Abstract
The transcriptional cofactor nascent polypeptide-associated complex and co-regulator α (NACA) regulates osteoblast maturation and activity. NACA functions, at least in part, by binding to Jun proto-oncogene, AP-1 transcription factor subunit (cJUN) and potentiating the transactivation of AP-1 targets such as osteocalcin (Bglap) and matrix metallopeptidase 9 (Mmp9). NACA activity is modulated by phosphorylation carried out by several kinases, but a phosphatase regulating NACA's activity remains to be identified. Here, we used affinity purification with MS in HEK293T cells to isolate NACA complexes and identified protein phosphatase 1 catalytic subunit α (PP1A) as a NACA-associated Ser/Thr phosphatase. NACA interacted with multiple components of the PP1A holoenzyme complex: the PPP1CA catalytic subunit and the regulatory subunits PPP1R9B, PPP1R12A and PPP1R18. MS analysis revealed that NACA co-expression with PPP1CA causes dephosphorylation of NACA at Thr-89, Ser-151, and Thr-174. NACA Ser/Thr-to-alanine variants displayed increased nuclear localization, and NACA dephosphorylation was associated with specific recruitment of novel NACA interactants, such as basic transcription factor 3 (BTF3) and its homolog BTF3L4. NACA and PP1A cooperatively potentiated cJUN transcriptional activity of the AP-1–responsive MMP9-luciferase reporter, which was abolished when Thr-89, Ser-151, or Thr-174 were substituted with phosphomimetic aspartate residues. We confirmed the NACA–PP1A interaction in MC3T3-E1 osteoblastic cells and observed that NACA phosphorylation status at PP1A-sensitive sites is important for the regulation of AP-1 pathway genes and for osteogenic differentiation and matrix mineralization. These results suggest that PP1A dephosphorylates NACA at specific residues, impacting cJUN transcriptional activity and osteoblast differentiation and function.
Keywords: phosphatase; c-Jun transcription factor; AP1 transcription factor (AP-1); cell signaling; cell differentiation; αNAC; nascent polypeptide-associated complex and co-regulator alpha (NACA); osteoblast; PP1A; Protein phosphatase 1 catalytic subunit alpha (PP1A); JUN, transcription
Introduction
Activator protein-1 (AP-1) is a dimeric transcription factor involved in the regulation of multiple cellular processes such as proliferation, differentiation and apoptosis (1–4). The AP-1 complex consists of members of the JUN (e.g. cJUN gene symbol: JUN, JUND) and FOS (e.g. FOSB, FRA-1) family of basic leucine zipper proteins, in several interfamilial combinations of JUN/JUN homodimers or JUN/FOS heterodimers (1–4). AP-1 characteristically mediates immediate early transcriptional responses to extracellular stimuli such as growth factors and cytokines by binding to sequence-specific cis-acting response elements in gene promotors (1–4). AP-1 factors canonically bind to the 12-O-tetradecanoylphorbol-13-acetate (TPA)–response element (TRE; TGA G/C TCA) leading to the activation of transcription (5, 6). The activity of AP-1 factors is enhanced by synergistic cooperation with general transcription factors and transcriptional coactivators (7, 8).
Nascent polypeptide-associated complex and co-regulator α (NACA, αNAC)3 is a 215–amino acid transcriptional cofactor involved in the regulation of AP-1 transcription (9–11). NACA was originally cloned as the α chain of the nascent polypeptide-associated complex (NAC), responsible for the sorting and translocation of polypeptides from the ribosome (12). NACA contains a DNA-binding domain, a calcium-binding EF-hand motif, and a NAC domain (13–15). The NAC domain, a region sharing homology with basic transcription factor 3 (BTF3), contains three α helices and six β-sheets, which are critical for nuclear localization and DNA binding (13, 14). NACA can bind to cJUN and stabilize the interaction of cJUN homodimers with chromatin, thereby enhancing AP-1 transcriptional activity (9, 10, 16). In addition to cJUN, NACA can also coactivate the activity of JUND (17). Physiologically, in osteoblast cells, we have previously shown that coactivation of AP-1–dependent transcription by NACA is important for the regulation of Type I collagen (Col1a1), Mmp9, osteocalcin (Bglap), and Lrp6 gene expression during differentiation (9, 11, 16–19). Furthermore, NACA also associates with TATA-binding protein and is thus hypothesized to act as a bridge between cJUN homodimers and the basal transcriptional machinery on target promoters (11).
An important mechanism for the regulation of protein function is the posttranslational modification of threonine, serine, and tyrosine residues by phosphorylation (20, 21). Phosphorylation events are fast and reversible, thus allowing for temporal modulation of protein activity. Phosphorylation of a single residue can alter protein localization, target proteins for degradation, enhance or inhibit protein–protein interactions, modify protein conformations, or stimulate or repress enzymatic activity (21–24). Despite there being more than 500 kinases and more than 100 phosphatases in the proteome, protein phosphorylation events are often highly specific (22–24). We, as well as others, have shown that the activity of NACA is extensively regulated by posttranslational modifications including phosphorylation. NACA is phosphorylated by casein kinase 2 (CK2) and integrin-linked kinase (ILK) at an N terminus cluster of phosphoacceptor sites (25, 26). CK2 phosphorylation of NACA at Ser-25, Thr-27, Ser-29, and Ser-34 leads to CRM1-mediated nuclear export of NACA (26). In osteoblasts, cell adhesion to fibronectin triggers ILK phosphorylation of NACA on Ser-43, leading to nuclear import of NACA (25). Nuclear import of NACA is also induced by protein kinase A phosphorylation of Ser-99 downstream of a parathyroid hormone signaling cascade in osteoblast cells (27). Additionally, NACA phosphorylation by glycogen synthase kinase 3β (GSK3β) on Thr-159 triggers the degradation of NACA by the 26S proteasome complex (28, 29). Nuclear accumulation of NACA as a result of ILK or PKA phosphorylation results in an increase of NACA coactivation of AP-1 transcriptional activity (17, 25, 27). Moreover, mutation of the CK2 export or the GSK3β degradation phosphoacceptor sites on NACA enhance its coactivation potency (25, 26, 29). To demonstrate the physiological relevance of NACA phosphorylation in vivo, we previously generated knock-in mice harboring a nonphosphorylatable serine-to-alanine mutation at NACA Ser-43 (19). NACA S43A mice were deficient in nuclear NACA and exhibited a reduction in bone formation and bone mass. Taken together, these data indicate that a balance between potentiating and inhibitory phosphorylation events of NACA alters the activation of AP-1 signaling pathways. Given the transient nature of phosphorylation events, NACA activity is likely to be fine-tuned by the dynamic removal of phosphate modifications. Despite our extensive understanding of NACA kinases, identification and characterization of a NACA phosphatase has remained elusive.
In the present study we used an unbiased proteomics approach to identify a NACA phosphatase. We report that NACA interacts with a major Ser/Thr phosphatase, PP1A (enzyme complex abbreviated as protein phosphatase 1 α; catalytic unit gene symbol PPP1CA). PP1A counteracts NACA phosphorylation on Thr-89, Ser-151, and Thr-174. Additionally, PP1A enhances NACA nuclear localization and coactivation activity.
Results
PP1A associates with NACA
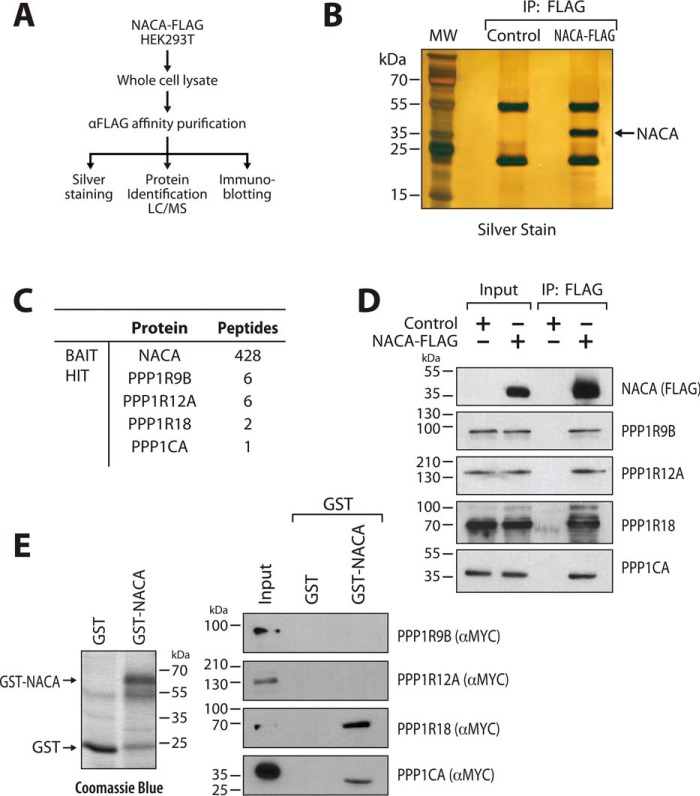
To identify potential NACA phosphatases, we utilized an unbiased affinity purification and proteomics approach (Fig. 1A). FLAG-tagged NACA protein complexes were immunopurified from HEK293T cells (Fig. 1B) and NACA-associated proteins identified by tandem MS analysis. Several components of the protein phosphatase 1 α holoenzyme complex (PP1A), including the regulatory subunits PPP1R9B, PPP1R12A, PPP1R18, and the catalytic subunit PPP1CA were identified (Fig. 1C). The catalytic subunit, PPP1CA was detected at a level lower than the standard threshold of two peptides. This low abundance most likely reflects the short-lived and transient nature of the catalytic unit/substrate reaction. PP1A components were the only phosphatase proteins found in the NACA complex. Co-immunoprecipitation confirmed the association of PPP1R9B, PPP1R12A, PPP1R18, and PPP1CA with NACA (Fig. 1D). Among these PP1A components, interaction with PPP1CA and PPP1R18 are likely to be direct, because a full-length GSH–S-transferase NACA fusion protein (GST-NACA) purified from Escherichia coli bacteria (Fig. 1E, left panel), bound to in vitro translated MYC-tagged PPP1CA and PPP1R18 protein (Fig. 1E, right panel).
Figure 1.
PP1A associates with NACA. A, schematic overview of the proteomics approach used to identify NACA-interacting proteins. B, silver-stained SDS-PAGE gel showing expression and purification of NACA protein. C, mass spectrometry (MS/MS) analysis identified numerous NACA-associated proteins, including several components of the PP1A holoenzyme complex. D, validation of interaction of NACA-FLAG with endogenous PP1A complex proteins. HEK293T cells expressing NACA-FLAG or FLAG empty vector were lysed and immunoprecipitated (IP) with anti-FLAG antibody beads and blotted with anti-FLAG antibody or the indicated PP1A-complex antibodies. E, GST-pulldown analysis showing direct interaction between NACA and PPP1CA as well as PPP1R18. GST-NACA fusion protein or GST alone expressed in bacteria (left panel) was incubated with in vitro translated PPP1R9B, PPP1R12A, PPP1R18, and PPP1CA, and then subjected to SDS-PAGE and Western blot analysis (right panel).
PP1A mediates the dephosphorylation of NACA
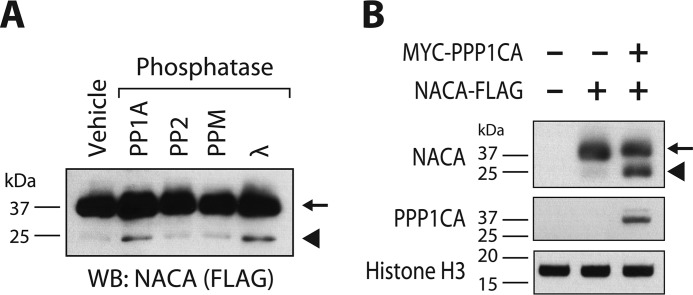
Increased mobility in SDS-PAGE is characteristic of hypo-phosphorylated protein forms. To determine whether PP1A could specifically dephosphorylate NACA, we treated NACA with recombinant serine/threonine phosphatases including PP1A, PP2, and PPM. As shown in Fig. 2A, PP1A activity increased the mobility of NACA, whereas incubation of NACA with PP2 or PPM did not result in a faster migrating hypo-phosphorylated form of NACA. The highly promiscuous Lambda phosphatase served as a positive control of NACA dephosphorylation. To determine whether PP1A could modify the phosphorylation status of NACA in vivo, we next overexpressed NACA together with or without the PP1A catalytic unit in HEK293T cells and examined NACA migration by Western blotting. Co-expression of NACA with PPP1CA resulted in a faster migrating hypo-phosphorylated NACA band indicative of NACA dephosphorylation in the presence of PPP1CA (Fig. 2B).
Figure 2.
PP1A dephosphorylates NACA in vitro and in vivo. A, recombinant NACA (arrow) was treated with the indicated recombinant enzymes for 90 min at 30 °C and then analyzed by Western blotting (WB). B, HEK293T cells were transfected with plasmid vectors expressing NACA or the PP1A catalytic unit (PPP1CA). Forty-eight h after transfection, cell lysates were separated by SDS-PAGE and analyzed by Western blotting. In vitro dephosphorylation of NACA (arrow) with recombinant phosphatase enzymes (A) or in vivo co-expression of NACA (B) with PP1A resulted in a faster migrating hypo-phosphorylated form of NACA (arrowhead).
PP1A mediates dephosphorylation of NACA at Thr-89, Ser-151, and Thr-174
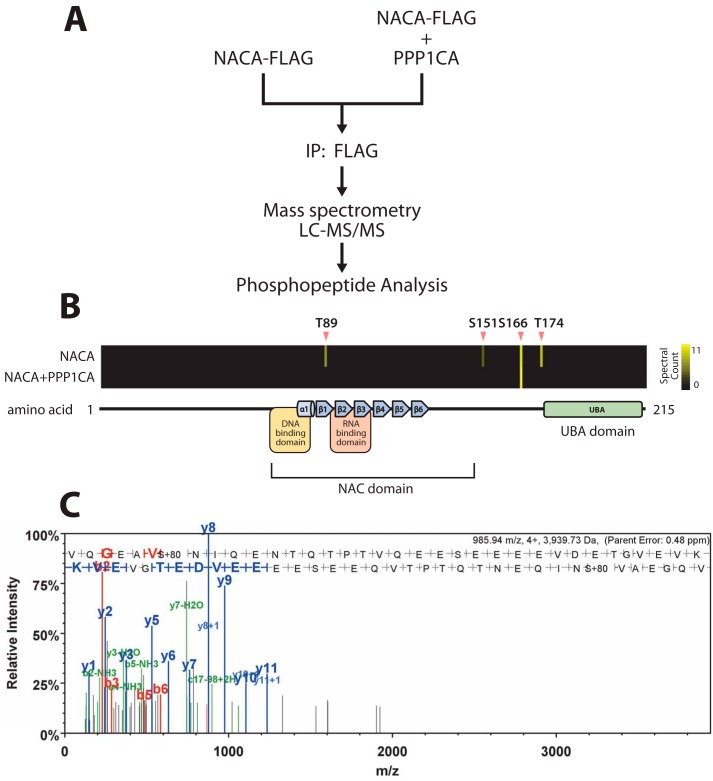
To identify NACA phosphorylation sites modified by PP1A, we performed a phosphoproteomic characterization of NACA from HEK293T cells expressing NACA alone or NACA together with PPP1CA (Fig. 3A). In cells expressing NACA alone, we detected four NACA phosphorylation sites at Thr-89, Ser-151, Ser-166, and Thr-174 (Fig. 3, B and C). Interestingly, PPP1CA co-expression led to a loss of phosphorylation at Thr-89, Ser-151, and Thr-174. Ser-166, however, was not regulated by PPP1CA overexpression (Fig. 3B).
Figure 3.
PP1A mediates dephosphorylation of NACA at specific residues. A, NACA was immunopurified from HEK293T cells expressing NACA with or without the PP1A catalytic unit (PPP1CA). Posttranslational modifications were identified by LC-tandem MS. B, heat maps indicating the differential abundance of phosphorylation sites detected on NACA in the presence or absence of PPP1CA. Black indicates zero spectral counts and increasing quantities of spectral counts are represented in yellow. C, a representative mass spectra of a NACA phosphopeptide modified at serine 151.
NACA dephosphorylation leads to recruitment of novel interactants
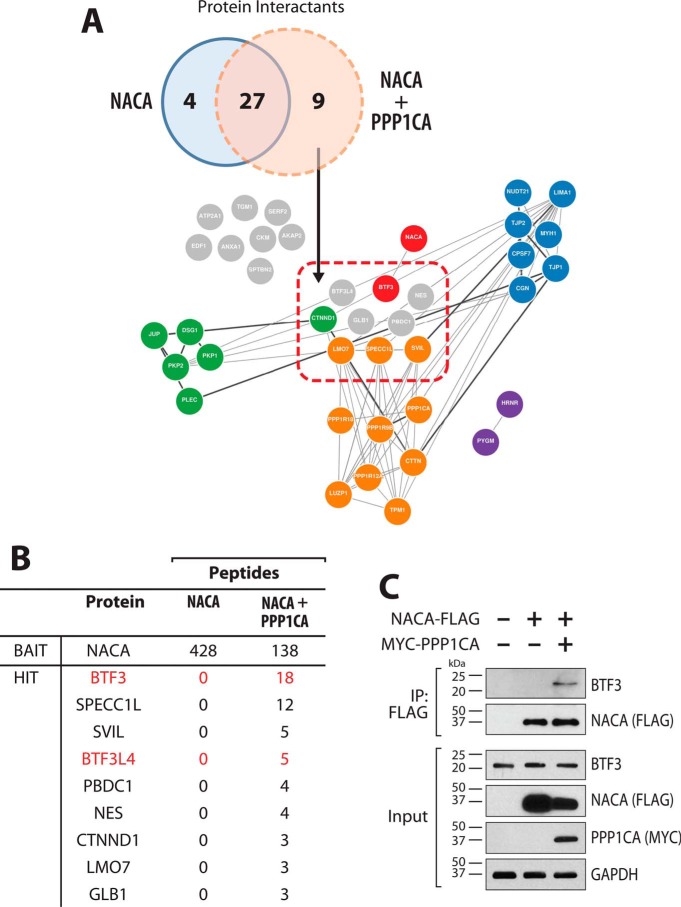
Protein phosphorylation is often linked to alterations in protein interactions. To better understand the molecular consequences of altered NACA phosphorylation status, we compared the NACA interactomes following affinity purification in the presence or absence of PPP1CA overexpression (Fig. 4A). Network analysis of the NACA interactomes revealed that in addition to the aforementioned PP1A complex, NACA is associated with five major functionally related protein clusters. Consistent with the diverse and multifunctional nature of NACA, we uncovered clustered enriched for components of the intermediate filaments, cajal bodies, cytoskeleton, mRNA processing, and transcriptional regulation. From the 40 high-confidence NACA-interacting proteins in both datasets, we found a total of 9 proteins that uniquely interacted with NACA following PPP1CA-mediated dephosphorylation (Fig. 4B). Of note was the induction of NACA interaction with BTF3 and BTF3L4 following PPP1CA co-expression. BTF3 is a general transcription factor shown to initiate transcription through interactions with TATA-binding protein and RNA polymerase II at proximal promoter regions (13, 14, 30–32). Furthermore, BTF3 has also previously been shown to form heterodimeric complexes with NACA (12, 33). BTF3-NACA complexes have been implicated in DNA binding as well as the sorting and targeting of cytosolic proteins from the ribosome. We further validated the PPP1CA-regulated association of NACA and BTF3 by immunoprecipitation and Western blotting (Fig. 4C). As shown in Fig. 4C, overexpression of PPP1CA significantly enhanced the interaction of BTF3 with NACA.
Figure 4.
NACA dephosphorylation leads to recruitment of novel interactants. A, Venn diagram showing overlap and comparison between the sets of NACA-associated proteins identified by MS. Network analysis and visualization of the identified NACA interactants based on curated protein– interaction databases (GeNets). Interactants unique to PP1A overexpression are delineated within the red rectangle. Closely connected protein clusters are color coded (green, intermediate filaments; purple, cajal bodies; orange, cytoskeleton; blue, mRNA processing; red, transcriptional regulation; interactants that are not part of defined clusters are shown in gray). B, table of the PP1A-dependent NACA interactome. NACA-associated proteins identified by MS in the presence or absence of PPP1CA. C, validation of NACA interaction with BTF3 in a PP1A-dependent manner. HEK293T cells expressing NACA-FLAG and/or MYC-PPP1CA were lysed and immunoprecipitated (IP) with anti-FLAG antibody beads and probed for endogenous BTF3. Overexpression of MYC-PPP1CA induces interaction of NACA and BTF3.
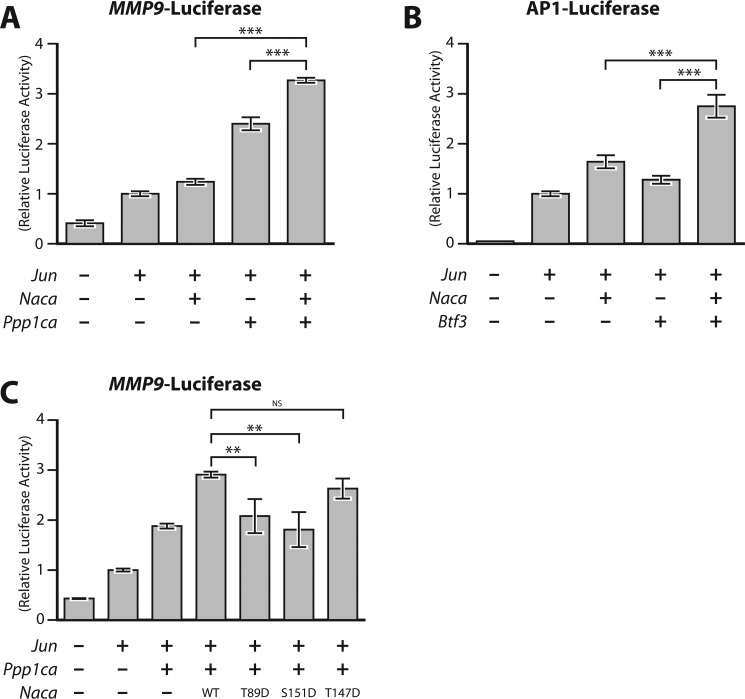
NACA cooperates with PPP1CA and BTF3 to coactivate cJUN transcriptional activity
To determine the functional consequences of NACA dephosphorylation by PP1A, we examined the effect of co-expression of PPP1CA and BTF3 on cJUN transcriptional activity. Luciferase reporter gene assays with the cJUN-responsive MMP9-promoter luciferase showed that NACA and PPP1CA cooperatively enhanced cJUN transcriptional activity (Fig. 5A). Similarly, BTF3 and NACA cooperatively stimulated transcriptional activity in the cJUN-mediated activation of an AP-1 luciferase reporter assay (Fig. 5B). Taken together, these data indicate that NACA can functionally interact with PPP1CA and BTF3 in the regulation of AP-1 transcriptional activity. Interestingly, PPP1CA was able to coactivate cJUN on its own. This may be a result of PPP1CA dephosphorylating endogenous NACA or another unknown AP-1 regulator. We next examined whether dephosphorylation of NACA was required for coactivation of transcription with PPP1CA. As shown in Fig. 5C, coactivation of NACA and PPP1CA was abolished when Thr-89 or Ser-151 were mutated into phosphatase-resistant phosphomimetic aspartate residues. This suggests that dephosphorylation of NACA on Thr-89 and Ser-151 contributes to coactivation of cJUN transcription.
Figure 5.
PPP1CA and BTF3 functionally cooperate with NACA to regulate cJUN transcription. A, transcriptional activity of a cJUN-responsive MMP9-promoter luciferase (MMP9-Luc) reporter construct in the presence of the indicated expression vectors. B, luciferase activity of a cJUN-responsive construct containing six tandem repeats of the AP-1 binding element (AP1-Luc) in cells transfected with the indicated expression vectors. C, effect of NACA serine/threonine-to-aspartate mutations on cJUN activity on the MMP9-Luc reporter. HEK293T cells were transiently transfected with reporter vectors together with cJUN, NACA, BTF3 and/or PPP1CA expression vectors alone or in combination and relative luciferase activity measured 24 h after transfection. Data are presented as mean ± S.D.; n ≥ 3. **, p < 0.01; ***, p < 0.001; ANOVA with post hoc test. NS, non significant.
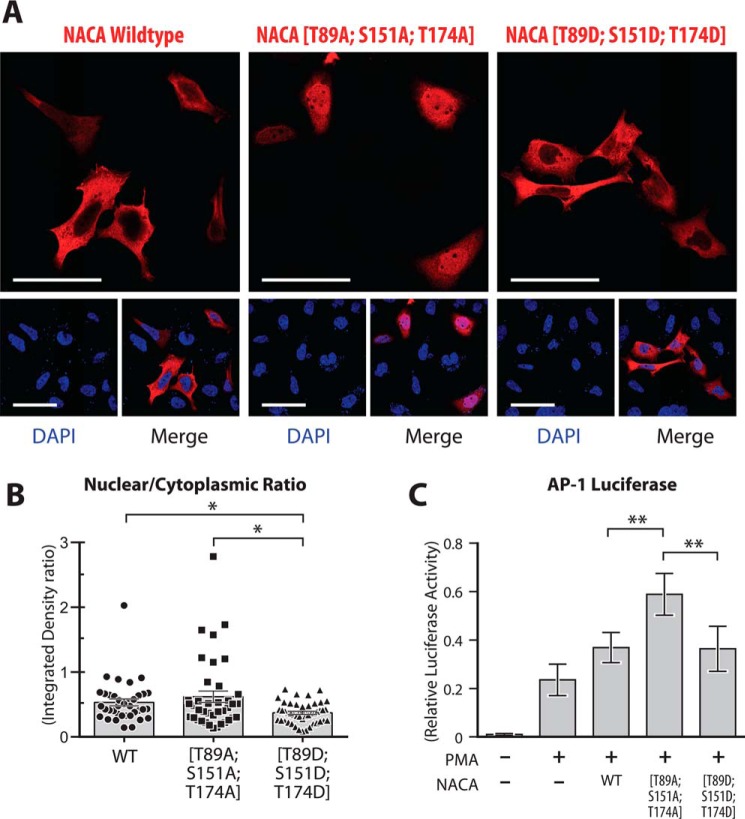
Regulation of NACA localization by PP1A-sensitive NACA residues
Changes in NACA phosphorylation are strongly linked to alterations in its subcellular localization. Because dephosphorylation of NACA by PPP1CA increased NACA coactivation of transcriptional activity, we examined the possibility that NACA phosphorylation at Thr-89, Ser-151, and Thr-174 may alter NACA subcellular localization. As shown in Fig. 6A and quantified in Fig. 6B, nuclear localization of NACA was increased in a mutant harboring phosphorylation-resistant threonine/serine-to-alanine mutations (T89A, S151A, T174A) compared with the phosphomimetic mutant (T89D, S151D, T174D). Furthermore, AP-1 luciferase reporter assays showed that phosphorylation-resistant NACA mutant had increased AP-1 coactivation activity relative to the phosphomimetic mutant (Fig. 6C). In conclusion, these findings suggest that dephosphorylation of NACA at multiple residues by PPP1CA enhances nuclear targeting of NACA, thereby leading to an increase in cJUN coactivation.
Figure 6.
Regulation of NACA localization by PP1A-sensitive NACA residues. A, HEK293T cells were transiently transfected with WT NACA or NACA mutant constructs and immunostained for NACA with anti-FLAG antibody. B, quantification of NACA distribution in cells treated as in (A) by fluorescence intensity analysis with ImageJ software. C, effect of NACA serine/threonine-to-aspartate or -alanine mutations on AP-1 activity using the AP1-Luc reporter. HEK293T cells were transiently transfected with reporter vectors together with WT NACA or the indicated mutants. Relative luciferase activity was measured 16 h after treatment with 200 nm PMA. Data are presented as mean ± S.D.; n ≥ 3. *, p < 0.05; **, p < 0.01; ANOVA with post hoc test. Magnification bars, 50 μm.
NACA cooperates with PPP1CA to coactivate cJUN activity in osteoblast cells
We have previously shown that in osteoblast cells, NACA is required for the potentiation of cJUN-mediated transcriptional activity during osteoblast differentiation (18). We next sought to determine whether NACA functionally interacts with PPP1CA in the context of osteoblast cell biology.
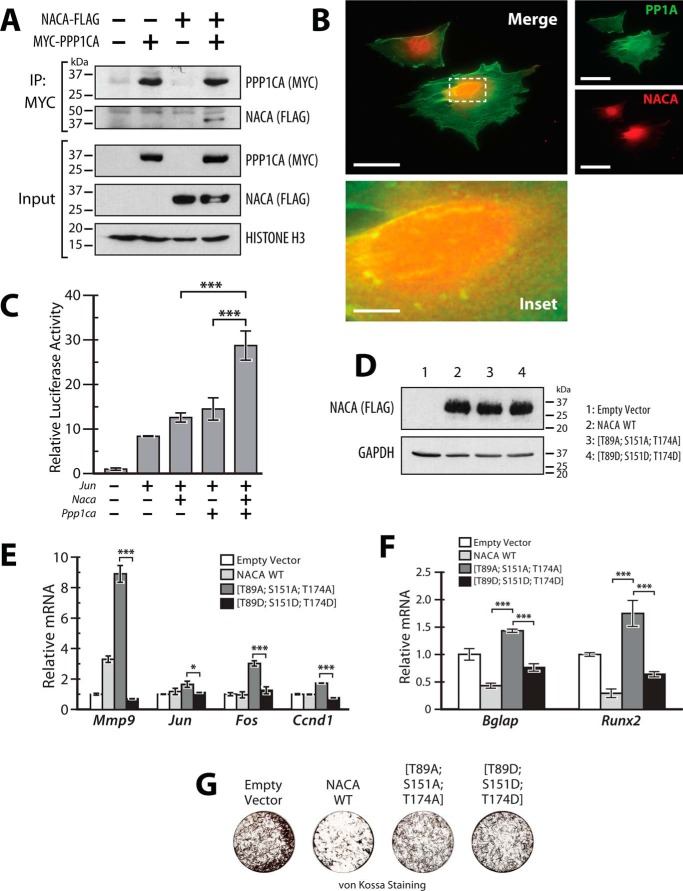
MC3T3-E1 cells are a calvaria-derived murine osteoblast cell line commonly used as a model for the differentiation and mineralization properties of osteoblast cells. To assess whether NACA interacts with PPP1CA in bone cells, MC3T3-E1 cells were cotransfected with constructs expressing NACA-FLAG and MYC-PPP1CA. Cell lysates were then immunoprecipitated with anti-MYC antibody and immunoblotted for NACA. As shown in Fig. 7A, a NACA-specific band was observed in immunoprecipitates from cotransfected cells but not from singly transfected cells, indicating that NACA interacts specifically with PPP1CA in osteoblast cells. To further verify the interaction of endogenous PPP1CA with endogenous NACA, we used immunofluorescence imaging to colocalize PP1A and NACA in untransfected MC3T3-E1 cells. We observed that a fraction of endogenous perinuclear NACA foci also contain PPP1CA (Fig. 7B). To investigate the role of NACA and PPP1CA in regulating cJUN function in osteoblast cells, we co-expressed NACA, cJUN, and PPP1CA together with the AP-1 luciferase reporter. Co-expression of NACA and PPP1CA led to a more significant activation of cJUN transcriptional activity compared with expression of either NACA or PPP1CA alone (Fig. 7C). These data indicate that NACA and PPP1CA physically interact and potentiate cJUN activity in osteoblast cells.
Figure 7.
NACA and PPP1CA regulate cJUN activity in osteoblast cells. A, interaction of NACA with PPP1CA. MC3T3-E1 cells expressing NACA-FLAG and/or MYC-PPP1CA were lysed and immunoprecipitated (IP) with anti-MYC antibody beads and blotted with the indicated antibodies. B, localization of NACA and PPP1CA in MC3T3-E1 cells was examined by immunofluorescence using antibodies against endogenous NACA (red) and endogenous PPP1CA (green). Areas of colocalization (yellow) were detected in the merged image. C, transcriptional activity of a cJUN-responsive luciferase reporter construct (AP1-Luc) in the presence of the indicated expression vectors in MC3T3-E1 cells. D, immunoblots of whole-cell lysates from MC3T3-E1 cells stably expressing WT NACA or NACA serine/threonine-to-aspartate or -alanine mutations. E, RT-qPCR analysis of AP-1 target gene expression in MC3T3-E1 cells stably expressing NACA or the indicated mutants. F, RT-qPCR analysis of osteoblast differentiation markers after 12 days of differentiation. G, MC3T3-E1 cells differentiated as in (F) were stained for mineral by von Kossa staining. Data are presented as mean ± S.D.; n ≥ 3. *, p < 0.05; ***, p < 0.001; ANOVA with post hoc test. Magnification bars, 50 μm except inset, 9 μm.
We next sought to determine whether phosphorylation of NACA at the PP1A-sensitive residues modulates AP-1 activity. To this end we stably expressed WT NACA, the phosphorylation-resistant mutant (T89A, S151A, T174A) and the phosphomimetic mutant (T89D, S151D, T174D) in MC3T3-E1 cells (Fig. 7D) and examined the expression levels of four direct AP-1/cJUN targets. Overexpression of NACA (T89A, S151A, T174A) led to a significantly enhanced expression of the AP-1 targets, Mmp9, Jun, Fos, and Ccnd1 compared with NACA (T89D, S151D, T174D) (Fig. 7E). This suggests that dephosphorylation of NACA at residues Thr-89, Ser-151, and Thr-174 is indeed important for regulation of native AP-1 target promoters in osteoblast cells. Osteocalcin (Bglap), a marker of terminal osteoblast differentiation, is also a well-characterized target of NACA/cJUN activity in osteoblasts (18). We examined whether the increased AP-1 activity induced by NACA mutant (T89A, S151A, T174A) also affects Bglap expression and/or osteoblast differentiation. MC3T3-E1 cells stably expressing NACA or phosphorylation mutants were differentiated in osteogenic media for 12 days after which gene expression and mineralization were assessed. RT-qPCR (reverse transcription-quantitative PCR) analysis of Bglap and Runx2 expression revealed that overexpression of NACA mutant (T89A, S151A, T174A) led to increased expression of these osteoblast differentiation markers compared with phosphomimetic mutant (T89D, S151D, T174D) or WT NACA (Fig. 7F). Interestingly, cells expressing WT NACA had lower levels of Bglap and Runx2 expression than control cells and behaved similarly to the phosphomimetic mutant (T89D, S151D, T174D). This suggests that in terminally differentiated osteoblasts, NACA may be highly phosphorylated under basal conditions. In addition to regulating Bglap expression and osteoblast differentiation via cJUN activity, NACA also influences osteoblast matrix mineralization by a separate distinct and as yet unclear mechanism (19). Examination of matrix mineralization by von Kossa staining revealed that cells expressing NACA mutant (T89A, S151A, T174A) deposited more mineral than cells expressing WT or mutant (T89D, S151D, T174D) (Fig. 7G). However, it must be noted that all forms of NACA displayed less mineral than control cells, perhaps indicative of the predictable involvement of other phosphorylation sites in the regulation of matrix mineralization by NACA. Nonetheless, taken together, these data indicate that the phosphorylation state of NACA at PPP1CA-sensitive residues is important for the regulation of AP-1-dependent transcription in a way that influences osteoblast differentiation and activity.
Discussion
Nuclear NACA functions as an important transcriptional coactivator of cJUN and AP-1 signaling (11, 16–19, 25, 27). Phosphorylation of NACA is known to alter its subcellular localization, transcriptional functions and its proteolytic degradation (25, 27, 29). Although several kinases are known to phosphorylate NACA, the process by which these modifications are reversed or negated has remained elusive. Here in this study, we have identified PP1A as the major serine/threonine phosphatase responsible for NACA dephosphorylation. We report that NACA associates with multiple PP1A holoenzyme constituents and that dephosphorylation of NACA is linked to functional changes in the localization and AP-1 coactivation potential of NACA.
The reversible phosphorylation of proteins is achieved by a balance between kinases and phosphatases. The human genome encodes for 428 known serine/threonine kinases but only about 30 serine/threonine phosphatases (23, 24). Serine/threonine phosphatases can be further divided into three major groups: phosphoprotein phosphatases (PPP), metal-dependent phosphatases (PPM), and the aspartate-based phosphatases. In our study, we found that NACA interacted with one major phosphatase: PP1, a PPP family member. PP1 is a ubiquitously expressed serine/threonine phosphatase with a diverse array of cellular functions (34, 35). In humans PP1 is encoded by three closely related genes (PP1α, PP1β, and PP1γ). These PP1 holoenzyme isoforms are combinatorial complexes of a catalytic subunit interacting with >90 distinct regulatory proteins that define enzyme localization, substrate specificity, and activity (34, 35). We demonstrated that NACA binds directly to the PP1A α-isoform catalytic subunit as well as the PPP1R18 regulatory subunit. Among the PP1 isoforms which can all be also found in the cytosol, the α isoform is unique in its localization to the nuclear matrix (23, 36, 37). It is easy to speculate that such close proximity to nuclear factors makes PP1A an important regulator of transcriptional events. Indeed, dephosphorylation of transcription factors such as cAMP-response element-binding protein (38) and RUNX2 (39) by PP1A is a major mechanism for the attenuation of their transcriptional activity. Here, we likewise show that dephosphorylation of NACA by PP1A has implications for AP-1 transcription. Our results suggest a model wherein PP1A controls the phosphorylation state of NACA, thereby altering the relative distribution of nuclear and cytoplasmic pools of NACA.
In addition to the catalytic subunit of PP1A, we also observed that NACA was associated with the regulatory subunits PPP1R9B, PPP1R18, and PPP1R12. This suggests that there are multiple PP1A holoenzymes interacting with NACA or that each of the specific residues are targeted by a particular PP1A complex. Regulatory subunits often target catalytic units to specific subcellular compartments (23, 40). Indeed, given that there are multiple pools of NACA it is understandable that specific regulatory subunits would be required to target the PPP1CA catalytic subunit to each compartment. Thus, it follows that regulatory subunits may also competitively inhibit each other. Further work would be required to dissect the function of these regulatory subunits in relation to NACA function. Withstanding some exceptions, PP1A usually interacts with regulatory subunits or substrates through two well-defined docking motifs: the RVXF motif ([KR][X]0–1[VI]{P}[FW]) and the SILK motif ([GS]IL[RK]) (41–43). Consistent with our findings that NACA associates with PP1A, NACA does indeed contain a putative RVXF motif at amino acids 100–104 (KNILF).
Several prior studies using in vitro kinase assays have demonstrated phosphorylation of NACA at multiple residues. We have used a MS-based proteomics approach to dynamically map NACA phosphorylation in the presence and absence of PP1A. We showed that NACA is indeed phosphorylated at Thr-89, Ser-151, Ser-166, and Thr-174. Among these sites, Thr-89, Ser-151, and Thr-174 are completely novel and have not been described previously. Given the varied expression of kinases and phosphatases across cell types it is highly likely that NACA phosphorylation sites and phosphatases could be cell-type specific. Phosphorylation of Ser-166 has previously been described in embryonic stem cells as an induced response to bone morphogenetic protein during differentiation (44). In our study, Ser-166 phosphorylation was resistant to PP1A, whereas Thr-89, Ser-151, and Thr-174 were dephosphorylated by PP1A. Thus, PP1A dephosphorylation of NACA is site specific and perhaps other phosphatases, in various cell types, might be responsible for dephosphorylation of Ser-166 and the other important NACA phosphorylation sites that we have previously described.
To our knowledge, this is the first comprehensive NACA interactome analysis performed in mammalian cells. NACA has previously been purified in yeast cells with a focus on its interaction with ribosomal proteins (45, 46). Consistent with this aspect of NACA function, we did observe, as part of the 31 NACA interactants that were not PPPICA-dependent (Fig. 4A), several ribosome-associated proteins interacting with NACA, including ribosomal protein L38 (RPL38). More importantly, among the previously known transcriptional partners of NACA, we observed interaction with TXLNG (FIAT) and TXLNA (47–52). Taken together these observations are supportive of the fidelity of our purification method. Our study focused on NACA associations that were differentially regulated by PPP1CA, namely BTF3. NACA has long been known to dimerize with BTF3 (also known as βNAC) in the formation of the nascent polypeptide-associated complex, a multifunctional protein complex involved in both protein translation and nuclear DNA binding (12–14, 51, 53). In the absence of the binding partner, NACA and BTF3 can also homodimerize and function independently. Until now there has been no study describing how heterodimerization and homodimerization of NACA and BTF3 are regulated. Here we show that phosphorylation of NACA is a possible mechanism regulating NACA/BTF3 dimerization. Crystal structures of NACA/BTF3 heterodimers have shown that the β-sheets between NACA amino acids 70 and 135 are important for dimerization (13, 14). Interestingly, Thr-89, which we have shown to be sensitive to PPP1CA, was a residue shown to be specifically involved in heterodimerization by the formation of intermolecular hydrogen bonds. The phosphorylation status of Thr-89 will therefore be important for NACA interaction with BTF3. BTF3 is one of several general transcription factors involved in TATA box–mediated proximal promoter transcription via interactions with RNA polymerase II (32, 54). Our own previous data demonstrated that NACA interacts with TATA-binding protein, and stabilizes cJUN heterodimers (9, 11). Here we add to this model, by showing that NACA and BTF3 can cooperatively enhance cJUN activity. BTF3 could therefore be the bridge between NACA, cJUN, TATA-binding protein, and RNA Pol II during cJUN-activated transcription.
Interestingly, our earlier attempts to understand the role of NACA/BTF3 dimers on transcription had suggested that BTF3 could be inhibitory to NACA transcriptional activity (11). In this system, NACA coactivated the transcriptional activity of a hybrid transcriptional activator (yeast GAL4 transcription factor fused to the activator domain of the herpes simplex virus VP16 protein) and this activity was inhibited by BTF3b (11). BTF3b is an alternative splice isoform of BTF3, lacking the first 44 amino acids (32). Indeed, Zheng et al. (32) demonstrated that BTF3b is transcriptionally inactive despite being able to interact with RNA Pol II. Thus, it is possible that BTF3b acts as a dominant negative inhibitor of NACA as we previously observed whereas full-length BTF3 coactivates NACA activity as demonstrated in this study. We also cannot exclude the possibility that NACA/BTF3 activity is promoter- and activator-dependent. In summary, we have identified PP1A as a major NACA phosphatase. We propose that dephosphorylation of NACA by PP1A is a mechanism for regulating AP-1 transcriptional activity. These findings extend to osteoblast biology where NACA and PP1A function cooperatively in the regulation of gene expression and osteoblast activity. Further studies into the cellular outcomes of PP1A and NACA interactions will permit us to understand the role of NACA in other signaling pathways.
Experimental procedures
Cell culture
HEK293T cells (ATCC) were cultured in Dulbecco's modified Eagle's medium (DMEM) (Life Technologies) supplemented with 10% fetal bovine serum (Hyclone) and 1% penicillin-streptomycin (Life Technologies). MC3T3-E1 cells (a generous gift from Dr. Hiroko Sudo, Tohoku Dental University, Japan) (55) were maintained in α-modified Eagle's minimum essential medium (αMEM) supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. Cells were transfected with X-tremeGENE HP (Roche) according to manufacturer's instructions. Stable cells lines were selected with 5 μg/ml puromycin (Life Technologies) for 3 days and thereafter maintained in complete medium with 2.5 μg/ml puromycin. Osteoblast differentiation was induced in MC3T3-E1 cells by culturing cells in αMEM supplemented with 10 mm β-glycerophosphate and 50 μg/ml ascorbic acid for 12 days. von Kossa staining for mineral was performed using 5% silver nitrate.
Plasmids
To generate pCMV6-Naca-FLAG, the mouse Naca coding sequence was PCR amplified from pLVX-Naca-FLAG (27) and inserted into EcoRI-linearized pCMV6-Kan/Neo (OriGene Technologies) by DNA assembly (In-Fusion; Takara). The Ppp1ca (pCMV6-MYC-Ppp1ca) and Btf3 (pCMV6-MYC-Btf3) expression vectors were constructed by PCR amplification from mouse MC3T3-E1 cell cDNA with MYC-containing primers and inserted into the EcoRI-linearized pCMV6-Kan/Neo. The MMP9 promoter reporter plasmid (MMP9-Luc), containing the 670-bp proximal promoter fragment in the pGL3 luciferase backbone was a kind gift from Dr. Shoukat Dedhar (University of British Columbia, Canada). The AP-1 reporter plasmid (pAP1(PMA)-TA-Luc) containing six tandem repeats of the AP-1 response element was obtained from Clontech. The cJUN expression vector as well as the GST-fusion constructs, pGEX-4T-3 and pGEX-4T-3-Naca have been described previously (11). pCMV6-Ppp1r9b, pCMV6-Ppp1r12a, pCMV6-Ppp1r18 were obtained from OriGene Technologies. Site-directed mutagenesis to obtain pCMV6-Naca(T89A, S151A, T174A)-FLAG and pCMV6-Naca(T89D, S151D, T174D)-FLAG was performed with the Q5 Site-Directed Mutagenesis Kit (New England Biolabs). All high-fidelity PCR amplifications were performed with CloneAMP HiFi Taq Polymerase (Takara). All expression vectors were verified by sequencing. Primer sequences for cloning and mutagenesis are available upon request.
Antibodies
Rabbit Anti-FLAG, Mouse Anti-FLAG, Rabbit Anti-MYC, Anti-Rabbit HRP, Anti-Rabbit (conformation-specific)-HRP, Anti-Mouse (light chain–specific)-HRP, Anti-Mouse HRP, Anti-Rabbit Alexa Fluor 594, Anti-Rabbit Alexa Fluor 488, Anti-Mouse Alexa Fluor 594, Anti-Mouse Alexa Fluor 488, Anti-PP1A, Anti-PPP1R9B, Anti-PPP1R12A, and Anti-GAPDH-HRP antibodies were from Cell Signaling Technology. Anti-BTF3 and Anti-PPP1CA antibody were obtained from Abcam. Anti-PPP1R18 was obtained from Santa Cruz Biotechnology. Anti-NACA antibody has been described previously (27).
Affinity purification, co-immunoprecipitation, and MS
HEK293T cells stably expressing NACA-FLAG, empty vector, or NACA-FLAG and MYC-PPP1CA were washed twice in cold PBS and scraped into PBS containing 0.2 mm phenylmethylsulfonyl fluoride (PMSF). After centrifugation, cells were resuspended in lysis buffer (25 mm Hepes, pH 7.4, 150 mm NaCl, 1% Triton X, 0.5% Nonidet P-40, and 10% glycerol), lysed with a Dounce homogenizer and cell debris pelleted by centrifugation. To purify NACA-FLAG complexes, 15 μl of Anti-FLAG M2 magnetic beads (Sigma) were added to clarified cell lysates (3 mg of protein) and incubated with gentle rotation for 3 h. Beads were then washed four times with a high-salt buffer (25 mm Hepes, pH 7.4, 900 mm NaCl, 1% Triton X, 0.5% Nonidet P-40, and 10% glycerol), twice with regular lysis buffer, and then twice more with a detergent-free buffer (25 mm Hepes, pH 7.4, 150 mm NaCl, and 10% glycerol). All buffers were supplemented with 1 mm DTT, 0.25 mm PMSF, and protease/phosphatase inhibitor mixture (Cell Signaling Technology). All steps were carried out in a 4 °C cold room. Beads were then subjected to LC coupled with tandem MS (LC-MS/MS) or immunoprecipitated proteins eluted with Laemmli buffer and separated by SDS-PAGE for silver staining (SilverQuest Silver Staining Kit; Thermo Fisher) and Western blot analysis. LC-MS/MS was performed at the Proteomics Core Facility of the Research Institute of the McGill University Health Centre using a Dionex Ultimate 3000 uHPLC and a Thermo Orbitrap Fusion mass spectrometer. Peptides were identified and analyzed using Mascot 2.3 and Scaffold Q+ Scaffold 4.7.5. Protein identifications with >99.9% probability by the Protein Prophet algorithm were considered valid. Common background proteins and contaminants such as keratin were removed (56, 57). Network visualization and ontology enrichment was performed with GeNets (58) (Broad Institute of MIT and Harvard) and Enrichr (59).
For co-immunoprecipitation assays, cells were lysed with HKMG lysis buffer (10 mm HEPES, pH 7.9, 100 mm KCl, 5 mm MgCl2, 10% glycerol, 1 mm DTT, and 0.5% Nonidet P-40) supplemented with protease/phosphatase inhibitor mixture (Cell Signaling Technology). After syringe homogenization with a 27-gauge needle, cell lysates were clarified by centrifugation and protein concentrations determined with the Bio-Rad Protein Assay reagent (Bio-Rad). One mg of protein was then incubated with 15 μl of Anti-FLAG M2 or Anti-MYC (Cell Signaling Technology) magnetic beads for 3 h at 4 °C. Beads were washed three times in HKMG lysis buffer, twice in high-salt buffer, and twice more in HKMG lysis buffer. Bound proteins were eluted with 2× SDS sample buffer and analyzed by Western blotting.
GST pulldown assay
Recombinant GST-NACA fusion protein and GST control protein were expressed and purified from T7 Express E. coli strain (New England Biolabs) using anti-GST magnetic beads (Cell Signaling Technology) according to standard manufacturer's protocols. Prey proteins were in vitro transcribed and expressed in rabbit reticulocytes with the TNT Quick Coupled Transcription/Translation system (Promega). GST-fusion protein–bound beads were incubated with prey protein in a GST pulldown buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.1% Nonidet P-40, 10% glycerol with protease/phosphatase inhibitor mixture) for 90 min at 4 °C. After washing five times with pulldown buffer, associated proteins were eluted in 2× SDS sample buffer and analyzed by SDS-PAGE and Western blotting.
In vitro phosphatase assays
Recombinant FLAG-tagged NACA was expressed and purified from HEK293T cells. Forty-eight h after transfection with Naca-FLAG cDNA plasmid, cells were lysed in a harsh lysis buffer (10 mm Tris-HCl, pH 8.0, 150 mm NaCl, 2% SDS, 1% Triton) supplemented with protease inhibitor mixture (Sigma). Following sonication and clarification by centrifugation, lysates were incubated with anti-FLAG M2 magnetic beads (Sigma) for 3 h at 4 °C. Beads were then washed five times with a wash buffer containing 10 mm Tris-HCl, pH 8.0, 1 m NaCl, 1% Nonidet P-40. NACA-FLAG fusion protein was then eluted with 200 μg/ml FLAG peptide (Sigma). In vitro phosphatase assays were carried out for 30 min at 30 °C in 50 mm HEPES, 100 mm NaCl, 2 mm DTT, 1 mm MnCl2 for PP1A (EMD Millipore), PPM (EMD Millipore), and Lambda Phosphatase (New England Biolabs). For PP2 (EMD Millipore), buffers were further supplemented with 1 mm MgCl2. Reaction products were separated by SDS-PAGE and visualized by Western blotting.
Luciferase reporter assays
Luciferase reporter plasmids together with expression plasmids were transfected into HEK293T or MC3T3-E1 cells using X-tremeGENE HP (Roche). Luciferase activity was measured 24 h after transfection with the Dual-Glo Assay System (Promega). Renilla luciferase (pRL-TK or pRL-SV40) was used as an internal control for transfection efficiency. For responses to PMA (phorbol 12-myristate 13-acetate, also known as 12-O-tetradecanoylphorbol-13-acetate (TPA)) (Cell Signaling Technology), cells were maintained in 0.5% FBS and treated with 200 nm PMA or vehicle 16 h prior to assay measurement.
Immunofluorescence and confocal microscopy
Cells cultured on coverslips or chamber slides were fixed with 3.7% formaldehyde and permeabilized with 0.1% Triton X in PBS. Cells were then blocked with 3% BSA for 30 min and then incubated with primary antibody in PBS with 1% BSA overnight at 4 °C. Cells were incubated with Alexa Fluor–labeled secondary antibodies for 1 h at room temperature and then mounted with Prolong Gold Antifade containing DAPI (Life Technologies). Images were acquired on a Zeiss LSM880 Laser Scanning Confocal or a Zeiss Spinning Disk Confocal Microscope at the Molecular Imaging Platform of the Research Institute of the McGill University Health Centre (Fig. 6) or a Leica DMR fluorescence microscope (Leica Microsystems) connected to a digital DP70 camera (Olympus) (Fig. 7). Image analysis and fluorescence intensity quantification was performed with ImageJ (National Institutes of Health) and the Zen Software Suite (Carl Zeiss Microscopy).
Statistical analysis
Data are presented as mean ± 1 S.D. Comparisons were made by analysis of variance (ANOVA) with Fisher's least significant difference post hoc test. Statistical significance relative to the specified control is represented as follows: *, p < 0.05; **, p < 0.01; ***, p < 0.001. Experiments were performed in triplicate.
Author contributions
W. N. A. and R. St-A. conceptualization; W. N. A. data curation; W. N. A. formal analysis; W. N. A. and M. P. methodology; W. N. A. writing-original draft; M. P. resources; R. St-A. supervision; R. St-A. funding acquisition; R. St-A. writing-review and editing.
Acknowledgments
We thank all members of the St-Arnaud Lab for insightful comments and assistance over the course of this work. Mark Lepik prepared the publication-ready final version of the figures.
This work was supported by Shriners Hospitals for Children Grants 86400 and 85100 (to R. St-A.). The authors declare that they have no conflicts of interest with the contents of this article.
- NACA
- nascent polypeptide-associated complex and co-regulator alpha
- NAC
- nascent polypeptide-associated complex
- ILK
- integrin-linked kinase
- ANOVA
- analysis of variance
- RT-qPCR
- reverse transcription-quantitative PCR.
References
- 1. Chinenov Y., and Kerppola T. K. (2001) Close encounters of many kinds: Fos-Jun interactions that mediate transcription regulatory specificity. Oncogene 20, 2438–2452 10.1038/sj.onc.1204385 [DOI] [PubMed] [Google Scholar]
- 2. Karin M., Liu Z.-G., and Zandi E. (1997) AP-1 function and regulation. Curr. Opin. Cell Biol. 9, 240–246 10.1016/S0955-0674(97)80068-3 [DOI] [PubMed] [Google Scholar]
- 3. Shaulian E., and Karin M. (2002) AP-1 as a regulator of cell life and death. Nat. Cell Biol. 4, E131–E136 10.1038/ncb0502-e131 [DOI] [PubMed] [Google Scholar]
- 4. Wagner E. F., and Eferl R. (2005) Fos/AP-1 proteins in bone and the immune system. Immunol. Rev. 208, 126–140 10.1111/j.0105-2896.2005.00332.x [DOI] [PubMed] [Google Scholar]
- 5. Bohmann D., Bos T. J., Admon A., Nishimura T., Vogt P. K., and Tjian R. (1987) Human proto-oncogene c-jun encodes a DNA binding protein with structural and functional properties of transcription factor AP-1. Science 238, 1386–1392 10.1126/science.2825349 [DOI] [PubMed] [Google Scholar]
- 6. Bohmann D., and Tjian R. (1989) Biochemical analysis of transcriptional activation by Jun: Differential activity of c- and v-Jun. Cell 59, 709–717 10.1016/0092-8674(89)90017-2 [DOI] [PubMed] [Google Scholar]
- 7. Eferl R., and Wagner E. F. (2003) AP-1: A double-edged sword in tumorigenesis. Nat. Rev. Cancer 3, 859–868 10.1038/nrc1209 [DOI] [PubMed] [Google Scholar]
- 8. St-Arnaud R., and Quelo I. (1998) Transcriptional coactivators potentiating AP-1 function in bone. Front. Biosci. 3, D838–D848 10.2741/A327 [DOI] [PubMed] [Google Scholar]
- 9. Moreau A., Yotov W. V., Glorieux F. H., and St-Arnaud R. (1998) Bone-specific expression of the α chain of the nascent polypeptide-associated complex, a coactivator potentiating c-Jun-mediated transcription. Mol. Cell Biol. 18, 1312–1321 10.1128/MCB.18.3.1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Quèlo I., Hurtubise M., and St-Arnaud R. (2002) αNAC requires an interaction with c-Jun to exert its transcriptional coactivation. Gene Expr. 10, 255–262 10.3727/000000002783992433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yotov W. V., Moreau A., and St-Arnaud R. (1998) The α chain of the nascent polypeptide-associated complex functions as a transcriptional coactivator. Mol. Cell Biol. 18, 1303–1311 10.1128/MCB.18.3.1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wiedmann B., Sakai H., Davis T. A., and Wiedmann M. (1994) A protein complex required for signal-sequence-specific sorting and translocation. Nature 370, 434–440 10.1038/370434a0 [DOI] [PubMed] [Google Scholar]
- 13. Liu Y., Hu Y., Li X., Niu L., and Teng M. (2010) The crystal structure of the human nascent polypeptide-associated complex domain reveals a nucleic acid-binding region on the NACA subunit. Biochemistry 49, 2890–2896 10.1021/bi902050p [DOI] [PubMed] [Google Scholar]
- 14. Wang L., Zhang W., Wang L., Zhang X. C., Li X., and Rao Z. (2010) Crystal structures of NAC domains of human nascent polypeptide-associated complex (NAC) and its αNAC subunit. Protein Cell 1, 406–416 10.1007/s13238-010-0049-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yotov W. V., and St-Arnaud R. (1996) Differential splicing-in of a proline-rich exon converts αNAC into a muscle-specific transcription factor. Genes Dev. 10, 1763–1772 10.1101/gad.10.14.1763 [DOI] [PubMed] [Google Scholar]
- 16. Akhouayri O., and St-Arnaud R. (2007) Differential mechanisms of transcriptional regulation of the mouse osteocalcin gene by Jun family members. Calcif. Tissue Int. 80, 123–131 10.1007/s00223-006-0102-7 [DOI] [PubMed] [Google Scholar]
- 17. Pellicelli M., Hariri H., Miller J. A., and St-Arnaud R. (2018) Lrp6 is a target of the PTH-activated αNAC transcriptional coregulator. Biochim. Biophys. Acta 1861, 61–71 10.1016/j.bbagrm.2018.01.008 [DOI] [PubMed] [Google Scholar]
- 18. Akhouayri O., Quélo I., and St-Arnaud R. (2005) Sequence-specific DNA binding by the αNAC coactivator is required for potentiation of c-Jun-dependent transcription of the osteocalcin gene. Mol. Cell Biol. 25, 3452–3460 10.1128/MCB.25.9.3452-3460.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Meury T., Akhouayri O., Jafarov T., Mandic V., and St-Arnaud R. (2010) Nuclear αNAC influences bone matrix mineralization and osteoblast maturation in vivo. Mol. Cell Biol. 30, 43–53 10.1128/MCB.00378-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cohen P. (2001) The role of protein phosphorylation in human health and disease. The Sir Hans Krebs Medal Lecture. Eur. J. Biochem. 268, 5001–5010 10.1046/j.0014-2956.2001.02473.x [DOI] [PubMed] [Google Scholar]
- 21. Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., and Mann M. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 10.1016/j.cell.2006.09.026 [DOI] [PubMed] [Google Scholar]
- 22. Manning G., Whyte D. B., Martinez R., Hunter T., and Sudarsanam S. (2002) The protein kinase complement of the human genome. Science 298, 1912–1934 10.1126/science.1075762 [DOI] [PubMed] [Google Scholar]
- 23. Moorhead G. B., Trinkle-Mulcahy L., and Ulke-Lemée A. (2007) Emerging roles of nuclear protein phosphatases. Nat. Rev. Mol. Cell Biol. 8, 234–244 10.1038/nrm2126 [DOI] [PubMed] [Google Scholar]
- 24. Shi Y. (2009) Serine/threonine phosphatases: Mechanism through structure. Cell 139, 468–484 10.1016/j.cell.2009.10.006 [DOI] [PubMed] [Google Scholar]
- 25. Quélo I., Gauthier C., Hannigan G. E., Dedhar S., and St-Arnaud R. (2004) Integrin-linked kinase regulates the nuclear entry of the c-Jun coactivator α-NAC and its coactivation potency. J. Biol. Chem. 279, 43893–43899 10.1074/jbc.M406310200 [DOI] [PubMed] [Google Scholar]
- 26. Quélo I., Gauthier C., and St-Arnaud R. (2005) Casein kinase II phosphorylation regulates αNAC subcellular localization and transcriptional coactivating activity. Gene Expr. 12, 151–163 10.3727/000000005783992070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pellicelli M., Miller J. A., Arabian A., Gauthier C., Akhouayri O., Wu J. Y., Kronenberg H. M., and St-Arnaud R. (2014) The PTH-gas-protein kinase A cascade controls αNAC localization to regulate bone mass. Mol. Cell Biol. 34, 1622–1633 10.1128/MCB.01434-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hotokezaka Y., van Leyen K., Lo E. H., Beatrix B., Katayama I., Jin G., and Nakamura T. (2009) αNAC depletion as an initiator of ER stress-induced apoptosis in hypoxia. Cell Death Differ. 16, 1505–1514 10.1038/cdd.2009.90 [DOI] [PubMed] [Google Scholar]
- 29. Quelo I., Akhouayri O., Prud'homme J., and St-Arnaud R. (2004) GSK3β-dependent phosphorylation of the αNAC coactivator regulates its nuclear translocation and proteasome-mediated degradation. Biochemistry 43, 2906–2914 10.1021/bi036256+ [DOI] [PubMed] [Google Scholar]
- 30. Hu G. Z., and Ronne H. (1994) Yeast BTF3 protein is encoded by duplicated genes and inhibits the expression of some genes in vivo. Nucleic Acids Res. 22, 2740–2743 10.1093/nar/22.14.2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Parthun M. R., Mangus D. A., and Jaehning J. A. (1992) The EGD1 product, a yeast homolog of human BTF3, may be involved in GAL4 DNA binding. Mol. Cell Biol. 12, 5683–5689 10.1128/MCB.12.12.5683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zheng X. M., Black D., Chambon P., and Egly J. M. (1990) Sequencing and expression of complementary DNA for the general transcription factor BTF3. Nature 344, 556–559 10.1038/344556a0 [DOI] [PubMed] [Google Scholar]
- 33. Beatrix B., Sakai H., and Wiedmann M. (2000) The α and β subunit of the nascent polypeptide-associated complex have distinct functions. J. Biol. Chem. 275, 37838–37845 10.1074/jbc.M006368200 [DOI] [PubMed] [Google Scholar]
- 34. Ceulemans H., and Bollen M. (2004) Functional diversity of protein phosphatase-1, a cellular economizer and reset button. Physiol. Rev. 84, 1–39 10.1152/physrev.00013.2003 [DOI] [PubMed] [Google Scholar]
- 35. Cohen P. T. (2002) Protein phosphatase 1–targeted in many directions. J. Cell Sci. 115, 241–256 [DOI] [PubMed] [Google Scholar]
- 36. Andreassen P. R., Lacroix F. B., Villa-Moruzzi E., and Margolis R. L. (1998) Differential subcellular localization of protein phosphatase-1 α, γ1, and δ isoforms during both interphase and mitosis in mammalian cells. J. Cell Biol. 141, 1207–1215 10.1083/jcb.141.5.1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kuret J., Bell H., and Cohen P. (1986) Identification of high levels of protein phosphatase-1 in rat liver nuclei. FEBS Lett. 203, 197–202 10.1016/0014-5793(86)80741-4 [DOI] [PubMed] [Google Scholar]
- 38. Hagiwara M., Alberts A., Brindle P., Meinkoth J., Feramisco J., Deng T., Karin M., Shenolikar S., and Montminy M. (1992) Transcriptional attenuation following cAMP induction requires PP-1-mediated dephosphorylation of CREB. Cell 70, 105–113 10.1016/0092-8674(92)90537-M [DOI] [PubMed] [Google Scholar]
- 39. Rajgopal A., Young D. W., Mujeeb K. A., Stein J. L., Lian J. B., van Wijnen A. J., and Stein G. S. (2007) Mitotic control of RUNX2 phosphorylation by both CDK1/cyclin B kinase and PP1/PP2A phosphatase in osteoblastic cells. J. Cell. Biochem. 100, 1509–1517 10.1002/jcb.21137 [DOI] [PubMed] [Google Scholar]
- 40. Trinkle-Mulcahy L., Sleeman J. E., and Lamond A. I. (2001) Dynamic targeting of protein phosphatase 1 within the nuclei of living mammalian cells. J. Cell Sci. 114, 4219–4228 [DOI] [PubMed] [Google Scholar]
- 41. Egloff M. P., Johnson D. F., Moorhead G., Cohen P. T., Cohen P., and Barford D. (1997) Structural basis for the recognition of regulatory subunits by the catalytic subunit of protein phosphatase 1. EMBO J. 16, 1876–1887 10.1093/emboj/16.8.1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hendrickx A., Beullens M., Ceulemans H., Den Abt T., Van Eynde A., Nicolaescu E., Lesage B., and Bollen M. (2009) Docking motif-guided mapping of the interactome of protein phosphatase-1. Chem. Biol. 16, 365–371 10.1016/j.chembiol.2009.02.012 [DOI] [PubMed] [Google Scholar]
- 43. Meiselbach H., Sticht H., and Enz R. (2006) Structural analysis of the protein phosphatase 1 docking motif: Molecular description of binding specificities identifies interacting proteins. Chem. Biol. 13, 49–59 10.1016/j.chembiol.2005.10.009 [DOI] [PubMed] [Google Scholar]
- 44. Van Hoof D., Muñoz J., Braam S. R., Pinkse M. W., Linding R., Heck A. J., Mummery C. L., and Krijgsveld J. (2009) Phosphorylation dynamics during early differentiation of human embryonic stem cells. Cell Stem Cell 5, 214–226 10.1016/j.stem.2009.05.021 [DOI] [PubMed] [Google Scholar]
- 45. Koplin A., Preissler S., Ilina Y., Koch M., Scior A., Erhardt M., and Deuerling E. (2010) A dual function for chaperones SSB-RAC and the NAC nascent polypeptide-associated complex on ribosomes. J. Cell Biol. 189, 57–68 10.1083/jcb.200910074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nyathi Y., and Pool M. R. (2015) Analysis of the interplay of protein biogenesis factors at the ribosome exit site reveals new role for NAC. J. Cell Biol. 210, 287–301 10.1083/jcb.201410086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hekmatnejad B., Akhouayri O., Jafarov T., and St-Arnaud R. (2014) SUMOylated αNAC potentiates transcriptional repression by FIAT. J. Cell. Biochem. 115, 866–873 10.1002/jcb.24729 [DOI] [PubMed] [Google Scholar]
- 48. Hekmatnejad B., Mandic V., Yu V. W., Akhouayri O., Arabian A., and St-Arnaud R. (2014) Altered gene dosage confirms the genetic interaction between FIAT and αNAC. Gene 538, 328–333 10.1016/j.gene.2014.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hotokezaka Y., Katayama I., van Leyen K., and Nakamura T. (2015) GSK-3β-dependent downregulation of γ-taxilin and αNAC merge to regulate ER stress responses. Cell Death Dis. 6, e1719 10.1038/cddis.2015.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. St-Arnaud R., and Hekmatnejad B. (2011) Combinatorial control of ATF4-dependent gene transcription in osteoblasts. Ann. N.Y. Acad. Sci. 1237, 11–18 10.1111/j.1749-6632.2011.06197.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yoshida K., Nogami S., Satoh S., Tanaka-Nakadate S., Hiraishi H., Terano A., and Shirataki H. (2005) Interaction of the taxilin family with the nascent polypeptide-associated complex that is involved in the transcriptional and translational processes. Genes Cells 10, 465–476 10.1111/j.1365-2443.2005.00848.x [DOI] [PubMed] [Google Scholar]
- 52. Yu V. W., Ambartsoumian G., Verlinden L., Moir J. M., Prud'homme J., Gauthier C., Roughley P. J., and St-Arnaud R. (2005) FIAT represses ATF4-mediated transcription to regulate bone mass in transgenic mice. J. Cell Biol. 169, 591–601 10.1083/jcb.200412139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Whitby M. C., and Dixon J. (2001) Fission yeast nascent polypeptide-associated complex binds to four-way DNA junctions. J. Mol. Biol. 306, 703–716 10.1006/jmbi.2000.4407 [DOI] [PubMed] [Google Scholar]
- 54. Zheng X. M., Moncollin V., Egly J. M., and Chambon P. (1987) A general transcription factor forms a stable complex with RNA polymerase B (II). Cell 50, 361–368 10.1016/0092-8674(87)90490-9 [DOI] [PubMed] [Google Scholar]
- 55. Sudo H., Kodama H. A., Amagai Y., Yamamoto S., and Kasai S. (1983) In vitro differentiation and calcification in a new clonal osteogenic cell line derived from newborn mouse calvaria. J. Cell Biol. 96, 191–198 10.1083/jcb.96.1.191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mellacheruvu D., Wright Z., Couzens A. L., Lambert J. P., St-Denis N. A., Li T., Miteva Y. V., Hauri S., Sardiu M. E., Low T. Y., Halim V. A., Bagshaw R. D., Hubner N. C., Al-Hakim A., Bouchard A., et al. (2013) The CRAPome: A contaminant repository for affinity purification-mass spectrometry data. Nat. Meth. 10, 730–736 10.1038/nmeth.2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Trinkle-Mulcahy L., Boulon S., Lam Y. W., Urcia R., Boisvert F. M., Vandermoere F., Morrice N. A., Swift S., Rothbauer U., Leonhardt H., and Lamond A. (2008) Identifying specific protein interaction partners using quantitative mass spectrometry and bead proteomes. J. Cell Biol. 183, 223–239 10.1083/jcb.200805092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Li T., Kim A., Rosenbluh J., Horn H., Greenfeld L., An D., Zimmer A., Liberzon A., Bistline J., Natoli T., Li Y., Tsherniak A., Narayan R., Subramanian A., Liefeld T., et al. (2018) GeNets: A unified web platform for network-based genomic analyses. Nat. Meth. 15, 543–546 10.1038/s41592-018-0039-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chen E. Y., Tan C. M., Kou Y., Duan Q., Wang Z., Meirelles G. V., Clark N. R., and Ma'ayan A. (2013) Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128 10.1186/1471-2105-14-128 [DOI] [PMC free article] [PubMed] [Google Scholar]