ABSTRACT
Innate immune defense against intracellular pathogens, like Salmonella, relies heavily on the autophagy machinery of the host. This response is studied intensively in epithelial cells, the target of Salmonella during gastrointestinal infections. However, little is known of the role that autophagy plays in macrophages, the predominant carriers of this pathogen during systemic disease. Here we utilize a zebrafish embryo model to study the interaction of S. enterica serovar Typhimurium with the macroautophagy/autophagy machinery of macrophages in vivo. We show that phagocytosis of live but not heat-killed Salmonella triggers recruitment of the autophagy marker GFP-Lc3 in a variety of patterns labeling tight or spacious bacteria-containing compartments, also revealed by electron microscopy. Neutrophils display similar GFP-Lc3 associations, but genetic modulation of the neutrophil/macrophage balance and ablation experiments show that macrophages are critical for the defense response. Deficiency of atg5 reduces GFP-Lc3 recruitment and impairs host resistance, in contrast to atg13 deficiency, indicating that Lc3-Salmonella association at this stage is independent of the autophagy preinitiation complex and that macrophages target Salmonella by Lc3-associated phagocytosis (LAP). In agreement, GFP-Lc3 recruitment and host resistance are impaired by deficiency of Rubcn/Rubicon, known as a negative regulator of canonical autophagy and an inducer of LAP. We also found strict dependency on NADPH oxidase, another essential factor for LAP. Both Rubcn and NADPH oxidase are required to activate a Salmonella biosensor for reactive oxygen species inside infected macrophages. These results identify LAP as the major host protective autophagy-related pathway responsible for macrophage defense against Salmonella during systemic infection.
Abbreviations: ATG: autophagy related gene; BECN1: Beclin 1; CFU: colony forming units; CYBA/P22PHOX: cytochrome b-245, alpha chain; CYBB/NOX2: cytochrome b-245 beta chain; dpf: days post fertilization; EGFP: enhanced green fluorescent protein; GFP: green fluorescent protein; hfp: hours post fertilization; hpi: hours post infection; IRF8: interferon regulatory factor 8; Lcp1/L-plastin: lymphocyte cytosolic protein 1; LAP: LC3-associated phagocytosis; MAP1LC3/LC3: microtubule-associated protein 1A/1B-light chain 3; mCherry: red fluorescent protein; mpeg1: macrophage expressed gene 1; mpx: myeloid specific peroxidase; NADPH oxidase: nicotinamide adenine dinucleotide phosphate oxidase; NCF4/P40PHOX: neutrophil cytosolic factor 4; NTR-mCherry: nitroreductase-mCherry fusion; PTU: phenylthiourea; PtdIns3K: class III phosphatidylinositol 3-kinase; PtdIns3P: phosphatidylinositol 3-phosphate; RB1CC1/FIP200: RB-1 inducible coiled coin 1; ROS: reactive oxygen species; RT-PCR: reverse transcriptase polymerase chain reaction; RUBCN/RUBICON: RUN and cysteine rich domain containing BECN1-interacting protein; SCV: Salmonella-containing vacuole; S. Typhimurium/S.T: Salmonella enterica serovar Typhimurium; TEM: transmission electron microscopy; Tg: transgenic; TSA: tyramide signal amplification; ULK1/2: unc-51-like autophagy activating kinase 1/2; UVRAG: UVRAG: UV radiation resistance associated; wt: wild type
KEYWORDS: Autophagy, LC3, LC3-associated phagocytosis (LAP), Salmonella Typhimurium, Rubicon, ATG5, NADPH oxidase, ROS, zebrafish
Introduction
Salmonella enterica serovar Typhimurium (S. Typhimurium) is a common cause of self-limiting gastrointestinal infections in human hosts, but can provoke a systemic typhoid-like disease upon infection of mice [1]. Therefore, this pathogen is widely used as surrogate model for S. enterica serovar Typhi (S. Typhi), the causative agent of typhoid fever, a life-threatening systemic human infectious disease. Salmonella can invade a variety of cell types owing to their ability to inject virulence effectors triggering phagocytosis by non-professional phagocytes, such as epithelial cells and fibroblasts [2]. Following this self-induced entry, Salmonella begins to replicate inside a growing compartment called the Salmonella-containing vacuole (SCV). Salmonella can also replicate inside professional phagocytes, including macrophages, which are the main carriers of this pathogen when it causes systemic disease [3].
The invasion of host cells by Salmonella or other intracellular pathogens triggers macroautophagy (hereafter autophagy), a cellular degradation pathway that delivers cytoplasmic content to lysosomes [4]. Many studies support that activation of the autophagy machinery functions to restrict cytosolic escape and intracellular replication of Salmonella [5–12]. Xenophagy is known as a selective autophagy process, wherein ubiquitin and galectin receptors target the membranes of damaged SCVs and bacteria that have escaped into the cytosol, and this is the main anti-Salmonella autophagy response in epithelial cells [8–11,13]. However, both the survival strategies of Salmonella and the host cell autophagy responses differ between cell types. For example, S. Typhimurium does not escape into the cytosol in fibroblasts, but in this cell type endosomal and lysosomal membranes accumulate around the SCV to form large aggregates that engage the autophagy machinery in an ATG9A-independent manner [14,15]. Another process dependent on components of the autophagy machinery is known as LC3-associated phagocytosis (LAP) [16]. During LAP, the autophagy marker microtubule-associated protein 1A/1B-light chain 3 (MAP1LC3, LC3) is recruited to the single membrane of the phagosome, whereas it marks the double membranes of autophagosomes during canonical autophagy [17,18]. It is likely that LAP contributes to the response of neutrophils and epithelial cells to Salmonella infection [7]. Studies of the autophagic response of macrophages to Salmonella infection are limited and point to roles in mediating programmed cell death as well as restricting bacterial replication [19].
The recruitment of LC3 to the phagosomal membrane of Salmonella-infected neutrophils and epithelial cells has been shown to depend on the function of the NADPH oxidase (nicotinamide adenine dinucleotide phosphate oxidase) [7]. NADPH oxidase is required for the generation of reactive oxygen species (ROS) with potentially damaging properties against intracellular pathogens. NADPH oxidase is inactive in resting macrophages and neutrophils and becomes activated when invading microbes trigger innate immunity signaling via the Toll-like receptor pathway [20]. Its importance in host defense is well exemplified by chronic granulomatous disease, a condition leading to life-threatening fungal and bacterial infections arising due to non-functional NADPH oxidase in phagocytes [21,22].
The RUN and cysteine rich domain containing BECN1/BECLIN1-interacting protein (RUBCN/RUBICON) has recently been shown to play an essential role in LAP and to be required for the activation of NADPH oxidase during this process [20,23,24]. RUBCN mediates the activation of NADPH oxidase in two ways, first by recruiting the NCF4/P40PHOX component of NADPH oxidase via activating Class III phosphatidylinositol 3-kinase (PtdIns3K) on phagosomes and generating phosphatidylinositol 3-phosphate (PtdIns3P), and second by stabilizing CYBA/P22PHOX to assemble the NADPH oxidase complex [20,23]. In contrast, RUBCN functions as a negative regulator of autophagosome formation, inhibiting the PtdIns3K complex and RAB7 guanosine triphosphatase activity [25]. Thus, RUBCN functions as a molecular switch that either suppresses canonical autophagy or promotes LAP. Most components of the autophagy machinery are required for the formation of autophagosomes as well as LAPosomes. However, LAP is independent of the preinitiation complex (ULK1 complex, including ULK1/2 [unc-51 like autophagy activating kinase 1/2], RB1CC1/FIP200, ATG13 and ATG101) [23,26], whereas this complex is essential for the induction of canonical autophagy [27]. Several studies have provided evidence for a role of LAP in restricting the growth of intracellular pathogens [20,23,28,29], while other studies suggest that pathogens might exploit LAPosomes as replication niches [30,31]. In addition, LAP has been proposed to promote MHC II class presentation of antigens [32,33]. Furthermore, LAP can be activated during internalization of dead or dying cells (efferocytosis) or of live cells (entosis), which is thought to ensure that cells are cleared effectively without triggering pathological inflammatory responses [26,34,35].
It is currently unknown how the different autophagy-related mechanisms, such as xenophagy and LAP, are involved in the interaction of macrophages with Salmonella during systemic infection. To study the encounter of macrophages with this pathogen in vivo, we took advantage of a zebrafish embryo model of S. Typhimurium infection, wherein bacteria are delivered by microinjection into the blood circulation or sub-cutaneously [36,37]. We have previously shown that zebrafish embryos respond to Salmonella infection by Toll-like receptor-mediated signaling inducing a strong proinflammatory gene expression signature similar as in mammalian hosts and human cells [38–41]. The zebrafish has become a widely utilized vertebrate model for human infection diseases, especially because microscopic imaging of infected zebrafish embryos provides new possibilities to gain insight into the interactions between pathogens and host innate immune cells in a living organism [42–44]. The zebrafish is also increasingly used to study autophagy and it has previously been demonstrated that zebrafish embryos can mount an autophagic defense response against Shigella flexneri and Mycobacterium marinum [45–48]. Here, by in vivo imaging of GFP-Lc3 transgenic zebrafish embryos we could dissect the role of macrophages and neutrophils in anti-Salmonella responses and expose LAP as the major pathway responsible for the macrophage-mediated defense against this pathogen.
Results
GFP-Lc3 recruitment is a dynamic response limited to live Salmonella cells
LC3, the vertebrate ortholog of yeast Atg8, is a widely accepted marker for autophagy-related processes, which can be visualized by fusion with fluorescent proteins [49]. We took this approach and used Tg(CMV:GFP-map1lc3b) transgenic zebrafish [50] to study the role of the autophagy machinery during Salmonella infection in vivo. In previous work, we had characterized the immune response of zebrafish embryos to intravenous infection with S. Typhimurium by microarray and RNA sequencing analysis. These studies showed that systemic Salmonella infection in the zebrafish host is characterized by a strong proinflammatory gene expression signature and that infection of embryos at 1 day post fertilization (dpf) causes lethality within one day [39–41,51–53]. In this study we modulated the infection model to investigate the host autophagic defense response over a longer period of infection. To this end, embryos were systemically infected at 2 dpf via caudal vein microinjection with a low dose (200–400 colony forming units (CFU)) of either live or heat-killed S. Typhimurium (SL1344). Both groups were scored for mortalities at 24 hour intervals up to 72 hours post infection (hpi). In contrast to heat-killed bacteria, live S. Typhimurium were able to kill approximately 50% of the infected zebrafish by 48 hpi, and the infection resulted in approximately 70% mortality at the endpoint of the survival experiment (72 hpi) (Figure 1(a)). To assess bacterial growth kinetics, CFU counts were determined by retrieving Salmonella cells from surviving embryos at 24, 48 and 72 hpi. As expected, the heat-killed S. Typhimurium could not establish infection within the host as no bacterial growth on agar plates was observed (Figure 1(b)). Conversely, live S. Typhimurium showed exponential growth inside the host reaching approximately 105 CFU per larva (Figure 1(b)). Of note, the above experiments were reproduced in the wild-type zebrafish line AB/TL (Figure S1); therefore, the observed response to S. Typhimurium is not affected by the GFP-Lc3 transgene expression.
Figure 1.
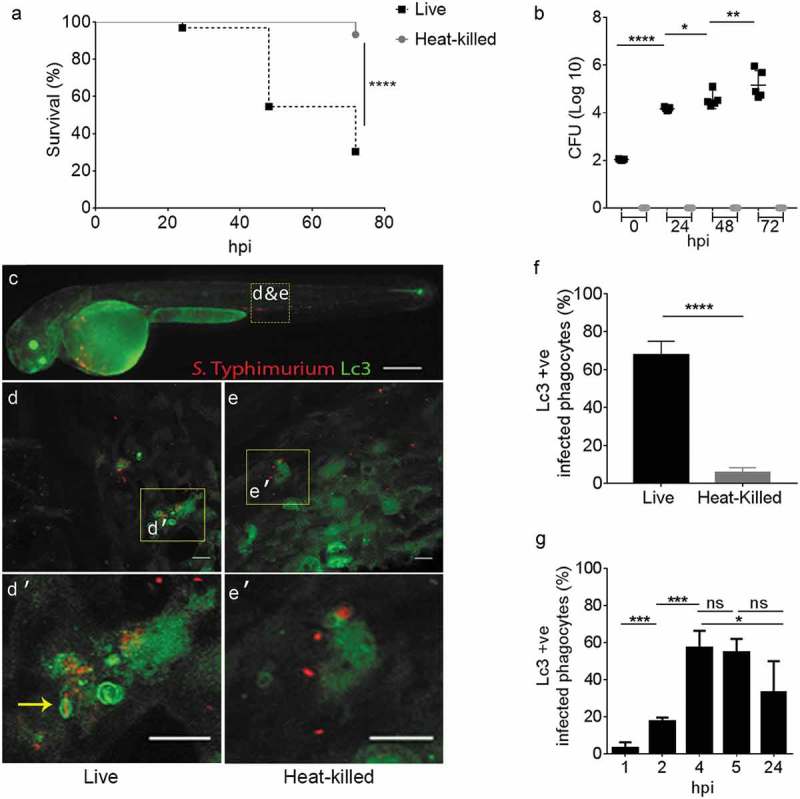
GFP-Lc3 recruitment to Salmonella during systemic infection of zebrafish. (a) Survival curves of Tg(CMV:GFP-map1lc3b) zebrafish embryos/larvae, systemically infected at 2 dpf by caudal vein injection with either live or heat-killed S. Typhimurium. One representative of three replicates is shown (n = 50 embryos per group). (b) CFU counts of the zebrafish infected with live or heat-killed S. Typhimurium. Five embryos/larvae per time point were used and the log-transformed CFU data are shown with the geometric mean per time point. One representative of three replicates is shown. (c) Stereo fluorescence image of a 2 dpf Tg(CMV:GFP-map1lc3b) embryo, systemically infected with live mCherry-expressing S. Typhimurium. The yellow dotted box shows the area in the tail used for Z-stack confocal imaging in d and e. (d and e) Representative confocal images at maximum projection taken at 4 hpi from time lapses recorded of embryos injected with live S. Typhimurium (e) or heat-killed S. Typhimurium (f) bacteria (see Movie M1 for live and M2 for heat-killed bacteria). Yellow boxes indicate the magnified regions shown in d’ and e’. The arrow in d’ indicates a ring-shaped structure of GFP-Lc3 signal enclosing an mCherry-expressing S. Typhimurium cell and several other GFP-Lc3-Salmonella associations can be observed in this image. (f) Quantification of GFP-Lc3-Salmonella associations at 4 hpi. Embryos injected with live or heat-killed S. Typhimurium were imaged over the yolk sac circulation valley region and quantified for percentages of phagocytes positive for GFP-Lc3-Salmonella associations (Lc3+ ve) over the total number of phagocytes with ingested bacteria. Error bars represent the SD. One representative of three replicates (each with n = 5 embryos per group) is shown. (g) Quantification of time course associations of GFP-Lc3-Salmonella during systemic infection with live S. Typhimurium. Five embryos per time point were imaged and quantified for percentages of phagocytes with GFP-Lc3-Salmonella associations as in f. Error bars represent the SD. Scale bars: c = 200 μm, d, e, d’ and e’ = 10 μm. ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05, ns = non-significant.
In order to visualize the dynamics of GFP-Lc3-associations to Salmonella cells, a high dose (2000–4000 CFU) of mCherry-expressing bacteria (either live or heat-killed) was injected and time-lapse confocal microscopy was performed. GFP-Lc3-Salmonella associations were found to be mostly limited to injection of live bacteria, where the bright GFP signal of Lc3 (Figure 1(c–e)) was observed associated to S. Typhimurium internalized by motile phagocytes (see Movies M1 and M2). Approximately 60% of phagocytes that had ingested live bacteria were positive for GFP-Lc3-Salmonella associations at 4 hpi, whereas the proportion of GFP-Lc3 positive phagocytes was close to zero in response to injection of heat-killed bacteria (Figure 1(f)). In addition, the dynamics of GFP-Lc3 associations to internalized S. Typhimurium was determined in a time course experiment where infected embryos with live S. Typhimurium were fixed at 1, 2, 4, 5 and 24 hpi and the proportion of infected phagocytes with Lc3 associations was quantified from confocal images. The highest percentage of GFP-Lc3 recruitment was observed at 4 hpi, which dropped to significantly lower levels at 24 hpi (Figure 1(g)).
GFP-Lc3 associates to Salmonella in different patterns of rings and puncta
To further describe the intracellular patterns of GFP-Lc3-Salmonella associations, we focused on the 4 hpi time point, where the highest observed Lc3 response occurred. We confirmed the presence of a proinflammatory immune response at this stage (Figure S2), in agreement with previous expression profiling studies [39–41,51–53]. Following intravenous infection, the majority of the S. Typhimurium bacteria contained by phagocytes residing in the yolk sac circulation valley or in blood vessels of the tail region (Figure 2(a)) could be classified as Lc3-positive associations (74% of intracellular Salmonella) (Figure 2(bi-vi,,c)), while the remaining fraction of intraphagocyte Salmonella (26%) was observed as Lc3-negative (Figure 2(bvii-ix)). The Lc3-positive associations were observed in different patterns, categorized as (i) a single punctum associated with a single bacterial cell (Figure 2(bi)), (ii) a spacious Lc3 ring around a single bacterial cell (Figure 2(bii)), (iii) a tight Lc3 ring around a single bacterial cell (Figure 2(biii)), (iv) a spacious Lc3 ring enclosing a cluster of loosely packed bacterial cells (Figure 2(biv)), (v) an Lc3 ring around a dense mCherry signal of bacteria where bacterial cell boundaries are not identifiable (Figure 2(bv)), and (vi) multiple Lc3 puncta associating with clusters of bacteria (Figure 2(bvi)). The four different Lc3 ring patterns (Figure 2(bii-v)) together constituted 45% of all Lc3-positive associations, and the Lc3 puncta were more frequently observed in association with clusters of bacteria (40% of Lc3-positive associations) than in association with single bacterial cells (15% of Lc3-positive associations) (Figure 2(c)). The Lc3-negative intracellular Salmonella were observed as single bacterial cells (Figure 2(bvii)), and loosely (Figure 2(bviii)) or tightly packed (Figure 2(bix)) clusters of bacteria that seemed contained inside a vesicular compartment. These different types of vesicles constituted 20%, 21%, and 9% of all intraphagocyte Salmonella observations, respectively (Figure 2(c)). In conclusion, Lc3 rings or puncta were observed in association with the majority of bacterial cells or clusters at 4 hpi, which prompted us to further investigate the relevance of this response for the host defense against Salmonella.
Figure 2.
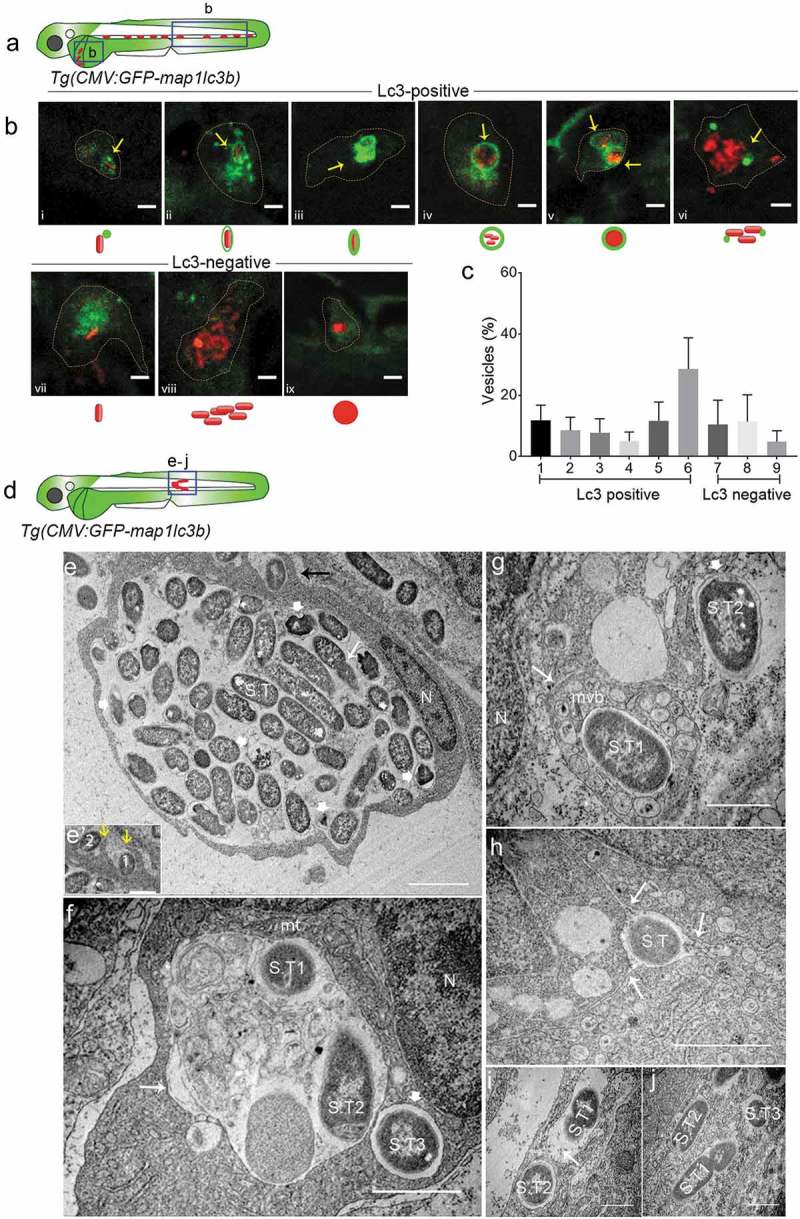
Types of Salmonella-containing vesicles in professional phagocytes. (a) Regions of interest (blue squares) for confocal images of infected phagocytes in the blood circulation in b. (b) Representative confocal micrographs for 6 distinctive patterns of GFP-Lc3 association with mCherry-expressing S. Typhimurium (bi-bvi) and 3 types of Lc3-negative cases (bvii-bix) observed in motile phagocytes in the tail and yolk sac regions at 4 hpi following systemic infection by caudal vein injection. Under each image the symbolic representation of the pattern is shown. Dotted lines outline the cellular boundaries of the phagocytes and yellow arrows point towards GFP-Lc3 association with bacterial cells. (c) Quantification of types of Lc3-positive and Lc3-negative Salmonella associations observed in b. Percentages of the 9 types of Lc3-positive and Lc3-negative vesicles were determined from images of Salmonella-infected phagocytes and averaged from 5 embryos (n > 30 Lc3+/Lc3− Salmonella associations per embryo, 196 in total). Error bars represent the SD. (d) Region of interest (blue square) in subcutaneously infected embryos for image acquisition through TEM in e to j. (e) TEM micrograph of a heavily infected macrophage with phagocytosed S. Typhimurium at 4 hpi, located in the lumen underneath the epidermis. Cytoplasm and nucleus (n) of the cell are pushed towards the cellular periphery and a large number of phagocytosed S. Typhimurium cells (S.T) are centrally contained in a large phagosomal compartment. The long white arrow indicates a dividing bacterial cell. The short thick white arrows indicate examples of dissociating Salmonella cells recognized by the retraction of the cytoplasm from the inner membrane. The black arrow indicates phagocytosing activity of the macrophage which is enlarged in e’. The yellow arrows in e’ point to membrane ruffles of the macrophage formed to enclose extracellular S. Typhimurium (1 and 2). Figure S1 shows a broader view of this macrophage in the tissue context. (f) TEM micrograph showing two S. Typhimurium-containing compartments. The larger compartment (long arrow) contains cellular debris in addition to bacterial cells (S.T1 and S.T2). The lumen of the smaller vesicle (short thick arrow), which holds a single bacterium (S.T3), is clean. The nucleus (n) and a mitochondrion (mt) in close vicinity of the bacterial compartments are indicated. (g) TEM micrograph of S. Typhimurium (S.T1) inside a multi-vesicular body (MVB, long arrow). The short thick arrow indicates a cytoplasmic Salmonella cell (S.T2) with intact outer and inner bacterial membrane. (h) TEM of S. Typhimurium (S.T1) inside a vesicle that forms tubular extensions into the cytoplasm (arrows). (i) TEM showing S. Typhimurium cells (S.T1 and S.T2) inside two single membrane-bound compartments. The presence of ribosomes (arrow) in the internal environment suggests membrane damage. (j) TEM of three S. Typhimurium (S.T) cells in the cytoplasm, one of which (S.T1) is replicating. Scale bars: b = 5 μm, e-j = 2 μm, e’ = 1 μm.
Ultrastructural analysis reveals the presence of Salmonella in phagosomes and a variety of other subcellular compartments
In an attempt to visualize S. Typhimurium inside the classical double membrane autophagosome compartments, a hallmark of canonical autophagy (xenophagy), we used transmission electron microscopy (TEM). In order to facilitate the localization of infected cells in TEM sections, a localized infection was established by inoculating a high dose of S. Typhimurium subcutaneously into the 2 dpf embryos (Figure 2(d)). In our limited number (n = 80) of TEM micrographs from multiple embryos (n = 20), we were not able to identify any S. Typhimurium cells residing within a double membrane structure, while diverse other types of Salmonella containing vesicles were observed (Figure 2(e-i)) as well as Salmonella residing freely in the cytoplasm (Figure 2(j)). For example, we observed a macrophage attached to the inner layer of the epidermis, where it centrally contains a phagosome with a large number of internalized S. Typhimurium (Figure 2(e)), also see Figure S3 showing the cell within the surrounding tissue). The membrane ruffles extending from the observed macrophage to extracellular bacteria in the sub-cutaneous space indicate that this phagocyte was involved in further phagocytosis of bacteria (Figure 2(e’)). Among the phagocytosed bacteria, some showed normal S. Typhimurium morphology where bacterial envelopes and cytoplasm were intact. In contrast, other ingested bacteria were identified with unusual morphology where the cytoplasm was found retracted away from the bacterial envelope. Additionally, the intraphagosome environment appeared cloudy and contained debris that could have resulted from dissociating bacteria. This large phagosome could be a possible representative for one of the larger types of Lc3-positive (Figure 2(biv-vi)) or Lc3-negative (Figure 2(bviii,ix)) Salmonella containing vesicles with multiple bacteria inside.
Another type of intracellular compartment containing S. Typhimurium was observed where a spacious vesicle held two bacterial cells (S.T1 and 2 in Figure 2(f)) but also contained cytoplasmic (membranous) material that might have resulted from intraluminal vesicle formation or from fusion with autophagic vesicles. In the same micrograph, another bacterial cell (S.T3) is observed inside a less expanded SCV, not containing cytoplasmic material, suggesting this structure to have originated from phagocytosis and not from xenophagy. Similar cases were observed in other micrographs (S.T2 in Figure. 2(g,i)).
We also noticed an S. Typhimurium cell centrally placed inside a multi-vesicular body (MVB) (S.T1 in Figure 2(g)) and enclosed inside a single membrane compartment. While no conclusions about the relationship between GFP-Lc3 and TEM structures can be drawn in the absence of correlative light and electron microscopy, it is tempting to speculate that the intense and tight ring of Lc3 around a single bacterium in Figure 2(biii) might correspond to such an MVB structure observed in the TEM analysis. A bacterial compartment forming tubular extensions into the cytoplasm was also observed (Figure 2(h)), which is reminiscent of the formation of Salmonella induced filaments, a process known to occur at later stages of SCV formation [2,54]. Finally, we spotted replicating S. Typhimurium cells inside vesicles (S.T1 in Figure 2(i)) as well as within the cytoplasm (S.T1 in Figure 2(j)).
In conclusion, TEM analysis did not provide direct evidence for targeting of Salmonella by autophagy, but in agreement with the results of confocal microscopy, it revealed a variety of Salmonella-containing compartments, including phagosomes, multivesicular compartments, and other compartments that could have arisen from intracellular vesicle fusion events.
Macrophages show more Lc3-Salmonella associations than neutrophils
In order to determine which phagocyte cell types are involved in the GFP-Lc3 response to S. Typhimurium, infected embryos were stained with tyramide signal amplification (TSA) and anti-Lcp1/L-plastin (lymphocyte cytosolic protein 1) immunolabeling, a method that is used to distinguish macrophages and neutrophils [55–58]. The fluorescent tyramide substrate is metabolized by the neutrophil-specific myeloperoxidase and the anti-Lcp1 antibody labels all phagocytes, such that macrophages can be identified as Lcp1-positive and TSA-negative cells. Lc3-Salmonella associations were not observed in Lcp1-negative cells, indicating that their presence is restricted to phagocytes at the time point of our analysis. It was observed that macrophages (Lcp1-positive/TSA-negative) in the blood circulation of systemically infected embryos (Figure 3(a)) show almost all types of Lc3-Salmonella associations recorded earlier (Figure 3(bi-vi) as compared with Figure 2(bi-vi)). Neutrophils (Lcp1/TSA double positive) were also observed with Lc3-Salmonella associations (Figure 3(bvii,viii)), however, not all types of Lc3-positive Salmonella containing vesicles could be observed in these cells.
Figure 3.
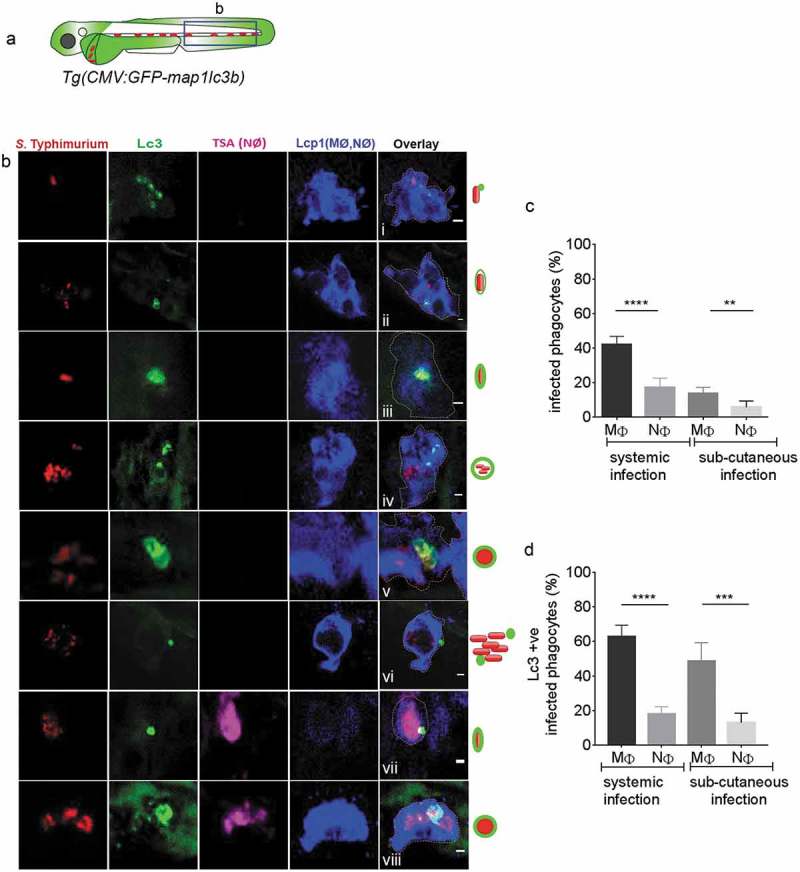
GFP-Lc3–Salmonella associations in specific phagocyte types. (a) Region of interest (blue square) for confocal images of infected phagocytes in the blood circulation in b. (b) Confocal micrographs in series (i–viii) of distinctive patterns of GFP-Lc3 associations with mCherry-expressing S. Typhimurium in macrophages (MΦ, TSA negative/Lcp1 positive; i-vi) and neutrophils (NΦ, TSA positive/Lcp1 positive; vii,viii). The dotted lines in the overlay image of the separate channels indicate the cellular boundaries. The symbolic presentations shown next to the images refer to the different types of GFP-Lc3-Salmonella associations presented in Figure 2(b). (c) Quantification of phagocytosing activity of macrophages and neutrophils in systemic and sub-cutaneous infections at 4 hpi. Numbers of infected and non-infected phagocytes were counted from confocal images and the percentages of infected over the total were averaged from five embryos per group. Error bars represent the SD. Macrophages (MΦ) and neutrophils (NΦ) were distinguished by anti-Lcp1 and TSA staining as in b. (d) Quantification of GFP-Lc3-Salmonella associations in macrophages and neutrophils at 4 hpi. Numbers of infected phagocytes positive or negative for GFP-Lc3-Salmonella associations were counted from confocal images and the percentages of Lc3-positive over the total were averaged from five embryos per group. Error bars represent the SD. Macrophages (MΦ) and neutrophils (NΦ) were distinguished by anti-Lcp1 and TSA staining as in b. Scale bars: b = 2 μm. ****P < 0.0001, ***P < 0.001, **P < 0.01.
Macrophages are known for readily phagocytosing intravenously injected Escherichia coli bacteria at higher rates than neutrophils, whilst neutrophils display a surface-associated phagocytic behavior and are reported to phagocytose bacteria more readily during a sub-cutaneous infection [59]. Despite this difference in phagocytic behavior between the cell types, we observed that, in both types of infection routes, macrophages phagocytose S. Typhimurium in 2–3-fold higher percentages when compared to neutrophils (Figure 3(c)). Furthermore, irrespective of the infection route, a significantly higher percentage of infected macrophages were scored positive for the presence of GFP-Lc3-Salmonella associations as compared to the percentage of infected neutrophils (Figure 3(d)). In conclusion, macrophages show all the above-described GFP-Lc3 ring or puncta patterns in response to Salmonella infection. Furthermore, independent of the route of infection, macrophages have higher phagocytic activity against Salmonella and more frequently display GFP-Lc3-Salmonella associations than neutrophils. These results suggest that macrophages are more critical for the defense response to Salmonella than neutrophils at this stage of the infection.
Macrophage depletion reduces Lc3-Salmonella associations and impairs host defense
The results showing that macrophages stand out as the predominant phagocyte cell type responsible for phagocytosing and Lc3 targeting of S. Typhimurium in our model, prompted us to investigate how the host would respond to Salmonella infection in a state without macrophages. To this end, we took advantage of the metronidazole/nitroreductase system, which has been successfully used for chemically induced ablation of either macrophages or neutrophils in zebrafish embryos [60–62]. Metronidazole treatment of embryos expressing a nitroreductase-mCherry fusion (NTR-mCherry) under control of the macrophage-specific mpeg1 promoter resulted in specific loss of fluorescent macrophages (Figure S4A,B). In Salmonella infection experiments, such ablation of macrophages resulted in 100% mortality of embryos by 24 hpi, while control groups (untreated NTR-mCherry, or untreated and treated siblings not expressing NTR-mCherry) showed over 40% survival at 48 hpi and over 15% survival at 72 hpi (Figure 4(a)). Ablation of neutrophils was achieved using embryos expressing NTR-mCherry under the mpx promoter (Figure S4C,D). Neutrophil ablation resulted in lower survival rates of Salmonella-infected embryos than observed for the control groups. However, compared to macrophage ablation the effect was relatively minor, with over 60% survival of neutrophil-ablated embryos at 24 hpi, the time point where all macrophage-ablated embryos had died (Figure 4(a)). In agreement with the survival rates, CFU counts were significantly higher in macrophage-ablated than in neutrophil-ablated embryos (Figure 4(b)).
Figure 4.
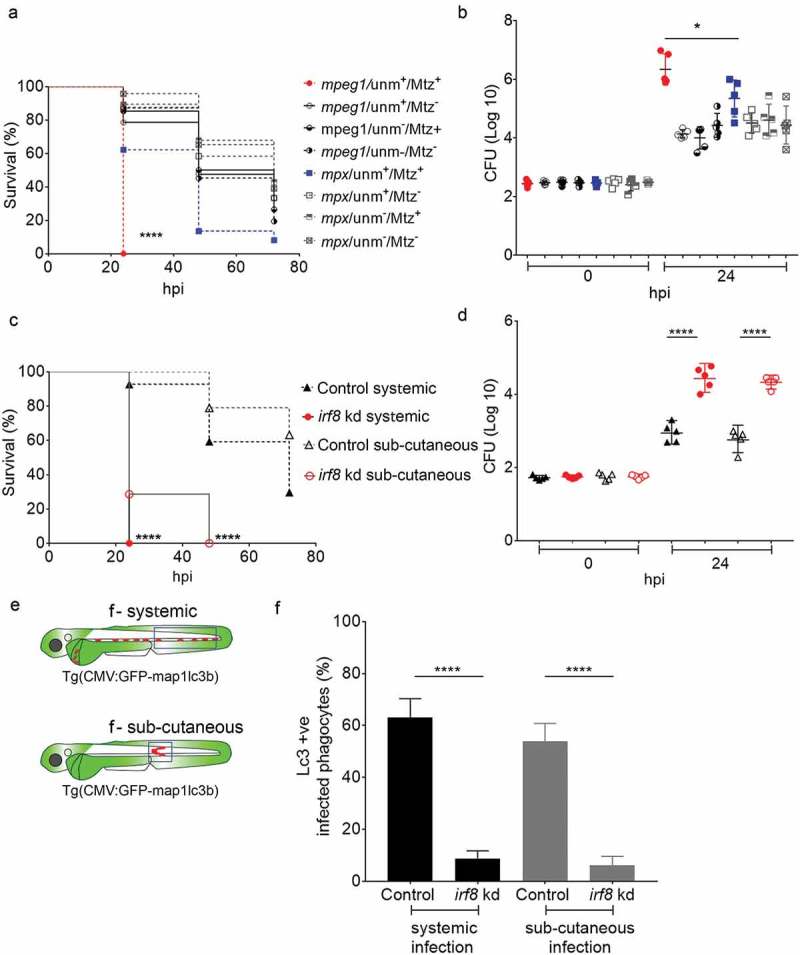
Essential role of macrophages in host defense against systemic Salmonella infection. (a) Survival curves of metronidazole-treated ablation groups for macrophages (mpeg1:unm+/Mtz+, red curve) and neutrophils (mpx:unm+/Mtz+, blue curve), and their respective controls (mpeg1:unm+/Mtz-, mpeg1:unm-/Mtz+, mpeg1:unm-/Mtz-, solid lines) and (mpx:unm+/Mtz-, mpx:unm-/Mtz+, mpx:unm-/Mtz-, dotted lines). Embryos were systemically infected by caudal vein injection. One representative of three replicates is shown. Significance is indicated for the difference in survival between the macrophage (mpeg1:unm+/Mtz+, red curve) and neutrophil (mpx:unm+/Mtz+, blue curve) ablation groups. (b) Representative CFU counts of controls and macrophage (red) and neutrophil (blue) ablation groups from a. Five embryos per time point were used and the log-transformed CFU data are shown with the geometric mean per time point. In case of the macrophage ablation group, dead embryos were used for CFU plating at the 24 hpi time point. One representative of three replicates is shown. (c) Survival curves of control and irf8 knockdown larvae in systemic and sub-cutaneous infection experiments. Significance is indicated for the difference between the systemically infected control and irf8 knockdown groups and for the sub-cutaneously infected control and irf8 knockdown groups. One representative of three replicates is shown. (d) Representative CFU counts of control and irf8 knockdown larvae in systemic and sub-cutaneous infection experiments. Five embryos/larvae per time point were used and the log transformed CFU data are shown with the geometric mean per time point. (e) Regions of interest (blue squares) for quantification of GFP-Lc3-Salmonella associations in infected phagocytes in the blood circulation or in the sub-cutaneous area in f. (f) Quantification of GFP-Lc3-Salmonella associations during systemic and sub-cutaneous infection in control and irf8 knockdown embryos at 4 hpi. Numbers of infected phagocytes positive or negative for GFP-Lc3-Salmonella associations were counted from confocal images and the percentages of Lc3-positive over the total were averaged from five embryos per group. Error bars represent the SD. ****P < 0.0001, *P < 0.1.
Next, we used a second strategy to further explore the impact of macrophage ablation. To this end, we used an interferon regulatory factor-8 (irf8) knockdown approach, where zebrafish myelopoiesis is reoriented and no macrophages are produced, whilst almost doubling the number of neutrophils [63]. This method has been successfully applied to zebrafish infection models [62,64,65]. The morpholino-mediated irf8 knockdown was validated by imaging macrophages and neutrophils in double transgenic Tg(mpeg1:mCherry) and Tg(mpx:egfp) zebrafish and resulted in embryos with almost no macrophages but expanded neutrophil numbers at 2 dpf, the time of S. Typhimurium infection, confirming the efficacy of this knockdown strategy (Figure S4E and F). We found that macrophage-depleted embryos were hypersusceptible to S. Typhimurium infection as most of the irf8 knockdown subjects died within 24 hpi regardless of the route of infection (Figure 4(c)). In addition, CFU counts revealed that in the absence of macrophages, the infection progresses faster as the bacterial numbers at 24 hpi were significantly higher in irf8 knockdown compared to the controls (Figure 4(d)). Subsequently, we investigated the effect of irf8 knockdown on GFP-Lc3-Salmonella associations. Because IRF8 has been reported to regulate autophagy [66], we checked that irf8 knockdown did not detectably affect GFP-Lc3 levels in uninfected embryos, neither in presence nor in the absence of bafilomycin A1, an inhibitor of autophagic flux (Figure S6B,C and B’,C’). In addition, confocal microscopy, combined with double staining with anti-Lcp1 and TSA was used to confirm the macrophage depletion and neutrophil expansion in GFP-Lc3 irf8 knockdown embryos (Figure S4G-L) in systemic and subcutaneous infection models. Both the intravenous and sub-cutaneous infection routes led to higher percentages of infected phagocytes with Lc3-Salmonella associations in control embryos compared with the irf8 knockdown groups, supporting that the Lc3 response observed is mainly driven by macrophages (Figure 4(e,f)).
Lc3-Salmonella associations are Atg5-dependent and Atg13-independent
A number of ATG proteins play essential roles during the process of autophagy. We wanted to determine the type of Lc3-Salmonella associations observed as evidence of either canonical autophagy or an autophagy-related process and therefore investigated the role of components of the ATG-machinery. ATG5 forms a complex with ATG12 and is required for autophagosome formation [67]. To determine the impact of zebrafish Atg5 in our Salmonella infection model, we took advantage of a previously described atg5 morpholino knockdown strategy [68,69]. Since atg5 depletion can cause lethality in zebrafish larvae from 4 dpf [69], we used a partial atg5 knockdown approach to avoid this early lethality such that survival rates of Salmonella-infected larvae could be studied until 5 dpf (72 hpi). To this end, we titrated the morpholino down to a level at which uninfected larvae showed no lethality or developmental aberrations over the time frame of our experiments. We verified the efficacy of the atg5 morpholino on autophagy inhibition by demonstrating a significant reduction of GFP-Lc3 levels in bafilomycin A1-treated atg5 knockdown embryos compared with control embryos (Figure S6D,E and D’,E’). Despite of studying a partial atg5 knockdown phenotype, it could be concluded that Atg5 is required for host defense against S. Typhimurium as control embryos had significantly higher survival rates during Salmonella infection when compared to the atg5 knockdown group (Figure 5(b)). In agreement, control embryos were able to curtail bacterial infection better than atg5 knockdown embryos as based on CFU counts (Figure 5(c)). Quantification of the GFP-Lc3-Salmonella associations (Figure 5(d,d’)) confirmed the importance of Atg5 for this response as controls showed a significantly higher percentage of GFP-Lc3-positive infected phagocytes when compared to atg5 knockdown embryos (Figure 5(e)).
Figure 5.
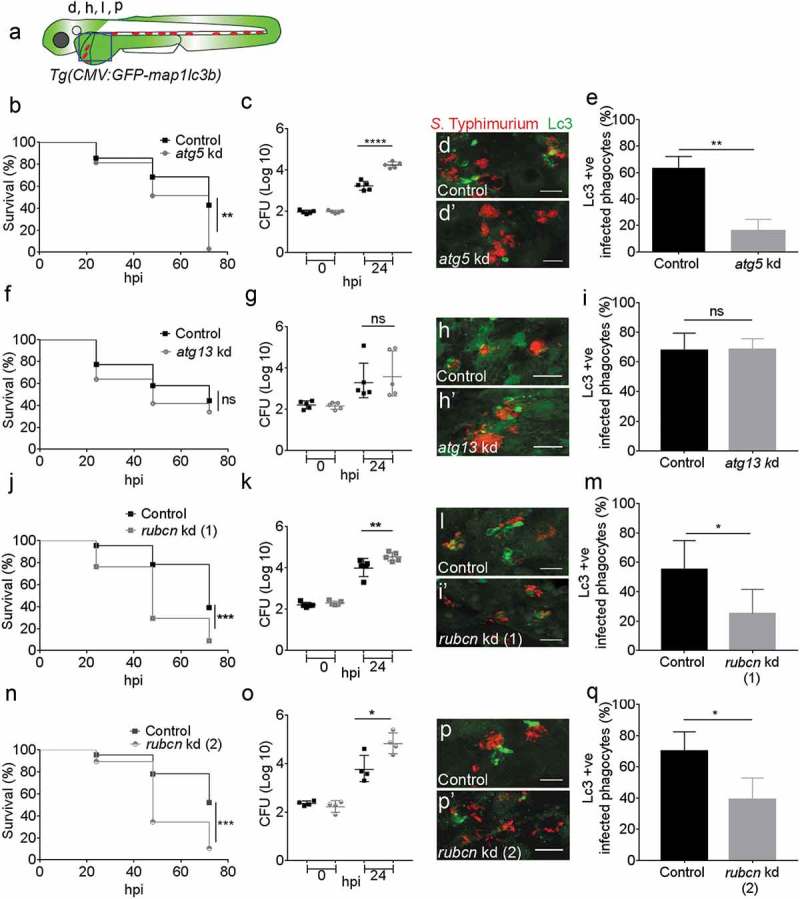
Roles of autophagy-related proteins in the host response to Salmonella infection. (a) Region of interest (blue square) for confocal image acquisition of infected phagocytes in the blood circulation in d, h, l and p. (b, f, j and n) Survival curves of systemic S. Typhimurium infection experiments of atg5 (b), atg13 (f), rubcn kd(1) (j) and rubcn kd(2) (n) morpholino knockdown groups with their respective control morpholino-injected groups. Kd1 and kd2 refer to use of translation blocking (MO1-rubcn) and splice blocking (MO2-rubcn) morpholinos, respectively. For each comparison one representative of three replicates is shown. Survival was scored at 24, 48 and 72 hpi, following infection at 48 hpf. (c, g, k and o) Representative CFU counts of the S. Typhimurium infections of atg5 (c), atg13 (g), rubcn kd(1) (k) and rubcn kd(2) (o) morpholino knockdown groups with their respective controls at 24 hpi. Five embryos/larvae per time point were used and the log transformed CFU data are shown with the geometric mean per time point. (d, h, l and p) Representative confocal images of atg5 (d’), atg13 (h’), rubcn kd(1) (l’) and rubcn kd(2)’ (p’) morpholino knockdown embryos with their respective controls (d, h, l and p) at 4 hpi. (e, i, m and q): Quantification of GFP-Lc3-Salmonella associations in atg5 (e), atg13 (i), rubcn kd(1) (m) and rubcn kd(2) (q) morpholino knockdown groups with their respective controls at 4 hpi. Numbers of infected phagocytes positive or negative for GFP-Lc3-Salmonella associations were counted from confocal images and the percentages of Lc3-positive over the total were averaged from five embryos per group. Error bars represent the SD. Scale bars: 10 μm. ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05, ns = non-significant.
Because Atg5 is required in both canonical autophagy [70] and autophagy-related processes [7,14,16], the loss of this protein mediated by atg5 knockdown did not reveal whether Lc3-Salmonella associations observed in our model are related to entrapment of Salmonella in autophagosomes (xenophagy) or could be due to Lc3 recruitment to phagosomes (LAP). Therefore, we knocked down atg13, encoding a component of the Ulk1 preinitiation complex, which is required only for canonical autophagy [26]. Similar as for atg5 knockdown, efficacy of the atg13 morpholino was validated with a functional analysis using bafilomycin A1 to block autophagic flux, wherein atg13 knockdown resulted in significantly lower GFP-Lc3 signal accumulation as compared to controls after bafilomycin A1 treatment at 2 dpf (Figure S6F,G and F’,G’). Knockdown of atg13 had no significant effect on survival rates during infection (Figure 5(f)), and bacterial burdens within controls and atg13 morphants did not differ from each other (Figure 5(g)). These results indicate that the Ulk1-Atg13-Rb1cc1/Fip200 complex is not required for the observed host defense response against Salmonella. Subsequent quantification of GFP-Lc3-Salmonella associations supported the non-essential role of the Ulk1 complex as both controls and atg13 morphants had similar numbers of Lc3-positive infected phagocytes (Figure 5(h-i)). Collectively, these data support the hypothesis that Lc3-Salmonella association is mediated by the Ulk1-independent LAP pathway in our model.
Rubcn plays a major role in the Lc3-mediated anti-Salmonella defense response
RUBCN is a BECN1 partner that negatively regulates canonical autophagy and has recently been identified as a molecular switch between autophagy and LAP [23]. Therefore, we investigated its importance in our model using a knockdown approach of the rubcn gene with two different morpholinos, targeting either the translation start site or an intron-exon boundary. This study revealed a significant role for Rubicon in host defense as Salmonella-infected control larvae had significantly higher survival rates and lower bacterial burdens as compared to both rubcn knockdown groups (Figure 5(j,k and n,o)). Furthermore, rubcn knockdown using either of the two morpholinos significantly reduced the percentage of Lc3-positive infected phagocytes as compared to control embryos (Figure 5(l,m and p,q)). In contrast to the reduction of Lc3-Salmonella associations, we observed no effect of the rubcn morpholinos on GFP-Lc3 levels in uninfected embryos in presence or absence of bafilomycin A1 (Figure S6H-K and H’-K’). The efficacy of rubcn knockdown with splice blocking morpholino was verified by reverse transcriptase PCR (RT-PCR), showing an alternatively spliced transcript and a near complete loss of the control transcript. (Figure S5A). Together, the Atg13 independency and the requirement of Rubcn point towards LAP as a host defense response towards S. Typhimurium infection.
The Lc3 response to phagocytosed Salmonella is strictly NADPH oxidase-dependent
To provide further evidence that Salmonella is targeted by LAP during systemic infection, we investigated the role of the ROS-producing enzyme complex NADPH oxidase, which has been shown to be stabilized on the phagosomal membrane by Rubcn [20]. To this end, we targeted the Cyba/p22phox (cytochrome b-245, alpha polypeptide) subunit of NADPH oxidase, using a published morpholino knockdown approach [71,72]. The knockdown of cyba was validated by RT-PCR (Figure S5B) and we verified that the morpholino did not affect GFP-Lc3 levels of uninfected embryos in the presence or absence of bafilomycin A1 (Figure S6L,M and L’,M’). In S. Typhimurium infection experiments, control embryos showed significantly higher survival rates and were able to curtail bacterial infection better than cyba-deficient embryos (Figure 6(b,c)). In addition, GFP-Lc3 recruitment to Salmonella compartments was almost completely abolished by cyba knockdown (Figure 6(d-e)).
Figure 6.
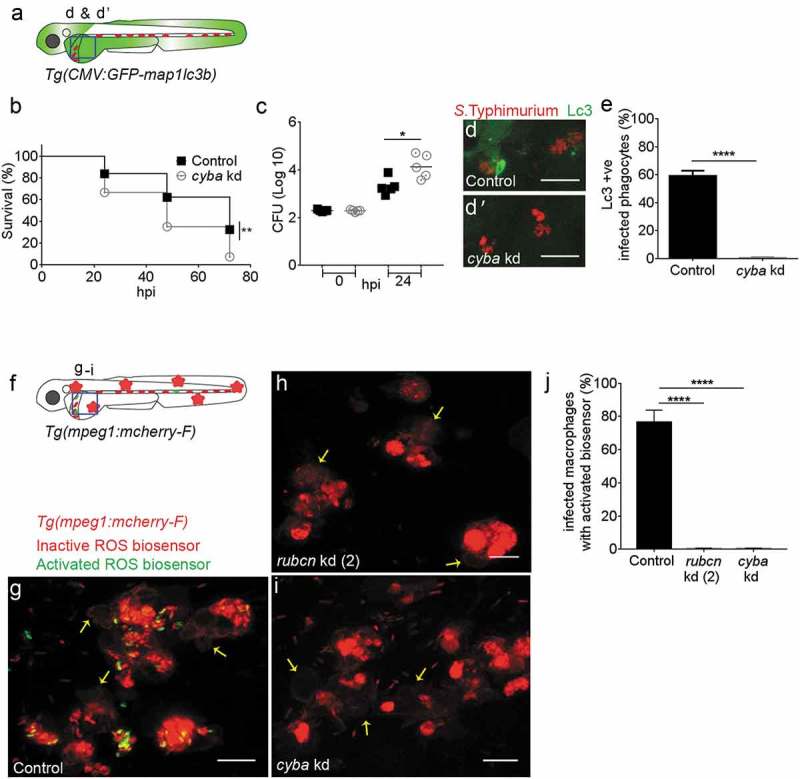
Requirement of NADPH oxidase for Lc3-mediated host defense against Salmonella.
(a) Region of interest (blue square) for confocal image acquisition of infected phagocytes in the blood circulation in d and d’. (b) Survival curves of S. Typhimurium-infected control and cyba knockdown embryos. One representative of three replicates is shown. (c) Representative CFU counts of S. Typhimurium- infected control and cyba knockdown embryos. Five embryos/larvae per time point were used and the log transformed CFU data are shown with the geometric mean per time point. (d) Representative confocal images of control (d) and cyba (d’) knockdown embryos at 4 hpi. (e) Quantification of GFP-Lc3-Salmonella associations in control and cyba knockdown embryos at 4 hpi. Numbers of infected phagocytes positive or negative for GFP-Lc3-Salmonella associations were counted from confocal images and the percentages of Lc3-positive over the total were averaged from five embryos per group. Error bars represent the SD. (f) Region of interest (blue square) for confocal image acquisition of infected phagocytes in the blood circulation of Tg(mpeg1:mCherry-F) embryos in g-i. Red stars symbolize the mCherry-expressing macrophages in the transgenic line used for this experiment. (g-i) Representative confocal images of Tg(mpeg1:mCherry-F)umsF001 embryos at 4 hpi following injection of a Salmonella ROS biosensor strain into control (g), and rubcn kd(2) (h) or cyba morpholino knockdown groups (i). The ROS biosensor strain expresses a constitutive mCherry marker and the GFP signal indicates ROS biosensor activation. Macrophages in the Tg(mpeg1::mCherry-F) line express a farnesylated mCherry protein localizing to membranes (arrows) and therefore the signal is separated from bacterial mCherry signal. (j) Quantification of ROS biosensor activation at 4 hpi in control, rubcn kd(2) and cyba morpholino knockdown groups of Tg(mpeg1::mCherryF) embryos at 4 hpi. Numbers of macrophages showing ROS biosensor activation (GFP and mCherry bacterial signals) or without ROS biosensor activation (mCherry bacterial signal only) were counted from confocal images and the percentages of ROS biosensor-positive over the total were averaged from five embryos per group. Error bars represent the SD. Scale bars: 10 μm. ****P < 0.0001, **P < 0.01, *P < 0.1.
To demonstrate that ingested Salmonella is targeted by intraphagocyte ROS, we used an S. Typhimurium biosensor strain that constitutively expresses mCherry and contains a GFP reporter under the control of the katGp promoter, which is activated when bacterial cells are exposed to ROS from the host [73]. Activation of this ROS biosensor was shown to occur inside mpeg1:mCherry-F labeled macrophages (Figure 6(f,g)), and this activation was strictly dependent on both rubcn (Figure 6(h,j)) and cyba (Figure 6(i,j)). The requirement of rubcn and cyba for activation of the ROS biosensor was confirmed in non-transgenic (AB/TL) zebrafish embryos (Figure S7). Collectively, our results show that Lc3 association with the Salmonella-containing compartments as well as ROS production in the infected cells occurs by a mechanism dependent on Rubcn and NADPH oxidase activity. The essential role of these proteins indicates that LAP is the process that targets Salmonella inside phagocytes, predominantly macrophages, in our model. Furthermore, we conclude that LAP is a host-beneficial response that restricts the intracellular replication of S. Typhimurium.
Discussion
Autophagy and related processes have emerged as important innate immune defense mechanisms against intracellular bacterial infections, and several pathogens, including Salmonella, are known to have evolved mechanisms to counteract the host autophagy response [4–6,8–12,74–76]. Macrophages are the key cell type responsible for dissemination of Salmonella during systemic typhoid disease. Yet, it is not known whether the autophagy machinery of macrophages plays a host protective role or might be exploited by pathogenic Salmonellae. In this study we used zebrafish embryos as a model host for S. Typhimurium to address this question on whole organism level. Using combined in vivo imaging and genetic approaches we demonstrate that macrophages target S. Typhimurium by an autophagy-related process known as LAP, where Lc3 is directly recruited to phagocytosed bacteria in a manner dependent on the activation of ROS production in the Salmonella-containing compartment. This conclusion is supported by data showing that GFP-Lc3-Salmonella association and restriction of bacterial growth required the functions of Atg5, Rubcn, and the Cyba component of NADPH oxidase, while independent of Atg13, a component of the Ulk1 preinitiation complex whose function is limited to canonical autophagy.
In addition to macrophages, neutrophils are also known to play an important role in host defense against Salmonella infection [73,77,78]. However, in our study macrophages stood out as the predominant cell type targeting Salmonella and showed higher phagocytic activity against blood borne as well as sub-cutaneous bacteria. Chemical ablation of macrophages strongly impaired the ability of zebrafish embryos to survive Salmonella infection, whereas neutrophil ablation led to comparatively minor increases in bacterial burden and embryo mortality. In addition, a genetically induced increase in neutrophil numbers at the expense of macrophages was detrimental to host defense against Salmonella infection. Furthermore, the majority of Lc3-Salmonella associations were observed in macrophages. It has previously been shown that S. Typhimurium causes programmed cell death of macrophages in an autophagy-dependent manner [19]. Whether this autophagy-induced macrophage cell death restricts Salmonella replication or promotes pathogenesis is not known. We did not study the effects of autophagy or LAP inhibition on the life span of infected macrophages, and it is well possible that Lc3 activation in our model could be linked with macrophage cell death when these cells reach the stage that they are no longer able to control Salmonella growth. However, it is clear from our data that Lc3 activation is an early response that is dependent on phagosomal ROS production, restricting bacterial replication at least for a limited period of time.
S. Typhimurium may defend itself against the observed defense responses of phagocytes by inhibiting Lc3 activation and NADPH oxidase or by expressing genes involved in detoxification of ROS, such as katG. Consistent with this hypothesis, Lc3 recruitment was not observed in all infected macrophages, and only a subset of bacteria in the infected macrophages expressed a katGp-driven ROS biosensor gene. The heterogeneous activation of this biosensor is consistent with studies in mice, which furthermore suggested that sub-lethal ROS production in resident macrophages allows Salmonella to spread and persist in the infected host [73]. In agreement with this work, we find that deficiency in autophagy genes and NADPH oxidase impairs zebrafish host defense, but autophagy and NADPH oxidase competent hosts are unable to fully control S. Typhimurium replication as 75% of infected individuals eventually succumb to infection. We also found that macrophage ablation led to more severe reduction of zebrafish host resistance than knockdown of autophagy genes or NADPH oxidase. Together, these results suggest that the Lc3/ROS response is important for host defense, but not the only mechanism that macrophages use to control Salmonella infection.
That LAP could be involved in the response of phagocytes against Salmonella infection follows from an early study, in which targeting of LC3 to phagosomes was observed in neutrophils isolated from wild-type but not from Cybb/Nox2−/- mice that lack NADPH oxidase activity [7]. Since then, LAP has been defined as a process wherein ROS production and LC3 recruitment are tightly coupled due to interaction between RUBCN and NADPH oxidase, and that is independent of the ULK1 preinitiation complex [20,23]. Lc3-Salmonella association in our model fulfilled all these criteria, consistent with LAP as the underlying mechanism. In agreement with other studies of RUBCN and LAP in human and mouse models, we found that rubcn depletion in Salmonella-infected zebrafish abolished the host Lc3 response as well as the ROS response [20,23,26]. In contrast, RUBCN knockdown led to enhanced ROS production in human neutrophils during phagocytosis of serum-opsonized zymosan [79]. These different effects of RUBCN can be explained by opposite effects on PtdIns3K activity dependent on different interaction partners, UVRAG (UV radiation resistance associated) or NADPH oxidase [24]. In the study of human neutrophils, it is proposed that binding of RUBCN to UVRAG prevents PtdIns3K activation on the phagosomal membrane, analogous to the described mechanism of autophagy inhibition by RUBCN [25]. In LAP, RUBCN is thought to interact with the NCF4 and CYBA subunits of NADPH oxidase and to stimulate LC3 recruitment by activating PtdIns3K [20,23,26]. We showed that deficiency in three essential components of LAP, Rubcn, Cyba and Atg5, impaired not only the Lc3/ROS responses but also the resistance of the zebrafish host against S. Typhimurium infection. A critical role for LAP in innate immunity was previously demonstrated in Aspergillus fumigatus and Listeria monocytogenes infection experiments using Rubcn-deficient or Rubcn-overexpressing mice [23]. Together, the studies in two different preclinical animal models, zebrafish and mice, highlight LAP as a critical and evolutionary conserved host defense mechanism against both fungal and bacterial infections.
The major role for LAP identified in our study does not exclude that canonical forms of autophagy are also involved in the response of the zebrafish host to S. Typhimurium. In fact, we observed a variety of GFP-Lc3 patterns associated with Salmonella bacteria in infected macrophages. Furthermore, besides bacteria residing in phagosomes, TEM analysis revealed Salmonella in multivesicular compartments and in other vesicles containing cytoplasmic material, suggesting the possible involvement of autophagy. Although this cytoplasmic content could be derived from intraluminal vesicles, it might also be the result of fusion between autophagosomes and phagocytic compartments, which generates amphisomes [80]. It is conceivable that the decoration of phagosomes with Lc3 could promote such fusion processes. We also noted that, at the time point of our analysis (4 hpi), some bacteria were already present freely in the cytosol of the infected phagocytes, and therefore xenophagy, a common process in Salmonella-infected epithelial cells, could become important at later stages of infection following the escape of Salmonella from LAPosomes [5,8–11,13]. Comparison between the cyba and rubcn knockdown effects in our study also provides an indication for different autophagy-related processes to be at play. Inhibition of NADPH oxidase activity by cyba knockdown was equally effective in abolishing ROS biosensor activation as knockdown of rubcn. However, despite that both factors are required for LAP, depleting cyba almost completely abolished GFP-Lc3-Salmonella associations, whereas GFP-Lc3-Salmonella associations were reduced but not absent under rubcn knockdown conditions. Since Rubcn is known to function as an inhibitor of canonical autophagy and an inducer of LAP [20,23,25,26], it is possible that depletion of Rubcn increases the escape of Salmonella from phagosomes and the subsequent targeting of cytosolic bacteria by xenophagy.
In conclusion, our study provides evidence that LAP serves as a host protective mechanism of macrophages limiting systemic infection with Salmonella. Together with the fact that LAP has also been shown to protect against Aspergillus and Listeria infections [23,81], this warrants further investigation into the development of therapeutic strategies promoting LAP as a host-directed approach to infectious disease treatment.
Materials and methods
Zebrafish lines and maintenance
Zebrafish adults and embryos were handled in compliance with local animal welfare regulations and maintained according to standard protocols (zfin.org). Breeding of zebrafish adults was approved by the local animal welfare committee (DEC) of the University of the Leiden, under license number 10612 and in compliance with international guidelines specified by the EU Animal Protective Directive 2010/63/EU. All studies in this work was performed on embryos/larvae before the free feeding stage, no adult fish were sacrificed, and experiments did not fall under animal experimentation law according to the EU Animal Protection Directive 2010/63/EU.
Fish lines used for the present work were wild-type (wt) strain AB/TL, transgenic lines Tg(CMV:GFP-map1lc3b)zf155 [50], Tg(mpeg1:mCherry-F)ump2 [82], Tg(mpx:egfp)i114 [83], Tg(mpeg1:GAL4gl24/UAS-E1b:nfsB-mCherryi149) [84,85] and Tg(mpx:GAL4i222/UAS-E1b:nfsB-mCherryi149) [85,86]. Embryos from adult fish were obtained by natural spawning and were kept at 28.5°C in egg water (Sera Marin salt, 05420; 60 μg/ml sera marin in distilled deionized water,). PTU (Sigma Aldrich, P7629;0.003% solution of 1-phenyl-2thiourea in PBS (0.8 NaCl%, 0.02% KCl, 0.02 M PO4, in distilled deionized water, pH 7.3)) was added to egg water to prevent melanization of embryos. For infection delivery and live imaging experiments embryos were anaesthetized in egg water with 0.02% buffered Tricaine (Sigma Aldrich, A-5040; 400 mg of 3-aminobenzoic acid ethyl ester in 97.9 ml of distilled deionized water, pH adjusted ~7 with 1 M Tris [pH 9]).
Bacterial cultures and infection experiments
Salmonella Typhimurium strains used for this study included wild type (wt) strain SL1344 [87] constitutively expressing mCherry, and a ROS biosensor strain (SL1344 sifBp::mCherry/pkatGp-gfpOVA) [73] constitutively expressing mCherry and expressing an unstable GFP variant (GFP-OVA) under an OxyR-activated promoter upon exposure to ROS from the host. The bacterial strains were plated from −80 stocks over LB agar plates (Sigma Aldrich, L2897) with appropriate antibiotics and were left overnight to grow at 37°C. Before the start of infection experiments colonies from LB agar plates were suspended into phosphate buffered saline (PBS) supplemented with polyvinylpyrrolidone-40 (Sigma Aldrich, PVP40; 2% in PBS) to obtain the low dose (200–400 CFU, for survival curves and CFU counts experiments) or high dose (2000–4000 CFU, for imaging experiments). Bacterial inoculum was injected systemically into the caudal vein of the anaesthetized embryos at 2 dpf or the local infections were established with infection delivery under the skin layers of a somite in the tail region. To check the inoculum size, the same dose was spotted onto agar plates, and bacterial counts determined following overnight incubation. After infection, embryos were kept individually in egg water in 48-well plates (Sigma Aldrich, SIAL0548) to score survival during larval development up to 5 dpf and to collect individuals for CFU counts at 24 h intervals.
Determination of in vivo bacterial (CFU) counts
Five embryos/larvae per time point were sacrificed and homogenized in PBS using the Bullet Blender Tissue Homogenizer (Next Advance Inc., USA). Homogenates were then serially diluted, and three technical replicates for each embryo/larva were plated on LB solid media with the appropriate antibiotics for S. Typhimurium. To determine the CFUs, the resulting colonies were counted manually after 24 h incubation at 37°C.
Gene expression analysis
Total RNA was extracted using TRIzol reagent (Thermo Fisher Scientific/Invitrogen, 15596026) according to the manufacturer’s instructions and extracted with RNeasy Min Elute Clean up kit (QIAGEN, 154015861,). Reverse transcription reaction was performed using iScript cDNA synthesis kit (Bio-Rad, 170–8891) with 0.5 µg of total RNA as input. The mRNA expression levels were determined by quantitative real-time PCR using iQSYBR Green Supermix (Bio-Rad, 170–8882) and Single color Real-Time PCR Detection System (Bio-Rad, U.S) as previously described [39]. The following primers were used to detect the expression level of tbp (TATA-box binding protein; reference gene), forward: CCTGCCCATTTCAGTC and reverse: TGTTGTTGCCTCTGTTGCTC; il1b (interleukin 1b), forward: TGTGTGTTTGGGAATCTCCA and reverse: TGATAAACCAACCGGGACA; tnfa (tumor necrosis factor a), forward: CAAAGACACCTGGCTGTAGAC and reverse: AGACCTTAGACGAGAGATGAC; mmp9 (matrix metallopeptidase 9), forward: CATTAAAGATGCCCTGATGTATCCC and reverse: AGTGGTGGTCCGTGGTTGAG.
Morpholino knockdowns, RT-PCR and pharmacological verification
Morpholino oligonucleotides (Gene Tools) were diluted in Danieau buffer (58 mM NaCl, 0.7 mM KCl, 0.4 mM MgSO4, 0.6 mM Ca(NO3)2, 5.0 mM HEPES; pH 7.6) to obtain the required concentrations. 1 nl volume of morpholino was injected into the yolk of 0 hours post fertilization (hpf) zebrafish embryos with microneedles and a Femtojet injector (Eppendorf, Germany) paired with a stereo-microscope (Leica, Germany). All morpholinos were used at concentrations that caused no developmental aberrations and did not affect survival compared with uninjected controls. As control 1 nl of the standard control morpholino by Gene Tools was injected in same concentration as the other morpholinos.
Knockdown of rubcn was achieved with a translation blocking morpholino (MO1-rubcn, ATCTTGATCCTCAGGTAATGCAGGT at 1 mM) and a splice blocking morpholino (MO2-rubcn, CGCTGTGAAATCTGCTGACCTGAGC at 0.25 mM). Knockdown of rubcn with MO2-rubcn was verified by RT-PCR with a pair of primers flanking the e6i6 boundary, Forward: TCTTATCAGCGCAGCTCAAAC and Reverse: GTGAAAATGGACCACAGCTCTT (Figure S5). Knockdown of cyba was performed using a published splice blocking morpholino (ATCATAGCATGTAAGGATACATCCC at 1 mM) [71] and verified by RT-PCR using described primer sequences (Forward: ATGGCGAAGATTGAGTGGGCGAT and Reverse: GCTGCAGCATGGAGGTTATCTGCT) [72]. Knockdown of irf8 was performed using a published splice blocking morpholino (AATGTTTCGCTTACTTTGAAAATGG at 1 mM) [63] and efficacy was verified by fluorescence imaging of macrophages and neutrophils (Figure S4).
Morpholino knockdown of atg5 (CATCCTTGTCATCTGCCATTATCAT and atg13 was achieved using published translation blocking morpholinos (atg5: CATCCTTGTCATCTGCCATTATCAT at 0.5 mM; atg13: GGCTCAGATCACTATCCATTGTCGC at 1 mM) [68,88] and was verified with functional analysis by quantification of GFP-Lc3 levels after incubation with bafilomycin A1 (Sigma Aldrich, B1793-10UG). Control and atg5 or atg13 knockdown embryos (n = 24) were bath treated with 100 μM bafilomycin A1 in egg water for 12 hours starting at 48 hpf. In parallel, control and atg5 or atg13 knockdown embryos were bathed only in egg water (without bafilomycin A1) as negative control groups of the treatment. Embryos from all groups were quickly washed with egg water and were immediately fixed in 4% PFA (Thermo Scientific, LE146786) for 24 hours. Subsequently 5 embryos per group were randomly selected for imaging in the tail fin region to detect reduction of GFP-Lc3 levels using confocal microscopy (Figure S6). The same procedure was applied to verify that the morpholinos for rubcn, irf8 and cyba did not affect GFP-Lc3 levels.
Macrophage and neutrophil ablation with the metronidazole/nitroreductase system
Zebrafish embryos from heterozygous transgenic lines Tg(mpeg1:GAL4/UAS-E1b:nfsB-mCherry) [84,85] and Tg(mpx:GAL4/UAS-E1b:nfsB-mCherry) [85,86] were screened at 24 hpf under a stereo fluorescence microscope for signal of the nitroreductase-mCherry fusion protein in macrophages (driven by mpeg1:GAL4) or neutrophils (driven by mpx:GAL4). Embryos with strong fluorescent signals expressed by the majority of their macrophages or neutrophils were selected and are referred to as mpeg1:unm+ and mpx:unm+, respectively. The embryos with complete absence of fluorescently-tagged macrophages or neutrophils were used as controls, referred to as mpeg1:unm- and mpx:unm-. The screened larvae of all groups, mpeg1:unm+, mpx:unm+, mpeg1:unm- and mpx:unm-, were further divided into two subgroups, one for treatment with metronidazole (Sigma Aldrich, M3761) and one to serve as untreated control. Metronidazole solution was freshly prepared just before the treatment at 5 mM strength in 0.2% DMSO in egg water [61]. For the macrophage ablation experiments, dechorionated 33 hpf old embryos of mpeg1:unm+ and mpeg1:unm- were incubated in metronidazole solution for 15 h in the dark at 28°C, and subsequently infected with S. Typhimurium at 48 hpf. Control groups were similarly treated with 0.2% DMSO in egg water. For the neutrophil ablation experiments, the same treatment was applied on mpx:unm+ and mpx:unm- embryos, but, for achieving complete neutrophil ablation, an additional 24 h treatment with metronidazole solution was applied following S. Typhimurium infection. Macrophage and neutrophil ablations were verified post treatment under a stereo fluorescence microscope (Figure S4A-D).
Whole mount immunohistochemistry and TSA staining
Identification of macrophages and neutrophils was facilitated by immunolabeling with leukocyte specific anti-Lcp1 antibody (paired with secondary antibody staining with Alexa Fluor 405, combined with neutrophil specific Tyramide Signal Amplification (TSA) paired with Cyanine-5 (PerkinElmer,01072016). Embryos were fixed at 4 hpi in 4% paraformaldehyde (PFA) in PBS-TX (1X PBS supplemented with 0.8% of Triton X-100 (Sigma Aldrich, X100) overnight and proceeded for anti-Lcp1 antibody staining and TSA as previously described [58].
Imaging and image analysis
Stereo Fluorescence images (Figure S4) were acquired with an MZ16FA microscope (Leica, Germany) equipped with a DFC420C digital color camera (Leica, Germany). Confocal laser scanning images and time lapse movies were acquired using a 63x water immersion objective (NA 1.2) with a Leica (TCS SPE system, Germany), (micrographs in Figure 1, 2, 5 and 6; Figsure S6 and S7, Movies M1 and M2) or a 63x oil immersion objective (NA 1.4) Zeiss Observer (6.5.32, Germany), (micrographs in Figure 3 and Figure S4).
For live imaging and recording of time lapses, embryos were mounted in 1% low-melting-point agarose (Sphaero Q,) and in the case of fixed sample imaging, embryos were fixed in 4% PFA and washed with PBS before image acquisitions. GFP-Lc3-Salmonella associations were determined by imaging the region of the tail just behind the yolk extension. Images were analyzed through Z-stacks in Leica LAS AF Lite software and bacterial clusters were observed and manually counted in the overlay channel. Max projections in the overlay channels were used for representative images. For images acquired under the Zeiss Observer, the Z-stack projection function of Fiji software at maximum intensity was used to make composite images of different channels acquired.
For quantification of GFP-Lc3 recruitment within infected phagocytes, for each embryo, the total number of observable phagocytes were manually counted through the Z-stacks of the acquired confocal image. Phagocytes were identified in the yolk sac circulation valley by bacterial clusters in the mCherry channel and cellular boundaries of phagocytes were determined in the light transmission channel. Among these total observable infected phagocytes, the numbers of cells with GFP-Lc3 signal in association with Salmonella bacteria were counted and the percentage of Lc3-positive phagocytes over the total observable phagocytes was determined for each embryo. The same approach was used to quantify the percentage of phagocytes showing activation of the Salmonella ROS biosensor strain.
For bafilomycin A1 treatment experiments, embryos from all four groups were washed with 1XPBS-Tx and imaged in the caudal fin region using a 63x water immersion objective (NA 1.2) with a Leica TCS SPE system. Five embryos per group were imaged in three separate replicates and GFP-Lc3 signal was quantified as fluorescent pixels with a described pixel count program [89] for five embryos per group in three replicates.
For transmission electron microscopy embryos were fixed in 2% glutaraldehyde (Polysciences, 00376–500) and 2% paraformaldehyde in sodium cacodylate buffer pH 7.2 (Agar scientific, R1103) at 4 hpi for 3 hours at room temperature followed by fixation at 4°C. Post-fixation was performed in 1% osmium tetraoxide (SPI supplies, 02604-AB) in sodium cacodylate buffer for 1 h at room temperature. After dehydration through a graded series of ethanol, specimens were kept in epoxy resin (Agar scientific, AGR 1043) for 16 h before embedding. Ultrathin sections were collected on Formvar coated 200 mesh or one hole copper grids (Agar scientific, AGS 162) stained with 2% uranyl acetate (BDH) in 50% ethanol and 0.2% lead citrate (BDH) for 10 min each. Electron microscopy images were acquired with a JEOL JEM-1010 (Japan) transmission electron microscope equipped with an Olympus Megaview camera (Japan). Transmission electron micrographs were viewed and cropped in Fiji software.
Statistics
All data sets were analyzed with Prism 7 software. Survival curves were analyzed with Log rank (Mantel-Cox) test. For CFU counts, one way ANOVA was performed on Log transformed data and was corrected for multiple comparisons using Sidak’s multiple comparisons test when required. Percentage GFP-Lc3-positive biosensor-positive phagocyte quantifications and pixel count means were analyzed for significance with unpaired parametric t-test between two groups, and for multiple groups the one way ANOVA test was performed and corrected for multiple comparisons.
Funding Statement
This work was supported by the Higher Education Commission of Pakistan and the Bahaudin Zakriya University, Multan with a fellowship to S.M.; the 7th Framework Programme of the European Commission under grants PIEF-GA-2013-625975 and PITN-GA-2011-289209; and the Netherlands Organization for Scientific Research (NWO) Domain Applied and Engineering Sciences under grant 13259.
Acknowledgements
We thank Dirk Bumann (University of Basel) for sharing of the Salmonella strains used in this study, Dan Klionsky (University of Michigan), Georges Lutfalla (University of Montpellier) Steve Renshaw (University of Sheffield) and Graham Lieschke (Monash University) for zebrafish lines, Yi Feng (University of Edinburgh) for anti-Lcp1 antibody, Annette Vergunst (INSERM, Nimes) for the rubcn translation blocking morpholino (MO1-rubcn), and Anna-Pavlina Haramis (Institute of Biology Leiden) for the atg5 morpholino. We are grateful to all members of the fish facility team for zebrafish care.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Fabrega A, Vila J.. Salmonella enterica serovar typhimurium skills to succeed in the host: virulence and regulation. Clin Microbiol Rev. 2013;26(2):308–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Finlay BB, Brumell JH.. Salmonella interactions with host cells: in vitro to in vivo. Philos Trans R Soc Lond B Biol Sci. 2000;355(1397):623–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Watson KG, Holden DW. Dynamics of growth and dissemination of salmonella in vivo. Cell Microbiol. 2010;12(10):1389–1397. [DOI] [PubMed] [Google Scholar]
- [4].Huang J, Brumell JH. Bacteria-autophagy interplay: a battle for survival. Nat Rev Microbiol. 2014;12(2):101–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Birmingham CL, Smith AC, Bakowski MA, et al. Autophagy controls salmonella infection in response to damage to the salmonella-containing vacuole. J Biol Chem. 2006;281(16):11374–11383. [DOI] [PubMed] [Google Scholar]
- [6].Cemma M, Kim PK, Brumell JH. The ubiquitin-binding adaptor proteins p62/SQSTM1 and NDP52 are recruited independently to bacteria-associated microdomains to target Salmonella to the autophagy pathway. Autophagy. 2011;7(3):341–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Huang J, Canadien V, Lam GY, et al. Activation of antibacterial autophagy by NADPH oxidases. Proc Natl Acad Sci U S A. 2009;106(15):6226–6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Thurston TL, Wandel MP, von Muhlinen N, et al. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature. 2012;482(7385):414–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Thurston TL, Ryzhakov G, Bloor S, et al. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol. 2009;10(11):1215–1221. [DOI] [PubMed] [Google Scholar]
- [10].Wild P, Farhan H, McEwan DG, et al. Phosphorylation of the autophagy receptor optineurin restricts salmonella growth. Science. 2011;333(6039):228–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zheng YT, Shahnazari S, Brech A, et al. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J Immunol. 2009;183(9):5909–5916. [DOI] [PubMed] [Google Scholar]
- [12].Thurston TL, Matthews SA, Jennings E, et al. Growth inhibition of cytosolic salmonella by caspase-1 and caspase-11 precedes host cell death. Nat Commun. 2016;7:13292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Shahnazari S, Namolovan A, Mogridge J, et al. Bacterial toxins can inhibit host cell autophagy through cAMP generation. Autophagy. 2011;7(9):957–965. [DOI] [PubMed] [Google Scholar]
- [14].Kageyama S, Omori H, Saitoh T, et al. The LC3 recruitment mechanism is separate from Atg9L1-dependent membrane formation in the autophagic response against Salmonella. Mol Biol Cell. 2011;22(13):2290–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lopez-Montero N, Ramos-Marquès E, Risco C, et al. Intracellular salmonella induces aggrephagy of host endomembranes in persistent infections. Autophagy. 2016;12(10):1886–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Sanjuan MA, Dillon CP, Tait SW. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450(7173):1253–1257. [DOI] [PubMed] [Google Scholar]
- [17].Galluzzi L, Baehrecke EH, Ballabio A, et al. Molecular definitions of autophagy and related processes. Embo J. 2017;36(13):1811–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Cunha LD, Martinez J.. Autophagy and lc3-associated phagocytosis mediate the innate immune response In: Hayat MA, editor. Autophagy: cancer, other pathologies, inflammation, immunity, infection, and aging. Cambridge (MA): Academic Press; 2017. p. 303–319. [Google Scholar]
- [19].Hernandez LD, Pypaert M, Flavell RA, et al. A Salmonella protein causes macrophage cell death by inducing autophagy. J Cell Biol. 2003;163(5):1123–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Yang CS, Lee J-S, Rodgers M, et al. Autophagy protein Rubicon mediates phagocytic NADPH oxidase activation in response to microbial infection or TLR stimulation. Cell Host Microbe. 2012;11(3):264–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Jones LB, McGrogan P, Flood TJ, et al. Special article: chronic granulomatous disease in the United Kingdom and Ireland: a comprehensive national patient-based registry. Clin Exp Immunol. 2008;152(2):211–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Martire B, Rondelli R, Soresina A, et al. Clinical features, long-term follow-up and outcome of a large cohort of patients with chronic granulomatous disease: an Italian multicenter study. Clin Immunol. 2008;126(2):155–164. [DOI] [PubMed] [Google Scholar]
- [23].Martinez J, Malireddi RKS, Lu Q, et al. Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins. Nat Cell Biol. 2015;17(7):893–906. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [24].Wong SW, Sil P, Martinez J. Rubicon: LC3-associated phagocytosis and beyond. Febs j. 2018;285(8):1379–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Matsunaga K, Saitoh T, Tabata K, et al. Two beclin 1-binding proteins, Atg14L and rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11(4):385–396. [DOI] [PubMed] [Google Scholar]
- [26].Martinez J, Almendinger J, Oberst A, et al. Microtubule-associated protein 1 light chain 3 alpha (LC3)-associated phagocytosis is required for the efficient clearance of dead cells. Proc Natl Acad Sci U S A. 2011;108(42):17396–17401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kim J, Kundu M, Viollet B, et al. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13(2):132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Gong L, Cullinane M, Treerat P, et al. The burkholderia pseudomallei type III secretion system and BopA are required for evasion of LC3-associated phagocytosis. PLoS One. 2011;6(3):e17852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Hubber A, Kubori T, Coban C, et al. Bacterial secretion system skews the fate of Legionella-containing vacuoles towards LC3-associated phagocytosis. Sci Rep. 2017;7:44795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lam GY, Cemma M, Muise AM, et al. Host and bacterial factors that regulate LC3 recruitment to listeria monocytogenes during the early stages of macrophage infection. Autophagy. 2013;9(7):985–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ligeon LA, Moreau K, Barois N, et al. Role of VAMP3 and VAMP7 in the commitment of yersinia pseudotuberculosis to LC3-associated pathways involving single- or double-membrane vacuoles. Autophagy. 2014;10(9):1588–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ma J, Becker C, Lowell CA, et al. Dectin-1-triggered recruitment of light chain 3 protein to phagosomes facilitates major histocompatibility complex class II presentation of fungal-derived antigens. J Biol Chem. 2012;287(41):34149–34156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Romao S, Gasser N, Becker AC, et al. Autophagy proteins stabilize pathogen-containing phagosomes for prolonged MHC II antigen processing. J Cell Biol. 2013;203(5):757–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Florey O, Kim SE, Sandoval CP, et al. Autophagy machinery mediates macroendocytic processing and entotic cell death by targeting single membranes. Nat Cell Biol. 2011;13(11):1335–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Martinez J, Cunha LD, Park S, et al. Noncanonical autophagy inhibits the autoinflammatory, lupus-like response to dying cells. Nature. 2016;533(7601):115–119. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [36].van der Sar AM, Musters RJP, van Eeden FJM, et al. Zebrafish embryos as a model host for the real time analysis of salmonella typhimurium infections. Cell Microbiol. 2003;5(9):601–611. [DOI] [PubMed] [Google Scholar]
- [37].Benard EL, van der Sar AM, Ellett F, et al. Infection of zebrafish embryos with intracellular bacterial pathogens. J Vis Exp. 2012;(61):e3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].van der Sar AM, Stockhammer OW, van der Laan C, et al. MyD88 innate immune function in a zebrafish embryo infection model. Infect Immun. 2006;74(4):2436–2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Stockhammer OW, Zakrzewska A, Hegedûs Z, et al. Transcriptome profiling and functional analyses of the zebrafish embryonic innate immune response to Salmonella infection. J Immunol. 2009;182(9):5641–5653. [DOI] [PubMed] [Google Scholar]
- [40].Stockhammer OW, Rauwerda H, Wittink FR, et al. Transcriptome analysis of Traf6 function in the innate immune response of zebrafish embryos. Mol Immunol. 2010;48(1–3):179–190. [DOI] [PubMed] [Google Scholar]
- [41].van der Vaart M, van Soest JJ, Spaink HP, et al. Functional analysis of a zebrafish myd88 mutant identifies key transcriptional components of the innate immune system. Dis Model Mech. 2013;6(3):841–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Tobin DM, May RC, Wheeler RT. Zebrafish: a see-through host and a fluorescent toolbox to probe host-pathogen interaction. PLoS Pathog. 2012;8(1):e1002349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Torraca V, Masud S, Spaink HP, et al. Macrophage-pathogen interactions in infectious diseases: new therapeutic insights from the zebrafish host model. Dis Model Mech. 2014;7(7):785–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Yoshida N, Frickel E-M, Mostowy S. Macrophage-microbe Interactions: lessons from the zebrafish model. Front Immunol. 2017;8:1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Mostowy S, Boucontet L, Mazon Moya MJ, et al. The zebrafish as a new model for the in vivo study of shigella flexneri interaction with phagocytes and bacterial autophagy. PLoS Pathog. 2013;9(9):e1003588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].van der Vaart M, Korbee CJ, Lamers GEM, et al. The DNA damage-regulated autophagy modulator DRAM1 links mycobacterial recognition via TLR-MYD88 to autophagic defense [corrected]. Cell Host Microbe. 2014;15(6):753–767. [DOI] [PubMed] [Google Scholar]
- [47].Mathai BJ, Meijer AH, Simonsen A. Studying autophagy in zebrafish. Cells. 2017;6(3):E21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Hosseini R, Lamers GE, Hodzic Z, et al. Correlative light and electron microscopy imaging of autophagy in a zebrafish infection model. Autophagy. 2014;10(10):1844–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140(3):313–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].He C, Bartholomew CR, Zhou W, et al. Assaying autophagic activity in transgenic GFP-Lc3 and GFP-gabarap zebrafish embryos. Autophagy. 2009;5(4):520–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Ordas A, Hegedus Z, Henkel CV, et al. Deep sequencing of the innate immune transcriptomic response of zebrafish embryos to salmonella infection. Fish Shellfish Immunol. 2011;31(5):716–724. [DOI] [PubMed] [Google Scholar]
- [52].Ordas A, Pereira DM, Amaral JD, et al. MicroRNA-146 function in the innate immune transcriptome response of zebrafish embryos to salmonella typhimurium infection. BMC Genomics. 2013;14:696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Kanwal Z, Zakrzewska A, Den Hertog J, et al. Deficiency in hematopoietic phosphatase ptpn6/Shp1 hyperactivates the innate immune system and impairs control of bacterial infections in zebrafish embryos. J Immunol. 2013;190(4):1631–1645. [DOI] [PubMed] [Google Scholar]
- [54].Rajashekar R, Liebl D, Chikkaballi D, et al. Live cell imaging reveals novel functions of salmonella enterica SPI2-T3SS effector proteins in remodeling of the host cell endosomal system. PLoS One. 2014;9(12):e115423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Le Guyader D, Redd MJ, Colucci-Guyon E, et al. Origins and unconventional behavior of neutrophils in developing zebrafish. Blood. 2008;111(1):132–141. [DOI] [PubMed] [Google Scholar]
- [56].Cui C, Benard EL, Kanwal Z. Infectious disease modeling and innate immune function in zebrafish embryos. Methods Cell Biol. 2011;105:273–308. [DOI] [PubMed] [Google Scholar]
- [57].Elks PM, Brizee S, van der Vaart M, et al. Hypoxia inducible factor signaling modulates susceptibility to mycobacterial infection via a nitric oxide dependent mechanism. PLoS Pathog. 2013;9(12):e1003789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Loynes CA, Martin JS, Robertson A, et al. Pivotal advance: pharmacological manipulation of inflammation resolution during spontaneously resolving tissue neutrophilia in the zebrafish. J Leukoc Biol. 2010;87(2):203–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Colucci-Guyon E, Tinevez JY, Renshaw SA, et al. Strategies of professional phagocytes in vivo: unlike macrophages, neutrophils engulf only surface-associated microbes. J Cell Sci. 2011;124(Pt 18):3053–3059. [DOI] [PubMed] [Google Scholar]
- [60].Curado S, Stainier DY, Anderson RM. Nitroreductase-mediated cell/tissue ablation in zebrafish: a spatially and temporally controlled ablation method with applications in developmental and regeneration studies. Nat Protoc. 2008;3(6):948–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Mesureur J, Feliciano JR, Wagner N, et al. Macrophages, but not neutrophils, are critical for proliferation of burkholderia cenocepacia and ensuing host-damaging inflammation. PLoS Pathog. 2017;13(6):e1006437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Prajsnar TK, Hamilton R, Garcia-Lara J, et al. A privileged intraphagocyte niche is responsible for disseminated infection of Staphylococcus aureus in a zebrafish model. Cell Microbiol. 2012;14(10):1600–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Li L, Jin H, Xu J, et al. Irf8 regulates macrophage versus neutrophil fate during zebrafish primitive myelopoiesis. Blood. 2011;117(4):1359–1369. [DOI] [PubMed] [Google Scholar]
- [64].Pagan AJ, Yang C-T, Cameron J, et al. Myeloid growth factors promote resistance to mycobacterial infection by curtailing granuloma necrosis through macrophage replenishment. Cell Host Microbe. 2015;18(1):15–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Mazon-Moya MJ, Willis AR, Torraca V, et al. Septins restrict inflammation and protect zebrafish larvae from shigella infection. PLoS Pathog. 2017;13(6):e1006467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Gupta M, Shin D-M, Ramakrishna L, et al. IRF8 directs stress-induced autophagy in macrophages and promotes clearance of Listeria monocytogenes. Nat Commun. 2015;6:6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Walczak M, Martens S. Dissecting the role of the Atg12-Atg5-Atg16 complex during autophagosome formation. Autophagy. 2013;9(3):424–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Hu Z, Zhang J, Zhang Q. Expression pattern and functions of autophagy-related gene atg5 in zebrafish organogenesis. Autophagy. 2011;7(12):1514–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Mans LA, Querol Cano L, van Pelt J, et al. The tumor suppressor LKB1 regulates starvation-induced autophagy under systemic metabolic stress. Sci Rep. 2017;7(1):7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Mizushima N, Noda T, Yoshimori T, et al. A protein conjugation system essential for autophagy. Nature. 1998;395(6700):395–398. [DOI] [PubMed] [Google Scholar]
- [71].Niethammer P, Grabher C, Look AT, et al. A tissue-scale gradient of hydrogen peroxide mediates rapid wound detection in zebrafish. Nature. 2009;459(7249):996–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Tauzin S, Starnes TW, Becker FB, et al. Redox and Src family kinase signaling control leukocyte wound attraction and neutrophil reverse migration. J Cell Biol. 2014;207(5):589–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Burton NA, Schürmann N, Casse O, et al. Disparate impact of oxidative host defenses determines the fate of salmonella during systemic infection in mice. Cell Host Microbe. 2014;15(1):72–83. [DOI] [PubMed] [Google Scholar]
- [74].Yu HB, Croxen MA, Marchiando AM, et al. Autophagy facilitates salmonella replication in hela cells. MBio. 2014;5(2):e00865–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Jia K, Thomas C, Akbar M, et al. Autophagy genes protect against salmonella typhimurium infection and mediate insulin signaling-regulated pathogen resistance. Proc Natl Acad Sci U S A. 2009;106(34):14564–14569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Riquelme S, Varas M, Valenzuela C, et al. Relevant genes linked to virulence are required for salmonella typhimurium to survive intracellularly in the social amoeba dictyostelium discoideum. Front Microbiol. 2016;7:1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Tyrkalska SD, Candel S, Angosto D, et al. Neutrophils mediate salmonella typhimurium clearance through the GBP4 inflammasome-dependent production of prostaglandins. Nat Commun. 2016;7:12077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Chen KW, Groß CJ, Sotomayor FV, et al. The neutrophil NLRC4 inflammasome selectively promotes IL-1beta maturation without pyroptosis during acute salmonella challenge. Cell Rep. 2014;8(2):570–582. [DOI] [PubMed] [Google Scholar]
- [79].Song ZM, Bouchab L, Hudik E, et al. Phosphoinositol 3-phosphate acts as a timer for reactive oxygen species production in the phagosome. J Leukoc Biol. 2017;101(5):1155–1168. [DOI] [PubMed] [Google Scholar]
- [80].Fader CM, Colombo MI. Autophagy and multivesicular bodies: two closely related partners. Cell Death Differ. 2009;16(1):70–78. [DOI] [PubMed] [Google Scholar]
- [81].Gluschko A, Herb M, Wiegmann K, et al. The beta2 integrin Mac-1 induces protective LC3-associated phagocytosis of listeria monocytogenes. Cell Host Microbe. 2018;23(3):324–337.e5. [DOI] [PubMed] [Google Scholar]
- [82].Bernut A, Herrmann J-L, Kissa K, et al. Mycobacterium abscessus cording prevents phagocytosis and promotes abscess formation. Proc Natl Acad Sci U S A. 2014;111(10):E943–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Renshaw SA, Loynes CA, Trushell DMI, et al. A transgenic zebrafish model of neutrophilic inflammation. Blood. 2006;108(13):3976–3978. [DOI] [PubMed] [Google Scholar]
- [84].Ellett F, Pase L, Hayman JW, et al. mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood. 2011;117(4):e49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Gray C, Loynes CA, Whyte MKB, et al. Simultaneous intravital imaging of macrophage and neutrophil behaviour during inflammation using a novel transgenic zebrafish. Thromb Haemost. 2011;105(5):811–819. [DOI] [PubMed] [Google Scholar]
- [86].Ellett F, Elks PM, Robertson AL, et al. Defining the phenotype of neutrophils following reverse migration in zebrafish. J Leukoc Biol. 2015;98(6):975–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Hoiseth SK, Stocker BA. Aromatic-dependent salmonella typhimurium are non-virulent and effective as live vaccines. Nature. 1981;291(5812):238–239. [DOI] [PubMed] [Google Scholar]
- [88].Musso G, Tasan M, Mosimann C, et al. Novel cardiovascular gene functions revealed via systematic phenotype prediction in zebrafish. Development. 2014;141(1):224–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Stoop EJ, Schipper T, Rosendahl Huber SK, et al. Zebrafish embryo screen for mycobacterial genes involved in the initiation of granuloma formation reveals a newly identified ESX-1 component. Dis Model Mech. 2011;4(4):526–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


