Abstract
Trypanosoma brucei, a protist parasite that causes African trypanosomiasis or sleeping sickness, relies mainly on glycolysis for ATP production when in its mammalian host. Glycolysis occurs within a peroxisome-like organelle named the glycosome. Previous work from our laboratory reported the presence of significant amounts of inorganic polyphosphate (polyP), a polymer of three to hundreds of orthophosphate units, in the glycosomes and nucleoli of T. brucei. In this work, we identified and characterized the activity of two Nudix hydrolases (NHs), T. brucei Nudix hydrolase (TbNH) 2 and TbNH4, one located in the glycosomes and the other in the cytosol and nucleus, respectively, which can degrade polyP. We found that TbNH2 is an exopolyphosphatase with higher activity on short chain polyP, while TbNH4 is an endo- and exopolyphosphatase that has similar activity on polyP of various chain sizes. Both enzymes have higher activity at around pH 8.0. We also found that only TbNH2 can dephosphorylate ATP and ADP but with lower affinity than for polyP. Our results suggest that NHs can participate in polyP homeostasis and therefore may help control polyP levels in glycosomes, cytosol and nuclei of T. brucei.
Keywords: glycosome, inorganic polyphosphates, nucleus, polyphosphatase, trypanosomes
Introduction
Inorganic polyphosphate (polyP) is a linear polymer of phosphate (orthophosphate [Pi]) that can range from three to hundreds of Pi units. PolyP has been found in most species investigated, from bacteria to animals [1]. In Trypanosoma brucei, one of the agents of African trypanosomiasis, polyP is synthesized by the vacuolar transporter chaperone (VTC) complex [2], which is located in the acidocalcisome [3], an acidic organelle that stores calcium and other cations together with Pi, inorganic pyrophosphate (PPi), and polyP [4]. Hydrolysis of polyP with release of Pi occurs by the activity of a cytosolic exopolyphosphatase (PPX) [5] and by the PPX activity of the acidocalcisome vacuolar soluble pyrophosphatase (VSP) [6–9]. No endopolyphosphatase (PPN) activity, which cleaves internal phosphoanhydride bonds generating shorter polyP molecules, like the yeast PPN1 (YDR452W) [10,11] and PPN2 (YNL217W) [12], has yet been reported in trypanosomatids.
A third endopolyphosphatase that has been described in yeast is diadenosine and diphosphoinositol polyphosphate phosphohydrolase (Ddp1) (YOR163W) [13], initially described as diadenosine hexaphosphate and diphosphoinositol polyphosphate hydrolase (DIPP) [14,15]. Ddp1 is localized in the cytosol and nucleus [16] and belongs to the Nudix (nucleoside diphosphate-linked moiety X) hydrolase family, which is characterized by a MuT motif or Nudix box of 23 amino acids (GX5EX7REUXEEXGU) where U is a bulky aliphatic residue and X is any amino acid [17]. The glutamic acid residues within the Nudix box binds to divalent cations cofactors like Mg2+ and Mn2+ [18]. Ddp1 and its human homologs DIPPs, DIPP1, DIPP2 and DIPP3, have polyP endopolyphosphatase activity [13]. Interestingly, the yeast and human enzymes also have 5-diphosphoinositol pentakisphosphate (5-IP7) hydrolase activity that helps to regulate inositol pyrophosphate signaling [13].
The Nudix superfamily (Pfam PF00293) is found in archaea, bacteria, eukaryotes and viruses and includes pyrophosphohydrolases of nucleotide sugars and alcohols, nucleoside and deoxynucleoside triphosphates ([d]NTPs), dinucleoside polyphosphates, dinucleotide coenzymes and capped RNAs [19]. Trypanosoma brucei has five Nudix proteins, of which two have been characterized. Nudix Hydrolase 1 (T. brucei Nudix hydrolase [TbNH] 1 or MERS1) binds to the RNA–editing complex and helps stabilizing edited mRNAs [20]. Nudix Hydrolase 4 (TbNH4 or TbDcp2) is a mRNA de-capping enzyme that removes the 5′ cap from processed mRNAs [21].
In this work, we investigated the ability of T. brucei Nudix hydrolases (NHs) to hydrolyze polyP and 5-IP7. We identified two polyphosphatases, TbNH2 and TbNH4, with polyP exopolyphosphatase and endopolyphosphatase activities, respectively. TbNH4 is the first trypanosome endopolyphosphatase class of enzyme described. TbNH2 localizes to the glycosomes while TbNH4 localizes to the cytosol and nucleus, results which are consistent with our recent demonstration of polyP in the glycosomes and nucleoli of these parasites [22]. None of the enzymes hydrolyzes 5-IP7.
Materials and methods
Materials
Chemically synthesized 5-diphosphoinositol pentakisphosphate [23] was provided by Dr. Henning Jessen, Albert-Ludwigs-University of Freiburg, Germany. PolyP60 was a gift from Dr. Toshikazu Shiba (RegeneTiss Inc., Tokyo, Japan). PolyP700 was purchased from Kerafast Inc. (Boston, MA, U.S.A.). The plasmid for expression of human DIPP was a gift from Dr. Dorothea Fiedler (Humboldt University of Berlin, Germany). Monoclonal antibody against phosphate pyruvate dikinase (PPDK) was a gift from Dr. Frédéric Bringaud (University of Bordeaux, France).
Cell cultures
Trypanosoma brucei procyclic form (PCF) Lister 427, 29-13 TetR/T7RNAP cell line was used. Procyclic cells were cultivated at 28°C in SDM-79 [24] supplemented with 10% heat-inactivated FBS and hemin (7.5 µg/ml). Drug concentrations used for selection and maintenance of procyclic cell lines were: hygromycin (50 µg/ml), G418 (15 µg/ml), and blasticidin S (5 µg/ml).
Methods
Cloning, primers, expression and SDS/PAGE
The sequences of TbNH1 (Tb927.11.15640), TbNH2 (Tb927.5.4350), TbNH3 (Tb927.11.9810), TbNH4 (Tb927.6.2670) and TbNH5 (Tb927.10.4680) were amplified from genomic DNA by PCR (Supplementary Table S1) and cloned in expression vector pET32 Ek/LIC (Novagen) following manufacturer instructions. Constructs inserts were verified by Sanger sequencing and then transformed in Escherichia coli BL21-CodonPlus (DE3). Protein expression was induced by addition of 0.1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) to bacterial cultures in Luria Bertani broth shaking for 2 h at 25°C. Culture was chilled on ice and bacteria harvested by centrifugation. Pellet was suspended in 20 mM Tris HCl, 150 mM NaCl, pH 7.4 with protease inhibitors (Sigma P8465) and sonicated on ice. Lysate was then centrifuged 15000 × g for 30 min and then filtered on 0.8 µm syringe filter units (Millipore). Protein purification was performed using Nickel column HIS-Select® Cartridges as recommended by manufacturer. Elution fractions with purified protein were dialyzed, replacing elution buffer for 300 mM NaCl, 200 mM Tris HCl, pH 7.4 with 20% glycerol. Expression was verified by SDS/PAGE followed by Coomassie blue staining and protein aliquots were stored at −80°C until further use. Protein concentration was determined using Pierce BCA Protein Assay Kit (Thermo Scientific) as instructed by manufacturer. Human DIPP was transformed, expressed and purified using same protocol described above.
Nudix hydrolases activity tests
Nudix activity assays were performed at 37°C using 50 mM NaCl, 40 mM Hepes buffer (pH 7.4, unless stated otherwise), 0.25 micromoles of polyP60 or 5 nanomoles of 5-diphosphoinositol pentakisphosphate (IP7), 6 mM MgCl2 or other specified cation and about 0.5 µg/ml of recombinant protein for 1 h or indicated time. For enzymatic reactions at different pHs, we used MES buffer for pH 5.5–6.5, Hepes for pH 7.0–8.0 and Tris-base for pH 8.5. Enzymatic reactions were stopped by addition of 3 µl of 100 mM EDTA and kept on ice or frozen until further use. Products were resolved by polyacrylamide gel electrophoresis using 30 or 35% acrylamide/bis-acrylamide 19:1 (National Diagnostics) gels in Tris/Borate/EDTA (TBE) buffer as previously described [25]. Gels were then stained with toluidine blue for 1 h and de-stained on 20% methanol for several hours until background staining was removed. For kinetic measurements, the same activity test was performed at pH 8.0 using various quantities of indicated substrate for 10 min. Then Pi released from substrates was quantified by malachite green assay. First, we prepared reagent mix (0.045% malachite green with 4.2% ammonium molybdate in 4 M HCl at a 1:3 ratio, respectively) and let it sit for at least 10 min, and then filtered the solution with 0.2 µm syringe filter units (Millipore). We then added 100 µl of reagent mix to 100 µl of reaction in a clear 96-well plate, mixed well and immediately read absorbance at 660 nm. We quantified Pi through comparison with a standard curve made by serial dilution of KH2PO4. Pi concentration obtained was used for kinetic calculations and plotted in GraphPad Prism 6 software.
Endogenous tagging
We generated cell lines with endogenous C-terminal tags using a one-step transfection method [26]. We amplified by PCR a cassette from pMOTag4H using primers that contained 80 nt homologous region of the 3′ end of CDS and 3′UTR of TbNH4 (Supplementary Table S1). The construct was verified by agarose gel electrophoresis, PCR purified using Minelute PCR purification kit (Qiagen), and transfected in T. brucei Lister 427 PCF cells. Transfection was performed as described before and cells were selected using hygromycin [27]. Preparation of culture lysates, and SDS/PAGE and western blot analyses using anti-HA antibody (Covance) were done to verify expression of tagged proteins [27]. We used microscopy to localize tagged proteins, sample preparation for immunofluorescence microscopy was done as described before [27].
Construct for overexpression
The sequence of TbNH2 (Tb427.05.4350) was amplified by PCR using Q5® high-fidelity polymerase (NED) and cloned in the vector pLEW100v5b1d-BSD using the Gibson Assembly® Cloning kit (NEB) (Supplementary Table S1). Sequence was verified by Sanger sequencing and plasmid transfected in T. brucei procyclic 29-13 TetR/T7RNAP cell line. Cells were cultured for 2 days with tetracycline (1 µg/ml) for induction of overexpression. To verify overexpression, we extracted RNA, synthesized cDNA and performed by qRT-PCR as described previously [27]. Relative gene expression data were obtained by comparison with actin expression levels. We also used western blot analysis with specific antibody to validate increase in TbNH2 protein translation.
Antibody production
We digested the recombinant protein construct of TbNH2 with thrombin (Sigma) to remove thioredoxin and the His-tag from the construct. We then applied products to a HIS-Select® Cartridge column to remove tag from mixture, allowing us to collect pure TbNH2 in the flow-through. This protein was quantified using Pierce BCA Protein Assay Kit (Thermo Scientific) and used for antibody production. The antigen was injected to six female CD-1 mice (Charles River Laboratories) intraperitoneally. The primary inoculation contained 100 µg purified protein mixed in equal parts with Freund’s complete adjuvant (Sigma). Subsequent boosts, spaced in 2-week intervals, contained 50 µg purified protein mixed in equal parts with Freund’s incomplete adjuvant (Sigma). Final bleeds were collected via cardiac puncture.
Fluorescence microscopy
Trypanosoma brucei PCF were centrifuged at 1000 × g for 10 min at 25°C; washed twice with PBS, pH 7.4; and fixed with 4% paraformaldehyde in PBS for 1 h at room temperature (RT). Afterward, cells were adhered to poly-L-lysine coated coverslips for 30 min; permeabilized with 0.1% Triton X-100 in PBS for 5 min, washed three times, and blocked with PBS containing 100 mM NH4Cl, 3% BSA, 1% fish gelatin, and 5% goat serum for 1 h. Cells were then incubated for 1 h, at RT, with primary antibodies: anti-HA tag monoclonal antibody (1:250), polyclonal mouse anti-NH2 antibody (1:1000), and polyclonal rabbit anti-PPDK antibody (1:30), as glycosomal marker. After washing three times with 3% BSA in PBS (pH 8.0), cells were incubated at RT in the dark with secondary antibodies: Alexa Fluor 488-conjugated goat anti-mouse (1:1000), or Alexa Fluor 546-conjugated goat anti-rabbit (1:1000). Then cells were counterstained with 5 µg/ml DAPI to label nuclei and kinetoplasts. Finally, all preparations were washed again three times with 3% BSA in PBS (pH 8.0) and mounted on glass slides with Fluoromount-G (Southern Biotechnology). Differential interference contrast (DIC) and fluorescence optical images were captured under non-saturating conditions and identical exposure times using an Olympus IX-71 inverted fluorescence microscope with a Photometrix Cool-SnapHQ charge-coupled device (CCD) camera driven by DeltaVision software (Applied Precision). Images were deconvolved for 15 cycles using Softwarx deconvolution software.
Results
Analysis of T. brucei Nudix hydrolase sequences
It has been reported [21] that five putative Nudix proteins are present in the T. brucei proteome: TbNH1 (Tb927.11.15640; MW: 44.4; isoelectric point [IP]: 5.49), TbNH2 (Tb927.5.4350; MW: 19.6; IP: 6.88), TbNH3 (Tb927.11.9810; MW: 27.5; IP: 4.83), TbNH4 (Tb927.6.2670; MW: 33.0; IP: 8.62) and TbNH5 (Tb927.10.4680; MW: 32.2; IP: 7.11). All of them have the Nudix box of 23 amino acids common to other NHs but have little identity with Ddp1 (16% for TbNH2 and 13% for TbNH4). TbNH1 (MERS) is a mitochondrial mRNA stability factor [20] and TbNH4 (TbNDcp2) has de-capping activity [21]. The activity of the other Nudix proteins has not been investigated. TbNH2 and TbNH3 has been localized to the glycosomes by proteomic studies [28].
PolyP polyphosphatase activity of NHs from T. brucei
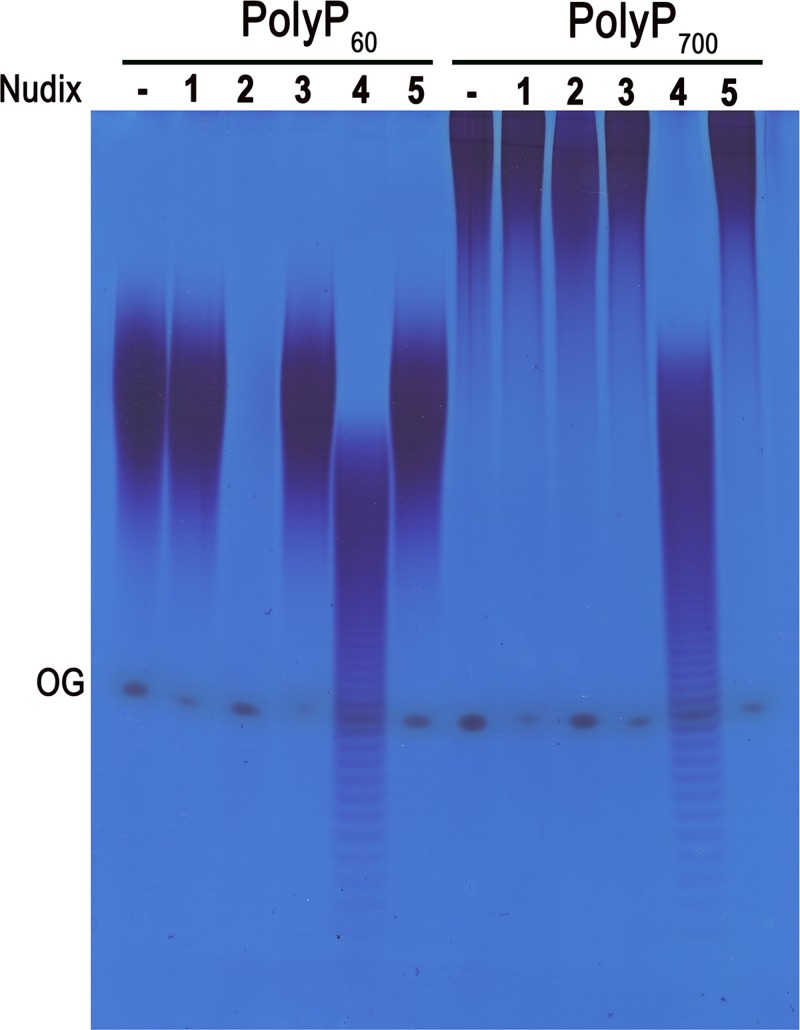
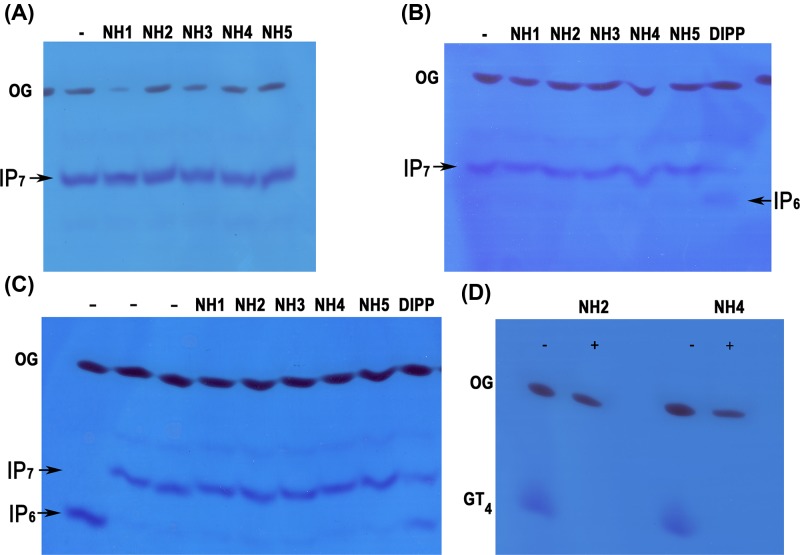
To test whether any of the five NHs from T. brucei has polyP polyphosphatase activity, we cloned and expressed them in bacteria with a polyhistidine tag, and purified the proteins using nickel columns, as described under Materials and Methods. We were able to obtain proteins of the expected size (Supplementary Figure S1), which were used on activity tests with polyPs of different sizes. Incubation of the enzymes with commercially available polyP60 showed a significant polyP hydrolyzing activity of TbNH2 and TbNH4 while neither TbNH1, TbNH3, nor TbNH5 was able to hydrolyze it. In contrast, only TbNH4 was able to hydrolyze polyP700 (Figure 1). None of the T. brucei NHs was able to hydrolyze 5-IP7 at either pH 6.0, 7.0 or 8.0 in contrast to human DIPP, used as positive control (Figure 2A–C). However, both TbNH2 and TbNH4 were able to degrade guanosine tetraphosphate (GP4) (Figure 2D).
Figure 1. Screening of Nudix hydrolase activities identifies two polyP phosphatases in T. brucei.
The ability of the five NHs to degrade short (polyP60) and long chain polyP (polyP700) was tested at 37°C for 1 h in medium containing 40 mM Hepes buffer, pH 7.4, 50 mM NaCl, 6 mM MgCl2, 0.25 micromoles of polyP60 or polyP700 and 5 µg/ml of recombinant protein. Controls (-) and enzymatic products from NH 1, 2, 3, 4 and 5 were resolved in 30% polyacrylamide gels. TbNH2 has high activity with polyP60 and apparently no activity with polyP700. TbNH4 has activity with both polyP60 and polyP700, as evidenced by the production of shorter chain polyP. Orange G (OG) dye was used as loading indicator.
Figure 2. Lack of hydrolytic activity of TbNH2 and TbNH4 against 5-diphosphoinositol pentakisphosphate (5-IP7) at different pHs and activity against GP4.
Phosphatase activity was assayed for 1 h at 37°C in medium containing 40 mM Hepes buffer, pH 7.4, 50 mM NaCl, 6 mM MgCl2, 5 nanomoles of 5-diphosphoinositol pentakisphosphate (5-IP7) and 5 µg/ml of recombinant protein, at pH 6 (A), 7 (B) or 8 (C). (D) Phosphatase activity against GT4. Assays were done as in (A–C), using 20 nanomoles GP4 instead of 5-IP7. The three first lanes in (C) do not have enzymes, and were loaded with IP6 (first lane) or IP7 (second and third lanes).
Characterization of TbNH2 activity
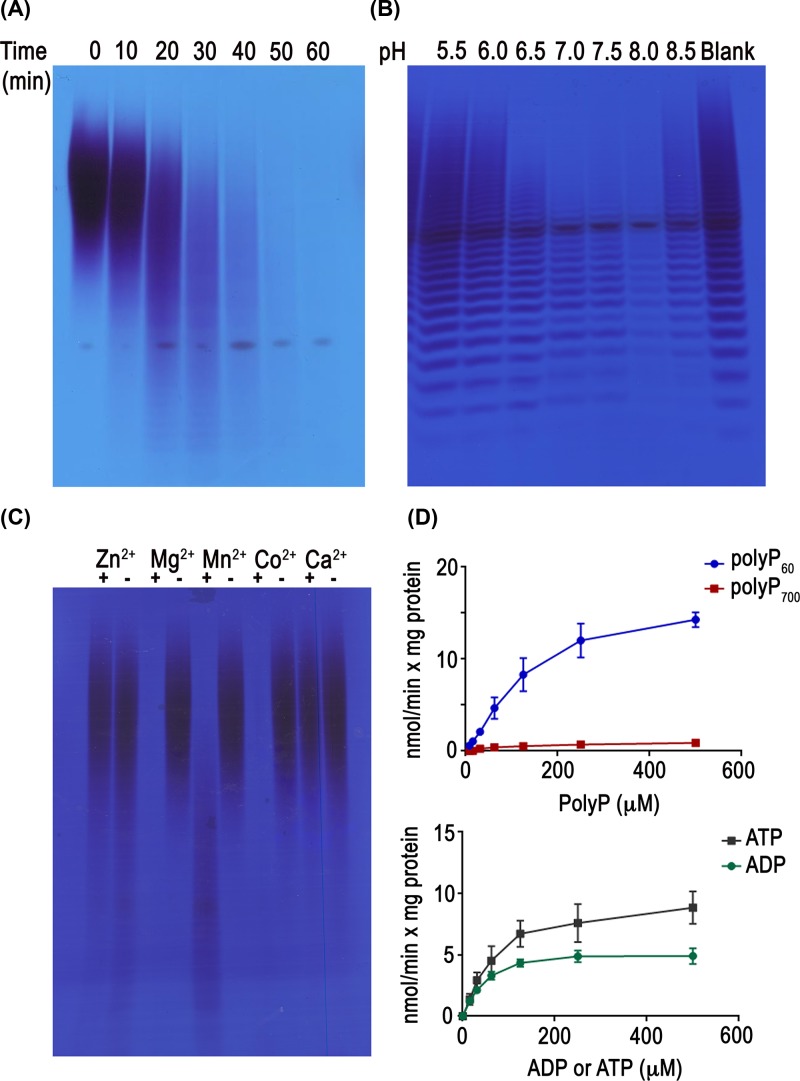
We tested the activity of TbNH2 on polyP60 over the course of 1 h and resolved the products by PAGE (Figure 3A). The progressive shortening of the polyP polymer with production of Pi (see below) demonstrates an PPX activity. Yeast Ddp1 has been reported to have higher activity at slightly acidic pH, while human DIPPs have preference for higher pH [13]. TbNH2 has higher activity at basic pH 8.0 (Figure 3B). We also tested the activity of TbNH2 with different cofactors. TbNH2 is able to hydrolyze polyP with Mg2+ and Co2+ as cofactors, but has lower activity with Mn2+ (Figure 3C). We used the malachite green assay to compare Pi release from polyP, ATP and ADP. TbNH2 has a higher affinity for polyP700 than for polyP60 (Km = 125.6 ± 45 µM versus 200.5 ± 36 µM with Vmax = 1.1 ± 0.1 nmol min−1 mg protein−1 versus 20.6 ± 1.7 nmol min−1mg protein−1) (Figure 3D, top and Table 1). This difference in affinity based on polyP size suggests that the enzyme works by removing the terminal phosphate from polyP chains exerting an PPX activity. Interestingly, using same method we detected TbNH2 also has activity to release the γ and β phosphates from ATP and ADP (Figure 3D, bottom).
Figure 3. Characterization of TbNH2 activity.
(A) Hydrolysis of polyP60 by TbNH2 over 1 h was tested at 37°C in medium containing 40 mM Hepes buffer, pH 7.4, 50 mM NaCl, 6 mM MgCl2, 0.25 micromoles of polyP60 and 5 µg/ml of recombinant protein. (B) TbNH2 activity on polyP60 at various pHs. (C) TbNH2 activity on polyP60 in the presence of divalent cations (6 mM). (D) Phosphatase activity of TbNH2 with different concentration of polyP60, polyP700, ATP and ADP. Note that the activity on polyP700, is about ten times lower than the activity on polyP60.
Table 1. Kinetic parameters of PPX activity of TbNH2 and TbNH4.
| Enzyme | Substrate | Vmax (nmol min−1 mg−1) | Km (µM) | kcat/Km (s−1M−1) |
|---|---|---|---|---|
| TbNH2 | PolyP60 | 20.6 ± 1.7 | 200.5 ± 36 | 2.2 × 105 |
| PolyP700 | 1.1 ± 0.1 | 125.6 ± 45 | 0.2 × 105 | |
| ADP | 5.6 ± 0.2 | 46.5 ± 6.3 | 2.7 × 105 | |
| ATP | 10.2 ± 0.7 | 78.1 ± 16.3 | 2.9 × 105 | |
| TbNH4 | PolyP60 | 9.1 ± 0.4 | 82.2 ± 13 | 3.4 × 105 |
| PolyP700 | 5.6 ± 0.2 | 149.4 ± 21 | 1.1 × 105 |
Characterization of TbNH4 activity
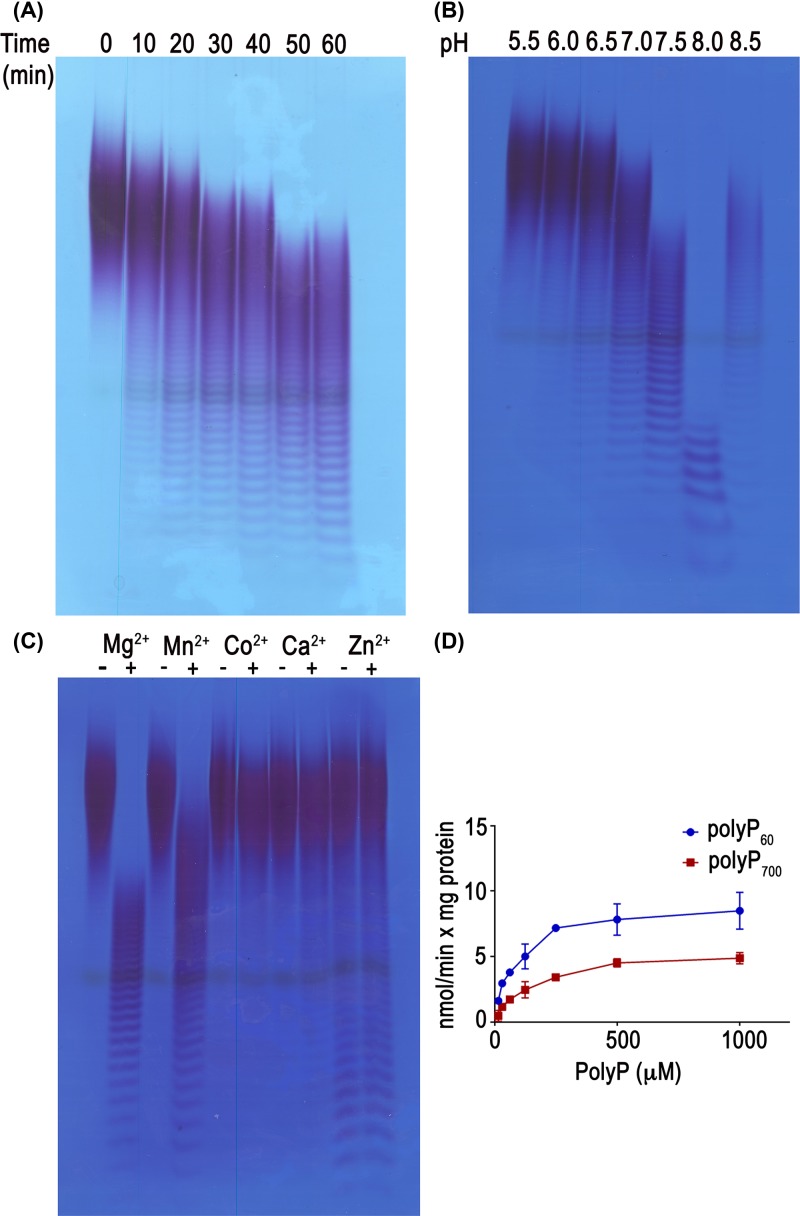
We also tested the activity of TbNH4 on polyP60 over the course of 1 h (Figure 4A). TbNH4 endopolyphosphatase activity was shown by the increase in the staining intensity of small polyP oligomers that were produced after incubation of polyP60 with the enzyme. TbNH4 has a slightly lower activity on polyP60 than TbNH2. Activity tests at various pHs showed TbNH4 has optimum activity at pH 8.0 (Figure 4B). Mg2+ or Mn2+ were the preferred cofactors while no activity was detected with Co2+ (Figure 4C). The malachite green assay for detection of Pi release showed that the enzyme also has exopolyposphatase activity and a higher affinity for polyP60 than for polyP700 (Figure 4D and Table 1) (Km = 82.1 ± 13 µM versus 149.4 ± 21 µM and Vmax = 9.1 ± 0.4 nmol min−1mg protein−1 versus = 5.6 ± 0.2 nmol min−1mg protein−1). TbNH4 does not have phosphatase activity against ATP or ADP.
Figure 4. Characterization of TbNH4 activity.
(A) Hydrolysis of polyP60 by TbNH4 over 1 h was tested at 37°C in medium containing 40 mM Hepes buffer, pH 7.4, 50 mM NaCl, 6 mM MgCl2, 0.25 micromoles of polyP60 and 5 µg/ml of recombinant protein. (B) TbNH4 activity on polyP60 at various pHs. (C) TbNH4 activity on polyP60 in the presence of divalent cations. (D) Phosphatase activity of TbNH4 with different concentration of polyP60 and polyP700.
Localization studies
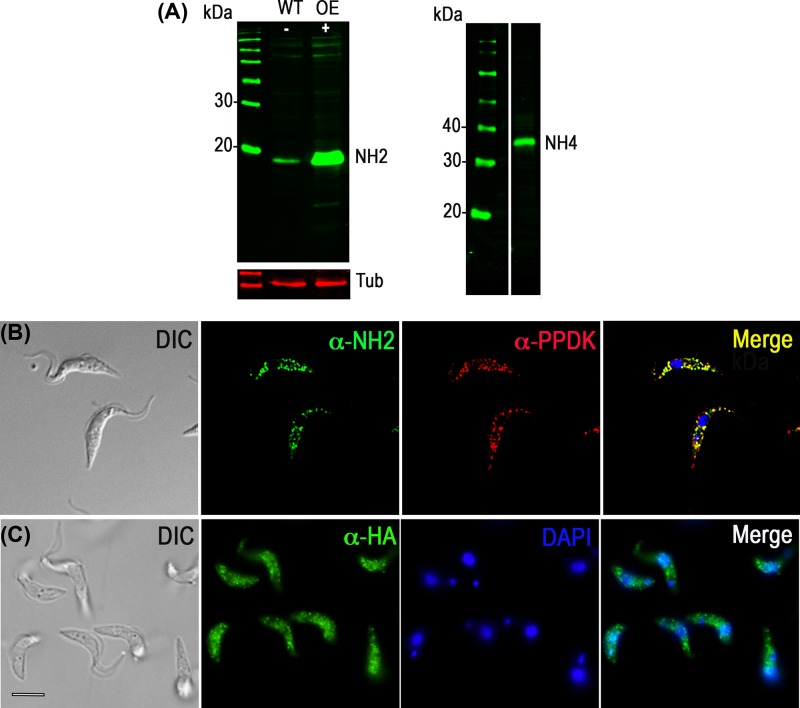
Trypanosomes accumulate large amounts of polyP in acidocalcisomes [4]. PolyP has also been found in glycosomes and nucleoli of T. brucei [22]. In order to determine the localization of TbNH4, we tagged the C-terminus of the gene with an HA tag using homologous recombination with the endogenous gene locus in procyclic trypomastigotes (PCF). Western blot analysis using anti-HA antibodies confirmed the expression of the protein of the expected size (37 kDa, Figure 5A right panel). Because the tag could interfere with the glycosomal localization signal of TbNH2, we prepared a specific polyclonal antibody and overexpressed the protein. Western blot analysis using this antibody labeled a protein of the expected size (19 kDa, Figure 5A, left panel). Immunofluorescence microscopy showed that overexpressed TbNH2 localized to the glycosomes, as demonstrated by co-localization with the glycosomal marker PPDK (Figure 5B). Immunofluorescence microscopy shows that tagged TbNH4 localizes to the cytosol and nuclei of the cells (Figure 5C). No fluorescence was observed in control parasites incubated only in the presence of secondary antibodies (not shown).
Figure 5. Subcellular localization of TbH2 and TbNH4.
(A) Left panel, western blot analysis of wild type or TbNH2 overexpressing cells using polyclonal anti-TbNH2 antibody showing a band of about 19 kDa. Antibodies against α-Tubulin (Tub) were used as loading control. Right panel, western blot analysis of endogenously tagged parasites using monoclonal anti-HA antibodies showing TbNH4 (37 kDa). Molecular weights are shown on the left. (B) TbNH2 co-localizes with PPDK in glycosomes of PCF. TbNH2 was detected with polyclonal anti-TbNH2 antibody in cells overexpressing the protein (green), and co-localized with antibodies against PPDK (red). The merge shows co-localization in yellow. (C) TbNH4 localizes in the cytosol and nucleus of PCF. TbNH4 was detected with monoclonal anti-HA antibodies in trypanosomes expressing TbNH4-HA (green). A punctate appearance could be the result of deconvolution of the images. Bar (for B and C) = 5 µm.
Discussion
The most important findings of this work are the identification of two NHs of T. brucei as polyP phosphatases, and evidence of their localization in subcellular organelles where polyP is or may be present. None of the other three NHs of the parasites has this activity, and none of the NHs can hydrolyze 5-IP7.
TbNH4 has polyP endopolyphosphatase activity, the ability to attack internal phosphoanhydride bonds hydrolyzing polyP molecules into smaller oligophosphates, and is the first such activity identified in trypanosomatids. TbNH4 has also PPX activity, the ability to remove Pi from the end of polyP chains, as the yeast PPN1 endopolyphosphatase previously described [29]. TbNH4 PPX has higher affinity for polyP60 than for polyP700. However, this could be attributed to the detection method. Although the enzyme can hydrolyze phosphoanhydride bonds at any position, only Pi is detected by the malachite green assay used and shorter polyP chains have more available ends to release Pi than polyP of longer chains. TbNH4 localizes to the cytosol and nucleus, as occurs with Ddp1 [16]. PolyP has been shown to be cytotoxic when in the yeast cytosol [30] and this enzyme, together with TbPPX [5], could help in controlling its cytosolic concentration. The cytosolic localization could also be important for its de-capping activity [21] and the nuclear localization could be relevant to regulate nucleolar polyP levels in procyclic forms [22].
TbNH2 has polyP PPX activity and has a preference for short chain polyP. PolyP is a linear polymer of phosphate, so there is no chemical difference among polyPs of different chain lengths except for the number of ends available for the PPX to hydrolyze. The convention is to quantify the amount of polyP by molarity of phosphate units. Therefore, polyP60 and polyP700 at the same molarity have the same amount of phosphate units, but not the same number of molecules. The activity of a PPX should be the same only if there is the same number of molecules in solution. Actually, the number of polyP molecules available on polyP60 is 11.7 lower than for polyP700, and our assays show about 18 times reduction in Vmax between polyP60 and polyP700. This result supports the activity of TbNH2 as a PPX.
TbNH2 was reported in two glycosome proteomes [28,31], has a peroxisomal targeting signal 2 (PTS2) and, as other peroxisomal NHs, has been proposed to have a role in destroying nucleotides damaged by reactive oxygen species [15]. Interestingly, expression of a yeast PPX in the glycosomes of T. brucei makes them more susceptible to oxidative stress [22] and polyP has been shown to have a role in protection against oxidative stress [32,33]. It would be interesting to test whether overexpression of an endogenous gene, which could be susceptible to regulatory mechanisms not present when overexpressing an exogenous gene, have the same effect.
Alkaline pH and divalent ions have been found before to be important for Nudix hydrolase activity [18]. Both TbNH2 and TbNH4 have similar preferences for alkaline pH and divalent cofactors (Mg2+ and Co2+ for TbNH2 and Mg2+ and Mn2+ for TbNH4).
In conclusion, we have identified that two of the five NHs present in T. brucei have polyP endo- and exopolyphosphatase activities but, in contrast to the yeast and mammalian NHs, are unable to hydrolyze 5-IP7. TbNH2 is a polyP PPX, and localizes in the glycosome. TbNH4 is a polyP endo- and exopolyphosphatase, and localizes in the cytosol and nucleus. Both enzymes could have a role in maintaining and regulating polyP levels in their respective localizations.
Supporting information
Supplementary Figure 1.
Supplementary Table S1. Primers used in this study.
Acknowledgments
The authors thank Dr Henning Jessen, Dr Toshikazu Shiba, Dr Dorothea Fiedler and Dr Frédéric Bringaud for reagents; Dr Adolfo Saiardi (University College London) for advice on detecting enzymatic activities by PAGE; and Melissa Storey for technical support with the antibody production in mice.
Abbreviations
- 5-IP7
5-Diphosphoinositol pentakisphosphate
- Ddp1
diphosphoinositol polyphosphate phosphohydrolase
- DIPP
diphosphoinositol polyphosphate hydrolase
- GP4
guanosine tetraphosphate
- IP
isoelectric point
- NH
nudix hydrolase
- Pi
orthophosphate
- polyP
inorganic polyphosphate
- PPi
inorganic pyrophosphate
- PPDK
phosphate pyruvate dikinase
- TbNH
T. brucei Nudix hydrolases
- RT
room temperature
- VTC
vacuolar transporter chaperone
Author Contribution
C.D.C. and R.D. were involved in the conception and design of the experiments and interpretation of the data. C.D.C., M.A.A. and B.W. performed the experiments. C.D.C. and R.D. wrote the final version of the manuscript.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was funded by a grant from the US National Institutes of Health [grant number AI-077358 to R.D.].
References
- 1.Rao N.N., Gomez-Garcia M.R. and Kornberg A. (2009) Inorganic polyphosphate: essential for growth and survival. Annu. Rev. Biochem. 78, 605–647 10.1146/annurev.biochem.77.083007.093039 [DOI] [PubMed] [Google Scholar]
- 2.Hothorn M., Neumann H., Lenherr E.D., Wehner M., Rybin V., Hassa P.O.. et al. (2009) Catalytic core of a membrane-associated eukaryotic polyphosphate polymerase. Science 324, 513–516 10.1126/science.1168120 [DOI] [PubMed] [Google Scholar]
- 3.Lander N., Ulrich P.N. and Docampo R. (2013) Trypanosoma brucei vacuolar transporter chaperone 4 (TbVtc4) is an acidocalcisome polyphosphate kinase required for in vivo infection. J. Biol. Chem. 288, 34205–34216 10.1074/jbc.M113.518993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Docampo R., de Souza W., Miranda K., Rohloff P. and Moreno S.N. (2005) Acidocalcisomes - conserved from bacteria to man. Nat. Rev. Microbiol. 3, 251–261 10.1038/nrmicro1097 [DOI] [PubMed] [Google Scholar]
- 5.Luginbuehl E., Kunz S., Wentzinger L., Freimoser F. and Seebeck T. (2011) The exopolyphosphatase TbrPPX1 of Trypanosoma brucei. BMC Microbiol. 11, 4 10.1186/1471-2180-11-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lemercier G., Espiau B., Ruiz F.A., Vieira M., Luo S., Baltz T.. et al. (2004) A pyrophosphatase regulating polyphosphate metabolism in acidocalcisomes is essential for Trypanosoma brucei virulence in mice. J. Biol. Chem. 279, 3420–3425 10.1074/jbc.M309974200 [DOI] [PubMed] [Google Scholar]
- 7.Kotsikorou E., Song Y., Chan J.M., Faelens S., Tovian Z., Broderick E.. et al. (2005) Bisphosphonate inhibition of the exopolyphosphatase activity of the Trypanosoma brucei soluble vacuolar pyrophosphatase. J. Med. Chem. 48, 6128–6139 10.1021/jm058220g [DOI] [PubMed] [Google Scholar]
- 8.Jamwal A., Round A.R., Bannwarth L., Venien-Bryan C., Belrhali H., Yogavel M.. et al. (2015) Structural and functional highlights of vacuolar soluble protein 1 from pathogen Trypanosoma brucei brucei. J. Biol. Chem. 290, 30498–30513 10.1074/jbc.M115.674176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Y., Ko T.P., Chen C.C., Huang G., Zheng Y., Liu W.. et al. (2016) Structures of trypanosome vacuolar soluble pyrophosphatases: antiparasitic drug targets. ACS Chem. Biol. 11, 1362–1371 10.1021/acschembio.5b00724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sethuraman A., Rao N.N and Kornberg A. (2001) The endopolyphosphatase gene: essential in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 98, 8542–8547 10.1073/pnas.151269398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi X. and Kornberg A. (2005) Endopolyphosphatase in Saccharomyces cerevisiae undergoes post-translational activations to produce short-chain polyphosphates. FEBS Lett. 579, 2014–2018 10.1016/j.febslet.2005.02.032 [DOI] [PubMed] [Google Scholar]
- 12.Gerasimaite R. and Mayer A. (2017) Ppn2, a novel Zn2+-dependent polyphosphatase in the acidocalcisome-like yeast vacuole. J. Cell Sci. 130, 1625–1636 10.1242/jcs.201061 [DOI] [PubMed] [Google Scholar]
- 13.Lonetti A., Szijgyarto Z., Bosch D., Loss O., Azevedo C. and Saiardi A. (2011) Identification of an evolutionarily conserved family of inorganic polyphosphate endopolyphosphatases. J. Biol. Chem. 286, 31966–31974 10.1074/jbc.M111.266320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caffrey J.J, Safrany S.T, Yang X. and Shears S.B (2000) Discovery of molecular and catalytic diversity among human diphosphoinositol-polyphosphate phosphohydrolases. An expanding Nudt family. J. Biol. Chem. 275, 12730–12736 10.1074/jbc.275.17.12730 [DOI] [PubMed] [Google Scholar]
- 15.Fisher D.I, Safrany S.T, Strike P., McLennan A.G and Cartwright J.L (2002) Nudix hydrolases that degrade dinucleoside and diphosphoinositol polyphosphates also have 5-phosphoribosyl 1-pyrophosphate (PRPP) pyrophosphatase activity that generates the glycolytic activator ribose 1,5-bisphosphate. J. Biol. Chem. 277, 47313–47317 10.1074/jbc.M209795200 [DOI] [PubMed] [Google Scholar]
- 16.Huh W.K, Falvo J.V, Gerke L.C, Carroll A.S, Howson R.W, Weissman J.S. et al. (2003) Global analysis of protein localization in budding yeast. Nature 425, 686–691 10.1038/nature02026 [DOI] [PubMed] [Google Scholar]
- 17.Srouji J.R, Xu A., Park A., Kirsch J.F and Brenner S.E (2017) The evolution of function within the Nudix homology clan. Proteins 85, 775–811 10.1002/prot.25223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoshimura K. and Shigeoka S. (2015) Versatile physiological functions of the Nudix hydrolase family in Arabidopsis. Biosci. Biotechnol. Biochem. 79, 354–366 10.1080/09168451.2014.987207 [DOI] [PubMed] [Google Scholar]
- 19.McLennan A.G. (2006) The Nudix hydrolase superfamily. Cell. Mol. Life Sci. 63, 123–143 10.1007/s00018-005-5386-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weng J., Aphasizheva I., Etheridge R.D, Huang L., Wang X., Falick A.M. et al. (2008) Guide RNA-binding complex from mitochondria of trypanosomatids. Mol. Cell 32, 198–209 10.1016/j.molcel.2008.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ignatochkina A.V, Takagi Y., Liu Y., Nagata K. and Ho C.K (2015) The messenger RNA decapping and recapping pathway in Trypanosoma. Proc. Natl. Acad. Sci. U.S.A. 112, 6967–6972 10.1073/pnas.1424909112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Negreiros R.S, Lander N., Huang G., Cordeiro C.D, Smith S.A, Morrissey J.H. et al. (2018) Inorganic polyphosphate interacts with nucleolar and glycosomal proteins in trypanosomatids. Mol. Microbiol. 110, 973–994 10.1111/mmi.14131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerasimaite R., Pavlovic I., Capolicchio S., Hofer A., Schmidt A., Jessen H.J. et al. (2017) Inositol pyrophosphate specificity of the SPX-dependent polyphosphate polymerase VTC. ACS Chem. Biol. 12, 648–653 10.1021/acschembio.7b00026 [DOI] [PubMed] [Google Scholar]
- 24.Cunningham I. and Honigberg B.M (1977) Infectivity reacquisition by Trypanosoma brucei brucei cultivated with tsetse salivary glands. Science 197, 1279–1282 10.1126/science.897667 [DOI] [PubMed] [Google Scholar]
- 25.Losito O., Szijgyarto Z., Resnick A.C and Saiardi A. (2009) Inositol pyrophosphates and their unique metabolic complexity: analysis by gel electrophoresis. PLoS ONE 4, e5580 10.1371/journal.pone.0005580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oberholzer M., Morand S., Kunz S. and Seebeck T. (2006) A vector series for rapid PCR-mediated C-terminal in situ tagging of Trypanosoma brucei genes. Mol. Biochem. Parasitol. 145, 117–120 10.1016/j.molbiopara.2005.09.002 [DOI] [PubMed] [Google Scholar]
- 27.Cordeiro C.D, Saiardi A. and Docampo R. (2017) The inositol pyrophosphate synthesis pathway in Trypanosoma brucei is linked to polyphosphate synthesis in acidocalcisomes. Mol. Microbiol. 106, 319–333 10.1111/mmi.13766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guther M.L, Urbaniak M.D, Tavendale A., Prescott A. and Ferguson M.A (2014) High-confidence glycosome proteome for procyclic form Trypanosoma brucei by epitope-tag organelle enrichment and SILAC proteomics. J. Proteome Res. 13, 2796–2806 10.1021/pr401209w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andreeva N., Trilisenko L., Eldarov M. and Kulakovskaya T. (2015) Polyphosphatase PPN1 of Saccharomyces cerevisiae: switching of exopolyphosphatase and endopolyphosphatase activities. PLoS ONE 10, e0119594 10.1371/journal.pone.0119594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gerasimaite R., Sharma S., Desfougeres Y., Schmidt A. and Mayer A. (2014) Coupled synthesis and translocation restrains polyphosphate to acidocalcisome-like vacuoles and prevents its toxicity. J. Cell Sci. 127, 5093–5104 10.1242/jcs.159772 [DOI] [PubMed] [Google Scholar]
- 31.Colasante C., Ellis M., Ruppert T. and Voncken F. (2006) Comparative proteomics of glycosomes from bloodstream form and procyclic culture form Trypanosoma brucei brucei. Proteomics 6, 3275–3293 10.1002/pmic.200500668 [DOI] [PubMed] [Google Scholar]
- 32.Dahl J.U, Gray M.J and Jakob U. (2015) Protein quality control under oxidative stress conditions. J. Mol. Biol. 427, 1549–1563 10.1016/j.jmb.2015.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gray M.J and Jakob U. (2015) Oxidative stress protection by polyphosphate–new roles for an old player. Curr. Opin. Microbiol. 24, 1–6 10.1016/j.mib.2014.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]