Abstract
Choroideremia (CHM) is an x-linked recessive chorioretinal dystrophy, with 30% caused by nonsense mutations in the CHM gene resulting in an in-frame premature termination codon (PTC). Nonsense-mediated mRNA decay (NMD) is the cell’s natural surveillance mechanism that detects and destroys PTC-containing transcripts, with UPF1 being the central NMD modulator. NMD efficiency can be variable amongst individuals with some transcripts escaping destruction, leading to the production of a truncated non-functional or partially functional protein. Nonsense suppression drugs, such as ataluren, target these transcripts and read-through the PTC, leading to the production of a full length functional protein. Patients with higher transcript levels are considered to respond better to these drugs, as more substrate is available for read-through. Using Quantitative reverse transcription PCR (RT-qPCR), we show that CHM mRNA expression in blood from nonsense mutation CHM patients is 2.8-fold lower than controls, and varies widely amongst patients, with 40% variation between those carrying the same UGA mutation [c.715 C>T; p.(R239*)]. These results indicate that although NMD machinery is at work, efficiency is highly variable and not wholly dependent on mutation position. No significant difference in CHM mRNA levels was seen between two patients’ fibroblasts and their induced pluripotent stem cell-derived retinal pigment epithelium. There was no correlation between CHM mRNA expression and genotype, phenotype or UPF1 transcript levels. NMD inhibition with caffeine was shown to restore CHM mRNA transcripts to near wild-type levels. Baseline mRNA levels may provide a prognostic indicator for response to nonsense suppression therapy, and caffeine may be a useful adjunct to enhance treatment efficacy where indicated.
Introduction
Choroideremia (CHM; MIM: 303100) is an x-linked recessive chorioretinal dystrophy that affects approximately 1 in 50 000–100 000 individuals (1). CHM is characterized by a progressive loss of vision, starting with night blindness in early childhood, followed by peripheral field loss and eventually leading to complete blindness in late middle age. CHM is caused by mutations in the CHM gene (MIM: 300390), located on chromosome Xq21.2, it spans ~150 kb and is composed of 15 exons. It encodes the ubiquitously expressed 653 amino acid protein, Rab Escort Protein 1 (REP1). REP1 is involved in intracellular trafficking of vesicles and post-translational modification of Rab-proteins.
Thirty percent of CHM cases are caused by nonsense mutations, resulting in an in-frame premature termination codon (PTC) (1). Nonsense-mediated mRNA decay (NMD) is the cell’s natural surveillance mechanism, which detects and destroys PTC-containing transcripts. Typically, PTCs found more than 50–55 nucleotides upstream of the last exon–exon junction are described as being marked for destruction (2). However, exceptions to this rule have been observed. For example, in T-cell receptor-β, PTCs located within 50 nucleotides of the last exon–exon junction are still degraded (3). In the case of the β-globin gene (MIM: 141900), PTCs in close proximity to an AUG codon evade NMD and trigger translation re-initiation (4). NMD is a complex multifactorial mechanism that is intrinsically linked to translation. UPF1 is the central NMD factor; it is an RNA dependent ATPase and an ATP-dependent RNA helicase that is recruited to mRNA and undergoes a cycle of phosphorylations and dephosphorylations (5). Although NMD is described primarily as a surveillance mechanism, it also plays an important role in the regulation of normal gene expression and response to cellular stress (5,6).
Some PTC-containing transcripts escape NMD, leading to the expression of a truncated partially functional or non-functional protein. Nonsense suppression drugs exploit these PTC-containing transcripts. They bind to the ribosomal subunit and increase the ability of a near cognate aminoacyl-tRNA to compete with the eukaryotic release factors for binding to the A-site. An amino acid is added to the growing polypeptide chain, effectively allowing ‘read-through’ of the PTC, leading to production of the full length functional protein (7). We have previously shown that small molecule drugs, PTC124 and PTC414, restore rep1/REP1 activity in the chmru848 zebrafish model and a patient CHMY42X fibroblast cell line (8), whereas it was less effective in CHMK248X fibroblasts and induced pluripotent stem cell (iPSC)-derived retinal pigment epithelium (RPE; 9).
It has been suggested that the response to nonsense suppression drugs is greater in patients with higher baseline transcripts, providing more substrate for drug action, as a result of lower NMD efficiency (10). NMD efficiency is known to be variable between individuals (11); however, it is not yet fully understood what governs these differences. Linde et al. (10) found patients with the same mutation, p.(W1282*), in the cystic fibrosis transmembrane conductance regulator gene (CFTR; MIM: 602421), had widely variable transcript levels, indicating that NMD is not entirely governed by PTC position alone.
Baseline mRNA levels may be used as prognostic indicators of treatment outcome and inhibition of the NMD pathway could be used as an adjunct to boost transcripts for nonsense suppression. Caffeine has been identified as an NMD inhibitor, due to its inhibitory action on SMG1 kinase activity (12). Ullrich’s disease (MIM: 254090) is a muscular dystrophy, caused by mutations in the collagen VI genes. Caffeine has been shown to rescue the phenotype in Ullrich’s disease fibroblasts, by increasing the level of collagen VI α2 mRNA and protein, resulting in efficient integration into the collagen VI triple helix (13). Co-administration of the NMD inhibitor NMDI-1 with the nonsense suppression drug, gentamicin, has been shown to restore full length protein in a model of Hurler syndrome (MIM: 607014; 14).
In preparation for a clinical trial with PTC124 (ataluren) for CHM, we examined NMD efficiency in nonsense mutation CHM patients, determining relative CHM and UPF1 mRNA transcript levels in blood, fibroblasts and iPSC-derived RPE. We have shown that NMD efficiency is variable in nonsense mutation CHM patients and does not correlate with genotype or phenotype. NMD inhibition increases CHM transcript levels and could be explored as an adjunct for the treatment of nonsense-mediated diseases.
Results
Variable CHM mRNA expression in patient whole blood
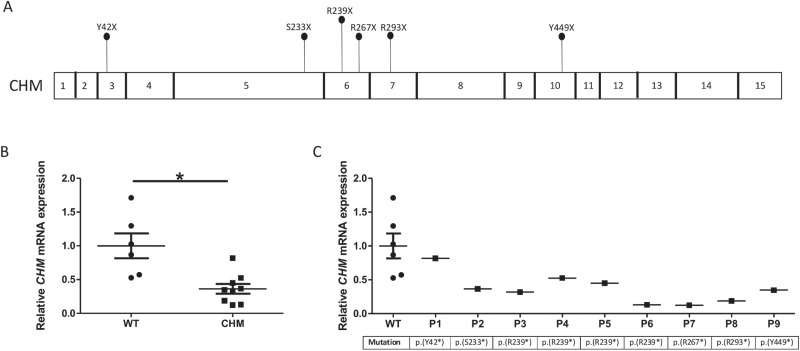
CHM transcript levels in whole blood from nine CHM male patients with nonsense mutations (mean age 49 ± 15 years) and six age- and sex-matched healthy controls (mean age 45 ± 15 years) were measured using Quantitative reverse transcription PCR (RT-qPCR). Patient mutations are shown in Figure 1A and Table 1. CHM transcript levels were reduced in all patients. Overall, mean CHM mRNA expression in patients was significantly reduced to 36.3 ± 7.3% of control (P = 0.002; Fig. 1B). A large variability in transcript levels amongst patients was seen, ranging from 12.5% to 81.2% of wild-type levels. These results indicate that a proportion of transcripts are escaping NMD. In our cohort, four patients have a c.715 C>T; p.(R239*) UGA mutation; in these patients, CHM transcript levels ranged from 13% to 52.6%. No correlation between CHM mRNA transcript level and genotype was found (Fig. 1C).
Figure 1.
CHM mRNA expression is significantly reduced in patients. (A) Schematic of the CHM gene. Patient mutations used in this study are labelled. (B) Relative CHM mRNA expression in patients analysed by RT-qPCR. Patients have a 2.8-fold lower expression compared to control (*P = 0.008). (C) Relative CHM mRNA expression in patients, ordered by mutation position. No correlation was found between CHM mRNA expression and genotype. Data expressed as mean ± SEM.
Table 1.
CHM male affected patients enrolled in this study
| Patient | Age | cDNA change | Amino acid change | Stop introduced | Exon | FAF area (mm2) |
|---|---|---|---|---|---|---|
| P1 | 28 | c.126 C>G | p.(Y42*) | UAG | 3 | 1.77 |
| P2 | 50 | c.698 C>G | p.(S233*) | UGA | 5 | 0.41 |
| P3 | 28 | c.715 C>T | p.(R239*) | UGA | 6 | 22.32 |
| P4 | 50 | c.715 C>T | p.(R239*) | UGA | 6 | 19.71 |
| P5 | 62 | c.715 C>T | p.(R239*) | UGA | 6 | 19.62 |
| P6 | 72 | c.715 C>T | p.(R239*) | UGA | 6 | 2.66 |
| P7 | 49 | c.799 C>T | p.(R267*) | UGA | 6 | 18.55 |
| P8 | 43 | c.877 C>T | p.(R293*) | UGA | 7 | 10.98 |
| P9 | 58 | c.1347 C>G | p.(Y449*) | UAG | 10 | 6.27 |
FAF analysed using the Heidelberg area tool, Heidelberg Engineering.
Variants correspond to RefSeq NM_000390.4.
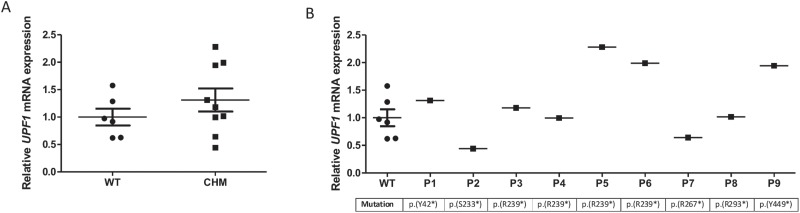
We next analysed the levels of UPF1 in patient blood to determine if there was a correlation between expression of genes encoding proteins involved in the NMD pathway and mRNA levels of CHM. There was no significant difference in UPF1 expression between patients and controls. However, there was a large variation in UPF1 mRNA expression amongst patients, ranging from 44.3% to 228.1%, compared to wild-type levels (Fig. 2). Except for P2 and P7, all other patients had higher UPF1 expression compared to controls. No correlation between CHM and UPF1 transcript levels was observed (r = 0.07).
Figure 2.
UPF1 mRNA expression is widely variable amongst patients. (A) Relative UPF1 mRNA expression in patients analysed by RT-qPCR. No significant difference was found between patients and controls. (B) Relative UPF1 mRNA expression in patients, ordered by mutation position. No correlation was found between UPF1 mRNA expression and genotype. Data expressed as mean ± SEM.
A genotype–phenotype correlation does not exist for CHM patients (15). In this population, the relationship between phenotype [age and fundus autofluorescence (FAF) size] and CHM transcript levels was investigated, but no statistically significant correlation was found (P = 0.21). Although it is important to note that in this population, there was also no correlation between age and FAF area (P = 0.53). So while the multivariate linear model did not suggest statistical significance, it did improve the correlation over any single factor correlation. Therefore, further investigation in a larger patient cohort may be needed to determine actual interaction.
Tissue-specific NMD variation
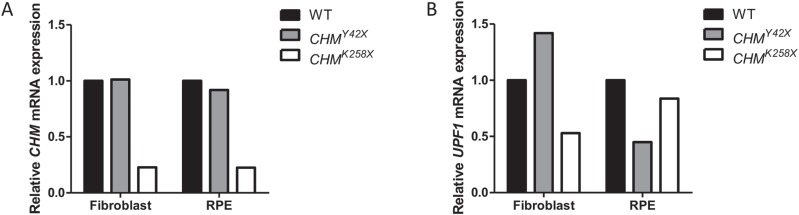
Previous studies have shown that NMD efficiency varies between different murine tissues (16). In order to investigate tissue-specific NMD variation, we have analysed CHM and UPF1 expression in fibroblasts and iPSC-derived RPE from two unrelated patients (i) p.(Y42*) and (ii) p.(K258*). CHM transcript levels in fibroblasts and iPSC-derived RPE for p.(Y42*) were 101 and 92% relative to an age-matched healthy control and for p.(K258*) were 22.8% and 22.6%, respectively (Fig. 3A). UPF1 transcript levels for p.(Y42*) were 142% and 45% and for p.(K258*) were 52.9% and 83.7%, respectively (Fig. 3B). Our results show that the CHM transcript levels in both cell types are similar for each patient; however, UPF1 expression varied considerably amongst the different tissues. CHM mRNA transcripts in p.(Y42*) are present at wild-type levels, indicating that this transcript is escaping NMD.
Figure 3.
No variation in NMD efficiency was found between cell types. Relative (A)CHM and (B)UPF1 mRNA expression in CHMY42X (grey) and CHMK258X (white) fibroblasts and iPSC-derived RPE.
NMD inhibition increases CHM mRNA expression in fibroblasts
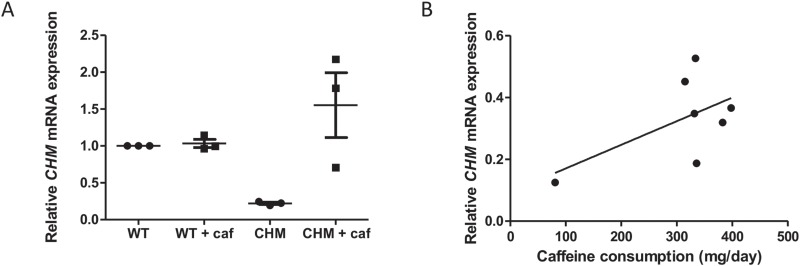
The effect of caffeine on CHM expression was tested in three independent and unrelated patient fibroblast cell lines, two with a p.(R270*) mutation and one with a p.(S190*) mutation. CHM transcript levels in the two p.(R270*) patients were 19.3% ± 3.9% and 24.6% ± 2.3%, and for p.(S190*) was 22.3% ± 2.1%. Overall, mean untreated CHM expression was 22.1% ± 1.5% (Fig. 4A). Treatment with 10 mm caffeine for 4 h increased CHM transcript levels in all cell lines, to a mean 155% ± 44% of wild-type levels, a 7-fold increase (P < 0.05; Fig. 4A). This confirms that active NMD is inhibited, leading to rescue of PTC-containing transcripts.
Figure 4.
NMD inhibition with caffeine increases CHM mRNA expression. (A) Effect of caffeine on CHM mRNA expression in three unrelated patient fibroblasts [p.(R270*), p.(R270*) and p.(S190*)]. Cells were treated with 10 mm caffeine for 4 h and mRNA expression analysed by RT-qPCR. Caffeine significantly increased CHM mRNA expression, compared to untreated cells (P < 0.05). Data expressed as mean ± SEM (n = 3). (B) Patients were asked to complete a food questionnaire, and the average daily caffeine intake was calculated. Correlation between caffeine consumption and relative CHM mRNA expression was analysed by Pearson’s correlation (r = 0.58). No significant correlation between patient caffeine consumption and CHM mRNA expression (P = 0.18) was observed.
To assess whether caffeine intake influenced whole blood CHM mRNA transcript levels, the average daily caffeine consumption from the FFQ was determined for each CHM patient. There was no significant difference in daily caffeine consumption between the CHM group (185.4 ± 28.9 mg/day) and the age-matched controls (177.1 ± 28.9 mg/day). The average caffeine intake for patients used in this study (excluding P1, who does not have an NMD-sensitive CHM variant) was 310.8 ± 40 mg/day. There was no sign of correlation between patient caffeine consumption and CHM mRNA expression in blood (r = 0.58, P = 0.18; Fig. 4B).
Discussion
In this study, we have shown that patients with nonsense mutations in the CHM gene have 2.8-fold lower levels of CHM transcripts compared to controls, indicating that the transcripts are subject to degradation by NMD, but also a proportion of transcripts are escaping destruction. All patient mutations are positioned at least 55 nucleotides upstream of the final exon–exon junction, and therefore likely substrates of NMD. A wide variation in CHM expression was observed amongst patients, which did not correlate with genotype, suggesting other factors may be responsible for NMD efficiency. UPF1 expression, a key NMD facilitator, was also highly variable with no correlation found with CHM transcript levels. Linde et al. (17) found NMD efficiency to be variable between different cell types transfected with the same PTC-containing genes, suggesting NMD efficiency to be an inherent characteristic of the cell. However, in this study similar levels of CHM transcripts were found between two different tissue-specific cell types in two unrelated patients. For this to be a useful patient screening tool for potential response to nonsense suppression, further validation with a greater number of tissues from more patients would be beneficial. In contrast, corresponding UPF1 transcript levels did vary between different cell types. This is consistent with the study by Zetoune et al. (16), which showed NMD efficiency varies between different murine tissues and does not correlate with UPF1 expression or expression of any other genes encoding proteins involved in NMD.
Our cohort of patients did not show a correlation between disease severity and mRNA expression, although investigation in a larger patient cohort would increase sensitivity. The role of NMD in the regulation of normal gene expression is becoming more apparent. Investigation in non-disease individuals may be valuable to elucidate the causes of variation in NMD efficiency.
In patient 1, [P1; p.(Y42*)], CHM mRNA levels are comparable to wild-type levels in all cell types, indicating that this transcript is escaping NMD. As this mutation is present near the start of the coding sequence, the AUG-proximity effect may be in play here. In a study of 10 000 human tumours, Lindeboom et al. (18) found that NMD efficiency is significantly reduced in transcripts with PTCs located within the first 200 nucleotides of the start codon. In P1, the PTC is located 126 nucleotides from the start codon. An AUG-proximal PTC transcript can evade NMD and trigger translation re-initiation at a downstream codon. Pereira et al. (4) showed the boundary for translation re-initiation in the β-globin mRNA is between codons 23 and 25. In the CHM transcript, the next AUG is present at codon 149, which is unlikely to trigger translation re-initiation. However, NMD efficiency is still lower in AUG-proximal PTC transcripts, even in the absence of a downstream start codon. An alternative mechanism suggests that the transcript is stabilized by interaction of cytoplasmic poly(A) binding protein 1 (PABPC1) and the termination complex. In the short open reading frame of an AUG-proximal PTC transcript, PABPC1 interacting with eukaryotic initiation factor 4G is bought into close proximity with the termination complex at the PTC, leading to an effective termination event, thereby suppressing NMD (19).
Linde et al. (10) showed in a group of cystic fibrosis patients with the same p.(W1282*) mutation, patients had varying levels of baseline CFTR transcripts, and those with higher levels responded better to the nonsense suppression drug gentamicin. A number of other studies have shown that response to nonsense suppression drugs is highly variable (10,20,21). We have previously shown that treatment with ataluren restores prenylation activity in CHMY42X fibroblasts (8). However, in the study by Torriano et al. (9), no significant rescue was observed in CHMK258X fibroblasts and iPSC-derived RPE, which had lower CHM transcript levels (~20%). In preparation for a phase 2 clinical trial with ataluren for CHM, levels of baseline mRNA may provide a prognostic indicator of response to treatment. Patients with lower CHM transcript levels may benefit from NMD inhibition to increase baseline levels, allowing for more effective read-through. Treatment with caffeine restored CHM mRNA expression to wild-type levels in treated cells. Further clinical studies assessing the direct effect of caffeine on NMD and resultant CHM mRNA levels following consumption would provide further evidence of therapeutic benefit. However, caution would be required due to the widespread side effects on the body and interactions with many other pathways, and so potentially local delivery would be more applicable. Keeling et al. (14) showed in the mucopolysaccharidosis I-Hurler mouse model, co-administration of the NMD specific inhibitor, NMDI-1, together with gentamicin increased enzyme activity compared to gentamicin alone. Recently, an analogue of NMDI-1 called VG1 has also been developed, using a more efficient process (22). The FDA-approved drug amlexanox has been shown to have a dual function by inhibiting NMD and promoting synthesis of full length protein though nonsense suppression (23).
Together, our results show that NMD efficiencies are highly variable in CHM patients, with no correlation with genotype or phenotype. Levels of transcripts did not vary between tissues; hence, measuring baseline mRNA levels in patients with nonsense mutations, if accessible, may act to guide choice of nonsense suppression therapy with or without NMD inhibitor adjuncts.
Materials and Methods
Clinical methods
The study protocol adhered to the tenets of the Declaration of Helsinki and received approval from the NRES Committee London Ethics Committee (REC12/LO/0489) and (REC12/LO/0141). Written, informed consent was obtained from all participants prior to their inclusion in this study.
Clinical data were collected from nine male subjects confirmed to have pathogenic variants in the CHM gene, (Table 1) including age, ethnicity and visual acuity. All retinal imaging was collected as part of an on-going natural history study of CHM patients for future gene augmentation therapies tailored to nonsense-mediated disease. Previous work has shown delineation of the central hyper-autofluorescent retinal island area to be the most repeatable metric to measure disease state (24). Images were acquired using short wavelength (488 nm) autofluorescence on the Heidelberg Spectralis confocal scanning laser ophthalmoscope (Heidelberg Engineering, Heidelberg, Germany), and area was measured using the vendor software (Heidelberg Eye Explorer Region Finder Version 2.4.3.0).
Blood collection and RNA extraction
Whole blood (2.5 ml) was collected from the nine CHM affected male patients and six age- and sex-matched healthy controls (Table 1) in PAXgene Blood RNA tubes (QIAGEN, Manchester, UK). These were incubated at room temperature for 2 h to lyse blood cells. Tubes were then transferred to −20°C until frozen and subsequently stored at −80°C, until further processing. Prior to RNA extraction, tubes were thawed to room temperature. RNA from blood was extracted using the PAXgene Blood RNA kit (QIAGEN), according to the manufacturer’s instructions.
Fibroblast cell culture and dosing
Three patient and one healthy control fibroblast lines were obtained from Coriell Biorepository or cultured from skin biopsies, as previously described (8; cell line details are listed in Table 2). Cells were maintained in Dulbecco’s Modified Eagles Medium, high glucose, supplemented with 15% FBS and Penicillin/Streptomycin (ThermoFisher Scientific, Paisley, UK). For cell collection, a T75 confluent flask was pelleted at 200g for 5 min; the pellet was washed in PBS at 200g for 5 min at 4°C twice. All liquid was removed and the pellet snap frozen in an alcohol/dry ice bath and stored at −80°C until further processing. RNA extraction from cells was carried out using RNAeasy Mini Kit (QIAGEN), following the manufacturer’s instructions. For dosing experiments, fibroblasts were plated in 24 well plates at a seeding density of 100 000 cells per well for 48 h. Media were replaced with antibiotic free growth medium, containing 10 mm caffeine (Sigma-Aldrich, Dorset, UK). Cells were incubated at 37°C for 4 h. RNA extraction was carried out using RNAeasy Mini Kit (QIAGEN), following the manufacturer’s instructions. Three independent experiments were performed.
Table 2.
Fibroblast cell lines used in this study
| Age | cDNA change | Amino acid change | Stop introduced | Exon | Coriell ID |
|---|---|---|---|---|---|
| 43 | Healthy control | GM23963 | |||
| 48 | c.569 C>G | p.(S190*) | UGA | 5 | GM25421 |
| 20 | c.808 C>T | p.(R270*) | UGA | 6 | GM25383 |
| 61 | c.808 C>T | p.(R270*) | UGA | 6 | GM25386 |
| 28 | c.126 C>G | p.(Y42*) | UAG | 3 | Patient skin biopsy |
| 10 | c.772 A>T | p.(K258*) | UAA | 6 | Torriano et al. (9) |
Cells were obtained from Coriell Institute for Medical Research or cultured from patient skin biopsies.
Fibroblast reprogramming to iPSCs and generation of RPE
Wild-type and CHMY42X fibroblasts were reprogrammed to iPSCs, using integration free episomal vectors, and subsequently differentiated into RPE, as previously described (25). RT-PCR of RPE-specific marker genes and immunohistochemistry are shown in supplementary data (Supplementary Material, Fig. S1). RNA extraction was carried out using RNAeasy Mini Kit (QIAGEN), following manufacturer’s instructions. CHMK258X fibroblasts and iPSC-derived RPE, as well as the corresponding RNA, were generated as previously described (9).
RT-qPCR
cDNA was synthesized from 500 ng of RNA using the Superscript III First Strand cDNA synthesis kit (Invitrogen), according to the manufacturer’s instructions. Transcript levels were analysed using SYBR Green MasterMix (ThermoFisher) on a StepOne Real-Time PCR system (Applied Biosystems, ThermoFisher, Paisley, UK), under standard cycling conditions. All samples were assayed in triplicate. Primers used for RT-qPCR are listed in Table 3. GAPDH was used as a reference gene. As the forward CHM primer overlapped the p.(Y42*) mutation in P1, the 5′ – CGTCAGACATCAGCAGGAGC (forward) and 5′ – GGATTTGGTGGAGGGGGACA (reverse) primers were used to analyse CHM transcript levels in P1 blood, fibroblasts and iPSC-derived RPE.
Table 3.
RT-qPCR primer sequences
| CHM forward | 5′ - AGAAGCTACTATGGAGGAAAC |
|---|---|
| CHM reverse | 5′ – TTCCTGGTATTCCTTTAGCC |
| UPF1 forward | 5′ – GCTGTCCCAGTATTAAAAGG |
| UPF1 reverse | 5′ - CAGTGGTGCTTCAGTTTTAG |
| GAPDH forward | 5′ - CTTTTGCGTCGCCAG |
| GAPDH reverse | 5′ - TTGATGGCAACAATATCCAC |
Patient caffeine consumption
A total of 25 CHM male patients and 25 age- and sex-matched control subjects were asked to complete a food frequency questionnaire (FFQ) on their average consumption of various foods and drinks over the past 12 months. The validated FFQ comprised a list of 147 food items and participants were asked to indicate their usual consumption from one of nine frequency categories ranging from “never or less than once per month” to “six or more times per day.” (26). Individuals would have been excluded if their answers to >10 items had been left blank, but this was not true for any of the participants. The amount of caffeine in food and drink items was calculated using a database with composition values obtained from the USDA Food Composition Databases (Accessed October 2018). Specifically, using data derived from the USDA National Nutrient Database for Standard Reference Legacy Release (April 2018) and USDA Branded Food Products Database to enable average caffeine consumption (mg/day) to be calculated for each patient.
Statistical analysis
All data are expressed as mean ± SEM. Differences between control and patient groups were analysed by Mann–Whitney test. Relationship between CHM and UPF1 transcript levels were analysed by Pearson’s correlation. To assess the relationship between CHM mRNA expression and clinical phenotype, multivariate regression analysis for subject age, FAF area and mRNA level was undertaken (JMP13, Marlow, Buckinghamshire, UK). The effect of caffeine treatment on cells was analysed by Kruskal-Wallis analysis, followed by Dunn’s multiple comparisons test. Correlation between caffeine consumption and CHM mRNA levels was analysed by Pearson’s correlation. A P-value of <0.05 was considered significant.
Supplementary Material
Acknowledgements
We gratefully acknowledge the support of the National Institute for Health Research (NIHR) Biomedical Research Centre based at Moorfields Eye Hospital NHS Foundation Trust and UCL Institute of Ophthalmology.
Conflict of Interest statement. The authors declare no competing interests.
Funding
Choroideremia Research Foundation US, France Choroideremia, Fight for Sight UK, National Eye Research Centre Charity, Moorfields Eye Charity, Retina UK and the Wellcome Trust.
References
- 1. Moosajee M., Ramsden S.C., Black G.C., Seabra M.C. and Webster A.R. (2014) Clinical utility gene card for: choroideremia. Eur. J. Hum. Genet., 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nagy E. and Maquat L.E. (1998) A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem. Sci., 23, 198–199. [DOI] [PubMed] [Google Scholar]
- 3. Carter M.S., Li S. and Wilkinson M.F. (1996) A splicing-dependent regulatory mechanism that detects translation signals. EMBO J., 15, 5965–5975. [PMC free article] [PubMed] [Google Scholar]
- 4. Pereira F.J., Teixeira A., Kong J., Barbosa C., Silva A.L., Marques-Ramos A., Liebhaber S.A. and Romao L. (2015) Resistance of mRNAs with AUG-proximal nonsense mutations to nonsense-mediated decay reflects variables of mRNA structure and translational activity. Nucleic Acids Res., 43, 6528–6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hug N., Longman D. and Caceres J.F. (2016) Mechanism and regulation of the nonsense-mediated decay pathway. Nucleic Acids Res., 44, 1483–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nickless A., Bailis J.M. and You Z. (2017) Control of gene expression through the nonsense-mediated RNA decay pathway. Cell Biosci., 7, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Richardson R., Smart M., Tracey-White D., Webster A.R. and Moosajee M. (2017) Mechanism and evidence of nonsense suppression therapy for genetic eye disorders. Exp. Eye Res., 155, 24–37. [DOI] [PubMed] [Google Scholar]
- 8. Moosajee M., Tracey-White D., Smart M., Weetall M., Torriano S., Kalatzis V., Cruz L., Coffey P., Webster A.R. and Welch E. (2016) Functional rescue of REP1 following treatment with PTC124 and novel derivative PTC-414 in human choroideremia fibroblasts and the nonsense-mediated zebrafish model. Hum. Mol. Genet., 25, 3416–3431. [DOI] [PubMed] [Google Scholar]
- 9. Torriano S., Erkilic N., Baux D., Cereso N., De Luca V., Meunier I., Moosajee M., Roux A.F., Hamel C.P. and Kalatzis V. (2018) The effect of PTC124 on choroideremia fibroblasts and iPSC-derived RPE raises considerations for therapy. Sci. Rep., 8, 8234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Linde L., Boelz S., Nissim-Rafinia M., Oren Y.S., Wilschanski M., Yaacov Y., Virgilis D., Neu-Yilik G., Kulozik A.E., Kerem E. et al. (2007) Nonsense-mediated mRNA decay affects nonsense transcript levels and governs response of cystic fibrosis patients to gentamicin. J. Clin. Invest., 117, 683–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nguyen L.S., Wilkinson M.F. and Gecz J. (2014) Nonsense-mediated mRNA decay: inter-individual variability and human disease. Neurosci. Biobehav. Rev., 46, 175–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yamashita A., Ohnishi T., Kashima I., Taya Y. and Ohno S. (2001) Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev., 15, 2215–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Usuki F., Yamashita A., Higuchi I., Ohnishi T., Shiraishi T., Osame M. and Ohno S. (2004) Inhibition of nonsense-mediated mRNA decay rescues the phenotype in Ullrich's disease. Ann. Neurol., 55, 740–744. [DOI] [PubMed] [Google Scholar]
- 14. Keeling K.M., Wang D., Dai Y., Murugesan S., Chenna B., Clark J., Belakhov V., Kandasamy J., Velu S.E., Baasov T. et al. (2013) Attenuation of nonsense-mediated mRNA decay enhances in vivo nonsense suppression. PLoS One, 8, 1–0.e60478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Freund P.R., Sergeev Y.V. and MacDonald I.M. (2016) Analysis of a large choroideremia dataset does not suggest a preference for inclusion of certain genotypes in future trials of gene therapy. Mol. Genet. Genomic. Med., 4, 344–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zetoune A.B., Fontaniere S., Magnin D., Anczukow O., Buisson M., Zhang C.X. and Mazoyer S. (2008) Comparison of nonsense-mediated mRNA decay efficiency in various murine tissues. BMC Genet., 9, 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Linde L., Boelz S., Neu-Yilik G., Kulozik A.E. and Kerem B. (2007) The efficiency of nonsense-mediated mRNA decay is an inherent character and varies among different cells. Eur. J. Hum. Genet., 15, 1156–1162. [DOI] [PubMed] [Google Scholar]
- 18. Lindeboom R.G., Supek F. and Lehner B. (2016) The rules and impact of nonsense-mediated mRNA decay in human cancers. Nat. Genet., 48, 1112–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Peixeiro I., Inacio A., Barbosa C., Silva A.L., Liebhaber S.A. and Romao L. (2012) Interaction of PABPC1 with the translation initiation complex is critical to the NMD resistance of AUG-proximal nonsense mutations. Nucleic Acids Res., 40, 1160–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Floquet C., Hatin I., Rousset J.P. and Bidou L. (2012) Statistical analysis of readthrough levels for nonsense mutations in mammalian cells reveals a major determinant of response to gentamicin. PLoS Genet., 8, e1002608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kerem E., Hirawat S., Armoni S., Yaakov Y., Shoseyov D., Cohen M., Nissim-Rafinia M., Blau H., Rivlin J., Aviram M. et al. (2008) Effectiveness of PTC124 treatment of cystic fibrosis caused by nonsense mutations: a prospective phase II trial. Lancet, 372, 719–727. [DOI] [PubMed] [Google Scholar]
- 22. Gotham V.J., Hobbs M.C., Burgin R., Turton D., Smythe C. and Coldham I. (2016) Synthesis and activity of a novel inhibitor of nonsense-mediated mRNA decay. Org. Biomol. Chem., 14, 1559–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gonzalez-Hilarion S., Beghyn T., Jia J., Debreuck N., Berte G., Mamchaoui K., Mouly V., Gruenert D.C., Deprez B. and Lejeune F. (2012) Rescue of nonsense mutations by amlexanox in human cells. Orphanet J. Rare Dis., 7, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jolly J.K., Edwards T.L., Moules J., Groppe M., Downes S.M. and MacLaren R.E. (2016) A qualitative and quantitative assessment of fundus autofluorescence patterns in patients with choroideremia. Invest. Ophthalmol. Vis. Sci., 57, 4498–4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schwarz N., Carr A.J., Lane A., Moeller F., Chen L.L., Aguila M., Nommiste B., Muthiah M.N., Kanuga N., Wolfrum U. et al. (2015) Translational read-through of the RP2 Arg120stop mutation in patient iPSC-derived retinal pigment epithelium cells. Hum. Mol. Genet., 24, 972–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Welch A.A., Luben R., Khaw K.T. and Bingham S.A. (2005) The CAFE computer program for nutritional analysis of the EPIC-Norfolk food frequency questionnaire and identification of extreme nutrient values. J. Hum. Nutr. Diet., 18, 99–116. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.