Abstract
FAM3C/Interleukin-like EMT Inducer (ILEI) is an oncogenic member of the FAM3 cytokine family and serves essential roles in both epithelial-mesenchymal transition (EMT) and breast cancer metastasis. ILEI expression levels are regulated through a non-canonical TGFβ signaling pathway by 3’-UTR-mediated translational silencing at the mRNA level by hnRNP E1. TGFβ stimulation or silencing of hnRNP E1 increases ILEI translation and induces an EMT program that correlates to enhanced invasion and migration. Recently, EMT has been linked to the formation of breast cancer stem cells (BCSCs) that confer both tumor cell heterogeneity as well as chemoresistant properties. Herein, we demonstrate that hnRNP E1 knockdown significantly shifts normal mammary epithelial cells to mesenchymal BCSCs in vitro and in vivo. We further validate that modulating ILEI protein levels results in the abrogation of these phenotypes, promoting further investigation into the unknown mechanism of ILEI signaling that drives tumor progression. We identify LIFR as the receptor for ILEI, which mediates signaling through STAT3 to drive both EMT and BCSC formation. Reduction of either ILEI or LIFR protein levels results in reduced tumor growth, fewer tumor initiating cells and reduced metastasis within the hnRNP E1 knock-down cell populations in vivo. These results reveal a novel ligand-receptor complex that drives the formation of BCSCs and represents a unique target for the development of metastatic breast cancer therapies.
Keywords: Breast cancer stem cell, epithelial-mesenchymal transition, tumor initiating cell, ILEI, Fam3C, hnRNP E1, PCBP1, LIFR, metastasis
Introduction
Breast cancer is the third leading cause of cancer-related deaths in the US with approximately 40,000 fatal outcomes each year. Breast cancer patients with late-stage metastasis have a 27% 5-year relative survival rate, a significant decrease in comparison to patients with localized disease (99% 5-year survival) 1. Metastases derived from primary breast lesions (commonly targeted to the bone, brain, liver, lung, and distant lymph nodes) require the coordination of several cellular and molecular processes, including the epithelial-mesenchymal transition (EMT) 1,2. Under normal conditions, EMT is utilized during embryonic development and tissue regeneration where cells undergo a highly dynamic switch between epithelial and mesenchymal phenotypes 3. Mammary epithelial cells acquire migratory and invasive properties upon EMT activation, thereby implicating EMT regulatory mechanisms in breast malignancies 4,5. EMT is facilitated by an array of transcriptional reprogramming, epigenetic modification, and differential expression patterns of epithelial/mesenchymal-specific cell adhesion and cytoskeletal proteins 4. It has also been suggested that the reverse process of EMT, the mesenchymal-epithelial transition (MET), is executed once cancer cells have reached their metastatic niche in order to promote secondary colonization and tumorigenesis 3,6.
The current understanding of EMT in cancer progression has recently shed light on the existence of undifferentiated mammary stem cells known as tumor initiating cells (TICs) or breast cancer stem cells (BCSCs) that are derived from basal mammary epithelial cells (MECs) undergoing EMT and subsequent self-renewal/differentiation, dissemination, and secondary tumorigenesis 7–9. The cells have been extensively characterized due to their ability to drive tumor progression and confer chemo-resistant properties 8,10. BCSCs are identified by both functional assays and differential regulation of established stem cell markers, including CD44/CD24, BMI1, aldehyde dehydrogenase activity, and Nanog 9,11–13.
The link between EMT and BCSC formation has been extensively investigated by groups focused on the pleotropic cytokine transforming growth factor β (TGFβ) and its signaling modalities 14–16. These studies collectively demonstrate the role of TGFβ-induced EMT and BCSC formation within the mammary epithelium. It is therefore important to characterize the molecular mechanisms defining TGFβ-induced BCSCs for the development of new therapies targeting breast cancer metastasis.
Studies from our lab uncovered a master regulator of the mesenchymal proteome during TGFβ-induced EMT. We identified a tumor suppressor ribonucleoprotein (mRNP) complex containing heterogeneous nuclear ribonucleoprotein E1 (hnRNP E1) and eukaryotic elongation factor 1A1 (eEF1a1) that binds a structurally conserved 3’-UTR or BAT element (TGFβ activated translation) and inhibits translation of several mRNAs involved in EMT 17,18. TGFβ signaling initiates a non-canonical kinase cascade that results in Akt2-mediated phosphorylation of hnRNP-E1 at serine 43. Subsequently, hnRNP E1 complexes are released from EMT related mRNA transcripts, translation is restored, and EMT progresses. Using a genome-wide combinatorial approach by expression profiling and RIP-Chip analysis, our lab revealed a cohort of over 30 mRNAs that are controlled by the 3’-UTR BAT mechanism of translational regulation by TGFβ 19. Knockdown of hnRNP E1 within normal mammary epithelial cells results in a transition to a mesenchymal, invasive and migratory state, and drives spontaneous tumor formation in a xenografts mouse model 18. Analysis of the mRNAs translationally regulated by hnRNP E1 revealed FAM3C, also known as interleukin-like EMT inducer (ILEI), a cytokine analyzed for its role in EMT and tumorigenesis.
ILEI is a member of the FAM3 family of secreted cytokines and was discovered through a sequence based search for predicted four-helical bundle cytokines 20. ILEI has been shown to play a role in a variety of biological processes including Alzheimer’s disease, retinal laminar formation, and bone density regulation 21–23. In the context of epithelial tumorigenesis, ILEI serves an essential role in EMT activation and metastatic progression after translational upregulation by TGFβ. Subsequent N-terminal processing and secretion results in the full-length 25kD molecule being reduced to a 18kD variant that functions within the extracellular space 24–26. Although these studies demonstrate an important role for ILEI in disease, mechanistic studies and identification of the extracellular receptor mediating ILEI function has yielded little progress. The previously described role of ILEI in EMT and tumorigenesis gives rise to its possible function in BCSC formation and self-renewal. Indeed, it has been demonstrated that secreted inflammatory cytokines such as interleukins 6 and 8 (IL-6/IL-8) play important roles in BCSC enrichment and chemoresistance 27–29. These findings present an interesting possibility that ILEI may also function in a similar capacity within the context of TGFβ-mediated EMT and BCSC formation.
The IL-6 family of secreted cytokines, that adopt four-helical bundle structures such as leukemia inhibitory factor (LIF), cardiotrophin-1 (CT-1), ciliary neurotrophic factor (CNTF), and oncostatin M (OSM), all utilize leukemia inhibitory factor receptor (LIFR) in complex with glycoprotein 130 (gp130) for downstream signal transduction 30,31. The gp130/LIFR signaling platform is involved in many important cell biological processes including inflammation, bone-remodeling, reproduction, cardiac function, neuromuscular function, the hematopoietic system, and embryonic stem cell (ESC) homeostasis 30. The wide realm of functionality for the gp130/LIFR axis is due to the activation of several major cell-regulatory pathways including the Jak/STAT, Erk/MAPK, and Akt/PI3K signaling pathways 30. In the context of breast cancer progression, LIFR and its ligands have been implicated in both the enhancement and repression of oncogenesis, metastatic progression, and dormancy phenotypes 32–34.
Herein, we aimed to understand the molecular mechanism underlying the TGFβ/hnRNP E1 pathway in the regulation of MEC self-renewal. We determined that loss of hnRNP E1 from normal MECs is sufficient to induce a transition to a BCSC state. Reduction of ILEI levels in hnRNP E1 knockdown cells reduces the tumor initiating properties of these cells both in vitro and in vivo. Additionally, leukemia inhibitory factor receptor (LIFR) and STAT3 were identified as effectors of ILEI signaling in BCSCs. Our data indicate that TGFβ induces the self-renewal capacity of BCSCs through hnRNP E1-dependent induction of ILEI and the subsequent activation of JAK-STAT signaling through LIFR.
Results
TGFβ/hnRNP E1 induces BCSC Self-Renewal through a Secreted Factor
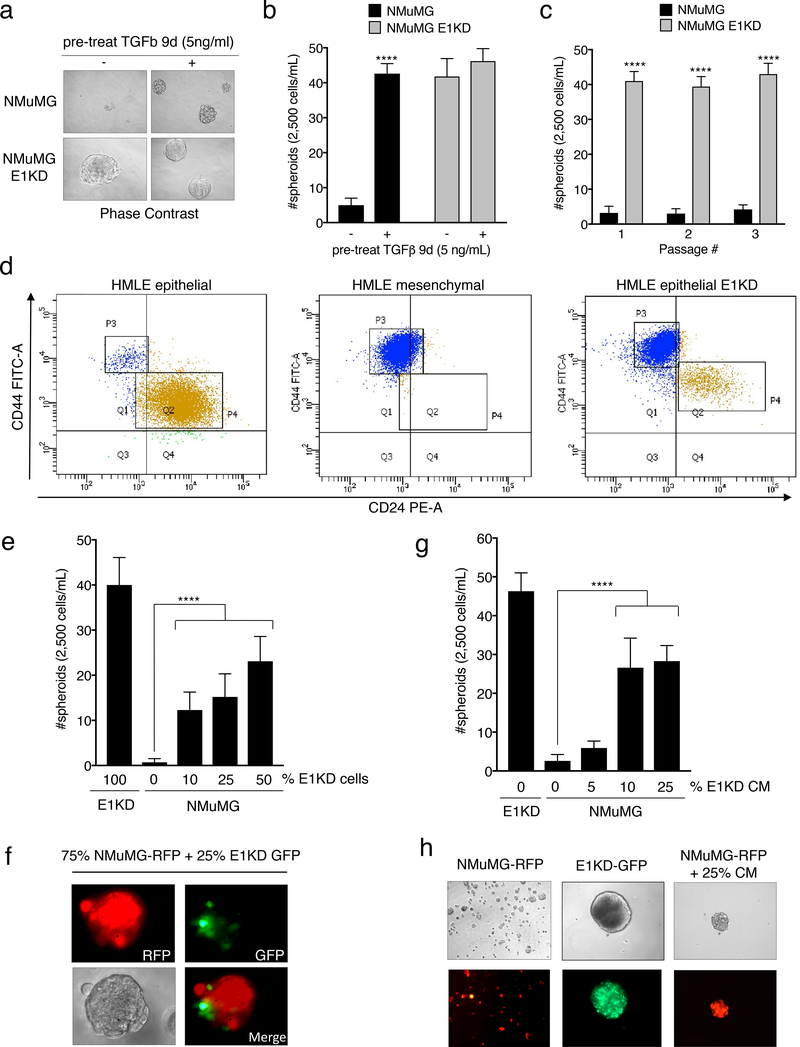
hnRNP E1 has been shown to regulate the migratory and invasive potential of cells undergoing TGFβ-mediated EMT 17. To determine whether hnRNP E1 regulates any of TGFβ’s effects in BCSC self-renewal, we utilized a normal murine mammary gland epithelial cell line (NMuMG) with or without shRNA-guided knockdown of hnRNP E1. These lines were passaged from two-dimensional cultures into non-adherent, serum-free conditions in the presence of limited growth factors to test their capacity to grow and self-renew as spheroid structures or mammospheres. Parental NMuMG cells do not form mammospheres without a 9-day pre-treatment with TGFβ. Upon attenuation of hnRNP E1 (E1KD cells), mammosphere formation is significantly enhanced even in the absence of TGFβ pre-treatment (Fig. 1a-1b; see Fig. S1a for relative expression levels of hnRNP E1 in NMuMG cells). To test self-renewal capacity, both NMuMG and E1KD mammospheres were trypsinized, resuspended, and passaged up to three times. E1KD mammospheres displayed increased self-renewal capacity sustained over multiple passages when compared to the NMuMG spheres, validating hnRNP E1 as an important factor mediating stem cell phenotypes in these lines (Fig. 1c). Interestingly, mammospheres generated from a single hnRNP E1 knockdown cell population stained for both basal (CK5 and CK14) and luminal (CK8 and CK18) cytokeratins, demonstrating that E1KD cells can exhibit both myoepithelial and luminal epithelial cell phenotypes (Fig. S1b) 35. Similarly, to test the contribution of hnRNP E1 to the self-renewal of human mammary cells in vitro, we performed FACS analysis of CD44/CD24 stem cell surface marker expression in epithelial and mesenchymal human mammary epithelial (HMLE) cells, and epithelial HMLE cells attenuated for hnRNP E1 (epithelial E1KD). Knockdown of hnRNP E1 in the HMLE epithelial population results in a switch to a more mesenchymal-like CD44high/CD24low profile (Fig. 1d; see Fig. S1c for relative expression levels of hnRNP E1 in HMLE cells) 14,36.
Figure 1. TGFβ/hnRNP E1 induces BCSC Self-Renewal through a Secreted Factor.
(a) Phase contrast images and (b) quantification of mammosphere formation by NMuMG and NMuMG shE1 (E1KD) cells with or without 9-day TGFβ pre-treatment (error bars represent mean +/− SD; n=5; ****p <0.0001, unpaired Student’s t-test). All mammospheres were counted at a minimum diameter of 100μm. (c) Quantification of sequentially passaged mammospheres from NMuMG and E1KD cells (error bars represent mean +/− SD; n=5; ****p <0.0001, unpaired Student’s t-test). (d) FACS analysis of CD44/CD24 surface marker expression in HMLE epithelial, mesenchymal, and epithelial E1KD populations (HMLE epi P3: 3.4%, P4: 92.7%; HMLE mes P3: 98.0% P4: 0.7%; HMLE epi E1KD P3: 80.8%, P4: 12.5%) (e) Mammosphere quantification and (f) representative images from co-cultured NMuMG-RFP and E1KD-GFP cells as well as (g & h) NMuMG cells supplied with increasing amounts of E1KD conditioned media (error bars represent mean +/− SD; n=10; ****p <0.0001, One-way Anova.
Additionally, Red Fluorescent Protein (RFP)-expressing NMuMG cells were capable of mammosphere growth upon co-culture with Green Fluorescent Protein (GFP)-expressing E1KD cells in a dose-dependent manner, with both cell types exhibiting co-localization within the spheroids (Fig. 1e quantification, Fig. 1f for RFP-NMuMG and GFP-E1KD co-culture images by immunofluorescence). To delineate between self-renewal induced by E1KD generated cell surface proteins and secreted factors, conditioned media (CM) collected from 7-day E1KD mammospheres was used to culture with the NMuMG parental cell line. The CM from E1KD cells was sufficient to increase, in a dose-dependent manner, the mammosphere growth of RFP-expressing NMuMG cells (Fig. 1g quantification; Fig. 1h for NMuMG-RFP mammosphere immunofluorescence upon E1KD conditioned media treatment). Therefore, a secreted factor derived from modulation of hnRNP E1 may play a critical role in TGFβ-induced BCSC formation and self-renewal.
The Secreted Cytokine ILEI is Necessary for TGFβ/hnRNP E1-Mediated EMT and BCSC formation
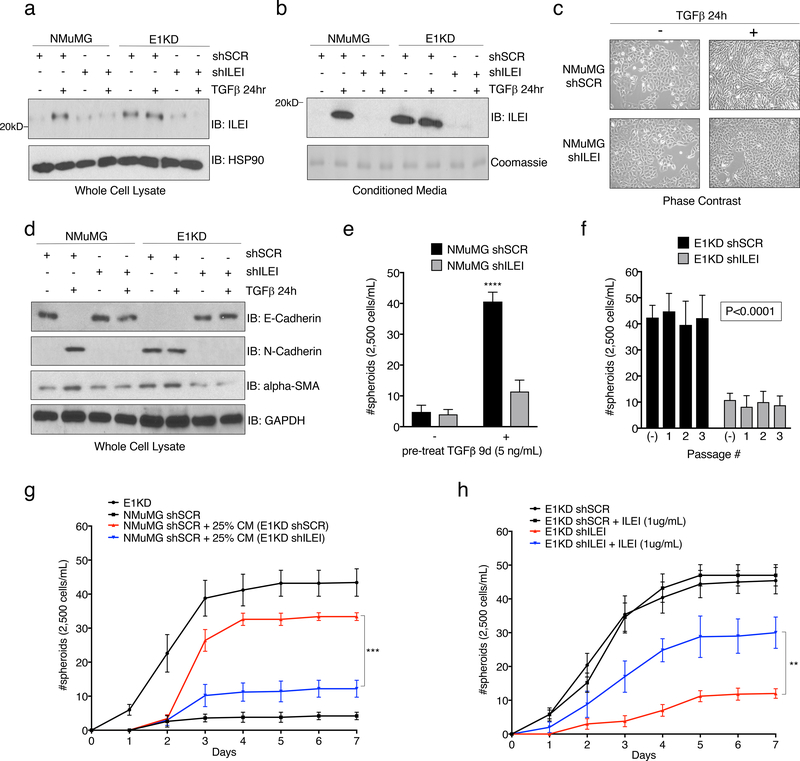
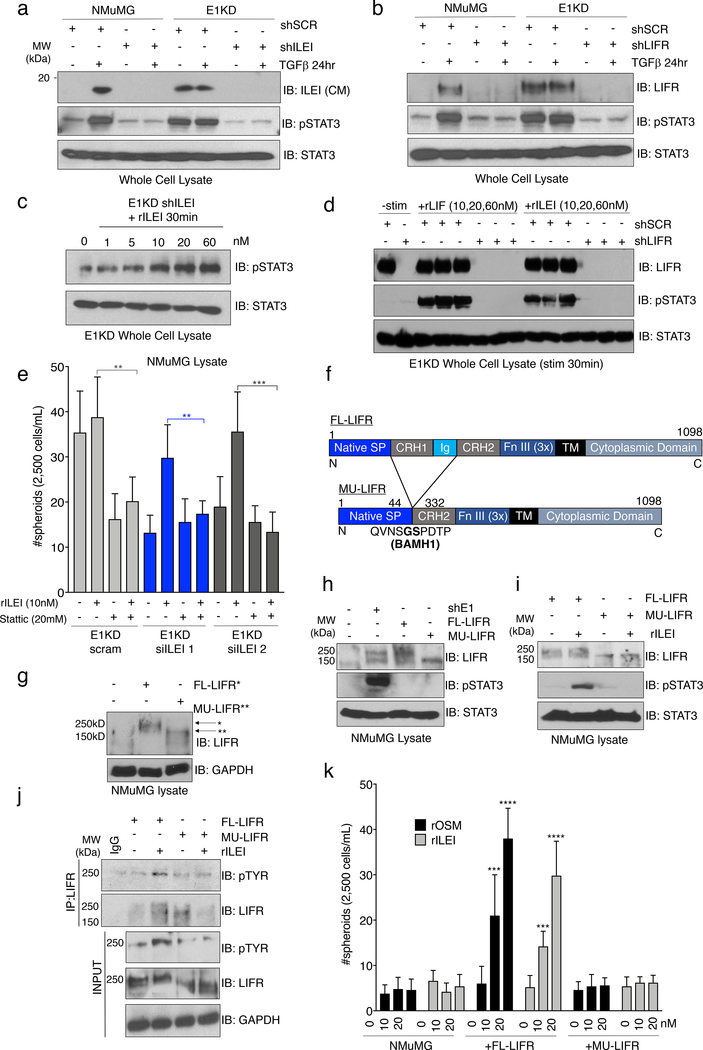
Previously, we identified the secreted cytokine interleukin-like EMT inducer (ILEI) as a member of a cohort of mRNAs involved in EMT that are regulated by TGFβ/hnRNP E1 through a 3’-UTR-mediated translational silencing mechanism 19. To examine whether TGFβ-induced expression of ILEI is necessary and/or sufficient to activate an EMT program, we introduced either scramble control or ILEI-specific shRNA constructs into the NMuMG and E1KD cell lines (referred to as NMuMG and E1KD shSCR/shILEI). Figure 2a demonstrates successful silencing of ILEI protein levels in NMuMG shILEI cells when compared to NMuMG shSCR lines after TGFβ stimulation for 24 hours, whereas E1KD shSCR cells that possess constitutive ILEI translation, lose this expression level after ILEI shRNA silencing (Fig. 2a). This effect is also observed in the secretion of the mature processed 18kD ILEI protein from the various cell lines after analysis of the conditioned media (Fig. 2b). To test TGFβ responsiveness in these cell lines, we assessed both morphology and EMT marker regulation after TGFβ stimulation for 24 hours. NMuMG shSCR cells undergo a morphological EMT after TGFβ treatment, indicated by elongated fibroblast-like cell morphology when examined by phase contrast microscopy, whereas NMuMG shILEI cells do not (Fig. 2c). The E1KD shSCR/shILEI cell lines display constitutive mesenchymal morphologies in both the presence and absence of TGFβ (Fig. S2a). These data suggest that ILEI knockdown is sufficient to block TGFβ-mediated EMT, but is insufficient to induce a MET program within the mesenchymal E1KD pool. Assessment of EMT markers in these cell lines shows that the epithelial cell adhesion marker E-cadherin is down-regulated in NMuMG shSCR cells after 24h TGFβ stimulation, but maintained in NMuMG shILEI cells after the same treatment (Fig. 2d). A change in expression pattern is also observed in the E1KD cell lines where E1KD shSCR cells that lack E-cadherin restore E-cadherin upon ILEI silencing (Fig. 2d). The mesenchymal markers N-cadherin and alpha-smooth muscle actin (α-SMA) show opposite trends compared to E-cadherin in the NMuMG and E1KD populations, whereby ILEI levels dictate the relative expression patterns of these markers (Fig. 2d). These data collectively demonstrate that ILEI translation/secretion through TGFβ/hnRNP E1 is important for induction of EMT in mammary epithelial cell populations.
Figure 2. The Secreted Cytokine ILEI is Necessary for TGFβ/hnRNP E1-Mediated EMT and BCSC formation.
Immunoblot analysis of either (a) whole cell lysates or (b) conditioned media from NMuMG and E1KD cells stably transduced with either scramble control shRNA (shSCR) or ILEI knock-down shRNA (shILEI) in the presence or absence of TGFβ (5ng/ml) for 24 hours. Coomassie staining was used as a loading control for conditioned media. (c) Phase contrast images of NMuMG shSCR and NMuMG shILEI cells in the presence and absence of TGFβ stimulation for 24 hours. (d) Immunoblot analysis of EMT markers in whole cell lysates derived from NMuMG/E1KD shSCR and NMuMG/E1KD shILEI cells in the presence and absence of TGFβ stimulation for 24 hours. (e) Quantification of mammosphere formation by TGFβ pre-treatment (5ng/mL) in NMuMG cells stably transduced with either scramble control shRNA (shSCR) or ILEI knock-down shRNA (shILEI) (error bars represent mean +/− SD; n=5; ****p<0.0001, unpaired Student’s t-test). (f) Quantification of mammosphere formation and self-renewal in E1KD cells stably transduced with either scramble control shRNA or ILEI knock-down through several passages (error bars represent mean +/− SD; n=5, p <0.0001 for all passages, paired Student’s t-tests between correlative passage numbers). (g) Mammosphere formation by NMuMG shSCR cells supplemented with conditioned media derived from either E1KD shSCR or E1KD shILEI cells (25% total volume) with E1KD mammosphere growth used as a positive control (error bars represent mean +/− SD, n=5; ***p=0.0002, 2-way ANOVA). (h) Quantification of mammosphere formation in E1KD shSCR/shILEI cells in the presence and absence of purified recombinant ILEI (error bars represent mean +/− SD; n=5; **p=0.0013, 2-way ANOVA).
Mammosphere assays were used to determine whether ILEI is responsible for BCSC self-renewal in E1KD cells. As shown, TGFβ pre-treatment induces mammosphere formation in NMuMG shSCR control cells, which is abolished in shRNA-ILEI-silenced NMuMG cells (Fig. 2e). Similarly, E1KD shSCR cells that display constitutive self-renewal and mammosphere formation properties partially lose this phenotype upon ILEI knockdown (Fig. 2f). To test the contribution of ILEI to stemness potential in a human cell line, we utilized shRNA-mediated silencing of ILEI in human basal epithelial A549 cells and performed oncosphere assays (Fig. S2b-d). We demonstrate that ILEI silencing significantly decreases the spheroid formation capacity of A549 cells, confirming ILEI as an important factor in both murine and human stem cell phenotypes.
To test the contribution of ILEI in the conditioned media derived from E1KD shSCR/ILEI cells on mammosphere formation, we assayed spheroid growth of NMuMG cells in the presence of either E1KD shSCR or E1KD shILEI conditioned media (Fig. 2g). We show that only the conditioned media derived from E1KD shSCR cells is able to induce mammosphere growth in the NMuMG cell lines relative to positive control E1KD cells. Analysis of the stemness markers BMI1 and Nanog support the mammosphere self-renewal experiments and reveal an upregulation of these factors in E1KD cells when compared to NMuMG cells (Fig. S2e). This observation in E1KD cells is ILEI-dependent, as siRNA-mediated silencing of ILEI induces the loss of BMI and Nanog levels (Fig. S2f). To further test the effect of ILEI on self-renewal, we performed rescue experiments with purified recombinant ILEI that lacks the N-terminal pro-peptide and signaling peptide sequences, resulting in an 18kD functional variant (rILEI; Fig. S3a-b). We show that supplementing pure rILEI in the mammosphere formation assay of E1KD shILEI cells is able to partially rescue their spheroid formation capacity relative to positive control E1KD shSCR cells (Fig. 2h). Collectively, these data suggest that ILEI secretion is necessary for inducing the self-renewal properties in BCSCs regulated by TGFβ/hnRNP E1.
ILEI Protein Levels Correlate with Mammary Tumor Progression
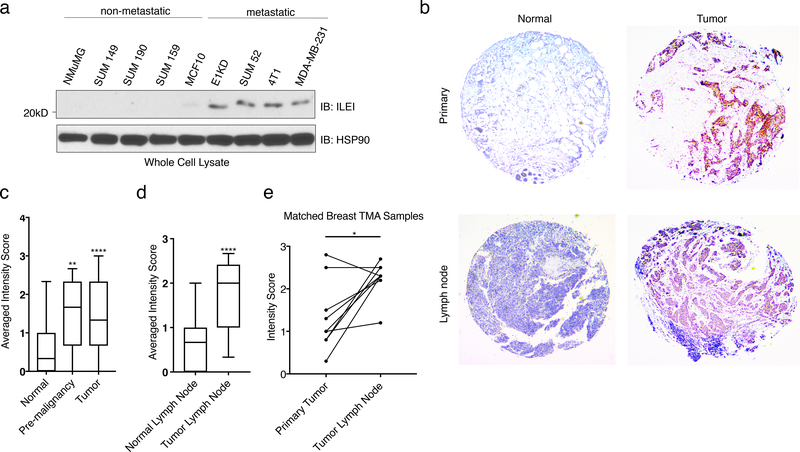
To further investigate the association between ILEI and human malignancy, we probed a series of cancer cell lines that represent different subtypes of breast carcinoma populations with various levels of metastatic potential. Human SUM cell lines were derived from patients with breast malignancies 37. Figure 3a demonstrates that ILEI is upregulated in a correlative manner in both human and murine metastatic populations (E1KD, 4T1, SUM44, MDA-MB-231) when compared to normal epithelial cells (NMuMG and MCF10) and non-metastatic tumor cell lines (SUM149, SUM190, and SUM 159). Furthermore, using human breast tissue microarrays from the Biorepository and Tissue Analysis core at MUSC, we correlated ILEI expression with tumorigenesis in human patients. Both pre-malignant and primary tumor samples demonstrate a significant increase in IHC score compared to normal breast tissue (Representative images Fig. 3b, Quantification Fig. 3c; for representative images of all conditions see Fig. S4a-b). Similarly, tumors found within the lymph node have significantly higher levels of ILEI compared to normal lymph node tissue (Fig. 3b and 3d). Using only patient samples that contained matched breast tissue tumors and lymph node tumors, we could see a significant increase in ILEI expression at the metastatic site compared to the primary tumor site (Fig. 3e). These data indicate that ILEI signaling is involved in tumorigenesis of mammary epithelial cells in human patients.
Figure 3. ILEI Protein Levels Correlate with Mammary Tumor Progression.
(a) Immunoblot analysis of basal ILEI levels in both murine and human cell lines that display altered metastatic potential. (b) Representative images taken at 5x magnification of normal breast, tumor breast, normal lymph node or tumor lymph node TMAs after immunohistochemistry analysis of ILEI. (c) Quantification of IHC scores for all normal breast tissue, pre-malignant tissue and tumor tissue samples (box plot represents intensity score distribution minimum to maximum; ****p<0.0001, **p<0.01, unpaired Student’s t-test). (d) Quantification of IHC scores for all normal lymph node tissue, and tumor lymph node tissue samples (box plot represents intensity score distribution minimum to maximum; ****p<0.0001, unpaired Student’s t-test). (e) Plot showing matched primary tumor and lymph node tumor patient samples (n=8, *p=0.013, paired Student’s t-test).
ILEI Binds to the Cytokine Receptor LIFR
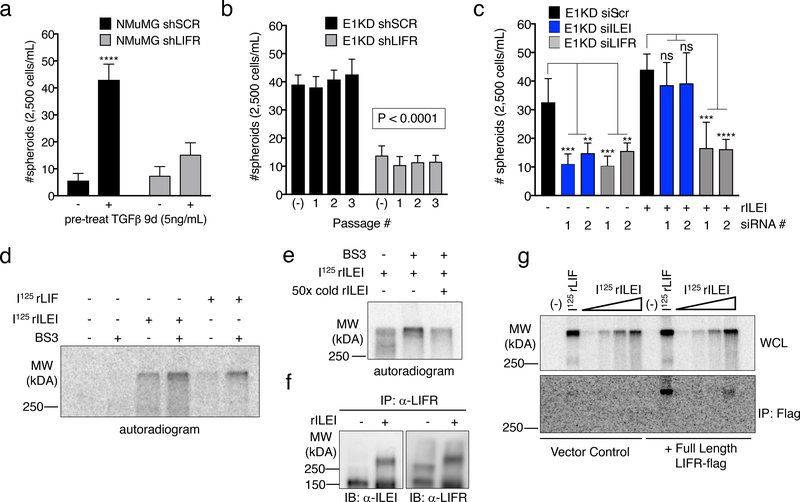
Little is known regarding ILEI’s mechanism of action. We utilized a yeast-two hybrid screen to identify potential ILEI binding partners using the ILEI coding sequence as bait and a HeLa cDNA library as prey. Among the 60 candidate interacting partners was the cytokine receptor LIFR precursor (Fig. S5a). Since LIFR serves as a receptor for several cytokines involved in self-renewal, we postulated that it may serve as the extracellular receptor mediating ILEI signaling. Indeed, TGFβ signaling or hnRNP E1 knockdown in NMuMG cells induces the upregulation of LIFR protein, suggesting a potential role of the receptor in E1KD/TGFβ-induced EMT/BCSC formation (Fig. S5b). We utilized shRNA-mediated silencing of LIFR in both NMuMG and E1KD cell lines in order to investigate any differential BCSC formation capabilities dependent on either TGFβ stimulation or hnRNP E1 knockdown (Fig. S5b). We observed attenuated mammosphere formation and self-renewal capacity with LIFR knockdown when compared to scramble control cells, phenocopying ILEI’s effect on mammosphere formation (Fig. 4a-b). Furthermore, we observed mammosphere rescue when rILEI was added to E1KD cells silenced for ILEI, as shown previously. Alternatively, mammosphere formation was not rescued when rILEI was incubated with E1KD cells stably silenced for LIFR (E1KD shLIFR), suggesting that ILEI is functioning through LIFR to mediate the self-renewal phenotype (Fig. S5c). We also transiently silenced ILEI and LIFR with targeted siRNAs in E1KD cells to test their mammosphere formation capacity both constitutively as well as after co-incubation with purified rILEI (Fig. 4c, see Fig. S5d for silencing of ILEI and LIFR using si molecules). As expected, either shRNA- or siRNA-mediated knockdown of ILEI and LIFR results in attenuated self-renewal properties. Only cells silenced for ILEI are able to regain this phenotype when supplied with recombinant rILEI. Another functional activity assay used to demonstrate stemness is monitoring aldehyde dehydrogenase (ALDH) activity. We observed an increase from 5.80% to 10.50% in aldehyde dehydrogenase activity in the E1KD cells when compared to NMuMG cells. This increase was respectively abolished and reduced upon downregulation of either ILEI or LIFR protein levels (Fig. S5e), further confirming their respective roles in the cancer stem cell phenotype.
Figure 4. ILEI Binds to the Cytokine Receptor LIFR.
(a) Mammosphere assay of NMuMG cells stably transduced with either scramble control or LIFR knock-down shRNA (shLIFR) in the presence and absence of 9-day TGFβ pre-treatment (5ng/mL) (error bars represent mean +/− SD; n=5; ****p<0.0001, unpaired Student’s t-test). (b) Quantification of mammosphere formation and self-renewal in E1KD cells stably transduced with either scramble control shRNA or LIFR knock-down (error bars represent mean +/− SD; n=5; p<0.0001 for all passages, paired Student’s t-tests between correlative passage numbers). (c) Quantification of mammosphere formation in E1KD siSCR/ siILEI/ siLIFR cells in the presence and absence of 10nM purified recombinant ILEI (error bars represent mean +/− SD; n=5; **p<0.01, ***p<0.001, ****p<0.0001, unpaired Student’s t-test). (d) BS3 cross-linking in HEK293 cells in the presence or absence of 625pM 125I-ILEI and 125I-LIF. Radiolabeled BSA was used in control lanes. (e) Cold-competition assay of radiolabeled 125I-ILEI with or without 50x cold recombinant ILEI. Indicated samples were BS3 cross-linked to the surface of HEK293 cells and separated by SDS-PAGE. (f) Immunoprecipitation of LIFR in HEK293 cells in the presence and absence of 10nM ILEI followed by immunoblot for either ILEI or LIFR. All lanes were crosslinked with BS3. (g) Overexpression of full-length FLAG-tagged LIFR or vector control followed by 125I-LIF (625pM) and 125I-ILEI (100pM-2nM) stimulation with BS3 crosslinking. Cell lysates were immunoprecipitated with Flag antibody and separated on an 8% SDS-PAGE gel.
To demonstrate ILEI binding to the cell surface, we [I125]-radiolabeled recombinant ILEI and performed crosslinking experiments using the non-permeable, non-reducible chemical cross-linker BS3. [I125]-rLIF was used as a positive control. In addition to binding the cell surface in a similar manner as rLIF (Fig. 4d), [I125]-rILEI binding could be out-competed using 50X-fold unlabeled rILEI protein, demonstrating the specificity of the interaction (Fig. 4e). The complexed radiolabeled LIF and ILEI were both observed at molecular weights in excess of 250kDa compared to the free ligand, which migrates at approximately 20kDa (Fig. S6a). Additionally, we immunoprecipitated LIFR from E1KD cells following rILEI treatment and BS3 crosslinking. Both ILEI and LIFR were detected by immunoblot analysis after LIFR immunoprecipitation. Due to the irreversibility of the BS3 cross-linker, both ILEI and LIFR were observed to shift to a molecular weight larger than LIFR alone and consistent with complex formation (Fig. 4f). Finally, transfection of either empty vector or a FLAG-tagged LIFR construct in HEK293 cells allows for radiographic detection of ILEI binding to LIFR after the addition of [I125]-rILEI and immunoprecipitation with a LIFR antibody. The positive control [I125]-rLIF was also observed to co-precipitate with LIFR (Fig. 4g; see Fig. S6b for FLAG-LIFR transfection levels in HEK293 cells).
ILEI Induces STAT3 Signaling through Activation of LIFR
Downstream of LIFR activation, STAT3 is known to contribute to maintenance and self-renewal of stem cells 38. To determine whether ILEI-induced LIFR activation resulted in phosphorylation of STAT3, we first monitored phospho-STAT3 at position Y705 (pSTAT3) in NMuMG and E1KD cells with and without TGFβ. Treatment with TGFβ for 24 hours induces pSTAT3 signaling in normal mammary epithelial cells (NMuMG). This activation is lost with both ILEI and LIFR protein reduction (Fig. 5a-b). As shown, E1KD cells, which exhibit basal stem cell properties, possess constitutive pSTAT3 in the absence of TGFβ, and silencing of ILEI or LIFR in these cells results in a decrease in constitutive pSTAT3 levels (Fig. 5a-b). Moreover, the addition of increasing concentrations of rILEI to E1KD shILEI cells for 30 minutes showed a corresponding increase in pSTAT3 (Fig. 5c). To test endogenous LIFR activation, we treated either control E1KD shSCR cells or shLIFR cells with ILEI and LIF. Only E1KD cells that contain LIFR are responsive to ILEI and LIF through the subsequent activation of STAT3, demonstrating that LIFR mediates ILEI signaling (Fig. 5d). In order to determine whether STAT3 activation is important for ILEI-mediated self-renewal phenotypes in E1KD cells, we utilized the small molecule specific inhibitor of STAT3 activation termed Stattic39. Stimulation of E1KD cells with rILEI while in the presence of Stattic significantly abrogates STAT3 signaling and spheroid formation capacity (Fig. S7a and 5e). The loss of this phenotype upon STAT3 inhibition cannot be rescued by rILEI treatment, suggesting that ILEI-mediated BCSC phenotypes are dependent upon LIFR and STAT3 (Fig. 5e). In addition to STAT3 signaling, we also observe constitutive activation of MAPK/Erk signaling in E1KD cells (Fig. S7b). Since MAPK/Erk signaling was observed to be linked to LIFR activation 27, we investigated whether ILEI also plays a role in constitutive levels of MAPK/Erk activity in E1KD cells. We first treated E1KD cells with the MEK1/2 inhibitor U0126 and observed potent inhibition of basal MAPK/Erk signaling (Fig. S7b). We then performed E1KD mammosphere assays in the presence or absence of U0126 and observed a slightly significant abrogation of E1KD spheroid forming capacity when MAPK/Erk signaling is inhibited (Fig. S7c). Furthermore, we observe an elevation of Erk phosphorylation after serum-starvation and stimulation of E1KD cells with purified rILEI (Fig. S7d).
Figure 5. ILEI Induces STAT3 Signaling through Activation of LIFR.
(a) Immunoblot analysis of basal pSTAT3 levels in NMuMG and E1KD cells stably transduced with control shSCR shRNA, as well as shRNA targeting either ILEI or (b) LIFR. Cells were stimulated with TGFβ (5ng/mL) for 24 hours where indicated. (c) Stimulation of E1KD shILEI cells with the indicated concentration of recombinant ILEI for 30 minutes. Lysates were probed for phosphorylated STAT3 protein. (d) E1KD cells or E1KD shLIFR cells treated with increasing concentration of rLIF and rILEI for 30 minutes. Lysates were probed for LIFR, pSTAT3, and total STAT3. (e) Mammosphere assay using E1KD cells treated with two siRNA molecules against ILEI and rescued with rILEI treatment at 10nM. The STAT3 inhibitor Stattic was added at 20μM where indicated (error bars represent mean +/− SD; n=5; **p<0.01, ***p<0.001, unpaired Student’s t-test). (f) Construct diagram of FL-LIFR and mutated LIFR (MU-LIFR) lacking the cytokine binding region. Regions include the native signal peptide, cytokine receptor homology domains 1 and 2, Ig-like domain, Fibronectin type III repeat, transmembrane and cytoplasmic domains. (g) Overexpression of FL-LIFR and MU-LIFR constructs in NMuMG cells. (h) Immunoblot analysis of basal STAT3 activation in NMuMG cells overexpressing either FL-LIFR or MU-LIFR compared to parental NMuMG and E1KD cells. (i) Immunoblot analysis of STAT3 activation in response to rILEI (20nM) for 30 minutes in either NMuMG FL-LIFR or MU-LIFR cells. (j) Immunoprecipitation of LIFR after rILEI stimulation (20nM) for 30 minutes in NMuMG cells overexpressing either FL-LIFR or MU-LIFR followed by immunoblot analysis. (k) Quantification of mammosphere formation in NMuMG FL/MU-LIFR cells in the presence or absence of rOSM/rILEI pre-treatment for 7 days (error bars represent mean +/− SD; n=5; ****p<0.0001, ***p<0.001, unpaired Student’s t-test).
In order to investigate ILEI/LIFR interactions in cells, we generated two constructs of LIFR containing either the full-length receptor (FL-LIFR) or a truncation of the extracellular cytokine binding region (MU-LIFR) (Fig. 5f). These constructs were overexpressed in NMuMG cells, which do not maintain high levels of endogenous LIFR (Fig. 5g). While we could not detect an increase in basal pSTAT3 levels in the transfectants compared to E1KD cells, most likely due to the absence of ILEI translation (Fig. 5h), we did observe activation of pSTAT3 in cells overexpressing the FL-LIFR, and not the MU-LIFR cells, following a 30 minute incubation with rILEI (Fig. 5i). Further, co-immunoprecipitation analyses revealed that a 30 minute stimulation of cells overexpressing the FL-LIFR, and not the MU-LIFR, with rILEI promoted tyrosine phosphorylation of LIFR (Fig. 5j). These results demonstrate that ILEI induces Jak/STAT signaling through the extracellular activation of the LIF receptor.
We performed mammosphere assays to observe ILEI/LIFR signaling as it relates to epithelial cell self-renewal with the NMuMG FL/MU-LIFR cell lines in the presence of long-term ILEI treatment. The LIFR ligand oncostatin M (OSM), recently shown to induce CSC phenotypes through pSTAT3 activation, was used as a positive control 40. Neither rILEI nor rOSM induce mammosphere formation in parental NMuMG cells (Fig. 5k). However, expression of FL-LIFR, but not the MU-LIFR, confers mammosphere formation capabilities to these cells, demonstrating that the cytokine-binding region of the receptor is necessary to elicit downstream biological phenotypes controlled by ILEI signaling (Fig. 5k). This observation was consistent when using recombinant LIF (Fig. S7e). Collectively, these data demonstrate that ILEI initiates STAT3 signaling through LIFR and this mechanism is necessary for BCSC phenotypes in mammary epithelial cells.
ILEI/LIFR Signaling Contributes to Mammary Stemness, Tumorigenesis and Metastasis in vivo.
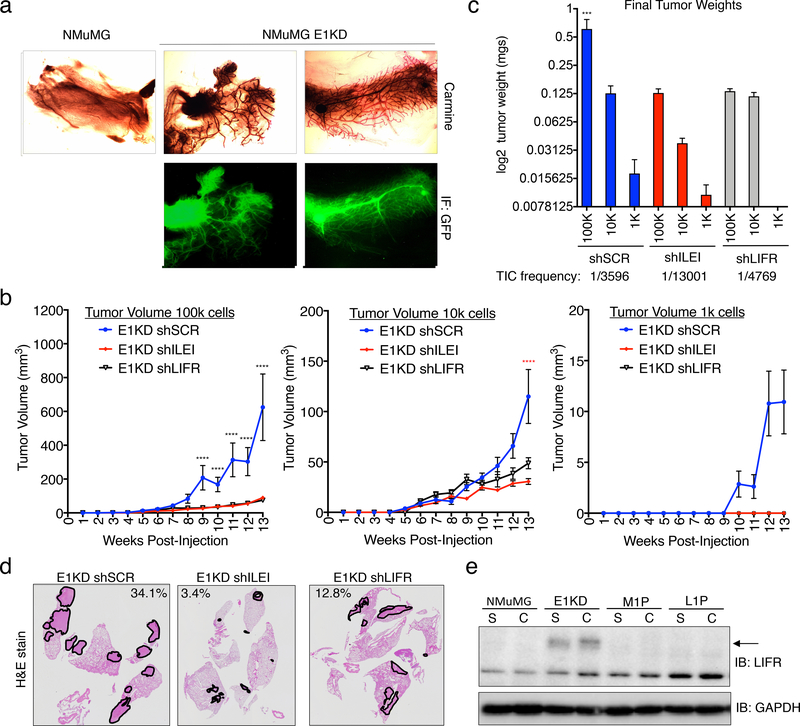
To elucidate the in vivo potential of E1KD cells, we performed mammary fat-pad reconstitution experiments. Cleared fat-pads from 3-week old NOD/SCID female mice, were injected with either NMuMG-RFP or E1KD-GFP cells. 6 weeks post-injection, fat pads were isolated and analyzed by Carmine staining and imaged for fluorescent signals. E1KD-GFP cells, but not the NMuMG-RFP parental cells, were able to reconstitute the ductal network of cleared fat pads as observed with Carmine stain and GFP fluorescence (Fig. 6a), demonstrating the pluripotent nature of the NMuMG cell line upon hnRNP E1 protein reduction. GFP expression in the ductal outgrowth validates that the de novo ductal tree growth is a result of E1KD cell injections and not failed clearance of the endogenous ductal tree. RFP signal was not observed in the NMuMG injected fat pads.
Figure 6. ILEI/LIFR Signaling Contributes to Mammary Stemness, Tumorigenesis and Metastasis in vivo.
. (a) Mammary fat-pad reconstitution assay of NMuMG-GFP and E1KD-GFP cells orthotopically implanted into the cleared fat-pad of 3-week old female NOD/SCID mice. (b) Quantification of tumor volume over time in female NOD/SCID mice injected with E1KD shSCR, shILEI, and shLIFR cells at concentrations of 1k, 10k, and 100k cells per injection into the mammary fat-pad region (error bars represent mean +/− SEM n≥4; ****p<0.0001, 2-way ANOVA; shSCR is significant compared to shILEI and shLIFR in 100k cell injections, shSCR is significant compared to shILEI in 10k cell injections). (c) Final mammary tumor weight quantification from female NOD/SCID mice injected with 1k, 10k, and 100k E1KD shSCR, shILEI, and shLIFR cells into the mammary fat-pad region with a TIC frequency displayed for each condition (error bars represent mean +/− SEM, n≥4, ***p=0.0002, One-way Anova; E1KD shSCR 100k condition is significant when compared to the shSCR 10k and 1k conditions, as well as all shILEI and shLIFR conditions). (d) H&E staining of metastatic area in lungs of female NOD/SCID mice injected with E1KD shSCR, shILEI, and shLIFR cells into the mammary fat-pad. Representative images show lungs with the largest metastatic burden for each cell condition. Metastatic area percentage is shown for these images. (e) Immunoblot analysis of LIFR in NMuMG, E1KD, and E1KD cells injected into NOD/SCID fat pads and cultured from mouse primary tumor formation (M1P) and lung metastases (L1P).
We have previously shown that E1KD cells, but not wild type NMuMG cells, are tumorigenic and metastatic when injected into the mammary fat-pad of NOD SCID mice17,18. To determine the contribution of ILEI/LIFR signaling to these results, we performed similar mammary fat pad injections with either control E1KD shSCR, shILEI, or shLIFR cells and monitored tumor formation and metastatic disease. Female NOD/SCID mice were injected in the mammary fat pad region with increasing concentrations ranging from 1k-100k cells. We demonstrate quantitatively that there is a significant alteration in tumor volume over time that is observed over multiple cell concentrations (Fig. 6b). Similarly, we detected significant alterations in final tumor weight and tumor initiating cell frequency (TIC, calculated using Extreme Limiting Dilution Analysis (ELDA) 41) between control E1KD shSCR cells and the E1KD shILEI/LIFR lines (Fig. 6c; see Fig. S8a for extracted tumor images).
To investigate whether there was a difference in metastatic spread to the lungs from these experiments, we harvested lungs at the time of tumor extraction and performed H&E staining to identify metastases. Our data indicate that there is an aberration of metastatic lesions between the control E1KD shSCR lungs and the E1KD shILEI/LIFR lungs. Representative images show lungs with the largest metastatic burden obtained from each cell line (Fig. 6d). Lungs extracted from mice injected with E1KD shSCR cells showed an average metastatic lesion area of 4.99% where the corresponding lungs from E1KD shILEI and shLIFR injected mice had an average metastatic area of 0.49% and 2.03%, respectively (Fig. S8b).
Previously, we observed changes in LIFR expression levels from low levels in the NMuMG cells to high levels in the BSCS E1KD cells (Fig. 5b). Corresponding to our NMuMG versus E1KD data, in HMLE cells, the mesenchymal cell population displays increased levels of LIFR when compared to the epithelial cell population (Fig. S8c). To further examine LIFR levels in tumorigenesis, we utilized a progression series developed in the lab and assessed the relative quantity of LIFR expression in our BCSC model (E1KDs) as compared to the resulting tumors from these cells 42. This model was initiated using the NMuMG normal mammary cell line after hnRNP E1 knockdown. E1KD cells injected into the mammary fat pad of NOD/SCID mice form both primary mammary tumors (M1P) and lung metastases (L1P), from which cells were isolated and cultured in puromycin selection 42. Interestingly, E1KD cells have significantly upregulated LIFR protein (Fig. 5b & 6e) compared to the wild type NMuMG cell line, yet tumor outgrowth at both the primary and metastatic sites have lost expression of the LIFR gene product (Fig. 6e). ILEI levels in the conditioned media of the progression series increase (Fig. S8d). As discussed below, LIFR expression could be important in tumor initiation and the breast cancer stem cell phenotype yet be downregulated during tumor outgrowth. Indeed, as observed in both primary tumors (Fig. S8e) and metastatic tumors (Fig. S8f) derived from the MMTV-PyMT breast cancer tumor model, LIFR levels are decreased overall in the tumor tissue. IHC staining in these tumors for LIFR and ILEI, in addition to H&E staining, indicate that despite low overall levels of LIFR, there are distinct cells within the cancer growth that maintain LIFR expression and could potentially act as BCSCs. ILEI expression is observed in all areas where LIFR expression is seen (Fig. S8e-f).
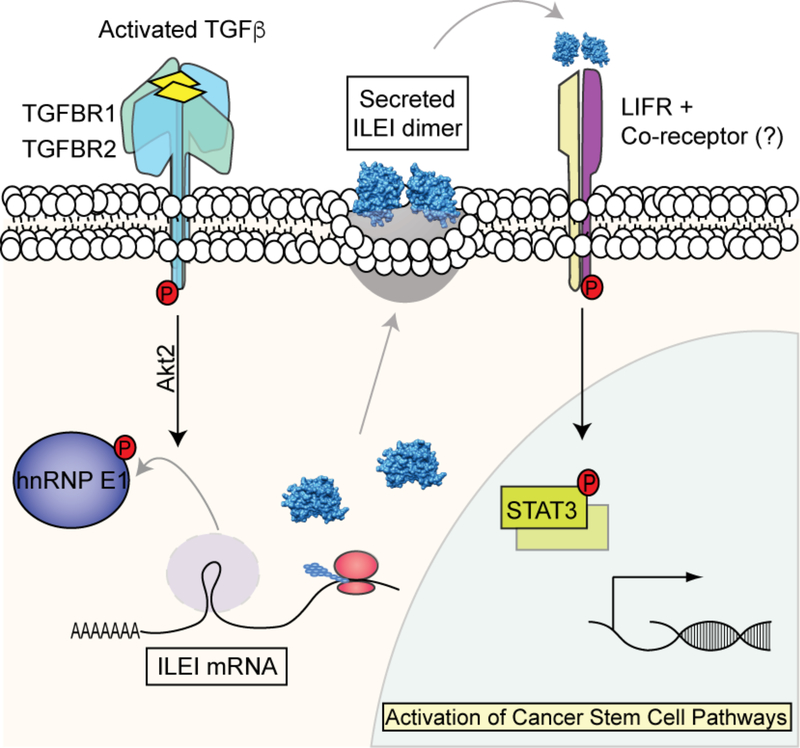
Our in vivo analyses indicate that hnRNP E1 is an important factor controlling in vivo stemness properties within the mammary epithelium. We further show that the tumorigenic properties harbored by cells lacking hnRNP E1, which has been established previously 17,18, is mediated by factors including ILEI and LIFR. The current model depicts an undiscovered signaling pathway that controls EMT and stemness properties regulated by TGFβ (Fig. 7). We collectively demonstrate that TGFβ-induced ILEI expression and activation of LIFR through the hnRNP-E1 mechanism of EMT induction is responsible for enhanced tumorigenesis and CSC formation in vitro and in vivo.
Figure 7.
Schematic of working model demonstrating ILEI-mediated CSC formation through LIFR.
Discussion
The dual roles of TGFβ signaling as a tumor-suppressor in normal epithelial cells and early-stage cancers, as well as tumor-promoter in advanced tumors stands as a complex paradigm of tumor development and spread 43,44. Given its role in EMT induction and CSC formation during late stages of tumorigenesis, well-defined insight into these TGFβ-regulated processes is vital for the development of new-targeted therapies. Here, we have delineated a non-canonical TGFβ/hnRNP-E1-driven mechanism of CSC formation and tumorigenesis through the progenitor factor ILEI and the cytokine receptor LIFR. We demonstrate that TGFβ induces the expression and secretion of ILEI, through an hnRNP E1 translational inhibitory mechanism, resulting in the activation of LIFR and downstream STAT3 signaling to mediate enhanced stemness and tumorigenic properties (Fig. 7).
BCSCs serve as valuable therapeutic targets due to their ability to self-renew and differentiate to generate heterogeneity in the bulk tumor population, as well as display resistance to chemotherapeutic agents 8,10. The emergence of tumor-initiating cancer stem cells (CSCs) has been identified across several cancer types including breast cancer, melanoma, and leukemia 8. Various lines of evidence indicate that cell transformation caused by TGFβ-mediated EMT may be partially involved in BCSC formation during epithelial tumorigenesis 14–16. Additionally, BCSCs rely on signaling pathways related to embryonic development and inflammatory signaling responses to maintain their stem-like properties, including Wnt/β-catenin, NOTCH, and importantly, Jak/STAT 38. Signaling through the Jak/STAT pathway promotes proliferation, invasion, angiogenesis, and metastasis 45. Several groups have demonstrated the role of STAT3 in aiding stem cell/mesenchymal properties in cell populations originated from epithelial derived cancers including self-renewal and tamoxifen resistance 46,47.
Our evaluation of TGFβ-mediated EMT and BCSC formation through hnRNP E1 regulation has revealed insightful evidence of STAT pathway activation through an ILEI signaling axis. Modulation of ILEI in epithelial NMuMG cells or mesenchymal NMuMG cells knocked down for hnRNP E1 results in abrogated EMT and self-renewal capacity upon TGFβ stimulation in vitro. We demonstrate that either silencing of hnRNP E1 or sustained translation of ILEI by TGFβ stimulation is both necessary for EMT induction as well as CSC formation in vitro. These observations prompted an investigation into candidate effector molecules that aid in the ILEI signaling pathway to further understand its role in BCSC formation. Utilizing yeast-two-hybrid screening with a cDNA library derived from HeLA cell mRNA, we identified an interaction between ILEI and a precursor of the cytokine receptor LIFR. LIFR serves as a signaling platform for several cytokines and functions together with its co-receptor gp130, and in some instances with other ligand-specific co-receptors 30. Activation of LIFR through ligand-binding induces a multitude of signaling pathways including JAK/STAT, AKT, and ERK 30. These modalities have been established to converge and orchestrate a complex signaling network that results in the maintenance, pluripotency and self-renewal properties observed in embryonic stem cells (ESCs) 48. Interestingly, it has been demonstrated that the assortment of ligands for LIFR elicit individual responses from this receptor 49. For example, OSM and LIF can induce similar, yet distinct, activation of STAT1, STAT3, ERK and JNK in T47D breast tumor cells 50. In neuronal cells, CNTF and LIF result in different activation of downstream pathways due to variances in localization of receptor subunits to lipid rafts in the membrane 51. Therefore, due to a complex network of co-receptors and regulators associated with the LIF receptor, further investigation comparing ILEI to previously identified LIFR ligands and the discovery of any co-receptors will be necessary to understand the ILEI/LIFR signaling pathway, and an important next step for targeted drug development.
The seeding of metastases and tumor heterogeneity requires intricate programming and over-activation/suppression of several signaling pathways 52. ILEI has been established as an oncogenic cytokine responsible for both EMT and tumorigenesis 53. Despite a clear function for STAT3 signaling in BCSCs, some reports describe LIFR as a metastasis suppressor 34,46,54,55. Conflicting studies show that high levels of LIFR in melanoma are associated with increased migration and poor prognosis in patients, and in prostate cancer epigenetic activation of LIFR is correlated with metastasis 56,57. Despite the apparent discrepancies, we believe that our data along with the findings from Johnson et al. provide an interesting explanation 33. Consistent with LIFR and STAT3’s role in cancer stem cell maintenance, migration and invasion, LIFR protein levels are low in the normal mammary epithelial cells (NMuMG cells), yet high in our hnRNP E1 knockdown cells with a BCSC phenotype (Fig. 5b and Fig. 6e). Most interestingly, when the primary tumors and metastatic lesions derived from NMuMG E1KD cells are extracted from NOD SCID mice and analyzed by western, LIFR levels are reduced in both the primary and metastatic tumor cells when compared to E1KD cells (Fig. 6e). This suggests that LIFR is playing important roles in maintenance of the cancer stem cells and in tumor formation yet may be inhibitory to tumor outgrowth both at the primary and secondary sites depending on the cellular context. We did observe cells that maintain LIFR expression within both the primary and metastatic tumors in a MMTV-PyMT model by IHC that could serve as BCSCs (Fig. S8e-f).
Similarly, Johnson et al. found that LIFR protein levels regulate the dormancy state of breast cancer cells disseminated to the bone 33. In particular, LIFR protein reduction induced proliferation of the metastatic lesions in bone from cells that would otherwise remain dormant. Upon downregulation of LIFR, specific reduction in the mRNA of several dormancy and cancer stem cell genes was observed. Hypoxia was also shown to reduce LIFR protein levels, thereby down-regulating stem cell associated genes to promote metastatic growth and an exit from the dormancy state. Conversely, valproic acid induced expression of LIFR and consistently induced dormancy and cancer stem cell associated genes 33. In our NMuMG E1KD cells, reducing LIFR inhibits the tumor initiating potential of our cells, and therefore reduces the tumorigenicity and metastatic potential of this system. Based on these findings, we believe that the ILEI/LIFR complex plays an important role in the maintenance of cancer stem cells at distant sites and may affect the switch from a dormant state to active metastatic growth. Herein, we have established a detailed mechanism by which TGFβ induces BCSC phenotypes during EMT induction.
Materials and Methods
Cell Culture, Treatments and Cell Manipulations
NMuMG (ATCC), HMLE, HEK293 (ATCC) and HEK293t (ATCC) cells were maintained at 37oC, 5% v/v CO2 in a humidified incubator. NMuMG, HEK293 and HEK293t cells were cultured in DMEM (Corning) high glucose supplemented with 10% fetal bovine serum (Atlanta Biologicals) and 1% antibiotic/antimycotic solution (penicillin G, streptomycin, amphotericin B) (ThermoFisher). HMLE cells were isolated for trypsin resistance and sorted by FACS as previously described 58. HMLE cells were cultured in DMEM:F12 supplemented with 5% calf serum (VWR), 0.5 μg/ml hydrocortisone, 10 μg/ml insulin, 20 ng/ml epidermal growth factor (Corning) and 1% antibiotic/antimycotic. TGFβ treatments were conducted using 5 ng/mL recombinant TGBβ2 (Genzyme) for the indicated durations. Phase contrast cell images were obtained using a Leica DM IL LED Inverted Phase Contrast microscope.
Lentiviral constructs were obtained from the shRNA core at MUSC or cloned into the pLKO.1-neo construct (Addgene) using EcoR1/Age1 sites. Sequences for the shRNA hairpins are listed in Table S1. HEK293t cells were grown to 60–70% confluence and transfected with the pLKO.1 shRNA plasmid containing the targeted hairpins, psPAX2 and pMD2.G packaging plasmids using Lipofectamine 3000 in OPTI-MEM. Media was changed after overnight incubation to fresh culture media. Virus was collected and filtered at 24 and 48 hours through a 0.22 μm sterile filter. For transduction, 1:5 to 1:2 ratios of virus containing media to culture media was incubated on the target cells with 8 μg/mL polybrene overnight. Culture media was changed to include either 1 μg/mL puromycin (ACROS Organics) or 2 mg/mL G418 (Invivogen) for NMuMG derivative cells. Resistant pools were collected for experimentation. For experiments utilizing siRNAs, cells were transfected at 60–70% confluence using Lipofectamine 3000 in OPTI-MEM for 24 hours before analyzing RNA and whole cell lysates for successful knock-down. Cells transfected with siRNA for mammosphere assays were monitored over the course of ten days post-transfection in order to observe steady knockdown during the experiments.
Mammosphere Assay
NMuMG wild-type and derivative cell lines were cultured either in the presence or absence of 5 ng/mL TGFβ for 9 days and were trypsinized from 2D adherent culture plates. Cell suspensions were diluted to 2,500 cells/mL and seeded into a 96-well non-adherent plate (500 cells/well) containing DMEM (Corning), 20 ng/mL epidermal growth factor (Corning), 20 ng/mL fibroblast growth factor (Corning) and B27 supplement (Gibco). After 7–9 days, spheroids from each cell line (n=5) were quantified under phase-contrast microscopy. For passaging, the mammospheres were pooled, trypsinized, and passed through a 0.22 micron filtered syringe before re-seeding into a fresh 96-well non-adherent plate. After an additional 7–9 days, spheroids that self-renewed were again quantified under phase-contrast microscopy with a Leica DM IL LED Inverted Phase Contrast microscope. Cell growth was considered a mammosphere at 100μm diameter for all mammosphere assays.
Human lung cancer cell line A549 shRNA constructs was cultured in adherent conditions in RPMI 1640 medium (HyClone) supplemented with 10% FBS (Atlanta Biological), 1X antibiotic–antimycotic (ThermoFisher), and prophylactic plasmocin (InvivoGen) at 37 °C and in 5% CO2. After 3–4 days, cells were detached using 1X trypsin-EDTA (ThermoFisher). Following a wash in 1X PBS (Fisher Scientific), cells were resuspended in serum free DMEM/F12 with 1x B-27 supplement (ThermoFisher), 20ng/ml human recombinant EGF, 20ng/ml mFGF, 1X antibiotic–antimycotic, and 0.25% methyl cellulose (Sigma-Aldrich) to reduce cell aggregation. Cells were plated at 1000 cells/ml in anchorage-independent conditions using a 6 well ultra-low attachment plate. Spheres were grown for 10 days at 37 °C and in 5% CO2 and then counted under light microscopy. Images were obtained at 5x magnification.
Semi-quantitative PCR
RNA was extracted from cells using Trizol (ThermoFisher) reagent according to manufacturer’s instructions. cDNA was synthesized using cDNA Supermix QScript (QuantaBio) with 1ug RNA. PCR reactions were run with Maxima HotStart Taq Polymerase Mix (ThermoFisher) and 10ng cDNA. Primers used for PCR were as follows: murine BMI1 F GGCTCGCATTCATTTTATGCTG, R ATGAAGTACCCTCCACACAG; Nanog F CTTCAGATAGGCTGATTTGGT, R AATTCGATGCTTCCTCAGAAC; ILEI F GAGCTGCAAAGTTGGTAGTG, R TAGAACGAATGGCTGAGTCC; GAPDH F GGTTGTCTCCTGCGACTTCA, R TAGGGCCTCTCTT GCTCAGT. PCR products were visualized using 9% Acrylamide gels in 1x TBE.
Western Blotting
Whole cell lysates were extracted as follows: Appropriate volumes of Tris-Triton lysis buffer (20 mM Tris pH 7.5, 1% Triton X-100, 10% glycerol, 137 mM NaCl, 2 mM EDTA, and Halt Protease and Phosphatase Inhibitor cocktail (ThermoFisher) was added to cell plates. Cells were immediately scraped, incubated on ice for 30 minutes, and cleared by centrifugation for 20 minutes at 16,000 x g. Protein concentrations were measured with Bradford Protein Assay (BioRad). For conditioned medium immunoblots, cells were serum starved overnight in serum-free DMEM. Medium was collected and precipitated using trichloroacetic acid/acetone. Protein samples were denatured by incubating at 95° for 5 minutes with 1x Laemmli reducing denaturing sample buffer (60 mM Tris-Cl pH 6.8, 1% SDS, 10% glycerol, 5% BME). 1–100 μg of whole cell lysate was resolved on a 8, 10, or 12% polyacrylamide SDS gels and transferred onto PVDF membrane. Membranes were blocked for 1h at RT in 5% skim milk/Tris-buffered saline with 0.01% Tween-20 (TBST) and incubated overnight at 4° on primary antibody + 5% skim milk/TBST. The following primary antibodies were used: ILEI (ab72182; Abcam; 1:1,000), hnRNP E1 (#M01, Abnova; 1: 1,000), E-Cadherin (#610181; BD biosciences; 1:5,000), N-Cadherin (#610920; BD Biosciences; 1:5,000), alpha smooth muscle Actin (ab5694; Abcam; 1:1,000), LIFR (sc-659, Santa Cruz; 1:1,000), LIFR (Santa Cruz sc-515337), FLAG (#2368, Cell Signaling Technologies; 1:1,000), pSTAT3 (#9145, Cell Signaling Technologies; 1:1,000), STAT3 (#4904, Cell Signaling Technologies; 1:5,000), gp130 (sc-376280, Santa Cruz; 1:1,000), pTYR (#9416, Cell Signaling Technologies; 1:5,000), GAPDH (sc-32233; Santa Cruz; 1:10,000), and HSP90 (sc-13119; Santa Cruz; 1:10,000). After primary antibody incubation, membranes were washed 4× 15 minutes in TBST and incubated for 1h at RT on secondary antibody + TBST. The following secondary antibodies were used: Goat anti-Mouse IgG (31430; ThermoFisher; 1:10,000) and Goat anti-Rabbit IgG (31460; ThermoFisher; 1:10,000). After secondary antibody incubation, membranes were washed 4× 15 minutes in TBST and detected using Luminata Forte Western HRP substrate (EMD Millipore) and HyBlot CL Autoradiography Film (Denville) or CCD camera (BioRad ChemiDoc System; BioRad).
Bacterial Protein Purification
Bacterial protein preparation was initiated using a construct that spans human ILEI amino acids 43–227 with an N-terminal TEV-cleavable hexahistidine tag for purification. E. coli BL21 transformed with ILEI cDNA were induced with IPTG for 4 hours, spun out and resuspended in 20mM Tris-HCl pH 8, 20% Sucrose. Cell suspensions were flash frozen and subsequently slow thawed in a circulating ice bath. Lysis Buffer (350mM NaCl (VWR), 10mM Imidazole, 100μg/L Lysozyme (MP Biomedicals), and 1mM BME) was added to the thawing cells suspension then spun at 35,000 rpm for 1 hour at 4°C in a SW40 rotor using a Beckman UltraCentrifuge (Optima LE-80K). His-tagged ILEI was separated from the clarified lysate using HisPur NiNTA resin (Thermo Scientific). The his tag was removed using a his tagged TEV protease. Cleavage was conducted overnight at 4°C and the digested his tag and TEV protein was separated from ILEI using HisPur NiNTA resin. rILEI was further purified by gel filtration on a SD75 16/60 column to greater than 95% purity (GE Healthcare).
Flow Cytometry
HMLE cells were cultured in normal growth media to 70% confluence in 6cm dishes (Denville). Cells were trypsinized, washed 2 times with PBS and resuspended in 1 mL 4% paraformaldehyde (Alfa Aesar) in PBS for 10 minutes at 37°C and one minute on ice. After fixation, cells were washed 3 times in PBS + 1% BSA (Fisher). Cells were incubated with PE conjugated anti-human CD24 antibody (Biolegend) and FITC conjugated anti-human CD44 primary antibody (Biolegend) in 100μL PBS + 1% BSA for 1 hour at room temperature. Cells were washed 3 times in PBS + 1% BSA, resuspended in 500μL PBS and filtered through a 40μm mesh strainer. Flow cytometry was performed on a BD Fortessa X-20 Analytic Flow Cytometry and analyzed using the BD Diva software. Gating on single cells was performed as shown in Supplemental Figure S1d.
The ALDEFLUOR assay was conducted according to manufacturer’s instructions (Stem Cell Technologies) in NMuMG shScr and E1KD cells modulated for ILEI and LIFR. Briefly, cells were grown to 70% confluence, trypsinized, washed in PBS, and resuspended at a concentration of 1*106 cells per mL in ALDEFLUOR buffer. 5μL ALDEFLUOR reagent was added to 1mL suspended cell solution. 500μL of the suspension was immediately transferred to a tube containing 5μL DEAB. All solutions were incubated for 45 minutes at room temperature. Subsequently, cells were pelleted, resuspended in 500μL ALDEFLUOR buffer and filtered through a 40μm mesh strainer. Flow cytometry was performed on 30,000 event counts using a BD Fortessa X-20 Analytic Flow Cytometry and analyzed using FlowJo software (FlowJo, LLC).
Yeast Two-hybrid Screening
ILEI ORF (corresponding to amino acid 42–227) was cloned into the yeast two-hybrid DNA-binding domain vector pGBT9 (Clontech) and transformed into the yeast strain PJ69–4A)59. The strain was mated with the yeast strain Y187 containing a normalized library of HeLa cell cDNAs cloned into a GAL4 AD vector (Clontech, Takara). The resulting library in the diploid strain was screened for activation of the ADE2 reporter gene on yeast minimal medium lacking leucine, tryptophan and adenine. The positive clones were confirmed for interaction by further tests in PJ69–4A strain and plasmid DNA sequenced to identify the interacting genes.
Protein Binding Experiments
Recombinant human ILEI protein (Sino Biologicals) and recombinant human LIF protein (R&D) were radiolabeled with 125 I (Perkin Elmer) using Iodobeads (Peirce) as described by the manufacturer. Briefly, Iodobeads were washed twice with PBS pH 7.2 and air dried. 5μg BSA (Fisher), rLIF, or rILEI was dissolved in 200μL PBS pH 7.2 with one washed Iodobead and 0.6mCi Na125I (Perkin Elmer). The reaction mixture was incubated for 15 minutes at room temperature. Solution was removed from the Iodobead to quench the reaction and radiolabeled protein was separated from free Na125I using PD-10 prepacked gravity columns (GE Healthcare).
HEK293 cells were transiently transfected with either empty pcDNA3.1 (Addgene) vector or pcDNA3.1-LIFR-flag using Lipofectamine 3000 (ThermoFisher) where indicated. Binding experiments were conducted 24 hours after transfection where applicable. Transiently transfected HEK293 or untransfected E1KD cells were cultured to 90% confluence in a 6 well dish, washed with PBS and incubated with radiolabeled protein or unlabeled protein for western blotting for 2 hours at 4°C with gentle rocking. 1mM bis(sulfosuccinimidyl)suberate (BS3) (ThermoFisher) was added to the well and incubated at room temperature for 30 minutes before quenching the crosslinking reaction with 20mM Tris pH 7.5 for 15 minutes at room temperature. Cell lysates were analyzed by 8% SDS-PAGE gel electrophoresis. Gels were dried, exposed to a phosphorImager screen and observed using a Typhoon FLA 1900 PhosphorImager or transferred to PVDF (BioRad) for western blotting analysis.
Tissue Microarray and immunohistochemistry
TMA sample identification is listed in Table S2. 5 μm FFPE sections were routinely deparaffinized in xylene, rehydrated in alcohol, and processed as follows: The sections were incubated with target retrieval solution (Dako S1699) in a steamer (Oster CKSTSTMD5-W) for 10 minutes and then 3% hydrogen peroxide solution for 10 minutes and protein block (Dako X0909) for 20 minutes at room temperature. FAM3C (Sigma HPA050548) 1:1,600 and LIFR 1:500 (Santa Cruz sc-515337) antibodies were incubated in a humid chamber at 4oC overnight followed by biotinylated secondary antibody for 30 minutes and ABC reagent (Vector PK-6101) for 30 minutes. Immunocomplexes of horseradish peroxidase was visualized by DAB (Dako K3468), and sections were counterstained with hematoxylin before mounting. Samples were imaged using a using Leica DM IL LED Inverted Phase Contrast microscope and scored blindly 0–3 for staining intensity by three independent researchers and averaged for the final IHC score. Images representing scores 0–3 are shown in Fig. S4a. Student’s t-test statistical analysis was performed using Prism 7 (Graphpad).
Mammary Fat Pad Clearance Assay
Mammary fat pad clearance surgery was performed on 3-week old NOD.CB17-Prkdcscid/J mice (Breeders obtained from Jackson Labs) according to the minimally invasive protocol 60. Briefly, mice were anesthetized using isoflurane and prepared for surgery by shaving the region around the 4th nipple, scrubbed the sight with betadine and alcohol wipes alternating three times each and applied topical lidocaine. A small incision around the 4th nipple was made, adjacent fat pad was separated from skin and surrounding muscle, and excised. 5,000 cells as indicated in 10μL of PBS suspension was injected into the visible cleared fat pad using a pointed Hamilton Syringe. Reconstituted fat pads were excised after a 6-week period. After extraction, fat pads were spread on a glass slide, allowed to air dry briefly to prevent future dissociation from the slide and placed in Carnoy’s Fixative (60% ethanol, 30% CHCl3, and 10% glacial acetic acid) overnight at 4°C. Slides were washed in 70% ethanol, 50% ethanol, 30% ethanol and water, each step for 20 minutes at room temperature. After hydration, slides were placed in Carmine stain overnight at room temperature. Carmine stain was prepared as follows: Place 1g carmine (Sigma C1022) and 2.5g aluminum potassium sulfate (Sigma A7167) in 500mL dH20 and boil 20 min. Adjusted final volume to 500mL with H20. Filtered and refrigerated. Once stained, fat pads were dehydrated in sequential ethanol washes at 70%, 95% and 100% for 15 minutes each at room temperature. Fat was cleared using toluene overnight and slides were mounted using Permount (Fisher). Slides were visualized for Carmine stain by bright field imaging, and RFP and GFP fluorescent signal using a 1.25x objective on an Olympus BX61 Fluorescent Microscope with an Olympus DP72 8-bit RGB camera and Cellsens software in the Laboratory Core in the Center for Oral Health Research at MUSC.
Serial Dilution Mammary Fat Pad Injection Studies
8 week old NOD.CB17-Prkdcscid/J mice (Jackson Labs) were injected with 100μL PBS cell suspension using E1KD shScr, E1KD shILEI or E1KD shLIFR cells at 1,000, 10,000 or 100,000 cells per fat pad. Mice were evaluated weekly using calipers to assess tumor volume. Mice were sacrificed and tumors and lungs were extracted at 13–14 weeks. Final tumor volumes were weighed and lungs were stained for H&E. Briefly, lungs were formalin-fixed and paraffin-embedded in the Biorepository and Tissue Analysis Core at MUSC. Paraffin-embedded lungs were cut into 5 μm sections and stained with hematoxylin and eosin. H&E stained lungs were imaged using a 1.25x objective on an Olympus BX61 Microscope with an Olympus DP72 8-bit RGB camera and Cellsens software in the Laboratory Core in the Center for Oral Health Research at MUSC. Two-way Anova statistical analysis for volume measurements over time and Student t-test analysis of final tumor weights and metastatic tumor burden were performed using Prism 7 (Graphpad).
PyMT Tumor Growth and Isolation
MMTV-PyMT mouse model was used to investigate tumor progression in vivo 61. This model permits the examination of in situ tumor progression from hyperplasia and MMTV-PyMT mice develop palpable mammary tumors as early as 5 weeks. In order to examine metastasis progression, the mice were scarified when tumor reached 1cm3 and both primary tumors and lungs were collected to perform IHC as described in the above section.
Supplementary Material
Figure S1. (a) Immunoblot analysis of hnRNP-E1 protein levels in parental NMuMG cells stably transduced with either scramble control shRNA or shRNA targeting hnRNP-E1. (b) E1KD mammospheres stained for basal and luminal cytokeratins. (c) Immunoblot analysis of trypsin-selected and FACS sorted (CD44/CD24) HMLE mesenchymal, HMLE epithelial, and HMLE epithelial cell lines silenced for hnRNP E1. (d) Example of gating technique used for HMLE flow experiments.
Figure S2. (a) Phase contrast images of NMuMG E1KD shSCR and NMuMG E1KD shILEI cells in the presence and absence of TGFβ stimulation for 24 hours. (b) Blot demonstrating ILEI knockdown with three shILEI constructs compared to shSCR in A549 cells. (c) Oncosphere quantification after 10 days of growth in minimal media for A549 shSCR and two shILEI cell lines. (d) Representative images for A549 oncospheres quantified in Fig. S2c. (e) Semi-quantitative PCR for BMI1 and Nanog in NMuMG and E1KD cells. (f) Semi-quantitative PCR for BMI1, Nanog, and ILEI RNA levels in E1KD cells silenced for ILEI.
Figure S3. (a) Schematic of construct design and purification of E.coli-derived rILEI through the incorporation of an N-terminal TEV-cleavable hexahistidine tag preceding the mature ILEI sequence comprising amino acids 43–227. (b) Purified rILEI analyzed by commassie and immunoblot in the presence and absence of beta-mercaptoethanol.
Figure S4. (a) TMA images representing 0–3 IHC scores. (b) Representative IHC images for each condition shown in Figure 4c-e.
Figure S5. (a) Yeast 2-hybrid of ILEI 43–227 probed against a HELA cDNA library demonstrating activation of adenine reporter and colony growth corresponding to mature ILEI interacting with LIFR precursor. (b) Immunoblot analysis of LIFR levels in TGFβ treated NMuMG and E1KD shSCR versus shLIFR cells. (c) Quantification of mammosphere formation in E1KD shSCR/ shILEI/ shLIFR cells in the presence and absence of 10nM purified recombinant ILEI (error bars represent mean +/− SD; n=5; ****p<0.0001, unpaired Student’s t-test). (d) Immunoblot analysis of ILEI and LIFR levels in E1KD cells transiently transfected with si molecules. (e) FACS analysis of NMuMG, E1KD shSCR, E1KD shILEI, and E1KD shLIFR cells using the ALDEFLUOR Assay as described by the manufacturer and analyzed using FlowJo Software.
Figure S6. (a) Full blot showing free ligand from whole cell lysate after 125I ligand incubation and BS3 crosslinking. (b) Immunoblot analysis of FLAG-LIFR overexpression in HEK293 cells.
Figure S7. (a) Immunoblot analysis of serum starved E1KD cells treated with ILEI or LIF in the presence and absence of the STAT3 inhibitor Stattic. (b) Immunoblot analysis of E1KD cells for pERK1/2 and total ERK levels treated with the MEK1/2 inhibitor U0126. (c) Quantification of E1KD mammospheres treated with the MEK1/2 inhibitor U0126 (error bars represent mean +/− SD; n=5; *p<0.05, unpaired Student’s t test). (d) Immunoblot analysis of pERK1/2 in serum starved E1KD cells treated with a partially purified ILEI or 10nM recombinant purified ILEI. (e) Quantification of mammosphere formation in the indicated cell lines supplemented with increasing concentrations of rLIF compared to NMuMG cells (error bars represent mean +/− SEM; n=5; ***p<0.001, unpaired Student’s t test).
Figure S8. (a) Images of tumors derived from female NOD/SCID mice injected with 100k E1KD shSCR, shILEI, and shLIFR cells. (b) Quantification of lung metastases in lungs derived from female mice injected with E1KD shSCR, shILEI, and shLIFR cells. (c) Immunoblot analysis of LIFR levels in epithelial and mesenchymal HMLE cells. (d) Immunoblot analysis of ILEI from conditioned media of E1KD, M1P, and L1P cells. (e) Sections of FFPE primary tumors from MMTV-PyMT mice stained for H&E or IHC for ILEI and LIFR and imaged at 10x or 40x magnification. (f) Sections of FFPE lungs from MMTV-PyMT mice stained for H&E or IHC for ILEI and LIFR and imaged at 10x or 40x magnification.
Acknowledgements
The authors would like to thank Ken Noguchi and the Howe laboratory members for their helpful discussions and critical reading of the manuscript. The SUM cell lines and HMLE cell lines were a generous gift from Dr. Steven Ethier and Dr. Juan Massague respectively. Funding for this project was provided by DOD BCRP BC170301 to A.C.D., NIH NCI T32 CA193201 to A.C.D., NIH NCI F31 CA213627 to A.N.W., and NIH NCI RO1CA154663 to P.H.H.. Cores at MUSC used for this project include the shRNA Shared Technology Resource, Flow Cytometry and Cell Sorting Unit, Laboratory Core in the Center for Oral Health Research, Cell and Molecular Imaging Core and the Biorepository and Tissue Analysis Shared Resource. The U.S. Army Medical Research Acquisition Activity, 820 Chandler Street, Fort Detrick MD 21702–5014 is the awarding and administering acquisition office for A.C.D.. This work was supported by the Office of the Assistant Secretary of Defense for Health Affairs through the Breast Cancer Research Program, under Award No. W81XWH-18–1-0003. Opinions, interpretations, conclusions and recommendations are those of the author and are not necessarily endorsed by the Department of Defense. In conducting research using animals, the investigators adhere to the laws of the United States and regulations of the Department of Agriculture.
Footnotes
Conflict of Interest
The authors declare no conflicts of interest.
References
- 1.Liu F, Gu L-N, Shan B-E, Geng C-Z & Sang M-X Biomarkers for EMT and MET in breast cancer: An update. Oncol. Lett 12, 4869–4876 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yates LR et al. Genomic Evolution of Breast Cancer Metastasis and Relapse. Cancer Cell 32, 169–184.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kalluri R & Weinberg RA The basics of epithelial-mesenchymal transition. J. Clin. Invest 119, 1420–8 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Larue L & Bellacosa A Epithelial-mesenchymal transition in development and cancer: role of phosphatidylinositol 3’ kinase/AKT pathways. Oncogene 24, 7443–54 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Yu M et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 339, 580–4 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsai JH, Donaher JL, Murphy DA, Chau S & Yang J Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell 22, 725–36 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ & Clarke MF Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci 100, 3983–3988 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kreso A & Dick JE Evolution of the cancer stem cell model. Cell Stem Cell 14, 275–91 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Ye X et al. Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature 525, 256–60 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Velasco-Velázquez MA, Homsi N, De La Fuente M & Pestell RG Breast cancer stem cells. Int. J. Biochem. Cell Biol 44, 573–7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lessard J & Sauvageau G Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature 423, 255–60 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Chiou S-H et al. Positive correlations of Oct-4 and Nanog in oral cancer stem-like cells and high-grade oral squamous cell carcinoma. Clin. Cancer Res. 14, 4085–95 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Tomita H, Tanaka K, Tanaka T & Hara A Aldehyde dehydrogenase 1A1 in stem cells and cancer. Oncotarget 7, 11018–32 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mani SA et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 133, 704–15 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Asiedu MK, Ingle JN, Behrens MD, Radisky DC & Knutson KL TGFbeta/TNF(alpha)-mediated epithelial-mesenchymal transition generates breast cancer stem cells with a claudin-low phenotype. Cancer Res. 71, 4707–19 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richards EJ et al. Long non-coding RNAs (LncRNA) regulated by transforming growth factor (TGF) β: LncRNA-hit-mediated TGFβ-induced epithelial to mesenchymal transition in mammary epithelia. J. Biol. Chem 290, 6857–67 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chaudhury A et al. TGF-beta-mediated phosphorylation of hnRNP E1 induces EMT via transcript-selective translational induction of Dab2 and ILEI. Nat. Cell Biol 12, 286–93 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hussey GS et al. Identification of an mRNP complex regulating tumorigenesis at the translational elongation step. Mol. Cell 41, 419–31 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hussey GS et al. Establishment of a TGFβ-induced post-transcriptional EMT gene signature. PLoS One 7, e52624 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu Y et al. Cloning, expression, and initial characterization of a novel cytokine-like gene family. Genomics 80, 144–50 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Hasegawa H, Liu L, Tooyama I, Murayama S & Nishimura M The FAM3 superfamily member ILEI ameliorates Alzheimer’s disease-like pathology by destabilizing the penultimate amyloid-β precursor. Nat. Commun 5, 3917 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Katahira T, Nakagiri S, Terada K & Furukawa T Secreted factor FAM3C (ILEI) is involved in retinal laminar formation. Biochem. Biophys. Res. Commun 392, 301–6 (2010). [DOI] [PubMed] [Google Scholar]
- 23.Määttä JA et al. Fam3c modulates osteogenic cell differentiation and affects bone volume and cortical bone mineral density. Bonekey Rep. 5, 787 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Waerner T et al. ILEI: a cytokine essential for EMT, tumor formation, and late events in metastasis in epithelial cells. Cancer Cell 10, 227–39 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Lahsnig C et al. ILEI requires oncogenic Ras for the epithelial to mesenchymal transition of hepatocytes and liver carcinoma progression. Oncogene 28, 638–50 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Csiszar A et al. Interleukin-like epithelial-to-mesenchymal transition inducer activity is controlled by proteolytic processing and plasminogen-urokinase plasminogen activator receptor system-regulated secretion during breast cancer progression. Breast Cancer Res. 16, 433 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Korkaya H et al. Activation of an IL6 inflammatory loop mediates trastuzumab resistance in HER2+ breast cancer by expanding the cancer stem cell population. Mol. Cell 47, 570–84 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim S-Y et al. Role of the IL-6-JAK1-STAT3-Oct-4 pathway in the conversion of non-stem cancer cells into cancer stem-like cells. Cell. Signal. 25, 961–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ginestier C et al. CXCR1 blockade selectively targets human breast cancer stem cells in vitro and in xenografts. J. Clin. Invest. 120, 485–97 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nicola NA & Babon JJ Leukemia inhibitory factor (LIF). Cytokine Growth Factor Rev. 26, 533–44 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yue X, Wu L & Hu W The regulation of leukemia inhibitory factor. Cancer cell Microenviron. 2, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X et al. LIF promotes tumorigenesis and metastasis of breast cancer through the AKT-mTOR pathway. Oncotarget 5, 788–801 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson RW et al. Induction of LIFR confers a dormancy phenotype in breast cancer cells disseminated to the bone marrow. Nat. Cell Biol 18, 1078–1089 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen D et al. LIFR is a breast cancer metastasis suppressor upstream of the Hippo-YAP pathway and a prognostic marker. Nat. Med 18, 1511–7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wahl GM & Spike BT Cell state plasticity, stem cells, EMT, and the generation of intra-tumoral heterogeneity. NPJ breast cancer 3, 14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grelet S et al. A regulated PNUTS mRNA to lncRNA splice switch mediates EMT and tumour progression. Nat. Cell Biol 19, 1105–1115 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Forozan F et al. Molecular cytogenetic analysis of 11 new breast cancer cell lines. Br. J. Cancer 81, 1328–34 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsui WH Cancer stem cell signaling pathways. Medicine (Baltimore). 95, S8–S19 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schust J, Sperl B, Hollis A, Mayer TU & Berg T Stattic: A Small-Molecule Inhibitor of STAT3 Activation and Dimerization. Chem. Biol 13, 1235–1242 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Junk DJ et al. Oncostatin M promotes cancer cell plasticity through cooperative STAT3-SMAD3 signaling. Oncogene 36, 4001–4013 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu Y & Smyth GK ELDA: Extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods 347, 70–78 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Howley BV, Link LA, Grelet S, El-Sabban M & Howe PH A CREB3-regulated ER-Golgi trafficking signature promotes metastatic progression in breast cancer. Oncogene 37, 1308–1325 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Akhurst RJ & Derynck R TGF-beta signaling in cancer--a double-edged sword. Trends Cell Biol. 11, S44–51 (2001). [DOI] [PubMed] [Google Scholar]
- 44.Lebrun J-J The Dual Role of TGFβ in Human Cancer: From Tumor Suppression to Cancer Metastasis. ISRN Mol. Biol 2012, 381428 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomas SJ, Snowden JA, Zeidler MP & Danson SJ The role of JAK/STAT signalling in the pathogenesis, prognosis and treatment of solid tumours. Br. J. Cancer 113, 365–71 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang T et al. JAK/STAT3-Regulated Fatty Acid β-Oxidation Is Critical for Breast Cancer Stem Cell Self-Renewal and Chemoresistance. Cell Metab. 27, 136–150.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang X et al. STAT3 mediates resistance of CD44(+)CD24(-/low) breast cancer stem cells to tamoxifen in vitro. J. Biomed. Res 26, 325–35 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hirai H, Karian P & Kikyo N Regulation of embryonic stem cell self-renewal and pluripotency by leukaemia inhibitory factor. Biochem. J 438, 11–23 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ishihara K & Hirano T Molecular basis of the cell specificity of cytokine action. Biochim. Biophys. Acta 1592, 281–96 (2002). [DOI] [PubMed] [Google Scholar]
- 50.Underhill-Day N & Heath JK Oncostatin M (OSM) cytostasis of breast tumor cells: characterization of an OSM receptor beta-specific kernel. Cancer Res. 66, 10891–901 (2006). [DOI] [PubMed] [Google Scholar]
- 51.Port MD, Gibson RM & Nathanson NM Differential stimulation-induced receptor localization in lipid rafts for interleukin-6 family cytokines signaling through the gp130/leukemia inhibitory factor receptor complex. J. Neurochem 101, 782–93 (2007). [DOI] [PubMed] [Google Scholar]
- 52.Gerlinger M et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med 366, 883–892 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Waerner T et al. ILEI: a cytokine essential for EMT, tumor formation, and late events in metastasis in epithelial cells. Cancer Cell 10, 227–39 (2006). [DOI] [PubMed] [Google Scholar]
- 54.Ma D et al. Leukemia inhibitory factor receptor negatively regulates the metastasis of pancreatic cancer cells in vitro and in vivo. Oncol. Rep 36, 827–36 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Luo Q et al. LIFR functions as a metastasis suppressor in hepatocellular carcinoma by negatively regulating phosphoinositide 3-kinase/AKT pathway. Carcinogenesis 36, 1201–12 (2015). [DOI] [PubMed] [Google Scholar]
- 56.Guo H, Cheng Y, Martinka M & McElwee K High LIFr expression stimulates melanoma cell migration and is associated with unfavorable prognosis in melanoma. Oncotarget 6, 25484–98 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lv S et al. Histone methyltransferase KMT2D sustains prostate carcinogenesis and metastasis via epigenetically activating LIFR and KLF4. Oncogene 37, 1354–1368 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Howley BV, Hussey GS, Link LA & Howe PH Translational regulation of inhibin βA by TGFβ via the RNA-binding protein hnRNP E1 enhances the invasiveness of epithelial-to-mesenchymal transitioned cells. Oncogene 35, 1725–35 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.James P, Halladay J & Craig EA Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425–36 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brill B, Boecher N, Groner B & Shemanko CS A sparing procedure to clear the mouse mammary fat pad of epithelial components for transplantation analysis. Lab. Anim 42, 104–10 (2008). [DOI] [PubMed] [Google Scholar]
- 61.Lin EY et al. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am. J. Pathol 163, 2113–26 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. (a) Immunoblot analysis of hnRNP-E1 protein levels in parental NMuMG cells stably transduced with either scramble control shRNA or shRNA targeting hnRNP-E1. (b) E1KD mammospheres stained for basal and luminal cytokeratins. (c) Immunoblot analysis of trypsin-selected and FACS sorted (CD44/CD24) HMLE mesenchymal, HMLE epithelial, and HMLE epithelial cell lines silenced for hnRNP E1. (d) Example of gating technique used for HMLE flow experiments.
Figure S2. (a) Phase contrast images of NMuMG E1KD shSCR and NMuMG E1KD shILEI cells in the presence and absence of TGFβ stimulation for 24 hours. (b) Blot demonstrating ILEI knockdown with three shILEI constructs compared to shSCR in A549 cells. (c) Oncosphere quantification after 10 days of growth in minimal media for A549 shSCR and two shILEI cell lines. (d) Representative images for A549 oncospheres quantified in Fig. S2c. (e) Semi-quantitative PCR for BMI1 and Nanog in NMuMG and E1KD cells. (f) Semi-quantitative PCR for BMI1, Nanog, and ILEI RNA levels in E1KD cells silenced for ILEI.
Figure S3. (a) Schematic of construct design and purification of E.coli-derived rILEI through the incorporation of an N-terminal TEV-cleavable hexahistidine tag preceding the mature ILEI sequence comprising amino acids 43–227. (b) Purified rILEI analyzed by commassie and immunoblot in the presence and absence of beta-mercaptoethanol.
Figure S4. (a) TMA images representing 0–3 IHC scores. (b) Representative IHC images for each condition shown in Figure 4c-e.
Figure S5. (a) Yeast 2-hybrid of ILEI 43–227 probed against a HELA cDNA library demonstrating activation of adenine reporter and colony growth corresponding to mature ILEI interacting with LIFR precursor. (b) Immunoblot analysis of LIFR levels in TGFβ treated NMuMG and E1KD shSCR versus shLIFR cells. (c) Quantification of mammosphere formation in E1KD shSCR/ shILEI/ shLIFR cells in the presence and absence of 10nM purified recombinant ILEI (error bars represent mean +/− SD; n=5; ****p<0.0001, unpaired Student’s t-test). (d) Immunoblot analysis of ILEI and LIFR levels in E1KD cells transiently transfected with si molecules. (e) FACS analysis of NMuMG, E1KD shSCR, E1KD shILEI, and E1KD shLIFR cells using the ALDEFLUOR Assay as described by the manufacturer and analyzed using FlowJo Software.
Figure S6. (a) Full blot showing free ligand from whole cell lysate after 125I ligand incubation and BS3 crosslinking. (b) Immunoblot analysis of FLAG-LIFR overexpression in HEK293 cells.
Figure S7. (a) Immunoblot analysis of serum starved E1KD cells treated with ILEI or LIF in the presence and absence of the STAT3 inhibitor Stattic. (b) Immunoblot analysis of E1KD cells for pERK1/2 and total ERK levels treated with the MEK1/2 inhibitor U0126. (c) Quantification of E1KD mammospheres treated with the MEK1/2 inhibitor U0126 (error bars represent mean +/− SD; n=5; *p<0.05, unpaired Student’s t test). (d) Immunoblot analysis of pERK1/2 in serum starved E1KD cells treated with a partially purified ILEI or 10nM recombinant purified ILEI. (e) Quantification of mammosphere formation in the indicated cell lines supplemented with increasing concentrations of rLIF compared to NMuMG cells (error bars represent mean +/− SEM; n=5; ***p<0.001, unpaired Student’s t test).
Figure S8. (a) Images of tumors derived from female NOD/SCID mice injected with 100k E1KD shSCR, shILEI, and shLIFR cells. (b) Quantification of lung metastases in lungs derived from female mice injected with E1KD shSCR, shILEI, and shLIFR cells. (c) Immunoblot analysis of LIFR levels in epithelial and mesenchymal HMLE cells. (d) Immunoblot analysis of ILEI from conditioned media of E1KD, M1P, and L1P cells. (e) Sections of FFPE primary tumors from MMTV-PyMT mice stained for H&E or IHC for ILEI and LIFR and imaged at 10x or 40x magnification. (f) Sections of FFPE lungs from MMTV-PyMT mice stained for H&E or IHC for ILEI and LIFR and imaged at 10x or 40x magnification.