Abstract
Dendritic cells (DCs) are professional antigen presenting cells located at mucosal surfaces and lymphoid tissues. Their main role is to present antigens to T cells and thus regulate the initiation of the acquired immune response and modulate tolerance mechanisms towards self-antigens. Despite their relevance, not many studies have addressed the identification and characterization of specific DC subsets in teleost fish. Previous studies in our group identified a DC subpopulation co-expressing CD8α and major histocompatibility complex II (MHC II) on the cell surface in rainbow trout (Oncorhynchus mykiss) skin and gills. A complete functional and phenotypical characterization of these cell subsets was then undertaken, unequivocally recognizing them as DCs (CD8+ DCs). In the current study, we report the identification of a homologous population in rainbow trout intestinal lamina propria (LP). We have studied the main features of these intestinal CD8+ DCs, comparing them to those of CD8+ DCs from another mucosal tissue (gills). Interestingly, intestinal CD8+ DCs exhibited significant phenotypical and functional differences when compared to gill CD8+ DCs, suggesting that the location of DCs strongly conditions their activation state. These results will contribute to further expand our knowledge on how intestinal immune responses are regulated in fish, helping us to rationally design oral vaccines in the future.
Keywords: Teleost fish, Rainbow trout, Dendritic cells (DCs), Cross-presentation, Intestine, Antigen presentation
Highlights
-
•
We have identified a CD8+ MHC II+ population in the intestine with characteristics of DCs.
-
•
These cells express many markers shared by different mammalian cross-presenting DC subsets.
-
•
Trout intestinal CD8+ DCs have strong phagocytic capacities and the ability to stimulate naïve T cells.
-
•
Trout intestinal CD8+ DCs seem to have a more tolerogenic profile than CD8+ DCs from gills.
1. Introduction
Oral vaccines are highly demanded by the aquaculture sector that requests alternatives to the labor-intensive injectable vaccines that require individual handling of fish, provoking stress-related immunosuppression and handling mortalities. Despite this, most previous attempts to obtain effective oral vaccines have failed both in fish as they have in mammals [1]. Although the fact that the antigen reaches the intestinal mucosa intact is an important issue when designing oral vaccines, recent advances in antigen protection through diverse encapsulating techniques have solved this issue [1]. Nevertheless oral vaccines are often not fully protective. Therefore, it seems that the lack of effectiveness of oral vaccines could be a consequence of the very strict tolerance mechanisms that regulate the intestine. Immune cells within this mucosa are at the frontline of antigen encounter, having to balance the delicate equilibrium between tolerance and immunity in a microbe-rich aquatic environment. Thus, the intestinal immune system is responsible for the maintenance of tolerance to self-antigens or innocuous antigens, such as food or commensal bacteria, while it has to be capable of triggering an effective immune response to potentially harmful antigens (reviewed in Ref. [2]). In this context, it seems essential to unravel how intestinal immunity is regulated in teleost fish.
Dendritic cells (DCs) are professional antigen presenting cells (APCs) that play a key role in regulating the onset of adaptive immune responses. Owing to high levels of expression of a wide range of pattern recognition receptors, they are able to rapidly sense invading microbes at mucosal surfaces, internalize them, and present them in the context of major histocompatibility complex (MHC) I (intracellular antigens) or MHC II (extracellular antigens) to T cells to start an adaptive immune response [3]. In this sense, intestinal DCs have been shown to be essential for both the induction of immunity and the maintenance of homeostasis, being this the reason why many novel oral vaccination strategies in mammals focused on directing antigens to DCs [4]. In these studies, it has been demonstrated that in order to stimulate an appropriate adaptive immune response within a mucosa, antigens need to be targeted to the correct DC subset through the use of specific adjuvants.
Despite the relevance of DCs in regulating adaptive immune responses, not many studies have been focused at studying these cells in teleost fish. The very first studies that dealt with DCs in teleost fish, indirectly demonstrated that these cells were also present in these species through the identification of cells containing Birbeck granules [5], cells with DC-like morphology [6] or cells expressing transcripts of specific DC markers [7]. Additional studies described the optimization of protocols to enrich leukocytes in DC-like cells [8,9]. However, it was not until 2015, that a specific DC subset was identified in the skin of rainbow trout (Oncorhynchus mykiss) [10]. This DC subpopulation was identified through flow cytometry combining specific antibodies against CD8α and MHC II. These cells transcribed high levels of specific DC markers and inappreciable levels of transcription of T cell markers. At a functional level, these skin CD8+ DCs were highly phagocytic, had the capacity to activate T cells and responded to antigenic stimulations both in vivo and in vitro [10]. Later on, a similar subpopulation was identified in rainbow trout gills [11]. Interestingly, the CD8+ DC populations described in rainbow trout skin and gills expressed a series of immune markers and transcription factors that are exclusive to DCs that have the capacity to cross-present antigens [10,11], pointing to these cells as a potential common ancestor of the diverse DC populations that have cross-presenting capacities in human and mice. Cross-presentation refers to the capacity of specific subsets of DCs to acquire extracellular antigens, process them and present them in the context of MHC I [12]. Murine cross-presenting DCs include resident CD8+ DCs found in secondary immune organs, mainly spleen and lymph nodes [13] and migratory CD103+ DCs present in mucosal tissues such as skin, lungs and intestine [14]. In humans, differently, cross-presenting DCs are characterized by the expression of CD141 [15]. Despite the expression of distinct surface markers, all DCs with cross-presenting capacities from both mice and human share common exclusive features. Among these, for instance, is the fact that these cells use TLR3 to respond to viral stimuli [[16], [17], [18]], whereas other non-cross-presenting DC subsets lack TLR3 expression. Additionally, the functionality of all these cross-presenting populations is regulated by the fms-like tyrosine kinase 3 (Flt3) ligand, IFN regulatory protein 8 (IRF8) [19,20] and Batf3 [21,22].
Although in the digestive tract of mammals a variety of subpopulations of DCs can be found throughout the lamina propria (LP) and associated lymph nodes [23], whether DCs are also found in the teleost intestinal mucosa has never been studied and this is what we have addressed in the current study. As performed in skin and gills, we have combined anti-CD8 and anti-MHC II antibodies to identify a CD8+ DC subset in the rainbow trout intestinal mucosa. We have performed some essential phenotypic and functional analysis to unequivocally establish that these cells correspond in fact to a DC subset. Interestingly, these CD8+ DCs showed some unique features when compared to the equivalent populations in gills and skin, pointing to distinctive traits of CD8+ DCs within the intestine. Interestingly, these specific features suggest a more tolerogenic profile of intestinal CD8+ DCs when compared to the corresponding DC subsets in gills. Expanding our knowledge on how DCs are regulated in the teleost intestine, will greatly contribute to understand how adaptive immunity can breach the strong tolerance mechanisms present in this tissue and thus develop effective oral vaccines.
2. Materials and Methods
2.1. Experimental fish
Female rainbow trout (Oncorhynchus mykiss) of ∼50 g were obtained from Piscifactoría Cifuentes (Guadalajara, Spain) and maintained at the animal facilities of the Animal Health Research Center (CISA-INIA) in an aerated recirculating water system at 16 °C, with 12:12 h light:dark photoperiod. Fish were fed twice a day with a commercial diet (Skretting, Spain). Prior to any experimental procedure, fish were acclimatized to laboratory conditions for at least 2 weeks. All of the experiments described comply with the Guidelines of the European Union Council (2010/63/EU) for use of laboratory animals and have been approved by the Instituto Nacional de Investigación Agraria y Alimentaria (INIA) Ethics Committee (PROEX002/17).
2.2. Leukocyte isolation
Rainbow trout were killed by benzocaine (Sigma) overdose. A transcardial perfusion of the rainbow trout was performed using Ringer solution pH 7.4 containing 0.1% procaine to remove blood from fish tissues. Single cell suspensions from spleen and gills were prepared using 100 μm nylon cell strainers (BD Biosciences) and Leibovitz medium (L-15, Life Technologies) supplemented with 100 I·U./ml penicillin and 100 μg/ml streptomycin (P/S) and 5% fetal calf serum (FCS) (all supplements from Life Technologies). Intestine cell suspensions were also prepared using the midgut and hindgut regions. For this, prior to cell extraction, small pieces of intestine were incubated for 30 min at 4 °C in L-15 medium with antibiotics (P/S) and 5% FCS, followed by agitation for 30 min in PBS containing 1 mM EDTA and 1 mM DTT. Tissue digestion was performed using 0.15 mg/ml collagenase type IV from Clostridium histolyticum (Sigma) in L-15 for 30 min at 20 °C. All cell suspensions were placed onto 30/51% Percoll density gradients and centrifuged at 500×g for 30 min at 4 °C. Cells at the interface were collected and washed twice in L-15 medium containing 5% FCS.
2.3. Flow cytometry
For the identification of DC populations, leukocytes were incubated for 30 min with an anti-trout CD8α (mAb rat IgG; 7 μg/ml) [24] antibody in L-15 media supplemented with 2% FCS (FACS staining buffer). Cells were then washed twice with FACS staining buffer and stained for 20 min with a secondary Ab for anti-CD8α [R-phycoerythrin F(ab')2 fragment of goat anti-rat IgG (H + L) (Life Technologies)] in FACS staining buffer. Thereafter, cells were washed and incubated with anti-trout MHC II [mAb mouse IgG1 coupled to allophycocyanin; 2 μg/ml] [10] in FACS staining buffer. After this incubation, cells were washed two times with FACS staining buffer and analyzed in a FACS Celesta flow cytometer (BD Biosciences) equipped with FACS DIVA software or on a FACSCalibur flow cytometer (BD Biosciences) equipped with CellQuest Pro software. Flow cytometry analysis was performed using Flow Jo 10 software package (Tree Star).
To determine the levels of expression of surface CCR7 in CD8+ DC populations, intestinal leukocytes were incubated with the anti-trout CD8α in combination with a specific anti-CCR7 polyclonal antibody (pAb rabbit IgG; 2 μg/ml) [25]. After 30 min, the cells were washed twice with FACS staining buffer and stained 20 min with secondary antibodies that included an R-phycoerythrin F(ab')2 fragment of goat anti-rat IgG (H + L) and an Alexa Fluor® 488 F(ab')2 fragment of goat anti-rabbit IgG (H + L) (Life Technologies). After this time, cells were washed again and stained with anti-MHC II coupled to allophycocyanin as described above.
In all cases, isotype controls for mouse and rat mAbs and rabbit pAb (BD Biosciences) were tested in parallel to discard unspecific binding of the Abs. All the incubations were performed at 4 °C. Cell viability was checked using YOPRO-1 (0.05 μM). Dead cells (YOPRO-1+ cells) and doublets were excluded from the analysis following the gating strategy described in Supplementary Fig. 1.
2.4. Confocal microscopy
Intestinal leukocyte suspensions were seeded on a poly-l-lysine–coated slide and incubated at 16 °C for 30 min. After gently washing with PBS, the slides were dried and fixed in ice-cold acetone for 15 min. The fixed samples were incubated for 1 h at room temperature (RT) with blocking solution (TBS, pH 7.5 containing 0.01% BSA and 0.5% saponin). The samples were then incubated with mAbs against trout CD8α (50 μg/ml) and trout MHC class II (coupled to allophycocyanin, 10 μg/ml) for 1 h at RT. Thereafter, slides were washed with TBS and incubated with an anti-rat IgG (H + L) Alexa Fluor 488 conjugate for 30 min at 4 °C. Slides were counterstained with 1 μg/ml DAPI (Sigma). Laser scanning confocal microscopy images (0.3 μm thickness) were acquired with an inverted Zeiss Axiovert LSM 880 microscope. Images were analyzed with Zen 2.0 (Carl Zeiss) and Fiji (NIH) software packages.
Rainbow trout intestine from anesthetized and exsanguinated rainbow trout were immediately fixed in 1:3 acetic acid: methanol overnight at RT, further incubated in a sucrose gradient for 48 h (15% and 30% sucrose in PBS), then embedded in PolyFreeze cryostat mounting medium (Sigma) and stored at −80○C until use. Cryostat sections with a thickness of 10 μm were prepared using a Leica CM3050 microtome and placed on SuperFrost glass slides (Menzel-Gläser). Dry sections were fixed in acetone at −20 °C for 20 min, air dried, encircled with a hydrophobic compound (ImmunoPen; Calbiochem), incubated for 1 h at RT with blocking solution (TBS buffer pH 7.5, containing 0.01% BSA, 0.5% saponin, 0.02% Tween-20) and then stained with mAbs against trout MHC II (coupled to allophycocyanin) and CD8α as described above. Slides were counterstained with 1 μg/ml DAPI. Laser scanning confocal microscopy image stacks (1 μm total thickness) acquired were analyzed with Zen 2.0 and Image J softwares.
2.5. Transcriptional analysis of FACS isolated populations
CD8+ DC populations were isolated by flow cytometry using a BD FACSAria III cell sorter (BD Biosciences) after staining intestinal leukocytes with anti-trout CD8α and anti-trout MHC II antibodies as described above. For this, CD8+ DCs were selected following their FSC/SSC profile (FSChighSSCmed/high) and their fluorescence characteristics (CD8+ MHC II+). In some experiments, intestinal leukocytes were treated with polyinosinic-polycytidylic acid (poly I:C; 50 μg/ml) for 16 h at 20 °C or with media alone prior to cell sorting. Splenic CD8+ cytotoxic T cells (lymphoid CD8+MHCII− cells) were also isolated by flow cytometry for comparative purposes. RTS11, a rainbow trout monocyte/macrophage cell line established from spleen [26] was also included in some transcriptional analysis.
Total cellular RNA was isolated from cell populations using the Power SYBR Green Cells-to-Ct Kit (Invitrogen) following manufacturer's instructions. RNA was treated with DNase during the process to remove genomic DNA that might interfere with the PCR reactions. Reverse transcription was also performed using the Power SYBR Green Cells-to-Ct Kit (Invitrogen) following manufacturer's instructions. To evaluate the levels of transcription of the different genes, real time PCR was performed with a LightCycler® 96 System instrument (Roche) using SYBR Green PCR core Reagents (Applied Biosystems) and specific primers previously described (Table 1) [10,11]. Each sample was measured in duplicate under the following conditions: 10 min at 95 °C, followed by 40 amplification cycles (15 s at 95 °C and 1 min at 60 °C). A melting curve for each primer set was obtained by reading fluorescence every degree between 60 °C and 95 °C to ensure only a single product had been amplified. The expression of individual genes was normalized to the relative expression of trout housekeeping gene elongation factor 1α (EF-1α), and the expression levels were calculated using the 2−ΔCt method, where ΔCt is determined by subtracting the EF-1α value from the target Ct. No template negative controls and minus reverse transcriptase controls were included in all the experiments.
Table 1.
List of primers used in this study.
| Gene | Forward Primer | Reverse Primer | Accession no |
|---|---|---|---|
| EF-1α | GATCCAGAAGGAGGTCACCA | TTACGTTCGACCTTCCATCC | AF498320 |
| APRIL | CACAGACATACACAATGGAATGGAA | TGTGATGACAGAGGAACAAGATGAA | EF451543.1 |
| BAFF | ATGTTTGATGCTTATTCTGGCAGG T | TGGGACTGTGTCTTGACTGTGTGTA | DQ218467 |
| BALM | TGGAGGTACAGTAGTTCAGCAGTCG | ACTATCCAAGGAATCACCGTCACAT | NM_001124566 |
| Batf3 | CACAGAGAGCAGATGAGTTGCATAA | TTGCTCCTCAGACAGAAACTGTACC | CDQ67442.1 |
| CCR7 | TTCACTGATTACCCCACAGACAATA | AAGCAGATGAGGGAGTAAAAGGTG | JX982103 |
| CD3 | CCTGATTGGAGTAGCTGTCTAC | GCTGTACTCAGATCTGTCCATGC | XM_021582112 |
| CD8β | GTTCAAGGCCAGTAAAAGGGACAT | GCCTCCACAACTCGTTCTCTTTCT | NM_001124008.1 |
| CD11b | GAAATTCACTGGGGATAGAGAAACAG | CTACTCCAACTCCTGTCCCTATTATC | AM713180.2 |
| CD80/86 | GTGTTTCCTGGTTCTGGTATCTA | AACTTGCTGCTCCCTTTCCTC | FJ467621 |
| CD83 | GCTGTTGATAGCGGGAGGTA | TGTGGACTCAAGGCAATCTG | AY263793.1 |
| CD103 | AGGAGTGATCTTAAAACACCCCAAG | TGGCAGACACAACACTGTAACCTAA | CDQ67442.1 |
| CD141 | CAGAATTCAGCAACTGGAAAGACAA | ACTTTTTCCTGACAAGGTCGTTCTG | KP203844 |
| DC-SIGN | GAGAAGGAAGGGGATTGGAG | CCCATGTGATCCTCCTGACT | NM_001124633 |
| IFN-γ | GAAGGCTCTGTCCGAGTTCA | TGTGTGATTTGAGCCTCTGG | NM_001124620 |
| IRF8 | CCGAGGAGGAGCAGAAGAGTAAAAG | GCGGCATTGAAAGAACCCAT | AJ829674 |
| MHC-I | GACAGTCCGTCCCTCAGTGT | CTGGAAGGTTCCATCATCGT | AF115522 |
| MHC-II | ACACCCTTATCTGCCACGTC | TCTGGGGTGAAGCTCAGACT | AF296384 |
| TCRα | ACGCACTTGGAATTATTCAACAAGA | GCTTCACATTTCTCTGAACCACCTA | AAA98477.1 |
| TLR1 | CAGACGCCCTGTTGATGTT C | CCTTCACAAGTTCCACCACG | NM_001166101 |
| TLR2 | GATCCAGAGCAACACTCTCAACAT | CTCCAGACCATGAAGTTGACAAAC | XM_021578334 |
| TLR3 | AGCCCTTTGCTGCCTTACAGAG | GTCTTCAGGTCATTTTTGGACACG | NM_001124578 |
| TLR5 | TTGACTTATCTTCCAACGGATTCA | CTTTGAAATTGCTGAAACCAAATG | NM_001124208 |
| TLR7 | TACAGCTTGGTAACATGACTCTCC | CAACTCTCTGAGACTTGTCGGTAA | GQ422119 |
| TLR8a | CATCTATGTTCTCATCCAGCAACC | GGTCCCCCTAATAGACAACCTCTT | GQ422120 |
| TLR9 | TCTTCATAGAGCTGAAGAGGCCTCA | GTTCCCACTGAGGAGAAGTGTTTT | NM_001129991 |
| TLR22 | TGGACAATGACGCTCTTTTACC | GAGCTGATGGTTGCAATGAGG | NM_001124412 |
2.6. Analysis of T cell activating capacity
To determine the capacity of intestinal CD8+ DCs to activate T cells, we used a protocol previously described [10,11] with minor modifications. Briefly, CD8+ DCs were isolated by cell sorting as described above and cultured for 12 h in L-15 medium supplemented with 5% FCS. Because no antibodies are available against extracellular pan-T cell markers in rainbow trout, we used T cell-enriched cultures as responder cells. These T cell-enriched cultures were obtained from isogeneic or allogeneic splenocytes by depleting all IgD+, IgM+ and MHC II+ cells through cell sorting. The resulting negative population, representing approximately 10% of splenocytes, was then labelled with Cell Trace™ CFSE Cell Proliferation Kit (Thermo Scientific). The enrichment in T cells was verified by stimulation with Concanavalin A (ConA, 4 μg/ml; Sigma), a typical T cell mitogen as previously described [11] and by intracellular staining with an anti-CD3ε Ab [27], which showed >90% of CD3+ cells in the cultures. To carry out the T cell-stimulating test, DCs were co-cultured with isogeneic or allogenic T cell-enriched splenocytes at a ratio of 1:30 (DCs:splenocytes). After 5 days of incubation at 20 °C, co-cultured samples were stained in some experiments with 7-AAD (BD Biosciences) at 2.5 μg/ml to check cell viability, and analyzed by flow cytometry to measure cell proliferation of the enriched T cell population through the degree of dilution of CellTrace Far Red vital marker. Flow cytometry analysis was performed using Flow Jo 10 software package.
2.7. Phagocytic activity
To analyze the phagocytic capacity of intestinal CD8+ DCs, leukocytes from the intestine were seeded in 24-well plates (Nunc) at a cell density of 1 × 106 cells per well and incubated for 16 h at 20 °C with fluorescent beads (FluoSpheres® Microspheres, 1.0 μm, Crimson Red Fluorescent 625/645, 2% solids; Life Technologies) at a cell:bead ratio of 1:10 or without beads in the case of negative controls. After the incubation period, cells were harvested by gently pipetting and non-ingested beads were removed by centrifugation (100×g for 10 min at 4 °C) over a cushion of 3% (weight/volume) BSA (Fraction V; Fisher Scientific) in PBS supplemented with 4.5% (weight/volume) d-glucose (Sigma). Cells were resuspended in L-15 with 5% FCS, labelled with the flow cytometry antibodies as described above and analyzed on a FACSCalibur flow cytometer. Flow cytometry analysis was performed using Flow Jo 10 software package.
2.8. Statistical analysis
Graphpad Prism software was used to carry out the statistical analyses. The analyses were performed using a two-tailed Student's t-test with Welch's correction when the F test indicated that the variances of both groups differed significantly. The differences between the mean values were considered significant on different degrees, where * means P ≤ 0.05, ** means P ≤ 0.01 and *** means P ≤ 0.005.
3. Results
3.1. Identification of CD8+ DCs in the rainbow trout intestine
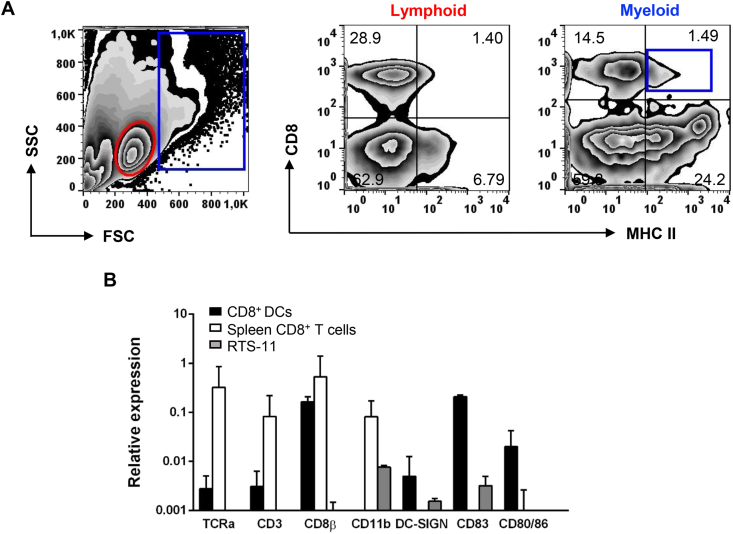
As previously described when identifying CD8+ DCs from rainbow trout skin and gills [10,11], we stained total leukocytes obtained from the midgut and hindgut segments of the intestine with anti-CD8α and anti-MHC-II to determine whether CD8+ DCs are also present in this tissue. Cells were first analyzed according to their FSC/SSC profile, and thus catalogued as cells included in the lymphoid gate (FSClowSSClow) mostly representing lymphocytes, or as cells that fall within what we have designated as the myeloid gate (FSChighSSCmed/high), representing larger and more complex cells, such as macrophages, neutrophils, and DCs (Fig. 1A). Within the lymphoid gate, a high percentage of cells expressed CD8α on the cell surface (Fig. 1A) (22.6% ± 8.9; n = 8). However these cells had no MHC II expression on the surface and therefore most probably correspond to cytotoxic CD8+ T cells, known to be present in high numbers in the teleost digestive tract [24]. On the other hand, a population of cells that co-expressed CD8α and MHC II was clearly visible within the myeloid gate (Fig. 1A). These cells represent 2.1% ± 1.1 (n = 8) of the cells included in the myeloid gate.
Fig. 1.
Identification and characterization of intestinal CD8+DCs. (A) Flow cytometry analysis of leukocytes from rainbow trout intestine labelled with anti-CD8α and anti-MHC II mAbs. A representative FSC/SSC profile is shown (left panel) and gates for lymphoid (red) and myeloid (blue) cells defined. Representative two-color CD8/MHC II dot plots of lymphoid and myeloid gated cells are also shown. The percentage of myeloid CD8+ MHC II+ (CD8+ DCs) cells among the total number of cells in the myeloid gate is shown in the upper right corner. (B) Intestinal CD8+ DCs and splenic CD8+ T cells were isolated by flow cytometry and RNA obtained as described in the Materials and Methods sections to study the transcription levels of different marker genes by real time PCR. The RTS11 monocytes-macrophage cell line was also included for comparative purposes. Results are shown as relative expression values to the endogenous control EF1α (mean ± SD; n = 3–7). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
To corroborate that these CD8+MHC II+ cells correspond in fact to a DC subset, we FACS isolated this population and analyzed the levels of transcription of different immune genes, comparing them to those obtained in RTS11 macrophages and splenic CD8+ T cells. CD8+MHC II+ cells from the intestine had very low mRNA levels of T cell markers such as TCRα or CD3, being these levels approximately 100 fold lower than those obtained in splenic CD8+ T cells (Fig. 1B). Interestingly, intestinal CD8+MHC II+ cells showed high expression levels of CD8β, strongly suggesting that this DC subset is CD8α/β (Fig. 1B) as the homologous population in gills [11]. Concerning the transcription of DC markers, these intestinal cells showed moderate levels of transcription of DC-SIGN and very high levels of CD83 and CD80/86 (Fig. 1B), confirming they correspond to a DC subset. On the other hand, no CD11b transcription was detected in intestinal CD8+ DCs (Fig. 1B).
3.2. Visualization of intestinal CD8+ DCs
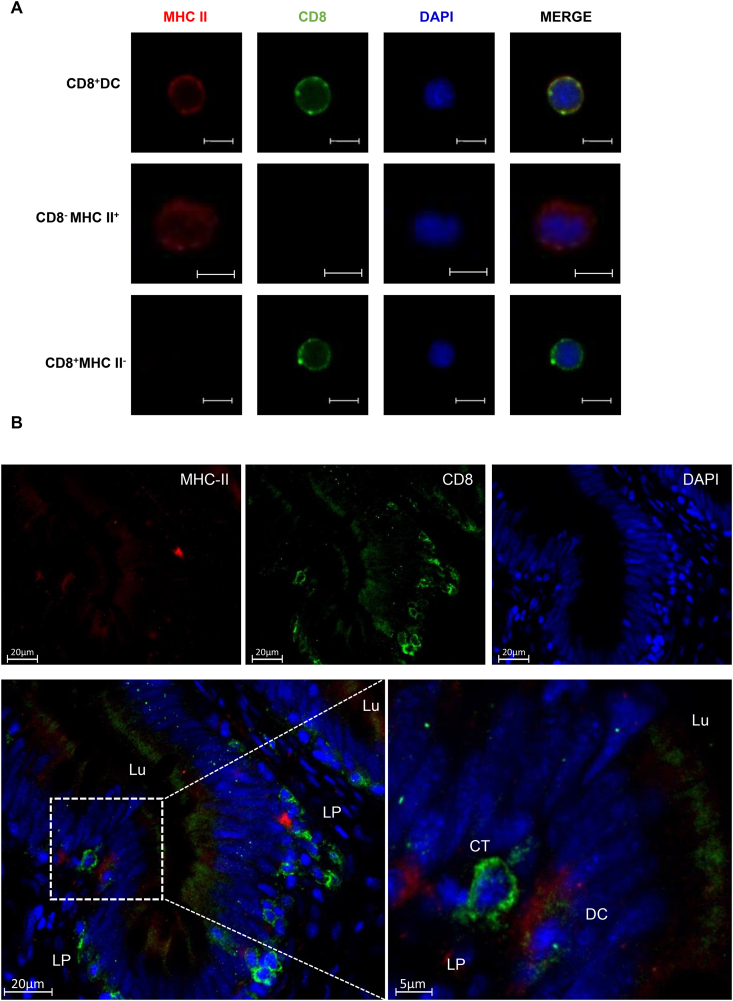
To further characterize the morphology of these cells, the intestinal leukocyte populations stained with anti-CD8α and anti-MHC-II were analyzed by confocal microscopy. By the use of this technique, we could confirm that CD8+MHC II+ cells, CD8−MHC II+ cells and CD8+MHC II− cells could all be identified among intestinal leukocytes (Fig. 2A) and that CD8+MHC II+ cells were larger than CD8+MHC II− cells (Fig. 2A). With an approximate size of 8 μm and a round morphology, these cells seem to correspond to immature DCs [28].
Fig. 2.
Visualization of intestinal CD8+DCs. (A) To confirm the presence of intestinal CD8+ DCs, total leukocytes from rainbow trout intestine were fixed and stained with anti-CD8α (green) and anti-MHC II (red) mAbs, counterstained with DAPI (blue) and analyzed by fluorescence microscopy. Scale bars, 5 μm. (B) In order to study the location of CD8+ DCs within the tissue, cryostat sections were prepared from rainbow trout intestine, fixed and labelled with anti-CD8α (green) and anti-MHC class II (red) Abs, counterstained with DAPI (blue), and analyzed by fluorescence microscopy. A representative image is shown (upper panels, scale bars, 20 μm), together with magnifications of an area containing a CD8+ DC (lower panels, scale bars 20 μm and 5 μm). LP = lamina propria; Lu = lumen; CT = Cytotoxic CD8+ T cell; DC = CD8+ MHC II+ dendritic cell. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
To visualize intestinal CD8+DCs within the intestinal mucosa, we also performed an immunofluorescence analysis through confocal microscopy in midgut and hindgut sections. Through this analysis we identified CD8+MHC II+ cells within the intestinal LP (Fig. 2B). Interestingly, within the tissue, these cells showed a characteristic DC morphology, including an irregular shape and multiple cytoplasmic extensions (Fig. 2B). Furthermore, as can be seen in the figure, these CD8+ DCs seem to be in close contact with CD8+ T cells (MHC II− cells) (Fig. 2B).
3.3. Transcription of markers of cross-presenting DCs in intestinal CD8+ DCs
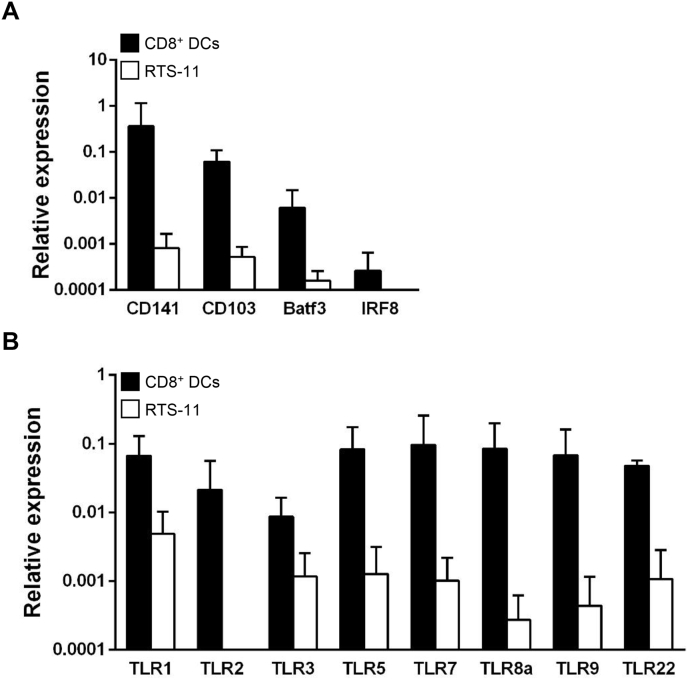
The skin and gills CD8+ DCs previously characterized in rainbow trout were shown to transcribe a wide range of specific markers that are exclusively expressed in mammalian DC populations with cross-presenting capacities [10,11]. Thus, we wanted to confirm whether intestinal CD8+ DCs showed a similar transcription pattern. We observed that rainbow trout intestinal CD8+ DCs transcribe CD141, CD103, Baft3 and IRF8 (Fig. 3A), all of them signature genes of different subsets of mammalian cross-presenting DCs [18,29,30]. Since TLR3 is also one of the immune genes that within mammalian DC populations are exclusively expressed in those with cross-presenting capacities [[16], [17], [18]], we next studied the levels of expression of all known rainbow trout TLRs in CD8+ DCs from the intestine, comparing them to the levels observed in rainbow trout RTS11 macrophages. We found that rainbow trout intestinal CD8+ DCs transcribed all TLR genes studied at very high levels, including TLR3 (Fig. 3B).
Fig. 3.
Transcription of mammalian cross-presenting markers and toll-like receptor (TLR) genes by rainbow trout intestinal CD8+ DCs. Intestinal CD8+ DCs were isolated by flow cytometry, total RNA extracted and used to study the levels of transcription of cross-presenting DCs markers (A) and TLRs (B) by real time PCR. RNA was also obtained from RTS-11 monocytes-macrophages for comparative reasons. Results are shown as relative expression levels to the endogenous control EF1α (mean + SD; n = 3–9).
3.4. Responsiveness of intestinal CD8+ DCs to a TLR3 agonist
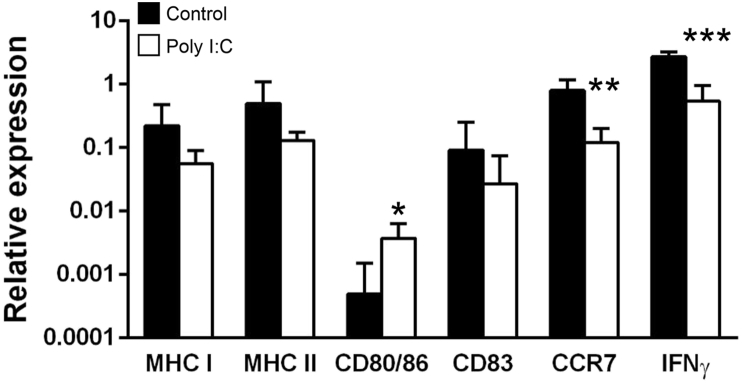
To further investigate the responsiveness of intestinal CD8+ DCs to TLR3 ligands, we incubated intestinal leukocytes with poly I:C (a known TLR3 agonist) and after 16 h of incubation, we FACS isolated the CD8+ DCs and analyzed the levels of transcription of a range of immune genes, comparing them to the levels obtained in CD8+ DCs from cultures not treated with poly I:C. Surprisingly, we found that poly I:C stimulation decreased the levels of transcription of CCR7 and IFNγ (Fig. 4). Conversely, poly I:C stimulation provoked a significant up-regulation of CD80/86 transcription levels (Fig. 4).
Fig. 4.
Transcriptional response of intestinal CD8+ DCs to poly I:C. Intestinal leukocytes in L-15 medium supplemented with 5% FCS were incubated with 50 μg/ml poly I:C for 16 h at 20 °C. Control cells incubated with media alone were also included. After this time, CD8+ DCs were FACS isolated from poly I:C-treated and control cultures. Total RNA was extracted from the FACS isolated cells and used to determine the levels of transcription of different immune genes by real time PCR. Data are shown as the gene expression relative to the expression of endogenous control EF-1α (mean + SD; n = 4). Asterisks denote statistical differences between values obtained in poly I:C-treated cells and cells from control cultures. Statistical analyses were performed, where * p ≤ 0.05 and **p ≤ 0.01.
3.5. T cell stimulating capacities of intestinal CD8+ DCs
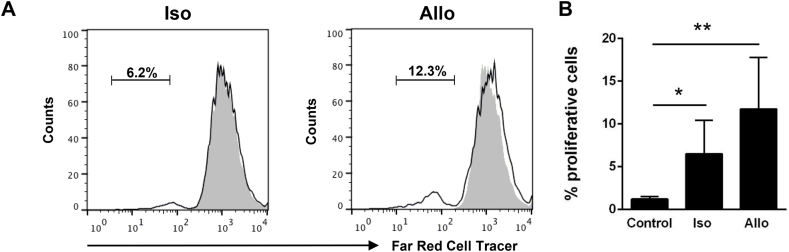
One of the main defining features of DCs is their capacity to stimulate T cells [31]. DCs can activate T cells in an antigen-dependent fashion but are also one of the few cell types that can directly activate naïve T cells [32]. Thus, to confirm this, we incubated isolated intestinal CD8+ DCs with either isogeneic (from the same fish) or allogeneic (from a different fish) T cell-enriched cultures. We observed that in both cases, the addition of intestinal CD8+ DCs to these cultures significantly activated the proliferation of T cells (Fig. 5), although the effect was stronger in allogeneic cultures.
Fig. 5.
Analysis of T cell activation by intestinal CD8+DCs. Intestinal CD8+ DCs were isolated by flow cytometry. Cells were then cultured for 12 h in L-15 medium supplemented with 5% FCS. Afterwards, they were co-cultured with isogenic (iso) or allogenic (allo) T cell-enriched splenocytes, previously labelled with CellTrace Far Red, as described in the Materials and Methods sections. Co-cultures of DCs and T cells were incubated for 4 days and then analyzed by flow cytometry. (A) A representative example from four individuals is shown. (B) Mean percentage (+SD) of proliferating cells (n = 4). Statistical analyses were performed and asterisks indicate levels of proliferation significantly higher than those observed in T cell-enriched splenocyte cultures not incubated with DCs (control). *p ≤ 0.05 and **p ≤ 0.01.
3.6. Phagocytic capacity of CD8+ DCs in the rainbow trout intestine
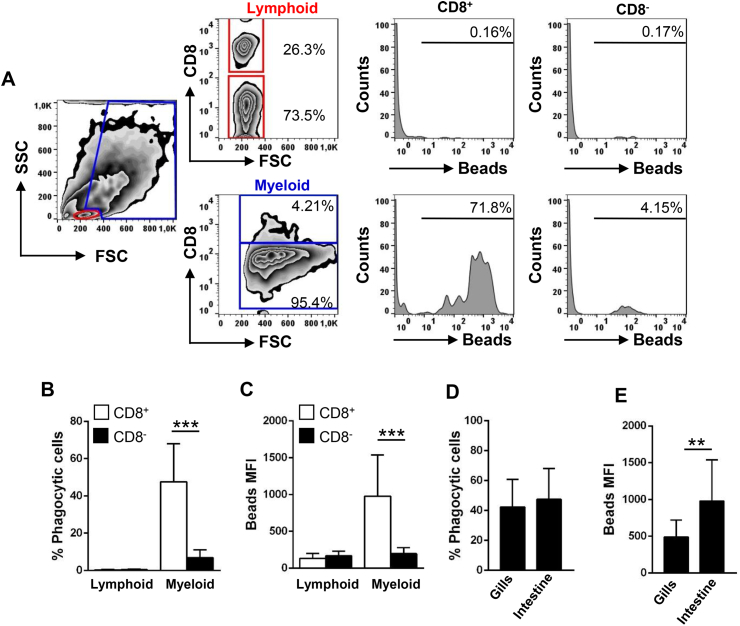
Although the main function of DCs is to present antigen and not to clear pathogens from the circulation, they exert a high phagocytic capacity, equivalent to that of other professional phagocytes, to uptake antigens [33]. In rainbow trout intestine, we found that none of the cells from the lymphoid gate showed significant phagocytic capacities (Fig. 6A–C). Within the myeloid gate, CD8+ DCs were the main cells with phagocytic activity, since 47.54% ± 20.48 of these cells exhibited a high phagocytic capacity, while only 6.86% ± 4.23 of the CD8− cells within this gate had the capacity to phagocytose beads (Fig. 6A–C). Furthermore, the mean fluorescence intensity (MFI) of internalized beads within CD8+ DCs was significantly higher than that observed in CD8− cells within the myeloid gate (Fig. 6C).
Fig. 6.
Phagocytic capacity of intestinal CD8+DCs. Intestinal leukocytes were incubated with Crimson Red fluorescent polystyrene beads (1 μm diameter) at a ratio 1:10 (cell/beads) for 16 h and then centrifuged through a 3% BSA 4.5% glucose gradient in order to remove non-ingested beads. Cells were then labelled with anti-CD8α and analyzed by flow cytometry. (A) Representative plots showing gated lymphoid cells (upper panels) and myeloid cell (lower panels) are shown. CD8− and CD8+ cells were further selected to analyze the fluorescence of internalized beads (shown in the histograms). The percentage of cells containing beads were determined using control samples without beads to set the positive threshold. Mean percentages of phagocytic cells (B) and median fluorescence intensity of the beads (C) in each subpopulation are shown as mean + SD (n = 12). The phagocytic capacity of intestinal CD8+ DCs was compared to that of gills CD8+ DCs, studying both the percentage of phagocytic cells (D) and MFI of beads (E). (n = 12). Statistical analyses were performed and asterisks denote significant differences between groups as indicated. **p ≤ 0.01, ***p ≤ 0.005. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
When the phagocytic capacity of intestinal CD8+ DCs was compared to that of the equivalent population in rainbow trout gills from the same animals, we found that while there were no differences between the percentages of cells with phagocytic potential between the two populations (Fig. 6D), the number of microparticles that these cells could ingest (measured as the MFI value) was significantly higher in the case of intestinal CD8+ DCs than that of gills CD8+ DCs (Fig. 6E).
3.7. CCR7 surface levels in intestinal CD8+ DCs
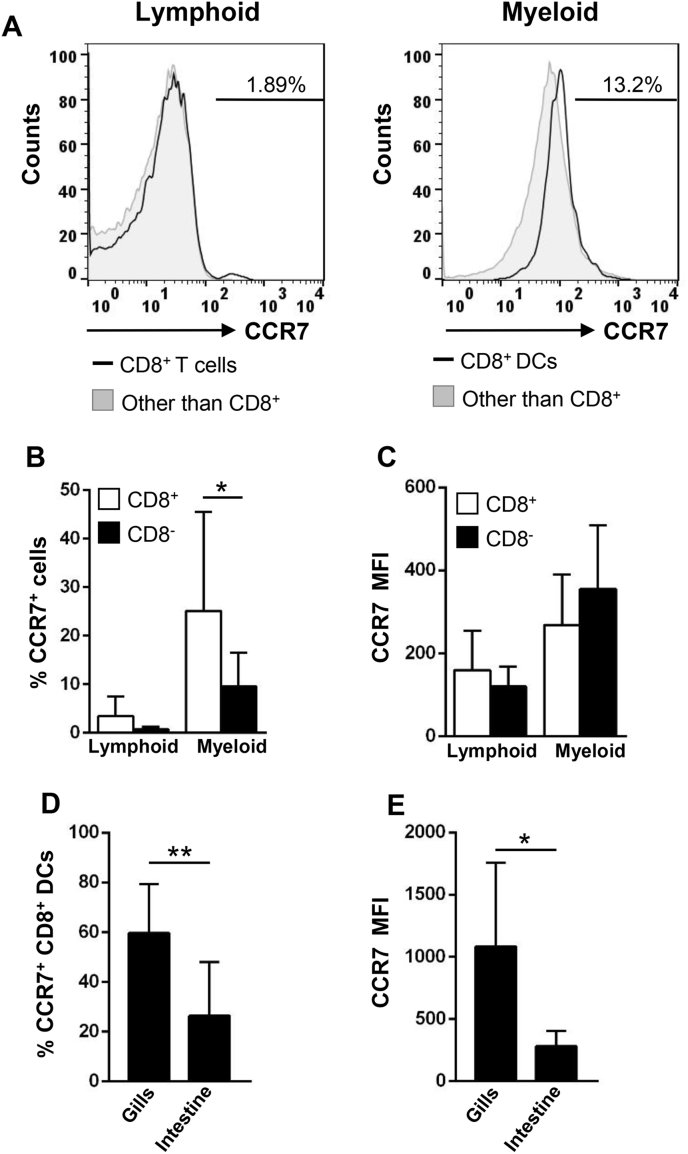
In mammals, CCR7 expression is up-regulated in activated DCs that consequently migrate to the lymph node following CCL19 and CCL21 ligands to encounter cognate T cells [34]. Thus, we also studied whether CD8+ DCs in the rainbow trout intestine expressed CCR7 on the cell surface using a specific anti-CCR7 polyclonal antibody previously characterized in our laboratory [25]. We found that although only a minor percentage of lymphoid cells (mainly CD8+ cells) expressed CCR7 on the cell membrane (Fig. 7A–C), 25.15% ± 20.38 of the CD8+ cells in the myeloid gate (CD8+ DCs) expressed CCR7 (Fig. 7A and B). Interestingly, although the percentage of myeloid CD8− cells with CCR7 on the membrane was significantly lower (Fig. 7A and B), the amount of CCR7 they exhibited on the cell membrane was similar among CCR7+ cells from the two myeloid subsets (Fig. 7C).
Fig. 7.
CCR7 expression levels in rainbow trout intestinal CD8+DCs. Trout leukocytes obtained from rainbow trout intestine were isolated, stained with Abs to CD8α, MHC II and CCR7 and analyzed by multicolour flow cytometry. (A) Cells were first gated as lymphoid and myeloid cell on the basis of their FSC and SSC profile. Then CD8+ cells were gated on those populations and CCR7 fluorescence determined in CD8+ MHC II+ cells (open line histogram) and compared against the CCR7 levels of all other cells in that gate (filled line histogram). Numbers on histograms correspond to the percentage of CCR7+ cells after exclusion of negative cell of the conjugate-only controls (data not shown). Average percentage of CCR7+ cells (B) and MFI of CCR7 expression (C) in the intestinal lymphoid and myeloid populations were calculated and shown as mean + SD (n = 7). The percentage of CD8+ DCs expressing CCR7 expression (D) and MFI of CCR7 (E) were measured in gills and intestine and shown as mean + SD (n = 7). Statistical analyses were performed and asterisks denote significant differences between groups as indicated. *p ≤ 0.05 and **p ≤ 0.01.
When CCR7 expression levels were compared between CD8+ DCs from gills and intestine, we found that a higher percentage of gill CD8+ DCs had CCR7 on the cell membrane (Fig. 7D) and the levels of expression per cell were also higher in gill CD8+ DCs than in intestinal CD8+ DCs (Fig. 7E).
3.8. Transcription of B-regulating cytokines by intestinal CD8+ DCs
DCs not only present antigens to T cells, but also regulate the function of neighboring cells through the secretion of cytokines. Among these cytokines, the production of cytokines from the tumor necrosis factor (TNF) superfamily, such as B cell activating factor (BAFF) and a proliferation-inducing ligand (APRIL), especially regulates the functionality of B cells [35]. In teleost fish, a third cytokine with close similarity to both BAFF and APRIL, designated as BALM (BAFF and APRIL-like molecule) is also known to regulate some functions of B cells [36]. Hence, we investigated whether intestinal CD8+ DCs transcribed these three cytokines, comparing the transcription levels to those of gills CD8+ DCs. We found that both gills and intestinal CD8+ DCs transcribed very high levels of BAFF and APRIL but much lower levels of BALM (Fig. 8).
Fig. 8.
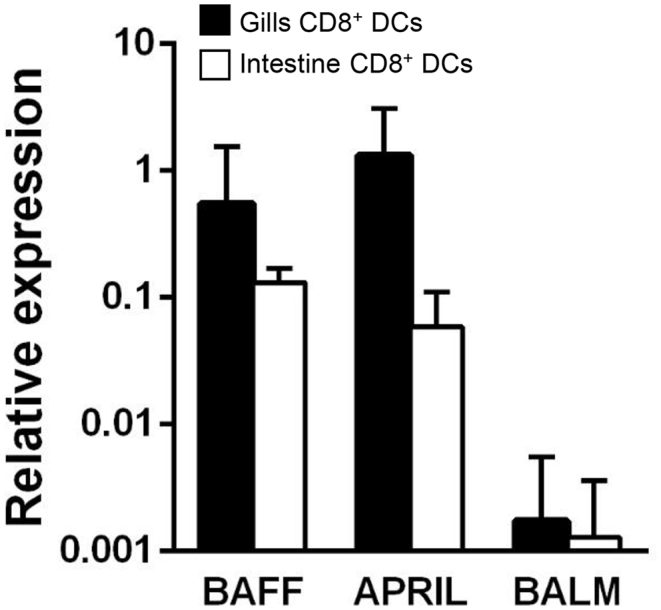
Transcription of B cell-stimulating TNF family ligands by CD8+DCs. Intestinal and gills CD8+DCs were isolated by flow cytometry and RNA obtained to determine the constitutive levels of transcription of TNF family ligands known to activate B cells by real time PCR. Relative expression levels of indicated genes to endogenous control EF1α were calculated (mean + SD, n = 4–9).
4. Discussion
Intestinal DCs are known to regulate tolerance mechanisms to harmless antigens that go through the intestinal tract, being also involved in mounting immune responses to pathogens [23]. Therefore, these cells are key factors in maintaining the balance between tolerance and immunity in this mucosal surface. In this context, after having identified a CD8+ DC subset in rainbow trout skin and gills, we thought of great relevance to establish if an equivalent DC subset was present in the intestinal mucosa. In the current work, we have verified that such subset exists. These cells that express both CD8α and MHC II on the cell surface, show insignificant transcription levels of T cell specific genes, while expressing genes characteristic of DCs such as DC-SIGN, CD83 and CD80/86. This transcriptional profile, together with a verification of their phagocytic and T stimulating capacities, unequivocally point to this population being a DC subset.
Many different subsets of DCs are found in mammals. These subsets are characterized by their location and the presence of specific surface markers. Interestingly, the profile of these markers is not the same in homologous populations from different species [18,37]. Among these subpopulations, some specific DC subsets have the capacity to cross-present antigens, that is, to uptake external antigens and present them in an MHC I context through a mechanism still not fully understood [12,38]. Murine cross-presenting DCs include lymphoid CD8+ DCs [13] and mucosal CD103+ DCs [14], whereas in humans, cross-presenting DCs are characterized by the expression of CD141 [15]. Despite these differences, all DCs with cross-presenting capacities from both mice and human share common exclusive features, such as TLR3 [[16], [17], [18]], Flt3 ligand, IFR8 [19,20] and Batf3 [21,22] expression. Interestingly, a similar molecular signature was found in cross-presenting DCs in sheep, which express CD26 as a specific surface marker [39]. Thus, the fact that all these diverse cross-presenting subsets shared these and other specific traits strongly pointed to the existence of a common ancestor. In this sense, the identification of subpopulation of DCs in teleost skin, gills and now in intestine co-expressing CD8, CD103, CD141, TLR3, IRF8 and Batf3 pointed to these cells as those potential common ancestors for mammalian cross-presenting DCs.
Concerning the expression of CD11b, intestinal CD8+ DCs similarly to gills CD8+DCs [11] and on the contrary to what happens in skin CD8+ DCs [10] do not transcribe CD11b. In mammals, CD103+ DCs, an important subset of intestinal DCs with migratory capacities can be divided into two populations according to whether they co-express CD11b or not on the cell membrane [23]. Interestingly, it is in fact the CD103+CD11b− population the one that is dependent on Baft3 and IRF8, while the CD103+CD11b+ subset develops independently of these transcription factors [19]. The dependence of these CD103+CD11b− DCs on transcription factors characteristic of cross-presenting DCs, induced researchers to hypothesize that this subset could be related to CD8+ DCs from lymphoid tissues. Supporting this idea, CD103+CD11b− DCs in the intestinal LP were shown to express CD8α [40]. Furthermore, in mammals, the distribution of these two subpopulations is not homogeneous along the digestive tract, as CD103+CD11b+ DCs comprise ∼70% of total CD103+ cDCs in the duodenum and CD103+CD11b− DCs constitute ∼75% of total CD103+ cDCs in the colon [41]. Given that immune cells have been identified all along the digestive tract of teleosts, whether CD8+ DCs present in other segments of the rainbow trout digestive tract express CD11b is an interesting issue to investigate in the future.
It has to be noted that as a consequence of their capacity to present external antigens in an MHC I context, cross-presenting DCs have a superior capacity to activate cytotoxic CD8+ T cells, and therefore are important elements in the activation of adaptive responses to intracellular pathogens or tumor cells [12,38]. Remarkably, when we studied the localization of CD8+ DCs in the intestinal mucosa, we visualized them in the LP, often in close contact with cytotoxic CD8+ T cells. This suggests that in fish, as in mammals, this DC subset is responsible for the activation of cytotoxic CD8+ T cells, being therefore, a key target to direct viral antigens to during the optimization of oral antiviral vaccines. In teleost fish, because organized secondary lymphoid structures, such as Peyer's patches or mesenteric lymph nodes are missing [42], the intestinal LP has been proposed as both an inductive and effector site [43], in which antigen sampling and presentation as well as effector functions could simultaneously take place. Although this hypothesis remains to be demonstrated, the fact that teleost intestine is a T cell rich tissue [24,27], and the identification of DCs that in some cases seemed in close contact with cytotoxic T cells supports this hypothesis. Indeed the low levels of surface CCR7 expression detected on intestinal CD8+ DCs when compared to the equivalent population in rainbow trout gills strongly suggests that in the intestine DCs can activate T cells locally without a need to migrate to external T cell-rich areas.
It is well known that local environmental conditions and maturation states will shape the functionality of the different DC lineages in different tissues. Therefore, we compared additional characteristics of intestinal CD8+ DCs to that of gills CD8+ DCs. We verified that although the percentages of DCs actively phagocyting microparticles were similar in both tissues, intestinal CD8+ DCs seemed to have a superior capacity to ingest microparticles (they could ingest a significantly higher number of particles). In mammals, a superior capacity to phagocytose is associated with an immature stage, given that as DCs mature they start to lose their capacity to uptake antigens [44]. Interestingly, mammalian steady state immature DCs exhibit a continuous endocytic activity through which they continuously present innocuous antigens to T cells, thus facilitating tolerance [45]. Therefore, it seems probable that a higher phagocytic capacity of intestinal DCs is associated with a more tolerogenic profile. Along this line, it was surprising to observe that intestinal CD8+ DCs responded to stimulation with a TLR3 agonist by down-regulating some immune genes, mainly CCR7 and IFNγ, while at the same time, the up-regulation of CD80/86 suggested that these cells were maturing in response to the stimulation [46]. This suggests that the activity of this population is tightly regulated in the intestine, given that poly I:C stimulation of rainbow trout CD8+ DCs from skin significantly increased the transcription of a wide range of immune genes [10]. On the other hand, the capacity of intestinal DCs to regulate B cells through the secretion of cytokines from the TNF family of ligands, is equivalent to that of gill DCs, pointing to the interaction between T cells and DCs as a critical factor in the regulation of oral tolerance.
In conclusion, we have identified for the first time DCs within the intestinal mucosa of teleost fish. This DC subset expresses a wide range of genes specific of mammalian DC subsets with cross-presenting capacities. The fact that rainbow trout CD8+ DCs in the intestine were found in some cases in close contact with CD8+ T cells suggests that as mammalian cross-presenting DC subsets, the DC population identified is responsible for the regulation of cytotoxic T cell activities. On the other hand, the functional and phenotypic characterization of this population points to a more immature state than that of the homologous populations in gills or skin. Thus, understanding how DCs interact with lymphoid cells in the LP seems to be quite relevant for the future design of vaccines/adjuvants that are able to circumvent oral tolerance.
Acknowledgements
The authors want to thank Lucía González and Laura Fernández for technical assistance. Dr. Uwe Fischer is greatly acknowledged for providing the anti-CD8α antibody used in this study. This work was supported by the European Research Council (ERC Consolidator Grant 2016 725061 TEMUBLYM) and by projects AGL2014-53061-R (Spanish Ministry of Economy and Competitiveness, MINECO) and AGL2017-85494-C2-1-R (Spanish Ministry of Science, Innovation and Universities).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.fsi.2019.04.001.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Embregts C.W., Forlenza M. Oral vaccination of fish: lessons from humans and veterinary species. Dev. Comp. Immunol. 2016;64:118–137. doi: 10.1016/j.dci.2016.03.024. [DOI] [PubMed] [Google Scholar]
- 2.Mowat A.M. Anatomical basis of tolerance and immunity to intestinal antigens. Nat. Rev. Immunol. 2003;3(4):331–341. doi: 10.1038/nri1057. [DOI] [PubMed] [Google Scholar]
- 3.Mellman I., Steinman R.M. Dendritic cells: specialized and regulated antigen processing machines. Cell. 2001;106(3):255–258. doi: 10.1016/s0092-8674(01)00449-4. [DOI] [PubMed] [Google Scholar]
- 4.Owen J.L., Sahay B., Mohamadzadeh M. New generation of oral mucosal vaccines targeting dendritic cells. Curr. Opin. Chem. Biol. 2013;17(6):918–924. doi: 10.1016/j.cbpa.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lovy J., Savidant G.P., Speare D.J., Wright G.M. Langerin/CD207 positive dendritic-like cells in the haemopoietic tissues of salmonids. Fish Shellfish Immunol. 2009;27(2):365–368. doi: 10.1016/j.fsi.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 6.Haugland G.T., Jordal A.E., Wergeland H.I. Characterization of small, mononuclear blood cells from salmon having high phagocytic capacity and ability to differentiate into dendritic like cells. PLoS One. 2012;7(11) doi: 10.1371/journal.pone.0049260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johansson P., Corripio-Miyar Y., Wang T., Collet B., Secombes C.J., Zou J. Characterisation and expression analysis of the rainbow trout (Oncorhynchus mykiss) homologue of the human dendritic cell marker CD208/lysosomal associated membrane protein 3. Dev. Comp. Immunol. 2012;37(3–4):402–413. doi: 10.1016/j.dci.2012.02.012. [DOI] [PubMed] [Google Scholar]
- 8.Lugo-Villarino G., Balla K.M., Stachura D.L., Banuelos K., Werneck M.B., Traver D. Identification of dendritic antigen-presenting cells in the zebrafish. Proc. Natl. Acad. Sci. U.S.A. 2010;107(36):15850–15855. doi: 10.1073/pnas.1000494107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bassity E., Clark T.G. Functional identification of dendritic cells in the teleost model, rainbow trout (Oncorhynchus mykiss) PLoS One. 2012;7(3) doi: 10.1371/journal.pone.0033196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Granja A.G., Leal E., Pignatelli J., Castro R., Abos B., Kato G., Fischer U., Tafalla C. Identification of teleost skin CD8alpha+ dendritic-like cells, representing a potential common ancestor for mammalian cross-presenting dendritic cells. J. Immunol. 2015;195(4):1825–1837. doi: 10.4049/jimmunol.1500322. [DOI] [PubMed] [Google Scholar]
- 11.Soleto I., Fischer U., Tafalla C., Granja A.G. Identification of a potential common ancestor for mammalian cross-presenting dendritic cells in teleost respiratory surfaces. Front. Immunol. 2018;9:59. doi: 10.3389/fimmu.2018.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joffre O.P., Segura E., Savina A., Amigorena S. Cross-presentation by dendritic cells. Nat. Rev. Immunol. 2012;12(8):557–569. doi: 10.1038/nri3254. [DOI] [PubMed] [Google Scholar]
- 13.den Haan J.M., Lehar S.M., Bevan M.J. CD8(+) but not CD8(-) dendritic cells cross-prime cytotoxic T cells in vivo. J. Exp. Med. 2000;192(12):1685–1696. doi: 10.1084/jem.192.12.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.del Rio M.L., Bernhardt G., Rodriguez-Barbosa J.I., Forster R. Development and functional specialization of CD103+ dendritic cells. Immunol. Rev. 2010;234(1):268–281. doi: 10.1111/j.0105-2896.2009.00874.x. [DOI] [PubMed] [Google Scholar]
- 15.Jongbloed S.L., Kassianos A.J., McDonald K.J., Clark G.J., Ju X., Angel C.E., Chen C.J., Dunbar P.R., Wadley R.B., Jeet V., Vulink A.J., Hart D.N., Radford K.J. Human CD141+ (BDCA-3)+ dendritic cells (DCs) represent a unique myeloid DC subset that cross-presents necrotic cell antigens. J. Exp. Med. 2010;207(6):1247–1260. doi: 10.1084/jem.20092140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poulin L.F., Salio M., Griessinger E., Anjos-Afonso F., Craciun L., Chen J.L., Keller A.M., Joffre O., Zelenay S., Nye E., Le Moine A., Faure F., Donckier V., Sancho D., Cerundolo V., Bonnet D., Reis e Sousa C. Characterization of human DNGR-1+ BDCA3+ leukocytes as putative equivalents of mouse CD8alpha+ dendritic cells. J. Exp. Med. 2010;207(6):1261–1271. doi: 10.1084/jem.20092618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jelinek I., Leonard J.N., Price G.E., Brown K.N., Meyer-Manlapat A., Goldsmith P.K., Wang Y., Venzon D., Epstein S.L., Segal D.M. TLR3-specific double-stranded RNA oligonucleotide adjuvants induce dendritic cell cross-presentation, CTL responses, and antiviral protection. J. Immunol. 2011;186(4):2422–2429. doi: 10.4049/jimmunol.1002845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haniffa M., Shin A., Bigley V., McGovern N., Teo P., See P., Wasan P.S., Wang X.N., Malinarich F., Malleret B., Larbi A., Tan P., Zhao H., Poidinger M., Pagan S., Cookson S., Dickinson R., Dimmick I., Jarrett R.F., Renia L., Tam J., Song C., Connolly J., Chan J.K., Gehring A., Bertoletti A., Collin M., Ginhoux F. Human tissues contain CD141hi cross-presenting dendritic cells with functional homology to mouse CD103+ nonlymphoid dendritic cells. Immunity. 2012;37(1):60–73. doi: 10.1016/j.immuni.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ginhoux F., Liu K., Helft J., Bogunovic M., Greter M., Hashimoto D., Price J., Yin N., Bromberg J., Lira S.A., Stanley E.R., Nussenzweig M., Merad M. The origin and development of nonlymphoid tissue CD103+ DCs. J. Exp. Med. 2009;206(13):3115–3130. doi: 10.1084/jem.20091756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu K., Victora G.D., Schwickert T.A., Guermonprez P., Meredith M.M., Yao K., Chu F.F., Randolph G.J., Rudensky A.Y., Nussenzweig M. In vivo analysis of dendritic cell development and homeostasis. Science. 2009;324(5925):392–397. doi: 10.1126/science.1170540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hildner K., Edelson B.T., Purtha W.E., Diamond M., Matsushita H., Kohyama M., Calderon B., Schraml B.U., Unanue E.R., Diamond M.S., Schreiber R.D., Murphy T.L., Murphy K.M. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. 2008;322(5904):1097–1100. doi: 10.1126/science.1164206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edelson B.T., Kc W., Juang R., Kohyama M., Benoit L.A., Klekotka P.A., Moon C., Albring J.C., Ise W., Michael D.G., Bhattacharya D., Stappenbeck T.S., Holtzman M.J., Sung S.S., Murphy T.L., Hildner K., Murphy K.M. Peripheral CD103+ dendritic cells form a unified subset developmentally related to CD8alpha+ conventional dendritic cells. J. Exp. Med. 2010;207(4):823–836. doi: 10.1084/jem.20091627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Persson E.K., Scott C.L., Mowat A.M., Agace W.W. Dendritic cell subsets in the intestinal lamina propria: ontogeny and function. Eur. J. Immunol. 2013;43(12):3098–3107. doi: 10.1002/eji.201343740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takizawa F., Dijkstra J.M., Kotterba P., Korytar T., Kock H., Kollner B., Jaureguiberry B., Nakanishi T., Fischer U. The expression of CD8alpha discriminates distinct T cell subsets in teleost fish. Dev. Comp. Immunol. 2011;35(7):752–763. doi: 10.1016/j.dci.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 25.Castro R., Bromage E., Abos B., Pignatelli J., Gonzalez Granja A., Luque A., Tafalla C. CCR7 is mainly expressed in teleost gills, where it defines an IgD+IgM- B lymphocyte subset. J. Immunol. 2014;192(3):1257–1266. doi: 10.4049/jimmunol.1302471. [DOI] [PubMed] [Google Scholar]
- 26.Ganassin R.C., Bols N.C. Development of a monocyte/macrophage-like cell line, RTS11, from rainbow trout spleen. Fish Shellfish Immunol. 1998;8:457–476. [Google Scholar]
- 27.Boardman T., Warner C., Ramirez-Gomez F., Matrisciano J., Bromage E. Characterization of an anti-rainbow trout (Oncorhynchus mykiss) CD3epsilon monoclonal antibody. Vet. Immunol. Immunopathol. 2012;145(1–2):511–515. doi: 10.1016/j.vetimm.2011.11.017. [DOI] [PubMed] [Google Scholar]
- 28.Dumortier H., van Mierlo G.J., Egan D., van Ewijk W., Toes R.E., Offringa R., Melief C.J. Antigen presentation by an immature myeloid dendritic cell line does not cause CTL deletion in vivo, but generates CD8+ central memory-like T cells that can be rescued for full effector function. J. Immunol. 2005;175(2):855–863. doi: 10.4049/jimmunol.175.2.855. [DOI] [PubMed] [Google Scholar]
- 29.Huysamen C., Willment J.A., Dennehy K.M., Brown G.D. CLEC9A is a novel activation C-type lectin-like receptor expressed on BDCA3+ dendritic cells and a subset of monocytes. J. Biol. Chem. 2008;283(24):16693–16701. doi: 10.1074/jbc.M709923200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sancho D., Joffre O.P., Keller A.M., Rogers N.C., Martinez D., Hernanz-Falcon P., Rosewell I., Reis e Sousa C. Identification of a dendritic cell receptor that couples sensing of necrosis to immunity. Nature. 2009;458(7240):899–903. doi: 10.1038/nature07750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steinman R.M., Gutchinov B., Witmer M.D., Nussenzweig M.C. Dendritic cells are the principal stimulators of the primary mixed leukocyte reaction in mice. J. Exp. Med. 1983;157(2):613–627. doi: 10.1084/jem.157.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sallusto F., Lanzavecchia A. The instructive role of dendritic cells on T-cell responses. Arthritis Res. 2002;4(3):S127–S132. doi: 10.1186/ar567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagl M., Kacani L., Mullauer B., Lemberger E.M., Stoiber H., Sprinzl G.M., Schennach H., Dierich M.P. Phagocytosis and killing of bacteria by professional phagocytes and dendritic cells. Clin. Diagn. Lab. Immunol. 2002;9(6):1165–1168. doi: 10.1128/CDLI.9.6.1165-1168.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Forster R., Schubel A., Breitfeld D., Kremmer E., Renner-Muller I., Wolf E., Lipp M. CCR7 coordinates the primary immune response by establishing functional microenvironments in secondary lymphoid organs. Cell. 1999;99(1):23–33. doi: 10.1016/s0092-8674(00)80059-8. [DOI] [PubMed] [Google Scholar]
- 35.Schneider P. The role of APRIL and BAFF in lymphocyte activation. Curr. Opin. Immunol. 2005;17(3):282–289. doi: 10.1016/j.coi.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 36.Soleto I., Abos B., Castro R., Gonzalez L., Tafalla C., Granja A.G. The BAFF/APRIL axis plays an important role in virus-induced peritoneal responses in rainbow trout. Fish Shellfish Immunol. 2017;64:210–217. doi: 10.1016/j.fsi.2017.03.023. [DOI] [PubMed] [Google Scholar]
- 37.Steinman R.M., Idoyaga J. Features of the dendritic cell lineage. Immunol. Rev. 2010;234(1):5–17. doi: 10.1111/j.0105-2896.2009.00888.x. [DOI] [PubMed] [Google Scholar]
- 38.Alloatti A., Kotsias F., Magalhaes J.G., Amigorena S. Dendritic cell maturation and cross-presentation: timing matters! Immunol. Rev. 2016;272(1):97–108. doi: 10.1111/imr.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Contreras V., Urien C., Guiton R., Alexandre Y., Vu Manh T.P., Andrieu T., Crozat K., Jouneau L., Bertho N., Epardaud M., Hope J., Savina A., Amigorena S., Bonneau M., Dalod M., Schwartz-Cornil I. Existence of CD8alpha-like dendritic cells with a conserved functional specialization and a common molecular signature in distant mammalian species. J. Immunol. 2010;185(6):3313–3325. doi: 10.4049/jimmunol.1000824. [DOI] [PubMed] [Google Scholar]
- 40.Fujimoto K., Karuppuchamy T., Takemura N., Shimohigoshi M., Machida T., Haseda Y., Aoshi T., Ishii K.J., Akira S., Uematsu S. A new subset of CD103+CD8alpha+ dendritic cells in the small intestine expresses TLR3, TLR7, and TLR9 and induces Th1 response and CTL activity. J. Immunol. 2011;186(11):6287–6295. doi: 10.4049/jimmunol.1004036. [DOI] [PubMed] [Google Scholar]
- 41.Denning T.L., Norris B.A., Medina-Contreras O., Manicassamy S., Geem D., Madan R., Karp C.L., Pulendran B. Functional specializations of intestinal dendritic cell and macrophage subsets that control Th17 and regulatory T cell responses are dependent on the T cell/APC ratio, source of mouse strain, and regional localization. J. Immunol. 2011;187(2):733–747. doi: 10.4049/jimmunol.1002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zapata A., Diez B., Cejalvo T., Gutierrez-de Frias C., Cortes A. Ontogeny of the immune system of fish. Fish Shellfish Immunol. 2006;20(2):126–136. doi: 10.1016/j.fsi.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 43.Parra D., Korytar T., Takizawa F., Sunyer J.O. B cells and their role in the teleost gut. Dev. Comp. Immunol. 2016;64:150–166. doi: 10.1016/j.dci.2016.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Platt C.D., Ma J.K., Chalouni C., Ebersold M., Bou-Reslan H., Carano R.A., Mellman I., Delamarre L. Mature dendritic cells use endocytic receptors to capture and present antigens. Proc. Natl. Acad. Sci. U.S.A. 2010;107(9):4287–4292. doi: 10.1073/pnas.0910609107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dudek A.M., Martin S., Garg A.D., Agostinis P. Immature, semi-mature, and fully mature dendritic cells: toward a DC-cancer cells interface that augments anticancer immunity. Front. Immunol. 2013;4:438. doi: 10.3389/fimmu.2013.00438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steinman R.M., Pack M., Inaba K. Dendritic cell development and maturation. Adv. Exp. Med. Biol. 1997;417:1–6. doi: 10.1007/978-1-4757-9966-8_1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.