Significance
Current antibiotics are increasingly ineffective due to rising resistance, and antibiotic discovery campaigns frequently fail. One key factor in effective antibiotic discovery is knowing whether a target’s function is essential in a bacterial species. We present a general paradigm for comprehensively identifying the core essential genome of clinical pathogens to identify candidate drug targets. We applied the paradigm to Pseudomonas aeruginosa, a priority antibiotic-resistant pathogen, by performing genome-wide genetic selection studies across a diverse set of clinical isolates and infection-relevant growth conditions (serum, sputum, and urine). We identified 321 core essential genes that constitute a high-priority list of candidate targets for drug discovery. The strategy should be applicable to define the core essential genome for most clinical pathogens.
Keywords: antibiotic discovery, gram-negative pathogens, ESKAPE, Tn-Seq
Abstract
Genomics offered the promise of transforming antibiotic discovery by revealing many new essential genes as good targets, but the results fell short of the promise. While numerous factors contributed to the disappointing yield, one factor was that essential genes for a bacterial species were often defined based on a single or limited number of strains grown under a single or limited number of in vitro laboratory conditions. In fact, the essentiality of a gene can depend on both the genetic background and growth condition. We thus developed a strategy for more rigorously defining the core essential genome of a bacterial species by studying many pathogen strains and growth conditions. We assessed how many strains must be examined to converge on a set of core essential genes for a species. We used transposon insertion sequencing (Tn-Seq) to define essential genes in nine strains of Pseudomonas aeruginosa on five different media and developed a statistical model, FiTnEss, to classify genes as essential versus nonessential across all strain–medium combinations. We defined a set of 321 core essential genes, representing 6.6% of the genome. We determined that analysis of four strains was typically sufficient in P. aeruginosa to converge on a set of core essential genes likely to be essential across the species across a wide range of conditions relevant to in vivo infection, and thus to represent attractive targets for novel drug discovery.
All current antibiotics to date target essential functions in the bacterial cell. The sequencing of the first bacterial genome in 1995 (1) offered the hope of revolutionizing antibiotic discovery by revealing the breadth of genes that could be mined for antibiotic targets, enabling genome-wide genetic screens to identify essential genes in a given bacterial species and paving the way for chemical screens to find new antibiotics inhibiting these essential targets. However, this revolution has, to date, failed to materialize.
Several factors contributed to the disappointing yield of new antibiotic candidates. These include the challenge of overcoming the impermeable membrane and efflux pumps in bacteria, which have made it difficult to translate inhibitors found in biochemical assays into compounds with whole-cell activity; the need for improved chemical libraries to provide better starting points for chemical optimization against bacteria; and the focus on searching for broad-spectrum agents with activity against a range of bacterial species (2, 3).
Another contributing factor, however, has been the erroneous determination of target essentiality, resulting in the pursuit of inhibitors of targets that are either not essential at all in the species or not essential in a subset of strains. Indeed, two major studies experienced this challenge, with one study describing “genomic blind spots” involving targets that were erroneously thought to be essential based on the limited pathogen genomic data available at the time, but were actually nonessential in additional subsequently tested strains (4, 5). As a result, some inhibitors of targets thought to be essential failed to have good activity against the full range of relevant pathogen strains (2, 3).
Another issue is that some targets may be essential only under certain growth conditions (i.e., conditional essentiality) (6). Given the variable environments encountered by bacterial pathogens in laboratory media and different infection types (i.e., blood, urine, lung, abscess infections), genes essential in artificial laboratory growth conditions need not be essential during human infection. This concept has been illustrated in the ongoing debate of whether fatty acid biosynthesis, specifically the type II fatty-acid synthesis (FASII) pathway, is essential in gram-positive bacteria during infection (7). Although the pathway is essential under conventional in vitro laboratory conditions, Brinster et al. (8) challenged its essentiality in vivo by showing that several gram-positive pathogens could be rescued in vitro by the addition of exogenous unsaturated fatty acids and that a mutant of Streptococcus agalactiae in which numerous FASII genes had been deleted could be grown in both human serum and septicemia mouse infection models, presumably because of its ability to scavenge host fatty acids. The concept of conditional essentiality is similarly illustrated in the notable example of an inhibitor series that was developed to have potent in vitro activity against Mycobacterium tuberculosis, but later found to have no activity in a mouse tuberculosis model because the inhibitor’s activity was dependent on growth in glycerol, the carbon source present in a standard laboratory medium but not the source utilized by M. tuberculosis during infection (9). These examples clearly highlight the value of defining targets relevant to in vivo infection, and not simply to in vitro conditions. At the same time, we note that targets that may be essential under some, but not all, relevant in vivo conditions may provide novel approaches to infection site-specific agents.
Given the challenges of genomic blind spots and conditional essentiality, we propose that antibiotic discovery could be improved by focusing on “core essential genes,” by which we mean genes that are essential across virtually all strains of a pathogen species and all relevant growth conditions. We therefore sought to develop a robust paradigm for defining the core essential genes of a bacterial species. We focused on Pseudomonas aeruginosa, a clinically significant pathogen that is a major cause of bacteremia as well as pulmonary and urinary tract infections, with high mortality rates (10–12) and for which there is the greatest need for new antibiotics. Due to its ability to evade current antibiotics or develop resistance, P. aeruginosa clinical strains are increasingly resistant to all current antibiotics (13, 14). The WHO has recently classified P. aeruginosa as a priority pathogen in need of research investment and new drugs (15). Alarmingly, only one in five antibacterial drugs succeed in clinical trials (16), and of the 42 potential antibacterials in development as of 2018, only two have expected activity against P. aeruginosa, with only one of these having a new mechanism of action (https://www.pewtrusts.org/en/research-and-analysis/articles/2016/12/tracking-the-pipeline-of-antibiotics-in-development).
In pursuit of a general strategy to define a pathogen’s core essential genome, we examined two fundamental questions. First, how accurately can the core essential genome be identified based on essentiality in one strain under one laboratory growth condition? Second, how many strains must be examined to converge on a set of core essential genes that are likely to be essential under conditions relevant for infection, and thus may be good drug targets? We addressed this question by using transposon insertion sequencing [Tn-Seq; also abbreviated as TIS, INseq, HITS, or TraDIS (17–21)] to perform genome-wide negative selection studies on libraries of transposon-insertion mutants under different growth conditions, with the distribution of transposon insertions determined by sequencing the pool of strains. Genes that are important for optimal growth under a specific growth condition can be identified, because the corresponding mutants containing disrupting transposon insertions in these genes will be significantly depleted from the pool of all possible mutants. These methods have been applied to the two commonly studied reference laboratory strains of P. aeruginosa, PA14 and PAO1, with varying numbers and identities of essential genes (22). Here, we applied this method to PA14 on Luria–Bertani (LB) medium and compared the essential genome determined from this single strain on a single laboratory-based medium with eight other diverse strains of P. aeruginosa under five different growth conditions. The strains comprised isolates from various human infections (including pulmonary, urinary, blood, wound, and ocular) and one environmentally isolated strain, while the growth conditions comprised three media intended to simulate the conditions of human infection (sputum, serum, and urine) and two laboratory-based media (LB and M9 minimal media). We further developed a simple statistical method, called FiTnEss (Finding Tn-Seq Essential genes), that maps measurements of fitness of individual transposon mutants onto a binary classification of essential or nonessential with user-defined levels of stringency. We applied FiTnEss to the Tn-Seq data from all strain and medium combinations and defined a set of 321 core essential genes, which represent 6.6% of the genome, that constitute a high-priority list of candidate targets for drug discovery against this important pathogen. Finally, we calculated that as few as four individual strains could be examined in combination to approach a plateau of core essential genes across a given species.
Results
Transposon Mutagenesis, Sequencing, and Mapping of Transposon Insertions.
We chose strains from a collection of 130 clinical P. aeruginosa isolates obtained from various sources (Materials and Methods). We performed whole-genome sequencing on the collection and mapped the isolates to a phylogenetic tree formed by 2,560 P. aeruginosa genomes in the National Center for Biotechnology Information (NCBI) Genome database. We also tested a subset for their ability to be efficiently mutagenized by the Himar1-derived transposon MAR2xT7 (23–25). We selected the MAR2xT7 transposon because it is engineered to reduce polar effects that can occur when transposon integration into a nonessential gene disrupts the transcription of a downstream essential gene (because it contains a gentamicin resistance cassette that lacks a transcriptional terminator downstream, and thus allows downstream transcription from the gentamicin resistance promoter).
Based on this information, we focused on nine strains that represented five different infection types (blood, urine, respiratory, ocular, and wound), with each strain representing a different branch of the dendrogram (NCBI; Fig. 1A). The genomes of these nine strains varied from 6.34 to 7.15 Mbp.
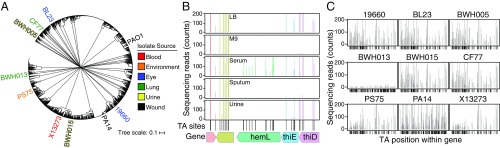
Fig. 1.
Tn-Seq of P. aeruginosa clinical isolates. (A) Phylogenetic dendrogram of 2,560 P. aeruginosa genomes (NCBI Genome database); PAO1 and strains selected for mutagenesis are indicated. (B) Variable sequencing reads that map to TA sites in an exemplary region of five genes in strain PA14 (including hemL, thiE, and thiD) under different growth conditions highlight the conditional essentiality of these genes. The gene arrow indicates gene direction from 5′ to 3′. (C) Normalized read counts mapping to the pilY1 gene (3,477 bp) in all nine strains in LB medium demonstrate the variable essentiality of pilY1 in different strains as an example of the genomic heterogeneity of P. aeruginosa isolates. Genes are shown 5′ (left) to 3′ (right); a black tick on the x axis indicates the TA insertion site location within the gene.
We constructed transposon libraries by performing tripartite matings of these nine P. aeruginosa strains with Escherichia coli donor strain SM10 carrying an episomal MAR2xT7 transposon (24) and E. coli strain SM10 carrying an episomal hyperactive transposase that results in efficient integration at the dinucleotide sequence “TA” (26) (SI Appendix, Fig. S1). Separating the transposase and transposon increased the efficiency of insertion sequencing and mapping, relative to the more common system of a single plasmid carrying both the transposase and the transposon. We obtained at least 5 × 106 distinct mutants for each strain from at least two independent conjugations, and selected mutants on each of five different agar media directly to avoid a bottleneck from preselecting the libraries on a given medium. To ensure saturating mutagenesis, a total of 1 × 106 mutants were selected on each medium in duplicate, yielding 10-fold more transposon mutants than possible insertion sites. The media types included rich (LB) and minimal (M9) laboratory media, to provide the boundaries (extremes of growth conditions) for essential gene identification, and three media intended to resemble infection site fluids: FBS, synthetic cystic fibrosis sputum (SCFM) (27), and urine. We mapped the transposon-insertion sites to the corresponding reference genomes for each strain.
In all, we created 90 Tn-Seq datasets (nine strains grown on five media, performed in duplicate), with an average number of mapped reads of ∼107. Reads at each TA site were highly concordant between replicates, with a mean R2 = 0.98 (Dataset S1).
Visual inspection readily identified examples of genes that were variably essential under different growth conditions for a certain strain, illustrating the conditional essentiality of some genes (Fig. 1B). For example, the thiamine synthesis genes thiD and thiE showed few insertions in M9 minimal medium, which lacks thiamine, but an abundance of insertions in rich LB medium, indicating their essentiality in M9 but not LB medium. Variability is also seen for the hemL gene under different growth conditions. We similarly saw examples of genes that were variably essential in different strains under the same growth condition. For example, the pilY1 gene did not tolerate insertions in strain BWH013, but readily tolerated insertions in the other eight strains, when grown on LB medium, highlighting the genomic plasticity of P. aeruginosa (Fig. 1C).
To optimize our accuracy in calling genes essential or nonessential, we removed from our analysis three classes of TA sites that can lead to technical errors. These classes include (i) nonpermissive insertion sites consisting of the sequence (GC)GNTANC(GC), which was recently reported to be intolerant to Himar1 transposon insertions in M. tuberculosis (28) and which we confirmed is also intolerant in P. aeruginosa; (ii) nondisruptive terminal insertions within 50 bp of the 5′- and 3′-gene termini (a distance we optimized empirically), which can nevertheless result in the expression of a functional, albeit truncated, version of the corresponding gene product (29); and (iii) insertion sequences at which genomic sequences flanking a TA site were not unique and could not be accurately mapped (Materials and Methods and SI Appendix, Fig. S2). In total, we removed 16,499 of 81,328 TA sites (20%) in PA14, which resulted in our inability to assess 150 genes in PA14 (2.5%). The inability to assess the essentiality of genes that contain zero TA sites (30) removed another 185 genes from analysis in PA14. In total, we were able to assess the essentiality of 5,708 of the 5,893 total genes in the PA14 genome (97%). The statistics were similar for the other eight strains (SI Appendix, Table S1).
FiTnEss: A Statistical Model to Identify Essential Genes.
We next sought to perform a comprehensive and quantitative analysis of the 90 Tn-Seq datasets. While various methods exist for analyzing Tn-Seq data (17, 31–33), they differ in their complexity and their stringency for calling a gene as essential. We thus developed a simple model and method (FiTnEss) for identifying essential genes from Tn-Seq data that required minimal assumptions and had good predictive power. The important features of FiTnEss are that (i) it evaluates genes (rather than individual TA sites or stretches of TA sites) and (ii) it uses a simple two-parameter model to capture all of the salient features of the data (with the simple model yielding greater statistical power).
FiTnEss assumes that the number of reads observed at a particular TA site in a gene depends on the fitness of loss-of-function mutant in the gene. We found that the distribution of the number of TA sites per gene is bimodal, with presumed nonessential genes on the right and essential genes on the left (SI Appendix, Fig. S3A). The distribution of nonessential genes can be well fit by a model in which the read counts at TA sites in a gene follow a geometric distribution with probability pg, with 1/pg drawn from a log-normal distribution. This model requires only two parameters: μ (the mean fitness for nonessential genes, as reflected in the mean reads per TA site in a gene) and σ (the variance of the fitness for nonessential genes). We calculate μ and σ for each individual Tn-Seq dataset, based on nonessential genes and then apply it to the entire dataset. Because the validity of the model depends on our estimates of μ and σ based on nonessential genes, we are conservative in including nonessential genes: We take only genes from the extreme right side of the distribution (top 75% of genes with the most reads, with 10 TA sites).
This model requires one other assumption, that the fitness distribution does not depend on gene size or number of TA sites contained in a gene. Rephrased, the model assumes that the number of reads at each TA site is independent of the numbers of TA sites in a gene. We tested the validity of this assumption. Because detection power for a gene depends on the number of TA sites in the gene (SI Appendix, Fig. S3B), we compared the distributions of reads at randomly sampled TA sites, allowing us to compare similar numbers of sites in genes of different size. We found that the numbers of reads at randomly sampled TA sites were independent of the numbers of TA sites in a gene (SI Appendix, Fig. S3D), confirming the validity of our assumption.
Using the two parameters (determined individually for each dataset), we then constructed a theoretical “nonessential” distribution for each gene size category within each corresponding dataset and calculated the probability (P value) of a given gene coming from this nonessential distribution. To vary the stringency with which we called essentiality, we applied two different levels of multiple testing adjustment: one with maximal stringency to offer the highest confidence set of essential genes [family-wise error rate (FWER)] to identify genes with no or very few sequencing reads and one with high stringency yet slightly relaxed [false discovery rate (FDR)] to identify genes that are statistically significant yet contain a low number of reads. Genes with an adjusted P value <0.05 in both replicates were predicted to be essential (Fig. 2A and SI Appendix, Additional Methods and Results). Virtually all maximal stringency calls are expected to be true essential genes, while among the high-stringency set, a small number of false-positive predictions are expected.
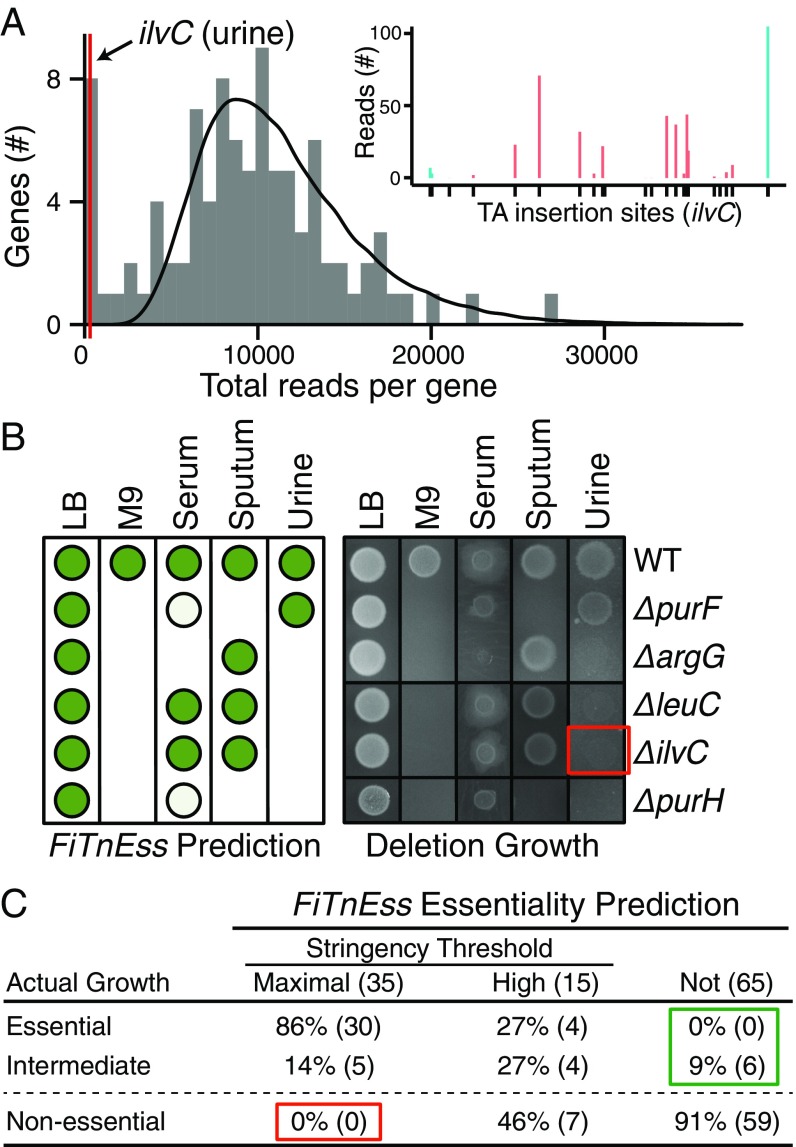
Fig. 2.
Validation of FiTnEss predictions on a set of conditionally essential gene deletions. (A) FiTnEss prediction for ilvC in urine. The gray histogram is the actual distribution of Tn-Seq reads for all genes with 18 insertion sites in PA14 grown in urine, the black line is the theoretical distribution calculated by FiTnEss, and the red line is where ilvC falls at the far left (i.e., essential) of the bimodal distribution. (Inset) Read numbers at useable (red) and removed (blue) TA sites are shown; a black tick on the x axis indicates the TA insertion site location within the gene, and the gene is presented 5′ (left) to 3′ (right). (B) FiTnEss essentiality predictions (Left; nonessential, high-stringency essential, and maximal stringency essential predictions are displayed as dark green circles, light green circles, and blank spaces, respectively) of five representative gene deletion mutants from PA14; actual mutant growth mirrored predicted growth on five media (Right). The red box identifies the absence of growth of ΔilvC (the deletion mutant highlighted in A), thus experimentally confirming its essentiality on urine. The full growth profiles of 23 gene deletions can be found in SI Appendix, Fig. S4. (C) Summary of FiTnEss performance based on actual deletion mutant growth profiles. Gene-medium instances are indicated in parentheses; and red and green boxes highlight false-positive and false-negative rates, respectively.
Validating FiTnEss Using Strain PA14.
To validate FiTnEss’s approach to predicting gene essentiality, we compared its predictions with actual viability and growth measurements for a set of PA14 mutants in which we cleanly deleted particular genes of interest. We created clean deletion mutants corresponding to 20 genes that FiTnEss identified as nonessential in LB medium, but were essential in one or more of the other media, as well as to three control genes that were predicted to be nonessential in all media. We determined the positive and negative predictive values of FiTnEss by growing the 23 mutants on the same five media as used in the original Tn-Seq experiments, for a total of 115 gene-medium combinations. Mutant strain viability was categorized as essential, intermediate, and nonessential using densitometry (<20%, 20–50%, and >50% relative to wild-type strain PA14 containing no transposons, respectively; Fig. 2 B and C and SI Appendix, Fig. S4). Of the 35 combinations predicted to be essential by the maximal stringency criteria, 30 were indeed found to be essential and five were of intermediate growth. Importantly, no strains within this criterion were, in fact, nonessential. By relaxing the stringency slightly to “highly stringent,” 15 additional strain-medium combinations were predicted to be essential, eight of which were truly essential or of intermediate growth and the remaining seven were nonessential, corroborating our prediction that some false-positive predictions would be expected in this category. Of the 65 combinations predicted to be nonessential, none were found to be essential, but six were found to be of intermediate growth. In this limited dataset, FiTnEss had a positive predictive value of 100% using maximal stringency and 86% using the high-stringency predictions (if intermediate genes are classified as essential). The negative predictive value of FiTnEss is 91% or 100% (depending on the classification of intermediate genes). Notably, all three control strains behaved as predicted, growing on all media, despite our having chosen these control strains with P values that fall at the boundary drawn to distinguish essential and nonessential genes, further reinforcing the accuracy of this binary classification. All together, these results support FiTnEss’s ability to accurately call essential and nonessential genes, and that FiTnEss’s stringency can be varied based on user tolerance of false-positive versus false-negative predictions. Importantly, FiTnEss correctly predicted gene essentiality despite the presence of a small number of mapped insertions in the primary Tn-Seq data of some genes, as exemplified in the case of the ilvC gene encoding ketol-acid reductoisomerase (Fig. 2A).
Defining the Core Genome.
We first defined the core genome consisting of 5,109 protein-coding genes (genes present in all nine strains) using the orthogroup clustering software Synerclust (34). The size of the core genome defined by these nine strains is comparable to what has been previously described for P. aeruginosa [5,316 total genes (35)]. Of these genes, 4,903 were present in a single copy with TA sites that allowed assessment by Tn-Seq; the remaining genes (86 multicopy genes in which reads could not be accurately mapped due to sequence homology and 120 small genes that do not have TA sites permissive to transposon insertion) could not be assessed (Dataset S2). The accessory genome within each strain (the genes that are not present in all nine strains) ranged from 655 to 1,369 genes.
Defining the Core Essential Genome.
We then examined the FiTnEss predictions for all 90 datasets to identify the core essential genes across the P. aeruginosa species (Table 1 and Dataset S3). If one examines only a single strain in a single medium, the number of essential genes varies widely between 354 and 727 genes, even when using the maximal stringency prediction (Table 1). If one examines only genes common to all strains (the core genome), however, the number of essential genes from strain to strain was much more tightly distributed (337–386 genes; SI Appendix, Fig. S5). In contrast, the number of essential genes in the accessory genome of each strain varied widely from 59 to 478 genes (SI Appendix, Fig. S5); interestingly, the number was roughly proportional to genome size (SI Appendix, Fig. S5).
Table 1.
Number of essential genes in each strain-medium combination
| Strain | Genome size* | Genes analyzed† | LB | M9 | Serum | Sputum | Urine | Strain essentials‡ |
| BL23 | 6,687 | 6,272 | 721 | 661 | 696 | 718 | 534 | 405 (794) |
| BWH005 | 6,307 | 5,956 | 547 | 592 | 495 | 508 | 453 | 379 (634) |
| BWH013 | 6,190 | 5,898 | 545 | 527 | 644 | 594 | 478 | 362 (715) |
| BWH015 | 5,907 | 5,622 | 388 | 393 | 395 | 390 | 354 | 300 (396) |
| CF77 | 6,480 | 6,122 | 556 | 518 | 626 | 737 | 529 | 410 (829) |
| PS75 | 5,812 | 5,558 | 441 | 417 | 445 | 468 | 404 | 324 (465) |
| PA14 | 5,893 | 5,708 | 437 | 519 | 430 | 489 | 475 | 364 (467) |
| X13273 | 6,594 | 6,261 | 590 | 458 | 643 | 566 | 437 | 356 (669) |
| 19660 | 6,271 | 5,891 | 508 | 442 | 512 | 552 | 429 | 334 (586) |
| Core‡ | 5,109 | 4,903 | 318 (424) | 329 (431) | 304 (412) | 338 (439) | 298 (390) | 249 (321) |
Total number of protein-coding genes.
Number of protein-coding genes analyzed in this study; absent genes were eliminated from study as described in the text.
Maximal stringency essential genes are shown; high-stringency essential genes are shown in parentheses.
When combining all nine strains across the five media and applying maximal stringency, we found that there are only 249 core essential genes (5.1% of the genome). This number is up to threefold fewer than the number found for a single strain and medium. If we apply the slightly lower standard of high stringency (to allow for the possibility of some false-negative predictions in the data), an additional 72 genes (1.5% of the genome) are included, resulting in 321 genes. We define this set as the core essential genome.
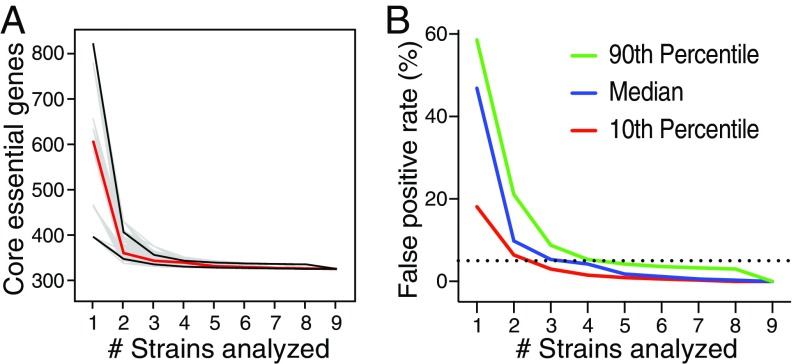
To assess whether the core essential genome had reached a plateau, we calculated how the number of core essential genes decreases with additional strains (with strains added in 10,000 different random orders) (Fig. 3A). We found that the median across these trajectories typically plateaued after four strains, but that five strains would ensure 90% of all trajectories reaching a plateau defined as a <5% false-positive rate (Fig. 3B). Beyond the four strains, the maximum number of core essential genes declined by only 13 genes. These genes just failed to reach the essential P value threshold, suggesting that they were false-negative predictions (Dataset S4). If we were to include these 13 genes, the core essential genome would reach 334 genes. We also examined how the number of essential genes varied under different growth conditions. The numbers of genes that were essential in all nine strains in a single medium ranged from 412 to 439 genes, versus 334 genes under all growth conditions (Table 1).
Fig. 3.
Core essential genome definition plateau. (A) Total of 10,000 random calculations (gray) of the trajectory of the number of core essential genes in all five media tested, determined upon the sequential introduction of additional strains, up to a total of nine strains. The 10th and 90th percentiles (black lines) and mean (red line) core essential genome sizes, as determined by the 10,000 calculations, are highlighted. (B) False-positive rate of core essential genes upon the introduction of strains as calculated in A. The dashed line represents a 5% false-positive rate, with the median (blue) number of strains to cross this threshold being four, and the 10th (red) and 90th (green) percentiles crossing the threshold at three and five strains, respectively.
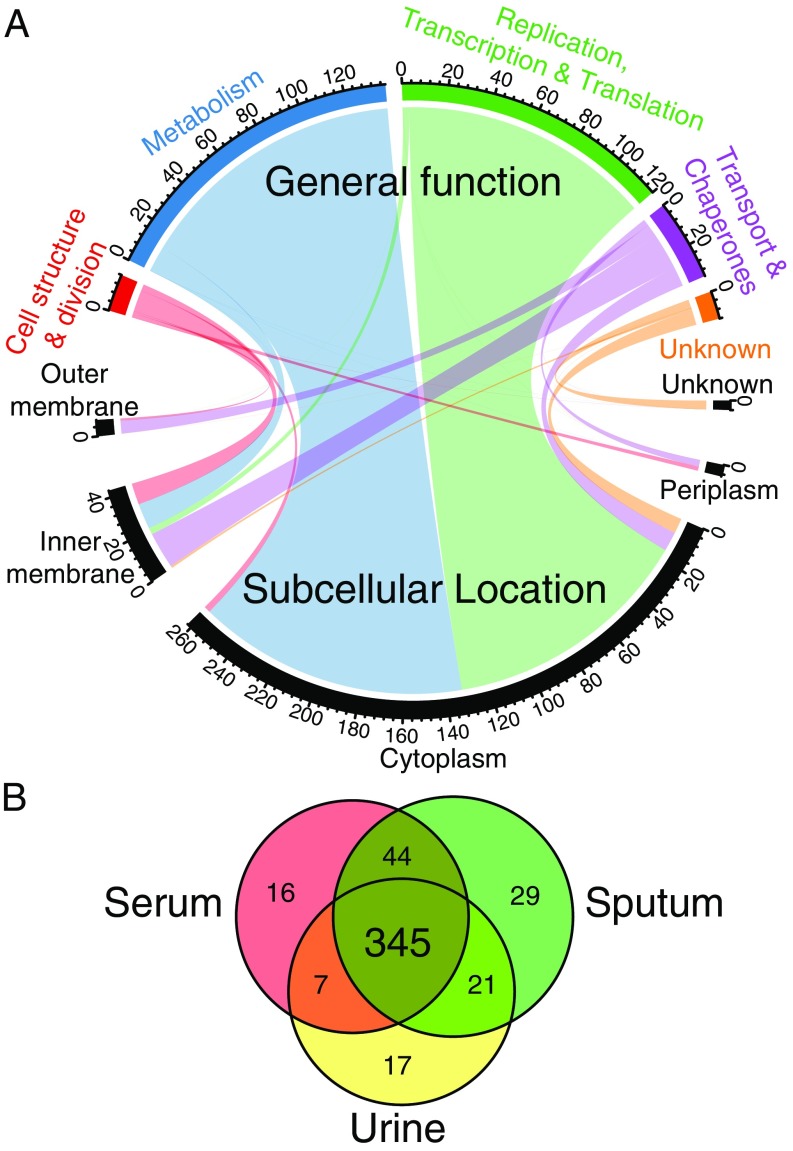
We examined the identities and functions of the core essential genes. Of the 321 core essential genes, 263 correspond to cytosolic proteins, with 132 involved in metabolic pathways (50%) and 119 involved in macromolecular synthesis, including DNA replication, transcription, or translation (45%). Another 56 correspond to cytoplasmic membrane, periplasmic, and outer membrane proteins, with the majority involved in cell structure and division, involved in metabolism, or acting as transporters/chaperones (13, 12, and 26 genes, respectively). The remaining 12 of the 321 genes are completely uncharacterized (Fig. 4A and Dataset S5).
Fig. 4.
Core and conditionally essential gene functions in P. aeruginosa. (A) Chord diagram of the 321 core essential genes showing the relationship between subcellular location (bottom) and general function (top), with the number of genes for each category indicated. (B) Venn diagram showing the number of essential genes in all strains across three infection-relevant media.
Conditionally Essential Genes.
In addition to the core essential genes, the core genome contains conditionally essential genes that are essential in one or more, but not all, media. Sputum and M9 media had the highest number of conditionally essential genes (118 and 110, respectively), consistent with these being the most nutritionally depleted media. LB medium had 103 conditionally essential genes, while urine and serum had the fewest (69 and 91, respectively) (Table 1). While the numbers of essential genes required in each growth condition did not vary significantly from condition to condition, the actual gene identities did vary (Dataset S5). Importantly, we identified an additional 24 conditionally essential genes required for growth in all three infection-relevant media (serum, sputum, and urine; Fig. 4B) but not in both of the laboratory-based media (LB and M9). Several of these genes are involved in pyrimidine and purine synthesis and are not required in LB medium, suggesting that sufficient nucleotide intermediates may be present in LB medium to sustain growth in vitro but that these genes may be valid targets during in vivo infection.
When we applied multiple correspondence analysis to all sets of essential genes for every strain-growth condition, we find that, indeed, the vast majority of strains formed distinct clusters based on growth condition (SI Appendix, Fig. S6). Interestingly, one strain, PA14, an extensively used laboratory strain, was an outlier under two conditions, M9 and urine. In these two media, we observed that, compared with other strains, PA14 has more essential genes involved in the TCA cycle and oxidative phosphorylation. This behavior could be a consequence of PA14 being a laboratory strain that has adapted to laboratory conditions over a long period of time, perhaps providing a slight cautionary flag if attempting to extrapolate PA14 behavior to the species in general.
Contained within the sets of conditionally essential genes for each growth condition are genes that are essential only in a single medium, termed “unique conditionally essential” genes. Considering only the three infection-relevant conditions, while ignoring the laboratory conditions, sputum had 29, serum had 16, and urine had 17 unique conditionally essential genes. These unique conditionally essential genes carry the intriguing potential of becoming infection site-specific targets for infection type-specific antibiotics (e.g., a urine-specific antipseudomonal antibiotic) as long as their essentiality is not dependent on factors that are variable from patient to patient.
The essential genes unique to sputum consist mainly of biosynthetic pathways such as thiamine, pyridoxine, and tryptophan synthesis, with the former two cofactors being required for multiple cellular processes, including synthesis and catabolism of sugars and amino acids, and the latter requirement suggesting that tryptophan levels in sputum may not be sufficient for growth (27). Similarly, urine-specific essential genes almost exclusively consist of genes involved in amino acid biosynthesis, specifically valine, leucine, and isoleucine pathways. Meanwhile, methionine and arginine pathways are essential in both urine and serum. The urine findings are consistent with the fact that these amino acids are among the least abundant in urine (30). However, despite the low abundance of proline and cysteine in urine, classical proline and cysteine biosynthesis genes are not essential, likely because alternative, functionally redundant synthesis pathways exist in P. aeruginosa for these amino acids (36, 37).
In contrast, most genes involved in amino acid biosynthesis are dispensable in serum, as are genes involved in heme biosynthesis (pdxA,H and hemA,B,C,D,E,F,H,J,L), likely due to the ability to scavenge amino acids (38) and heme from free hemoglobin (39, 40) from serum. Interestingly, despite the nonessentiality of porphyrin genes in serum, genes involved in the formation and utilization of porphyrin-containing cytochrome c were uniquely essential in serum and no other media, including the cytochrome c biogenesis protein CcmH (PA14_57540), cytochrome c1 (Cyt1; PA14_57540), the ubiquinol cytochrome c reductase (PA14_57570), and cytochrome c oxidase cbb3-type subunit I (CcoN; PA14_44370). P. aeruginosa’s respiratory chain is highly branched and able to use diverse electron donors and acceptors under different environments (41). Here, we find that in an environment containing high concentrations of heme, P. aeruginosa’s respiratory flexibility is lost, as it becomes dependent on a single pathway.
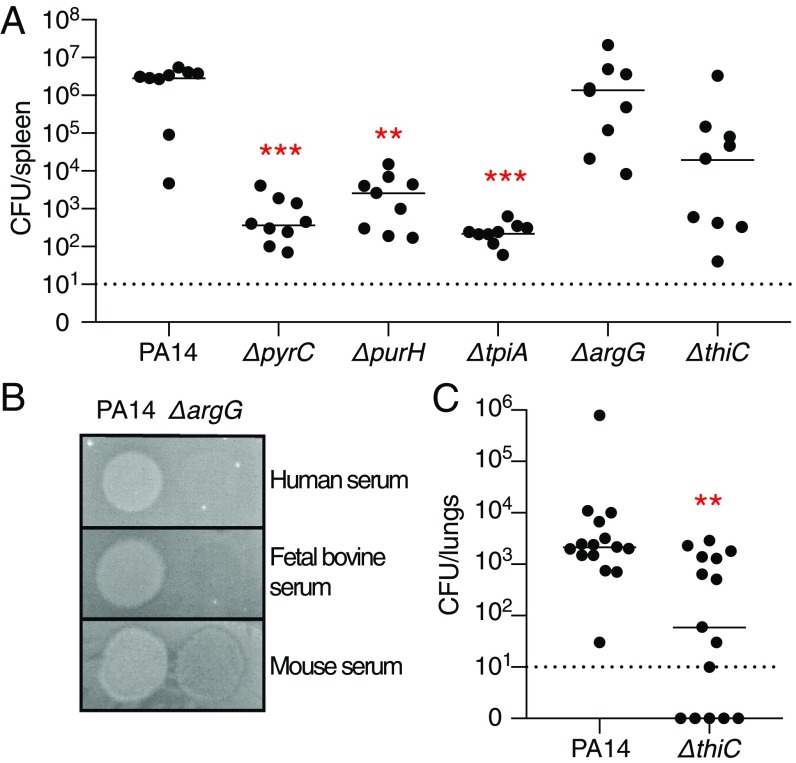
To validate in vivo a strategy of targeting conditionally essential genes identified from our in vitro Tn-Seq experiments, we tested a set of PA14 deletion mutants for their ability to survive in different in vivo mouse models of P. aeruginosa infection. Using a neutropenic mouse model in which the bacteria are administered i.v. to test translation of the in vitro serum growth condition, we infected mice with six strains: wild-type PA14; three mutants containing deletions of metabolic genes predicted to be essential in serum, sputum, and urine but not LB medium (pyrC, pyrimidine biosynthesis; tpiA, glycolysis; and purH, purine biosynthesis); one mutant containing a deletion in a gene predicted be essential in serum and urine but not sputum (argG, arginine biosynthesis); and one mutant containing a deletion in a gene predicted to be conditionally essential in sputum alone but not serum or urine (thiC, thiamine biosynthesis; Fig. 5A). In concordance with their predicted conditional essentiality, the pyrC, tpiA, and purH mutants were significantly attenuated in the neutropenic mouse model with a three- to four-log reduction in total colony-forming units in the spleen 16 h postinfection. Interestingly, the argG mutant was not attenuated as predicted. To understand this discrepancy, we compared its growth on agar plates supplemented with mouse, bovine (which was used in the Tn-Seq experiments), or human serum. Indeed, the mutant was able to grow on mouse serum, but not on bovine or human serum, thereby explaining the lack of mutant attenuation in the mouse model, likely due to the higher levels of arginine available in mouse serum (42–44) (Fig. 5B). Finally, as predicted, the thiC mutant was not significantly attenuated in the neutropenic mouse model; it was, however, attenuated in an acute pneumonia mouse model, where the inoculum is delivered intranasally followed by colonization of the lungs, which is consistent with FiTnEss predictions (Fig. 5C). Together, these datasets highlighted the tremendous differences required by P. aeruginosa in different microenvironments that could be exploited in the development of infection condition-specific therapeutics.
Fig. 5.
In vivo murine infection with gene deletion mutants of conditionally essential genes identified by FiTnEss. (A) Bacterial burden in the spleen of neutropenic mice infected i.v. with wild-type (WT) PA14 and ΔpyrC, ΔpurH, ΔtpiA, ΔargG, and ΔthiC deletion mutants; statistical significance was determined with a Kruskal–Wallis test (n = 9). (B) Growth of WT PA14 and the ΔargG deletion mutant in human, fetal bovine, and mouse sera. (C) Bacterial burden in the lungs from mice infected intranasally with PA14 or the ΔthiC deletion mutant; statistical significance was determined with a Mann–Whitney U test (n = 15). (A and C) Bacterial burden determined at 16 h postinfection. Each dot represents a single mouse, with a line indicating the median; significance is displayed at ***P < 0.001 or **P < 0.01; the dashed line indicates the limit of detection of the assay; and data are a combination of two biological replicates.
Discussion
Target-informed antibiotic discovery and development have been predicated on knowing which genes within a given species constitute good targets, with genomic technologies such as Tn-Seq paving the way for more comprehensive definition of essential targets. However, comprehensive genomic methods for defining essential targets, such as Tn-Seq, have largely been applied to only a single or a few bacterial strains under a single or a few growth conditions, with the implicit assumption that these results will apply to the entire species under infection-relevant conditions. Here, we sought to develop a general strategy for determining the core essential genome of a bacterial species and to examine how the number of essential genes varies among strains and growth conditions. We empirically determined that the analysis of essential genes among four diverse strains was sufficient to define the core essential genes in P. aeruginosa.
Using Tn-Seq and a method of analysis, FiTnEss, to establish the core essential genome of P. aeruginosa, we determined that while a single strain has ∼400–800 essential genes, the core essential genome across all strains analyzed is ∼321 genes, thus demonstrating the limitations in determining species essentiality based on a single strain and/or single medium condition. Further, there are an additional 24 essential genes required for growth in the three infection-relevant media examined, which are nonessential in LB and M9 media. Finally, we find that there are ∼15–30 unique, conditionally essential genes for each of the infection-relevant media examined, with their corresponding biological pathways important for survival only within a particular host tissue environment; these genes may represent a unique set of targets for infection type-specific therapeutics, with the obvious caveat that their essentiality cannot be dependent on microenvironmental factors that vary widely from patient to patient. Interestingly, our study provides insight into differences in gene essentiality between P. aeruginosa and other characterized bacterial species such as E. coli, which differ, for example, in genes required for ATP or fatty acid biosynthesis (45, 46).
Previous transposon mutagenesis studies of two common laboratory strains, PA14 and PAO1, have found varying numbers of essential genes, as reviewed by Juhas (22). These studies have predominantly focused on identifying genes refractory to transposon mutagenesis when bacteria are selected for growth on laboratory media, including LB, brain–heart infusion (BHI), and minimal media (24, 47–50), although recent studies have extended growth conditions to include sputum or sputum-like media (49, 50) in vitro. A comparison of all of these datasets combined revealed an intersection of only 109 essential genes common to all studies (Dataset S6). This low concordance is likely due to methodological or analytical differences between the studies. One way in which our study differs from the majority of previous studies is that we did not initially select transposon mutants on a rich (isolation) medium (i.e., LB medium) before selection on the condition of interest, thereby eliminating a bottleneck that prevents evaluation of genes essential in the isolation medium. By omitting the initial isolation step, we were able to identify 103 conditionally essential genes that are required in LB medium, but not in at least one of the other four media. In addition to these in vitro studies, in vivo Tn-Seq studies can be valuable in determining what genes are required for fitness in the context of an active infection (48); however, they also suffer from the problem of experimental bottlenecks that practically limit the ability to truly interrogate essentiality on a genome-wide scale. These bottlenecks include the required isolation step before inoculating into an animal and the depletion of mutants in vivo due to stochastic loss rather than a true fitness loss of the mutant itself (17).
Experimental methods for identifying gene essentiality have varied greatly through the years. Despite significant advances for defining fitness costs of gene disruptions on a genome-wide scale using sequencing (Tn-Seq), limitations persist. They include (i) analysis of mutant behavior in pools where there can be both competition as well as in trans complementation, factors that can cause mutant growth in a pool to diverge from growth in isolation; (ii) transposon sequence insertion biases (28, 51); and (iii) polar effects on adjacent genes conferred by transposon insertion. These limitations prevented us from evaluating the essentiality of ∼5% of the genome and can technically lead to errors in assessing fitness and essentiality. We indeed found examples of discordance where our Tn-Seq data classified genes as essential even though mutants of these genes are available, albeit growth-defective, such as hfq, rpoN, and gidA (52–54). These cases could be due to FiTnEss errors; alternatively, one must consider the possibility that these mutants have reduced fitness when grown in competition but not in isolation, or that reported mutants could contain compensatory mutants acquired in their construction that allow deletion of the gene of interest.
A major challenge to analysis is translating measurements that quantify a continuum of fitness to a binary classification of essentiality versus nonessentiality to define the best antibiotic targets. Approaches can vary substantially, with different systematic errors and different tolerances for false-positive versus false-negative predictions (SI Appendix, Additional Methods and Results and Fig. S7). We therefore developed a Tn-Seq analysis pipeline, FiTnEss, that balances false-positive and false-negative rates with the aim of accurately classifying gene essentiality, while providing two levels of stringency depending on one’s tolerance for false-positive versus false-negative predictions. We validated FiTnEss using clean gene deletion mutants (Fig. 2 and SI Appendix, Fig. S4).
We used FiTnEss to perform the binary classification of the 4,903 genes in the core genome that could be assessed across nine P. aeruginosa isolates. The great majority of core essential genes can be broadly categorized as being involved in metabolic pathways or macromolecular synthesis such as DNA replication, transcription, or translation. That the core essential genome is dominated by genes involved in macromolecular synthesis (i.e., nucleic acids, protein, cell wall) may explain, in part, why most current antibiotics seem to target this limited set of functions (i.e., fluoroquinolones, aminoglycosides, beta-lactams, respectively). Three recent gram-negative antibiotic candidates reported in the literature also target essential genes identified in this study [i.e., lptD, lepB, msbA (55–58)]. There has been greater reticence to target metabolic pathways, given concern over the ability of bacteria to scavenge nutrients from the host, thereby rendering their biosynthesis nonessential during infection. Indeed, we see variable requirements for metabolic genes in our identification of conditionally essential genes, such as a greater dependence on amino acid biosynthesis in urine than other growth conditions. We have, however, demonstrated that this conditional essentiality can, in fact, be exploited in vivo, as mice infected with the pyrC, tpiA, and purH mutants that are significantly growth-impaired on infection-relevant media have a dramatically reduced bacterial burden in a systemic infection model. Further, the thiC mutant, found to be essential in M9 medium and sputum alone, was attenuated in an acute pneumonia model, yet was still virulent if introduced systemically, demonstrating variable metabolite levels in different infection sites in vivo and raising the possibility of infection site-specific therapeutics.
In summary, we suggest that a major factor in the failure of genomics to transform antibiotic discovery in the late 1990s to early 2000s was due not to a fundamental flaw with the concept of defining essential genes, but rather to challenges in implementing the approach, namely, defining essential genes based on limited information. Advances in genomic technologies now make possible studies on a much greater scale, allowing us to define essential genes in a way that overcomes previous shortcomings. Our work describes a general approach applicable to other pathogens, given the explosion in available bacterial genomes. While the number of strains required to reach a plateau in essential genes for different species may vary based on the genomic diversity of a species, the basic paradigm should apply broadly. Importantly, defining the core essential genome for different species will enable comparative genomics studies to understand differing evolutionary or adaptive programs adopted by the different species, and to distinguish targets that may be ideal for species-specific versus broad-spectrum targeting. For important pathogens such as P. aeruginosa, our hope is that defining a core essential genome by selecting diverse strains across its phylogenetic tree will enable more effective discovery and development of much needed antibacterial therapeutics.
Materials and Methods
Strain Selection and Plasmid Construction.
A genome tree report of 2,560 sequenced P. aerguinosa strains was downloaded from the NCBI (organism ID 187) and visualized with the Interactive Tree of Life (iTOL) tool (59). Nine strains were selected for genetic diversity and graciously gifted from various sources: PA14, 19660, and X13273 were obtained from Frederick M. Ausubel, Massachusetts General Hospital, Boston (60); BWH005, BWH013, and BWH015 were collected through the Brigham and Women’s Hospital Specimen Bank per a protocol previously described (61); BL23 was obtained from Bausch & Lomb (62); PS75 was obtained from Paula Suarez, Simon Bolivar University, Sartenejas, Miranda State, Venezuela; and CF77 was obtained from the Boston Children’s Hospital (63). The pC9 containing a hyperactive transposase was derived from pSAM-DGm (48), and the pMAR containing the Himar1 transposon was derived from pMAR2xT7 (24).
Transposon Library Construction and Sequencing.
Recipient P. aeruginosa strains were prepared for mating as previously described (64). P. aeruginosa and midlog cultures of E. coli SM10(pC9) and E. coli SM10(pMAR) were collected by centrifugation, washed, and resuspended in LB medium. A total of 3 × 1011 cfu were mixed in a 2:2:1 ratio of pC9/pMAR/recipient and collected by centrifugation. The cell mating mixture was resuspended to a concentration of 1011 cfu/mL, and 30-μL spots were dispensed to a dry LB agar plate. Mating plates were incubated at 37 °C for 1.5 h before cells were scraped, resuspended in PBS, mixed with glycerol to a final concentration of 40%, aliquoted, and flash-frozen before storage at −80 °C. Matings were performed at least twice for each recipient strain, and efficiencies were quantified by plating to LB-selective agar. Two hundred fifty milliliters of each medium containing 1.5% agar, 5 μg/mL irgasan, and 30 μg/mL gentamicin was prepared in a Biodish XL (Nunc). LB and M9 minimal agar (US Biologicals) and synthetic cystic fibrosis medium agar (SCFM) (27) were prepared. Pooled filter-sterilized urine and FBS (Thermo Fisher Scientific) were warmed to 55 °C and mixed with a 5% agar solution (Teknova) to achieve a 1.5% final agar concentration. Five hundred thousand colony-forming units of each transposon-integrated strain was plated to each medium in duplicate and grown at 37 °C for 24 h (LB medium, FBS, and SCFM) or 48 h (urine and M9) before scraping and resuspending cells in PBS. Genomic DNA was isolated, and Illumina libraries were prepared using a custom method and primers described in SI Appendix, Fig. S1 and Dataset S7. Sequencing was performed with an Illumina NextSeq platform to obtain 50-bp genomic DNA reads.
Determining Essential Genes from Tn-Seq Data Using FiTnEss.
All sequencing read count data and analysis files are available (https://data.broadinstitute.org/fitness/) and on the NCBI Sequence Read Archive (65). The analysis pipeline, including sequencing summarization and the FiTnEss software, is freely available (https://github.com/broadinstitute/FiTnEss) (66). Genomes and annotations for each strain were obtained from www.pseudomonas.com and https://patricbrc.org (67). The core and accessory genomes were determined by gene clustering analysis across the strains tested using Synerclust (33). Illumina reads were mapped to each respective genome using Bowtie (68), utilizing the options for exact and unique read mapping. Reads potentially mapping to more than one location in a genome were discarded, and homologous TA sites were removed from analysis by searching the genome using custom scripts. TA insertion sites at the distal 50 bp from each end of the gene and nonpermissive insertion sites containing the sequence (GC)GNTANC(GC) were removed using custom scripts. Reads mapped to each TA site were tallied using scripts modified from a study by DeJesus et al. (32). For each Tn-Seq dataset, a lognormal − negative binomial distribution was conservatively fit using genes with a median number of TA sites and the top 75% of reads per gene (nonessential genes) to identify parameters . Then, a theoretical distribution was constructed using these two parameters for each gene size category based on the number of TA sites per gene . Background distributions for these categories were obtained from numerical sampling of the theoretical distribution. The actual number of reads for each gene was compared with the background distribution for the corresponding category, and a P value was calculated as the probability of obtaining the number of reads or less by chance. Two-layer multiple comparison adjustments were conducted. First, to obtain a maximally stringent essential set, we adjusted for the FWER using the Holm–Bonferroni method. Second, to reduce the risk of losing targets, we relaxed the stringency slightly to obtain a highly stringent essential set, by adjusting for the FDR using the Benjamini–Hochberg method. After either correction process, genes with an adjusted P value smaller than 0.05 in both replicates were identified as essential. A full description and calculations are provided in SI Appendix.
Method Validation with Clean Gene Deletions.
Gene deletions were performed as previously described in strain PA14 (64). Gene deletions were confirmed by PCR amplification and sequencing. Successful gene deletion strains were grown in duplicate in LB medium at 37 °C for 16 h before diluting 10−4 in PBS. Five microliters of diluted culture was spotted to the five solid media used in this study and grown at 37 °C for 24 h. Images were captured, densitometry was performed using ImageJ (NIH), and growth was categorized relative to 10 wild-type replicates: essential (0–20%), growth-defective (21–50%), and nonessential (>50%).
In Vivo Mouse Models.
All vertebrate animal experiments were done with the approval of the Massachusetts General Hospital’s Institutional Animal Care and Use Committee. Bacteria were grown to midlog, collected by centrifugation, washed, and resuspended in PBS. For the systemic infection model, 9-wk-old female BALB/c mice (The Jackson Laboratory) were injected i.p. with 4 mg of cyclophosphamide 3 d before infection. Mice were infected i.v. with 5 × 105 cfu per mouse. For the acute pneumonia model, mice were infected intranasally with 1 × 106 cfu per mouse. For both infection models, mice were euthanized 16 h postinfection and spleens (systemic) or lungs (pneumonia) were harvested and homogenized in 1 mL of PBS + 0.1% Triton X-100 using a TissueLyser LT (Qiagen) before plating to LB agar + 5 μg/mL irgasan to enumerate the bacterial burden.
Supplementary Material
Acknowledgments
This work was supported by a generous gift from Anita and Josh Bekenstein, NIH Grant R33AI098705 (to D.T.H.), and a Cystic Fibrosis Canada Fellowship (to B.E.P.).
Footnotes
Conflict of interest statement: Eric Lander serves on the Board of Directors for and holds equity in Codiak BioSciences and Neon Therapeutics, and serves on the Scientific Advisory Board of F-Prime Capital Partners and Third Rock Ventures; he is also affiliated with several nonprofit organizations, including serving on the Board of Directors of the Innocence Project, Count Me In, and Biden Cancer Initiative, and the Board of Trustees for the Parker Institute for Cancer Immunotherapy. He has served and continues to serve on various federal advisory committees.
Data deposition: All sequencing read count data and analysis files are available at https://data.broadinstitute.org/fitness/ and on the National Center for Biotechnology Information Sequence Read Archive Sequence Read Archive (https://www.ncbi.nlm.nih.gov/bioproject/533044). The analysis pipeline, including sequencing summarization and the FiTnEss software, is freely available at https://github.com/broadinstitute/FiTnEss.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1900570116/-/DCSupplemental.
References
- 1.Fleischmann RD, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 2.Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. Drugs for bad bugs: Confronting the challenges of antibacterial discovery. Nat Rev Drug Discov. 2007;6:29–40. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- 3.Tommasi R, Brown DG, Walkup GK, Manchester JI, Miller AA. ESKAPEing the labyrinth of antibacterial discovery. Nat Rev Drug Discov. 2015;14:529–542. doi: 10.1038/nrd4572. [DOI] [PubMed] [Google Scholar]
- 4.Gentry DR, et al. Variable sensitivity to bacterial methionyl-tRNA synthetase inhibitors reveals subpopulations of Streptococcus pneumoniae with two distinct methionyl-tRNA synthetase genes. Antimicrob Agents Chemother. 2003;47:1784–1789. doi: 10.1128/AAC.47.6.1784-1789.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yao Z, Davis RM, Kishony R, Kahne D, Ruiz N. Regulation of cell size in response to nutrient availability by fatty acid biosynthesis in Escherichia coli. Proc Natl Acad Sci USA. 2012;109:E2561–E2568. doi: 10.1073/pnas.1209742109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown ED, Wright GD. Antibacterial drug discovery in the resistance era. Nature. 2016;529:336–343. doi: 10.1038/nature17042. [DOI] [PubMed] [Google Scholar]
- 7.Parsons JB, Rock CO. Is bacterial fatty acid synthesis a valid target for antibacterial drug discovery? Curr Opin Microbiol. 2011;14:544–549. doi: 10.1016/j.mib.2011.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brinster S, et al. Type II fatty acid synthesis is not a suitable antibiotic target for gram-positive pathogens. Nature. 2009;458:83–86. doi: 10.1038/nature07772. [DOI] [PubMed] [Google Scholar]
- 9.Pethe K, et al. A chemical genetic screen in Mycobacterium tuberculosis identifies carbon-source-dependent growth inhibitors devoid of in vivo efficacy. Nat Commun. 2010;1:57. doi: 10.1038/ncomms1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Folkesson A, et al. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: An evolutionary perspective. Nat Rev Microbiol. 2012;10:841–851. doi: 10.1038/nrmicro2907. [DOI] [PubMed] [Google Scholar]
- 11.Gellatly SL, Hancock RE. Pseudomonas aeruginosa: New insights into pathogenesis and host defenses. Pathog Dis. 2013;67:159–173. doi: 10.1111/2049-632X.12033. [DOI] [PubMed] [Google Scholar]
- 12.Klevens RM, et al. Estimating health care-associated infections and deaths in U.S. hospitals, 2002. Public Health Rep. 2007;122:160–166. doi: 10.1177/003335490712200205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Del Barrio-Tofiño E, et al. Genomics and susceptibility profiles of extensively drug-resistant Pseudomonas aeruginosa isolates from Spain. Antimicrob Agents Chemother. 2017;61:e01589-17. doi: 10.1128/AAC.01589-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strateva T, Yordanov D. Pseudomonas aeruginosa–A phenomenon of bacterial resistance. J Med Microbiol. 2009;58:1133–1148. doi: 10.1099/jmm.0.009142-0. [DOI] [PubMed] [Google Scholar]
- 15.Tacconelli E, et al. WHO Pathogens Priority List Working Group Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018;18:318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 16.Hay M, Thomas DW, Craighead JL, Economides C, Rosenthal J. Clinical development success rates for investigational drugs. Nat Biotechnol. 2014;32:40–51. doi: 10.1038/nbt.2786. [DOI] [PubMed] [Google Scholar]
- 17.Chao MC, Abel S, Davis BM, Waldor MK. The design and analysis of transposon insertion sequencing experiments. Nat Rev Microbiol. 2016;14:119–128. doi: 10.1038/nrmicro.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gawronski JD, Wong SM, Giannoukos G, Ward DV, Akerley BJ. Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung. Proc Natl Acad Sci USA. 2009;106:16422–16427. doi: 10.1073/pnas.0906627106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodman AL, et al. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe. 2009;6:279–289. doi: 10.1016/j.chom.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langridge GC, et al. Simultaneous assay of every Salmonella typhi gene using one million transposon mutants. Genome Res. 2009;19:2308–2316. doi: 10.1101/gr.097097.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Opijnen T, Bodi KL, Camilli A. Tn-seq: High-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods. 2009;6:767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Juhas M. Pseudomonas aeruginosa essentials: An update on investigation of essential genes. Microbiology. 2015;161:2053–2060. doi: 10.1099/mic.0.000161. [DOI] [PubMed] [Google Scholar]
- 23.Lampe DJ, Grant TE, Robertson HM. Factors affecting transposition of the Himar1 mariner transposon in vitro. Genetics. 1998;149:179–187. doi: 10.1093/genetics/149.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liberati NT, et al. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci USA. 2006;103:2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rubin EJ, et al. In vivo transposition of mariner-based elements in enteric bacteria and mycobacteria. Proc Natl Acad Sci USA. 1999;96:1645–1650. doi: 10.1073/pnas.96.4.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lampe DJ, Akerley BJ, Rubin EJ, Mekalanos JJ, Robertson HM. Hyperactive transposase mutants of the Himar1 mariner transposon. Proc Natl Acad Sci USA. 1999;96:11428–11433. doi: 10.1073/pnas.96.20.11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palmer KL, Aye LM, Whiteley M. Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J Bacteriol. 2007;189:8079–8087. doi: 10.1128/JB.01138-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeJesus MA, et al. Comprehensive essentiality analysis of the Mycobacterium tuberculosis genome via saturating transposon mutagenesis. MBio. 2017;8:e02133-16. doi: 10.1128/mBio.02133-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Griffin JE, et al. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog. 2011;7:e1002251. doi: 10.1371/journal.ppat.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Venta R. Year-long validation study and reference values for urinary amino acids using a reversed-phase HPLC method. Clin Chem. 2001;47:575–583. [PubMed] [Google Scholar]
- 31.Zomer A, Burghout P, Bootsma HJ, Hermans PW, van Hijum SA. ESSENTIALS: Software for rapid analysis of high throughput transposon insertion sequencing data. PLoS One. 2012;7:e43012. doi: 10.1371/journal.pone.0043012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeJesus MA, Ambadipudi C, Baker R, Sassetti C, Ioerger TR. TRANSIT–A software tool for Himar1 TnSeq analysis. PLOS Comput Biol. 2015;11:e1004401. doi: 10.1371/journal.pcbi.1004401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pritchard JR, et al. ARTIST: High-resolution genome-wide assessment of fitness using transposon-insertion sequencing. PLoS Genet. 2014;10:e1004782. doi: 10.1371/journal.pgen.1004782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Georgescu CH, et al. SynerClust: A highly scalable, synteny-aware orthologue clustering tool. Microb Genom. 2018;4 doi: 10.1099/mgen.0.000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ozer EA, Allen JP, Hauser AR. Characterization of the core and accessory genomes of Pseudomonas aeruginosa using bioinformatic tools Spine and AGEnt. BMC Genomics. 2014;15:737. doi: 10.1186/1471-2164-15-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stalon V, Vander Wauven C, Momin P, Legrain C. Catabolism of arginine, citrulline and ornithine by Pseudomonas and related bacteria. J Gen Microbiol. 1987;133:2487–2495. doi: 10.1099/00221287-133-9-2487. [DOI] [PubMed] [Google Scholar]
- 37.Vermeij P, Kertesz MA. Pathways of assimilative sulfur metabolism in Pseudomonas putida. J Bacteriol. 1999;181:5833–5837. doi: 10.1128/jb.181.18.5833-5837.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stein WH, Moore S. The free amino acids of human blood plasma. J Biol Chem. 1954;211:915–926. [PubMed] [Google Scholar]
- 39.Choby JE, Skaar EP. Heme synthesis and acquisition in bacterial pathogens. J Mol Biol. 2016;428:3408–3428. doi: 10.1016/j.jmb.2016.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Na N, Ouyang J, Taes YE, Delanghe JR. Serum free hemoglobin concentrations in healthy individuals are related to haptoglobin type. Clin Chem. 2005;51:1754–1755. doi: 10.1373/clinchem.2005.055657. [DOI] [PubMed] [Google Scholar]
- 41.Arai H. Regulation and function of versatile aerobic and anaerobic respiratory metabolism in Pseudomonas aeruginosa. Front Microbiol. 2011;2:103. doi: 10.3389/fmicb.2011.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baetz AL, Hubbert WT, Graham CK. Developmental changes of free amino acids in bovine fetal fluids with gestational age and the interrelationships between the amino acid concentrations in the fluid compartments. J Reprod Fertil. 1975;44:437–477. doi: 10.1530/jrf.0.0440437. [DOI] [PubMed] [Google Scholar]
- 43.Lüneburg N, et al. Reference intervals for plasma L-arginine and the L-arginine:asymmetric dimethylarginine ratio in the Framingham Offspring Cohort. J Nutr. 2011;141:2186–2190. doi: 10.3945/jn.111.148197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rivera S, López-Soriano FJ, Azcón-Bieto J, Argilés JM. Blood amino acid compartmentation in mice bearing Lewis lung carcinoma. Cancer Res. 1987;47:5644–5646. [PubMed] [Google Scholar]
- 45.Martínez-Carranza E, et al. Variability of bacterial essential genes among closely related bacteria: The case of Escherichia coli. Front Microbiol. 2018;9:1059. doi: 10.3389/fmicb.2018.01059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goodall ECA, et al. The essential genome of Escherichia coli K-12. MBio. 2018;9:e02096-17. doi: 10.1128/mBio.02096-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jacobs MA, et al. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2003;100:14339–14344. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Skurnik D, et al. A comprehensive analysis of in vitro and in vivo genetic fitness of Pseudomonas aeruginosa using high-throughput sequencing of transposon libraries. PLoS Pathog. 2013;9:e1003582. doi: 10.1371/journal.ppat.1003582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee SA, et al. General and condition-specific essential functions of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2015;112:5189–5194. doi: 10.1073/pnas.1422186112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Turner KH, Wessel AK, Palmer GC, Murray JL, Whiteley M. Essential genome of Pseudomonas aeruginosa in cystic fibrosis sputum. Proc Natl Acad Sci USA. 2015;112:4110–4115. doi: 10.1073/pnas.1419677112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ason B, Reznikoff WS. DNA sequence bias during Tn5 transposition. J Mol Biol. 2004;335:1213–1225. doi: 10.1016/j.jmb.2003.11.039. [DOI] [PubMed] [Google Scholar]
- 52.Gupta R, Gobble TR, Schuster M. GidA posttranscriptionally regulates rhl quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 2009;191:5785–5792. doi: 10.1128/JB.00335-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hendrickson EL, Plotnikova J, Mahajan-Miklos S, Rahme LG, Ausubel FM. Differential roles of the Pseudomonas aeruginosa PA14 rpoN gene in pathogenicity in plants, nematodes, insects, and mice. J Bacteriol. 2001;183:7126–7134. doi: 10.1128/JB.183.24.7126-7134.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sonnleitner E, et al. Reduced virulence of a hfq mutant of Pseudomonas aeruginosa O1. Microb Pathog. 2003;35:217–228. doi: 10.1016/s0882-4010(03)00149-9. [DOI] [PubMed] [Google Scholar]
- 55.Zhang G, et al. Cell-based screen for discovering lipopolysaccharide biogenesis inhibitors. Proc Natl Acad Sci USA. 2018;115:6834–6839. doi: 10.1073/pnas.1804670115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Srinivas N, et al. Peptidomimetic antibiotics target outer-membrane biogenesis in Pseudomonas aeruginosa. Science. 2010;327:1010–1013. doi: 10.1126/science.1182749. [DOI] [PubMed] [Google Scholar]
- 57.Ho H, et al. Structural basis for dual-mode inhibition of the ABC transporter MsbA. Nature. 2018;557:196–201. doi: 10.1038/s41586-018-0083-5. [DOI] [PubMed] [Google Scholar]
- 58.Craney A, Romesberg FE. The inhibition of type I bacterial signal peptidase: Biological consequences and therapeutic potential. Bioorg Med Chem Lett. 2015;25:4761–4766. doi: 10.1016/j.bmcl.2015.07.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Letunic I, Bork P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee DG, et al. Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial. Genome Biol. 2006;7:R90. doi: 10.1186/gb-2006-7-10-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pecora ND, et al. Genomically informed surveillance for carbapenem-resistant Enterobacteriaceae in a health care system. MBio. 2015;6:e01030. doi: 10.1128/mBio.01030-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Haas W, Gearinger LS, Usner DW, Decory HH, Morris TW. Integrated analysis of three bacterial conjunctivitis trials of besifloxacin ophthalmic suspension, 0.6%: Etiology of bacterial conjunctivitis and antibacterial susceptibility profile. Clin Ophthalmol. 2011;5:1369–1379. doi: 10.2147/OPTH.S23519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ordoñez CL, et al. Inflammatory and microbiologic markers in induced sputum after intravenous antibiotics in cystic fibrosis. Am J Respir Crit Care Med. 2003;168:1471–1475. doi: 10.1164/rccm.200306-731OC. [DOI] [PubMed] [Google Scholar]
- 64.Hmelo LR, et al. Precision-engineering the Pseudomonas aeruginosa genome with two-step allelic exchange. Nat Protoc. 2015;10:1820–1841. doi: 10.1038/nprot.2015.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Poulsen BE, Hung DT. 2019 Data from “Defining the core essential genome of Pseudomonas aeruginosa.” BioProject. Available at https://www.ncbi.nlm.nih.gov/bioproject/533044. Deposited April 12, 2019.
- 66.Poulsen BE, Yang R. 2019 Data from “broadinstitute/FiTnEss: First release of FiTnEss.” Zenodo. Available at https://zenodo.org/record/2640897. Deposited April 15, 2019.
- 67.Winsor GL, et al. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 2016;44:D646–D653. doi: 10.1093/nar/gkv1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.