Abstract
Gene editing constitutes a novel approach for precisely correcting disease-causing gene mutations. Frameshift mutations in COL7A1 causing recessive dystrophic epidermolysis bullosa are amenable to open reading frame restoration by non-homologous end joining repair-based approaches. Efficient targeted deletion of faulty COL7A1 exons in polyclonal patient keratinocytes would enable the translation of this therapeutic strategy to the clinic. In this study, using a dual single-guide RNA (sgRNA)-guided Cas9 nuclease delivered as a ribonucleoprotein complex through electroporation, we have achieved very efficient targeted deletion of COL7A1 exon 80 in recessive dystrophic epidermolysis bullosa (RDEB) patient keratinocytes carrying a highly prevalent frameshift mutation. This ex vivo non-viral approach rendered a large proportion of corrected cells producing a functional collagen VII variant. The effective targeting of the epidermal stem cell population enabled long-term regeneration of a properly adhesive skin upon grafting onto immunodeficient mice. A safety assessment by next-generation sequencing (NGS) analysis of potential off-target sites did not reveal any unintended nuclease activity. Our strategy could potentially be extended to a large number of COL7A1 mutation-bearing exons within the long collagenous domain of this gene, opening the way to precision medicine for RDEB.
Keywords: CRISPR/Cas9, epidermal stem cells, epidermolysis bullosa, gene therapy
Highly efficient and safe approaches for precise correction of pathogenic mutations are required to realize the full potential of gene-editing protocols for clinical practice. Here we describe a single-step NHEJ-based CRISPR/Cas9 strategy of COL7A1 mutation-bearing exon deletion with unprecedented efficacy for ex vivo correction of recessive dystrophic epidermolysis bullosa.
Introduction
Recessive dystrophic epidermolysis bullosa (RDEB) is a severe skin fragility genodermatosis caused by loss-of-function mutations in the COL7A1 gene, encoding type VII collagen (C7). C7 deficiency results in generalized blistering of the skin and other stratified epithelia, scarring, fibrosis, mitten-like deformities of hands and feet, and a high risk of developing metastatic squamous cell carcinoma.1
Ex vivo gene addition therapies based on transplantation of keratinocyte sheets modified by retroviral vectors are already at the clinical stage for forms of epidermolysis bullosa, including junctional epidermolysis bullosa (JEB) and RDEB, with encouraging results.2, 3, 4, 5 However, significant hurdles face the gene addition approach that include suboptimal viral gene delivery to the stem cell population, inaccurate spatial-temporal gene expression, and potential insertional mutagenesis-derived adverse events, which are particularly relevant for RDEB patients given their high proneness to carcinoma development. Therefore, the implementation of gene therapy approaches for RDEB based on highly precise gene-editing technologies is warranted and has been pursued by employing different types of nucleases, i.e., meganucleases, transcription activator-like effector nucleases (TALENs), and CRISPR/Cas9, and target cells.6, 7, 8, 9, 10
Since neither homology-directed repair (HDR)- nor non-homologous end joining (NHEJ)-mediated gene-editing strategies tested so far in patient cells have reached a sufficient level of efficacy to enable therapeutic C7 replacement by direct transplantation of cells treated in bulk, isolation of corrected cell clones has been necessary, either from patient-derived induced pluripotent stem cells (iPSCs) and subsequent target cell derivation6, 7 or from patient keratinocytes.9, 10, 11 Our laboratory previously demonstrated long-term skin regeneration from single, gene-edited epidermal stem cell clones of primary RDEB patient keratinocytes.11 Our original approach involved the use of TALENs delivered by adenoviral vectors to induce NHEJ-mediated insertions or deletions (indels) able to restore the reading frame of COL7A1 in patient cells carrying the frameshift mutation c.6527insC12 located at exon 80, which is highly prevalent in the cohort of Spanish RDEB patients.13, 14 One of the edited patient keratinocyte clones described in this study carried a long deletion encompassing the whole exon 80, and it showed restoration of C7 expression and persistent phenotypic correction in vivo upon transplantation onto immunocompromised mice.11 The functionality of C7 variants lacking the amino acids encoded by specific exons within the collagenous domain (i.e., exons 73, 80, and 105) has been previously demonstrated.15, 16 Further, Wu et al.17 also showed that Col7a1 exon 80-skipped mice generated with the CRISPR/Cas9 system were indistinguishable from their wild-type littermates. Building on these results, we sought more efficient and safe methods to achieve targeted deletion of mutation-carrying COL7A1 exon 80 using the CRISPR/Cas9 system in RDEB patient keratinocytes.
In this study, we show the remarkable efficacy and safety of a non-viral strategy employing a dual single-guide RNA (sgRNA)-guided Cas9 nuclease delivered as a ribonucleoprotein (RNP) complex by electroporation to precisely excise COL7A1 exon 80 carrying the c.6527insC mutation in RDEB patient keratinocytes. Moreover, we demonstrate the long-term skin regeneration ability of the corrected cells upon grafting of polyclonal and monoclonal populations of edited cells to immunodeficient mice, which is indicative of epidermal stem cell correction. This highly efficacious and one-step strategy would enable quick translation to clinical application.
Results
Delivery of Dual sgRNA-Guided Cas9 Nuclease to RDEB Keratinocytes as an RNP Complex Enables Highly Efficient Targeted Deletion of COL7A1 Exon 80
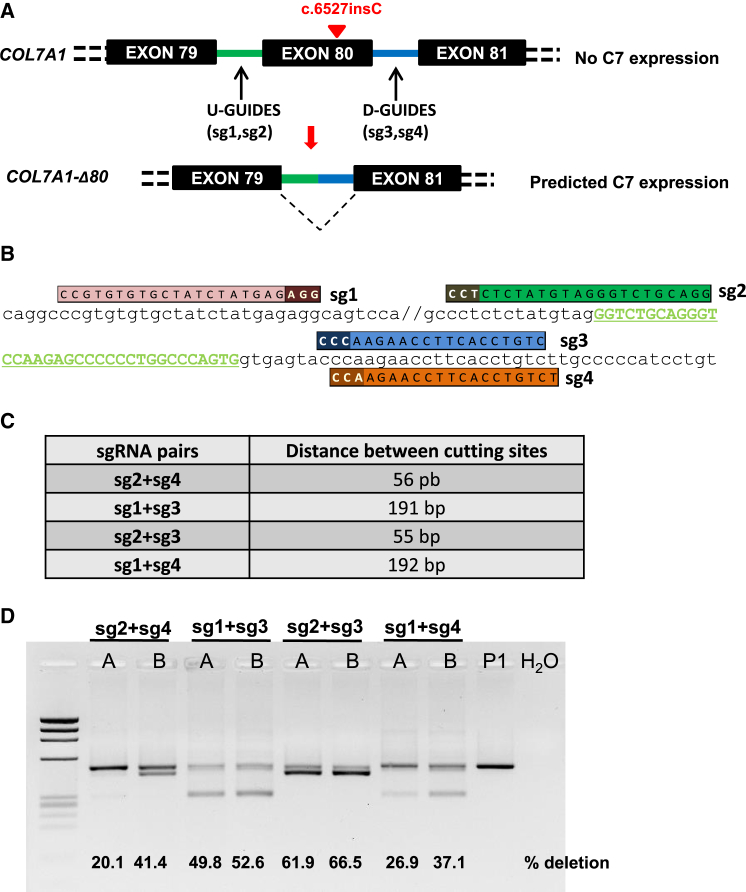
The dual sgRNA CRISPR/Cas9 deletion strategy of COL7A1 exon 80 (E80) is shown in the scheme (Figure 1A). Four different sgRNA pairs targeted to DNA sequences within introns 79 and 80 were designed (Figure 1B), with the aim of generating deletions of different sizes covering E80 (Figure 1C). RDEB keratinocytes from a homozygous carrier of the c.6527insC mutation were nucleofected with the CRISPR/Cas9 RNP complexes at two different electroporation pulse conditions (designated as A and B, Figure 1D). Once treated, keratinocytes reached confluency, genomic DNA was extracted, and E80 deletion (ΔE80) was assessed by PCR amplification of a fragment spanning the sgRNA target sites (Figure 1D). All sgRNA pairs led to E80 deletion, as shown by the presence of smaller molecular weight bands with sizes consistent with the distances between Cas9 cutting sites. Electroporation condition B performed better for all sgRNA pairs. Differences in deletion efficacy were found among the different sgRNA pairs (i.e., sg2 + sg3 > sg1 + sg3 > sg2 + sg4 > sg1 + sg4) (Figure 1D). In keratinocytes treated with sg2 + sg3, the most efficacious pair of guides, E80 deletions accounted for 66.5% of alleles, as determined by densitometric quantitation of the PCR products (Figure 1D). The presence of ΔE80 alleles in a ratio higher than 50% indicated that homozygous deletion of E80 was a frequent event.
Figure 1.
COL7A1 E80 Dual RNA Guide CRISPR/Cas9 Deletion Strategy
(A) Scheme of the strategy. Single CRISPR RNA guides were designed to induce Cas9-mediated DNA double-strand breaks within selected COL7A1 intron sequences flanking exon 80 (U-Guides and D-Guides). NHEJ repair leads to intron-intron rejoining with concomitant E80 deletion and restoration of COL7A1 reading frame and truncated C7 expression. (B) CRISPR guide design. sgRNA (sgRNAs 1, 2, 3, and 4) sequences and alignment to E80-flanking sequences E80 are shown. The protospacer adjacent motif (PAM) is indicated in a darker color. (C) Predicted amplicon sizes for E80 deletions corresponding to each sgRNA pair combination. (D) PCR analysis of genomic DNA from RDEB keratinocytes from patient P1 treated with the RNP complexes containing the different sgRNA pair combinations. The sg2 + sg3 pair yielded the highest proportion of excised E80 according to the intensity of the lower 440-bp band. Deletion ratios assessed by densitometry are shown in each lane. “A” and “B” refer to the two different electroporation conditions tested.
Targeted COL7A1 E80 Deletion Produced a Diversity of Spliceable Chimerical Intronic Variants
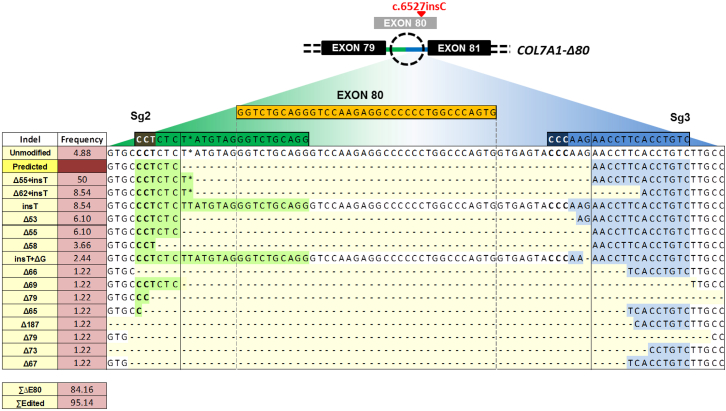
A variety of intron 79-intron 80 joining events after Cas9-mediated cleavage at sequences flanking E80 and subsequent NHEJ repair were expected to occur in a polyclonal cell population. The characterization of the DNA repair outcomes in RDEB keratinocytes from patient P1 treated with the most effective sgRNA pair (sg2 + sg3) was initially performed by thymidine-adenine (TA) cloning of PCR products spanning both Cas9 cutting sites and Sanger sequencing of individual colonies (n = 82). This analysis detected a spectrum of end joining repair variants, the majority of which (41 of 82) corresponded to the fusion of the predicted sgRNA-targeted Cas9 cleavage sites plus the insertion of a T (Figure 2). Other fusion events included small indels (both deletions and insertions) not affecting intron-splicing signals and, therefore, not likely to disrupt the splicing of the resulting chimerical intron. The analysis also showed that, in addition to dual Cas9 cuts leading to the intended E80 deletion, which accounted for 84% of alleles, indels corresponding to DNA repair after single cuts (i.e., at either intron 79 or 80) were also present. Taking these into account, 95% of alleles had been edited (Figure 2).
Figure 2.
Indel Spectrum in COL7A1 E80 Region
Sequences (Sanger) listed from higher to lower frequencies. 50% of rejoining events corresponded to the fusion between E80-flanking cutting sites plus the addition of a T (Δ55 + insT). 95.14% of all alleles had been edited and 84.16% presented E80 deletion.
Next-Generation Sequencing Analysis of On-Target and Off-Target CRISPR-Generated Variants
For in-depth characterization of repair events at the on-target region and to analyze potential off-target cleavage activity at in silico-predicted sites, we performed next-generation sequencing (NGS) of PCR amplicons spanning the corresponding sites. Genomic DNA samples from keratinocytes from two homozygous c.6527insC carrier patients, P1 and P2, treated in duplicate with RNPs (biological replicates BR1 and BR2) as well as control DNA samples from both patients were subjected to this analysis. On-target NGS analysis confirmed highly efficient deletion of E80 in Cas9 RNP-treated patient cells. Sequence variations within a 60-bp window centered on the midpoint between both cut sites were considered. The two most frequent repair variants, i.e., fusion of the Cas9 cleavage sites with (62.2%) or without (9.7%) the insertion of a T (Figure S1), found in P1 (BR1), were also among the most frequently represented in the Sanger sequencing characterization (50.0% and 6.1%, respectively) for this sample (Figure 2). For less frequent repair variants, higher diversity was detected with NGS than with Sanger sequencing. Taking into account all variants resulting in E80 deletion, similar frequencies of deletion were found with either technique (84% versus 87% for Sanger and NGS, respectively) in P1 (BR1) patient keratinocytes. Patient P2 cells, analyzed by NGS only, showed 95% E80 deletion with a similar spectrum of allelic variants (Figure S1). The lower frequency of E80 deletion estimated by PCR genotyping in patient P1 (66%), as compared with Sanger or NGS sequencing estimates, might be explained by heteroduplex DNA formation between deleted and undeleted alleles that results in decreased intensity of the lower molecular weight PCR band.18
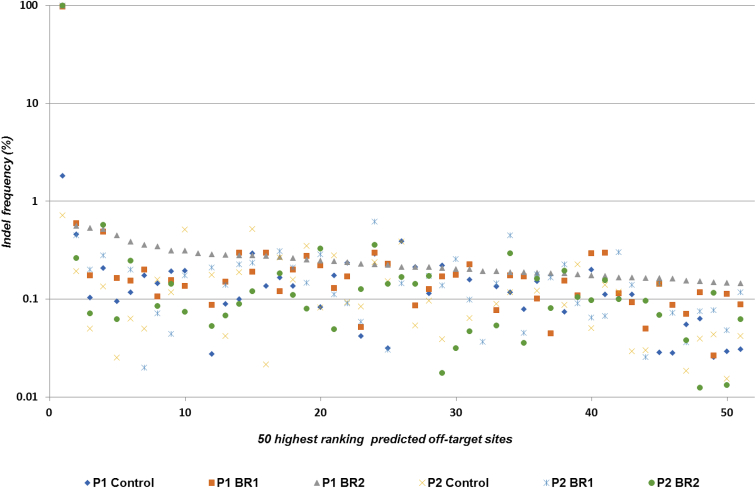
To assess for potential off-target Cas9 cleavage activity, NGS was used to analyze PCR amplicons covering 278 in silico-predicted sites, which included all off-target sites up to 3 mismatches, all sites with 1- or 2-bp bulges, and the top 53 coding region sites with 4 mismatches for each guide. After establishing a 1,000-read cutoff for sequencing depth, 244 amplicons were studied. The only sequence variations considered were indels within a 6-bp window centered on the target site for each sgRNA.
The threshold percentage for off-target activity was set at 0.52%, since this was the highest percentage of indel-containing reads (within the 6-bp window considered for the off-target evaluation) in unedited control samples. Thus, for each site, indel-containing reads below this percentage were considered as noise. For every off-target site analyzed, indel-containing reads represented less than 0.52% of the total (Figure 3), except for five predicted sites. Although these sites showed indel-containing reads at a slightly higher frequency (0.52%–0.6%) above the 0.52% threshold (Figure S2), the sequence variations found were also present in both controls and edited samples, suggesting that these were not bona fide Cas9 off-targets. We therefore concluded that our NGS analysis of 244 predicted off-target sites did not reveal any off-target events above the threshold of detection.
Figure 3.
NGS Assessment for the Presence of Indels in Potential Off-Target Sites
PCR amplicons for 244 predicted off-target sites sequenced with at least 1,000× depth, as well as the on-target region, were analyzed (x axis, putative off-target sites arranged according to the frequency of sequence variation). The 50 highest ranking sites are represented. The first point corresponds to the on-target site (y axis, percentage of reads containing sequence variations within a 6-bp window centered on the target site for each sgRNA).
COL7A1 Transcription Analysis after E80 Deletion in c.6527insC Patient Cells
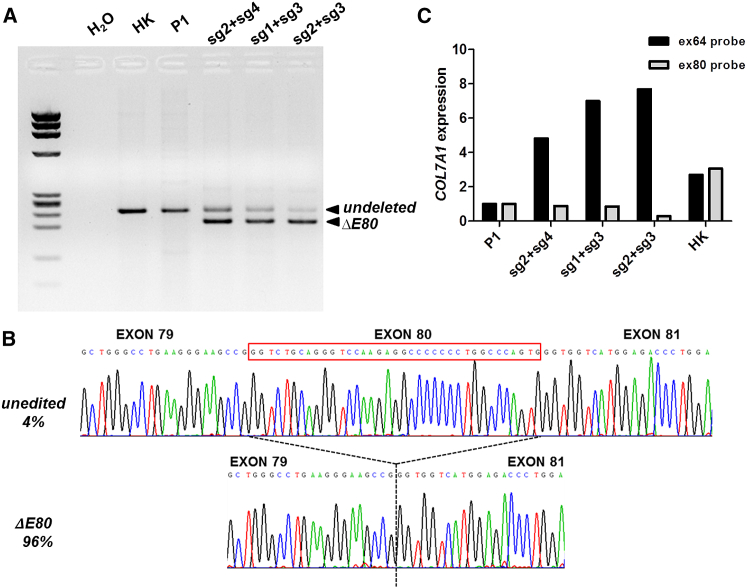
Our NHEJ correction strategy was designed to generate, upon E80 deletion, new functional introns with donor and acceptor splicing sequences corresponding to those from exons 79 and 81, respectively. Still, the potential generation of cryptic splicing sites could result in inappropriately spliced transcripts. To confirm proper reading frame restoration derived from the precise in-frame exon 79-exon 81 junction, we studied COL7A1 transcription by performing RT-PCR analysis. For all three sgRNA pairs suitable for E80 deletion, a smaller band consistent with amplification of transcripts lacking E80 was found (Figure 4A). The intensity of the smaller band (Figure 4A) was proportional to the efficiency of the deletion, as detected by PCR analysis of genomic DNA (Figure 1D). The RT-PCR products corresponding to cells treated with the best-performing sgRNA pair (sg2 + sg3) were TA cloned (n = 49) and Sanger sequenced. This analysis revealed only two types of transcripts: 47 colonies contained the proper exon 79-exon 81 junction (96% of transcripts), and 2 colonies (4%) contained the E80 sequence originating from the unedited (c.6527insC) allele (Figure 4B).
Figure 4.
Collagen VII mRNA Expression of ΔE80 Gene-Edited RDEB Keratinocytes
(A) RT-PCR analysis of COL7A1 transcripts amplified with primers in exons 78–84. Wild-type/c.6527insC unedited transcripts produced a 240/241-bp band (non-modified, n-m) found in all RNA samples. A smaller 205-bp band corresponding to transcripts lacking exon 80 was detected in samples from edited cells. M, DNA Molecular Weight Marker IX (Sigma-Aldrich) molecular weight marker; HK, healthy human keratinocytes; P1, patient keratinocytes; H2O, negative control without cDNA. (B) Representative sequence chromatograms showing the two different resulting transcripts. Transcript frequencies are shown on the left. (C) COL7A1 expression quantification by real-time qPCR using Taqman probes for two different COL7A1 regions (ex64, specific for all COL7A1 transcripts; and Ex80 probe, for exon 80-containing transcripts).
To further quantify the presence of E80-deleted and E80-containing transcripts in gene-edited cells, qRT-PCR was performed using Taqman probes specific for exon 80- and exon 64-encoded sequences. The exon 64-specific probe detects transcripts originating from both edited and unedited alleles, and the exon 80-specific probe detects only transcription from unedited E80 sequence-containing alleles. While in healthy donor control keratinocytes both probes detected similar expression levels, in the different RNP-treated pools of cells we observed higher expression with the exon 64 probe than with the exon 80 probe. The increased COL7A1 transcription detected with the exon 64 probe as compared to the exon 80 probe was proportional to the deletion efficacy of the guide combinations. This analysis was consistent with the RT-PCR product quantification, and it clearly demonstrated the prevalence of E80-lacking transcripts in edited cells, confirming sg2 + sg3 guides as the best performing pair for E80 deletion (Figure 4C).
De Novo C7 Expression after Targeted Deletion of E80 in RDEB Keratinocytes
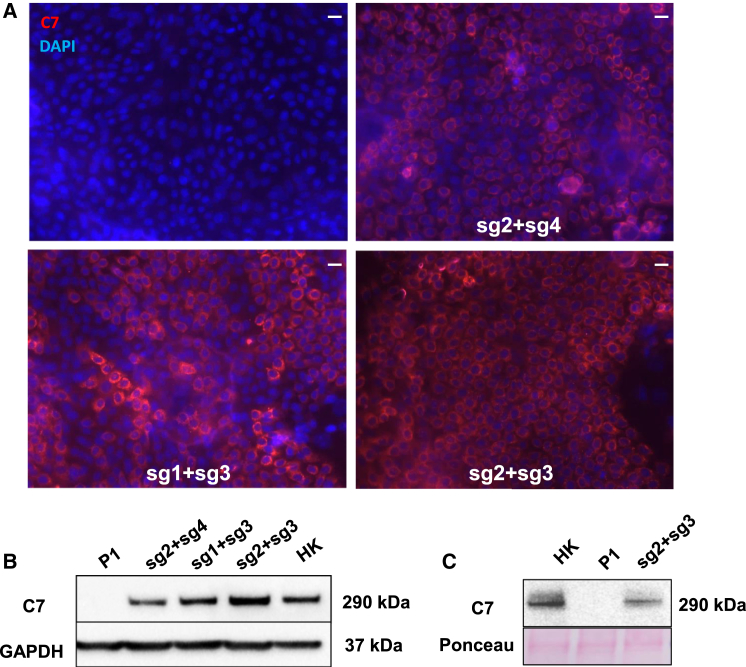
COL7A1 reading frame restoration by targeted deletion of mutant E80 and splicing of the resulting chimerical intron should result in C7 expression, as we previously observed in a clone of RDEB keratinocytes containing a deletion that encompassed COL7A1 E80 entirely.11 Previous studies from others have also shown that C7-null cells expressing a retrovirally transferred ΔE80 COL7A1 cDNA construct were able to produce a functional C7 variant.16 We therefore assessed C7 expression by immunofluorescence and western blot in RDEB keratinocytes nucleofected with the three most efficient E80-deleting RNP combinations (sg2 + sg4, sg1 + sg3, and sg2 + sg3). The number of C7-expressing cells detected by immunofluorescence analysis (Figures 5A and S3A) matched the E80 deletion efficacy found by PCR analysis (Figure 1D) and the COL7A1 mRNA transcription levels (Figures 4A and 4C). In fact, quantification of C7-positive cells by flow cytometry using a specific anti-C7 antibody showed that 81% of patient keratinocytes treated with the sg2 + sg3 RNPs expressed C7 (Figure S3B). Accordingly, western blot analysis performed on cellular extracts further demonstrated the highest expression of C7 in patient keratinocyte samples treated with sg2 + sg3 RNPs. The mobility of the C7 band detected in these samples was indistinguishable from that of the wild-type (WT) protein, as expected since the size difference between WT C7 and the variant lacking the 12 amino acids encoded by E80 cannot be resolved at the molecular weight range of C7 (290 kDa) (Figure 5B). In addition, to study C7 secretion, we performed western blot analysis of proteins precipitated from keratinocyte culture supernatants. As shown, C7 was present in the media of sg2 + sg3 RNP-treated keratinocytes (Figure 5C), confirming that removal of E80 does not impair C7 secretion.
Figure 5.
Collagen VII Expression in ΔE80 RDEB Polyclonal Keratinocytes
Keratinocytes were treated with the RNP complex containing different CRISPR dual sgRNA combinations, and C7 expression was assessed by immunofluorescence (IF) and western blot. (A) C7 IF analysis. Left top: control, untreated RDEB (c.6527insC) keratinocytes are shown. Right top: RDEB keratinocytes were treated with the sg2 + sg4 pair. Left bottom: RDEB keratinocytes were treated with the sg1 + sg3 pair. Right bottom: RDEB keratinocytes were treated with the sg2 + sg3 pair. DAPI was used to stain nuclei. Scale bars, 50 μm. (B) Western blot analysis of C7 expression in unedited and edited RDEB keratinocytes, showing C7 bands intensities consistent with the IF data. (C) Western blot analysis of secreted C7 from culture supernatant of normal, untreated RDEB and the sg2 + sg3 RNP-treated RDEB keratinocytes. Protein loading was assessed by Ponceau red staining of the membrane (bottom). The ΔE80 C7 in edited RDEB cells is indistinguishable from that of normal human keratinocytes.
Long-Term Engraftment of Polyclonal Populations of ΔE80 RDEB Keratinocytes
The high proportion of C7-expressing keratinocytes obtained with the non-viral CRISPR/Cas9 approach indicated that a C7 amount sufficient to restore epidermal-dermal adhesion would be attainable. To test this, polyclonal populations of edited RDEB cells, obtained with the different combinations of sgRNAs, and control unedited cells were used to produce bioengineered skin constructs that were subsequently grafted onto immunodeficient mice. Animals were monitored for engraftment, and biopsies were obtained at different time points for histopathological analysis of regenerated skin and assessment of C7 expression.
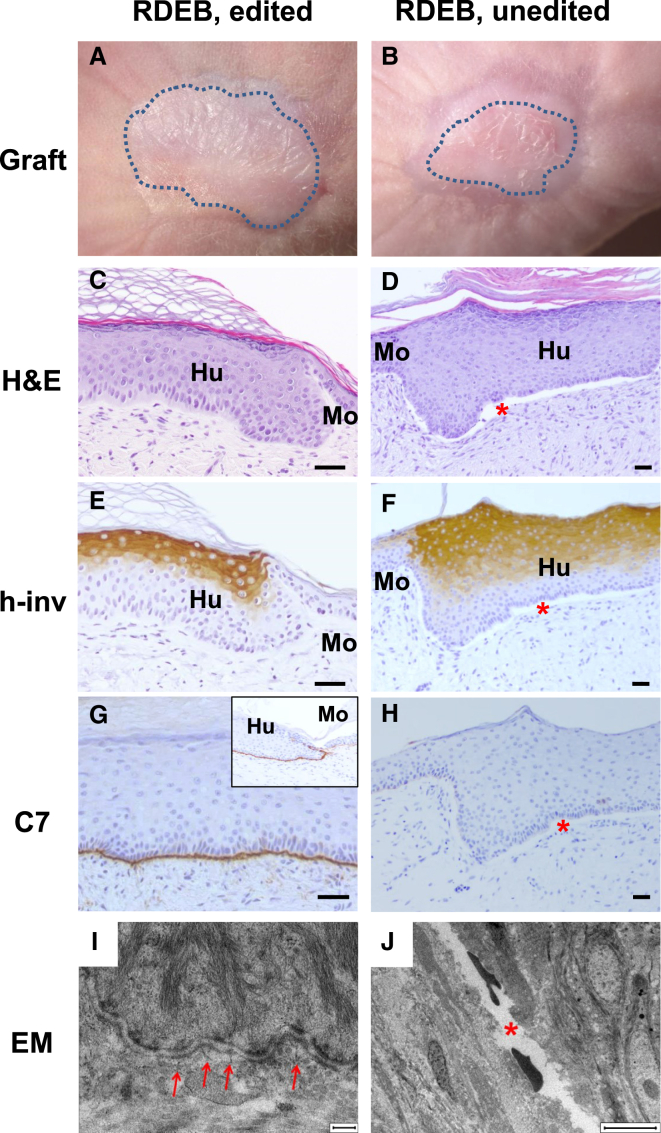
Macroscopic examination clearly showed human skin engraftment for the most efficient guide combination (sg2 + sg3) (Figures 6A and 6B). Routine histological analysis (H&E staining) of 12-week-old grafts showed normal skin architecture and uninterrupted dermal-epidermal attachment in grafts from these RDEB-edited cells (Figure 6C). On the contrary, epidermal-dermal separation was evident in grafts from non-edited cells (Figure 6D). Both types of grafts showed correct suprabasal human involucrin expression indicating normal epidermal differentiation (Figures 6E and 6F). Immunoperoxidase staining clearly exhibited C7 expression with appropriate localization at the basement membrane zone (BMZ) exclusively in grafts from edited patient cells (Figures 6G and 6H). Ultrastructural analysis by electron microscopy accordingly showed the presence of abundant anchoring fibrils in edited grafts (Figure 6I), but not in control unedited grafts where dermal-epidermal separation was evident (Figure 6J).
Figure 6.
Skin Regeneration, Collagen VII Expression, and Ultrastructural Analysis of Grafts from CRISPR/Cas9-Edited (ΔE80) Polyclonal Keratinocytes
(A and B) Macroscopic view of engrafted areas 12 weeks after grafting of bioengineered skins containing gene-edited (ΔE80) (A) or unedited RDEB keratinocytes (B). (C and D) Histological analysis (H&E staining) of grafts from gene-edited (ΔE80) (C) or unedited RDEB keratinocytes (D). The dermal-epidermal separation in grafts from control, unedited cells is indicated by the red asterisk. (E and F) Human involucrin (h-inv) immunostaining (suprabasal expression) showing normal epidermal differentiation in grafts from ΔE80 keratinocytes (E) or unedited RDEB keratinocytes (F). (G and H) C7 immunoperoxidase expression analysis showing the continuous and correct deposition of C7 at the BMZ in ΔE80-edited grafts (G) and its complete absence in unedited RDEB keratinocyte grafts (H). Scale bars, 100 μm. (I and J) Electron microscopy analysis shows the presence of mature anchoring fibrils (arrows) at the dermal-epidermal junction of the gene-edited RDEB skin (I) and an empty electron lucent split area, corresponding to a blister (red asterisk) in the unedited graft (J). Scale bars, 200 nm (I) and 6 μm (J).
Patient keratinocytes modified with the other sgRNA pairs able to induce alternative E80 deletions, albeit at lower efficiencies, were also tested for skin regeneration. Histological and immunohistochemical analysis 12 weeks after grafting showed full dermal-epidermal adhesion (Figure S4D) and continuous C7 expression (Figure S4F) at the BMZ in grafts of keratinocytes edited with sg1 + sg3. In contrast, microblisters (Figure S4A) and reduced and patchy C7 expression (Figure S4C) were observed in grafts generated from cells treated with the less efficient sg2 + sg4 RNP combination, suggesting that the C7 amount provided by these cells was not sufficient to sustain continuous dermal-epidermal adhesion. Human involucrin expression demonstrated normal epidermal differentiation of these grafts (Figures S4B and S4E).
Non-viral CRISPR/Cas9 Delivery Targets the Epidermal Stem Compartment
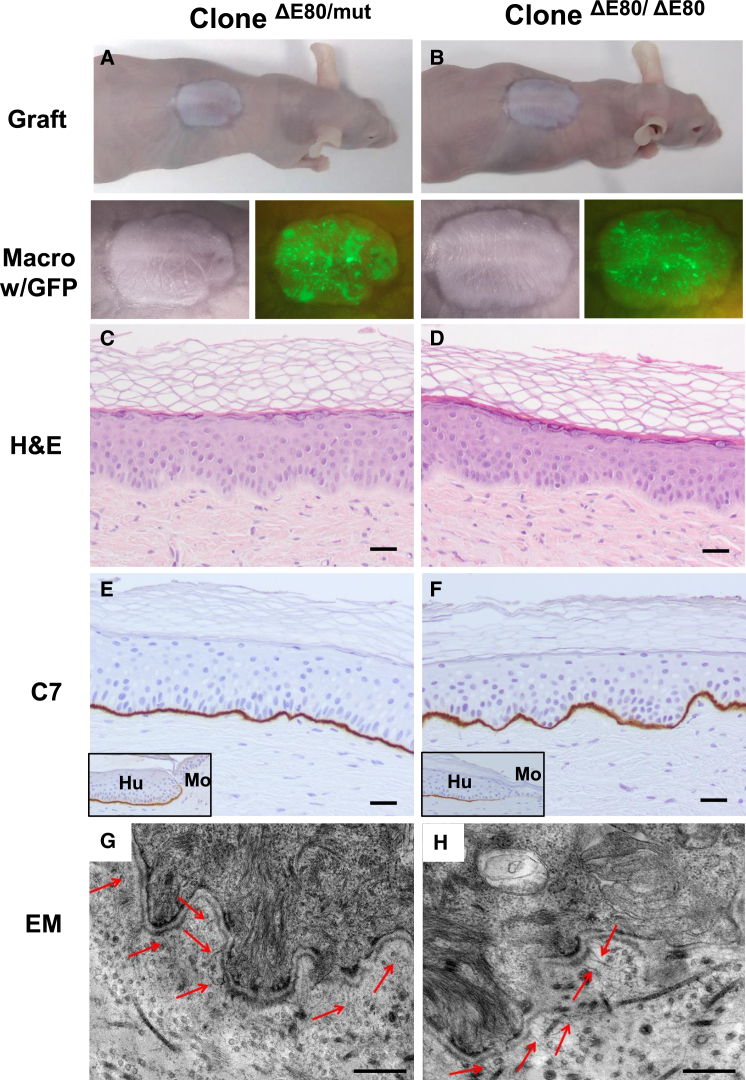
The very high efficiency of E80 deletion attained through our non-viral CRISPR/Cas9 RNP complex delivery and the long-term skin regeneration achieved with a polyclonal population of corrected cells suggested that targeting of the epidermal stem cell compartment had occurred. To confirm this, 11 clones from the bulk RDEB keratinocyte population edited with sg2 + sg3 RNPs were isolated by limiting dilution. Genotype analysis revealed that all of these clones had been edited (Figure S5A). Two clones, one monoallelic and the other biallelic for the E80 deletion (named as ΔE80/ΔE80 and ΔE80/mut), were selected for in vivo skin regeneration performance. These clones were accurately genotyped by sequencing (Figure S5C) and COL7A1 expression was precisely assessed (Figures S5B and S5D–S5F). Keratinocytes from these clones were labeled with GFP by lentiviral transduction to facilitate the monitoring of graft persistence over time.
Macroscopic analysis under white and blue (GFP) light illumination of grafts 20 weeks post-grafting, when several epidermal turnover cycles had occurred, revealed clearly distinguishable human skin morphological characteristics (Figures 7A and 7B). Grafts from both clones displayed normal histopathological features with continuous dermal-epidermal attachment (Figures 7C and 7D). As shown with the polyclonal ΔE80 RDEB keratinocyte population and consistent with the epidermal-dermal attachment observed histologically, both C7 expression (Figures 7E and 7F) and anchoring fibrils (Figures 7G and 7H) were also clearly detectable in the monoallelic and biallelic ΔE80 clonal grafts. Appropriate epidermal differentiation was confirmed by human involucrin expression immunodetection (Figures S6B and S6E). In addition to graft endurance over time, a robust p63 immunostaining also indicated that a pool of keratinocytes with persistent regenerative functionality was maintained19 (Figures S6C and S6F). Long-term clonal skin regeneration and persistence of the corrective effect, as determined by this stringent stemness in vivo test, indicated that the epidermal stem cell compartment had been effectively targeted.
Figure 7.
Skin Regeneration, C7 Expression, and Ultrastructural Analysis of Grafts from CRISPR/Cas9-Edited (ΔE80) Stem Cell Clones
Keratinocyte clones, derived from the polyclonal population of cells treated with the sg2 + sg3 combination, with monoallelic (ΔE80/mut) and biallelic (ΔE80/ΔE80) E80 deletions being used to generate bioengineered skin equivalents and grafted onto immunodeficient mice. (A and B) Top: macroscopic view of an engrafted mouse 20 weeks after grafting of ΔE80/mut clone (A) and ΔE80/ΔE80 clone (B). Bottom: close-up view of the grafts under white and blue (GFP) light show the engrafted areas. (C and D) Histological analysis (H&E) of regenerated skin from ΔE80/mut clone (C) and ΔE80/ΔE80 clone (D). Note the clear dermal-epidermal adhesion in both types of grafts. (E and F) C7 immunoperoxidase expression analysis in ΔE80/mut clone (E) and ΔE80/ΔE80 clone (F) grafts. Both types of grafts display robust and continuous C7 expression. Insets show C7 expression at the human-mouse skin boundary. Note that mouse C7 is not recognized at the dilution of the antibody used. Scale bars, 100 μm. (G and H) Electron microscopy analysis shows the presence of well-developed anchoring fibrils (arrowheads) at the dermal-epidermal junction in grafts from both types of gene-edited clones. Scale bars, 500 nm.
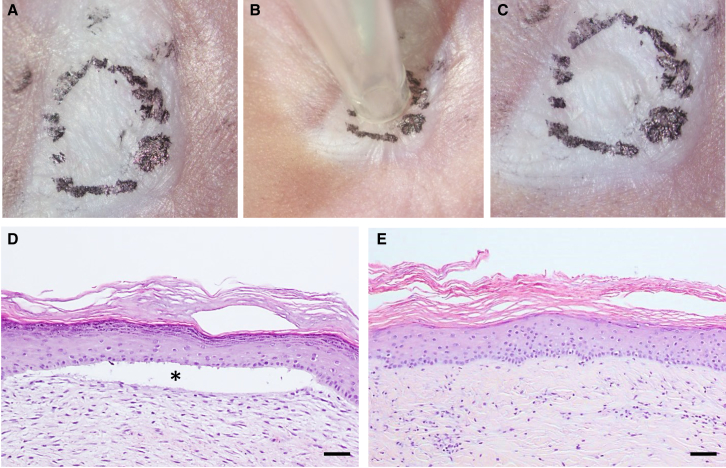
Blistering Resistance Assay of Gene-Edited Skin Grafts
Mechanical strength of human skin grafts regenerated from patient P1 cells was assessed by using a suction device able to exert precisely controlled negative pressure on circular areas of a 3-mm diameter. A negative pressure of 10 ± 2 kPa20 was applied for 5 min on two different points of each graft. No blistering was apparent on 12-week grafts from sg2 + sg3 RNP-treated patient keratinocytes (Figures 8A–8C). Histological analysis showed dermal-epidermal detachment after suction in grafts from unedited cells (Figure 8D). In contrast, uninterrupted dermal-epidermal adhesion (Figure 8E) was found after suction in ΔE80 grafts. To characterize the basement membrane integrity upon C7 expression restoration in edited RDEB grafts, immunofluorescence detection localization of type IV collagen (C4) and C7 was compared in blisters induced in ΔE80 RDEB and in normal human keratinocyte grafts. We found that, in ΔE80 RDEB grafts, C4 is localized at the floor of the blister, indicating that suction-induced blistering creates a separation between the basal keratinocytes and the lamina lucida, as observed in normal keratinocyte grafts (Figure S7).
Figure 8.
Blister Formation Resistance Assay
(A–C) Macroscopic view of a representative ΔE80 skin graft regenerated from sg2 + sg3 RNP-treated RDEB (P1) cells before (A), during (B), and after (C) the suction procedure. Black dotted circle line shows the human skin area where suction was applied. (D and E) Histological (H&E staining) characterization of skin sections corresponding to suctioned areas from unedited (D) and ΔE80 (E) grafts. Asterisk shows blistered area. Scale bars, 100 μm.
Discussion
To date, more than 650 different mutations causing the different subtypes of RDEB have been identified within COL7A1.21, 22 Severe generalized (sev-gen) RDEB, presenting complete absence or marked deficiency of C7, is the most devastating subtype. Mutations leading to a premature termination codon (PTC), that is, nonsense mutations or out-of-frame indels, cause sev-gen RDEB. A large proportion of these mutations occur within the coding region of the C7 collagenous domain encompassing exons 29–112.23 Although the majority of COL7A1 mutations are private, a limited number have been shown to be highly prevalent. This is the case of c.6527insC at E80, which is characteristic of the Spanish and some Latin American RDEB cohorts.14, 24 To our knowledge, this is the most prevalent COL7A1 mutation. Thus, an advanced gene-editing strategy tailored to this mutation would be beneficial for a large patient cohort.
All COL7A1 exons coding for the collagenous domain are in frame, and they might be amenable to an exon-skipping approach aimed at producing internally truncated, potentially functional C7. However, most of this domain is constituted by Gly-X-Y repeats that form the collagen triple helix, and the preservation of this repeat pattern must be taken into account when considering exon removal-based corrections. Since deletion of particular exons might result in structural alterations of C7, the functionality of each variant derived from exon removal needs to be assessed. Thus, production of functional C7 able to form anchoring fibrils has been demonstrated after transient or permanent removal of several different exons within the triple-helix-forming domain.11, 15, 16 Previous attempts by our laboratory using TALE nucleases designed to permanently restore the COL7A1 reading frame in homozygous c.6527insC carrier keratinocytes by NHEJ-mediated repair clearly proved that a deletion encompassing the whole of exon 80 restored the COL7A1 reading frame, resulting in a functional C7 variant10, 11 with healthy skin regeneration capacity. In spite of the increased efficacy with respect to HDR approaches, COL7A1 frame restoration by NHEJ with TALENs was only found in approximately 1% of the isolated, expanded, and genotyped epidermal stem cell clones. Although this gene-editing efficacy was yet not ready for clinical application, this proof-of-principle correction of primary patient cells by NHEJ, together with additional previous evidence from other laboratories showing the functionality of ΔE80 C7,15, 16, 25, 26 led us to seek a more efficient gene-editing strategy based on precise targeted excision of E80.
Our non-viral CRISPR/Cas9 delivery protocol resulted in C7 restoration in a very large percentage (close to 85%) of RDEB keratinocytes without noticeable toxicity, allowing us to overcome limitations in the use of gene-editing tools in hard-to-transfect primary cells. CRISPR/Cas9 RNP delivery systems have previously been shown to provide fast action and short permanence in the nucleus, as well as increased efficiency derived from robust nuclease activity, leading to the introduction of the desired modification in a high percentage of cells.27, 28, 29 This technology therefore demonstrates a superior safety profile by not using long-lasting expression from viral vectors (e.g., adenoviral or non-integrative lentiviral), which could maintain Cas9 cleavage activity for longer than desired, thus increasing the probability of off-target indel generation. Accordingly, assessment of the safety of the strategy by deep sequencing analysis of a large number of in silico-predicted off-target sites did not reveal any undesired cleavage. Concurrently, the intended DNA deletion to remove the faulty exon 80 sequence while preserving appropriate splicing motifs in adjacent introns was effected in a precise and efficient fashion, as evidenced by the very high percentage of transcripts lacking exon 80 and the absence of aberrant transcripts derived from undesired splicing events.
Gene correction protocols for RDEB cells using CRISPR/Cas9 have been tried before. Delivery of CRISPR/Cas9 components by plasmid electroporation into primary fibroblasts7 or by polymer-mediated transfection of an immortalized keratinocyte cell line9 required drug or marker selection to achieve mutation correction in a significant percentage of cells. Wu et al.17 used CRISPR/Cas9 RNPs to generate an E80-deleted mouse model, proving the functionality of a C7 variant lacking Col7a1 exon 80. In addition, they explored the in vivo delivery of CRISPR/Cas9 by direct mouse skin electroporation as a therapeutic avenue, demonstrating that the skin-blistering phenotype was significantly ameliorated after treatment. However, additional experimentation using skin-humanized preclinical models will be necessary before clinical translation of this approach can be envisioned. Furthermore, even if the challenge of efficient delivery of CRISPR/Cas9 complexes to human skin is overcome, preexisting immunity to Cas9 might still pose an additional hurdle for in vivo therapies.30 Hence, current protocols based on ex vivo manipulation of epidermal stem cells, already validated in the clinic, represent a safe and feasible therapeutic scenario.3 However, gene correction protocols based on CRISPR/Cas9 RNPs delivered by electroporation to primary patient keratinocytes, the main source of C7 in the skin, have not been previously reported. We have now demonstrated here that one such efficient protocol affords the possibility of using the whole polyclonal gene-edited keratinocyte population capable of producing therapeutic C7 levels, without the need for drug selection and isolation of corrected clones, which will facilitate its translation to the clinic. Moreover, we have proven that functional correction of patient cells has been achieved with this protocol, not only by histological analysis showing the structural integrity of the gene-edited skin grafts and ultrastructural analysis demonstrating the presence of anchoring fibrils at the BMZ but also by showing resistance to blister formation upon negative pressure application on these grafts.
In the present study, we provide evidence, through a rigorous in vivo skin regeneration test, that the epidermal stem cell population has been targeted, providing a long-term effect. A recent study involving the replacement of a large area of skin affected by JEB showed that engraftment occurred through a limited number of holoclones not isolated beforehand.3 Conceivably, a similar clinical outcome could be anticipated using isolated and characterized bona fide stem cells. Although the feasibility of epidermal stem clonal-based therapies with either added or edited genes has been previously established preclinically by us and others11, 31, 32, 33 and herein accomplished again, in terms of clinical applicability, the use of the RNP-edited bulk population containing long-term corrected stem cells represents a clear practical advantage.
As discussed above, the majority of RDEB-causing mutations occur within small and in-frame exons encoding the central helical collagenous domain and thus are ideal for an exon-skipping-mediated approach. Although in this study we have focused on E80 as the host of a highly recurrent mutation in RDEB patients, our data provide strong proof of principle for a strategy that should be easily transferrable to other potentially dispensable exons. Furthermore, we establish CRISPR/Cas9 delivered as RNP by electroporation as the state-of-the-art technology for genome editing in primary keratinocytes.
Overall, our results provide compelling preclinical evidence of the efficacy and safety of a novel CRISPR/Cas9-based ex vivo polyclonal gene-editing approach capable of correcting a highly prevalent mutation, opening up the way to feasible gene-editing-based therapy for RDEB patients that can be easily translated to the clinic.
Materials and Methods
Keratinocyte Culture and Isolation of Clones
Patient keratinocytes were originally obtained from skin biopsies of two RDEB (sev-gen RDEB) patients carrying the c.6527insC homozygous mutation in the COL7A1 gene. Clinical features of these patients have been previously described (patients 4 and 9).14 Skin biopsies were obtained from patients after approval from the Ethics Committee of the collaborating hospital upon informed consent.
Primary human RDEB and healthy donor keratinocytes were cultured as previously described.11, 32 Human primary RDEB keratinocytes from patients P1 and P211 were plated onto lethally irradiated 3T3-J2 cells and cultured in keratinocyte growth complete FAD (cFAD) medium (KCa), a 3:1 mix of DMEM and Ham’s F12 medium (Gibco-BRL, Barcelona, Spain) containing fetal bovine calf serum (Hyclone, GE Healthcare, Logan, UT) (10%), penicillin-streptomycin (1%), glutamine (2%), insulin (5 μg/mL; Sigma-Aldrich), adenine (0.18 mmol/L; Sigma-Aldrich), hydrocortisone (0.4 μg/mL; Sigma-Aldrich), cholera toxin (0.1 nmol/L; Sigma-Aldrich), triiodothyronine (2 nmol/L; Sigma-Aldrich), epidermal growth factor (EGF) (10 ng/mL; Sigma-Aldrich), and Y-27632 Rho-associated protein kinase (ROCK) inhibitor (Sigma-Aldrich) at 10 μM. To obtain isolated clones, cells were then trypsinized and plated at low density in 100-mm plates (103cells/plate) with 2 × 106 lethally irradiated 3T3 feeder cells per plate. Cell clones were then collected using polystyrene cloning cylinders (Sigma, St. Louis, MO) and expanded for further analysis. Clonal keratinocytes were GFP labeled using a phosphoglycerate kinase (PGK)-EGFP lentiviral vector, as previously described,34 to allow better visualization and follow-up of regenerated human skin after grafting.
CRISPR/Cas9 Delivery
sgRNAs were designed using a CRISPR design online tool (https://zlab.bio/guide-design-resources). Synthetic RNAs and recombinant Cas9 were purchased from Integrated DNA Technologies (IDT, IL), and RNP complexes were reconstituted according to the manufacturer’s instructions and delivered to the RDEB patient primary keratinocytes by nucleofection using the Neon Transfection System 10 μL Kit (Thermo Fisher Scientific). Primary cells were trypsinized and washed with PBS. 1.5 × 105 primary keratinocytes were resuspended in 10 μL Resuspension Buffer R for each reaction, and RNP complexes were added to each sample (72.7 pmol CRISPR RNA [crRNA][sgRNA]:trans-activating crRNA [tracrRNA], 10.9 pmol Cas9, 6.6:1 molar ratio). Two electroporation conditions were tested: 1,150 V/30 ms/2 pulses (condition A) and 1,700 V/20 ms/1 pulse (condition B).35 After electroporation, cells were seeded into 6-well plates containing a feeder layer and pre-warmed KCa medium.
Genotyping of Gene-Edited Keratinocytes
Genomic DNA was isolated by isopropanol precipitation of keratinocyte lysates (lysis buffer was Tris [pH 8] 100 mM, EDTA 5 mM, SDS 0.2%, NaCl 200 mM, and 1 mg/mL proteinase K [Roche Diagnostics, Mannheim, Germany]) and resuspended in Tris/EDTA (TE) buffer. Approximately 20–50 ng genomic DNA was used for PCR amplification. PCR fragments spanning the nuclease target sites were generated with primers F1/R (F1, 5′-gtgagtggtggctgaagcac-3′; and R, 5′-accccaccaaggaaactga-3′). PCR program TD 68-63 was as follows: 94°C for 5 min; 5 cycles of 94°C for 30 s, 68°C for 30 s, and 72°C for 30 s, decreasing annealing temperature 1°C every cycle; followed by 30 cycles of 94°C for 30 s, 63°C for 30 s, and 72°C for 30 s; then 72°C for 7 min. PCR products were analyzed in 1.5% agarose gel. Molecular weight marker was IX (Sigma-Aldrich). For sequencing, PCR products were treated with Illustra ExoProStar (GE Healthcare, UK), sequenced using Big Dye Terminator version (v.)1.1 Cycle Sequencing kit (Thermo Fisher Scientific, Waltham, MA) and examined on a 3730 DNA Analyzer (Life Technologies, Carlsbad, CA). Chromatograms were analyzed using Sequencher (Gene Codes, Ann Harbor, MI) and Chromas (Technelysium, Australia) software. Bio-Rad Image Lab Software 6.0 was used for PCR band densitometry.
Genome Editing Off-Target Analysis by NGS
On-target and potential off-target sites were designed, sequenced, and analyzed using the Thera-Seq genotyping services (Desktop Genetics). Sequencing libraries were prepared and subjected to paired-end sequencing on a MiSeq using Reagent Kit v.2 (500 cycles) (Illumina).
COL7A1 Transcription Analysis by RT-PCR and qPCR
Total RNA was extracted from keratinocytes with the miRNeasy Mini Kit (QIAGEN, Hilden, Germany), and cDNA was synthesized using the SuperScript III First-Strand Synthesis System (Invitrogen, Carlsbad, CA). The primers (forward) 5′-aggggtcaggacggcaac-3′ and (reverse) 5′-cagctccagtaggtccagtcag-3′ were used to amplify 241-bp (unedited) and 205-bp (exon 80-deleted) fragments spanning exons 78–84 of COL7A1. The PCR program was 94°C for 5 min; 5 cycles of 94°C for 30 s, 68°C for 30 s, and 72°C for 30 s, decreasing annealing temperature 1°C every cycle; followed by 25 cycles of 94°C for 30 s, 63°C for 30 s, and 72°C for 30 s; then 72°C for 7 min. The human glyceraldehyde-3-phosphate dehydrogenase gene (GAPDH) was analyzed as a loading control with GAPDH- specific primers (forward, 5′-accacagtccatgccatcac-3′; and reverse, 5′-tccaccaccctgttgctgt-3′). For the qPCR analysis, 1:20 dilutions of each cDNA synthesis reaction were analyzed in triplicate using Taqman gene expression assays Hs00164310_m1 (COL7A1 exon64 probe), Hs01574801_g1 (COL7A1 exon80 probe), and Hs02758991_g1 (GAPDH probe, control). Amplification was performed using a QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems).
Western Blot Analysis
Keratinocytes were lysed in protein extraction buffer (50 mM Tris-HCl [pH 7.5], 100 mM NaCl, 1% Nonidet P-40, and 4 mM EDTA) containing proteinase inhibitors cocktail (Complete Mini, EDTA-free; Roche Diagnostics, Mannheim, Germany). Lysates were incubated for 30 min on ice and centrifuged at 15,000 × g for 30 min at 4°C. Supernatants were collected and concentrated by ultrafiltration using Amicon Ultra columns (10 kDa; Millipore, Ireland). Protein concentrations were measured using the Bradford assay (Bio-Rad, Hercules, CA). For each sample, 40 μg total protein was resolved on NuPAGE Novex 3%–8% Tris-Acetate gel electrophoresis (Invitrogen, Carlsbad, CA) and electrotransferred to nitrocellulose membranes (Invitrogen, Carlsbad, CA). For type VII collagen analysis, blots were probed with a monospecific polyclonal anti-C7 antibody (a generous gift from Dr. A. Nystrom, University of Freiburg). An antibody against GAPDH was used as a loading control. Visualization was performed by incubating with horseradish peroxidase (HRP)-conjugated anti-rabbit antibody (Amersham, Burlington, MA) and West Pico Chemiluminescent Substrate (Pierce, Rockford, IL).
Immunofluorescence and Immunohistochemical Staining
For immunofluorescence detection of C7 in keratinocytes, cells grown on glass coverslips were fixed in methanol/acetone (1:1) for 10 min at −20°C. After washing three times in PBS and once in PBS with 3% BSA (Sigma-Aldrich, St. Louis, MO) for 30 min, cells were incubated with monospecific polyclonal anti-C7 antibody at 1:5,000 dilution. Secondary antibody (AlexaFluor488, Invitrogen, Carlsbad, CA) was used at 1:1,000 dilution. After the final washing step in PBS, preparations were mounted using Mowiol (Hoechst, Somerville, NJ) mounting medium and DAPI 20 μg/mL (Sigma-Aldrich, St Louis, MO) for nuclei visualization. Immunoperoxidase detection of C7 in paraffin-embedded, formalin-fixed sections was carried out with proteinase K antigen retrieval as described.36 Immunoperoxidase staining for human involucrin and p63 was performed using rabbit SY5 monoclonal antibody (Sigma) and 4A4 monoclonal antibody, respectively, on paraffin sections without antigen retrieval. The ABC peroxidase kit (Vector Laboratories) was used for immunohistochemical detection. Immunofluorescence detection of C4 in regenerated skin tissue was carried out using C4 monoclonal antibody (Clone CIV 22; Dako, M0785). Secondary antibody (AlexaFluor 488, Invitrogen, Carlsbad, CA) was used at 1:1,000 dilution.
Electron Microscopy
Specimens of approximately 0.4 × 0.3 cm were fixed for at least 2 h at room temperature in 3% glutaraldehyde solution in 0.1 M cacodylate buffer (pH 7.4), cut into pieces of approximately 1 mm3, washed in buffer, post-fixed for 1 h at 4°C in 1% osmium tetroxide, rinsed in water, dehydrated through graded ethanol solutions, transferred into propylene oxide, and embedded in epoxy resin (glycidether 100). Semi-thin and ultrathin sections were cut with an ultramicrotome (Reichert Ultracut E). Ultrathin sections were treated with uranyl acetate and lead citrate, and they were examined with an electron microscope (JEM 1400) equipped with a 2k charge-coupled device (CCD) camera (TVIPS).
Generation of Skin Equivalents, Grafting onto Immunodeficient Mice, and Graft Analysis
Animal studies were approved by our institutional animal care and use committee according to national and European legal regulations.
CRISPR/Cas9 RNP-treated keratinocytes (polyclonal or monoclonal populations) were seeded on fibrin dermal equivalents containing RDEB fibroblasts null for C7 expression, prepared as previously described.37 Bioengineered skin equivalents were grafted onto the back of 7-week-old female immunodeficient mice (nu/nu, NMRI background) purchased from Elevage-Janvier (France), as previously described.32 Grafting was performed under sterile conditions, and mice were housed in pathogen-free conditions for the duration of the experiment at the CIEMAT Laboratory Animals Facility (Spanish registration 28079-21 A). Animals were housed in individually ventilated type II cages, with 25 air changes/h and 10 kGy gamma-irradiated soft wood pellets as bedding. All handling was carried out under sterile conditions, and all experimental procedures were according to European and Spanish laws and regulations. Grafts were monitored using a magnifying lens equipped with white and blue light to allow EGFP visualization. Mice were sacrificed at different time points post-grafting, and grafts were harvested for skin histology, immunohistochemistry analyses, and electron microscopy studies.
In Vivo Skin Fragility Test
A suction device developed in our laboratory was set up to exert a negative pressure of 10 ± 2 kPa on a 3-mm diameter area for 5 min to induce blister formation onto human skin grafts regenerated in immunodeficient mice 12 weeks after grafting. Two mice bearing grafts from unedited and two from sg2 + sg3 RNP-treated keratinocytes were used. Suction was applied on two different sites for each graft. Before applying suction, to promote blister formation, an incandescent light bulb was set on top of the graft area approximately 2 cm away for 2 min.38 After that, the bulb was kept on for the entire duration of the experiment. The suctioned area was photographed 10 min after suctioning and excised for histological analysis.
Author Contributions
J.B., A.M., R.M., and F.L. designed the experiments. J.B., A.M., M.G., R.T., S.R., E.C.-S., and S.M.-H. performed molecular and cellular studies. M.D.R. contributed to blistering tests and M.J.E. contributed to the discussion of experimental results and provided reagents and ethical approval for the studies. M.G., J.B., and F.L. performed animal experiments and histological analysis. M.C. and C.L. contributed to data analysis and critical review of the manuscript. I.H. performed the electron microscopy analysis. L.M. and M.G. developed the suction device and performed the in vivo skin fragility test. J.B., A.M., F.L., and R.M. wrote the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
The study was mainly supported by DEBRA International, funded by DEBRA Austria (grant termed as Larcher 1). Additional funds came from Spanish grants SAF2017-86810-R (to M.D.R.) and PI17/01747 (to F.L.) from the Ministry of Economy and Competitiveness and Instituto de Salud Carlos III, respectively, both co-funded with European Regional Development Funds (ERDF) ERA-NET E-RARE JTC 2017 (MutaEB) and CIBERER (grant termed as Murillas-TERAPIAS ER2017). Authors are indebted to Blanca Duarte, Almudena Holguín, and Nuria Illera for grafting experiments and to Jesus Martínez and Edilia De Almeida for animal maintenance and care. We also thank Juan Manuel Ruibal Mera for the suction blister device manufacturing and Jonathan O’Keeffe for professional language examination of the manuscript.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ymthe.2019.03.007.
Contributor Information
Rodolfo Murillas, Email: rodolfo.murillas@ciemat.es.
Fernando Larcher, Email: fernando.larcher@ciemat.es.
Supplemental Information
References
- 1.Fine J.D., Bruckner-Tuderman L., Eady R.A., Bauer E.A., Bauer J.W., Has C., Heagerty A., Hintner H., Hovnanian A., Jonkman M.F. Inherited epidermolysis bullosa: updated recommendations on diagnosis and classification. J. Am. Acad. Dermatol. 2014;70:1103–1126. doi: 10.1016/j.jaad.2014.01.903. [DOI] [PubMed] [Google Scholar]
- 2.Bauer J.W., Koller J., Murauer E.M., De Rosa L., Enzo E., Carulli S., Bondanza S., Recchia A., Muss W., Diem A. Closure of a Large Chronic Wound through Transplantation of Gene-Corrected Epidermal Stem Cells. J. Invest. Dermatol. 2017;137:778–781. doi: 10.1016/j.jid.2016.10.038. [DOI] [PubMed] [Google Scholar]
- 3.Hirsch T., Rothoeft T., Teig N., Bauer J.W., Pellegrini G., De Rosa L., Scaglione D., Reichelt J., Klausegger A., Kneisz D. Regeneration of the entire human epidermis using transgenic stem cells. Nature. 2017;551:327–332. doi: 10.1038/nature24487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mavilio F., Pellegrini G., Ferrari S., Di Nunzio F., Di Iorio E., Recchia A., Maruggi G., Ferrari G., Provasi E., Bonini C. Correction of junctional epidermolysis bullosa by transplantation of genetically modified epidermal stem cells. Nat. Med. 2006;12:1397–1402. doi: 10.1038/nm1504. [DOI] [PubMed] [Google Scholar]
- 5.Siprashvili Z., Nguyen N.T., Gorell E.S., Loutit K., Khuu P., Furukawa L.K., Lorenz H.P., Leung T.H., Keene D.R., Rieger K.E. Safety and Wound Outcomes Following Genetically Corrected Autologous Epidermal Grafts in Patients With Recessive Dystrophic Epidermolysis Bullosa. JAMA. 2016;316:1808–1817. doi: 10.1001/jama.2016.15588. [DOI] [PubMed] [Google Scholar]
- 6.Osborn M.J., Starker C.G., McElroy A.N., Webber B.R., Riddle M.J., Xia L., DeFeo A.P., Gabriel R., Schmidt M., von Kalle C. TALEN-based gene correction for epidermolysis bullosa. Mol. Ther. 2013;21:1151–1159. doi: 10.1038/mt.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Webber B.R., Osborn M.J., McElroy A.N., Twaroski K., Lonetree C.L., DeFeo A.P., Xia L., Eide C., Lees C.J., McElmurry R.T. CRISPR/Cas9-based genetic correction for recessive dystrophic epidermolysis bullosa. NPJ Regen. Med. 2016;1:16014. doi: 10.1038/npjregenmed.2016.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Izmiryan A., Danos O., Hovnanian A. Meganuclease-Mediated COL7A1 Gene Correction for Recessive Dystrophic Epidermolysis Bullosa. J. Invest. Dermatol. 2016;136:872–875. doi: 10.1016/j.jid.2015.11.028. [DOI] [PubMed] [Google Scholar]
- 9.Hainzl S., Peking P., Kocher T., Murauer E.M., Larcher F., Del Rio M., Duarte B., Steiner M., Klausegger A., Bauer J.W. COL7A1 Editing via CRISPR/Cas9 in Recessive Dystrophic Epidermolysis Bullosa. Mol. Ther. 2017;25:2573–2584. doi: 10.1016/j.ymthe.2017.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chamorro C., Mencía A., Almarza D., Duarte B., Büning H., Sallach J., Hausser I., Del Río M., Larcher F., Murillas R. Gene Editing for the Efficient Correction of a Recurrent COL7A1 Mutation in Recessive Dystrophic Epidermolysis Bullosa Keratinocytes. Mol. Ther. Nucleic Acids. 2016;5:e307. doi: 10.1038/mtna.2016.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mencía Á., Chamorro C., Bonafont J., Duarte B., Holguin A., Illera N., Llames S.G., Escámez M.J., Hausser I., Del Río M. Deletion of a Pathogenic Mutation-Containing Exon of COL7A1 Allows Clonal Gene Editing Correction of RDEB Patient Epidermal Stem Cells. Mol. Ther. Nucleic Acids. 2018;11:68–78. doi: 10.1016/j.omtn.2018.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hovnanian A., Rochat A., Bodemer C., Petit E., Rivers C.A., Prost C., Fraitag S., Christiano A.M., Uitto J., Lathrop M. Characterization of 18 new mutations in COL7A1 in recessive dystrophic epidermolysis bullosa provides evidence for distinct molecular mechanisms underlying defective anchoring fibril formation. Am. J. Hum. Genet. 1997;61:599–610. doi: 10.1086/515495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cuadrado-Corrales N., Sánchez-Jimeno C., García M., Escámez M.J., Illera N., Hernández-Martín A., Trujillo-Tiebas M.J., Ayuso C., Del Rio M. A prevalent mutation with founder effect in Spanish Recessive Dystrophic Epidermolysis Bullosa families. BMC Med. Genet. 2010;11:139. doi: 10.1186/1471-2350-11-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Escámez M.J., García M., Cuadrado-Corrales N., Llames S.G., Charlesworth A., De Luca N., Illera N., Sánchez-Jimeno C., Holguín A., Duarte B. The first COL7A1 mutation survey in a large Spanish dystrophic epidermolysis bullosa cohort: c.6527insC disclosed as an unusually recurrent mutation. Br. J. Dermatol. 2010;163:155–161. doi: 10.1111/j.1365-2133.2010.09713.x. [DOI] [PubMed] [Google Scholar]
- 15.Bornert O., Kühl T., Bremer J., van den Akker P.C., Pasmooij A.M., Nyström A. Analysis of the functional consequences of targeted exon deletion in COL7A1 reveals prospects for dystrophic epidermolysis bullosa therapy. Mol. Ther. 2016;24:1302–1311. doi: 10.1038/mt.2016.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turczynski S., Titeux M., Tonasso L., Décha A., Ishida-Yamamoto A., Hovnanian A. Targeted Exon Skipping Restores Type VII Collagen Expression and Anchoring Fibril Formation in an In Vivo RDEB Model. J. Invest. Dermatol. 2016;136:2387–2395. doi: 10.1016/j.jid.2016.07.029. [DOI] [PubMed] [Google Scholar]
- 17.Wu W., Lu Z., Li F., Wang W., Qian N., Duan J., Zhang Y., Wang F., Chen T. Efficient in vivo gene editing using ribonucleoproteins in skin stem cells of recessive dystrophic epidermolysis bullosa mouse model. Proc. Natl. Acad. Sci. USA. 2017;114:1660–1665. doi: 10.1073/pnas.1614775114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ousterout D.G., Kabadi A.M., Thakore P.I., Majoros W.H., Reddy T.E., Gersbach C.A. Multiplex CRISPR/Cas9-based genome editing for correction of dystrophin mutations that cause Duchenne muscular dystrophy. Nat. Commun. 2015;6:6244. doi: 10.1038/ncomms7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pellegrini G., Dellambra E., Golisano O., Martinelli E., Fantozzi I., Bondanza S., Ponzin D., McKeon F., De Luca M. p63 identifies keratinocyte stem cells. Proc. Natl. Acad. Sci. USA. 2001;98:3156–3161. doi: 10.1073/pnas.061032098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petrof G., Lwin S.M., Martinez-Queipo M., Abdul-Wahab A., Tso S., Mellerio J.E., Slaper-Cortenbach I., Boelens J.J., Tolar J., Veys P. Potential of Systemic Allogeneic Mesenchymal Stromal Cell Therapy for Children with Recessive Dystrophic Epidermolysis Bullosa. J. Invest. Dermatol. 2015;135:2319–2321. doi: 10.1038/jid.2015.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van den Akker P.C., Jonkman M.F., Rengaw T., Bruckner-Tuderman L., Has C., Bauer J.W., Klausegger A., Zambruno G., Castiglia D., Mellerio J.E. The international dystrophic epidermolysis bullosa patient registry: an online database of dystrophic epidermolysis bullosa patients and their COL7A1 mutations. Hum. Mutat. 2011;32:1100–1107. doi: 10.1002/humu.21551. [DOI] [PubMed] [Google Scholar]
- 22.Wertheim-Tysarowska K., Sobczyńska-Tomaszewska A., Kowalewski C., Skroński M., Swięćkowski G., Kutkowska-Kaźmierczak A., Woźniak K., Bal J. The COL7A1 mutation database. Hum. Mutat. 2012;33:327–331. doi: 10.1002/humu.21651. [DOI] [PubMed] [Google Scholar]
- 23.Stenson P.D., Mort M., Ball E.V., Evans K., Hayden M., Heywood S., Hussain M., Phillips A.D., Cooper D.N. The Human Gene Mutation Database: towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum. Genet. 2017;136:665–677. doi: 10.1007/s00439-017-1779-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodríguez F.A., Gana M.J., Yubero M.J., Zillmann G., Krämer S.M., Catalán J., Rubio-Astudillo J., González S., Liu L., Ozoemena L. Novel and recurrent COL7A1 mutations in Chilean patients with dystrophic epidermolysis bullosa. J. Dermatol. Sci. 2012;65:149–152. doi: 10.1016/j.jdermsci.2011.11.010. [DOI] [PubMed] [Google Scholar]
- 25.Turczynski S., Titeux M., Pironon N., Hovnanian A. Antisense-mediated exon skipping to reframe transcripts. Methods Mol. Biol. 2012;867:221–238. doi: 10.1007/978-1-61779-767-5_15. [DOI] [PubMed] [Google Scholar]
- 26.Bremer J., Bornert O., Nyström A., Gostynski A., Jonkman M.F., Aartsma-Rus A., van den Akker P.C., Pasmooij A.M. Antisense Oligonucleotide-mediated Exon Skipping as a Systemic Therapeutic Approach for Recessive Dystrophic Epidermolysis Bullosa. Mol. Ther. Nucleic Acids. 2016;5:e379. doi: 10.1038/mtna.2016.87. [DOI] [PubMed] [Google Scholar]
- 27.Kim S., Kim D., Cho S.W., Kim J., Kim J.S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24:1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seki A., Rutz S. Optimized RNP transfection for highly efficient CRISPR/Cas9-mediated gene knockout in primary T cells. J. Exp. Med. 2018;215:985–997. doi: 10.1084/jem.20171626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dever D.P., Bak R.O., Reinisch A., Camarena J., Washington G., Nicolas C.E., Pavel-Dinu M., Saxena N., Wilkens A.B., Mantri S. CRISPR/Cas9 β-globin gene targeting in human haematopoietic stem cells. Nature. 2016;539:384–389. doi: 10.1038/nature20134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Charlesworth C.T., Deshpande P.S., Dever D.P., Camarena J., Lemgart V.T., Cromer M.K., Vakulskas C.A., Collingwood M.A., Zhang L., Bode N.M. Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat. Med. 2019;25:249–254. doi: 10.1038/s41591-018-0326-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Droz-Georget Lathion S., Rochat A., Knott G., Recchia A., Martinet D., Benmohammed S., Grasset N., Zaffalon A., Besuchet Schmutz N., Savioz-Dayer E. A single epidermal stem cell strategy for safe ex vivo gene therapy. EMBO Mol. Med. 2015;7:380–393. doi: 10.15252/emmm.201404353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duarte B., Miselli F., Murillas R., Espinosa-Hevia L., Cigudosa J.C., Recchia A., Del Río M., Larcher F. Long-term skin regeneration from a gene-targeted human epidermal stem cell clone. Mol. Ther. 2014;22:1878–1880. doi: 10.1038/mt.2014.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Larcher F., Dellambra E., Rico L., Bondanza S., Murillas R., Cattoglio C., Mavilio F., Jorcano J.L., Zambruno G., Del Rio M. Long-term engraftment of single genetically modified human epidermal holoclones enables safety pre-assessment of cutaneous gene therapy. Mol. Ther. 2007;15:1670–1676. doi: 10.1038/sj.mt.6300238. [DOI] [PubMed] [Google Scholar]
- 34.Di Nunzio F., Maruggi G., Ferrari S., Di Iorio E., Poletti V., Garcia M., Del Rio M., De Luca M., Larcher F., Pellegrini G., Mavilio F. Correction of laminin-5 deficiency in human epidermal stem cells by transcriptionally targeted lentiviral vectors. Mol. Ther. 2008;16:1977–1985. doi: 10.1038/mt.2008.204. [DOI] [PubMed] [Google Scholar]
- 35.Liang X., Potter J., Kumar S., Zou Y., Quintanilla R., Sridharan M., Carte J., Chen W., Roark N., Ranganathan S. Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J. Biotechnol. 2015;208:44–53. doi: 10.1016/j.jbiotec.2015.04.024. [DOI] [PubMed] [Google Scholar]
- 36.Niskanen J., Dillard K., Arumilli M., Salmela E., Anttila M., Lohi H., Hytönen M.K. Nonsense variant in COL7A1 causes recessive dystrophic epidermolysis bullosa in Central Asian Shepherd dogs. PLoS ONE. 2017;12:e0177527. doi: 10.1371/journal.pone.0177527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Llames S.G., Del Rio M., Larcher F., García E., García M., Escamez M.J., Jorcano J.L., Holguín P., Meana A. Human plasma as a dermal scaffold for the generation of a completely autologous bioengineered skin. Transplantation. 2004;77:350–355. doi: 10.1097/01.TP.0000112381.80964.85. [DOI] [PubMed] [Google Scholar]
- 38.Hatje L.K., Richter C., Blume-Peytavi U., Kottner J. Blistering time as a parameter for the strength of dermoepidermal adhesion: a systematic review and meta-analysis. Br. J. Dermatol. 2015;172:323–330. doi: 10.1111/bjd.13298. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.