Abstract
DNA-encoded libraries represent an exciting and powerful modality for high-throughput screening. In this article, we highlight recent important advances in this field and also suggest some important directions that would make the technology even more powerful.
Introduction
Most bioactive small molecules are now discovered via some type of high-throughput screening (HTS) campaign. Traditional HTS involves dispensing tens to hundreds of thousands of individual compounds into the wells of microtiter plates, along with the target enzyme and appropriate reagents to monitor enzyme activity. The effect of each compound on the activity of the enzyme is then assessed. This screening format has also been utilized to carry out other types of biochemical screens, for example the disruption of protein-protein interactions, as well as phenotypic assays using cells engineered to report an event of interest. All such plate-based screens rely on a sophisticated robotic infrastructure, since it is impractical to carry out thousands of assays manually.
In sharp contrast, peptide ligands for a target protein of interest can be identified by screening peptide libraries in batch mode (i.e. all of the molecules are present in a single tube or well) using techniques such as phage display1 or ribosome display2–4. The critical feature of these screening schemes is that each peptide is linked physically to an encoding DNA. When peptides that bind a target protein are separated from those that do not, the encoding DNA is carried along. These tags are then amplified and sequenced. This process can be repeated several times in order to “evolve” a peptide to bind selectively to the target protein, yielding high affinity ligands in a far cheaper and operationally simpler format than standard HTS.
For many years it has been the dream of chemical biologists to apply this type of screening methodology to synthetic, rather than genetically encoded, libraries. The idea was first introduced in 1992 by Brenner and Lerner5 and accomplished experimentally by Gallop and colleagues6 in the following year. However, this pioneering work was severely limited by the crude DNA sequencing technology of the time, which involved Sanger analysis of one DNA sequence at a time. The advent of deep sequencing (also called next generation sequencing)7, which allows millions of individual DNAs to be sequenced simultaneously, changed everything. Once a large library of DNA-small molecule conjugates was created, it became feasible to simply incubate it with a protein of interest, pull down that protein and sequence the entire population of encoding tags that co-precipitate due to a small molecule-protein binding event. Truly a synthetic analogue of phage display.
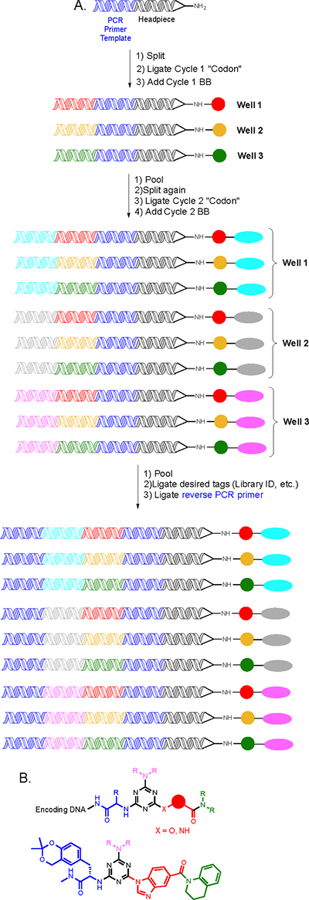
Several novel platforms have been described for the synthesis of DNA-encoded libraries (DELs). In most, the DNA is merely a bystander. But in others, the DNA tag actually templates the reactions used to couple building blocks8. In others, single-stranded DNA is tethered to small molecular weight fragments. Association of the DNA strands allows the fragments to engage protein targets as bivalent ligands (the DNA “bridge” is later replaced with a suitable synthetic linker)9. Perhaps the most popular platform for DNA-encoded library (DEL) synthesis was first described by workers at Praceis (now GSK)10. Synthesis commences from a “headpiece DNA” with a tuning fork-like structure (Fig. 1). At the bottom of the “handle” is an amino group or some other chemical functionality that serves as the starting point for library synthesis. The forks are two covalently linked complementary DNA strands with a single-stranded overhang onto which a PCR primer template site is ligated. The venerable split and pool synthesis scheme11, 12 (Fig. 1) is then utilized to create a combinatorial library of DNA-encoded small molecules. While the split and pool procedure is most often associated with solid-phase synthesis to make one bead one compound (OBOC) libraries, it can be used here because the DNA conjugates are readily precipitated in ethanol, allowing excess chemical reagents to be removed after each step. In other words, the DNA also serves the function of a solid support. After the split, both a chemical step and a ligation event are carried out prior to pooling. The encoding DNAs are small duplex oligonucleotides with single-stranded overhangs to facilitate ligations. The duplex regions contain a unique sequence that encodes the chemical unit added in that tube. Once the desired library is created, certain barcodes are added, and finally a reverse PCR primer template. Split and pool synthesis facilitates the creation of large libraries with only a few chemical steps. For example, building a molecule from three variable building blocks using 100 different units at each position would yield one million (1003) unique molecules. The first library reported by the Praceis/GSK group contained over seven million molecules, significantly exceeding the size of any existing screening collection. In the same paper, they reported a more elaborate library putatively containing about 800 million unique compounds (Fig. 1B). A screen of this library against p38 MAPK yielded several high affinity ligands (Fig. 1B).
Figure 1. Split and pool synthesis of a DNA-encoded library.
A. Schematic depiction of a two cycle DEL using three building blocks at each position to create nine unique species. In general, the size of libraries created by split and pool chemistry will be xy, where x = the number of unique building blocks used at that position and y = the number of cycles used to create the library. B. The general structure of an 800 million compound library containing a central triazine core. The structure of a screening hit against p38 MAP kinase (the identity of the unit shown in pink was not relevant to binding).
What DELs have been made and what DELs should be made?
One of the most appealing features of encoding a library constructed by split and pool synthesis11, whether with DNA or using some other strategy13, is the vastly expanded chemical space that can practically be explored. Since it impossible to keep track of individual beads (or molecules in the case of solution-phase synthesis of DELs) in the split and pool process, the molecular structure of screening hits from a non-encoded library must be determined de novo, usually by tandem mass spectrometry (MS/MS). This limits the nature of the molecules that can be practically used on the OBOC platform to peptides11 and other oligoamides such as peptoids14, 15, which fragment in a logical fashion. Encoding removes this constraint.
Given this freedom to explore almost any region of “chemical space”, as long as it can be accessed with DNA-tolerant reactions (vide infra) what type of library should one make? Different chemists would give justifiably different answers, depending on their priorities. But most researchers in the field agree upon a few general guidelines. First, “stiffness” is good, at least to a point. “Floppy” molecules tend to bind to proteins with only modest affinity, in part because of the high entropic cost for a floppy molecule to assume a single bound conformation. Second, “three-dimensionality” is desirable. In other words, one should avoid achieving conformational restraint simply by including mostly sp2 centers and fused aromatic rings in the molecules. This is known colloquially as “escaping flatland”16, 17. While flat, hydrophobic, aromatic molecules can bind proteins in aqueous solution with high affinity, selectivity can be a major problem. Third, scaffold diversity is highly desirable17. If one creates many millions of molecules all containing a single scaffold, a high rate of screening failure is likely since only a small number of bindable surfaces on a protein are likely to be suitable partners for that scaffold. Finally, if one wishes to target an intracellular biomolecule, then the library should be comprised of mostly cell permeable molecules.
This review focuses on the chemical composition of DELs. Several other reviews are available that focus on different drug leads that have been found using DELs and strategies for employing DELs18–21. First, we consider, in general terms, the strengths and weaknesses of DELs that have been reported to date with respect to the features described immediately above. Next we attempt to predict what types of libraries will be made in the near future as library developers attempt to include more and more of these desirable characteristics into their designs. In particular, we focus on the important issues of achieving stereochemical complexity and scaffold diversity. Finally, we argue that drugging certain classes of targets is likely to require higher molecular weight molecules, which has not been a major focus of the field so far. The creation of high quality libraries of such molecules is not trivial because of the sensitivity of DNA to many of the reactions commonly used to splice together building blocks. Larger molecules requiring many steps (“cycles” in DEL parlance) to construct will force researchers pay greater attention to the number of tags that remain amplifiable after all of the chemistry is complete. Therefore, a brief primer on DNA sensitivity and experimental methods to monitor the integrity of the encoding material is presented towards the end of this review.
What Kind of DELs Have Been Made So Far?
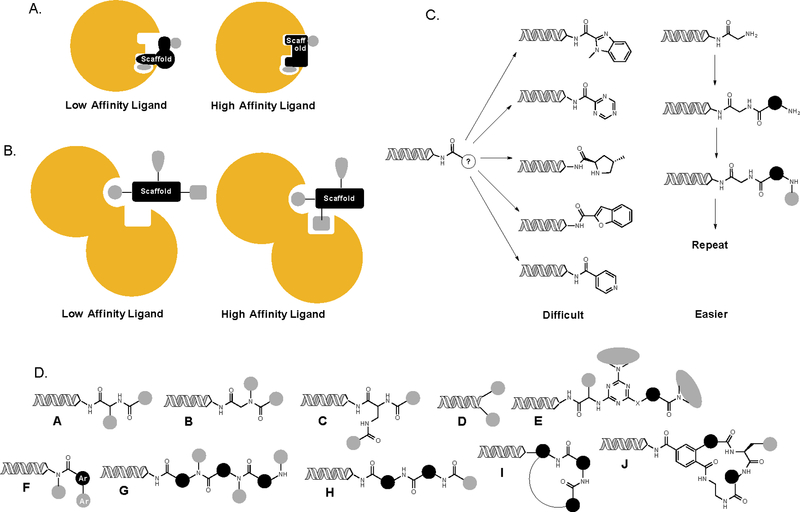
Most DELs reported to date reflect some of the desirable features mentioned above, but certainly not all. Libraries of low molecular weight (MW) molecules dominate DEL chemical space so far. This is in part to ensure compliance with the usual guidelines for drug likeness22, 23, especially since much of the activity in this arena has taken place in industry. It also largely avoids the problem of DNA damage, since only a few steps (known as “cycles” in DEL parlance) are required to complete the synthesis. However, low MW DELs are not at all like traditional HTS screening collections. Most DELs are “scaffold-poor” and “appendage-rich”, built around a small number of core scaffolds, often only one, as was the case with the first Praceis library10. Before proceeding further, let us define carefully what we mean when we use the terms scaffold and appendage (see Fig. 2), since these are core concepts in this review. A medicinal chemist who deals largely with low MW molecules would say that a compound collection is scaffold-diverse if it contains many different types of ring structures (pyridines, furans, benzimidazoles, oxazoles, etc.). These scaffolds are decorated with appendages. This type of scaffold diversity is important because in low MW ligands, the scaffold almost invariably makes critical contacts with the target biomolecule (Fig. 2A). In this view, it would foolish to assemble a screening collection containing only a single scaffold. Any particular scaffold might be good for recognizing a particular type of protein pocket (for example adenine-like heterocycles binding to kinase active sites), but would fail to engage with a plethora of other types of pockets. Indeed, traditional HTS collections recognize an implicit compromise in which appendage diversity on any given scaffold is sacrificed for scaffold diversity. The goal is generally to first identify what scaffold is right for the particular target of interest by identifying a moderate potency primary hit, then to carry out a medicinal chemistry campaign to explore the effect of many different appendages on that scaffold. This is now often done by creating focused libraries that explore chemical space in the immediate vicinity of a primary screening hit. Most current DELs can thus be thought of as the equivalent of a “focused library on steroids”. They exhaustively cover essentially all appendage space around one or a very few core scaffolds. There is good reason for this. It is quite difficult to imagine creating many different heterocyclic scaffolds from a common precursor during the course of a split and pool library synthesis (Fig. 2C). Moreover, many methods to make heterocycles employ harsh conditions that would be incompatible with DNA. It is much easier to add many different appendages to a single scaffold, for example coupling a wealth of different amines to the triazine core in the Praceis library. Larger companies ameliorate this problem to some extent by creating many different libraries containing different scaffolds. These are screened all at once against a given target by assigning each library a unique barcode. However, this approach is likely impractical for academic laboratories and smaller commercial facilities that practice DEL technology. Therefore, it can be safely said that the development of strategies to create much more scaffold-rich low MW DELs is an important goal. Some promising approaches to this problem will be discussed below.
Fig. 2. Scaffold diversity.
A. Scaffolds (black) in low molecular weight (MW) molecules usually make critical contacts with the target biomolecule in addition to positioning appendages in a given three-dimensional orientation. Thus, it is important to have chemically diverse core scaffolds in libraries of low MW molecules. B. In higher MW foldamer-type molecules, the scaffold is not expected to engage the target biomolecule directly. Here, the critical issue is to hold appendages in relatively inflexible and distinct three-dimensional orientations. C. Scaffold diversity, especially with respect to heterocyclic cores, is difficult to achieve in split and pool chemistry due to chemical limitations. It is much easier to construct stiff building blocks onto which appendages can be grafted and oligomerize these into diverse scaffolds. D. A graphical representation of several existing DELs. Many of the lower MW libraries (A-D) have essentially no scaffold diversity, whereas some contain several (E,F). Some scaffold-diverse oligomer libraries (G, H) have been reported, including macrocycles (I,J). Ar = aryl.
A chemist working in the peptidomimetic “foldamer”24 arena would have a rather different view of scaffold diversity. In this field, the goal is to construct a non-natural molecule that mimics the secondary structure of a region of a protein that interacts with a partner protein. For example, several innovative designs have been put forth that display 2–3 appendages in a similar orientation as the side chains on one face of a protein α-helix 25–27. In the foldamer arena, the scaffold is usually not expected to make important contacts with the target, but is simply asked to display several appendages in a particular three-dimensional orientation (Fig. 2B). Achieving scaffold diversity in this sense is more straightforward (Fig. 2C). One would simply have to string together conformationally constrained units, each bearing one or more appendage, into an oligomer. The scaffold diversity would equal xy, where x = the number of building blocks used at each variable position and y is the number of units in the oligomer. For example a trimer made using 10 building blocks at each site would have a scaffold diversity of 1000. Again, this view of scaffold diversity only holds if the “main chain” of the molecule is quite stiff. Libraries of short peptides, peptoids and indeed most short oligomers do not have high scaffold diversity because they are floppy. As will be discussed below, libraries of foldamer-type libraries have been used broadly already and are just now coming into use for DELs.
Fig. 2D provides a graphical representation of representative DELs reported in the literature28,10, 29–35,36. The black circles represent main chain elements and the gray circles appendages. Library designs A-D have low scaffold diversity, library designs E and F have moderate scaffold diversity and G-J have higher scaffold diversity. Again, this assumes that the main chain scaffolding units (black in Fig. 2) have little conformational flexibility.
What kind of DELs will be made in the near future?
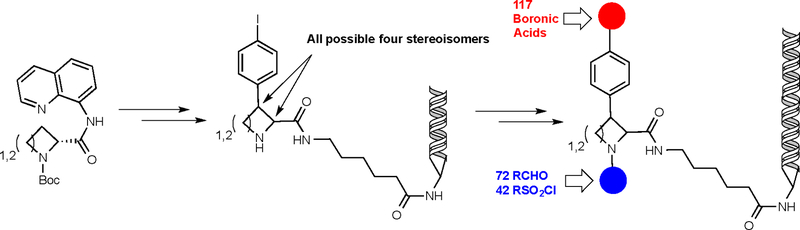
While it is always dangerous to predict the future, one safe answer to the above question is that DELs containing molecules resembling natural products will constitute a major goal for chemists working in this area. Natural products have many of the desirable traits mentioned above. Along these lines, a particular focus will be to increase the stereochemical complexity of molecules in DELs as a way of “escaping flatland”16. One way to do this is to create all of the isomers of a given scaffold with one or more stereocenters (each of which represents a unique scaffold in the Fig. 2B sense), attach each of them to a DNA headpiece and then elaborate these cores in an encoded synthesis. This strategy is applicable to the creation of classically drug-like, Lipinski-compliant23 molecules that are able to target enzyme active sites. A recent illustrative example of this strategy was reported by Clemons, Schreiber, and co-workers37. They used a stereospecific C-H arylation route to produce all of the stereoisomers of the azetidine and pyrrolidine core scaffolds shown in Fig. 3. A DEL of just over 100,000 molecules was then produced by attaching these scaffolds to DNA and elaborating them by either sulfonylation or reductive amination of the amine, followed by Pd-catalyzed displacement of the iodide with a boronic acid. The library was screened against carbonic anhydrase IX, yielding several compounds that inhibit the enzyme with mid- nM Kis.
Figure 3.
A DEL based on eight stereochemically-rich scaffolds. These were synthesized using DNA-intolerant chemistry. Once the scaffolds were complete, each was connected to DNA and the substituents shown in blue and red were attached in an encoded fashion using DNA-tolerant coupling reactions.
This general strategy acknowledges that many of the tools of modern organic synthesis, in this case the quinoline-directed C-H arylation reaction used to add the phenyl iodide to the ring, are incompatible with DNA. As will be discussed in more detail below, this is a defining issue in the synthesis of DELs. Therefore the chemistry to generate stereochemical diversity and the core scaffolds is done “off-line”.
In another example, the DEC-TEC Core at Baylor College of Medicine synthesized a set of enantiomerically pure substituted, piperazine acetic acid esters that are well-poised for subsequent diversification38, 39. These units could be easily installed into traditional DNA-encoded libraries and be transformed using already-established chemistries to afford skeletally diverse, high diversity libraries. However, they are similarly restricted by their laborious building-block preparation.
The DEL shown in Fig. 3 could be given a thumbs up or a thumbs down with respect to scaffold diversity depending on one’s point of view. While every molecule in the library contains an azetidine or pyrrolidine core, the inclusion of all possible stereoisomers of the rigid scaffold provides eight, rather than two, distinct scaffolds. This is clearly far better than one, which is typical of many DELs, so in that sense it is an excellent design. On the other hand, eight is a modest number compared to the scaffold diversity of typical classical HTS screening collections.
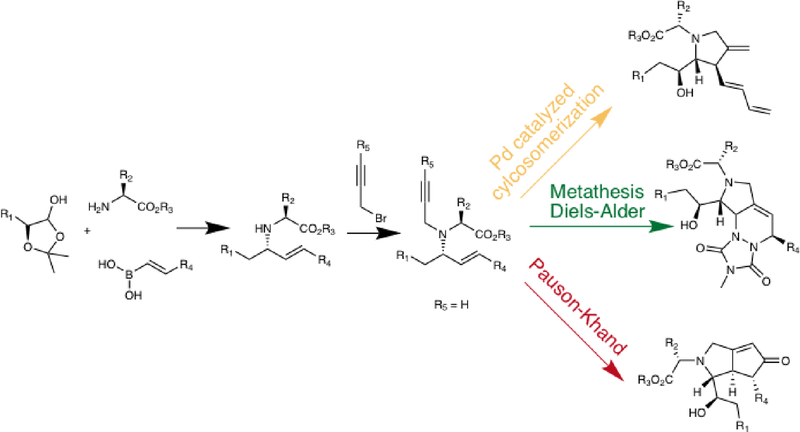
One way to begin to bridge this gap might be to adapt the so-called build-couple-pair (BCP) strategy40 to DEL synthesis. The BCP procedure is capable of creating dozens of different scaffolds in a single library (Fig. 4). In the build phase, stereochemically diverse building blocks with appropriate functional groups to support linkages are created. In the couple phase, they are linked together. Finally, in the pair phase, the coupled products are subjected to some type of intramolecular reaction, usually a ring-closing process, to yield highly rigidified and complex molecular scaffolds. For example, Fig. 4 shows work from the Schreiber laboratory in which a scaffold-diverse screening collection was created through a BCP strategy that included a Petasis condensation and an amine alkylation in the couple phase, then subjected these products to various ring-closing reactions (Fig. 4)41. For example, Pd-catalyzed cycloisomerization reactions provide complex pyrrolidines. Alternatively, the Grubbs catalyst was employed to transform the coupled products into dienes that entered into Diels-Alder reactions with suitable dienophiles. Fused bicyclic compounds were created using the Pauson-Khand reaction.
Figure 4. Schematic illustration of the Build Couple Pair strategy.
Adapted from ref. 32 DOI: 10.1002/anie.200600497.
To the best of our knowledge, the BCP strategy has not been used in the context of split and pool synthesis of DELs. This will not be without challenges, for example developing more pairing reactions that are DNA compatible, but we believe that this approach has a bright future.
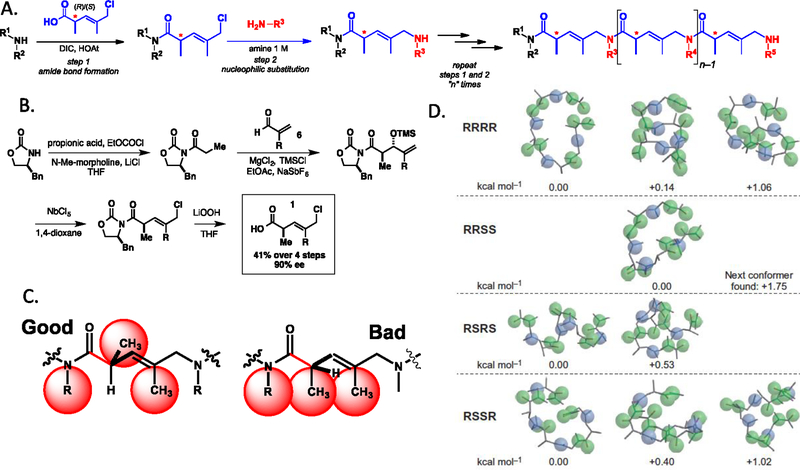
The creation of DELs comprised of foldamer-type, conformationally-constrained, non-peptidic oligomers is likewise at an early stage. Like the library shown in Fig. 3, those that incorporate some level of stereochemical diversity do so by employing pre-formed chiral sub-units. A good example is the class of compounds called COPAs (chiral oligomers of pentenoic acids, Fig. 5). COPAs are constructed via a simple, peptoid-like42 synthetic scheme (Fig. 5) using either enantiomer of a chiral pentenoic acid and diverse amines. Avoidance of allylic 1,3 (A1,3) strain strongly encourages the hydrogen atom at the chiral center in each unit of the oligomer to sit in the plane occupied by the allylic methyl group and the N-substituent just upstream in the chain. Thus, the chain takes a “right turn” or “left turn” at the chiral center depending on the absolute stereochemistry at that position (Fig. 5). Calculations suggest that only a few conformers of even short COPAs can be populated, making them true foldamers.
Figure 5. COPAs, a class of peptoid-inspired, conformationally constrained oligomers (PICCOs)43.
A. Synthesis of COPA oligomers follows the “sub-monomer” scheme developed by Zuckermann and colleagues for peptoids42. B. Scheme for the asymmetric synthesis of COPA sub-monomers. C. Cartoon illustrating the conformational preferences that cause COPAs to be relatively stiff foldamers. D. Predicted low energy conformers of a COPA tetramer depending on the absolute stereochemistry of each sub-unit. The blue balls represent the alkene atoms and the green balls heteroatoms. Reprinted with permission form ref. 44.
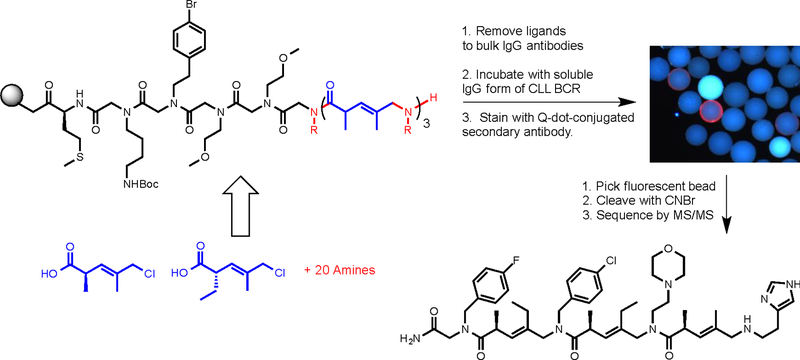
While DELs comprised solely of COPA sub-units have not yet been made, this chemistry has been used to make non-encoded libraries by split and pool synthesis, since COPA structures can be determined by MS/MS45. Specifically, Sarkar, et al. reported a library of about 1.3 million tetramers in which the first unit was a peptoid and the next three were either peptoids or COPAs (using both enantiomeric building block and mass-encoding the absolute stereochemistry)46. 20 amines were used as variable elements at each position. This library was made by solid-phase synthesis, producing a one bead one compound (OBOC) library that was screened on-bead under demanding conditions for ligands to a B cell receptor (BCR) cloned from a chronic lymphocytic leukemia (CLL) patient. 16 high quality hits ligands were identified from this screen, with the highest affinity compound (2; Fig. 6) binding to the CLL BCR with a KD of about 90 nM. Notably, all of the 16 screening hits contained a COPA unit at positions 2–4. No peptoids were pulled out as hits even though they were in the library. Moreover, changing the absolute stereochemistry of any of the chiral centers in hit 2 essentially abolished binding46. These data argue that the conformation of the oligomer is critical to binding, though the structure of the complex has not been solved.
Figure 6. A COPA library screen.
The library was constructed by split and pool solid-phase synthesis using the COPA building blocks shown and 20 different amines at each variable position. The general structure of the library is shown at the top left. The OBOC library was first incubated with bulk IgG antibodies and a fluorescently labeled secondary antibody. Beads that retained the label were removed to eliminate ligands from the library that recognize the conserved regions of an IgG antibody. The remainder of the library was then incubated with an antigen-specific CLL BCR and a fluorescently labeled second antibody. Beads that displayed a bright red halo (top right) were harvested. After release from the bead, the structure of the ligand was determined by mass spectrometry. One of the highest affinity hits from this screen is shown at the bottom right.
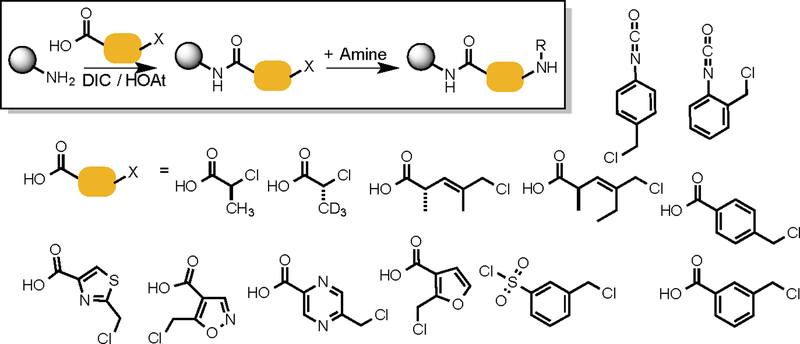
A library including three chiral pentenoic acid elements can be considered to have eight (23) different scaffolds. Again, eight scaffolds are better than one, but much less desirable than dozens or even hundreds. In order to achieve much greater levels of scaffold diversity, we have begun to develop DELs of oligomers that we call PICCOs43 (peptoid-inspired, conformationally constrained oligomers, Fig. 7), of which COPAs are a specific subset. Like COPAs, PICCOs are made using the sub-monomer scheme developed for peptoids42, but employ a variety of conformationally constrained building blocks in place of chloroacetic acid, not simply the two chloropentenoic acid enantiomers. By “blowing up” the number of different backbone units used to make the oligomer, extremely scaffold-diverse libraries of oligomers can be created, again using this term in the foldamer sense.. Of course, the backbone can also enter into interactions with the protein target directly, rather than merely acting as a scaffold (Fig. 2A). Given that PICCOs contain several heterocyclic main chain units 47, this is likely to occur. Thus PICCOs fit both definitions of scaffold diversity shown in Fig. 2A, the more likely this is to occur. It should be noted that other laboratories have developed distinct strategies to produce libraries of relatively stiff synthetic oligomers, for example the elegant α-helix mimics reported by Lim and co-workers48. But these lack the scaffold diversity of PICCO libraries.
Fig. 7. Synthesis of PICCOs.
Top box: The peptoid-like protocol employed to synthesize PICCOs. The Structures of the sub-monomers used (along with diverse amines) to date to make PICCOs.
An enormous limitation to using the full range of PICCO sub-units in a non-encoded library is the widely different efficiencies of fragmentation between different main chain units in the tandem mass spectrometer. This makes the de novo characterization of screening hits almost impossible. Encoding is essential49. Thus, an important development was the adaptation of the Praceis DNA-encoding technology to split and pool solid-phase synthesis by Paegel and co-workers50. In this scheme, only a small percentage (<1%) of the chains on each bead are tethered to an encoding DNA, allowing the library to be screened on-bead without interference from a “forest” of DNA chains on the bead surface. DNA-encoded OBOC library synthesis enables the use of the full PICCO building block set, since MS-based hit characterization is no longer necessary. Moreover, since the encoding tags of screening hits will be amplified by PCR prior to deep sequencing, only a small amount of material is required, allowing the libraries to be constructed on tiny (10 µm) TentaGel beads, which pass easily through a standard flow cytometer51. Thus, screens can be done by incubating the library with a fluorescently labeled target protein doped into a large excess of diverse, unlabeled competitor proteins. After washing, the beads are passed through the flow cytometer and those that capture a high level of fluorescent protein on their surface are collected34, 51. Since we wish to focus on the chemical composition of DELs here, a discussion of the details of this screening modality is beyond the scope of this article, except to say that the ability of flow cytometers to monitor multiple wavelengths opens up new possibilities for how DELs are screened. For example, one could easily incorporate a stringent selectivity filter into a screen by labeling the target with a red dye and off-targets with a green dye, then gating the flow cytometer to collect beads that exhibit a high level of red, but a low level of green, fluorescence (see ref. 34 for an example of this approach). It is possible that the same type of selectivity filter could be achieved by parallel or sequential screening of soluble DELs against a target and off-targets, then comparing the DNA sequences that co-precipitate with the protein in each case. But, to the best of our knowledge, this remains to be determined.
DELs of macrocycles
Macrocyclization of linear oligomers greatly reduces the degrees of freedom available to the molecule, especially when the linear compound is inherently floppy like a peptide or peptoid. Even stiffer oligomers such as PICCOs contain some bonds with little restriction to rotation and therefore macrocyclization of these species would be of interest as well.
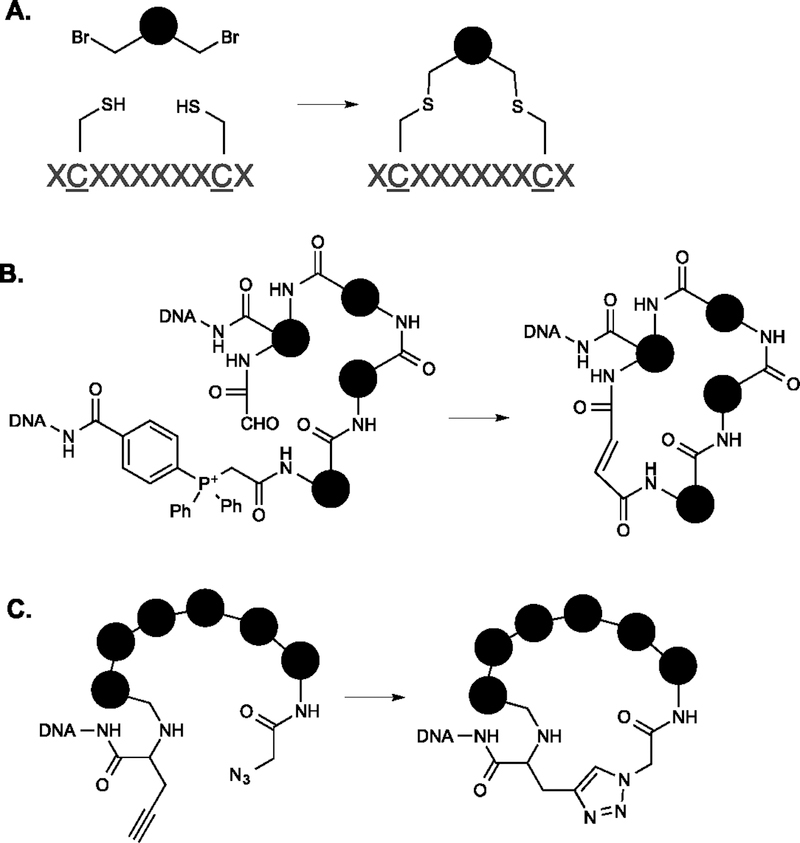
An abundance of work on DNA-encoded macrocyclic peptide libraries made by the ribosome has amply demonstrated the utility of this class of molecules as protein ligands. Many selective, high affinity protein binders have resulted from such screens2, 4. Clever schemes have been developed for cyclizing the linear precursors. One that merits particular mention is the strategy of Heinis and co-workers in which macrocyclic mono- or bi-cycles are constructed from linear peptides via multiple alkylation reactions Fig. 8A)52. By varying the nature of the electrophilic moiety, different macrocyclic scaffolds can be generated from the same linear precursor53. Because of the constraints of the chemical matter present in genetically encoded peptides, this strategy has been largely limited to multiple thioether bond-forming reactions. But in fully synthetic macrocycles, one can imagine using a similar strategy with many other types of reactions.
Fig. 8. DELs of macrocycles.
A. Peptide macrocycles formed by multiple thioether linkages. B. Synthetic macrocycles formed via a Wittig reaction. C. Synthetic peptide macrocycles formed via Click chemistry.
The first synthetic macrocyclic DELs were reported by Liu and co-workers using their DNA-templated synthesis platform. They constructed a library of 13,824 compounds, using a Wittig reaction to close the macrocyclic ring (Fig. 8B)8, 54. They were able to identify high quality ligands for several kinases, including some with nM affinity. An insulin-degrading enzyme (IDE) inhibitor was also isolated from this library35. More recently, the same group described a second generation library of similar design, but with many more compounds (256,000)55. These pioneering efforts in the DEL macrocycle space highlight the potential utility of such libraries.
More work in this area has been forthcoming. Zhu, et al. reported the synthesis of a macrocyclic peptide DEL in which the ring was closed by a copper-catalyzed Huisgen cycloaddition (Click reaction). This library (Fig. 8C) contained, theoretically, 2.4 × 1012 unique molecules of varying scaffold size from 4 to 20 amino acids56. They reported good ligands for the RSV N protein and the VHL E3 ubiquitin ligase. A different DEL of peptide macrocycles was reported by Neri and co-workers that contains a single macrocyclic scaffold and a high level of appendage diversity57. Very recently, Gillingham and colleagues reported a macrocyclic DEL (design J in Fig. 2D) containing over one million compounds with good scaffold diversity 36.
We believe that DELs of macrocycles have an extremely bright future. In addition to the ample evidence that they are capable of targeting protein-protein interaction surfaces and other difficult-to-drug targets, Heath and co-workers (using non-encoded macrocyclic peptide libraries) have demonstrated that macrocycles are even able to bind linear peptide epitopes58, 59.
In the future, however, the further advancement of this technology will require meeting two significant challenges. The first is to develop classes of macrocycles that are more likely to be cell permeable than peptides. The secondary amide bonds of peptides are highly hydrated. Shedding this shell of water represents a formidable barrier to crossing a membrane60, 61. Macrocyclic peptide natural products that penetrate cells tend to have multiple tertiary amide bonds (i.e. N-methyl amino acid units) and can adopt conformations that promote intracellular hydrogen bonding, thus reducing the hydration shell62. However, the conditions required to acylate N-alkylated amino acids are quite harsh and unlikely to be compatible with the synthesis of DELs49, 63, 64, so simply mimicking Nature’s strategy of creating libraries of macrocycles with multiple N-methyl amino acids will be difficult.
Second, most libraries of macrocycles reported to date were built by cyclizing collections of quite floppy linear precursors, such as peptides. One can imagine that most of the molecules in a library of this type will cyclize with reasonable efficiency. Unfortunately, the lack of conformational restraint will persist to some degree even in the macrocycle. Indeed, in the extreme case of peptoids, where the amide bonds explore both the cis and trans conformations, we have shown that macrocyclization makes little difference when the ring is relatively large (20 or more atoms)65. In other words, libraries of linear peptoids and the analogous large ring macrocycles provide protein ligands of approximately the same quality when screened under identical conditions. It is only when the ring becomes small (17 atoms or less) that a macrocyclic peptoid library yields higher affinity ligands than the analogous library of linear peptoids. Presumably, this ring size eliminates some of the conformers that the larger rings can explore. It stands to reason that it would be desirable to make libraries of macrocycles from linear precursors that have substantial conformational restraints to begin with, such as PICCOs. But this will inevitably create issues with macrocyclization yields for molecules where the most stable conformation prevents the reactive ends from coming into close proximity. In a scaffold-diverse library, some linear precursors of this type will cyclize well and some will not. So it will be important to develop strategies to either identify these problematic cases at the stage of library design and exclude them from the synthetic scheme, or come up with some way to remove linear molecules from the library prior to screening. Alternatively, one could simply live with the fact the library will contain many linear molecules.
Compromises in the construction of DELs: Scaffold-rich or appendage-rich libraries?
If a high level of stereochemical and chemotype diversity in the core scaffolding of a DEL is desirable, does this require that compromises be made in library design with respect to the level of appendage diversity? If one creates OBOC DELs by solid-phase synthesis, the answer is clearly yes. The number of unique molecules in the library cannot exceed the number of beads used for the synthesis. There are about 300 million particles in 100 mg of 10 µm TentaGel resin, which is the scale on which we routinely make libraries, so this represents the ceiling of library size. In practice, it must be at least 10- to 100-fold smaller, since multiple copies have to be made if one wishes to screen on resin66. While 3–30 million is still a large number, it does not cover the theoretical diversity one might ideally like to achieve. For example, a PICCO library comprised of four units made from ten different chloro acids and twenty amines (Fig. 7) would have a theoretical diversity of two billion molecules (104 × 204), exceeding the number of beads by a large margin.
For libraries created by solution-phase split and pool synthesis, the answer is less clear. These DELs are limited only by the number of molecules one begins with, and thus can be enormous. However, even for DELs created by solution-phase synthesis, compelling arguments are emerging to hold the line at 10–100 million molecules at most67, 68. While a detailed discussion of these data is beyond the scope of this article, the basic argument is that when the theoretical diversity of the library approaches the number of reads one can achieve in a deep sequencing run (currently 100–1000 million, depending on the instrument), the more difficult it is to distinguish real hits from noise in a screening experiment. A much better situation is to be able to cover the entire library several times over with a single sequencing run. This allows one to assess the enrichment of encoding units in screening hits relative to the naive library in a statistically significant manner37. So even for DELs made by solution-phase synthesis, there is a practical size limit, though what that number is remains a matter of debate.
If a compromise must be made, we would like to make the argument that it is much better to strive for maximum scaffold diversity in a primary screening library, even if this means reducing appendage diversity significantly (see ref. 17 for DEL screening data supporting this view). In classical HTS, it is a given that primary screening hits will usually display modest activity. The goal is to identify a scaffold that can be elaborated in a follow-up medicinal chemistry campaign to develop more potent leads. We believe it would be valuable to adopt this mentality in the DEL world as well, but this is not common. As mentioned above, most of the DELs in existence are appendage-rich and scaffold-poor. These are essentially state of the art versions of the focused libraries a medicinal chemist would make to exhaustively examine chemical space in the immediate neighborhood of a primary screening hit.
Building stereochemical complexity during DEL synthesis
Returning to the topic of making more stereochemically complex libraries, to the best of our knowledge, all combinatorial libraries constructed to date have employed a “pre-fab” strategy to incorporate chiral elements. In other words, the chiral centers come from chiral building blocks, for example the azetidine and pyrrolidine scaffolds of the Fig. 3 library or the pentenoic acid precursors of COPAs (Fig. 5). Keeping in mind the desirability of making scaffold-rich libraries, this approach will necessitate the maintenance of gram quantity stocks of many different chiral building blocks, each of which might require several steps to make. This is tolerable for large, well-funded organizations like pharma companies, but will strain most academic laboratories. Therefore, a major goal in the future must be to develop more asymmetric reaction that are DNA-compatible, allowing the generation of chirality during synthesis of the library from inexpensive achiral building blocks.
One likely source of DEL-relevant asymmetric reactions is the field of organocatalysis69, which has exploded over the last several years. These reactions generally proceed under mild conditions that are unlikely to compromise the amplification of DNA encoding tags. Indeed we have recently shown that the proline-catalyzed aldol or Mannich addition 70, 71 of certain ketones to bead-displayed aldehydes proceeds in good enantiomeric excess and high chemical yield72, 73. This chemistry incurs no detectable DNA damage. To date these methods have not been used to create DELs, but work towards this goal is in progress.
Several other organocatalytic reactions have been reported in the literature that, to our knowledge, have yet to be applied to DEL synthesis, but would appear to be promising. For example, catalysts have been reported for the asymmetric Henry reaction under conditions that are highly likely to be DNA-tolerant74. A number of synthetically useful transformations are catalyzed by thiourea-containing molecules under mild conditions that would almost certainly be useful for DELs75–77. The list goes on. Therefore, another safe prediction is that organocatalytic reactions will play a major, perhaps even dominant, role in the synthesis of DELs in the near future. This is likely to be the simplest path to the first generation of DELs that truly resemble natural products.
DNA is not Teflon: The importance of quantitative assessment of DNA damage
We have mentioned several times above the rather obvious fact that chemistry used to build DELs must be DNA tolerant. A compound tethered to an encoding tag that cannot be amplified by the PCR is effectively absent from the library. Methods that employ harsh acidic or basic conditions, in particular, must be avoided. Unfortunately, many of the workhorse methods for asymmetric synthesis, for example the Evans aldol condensation, are thus out of bounds. We are unaware of reports of the DNA tolerance of other standout asymmetric reactions such as Jacobsen78 or Sharpless79 epoxidations, or Sharpless dihydroxylation of alkenes80, but it would not be surprising if they damage DNA. Many functional group transformations that are conducted routinely in total synthesis, such as the oxidation of a primary alcohol to an aldehyde, are almost likely problematic.
However, while it is clear to everyone in the field that there exist “black and white” dividing lines of reactions that can and cannot be used for DEL synthesis, perhaps less appreciated is the grey zone of reactions that employ conditions that wipe out some, but not all, of the encoding tags. A good example is the acylation of amines, probably the most commonly employed reaction in the construction of DELs. Using a quantitative PCR assay comparing DNA tags that had or had not been exposed to reaction conditions, Malone and Paegel81 showed that between 20–80% of the DNA population was rendered non-amplifiable depending on the acylation conditions employed. Typical Suzuki conditions damaged 50%−70% of the tags. So while these reactions are used commonly in the construction of DELs and are “DNA-tolerant”, they do not leave the nucleic acid unscathed. As chemists attempt to deploy more and more new reactions to DEL synthesis that are likely to have some impact on DNA tags, such as photoredox reactions and other processes involving radical intermediates82,83 quantitative evaluations of DNA damage will become increasingly important. The likely interest in more elaborate DEL designs, necessitating a larger number of cycles36, will also create pressure to get a better handle on DNA damage levels for each component reaction. Quantitative damage assays will allow the designer to make an educated guess as to whether a given series of reactions will allow enough tags to survive to support a robust screening campaign. This issue will also have an effect on interpretation of screening results, if one relies on relative levels of enrichment of a given building block at a given position as an indication of a true hit. Depending on the level of tag damage incurred in a particular step, different molecules in the library could be seen at different frequencies in the naïve library. Therefore it is critical to deep sequence the naïve library and empirically determine the representation of each molecule rather than assume that this will match a theoretical prediction. This, in turn, goes back to the point of the desirability of achieving a high level of sequencing depth by limiting the theoretical diversity.
These considerations lead us to predict that to access more elaborate, higher cycle number DELs, developers will begin to deemphasize current workhorse reactions such as peptide bond formation (at least acylation of secondary amines) in favor of true “freebies” that incur little or no DNA damage. These would include reductive aminations,81 pericyclic reactions84 and organocatalytic processes72, 73, amongst others. Another exciting possibility is to adapt enzyme-catalyzed transformations to DEL synthesis.85
Conclusions
In summary, it is clear that DEL technology represents an exciting option for bioactive molecule discovery. The technology has already had significant impact, which will only grow as more and more reactions are adapted to this platform. As discussed above, seminal challenges in the field include the development of better strategies to construct scaffold-rich and stereochemically diverse libraries that more closely resemble natural products. Especially exciting would be the introduction of more and better reactions for the generation of stereochemical diversity during DEL synthesis from cheap, achiral precursors, thus reducing the burden of maintaining large stocks of chiral building blocks.
Acknowledgements
Some of the work described in this article from our laboratory was supported by NIH grants AG054892 and GM 133041.
Footnotes
Conflict of Interest Statement
T.K. is a co-founder of Deluge Biotechnologies, which employs DEL technology.
Literature Cited
- 1.Smith GP and Petrenko VA, Chem. Rev, 1997, 97, 391–410. [DOI] [PubMed] [Google Scholar]
- 2.Roberts RW and Szostak JW, Proc. Natl. Acad. Sci. USA, 1997, 94, 12297–12302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanes J, Jermutus L, Weber-Bornhauser S, Bosshard HR and Plückthun A, Proc. Natl. Acad. Sci. USA, 1998, 95, 14130–14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ohuchi M, Murakami H and Suga H, Current opinion in chemical biology, 2007, 11, 537–542. [DOI] [PubMed] [Google Scholar]
- 5.Brenner S and Lerner RA, Proc Natl Acad Sci U S A, 1992, 89, 5381–5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Needels MC, Jones DG, Tate EH, Heinkel GL, Kochersperger LM, Dower WJ, Barrett RW and Gallop MA, Proc. Natl. Acad. Sci. USA, 1993, 90, 10700–10704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shendure J and Ji H, Nature biotechnology, 2008, 26, 1135–1145. [DOI] [PubMed] [Google Scholar]
- 8.Li X and Liu DR, Angewandte Chemie (International ed, 2004, 43, 4848–4870. [DOI] [PubMed] [Google Scholar]
- 9.Melkko S, Scheuermann J, Dumelin CE and Neri D, Nature biotechnology, 2004, 22, 568–574. [DOI] [PubMed] [Google Scholar]
- 10.Clark MA, Acharya RA, Arico-Muendel CC, Belyanskaya SL, Benjamin DR, Carlson NR, Centrella PA, Chiu CH, Creaser SP, Cuozzo JW, Davie CP, Ding Y, Franklin GJ, Franzen KD, Gefter ML, Hale SP, Hansen NJ, Israel DI, Jiang J, Kavarana MJ, Kelley MS, Kollmann CS, Li F, Lind K, Mataruse S, Medeiros PF, Messer JA, Myers P, O’Keefe H, Oliff MC, Rise CE, Satz AL, Skinner SR, Svendsen JL, Tang L, van Vloten K, Wagner RW, Yao G, Zhao B and Morgan BA, Nature Chem Biol, 2009, 5, 647–654. [DOI] [PubMed] [Google Scholar]
- 11.Lam KS, Salmon SE, Hersh EM, Hruby VJ, Kazmierski WM and Knapp RJ, Nature, 1991, 354, 82–84. [DOI] [PubMed] [Google Scholar]
- 12.Houghten RA, Pinilla C, Blondelle SE, Appel JR, Dooley CT and Cuervo JH, Nature, 1991, 354, 84–86. [DOI] [PubMed] [Google Scholar]
- 13.Liu R, Marik J and Lam KS, J. Amer. Chem. Soc, 2002, 124, 7678–7680. [DOI] [PubMed] [Google Scholar]
- 14.Figliozzi GM, Goldsmith R, Ng SC, Banville SC and Zuckermann RN, Methods Enzymol, 1996, 267, 437–447. [DOI] [PubMed] [Google Scholar]
- 15.Alluri PG, Reddy MM, Bachhawat-Sikder K, Olivos HJ and Kodadek T, Journal of the American Chemical Society, 2003, 125, 13995–14004. [DOI] [PubMed] [Google Scholar]
- 16.Lovering F, Bikker J and Humblet C, Journal of medicinal chemistry, 2009, 52, 6752–6756. [DOI] [PubMed] [Google Scholar]
- 17.Favalli N, Biendl S, Hartmann M, Piazzi J, Sladojevich F, Graslund S, Brown PJ, Nareoja K, Schuler H, Scheuermann J, Franzini R and Neri D, ChemMedChem, 2018, 13, 1303–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scheuermann J and Neri D, Chembiochem, 2010, 11, 931–937. [DOI] [PubMed] [Google Scholar]
- 19.Neri D and Lerner RA, Annual review of biochemistry, 2018, 87, 479–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuen LH and Franzini RM, Chembiochem, 2017, 18, 829–836. [DOI] [PubMed] [Google Scholar]
- 21.Clark MA, Current opinion in chemical biology, 2010, 14, 396–403. [DOI] [PubMed] [Google Scholar]
- 22.Lipinski C and Hopkins A, Nature, 2004, 432, 855–861. [DOI] [PubMed] [Google Scholar]
- 23.Lipinski CA, Journal of pharmacological and toxicological methods, 2000, 44, 235–249. [DOI] [PubMed] [Google Scholar]
- 24.Gellman SH, Acc. Chem. Res, 1998, 31, 173–180. [Google Scholar]
- 25.Kutzki O, Park HS, Ernst JT, Orner BP, Yin H and Hamilton AD, J. Amer. Chem. Soc, 2002, 124, 11838–11839. [DOI] [PubMed] [Google Scholar]
- 26.Shaginian A, Whitby LR, Hong S, Hwang I, Farooqi B, Searcey M, Chen J, Vogt PK and Boger DL, Journal of the American Chemical Society, 2009, 131, 5564–5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moon H, Lee WS, Oh M, Lee H, Lee JH, Im W and Lim HS, ACS Comb Sci, 2014, 16, 695–701. [DOI] [PubMed] [Google Scholar]
- 28.Leimbacher M, Zhang Y, Mannocci L, Stravs M, Geppert T, Scheuermann J, Schneider G and Neri D, Chemistry, 2012, 18, 7729–7737. [DOI] [PubMed] [Google Scholar]
- 29.Petersen LK, Blaksajaer P, Chaikuad A, Christensen AB, Dietvorst J, Holmkvist J, Knapp S, Korinek M, Larsen LK, Pedersen AE, Rohm S, Slok FA and Hansen NJV, MedChemComm, 2016, 7, 1332–1339. [Google Scholar]
- 30.Samain F, Ekblad T, Mikutis G, Zhong N, Zimmermann M, Nauer A, Bajic D, Decurtins W, Scheuermann J, Brown PJ, Hall J, Graslund S, Schuler H, Neri D and Franzini RM, Journal of medicinal chemistry, 2015, 58, 5143–5149. [DOI] [PubMed] [Google Scholar]
- 31.Litovchick A, Dumelin CE, Habeshian S, Gikunju D, Guie MA, Centrella P, Zhang Y, Sigel EA, Cuozzo JW, Keefe AD and Clark MA, Sci Rep, 2015, 5, 10916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harris PA, King BW, Bandyopadhyay D, Berger SB, Campobasso N, Capriotti CA, Cox JA, Dare L, Dong X, Finger JN, Grady LC, Hoffman SJ, Jeong JU, Kang J, Kasparcova V, Lakdawala AS, Lehr R, McNulty DE, Nagilla R, Ouellette MT, Pao CS, Rendina AR, Schaeffer MC, Summerfield JD, Swift BA, Totoritis RD, Ward P, Zhang A, Zhang D, Marquis RW, Bertin J and Gough PJ, Journal of medicinal chemistry, 2016, 59, 2163–2178. [DOI] [PubMed] [Google Scholar]
- 33.Wichert M, Krall N, Decurtins W, Franzini RM, Pretto F, Schneider P, Neri D and Scheuermann J, Nat Chem, 2015, 7, 241–249. [DOI] [PubMed] [Google Scholar]
- 34.Mendes K, Malone ML, Ndungu JM, Suponitsky-Kroyter I, Cavett VJ, McEnaney PJ, MacConnell AB, Doran TM, Ronacher K, Stanley K, Utset O, Walzl G, Paegel BM and Kodadek T, ACS Chem. Biol, 2017, 19, 234–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maianti JP, McFedries A, Foda ZH, Kleiner RE, Du XQ, Leissring MA, Tang WJ, Charron MJ, Seeliger MA, Saghatelian A and Liu DR, Nature, 2014, 511, 94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stress C, Sauter B, Schneider L, Sharpe T and Gillingham D, Angewandte Chemie (International ed, 2019, DOI: 10.1002/anie.201902513. [DOI] [PubMed] [Google Scholar]
- 37.Gerry C, Wawer M, Clemons P and Schreiber SL, ChemRxiv, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reddy Guduru SK, Chamakuri S, Raji IO, MacKenzie KR, Santini C and Young DW, J Org Chem, 2018, 83, 11777–11793. [DOI] [PubMed] [Google Scholar]
- 39.Chamakuri S, Jain P, Reddy Guduru SK, Arney JW, MacKenzie KR, Santini C and Young DW, J Org Chem, 2018, 83, 6541–6555. [DOI] [PubMed] [Google Scholar]
- 40.Nielsen TE and Schreiber SL, Angewandte Chemie (International ed, 2008, 47, 48–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumagai N, Muncipinto G and Schreiber SL, Angewandte Chemie (International ed, 2006, 45, 3635–3638. [DOI] [PubMed] [Google Scholar]
- 42.Zuckermann RN, Kerr JM, Kent SBH and Moos WH, J. Amer. Chem. Soc, 1992, 114, 10646–10647. [Google Scholar]
- 43.Kodadek T and McEnaney PJ, Chemical communications (Cambridge, England), 2016, 52, 6038–6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aquino C, Sarkar M, CHalmers MJ, Mendes K, Kodadek T and Micalizio G, Nature Chem, 2011, 4, 99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sarkar M, Pascal BD, Steckler C, Micalizio GC, Kodadek T and Chalmers MJ, J. Amer. Soc. Mass Spec, 2013, 24, 1026–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sarkar M, Liu Y, Morimoto J, Peng H, Aquino C, Rader C, Chiorazzi N and Kodadek T, Chem. & Biol, 2014, 111, 1670–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aditya A and Kodadek T, ACS Comb Sci, 2012, 14, 164–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moon H and Lim HS, Current opinion in chemical biology, 2015, 24, 38–47. [DOI] [PubMed] [Google Scholar]
- 49.Morimoto J and Kodadek T, Molecular bioSystems, 2015, 11, 2770–2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.MacConnell AB, McEnaney PJ, Cavett VJ and Paegel BM, ACS Comb Sci, 2015, 17, 518–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Erharuyi O, Simanski S, McEnaney PJ and Kodadek T, Bioorganic & medicinal chemistry letters, 2018, 28, 2773–2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen S, Morales-Sanfrutos J, Angelini A, Cutting B and Heinis C, Chembiochem, 2012, 13, 1032–1038. [DOI] [PubMed] [Google Scholar]
- 53.Kale SS, Villequey C, Kong XD, Zorzi A, Deyle K and Heinis C, Nat Chem, 2018, 10, 715–723. [DOI] [PubMed] [Google Scholar]
- 54.Kleiner RE, Dumelin CE, Tiu GC, Sakurai K and Liu DR, Journal of the American Chemical Society, 2010, 132, 11779–11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Usanov DL, Chan AI, Maianti JP and Liu DR, Nat Chem, 2018, 10, 704–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu Z, Shaginian A, Grady LC, O’Keeffe T, Shi XE, Davie CP, Simpson GL, Messer JA, Evindar G, Bream RN, Thansandote PP, Prentice NR, Mason AM and Pal S, ACS Chem Biol, 2018, 13, 53–59. [DOI] [PubMed] [Google Scholar]
- 57.Li Y, De Luca R, Cazzamalli S, Pretto F, Bajic D, Scheuermann J and Neri D, Nat Chem, 2018, 10, 441–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Farrow B, Wong M, Malette J, Lai B, Deyle KM, Das S, Nag A, Agnew HD and Heath JR, Angewandte Chemie (International ed, 2015, 54, 7114–7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lai BT, Wilson JA, Malette Loredo J, Pitram SM, LaBerge NA, Heath JR and Agnew H, Chemistry, 2018, 24, 3760–3767. [DOI] [PubMed] [Google Scholar]
- 60.Yu P, Liu B and Kodadek T, Nature Biotech, 2005, 23, 746–751. [DOI] [PubMed] [Google Scholar]
- 61.Kwon YU and Kodadek T, J. Amer. Chem. Soc, 2007, 129, 1508–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ahlbach CL, Lexa KW, Bockus AT, Chen V, Crews P, Jacobson MP and Lokey RS, Future Med Chem, 2015, 7, 2121–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gao Y and Kodadek T, Chem. & Biol, 2013, 20, 360–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Falb E, Yechezkel T, Salitra Y and Gilon C, J Pept Res, 1999, 53, 507–517. [DOI] [PubMed] [Google Scholar]
- 65.Gao Y and Kodadek T, ACS Comb Sci, 2015, 17, 190–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Doran TM, Gao Y, Mendes K, Dean S, Simanski S and Kodadek T, ACS Comb. Sci, 2014, 16, 259–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Eidam O and Satz AL, MedChemComm, 2016, 7, 1323–1331. [Google Scholar]
- 68.Satz AL, Hochstrasser R and Petersen AC, ACS Comb Sci, 2017, 19, 234–238. [DOI] [PubMed] [Google Scholar]
- 69.Jacobsen EN and MacMillan DW, Proc Natl Acad Sci U S A, 2010, 107, 20618–20619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Notz W, Tanaka F and Barbas CF 3rd, Acc Chem Res, 2004, 37, 580–591. [DOI] [PubMed] [Google Scholar]
- 71.List B, J. Amer. Chem. Soc, 2000, 122, 9336–9337. [Google Scholar]
- 72.Shu K and Kodadek T, ACS Comb Sci, 2018, 20, 277–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tran NH and Kodadek T, ACS Comb. Sci, 2018, 20, 55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Marcelli T, van der Haas RN, van Maarseveen JH and Hiemstra H, Angewandte Chemie (International ed, 2006, 45, 929–931. [DOI] [PubMed] [Google Scholar]
- 75.Klausen RS, Kennedy CR, Hyde AM and Jacobsen EN, Journal of the American Chemical Society, 2017, 139, 12299–12309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kennedy CR, Guidera JA and Jacobsen EN, ACS Cent Sci, 2016, 2, 416–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yeung CS, Ziegler RE, Porco JA Jr. and Jacobsen EN, Journal of the American Chemical Society, 2014, 136, 13614–13617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang W, LOebach JL, Wilson SR and Jacobsen EN, J. Amer. Chem. Soc, 1990, 112, 2801–2803. [Google Scholar]
- 79.Katsuki T and Sharpless KB, J. Amer. Chem. Soc, 1980, 102, 5974–5976. [Google Scholar]
- 80.Kolb HC, Van Nieuwenhze MS and Sharpless KB, Chem. Reviews, 1994, 94, 2483–2547. [Google Scholar]
- 81.Malone ML and Paegel BM, ACS Comb Sci, 2016, 18, 182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Phelan JP, Lang SB, Sim J, Berritt S, Peat AJ, Billings K, Fan L and Molander GA, Journal of the American Chemical Society, 2019, 141, 3723–3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kolmel DK, Loach RP, Knauber T and Flanagan ME, ChemMedChem, 2018, 13, 2159–2165. [DOI] [PubMed] [Google Scholar]
- 84.Gerry CJ, Yang Z, Stasi M and Schreiber SL, Organic letters, 2019, DOI: 10.1021/acs.orglett.9b00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Thomas B, Lu X, Birmingham WR, Huang K, Both P, Reyes Martinez JE, Young RJ, Davie CP and Flitsch SL, Chembiochem, 2017, 18, 858–863. [DOI] [PubMed] [Google Scholar]