ABSTRACT
Plants possess the structural maintenance of chromosome (SMC) protein complexes cohesin, condensin, and SMC5/6, which function in fundamental biological processes such as sister chromatid cohesion, chromosome condensation and segregation, and damaged DNA repair. Recently, increasing evidence in several organisms has suggested that condensin is involved in chromatin organizations during interphase. In Arabidopsis thaliana, condensin II is localized in the nucleus throughout interphase and is suggested to be required for keeping centromeres apart and the assembly of euchromatic chromosome arms. However, it remains unclear how condensin II organizes chromatin associations. Here, we first showed the high possibility that the function of condensin II as a complex is required for the disassociation of centromeres. Analysis of the rDNA array distribution revealed that condensin II is also indispensable for the association of centromeres with rDNA arrays. Reduced axial compaction of chromosomes and impaired genome integrity in condensin II mutants are not related to the disruption of chromatin organization. In contrast, the axial compaction of chromosomes by condensin II produces the force leading to the disassociation of heterologous centromeres in Drosophila melanogaster. Taken together, our data imply that the condensin II function in chromatin organization differs among eukaryotes.
KEYWORDS: Condensin II, centromere, rDNA, histone hyperacetylation, genome integrity
Introduction
Eukaryotes possess evolutionarily conserved protein complexes composed of chromosomal ATPases that belong to the structural maintenance of chromosomes (SMC) family. There are three types of SMC protein complexes distinguished by different combinations of SMC proteins. SMC1 and SMC3 comprise the core subunits of cohesin, which is involved in sister chromatid cohesion [1,2]. Similarly, SMC2 and SMC4 are the core subunits of condensin, which is required for chromosome condensation and segregation [1–5]. The remaining two, SMC5 and SMC6, interact with each other and are crucial for damaged DNA repair and recombination [1,2].
In animals and plants that have been investigated including Arabidopsis thaliana, condensin is subdivided into two types, I and II, by differences in regulatory subunits. In association with SMC2 and SMC4, chromosomal associated protein (CAP)-H, CAP-G, and CAP-D3 form condensin I, while CAP-H2, CAP-G2, and CAP-D3 form condensin II [3]. In vertebrates, repression of condensin I or II causes misregulation of both mitotic and meiotic chromosome condensation and segregation [4,5]. Likewise, defects in SMC2 or SMC4 cause missegregation of mitotic and meiotic chromosomes in A. thaliana [6–9]. However, A. thaliana condensin II is unlikely to participate in the regulation of mitotic chromosomes [10]. This is also considered to be the case in Drosophila melanogaster [11] and the primitive red alga Cyanidioschyzon merolae [12].
Unlike condensin I, which is sequestrated in the cytoplasm, condensin II stays in the nucleoplasm and the nucleolus during interphase in vertebrates [13]. In plants, although condensin II subunit CAP-D3 is not expressed in the nucleolus [14], the expressions of other subunits CAP-H2 and CAP-G2 indicate that the plant condensin II during interphase shows similar localization pattern to the vertebrate condensin II [10,15]. Therefore, it is considered that condensin II plays important roles in the interphase nucleus, and conversely, that condensin I has a minimal function during interphase [4]. In fact, recent studies on animal, fly and plant cells have shown that condensin II contributes to interphase chromosome functions such as damaged DNA repair [10,16,17] and local chromatin organization associated with gene transcription [18–22].
Condensin II affects a wide range of chromosome structures. In D. melanogaster, condensin II promotes the axial compaction of chromosomes, narrowing the distance between two distinct loci on the same chromosome arm, which drives the force that disperses heterologous centromeres and forms chromosome territories [23]. Condensin II also contributes to suppressing the association of centromeric regions in mouse neuronal stem cells and neurons [24]. A lack of CAP-D3 function in A. thaliana causes increased association of centromeres and disassembly of euchromatic chromosome arms [14]. Condensin I in Schizosaccharomyces pombe is recruited to RNA Pol III-transcribed gene loci including 5S rDNA arrays during interphase. It promotes centromere association and facilitates faithful chromosome segregation during mitosis [25,26].
In this study, we established that the function of condensin II as a complex is required for the regulation of centromere and rDNA array association using condensin II mutants. We also found that defects in condensin II enhanced chromatin loosening and impaired genome integrity. However, the abnormal chromatin association between centromeres and rDNA arrays was not attributable to those impairments. Therefore, we propose a possibility that condensin II mediates the recruitment of rDNA arrays to centromeres in plants, as is the case for condensin I in S. pombe [25,26].
Results
Condensin II mutants show an increase in centromere association
CAP-D3, a subunit of condensin II, is required for keeping centromeres apart [14]. However, because only CAP-D3 has been analyzed among the five subunits of condensin II, it remains possible that CAP-D3 functions independently of the condensin II complex. To clarify this, we elucidated the functions of the other condensin II-specific subunits, CAP-H2 and CAP-G2, in centromere association. The lack of CAP-D3 is known to cause an increase in centromere association into a few clusters in nuclei from rosette leaves [14]. Similarly, our FISH analysis using nuclei from flower buds not only in cap-d3-1 but also in cap-h2-2 and cap-g2-1 showed a reduction in the number of centromeric signals indicated by a probe for 180 bp repeats compared with the wild-type (Figure 1(a)). Wild-type 2–8C nuclei of A. thaliana contain ~4 to 10 centromere clusters [14]. In our analysis, nuclei containing ~7 to 10 centromere clusters were most frequent in the wild-type, while nuclei containing ~1 to 4 centromere clusters were most frequent in cap-h2-2 and cap-d3-1 (Figure 1(b)). Although it did not occur frequently in cap-g2-1 compared with cap-h2-2 and cap-d3-1, centromere association was significantly increased in cap-g2-1 compared with the wild-type (Figure 1(b)). Taken together, these data strongly suggested that CAP-H2, CAP-G2, and CAP-D3 act in a condensin II complex to maintain the correct centromere association in A. thaliana.
Figure 1.
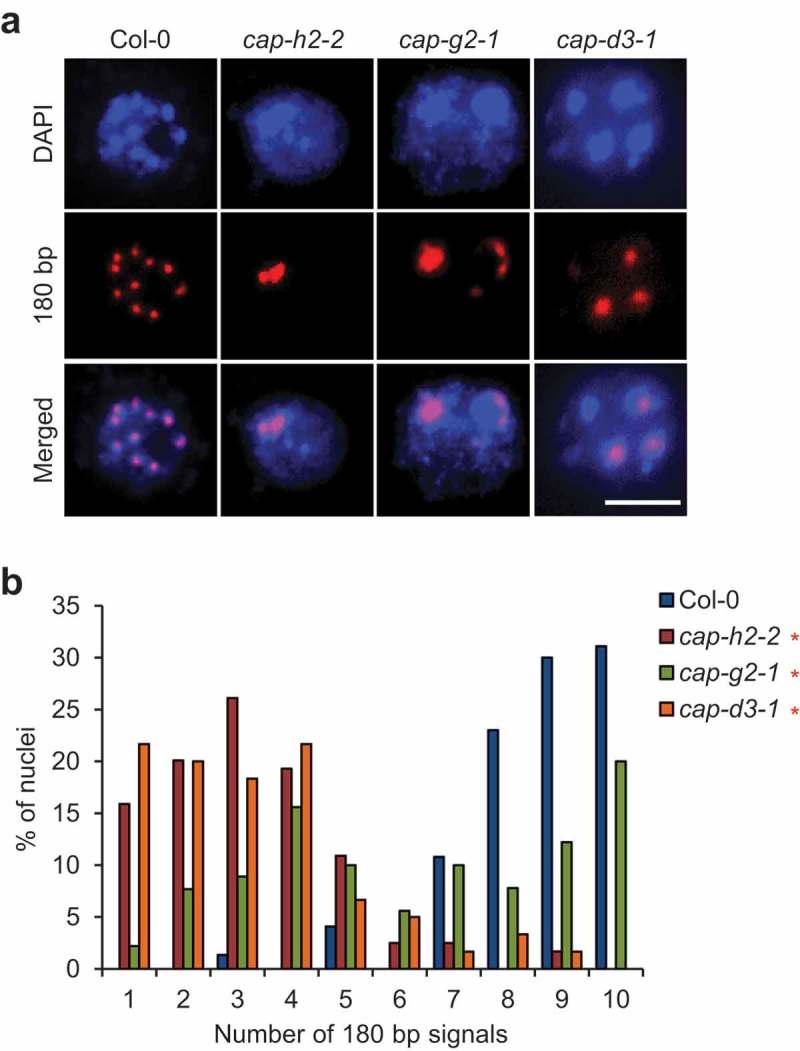
An increase in centromere association is observed in all mutants of condensin II-specific subunits. (a) Representative images of FISH detection of pericentromeric 180 bp signals in nuclei from flower buds of Col-0, cap-h2-2, cap-g2-1, and cap-d3-1. Bar = 5 μm. (b) Number of 180 bp signals in nuclei from flower buds of Col-0, cap-h2-2, cap-g2-1, and cap-d3-1. The frequency distribution of the number of 180 bp signals in each mutant was statistically compared with that in Col-0 by chi-squared test (*, p < 0.05, n = 74 for Col-0, n = 119 for cap-h2-2, n = 90 for cap-g2-1, n = 60 for cap-d3-1).
On the other hand, FISH analysis of telomere repeats revealed that condensin II mutants show nucleolar peripheral localization of telomeres without clustering as the wild-type does (Supplementary Fig. 1), suggesting that condensin II is not involved in the maintenance of telomere organization.
Condensin II mutants show disassociation of rDNA arrays from centromeres
Condensins are reported to function in axial compaction of chromosomes and recruitment of rDNA arrays to centromeres in D. melanogaster and S. pombe, respectively [23,25,26]. These facts led us to speculate that one or both functions would be conserved in A. thaliana condensin II. To investigate this possibility, we simultaneously visualized rDNA arrays and centromeres in nuclei of flower buds by FISH. Because in interphase nuclei of A. thaliana, condensin II subunits, CAP-H2 and CAP-G2, are localized in the nucleoplasm and the nucleolus [10,15] that is formed around nucleolar organizer regions (NORs) containing 45S rDNA arrays [27], we focused on the distribution of both 5S and 45S rDNA arrays. In A. thaliana ecotype Columbia, 5S rDNA arrays are located in close proximity to centromeres on chromosomes 3, 4 and 5; in contrast, 45S rDNA arrays are located away from centromeres on chromosomes 2 and 4 [27] (Figure 2(a)). However, similarly to 5S rDNA, the spatial location of 45S rDNA arrays basically shows an association with centromeres [28] (Figure 2(b)). Our FISH analysis revealed that more than 40% of nuclei showed the separation of 45S rDNA from centromeric regions in all condensin II mutants tested, while less than 7% of nuclei did in wild-type (Figure 2(b,c)). The differences between each mutant and wild-type were statistically significant (Figure 2(c)). We also found a reduction in the association of 5S rDNA arrays with the centromeric regions in both cap-h2-2 and cap-g2-1 (Figure 2(d)). The distance between the 5S rDNA signal and the nearest centromeric signal was more than twofold longer in both condensin II mutants compared with wild-type (Figure 2(e)). These results indicate that condensin II is required for the association of rDNA arrays with centromeres during interphase in A. thaliana, possibly through the axial compaction of chromosomes and/or direct recruitment of rDNA arrays to centromeres (Figure 2(f)).
Figure 2.
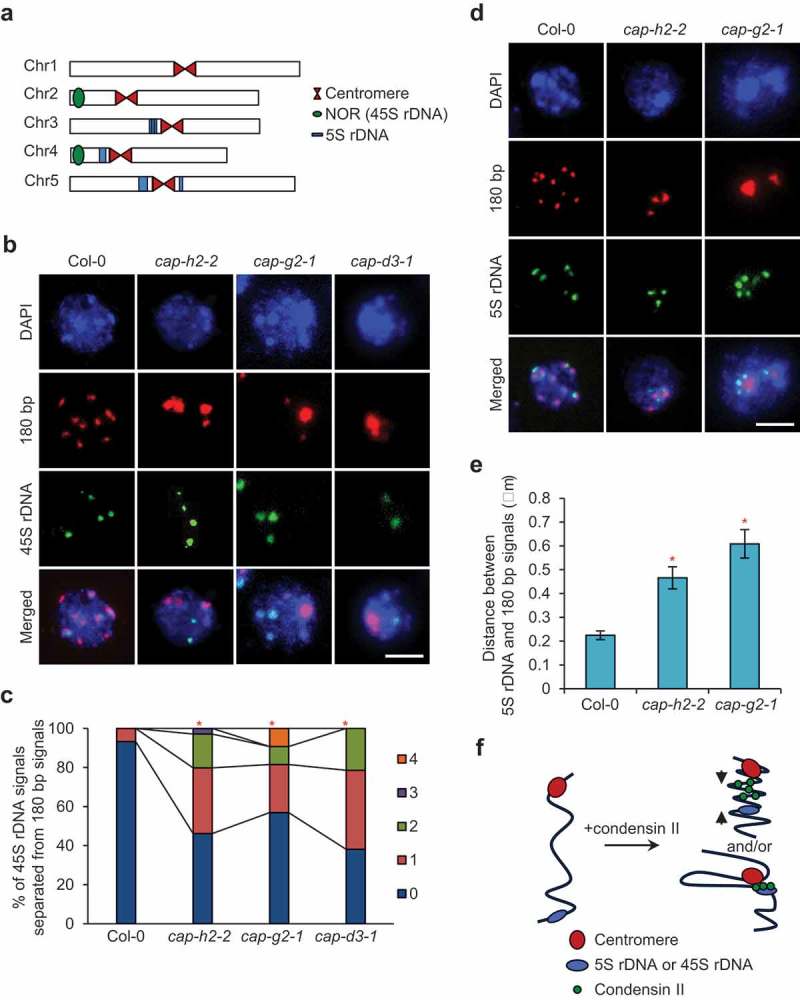
Defects in condensin II cause dissociation of centromeres from rDNA arrays. (a) Localization of rDNA arrays in the A. thaliana genome. 45S rDNA arrays are contained in nucleolar organizing regions (NORs) located on the short arms of chromosomes 2 and 4. 5S rDNA arrays are located in close proximity to the centromeres of chromosomes 3, 4, and 5. (b) Representative images of simultaneous FISH detection of pericentromeric 180 bp signals and 45S rDNA signals in nuclei from flower buds of Col-0, cap-h2-2, cap-g2-1, and cap-d3-1. Bar = 5 μm. (c) Number of 45S rDNA signals separated from 180 bp signals in nuclei from flower buds of Col-0, cap-h2-2, cap-g2-1, and cap-d3-1. The frequency distribution of the number of separated 45S rDNA signals in each mutant was statistically compared with that in Col-0 by chi-squared test (*, p < 0.05, n = 52 for Col-0, n = 104 for cap-h2-2, n = 65 for cap-g2-1, n = 42 for cap-d3-1). (d) Representative images of simultaneous FISH detection of pericentromeric 180 bp signals and 5S rDNA signals in nuclei from flower buds of Col-0, cap-h2-2, and cap-g2-1. Bar = 5 μm. (e) Quantification of the distance between 5S rDNA signals and 180 bp signals in nuclei from flower buds of Col-0, cap-h2-2, and cap-g2-1. Means ± SD are shown. The distance in each mutant was statistically compared with that in Col-0 by Student’s t-test (*, p < 0.01, n = 61 for Col-0, n = 51 for cap-h2-2, n = 61 for cap-g2-1). (f) Two possible models for condensin II-mediated association of centromeres with rDNA arrays: i) axial compaction of chromatin located between two loci (upper), ii) direct recruitment of an rDNA array to a centromeric region (lower).
The abnormal arrangement of 45S rDNA association with centromeres in condensin II mutants led us to speculate the function of condensin II in controlling rRNA gene expression. Therefore, we analyzed the expression levels of 5S rRNA and 45S rRNA (5.8S, 18S and 25S rRNA) genes and found that the defects in condensin II mutant showed similar expression levels of rRNA genes to the wild-type (Supplementary Fig. 2), suggesting that condensin II is not involved in the regulation of rRNA genes.
Chromatin loosening does not cause disassociation of rDNA arrays from centromeres
If we assume that axial compaction of chromosomes is required for the association of rDNA array with centromeres, defects in condensin II would cause the loosening of chromatin, at least which resides between the two loci. To verify this hypothesis, we analyzed histone acetylation, which is an epigenetic mark for loosened chromatin [29]. We found that the levels of both acetylated histone H3 and H4 in the whole nucleus were significantly higher in cap-h2-2 (Figure 3(a–c)), suggesting that chromatin loosening is enhanced by defects in condensin II function. Based on these results, we hypothesized that chromatin loosening would cause disassociation of rDNA arrays from centromeres. This hypothesis was assessed by analyzing the spatial relationship between 45S rDNA arrays and centromeres in plants showing increased genome-wide histone acetylation caused by treatment with a histone deacetylase inhibitor, trichostatin A (TSA) [30] (Figure 3(d)). We found that TSA treatment did not affect the association between centromeres or between 45S rDNA arrays and centromeres (Figure 3(e,f)). Taken together, these results suggest that although condensin II is involved in the maintenance of the epigenetic state to prevent an increase in chromatin loosening, unlike the case in D. melanogaster axial compaction of chromosomes is likely not required for the association of rDNA arrays and centromeres. In other words, it is plausible that the association of rDNA arrays and centromeres results from condensin II-mediated recruitment of rDNA arryas to centromeres as is the case for condensin I in S. pombe [25,26].
Figure 3.
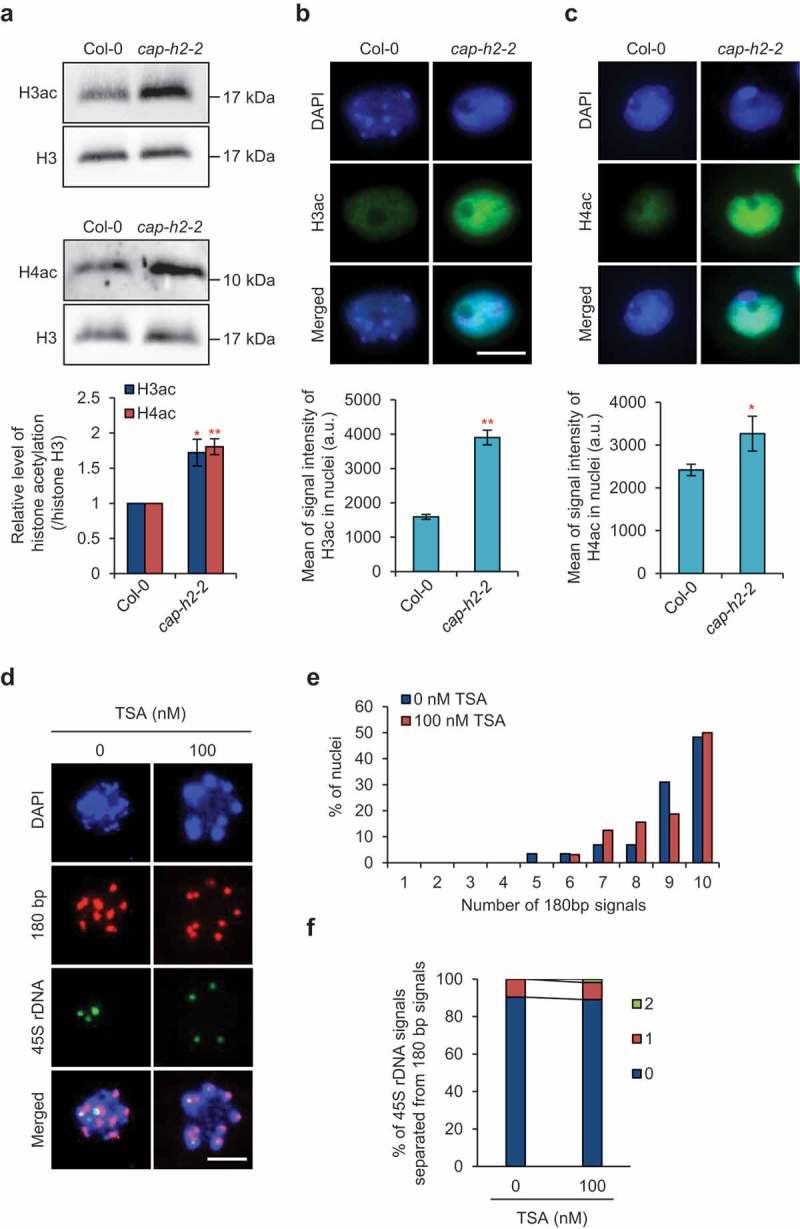
Defects in condensin II cause histone hyperacetylation, but this is unlikely to cause disruption of chromatin associations. (a) Immunoblotting of acetylated histone H3 and H4 in nuclei from 7-day-old seedlings of Col-0 and cap-h2-2. The graph shows the acetylated histone levels normalized by histone H3 levels. Error bars indicate SE. The level in cap-h2-2 was statistically compared with that in Col-0 by Student’s t-test (*, p < 0.05, **, p < 0.01, n = 3). (b,c) Immunostaining of acetylated histone H3 (b) and H4 (c) in nuclei from flower buds of Col-0 and cap-h2-2. Representative images of immunostained nuclei are shown. Bar = 5 μm. Each graph shows the acetylated histone levels represented by the mean intensity of immunofluorescence. Error bars indicate SE. The level in cap-h2-2 was statistically compared with that in Col-0 by Student’s t-test (*, p < 0.05, **, p < 0.01, n = 53 and 68 for H3ac, n = 41 and 65 for H4ac). (d) Representative images of simultaneous FISH detection of pericentromeric 180 bp signals and 45S rDNA signals in nuclei from 0 and 100 nM TSA-treated flower buds. Bar = 5 μm. (e) Number of 180 bp signals in nuclei from 0 and 100 nM TSA-treated flower buds. No significant differences in the frequency distribution of the number of 180 bp signals were detected in 100 nM TSA-treated plants compared with each control by chi-squared test (n = 29 for 0 nM TSA, n = 32 for 100 nM TSA). (f) Number of 45S rDNA signals separated from 180 bp signals in nuclei from 0 and 100 nM TSA-treated flower buds. No significant differences in the frequency distribution of the number of separated 45S rDNA signals were detected in 100 nM TSA-treated plants compared with each control by chi-squared test (n = 46).
Impaired genome integrity does not cause disassociation of heterochromatic loci
Condensin II is known to be crucial for the maintenance of genome integrity to alleviate DNA damage accumulation caused by genotoxic stresses, especially to high-boron stress, although the molecular mechanism is still unknown [10]. Thus, we next questioned whether impaired genome integrity would result in the disassociation of heterochromatic loci. To address this question, we focused on the mutants lig4-4 and rpt5a-4. lig4-4 is a mutant of DNA ligase IV, which functions in DNA repair [31], and rpt5a-4 is a mutant of RPT5a, which encodes a subunit of the 26S proteasome that functions in protecting the genome from attack by DNA-damaging factors [30]. Both mutants are known to show the hypersensitivity to high-B stress and accumulate DNA damage even under the normal growth condition [10,30,32], which are common features to condensin II mutants [10]. The reduced genome integrity in cap-h2-2, lig4-4 or rpt5a-4 was also confirmed by the hypersensitivity of root elongation through the treatment with a DNA-damaging reagent zeocin (Supplementary Fig. 3A). We visualized both 45S rDNA arrays and centromeric regions of these two mutants by FISH (Supplementary Fig. 3B) and found that both mutants showed a normal association of centromeres and a normal spatial relationship between 45S rDNA arrays and centromeres (Supplementary Fig. 3C, D). These results suggested that the impairment of genome integrity does not cause the disassociation of heterochromatic loci.
Discussion
In this study, it is strongly suggested that the function of condensin II as a complex is required for the proper organization of centromere distribution in interphase nuclei, rather than the independent function of individual condensin II-specific subunits, although we cannot completely show the evidence that CAP-H2, -G2, and -D3 form a complex. Considering that condensin II also prevents centromere clustering in mouse and D. melanogaster [23,24], its role in the separation of heterologous centromeres seems to be highly conserved among eukaryotes. However, the molecular mechanism by which the correct centromere distribution is maintained seems to differ among organisms. Our findings showed that the enforced chromatin loosening attributable to histone hyperacetylation induced by TSA did not affect the centromere distribution. Considering that histone hyperacetylation interferes with the axial compaction of meiotic chromosomes in mouse oocytes [33], unlike the case in D. melanogaster, A. thaliana condensin II acts in a different way to ensure the correct centromere distribution. Interestingly, a similar phenotype regarding the increase in centromere association is also seen in A. thaliana mutants of CROWDED NUCLEI (CRWN) proteins [34], which reside at the inner side of the nuclear periphery and play important roles in specifying nuclear shape and size [35,36]. This led us to speculate that condensin II and CRWNs act concertedly to ensure the correct centromere distribution during interphase in A. thaliana.
In yeast, as condensin II is absent, condensin I is considered to perform the job of condensin II [5]. In S. pombe, condensin mediates the interactions between centromeres and RNA Pol III-transcribed genes including 5S rDNA arrays and retrotransposons, which are crucial for the construction of 3D chromatin structure and contribute to gene silencing at these loci [25]. In A. thaliana, we found that in addition to 5S rDNA arrays, condensin II is involved in the association of 45S rDNA arrays with centromeres despite these locations being far apart on the same chromosome. The core subunit of condensin I and II, SMC4, was found to be involved in the silencing of transposons in pericentromeric heterochromatin of A. thaliana in conjunction with the enrichment of DNA methylation and the repressive histone modifications, H3K9me2 and H3K27me1 [37]. However, in A. thaliana, condensin II is not involved in repressing rRNA gene expression (Supplementary Fig. 2), unlike the case of condensin I in S. pombe [25].
Besides heterologous centromere associations, our results suggest that the association between centromeres and rDNA arrays does not require the axial compaction of chromosomes. The genome-wide distribution of condensins has now been unveiled in several species, and it has been found that condensin is enriched at promoters of highly expressed RNA Pol II genes, tRNA, and heterochromatic regions such as centromeres and rDNA arrays [25,38,39]. Therefore, it is tempting to speculate that direct binding of condensin II to rDNA array regions is necessary for their association with centromeres in A. thaliana. An understanding of the genome-wide distribution of condensin II and chromatin interactions between rDNA arrays and other loci provided by the genome-wide chromosome conformation capture method called Hi-C [40] will help to clarify condensin II’s function in the construction of 3D chromatin organization in A. thaliana.
Previously, we showed that condensin II is essential for maintaining genome integrity [10]. Consistent with this, we found that condensin II suppresses hyperacetylation of histone H3 and H4, which would impair genome integrity and increase susceptibility to genotoxic factors. Our mutant analysis suggests that the disruption of chromatin associations is not related to impaired genome stability. This is also supported by the fact that the fas2 and bru1 mutants defective in DNA damage responses maintain a normal centromere distribution [41,42]. Further analysis is needed to clarify whether the maintenance of genome integrity by condensin II is merely brought about by the correct organization of chromatin associations.
Materials and methods
Plant materials and growth condition
The cap-h2-2, cap-g2-1, lig4-4, and rpt5a-4 mutants have been described previously [10,30]. The cap-d3-1 mutant (SALK_094776, background Col-0) was obtained from the Arabidopsis Biological Resource Center. Plants were grown on vermiculite for ~1.5–2 months to obtain flower buds for FISH analysis. TSA treatment was conducted by soaking the flower buds in TSA solution diluted with water for 24 h. For the root elongation assay, seeds were sown on media containing MGRL solution, 1% (w/v) sucrose and 1.5% (w/v) gellan gum [10]. After 5 days incubation, the plants were transferred to MGRL medium containing the indicated concentrations of zeocin. After an additional 4 days incubation, the lengths of the newly elongated primary roots from the marked positions were determined using the ImageJ software (ver.1.51, http://rsb.info.nih.gov/ij/).
FISH analysis
Flower buds were fixed in Farmer’s fixative at 25°C for 12 h. The preparation of nuclei and hybridization of DNA with FISH probes were performed as described previously [43]. The DNA probes were synthesized as follows. Probes recognizing the centromeric 180 bp repeats (FP; 5ʹ-GATCAAGTCATATTCGACTC-3ʹ, RP; 5ʹ-GTTGTCATGTGTATGATTGA-3ʹ) was synthesized by nick translation using Biotin Nick Translation Mix (Roche, Basel, Switzerland). Probes recognizing 5S rDNA (FP; 5ʹ-GCGGAGCTCCCCAAATTTTGAC-3ʹ, RP; 5ʹ-GACCACGTGGTCGACAAAAAGTC-3ʹ), 45S rDNA (FP; 5ʹ-CAAGCAAGCCCATTCTCCTC-3ʹ, RP; 5ʹ-CAACTAGACCATGAAAATCC-3ʹ), and telomere repeats (FP; 5ʹ- TAAACCCTAAACCCTAAACCCTAAACCCTAAACCCTAAACCCTAAACCC −3ʹ, RP; 5ʹ- GGGTTTAGGGTTTAGGGTTTAGGGTTTAGGGTTTAGGGTTTAGGGTTTA-3ʹ) were synthesized by nick translation using DIG Nick Translation Mix (Roche). The hybridized nuclei were mounted with VECTASHIELD™ (Vector Laboratories, Burlingame, CA). The nuclei were observed under a light microscope (BX53m, Olympus, Tokyo, Japan) equipped with a CCD camera (DOC CAM U3-50S5M-C, Molecular Devices, Tokyo, Japan). The overlapping of the 180 bp signals and the 45S rDNA signals was analyzed with the ImageJ software. The distances between the position showing the maximum intensity of 5S rDNA signals and that of the nearest 180 bp signals were analyzed with the ImageJ software.
Immunoblotting
To detect histones from 7-day-old seedlings of Col-0 or cap-h2-2, proteins were extracted using 1xSDS sample buffer (without Bromophenol blue). The extract was filtered using two layers of Miracloth and then boiled for 10 min. The antibodies used in this study were anti-histone H3 (MABI0301, MBL) (1:2000 dilution), anti-acetylated histone H3 (06–599, Millipore) (1:1000 dilution), and anti-acetylated histone H4 (06–866, Millipore) (1:1000 dilution). The target proteins of each antibody were visualized with ImmunoStar (Wako, Osaka, Japan) on a Fusion Pulse system (Vilber Lourmat, Osaka, Japan). To quantify the protein levels, the signal intensities of target bands were measured using Gel Analyzer in the Image J software.
Immunostaining
Flower buds were fixed with 4% paraformaldehyde/1xPBS for 40 min and then washed with 1xPBS three times. The fixed flower buds were digested in solution {1% Doriselase (Sigma-Aldrich Japan, Tokyo, Japan), 0.5% Cellulase R-10 (Yakult Pharmaceutical, Tokyo, Japan), 0.025% pectolyase (Seishin Pharmaceutical, Tokyo, Japan) in water} for 30 min at 37°C. After washing with 1xPBS twice, the fixed flower buds were squashed using a coverslip on a slide coated with poly-l-lysine. The slides were immediately placed in liquid N2 and the cover glass was removed. After the slides were dried, they were washed with 1xPBS once. Then, each sample was treated with 4% (w/v) bovine serum albumin (BSA)/1xPBS on the slide for 30 min at room temperature. After the solution was removed from the slide, 100 μl of a solution of primary antibodies diluted 100 times with 1% (w/v) BSA/1xPBS was added to the sample. Anti-H3ac rabbit IgG (Millipore, Temecula, CA, USA) and anti-H4ac rabbit IgG (Millipore) were used as the primary antibodies. The slides were incubated overnight at 4°C and then washed with 0.05% Tween20/1xPBS twice. Then, each sample was treated with 4% (w/v) bovine serum albumin (BSA)/1xPBS on the slide for 30 min at room temperature. After washing with 1xPBS twice, 50 μl of a solution of anti-rabbit IgG Alexa Fluor® 488 Fab (Invitrogen, Carlsbad, CA) diluted 200 times with 1% (w/v) BSA/1xPBS was added to the sample. The slides were incubated at 37°C for 1 h in the dark and then treated with 0.05% Tween20/1xPBS containing 1 µg/mL DAPI for 10 min. The slides were then washed with 0.05% Tween20/1xPBS and subsequently with water three times. After drying the slides at 37°C in the dark, the hybridized nuclei were mounted with VECTASHIELD™. The nuclei were observed under the same light microscope (BX53m). The mean intensity of the fluorescence in the entire nuclei was analyzed using the ImageJ software.
rRNA gene expression
Total RNA from shoots and roots of 7-day-old seedlings was extracted as described previously [30]. Approximately 1 μg of total RNA was reverse-transcribed with a Verso cDNA Synthesis Kit (Thermo Fisher Scientific, Yokohama, Japan) following the manufacturer’s protocol. Real-time PCR was performed with Thermal Cycler Dice Real Time System II (Takara Bio, Shiga, Japan) and Luna qPCR Master Mix (New England Biolabs, Tokyo, Japan). Actin2 was used for the normalization of cDNA concentration. The primers for the detection of used were as follows: 5ʹ-GATGCGATCATACCAGCACT-3ʹ (FP) and 5ʹ-GGATGCAACACGAGGACTTC-3ʹ (RP) for 5S rRNA, 5ʹ-CCTGCGGCTTAATTTGACTC-3ʹ (FP) and 5ʹ-GACAAATCGCTCCACCAACT-3ʹ (RP) for 18S rRNA, 5ʹ-CGCGAGTTCTATCGGGTAAA-3ʹ (FP) and 5ʹ-CACTTGGAGCTCTCGATTCC-3ʹ (RP) for 25S rRNA, and 5ʹ-AAGTCATAACCATCGGAGCTG-3ʹ (FP) and 5ʹ-ACCAGATAAGACAAGACACAC-3ʹ (RP) for ACT2.
Funding Statement
This work was supported by Japan Society for the Promotion of Science [JP15H05955]; Japan Society for the Promotion of Science [JP15H05962] to S.M.
Acknowledgments
We thank Robbie Lewis, MSc, from Edanz Group (www.edanzediting.com/ac) for editing a draft of this manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Jeppsson K, Kanno T, Shirahige K, et al. The maintenance of chromosome structure: positioning and functioning of SMC complexes. Nat Rev Mol Cell Biol. 2014;15:601–614. [DOI] [PubMed] [Google Scholar]
- [2].Uhlmann F. SMC complexes: from DNA to chromosomes. Nat Rev Mol Cell Biol. 2016;17:399–412. [DOI] [PubMed] [Google Scholar]
- [3].Hirano T. Condensins: organizing and segregating the genome. Curr Biol. 2005;15:R265–275. [DOI] [PubMed] [Google Scholar]
- [4].Hirano T. Chromosome territories meet a condensin. PLoS Genet. 2012;8:e1002939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hirano T. Condensin-based chromosome organization from bacteria to vertebrates. Cell. 2016;164:847–857. [DOI] [PubMed] [Google Scholar]
- [6].Liu CC, McElver J, Tzafrir I, et al. Condensin and cohesin knockouts in Arabidopsis exhibit a titan seed phenotype. Plant J. 2002;29:405–415. [DOI] [PubMed] [Google Scholar]
- [7].Tzafrir I, McElver JA, Liu CC, et al. Diversity of TITAN functions in Arabidopsis seed development. Plant Physiol. 2002;128:38–51. [PMC free article] [PubMed] [Google Scholar]
- [8].Siddiqui NU, Stronghill PE, Dengler RE, et al. Mutations in Arabidopsis condensin genes disrupt embryogenesis, meristem organization and segregation of homologous chromosomes during meiosis. Development. 2003;130:3283–3295. [DOI] [PubMed] [Google Scholar]
- [9].Siddiqui NU, Rusyniak S, Hasenkampf CA, et al. Disruption of the Arabidopsis SMC4 gene, AtCAP-C, compromises gametogenesis and embryogenesis. Planta. 2006;223:990–997. [DOI] [PubMed] [Google Scholar]
- [10].Sakamoto T, Inui YT, Uraguchi S, et al. Condensin II alleviates DNA damage and is essential for tolerance of boron overload stress in Arabidopsis. Plant Cell. 2011;23:3533–3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Oliveira RA, Heidmann S, Sunkel CE. Condensin I binds chromatin early in prophase and displays a highly dynamic association with Drosophila mitotic chromosomes. Chromosoma. 2007;116:259–274. [DOI] [PubMed] [Google Scholar]
- [12].Fujiwara T, Tanaka K, Kuroiwa T, et al. Spatiotemporal dynamics of condensins I and II: evolutionary insights from the primitive red alga Cyanidioschyzon merolae. Mol Biol Cell. 2013;24:2515–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ono T, Fang Y, Spector DL, et al. Spatial and temporal regulation of Condensins I and II in mitotic chromosome assembly in human cells. Mol Biol Cell. 2004;15:3296–3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Schubert V, Lermontova I, Schubert I. The Arabidopsis CAP-D proteins are required for correct chromatin organisation, growth and fertility. Chromosoma. 2013;122:517–533. [DOI] [PubMed] [Google Scholar]
- [15].Fujimoto S, Yonemura M, Matsunaga S, et al. Characterization and dynamic analysis of Arabidopsis condensin subunits, AtCAP-H and AtCAP-H2. Planta. 2005;222:293–300. [DOI] [PubMed] [Google Scholar]
- [16].Heale JT, Ball AR, Schmiesing JA, et al. Condensin I interacts with the PARP-1-XRCC1 complex and functions in DNA single-strand break repair. Mol Cell. 2006;21:837–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Wood JL, Liang Y, Li K, et al. Microcephalin/MCPH1 associates with the Condensin II complex to function in homologous recombination repair. J Biol Chem. 2008;283:29586–29592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Xu Y, Leung CG, Lee DC, et al. MTB, the murine homolog of condensin II subunit CAP-G2, represses transcription and promotes erythroid cell differentiation. Leukemia. 2006;20:1261–1269. [DOI] [PubMed] [Google Scholar]
- [19].Hartl TA, Smith HF, Bosco G. Chromosome alignment and transvection are antagonized by condensin II. Science. 2008;322:1384–1387. [DOI] [PubMed] [Google Scholar]
- [20].Joyce EF, Williams BR, Xie T, et al. Identification of genes that promote or antagonize somatic homolog pairing using a high-throughput FISH-based screen. PLoS Genet. 2012;8:e1002667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Schuster AT, Sarvepalli K, Murphy EA, et al. Condensin II subunit dCAP-D3 restricts retrotransposon mobilization in Drosophila somatic cells. PLoS Genet. 2013;9:e1003879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wallace HA, Klebba JE, Kusch T, et al. Condensin II regulates interphase chromatin organization through the Mrg-Binding motif of Cap-H2. G3 (Bethesda). 2015;5:803–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Bauer CR, Hartl TA, Bosco G. Condensin II promotes the formation of chromosome territories by inducing axial compaction of polyploid interphase chromosomes. PLoS Genet. 2012;8:e1002873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Nishide K, Hirano T. Overlapping and non-overlapping functions of condensins I and II in neural stem cell divisions. PLoS Genet. 2014;10:e1004847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Iwasaki O, Tanizawa H, Kim K-D, et al. Interaction between TBP and condensin drives the organization and faithful segregation of mitotic chromosomes. Mol Cell. 2015;59:755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Iwasaki O, Corcoran CJ, Noma K-I. Involvement of condensin-directed gene associations in the organization and regulation of chromosome territories during the cell cycle. Nucleic Acids Res. 2016;44:3618–3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Layat E, Sáez-Vásquez J, Tourmente S. Regulation of Pol I-transcribed 45S rDNA and Pol III-transcribed 5S rDNA in Arabidopsis. Plant Cell Physiol. 2012;53:267–276. [DOI] [PubMed] [Google Scholar]
- [28].Fransz PF, de Jong JH. Chromatin dynamics in plants. Curr Opin Plant Biol. 2002;5:560–567. [DOI] [PubMed] [Google Scholar]
- [29].Takata H, Hanafusa T, Mori T, et al. Chromatin compaction protects genomic DNA from radiation damage. PLoS ONE. 2013;8:e75622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Sakamoto T, Tsujimoto-Inui Y, Sotta N, et al. Proteasomal degradation of BRAHMA promotes Boron tolerance in Arabidopsis. Nat Commun. 2018;9:5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].West CE, Waterworth WM, Jiang Q, et al. Arabidopsis DNA ligase IV is induced by gamma-irradiation and interacts with an Arabidopsis homologue of the double strand break repair protein XRCC4. Plant J. 2000;24:67–78. [DOI] [PubMed] [Google Scholar]
- [32].Fulcher N, Sablowski R. Hypersensitivity to DNA damage in plant stem cell niches. Pnas. 2009;106:20984–20988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Yang F, Baumann C, Viveiros MM. De La Fuente R.. Histone hyperacetylation during meiosis interferes with large-scale chromatin remodeling, axial chromatid condensation and sister chromatid separation in the mammalian oocyte. Int J Dev Biol. 2012;56:889–899. [DOI] [PubMed] [Google Scholar]
- [34].Wang H, Dittmer TA, Richards EJ. Arabidopsis CROWDED NUCLEI (CRWN) proteins are required for nuclear size control and heterochromatin organization. BMC Plant Biol. 2013;13:200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Dittmer TA, Stacey NJ, Sugimoto-Shirasu K, et al. LITTLE NUCLEI genes affecting nuclear morphology in Arabidopsis thaliana. Plant Cell. 2007;19:2793–2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Sakamoto Y, Takagi S. LITTLE NUCLEI 1 and 4 regulate nuclear morphology in Arabidopsis thaliana. Plant Cell Physiol. 2013;54:622–633. [DOI] [PubMed] [Google Scholar]
- [37].Wang J, Blevins T, Podicheti R, et al. Mutation of Arabidopsis SMC4 identifies condensin as a corepressor of pericentromeric transposons and conditionally expressed genes. Genes Dev. 2017;31:1601–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kranz A-L, Jiao C-Y, Winterkorn LH, et al. Genome-wide analysis of condensin binding in Caenorhabditis elegans. Genome Biol. 2013;14:R112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Sutani T, Sakata T, Nakato R, et al. Condensin targets and reduces unwound DNA structures associated with transcription in mitotic chromosome condensation. Nat Commun. 2015;6:7815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Yu S, Lemos BA. Portrait of ribosomal DNA contacts with Hi-C reveals 5S and 45S rDNA anchoring points in the folded human genome. Genome Biol Evol. 2016;8:3545–3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Takeda S, Tadele Z, Hofmann I, et al. BRU1, a novel link between responses to DNA damage and epigenetic gene silencing in Arabidopsis. Genes Dev. 2004;18:782–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Endo M, Ishikawa Y, Osakabe K, et al. Increased frequency of homologous recombination and T-DNA integration in Arabidopsis CAF-1 mutants. Embo J. 2006;25:5579–5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Hasegawa J, Sakamoto T, Fujimoto S, et al. Auxin decreases chromatin accessibility through the TIR1/AFBs auxin signaling pathway in proliferative cells. Sci Rep. 2018;8:7773. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


