Abstract
Mitochondrial DNA encodes 13 proteins that comprise components of the respiratory chain that maintain oxidative phosphorylation. The replication of mitochondrial DNA is performed by the sole mitochondrial DNA polymerase γ. As acute myeloid leukemia (AML) cells and stem cells have an increased reliance on oxidative phosphorylation, we sought to evaluate polymerase γ inhibitors in AML. The thymidine dideoxynucleoside analog, alovudine, is an inhibitor of polymerase γ. In AML cells, alovudine depleted mitochondrial DNA, reduced mitochondrial encoded proteins, decreased basal oxygen consumption, and decreased cell proliferation and viability. To evaluate the effects of polymerase γ inhibition with alovudine in vivo, mice were xenografted with OCI-AML2 cells and then treated with alovudine. Systemic administration of alovudine reduced leukemic growth without evidence of toxicity and decreased levels of mitochondrial DNA in the leukemic cells. We also showed that alovudine increased the monocytic differentiation of AML cells. Genetic knockdown and other chemical inhibitors of polymerase γ also promoted AML differentiation, but the effects on AML differentiation were independent of reductions in oxidative phosphorylation or respiratory chain proteins. Thus, we have identified a novel mechanism by which mitochondria regulate AML fate and differentiation independent of oxidative phosphorylation. Moreover, we highlight polymerase γ inhibitors, such as alovudine, as novel therapeutic agents for AML.
Introduction
Acute myeloid leukemia (AML) is a hematologic malignancy with a poor prognosis, characterized by clonal, pathological and often poorly differentiated hematopoietic cells that infiltrate the bone marrow (BM), blood and extra-medullary tissues. In spite of some recent new therapies for AML, the disease is curable in only up to 40% of adults under 60 years of age, and older patients and those with high-risk cytogenetics have a dismal outcome, with a 2-year survival rate of <10%.1,2 Thus, new treatment options for this disease are required.
As previously described by us and others, a subset of AML cells present unique features that make them more vulnerable to impairment of mitochondrial function, such as increased mitochondrial biogenesis, decreased spare reserve capacity, and increased dependence on oxidative phosphorylation compared to normal hematopoietic progenitor cells.3–6 Based on these data, our current study focuses on inhibiting polymerase gamma (POLG), the sole mitochondrial DNA polymerase, as a new therapeutic target for this disease.
Eukaryotic cells have two separate genomes: nuclear DNA (nuDNA), organized in chromosomes, and the circular mitochondrial DNA (mtDNA), organized within nucleoids located within the mitochondria. Mitochondrial DNA is comprised of a double-stranded circular genome that is 16.6 kb in length and lacks introns. It encodes two rRNAs, 22 t-RNAs, and 13 of the 90 proteins in the mitochondrial respiratory chain, the cascade of enzymes central to ATP production in the mitochondria via oxidative phosphorylation.2 Synthesis of the 13 proteins encoded by the mtDNA occurs in the mitochondrial matrix on mitochondrial ribosomes using mitochondrial-specific protein synthesis machinery.7,8 The remaining approximately 1300 mitochondrial proteins are encoded by nuclear genes and translated in the cytoplasm.
Contrary to nuDNA, that replicates once during cell division, mtDNA is continuously replicated independently from the cell cycle.9 Mitochondria contain their own specialized machinery for DNA replication, transcription and translation of the mitochondrial genome. Loss of integrity of mtDNA results in dysfunctional respiratory complexes and negatively affects the production of ATP.
In humans, the nuclear-encoded mitochondrial POLG is the sole polymerase responsible for mitochondrial DNA replication. Human POLG is comprised of a catalytic polymerase domain at the C-terminus and an exonuclease domain separated by a linker region at the N-terminus. The holoenzyme consists of the primary subunit POLG and a homodimeric form of its accessory subunit POLG2.10 POLG forms a multi-protein-DNA complex, termed nucleoid, which acts as a hub for mtDNA replication, transcription and translation.11
Mitochondrial and cytoplasmic pathways support mtDNA biosynthesis by supplying it with sufficient nucleotide pools. The former, the mitochondrial nucleotide salvage pathway, converts nucleoside precursors to nucleotides by a cascade of kinases within the mitochondria,12 while in the latter, the cytoplasmic pathway, kinases catalyze the phosphorylation of nucleosides to nucleotides in the cytoplasm. Nucleosides in this cytoplasmic pathway are synthesized from de novo biosynthesis,13,14 and following their phosphorylation to nucleotides they are imported into the mitochondria by specific nucleotide transporters.15,16
In this current study, we investigated the POLG inhibitor, dideoxynucleoside analog of thymidine, alovudine (3′-deoxy-3′-fluorothymidine, FLT), in AML. We evaluated the effects of alovudine on mitochondrial function as well as on stemness and differentiation in AML.
Methods
Cell lines and primary samples
OCI-AML2, MV4-11 and K562 cells were cultured in Iscove’s modified Dulbecco’s medium (IMDM) augmented with 10% fetal bovine serum (FBS) and antibiotics. TEX cells (a gift from Dr. J. Dick) were cultured in IMDM augmented with 20% FBS, 2 mM L-glutamine, 2 ng/mL human Interleukin-3 (IL-3), 20 ng/mL human stem cell factor (SCF) (R&D Systems) and antibiotics. NB4 cells were cultured in RPMI 1640 medium augmented with 10% FBS. A total of 8227 cells (a gift from Dr. J. Dick) were cultured in X-VIVO 10 supplemented with 20% BIT (StemCell Technologies, Vancouver, Canada), 10 ng/mL hIL-3, 50 ng/mL hSCF, 10 ng/mL G-CSF, and 25 ng/mL TPO (Pepro Tech).
Peripheral blood was collected from consenting patients with AML. Samples with at least 80% leukemic blasts among low-density cells isolated by Ficoll density gradient centrifugation were included in this analysis. Normal hematopoietic cells were derived from healthy volunteers donating peripheral blood stem cells (PBSCs) for allogeneic stem cell transplantation following granulocyte colony-stimulating factor (G-CSF) mobilization. Primary AML and normal hematopoietic mononuclear cells were cultured in Iscove-modified Dulbecco medium (IMDM) supplemented with 20% FBS, 2 mM L-glutamine, 2 ng/mL human IL-3, and 20 ng/mL human SCF. Sample collection and the use of human tissue were approved by the University Health Network institutional review.
All cell lines and primary samples used in our experiments were incubated at 37°C and 5% CO2 in humidified atmosphere.
Xenograft models of human acute myeloid leukemia
For in vivo studies, alovudine was supplied by Medivir AB (Huddinge, Sweden). OCI-AML2 leukemia cells (1x106) were injected subcutaneously into the flanks of severe combined immune deficient (SCID) mice (Ontario Cancer Institute, Toronto, ON, Canada). After the appearance of a palpable tumor (9-11 days), the mice were treated orally with alovudine (50 mg/kg) twice daily or vehicle (saline) control (n=10 per group) at a treatment schedule of 5 out of 7 days for a total of 21 days (total number of 27 doses). Tumors were measured 3 times a week based on caliper measurements of tumor length and width (volume=tumor length × width2 × 0.5236). At the end of treatment, mice were sacrificed and tumor volumes and mass were measured from excised tumors. Mitochondrial DNA (mtDNA) assessment was also carried out from excised tumors, using qRT-PCR.
To test alovudine efficacy in a primary AML engraftment mouse model, a frozen aliquot of primary AML cells was thawed, counted, and re-suspended in phosphate-buffered saline. Viable trypan blue-negative cells (2.5×106) were injected into the right femur of 10-week old female NOD-SCID mice that were sublethally irradiated (2 Gy), and pretreated with 200 μg of anti-mouse CD-122. Mice were treated once daily with oral alovudine at 25 mg/kg or vehicle (saline) control (n=10 per group) 5 out of 7 days for 24 days (total number of 17 doses). Mice were then sacrificed, femurs flushed, and primary AML engraftment (CD45+CD33+CD19− cells) in the left femur was determined by flow cytometry.
In vivo studies were performed according to the regulations of the Canadian Council on Animal Care and with the approval of the Ontario Cancer Institute Animal Ethics Review Board.
Results
The nucleoside analog alovudine depletes mitochondrial DNA and reduces cell growth and viability in acute myeloid leukemia cells
Alovudine (Figure 1A) is a dideoxynucleoside analog of thymidine, originally developed as a reverse transcriptase inhibitor and evaluated in patients for the treatment of HIV.17–19 In addition to inhibiting viral replication, alovudine triphosphate is incorporated into DNA by POLG preferentially over nuclear polymerases, resulting in chain termination and inhibition of its enzymatic activity in cell-free assays.20 However, its anti-leukemic activity has not been previously described. Therefore, we focused our further investigations on this compound.
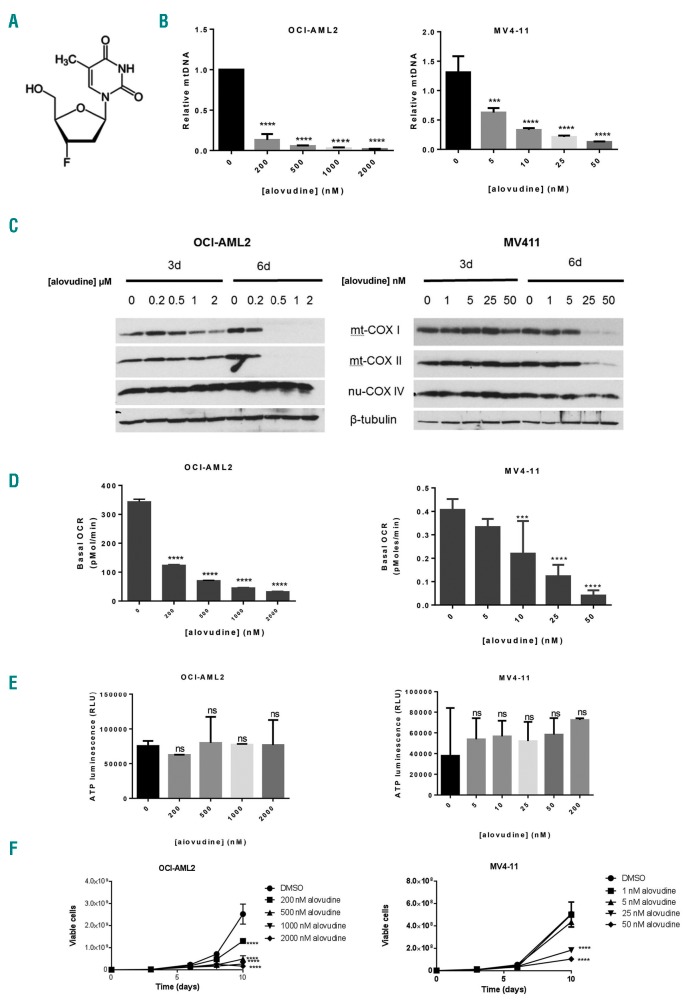
Figure 1.
Alovudine inhibits mitochondrial DNA biosynthesis and oxidative phosphorylation in acute myeloid leukemia (AML). (A) Alovudine’s chemical structure. (B) OCI-AML2 and MV4-11 cells were treated with increasing concentrations of alovudine for 6 days. Relative mitochondrial DNA (mtDNA) content was assessed by qRT-PCR as described in the Methods section. Data represent mean+Standard Deviation (SD) mtDNA relative to untreated controls from one of three representative experiments. (C) OCI-AML2 and MV411 were treated with increasing concentrations of alovudine. Three and 6 days after treatment, cells were harvested, lysed and levels of cytochrome C oxidase subunits: mitochondrial COXI (mt-COX1), mitochondrial COXII (mt-COXII), nuclear COX IV (nu-COX IV), and ®-tubulin were measured by immunoblotting. The immunoblot from one of three representative experiments is shown. (D) Basal oxygen consumption rate (OCR) was assessed in OCI-AML2 and MV411 cells following 6 days of alovudine treatment, using the Seahorse XF96 Metabolic Flux Assay. Data represent the mean±SD basal OCR from one of three representative experiments. n=6. (E) OCI-AML2 and MV411 cells were treated with increasing concentrations of alovudine for 6 days. ATP production was assessed by CellTitre-Glo Luminescent Cell Viability Assay. Data represent the mean±SD from two independent experiments in triplicate. (F) OCI-AML2 and MV411 cells were treated with increasing concentrations of alovudine. Cell growth and viability was assessed by trypan blue exclusion staining at increasing times after incubation. Data represent the mean±SEM from one of three representative experiments. For all experiments, ***P<0.001 and ****P<0.0001 using Dunnett’s multiple comparisons test after one-way ANOVA (B, D and E). (F) Two-way ANOVA.
Alovudine reduces mtDNA and impairs mitochondrial function in acute myeloid leukemia cells
To investigate the effects of alovudine on mitochondrial function, OCI-AML2 and MV4-11 leukemia cells were treated with increasing concentrations of alovudine. Six days after incubation, changes in mtDNA and bioenergetics were measured. Alovudine decreased mtDNA in both OCI-AML2 and MV4-11 cells, although MV4-11 cells were more sensitive with reductions in mtDNA observed at low nM concentrations and >80% reductions in mtDNA observed with 25 nM of the drug (Figure 1B). In contrast to the large reduction in levels of mtDNA, there was only a small reduction in mitochondrial mass after 6 days of alovudine treatment (Online Supplementary Figure S1) and no change in nuclear DNA (Online Supplementary Figure S2). In keeping with reductions in mtDNA, we noted prominent reductions in protein levels of the mtDNA-encoded respiratory chain IV complex subunits, mt-COXI and mt-COX II (Figure 1C). In contrast, no changes were seen in levels of nu-COX IV, a subunit of the same respiratory chain complex IV, but encoded by nuclear DNA (Figure 1C). mt-COX I and mt-COX II mRNA were also decreased >95%, but little change in the mRNA expression of nu-COX IV was seen (Online Supplementary Figure S3). We also observed reductions in basal oxygen consumption at concentrations associated with reductions in mtDNA and respiratory chain proteins (Figure 1D). Of note, alovudine did not reduce ATP level in treated cells (Figure 1E).
Finally, we observed reductions in cell proliferation and viability at concentrations of alovudine that reduced mtDNA and inhibited oxidative phosphorylation. Of the tested AML cell lines (MV4-11, OCI-AML2, TEX, NB4, and K562), MV4-11 was the most sensitive to alovudine (Figure 1F and Online Supplementary Figure S4A-C). Compared to the other tested cell lines, MV4-11 had highest expression of POLG (Online Supplementary Figure S4D).
There was no increase in apoptosis as measured by Annexin V staining 6 days after alovudine treatment, a time at which reductions in cell viability and mtDNA biosynthesis were already evident. However, cell death was observed after more prolonged (11-day) incubation with alovudine (Online Supplementary Figure S4E). Furthermore, analysis of the cell cycle in alovudine-treated AML cells showed little or no change (Online Supplementary Figure S5).
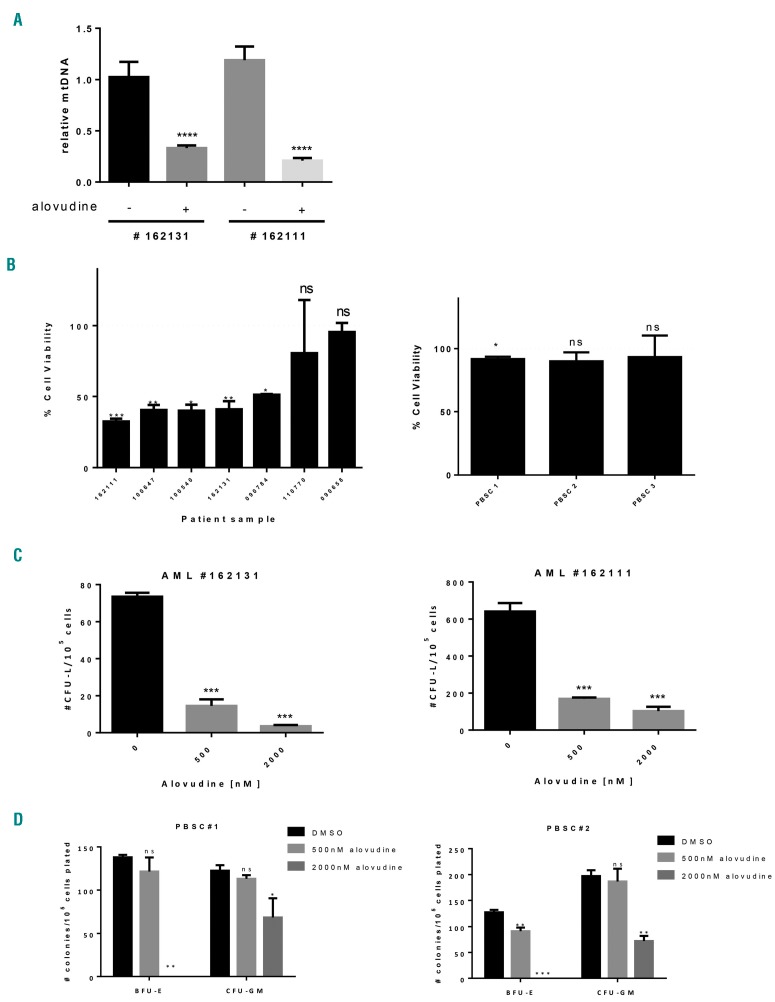
Next, we investigated the effects of alovudine on primary AML cells and normal hematopoietic cells. Primary AML and normal hematopoietic cells were treated for 6 days with increasing concentrations of alovudine. After incubation, we measured cell viability and levels of mtDNA. The number of viable primary cells treated with the vehicle control did not increase during the period of incubation, suggesting that the primary cells were quiescent and did not proliferate. Alovudine reduced levels of mtDNA in primary AML cells at concentrations that also reduced cell viability (Figure 2A and B and Online Supplementary Table S1). In contrast, alovudine did not reduce the viability of the normal hematopoietic cells (Figure 2B). Furthermore, alovudine reduced the clonogenic growth of primary AML cells. We also tested the effects of alovudine on the clonogeneic growth of normal hematopoietic cells. Compared to AML cells, normal hematopoietic cells were more resistant to alovudine. However, at the highest tested dose (2000 nM), alovudine partially reduced the clonogenic growth of granulocyte precursors (CFU-GM) and completely inhibited the growth of primitive erythroid progenitor cells (BFU-E) (Figure 2C and D and Online Supplementary Table S1).
Figure 2.
Effects of alovudine on primary acute myeloid leukemia (AML) and normal hematopoietic cells. Primary AML samples and normal peripheral blood stem cells (PBSCs) were treated with 2000 nM alovudine for 6 days. (A) Relative mitochondrial DNA (mtDNA) was assessed by qRT-PCR in AML patients (n=2). (B) Primary AML and normal hematopoietic cells (PBSCs) were treated with 2000 nM of alovudine for 6 days. Cell viability was assessed by trypan blue exclusion staining in primary AML cells (n=7) and CellTiter-Fluor for PBSCs (n=3). The dotted line represents cell viability of DMSO control. (C and D) Primary AML and normal hematopoietic cells (PBSCs) were treated with increasing concentrations of alovudine for 6 days. Colony forming abilities were assessed in primary AML cells and PBSCs (n=2 of each) as described above in the Online Supplementary Methods. PBSC samples 1 and 2 were the same samples as used in (B). For all experiments, ns: non-significant; *P<0.5, **P<0.01, ***P<0.001, and ****P<0.0001 using Sidak’s (A) or Dunnett’s (B-D) multiple comparisons test after one-way ANOVA. BFU-E: primitive erythroid progenitor cells; CFU-GM: granulocyte precursors.
Thus, alovudine inhibits mtDNA biosynthesis in primary AML cells and selectively targets a subset of AML cells in vitro.
Alovudine displays efficacy in mouse models of human leukemia
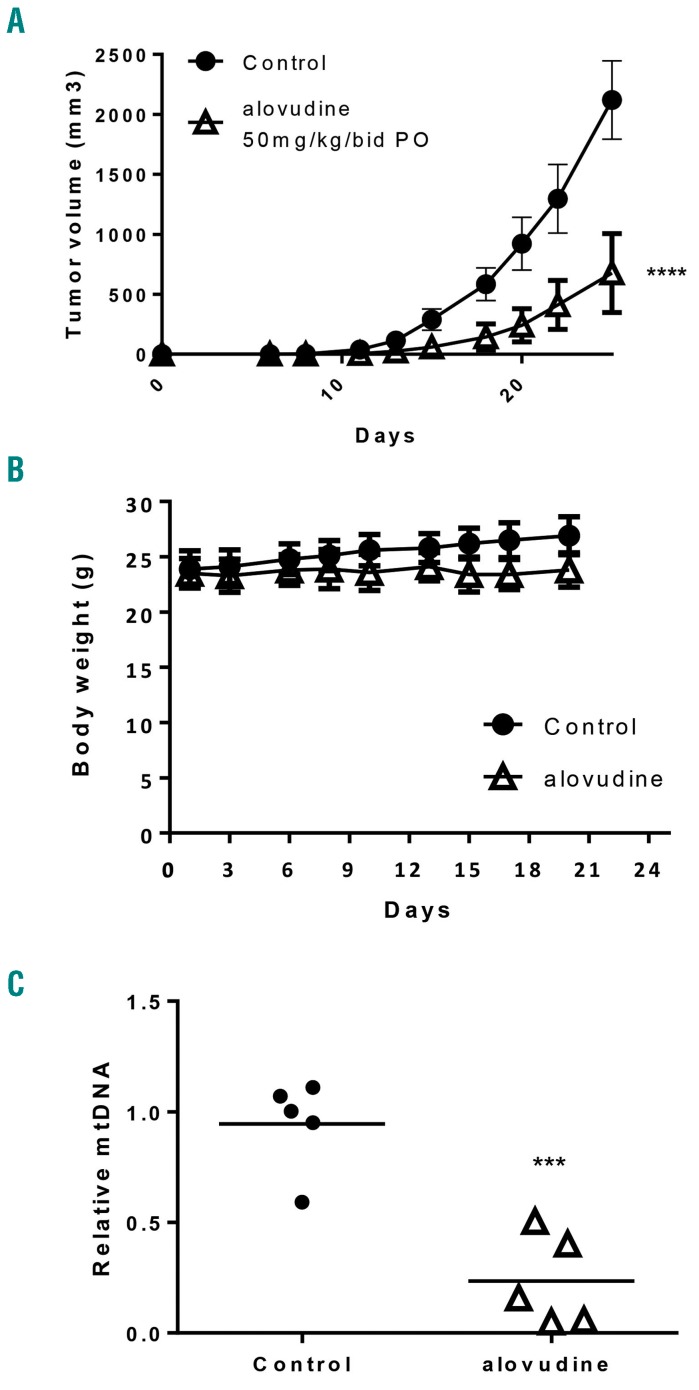
Given the anti-leukemic effects of alovudine in vitro, we examined the efficacy and toxicity of alovudine in a mouse model of leukemia. Severe combined immune deficient (SCID) mice xenografted with OCI-AML2 cells were treated with alovudine (50 mg/kg bid) or vehicle control by oral gavage. Alovudine reduced the growth of leukemia in vivo by approximately 70% without evidence of toxicity (Figure 3A). Specifically, doses of alovudine that reduced tumor growth did not alter mouse body weight, behavior, serum chemistries, or organ histology (Figure 3B and Online Supplementary Figure S6). Finally, we conducted correlative studies and measured mtDNA in leukemic cells isolated from mice treated with alovudine. We observed reductions in mtDNA by greater than 75% in OCI-AML2 tumors excised from mice treated with alovudine (Figure 3C).
Figure 3.
Alovudine displays efficacy in mouse models of human acute myeloid leukemia. OCI-AML2 cells were injected subcutaneously into the flank of SCID mice. Once tumors were palpable, mice were treated with oral (PO) alovudine (50 mg/kg bid) or vehicle control for 14 days. (A) Tumor volume was assessed from excised tumors. The error bars represent Standard Deviation (SD) (n=10 per group). (B) Body weight was assessed every 2-3 days. The error bars represent SD (n=10 per group). (C) Relative mitochondrial DNA (mtDNA) from xenograft tumors excised in (B) (n=5 per group). For all experiments, ***P<0.001 and ****P<0.0001 using two-way ANOVA (A and B) and t-test (C).
Next, we tested whether alovudine targets primary AML cells in vivo. Sublethally irradiated NOD-SCID mice preconditioned with anti-CD122 received intrafemural injection of primary AML cells. In this mouse model, the maximum tolerated dose of alovudine was only 25 mg/kg/day and less than the dose tolerated by the SCID mice engrafted with OCI-AML2 cells. Therefore, mice were treated with 25 mg/kg alovudine for 5 of 7 days for 17 days. A small, but statistically significant reduction in primary AML engraftment was observed (Online Supplementary Figure S7), potentially reflecting the reduced treatment dose, an alovudine-resistant primary AML sample, or a protective effect from the marrow niche.
Alovudine promotes monocytic differentiation in acute myeloid leukemia
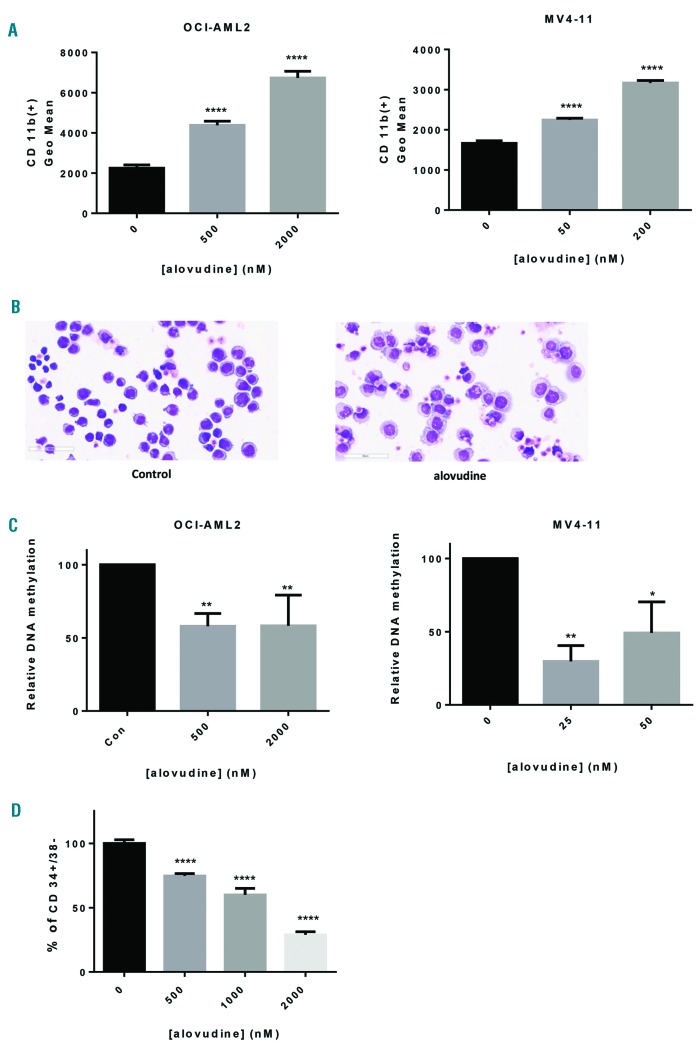
Recent studies reported that mitochondrial pathways and metabolism can regulate the differentiation of malignant cells.21–24 Therefore, we explored the effects of alovudine on the differentiation of AML cells. OCI-AML2 and MV4-11 cells were treated with concentrations of alovudine that depleted mtDNA for 10 days. Alovudine increased expression of CD11b, a cell surface marker associated with monocytic differentiation (Figure 4A). It also induced morphological changes typical of monocytic differentiation by Giemsa stain (Figure 4B). Finally, alovudine decreased global DNA methylation (Figure 4C), in keeping with prior studies showing a reduction in methylation correlates with differentiation.25–27
Figure 4.
Alovudine promotes monocytic differentiation in acute myeloid leukemia. (A) OCI-AML2 and MV4-11 cells were treated with increasing concentrations of alovudine for 10 days. CD11b expression was assessed by flow cytometry. (B) MV4-11 cells were treated with alovudine (200 nM) for 9 days. Morphology was assessed by Giemsa stain (magnification 40X, scale bar=60 μm). (C) OCI-AML2 and MV4-11 cells were treated with alovudine for 6 days. Methylation was assessed by dot-blot assay. (D) 8227 cells were treated with increasing concentrations of alovudine for 6 days. CD34+/CD38− expression was assessed by flow cytometry. For all experiments, *P<0.5, **P<0.01, ***P<0.001, and ****P<0.0001 using Dunnett’s multiple comparisons test after one-way ANOVA. Data represent the mean+Standard Deviaiton (SD) from one of three representative experiments, except (C) which represent the mean+SEM of average of 3 (MV4-11) or 4 experiments (OCI-AML2).
Next, we evaluated alovudine in the 8227 primary AML culture model; 8227 leukemia cells are patient-derived cells that are organized into a hierarchy of stem and bulk cells with the stem cells residing in the CD34+CD38− com partment.28 Treatment of 8227 cells with alovudine decreased the CD34+CD38− stem cells (Figure 4D).
Inhibition of polymerase gamma but not reductions in oxidative phosphorylation or respiratory chain proteins influence acute myeloid leukemia differentiation
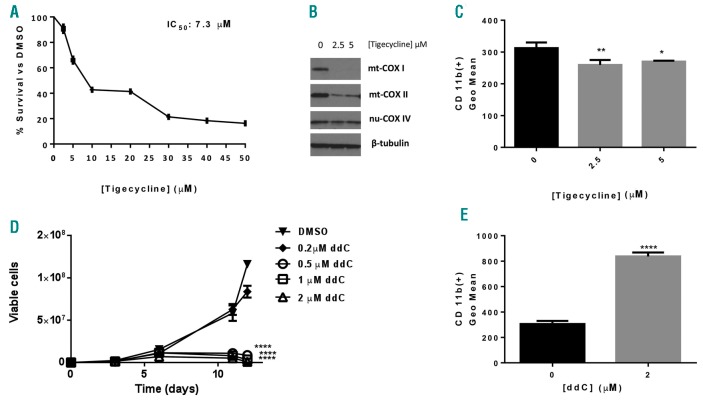
We then explored the mechanism by which alovudine promoted AML differentiation. We tested whether reducing levels of mitochondrial encoded respiratory chain proteins and basal oxygen consumption without altering levels of mtDNA would be sufficient to mimic the effects of alovudine and induce AML differentiation. We treated OCI-AML2 cells with tigecycline that we previously showed inhibits mitochondrial protein translation by targeting mitochondrial ribosomes. Consistent with our previous studies,4 tigecycline reduced levels of mtDNA-encoded proteins but not nuclear-DNA-encoded proteins and reduced the proliferation and viability of AML cells (Figure 5A and B). However, there was no evidence of differentiation (Figure 5C).
Figure 5.
Effects of inhibiting mitochondrial translation and polymerase gamma (POLG) on acute myeloid leukemia growth and differentiation. (A-C) OCI-AML2 cells were treated with increasing concentrations of Tigecycline for 72 hours (h). (A) Cell growth and viability was assessed by Alamar Blue assay [Mean+Standard Deviation (SD)]. (B) Cells were harvested 72 h after treatment, lysed, and levels of cytochrome C oxidase subunits: mitochondrial COXI (mt-COXI), mitochondrial COXII (mt-COXII), nuclear COX IV (nu-COX IV), and ®-tubulin measured by immunoblotting. The immunoblot from a representative experiment is shown. (C) CD11b expression was assessed by flow cytometry. (D) OCI-AML2 cells were treated with increasing concentrations of antiviral ddC. Cell growth and viability was assessed by trypan blue exclusion staining at increasing times after incubation. Data represent the mean±Standard Error of Mean (SEM) from one of three representative experiments. (E) OCI-AML2 cells were treated with increasing concentrations of 2′3′-dideoxycytidine (ddC) for 10 days. CD11b expression was assessed by flow cytometry. For all experiments, *P<0.5, **P<0.01, ***P<0.001, and ****P<0.0001 using Dunnett’s multiple comparisons test after one-way ANOVA (C) and two-way ANOVA (D). (E) Unpaired t-test.
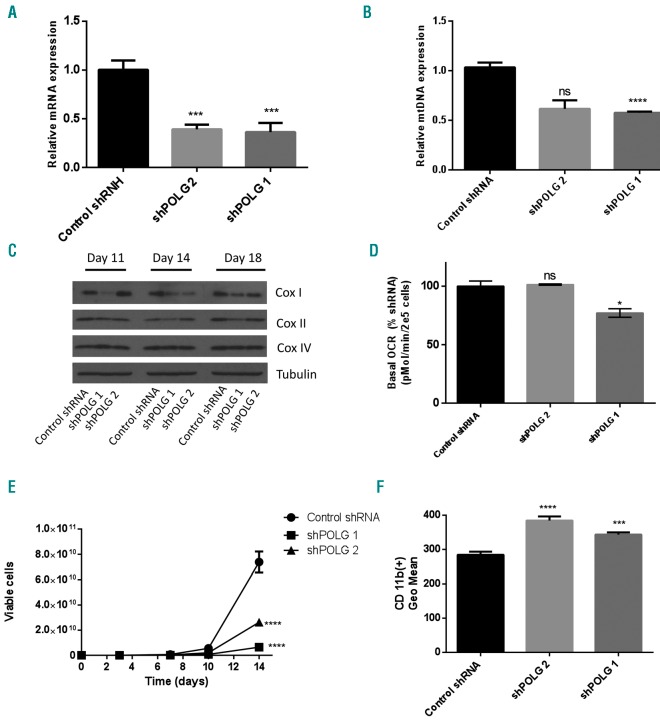
Then, we asked whether other chemical or genetic inhibitors of POLG would also promote AML differentiation. We treated OCI-AML2 cells with the antiviral 2′3′-dideoxycytidine (ddC) that we and others previously showed inhibited POLG.6,20 Similar to alovudine, ddC reduced mtDNA and reduced levels of mitochondrial encoded respiratory chain proteins.6 Like alovudine, ddC also reduced AML growth (Figure 5D) and promoted AML differentiation (Figure 5E). As a genetic approach, we knocked down POLG with shRNA in lentiviral vectors. Partial POLG knockdown reduced mtDNA, but the reductions in mtDNA were not sufficient to significantly reduce levels of respiratory chain proteins or impair oxidative phosphorylation. However, partial POLG knockdown mimicked the effects of alovudine and reduced AML proliferation, and induced differentiation of AML cells (Figure 6A–F).
Figure 6.
Polymerase gamma (POLG) knockdown promotes acute myeloid leukemia differentiation. OCI-AML2 cells were infected with shRNA targeting POLG or a control sequenced in lentiviral vectors. (A) POLG knockdown at 14 days post transduction was assessed by qRT-PCR. Data represent the mean+Standard Deviation (SD) mtDNA from one of three representative experiments. (B) Relative mitochondrial DNA (mtDNA) content was assessed by qRT-PCR 14 days post transduction with POLG shRNA, as described in the “Methods” section. Data represent the mean+Standard Error of Mean (SEM) from two independent experiments. (C) Cells were harvested 11, 14 and 18 days post POLG knockdown and levels of cytochrome C oxidase subunits: mitochondrial COXI (mt-COX1), nuclear COX IV (nu-COX IV), and β-tubulin in whole-cell extracts were measured by immunoblotting. The immunoblot from one of three representative experiments is shown. (D) Basal oxygen consumption rate (OCR) was assessed 10 days following POLG knockdown, using the Seahorse XF96 Metabolic Flux Assay. Data represent Mean±SEM basal OCR from two independent experiments; n=6. (E) Cell growth and viability were assessed by trypan blue exclusion staining. Data represent the mean±SD from one of three representative experiments. (F) CD11b expression was assessed by flow cytometry. Data represent the mean±SD from one of three representative experiments. For all experiments, *P<0.5, **P<0.01, ***P<0.001, and ****P<0.0001 using Dunnett’s multiple comparison following one-way ANOVA (A and F) or two-way ANOVA (B and D). (E) Two-way ANOVA test.
Thus, taken together, our data suggest that inhibition of POLG promotes AML differentiation through effects that are independent of reductions in oxidative phosphorylation or respiratory chain proteins.
Discussion
Inhibiting nuclear DNA replication by targeting nuclear DNA polymerases is the primary mechanism of many standard anti-leukemic agents, including cytarabine, clofarabine and cladribine. Some nucleoside analogs also cross react with the mitochondrial polymerase, POLG, and thereby inhibit mitochondrial DNA replication. Given that a subset of AML cells and stem cells have increased mitochondrial mass, mitochondrial DNA, and a greater reliance on oxidative phosphorylation,3,5,29 we sought to identify new nucleoside analogs that deplete mitochondrial DNA. In this current study, we evaluated alovudine as a potent nucleoside analog targeting mtDNA in AML. Alovudine is a dideoxynucleoside analog of thymidine, which inhibits mtDNA polymerase activity through its 5’-triphosphate metabolite. First introduced in the early 90s, this drug was originally developed as a reverse transcriptase inhibitor and evaluated in patients for the treatment of HIV.30–32 More recently, it has been labeled with 18F and used as a reagent in PET imaging to visualize malignancy.33–35
We demonstrated that inhibition of POLG with alovudine inhibits the viability and proliferation of AML cells in vitro and in vivo. Human mitochondrial DNA is replicated exclusively by the mitochondrial DNA polymerase gamma. The holoenzyme is a heterotrimer which consists of the primary subunit POLG and a homodimeric form of its accessory subunit POLG2.10 The nuclear-encoded primary subunit contains a C-terminal catalytic polymerase domain and N-terminal exonuclease domain separated by a linker/spacer region.36 The polymerase domain is responsible for extension of the DNA strand, and similar to nuclear-DNA polymerase, utilizes deoxynucleotide triphosphates (dNTPs) as its substrate to the growing chain.37 Any alteration of dNTP (i.e. analogs of dNTPs, called ddNTPs) can inhibit POLG function through chain termination. We showed that alovudine decreased the growth of OCI-AML2 cells xenografted into SCID mice. However, in sublethally irradiated NOD/SCID mice engrafted by intrafemural injection with a primary AML patient sample, alovudine produced only a small reduction in primary AML engraftment. Unfortunately, in this mouse model, the maximum tolerated dose of alovudine was only 25 mg/kg/day. In contrast, in SCID mice engrafted with OCI-AML2 cells, the maximum tolerated dose was over 50 mg/kg/day. We suspect that the difference in the maximal tolerated dose is due to differences in the mouse strains or the sublethal irradiation that the NOD/SCID mice received prior to engraftment with primary AML cells. Alternatively, the reduced efficacy in the primary AML model may reflect protection from the BM niche, or that the tested sample was resistant to alovudine. Further studies will be needed to clarify the basis for this sensitivity and resistance.
In clinical trials with patients with AIDS and HIV, alovudine reduced viral load but its development for HIV did not progress beyond phase II and small scale phase III trials.17,30,36 In these trials, alovudine was associated with dose-dependent reversible leukopenia and anemia.19 Although myelosuppression was not an acceptable toxicity for HIV, it may be a more manageable side effect for an anti-leukemic therapy. Moreover, our data suggest that alovudine can preferentially target AML cells over normal hematopoeitic cells. Thus, given the known toxicology and pharmacology of alovudine, along with the prior experience of its clinical use, it could potentially be rapidly repositioned for the treatment of AML.
Using a combination of pharmacological and genetic approaches, we discovered that inhibition of POLG induces the monocytic differentiation of AML cells. Thus, we identified new mechanisms by which mitochondrial pathways control differentiation in AML and highlight alovudine as a novel potential therapeutic agent for AML. Most therapies currently in use or under investigation for AML are cytotoxic and induce cell death. A less explored therapeutic strategy is to promote the differentiation of AML cells into more mature progeny. Upon differentiation, the leukemic cells will cease to proliferate or die. Targeting the block in differentiation is the standard therapy for acute promyelocytic leukemia (APL)-AML.39 There is growing evidence that targeting mitochondrial pathways can impact cell fate and differentiation of AML beyond the subset of APL.22–24 For example, the cytosolic isocitrate dehydrogenase 1 (IDH1) and its mitochondrial homolog IDH2 encode enzymes that convert isocitrate to α-ketoglutarate (αKG), a co-factor for TET2 that demethylates DNA. Mutations in IDH1 and IDH2 modify the affinity between the encoded enzymes and their substrates, resulting in conversion of αKG to R-2-hydroxyglutarate (R-2-HG) which inhibits both TET2 and other αKG-dependent enzymes.40 The result is an increase in DNA methylation, altered gene expression, and a block in differentiation. Inhibiting mutant IDH1/2 restores normal IDH function, decreases DNA methylation, and promotes the differentiation of AML cells in vitro and in vivo.21,41
In this study, we report a new mechanism by which mitochondrial pathways control differentiation. We demonstrated that inhibition of POLG and/or reductions in levels of mitochondrial DNA that were not sufficient to impair oxidative phosphorylation led to increased monocytoid differentiation. Previous studies have reported that mitochondrial stress such as inhibition of the mitochondrial protease ClpP42,43 or oxidative stress44 can result in translocation of mitochondrial proteins to the nucleus and alter gene expression. Potentially similar mechanisms could occur upon inhibition of POLG and lead to translocation of proteins from the mitochondria to the nucleus, impacting gene expression to promote differentiation.
Thus, we have identified a novel mechanism by which mitochondria regulate AML fate and differentiation independently of oxidative phosphorylation. Moreover, we highlight POLG inhibitors such as alovudine as potential therapeutic agents for AML.
Supplementary Material
Acknowledgments
We thank Jill Flewelling (Princess Margaret Cancer Center) for administrative assistance.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/104/5/963
Funding
This work was supported by Medivir AB, the Leukemia and Lymphoma Society, the Canadian Institutes of Health Research, the Princess Margaret Cancer Centre Foundation, and the Ministry of Long Term Health and Planning in the Province of Ontario. ADS holds the Barbara Baker Chair in Leukemia and Related Diseases
References
- 1.Döhner H, Weisdorf DJ, Bloomfield CD. Acute Myeloid Leukemia. N Engl J Med. 2015;373(12):1136–1152. [DOI] [PubMed] [Google Scholar]
- 2.Lang BF, Gray MW, Burger G. Mitochondrial genome evolution and the origin of eukaryotes. Annu Rev Genet. 1999;33(1):351–397. [DOI] [PubMed] [Google Scholar]
- 3.Lagadinou ED, Sach A, Callahan K, et al. BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell Stem Cell. 2013;12(3):329–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Skrtić M, Sriskanthadevan S, Jhas B, et al. Inhibition of mitochondrial translation as a therapeutic strategy for human acute myeloid leukemia. Cancer Cell. 2011; 20(5):674–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sriskanthadevan S, Jeyaraju DV, Chung TE, et al. AML cells have low spare reserve capacity in their respiratory chain that renders them susceptible to oxidative metabolic stress. Blood. 2015;125(13):2120–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liyanage SU, Hurren R, Voisin V, et al. Leveraging increased cytoplasmic nucleoside kinase activity to target mtDNA and oxidative phosphorylation in AML. Blood. 2017;129(19):2657–2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaur R, Grasso D, Datta PP, et al. A single mammalian mitochondrial translation initiation factor functionally replaces two bacterial factors. Mol Cell. 2008;29(2):180–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christian BE, Spremulli LL. Mechanism of protein biosynthesis in mammalian mitochondria. Biochim Biophys Acta. 2012;1819: 1035–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Birky CW. Eukaryotic Origins. Science. 1994;266(5183):309–310. [DOI] [PubMed] [Google Scholar]
- 10.Chan SSL, Copeland WC. DNA polymerase gamma and mitochondrial disease: Understanding the consequence of POLG mutations. Biochim Biophys Acta -Bioenerg. 2009;1787(5):312–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bogenhagen DF. Mitochondrial DNA nucleoid structure. Biochim Biophys Acta. 2012;1819(9-10):914–920. [DOI] [PubMed] [Google Scholar]
- 12.Carling PJ, Cree LM, Chinnery PF. The implications of mitochondrial DNA copy number regulation during embryogenesis. Mitochondrion. 2011;11(5):686–692. [DOI] [PubMed] [Google Scholar]
- 13.Lane AN, Fan TW-M. Regulation of mammalian nucleotide metabolism and biosynthesis. Nucleic Acids Res. 2015;43(4):2466–2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mathews CK. Deoxyribonucleotide metabolism, mutagenesis and cancer. Nat Rev Cancer. 2015;15(9):528–539. [DOI] [PubMed] [Google Scholar]
- 15.Kakuda TN. Pharmacology of nucleoside and nucleotide reverse transcriptase inhibitor-induced mitochondrial toxicity. Clin Ther. 2000;22(6):685–708. [DOI] [PubMed] [Google Scholar]
- 16.Gandhi VV, Samuels DC. A review comparing deoxyribonucleoside triphosphate (dNTP) concentrations in the mitochondrial and cytoplasmic compartments of normal and transformed cells. Nucleosides Nucleotides Nucleic Acids. 2011;30(5):317–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katlama C, Ghosn J, Tubiana R, et al. MIV-310 reduces HIV viral load in patients failing multiple antiretroviral therapy: results from a 4-week phase II study. AIDS. 2004;18(9):1299–1304. [DOI] [PubMed] [Google Scholar]
- 18.Rusconi S. Alovudine Medivir. Curr Opin Investig Drugs. 2003;4(2):219–223. [PubMed] [Google Scholar]
- 19.Flexner C, van der Horst C, Jacobson MA, et al. Relationship between plasma concentrations of 3’-deoxy-3’-fluorothymidine (alovudine) and antiretroviral activity in two concentration-controlled trials. J Infect Dis. 1994;170(6):1394–1403. [DOI] [PubMed] [Google Scholar]
- 20.Martin JL, Brown CE, Matthews-Davis N, Reardon JE. Effects of antiviral nucleoside analogs on human DNA polymerases and mitochondrial DNA synthesis. Antimicrob Agents Chemother. 1994;38(12):2743–2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Birendra KC, DiNardo CD. Evidence for Clinical Differentiation and Differentiation Syndrome in Patients With Acute Myeloid Leukemia and IDH1 Mutations Treated With the Targeted Mutant IDH1 Inhibitor, AG-120. Clin Lymphoma Myeloma Leuk. 2016;16(8):460–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller WH, Waxman S. Differentiation induction as a treatment for hematologic malignancies. Oncogene. 2002;21(21):3496–3506. [DOI] [PubMed] [Google Scholar]
- 23.Stegmaier K, Corsello SM, Ross KN, Wong JS, Deangelo DJ, Golub TR. Gefitinib induces myeloid differentiation of acute myeloid leukemia. Blood. 2005; 106(8): 2841–2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Waxman S. Differentiation therapy in acute myelogenous leukemia (non-APL). Leukemia. 2000;14:491–496. [DOI] [PubMed] [Google Scholar]
- 25.Bröske A-M, Vockentanz L, Kharazi S, et al. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat Genet. 2009;41(11): 1207–1215. [DOI] [PubMed] [Google Scholar]
- 26.Gereige L-M, Mikkola HKA. DNA methylation is a guardian of stem cell self-renewal and multipotency. Nat Genet. 2009;41(11): 1164–1166. [DOI] [PubMed] [Google Scholar]
- 27.Challen GA, Sun D, Jeong M, et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet. 2012;44(1):23–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lechman ER, Gentner B, Ng SWK, et al. miR-126 Regulates Distinct Self-Renewal Outcomes in Normal and Malignant Hematopoietic Stem Cells. Cancer Cell. 2016;29(4):602–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Škrtić M, Sriskanthadevan S, Jhas B, et al. Inhibition of Mitochondrial Translation as a Therapeutic Strategy for Human Acute Myeloid Leukemia. Cancer Cell. 2011; 20(5):674–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ghosn J, Quinson A-M, Sabo N, et al. Antiviral activity of low-dose alovudine in antiretroviral-experienced patients: results from a 4-week randomized, double-blind, placebo-controlled dose-ranging trial. HIV Med. 2007;8(3):142–147. [DOI] [PubMed] [Google Scholar]
- 31.Otto MJ. New nucleoside reverse transcriptase inhibitors for the treatment of HIV infections. Curr Opin Pharmacol. 2004; 4(5):431–436. [DOI] [PubMed] [Google Scholar]
- 32.Kim E-Y, Vrang L, Öberg B, Merigan TC. Anti-HIV Type 1 Activity of 3′-Fluoro-3′-Deoxythymidine for Several Different Multidrug-Resistant Mutants. AIDS Res Hum Retroviruses. 2001;17(5):401–407. [DOI] [PubMed] [Google Scholar]
- 33.Shen G, Ma H, Pang F, Ren P, Kuang A. Correlations of 18F-FDG and 18F-FLT uptake on PET with Ki-67 expression in patients with lung cancer: a meta-analysis. Acta Radiol. 2018;59(2):188–195. [DOI] [PubMed] [Google Scholar]
- 34.Bahri H, Laurence L, Edeline J, et al. High Prognostic Value of 18F-FDG PET for Metastatic Gastroenteropancreatic Neuroendocrine Tumors: A Long-Term Evaluation. J Nucl Med. 2014;55(11):1786–1790. [DOI] [PubMed] [Google Scholar]
- 35.Gerbaudo VH, Killoran JH, Kim CK, et al. Pilot study of serial FLT and FDG-PET/CT imaging to monitor response to neoadjuvant chemoradiotherapy of esophageal adenocarcinoma: correlation with histopathologic response. Ann Nucl Med. 2018;32(3):165–174. [DOI] [PubMed] [Google Scholar]
- 36.Lee Y-S, Kennedy WD, Yin YW. Structural Insight into Processive Human Mitochondrial DNA Synthesis and Disease-Related Polymerase Mutations. Cell. 2009;139(2):312–324.2018;32(3): 165–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang TS. Eukaryotic DNA polymerases. Annu Rev Biochem. 1991;60(1):513–552. [DOI] [PubMed] [Google Scholar]
- 38.Martin JL, Brown CE, Matthews-Davis N, Reardon JE. Effects of antiviral nucleoside analogs on human DNA polymerases and mitochondrial DNA synthesis. Antimicrob Agents Chemother. 1994;38(12):2743–2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Warrell RP, Frankel SR, Miller WH, et al. Differentiation therapy of acute promyelocytic leukemia with tretinoin (all-trans-retinoic acid). N Engl J Med. 1991; 324(20):1385–1393. [DOI] [PubMed] [Google Scholar]
- 40.Figueroa ME, Abdel-Wahab O, Lu C, et al. Leukemic IDH1 and IDH2 Mutations Result in a Hypermethylation Phenotype, Disrupt TET2 Function, and Impair Hematopoietic Differentiation. Cancer Cell. 2010;18(6):553–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stein EM, DiNardo CD, Pollyea DA, et al. Enasidenib in mutantIDH2relapsed or refractory acute myeloid leukemia. Blood. 2017;130(6):722–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fiorese CJ, Schulz AM, Lin Y-F, Rosin N, Pellegrino MW, Haynes CM. The Transcription Factor ATF5 Mediates a Mammalian Mitochondrial UPR. Curr Biol. 2016;26(15):2037–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haynes CM, Yang Y, Blais SP, Neubert TA, Ron D. The matrix peptide exporter HAF-1 signals a mitochondrial UPR by activating the transcription factor ZC376.7 in C. elegans. Mol Cell. 2010;37(4):529–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nagaraj R, Sharpley MS, Chi F, et al. Nuclear Localization of Mitochondrial TCA Cycle Enzymes as a Critical Step in Mammalian Zygotic Genome Activation. Cell. 2017;168:210–223.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.