Abstract
Quorum sensing (QS) is a cell-to-cell communication system that uses autoinducers as signaling molecules to enable inter-species and intra-species interactions in response to external stimuli according to the population density. QS allows bacteria such as Acinetobacter baumannii to react rapidly in response to environmental changes and hence, increase the chances of survival. A. baumannii is one of the causative agents in hospital-acquired infections and the number of cases has increased remarkably in the past decade. In this study, A. baumannii strain 863, a multidrug-resistant pathogen, was found to exhibit QS activity by producing N-acyl homoserine lactone. We identified the autoinducer synthase gene, which we named abaI, by performing whole genome sequencing analysis of A. baumannii strain 863. Using high resolution tandem triple quadrupole mass spectrometry, we reported that abaI of A. baumannii strain 863 produced 3-hydroxy-dodecanoyl-homoserine lactone. A gene deletion mutant was constructed, which confirmed the functionality of abaI. A growth defect was observed in the QS-deficient mutant strain. Transcriptome profiling was performed to determine the possible genes regulated by QS. Four groups of genes that showed differential expression were discovered, namely those involved in carbon source metabolism, energy production, stress response and the translation process.
Keywords: Acinetobacter baumannii, quorum sensing, transcriptomic, RNA-Seq, recombineering, mutagenesis
1. Introduction
Quorum sensing (QS) is a sophisticated cell-to-cell communication system that involves the production, detection and response to signaling molecules known as autoinducers. QS system allows bacteria to detect its populations density in response to the concentration of autoinducers [1]. There are three steps involved in the QS system [1]. First, the autoinducers are produced in the cells. Second, these autoinducers either diffuse or are actively transported to the extracellular environment and their concentration increases as the population density increases. Third, when the concentration of the autoinducers goes beyond the minimal threshold for detection, the autoinducers bind to the intracellular cognate receptors in the cells. Autoinducer-bound receptors act as transcriptional factors and induce cell signaling cascades resulting in a change in gene expression on a population basis [2]. Alterations in gene expression can trigger phenotypic changes as a response to the environmental changes. These phenotypic changes include biofilm formation [3], antibiotic resistance [4], toxin secretion [5] and conjugation [6,7]. There are various autoinducers and their respective receptors are used by different bacterial species. For instance, the QS system in Vibrio fischeri involves two genes, autoinducer synthase, luxI and autoinducer receptor, luxR and autoinducer, and N-acyl homoserine lactones (AHLs) [8]. Many Gram-negative bacteria including Acinetobacter baumannii possess luxIR homologues and utilize AHLs as autoinducers [9].
A. baumannii is a Gram-negative, catalase producing, non-fermenting and strictly aerobic coccobacilli [10]. This bacterium has become increasingly important because of its frequent association with nosocomial infections, especially among immunocompromised patients [11]. It was once coined the nickname “Iraqibacter” due to many infections found in wounded soldiers that served in Iraq [10]. There was a misconception that A. baumannii can be isolated from various environments [12]. While this is true for the genus which contributed to the misconception, the natural reservoir of A. baumannii is still remained to be identified [13,14,15]. However, the hospital environment has become an important reservoir [13,14,16]. In addition, A. baumannii has acquired resistance towards a wide range of antibiotics and recent studies suggested that the prevalence of multidrug-resistant A. baumannii is still on the rise in many countries [17]. In 2017, the World Health Organization (WHO) has included A. baumannii as one of the most dangerous pathogens hit-list and hence, the highest priority in new antibiotic development [18]. Being able to persist in hospital environment and to defend themselves against multiple antibiotics show that A. baumannii has an amazing capability to survive and adapt in a harsh environment.
Since the QS system is used by various bacterial species to react against changes in the environment, it is of interest to elucidate the genes regulated by this cell-to-cell communication system. Transcriptomic profiling fits well in this study as QS controls a wide range of genes. Transcriptomic profiling previously employs hybridization-based microarray. However, as next-generation sequencing (NGS) technology has improved, RNA-sequencing (RNA-Seq) has become more common in transcriptomic studies. This is because RNA-Seq provides three advantages over hybridization-based microarray platforms: (i) detection of novel transcripts, (ii) higher resolution data, and (iii) a higher dynamic range [19,20]. Hence, whole genome sequencing and RNA-Seq could work hand in hand to provide a more comprehensive way to elucidate the expression network revolved around QS in A. baumannii.
In this study, we characterized the autoinducer synthase gene, abaI, in a multidrug-resistant A. baumannii strain 863 and the genes regulated by QS using mutagenesis and transcriptome analyses. With verification of the synthase activity, it provides a platform to study the regulatory role of the AHLs on the virulence and unknown genetic traits of this pathogenic isolate. With further insights of the role of the secreted AHLs, it facilitates the antimicrobial strategies to attenuate bacterial virulence among Acinetobacter sp.
2. Materials and Methods
2.1. Bacterial Strain and Growth Conditions
A. baumannii strain 863 was obtained from the culture collection of a local diagnostic lab. The strain was cultured in Luria-Bertani (LB) medium. Table 1 shows the bacteria strains and plasmids used in this study.
Table 1.
Bacteria strains and plasmids used in this study.
| Strain/Plasmid | Genotype and Property | Source |
|---|---|---|
| Strains | ||
| Acinetobacter baumannii 863 | Wild type | This study |
| A. baumannii 863 ΔabaI:Km | Quorum sensing (QS)-deficient (abaI deleted) mutant | This study |
| Plasmids | ||
| pKD4 | Kanamycin resistant gene (Km), used as a template for selectable marker in knockout (KO) cassette | [21] |
| pAT04 | Recombineering system (RecAb), tetracycline resistant | [21] |
2.2. Whole Genome Sequencing (WGS), Assembly and Annotation
The genomic DNA from strain 863 was extracted using the Masterpure DNA Purification Kit (Epicentre, Madison, WI, USA) according to the manufacturer’s protocol. The quality of the genomic DNA was assessed using a Nanodrop Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). The extracted DNA was used in Nextera Library Preparation Kit (Illumina San Diego, CA, USA). The prepared library was sequenced using 100 bp × 2 cartridge in HiSeq 2500 High Throughput Sequencer (Illumina) on rapid run mode. The quality of the sequenced data was assessed using FastQC software [22]. The sequenced data were then trimmed and assembled using CLC Genomic Workbench (V7.5; Qiagen, Hilden, Germany). Following this, the assembled sequence was annotated by the National Center for Biotechnology Information (NCBI) Prokaryotic Genome Automatic Annotation Pipeline (PGAAP) [23] and RAST [24].
2.3. Autoinducer Synthase Identification and Bioinformatics Analysis
The nucleotide sequence generated from the WGS which was annotated as autoinducer synthase (abaI) was used to compare with GenBank databases via BLASTX. Eleven protein sequences of autoinducer synthases from Acinetobacter species were selected. These sequences were aligned using ClustalW and a phylogenetic tree was generated via Maximum Likelihood method using MEGA 7 [25]. The algorithm applied the Jones-Taylor-Thornton (JTT) matrix-based model [26] and to provide confidence estimation, 1000 bootstrap replications were used to construct the phylogenetic tree. On the other hand, the putative promoter sequences were identified using BPROM webtool [27]. All parameters were set to default settings, unless specified.
2.4. AHL Extraction and Its Identification Using Mass Spectrometry (MS)
One millimeter of an overnight culture of strain 863 was subcultured into 100 mL LB broth buffered to pH 6.5 with 50 mM of 3-[N-morpholino] propanesulfonic acid (MOPS) to prevent the AHLs from being degraded in a basic medium [28]. The culture was then incubated overnight at 37 °C with agitation at 230 rpm. The culture supernatant was obtained by centrifugation and subsequently extracted three times with equal volume of acidified ethyl acetate (0.1% v/v glacial acetic acid in ethyl acetate). The extracts were dried in a fume hood before resuspended with 1 mL of acidified ethyl acetate and dried again inside fume hood. The final dried extracts were reconstituted in 1 mL of acetonitrile. The insoluble solids in the extracts were removed using a syringe filter. AHL identification was done using high-resolution tandem triple quadrupole mass spectrometry (LCMS/MS; Agilent 1290 Infinity LC coupled with Agilent 6490 Triple Quadrupole LC/MS system, Agilent Technologies Inc., Santa Clara, CA, USA).
2.5. Construction of Abai Deletion (ΔabaI:Km) Strain
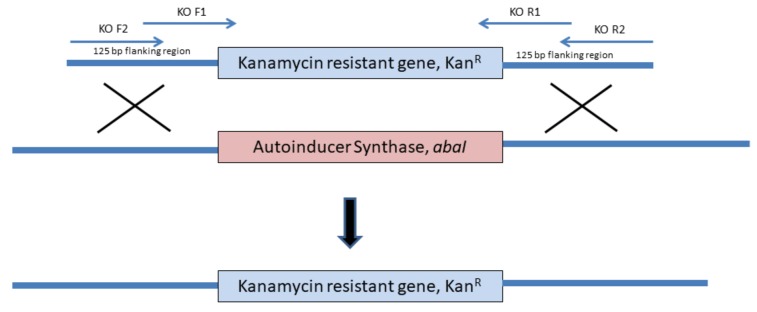
An autoinducer synthase (abaI) deletion mutant was constructed as described by Tucker et al. [21]. In brief, plasmid pKD4 (Table 1) was used as a template to amplify the kanamycin resistance gene (Km) using polymerase chain reaction (PCR). Two pairs of primers were used to construct the knockout (KO) cassette Table 2. KO F1 and KO R1 primers were used to amplify the intermediate KO cassette. The amplicon was gel-purified and used as a template for a second PCR. In the second PCR, KO F2 and KO R2 primers were used to construct the final KO cassette with Km and the resultant amplicon was flanked by 125 bp upstream and downstream of abaI Figure 1) This PCR product (KO cassette) was gel purified and used in transformation. The plasmid pAT04 Table 1 was transformed into strain 863 through electroporation and selected via LB agar supplemented with tetracycline (10 µg/mL, final concentration). Strain 863 harboring pAT04 was confirmed with PCR using primer Screen pAT04 F and Screen pAT04 R. A. baumannii strain 863 harboring pAT04 was cultured in 50 mL LB broth containing 10 µg/mL of tetracycline to maintain the plasmid for 45 min before induction with isopropyl β-D-thiogalactopyranoside (IPTG (2 mM, final concentration). The induced culture was grown until the OD600 reached 0.4. Next, the culture was incubated on ice for 30 min before washing three times with ice-cold 10% (v/v) glycerol and concentrated 250-fold. Twenty microliters of these washed cells were transformed with 5 µg of KO cassette via electroporation. The transformants were then incubated for 4 h at 37 °C and plated on LB agar supplemented with 30 µg/mL kanamycin. The transformant colonies (mutant strain, ΔabaI:Km) were verified using PCR (Screen KO F and Screen KO R as primers) and the resulting mutant loss of its AHL production was confirmed with LCMS/MS. Finally, the desired mutant was sub-cultured sequentially to enable the loss of pAT04. A PCR was then performed to confirm the loss of pAT04.
Table 2.
Primers used in KO cassette construction.
| Primer Name | Sequence |
|---|---|
| KO F1 | AGTTACCGCCTTTCCAGCTGTTCAGTAAAGTCACTTGAAAAATAAGTGGAAGCACTTGTAATGTATGGACAGCAAGCGAACCG |
| KO R1 | GATATGTAAAAATTAGGACTCATACCCCACGGATAGGCATGAGTCCTATATAAGAAATTATCAGAAGAACTCGTCAAGAAG |
| KO F2 | ATACGTCATTAACCAAGTCTTCATTAAGTCAAACCTTCTCTTAGAAACCTGTAAATTCTTACAGTTACCGCCTTTCCAGC |
| KO R2 | GAGCTATAAAAAGGATGAGACTTATTATAAGAAAGCAAACCTAAATCTAAAAACCAAGATTGATTGATATGTAAAAATTAGGAC |
| Screen KO F | ATACGTCATTAACCAAGTCTTCA |
| Screen KO R | GAGCTATAAAAAGGATGAGACT |
| Screen pAT04 F | GGTCTCCCCATGCGAGAGTA |
| Screen pAT04 R | CTCTTGCCCGGCGTCAACAC |
Figure 1.
Schematic diagram of the KO cassette construction and abaI deletion process in the cell. Two pair of primers were used to create a selectable marker with 125 bp flanking regions from upstream and downstream of abaI. The gene abaI was replaced with kanamycin resistant gene as a selectable marker with ReAB system from pAT04 transformed into A. baumannii 863 earlier.
2.6. Growth Curve Analysis
Both overnight cultures of the wild type and the mutant strains were inoculated in a 100 mL fresh LB broth, and the cell density was adjusted to OD600 = 0.001. The cultures were grown in a shaking incubator at 37 °C with shaking at 230 rpm. The cell density (OD600) was measured every 30 min until at least three readings indicated stationary phase. The experiment was performed with three biological replicates.
2.7. RNA-Seq Library Preparation
Both overnight bacterial cultures of the wild type and the mutant strains were sub-cultured into 100 mL LB broth at OD600 = 0.001 and grown at 37 °C until late log phase (OD600 reached 2.75 for wild type, and 2.25 for ΔabaI:Km) was achieved. The harvested cells were stored in RNAprotect Bacteria Reagent (Qiagen) according to the manufacturer’s instructions until RNA extraction was performed. The total RNA extraction was performed using Nucleospin RNA kit (Macherey-Nagel, Düren, Germany). The ribosomal RNA (rRNA) was removed from total RNA using Ribo-Zero rRNA Removal Kit (Bacteria) (Illumina). Finally, the rRNA depleted RNA was used as the input for ScriptSeq v2 RNA-Seq Library Preparation Kit (Illumina). The libraries obtained were sequenced using 75 × 2 cartridge on MiSeq Sequencer (Illumina). This analysis was performed with three biological replicates.
2.8. RNA-Seq Bioinformatics Analysis
The quality of the sequencing data was assessed with FastQC [22]. The assessed RNA-Seq sequencing data were trimmed, assembled and mapped using CLC Genomics Workbench (V 7.5; Qiagen). Then, the assembled reads were converted into BAM format and counted using HTSeq-count script from HTSeq [29]. Following this, a differential expression (DE) analysis was performed using DESeq2 [30] and a heatmap was generated. Genes with Log2 fold change (Log2FC) ≥ and the adjusted p-value of < 0.05 were considered as genes with DE.
2.9. Accession Numbers
The whole genome sequence of A. baumannii strain 863 was submitted to GenBank under accession number LZTF00000000. The RNA-Seq dataset has been deposited to Gene Expression Omnibus (GEO) under accession number GSE120346.
3. Results
3.1. Identification and Phylogenetic Analysis of Autoinducer Synthase Gene
The whole-genome sequencing of strain 863 was performed using a HiSeq 2500 High Throughput Sequencer. The genome sequence was assembled into 65 contigs consisting of 3.812 Mb. A total of 3495 coding DNA sequences (CDS) was predicted by PGAAP. The sequence has been deposited at GenBank under accession no. LZTF00000000.
From the annotations by PGAAP, an open reading frame of 552 bp (A9801_RS12630) was predicted to encode the AHL synthase protein (WP_029424600), which is 183 amino acids in length. To further verify the identity of this gene, its nucleotide sequence was searched against NCBI database using BLASTX program. The result revealed that the sequence is a homologue to AHL synthase and autoinducer synthase from other A. baumannii strains with similarity exceeded 95% identical residues. Multiple sequence alignment analysis further showed that these sequences exhibit a high degree of similarity between various Acinetobacter species, particularly the 10 conserved amino acids signature to autoinducer synthase homologue protein (Figure 2). A phylogenetic tree was constructed based on the protein sequences of autoinducer synthase from 11 Acinetobacter strains (Figure 3). The putative abaI gene from strain 863 was grouped with other A. baumannii strains with a bootstrap value of 97%. The protein sequence from the putative abaI was analyzed using InterProScan [31]. Results showed that the amino acid sequence matches with protein family from autoinducer synthase and possessed acyl-CoA-N-acyltransferase domain, a signature structural domain for autoinducer synthase.
Figure 2.
Multiple sequence alignment of autoinducer synthase protein sequence of strain 863 and other Acinetobacter species. Identical residues are highlighted in grey. All 10 invariant residues in autoinducer synthase homologues are labelled with asterisks. National Center for Biotechnology Information (NCBI) accession numbers: A. baumannii SIPA14 (OXU71347), A. baumannii OIFC110 (EKL58643), A. baumannii IS-116 (EKA70172), A. baumannii 118362 (EXA85498), A. baumannii WC-348 (EKU52127), Acinetobacter sp. WC-136 (EKU68177), A. pittii UKK-0540 (ODI99480), A. calcoaceticus EGD_AQ_BF14 (OBA11519), Acinetobacter sp. 1245249 (EXH33584), Acinetobacter genomosp. 33YU A362 (ONN53459), A. oleivorans DR1 (ADB80056), A. baumannii 863 (WP_029424600).
Figure 3.
Phylogenetic tree of autoinducer synthase from strain 863 and other Acinetobacter species. The tree was generated using maximum likelihood method based on the Jones-Taylor-Thornton (JTT) matrix-based model. The numbers at the node show the bootstrap values as a percentage of 1000 bootstrap replications. Autoinducer synthase from Janthinobacterium sp. ROICE36 (WP_102124182) was used as the outgroup in this tree. NCBI accession numbers: A. baumannii SIPA14 (OXU71347), A. baumannii OIFC110 (EKL58643), A. baumannii IS-116 (EKA70172), A. baumannii 118362 (EXA85498), A. baumannii WC-348 (EKU52127), Acinetobacter sp. WC-136 (EKU68177), A. pittii UKK-0540 (ODI99480), A. calcoaceticus EGD_AQ_BF14 (OBA11519), Acinetobacter sp. 1245249 (EXH33584), Acinetobacter genomosp. 33YU A362 (ONN53459), A. oleivorans DR1 (ADB80056), A. baumannii 863 (WP_029424600).
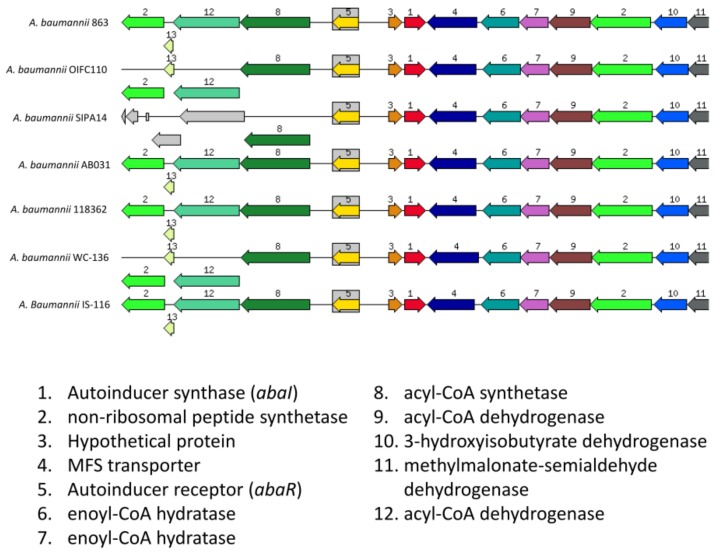
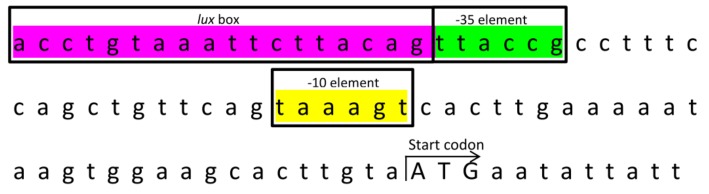
When we analyzed the organization of gene cluster of abaI, we found that the gene clusters demonstrated conserved variation when compared to other A. baumannii strains except A. baumannii SIPA14 (Figure 4). To further analyze the flanking regions of this abaI gene, we first searched 200 bp upstream of abaI using BPROM to determine the presence of a promoter in this region. BPROM analysis predicted a putative −10 element (Pribnow box) sequence, TAAAGT and a −35 element sequence, TTACCG. These sequences are located at 30 and 53 nucleotides upstream of abaI, respectively (Figure 5). Besides, both element sequences are 17 nucleotides apart which are found to be an optimum spacing proposed by Hawley and McClure [32]. Subsequently, we aimed to identify the “lux box” which is palindromic in nature and found in other AHL producing bacteria [33,34,35,36]. We found a palindromic sequence (CTGTAAATTCTTACAG) at 59 nucleotides upstream of start codon site could be the putative lux box. This sequence is the binding site for LuxR homologue protein, designated AbaR.
Figure 4.
The organization of gene cluster of abaI. The organization of abaI in strain 863 and other A. baumannii strains.
Figure 5.
Schematic diagram of the upstream region of abaI. The start codon of the gene is labelled in uppercase. Promoter elements were highlighted in colors. Yellow, −10 (Pribnow box); Green, −35; Magenta, lux box.
3.2. Verification of Autoinducer Synthase Gene through Mutagenesis
Next, we determined the authenticity of the gene with a two-part functional study experiment. First, we determined the production of AHL in strain 863. This involved AHL extraction from an overnight culture of strain 863 and these extracts were subjected to high-resolution mass spectrometry analysis to detect the presence of AHL. The LCMS/MS mass spectra showed the presence of a long chain AHL (3-hydroxy-dodecanoyl-homoserine lactone, hydroxyl C12 in short) in the spent culture supernatant of strain 863 (Figure 6). We then proceeded to confirm the putative AHL synthase gene is indeed the gene responsible for the AHL production in strain 863. This was done by replacing the putative abaI gene with a kanamycin resistant cassette via a recombineering method. Cell-free extracts from overnight cultures of the mutant strains were then again subjected to LCMS/MS analysis. Mass spectrometry analysis confirmed the absence of AHL in the mutant strain, therefore verified that the putative abaI gene was responsible for the production of AHL in strain 863.
Figure 6.
Mass spectrometry analysis shows the presence of a long chain AHL, hydroxy-C12 in the spent culture supernatant of strain 863. (A) Mass spectra of synthetic hydroxyl-C12 and (B) hydroxyl-C12 from strain 863. A peak at m/z 300.3 was detected which indicated the presence of hydroxyl-C12.
3.3. Growth Curve Analysis
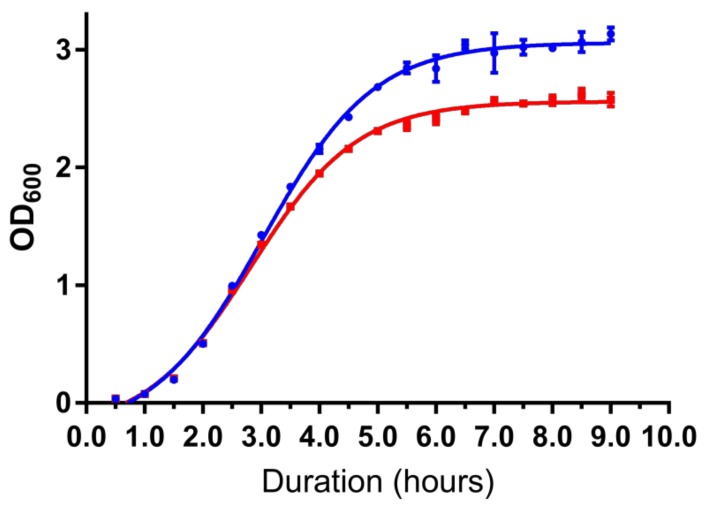
A growth curve analysis was performed to determine the difference in growth rate between the wild type and the mutant strains (A. baumannii strain 863 ΔabaI:Km). We observed a difference in cell density between the wild type and QS-deficient mutant at late log phase. This trend continued when the cultures reached the stationary phase in which the OD600 of the wild type was higher than 3.0 while the mutant stayed around 2.5 Figure 7.
Figure 7.
Growth curve analysis shows QS-deficient mutant suffered growth defect. Mutant strain (red line) has significantly lower cell density compared to the wild type (blue line). Data are presented as the mean (± S.E.) of three independent biological replicates.
3.4. Transciptome Profiling Study
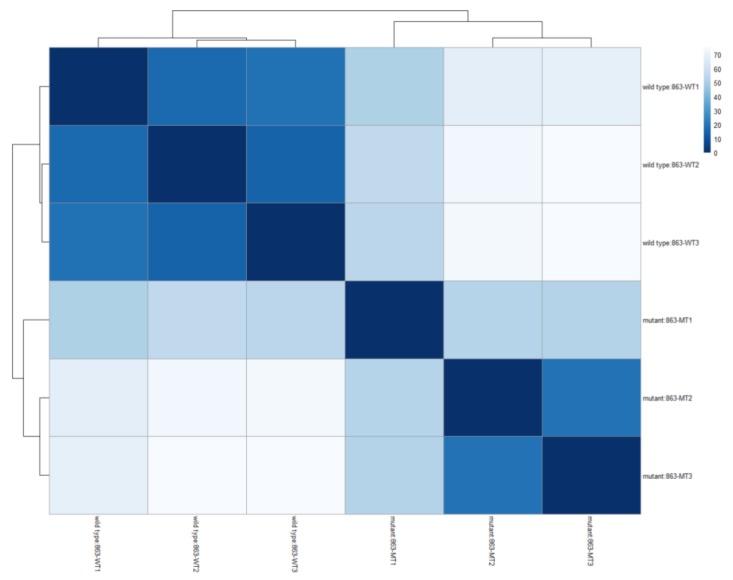
Transcriptome profiling was conducted via RNA-Seq to identify the genes regulated by QS in strain 863 and generated a total of around 43.5 million raw reads. From these total raw reads, 33.1 million (76.2%) passed the filter reads (PF read) and 98.5% of these reads were identified. The composition of every individual sample ranged from 14.6 to 19.3% of the total identified read. The heat map in Figure 8 shows both wild type and mutant strains were grouped into 2 clusters, indicating a good replication of raw data among samples.
Figure 8.
A heatmap showing sample-to-sample Euclidean distances computed from the regularized log transformation. This heat map shows that the wild type and QS-deficient mutant were clustered in respective groups, which indicates a distinct difference in the expression of various genes.
There are a total of 352 genes found to have a differential in expression (Log2 |fold change| ≥ 2) between the wild type and ΔabaI:Km strains Table S1. Out of the 352 genes, 175 genes are downregulated while 177 genes are upregulated in the mutant strain. From this list, we found that the average magnitude of DE is significantly higher in the downregulated genes (mean Log2FC = −4.69) compared to the upregulated genes (mean Log2FC = 2.28). Table 3 shows the 10 genes with the highest magnitude in DE for both downregulated and upregulated genes.
Table 3.
Ten Highest Fold Changes of Downregulated and Upregulated Genes in QS-Deficient Mutant.
| Locus tag | Gene | Log2 FC |
|---|---|---|
| Down-regulated genes | ||
| A9801_RS14870 | 3-methylcrotonyl-CoA carboxylase subunit alpha, liuD | −9.61393 |
| A9801_RS14965 | NAD(P)-dependent oxidoreductase | −9.42986 |
| A9801_RS15335 | phenylacetate-CoA ligase, paaK | −9.37778 |
| A9801_RS14955 | hypothetical protein | −9.30532 |
| A9801_RS16540 | aminoglycoside N-acetyltransferase AAC(3)-IId | −9.20034 |
| A9801_RS15345 | protein paaH | −9.18939 |
| A9801_RS14890 | TetR family transcriptional regulator | −9.1678 |
| A9801_RS14960 | catalase HPII | −9.1216 |
| A9801_RS14875 | enoyl-CoA hydratase | −9.00212 |
| A9801_RS14855 | MFS transporter | −8.94764 |
| Up-regulated genes | ||
| A9801_RS10685 | UmuC | 3.880526 |
| A9801_RS13285 | hypothetical protein | 2.842797 |
| A9801_RS13465 | GntR family transcriptional regulator | 2.737668 |
| A9801_RS06690 | 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline decarboxylase | 2.725173 |
| A9801_RS13170 | alpha/beta hydrolase | 2.641788 |
| A9801_RS01690 | terminase | 2.622548 |
| A9801_RS10575 | ammonium transporter | 2.621587 |
| A9801_RS16845 | allantoin permease | 2.61855 |
| A9801_RS05955 | MFS transporter | 2.613142 |
| A9801_RS12660 | non-ribosomal peptide synthetase | 2.608198 |
A number of genes involved in carbon source metabolism were down-regulated in the ΔabaI:Km strain. The genes (A9801_RS14885, liuA; A9801_RS14880, liuB; A9801_RS14875, liuC; A9801_RS14870, liuD; A9801_RS14865, liuE) from the leucine/isovalerate utilization (Liu) operon were highly repressed in the mutant strain compared to the wild type. These genes had a Log2FC of less than −8.22 and liuD was the gene with the highest degree of down-regulation (Log2FC = −9.61) among other DE genes (Table 3). Besides, the phenylacetic acid degradation pathway was found to be affected as well. The genes (paaH, A9801_RS15345; paaJ, A9801_RS15340; paaK, A9801_RS15335) involved in this process were heavily down-regulated in ΔabaI:Km strain (Log2FC < −8.75).
Other than carbon source metabolism, it was found that a protein family involved in energy production was affected in the mutant strain (A. baumannii strain 863 ΔabaI:Km). The expression of a few cytochrome proteins or the components of them which are vital in the redox reaction of oxidative phosphorylation, were reduced in the mutant strain compared to the wild type. These genes included cytochrome b (A9801_RS14635), cytochrome o ubiquinol oxidase (cyoA, A9801_RS16360; cyoB, A9801_RS16365) and cytochrome bd complex (cydA, A9801_RS11155; cydB, A9801_RS11150). In addition, another vital component of the energy production process, F-ATPase, was also found to be affected in the mutant strain. This is because the majority of the components that constitute this protein complex were down-regulated (atpA, A9801_RS12840; atpB, A9801_RS12820; atpD, A9801_RS12850; atpF, A9801_RS12830; atpG, A9801_RS12845 & atpH, A9801_RS12835).
Interestingly, a number of genes responsible in stress response were expressed differently in the QS mutant strain. From an oxidative stress perspective, we found that two catalases (A9801_RS14960 & A9801_RS14630) were heavily repressed in the mutant strain. We also discovered that the expression of umuC (A9801_RS10685) and umuD (A9801_RS14985) are different from the wild type. These two genes encode the subunit of DNA polymerase V which is involved in a DNA repair mechanism [37]. However, both genes have an opposite pattern on one another. The first subunit, umuD, was down-regulated in the mutant strain while umuC was upregulated. In fact, umuC was the gene with the highest degree of differential expression among the upregulated genes (Log2 FC = 3.88) (Table 3).
Another group of genes worth noting is the genes associated with the ribosomes. There were 16 genes encoding for ribosomal proteins that exhibited reduced expression in the mutant strain. Seven of them (S2, A9801_RS17795; S4, A9801_RS14500; S6, A9801_RS16385; S10, A9801_RS14375; S13, A9801_RS14490; S16, A9801_RS13965 & S18, A9801_RS16390) of these genes are associated with the small subunit while the remaining 9 (L2, A9801_RS14395; L4, A9801_RS14385; L11, A9801_RS03460; L13, A9801_RS15160; L14, A9801_RS14430; L17, A9801_RS14510; L23, A9801_RS14390; L25, A9801_RS05355 & L28, A9801_RS02635) were associated with the large subunit. Besides ribosomal proteins, ribosome maturation factor, rimM (A9801_RS13970) and ribosome recycling factor, rrf (A9801_RS10865) were also repressed in A. baumannii strain 863 ΔabaI:Km.
4. Discussion
Whole genome sequencing was performed on strain 863 to decipher its draft genome. Raw sequencing data were processed and annotated by annotation pipelines with respective databases. From annotated data, a pair of luxIR homologues which consist of autoinducer synthase, abaI and autoinducer-binding receptor, abaR, was found. In this work, we aimed to characterize the AHL synthase, AbaI. From the multiple sequence alignment result, we observed that the AHL synthase protein from strain 863 possesses 10 amino acid residues which are conserved in autoinducer synthases among many bacterial species [38], and has high similarity with other Acinetobacter species. In addition, the phylogenetic tree shows that the autoinducer synthase protein from strain 863 was clustered with other homologues of A. baumannii strains. The phylogenetic tree reveals how closely related is AbaI with other autoinducer synthases in terms of molecular evolutionary distance. AbaI is clearly shown to be clustered closely among LuxI homologues of other A. baumanii strains, hence, strongly indicates a low rate of random mutation for this autoinducer gene. On the other hand, AbaI is the least phylogenetically related to AHL synthase from A. oleivorans DR1, possibly indicating that the latter was diverted in evolution. Besides, the amino acid analysis from Interproscan also showed the presence of acyl-CoA-N-acyltransferase domain in the putative AbaI as well as autoinducer synthase from various bacterial species that use AHLs as autoinducers.
Further studies on its upstream sequence, we managed to identify a few important promoter sequences, −10 and −35 elements. The presence of both promoter regions meets the requirement of a typical Escherichia coli RNA polymerase σ70 consensus promoter sequences. We also found a palindromic sequence that has a high possibility to be a putative lux box, which is associated with LuxR transcriptional binding factor, at 59 bp upstream of abaI. This 16-bp palindromic sequence is identical to the lux box found in other Acinetobacter species [39,40,41], indicating those species may possibly possess similar regulation of AHLs. In fact, many of these A. baumannii strains share a similar cluster of genes in the vicinity of LuxI/R genes. An interesting point is the presence of fatty acid synthesis related genes which are found at the downstream and upstream of luxI/R genes (Figure 4). It is highly suggested that the metabolites from these fatty acid biosynthetic machinery are used as the precursors to form the lactone ring and the acyl group of AHLs.
Following this analysis, we then determined whether strain 863 is an AHL-producing bacterium. LCMS/MS mass spectra showed the presence of hydroxyl C12 in the cell-free extract. This result correlates with other studies in which hydroxyl C12 is detected in the cultures from other Acinetobacter species [39,40], but not the mutant ΔabaI:Km strain, hence the functional role of abaI was confirmed in this work. Even though hydroxyl C12 is commonly produced by Acinetobacter species, there are exceptional cases. A study by Anbazhagan et al. [3] on multiple clinical strains of Acinetobacter sp. found that none of them produced hydroxyl C12. Chan et al. [9] and John et al. [42] who worked on A. baumannii 4KT and A. baumannii AB 14, respectively, also did not detect hydroxyl C12 using LCMS analysis.
As RNA-Seq provides a better result than the microarray approach, we used high throughput transcriptomic studies to investigate gene regulation on both wildtype and mutant strains [19,20,43]. In this work, we studied the transcriptome profiling of late log phase of the bacterial growth curve because we hope this phase could provide more information on QS in strain 863 during rapid growth and in preparation of the stationary phase. This is because there are studies that found that QS helps optimize growth in bacteria [44,45]. We also hope that by studying this phase, we could unlock some insights related to the differences in the growth rate and cell density in stationary phase between the wild type and QS-deficient mutant. As clearly illustrated in Figure 7, the mutant strain achieved a lower cell density at stationary phase as compared to its wildtype counterpart. This suggests that a paralyzed QS system in the mutant strain affects the basic cellular mechanism and metabolic pathways, hence producing a much slower growth rate. From the analyzed transcriptome data, we discovered that there are four distinct groups of genes which were expressed differently in the mutant strain compared to the wild type. These groups are related to carbon source metabolism, energy production, stress and ribosomes.
There are a few studies that associate QS with bacterial metabolism [46,47,48]. In this study, we observed that the expression of a number of genes related to carbon source metabolism was downregulated. The Liu operon is found to be affected the most as most of the genes in this operon were downregulated significantly in the mutant strain. The Liu operon is important in the leucine catabolism pathway [49,50]. Leucine is an alternative carbon source for metabolic activity during starvation, hence, increases the chance of survival in multiple bacterial species [51,52,53]. Therefore, in this study, we speculated that the mutant strain’s ability to survive in a nutrient-limiting environment could be hampered if the QS system was paralyzed. Nevertheless, further investigation is needed to validate such hypothesis. Besides Liu operon, a few genes in the Paa operon which are involved in phenylacetic acid degradation were also repressed in the QS-deficient mutant strain. This operon enables the bacteria to utilize phenylalanine as a carbon source and break it down into other aromatic compounds [54]. According to Bhuiyan et al. [12], phenylacetate accumulation in the infected site of zebrafish embryos could induce neutrophil chemotaxis. In our study, we found that paaK was greatly repressed in the mutant strain. The gene product of paaK, phenylacetate-CoA ligase, catalyzes the conversion of phenylacetate to phenylacetyl-CoA [55]. In fact, a paaK homologue which was found in Phaeobacter gallaeciensis DSM 17395 is essential for phenylacetate degradation [56]. Hence, we predicted that the downregulation of paaK could lead to phenylacetate accumulation in the infected site. However, Bhuiyan and co-workers [12] found that there was no difference in neutrophil migration pattern when the QS-deficient mutant of A. baumannii M2 was inoculated into zebrafish embryos. Hence, it is of high interest to further investigate the effect of downregulation of paaK in the mutant strain ΔabaI:Km.
In addition, several genes associated with energy production were affected in the QS-deficient mutant, ΔabaI:Km. For instance, a few genes related to cytochrome were downregulated in the QS-deficient mutant. This included cytochrome b, cytochrome o ubiquinol oxidase (cyoA & cyoB) and cytochrome bd complex (cydA & cydB). A study showed that the extracellular ATP production of the ∆cyoA mutants was lowered by more than two fold compared to the wild type strains in both E. coli and Salmonella enterica. Meanwhile, the production of extracellular ATP of the ∆cyoB mutant in S. enterica was also altered less than two fold, hence, suggesting that cytochrome o ubiquinol oxidase is responsible for extracellular ATP production [57]. The same study also reported that extracellular ATP could improve bacterial survivability. The cytochrome bd complex is a terminal oxidase that helps in establishing proton motive force. Although the biochemical reaction is less efficient, the protein’s function becomes more significant under stress conditions which are unfavorable to other terminal oxidases [58]. Boot et al. [58] also observed a growth defect in the ∆cydB mutant of E. coli, which could be a factor contributing to the slower growth in the QS-deficient mutant strain 863 ΔabaI:Km. In E. coli, the cytochrome bd complex is important against oxidative stress from hydrogen peroxide [59]. Furthermore, multiple in vitro and in vivo studies suggested that the cytochrome bd complex in various bacterial species could play a role in bacterial survivability during infection in a number of vertebrate hosts [58,60,61,62,63,64,65,66].
Another important component in energy production is F-ATPase. F-ATPase generates energy in the form of ATP by transporting hydrogen ions across the cell membrane through a proton motive force [67]. Besides, F-ATPase is also reported to help bacteria to have a higher tolerance to acidic environments [68,69]. For example, Streptococcus mutans was found to be able to increase the expression of F-ATPase to pump hydrogen ions out of the cell to maintain its cytoplasmic pH in order to adapt to an acidic environment [70,71]. Moreover, F-ATPase can also act as ATP synthase when bacteria under starvation are exposed to an acidic environment [72]. Most of the subunits associated with the F-ATPase complex were downregulated in the QS-deficient mutant, ΔabaI:Km suggesting that QS could have a role in regulating these genes.
Bacteria are constantly experiencing stress in a harsh environment. Our result showed that QS is involved in some stress responses in strain 863. The synthesis of catalases was drastically downregulated in the mutant strain. This is in agreement with a report from Bhargava et al. [73] in which the QS-deficient mutant of A. baumannii M2 showed a reduction in catalase production and was more sensitive to hydrogen peroxide. A decrease in catalase and cytochrome bd complex production in the QS-deficient mutant could signify a potential disinfection approach in healthcare environments using anti-QS compounds followed by hydrogen peroxide. Abiotic stress such as oxidative stress could often lead to DNA damage [74,75]. When DNA is damaged beyond repair by other cellular mechanisms, an error-prone repair mechanism known as the “SOS response” will be triggered [37]. When the SOS response is triggered, the cell cycle is arrested and there is a marked increase in the mutation rate [76]. One of the crucial proteins involved in this mechanism is DNA Polymerase V, which consists of subunits encoded by umuCD [37]. The formation of DNA Polymerase V requires the binding of two RecA-mediated cleavage of UmuD and one UmuC protein [37]. Both UmuC and UmuD proteins are involved in translesion repair of DNA and are essential in induced mutagenesis. They are able to replicate DNA across DNA lesions in the presence of activated RecA [77]. In our RNA-Seq data, we discovered that the umuC gene was upregulated the most compared to other upregulated genes. Studies have shown that elevated umuC expression is generally associated with mutagenic agents such as ultraviolet rays and methyl methanesulfonate [78,79]. The action of both UmuC and UmuD proteins are needed for cell cycle checkpoint control and they are expressed in the late SOS response [80]. However, we were intrigued by our results as both umuD and umuC expression level was contradictory to each other. This phenomenon could be explained by a study done by Aranda et al. [37], which suggested that the expression of UmuC is probably regulated by UmuD which can act as a repressor or an activator. Nevertheless, further investigations are required to determine if DNA repair systems were impaired in the QS-deficient mutant.
Our result suggested that QS may play a role in strain 863 ribosome biogenesis. Multiple genes encoded for ribosomal protein were downregulated in the QS-deficient mutant. Though many ribosomal proteins are not essential in translation, these proteins could increase the integrity of ribosomes [81]. Other than protein synthesis, ribosomal proteins also act as transcriptional factors to regulate their own or other ribosomal proteins’ transcriptional level [81]. Additionally, they were also found to control transcription termination [82]. Other than ribosomal proteins, we also discovered two interesting proteins related to translation, namely, ribosome maturation factor M, RimM and ribosome recycling factor, RRF which were also downregulated in the mutant strain. Bylund et al. [83] reported that RimM plays an important role in 30S subunit maturation in E. coli. During the maturation process, RimM is involved in the stability of 16S 3′-domain [84,85]. Therefore, E. coli with deleted rimM had been demonstrated to have retarded growth and a lower cell density in stationary phase compared to the wild type strain [83]. In fact, it was demonstrated that E. coli strains from which the rimM gene has been deleted, have a sevenfold-reduced growth rate and a reduced translational efficiency [85]. Additionally, RimM was found to be critical for efficient processing of 16S rRNA. RRF encoded by frr is involved in the separation of ribosomes from mRNA after stop codon is detected to allow the next translation to take place [86]. A study on E. coli found that RRF is crucial for bacterial viability as no colony with a mutated ffr gene was able to be isolated unless it was complemented with a plasmid harboring intact ffr [86]. This could be a possible factor which contributes to the growth defect in the QS-deficient strain, as both rimM and ffr were downregulated compared to the wild type strain.
Surprisingly, abaI gene was not found in the top ten downregulated genes. This could be due to a very low transcription level in the wild type strain. This can be validated by the count table generated by HTSeq-count script. We found that the raw count for abaI gene was extremely low in the wild types among the three replicates (9, 3 and 6) while no read was found in the mutant replicates (A. baumannii strain 863 ΔabaI:Km). We postulated this result could be due to culture conditions. A recent study found that static culture condition increases the production of AHL and abaI expression in A. baumannii ATCC17978 [87]. Hence, Mayer et al. [87] speculated that the expression of QS genes in Acinetobacter species requires cell-to-cell or surface attachment. Nonetheless, we should not discount the differences in gene expression of other genes even with the low expression of abaI as demonstrated in the RNA-Seq data.
5. Conclusions
In this study, we had identified and characterized a luxI homologue, abaI, in multidrug-resistant strain 863. Through transcriptomic study, this paper reported that QS in strain 863 could possibly regulate more than 300 genes, and of these, four main groups of genes were significantly differentially expressed in the QS-deficient mutant, each with respective roles in bacterial fitness. We hope the findings in this study enable us to unlock more insights about the QS system in A. baumannii, and at the same time incorporate this knowledge in antimicrobial approaches against A. baumannii.
Acknowledgments
We would like to thank Cheong Yuet Meng for supplying A. baumannii strain 863. We also thank Bryan W. Davies for providing us plasmids pAT04 and pKD4.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/10/4/282/s1, Table S1: List of Genes with differential expression.
Author Contributions
Conceptualization, K.-Y.H. and K.-G.C.; Data curation, C.-K.N.; Formal analysis, C.-K.N.; Funding acquisition, K.-G.C.; Investigation, C.-K.N. and K.-Y.H.; Methodology, C.-K.N. and K.-Y.H.; Project administration, K.-G.C.; Resources, K.-G.C.; Software, C.-K.N.; Supervision, K.-K.T. and K.-G.C.; Writing—original draft, C.-K.N.; Writing—review & editing, K.-Y.H. and K.-K.T.
Funding
This study was funded by the University of Malaya (Grant No. A0001-500001), grant awarded to KGC. CKN gives thanks for the PPP Grant (Grant No. PG134-2016A) from the University of Malaya.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Ng W.-L., Bassler B.L. Bacterial quorum-sensing network architectures. Annu. Rev. Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yin W.-F., Purmal K., Chin S., Chan X.-Y., Koh C.-L., Sam C.-K., Chan K.-G. N-acyl homoserine lactone production by Klebsiella pneumoniae isolated from human tongue surface. Sensors. 2012;12:3472–3483. doi: 10.3390/s120303472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anbazhagan D., Mansor M., Yan G.O.S., Yusof M.Y.M., Hassan H., Sekaran S.D. Detection of quorum sensing signal molecules and identification of an autoinducer synthase gene among biofilm forming clinical isolates of Acinetobacter spp. PLoS ONE. 2012;7:e36696. doi: 10.1371/journal.pone.0036696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang D., Seeve C., Pierson L.S., Pierson E.A. Transcriptome profiling reveals links between ParS/ParR, MexEF-OprN, and quorum sensing in the regulation of adaptation and virulence in Pseudomonas aeruginosa. Bmc Genom. 2013;14:618. doi: 10.1186/1471-2164-14-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khajanchi B.K., Sha J., Kozlova E.V., Erova T.E., Suarez G., Sierra J.C., Popov V.L., Horneman A.J., Chopra A.K. N-acylhomoserine lactones involved in quorum sensing control the type VI secretion system, biofilm formation, protease production, and in vivo virulence in a clinical isolate of Aeromonas hydrophila. Microbiology. 2009;155:3518–3531. doi: 10.1099/mic.0.031575-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He X., Chang W., Pierce D.L., Seib L.O., Wagner J., Fuqua C. Quorum sensing in Rhizobium sp. strain NGR234 regulates conjugal transfer (tra) gene expression and influences growth rate. J. Bacteriol. 2003;185:809–822. doi: 10.1128/JB.185.3.809-822.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Piper Kevin R., von Bodman S.B., Hwang I., Stephen K.F. Hierarchical gene regulatory systems arising from fortuitous gene associations: Controlling quorum sensing by the opine regulon in Agrobacterium. Mol. Microbiol. 1999;32:1077–1089. doi: 10.1046/j.1365-2958.1999.01422.x. [DOI] [PubMed] [Google Scholar]
- 8.Boettcher K.J., Ruby E.G. Detection and quantification of Vibrio fischeri autoinducer from symbiotic squid light organs. J. Bacteriol. 1995;177:1053–1058. doi: 10.1128/jb.177.4.1053-1058.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan K.-G., Cheng H.J., Chen J.W., Yin W.-F., Ngeow Y.F. Tandem mass spectrometry detection of quorum sensing activity in multidrug resistant clinical isolate Acinetobacter baumannii. Sci. World J. 2014;2014:6. doi: 10.1155/2014/891041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peleg A.Y., Seifert H., Paterson D.L. Acinetobacter baumannii: Emergence of a successful pathogen. Clin. Microbiol. Rev. 2008;21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoon E.-J., Chabane Y.N., Goussard S., Snesrud E., Courvalin P., Dé E., Grillot-Courvalin C. Contribution of resistance-nodulation-cell division efflux systems to antibiotic resistance and biofilm formation in Acinetobacter baumannii. mBio. 2015:6. doi: 10.1128/mBio.00309-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhuiyan M.S., Ellett F., Murray G.L., Kostoulias X., Cerqueira G.M., Schulze K.E., Maifiah M.H.M., Li J., Creek D.J., Lieschke G.J., et al. Acinetobacter baumannii phenylacetic acid metabolism influences infection outcome through a direct effect on neutrophil chemotaxis. Proc. Natl. Acad. Sci. USA. 2016;113:9599–9604. doi: 10.1073/pnas.1523116113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Antunes L.C.S., Visca P., Towner K.J. Acinetobacter baumannii: Evolution of a global pathogen. Pathog. Dis. 2014;71:292–301. doi: 10.1111/2049-632X.12125. [DOI] [PubMed] [Google Scholar]
- 14.Eveillard M., Kempf M., Belmonte O., Pailhoriès H., Joly-Guillou M.-L. Reservoirs of Acinetobacter baumannii outside the hospital and potential involvement in emerging human community-acquired infections. Int. J. Infectious Dis. 2013;17:e802–e805. doi: 10.1016/j.ijid.2013.03.021. [DOI] [PubMed] [Google Scholar]
- 15.Seifert H., Dijkshoorn L., Gerner-Smidt P., Pelzer N., Tjernberg I., Vaneechoutte M. Distribution of Acinetobacter species on human skin: Comparison of phenotypic and genotypic identification methods. J. Clin. Microbiol. 1997;35:2819–2825. doi: 10.1128/jcm.35.11.2819-2825.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joly-Guillou M.L. Clinical impact and pathogenicity of Acinetobacter. Clin. Microbiol. Infect. 2005;11:868–873. doi: 10.1111/j.1469-0691.2005.01227.x. [DOI] [PubMed] [Google Scholar]
- 17.Xie R., Zhang X.D., Zhao Q., Peng B., Zheng J. Analysis of global prevalence of antibiotic resistance in Acinetobacter baumannii infections disclosed a faster increase in OECD countries. Emerg. Microbes Infect. 2018;7:1–10. doi: 10.1038/s41426-018-0038-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.World Health Organization (WHO) WHO Publishes List of Bacteria for Which New Antibiotics are Urgently Needed. WHO; Geneva, Switzerland: 2017. [Google Scholar]
- 19.Croucher N.J., Thomson N.R. Studying bacterial transcriptomes using RNA-Seq. Curr. Opin. Microbiol. 2010;13:619–624. doi: 10.1016/j.mib.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan K.-G. Whole-genome sequencing in the prediction of antimicrobial resistance. Expert Rev. Anti-Infect. Ther. 2016;14:617–619. doi: 10.1080/14787210.2016.1193005. [DOI] [PubMed] [Google Scholar]
- 21.Tucker A.T., Nowicki E.M., Boll J.M., Knauf G.A., Burdis N.C., Trent M.S., Davies B.W. Defining gene-phenotype relationships in Acinetobacter baumannii through one-step chromosomal gene inactivation. mBio. 2014;5:e01313-14. doi: 10.1128/mBio.01313-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Babraham Bioinformatics-FastQC, A quality control tool for high throughput sequence data. [(accessed on 8 August 2016)]; Available online: http://www.Bioinformatics.babraham.ac.uk/projects/fastqc.
- 23.Tatusova T., DiCuccio M., Badretdin A., Chetvernin V., Nawrocki E.P., Zaslavsky L., Lomsadze A., Pruitt K.D., Borodovsky M., Ostell J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016;44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aziz R.K., Bartels D., Best A.A., DeJongh M., Disz T., Edwards R.A., Formsma K., Gerdes S., Glass E.M., Kubal M., et al. The RAST server: Rapid Annotations using Subsystems Technology. BMC Genom. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar S., Stecher G., Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thornton J.M., Jones D.T., Taylor W.R. The Rapid Generation of Mutation Data Matrices from Protein Sequences 8, no. Bioinformatics. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 27.Solovyev V., Salamov A. Automatic annotation of microbial genomes and metagenomic sequences. In: Robert W.L., editor. Metagenomics and Its Applications in Agriculture, Biomedicine and Environmental Studies. Nova Science Publishers; Hauppauge, NY, USA: 2011. pp. 61–78. [Google Scholar]
- 28.Chan K.-G., Atkinson S., Mathee K., Sam C.-K., Chhabra S.R., Cámara M., Koh C.-L., Williams P. Characterization of N-acylhomoserine lactone-degrading bacteria associated with the Zingiber officinale (ginger) rhizosphere: Co-existence of quorum quenching and quorum sensing in Acinetobacter and Burkholderia. BMC Microbiol. 2011;11:51. doi: 10.1186/1471-2180-11-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anders S., Pyl P.T., Huber W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:1–21. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mitchell A., Chang H.Y., Daugherty L., Fraser M., Hunter S., Lopez R., McAnulla C., McMenamin C., Nuka G., Pesseat S., et al. The InterPro protein families database: The classification resource after 15 years. Nucleic Acids Res. 2015;43:D213–D221. doi: 10.1093/nar/gku1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hawley D.K., McClure W.R. Compilation and analysis of Escherichia coli promoter DNA Sequences. Nucleic Acids Res. 1983;11:2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Devine J.H., Shadel G.S., Baldwin T.O. Identification of the operator of the lux regulon from the Vibrio fischeri strain ATCC7744. Proc. Natl. Acad. Sci. USA. 1989;86:5688–5692. doi: 10.1073/pnas.86.15.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Farah C., Vera M., Morin D., Haras D., Jerez C.A., Guiliani N. Evidence for a functional quorum-sensing type AI-1 system in the extremophilic bacterium Acidithiobacillus ferrooxidans. Appl. Environ. Microbiol. 2005;71:7033–7040. doi: 10.1128/AEM.71.11.7033-7040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fuqua C., Winans S.C. Conserved cis-acting promoter elements are required for density-dependent transcription of Agrobacterium tumefaciens conjugal transfer genes. J. Bacteriol. 1996;178:435–440. doi: 10.1128/jb.178.2.435-440.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gray K.M., Passador L., Iglewski B.H., Greenberg E.P. Interchangeability and specificity of components from the quorum-sensing regulatory systems of Vibrio fischeri and Pseudomonas aeruginosa. J. Bacteriol. 1994;176:3076–3080. doi: 10.1128/jb.176.10.3076-3080.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aranda J., Poza M., Shingu-Vázquez M., Cortés P., Boyce J.D., Adler B., Barbé J., Bou G. Identification of a DNA-damage-inducible regulon in Acinetobacter baumannii. J. Bacteriol. 2013;195:5577–5582. doi: 10.1128/JB.00853-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parsek M.R., Schaefer A.L., Greenberg E.P. Analysis of random and site--directed mutations in rhlI, a Pseudomonas aeruginosa gene encoding an acylhomoserine lactone synthase. Mol. Microbiol. 1997;26:301–310. doi: 10.1046/j.1365-2958.1997.5741935.x. [DOI] [PubMed] [Google Scholar]
- 39.How K.Y., Hong K.-W., Sam C.-K., Koh C.-L., Yin W.-F., Chan K.-G. Unravelling the genome of long chain N-acylhomoserine lactone-producing Acinetobacter sp. strain GG2 and identification of its quorum sensing synthase gene. Front. Microbiol. 2015;6:240. doi: 10.3389/fmicb.2015.00240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niu C., Clemmer K.M., Bonomo R.A., Rather P.N. Isolation and characterization of an autoinducer synthase from Acinetobacter baumannii. J. Bacteriol. 2008;190:3386–3392. doi: 10.1128/JB.01929-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oh M.H., Choi C.H. Role of LuxIR homologue AnoIR in Acinetobacter nosocomialis and the effect of virstatin on the expression of anoR gene. J. Microbiol. Biotechnol. 2015;25:1390–1400. doi: 10.4014/jmb.1504.04069. [DOI] [PubMed] [Google Scholar]
- 42.John J., Saranathan R., Adigopula L.N., Thamodharan V., Singh S.P., Lakshmi T.P., CharanTej M.A., Rao R.S., Krishna R., Rao H.S.P., et al. The quorum sensing molecule N-acyl homoserine lactone produced by Acinetobacter baumannii displays antibacterial and anticancer properties. Biofouling. 2016;32:1029–1047. doi: 10.1080/08927014.2016.1221946. [DOI] [PubMed] [Google Scholar]
- 43.Mutz K.-O., Heilkenbrinker A., Lönne M., Walter J.-G., Stahl F. Transcriptome analysis using next-generation sequencing. Curr. Opin. Biotechnol. 2013;24:22–30. doi: 10.1016/j.copbio.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 44.Bruger E.L., Waters C.M. Bacterial quorum sensing stabilizes cooperation by optimizing growth strategies. Appl. Environ. Microbiol. 2016;82:6498–6506. doi: 10.1128/AEM.01945-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nickzad A., Déziel E. Adaptive significance of quorum sensing-dependent regulation of rhamnolipids by integration of growth rate in Burkholderia glumae: A trade-off between survival and efficiency. Front. Microbiol. 2016;7:1215. doi: 10.3389/fmicb.2016.01215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davenport P.W., Griffin J.L., Welch M. Quorum sensing is accompanied by global metabolic changes in the opportunistic human pathogen Pseudomonas aeruginosa. J. Bacteriol. 2015;197:2072–2082. doi: 10.1128/JB.02557-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goo E., Majerczyk C.D., An J.H., Chandler J.R., Seo Y.-S., Ham H., Lim J.Y., Kim H., Lee B., Jang M.S., et al. Bacterial quorum sensing, cooperativity, and anticipation of stationary-phase stress. Proc. Natl. Acad. Sci. USA. 2012;109:19775–19780. doi: 10.1073/pnas.1218092109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hawver L.A., Giulietti J.M., Baleja J.D., Ng W.-L. Quorum sensing coordinates cooperative expression of pyruvate metabolism genes to maintain a sustainable environment for population stability. mBio. 2016;7:e01863-16. doi: 10.1128/mBio.01863-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aguilar J.A., Zavala A.N., Díaz-Pérez C., Cervantes C., Díaz-Pérez A.L., Campos-García J. The atu and liu clusters are involved in the catabolic pathways for acyclic monoterpenes and leucine in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2006;72:2070–2079. doi: 10.1128/AEM.72.3.2070-2079.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Förster-Fromme K., Höschle B., Mack C., Bott M., Armbruster W., Jendrossek D. Identification of genes and proteins necessary for catabolism of acyclic terpenes and leucine/isovalerate in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2006;72:4819–4828. doi: 10.1128/AEM.00853-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beck H.C., Hansen A.M., Lauritsen F.R. Catabolism of leucine to branched-chain fatty acids in Staphylococcus xylosus. J. Appl. Microbiol. 2004;96:1185–1193. doi: 10.1111/j.1365-2672.2004.02253.x. [DOI] [PubMed] [Google Scholar]
- 52.Dherbécourt J., Maillard M.-B., Catheline D., Thierry A. Production of branched-chain aroma compounds by Propionibacterium freudenreichii: Links with the biosynthesis of membrane fatty acids. J. Appl. Microbiol. 2008;105:977–985. doi: 10.1111/j.1365-2672.2008.03830.x. [DOI] [PubMed] [Google Scholar]
- 53.Serrazanetti D.I., Ndagijimana M., Sado-Kamdem S.L., Corsetti A., Vogel R.F., Ehrmann M., Guerzoni M.E. Acid stress-mediated metabolic shift in Lactobacillus sanfranciscensis LSCE1. Appl. Environ. Microbiol. 2011;77:2656–2666. doi: 10.1128/AEM.01826-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cerqueira G.M., Kostoulias X., Khoo C., Aibinu I., Qu Y., Traven A., Peleg A.Y. A global virulence regulator in Acinetobacter baumannii and its control of the phenylacetic acid catabolic pathway. J. Infect. Dis. 2014;210:46–55. doi: 10.1093/infdis/jiu024. [DOI] [PubMed] [Google Scholar]
- 55.Teufel R., Mascaraque V., Ismail W., Voss M., Perera J., Eisenreich W., Haehnel W., Fuchs G. Bacterial phenylalanine and phenylacetate catabolic pathway revealed. Proc. Natl. Acad. Sci. USA. 2010;107:14390–14395. doi: 10.1073/pnas.1005399107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Berger M., Brock N.L., Liesegang H., Dogs M., Preuth I., Simon M., Dickschat J.S., Brinkhoff T. Genetic analysis of the upper phenylacetate catabolic pathway in the production of tropodithietic acid by Phaeobacter gallaeciensis. Appl. Environ. Microbiol. 2012;78:3539–3551. doi: 10.1128/AEM.07657-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mempin R., Tran H., Chen C., Gong H., Ho K.K., Lu S. Release of extracellular ATP by bacteria during growth. BMC Microbiol. 2013;13:301. doi: 10.1186/1471-2180-13-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boot M., Jim K.K., Liu T., Commandeur S., Lu P., Verboom T., Lill H., Bitter W., Bald D. A fluorescence-based reporter for monitoring expression of mycobacterial cytochrome bd in response to antibacterials and during infection. Sci. Rep. 2017;7:10665. doi: 10.1038/s41598-017-10944-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wall D., Delaney J.M., Fayet O., Lipinska B., Yamamoto T., Georgopoulos C. arc-dependent thermal regulation and extragenic suppression of the Escherichia coli cytochrome d operon. J. Bacteriol. 1992;174:6554–6562. doi: 10.1128/jb.174.20.6554-6562.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Endley S., McMurray D., Ficht T.A. Interruption of the cydB locus in Brucella abortus attenuates intracellular survival and virulence in the mouse model of infection. J. Bacteriol. 2001;183:2454–2462. doi: 10.1128/JB.183.8.2454-2462.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Turner A.K., Barber L.Z., Wigley P., Muhammad S., Jones M.A., Lovell M.A., Hulme S., Barrow P.A. Contribution of proton-translocating proteins to the virulence of Salmonella enterica serovars Typhimurium, Gallinarum, and Dublin in chickens and mice. Infect. Immun. 2003;71:3392–3401. doi: 10.1128/IAI.71.6.3392-3401.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jones-Carson J., Husain M., Liu L., Orlicky D.J., Vázquez-Torres A. Cytochrome bd-dependent bioenergetics and antinitrosative defenses in Salmonella pathogenesis. mBio. 2016;7:e02052-16. doi: 10.1128/mBio.02052-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shepherd M., Achard M.E.S., Idris A., Totsika M., Phan M.-D., Peters K.M., Sarkar S., Ribeiro C.A., Holyoake L.V., Ladakis D., et al. The cytochrome bd-I respiratory oxidase augments survival of multidrug-resistant Escherichia coli during infection. Sci. Rep. 2016;6:35285. doi: 10.1038/srep35285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Way S.S., Sallustio S., Magliozzo R.S., Goldberg M.B. Impact of either elevated or decreased levels of cytochrome bd expression on Shigella flexneri virulence. J. Bacteriol. 1999;181:1229–1237. doi: 10.1128/jb.181.4.1229-1237.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hammer N.D., Reniere M.L., Cassat J.E., Zhang Y., Hirsch A.O., Hood M.I., Skaar E.P. Two heme-dependent terminal oxidases power Staphylococcus aureus organ-specific colonization of the vertebrate host. mBio. 2013;4:e00241-13. doi: 10.1128/mBio.00241-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yamamoto Y., Poyart C., Trieu-Cuot P., Lamberet G., Gruss A., Gaudu P. Respiration metabolism of Group B Streptococcus is activated by environmental haem and quinone and contributes to virulence. Mol. Microbiol. 2005;56:525–534. doi: 10.1111/j.1365-2958.2005.04555.x. [DOI] [PubMed] [Google Scholar]
- 67.Mitchell P. Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature. 1961;191:144–148. doi: 10.1038/191144a0. [DOI] [PubMed] [Google Scholar]
- 68.Kuhnert W.L., Zheng G., Faustoferri R.C., Quivey R.G. The F-ATPase operon promoter of Streptococcus mutans is transcriptionally regulated in response to external pH. J. Bacteriol. 2004;186:8524–8528. doi: 10.1128/JB.186.24.8524-8528.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sturr M.G., Marquis R.E. Comparative acid tolerances and inhibitor sensitivities of isolated F-ATPases of oral lactic acid bacteria. Appl. Environ. Microbiol. 1992;58:2287–2291. doi: 10.1128/aem.58.7.2287-2291.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Belli W.A., Marquis R.E. Adaptation of Streptococcus mutans and Enterococcus hirae to acid stress in continuous culture. Appl. Environ. Microbiol. 1991;57:1134–1138. doi: 10.1128/aem.57.4.1134-1138.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bender G.R., Sutton S.V., Marquis R.E. Acid tolerance, proton permeabilities, and membrane ATPases of oral Streptococci. Infect. Immun. 1986;53:331–338. doi: 10.1128/iai.53.2.331-338.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sheng J., Marquis R.E. Enhanced acid resistance of oral Streptococci at lethal pH values associated with acid-tolerant catabolism and with ATP synthase activity. FEMS Microbiol. Lett. 2006;262:93–98. doi: 10.1111/j.1574-6968.2006.00374.x. [DOI] [PubMed] [Google Scholar]
- 73.Bhargava N., Sharma P., Capalash N. Pyocyanin stimulates quorum sensing-mediated tolerance to oxidative stress and increases persister cell populations in Acinetobacter baumannii. Infect. Immun. 2014;82:3417–3425. doi: 10.1128/IAI.01600-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dukan S., Touati D. Hypochlorous acid stress in Escherichia coli: Resistance, DNA damage, and comparison with hydrogen peroxide stress. J. Bacteriol. 1996;178:6145–6150. doi: 10.1128/jb.178.21.6145-6150.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Martinez A., Kolter R. Protection of DNA during oxidative stress by the nonspecific DNA-binding protein Dps. J. Bacteriol. 1997;179:5188–5194. doi: 10.1128/jb.179.16.5188-5194.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rajagopalan M., Lu C., Woodgate R., Donnell M., Goodman M.F., Echols H. Activity of the purified mutagenesis proteins UmuC, UmuD‘, and RecA in replicative bypass of an abasic DNA lesion by DNA polymerase III. Proc. Natl. Acad. Sci. USA. 1992;89:10777–10781. doi: 10.1073/pnas.89.22.10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Simmons L., Foti J., Cohen S., Walker G. The SOS regulatory network. Ecosal Plus. 2008 doi: 10.1128/ecosalplus.5.4.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bagg A., Kenyon C.J., Walker G.C. Inducibility of a gene product required for UV and chemical mutagenesis in Escherichia coli. Proc. Natl. Acad. Sci. USA. 1981;78:5749–5753. doi: 10.1073/pnas.78.9.5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mesak L.R., Miao V., Davies J. Effects of subinhibitory concentrations of antibiotics on SOS and DNA repair gene expression in Staphylococcus aureus. Antimicrob. Agents Chemother. 2008;52:3394–3397. doi: 10.1128/AAC.01599-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sutton M.D., Walker G.C. umuDC-mediated cold sensitivity is a manifestation of functions of the UmuD2C complex involved in a DNA damage checkpoint control. J. Bacteriol. 2001;183:1215–1224. doi: 10.1128/JB.183.4.1215-1224.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nikolay R., van den Bruck D., Achenbach J., Knud H.N. eLS. Hoboken, NJ, USA: 2015. Ribosomal proteins: Role in ribosomal functions. [DOI] [Google Scholar]
- 82.Friedman D.I., Schauer A.T., Baumann M.R., Baron L.S., Adhya S.L. Evidence that ribosomal protein S10 participates in control of transcription termination. Proc. Natl. Acad. Sci. USA. 1981;78:1115–1118. doi: 10.1073/pnas.78.2.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bylund G.O., Persson B.C., Lundberg L.A., Wikström P.M. A novel ribosome-associated protein is important for efficient translation in Escherichia coli. J. Bacteriol. 1997;179:4567–4574. doi: 10.1128/jb.179.14.4567-4574.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bylund G.O., Wipemo L.C., Lundberg L.C., Wikström P.M. RimM and RbfA are essential for efficient processing of 16S rRNA in Escherichia coli. J. Bacteriol. 1998;180:73–82. doi: 10.1128/jb.180.1.73-82.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guo Q., Goto S., Chen Y., Feng B., Xu Y., Muto A., Himeno H., Deng H., Lei J., Gao N. Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process. Nucleic Acids Res. 2013;41:2609–2620. doi: 10.1093/nar/gks1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Janosi L., Shimizu I., Kaji A. Ribosome recycling factor (ribosome releasing factor) is essential for bacterial growth. Proc. Natl. Acad. Sci. USA. 1994;91:4249–4253. doi: 10.1073/pnas.91.10.4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mayer C., Muras A., Romero M., López M., Tomás M., Otero A. Multiple quorum quenching enzymes are active in the nosocomial pathogen Acinetobacter baumannii ATCC17978. Front. Cell. Infect. Microbiol. 2018;8:310. doi: 10.3389/fcimb.2018.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.