Significance
Complement regulation is critical for protecting the body’s own cells from homologous complement. This function is primarily mediated by the regulators of complement activation (RCA) proteins through two functional mechanisms termed decay-accelerating activity (DAA) and cofactor activity (CFA). Here, we show the presence of functional modularity in the RCA proteins. We demonstrate that the basic functional blocks for DAA and CFA in these proteins are formed by only two domains and that these are arranged in a specific order. Furthermore, we show that linking functional blocks of the decay-accelerating factor and membrane cofactor protein—which possess DAA and CFA, respectively—and adding C-terminal domains for enhancing avidity toward C3b/C4b results in a regulator molecule with dual activity.
Keywords: complement, complement regulation, RCA proteins, decay-accelerating factor, membrane cofactor protein
Abstract
The complement system is highly efficient in targeting pathogens, but lack of its apposite regulation results in host-cell damage, which is linked to diseases. Thus, complement activation is tightly regulated by a series of proteins, which primarily belong to the regulators of complement activation (RCA) family. Structurally, these proteins are composed of repeating complement control protein (CCP) domains where two to four successive domains contribute to the regulatory functions termed decay-accelerating activity (DAA) and cofactor activity (CFA). However, the precise constitution of the functional units and whether these units can be joined to form a larger composition with dual function have not been demonstrated. Herein, we have parsed the functional units for DAA and CFA by constructing chimeras of the decay-accelerating factor (DAF) that exhibits DAA and membrane cofactor protein (MCP) that exhibits CFA. We show that in a four-CCP framework, a functional unit for each of the regulatory activities is formed by only two successive CCPs wherein each participates in the function, albeit CCP2 has a bipartite function. Additionally, optimal activity requires C-terminal domains that enhance the avidity of the molecule for C3b/C4b. Furthermore, by composing a four-CCP DAF-MCP chimera with robust CFA (for C3b and C4b) and DAA (for classical and alternative pathway C3 convertases), named decay cofactor protein, we show that CCP functional units can be linked to design a dual-activity regulator. These data indicate that the regulatory determinants for these two biological processes are distinct and modular in nature.
The complement system is an essential effector system of innate immunity. Although classically it is known for its direct action on pathogens (1), it is also critically involved in boosting the pathogen-specific adaptive immune responses (2) as well as other processes such as cell differentiation and polarization, tissue regeneration, lipid metabolism, and removal of immune complexes and apoptotic cells (3). The system is tightly regulated by multiple regulatory proteins, as inappropriate activation is known to result in host tissue destruction (4, 5). The major proteins that regulate complement activation belong to a family dubbed as regulators of complement activation (RCA). These proteins are exclusively composed of complement control protein (CCP) modules and are located on the cell surface [e.g., decay-accelerating factor (DAF; CD55), membrane cofactor protein (MCP; CD46), and complement receptor 1 (CR1; CD35)] as well as in the fluid phase [e.g., factor H (FH) and C4b-binding protein (C4BP)] (6). Notably, defects in the functioning of these RCA proteins are linked to various diseases such as age-related macular degeneration, atypical hemolytic uremic syndrome, and dense deposit disease (4, 5, 7). Intriguingly, mimics of RCA proteins are also encoded by human orthopox and gamma herpes viruses (e.g., variola and HHV-8) (8).
Activation of the complement system is initiated by three major pathways (lectin, classical, and alternative) that converge at the C3 convertase (C4b2a or C3bBb) formation—the enzymes that cleave complement component C3, which is necessary for initiation of all of the downstream effector functions of the complement. The C3 convertases are composed of two subunits wherein a catalytic subunit (Bb/C2a) is bound to a noncatalytic subunit (C3b/C4b) in a Mg2+-dependent manner (9, 10). The RCA proteins bind to these convertases or their noncatalytic subunits and inactivate them. These regulatory activities of RCA are termed decay-accelerating activity (DAA) and cofactor activity (CFA). In DAA, the RCA protein binds to the convertase and irreversibly dissociates it into its subunits, while, in CFA, the RCA protein binds to the noncatalytic subunit of the convertase (C3b/C4b) and recruits serine protease factor I (FI) to cleave and inactivate it, thus ending its ability to form C3 convertase (6, 11).
Among the RCA proteins, CR1, DAF, and MCP, the membrane-tethered complement regulators, are critically involved in the protection of the host cells from autologous complement attack (12, 13). The most abundant form of CR1 is composed of 30 CCP domains, which are organized into four long homologous repeats (LHRs) A–D. Intriguingly, the first three CCPs of LHRs A–C have the regulatory activities; LHR-A (CCP1–3) primarily harbors DAA, while LHR-B/C (CCP8–10/CCP15–17) mainly harbors CFA (14). Importantly, the LHR-A (CCP1–3) is in clinical development, and its improved version (CCP1–3, D109N/E116K) has also been designed (14).
Unlike CR1, however, DAF and MCP harbor only one activity; DAF has DAA, while MCP has CFA. Both these proteins are much shorter than CR1 and are composed of only four CCP modules, which are arranged in extended arrangements (15–17). Of note, CCP2–4 domains of DAF are similar to CCP1–3 of MCP. Domain-deletion and site-directed mutagenesis in DAF demonstrated that CCP2–3 of DAF are required for its ability to decay the classical/lectin pathway C3 convertase (CP-DAA), and CCP2–4 are essential for the decay of the alternative pathway C3 convertase (AP-DAA) (18, 19). In MCP, domain deletion and site-directed mutagenesis data implicated CCP2–4 in CFA against C3b (C3b-CFA) and C4b (C4b-CFA) (20, 21). The recent crystal structures of DAF and MCP in complex with C3b showed that these RCA proteins bind through a common binding mode and that CCP3–4 of these proteins form contact with C3b (22). However, the questions that remained unanswered are the following: (i) What is the smallest structural unit of DAF required for imparting DAA? (ii) What is the functional significance of each of DAF’s domains in DAA? (iii) What represents the minimal FI interaction sites on MCP that are essential for imparting CFA against C3b and C4b?
Although RCA proteins are modular in structure, what largely remains unanswered is whether functional modularity exists in these proteins, i.e., whether individual CCPs or multi-CCP units are capable of imparting a specific function to the protein and whether joining them would add new functional capabilities. Thus, to test this, we have generated four CCP DAF-MCP chimeras and biochemically characterized them and their site-directed mutants. These data demonstrate the functional role of individual modules of DAF and MCP in a structural framework of four contiguous CCPs. Additionally, construction of the DAF-MCP chimeric mutant that contains DAA and CFA equivalent to that of the parent proteins illustrates the existence of functional modularity in RCA proteins. Our data also provide mechanistic insight into both the regulatory activities.
Results
CCP2–3 of DAF Induce Decay, While CCP4 Aids by Serving as a C3b-Binding Domain.
DAF is composed of four CCP modules, wherein CCP2–4 are required for DAA (18). Thus, to determine the role of each of its domains in DAA, we utilized the domain swap strategy. Because MCP completely lacks DAA, we swapped one or more of its domains (M1–M3) and the linkers with the homologous domains of DAF (D2–D4) and the associated linkers (Fig. 1) and tested these mutants for gain of function (Fig. 2). First, we swapped M1 and the attached intermodular linker with D2 and the associated linker (mutant D2M2–4). Intriguingly, this substitution alone was sufficient to impart CP-DAA, although the mutant was ∼30-fold less active compared with DAF. The mutant, however, displayed diminished AP-DAA highlighting the crucial role of other modules in the activity (Fig. 2 A and B, Left).
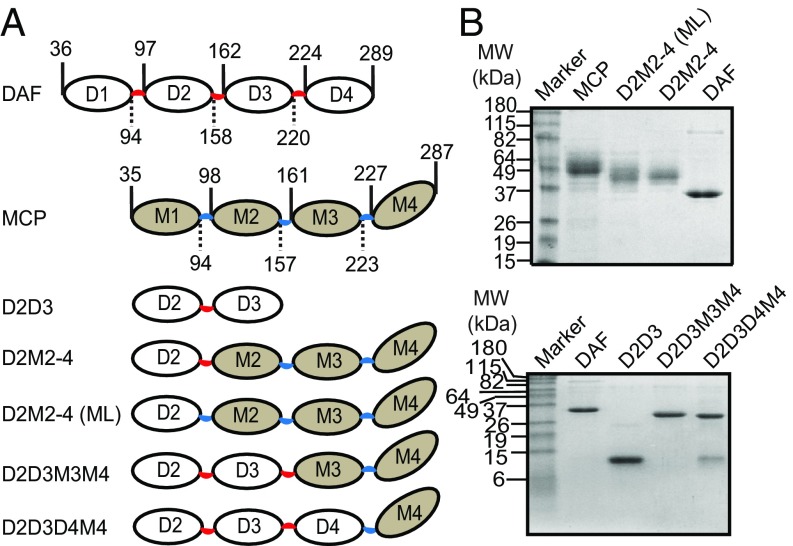
Fig. 1.
Construction of various DAF-MCP chimeras. (A) Diagrammatic representation of DAF, MCP, and the DAF-MCP chimeras. The domains are numbered and the boundaries are marked by the vertical lines. The numbers associated with the vertical lines represent the boundary residues and are according to UniProt numbering. The linkers connecting the DAF domains are in red, while the linkers connecting the MCP domains are in blue; the same color scheme is used to depict the linkers in the DAF-MCP chimeras. D2M2–4 (ML) represents the chimera with the M1–M2 interdomain linker. (B, Upper) SDS/PAGE analysis of DAF, MCP, and the DAF-MCP chimeras expressed in Pichia. (B, Lower) SDS/PAGE analysis of DAF, DAF mutant D2D3, and the DAF-MCP chimeras D2D3M3M4 and D2D3D4M4 expressed in E. coli. MW, molecular weight.
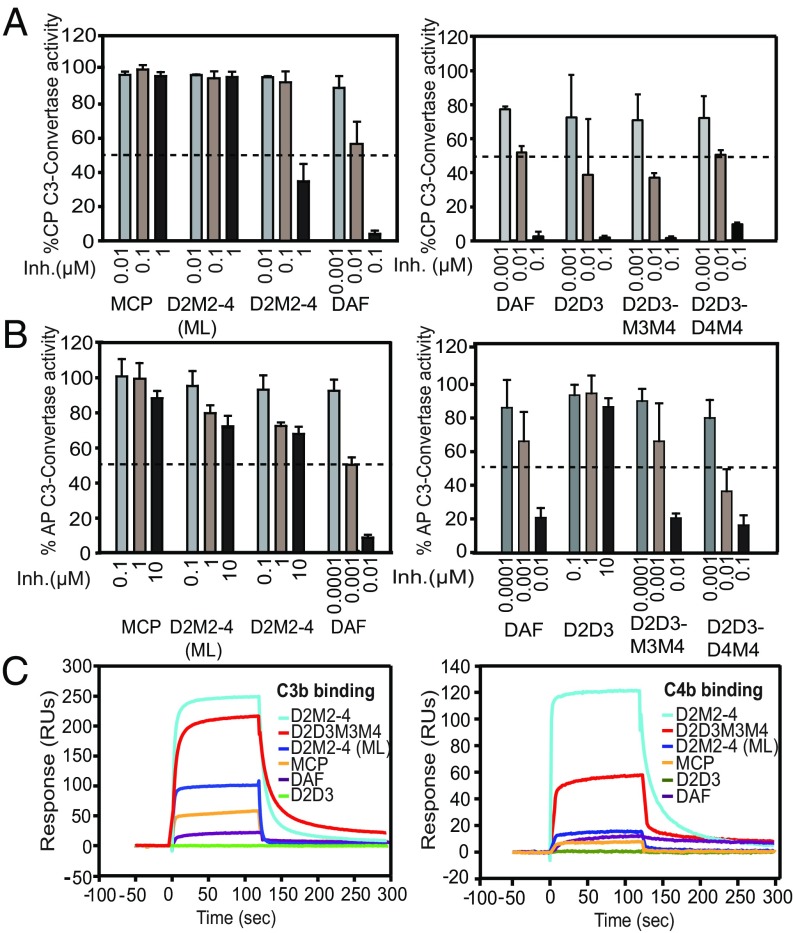
Fig. 2.
Decay-accelerating activity and binding analysis of DAF-MCP chimeras. (A) CP-DAA and (B) AP-DAA of DAF, MCP, D2D3 mutant, and the indicated DAF-MCP chimeras. “Inh.” denotes inhibitor concentration. Data shown are mean ± SD of three independent experiments summarized in SI Appendix, Table S1. The dashed line denotes 50% activity. (C) Binding analysis of DAF, MCP, D2D3 mutant, and the DAF-MCP chimeras to C3b (Left) and C4b (Right). The graph represents the sensogram overlays of binding interactions shown as response units on the y axis. Binding was measured by flowing 1 µM of the respective proteins over the C3b- (Flow cell-2) and C4b-biotin (Flow cell-3) immobilized on a streptavidin chip. Data shown here are from one of the three independent experiments shown in SI Appendix, Fig. S1.
Next, before swapping the successive domain, we tested the importance of the linker between D2–D3 (i.e., KKKS; net positive charge +3) in DAA as earlier studies have highlighted the role of the positively charged residues of this linker in DAA (19). Moreover, the recent DAF-C3b complex structure also illustrated that the linker residues interact with C3b (22). In addition, positive electrostatic potential around the N terminus CCP has been shown to enhance the initial recognition of C3b/C4b (23). Thus, we replaced the D2–D3 linker in D2M2–4 with the neutral linker (YRET) of MCP present at the homologous position. The resultant mutant (D2M2–4-ML), unlike that of D2M2–4, showed no gain in CP-DAA (Fig. 2A, Left). The D2M2-4 mutant with the MCP linker (D2M2–4-ML) also showed a considerable loss in binding to C3b and C4b compared with the D2M2–4 mutant with the DAF linker (D2M2–4) (Fig. 2C and SI Appendix, Fig. S1A).
We next substituted two N-terminal modules of DAF along with the associated linkers in MCP (D2D3M3M4) and assessed the DAA of this mutant. We observed that the mutant demonstrated CP-DAA and AP-DAA equal to that of DAF. The substitution of three domains of DAF along with the linkers in MCP (D2D3D4M4), however, did not result in any further increase in DAA. If any, there was an approximate 10-fold reduction in AP-DAA compared with DAF (Fig. 2 A and B, Right). The surface plasmon resonance (SPR) data showed that the off-rate of this mutant from C3b is much slower than D2D3M3M4; its off-rate did not change for C4b (SI Appendix, Fig. S1B). These data thus suggest that the slower off-rate affects the regulator’s recycle rate for AP C3 convertase complexes, which results in its reduced AP-DAA. Together, the above results indicate that decay is induced only by CCP2–3 of DAF. It should, however, be pointed out that earlier studies have implicated the role of D4 in addition to that of D2–D3 in inducing AP-DAA (18, 24), which is also apparent from our data as D2–D3 alone do not show any AP-DAA (Fig. 2B, Right). It is therefore apparent that the D4 module in DAF merely plays a role of C3b-binding domain, and its function can be substituted by M3–M4.
To gain further insight into the possible interactions of D2D3 modules in the D2D3M3M4 chimera with the AP C3 convertase subunits (i.e., C3b and Bb), we modeled C3b-D2D3M3M4-Bb complex (SI Appendix, SI Materials and Methods). Upon interface analysis, we found that D2 and D3 interact with both C3b and Bb. Namely, D2 and D3 formed interfaces with an α′ N-terminal region and macroglobulin-6 domain of C3b (SI Appendix, Fig. S2A). In addition, D2 and the D2–D3 linker formed contacts with the α7-helix of the Von Willebrand factor type-A (VWA) domain in Bb, while D3 formed contacts with the α7-helix, and α6-βF and α5-βE loops of the VWA domain in Bb (SI Appendix, Fig. S2B). Previous studies have suggested that a stable conformation of the metal ion-dependent adhesion site (MIDAS) (formed by βA-α1,α3–α4 and βD-α5 loops) is critical for maintenance of a high-affinity conformation of VWA and the stability of the AP C3 convertase (25, 26). Furthermore, it was also proposed that the α7-helix is allosterically coupled to the MIDAS site (26). Our modeling data show that D2 and D3 majorly interact with α6-βF and α5-βE loops in addition to the α7-helix. It is therefore likely that interaction of D2–D3 with these loops is important for allosteric changes in the MIDAS site leading to decay of Bb from C3b.
Factor I Interaction Sites on M2 and M3 Are Required for Optimum CFA of MCP.
The recently solved crystal structure of the complex of human C3b, mini FH, and FI showed that, along with C3b, both CCP2 and CCP3 of FH (homologous to M2 and M3 modules of MCP) make contact with FI (27). Furthermore, CCP2–3 of viral RCA regulators (Kaposica and CCPH) have been shown to drive CFA (8). These data suggested that FI interaction sites are likely conserved in CCP2 and CCP3 of human and viral RCA proteins. The relative importance of these sites in CFA, however, was not clear. We, therefore, asked whether the FI site on M3 alone is sufficient to drive CFA in MCP. Hence, we examined the CFA of the D2D3M3M4 chimera, which lacks the M2 domain of MCP. Remarkably, the chimera displayed CFA for C3b and C4b, although the CFA for C3b and C4b was 34- and 35-fold less, respectively, compared with that of MCP (Fig. 3A and SI Appendix, Fig. S3). These results, therefore, show that the FI interaction site present on M3 alone can contribute functionally to CFA.
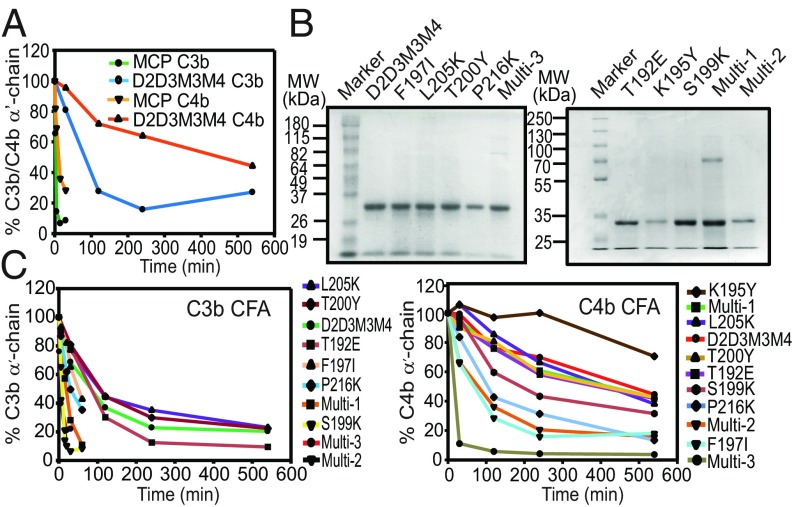
Fig. 3.
Cofactor activity of D2D3M3M4 and its substitution mutants. (A) Relative C3b-CFA and C4b-CFA of MCP and D2D3M3M4. Data shown are mean of three independent experiments summarized in SI Appendix, Table S1. (B) SDS/PAGE analysis of D2D3M3M4 and its single and multiresidue mutants expressed in E. coli. (C) Relative C3b-CFA and C4b-CFA of D2D3M3M4 and its single and multiresidue mutants. The order of the symbol key from top to bottom corresponds to the order of the lines (i.e., least to most active). Data shown in the graphs are mean of three independent experiments summarized in SI Appendix, Table S1.
Having seen CFA in the D2D3M3M4 chimera, we next asked whether we can enhance the CFA of this chimera by substituting the FI interaction site in the D3 domain (homologous to M2), but without affecting its DAA. In other words, we asked, can potent DAA and CFA toward both the classical and alternative pathway C3 convertases coexist in a four-CCP regulator? To answer this, we performed the substitution of putative FI-interacting residues in D3 and the attached linker of the D2D3M3M4 mutant based on the earlier mutagenesis studies of the viral and human RCA proteins (SI Appendix, Fig. S4). Herein, we generated a total of seven single and three multiresidue mutants of D2D3M3M4 (Fig. 3B). These mutations reside between the Cys2–Cys4 region of D3 and the D3–M3 linker (ECREIY) as the earlier swapping of this region of DAF with the homologous region of MCP resulted in the incorporation of CFA into DAF (28). Biochemical analysis of the single and multiresidue mutants showed varying results – a complete loss to 28-fold gain in CFA. Substitutions that showed more than twofold gain in C3b CFA included single amino acid mutants such as S199K and P216K and multiresidue mutants such as multi-1 (linker substitution mutants 219ECREIY224 to ICEKVL), multi-2 (linker substitution + S199K), and multi-3 (215DPL217 to PKA). Likewise, mutations that showed more than twofold gain in C4b CFA included F197I, S199K, P216K, multi-2, and multi-3 (Fig. 3C and SI Appendix, Figs. S5 A and B and Table S1).
Next, we evaluated the DAA of these D2D3M3M4 mutants and looked for the loss in DAA, particularly in the mutants that showed a gain in CFA (SI Appendix, Fig. S6 and Table S1). Our results demonstrated that three mutants that exhibited a gain in CFA showed a loss in DAA. For example, an approximately twofold loss in AP-DAA was observed in S199K and multi-2 and muti-3 mutants. The other three mutants (F197I, P216K, and multi-1) showed little or no loss in AP-DAA. Additionally, none of the gain in CFA mutants showed an approximately twofold loss in CP-DAA. These data, therefore, support the premise that coexistence of strong CFA and DAA in a structural framework of four CCPs is achievable.
To examine whether residues that provided gain in CFA are conserved in other RCA proteins that show CFA, we performed the sequence alignment of various RCA proteins. We observed that 5 of 10 residues that are associated with a gain in activity are conserved in position in other proteins with CFA. For example, a conserved isoleucine was seen at positions comparable with F197 and E219 of DAF. Similarly, positively charged residues were present at positions comparable with P216 and E222 of DAF, and negatively charged residues were present at the position analogous to R221 of DAF. Among these residues, mutation of isoleucine in MCP (21) at a position collinear to E219 of DAF, and of positively charged residues in CR1, SPICE, Kaposica, and CCPH (8, 28, 29) at positions corresponding to P216 and/or E222 of DAF have shown a loss in CFA (SI Appendix, Fig. S4).
Designing of a Four-CCP DAF-MCP Chimera with Potent DAA and CFA.
In the above exercise, three single-residue mutants (F197I, S199K, P216K) and one multiresidue mutant (multi-1, i.e., linker substitution mutant) showed moderate to a considerable increase in CFA (Fig. 3C and SI Appendix, Table S1). It was therefore conceivable that collective substitution of these residues in D2D3M3M4 chimera is likely to result in the generation of a molecule with DAA and CFA as strong as DAF and MCP. We thus generated two multivariants of D2D3M3M4: one where all of the above mutations were incorporated into D2D3M3M4 (multi-4) and the other where all, except S199K, were incorporated into D2D3M3M4 (multi-5) as this mutation increases CFA, but decreases AP-DAA. Examination of the CFA of these two mutants showed that both the mutants have good C3b CFA, but multi-5 was equivalent to MCP (Fig. 4A and SI Appendix, Fig. S7). Both the mutants, however, displayed better C4b CFA (approximately two- to threefold) compared with MCP (Fig. 4B and SI Appendix, Fig. S7). We also determined if these mutations have any effect on the DAA of these mutants. The multi-4 mutant showed a complete loss in AP-DAA and an ∼2.5-fold loss in CP-DAA, while multi-5 displayed AP- and CP-DAA equivalent to DAF (Fig. 4 C and D and SI Appendix, Table S1). Consistent with the loss in AP-DAA of multi-4 mutant, its binding to C3b but not C4b, was found to be perturbed in comparison with D2D3M3M4 (SI Appendix, Fig. S1C).
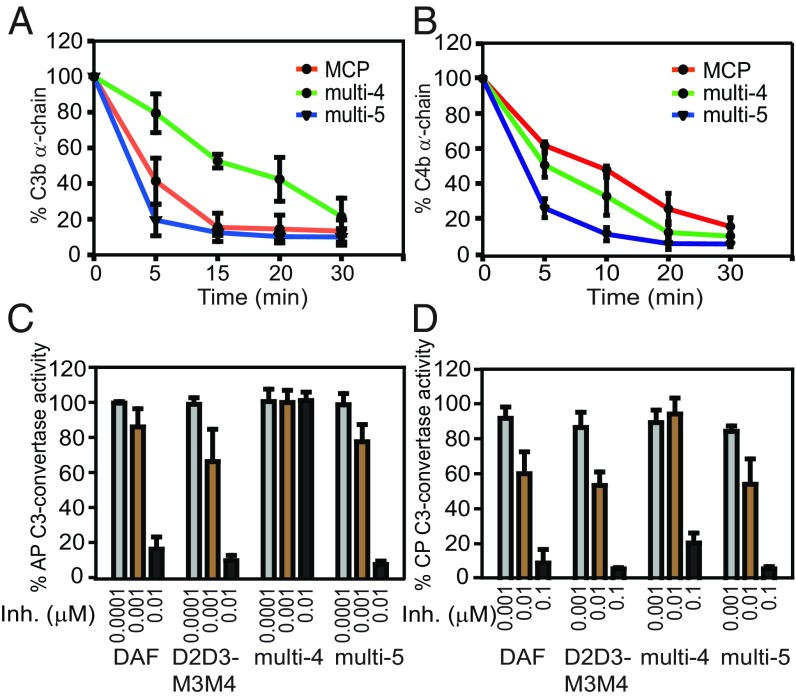
Fig. 4.
Substitution of the putative factor I interaction sites in the D3 domain of D2D3M3M4 generates a molecule with CP- and AP-DAA and C3b- and C4b-CFA. (A) C3b-CFA and (B) C4b-CFA of MCP and the D2D3M3M4 substitution mutants multi-4 and multi-5. (C) AP-DAA and (D) CP-DAA of DAF, D2D3M3M4, and its substitution mutants multi-4 and multi-5. “Inh.” denotes inhibitor concentration.
Next, we tested the regulatory activity of the multi-5 mutant on all of the three major pathways of complement using the Wieslab complement screen ELISA and compared it to DAF and MCP. The multi-5 was 2- to 7.5-fold more active than DAF and 35- to 225-fold more active than MCP in inhibiting the various pathways (SI Appendix, Fig. S7 and Table S2A). Thus, we describe the successful design of a four-CCP molecule with efficient CFA and DAA. Based on the strong CFA and DAA of multi-5, we name this molecule DCP (decay-cofactor protein). A comparison of the inhibitory potential of DCP with CR1 inhibitors such as LHR-A (CCP1–3) and its improved variant LHR-Amut (CCP1–3, D109N/E116K) showed that DCP was more potent than LHR-A (CCP1–3) in inhibiting the alternative and lectin pathways and more potent than LHR-Amut in inhibiting the alternative pathway (SI Appendix, Fig. S7 and Table S2B). DCP, however, was more similar to LHR-Amut in inhibiting the classical and lectin pathways.
Gain of CFA Mutations in the DAF-MCP Chimera D2D3M3M4 Enhance Its Interaction with FI.
The exercise detailed above clearly identified the residues that impart gain in CFA when substituted in the D2D3M3M4 mutant. To decipher the influence of these residues on molecular interactions with FI and C3b, a structural model of a ternary complex of C3b-multi-4 mutant-FI was generated by replacing FH with the chimera in the C3b-FH-FI structure (SI Appendix, SI Materials and Methods). The predicted ternary complex was subjected to molecular dynamics simulations (for 50 ns) to highlight the effects of these mutations on the stability and their role in interactions with FI and C3b (Fig. 5A). The rmsd was calculated to characterize structural variations in the protein for the entire simulation period. The rmsd was found to be stable for the entire simulation period with small drifts and plateaus. The root mean square fluctuation (RMSF) plot was calculated for the chimera to understand the residue-wise fluctuation. The RMSF plot revealed a similar fluctuation pattern between the D2D3M3M4 chimera and multi-4 mutant structure (SI Appendix, Fig. S8A).
Fig. 5.
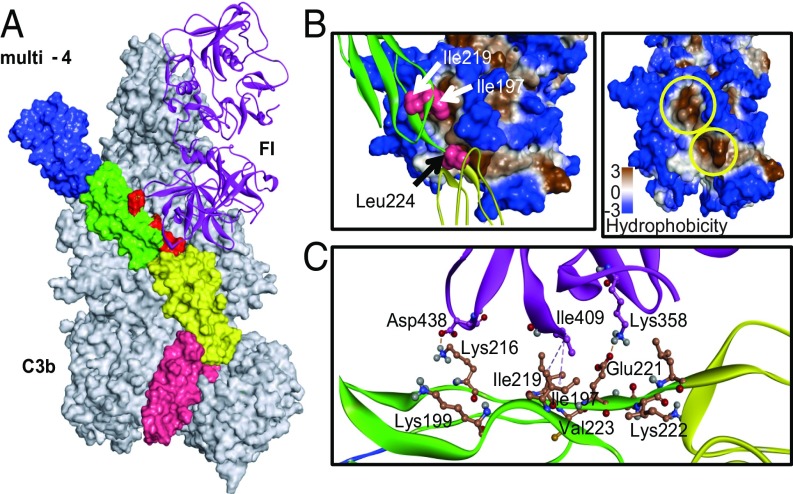
Mapping of factor I interaction sites in the C3b-multi-4 mutant-FI trimolecular complex. (A) Model of the C3b-multi-4 mutant-FI trimolecular complex. The D2D3M3M4 chimera having all of the CFA gain-of-function residues (multi-4 mutant) was superimposed with the coordinates of FH in the C3b-FH-FI crystal structure (Protein Data Bank ID code 5O35). The model was then subjected to MD simulations for 50 ns and analyzed for the interactions of gain-of-function residues with FI. Each of the four domains of the multi-4 mutant (shown as surface) was labeled with different colors (CCP1-blue, CCP2-green, CCP3- yellow, CCP4-pink). C3b (shown as surface) and FI (ribbon representation) are in gray and magenta, respectively. Gain-of-function residues are in red. (B) Zoomed view of the interactions shown by the gain-of-function residues with FI. (Left) I197 and I219 sit in the upper hydrophobic pocket formed by the residues W393, P402, L404, I407, V408, I409, and Y411, while L224 sits in a lower hydrophobic pocket formed by residues I357, G362, I363, A360, V396, V397, W399, and I400. (Right) The hydrophobic pockets shown on the Left are shown again and marked with yellow circles. (C) Charge and hydrophobic interactions of gain-of-function residues with FI.
The interactions of the chimera with C3b and FI evaluated in the simulated structure showed that I197 and I219 of the chimera accommodate in a hydrophobic pocket of FI formed by residues W393, P402, L404, I407, V408, I409, and Y411. Similarly, L224 (linker residue) sits in the adjacent hydrophobic pocket of FI formed by residues I357, G362, I363, A360, V396, V397, W399, and I400 (Fig. 5B). Thus, the hydrophobic interactions of these residues seem crucial to enhance the stability of the chimera and FI, as reported previously for FH-FI (27). Furthermore, residues K216 and E221 located in the linker region interact with residues D438 and K358 of FI through salt bridge interactions. The mutant residues K199 and K222 do not show any interaction as side-chain orientations are away from the FI interface. This explains why the reversion of K199 to Ser (in multi-5) has no significant effect on CFA. The S199 residue, however, has a high BSA score with respect to contact with α7 and α6-βF loops of the VWA domain of Bb (SI Appendix, Fig. S2B) and, therefore, possibly accounts for a major gain in AP-DAA following K199S reversion. Interestingly, D403 of FI acts as bridging residue by forming hydrogen bonds with K195 of D3 and S214 of M3 and might be involved in D3-M3 domain coordination of the chimera. Additionally, residues E179 and E177 form a salt bridge network with residue R480 of FI. The residue V178 of the chimera forms a pi-alkyl interaction with W399 of FI (SI Appendix, Fig. S9C). Overall, the interaction patterns of the linker and associated regions of the chimera with FI are congruent with our experimental study. The interaction of the multi-4 mutant with the C3b domain is depicted in SI Appendix, Fig. S9 A and B. The domain-specific interactions were found to be similar to DAF and MCP (22).
Discussion
The members of the RCA family of proteins are majorly responsible for preventing the indiscriminate damage of host cells from homologous complement (4, 5). These modular proteins are formed solely by CCP domains, which vary from 4 to 59, and are linked by short linkers of three to eight amino acids. Although overall it appears that the regulatory activities (DAA and CFA) of these proteins are primarily driven by two to four CCPs (18, 20, 30), what constitutes the optimal functional unit, what role individual domains of the unit play, and is the arrangement of such units orderly in the context of four CCPs, has not yet been resolved.
Earlier domain deletion and mutagenesis studies have clearly pointed out the role of DAF domains 2–3 (D2–D3) and 2–4 (D2–D4) in CP-DAA and AP-DAA, respectively (18, 19). It is also known that these domains bind to both the subunits of C3 convertases, i.e., C3b/C4b as well as Bb/C2a (24, 31). However, the exact role of these domains in DAA is not clear. For example, even though mutagenesis studies have identified multiple residues in D2 and D3 as critical for AP- and CP-DAA (17, 19), it was proposed that these domains do not mediate AP-DAA. Rather, D2 binds to the Bb subunit of the convertase to increase its avidity and D3 serves as an essential linker. And, it is D4, which mediates decay of C3bBb owing to its interaction with C3b, which results in its conformational transition leading to destabilization of Bb and therefore its release (31). This model, however, could not explain how D2–D3 alone mediate CP-DAA. Moreover, swapping of D3 in the vaccinia virus complement control protein was shown to significantly enhance AP-DAA (32). Thus, to clarify the role of individual functional domains of DAF (D2–D4) and its interdomain linkers in DAA, we swapped them in place of homologous domains/linkers of MCP, which lack DAA, but binds to the noncatalytic subunit of the convertase (C3b and likely C4b) in a similar manner (22). Our data show that D2 along with its linker (KKKS) are enough to mediate moderate CP-DAA, although not AP-DAA when linked in an MCP framework (D2M2–4). Furthermore, our data also show that swapping of D2–D3 and the linkers in MCP (D2D3M3M4) is enough to drive optimum CP- as well as AP-DAA. We therefore unequivocally show that D2–D3 constitutes the functional unit in DAF that imparts DAA. The question that remains then is, why is D4 required for AP-DAA? We propose that it purely serves the purpose of increasing the avidity to the convertase through its interaction with the noncatalytic subunit C3b, as M3–M4 domains that are known to bind to C3b (20, 22, 32) serve this purpose in the D2D3M3M4 chimera. Having said the above, we add that such binding to C3b must have a fast-to-moderate off-rate to achieve efficient recycling over the AP C3 convertase because a decrease in the off-rate, as seen in D2D3D4M4, significantly reduces AP-DAA.
Diminished DAF-induced decay of the C3 convertases (C3bBb and C4b2a) assembled using factors B and C2 mutated in their respective VWA domains suggested that DAF mediates decay by interacting with the VWA domains of Bb and C2a (33, 34). The VWA domain is known to exist in high- and low-affinity forms, which are dependent on the presence of metal ions (35). Moreover, DAF has been shown to bind to VWA in the presence of Mg2+ (31). It was therefore proposed that binding of DAF to VWA induces its low-affinity conformation, which results in displacement of Bb/C2a from the respective convertases (31). Modeling of D2D3M3M4 in complex with C3bBb shows that D2 interacts with α7 helix of Bb, while D3 interacts with α7 helix and α6-βF and α5-βE loops of Bb, which appear to be conformationally connected to MIDAS. Because the presence of D2 is enough for moderate CP-DAA, and D2–D3 are required for optimum AP- and CP-DAA, we suggest that interaction of DAF with the α7 helix of C2a and C4b is enough to initiate CP-DAA, but not AP-DAA, which requires additional interaction with α6-βF as well as α5-βE loops of Bb in addition to C3b. Earlier mutagenesis studies in the VWA domain of Bb pointed out that mutations in the α4/α5 helices (Q335A, Y338A, S339A, and D382A) renders the convertase resistant to decay by DAF/CR1, but not by factor H (34). Mapping of these mutations in our model shows that these residues are away from the D2D3-Bb interface. It is therefore likely that the α4/5 region participates in the propagation of an allosteric signal from the D2D3-Bb interface to the C4b2a/C3bBb interface, resulting in the decay, as suggested earlier (33).
Mapping of RCA proteins (e.g., FH, MCP, CR1 C4BP, and viral regulators Kaposica and SPICE) for CFA revealed that three to four CCPs are required for optimum activity. What, however, remained unresolved was what role each of the domains play in CFA. The current model of CFA based on the structural and biochemical data suggests that during this process the RCA protein binds to the target protein (C3b/C4b) and provides a platform for docking of the protease FI, which then cleaves the target protein (C3b/C4b), resulting in its inactivation. The structures of C3b in complex with the RCA proteins that mediate CFA (FH, MCP, and the viral regulator SPICE) showed that their binding results in ∼6–9 Å of upward movement of the CUB and TED domains (22). Such movement and rearrangements of domains seem important for the activity as bridging of CUB and MG2 is critical for potent CFA (28). The structure of free FI and its docking onto the C3b-FH structure showed that FI docks into a niche formed by the RCA and the CTC domain of C3b (36). More recently, the structure of C3b-miniFH-FI has been solved, and it showed that FI interacts with CCPs 2 and 3 of miniFH, as well as the CTC, MG2, and CUB domains of C3b (27). The functional significance of the interaction of FI with each of the CCPs, however, was not determined in this study, and therefore it is not clear whether interaction of FI with both CCPs is needed for strong CFA.
In the present study, the DAF-MCP chimera D2D3M3M4 was shown to possess robust CP- and AP-DAA. We thus tested whether strong CFA can be incorporated in this molecule without affecting DAA. We expected the molecule to display CFA as it showed good binding to C3b/C4b and also had the M3 domain, which is expected to harbor a conserved FI-binding site [based on the earlier biochemical and structural data (21, 27)] and bridge CUB-MG2 domains (28). The molecule did show CFA, although weak compared with MCP, suggesting that the FI interaction site present on M3 (CCP3) contributes to CFA. Next, based on the earlier mutagenesis data from our and other laboratories, we successfully introduced FI interaction sites in D3 (CCP2) without affecting AP- or CP-DAA. Thus, we established that the interaction of FI with both CCP2 and CCP3 is essential for strong CFA. Additionally, in the process, we generated a molecule (now named DCP) that displays robust CFA (C3b- and C4b-CFA) as well as DAA (CP- and AP-DAA). A CR1-derived molecule, LHR-A (CCP1–3), is being developed for clinical use (14). We thus compared the activity of DCP with this molecule as well as its more potent variant LHR-Amut (CCP1–3, D109N/E116K). In general, the DCP molecule displayed inhibitory potential more comparable to that of the mutant; however, its ability to inhibit the alternative pathway was even better than LHR-Amut, suggesting that DCP is indeed a potent inhibitor of complement activation.
In summary, the present study identifies the role of individual CCPs of DAF and MCP in mediating DAA and CFA, respectively. Furthermore, it also provides the structural definition of a “functional unit” of RCA for DAA and CFA and their spatial arrangement in a structural framework of four CCPs. Based on the present and earlier data, we suggest that DAA is primarily mediated by the interaction of a pair of RCA CCPs with Bb/C2a, which induces low-affinity conformation of the VWA domain of Bb/C2a, leading to their release from the convertase. However, optimal interaction of RCA with C3b/C4b is essential for its proper docking onto the convertase. The CFA, on the other hand, requires a pair of RCA CCPs, which on one face interact with C3b/C4b and bridge its CUB-MG2 domain, while on the other face act as a platform for recruitment and reorientation of FI to induce cleavage in the scissile bonds in the CUB domain. Together, the data presented here suggest that an optimal dual-activity regulator having robust CP- and AP-DAA as well as C3b- and C4b-CFA requires four CCPs. Additionally, the dual-activity regulator generated here (DCP) may serve as a lead molecule for developing RCA-based therapeutics for treating pathological conditions involving the complement system.
Materials and Methods
Construction, Expression, and Purification of DAF, MCP, DAF-MCP Chimeras, and the Substitution Mutants of D2D3M3M4.
The CCP1–4 portion of human DAF and MCP cloned in pGEM-T Easy vector was used as the template for cloning and generation of chimeric mutants. The construction, expression, and purification of DAF, MCP, DAF-MCP chimeras, and the point mutants of D2D3M3M4 are detailed in SI Appendix, SI Materials and Methods. Both the parent proteins were expressed in Pichia pastoris and in Escherichia coli. Some of the domain swap mutants, namely D2M2–4 and D2M2–4 (ML), were expressed in P. pastoris, whereas other domain swap mutants (D2D3M3M4, D2D3D4M4, D2D3) and the substitution mutants of D2D3M3M4 chimera were expressed in E. coli BL21 (DE3) cells.
Complement Assays and SPR Measurements.
Details of CP and AP C3 convertase DAA assay, C3b and C4b CFA assay, Wieslab complement screen ELISA, and SPR measurements are in SI Appendix, SI Materials and Methods.
Molecular Modeling and MD Simulation.
The details of model building and validation and MD simulations are provided in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Prof. Piet Gros (Utrecht University) and Dr. Jayati Mullick (National Institute of Virology, Pune) for valuable suggestions and critical reading of the manuscript; Ankita Vaishampayan for assistance with DNA sequencing; and Avneesh Gautam, Rajashri Shende, and Jitendra Kumar for help during the experimental work. This work was done in partial fulfillment of the Ph.D. thesis of H.S.P. to be submitted to the S. P. Pune University. We acknowledge financial assistance from the Council of Scientific and Industrial Research, New Delhi (H.S.P.) and the Department of Biotechnology, New Delhi (H.O.). This work was supported by the Department of Biotechnology, India Project Grant BT/PR28506/MED/29/1307/2018 (to A.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1818573116/-/DCSupplemental.
References
- 1.Walport MJ. Complement. First of two parts. N Engl J Med. 2001;344:1058–1066. doi: 10.1056/NEJM200104053441406. [DOI] [PubMed] [Google Scholar]
- 2.Carroll MC, Isenman DE. Regulation of humoral immunity by complement. Immunity. 2012;37:199–207. doi: 10.1016/j.immuni.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ricklin D, Hajishengallis G, Yang K, Lambris JD. Complement: A key system for immune surveillance and homeostasis. Nat Immunol. 2010;11:785–797. doi: 10.1038/ni.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liszewski MK, Atkinson JP. Complement regulators in human disease: Lessons from modern genetics. J Intern Med. 2015;277:294–305. doi: 10.1111/joim.12338. [DOI] [PubMed] [Google Scholar]
- 5.Martínez-Barricarte R, et al. The molecular and structural bases for the association of complement C3 mutations with atypical hemolytic uremic syndrome. Mol Immunol. 2015;66:263–273. doi: 10.1016/j.molimm.2015.03.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hourcade D, Liszewski MK, Krych-Goldberg M, Atkinson JP. Functional domains, structural variations and pathogen interactions of MCP, DAF and CR1. Immunopharmacology. 2000;49:103–116. doi: 10.1016/s0162-3109(00)80296-9. [DOI] [PubMed] [Google Scholar]
- 7.Holers VM. Complement and its receptors: New insights into human disease. Annu Rev Immunol. 2014;32:433–459. doi: 10.1146/annurev-immunol-032713-120154. [DOI] [PubMed] [Google Scholar]
- 8.Ojha H, Panwar HS, Gorham RD, Jr, Morikis D, Sahu A. Viral regulators of complement activation: Structure, function and evolution. Mol Immunol. 2014;61:89–99. doi: 10.1016/j.molimm.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 9.Kerr MA. The human complement system: Assembly of the classical pathway C3 convertase. Biochem J. 1980;189:173–181. doi: 10.1042/bj1890173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nicholson A, et al. Interaction of C3b, B, and D in the alternative pathway of complement activation. J Immunol. 1975;115:1108–1113. [PubMed] [Google Scholar]
- 11.Zipfel PF, Skerka C. Complement regulators and inhibitory proteins. Nat Rev Immunol. 2009;9:729–740. doi: 10.1038/nri2620. [DOI] [PubMed] [Google Scholar]
- 12.Seya T, Turner JR, Atkinson JP. Purification and characterization of a membrane protein (gp45-70) that is a cofactor for cleavage of C3b and C4b. J Exp Med. 1986;163:837–855. doi: 10.1084/jem.163.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicholson-Weller A, Burge J, Fearon DT, Weller PF, Austen KF. Isolation of a human erythrocyte membrane glycoprotein with decay-accelerating activity for C3 convertases of the complement system. J Immunol. 1982;129:184–189. [PubMed] [Google Scholar]
- 14.Krych-Goldberg M, Atkinson JP. Structure-function relationships of complement receptor type 1. Immunol Rev. 2001;180:112–122. doi: 10.1034/j.1600-065x.2001.1800110.x. [DOI] [PubMed] [Google Scholar]
- 15.Lukacik P, et al. Complement regulation at the molecular level: The structure of decay-accelerating factor. Proc Natl Acad Sci USA. 2004;101:1279–1284. doi: 10.1073/pnas.0307200101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Persson BD, et al. Structure of the extracellular portion of CD46 provides insights into its interactions with complement proteins and pathogens. PLoS Pathog. 2010;6:e1001122. doi: 10.1371/journal.ppat.1001122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uhrinova S, et al. Solution structure of a functionally active fragment of decay-accelerating factor. Proc Natl Acad Sci USA. 2003;100:4718–4723. doi: 10.1073/pnas.0730844100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brodbeck WG, Liu D, Sperry J, Mold C, Medof ME. Localization of classical and alternative pathway regulatory activity within the decay-accelerating factor. J Immunol. 1996;156:2528–2533. [PubMed] [Google Scholar]
- 19.Kuttner-Kondo L, et al. Structure-based mapping of DAF active site residues that accelerate the decay of C3 convertases. J Biol Chem. 2007;282:18552–18562. doi: 10.1074/jbc.M611650200. [DOI] [PubMed] [Google Scholar]
- 20.Adams EM, Brown MC, Nunge M, Krych M, Atkinson JP. Contribution of the repeating domains of membrane cofactor protein (CD46) of the complement system to ligand binding and cofactor activity. J Immunol. 1991;147:3005–3011. [PubMed] [Google Scholar]
- 21.Liszewski MK, et al. Dissecting sites important for complement regulatory activity in membrane cofactor protein (MCP; CD46) J Biol Chem. 2000;275:37692–37701. doi: 10.1074/jbc.M004650200. [DOI] [PubMed] [Google Scholar]
- 22.Forneris F, et al. Regulators of complement activity mediate inhibitory mechanisms through a common C3b-binding mode. EMBO J. 2016;35:1133–1149. doi: 10.15252/embj.201593673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pyaram K, Kieslich CA, Yadav VN, Morikis D, Sahu A. Influence of electrostatics on the complement regulatory functions of Kaposica, the complement inhibitor of Kaposi’s sarcoma-associated herpesvirus. J Immunol. 2010;184:1956–1967. doi: 10.4049/jimmunol.0903261. [DOI] [PubMed] [Google Scholar]
- 24.Harris CL, Pettigrew DM, Lea SM, Morgan BP. Decay-accelerating factor must bind both components of the complement alternative pathway C3 convertase to mediate efficient decay. J Immunol. 2007;178:352–359. doi: 10.4049/jimmunol.178.1.352. [DOI] [PubMed] [Google Scholar]
- 25.Milder FJ, et al. Structure of complement component C2A: Implications for convertase formation and substrate binding. Structure. 2006;14:1587–1597. doi: 10.1016/j.str.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 26.Forneris F, et al. Structures of C3b in complex with factors B and D give insight into complement convertase formation. Science. 2010;330:1816–1820. doi: 10.1126/science.1195821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xue X, et al. Regulator-dependent mechanisms of C3b processing by factor I allow differentiation of immune responses. Nat Struct Mol Biol. 2017;24:643–651. doi: 10.1038/nsmb.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gautam AK, et al. Mutational analysis of Kaposica reveals that bridging of MG2 and CUB domains of target protein is crucial for the cofactor activity of RCA proteins. Proc Natl Acad Sci USA. 2015;112:12794–12799. doi: 10.1073/pnas.1506449112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krych M, Hauhart R, Atkinson JP. Structure-function analysis of the active sites of complement receptor type 1. J Biol Chem. 1998;273:8623–8629. doi: 10.1074/jbc.273.15.8623. [DOI] [PubMed] [Google Scholar]
- 30.Klickstein LB, et al. Identification of distinct C3b and C4b recognition sites in the human C3b/C4b receptor (CR1, CD35) by deletion mutagenesis. J Exp Med. 1988;168:1699–1717. doi: 10.1084/jem.168.5.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harris CL, Abbott RJ, Smith RA, Morgan BP, Lea SM. Molecular dissection of interactions between components of the alternative pathway of complement and decay accelerating factor (CD55) J Biol Chem. 2005;280:2569–2578. doi: 10.1074/jbc.M410179200. [DOI] [PubMed] [Google Scholar]
- 32.Ahmad M, et al. Domain swapping reveals complement control protein modules critical for imparting cofactor and decay-accelerating activities in vaccinia virus complement control protein. J Immunol. 2010;185:6128–6137. doi: 10.4049/jimmunol.1001617. [DOI] [PubMed] [Google Scholar]
- 33.Kuttner-Kondo LA, et al. A corresponding tyrosine residue in the C2/factor B type A domain is a hot spot in the decay acceleration of the complement C3 convertases. J Biol Chem. 2003;278:52386–52391. doi: 10.1074/jbc.M304620200. [DOI] [PubMed] [Google Scholar]
- 34.Hourcade DE, Mitchell L, Kuttner-Kondo LA, Atkinson JP, Medof ME. Decay-accelerating factor (DAF), complement receptor 1 (CR1), and factor H dissociate the complement AP C3 convertase (C3bBb) via sites on the type A domain of Bb. J Biol Chem. 2002;277:1107–1112. doi: 10.1074/jbc.M109322200. [DOI] [PubMed] [Google Scholar]
- 35.Bhattacharya AA, Lupher ML, Jr, Staunton DE, Liddington RC. Crystal structure of the A domain from complement factor B reveals an integrin-like open conformation. Structure. 2004;12:371–378. doi: 10.1016/j.str.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 36.Roversi P, et al. Structural basis for complement factor I control and its disease-associated sequence polymorphisms. Proc Natl Acad Sci USA. 2011;108:12839–12844. doi: 10.1073/pnas.1102167108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.