Abstract
Purkinje cells (PCs) are primarily affected in neurodegenerative spinocerebellar ataxias (SCAs). For generating animal models for SCAs, genetic regulatory elements specifically targeting PCs are required, thereby linking pathological molecular effects with impaired function and organismic behavior. Because cerebellar anatomy and function are evolutionary conserved, zebrafish represent an excellent model to study SCAs in vivo. We have isolated a 258 bp cross-species PC-specific enhancer element that can be used in a bidirectional manner for bioimaging of transgene-expressing PCs in zebrafish (both sexes) with variable copy numbers for tuning expression strength. Emerging ectopic expression at high copy numbers can be further eliminated by repurposing microRNA-mediated posttranslational mRNA regulation.
Subsequently, we generated a transgenic SCA type 13 (SCA13) model, using a zebrafish-variant mimicking a human pathological SCA13R420H mutation, resulting in cell-autonomous progressive PC degeneration linked to cerebellum-driven eye-movement deficits as observed in SCA patients. This underscores that investigating PC-specific cerebellar neuropathologies in zebrafish allows for interconnecting bioimaging of disease mechanisms with behavioral analysis suitable for therapeutic compound testing.
SIGNIFICANCE STATEMENT SCA13 patients carrying a KCNC3R420H allele have been shown to display mid-onset progressive cerebellar atrophy, but genetic modeling of SCA13 by expressing this pathogenic mutant in different animal models has not resulted in neuronal degeneration so far; likely because the transgene was expressed in heterologous cell types. We developed a genetic system for tunable PC-specific coexpression of several transgenes to manipulate and simultaneously monitor cerebellar PCs. We modeled a SCA13 zebrafish accessible for bioimaging to investigate disease progression, revealing robust PC degeneration, resulting in impaired eye movement. Our transgenic zebrafish mimicking both neuropathological and behavioral changes manifested in SCA-affected patients will be suitable for investigating causes of cerebellar diseases in vivo from the molecular to the behavioral level.
Keywords: bioimaging, cerebellum, neuronal degeneration, Purkinje cells, spinocerebellar ataxia, zebrafish
Introduction
One of the best understood neural circuits is the connectivity of neurons of the vertebrate cerebellum, which is responsible for coordinating body posture, balance, and locomotive control among others. Therefore, the cerebellum provides a highly valuable model to gain insight into how neuronal networks integrate external sensory input and generate an appropriate behavioral response (Ito, 2006; Jörntell, 2017).
Purkinje cells (PCs) play a central role in the cerebellar circuitry because these key neurons integrate and process the sensory information they receive from the periphery via synapses from the parallel fibers and climbing fibers, respectively, and generate motor commands to mediate smooth and skillful movements (Ito, 2006; Jörntell, 2017). Because of this central role of PCs in the cerebellar cortex, their dysfunction results in many cerebellar diseases of which the clinical manifestations include motor incoordination, ataxia, tremor, dysarthria, and oculomotor disturbances such as nystagmus (Rossi et al., 2014; Hekman and Gomez, 2015).
The cytoarchitecture and connectivity of cerebellar neurons are highly conserved between teleost fish and mammals (Hashimoto and Hibi, 2012; Matsui et al., 2014). Hence, the almost transparent zebrafish larva is increasingly used for non-invasive optical imaging to disclose developmental programs organizing cerebellar development including cell biological mechanisms of PC differentiation (Köster and Fraser, 2001a; Distel et al., 2010; Tanabe et al., 2010; Theisen et al., 2018). In addition, it was shown that the cerebellum of larval zebrafish is well suited to dissect physiology and function of PCs controlling motor behaviors and learning (Hsieh et al., 2014; Matsui et al., 2014; Sengupta and Thirumalai, 2015; Harmon et al., 2017).
Despite the increasing knowledge about the development and function of the zebrafish cerebellum including underlying genetic and cell biological mechanisms, little effort has been made to exploit zebrafish for modeling human cerebellar diseases, especially caused by PC dysfunction. We therefore sought to develop a gain-of-function neurodegeneration model by overexpression of a pathogenic gene typically affecting cerebellar PC survival. Ideally, these neurons should be the only ones targeted by transgene expression to attribute the cause of the observed morphological, cell biological, molecular, physiological and behavioral phenotypes unambiguously to PC dysfunction.
Thus, we first isolated a novel PC regulatory element derived from the upstream region of zebrafish carbonic anhydrase VIII (ca8) gene whose expression is detected almost exclusively in zebrafish PCs (Bae et al., 2009). By identifying a small enhancer of only 258 bp, we were able to develop a bidirectional PC-specific system of two transgenes whose expression strength could be tuned at will. The established expression system is suitable for monitoring PCs expressing a pathogenic mutant gene using a fluorescent reporter protein targeted to cellular structures different from the pathogenic protein.
As a proof-of-concept for this expression system, we modeled spinocerebellar ataxia type 13 (SCA13). SCA13 represents an autosomal dominant neurological disease, which can be accompanied by prominent cerebellar atrophy (Kaczmarek and Zhang, 2017). This disease is caused by the expression of mutant variants of the voltage-gated potassium channel Kv3.3 encoded by the KCNC3 gene (Waters et al., 2006) resulting in ataxia with prominent cerebellar signs and symptoms as well as often accompanied by mild mental retardation and epilepsy in heterozygous carriers (Waters and Pulst, 2008; Stevanin and Dürr, 2012). However, the biological consequence of its mutant allele in terms of PC degeneration is poorly understood. With the help of the bidirectional PC-specific expression system, we genetically modeled SCA13 in zebrafish, which indeed resulted in robust and progressive degeneration of PCs already at larval stages and caused eye movement deficits as behavioral symptoms. These phenotypes will allow for bioimaging approaches addressing disease-causing mechanisms as well as therapeutic compound testing in vivo.
Materials and Methods
Animal husbandry.
AB wild-type fish and the less pigmented brass mutant were used throughout the study. Fish were maintained according to standard protocols (Westerfield, 2007). The stable transgenic lines established in this work (Tg(-7.5ca8:GFP)bz12, Tg(cpce-E1b:GFP)bz13, and Tg(2xcpce-E1b:KALNFB, he1.1:mTagBFP2)bz14) are characterized in Figures 3, 5, and 7, respectively. In addition, the following two UAS transgenic lines were used; Tg(4xUAS:GFP)hzm3 (Distel et al., 2009), and Tg(5xUAS:FMAVenus-2A-3xNLS-HA-mseCFP)bz8 (Matsui et al., 2014). Zebrafish embryos and larvae were maintained in zebrafish rearing medium (30% Danieau: 58 mm NaCl, 0.7 mm KCl, 0.4 mm MgSO4, 0.6 mm Ca(NO3)2, and 5 mm HEPES, pH7.2) at 28°C. To inhibit development of pigmentation, the rearing medium was supplemented with 1-phenyl-2-thiourea (PTU; Sigma-Aldrich) starting from 24 h postfertilization (hpf) at concentrations of 200 μm for microscopic observations or 70 μm for optokinetic response (OKR) experiments, respectively. All procedures involving zebrafish were performed according to EU guidelines and German legislation (EU Directive 2010_63, license AZ 325.1.53/56.1-TU-BS). Mouse experiments were approved by the animal welfare representative of the TU Braunschweig and the LAVES [Az. §4 (02.05) TSchB TU BS].
Figure 3.
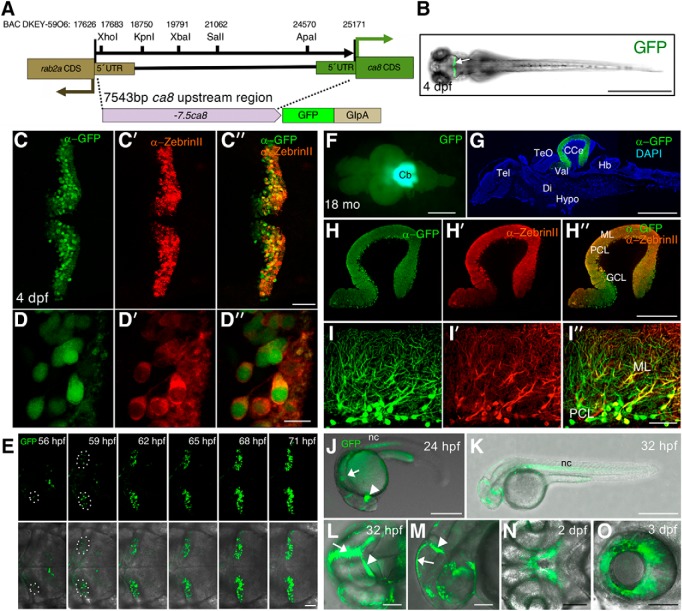
Characterization of GFP expression pattern of Tg (-7.5ca8:GFP)bz12. The structure of the −7.5ca8:GFP transgene and its expression in Tg(-7.5ca8:GFP)bz12 fish. A, Schematic map of the −7.5ca8:GFP-globin intron-SV40 polyA (GIpA) transgene representing the 5′-upstream sequence of the ca8 gene, which corresponds to the sequence flanked by ca8 and rab2a coding regions (17626–25171 of BAC DKEY-59O6, GenBank accession number: CR847864). Unique restriction enzyme sites in this fragment are shown. B, Transgenic F2 larva at 4 dpf displays fluorescent GFP expression (arrow) in the cerebellum. PC-specific GFP expression of the transgene in the cerebellum of 4 dpf larvae was confirmed by co-immunostaining with anti-GFP (C, green) and anti-ZebrinII (C′, red) antibodies, a merged image demonstrates coexpression (C″, green and red channels), magnified views are shown in (D, D′, D″). E, Time-lapse imaging analysis of the cerebellum of Tg(-7.5ca8:GFP)bz12 larva between 56 and 71 hpf. The areas encircled by white dotted ellipses indicate emerging GFP-expressing PCs. Upper image row shows GFP fluorescence (green) recordings only that were overlaid in the lower row by transmitted light field images for anatomical orientation. Dorsal view (F), and a sagittal section (G) of an 18-month-old adult brain of Tg(-7.5ca8:GFP)bz12 fish expressing GFP (green) in the cerebellar region. Immunostaining was performed with anti-GFP antibody and counterstained with DAPI (G, blue). Costaining with anti-GFP (H) and anti-ZebrinII (H′) antibodies, that were merged to reveal coexpression in Purkinje cells (H″), magnified views are shown in (I, I′, I″). J–O, GFP expression pattern in Tg(-7.5ca8:GFP)bz12 larvae during embryonic stages. J, Lateral view of a 24 hpf embryo showing GFP (green) expression in the diencephalon (white arrowhead), the hindbrain (white arrow) and the notochord (nc). K, Lateral view of a 32 hpf embryo showing GFP expression in the notochord. Higher magnified dorsal (L) and lateral (M) views of the head regions of a 32 hpf embryo showing green fluorescent epithelia in the midline seam (white arrows), and the midbrain-hindbrain boundary (white arrowheads). N, Ventral view of a 2 dpf embryo showing GFP expression in the stomodeum. O, Lateral view of a 3 dpf larva showing GFP expression in the retinal pigment epithelium. Scale bars: B, G, J, K, 1 mm; H″, 400 μm; L–O, 100 μm; C″, E, I″, 50 μm; D″, 10 μm. CCe, Corpus cerebelli; CDS, coding sequence; Di, diencephalon; GCL, granule cell layer; Hb, hindbrain; Hypo, hypothalamus; ML, molecular layer; PCL, PC layer; Tel, telencephalon; TeO, optic tectum; UTR, untranslated region; Val, lateral division of the valvula cerebelli.
Figure 5.
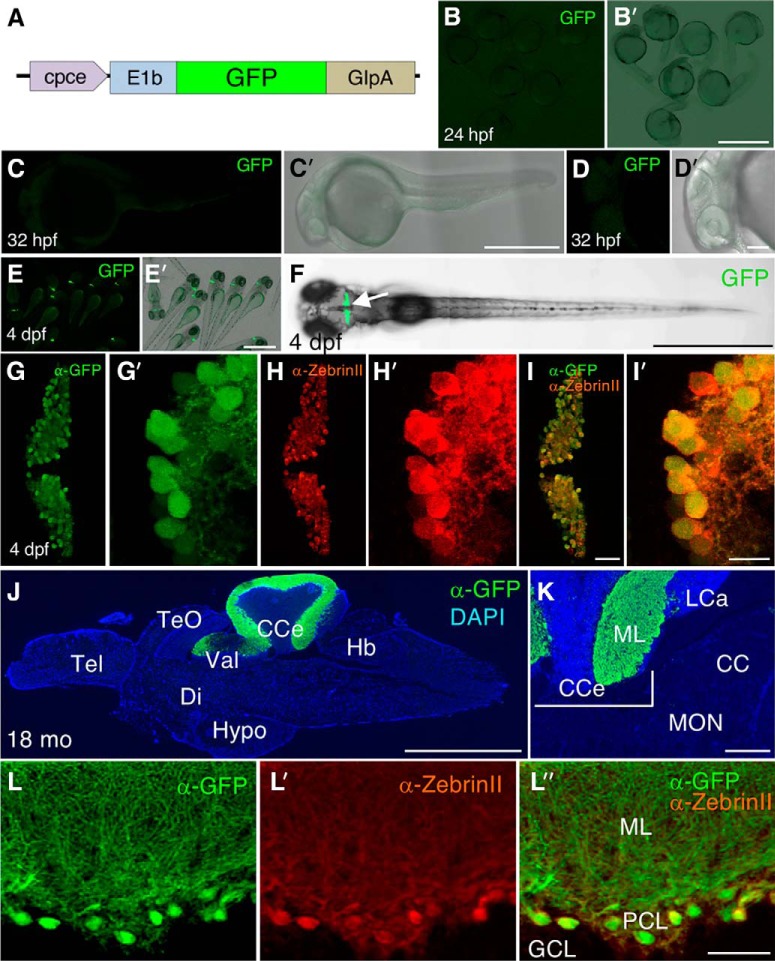
Characterization of GFP expression pattern of Tg(cpce-E1b:GFP)bz13. A, Schematic drawing of construct carrying the cpce-E1b promoter and driving GFP expression used to establish the Tg(cpce-E1b:GFP)bz13 transgenic strain. GFP reporter expression is not detectable in embryos at 24 (B, B′) and 32 hpf (C, C′, D, D′). In contrast to Tg(-7.5ca8:GFP)bz12 larvae that show ectopic expression (Fig. 3), GFP expression is restricted to the cerebellum (F, arrow) in 4 dpf larval Tg(cpce-E1b:GFP)bz13 zebrafish (E, E′, F). Co-immunostaining with anti-GFP (G, G′, green) and anti-ZebrinII (H, H′, red) antibodies reveals PC-specific expression of GFP in PCs at 4 dpf in merged images (I, I′). J, Sagittal section of an adult 18-month-old brain from Tg(cpce-E1b:GFP)bz13 fish showing immunostaining of GFP (green) in the cerebellum and DAPI counterstaining (blue). K, Higher-magnification of the MON reveals lack of GFP expression in this cerebellar-like structure compared with the PCL in the adjacent CCe. L, L′, L″, GFP-positive (green) cells are detected as ZebrinII-positive PCs (red). Scale bars: B′, E′, F, J, 1 mm; C′, 500 μm; K, 200 μm; D′, 100 μm; I, L″, 50 μm; I′, 10 μm. CC, Crista cerebellaris; CCe, corpus cerebelli; Di, diencephalon; GCL, granule cell layer; Hb, hindbrain; Hypo, hypothalamus; LCa, lobus caudalis cerebelli; ML, molecular layer; PCL, PC layer; Tel, telencephalon; TeO, optic tectum; Val, lateral division of the valvula cerebelli.
Figure 7.
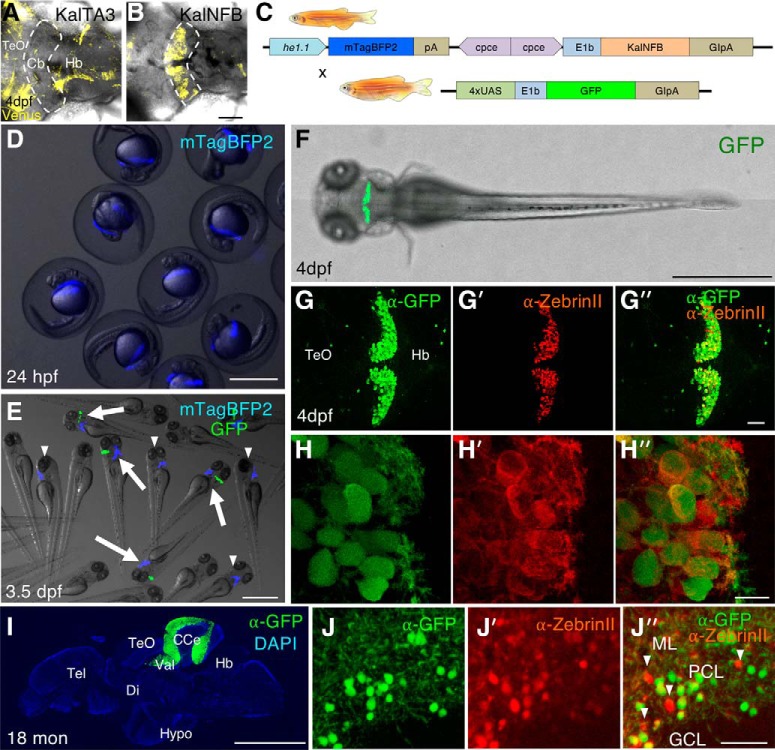
Characterization of 2xcpce-E1b:Gal4 driver line. A, B, An expression construct with KalTA3 (equivalent to GalFF) or KalNFB under control of the cpce-E1b promoter was injected into heterozygous Tg(5xUAS:FMAVenus-2A-3xNLS-HA-mseCFP)bz8 embryos harboring a Gal4-dependent 5xUAS:membrane tagged Fyn-Venus reporter transgene in their genome. Images show a dorsal view of the cerebellar region (enclosed by the white dashed line) of 4 dpf zebrafish injected with each construct at the one cell stage. KalTA3 induces Venus expression only marginally in the cerebellum (A); in contrast, KalNFB strongly activates Fyn-Venus expression in a cerebellum-specific manner (B). C, Top row, Schematic representation of transgenes of the cpce-E1b:Gal4 driver line Tg(2xcpce-E1b:KALNFB,he1.1:mTagBFP2)bz14 carrying two expression cassettes composed of KalNFB under the control of 2xcpce, and mTagBFP2 regulated by the hatching enzyme 1.1 promoter (he1.1). C, Bottom row, Scheme of transgene of UAS effector line Tg(4xUAS:GFP)hzm3. Mating the 2xcpce-E1b:Gal4 driver line with Tg(4xUAS:GFP)hzm3 generates embryos displaying blue fluorescent mTagBFP2 expression in the hatching gland at 24 hpf (D). E, At 3.5 dpf, larvae carrying both driver and effector transgenes display in addition GFP expression in the cerebellum (white arrows), whereas Tg(2xcpce-E1b:KALNFB, he1.1:mTagBFP2)bz14 larvae only reveal mTagBFP2 expression in their hatching glands allowing for easy identification of Gal4 driver carriers (white arrowheads). Four days postfertilization larva heterozygous for both KalNFB and UAS:GFP transgenes displaying GFP expression in cerebellar PCs (F), as demonstrated by co-immunostaining of GFP (G, H, green) and ZebrinII (G′, H′). Merged images reveal coexpression (G″, H″). A few scattered GFP-positive cells are also detectable in the optic tectum (TeO) and hindbrain (Hb) regions (G, G″, green). I, Immunostaining of sagittal brain section from an 18-months-old heterozygous cpce-E1b:KalNFB/4xUAS:GFP fish depicting GFP expression (green) in the cerebellum counterstained with DAPI (blue). J, J′, J″, Most GFP-positive cells (J, J″, green) also express ZebrinII (J′, J″, red), with a few exceptions (J″, white arrowheads). Scale bars: B, D, E, F, I, 1 mm; G″, J″, 50 μm; H″, 10 μm. Cb, Cerebellum; CCe, corpus cerebelli; Di, diencephalon; GCL, granule cell layer; Hypo, hypothalamus; ML, molecular layer; PCL, PC layer; Tel, telencephalon; Val, lateral division of the valvula cerebelli.
In situ hybridization.
Whole-mount in situ hybridization on embryonic and larval zebrafish was performed according to the protocols as described previously (Lauter et al., 2011a,b; Matsui et al., 2014). The plasmids carrying the following cDNAs were used to prepare in vitro transcribed complementary RNA (cRNA) probes incorporated with Digoxigenin (DIG)-UTP, or fluorescein (FLUO)-UTP (Roche): ca8: (1-661 of NM_001017571), krox20: (1–1901 of X70322; a gift from Matthias Hammerschmidt, University of Cologne, Germany), pitx3 (39–1773 of XM_021480551; a gift from Wolfgang Driever, University of Freiburg), pvalb7 (140–965 of NM_205574), kcnc3a (for detecting all splice variants, 1–1009 of NM_001195240.1), kcnc3b (999–1944 of NM_001195241.1), kcnc3a-X5 (3060–4183 of XM_021470855), kcnc3a-X6,7,8,9 (3061–3374 of XM_009299545), kcnc3a-X11 (3058–4129 of XM_021470866), and kcnc3a-X12 (3058–4093 of XM_009299549).
For a single transcript detection, hybridized probes labeled with DIG-UTP were visualized by a chromogenic reaction using NBT and BCIP (Roche), or detected by fluorescent staining using fast red (Roche; see Fig. 2R) as an alkaline phosphatase (AP) substrate. The detection of two transcripts by two-color fluorescence in situ hybridization was performed according to protocols using tyramide signal amplification as described previously (Lauter et al., 2011a,b). Alternatively, hybridized DIG-labeled probes were brought to reaction with anti-DIG-AP, followed by the visualization with fast red (see Fig. 2C). In situ hybridization analysis on adult (18 months old) female brain sections was performed according to the protocol optimized for mouse/rat brain sections (Matsumoto et al., 2012), except for skipping a de-lipidation step before pre-hybridization.
Figure 2.
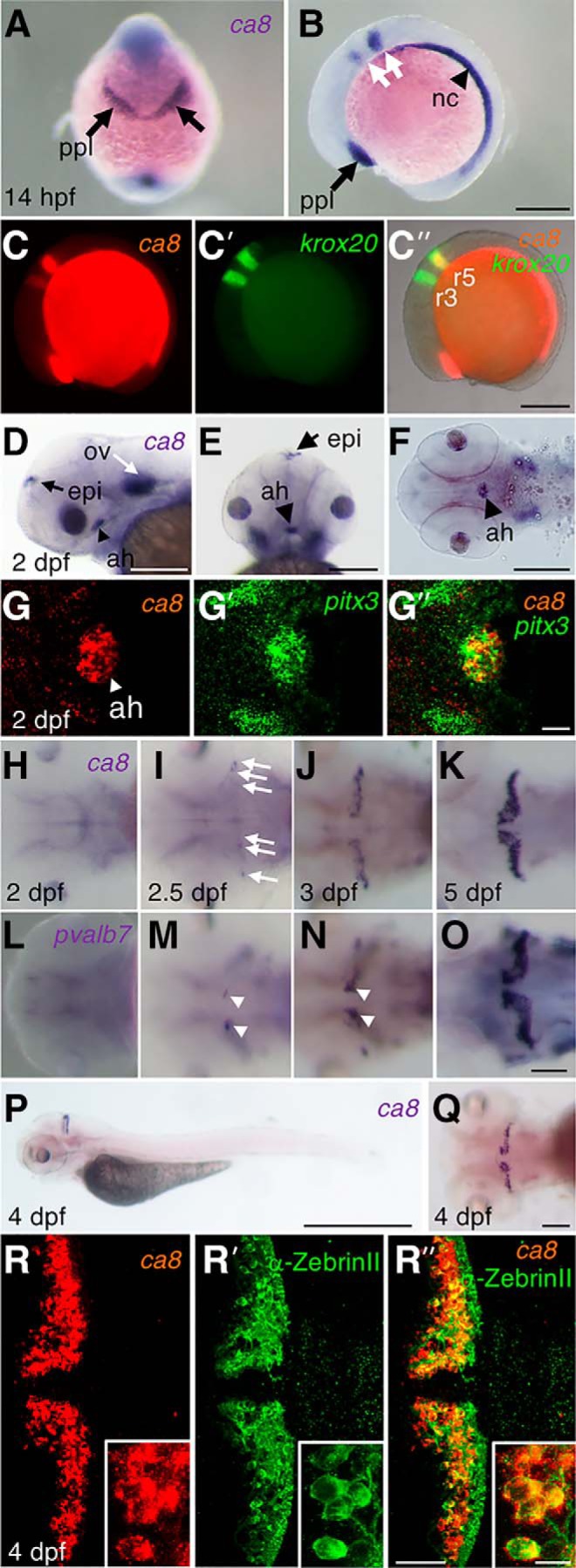
ca8 mRNA expression on embryonic and larval zebrafish. A, B, In situ hybridization analysis of ca8 in 14 hpf embryos (A, frontal view, dorsal upward; B, lateral view, anterior is to the left) reveals expression in the prechordal plate (ppl) (black arrows) and the notochord (nc; black arrowhead) as well as within the hindbrain as two stripes (white arrows). C, C′, C″, Two-color fluorescent in situ hybridization (FISH) against ca8 and krox-20 reveals coexpression (C″, merged images) of ca8 (C, red) and krox20 (C′, green) in rhombomeres 3 and 5 (labeled r3 and r5). D–F, At 2 dpf, the expression of ca8 is detected in the otic vesicles (D, ov, white arrow), epiphysis (D, E, epi, black arrow), and the adenohypophysis (D–F, ah, black arrowhead; D, lateral view; E, frontal view, dorsal upward; F, ventral view). G, G′, G″, FISH of ca8 (G, red) and pitx3 (G′, green), a marker gene for the adenohypophysis (ah, arrowhead), shows colocalization of ca8 and pitx3 expression (G″, merged images), (right, merged; anterior is to the left). H–O, In situ hybridization analysis with ca8 (H–K), or pvalb7 (L–O) probe on 2 (H, L), 2.5 (I, M), 3 (J, N), and 5 dpf larvae (K, O), respectively. In 2.5 dpf larvae, ca8 expression is faintly scattered (white arrows) in the cerebellar plate, and then expands and increases gradually from 3 to 5dpf, whereas pvalb7 (white arrowheads) is initially confined close to the cerebellar dorsal midline of larvae at 2.5 and 3 dpf. P, Q, Whole-mount in situ hybridization showing ca8 expression in lateral (P) and dorsal (Q) views of 4 dpf larval zebrafish. R, R′, R″, Fluorescent in situ hybridization together with immunohistochemistry in 4 dpf larvae depicts ca8 expression (R, red), anti-ZebrinII antibody staining (R′, green), and coexpression (R″). R, R′, R″, Insets depict colocalization of ca8 and ZebrinII in PCs at higher-magnification. Scale bars: P, 1 mm; B, C″, D–F, 200 μm; O, Q, R″, 50 μm; G″, 20 μm; R″ (inset), 10 μm.
Immunohistochemistry.
Whole-mount immunohistochemistry on larval zebrafish and fluorescent immunohistochemical detection on brain sections of adult (18 months old) female zebrafish were performed according to the protocol described in a previous report (Kani et al., 2010). Immunodetection after in situ hybridization on adult (18 months old) female brain sections was performed using 3,3′ diaminobenzidine (DAB)-chromogen (Sigma-Aldrich) to visualize signals as brown precipitates (Fig. 1C) according to the procedure as previously described (Matsui et al., 2014). For the immunostaining of mouse organotypic cerebellar slices 6 d after transfection [8 d in vitro (DIV)], the slices were treated as described previously (Zagrebelsky et al., 1998).
Figure 1.
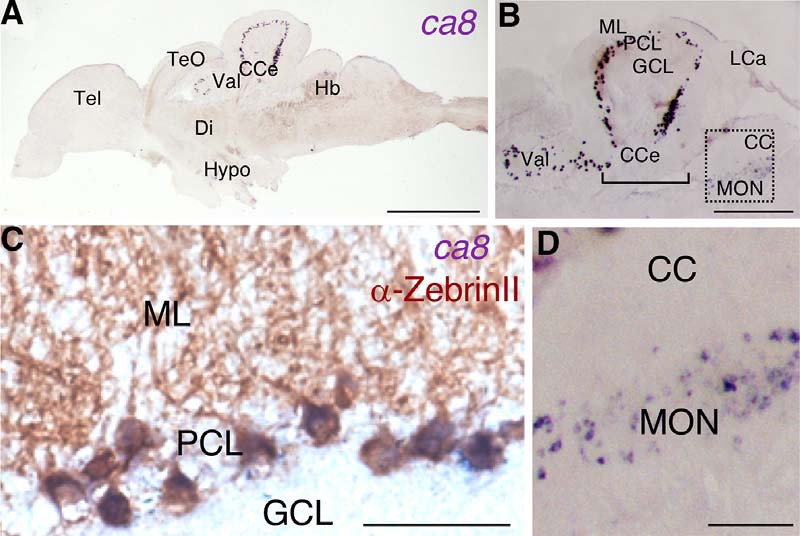
ca8 mRNA expression in adult zebrafish brain. In situ hybridization of ca8 mRNA using a DIG-labeled antisense probe. A, Overview of ca8 mRNA expression (purple) in a sagittal brain section of an 18-month-old zebrafish. B, Magnified view depicting intense expression of ca8 mRNA in the PC layer (PCL) of the lateral division of the valvula cerebelli (Val) and the corpus cerebelli (CCe), whereas weak expression is observed in the MON. C, Costaining of ZebrinII antigen (brown) and ca8 mRNA (purple) in the PCL. D, Higher-magnification of the boxed region in B showing ca8 mRNA-positive crest cells in the MON. Scale bars: A, 1 mm; B, 400 μm; D, 100 μm; C, 50 μm. CC, Crista cerebellaris; Di, diencephalon; GCL, granule cell layer; Hb, hindbrain; Hypo, hypothalamus; LCa, lobus caudalis cerebelli; ML, molecular layer; Tel, telencephalon; TeO, optic tectum.
Antibodies.
The following primary and secondary antibodies were used: polyclonal chicken anti-GFP IgY (1:1000; Aves Labs), monoclonal mouse anti-ZebrinII (1:500; a gift from Richard Hawkes, University of Calgary, Canada), donkey anti-chicken IgY-FITC (1:1000; Jackson ImmunoResearch/Dianova), mouse anti-calbindin antibodies (monoclonal, 1:5000; Swant), goat anti-mouse IgG AlexaFluor-488 (1:1000) and goat anti-mouse IgG AlexaFluor-546 (1:1000; Invitrogen/Life Technologies), goat anti-mouse IgG Cy5 (1:400; Dianova), biotinylated anti-mouse secondary antibody (1:500; Vectastain ABC HRP Kit, Vector Laboratories/Biozol), sheep anti-DIG-AP antibody (1:5000; Roche), sheep anti-DIG-POD antibody (1:300; Roche), and sheep anti-FLUO-POD antibody (1:300; Roche).
Isolation of a genomic ca8 regulatory element.
The 7543 bp upstream genomic region extending from the translational start site of the ca8 gene (−7543 to −1, named as −7.5ca8) was subcloned from BAC DKEY-59O6 (RZPD-German Science Centre for Genome Research, Berlin, Germany), which encompasses an ∼180 kb zebrafish genomic fragment, including rab2a and ca8 genes by using a prophage-recombination system as previously described (Lee et al., 2001).
Constructs for a reporter gene assay and generation of PC-specific GFP reporter line.
The isolated −7.5ca8 fragment was inserted in front of the GFP-globin intron-SV40pA (pA) transgene flanked by Tol2 transposase recognition sites whose backbone was pBluescriptII-SK (−7.5ca8:GFP). Stepwise deletions of −7.5ca8 were performed by digesting −7.5ca8:GFP with each restriction enzyme shown in Figure 4A to generate each ca8 promoter deletion construct whose green fluorescent protein (GFP) expression is under the control of −4.1ca8, −2.24ca8, −1.98ca8, or −0.8ca8, respectively. Finally, PCR amplified cpce (ca8 promoter-derived PC-specific enhancer element) with or without CMV minimal (CMVmini; Bajoghli et al., 2004) or E1b promoter (E1b; Köster and Fraser, 2001b) was cloned into the GFP reporter vector in orientations as illustrated in Figures 4B and 5A. The nucleotide sequence of cpce is as follows: 5′-GCTAATTCAGATTTTTTAATATGTCGATTAAAGATGC TTAGGTCATGTGACAGCTCTCGTTATATCCTTGAGCTTTGGGGA TTAAATCAACCTACTTTTACTTAATTAGATAGTGATTTGTCTTT TGTGTAATGTGCTATCTTTAATTTTAGACAATCAGGGTCAAATT AGTTTCTTAGTATAGCATATATGGCAATACGCAATAGAGTTCAG TGCCTTTGGTTGTTACTTTTTTTTTATCTAGGACAGTCAGTGCAT-3′. The constructs inducing GFP under the control of cpce linked to CMVmini, or E1b, which were used for transgenesis experiments, are listed as no. 2045 or 2414, respectively, in the laboratory's internal database.
Figure 4.
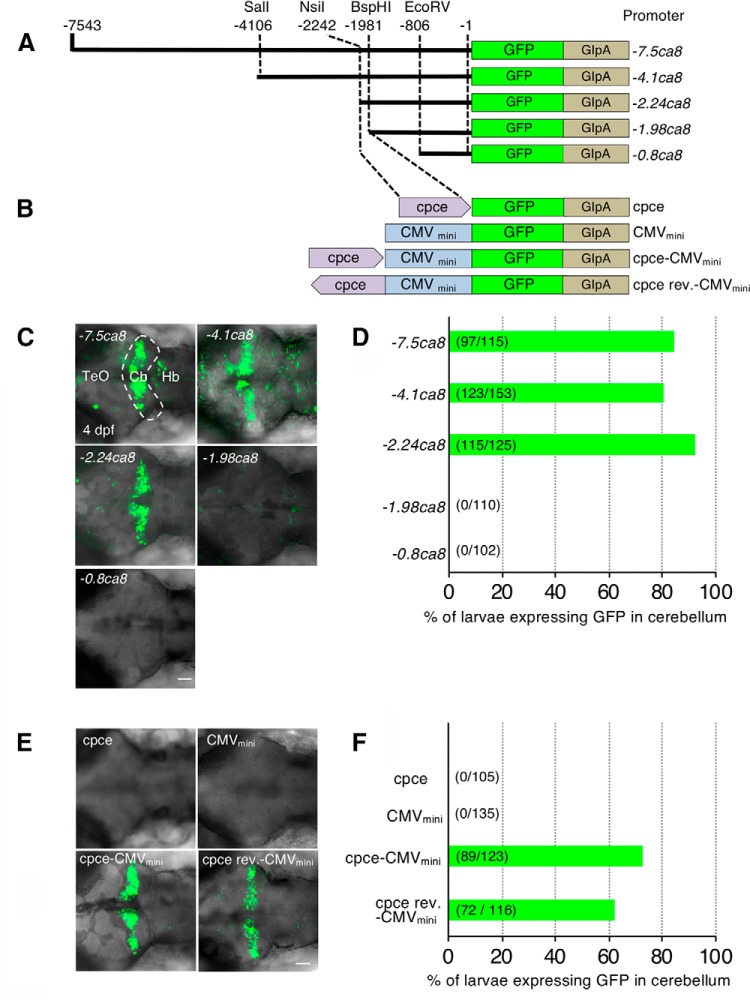
In vivo reporter gene assay to identify cpce. A, B, Schematic representations of GFP reporter constructs carrying deleted ca8 5′-upstream regions [7543 (−7.5ca8), 4106 (−4.1ca8), 2242 (−2.24ca8), 1981(−1.98ca8), and 806 bp (-0.8ca8) fragments, respectively; A], and cpce or cpce cloned in reverse orientation (cpce rev) with or without CMVmini promoter (B). The translational initiation start site is designated as +1. C, E, Representative images showing a dorsal view of the GFP expressing cerebellar region (C, white dashed line) of 4 dpf zebrafish injected with each construct shown in A and B, respectively. D, F, Bars in each diagram indicate the percentage of larvae displaying GFP expression, driven by constructs shown in A and B, respectively, in the cerebellum of larval zebrafish at 4 dpf (total numbers in parentheses). Three independent injection experiments were performed for each group. Cb, Cerebellum; Hb; hindbrain; TeO, optic tectum. Scale bars: C, E, 50 μm.
Generation of transient and stable transgenic fish.
Plasmid DNA was injected into one cell stage zebrafish brass embryos together with Tol2 or Tol1 transposase mRNA (25 ng/μl DNA together with 25 ng/μl mRNA in 1.5 nl of injection mix; Kawakami, 2007; Koga et al., 2008). Fluorescent larvae were selected and raised to adulthood. To identify transgenic F0 carriers, injected fish were crossed with brass fish, and larvae with tissue-specific fluorescent reporter expression were selected and raised to adulthood (F1 generation) and maintained by raising further generations (F2–F8).
Preparation of cerebellar organotypic cultures and particle-mediated gene transfer.
Cerebellar organotypic cultures (thickness of 400 μm) were prepared from postnatal day (P)10 C57BL/ 6 mice of either sex as previously described (Stoppini et al., 1991; Zagrebelsky et al., 1998). Individual neurons were biolistically transfected after 2 DIV using the Helios Gene Gun System (Bio-Rad) with the cpce-CMVmini:GFP and CMV enhancer/promoter (CMV):mCherry constructs. Gold beads (1 μm) coated with expression plasmids were shot with a pressure of 80–100 psi, and a filter with a pore size of 3 μm was used to prevent clusters from reaching the slices.
Construct for generation of a stable transgenic PC-specific Gal4 driver strain.
For transient transgenic assays to examine the activity of Gal4 constructs (see Fig. 7A,B), a fragment encoding the Gal4-DNA binding domain with translation optimization for zebrafish expression (Kal; Distel et al., 2009) was fused to either a two tandem repeat minimal transactivation domain (PADALDDFDLDML PADALDDFDLDMLPGN) from herpes simplex virus type 1 (HSV-1) VP16 (Asakawa and Kawakami, 2008; Distel et al., 2009) or a human NFκB p65 transactivation domain (generous gift from Darren Gilmour, EMBL; (Emelyanov and Parinov, 2008), generating KalTA3 or KalNFB, respectively (no. 2455 or 2777, respectively).
Each Gal4-transcriptional activator cDNA was flanked by cpce-E1b and pA and subsequently cloned into pDon [Tol1] (generous gift from Akihiko Koga, Kyoto University, Japan; Koga et al., 2008) carrying recognition sites of the Tol1 transposase. For generating a stable transgenic strain expressing PC-specific KalNFB, we used a promoter composed of two tandem cpce fused to E1b (2xcpce-E1b; Matsui et al., 2014). mTagBFP cDNA was converted to mTagBFP2 (I174A substitution by site directed mutagenesis; Subach et al., 2011). The final expression construct (see Fig. 7C) for generating a stable transgenic line carries mTagBFP2-pA under the control of the hatching enzyme 1, tandem duplicate 1 promoter (he1.1) (a gift from Jeffrey Mumm, Johns Hopkins Medicine; Xie et al., 2012) for visualization of transgenic carriers (see Fig. 7D,E). These two expression cassettes were flanked by Tol2 transposon recognition arms in the pBluescript II-SK backbone (no. 2973).
Generation of PC-specific bidirectional expression vectors.
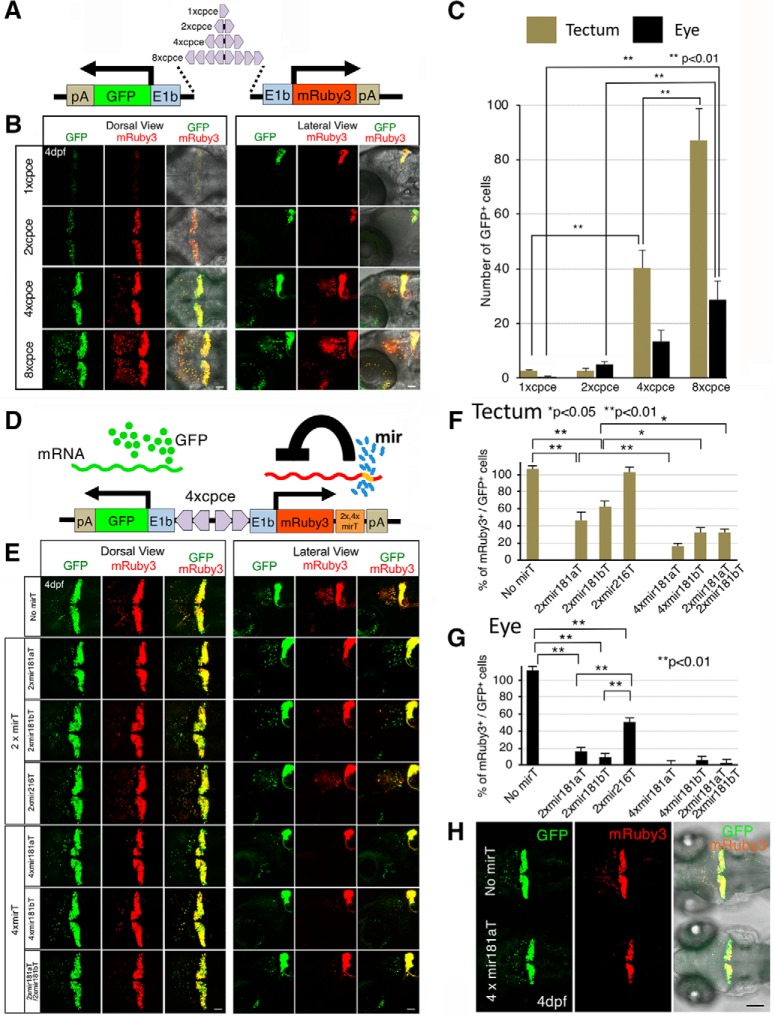
The cpce sequence was isolated by PCR from −7.5ca8 with the following primer pair: 5′-CCGCGGCCGCGGATCCGCTAATTCAGATTTTTTAATATGTCG-3′ (NotI-BamHI sites underlined). 5′-CCGAATTCAGATCTATGCACTGACTGTCCTAG-3′ (EcoRI-BglII sites underlined). The amplified product was digested with NotI/EcoRI, and cloned into pBluescript II-SK (pB-cpce). To create 2× tandem cpce, cpce was released from pB-cpce by BglII/BamHI digestion and inserted into the BamHI site of pB-cpce (pB-2xcpce). Further generation of 4× tandem cpce was performed by a similar strategy and 2× cpce was cloned into pB-2xcpce (pB-4xcpce). Each 1×, 2×, or 4× cpce was fused to the E1b minimal promoter sequence, followed by placing GFP-pA or mRuby3-pA behind each promoter. mRuby3 (Addgene, plasmid 74252) was a gift from Michael Lin, Stanford University (Bajar et al., 2016). Because joining 5′ and 3′ end of cpce in a tail to tail direction proved difficult, an ∼110 bp spacer sequence devoid of DNA methylation was PCR-amplified from a CpG free vector backbone (pCpGL-basic: a gift from Michael Rehli, University of Regensburg; Klug and Rehli, 2006) and inserted between each cpce. This allowed us to create pA-GFP-E1b-1/2/4xcpce-spacer-1/2/4xcpce-E1b-mRuby3-pA. These fragments were cloned into the KpnI/NotI sites of pTol2-MCS (no. 3822) carrying an MCS derived from pBluescript II-SK with T3 and T7 promoters, flanked by minimal Tol2 transposase recognition sites in a pUC19 backbone, resulting in the generation of p-GFP:2/4/8xcpce:Ruby. Finally, 2xcpce in p-GFP:2xcpce:Ruby was replaced by 1xcpce (p-GFP:1xcpce:Ruby; see Fig. 9A). To find a miRNA target sequence (mirT) to specifically reduce ectopic expression in cells in the optic tectum and the eyes, a miRNA expression image library (http://science.sciencemag.org/content/suppl/2005/07/05/1114519.DC1; Wienholds et al., 2005) was searched for miRNAs predominantly expressed in tectal and retinal cells at larval stages.
Figure 9.
PC-selective bidirectional expression system. A, Schematic view of the bidirectional PC-specific expression system carrying 1×, 2×, 4×, or 8× cpce flanked by two E1b promoters to drive both GFP and mRuby3 reporter genes. The 2×, 4×, or 8× tandem cpce was generated by flanking a spacer sequence on each side with 1×, 2×, 4× cpce sequences in a mirror-symmetric orientation. B, Four days postfertilization larvae displayed from dorsal (left) or lateral side (right) injected with 1×, 2×, 4×, or 8× cpce constructs. The expression levels of both GFP and mRuby3 gradually increase with a higher number of cpce. Ectopic expression in the optic tectum and eyes becomes first visible in larvae injected with 4xcpce-construct, which is further increased with 8xcpce-construct. Images were recorded by confocal microscopy with identical illumination and detection settings and the same z-stack thickness (see Materials and Methods). C, Bar diagram showing the number of GFP-positive cells ectopically expressing GFP in the optic tectum (khaki bars) and the eyes (black bars). D, Schematic drawing of the bidirectional expression system containing mirT-sites to reduce the ectopic expression of mRuby3. mirT-sites are inserted between the coding sequence of mRuby3 and the polyA-sequence. E, Embryos were injected with the PC-selective bidirectional expression construct with or without different mirTs between the mRuby3 coding sequence and the polyA-sequence. Four days postfertilization larvae are depicted from a dorsal (left) or lateral (right) perspective of the cerebellar region to visualize ectopic expression of GFP and mRuby3 in the optic tectum and the eye, but also PC-specific expression in the cerebellum. Note that 4xmir181aT almost completely eliminates ectopic mRuby3 expression, but not GFP expression outside of the cerebellum. The confocal images were recorded with the same excitation and detection settings as well as, the thickness of the z-stacks was maintained (see Materials and Methods). F, G, Quantification of the silencing effect of different mirTs revealed by the percentage of mRuby3- relative to GFP-positive cells in the optic tectum (F) and eye (G) in 4dpf larvae. H, Comparison of ectopically expressed GFP to mRuby3 driven by the plasmids carrying 4xcpce without (top) or with 4xmir181aT (bottom larva). Whereas ectopic GFP expression is observed in the optic tectum in addition to expression in cerebellum with or without 4xmir181aT (left, green), ectopic mRuby3 expression in the optic tectum is significantly repressed in the presence of 4xmir181aT without changing the cerebellar expression (middle, red). Merged images (left) reveal GFP-only expressing cells outside of the cerebellum (bottom larva). N values of zebrafish in each group are 6–10 for C, G, and H. p values were determined by one-way ANOVA with post hoc Tukey HSD test. C, 1xcpce versus 4xcpce (tectum), p = 0.0025945; 4xcpce versus 8xcpce (tectum), p = 0.0010053; 2xcpce versus 8xcpce (eye), p = 0.0010053; 2xcpce versus 8xcpce (eye), p = 0.0010053. G, No mirT versus 2xmir181aT, p = 0.0010053; No mirT versus 2xmir181bT, p = 0.0010053; 2xmir181aT versus 4xmir181aT, p = 0.0078459; 2xmir181bT versus 4xmir181bT, p = 0.0126719; 2xmir181bT versus 2xmir181aT/ 2xmir181aT, p = 0.0244631. H, No mirT versus 2xmir181aT, p = 0.0010053; No mirT versus 2xmir181bT, p = 0.0010053; No mirT versus 2x216T, p = 0.0010053; 2xmir181aT versus 2xmir216T, p = 0.0010053; 2xmir181bT versus 2xmir216T, p = 0.0010053. Scale bars: B, E, H, 50 μm.
To examine the effect of each mirT on specific silencing in tectal and retinal cells, bidirectional expression vectors were generated by inserting each mirT fragment between the mRuby3 open reading frame and the following polyA-sequence. The following oligos each creating 2×/4× mirT (each mirT sequence is underlined) flanked by XhoI/NheI sites were annealed, and ligated into XhoI/XbaI sites of p-GFP:4xcpce:Ruby.
mir2x181aT: 5′-CTCGAGGACTAGCTCTAGAACTCACCGACAGCGTTGAATGTTGCTAGAACTCACCGACAGCGTTGAATGTTGCTAGC-3′
mir2x181bT: 5′-CTCGAGGACTAGCTCTAGAAACCCACCGACAGCAATGAATGTTGCTAGAAACCCACCGACAGCAATGAATGTTGCTAGC-3′
mir2x216T: 5′-CTCGAGGACTAGCTCTAGACACAGTTGCCAGCTGAGATTAGCTAGACACAGTTGCCAGCTGAGATTAGCTAGC-3′
mir4x181aT: 5′-CTCGAGGACTAGCTCTAGAACTCACCGACAGCGTTGAATGTTGCTAGAACTCACCGACAGCGTTGAATGTTGCTAGAACTCACCGACAGCGTTGAATGTTGCTAGAACTCACCGACAGCGTTGAATGTTGCTAGC-3′
mir4x181bT: 5′-CTCGAGGACTAGCTCTAGAAACCCACCGACAGCAATGAATGTTGCTAGAAACCCACCGACAGCAATGAATGTTGCTAGAAACCCACCGACAGCAATGAATGTTGCTAGAAACCCACCGACAGCAATGAATGTTGCTAGC-3′
mir2x181aT/mir2x181bT: 5′-CTCGAGCCTCTAGAAACCCACCGACAGCAATGAATGTTGCTAGAAACCCACCGACAGCAATGAATGTTGCTAGAACTCACCGACAGCGTTGAATGTTGCTAGAACTCACCGACAGCGTTGAATGTTGCTAGC-3′
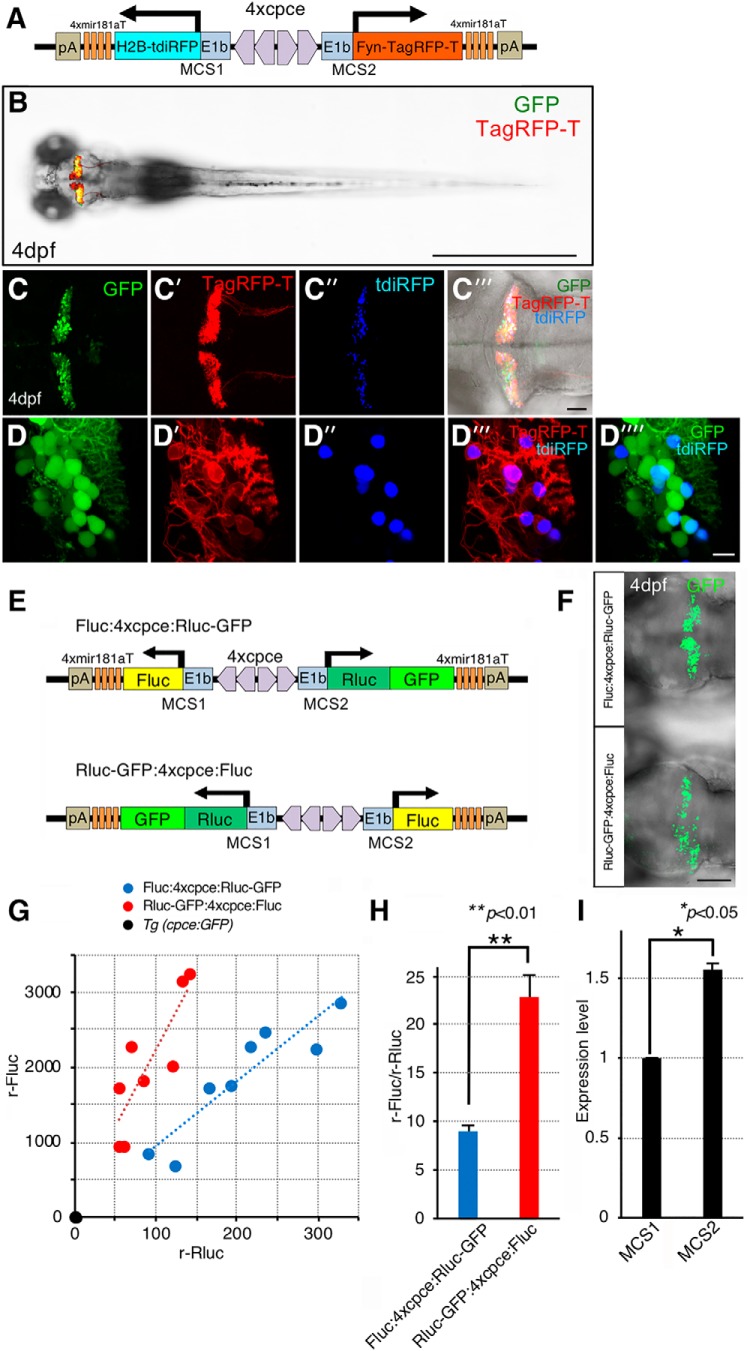
Next, we generated a PC-selective expression cassette carrying 4xcpce by replacing reporter genes with two multiple cloning sites (MCS1 and MCS2) linked to 4× mir181aT that were flanked by Tol2 minimal arms in pTol2-MCS (no. 3822). The resulting construct was designated pTol2-4xcpce-MCS (no. 4794) which will be useful for inserting any two expression units-of-interest. To achieve PC selective subcellular labeling for the nucleus and the plasma membrane, H2B fused to tandem iRFP (tdiRFP), a gift from Yusuke Miyanari (Okazaki Institute for Integrative Bioscience, Japan; Miyanari et al., 2013), and Fyn-TagRFP-T (Matsui et al., 2014) were cloned into MCS1 and MCS2 of the pTol2-4xcpce-MCS, respectively (see Fig. 10A; no. 5162). This expression cassette was also used for generating PC-specific bidirectional reporter constructs expressing dual luciferases [firefly luciferase2 (Fluc) and Renilla luciferase fused to GFP (Rluc-GFP), Promega; see Fig. 10E; Fluc:4xcpce:Rluc-GFP and Fluc:4xcpce:Rluc-GFP; no. 5533 and 5534, respectively]. A PC selective coexpression cassette carrying 1× or 2× cpce without mir181aT insertions (pTol2-1xcpce-MCS, or pTol2-2xcpce-MCS) is also available upon request (no. 5194 or 5163, respectively).
Figure 10.
pTol2-4xcpce allows for dual subcellular reporter incorporation with slightly different expression levels in each direction. A, Schematic drawing of the construct containing two subcellular markers, membrane targeted Fyn-TagRFP-T and nuclear localized H2B-tdiRFP, which are inserted into each multiple cloning site (MCS1, or MCS2) of pTol2-cpce-MCS, respectively. This vector contains insertions of 4× mir181aT fragments immediately upstream to both pA-sequences to restrict expression of transgenes in PCs. B, Dorsal view of 4 dpf larva expressing membrane targeted Fyn-TagRFP-T (red) and GFP (green) in the cerebellum. Tg(-7.5ca8:GFP)bz12 embryos were injected with the plasmid shown in A. C–C⁗, Cerebellar overview of injected Tg(-7.5ca8:GFP)bz12 larva expressing GFP (C, green) together with Fyn-TagRFP-T (C′, red) and tdiRFP (C″, blue). C‴, A merged image of C, C″, C‴ under the bright field. D–D⁗, Higher-magnified images showing coexpression of membrane tagged Fyn-TagRFP-T (D′, D‴, red) and nuclear localized H2B-tdiRFP (blue, D″, D‴, D⁗) in PCs marked by GFP-mediated green fluorescence (D, D⁗, green). The two subcellular markers (D‴, red and blue) are coexpressed in a mosaic manner by the injected plasmid in GFP-positive PCs (D⁗). E, Schematic representations of the bidirectional 4xcpce-dual luciferase (luc) constructs expressing firefly luc (Fluc) and Renilla luc fused to GFP (Rluc-GFP), which are cloned into MCS1 and MCS2, respectively, and vice versa (Fluc:4xcpce: Rluc-GFP and Rluc-GFP:4xcpce:Fluc). Larval fish injected with either construct, showing GFP expression throughout the entire PC plate (F), were sorted for luciferase assay. G, Scatter plot depicting the values of relative Fluc (y-axis), and Rluc activity (x-axis; r-Fluc and r-Rluc, respectively) measured from a head lysate of a 4 dpf Tg(cpce-E1b:GFP) (black circle), or injected larva with Fluc:4xcpce:Rluc-GFP (blue circle) Rluc-GFP:4xcpce:Fluc (red circle), respectively, with a linear trend line (a blue or red dotted line). r-Fluc and r-Rluc for each lysate was obtained as the ratio of an obtained luciferase activity by a mean background activity in lysate from a head of a brass larva. H, The mean ratio of r-Fluc activity by r-Rluc (r-Fluc/r-Rluc) in an individual head lysate of a larva injected with each construct. N values of zebrafish in each group in G and H are 8. I, The mean value indicating expression level of a reporter gene cloned into MCS1 (Fig. 9E, facing left), or MCS2 (Fig. 9E, facing right), was calculated by the square root of the mean ratio of (r-Fluc/r-Rluc) for each group (see Materials and Methods), which were obtained from two individual experiment. p values were determined by unpaired Student's t test. H, Fluc:4xcpce:Rluc-GFP versus Rluc-GFP:4xcpce:Fluc p = 0.0036; I, MCS1 versus MCS2 p = 0.0438. Scale bars: B, 1 mm; F, 100 μm; C‴, 50 μm; D⁗, 10 μm.
Luciferase assay.
After injection with either Fluc:4xcpce:Rluc-GFP, or Rluc-GFP:4xcpce:Fluc construct, 4 d postfertilization (dpf) zebrafish showing GFP fluorescence throughout the entire PC layer were collected, followed by preparation of a head lysate from each individual with 100 μl of 1xpassive lysis buffer (Promega). To prepare a control head lysate, a non-injected brass, or Tg(cpce-E1b:GFP)bz13 larva was processed in the same manner. Luciferase activities for Fluc and Rluc in 20 μl of each lysate were measured using Beetle Juice and Renilla Glow Juice kits (p.j.k.), respectively, according to the manufacturer's protocol. The relative luciferase activity for each luciferase (r-Fluc, or r-Rluc) in each lysate was determined as the ratio of a measured value by a mean value of background luciferase activity in brass head lysate (see Fig. 10G), followed by the calculation of the ratio of r-Fluc by r-Rluc: (r-FlucFR/r-RlucFR), or (r-FlucRF/r-RlucRF) obtained from each head lysate expressing Fluc:4xcpce:Rluc-GFP, or Rluc-GFP:4xcpce:Fluc, respectively (see Fig. 10H). To demonstrate the difference in the bidirectional vector induced expression level in each direction, we assumed that κ times higher level of reporter (cloned into MCS2) is expressed in the one direction than a second reporter (cloned into MCS1) expressed in the other direction. To calculate the factor κ, r-RlucFR and r-FlucRF, which are the resultants of reporters (cloned into MCS2) expressing in the one direction, were normalized by dividing these values by the factor κ to obtain a formula: [r-FlucFR/(r-RlucFR/κ)] = [(r-FlucRF/κ)/r-RlucRF], indicating that the ratio of r-Fluc by r-Rluc are equally calculated after the normalization by the factor κ independent of injected vectors. Thus, the factor κ could be obtained as the square root of (r-FlucRF/r-RlucRF)/(r-FlucFR/r-RlucFR). The mean values of (r-FlucRF/r-RlucRF) and (r-FlucFR/r-RlucFR) (see Fig. 10H) calculated from two independent assay were used for this calculation to obtain the factor κ as a value of difference in expression level of one reporter cloned into MCS1 (facing in left direction) compared with the other cloned into MCS2 (facing in right direction; see Fig. 10I) according to vector scheme shown in Figure 10E.
Zebrafish larvae dissociation and FACS to isolate single PCs.
Heads of GFP-expressing Tg (cpce-E1b:GFP)bz13 and brass zebrafish larvae at 4 dpf were placed under a sterile workbench directly after decapitation and processed as described previously (Sassen et al., 2017). Completed dissociation into single-cell suspension was followed by filtration through a sterile cell strainer (EasyStrainer 40 μm; Greiner Bio-One). Cells were pelleted by centrifugation (250 × g, 5 min) and resuspended in 3 ml of fluorescence activated cell sorting (FACS)-buffer (0.5% BSA, 5 mm EDTA in 1× PBS solution). The filtration step was repeated also directly before FACS. Subsequently cell sorting was performed using an SH-800 cell sorter (Sony) according to the manufacturer's protocol. To evaluate cell size and granularity, cells were pre-gated using forward scatter (FSC) and backward scatter (BSC), respectively. Since living cells are larger and less granular compared with dead cells, they can be separated by appropriate gating (see Fig. 11G,H). Single cells were subsequently selected by plotting the FSC signal height (FSC-H) against its width (FSC-W), thus discriminating against cell doublets with double or more of the signal width (see Fig. 11I,J). In a third gating step, round cells were selected by plotting BSC height (BSC-H) against its width (BSC-W), thus further discriminating against cell aggregates and cells containing a high granularity index (Fig. 11K,L) thereby enriching the PC population as described previously (Takeuchi et al., 2017). For single living cells, GFP fluorescence was excited by 488 nm laser illumination and detected by a 525 ± 25 nm optical bandpass filter (see Fig. 11M,N). PCs were sorted directly into PCR tubes containing 15 μl reverse transcription reaction mix (500 μm dNTP, 150 ng random hexamer primer, 2U RNase inhibitor; Qiagen), 40 U RNase inhibitor, 7 mm DTT, 0.1% Tween 20, 25 U Superscript IV, and 1× Superscript IV buffer (ThermoFisher Scientific, Life Technologies). Predefined “single cell” sorting mode was adopted.
Figure 11.
Expression of kcnc3a splice isoforms in PCs. A, Whole-mount in situ hybridization of kcnc3a (A, B) or kcnc3b (C, D) in 4 dpf larvae. Dorsal (A, C) and lateral views (B, D). mRNA for kcnc3a is strongly expressed in the cerebellum reminiscent of the PC layer (A, B), whereas expression of kcnc3b is much weaker and not well confined to any cerebellar cell population (C, D). Images showing two-color fluorescent in situ hybridization of kcnc3a (E, F, green), ca8 (E′, F′, red), in 4 dpf larvae. Merged images (E″, F″, green and red). Images recorded at higher-magnification (F, F′, F″) reveal mRNA coexpression of kcnc3a and ca8 in PCs. Green fluorescent GFP-expressing PCs were isolated by FACS from dissociated heads of 4 dpf Tg(cpce-E1b:GFP)bz13 larval zebrafish through four sequential gatings (H, J, L, N). The same gatings applied to samples from dissociated heads of 4 dpf larval zebrafish of the brass strain (G, I, K, M). This allowed for discrimination of GFP-negative from GFP-positive cells of Tg(cpce-E1b:GFP)bz13 samples. G, H, Alive cells gate. BSC-A (area) and FSC-A (area). I, J, Single cells gate. FSC-W and FSC-H. K, L, Round cells gate. BSC-W and BSC-H. M–O, GFP-positive cells gate. No GFP-positive cells were detected within the GFP-positive cells gate in the brass sample, but only in the Tg(cpce-E1b:GFP)bz13 sample. Plots in G–N show 100,000 cells each, while plot in O shows 1,256,387 cells to allow for better visualization of the small percentage of GFP-positive PCs. P, Schematic drawings outlining annealing sites of specific primer pairs (red arrowheads) to detect the expression of groups of splice variants which encode different C-terminal amino acid sequences terminating with -WIPK, -PSIL, -VTNV, or -VLSF, respectively. Also see Figure 11-1 and Figure 11-2 supporting Figure 11P. Q, The top gel image shows single-cell RT-PCR analysis subsequent to FACS isolation of single PCs for detecting kcnc3a splice variants. Subsequent cloning of amplified PCR products, followed by sequencing failed to obtain expression data for isoforms containing exon 5 (X1, 2, 3, 4, 10), whereas the other variants shown in P were found to be expressed in single PCs purified from 4dpf Tg(cpce-E1b:GFP)bz13 larvae. RT-PCR on a plasmid containing each isoform (Q, bottom image) confirms the specificity of the PCR. The detection of ca8 confirms that the isolated single cell was a PC. R–U, mRNA expression of different kcnc3a splice variants in 4 dpf zebrafish larvae detected by whole-mount in situ hybridization. The different probes recognize isoforms X6/7/8/9 (R), X5 (S), X11 (T), or X12 (U), respectively. Colocalization (V″, W″, merged images) of kcnc3a-X12 (V, W, green), and ca8 (V′, W′, red) in PCs revealed by fluorescent in situ hybridization. Scale bars: D, U, 100 μm; E″, V″, 50 μm; F″, W″, 10 μm. Cb, Cerebellum; Hb, hindbrain; TeO, optic tectum.
Reverse-transcription PCR analysis.
The reverse-transcription reaction mix containing FACS sorted single PC was subjected to cDNA synthesis. PCRs were performed at 40 cycles with the specific primer pairs spanning exon3 to varying exons downstream (exon 4, 6, 7, or 8) of zebrafish kcnc3a to amplify ∼300 bp fragments containing isoform specific C-terminal amino acid sequences. The primers used to amplify the different kcnc3a transcripts were as follows: exon 3 forward, 5′-TCGAAGCAGAATGGGGATGC-3′, exon 4 reverse, 5′-GTTTGATCCAGGAAGAGG-3′ (313 bp); exon 6 reverse, 5′-TTGTTGATGGGGGTGATGGG-3′ (296 bp); exon 7 reverse, 5′-TCGGGAATGTGTTTGTGAAGGA-3′ (316 bp); exon 8 reverse 5′-TCTCAAATCCTGCGGTGACTC-3′ (295 bp). In addition, primers spanning exon1 and exon2 of ca8 were used to confirm that the synthesized single-cell cDNA was PC-specific: ca8 forward, 5′-TGTGGTTGGTGTGCCTTTGA-3′, ca8 reverse, 5′-AGGGGACTGGTACTCTCCAT-3′ (301 bp). The reaction aliquots were separated by electrophoresis in 1.5% agarose gels, and visualized. Each amplified fragment was further cloned to confirm the identity of each fragment by sequencing. These cloned DNAs were also used in further PCRs as templates for control PCRs with 35 cycles to ensure that each primer pair could efficiently amplify its target product. Three different cDNAs prepared from three different, single-sorted PCs were used to confirm the reproducibility of the PCR data.
PC-specific expression constructs for neurodegenerative disease modeling.
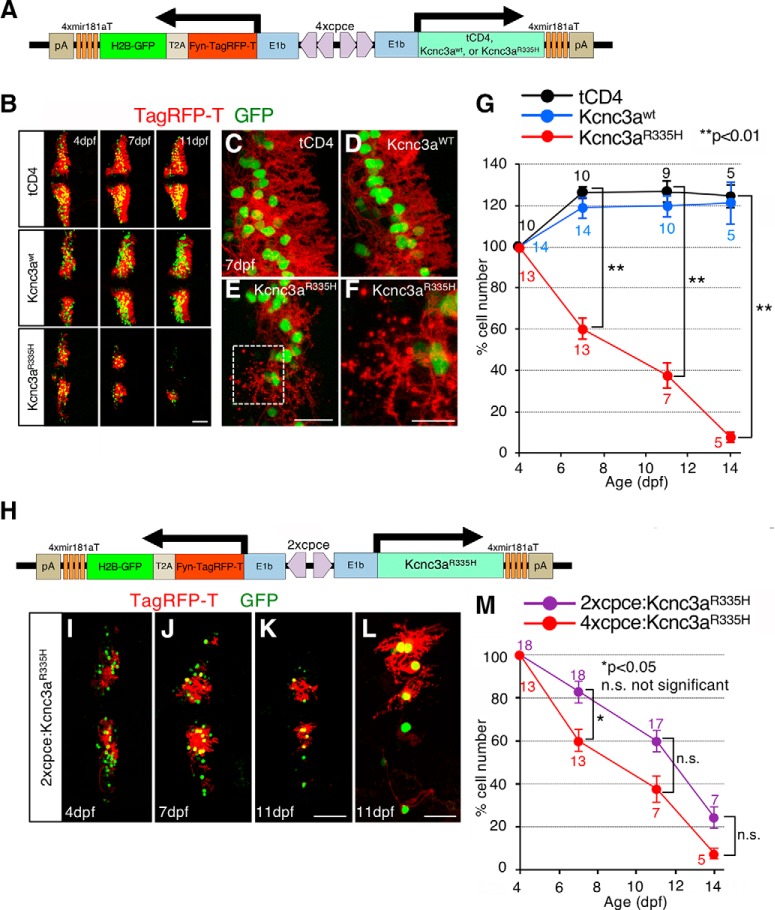
Zebrafish cDNA containing the coding sequence for kcnc3awt and kcnc3aR335H, and terminating with -WIKP (Mock et al., 2010), were gifts from Diane Papazian (University of California, Los Angeles). As we found that the isoform kcnc3a-X12 was the most abundantly expressed transcript in larval PCs (see Fig. 11R–W″) the C-terminal sequence was replaced by exon 8-derived sequences generating kcnc3awt-X12 and kcnc3aR335H-X12 (referred as kcnc3awt and kcnc3aR335H hereafter). The bicistronic expression unit coding for the two subcellular fluorescent reporters Fyn-TagRFP-T and H2B-GFP linked by a T2A self-cleaving peptide was inserted into MCS1 of pTol2-4xcpce-MCS. MCS2 was used to insert either tCD4 (a truncated form of human CD4 encoding a nonfunctional membrane surface protein; a gift from Olivier Gandrillon, Université de Lyon, France), kcnc3awt, or kcnc3aR335H. The resulting constructs drive expression of tCD4, kcnc3awt, or kcnc3aR335H together with both Fyn-TagRFP-T and H2B-GFP in a PC-specific manner (see Fig. 12A; no. 4815, 4813, and 4814, respectively). Furthermore, Fyn-TagRFP-T-T2A-H2B-GFP and kcnc3aR335H were cloned into MSC1 and MSC2 of pTol2-2xcpce-MCS, respectively, to generate the construct expressing these genes under control of 2xcpce (Fig. 12H; no. 5503).
Figure 12.
Cerebellar neurodegenerative disease modeling for human SCA13. A, Schematic drawing of a bidirectional PC-specific expression system using 4xpce to express tCD4, Kcnc3awt, or Kcnc3aR335H together with nuclear H2B-GFP and membrane targeted Fyn-TagRFP-T reporter genes linked to a self-cleaving T2A peptide. B–G, Time course of Kcnc3aR335H-induced PC degeneration. B, Larvae injected with a construct expressing tCD4, Kcnc3awt, or Kcnc3aR335H (upper, middle or lower rows, respectively) and monitored in vivo at 4, 7, and 11 dpf by laser scanning confocal microscopy (note images of consecutive time points have been recorded from the same larva). C, Cerebellar PCs expressing tCD4 (C) or Kcnc3awt (D) exhibit highly arborized dendrites in 7 dpf larva, whereas PCs expressing Kcnc3aR335H (E, F) display atrophic and degenerative changes with fragmented red labeled puncta from dendritic or axonal structures. A higher-magnification image of the region marked by the white dashed rectangle (E) is presented in F. G, Percentage of surviving PCs in each larva was calculated at each time point in relation to the numbers of PCs of the same larva at 4 dpf. H, Schematic representation of the 2xcepc:Kcnc3aR335H construct expressing Kcnc3aR335H together with subcellular reporters (Fyn-TagRFP-T, red and H2B-GFP, green). I–K, The cerebellar region of the same larva injected with the construct shown in H was monitored in vivo at 4, 7, and 11 dpf, respectively, by laser scanning confocal microscopy. L, A higher magnified image of K displays degenerating PCs with atrophic and fragmented dendritic/or axonal structures. M, Percentage of surviving PCs in each larva. N values at each time point in G are reported in three different colors [tCD4 (black), Kcnc3awt (blue), Kcnc3aR335H (red)], and those in M are two [2xcpce:Kcnc3a R335H (purple), 4xcpce:Kcnc3aR335H (red)]. p values were determined by one-way ANOVA with post hoc Tukey HSD test. G, tCD4 versus Kcnc3aR335H (7 dpf) p = 0.0010053; tCD4 versus Kcnc3aR335H (11 dpf) p = 0.0010053; tCD4 versus Kcnc3aR335H (14 dpf) p = 0.0010053. M, 2xcpce:Kcnc3aR335H versus 4xcpce:Kcnc3aR335H (7 dpf) p = 0.0368766; 2xcpe:Kcnc3aR335H versus 4xcpce:Kcnc3aR335H (11 dpf) p = 0.165158; 2xcpce: Kcnc3aR335H versus 4xcpce:Kcnc3aR335H (14 dpf) p = 0.3135167. Scale bars: B, K, 50 μm; E, L, 20 μm; F, 10 μm.
Imaging.
Fluorescent image recording was performed using a laser scanning confocal microscope (TCS SP8, Leica Microsystems); reconstructions and projections from z-stacks of images were generated with the 3D-projection program included in the LAS X software (Leica Microsystems).
Alternatively, fluorescent images and images of DAB and NBT/BCIP costainings were captured with a fluorescent stereomicroscope (MZ205FA, Leica Microsystems), or a transmitted light fluorescence microscope (DM5000B, Leica Microsystems) equipped with cameras (DMZ2900 and DFC3000G, Leica Microsystems). For recording in situ hybridization signals stained with NBT/BCIP, a camera- (Digital Sight DS-Vi1, Nikon) equipped stereo microscope (MZ205FA, Leica Microsystems) was used. Acquired images were further processed with Adobe Photoshop (Adobe Systems) to prepare figures and movies. For recording LSM confocal images shown in Figure 9, B and E, the same laser and detection settings as well as z-stack thickness were applied. For quantification purposes the number of fluorescent cells (see Figs. 8H, 9C,F,G, 12G,M) was counted automatically using Volocity 6.3 software (PerkinElmer). For microscopy of organotypic mouse cerebellar cultures, the stained slices were imaged using an Axioplan fluorescence microscope (Carl Zeiss) for quantification, and a Fluoview1000 confocal microscope (Olympus) for providing higher-magnification images of PCs.
Figure 8.
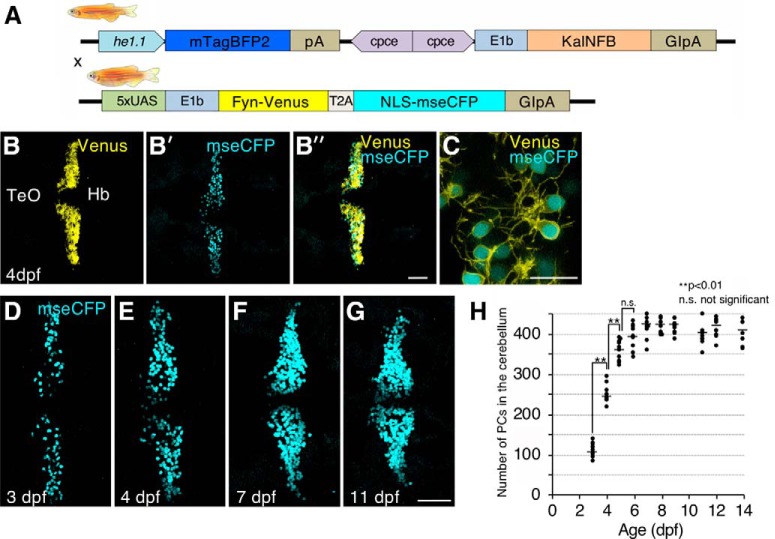
PC-specific Gal4/UAS system mediated dual reporter expression allows the quantification of developmental growth dynamics of the zebrafish PCs. A, Schematic representation of the Tg(2xcpce-E1b:KALNFB, he1.1:mTagBFP2)bz14 driver (top row) and effector line Tg(5xUAS:FMAVenus-2A-3xNLS-HA-mseCFP)bz8 carrying 5xUAS driving Gal4-dependent expression of membrane targeted Fyn-Venus and nuclear localized NLS-mseCFP linked by a T2A peptide (bottom row). B, B′, B″, C, Four days postfertilization larva heterozygous for both KalNFB and UAS:reporter transgenes displaying membrane tagged Fyn-Venus (yellow) together with nuclear localized NLS-mseCFP expression (cyan) in PCs. A higher magnified image from (B″) is presented in C. D–G, Nuclear localized NLS-mseCFP expression in PCs from larva at 3, 4, 7, and 11 dpf, respectively. H, Scatter plot depicting the mean values of the number of PCs in 3–14 dpf zebrafish. N values of zebrafish in each group are 9–10 for H. p values were determined by one-way ANOVA with post hoc Tukey HSD test; 3 vs 4 dpf, p = 0.0010053; 4 vs 5 dpf, p = 0.0010053; 5 vs 6dpf, p = 0.0830332. Scale bars: B″, G, 50 μm; C, 20 μm. Hb, Hindbrain; TeO, optic tectum.
OKR experimental setup and data analysis.
Larval zebrafish injected with a bidirectional vector carrying kcnc3awt or kcnc3aR335H were reared in 30% Danieau supplemented with PTU at a concentration of 70 μm until 4 dpf, allowing for sorting fluorescent-positive larvae throughout the entire PC layer. This PTU treatment does not affect OKR observed in larval fish (Matsui et al., 2014). At 6.3 dpf, OKR was triggered (see Fig. 13A) as described in our previous work (Matsui et al., 2014). OKR of larvae illuminated with white light from the bottom was recorded for 45 s with the speed of 2 fps using a stereomicroscope (MZ205FA, Leica Microsystems) equipped with a digital camera (DFC3000G, Leica Microsystems). Each time frame was analyzed with Fiji software (http://fiji.sc) to determine the eye orientation, which was performed by measuring the angle of the long axis (white arrows) of the fitted ellipse of each eye relative to the body axis (dashed white lines). The angle was defined as positive value when it opened counterclockwise and as negative value when it opened clockwise with respect to the vertical line (see Fig. 13B). Images were processed in Adobe Photoshop (Adobe Systems) followed by their animation using the QuickTime player (Apple).
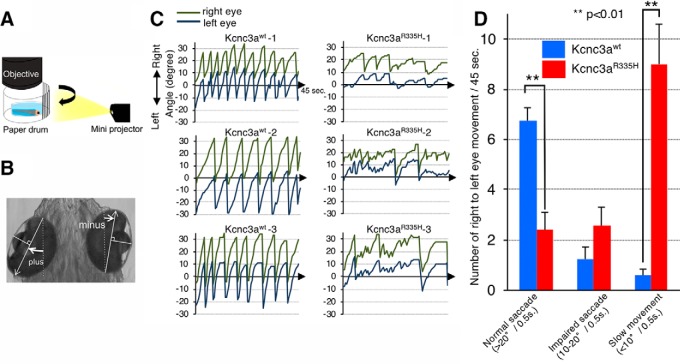
Figure 13.
Expression of Kcnc3aR335H pathological variant in PCs causes eye movement deficits. A, Schematic drawing of the experimental set up for OKR monitoring. Moving black and white stripes from left to right were projected onto a white paper drum while the larva was immobilized in agarose with only its eyes allowed to move freely. B, Measurement of eye position. The angle of each eye was measured with respect to the external vertical axis parallel to the midline; angles of eye position deflected counterclockwise were defined as positive values, whereas clockwise deflected ones were defined as negative values. White arrows, The long axis of an ellipse; dotted line, external vertical axis. C, The graphs show left and right eye position tracking by angular eye displacement during visually triggered OKR from three representative 6.3 dpf larvae expressing Kcnc3awt (left) or Kcnc3aR335H (right) in PCs. D, Each bar in the graph indicates the average number of saccades observed during individual OKR trials (45 s) for Kcnc3awt (blue) or Kcnc3aR335H (red) larvae. Normal saccades (angle change larger than 20°/0.5 s right to left); impaired saccades (angle change at 10–20°/0.5 s right to left); or slow eye movements (angle change <10°/0.5 s right to left) were distinguished. N values for D are 6 OKR responses corresponding to 12 eye movements for 6 zebrafish larvae each. p values were determined by one-way ANOVA with post hoc Tukey HSD test. D, Kcnc3awt versus Kcnc3aR335H (Normal saccade), p = 0.0010053; Kcnc3awt versus Kcnc3aR335H (Slow movement), p = 0.0010053.
Statistical analysis.
N values for each group are reported in each figure legend, or directly described in Figures 4, D and F, and 12, G and M. All data illustrated are provided as mean ± SEM. One-way ANOVA with post hoc Tukey HSD test (see Figs. 8H, 9C,F,G, 12G,M, 13D), or unpaired Student's t test (see Fig. 10H,I) was used for the evaluation of the statistical significance between each experimental group. p values are indicated as *p < 0.05 or **p < 0.01 and were considered significant. Exact p values are additionally described in the Results and each figure legend.
Results
mRNA of ca8 is predominantly expressed in larval and adult cerebellar PCs in zebrafish
Ca8 represents an evolutionary conserved gene throughout vertebrates (Aspatwar et al., 2010) with strong expression observed in cerebellar PCs (Kato, 1990; Nógrádi et al., 1997; Bae et al., 2009). mRNA in situ hybridization on adult zebrafish brain sections confirmed this strong PC-specific expression (Fig. 1A,B) based on colocalization with the PC-specific ZebrinII antigen (Fig. 1C). Faint ca8 expression was also detectable in crest cells of the medial octavolateralis nucleus (MON; Fig. 1D), which function like PCs in this cerebellar-like structure (Bell et al., 2008).
The embryonic and larval ca8 expression during zebrafish development, which has not been systematically described so far (Aspatwar et al., 2013; Huang et al., 2014; Takeuchi et al., 2017), could be detected first at 14 hpf throughout the notochord and prechordal plate as well as in rhombomere 3 and 5 marked by krox20 coexpression (Fig. 2A–C″).
At 48 hpf, restricted ca8 mRNA expression was detected in the otic vesicles, the epiphysis, and the adenohypophysis (Fig. 2D–G″). Starting at 2.5 dpf, ca8 expression appeared in the larval cerebellum coincident with the onset of PC differentiation (Hamling et al., 2015) and increases in strength (Fig. 2H–K) marking ZebrinII-positive PCs (Fig. 2P–R″) similar to the expression dynamics of the zebrafish PC marker parvalbumin7 (pvalb7; Fig. 2L–O; Bae et al., 2009). Early expression of ca8 appears more uniformly distributed throughout the developing PC layer compared with pvalb7 of which strong expression is restricted to cells near the dorsal cerebellar midline (Fig. 2I,J,M,N, compare signals at 2.5 and 3 dpf, white arrows and arrowheads).
In summary, ca8 mRNA is distributed in few cell types (prechordal plate, notochord, rhombomere 3/5, otic vesicle, epiphysis, adenohypophysis) during early embryonic stages, but becomes restricted to PCs during larval stages, in which it is maintained throughout adulthood.
The zebrafish ca8 upstream genomic region contains a regulatory element to induce gene expression in PCs
The restricted expression profile of the ca8 mRNA including PCs indicated ca8 as a good candidate gene for isolating a regulatory element driving specific expression in zebrafish PCs. We first generated a stable reporter line in which enhanced GFP expression was placed under control of a 7543 bp genomic region upstream of the translational initiation site of the ca8 gene (−7543 to −1; −7.5ca8; Fig. 3A). Of 36 injected and raised P0 fishes, offspring of five of them exhibited intense GFP expression in PCs, although some ectopic GFP expression outside of the cerebellum could be observed. F2 offspring obtained from the F1 founders, designated as Tg(-7.5ca8:GFP)bz12, with most restricted PC expression again displayed strong GFP expression in larval PCs in Mendelian ratio (Fig. 3B–D″). Whole-mount immunostaining on 4 dpf larvae confirmed that ZebrinII-positive PCs coexpressed GFP (Fig. 3C,D″). Time-lapse analysis of the developing cerebellum revealed that GFP-positive cerebellar cells emerged at 56 hpf (2.3 dpf) in Tg(-7.5ca8:GFP)bz12 larva, increasing in number along with cerebellar differentiation reminiscent to the appearance and expansion of differentiating PCs (Hamling et al., 2015; Fig. 3E; Movie 1). Restricted GFP expression in PCs lasted until adulthood (Fig. 3F–I″). These Tg(-7.5ca8:GFP)bz12 embryos also showed GFP fluorescence at 24 hpf in the notochord, the diencephalic region, and the hindbrain likely corresponding to rhombomere 3 and 5 domains (Fig. 3J,K) equivalent to the distribution of endogenous ca8 mRNA (Fig. 2A–C″). In addition, transient GFP expression was observed in epithelia along the midline seam, the stomodeum and the eye retinal pigment epithelia (Fig. 3L–O). These results indicate that the isolated 7543 bp genomic fragment upstream of ca8 harbors among others a regulatory element driving expression in PCs.
Time-lapse imaging of the cerebellar region of larval zebrafish from 56 to 71.5 hpf.
In Tg(−7.5:GFP)bz12 cerebellum, initial appearance of green fluorescent PCs (green) occurs at 56 hpf. PCs increase in number and expand across the cerebellar surface. Imaging was carried out by recording z-stacks encompassing the same cerebellar region (177.22 μm thickness, 233 images in 13.027 μm steps) at 30 minutes intervals, and captured z-stacks were projected as maximum projection into a single plane for each time point and animated to replay at a speed of 2 frames per second.
Identification of a compact regulatory element to activate gene expression exclusively in PCs
To identify a minimal regulatory element for PC-exclusive gene expression, stepwise deleted −7.5ca8 fragments were cloned into a construct carrying a GFP reporter (Fig. 4A) flanked by Tol2-transposon sites (Kawakami, 2007). This was followed by reporter plasmid injection into one cell stage embryos and subsequent observation of GFP fluorescence in the cerebellum of injected larvae at 4 dpf. Tol2-transposon-mediated early genomic integration of the transgene resulted in broader expression throughout the PC population (Fig. 4C).
This transient reporter construct analysis revealed that a 2.24 kb fragment (−2242 to −1: −2.24ca8) clearly drove reporter expression in the cerebellum, whereas a 1.98kb fragment (−1981 to −1: −1.98ca8) completely lost this activity (Fig. 4C,D), suggesting the existence of a PC-specific regulatory sequence of 258 bp between the two breakpoints (−2240 to −1983). Thus, we subcloned this element with or without a CMVmini basal promoter (Bajoghli et al., 2004) into the GFP reporter vector for further analysis (Fig. 4B). Injections revealed that the isolated element alone was able to efficiently induce cerebellar GFP expression in an orientation-independent manner but required the CMVmini (Fig. 4E,F). The identified 258 bp regulatory element thus fulfills the characteristics of a cell-type-specific enhancer element which we named cpce (ca8 promoter derived PC-specific enhancer element).
The CMVmini is prone to induce ectopic expression especially in the spinal cord and suffers from silencing along with successive generations (own observations). To examine cpce activity in stable transgenics we combined it with the minimal E1b promoter (Köster and Fraser, 2001b), which is widely used in zebrafish as a basal promoter lacking background expression, followed by the GFP reporter (Fig. 5A) and flanked by Tol2-transposon sites (Kawakami, 2007). Injected GFP-positive larvae were raised to adult fish to generate stable transgenic lines. All of the F1 offspring from P0 founders (20%; 6 of 30 injected fish screened), showed PC restrictive GFP expression at larval stages. One of these lines was raised to adulthood to generate the transgenic strain Tg(cpce-E1b:GFP)bz13 with Mendelian inheritance of the fluorescent reporter in cerebellar cells (Fig. 5E,E′,F) that were identified as PCs by ZebrinII immunohistochemistry (Fig. 5G–I′). Noteworthy, unlike Tg(-7.5ca8:GFP)bz12, Tg(cpce-E1b:GFP)bz13 embryos did not show GFP expression outside of the cerebellum during embryonic stages (Fig. 5B–D′). In addition, adult brain sections stained with an anti-GFP antibody revealed that GFP expression was confined only to PCs without detectable expression in MON crest cells (Fig. 5J–L″), which faintly expresses endogenous ca8 mRNA (Fig. 1D). Intriguingly, gene gun mediated transfection of mouse cerebellar organotypic slices with the cpce-CMVmini:GFP construct also resulted in PC restricted GFP expression (Fig. 6). These results indicate that the cpce represents an evolutionary conserved compact enhancer element mediating PC-specific expression not only limited to transgenic zebrafish.
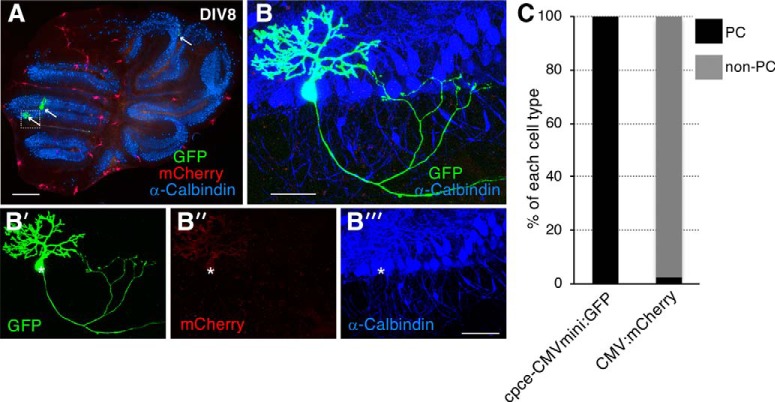
Figure 6.
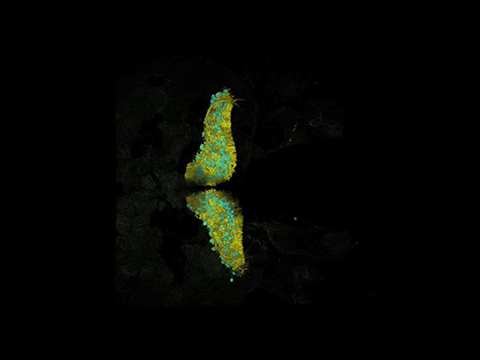
cpce induces GFP expression exclusively in PCs in transfected mouse organotypic cerebellar slice culture. A, Overview of an 8 d in vitro (DIV8) cerebellar slice culture cotransfected with cpce-CMVmini:GFP (green) and CMV enhancer/promoter (CMV): mCherry (red), cultured for 6 d and stained for calbindin (blue). Arrows indicate strongly GFP-expressing PCs. The white dotted box identifies the area magnified in B. B′, B″, B‴, Higher-magnification image of the cerebellar region shown by the white dotted box in A containing one calbindin-expressing PC (B‴, white star) strongly labeled with GFP expression (B′) and very faint mCherry fluorescence (B″). Throughout the cerebellum strongly expressing mCherry cells are found belonging to cell types different from PCs based on the absence of calbindin coexpression (A). The very weak expression of mCherry in PCs (B″), indicates that the CMV has a low preference for driving gene expression in mouse PCs. This is opposite for the cpce which induces strong GFP expression in PCs, but not elsewhere throughout a cerebellar slice, neither strongly or weakly. C, To quantify the reporter gene expression driven by each construct in PCs and other cell types (non-PCs), fluorescent cell counting was performed on overview images from 4 cerebellar slices cotransfected with cpce-CMVmini:GFP and CMV:mCherry constructs. All GFP-positive cells (8/8) are calbindin-positive PCs, whereas 97.2% (74/76) of cells strongly expressing mCherry are non-PCs lacking calbindin expression. Sale bars: A, 1 mm; B, B‴, 50 μm.
Establishment of stable transgenic PC-specific Gal4-driver zebrafish strain
The Gal4/UAS system is a versatile combinatorial genetic system widely used in zebrafish (Asakawa and Kawakami, 2008; Distel et al., 2009). To establish a PC-specific Gal4 driver line, we generated two cpce expression constructs with different Gal4 activators; cpce-E1b:KalTA3 (equivalent to Gal4FF), and KalNFB, in which the transcriptional activation domains are derived from HSV1-VP16 (Asakawa and Kawakami, 2008; Distel et al., 2009), or the human NFκB p65 subunit (Emelyanov and Parinov, 2008), respectively. Injection of each cpce-E1b:Gal4 construct into transgenic UAS reporter embryos Tg(5xUAS:FMAVenus-2A-3xNLS-HA-mseCFP)bz8 (Matsui et al., 2014) revealed that KalNFB in contrast to KalTA3 induced prominent cerebellar expression (Fig. 7A,B). Thus, we used KalNFB under control of a tandem cpce repeat for stronger expression to generate stable transgenics. In addition, mTagBFP2 expression driven by the hatching enzyme 1.1 promoter (he1.1; Xie et al., 2012) was included in the construct (Fig. 7C, top) to identify transgenic KalNFB carriers. All identified P0 founders (10/56 screened adults, 17.9%) produced offspring with mTagBFP2 labeled hatching glands. When crossed against heterozygous Tg(4xUAS:GFP)hzm3 carriers (Fig. 7C, bottom; Distel et al., 2009), F1-larvae displayed GFP expression in the cerebellum, but also in cells of the optic tectum to a varying extent. Nevertheless, F1 offspring showing mTagBFP2 in hatching glands at 24 dpf (Fig. 7D), followed by inducing almost exclusive PC expression of GFP (Fig. 7E,F) were raised to generate F2 offspring of the transgenic strain Tg(2xcpce-E1b:KALNFB, he1.1:mTagBFP2)bz14. PC-specific expression was confirmed by ZebrinII immunohistochemistry using larvae carrying both KalNFB and UAS:GFP transgenes (Fig. 7G,H″). Sparse and faint ectopic expression could be found in a few cells of the optic tectum, and hindbrain (Fig. 7G,G″). On adult sagittal brain sections, GFP fluorescence was still confined to ZebrinII-positive PCs (Fig. 7I,J″). In contrast to ubiquitous PC expression during larval stages, the absence of GFP-expression in some ZebrinII-positive neurons though (Fig. 7J″) may reflect the inactivation of repetitive UAS sequences by DNA methylation (Goll et al., 2009).
Quantification of developmental growth dynamics of the zebrafish PC population
Previous quantifications of differentiated PCs in the developing zebrafish cerebellum during 3 and 14 dpf using anti-Parvalbumin immunohistochemistry showed significant differences in cell numbers as well as the temporal profile of their appearance (Kaslin et al., 2013; Hamling et al., 2015). We could resolve these discrepancies using double-transgenic larval fish expressing PC-specific KalNFB in Tg(5xUAS:FMAVenus-2A-3xNLS-HA-mseCFP)bz8 background (Fig. 8A). In these larvae individual PCs are labeled with two different fluorescent reporters allowing one to detect blue labeled PC nuclei clearly separated from the coexpressed yellow fluorescent membrane staining using high resolution confocal microscopy (Fig. 8B–G; Movie 2; Movie 3), followed by automated segmentation and counting of cells. The largest increase in PC number occurred between 3 and 5 dpf, with PC generation reaching a plateau at ∼6–7 dpf (Fig. 8H; 3 vs 4 dpf, p = 0.0010053; 4 vs 5 dpf p = 0.0010053; 5 vs 6 dpf p = 0.0830332, one-way ANOVA with post hoc Tukey HSD test) similar to the temporal profile of PC differentiation described by Hamling et al. (2015). The average number of PCs though was significantly higher than those previously reported deviating by twofold to fourfold at each developmental time point analyzed (Fig. 8H; e.g., at 7 dpf: 100 PCs: Kaslin et al., 2013; 250 PCs: Hamling et al., 2015; compared to 420 PCs in this study). Thus based on Gal4-mediated cell type-specific activation of several transgenic reporters, PCs can be analyzed at higher detail, increased precision and with less effort compared with antibody-based methods.
A 3D reconstruction of the cerebellum of a double transgenic zebrafish labeled with membrane tagged Venus and nuclear localized mseCFP.
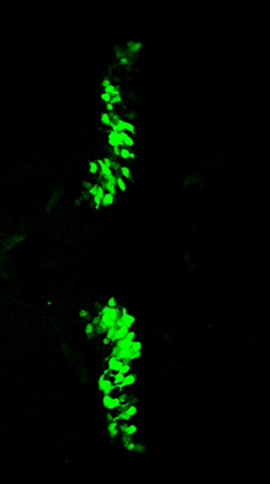
In Tg(2xcpce-E1b:KALNFB, he1.1:mTagBFP2)bz14; Tg(5xUAS:FMAVenus-2A-3xNLS-HA-mseCFP)bz8 larva at 7dpf, PCs are labelled with differently colored fluorescent proteins in the nucleus (cyan) and the cytoplasmic membrane (yellow) respectively. For 3D reconstruction of the cerebellum laser scanning confocal microscope z-stack imaging was performed by recording 197 images in 13.027 μm steps (total thickness: 149.63 μm). Subsequent reconstruction and animation were performed using the Leica LAS-AF software.
A 3D reconstruction of the cerebellum of a double transgenic zebrafish labeled with membrane tagged Venus and nuclear localized mseCFP.
In Tg(2xcpce-E1b:KALNFB, he1.1:mTagBFP2)bz14; Tg(5xUAS:FMAVenus-2A-3xNLS-HA-mseCFP)bz8 larva at 7dpf, PCs are labelled with differently colored fluorescent proteins in the nucleus (cyan) and the cytoplasmic membrane (yellow) respectively. For 3D reconstruction of the cerebellum laser scanning confocal microscope z-stack imaging was performed by recording 197 images in 13.027 μm steps (total thickness: 149.63 μm). Subsequent reconstruction and animation were performed using the Leica LAS-AF software.
Generation of a PC selective multi-cistronic coexpression system tunable in expression strength
A cell-type-specific single-vector coexpression system independent of combinatorial genetics such as the Gal4, LexA, or Q system (Asakawa and Kawakami, 2008; Emelyanov and Parinov, 2008; Ghosh and Halpern, 2016), is a tremendously versatile tool. It allows one to monitor specific cells expressing a gene-of-interest together with the help of a fluorescent reporter protein targeted to cellular structures independently of the protein encoded by the gene-of-interest (Distel et al., 2011; Hocking et al., 2013). In addition, without having to use combinatorial genetics, only single transgenic lines need to be generated, which propagate the cistrons like a single transgene and do not suffer from inactivation of host-unspecific repetitive elements, as observed for example for UAS sequences of the Gal4 system (Goll et al., 2009). As the cpce is of very small size, we sought to establish such a PC-specific coexpression system in analogy to bidirectional Janus and Medusa UAS expression constructs that we reported before (Distel et al., 2010).
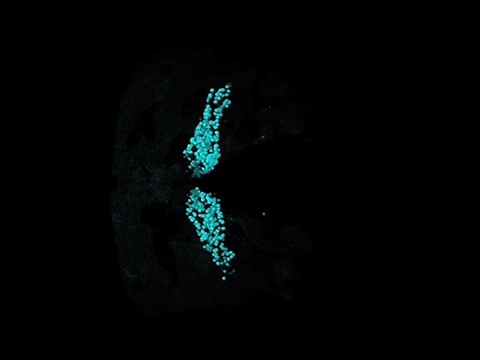
Initially, a single cpce was flanked on both sides by an E1b basal promoter to drive expression of a red fluorescent protein, mRuby3, and GFP in a bidirectional manner (p-GFP:cpce:Ruby; Fig. 9A). Larvae injected with this construct at the one cell stage showed tight coexpression of both reporters in PCs albeit at low levels (Fig. 9B), useful for low-level coexpression of highly effective transgenes such as enzymes. By stepwise increasing cpce numbers in the coexpression vector (p-GFP:2/4/8xcpce:Ruby; Fig. 9A) expression of both cistrons successively enhanced from moderate to strong and very strong, while their tight coexpression was kept (Fig. 9B). Starting with vectors carrying 4× repeats of cpce, moderate ectopic expression outside of the cerebellum in cells of the optic tectum and the retina was observed, which further increased in extent and strength by using 8xcpce (Fig. 9B,C; 1xcpce vs 4xcpce (tectum) p = 0.0025945; 4xcpce vs 8xcpce (tectum) p = 0.0010053; 2xcpce vs 8xcpce (eye) p = 0.0010053; 2xcpce vs 8xcpce (eye) p = 0.0010053, one-way ANOVA with post hoc Tukey HSD test).
To also allow for strong PC-specific transgene coexpression we further refined p-GFP:4x cpce:Ruby by eliminating unwanted expression outside of the cerebellum with the help of a cell-type-specific microRNA (mir)-mediated gene silencing strategy (Brown et al., 2007). mir181a, mir181b, and mir216 are strongly expressed in tectal and retinal ganglion cells but are devoid of PC expression (Wienholds et al., 2005). Each mir target sequence (mirT) was inserted in tandem immediately behind mRuby3 (Fig. 9D). Constructs carrying 2xmir181aT, or 2xmir181bT but not 2xmir216T effectively repressed ectopic tectal and retinal mRuby3 expression with keeping high expression levels in PCs at levels comparable to p-GFP:4xcpce:Ruby (Fig. 9E–G). As expected, the inserted 2xmirT-sequences did not change the expression of GFP, which served as control for monitoring the inhibitory effects of the respective 2xmirT-sequences on mRuby3 expression. Ectopic expression was further decreased when 4xmir181aT, 4xmir181bT, or their combination (2xmir181aT/2xmir181bT) were used, again leaving strong PC-specific coexpression unaffected (Fig. 9E–H; F: No mirT vs 2xmir181aT p = 0.0010053; No mirT vs 2xmir181bT p = 0.0010053; 2xmir181aT vs 4xmir181aT p = 0.0078459; 2xmir181bT vs 4xmir181bT p = 0.0126719; 2xmir181bT vs 2xmir181aT/ 2xmir181aT p = 0.0244631. G: No mirT vs 2xmir181aT p = 0.0010053; No mirT vs 2xmir181bT p = 0.0010053; No mirT vs 2x216T p = 0.0010053; 2xmir181aT vs 2xmir216T p = 0.0010053; 2xmir181bT vs 2xmir216T p = 0.0010053, one-way ANOVA with post hoc Tukey HSD test). Based on these results we finally generated a universal bidirectional strong PC-specific 4xcpce coexpression construct, named pTol2-4x cpce-MCS, containing two multiple cloning sites (MCS1 and MCS2) each of which is followed by a 4xmir181aT-polyA sequence. This vector allows one to insert two genes-of-interest into these MCSs, which we demonstrated by cloning membrane-targeted Fyn-TagRFP-T and nuclear localized H2B-tdiRFP into this cassette (Fig. 10A). When the resulting construct was injected into Tg(-7.5ca8:GFP)bz12 embryos, strong PC-specific coexpression of both subcellular reporters with negligible ectopic expression was observed (Fig. 10B–D⁗). We further revealed by a luciferase-based quantitative assay (Fig. 10E–I) that pTol2–4x cpce construct drove a slightly higher expression of one transgene cloned into MCS2 (Fig. 10E, facing right) compared with expression of another cloned into MCS1 (Fig. 10E, facing left; expression from MCS1:expression from MCS2 = 1:1.55 ± 0.038; H: Fluc:4xcpce:Rluc-GFP vs Rluc-GFP:4xcpce:Fluc p= 0.0036; I: MCS1 vs MCS2 p = 0.0438, unpaired Student's t test). An asymmetric sequence contained in this vector, a spacer sequence positioned in the middle of 2xcpec and/or Tol2 recognition sites flanking the whole expression units, may slightly affect expression level in each direction, the PC-specific enhancer as well as the mirT-pA fragments are constituted by a completely mirror symmetric sequence though.
In summary we provide a series of PC-specific constructs to tune transgene coexpression from low to moderate to strong levels depending on the experimental needs.
Genetic modeling of a PC degeneration disease in zebrafish
With these established genetic tools in hand we aimed to genetically model and monitor a human PC neurodegenerative disease in zebrafish. SCA13 is an autosomal dominant disease caused by a missense mutation allele of the KCNC3 gene encoding a voltage-dependent potassium channel KCNC3/Kv3.3 (Waters et al., 2006; Zhang and Kaczmarek, 2016). Fast firing neurons such as cerebellar PCs are crucially dependent on this channel as it mediates a rapid repolarization phase of an action potential, generating the repetitive spikelets of the complex spike (Kaczmarek and Zhang, 2017). Patients heterozygous for the missense mutant allele KCNC3R420H suffer from progressive cerebellar atrophy (Waters et al., 2006), although the neurodegenerative changes have not been investigated in detail in postmortem cerebellar tissue. Because rodent KCNC3 mRNA and protein expression were detected most strongly in cerebellar PCs (Weiser et al., 1994; Chang et al., 2007), it is believed that PCs are the primarily affected neurons in SCA13 patients. So far only few animal models have been generated for SCA13 (Issa et al., 2011, 2012; Gallego-Iradi et al., 2014) using the expression of human KCNC3R420H in Drosophila and expression of its zebrafish pathological counterpart kcnc3aR335H in zebrafish motoneurons, respectively, but these models failed to induce apparent neuronal degeneration (Issa et al., 2012; Gallego-Iradi et al., 2014). In addition to strong expression in PCs, moderate expression of rodent KCNC3 mRNA and protein were found in cerebellar granule cells, neurons of the deep cerebellar nuclei as well as in the brainstem (Weiser et al., 1994; Chang et al., 2007), providing an alternative hypothesis that SCA13 neuropathologies initiate from these regions, and only secondarily affect PC integrity and survival thereby leaving the origin of observed cerebellar shrinkage, manifested in SCA13R420H patients uncertain. Our PC-specific coexpression system is well suited to address these open questions, whether expression of the pathogenic SCA13R420H allele induces PC degeneration in a cell-autonomous manner and causes behavioral coordination deficits such as impaired eye movement control.
We first confirmed a previous finding that the transcript of kcnc3a, one of two zebrafish genes homologous to human KCNC3 (Mock et al., 2010), is highly expressed in PCs at larval stages (Fig. 11A,B,E,F″) as reported previously (Hsieh et al., 2014), whereas mRNA for kcnc3b, was not clearly detected in the cerebellum (Fig. 11C,D). Indeed, it has been previously shown that the zebrafish kcnc3a gene product harbors unique gating properties similar to the human KCNC3 channel (Mock et al., 2010). In addition, a Kcnc3aR335H mutant channel corresponding to the human pathogenic KCNC3R420H variant has been shown to be nonfunctional when expressed alone, but acts as a dominant-negative form in the presence of the wild-type subunit (Mock et al., 2010), causing similar electrophysiological signatures compared with the expression of the human KCNC3R420H variant (Waters et al., 2006). These results indicate that zebrafish kcnc3aR335H is equivalent to the KCNC3R420H pathogenic variant, suitable for modeling human SCA13R420H in zebrafish.
kcnc3a gene structure and alternative splicing. Exon-intron structure of the zebrafish kcnc3a gene. Boxes represent exons, and black boxes indicate exons used to generate each splice variant. Currently, twelve predicted variants are deposited in GenBank as variants as follows: X1; XM_021470843, X2; XM_021470845, X3; XM_021470847, X4; XM_009299543, X5; XM_021470855, X6; XM_009299545, X7; XM_021470856, X8; XM_021470857, X9; XM_005164037, X10; XM_009299548, X11; XM_021470866, and X12; XM_009299549. The translation initiation codon for all variants is located in exon1. Exons 1, 2, and 3 are present in all splice isoforms to encode most of the protein coding sequence, whereas the subsequent exons 4, 6, 7, or 8 determine the C-terminal intracellular coding sequence of kcnc3a terminating with -WIPK, -PSIL, -VTNV, or –VLSF, respectively. Note that variants X2 and X3 partially lack exon7, whereas variant X10 partially lacks exon2. Download Figure 11-1, TIF file (459.3KB, tif)
An amino acid sequence represents a C-terminal intracellular domain of a zebrafish Kcnc3a protein generated from each splicing variant. The black line indicates a position of nucleotide sequence corresponding to exon3 shared by all transcripts, whereas red shows each exon sequence translated into a specific C-terminal intracellular domain. Only nucleotide sequences (indicated by small characters) close to the junction between exon3 and each following exon are presented. Note that an exon 6 used in isoform X1, 2, 3, 4, 5, and 10 encodes a C-terminal sequence terminating with PSIL (enclosed in boxes) conserved among vertebrate. This sequence matches the potential class I PDZ (PSD-95/Discs Large/ZO-1) binding motif (X-S/T-X-L/V) (Sheng, and Sala, 2001). * indicates a C-terminal amino acid. Download Figure 11-2, TIF file (509.8KB, tif)
kcnc3a is expressed in many alternative isoforms in zebrafish PCs
There are multiple splice isoforms of mammalian KCNC channel family members, resulting in channels with different C-terminal intracellular regions (Rudy et al., 1992; Weiser et al., 1994), but the functional significance of this diversity is not fully investigated yet. In zebrafish, there are 12 predicted kcnc3a transcripts deposited in GenBank (Fig. 11-1), encoding five different C-terminal intracellular domains (Fig. 11-2; Sheng and Sala, 2001). All of the splicing variants share exon 2 encoding arginine 335 that is substituted to histidine in the pathogenic SCA13 variant (Mock et al., 2010). Thus, we exploited the exclusive GFP expression in PCs in 4dpf Tg(cpce-E1b:GFP)bz13 larvae for single cell FACS from dissociated head regions to identify the kcnc3a isoforms predominantly expressed in PCs (Fig. 11G–O; Table 1). This was followed by single cell reverse-transcription (RT) PCR analysis with specific primer pairs to amplify each of the five possible different C-termini generated by alternative splicing (Fig. 11P,Q). Although isoforms for four C-termini were found to be expressed in PCs by this analysis (Fig. 11Q), further in situ hybridization using a specific probe against each transcript revealed that isoform X12 encoding a protein terminating with −VLSF is the most abundantly expressed kcnc3a mRNA in larval PCs (see Fig. 11R–W″).
Table 1.
FACS statistics
| Brass | Tg(cpce-E1b:GFP)bz13 | |
|---|---|---|
| No. of events | 100,000 | 1,256,387 |
| Alive, % | 32.32 on all events | 40.17 on all events |
| Alive-Singlets, % | 61.05 on Alive | 67.73 on Alive |
| Alive-Singlets-Round, % | 59.79 on Alive | 66.66 on Alive |
| Alive-Singlets-Round-GFP+, % | 0.0000 on all the events | 0.0451 on all the events |
| 0.0000 on Alive | 0.1123 on Alive | |
| 0.0000 on Alive-Singlets-Round | 0.1685 on Alive-Singlets-Round |
FACS of Tg(cpce-E1b:GFP)bz13 zebrafish was used to isolate GFP-expressing PCs, whereas nontransgenic cells from brass larvae were used to distinguish GFP-positive from GFP-negative cells. The table shows the percentage of living cells found in each sample, the percentage of Singlets, Round-Singlets, and GFP-positive Round-Singlets among the alive cells. To provide a better understanding of the initial number of cells required to isolate a significant number of single PCs, the percentage of GFP-positive PCs is expressed with respect to the overall number of events. To show the enrichment factor introduced by the sequential gatings, which precede the selection of single PCs, the percentage of GFP-positive cells is also expressed with respect to the live-Singlets-Round cell population.
Expression of SCA13 causing kcnc3aR355H results in prominent progressive degeneration of zebrafish PCs
To express this kcnc3a X12 isoform specifically in PCs, we inserted the cDNA of this variant (referred to hereafter as kcnc3awt) as well as its pathogenic allele carrying the R355H mutation (kcnc3aR335H; Mock et al., 2010) into one of the MCS of our established pTol2-4xcpce-MCS vector. As additional control for transgene expression a cDNA encoding a nonfunctional truncated receptor of human CD4 (tCD4) lacking its cytoplasmic domain (Mejia-Pous et al., 2009) was used. Into the other MCS, a bicistronic transgene was inserted encoding two subcellular fluorescent reporters: membrane targeted Fyn-TagRFP-T to visualize Kcnc3aR335H affected PC morphologies, and nuclear localized H2B-GFP to quantify PC numbers. Both reporters were linked by a self-cleaving T2A-peptide (Fig. 12A; Kim et al., 2011). The PC layer of the same individuals injected with each construct at the one cell stage were subjected to confocal microscopy characterization at 4, 7, 11, and 14 dpf.
This analysis revealed that red fluorescent Kcnc3aR335H-expressing PCs displayed clear signs of neuronal degeneration, which was progressive with ongoing larval differentiation. These signs included a shrinkage of PCs and their extensive dendritic tree with dying cells giving rise to fragmented red labeled debris likely originating from degenerating dendritic and/or axonal structures (Fig. 12B,E,F). In contrast, PCs expressing either tCD4 or Kcnc3awt did not show such signs of neuronal damage and maintained highly branched dendritic structures (Fig. 12B–D).
Confocal analysis was further used to quantify the progression of Kcnc3aR335H induced neurodegeneration by counting the number of H2B-GFP labeled PCs for each individual larva at 4, 7, 11, and 14 dpf. Larvae expressing tCD4 or Kcnc3awt showed an increase in their PC number, which reached a plateau ∼7 dpf (average 126.3 and 118.7%, respectively, compared with those at 4 dpf that were maintained until 14 dpf; Fig. 12G). This temporal profile of the expansion of the PC population is in good agreement with our quantitative analysis in stable transgenic Tg(2xcpce-E1b:KALNFB,he1.1:mTagBFP2)bz14; Tg(5xUAS:FMAVenus-2A-3xNLS-HA-mseCFP)bz8 specimens (Fig. 8H). In contrast, larval fish expressing Kcnc3aR335H displayed a continuous, gradually increasing reduction in their PC number (average; 60.3 and 37.6% at 7 and 11 dpf, respectively, compared with those at 4 dpf) with nearly all of Kcnc3aR335H expressing PCs having disappeared by 14 dpf [Fig. 12G; tCD4 vs Kcnc3aR335H (7 dpf) p = 0.0010053; tCD4 vs Kcnc3aR335H (11 dpf) p = 0.0010053; tCD4 vs Kcnc3aR335H (14 dpf) p = 0.0010053, one-way ANOVA with post hoc Tukey HSD test]. A reduced Kcnc3aR335H expression driven by 2xcpce (Fig. 12H) still induced PC cell death (Fig. 12I–L), the progression though was slower especially in the initial degeneration phase at 7 dpf [Fig. 12M; 2xcpce:Kcnc3aR335H vs 4xcpce:Kcnc3aR335H (7 dpf) p= 0.0368766, one-way ANOVA with post hoc Tukey HSD test], indicating a dose-dependent cytotoxic effect of Kcnc3aR335H expression on PCs.
These results demonstrate that clear, comprehensive and progressive neurodegeneration can be obtained if a well characterized pathogenic allele of kcnc3a is expressed in a targeted manner in the primarily affected neuronal cell type. Furthermore, our PC-specific coexpression system reveals that Kcnc3aR335H expression exerts a direct and cell autonomous cytotoxic effect on PCs.
Expression of kcnc3aR335H pathological variant in PCs impairs eye movement behaviors
Patients affected by SCAs suffer among other symptoms from eye-movement deficits (Rossi et al., 2014; Hekman and Gomez, 2015). Zebrafish larvae respond to patterns of vertical, alternating black and white stripes that move laterally in front of their field-of-view (Fig. 13A) with a robust OKR starting at 5 dpf (Huang and Neuhauss, 2008). This OKR is composed of a serial combination of smooth pursuits and quick saccadic eye movements (Huang and Neuhauss, 2008). As we recently demonstrated that zebrafish PCs control saccadic eye movements during OKR (Matsui et al., 2014), we addressed whether Kcnc3aR335H-induced PC neuropathologies in zebrafish would cause similar behavioral changes by using larvae injected with Kcnc3a constructs in the pTol2-4xcpce-MCS backbone (Fig. 12A).
When the angles of the eyes (Fig. 13B) in Kcnc3awt expressing larvae were plotted during the trial period, regular waves indicating stereotypic repetitive pursuit-saccade eye movements were induced (Fig. 13C; Movie 4). In contrast, Kcnc3aR335H affected larvae displayed a significant reduction in numbers of saccade eye movements (changes in angle larger than 20°/0.5 s right to left) compared with Kcnc3awt larvae (Fig. 13C,D; Movie 4; Kcnc3awt vs Kcnc3aR335H, p = 0.0010053, one-way ANOVA with post hoc Tukey HSD test). In addition, slow eye movements (changes in angle smaller than 10°/0.5 s right to left) were significantly increased in number, whereas such abnormal eye movements were barely detected in Kcnc3awt-expressing larvae (Fig. 13C,D; Movie 4; Kcnc3awt vs Kcnc3aR335H, p = 0.0010053, one-way ANOVA with post hoc Tukey HSD test). Together, these results confirmed that PCs are involved in the regulation of saccade eye movements in zebrafish, supporting our previous results obtained by optogenetic silencing of PC activity (Matsui et al., 2014). Moreover, the selective expression of the pathogenic SCA13 allele Kcnc3aR335H restricted to PCs allows for directly connecting behavioral symptoms to an individual neuronal population. Thus, our PC-specific SCA13 model in zebrafish displays behavioral symptoms and anatomical alterations at the cellular level reminiscent of the clinical manifestations, a widely shared symptom in progressive SCA neurodegenerative patients (Rossi et al., 2014; Hekman and Gomez, 2015).
Optokinetic response (OKR) of a 6.3 dpf larva expressing Kcnc3awt or Kcnc3aR335H in PCs.
This movie displays a representative optokinetic response (OKR) of a 6.3 dpf larva expressing Kcnc3awt or Kcnc3aR335H in PCs. The movie was recorded at video rate of 2 frames per second and replays in real time.
Discussion
The cpce as a versatile tool to study PC-specific molecule-structure–function-behavior relationships
The rapid appearance of a fully functional cerebellum in zebrafish larvae at 5–7 dpf (Kani et al., 2010; Hsieh et al., 2014) makes it an attractive brain compartment for non-invasive high-resolution bioimaging studies, as it is easily accessible just underneath the skin below the anterodorsal hindbrain. Moreover, the cerebellum is highly conserved during vertebrate evolution consisting only of several neuronal cell types of clear connectivity organized in distinct layers and controlling locomotive behavior, that can be easily monitored in fish (Ahrens et al., 2012; Matsui et al., 2014; Portugues et al., 2014; Knogler et al., 2017).
A prerequisite though for studying cell biological and molecular mechanisms of cerebellar neuronal differentiation, function, homeostasis, plasticity, degeneration, and regeneration, is the specific accessibility of distinct cerebellar neuronal cell types for genetic manipulation (Distel et al., 2010; Matsui et al., 2014; Weber et al., 2016; Theisen et al., 2018). In zebrafish, a PC specific regulatory element of the zebrafish aldolase c gene encoding the ZebrinII antigen has been identified previously, which has been very useful in establishing PC reporter strains (Tanabe et al., 2010). Its size >5 kb though restricts tuning of expression and a bidirectional use. Although the expression of two transgenes mediated by a large enhancer fragment could also be achieved with the help of 2A-peptides, their self-cleaving reaction results in addition of amino acid sequences to the C- and N-terminal ends of the expressed transgenes (Kim et al., 2011) and does not permit expression of native pathogenic protein variants. In general, bidirectional enhancers allow for expressing twice as many transgenes/2A-peptide cassettes as could be achieved with a unidirectional promoter. We have therefore isolated a compact regulatory element of 258 bp, cpce, which specifically drives expression in PCs, the central neuronal population of the cerebellar cortex. We rendered cpce into a bidirectional expression unit and tuned its expression strength to low, moderate, and strong levels by combining it with varying copy numbers of the enhancer. Thus, this specific regulatory element of compact size driving expression exclusively in cerebellar PCs allows for linking genetic manipulations to morphological, cell biological and physiological changes in a single neuronal cell type and to functional and behavioral consequences for a vertebrate organism.
A specific advantage of the cpce is that variations of the copy number of multimerized cpce result in different expression strength. As all these multimers still work in a bidirectional manner, expression strength of two transgene cassettes can be adjusted similarly, again an advantage that cannot be achieved with 2A-peptide-mediated single transgene cassettes driven by a long promoter fragment. Low-level expression can be an advantage for expressing fluorescent proteins as organelle markers that become quickly saturating or deleterious at higher concentration, such as centrosome or microtubule reporters (own observation). In contrast, expression at higher levels is useful to establish sufficient expression levels for pathogenic mutant genes in genetic models of neurological diseases. Moreover, the availability of a cpce-Gal4 driver (Fig. 7) allows for further adjustments of expression by using one or several bidirectional UAS-cassettes (Distel et al., 2010) with varying repeats of Gal4-binding sites.
Finally, the ready to use pTol2-4xcpce-MCS construct has several advantages to drive targeted expression in PCs. The Tol2 transposon mediates insertions of DNA efficiently into the zebrafish genome, if the size is <10 kb (Urasaki et al., 2006), whereas larger fragments are less efficiently integrated into the genome, resulting in reduced numbers of stable transgenic F1 offspring (Suster et al., 2009). This vector carrying a relatively short promoter of ∼1.35 kb, will facilitate the investigation of physiological and pathophysiological states of PCs at the subcellular level, in which transgene expression to alter molecular events is combined with the expression of several suitable subcellular markers and fluorescent sensor proteins (Hocking et al., 2013). This enables elucidating the interdependence of several subcellular structures upon altering the molecular composition of PCs in vivo in real time.
Cross-species cpce activity in PCs
It is worth mentioning that the regulatory cpce element induced PC restricted gene expression not only in zebrafish (Fig. 5), but also in mice at least in transfected organotypic cerebellar slices (Fig. 6). This result suggests that transcriptional regulatory processes for the ca8 gene are highly conserved between zebrafish and mammals. Although our search in the mouse genome could not identify a cpce homologous sequence in the upstream region of the mouse ca8 gene, our findings suggest a conserved transcriptional regulation of this element. Further studies will be needed to clarify the transcriptional mechanism leading to cpce-mediated PC selective gene expression. The compact size of the cpce promises to help disclosing the minimal core transcriptional events mediating PC selective gene expression. Currently, a widely used PC-specific promoter in mouse transgenic research is represented by a 3 kb genomic fragment encompassing exons 1–4 of the L7/pcp2 gene, and its upstream promoter region with destroyed translational initiation sites (Sługocka et al., 2017). Especially for establishing PC-specific viral vectors, only genetic elements of compact size can be used to not interfere with packaging of viral vectors into infectious particles (Hirai and Iizuka, 2011). Thus, the cpce has a great potential for PC-specific multigene expression in rodents as well as in primates using viral transfections (Hirai and Iizuka, 2011; Nitta et al., 2017).
Disease modeling in zebrafish PCs revealed cell autonomous cytotoxicity/degenerative activity of the kcnc3aR335H mutant in PCs
With the help of our established PC-specific expression system, we attempted to model human SCA13, in particular for the SCA13R420H mutant allele, which causes a slowly progressing cerebellar atrophy (Waters et al., 2006). The cellular changes and cytotoxic mechanisms associated with this atrophy have not been clarified in humans so far, as investigations on postmortem tissue are missing. A previous in vitro culture study described mouse PC death caused by the expression of the mouse KCNC3R424H variant corresponding to human pathogenic SCA13R423H (Irie et al., 2014). There are no data available though about neurodegenerative consequences of expressing any of the SCA13 pathogenic alleles in vivo in whole animal models. Expression of the SCA13R420H pathogenic gene in Drosophila resulted in perturbed patterning of photoreceptor cells and wing veins (Gallego-Iradi et al., 2014), whereas expression of the zebrafish Kcnc3aR335H variant targeted to motoneurons increased the complexity of distal axonal branches of primary caudal zebrafish motoneurons (Issa et al., 2012). In both models though, no apparent cellular degeneration was observed. Instead cpce-specific expression of this pathogenic allele in cerebellar PCs elicited a robust degenerative phenotype, which is reminiscent of cerebellar atrophy observed in SCA13 patients, indicating that PCs represent a major vulnerable cell type for malfunctioning KCNC3. This underscores the importance of targeting pathogenic allele expression to the specific type of cell affected in a certain disease. Physiological studies in Xenopus oocytes and zebrafish motoneurons demonstrated a dominant-negative effect of both human KCNC3R420H and zebrafish Kcnc3aR335H on wild-type KCNC3 subunits that, like other KCNC/Kv3 family members, form tetrameric channels (Waters et al., 2006; Mock et al., 2010; Minassian et al., 2012). Indeed, incorporation of KCNC3R420H into the channel reduced the excitability of fast-spiking motoneurons and impaired startle reflex in zebrafish (Issa et al., 2011). The design of our PC-specific vector will allow the investigation of the cell biological mechanisms underlying the cytotoxicity of this mutant allele, including the change of electrophysiological properties (Issa et al., 2011) as well as an impaired intracellular trafficking as has been recently suggested (Gallego-Iradi et al., 2014; Khare et al., 2017).
A slight drawback of zebrafish cerebellar disease modeling though may arise from only a few differences in cerebellar cytoarchitecture compared with mammals. The major population of zebrafish PCs extends only short axons innervating eurydendroid cells distributed just underneath them or within the PC layer (Bae et al., 2009; Matsui et al., 2014). The equivalent efferent neurons in mammals are located in deep cerebellar nuclei and receive relatively long projections from PCs through the cerebellar white matter. Thus, for addressing diseases caused by axonal pathologies such as dying-back axonal degeneration, subpopulations of zebrafish PCs projecting long axons to the octaval nuclei (Bae et al., 2009; Matsui et al., 2014) are to be selected. In addition, the role of GABAergic inhibitory interneurons in zebrafish cerebellum remains to be elucidated. Especially, the basket cells synapsing onto PC somata and proximal dendrites have not been identified in zebrafish yet (Hashimoto and Hibi, 2012), the enhanced basket cell–PC connectivity though was reported to be causative of PC dysfunction, at least, in a SCA type 1 mice model (Edamakanti et al., 2018).
Even considering the limitations mentioned above, zebrafish larvae coexpressing fluorescent reporters or biosensors together with SCA causing alleles in PCs provide a powerful tool to achieve in vivo analysis of pathogenic mechanisms at high subcellular and temporal resolution in the native context of the cerebellar circuitry. Moreover, such transgenic zebrafish offer to validate compounds for mitigating the progression of neuronal degeneration thereby advancing basic/translational research to therapeutic approaches in a living vertebrate disease model.
Footnotes
This work was supported in part by funding from Deutsche Forschungsgemeinschaft Grants (KO1949/4-1 and 1949/7-1) to R.W.K. We thank Matthias Hammerschmidt, Wolfgang Driever, Darren Gilmour, Akihiko Koga, Jeffrey Mumm, Michael Rehli, Michael Lin, Yusuke Miyanari, Diane Papazian, and Olivier Gandrillon for generous gifts of plasmids, Richard Hawkes for anti-ZebrinII antibody, Jakob von Trotha and Thomas Weber for critical reading of the paper, Timo Fritsch for zebrafish care and mating, and all of the Köster Laboratory members for helpful discussions and technical assistance.
The authors declare no competing financial interests.
References
- Ahrens MB, Li JM, Orger MB, Robson DN, Schier AF, Engert F, Portugues R (2012) Brain-wide neuronal dynamics during motor adaptation in zebrafish. Nature 485:471–477. 10.1038/nature11057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakawa K, Kawakami K (2008) Targeted gene expression by the Gal4-UAS system in zebrafish. Dev Growth Differ 50:391–399. 10.1111/j.1440-169X.2008.01044.x [DOI] [PubMed] [Google Scholar]
- Aspatwar A, Tolvanen ME, Parkkila S (2010) Phylogeny and expression of carbonic anhydrase-related proteins. BMC Mol Biol 11:25. 10.1186/1471-2199-11-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspatwar A, Tolvanen ME, Jokitalo E, Parikka M, Ortutay C, Harjula SK, Rämet M, Vihinen M, Parkkila S (2013) Abnormal cerebellar development and ataxia in CARP VIII morphant zebrafish. Hum Mol Genet 22:417–432. 10.1093/hmg/dds438 [DOI] [PubMed] [Google Scholar]
- Bae YK, Kani S, Shimizu T, Tanabe K, Nojima H, Kimura Y, Higashijima S, Hibi M (2009) Anatomy of zebrafish cerebellum and screen for mutations affecting its development. Dev Biol 330:406–426. 10.1016/j.ydbio.2009.04.013 [DOI] [PubMed] [Google Scholar]
- Bajar BT, Wang ES, Lam AJ, Kim BB, Jacobs CL, Howe ES, Davidson MW, Lin MZ, Chu J (2016) Improving brightness and photostability of green and red fluorescent proteins for live cell imaging and FRET reporting. Sci Rep 6:20889. 10.1038/srep20889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajoghli B, Aghaallaei N, Heimbucher T, Czerny T (2004) An artificial promoter construct for heat-inducible misexpression during fish embryogenesis. Dev Biol 271:416–430. 10.1016/j.ydbio.2004.04.006 [DOI] [PubMed] [Google Scholar]
- Bell CC, Han V, Sawtell NB (2008) Cerebellum-like structures and their implications for cerebellar function. Annu Rev Neurosci 31:1–24. 10.1146/annurev.neuro.30.051606.094225 [DOI] [PubMed] [Google Scholar]
- Brown BD, Gentner B, Cantore A, Colleoni S, Amendola M, Zingale A, Baccarini A, Lazzari G, Galli C, Naldini L (2007) Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat Biotechnol 25:1457–1467. 10.1038/nbt1372 [DOI] [PubMed] [Google Scholar]
- Chang SY, Zagha E, Kwon ES, Ozaita A, Bobik M, Martone ME, Ellisman MH, Heintz N, Rudy B (2007) Distribution of Kv3.3 potassium channel subunits in distinct neuronal populations of mouse brain. J Comp Neurol 502:953–972. 10.1002/cne.21353 [DOI] [PubMed] [Google Scholar]
- Distel M, Wullimann MF, Köster RW (2009) Optimized Gal4 genetics for permanent gene expression mapping in zebrafish. Proc Natl Acad Sci U S A 106:13365–13370. 10.1073/pnas.0903060106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distel M, Hocking JC, Volkmann K, Köster RW (2010) The centrosome neither persistently leads migration nor determines the site of axonogenesis in migrating neurons in vivo. J Cell Biol 191:875–890. 10.1083/jcb.201004154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distel M, Hocking JC, Köster RW (2011) In vivo cell biology using Gal4-mediated multicolor subcellular labeling in zebrafish. Commun Integr Biol 4:336–339. 10.4161/cib.4.3.15037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edamakanti CR, Do J, Didonna A, Martina M, Opal P (2018) Mutant ataxin1 disrupts cerebellar development in spinocerebellar ataxia type 1. J Clin Invest 128:2252–2265. 10.1172/JCI96765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emelyanov A, Parinov S (2008) Mifepristone-inducible LexPR system to drive and control gene expression in transgenic zebrafish. Dev Biol 320:113–121. 10.1016/j.ydbio.2008.04.042 [DOI] [PubMed] [Google Scholar]
- Gallego-Iradi C, Bickford JS, Khare S, Hall A, Nick JA, Salmasinia D, Wawrowsky K, Bannykh S, Huynh DP, Rincon-Limas DE, Pulst SM, Nick HS, Fernandez-Funez P, Waters MF (2014) KCNC3R420H, a K+channel mutation causative in spinocerebellar ataxia 13 displays aberrant intracellular trafficking. Neurobiol Dis 71:270–279. 10.1016/j.nbd.2014.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A, Halpern ME (2016) Transcriptional regulation using the Q system in transgenic zebrafish. In: The zebrafish: genetics, genomics, and transcriptomics (Detrich HW, Westerfield M, Zon L, eds), pp 205–218. Amsterdam; Boston; Heidelberg: Elsevier. [Google Scholar]
- Goll MG, Anderson R, Stainier DY, Spradling AC, Halpern ME (2009) Transcriptional silencing and reactivation in transgenic zebrafish. Genetics 182:747–755. 10.1534/genetics.109.102079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamling KR, Tobias ZJ, Weissman TA (2015) Mapping the development of cerebellar Purkinje cells in zebrafish. Dev Neurobiol 75:1174–1188. 10.1002/dneu.22275 [DOI] [PubMed] [Google Scholar]
- Harmon TC, Magaram U, McLean DL, Raman IM (2017) Distinct responses of Purkinje neurons and roles of simple spikes during associative motor learning in larval zebrafish. eLife 6:e22537. 10.7554/eLife.22537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M, Hibi M (2012) Development and evolution of cerebellar neural circuits. Dev Growth Differ 54:373–389. 10.1111/j.1440-169X.2012.01348.x [DOI] [PubMed] [Google Scholar]
- Hekman KE, Gomez CM (2015) The autosomal dominant spinocerebellar ataxias: emerging mechanistic themes suggest pervasive Purkinje cell vulnerability. J Neurol Neurosurg Psychiatry 86:554–561. 10.1136/jnnp-2014-308421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai H, Iizuka A (2011) Recent developments in gene therapy research targeted to cerebellar disorders. In: Gene therapy applications (Kang C, ed), pp 401–422. Rijeka, Croatia: InTech. [Google Scholar]
- Hocking JC, Distel M, Köster RW (2013) Studying cellular and subcellular dynamics in the developing zebrafish nervous system. Exp Neurol 242:1–10. 10.1016/j.expneurol.2012.03.009 [DOI] [PubMed] [Google Scholar]
- Hsieh JY, Ulrich B, Issa FA, Wan J, Papazian DM (2014) Rapid development of Purkinje cell excitability, functional cerebellar circuit, and afferent sensory input to cerebellum in zebrafish. Front Neural Circuits 8:147. 10.3389/fncir.2014.00147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang MS, Wang TK, Liu YW, Li YT, Chi TH, Chou CW, Hsieh M (2014) Roles of carbonic anhydrase 8 in neuronal cells and zebrafish. Biochim Biophys Acta 1840:2829–2842. 10.1016/j.bbagen.2014.04.017 [DOI] [PubMed] [Google Scholar]
- Huang YY, Neuhauss SC (2008) The optokinetic response in zebrafish and its applications. Front Biosci 13:1899–1916. 10.2741/2810 [DOI] [PubMed] [Google Scholar]
- Irie T, Matsuzaki Y, Sekino Y, Hirai H (2014) Kv3.3 channels harbouring a mutation of spinocerebellar ataxia type 13 alter excitability and induce cell death in cultured cerebellar Purkinje cells. J Physiol 592:229–247. 10.1113/jphysiol.2013.264309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa FA, Mazzochi C, Mock AF, Papazian DM (2011) Spinocerebellar ataxia type 13 mutant potassium channel alters neuronal excitability and causes locomotor deficits in zebrafish. J Neurosci 31:6831–6841. 10.1523/JNEUROSCI.6572-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa FA, Mock AF, Sagasti A, Papazian DM (2012) Spinocerebellar ataxia type 13 mutation that is associated with disease onset in infancy disrupts axonal pathfinding during neuronal development. Dis Model Mech 5:921–929. 10.1242/dmm.010157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M. (2006) Cerebellar circuitry as a neuronal machine. Prog Neurobiol 78:272–303. 10.1016/j.pneurobio.2006.02.006 [DOI] [PubMed] [Google Scholar]
- Jörntell H. (2017) Cerebellar physiology: links between microcircuitry properties and sensorimotor functions. J Physiol 595:11–27. 10.1113/JP272769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczmarek LK, Zhang Y (2017) Kv3 channels: enablers of rapid firing, neurotransmitter release, and neuronal endurance. Physiol Rev 97:1431–1468. 10.1152/physrev.00002.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kani S, Bae YK, Shimizu T, Tanabe K, Satou C, Parsons MJ, Scott E, Higashijima SI, Hibi M (2010) Proneural gene-linked neurogenesis in zebrafish cerebellum. Dev Biol 343:1–17. 10.1016/j.ydbio.2010.03.024 [DOI] [PubMed] [Google Scholar]
- Kaslin J, Kroehne V, Benato F, Argenton F, Brand M (2013) Development and specification of cerebellar stem and progenitor cells in zebrafish: from embryo to adult. Neural Dev 8:9. 10.1186/1749-8104-8-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K. (1990) Sequence of a novel carbonic anhydrase-related polypeptide and its exclusive presence in Purkinje cells. FEBS Lett 271:137–140. 10.1016/0014-5793(90)80390-5 [DOI] [PubMed] [Google Scholar]
- Kawakami K. (2007) Tol2: a versatile gene transfer vector in vertebrates. Genome Biol 8:S7. 10.1186/gb-2007-8-s1-s7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare S, Nick JA, Zhang Y, Galeano K, Butler B, Khoshbouei H, Rayaprolu S, Hathorn T, Ranum LPW, Smithson L, Golde TE, Paucar M, Morse R, Raff M, Simon J, Nordenskjöld M, Wirdefeldt K, Rincon-Limas DE, Lewis J, Kaczmarek LK, et al. (2017) A KCNC3 mutation causes a neurodevelopmental, non-progressive SCA13 subtype associated with dominant negative effects and aberrant EGFR trafficking. PLoS One 12:e0173565. 10.1371/journal.pone.0173565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Lee SR, Li LH, Park HJ, Park JH, Lee KY, Kim MK, Shin BA, Choi SY (2011) High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PloS One 6:e18556. 10.1371/journal.pone.0018556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug M, Rehli M (2006) Functional analysis of promoter CpG methylation using a CpG-free luciferase reporter vector. Epigenetics 1:127–130. 10.4161/epi.1.3.3327 [DOI] [PubMed] [Google Scholar]
- Knogler LD, Markov DA, Dragomir EI, Štih V, Portugues R (2017) Sensorimotor representations in cerebellar granule cells in larval zebrafish are dense, spatially organized, and non-temporally patterned. Curr Biol 27:1288–1302. 10.1016/j.cub.2017.03.029 [DOI] [PubMed] [Google Scholar]
- Koga A, Cheah FS, Hamaguchi S, Yeo GH, Chong SS (2008) Germline transgenesis of zebrafish using the medaka Tol1 transposon system. Dev Dyn 237:2466–2474. 10.1002/dvdy.21688 [DOI] [PubMed] [Google Scholar]
- Köster RW, Fraser SE (2001a) Direct imaging of in vivo neuronal migration in the developing cerebellum. Curr Biol 11:1858–1863. 10.1016/S0960-9822(01)00585-1 [DOI] [PubMed] [Google Scholar]
- Köster RW, Fraser SE (2001b) Tracing transgene expression in living zebrafish embryos. Dev Biol 233:329–346. 10.1006/dbio.2001.0242 [DOI] [PubMed] [Google Scholar]
- Lauter G, Söll I, Hauptmann G (2011a) Multicolor fluorescent in situ hybridization to define abutting and overlapping gene expression in the embryonic zebrafish brain. Neural Dev 6:10. 10.1186/1749-8104-6-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauter G, Söll I, Hauptmann G (2011b) Two-color fluorescent in situ hybridization in the embryonic zebrafish brain using differential detection systems. BMC Dev Biol 11:43. 10.1186/1471-213X-11-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG (2001) A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73:56–65. 10.1006/geno.2000.6451 [DOI] [PubMed] [Google Scholar]
- Matsui H, Namikawa K, Babaryka A, Köster RW (2014) Functional regionalization of the teleost cerebellum analyzed in vivo. Proc Natl Acad Sci U S A 111:11846–11851. 10.1073/pnas.1403105111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto S, Konishi H, Maeda R, Kiryu-Seo S, Kiyama H (2012) Expression analysis of the regenerating gene (Reg) family members Reg-IIIβ and Reg-IIIγ in the mouse during development. J Comp Neurol 520:479–494. 10.1002/cne.22705 [DOI] [PubMed] [Google Scholar]
- Mejia-Pous C, Viñuelas J, Faure C, Koszela J, Kawakami K, Takahashi Y, Gandrillon O (2009) A combination of transposable elements and magnetic cell sorting provides a very efficient transgenesis system for chicken primary erythroid progenitors. BMC Biotechnol 9:81. 10.1186/1472-6750-9-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minassian NA, Lin MC, Papazian DM (2012) Altered Kv3.3 channel gating in early-onset spinocerebellar ataxia type 13. J Physiol 590:1599–1614. 10.1113/jphysiol.2012.228205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyanari Y, Ziegler-Birling C, Torres-Padilla ME (2013) Live visualization of chromatin dynamics with fluorescent TALEs. Nat Struct Mol Biol 20:1321–1324. 10.1038/nsmb.2680 [DOI] [PubMed] [Google Scholar]
- Mock AF, Richardson JL, Hsieh JY, Rinetti G, Papazian DM (2010) Functional effects of spinocerebellar ataxia type 13 mutations are conserved in zebrafish Kv3.3 channels. BMC Neurosci 11:99. 10.1186/1471-2202-11-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta K, Matsuzaki Y, Konno A, Hirai H (2017) Minimal Purkinje cell-specific PCP2/L7 promoter virally available for rodents and non-human primates. Mol Ther Methods Clin Dev 6:159–170. 10.1016/j.omtm.2017.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nógrádi A, Jonsson N, Walker R, Caddy K, Carter N, Kelly C (1997) Carbonic anhydrase II and carbonic anhydrase-related protein in the cerebellar cortex of normal and lurcher mice. Brain Res Dev Brain Res 98:91–101. 10.1016/S0165-3806(96)00177-0 [DOI] [PubMed] [Google Scholar]
- Portugues R, Feierstein CE, Engert F, Orger MB (2014) Whole-brain activity maps reveal stereotyped, distributed networks for visuomotor behavior. Neuron 81:1328–1343. 10.1016/j.neuron.2014.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi M, Perez-Lloret S, Doldan L, Cerquetti D, Balej J, Millar Vernetti P, Hawkes H, Cammarota A, Merello M (2014) Autosomal dominant cerebellar ataxias: a systematic review of clinical features. Eur J Neurol 21:607–615. 10.1111/ene.12350 [DOI] [PubMed] [Google Scholar]
- Rudy B, Kentros C, Weiser M, Fruhling D, Serodio P, Vega-Saenz de Miera E, Ellisman MH, Pollock JA, Baker H (1992) Region-specific expression of a K+ channel gene in brain. Proc Natl Acad Sci U S A 89:4603–4607. 10.1073/pnas.89.10.4603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassen WA, Lehne F, Russo G, Wargenau S, Dübel S, Köster RW (2017) Embryonic zebrafish primary cell culture for transfection and live cellular and subcellular imaging. Dev Biol 430:18–31. 10.1016/j.ydbio.2017.07.014 [DOI] [PubMed] [Google Scholar]
- Sengupta M, Thirumalai V (2015) AMPA receptor mediated synaptic excitation drives state-dependent bursting in Purkinje neurons of zebrafish larvae. eLife 4:e09158. 10.7554/eLife.09158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng M, Sala C (2001) PDZ domains and the organization of supramolecular complexes. Annu Rev Neurosci 24:1–29. 10.1146/annurev.neuro.24.1.1 [DOI] [PubMed] [Google Scholar]
- Sługocka A, Wiaderkiewicz J, Barski JJ (2017) Genetic targeting in cerebellar Purkinje cells: an update. Cerebellum 16:191–202. 10.1007/s12311-016-0770-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevanin G, Dürr A (2012) Spinocerebellar ataxia 13 and 25. Handb Clin Neurol 103:549–553. 10.1016/B978-0-444-51892-7.00035-8 [DOI] [PubMed] [Google Scholar]
- Stoppini L, Buchs PA, Muller D (1991) A simple method for organotypic cultures of nervous tissue. J Neurosci Methods 37:173–182. 10.1016/0165-0270(91)90128-M [DOI] [PubMed] [Google Scholar]
- Subach OM, Cranfill PJ, Davidson MW, Verkhusha VV (2011) An enhanced monomeric blue fluorescent protein with the high chemical stability of the chromophore. PLoS One 6:e28674. 10.1371/journal.pone.0028674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suster ML, Sumiyama K, Kawakami K (2009) Transposon-mediated BAC transgenesis in zebrafish and mice. BMC Genomics 10:477. 10.1186/1471-2164-10-477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi M, Yamaguchi S, Sakakibara Y, Hayashi T, Matsuda K, Hara Y, Tanegashima C, Shimizu T, Kuraku S, Hibi M (2017) Gene expression profiling of granule cells and Purkinje cells in the zebrafish cerebellum. J Comp Neurol 525:1558–1585. 10.1002/cne.24114 [DOI] [PubMed] [Google Scholar]
- Tanabe K, Kani S, Shimizu T, Bae YK, Abe T, Hibi M (2010) Atypical protein kinase C regulates primary dendrite specification of cerebellar Purkinje cells by localizing Golgi apparatus. J Neurosci 30:16983–16992. 10.1523/JNEUROSCI.3352-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theisen U, Hennig C, Ring T, Schnabel R, Köster RW (2018) Neurotransmitter-mediated activity spatially controls neuronal migration in the zebrafish cerebellum. PLoS Biol 16:e2002226. 10.1371/journal.pbio.2002226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urasaki A, Morvan G, Kawakami K (2006) Functional dissection of the Tol2 transposable element identified the minimal cis-sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics 174:639–649. 10.1534/genetics.106.060244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MF, Pulst SM (2008) Sca13. Cerebellum 7:165–169. 10.1007/s12311-008-0039-7 [DOI] [PubMed] [Google Scholar]
- Waters MF, Minassian NA, Stevanin G, Figueroa KP, Bannister JP, Nolte D, Mock AF, Evidente VG, Fee DB, Müller U, Dürr A, Brice A, Papazian DM, Pulst SM (2006) Mutations in voltage-gated potassium channel KCNC3 cause degenerative and developmental central nervous system phenotypes. Nat Genet 38:447–451. 10.1038/ng1758 [DOI] [PubMed] [Google Scholar]
- Weber T, Namikawa K, Winter B, Müller-Brown K, Kühn R, Wurst W, Köster RW (2016) Caspase-mediated apoptosis induction in zebrafish cerebellar Purkinje neurons. Development 143:4279–4287. 10.1242/dev.122721 [DOI] [PubMed] [Google Scholar]
- Weiser M, Vega-Saenz de Miera E, Kentros C, Moreno H, Franzen L, Hillman D, Baker H, Rudy B (1994) Differential expression of shaw-related K+ channels in the rat central nervous system. J Neurosci 14:949–972. 10.1523/JNEUROSCI.14-03-00949.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. (2007) The zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio), Ed 5 Eugene: University of Oregon. [Google Scholar]
- Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH (2005) MicroRNA expression in zebrafish embryonic development. Science 309:310–311. 10.1126/science.1114519 [DOI] [PubMed] [Google Scholar]
- Xie X, Mathias JR, Smith MA, Walker SL, Teng Y, Distel M, Köster RW, Sirotkin HI, Saxena MT, Mumm JS (2012) Silencer-delimited transgenesis: NRSE/RE1 sequences promote neural-specific transgene expression in a NRSF/REST-dependent manner. BMC Biol 10:93. 10.1186/1741-7007-10-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagrebelsky M, Buffo A, Skerra A, Schwab ME, Strata P, Rossi F (1998) Retrograde regulation of growth-associated gene expression in adult rat Purkinje cells by myelin-associated neurite growth inhibitory proteins. J Neurosci 18:7912–7929. 10.1523/JNEUROSCI.18-19-07912.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Kaczmarek LK (2016) Kv3.3 potassium channels and spinocerebellar ataxia. J Physiol 594:4677–4684. 10.1113/JP271343 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Time-lapse imaging of the cerebellar region of larval zebrafish from 56 to 71.5 hpf.
In Tg(−7.5:GFP)bz12 cerebellum, initial appearance of green fluorescent PCs (green) occurs at 56 hpf. PCs increase in number and expand across the cerebellar surface. Imaging was carried out by recording z-stacks encompassing the same cerebellar region (177.22 μm thickness, 233 images in 13.027 μm steps) at 30 minutes intervals, and captured z-stacks were projected as maximum projection into a single plane for each time point and animated to replay at a speed of 2 frames per second.

A 3D reconstruction of the cerebellum of a double transgenic zebrafish labeled with membrane tagged Venus and nuclear localized mseCFP.
In Tg(2xcpce-E1b:KALNFB, he1.1:mTagBFP2)bz14; Tg(5xUAS:FMAVenus-2A-3xNLS-HA-mseCFP)bz8 larva at 7dpf, PCs are labelled with differently colored fluorescent proteins in the nucleus (cyan) and the cytoplasmic membrane (yellow) respectively. For 3D reconstruction of the cerebellum laser scanning confocal microscope z-stack imaging was performed by recording 197 images in 13.027 μm steps (total thickness: 149.63 μm). Subsequent reconstruction and animation were performed using the Leica LAS-AF software.

A 3D reconstruction of the cerebellum of a double transgenic zebrafish labeled with membrane tagged Venus and nuclear localized mseCFP.
In Tg(2xcpce-E1b:KALNFB, he1.1:mTagBFP2)bz14; Tg(5xUAS:FMAVenus-2A-3xNLS-HA-mseCFP)bz8 larva at 7dpf, PCs are labelled with differently colored fluorescent proteins in the nucleus (cyan) and the cytoplasmic membrane (yellow) respectively. For 3D reconstruction of the cerebellum laser scanning confocal microscope z-stack imaging was performed by recording 197 images in 13.027 μm steps (total thickness: 149.63 μm). Subsequent reconstruction and animation were performed using the Leica LAS-AF software.

kcnc3a gene structure and alternative splicing. Exon-intron structure of the zebrafish kcnc3a gene. Boxes represent exons, and black boxes indicate exons used to generate each splice variant. Currently, twelve predicted variants are deposited in GenBank as variants as follows: X1; XM_021470843, X2; XM_021470845, X3; XM_021470847, X4; XM_009299543, X5; XM_021470855, X6; XM_009299545, X7; XM_021470856, X8; XM_021470857, X9; XM_005164037, X10; XM_009299548, X11; XM_021470866, and X12; XM_009299549. The translation initiation codon for all variants is located in exon1. Exons 1, 2, and 3 are present in all splice isoforms to encode most of the protein coding sequence, whereas the subsequent exons 4, 6, 7, or 8 determine the C-terminal intracellular coding sequence of kcnc3a terminating with -WIPK, -PSIL, -VTNV, or –VLSF, respectively. Note that variants X2 and X3 partially lack exon7, whereas variant X10 partially lacks exon2. Download Figure 11-1, TIF file (459.3KB, tif)
An amino acid sequence represents a C-terminal intracellular domain of a zebrafish Kcnc3a protein generated from each splicing variant. The black line indicates a position of nucleotide sequence corresponding to exon3 shared by all transcripts, whereas red shows each exon sequence translated into a specific C-terminal intracellular domain. Only nucleotide sequences (indicated by small characters) close to the junction between exon3 and each following exon are presented. Note that an exon 6 used in isoform X1, 2, 3, 4, 5, and 10 encodes a C-terminal sequence terminating with PSIL (enclosed in boxes) conserved among vertebrate. This sequence matches the potential class I PDZ (PSD-95/Discs Large/ZO-1) binding motif (X-S/T-X-L/V) (Sheng, and Sala, 2001). * indicates a C-terminal amino acid. Download Figure 11-2, TIF file (509.8KB, tif)
Optokinetic response (OKR) of a 6.3 dpf larva expressing Kcnc3awt or Kcnc3aR335H in PCs.
This movie displays a representative optokinetic response (OKR) of a 6.3 dpf larva expressing Kcnc3awt or Kcnc3aR335H in PCs. The movie was recorded at video rate of 2 frames per second and replays in real time.