Abstract
The prospection of bioherbicides has been an alternative to weed control, aiming at mitigating chemical risks to human, animal and environmental health due to extreme use of synthetic herbicides. In the present study, various fungi were isolated from plants with symptoms of fungal diseases for bioherbicide purposes against weeds (Urochloa plantaginea, Euphorbia heterophylla and Bidens pilosa). Fungi isolated were identified by molecular methods and enzymatic products obtained by fungi fermentation (cellulase, lipase, peroxidase, and amylase) were quantified. Bioherbicide selectivity study was performed on crops (soybean and corn), as well as on resistant weeds. Among the isolated fungi, Fusarium oxysporum, Fusarium ploriferatum, and Trichoderma koningiopsis presented bioherbicide potential. T. koningiopsis, in particular, presented the highest effect on Euphorbia heterophylla (popular name - Mexican fire plant), causing up to 60% of foliar damage, without presenting phytotoxicity against corn crop. New perspectives for weeds control and their use in corn crops were prospected, considering the bioherbicide selectivity described in this study.
Keywords: Biotechnology, Plant biology, Systems biology, Microbiology
1. Introduction
Weeds have traits, which confer them great aggressiveness even in adverse environments. Weeds may rapidly capture resources and occupy space. This is often linked to their competitive ability because rapid growth requires the prompt and efficient conversion of resources into biomass. Thus, the yield crops are reduced and the agriculture costs increased, resulting in a decrease in farmer's income (Trognitz et al., 2016; Silva et al., 2013).
The high diversity of fungi establishes them in diverse areas of the human activity, from those for culinary use like for control of other organisms. For the biological control of weeds, the focus is on the use of phytopathogenic fungi, which are called bioherbicides (Charudattan, 2000). The first record of the use of fungi for weed management in the form of a biocontrol occurred in 1971, with Puccinia chondrillina, introduced in Australia for the control of Chondrilla juncea (Barton, 2004).
In 2009 only eleven bioherbicides were available in agriculture; this shows the need for the development of new products. However, when considering the investments of chemical industries in the development of bioherbicides these have a success rate below 1% against a success rate of 5% in the development of conventional herbicides (Ash, 2010; Klaic et al., 2015).
The bioherbicidal activity of microorganisms was related to the compatibility between plant and microorganism. The different virulence factors are directly involved in this infection process. First, the microorganism infection and phytotoxic efficiency may be through enzymes that degrade the cell wall thereby facilitating the penetration of such compounds. Enzymes such as cellulases and lipases degrade the cell walls and lipid membranes, being the main means of entry into the plant (Ghorbani et al., 2005; Cordeau et al., 2016).
Development studies and selection of promising microorganisms for weed control becomes important. The present work aimed to select and evaluate phytopathogenic microorganisms isolated from weeds regarding their potential to control these plants and their effect on summer crops.
2. Material and methods
2.1. Infected plants collection
Digitaria ciliares, Bidens pilosa and Euphorbia heterophylla were infected with fungi, showing typical symptoms of fungal diseases. These plants were collected from soybean and corn crop areas in Southern Brazil, in the period of 2015 and 2016.
The collected samples were conditioned in individual bags and, kept under cooling in polystyrene boxes. Samples were transported to the Agroecology Laboratory of the Federal University of Fronteira Sul - campus Erechim, and isolations of endophytic phytopathogenic microorganisms were carried.
2.2. Isolation of phytopathogenic microorganisms
For endophytic phytopathogenic fungi, isolation was used the scraping method on the plant's lesions, where small samples of infected material were placed in petri dishes containing Potato Dextrose Agar (PDA) culture medium, which was incubated at 28 °C for 7 days, as described by De Souza et al. (2017). After this period, successive replications were carried in PDA, until that pure culture was obtained for fermentative process.
2.3. Fermentative process
The fermented extract contain mycelium and culture medium was obtained by submerged fermentation, using shaker flasks containing 350 mL of culture medium. For this, culture medium were prepared by addition of 10 g L−1 glucose (C6H12O6); 7.5 g L−1 yeast extract; 10 g L−1 peptone; 2 g L−1 of ammonium sulfate ((NH4)2SO4); 0.5 g L−1 of magnesium sulfate (MgSO4 .7H2O); 1 g ferrous sulfate L−1 (FeSO4 .7H2O) and 1 g L−1 manganese sulfate (MnSO4 .H2O) (De Souza et al., 2017).
Fermentation experiments were performed at 28 °C in a shaker (120 rpm) during 3 days. The total fermentative extracts was evaluated as potential bioherbicide in crops and weeds.
2.4. Evaluation of bioherbicide potential
The fermented extracts obtained from the studied fungi were used for the bioherbicide activity assays. For this, Aureo® Emulsifiable concentrate (EC) vegetable oil at the recommended dose of 0.1% v/v (100 mL in 100 liters of water) was added to the extracts, and in order to test the effect of a greater number of applications, the pure extract with the adjuvant was applied sequentially and applied 1, 2, 4 and 8 times. For the applications 2 to 8, a 5-minute interval was established between one application and another. The evaluations were carried out 1, 7, 15, 20 and 30 days after the application of the treatments, evaluating the percentage of injury on the plants according to the aforementioned diagrammatic scale.
In order to evaluate the effect of fermented fungi on crop cultivation (corn and soybean), the extract was applied with the vegetable oil at the same concentrations, and the same weed evaluations were performed. For comparison purposes, two controls were used, one only with the application of water and another only with the application of water and adjuvant. For the bioherbicide potential test, the design was completely randomized (DIC), with 3 replicates and 12 treatments.
This experiment aimed to identify the pure cultures with the capacity to inhibit the development of seedlings before a great diversity of isolates. The bioassay was conducted in a greenhouse, in which weeds hairy beggarticks, alexandergrass and mexican fire plant were used to evaluate the effectiveness of the extracts. Seeding of weeds was carried out in plastic trays containing organic substrate (30%) and sieved earth (70%). The application of bioherbicides occurred by sprinkling on the plants.
A completely randomized design (CRD) with 3 replicates was used. The treatments consisted of the application of the extracts obtained in the fermentation. The different extracts used in the plants were considered treatments applying the same direction on the aerial part of the plant until the drainage with a garden sprayer.
Visual analyzes were performed after 1, 7, 15, 20 and 30 days of application of the treatments (DAT), evaluating the percentage of injury from a diagrammatic scale proposed by Nunes & Alvez (Nunes and Alves, 2012).
2.5. Enzymatic activity of the extracts
To evaluate the enzymatic activity of the extracts obtained, follow enzymes were selected in this study: (1) cellulase, (2) peroxidase, (3) lipase and (4) amylase.
2.5.1. Cellulase activity
Following the proposal of Ghose (1987), with some modifications, 50 mg of Whatman #1 filter paper (cellulose source) were weighed and added to test tubes containing 2 mL of 0.2 M acetate buffer pH 5.5. A volume of 1 ml of the enzyme solution was added to the tube in a thermostatic bath at 50 °C for 1 hour. The release of total reducing sugars was measured by the DNS method. A unit of enzymatic activity (U) was defined as the amount of cellulase capable of releasing 1 μmol of glucose per minute under the reaction conditions.
2.5.2. Peroxidase activity
The activity of the peroxidase enzyme was analyzed by its oxidative action on the guaiacol substrate in the presence of hydrogen peroxide in buffering medium. The reaction medium was composed of 1.5 mL of 5 mM phosphate buffer pH 5.0 and 2 mL of distilled water over 0.5 mL of the 1% guaiacol substrate and 1 mL of 8% hydrogen peroxide. The medium was kept in a thermostatic bath until the temperature reached 35 °C. After temperature stabilization, 1 mL of the enzyme extract was added, and the reaction medium was maintained in the thermostatic bath for 20 minutes. The transmittance of the oxidized compounds was obtained in UV-VIS spectrophotometer at 470 nm (Khan and Robinson, 1994).
2.5.3. Lipase activity
A 10% (w/v) olive oil emulsion and 5% (w/v) Arabic gum extract, diluted in 90% (v/v) sodium phosphate buffer 100 mM pH 6, were used to determine the lipase activity present in the fermentative extract. For the hydrolysis reaction process, 18 mL of emulsion and 2 mL of the extract were used, the solution of which was incubated for 32 minutes at 35 °C in a shaker with shaking of 165 rpm (Treichel et al., 2017). After this period, the reaction was stopped by adding 20 mL of acetone-ethanol solution (1:1, v/v). The fatty acids released during the reaction were titrated to pH 11 with 0.05 M sodium hydroxide solution. For purposes of comparison of the initial amount of free fatty acids with the amount after the reaction, a control sample (reaction blank) was used and the acetone-ethanol solution was added before the addition of the extract to the emulsion. For the calculation of the activity, Eq. (1) was used.
| (1) |
where Va is the mean of the volumes of NaOH spent in the titration of the samples; Vb is the volume of NaOH spent by the control; MNaOH is the molarity value of sodium hydroxide; t is the reaction time and Vc is the volume of enzyme extract used (Treichel et al., 2017). A unit of enzymatic activity (U) was defined as the amount of lipase capable of releasing 1 μmol of fatty acids per minute under the reaction conditions (Cavalcanti et al., 2005).
2.5.4. Amylase activity
The method described by Fuwa (1954) and Pongsawasdi and Yagisawa (1987) was used, with some modifications. Starch was diluted in 100 mM acetate buffer pH 5.0 in the ratio of 1:100. After, 1 ml of the diluted starch was added to 1 ml of the extract, which was incubated at 38 °C for 10 minutes. The enzymatic activity was determined by the DNS method, the same methodology used for cellulase activity.
2.6. Molecular identification of fungi
The DNA of the isolates fungi was extracted according to the method described by Doyle and Doyle (1987), from the mycelium grown in culture medium. The extracted genomic DNA sample was submitted to the polymerase chain reaction (PCR) for amplification of the rDNA internal transcribed spacer (ITS) region. The oligonucleotide primers for the ITS region were ITS1 (5′-TCCGTAGGTGAACCTGCGG-3 ′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) (White et al., 1990). The PCR mixture consisted of 1 μl of DNA, 1 μl of each primer at 10 μM, 10 μl of 5X PCR buffer, 1 μl of 10 mM dNTPs, 0.2 μl of GoTaq DNA polymerase 5U/μL (Promega) and autoclaved 35.8 μL MilliQ H2O, to a final volume of 50 μL. The amplification program consisted of initial denaturation at 94 °C/2 min followed by 40 denaturation cycles at 94 °C/10 s, annealing at 54 °C/30 s, extension at 72 °C/45 s, and a final extension at 72 °C/4 min. The amplified products were verified by means of 0.8% agarose gel electrophoresis, stained with ethidium bromide. The amplified products were purified by precipitation with polyethyleneglycol, subjected to the chain termination sequencing reaction using the Big Dye 3.1 reagent (Applied Biosystems) and analyzed in a 3,500 XL automatic capillary sequencer (Applied Biosystems). Sequences similar to those obtained for the isolates of the present study were found in GenBank through the Blastn tool (Schmitz and Riesner, 2006).
3. Results and discussion
3.1. Microorganisms isolation and screening
Thirty fungi were isolated and stored in the Agroecology Laboratory of the Federal University of Fronteira Sul - campus Erechim. These microorganisms were submitted to screening, to identify strains with bioherbicidal potential. From the tests, three microorganisms presented bioherbicide potential and, were selected when showed damage to at least one of the test plants.
The fungi were identified as Fusarium oxysporum – GenBank sequence number MK860712 (99% query coverage and 99% identity), Fusarium ploriferatum - GenBank sequence number MK860713 (99% query coverage and 99% identity), and Trichoderma koningiopsis MK860714 (100% query coverage and 100% identity), respectively.
As demonstrated in Fig. 1, in the initial tests, the species that presented the greatest susceptibility to the selected fungi was the Mexican fire plant (Euphorbia heterophylla), with damage of up to 10% in the leaf with T. koningiopsis no effect was observed on Bidens sp. and Urochloa plantaginea. From the 7 days after the application, the Mexican fire plant that presented damages by the bioherbicides began the emission of new shoots, suggesting that the damage was only of contact and not of a systemic form.
Fig. 1.
Mexican fire plant (Euphorbia heterophylla) control untreated (a) and, with damage after aplication bioherbicide from T. koningiopsis (b).
Damage suppression may be related to the hypersensitive response, which is one of the plant's main defense responses against pathogen attack, being a rapid and localized response in the pathogen infection region. This phenomenon is characterized by a rapid and localized collapse of the plant tissue around the infection, preventing the progression of disease and circulation of toxic compounds (Agrios, 2004). New methodologies are needed that aim at the biocontrol of weeds based on microorganisms associated with soil and plant (Trognitz et al., 2016).
Species of Fusarium fungi have a well-established phytopathogenicity and awere used in several studies as a source of biological control of weeds (Hodosy, 1981; Bouzoukov and Kouzmanova, 1994; Logrieco and Moretti, 1995; Boari and Vurro, 2003; Saremi and Okhovvat, 2008). These fungi cause various types of diseases in a wide range of plant species.
T. koningiopsis is well established as a biological control of insects and fungi (Samuels et al., 2006; You et al., 2016), including in the control of fusariosis in pineapple (Souza et al., 2016). However, there are no reports of this microorganism being used as a source for weed control. To date, only in the work of Qian et al. (2013) this fungus was identified as the causal agent of leaf spot in Curcuma wenyujin, suggesting that it may be phytopathogenic. Except for the work described by Qian et al. (2013), there are no more reports in the literature on the possibility of using T. koningiopsis as a weed control agent. Positive identification in this work indicates a new possibility of use for this fungus.
3.2. Enzymatic activity of the extracts
Table 1 show the enzymatic activity results of the fungi, considering cellulase, amylase, peroxidase and lipase enzymes.
Table 1.
Enzymatic activity of cellulase, amylase, peroxidase and lipase, present in the fermentative extract of the fungi Fusarium oxysporum, F. ploriferatum and Trichoderma koningiopsis.
| Fungus | Enzymatic activity (U/mL) |
|||
|---|---|---|---|---|
| Cellulase | Amylase | Peroxidase | Lipase | |
| Fusarium oxysporum | 0.950 | 6.433 | 479.33 | 0 |
| Fusarium ploriferatum | 0.416 | 0.647 | 285.33 | 0 |
| Trichoderma koningiopsis | 11.455 | 1.320 | 261.33 | 9.84 |
T. koningiopsis showed enzymatic activity for cellulase and lipase, is also the most damaging fungus to the Mexican fire plant. This increased enzymatic activity may have facilitated the entry of the microorganism along with its compounds, thus enhancing phytotoxicity.
The mode of action of bioherbicides is similar to the mechanisms of plant-pathogen interaction. In the case of plant-pathogen interaction, the microorganism must bypass the plant defense mechanisms and the relationship between the two individuals must be compatible so that the biocontrol agent can infect the target plant. Virulence factors are related directly or indirectly to the infection process (Trognitz et al., 2016). Thus, virulence factors could be enzymes that degrade cell walls, facilitating the entry of the microorganism and the phytotoxic components and phytotoxins or secondary metabolites, that can interfere in the metabolism of the plant (Ghorbani et al., 2005; Stergiopoulos et al., 2013; Harding and Raizada, 2015; Cordeau et al., 2016).
Among the enzymes studied, the most important are cellulase and lipase, the first one since its role is the degradation of cellulose, the main component of the vegetal cell wall, and the second to assist in the degradation of lipid membranes. Other enzymes such as proteases, peptidases, peroxidases, amylases, and phospholipases could help in the degradation of proteins and other structures (Ghorbani et al., 2005). Studies aiming at the addition of extracellular cellulase in bioherbicides showed that the addition of this enzyme generated the death of 58% of Orobanche aegyptiaca plants when compared to the extract without the addition of the enzyme, which resulted in approximately 38% mortality (Babalola, 2010).
3.3. Bioherbicidal potential
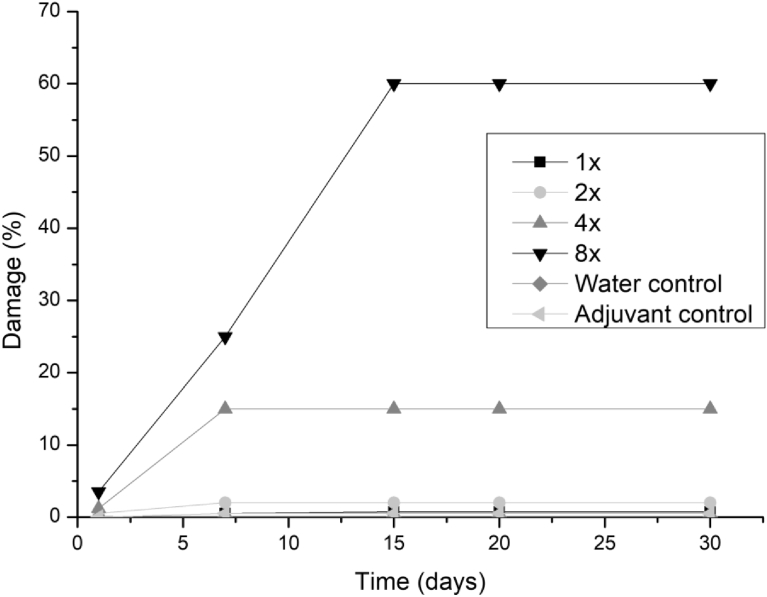
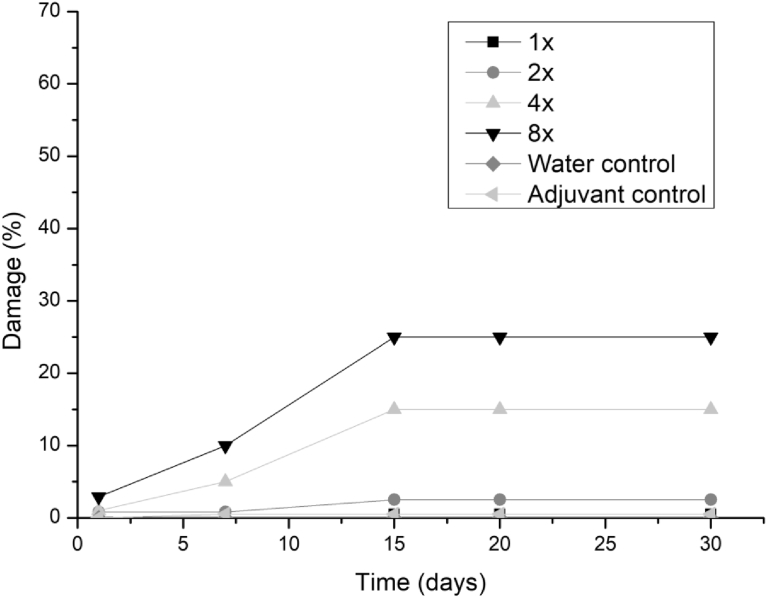
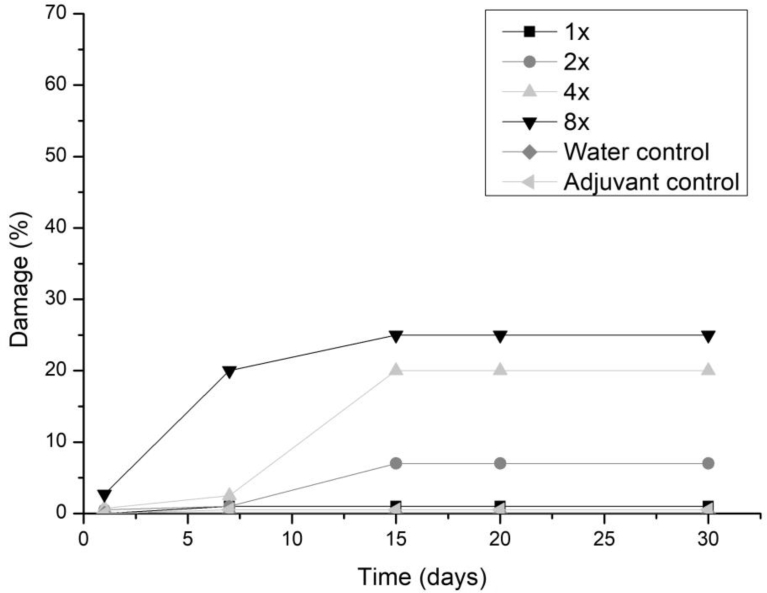
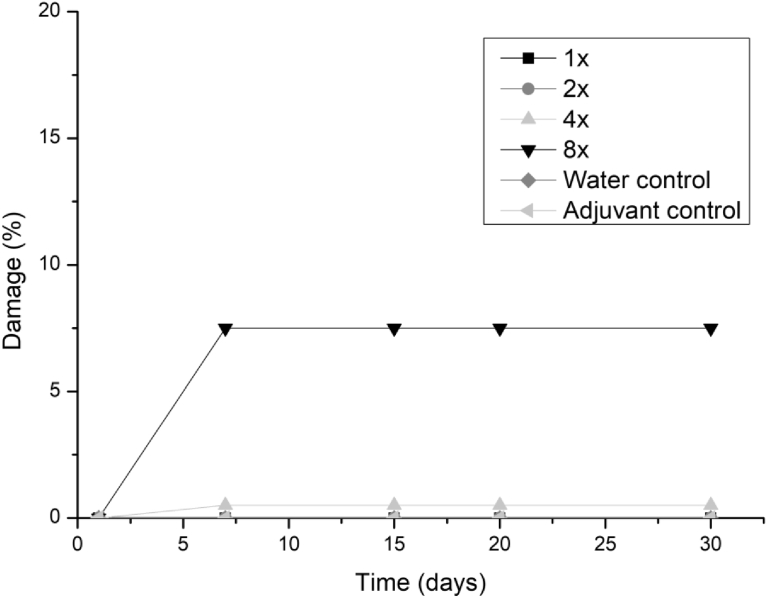
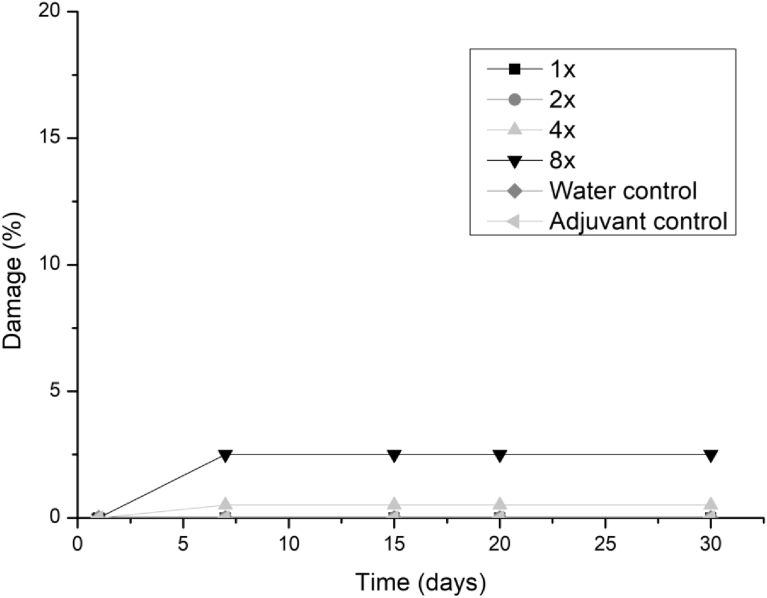
All three fungi presented the same symptoms for the dicotyledonous soybean and Mexican fire plant (necrotic areas on the leaf and shriveling). Hairy beggarticks and alexander grass did not present susceptibility to any of the microorganisms, concentrations and times evaluated, as did corn. The Mexican fire plant was the weed that showed greater sensitivity to the increase of the bioherbicide concentration for all the fungi (Fig. 2). Phytotoxicity was 60% at the highest adjuvant concentration for the fungus T. koningiopsis. For F. oxysporum and F. ploriferatum, phytotoxicity increased up to 25% for 8-times applications (Figs. 3 and 4).
Fig. 2.
Damage percentage of T. koningiopsis extract on Mexican fire plant with addition of Aureo® adjuvant in 1, 2, 4 and 8 applications up to 30 days after application.
Fig. 3.
Damage percentage of the F. ploriferatum extract on Mexican fire plant with addition of the Aureo® adjuvant, in 1, 2, 4 and 8 applications up to 30 days after application.
Fig. 4.
Damage percentage of F. oxysporum extract on Mexican fire plant with addition of Aureo® adjuvant in 1, 2, 4 and 8 applications up to 30 days after application.
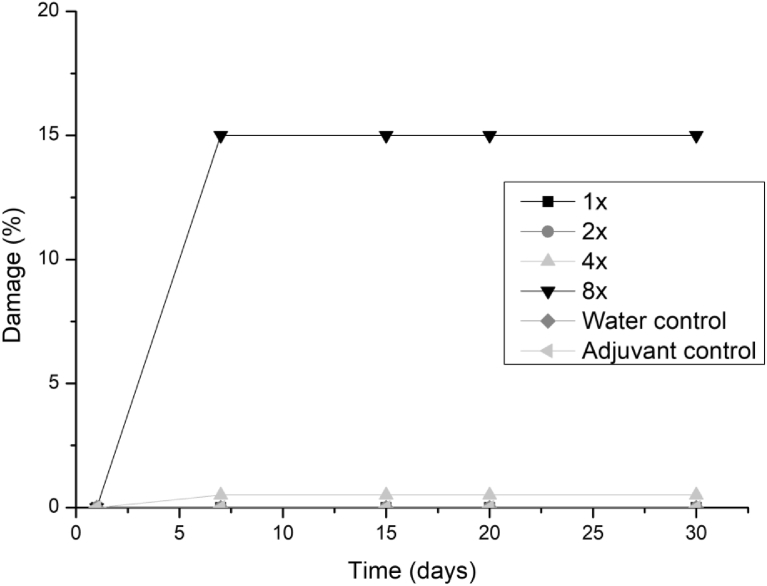
Some symptoms could be observed from 7 days for alexander grass and corn plants at concentrations of 4 and 8 times, but these symptoms are associated with larger droplets of extract that accumulated in some points of the leaf, presenting necrosis in the evaluated times. However, there was no increase in symptoms after 7 days of evaluation. The damages observed for these species did not exceed 1% even with the addition of adjuvant. The soybean showed sensitivity to the extracts only for 4 and 8 applications for the three fungi, reaching damages of up to 15% (Figs. 5, 6, and 7).
Fig. 5.
Damage percentage of T. koningiopsis extract on soybean with addition of Aureo® adjuvant in 1, 2, 4 and 8 applications up to 30 days after application.
Fig. 6.
Damage percentage of F. ploriferatum extract on soybean with addition of Aureo® adjuvant in 1, 2, 4 and 8 applications up to 30 days after application.
Fig. 7.
Damage percentage caused by F. oxysporum extract on soybean with addition of Aureo® adjuvant in 1, 2, 4 and 8 applications up to 30 days after application.
4. Conclusion
Considering our studies, was possible to affirm that the tested fungi presented phytotoxicit, being that the T. koningiopsis presented interesting bioherbicidal potential. The phytotoxic activity of T. koningiopsis may be a relation with enzymatic activity, suggesting that these enzymes participate in the pathogens infection process.
Declarations
Author contribution statement
Francisco Wilson Reichert Júnior, Maurício Albertoni Scariot, César Tiago Forte, Leonardo Pandolfi, Jacqueline Mara Dil, Sabrina Weirich, Carine Carezia, Jéssica Mulinari, Marcio Antônio Mazutti, Gislaine Fongaro, Leandro Galon, Helen Treichel, Altemir José Mossi: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This work was supported by CNPq, CAPES-PNPD and FAPERGS.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
The authors thank Laboratory of Applied Molecular Biology of the Biological Institute - Brazil (Dr. Ricardo Harakava) by molecular sequencing, and the CNPq, CAPES-PNPD and FAPERGS by financial support of this work.
References
- Agrios G.N. fifth ed. San Diego Press; CA: 2004. Plant Pathology. [Google Scholar]
- Ash G.J. The science art business of succeful bioherbicides. Biol. Control. 2010;52:230–240. [Google Scholar]
- Babalola O.O. Exogenous cellulose contributes to mycoherbicidal acivity of Fusarium arthrosporioides on Orobanche aegyptiaca. Int. J. Agron. 2010;3:1–4. [Google Scholar]
- Barton J. How good are we at predicting the Field host-range of fungal pathogens used for classical biological control of weeds? Biol. Control. 2004;31:99–112. [Google Scholar]
- Boari A., Vurro M. Evaluation of Fusarium spp. and other fungi as biological control agents of broomrape (orobanche ramose) Biol. Control. 2003;30:212–219. [Google Scholar]
- Bouzoukov H., Kouzmanova I. Biological control of tobacco broomrape (Orobanche spp) by means of some fungi of the genus Fusarium. Biology and management of Orobanche. In: Pieterse A.H., Verkleig J.A.C., Borg S.J.T.E.R., editors. Proceedings of the Third International Workshop on Orobanche and Related Striga Research. Royal Tropical Institute; Amsterdam: 1994. [Google Scholar]
- Cavalcanti E.A.C., Gutarra M.L.E., Freire D.M.G., Castilho L.R., Sant'Anna Júnior G.L. Lipase production by solid-state fermentation in fixed-bed bioreactors. Braz. Arch. Biol. Technol. 2005;48:79–84. doi: 10.1385/abab:121:1-3:0105. [DOI] [PubMed] [Google Scholar]
- Charudattan R. Current status of biological of weeds. In: Kennedy C.G., Sutton T.B., editors. Emerging Technologies for Integrated Pest Management: Concepts, Research, and Implementation. APS; Kenya: 2000. [Google Scholar]
- Cordeau S., Triolet M., Wayman S., Steinberg C., Guillemin J. Bioherbicides: dead in the water? A review of the existing products for integrated weed management. Crop Protect. 2016;87:44–49. [Google Scholar]
- De Souza A.R.C., Baldoni D.B., Lima J., Porto V., Marcuz C., Machado C., Ferraz R.C., Kuhn R.C., Jacques R.J.S., Guedes J.V.C., Mazutti M.A. Selection, isolation, and identification of fungi for bioherbicide production. Braz. J. Microbiol. 2017;48:101–108. doi: 10.1016/j.bjm.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J.J., Doyle J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987;19:11–15. [Google Scholar]
- Fuwa H. A new method for microdetermination of amylase activity by the use of amylose as the substrate. J. Biochem. 1954;41:583–603. [Google Scholar]
- Ghorbani R., Leifert C., Seel W. Biological control of weeds with antagonistic plant pathogens. Adv. Agron. 2005;86:191–225. [Google Scholar]
- Ghose T.K. Measurement of cellulase activities. Pure Appl. Chem. 1987;59:257–268. [Google Scholar]
- Harding D.P., Raizada M.N. Controlling weeds with fungi, bacteria and viruses: a review. Front. Plant Sci. 2015;6:659. doi: 10.3389/fpls.2015.00659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodosy S. Biological control of broomrape, Orobanche ramosa, a tomato parasite. In: Hadj M., Hadi S., Fazeli F., Taghi M., Darzi, Shahmoradi R., editors. Occurrence and Adaptability of Fusarium Species to Control Broomrape in Hungary. Roodhen Branch; Iran: 1981. [Google Scholar]
- Khan A.A., Robinson D.S. Hydrogen donor specificity of mango isoperoxidases. Food Chem. 1994;49:407–410. [Google Scholar]
- Klaic R., Kuhn R.C., Foletto E.L., Dal Prá V., Jacques R.J.S., Guedes J. first ed. John Wiley & Sons; Ltda: 2015. An Overview Regarding Bioherbicide and Their Production Methods by fermentation. Fungal Biomolecules. [Google Scholar]
- Logrieco A., Moretti A. Ocurrence and toxigenicity of Fusarium proliferatum from preharvest maize ear rot, and associated mycotoxins, in Italy. Plant Dis. 1995;79:727–731. [Google Scholar]
- Nunes C.C., Alves S.A.M. Development and validation of a diagrammatic scale to quantify the severity of Fabraea leaf spot of pear. Summa Phytopathol. 2012;38:239–244. [Google Scholar]
- Pongsawasdi P., Yagisawa M. Screening and indentification of a cyclomaltodextrin glucanotransferase-producing bacteria. J. Ferment. Technol. 1987;65:463–467. [Google Scholar]
- Qian Y.S., Cai S., Huo Y.N., Mao P.P., Wang H.Z., Wuet J.B. First report of leaf blight disease of Curcuma wenyujin caused by Trichoderma koningiopsis in China. J. Plant Pathol. 2013;95:77. [Google Scholar]
- Samuels G.J., Dodd S.L., Lu B.S.P., Petrini O., Schroers H.J., Druzhinina I.S. The Trichoderma koningii aggregate species. Stud. Mycol. 2006;56:67–113. doi: 10.3114/sim.2006.56.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saremi H., Okhovvat S.M. Biological control of Orobanche aegyotiaca by Fusarium oxysporum f. sp. Orobanchein northwest Iran. Commun. Agric. Appl. Biol. Sci. 2008;73:931–938. [PubMed] [Google Scholar]
- Schmitz A., Riesner D. Purification of nucleic acids by selective precipitation with polyethylene glycol 6000. Anal. Biochem. 2006;354:311–313. doi: 10.1016/j.ab.2006.03.014. [DOI] [PubMed] [Google Scholar]
- Silva A. F. da, Galon L., Aspiazú I., Ferreira E.A., Concenco G., Ramos Junior E.U., Rocha P.R.R. Weed management in the soybean crop. In: EL-Shemy H.A., editor. Soybean: Pest Resistance. In Tech; Rijeka: 2013. [Google Scholar]
- Souza J.T., Trocoli R.O., Monteiro F.P. Plants from the caatinga biome harbor endophytic Trichoderma species active in the biocontrol of pineapple fusarioses. Biol. Control. 2016;94:25–32. [Google Scholar]
- Stergiopoulos I., Collemare J., Mehrabi R., De Wit P. Phytotoxic secondary metabolites and peptides produced by plant pathogenic Dothideomycete fungi. FEMS Microbiol. Rev. 2013;37:67–93. doi: 10.1111/j.1574-6976.2012.00349.x. [DOI] [PubMed] [Google Scholar]
- Treichel H., Sbardelotto M., Venturin B., Dall Agnol A., Mulinari J., Golunski S.M., Baldoni D.B., Bevilacqua C.B., Jacques R.J.S., Vargas G.D.L.P., Mossi A.J. Lipase production from a newly isolated Aspergillus niger by solid state fermentation using canola cake as substrate. Curr. Biotechnol. 2017;6:295–300. [Google Scholar]
- Trognitz F., Hackl E., Widhalm S., Sessitsch A. The role of plant–microbiome interactions in weed establishment and control. FEMS Microbiol. Ecol. 2016;92(10) doi: 10.1093/femsec/fiw138. pii: fiw138. [DOI] [PubMed] [Google Scholar]
- White T.J., Bruns T., Lee S., Taylor J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M.A., Gelfand D.H., Sninsky J.J., White T.J., editors. Protocols: A Guide to Methods and Applications. Academic Press, Inc.; New York: 1990. [Google Scholar]
- You J., Zhang J., Wu M., Yang L., Chen W., Li G. Multiple criteria-based screening of Trichoderma isolates for biological control of Botrytis cinereal on tomato. Biol. Control. 2016;101:31–38. [Google Scholar]