hypercholesterolaemia in African Americans. NPJ Genom Med. 2016;1:16012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hopkins PN, et al. Familial hypercholesterolemias: prevalence, genetics, diagnosis and screening recommendations from the National Lipid Association Expert Panel on Familial Hypercholesterolemia. J Clin Lipidol. 2011;5:S9–17. [DOI] [PubMed] [Google Scholar]
- 15.Sjouke B, et al. Homozygous autosomal dominant hypercholesterolaemia in the Netherlands: prevalence, genotype-phenotype relationship, and clinical outcome. Eur Heart J. 2015;36:560–5. [DOI] [PubMed] [Google Scholar]
- 16.Watts GF, et al. Prevalence and treatment of familial hypercholesterolaemia in Australian communities. Int J Cardiol. 2015;185:69–71. [DOI] [PubMed] [Google Scholar]
- 17.Benn M, et al. Mutations causative of familial hypercholesterolaemia: screening of 98 098 individuals from the Copenhagen General Population Study estimated a prevalence of 1 in 217. Eur Heart J. 2016;37:1384–94. [DOI] [PubMed] [Google Scholar]
- 18.Klancar G, et al. Universal Screening for Familial Hypercholesterolemia in Children. J Am Coll Cardiol. 2015;66:1250–7. [DOI] [PubMed] [Google Scholar]
- 19.Ademi Z, et al. Cascade screening based on genetic testing is cost-effective: evidence for the implementation of models of care for familial hypercholesterolemia. J Clin Lipidol. 2014;8:390–400. [DOI] [PubMed] [Google Scholar]
- 20.Wald DS, et al. Child-parent screening for familial hypercholesterolemia. J Pediatr. 2011;159:865–7. [DOI] [PubMed] [Google Scholar]
- 21.Wald DS, et al. Child-Parent Familial Hypercholesterolemia Screening in Primary Care. N Engl J Med. 2016;375:1628–1637. [DOI] [PubMed] [Google Scholar]
- 22.Hadfield SG, et al. Implementation of cascade testing for the detection of familial hypercholesterolaemia. Curr Opin Lipidol. 2005;16:428–33. [DOI] [PubMed] [Google Scholar]
- 23.Haralambos K, et al. Diagnostic scoring for familial hypercholesterolaemia in practice. Curr Opin Lipidol. 2016;27:367–74. [DOI] [PubMed] [Google Scholar]
- 24.Sturm AC, et al. Clinical Genetic Testing for Familial Hypercholesterolemia: JACC Scientific Expert Panel. J Am Coll Cardiol. 2018;72:662–680. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.