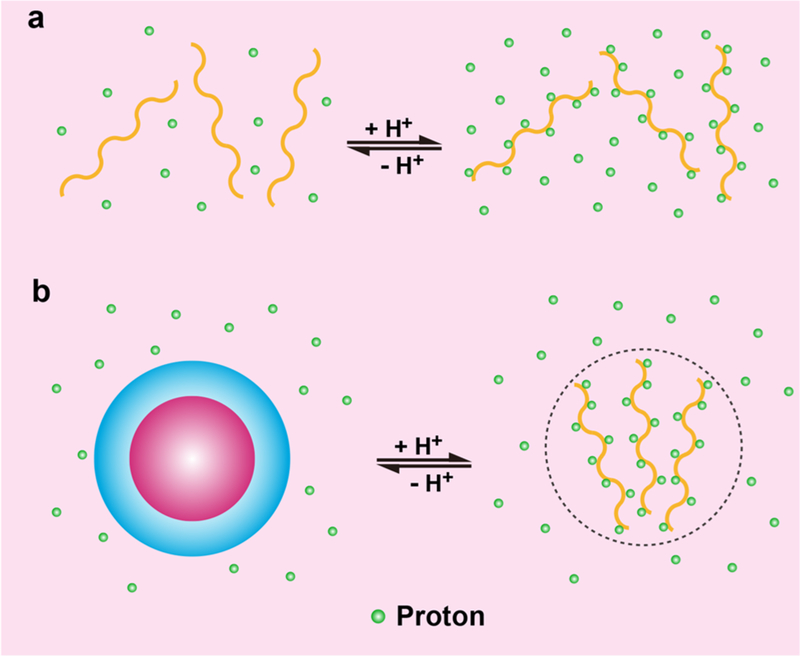
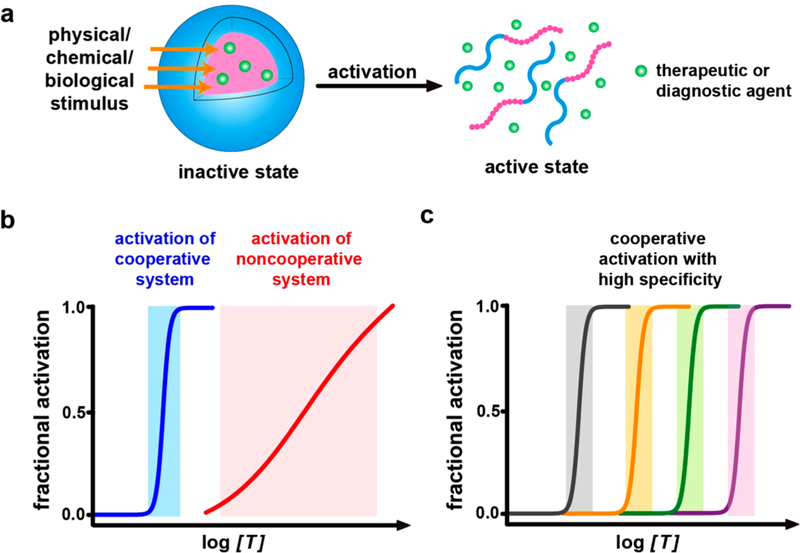
Abstract
Nanomedicine is a discipline that applies nanoscience and nanotechnology principles to the prevention, diagnosis, and treatment of human diseases. Self-assembly of molecular components is becoming a common strategy in the design and syntheses of nanomaterials for biomedical applications. In both natural and synthetic self-assembled nanostructures, molecular cooperativity is emerging as an important hallmark. In many cases, interplay of many types of noncovalent interactions leads to dynamic nanosystems with emergent properties where the whole is bigger than the sum of the parts. In this review, we provide a comprehensive analysis of the cooperativity principles in multiple self-assembled nanostructures. We discuss the molecular origin and quantitative modeling of cooperative behaviors. In selected systems, we describe the examples on how to leverage molecular cooperativity to design nanomedicine with improved diagnostic precision and therapeutic efficacy in medicine.
Graphical Abstract

1. INTRODUCTION
Nanomaterials are rapidly evolving and impact a broad range of applications in photonics, electronics, and medicine.1–6 In particular, they play an increasingly important role in medicine, where numerous nanosystems have been developed for biochemical sensing, molecular imaging, disease diagnosis, and treatment.7–12 Various nanoplatforms have been extensively investigated to address challenges in medicine to overcome deficiencies in conventional small molecular sensors and drugs, resulting in the rapid growth of nanomedicine as a new discipline.13–15
In contrast to “top-down” methods like lithography, a “bottom-up” approach allows the formation of nanoscopic architectures driven by noncovalent self-assembly of molecular components.16–18 Self-assembly, which bridges the structures of individual building blocks and the function of the obtained nanocomplex, is an essential part of nanotechnology.19 The underlying supramolecular chemistry principles were described by Lehn and Whitesides over two decades ago.20,21 Compared to covalent chemistry, noncovalent self-assembly employs weak and polyvalent interactions to achieve a thermodynamically stable nanostructure. This strategy can produce nanoscopic structures (104−1010 Da) that are not easily synthesizable by covalent chemistry. The resulting system often has a faster temporal response to environmental stimuli due to the lower energy barrier (e.g., dissociation of noncovalent complexes requires lower energy than breaking of covalent bonds).22,23
A hallmark of self-assembled systems is molecular cooperativity,21 where the system behaves quite differently as a whole from the sum of parts acting in isolation. Positive cooperativity has been identified in many biological and physiological processes (e.g., oxygen transport by hemoglobin).24 Mechanistic investigations on several established self-assembled nanosystems also suggest that positive cooperativity contributes to enhanced detection sensitivity and specificity in chemical and biological sensing.5 Understanding the supramolecular self-assembly process and associated cooperativity offers a new paradigm for the design and development of nanomaterials in medicine.
In this article, we highlight the recent advances in the investigation of cooperativity principles underlying the design of self-assembled nanomedicine (Figure 1). The current review focuses on the bottom-up chemistry and material science considerations of nanomedicine. Implementation of a top-down method for nanomedicine development is beyond the scope of the current review.
Figure 1.
Supramolecular self-assembly for the development of cooperative nanomedicine.
2. CONFORMATION CHANGE-INDUCED COOPERATIVITY IN NATURAL SELF-ASSEMBLED NANOSTRUCTURES
Cooperativity is frequently employed in biology to modulate molecular recognition through sequential binding events, usually operated by the conformational changes of the macromolecules.25 The binding may display either positive or negative cooperativity. Positive cooperativity is described as synergistic (whole is bigger than the sum of the parts) and negative cooperativity as interfering.26 In this section, we begin the discussion of biological cooperativity using well-established protein/RNA folding and allosteric examples (e.g., hemoglobin-O2 interactions), then move on to more complex multivalent cell surface interactions, and finally present the emerging microphase separations of large protein signaling complexes.
2.1. Self-Organization of Protein and RNA
2.1.1. Protein Folding.
A defining characteristic of biological systems is their capability to organize small molecular components into supramolecular structures with extraordinary precision and fidelity. Protein folding, where a polypeptide chain self-organizes into a perfectly folded three-dimensional structure, is a great example.27 A newly synthesized chain of amino acids can form multiple structured assemblies, such as secondary/tertiary structures and macromolecular complexes. The populations and interconversion of different assemblies are governed by thermodynamic and kinetic stabilities.
Cooperativity is observed in the final folding step of proteins when the side-chains are locked in the native state and water molecules are squeezed out of the hydrophobic protein core.28,29 The Onuchic group used a minimalist model to search for the intermediate states leading to the native structure in parallel with desolvation during protein folding.29 Their results suggested that the majority of the structural formation is accomplished before water is expelled from the hydrophobic core. In another study, the Bustamante group reported that chain topology impacted the cooperative folding of proteins.30 They used an optical tweezer method to selectively unfold specific regions of T4 lysozymes and monitored its perturbation on other regions. Results showed the topological arrangement of the polypeptide chain is critical in determining the folding cooperativity. Data indicated that cooperative interactions among protein domains depend not only on the local interactions between amino acids but also on the degree of complementary shape and topography of the polypeptide chains.
2.1.2. Nucleic Acid Folding.
Ribonucleic acid (RNA) is essential in various biological roles such as coding, regulation, and transcription of genes. Some RNA molecules also catalyze biological reactions and sense or communicate responses to cellular machineries. Similar to polypeptides, a linear ribonucleic acid sequence can also fold into an active conformation with well-defined secondary or tertiary structure. The folded RNA motif often binds to proteins to form specific RNA−protein complexes, where proteins help RNAs reach their native state by stabilizing the assembled structures or by chaperoning the folding process.31
The Woodson group reported that cooperative assembly of RNA helices reduces the misfolding of Tetrahymena group I ribozyme.32 Disruption of the tetraloop structure destabilizes the free energy of RNA folding by 2−3 kcal/mol. The same group also reported that the cooperative tertiary interaction guides RNA folding (Figure 2).33 Interaction between tertiary structures increases the free energy gap between the native state and the intermediate state, thereby facilitating the RNA folding to the native state.34 Daniel and co-workers further quantified tertiary contact interactions in RNA folding using single-molecule Förster resonance energy transfer method.35
Figure 2.
Cooperative folding of wildtype ribozyme leads to lowered free energy of the native-like Ic intermediate and the native state (N). Reproduced with permission from ref 33. Copyright 2012 Elsevier Ltd.
2.2. Allosteric Cooperativity
Allosteric cooperativity is extensively investigated and describes the process where ligand binding at one site regulates the binding or function at another site.36 Goodey suggests that the conformational mobility is a common mechanism that underlies allosteric regulation and catalysis in biological systems.37 Intrinsic flexibility of proteins contributes to multiple conformations that can interconvert at different time scales. The binding of an allosteric effector may lead to the conformational change with modulated binding site geometries and activity. As a result, allostery is used by Nature to regulate the catalytic function of proteins.38–40
2.2.1. Hemoglobin-Oxygen Binding.
All cells in our body use oxygen to make ATP, which provides the energy for many physiological functions. Oxygen molecules are transported from the lung to individual cells. After oxygen is breathed into the lung, it first diffuses to the blood. Its low solubility in water (40 mg/L) makes it impossible to meet the metabolic needs of our tissues and cells. Hemoglobin, a tetrameric protein residing in the red blood cells, serves as a carrier for the transportation of oxygen.
Allosteric oxygen binding is associated with conformational changes of hemoglobin triggered by the oxygen−iron(II) interactions.24 Perutz first reported the structure of hemoglobin in various forms.41 Each hemoglobin molecule consists of two α subunits and two β subunits with similar 3D structures. The binding affinity of hemoglobin to oxygen molecules depends on the heme cofactor, responsible for the red color of blood.42 Each heme group has a central iron atom chelated by protoporphyrin. Each iron within the heme group can serve as a single binding site to an oxygen molecule, and one hemoglobin protein can bind to four oxygen molecules. Under normal physiology, the iron is in the ferrous (Fe2+) oxidation state. The binding of the oxygen molecule to the ferrous ion results in a smaller ferrous ion, allowing it to move into the plane of the porphyrin. Such oxygenation-driven conformation change leads to a transition from deoxy T state to oxy R state of the quaternary structure of hemoglobin, where one pair of αβ subunits rotates relative to the other by 15 degrees.24,43
The structural alteration in hemoglobin significantly changes the oxygen binding affinity to hemoglobin. Initial oxygen binding to hemoglobin facilitates the binding of the second and ensuing oxygen molecules (Figure 3). When three binding sites of hemoglobin are occupied, the binding affinity of the last free site for oxygen is 20-fold higher than that for the first oxygen molecule. The cooperative binding improves the oxygen transport efficiency. The oxygen−hemoglobin saturation curve displays a sigmoid shape, typical for a cooperative binding process.
Figure 3.
Cooperative binding of oxygen to hemoglobin. Initial oxygen binding to hemoglobin makes it easier for the subsequent binding events. The transition of hemoglobin from oxygen-free state (T state) to occupied state (R state) displays strong allosteric cooperativity.
2.2.2. Cooperative Enzyme Catalysis.
Enzymes can dramatically accelerate the rate of biochemical reactions by reduction of activation energy barriers. Many enzymes function as oligomeric complexes of multiple subunits, and each subunit contains an active site for ligand binding and/or catalysis.44 Figure 4 summarizes representative cooperative activation processes of enzymes.37 An “induced fit model” has often been used to describe the enzyme−substrate interactions. The initial interaction is capable of inducing conformational changes of enzymes to increase the strength of subsequent binding events. The conformational changes are described as a key mechanism of enzyme catalysis. It is worth pointing out that the initiation of the conformation change is usually the rate-limiting step instead of the ensuing steps.
Figure 4.
Representative types of cooperative activation of enzymes. (a) The binding of an allosteric effector leads to increased affinity in the adjacent site. (b) The binding of an allosteric effector introduces a new active site. Reproduced with permission from ref 37. Copyright 2008 Nature Publishing Group.
Dihydrofolate reductase (DHFR) is an enzyme responsible for the syntheses of purine, thymine, and several amino acids. It catalyzes the hydride transfer reaction to convert dihydrofolic acid to tetrahydrofolic acid, using NADPH as an electron donor. Conformational changes are observed in the DHFR catalytic cycle. The Hammes-Schiffer group reported that such conformational changes facilitated the hydride transfer by associating and aligning the substrates and cofactors properly with a more favorable electrostatic environment.45–47
2.3. Multivalent Cooperativity and Molecular Recognition
Cell adhesion is the process where cells form contacts to a surface, substrate, or another cell through multivalent interactions. Cell adhesion is critical for many cellular functions such as proliferation, migration, and apoptosis. Cell adhesion is carried out by interaction of transmembrane glycoproteins, which include selectins, integrins, syndecans, and cell surface receptors.48 Cell surface proteins can diffuse and rotate on the membrane surface and sometimes preorganize before binding events. Constraining these proteins to the membrane surface dramatically reduces the entropy and contributes to increased binding affinity compared to the same proteins interacting in solution. Cooperativity through multivalent interactions has been proposed as a physiological mechanism for modulating the strength of cell adhesion.49–51
Epidermal growth factor receptors (EGFRs) have been shown to assume a predimerized (ligand free and inactive) state on the cell surface.52 Binding of an EGF ligand to the predimerized receptor occurs with positive cooperativity.53 Integrin-induced clustering of growth factor receptors facilitates binding of EGF and triggers receptor activation.53,54 Cooperativity due to protein colocalization on the cell surface was also observed in the immunological synapse, an orchestrated interaction between T cells and antigen-presenting cells (Figure 5).55–58 The cooperativity arises from the polyvalent interactions of protein−protein interactions and spatial constraint of the binding partners in the contact region at the cell−cell interface.59
Figure 5.
Key ligand pairs and signaling molecules in an immunological synapse. This process is mediated by a series of cooperative bindings of a complementary array of adhesion and costimulatory molecules. Orchestration of (1) antigen presentation by MHC molecule to the T-cell receptor, (2) CD80/86 costimulation, and (3) cytokine signals is necessary to achieve antigen-specific T cell activation. Reprinted with permission from ref 57. Copyright 2017 Elsevier Ltd.
Another cooperativity example resides in the sequential assembly of weak binding components into a stable multi-molecular complex.60 One such example is the nucleosome-mediated cooperativity between transcription factors.61 Sequential binding of different transcription factor proteins to the promoter region is critical for precise control of gene expression. DNA regions depending on histone binding status can be classified as nucleosomal (N) with low binding affinity or open (O) state with high binding affinity. Binding of transcription factors evicts a nucleosome and frees up new distant binding sites for transcription factors with significantly increased binding affinity. Displacement of nucleosome and generation of new open sites contribute to observed strong cooperative binding of transcription factors.
Such multicomponent cooperativity is also seen in the formation of interferon-β (IFN-β) “enhanceosome” complex,62 a multiprotein complex that binds to the IFN-β enhancer site on the DNA. This multiprotein complex contains more than five proteins, and these proteins assemble cooperatively on a chromatin template with the help of an architectural factor. Preorganization of some proteins generates a new binding site for others with additional stabilization. The absence of any individual component will destabilize the eventual nanocomplex, which suggests strong multivalent cooperativity among individual components.
2.4. Biomolecular Condensation and Phase Transition-Induced Cooperativity
Biological macromolecules are spatially organized within the cells. Membrane-bound subcellular organelles offer the physical separation needed for biochemical reactions in optimized compartments within a cell. Hyman and co-workers first reported subcellular structures consisting of heterogeneous mixtures of proteins and nucleic acids in membrane-less organelles. The formation of these nonmembraned organelles is driven by phase separation similar to polymer condensation.63,64 Living cells contain many such types of nanoscopic droplet-like structures from different compositions of biological molecules (Figure 6).65 Phase separation and condensation of biomacromolecules also display supramolecular cooperativity.66–68
Figure 6.
Schematic illustration of regulated liquid phase separation in cells. Reprinted from ref 65. Copyright 2017 American Association for the Advancement of Science.
One example is the phase separation of proteins with intrinsically disordered regions.69,70 Intrinsically disordered proteins (IDPs) are crucial components of the cellular signaling machinery. They participate in the dynamic assembly of signaling complexes and membrane-less nuclear and cytoplasmic organelles.71 IDPs are found in many biomolecular condensates such as stress granules, germ granules, and nuclear ultrastructures.72,73 Many intrinsically disordered proteins undergo similar phase separation in vitro under solution conditions.74 IDPs display complex allosteric cooperativity that is responsible for their tunable regulatory interactions.36,70
Another example is the formation of micrometer-sized droplets from multivalent protein complexes (e.g., 2 + 3 systems).75 Rosen and co-workers reported the nephrin/Nck/N-WASP system constituting a three-component interaction with the formation of phase separated liquid droplets (Figure 7).76–79 The cooperative association is controlled by the phosphorylation status of the nephrin protein and consequently shifted the phase boundary of the complex.79
Figure 7.
Representative multivalent self-assembly process and microscopic images of liquid droplets (scale bar = 20 μm). Reproduced with permission from ref 79. Copyright 2012 Nature Publishing Group.
3. NONCOVALENT INTERACTIONS: MOLECULAR BASIS OF SUPRAMOLECULAR COOPERATIVITY
Noncovalent self-assembly of molecular modules can form thermodynamically stable nanocomplexes in biological systems. They determine the higher order structures of proteins, DNA, and RNA as well as molecular recognition between biomacromolecules. Through multivalent interactions, molecules or groups of molecules associate into organized structures with increasing complexity. A hallmark of these nanoscale structures and architectures is positive cooperativity, which arises from subtle interplay of two or more noncovalent interactions.80–82
Compared to noncovalent interactions, the length of a covalent bond is short with an average distance less than 0.2 nm between pairing atoms.83–85 The strength of the covalent bond is strong varying from 149 kJ/mol for breaking an I−I bond to 411 kJ/mol for a C−H bond.86 Covalent synthesis alone is incapable of generating well-defined, functional structures with dimensions from tens of nanometers to hundreds of nanometers in size, which covers biological structures from protein complexes to viruses to subcellular organelles.
Noncovalent interactions can occur at longer distances than covalent bonds. Interaction of hydrophobic surfaces or electrostatic interactions between charged species can happen over tens of nanometers. Compared to covalent bonds, noncovalent bonds are 10−100 times weaker (Table 1). Polyvalent interactions involving multiple types of noncovalent bonds through contact of large surface areas compensate for the weaker bond strengths, while allowing the formation, disintegration, and reformation of large scale structures that are not easily attainable by covalent chemistry. Numerous reviews have discussed the nature and strengths of noncovalent interactions.87–90 In this section, we offer a brief summary of several key types of noncovalent interactions that impact molecular cooperativity in biological environments.
Table 1.
Types and Strengths of Typical Noncovalent Interactions
| Type of interaction | Example | Strengthb (kJ mol-1) |
|---|---|---|
| electrostaic interaction |  |
5–100 |
| hydrogen bond |  |
5–150 |
| dipole-dipole interaction |  |
5–50 |
| π-π interaction |  |
0–50 |
| hydrophobic interaction |  |
0–50 |
3.1. Hydrophobic Interaction
The mixture of oil and water tends to segregate into two independent phases: an aqueous phase and an oil phase with well-defined boundaries. The noncovalent interactions that are responsible for aggregation of hydrophobic structures are termed the hydrophobic effect.91,92 Hydrophobic interactions tend to minimize the energy penalty in order to insert a nonpolar molecule into water.93–98 Solvation of nonpolar substances in water can disrupt the hydrogen bonding network of water. A large hydrophobic solute is able to force the water into a rigid cage. The cages restrict the motion and increase the structural organization of water molecules, which facilitates hydrogen bonding interactions and gains in enthalpy. Meanwhile, the randomness (entropy) of the water molecules decreases and causes an overall penalty in free energy. To minimize such penalty, nonpolar molecules tend to come together and aggregate in aqueous solution to exclude water molecules (Figure 8).
Figure 8.
Schematic illustration of the hydrophobic effect, where aggregation of a hydrophobic substance reduces the number of water molecules in the rigid cage surrounding the hydrophobic surface.
Hydrophobic interaction contributes to a multitude of biological structures and processes such as cell membranes, protein folding, formation of subcellular vesicles, and insertion of membrane proteins into the nonpolar lipid environment. Chemists have learned to use the hydrophobic effect as a strategy to generate well-defined structures.99–104 Numerous drug delivery carriers such as polymer- or lipid-based nanoparticles have been developed to improve the pharmacological properties of encapsulated drugs.105–114 Amphiphilic block copolymers have been synthesized to form micellar nanoparticles for the delivery of hydrophobic therapeutics.115 Liposome was first demonstrated in the 1960s and is one of the few nanoparticle-based drug carriers that were translated into the clinic successfully.116–118
3.2. Hydrogen Bonding
A hydrogen bond describes attractive interactions between a hydrogen donor and an acceptor (most often an electron rich atom such as oxygen or nitrogen). Although the interaction is relatively weak, multiplication of hydrogen bonds can drive the self-assembly of individual building blocks to well-defined nano or macrostructures.108,119–123 Formation of a protein α-helix and DNA base pairs are well-known examples of hydrogen bond-mediated complexes. Noncovalent hydrogen bond interactions have also been used to form higher order complexes from synthetic molecules.109 Whitesides and co-workers reported a stable supramolecular complex from cyanuric acid (CA) and melamine (M) based on the hydrogen bond interactions.124–126 The Rotello group reported a polymer-mediated` bricks and mortar’ strategy to order surface functionalized gold particles into aggregated assemblies via intermolecular hydrogen bonding interactions.127–129
3.3. Electrostatic Interaction
Breaking of ionic bonds in vacuum requires higher energy (e.g., 788 kJ/mol for separating Na+Cl−ion pairs) than the breaking of covalent bonds (e.g., 411 kJ/mol for C−H bonds).86 In aqueous environments, solvation of ions by water molecules dramatically reduces the energy cost to separate oppositely charged species. Coulomb interactions between two point-charges are shielded by a factor of relative permittivity (εr, also known as dielectric constant) of the medium. For water, the value of εr, is 78.3 at 25°C, which places the electrostatic interactions at the same energy scale as other noncovalent interactions (Table 1). In biological systems, electrostatic interactions between charged macromolecules are important in nucleic acid condensation,130 ligand−receptor binding,131 and cell−cell interactions.132
Layer-by-layer self-assembly represents a common strategy to construct nanoparticles based on electrostatic interactions.133–136 The film architecture and composition can be precisely controlled at the nanoscale.135 This capability has spawned the development of artificial cells and drug delivery systems.137–146 Electrostatic interaction-mediated condensation between polycations and the phosphate backbone of nucleic acids has been investigated for the development of gene delivery systems over the past several decades.147–152
3.4. π−π Stacking
In chemistry, π−π stacking describes the noncovalent, attractive interactions between neighboring aromatic residues. The stacking effect is critical in multiple biological processes, such as protein folding,28 molecular recognition153 and template-directed synthesis.154 Many groups have reported noncovalent complexes based on π−π interactions.155–161 Stoddart and co-workers have designed several generations of rotaxanes and catenanes functionalized with electron-rich and electron-deficient aromatic units.162 Meijer and co-workers have developed a hierarchical self-assembly strategy to produce molecular nanostructures.163,164 A nucleation−growth strategy is conceptualized that yields a high degree of cooperativity from π−π stacking interactions between adjacent repeating units.163
3.5. Cooperativity: Interplay of Noncovalent Interactions
In macromolecular self-assembly systems, multiple types of noncovalent interactions as described above can simultaneously occur, with compensating energetics leading to highly complex architectures and interacting dynamics.20,21,80,165 Such examples include the association of hydrophobic side chains with H-bonding of polypeptide backbones and salt-bridge formation during protein folding; the interplay of electrostatic and hydrophobic interactions in the chaotropic anion-induced micelle self-assembly; and predominantly H-bonding and hydrophobic interactions in the thermosensitive properties of elastin-like polymers. A hallmark of complex and dynamic systems is the emergence of cooperativity (Figure 9). Below we summarize a few well-established cooperative systems, with the hope of deciphering the underlying mechanism to help predict and program new systems in the future.
Figure 9.
Supramolecular cooperativity arises from interplay of multiple types of noncovalent interactions acting in coordination.
4. PHASE TRANSITION-INDUCED COOPERATIVITY IN SYNTHETIC SELF-ASSEMBLED NANOSTRUCTURES
4.1. Oligonucleotides-Conjugated Gold Nanoparticles
Spherical nucleic acids (SNAs) are three-dimensional nanostructures with densely packed nucleic acids covalently conjugated to the nanoparticle surface (Figure 10).166 These constructs were originally created using gold nanoparticles (AuNPs) by the Mirkin group.167 The unique three-dimensional framework introduces new physical, chemical, and biological properties over one-dimensional linear nucleic acids, which found broad uses in biological sensing, molecular diagnostics, and intracellular gene regulation.5
Figure 10.
Distinctive structural forms of nucleic acids. Reproduced with permission from ref 166. Copyright 2012 American Chemical Society.
The linear nucleic acid chains are typically functionalized with a headgroup to improve the stability of the nanocomplex in aqueous environments. The first SNA conjugates were prepared by covalent attachment of the alkanethiol-terminated, single-stranded oligonucleotides to the surface of gold nanoparticles.167 A dense layer of nucleic acids can be achieved through salt additions, where positively charged counterions are necessary to minimize electrostatic repulsion between adjacent negatively charged DNA strands. In living systems, the nucleic acids usually exist in the hybridized duplex structure. In contrast, the SNAs adopt their morphology to the shape of the inorganic cores.
Besides SNA-NPs, several nucleic acid−based assemblies have been developed for biological sensing or catalysis applications. The Willner group reported improved specificity in the sensing of DNA or selected sequence of aptamers.168 The nucleic acid structures activated the DNAzyme cascades that catalyzed the oxidation of ABTS2− by H2O2. The Kolpashchikov group developed a binary DNA probe for nucleic acid detection.169,170 Two short DNA hairpin cooperativities to the targeted sequence enabled the molecular recognition with high sensitivity and selectivity.
4.1.1. Cooperativity in Aggregation of Oligonucleotides-Conjugated Nanoparticles.
The hybridization of complementary nucleic acid sequences enables the binding interactions between SNA particles with matched DNA sequences. These interactions lead to the aggregation of gold-nanoparticles. The SNA nanoparticles can be released from the aggregates through dehybridization upon heating that disrupts noncovalent base pairing interactions. DNA duplexes and SNAs have characteristic melting temperatures (Tm) when dehybridization occurs.
Jin and co-workers reported a striking sharp melting curve for the dehybridization of SNA-NP aggregates.171 Typically, the melting of the linear DNA duplex happens over a broad temperature range (~20 °C). In contrast, the thermal transition of the SNA−Au NPs from the aggregate state to the individual particle state occurs over a narrower temperature range of 2−8°C. In addition, the phase transition temperature of SNA is higher than that of the corresponding free DNA duplex. The sharp phase transition was observed in both SNA-NPs and a chip-based assay.171,172 Importantly, a single oligonucleotide base-pair mismatch can be differentiated by melting behavior from those with fully complementary sequences.
Mechanistic investigation suggests that high surface density of oligonucleotides on SNA-NPs contributes to polyvalent interparticle connections that are collectively stronger in binding compared to free DNA duplexes in aqueous solution.171 High salt concentration is necessary to achieve the melting cooperativity and sharp phase transition. The Schatz group proposed a “shared ion cloud” model to describe the cooperative melting transition in SNA-NPs, which is supported by experimental evidence (Figure 11).173 The established thermodynamic model also enabled the quantitative assessment of the contributions from the neighboring-duplex effect. Nguyen and co-workers reported as little as two DNA duplexes were necessary to elicit cooperative melting behavior.174 The experimental data fit well into a coarse-grain dynamic stimulation-based model. The oligonucleotides assumed an orientation that enabled the sharing of counterions for the cooperative response.
Figure 11.
Schematic of the “shared ion cloud” model in the cooperative melting behavior of SNA-NPs. Reprinted with permission from ref 173. Copyright 2007 American Chemical Society.
4.1.2. Tunable Phase Transition Temperature.
In contrast to natural biomacromolecules, the physicochemical properties of synthetic nanomaterials can be easily modified by tailoring their structure and composition. For example, controlling the surface nucleic acid density of AuNPs can affect the hybridization efficiency and cooperative melting response (Figure 12).171 The thermal transition temperature was found proportional to the surface DNA density while keeping nanoparticle and target concentration unaltered.
Figure 12.
Cooperative melting response depending on surface DNA density of gold nanoparticles. B and C refer to data from solution and glass surface, respectively. Reprinted with permission from ref 171. Copyright 2003 American Chemical Society.
One unique feature of nanomaterials is the large surface-to-volume ratio due to the small nanoparticle size. The nanoparticle size is expected to affect the phase transition behaviors of SNAs. The melting transition temperature decreased from 50 to 47 °C when the size of gold particles increased from 13 to 50 nm, respectively. Interestingly, larger SNAs generally exhibited sharper melting transitions compared to smaller ones.
The melting curves of natural single strand DNA exhibit a salt concentration dependence.175 The transition temperature of SNAs increased from 41 to 61.5 °C when the NaCl concentration went from 0.05 to 1.0 M. In addition, the increase in salt concentration also led to the formation of larger aggregates.176 This can be attributed to a charge shielding effect by the salt, which can reduce electrostatic repulsions between the oligonucleotide-modified gold nanoparticles and permit further hybridization between nanoparticles.
Aggregation of SNAs in DNA sensing can result in a distinct color change from red to purple by visual inspection. The electromagnetic coupling between nanoparticles that affects the surface plasmon resonance is distance dependent, which also impacts the van der Waals and electrostatic interactions between particles. The melting analysis showed that longer interparticle distance resulted in higher transition temperature of SNAs. Further mechanistic investigation suggested that the electrostatic interaction was expected to be the dominant factor in regulating distance-dependent melting behaviors.
Besides DNAs, ribonucleic acids (RNAs) have also shown a promising therapeutic effect.177 RNAs were also introduced onto AuNPs surface to generate the RNA SNAs. In a recent study,178 Barnaby et al. reported a systematic investigation on the structure−function relationships in RNA SNAs, which would help elucidate the interactions of RNAs with a specific type of serum nucleases. A combined experimental and theoretical study investigated the impact of several key parameters (i.e., RNA sequence, density, linker, etc.) of RNA-SNAs for rational design of SNAs in biomedical applications.
4.2. Poly(acrylamide)-Based Thermoresponsive Hydrogels
Stimuli-responsive polymers often display a sharp change in physical or chemical properties upon a small perturbation in environmental conditions, which is used for the design of “smart” nanomaterials for the controlled release of therapeutics. 179–181 Thermoresponsiveness is usually measured as a change of light transmittance or solubility of polymeric materials.182,183 Thermoresponsive nanomaterials are among the most investigated systems in drug delivery and cancer therapy. The sharp thermal response was exploited for the triggered-release of drugs in response to change in the surrounding temperature. For biomedical applications, thermosensitive nanocarriers are expected to retain their therapeutic load at normal physiological temperature (i.e., 37 °C); upon local heating by an external source, the nanocarriers can rapidly release the drug in the desired location. Thermoresponsive systems include liposomes or polymeric micelles that undergo phase transitions at specific temperatures.184,185
Poly(N-isopropylacrylamide) (PNIPAM) was first synthesized in the 1950s, and it is widely adopted for use as a thermosensitive polymeric drug carrier.186,187 It is typically prepared by polymerization of commercially available N-isopropylacrylamide monomer. When heated above 32 °C in water, PNIPAM undergoes conformation changes from a hydrated gel to an aggregated solid across the lower critical solution temperature (LCST). The gel will lose about 90% of its original volume. This LCST temperature has close proximity to physiological temperatures that can trigger a reversible phase transition without causing damage to surrounding tissues. Considerable efforts have been dedicated to the design of PNIPAM-based thermosensitive nanomaterials as delivery vehicles for controlled drug release.187–190
PNIPAM polymer stays in the gel state below the LCST, where water molecules form a hydrated cage around the hydrophobic moieties along the polymer chain. When temperatures are raised above the phase transition temperature, hydrogen bonds between the polymers and water molecules become more favorable in comparison to polymer−polymer or water−water interactions. Such destabilization results in the desolvation of the hydrophobic groups of polymer chains. The increase in entropy of the released water molecules and hydrophobic interactions drives the collapse of polymer chains. The LCST can be controlled by adjusting the hydrophobicity−hydrophilicity ratio of the polymer chains. An increase in hydrophilic groups increases the LCST, and an increase in hydrophobic groups has the opposite effect.186 Studies show that the concentration or molecular weight of the polymer has little effect on the phase transition temperature of PNIPAM.186,191 It is notable that some thermoresponsive systems do display molecular weight or size dependence in LCST transitions.192,193
4.2.1. Cooperativity in Gelation of Thermoresponsive Polymers.
The first detailed study of thermosensitive PNIPAM in aqueous solution was reported in 1969 by Heskins and Guillet.194 They observed the change in turbidity of a solution upon heating at 32 °C. Since then, continuous efforts have been made to investigate the phase transition properties of PNIPAM and its derivatives. Extensive mechanistic investigation suggests that the driving force for this phase transition is the balance of hydrophilic and hydrophobic moieties.195–197 PNIPAM chains carry two types of bound water molecules with one around the hydrophobic isopropyl moiety and the other associated with the amide group.198 Change in the hydration status of the hydrophobic side chains results in association of the PNIPAM chains.
Tanaka and co-workers first reported cooperative dehydration of the PNIPAM chains in the temperature-induced phase separation.199 They concluded that dehydration of the neighboring water molecules around the polymer chains was responsible for the sharp phase transition with little dependence on molecular weight or chain length. The degree of hydration versus temperature by theoretical calculation correlated well with the experimental data reported by Fujishige et al.200–202
The Winnik group also investigated the phase transition behavior of cyclic PNIPAM in aqueous solution.203 They found that the melting curves of cyclic PNIPAM solutions occurred over a much wider temperature range over the linear counterpart, indicating the importance of side chain geometry on cooperative response. A recent study by Muller-Buschbaum reported how partial dehydration affected the volume changes in the phase separations of PNIPAM hydrogel.204
4.2.2. Tunable Sol−Gel Transition Temperature.
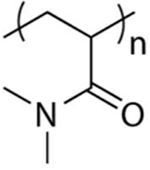
Fujishige et al. reported that neither molecular weight (5 × 104 to 8.4 × 106 Da) nor concentration (0.01 to 1 wt %) greatly impact the thermal transition temperature of PNIPAM.202 In contrast, many studies show that the LCST is tunable by shifting the hydrophilic/hydrophobic balance.186 Different types of N-alkyl-substituted poly(meth) acrylamides have been synthesized, and their LCST values were investigated.185 Poly(N-n-propylacrylamide) (PNNPAM)205,206 had a LCST of 10 °C compared to 32 °C of PNIPAM, suggesting that hydrophobic geometry affects the transition temperature (Table 2). LogP is the octanol−water partition coefficient of a molecule, which is commonly used as a quantitative measure of molecular hydrophobicity.207 A higher LogP indicates stronger hydrophobicity. The LCST of poly(N-cyclopropylacrylamide) (PNCPAM), in which iso-propyl of PINPAM is replaced by the less hydrophobic cyclo-propyl group, occurs around 53 °C. Poly(N,N-diethylacrylamide) (PDEAM) displayed a phase transition temperature similar to that of PNIPAM at 33 °C.208 The LCST of poly(N,N-ethylmethyl acrylamide) (PNEMAM) shifted to a much higher temperature of 70 °C. It is worth noting that the poly(N,N-dimethyl acrylamide) did not show phase transition behavior below the boiling point of water.209 The hydrophobicity of PNIPAM can also be controlled by incorporating an additional alkyl group in the backbone instead of side chains. Poly(N-isopropyl methacrylamide) has a LCST at 45 °C, which indicates that restricting the rotation freedom of the polymer backbone can increase the transition temperature and is opposite to that in the side chain.
Table 2.
Structures, Hydrophobicity of Repeating Units and Phase Transition Temperatures of Several Representative N-Alkyl- Substituted Poly(acrylamide)s
| polymer | structure | LogP | LCST (°C) |
|---|---|---|---|
| Poly(N-n-propylacrylamide) (PNNPAM) |
 |
1.74 | 10 |
| Poly(N-isopropylacrylamide) (PNIPAM) |
 |
1.57 | 32 |
| Poly(N-cyclopropylacrylamide) (PNCPAM) |
 |
1.21 | 53 |
| Poly(N,N-diethylacrylamide) (PNDEAM) |
 |
1.83 | 33 |
| Poly(N,N-ethylmethylacrylamide) (PNEMAM) |
 |
1.49 | 70 |
| Poly(N,N-dimethylacrylamide) (PNDMAM) |
 |
1.15 | - |
Salt can greatly impact the solubility of proteins.210 The structure of water in the vicinity of different solute ions has been studied for many decades.211 Hofmeister initially observed that different salts have contrasting effects on protein solubilities.212 The ions are divided into kosmotropes or chaotropes depending on their ability to make or break water network structures, respectively. Kosmotropes decrease protein solubility in water whereas chaotropes increase the solubility.213 Salt also critically affects the physicochemical properties of synthetic polymers. The Cremer group reported the salt effect on the thermoresponsive behavior of PNIPAM. They found that increasing the concentration of NaCl led to a decrease of LCST. They then expanded the ion effect on PNIPAM to the entire Hofmeister series.214 Specific anions’ ability to lower the LCST of PNIPAM followed the Hofmeister trend in protein solubility (Figure 13).215 Mechanistic investigation indicates that chaotropic species lowered the LCST via change of surface-tension, which triggers hydrophobic collapse. For kosmotropic anions, the surface-tension and polarization of hydrated water molecules are both important in regulating the transition temperature of PINPAM. In a follow-up study, Zhang et al. found that the effect of Hofmeister anions on the LCST of PINPAM was molecular weight-dependent.216
Figure 13.
LCST values of PNIPAM over a broad range of anion concentrations in the Hofmeister ion series. Reprinted with permission from ref 215. Copyright 2005 American Chemical Society.
The promise of PNIPAM in biomedical applications has inspired further development of other thermoresponsive polymers. The Gibson group synthesized a series of poly(acrylamide)-based polymers with cyclic alkyl groups as N-substituents.193 Poly(N-vinylpiperidone) (PVPip), with a six-member-ring side chain, showed a LCST between 65 and 90°C.217 The phase transition temperature of poly(N-vinylcaprolactam) (PVCap) shifted to a lower 40 °C with a seven-member-ring side chain.218,219 Although PVPip and PVCap demonstrated similar hydrophobicity-dependent phase transition behavior, they also showed significant molecular weight-dependent LCST shift, which is different from the case of PNIPAM.
The Zhang group synthesized a series of N-ester-substituted poly(acrylamide)s and systematically investigated their LCST behavior.220 In one of their polymer series, poly(N-acryloylglycine methyl ester) (PNAGME), the melting temperature displayed strong molecular weight dependence. The LCST decreased from 57 to 42 °C when the polymerization degree increased from 20 to 180. They also observed that increasing the concentration of NaCl shifted the LCST to a lower temperature.
New thermoresponsive nanomaterials have also been developed by coating these polymers onto the surface of different solid nanoparticles. Edwards et al. demonstrated that poly(ethylene glycol) methacrylate (PEGMA) coated gold nanoparticles facilitates their transport across an oil/water interface above the LCST.221 Boyer et al. prepared a series of thermoresponsive block copolymers with tunable phase transition temperature by altering monomer compositions.222 The Tenhu group reported the development of thermoresponsive nanoparticles by grafting PNIPAM brushes on the surface of gold nanoparticles.223 Increasing the molecular weight of PNIPAM resulted in the decrease of LCST. They also found that a decrease in gold nanoparticle size resulted in a small increase of phase transition temperature. Klok and co-workers observed similar size-dependent thermotransitions in their system.192
4.3. Thermoresponsive Elastin Like Polypeptides (ELPs)
Elastin is an elastic protein that allows tissues to resume their original shape after stretching or contracting.224 Elastin-like polypeptides (ELPs) are synthetic polymers inspired from mammalian elastin. Early pioneering work by Urry and co-workers identified a pentapeptide repeat, VPGXG, where X refers to any natural amino acid except proline for the development of ELPs.225 These polymers display thermal transitions with an LCST similar to that of PNIPAM. The thermoresponse and biocompatibility make ELPs ideal materials for different biomedical applications (Figure 14).226
Figure 14.
Different methods for drug delivery using elastin-like polypeptides (ELPs) in vivo. Reprinted with permission from ref 226. Copyright 2010 Elsevier Ltd.
4.3.1. Cooperativity in Supramolecular Self-Assembly of ELPs.
Elastin-like polypeptides can be genetically engineered with precise control of peptide sequence and chain length. Mechanistic investigation of ELPs elucidates the key factors that impact their phase transition temperature. The ELPs have also been used to investigate the physical behavior of intrinsically disordered proteins (IDPs).227 The Hinderberger group pioneered the study of the temperature-triggered reversible phase transition behavior of ELPs (Figure 15).228 They showed that the hydration layers can vary depending on the composition of hydrophobic side chains and amide backbones. A strongly coupled hydration state can lead to a cooperative dehydration of both segments. Chilkoti and co-workers established an empirical model to predict the transition temperature of ELPs from amino acid composition, peptide chain length, and concentration in phosphate buffered saline.229 They also performed molecular simulation of the LCST behavior of ELPs.230 Increase of temperature can lead to a gradual conformational change of ELPs, arising from the formation of more ordered secondary structures. Higher temperature also exposed the hydrophobic side chains of valine to water, contributing to the collapse of polypeptide chains.
Figure 15.
Coupled hydration layers (left to right) lead to cooperative dehydration of water molecules surrounding the ELP chains. Reprinted with permission from ref 228. Copyright 2013 American Chemical Society.
4.3.2. Tunable Phase Transition Temperature of ELPs.
It is generally accepted that the folding and phase transition behavior of proteins is encoded in its amino acid sequence. Mutation of key residues in a protein can result in dramatic alteration of property and function.231,232 Elastin-like polymers contain a specific amino acid sequence that is critical for their thermosensitivity. The temperature-induced phase transition of ELPs is affected by pH, ionic strength, and polymer molecular weight. It is also expected that alteration of key residues may also significantly impact their phase transition behavior.
The Rodriguez-Cabello group reported a series of model elastin-like polymers to investigate how amino acid sequence affects solution−gel transition temperature.233 They synthesized three types of ELPs with the same molecular weight and composition but different sequences. The LCST values of the three ELP structural isomers (E100A40, E50A40E50, and E50A20E50A20) were 43.8, 47.1, and 60.1 °C, respectively. These data showed that the arrangement of polymer blocks greatly impacted the LCST of ELPs. In another systematic comparison,234 they found that the transition temperature decreased with the increase in polymer chain length.
Quiroz et al. recently reported a number of elastin-like polymers with variable lower or upper critical solution temperatures.235 They screened a library of ELPs offering heuristic evidence to identify proteins that may display thermal responsiveness and established the foundation for encoding their phase transition behavior at the sequence level. Mutation of key residues or insertion of additional amino acids in each repeating unit drastically shifted the phase transition temperature (left panel, Figure 16).235 Further examination of different sequence parameters showed the number of repeating units or molecular weight played an important role in controlling the LCST (right panel, Figure 16).235 The phase transition temperature also showed linear correlation with polymer concentration in one specific example. A change of environmental conditions such as pH or salt concentration also resulted in the shift of phase transition temperature.
Figure 16.
Effect of amino acid composition (left panel) and number of repeating units (right panel) on the LCST values of elastin-like polymers. Reproduced from ref 235. Copyright 2015 Nature Publishing Group.
Cho et al. investigated the effect of ion species and concentration on the reversible gelation temperature of ELPs.236 Temperature-triggered hydrophobic self-assembly of ELPs generally followed the Hofmeister trend. Mechanistic investigation suggested that kosmotropes increased the LCST by polarizing interfacial water molecules in the hydration shell of ELPs. Chaotropic anions lowered the gelation−solution transition temperature via reduced surface tension. These observations were in agreement with previous conclusions in the poly(N-isopropylacrylamide) (PNIPAM) system.
The Holland group reported that the molecular architecture also impacted the thermal phase transition of ELPs.237 They designed a star-shaped elastin-like polypeptide and compared its LCST behavior with the linear analogs. Melting curve analysis showed that the transition temperature of both the linear and star ELPs decreased with the increase in concentration. Their results also showed that the molecular architecture and morphology also contributed to the folding of ELPs chains.
4.4. Ultra-pH Sensitive (UPS) Nanoparticles
Dysregulated pH is considered to be a distinct characteristic of tumors as described by Barber and co-workers.238 Cancer cells have increased intracellular pH (pHi) and decreased extracellular pH (pHe) compared to normal tissues.238 The increased pHi protects the cancer cells from apoptotic cell death, facilitates cell proliferation, and is necessary for cell migration. The decreased extracellular pH,239–242 or tumor acidosis, activates proteases for matrix remodeling and cancer metastasis. Highly glycolytic tumors are shown to have an acidic extracellular pH by Gillies and others.243 In addition to metabolic abnormality, impaired lymphatic drainage may further contribute to the accumulation of acidic metabolites inside the tumors. Extensive investigations suggest that regardless of the bioenergetic types of tumors, tumor acidosis is a persistent characteristic of solid cancers.244–248
Targeting tumor extracellular acidity offers a viable strategy for cancer staging and drug delivery.183,184,249–252 These pH-responsive nanocarriers can be classified into two categories: polymers with ionizable moieties and polymers with cleavable covalent linkages.251,253 Ionizable polymers employ a non-covalent strategy to achieve pH sensitivity, where dissociation of carboxylic acids or protonation of amine groups occurs at different pH values.
Carboxylic acid-based hydrogels and their pH-triggered drug release behavior were first reported in the 1950s.254 These hydrogels undergo pH-driven swelling upon ionization in aqueous medium, and the apparent pKa of carboxylic acids in these hydrogels varies with the monomer structure, copolymer composition, and surrounding environment. Philippova et al. reported the impact of hydrophobic groups on the pH response in the poly(acrylic acid) hydrogels.255 Hydrophobic n-alkyl acrylates were blended in the poly(acrylic acid) network. Data showed that hydrophobic modification increased the apparent pKa of poly(acrylic acid)-based pH-responsive hydrogels.
The Bae group developed polymeric sulfonamides for cytosolic delivery of nucleic acids.256 These polymers had a reversible phase transition at pH 7.4. The research group was able to lower the pKa to the endosomal pH range by copolymerization with N,N-dimethylacrylamide (DMAAm) monomers. The pKa of the obtained copolymers shifted from 6.9 to 6.1 as the feeding ratio of hydrophilic DMAAm increased from 50% to 90%.257 In another study, Kang et al. reported that the pKa and buffering effect of oligomeric sulfonamides (OSAs) are influenced by the hydrophobicity of the sulfonamide monomer.258 Park et al. also found that the pKa of polymers containing sulfonamide groups showed polymer concentration dependence where higher concentrations led to an increased transition pH.259
Gao and co-workers reported a library of ultra-pH sensitive (UPS) nanoparticles for tumor-targeted imaging and drug delivery applications.260–266 The UPS nanoparticles are composed of block copolymers of PEO-b-PR, where PEO is poly(ethylene oxide) and PR is a hydrophobic block with multiple ionizable tertiary amines (Figure 17).262 At pH below the apparent pKa, the copolymers with protonated ammonium groups stay in solution as unimers. Upon pH increase, the PR segments become neutral and associate into core−shell micelles. When fluorescent dyes are conjugated onto the hydrophobic PR segment, the UPS systems display a sharp pH transition with over a 100-fold increase in fluorescence intensity within 0.25 pH unit, which allows precise imaging of acidification of tumors or endocytic organelles.260,265
Figure 17.
Schematic design of ultra-pH sensitive (UPS) nanoparticles. Different hydrophobic side chains were used to fine-tune the pH transition of the resulting copolymers. Reprinted with permission from ref 262. Copyright 2011 Wiley-VCH.
4.4.1. Cooperativity in Reversible Protonation of UPS Block Copolymers.
The sharp pH transition, absent in commonly used small molecular and polymeric pH sensors, inspired the mechanistic investigation on the molecular basis of the cooperative response. The authors first compared the pH responsive behavior of pH-sensitive small molecules or polymers. pH titration results showed that NH4Cl (pKa = 10.5) and chloroquine (pKa = 8.3), commonly used lysosomotropic agents to manipulate the pH of endocytic organelles, had typical broad pH responses in the range of pH 7 to 11. pH titrations of several extensively investigated pH-sensitive polymers including polyethylenimine (PEI),267 poly(L-Lysine) (PLL),113 chitosan,268 and poly(L-Histidine) (PLH)114,269 showed different degrees of broad pH response compared to small molecular bases. In contrast, pH titration of three UPS polymers (PDPA, PDBA, and PD5A with propyl, butyl, and pentyl side chains, respectively) showed initial pH decrease after HCl addition followed by a remarkable plateau, indicating a strong buffer effect and ultra-pH response. A plot of pH transition sharpness as a function of the octanol−water partition coefficient (LogP) of the repeating unit from different polymers suggested that the hydrophobic micellization contributed to the sharp pH transition of UPS nanoparticles.
Dialysis and 1H NMR experiments were used to investigate the pH-triggered self-assembly process. Collective evidence indicated that the micelle phase transition is responsible for the bistable protonation states along the titration coordinate. This all-or-nothing divergent proton distribution between the unimer and micelle states is a hallmark of positive cooperativity (Figure 18).270
Figure 18.
Schematic illustration of distinctive deprotonation pathways by two structurally related copolymers, PEO-b-PDMA and PEO-b-PDPA. Increase in hydrophobicity of the PDPA copolymers led to an “all or nothing” cooperative deprotonation phenotype but not in PDMA copolymers. Reprinted with permission from ref 270. Copyright 2016 Nature Publishing Group.
4.4.2. Tunable pKa and pH Transition Sharpness.
Ma et al. reported a copolymerization strategy to fine-tune the pKa of UPS block copolymers (Figure 19).261 A library of UPS nanoprobes was established to cover a broad physiological pH range from 4 to 7.4, where polymers with more hydrophobic repeating units displayed lower pKa. Readily tunable pKa may offer exciting opportunities to target endosomes for the cytosolic delivery of diagnostic and therapeutic agents before reaching lysosomes.271–276
Figure 19.
UPS library consisting of sharp pH threshold nanosensors spanning a wide physiological pH range from 4 to 7.4. Reprinted with permission from ref 261. Copyright 2014 American Chemical Society.
In a recent study, Li et al. reported a quantitative correlation between the hydrophobicity of repeating units of UPS block copolymers and their pKa values.277 They also expanded the composition of UPS nanoprobes to polymers with aromatic side chains. In the same study, they showed that both anionic species and salt concentration affect the apparent pKa of UPS copolymers. Higher salt concentration led to the increase of apparent pKa (Figure 20).277 When sodium chloride concentration increased from almost zero to 0.15 M, the pKa values of a representative polymer increased by 1.1 pH unit. Moreover, chaotropic anions (ClO4−) had the most impact whereas kosmotropic anions (SO42−) had the least effect on the apparent pKa.
Figure 20.
Chaotropic anions and concentration impacted the pH transition of UPS micelles. Using PEO-b-nPDPA as an example, the effects of NaCl (a), Na2SO4 (b), and NaClO4 (c) at different salt concentrations on pH titration are presented. (d) The apparent pKa is ion species and ionic strength dependent. Reprinted with permission from ref 277. Copyright 2016 Royal Society of Chemistry.
4.5. pH-(Low) Insertion Peptides (pHLIPs)
Many proteins in living systems display a unique pH-dependent membrane insertion property.278–282 pHLIP peptides consist of about 36 amino acids. In acidic environments, pHLIPs can insert across the cell membrane with increased accumulation in acidic tissues (Figure 21).283 In the acidic tumor environment, the low pH-driven insertion characteristics were exploited for the development of tumor-targeted imaging agents and drug delivery systems.284
Figure 21.
Schematic illustration of pHLIP’s interaction with lipid bilayers at neutral and acidic pH. State I refers to the free peptide conformation at high pH. State II describes the adsorption of the unstructured peptide on the membrane surface. At state III, acidification allows the protonation of Asp residues with increased hydrophobicity and results in the formation of a transmembrane α-helix. Reprinted with permission from ref 283. Copyright 2010 National Academy of Sciences.
In 2007, Engelman and co-workers reported a fluorescently labeled pHLIP for tumor imaging.285 The imaging probe identified solid tumors with good signal-to-noise ratio (3−5 times higher in tumors than adjacent normal tissues) and was stable over 4 days. A pHLIP-based delivery system was also reported for the transport of phalloidin, a cell-impermeable toxin, into the cytoplasm of cancer cells.286 The pHLIP peptide inserted its C terminus across the cell membrane at lower pH, which allowed for triggered release of toxin through the cleavage of a disulfide bond. Proliferation of multiple cancer cell types was inhibited. Nitin and co-workers designed an Alexa-647 labeled pHLIP for the imaging of variations in extracellular pH in head and neck squamous cell carcinoma. The fluorescence intensity is 4−8-fold higher in cancer tissues over healthy tissues.
The molecular mechanism and principles of pKa control were extensively studied. Weerakkody et al. reported the structure−property correlations in altering the transition pKa of pHLIPs by screening a library of 16 rationally designed peptides.287 It was found that the pKa values of pHLIP variants were sequence-dependent. For example, the variants with Asp residue generally displayed a lower pKa while the analogs containing Glu residues usually had a higher pKa. In another study, they showed that the pKa of pHLIPs shifted to a lower pH as the hydrophobic thickness of the membrane increased.288 Both the composition of the peptides and the physical properties of the lipid bilayers impacted the pH-triggered membrane insertion process and ensuing tumor targeting, organ distribution, and blood clearance outcomes.
It was postulated that pHLIPs can exist in three distinctive states: unstructured and soluble state in aqueous solution, unstructured and binding state to the outer leaflet of the cell membrane, and α-helical state after membrane insertion in response to acidic pH signal.289 The two aspartic acid residues were critical for the observed pH-induced membrane insertion behavior.290 These residues are negatively charged at neutral or basic pH, which prevents insertion into the phospholipid bilayers due to electrostatic repulsion. At low pH, the protonated carboxylate groups enable the reduction in polarity leading to the conformation change and membrane insertion. Previous investigations have shown that the formation of an α-helix is a cooperative process.291 Engelman and co-workers reported that increasing the number of ionizable residues can promote the pH-dependent cooperative membrane insertion process (Figure 22).292,293 Protonation of the initial Asp allowed peptides to insert into the cell membrane partially. Consequently, exposure in the membrane environment drives further protonation of the adjacent Asp, leading to a positive feedback and complete membrane insertion. The cooperative insertion process was further validated by the mutation of conformation-restrained proline residue by glycine.288 Data show that the proline at position 20, midway through the transmembrane region, is crucial for pH-induced insertion activity. Replacement of proline-20 by glycine resulted in variable insertion over a broader pH range, suggesting reduced cooperativity compared to wide type pHLIPs.
Figure 22.
Molecular description of membrane insertion by pHLIPs. The insertion and folding of peptide chains appear without intermediate states, indicating a positive cooperative process. Reprinted with permission from ref 293. Copy right 2012 Elsevier Ltd.
5. MOLECULAR MECHANISM OF SUPRAMOLECULAR COOPERATIVITY
Cooperativity is universally found in the nanoscale systems where identical or near-identical components self-assemble into multicomponent structures through a multitude of noncovalent interactions. Cooperativity can be described as the synergistic process in which individual components interact with each other to accelerate or facilitate the formation of a multicomponent complex, which is usually the most thermodynamically favorable state. Cooperativity can be manifested in either intramolecular (e.g., protein folding) or intermolecular (e.g., micellization) processes.
Mechanistically, nanoscale cooperativity can be broadly categorized into two types: allostery and preorganization.25,60 In allosteric cooperativity, binding between A and B induces conformational change of A, which results in increased binding affinity for component C (Figure 23a). Compared to the free state A, the formation of AB complex opens a new binding site on A with enhanced binding affinity for C. In the preorganization model (Figure 23b), the initial complexation of A and B decreases the number of nonproductive configurations and thereby reduces the entropic cost of bringing C into the bound state from its free state. Preorganization promoted cooperativity can further be augmented by the additional interactions (Figure 23c). For example, initial formation of complex AB not only facilitates the binding between A and C, additional interactions between B and C render gains in free energy of binding that further drive the formation of complex ABC. It should be noted that these two types of cooperativity are not mutually exclusive and can occur concurrently in the same nanosystem.
Figure 23.
Different types of molecular cooperativity. (a) Allosteric cooperativity: binding of component B changes the conformation of component A and opens a new binding site for component C. (b) Preorganization of complex AB renders intramolecular binding between A and C to facilitate the formation of complex ABC. (c) Preorganization of complex AB provides additional stabilization between B and C to drive the formation of complex ABC.
5.1. Origin of Cooperativity
5.1.1. Cooperative Folding of Proteins.
Scientists have long studied the impact of amino acid sequence on a protein’s native structure and the stochastic nature of the folding process. A “folding funnel” hypothesis is proposed in an energy landscape model (Figure 24).294,295 The transition states, the energy barrier that denatured conformations must overcome in order to fold into the native state, are represented by the saddle points on the surface of the above landscape. Superimposed on the surface are intermediate states that represent different stages of the progressive folding process. The folding funnel theory assumes the existence of many non-native local minima of free energy, where partially folded proteins are trapped.
Figure 24.
Folding funnel model to describe protein folding. Intermediate structures collapse into the native state mainly driven by hydrophobic interactions. Reprinted with permission from ref 295. Copyright 2003 Nature Publishing Group.
The folding funnel theory hypothesizes that hydrophobic collapse plays an essential role in the folding of proteins. Hydrophobic interaction between amino acids’ side chains stabilizes the intermediate states and in the folded domains. The free energy of folded structures can be further lowered by the relocation of charged side chains on the surface of proteins or the formation of salt bridges to balance the charges in the core. The interplay of many types of noncovalent interactions contributes to the observed positive cooperativity, a hallmark of protein folding.296
The dynamic coupling between the interactions which stabilizes a packed natural state determines the cooperativity of the folding landscape. In other words, cooperativity implies a favored protein folding pathway to the native state. Strong coupling between the stabilization forces will lead to a cooperative two-state transition in protein folding as observed in the self-assembly of small globular proteins.297,298
5.1.2. Cooperative Activation of Ion Channels.
In voltage-gated channels (e.g., Na+, K+ and Ca2+), separate protein domains are responsible for ion conduction and voltage sensing. Isacoff showed that the two subunits of the human hydrogen voltage-gated channel 1 (Hv1) affect one another during gating with positive cooperativity.299 Opening of either subunit favors the opening of the other one dramatically. This model correlated with the experimental observation that the two pores of Hv1 tended to stay either both open or closed (all or nothing) (Figure 25).299
Figure 25.
Cooperative gating of the Hv1 channel where both subunit channels stay either open or closed. Reprinted with permission from ref 299. Copyright 2010 Nature Publishing Group.
5.1.3. Cooperative Dehydration of Thermoresponsive Polymers.
Tanaka and co-workers proposed a “pearl-necklace” model to describe the cooperative hydration process in the solvation of PNIPAM polymers (Figure 26).199,300 When a water molecule initiates a hydrogen bond with an amide group in the backbone, it results in displacement of the isopropyl group to enable the second water molecule to form another hydrogen bond. Consecutive hydration of water molecules behaves like a pearl-necklace type along the polymer chain. When temperature increases, each sequence can be dehydrated cooperatively, leading to the collective collapse of the polymer chain and observed sharp melting curve. In a separate study, Wu and co-workers have discovered the presence of a molten globule state along the thermal transition coordinate of a single PNIPAM chain, which resembles that in protein folding.301 The molten globule state is characterized by a dense core and a molten shell, which suggests a heterogeneous assembly process during phase transition.
Figure 26.
Pearl-necklace model to describe conformation change of polymer chains by cooperative hydration. Reprinted with permission from ref 199. Copyright 2005 American Chemical Society.
5.1.4. Hydrophobic Micellization-Driven Cooperative Protonation.
A polymeric allosteric model was proposed by Li et al. to describe the pH-triggered phase transition of ultra-pH sensitive block copolymers.270 The polymer chains with multiple ionizable tertiary amines were considered as a multisite receptor and the protons as monovalent ligands. Experimental data showed that the copolymers in the micelle state were mostly neutral, whereas the majority of the tertiary amines were protonated in the unimer state in solution. In the protonation of UPS polymers, the micelles initially created a hydrophobic core to prevent the protons from ionizing the tertiary amines (Figure 27). Protons cannot break through the hydrophobic barrier until a critical pH threshold (or a critical proton concentration) is reached. Once protonation started, the ionized ammonium groups are hypothesized to expose the hydrophobic chains to the aqueous environment, which facilitates the protonation of the remaining tertiary amines. The reversed deprotonation process also displayed strong pH cooperativity following the “loss of protons—increase of hydrophobicity—polymer condensation” cycle. The hydrophobic micellization-driven cooperativity leads to a Hill coefficient of 51 and shifts the pKa from alkaline pH to acidic pH (e.g., 9 to 5).
Figure 27.
(a) No positive cooperativity in the protonation of hydrophilic polymers. (b) Hydrophobic phase separation (micellization) drives cooperative protonation or deprotonation of ionizable groups at a threshold proton concentration. Free proton concentration remains the same during pH titration.
5.2. Quantitative Analysis of Cooperativity
Hunter and Anderson described different kinds of cooperative behaviors in multicomponent complexes.302 Among these, allosteric cooperativity is best understood, where binding a ligand to a multisite receptor will affect the binding affinity of the next ligand as a result of conformational changes (Figure 28a, b). Allosteric enzymes change conformation upon the binding of the first substrate, which affects the binding of molecules at other sites. Cooperativity is also commonly found in bivalent binding processes such as cell adhesion and chelation (Figure 28c, d). A bivalent ligand may bind to a bivalent receptor at either site. After the first binding, subsequent binding becomes an intramolecular event with reduced entropic cost.60 Polyvalent ligand may pertain to multiple distinct binding elements, which can be identical or dissimilar.303 For multivalent interactions, valence over [3 + 2] can lead to physical cross-links and phase condensation as shown previously in the nephrin/Nck/N-WASP system (Figure 7).76–78,193 The third type of cooperativity is found (Figure 28 e) in the oligomerization or polymerization of amyloid peptides,304 actin strands,305 and other polymer systems.163 Similar to the allosteric scenario, initial organization of repeating units such as nucleation makes the subsequent binding more favorable and triggers cooperative self-assembly. Recently, Cheng and co-workers reported a cooperative synthetic polymer system.306 This polymer can catalyze its own chain elongation. Initial formation of α-helices accelerates the polymerization rate due to cooperative interactions of macrodipoles between neighboring α-helices.
Figure 28.
Cooperative associations in ligands (pink) and receptors with multiple binding sites (blue). Allosteric cooperativity: initial binding of ligand induces conformational change of receptors and increases the binding affinity of the same ligand (a) or a secondary ligand (b). Multivalence cooperativity: anchoring of first ligand brings the unoccupied binding site closer to free ligand and increases binding affinity of ensuing the same (c) or different ligand (d). (e) Cooperative oligomerization or polymerization triggered by initial self-organization of several repeating units.
To determine whether a protein−ligand binding process exhibits any cooperativity, binding parameters of the ligand to the protein are first quantified at varying concentrations of the ligand. In a representative case, θA is defined as the molar fraction of protein binding sites that are occupied by the ligand of interest. For a process with no cooperativity, it takes about 100-fold change in ligand concentration to increase the site-occupancy from 10% to 90%. If a system displays positive cooperativity, it takes smaller changes in concentration for the same increase in occupation percentage. For allosteric systems such as in protein−ligand interactions, a Hill plot is often used to quantify cooperativity.307–309 In practice, the Hill plot is obtained by plotting log(θ/(1 − θ)) versus logarithmic concentration of ligands (eq 1).
| (1) |
The Hill coefficient nH, corresponding to the slope of this plot measured at 50% saturation, is used to quantify the cooperativity strength experimentally. A Hill coefficient of one suggests no cooperativity in the binding process. A Hill coefficient of greater or less than one indicates positive or negativecooperativity, respectively.
The Hill coefficient is widely used in allosteric binding studies.310 Many pharmacokinetic−pharmacodynamic models reported the use of the Hill equation to quantify the nonlinear drug dose−response relationships. Other quantification methods have also been developed in different self-assembly systems where a Hill plot is not applicable or not very accurate. Yifrach showed that a modified Boltzmann equation can estimate the degree of cooperativity in voltage-dependent ion channels.311 This approach allowed the quantification of the steady-state cooperativity of ion channels and enzymes.312 Camara-Campos et al. reported the use of double mutant cycles to investigate chelate cooperativity in multiple hydrogen-bonded complexes.313 This method allowed for the delineation of the free energy contribution associated with the intramolecular non-covalent interactions. Ercolani proposed a method to quantitatively evaluate the cooperativity in helicate and porphyrin ladders.314 He defined a new parameter, statistical stability constant, to evaluate the cooperativity.
A binding isotherm from a receptor−ligand titration study (e.g., fluorescence anisotropy315 and isothermal titration calorimetry316) is another common methodology to analyze cooperativity. Saykally reported the use of far-infrared vibration−rotation tunneling (VRT) spectroscopy to quantify hydrogen bond cooperativity.317 The Mariuzza group employed a surface plasmon resonance method to quantify the strength of binding cooperativity in a three-component complex.318 Their method for the quantification of cooperativity strength may probably be applicable in modeling more complicated protein assemblies.
The Hill coefficient of oxygen binding to hemoglobin is in the range 1.7−3.2.319 Berg group’s investigation in E. coli indicated that assemblies of bacterial chemoreceptors work cooperatively with a Hill coefficient ranging from 1.4 to 3.8.320 Their results were consistent with several previous reports that long-range cooperative interactions can serve as a general mechanism for signal amplification.321–323 The maturation of Xenopus oocytes with hormone progesterone operates in an all-or-nothing manner. The cooperative response is generated by the mitogen-activated protein kinase (MAPK) cascade.324 Analysis of individual oocytes suggested that the response of MAPK to progesterone was equivalent to that of a cooperative enzyme with a Hill coefficient of 35.
Li et al. quantified the pH cooperativity of UPS polymers (Figure 29).270 The Hill coefficients of ultra-pH sensitive block copolymers were around 51, compared to 1 of commonly used small molecular bases. They showed that the cooperativity can be further strengthened by increasing the hydrophobic chain length. The cooperativities in similar anion-induced self-assembly systems were also investigated.270,325 The Hill coefficient ranged from 5 to 30 depending on the anion species. The self-assembly process was driven by a novel micellization process induced by the chaotropic anions.325,326
Figure 29.
(a) Binding isotherm and (b) Hill plot of small molecular base DPA (dipropylaminoethanol), polymeric bases of PEI (polyethylenimine), PEO-b-PDMA (poly(ethylene oxide)-b-poly(2-(dipropylamino) ethyl methacrylate)), and PEO-b-PDPA (poly(ethylene oxide)-b-poly(2-(dipropylamino) ethyl methacrylate)). DPA and PEO-b-PDMA showed no cooperativity. PEI displayed negative pH cooperativity, and PEO-b- PDPA showed strong positive cooperativity. (c) Binding isotherm and (d) Hill plot of PEO-b-PDPA copolymers with different numbers of repeating units in the hydrophobic segment. Increase of hydrophobic chain length led to stronger positive cooperativity and sharper pH response. Reprinted with permission from ref 270. Copyright 2016 Nature Publishing Group.
6. SUPRAMOLECULAR COOPERATIVITY IN ADDRESSING THE CHALLENGES IN MEDICINE
Nanoscale cooperativity can be exploited in the design of activatable nanomedicine with increased biological precision and specificity. These nanostructures can be designed to stay in the inactive state at normal physiological conditions but become activated at the site of disease to achieve diagnostic and therapeutic functions (Figure 30a). The release of imaging signals or payloads can be triggered by physical (ultrasound, heat, light), chemical (pH, redox potential), or biological (enzyme, DNA) stimuli. Compared to noncooperative systems, cooperative nanostructures can respond to stimuli more rapidly and efficiently (Figure 30b). Small changes in the amount/ concentration of target signals ([T]) are able to elicit large signal changes in diagnostic or therapeutic outcomes. Another benefit of the cooperative system is the ability to fine-tune the threshold of stimuli response, which can be used to target selective oxygen pressure, pH, or temaperature (Figure 30c) to enlarge the therapeutic window. The precise spatiotemporal control of the activation of functionalized nanoparticles will be further discussed in this section with selected cooperative nanomedicine systems.
Figure 30.
Cooperative nanomedicine with improved precision and specificity. (a) Schematic illustration of stimuli-responsive nanomedicine. (b) Compared to noncooperative systems, small changes in target signals can lead to amplified response in cooperative systems. T refers to the target species (e.g., proton) or signals (e.g., heat). (c) Tunable transition of cooperative systems enables precise control of signal activation at a predetermined threshold to enlarge the therapeutic window.
6.1. Targeted Drug Delivery
A fundamental challenge in medicine is the efficient delivery of therapeutic cargos into the targeted cells. In cancer, solid tumors usually show anatomical and pathophysiological properties different from those of normal tissues. For example, tumors have leaky vasculature and impaired lymphatic systems that can lead to accumulation of nanoparticulates, a phenomenon termed as the enhanced permeability and retention (EPR) effect.327,328
Although drug-loaded nanoparticles with optimized diameter and surface chemistry have been designed to take advantage of the EPR effect to accumulate in tumor sites, cancer cells develop drug resistance over time.329,330 To overcome drug resistance, active targeting strategies have been developed.331 Ligands are functionalized onto the surface of nanoparticles by various conjugation chemistries. These ligand-encoded nanoparticles can specifically bind to the receptors on the surfaces of targeted tumor cells after extravasation. The bound nanocarriers can be internalized via endocytosis, enabling the intracellular release of drugs. Such active targeting methods can overcome the efflux-pump mediated drug resistance with increased intracellular drug concentration.332
The internalization of drug-loaded nanoparticles is dependent on receptor-mediated endocytosis. Both the initial binding events and ensuing cell uptake can be enhanced by multivalent binding (Figure 31). The multivalent binding process may display positive cooperativity, where binding of one ligand on the nanoparticles will facilitate further binding events for the neighboring ligands.333,334 For example, in multiple folate receptor binding, the nanoparticle-cell association is enhanced by more than 2,500-fold.335 Multivalent antiviral and anti-inflammation therapeutics also showed significantly improved potencies compared to corresponding monovalent counterparts.336 The overall binding affinity of ligand-modified nanoparticles to targeted cells generally increases with an increasing ligand density. However, too high a ligand density may result in the decrease of binding affinity due to unfavorable steric crowding, where the ligand may have limited conformational freedom to effectively bind to the target molecules.
Figure 31.
Cooperative multivalent interactions increase the binding avidity of ligand-conjugated nanoparticles to the cell surface.
Besides naturally existing cell surface receptors (e.g., folate receptor),337 rapid advances in bioorthogonal chemistry have inspired the metabolic labeling of cancer cells for the targeted delivery of nanomedicine.338 Metabolic labeling artificially introduces chemical receptors onto the cell surfaces and enables a “two-step” targeting strategy. This strategy is especially useful for delivering therapeutics without nascent biomarkers. Recently, Kim and co-workers developed an active targeting strategy through Cu2+ free bioorthogonal chemistry.339 Unnatural sialic acids with azide groups were introduced on the cell surface of tumors via metabolic glycoengineering, which effectively enhanced the accumulation of nanoparticles by multivalent interactions.
6.2. Biological Sensing and Molecular Imaging
Biosensors usually rely on biological recognition of disease-specific biomarkers where the signals are further processed by a transducer.340 There are different categories of biosensor designs including small molecules, peptides, aptamers, antibodies, proteins, and different types of nanoparticles.341 One significant drawback of conventional biosensing technology is that most biosensors are analog sensors, where noise can be propagated without signal amplification, leading to degraded signal-to-noise ratios. Use of nanomaterial biosensors has the potential to overcome the deficiencies of commonly used biosensors.342–344
One such example is spherical nucleic acid (SNAs)-based nanoflares.5 These nanosensors have been used in the molecular sensing of a range of analytes including nucleic acids,345 proteins,346 small molecules,347 and metal ions.348 The combination of an inorganic core and polyvalent oligonucleotide shell offers advantages over unimolecular counterparts. A target analyte such as nucleic acids can be recognized by two different designs of SNAs. Subsequent binding of target sequences will trigger the aggregation of the SNAs nanoparticles, which is accompanied by a visible color transition. The aggregates exhibit a narrow melting transition compared to duplex DNAs.171 A single base pair mismatch, insertion, or deletion will result in a shift of transition temperature that leads to the detection of target nucleic acids with high specificity. Moreover, the high extinction coefficient of gold nanoparticles allows for sensitive detection of target molecules at lower concentrations than with conventional dyes.
Recently, the Mirkin team applied the nanoflare technology to detect circulating tumor cells (CTCs) in blood (Figure 32).349 Detection of CTCs offers early opportunities for metastatic risk assessment. The nanoflares were designed to target mRNAs (mRNAs) that code for protein biomarkers for breast cancer cells. They were able to detect the genetic markers of CTCs in blood with less than one percent false positive results. This technique also successfully detected CTCs in a murine model of metastatic breast cancer. This nano cooperativity-enabled approach offers a new paradigm for tumor diagnosis and personalized treatment.
Figure 32.
Nanoflares for mRNA detection in circulating tumor cells. Reprinted with permission form ref 349. Copyright 2007 American Chemical Society.
6.3. Digitization of pH Signals by Threshold Sensors
Cancer is a heterogeneous disease that makes it challenging for universal, cancer-specific detection.350 Most common strategies in the development of tumor-targeted imaging agents focus on cell surface proteins such as the folate receptor,351 chlorotoxin,352 epidermal growth factor receptor,353 and some tumor-associated antigens.354 Although various preclinical studies have shown some success, the ability to detect a broad range of cancer types is often not possible because of genetic or phenotypic variability among different tumors.355,356 Tumor acidosis, which is well recognized as a hallmark of cancer regardless of genotypes and phenotypes, can be used as a universal target for cancer-specific imaging and drug delivery.357–360 However, commonly used small molecular pH sensors display broad pH response (2 pH units) and are not capable to differentiate subtle pH variation between tumor and surrounding normal tissues.
To overcome these deficiencies, Zhao et al. reported a transistor-like pH threshold sensor for the tumor-specific fluorescent imaging of different types of cancers (Figure 33).361 The fluorescent nanosensor amplified the tumor acidosis signals with discretized output while remaining silent in the normal tissue. The binary on/off digitization of tumor pH and surrounding normal tissue pH allows for clear tumor margin depiction with high sensitivity and specificity. The real-time image-guided surgery of primary tumors and occult nodules (<1 mm3) in mice bearing head and neck or breast tumors significantly improved the long-term survival over white light controls.
Figure 33.
Tumor margin delineation by a pH threshold sensor. Representative frozen section of HN5 tumor with surrounding tissues showed excellent matching of fluorescence signal with H&E tumor histology; scale bar = 2 mm. Dashed line indicates the tumor margin. Reprinted with permission from ref 361. Copyright 2017 Nature Publishing Group.
In a separate study, Wang et al. reported a hybrid ultra-pH-sensitive (HyUPS) nanosensor design to digitize the luminal pH of endocytic organelles in live cells (Figure 34).362 The HyUPS nanosensor consisted of a mixture of three different copolymers with each exhibiting a sharp (<0.25 pH) response at different pH thresholds (pHt). HyUPS allowed for the quantification of acidification kinetics of endocytic organelles at a single-organelle resolution. Compared to a conventional analog pH sensor (e.g., Lysosensor), the HyUPS design does not require a calibration curve before pH measurement and is less sensitive to photobleaching. A digital barcode (e.g., 000, 001, etc.) can be easily assigned based on the binary on/off signal output in each fluorescence channel for each organelle, whereas Lysosensor only measures the average endocytic pH from all the acidic organelles (including Golgi) within each cell. This simple analog to digital signal conversion allowed for fast quantification of organelle pH and permitted identification of mutant Kras as an oncologic driver for the accelerated acidification of endocytic organelles in cancer cells.
Figure 34.
Multispectral hybrid ultra-pH sensitive (HyUPS) nanosensor to digitize organelle pH after receptor-mediated endocytosis. Reprinted with permission from ref 362. Copyright 2017 Wiley-VCH.
6.4. On Demand Drug Release
Nanostructures that can be externally triggered to release drugs on demand have the potential to improve therapeutic efficacy with reduced toxicity.363 One major challenge in the design of externally triggered drug release systems is the low sensitivity and poor response. Long time exposure to external energy sources may result in serious tissue damage and side effects. Nanosystems with cooperative response to external stimuli can deliver therapeutics more effectively and precisely.
Elastin-like polypeptides (ELPs) have been developed as thermoresponsive micelles and liposomes for drug delivery.364 Liu et al. reported a local cancer radiotherapy consisting of ELPs conjugated to a therapeutic radionuclide.365 This injectable depot successfully delayed the tumor progression and showed controlled advanced-stage cancers. Chilkoti and co-workers also reported “heat activatable” drug-loaded ELP nanoparticles to target solid tumors.366 The Kostarelos group reported a lipid-peptide nanoplatform for sustained release of therapeutics triggered by hyperthermia.367 The lipid-based nanoparticles showed extraordinary stability in blood circulation at physiological temperature. In vivo data by 14C-doxorubicin quantitation illustrated significantly increased tumor accumulation at 24 h after intravenous administration with hyperthermia. ELP-functionalized plasmonic nanoparticles,368 liposomes,369 and dendrimers370 were also developed. Besides cancer, ELP drug depot has also been investigated in applications for joint degeneration,371,372 neuro-inflammation,373 and diabetes.374
The backbone of poly(β-amino esters) (PAEs) can be degraded through hydrolysis under physiological conditions, which improves their safety profiles in biomedical applications. PAEs were employed in the design of gene/drug delivery systems.375–377 However, the thermoresponsive behavior of these polymers was less known and their phase transition behavior was largely unexplored. Recently, Wang and co-workers reported the temperature-induced phase transition behavior of hyperbranched PAEs (HPAEs) (Figure 35).378 By varying the length of the ethylene glycol spacers and the molecular weight of polymers, the LCST of HPAEs was successfully fine-tuned in aqueous environment. The sharp melting curves suggest strong cooperativity in the reversible dehydration of these polymers, which makes them another model system for the investigation of molecular cooperativity in aqueous environment.
Figure 35.
Thermoresponsive hyperbranched poly(β-amino ester) with tunable phase transition temperature. Reprinted with permission from ref 378. Copyright 2017 American Chemical Society.
7. SUMMARY AND FUTURE PERSPECTIVE
Nanomedicine is an interdisciplinary field that integrates physics, chemistry, materials science, biology, and pathophysiology principles toward prevention, diagnosis, and treatment of diseases.379 Over the past decade, the field has advanced rapidly as a result of the push by the medical needs to improve patient care and the pull of novel science at the nanoscale that is absent in the traditional single molecular arena.380–388 Multiple therapeutic nanomedicines have progressed into the clinical stages, with some successes (e.g., Doxil) as well as unsuccessful attempts.389,390 The lack of success has spurred heated debates on the potential promise of nanomedicine and allowed for a healthy introspection and reality check by many in the field.391
In contrast to small molecule-based diagnostics and therapeutics, nanomedicine represents a new paradigm that employs a system-based approach toward problem solving. A clear advantage is the ability to incorporate multiple functions and tools within a small size confinement to address multiple challenges simultaneously. One such example is the multilayered liposomal carriers with sequential delivery of two therapeutic drugs and surface functionalization by folate and Cy5.5 dye (Figure 36).392 The elaborate engineering design synergizes cell surface receptor targeting by the folate ligand for cell uptake, combined with therapeutic targeting by erlotinib and doxorubicin to exploit different vulnerable molecular pathways inside cancer cells. It also allows for the tracking of the nanoparticles in cancer cells by fluorescence imaging. Such system-based combination of multiple therapeutic and imaging modalities is beneficial over single molecular drug therapy where adaptive resistance arises over time in cancer patients.393–403 Despite the therapeutic promise, nanomedicine also introduces exponentially increased complexity inherent to the multiple interacting components within the system. Although synergistic outcome is desirable, chaos, where subtle changes in the initial conditions can result in widely divergent outcomes (aka the butterfly effect),404,405 can also occur and introduce uncertainties in data irreproducibility, increase in the cost of production, and challenges in quality control. How to achieve robustness in action from complex nanomedicine systems is a paramount but perplexing challenge.
Figure 36.
Design of a folate-targeted, Cy5.5-encoded multilayered liposomal system for sequential delivery of hydrophobic erlotinib and hydrophilic doxorubicin drugs. Reprinted with permission from ref 392. Copyright 2014 American Association for the Advancement of Science.
Biology may provide the answer to many of these challenges. Biological systems are complex, dynamic systems that have held the imprint of evolution for over four billion years of history on Earth. Although chaos does occur (e.g., formation of cancer, memory loss), life has a plethora of high fidelity processes such as maintenance of DNA identity and hereditary traits. In aqueous solution, the unique property of water molecules creates an exceptional environment where noncovalent interactions (electrostatic, hydrogen bonding, hydrophobic interactions, etc.) can interact and compensate each other at relatively low energy levels (5−100 kJ/mol, Table 1) comparable to the random thermal energy (~5 kJ/mol for each degree of freedom) in the environment. This creates a dynamic system with high entropy and low enthalpy exchange processes (e.g., in contrast to covalent bond formation or breaking). Supramolecular self-assembly, whether in protein folding, biomolecular condensation, or gene transcription, introduces cooperativity and nonlinear dynamics to amplify signals over background noise to achieve the intended biological specificity. Cooperativity has the potential to overcome chaos to achieve robustness in function.
Cooperativity principles, which have not yet drawn significant attention and are not necessarily the mainstream concept in nanomedicine design, appear to be critical in several biomedical applications such as tumor imaging (UPS nanoparticles), drug delivery (ELPs), and DNA sensing (SNAs). These systems manifest a “controlled chaos” phenotype where precipitive phase transitions occur that amplify the signal response to an external stimulus. The underlying cooperative process resembles that of protein folding (hydrophobic collapse) or biomolecular condensation in nature. In the engineering systems, new phenomena are further discovered as in the case of UPS nanoparticles, where an all-or-nothing proton distribution phenotype was uncovered between the unimer and micelle states of the UPS polymers, respectively. This bistable state solution along the pH titration coordinate is responsible for the threshold fluorescence response to subtle pH changes (e.g., < 0.3 pH unit) in the surrounding environment, which allowed a binary delineation of tumor margins (Figure 33) and improved accuracy in image-guided surgery. For the ELP and SNA systems, molecular cooperativity allowed efficient on-demand drug release and ultrasensitive detection of DNA strands, respectively. In these examples, molecular cooperativity offers a useful strategy to transform random chaotic events into directed, synchronized outcomes for signal amplification and enlargement of therapeutic windows, which are essential to achieve precision in function for medical applications.
This review article aims to highlight the cooperativity principles and their potential values in self-assembled nanomedicine design. Although Nature might have learned how to engage cooperativity in managing complex biological functions, we are only touching the tip of the iceberg to implement it in medicine. Moving forward, several questions may warrant future considerations. First, the fundamental nature of cooperativity requires mechanistic clarity. Nanoscale may represent the smallest size scale at which cooperativity manifests itself. Despite decades of biophysical research, we have not fully understood the molecular mechanism of protein folding or even the hydrophobic effect. The energy costs of water molecules solvating extended areas of hydrophobic surfaces, the contribution of hydrogen bonding and/or electrostatic interactions in the changing dielectric environment, and the effects of different ionic species remain to be elucidated. Related to the first question is how to model cooperative behaviors. Cooperativity is an emergent property arising from the system as a whole bigger than the sum of the parts. By definition, this may call for system-based models over traditional molecular based simulations. Chaos theory, originally from the study of weather and climate change, applies probability theory to study dynamic and complex systems. Similar treatment may be warranted to investigate the cooperativity behavior of nanomedicine systems. Lastly, is rational prediction of cooperative response feasible in nanomedicine? Answers to the first two questions may provide insights for the design of cooperative systems in response to any biological stimuli of interest. In the short term, attempts can be made toward the understanding of the structure−property relationships of existing cooperative systems. Such examples include the effect of hydrophobicity of the polymer matrix on the LCST of PNIPAM185 and apparent pKa of UPS polymers.277 The latter example draws a different molecular strategy (i.e., controlling hydrophobicity of polymer segment) from the use of electron withdrawing/donating groups to modulate pKa in small ionizable molecules.406 Additional mechanistic studies are necessary to build a systematic set of independent evidence to help elucidate nonlinear dynamics of interacting components in coordination. Although many critical questions remain, we anticipate the field of nanomedicine is at an exciting juncture to make a major impact in the implementation of cooperativity principles in medicine.
ACKNOWLEDGMENTS
We appreciate the funding support from the National Institutes of Health (R01CA192221, R01CA211930, U01CA218422 to JG, and pilot grant from SCCC P30CA142543 to GH) and the Cancer Prevention Research Institute of Texas (RP140140 to JG, and cyclotron pilot award RP110771 to GH).
Biographies
Yang Li obtained his B.S. (2011) in Polymer Chemistry from the University of Science and Technology of China. He earned his Ph.D. (2017) in Biomedical Science from the University of Texas Southwestern Medical Center under the supervision of Dr. Jinming Gao. Currently, he works as a postdoctoral fellow in the research group of Dr. Daniel Kohane at Boston Children’s Hospital and Harvard Medical School. His research interest focuses on developing stimuli-responsive nanomaterials for biological sensing and drug delivery. He is also interested in mechanistic investigation of supramolecular self-assembly.
Yiguang Wang received his Ph.D. in Pharmaceutics from Peking University in 2008 under the supervision of Dr. Qiang Zhang. Then he worked as a postdoctoral fellow in Institute of Biophysics, Chinese Academy of Sciences. In 2010, he moved to University of Texas Southwestern Medical Center at Dallas and worked as a postdoctoral fellow under the supervision of Dr. Jinming Gao. In 2015, he joined Peking University School of Pharmaceutical Sciences as a Professor. His research focuses on targeted drug delivery, stimuli-responsive nanoparticles for cancer imaging and therapy, as well as intracellular trafficking of nanodrug delivery systems.
Gang Huang is an assistant professor in the Simmons Comprehensive Cancer Center and Department of Pharmacology at UT Southwestern Medical Center. He received his Ph.D. in Chemistry from the University of North Texas in 2004. His postgraduate training was performed at University of Utah under Dr. Russell Stewart and at UT Southwestern Medical Center under Dr. Jinming Gao. His current research interest focuses on the design and development of novel nanomaterials and nanoarchitectures for cancer diagnosis and targeted therapeutic applications.
Jinming Gao received his B.S. in chemistry from Peking University in 1991. He obtained his Ph.D. under Dr. George M. Whitesides in physical organic chemistry at Harvard University in 1996. His postdoctoral training was under Dr. Robert S. Langer in biomedical engineering at MIT. In 1998, he started his lab in the Department of Biomedical Engineering at Case Western Reserve University. In 2005, he moved to UT Southwestern Medical Center at Dallas, Texas. His current research interests focus on the understanding of nanoscale cooperativity and its implementation in niche biomedical applications. The lab-invented pH transistor nanosensor is entering first-in-human trials for image-guided cancer surgery in 2018.
Footnotes
The authors declare no competing financial interest.
REFERENCES
- (1).Liu X; Yan C-H; Capobianco JA Photon Upconversion Nanomaterials. Chem. Soc. Rev 2015, 44, 1299–1301. [DOI] [PubMed] [Google Scholar]
- (2).Pumera M Graphene-based Nanomaterials and Their Electrochemistry. Chem. Soc. Rev 2010, 39, 4146–4157. [DOI] [PubMed] [Google Scholar]
- (3).Wang J Nanomaterial-based Electrochemical Biosensors. Analyst 2005, 130, 421–426. [DOI] [PubMed] [Google Scholar]
- (4).Chen X; Li C; Grätzel M; Kostecki R; Mao SS Nanomaterials for Renewable Energy Production and Storage. Chem. Soc. Rev 2012, 41, 7909–7937. [DOI] [PubMed] [Google Scholar]
- (5).Rosi NL; Mirkin CA Nanostructures in Biodiagnostics. Chem. Rev 2005, 105, 1547–1562. [DOI] [PubMed] [Google Scholar]
- (6).Sun T; Zhang YS; Pang B; Hyun DC; Yang M; Xia Y Engineered Nanoparticles for Drug Delivery in Cancer Therapy. Angew. Chem., Int. Ed 2014, 53, 12320–12364. [DOI] [PubMed] [Google Scholar]
- (7).Langer R; Tirrell DA Designing Materials for Biology and Medicine. Nature 2004, 428, 487–492. [DOI] [PubMed] [Google Scholar]
- (8).Gatenby RA A Change of Strategy in the War on Cancer. Nature 2009, 459, 508–509. [DOI] [PubMed] [Google Scholar]
- (9).Brannon-Peppas L; Blanchette JO Nanoparticle and Targeted Systems for Cancer Therapy. Adv. Drug Delivery Rev 2004, 56, 1649–1659. [DOI] [PubMed] [Google Scholar]
- (10).Chauhan VP; Jain RK Strategies for Advancing Cancer Nanomedicine. Nat. Mater 2013, 12, 958–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Webber MJ; Appel EA; Meijer E; Langer R Supramolecular Biomaterials. Nat. Mater 2016, 15, 13–26. [DOI] [PubMed] [Google Scholar]
- (12).Chabner BA; Roberts TG Chemotherapy and the War on Cancer. Nat. Rev. Cancer 2005, 5, 65–72. [DOI] [PubMed] [Google Scholar]
- (13).Jain RK; Stylianopoulos T Delivering Nanomedicine to Solid Tumors. Nat. Rev. Clin. Oncol 2010, 7, 653–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Misra R; Acharya S; Sahoo SK Cancer Nanotechnology: Application of Nanotechnology in Cancer Therapy. Drug Discovery Today 2010, 15, 842–850. [DOI] [PubMed] [Google Scholar]
- (15).Farokhzad OC; Langer R Impact of Nanotechnology on Drug Delivery. ACS Nano 2009, 3, 16–20. [DOI] [PubMed] [Google Scholar]
- (16).Stupp SI; Palmer LC Supramolecular Chemistry and Self-Assembly in Organic Materials Design. Chem. Mater 2014, 26, 507–518. [Google Scholar]
- (17).Lehn J-M Supramolecular Chemistry. Science 1993, 260, 1762–1764. [DOI] [PubMed] [Google Scholar]
- (18).Mattia E; Otto S Supramolecular Systems Chemistry. Nat. Nanotechnol 2015, 10, 111–119. [DOI] [PubMed] [Google Scholar]
- (19).Lehn JM Perspectives in Chemistry—Steps Towards Complex Matter. Angew. Chem., Int. Ed 2013, 52, 2836–2850. [DOI] [PubMed] [Google Scholar]
- (20).Lehn JM Supramolecular Chemistry—Scope and Perspectives Molecules, Supermolecules, and Molecular Devices (Nobel Lecture). Angew. Chem., Int. Ed. Engl 1988, 27, 89–112. [Google Scholar]
- (21).Whitesides G; Mathias J; Seto C Molecular Self-Assembly and Nanochemistry- A Chemical Strategy for the Synthesis of Nanostructures. Science 1991, 254, 1312–1319. [DOI] [PubMed] [Google Scholar]
- (22).Stuart MAC; Huck WT; Genzer J; Müller M; Ober C; Stamm M; Sukhorukov GB; Szleifer I; Tsukruk VV; Urban M; et al. Emerging Applications of Stimuli-Responsive Polymer Materials. Nat. Mater 2010, 9, 101–113. [DOI] [PubMed] [Google Scholar]
- (23).de las Heras Alarcón C; Pennadam S; Alexander C Stimuli Responsive Polymers for Biomedical Applications. Chem. Soc. Rev 2005, 34, 276–285. [DOI] [PubMed] [Google Scholar]
- (24).Eaton WA; Henry ER; Hofrichter J; Mozzarelli A Is Cooperative Oxygen Binding by Hemoglobin Really Understood? Nat. Struct. Biol 1999, 6, 351–358. [DOI] [PubMed] [Google Scholar]
- (25).Williamson JR Cooperativity in Macromolecular Assembly. Nat. Chem. Biol 2008, 4, 458–465. [DOI] [PubMed] [Google Scholar]
- (26).Purich DL Enzyme Kinetics: Catalysis and Control: A Reference of Theory and Best-Practice Methods; Elsevier, 2010; p 703. [Google Scholar]
- (27).Dill KA; MacCallum JL The Protein-Folding Problem, 50 years on. Science 2012, 338, 1042–1046. [DOI] [PubMed] [Google Scholar]
- (28).Levy Y; Onuchic JN Water Mediation in Protein Folding and Molecular Recognition. Annu. Rev. Biophys. Biomol. Struct 2006, 35, 389–415. [DOI] [PubMed] [Google Scholar]
- (29).Cheung MS; García AE; Onuchic JN Protein Folding Mediated by Solvation: Water Expulsion and Formation of the Hydrophobic Core Occur After the Structural Collapse. Proc. Natl. Acad. Sci. U. S. A 2002, 99, 685–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Shank EA; Cecconi C; Dill JW; Marqusee S; Bustamante C The Folding Cooperativity of a Protein is Controlled by Its Chain Topology. Nature 2010, 465, 637–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Herschlag D RNA Chaperones and the RNA Folding Problem. J. Biol. Chem 1995, 270, 20871–20874. [DOI] [PubMed] [Google Scholar]
- (32).Chauhan S; Woodson SA Tertiary Interactions Determine the Accuracy of RNA Folding. J. Am. Chem. Soc 2008, 130, 1296–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Behrouzi R; Roh JH; Kilburn D; Briber RM; Woodson SA Cooperative Tertiary Interaction Network Guides RNA Folding. Cell 2012, 149, 348–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Gonzalez RL Navigating the RNA Folding Landscape. Nat. Chem. Biol 2008, 4, 451–452. [DOI] [PubMed] [Google Scholar]
- (35).Sattin BD; Zhao W; Travers K; Chu S; Herschlag D Direct Measurement of Tertiary Contact Cooperativity in RNA Folding. J. Am. Chem. Soc 2008, 130, 6085–6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Motlagh HN; Wrabl JO; Li J; Hilser VJ The Ensemble Nature of Allostery. Nature 2014, 508, 331–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Goodey NM; Benkovic SJ Allosteric Regulation and Catalysis Emerge via a Common Route. Nat. Chem. Biol 2008, 4, 474–482. [DOI] [PubMed] [Google Scholar]
- (38).Kuriyan J; Eisenberg D The Origin of Protein Interactions and Allostery in Colocalization. Nature 2007, 450, 983–990. [DOI] [PubMed] [Google Scholar]
- (39).Tzeng S-R; Kalodimos CG Protein Activity Regulation by Conformational Entropy. Nature 2012, 488, 236–240. [DOI] [PubMed] [Google Scholar]
- (40).Masterson LR; Mascioni A; Traaseth NJ; Taylor SS; Veglia G Allosteric Cooperativity in Protein Kinase A. Proc. Natl. Acad. Sci. U. S. A 2008, 105, 506–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Perutz MF; Rossmann MG; Cullis AF; Muirhead H; Will G; North A Structure of Haemoglobin: A Three-Dimensional Fourier Synthesis at 5.5-A. Resolution, Obtained by X-Ray Analysis. Nature 1960, 185, 416–422. [DOI] [PubMed] [Google Scholar]
- (42).Perutz MF Mechanisms of Cooperativity and Allosteric Regulation in Proteins. Q. Rev. Biophys 1989, 22, 139–237. [DOI] [PubMed] [Google Scholar]
- (43).Lukin JA; Kontaxis G; Simplaceanu V; Yuan Y; Bax A; Ho C Quaternary Structure of Hemoglobin in Solution. Proc. Natl. Acad. Sci. U. S. A 2003, 100, 517–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Laskowski RA; Gerick F; Thornton JM The Structural Basis of Allosteric Regulation in Proteins. FEBS Lett 2009, 583, 1692–1698. [DOI] [PubMed] [Google Scholar]
- (45).Hammes-Schiffer S Hydrogen Tunneling and Protein Motion in Enzyme Reactions. Acc. Chem. Res 2006, 39, 93–100. [DOI] [PubMed] [Google Scholar]
- (46).Hammes-Schiffer S; Benkovic SJ Relating Protein Motion to Catalysis. Annu. Rev. Biochem 2006, 75, 519–541. [DOI] [PubMed] [Google Scholar]
- (47).Agarwal PK; Billeter SR; Rajagopalan PR; Benkovic SJ; Hammes-Schiffer S Network of Coupled Promoting Motions in Enzyme Aatalysis. Proc. Natl. Acad. Sci. U. S. A 2002, 99, 2794–2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Gumbiner BM Cell Adhesion: the Molecular Basis of Tissue Architecture and Morphogenesis. Cell 1996, 84, 345–357. [DOI] [PubMed] [Google Scholar]
- (49).Zhang X; Moy VT Cooperative Adhesion of Ligand−Receptor Bonds. Biophys. Chem 2003, 104, 271–278. [DOI] [PubMed] [Google Scholar]
- (50).Selhuber-Unkel C; López-García M; Kessler H; Spatz JP Cooperativity in Adhesion Cluster Formation During Initial Cell Adhesion. Biophys. J 2008, 95, 5424–5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Chen A; Moy VT Cross-Linking of Cell Surface Receptors Enhances Cooperativity of Molecular Adhesion. Biophys. J 2000, 78, 2814–2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Moriki T; Maruyama H; Maruyama IN Activation of Preformed EGF Receptor Dimers by Ligand-Induced Rotation of the Transmembrane Domain. J. Mol. Biol 2001, 311, 1011–1026. [DOI] [PubMed] [Google Scholar]
- (53).Teramura Y; Ichinose J; Takagi H; Nishida K; Yanagida T; Sako Y Single-Molecule Analysis of Epidermal Growth Factor Binding on the Surface of Living Cells. EMBO J 2006, 25, 4215–4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Yamada KM; Even-Ram S Integrin Regulation of Growth Factor Receptors. Nat. Cell Biol 2002, 4, E75–E76. [DOI] [PubMed] [Google Scholar]
- (55).Grakoui A; Bromley SK; Sumen C; Davis MM; Shaw AS; Allen PM; Dustin ML The Immunological Synapse: A Molecular Machine Controlling T Cell Activation. Science 1999, 285, 221–227. [DOI] [PubMed] [Google Scholar]
- (56).Huppa JB; Davis MM T-Cell-Antigen Recognition and the Immunological Synapse. Nat. Rev. Immunol 2003, 3, 973–983. [DOI] [PubMed] [Google Scholar]
- (57).Luo M; Samandi LZ; Wang Z; Chen ZJ; Gao J Synthetic Nanovaccines for Immunotherapy. J. Controlled Release 2017, 263, 200–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Luo M; Wang H; Wang Z; Cai H; Lu Z; Li Y; Du M; Huang G; Wang C; Chen X A STING-Activating Nanovaccine for Cancer Immunotherapy. Nat. Nanotechnol 2017, 12, 648–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Kaizuka Y; Douglass AD; Varma R; Dustin ML; Vale RD Mechanisms for Segregating T Cell Receptor and Adhesion Molecules during Immunological Synapse Formation in Jurkat T Cells. Proc. Natl. Acad. Sci. U. S. A 2007, 104, 20296–20301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Whitty A Cooperativity and Biological Complexity. Nat. Chem. Biol 2008, 4, 435–439. [DOI] [PubMed] [Google Scholar]
- (61).Mirny LA Nucleosome-Mediated Cooperativity between Transcription Factors. Proc. Natl. Acad. Sci. U. S. A 2010, 107, 22534–22539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Courey AJ Cooperativity in Transcriptional Control. Curr. Biol 2001, 11, R250–R252. [DOI] [PubMed] [Google Scholar]
- (63).Mitrea DM; Kriwacki RW Phase Separation in Biology; Functional Organization of a Higher Order. Cell Commun. Signaling 2016, 14, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Brangwynne CP; Eckmann CR; Courson DS; Rybarska A; Hoege C; Gharakhani J; Jülicher F; Hyman AA Germline P Granules Are Liquid Droplets That Localize by Controlled Dissolution/ Condensation. Science 2009, 324, 1729– 1732. [DOI] [PubMed] [Google Scholar]
- (65).Shin Y; Brangwynne CP Liquid Phase Condensation in Cell Physiology and Disease. Science 2017, 357, eaaf4382. [DOI] [PubMed] [Google Scholar]
- (66).Banani SF; Lee HO; Hyman AA; Rosen MK Biomolecular Condensates: Organizers of Cellular Biochemistry. Nat. Rev. Mol. Cell Biol 2017, 18, 285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Hyman AA; Simons K Beyond Oil and Water—Phase Transitions in Cells. Science 2012, 337, 1047–1049. [DOI] [PubMed] [Google Scholar]
- (68).Hyman AA; Weber CA; Jülicher F Liquid-Liquid Phase Separation in Biology. Annu. Rev. Cell Dev. Biol 2014, 30, 39–58. [DOI] [PubMed] [Google Scholar]
- (69).Habchi J; Tompa P; Longhi S; Uversky VN Introducing Protein Intrinsic Disorder. Chem. Rev 2014, 114, 6561–6588. [DOI] [PubMed] [Google Scholar]
- (70).Wright PE; Dyson HJ Intrinsically Disordered Proteins in Cellular Signalling and Regulation. Nat. Rev. Mol. Cell Biol 2015, 16, 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Dunker AK; Lawson JD; Brown CJ; Williams RM; Romero P; Oh JS; Oldfield CJ; Campen AM; Ratliff CM; Hipps KW Intrinsically Disordered Protein. J. Mol. Graphics Modell 2001, 19, 26–59. [DOI] [PubMed] [Google Scholar]
- (72).Molliex A; Temirov J; Lee J; Coughlin M; Kanagaraj AP; Kim HJ; Mittag T; Taylor JP Phase Separation by Low Complexity Domains Promotes Stress Granule Assembly and Drives Pathological Fibrillization. Cell 2015, 163, 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Elbaum-Garfinkle S; Kim Y; Szczepaniak K; Chen CC-H; Eckmann CR; Myong S; Brangwynne CP The Disordered P Granule Protein LAF-1 Drives Phase Separation into Droplets with Tunable Viscosity and Dynamics. Proc. Natl. Acad. Sci. U. S. A 2015, 112, 7189–7194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Burke KA; Janke AM; Rhine CL; Fawzi NL Residue-by-Residue View of in vitro FUS Granules that Bind the C-Terminal Domain of RNA Polymerase II. Mol. Cell 2015, 60, 231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Brangwynne CP Phase Transitions and Size Scaling of Membrane-Less Organelles. J. Cell Biol 2013, 203, 875–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Banjade S; Rosen MK Phase Transitions of Multivalent Proteins Can Promote Clustering of Membrane Receptors. eLife 2014, 3, e04123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Lin Y; Protter DS; Rosen MK; Parker R Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Mol. Cell 2015, 60, 208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Su X; Ditlev JA; Hui E; Xing W; Banjade S; Okrut J; King DS; Taunton J; Rosen MK; Vale RD Phase Separation of Signaling Molecules Promotes T Cell Receptor Signal Transduction. Science 2016, 352, 595–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Li P; Banjade S; Cheng H-C; Kim S; Chen B; Guo L; Llaguno M; Hollingsworth JV; King DS; Banani SF Phase Transitions in the Assembly of Multivalent Signalling Proteins. Nature 2012, 483, 336–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Whitesides GM; Grzybowski B Self-Assembly at All Scales. Science 2002, 295, 2418–2421. [DOI] [PubMed] [Google Scholar]
- (81).Lehn J-M Toward Complex Matter: Supramolecular Chemistry and Self-Organization. Proc. Natl. Acad. Sci. U. S. A 2002, 99, 4763–4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Whitesides GM; Boncheva M Beyond molecules: Self- Assembly of Mesoscopic and Macroscopic Components. Proc. Natl. Acad. Sci. U. S. A 2002, 99, 4769–4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Beyer MK The Mechanical Strength of A Covalent Bond Calculated by Density Functional Theory. J. Chem. Phys 2000, 112, 7307–7312. [Google Scholar]
- (84).Gilli P; Bertolasi V; Pretto L; Ferretti V; Gilli G Covalent versus Electrostatic Nature of the Strong Hydrogen Bond: Discrimination among Single, Double, and Asymmetric Single-Well Hydrogen Bonds by Variable-Temperature X-Ray Crystallographic Methods in β-Diketone Enol RAHB Systems. J. Am. Chem. Soc 2004, 126, 3845–3855. [DOI] [PubMed] [Google Scholar]
- (85).Grandbois M; Beyer M; Rief M; Clausen-Schaumann H; Gaub HE How Strong is a Covalent Bond? Science 1999, 283, 1727–1730. [DOI] [PubMed] [Google Scholar]
- (86).Blanksby SJ; Ellison GB Bond Dissociation Energies of Organic Molecules. Acc. Chem. Res 2003, 36, 255–263. [DOI] [PubMed] [Google Scholar]
- (87).Müller-Dethlefs K; Hobza P Noncovalent Interactions: A Challenge for Experiment and Theory. Chem. Rev 2000, 100, 143–168. [DOI] [PubMed] [Google Scholar]
- (88).Johnson ER; Keinan S; Mori-Sanchez P; Contreras-Garcia J; Cohen AJ; Yang W Revealing Noncovalent Interactions. J. Am. Chem. Soc 2010, 132, 6498–6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Frieden E Non-covalent Interactions: Key to Biological Flexibility and Specificity. J. Chem. Educ 1975, 52, 754–761. [DOI] [PubMed] [Google Scholar]
- (90).Černý J; Hobza P Non-Covalent Interactions in Biomacromolecules. Phys. Chem. Chem. Phys 2007, 9, 5291–5303. [DOI] [PubMed] [Google Scholar]
- (91).Tanford C The Hydrophobic Effect and the Organization of Living Matter. Science 1978, 200, 1012–1018. [DOI] [PubMed] [Google Scholar]
- (92).Lodge TP; Bang J; Park MJ; Char K Origin of the Thermoreversible Fcc-BccTransition in Block Copolymer Solutions. Phys. Rev. Lett 2004, 92, 145501. [DOI] [PubMed] [Google Scholar]
- (93).Dill KA; Truskett TM; Vlachy V; Hribar-Lee B Modeling Water, the Hydrophobic Effect, and Ion Solvation. Annu. Rev. Biophys. Biomol. Struct 2005, 34, 173–199. [DOI] [PubMed] [Google Scholar]
- (94).Biedermann F; Nau WM; Schneider HJ The Hydrophobic Effect Revisited—Studies with Supramolecular Complexes Imply High-Energy Water as A Noncovalent Driving Force. Angew. Chem., Int. Ed 2014, 53, 11158–11171. [DOI] [PubMed] [Google Scholar]
- (95).Davis JG; Gierszal KP; Wang P; Ben-Amotz D Water Structural Transformation at Molecular Hydrophobic Interfaces. Nature 2012, 491, 582–585. [DOI] [PubMed] [Google Scholar]
- (96).Chandler D Interfaces and the Driving Force of Hydrophobic Assembly. Nature 2005, 437, 640–647. [DOI] [PubMed] [Google Scholar]
- (97).Israelachvili J; Pashley R The Hydrophobic Interaction is Long Range, Decaying Exponentially with Distance. Nature 1982, 300, 341–342. [DOI] [PubMed] [Google Scholar]
- (98).Southall NT; Dill KA; Haymet A A View of the Hydrophobic Effect. J. Phys. Chem. B 2002, 106, 521–533. [Google Scholar]
- (99).Shimizu T; Masuda M; Minamikawa H Supramolecular Nanotube Architectures based on Amphiphilic Molecules. Chem. Rev 2005, 105, 1401–1444. [DOI] [PubMed] [Google Scholar]
- (100).Lee JS; Feijen J Polymersomes for Drug Delivery: Design, Formation and Characterization. J. Controlled Release 2012, 161, 473–483. [DOI] [PubMed] [Google Scholar]
- (101).Discher DE; Ahmed F Polymersomes. Annu. Rev. Biomed. Eng 2006, 8, 323–341. [DOI] [PubMed] [Google Scholar]
- (102).Chen XM; Liu GF Double-Stranded Helices and Molecular Zippers Assembled from Single-Stranded Coordination Polymers Directed by Supramolecular Interactions. Chem. - Eur. J 2002, 8, 4811–4817. [DOI] [PubMed] [Google Scholar]
- (103).Immordino ML; Dosio F; Cattel L Stealth Liposomes: Review of the Basic Science, Rationale, and Clinical Applications, Existing and Potential. Int. J. Nanomedicine 2006, 1, 297–315. [PMC free article] [PubMed] [Google Scholar]
- (104).Chang H-I; Yeh M-K Clinical Development of Liposome-based Drugs: Formulation, Characterization, and Therapeutic efficacy. J. Nanomed. Biother. Discovery 2012, 7, 49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (105).Allen TM; Cullis PR Drug Delivery Systems: Entering the Mainstream. Science 2004, 303, 1818–1822. [DOI] [PubMed] [Google Scholar]
- (106).Gaucher G; Dufresne M-H; Sant VP; Kang N; Maysinger D; Leroux J-C Block Copolymer Micelles: Preparation, Characterization and Application in Drug Delivery. J. Controlled Release 2005, 109, 169–188. [DOI] [PubMed] [Google Scholar]
- (107).Kedar U; Phutane P; Shidhaye S; Kadam V Advances in Polymeric Micelles for Drug Delivery and Tumor Targeting. Nanomedicine 2010, 6, 714–729. [DOI] [PubMed] [Google Scholar]
- (108).Stoffelen C; Huskens J Soft Supramolecular Nanoparticles by Noncovalent and Host−Guest Interactions. Small 2016, 12, 96–119. [DOI] [PubMed] [Google Scholar]
- (109).Sherrington DC; Taskinen KA Self-Assembly in Synthetic Macromolecular Systems via Multiple Hydrogen Bonding Interactions. Chem. Soc. Rev 2001, 30, 83–93. [Google Scholar]
- (110).Bangham A; Standish M; Weissmann G The Action of Steroids and Streptolysin S on the Permeability of Phospholipid Structures to Cations. J. Mol. Biol 1965, 13, 253–259. [DOI] [PubMed] [Google Scholar]
- (111).Padilla De Jesús OL; Ihre HR; Gagne L; Frechet JM; Szoka FC Polyester Dendritic Systems for Drug Delivery Applications: in vitro and in vivo Evaluation. Bioconjugate Chem 2002, 13, 453–461. [DOI] [PubMed] [Google Scholar]
- (112).Li C Poly (L-Glutamic Acid)−Anticancer Drug Conjugates. Adv. Drug Delivery Rev 2002, 54, 695–713. [DOI] [PubMed] [Google Scholar]
- (113).Choi YH; Liu F; Kim J-S; Choi YK; Park JS; Kim SW Polyethylene Glycol-Grafted Poly-L-Lysine as Polymeric Gene Carrier. J. Controlled Release 1998, 54, 39–48. [DOI] [PubMed] [Google Scholar]
- (114).Lee ES; Shin HJ; Na K; Bae YH Poly (L-Histidine)−PEG Block Copolymer Micelles and pH-Induced Destabilization. J. Controlled Release 2003, 90, 363–374. [DOI] [PubMed] [Google Scholar]
- (115).Miyata K; Christie RJ; Kataoka K Polymeric Micelles for Nano-Scale Drug Delivery. React. Funct. Polym 2011, 71, 227–234. [Google Scholar]
- (116).Pattni BS; Chupin VV; Torchilin VP New Developments in Liposomal Drug Delivery. Chem. Rev 2015, 115, 10938–10966. [DOI] [PubMed] [Google Scholar]
- (117).Barenholz YC Doxil®—the First FDA-Approved Nano-Drug: Lessons Learned. J. Controlled Release 2012, 160, 117–134. [DOI] [PubMed] [Google Scholar]
- (118).Petre CE; Dittmer DP Liposomal Daunorubicin as Treatment for Kaposi’s Sarcoma. Int. J. Nanomedicine 2007, 2, 277–288. [PMC free article] [PubMed] [Google Scholar]
- (119).Peppas NA Hydrogels and Drug Delivery. Curr. Opin. Colloid Interface Sci 1997, 2, 531–537. [Google Scholar]
- (120).Kozlovskaya V; Sukhishvili SA pH-Controlled Permeability of Layered Hydrogen-Bonded Polymer Capsules. Macromolecules 2006, 39, 5569–5572. [Google Scholar]
- (121).Kim SH; Tan JP; Nederberg F; Fukushima K; Colson J; Yang C; Nelson A; Yang Y-Y; Hedrick JL Hydrogen Bonding-Enhanced Micelle Assemblies for Drug Delivery. Biomaterials 2010, 31, 8063–8071. [DOI] [PubMed] [Google Scholar]
- (122).Kim B-S; Park SW; Hammond PT Hydrogen-Bonding Layer-by-Layer-Assembled Biodegradable Polymeric Micelles as Drug Delivery Vehicles From Surfaces. ACS Nano 2008, 2, 386–392. [DOI] [PubMed] [Google Scholar]
- (123).Kharlampieva E; Kozlovskaya V; Sukhishvili SA Layer-by-Layer Hydrogen-Bonded Polymer Films: from Fundamentals to Applications. Adv. Mater 2009, 21, 3053–3065. [Google Scholar]
- (124).Seto CT; Whitesides GM Self-Assembly based on the Cyanuric Acid-Melamine Lattice. J. Am. Chem. Soc 1990, 112, 6409–6411. [Google Scholar]
- (125).Mathias JP; Simanek EE; Zerkowski JA; Seto CT; Whitesides GM Structural Preferences of Hydrogen-Bonded Networks in Organic Solution-the Cyclic CA3. cntdot. M3″ Rosette″. J. Am. Chem. Soc 1994, 116, 4316–4325. [Google Scholar]
- (126).Whitesides GM; Simanek EE; Mathias JP; Seto CT; Chin D; Mammen M; Gordon DM Noncovalent Synthesis: Using Physical-Organic Chemistry to Make Aggregates. Acc. Chem. Res 1995, 28, 37–44. [Google Scholar]
- (127).Boal AK; Ilhan F; DeRouchey JE; Thurn-Albrecht T; Russell TP; Rotello VM Self-Assembly of Nanoparticles into Structured Spherical and Network Aggregates. Nature 2000, 404, 746–748. [DOI] [PubMed] [Google Scholar]
- (128).Frankamp BL; Uzun O; Ilhan F; Boal AK; Rotello VM Recognition-Mediated Assembly of Nanoparticles into Micellar Structures with Diblock Copolymers. J. Am. Chem. Soc 2002, 124, 892–893. [DOI] [PubMed] [Google Scholar]
- (129).Ilhan F; Galow TH; Gray M; Clavier G; Rotello VM Giant Vesicle Formation through Self-Assembly of Complementary Random Copolymers. J. Am. Chem. Soc 2000, 122, 5895–5896. [Google Scholar]
- (130).Bloomfield VA DNA Condensation. Curr. Opin. Struct. Biol 1996, 6, 334–341. [DOI] [PubMed] [Google Scholar]
- (131).Ananthanarayanan VS; Kerman A Role of Metal Ions in Ligand−Receptor Interaction: Insights from Structural Studies. Mol. Cell. Endocrinol 2006, 246, 53–59. [DOI] [PubMed] [Google Scholar]
- (132).Fuqua C; Parsek MR; Greenberg EP Regulation of Gene Expression by Cell-to-Cell Communication: Acyl-Homoserine Lactone Quorum Sensing. Annu. Rev. Genet 2001, 35, 439–468. [DOI] [PubMed] [Google Scholar]
- (133).Iler R Multilayers of Colloidal Particles. J. Colloid Interface Sci 1966, 21, 569–594. [Google Scholar]
- (134).Decher G; Hong J; Schmitt J Buildup of Ultrathin Multilayer Films by A Self-Assembly Process: III. Consecutively Alternating Adsorption of Anionic and Cationic Polyelectrolytes on Charged sSurfaces. Thin Solid Films 1992, 210, 831–835. [Google Scholar]
- (135).De Villiers MM; Otto DP; Strydom SJ; Lvov YM Introduction to Nanocoatings Produced by Layer-by-Layer (LbL) Self- Assembly. Adv. Drug Delivery Rev 2011, 63, 701–715. [DOI] [PubMed] [Google Scholar]
- (136).Zhang X; Chen H; Zhang H Layer-by-Layer Assembly: from Conventional to Unconventional Methods. Chem. Commun 2007, 43, 1395–1405. [DOI] [PubMed] [Google Scholar]
- (137).Ai H; Jones SA; Lvov YM Biomedical Applications of Electrostatic Layer-by-Layer Nano-Assembly of Polymers, Enzymes, and Nanoparticles. Cell Biochem. Biophys 2003, 39, 23–43. [DOI] [PubMed] [Google Scholar]
- (138).Sondjaja HR; Hatton TA; Tam K Self-Assembly of Poly (Ethylene Oxide)-Block-Poly (Acrylic Acid) Induced by CaCl2: Mechanistic Study. Langmuir 2008, 24, 8501–8506. [DOI] [PubMed] [Google Scholar]
- (139).Harada A; Kataoka K Formation of Polyion Complex Micelles in An Aqueous Milieu from a Pair of Oppositely-Charged Block Copolymers with Poly(Ethylene Glycol) Segments. Macromolecules 1995, 28, 5294–5299. [Google Scholar]
- (140).Kabanov AV; Bronich TK; Kabanov VA; Yu K; Eisenberg A Soluble Stoichiometric Complexes from Poly (N-Ethyl-4-Vinylpyridinium) Cations and Poly (Ethylene Oxide)-Block-Polymethacrylate Anions. Macromolecules 1996, 29, 6797–6802. [Google Scholar]
- (141).Cohen Stuart MA; Besseling N; Fokkink R Formation of Micelles with Complex Coacervate Cores. Langmuir 1998, 14, 6846–6849. [Google Scholar]
- (142).Lee Y; Ishii T; Cabral H; Kim HJ; Seo JH; Nishiyama N; Oshima H; Osada K; Kataoka K Charge-Conversional Polyionic Complex Micelles—Efficient Nanocarriers for Protein Delivery into Cytoplasm. Angew. Chem 2009, 121, 5413–5416. [DOI] [PubMed] [Google Scholar]
- (143).Chen J; Ding J; Zhang Y; Xiao C; Zhuang X; Chen X Polyion Complex Micelles with Gradient pH-Sensitivity for Adjustable Intracellular Drug Delivery. Polym. Chem 2015, 6, 397–405. [Google Scholar]
- (144).Lee Y; Kataoka K Biosignal-Sensitive Polyion Complex Micelles for the Delivery of Biopharmaceuticals. Soft Matter 2009, 5, 3810–3817. [Google Scholar]
- (145).Insua I; Wilkinson A; Fernandez-Trillo F Polyion Complex (PIC) Particles: Preparation and Biomedical Applications. Eur. Polym. J 2016, 81, 198–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (146).Harada A; Kataoka K Chain Length Recognition: Core-Shell Supramolecular Assembly from Oppositely Charged Block Copolymers. Science 1999, 283, 65–67. [DOI] [PubMed] [Google Scholar]
- (147).Green JJ; Chiu E; Leshchiner ES; Shi J; Langer R; Anderson DG Electrostatic Ligand Coatings of Nanoparticles Enable Ligand-Specific Gene Delivery to Human Primary Cells. Nano Lett 2007, 7, 874–879. [DOI] [PubMed] [Google Scholar]
- (148).Waehler R; Russell SJ; Curiel DT Engineering Targeted Viral Vectors for Gene Therapy. Nat. Rev. Genet 2007, 8, 573–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (149).Mintzer MA; Simanek EE Nonviral Vectors for Gene Delivery. Chem. Rev 2009, 109, 259–302. [DOI] [PubMed] [Google Scholar]
- (150).Morille M; Passirani C; Vonarbourg A; Clavreul A; Benoit J-P Progress in Developing Cationic Vectors for Non-Viral Systemic Gene Therapy Against Cancer. Biomaterials 2008, 29, 3477–3496. [DOI] [PubMed] [Google Scholar]
- (151).Pack DW; Hoffman AS; Pun S; Stayton PS Design and Development of Polymers for Gene Delivery. Nat. Rev. Drug Discovery 2005, 4, 581–593. [DOI] [PubMed] [Google Scholar]
- (152).Shmueli RB; Anderson DG; Green JJ Electrostatic Surface Modifications to Improve Gene Delivery. Expert Opin. Drug Delivery 2010, 7, 535–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (153).Hunter CA Meldola lecture. The Role of Aromatic Interactions in Molecular Recognition. Chem. Soc. Rev 1994, 23, 101–109. [Google Scholar]
- (154).Crowley JD; Goldup SM; Lee A-L; Leigh DA; McBurney RT Active Metal Template Synthesis of Rotaxanes, Catenanes and Molecular Shuttles. Chem. Soc. Rev 2009, 38, 1530–1541. [DOI] [PubMed] [Google Scholar]
- (155).Robinson JT; Tabakman SM; Liang Y; Wang H; Sanchez Casalongue H; Vinh D; Dai H Ultrasmall Reduced Graphene Oxide with High Near-Infrared Absorbance for Photothermal Therapy. J. Am. Chem. Soc 2011, 133, 6825–6831. [DOI] [PubMed] [Google Scholar]
- (156).Liu Z; Robinson JT; Sun X; Dai H PEGylated Nanographene Oxide for Delivery of Water-Insoluble Cancer Drugs. J. Am. Chem. Soc 2008, 130, 10876–10877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (157).Liu Z; Fan AC; Rakhra K; Sherlock S; Goodwin A; Chen X; Yang Q; Felsher DW; Dai H Supramolecular Stacking of Doxorubicin on Carbon Nanotubes for in vivo Cancer Therapy. Angew. Chem., Int. Ed 2009, 48, 7668–7672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (158).Sun X; Liu Z; Welsher K; Robinson JT; Goodwin A; Zaric S; Dai H Nano-Graphene Oxide for Cellular Imaging and Drug Delivery. Nano Res 2008, 1, 203–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (159).Liu Z; Robinson JT; Tabakman SM; Yang K; Dai H Carbon Materials for Drug Delivery & Cancer Therapy. Mater. Today 2011, 14, 316–323. [Google Scholar]
- (160).Liu Z; Sun X; Nakayama-Ratchford N; Dai H Supramolecular Chemistry on Water-Soluble Carbon Nanotubes for Drug Loading and Delivery. ACS Nano 2007, 1, 50–56. [DOI] [PubMed] [Google Scholar]
- (161).Vintiloiu A; Leroux J-C Organogels and Their Use in Drug Delivery—A Review. J. Controlled Release 2008, 125, 179–192. [DOI] [PubMed] [Google Scholar]
- (162).Stoddart JF The Chemistry of the Mechanical Bond. Chem. Soc. Rev 2009, 38, 1802–1820. [DOI] [PubMed] [Google Scholar]
- (163).Jonkheijm P; van der Schoot P; Schenning AP; Meijer E Probing the Solvent-Assisted Nucleation Pathway in Chemical Self-Assembly. Science 2006, 313, 80–83. [DOI] [PubMed] [Google Scholar]
- (164).Aida T; Meijer E; Stupp S Functional Supramolecular Polymers. Science 2012, 335, 813–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (165).Mahadevi AS; Sastry GN Cooperativity in Noncovalent Interactions. Chem. Rev 2016, 116, 2775–2825. [DOI] [PubMed] [Google Scholar]
- (166).Cutler JI; Auyeung E; Mirkin CA Spherical Nucleic Acids. J. Am. Chem. Soc 2012, 134, 1376–1391. [DOI] [PubMed] [Google Scholar]
- (167).Mirkin CA; Letsinger RL; Mucic RC; Storhoff JJ A DNA-based Method for Rationally Assembling Nanoparticles into Macroscopic Materials. Nature 1996, 382, 607–609. [DOI] [PubMed] [Google Scholar]
- (168).Elbaz J; Moshe M; Shlyahovsky B; Willner I Cooperative Multicomponent Self-Assembly of Nucleic Acid Structures for the Activation of DNAzyme Cascades: A Paradigm for DNA Sensors and Aptasensors. Chem. - Eur. J 2009, 15, 3411–3418. [DOI] [PubMed] [Google Scholar]
- (169).Kolpashchikov DM Split DNA Enzyme for Visual Single Nucleotide Polymorphism Typing. J. Am. Chem. Soc 2008, 130, 2934–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (170).Kolpashchikov DM A Binary DNA Probe for Highly Specific Nucleic Acid Recognition. J. Am. Chem. Soc 2006, 128, 10625–10628. [DOI] [PubMed] [Google Scholar]
- (171).Jin R; Wu G; Li Z; Mirkin CA; Schatz GC What Controls the Melting Properties of DNA-Linked Gold Nanoparticle Assemblies? J. Am. Chem. Soc 2003, 125, 1643–1654. [DOI] [PubMed] [Google Scholar]
- (172).Taton TA; Mucic RC; Mirkin CA; Letsinger RL The DNA-Mediated Formation of Supramolecular Mono-and Multilayered Nanoparticle Structures. J. Am. Chem. Soc 2000, 122, 6305–6306. [Google Scholar]
- (173).Gibbs-Davis JM; Schatz GC; Nguyen ST Sharp Melting Transitions in DNA Hybrids without Aggregate Dissolution: Proof of Neighboring-Duplex Cooperativity. J. Am. Chem. Soc 2007, 129, 15535–15540. [DOI] [PubMed] [Google Scholar]
- (174).Eryazici I; Prytkova TR; Schatz GC; Nguyen ST Cooperative Melting in Caged Dimers with Only Two DNA Duplexes. J. Am. Chem. Soc 2010, 132, 17068–17070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (175).Pohl FM; Jovin TM Salt-Induced Cooperative Conformational Change of a Synthetic DNA: Equilibrium and Kinetic Studies with Poly (dG-dC). J. Mol. Biol 1972, 67, 375–396. [DOI] [PubMed] [Google Scholar]
- (176).Storhoff JJ; Lazarides AA; Mucic RC; Mirkin CA; Letsinger RL; Schatz GC What Controls the Optical Properties of DNA-Linked Gold Nanoparticle Assemblies? J. Am. Chem. Soc 2000, 122, 4640–4650. [Google Scholar]
- (177).Burnett JC; Rossi JJ RNA-based Therapeutics: Current Progress and Future Prospects. Chem. Biol 2012, 19, 60–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (178).Barnaby SN; Perelman GA; Kohlstedt KL; Chinen AB; Schatz GC; Mirkin CA Design Considerations for RNA Spherical Nucleic Acids (SNAs). Bioconjugate Chem 2016, 27, 2124–2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (179).Yoshida M; Lahann J Smart Nanomaterials. ACS Nano 2008, 2, 1101–1107. [DOI] [PubMed] [Google Scholar]
- (180).Dvir T; Timko BP; Kohane DS; Langer R Nanotechnological Strategies for Engineering Complex Tissues. Nat. Nanotechnol 2011, 6, 13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (181).Lu Y; Aimetti AA; Langer R; Gu Z Bioresponsive Materials. Nat. Rev. Mater 2016, 2, 16075. [Google Scholar]
- (182).Dai S; Ravi P; Tam KC Thermo-and Photo-Responsive Polymeric Systems. Soft Matter 2009, 5, 2513–2533. [Google Scholar]
- (183).Schmaljohann D Thermo-and pH-Responsive Polymers in Drug Delivery. Adv. Drug Delivery Rev 2006, 58, 1655–1670. [DOI] [PubMed] [Google Scholar]
- (184).Mura S; Nicolas J; Couvreur P Stimuli-Responsive Nanocarriers for Drug Delivery. Nat. Mater 2013, 12, 991–1003. [DOI] [PubMed] [Google Scholar]
- (185).Roy D; Brooks WL; Sumerlin BS New Directions in Thermoresponsive Polymers. Chem. Soc. Rev 2013, 42, 7214–7243. [DOI] [PubMed] [Google Scholar]
- (186).Schild HG Poly (N-Isopropylacrylamide): Experiment, Theory and Application. Prog. Polym. Sci 1992, 17, 163–249. [Google Scholar]
- (187).Bikram M; West JL Thermo-Responsive Systems for Controlled Drug Delivery. Expert Opin. Drug Delivery 2008, 5, 1077–1091. [DOI] [PubMed] [Google Scholar]
- (188).Zhang J; Chen H; Xu L; Gu Y The Targeted Behavior of Thermally Responsive Nanohydrogel Evaluated by NIR System in Mouse Model. J. Controlled Release 2008, 131, 34–40. [DOI] [PubMed] [Google Scholar]
- (189).Serres A; Baudyš M; Kim SW Temperature and pH Polymers for Human Calcitonin Delivery. Pharm. Res 1996, 13, 196–201. [DOI] [PubMed] [Google Scholar]
- (190).Yong-Hee K; Bae YH; Kim SW pH/Temperature-Sensitive Polymers for Macromolecular Drug Loading and Release. J. Controlled Release 1994, 28, 143–152. [Google Scholar]
- (191).Shibayama M; Norisuye T; Nomura S Cross-Link Density Dependence of Spatial Inhomogeneities and Dynamic Fluctuations of poly (N-Isopropylacrylamide) Gels. Macromolecules 1996, 29, 8746–8750. [Google Scholar]
- (192).Gibson MI; Paripovic D; Klok HA Size-Dependent LCST Transitions of Polymer-Coated Gold Nanoparticles: Cooperative Aggregation and Surface Assembly. Adv. Mater 2010, 22, 4721–4725. [DOI] [PubMed] [Google Scholar]
- (193).Ieong NS; Hasan M; Phillips DJ; Saaka Y; O’Reilly RK; Gibson MI Polymers with Molecular Weight Dependent LCSTs are Essential for Cooperative Behaviour. Polym. Chem 2012, 3, 794–799. [Google Scholar]
- (194).Heskins M; Guillet JE Solution Properties of Poly (N-Isopropylacrylamide). J. Macromol. Sci., Chem 1968, 2, 1441–1455. [Google Scholar]
- (195).Tanaka N; Matsukawa S; Kurosu H; Ando I A Study on Dynamics of Water in Crosslinked Poly (N-Isopropylacrylamide) Gel by NMR Spectroscopy. Polymer 1998, 39, 4703–4706. [Google Scholar]
- (196).Sasaki S; Kawasaki H; Maeda H Volume Phase Transition Behavior of N-Isopropylacrylamide Gels as a Function of the Chemical Potential of Water Molecules. Macromolecules 1997, 30, 1847–1848. [Google Scholar]
- (197).Grinberg NV; Dubovik AS; Grinberg VY; Kuznetsov DV; Makhaeva EE; Grosberg AY; Tanaka T Studies of the Thermal Volume Transition of Poly (N-Isopropylacrylamide) Hydrogels by High-Sensitivity Differential Scanning Microcalorimetry. 1. Dynamic effects. Macromolecules 1999, 32, 1471–1475. [Google Scholar]
- (198).Otake K; Inomata H; Konno M; Saito S Thermal Analysis of the Volume Phase Transition with N-Isopropylacrylamide Gels. Macromolecules 1990, 23, 283–289. [Google Scholar]
- (199).Okada Y; Tanaka F Cooperative Hydration, Chain Collapse, and Flat LCST Behavior in Aqueous Poly (N-Isopropylacrylamide) Solutions. Macromolecules 2005, 38, 4465–4471. [Google Scholar]
- (200).Tanaka F; Koga T; Winnik FM Temperature-Responsive Polymers in Mixed Solvents: Competitive Hydrogen Bonds Cause Cononsolvency. Phys. Rev. Lett 2008, 101, 028302. [DOI] [PubMed] [Google Scholar]
- (201).Tanaka F; Koga T; Kojima H; Winnik FM Temperature- and Tension-Induced Coil− Globule Transition of Poly (N-Isopropylacrylamide) Chains in Water and Mixed Solvent of Water/Methanol. Macromolecules 2009, 42, 1321–1330. [Google Scholar]
- (202).Fujishige S; Kubota K; Ando I Phase Transition of Aqueous Solutions of Poly (N-Isopropylacrylamide) and Poly (N-Isopropylmethacrylamide). J. Phys. Chem 1989, 93, 3311–3313. [Google Scholar]
- (203).Qiu X-P; Tanaka F; Winnik FM Temperature-Induced Phase Transition of Well-Define d Cyclic Poly (N-Isopropylacrylamide)s in Aqueous Solution. Macromolecules 2007, 40, 7069–7071. [Google Scholar]
- (204).Philipp M; Kyriakos K; Silvi L; Lohstroh W; Petry W; Krüger JK; Papadakis CM; Müller-Buschbaum P From Molecular Dehydration to Excess Volumes of Phase-Separating PNIPAM Solutions. J. Phys. Chem. B 2014, 118, 4253–4260. [DOI] [PubMed] [Google Scholar]
- (205).Ito D; Kubota K Solution Properties and Thermal Behavior of Poly (N-n-Propylacrylamide) in Water. Macromolecules 1997, 30, 7828–7834. [Google Scholar]
- (206).Feil H; Bae YH; Feijen J; Kim SW Effect of Comonomer Hydrophilicity and Ionization on the Lower Critical Solution Temperature of N-Isopropylacrylamide Copolymers. Macromolecules 1993, 26, 2496–2500. [Google Scholar]
- (207).Leo A; Jow P; Silipo C; Hansch C Calculation of Hydrophobic Constant (log P) from. pi. and f Constants. J. Med. Chem 1975, 18, 865–868. [DOI] [PubMed] [Google Scholar]
- (208).Idziak I; Avoce D; Lessard D; Gravel D; Zhu X Thermosensitivity of Aqueous Solutions of Poly (N, N-Diethylacrylamide). Macromolecules 1999, 32, 1260–1263. [Google Scholar]
- (209).Cao Y; Zhu X; Luo J; Liu H Effects of Substitution Groups on the RAFT Polymerization of N-Alkylacrylamides in the Preparation of Thermosensitive Block Copolymers. Macromolecules 2007, 40, 6481–6488. [Google Scholar]
- (210).Arakawa T; Timasheff SN Mechanism of Protein Salting In and Salting Out by Divalent Cation Salts: Balance between Hydration and Salt Binding. Biochemistry 1984, 23, 5912–5923. [DOI] [PubMed] [Google Scholar]
- (211).Hofmeister F Zur Lehre von der Wirkung der Salze. Naunyn-Schmiedeberg’s Arch. Pharmacol 1888, 25, 1–30. [Google Scholar]
- (212).Parsegian VA Hopes for Hofmeister. Nature 1995, 378, 335–336. [Google Scholar]
- (213).Jungwirth P; Cremer PS Beyond Hofmeister. Nat. Chem 2014, 6, 261–263. [DOI] [PubMed] [Google Scholar]
- (214).Zhang Y; Cremer PS Chemistry of Hofmeister Anions and Osmolytes. Annu. Rev. Phys. Chem 2010, 61, 63–83. [DOI] [PubMed] [Google Scholar]
- (215).Zhang Y; Furyk S; Bergbreiter DE; Cremer PS Specific Ion Effects on the Water Solubility of Macromolecules: PNIPAM and the Hofmeister Series. J. Am. Chem. Soc 2005, 127, 14505–14510. [DOI] [PubMed] [Google Scholar]
- (216).Zhang Y; Furyk S; Sagle LB; Cho Y; Bergbreiter DE; Cremer PS Effects of Hofmeister Anions on the LCST of PNIPAM as A Function of Molecular Weight. J. Phys. Chem. C 2007, 111, 8916–8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (217).Ieong NS; Redhead M; Bosquillon C; Alexander C; Kelland M; O’Reilly RK The Missing Lactam-Thermoresponsive and Biocompatible Poly (N-Vinylpiperidone) Polymers by Xanthate- Mediated RAFT Polymerization. Macromolecules 2011, 44, 886–893. [Google Scholar]
- (218).Weaver J; Bannister I; Robinson K; Bories-Azeau X; Armes S; Smallridge M; McKenna P Stimulus-Responsive Water-Soluble Polymers based on 2-Hydroxyethyl Methacrylate. Macromolecules 2004, 37, 2395–2403. [Google Scholar]
- (219).Phillips DJ; Gibson MI Degradable Thermoresponsive Polymers Which Display Redox-Responsive LCST Behaviour. Chem. Commun 2012, 48, 1054–1056. [DOI] [PubMed] [Google Scholar]
- (220).Chen S; Zhang Y; Wang K; Zhou H; Zhang W N-Ester-Substituted Polyacrylamides with a Tunable Lower Critical Solution Temperature (LCST): the N-Ester-Substitute Dependent Thermoresponse. Polym. Chem 2016, 7, 3509–3519. [Google Scholar]
- (221).Edwards EW; Chanana M; Wang D; Möhwald H Stimuli-Responsive Reversible Transport of Nanoparticles Across Water/Oil Interfaces. Angew. Chem., Int. Ed 2008, 47, 320–323. [DOI] [PubMed] [Google Scholar]
- (222).Boyer C; Whittaker MR; Luzon M; Davis TP Design and Synthesis of Dual Thermoresponsive and Antifouling Hybrid Polymer/ Gold Nanoparticles. Macromolecules 2009, 42, 6917–6926. [Google Scholar]
- (223).Shan J; Zhao Y; Granqvist N; Tenhu H Thermoresponsive Properties of N-Isopropylacrylamide Oligomer Brushes Grafted to Gold Nanoparticles: Effects of Molar Mass and Gold Core Size. Macromolecules 2009, 42, 2696–2701. [Google Scholar]
- (224).Keeley FW; Bellingham CM; Woodhouse KA Elastin as A Self−Organizing Biomaterial: Use of Recombinantly Expressed Human Elastin Polypeptides as A Model for Investigations of Structure and Self−Assembly of Elastin. Philos. Trans. R. Soc., B 2002, 357, 185–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (225).Urry DW Free Energy Transduction in Polypeptides and Proteins based on Inverse Temperature Transitions. Prog. Biophys. Mol. Biol 1992, 57, 23–57. [DOI] [PubMed] [Google Scholar]
- (226).McDaniel JR; Callahan DJ; Chilkoti A Drug Delivery to Solid Tumors by Elastin-Like Polypeptides. Adv. Drug Delivery Rev 2010, 62, 1456–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (227).Dyson HJ; Wright PE Coupling of Folding and Binding for Unstructured Proteins. Curr. Opin. Struct. Biol 2002, 12, 54–60. [DOI] [PubMed] [Google Scholar]
- (228).Kurzbach D; Hassouneh W; McDaniel JR; Jaumann EA; Chilkoti A; Hinderberger D Hydration Layer Coupling and Cooperativity in Phase Behavior of Stimulus Responsive Peptide Polymers. J. Am. Chem. Soc 2013, 135, 11299–11308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (229).McDaniel JR; Radford DC; Chilkoti A A Unified Model for de novo Design of Elastin-Like Polypeptides with Tunable Inverse Transition Temperatures. Biomacromolecules 2013, 14, 2866–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (230).Li NK; Quiroz FG; Hall CK; Chilkoti A; Yingling YG Molecular Description of the LCST Behavior of An Elastin-Like Polypeptide. Biomacromolecules 2014, 15, 3522–3530. [DOI] [PubMed] [Google Scholar]
- (231).Kato S; Han S-Y; Liu W; Otsuka K; Shibata H; Kanamaru R; Ishioka C Understanding the Function−Structure and Function−Mutation Relationships of p53 Tumor Suppressor Protein by High- Resolution Missense Mutation Analysis. Proc. Natl. Acad. Sci. U. S. A 2003, 100, 8424–8429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (232).Ng PC; Henikoff S SIFT: Predicting Amino Acid Changes that Affect Protein Function. Nucleic Acids Res 2003, 31, 3812–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (233).Ribeiro A; Arias FJ; Reguera J; Alonso M; Rodríguez-Cabello JC Influence of the Amino-Acid Sequence on the Inverse Temperature Transition of Elastin-Like Polymers. Biophys. J 2009, 97, 312–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (234).Girotti A; Reguera J; Arias FJ; Alonso M; Testera AM; Rodríguez-Cabello JC Influence of the Molecular Weight on the Inverse Temperature Transition of A Model Genetically Engineered Elastin-Like pH-Responsive Polymer. Macromolecules 2004, 37, 3396–3400. [Google Scholar]
- (235).Quiroz FG; Chilkoti A Sequence Heuristics to Encode Phase Behaviour in Intrinsically Disordered Protein Polymers. Nat. Mater 2015, 14, 1164–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (236).Cho Y; Zhang Y; Christensen T; Sagle LB; Chilkoti A; Cremer PS Effects of Hofmeister Anions on the Phase Transition Temperature of Elastin-Like Polypeptides. J. Phys. Chem. B 2008, 112, 13765–13771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (237).Ghoorchian A; Holland NB Molecular Architecture Influences the Thermally Induced Aggregation Behavior of Elastin- Like Polypeptides. Biomacromolecules 2011, 12, 4022–4029. [DOI] [PubMed] [Google Scholar]
- (238).Webb BA; Chimenti M; Jacobson MP; Barber DL Dysregulated pH: A Perfect Storm for Cancer Progression. Nat. Rev. Cancer 2011, 11, 671–677. [DOI] [PubMed] [Google Scholar]
- (239).Gillies RJ; Liu Z; Bhujwalla Z P-31-Mrs Measurements of Extracellular pH of Tumors Using 3-Aminopropylphosphonate. Am. J. Physiol 1994, 267, C195–C203. [DOI] [PubMed] [Google Scholar]
- (240).Gillies RJ; Raghunand N; Garcia-Martin ML; Gatenby RA pH Imaging. IEEE Eng. Med. Biol. Mag 2004, 23, 57–64. [DOI] [PubMed] [Google Scholar]
- (241).van Sluis R; Bhujwalla ZM; Raghunand N; Ballesteros P; Alvarez J; Cerdan S; Galons JP; Gillies RJ In vivo Imaging of Extracellular pH Using (1)H MRSI. Magn. Reson. Med 1999, 41, 743–750. [DOI] [PubMed] [Google Scholar]
- (242).Volk T; Jahde E; Fortmeyer HP; Glusenkamp KH; Rajewsky MF pH in Human Tumour Xenografts: Effect of Intravenous Administration of Glucose. Br. J. Cancer 1993, 68, 492–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (243).Gatenby RA; Gillies RJ Why Do Cancers Have High Aerobic Glycolysis? Nat. Rev. Cancer 2004, 4, 891–899. [DOI] [PubMed] [Google Scholar]
- (244).Helmlinger G; Sckell A; Dellian M; Forbes NS; Jain RK Acid Production in Glycolysis-Impaired Tumors Provides New Insights into Tumor Metabolism. Clin. Cancer Res 2002, 8, 1284–1291. [PubMed] [Google Scholar]
- (245).Newell K; Franchi A; Pouyssegur J; Tannock I Studies with Glycolysis-Deficient Cells Suggest that Production of Lactic Acid is Not the Only Cause of Tumor Acidity. Proc. Natl. Acad. Sci. U. S. A 1993, 90, 1127–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (246).Yamagata M; Hasuda K; Stamato T; Tannock IF The Contribution of Lactic Acid to Acidification of Tumours: Studies of Variant Cells Lacking Lactate Dehydrogenase. Br. J. Cancer 1998, 77, 1726–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (247).Neri D; Supuran CT Interfering with pH Regulation in Tumours as A Therapeutic Strategy. Nat. Rev. Drug Discovery 2011, 10, 767–777. [DOI] [PubMed] [Google Scholar]
- (248).Jose C; Bellance N; Rossignol R Choosing between Glycolysis and Oxidative Phosphorylation: A Tumor’s Dilemma? Biochim. Biophys. Acta, Bioenerg 2011, 1807, 552–561. [DOI] [PubMed] [Google Scholar]
- (249).Blum AP; Kammeyer JK; Rush AM; Callmann CE; Hahn ME; Gianneschi NC Stimuli-Responsive Nanomaterials for Biomedical Applications. J. Am. Chem. Soc 2015, 137, 2140–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (250).Li H-J; Du J-Z; Liu J; Du X-J; Shen S; Zhu Y-H; Wang X; Ye X; Nie S; Wang J Smart Superstructures with Ultrahigh pH-Sensitivity for Targeting Acidic Tumor Microenvironment: Instantaneous Size Switching and Improved Tumor Penetration. ACS Nano 2016, 10, 6753–6761. [DOI] [PubMed] [Google Scholar]
- (251).Gao W; Chan JM; Farokhzad OC pH-Responsive Nanoparticles for Drug Delivery. Mol. Pharmaceutics 2010, 7, 1913–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (252).Hu J; Zhang G; Ge Z; Liu S Stimuli-Responsive Tertiary Amine Methacrylate-based Block Copolymers: Synthesis, Supramolecular Self-Assembly and Functional Applications. Prog. Polym. Sci 2014, 39, 1096–1143. [Google Scholar]
- (253).Binauld S; Stenzel MH Acid-Degradable Polymers for Drug Delivery: a Decade of Innovation. Chem. Commun 2013, 49, 2082–2102. [DOI] [PubMed] [Google Scholar]
- (254).Kuhn W; Hargitay B; Katchalsky A; Eisenberg H Reversible Dilation and Contraction by Changing the State of Ionization of High-Polymer Acid Networks. Nature 1950, 165, 514–516. [Google Scholar]
- (255).Philippova OE; Hourdet D; Audebert R; Khokhlov AR pH-Responsive Gels of Hydrophobically Modified Poly (Acrylic Acid). Macromolecules 1997, 30, 8278–8285. [Google Scholar]
- (256).Kang SI; Bae YH pH-Induced Volume-Phase Transition of Hydrogels Containing Sulfonamide Side Group by Reversible Crystal Formation. Macromolecules 2001, 34, 8173–8178. [Google Scholar]
- (257).Kang SI; Bae YH pH-Induced Solubility Transition of Sulfonamide-based Polymers. J. Controlled Release 2002, 80, 145–155. [DOI] [PubMed] [Google Scholar]
- (258).Kang HC; Bae YH pH-Tunable Endosomolytic Oligomers for Enhanced Nucleic Acid Delivery. Adv. Funct. Mater 2007, 17, 1263–1272. [Google Scholar]
- (259).Park SY; Bae YH Novel pH-Sensitive Polymers Containing Sulfonamide Groups. Macromol. Rapid Commun 1999, 20, 269–273. [Google Scholar]
- (260).Wang Y; Zhou K; Huang G; Hensley C; Huang X; Ma X; Zhao T; Sumer BD; DeBerardinis RJ; Gao J A Nanoparticle-based Strategy for the Imaging of A Broad Range of Tumours by Nonlinear Amplification of Microenvironment Signals. Nat. Mater 2014, 13, 204–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (261).Ma X; Wang Y; Zhao T; Li Y; Su L-C; Wang Z; Huang G; Sumer BD; Gao J Ultra-pH-Sensitive Nanoprobe Library with Broad pH Tunability and Fluorescence Emissions. J. Am. Chem. Soc 2014, 136, 11085–11092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (262).Zhou K; Wang Y; Huang X; Luby-Phelps K; Sumer BD; Gao J Tunable, Ultrasensitive pH-Responsive Nanoparticles Targeting Specific Endocytic Organelles in Living Cells. Angew. Chem., Int. Ed 2011, 50, 6109–6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (263).Wang C; Niederstrasser H; Douglas PM; Lin R; Jaramillo J; Li Y; Olswald NW; Zhou A; McMillan EA; Mendiratta S; et al. Small-Molecule TFEB Pathway Agonists that Ameliorate Metabolic Syndrome in Mice and Extend C. Elegans Lifespan. Nat. Commun 2017, 8, 2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (264).Huang X; Huang G; Zhang S; Sagiyama K; Togao O; Ma X; Wang Y; Li Y; Soesbe TC; Sumer BD; et al. Multi-Chromatic pH-Activatable 19F-MRI Nanoprobes with Binary ON/OFF pH Transitions and Chemical-Shift Barcodes. Angew. Chem., Int. Ed 2013, 52, 8074–8078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (265).Wang C; Wang Y; Li Y; Bodemann B; Zhao T; Ma X; Huang G; Hu Z; DeBerardinis RJ; White MA; et al. A Nnanobuffer Reporter Library for Fine-Scale Imaging and Perturbation of Endocytic Organelles. Nat. Commun 2015, 6, 8524–8534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (266).Wang C; Zhao T; Li Y; Huang G; White MA; Gao J Investigation of Endosome and Lysosome Biology by Ultra pH-Sensitive Nanoprobes. Adv. Drug Delivery Rev 2017, 113, 87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (267).Boussif O; Lezoualc’h F; Zanta MA; Mergny MD; Scherman D; Demeneix B; Behr J-P A Versatile Vector for Gene and Oligonucleotide Transfer into Cells in Culture and in vivo: Polyethylenimine. Proc. Natl. Acad. Sci. U. S. A 1995, 92, 7297–7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (268).Roy K; Mao H-Q; Huang S-K; Leong KW Oral Gene Delivery with Chitosan−DNA Nanoparticles Generates Immunologic Protection in A Murine Model of Peanut Allergy. Nat. Med 1999, 5, 387–391. [DOI] [PubMed] [Google Scholar]
- (269).Kim D; Lee ES; Oh KT; Gao ZG; Bae YH Doxorubicin-Loaded Polymeric Micelle Overcomes Multidrug Resistance of Cancer by Double-Targeting Folate Receptor and Early Endosomal pH. Small 2008, 4, 2043–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (270).Li Y; Zhao T; Wang C; Lin Z; Huang G; Sumer BD; Gao J Molecular Basis of Cooperativity in pH-Triggered Supramolecular Self-assembly. Nat. Commun 2016, 7, 13214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (271).Evans WH; Hardison W Phospholipid, Cholesterol, Polypeptide and Glycoprotein Composition of Hepatic Endosome Subfractions. Biochem. J 1985, 232, 33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (272).Cho YW; Kim JD; Park K Polycation Ggene Delivery Systems: Escape from Endosomes to Cytosol. J. Pharm. Pharmacol 2003, 55, 721–734. [DOI] [PubMed] [Google Scholar]
- (273).Søndergaard RV; Christensen NM; Henriksen JR; Kumar EP; Almdal K; Andresen TL Facing the Design Challenges of Particle-based Nanosensors for Metabolite Quantification in Living Cells. Chem. Rev 2015, 115, 8344–8378. [DOI] [PubMed] [Google Scholar]
- (274).Wagner E Strategies to Improve DNA Polyplexes for in vivo Gene Transfer: Will “Artificial Viruses” Be the Answer? Pharm. Res 2004, 21, 8–14. [DOI] [PubMed] [Google Scholar]
- (275).Wang Z; Luo M; Mao C; Wei Q; Zhao T; Li Y; Huang G; Gao J A Redox-Activatable Fluorescent Sensor for the High-Throughput Quantification of Cytosolic Delivery of Macromolecules. Angew. Chem., Int. Ed 2017, 56, 1319–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (276).Fu L; Yuan P; Ruan Z; Liu L; Li T; Yan L Ultra-pH-Sensitive Polypeptide Micelles with Large Fluorescence Off/On Ratio in Near Infrared Range. Polym. Chem 2017, 8, 1028–1038. [Google Scholar]
- (277).Li Y; Wang Z; Wei Q; Luo M; Huang G; Sumer BD; Gao J Non-Covalent Interactions in Controlling pH-Responsive Behaviors of Self-Assembled Nanosystems. Polym. Chem 2016, 7, 5949–5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (278).Vargas-Uribe M; Rodnin MV; Ladokhin AS Comparison of Membrane Insertion Pathways of the Apoptotic Regulator Bcl-xL and the Diphtheria Toxin Translocation Domain. Biochemistry 2013, 52, 7901–7909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (279).Jakes KS; Cramer WA Border Crossings: Colicins and Transporters. Annu. Rev. Genet 2012, 46, 209–231. [DOI] [PubMed] [Google Scholar]
- (280).Zakharov SD; Lindeberg M; Cramer WA Kinetic Description of Structural Changes Linked to Membrane Import of the Colicin E1 Channel Protein. Biochemistry 1999, 38, 11325–11332. [DOI] [PubMed] [Google Scholar]
- (281).Zhan H; Oh KJ; Shin Y-K; Hubbell WL; Collier RJ Interaction of the Isolated Transmembrane Domain of Diphtheria Toxin with Membranes. Biochemistry 1995, 34, 4856–4863. [DOI] [PubMed] [Google Scholar]
- (282).Ladokhin AS; Legmann R; Collier RJ; White SH Reversible Refolding of the Diphtheria Toxin T-Domain on Lipid Membranes. Biochemistry 2004, 43, 7451–7458. [DOI] [PubMed] [Google Scholar]
- (283).Andreev OA; Karabadzhak AG; Weerakkody D; Andreev GO; Engelman DM; Reshetnyak YK pH (Low) Insertion Peptide (pHLIP) Inserts across a Lipid Bilayer as A Helix and Exits by a Different Path. Proc. Natl. Acad. Sci. U. S. A 2010, 107, 4081–4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (284).Deacon JC; Engelman DM; Barrera FN Targeting Acidity in Diseased Tissues: Mechanism and Applications of the Membrane- Inserting Peptide, pHLIP. Arch. Biochem. Biophys 2015, 565, 40–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (285).Andreev OA; Dupuy AD; Segala M; Sandugu S; Serra DA; Chichester CO; Engelman DM; Reshetnyak YK Mechanism and Uses of A Membrane Peptide That Targets Tumors and Other Acidic Tissues in vivo. Proc. Natl. Acad. Sci. U. S. A 2007, 104, 7893–7898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (286).An M; Wijesinghe D; Andreev OA; Reshetnyak YK; Engelman DM pH-(Low)-Insertion-Peptide (pHLIP) Translocation of Membrane Impermeable Phalloidin Toxin Inhibits Cancer Cell Proliferation. Proc. Natl. Acad. Sci. U. S. A 2010, 107, 20246–20250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (287).Weerakkody D; Moshnikova A; Thakur MS; Moshnikova V; Daniels J; Engelman DM; Andreev OA; Reshetnyak YK Family of pH (Low) Insertion Peptides for Tumor Targeting. Proc. Natl. Acad. Sci. U. S. A 2013, 110, 5834–5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (288).Barrera FN; Fendos J; Engelman DM Membrane Physical Properties Influence Transmembrane Helix Formation. Proc. Natl. Acad. Sci. U. S. A 2012, 109, 14422–14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (289).Reshetnyak YK; Andreev OA; Segala M; Markin VS; Engelman DM Energetics of Peptide (pHLIP) Binding to and Folding Across A Lipid Bilayer Membrane. Proc. Natl. Acad. Sci. U. S. A 2008, 105, 15340–15345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (290).Hunt JF; Rath P; Rothschild KJ; Engelman DM Spontaneous, pH-Dependent Membrane Insertion of A Transbilayer α-Helix. Biochemistry 1997, 36, 15177–15192. [DOI] [PubMed] [Google Scholar]
- (291).Scholtz JM; Baldwin RL The Mechanism of Alpha-Helix Formation by Peptides. Annu. Rev. Biophys. Biomol. Struct 1992, 21, 95–118. [DOI] [PubMed] [Google Scholar]
- (292).Barrera FN; Weerakkody D; Anderson M; Andreev OA; Reshetnyak YK; Engelman DM Roles of Carboxyl Groups in The Transmembrane Insertion of Peptides. J. Mol. Biol 2011, 413, 359–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (293).Karabadzhak AG; Weerakkody D; Wijesinghe D; Thakur MS; Engelman DM; Andreev OA; Markin VS; Reshetnyak YK Modulation of the pHLIP Transmembrane Helix Insertion Pathway. Biophys. J 2012, 102, 1846–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (294).Dinner AR; Šali A; Smith LJ; Dobson CM; Karplus M Understanding Protein Folding via Free-Energy Surfaces from Theory and Experiment. Trends Biochem. Sci 2000, 25, 331–339. [DOI] [PubMed] [Google Scholar]
- (295).Dobson CM Protein Folding and Misfolding. Nature 2003, 426, 884–890. [DOI] [PubMed] [Google Scholar]
- (296).Keskin O; Gursoy A; Ma B; Nussinov R Principles of Protein−Protein Interactions: What Are the Preferred Ways for Proteins to Interact? Chem. Rev 2008, 108, 1225–1244. [DOI] [PubMed] [Google Scholar]
- (297).Jackson SE; Fersht AR Folding of Chymotrypsin Inhibitor 2. 1. Evidence for A Two-State Transition. Biochemistry 1991, 30, 10428–10435. [DOI] [PubMed] [Google Scholar]
- (298).Tsai CJ; Lin SL; Wolfson HJ; Nussinov R Studies of Protein-Protein Interfaces: A Statistical Analysis of the Hydrophobic Effect. Protein Sci 1997, 6, 53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (299).Tombola F; Ulbrich MH; Kohout SC; Isacoff EY The Opening of the Two Pores of the Hv1 Voltage-Gated Proton Channel is Tuned by Cooperativity. Nat. Struct. Mol. Biol 2010, 17, 44–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (300).Tanaka F; Koga T; Kaneda I; Winnik FM Hydration, Phase Separation and Nonlinear Rheology of Temperature-Sensitive Water-Soluble Polymers. J. Phys.: Condens. Matter 2011, 23, 284105. [DOI] [PubMed] [Google Scholar]
- (301).Wu C; Zhou S First Observation of the Molten Globule State of A Single Homopolymer Chain. Phys. Rev. Lett 1996, 77, 3053–3055. [DOI] [PubMed] [Google Scholar]
- (302).Hunter CA; Anderson HL What is Cooperativity? Angew. Chem., Int. Ed 2009, 48, 7488–7499. [DOI] [PubMed] [Google Scholar]
- (303).Zhang B; Breslow R Enthalpic Domination of the Chelate Effect in Cyclodextrin Dimers. J. Am. Chem. Soc 1993, 115, 9353–9354. [Google Scholar]
- (304).Terzi E; Hölzemann G; Seelig J Self-Association of β-Amyloid Peptide (1−40) in Solution and Binding to Lipid Membranes. J. Mol. Biol 1995, 252, 633–642. [DOI] [PubMed] [Google Scholar]
- (305).Zhao D; Moore JS Nucleation−Elongation: A Mechanism for Cooperative Supramolecular Polymerization. Org. Biomol. Chem 2003, 1, 3471–3491. [DOI] [PubMed] [Google Scholar]
- (306).Baumgartner R; Fu H; Song Z; Lin Y; Cheng J Cooperative Polymerization of α-Helices Induced by Macromolecular Architecture. Nat. Chem 2017, 9, 614–622. [DOI] [PubMed] [Google Scholar]
- (307).Weiss JN The Hill Equation Revisited: Uses and Misuses. FASEB J 1997, 11, 835–841. [PubMed] [Google Scholar]
- (308).Lopez-Fontal E; Milanesi L; Tomas S Multivalence Cooperativity Leading to “All-or-Nothing” Assembly: the Case of Nucleation-Growth in Supramolecular Polymers. Chem. Sci 2016, 7, 4468–4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (309).Filot IA; Palmans AR; Hilbers PA; van Santen RA; Pidko EA; de Greef TF Understanding Cooperativity in Hydrogen- Bond-Induced Supramolecular Polymerization: A Density Functional Theory Study. J. Phys. Chem. B 2010, 114, 13667–13674. [DOI] [PubMed] [Google Scholar]
- (310).Goutelle S; Maurin M; Rougier F; Barbaut X; Bourguignon L; Ducher M; Maire P The Hill Equation: A Review of Its Capabilities in Pharmacological Modelling. Fundam. Clin. Pharmacol 2008, 22, 633–648. [DOI] [PubMed] [Google Scholar]
- (311).Yifrach O Hill Coefficient for Estimating the Magnitude of Cooperativity in Gating Transitions of Voltage-Dependent Ion Channels. Biophys. J 2004, 87, 822–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (312).Williams DH; Stephens E; O’Brien DP; Zhou M Understanding Noncovalent Interactions: Ligand Binding Energy and Catalytic Efficiency from Ligand-Induced Reductions in Motion within Receptors and Enzymes. Angew. Chem., Int. Ed 2004, 43, 6596–6616. [DOI] [PubMed] [Google Scholar]
- (313).Camara-Campos A; Musumeci D; Hunter CA; Turega S Chemical Double Mutant Cycles for the Quantification of Cooperativity in H-Bonded Complexes. J. Am. Chem. Soc 2009, 131, 18518–18524. [DOI] [PubMed] [Google Scholar]
- (314).Ercolani G Assessment of Cooperativity in Self-Assembly. J. Am. Chem. Soc 2003, 125, 16097–16103. [DOI] [PubMed] [Google Scholar]
- (315).Valeur B; Berberan-Santos MN Molecular Fluorescence: Principles and Applications; John Wiley & Sons, 2012; p 210. [Google Scholar]
- (316).Brown A Analysis of Cooperativity by Isothermal Titration Calorimetry. Int. J. Mol. Sci 2009, 10, 3457–3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (317).Cruzan J; Braly L; Liu K; Brown M; Loeser J Quantifying Hydrogen Bond Cooperativity in Water: VRT Spectroscopy of the Water Tetramer. Science 1996, 271, 59–62. [DOI] [PubMed] [Google Scholar]
- (318).Andersen PS; Schuck P; Sundberg EJ; Geisler C; Karjalainen K; Mariuzza RA Quantifying the Energetics of Cooperativity in A Ternary Protein Complex. Biochemistry 2002, 41, 5177–5184. [DOI] [PubMed] [Google Scholar]
- (319).Dexter F; Hindman BJ Theoretical Analysis of Cerebral Venous Blood Hemoglobin Oxygen Saturation as an Index of Cerebral Oxygenation during Hypothermic Cardiopulmonary Bypass A Counter-proposal to the Luxury Perfusion Hypothesis. Anesthesiology 1995, 83, 405–412. [DOI] [PubMed] [Google Scholar]
- (320).Sourjik V; Berg HC Functional Interactions between Receptors in Bacterial Chemotaxis. Nature 2004, 428, 437–441. [DOI] [PubMed] [Google Scholar]
- (321).Mello BA; Tu Y Quantitative Modeling of Sensitivity in Bacterial Chemotaxis: the Role of Coupling Among Different Chemoreceptor Species. Proc. Natl. Acad. Sci. U. S. A 2003, 100, 8223–8228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (322).Duke T; Bray D Heightened Sensitivity of A Lattice of Membrane Receptors. Proc. Natl. Acad. Sci. U. S. A 1999, 96, 10104–10108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (323).Bray D; Levin MD; Morton-Firth CJ Receptor Clustering As A Cellular Mechanism to Control Sensitivity. Nature 1998, 393, 85–88. [DOI] [PubMed] [Google Scholar]
- (324).Ferrell JE Jr.; Machleder EM The Biochemical Basis of An All-or-None Cell Fate Switch in Xenopus Oocytes. Science 1998, 280, 895–898. [DOI] [PubMed] [Google Scholar]
- (325).Li Y; Wang Y; Huang G; Ma X; Zhou K; Gao J Chaotropic-Anion-Induced Supramolecular Self-Assembly of Ionic Polymeric Micelles. Angew. Chem., Int. Ed 2014, 53, 8074–8078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (326).Christian DA; Tian A; Ellenbroek WG; Levental I; Rajagopal K; Janmey PA; Liu AJ; Baumgart T; Discher DE Spotted Vesicles, Striped Micelles and Janus Assemblies Induced by Ligand Binding. Nat. Mater 2009, 8, 843–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (327).Fang J; Nakamura H; Maeda H The EPR Effect: Unique Features of Tumor Blood Vessels for Drug Delivery, Factors Involved, and Limitations and Augmentation of the Effect. Adv. Drug Delivery Rev 2011, 63, 136–151. [DOI] [PubMed] [Google Scholar]
- (328).Maeda H; Wu J; Sawa T; Matsumura Y; Hori K Tumor Vascular Permeability and the EPR Effect in Macromolecular Therapeutics: A Review. J. Controlled Release 2000, 65, 271–284. [DOI] [PubMed] [Google Scholar]
- (329).Gottesman MM; Fojo T; Bates SE Multidrug Resistance in Cancer: Role of ATP−Dependent Transporters. Nat. Rev. Cancer 2002, 2, 48–58. [DOI] [PubMed] [Google Scholar]
- (330).Patel NR; Pattni BS; Abouzeid AH; Torchilin VP Nanopreparations to Overcome Multidrug Resistance in Cancer. Adv. Drug Delivery Rev 2013, 65, 1748–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (331).Peer D; Karp JM; Hong S; Farokhzad OC; Margalit R; Langer R Nanocarriers as An Emerging Platform for Cancer Therapy. Nat. Nanotechnol 2007, 2, 751–760. [DOI] [PubMed] [Google Scholar]
- (332).Schluep T; Hwang J; Cheng J; Heidel JD; Bartlett DW; Hollister B; Davis ME Preclinical Efficacy of the Camptothecin- Polymer Conjugate IT-101 in Multiple Cancer Models. Clin. Cancer Res 2006, 12, 1606–1614. [DOI] [PubMed] [Google Scholar]
- (333).Ho K; Lapitsky Y; Shi M; Shoichet MS Tunable Immunonanoparticle Binding to Cancer Cells: Thermodynamic Analysis of Targeted Drug Delivery Vehicles. Soft Matter 2009, 5, 1074–1080. [Google Scholar]
- (334).Lane LA; Qian X; Smith AM; Nie S Physical Chemistry of Nanomedicine: Understanding the Complex Behaviors of Nanoparticles in vivo. Annu. Rev. Phys. Chem 2015, 66, 521–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (335).Hong S; Leroueil PR; Majoros IJ; Orr BG; Baker JR; Holl MMB The Binding Avidity of A Nanoparticle-based Multivalent Targeted Drug Delivery Platform. Chem. Biol 2007, 14, 107–115. [DOI] [PubMed] [Google Scholar]
- (336).Mammen M; Choi S-K; Whitesides GM Polyvalent Interactions in Biological Systems: Implications for Design and Use of Multivalent Ligands and Inhibitors. Angew. Chem., Int. Ed 1998, 37, 2754–2794. [DOI] [PubMed] [Google Scholar]
- (337).Zwicke GL; Ali Mansoori G; Jeffery CJ Utilizing the Folate Receptor for Active Targeting of Cancer Nanotherapeutics. Nano Rev 2012, 3, 18496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (338).Prescher JA; Bertozzi CR Chemistry in Living Systems. Nat. Chem. Biol 2005, 1, 13–21. [DOI] [PubMed] [Google Scholar]
- (339).Koo H; Lee S; Na JH; Kim SH; Hahn SK; Choi K; Kwon IC; Jeong SY; Kim K Bioorthogonal Copper-Free Click Chemistry In Vivo for Tumor-Targeted Delivery of Nanoparticles. Angew. Chem., Int. Ed 2012, 51, 11836–11840. [DOI] [PubMed] [Google Scholar]
- (340).Luong JH; Male KB; Glennon JD Biosensor Technology: Technology Push versus Market Pull. Biotechnol. Adv 2008, 26, 492–500. [DOI] [PubMed] [Google Scholar]
- (341).James ML; Gambhir SS A Molecular Imaging Primer: Modalities, Imaging Agents, and Applications. Physiol. Rev 2012, 92, 897–965. [DOI] [PubMed] [Google Scholar]
- (342).Holzinger M; Le Goff A; Cosnier S Nanomaterials for Biosensing Applications: A Review. Front. Chem 2014, 2, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (343).Davis ME; Shin DM Nanoparticle Therapeutics: An Emerging Treatment Modality for Cancer. Nat. Rev. Drug Discovery 2008, 7, 771–782. [DOI] [PubMed] [Google Scholar]
- (344).Lei J; Ju H Signal Amplification Using Functional Nanomaterials for Biosensing. Chem. Soc. Rev 2012, 41, 2122–2134. [DOI] [PubMed] [Google Scholar]
- (345).Taton TA; Mirkin CA; Letsinger RL Scanometric DNA Array Detection with Nanoparticle Probes. Science 2000, 289, 1757–1760. [DOI] [PubMed] [Google Scholar]
- (346).Nam J-M; Thaxton CS; Mirkin CA Nanoparticle-based Bio-Bar Codes for the Ultrasensitive Detection of Proteins. Science 2003, 301, 1884–1886. [DOI] [PubMed] [Google Scholar]
- (347).Han MS; Lytton-Jean AK; Oh BK; Heo J; Mirkin CA Colorimetric Screening of DNA-Binding Molecules with Gold Nanoparticle Probes. Angew. Chem., Int. Ed 2006, 45, 1807–1810. [DOI] [PubMed] [Google Scholar]
- (348).Liu C-W; Hsieh Y-T; Huang C-C; Lin Z-H; Chang H-T Detection of Mercury (II) based on Hg 2+−DNA Complexes Inducing the Aggregation of Gold Nanoparticles. Chem. Commun 2008, 44, 2242–2244. [DOI] [PubMed] [Google Scholar]
- (349).Seferos DS; Giljohann DA; Hill HD; Prigodich AE; Mirkin CA Nano-Flares: Probes for Transfection and mRNA Detection in Living Cells. J. Am. Chem. Soc 2007, 129, 15477–15479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (350).Vogelstein B; Papadopoulos N; Velculescu VE; Zhou S; Diaz LA; Kinzler KW Cancer Genome Landscapes. Science 2013, 339, 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (351).Van Dam GM; Themelis G; Crane LM; Harlaar NJ; Pleijhuis RG; Kelder W; Sarantopoulos A; De Jong JS; Arts HJ; Van Der Zee AG; et al. Intraoperative Tumor-Specific Fluorescence Imaging in Ovarian Cancer by Folate Receptor-[Alpha] Targeting: First in-Human Results. Nat. Med 2011, 17, 1315–1319. [DOI] [PubMed] [Google Scholar]
- (352).Veiseh M; Gabikian P; Bahrami S-B; Veiseh O; Zhang M; Hackman RC; Ravanpay AC; Stroud MR; Kusuma Y; Hansen SJ; et al. Tumor Paint: A Chlorotoxin: Cy5. 5 Bioconjugate for Intraoperative Visualization of Cancer Foci. Cancer Res 2007, 67, 6882–6888. [DOI] [PubMed] [Google Scholar]
- (353).Ke S; Wen X; Gurfinkel M; Charnsangavej C; Wallace S; Sevick-Muraca EM; Li C Near-Infrared Optical Imaging of Epidermal Growth Factor Receptor in Breast Cancer Xenografts. Cancer Res 2003, 63, 7870–7875. [PubMed] [Google Scholar]
- (354).Nakajima T; Mitsunaga M; Bander NH; Heston WD; Choyke PL; Kobayashi H Targeted, Activatable, in vivo Fluorescence Imaging of Prostate-Specific Membrane Antigen (PSMA) Positive Tumors Using the Quenched Humanized J591 Antibody−Indocyanine Green (ICG) Conjugate. Bioconjugate Chem 2011, 22, 1700–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (355).Paik S; Bryant J; Tan-Chiu E; Yothers G; Park C; Wickerham DL; Wolmark N HER2 and Choice of Adjuvant Chemotherapy for Invasive Breast Cancer: National Surgical Adjuvant Breast and Bowel Project Protocol B-15. J. Natl. Cancer Inst 2000, 92, 1991–1998. [DOI] [PubMed] [Google Scholar]
- (356).Jacobs TW; Gown AM; Yaziji H; Barnes MJ; Schnitt SJ HER-2/neu Protein Expression in Breast Cancer Evaluated by Immunohistochemistry: A Study of Interlaboratory Agreement. Am. J. Clin. Pathol 2000, 113, 251–258. [DOI] [PubMed] [Google Scholar]
- (357).Švastová E; Hulíková A; Rafajová M; Zat’ovičová M; Gibadulinová A; Casini A; Cecchi A; Scozzafava A; Supuran CT; Pastorek J; et al. Hypoxia Activates the Capacity of Tumor-Associated Carbonic Anhydrase IX to Acidify Extracellular pH. FEBS Lett 2004, 577, 439–445. [DOI] [PubMed] [Google Scholar]
- (358).Xu X; Wu J; Liu Y; Yu M; Zhao L; Zhu X; Bhasin S; Li Q; Ha E; Shi J; et al. Ultra-pH-Responsive and Tumor-Penetrating Nanoplatform for Targeted siRNA Delivery with Robust Anti-Cancer Efficacy. Angew. Chem., Int. Ed 2016, 55, 7091–7094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (359).Yu H; Guo C; Feng B; Liu J; Chen X; Wang D; Teng L; Li Y; Yin Q; Zhang Z; et al. Triple-Layered pH-Responsive Micelleplexes Loaded with siRNA and Cisplatin Prodrug for NF-Kappa B Targeted Treatment of Metastatic Breast Cancer. Theranostics 2016, 6, 14–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (360).Cho K; Wang X; Nie S; Shin DM Therapeutic Nanoparticles for Drug Delivery in Cancer. Clin. Cancer Res 2008, 14, 1310–1316. [DOI] [PubMed] [Google Scholar]
- (361).Zhao T; Huang G; Li Y; Yang S; Ramezani S; Lin Z; Wang Y; Ma X; Zeng Z; Luo M A Transistor-like pH Nanoprobe for Tumour Detection and Image-Guided Surgery. Nat. Biomed. Eng 2017, 1, 0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (362).Wang Y; Wang C; Li Y; Huang G; Zhao T; Ma X; Wang Z; Sumer BD; White MA; Gao J Digitization of Endocytic pH by Hybrid Ultra-pH-Sensitive Nanoprobes at Single-Organelle Resolution. Adv. Mater 2017, 29, 1603794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (363).Wang Y; Kohane DS External Triggering and Triggered Targeting Strategies for Drug Delivery. Nat. Rev. Mater 2017, 2, 17020. [Google Scholar]
- (364).Kono K Thermosensitive Polymer-Modified Liposomes. Adv. Drug Delivery Rev 2001, 53, 307–319. [DOI] [PubMed] [Google Scholar]
- (365).Liu W; MacKay JA; Dreher MR; Chen M; McDaniel JR; Simnick AJ; Callahan DJ; Zalutsky MR; Chilkoti A Injectable Intratumoral Depot of Thermally Responsive Polypeptide−Radionuclide Conjugates Delays Tumor Progression in A Mouse Model. J. Controlled Release 2010, 144, 2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (366).McDaniel JR; MacEwan SR; Li X; Radford DC; Landon CD; Dewhirst M; Chilkoti A Rational Design of “Heat Seeking” Drug Loaded Polypeptide Nanoparticles that Thermally Target Solid Tumors. Nano Lett 2014, 14, 2890–2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (367).Al-Ahmady ZS; Al-Jamal WT; Bossche JV; Bui TT; Drake AF; Mason AJ; Kostarelos K Lipid−Peptide Vesicle Nanoscale Hybrids for Triggered Drug Release by Mild Hyperthermia in vitro and in vivo. ACS Nano 2012, 6, 9335–9346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (368).Nath N; Chilkoti A Interfacial Phase Transition of An Environmentally Responsive Elastin Biopolymer Adsorbed on Functionalized Gold Nanoparticles Studied by Colloidal Surface Plasmon Resonance. J. Am. Chem. Soc 2001, 123, 8197–8202. [DOI] [PubMed] [Google Scholar]
- (369).Park SM; Kim MS; Park S-J; Park ES; Choi K-S; Kim Y -s.; Kim, H. R. Novel Temperature-Triggered Liposome with High Stability: Formulation, in vitro Evaluation, and in vivo Study Combined with High-Intensity Focused Ultrasound (HIFU). J. Controlled Release 2013, 170, 373–379. [DOI] [PubMed] [Google Scholar]
- (370).Kojima C; Irie K Synthesis of Temperature-Dependent Elastin-Like Peptide-Modified Dendrimer for Drug Delivery. Biopolymers 2013, 100, 714–721. [DOI] [PubMed] [Google Scholar]
- (371).Shamji MF; Betre H; Kraus VB; Chen J; Chilkoti A; Pichika R; Masuda K; Setton LA Development and Characterization of A Fusion Protein between Thermally Responsive Elastin-Like Polypeptide and Interleukin-1 Receptor Antagonist: Sustained Release of A Local Antiinflammatory Therapeutic. Arthritis Rheum 2007, 56, 3650–3661. [DOI] [PubMed] [Google Scholar]
- (372).Betre H; Liu W; Zalutsky MR; Chilkoti A; Kraus VB; Setton LA A Thermally Responsive Biopolymer for Intra-Articular Drug Delivery. J. Controlled Release 2006, 115, 175–182. [DOI] [PubMed] [Google Scholar]
- (373).Shamji MF; Chen J; Friedman AH; Richardson WJ; Chilkoti A; Setton LA Synthesis and Characterization of A Thermally-Responsive Tumor Necrosis Factor Antagonist. J. Controlled Release 2008, 129, 179–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (374).Amiram M; Luginbuhl KM; Li X; Feinglos MN; Chilkoti A Injectable Protease-Operated Depots of glucagon-like peptide-1 provide extended and tunable glucose control. Proc. Natl. Acad. Sci. U. S. A 2013, 110, 2792–2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (375).Lynn DM; Langer R Degradable poly (β-amino esters): synthesis, characterization, and self-assembly with plasmid DNA. J. Am. Chem. Soc 2000, 122, 10761–10768. [Google Scholar]
- (376).Zhou D; Cutlar L; Gao Y; Wang W; O’Keeffe-Ahern J; McMahon S; Duarte B; Larcher F; Rodriguez BJ; Greiser U The Transition from Linear to Highly Branched Poly (β-Amino Ester)s: Branching Matters for Gene Delivery. Sci. Adv 2016, 2, e1600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (377).Newland B; Zheng Y; Jin Y; Abu-Rub M; Cao H; Wang W; Pandit A Single Cyclized Molecule versus Single Branched Molecule: A Simple and Efficient 3D “Knot” Polymer Structure for Nonviral Gene Delivery. J. Am. Chem. Soc 2012, 134, 4782–4789. [DOI] [PubMed] [Google Scholar]
- (378).Zhou D; Pierucci L; Gao Y; O’Keeffe Ahern J; Huang X; Sigen A; Wang W Thermo-and pH-Responsive, Coacervate-Forming Hyperbranched Poly (β-amino ester) s for Selective Cell Binding. ACS Appl. Mater. Interfaces 2017, 9, 5793–5802. [DOI] [PubMed] [Google Scholar]
- (379).Kim B; Rutka JT; Chan WC Nanomedicine. N. Engl. J. Med 2010, 363, 2434–2443. [DOI] [PubMed] [Google Scholar]
- (380).Bertrand N; Wu J; Xu X; Kamaly N; Farokhzad OC Cancer Nanotechnology: the Impact of Passive and Active Targeting in the Era of Modern Cancer Biology. Adv. Drug Delivery Rev 2014, 66, 2–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (381).Kim BY; Rutka JT; Chan WC Nanomedicine. N. Engl. J. Med 2010, 363, 2434–2443. [DOI] [PubMed] [Google Scholar]
- (382).Keereweer S; Kerrebijn JD; Van Driel PB; Xie B; Kaijzel EL; Snoeks TJ; Que I; Hutteman M; Van der Vorst JR; Mieog JSD; et al. Optical Image-Guided Surgery—Where Do We Stand? Mol. Imaging Biol 2011, 13, 199–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (383).Doane TL; Burda C The Unique Role of Nanoparticles in Nanomedicine: Imaging, Drug Delivery and Therapy. Chem. Soc. Rev 2012, 41, 2885–2911. [DOI] [PubMed] [Google Scholar]
- (384).Blanco E; Shen H; Ferrari M Principles of Nanoparticle Design for Overcoming Biological Barriers to Drug Delivery. Nat. Biotechnol 2015, 33, 941–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (385).Sumer B; Gao J Theranostic nanomedicine for Cancer. Nanomedicine 2008, 3, 137–140. [DOI] [PubMed] [Google Scholar]
- (386).Ferrari M Cancer Nanotechnology: Opportunities and Challenges. Nat. Rev. Cancer 2005, 5, 161–171. [DOI] [PubMed] [Google Scholar]
- (387).Freitas RA What Is Nanomedicine? Nanomedicine 2005, 1, 2–9. [DOI] [PubMed] [Google Scholar]
- (388).Etheridge ML; Campbell SA; Erdman AG; Haynes CL; Wolf SM; McCullough J The Big Picture on Nanomedicine: the State of Investigational and Approved Nanomedicine Products. Nanomedicine 2013, 9, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (389).Bawa R Regulating Nanomedicine-Can the FDA Handle It? Curr. Drug Delivery 2011, 8, 227–234. [DOI] [PubMed] [Google Scholar]
- (390).Nie S Understanding and Overcoming Major Barriers in Cancer Nanomedicine. Nanomedicine 2010, 5, 523–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (391).Wilhelm S; Tavares AJ; Dai Q; Ohta S; Audet J; Dvorak HF; Chan WC Analysis of Nanoparticle Delivery to Tumours. Nat. Rev. Mater 2016, 1, 16014. [Google Scholar]
- (392).Morton SW; Lee MJ; Deng ZJ; Dreaden EC; Siouve E; Shopsowitz KE; Shah NJ; Yaffe MB; Hammond PT A Nanoparticle-based Combination Chemotherapy Delivery System for Enhanced Tumor Killing by Dynamic Rewiring of Signaling Pathways. Sci. Signaling 2014, 7, ra44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (393).Conde J; Oliva N; Artzi N Revisiting the ‘One Material Fits All’Rule for Cancer Nanotherapy. Trends Biotechnol 2016, 34, 618–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (394).Chauhan VP; Stylianopoulos T; Martin JD; Popović Z; Chen O; Kamoun WS; Bawendi MG; Fukumura D; Jain RK Normalization of Tumour Blood Bessels Improves the Delivery of Nanomedicines in A Size-Dependent Manner. Nat. Nanotechnol 2012, 7, 383–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (395).Wang H; Gauthier M; Kelly JR; Miller RJ; Xu M; O’Brien WD; Cheng J Targeted Ultrasound-Assisted Cancer- Selective Chemical Labeling and Subsequent Cancer Imaging using Click Chemistry. Angew. Chem., Int. Ed 2016, 55, 5452–5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (396).Jain RK Normalizing Tumor Microenvironment to Treat Cancer: Bench to Bedside to Biomarkers. J. Clin. Oncol 2013, 31, 2205–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (397).Conde J; Oliva N; Zhang Y; Artzi N Local Triple- Combination Therapy Results in Tumour Regression and Prevents Recurrence in A Colon Cancer Model. Nat. Mater 2016, 15, 1128–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (398).Xu X; Xie K; Zhang X-Q; Pridgen EM; Park GY; Cui DS; Shi J; Wu J; Kantoff PW; Lippard SJ Enhancing Tumor Cell Response to Chemotherapy through Nanoparticle-Mediated Codelivery of siRNA and Cisplatin Prodrug. Proc. Natl. Acad. Sci. U. S. A 2013, 110, 18638–18643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (399).Sengupta S; Eavarone D; Capila I; Zhao G; Watson N; Kiziltepe T; Sasisekharan R Temporal Targeting of Tumour Cells and Neovasculature with A Nanoscale Delivery System. Nature 2005, 436, 568–572. [DOI] [PubMed] [Google Scholar]
- (400).Kemp JA; Shim MS; Heo CY; Kwon YJ Combo” Nanomedicine: Co-Delivery of Multi-Modal Therapeutics for Efficient, Targeted, and Safe Cancer Therapy. Adv. Drug Delivery Rev 2016, 98, 3–18. [DOI] [PubMed] [Google Scholar]
- (401).Pacardo DB; Ligler FS; Gu Z Programmable Nanomedicine: Synergistic and Sequential Drug Delivery Systems. Nanoscale 2015, 7, 3381–3391. [DOI] [PubMed] [Google Scholar]
- (402).Goren D; Horowitz AT; Tzemach D; Tarshish M; Zalipsky S; Gabizon A Nuclear Delivery of Doxorubicin via Folate-Targeted Liposomes with Bypass of Multidrug-Resistance Efflux Pump. Clin. Cancer Res 2000, 6, 1949–1957. [PubMed] [Google Scholar]
- (403).Ashley CE; Carnes EC; Phillips GK; Padilla D; Durfee PN; Brown PA; Hanna TN; Liu J; Phillips B; Carter MB; et al. The Targeted Delivery of Multicomponent Cargos to Cancer Cells by Nanoporous Particle-Supported Lipid Bilayers. Nat. Mater 2011, 10, 389–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (404).Karkuszewski ZP; Jarzynski C; Zurek WH Quantum Chaotic Environments, the Butterfly Effect, and Decoherence. Phys. Rev. Lett 2002, 89, 170405. [DOI] [PubMed] [Google Scholar]
- (405).Shinbrot T; Grebogi C; Yorke JA; Ott E Using Small Perturbations to Control Chaos. Nature 1993, 363, 411–417. [Google Scholar]
- (406).Atkins P; De Paula J Elements of physical chemistry; Oxford University Press: New York, 2009; p 175. [Google Scholar]