Abstract
Long non-coding RNAs (lncRNAs) are a major class of non-coding RNAs, and the functional deregulations of lncRNAs have been shown to be associated with the development and progression of BC. In this work, we conduct an integrative analysis on five re-annotated lncRNA expression datasets from the Gene Expression Omnibus (GEO) which included a total of 891 BC samples. We identified a five-lncRNA signature that was significantly associated with DFS in the training cohort of 327 patients. We found the five-lncRNA signature could effectively stratify patients in the training dataset into high- and low-risk groups with significantly different DFS (p = 3.29×10−5, log-rank test). The five-lncRNA signature was effectively validated in four independent cohorts, and prognostic analysis results showed that the five-lncRNA signature was independent of clinical prognostic factors, such as BC subtypes and adjuvant treatments. Furthermore, GSEA suggested that the five-lncRNA signature was involved in BC metastasis-related pathways. Our findings indicate that these five lncRNAs may be implicated in BC pathogenesis, and further, these lncRNAs may potentially serve as novel candidate biomarkers for the identification of BC patients at high risk for tumor recurrence.
Keywords: lncRNA signature, prognosis, breast cancer, array re-annotation, tumor recurrence
Introduction
Long non-coding RNA (lncRNA), defined as RNA transcripts of more than 200 base pairs in length, is a major class of ncRNA.1 Recently, several studies have demonstrated that abnormal expression of lncRNAs is closely associated with human complex diseases, especially in cancers.2 A growing number of lncRNAs have been identified and recognized as “oncogenes” or “tumor suppressors”, and functional dysregulations of lncRNAs have been shown to contribute to cancer development, progression, and metastasis.3 For instance, metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) has been shown to play a key role in oral squamous cell carcinoma, and high expression of MALAT1 was related to tumor metastasis.4 Another lncRNA, HOX Transcript Antisense RNA (HOTAIR), has been identified as an “oncogene” in BC-its high expression was associated with metastasis and death in primary breast tumors.5 Growth arrest-specific 5 (GAS5) was found to be downregulated in BC tissues, and its overexpression in the MCF-7 BC cell line furthered growth arrest and apoptosis.6 Expression profiling has revealed highly aberrant lncRNA expression in cancers, which may indicate their potential as possible biomarkers predictive of clinical outcome.7
BC is the most frequent malignancy in women, affecting more than 10% of women in western countries.8 Early diagnosis improvements have been made in BC prognosis by mammographic screening. However, tumor recurrence with local recurrence or distant recurrence following conventional therapies is still a major cause of morbidity and mortality for BC patients.9 To improve BC prognosis analysis, several systems biology approaches have been developed to identify BC prognosis-related lncRNA biomarkers and to construct lncRNA signatures.10, 11 Meng et al. identified a four-lncRNA set through analysis of lncRNA expression profiling in 887 BC patients from GEO datasets using the random survival forest algorithm.10 Zhou et al. discovered a 12-lncRNA biomarker to predict the risk of tumor recurrence in BC patients.11 Both sets of authors tested their lncRNA signatures in three independent datasets. The log-rank p-value of the 12-lncRNA-based signature from Zhou et al. was not significant in one testing dataset (GSE20711, p = 0.289), implying that identification of robust lncRNA signatures remains a challenge. More patient cohorts are needed to validate signatures.
Because lncRNAs related to survival are generally associated with the development and metastasis of cancers, it is critical to identify BC prognosis-related lncRNA signatures through analysis of their biological functions. Therefore, to efficiently identify a robust BC prognosis-related lncRNA signature, we present a systematic pipeline to identify BC-related lncRNA biomarkers through analyzing five lncRNA expression datasets with a total of 891 BC patient samples. A five-lncRNA signature associated with BC both in function and prognosis was identified and successfully validated in four independent validation cohorts. We found that the five-lncRNA signature was involved in important biological processes and pathways of BC. The GSEA analysis also showed similar results, which suggested that the five-lncRNA signature could effectively predict recurrence risk in BC patients and provides a better understanding of the molecular mechanisms underlying BC prognosis.
Methods
Breast cancer patients
For consistency in microarray platforms, BC-related gene expression datasets were measured by the Affymetrix HU133 Plus 2.0 microarray. Corresponding clinical data were obtained from the GEO database (https://www.ncbi.nlm.nih.gov/geo/).12 To analyze the correlations of lncRNA signatures with survival endpoints for BC patients, we selected those datasets that included more than 80 patients with DFS or overall survival (OS) in this study. In total, 891 samples (327 from GSE20685,13 88 from GSE20711,14 252 from GSE21653,15 117 from GSE88770,16 and 107 from GSE5881217) were obtained (Table 1). All selected expression datasets were log2-transformed, then standardized.
Table 1.
Summary of BC-related lncRNA expression datasets and corresponding clinical characteristics.
| Characteristic | GSE20685 (n=327) | GSE20711 (n=88) | GSE21653 (n=252) | GSE88770 (n=117) | GSE58812 (n=107) | |
|---|---|---|---|---|---|---|
| Age(years) | <=50 | 209 | 107 | 43 | ||
| >50 | 118 | 145 | 64 | |||
| Survival status | Living | 244 | 49 | 169 | 89 | 78 |
| Dead | 83 | 39 | 83 | 28 | 29 | |
| Metastasis stage | M0 (Living) | 244 (233) | 76 (73) | |||
| M1 (dead) | 83 (72) | 31 (26) | ||||
| Grade | 1 | 13 | 43 | 13 | ||
| 2 | 5 | 84 | 96 | |||
| 3 | 70 | 119 | 7 | |||
| ER status | Negative | 45 | 110 | 11 | 107 | |
| Positive | 42 | 140 | 106 | 0 | ||
| PR status | Negative | 123 | 37 | 107 | ||
| Positive | 126 | 79 | 0 | |||
| HER2 status | Negative | 62 | 206 | 108 | 107 | |
| Positive | 26 | 27 | 7 | 0 | ||
| Tumor stage | T1 | 101 | ||||
| T2 | 188 | |||||
| T3 | 26 | |||||
| T4 | 12 | |||||
| Lymph node stage | N0 | 137 | ||||
| N1 | 87 | |||||
| N2 | 63 | |||||
| N3 | 40 | |||||
| Subtype | Basal-like | 43 | 24 | 70 | ||
| Her2-enriched | 75 | 20 | 41 | |||
| Luminal A | 89 | 15 | 63 | |||
| Luminal B | 97 | 28 | 68 | |||
| Normal-like | 23 | 1 | 10 | |||
| Chemotherapy | No treatment | 59 | ||||
| CAF | 88 | |||||
| CMF | 61 | |||||
| Paclitaxel | 119 |
Construction of lncRNA expression through re-annotation
Based on the gene expression datasets obtained above, we applied a custom pipeline to re-annotate Affymetrix HU133 Plus 2.0 microarrays by taking advantage of its large amount of probes annotated to thousands of lncRNAs as follows:
All the microarray raw data (CEL files) of five breast cancer cohorts were obtained from the GEO database. The Affymetrix probe sequences were downloaded from the manufacturer’s website (http://www.affymetrix.com) and uniquely mapped to the human genome (hg19) by Bowtie without mismatch. Specific probes of lncRNAs were obtained by matching the chromosomal position of probes to the chromosomal position of lncRNA genes based on annotations from GENCODE (Release 23).18 Through using BEDTools (http://code.google.com/p/bedtools),19 probes completely falling into exons of lncRNAs but without overlapping with protein-coding genes were selected. Expression values of one lncRNA gene detected by at least four probes were retained. The median expression value of multiple probes mapping to the same lncRNA was used to represent the expression level of the lncRNA. For each cohort, the expression data was log2 transformed and normalized by the quantile-normalization approach. Finally, five corresponding lncRNA expression datasets, containing 2673 lncRNAs, were constructed.
Identification of a survival-related lncRNA signature set associated with breast cancer
The largest lncRNA expression dataset, GSE20685, was used to construct the training cohort and analyze the correlation of lncRNAs’ expression with the DFS time based on univariate Cox proportional hazards regression (PHR) analysis. The lncRNAs with the most significant p values (p < 0.05) were selected as candidates. To further help create the predictive signature, we analyzed the co-expression relations between lncRNAs and protein coding genes and inferred the functions of lncRNAs based on the “guilt by association” hypothesis: genes or lncRNAs with similar expression patterns under multiple experimental conditions have high probabilities of sharing similar functions or being involved in common biological pathways. Such co-expression-based approaches have been widely used in previous studies.20, 21, 22 Thus, using the highly co-expressed genes associated with lncRNAs (Pearson correlation coefficients ≥ 0.8 and multiple testing correction p < 0.05), we investigated lncRNAs’ functions through GO and KEGG annotation or enrichment analysis through the Database for Annotation, Visualization and Integrated Discovery (DAVID, https://david.ncifcrf.gov/).23 Through manually searching literature by Google or PubMed, we obtained GO terms or KEGG pathways associated with BC with high confidence. We extracted the functions that play important roles in BC, which we termed “BC-related functions” (Supplementary Table 1). Subsequently, the intersection of the survival-related lncRNA and function-related lncRNA sets was generated. Notably, given more lncRNAs meant more redundancy and a smaller number of lncRNAs would make the model more practical and parsimonious, so we used the forward stepwise approach to find a minimal set of lncRNAs. Forward stepwise, which involves starting with no variables in the model, adding the variable whose inclusion gives the most statistically significant improvement of the fit (if any), and repeating this process until including no variable improves the model to a statistically significant extent. Finally the five-lncRNA signature recognized by forward stepwise was tested in the validation cohort, including the GSE20711, GSE21653, GSE88770 and GSE58812 data sets. The detailed workflow is shown in Figure 1.
Figure 1.
The workflow of identification of BC survival-related five-lncRNA signature.
Subtypes and adjuvant treatments of breast cancer patients
To stratify all BC patients into different BC subtypes, the PAM50 classifier was used by the R package geneFu to divide all five BC patients datasets into Basal-like, Luminal A, Luminal B, Her2 and Normal-like intrinsic subtypes.24 Moreover, the clinical information about treatment of BC patients in the five expression datasets was obtained from the GEO database. In the GSE20685 dataset, 268 patients were treated with three different drug therapies, including CAF (cyclophosphamide, doxorubicin and fluorouracil), CMF (cyclophosphamide, methotrexate and fluorouracil), and paclitaxel alone.
Statistical analysis
We fit a multivariate Cox PHR model to a training cohort in order to construct a lncRNA signature. This signature was then used to identify the best cutoff value for BC patient stratification. The time-dependent receiver operating characteristic (ROC) curve was then calculated to compare the sensitivity and specificity of risk prediction of the lncRNA signature set for recurrence-free survival.25 BC patients in the training cohort were classified into high- and low-risk groups according to the stratification cutoff obtained above. Subsequently, Kaplan-Meier survival curve analysis and a log-rank test were used to compare the difference of recurrence-free survival between high- and low-risk groups in each set of the validation cohort. Multivariate analyses with Cox PHR were conducted to test whether the lncRNA signature set is independent of other clinicopathological factors, including age, survival status, grade, clinical stage, estrogen receptor (ER) status, subtype, and adjuvant treatment. The hazard ratio (HR) and its 95% confidence intervals (CI) were calculated with the Cox PHR model.26
Functional enrichment analysis
Based on lncRNA and gene relationships calculated by the Pearson correlation coefficients, functional enrichment analysis for lncRNAs in the lncRNA signature set was performed using DAVID. GSEA was performed by the JAVA program (http://software.broadinstitute.org/gsea/downloads.jsp) using the MSigDB C2 Canonical pathways gene set collection, which contains 1320 gene sets. Gene sets with a false discovery rate (FDR) value less than 0.05 after performing 1000 permutations were considered to be significantly enriched.27 Statistical analysis was implemented using the R software (www.rproject.org).26
Results
Acquisition of lncRNA expression datasets through array re-annotation
Because a number of large-scale gene expression datasets are available in the GEO database, an integrative analysis of re-annotated lncRNA expression datasets would provide good statistical power to capture the expression changes of lncRNAs in disease condition. To this end, we collected BC expression datasets measured on the Affymetrix HU133 Plus 2.0 microarray platform to identify potential lncRNA prognostic biomarkers. After a thorough search of the GEO database, we identified four gene expression datasets with DFS time (GSE20685, GSE21653, GSE20711, and GSE88770) and one with OS time (GSE58812). Together, these data sets include a total of 891 BC patient samples (Table 1). Next, the corresponding lncRNA expression datasets were constructed using array re-annotation analysis as described in the Methods section. Relevant clinical information including age, grade, Estrogen receptor (ER) status, progesterone receptor (PR) status, HER2 status, tumor stage, lymph node stage, metastasis status, subtype, chemotherapy information and survival status (if patients died because of the occurrence of metastasis) for the five lncRNA expression datasets are summarized in Table 1.
Identification of a five-lncRNA signature for breast cancer survival
In the five BC-related lncRNA expression datasets, the GSE20685 data set (with 327 patients-the largest sample size of the selected data sets) was used to construct the training cohort. First, survival-related lncRNAs were identified based on univariate Cox PHR analysis, and the 384 lncRNAs with the most significant p values were selected (p < 0.05, shown in Supplementary Table 2). Next, we analyzed co-expression associations of lncRNAs and protein coding genes and inferred the functions of lncRNAs based on the guilt by association hypothesis (Supplementary Table 3). Then, 46 lncRNAs and 193 genes with high co-expression relations (Pearson correlation coefficients ≥ 0.8 and multiple testing correction p < 0.05) were obtained. Using the highly co-expressed genes associated with lncRNAs, we investigate the lncRNAs’ functions through GO and KEGG annotation or enrichment analysis by DAVID. To obtain GO terms or KEGG pathways associated with BC with high confidence, we manually searched literature by Google and PubMed. Through functional annotation, we found that all of the 46 lncRNAs were involved with BC-related biological functions or pathways, and thus we retained these lncRNAs for subsequent analysis (Supplementary Table 1). By the intersecting of these two lncRNA sets (those associated with survival and those have significant function), a 15 lncRNA signature set (Supplementary Table 4) was then generated for subsequent analysis. The detailed workflow is described in the Methods section and shown Figure 1.
The univariate z-score for each lncRNA was calculated to characterize its predictive power. More negative (positive) z-scores indicate longer (shorter) OS time.28 Considering the issue that some of the lncRNA predictors may not be independent, we applied forward stepwise approach based on the 15-lncRNAs in Supplementary Table 4 to select a more effective signature. Starting with the lncRNA “RP11-13L2.4” (with the largest univariate z-score), we added one lncRNA that showed the most association with longer or shorter OS at each step and evaluated if the prognostic performance could be improved. We repeated this process until no improvement could be achieved. Finally, five lncRNAs-including ENSG00000261295 (RP11-524D16_A.3), ENSG00000228630 (HOTAIR), ENSG00000223764 (AL645608.1), ENSG00000265148 (TSPOAP1-AS1) and ENSG00000261179 (RP11-13L2.4) were selected. Moreover, to systematically evaluate the prognostic performance of the five-lncRNA signature, we compared the survival result with those from another lncRNA set with different sizes based on the absolute z-scores from the 15-lncRNAs. We found good performance of the five-lncRNA signature in the five independent datasets (Supplementary Figure 1). Among these lncRNAs, HOTAIR has been validated to be associated with BC. Several studies have demonstrated that this lncRNA contributes to BC development.5, 29, 30 Overexpression of HOTAIR predicts a poor prognosis in BC patients.
Determination and analysis of the five-lncRNA signature in the training cohort
We hypothesized that the identified five-lncRNA signature strongly contributes to the survival of BC. Therefore, we took this lncRNA set as an independent predictive signature to predict the risk of tumor recurrence for BC patients. A multivariate Cox PHR analysis was performed and the estimated regression coefficients (see the Methods section) are as follows:
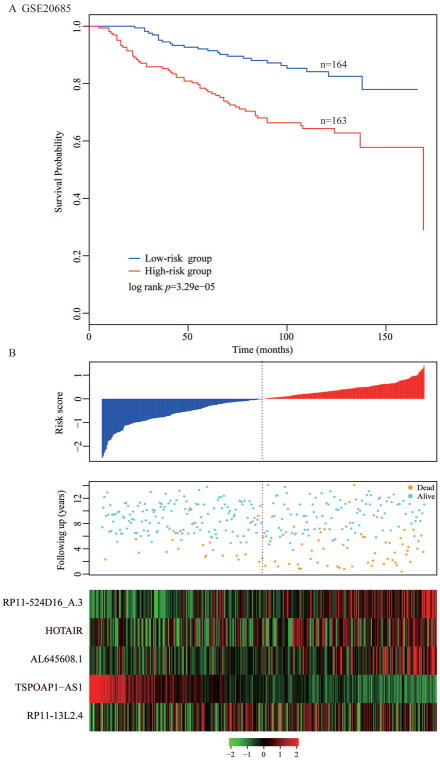
where x1 represents the expression of RP11-524D16_A.3, x2 represents HOTAIR, x3 AL645608.1, x4 TSPOAP1-AS1, and x5 RP11-13L2.4. In the training cohort, the time-dependent ROC curves analysis for the five-lncRNA signature achieved an area under the ROC curve (AUC) of 0.69 at five years of recurrence-free survival. We calculated a five-lncRNA expression-based risk score for each patient and classified all BC patients in the training cohort into high-risk group (n = 163) and low-risk group (n = 164) by using the median risk score (0.1121) as the cutoff point. We found that BC patients with low-risk scores have the better recurrence-free survival outcome. Further, the survival time of the high-risk group was significantly shorter than the low risk group (p = 3.29×10−05, log-rank test, Figure 2A).
Figure 2. Determination and analysis of the five-lncRNA signature in the training cohort.
(A) Kaplan–Meier survival curves of DFS survival between high-risk and low-risk patients in the training cohort. (B) The distribution of patients’ risk score and recurrence status, and the expression pattern of the five-lncRNA signature.
Distribution of the five-lncRNA expression-based risk scores, the survival status, and the expression pattern of five lncRNA biomarkers in the 327 breast patients belonging to the training cohort is shown in Figure 2B. Of these five lncRNAs, two were protective lncRNAs (TSPOAP1-AS1 and RP11-13L2.4) whose high expressions were associated with better prognosis. In contrast, high expressions of the remaining three (RP11-524D16_A.3, HOTAIR, and AL645608.1) were associated with poor prognosis.
Validation of the five-lncRNA signature in four independent cohorts
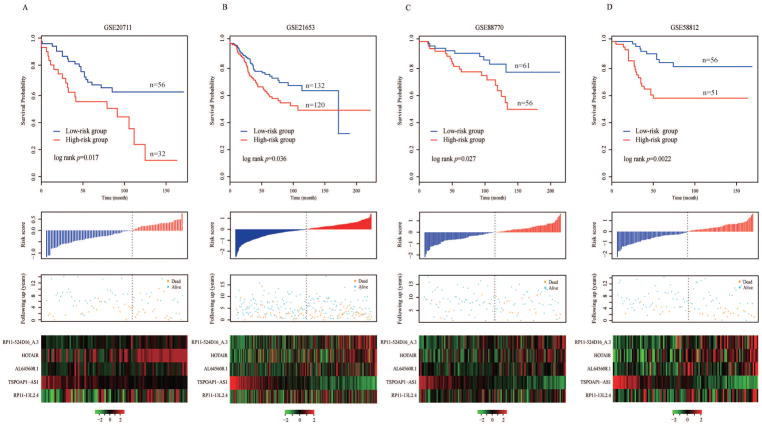
To evaluate the robustness of the five-lncRNA signature in predicting the risk of tumor recurrence for BC patients, the five-lncRNA signature was then tested for its predictive power in the validation cohort, which consisted of the remaining GSE20711, GSE21653, GSE88770 and GSE58812 data sets. By using the same model and the cutoff point derived from the training cohort, 88 BC patients in GSE20711 with DFS information were stratified into the high- and low-risk groups (n = 32 and 56, respectively). As shown in Figure 3A, we found a significant difference between the high-risk group and low-risk group (p = 0.017, log-rank test) by using five lncRNA risk scores. The HR was 2.11 (95% CI: 1.13–3.96; p = 0.02) based on univariate analysis. The AUC was 0.59 at the survival time of five years in GSE20711.
Figure 3. Validation of the five-lncRNA signature in four independent cohorts.
Kaplan–Meier survival curves of DFS or OS between high-risk and low-risk patients, the distributions of the risk score, survival status and lncRNA expression values associated with breast cancer patients in (A) GSE20711, (B) GSE21653, (C) GSE88770 and (D) GSE58812.
Further validation of the five-lncRNA signature was conducted in GSE21653, which included 252 samples. Similar to the results generated by the training cohort and GSE20711, the five-lncRNA signature was used to efficiently predict the risk of tumor recurrence. Utilizing the risk score formula estimated from the training cohort, the five-lncRNA signature was able to classify 252 BC patients into the high-risk group (n = 120) and low-risk group (n = 132) with significantly different recurrence-free survival (p = 0.036, log-rank test). The recurrence-free survival time was significantly shorter in the high-risk group when compared with the low-risk group (Figure 3B). At five years, the respective absolute difference in DFS between the low-risk group and high-risk group was 15% (75.5% versus 60.5%). The HR of high-risk group versus low-risk group for DFS was 1.59 (95% CI: 1.03–2.46; p = 0.037). The AUC of time-dependent ROC curves for the five-lncRNA signature in GSE21653 was 0.64 at five years.
Another validation of the five-lncRNA signature in GSE88770 with 117 BC patients again showed its good performance when predicting the risk of tumor recurrence (Figure 3C). The five-lncRNA signature stratified all patients into the high- and low-risk groups (n = 56 and 61, respectively) by using the same risk score formula and cutoff as above. A significant difference between the high-risk group and the low-risk group (p = 0.027, log-rank test) was found. The HR for recurrence-free survival of high-group versus low-group was 2.39 (95% CI: 1.08–5.27; p = 0.032). The corresponding AUC was 0.63 at five-year DFS time in GSE88770.
Final validation was performed in GSE58812, which contains data on 107 BC patients. Similar to the results above, the five-lncRNA signature enabled us to successfully stratify all patients into high- and low-risk groups (n = 51 and 56, respectively). As we can see in Figure 3D, BC patients in the high-risk group had significantly shorter recurrence-free survival time than those in the low-risk group (p = 0.0022, log-rank test). The five-year recurrence-free survival rate of the high-risk group was 58.1% but 83.6% for the low-risk group. The HR of the high-risk versus low-risk group for OS was 3.19 (95% CI: 1.45–7.02; p = 0.0039). The AUC for the GSE58812 data set was 0.72 at five years of recurrence-free survival.
The distribution of risk score, survival status and expression pattern of five lncRNA biomarkers in the validation cohort are shown in Figure 3. In addition to this, sensitivity and specificity of classification performance generated by the five-lncRNA signature are also presented in Supplementary Table 5. These results suggest that the five-lncRNA signature may potentially play an essential role in BC prognostic prediction.
Independent predictive power of the five-lncRNA signature from clinicopathological factors
To further investigate whether predictive power of the five-lncRNA signature was independent of clinicopathological factors in the training cohort, such as age, tumor grade (or stage), Estrogen receptor (ER) status, progesterone receptor (PR) status, HER2 status, lymph node stage and metastasis status, a multivariate Cox PHR analysis was performed. The analysis results from the training cohort suggested that the five-lncRNA signature (HR = 2.38; 95% CI: 1.24–4.54; p = 0.009), metastasis status, Her2-enriched, Luminal A and Luminal B were five independent prognostic factors for BC patients (Supplementary Table 6). In GSE20711, the five-lncRNA signature (HR = 2.65; 95% CI: 1.17–6.02; p = 0.02), Her2-enriched and normal-like were three independent prognostic factors for BC patients. In GSE21653 only the five-lncRNA signature (HR = 1.8; 95% CI: 1.10–2.96; p = 0.02) and Luminal A were significantly correlated with recurrence-free survival of BC patients based on the multivariate analysis. In GSE88770, only the five-lncRNA signature was correlated with recurrence-free survival of BC patients (HR = 2.03; 95% CI: 0.90–4.56; p = 0.086). In another validation dataset, GSE58812, the result showed both the five-lncRNA signature (HR = 2.31; 95% CI: 1.00–5.28; p = 0.047) and metastasis status were correlated with the OS of BC patients.
The impact of subtypes and adjuvant treatments on the survival of breast cancer patients
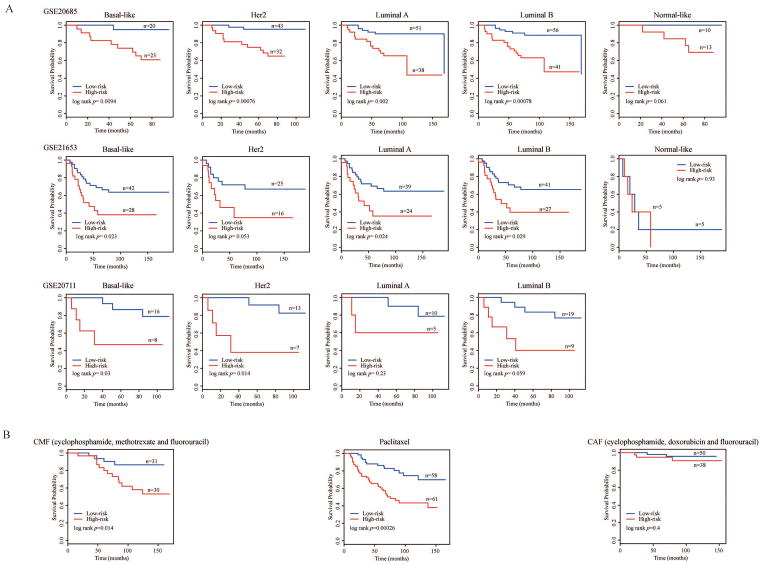
From a clinical point of view, patients with different subtypes are managed and treated as different diseases.31 Therefore, we investigated the impact of patient subtypes on patient survival. We used the PAM50 classifier to stratify BC patients from three mRNA expression datasets (GSE20685, GSE21653 and GSE20711) into Basal-like, Luminal A, Luminal B, Her2 and Normal-like subtypes (Table 1). We observed poor subtype classification in the other two expression datasets (GSE58812 and GSE88770) that were recorded as triple-negative and invasive lobular carcinoma respectively, both of which substantially belonged to the intrinsic subtypes of BC.16, 17 The multivariate Cox PHR analysis results showed that the five-lncRNA signature was still an independent prognosis factor (p values were 0.009, 0.019 and 0.020 in the GSE20685, GSE21653 and GSE20711 data sets, respectively; see Supplementary Table 6). Moreover, we found that the five-lncRNA signature generated good performance for predicting the survival benefit in specific BC subtypes (Figure 4A).
Figure 4. The impact of subtypes and chemotherapy treatments on the survival of BC patients.
Kaplan–Meier survival curves of DFS between high-risk and low-risk patients in different intrinsic subtypes of GSE20685, GSE21653 and GSE20711 (A), and adjuvant treatments (B).
We further investigated the impact of the three specified treatment regimens on patient survival. Within the five expression datasets, GSE20685 contained the adjuvant treatment information, in which 268 patients were treated with three different drug therapies: CAF (cyclophosphamide, doxorubicin and fluorouracil), CMF (cyclophosphamide, methotrexate and fluorouracil), and paclitaxel alone. After adding the treatment effects, the multivariate Cox PHR analysis showed that the five-lncRNA signature was still an independent prognostic factor (p = 0.009, Supplementary Table 6). Furthermore, the patients treated with CMF and paclitaxel could be successfully divided into high- or low-risk groups (log-rank p = 0.014 and 0.00026, respectively, Figure 4B) using the five-lncRNA signature. However, patients treated with CAF could not be successfully grouped (log-rank p = 0.4, Figure 4B).
Identification of the five-lncRNA-signature associated biological pathways
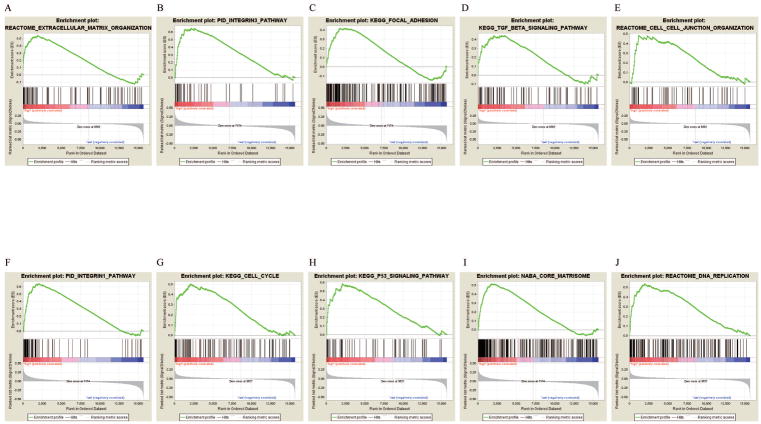
The strong stratification power of the five-lncRNA signature in predicting recurrence risk of BC patients could be attributed to their crucial roles in tumor development or metastasis. Therefore, GSEA analysis was performed to identify lncRNA associated pathways in all five gene expression datasets. For each gene expression dataset, genes were ranked on the basis of differential significance between the high- and low-risk groups, which were classified by the five-lncRNA signature in the lncRNA expression dataset. Then, gene sets were considered significantly enriched if the nominal p-value was less than 0.005 and the FDR less than 0.05 based on a canonical pathways gene set from the MSigDB database. In the GSEA enrichment results, we observed that the “Extracellular matrix organization” pathway was enriched in the high risk groups of all five cohorts. Several studies have demonstrated that this pathway is associated with the development of BC. Furthermore, several cancer related pathways, such as the “Integrin1 pathway”, “Integrin3 pathway”, “Cell cycle”, “Focal adhesion”, “P53 signaling pathway”, “TGF-beta signaling pathway”, “Core matrisome”, “Cell-cell junction organization” and “DNA replication” pathways, were enriched in the high-risk groups in most of the five test cohorts.32, 33 In summary, the GSEA analysis results implied that the five-lncRNA signature was associated with the BC development and progress (Supplementary Table 7 and Figure 5).
Figure 5. The GSEA analysis results in five datasets.
GSEA validated enhanced activity of (A) “Extracellular matrix organization”, (B) “Integrin3 pathway”, (C) “Focal adhesion”, (D) “TGF-beta signaling pathway”, (E) “Cell-cell junction organization”, (F) “Integrin1 pathway”, (G) “Cell cycle”, (H) “P53 signaling pathway”, (I) “Core matrisome” and (J) “DNA replication” in high risk score group.
Comparison of the five-lncRNA signature with other breast cancer prognostic signatures
We compared the prognostic value of the five-lncRNA signature to that of different gene sets and lncRNA sets used for risk stratification of BC: the genes used in the 70-gene MammaPrint profile,28 the genes of Oncotype DX,29 the 12-lncRNAs published by Zhou et al.,11 the 4-lncRNAs published by Meng et al.,10 “rank-based” five-lncRNA signature, and five-lncRNAs related with function. For the “rank-based” five-lncRNA signature, the univariate Cox PHR analysis generated 384 lncRNAs that are significantly associated with survival (p < 0.05). We directly extracted the five lncRNAs with the most significant Cox PHR p values as the “rank-based” five lncRNA signature. For the functionally-related five-lncRNA signature, we selected the five lncRNAs showing the highest expression correlations with the 193 genes that were associated with BC-related biological functions.
We calculated the area under the ROC curves and p-values for each possible signature in the five cohorts (Supplementary Table 8). In the GSE20685 cohort, all signatures tested were successful with similar performances. The 70-gene classifier had poor prediction performances on the GSE58812, GSE88770, and GSE20711 data sets. Moreover, prognostic results generated by the established classifier Oncotype DX showed poor prognostic value when applied to the GSE58812 and GSE88770 data sets (Supplementary Table 8). Subsequently, we used the 12-lncRNA prognostic model published by Zhou et al. and the four-lncRNA prognostic model published by Meng et al. in five independent cohorts. We found that the Zhou et al. classifier had poor prediction performances in two datasets (GSE58812 and GSE20711) while the Meng et al. classifier exhibited poor performance on three datasets (GSE21653, GSE58812, and GSE20711) respectively.10, 11 The rank-based five-lncRNA signature and five-lncRNAs related to function only had good performance on the GSE20685 data set. This result showed that our approach using both survival and functional information has better performance in predicting recurrent risk in BC patients.
Discussion
With the recent development of clinical management and treatment of BC, some prognostic factors, including stage, lymph node status, tumor size, tumor grade, lymphatic and vascular invasion were considered to be associated with the mortality rate in BC. Simultaneously, genes or miRNAs detected by high-throughput biological technologies have been widely used to predict tumor metastasis and cancer recurrence and ultimately survival of BC patients.34, 35 A major class of non-coding RNAs, lncRNAs, provide a promising opportunity to predict the BC recurrence risk as a supplement to genes or miRNAs.36 However, few lncRNA signatures constructed by multiple lncRNAs have been verified and linked to BC prognosis, and patterns of recurrence at lncRNA levels in BC have remained poor. Therefore, it is necessary to develop a systematic pipeline to identify lncRNA signature sets capable of predicting the survival of BC patients.
In this study, we developed a lncRNA function-based prognosis model to systematically identify a five-lncRNA signature through integration analysis of five gene expression profiles with 891 BC samples from the GEO, including GSE20685, GSE21653, GSE20711, GSE88770, and GSE58812. Through re-annotation, five lncRNA expression datasets with matched disease samples were generated. We chose the largest dataset (GSE20685) as the training cohort, while the other four we kept aside for validation. Fifteen lncRNAs associated with BC prognosis and biological functions were generated through integration analysis. Using this 15-lncRNA subset, we further applied a forward stepwise approach to find the minimal set of five lncRNAs and used these to construct a five-lncRNA signature to predict recurrence risk in BC patients.28 Based on the risk score calculated by the lncRNA expression data of five lncRNAs, we efficiently separated the training cohort into high-risk and low-risk groups. Additionally, five lncRNAs’ expression levels were significantly different between the two groups. Validation results showed that the five-lncRNA signature was reliable and robust in the prediction of BC recurrence risk.
Furthermore, a multivariate Cox PHR analysis was performed to investigate predictive power of the five-lncRNA signature. This analysis demonstrated that the five-lncRNA signature was an independent predictor (in addition to clinicopathological factors) in both the training and validation cohorts. We also found the five-lncRNA signature could generate good performances for predicting the survival benefit in most specific BC subtypes and chemotherapy treatments. However, the five-lncRNA signature failed to distinguish high-risk from low-risk groups within “normal-like” BC subtype for the GSE20685 and GSE21653 data sets; we believe the comparatively poor performance on these two data sets is due to the high consistency between the “normal-like” BC subtype and normal breast tissue.37
Moreover, GSEA analysis for gene expression data in all five datasets analyzed the difference between high- and low-risk groups as stratified by the five-lncRNA signature. Several BC-related pathways, such as “Extracellular matrix organization”, “Integrin1 pathway”, “Integrin3 pathway”, “Cell cycle”, “Focal adhesion”, “P53 signaling pathway”, “TGF-beta signaling pathway”, “Core matrisome”, “Cell-cell junction organization” and “DNA replication”, were significantly enriched in the high risk group. The GSEA enrichment result also suggested that the five-lncRNA signature may be involved in BC-related biological pathways and their functional dysregulations may contribute to BC recurrence.
We now discuss several limitations to this study. First, highly heterogeneous patient cohorts were used in the evaluation of prognostic performance: several clinical factors, such as subtype, grade, ER status, and HER2 status showed large between-cohort variability. Second, only limited clinical information was available. Adjuvant treatment information only in GSE20685 could be used, the metastasis stage information was recorded only in GSE20685 and GSE58812. These limitations potentially hamper capturing an accurate prognostic characterization of the five-lncRNA signature and weaken potential comparisons with other published signatures in BC. Indeed, only moderate sensitivity and specificity generated by the five-lncRNA signature were observed in our work.
Also, we note that some published signatures were developed for patients associated with specific clinical conditions. For example, the MammaPrint profile was developed in solely untreated patients to classify patients which may or may not benefit from endocrine treatments.38 It comes as no surprise then that these specific signatures cannot achieve optimal prognostic performance using the five heterogeneous datasets in our study. Therefore, more comprehensive and homogeneous datasets are needed assess the five-lncRNA signature before clinical applications. Finally, nearly most of cellular mechanisms and numerous pathways were associated with BC development and progress. Rigorous definition of “BC-related functions” should be applied in patients of distinct BC subtypes or specific clinical conditions. Then, the BC-related functions generated by this strict definition should provide more accurate and representative information for identification of functional roles of these five lncRNAs.
In summary, we have presented a systematic approach for BC prognostic analysis and identified a robust five-lncRNA signature. We expect this robust signature to provide clues on biological behaviors as well as prognostic characteristics of breast tumors in clinical tests. Further, the research framework used in our study for identifying prognostic markers may help provide guidance in prognostic analysis for future cancer-related lncRNA expression profile studies. The five-lncRNA signature may indicate the potential roles of lncRNAs in BC pathogenesis and have clinical implications as molecular diagnostic markers and therapeutic targets in BC patients.
Supplementary Material
The comparison results with other lncRNA sets with different sizes based on the absolute z-scores from 15 lncRNAs.
Novelty and Impact.
Identification of a survival-related lncRNA signature has been a big challenge in breast cancer (BC) research. We have developed a systematic approach to identify a five-lncRNA signature, and we have shown this signature to be significantly associated with disease-free survival (DFS). We found the five-lncRNA signature could classify patients into high- and low-risk groups with significantly different DFS in a training dataset. Subsequently, the five-lncRNA signature was effectively validated in four cohorts. Prognostic results demonstrated that the signature was independent of subtype classification and adjuvant treatment. In addition, Gene Set Enrichment Analysis (GSEA) results suggested that the five-lncRNA signature was involved in BC metastasis-related pathways and indicated that these five lncRNAs may potentially serve as novel prognostic biomarkers for BC patients.
Acknowledgments
This work was supported by the National High Technology Research and Development Program of China [863 Program, Grant No. 2014AA021102], the National Program on Key Basic ResearchProject [973 Program, Grant Nos. 2014CB910504], the National Natural Science Foundation of China [Grant Nos. 91439117, 61473106, 61573122, 31601076], Wu lien-teh youth science fund project of Harbin medical university [Grant Nos. WLD-QN1407]. The Postdoctoral project of Heilongjiang Province (Grant Nos. LBH-Z14130), and the National Cancer Institute (USA) [Grant Nos. R01CA200987, R01CA158472, and U24CA210954]. We would like to thank Dr. Gabriel Odom for revising the manuscript.
Abbreviations
- LncRNAs
Long non-coding RNAs
- BC
Breast cancer
- GEO
Gene expression omnibus
- DFS
Disease-free survival
- OS
overall survival
- GSEA
Gene set enrichment analysis
- MALAT1
Metastasis-associated lung adenocarcinoma transcript 1
- HOTAIR
HOX Transcript Antisense RNA
- GAS5
Growth arrest-specific 5
- ROC
Receiver operating characteristic
- AUC
an area under the ROC curve
- ER
Estrogen receptor
- PR
progesterone receptor
- FDR
false discovery rate
- Cox PHR
Cox proportional hazards regression
- HR
hazard ratio
Footnotes
Conflict of interest
No potential conflicts of interest were disclosed.
References
- 1.Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene. 2012;31:4577–87. doi: 10.1038/onc.2011.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hauptman N, Glavac D. Long non-coding RNA in cancer. International journal of molecular sciences. 2013;14:4655–69. doi: 10.3390/ijms14034655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nature reviews Genetics. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 4.Zhou X, Liu S, Cai G, Kong L, Zhang T, Ren Y, Wu Y, Mei M, Zhang L, Wang X. Long Non Coding RNA MALAT1 Promotes Tumor Growth and Metastasis by inducing Epithelial-Mesenchymal Transition in Oral Squamous Cell Carcinoma. Scientific reports. 2015;5:15972. doi: 10.1038/srep15972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28:195–208. doi: 10.1038/onc.2008.373. [DOI] [PubMed] [Google Scholar]
- 7.Gibb EA, Vucic EA, Enfield KS, Stewart GL, Lonergan KM, Kennett JY, Becker-Santos DD, MacAulay CE, Lam S, Brown CJ, Lam WL. Human cancer long non-coding RNA transcriptomes. PloS one. 2011;6:e25915. doi: 10.1371/journal.pone.0025915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sorensen KP, Thomassen M, Tan Q, Bak M, Cold S, Burton M, Larsen MJ, Kruse TA. Long non-coding RNA HOTAIR is an independent prognostic marker of metastasis in estrogen receptor-positive primary breast cancer. Breast cancer research and treatment. 2013;142:529–36. doi: 10.1007/s10549-013-2776-7. [DOI] [PubMed] [Google Scholar]
- 9.Lin PH, Yeh MH, Liu LC, Chen CJ, Tsui YC, Su CH, Wang HC, Liang JA, Chang HW, Wu HS, Yeh SP, Li LY, et al. Clinical and pathologic risk factors of tumor recurrence in patients with node-negative early breast cancer after mastectomy. Journal of surgical oncology. 2013;108:352–7. doi: 10.1002/jso.23403. [DOI] [PubMed] [Google Scholar]
- 10.Meng J, Li P, Zhang Q, Yang Z, Fu S. A four-long non-coding RNA signature in predicting breast cancer survival. Journal of experimental & clinical cancer research : CR. 2014;33:84. doi: 10.1186/s13046-014-0084-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou M, Zhong L, Xu W, Sun Y, Zhang Z, Zhao H, Yang L, Sun J. Discovery of potential prognostic long non-coding RNA biomarkers for predicting the risk of tumor recurrence of breast cancer patients. Scientific reports. 2016;6:31038. doi: 10.1038/srep31038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, Yefanov A, Lee H, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic acids research. 2013;41:D991–5. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kao KJ, Chang KM, Hsu HC, Huang AT. Correlation of microarray-based breast cancer molecular subtypes and clinical outcomes: implications for treatment optimization. BMC cancer. 2011;11:143. doi: 10.1186/1471-2407-11-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dedeurwaerder S, Desmedt C, Calonne E, Singhal SK, Haibe-Kains B, Defrance M, Michiels S, Volkmar M, Deplus R, Luciani J, Lallemand F, Larsimont D, et al. DNA methylation profiling reveals a predominant immune component in breast cancers. EMBO molecular medicine. 2011;3:726–41. doi: 10.1002/emmm.201100801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sabatier R, Finetti P, Cervera N, Lambaudie E, Esterni B, Mamessier E, Tallet A, Chabannon C, Extra JM, Jacquemier J, Viens P, Birnbaum D, et al. A gene expression signature identifies two prognostic subgroups of basal breast cancer. Breast cancer research and treatment. 2011;126:407–20. doi: 10.1007/s10549-010-0897-9. [DOI] [PubMed] [Google Scholar]
- 16.Metzger-Filho O, Michiels S, Bertucci F, Catteau A, Salgado R, Galant C, Fumagalli D, Singhal SK, Desmedt C, Ignatiadis M, Haussy S, Finetti P, et al. Genomic grade adds prognostic value in invasive lobular carcinoma. Annals of oncology : official journal of the European Society for Medical Oncology. 2013;24:377–84. doi: 10.1093/annonc/mds280. [DOI] [PubMed] [Google Scholar]
- 17.Jezequel P, Loussouarn D, Guerin-Charbonnel C, Campion L, Vanier A, Gouraud W, Lasla H, Guette C, Valo I, Verriele V, Campone M. Gene-expression molecular subtyping of triple-negative breast cancer tumours: importance of immune response. Breast cancer research : BCR. 2015;17:43. doi: 10.1186/s13058-015-0550-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrow J, Denoeud F, Frankish A, Reymond A, Chen CK, Chrast J, Lagarde J, Gilbert JG, Storey R, Swarbreck D, Rossier C, Ucla C, et al. GENCODE: producing a reference annotation for ENCODE. Genome biology. 2006;7(Suppl 1):S41–9. doi: 10.1186/gb-2006-7-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, Attardi LD, Regev A, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–19. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan X, Hu Z, Feng Y, Hu X, Yuan J, Zhao SD, Zhang Y, Yang L, Shan W, He Q, Fan L, Kandalaft LE, et al. Comprehensive Genomic Characterization of Long Non-coding RNAs across Human Cancers. Cancer cell. 2015;28:529–40. doi: 10.1016/j.ccell.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic acids research. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, Quackenbush JF, Stijleman IJ, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009;27:1160–7. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heagerty PJ, Lumley T, Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000;56:337–44. doi: 10.1111/j.0006-341x.2000.00337.x. [DOI] [PubMed] [Google Scholar]
- 26.Moreno-Betancur M, Sadaoui H, Piffaretti C, Rey G. Survival Analysis with Multiple Causes of Death: Extending the Competing Risks Model. Epidemiology. 2017;28:12–9. doi: 10.1097/EDE.0000000000000531. [DOI] [PubMed] [Google Scholar]
- 27.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lossos IS, Czerwinski DK, Alizadeh AA, Wechser MA, Tibshirani R, Botstein D, Levy R. Prediction of survival in diffuse large-B-cell lymphoma based on the expression of six genes. The New England journal of medicine. 2004;350:1828–37. doi: 10.1056/NEJMoa032520. [DOI] [PubMed] [Google Scholar]
- 29.Avazpour N, Hajjari M, Tahmasebi Birgani M. HOTAIR: A Promising Long Non-coding RNA with Potential Role in Breast Invasive Carcinoma. Frontiers in genetics. 2017;8:170. doi: 10.3389/fgene.2017.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xue X, Yang YA, Zhang A, Fong KW, Kim J, Song B, Li S, Zhao JC, Yu J. LncRNA HOTAIR enhances ER signaling and confers tamoxifen resistance in breast cancer. Oncogene. 2016;35:2746–55. doi: 10.1038/onc.2015.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheang MC, Voduc KD, Tu D, Jiang S, Leung S, Chia SK, Shepherd LE, Levine MN, Pritchard KI, Davies S, Stijleman IJ, Davis C, et al. Responsiveness of intrinsic subtypes to adjuvant anthracycline substitution in the NCIC.CTG MA. 5 randomized trial. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:2402–12. doi: 10.1158/1078-0432.CCR-11-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Felding-Habermann B, O’Toole TE, Smith JW, Fransvea E, Ruggeri ZM, Ginsberg MH, Hughes PE, Pampori N, Shattil SJ, Saven A, Mueller BM. Integrin activation controls metastasis in human breast cancer. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:1853–8. doi: 10.1073/pnas.98.4.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naba A, Clauser KR, Hoersch S, Liu H, Carr SA, Hynes RO. The matrisome: in silico definition and in vivo characterization by proteomics of normal and tumor extracellular matrices. Molecular & cellular proteomics : MCP. 2012;11:M111 014647. doi: 10.1074/mcp.M111.014647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thewes B, Prins J, Friedlander M. 70-Gene Signature in Early-Stage Breast Cancer. The New England journal of medicine. 2016;375:2199–200. doi: 10.1056/NEJMc1612048. [DOI] [PubMed] [Google Scholar]
- 35.Lerebours F, Cizeron-Clairac G, Susini A, Vacher S, Mouret-Fourme E, Belichard C, Brain E, Alberini JL, Spyratos F, Lidereau R, Bieche I. miRNA expression profiling of inflammatory breast cancer identifies a 5-miRNA signature predictive of breast tumor aggressiveness. International journal of cancer. 2013;133:1614–23. doi: 10.1002/ijc.28171. [DOI] [PubMed] [Google Scholar]
- 36.Niknafs YS, Han S, Ma T, Speers C, Zhang C, Wilder-Romans K, Iyer MK, Pitchiaya S, Malik R, Hosono Y, Prensner JR, Poliakov A, et al. The lncRNA landscape of breast cancer reveals a role for DSCAM-AS1 in breast cancer progression. Nature communications. 2016;7:12791. doi: 10.1038/ncomms12791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dennison JB, Shahmoradgoli M, Liu W, Ju Z, Meric-Bernstam F, Perou CM, Sahin AA, Welm A, Oesterreich S, Sikora MJ, Brown RE, Mills GB. High Intratumoral Stromal Content Defines Reactive Breast Cancer as a Low-risk Breast Cancer Subtype. Clinical cancer research : an official journal of the American Association for Cancer Research. 2016;22:5068–78. doi: 10.1158/1078-0432.CCR-16-0171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bedard PL, Mook S, Piccart-Gebhart MJ, Rutgers ET, Van’t Veer LJ, Cardoso F. MammaPrint 70-gene profile quantifies the likelihood of recurrence for early breast cancer. Expert opinion on medical diagnostics. 2009;3:193–205. doi: 10.1517/17530050902751618. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The comparison results with other lncRNA sets with different sizes based on the absolute z-scores from 15 lncRNAs.