ABSTRACT
The mammalian nucleolar proteins nucleostemin and GNL3-like (GNL3L) are encoded by paralogous genes that arose from an ancestral invertebrate gene, GNL3. Invertebrate GNL3 has been implicated in ribosome biosynthesis, as has its mammalian descendent, GNL3L. The paralogous mammalian nucleostemin protein has, instead, been implicated in cell renewal. Here, we found that depletion of nucleostemin in a human breast carcinoma cell line triggers prompt and significant DNA damage in S-phase cells without perturbing the initial step of ribosomal (r)RNA synthesis and only mildly affects the total ribosome production. By contrast, GNL3L depletion markedly impairs ribosome production without inducing appreciable DNA damage. These results indicate that, during vertebrate evolution, GNL3L retained the role of the ancestral gene in ribosome biosynthesis, whereas the paralogous nucleostemin acquired a novel genome-protective function. Our results provide a coherent explanation for what had seemed to be contradictory findings about the functions of the invertebrate versus vertebrate genes and are suggestive of how the nucleolus was fine-tuned for a role in genome protection and cell-cycle control as the vertebrates evolved.
KEY WORDS: Cell cycle, DNA damage, GNL3L, Nucleolus, Nucleostemin, Ribosomal synthesis
INTRODUCTION
The mammalian proteins nucleostemin, GNL3L and GNL2 (also known as NGP1) constitute a newly recognized family of GTP-binding proteins defined by their unique MMR-HSR1 motif and nucleolar localization. Nucleostemin, the most well-known member of this family, was discovered as a gene preferentially expressed in rat embryonic neural stem cells compared with their differentiated progeny and was also found to be expressed at elevated levels in many cancer cells (Tsai and McKay, 2002). Nucleostemin plays an indispensable role in early embryogenesis and the maintenance of the self-renewal of stem or progenitor cells (Qu and Bishop, 2012; Tsai, 2011; Zhu et al., 2006). GNL3L was discovered in a screen for nucleostemin-related genes (Du et al., 2006; Yasumoto et al., 2007). Later studies revealed that nucleostemin and GNL3L exist as separate genes only in vertebrate species (Tsai and Meng, 2009). The invertebrate genomes for which sequence information is available contain a common ortholog of nucleostemin and GNL3L, termed GNL3 collectively or specifically named NS1 in Drosophila melanogaster (CG3983), NST-1 in Caenorhabditis elegans (K01C8.9), Nug1 in Saccharomyces cerevisiae and Grn1 in Schizosaccharomyces pombe (NM_001022573). By contrast, GNL2 represents a single gene product that is highly conserved from yeast to human. Although many members of the MMR-HSR1 family, including nucleostemin, GNL3L and GNL2 (Meng et al., 2006), are capable of binding to GTP, most of them do not possess intrinsic GTPase activity. For the few that do [i.e. YjeQ (Daigle et al., 2002), Lsg1 (Reynaud et al., 2005) and Drosophila GNL3 (Rosby et al., 2009)], the detected GTPase activity is relatively weak.
Nucleostemin, GNL3L and GNL2 proteins are conspicuously localized in the nucleolus but, like many nucleolus-concentrated proteins, also shuttle between the nucleolus and the nucleoplasm (Meng et al., 2007). Because of the nucleolar presence of nucleostemin, it has always been thought to be involved in ribosome biogenesis. Of course, such a hypothesis assumes that proteins stationed in the nucleolus at higher concentration than in the nucleoplasm are involved in the canonical function of this nuclear domain (i.e. ribosome synthesis), but we now know that not all nucleolar proteins serve such a role (Andersen et al., 2005; Ma and Pederson, 2008; Pederson, 1998; Pederson and Tsai, 2009; Scherl et al., 2002). To date, most of the studies showing a ribosomal effect of nucleostemin have been performed on invertebrate GNL3 (i.e. Grn1, NST-1 and NS1). It has been reported that deletion of Grn1 in S. pombe perturbs 35S preribosomal (pre-r)RNA processing and nucleolar export of the Rpl25a (60S) complex (Du et al., 2006). In C. elegans, loss of NST-1 decreases the levels of 18S and 26S rRNAs (Kudron and Reinke, 2008), and in Drosophila depletion of NS1 protein results in nucleolar accumulation of the large ribosomal subunit proteins L11 and L26 (Rosby et al., 2009). In mammalian cells, a potential role of nucleostemin in ribosomal synthesis was suggested by a study showing that prolonged knockdown of nucleostemin delayed the processing of 32S pre-rRNA to 28S ribosomal (r)RNA (Romanova et al., 2009a). Although these studies indicate that the loss of nucleostemin might eventually lead to the perturbation of ribosomes, they fail to establish a coherent mechanism or a direct target of nucleostemin action in the ribosomal-synthetic pathway. Indeed, a direct role of mammalian nucleostemin in pre-rRNA processing is contradicted by a study showing that the impaired 35S pre-rRNA processing and Rpl25a nucleolar export phenotypes of Grn1-null yeast can be restored by human GNL3L, but not by human nucleostemin (Du et al., 2006). Moreover, mammalian nucleostemin fails to rescue the growth phenotype in NST-1-deficient C. elegans, and NST-1 is absent from nucleolar regions where rRNA transcription and processing occur (Kudron and Reinke, 2008). Furthermore, mammalian nucleostemin is localized in subnucleolar regions that are deficient in nascent 28S rRNA (Politz et al., 2005). Because of the paralogous descent of mammalian nucleostemin and GNL3L, and given the evidence that only GNL3L retains the ribosome-biosynthesis function of invertebrate GNL3, it is possible that mammalian nucleostemin might have functionally diverged from mammalian GNL3L and invertebrate GNL3.
These apparent discrepancies raise the issue that perturbations in ribosome synthesis observed upon nucleostemin depletion might not represent a direct effect. In this respect, it is noteworthy that effects on ribosome synthesis have been observed following two rounds of RNAi-mediated nucleostemin knockdown over a period of 5 days (Romanova et al., 2009a; Romanova et al., 2009b). Motivated by the recent discovery of a role for nucleostemin in protecting telomeres and the genome integrity of stem and progenitor cells (Hsu et al., 2012; Lin et al., 2013; Meng et al., 2013; Yamashita et al., 2013), we began to consider whether the accumulation of DNA damage, rather than impaired ribosome production, is actually the initial event following nucleostemin depletion. Using human breast cancer MDA-MB-231 cells, we found that DNA damage arises as early as 12 hours after the initiation of nucleostemin depletion, without incurring a significant effect on rRNA synthesis or nucleolar structure for up to 48 hours. By striking contrast, depletion of GNL3L perturbs the processing of pre-rRNA while evoking little, if any, DNA damage. Our results thus indicate that, during vertebrate evolution, nucleostemin acquired a unique role in genome protection, whereas GNL3L retained a conserved function in ribosome synthesis.
RESULTS
Loss of nucleostemin elicits a prompt DNA damage response
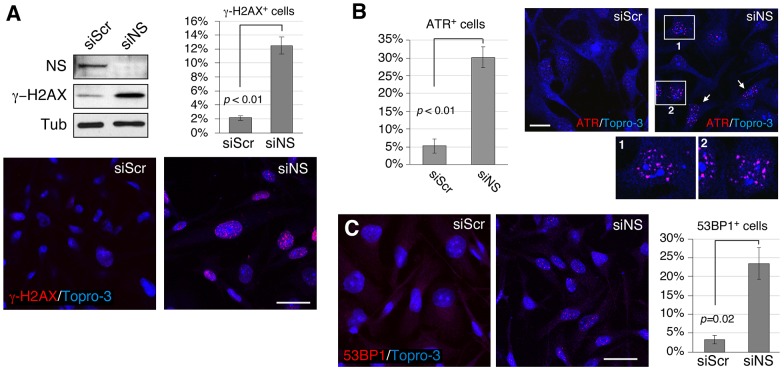
MDA-MB-231 cells, a human breast carcinoma cell line, were transfected with nucleostemin-specific (siNS) or with control scrambled (siScr) siRNA duplexes and were collected for analysis at 48 hours after transfection. The target specificity of siNS has been demonstrated previously (Meng et al., 2008), and the knockdown efficiency in MDA-MB-231 cells is confirmed in Fig. 1A (by western blotting). Notably, there was a pronounced elevation in the amount of phosphorylated H2AX (γ-H2AX, a histone modification associated with DNA damage), as shown by analysis of western blots, and an increased percentage of cells with γ-H2AX foci (from 2.3% to 12.2%, P<0.01) was revealed by using immunofluorescence (Fig. 1A). Parallel immunostaining also revealed an increase in the percentage of cells with foci of the protein ataxia telangiectasia and rad3-related (ATR) (Fig. 1B, from 5.3% to 30.2%, P<0.01) or with 53BP1 foci (Fig. 1C, from 3.3% to 23.5%, P = 0.02), both of which are indicative of a DNA damage response.
Fig. 1.
Loss of nucleostemin increases the number of DNA damage foci as visualized by staining for multiple DNA-damage-response proteins. MDA-MB-231 cells were treated with scrambled (siScr) or nucleostemin-specific (siNS) siRNA duplexes for 48 hours at 100 nM. (A) Upper left panel, western blots of nucleostemin and γ-H2AX. Protein loading was controlled by the amount of α-tubulin (Tub). NS, nucleostemin. Lower panels, immunofluorescence with an anti-γ-H2AX antibody. Upper-right panel, quantitative measurement of the percentage of γ-H2AX+ cells in siScr- and siNS-treated samples. (B) Percentage of ATR+ cells (left) as determined by using immunofluorescence (right). Panels 1 and 2 show magnified views of the regions outlined in white. White arrows, examples of ATR+ cells. (C) The percentage of 53BP1+ cells (right) as determined by using immunofluorescence (left). Scale bars: 50 µm (A and C), 20 µm (B). Bar graphs represent mean±s.e.m.
DNA damage in S-phase cells is an early event after nucleostemin depletion
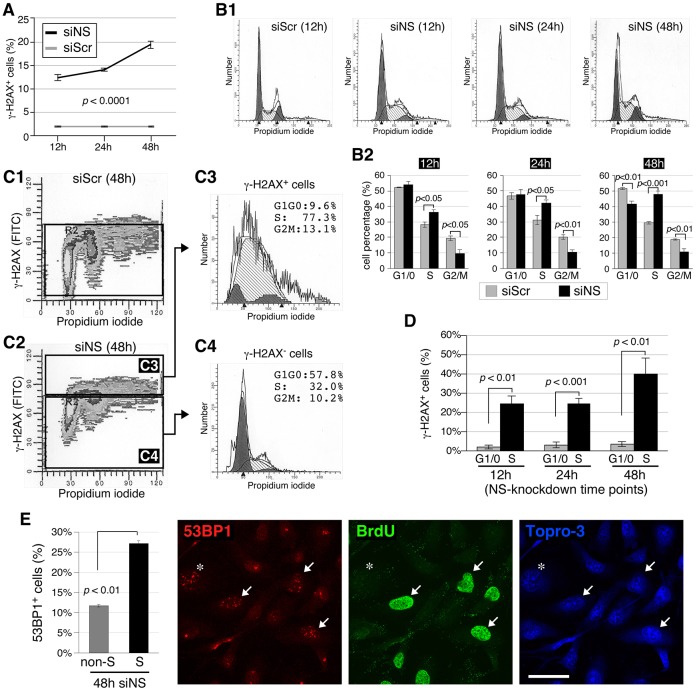
To determine the immediacy of the DNA damage effect and also to assess whether a particular stage of the cell cycle was impacted, we quantified the increase in the percentage of γ-H2AX+ cells over 12–48 hours by using FACS. As shown in Fig. 2A, the percentage of γ-H2AX+ cells increased as early as 12 hours after the initiation of nucleostemin knockdown and continued to rise within the first 48 hours. FACS analysis of propidium-iodide-stained cells revealed that nucleostemin depletion resulted in an increase in the percentage of S-phase cells and a decrease in the percentage of G2/M-phase cells at the 12, 24 and 48-hour time-points, and that there was a decrease in the percentage of G1/G0 cells only at the 48-hour time-point (Fig. 2B1,2). This suggested that nucleostemin depletion triggers replication-dependent DNA damage. To address this possibility, double-label FACS was performed to measure the cell-cycle distributions in the γ-H2AX+ versus γ-H2AX− subpopulations of siNS-treated cells (Fig. 2C). The γ-H2AX+ nucleostemin-depleted population contained significantly more S-phase cells and fewer cells in G1/G0 compared with the γ-H2AX− nucleostemin-depleted population (Fig. 2C3,4). Fig. 2D shows the percentages of G1-phase versus S-phase cells with γ-H2AX foci as a function of the duration of nucleostemin knockdown. We found that 24.5% (12 hours) to 40.2% (48 hours) of the S-phase cells showed γ-H2AX+ signals in response to nucleostemin depletion, compared with only 1.8% (12 hours) to 3.5% (48 hours) of the G1/G0 cells (Fig. 2D, P<0.01). These results demonstrate that DNA damage is induced in S-phase cells as early as 12 hours after nucleostemin depletion commences, triggering an intra-S-phase cell-cycle arrest. To substantiate the result that nucleostemin-knockdown-triggered DNA damage occurs preferentially in S-phase cells, we performed a 30-minute BrdU pulse on siNS-treated cells, followed by immunofluorescence with antibodies against 53BP1 and BrdU. At the 48-hour time-point, 27% of the S-phase (BrdU-labeled) cells showed an increased number of 53BP1 foci (≥5 foci per cell), whereas only 11% of the non-S-phase cells showed an increased number of 53BP1 foci (Fig. 2E, P<0.01). The higher percentage of cells with DNA damage in the non-BrdU-labeled cell population (Fig. 2E, gray bar) compared with that in the G0/G1 population as measured by FACS of cells labeled with γ-H2AX and PI (Fig. 2D, gray bar) might be due to the inclusion of G2/M-phase cells as well as some S-phase cells that were already arrested at the time of BrdU labeling in the non-BrdU-labeled population. It might also be due to the different sensitivities of immunofluorescence versus FACS.
Fig. 2.
S-phase DNA damage is an early event during knockdown of nucleostemin. (A) FACS-based quantification of the percentage of γ-H2AX+ cells in MDA-MB-231 cell samples treated with siScr or siNS (100 nM) for 12, 24 or 48 hours. Data show the mean±s.e.m. (B) Cell-cycle profiles (B1) and quantitative analyses (n = 4, B2) of siScr- and siNS-treated MDA-MB-231 cells at different knockdown time-points. The fitted profiles shaded in gray represent cells with more than 2N copies of chromosomes. (C) Cell-cycle profiles of the γ-H2AX+ versus γ-H2AX− cells in nucleostemin-knockdown samples were quantitatively analyzed by using FACS with anti-γ-H2AX and propidium iodide (PI). (D) Based on the analyses shown in C, the percentages of γ-H2AX+ cells in S-phase versus G1/G0-phase in the nucleostemin-knockdown sample were compared at the 12, 24 and 48-hour time-points. Data show the mean±s.e.m. (E) Immunofluorescent imaging (right) using anti-53BP1 and anti-BrdU antibodies in MDA-MB-231 cells treated with siNS (100 nM) for 48 hours. Quantitative data (left) represent the mean±s.e.m. of the percentage of 53BP1+ cells in the non-BrdU-labeled (non-S-phase, asterisk) versus BrdU-labeled (S-phase, arrows) cell populations. Scale bar: 50 µm.
Nucleostemin depletion does not significantly perturb rRNA biogenesis or nucleolar structure coincident with the activation of DNA damage
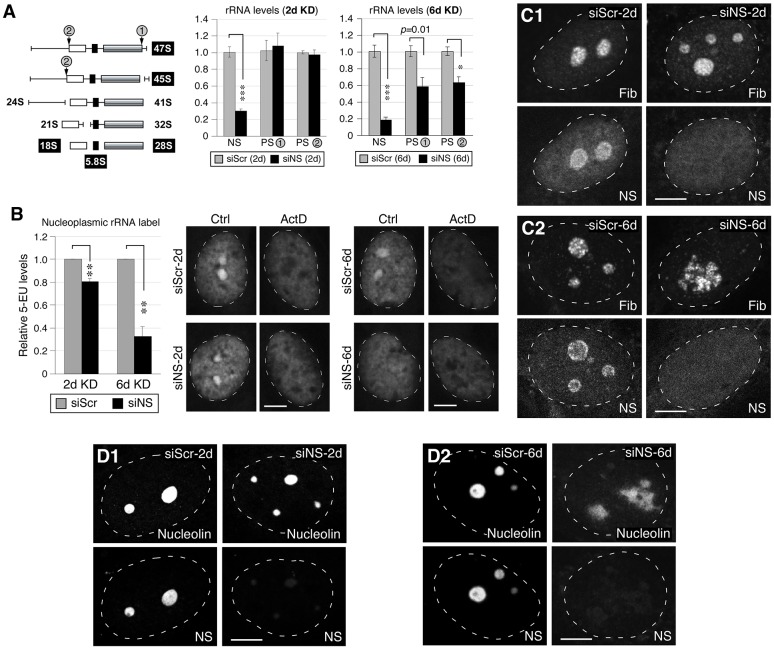
Given the conflicting reports on whether nucleostemin depletion affects rRNA synthesis (see Introduction), we next measured the effects of nucleostemin knockdown on the levels of 47S and 45S pre-rRNAs by real-time RT-PCR. As shown in Fig. 3A (left panel), the primer pair for Processing Site 1 (PS-1) spans the junction between the 28S region and the 3′ external transcribed spacer (ETS), and therefore its PCR product reflects only the 47S primary transcript, whereas the primer pair for PS-2 spans the junction between the 18S region and the 5′ ETS, and therefore it detects both the 47S and 45S precursors. At 48 hours of nucleostemin depletion, there was no effect on the level of the 47S primary transcript or on the combined levels of the 47S primary transcript and the 45S pre-rRNA intermediate (Fig. 3A, center panel). However, when subjected to two rounds of RNAi-mediated knockdown, lasting for a total of 6 days (see Materials and Methods), a significant impairment of rRNA synthesis was observed (Fig. 3A, right panel).
Fig. 3.
Knockdown of nucleostemin has minimal effects on rRNA synthesis or nucleolar integrity at the 48-hour time-point. (A) Left, diagram showing the pre-rRNA-processing step and the primer positions flanking the processing sites 1 and 2 (PS-1, PS-2). White, black and grey boxes represent 18S, 5.8S and 28S rRNAs, respectively. The PS-1 and PS-2 primer recognition sites are indicated by gray circles. Middle and left panels, qRT-PCR assays of nucleostemin (NS), PS-1 and PS-2 levels in MDA-MB-231 cells treated with siScr or siNS for 2 days (2d) or 6 days (6d). The expression levels of target transcripts were quantified by normalizing to the expression of HMG-14. The expression levels of each transcript in the corresponding siScr samples were arbitrarily set as 1. (B) A modified 5-EU click-chemistry assay was designed to measure the amount of newly synthesized mature rRNA present in the nucleoplasm. The differential 5-EU signals [control (Ctrl) minus actinomycin (ActD)-treated samples] in the nucleoplasm were quantified in MDA-MB-231 cells treated with siScr or siNS for 2 days or 6 days. Data show the mean±s.e.m.; *P<0.01, **P<0.001, ***P<0.0001. (C) Confocal immunofluorescence of the fibrillarin (Fib)-labeled dense fibrillar component and nucleostemin-labeled structure in siRNA-treated cells at the 2-day and 6-day time-points. (D) Confocal immunofluorescence of nucleolin- and nucleostemin-labeled nucleolar structures in siRNA-treated cells at the 2-day and 6-day time-points. White dashed lines show nucleus outlines. Scale bars: 10 µm.
We then sought to determine whether nucleostemin knockdown affected the accumulation of processed rRNA in the nucleus, as would be expected if the ribosomal-synthetic pathway was inhibited. The experimental design was based on the uptake of the uridine analog 5-ethynyl-uridine (5-EU), and its incorporation into RNA, followed by detection of the newly synthesized RNA in fixed cells by a click-chemistry-mediated dye-coupling reaction (Jao and Salic, 2008). Moreover, we reasoned that, because rRNA synthesis can be selectively inhibited in mammalian cells with a low concentration of actinomycin (Perry, 1962), it would be possible to assess the amount of dye label in rRNA versus non-rRNA transcripts in control versus knockdown cells, based on the presence or absence of the inhibitor. The cells were incubated with 5-EU for 2 hours, by which time the nuclear rRNA labeling has attained steady state (Penman, 1966). As shown in Fig. 3B, the nucleoplasmic 5-EU differential (calculated as the dye signal level in actinomycin-treated cells subtracted from that of untreated cells, thus representing mature rRNAs) showed a small decrease (20%) after 2 days of nucleostemin knockdown compared with siScr-treated cells. However, cells in which nucleostemin had been knocked down for 6 days displayed a substantial reduction (75%) in their nucleoplasmic 5-EU differential signal compared with siScr-treated cells. Finally, in contrast to a previous report (Romanova et al., 2009b) showing that nucleostemin depletion causes a dispersion of the dense fibrillar component (DFC) of nucleolar structure, which is defined by the protein fibrillarin, we found no discernible changes in the fibrillarin-labeled DFC structure in cells in which nucleostemin was knocked down for 2 days (Fig. 3C1), but a disorganized DFC structure was apparent in some cells after a 6-day knockdown (Fig. 3C2). In support, we found no changes in the nucleolin-labeled nucleolar structure in cells treated with siNS for 2 days (Fig. 3D1), but a disorganized nucleolin-labeled nucleolar structure in some of the cells treated with siNS for 6 days (Fig. 3D2). These results indicate that prolonged nucleostemin depletion might perturb rRNA transcription and processing secondarily to the induction of the DNA damage response described in the previous section.
Partial nucleostemin depletion induces a G2/M arrest
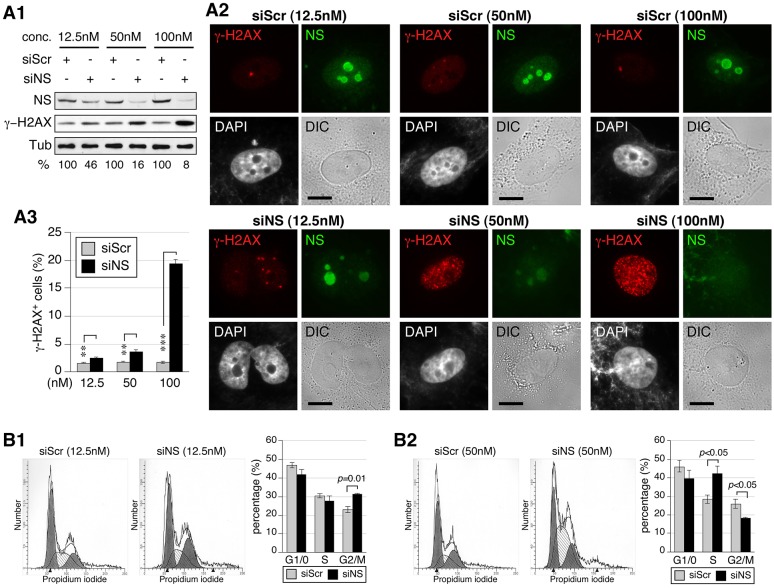
We previously found that nucleostemin-haploinsufficient mouse embryonic fibroblasts and shRNAmir-mediated nucleostemin-depleted human osteosarcoma cells both display a G2/M-phase arrest (Meng et al., 2008; Zhu et al., 2006). Because the nucleostemin depletion in both cases was in the range of 50–60%, and based on the results of the present study, we speculated that partial nucleostemin depletion is permissive for S-phase transit until the G2/M checkpoint. To investigate this possibility, we tested a range of siRNA concentrations that led to nucleostemin depletions of 54%, 84% and 92% at the 48-hour time-point (Fig. 4A1,2). FACS analysis confirmed that the percentage of γ-H2AX+ cells increased in proportion to nucleostemin depletion (Fig. 4A3). Notably, there was an increase in the percentage of G2/M-phase cells in samples with the least nucleostemin depletion (Fig. 4B1). When the nucleostemin depletion was 84%, S-phase arrest became apparent, with a concomitant decrease in the G2/M-phase population (Fig. 4B2). These results indicate that cells with a partial loss of nucleostemin progress through S phase until the G2/M checkpoint and thus offer an explanation of the seemingly conflicting findings on the effect of nucleostemin knockdown on the cell cycle.
Fig. 4.
The knockdown efficiency dictates the nature of the cell-cycle arrest of nucleostemin-depleted cells. (A1) Western blots to show the knockdown efficiency in MDA-MB-231 cells treated with different concentrations (conc.) of siScr and siNS. Samples were collected at the 48-hour time-point. Relative nucleostemin intensities (%) between paired siScr and siNS samples are shown below the blots. α-tubulin (Tub) is shown as a loading control. NS, nucleostemin. (A2) Immunofluorescence performed using anti-nucleostemin and γ-H2AX antibodies in siScr- and siNS-treated MDA-MB-231 cells. Nuclear structure and cell morphology are shown by DAPI staining and differential interference contrast (DIC) imaging, respectively. Scale bars: 10 µm. (A3) FACS-based quantification of the percentage of γ-H2AX+ cells in siScr- and siNS-treated samples. **P<0.001, ***P<0.0001. (B) Cell-cycle profiles and quantitative analyses (n = 4) of MDA-MB-231 cells treated with 12.5 nM (B1) or 50 nM (B2) siRNAs for 48 hours. Data show the mean±s.e.m. of three independent experiments.
GNL3L or GNL2 knockdown contrasts with nucleostemin depletion
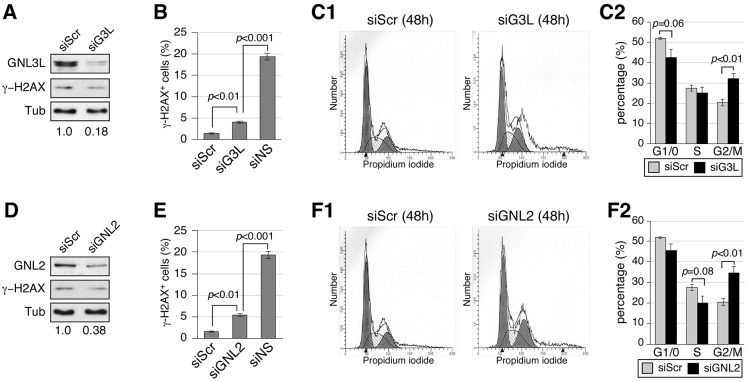
Because human cells contain two other nucleostemin homologs, GNL3L and GNL2, it was of interest to determine how their depletion might impact on DNA damage and cell-cycle progression. Cells were transfected with GNL3L-specific (siG3L) or GNL2-specific (siGNL2) siRNA duplexes and analyzed 48 hours later. To detect GNL3L and GNL2 proteins, antibodies were generated to specifically recognize either human GNL3L (Ab134) or human GNL2 (Ab136). Analysis of western blots confirmed that siG3L treatment reduced the amount of GNL3L protein by >82% but did not increase the amount of γ-H2AX protein (Fig. 5A). FACS analysis showed that only 3.9% of GNL3L-depleted cells were γ-H2AX+, which is slightly higher than the 1.5% of γ-H2AX+ cells in the siScr-treated sample but distinctly lower than the 19.3% of γ-H2AX+ cells in the nucleostemin-depleted sample (Fig. 5B). In addition, GNL3L-depleted cells differ from nucleostemin-depleted cells in showing an increase in the percentage of G2/M-phase cells and a slight decrease in the percentage of G1/G0 cells (Fig. 5C1,2). Western blotting showed that a 62% reduction in the amount of GNL2 elicited no change in γ-H2AX protein (Fig. 5D). FACS analysis revealed that 5.4% of GNL2-depleted cells were γ-H2AX+, which is still significantly lower than for nucleostemin-depleted cells (P<0.001) (Fig. 5E). In addition, GNL2 depletion led to an increase in the percentage of G2/M-phase cells and a slight decrease in the percentage of S-phase cells, indicative of a G2/M arrest (Fig. 5F). These results demonstrate that depletion of neither GNL3L nor GNL2 elicits as extensive a DNA damage response as does nucleostemin depletion, and that depletion of either of these proteins does not trigger the S-phase arrest seen upon nucleostemin depletion.
Fig. 5.
Loss of GNL3L or GNL2 causes G2/M arrest with minimal DNA damage. MDA-MB-231 cells were treated with GNL3L-specific (siG3L, A–C) or GNL2-specific (siGNL2, D–F) siRNA duplexes at 100 nM for 48 hours. (A,D) Western blots of GNL3L, GNL2 and γ-H2AX. Protein loading was controlled by the amount of α-tubulin (Tub). The relative amount of the siRNA-targeted protein is indicated beneath the blots. (B,E) FACS-based quantification of the percentage of γ-H2AX+ cells in siRNA-treated samples. (C,F) Cell-cycle profiles and quantitative analyses (n = 4) of siRNA-treated MDA-MB-231 cells. Data represent the mean±s.e.m.
Loss of GNL3L perturbs pre-rRNA processing
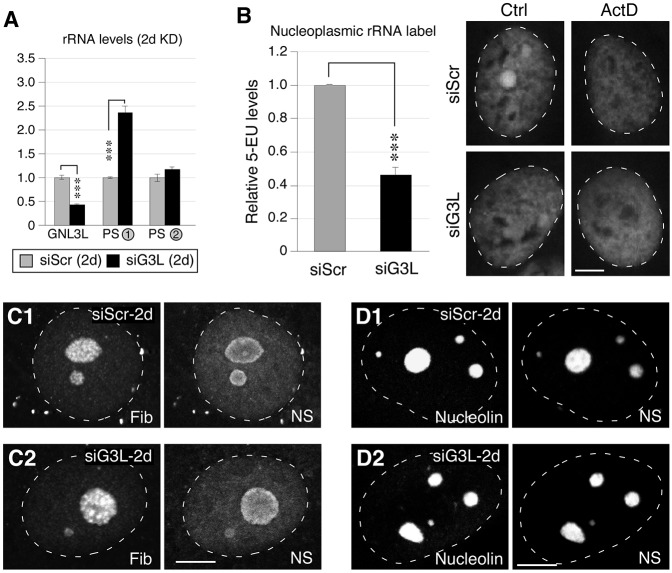
Given that GNL3L is paralogous to nucleostemin but elicits minimal DNA damage when depleted, we reasoned that GNL3L is likely to be the member of this protein pair that has retained the ribosome synthesis function of the ancestral protein, GNL3. To test this hypothesis, we determined the levels of rRNA transcripts in GNL3L-depleted cells (Fig. 6A). GNL3L depletion led to a substantial increase in the level of the 47S primary transcript, indicating an inhibition of the 47S-to-45S processing step. In addition, the differential 5-EU assay revealed a significant (57%) reduction in nucleoplasmic rRNA after GNL3L knockdown (Fig. 6B). However, as with short-term (2-day) nucleostemin knockdown, GNL3L depletion did not alter the fibrillarin-, nucleolin- or nucleostemin-labeled subnucleolar structures (Fig. 6C,D), indicating that the effect of the 2-day GNL3L knockdown on 47S-to-45S processing does not manifest as a change in nucleolar structure such as that known to occur when transcription of the rDNA is perturbed (Dousset et al., 2000; Ma and Pederson, 2013).
Fig. 6.
Loss of GNL3L perturbs the processing of pre-rRNAs. (A) qRT-PCR assays of GNL3L, PS-1 and PS-2 levels in MDA-MB-231 cells treated with siScr or siG3L for 2 days (2d). The expression levels of target transcripts were quantified by normalizing to the expression of HMG-14. The expression levels of each transcript in the siScr samples were arbitrarily set as 1. (B) The differential 5-EU signals [control (Ctrl) minus actinomycin (ActD)-treated samples] in the nucleoplasm were quantified in MDA-MB-231 cells treated with siScr or siG3L for 2 days (left). Right, immunofluorescence of 5-EU signals in control (siScr) and GNL3L-knockdown (siG3L) cells with (ActD) or without (Ctrl) treatment with a low dose of ActD. Data show the mean±s.e.m. ***P<0.0001. (C) Confocal immunofluorescence to show the fibrillarin (Fib)- and nucleostemin (NS)-labeled structures in cells treated with siScr or siG3L for 2 days. (D) Confocal immunofluorescence of the nucleolin- and nucleostemin-labeled structures in cells treated with siScr or siG3L for 2 days. White dashed lines show nucleus outlines. Scale bars: 10 um.
Overexpression of nucleostemin, but not of GNL3L or GNL2, confers protection against replication-induced DNA damage
Spontaneous DNA damage can be caused by stalling of the replication fork, oxidative stress or DNA base hydrolysis. To determine the type of DNA damage involved, we measured the sensitivities of control and nucleostemin-depleted cells to hydoxyurea, H2O2 or UV perturbations that introduce replication fork stalling, reactive oxygen species and thymine dimers, respectively. Nucleostemin depletion increased the sensitivity of MDA-MB-231 cells to HU-induced replication stalling (P<0.001, repeated measures ANOVA) but not to UV- or H2O2-induced DNA damage (Fig. 7A). To further investigate the function of nucleostemin in replication-linked DNA damage, we performed rescue experiments. Nucleostemin–GFP expression significantly protected cells against replication-linked DNA damage, whereas neither GNL3L–GFP nor GNL2–GFP was able to do so (Fig. 7B, upper panel). This rescue effect was seen only in transfected (GFP-positive) cells in the nucleostemin–GFP-transfected samples (Fig. 7B, compare upper and lower panels). Given the high concentration of nucleostemin protein in the nucleolus, we investigated whether the protective effect of nucleostemin against replication-linked DNA damage might occur through some indirect mechanism requiring the nucleolar localization of nucleostemin. However, we found that the expression of a version of nucleostemin that lacked the nucleolus-localization signal (NSdB–GFP) – and thus accumulated in the nucleoplasm – also conferred protection against DNA damage (Fig. 7B, upper panel). Thus, this property of nucleostemin does not require nucleolar localization and is likely to be a direct effect, or at least one that occurs in proximity to replication sites. Taken together, the results of this investigation show that nucleostemin has a unique role in maintaining genomic stability by protecting against replication-induced DNA damage, and that it is the paralogous protein, GNL3L, that has retained the role in ribosome synthesis established in the ancestral pre-vertebrate protein.
Fig. 7.
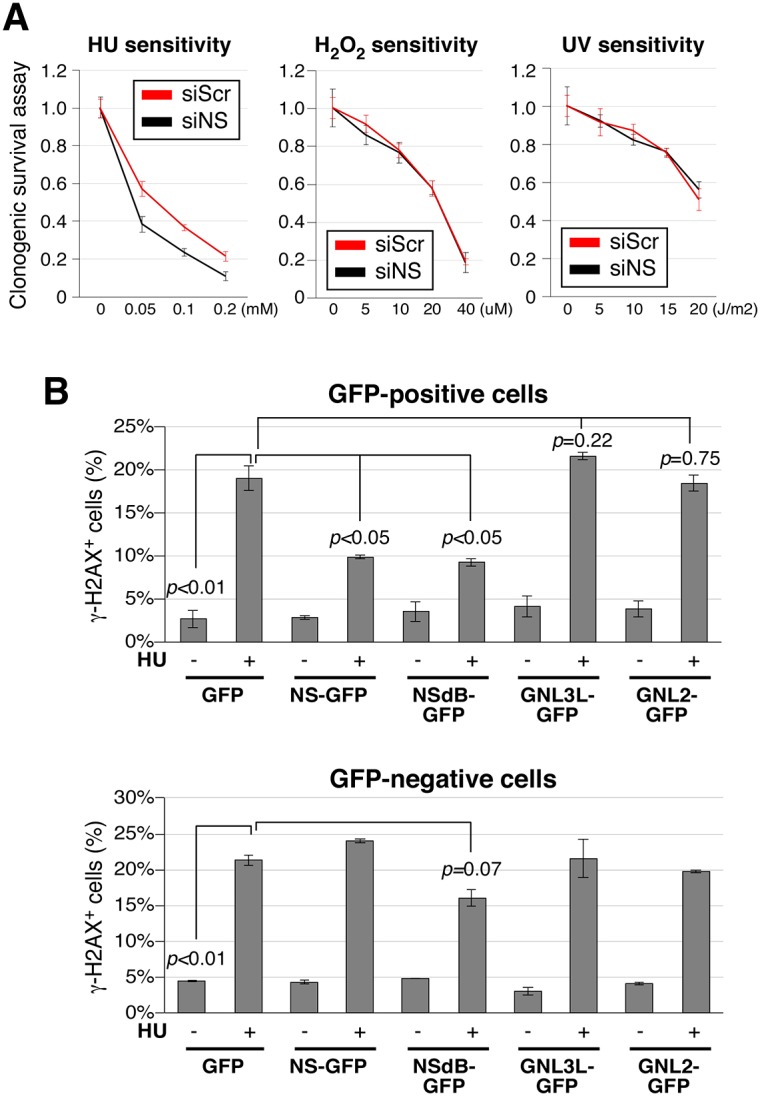
Overexpression of nucleostemin, but not of GNL3L or GNL2, protects against hydroxyurea (HU)-induced DNA damage. (A) MDA-MB-231 cells treated with siScr or siNS were tested for their clonogenic survival following treatment with HU, H2O2 or UV. (B) Upper panel, the percentage of γ-H2AX+ cells in the transfected (GFP-positive) populations of MDA-MB-231 cells transiently overexpressing GFP, nucleostemin (NS)–GFP, NSdB–GFP (a nucleoplasmic nucleostemin mutant), GNL3L–GFP or GNL2–GFP, with or without a 24-hour HU treatment. Lower panel, the percentage of γ-H2AX+ cells in the non-transfected (GFP-negative) populations. Differences were analyzed by repeated measures ANOVA. Data represent the mean±s.e.m.
DISCUSSION
The hypothesis
When nucleostemin was first discovered in rat embryonic neural stem cells (Tsai and McKay, 2002), the focus was, of course, on its role as a determinant in the cells, not its evolutionary origin. The subsequent identification of GNL3L, the mammalian paralog of nucleostemin, broadened the landscape of possible functions of these two nucleolar GTP-binding proteins. Subsequent work on both nucleostemin and GNL3L (Lin et al., 2010; Ma and Pederson, 2007; Meng et al., 2011a; Meng et al., 2011b; Meng et al., 2008; Zhu et al., 2009; Zhu et al., 2006) drew attention to the involvement of both proteins in cell proliferation, although the direct roles of the proteins were not rigorously proven. At the same time, evidence appeared in Drosophila linking the invertebrate protein, GNL3, to ribosome biosynthesis (Rosby et al., 2009), and another report implicated mammalian nucleostemin in ribosome biosynthesis (Romanova et al., 2009a). It was against this background that we launched the present study. Our hypothesis was that mammalian GNL3L has retained the role of the ancestral protein in ribosome biosynthesis, whereas the paralogous nucleostemin acquired a different function or functions. Our findings reveal distinct activities of mammalian nucleostemin and GNL3L in genome protection and ribosome biosynthesis, respectively, and strongly support the hypothesis that nucleostemin functionally diverged from its vertebrate paralog, GNL3L, and the invertebrate ortholog, GNL3.
DNA damage, not impairment of ribosome biosynthesis, is an early event following nucleostemin depletion
As discussed above, whether nucleostemin plays a direct role in ribosome biogenesis has not been clear. Most previous studies examined only the terminal results of nucleostemin gene knockout or knockdown, without resolving the temporal relationship of the events. This issue applies to both whole-organism studies (Kudron and Reinke, 2008; Rosby et al., 2009) and to the nucleostemin-knockdown study of Romanova et al. (Romanova et al., 2009a) in HeLa cells, in which cells were analyzed on the 5th day after two rounds of knockdown. Our timecourse analyses show that nucleostemin depletion triggers DNA damage and cell-cycle arrest shortly after the initiation of knockdown (<12 hours). By contrast, the biosynthesis of 47S and 45S rRNA precursors is not appreciably perturbed within 48 hours of nucleostemin knockdown. Moreover, the differential 5-EU labeling assay reveals only a minor reduction in the steady-state labeled rRNA species in the nucleoplasm after a 2-day nucleostemin knockdown. Most relevant to the Romanova et al. study, we found that the transcription and maturation of rRNAs are both severely inhibited after 6 days of nucleostemin knockdown, a period of time that is comparable to their 5-day knockdown. We also noted that the rRNA labeling kinetics reported in the study by Romanova et al. were much slower than those reported in numerous other studies (e.g. Penman, 1966), suggesting that these 5-day-knockdown cells succumb to pronounced cytopathic events. This interpretation of their findings gains further traction given the fact that the 5-day-knockdown cells display a ∼50% decrease in histone 2B, which substantial literature indicates would elicit a cell-cycle arrest. There is abundant evidence in the literature showing that cell-cycle arrest of transformed mammalian cells leads to an inhibition of rRNA synthesis. Thus, we believe that the conclusion reached by Romanova et al. might be confounded by unsuspected issues surrounding the immediate versus long-term effects of nucleostemin depletion, as well as the interconnecting relationship between nucleolar function and cell cycle. Furthermore, while the present investigation was under way, a parallel study suggested that there is a direct molecular recruitment of nucleostemin to DNA damage sites, where it interacts with the DNA repair protein RAD51 (Meng et al., 2013). This also alerted us to the possibility that nucleostemin is involved in genome protection and not rRNA synthesis.
The effect of nucleostemin depletion on cell-cycle progression
The exact effects of nucleostemin depletion on the progression of the cell cycle vary among different studies. Effects shown include a G1/S arrest (Dai et al., 2008; Ma and Pederson, 2007; Nikpour et al., 2009) or a G2/M arrest (Meng et al., 2008; Nikpour et al., 2009; Romanova et al., 2009a; Zhu et al., 2006). This discrepancy cannot be explained by the status of p53, Rb or p16 in the cell models used. The present study reveals that the primary cell-cycle event of nucleostemin depletion is an S-phase arrest and that a less-efficient knockdown of nucleostemin produces a G2/M-phase arrest (Fig. 4). This provides a potential resolution to the contradicting results on cell-cycle arrest reported previously, in which a near-complete or partial nucleostemin knockdown might result in an early S-phase (apparent G1/S) arrest, an intra-S-phase arrest or a late S and G2/M arrest.
The role of nucleostemin in DNA damage protection
When DNA damage occurs as a result of replication stalling, formation of reactive oxygen species or base hydrolysis, cells respond by mounting distinct repair mechanisms. The homologous recombination machinery is the primary mechanism responsible for the restart and repair of stalled or collapsed replication forks (Helleday, 2008). The base excision repair (BER) machinery repairs non-helix-distorting base lesions caused by reactive oxygen species, alkylation or deamination, and thus BER impairment leads, experimentally, to sensitivity to H2O2. The nucleotide excision repair (NER) machinery removes depurinated bases and intra-strand crosslinks. Cells with dysfunctional NER show an increased sensitivity to UV-induced damage. Our data show that DNA damage induced by nucleostemin knockdown preferentially occurs in S-phase cells. Compared with control cells, nucleostemin-depleted cells are more sensitive to HU-induced damage but not to H2O2- or UV-induced damage, and overexpression of nucleostemin is able to rescue HU-induced DNA damage. These findings suggest that nucleostemin might safeguard the genomic integrity of actively dividing cells by promoting the repair of DNA damage that takes place during DNA replication. Finally, because MDA-MB-231 cells are p53 mutated, the observed genome-protective activity of nucleostemin is expected to act independently of the MDM2–p53 pathway and is more in synchrony with the obligatory function of nucleostemin documented in p53-null embryos (Beekman et al., 2006). This study, and a previous one (Meng et al., 2008), emphasize that the genome-protective activity of nucleostemin occurs constitutively, whereas its MDM2-regulatory function takes place primarily when the protein is mobilized from the nucleolus to the nucleoplasm under conditions of nucleolar stress or mitotic transit.
An evolving view of nucleostemin family proteins
The conflicting reports on the role of nucleostemin in cell proliferation, MDM2–p53 regulation and ribosome biosynthesis gave rise to the notion that this is a multitasking protein. Our results suggest that this might not be the case. Instead, the primary role of nucleostemin might be a genome-protective function, and the effect of its knockdown on ribosome biosynthesis might be a secondary consequence. The present findings can aid in the interpretation of earlier studies in which invertebrate GNL3 and mammalian nucleostemin have been investigated. Thus, because genetic deletion of the invertebrate ortholog, GNL3, was observed to impair ribosome biosynthesis (Du et al., 2006; Kudron and Reinke, 2008; Rosby et al., 2009), reports of similar effects of mammalian nucleostemin knockdown naturally led some investigators to regard nucleostemin as the mammalian equivalent of invertebrate GNL3. Here, we show that a significant impairment of ribosome biosynthesis occurs only after prolonged nucleostemin knockdown, and that it is in fact GNL3L that plays a major role in rRNA biosynthesis. Thus, the entire phyletic situation of the nucleostemin family proteins becomes clear – nucleostemin is the paralog of GNL3L in mammalian cells, and the latter is the direct descendant of the invertebrate gene. It is not yet known whether the functional divergence of nucleostemin from GNL3L and GNL3 is an on-going event that differs in lower versus higher vertebrate species. One recent study by Essers and colleagues reported that nucleostemin mutation in zebrafish (Danio rerio) causes a reduction in the number of 60S ribosomes as well as p53 stabilization (Essers et al., 2014). Interestingly, they also found that deleting p53 can rescue the reduced number of 60S ribosomes in nucleostemin-mutant fish and elevate the number of 60S ribosomes in wild-type fish, which indicates that the ribosomal effect seen in nucleostemin-mutant fish might be explained by the stabilization of p53 as a result of nucleostemin mutation. By contrast, GNL3L-mutant fish show a decrease in the number of 80S ribosomes (by 38.5%) that is comparable to that of nucleostemin-mutant (by 41.5%) and GNL2-mutant (by 45.7%) fish, as well as a prominent increase in the amount of 47S rRNA precursor, but without an apparent effect on the amount of newly synthesized proteins.
In conclusion, this study reports that the DNA damage induced by nucleostemin knockdown occurs significantly earlier than the ribosomal effect reported previously, that the cell-cycle profile of nucleostemin-depleted cells (intra-S versus G2/M arrest) depends on the level of nucleostemin knockdown and that, in contrast to nucleostemin knockdown, the loss of human GNL3L causes ribosomal defects without introducing DNA damage. On the basis of these new findings, we suggest that, during the evolution of vertebrate species, GNL3L might have retained the role of its ancestral gene in ribosome biosynthesis, whereas the paralog, nucleostemin, acquired a new and unique function in genome protection.
MATERIALS AND METHODS
Cell culture and siRNA knockdown
The human breast carcinoma cell line MDA-MB-231 was obtained from the American Type Culture Collection and maintained in monolayer culture in DMEM plus 10% fetal bovine serum as described previously (Lin et al., 2010). Target-specific siRNA duplexes were introduced by transfection using Oligofectamine (Invitrogen, Carlsbad, CA) at 24 hours after replating. Timecourse studies were performed by incubating cells in medium containing siRNA (100 nM) for 12, 24 or 48 hours. Dose-dependent studies were carried out by incubating cells in medium containing siRNA (12.5 nM, 50 nM or 100 nM) for 48 hours. For the long-term (6-day) knockdown study, cells were incubated with siRNA (100 nM) for 24 hours, allowed to recover for 48 hours, treated with another round of siRNA (100 nM) for 24 hours and allowed to recover for 48 hours before harvesting. The RNA sequences targeted by the control (siScr), nucleostemin-specific (siNS), GNL3L-specific (siG3L) and GNL2-specific (siGNL2) siRNA duplexes were; 5′-UGACGAUCAGAAUGCGACU-3′ (siScr), 5′-GAACUAAAACAGCAGCAGA-3′ (siNS), 5′-CUAUUGCCGCCUUGGUGA A-3′ (siG3L) and 5′-ACAAAGGUCUGGCAGUAUA-3′ (siGNL2).
Western blotting
Western blot analyses were performed as described previously and were repeated twice (Meng et al., 2006; Tsai and McKay, 2005). The primary antibodies used in this study were rabbit polyclonal antibodies against human nucleostemin (Ab138), GNL3L (Ab134) and GNL2 (Ab136) (all generated in the Tsai laboratory) γ-H2AX (JBW301, Millipore, Billerica, MA) and α-tubulin (Sigma, St Louis, MO). Secondary antibodies were conjugated to peroxidase (Jackson ImmunoResearch, West Grove, PA).
Immunofluorescence
DNA damage foci were detected by staining with antibodies against γ-H2AX, ATR (N-19, Santa Cruz, Dallas, TX) and 53BP1 (#4937, Cell Signaling Technology, Danvers, MA). For γ-H2AX staining, 1696 and 1113 cells were counted for the siScr and siNS groups, respectively. For ATR staining, 580 and 592 cells were counted for the siScr and siNS groups, respectively. For 53BP1 staining, 1572 and 1615 cells were counted for the siScr and siNS groups, respectively. Nucleoli were labeled by using anti-fibrillarin antibody (38F3, EnCor, Gainesville, FL) and anti-nucleolin antibody (4E2, Research Diagnostics, CA). Secondary antibodies were conjugated with Rhodamine-X or FITC. After fixation in 4% formaldehyde (in all cases except for anti-nucleolin staining) or ice-cold methanol (nucleolin staining) for 15 minutes at room temperature, cells were permeabilized with 0.3% Triton X-100 and incubated with primary and secondary antibodies. To detect S-phase cells, cultures were treated with bromodeoxyuridine (BrdU, 10µM, Accurate, OBT0030) 30 minutes before harvest and stained with rat anti-BrdU antibody (BU1/75, Accurate) following treatment with 4N HCl. A total of 780 cells were randomly selected and analyzed. Fluorescence images were acquired on a Zeiss LSM510 confocal microscope using the 40× or 63× plan-apochromat objectives. For analysis of nucleolar structure, images were captured using the 63× plan-apochromat oil objective and scanned with a 512×512 pixel frame size, 5× zoom and an optical thickness of <1.0 µm. Detector gain and amplifier offset were adjusted to ensure that all signals were displayed within the linear range. Quantification of γ-H2AX+, ATR+ and 53BP1+ cells was performed using ImageJ software with a set threshold level, and positive cells were defined as those showing five or more visible foci. Final data represent the average of three independent experiments.
Flow cytometry
Cells were washed with PBS, trypsinized, fixed in 72% ice-cold ethanol for 30 minutes and sequentially stained with γ-H2AX antibody (1∶2000) and propidium iodide (PI, 50 ug/ml, Sigma). PI staining was performed in the presence of 100 µg/ml DNase-free RNase A. Flow cytometry analyses were conducted using a COULTER EPICS XL flow cytometer and the XL System II software in the Flow Cytometry Core Facility at the Texas Children's Hospital. Cell-cycle profiles were compiled from 2×104 gated events and were analyzed using Multi Cycle AV software. All data represent the average of four independent experiments.
qRT-PCR analysis of rRNA transcripts
Total cell RNA samples (5 µg) were reversed transcribed into first strand cDNAs using random hexanucleotide primers and Moloney murine leukemia virus (Mo-MLV) reverse transcriptase. For quantitative (q)PCR, the ΔCt values between the target RNA and a reference mRNA transcript, HMG-14 (high mobility group-14), were determined by using the MyiQ single-color real-time PCR detection system and supermix SYBR green reagent. The ΔΔCt values were measured from three repeats to compare the relative expression levels of target sequences. The PCR primer sequences were: nucleostemin (annealing temperature 56°C), 5′-GAACAAAGCCAAGTCGGG-3′ and 5′-GTCCACTCTGGACAATGG-3′; GNL3L (56°C), 5′-TACCTTCGGAATGAGTTG-3′ and 5′-TTCAGCTCCAAAGCAGGC-3′; PS-1 (62°C), 5′-TGCCCTTCGTCCTGGGAAAC-3′ and 5′-CGCGCGCGGACAAACCCTT-3′; PS-2 (62°C), 5′-GCGCTCTACCTTACCTACCTG-3′ and 5′-CCGTCGGCATGTATTAGCTCT-3′; and HMG-14 (56–62°C), 5′-GGCAGCAGCGAAGGATAAATC-3′ and 5′-TTCATCAGAGGCTGGACTCTC-3′. The final data represent the average of two independent knockdown experiments with two qPCR repeats.
Imaging of rRNA synthesis by click chemistry
Synthesis of rRNAs was detected by using the Click-iT™ RNA Alexa Fluor 488 imaging kit (C10329, Invitrogen) in experiments in which the synthesis of specifically rRNA was measured by using actinomycin to selectively inhibit rRNA synthesis (see Results for details of this experimental strategy). To detect the synthesis of mature rRNAs in the nucleoplasm, control or knockdown cells were treated with 0.04 µg/ml of actinomycin for 20 minutes and then incubated for 2 hours with 5-ethynyl uridine (5-EU) at 1 mM in the continued presence or absence of actinomycin. The cells were fixed with 4% formaldehyde in PBS for 15 minutes, permeabilized with 0.5% Triton X-100 in PBS for 15 minutes and then incubated with the Click-iT™ reaction cocktail for 1 hour. The cells were then washed with rinse buffer and DNA was counterstained with TO-PRO®-3 for 30 minutes. The average 5-EU signal intensity in the nucleoplasm was measured from 27 cells per experiment and three independent repeats for each condition using the ImageJ program. The level of nuclear rRNA was determined by the difference in nucleoplasmic intensity between the actinomycin-treated and non-treated groups. With 5-EU label times of 2 hours, little label was seen in the nucleoli, owing to the fact that, after this labeling period, the nuclear rRNA has been labeled to equilibrium, reflecting RNA mass, and the minute amounts of pre-rRNA in the nucleoli are only present at background levels.
Clonogenic survival assay
Cells were treated with the desired siRNA (100 nM) duplexes for 48 hours. After recovering for 24 hours, cells were replated at low density (55 cells/cm2). At 24 hours, cells were treated for 24 hours with different concentrations of hydroxyurea or H2O2. For UV irradiation, cells were washed with PBS, supernatant was removed and cells were exposed to 254-nm light at doses of 0–20 J/m2. All cultures were then maintained in normal growth medium for 2 weeks to allow colony formation. Colonies were visualized by fixing with 3.7% formaldehyde for 15 minutes at room temperature and staining with 0.05% Crystal Violet for 30 minutes. Colonies containing 50 or more cells were counted. Final data represent the average of three independent experiments and are analyzed by repeated measures ANOVA.
Acknowledgments
We thank Hanhui Ma in the T.P. laboratory for advice on the 5-EU assay.
Footnotes
Competing interests
The authors declare no competing interests.
Author contributions
T.L., L.M., T.P. and R.Y.L.T. designed research; T.L., L.M., T.-C.L. and L.J.W performed research; T.L., T.P. and R.Y.L.T. analyzed data; T.P. and R.Y.L.T. wrote the paper.
Funding
This work was supported in part by Texas A & M Cancer Research Council Incentive Award to R.Y.L.T., National Science Council (Taiwan R.O.C.) Postdoctoral Fellowship Award to T.-C. L. and National Science Foundation [grant number MCB-1051398] to T.P.
References
- Andersen J. S., Lam Y. W., Leung A. K., Ong S. E., Lyon C. E., Lamond A. I., Mann M. (2005). Nucleolar proteome dynamics. Nature 433, 77–83. 10.1038/nature03207 [DOI] [PubMed] [Google Scholar]
- Beekman C., Nichane M., De Clercq S., Maetens M., Floss T., Wurst W., Bellefroid E., Marine J. C. (2006). Evolutionarily conserved role of nucleostemin: controlling proliferation of stem/progenitor cells during early vertebrate development. Mol. Cell. Biol. 26, 9291–9301. 10.1128/MCB.01183--06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai M. S., Sun X. X., Lu H. (2008). Aberrant expression of nucleostemin activates p53 and induces cell cycle arrest via inhibition of MDM2. Mol. Cell. Biol. 28, 4365–4376. 10.1128/MCB.01662--07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle D. M., Rossi L., Berghuis A. M., Aravind L., Koonin E. V., Brown E. D. (2002). YjeQ, an essential, conserved, uncharacterized protein from Escherichia coli, is an unusual GTPase with circularly permuted G-motifs and marked burst kinetics. Biochemistry 41, 11109–11117. 10.1021/bi020355q [DOI] [PubMed] [Google Scholar]
- Dousset T., Wang C., Verheggen C., Chen D., Hernandez-Verdun D., Huang S. (2000). Initiation of nucleolar assembly is independent of RNA polymerase I transcription. Mol. Biol. Cell 11, 2705–2717. 10.1091/mbc.11.8.2705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X., Rao M. R., Chen X. Q., Wu W., Mahalingam S., Balasundaram D. (2006). The homologous putative GTPases Grn1p from fission yeast and the human GNL3L are required for growth and play a role in processing of nucleolar pre-rRNA. Mol. Biol. Cell 17, 460–474. 10.1091/mbc.E05--09--0848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers P. B., Pereboom T. C., Goos Y. J., Paridaen J., Macinnes A. W. (2014). A comparative study of nucleostemin family members in zebrafish reveals specific roles in ribosome biogenesis. Dev. Biol. 385, 304–315. [DOI] [PubMed] [Google Scholar]
- Helleday T. (2008). Amplifying tumour-specific replication lesions by DNA repair inhibitors - a new era in targeted cancer therapy. Eur. J. Cancer 44, 921–927. 10.1016/j.ejca.2008.02.044 [DOI] [PubMed] [Google Scholar]
- Hsu J. K., Lin T., Tsai R. Y. (2012). Nucleostemin prevents telomere damage by promoting PML-IV recruitment to SUMOylated TRF1. J. Cell Biol. 197, 613–624. 10.1083/jcb.201109038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao C. Y., Salic A. (2008). Exploring RNA transcription and turnover in vivo by using click chemistry. Proc. Natl. Acad. Sci. USA 105, 15779–15784. 10.1073/pnas.0808480105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudron M. M., Reinke V. (2008). C. elegans nucleostemin is required for larval growth and germline stem cell division. PLoS Genet. 4, e1000181 10.1371/journal.pgen.1000181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T., Meng L., Li Y., Tsai R. Y. (2010). Tumor-initiating function of nucleostemin-enriched mammary tumor cells. Cancer Res. 70, 9444–9452. 10.1158/0008--5472.CAN--10--2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T., Ibrahim W., Peng C-Y., Finegold M. J., Tsai R. Y. (2013). A novel role of nucleostemin in maintaining the genome integrity of dividing hepatocytes during mouse liver development and regeneration. Hepatology 58, 2176–2187. 10.1002/hep.26600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Pederson T. (2007). Depletion of the nucleolar protein nucleostemin causes G1 cell cycle arrest via the p53 pathway. Mol. Biol. Cell 18, 2630–2635. 10.1091/mbc.E07--03--0244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Pederson T. (2008). Nucleostemin: a multiplex regulator of cell-cycle progression. Trends Cell Biol. 18, 575–579. 10.1016/j.tcb.2008.09.003 [DOI] [PubMed] [Google Scholar]
- Ma H., Pederson T. (2013). The nucleolus stress response is coupled to an ATR-Chk1-mediated G2 arrest. Mol. Biol. Cell 24, 1334–1342. 10.1091/mbc.E12--12--0881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L., Yasumoto H., Tsai R. Y. (2006). Multiple controls regulate nucleostemin partitioning between nucleolus and nucleoplasm. J. Cell Sci. 119, 5124–5136. 10.1242/jcs.03292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L., Zhu Q., Tsai R. Y. (2007). Nucleolar trafficking of nucleostemin family proteins: common versus protein-specific mechanisms. Mol. Cell. Biol. 27, 8670–8682. 10.1128/MCB.00635--07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L., Lin T., Tsai R. Y. (2008). Nucleoplasmic mobilization of nucleostemin stabilizes MDM2 and promotes G2-M progression and cell survival. J. Cell Sci. 121, 4037–4046. 10.1242/jcs.037952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L., Hsu J. K., Tsai R. Y. (2011a). GNL3L depletion destabilizes MDM2 and induces p53-dependent G2/M arrest. Oncogene 30, 1716–1726. 10.1038/onc.2010.550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L., Hsu J. K., Zhu Q., Lin T., Tsai R. Y. (2011b). Nucleostemin inhibits TRF1 dimerization and shortens its dynamic association with the telomere. J. Cell Sci. 124, 3706–3714. 10.1242/jcs.089672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L., Lin T., Peng G., Hsu J. K., Lee S., Lin S-Y., Tsai R. Y. (2013). Nucleostemin deletion reveals an essential mechanism that maintains the genomic stability of stem and progenitor cells. Proc. Natl. Acad. Sci. USA 110, 11415–11420. 10.1073/pnas.1301672110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikpour P., Mowla S. J., Jafarnejad S. M., Fischer U., Schulz W. A. (2009). Differential effects of Nucleostemin suppression on cell cycle arrest and apoptosis in the bladder cancer cell lines 5637 and SW1710. Cell Prolif. 42, 762–769. 10.1111/j.1365--2184.2009.00635.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T. (1998). The plurifunctional nucleolus. Nucleic Acids Res. 26, 3871–3876. 10.1093/nar/26.17.3871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T., Tsai R. Y. (2009). In search of nonribosomal nucleolar protein function and regulation. J. Cell Biol. 184, 771–776. 10.1083/jcb.200812014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penman S. (1966). RNA metabolism in the HeLa cell nucleus. J. Mol. Biol. 17, 117–130, IN4. 10.1016/S0022--2836(66)80098--0 [DOI] [PubMed] [Google Scholar]
- Perry R. P. (1962). The cellular sites of synthesis of ribosomal and 4S RNA. Proc. Natl. Acad. Sci. USA 48, 2179–2186. 10.1073/pnas.48.12.2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz J. C., Polena I., Trask I., Bazett-Jones D. P., Pederson T. (2005). A nonribosomal landscape in the nucleolus revealed by the stem cell protein nucleostemin. Mol. Biol. Cell 16, 3401–3410. 10.1091/mbc.E05--02--0106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu J., Bishop J. M. (2012). Nucleostemin maintains self-renewal of embryonic stem cells and promotes reprogramming of somatic cells to pluripotency. J. Cell Biol. 197, 731–745. 10.1083/jcb.201103071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynaud E. G., Andrade M. A., Bonneau F., Ly T. B., Knop M., Scheffzek K., Pepperkok R. (2005). Human Lsg1 defines a family of essential GTPases that correlates with the evolution of compartmentalization. BMC Biol. 3, 21 10.1186/1741--7007--3--21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanova L., Grand A., Zhang L., Rayner S., Katoku-Kikyo N., Kellner S., Kikyo N. (2009a). Critical role of nucleostemin in pre-rRNA processing. J. Biol. Chem. 284, 4968–4977. 10.1074/jbc.M804594200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanova L., Kellner S., Katoku-Kikyo N., Kikyo N. (2009b). Novel role of nucleostemin in the maintenance of nucleolar architecture and integrity of small nucleolar ribonucleoproteins and the telomerase complex. J. Biol. Chem. 284, 26685–26694. 10.1074/jbc.M109.013342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosby R., Cui Z., Rogers E., deLivron M. A., Robinson V. L., DiMario P. J. (2009). Knockdown of the Drosophila GTPase nucleostemin 1 impairs large ribosomal subunit biogenesis, cell growth, and midgut precursor cell maintenance. Mol. Biol. Cell 20, 4424–4434. 10.1091/mbc.E08--06--0592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherl A., Couté Y., Déon C., Callé A., Kindbeiter K., Sanchez J. C., Greco A., Hochstrasser D., Diaz J. J. (2002). Functional proteomic analysis of human nucleolus. Mol. Biol. Cell 13, 4100–4109. 10.1091/mbc.E02--05--0271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai R. Y. (2011). New frontiers in nucleolar research: nucleostemin and related proteins. The Nucleolus: Protein Reviews 15, 301–320. 10.1007/978--1--4614--0514--6_13 [DOI] [Google Scholar]
- Tsai R. Y., McKay R. D. (2002). A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes Dev. 16, 2991–3003. 10.1101/gad.55671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai R. Y., McKay R. D. (2005). A multistep, GTP-driven mechanism controlling the dynamic cycling of nucleostemin. J. Cell Biol. 168, 179–184. 10.1083/jcb.200409053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai R. Y., Meng L. (2009). Nucleostemin: a latecomer with new tricks. Int. J. Biochem. Cell Biol. 41, 2122–2124. 10.1016/j.biocel.2009.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita M., Nitta E., Nagamatsu G., Ikushima Y. M., Hosokawa K., Arai F., Suda T. (2013). Nucleostemin is indispensable for the maintenance and genetic stability of hematopoietic stem cells. Biochem. Biophys. Res. Commun. 441, 196–201. 10.1016/j.bbrc.2013.10.032 [DOI] [PubMed] [Google Scholar]
- Yasumoto H., Meng L., Lin T., Zhu Q., Tsai R. Y. (2007). GNL3L inhibits activity of estrogen-related receptor gamma by competing for coactivator binding. J. Cell Sci. 120, 2532–2543. 10.1242/jcs.009878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q., Yasumoto H., Tsai R. Y. (2006). Nucleostemin delays cellular senescence and negatively regulates TRF1 protein stability. Mol. Cell. Biol. 26, 9279–9290. 10.1128/MCB.00724--06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q., Meng L., Hsu J. K., Lin T., Teishima J., Tsai R. Y. (2009). GNL3L stabilizes the TRF1 complex and promotes mitotic transition. J. Cell Biol. 185, 827–839. 10.1083/jcb.200812121 [DOI] [PMC free article] [PubMed] [Google Scholar]