ABSTRACT
Sessile plants employ a diverse array of plasma membrane-bound receptors to perceive endogenous and exogenous signals for regulation of plant growth, development and immunity. These cell surface receptors include receptor-like kinases (RLKs) and receptor-like proteins (RLPs) that harbor different extracellular domains for perception of distinct ligands. Several RLK and RLP signaling pathways converge at the somatic embryogenesis receptor kinases (SERKs), which function as shared co-receptors. A repertoire of receptor-like cytoplasmic kinases (RLCKs) associate with the receptor complexes to relay intracellular signaling. Downstream of the receptor complexes, mitogen-activated protein kinase (MAPK) cascades are among the key signaling modules at which the signals converge, and these cascades regulate diverse cellular and physiological responses through phosphorylation of different downstream substrates. In this Review, we summarize the emerging common theme that underlies cell surface receptor-mediated signaling pathways in Arabidopsis thaliana: the dynamic association of RLKs and RLPs with specific co-receptors and RLCKs for signal transduction. We further discuss how signaling specificities are maintained through modules at which signals converge, with a focus on SERK-mediated receptor signaling.
KEY WORDS: Receptor-like kinase, Receptor-like protein, Receptor-like cytoplasmic kinase, Mitogen-activated protein kinase cascade, Signal transduction
Summary: Here, we review the emerging common theme of cell surface receptor-mediated signaling in Arabidopsis, and discuss how the convergent modules regulate divergent signaling outputs while maintaining specificity.
Introduction
Plants are multicellular organisms that are sessile but live in fluctuating environments. Correct communications between cells, and between a cell and its environment are therefore critical to plant growth, development and adaptation to the environment. Compared to animals, plants have evolved a largely expanded number of receptor-like kinases (RLKs) and receptor-like proteins (RLPs), with over 600 members in Arabidopsis thaliana and over 1000 in rice (Fritz-Laylin et al., 2005; Shiu and Bleecker, 2003; Shiu et al., 2004). RLKs and RLPs have been implicated in sensing extrinsic and intrinsic signals to regulate plant growth and development (De Smet et al., 2009), and their responses to pathogen attack (Couto and Zipfel, 2016; Tang et al., 2017). A typical RLK contains a unique extracellular domain, a single transmembrane domain and a cytoplasmic kinase domain, whereas a RLP lacks the kinase domain and possesses a short cytoplasmic domain. Various extracellular domains of certain RLKs and RLPs perceive the cognate ligands, often leading to hetero-dimerization (Fig. 1). The cytoplasmic kinase domain of RLKs usually transduces the signal to intracellular signaling networks through phosphorylation events, and eventually establishes a cellular response to a specific ligand (Couto and Zipfel, 2016; Hohmann et al., 2017; Tang et al., 2017) (Fig. 1). For this, RLPs rely on the cytoplasmic kinase domain of interacting RLKs.
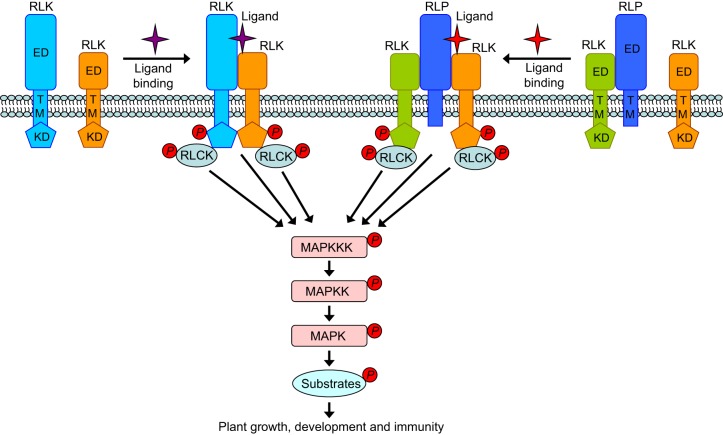
Fig. 1.
A model of ligand-induced receptor activation and signal transduction in plants. Plasma membrane-localized RLKs and RLPs perceive endogenous or exogenous ligands. A typical RLK contains an extracellular domain (ED), a single transmembrane domain (TM), and a cytoplasmic kinase domain (KD). Ligand binding to a RLK-type receptor often induces hetero-dimerization of the receptor and a regulatory RLK, which further leads to phosphorylation and activation of both RLKs. In contrast, an RLP lacks the kinase domain and often associates with a RLK for signal transduction. Similar to what occurs with a RLK-type receptor, activation of a RLP–RLK receptor complex also usually requires a ligand-dependent interaction with another regulatory RLK. Downstream of the receptor complexes, RLCKs associate with different RLKs to transduce intracellular signaling through protein phosphorylation. Among the intracellular pathways, MAPK cascades represent a key convergent module. A MAPK cascade contains three sequentially activated kinases: a MAPK kinase kinase (MAPKKK), a MAPK kinase (MAPKK) and a MAPK, which further phosphorylates diverse protein substrates to regulate plant growth, development and immunity.
The largest subfamily of RLKs and RLPs contains an extracellular leucine-rich repeat (LRR) domain, named LRR-RLKs or LRR-RLPs. In Arabidopsis, there are more than 200 LRR-RLK and LRR-RLP members (Shiu and Bleecker, 2001), many of which function as bona fide receptors for phytohormones, endogenous peptides or pathogen-derived molecules in order to regulate plant growth, development and defense responses (Couto and Zipfel, 2016; De Smet et al., 2009; Tang et al., 2017). A subgroup of LRR-RLKs, known as the somatic embryogenesis receptor kinases (SERKs), function as shared co-receptors and modules at which various signals converge (hereafter ‘convergent modules’) in multiple signaling pathways through hetero-dimerization with distinct LRR-RLK-type receptors (Ma et al., 2016). Receptor-like cytoplasmic kinases (RLCKs) associate with various RLK complexes to transduce intracellular signaling (Fig. 1), which provides RLKs with the capacity to activate divergent signaling pathways (Lin et al., 2013b). Downstream of the receptor complexes, mitogen-activated protein kinase (MAPK) cascades are emerging as convergent intracellular hubs that regulate diverse developmental or immune processes through phosphorylation of different downstream substrates (Fig. 1) (Meng and Zhang, 2013; Xu and Zhang, 2015). Here, we review the diverse functions and pathways of RLK- and RLP-mediated signaling in Arabidopsis and highlight the convergent signaling modules in these receptor-mediated pathways. Furthermore, we summarize the emerging common theme of receptor-mediated signaling, which is the dynamic association of receptors with different co-receptors and RLCKs in order to relay intracellular signaling. We also discuss how the convergent modules regulate divergent signaling outputs while maintaining specificity.
RLKs and RLPs – sensors and regulators for plant growth, development and immunity
Based on their varied extracellular motifs, plant RLKs and RLPs can be phylogenetically classified into multiple subfamilies, among which LRR-RLKs and LRR-RLPs represent the largest (Shiu and Bleecker, 2001). According to their biological roles, most plant RLKs and RLPs can be categorized into two functional groups: those that regulate growth and/or development, and those that mediate immunity.
RLKs and RLPs in plant growth and development
In Arabidopsis, a number of LRR-RLKs have been shown to recognize endogenous hormone or peptide signals to mediate the regulation of a wide range of plant growth or developmental processes. For instance, CLAVATA1 (CLV1) perceives the signaling peptide CLAVATA3 (CLV3) to control shoot apical meristem maintenance (Brand et al., 2000; Ogawa et al., 2008), BRASSINOSTEROID INSENSITIVE 1 (BRI1) acts as the receptor for brassinosteroids (BRs) to promote plant growth (Li and Chory, 1997), and ERECTA and ERECTA-LIKE 1 (ERL1) recognize the peptides EPIDERMAL PATTERNING FACTOR 1 (EPF1) and EPF2 to regulate stomatal development and patterning (Lee et al., 2012) (Fig. 2). Moreover, HAESA (HAE) and HAESA-LIKE 2 (HSL2) control cell separation during floral organ abscission and lateral root emergence in response to the peptide INFLORESCENCE DEFICIENT IN ABSCISSION (IDA) (Cho et al., 2008; Kumpf et al., 2013), and EXCESS MICROSPOROCYTES 1 (EMS1) perceives the peptide TAPETUM DETERMINANT 1 (TPD1) to regulate male gametophyte development (Jia et al., 2008) (Fig. 2). In addition, xylem differentiation is controlled by PHLOEM INTERCALATED WITH XYLEM [PXY; also known as TDIF RECEPTOR (TDR)], which functions as the receptor for the peptide TRACHEARY ELEMENT DIFFERENTIATION INHIBITORY FACTOR (TDIF) (Hirakawa et al., 2008) (Fig. 2). Furthermore, PHYTOSULFOKINE (PSK) RECEPTOR 1 (PSKR1) recognizes the sulfated pentapeptide PSK to stimulate plant growth (Wang et al., 2015), and ROOT GROWTH FACTOR (RGF) RECEPTOR 1 (RGFR1), through RGFR5, perceives RGF peptides to mediate root meristem development (Ou et al., 2016; Shinohara et al., 2016) (Fig. 2).
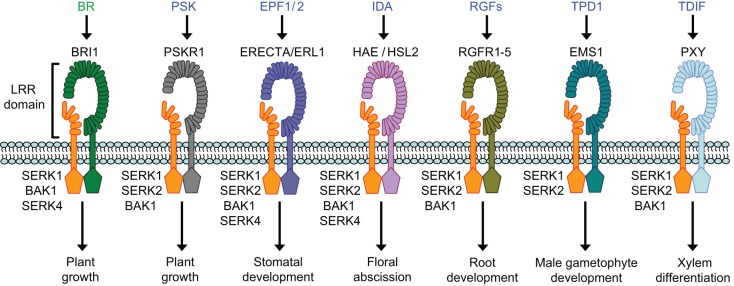
Fig. 2.
SERK-associated receptor complexes in plant growth and development. Plant hormones (green) or endogenous signaling peptides (blue) are perceived by plasma membrane-localized receptors. In Arabidopsis, the SERK-family RLKs form ligand-induced complexes with the LRR-RLK-type receptors BRI1, PSKR1, ERECTA and ERL1, HAE and HSL2, RGFR-family RLKs, EMS1, and PXY to regulate a variety of plant growth and developmental processes. Individual SERKs have differential or specific contributions to a particular pathway. For example, SERK1 and SERK2, but not BAK1 or SERK4, are involved in male gametophyte development, whereas SERK1, BAK1, and SERK4, but not SERK2, contribute to BR signaling.
As they lack an intracellular kinase domain, some LRR-RLPs associate with LRR-RLKs to regulate plant development in Arabidopsis. For example, the LRR-RLP TOO MANY MOUTHS (TMM) forms complexes with the LRR-RLKs ERECTA and ERL1 to perceive the EPF1 and EPF2 peptides for regulation of stomatal patterning (Lin et al., 2017).
Besides LRR-RLKs, some RLKs from very distinct subfamilies have also been found to regulate plant development in response to endogenous peptides. For instance, Arabidopsis FERONIA (FER), which is from the Catharanthus roseus RLK1-LIKE (CrRLK1L) subfamily and contains an extracellular malectin-like domain, has been shown to perceive the peptide RAPID ALKALINIZATION FACTOR 1 (RALF1) in order to regulate root growth (Haruta et al., 2014), besides its functions in female fertility and hormone signaling (Li et al., 2016). In addition, there are still many other plant-secreted peptides that could be potential ligands for RLKs in regulating plant growth and/or development. For instance, the endosperm-derived EMBRYO SURROUNDING FACTOR 1 (ESF1) peptides regulate early embryo patterning, and their cognate embryo-expressed RLK(s) have been proposed to sense the ESF1 peptides for regulation of embryo development (Costa et al., 2014). However, the RLK(s) that are responsible for this perception have yet to be identified.
RLKs and RLPs in plant immunity
Members of plant RLKs and RLPs are known to recognize pathogen-associated molecular patterns (PAMPs), as well as damage-associated molecular patterns (DAMPs) that are produced by plants during pathogen attacks (Fig. 3). RLKs and RLPs that perceive PAMPs or DAMPs are collectively referred to as pattern recognition receptors (PRRs). The stimulation of PRRs by PAMPs or DAMPs induces convergent intracellular signaling events, including MAPK activation and reactive oxygen species (ROS) bursts, which eventually result in the so-called pattern-triggered immunity (PTI). Among the well-characterized PRRs in Arabidopsis, FLAGELLIN-SENSING 2 (FLS2) perceives a conserved 22-amino-acid peptide (flg22) from bacterial flagellin (Gómez-Gómez and Boller, 2000), ELONGATION FACTOR-TU (EF-Tu) RECEPTOR (EFR) senses a conserved 18-amino-acid epitope (elf18) from the bacterial EF-Tu (Zipfel et al., 2006), and LYSIN MOTIF RECEPTOR KINASE 5 (LYK5) and CHITIN ELICITOR RECEPTOR KINASE 1 (CERK1) recognize the fungal chitin oligomers (Cao et al., 2014) (Fig. 3). Moreover, PEP1 RECEPTOR 1 (PEPR1) and PEPR2 perceive the endogenous plant elicitor peptides (Peps) (Yamaguchi et al., 2006, 2010) (Fig. 3). Together with most RLKs that are involved in plant growth and development, FLS2, EFR, PEPR1 and PEPR2 all belong to the subfamily of LRR-RLKs. Their extracellular LRR domain determines the binding specificity for their corresponding peptide ligands (Sun et al., 2013b; Tang et al., 2015, 2017), which suggests that LRR-RLKs preferentially bind proteins or peptides. In contrast, LYK5 and CERK1 both belong to another type of RLK with extracellular lysine motifs (LysM-RLKs), which are involved in perceiving microbial glycans, such as fungal chitin and bacterial peptidoglycans (Cao et al., 2014; Couto and Zipfel, 2016; Tang et al., 2017) (Fig. 3). Aside from LRR- and LysM-RLKs, there are still other RLKs with distinct extracellular domains, which also function as PRRs to perceive PAMPs in Arabidopsis. For instance, the lectin S-domain RLK LIPOOLIGOSACCHARIDE-SPECIFIC REDUCED ELICITATION (LORE) recognizes the bacterial lipopolysaccharide (LPS) (Ranf et al., 2015).
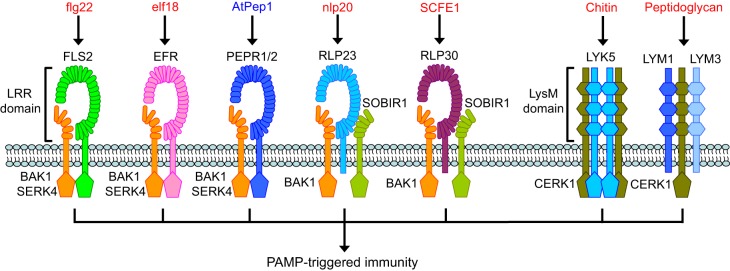
Fig. 3.
PAMP- or DAMP-induced receptor complexes in plant immunity. PAMPs (red) or DAMPs (blue) are perceived by RLK- or RLP-type receptors in Arabidopsis. The SERK-family LRR-RLKs and the LysM-RLK CERK1 form ligand-induced complexes with LRR- and LysM-containing receptors, respectively, to mediate PTI. BAK1 and SERK4 are recruited upon ligand perception by the LRR-RLK-type receptors FLS2, EFR, PEPR1 and PEPR2. The LRR-RLP-type receptor RLP23, which constitutively forms a complex with the LRR-RLK SOBIR1, also recruits BAK1 upon ligand perception. Both BAK1 and SOBIR1 are required for RLP30-mediated immunity, which implies that RLP30 may also be in a complex with both BAK1 and SOBIR1. CERK1 forms ligand-induced complexes with the LysM-RLK-type receptor LYK5, and the LysM-RLP-type receptors LYM1 and LYM3, respectively.
In addition, several Arabidopsis RLPs have been shown to function together with RLKs to regulate plant immunity. The LysM-RLPs LYSIN-MOTIF 1 (LYM1) and LYM3 act in concert with the LysM-RLK CERK1 to recognize bacterial peptidoglycans to trigger PTI signaling (Willmann et al., 2011) (Fig. 3). Similar to LRR-RLPs from other plant species (Gust and Felix, 2014), the Arabidopsis LRR-RLP RLP23 associates with the LRR-RLK SUPPRESSOR OF BIR1-1 (SOBIR1) to mediate PTI signaling that is triggered by NECROSIS- AND ETHYLENE-INDUCING PEPTIDE 1 (NEP1)-LIKE PROTEINS (NLPs) (Albert et al., 2015), and SOBIR1 is also required for the RLP30-mediated immunity in response to a fungal elicitor called SCLEROTINIA CULTURE FILTRATE ELICITOR1 (SCFE1) (Zhang et al., 2013) (Fig. 3). These data suggest that RLPs depend on regulatory RLKs to transduce ligand perception into intracellular signaling.
Interestingly, several RLKs, such as BRI1 (Belkhadir et al., 2012), ERECTA (Jordá et al., 2016), PSKR1 (Igarashi et al., 2012) and FER (Kessler et al., 2010), are not only involved in regulating plant growth and development, but also in modulating plant immunity. Specifically, it has been recently proposed that FER functions as a scaffold that modulates PRR complex assembly (Stegmann et al., 2017). Such a dual function for RLKs may help to mediate the interplay of plant growth, development and immunity.
Overall, plants deploy a large family of plasma membrane-localized RLKs and RLPs to perceive various endogenous and exogenous signals for regulation of a diverse range of plant growth, developmental or immune processes. Strikingly, despite inducing distinct signaling outputs, these cell surface receptors often employ common downstream signaling components, including the SERK-family RLKs as co-receptors, and RLCKs and MAPK cascades for intracellular signal transduction.
SERKs – co-receptors for multiple receptors
RLK- and RLP-mediated signaling is usually initiated by phosphorylation events that occur between proteins in the receptor complexes. Thus, the rapid formation or stabilization of the receptor complex upon ligand binding represents a common theme of cell surface receptor-mediated signaling (Fig. 1). The SERK-family RLKs associate with multiple LRR-containing receptors and play crucial roles in regulating a variety of plant growth, developmental and immune processes (Figs 2 and 3).
SERKs in plant growth and development
SERKs belong to the subgroup II of LRR-RLKs and contain a relatively short extracellular domain with five LRRs. Among the five SERK members in Arabidopsis, SERK3 was initially identified as a BRI1-associated receptor kinase (BAK1) that is involved in BR perception (Li et al., 2002; Nam and Li, 2002). Subsequently, SERK1 and SERK4 (also known as BAK1-LIKE 1, BKK1) were found to function redundantly with BAK1 in associating with BRI1 to mediate BR signaling and regulate plant growth (Gou et al., 2012). Recent studies have revealed strikingly diverse functions for SERKs in regulating plant growth and development (Fig. 2). Arabidopsis SERKs form ligand-induced complexes with the ERECTA-family RLKs (Meng et al., 2015), the RGFR-family RLKs (Song et al., 2016), and HAE and HSL2 (Meng et al., 2016) to regulate stomatal patterning, root meristem development, and floral organ abscission, respectively (Fig. 2). Furthermore, SERKs also regulate plant growth, xylem differentiation and male gametophyte development through ligand-induced association with the receptors PSKR1 (Wang et al., 2015), PXY (Zhang et al., 2016), and EMS1 (Li et al., 2017), respectively (Fig. 2).
SERKs in plant immunity
SERKs also form complexes with various members of LRR-containing immune receptors (Fig. 3). The Arabidopsis LRR-RLK-type immune receptors FLS2, EFR, PEPR1 and PEPR2 all associate with BAK1 and related SERKs in a ligand-dependent manner to mediate PTI signaling (Chinchilla et al., 2007; Heese et al., 2007; Roux et al., 2011; Schulze et al., 2010). The LRR-RLP-type immune receptor RLP23, which constitutively interacts with SOBIR1, also requires a ligand-dependent association with BAK1 for Arabidopsis to perceive nlp20, a conserved 20-amino-acid fragment found in most NECROSIS- AND ETHYLENE-INDUCING PEPTIDE 1-LIKE PROTEINS (NLPs) from multiple pathogen species (Albert et al., 2015). Furthermore, Arabidopsis BAK1 and SOBIR1 are both required for RLP30-mediated perception of the fungal elicitor SCFE1, which implies that RLP30 may also be in a complex with both BAK1 and SOBIR1 for the perception of SCFE1 (Zhang et al., 2013). Therefore, it has been hypothesized that LRR-RLPs constitutively form a complex with SOBIR1, and that this complex resembles a functional LRR-RLK module, which then further recruits the SERK-family RLKs upon ligand perception (Gust and Felix, 2014). Together, the SERK-family RLKs form complexes with multiple LRR-RLK- or LRR-RLP-type receptors in a ligand-dependent manner, and function as convergent modules in diverse signaling pathways (Figs 2 and 3).
SERKs as co-receptors
Structural studies have provided insights into the mechanisms of ligand-induced hetero-dimerization of SERKs with the cognate receptors. The crystal structures of the extracellular domains of the BR–BRI1–BAK1 and flg22–FLS2–BAK1 complexes revealed that both BR and flg22 ligands not only bind to their cognate receptors, but also clench onto the N-terminus of the LRR domain of BAK1. This suggests that BR and flg22 act as molecular glues to induce the hetero-dimerization of BRI1 with BAK1, and FLS2 with BAK1, respectively (Santiago et al., 2013; Sun et al., 2013a,b). A similar mechanism also extends to the ligand-induced hetero-dimerization of HAE with SERK1 (Santiago et al., 2016), PXY with SERK2 (Zhang et al., 2016), and RGFR1 with BAK1 (Song et al., 2016). However, SERKs are not always engaged in ligand binding during their dimerization with receptors upon ligand perception. The crystal structure of the extracellular domains of the PSK–PSKR1–SERK1 complex revealed that SERK1 does not participate in PSK binding. Instead, PSK induces allosteric modifications on the surface of PSKR1 that enable subsequent recruitment of SERK1 (Wang et al., 2015).
Uncoupled functions of SERKs in diverse pathways
SERKs function in diverse signaling pathways, and these functions appear to be uncoupled (Figs 2 and 3). For example, the function of BAK1 in plant immunity can be separated from its involvement in BR signaling, as indicated by the phenotype of the bak1-5 mutant, which displays severely compromised immunity, but normal BR signaling (Schwessinger et al., 2011). In addition, the involvement of SERKs in stomatal patterning appears to be independent of BR signaling, since high-order mutations of certain SERK members confers defects only in one signaling pathway, but not the other (Meng et al., 2015).
Moreover, it appears that individual SERKs function differentially during distinct physiological responses (Figs 2 and 3). For example, BAK1 and SERK4, but not SERK1 or SERK2, are required for plant immunity (Roux et al., 2011). In contrast, SERK1 and SERK2, but not BAK1 or SERK4, are essential for male gametophyte development (Li et al., 2017), whereas BAK1, SERK1 and SERK4, but not SERK2, are involved in BR signaling (Gou et al., 2012). In addition, the functionally redundant SERKs often contribute unequally to the same physiological response, which has been commonly observed in plant immune signaling, BR signaling and stomatal patterning pathways (Gou et al., 2012; Meng et al., 2015; Roux et al., 2011). How the functional specificity of individual SERKs is governed has largely not been investigated. The differences in protein structure and abundance, as well as the spatial and temporal expression patterns of individual SERKs, potentially contribute to their functional specificity in different signaling pathways.
Since multiple receptors employ the same group of SERKs as co-receptors to induce distinct signaling outputs, another important question is therefore how SERKs act in concert with these different receptors to initiate distinct and even antagonistic signaling pathways. Differential phosphorylation events between SERKs and various receptors may contribute to this specificity. For example, the bak1-5 mutation, which results in reduced BAK1 kinase activity owing to a point mutation in the kinase domain, specifically blocks immune responses without affecting BR signaling. This suggests a phosphorylation-dependent differential regulation of signaling outputs in distinct SERK-mediated pathways (Schwessinger et al., 2011). The recruitment of specific downstream RLCKs to different SERK-associated receptor complexes may also contribute to the initiation of specific signaling outputs (see below). Recently, single-particle tracking has revealed that FLS2 and BRI form spatially separated nanoclusters within the plasma membrane (Bücherl et al., 2017). This suggests that the signaling specificity between FLS2- and BRI1-mediated pathways may be explained by the spatial separation of the immune and growth receptor complexes, which contain the associated signaling components, including SERKs and RLCKs (Bücherl et al., 2017). It remains unknown how these functional nanoclusters of signaling components are assembled and located within separated nanodomains. Thus, a combination of spatial separation, post-translational modification and tailored gene expression programs might ensure the functional specificity of SERKs in regulating various plant growth, developmental and immune processes.
RLCKs – signal transmitters in the receptor complexes
Upon formation of the ligand-induced receptor complexes, auto- and/or trans-phosphorylation events usually occur within the complexes. This is followed by the activation of receptor complexes, which then transduce signals to intracellular networks (Couto and Zipfel, 2016; Ma et al., 2016). Accumulating evidence suggests that a large repertoire of RLCKs associate with diverse receptor complexes to transduce intracellular signaling (Lin et al., 2013b) (Figs 1, 4 and 5).
Fig. 4.
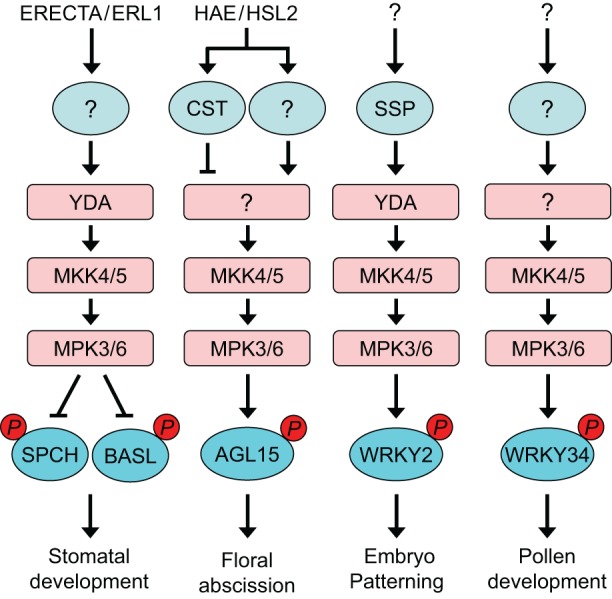
Diverse roles of RLCKs and the MKK4/MKK5–MPK3/MPK6 cascade in plant development. RLCKs (light blue) associate with various cell surface receptors to transduce intracellular signaling. In Arabidopsis, the RLCK CST interacts with the receptor HAE to negatively regulate floral abscission, and the RLCK SSP has been proposed to function downstream of the unidentified ESF1 receptor to regulate embryo development. The RLCK(s) downstream of the receptors ERECTA and ERL1 remain unknown. Downstream of multiple RLK and RLCK complexes, the MKK4/MKK5–MPK3/MPK6 cascade regulates diverse plant developmental processes through phosphorylation of the substrate proteins BASL, SPCH, AGL15, WRKY2 and WRKY34. YDA functions upstream of MKK4/MKK5 to regulate stomatal development and embryo patterning; the MAPKKK(s) that regulate floral abscission and pollen development upstream of MKK4/MKK5 remain unknown.
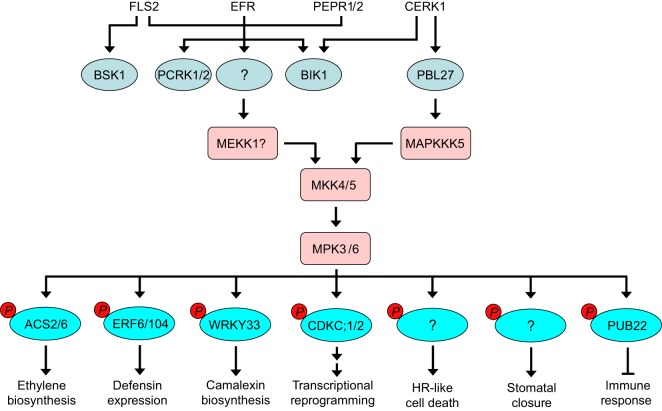
Fig. 5.
Diverse roles of RLCKs and the MKK4/MKK5–MPK3/MPK6 cascade in plant immunity. The RLCKs BSK1, PCRK1, PCRK2, BIK1 and PBL27 associate with the immune receptors FLS2, EFR, PEPR1, PEPR2 or CERK1 to regulate PTI responses, including MAPK activation. Downstream of multiple immune receptors, the MKK4/MKK5–MPK3/MPK6 cascade regulates various immune responses through phosphorylation of ACS2, ACS6, ERF6, ERF104, WRKY33, CDKC;1, CDKC;2, PUB22, and some unknown substrate proteins. MAPKKK5 functions downstream of CERK1 in mediating MPK3/MPK6 activation, and the CERK1-associated RLCK PBL27 interacts with MAPKKK5 to transduce the immune signal. However, downstream of the FLS2, EFR, PEPR1 and PEPR2 receptors, both of the RLCK and the MAPKKK that mediate MPK3/MPK6 activation still remain elusive.
RLCKs are members of the RLK family, which lack extracellular and transmembrane domains, but contain kinase domains that are closely related to those of the RLKs (Shiu and Bleecker, 2001). Similar to what has occurred for the RLKs, plants have also evolved a significant number of RLCKs, with 147 members in Arabidopsis, which can be further grouped into 11 subfamilies (Shiu and Bleecker, 2001). Of all the RLCK subfamilies in Arabidopsis, the VII subfamily is the largest, with 46 members.
RLCKs in plant growth and development
In Arabidopsis, a number of RLCKs have been shown to act in concert with RLKs to regulate plant growth and development (Fig. 4). BR-SIGNALING KINASE1 (BSK1) and CONSTITUTIVE DIFFERENTIAL GROWTH 1 (CDG1), two RLCKs from the XII and VII subfamily, respectively, function in parallel to positively regulate BR signaling and plant growth (Kim et al., 2011; Tang et al., 2008). The BR-activated BRI1 interacts with and phosphorylates BSK1 and CDG1, which then dissociate from BRI1 to relay the signal to the downstream components (Kim et al., 2011; Tang et al., 2008). Intriguingly, BOTRYTIS-INDUCED KINASE 1 (BIK1), another RLCK VII subfamily member, which also interacts with and is phosphorylated by BRI1, negatively regulates BR signaling (Lin et al., 2013a). The RLCK CAST AWAY (CST), also from the VII subfamily, interacts with HAE and negatively regulates floral organ abscission (Burr et al., 2011). SHORT SUSPENSOR (SSP), a RLCK II subfamily member, has been proposed to function downstream of the unidentified ESF1 receptor in regulating early embryo development (Bayer et al., 2009; Costa et al., 2014). In addition, RPM1-INDUCED PROTEIN KINASE (RIPK) and MARIS (MRI), two RLCKs from the VII and VIII subfamily, respectively, have been found to act in concert with FER in regulating root hair growth (Boisson-Dernier et al., 2015; Du et al., 2016). Most RLCKs that are involved in plant growth and development belong to the VII subfamily, which suggests that this RLCK subfamily has a general role in RLK signaling transduction.
RLCKs in plant immunity
Several RLCK VII subfamily members also associate with immune receptors to transduce defense signals in Arabidopsis (Fig. 5). BIK1 and its closely related subfamily member PBS1-LIKE 1 (PBL1) interact directly with the PRRs FLS2, EFR, CERK1 and PEPR1, and the co-receptor BAK1, and are required to activate PTI responses (Liu et al., 2013; Lu et al., 2010; Zhang et al., 2010). Activation of these PRRs results in a rapid phosphorylation of BIK1 by BAK1, which leads to subsequent dissociation of BIK1 from the immune receptors to activate downstream signaling. PATTERN-TRIGGERED IMMUNITY COMPROMISED RLCK 1 (PCRK1) and PCRK2, two RLCKs from the VII subfamily, also interact with FLS2 to mediate PTI responses (Kong et al., 2016; Sreekanta et al., 2015). Interestingly, BSK1 also associates with FLS2 to regulate specific subsets of flg22-induced immune responses, in addition to its role in BR signaling through the interaction with BRI1 (Shi et al., 2013). Furthermore, PBS1-LIKE 27 (PBL27), another RLCK from the VII subfamily, mediates chitin-induced immune responses through interaction with CERK1 (Shinya et al., 2014; Yamada et al., 2016). Additionally, RLCKs, particularly members of subfamily VII, are overrepresented among genes that are induced in response to biotic stress (Lehti-Shiu et al., 2009). Moreover, phosphoproteomics studies have revealed that many RLCKs are phosphorylated within minutes of PAMP perception (Benschop et al., 2007; Nühse et al., 2007). These observations further suggest that RLCKs play important roles in mediating immune signal transduction.
Signaling specificity through RLCKs
Recruitment of distinct RLCKs into specific receptor complexes may contribute to signaling specificity by activating different downstream components. RLCKs exhibit certain degrees of specificity in their interaction with upstream ligand receptors (Figs 4 and 5). For example, RIPK interacts with FER, but not with its closely related family members ANXUR1 (ANX1) or ANX2. In addition, FER does not interact with four RLCK members that are closely related to RIPK (Du et al., 2016). PBL27 mediates chitin- but not flg22-triggered immune responses (Shinya et al., 2014; Yamada et al., 2016), whereas BSK1 regulates subsets of flg22-, but not elf18-induced PTI responses (Shi et al., 2013). This indicates a specific involvement of PBL27 and BSK1 with defined immune receptors. RLCKs also show specificity in the activation of downstream signaling events. For instance, BIK1 phosphorylates the plasma membrane RESPIRATORY BURST OXIDASE HOMOLOG D (RBOHD) (Kadota et al., 2014; Li et al., 2014b), and is required for PAMP-induced ROS burst, but not MAPK activation (Feng et al., 2012). PBL27, by contrast, specifically interacts with and phosphorylates the MAPK kinase kinase 5 (MAPKKK5) to mediate chitin-induced MAPK activation (Yamada et al., 2016). However, there is also evidence that one RLCK can associate with several RLKs to mediate the same or different downstream signaling outputs. For example, BIK1 interacts with multiple PRRs and BAK1 to regulate the ROS burst (Liu et al., 2013; Zhang et al., 2010), whereas BSK1 associates with BRI1 and FLS2 to regulate BR signaling and immune responses, respectively (Shi et al., 2013; Tang et al., 2008). Similarly, one RLK can also interact with multiple RLCKs to regulate the same or different downstream responses. For instance, BRI associates with both BSK1 and CDG1 to mediate BR signaling (Kim et al., 2011; Tang et al., 2008), and CERK1 interacts with BIK1 and PBL27 to regulate the ROS burst and MAPK activation, respectively (Yamada et al., 2016; Zhang et al., 2010). Taken together, RLCKs contribute to receptor signaling specificity through distinct interactions with different upstream receptors, as well as through activation of diverse downstream signaling pathways (Figs 4 and 5).
MAPK cascades – an intracellular hub downstream of receptor complexes
Downstream of the receptor complexes, MAPK cascades represent another convergent module. Typically, a MAPK cascade contains three sequentially activated kinases, the first being a MAPK kinase kinase (MAPKKK or MEKK), the second a MAPK kinase (MAPKK or MKK), and finally a MAPK (MPK), which further phosphorylates diverse protein substrates to transduce signals (Figs 1, 4 and 5).
MAPK cascades in plant growth and development
In Arabidopsis, a MAPK cascade that is composed of YDA (MAPKKK), MKK4 and MKK5 (hereafter, MKK4/MKK5), and MPK3 and MPK6 (MPK3/MPK6) functions downstream of the ERECTA-family receptors to regulate both stomatal development and plant organ growth in response to the EPF1 and EPF2 peptides, or EPF-LIKE 4 (EPFL4) and EPFL6 peptides, respectively (Bemis et al., 2013; Lampard et al., 2009; Lee et al., 2012; Meng et al., 2012; Uchida et al., 2012; Wang et al., 2007). Moreover, MKK4/MKK5 and MPK3/MPK6 also function downstream of the IDA peptide receptors HAE and HSL2 in controlling floral organ abscission (Cho et al., 2008), although the MAPKKK upstream of MKK4/MKK5 in this case remains unknown. In addition, the YDA–MKK4/MKK5–MPK3/MPK6 cascade was recently found to act downstream of the ESF1 peptides and the RLCK SSP in regulating early embryo patterning (Costa et al., 2014; Ueda et al., 2017). Here, an unidentified receptor that is associated with SSP was proposed to perceive the ESF1 peptides and transduce signals to the MAPK cascade. Thus, the Arabidopsis MKK4/MKK5–MPK3/MPK6 cascade functions as a key module that is downstream of multiple cell surface receptors and regulates a diverse array of plant growth and developmental processes (Fig. 4).
MAPK cascades in plant immunity
In Arabidopsis, the perception of various PAMPs or DAMPs, such as bacterial flg22 and elf18, fungal chitin, and Arabidopsis Pep1, rapidly and transiently activates two major MAPK cascades: MKK4/MKK5–MPK3/MPK6 and MEKK1–MKK1/MKK2–MPK4, indicating that these MAPK cascades are the points at which separate PTI signaling pathways converge (Fig. 5) (Meng and Zhang, 2013). Several lines of evidence support that MEKK1 also acts upstream of the MKK4/MKK5–MPK3/MPK6 cascade and positively regulates immune gene expression and defense responses (Asai et al., 2002; Cheng et al., 2015; Ren et al., 2008). Surprisingly, mekk1 mutant plants show unaltered MPK3/MPK6 activation in response to flg22 or the bacterial pathogen-secreted protease PrpL (Cheng et al., 2015; Suarez-Rodriguez et al., 2007). This seeming inconsistency is likely caused by the redundant function of multiple MAPKKKs in the PAMP-triggered activation of MPK3/MPK6. Indeed, a recent study has found that MAPKKK5 functions upstream of MKK4/MKK5 during chitin-induced activation of MPK3/MPK6, and the CERK1-associated RLCK PBL27 interacts with and phosphorylates MAPKKK5 to transduce immune signals (Yamada et al., 2016) (Fig. 5). These findings provide a molecular link that connects an activated PRR to the intracellular MAPK cascade in plants. However, MAPKKK5 is not required for flg22-induced activation of MPK3/MPK6. Taken together, it is likely that multiple MAPKKKs function redundantly or specifically in the regulation of MPK3/MPK6 activation in response to different PAMPs (Fig. 5).
MPK3 and MPK6 are redundant positive regulators of plant immunity. Through phosphorylation of diverse downstream substrates (see below), MPK3 and MPK6 control a wide array of defense responses, including immune gene expression, ethylene induction, camalexin biosynthesis, ROS production, stomatal closure and hypersensitive cell death (the localized death of plant cells at the site of pathogen infection) (Meng and Zhang, 2013). MPK4 was originally considered to be a negative regulator of plant immunity, because mpk4 mutant plants exhibit autoimmune phenotypes that are characterized by spontaneous cell death, constitutive defense responses and a dwarf morphology (Gao et al., 2008; Petersen et al., 2000). Later, it was discovered that the autoimmune responses in mpk4 mutants are mediated by the disease resistance protein SUPPRESSOR OF MKK1 MKK2 2 (SUMM2) (Zhang et al., 2012). Therefore, the basal activity of MPK4 is required for suppression of SUMM2-mediated immunity (Zhang et al., 2012). In contrast, PAMP- or pathogen-activated MPK4 positively regulates immune gene expression and plant basal defense (Frei dit Frey et al., 2014; Qiu et al., 2008; Zhang et al., 2012). Thus, the basal MPK4 activity and the PAMP-triggered activation of MPK4 play distinct roles in different layers of plant immune responses. However, the molecular mechanisms underlying MPK4-mediated regulation of PTI signaling remain to be further elucidated.
Signaling specificity of MAPK cascades
How the signaling specificities of the MKK4/MKK5–MPK3/MPK6 cascade are maintained in distinct pathways is an intriguing question. The signaling specificity may be conferred by pathway-specific MAPKKKs upstream of MKK4/MKK5 (Figs 4 and 5), as exemplified by MAPKKK5 and YDA, which act in different RLK pathways to regulate distinct signaling outputs (Bergmann et al., 2004; Lukowitz et al., 2004; Ueda et al., 2017; Wang et al., 2007). Tissue- or cell-specific expression of upstream RLKs and/or peptide ligands can also provide a mechanism for the signaling specificity of the MKK4/MKK5–MPK3/MPK6 cascade in regulating distinct pathways. For instance, the abscission zone-specific expression of HAE and HSL2 and their ligand IDA specifies the function of this MAPK cascade in flower organ abscission (Butenko et al., 2003; Cho et al., 2008). Furthermore, the specific expression of EPF1 and EPF2 peptides in stomatal lineage cells, and the preferential expression of EPFL4 and EPFL6 peptides in inflorescence stems define the roles of this MAPK module in stomatal development and inflorescence growth, respectively (Hara et al., 2007; Hunt and Gray, 2009; Uchida et al., 2012).
MAPKs regulate plant development and immunity through the phosphorylation of diverse downstream protein substrates, which include transcription factors and enzymes (Figs 4 and 5). The multifunctionality of MAPKs is thereby largely conferred by their ability to phosphorylate different substrates.
MAPK substrates in plant growth and development
Several substrates of Arabidopsis MPK3 and MPK6 have been shown to play important roles in regulating plant development (Fig. 4). One of the best-characterized MPK3/MPK6 substrates is SPEECHLESS (SPCH), a basic helix-loop-helix transcription factor, which promotes asymmetric cell division for stomatal initiation (Lampard et al., 2008). Phosphorylation of SPCH by MPK3/MPK6 likely destabilizes the SPCH protein, thereby inhibiting stomatal initiation (Lampard et al., 2008). During stomatal development, MPK6 is also involved in the differentiation of asymmetric cell fate through phosphorylation of the polarity protein BREAKING OF ASYMMETRY IN THE STOMATAL LINEAGE (BASL) (Zhang et al., 2015). Recently, the transcription factor WRKY2 was identified as the MPK3/MPK6 substrate that regulates early embryo patterning (Ueda et al., 2017), whereas MPK3/MPK6-mediated phosphorylation of WRKY34, a close homolog of WRKY2, was found to be required for pollen development (Guan et al., 2014). In addition, phosphorylation of the transcription factor AGL15 by MPK3/MPK6 controls floral abscission through regulation of HAE gene expression (Patharkar and Walker, 2015).
MAPK substrates in plant immunity
Arabidopsis MPK3 and MPK6 also regulate a wide array of immune responses by phosphorylating diverse downstream substrates (Fig. 5). MPK3/MPK6-mediated phosphorylation and stabilization of the 1-aminocyclopropane-1-carboxylic acid synthase (ACS) isoforms ACS2 and ACS6 promotes pathogen-induced ethylene production (Han et al., 2010; Liu and Zhang, 2004). MPK3/MPK6 also positively regulate camalexin biosynthesis through phosphorylation of the WRKY33 transcription factor, which activates the expression of camalexin biosynthetic genes upon pathogen infection (Mao et al., 2011). In addition, phosphorylation and stabilization of the ERF6 and ERF104 transcription factors by MPK3/MPK6 drives defensin gene expression, which is required for fungal resistance (Bethke et al., 2009; Meng et al., 2013). MPK3/MPK6 also phosphorylates and activates the cyclin-dependent kinases CDKC;1 and CDKC;2, which further regulate transcription dynamics in plant immunity through phosphorylation of the C-terminal domain of RNA polymerase II (Li et al., 2014a). Interestingly, MPK3 is also involved in the negative regulation of plant immunity through phosphorylation and stabilization of the E3 ubiquitin ligase PUB22, which dampens the immune response (Furlan et al., 2017). These results highlight the variety of plant immune processes that are controlled by MPK3/MPK6-mediated phosphorylation events.
Specificity of substrate phosphorylation
The signaling specificity of MAPKs is largely mediated by their ability to spatially or temporally phosphorylate specific substrates (Figs 4 and 5). For example, MPK3/MPK6 regulate stomatal development through phosphorylation of SPCH, which is expressed specifically in stomatal lineage cells (MacAlister et al., 2007), whereas WKRY2, which is expressed in the basal embryo, functions as a substrate of MPK3/MPK6 to regulate embryo patterning (Ueda et al., 2017). Pollen-specific expression of WRKY34 allows it to function downstream of MPK3/MPK6 in the regulation of pollen development (Guan et al., 2014). In contrast, PAMP- or pathogen-induced expression of ACS2, ACS6, WRKY33, ERF6 and PUB22 allows these substrates of MPK3/MPK6 to regulate a diverse array of immune responses upon pathogen infection (Li et al., 2012; Mao et al., 2011; Meng et al., 2013; Trujillo et al., 2008).
In summary, MAPK cascades are a key converging module that act downstream of ligand-activated cell surface receptors and their co-receptors. The specificity of the MAPK cascade is largely conferred by the spatially and temporally fine-tuned targeting of phosphorylation substrates, which in turn control a variety of plant developmental and immune processes, such as those described above.
Concluding remarks and future perspectives
Plants have evolved diverse plasma membrane-bound receptors to regulate plant growth, development and immunity in response to various extrinsic and intrinsic stimuli (De Smet et al., 2009; Tang et al., 2017). Although hundreds of RLKs and RLPs have been identified in many plant species, most of them have not been functionally characterized in detail. Despite the diversity in each module, a common theme of cell surface receptor-mediated signal transduction is emerging: RLKs and RLPs dynamically associate with different co-receptors and RLCKs to initiate distinct intracellular signaling. The SERK-family RLKs are hereby emerging as shared co-receptors for LRR-RLKs and LRR-RLPs (Ma et al., 2016). How SERKs coordinate their activity and association with different receptors at the plasma membrane to initiate specific signaling outputs is not yet resolved. The understanding of how specific receptor signaling is generated will likely require multi-disciplinary approaches to decipher the composition, organization and phosphorylation dynamics of receptor complexes at the plasma membrane.
MAPK cascades are another convergent module downstream of receptor complexes that regulate diverse plant developmental and immune pathways (Meng and Zhang, 2013; Xu and Zhang, 2015). Another major gap in our understanding of plant receptor signaling is what links the receptor complexes and the downstream MAPK cascades. RLCKs likely function as a direct link between activated receptors and intracellular MAPK cascades, as indicated by the recently identified chitin signaling pathway (Yamada et al., 2016). However, the RLCKs upstream of MAPK cascades in many other receptor signaling pathways still need to be identified. In order to understand the specific function of a MAPK cascade, we also need to identify specific downstream substrates. High-throughput genomic and proteomic approaches have enabled the genome-wide identification of potential MAPK substrates in Arabidopsis (Hoehenwarter et al., 2013; Popescu et al., 2009). Further plant-based analyses will reveal details of their involvement in plant receptor signaling.
Acknowledgements
We would like to thank the budding scientist Pierce A. Jamieson of Texas A&M University for discussion and proofreading of this manuscript prior to publication. We apologize for not being able to cite all publications in the field due to page limitations.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
The work of our laboratories is supported by the National Natural Science Foundation of China (grant 31671515) and the Chinese Thousand Talents Program (grant to X.M.), and the Robert A. Welch foundation (A-1795 to L.S.).
References
- Albert I., Böhm H., Albert M., Feiler C. E., Imkampe J., Wallmeroth N., Brancato C., Raaymakers T. M., Oome S., Zhang H. et al. (2015). An RLP23-SOBIR1-BAK1 complex mediates NLP-triggered immunity. Nat. Plants 1, 15140 10.1038/nplants.2015.140 [DOI] [PubMed] [Google Scholar]
- Asai T., Tena G., Plotnikova J., Willmann M. R., Chiu W.-L., Gomez-Gomez L., Boller T., Ausubel F. M. and Sheen J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415, 977-983. 10.1038/415977a [DOI] [PubMed] [Google Scholar]
- Bayer M., Nawy T., Giglione C., Galli M., Meinnel T. and Lukowitz W. (2009). Paternal control of embryonic patterning in Arabidopsis thaliana. Science 323, 1485-1488. 10.1126/science.1167784 [DOI] [PubMed] [Google Scholar]
- Belkhadir Y., Jaillais Y., Epple P., Balsemao-Pires E., Dangl J. L. and Chory J. (2012). Brassinosteroids modulate the efficiency of plant immune responses to microbe-associated molecular patterns. Proc. Natl. Acad. Sci. USA 109, 297-302. 10.1073/pnas.1112840108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemis S. M., Lee J. S., Shpak E. D. and Torii K. U. (2013). Regulation of floral patterning and organ identity by Arabidopsis ERECTA-family receptor kinase genes. J. Exp. Bot. 64, 5323-5333. 10.1093/jxb/ert270 [DOI] [PubMed] [Google Scholar]
- Benschop J. J., Mohammed S., O'Flaherty M., Heck A. J. R., Slijper M. and Menke F. L. H. (2007). Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol. Cell. Proteomics 6, 1198-1214. 10.1074/mcp.M600429-MCP200 [DOI] [PubMed] [Google Scholar]
- Bergmann D. C., Lukowitz W. and Somerville C. R. (2004). Stomatal development and pattern controlled by a MAPKK kinase. Science 304, 1494-1497. 10.1126/science.1096014 [DOI] [PubMed] [Google Scholar]
- Bethke G., Unthan T., Uhrig J. F., Poschl Y., Gust A. A., Scheel D. and Lee J. (2009). Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proc. Natl. Acad. Sci. USA 106, 8067-8072. 10.1073/pnas.0810206106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson-Dernier A., Franck C. M., Lituiev D. S. and Grossniklaus U. (2015). Receptor-like cytoplasmic kinase MARIS functions downstream of CrRLK1L-dependent signaling during tip growth. Proc. Natl. Acad. Sci. USA 112, 12211-12216. 10.1073/pnas.1512375112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand U., Fletcher J. C., Hobe M., Meyerowitz E. M. and Simon R. (2000). Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289, 617-619. 10.1126/science.289.5479.617 [DOI] [PubMed] [Google Scholar]
- Bücherl C. A., Jarsch I. K., Schudoma C., Segonzac C., Mbengue M., Robatzek S., MacLean D., Ott T. and Zipfel C. (2017). Plant immune and growth receptors share common signalling components but localise to distinct plasma membrane nanodomains. Elife 6, e25114 10.7554/eLife.25114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burr C. A., Leslie M. E., Orlowski S. K., Chen I., Wright C. E., Daniels M. J. and Liljegren S. J. (2011). CAST AWAY, a membrane-associated receptor-like kinase, inhibits organ abscission in Arabidopsis. Plant Physiol. 156, 1837-1850. 10.1104/pp.111.175224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butenko M. A., Patterson S. E., Grini P. E., Stenvik G. E., Amundsen S. S., Mandal A. and Aalen R. B. (2003). Inflorescence deficient in abscission controls floral organ abscission in Arabidopsis and identifies a novel family of putative ligands in plants. Plant Cell 15, 2296-2307. 10.1105/tpc.014365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Liang Y., Tanaka K., Nguyen C. T., Jedrzejczak R. P., Joachimiak A. and Stacey G. (2014). The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. Elife 3, e03766 10.7554/eLife.03766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z., Li J.-F., Niu Y., Zhang X.-C., Woody O. Z., Xiong Y., Djonović S., Millet Y., Bush J., McConkey B. J. et al. (2015). Pathogen-secreted proteases activate a novel plant immune pathway. Nature 521, 213-216. 10.1038/nature14243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D., Zipfel C., Robatzek S., Kemmerling B., Nurnberger T., Jones J. D. G., Felix G. and Boller T. (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448, 497-500. 10.1038/nature05999 [DOI] [PubMed] [Google Scholar]
- Cho S. K., Larue C. T., Chevalier D., Wang H., Jinn T.-L., Zhang S. and Walker J. C. (2008). Regulation of floral organ abscission in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 105, 15629-15634. 10.1073/pnas.0805539105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa L. M., Marshall E., Tesfaye M., Silverstein K. A., Mori M., Umetsu Y., Otterbach S. L., Papareddy R., Dickinson H. G., Boutiller K. et al. (2014). Central cell-derived peptides regulate early embryo patterning in flowering plants. Science 344, 168-172. 10.1126/science.1243005 [DOI] [PubMed] [Google Scholar]
- Couto D. and Zipfel C. (2016). Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 16, 537-552. 10.1038/nri.2016.77 [DOI] [PubMed] [Google Scholar]
- De Smet I., Voß U., Jurgens G. and Beeckman T. (2009). Receptor-like kinases shape the plant. Nat. Cell Biol. 11, 1166-1173. 10.1038/ncb1009-1166 [DOI] [PubMed] [Google Scholar]
- Du C., Li X., Chen J., Chen W., Li B., Li C., Wang L., Li J., Zhao X., Lin J. et al. (2016). Receptor kinase complex transmits RALF peptide signal to inhibit root growth in Arabidopsis. Proc. Natl. Acad. Sci. USA 113, E8326-E8334. 10.1073/pnas.1609626113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng F., Yang F., Rong W., Wu X., Zhang J., Chen S., He C. and Zhou J.-M. (2012). A Xanthomonas uridine 5'-monophosphate transferase inhibits plant immune kinases. Nature 485, 114-118. 10.1038/nature10962 [DOI] [PubMed] [Google Scholar]
- Frei dit Frey N., Garcia A. V., Bigeard J., Zaag R., Bueso E., Garmier M., Pateyron S., de Tauzia-Moreau M.-L., Brunaud V., Balzergue S. et al. (2014). Functional analysis of Arabidopsis immune-related MAPKs uncovers a role for MPK3 as negative regulator of inducible defences. Genome Biol. 15, R87 10.1186/gb-2014-15-6-r87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz-Laylin L. K., Krishnamurthy N., Tör M., Sjolander K. V. and Jones J. D. (2005). Phylogenomic analysis of the receptor-like proteins of rice and Arabidopsis. Plant Physiol. 138, 611-623. 10.1104/pp.104.054452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlan G., Nakagami H., Eschen-Lippold L., Jiang X., Majovsky P., Kowarschik K., Hoehenwarter W., Lee J. and Trujillo M. (2017). Changes in PUB22 ubiquitination modes triggered by MITOGEN-ACTIVATED PROTEIN KINASE3 dampen the immune response. Plant Cell 29, 726-745. 10.1105/tpc.16.00654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M., Liu J., Bi D., Zhang Z., Cheng F., Chen S. and Zhang Y. (2008). MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen-activated protein kinase cascade to regulate innate immunity in plants. Cell Res. 18, 1190-1198. 10.1038/cr.2008.300 [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L. and Boller T. (2000). FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5, 1003-1011. 10.1016/S1097-2765(00)80265-8 [DOI] [PubMed] [Google Scholar]
- Gou X., Yin H., He K., Du J., Yi J., Xu S., Lin H., Clouse S. D. and Li J. (2012). Genetic evidence for an indispensable role of somatic embryogenesis receptor kinases in brassinosteroid signaling. PLoS Genet. 8, e1002452 10.1371/journal.pgen.1002452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Meng X., Khanna R., LaMontagne E., Liu Y. and Zhang S. (2014). Phosphorylation of a WRKY transcription factor by MAPKs is required for pollen development and function in Arabidopsis. PLoS Genet. 10, e1004384 10.1371/journal.pgen.1004384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gust A. A. and Felix G. (2014). Receptor like proteins associate with SOBIR1-type of adaptors to form bimolecular receptor kinases. Curr. Opin. Plant Biol. 21, 104-111. 10.1016/j.pbi.2014.07.007 [DOI] [PubMed] [Google Scholar]
- Han L., Li G. J., Yang K. Y., Mao G., Wang R., Liu Y. and Zhang S. (2010). Mitogen-activated protein kinase 3 and 6 regulate Botrytis cinerea-induced ethylene production in Arabidopsis. Plant J. 64, 114-127. 10.1111/j.1365-313X.2010.04318.x [DOI] [PubMed] [Google Scholar]
- Hara K., Kajita R., Torii K. U., Bergmann D. C. and Kakimoto T. (2007). The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes Dev. 21, 1720-1725. 10.1101/gad.1550707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta M., Sabat G., Stecker K., Minkoff B. B. and Sussman M. R. (2014). A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 343, 408-411. 10.1126/science.1244454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heese A., Hann D. R., Gimenez-Ibanez S., Jones A. M. E., He K., Li J., Schroeder J. I., Peck S. C. and Rathjen J. P. (2007). The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl. Acad. Sci. USA 104, 12217-12222. 10.1073/pnas.0705306104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa Y., Shinohara H., Kondo Y., Inoue A., Nakanomyo I., Ogawa M., Sawa S., Ohashi-Ito K., Matsubayashi Y. and Fukuda H. (2008). Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc. Natl. Acad. Sci. USA 105, 15208-15213. 10.1073/pnas.0808444105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoehenwarter W., Thomas M., Nukarinen E., Egelhofer V., Röhrig H., Weckwerth W., Conrath U. and Beckers G. J. (2013). Identification of novel in vivo MAP kinase substrates in Arabidopsis thaliana through use of tandem metal oxide affinity chromatography. Mol. Cell. Proteomics 12, 369-380. 10.1074/mcp.M112.020560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann U., Lau K. and Hothorn M. (2017). The structural basis of ligand perception and signal activation by receptor kinases. Annu. Rev. Plant Biol. 68, 109-137. 10.1146/annurev-arplant-042916-040957 [DOI] [PubMed] [Google Scholar]
- Hunt L. and Gray J. E. (2009). The signaling peptide EPF2 controls asymmetric cell divisions during stomatal development. Curr. Biol. 19, 864-869. 10.1016/j.cub.2009.03.069 [DOI] [PubMed] [Google Scholar]
- Igarashi D., Tsuda K. and Katagiri F. (2012). The peptide growth factor, phytosulfokine, attenuates pattern-triggered immunity. Plant J. 71, 194-204. 10.1111/j.1365-313X.2012.04950.x [DOI] [PubMed] [Google Scholar]
- Jia G., Liu X., Owen H. A. and Zhao D. (2008). Signaling of cell fate determination by the TPD1 small protein and EMS1 receptor kinase. Proc. Natl. Acad. Sci. USA 105, 2220-2225. 10.1073/pnas.0708795105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordá L., Sopeña-Torres S., Escudero V., Nuñez-Corcuera B., Delgado-Cerezo M., Torii K. U. and Molina A. (2016). ERECTA and BAK1 receptor like kinases interact to regulate immune responses in arabidopsis. Front Plant. Sci 7, 897 10.3389/fpls.2016.00897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota Y., Sklenar J., Derbyshire P., Stransfeld L., Asai S., Ntoukakis V., Jones J. D. G., Shirasu K., Menke F., Jones A. et al. (2014). Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol. Cell 54, 43-55. 10.1016/j.molcel.2014.02.021 [DOI] [PubMed] [Google Scholar]
- Kessler S. A., Shimosato-Asano H., Keinath N. F., Wuest S. E., Ingram G., Panstruga R. and Grossniklaus U. (2010). Conserved molecular components for pollen tube reception and fungal invasion. Science 330, 968-971. 10.1126/science.1195211 [DOI] [PubMed] [Google Scholar]
- Kim T.-W., Guan S., Burlingame A. L. and Wang Z.-Y. (2011). The CDG1 kinase mediates brassinosteroid signal transduction from BRI1 receptor kinase to BSU1 phosphatase and GSK3-like kinase BIN2. Mol. Cell 43, 561-571. 10.1016/j.molcel.2011.05.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Q., Sun T., Qu N., Ma J., Li M., Cheng Y. T., Zhang Q., Wu D., Zhang Z. and Zhang Y. (2016). Two redundant receptor-like cytoplasmic kinases function downstream of pattern recognition receptors to regulate activation of SA biosynthesis. Plant Physiol. 171, 1344-1354. 10.1104/pp.15.01954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumpf R. P., Shi C.-L., Larrieu A., Sto I. M., Butenko M. A., Peret B., Riiser E. S., Bennett M. J. and Aalen R. B. (2013). Floral organ abscission peptide IDA and its HAE/HSL2 receptors control cell separation during lateral root emergence. Proc. Natl. Acad. Sci. USA 110, 5235-5240. 10.1073/pnas.1210835110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampard G. R., Macalister C. A. and Bergmann D. C. (2008). Arabidopsis stomatal initiation is controlled by MAPK-mediated regulation of the bHLH SPEECHLESS. Science 322, 1113-1116. 10.1126/science.1162263 [DOI] [PubMed] [Google Scholar]
- Lampard G. R., Lukowitz W., Ellis B. E. and Bergmann D. C. (2009). Novel and expanded roles for MAPK signaling in Arabidopsis stomatal cell fate revealed by cell type-specific manipulations. Plant Cell 21, 3506-3517. 10.1105/tpc.109.070110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. S., Kuroha T., Hnilova M., Khatayevich D., Kanaoka M. M., McAbee J. M., Sarikaya M., Tamerler C. and Torii K. U. (2012). Direct interaction of ligand-receptor pairs specifying stomatal patterning. Genes Dev. 26, 126-136. 10.1101/gad.179895.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehti-Shiu M. D., Zou C., Hanada K. and Shiu S.-H. (2009). Evolutionary history and stress regulation of plant receptor-like kinase/pelle genes. Plant Physiol. 150, 12-26. 10.1104/pp.108.134353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. and Chory J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90, 929-938. 10.1016/S0092-8674(00)80357-8 [DOI] [PubMed] [Google Scholar]
- Li J., Wen J., Lease K. A., Doke J. T., Tax F. E. and Walker J. C. (2002). BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110, 213-222. 10.1016/S0092-8674(02)00812-7 [DOI] [PubMed] [Google Scholar]
- Li G., Meng X., Wang R., Mao G., Han L., Liu Y. and Zhang S. (2012). Dual-level regulation of ACC synthase activity by MPK3/MPK6 cascade and its downstream WRKY transcription factor during ethylene induction in Arabidopsis. PLoS Genet. 8, e1002767 10.1371/journal.pgen.1002767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Cheng C., Cui F., de Oliveira M. V. V., Yu X., Meng X., Intorne A. C., Babilonia K., Li M., Li B. et al. (2014a). Modulation of RNA polymerase II phosphorylation downstream of pathogen perception orchestrates plant immunity. Cell Host Microbe 16, 748-758. 10.1016/j.chom.2014.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Li M., Yu L., Zhou Z., Liang X., Liu Z., Cai G., Gao L., Zhang X., Wang Y. et al. (2014b). The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe 15, 329-338. 10.1016/j.chom.2014.02.009 [DOI] [PubMed] [Google Scholar]
- Li C., Wu H. M. and Cheung A. Y. (2016). FERONIA and her pals: functions and mechanisms. Plant Physiol. 171, 2379-2392. 10.1104/pp.16.00667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Wang Y., Huang J., Ahsan N., Biener G., Paprocki J., Thelen J. J., Raicu V. and Zhao D. (2017). Two SERK receptor-like kinases interact with EMS1 to control anther cell fate determination. Plant Physiol. 173, 326-337. 10.1104/pp.16.01219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W., Lu D., Gao X., Jiang S., Ma X., Wang Z., Mengiste T., He P. and Shan L. (2013a). Inverse modulation of plant immune and brassinosteroid signaling pathways by the receptor-like cytoplasmic kinase BIK1. Proc. Natl. Acad. Sci. USA 110, 12114-12119. 10.1073/pnas.1302154110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W., Ma X., Shan L. and He P. (2013b). Big roles of small kinases: the complex functions of receptor-like cytoplasmic kinases in plant immunity and development. J. Integr. Plant. Biol. 55, 1188-1197. 10.1111/jipb.12071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin G., Zhang L., Han Z., Yang X., Liu W., Li E., Chang J., Qi Y., Shpak E. D. and Chai J. (2017). A receptor-like protein acts as a specificity switch for the regulation of stomatal development. Genes Dev. 31, 927-938. 10.1101/gad.297580.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. and Zhang S. (2004). Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 16, 3386-3399. 10.1105/tpc.104.026609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Wu Y., Yang F., Zhang Y., Chen S., Xie Q., Tian X. and Zhou J.-M. (2013). BIK1 interacts with PEPRs to mediate ethylene-induced immunity. Proc. Natl. Acad. Sci. USA 110, 6205-6210. 10.1073/pnas.1215543110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D., Wu S., Gao X., Zhang Y., Shan L. and He P. (2010). A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl. Acad. Sci. USA 107, 496-501. 10.1073/pnas.0909705107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz W., Roeder A., Parmenter D. and Somerville C. (2004). A MAPKK kinase gene regulates extra-embryonic cell fate in Arabidopsis. Cell 116, 109-119. 10.1016/S0092-8674(03)01067-5 [DOI] [PubMed] [Google Scholar]
- Ma X., Xu G., He P. and Shan L. (2016). SERKing coreceptors for receptors. Trends Plant Sci. 21, 1017-1033. 10.1016/j.tplants.2016.08.014 [DOI] [PubMed] [Google Scholar]
- MacAlister C. A., Ohashi-Ito K. and Bergmann D. C. (2007). Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature 445, 537-540. 10.1038/nature05491 [DOI] [PubMed] [Google Scholar]
- Mao G., Meng X., Liu Y., Zheng Z., Chen Z. and Zhang S. (2011). Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23, 1639-1653. 10.1105/tpc.111.084996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X. and Zhang S. (2013). MAPK cascades in plant disease resistance signaling. Annu. Rev. Phytopathol. 51, 245-266. 10.1146/annurev-phyto-082712-102314 [DOI] [PubMed] [Google Scholar]
- Meng X., Wang H., He Y., Liu Y., Walker J. C., Torii K. U. and Zhang S. (2012). A MAPK cascade downstream of ERECTA receptor-like protein kinase regulates Arabidopsis inflorescence architecture by promoting localized cell proliferation. Plant Cell 24, 4948-4960. 10.1105/tpc.112.104695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Xu J., He Y., Yang K.-Y., Mordorski B., Liu Y. and Zhang S. (2013). Phosphorylation of an ERF transcription factor by Arabidopsis MPK3/MPK6 regulates plant defense gene induction and fungal resistance. Plant Cell 25, 1126-1142. 10.1105/tpc.112.109074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Chen X., Mang H., Liu C., Yu X., Gao X., Torii K. U., He P. and Shan L. (2015). Differential function of arabidopsis SERK family receptor-like kinases in stomatal patterning. Curr. Biol. 25, 2361-2372. 10.1016/j.cub.2015.07.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Zhou J., Tang J., Li B., de Oliveira M. V. V., Chai J., He P. and Shan L. (2016). Ligand-induced receptor-like kinase complex regulates floral organ abscission in arabidopsis. Cell Rep. 14, 1330-1338. 10.1016/j.celrep.2016.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam K. H. and Li J. (2002). BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110, 203-212. 10.1016/S0092-8674(02)00814-0 [DOI] [PubMed] [Google Scholar]
- Nühse T. S., Bottrill A. R., Jones A. M. E. and Peck S. C. (2007). Quantitative phosphoproteomic analysis of plasma membrane proteins reveals regulatory mechanisms of plant innate immune responses. Plant J. 51, 931-940. 10.1111/j.1365-313X.2007.03192.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M., Shinohara H., Sakagami Y. and Matsubayashi Y. (2008). Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science 319, 294 10.1126/science.1150083 [DOI] [PubMed] [Google Scholar]
- Ou Y., Lu X., Zi Q., Xun Q., Zhang J., Wu Y., Shi H., Wei Z., Zhao B., Zhang X. et al. (2016). RGF1 INSENSITIVE 1 to 5, a group of LRR receptor-like kinases, are essential for the perception of root meristem growth factor 1 in Arabidopsis thaliana. Cell Res. 26, 686-698. 10.1038/cr.2016.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patharkar O. R. and Walker J. C. (2015). Floral organ abscission is regulated by a positive feedback loop. Proc. Natl. Acad. Sci. USA 112, 2906-2911. 10.1073/pnas.1423595112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen M., Brodersen P., Naested H., Andreasson E., Lindhart U., Johansen B., Nielsen H. B., Lacy M., Austin M. J., Parker J. E. et al. (2000). Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell 103, 1111-1120. 10.1016/S0092-8674(00)00213-0 [DOI] [PubMed] [Google Scholar]
- Popescu S. C., Popescu G. V., Bachan S., Zhang Z., Gerstein M., Snyder M. and Dinesh-Kumar S. P. (2009). MAPK target networks in Arabidopsis thaliana revealed using functional protein microarrays. Genes Dev. 23, 80-92. 10.1101/gad.1740009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J.-L., Fiil B. K., Petersen K., Nielsen H. B., Botanga C. J., Thorgrimsen S., Palma K., Suarez-Rodriguez M. C., Sandbech-Clausen S., Lichota J. et al. (2008). Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO J. 27, 2214-2221. 10.1038/emboj.2008.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranf S., Gisch N., Schäffer M., Illig T., Westphal L., Knirel Y. A., Sánchez-Carballo P. M., Zähringer U., Hückelhoven R., Lee J. et al. (2015). A lectin S-domain receptor kinase mediates lipopolysaccharide sensing in Arabidopsis thaliana. Nat. Immunol. 16, 426-433. 10.1038/ni.3124 [DOI] [PubMed] [Google Scholar]
- Ren D., Liu Y., Yang K.-Y., Han L., Mao G., Glazebrook J. and Zhang S. (2008). A fungal-responsive MAPK cascade regulates phytoalexin biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 105, 5638-5643. 10.1073/pnas.0711301105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux M., Schwessinger B., Albrecht C., Chinchilla D., Jones A., Holton N., Malinovsky F. G., Tor M., de Vries S. and Zipfel C. (2011). The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 23, 2440-2455. 10.1105/tpc.111.084301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago J., Henzler C. and Hothorn M. (2013). Molecular mechanism for plant steroid receptor activation by somatic embryogenesis co-receptor kinases. Science 341, 889-892. 10.1126/science.1242468 [DOI] [PubMed] [Google Scholar]
- Santiago J., Brandt B., Wildhagen M., Hohmann U., Hothorn L. A., Butenko M. A. and Hothorn M. (2016). Mechanistic insight into a peptide hormone signaling complex mediating floral organ abscission. Elife 5, e15075 10.7554/eLife.15075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze B., Mentzel T., Jehle A. K., Mueller K., Beeler S., Boller T., Felix G. and Chinchilla D. (2010). Rapid heteromerization and phosphorylation of ligand-activated plant transmembrane receptors and their associated kinase BAK1. J. Biol. Chem. 285, 9444-9451. 10.1074/jbc.M109.096842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwessinger B., Roux M., Kadota Y., Ntoukakis V., Sklenar J., Jones A. and Zipfel C. (2011). Phosphorylation-dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor-like kinase BAK1. PLoS Genet. 7, e1002046 10.1371/journal.pgen.1002046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Shen Q., Qi Y., Yan H., Nie H., Chen Y., Zhao T., Katagiri F. and Tang D. (2013). BR-SIGNALING KINASE1 physically associates with FLAGELLIN SENSING2 and regulates plant innate immunity in Arabidopsis. Plant Cell 25, 1143-1157. 10.1105/tpc.112.107904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara H., Mori A., Yasue N., Sumida K. and Matsubayashi Y. (2016). Identification of three LRR-RKs involved in perception of root meristem growth factor in Arabidopsis. Proc. Natl. Acad. Sci. USA 113, 3897-3902. 10.1073/pnas.1522639113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinya T., Yamaguchi K., Desaki Y., Yamada K., Narisawa T., Kobayashi Y., Maeda K., Suzuki M., Tanimoto T., Takeda J. et al. (2014). Selective regulation of the chitin-induced defense response by the Arabidopsis receptor-like cytoplasmic kinase PBL27. Plant J. 79, 56-66. 10.1111/tpj.12535 [DOI] [PubMed] [Google Scholar]
- Shiu S.-H. and Bleecker A. B. (2001). Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. USA 98, 10763-10768. 10.1073/pnas.181141598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu S.-H. and Bleecker A. B. (2003). Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol. 132, 530-543. 10.1104/pp.103.021964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu S.-H., Karlowski W. M., Pan R., Tzeng Y.-H., Mayer K. F. X. and Li W.-H. (2004). Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16, 1220-1234. 10.1105/tpc.020834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W., Liu L., Wang J., Wu Z., Zhang H., Tang J., Lin G., Wang Y., Wen X., Li W. et al. (2016). Signature motif-guided identification of receptors for peptide hormones essential for root meristem growth. Cell Res. 26, 674-685. 10.1038/cr.2016.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreekanta S., Bethke G., Hatsugai N., Tsuda K., Thao A., Wang L., Katagiri F. and Glazebrook J. (2015). The receptor-like cytoplasmic kinase PCRK1 contributes to pattern-triggered immunity against Pseudomonas syringae in Arabidopsis thaliana. New Phytol. 207, 78-90. 10.1111/nph.13345 [DOI] [PubMed] [Google Scholar]
- Stegmann M., Monaghan J., Smakowska-Luzan E., Rovenich H., Lehner A., Holton N., Belkhadir Y. and Zipfel C. (2017). The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 355, 287-289. 10.1126/science.aal2541 [DOI] [PubMed] [Google Scholar]
- Suarez-Rodriguez M. C., Adams-Phillips L., Liu Y., Wang H., Su S.-H., Jester P. J., Zhang S., Bent A. F. and Krysan P. J. (2007). MEKK1 is required for flg22-induced MPK4 activation in Arabidopsis plants. Plant Physiol. 143, 661-669. 10.1104/pp.106.091389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Han Z., Tang J., Hu Z., Chai C., Zhou B. and Chai J. (2013a). Structure reveals that BAK1 as a co-receptor recognizes the BRI1-bound brassinolide. Cell Res. 23, 1326-1329. 10.1038/cr.2013.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Li L., Macho A. P., Han Z., Hu Z., Zipfel C., Zhou J.-M. and Chai J. (2013b). Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science 342, 624-628. 10.1126/science.1243825 [DOI] [PubMed] [Google Scholar]
- Tang W., Kim T.-W., Oses-Prieto J. A., Sun Y., Deng Z., Zhu S., Wang R., Burlingame A. L. and Wang Z. Y. (2008). BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321, 557-560. 10.1126/science.1156973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J., Han Z., Sun Y., Zhang H., Gong X. and Chai J. (2015). Structural basis for recognition of an endogenous peptide by the plant receptor kinase PEPR1. Cell Res. 25, 110-120. 10.1038/cr.2014.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D., Wang G. and Zhou J.-M. (2017). Receptor kinases in plant-pathogen interactions: more than pattern recognition. Plant Cell 29, 618-637. 10.1105/tpc.16.00891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo M., Ichimura K., Casais C. and Shirasu K. (2008). Negative regulation of PAMP-triggered immunity by an E3 ubiquitin ligase triplet in Arabidopsis. Curr. Biol. 18, 1396-1401. 10.1016/j.cub.2008.07.085 [DOI] [PubMed] [Google Scholar]
- Uchida N., Lee J. S., Horst R. J., Lai H.-H., Kajita R., Kakimoto T., Tasaka M. and Torii K. U. (2012). Regulation of inflorescence architecture by intertissue layer ligand-receptor communication between endodermis and phloem. Proc. Natl. Acad. Sci. USA 109, 6337-6342. 10.1073/pnas.1117537109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda M., Aichinger E., Gong W., Groot E., Verstraeten I., Vu L. D., De Smet I., Higashiyama T., Umeda M. and Laux T. (2017). Transcriptional integration of paternal and maternal factors in the Arabidopsis zygote. Genes Dev. 31, 617-627. 10.1101/gad.292409.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Ngwenyama N., Liu Y., Walker J. C. and Zhang S. (2007). Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19, 63-73. 10.1105/tpc.106.048298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Li H., Han Z., Zhang H., Wang T., Lin G., Chang J., Yang W. and Chai J. (2015). Allosteric receptor activation by the plant peptide hormone phytosulfokine. Nature 525, 265-268. 10.1038/nature14858 [DOI] [PubMed] [Google Scholar]
- Willmann R., Lajunen H. M., Erbs G., Newman M.-A., Kolb D., Tsuda K., Katagiri F., Fliegmann J., Bono J.-J., Cullimore J. V. et al. (2011). Arabidopsis lysin-motif proteins LYM1 LYM3 CERK1 mediate bacterial peptidoglycan sensing and immunity to bacterial infection. Proc. Natl. Acad. Sci. USA 108, 19824-19829. 10.1073/pnas.1112862108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J. and Zhang S. (2015). Mitogen-activated protein kinase cascades in signaling plant growth and development. Trends Plant Sci. 20, 56-64. 10.1016/j.tplants.2014.10.001 [DOI] [PubMed] [Google Scholar]
- Yamada K., Yamaguchi K., Shirakawa T., Nakagami H., Mine A., Ishikawa K., Fujiwara M., Narusaka M., Narusaka Y., Ichimura K. et al. (2016). The Arabidopsis CERK1-associated kinase PBL27 connects chitin perception to MAPK activation. EMBO J. 35, 2468-2483. 10.15252/embj.201694248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y., Pearce G. and Ryan C. A. (2006). The cell surface leucine-rich repeat receptor for AtPep1, an endogenous peptide elicitor in Arabidopsis, is functional in transgenic tobacco cells. Proc. Natl. Acad. Sci. USA 103, 10104-10109. 10.1073/pnas.0603729103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y., Huffaker A., Bryan A. C., Tax F. E. and Ryan C. A. (2010). PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis. Plant Cell 22, 508-522. 10.1105/tpc.109.068874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Li W., Xiang T., Liu Z., Laluk K., Ding X., Zou Y., Gao M., Zhang X., Chen S. et al. (2010). Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 7, 290-301. 10.1016/j.chom.2010.03.007 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Wu Y., Gao M., Zhang J., Kong Q., Liu Y., Ba H., Zhou J. and Zhang Y. (2012). Disruption of PAMP-induced MAP kinase cascade by a Pseudomonas syringae effector activates plant immunity mediated by the NB-LRR protein SUMM2. Cell Host Microbe 11, 253-263. 10.1016/j.chom.2012.01.015 [DOI] [PubMed] [Google Scholar]
- Zhang W., Fraiture M., Kolb D., Loffelhardt B., Desaki Y., Boutrot F. F. G., Tor M., Zipfel C., Gust A. A. and Brunner F. (2013). Arabidopsis receptor-like protein30 and receptor-like kinase suppressor of BIR1-1/EVERSHED mediate innate immunity to necrotrophic fungi. Plant Cell 25, 4227-4241. 10.1105/tpc.113.117010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wang P., Shao W., Zhu J.-K. and Dong J. (2015). The BASL polarity protein controls a MAPK signaling feedback loop in asymmetric cell division. Dev. Cell 33, 136-149. 10.1016/j.devcel.2015.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Lin X., Han Z., Wang J., Qu L.-J. and Chai J. (2016). SERK family receptor-like kinases function as co-receptors with PXY for plant vascular development. Mol. Plant. 9, 1406-1414. 10.1016/j.molp.2016.07.004 [DOI] [PubMed] [Google Scholar]
- Zipfel C., Kunze G., Chinchilla D., Caniard A., Jones J. D. G., Boller T. and Felix G. (2006). Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125, 749-760. 10.1016/j.cell.2006.03.037 [DOI] [PubMed] [Google Scholar]