Summary
Limitation for amino acids is thought to regulate translation in mammalian cells primarily by signaling through the kinases mTORC1 and GCN2. We find that a selective loss of arginine tRNA charging during limitation for the amino acid arginine regulates translation through ribosome pausing at two of six arginine codons. Surprisingly, limitation for leucine, an essential and abundant amino acid in protein, results in little or no ribosome pausing. Chemical and genetic perturbation of mTORC1 and GCN2 signaling revealed that their robust response to leucine limitation prevents ribosome pausing, while an insufficient response to arginine limitation led to loss of tRNA charging and ribosome pausing. Codon-specific ribosome pausing decreased protein production and triggered premature ribosome termination without reducing mRNA levels. Together, our results suggest that amino acids which are not optimally sensed by the mTORC1 and GCN2 pathways still regulate translation through an evolutionarily conserved mechanism based on codon-specific ribosome pausing.
Introduction
Protein synthesis consumes the highest fraction of nutrients and energy stores in proliferating cells (Buttgereit and Brand, 1995), and is therefore tightly controlled in response the levels of its amino acid substrates. In eukaryotic cells, amino acid limitation is sensed by two conserved signaling pathways anchored around the kinases mechanistic Target Of Rapamycin in Complex 1 (mTORC1) and General Control Nonderepressible 2 (GCN2) (Kimball, 2002). Amino acid limitation inhibits mTORC1 signaling and activates GCN2 signaling, reducing overall protein synthesis rate through a decrease in the rate of ribosome initiation on mRNA transcripts (Sonenberg and Hinnebusch, 2009). The failure of either pathway to respond to amino acid limitation can lead to cell death, particularly in nutrient-challenged contexts such as tumors (Nofal et al., 2017; Ye et al., 2010), underscoring the importance of regulatory control over protein synthesis in maintaining cellular homeostasis. The mTORC1 and GCN2 pathways respond strongly to simultaneous limitation for all 20 amino acids (Kimball, 2002), yet their responses to individual amino acid limitation are markedly different. mTORC1 signaling is highly sensitive to leucine levels, and to a lesser extent, to arginine and glutamine levels (Hara et al., 1998). GCN2, which senses amino acid limitation by binding uncharged tRNAs, has a similar affinity for different tRNAs in vitro (Dong et al., 2000; Zaborske et al., 2010), but variable activation of its effectors upon limitation for individual amino acids is nonetheless detected (Jousse et al., 2000; Tang et al., 2015). As many cancers exhibit dependence on specific amino acids (Hattori et al., 2017; Jain et al., 2012; Knott et al., 2018; Krall et al., 2016; Loayza-Puch et al., 2016; Possemato et al., 2011; Scott et al., 2000; Wise and Thompson, 2010), it is crucial to understand how these variegated mTORC1 and GCN2 responses are integrated, and whether they are sufficient, to regulate protein synthesis during individual amino acid limitation.
In addition to mTORC1- and GCN2-mediated regulation of translation initiation, amino acid limitation can affect protein synthesis by reducing the elongation rate of ribosomes. In bacteria, limitation for single auxotrophic amino acids causes loss of tRNA charging and ribosome pausing at a subset of synonymous codons cognate to the limiting amino acid (Dittmar et al., 2005; Subramaniam et al., 2013a). This ribosome pausing results in abortive termination and a consequent decrease in protein expression (Ferrin and Subramaniam, 2017; Subramaniam et al., 2013b, 2014). Notably, the codons at which ribosomes pause during amino acid limitation are not necessarily rare codons or decoded by low abundance tRNAs (Subramaniam et al., 2013b, 2013a, 2014). Ribosome pausing has also been observed in pathological mammalian states, including in a mouse model of neurodegeneration (Ishimura et al., 2014), and in patient-derived cancer tissue (Loayza-Puch et al., 2016), although the cause of steady-state ribosome pausing in tumors is unclear. Further, the codon-specificity and effect of ribosome pausing on protein expression have not been studied in mammalian systems, though codon usage frequency and tRNA levels have been implicated in the regulation of ribosome elongation rate and protein production during metastasis, differentiation, and amino acid limitation (Gingold et al., 2014; Goodarzi et al., 2016; Saikia et al., 2016). However, ribosome profiling studies have failed to find evidence for a simple relationship between codon usage, tRNA levels and ribosome density in mammalian cells (Ingolia et al., 2011; Qian et al., 2012).
Here, we investigated how amino acid signaling pathways and codon usage interact to regulate protein synthesis in response to limitation for single amino acids across multiple human cell lines. We focused on two amino acids, leucine and arginine, which can both regulate protein synthesis by acting as direct signals to mTORC1 (Chantranupong et al., 2016; Wang et al., 2015; Wolfson et al., 2016). Upon arginine limitation, we found that a stereotypical pattern of ribosome pausing emerges at two out of six arginine codons across cell lines, suggesting that arginine becomes a rate-limiting substrate for protein synthesis. Intriguingly, there was little to no ribosome slow-down at any leucine codon upon its limitation, though it is an essential amino acid. Perturbing amino acid signaling revealed that tRNA charging loss and ribosome pausing are driven by an inadequate response to amino acid limitation through the mTORC1 and GCN2 pathways. By establishing a molecular framework relating tRNA charging, ribosome elongation, and protein expression during amino acid depletion, our work provides a rational starting point from which to dissect disease states, such as cancers, that experience nutrient limitation and exhibit dysregulated ribosome dynamics.
Results
1.1. Ribosomes pause at specific synonymous codons upon limitation for arginine but not leucine
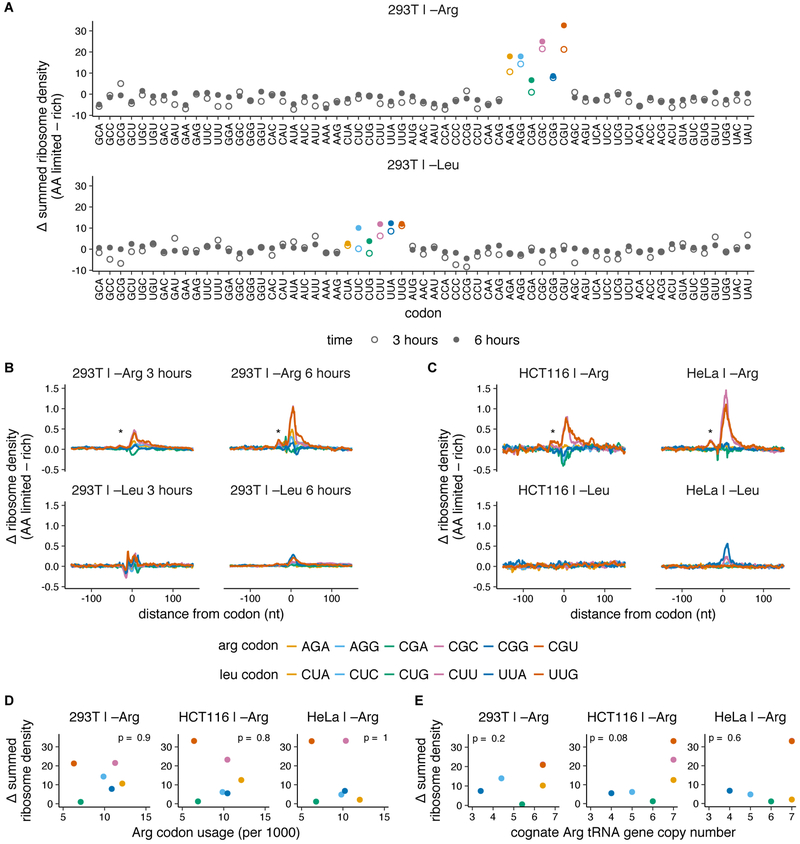
To systematically explore the effect of individual amino acid depletion on translation in mammalian cells, we performed ribosome profiling (Ingolia et al., 2009, 2012) in three human cell lines – HEK293T, HeLa and HCT116 – during limitation for either leucine or arginine. Although ribonuclease I (RNaseI) is typically used to generate RNA footprints for ribosome profiling, micrococcal nuclease (MNase) better preserved monosome integrity (Fig. S1A-C, Methods), and sequencing the resulting footprints (Fig. S1D) yielded reads with three nucleotide periodicity enriched in coding regions, despite a broader length distribution (Dunn et al., 2013; Reid et al., 2015) (Fig. S1E-G). To assess the extent of ribosome pausing upon amino acid limitation, we quantified the net increase in normalized average ribosome footprint density in the window around each of the 61 sense codons (Fig. 1A; Fig. S1H, Methods).
Fig. 1. Codon-specific ribosome pausing emerges during limitation for arginine, but not leucine.
(A-C) Changes in codon-specific ribosome density in HEK293T cells, HCT116, and HeLa cells upon 3 or 6 hours of leucine or arginine limitation. Ribosome density for each codon is calculated relative to the mean footprint density for each coding sequence, and is averaged over all occurrences of the codon across detectably expressed transcripts (see Methods for details). The difference in ribosome density between amino acid limited and rich conditions across a 150 nt window around each codon is summed (A) or shown as such (B,C) (* = trailing ribosome stalled behind the paused ribosome). Arg and Leu codons are colored according to legend in B,C (D-E) The summed change in ribosome density at arginine codons following 3 hours of arginine limitation in each cell line (see A, S1H) is compared to the transcriptome usage frequency of Arg codons (see Fig. S1J) (D) or genomic copy number of the cognate tRNA for each Arg codon (see Fig. S1K) (E). Arg and Leu codons are colored according to legend in B,C. p indicates p-value of Spearman’s rank coefficient, ρ and is shown at the top of each plot (in D: HEK293T, ρ = −0.1; HCT116, ρ = −0.14; HeLa, ρ = 0.03. In E: HEK293T, ρ = 0.58; HCT116, ρ = 0.76; HeLa, ρ = 0.27).
Upon arginine limitation for three hours, two of the six arginine codons–CGC and CGU–had a substantial increase in ribosome density across all three cell lines (Fig. 1A,C; Fig. S1H). Ribosome pausing at these codons increased after arginine limitation for six hours (Fig. 1B). None of the codons encoding the other 19 amino acids had increased ribosome density upon arginine limitation (Fig. 1A, Fig. S1H). We also observed smaller peaks in ribosome density approximately one ribosome footprint length (~ 30 nucleotides) behind the major peaks at CGC and CGU codons (Fig. 1 B,C; asterisks). Similar satellite peaks, presumably caused by collision of the trailing ribosome with the paused ribosome, have been previously observed during limitation for single amino acids in E. coli (Subramaniam et al., 2014) and S. cerevisiae (Guydosh and Green, 2014).
In contrast, none of the six leucine codons displayed a consistent increase in ribosome density in response to leucine limitation (Fig. 1A-C; Fig. S1H). Since leucine cannot be synthesized, we were surprised to find that ribosome elongation at leucine codons is largely unperturbed by leucine limitation. We considered the possibility that cells do not experience major changes in intracellular leucine levels upon its external limitation. However, intracellular arginine and leucine levels fell close to the detection limit of our measurement when they were each removed from the growth medium, suggesting that cells are effectively starved for both amino acids in these conditions (Fig. S1I).
We then tested whether the increase in ribosome density at specific codons upon arginine limitation correlated with simple measures of codon optimality or tRNA abundance, as hypothesized previously (Gingold et al., 2014; Goodarzi et al., 2016; Saikia et al., 2016). The pausing hierarchy did not correlate significantly in any cell line with either transcriptomic codon usage (Fig. 1D, Fig. S1J) or genomic copy number of the cognate tRNA (Fig. 1E, Fig. S1K,L) (Spearman’s rank correlation p-values displayed on plots). Nevertheless, the consistent hierarchy of codon-specific ribosome pausing upon arginine limitation, and its absence during leucine limitation, suggests a common principle underlying the emergence of ribosome pausing.
1.2. Cognate tRNA charging loss upon amino acid limitation sets the hierarchy of ribosome pausing at synonymous codons
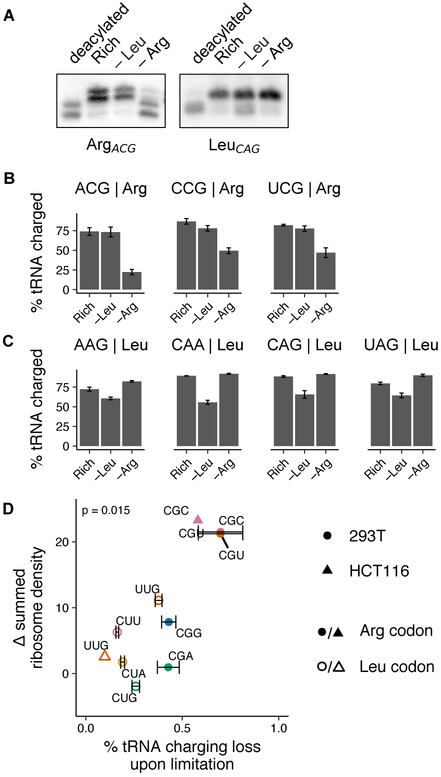
As ribosome elongation rate at a codon depends on recruitment of the cognate charged tRNA, we expected that the arginine tRNA which decodes the pause-site codons CGC and CGU, with the anticodon ACG (tRNAArgACG), would exhibit a greater charging loss upon arginine limitation than the isoacceptor arginine tRNAs that decode the remaining four arginine codons. In line with this expectation, tRNAArgACG lost 70% of its charging upon arginine limitation in HEK293T cells (Fig. 2A, Fig. S2A). By contrast, tRNAArgCCG and tRNAArgUCG, which decode the arginine codons CGG and CGA at which we did not observe strong pausing, lost less than 45% of their charging (Fig. 2A, Fig. S2A). All leucine tRNAs tested lost less than 40% of their charging upon leucine limitation, consistent with the observation that there is no ribosome pausing at leucine codons (Fig. 2B, Fig. S2B). Charging loss was also more severe for tRNAArgACG than a leucine tRNA in the HCT116 cell line (Fig. S2C). As expected, arginine and leucine tRNAs were between 75% to 90% charged during growth in rich conditions, and upon limitation for a non-cognate amino acid (Fig. 2A,B). Charging loss was also more severe for tRNAArgACG than a leucine tRNA in the HCT116 cell line (Supp. Fig. 2C). Overall we found a positive correlation between the change in ribosome density at a codon and the loss in charging of its cognate tRNA upon limitation for an amino acid (Fig. 2C; Spearman’s rank correlation coefficient p-value = 0.015). Further, our results suggest that ribosomes begin to pause at a codon only when a majority of the cognate charged tRNA is depleted.
Fig. 2. Selective loss of cognate tRNA charging during arginine limitation.
(A) Representative northern blots for determination of Arg and Leu tRNA charging levels (as shown in B) in HEK293T cells following 3 hours of limitation for leucine or arginine or growth in rich medium. A control deacylated total RNA sample is used to identify uncharged tRNA species. tRNA probe is indicated below each blot (see Methods for details on interpretation, quantification, and probe design; see Fig. S2 for additional blots). (B-C) tRNA charging levels for 3 Arg (B) and 4 Leu tRNAs (C) in HEK293T cells following 3 hours of leucine or arginine limitation or growth in rich medium (calculated as described in Methods). tRNA anticodon and isotype are indicated above plots; error bars represent the standard error of the mean from three technical replicates (see A and Fig. S2 for additional representative blots and Fig. S1L for codon-tRNA pairs). (D) The summed change in ribosome density at arginine and leucine codons following 3 hours of leucine or arginine limitation in HEK293T and HCT116 cells (see Fig. 1A) is plotted against the loss in charging for the cognate tRNA (for those measured) in the same condition. p indicates p-value of Spearman’s rank coefficient, ρ and is shown at the top of the plot (ρ = 0.7).
1.3. Differential mTORC1 and GCN2 responses to arginine and leucine limitation
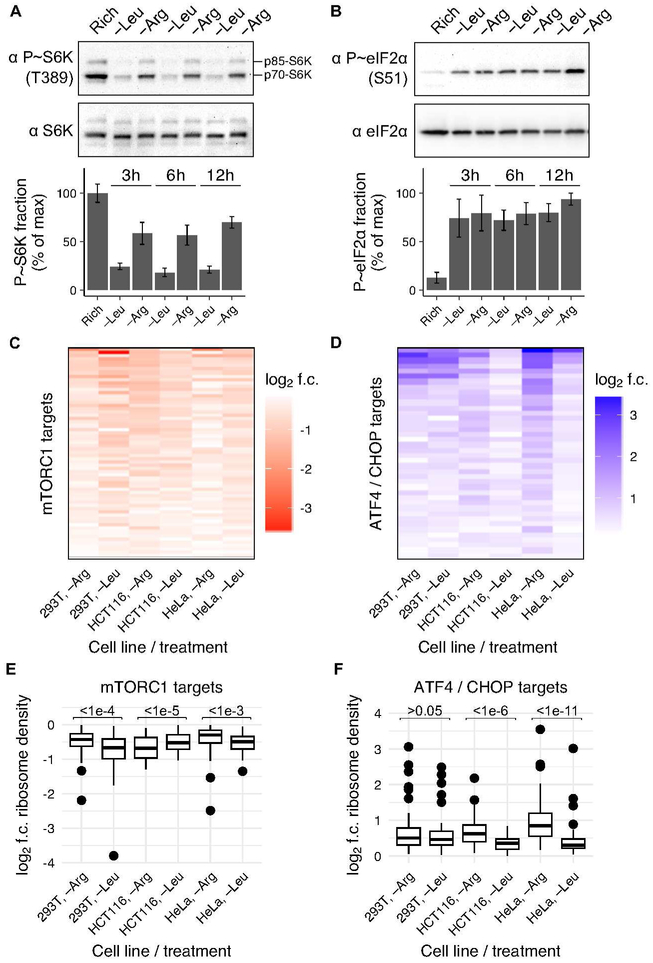
We next examined whether the loss of charged tRNA and emergence of ribosome pausing during arginine but not during leucine limitation might be related to the amino acid signaling response through the GCN2 and mTORC1 kinases, given that they are presumed to sense amino acid levels and co-ordinately regulate protein synthesis in order to maintain intracellular amino acid homeostasis (Park et al., 2017; Zhang et al., 2002). Consistent with previous reports (Hara et al., 1998), we observed greater mTORC1 inhibition during limitation for leucine in comparison to arginine – levels of the mTORC1 target phosphorylated ribosomal protein S6 kinase 1 (P~S6K) fell by 75% during leucine limitation, but only 45% during arginine limitation in HEK293T cells (Fig. 3A). Levels of the S6K target phosphorylated ribosomal protein S6 (P~RPS6) reflected the same differential response (Fig. S3A,B). GCN2 signaling was activated during limitation for both amino acids in these cells, as levels of the GCN2 target phosphorylated eIF2 ✔ (P~eIF2 ✔) increased to a similar extent (Fig. 3B).
Fig. 3. Differential mTORC1 and GCN2 responses to arginine and leucine limitation.
(A,B) Representative western blots for phosphorylated and total levels of ribosomal protein S6 kinase 1 (S6K) (A) or eIF2α (B) in HEK293T cells after growth in rich medium or after 3, 6, or 12 hours of leucine or arginine limitation. Bar graph shows percent of protein that is phosphorylated in each condition, relative to the maximum. Error bars represent the standard error of the mean from three technical replicates. (C,D) Heatmap of log2 fold-change (f.c.) in ribosome density for mRNA targets of mTORC1 inhibition (Hsieh et al., 2012) (C) or GCN2 activation via ATF4/CHOP (Han et al., 2013) (D), following 3 hours of leucine or arginine limitation relative to growth in rich medium for HEK293T, HCT116, and HeLa cells. Only targets with a log2 fold change of < 0, for mTORC1 targets, or > 0, for ATF4/CHOP targets, were considered. In HEK293T, HCT116, and HeLa cells, 46/63 (73%), 14/63 (22%), and 45/63 (71%) of mTORC1 targets had higher ribosome density upon arginine than leucine limitation, respectively (C) and 26/40 (65%), 35/40 (88%), and 40/40 (100%) of GCN2 targets had higher ribosome density upon arginine than leucine limitation, respectively (D). (E,F) Box plot of the log2 fold change for each mTORC1 (E) or ATF4/CHOP (F) target upon amino acid limitation (as shown in C,D). A two-sided Wilcoxon signed rank test with continuity correction was performed with μ = 0; the resulting p-value is shown above each comparison (see Methods for details). In HEK293T, HCT116, and HeLa cells, the mTORC1 signaling response was 1.2-, 0.9-, and 1.1-fold higher during limitation for arginine, respectively (E) and the GCN2 signaling response was 1-, 1,2, and 1.5-fold higher during limitation for arginine, respectively (F).
mTORC1 kinase activity in HEK293T cells mirrored downstream changes in ribosome density on mRNA targets of the pathway. 46 of 63 mRNAs that are translationally repressed by mTORC1 inhibition (Hsieh et al., 2012; Thoreen et al., 2012) had lower ribosome density during limitation for leucine than arginine (Fig. 3C,E; Fig. S3C,E,G; Wilcoxon signed rank test p = 1.2e-05). mTORC1 signaling was also more repressed during limitation for leucine in HeLa cells (Fig. 3C,E; Fig. S3G; p = 0.0003). There was little mTORC1 or GCN2 signaling response to leucine limitation in HCT116 cells (Fig. 3C-F; Fig. S3G), consistent with our observation that leucine tRNA charging is largely unaffected by leucine limitation in this cell line (Fig. S2C).
Comparing downstream changes in ribosome density on mRNA targets of ATF4 and CHOP, transcriptional effectors downstream of GCN2 (Han et al., 2013), during arginine versus leucine limitation revealed subtle but consistent differential GCN2 responses. In HEK293T cells, the GCN2 transcriptional response was similar during limitation for leucine and arginine (Fig. 3D,F; Fig. S3D,F,G; Wilcoxon signed rank test p = 0.33). However, GCN2 became significantly more activated during arginine limitation after a longer duration of amino acid limitation (Fig. S3D,F; p = 5.7e-4), which also increased ribosome pausing (Fig. 1A,B). GCN2 was also more activated during limitation for arginine in the HCT116 and HeLa cell lines (Fig. 3D,F; Fig. S3G; p = 9.3e-07 and 1.8e-12, respectively). The GCN2 response was generally most robust in the conditions and cell lines in which ribosome pausing was most severe, consistent with the recent observation that GCN2 may be activated downstream of ribosome pausing (Ishimura et al., 2016).
The variability of the signaling responses across all three cell lines was surprising, given that we observed a conserved signature of ribosome pausing. However, if pausing is determined by the extent to which the amino acid supply and demand are matched under each condition, it may be the totality of the signaling response, rather than the activity of each single pathway, that regulates this balance. We sought to test this idea in the HEK293T cell line, in which ribosome pausing emerges only during arginine limitation, in the context of a relatively weaker overall signaling response than leucine limitation.
1.4. An insufficient mTORC1 and GCN2 response to amino acid limitation induces ribosome pausing
The mTORC1 and GCN2 pathways inhibit the initiation phase of protein synthesis in response to amino acid limitation (Sonenberg and Hinnebusch, 2009). This lowers the number of elongating ribosomes, a major demand source for the cytosolic amino acid pool, and thereby reduces the consumption rate of a limiting amino acid. If this combined signaling response does not sufficiently reduce arginine consumption during its limitation, tRNA charging loss and ribosome pausing could result. Specifically, if residual mTORC1 activity and/or inadequate activation of GCN2 drives loss of tRNA charging and ribosome pausing, we hypothesized that increasing the response of these pathways would reduce pausing upon arginine limitation, and conversely, that decreasing their response would induce pausing upon leucine limitation. To test this hypothesis, we employed chemical and genetic methods to perturb the mTORC1 and GCN2 responses to arginine and leucine limitation in HEK293T cells, and determined the resulting changes to tRNA charging and ribosome pausing.
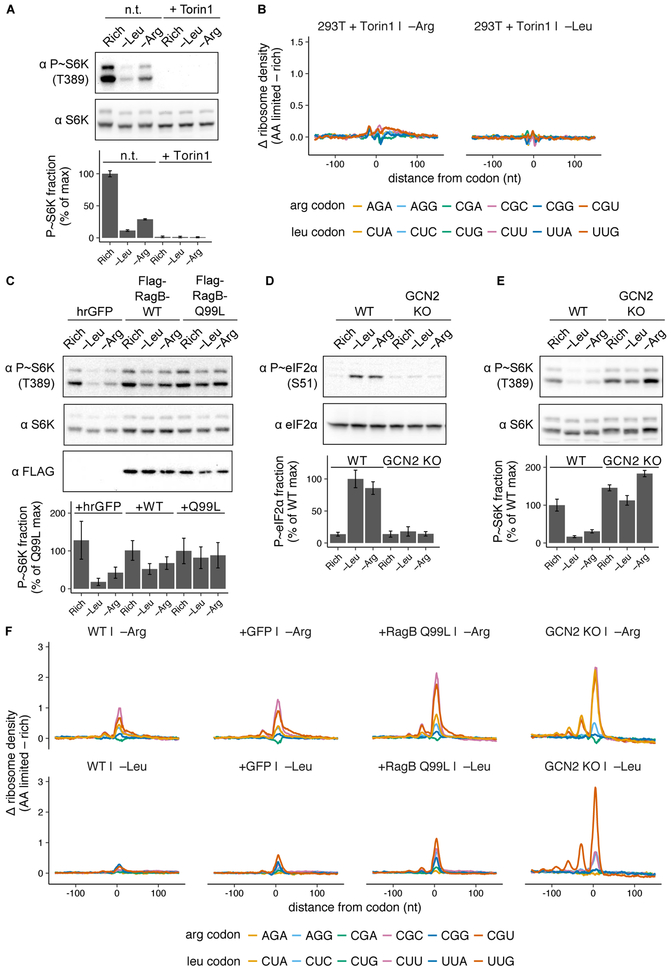
We first inhibited mTOR kinase activity using the catalytic site inhibitor Torin1 (Thoreen et al., 2009) (Fig. 4A) during both arginine and leucine limitation, and found that charging of all arginine and leucine tRNAs tested increased back to baseline rich condition levels (Fig. S4A). Torin1 treatment also prevented an increase in ribosome density at any codon upon leucine or arginine limitation (Fig. 4B, Fig. S4B), demonstrating that mTORC1 inhibition during amino acid limitation is sufficient to block depletion of the cognate charged tRNA fraction and ribosome pausing.
Fig. 4. Signaling through the mTORC1 and GCN2 pathways regulates the magnitude of ribosome pausing during amino acid limitation.
(A) Representative western blots for phosphorylated and total S6K in HEK293T cells after growth in rich medium or limitation for leucine or arginine for 3 hours, with or without (n.t.) 250 nM Torin1. Bar graph shows percent of protein that is phosphorylated, relative to the maximum; error bars represent the standard error of the mean from three technical replicates. (B) Changes in codon-specific ribosome density in the hrGFP cell line (as shown in C) after 3 hours of leucine or arginine limitation with 250 nM Torin1, relative to rich medium. (C) Representative western blots for phosphorylated S6K, total S6K, and FLAG after growth in rich medium, or 3 hours of leucine or arginine limitation in HEK293T cells stably expressing either hrGFP, FLAG-RagB-WT (RagB-WT), or FLAG-RagB-Q99L (RagB-Q99L). Bar graph shows percent of protein that is phosphorylated, relative to the maximum in the RagB-Q99L cell line; error bars represent the standard error of the mean from three technical replicates. (D,E) Representative western blots for phosphorylated and total eIF2α (D) or S6K (E) after growth in rich medium, or 3 hours of leucine or arginine limitation in the HEK293T (WT) or GCN2 KO cell lines. Bar graphs show percent of protein that is phosphorylated, relative to the maximum in WT cells; error bars represent the standard error of the mean from three technical replicates. (F) Changes in codon-specific ribosome density for WT, hrGFP, FLAG-RagB-Q99L, and GCN2 KO cell lines following 6 hours of limitation for leucine or arginine, relative to rich medium.
Next, we tested whether loss of the mTORC1 response to amino acid limitation would exacerbate tRNA charging loss and ribosome pausing. We rendered mTORC1 insensitive to amino acid levels by stable overexpression of a constitutively active form of its upstream regulator, RagB GTPase (RagB-Q99L) (Sancak et al., 2008) (Fig. 4C). The RagB-Q99L cell line exhibited reduced leucine tRNA charging during leucine limitation; charging fell to 22% for tRNALeuCAA, which decodes the codon UUG (Fig. S4C). Compared with charging in a control line that over-expressed humanized R. reniformis fluorescent protein (hrGFP), constitutive mTORC1 activity increased charging loss due to leucine limitation by 50%. Charging was also reduced by 36% due to constitutive mTORC1 activity for tRNALeuAAG, which decodes CUU (Fig. S4C). Concordantly, minor ribosome pausing emerged at the leucine codons UUG and CUU (Fig. S4D). Little difference was detected in arginine tRNA charging or ribosome pausing at arginine codons upon arginine limitation (Fig. S4C,D), and we thus repeated these measurements after 6 hours, rather than 3 hours, of amino acid limitation to reveal any effects that might become more pronounced over time.
After prolonged amino acid limitation, the RagB-Q99L cell line exhibited further increased tRNA charging loss and ribosome pausing compared to control cell lines. Charging fell as low as 18% for tRNALeuCAA (Fig. S4E) and ribosome pausing emerged at the cognate leucine codon UUG as well as the CUC and CUU codons (Fig. 4F; Fig. S4F). Similarly, the proportion of charged tRNAArgACG fell to 19% (Fig. S4E) and ribosome pausing increased at the cognate arginine codons CGC and CGU (Fig. 4F; Fig. S4F). Ribosome pausing was also increased slightly in the hrGFP control cell line relative to unperturbed HEK293T cells (WT) (Fig. 4F, Fig. S4F), possibly due to the translational burden of transgene overexpression (Elf et al., 2003). In summary, constitutive mTORC1 activation significantly worsened tRNA charging loss and exacerbated ribosome pausing during both leucine and arginine limitation.
We next investigated the role of GCN2 in ribosome pausing. We constructed a GCN2 knockout (GCN2 KO) cell line by CRISPR/Cas9 targeting (Cong et al., 2013) (Fig. S4G) in which eIF2α was not phosphorylated in response to amino acid limitation (Fig. 4D). GCN2 activation is reported to be necessary for inhibition of mTORC1 signaling upon leucine or arginine limitation (Averous et al., 2016); we confirmed that there is no significant mTORC1 response to those conditions in our GCN2 KO cell line (Fig. 4E).
tRNA charging loss and ribosome pausing were greatly amplified in the GCN2 KO cell line. tRNALeuCAA charging fell to only 14% upon leucine limitation (Fig. S4E) and ribosome density at the UUG leucine codon rose substantially, with an average of 4 ribosomes stacked behind the paused ribosome (Fig. 4F, Fig. S4F). Pausing increased only slightly at the arginine CGC and CGU codons (Fig. 4F, Fig. S4F), although tRNAArgACG charging continued to drop (Fig. S4E), indicating that pause duration is approaching an upper limit at these codons. Indeed, significant ribosome pausing emerged at the AGA arginine codon (Fig. 4F, top panel; Fig. S4F), suggesting that charging of a second arginine isoacceptor, tRNAArgUCU, is exhausted upon arginine limitation in the GCN2 KO cell line. Together, these results indicate that the absence of a response through the GCN2 or mTORC1 pathways during amino acid limitation is sufficient to deplete charged tRNA pools and induce extensive genome-wide ribosome pausing at cognate codons, consistent with our hypothesis that an insufficient signaling response to amino acid limitation can drive consumption of the limiting amino acid into a substrate-limiting regime for protein synthesis.
In addition to their control over translation, mTORC1 and GCN2 regulate other critical functions, such as metabolism and autophagy (Sonenberg and Hinnebusch, 2009), which could affect intracellular amino acid levels. To test our hypothesis that amino acid levels during their respective limitation are primarily set by the demand from translation elongation, we briefly exposed cells limited for leucine or arginine to the elongation inhibitor cycloheximide. This was sufficient to significantly restore tRNALeuCAA and tRNAArgACG charging (Fig. S4H), indicating that the flux of arginine and leucine into translation is a key determinant of the cytosolic levels of these amino acids upon their limitation. Thus, the ribosome pausing outcome during limitation for an amino acid is likely determined by the translational control imposed by mTORC1 and GCN2.
1.5. Genome-wide ribosome pausing reduces global protein synthesis rate during arginine limitation
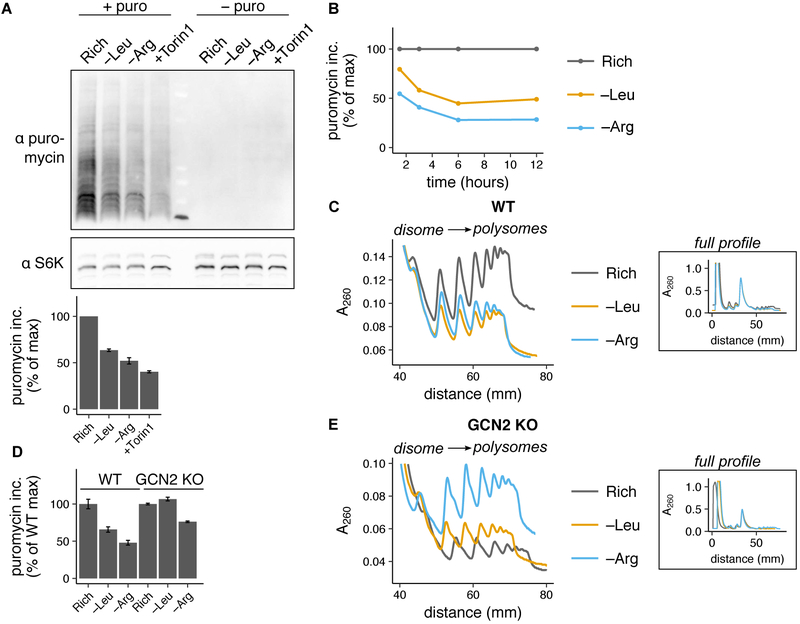
Having examined the upstream determinants of ribosome pausing, we next sought to investigate its impact on cellular translation. We measured global protein synthesis rate during limitation for leucine or arginine by quantifying incorporation of the antibiotic puromycin into nascent polypeptides (Schmidt et al., 2009), and found that global protein synthesis rate was consistently lower during limitation for arginine than leucine (Fig. 5A,B; Fig. S5A). This is consistent with previous measurements following protracted amino acid limitation (Scott et al., 2000).
Fig. 5. Ribosome pausing reduces global protein synthesis rate during amino acid limitation.
(A) Representative western blots for puromycin and S6K in HEK293T cells after (+ puro) or without (− puro) a pulse of 10 μg/mL puromycin following 3 hours of leucine or arginine limitation, treatment with 250 nM Torin1, or growth in rich medium. Bar graph shows puromycin incorporation relative to rich medium (calculated as described in Methods); error bars represent the standard error of the mean from three technical replicate experiments. (B) Puromycin incorporation in HEK293T cells following 1.5, 3, 6, or 12 hours of leucine or arginine limitation, relative to rich medium. (C) Polysome profiles from HEK293T (WT) cells following 6 hours of leucine or arginine limitation or growth in rich medium. The main plot shows overlaid polysome profiles starting at the disome (2 ribosome) peak and the inset plots show the entire profile, aligned with respect to the monosome peak height along the y-axis and position along the x-axis. (D) Puromycin incorporation in WT or GCN2 KO cell lines following 3 hours of leucine or arginine limitation, relative to rich medium (calculated as in described in Methods, see S5G for representative blots). Error bars represent the standard error of the mean for three technical replicates. (E) Polysome profiles (as described in C) from the GCN2 KO cell line following 6 hours of limitation for leucine or arginine or growth in rich medium.
We reasoned that three processes could contribute to the regulation of translation during amino acid limitation: mTORC1 inhibition, GCN2 activation, or ribosome pausing. Given that mTORC1 activity, which stimulates translation initiation, is higher during arginine limitation than leucine limitation (Fig. 3A,C,E; Fig. S3C,E), mTORC1 signaling cannot account for lower global protein synthesis during arginine relative to leucine limitation. The principal difference between GCN2- and ribosome pausing-mediated control over translation is that GCN2 regulates initiation, while ribosome pausing regulates elongation. To assess whether initiation or elongation rate control accounts for the greater reduction of global protein synthesis rate upon arginine limitation versus leucine limitation, we used polysome profiling to determine the average number of ribosomes per transcript in each condition. If global protein synthesis rate is lower during arginine limitation due to inhibition of initiation, there would be fewer ribosomes per transcript upon limitation for arginine compared to leucine. Instead, if global protein synthesis rate is reduced by slow elongation, we would find relatively more ribosomes per transcript upon arginine limitation. While the polysome fraction was reduced by limitation for either leucine or arginine, it was higher during arginine than leucine limitation in HEK293T cells (Fig. 5C, Fig. S5B), indicating that there are more ribosomes per transcript during arginine limitation despite a lower global protein synthesis rate. Thus, elongation rate control must account for the greater repression of global protein synthesis rate upon arginine limitation.
Elongation rate could be reduced by a global mechanism or by ribosome pausing during arginine limitation. To assess the role of reduced eukaryotic elongation factor 2 (EEF2) activity due to phosphorylation by EEF2 kinase (EEF2K) downstream of mTORC1 inhibition (Leprivier et al., 2013), we generated an EEF2K knockout cell line (Fig. S5C,D). Loss of general elongation factor regulation by EEF2K increased global protein synthesis rate upon arginine and leucine limitation by a similar, small margin (Fig. S5E). Therefore, downregulation of global elongation factor activity cannot explain the greater reduction of protein synthesis upon arginine than leucine limitation, and we instead attribute this difference to elongation rate control by ribosome pausing.
To isolate and quantify the contribution of ribosome pausing to global protein synthesis rate reduction, we made use of the GCN2 KO cell line, which lacks an initiation rate control response to amino acid limitation through both the GCN2 and mTORC1 pathways (Harding et al., 2000) (Fig. 4D,E). We reasoned that any residual inhibition of global protein synthesis rate during leucine or arginine limitation in the GCN2 KO cell line would be due to ribosome pausing. Global protein synthesis rate was reduced by 25% during arginine limitation (Fig. 5D, Fig. S5F). Strikingly, despite this lower global protein synthesis rate, there was a higher polysome fraction during arginine limitation than in rich conditions in this cell line (Fig. 5E, Fig. S5B), consistent with our observation of strong ribosome pausing under these conditions (Fig. 4F). Ribosome pausing also develops at a leucine codon in the GCN2 KO cell line, and accordingly the polysome fraction was higher upon limitation for leucine than in rich conditions as well (Fig. 5E). However, there was no change to global protein synthesis rate upon limitation for leucine in the GCN2 KO cell line (Fig. 5D, Fig. S5F), suggesting that global protein synthesis rate reduction in this condition in wild-type cells is primarily mediated by the mTORC1 and/or GCN2 responses. In sum, the inverse relationship between global protein synthesis rate and ribosome loading per transcript upon arginine limitation supports a model in which ribosome pausing reduces global protein synthesis rate.
1.6. Pause-site codons in mRNAs reduce protein expression and induce premature termination of translation
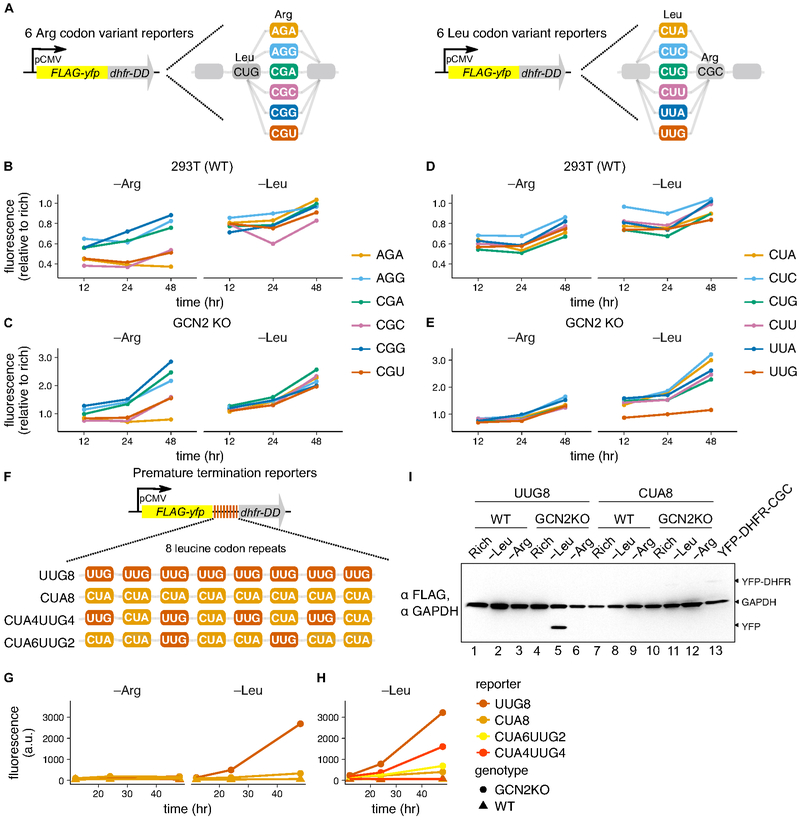
We next investigated whether pausing on mRNAs specifically inhibits production of the encoded protein. Towards this goal, we adapted a protein synthesis reporter in which YFP is fused to an engineered unstable E. coli dihydrofolate reductase (DHFR) domain (Han et al., 2014). In this reporter system, the unstable reporter is rapidly degraded and fluorescence only accumulates upon addition of a stabilizing ligand, trimethoprim (TMP). Fluorescence upon arginine and leucine limitation correlated well with the global protein synthesis rates we measured in those conditions (Fig. S6A, left plot, versus Fig. 5A,B), suggesting that it faithfully reflects the protein synthesis rate of the reporter. To determine the specific effect of ribosome pausing on reporter protein synthesis rate, we constructed a set of codon variant reporters in which either all arginine codons or all leucine codons were swapped to each of the six leucine or arginine codons, respectively (Fig. 6A).
Fig. 6. Ribosome pausing reduces protein expression from reporter mRNAs and induces premature termination of translation.
(A) Arginine and leucine YFP codon variant reporter design (see Methods for details). (B-E) Mean YFP fluorescence in the HEK293T (WT) (B,D) or GCN2 KO cell lines (C,E) stably expressing the arginine (B,C) or leucine (D,E) YFP codon variant reporters, following limitation for leucine or arginine with 10 μM trimethoprim (+TMP) for 12, 24, or 48 hours, relative to rich medium +TMP. (F) Premature termination reporter design. A short linker of 8 tandem CUA or UUG leucine codons was added to the YFP-CUA reporter (as shown in A). (G,H) Mean YFP fluorescence in the WT or GCN2 KO cell lines stably expressing the UUG8, CUA8 (G,H) CUA6UUG2, or CUA4UUG4 (H) reporters following limitation for leucine or arginine for 12, 24, or 48 hours without TMP. (I) Western blot for FLAG epitope and GAPDH in the WT or GCN2 KO cell lines stably expressing the UUG8 or CUA8 reporters after growth in rich medium or 48 hours of leucine or arginine limitation. Lane 13 contains lysate from the YFP-WT reporter cell line for a full-length reporter size reference; GAPDH provides an intermediate size reference (see Fig. S6F for overexpressed image).
We first determined whether the pause-site arginine codons, CGC and CGU, would reduce reporter protein synthesis rate during arginine limitation. In all three cell lines in which we detected ribosome pausing during arginine limitation (Fig. 1A-C), YFP-DHFR synthesis rate was reduced during arginine limitation when the pause-site codons CGC or CGU were used to encode arginine (YFP-CGC or YFP-CGU, respectively) (Fig. 6B, left plot; Fig. S6B,C). YFP-AGA synthesis rate was also reduced in HEK293T cells (Fig. 6B, left plot; Fig. S6B), suggesting that pausing might emerge at this codon after the extended duration of arginine limitation that was necessary for detectable accumulation of reporter fluorescence. In the GCN2 KO cell line, in which ribosomes pause at CGC, CGU, and AGA codons (Fig. 4F), use of each of these codons also reduced YFP synthesis rate upon arginine limitation (Fig. 6C). Importantly, there was little difference in the measured protein synthesis rates between the arginine codon variants upon leucine limitation (Fig. 6B,C; Fig. S6B). Similarly, the six leucine codon variants had comparable reductions in YFP synthesis rate upon leucine or arginine limitation (Fig. 6D; Fig. S6B), consistent with the absence of ribosome pausing at these codons in wild-type cells (Fig. 1A-C). However, YFP-UUG synthesis rate was strongly reduced in the GCN2 KO cell line upon limitation for leucine, reflecting the emergence of ribosome pausing at this codon in this condition (Fig. 6E; Fig. 4F). In all cases, ribosome pausing upon amino acid limitation was sufficient to inhibit reporter protein synthesis.
Recent work suggests a role for mRNA degradation in the reduction of protein synthesis rates downstream of slow translation of rare codons in yeast (Presnyak et al., 2015; Radhakrishnan et al., 2016). To determine whether lower YFP production rates could be explained by reporter mRNA degradation downstream of ribosome pausing, we measured changes to YFP-CGC and YFP-CGG reporter mRNA levels during arginine and leucine limitation. Levels of YFP-CGC, which contains pause sites, were 2-fold higher than levels of YFP-CGG, which does not contain pause sites, in all conditions (Fig. S6D). The addition of the reporter stabilizing ligand TMP did not affect mRNA levels (Fig. S6D). YFP-CGC and YFP-CGG levels were similarly reduced by 50% upon arginine limitation, and unaffected by leucine limitation (Fig. S6D). Thus, pausing is not clearly linked to a reduction in mRNA levels, and such an effect cannot explain why less protein is produced from the YFP-CGC reporter specifically upon arginine limitation.
To determine whether premature abortive termination of translation might instead account for the reduction of protein synthesis rate by ribosome pausing, as previously described in bacteria (Subramaniam et al., 2014), we modified our protein synthesis rate reporter to detect premature termination at pause-site codons. We inserted a tandem repeat of 8 pause-site or non-pause-site codons in between the YFP and DHFR domains (Fig. 6F). The full-length YFP-DHFR protein is degraded efficiently and results in little fluorescence signal (Fig S6A). However, abortive termination at the pause-site codons would prevent synthesis of the DHFR degron and therefore generate stable YFP. Indeed, we observed a 100-fold increase in YFP fluorescence signal specifically upon leucine limitation when 8 tandem pause-site UUG leucine codons were inserted (UUG8) and this reporter was expressed in the GCN2 KO cell line (Fig. 6G). We confirmed that the size of the stable UUG8 reporter protein corresponded to the predicted size for the premature truncation product in this condition (Fig. 6I). By contrast, we detected only a minor fluorescence increase for the CUA8 reporter upon leucine limitation (Fig. 6G), and the size of the polypeptide produced in this case corresponded to the full length reporter (Fig. 6I, Fig. S6F). There was no evidence for premature termination of UUG reporter translation in any condition in which pausing does not occur at UUG codons (Fig. 1A-C), such as in wild-type HEK293T cells, during limitation for a non-cognate amino acid, or in rich conditions. Premature termination correlated positively with the number of pause-site codons in the reporter, was detectable when as few as 2 pause sites were present (Fig. 6H), and did not reduce mRNA levels (Fig. S6E). In fact, premature termination was associated with increased mRNA level, which may be explained by increased ribosome loading due to stalling upstream of tandem pause sites. We did not find evidence for similar levels of premature termination at arginine codons during arginine limitation in wild-type cells. This may be because premature termination products are rapidly degraded in these conditions, as polyarginine tracts can trigger ribosome quality control responses (Brandman and Hegde, 2016).
Based on our observation that ribosome pausing reduces protein expression, we sought to identify endogenous proteins whose levels might be regulated by pause-site codons during arginine limitation. Towards this goal, we calculated the bias in genomic usage of the pause-site arginine codons CGC and CGT for 18,660 coding sequences based on the genome-wide average usage frequency of these arginine codons (Fig. S6G). Among coding sequences biased against use of pause-site arginine codons, we found significant enrichment for gene ontology (GO) terms broadly related to organelle organization, macromolecule and nitrogen-compound metabolism, RNA processing, and positive regulation of GTPase activity (Fig. S6H, left plot). Conversely, genes with bias in favor of CGC and CGT codons were significantly enriched for GO terms related to nucleosomes, intermediate filaments, and ion channels involved in neuronal signal transduction (Fig. S6H, right plot). Given our evidence that ribosome pausing can regulate protein production rates and stimulate premature termination, the genes we identified as being enriched in pause sites are likely to be more translationally repressed upon a shift to arginine-limiting conditions than those depleted of pause sites.
Discussion
Our study provides a mechanistic dissection of the cause and consequences of ribosome pausing due to amino acid limitation in mammalian cells. We reveal an evolutionarily conserved role for synonymous codon-specific ribosome pausing in the regulation of protein synthesis during amino acid limitation, a phenomenon which has been previously observed only in bacteria (Subramaniam et al., 2013b, 2014). We also discovered a layer of complexity in this process that is unique to mammalian cells – quantitative differences in the activity of amino acid signaling pathways upon limitation for two amino acids result in qualitative differences in ribosome pausing.
Despite recent evidence that tRNA level and synonymous codon usage influence translation in mammalian systems (Gingold et al., 2014; Goodarzi et al., 2016; Saikia et al., 2016), we did not find a correlation between ribosome pausing upon arginine limitation and these quantities. Ribosome pausing observed in bacteria is also not explained by these measures (Subramaniam et al., 2013b, 2013a, 2014). However, an exact accounting of the tRNA supply for each codon is challenging given the degeneracy introduced by wobble decoding, extensive tRNA modifications that influence codon reading, and multiple codons that compete for a single tRNA species. Furthermore, we did not measure tRNA levels, and cannot exclude the possibility that a more accurate accounting of tRNA supply would explain the observed hierarchy of ribosome pausing. However, it is more likely that the balance between tRNA supply and codon usage demand determines differential isoacceptor sensitivity to changes in arginine levels, as observed in bacteria (Dittmar et al., 2005; Elf et al., 2003). We propose that a consideration of nutrient context is critical for defining which codons or tRNAs are functionally “optimal”.
Our measurements of tRNA charging loss upstream of ribosome pausing suggest that even a 50% charging level for many tRNAs upon amino acid limitation was insufficient to cause ribosome pausing at the cognate codons. This reflects a robustness of ribosome elongation rate to fluctuations in charged tRNA concentrations, and thus changes in charged tRNA concentrations (Saikia et al., 2016) might not always cause changes in translation elongation rate. This finding is also consistent with the proposal that tRNA abundance in mammals is unlikely to be evolutionarily optimized for globally efficient translation (Galtier et al., 2017). Instead, an understanding of what underlies the sensitivity of charging for specific isoacceptor to amino acid levels may reveal the evolutionary forces shaping translation elongation.
Our finding that the mTORC1 and GCN2 pathways respond more potently to limitation for different single amino acids highlights an unusual divergence in their roles, challenging the idea that both pathways act co-ordinately to sense amino acid limitation and appropriately regulate translation rate (Park et al., 2017). The mTORC1 response is clearly non-optimal with respect to preserving arginine homeostasis for protein synthesis: mTORC1 responds more weakly to arginine than leucine limitation, even though arginine becomes more rate-limiting for translation than leucine. Given that direct sensors for arginine (Chantranupong et al., 2016; Wang et al., 2015) and leucine (Wolfson et al., 2016) have been identified in the mTORC1 pathway, this observation is surprising. One possibility is that in the context of a tissue or a whole organism, arginine limitation might be typically accompanied by additional cue(s) to stimulate an optimal mTORC1 response, and limitation for only arginine in vitro might be insufficient to evoke this response. Investigating the response to arginine limitation in vivo will shed light on the role of mTORC1 in regulating arginine consumption.
In contrast to mTORC1, GCN2 – which senses uncharged tRNA – appears to respond optimally; it is equally or more strongly activated during arginine than leucine limitation across all three cell lines. This raises the question of why this robust GCN2 response is insufficient to prevent pausing. It has been recently shown that GCN2 can also sense ribosome pausing, creating a feedback regulation loop between elongation and initiation rates (Ishimura et al., 2016). Therefore, GCN2 activation may in part be downstream of the emergence of ribosome pausing. Dissecting the dynamics of the GCN2 response to amino acid limitation with respect to the emergence of ribosome pausing will clarify whether its role is primarily to prevent, or to respond to, such a loss of amino acid homeostasis.
Although we found that the signaling response to amino acid limitation was necessary to prevent ribosome pausing in HEK293T cells, we note that other mechanisms may exert control over ribosome pausing in distinct cell types. For example, rates of protein catabolism, proliferation, or lysosomal amino acid content could affect intracellular amino acid supply and demand. Indeed, we found no ribosome pausing at leucine codons upon limitation for leucine in HCT116 cells despite a weak amino acid signaling response (Fig. 3C-F, Fig. S3G). A mechanistic investigation in multiple cell types will clarify the range of cellular processes that exert control over ribosome pausing.
We find that ribosome pausing reduces both global and gene-specific protein synthesis rates. The effects of slow translation at specific codons on protein production have been widely linked to mRNA decay: recent work in yeast has suggested that ribosome stalling at non-optimal codons represses protein synthesis rates by increasing mRNA decay rates (Presnyak et al., 2015; Radhakrishnan et al., 2016; Simms et al., 2017). We did not find evidence for a reduction in mRNA levels due to pausing, although we cannot exclude an increase in mRNA decay rate balanced by an increased synthesis rate. In addition, significant changes in mRNA levels have not been observed in cases where protein production is altered by ribosome pausing at specific codons during amino acid limitation (Saikia et al., 2016; Subramaniam et al., 2013b, 2013a, 2014). Perhaps pausing during limitation in the presence of excess uncharged tRNA is qualitatively different from typical “no-go” pauses that result from overall tRNA scarcity, and thus might not stimulate NGD (Buskirk and Green, 2017). We did find evidence for truncated nascent peptides upon ribosome pausing at leucine codons in GCN2 KO cells, suggesting that pausing due to limiting charged tRNA can trigger abortive termination of translation, although the factors involved remain to be elucidated.
Dysregulated amino acid signaling and elevated proliferative demand for amino acids are characteristic features of many cancers (Saxton and Sabatini, 2017; Vander Heiden and DeBerardinis, 2017). Ribosome pausing may thus occur across a range of malignant states in vivo. This raises the important question of whether cell-autonomous ribosome pausing is a deleterious, neutral, or an adaptive response. In bacteria, ribosome stalling during amino acid limitation can act as a sensor for upregulating amino acid biosynthesis genes and for entering into a biofilm state, suggesting an adaptive role (Dittmar et al., 2005; Subramaniam et al., 2013b). Our finding that genes involved in nucleotide metabolism are biased against the use of arginine pause site codons is intriguing, as arginine is a substrate for nucleotide synthesis, which can be limiting for cancer cell proliferation (Rabinovich et al., 2015). Histone genes are biased towards the use of pause sites, and reduced nucleosome levels could underlie the S-phase cell cycle arrest that accompanies arginine limitation (Scott et al., 2000)– though it is unclear whether this would be adaptive. We find that ribosome pausing is linked to reduced cell viability (Fig. S6I), consistent with previous reports that arginine limitation induces cell death in multiple cancer cell lines (Lind, 2004). Therefore, pausing may have a deleterious effect on the cell, for example via protein misfolding or mistranslation stress (Drummond and Wilke, 2008). If not directly harmful, pausing might be a symptom of loss of metabolic homeostasis. To assess whether ribosome pausing is a targetable metabolic vulnerability, it will be important to determine in these and other contexts whether it adapts cellular metabolism and gene expression to amino acid limitation, or increases cellular stress.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Arvind Subramaniam (rasi@fredhutch.org).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
See key resources table for catalog numbers for commercial reagents.
KEY RESOURCES TABLE :
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti GCN2 | Cell Signaling Technology (CST) | 3302S |
| Rabbit polyclonal anti eEF2K | CST | 3692S |
| Rabbit polyclonal anti EEF2 | CST | 2332S |
| Rabbit polyclonal anti P~T56 EEF2 | CST | 2331S |
| Rabbit monoclonal anti eIF2α | CST | 5324P |
| Rabbit monoclonal anti P~S51 eIF2α | CST | 3398P |
| Rabbit polyclonal anti S6K | CST | 9202S |
| Rabbit polyclonal anti P~T389 S6K | CST | 9205S |
| Rabbit monoclonal anti S6 ribosomal protein (RPS6) | CST | 2217S |
| Rabbit monoclonal anti P~S235/6 RPS6 | CST | 4858S |
| Rabbit monoclonal anti GAPDH | CST | 2118S |
| Rabbit monoclonal anti puromycin | Sigma-Aldrich | MABE343 |
| Mouse monoclonal anti FLAG | Sigma-Aldrich | F3165 |
| Goat polyclonal anti rabbit IgG, HRP-linked | CST | 7074S |
| Goat polyclonal anti mouse IgG, HRP-linked | Sigma-Aldrich | 12-349 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| High-glucose DMEM without pyruvate | Gibco | 11995065 |
| Fetal bovine serum | ATCC | 30-2020 |
| Low glucose DMEM powder without amino acids | US Bio | D9800-13 |
| Dialyzed FBS | Invitrogen | 26400-044 |
| Lipofectamine 3000 | Thermo Fisher Scientific (TFS) | L3000015 |
| Cycloheximide | Sigma-Aldrich | C7698-1G |
| Turbo DNase | TFS | AM2238 |
| Micrococcal nuclease | Worthington Biochemical | LS004798 |
| SYBR gold nucleic acid gel stain | TFS | S11494 |
| Superase-In RNase inhibitor | TFS | AM2696 |
| T4 PNK | NEB | M0201S |
| E. coli poly A polymerase | NEB | M0276S |
| Superscript III | TFS | 18080093 |
| Circligase | Epicentre | CL4111K |
| Acid-Phenol:Chloroform pH 4.5 with isoamyl alcohol at 25:24:1 | TFS | AM9720 |
| MyOne Streptavidin Dynabeads | TFS | 65001 |
| Phusion Flash High-Fidelity PCR master mix | TFS | F548S |
| RNase I | Invitrogen | AM2294 |
| Sequagel Urea gel system | National Diagnostics | EC-833 |
| BSA | CST | 9998S |
| SuperSignal West Femto Substrate | TFS | 34095 |
| Restore Western Blot stripping buffer | TFS | 21059 |
| PerfectHyb Plus Hybridization buffer | Sigma-Aldrich | H-7033 |
| [γ-P32]-ATP EasyTide | Perkin Elmer | NEG502A250UC |
| Puromycin | Sigma-Aldrich | P8833 |
| dT-20 primer | TFS | 18418020 |
| PowerUp SYBR Green PCR master mix | TFS | A25742 |
| Glycoblue | TFS | AM9516 |
| Critical Commercial Assays | ||
| CellTiter-Glo | Promega | G7570 |
| Quick RNA Miniprep | Zymo | R1054 |
| Deposited Data | ||
| Ribosome profiling | this study | GSE113751 |
| Raw/processed data on Github | this study | https://github.com/rasilab/adarnell_2018 |
| Experimental Models: Cell Lines | ||
| HEK293T | ATCC | CRL-3216 |
| HeLa | ATCC | CCL2 |
| HCT116 | NIH, National Cancer Institute (NCI) | NCI-60 cancer cell line panel |
| hsAD1: HEK293T AAVS1-hrGFP | Donor pADHS1, targeting pADHS4 | |
| hsAD2: HEK293T AAVS1-RagB-WT | Donor pADHS2, targeting pADHS4 | |
| hsAD3: HEK293T AAVS1-RagB-Q99L | Donor pADHS3, targeting pADHS4 | |
| hsAD4: HEK293T GCN2 KO (clones 1,2,3) | Targeting pADHS7,8 | |
| hsAD5: HEK293T EEF2K KO (clones 1,4,5) | Targeting pADHS9,10 | |
| hsAD6: HEK293T YFP-CGC | Donor pADHS15, pack/env pADHS11,12 | |
| hsAD7: HEK293T YFP-CGG | Donor pADHS16, pack/env pADHS11,12 | |
| hsAD8: HEK293T YFP-CGA | Donor pADHS17, pack/env pADHS11,12 | |
| hsAD9: : HEK293T YFP-CGU | Donor pADHS18, pack/env pADHS11,12 | |
| hsAD10: HEK293T YFP-AGA | Donor pADHS19, pack/env pADHS11,12 | |
| hsAD11: HEK293T YFP-AGG | Donor pADHS20, pack/env pADHS11,12 | |
| hsAD12: HEK293T GCN2 KO YFP-CGC | Donor pADHS15, pack/env pADHS11,12 | |
| hsAD13: HEK293T GCN2 KO YFP-CGG | Donor pADHS16, pack/env pADHS11,12 | |
| hsAD14: HEK293T GCN2 KO YFP-CGA | Donor pADHS17, pack/env pADHS11,12 | |
| hsAD15: HEK293T GCN2 KO YFP-CGU | Donor pADHS18, pack/env pADHS11,12 | |
| hsAD16: HEK293T GCN2 KO YFP-AGA | Donor pADHS19, pack/env pADHS11,12 | |
| hsAD17: HEK293T GCN2 KO YFP-AGG | Donor pADHS20, pack/env pADHS11,12 | |
| hsAD18: HEK293T YFP-CUA | Donor pADHS21, pack/env pADHS11,12 | |
| hsAD19: HEK293T YFP-CUC | Donor pADHS22, pack/env pADHS11,12 | |
| hsAD20: HEK293T YFP-CUU | Donor pADHS23, pack/env pADHS11,12 | |
| hsAD21: HEK293T YFP-UUA | Donor pADHS24, pack/env pADHS11,12 | |
| hsAD22: HEK293T YFP-UUG | Donor pADHS25, pack/env pADHS11,12 | |
| hsAD23: HEK293T GCN2 KO YFP-CUA | Donor pADHS21, pack/env pADHS11,12 | |
| hsAD24: HEK293T GCN2 KO YFP-CUC | Donor pADHS22, pack/env pADHS11,12 | |
| hsAD25: HEK293T GCN2 KO YFP-CUU | Donor pADHS23, pack/env pADHS11,12 | |
| hsAD26: HEK293T GCN2 KO YFP-UUA | Donor pADHS24, pack/env pADHS11,12 | |
| hsAD27: HEK293T GCN2 KO YFP-UUG | Donor pADHS25, pack/env pADHS11,12 | |
| hsAD28: HEK293T UUG8 | Donor pADHS26, pack/env pADHS11,12 | |
| hsAD29: HEK293T CUA8 | Donor pADHS27, pack/env pADHS11,12 | |
| hsAD30: HEK293T GCN2 KO UUG8 | Donor pADHS26, pack/env pADHS11,12 | |
| hsAD31: HEK293T GCN2 KO CUA8 | Donor pADHS27, pack/env pADHS11,12 | |
| hsAD32: HEK293T CUA4UUG4 | Donor pADHS28, pack/env pADHS11,12 | |
| hsAD33: HEK293T CUA6UUG2 | Donor pADHS29, pack/env pADHS11,12 | |
| hsAD34: HEK293T GCN2 KO CUA4UUG4 | Donor pADHS28, pack/env pADHS11,12 | |
| hsAD35: HEK293T GCN2 KO CUA6UUG2 | Donor pADHS29, pack/env pADHS11,12 | |
| hsAD39: HEK293T pAAVS1P-iCAG.FlagYFP-DHFR-CGC | Donor pADHS34, targeting pADHS4 | |
| hsAD40: HEK293T pAAVS1P-iCAG.FlagYFP-DHFR-CGG | Donor pADHS35, targeting pADHS4 | |
| hsAD41: HCT116 pAAVS1P-iCAG.FlagYFP-DHFR-CGC | Donor pADHS34, targeting pADHS4 | |
| hsAD42: HCT116 pAAVS1P-iCAG.FlagYFP-DHFR-CGG | Donor pADHS34, targeting pADHS4 | |
| hsAD43: HCT116 YFP-CGC | Donor pADHS15, pack/env pADHS11,12 | |
| hsAD44: HCT116 YFP-CGG | Donor pADHS16, pack/env pADHS11,12 | |
| hsAD45: HeLa YFP-CGC | Donor pADHS15, pack/env pADHS11,12 | |
| hsAD46: HeLa YFP-CGG | Donor pADHS16, pack/env pADHS11,12 | |
| Oligonucleotides | ||
| tRNA northern blot probe, Arg-ACG1: CCAGGAGTCGAACCTRGAATCTTCTGATCCGTAGTCAGACGCG |
this study | |
| tRNA northern blot probe, Arg-CCG3: ACTCGAACCCTCAATCTTCTGATCCGGAATCAGACGCCTT |
this study | |
| tRNA northern blot probe, Arg-TCG2: GGATTCGAACCCTCAATCTTCTGATCCGAAGTCAGACGCC |
this study | |
| tRNA northern blot probe, Leu-TAG3: AAGAGACTGGAGCCTAAATCCAGCGCCTTAGACCGCTCGGCCACACTACC |
this study | |
| tRNA northern blot probe, Leu-AAG3: AGTCTTAATACAGTGCCTTAGACCGCTCGGCCACCCTACC |
this study | |
| tRNA northern blot probe, Leu-CAG2: CACGCCTCCAGGGGAGACTGCGACCTGAACGCAGCGCCTT |
this study | |
| tRNA northern blot probe, Leu-CAA5: CCACGCCTCCATTGGAGACCACAAGCTTGAGTCTGGCGCC |
this study | |
| o3285_123:154_rp rRNA subtraction oligo: [Biotin-5]CCTCCCGGGGCTACGCCTGTCTGAGCGTCGC | this study | |
| o3287_4990:5022_rp rRNA subtraction oligo: [Biotin-5]TCGCTGCGATCTATTGAAAGTCAGCCCTCGAC | this study | |
| YFP-DHFR reporter qPCR primer F (to DHFR C terminus): ATATCGACGCAGAAGTGGAAGG |
this study | |
| YFP-DHFR reporter qPCR primer R (to DHFR C terminus): ATCAGCATCGTGGAATTCGC |
this study | |
| Premature termination reporter qPCR primer F (spans tandem Leucine codons): GAGTTCGTGACCGCCGC |
this study | |
| Premature termination reporter qPCR primer R (spans tandem Leucine codons): CCATGCCGATAACGTGATCTACCG |
this study | |
| Homologous recombination at AAVS1 locus PCR check, F (in AAVS1 locus): CTCTCTCCTGAGTCCGGACCACTTTGAGCTC | this study | |
| Homologous recombination at AAVS1 locus PCR check, R (in puroR): CGCACCGTGGGCTTGTACTCGGTCAT | ||
| Recombinant DNA | ||
| pADHS1: AAVS1-CAG-hrGFP | Qian et al., 2014 | Addgene #52344 |
| pADHS2: AAVS1-CAG-RagBWT | Cloned from Qian et al., 2014 and Sancak et al. 2008 | Cloned Flag-RagB-WT from Flag pLJM1 RagB wt, Addgene #19313, into AAVS1-CAG-hrGFP |
| pADHS3: AAVS1-CAG-RagBQ99L | Cloned from Qian et al., 2014 and Sancak et al. 2008 | Cloned Flag-RagB-Q99L from Flag pLJM1 RagB 99L, Addgene #19315, into AAVS1-CAG-hrGFP |
| pADHS4: px330-AAVS1-T2 | Cloned from Cong et al., 2013 | Cloned AAVS1 T2 guide sequence in px330-U6-Chimeric-BB-CBh-hSpCas9, Addgene #42230 |
| pADHS5: pU6-(BbsI)_CBh-Cas9-T2A-BFP | Chu et al., 2015 | Addgene #64323 |
| pADHS6: pSpCas9(BB)-2A-GFP (PX458) | Ran et al., 2013 | Addgene #48138 |
| pADHS7: pU6-GCN2-1-Cas9-2A-BFP | Cloned from Chu et al., 2015 | Cloned GCN2 (EIF2AK4) guide RNA sequence 2 (Doench et al.2016, Addgene #75876) into pU6-(BbsI)_CBh-Cas9-T2A-BFP |
| pADHS8: pU6-GCN2-2-Cas9-2A-GFP | Cloned from Ran et al., 2013 | Cloned GCN2 (EIF2AK4) guide RNA sequence 3 (Doench et al. 2016, Addgene #75877) into pSpCas9(BB)-2A-GFP (PX458) |
| pADHS9: pU6-EEF2K-1-Cas9-2A-BFP | Cloned from Chu et al., 2015 | Cloned EEF2K guide RNA sequence 2 (Doench et al.2016, Addgene #77855) into pU6-(BbsI)_CBh-Cas9-T2A-BFP |
| pADHS10: pU6-EEF2K-2-Cas9-2A-GFP | Cloned from Ran et al., 2013 | Cloned EEF2K guide RNA sequence 3 (Doench et al.2016, Addgene #77856) into pU6-EEF2K-2-Cas9-2A-GFP |
| pADHS11: psPAX2 | Addgene #12260 | |
| pADHS12: pCMV-VSV-G | Steward et al., 2003 | Addgene #8454 |
| pADHS13: pLJM1-EGFP | Sancak et al., 2008 | Addgene #19319 |
| pADHS14: KHT61-Unreg-YFP-DD | Han et al., 2014 | A gift from KyuhoHan |
| pADHS15: pLJM1-Flag-YFP-DHFR (YFP-CGC) | Cloned from Sancak et al., 2008 and Han et al., 2014 | Cloned YFP-DD from KHT61-Unreg-YFP-DD into pLJM1-EGFP with a Flag Tag |
| pADHS16: pLJM1-Flag-YFP-DHFR (YFP-CGG) | Cloned from YFP-CGC | Cloned 2 gBlocks with all YFP and DHFR arginine codons swapped to CGG into YFP-CGC |
| pADHS17: pLJM1-Flag-YFP-DHFR (YFP-CGA) | Cloned from YFP-CGC | Cloned 2 gBlocks with all YFP and DHFR arginine codons swapped to CGA into YFP-CGC |
| pADHS18: pLJM1-Flag-YFP-DHFR (YFP-CGU) | Cloned from YFP-CGC | Cloned 2 gBlocks with all YFP and DHFR arginine codons swapped to CGU into YFP-CGC |
| pADHS19: pLJM1-Flag-YFP-DHFR (YFP-AGA) | Cloned from YFP-CGC | Cloned 2 gBlocks with all YFP and DHFR arginine codons swapped to AGA into YFP-CGC |
| pADHS20: pLJM1-Flag-YFP-DHFR (YFP-AGG) | Cloned from YFP-CGC | Cloned 2 gBlocks with all YFP and DHFR arginine codons swapped to AGG into YFP-CGC |
| pADHS21: pLJM1-Flag-YFP-DHFR (YFP-CUA) | Cloned from YFP-CGC | Cloned 1 gBlock with all YFP leucine codons swapped to CUA into YFP-CGC |
| pADHS22: pLJM1-Flag-YFP-DHFR (YFP-CUC) | Cloned from YFP-CGC | Cloned 1 gBlock with all YFP leucine codons swapped to CUC into YFP-CGC |
| pADHS23: pLJM1-Flag-YFP-DHFR (YFP-CUU) | Cloned from YFP-CGC | Cloned 1 gBlock with all YFP leucine codons swapped to CUU into YFP-CGC |
| pADHS24: pLJM1-Flag-YFP-DHFR (YFP-UUA) | Cloned from YFP-CGC | Cloned 1 gBlock with all YFP leucine codons swapped to UUA into YFP-CGC |
| pADHS25: pLJM1-Flag-YFP-DHFR (YFP-UUG) | Cloned from YFP-CGC | Cloned 1 gBlock with all YFP leucine codons swapped to UUG into YFP-CUA |
| pADHS26: pLJM1-Flag-YFP-UUG8-DHFR (UUG8) | Cloned from YFP-CUA | Cloned 8 tandem UUG codons into YFP-CUA |
| pADHS27: pLJM1-Flag-YFP-CUA8-DHFR (CUA8) | Cloned from YFP-CUA | Cloned 8 tandem CUA codons into YFP-CUA |
| pADHS28: pLJM1-Flag-YFP-CUA4UUG4-DHFR (CUA4UUG4) | Cloned from YFP-CUA | Cloned UUG-CUA-UUG-CUA-UUG-CUA-UUG-CUA into YFP-CUA |
| pADHS29: pLJM1-Flag-YFP-CUA6UUG2-DHFR (CUA6UUG2) | Cloned from YFP-CUA | Cloned CUA-CUA-UUG-CUA-CUA-UUG-CUA-CUA into YFP-CUA |
| pADHS33: pAAVS1P-iCAG.copGFP | Cerbini et al., 2015 | Addgene #66577 |
| pADHS34: pAAVS1P-iCAG.FlagYFP-DHFR-CGC | Cloned fromCerbini et al., 2015 and YFP-CGC | Cloned YFP-DHFR from YFP-CGC into pAAVS1P-iCAG.copGFP |
| pADHS35: pAAVS1P-iCAG.FlagYFP-DHFR-CGG | Cloned from Cerbini et al., 2015 and YFP-CGG | Cloned YFP-DHFR from YFP-CGG into pAAVS1P-iCAG.copGFP |
| Software and Algorithms | ||
| cutadapt | (Martin, 2011) | |
| bowtie | (Langmead et al., 2009) | |
| rsem | (Li and Dewey, 2011) | |
| DESeq2 | (Love et al., 2014) | |
| samtools | (Li, 2011) | |
| tophat | (Trapnell, 2009) | |
| topGO | (Alexa and Rahnenfuhrer, 2016) | |
| REVIGO | (Supek et al., 2011) | |
Human cell line culture
The HEK293T (RRID:CVCL_0063) and HeLa cell lines (RRID:CVCL_0030) were obtained from ATCC, catalog numbers CRL-3216 and CCL-2. The HCT116 cell line (RRID:CVCL_0291) was obtained from the National Cancer Institute (NCI) panel of 60 cancer lines. Cells lines from ATCC and the NCI-60 panel are authenticated. All cell lines were passaged in high-glucose DMEM without pyruvate, with penicillin/streptomycin, and with 10% fetal bovine serum at 37°C and 5% CO2. The HEK293T cell line, used as the primary model and parental cell line for CRISPR/lentiviral genome editing in the paper, and the HCT116 cell line tested negative for mycoplasma contamination. The HeLa cell line was not tested, but all experiments were performed with low passage number stocks from ATCC.
Limitation for single amino acids
Amino acid limitation media were prepared from low glucose DMEM powder without amino acids; all amino acids except leucine and arginine, and glucose were supplemented according to this recipe: 3 g/L additional glucose, 30 mg/L glycine, 63 mg/L cysteine 2·HCl, 580 mg/L glutamine, 42 mg/L histidine HCl·H2O, 105 mg/L isoleucine, 146 mg/L lysine HCl, 30 mg/L methionine, 66 mg/L phenylalanine, 42 mg/L serine, 95 mg/L threonine, 16 mg/L tryptophan, 64 mg/L tyrosine 2·Na 2·H2O, and 94 mg/L valine. Medium was prepared in batches of 2 liters; the pH was adjusted to 7.2 – 7.4 with HCl, and medium was vacuum filtered and supplemented with 10% dialyzed FBS. For all amino acid limitation assays – except time course experiments over multiple days – cells were expanded to 60-70% confluency in amino acid limitation medium supplemented with 105 mg/L leucine and 84 mg/L arginine HCl. Cells were then washed once in PBS, and transferred to limitation medium supplemented with either 105 mg/L leucine (for arginine limitation) or 84 mg/L arginine HCl (for leucine limitation), or both (for rich medium). To account for different proliferation rates for cells in the rich and amino acid limited conditions in time course experiments over multiple days, cells for the rich medium condition were expanded to 10-20% confluency and cells for the leucine / arginine limitation conditions were expanded to 60-70% confluency in the same arginine and leucine supplemented amino acid limitation medium before beginning the experiment.
METHODS DETAILS
See key resources table for catalog numbers for commercial reagents. Unless otherwise indicated, commercial reagents were used according to the manufacturer’s instructions.
Construction of plasmids
AAVS1-CAG-hrGFP was from Su-Chun Zhang (Addgene # 52344) (Qian et al., 2014). We cloned sequences for FLAG-RagB-WT and FLAG-RagB-Q99L into this plasmid in place of hrGFP, from sequences in FLAG pLJM1 RagB wt (Addgene # 19313) and FLAG pLJM1 RagB 99L (Addgene # 19315) from David Sabatini (Sancak et al., 2008). The resulting CRISPR homology donor plasmids AAVS1-CAG-hrGFP, AAVS1-CAG-RagBWT, and AAVS1-CAG-RagBQ99L were then introduced into HEK293T cells by CRISPR/Cas9 mediated homologous recombination with the AAVS1 sgRNA and Cas9 expression plasmid px330-AAVS1-T2 (see Stable overexpression cell line generation by CRISPR/Cas9 genome editing section), which was cloned by inserting the AAVS1-T2 target sequence GGGGCCACTAGGGACAGGAT (Mali et al., 2013) into the px330-U6-Chimeric-BB-CBh-hSpCas9 plasmid, from Feng Zhang (Addgene # 42230) (Cong et al., 2013) (see Fig. 4C).
To generate plasmids for targeting endogenous GCN2 (alias EIF2AK4) and EEF2K (see Fig. S4G and S5D), sgRNA sequences were obtained from the lentiGuide-Puro library (Doench et al., 2016). Two sgRNA sequences each targeting exonic sequences ~790 bp apart in GCN2 (from Addgene #75876 and 75877), and ~230 bp apart in EEF2K (from Addgene #77855 and 77856), were selected. For each pair, one sgRNA was cloned into pU6-(BbsI)_CBh-Cas9-T2A-BFP, from Ralf Kuehn (Addgene # 64323) (Chu et al., 2015), and the other into pSpCas9(BB)-2A-GFP (PX458), from Feng Zhang (Addgene # 48138) (Ran et al., 2013). This produced the targeting plasmids pU6-GCN2-1-Cas9-2A-BFP, pU6-GCN2-2-Cas9-2A-GFP, pU6-EEF2K-1-Cas9-2A-BFP, and pU6-EEF2K-2-Cas9-2A-GFP (see also Knockout cell line generation by CRISPR/Cas9 genome editing section).
Our YFP-DHFR protein synthesis rate reporters (see Fig. 6A) were cloned into pLJM1-EGFP, from David Sabatini (Addgene # 19319) (Sancak et al., 2008). The EGFP coding sequence in this vector was replaced by the YFP-DHFR sequence from KHT61-Unreg-YFP-DD, a gift from Kyuho Han (Han et al., 2014), along with an N-terminal FLAG tag to generate the pLJM1-FLAG-YFP-DHFR reporter (YFP-CGC). The YFP-CGC reporter has 13 CGC and 1 CGU arginine codons, and 23 CUG, 5 CUC, 2 UUA, and 2 UUG leucine codons. To generate codon variants, gBlocks (IDT) were ordered in which all 14 arginine codons in YFP and DHFR, or all 21 leucine codons in YFP, were swapped to one out of each of the six synonymous leucine or arginine codons; these gBlocks were cloned in place of the YFP-CGC sequence in the pLJM1 plasmid backbone. The following library of FLAG-tagged codon variant reporter lentiviral donor plasmids was generated: YFP-CGC, YFP-CGG, YFP-CGA, YFP-CGU, YFP-AGA, YFP-AGG, YFP-CUA, YFP-CUC, YFP-CUU, YFP-UUA, and YFP-UUG (see Stable overexpression cell line generation by lentiviral transduction section).
These reporters were modified to generate premature termination reporters (see Fig. 6F) by cloning in eight tandem leucine codons into the pLJM1-YFP-CUA lentiviral donor plasmid in between the YFP and DHFR sequences. The following library of four FLAG-tagged premature termination reporter lentiviral donor plasmids was generated, in which the numbers refer to the composition of the eight leucine codon repeat: UUG8, CUA8, CUA4UUG4, CUA6UUG2. These plasmids were used to generate stable reporter cell lines by lentiviral transduction into HEK293T and the HEK293T GCN2 KO cell line (see Stable overexpression cell line generation by lentiviral transduction section).
A variant YFP-DHFR protein synthesis rate reporter (see Fig. S6C) was built by cloning from pAAVS1P-iCAG.copGFP, from Jizhong Zou (Addgene # 66577) (Cerbini et al., 2015). To generate pAAVS1P-iCAG.FLAG-YFP-DHFR-CGC and –CGG codon variant reporters, reporter sequences from YFP-CGC and YFP-CGG were cloned in place of copGFP. These plasmids were used to generate stable reporter cell lines in HEK293T and HCT116 cells by CRISPR/Cas9-mediated homologous recombination with px330-AAVS1-T2 (see Stable overexpression cell line generation by CRISPR/Cas9 genome editing section).
Stable cell line generation
All transfections were performed at 75% confluency using Lipofectamine 3000. Selection was performed with puromycin: 2μg/ml for HEK293T and GCN2 KO cells, 1 μg/ml for HCT116/HeLa cells.
I. Lentiviral transduction, overexpression
HEK293T cells were transfected in a 10 cm plate with donor expression plasmid pLJM1 containing the desired insert, psPAX2, from Didier Trono (Addgene # 12260), and pCMV-VSV-G, from Bob Weinberg (Addgene # 8454) in a 10:9:1 ratio (by weight). The media was replaced after 12-16 hours, and lentivirus was harvested at 48 hours by passing culture supernatant through a low-protein binding filter with 0.45 μm pore size. 1 mL of virus was then used to transduce 50-60% confluent HEK293T, HeLa, HCT116, or GCN2 KO cells in a 6 cm plate. Transduced cells were passaged to a 10 cm plate after 24 hours, and puromycin selection was initiated after 48 hours (see Fig. 6B-E, S6A-C).
II. CRISPR/Cas9 genome editing, overexpression
To generate hrGFP, RagB-WT, and RagB-Q99L cell lines (see Fig. 4C): HEK293T cells in a 6 well plate were transfected with homology donor plasmid (pAAVS1-CAG-hrGFP, pAAVS1-CAG-RagBWT, or pAAVS1-CAG-RagBQ99L) and px330-AAVS1-T2 at a ratio of 4:1 (2 μg donor : 500 ng guide). Homologous recombination and expression of transgenes were confirmed in the resulting polyclonal population by PCR, flow cytometry, and western blotting after puromycin selection.
To generate arginine/leucine codon variant YFP-DHFR reporter cells lines (see Fig. S6B): HEK293T or HCT116 cells were transfected with homology donor plasmid (for YFP reporter lines: pAAVS1P-iCAG.copGFP, pAAVS1P-iCAG.FLAG-YFP-DHFR-CGC, pAAVS1P-iCAG.FLAG-YFP-DHFR-CGG) and px330-AAVS1-T2 at a ratio of 2:1 (10 μg donor : 5 μg guide). Homologous recombination and TMP-inducible YFP fluorescence were confirmed in the resulting polyclonal population by PCR, flow cytometry, and western blotting after puromycin selection.
III. CRISPR/Cas9 genome editing, knockout
HEK293T cells in a 12 well plate were transfected with 500 ng of each targeting RNA plasmid, in the following four combinations: 1) both pU6-GCN2-1-Cas9-2A-BFP and pU6-GCN2-2-Cas9-2A-GFP, 2) both pU6-EEF2K-1-Cas9-2A-BFP and pU6-EEF2K-2-Cas9-2A-GFP, 3) pU6-GCN2-1-Cas9-2A-BFP only, and 4) pU6-GCN2-2-Cas9-2A-GFP only. Cells were transferred to a 6 well plate 24 hours post transfection. After 48 hours, dual-fluorescing BFP+ and GFP+ single cells were sorted into individual wells of a 96 well plate; samples 3) and 4) were used to define individual gates. Plates were spun 100xG, 1 minute to sediment cells. After expansion, western blotting confirmed complete knockout. 92% of clones tested were positive for complete GCN2 KO (11/12), and 83% for EEF2K KO (10/12) (see Fig. S4G and S5D).
Ribosome profiling
To detect codon-specific ribosome pausing, ribosome profiling was performed according to (Ingolia et al., 2009), with modifications detailed below (see Fig. 1,4 and Fig. S1,4).
Cells were expanded to 75% confluency in two 15 cm plates. Cells were washed once, briefly, in ice cold PBS. PBS was thoroughly drained, and plates were immersed in liquid nitrogen for flash freezing and then transferred to −80°C. Frozen cells were lysed on each plate by scraping into 300 μL lysis buffer (20 mM T ris pH 7.5, 15 mM MgCl2, 150 mM NaCl, 100 μg/mL cycloheximide, 5 mM CaCl2, 1% Triton X-100, 50 U/mL Turbo DNase), and lysates from the two 15 cm plates were combined to yield ~1 mL of lysate. Ribosome footprints were generated from 450 μL of lysate by 1 hour of digestion with 800 U micrococcal nuclease at room temperature (25°C) with nutation, and quenched by addition of 4.5 μL 0.5 M EGTA. Footprints were purified by sucrose density gradient fractionation; a BioComp Gradient Station was used to generate 10-50% sucrose density gradients in 1X polysome resuspension buffer (20 mM Tris pH 7.5, 15 mM MgCl2, 150 mM NaCl, 100 μg/mL cycloheximide). 400 μL digested lysate was loaded onto gradients in SW41 rotor buckets and samples were spun for 2.5 hours at 35,000 RPM and 4°C. Fractionation was performed at 0.22 mm/sec with UV absorbance monitoring at 254 nm (EconoUV Monitor) and the monosome fraction was collected in addition to the contiguous disome “shoulder”. Total RNA was purified by addition of 7 mM EDTA and 1% SDS, extraction in acid-phenol:chloroform pH 4.5 with isoamyl alcohol at 25:24:1 at 65°C, and precipitated.
8 μg of the monosome fraction RNA was run on a 15% TBE-urea gel (Bio-Rad) and footprints were excised from ~26-40 nt (see Fig. S1D). RNA was extracted in 0.3 M NaOAc pH 5.5, 1 mM EDTA, and 100 U/mL Superase-In overnight at room temperature with rotating and then precipitated.
Footprints were dephosphorylated with T4 PNK, then precipitated. Footprints were then polyA-tailed with E. coli polyA polymerase for 10 minutes, then precipitated. Reverse transcription was performed using SuperScript III and 0.5 μM oNTI19pA oligo primer (Ingolia et al., 2009) for 30 minutes at 48°C, RT products were run on a 10% TBE-urea gel (Bio-Rad), extracted from gel slices and precipitated. RT products were circularized with CircLigase, then precipitated. rRNA was removed by subtractive hybridization with MyOne Streptavidin Dynabeads. Two biotinylated reverse complement oligos to discrete rRNA sequences that were recovered extremely abundantly in our test ribosome profiling libraries (o3285, o3287; used in a 1:3 ratio) were annealed to circularized libraries, and an equal volume of washed beads was added to annealed oligo/libraries for 15 minutes at 37°C. Supernatant was recovered and precipitated. Resulting libraries were amplified by 6-12 cycles of PCR with 2X Phusion Flash master mix with common reverse and unique 6nt index forward library primers and purified after running on a 10% TBE gel (Bio-Rad). Libraries were extracted from gel slices, precipitated, resuspended in 10 μL Tris 10 mM pH 7, and quantified using an Agilent TapeStation or Bioanalyzer. Up to 15 multiplexed libraries were submitted for sequencing on both lanes of an Illumina HiSeq 2500 Rapid Flow Cell. Sequencing runs yielded approximately 150 million reads per lane.
Notably, two ribonucleases, RNase I and micrococcal nuclease (MNase), are commonly used for ribosome profiling. We observed near-complete degradation of the 60S ribosomal subunit and ribosome-bound mRNA fractions by RNase I in buffers with either high (Ingolia et al., 2012) or low magnesium (Andreev et al., 2015) and across a broad range of RNase I concentrations (Fig. S1A,B). The 60S and monosome fractions were largely intact after digestion with MNase (Fig. S1C), and therefore we used this nuclease. As previously reported (Dunn et al., 2013; Reid et al., 2015), MNase results in slightly longer reads and a broader read length distribution (Fig. S1E) as it does not digest completely around bound ribosomes. However, read density exhibited robust three nucleotide periodicity, is clearly enriched in the coding region, and exhibits peaks at start and stop codons (Fig. S1F,G), allowing resolution of codon-level changes in translation elongation.
Polysome profiling
The same procedure as in the “Ribosome profiling” section was used, with the following modifications. 150 μL of clarified lysate was loaded directly onto sucrose density gradients. Gradients were centrifuged in a SW41 rotor at 35,000 RPM for 3 hours at 4°C with the “slow” brake setting. After fractionation, the relative polysome to monosome fraction area was calculated for each profile by 1) manual definition of the fraction boundaries, 2) subtracting the lowest value in the profile from all points along the profile, and 3) manual integration using the trapezoid rule (see Fig. S5B).
Intracellular amino acid quantitation
HEK293T cells in a 10 cm dish were washed twice with PBS on ice. Ice cold HPLC-grade 80:20 MeOH:H2O was added to cells to extract polar metabolites. After vortexing, 80% methanol extracts were dried under vacuum and resuspended in water for analysis on an Agilent 6460 LC-MS/MS.
tRNA charging analysis
tRNA charging analysis was performed according to (Varshney et al., 1991) with the following modifications (see Fig. 2 and Fig. S2,4). 75% confluent cells in a 10 or 6 cm plate were washed once in PBS and flash frozen. Cells were scraped into ice cold 500 μL AE buffer (0.3 M NaOAc pH 4.5, 10 mM EDTA) on plates and added to 500 μL ice cold acid-saturated phenol:chloroform pH 4.5. Extractions were vortexed for 10 minutes, rested on ice for 3 minutes, and spun for 10 minutes at 20,000xG at 4°C. Aqueous supernatant was precipitated and resuspended in 10 mM NaOAc pH 4.5, 1 mM EDTA. To mark electrophoretic mobility of uncharged tRNA, RNA was deacylated in 100 mM Tris pH 9 at 37°C for 30 minutes, following by quenching with addition of an equal volume of 50 mM NaOAc, 100mM NaCl and precipitation.
For acid urea gel electrophoresis, 500 ng – 1 μg RNA and deacylated control in 0.1 M NaOAc pH 4.5, 8 M urea, 0.05% bromphenol blue, and 0.05% xylene cyanol were electrophoresed on a 0.4 mm 6.5% polyacrylamide gel (SequaGel) with 8M urea in 0.1M NaOAc pH 4.5 at 450V and 4°C for 18-20 hours. The gel region between the loading dye bands was excised and transferred according to “Northern blotting” section. Absolute charging level was calculated by dividing the intensity of the charged band(s) by the sum of all band intensities (quantified using ImageJ).
Probes for northern blotting were designed to hybridize uniquely to tRNA isoacceptors, where possible, or isodecoders after alignment of all arginine and leucine tRNAs. tRNAs with introns and psueo-tRNAs were identified using tRNAscan-SE (Lowe and Eddy, 1997). All probes were validated for specificity by Northern blotting against in vitro transcribed target tRNAs and candidates for cross-hybridization identified by a genomic tRNA BLAST. We were not able to find uniquely hybridizing probe for tRNALeuAAG and tRNALeuUAG as these isoacceptor gene families have a great degree of sequence homology; however, the major species detected for these probes is the indicated tRNA. We also note that two charged and uncharged species of tRNAArgACG are detected (see Fig. S2A,C), possibly due to covalent modification of this tRNA.
Western and dot-blotting
75% confluent cells in a 10 cm plate were lysed by scraping and pooling in 300 μL of 50 mM HEPES pH 7.4, 40 mM NaCl, 2 mM EDTA, 1 mM sodium orthovandate, 10 mM sodium glycerophosphate, 10 mM sodium pyrophosphate, 50 mM sodium fluoride, 1% Triton X-100. After 10 minutes at 4°C, the insoluble fraction was cleared by centrifugation for 10 minutes at 4°C and 20,000g. Lysate was electrophoresed in 1X Laemmli buffer on a 4-20% Tris-glycine gel (Novex) and blotted onto 0.45 μm nitrocellulose. Primary antibodies are listed in key resources table and were used at 1: 1000 final dilution. The primary antibody from rabbit against puromycin was used at 1:25,000. HRP-conjugated secondary antibodies were used at 1:5000. 5% BSA in TBST was used for all blocking and antibody solutions for phospho-antibody blots, and 5% milk in TBST was used for all others. SuperSignal West Femto Substrate was used for developing, and Restore Western Blot stripping buffer was used to strip blots.
To calculate a relative phosphorylation index for mTORC1 and GCN2 targets to evaluate kinase activity, phospho-protein band intensity was divided by total protein band intensity and normalized to the appropriate maximum, minimum, or untreated ratio (see individual Fig. legends). This normalized phosphorylation index was first calculated for each sample on one technical replicate blot and then averaged between blots from replicate experiments although similar results were obtained using different normalization methods.
For dot-blotting, 2 μL of lysate was spotted onto a dry 0.45 μm nitrocellulose membrane, allowed to dry for 15 minutes, and then blots were processed as described above for western blotting (see Fig. and Fig. S5).
Northern blotting
After electrophoresis, gels were rinsed thoroughly in 0.5X TBE and transferred to HyBond Nylon+ membrane in 0.5X TBE using a semi-dry transfer apparatus (Bio-Rad Transblot) at 3 mA/cm2 for 1 hour. The blot was crosslinked using the Stratalinker “auto-crosslink” setting once on each side, prehybridized in PerfectHyb buffer for 1 hour at 64°C, and hybridized at 64°C with 5 pmol probe (listed in key resources table). Probes were end-labelled with [γ-P32]-ATP using T4 PNK and purified with G25 sepharose columns (GE Healthcare Life Sciences). The blot was washed 2x in a low-stringency wash buffer (2X SSC, 0.1% SDS) and 1X in a high stringency wash buffer (0.5X SSC, 0.1% SDS) at 64°C, exposed to a Phosphor-Imaging screen for 12 – 24 hours, and imaged using a Typhoon scanner.
Flow cytometry
Cells were trypsinized from a 6 or 12 well plate, quenched with DMEM + 10% FBS, and cells were pelleted by centrifugation at 125g for 5 minutes. Pellets were resuspended in 500 μL (for a 12 well plate well) to 1 mL (for a 6 well plate well) of PBS and the cell suspension was passed through a 0.35 μm nylon mesh strainer-top tube (Corning). 10,000 – 30,000 events were collected for all experiments. YFP fluorescence measurements were log-transformed and the mean and standard deviation of all events was calculated from the population (see Fig. and Fig. S6).
Puromycin incorporation assays
75% confluent cells in a 6 cm plate were limited for leucine or arginine or grown in nutrient rich conditions for the desired time, followed by addition of puromycin (Sigma-Aldrich, P8833) to the culture medium at 10 μg/mL for exactly 5 or 10 minutes at 37°C. Cells were then washed once in ice-cold PBS and flash frozen in liquid nitrogen. Western blots or dot blots were performed to quantify puromycin incorporation into nascent polypeptide chains. To quantify blots, the total puromycin signal is integrated from each lane or dot and normalized to the signal intensity of a loading control (see Fig. and Fig. S5).
Reverse transcription & qPCR
RNA was extracted from cells in a 6-well plate with a Quick-RNA Miniprep kit (Zymo). Reverse transcription using a dT-20 primer or gene-specific primers was performed using Superscript III. cDNA template was diluted in water and qPCR was performed in 10 μL reaction volumes in 96 well plates, using the PowerUp SYBR Green PCR master mix (Thermo Fisher Scientific). To calculate relative YFP reporter mRNA levels, the YFP Ct value from qPCR analysis in each condition was normalized to the GAPDH Ct value to find ΔCt, and then to the ΔCt for the normalization sample indicated in figure legend to find ΔΔCt, which was converted to a normalized mRNA level by taking 2−ΔΔCt (see Fig. S6D).
Cell viability assays
20,000 cells were seeded in individual wells of 96-well plates (1 plate per assay time point, 5 technical replicate wells per plate) in amino acid limitation medium or rich medium. At desired time points, CellTiterGlo (CTG) assay was performed with the following modifications. Cells were lysed by adding 1 volume of CTG reagent and then transferred to an opaque black 96 well plate for luminescence reading. Luminesence was measured immediately on a TopCount instrument (Perkin Elmer) at 30°C. All viability measurements were normalized to an initial reading for each well taken 1.5 hours after seeding adherent cells (see Fig. S6I).
QUANTIFICATION AND STATISTICAL ANALYSIS
Databases utilized
A subset of unique canonical transcripts used for mapping aligned ribosome profiling sequencing reads was defined based on the Gencode v24 database annotation file (gencode.v24.annotation.gff3). For each gene, only transcripts annotated as both CCDS in the APPRIS principal splice isoform database (Rodriguez et al., 2013) were included; of this subset, the transcript with the lowest CCDS number for each gene was selected to generate a unique set.
tRNA gene numbers (see Fig. S1K) were obtained from the genomic tRNA database (Chan and Lowe, 2016).
Ribosome profiling data analysis
Analysis was performed using R and Bash programming languages. For analysis code and more details see https://github.com/rasilab/adarnell_2018. The polyA tail was trimmed from 50 nt single-end raw sequencing reads using cutadapt (Martin, 2011) with a minimum length cutoff of 13 nt. A subtractive alignment was performed against ribosomal RNA using bowtie (Langmead et al., 2009), and the remaining reads were aligned to the transcriptome using rsem and bowtie (Li and Dewey, 2011). To calculate the preprocessing statistics and assess library quality (see Fig. S1E-G), we used 3' trimming of 12 nt for reads <= 32 nt and 13 nt trimming for reads > 32 nt to demonstrate 3 nt periodicity. For the rest of the analyses, as we were interested in the overall increase in ribosome density at codons and frame information was not required, we trimmed 12 nt from both sides to smooth our ribosome density profiles as previously described (Li et al., 2012; Oh et al., 2011; Subramaniam et al., 2014). To calculate reads counts for each transcript, each transcript position aligning to the trimmed read was assigned a count of the inverse value of the read length. The DESeq2 package was used to normalize each sample and then calculate gene fold changes (see Fig. 3C,D) (Love et al., 2014).
To calculate the average ribosome density around each codon, only transcripts with a minimum average read density of 1 read per codon were considered. Reads at each transcript position were first normalized to the mean read count for that transcript. For each codon, the average read coverage was found for each position in a 150 nt window on either side of all occurrences of that codon. To calculate the change in average ribosome density around each codon upon amino acid limitation (see for example Fig. 1B), the average ribosome density at each position in the 150 nt window around the codon in rich conditions was subtracted from that in an amino acid limited condition. To calculate the summed ribosome density at each codon (see for example Fig. 1A), this 300 nt average ribosome density vector for each codon was summed.
Estimation of usage bias for pause-site arginine codons and GO analysis
We employed a binomial probability distribution to estimate the probability, for each gene, of having the observed number of CGC and CGU codons given the genome-wide average arginine codon usage frequencies (see Fig. S1J). To avoid skew due to local GC bias in our analysis, we only considered sets of pause-site or non-pause-site arginine codons with equivalent GC content (CGC/CGU vs. CGA/CGG, respectively; “CGN codons”). We calculated the average expected number of pause-site codons for each gene as the mean of a theoretical binomial probability distribution (μ); n*p, where n is the total number of arginine codons and p is the average frequency of stall sites relative to other CGN codons (p = 0.46). We calculated the standard deviation of that theoretical binomial probability distribution (σ) for each gene as the square root of n*p*(1-p). To then calculate a Z-score, we subtracted μ from the observed number of pause-site codons in that gene, and normalized by σ. When ranked, the resulting Z-scores range from −4.7 to 8.4 and represent bias towards (high scores) or against (low scores) the use of pause-site arginine codons to encode arginine in each gene (see Fig. S6G).
Gene ontology (GO) analysis to detect enrichment for GO terms in genes with biased usage of pause-site arginine codons was performed in R using the topGO library (Alexa and Rahnenfuhrer, 2016). GO terms with a false-discovery rate adjusted p-value of < 0.05 were visualized using R scripts to plot generated by REVIGO (Supek et al., 2011) (see Fig. S6H).
Statistics
For all experiments, technical replicates refer to the repetition an entire experiment with a separate dish of cells split off from the same parental cell line (i.e. produced from the same lentiviral transduction or CRISPR editing process). Unless otherwise indicated in figure legends, three replicates were performed.
To assess whether summed differences in ribosome density at arginine and leucine codons correlated significantly with measures of codon usage frequency (Fig. 1D; S1J,M), cognate tRNA copy number (Fig. 1E; S1K,N), or cognate tRNA charging loss (Fig. 2C) upon amino acid limitation, we performed a Spearman’s rank-order correlation analysis. For this test, we considered p < 0.05 to indicate a significant correlation between the variables compared.
To assess whether the expression of mTORC1 or GCN2 target genes was significantly different between arginine and leucine limitation, we used a two-sided Wilcoxon signed rank test with continuity correction. For this test, the null hypothesis was that the median difference (μ) in the log2 fold change for each target between arginine and leucine limitation was equal to zero, and we considered p < 0.05 to indicate a significant differential mTORC1 or GCN2 response to arginine versus leucine limitation (Fig. 3 & S3).
Fisher’s exact test was used to determine significance in enrichment of GO terms in genes with the highest and lowest 5% of Z-scores (Fig. S6G,H). GO terms with a false-discovery rate adjusted p-value of < 0.05 were considered significantly enriched in genes with the strongest bias against (low Z-scores) or towards (high Z-scores) arginine pause-site codon usage.
DATA AND SOFTWARE AVAILABILITY
Full code and detailed instructions for generating the final figures in our paper starting from raw sequencing data is provided as a README.md file and an interactive Jupyter notebook and static HTML files in the following Github repository (https://github.com/rasilab/adarnell_2018). Raw and processed high throughput sequencing data is available at GEO, accession number GSE113751.
Supplementary Material
Fig. S1 Codon-specific ribosome pausing emerges during limitation for arginine, but not leucine. (A-C) HEK293T cell polysome digestion into monosomes with varying amounts of RNaseI (B) in buffers with varying Mg2+ concentrations (A), or with MNase (C), was assessed by sucrose density gradient fractionation. (* = monosome fraction). (D) Representative size selection gel for RNA footprint extraction for library preparation. Box indicates the region excised. (E-G) Aligned read length distribution (E) and genome-wide read density profiles around annotated start (F) and stop codons (G) for data in Fig. 1A-C & S1H. After 3’ end trimming, normalized read density is calculated as described in Fig. 1A & Methods. A region of the read density profile in G is magnified in a second (right) panel. (H) Overlaid summed changes in codon-specific ribosome density for HCT116 and HeLa cells following 3 hours of leucine or arginine limitation, calculated as described in Fig. 1A & Methods. Arg and Leu codons are colored according to the legend in Fig. 1. (I) Intracellular arginine, isoleucine, leucine, and serine levels in HEK293T cells following limitation for leucine or arginine for 3 hours or growth in rich medium. Error bars represent the standard error of the mean from three technical replicate measurements. Intracellular leucine level was below the detection limit (n.d.) upon its limitation. (J) Usage frequencies for Arg codons in the transcriptome in HEK293T, HCT116, and HeLa cells following 3 hours of limitation for arginine or growth in rich medium. (K) Genomic copy number of Arg isoacceptor tRNAs (Chan and Lowe, 2016). (L) Arg and Leu codons matched with their cognate tRNA(s). Decoding by multiple tRNAs is indicated with a slash, I = inosine.
Fig. S2 Selective loss of cognate tRNA charging during arginine limitation. (A-C) Representative northern blots for determination of Arg and Leu tRNA charging levels (as shown in Fig. 2A,B) in HEK293T (A,B) cells or HCT116 cells (C) following 3 hours of limitation for leucine or arginine or growth in rich medium. A control deacylated total RNA sample is used to identify uncharged tRNA species. tRNA probe is indicated below each blot (see Methods for details of blot interpretation, quantification, and probe design).
Fig. S3 Differential mTORC1 and GCN2 responses to arginine and leucine limitation. (A,B) Representative western blots for phosphorylated and total levels of the S6K target, RPS6, in HEK293T cells after growth in rich medium or limitation for leucine or arginine for 3 hours + / − 250 nM Torin1 (A) or limitation for leucine or arginine for 3, 6 or 12 hours, or for all amino acids for 6 hours (B). Bar graphs show the fraction of protein that is phosphorylated in each condition, relative to rich medium; error bars represent the standard error of the mean from three technical replicate experiments. (C,D) Heatmap of log2 fold-changes (f.c.) in ribosome density for mRNA targets of mTORC1 inhibition (Hsieh et al., 2012) (C) or GCN2 activation via ATF4/CHOP (Han et al., 2013) (D) following 3 or 6 hours of leucine or arginine limitation, relative to rich medium, in HEK293T cells. Only targets with a log2 fold change of < 0, for mTORC1 targets, or > 0, for ATF4/CHOP targets, were considered. At 3 versus 6 hours, 43/73 (59%) versus 47/73 (64%) of mTORC1 targets (C), and 67/87 (77%) versus 77/87 (89%) of ATF4/CHOP targets (D) had higher ribosome density upon arginine than leucine limitation, respectively. (E,F) Box plot of the log2 fold change for each mTORC1 (E) or GCN2 (F) target upon amino acid limitation (as shown in C,D). A two-sided Wilcoxon signed rank test with continuity correction was performed with μ = 0; the resulting p-value is shown above each comparison (see Methods for details). At 3 hours versus 6 hours, the mTORC1 signaling response was 1.3- versus 1.4-fold higher (E), and the GCN2 signaling response was 1- versus 1.1-fold higher during arginine than leucine limitation, respectively (F). (G) Box plot of the difference in the log2 fold change between each mTORC1 or GCN2 target following 3 hours of limitation for arginine versus leucine in HEK293T, HCT116, and HeLa cells.
Fig. S4 Signaling through the mTORC1 and GCN2 pathways regulates the magnitude of ribosome pausing during amino acid limitation. (A) tRNA charging levels for 2 Arg tRNAs and 1 Leu tRNA in HEK293T cells following 3 hours of leucine or arginine limitation or growth in rich medium, + / − 250 nM Torin1 (calculated as described in Methods). Error bars represent the standard error of the mean from three technical replicate experiments. (B) Summed changes in codon-specific ribosome density in the hrGFP cell line following 3 hours of limitation for leucine or arginine with 250 nM Torin1, relative to rich medium. Arg and Leu codons are colored according to the legend in D. (C) tRNA charging levels for 3 Arg and 4 Leu tRNAs in the hrGFP, RagB-WT, or RagB-Q99L cell lines following limitation for leucine or arginine for 3 hours or growth in rich medium (calculated as described in Methods). Error bars represent the standard error of the mean from three technical replicate experiments. (D) Changes in codon-specific ribosome density for the hrGFP, RagB-WT, and RagB-Q99L cell lines following limitation for leucine or arginine for 3 hours, relative to rich medium. (E) tRNA charging levels for 1 Arg and 1 Leu tRNA in the WT, hrGFP, RagB-Q99L, or GCN2 KO cell lines following limitation for leucine or arginine for 3 hours or growth in rich medium. (F) Overlaid summed changes in codon-specific ribosome density for the WT, hrGFP, RagB-Q99L, and GCN2 KO cell lines following 6 hours of leucine or arginine limitation, relative to rich medium. Arg and Leu codons are colored according to the legend in D. (G) Western blots for GCN2 and GAPDH proteins in HEK293T and 3 clonal replicate GCN2 KO cell lines. (H) tRNA charging levels for 1 Arg and 1 Leu tRNA in the hrGFP cell line + / − 250 nM Torin1, the RagB-WT, and the RagB-Q99L cell lines after exposure for <1 minute to 100 μg/mL cycloheximide in ice-cold PBS following limitation for leucine or arginine for 3 hours or growth in rich medium.
Fig. S5 Ribosome pausing reduces global protein synthesis rate during amino acid limitation. (A) Representative western blots for puromycin and S6K in HCT116 cells after (+ puro) or without (− puro) a pulse of 10 μg/mL puromycin following 3 hours of leucine or arginine limitation, or growth in rich medium. Bar graph shows puromycin incorporation relative to rich medium (calculated as described in Methods); error bars represent the standard error of the mean from three technical replicate experiments. (B) Polysome profiles from the hrGFP and RagB-Q99L cell lines following 6 hours of leucine or arginine limitation or growth in rich medium. The main plot shows overlaid polysome profiles from the disome (2 ribosome) peak to the end of the polysomes, the inset plots show the entire profile. Bar graph shows the area in the polysome to monosome fraction, relative to that in rich medium (calculated as described in Methods). (C) Representative western blots for EEF2K and GAPDH in HEK293T and 3 clonal replicate EEF2K KO cell lines. (D) Western blots for phosphorylated and total EEF2 in WT and EEF2K KO cell lines following 3 hours of growth in rich medium, leucine limitation, or arginine limitation. (E) Puromycin incorporation in the WT (same data as Fig. 5D) or EEF2K KO cell lines following 3 hours of leucine or arginine limitation, relative to rich medium. Error bars represent the standard error of the mean for three technical replicate measurements. (F) Representative dot blots for puromycin and GAPDH in WT cells and the GCN2 KO cell line following 3 hours of leucine or arginine limitation or growth in rich medium.
Fig. S6 Ribosome pausing reduces protein expression from reporter mRNAs and induces premature termination of translation. (A-C) YFP codon variant reporter fluorescence measurements across multiple time points, cell lines, and reporter constructs (see Methods for details). Error bars represent the standard error of the mean from three technical replicate experiments. (A) Mean increase in YFP fluorescence per hour, + / − 10 μM TMP in HEK293T cells stably expressing the YFP-CGC reporter, following 24 or 38 hours of leucine or arginine limitation or growth in rich medium. (B) Mean YFP fluorescence in the HEK293T cells stably expressing the leucine or arginine YFP codon variant reporters following limitation for leucine or arginine with 10 μM trimethoprim for 24 hours, relative to rich medium +TMP. Arg and Leu codons are colored according to the legend in Fig. 6. (C) Mean YFP fluorescence in the HCT116, HeLa, and HEK293T cell lines stably expressing the YFP-CGC and -CGG reporters, following limitation for arginine, leucine or serum +TMP for 12, 24, or 48 hours, relative to rich medium +TMP. (D) YFP-CGC and -CGG CRISPR-AAVS1 reporter mRNA levels in HEK293T cells following 24 hours of limitation for leucine or arginine + / − TMP (calculated as described in Methods). From the top to bottom panel, the data is plotted 1) as the ratio of the YFP-CGG variant to the YFP-CGC variant for each condition, 2) normalized to the rich condition for the YFP-CGC reporter, 3) normalized to the rich condition for each YFP variant separately. Error bars represent the standard error of the mean for three technical replicate experiments. (E) CUA8 and UUG8 premature termination YFP reporter mRNA levels in the WT and GCN2 KO cell lines following 48 hours of limitation for leucine or arginine −TMP, relative to rich medium −TMP (see Methods section for details of calculation). Error bars represent the standard error of the mean for three technical replicate experiments. (F) Overexpressed blot image from Fig. 6I, for FLAG and GAPDH in the WT or GCN2 KO cell lines stably expressing the UUG8 or CUA8 reporters after growth in rich medium or 48 hours of leucine or arginine limitation. (G) Distribution of Z-scores reflecting pause-site arginine codon usage bias in coding sequences (see Methods section for details of calculation). (H) Biological process (BP) or cellular component (CC) gene ontology (GO) categories enriched in genes with bias against (left plot) or in favor of (right plot) usage of pause-site codons to encode arginine; visualized using REVIGO (Supek et al., 2011). Each bubble represents a significantly enriched GO term; color represents log10 of the false-discovery rate adjusted p-value, and size scales with the number of genes for a term. (I) Cell viability in the HEK293T or GCN2 KO cell lines following 1 to 13 days of leucine or arginine limitation, or growth in rich medium, relative to day 0. Error bars represent the standard error of the mean from five technical replicate measurements. Both cell lines reached confluency in rich medium after 4 days.
Acknowledgments
We thank B. Zid, C. Chidley, T. Pan, A. Murray, V. Denic, V. Mootha, and the O’Shea lab for helpful discussions, J. Piechura and J. Darnell for comments on the manuscript, C. Shoemaker for reagents and advice regarding CRISPR/Cas9, and K. Han for reagents used to build the YFP reporters. From the Harvard Bauer Core Facilities we thank C. Daly for high-throughput sequencing data, K. Chatman for LC-MS/MS data, and the Flow Cytometry Core. The sequencing data analysis was run on the Odyssey cluster, supported by the Research Computing Group at Harvard, and on the Rhino cluster at the Fred Hutchinson Cancer Research Center. This research was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award numbers, R00GM107113 and R35GM119835 (A.R.S.). EKO is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Declaration of Interests
The authors declare no competing interests.
References
- Alexa A, and Rahnenfuhrer J (2016). topGO: Enrichment Analysis for Gene Ontology. [Google Scholar]
- Andreev DE, O’Connor PB, Fahey C, Kenny EM, Terenin IM, Dmitriev SE, Cormican P, Morris DW, Shatsky IN, and Baranov PV (2015). Translation of 5’ leaders is pervasive in genes resistant to eIF2 repression. ELife e03971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Averous J, Lambert-Langlais S, Mesclon F, Carraro V, Parry L, Jousse C, Bruhat A, Maurin A-C, Pierre P, Proud CG, et al. (2016). GCN2 contributes to mTORC1 inhibition by leucine deprivation through an ATF4 independent mechanism. Sci. Rep 6, 27698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandman O, and Hegde RS (2016). Ribosome-associated protein quality control. Nat. Struct. Mol. Biol 23, 7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buskirk AR, and Green R (2017). Ribosome pausing, arrest and rescue in bacteria and eukaryotes. Philos. Trans. R. Soc. Lond. B. Biol. Sci 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttgereit F, and Brand MD (1995). A hierarchy of ATP-consuming processes in mammalian cells. Biochem. J. 312, 163–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerbini T, Funahashi R, Luo Y, Liu C, Park K, Rao M, Malik N, and Zou J (2015). Transcription Activator-Like Effector Nuclease (TALEN)-Mediated CLYBL Targeting Enables Enhanced Transgene Expression and One-Step Generation of Dual Reporter Human Induced Pluripotent Stem Cell (iPSC) and Neural Stem Cell (NSC) Lines. PLoS ONE 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PP, and Lowe TM (2016). GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 44, D184–D189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantranupong L, Scaria SM, Saxton RA, Gygi MP, Shen K, Wyant GA, Wang T, Harper JW, Gygi SP, and Sabatini DM (2016). The CASTOR proteins are arginine sensors for the mTORC1 pathway. Cell 165, 153–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu VT, Weber T, Wefers B, Wurst W, Sander S, Rajewsky K, and Kühn R (2015). Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol. 33, 543–548. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. (2013). Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmar KA, Sørensen MA, Elf J, Ehrenberg M, and Pan T (2005). Selective charging of tRNA isoacceptors induced by amino-acid starvation. EMBO Rep. 6, 151–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, et al. (2016). Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Qiu H, Garcia-Barrio M, Anderson J, and Hinnebusch AG (2000). Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Mol. Cell 6, 269–279. [DOI] [PubMed] [Google Scholar]
- Drummond DA, and Wilke CO (2008). Mistranslation-Induced Protein Misfolding as a Dominant Constraint on Coding-Sequence Evolution. Cell 134, 341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn JG, Foo CK, Belletier NG, Gavis ER, and Weissman JS (2013). Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. ELife 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edri S, and Tuller T (2014). Quantifying the Effect of Ribosomal Density on mRNA Stability. PLOS ONE 9, e102308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efeyan A, Zoncu R, Chang S, Gumper I, Snitkin H, Wolfson RL, Kirak O, Sabatini DD, and Sabatini DM (2013). Regulation of mTORC1 by the Rag GTPases is necessary for neonatal autophagy and survival. Nature 493, 679–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elf J, Nilsson D, Tenson T, and Ehrenberg M (2003). Selective charging of tRNA isoacceptors explains patterns of codon usage. Science 300, 1718–1722. [DOI] [PubMed] [Google Scholar]
- Ferrin MA, and Subramaniam AR (2017). Kinetic modeling predicts a stimulatory role for ribosome collisions at elongation stall sites in bacteria. ELife 6, e23629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N, Roux C, Rousselle M, Romiguier J, Figuet E, Glemin S, Bierne N, and Duret L (2017). Codon usage bias in animals: disentangling the effects of natural selection, effective population size and GC-biased gene conversion. BioRxiv 184283. [DOI] [PubMed] [Google Scholar]
- Gingold H, Tehler D, Christoffersen NR, Nielsen MM, Asmar F, Kooistra SM, Christophersen NS, Christensen LL, Borre M, Sørensen KD, et al. (2014). A Dual Program for Translation Regulation in Cellular Proliferation and Differentiation. Cell 158, 1281–1292. [DOI] [PubMed] [Google Scholar]
- Goodarzi H, Nguyen HCB, Zhang S, Dill BD, Molina H, and Tavazoie SF (2016). Modulated Expression of Specific tRNAs Drives Gene Expression and Cancer Progression. Cell 165, 1416–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossmann S, Bauer S, Robinson PN, and Vingron M (2007). Improved detection of overrepresentation of Gene-Ontology annotations with parent child analysis. Bioinforma. Oxf. Engl 23, 3024–3031. [DOI] [PubMed] [Google Scholar]
- Guydosh NR, and Green R (2014). Dom34 Rescues Ribosomes in 3’ Untranslated Regions. Cell 156, 950–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Back SH, Hur J, Lin Y-H, Gildersleeve R, Shan J, Yuan CL, Krokowski D, Wang S, Hatzoglou M, et al. (2013). ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat. Cell Biol 15, 481–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K, Jaimovich A, Dey G, Ruggero D, Meyuhas O, Sonenberg N, and Meyer T (2014). Parallel measurement of dynamic changes in translation rates in single cells. Nat. Methods 11, 86–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K, Yonezawa K, Weng Q-P, Kozlowski MT, Belham C, and Avruch J (1998). Amino Acid Sufficiency and mTOR Regulate p70 S6 Kinase and eIF-4E BP1 through a Common Effector Mechanism. J. Biol. Chem 273, 14484–14494. [DOI] [PubMed] [Google Scholar]
- Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, and Ron D (2000). Regulated Translation Initiation Controls Stress-Induced Gene Expression in Mammalian Cells. Mol. Cell 6, 1099–1108. [DOI] [PubMed] [Google Scholar]
- Hattori A, Tsunoda M, Konuma T, Kobayashi M, Nagy T, Glushka J, Tayyari F, McSkimming D, Kannan N, Tojo A, et al. (2017). Cancer progression by reprogrammed BCAA metabolism in myeloid leukemia. Nature 545, 500–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, Shi EY, Stumpf CR, Christensen C, Bonham MJ, et al. (2012). The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 485, 55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JRS, and Weissman JS (2009). Genome-Wide Analysis in Vivo of Translation with Nucleotide Resolution Using Ribosome Profiling. Science 324, 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, and Weissman JS (2011). Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147, 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Rouskin S, McGeachy AM, and Weissman JS (2012). The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc 7, 1534–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimura R, Nagy G, Dotu I, Zhou H, Yang X-L, Schimmel P, Senju S, Nishimura Y, Chuang JH, and Ackerman SL (2014). Ribosome stalling induced by mutation of a CNS-specific tRNA causes neurodegeneration. Science 345, 455–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimura R, Nagy G, Dotu I, Chuang JH, and Ackerman SL (2016). Activation of GCN2 kinase by ribosome stalling links translation elongation with translation initiation. ELife 5, e14295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Nilsson R, Sharma S, Madhusudhan N, Kitami T, Souza AL, Kafri R, Kirschner MW, Clish CB, and Mootha VK (2012). Metabolite Profiling Identifies a Key Role for Glycine in Rapid Cancer Cell Proliferation. Science 336, 1040–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jousse C, Bruhat A, Ferrara M, and Fafournoux P (2000). Evidence for Multiple Signaling Pathways in the Regulation of Gene Expression by Amino Acids in Human Cell Lines. J. Nutr 130, 1555–1560. [DOI] [PubMed] [Google Scholar]
- Kanaya S, Yamada Y, Kudo Y, and Ikemura T (1999). Studies of codon usage and tRNA genes of 18 unicellular organisms and quantification of Bacillus subtilis tRNAs: gene expression level and species-specific diversity of codon usage based on multivariate analysis. Gene 238, 143–155. [DOI] [PubMed] [Google Scholar]
- Kimball SR (2002). Regulation of Global and Specific mRNA Translation by Amino Acids. J. Nutr 132, 883–886. [DOI] [PubMed] [Google Scholar]
- Knott SRV, Wagenblast E, Khan S, Kim SY, Soto M, Wagner M, Turgeon M-O, Fish L, Erard N, Gable AL, et al. (2018). Asparagine bioavailability governs metastasis in a model of breast cancer. Nature 554, 378–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krall AS, Xu S, Graeber TG, Braas D, and Christofk HR (2016). Asparagine promotes cancer cell proliferation through use as an amino acid exchange factor. Nat. Commun 7, 11457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, and Salzberg SL (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leprivier G, Remke M, Rotblat B, Dubuc A, Mateo A-RF, Kool M, Agnihotri S, El-Naggar A, Yu B, Somasekharan SP, et al. (2013). The eEF2 Kinase Confers Resistance to Nutrient Deprivation by Blocking Translation Elongation. Cell 153, 1064–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, and Dewey CN (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G-W, Oh E, and Weissman JS (2012). The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature 484, 538–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind DS (2004). Arginine and Cancer. J. Nutr. 134, 2837S–2841S. [DOI] [PubMed] [Google Scholar]
- Loayza-Puch F, Rooijers K, Buil LCM, Zijlstra J, Vrielink F. Oude J., Lopes R, Ugalde AP, van Breugel P, Hofland I, Wesseling J, et al. (2016). Tumour-specific proline vulnerability uncovered by differential ribosome codon reading. Nature 530, 490–494. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, and Eddy SR (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, and Church GM (2013). RNA-Guided Human Genome Engineering via Cas9. Science 339, 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.Journal 17, 10–12. [Google Scholar]
- MGlincy NJ, and Ingolia NT (2017). Transcriptome-wide measurement of translation by ribosome profiling. Methods 126, 112–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nofal M, Zhang K, Han S, and Rabinowitz JD (2017). mTOR Inhibition Restores Amino Acid Balance in Cells Dependent on Catabolism of Extracellular Protein. Mol. Cell 67, 936–946.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Becker AH, Sandikci A, Huber D, Chaba R, Gloge F, Nichols RJ, Typas A, Gross CA, Kramer G, et al. (2011). Selective ribosome profiling reveals the co-translational chaperone action of trigger factor in vivo. Cell 147, 1295–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y, Reyna-Neyra A, Philippe L, and Thoreen CC (2017). mTORC1 Balances Cellular Amino Acid Supply with Demand for Protein Synthesis through Post-transcriptional Control of ATF4. Cell Rep. 19, 1083–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, Sethumadhavan S, Woo H-K, Jang HG, Jha AK, et al. (2011). Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature 476, 346–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presnyak V, Alhusaini N, Chen Y-H, Martin S, Morris N, Kline N, Olson S, Weinberg D, Baker KE, Graveley BR, et al. (2015). Codon optimality is a major determinant of mRNA stability. Cell 160, 1111–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian K, Huang C-L, Chen H, Blackbourn LW, Chen Y, Cao J, Yao L, Sauvey C, Du Z, and Zhang S-C (2014). A Simple and Efficient System for Regulating Gene Expression in Human Pluripotent Stem Cells and Derivatives. Stem Cells Dayt. Ohio 32, 1230–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian W, Yang J-R, Pearson NM, Maclean C, and Zhang J (2012). Balanced Codon Usage Optimizes Eukaryotic Translational Efficiency. PLOS Genet. 8, e1002603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovich S, Adler L, Yizhak K, Sarver A, Silberman A, Agron S, Stettner N, Sun Q, Brandis A, Helbling D, et al. (2015). Diversion of aspartate in ASS1-deficient tumours fosters de novo pyrimidine synthesis. Nature 527, 379–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan A, Chen Y-H, Martin S, Alhusaini N, Green R, and Coller J (2016). The DEAD-Box Protein Dhh1p Couples mRNA Decay and Translation by Monitoring Codon Optimality. Cell 167, 122–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, and Zhang F (2013). Genome engineering using the CRISPR-Cas9 system. Nat. Protoc 8, 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid DW, Shenolikar S, and Nicchitta CV (2015). Simple and inexpensive ribosome profiling analysis of mRNA translation. Methods San Diego Calif 91, 69–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez JM, Maietta P, Ezkurdia I, Pietrelli A, Wesselink J-J, Lopez G, Valencia A, and Tress ML (2013). APPRIS: annotation of principal and alternative splice isoforms. Nucleic Acids Res. 41, D110–D117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikia M, Wang X, Mao Y, Wan J, Pan T, and Qian S-B (2016). Codon optimality controls differential mRNA translation during amino acid starvation. RNA 22, 1719–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, and Sabatini DM (2008). The Rag GTPases Bind Raptor and Mediate Amino Acid Signaling to mTORC1. Science 320, 1496–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton RA, and Sabatini DM (2017). mTOR Signaling in Growth, Metabolism, and Disease. Cell 168, 960–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt EK, Clavarino G, Ceppi M, and Pierre P (2009). SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods 6, 275–277. [DOI] [PubMed] [Google Scholar]
- Scott L, Lamb J, Smith S, and Wheatley DN (2000). Single amino acid (arginine) deprivation: rapid and selective death of cultured transformed and malignant cells. Br. J. Cancer 83, 800–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms CL, Yan LL, and Zaher HS (2017). Ribosome Collision Is Critical for Quality Control during No-Go Decay. Mol. Cell 68, 361–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, and Hinnebusch AG (2009). Regulation of Translation Initiation in Eukaryotes: Mechanisms and Biological Targets. Cell 136, 731–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS, Sabatini DM, Chen ISY, Hahn WC, Sharp PA, et al. (2003). Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA 9, 493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam AR, Pan T, and Cluzel P (2013a). Environmental perturbations lift the degeneracy of the genetic code to regulate protein levels in bacteria. Proc. Natl. Acad. Sci. 110, 2419–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam AR, DeLoughery A, Bradshaw N, Chen Y, O’Shea E, Losick R, and Chai Y (2013b). A serine sensor for multicellularity in a bacterium. ELife 2, e01501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam AR, Zid BM, and O’Shea EK (2014). An Integrated Approach Reveals Regulatory Controls on Bacterial Translation Elongation. Cell 159, 1200–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F, Bošnjak M, Škunca N, and Šmuc T (2011). REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLoS One 6, e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Keenan MM, Wu J, Lin C-A, Dubois L, Thompson JW, Freedland SJ, Murphy SK, and Chi J-T (2015). Comprehensive Profiling of Amino Acid Response Uncovers Unique Methionine-Deprived Response Dependent on Intact Creatine Biosynthesis. PLoS Genet 11, e1005158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, and Gray NS (2009). An ATP-competitive Mammalian Target of Rapamycin Inhibitor Reveals Rapamycin-resistant Functions of mTORC1. J. Biol. Chem 284, 8023–8032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, and Sabatini DM (2012). A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485, 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, and DeBerardinis RJ (2017). Understanding the Intersections between Metabolism and Cancer Biology. Cell 168, 657–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney U, Lee CP, and RajBhandary UL (1991). Direct analysis of aminoacylation levels of tRNAs in vivo. Application to studying recognition of Escherichia coli initiator tRNA mutants by glutaminyl-tRNA synthetase. J. Biol. Chem 266, 24712–24718. [PubMed] [Google Scholar]
- Wang S, Tsun Z-Y, Wolfson RL, Shen K, Wyant GA, Plovanich ME, Yuan ED, Jones TD, Chantranupong L, Comb W, et al. (2015). Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science 347, 188–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise DR, and Thompson CB (2010). Glutamine addiction: a new therapeutic target in cancer. Trends Biochem. Sci 35, 427–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfson RL, Chantranupong L, Saxton RA, Shen K, Scaria SM, Cantor JR, and Sabatini DM (2016). Sestrin2 is a leucine sensor for the mTORC1 pathway. Science 351, 43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Kumanova M, Hart LS, Sloane K, Zhang H, De Panis DN, Bobrovnikova-Marjon E, Diehl JA, Ron D, and Koumenis C (2010). The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. EMBO J. 29, 2082–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaborske JM, Wu X, Wek RC, and Pan T (2010). Selective control of amino acid metabolism by the GCN2 eIF2 kinase pathway in Saccharomyces cerevisiae. BMC Biochem. 11, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, McGrath BC, Reinert J, Olsen DS, Lei L, Gill S, Wek SA, Vattem KM, Wek RC, Kimball SR, et al. (2002). The GCN2 eIF2α Kinase Is Required for Adaptation to Amino Acid Deprivation in Mice. Mol. Cell. Biol 22, 6681–6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Codon-specific ribosome pausing emerges during limitation for arginine, but not leucine. (A-C) HEK293T cell polysome digestion into monosomes with varying amounts of RNaseI (B) in buffers with varying Mg2+ concentrations (A), or with MNase (C), was assessed by sucrose density gradient fractionation. (* = monosome fraction). (D) Representative size selection gel for RNA footprint extraction for library preparation. Box indicates the region excised. (E-G) Aligned read length distribution (E) and genome-wide read density profiles around annotated start (F) and stop codons (G) for data in Fig. 1A-C & S1H. After 3’ end trimming, normalized read density is calculated as described in Fig. 1A & Methods. A region of the read density profile in G is magnified in a second (right) panel. (H) Overlaid summed changes in codon-specific ribosome density for HCT116 and HeLa cells following 3 hours of leucine or arginine limitation, calculated as described in Fig. 1A & Methods. Arg and Leu codons are colored according to the legend in Fig. 1. (I) Intracellular arginine, isoleucine, leucine, and serine levels in HEK293T cells following limitation for leucine or arginine for 3 hours or growth in rich medium. Error bars represent the standard error of the mean from three technical replicate measurements. Intracellular leucine level was below the detection limit (n.d.) upon its limitation. (J) Usage frequencies for Arg codons in the transcriptome in HEK293T, HCT116, and HeLa cells following 3 hours of limitation for arginine or growth in rich medium. (K) Genomic copy number of Arg isoacceptor tRNAs (Chan and Lowe, 2016). (L) Arg and Leu codons matched with their cognate tRNA(s). Decoding by multiple tRNAs is indicated with a slash, I = inosine.
Fig. S2 Selective loss of cognate tRNA charging during arginine limitation. (A-C) Representative northern blots for determination of Arg and Leu tRNA charging levels (as shown in Fig. 2A,B) in HEK293T (A,B) cells or HCT116 cells (C) following 3 hours of limitation for leucine or arginine or growth in rich medium. A control deacylated total RNA sample is used to identify uncharged tRNA species. tRNA probe is indicated below each blot (see Methods for details of blot interpretation, quantification, and probe design).
Fig. S3 Differential mTORC1 and GCN2 responses to arginine and leucine limitation. (A,B) Representative western blots for phosphorylated and total levels of the S6K target, RPS6, in HEK293T cells after growth in rich medium or limitation for leucine or arginine for 3 hours + / − 250 nM Torin1 (A) or limitation for leucine or arginine for 3, 6 or 12 hours, or for all amino acids for 6 hours (B). Bar graphs show the fraction of protein that is phosphorylated in each condition, relative to rich medium; error bars represent the standard error of the mean from three technical replicate experiments. (C,D) Heatmap of log2 fold-changes (f.c.) in ribosome density for mRNA targets of mTORC1 inhibition (Hsieh et al., 2012) (C) or GCN2 activation via ATF4/CHOP (Han et al., 2013) (D) following 3 or 6 hours of leucine or arginine limitation, relative to rich medium, in HEK293T cells. Only targets with a log2 fold change of < 0, for mTORC1 targets, or > 0, for ATF4/CHOP targets, were considered. At 3 versus 6 hours, 43/73 (59%) versus 47/73 (64%) of mTORC1 targets (C), and 67/87 (77%) versus 77/87 (89%) of ATF4/CHOP targets (D) had higher ribosome density upon arginine than leucine limitation, respectively. (E,F) Box plot of the log2 fold change for each mTORC1 (E) or GCN2 (F) target upon amino acid limitation (as shown in C,D). A two-sided Wilcoxon signed rank test with continuity correction was performed with μ = 0; the resulting p-value is shown above each comparison (see Methods for details). At 3 hours versus 6 hours, the mTORC1 signaling response was 1.3- versus 1.4-fold higher (E), and the GCN2 signaling response was 1- versus 1.1-fold higher during arginine than leucine limitation, respectively (F). (G) Box plot of the difference in the log2 fold change between each mTORC1 or GCN2 target following 3 hours of limitation for arginine versus leucine in HEK293T, HCT116, and HeLa cells.
Fig. S4 Signaling through the mTORC1 and GCN2 pathways regulates the magnitude of ribosome pausing during amino acid limitation. (A) tRNA charging levels for 2 Arg tRNAs and 1 Leu tRNA in HEK293T cells following 3 hours of leucine or arginine limitation or growth in rich medium, + / − 250 nM Torin1 (calculated as described in Methods). Error bars represent the standard error of the mean from three technical replicate experiments. (B) Summed changes in codon-specific ribosome density in the hrGFP cell line following 3 hours of limitation for leucine or arginine with 250 nM Torin1, relative to rich medium. Arg and Leu codons are colored according to the legend in D. (C) tRNA charging levels for 3 Arg and 4 Leu tRNAs in the hrGFP, RagB-WT, or RagB-Q99L cell lines following limitation for leucine or arginine for 3 hours or growth in rich medium (calculated as described in Methods). Error bars represent the standard error of the mean from three technical replicate experiments. (D) Changes in codon-specific ribosome density for the hrGFP, RagB-WT, and RagB-Q99L cell lines following limitation for leucine or arginine for 3 hours, relative to rich medium. (E) tRNA charging levels for 1 Arg and 1 Leu tRNA in the WT, hrGFP, RagB-Q99L, or GCN2 KO cell lines following limitation for leucine or arginine for 3 hours or growth in rich medium. (F) Overlaid summed changes in codon-specific ribosome density for the WT, hrGFP, RagB-Q99L, and GCN2 KO cell lines following 6 hours of leucine or arginine limitation, relative to rich medium. Arg and Leu codons are colored according to the legend in D. (G) Western blots for GCN2 and GAPDH proteins in HEK293T and 3 clonal replicate GCN2 KO cell lines. (H) tRNA charging levels for 1 Arg and 1 Leu tRNA in the hrGFP cell line + / − 250 nM Torin1, the RagB-WT, and the RagB-Q99L cell lines after exposure for <1 minute to 100 μg/mL cycloheximide in ice-cold PBS following limitation for leucine or arginine for 3 hours or growth in rich medium.
Fig. S5 Ribosome pausing reduces global protein synthesis rate during amino acid limitation. (A) Representative western blots for puromycin and S6K in HCT116 cells after (+ puro) or without (− puro) a pulse of 10 μg/mL puromycin following 3 hours of leucine or arginine limitation, or growth in rich medium. Bar graph shows puromycin incorporation relative to rich medium (calculated as described in Methods); error bars represent the standard error of the mean from three technical replicate experiments. (B) Polysome profiles from the hrGFP and RagB-Q99L cell lines following 6 hours of leucine or arginine limitation or growth in rich medium. The main plot shows overlaid polysome profiles from the disome (2 ribosome) peak to the end of the polysomes, the inset plots show the entire profile. Bar graph shows the area in the polysome to monosome fraction, relative to that in rich medium (calculated as described in Methods). (C) Representative western blots for EEF2K and GAPDH in HEK293T and 3 clonal replicate EEF2K KO cell lines. (D) Western blots for phosphorylated and total EEF2 in WT and EEF2K KO cell lines following 3 hours of growth in rich medium, leucine limitation, or arginine limitation. (E) Puromycin incorporation in the WT (same data as Fig. 5D) or EEF2K KO cell lines following 3 hours of leucine or arginine limitation, relative to rich medium. Error bars represent the standard error of the mean for three technical replicate measurements. (F) Representative dot blots for puromycin and GAPDH in WT cells and the GCN2 KO cell line following 3 hours of leucine or arginine limitation or growth in rich medium.
Fig. S6 Ribosome pausing reduces protein expression from reporter mRNAs and induces premature termination of translation. (A-C) YFP codon variant reporter fluorescence measurements across multiple time points, cell lines, and reporter constructs (see Methods for details). Error bars represent the standard error of the mean from three technical replicate experiments. (A) Mean increase in YFP fluorescence per hour, + / − 10 μM TMP in HEK293T cells stably expressing the YFP-CGC reporter, following 24 or 38 hours of leucine or arginine limitation or growth in rich medium. (B) Mean YFP fluorescence in the HEK293T cells stably expressing the leucine or arginine YFP codon variant reporters following limitation for leucine or arginine with 10 μM trimethoprim for 24 hours, relative to rich medium +TMP. Arg and Leu codons are colored according to the legend in Fig. 6. (C) Mean YFP fluorescence in the HCT116, HeLa, and HEK293T cell lines stably expressing the YFP-CGC and -CGG reporters, following limitation for arginine, leucine or serum +TMP for 12, 24, or 48 hours, relative to rich medium +TMP. (D) YFP-CGC and -CGG CRISPR-AAVS1 reporter mRNA levels in HEK293T cells following 24 hours of limitation for leucine or arginine + / − TMP (calculated as described in Methods). From the top to bottom panel, the data is plotted 1) as the ratio of the YFP-CGG variant to the YFP-CGC variant for each condition, 2) normalized to the rich condition for the YFP-CGC reporter, 3) normalized to the rich condition for each YFP variant separately. Error bars represent the standard error of the mean for three technical replicate experiments. (E) CUA8 and UUG8 premature termination YFP reporter mRNA levels in the WT and GCN2 KO cell lines following 48 hours of limitation for leucine or arginine −TMP, relative to rich medium −TMP (see Methods section for details of calculation). Error bars represent the standard error of the mean for three technical replicate experiments. (F) Overexpressed blot image from Fig. 6I, for FLAG and GAPDH in the WT or GCN2 KO cell lines stably expressing the UUG8 or CUA8 reporters after growth in rich medium or 48 hours of leucine or arginine limitation. (G) Distribution of Z-scores reflecting pause-site arginine codon usage bias in coding sequences (see Methods section for details of calculation). (H) Biological process (BP) or cellular component (CC) gene ontology (GO) categories enriched in genes with bias against (left plot) or in favor of (right plot) usage of pause-site codons to encode arginine; visualized using REVIGO (Supek et al., 2011). Each bubble represents a significantly enriched GO term; color represents log10 of the false-discovery rate adjusted p-value, and size scales with the number of genes for a term. (I) Cell viability in the HEK293T or GCN2 KO cell lines following 1 to 13 days of leucine or arginine limitation, or growth in rich medium, relative to day 0. Error bars represent the standard error of the mean from five technical replicate measurements. Both cell lines reached confluency in rich medium after 4 days.
Data Availability Statement
Full code and detailed instructions for generating the final figures in our paper starting from raw sequencing data is provided as a README.md file and an interactive Jupyter notebook and static HTML files in the following Github repository (https://github.com/rasilab/adarnell_2018). Raw and processed high throughput sequencing data is available at GEO, accession number GSE113751.