Abstract
Cellular adaptation to environmental stress relies on a wide range of tightly controlled regulatory mechanisms, including transcription. Changes in chromatin structure and organization accompany the transcriptional response to stress, and in some cases, can impart memory of stress exposure to subsequent generations through mechanisms of epigenetic inheritance. In the budding yeast Saccharomyces cerevisiae, histone post-translational modifications, and in particular histone methylation, have been shown to confer transcriptional memory of exposure to environmental stress conditions through mitotic divisions. Recent evidence from Caenorhabditis elegans also implicates histone methylation in transgenerational inheritance of stress responses, suggesting a more widely conserved role in epigenetic memory.
Keywords: chromatin, histone methylation, transcriptional memory, epigenetic inheritance, stress
1. Introduction
Here we review recent studies showing how histone methylation contributes to the mitotic inheritance of transcriptional changes induced by stress in S. cerevisiae, and specific cases where exposure to high temperature, chemicals, and starvation have been associated with changes in histone methylation in C. elegans, a model system widely used for transgenerational studies. The examples described in C. elegans suggest that histone modifications may also play a role in transgenerational inheritance of environmental exposure, although most of the observed effects extend across a few generations at most, and the underlying mechanisms remain to be described. Nonetheless, the tractability of C. elegans continues to provide valuable insights into the complex and varied mechanisms of transgenerational inheritance.
Histone post-translational modifications (PTMs) are covalent modifications of histones that contribute to chromosome structure and function. In most species, repressive chromatin is generally enriched in H3K9 and H3K27 trimethylation (H3K9me3 and H3K27me3), while H3K4me3 is enriched at promoter regions and associated with more accessible chromatin [1]. In S. cerevisiae H3K9 methylation is absent, but other features of both repressive and active chromatin are conserved. In C. elegans, transcriptionally silent regions marked by H3K9me3/H3K27me3 and enriched in transposons and other repetitive sequences, are found predominantly on the arms of chromosomes, while the distribution of H3K4me3 is similar to that found in other organisms [2]. PTMs and their readers influence fundamental processes, including transcription, replication, and the maintenance of genome integrity. In a developmental context, regulation of the chromatin landscape is pivotal in the establishment and maintenance of the epigenetic programs of development and differentiation, and in the response to environmental stress [3].
2. Histone Methylation and Transcriptional Memory of Stress Induced Gene Activation in Yeast
Unicellular organisms such as yeast survive environmental stress by responding rapidly to changes in temperature, nutrients, and osmotic pressure, among others. In specific cases, survival is improved when yeast cells “remember” a previous stressful experience, and are capable not only of a more rapid reactivation of the appropriate transcriptional stress response, but also of transmitting this response to their descendants. Here, we focus on examples of epigenetic transcriptional memory implicating chromatin modifications, although alternative mechanisms have been reported [4].
2.1. INO1 Transcriptional Memory
One of the best-studied examples of epigenetic transcriptional memory has been described for yeast cells that have previously experienced inositol deprivation. Lack of inositol triggers the transcription of INO1, encoding inositol-1-phosphate synthase (the rate limiting enzyme for synthesis of inositol-containing phospholipids), and its relocalization to the nuclear periphery where it interacts with the Nuclear Pore Complex (NPC). INO1 association with the NPC is mediated by the transcription factors Put3 and Cbf1 that bind two recruitment sequences (GRS I and GRS II) present in the INO1 promoter [5,6]. Addition of inositol triggers transcriptional repression of INO1, but the gene remains anchored to the NPC through a different mechanism that relies on binding of the transcription factor Sfl1 to the Memory Recruitment Sequence (MRSINO1), and on interactions with different components of the NPC, e.g. Nup100 [7,8,9]. Persistent localization of INO1 at the nuclear periphery results in faster transcriptional reactivation when inositol depletion reoccurs, and is inherited mitotically for up to four generations. Deposition of chromatin modifications at the INO1 promoter is another component essential for memory. More specifically, incorporation of the histone variant H2A.Z and dimethylation of H3K4 (H3K4me2) are required for the ultimate output of memory: the recruitment of a poised RNA polymerase II (RNAPII) allowing faster transcriptional reactivation [8,9,10]. This final step also relies on the interaction of RNAPII with a specific form of the Mediator Complex containing the Cdk8 kinase [9].
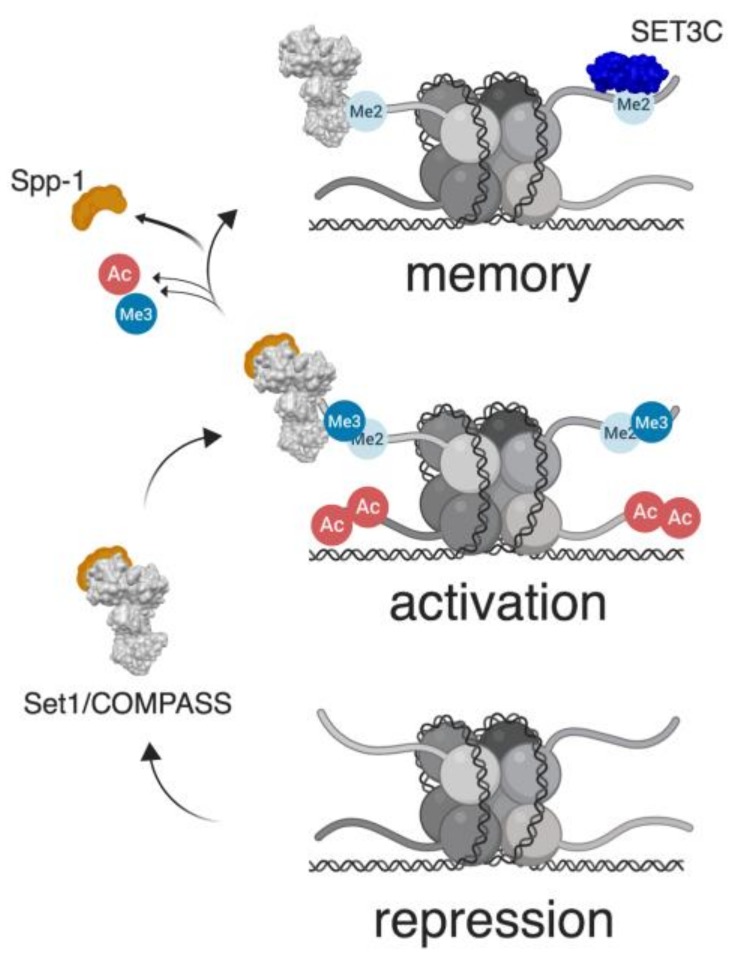
Importantly, characterization of PTMs at the INO1 promoter during the different transcriptional states has emphasized the pivotal role played by Set1/COMPASS, the only H3K4 methyltransferase present in yeast [11], in the regulation of transcriptional memory (Figure 1). During transcriptional repression, the INO1 promoter is hypoacetylated and hypomethylated on H3K4. Upon activation, H3/H4 acetylation at the promoter is accompanied by Set1/COMPASS dependent H3K4 di- and tri-methylation [9]. Finally, as transcriptional memory is induced, histone H3 and H4 are deacetylated, and H3K4me2 prevails over H3K4me3. This shift between methylation states depends on the activity of a remodeled version of the Set1/COMPASS complex lacking the Spp1 subunit, which in yeast has H3K4 mono- and dimethylation activities, but is incapable of H3K4 trimethylation [12,13,14,15,16,17]. H3K4me2 enrichment at the INO1 promoter requires both Sfl1 and MRSINO1, and is maintained during memory through an interaction with the histone deacetylase complex SET3C by mechanisms yet to be identified [9] (Figure 1).
Figure 1.
Model for S. cerevisiae INO1 transcriptional memory. When INO1 transcription is repressed (lower panel), nucleosomes associated with the INO1 promoter are hypoacetylated and hypomethylated. Upon activation (middle panel), INO1 relocalizes to the nuclear periphery, promoter nucleosomes are acetylated, and Set1/COMPASS di- and tri-methylates H3K4. When memory is induced (upper panel), INO1 remains at the nuclear periphery, nucleosomes are deacetylated, and a remodeled version of Set1/COMPASS (lacking the Spp1 component required for H3K4 tri-methylation) leads to the accumulation of H3K4me2, which is maintained through its interaction with the SET3C histone deacetylase complex.
2.2. GAL Transcriptional Memory
Another well-characterized example of yeast transcriptional memory is activated by galactose, and applies to several inducible genes (GAL1, GAL2, GAL7, GAL10) required for galactose catabolism. The expression of GAL genes depends on the Gal4 transcription factor, which binds to the promoter of each gene regardless of the carbon source available. When galactose is absent, the transcriptional repressor Gal80 interacts with Gal4 and inhibits its transcriptional activity. Conversely, when galactose is present Gal80 is retained in the cytosol, allowing Gal4 to activate the GAL genes [18]. When naive yeast cells are switched from glucose to galactose medium, derepression of GAL genes occurs very slowly. However, when the same population is exposed to galactose for a second time after growing for several hours in repressive glucose medium, activation of the GAL genes occurs much faster. Memory of GAL transcription persists through up to seven cell divisions, the longest so far reported in S. cerevisiae [7]. Gal1, a regulatory protein capable of preventing Gal80 nuclear translocation, is instrumental for the establishment of memory. Accumulation of Gal1 during galactose exposure, followed by its cytoplasmic inheritance through mitosis upon glucose repression, allows faster removal of Gal80 inhibition and, consequently, faster GAL gene reactivation once cells re-experience galactose [19].
Interestingly, Gal1 is responsible for GAL gene targeting to the nuclear periphery both under activating conditions (galactose) and during GAL memory [7]. Similarly to what is observed for INO1, GAL1 localization to the NPC during memory requires Nup100 and a specific MRS in the promoter (MRSGAL1). However, in contrast to the INO1 memory mechanism, disruption of GAL1 localization at the nuclear periphery does not affect GAL1 transcriptional memory [20]. An additional regulator of GAL memory is Tup1 (both a transcriptional activator and repressor), which mediates the effect of Gal1 on GAL1 peripheral localization as well as the binding of a poised RNAPII to the GAL1 promoter. Importantly, Tup1 is also required for incorporation of H2A.Z and dimethylation of H3K4 at the GAL1 promoter [20]. Similarly to what is observed for INO1, H2A.Z, and most likely H3K4me2, mediate the recruitment of RNAPII, which contributes to faster GAL1 reactivation during memory. Therefore, both the accumulation of long-lived Gal1 and the chromatin-driven binding of RNAPII to the GAL1 promoter are essential to establish memory.
2.3. Hormesis-Based Transcriptional Memory
Transcriptional memory is also associated with hormesis, an adaptive response to mild environmental stress through which the system improves its tolerance to more severe challenges [21]. Notably, yeast cells previously exposed to mild osmotic stress (0.7M NaCl) acquire both H2O2 resistance and the ability to mount a faster transcriptional response to H2O2 treatment. Both effects last for up to four generations, although they depend on different mechanisms. H2O2 resistance is due to the accumulation of a stable catalase (Ctt1, a H2O2 -detoxifying enzyme) that, by analogy with Gal1, is slowly diluted through cell division in the post-salt-stress period. The second effect, faster response to H2O2, represents a form of transcriptional memory, the first reported to include a large set of genes, both activated and repressed [22]. Intriguingly, salt-stress memory requires Nup42 but not Nup100, suggesting that it may rely on an interaction with the NPC through a mechanism different from those described for INO1 and the GAL genes [22]. Nonetheless, similarities have also been reported between salt-induced and other transcriptional memories. For instance, H3K4me2 and RNAPII are detected at the promoters of several genes induced by salt. Furthermore, a role for the SET3C deacetylase complex in maintaining H3K4me2 and promoting RNAPII recruitment appear to be conserved in both INO1 and salt-stress memories [9]. Lastly, an enrichment of Cdk8 is also observed at the promoters of genes displaying salt-stress memory, suggesting that, as for INO1 memory, the presence of Cdk8 on Mediator is required to activate transcriptional memory [9].
Strikingly, several aspects of yeast transcriptional memory are conserved in human cells. In HeLa cells, treatment with interferon-γ (IFN-γ) promotes transcriptional memory of a large number of genes that are more rapidly reactivated when cells are exposed to IFN-γ for a second time [23]. This memory is inherited mitotically for at least four generations and, as in yeast, is associated with H3K4me2 and RNAPII recruitment at the promoters of the affected genes. Moreover, Cdk8 is detected at these promoters, and Nup98 (orthologue of yeast Nup100) is essential for memory, although most likely through a distinct mechanism occurring in the cytoplasm rather than at the nuclear pore [8,9]. Taken together, evidence from yeast and mammalian cells suggests important roles for nuclear pore proteins and chromatin modifications, and in particular H3K4 methylation, in the establishment of transcriptional memory through the Cdk8+/Mediator-dependent recruitment of poised RNAPII.
3. Transcriptional Memory of Repression in Yeast
The vast majority of examples of transcriptional memory in yeast concerns activation of genes. However, hormetic memory induced by osmotic stress also includes numerous repressed genes [22]. Recently, another example of transcriptional repression memory (TREM) has been reported in response to carbon source shifts [24]. More specifically, galactose-driven repression of hundreds of genes occurs more rapidly in if yeast cells previously exposed to galactose. Notably, TREM requires the histone deacetylase complex Rpd3L, which is targeted to the promoter of TREM genes by the interaction of its Pho23 subunit with H3K4me3. Based on these results, the authors propose that TREM occurs as a consequence of histone deacetylation targeted by H3K4me3, in turn provoking loss of RNAPII at the promoter of TREM genes [24]. Of note, in this particular setting promoter H3K4me3 plays a repressive role, as already described in other contexts [25]. Therefore, yeast Set1/COMPASS plays key roles in both memory of transcriptional activation and repression through its ability to generate either H3K4me2 or H3K4me3, depending on the presence of the Spp1 component.
4. Transgenerational Inheritance in C. elegans
Transgenerational memory in mammals has been mostly correlated with changes in DNA methylation, an established carrier of epigenetic information [26]. However, the causality of these effects has been challenged because of the global epigenetic reprogramming of primordial germ cells (PGCs) during early embryogenesis, and transgenerational inheritance in mammalian species remains an area of extensive debate (see references [27,28] for recent reviews on the subject). C. elegans lacks DNA methylation on cytosines [29], and recent evidence suggests that histone modifications may play an important role in the transmission of epigenetic information in this model organism. Data from several labs has shown that gene silencing initiated by exogenous double-stranded RNA (dsRNA) or piwi-interacting RNAs (piRNAs) can be stably inherited, with transgenerational effects lasting more than 20 generations [30]. Transmission of these small RNAs has also been implicated in imparting memory of the response to environmental stressors, including starvation and high temperature. However, in most of these cases memory of the inherited state is observed for only a few generations [31,32,33,34]. Here we will discuss recent examples of epigenetic inheritance of environmental exposure mediated by H3K9me3, H3K27me3 and H3K4me3. Importantly, it should be noted that in order to be considered transgenerational, and to exclude the possibility of effects due to direct exposure to the trigger, the trait observed should be transmitted at least to the F3 generation following exposure of P0 mothers, and the F2 generation following exposure of P0 males [35].
4.1. Repressive H3K9 and H3K27 Tri-Methylation and Transgenerational Transgene Desilencing
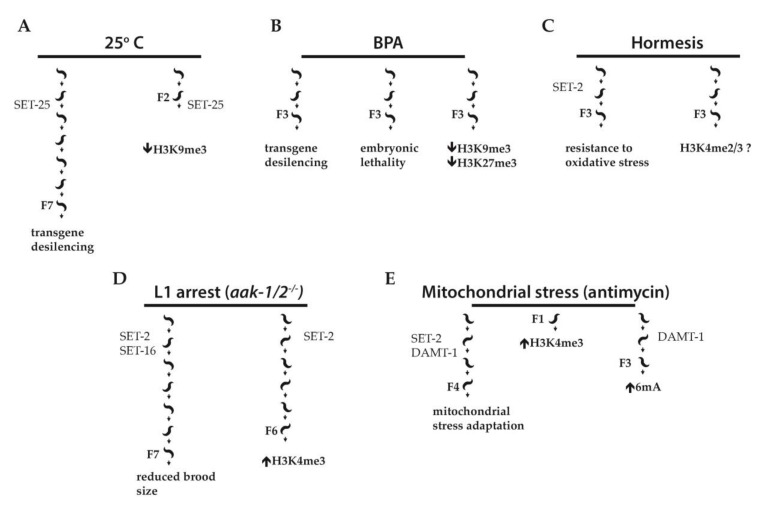
H3K9 and H3K27 tri-methylation have been implicated in heritability by exogenous dsRNA [30], although recent data suggest that their role may be limited to the establishment of heritable transgenerational silencing rather than its inheritance [36,37]. However, in several models of environmental stress under laboratory conditions, H3K9me3 was shown to mediate inheritance of phenotypes over several generations. The longest lasting effect was observed in animals carrying an integrated multicopy array consisting of GFP under control of the heat inducible daf-21/Hsp90 promoter (pdaf-21::GFP). Following exposure to high temperature (25 °C), GFP expression from the array strongly increased, and 14 generations were required for it to return to basal levels when animals were transferred to 20 °C after five generations at 25 °C. Even more surprising, exposure to the higher temperature for a single generation was sufficient to impart memory for 7 generations following a shift to 20 °C (Figure 2A). This memory effect was not observed for a single copy transgene, suggesting that inheritance depends on the repetitive nature of the transgene [38].
Figure 2.
Examples of stress-induced transgenerational inheritance in C. elegans. (A) Worms exposed to high temperature (25 °C) for a single generation show desilencing of a multicopy pdaf-21p::GFP transgene for seven generations, dependent on the histone methyltransferase SET-25. F2 embryos derived from ancestors developed at 25 °C show decreased levels of H3K9me3 on the multicopy array as compared to controls developed at 16 °C. (B) A germline heterochromatic transgene array is desilenced by Bisphenol A (BPA). F3 progeny of treated worms show transgene desilencing, embryonic lethality, and a global reduction in H3K9me3 and H3K27me3. (C) Hormesis triggered by low doses of heavy metals, hyperosmosis, or fasting during embryonic development promotes resistance to oxidative stress in adult worms. The hormetic effect is transmitted to the F3 generation and, at least in the F1 generation, its presence requires the activity of SET-2. (D) In worms lacking AMPK activity (aak-1/2−/−), prolonged starvation-induced L1 arrest results in reduced brood size for up to seven generations. H3K4me3 levels are increased in PGCs of post L1 diapause aak-1/2−/− animals for up to at least six generations. SET-2 and the MLL related H3K4 methyltransferase SET-16 both contribute to the reduced brood size phenotype while SET-2 is also implicated in the post L1 diapause accumulation of H3K4me3 in aak-1/2−/− mutants. (E) Exposure of worms to the electron transport chain inhibitor antimycin induces developmental delay; antimycin treatment of parental worms protects future generations (up to F4) from developmental delay (mitochondrial stress adaptation). Loss of N6-methyldeoxyadenine methyltransferase DAMT-1 prevents 6mA accumulation. Both SET-2 and DAMT-1 are required for mitochondrial stress adaptation.
Immunofluorescence experiments combined with DNA fluorescence in situ hybridization (DNA FISH) showed that at 20 °C, F2 embryos derived from P0 ancestors developed at 25° C accumulated less repressive H3K9me3 on the pdaf-21::GFP array than F2 embryos derived from control P0 developed at 16 °C, consistent with these arrays having features of heterochromatin that are sensitive to temperature (Figure 2A). No differences were observed in Polycomb mediated repressive H3K27me3, or in either H3K36me3 or H3K4me2, associated with active chromatin. Importantly, this difference was apparent in early embryos before the onset of zygotic transcription, and was inherited through both oocytes and sperm, indicating that the altered chromatin is not a secondary response to transcriptional changes in the embryo following heat shock. Although it is not known whether the histone methylation patterns themselves are responsible for transmitting memory of daf-21/Hsp-90 expression, memory was found to be dependent on the SET-25 histone methyltransferase required for deposition of H3K9me3, while inactivation of small RNAs had no effect [38]. Transcription profiling showed increased expression of repetitive sequences in the absence of set-25 up to three generations after a return of heat exposed animals to low temperature. These results suggest that changes in heterochromatin structure triggered by environmental stress may transmit epigenetic information between generations. How the increase in temperature leads to the loss of histone methylation marks deposited by SET-25 remains to be addressed. Interestingly, temperature-dependent phase separation of chromatin domains into liquid-like foci with physical properties critical for silencing has been described [39]. It is tempting to speculate that a related phenomenon may be implicated.
Global changes in H3K9me3 were also found to be associated with transgenerational germline defects in progeny of animals exposed to Bisphenol A (BPA) [40], a widely used plastic chemical highly prevalent in human samples and associated with endocrine disruption [41]. The authors showed that a heterochromatic transgene array transcriptionally silenced in the germline was desilenced in animals exposed to BPA, and in their F2 and F3 progeny (Figure 2B). The authors also showed increased embryonic lethality and germline apoptosis in F3 progeny of exposed animals, consistent with compromised germline function (Figure 2B). Mating of F1 progeny derived from exposed P0 mothers with unexposed males did not rescue germline desilencing, indicating that the primary mode of inheritance of the BPA effect is through the female germline. Perdurance of the effects in the F3 generation is consistent with an epigenetic, rather than a maternally inherited effect. Chromatin immunoprecipitation sequencing (ChIP-seq) of whole adult worms at the F3 generation revealed a minor decrease in H3K27me3 for genes upregulated in F3 progeny of BPA treated mothers, as well as reductions in both H3K9me3 and H3K27me3 from the distal, largely heterochromatic chromosomal regions (Figure 2B). Immunofluorescence analysis suggested that at least some of this global reduction is observed in germ cells. This was supported by experiments showing that inactivation of the H3K9me3/H3K36me3 demethylases JMJD-2 and the H3K27me3 demethylase JMJD-3/UTX-1 re-established transgene silencing as well as H3K9me3 and H3K27me3 in F3 animals [40].
4.2. H3K4 Methylation in the Inheritance of Stress Responses and Life History Traits
The above examples of epigenetic memory of exposure to stress rely on largely artificial systems based on the expression of a repetitive transgene. Kishimoto et al. [42] showed that, as in yeast, effects induced by hormesis could be passed transgenerationally to descendants. Animals exposed to various stressors, including heavy metal (arsenite), hyperosmosis (NaCl) and fasting during embryonic development exhibited increased resistance to oxidative stress and proteotoxicity as adults, and increased resistance was transmitted to the subsequent F2 and F3 generations grown under unstressed conditions (Figure 2C). Exposure of only male parents to stressors during development resulted in increased oxidative stress resistance and lifespan extension in the F1 descendants, showing that epigenetic information could be transmitted through sperm for at least one generation.
Because H3K4 methylation and COMPASS complex components required for its deposition mediate transgenerational effects on longevity in C. elegans [43], the authors investigated the role of COMPASS in regulating the transgenerational hormetic response. Knockdown of COMPASS components wdr-5.1, ash-2 and the SET1 homologue set-2 in the P0 generation had no effect on the increased stress resistance of the parent, but abrogated it in F1 descendants. This suggests that H3K4me3 is not required for stress-induced hormesis in the exposed animals, but is required for its inheritance at least in the F1 offspring. Knock-down of wdr-5.1 in the germline, but not intestinal or neuronal cells of exposed parental animals or their F1 offspring, led to suppression of increased resistance in F1 descendants. However, no significant difference in global H3K4me3 levels between stressed and unstressed animals was observed, and whether H3K4 methylation is able to transmit information through the male germline was not investigated. Additional factors were also identified as contributors to transgenerational inheritance, including known mediators of the stress response such as the insulin-like growth factor (IGF) signaling effector DAF-16/FOXO, the heat-shock factor HSF-1, and the transcription factor SKN-1/Nrf in the parental somatic cells, an interesting example of germ-to-soma communication.
In a related study, exposing C. elegans to high glucose concentrations was shown to have deleterious consequences on the germline of the F1 and to a lesser extent F2 progeny of exposed animals, resulting in a small decrease in the number of progeny [44]. Decreased brood size in F1 progeny was accompanied by increased resistance to oxidative stress and conveyed protection against neurodegeneration. As in the previous example, this effect was shown to depend on the COMPASS components SET-2 and WDR-5, but H3K4 was unaltered in affected F1 progeny, so that an indirect effect could not be ruled out. Furthermore, effects on stress resistance were not inherited beyond the non-exposed F1-F2 generation, showing that they reflect maternal inheritance rather than a transgenerational inheritance mechanism.
Recent evidence suggests that H3K4 methylation may also play a role in transgenerational memory of life history traits. C. elegans has a variety of developmental responses to nutrient availability, and two of the most well studied ones are dauer diapause and L1 developmental arrest, also known as L1 diapause [45,46]. Interestingly, examples of epigenetic inheritance have been observed in both [31,47,48]. In the absence of food, L1 larvae survive starvation up to two weeks by remaining arrested in the first larval stage, L1, and this is reversible upon feeding [49,50,51]. Jobson et al. [47] characterized the long-term phenotypic consequences of L1 larval arrest. They found that animals recovering from extended periods of starvation (8 days) had reduced growth and fertility compared to animals starved only 1 day. These animals were also more sensitive to subsequent starvation, suggesting that the experience of extended starvation does not protect against subsequent starvation. Curiously however, the F1, F2 and to a lesser extent F3 progeny of individuals most severely affected by extended L1 arrest were more resistant to heat, suggesting some form of epigenetic inheritance of stress resistance.
Subsequent work suggests that histone methylation may play a role in the transgenerational effect described above [48]. Activation of AMP-activated protein kinase (AMPK) in response to starvation is a conserved mechanism that facilitates metabolic adjustment [52]. In C. elegans, disruption of the genes encoding the two AMPK catalytic subunits, AAK-1 and -2, causes starved L1 larvae to die prematurely following prolonged starvation [53]. The authors found that when surviving post-L1 diapause mutant animals reached the adult stage, they showed both somatic and reproductive defects, including a reduced brood size. Remarkably, the reduced brood size was observed for up to 7 generations (Figure 2D). Levels of H3K4me3 were increased in the precursor germline cells (PGCs) of emergent post L1 diapause aak-1/2−/− mutant animals up to at least 6 generations (Figure 2D). RNAi knock down of the genes coding for the COMPASS subunits SET-2, ASH-2 or WDR-5.1, but also of set-16, encoding a MLL related H3K4 methyltransferase, partially suppressed both sterility and the reduced brood size in the F1 generation of post L1 diapause aak-1/2−/− mutant larvae. A set-2 knock-out also reduced global H3K4me3 levels of post L1 diapause aak-1/2−/− mutant larvae (Figure 2D). Interestingly, SET-2 protein levels appeared to be slightly higher in the PGCs of aak-1/2−/− mutant larvae, suggesting that AMPK may influence H3K4 levels through regulation of SET-2 protein stability. The transgenerational signal responsible for this increase in H3K4me3 and the genetic consequences of increased H3K4me3 in starved aak-1/2−/− worms and their progeny remain unknown. However, recent results showing that SET-2 also plays a role in the inheritance of another AMPK-dependent response, namely the mitochondrial stress response [54,55], suggests the intriguing possibility that AMPK signaling and changes in H3K4me3 may be interconnected processes contributing to transgenerational inheritance.
Liu et al [55] showed that C. elegans can adapt to mitochondrial stress induced by electron transport chain (ETC) inhibitors, and this response depends on N6-methyladenine (6mA) and SET-2 (Figure 2E). Animals exposed to antimycin, an ETC complex III inhibitor, showed dose-dependent developmental delays. Exposure of P0 parents protected progeny from developmental delay defects up to the F4 generation (Figure 2E). Similar results were obtained with other mitochondrial inhibitors. Crossing antimycin treated males with untreated hermaphrodites resulted in F1 progeny with increased resistance compared to progeny from untreated males, showing that adaptation is not due to a maternal effect. The ATP content and oxygen consumption levels of these F1 animals, two parameters of mitochondrial function, were also comparable to those of the F1 progeny from untreated animals.
Most importantly, reduced activity of SET-2, but not other methyltransferases tested, severely impaired transmission of mitochondrial stress adaptation. ChIP-seq analysis of antimycin treated animals showed a global increase in H3K4me3 on promoters of F1 progeny. Mitochondrial stress adaptation was also shown to depend on N6-methyldeoxyadenine (6 mA), and genetic analysis suggested that 6mA may act downstream of H3K4me3 in this response. The mechanism responsible for the increase in H3K4me3 following the induction of mitochondrial stress was not explored in this study, and it is not known whether this increase is maintained across generations. Nonetheless, it is intriguing that under starvation conditions, AMPK activation dampens germline H3K4 methylation to preserve germline function [48], while adaptation to mitochondrial stress is associated with a global increase of this same mark [55]. These results suggest the interesting possibility that in both cases AMPK activation impacts H3K4me3 methylation with different consequences on the germline and soma, possibly constituting an interesting example of germline to soma communication impacting inheritance of environmental exposure.
5. Perspectives
The yeast S. cerevisiae, in which examples of transcriptional memory through mitosis are well established, continues to provide useful insight into the molecular mechanisms responsible for transcriptional inheritance of environmental stress responses. Histone post-translational modifications, and in particular H3K4 methylation, have emerged as primary actors in inheritance, and at least some features of this mitotic memory mechanism may be conserved in mammals. More controversial is whether transcriptional memory of exposure to environmental stress can be inherited across multiple generations through the germline. This has become an area of intense research with potential impacts on human health and implications in evolutionary biology. Key to this question is how acquired information can be inherited given the extensive reprogramming of the genome accompanying germline development. Work on model systems such as C. elegans, in which environmental conditions can be tightly controlled and robust assays can be used to monitor inheritance, will continue to provide useful insights in understanding how epigenetic marks are established and maintained transgenerationally, and how cellular metabolism impacts these processes. In this context, the development of techniques allowing chromatin profiling to be carried out specifically on purified germline tissue will allow a more accurate description of histone modification profiles across generations and contribute to the identification of the molecular signatures accompanying transgenerational memory.
Acknowledgments
We thank members of the Palladino Lab for helpful discussions.
Funding
This research was funded by support from the ANR (N° 15-CE12-0018-01), the CNRS, and the Foundation ARC.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Black J.C., Van Rechem C., Whetstine J.R. Histone lysine methylation dynamics: Establishment, regulation, and biological impact. Mol. Cell. 2012;48:491–507. doi: 10.1016/j.molcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahringer J., Gasser S.M. Repressive chromatin in Caenorhabditis elegans: Establishment, composition, and function. Genetics. 2018;208:491–511. doi: 10.1534/genetics.117.300386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suganuma T., Workman J.L. Signals and combinatorial functions of histone modifications. Annu. Rev. Biochem. 2011;80:473–499. doi: 10.1146/annurev-biochem-061809-175347. [DOI] [PubMed] [Google Scholar]
- 4.Xue Y., Acar M. Mechanisms for the epigenetic inheritance of stress response in single cells. Curr. Genet. 2018;64:1221–1228. doi: 10.1007/s00294-018-0849-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahmed S., Brickner D.G., Light W.H., Cajigas I., McDonough M., Froyshteter A.B., Volpe T., Brickner J.H. DNA zip codes control an ancient mechanism for gene targeting to the nuclear periphery. Nat. Cell Biol. 2010;12:111–118. doi: 10.1038/ncb2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Randise-Hinchliff C., Coukos R., Sood V., Sumner M.C., Zdraljevic S., Meldi Sholl L., Garvey Brickner D., Ahmed S., Watchmaker L., Brickner J.H. Strategies to regulate transcription factor-mediated gene positioning and interchromosomal clustering at the nuclear periphery. J. Cell Biol. 2016;212:633–646. doi: 10.1083/jcb.201508068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brickner D.G., Cajigas I., Fondufe-Mittendorf Y., Ahmed S., Lee P.C., Widom J., Brickner J.H. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 2007;5:e81. doi: 10.1371/journal.pbio.0050081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Light W.H., Freaney J., Sood V., Thompson A., D’Urso A., Horvath C.M., Brickner J.H. A conserved role for human Nup98 in altering chromatin structure and promoting epigenetic transcriptional memory. PLoS Biol. 2013;11:e1001524. doi: 10.1371/journal.pbio.1001524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D’Urso A., Takahashi Y.H., Xiong B., Marone J., Coukos R., Randise-Hinchliff C., Wang J.P., Shilatifard A., Brickner J.H. Set1/COMPASS and Mediator are repurposed to promote epigenetic transcriptional memory. Elife. 2016;5:e16691G. doi: 10.7554/eLife.16691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Light W.H., Brickner D.G., Brand V.R., Brickner J.H. Interaction of a DNA zip code with the nuclear pore complex promotes H2A.Z incorporation and INO1 transcriptional memory. Mol. Cell. 2010;40:112–125. doi: 10.1016/j.molcel.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shilatifard A. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr. Opin. Cell Biol. 2008;20:341–348. doi: 10.1016/j.ceb.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takahashi Y.H., Lee J.S., Swanson S.K., Saraf A., Florens L., Washburn M.P., Trievel R.C., Shilatifard A. Regulation of H3K4 trimethylation via Cps40 (Spp1) of COMPASS is monoubiquitination independent: Implication for a Phe/Tyr switch by the catalytic domain of Set1. Mol. Cell. Biol. 2009;29:3478–3486. doi: 10.1128/MCB.00013-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dehe P.M., Dichtl B., Schaft D., Roguev A., Pamblanco M., Lebrun R., Rodriguez-Gil A., Mkandawire M., Landsberg K., Shevchenko A., et al. Protein interactions within the Set1 complex and their roles in the regulation of histone 3 lysine 4 methylation. J. Biol. Chem. 2006;281:35404–35412. doi: 10.1074/jbc.M603099200. [DOI] [PubMed] [Google Scholar]
- 14.Miller T., Krogan N.J., Dover J., Erdjument-Bromage H., Tempst P., Johnston M., Greenblatt J.F., Shilatifard A. COMPASS: A complex of proteins associated with a trithorax-related SET domain protein. Proc. Natl. Acad. Sci. USA. 2001;98:12902–12907. doi: 10.1073/pnas.231473398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roguev A., Schaft D., Shevchenko A., Pijnappel W.W., Wilm M., Aasland R., Stewart A.F. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. Embo. J. 2001;20:7137–7148. doi: 10.1093/emboj/20.24.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morillon A., Karabetsou N., Nair A., Mellor J. Dynamic lysine methylation on histone H3 defines the regulatory phase of gene transcription. Mol. Cell. 2005;18:723–734. doi: 10.1016/j.molcel.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 17.Schneider J., Wood A., Lee J.S., Schuster R., Dueker J., Maguire C., Swanson S.K., Florens L., Washburn M.P., Shilatifard A. Molecular regulation of histone H3 trimethylation by COMPASS and the regulation of gene expression. Mol. Cell. 2005;19:849–856. doi: 10.1016/j.molcel.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 18.Peng G., Hopper J.E. Gene activation by interaction of an inhibitor with a cytoplasmic signaling protein. Proc. Natl. Acad. Sci. USA. 2002;99:8548–8553. doi: 10.1073/pnas.142100099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kundu S., Peterson C.L. Dominant role for signal transduction in the transcriptional memory of yeast GAL genes. Mol. Cell Biol. 2010;30:2330–2340. doi: 10.1128/MCB.01675-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sood V., Cajigas I., D’Urso A., Light W.H., Brickner J.H. Epigenetic transcriptional memory of GAL genes depends on growth in glucose and the Tup1 transcription factor in Saccharomyces cerevisiae. Genetics. 2017;206:1895–1907. doi: 10.1534/genetics.117.201632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zimmermann A., Bauer M.A., Kroemer G., Madeo F., Carmona-Gutierrez D. When less is more: Hormesis against stress and disease. Microb. Cell. 2014;1:150–153. doi: 10.15698/mic2014.05.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guan Q., Haroon S., Bravo D.G., Will J.L., Gasch A.P. Cellular memory of acquired stress resistance in Saccharomyces cerevisiae. Genetics. 2012;192:495–505. doi: 10.1534/genetics.112.143016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gialitakis M., Arampatzi P., Makatounakis T., Papamatheakis J. Gamma interferon-dependent transcriptional memory via relocalization of a gene locus to PML nuclear bodies. Mol. Cell Biol. 2010;30:2046–2056. doi: 10.1128/MCB.00906-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee B.B., Choi A., Kim J.H., Jun Y., Woo H., Ha S.D., Yoon C.Y., Hwang J.T., Steinmetz L., Buratowski S., et al. Rpd3L HDAC links H3K4me3 to transcriptional repression memory. Nucleic Acids Res. 2018;46:8261–8274. doi: 10.1093/nar/gky573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Howe F.S., Fischl H., Murray S.C., Mellor J. Is H3K4me3 instructive for transcription activation? BioEssays News Rev. Mol. Cell. Dev. Biol. 2017;39:1–12. doi: 10.1002/bies.201600095. [DOI] [PubMed] [Google Scholar]
- 26.Seisenberger S., Peat J.R., Reik W. Conceptual links between DNA methylation reprogramming in the early embryo and primordial germ cells. Curr. Opin. Cell Biol. 2013;25:281–288. doi: 10.1016/j.ceb.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 27.Heard E., Martienssen R.A. Transgenerational epigenetic inheritance: Myths and mechanisms. Cell. 2014;157:95–109. doi: 10.1016/j.cell.2014.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Otterdijk S.D., Michels K.B. Transgenerational epigenetic inheritance in mammals: How good is the evidence? FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2016;30:2457–2465. doi: 10.1096/fj.201500083. [DOI] [PubMed] [Google Scholar]
- 29.Simpson V.J., Johnson T.E., Hammen R.F. Caenorhabditis elegans DNA does not contain 5-methylcytosine at any time during development or aging. Nucleic Acids Res. 1986;14:6711–6719. doi: 10.1093/nar/14.16.6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rechavi O., Lev I. Principles of transgenerational small RNA inheritance in Caenorhabditis elegans. Curr. Biol. CB. 2017;27:R720–R730. doi: 10.1016/j.cub.2017.05.043. [DOI] [PubMed] [Google Scholar]
- 31.Webster A.K., Jordan J.M., Hibshman J.D., Chitrakar R., Baugh L.R. Transgenerational effects of extended dauer diapause on starvation survival and gene expression plasticity in Caenorhabditis elegans. Genetics. 2018;210:263–274. doi: 10.1534/genetics.118.301250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rechavi O., Houri-Ze’evi L., Anava S., Goh W.S.S., Kerk S.Y., Hannon G.J., Hobert O. Starvation-induced transgenerational inheritance of small RNAs in C. elegans. Cell. 2014;158:277–287. doi: 10.1016/j.cell.2014.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schott D., Yanai I., Hunter C.P. Natural RNA interference directs a heritable response to the environment. Sci. Rep. 2014;4:7387. doi: 10.1038/srep07387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ni J.Z., Kalinava N., Chen E., Huang A., Trinh T., Gu S.G. A transgenerational role of the germline nuclear RNAi pathway in repressing heat stress-induced transcriptional activation in C. elegans. Epigenetics Chromatin. 2016;9:3. doi: 10.1186/s13072-016-0052-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skinner M.K. What is an epigenetic transgenerational phenotype? Reprod. Toxicol. 2008;25:2–6. doi: 10.1016/j.reprotox.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woodhouse R.M., Buchmann G., Hoe M., Harney D.J., Low J.K.K., Larance M., Boag P.R., Ashe A. Chromatin modifiers SET-25 and SET-32 are required for establishment but not long-term maintenance of transgenerational epigenetic inheritance. Cell Rep. 2018;25:2259–2272. doi: 10.1016/j.celrep.2018.10.085. [DOI] [PubMed] [Google Scholar]
- 37.Kalinava N., Ni J.Z., Gajic Z., Kim M., Ushakov H., Gu S.G. C. elegans heterochromatin factor SET-32 plays an essential role in transgenerational establishment of nuclear RNAi-mediated epigenetic silencing. Cell Rep. 2018;25:2273–2284. doi: 10.1016/j.celrep.2018.10.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klosin A., Casas E., Hidalgo-Carcedo C., Vavouri T., Lehner B. Transgenerational transmission of environmental in formation in C. elegans. Science. 2017;356:320–323. doi: 10.1126/science.aah6412. [DOI] [PubMed] [Google Scholar]
- 39.Strom A.R., Emelyanov A.V., Mir M., Fyodorov D.V., Darzacq X., Karpen G.H. Phase separation drives heterochromatin domain formation. Nature. 2017;547:241–245. doi: 10.1038/nature22989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Camacho J., Truong L., Kurt Z., Chen Y.-W., Morselli M., Gutierrez G., Pellegrini M., Yang X., Allard P. The memory of environmental chemical exposure in C. elegans is dependent on the Jumonji demethylases jmjd-2 and jmjd-3/utx-1. Cell Rep. 2018;23:2392–2404. doi: 10.1016/j.celrep.2018.04.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vandenberg L.N., Chahoud I., Heindel J.J., Padmanabhan V., Paumgartten F.J.R., Schoenfelder G. Urinary, circulating, and tissue biomonitoring studies indicate widespread exposure to bisphenol A. Environ. Health Perspect. 2010;118:1055–1070. doi: 10.1289/ehp.0901716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kishimoto S., Uno M., Okabe E., Nono M., Nishida E. Environmental stresses induce transgenerationally inheritable survival advantages via germline-to-soma communication in Caenorhabditis elegans. Nat. Commun. 2017;8:14031. doi: 10.1038/ncomms14031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Greer E.L., Maures T.J., Ucar D., Hauswirth A.G., Mancini E., Lim J.P., Benayoun B.A., Shi Y., Brunet A. Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans. Nature. 2011;479:365–371. doi: 10.1038/nature10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tauffenberger A., Parker J.A. Heritable transmission of stress resistance by high dietary glucose in Caenorhabditis elegans. PLoS Genet. 2014;10:e1004346. doi: 10.1371/journal.pgen.1004346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Riddle D.L., Albert P.S. Genetic and environmental regulation of dauer larva development. In: Riddle D.L., Blumenthal T., Meyer B.J., Priess J.R., editors. C. elegans II. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY, USA: 1997. pp. 739–768. [PubMed] [Google Scholar]
- 46.Baugh L.R. To grow or not to grow: Nutritional control of development during Caenorhabditis elegans L1 arrest. Genetics. 2013;194:539–555. doi: 10.1534/genetics.113.150847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jobson M.A., Jordan J.M., Sandrof M.A., Hibshman J.D., Lennox A.L., Baugh L.R. Transgenerational effects of early life starvation on growth, reproduction, and stress resistance in Caenorhabditis elegans. Genetics. 2015;201:201–212. doi: 10.1534/genetics.115.178699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Demoinet E., Li S., Roy R. AMPK blocks starvation-inducible transgenerational defects in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2017;114:E2689–E2698. doi: 10.1073/pnas.1616171114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hong Y., Roy R., Ambros V. Developmental regulation of a cyclin-dependent kinase inhibitor controls postembryonic cell cycle progression in Caenorhabditis elegans. Dev. Camb. Engl. 1998;125:3585–3597. doi: 10.1242/dev.125.18.3585. [DOI] [PubMed] [Google Scholar]
- 50.Baugh L.R., Sternberg P.W. DAF-16/FOXO regulates transcription of cki-1/Cip/Kip and repression of lin-4 during C. elegans L1 arrest. Curr. Biol. 2006;16:780–785. doi: 10.1016/j.cub.2006.03.021. [DOI] [PubMed] [Google Scholar]
- 51.Johnson T.E., Mitchell D.H., Kline S., Kemal R., Foy J. Arresting development arrests aging in the nematode Caenorhabditis elegans. Mech. Ageing Dev. 1984;28:23–40. doi: 10.1016/0047-6374(84)90150-7. [DOI] [PubMed] [Google Scholar]
- 52.Hardie D.G., Ross F.A., Hawley S.A. AMPK: A nutrient and energy sensor that maintains energy homeostasis. Nat. Rev. Mol. Cell Biol. 2012;13:251–262. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fukuyama M., Sakuma K., Park R., Kasuga H., Nagaya R., Atsumi Y., Shimomura Y., Takahashi S., Kajiho H., Rougvie A., et al. C. elegans AMPKs promote survival and arrest germline development during nutrient stress. Biol. Open. 2012;1:929–936. doi: 10.1242/bio.2012836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Herzig S., Shaw R.J. AMPK: Guardian of metabolism and mitochondrial homeostasis. Nat. Rev. Mol. Cell Biol. 2018;19:121–135. doi: 10.1038/nrm.2017.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ma C., Niu R., Huang T., Shao L.-W., Peng Y., Ding W., Wang Y., Jia G., He C., Li C.-Y., et al. N6-methyldeoxyadenine is a transgenerational epigenetic signal for mitochondrial stress adaptation. Nat. Cell Biol. 2019;21:319–327. doi: 10.1038/s41556-018-0238-5. [DOI] [PubMed] [Google Scholar]