Abstract
Previous studies demonstrated that miRNA-1 (miR-1) is downregulated in certain human cancer and serves a crucial role in the progression of cancer. However, there are only a few previous studies examining the association between miR-1 and lung squamous cell carcinoma (LUSC) and the regulatory mechanism of miR-1 in LUSC remains unclear. Therefore, the present study investigated the clinical significance and determined the potential molecular mechanism of miR-1 in LUSC. The expression of miR-1 and its clinical significance in LUSC was examined by conducting a meta-analysis of 12 studies using Stata 14, MetaDiSc1.4 and SPSS version 23. In addition, Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analyses were performed using the potential target genes of miR-1 gathered from Gene Expression Omnibus and ArrayExpress. Meta-analysis demonstrated that miR-1 was significantly downregulated in LUSC [standardized mean difference: −1.44; 95% confidence interval (CI): −2.08, −0.81], and the area under the curve was 0.9096 (Q*=0.8416) with sensitivity of 0.71 (95% CI: 0.66, 0.76) and specificity of 0.88 (95% CI: 0.86, 0.90). The pooled positive likelihood ratio and negative likelihood ratio were 4.93 (95% CI: 2.54, 9.55) and 0.24 (95% CI: 0.10, 0.54), respectively. Bioinformatics analysis demonstrated that miR-1 may be involved in the progression of LUSC via the ‘cell cycle’, ‘p53 signaling pathway’, ‘Fanconi anemia pathway’, ‘homologous recombination’, ‘glycine, serine and threonine metabolism’ and ‘oocyte meiosis’. In summary, miR-1 was significantly downregulated in LUSC, suggesting a novel and promising non-invasive biomarker for diagnosing LUSC, and miR-1 was involved in LUSC progression via a number of significant pathways.
Keywords: microRNA-1-3p, lung squamous cell carcinoma, microRNA, bioinformatics, microarray
Introduction
Non-small cell lung cancer (NSCLC) accounts for 85–90% lung cancer, and the most common subtypes are lung squamous cell carcinoma (LUSC) and lung adenocarcinoma (LUAD) (1–4). LUSC typically occurs in men and is associated with smoking, accounting for ~25–30% total lung cancer cases (5–7). Therapies for LUAD are frequently ineffective for LUSC, reflecting differences between LUAD and LUSC; therefore, distinguishing LUSC from other types of lung cancer and investigating the molecular mechanism of LUSC are crucial.
MicroRNAs (miRNAs) are a class of small non-coding RNAs with lengths of 19–25 nucleotides (8–13). As post-transcriptional regulators of gene expression, these molecules are involved in the regulation of a number of biological processes, including differentiation, proliferation, migration and apoptosis (14–20). miRNAs may be used as diagnostic biomarkers and treatment targets for human cancer (21–25). The miR-1 family is comprised of miR-1-1, miR-1-2 and miR-206. In the family, miR-1-1 and miR-1-2 locate on chromosomes 20 and 18, respectively (26,27). The upregulation of miR-1 inhibits proliferation, promotes apoptosis and reverses the drug resistance of various types of cancer in vivo and in vitro, serving a role as a cancer suppressor gene (27). Previous studies demonstrated that miR-1 is downregulated in approximately all human cancer types and miR-1 serves a crucial role in the progression of cancer. Han et al (28) demonstrated that miR-1 is downregulated in gastric cancer, and inhibits gastric cancer cell proliferation and migration by targeting MET proto-oncogene, receptor tyrosine kinase. Wang et al (29) demonstrated that miR-1-3p, the mature miRNA of miR-1, suppresses the proliferation and invasion of bladder cancer cells by inhibiting C-C motif chemokine ligand 2 expression. Wang et al (30) observed that microRNA-1-3p is downregulated in oral squamous cell carcinoma (OSCC) tissues and cells, and serves as a suppressor of OSCC progression. There are a few previous studies that address the roles of miRNA in lung cancer progression (31–33). Meta-analyses have additionally been conducted to confirm the association between miRNA expression and prognosis in NSCLC (34). Nevertheless, no consistent conclusion has been reached and direct evidence of the association between miR-1 and LUSC is limited. Therefore, the regulatory mechanism of miR-1 in LUSC requires further investigation.
The present study conducted a meta-analysis to evaluate the clinical significance of miR-1 in LUSC. Potential target genes of miR-1 in LUSC were obtained from the gene chip analysis of LUSC cells transfected with miR-1-3p, combined with target gene prediction and differential gene expression in The Cancer Genome Atlas (TCGA). Subsequently, a signaling pathway analysis was conducted to determine the potential molecular mechanism of miR-1 in LUSC.
Materials and methods
Collection of microarray datasets from Gene Expression Omnibus (GEO) and ArrayExpress
To examine the level of miR-1 expression in LUSC and adjacent non-cancer tissues, retrieval in GEO (http://www.ncbi.nlm.nih.gov/geo/) and ArrayExpress (http://www.ebi.ac.uk/arrayexpress/) was performed using the following key words: [‘lung’ OR ‘pulmonary’ OR ‘respiratory’ OR ‘bronchioles’ OR ‘bronchi’ OR ‘alveoli’ OR ‘pneumocytes’ OR ‘air way’ (MeSH)] AND (‘cancer’ OR ‘carcinoma’ OR ‘tumor’ OR ‘neoplas* OR malignan* squamous cell carcinoma’). ‘Series’ and ‘Homo sapiens’ were filtered. Studies with sample sizes ≥3 and miR-1 expression measured in LUSC and control groups were included. To identify promising miR-1 target genes, GEO and ArrayExpress were searched again using the following terms: [‘lung’ OR ‘pulmonary’ OR ‘respiratory’ OR ‘bronchioles’ OR ‘bronchi’ OR ‘alveoli’ OR ‘pneumocytes’ OR ‘air way’ (MeSH)] AND (‘cancer’ OR ‘carcinoma’ OR ‘tumor’ OR ‘neoplas* OR malignan* squamous cell carcinoma’) AND (‘miR-1’ OR ‘miRNA-1’ OR ‘microRNA-1’ OR ‘miR-1’ OR ‘miRNA-1’ OR ‘microRNA1’ OR ‘miR-1’ OR ‘miRNA-1’ OR ‘microRNA-1’ OR ‘miR-1-3p’ OR ‘miRNA-1-3p’ OR ‘microRNA-1-3p’ OR ‘miR-1-1’ OR ‘miR-1-2’ OR ‘miR1-1’ OR ‘miR1-2’). Gene chips intervened with miR-1 in LUSC cell lines, knockdown or transfection, were included in the present analysis. Datasets are presented in Table I.
Table I.
Forest plot of studies evaluating the SMD of microRNA-1 expression between patients with lung squamous cell carcinoma and the control group (a random-effects model).
| Experimental | Control | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Author, year | Study | Mean | SD | Total | Mean | SD | Total | SMD (95% CI) | (Refs.) |
| Seike et al, 2009 | GSE14936 | 7.06532 | 0.248859 | 3 | 7.384533 | 0.365138 | 21 | −0.90 (−2.13, 0.34) | (48) |
| Raponi et al, 2009 | GSE16025 | 4.811725 | 0.183384 | 61 | 4.9321 | 0.158178 | 10 | −0.67 (−1.35, 0.01) | (49) |
| Ohba et al, 2013 | GSE19945 | −1.06547 | 2.263498 | 5 | 2.027512 | 0.642366 | 8 | −2.12 (−3.55, −0.70) | (50) |
| Nymark et al, 2011 | GSE25508 | 6.154964 | 0.23209 | 8 | 6.334855 | 0.624779 | 34 | −0.31 (−1.09, −0.46) | (51) |
| Patnaik et al, 2017 | GSE40738 | −4.65092 | 0.333811 | 33 | −4.45357 | 0.468942 | 56 | −0.47 (−0.90, −0.03) | (52) |
| van Jaarsveld et al, 2014 | GSE47525 | 5.378686 | 2.603978 | 5 | 3.84297 | 1.091469 | 14 | 0.97 (−0.10, 2.04) | (53) |
| Arima et al, 2014 | GSE51853 | −5.83195 | 1.588947 | 29 | −2.78567 | 2.077722 | 5 | −1.84 (−2.89, −0.79) | (54) |
| Jin et al, 2015 | GSE74190 | 0.321576 | 0.861687 | 30 | 3.414288 | 1.172088 | 44 | −2.92 (−3.59, −2.26) | (55) |
| Seike et al, 2009 | PMID:18818206-1 | 0.00442 | 0.00416 | 7 | 0.031476 | 0.021179 | 7 | −1.77 (−3.04, −0.51) | (48) |
| PMID:18818206-2 | 0.015752 | 0.007522 | 7 | 0.172871 | 0.074251 | 7 | −2.98 (−4.56, −1.39) | ||
| TCGA-1 | 2.861394 | 1.672886 | 444 | 6.550327 | 1.177112 | 45 | −2.26 (−2.60, −1.92) | ||
| TCGA-2 | 2.876746 | 1.652493 | 458 | 6.590287 | 1.154916 | 45 | −2.30 (−2.64, −1.96) | ||
| Total (95% CI) | 1,090 | 296 | −1.70 (−1.87, −1.52) | ||||||
SD, standard deviation; GSE, Genomic Spatial Event; PMID, PubMed ID; TCGA, The Cancer Genome Atlas; TCGA, The Cancer Genome Atlas; CI, confidence interval; SMD, standard mean difference.
Acquisition of TCGA miRNA data
The LUSC miRNA matrix was downloaded from TCGA (http://cancergenome.nih.gov/). Data were normalized to a log2 scale. The values of miR-1-1 and miR-1-2 were included in the present study. Samples missing values for miR-1-1 or miR-1-2 were removed from the analysis. The data from TCGA were additionally included in the continuous variable meta-analysis and diagnostic meta-analysis.
Literature reviewing and selecting
PubMed (https://www.ncbi.nlm.nih.gov/pubmed), Science Direct (https://www.sciencedirect.com/), Google Scholar (https://scholar.google.com/), Ovid (https://ovidsp.ovid.com/), Wiley Online Library (https://onlinelibrary.wiley.com/), Embase (https://www.embase.com/), Web of Science (http://www.webofknowledge.com), Chong Qing VIP (http://www.cqvip.com), CNKI (http://www.cnki.net/), Wan Fang (http://www.wanfangdata.com.cn/) and China Biology Medicine Disc (http://http://www.sinomed.ac.cn/) were searched to identify all studies associated with miR-1-3p in LUSC using the following keywords: [‘lung’ OR ‘pulmonary’ OR ‘respiratory’ OR ‘bronchioles’ OR ‘bronchi’ OR ‘alveoli’ OR ‘pneumocytes’ OR ‘air way’ (MeSH)] AND (‘cancer’ OR ‘carcinoma’ OR ‘tumor’ OR ‘neoplas* OR malignan* squamous cell carcinoma’) AND (‘miR-1’ OR ‘miRNA-1’ OR ‘microRNA-1’ OR ‘miR-1’ OR ‘miRNA-1’ OR ‘microRNA-1’ OR ‘miR-1’ OR ‘miRNA-1’ OR ‘microRNA-1’ OR ‘miR-1-3p’ OR ‘miRNA-1-3p’ OR ‘microRNA-1-3p’ OR ‘miR-1-1’ OR ‘miR-1-2’ OR ‘miR1-1’ OR ‘miR1-2’). The cut-off point for the search was July 20, 2017, so no studies later than this date were included. Subsequently, the eligible literature was independently evaluated in the present study using the same multi-step process. Eligible studies had to meet the following criteria: i) They detected miR-1 expression in human tissue or serum in LUSC and control groups; ii) the study offered sufficient data to calculate the diagnostic accuracy; and iii) the study was published in Chinese or English. The following exclusion criteria were used: i) Duplicated studies, case reports, letters, reviews and conference articles; ii) studies with unavailable data; and iii) animal studies.
Data extraction
Information was independently extracted, including the mean, standard deviation, sample size, true positive, false positive, true negative and false negative, from the included studies.
Statistical analysis
All data from the included microarray datasets were converted to a log2 scale, and the mean, standard deviation and case numbers of LUSC and control groups were calculated. Subsequently, Stata 14 (StataCorp LLC, College Station, TX, USA) was used to combine the standard mean difference (SMD) value and its 95% confidence interval (CI). The pooled sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio (DOR) and 95% CI values from the accuracy data of each study were calculated using MetaDiSc 1.4 software (http://www.hrc.es/investigacion/metadisc_en.htm). The DOR value ranged between 0 and ∞, with higher values suggesting improved discriminatory performance (35–39). To assess the clinical significance of miR-1 in LUSC, the summarized receiver operating characteristic (SROC) curves were obtained using SPSS version 23 (IBM Corp., Armonk, NY, USA). An area under the curve (AUC) value near 1.0 indicates that the test has perfect discrimination, whereas, a value close to 0.5 implies poor discrimination. Potential heterogeneity among the included studies was estimated using Cochran's Q test and the I2 index, and heterogeneity significantly existed if P<0.05 or I2>50%. If there was no distinct heterogeneity in the analysis, a fixed-effect model was used; otherwise, a random-effects model was employed. Heterogeneity was explained using the threshold effect analysis and meta-regression analysis. A sensitivity analysis was conducted to identify sources of heterogeneity. Finally, a funnel plot was applied to assess the publication bias among the included studies. P<0.05 was considered to indicate a statistically significant difference.
Acquisition of potential target genes of miR-1 in LUSC
The downregulated genes recorded in Genomic Spatial Event (GSE)56243 were cross-referenced based on two cell lines. The mRNA data were additionally downloaded at the entry of LUSC in TCGA. DESeq data R package was applied to obtain the differentially expressed genes, log2 fold change >2 was considered a criterion of the differentially expressed genes (40). miRWalk version 2.0 (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/custom.html) was used to predict the target genes of miR-1 based on 12 different online miRNA prediction tools: miRWalk; Microt4; miRanda; miRBridge; miRDB; miRMap; miRNAMap; PicTar; PITA; RNA22; RNAhybrid; and TargetScan (https://bioconductor.org/biocLite/). Subsequently, the downregulated genes in GSE56243, TCGA differentially expressed genes and predicted target genes of miR-1 were cross-referenced. The overlapping genes selected by VENNY diagram (https://omictools.com/venny-tool) were considered promising target genes of miR-1 in LUSC.
Bioinformatics based on the promising target genes of miR-1 in LUSC
Gene Ontology (GO) (41,42) annotation, comprising three parts (biological process, cellular component and molecular function) and Kyoto Encyclopedia of Genes and Genomes (KEGG) (43,44) pathway analysis were performed in Database for the Annotation, Visualization and Integrated Discovery version 6.8 (https://david-d.ncifcrf.gov/) based on the promising target genes of miR-1 in LUSC. The BINGO and Enrichment Map plug-ins of Cytoscape (version 3.6.1) were used to visualize the GO annotation and KEGG pathways (45–47). Search Tool for the Retrieval of Interacting Genes/Proteins (http://www.string-db.org), an online tool, was used to construct the protein-protein interaction (PPI) network with the disconnected nodes hidden.
Results
Data selection
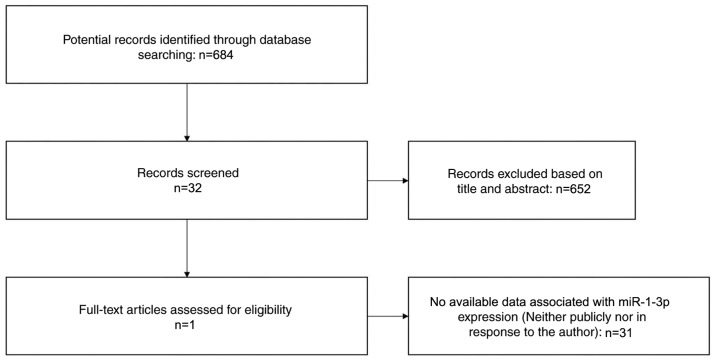
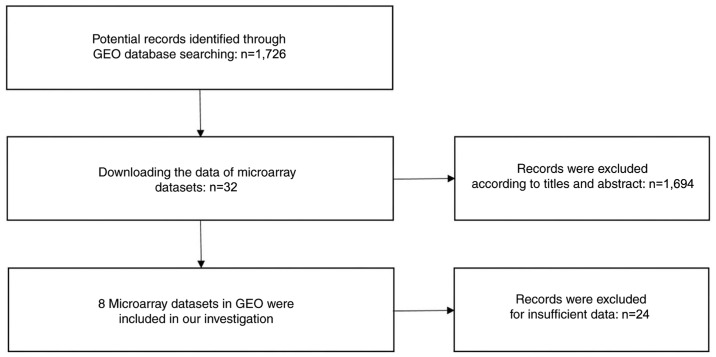
A search for literature, and GEO, ArrayExpress and TCGA datasets was performed to investigate the level of miRNA expression in LUSC. A total of 684 potential studies were identified following preliminary literature searches and finally a study containing two studies was deemed eligible for the present analysis following abstract screening and examination of the full text (41) (Fig. 1). A total of eight GEO datasets (GSE14936, GSE16025, GSE19945, GSE25508, GSE40738, GSE47525, GSE51853 and GSE74190) were selected (48–55) (Fig. 2), and two studies from TCGA were eligible for the present analysis. Eventually 12 studies with 1,386 cases were included. The GSE40738 samples were derived from serum, while the remaining samples were derived from tissue.
Figure 1.
Flow chart of the study selection process used in the present meta-analysis. miR, microRNA.
Figure 2.
Flow diagram of the GEO dataset selection process used in the present meta-analysis. GEO, Gene Expression Omnibus.
miR-1 expression in LUSC
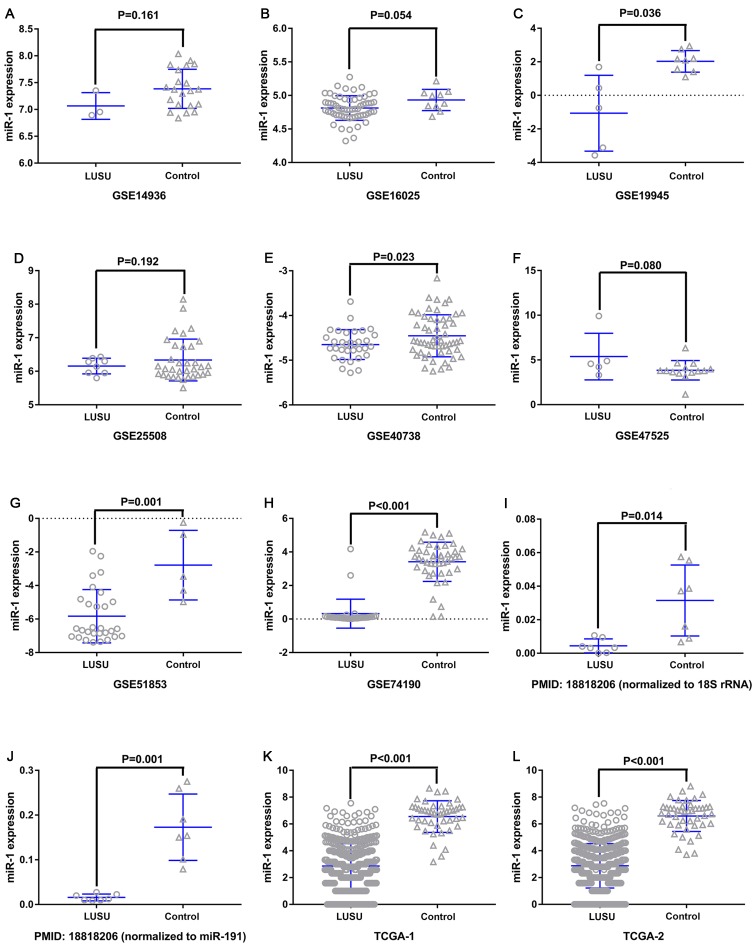
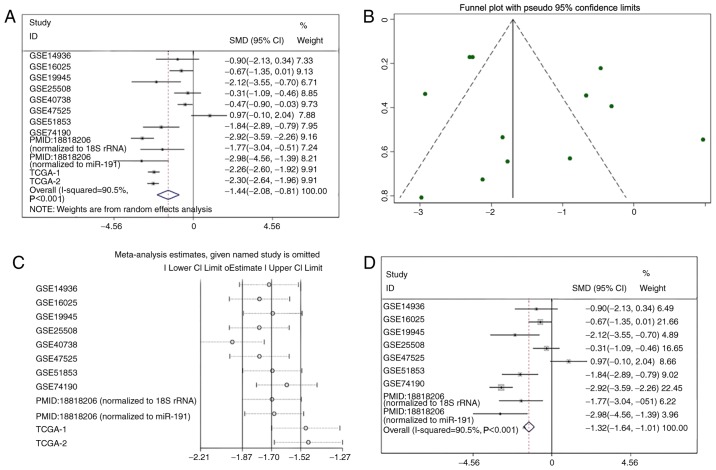
miR-1 was upregulated in the GSE47525 study and downregulated in other previous studies (Table I; Fig. 3). A meta-analysis was performed to examine the level of miR-1 expression in LUSC. A random effects model was implied due to the high heterogeneity (I2=90.5%), and the combined SMD was −1.44 (95% CI: −2.08, −0.81; Fig. 4A), demonstrating that miR-1 was significantly downregulated in LUSC. Publication bias was not observed in the present study (P>0.05; Fig. 4B). Sensitivity analysis was additionally performed to detect the source of heterogeneity (Fig. 4C), and the combined SMD was −1.32 (95% CI: −1.64, −1.01; I2=95.5%) when the GSE40738 studies and TCGA data were removed (Fig. 4D). The consistent conclusions obtained confirmed that these results were statistically stable.
Figure 3.
miR-1 expression in the LUSC and control groups. (A) GSE14936, (B) GSE16025, (C) GSE19945, (D) GSE25508, (E) GSE40738, (F) GSE47525, (G) GSE51853, (H) GSE74190, (I) PMID18818206-1, (J) PMID18818206-2, (K) TCGA-1 and (L) TCGA-2. The GSE40738 samples were derived from serum, and the remaining samples were derived from tissue. miR, microRNA; LUSC, lung squamous cell carcinoma; GSE, Genomic Spatial Event.
Figure 4.
Summary of the included studies. (A) Forest plots of all included studies. (B) Funnel plot. (C) Sensitivity analysis. (D) Forest plots of the studies following removal of the GSE40738 and TCGA data. GSE, Genomic Spatial Event; PMID, PubMed ID; TCGA, The Cancer Genome Atlas; SMD, standard mean difference; CI, confidence interval.
Clinical significance of miR-1 in LUSC
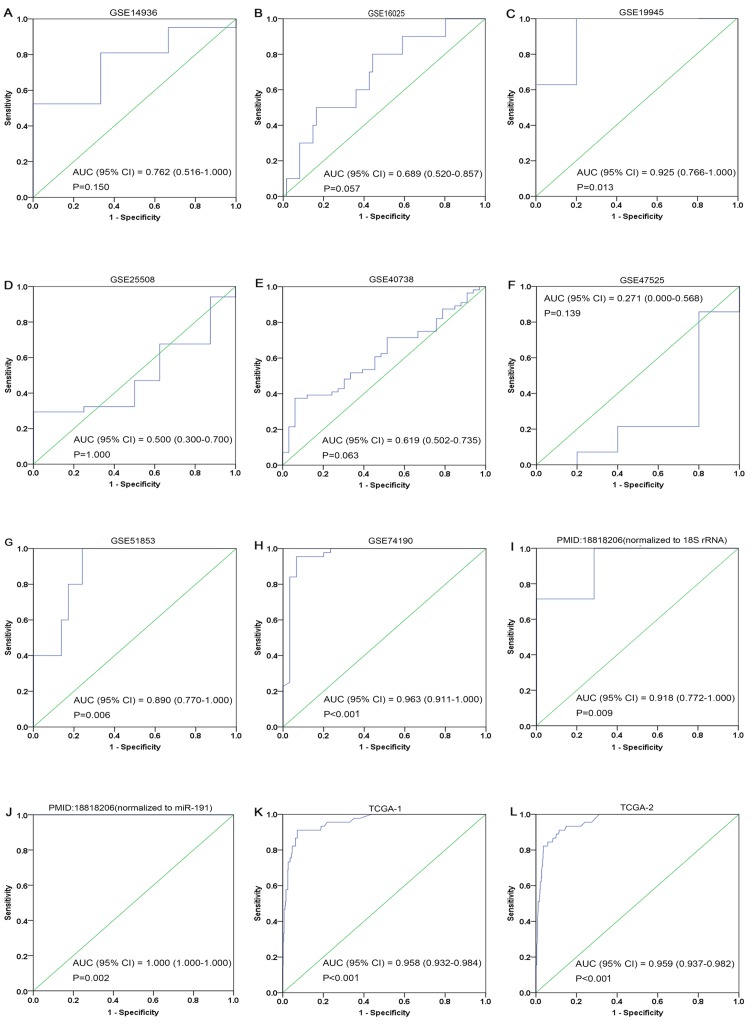
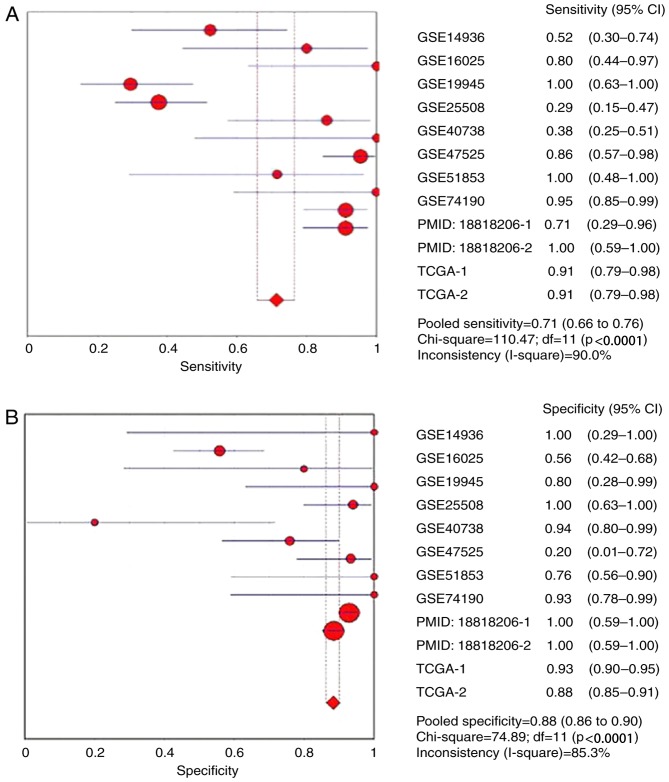
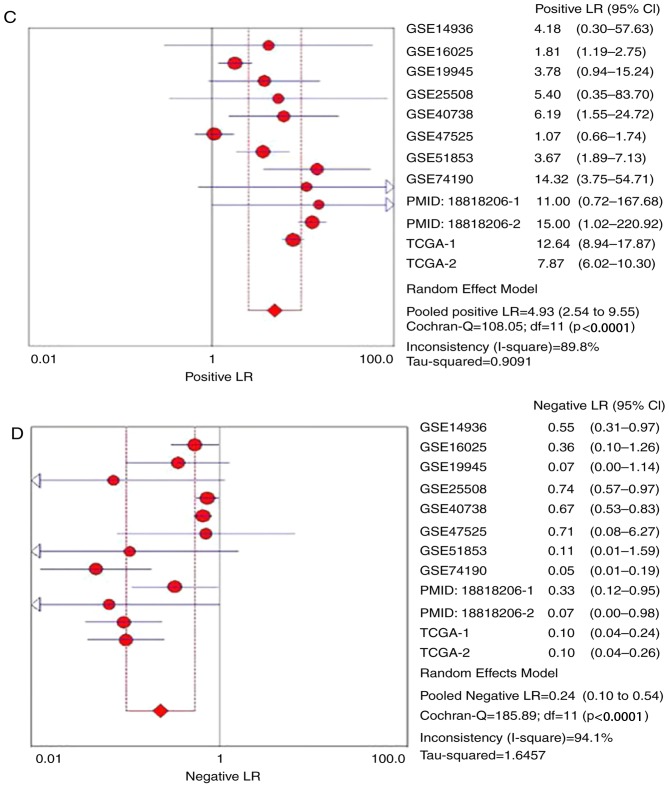
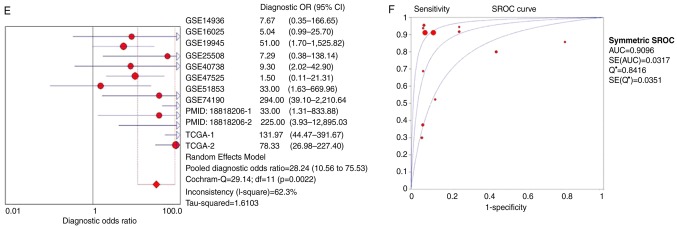
The ROC curves of miR-1 in LUSC for each study are presented in Fig. 5. It is evident that there was significant heterogeneity in these datasets, with sensitivity ranging from 0.38–1 and specificity ranging from 0.2–1. To circumvent this heterogeneity, a random effects model was adopted in the combined analyses (Fig. 6A-E). The combined sensitivity, specificity, PLR, NLR and DOR were 0.71 (95% CI: 0.66, 0.76; P<0.01), 0.88 (95% CI: 0.86, 0.90; P<0.01), 4.93 (95% CI: 2.54, 9.55; P<0.01), 0.24 (95% CI: 0.10, 0.54; P<0.01) and 28.24 (95% CI: 10.56, 75.53; P=0.0022), respectively. The SROC curve is presented in Fig. 6F, with an AUC of 0.9096 (Q*=0.8416), suggesting a good accuracy of miR-1 to distinguish patients with LUSC from control subjects.
Figure 5.
ROC curves of microRNA-1 in lung squamous cell carcinoma. (A) GSE14936, (B) GSE16025, (C) GSE19945, (D) GSE25508, (E) GSE40738, (F) GSE47525, (G) GSE51853, (H) GSE74190, (I) 18818206-1, (J) 18818206-2, (K) TCGA-1 and (L) TCGA-2. Blue represents a sensitive curve and green indicates the identifying line. The X-axis, presented as ‘1-Specificity’, indicates the false positive rate. The Y-axis, presented as ‘Sensitivity’, indicates the true positive rate. ROC, receiver operating characteristic.
Figure 6.
Forest plots of pooled miR-1 in the diagnosis of lung squamous cell carcinoma. (A) Sensitivity, (B) specificity, (C) positive LR, (D) negative LR, indicating the sensitivity and specificity of all included studies. miR, microRNA; SROC, summarized receiver operating characteristic; GSE, Genomic Spatial Event; PMID, PubMed ID; TCGA, The Cancer Genome Atlas; TCGA, The Cancer Genome Atlas; CI, confidence interval; AUC, area under the curve; LR, likelihood ratio; OR, odds ratio. Forest plots of pooled miR-1 in the diagnosis of lung squamous cell carcinoma. (E) Diagnostic OR and (F) SROC graphs indicating the sensitivity and specificity of all included studies.
Function analysis of miR-1-associated genes in LUSC
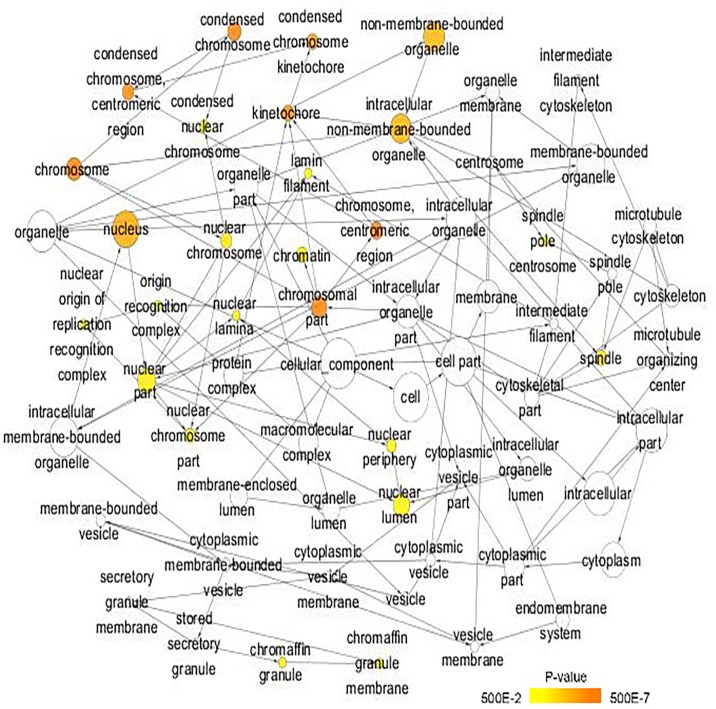
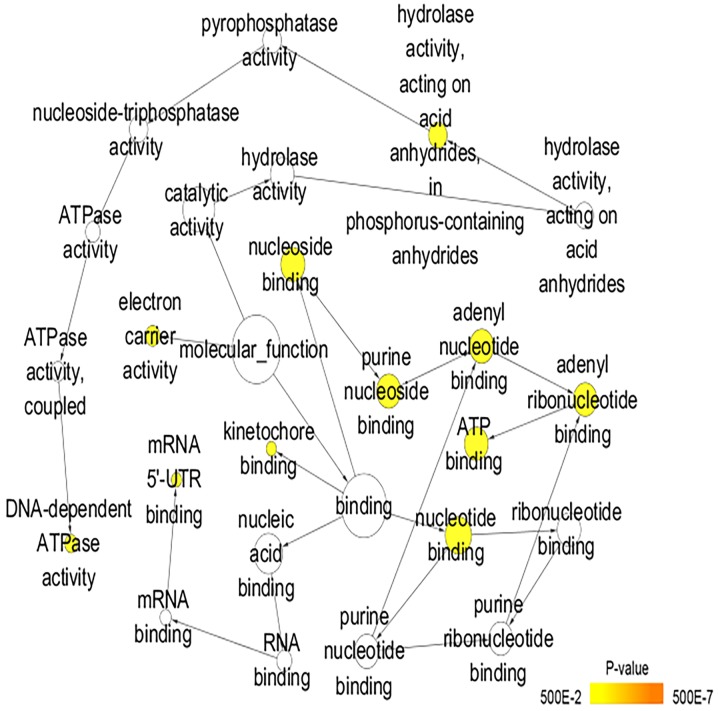
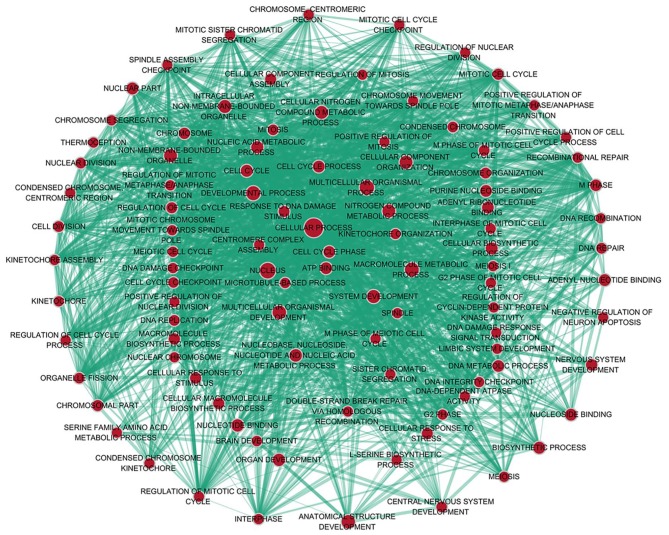
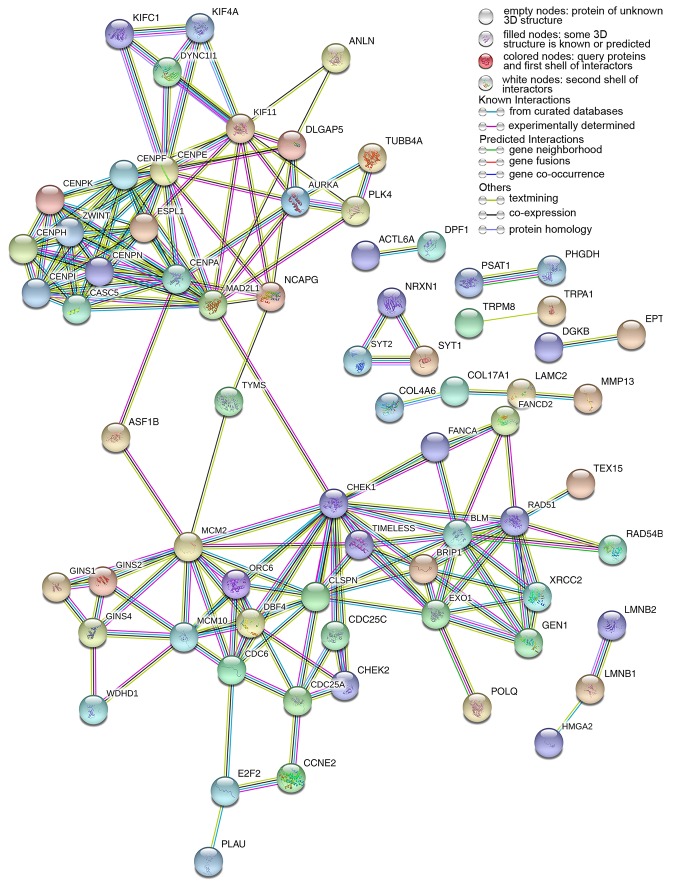
A total of 222 overlapping potential target genes of miR-1 in LUSC were obtained. Subsequently, GO and KEGG analyses were performed to examine the mechanism of action of miRNA in LUSC. The GO biological process annotation demonstrated that the promising target genes were primarily enriched in ‘DNA replication’, ‘cell division’, ‘DNA repair’, ‘G1/S transition of mitotic cell cycle’ and ‘sister chromatid cohesion’ (Table II; Fig. 7). In cellular component, the promising target genes were closely associated with ‘nucleoplasm’, ‘chromatin’, ‘kinetochore’, ‘GINS complex’ and ‘chromosome, centromeric region’ (Table II; Fig. 8). For molecular function, five of the most enriched items were ‘chromatin binding’, ‘ATP binding’, ‘3’-5’ DNA helicase activity’, ‘DNA replication origin binding’ and ‘microtubule motor activity’ (Table II; Fig. 9). In addition, the significant KEGG pathways included ‘cell cycle’, ‘p53 signaling pathway’, ‘Fanconi anemia pathway’, ‘homologous recombination’, ‘glycine, serine and threonine metabolism’ and ‘oocyte meiosis’ (Table III; Fig. 10). These results suggested that miR-1 may serve an important role in LUSC via multiple pathways. There were 222 nodes and 1,004 edges in the PPI network. In terms of the PPI network, in the current study, GINS complex subunit 4, DBF4 zinc finger, GINS complex subunit 1, phosphoserine aminotransferase 1, minichromosome maintenance complex component 2, minichromosome maintenance complex component 10, checkpoint kinase 1, GINS complex subunit 2, 3-phosphoglycerate dehydrogenase, cell division cycle 6 and cell division cycle 25C exhibited the highest degrees (Table IV; Fig. 11).
Table II.
GO analysis for the predicted target genes of microRNA-1 (only the top 10 pathways are presented).
| Category | Count | P-value | Target genes |
|---|---|---|---|
| A, Biological process | |||
| GO:0051301~cell division | 17 | 4.73×10−6 | CDC6, KIFC1, KIF11, CENPF, AURKA, CENPE, CHEK2, etc. |
| GO:0006281~DNA repair | 14 | 4.97×10−6 | EXO1, CLSPN, XRCC2, BLM, GEN1, CHEK1, RAD51, etc. |
| GO:0000082~G1/S transition of mitotic cell cycle | 9 | 2.96×10−5 | CCNE2, CDC6, TYMS, DBF4, ORC6, MCM2, MCM10, etc. |
| GO:0007062~sister chromatid cohesion | 9 | 3.18×10−5 | CENPN, MAD2L1, CENPA, ZWINT, CENPF, CENPE, etc. |
| GO:0006270~DNA replication initiation | 6 | 3.53×10−5 | CNE2, CDC6, GINS4, ORC6, MCM2, MCM10 |
| GO:0032508~DNA duplex unwinding | 6 | 1.70×10−4 | GINS1, GINS2, BLM, GINS4, BRIP1, RAD54B |
| GO:0007067~mitotic nuclear division | 12 | 1.97×10−4 | CENPN, CDC6, FAM64A, KIF11, TIMELESS, CENPF, etc. |
| GO:0000070~mitotic sister chromatid segregation | 5 | 2.03×10−4 | KIFC1, MAD2L1, CENPA, ZWINT, ESPL1 |
| GO:0000732~strand displacement | 5 | 2.38×10−4 | EXO1, XRCC2, BLM, BRIP1, RAD51 |
| B, Cellular component | |||
| GO:0005654~nucleoplasm | 59 | 3.45×10−6 | E2F2, CLSPN, XRCC2, DBF4, AURKA, CBX2, MCM10, etc. |
| GO:0000785~chromatin | 8 | 7.79×10−5 | CENPF, CHEK1, MCM2, ASF1B, HMGA2, ESCO2, etc. |
| GO:0000776~kinetochore | 7 | 3.54×10−4 | DYNC1I1, MAD2L1, ZWINT, CENPF, CENPE, CENPI, CENPH |
| GO:0000811~GINS complex | 3 | 3.97×10−4 | GINS1, GINS2, GINS4 |
| GO:0000775~chromosome, centromeric region | 6 | 5.09×10−4 | CENPN, MKI67, CENPA, CENPF, CENPE, HELLS |
| GO:0000777~condensed chromosome kinetochore | 7 | 5.20×10−4 | DYNC1I1, CENPN, MAD2L1, ZWINT, CENPE, CENPK, CENPH |
| GO:0031298~replication fork protection complex | 3 | 2.70×10−3 | GINS2, GINS4, MCM10 |
| GO:0000922~spindle pole | 6 | 8.77×10−3 | DYNC1I1, CENPN, MAD2L1, ZWINT, CENPE, CENPK, CENPH |
| GO:0005634~nucleus | 80 | 9.28×10−3 | KIFC1, FIGNL1, AURKA, CBX2, MCM10, ZIC1, GLDC, etc. |
| GO:0005829~cytosol | 53 | 1.12×10−2 | TFERMT1, AURKA, GTSE1, GLDC, CCNE2, PCP4, etc. |
| C, Molecular function | |||
| GO:0003682~chromatin binding | 14 | 8.89×10−4 | EXO1, CENPF, ATAD2, CBX2, VSX1, TP73, RAD51, DLX1, etc. |
| GO:0005524~ATP binding | 32 | 1.78×10−3 | KIFC1, KIF4A, XRCC2, GCLC, ATP10B, BLM, FIGNL1,etc. |
| GO:0043138~3′-5′; DNA helicase activity | 3 | 2.88×10−3 | GINS1, GINS2, GINS4 |
| GO:0003688~DNA replication origin binding | 3 | 7.30×10−3 | ORC6, MCM2, MCM10 |
| GO:0003777~microtubule motor activity | 5 | 1.55×10−2 | DYNC1I1, KIFC1, KIF4A, KIF11, CENPE |
| GO:0030276~clathrin binding | 4 | 2.43×10−2 | SYT1, SYT2, SYT13, SYT16 |
| GO:0003697~single-stranded DNA binding | 5 | 2.55×10−2 | XRCC2, BLM, NEIL3, MCM10, RAD51 |
| GO:0004520~endodeoxyribonuclease activity | 3 | 2.81×10−2 | XRCC2, GEN1, RAD51 |
| GO:0005544~calcium-dependent phospholipid binding | 4 | 3.22×10−2 | SYT1, SYT2, SYT13, SYT16 |
| GO:0008395~steroid hydroxylase activity | 3 | 3.84×10−2 | CYP2B6, CYP2S1, CYP2W1 |
GO, Gene Ontology.
Figure 7.
GO analysis of the biological process category. Nodes represent GO terms and arrows represent interactions. Orange nodes imply that the items are statistically significant (P<0.01). White nodes imply that the items only take part in connecting items but are not statistically significant. GO, Gene Ontology.
Figure 8.
Gene Ontology analysis of the cellular component category. Nodes represent GO terms and arrows represent interactions. Orange nodes imply that the items are statistically significant (P<0.05). White nodes imply that the items only take part in connecting items but are not statistically significant. GO, Gene Ontology.
Figure 9.
Gene Ontology analysis of the molecular function category. Nodes represent GO terms and arrows represent interactions. Orange nodes imply that the items are statistically significant (P<0.05). White nodes imply that the items only take part in connecting items but are not statistically significant. GO, Gene Ontology.
Table III.
KEGG pathway of validated target genes of microRNA-1.
| KEGG Pathway | Count | P-value | Target genes |
|---|---|---|---|
| hsa04110: Cell cycle | 12 | 3.52×10−7 | CCNE2, E2F2, CDC6, MAD2L1, DBF4, etc. |
| hsa04115: p53 signaling pathway | 6 | 1.47×10−3 | CCNE2, SERPINB5, CHEK1, CHEK2, GTSE1, etc. |
| hsa03460: Fanconi anemia pathway | 5 | 4.29×10−3 | BLM, FANCD2, BRIP1, FANCA, RAD51 |
| hsa03440: Homologous recombination | 4 | 5.56×10−3 | XRCC2, BLM, RAD54B, RAD51 |
| hsa00260: Glycine, serine and threonine metabolism | 4 | 1.27×10−2 | BHMT, PHGDH, PSAT1, GLDC |
| hsa04114: Oocyte meiosis | 5 | 4.81×10−2 | CCNE2, MAD2L1, AURKA, ESPL1, CDC25C |
| hsa05206: MicroRNAs in cancer | 8 | 6.69×10−2 | CCNE2, E2F2, SERPINB5, HMGA2, CDC25C, etc. |
A total of eight pathways were available, four of which were significant (P<0.01). KEGG, Kyoto Encyclopedia of Genes and Genomes.
Figure 10.
Kyoto Encyclopedia of Genes and Genomes pathway enrichment analyses. Nodes represent proteins and edges represent interactions.
Table IV.
Top 10 genes with combined scores in the protein-protein interaction network of potential target genes of microRNA-1.
| Node1 | Node2 | Node1 string internal ID | Co-expression | Experimentally determined interaction | Database annotated | Automated text mining | Combined score |
|---|---|---|---|---|---|---|---|
| GINS4 | GINS2 | 1846499 | 0.367 | 0.997 | 0.9 | 0.953 | 0.999 |
| DBF4 | MCM2 | 1845796 | 0.164 | 0.922 | 0.9 | 0.875 | 0.999 |
| GINS1 | GINS2 | 1845151 | 0.764 | 0.997 | 0.9 | 0.953 | 0.999 |
| PSAT1 | PHGDH | 1856523 | 0.971 | 0.417 | 0.941 | 0.743 | 0.999 |
| MCM2 | GINS2 | 1845676 | 0.859 | 0.898 | 0.9 | 0.484 | 0.999 |
| MCM2 | CDC6 | 1845676 | 0.582 | 0.957 | 0.9 | 0.923 | 0.999 |
| MCM10 | MCM2 | 1854261 | 0.769 | 0.934 | 0.9 | 0.842 | 0.999 |
| GINS4 | GINS1 | 1846499 | 0.371 | 0.996 | 0.9 | 0.953 | 0.999 |
| CHEK1 | CDC25C | 1859362 | 0.448 | 0.528 | 0.9 | 0.956 | 0.998 |
Figure 11.
Protein interactions of correlative genes for miR-1; the protein-to-protein network analysis was performed using Search Tool for the Retrieval of Interacting Genes/Proteins version 10.5. miR, microRNA.
Discussion
Recently, numerous studies demonstrated that miR-1 is predominantly downregulated in multiple human tumors, including lung cancer, prostate cancer, breast cancer and sarcomas (28,56), and serves as a tumor suppressor gene involved in cancer progression. However, studies on the expression of miR-1 and its clinical significance in LUSC are rare, and the underlying mechanism of miR-1 in LUSC remains to be elucidated. The present study investigated miR-1 expression, clinical significance and the potential molecular mechanisms of miR-1 in LUSC.
To the best of the authors' knowledge, the present study is the first meta-analysis to examine the expression of miR-1 and its clinical significance in LUSC. The present meta-analysis involved 1,386 cases from 12 eligible studies. The combined SMD was −1.70 (95% CI: −1.857, 1.52) with high heterogeneity (I2=90.5%; P=0.001), demonstrating that miR-1 was significantly downregulated in LUSC. The AUC was 0.9096 (Q*=0.8416) with a sensitivity of 0.71 (95% CI: 0.66, 0.76, P<0.01) and a specificity of 0.88 (95% CI: 0.86, 0.90, P<0.01). The pooled PLR and NLR were 4.93 (95% CI: 2.54, 9.55) and 0.24 (95% CI: 0.10, 0.54), respectively. A PLR value of 4.00 suggested that patients with LUSC possessed an ~4-fold higher probability of exhibiting downregulated miR-1 compared with control groups. An NLR value of 0.34 suggested that the probability of having LUSC was 34% when miR-1 is abnormal. In addition, the pooled DOR was 28.24 (95% CI: 10.56, 75.53). According to these results, miR-1 may better differentiate between LUSC and patients without cancer. Sensitivity analysis demonstrated that high heterogeneity may reflect the GSE40738 studies and TCGA data. To obtain more convincing conclusions, additional studies using a higher quality, larger sample size and a consistent standard procedure are required.
Accumulating evidence demonstrated that miR-1 serves an important role in the development of multiple tumors (57,58). Mataki et al (59) demonstrated that miR-1/133a was significantly downregulated in LUSC tissues and enhanced cancer cell invasion and migration via the regulation of Coronin1C. However, little is known of the potential molecular mechanisms of miR-1 in LUSC. Therefore, 222 validated targeting genes of miR-1 were collected and a comprehensive target genes network analysis performed. GO analysis demonstrated that miR-1 may be involved in multiple biological processes, including ‘DNA replication’, ‘cell division’, ‘DNA repair’ and the ‘G1/S transition of the mitotic cell cycle’. KEGG pathway analysis identified that miR-1 may serve a pivotal role in LUSC via different pathways, including ‘cell cycle’, ‘p53 signaling pathway’, ‘Fanconi anemia pathway’, ‘homologous recombination’, ‘glycine, serine and threonine metabolism’ and ‘oocyte meiosis’. The p53 gene, a key tumor suppressor located on chromosome 17p13, is the most frequently mutated gene in cancer and is involved in in a variety of biological processes to prevent tumorigenesis through the transcriptional regulation of downstream target genes (60). A previous study demonstrated that p53 is associated with cell cycle arrest, apoptosis and drug resistance in lung cancer cells (61). Zhang et al (62) demonstrated that GluA2 induces apoptosis in non-small cell lung cancer A549 cells through the p53 signaling pathway; the p53 signaling pathway was a significant pathway (Table III; P=0.001) that ranked second in the biological process of LUSC. The p53 pathway may serve an important role in radio sensitivity in non-small cell lung cancer H460 cells via the upregulation of phosphatase and tensin homolog expression level (63). A number of previous studies (64–66) demonstrated that miRNAs may be a key effector of p53 tumor-suppressor function; mediating the biological effects of p53 and inactivating this molecule may contribute to specific cancer types (67), suggesting that miRNAs may serve a vital role in the p53 gene signaling pathway. Therefore, it is hypothesized that miR-1 may be involved in the progression of LUSC through the p53 signaling pathway.
There are a number of limitations to the present study. The study size included in this meta-analysis was relatively small. The prognostic value of miR-1 was not discussed in this analysis, reflecting that there are no studies regarding the association between miR-1 and prognosis and clinic pathological parameters in LUSC. Only studies reported in Chinese and English were included in this meta-analysis, which may result in the omission of eligible studies due to language criteria. Furthermore, among the 12 studies included in the present study, only one was derived from serum, and the remaining 11 studies used samples derived from tissue. For a better understanding of the role of miR-1, large cohort and independent studies are required to examine the clinical significance of miRNA and its potential mechanism in LUSC.
In conclusion, the present study demonstrated that miR-1 is significantly downregulated in LUSC and may be involved in cancer progression via multiple, crucial pathways. Therefore, miR-1 may be used as a screening tool for LUSC in the future. Large-scale studies are required to further investigate the clinical significance of miR-1.
Acknowledgements
The authors would like to acknowledge the public data provided by The Cancer Genome Atlas and Gene Expression Omnibus datasets.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
XL, MQ and JH contributed to the design of the study, data collection, analysis and drafting of the manuscript. JM and XH contributed to the design of the study, interpretation of the data and drafting of the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Chen M, Liu X, Du J, Wang XJ, Xia L. Differentiated regulation of immune-response related genes between LUAD and LUSC subtypes of lung cancers. Oncotarget. 2017;8:133–144. doi: 10.18632/oncotarget.13346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Freeman JR, Chu S, Hsu T, Huang YT. Epigenome-wide association study of smoking and DNA methylation in non-small cell lung neoplasms. Oncotarget. 2016;7:69579–69591. doi: 10.18632/oncotarget.11831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peng L, Bian XW, Li DK, Xu C, Wang GM, Xia QY, Xiong Q. Large-scale RNA-Seq transcriptome analysis of 4043 cancers and 548 normal tissue controls across 12 TCGA cancer types. Sci Rep. 2015;5:13413. doi: 10.1038/srep13413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Czarnecka KH, Migdalska-Sęk M, Domańska D, Pastusza-Lewandoska D, Dutkowska A, Kordiak J, Nawrot E, Kiszałkiewicz J, Antczak A, Brzeziańska-Lasota E. FHIT promoter methylation status, low protein and high mRNA levels in patients with non-small cell lung cancer. Int J Oncol. 2016;49:1175–1184. doi: 10.3892/ijo.2016.3610. [DOI] [PubMed] [Google Scholar]
- 5.Chang JT, Lee YM, Huang RS. The impact of the cancer genome atlas on lung cancer. Transl Res. 2015;166:568–585. doi: 10.1016/j.trsl.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang L, Chen Z, An L, Wang Y, Zhang Z, Guo Y, Liu C. Analysis of long non-coding RNA expression profiles in non-small cell lung cancer. Cell Physiol Biochem. 2016;38:2389–2400. doi: 10.1159/000445591. [DOI] [PubMed] [Google Scholar]
- 7.Bittoni MA, Focht BC, Clinton SK, Buckworth J, Harris RE. Prospective evaluation of C-reactive protein, smoking and lung cancer death in the Third National Health and Nutrition Examination Survey. Int J Oncol. 2015;47:1537–1544. doi: 10.3892/ijo.2015.3141. [DOI] [PubMed] [Google Scholar]
- 8.Eldem V, Çelikkol Akçay U, Ozhuner E, Bakır Y, Uranbey S, Unver T. Genome-wide identification of miRNAs responsive to drought in peach (Prunus persica) by high-throughput deep sequencing. PLoS One. 2012;7:e50298. doi: 10.1371/journal.pone.0050298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma N, Zhang W, Qiao C, Luo H, Zhang X, Liu D, Zang S, Zhang L, Bai J. The tumor suppressive role of MiRNA-509-5p by targeting FOXM1 in non-small cell lung cancer. Cell Physiol Biochem. 2016;38:1435–1446. doi: 10.1159/000443086. [DOI] [PubMed] [Google Scholar]
- 10.Li P, Liu H, Wang Z, He F, Wang H, Shi Z, Yang A, Ye J. MicroRNAs in laryngeal cancer: Implications for diagnosis, prognosis and therapy. Am J Transl Res. 2016;8:1935–1944. [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J, Li Y, Ding M, Zhang H, Xu X, Tang J. Molecular mechanisms and clinical applications of miR-22 in regulating malignant progression in human cancer (Review) Int J Oncol. 2017;50:345–355. doi: 10.3892/ijo.2016.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gambari R, Brognara E, Spandidos DA, Fabbri E. Targeting oncomiRNAs and mimicking tumor suppressor miRNAs: Νew trends in the development of miRNA therapeutic strategies in oncology (Review) Int J Oncol. 2016;49:5–32. doi: 10.3892/ijo.2016.3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Subramani R, Gangwani L, Nandy SB, Arumugam A, Chattopadhyay M, Lakshmanaswamy R. Emerging roles of microRNAs in pancreatic cancer diagnosis, therapy and prognosis (Review) Int J Oncol. 2015;47:1203–1210. doi: 10.3892/ijo.2015.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang P, Yang D, Zhang H, Wei X, Ma T, Cheng Z, Hong Q, Hu J, Zhuo H, Song Y, et al. Early detection of lung cancer in serum by a panel of MicroRNA biomarkers. Clin Lung Cancer. 2015;16:313–319.e1. doi: 10.1016/j.cllc.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 15.Ma T, Zhao Y, Wei K, Yao G, Pan C, Liu B, Xia Y, He Z, Qi X, Li Z, et al. MicroRNA-124 functions as a tumor suppressor by regulating CDH2 and epithelial-mesenchymal transition in non-small cell lung cancer. Cell Physiol Biochem. 2016;38:1563–1574. doi: 10.1159/000443097. [DOI] [PubMed] [Google Scholar]
- 16.Feng X, Jiang J, Shi S, Xie H, Zhou L, Zheng S. Knockdown of miR-25 increases the sensitivity of liver cancer stem cells to TRAIL-induced apoptosis via PTEN/PI3K/Akt/Bad signaling pathway. Int J Oncol. 2016;49:2600–2610. doi: 10.3892/ijo.2016.3751. [DOI] [PubMed] [Google Scholar]
- 17.Meerson A, Yehuda H. Leptin and insulin up-regulate miR-4443 to suppress NCOA1 and TRAF4, and decrease the invasiveness of human colon cancer cells. BMC Cancer. 2016;16:882. doi: 10.1186/s12885-016-2938-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumamoto T, Seki N, Mataki H, Mizuno K, Kamikawaji K, Samukawa T, Koshizuka K, Goto Y, Inoue H. Regulation of TPD52 by antitumor microRNA-218 suppresses cancer cell migration and invasion in lung squamous cell carcinoma. Int J Oncol. 2016;49:1870–1880. doi: 10.3892/ijo.2016.3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Chen L, Wu Z, Wang M, Jin F, Wang N, Hu X, Liu Z, Zhang CY, Zen K, et al. miR-124-3p functions as a tumor suppressor in breast cancer by targeting CBL. BMC Cancer. 2016;16:826. doi: 10.1186/s12885-016-2862-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu X, Li Z. New insights into MicroRNAs involves in drug resistance in diffuse large B cell lymphoma. Am J Transl Res. 2015;7:2536–2542. [PMC free article] [PubMed] [Google Scholar]
- 21.Li Z, Yu X, Shen J, Law PT, Chan MT, Wu WK. MicroRNA expression and its implications for diagnosis and therapy of gallbladder cancer. Oncotarget. 2015;6:13914–13921. doi: 10.18632/oncotarget.4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cui L, Li Y, Lv X, Li J, Wang X, Lei Z, Li X. Expression of MicroRNA-301a and its functional roles in malignant melanoma. Cell Physiol Biochem. 2016;40:230–244. doi: 10.1159/000452540. [DOI] [PubMed] [Google Scholar]
- 23.Zhang X, Zhang Y, Liu X, Fang A, Wang J, Yang Y, Wang L, Du L, Wang C. Direct quantitative detection for cell-free miR-155 in urine: A potential role in diagnosis and prognosis for non-muscle invasive bladder cancer. Oncotarget. 2016;7:3255–3266. doi: 10.18632/oncotarget.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao Y, Feng B, Han S, Lu L, Chen Y, Chu X, Wang R, Chen L. MicroRNA-129 in human cancers: From tumorigenesis to clinical treatment. Cell Physiol Biochem. 2016;39:2186–2202. doi: 10.1159/000447913. [DOI] [PubMed] [Google Scholar]
- 25.Deng T, Yuan Y, Zhang C, Zhang C, Yao W, Wang C, Liu R, Ba Y. Identification of circulating MiR-25 as a potential biomarker for pancreatic cancer diagnosis. Cell Physiol Biochem. 2016;39:1716–1722. doi: 10.1159/000447872. [DOI] [PubMed] [Google Scholar]
- 26.Lu L, Zhou L, Chen EZ, Sun K, Jiang P, Wang L, Su X, Sun H, Wang H. A Novel YY1-miR-1 regulatory circuit in skeletal myogenesis revealed by genome-wide prediction of YY1-miRNA network. PLoS One. 2012;7:e27596. doi: 10.1371/journal.pone.0027596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han C, Shen JK, Hornicek FJ, Kan Q, Duan Z. Regulation of microRNA-1 (miR-1) expression in human cancer. Biochim Biophys Acta. 2017;1860:227–232. doi: 10.1016/j.bbagrm.2016.12.004. [DOI] [PubMed] [Google Scholar]
- 28.Han C, Zhou Y, An Q, Li F, Li D, Zhang X, Yu Z, Zheng L, Duan Z, Kan Q. MicroRNA-1 (miR-1) inhibits gastric cancer cell proliferation and migration by targeting MET. Tumour Biol. 2015;36:6715–6723. doi: 10.1007/s13277-015-3358-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang W, Shen F, Wang C, Lu W, Wei J, Shang A, Wang C. MiR-1-3p inhibits the proliferation and invasion of bladder cancer cells by suppressing CCL2 expression. Tumour Biol. 2017;39:1010428317698383. doi: 10.1177/1010428317698383. [DOI] [PubMed] [Google Scholar]
- 30.Wang Z, Wang J, Chen Z, Wang K, Shi L. MicroRNA-1-3p inhibits proliferation and migration of oral squamous cell carcinoma cells by targeting DKK1. Biochem Cell Biol. 2018;96:355–364. doi: 10.1139/bcb-2017-0015. [DOI] [PubMed] [Google Scholar]
- 31.Cui R, Meng W, Sun HL, Kim T, Ye Z, Fassan M, Jeon YJ, Li B, Vicentini C, Peng Y, et al. MicroRNA-224 promotes tumor progression in nonsmall cell lung cancer. Proc Natl Acad Sci USA. 2015;112:E4288–E4297. doi: 10.1073/pnas.1502068112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang WC, Chin TM, Yang H, Nga ME, Lunny DP, Lim EK, Sun LL, Pang YH, Leow YN, Malusay SR, et al. Tumour-initiating cell-specific miR-1246 and miR-1290 expression converge to promote non-small cell lung cancer progression. Nat Commun. 2016;7:11702. doi: 10.1038/ncomms11702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang X, Wang C, Shan S, Liu X, Jiang Z, Ren T. TLR4/ROS/miRNA-21 pathway underlies lipopolysaccharide instructed primary tumor outgrowth in lung cancer patients. Oncotarget. 2016;7:42172–42182. doi: 10.18632/oncotarget.9902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu N, Zhang Q, Liu Q, Yang J, Zhang S. A meta-analysis: microRNAs' prognostic function in patients with nonsmall cell lung cancer. Cancer Med. 2017;6:2098–2105. doi: 10.1002/cam4.1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao L, Li SH, Tian YX, Zhu QQ, Chen G, Pang YY, Hu XH. Role of downregulated miR-133a-3p expression in bladder cancer: A bioinformatics study. Onco Targets Ther. 2017;10:3667–3683. doi: 10.2147/OTT.S137433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Y, Xing Z, Zhan P, Liu H, Ye W, Lv T, Song Y. Is it feasible to detect epidermal growth factor receptor mutations in circulating tumor cells in nonsmall cell lung cancer?: A meta-analysis. Medicine (Baltimore) 2016;95:e5115. doi: 10.1097/MD.0000000000005115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang J, Yu Y, Li Y, Wei L. Diagnostic value of contrast-enhanced ultrasound in hepatocellular carcinoma: A meta-analysis with evidence from 1998 to 2016. Oncotarget. 2017;8:75418–75426. doi: 10.18632/oncotarget.20049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen WS, Li JJ, Hong L, Xing ZB, Wang F, Li CQ. Comparison of MRI, CT and 18F-FDG PET/CT in the diagnosis of local and metastatic of nasopharyngeal carcinomas: An updated meta analysis of clinical studies. Am J Transl Res. 2016;8:4532–4547. [PMC free article] [PubMed] [Google Scholar]
- 39.Ma X, Wang L, Wu H, Feng Y, Han X, Bu H, Zhu Q. Spleen stiffness is superior to liver stiffness for predicting esophageal varices in chronic liver disease: A meta-analysis. PLoS One. 2016;11:e0165786. doi: 10.1371/journal.pone.0165786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y, Dang YW, Wang X, Yang X, Zhang R, Lv ZL, Chen G. Comprehensive analysis of long non-coding RNA PVT1 gene interaction regulatory network in hepatocellular carcinoma using gene microarray and bioinformatics. Am J Transl Res. 2017;9:3904–3917. [PMC free article] [PubMed] [Google Scholar]
- 42.Zeng JH, Xiong DD, Pang YY, Zhang Y, Tang RX, Luo DZ, Chen G. Identification of molecular targets for esophageal carcinoma diagnosis using miRNA-seq and RNA-seq data from The Cancer Genome Atlas: A study of 187 cases. Oncotarget. 2017;8:35681–35699. doi: 10.18632/oncotarget.16051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y, He RQ, Dang YW, Zhang XL, Wang X, Huang SN, Huang WT, Jiang MT, Gan XN, Xie Y, et al. Comprehensive analysis of the long noncoding RNA HOXA11-AS gene interaction regulatory network in NSCLC cells. Cancer Cell Int. 2016;16:89. doi: 10.1186/s12935-016-0366-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, Huang JC, Cai KT, Yu XB, Chen YR, Pan WY, He ZL, Lv J, Feng ZB, Chen G. Long non-coding RNA HOTTIP promotes hepatocellular carcinoma tumorigenesis and development: A comprehensive investigation based on bioinformatics, qRT-PCR and meta-analysis of 393 cases. Int J Oncol. 2017;51:1705–1721. doi: 10.3892/ijo.2017.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maere S, Heymans K, Kuiper M. BiNGO: A Cytoscape plugin to assess overrepresentation of Gene Ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 46.Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: A network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010;5:e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seike M, Goto A, Okano T, Bowman ED, Schetter AJ, Horikawa I, Mathe EA, Jen J, Yang P, Sugimura H, et al. MiR-21 is an EGFR-regulated anti-apoptotic factor in lung cancer in never-smokers. Proc Natl Acad Sci USA. 2009;106:12085–12090. doi: 10.1073/pnas.0905234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raponi M, Dossey L, Jatkoe T, Wu X, Chen G, Fan H, Beer DG. MicroRNA classifiers for predicting prognosis of squamous cell lung cancer. Cancer Res. 2009;69:5776–5783. doi: 10.1158/0008-5472.CAN-09-0587. [DOI] [PubMed] [Google Scholar]
- 50.Ohba T, Nagano H. A small-cell lung cancer subtype with good prognosis found by a three miRNA signature. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE19945. [Oct 1st;2017 ];
- 51.Nymark P, Guled M, Borze I, Faisal A, Lahti L, Salmenkivi K, Kettunen E, Anttila S, Knuutila S. Integrative analysis of microRNA, mRNA and aCGH data reveals asbestos- and histology-related changes in lung cancer. Genes Chromosomes Cancer. 2011;50:585–597. doi: 10.1002/gcc.20880. [DOI] [PubMed] [Google Scholar]
- 52.Patnaik SK, Kannisto ED, Mallick R, Vachani A, Yendamuri S. Whole blood microRNA expression may not be useful for screening non-small cell lung cancer. PLoS One. 2017;12:e0181926. doi: 10.1371/journal.pone.0181926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Jaarsveld MT, Wouters MD, Boersma AW, Smid M, van Ijcken WF, Mathijssen RH, Hoeijmakers JH, Martens JW, van Laere S, Wiemer EA, Pothof J. DNA damage responsive microRNAs misexpressed in human cancer modulate therapy sensitivity. Mol Oncol. 2014;8:458–468. doi: 10.1016/j.molonc.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arima C, Kajino T, Tamada Y, Imoto S, Shimada Y, Nakatochi M, Suzuki M, Isomura H, Yatabe Y, Yamaguchi T, et al. Lung adenocarcinoma subtypes definable by lung development-related miRNA expression profiles in association with clinicopathologic features. Carcinogenesis. 2014;35:2224–2231. doi: 10.1093/carcin/bgu127. [DOI] [PubMed] [Google Scholar]
- 55.Jin Y, Liu YL, Lu SH. The miRNA expression profiles in three subtypes of lung carcinomas. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE74190. [Oct 12th;2017 ];
- 56.Liu T, Hu K, Zhao Z, Chen G, Ou X, Zhang H, Zhang X, Wei X, Wang D, Cui M, Liu C. MicroRNA-1 down-regulates proliferation and migration of breast cancer stem cells by inhibiting the Wnt/β-catenin pathway. Oncotarget. 2015;6:41638–41649. doi: 10.18632/oncotarget.5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xie M, Dart DA, Guo T, Xing XF, Cheng XJ, Du H, Jiang WG, Wen XZ, Ji JF. MicroRNA-1 acts as a tumor suppressor microRNA by inhibiting angiogenesis-related growth factors in human gastric cancer. Gastric Cancer. 2018;21:41–54. doi: 10.1007/s10120-017-0721-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu X, Wu X, Jiang Q, Sun Y, Liu H, Chen R, Wu S. Downregulation of microRNA-1 and microRNA-145 contributes synergistically to the development of colon cancer. Int J Mol Med. 2015;36:1630–1638. doi: 10.3892/ijmm.2015.2364. [DOI] [PubMed] [Google Scholar]
- 59.Mataki H, Enokida H, Chiyomaru T, Mizuno K, Matsushita R, Goto Y, Nishikawa R, Higashimoto I, Samukawa T, Nakagawa M, et al. Downregulation of the microRNA-1/133a cluster enhances cancer cell migration and invasion in lung-squamous cell carcinoma via regulation of Coronin1C. J Hum Genet. 2015;60:53–61. doi: 10.1038/jhg.2014.111. [DOI] [PubMed] [Google Scholar]
- 60.Liu J, Zhang C, Feng Z. Tumor suppressor p53 and its gain-of-function mutants in cancer. Acta Biochim Biophys Sin (Shanghai) 2014;46:170–179. doi: 10.1093/abbs/gmt144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhong G, Chen X, Fang X, Wang D, Xie M, Chen Q. Fra-1 is upregulated in lung cancer tissues and inhibits the apoptosis of lung cancer cells by the P53 signaling pathway. Oncol Rep. 2016;35:447–453. doi: 10.3892/or.2015.4395. [DOI] [PubMed] [Google Scholar]
- 62.Zhang HY, Yang W, Lu JB. Knockdown of GluA2 induces apoptosis in non-small-cell lung cancer A549 cells through the p53 signaling pathway. Oncol Lett. 2017;14:1005–1010. doi: 10.3892/ol.2017.6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jung IL, Kang HJ, Kim KC, Kim IG. PTEN/pAkt/p53 signaling pathway correlates with the radioresponse of non-small cell lung cancer. Int J Mol Med. 2010;25:517–523. doi: 10.3892/ijmm_00000372. [DOI] [PubMed] [Google Scholar]
- 64.Zhang C, Liu J, Tan C, Yue X, Zhao Y, Peng J, Wang X, Laddha SV, Chan CS, Zheng S, et al. microRNA-1827 represses MDM2 to positively regulate tumor suppressor p53 and suppress tumorigenesis. Oncotarget. 2016;7:8783–8796. doi: 10.18632/oncotarget.7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Perdas E, Stawski R, Nowak D, Zubrzycka M. Potential of liquid biopsy in papillary thyroid carcinoma in context of miRNA, BRAF and p53 mutation. Curr Drug Targets. 2018;19:1721–1729. doi: 10.2174/1389450119666180226124349. [DOI] [PubMed] [Google Scholar]
- 66.Xiao S, Wang R, Wu X, Liu W, Ma S. The long noncoding RNA TP73-AS1 interacted with miR-124 to modulate glioma growth by targeting inhibitor of apoptosis-stimulating protein of p53. DNA Cell Biol. 2018;37:117–125. doi: 10.1089/dna.2017.3941. [DOI] [PubMed] [Google Scholar]
- 67.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.