Abstract
DNA in the human Herpes simplex virus type 1 (HSV-1) capsid is packaged to a tight density. This leads to tens of atmospheres of internal pressure responsible for the delivery of the herpes genome into the cell nucleus. In this study we show that, despite its liquid crystalline state inside the capsid, the DNA is fluid-like, which facilitates its ejection into the cell nucleus during infection. We found that the sliding friction between closely packaged DNA strands, caused by interstrand repulsive interactions, is reduced by the ionic environment of epithelial cells and neurons susceptible to herpes infection. However, variations in the ionic conditions corresponding to neuronal activity can restrict DNA mobility in the capsid, making it more solid-like. This can inhibit intranuclear DNA release and interfere with viral replication. In addition, the temperature of the human host (37 °C) induces a disordering transition of the encapsidated herpes genome, which reduces interstrand interactions and provides genome mobility required for infection.
Herpesviruses are a leading cause of human viral disease, second only to influenza and cold viruses1. Herpesvirus infections are life-long with extended latency periods between recurrent reactivations, making treatment of herpesvirus infections more difficult2. Current anti-herpes drugs consist primarily of viral DNA polymerase inhibitors, which can be effective. Unfortunately, high mutation rates during viral genome replication lead to drug resistance upon extended use, which is problematic especially in the immunocompromised patients in which they are most needed3,4. This problem has led to a search for the less specific physical properties of the viral particles regulating replication.
HSV-1 is a prototypical model system used to investigate the infection pathways of herpesviruses and other viruses that deliver their genome into a cell nucleus without capsid disassembly5. Analogous to double-stranded (ds) DNA bacteriophages6–9, herpesviruses package their micrometer-long dsDNA into a nanometer-scale protein shell called the capsid10. The infectivity of HSV-1 is facilitated by its ability to eject its genome into the host cell nucleus upon binding of the capsid portal to the nuclear pore complex (NPC)5. We recently discovered and measured a high internal DNA pressure of 20 atm in HSV-1 capsids11. This internal pressure results from repulsive DNADNA interactions and bending stress acting on the tightly confined genome12. This pressure is capable of driving ejection of the entire viral genome from the capsid, suggesting a pressure-dependent infection mechanism in herpesviruses. At the same time, DNA confined in the HSV-1 capsid is at the extreme end of the packaging limit, where the genome is hexagonally ordered with interstrand surface separations of only ~11 Å or less13. It has been experimentally shown in vitro that DNA confined inside viral capsids at comparable packing densities is trapped in a glassy state with restricted molecular motion14. Despite decades of investigations of encapsidated genome structures15,16, it remains unknown what provides the mobility of closely packaged DNA required for the initiation of genome ejection through a single portal opening in the capsid.
In this work, we demonstrate how the ionic conditions and temperature mimicking those of the host cytoplasm regulate the structure, and the resulting mobility, of the encapsidated HSV-1 genome. We show that, close to the optimum temperature for infection (37 °C), the HSV-1 DNA in the capsid becomes more disordered and locally less densely packed with increased interstrand separations, which reduces DNA-DNA repulsions. This leads to a major increase in genome mobility or fluidity, which facilitates infection. Temperature is a parameter that is rarely varied in biophysical measurements on viruses but is pertinent to viral replication. The intracapsid DNA structure is determined using solution small-angle X-ray scattering (SAXS). The relative mobility of the packaged herpes genome is measured with atomic force microscopy (AFM) nano-indentation of HSV-1 capsids. Our results suggest a remarkable physical adaptation of herpesviruses to the environment of their human host and describe favorable as well as inhibiting in vivo conditions for HSV-1 infectivity and replication.
RESULTS
Tightly packaged DNA inside the HSV-1 capsid
Herpesvirus capsids assemble from many copies of just a few subunits in a multistage process. During the final stages of HSV-1 genome packaging, the terminase motor cleaves concatameric viral DNA, followed by sealing of the portal vertex to ensure stable retention of the encapsidated genome. The scaffold protein around which the capsid is initially self-assembled is proteolytically cleaved and removed, and DNA containing C-capsids is formed. These represent properly packaged nucleocapsids that mature into infectious virus particles17. Along with C-capsids, a number of empty A- and B-capsids are produced during viral replication. B-capsids, which retain scaffolding proteins, do not complete the maturation pathway and are a dead-end product17. A-capsids lack scaffolding proteins and are thought to arise from aborted packaging and loss of packaged DNA17. The shells of both A- and C-capsids are essentially identical in structure. This allows for a direct comparison of capsid stiffness between empty and DNA-filled capsids measured with AFM nano-indentation.
The stiffness of DNA packaged inside the HSV-1 capsid is measured by recording the force resisting the capsid indentation when the AFM tip is brought into contact with the DNA-filled capsid in solution (Online Methods). As viral capsids are permeable to water, capsid deformation leads to displacement of water molecules hydrating the DNA18, which occurs with compression of the packaged DNA strands. The resisting force attributed to the deformation of the empty capsid shell can be determined separately by AFM indentation of A-capsids. Hence, the force resisting C-capsid AFM indentation (in addition to the stiffness of the empty capsid) is associated with the free energy required to remove water molecules hydrating the closely packaged DNA strands and the change in the interhelical phosphate-phosphate correlations18. This energy is described by DNA-DNA repulsive interactions and DNA bending stress12,19. Furthermore, if the AFM tip rate of indentation is faster than the relaxation dynamics of DNA during deformation, there can be an additional contribution to the measured DNA stiffness from the interstrand sliding friction. The sliding friction occurs from dragging closely packed negatively charged DNA helices past other helices. It has been proposed that the so-called Coulomb sliding friction between neighboring DNA helices has a major role in mobility of DNA at high packing densities in the viral capsids20,21. Indeed, it was recently shown that interhelical sliding friction leads to a kinetically trapped, glassy DNA state inside the capsid. This high friction genome state was found to strongly affect the rates of DNA packaging in vitro14.
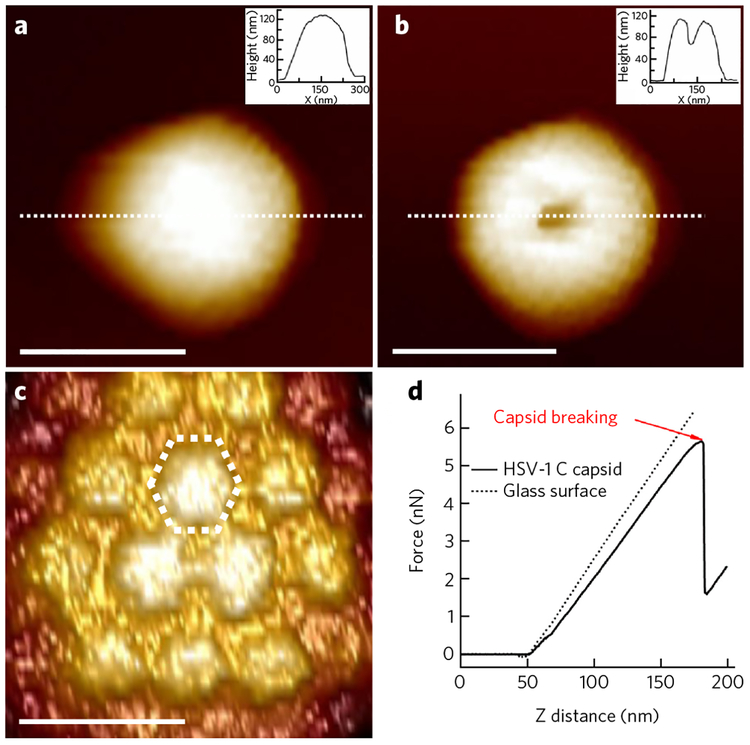
Figure 1 shows this single-molecule approach for analyzing intracapsid DNA stiffness. The force-distance curve in Figure 1c is linear, suggesting an elastic deformation of the HSV-1 capsid18,22. An abrupt drop in the force-distance curve signifies breaking of the capsid. The slope of the force-distance curve is the spring constant k, which describes the stiffness of the viral particle. Unless specified otherwise, the rate of indentation in all of the measurements was 60 nm s−1, which we have verified is slower than the relaxation rates for AFM-induced capsid and DNA deformations. Thus, measured stiffness reflects the free energy of the capsid and the encapsi-dated genome. The stiffness of the packaged DNA is measured as k(DNA) = k(C-capsid) − k(A-capsid) (assuming that capsid and DNA deformations are independent of each other). This k(DNA) value allows us to determine the change in the stiffness attributed only to the DNA state in the capsid. A decrease in capsid stiffness attributable to DNA indicates an increase in genome fluidity or, equivalently, mobility. The observed variation in the stiffness and the relative compressibility of the encapsidated herpes genome is explained by the structural DNA transitions determined with SAXS measurements16,23.
Figure 1 |. AFM nano-indentation analysis of encapsidated DNA mobility.
(a) AFM image of HSV-1 capsid in buffer. The inset shows a cross-section profile along the dashed line. Scale bar, 150 nm. (b) AFM image of HSV-1 capsid after breaking. Scale bar, 150 nm. (c) Zoomed-in image of HSV-1 C-capsid with individual hexons observed on the capsid surface. Scale bar, 30 nm. (d) Force-distance curves for glass substrate and for HSV-1 C-capsid at 24 °C in 10 mM Mg-Tris buffer. An abrupt drop in the force-distance curve signifies breaking of the capsid.
As mentioned above, both ionic conditions and temperature influence the intracapsid DNA structure and determine its mobility. dsDNA is a stiff polymer chain with a high negative charge density. The interplay between the bending stress and the DNA-DNA repulsive interactions determines the structure and interstrand separation for the encapsidated genome23,24. First we investigated the effect of ionic conditions approximating those in the cellular cytoplasm on the mobility of the encapsidated HSV-1 genome. As all viral capsids are permeable to water and ions, the ionic conditions of the cellular cytoplasm directly affect the DNA-DNA repulsive interactions by screening the negative charges on the tightly packaged DNA strands24. We showed that weaker DNA-DNA interactions lead to greater DNA fluidity as well as lower interhelical sliding friction. Next we showed how the encapsidated HSV-1 genome undergoes a structural transition close to the temperature of infection. The DNA bending stress is controlled by variation of temperature25. With increasing temperature, DNA becomes more flexible, leading to less bending stress. This results in larger DNA-DNA spacings and therefore weaker inter-strand interactions and increased DNA mobility. Our data reveal a solid-to-fluid–like transition of the encapsidated HSV-1 genome occurring close to the temperature of infection. This observed DNA mobility switch occurring in vitro is likely to contribute to efficient delivery of the viral genome into a cell nucleus during infection.
Effect of ionic conditions on the intracapsid DNA mobility
The stiffness of DNA packaged in HSV-1 capsids was determined with AFM nano-indentation under various ionic conditions. The ionic conditions were chosen to mimic the ionic environment of epithelial and neuronal cell cytoplasms (with similar ionic strength and ion species). As cations interact with negatively charged DNA in the capsid, we used an effective ionic strength value based only on the cation concentration to define our buffer conditions. HSV-1 replicates mainly in epithelial cells and establishes a latent infection in neurons2. In the cytoplasm of epithelial cells, the effective ionic strength from the cations is 0.1 M and is similar to that in the mammalian neuronal cell at its resting potential26–29. However, the cation ionic strength in neurons varies by nearly tenfold between 0.02 M and 0.2 M, depending on the phase of the action potential26. In addition to the monovalent cations in the cytoplasm of both cell types (for example, Na+, K+), the epithelial cells also contain Mg2+ at 10 mM27–29. The neuronal cytoplasm has, in contrast, only trace amounts of Ca2+ at 0.0001 mM26.
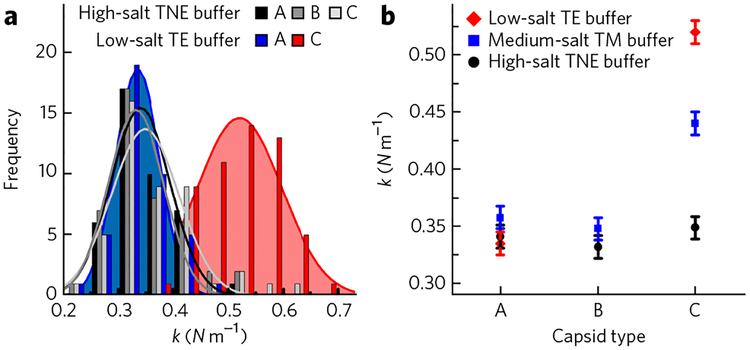
As a reference cation ionic condition for our measurements, we chose the lower ionic strength limit in neuronal cytoplasm (close to 0.02 M). This choice is motivated by our previous AFM nano-indentation measurements, which demonstrated that intracapsid DNA stiffness decreases with increasing ionic strength22. Thus, if DNA packaged in the HSV-1 capsid has low stiffness and fluid-like behavior at the lower ionic strength limit, it will remain fluid-like also at the higher ionic strength. Figure 2 shows measured spring constants for HSV-1 A-, B- and C-capsids at 24 °C in three different buffers: low-salt TE buffer (7.5 mM MgCl2, 15 mM EDTA, 50 mM Tris-HCl), medium-salt TM buffer (50 mM Tris-HCl, 10 mM MgCl2) and high-salt TNE buffer (500 mM NaCl, 1 mM EDTA, 10 mM Tris-HCl). Figure 2 shows that empty A- and B-capsids have identical spring constants that are not influenced by the ionic conditions (k ~ 0.34 ± 0.01 N m−1; error represents s.e., obtained from Gaussian curve fitting to the distribution of the spring constants).
Figure 2 |. Effect of cellular ionic conditions on the mobility of encapsidated DNA.
(a) Histograms of spring constants for HSV-1 A-, B- and C-capsids at 24 °C in different buffer conditions. In high-salt TNE buffer (TNE), no difference in spring constants is observed for A-, B- and C-capsids. On the contrary, in low-salt TE buffer, the spring constant of C-capsids is much larger compared to that of the A-capsids. (b) Spring constants for HSV-1 A-, B- and C-capsids in different ionic conditions. Forty to sixty force-distance curves were analyzed for each case. The Gaussian curve fitting to the distribution of the spring constants yields the s.d., which is converted to the s.e. The error bars show the s.e.
Because mammalian neuronal cytoplasm has only trace amounts of divalent ions, we removed Mg ions from the HSV-1 solution in TM buffer by adding an excess of EDTA. The resulting TE buffer has a cation ionic strength of 0.015 M and mimics the ionic environment in the neurons at the lowest salt concentration. The DNA-filled C-capsid in TE buffer at 24 °C shows a large increase in stiffness of 53% compared to the A-capsid (k(A-capsid) ~0.34 ± 0.01 N m−1 versus k(C-capsid) ~0.52 ± 0.01 N m−1; Fig. 2b). This observation suggests that DNA is more restricted or more solid-like in the HSV-1 capsid at the low-salt limit. At the same time, the stiffness of the C-capsid in the medium-salt TM buffer, which has a higher cation ionic strength than TE buffer (0.015 M versus 0.02 M), is reduced by 15% compared to the capsid stiffness in TE buffer (Fig. 2b). The presence of Mg ions resulted in a decrease in the DNA-DNA repulsive interactions and consequently improved intracapsid DNA mobility. Finally, in the high-salt conditions set by TNE buffer with a cation ionic strength of 0.25 M (similar to that in neurons at the peak of an action potential26), which is typically used for storage of HSV-1 capsids, there is no measurable contribution to the overall capsid stiffness from the packaged DNA. A-, B- and C-capsids in TNE buffer have the same stiffness (k ~ 0.34 ± 0.01 N m−1), which was also observed in ref. 30. The DNA-DNA repulsive interactions are now considerably reduced by the counterion screening, making the encapsidated genome even more fluid.
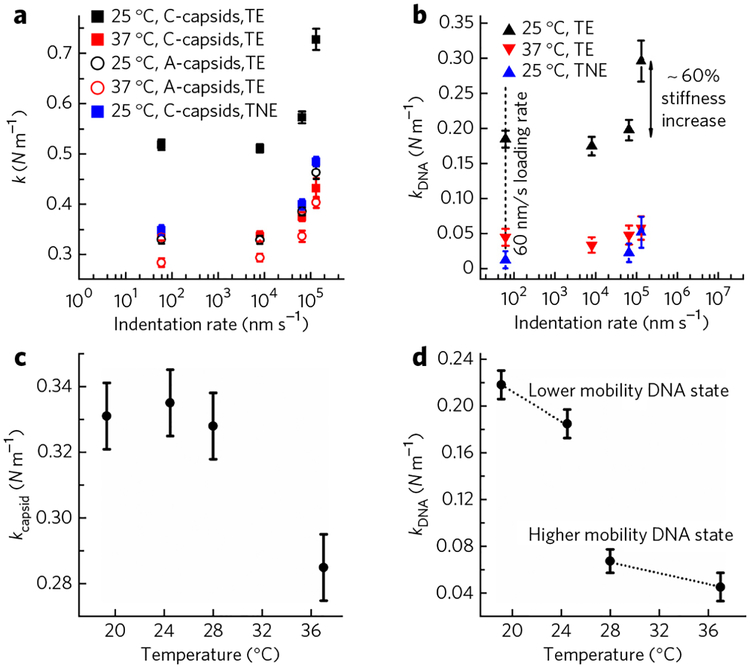
It has been shown for several bacteriophages31,32 with DNA packing densities similar to that of HSV-1 that DNA ejection rates in vitro could be as high as 60,000–75,000 base pairs per second. For the Archaeal virus His1, the ejection rate in vitro is 140,000 base pairs per second33. Assuming that DNA strands are sliding past each other during the ejection process20,21, these ejection velocities correspond to an interhelical sliding rate of 104–105 nm s−1 (using 0.34 nm per base pair). As mentioned above, when a C-capsid is compressed with an AFM tip, the DNA strands inside the capsid slide past each other as well. By choosing the AFM indentation rates corresponding to the DNA sliding rates during ejection, we attempted to simulate the friction effects occurring during the genome ejection process. Therefore, in addition to the DNA stiffness associated with the free energy of genome confined inside the capsid measured above, we also investigated whether the DNA sliding friction contributes to the restricted genome mobility at the higher rates of indentation. The intracapsid DNA stiffness, k(DNA), was measured as a function of AFM tip indentation rate. Figure 3a shows the spring constant, k, versus the indentation rate for A- and C-capsids in both low-salt TE buffer and high-salt TNE buffer at 24 °C. Figure 3b shows the k(DNA) versus indentation rate derived from k(C-capsid) –k(A-capsid), where the spring constants for C- and A-capsids were measured separately for each rate of indentation. At the indentation rate of 60 nm s−1 used for all of the spring constant measurements above, the measured DNA stiffness does not depend on the rate of indentation in both low- and high-salt buffers, as the spring constants measured at indentation rates of 60 nm s−1 and 7.8 × 103 nm s−1 are the same (Fig. 3b). However, as the indentation rates approach 105 nm s−1 (corresponding to the DNA sliding rates during ejection), the DNA stiffness rises by 60% in the low-salt TE buffer. At the same time, in the high-salt TNE buffer, the DNA stiffness remains essentially unchanged within the entire indentation rate interval (102–105 nm s−1). These observations suggest that the increase in the DNA stiffness in the low-salt conditions is accompanied by interhelical sliding friction caused by the strong repulsive interactions between the DNA strands. When the DNA-DNA repulsions are reduced in the high-salt conditions, the genome remains fluid-like, with little frictional contribution even at very high indentation rates.
Figure 3 |. Solid-to-fluid like DNA transition inside the HSV-1 capsid close to the temperature of infection.
(a) Spring constants of HSV-1 C- and A-capsids as a function of indentation rates at 24 °C and 37 °C in low-salt TE buffer. We also performed measurements in high-salt TNE buffer at 24 °C. Note that the A-capsid spring constants are the same in both TE- and TNE buffers. The error bars show the s.e. (b) Spring constants of encapsidated DNA obtained from subtracting spring constants for A-capsids from that of C-capsids at each indentation rate. k(DNA) is shown at 24 °C and 37 °C in low-salt TE buffer and at 24 °C in high-salt TNE buffer. The error bars show s.e. The dashed line shows the indentation rate of 60 nm s−1, at which the DNA relaxation rate during the indentation is faster than the AFM tip indentation rate (i.e., indentation occurs at equilibrium). (c) Spring constants for HSV-1 A-capsid in TE buffer as a function of temperature. (d) Spring constants for encapsidated DNA alone in C-capsid in TE buffer as a function of temperature. kDNA is derived by subtracting the spring constant for A-capsid from that of the C-capsid. Forty to sixty force-distance curves were analyzed for each case. The Gaussian curve fitting to the distribution of the spring constants yields the s.d., which is converted to the s.e. The error bars show the s.e.
These results demonstrate that DNA-DNA repulsive interactions determine the stiffness of DNA packaged inside the capsid. AFM data suggest that both repulsive inter-strand interactions and interhelical sliding friction restrict the mobility of the intracapsid genome. This is likely to cause an energy barrier for initiation of DNA ejection from the capsid in the low-salt conditions. Given the capsid permeability, the ionic environment of the host cytoplasm will therefore affect the ejection mobility of the encapsidated genome. Although our data show that intracapsid DNA is fluid-like in salt conditions similar to those of epithelial cells, its mobility seems to be restricted at room temperature in the low-salt conditions similar to those in neurons. The results in the next section provide an explanation to the fundamentally important question of how the HSV-1 genome is successfully released in the cell nucleus under the low-salt conditions of the neuronal cytoplasm.
Using AFM indentation measurements combined with SAXS, we demonstrate that DNA-DNA separation is increased when the temperature is raised from 20 °C to 37 °C as a result of decreased DNA bending stress inside the capsid. This structural transition leads to decreased interstrand repulsive interactions and a major increase in the fluidity of the encapsidated genome at the temperature of infection.
Solid-to-fluid DNA transition inside the HSV-1 capsid
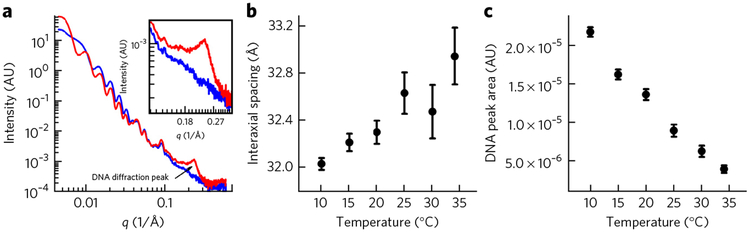
Solution SAXS provides direct structural information about the encapsidated viral genome16,23. Capsid proteins and packaged DNA have well-resolved scattering profiles23. Figure 4a shows SAXS scattering data for HSV-1 A- and C-capsids displaying a distinct diffraction peak associated only with DNA packaged in the capsid. The short-range DNA interaxial spacings determine the position of the DNA diffraction peak, whereas the area of this peak provides information on the total number of ordered DNA base pairs of the encapsidated genome16. When DNA inside the capsid becomes less ordered, the DNA peak area decreases as a result of less coherent diffraction. We collected SAXS scattering data for HSV-1 C-capsids in TE buffer between temperatures of 10 °C and 34 °C to investigate the effect of temperature on the DNA bending stress and the resulting DNA structure in the capsid (Fig. 4).
Figure 4 |. Solution SAXS analysis of intracapsid DNA structure.
(a) Background subtracted SAXS data, I(q) versus q, for HSV-1 A-capsid (blue curve) and C-capsid (red curve). The inset figure shows a zoomed-in region of the DNA diffraction peak from C-capsid. AU, arbitrary units. (b) DNA-DNA interaxial spacing d as a function of temperature for DNA packaged in HSV-1 capsid in TE buffer. (c) Area of the DNA diffraction peak in arbitrary units as a function of temperature for HSV-1 C-capsid in TE buffer. The error bars in b and c are calculated from the Gaussian curve fitting of the DNA diffraction peak.
Cryo-EM single-particle reconstructions of HSV-1 particles reveal that the entire capsid volume is filled with DNA13. Starting from the capsid walls, there are multiple, well-ordered concentric DNA layers. The layers are evenly spaced, indicating that DNA has adapted an ordered repetitive structure characteristic of a liquid crystalline state. Intracapsid confinement requires DNA to bend along radii that are energetically unfavorable given the stiffness of dsDNA12,34. To relieve the bending stress, helices are packed closer to the capsid wall, increasing the bending radius and also decreasing DNA-DNA spacing. At the same time, the repulsive DNA-DNA interactions attempt to push DNA strands as far from each other as possible, maximizing the interstrand separations35 and filling the entire capsid volume. The balance between the bending stress and the interstrand interactions determines the interaxial distance between the ordered DNA strands23,35. With increasing temperature, DNA becomes more flexible25, leading to less bending stress. Reduced bending stress causes an increase in DNA-DNA spacings, resulting in decreased repulsive interactions and enhanced mobility of the packaged genome. Indeed, our SAXS data show that the DNA interaxial distance in an HSV-1 capsid increases when the temperature is increased (Fig. 4b). Simultaneously, the area of the DNA scattering peak undergoes a marked ~5.6-fold decrease with increasing temperature (Fig. 4c). This area drop signifies a loss in the amount of ordered DNA inside the capsid. This increase in the amount of disordered DNA allows less dense packing and a larger interaxial distance of the DNA remaining in ordered domains. It is reasonable to infer that less densely packaged DNA and weaker DNA-DNA repulsive interactions lead to greater positional fluctuations and a more mobile DNA state.
Furthermore, with the help of osmotic stress measurements on bulk DNA arrays condensed in solution by an osmotic stress polymer (polyethylene glycol; see details in Supplementary Fig. 1), we also analyzed the effect of temperature on the DNA-DNA inter action energy alone without the DNA bending contribution induced by the capsid confinement35,36,23,24. DNA-DNA repulsive interactions and, therefore, compressibility for linearly packaged DNA at comparable interaxial distances as in the capsid are almost unaffected by the temperature increase from 5 °C to 50 °C (Supplementary Results, Supplementary Fig. 1).
To correlate the effect of these structural DNA transitions with the intracapsid DNA mobility, we performed AFM nano-indentation measurements. Figure 3 shows spring constants for empty A-capsids (Fig. 3c) and DNA stiffness alone in C-capsids (obtained from k(DNA) = k(C-capsid) –k(A-capsid)) in TE buffer in the temperature range 20–37 °C (Fig. 3d). The temperature dependence of both k(C-capsid) and k(A-capsid) was measured separately to deduce the value k(DNA) at each temperature. In correlation with the SAXS observations, the AFM measurements show that, as the temperature increases, the stiffness of the encapsidated herpes genome markedly decreases, suggesting an increase in fluidity. Figure 3d shows an abrupt drop in the intracapsid DNA stiffness by ~4.8 times (from 0.22 N m−1 at 20 °C to 0.046 N−1 at 37 °C), occurring close to the optimum temperature for infection. It is interesting to note that the DNA interaxial distance in HSV-1 increases from ~32 Å to ~33 Å in the measured temperature interval (Fig. 4b). It has been shown for bulk DNA condensed in solution that a hexagonal-to-cholesteric DNA structural transition occurs at the same characteristic interaxial distance of 32–33 Å35,37,38. Unlike hexagonally ordered DNA, the cholesteric DNA structure is more fluid-like or compressible and can be more readily released from the capsid during infection. The cholesteric DNA state is also found in eukaryotic chromosomes and in bacterial nucleoids39, where the existence of a fluid-like DNA state is required for biological functions40,41. Such a potential hexagonal-to-cholesteric transition of the encapsidated HSV-1 genome can contribute additionally to the abrupt increase in genome mobility.
At the temperature of infection, 37 °C, the DNA stiffness in TE buffer is small, kDNA ~ 0.046 ± 0.01 N m−1. However, its nonzero value suggests that mobility of the packaged DNA remains somewhat restricted. As we have shown before, the osmotic pressure of the cellular cytoplasm (caused by molecular crowding) further decreases the DNA-DNA repulsive interactions in the capsid11. This additionally increases the mobility of the encapsidated genome. Furthermore, the ionic strength in epithelial cells, where HSV-1 mainly replicates, is higher than the ionic strength in TE buffer (selected to mimic the lowest ion concentration in neurons). Also, cellular polyamines (for example, spermine and spermidine)42,43 can further contribute to the weakening of the DNA-DNA repulsions in the capsid22,24. Yet, it is important to emphasize that DNA mobility would increase only at low polyamine concentrations, where the net DNA interactions remain repulsive35. Conversely, at higher polyamine concentrations, where net DNA interactions are positive, the DNA inside the capsid would be completely condensed and trapped in a more solid-like state14. However, most of the polyamines in the cell are bound to the cellular DNA, RNA and other molecular components42–44. Therefore, the free polyamine concentration in the cell is low and insufficient for condensation of the herpes genome.
We also investigated the effect of temperature on interhelical sliding friction at higher AFM tip indentation rates to approximate DNA-DNA sliding during its ejection from the capsid. Analogous to the experiment above, the spring constants were measured for C- and A-capsids in low-salt TE buffer at 37 °C at indentation rates up to 104–105 nm s−1, corresponding to DNA sliding rates of 10,000–100,000 base pairs s−1 (Fig. 3c). Figure 3d shows DNA stiffness, k(DNA), versus the indentation rate in the low-salt TE buffer at 24 °C and 37 °C. When the indentation rate is increased to 104–105 nm s−1, we have shown that k(DNA) sharply increases at 24 °C. At the same time, k(DNA) remains close to zero and is not changed at 37 °C even at the highest indentation rates (Fig. 3d). This observation shows that intracapsid DNA is mobile with low sliding friction even under low-salt conditions at the temperature of infection.
In summary, our data demonstrate that, under most ionic conditions in epithelial and neuronal cells, the DNA in HSV-1 capsids is fluid-like at the temperature of infection, setting up conditions that would allow for efficient translocation of the viral genome into the cell nucleus.
In addition to showing encapsidated DNA gaining mobility, Figure 3a also shows that empty A-capsids become softer with increasing temperature. The AFM-measured spring constant decreases by ~18% when the temperature is increased from 20 °C to 37 °C, suggesting an increased flexibility in the capsid structure. Notably, we have not observed this effect of temperature on the stiffness of empty phage λ capsids within the same temperature interval18. This shows that capsomer-capsomer interactions in the HSV-1 capsid are more flexible than in phage λ, leading to larger fluctuations with temperature increase and resulting in softening of the capsids. These observations suggest a correlation with the differences in the genome delivery pathways between tailed phage and herpesviruses. A phage λ capsid does not directly interact with its receptor but releases its genome through the tail, which binds the LamB receptor on the Escherichia coli surface45. On the contrary, the portal structure within the HSV-1 capsid binds directly to the NPC in the nuclear membrane, which leads to DNA release46. Thus, the increased flexibility of the herpes capsid will increase its binding probability to the NPC at the temperature favorable for infection. Similarly, in the case of dengue virus47 and HIV48, it has been shown that either temperature or capsid maturation increase capsid flexibility, which facilitates more efficient capsid binding to membrane receptors and increases infectivity.
DISCUSSION
The HSV-1 genome is packaged in a capsid to a crystalline density10. The encapsidated DNA structure is trapped in a glassy state with restricted molecular motion between closely packed negatively charged DNA strands14. This can strongly interfere with the initiation of genome ejection into the nucleus, affecting the rate of herpes replication. In this work, we reveal that the mobility of the DNA in the HSV-1 capsid is increased and the interstrand sliding friction is minimized when herpesvirus is in ionic conditions similar to those of the epithelial or neuronal cell cytoplasm. We show that, despite its tight packaging, the encapsidated herpes genome has a fluid-like structure. This is explained by the fact that mono- and divalent cations present in the cytoplasm provide sufficient screening of negative charges on neighboring DNA helices, permitting less restrained DNA motion. Furthermore, DNA strands can slide past each other without considerable electrostatic friction20,49. Only in the low-salt conditions similar to those of the hyperpolarization phase in neurons did we find that intracapsid DNA is rigid at room temperature, indicating restricted mobility. However, at the favorable temperature for infection of the human host, the DNA bending stress of the packaged genome is decreased, leading to an increase in DNA interaxial spacings and less ordered DNA structure. This structural transition provides a major increase in the genome mobility required for efficient release from the HSV-1 capsid.
These observations provide new insight into physical conditions in vivo required for successful delivery of the herpes genome into the cell nucleus. Our data suggest that variations in the temperature and ionic conditions in the cellular cytoplasm could affect the infectivity of HSV-1 and rate of spread of infection. Indeed, it was previously shown that neuronal activity regulates the replication rate of HSV-1 in the nervous system50. Viral replication rates show strong dependence on the ionic conditions in the neuronal cytoplasm. HSV-1 replication in neurons varies with the cell type and location in the nervous system, which determine the pattern of neuronal cell activity. Although variations in cytoplasmic ionic conditions can affect the expression of viral immediate early genes50, our observation of limited DNA mobility in the capsid also supports the data.
Notably, using plaque assays, we found that the average area of HSV-1 plaques formed on a fixed layer of cells during the same incubation time has strong temperature dependence. Within the measured temperature interval of 33–39 °C, the average plaque area associated with the rate of spread of virions in the cells is reduced by a factor of ~2 for each 2-°C decrease in incubation temperature (for example, from 37 °C to 35 °C; Supplementary Fig. 2 and Table 1). Likewise, many factors can contribute to this behavior51,52, but the temperature-sensitive variation in the intracapsid DNA mobility could also potentially have a role.
Table 1 |.
Plaque area temperature dependence.
| Temperature (°C) | Plaque area (× 105 μm2) |
|---|---|
| 30 | No detectable plaques |
| 33 | 3.00 ± 0.7 |
| 35 | 5.52 ± 1.2 |
| 37 | 9.15 ± 1.6 |
| 39 | 16.0 ± 2.5 |
A fixed layer of Vero cells was infected with HSV-1 and incubated at each temperature for 75 h.
The error shows the s.d.
Furthermore, this work establishes the new concept of metastability of the viral genome packaged in HSV-1 and suggests a physical adaptation of herpesviruses to the environment of their host. At lower temperatures outside the host, the DNA in the herpes capsid is rigid, which helps to prevent its spontaneous release. Once the capsid is in the cell cytoplasm, the increased temperature induces mobility of the viral genome, facilitating infection. Preventing the solid-to-fluid–like transition of the tightly confined herpes genome offers a new antiviral strategy and an evolutionary static target. This limits the potential for development of drug resistance that can occur because of rapid adaptive mutations of viral genomes.
METHODS
Methods and any associated references are available in the online version of the paper.
ONLINE METHODS
HSV-1 capsid isolation.
African green monkey kidney cells (Vero) grown in Dulbecco’s Modified Eagle’s Medium (Cellgro) with 5% FCS (GeneMate) and 5% penicillin/streptomycin (Cellgro) were infected with HSV-1 KOS strain at a multiplicity of infection (MOI) of 5 PFU/cell for 20 h at 37 °C. Cells were scraped into solution and centrifuged at 3,500 r.p.m. for 10 min in a JLA-16.250 rotor. The cell pellet was resuspended in 20 mM Tris buffer (pH 7.5) on ice for 20 min and lysed by addition of 1.25% (v/v) Triton X-100 (Alfa Aesar) for 30 min on ice. Samples were centrifuged at 2,000 r.p.m. for 10 min, and the nuclei pellet was resuspended in TNE (10 mM Tris, 0.5 M NaCl, 1 mM EDTA) buffer with protease inhibitor cocktail (Complete; Roche). Nuclei were disrupted by sonication for 30 s. Large debris were cleared by brief centrifugation, and the supernatant was spun in a 20–50% (w/w) TNE sucrose gradient at 24,000 r.p.m. in a SW41 rotor for 1 h. The C-capsid band was isolated by side puncture, diluted in TNE buffer and centrifuged at 24,000 r.p.m. for an additional 1 h. Capsids were resuspended in TNE and stored at 4 °C. Prior to the measurements, capsids were dialyzed against the desired buffer solution used in this study.
AFM.
All of the AFM measurements were performed on a MultiMode 8 AFM with NanoScope V controller, NanoScope software and NanoScope Analysis software (Bruker AXS Corporation, Santa Barbara, CA). Images were acquired in Peak Force Tapping mode. All data (images and force-distance curves) were collected in liquid. A droplet of 40-μl sample was deposited on a glass coverslip. The details of substrate and sample preparations can be found elsewhere18,53. After 30 min, the sample was ready for the experiments. Rectangular gold-coated cantilevers (Olympus, Tokyo, Japan) were used. The measured average stiffness of cantilevers was 0.05 N/m, determined by the thermal fluctuations method54. Temperature was controlled with the Thermal Applications Controller (TAC). The heating stage connected to the TAC is inserted between the liquid sample and the Z piezo scanner. Our experimental temperature range was 20 °C to 40 °C. AFM records the vertical deflection of the cantilever in voltage. To convert the voltage to a force, we have to know two values from the calibration. The first value is the deflection sensitivity, which is how much a piezo can vertically move given the voltage supply. The value is measured in nm/V. The other value is the spring constant of the cantilever. This can be determined using the technique called thermal tune54. By multiplying these three numbers (deflection in V, deflection sensitivity in nm/V, and the spring constant in N/m), we obtain the force in nN. This applied force on the surface of the viral particle is then plotted as a function of the vertical displacement of the piezo (displayed in nm). This yields a force-distance curve. The spring constant, describing the stiffness, is determined as a slope, k (expressed in units N/m), of the linear deformation region on the force-distance curve. The AFM records both vertical and horizontal deflection of the cantilever simultaneously. The vertical deflection is converted to the force applied on the surface of the viral particle, which yields the F-z curves shown in this work. Furthermore, we also record the lateral force to detect any horizontal displacement of the cantilever during the indentation. As the cantilever is parallel to the surface of the viral capsid at all times during the indentation, the lateral force is essentially equal to zero throughout the indentation. We check the lateral force signal for each particle indentation to confirm that the particle does not slip on the surface as it is being indented. When the virus particle is indented, we measure the effective spring constant of the virus particle and the cantilever combined (keff), obtained as a slope of the linear force-distance curve. To obtain the spring constant of the viral particle alone (kv), we separately measure the spring constant of the cantilever by indenting the glass surface next to the viral particle (kc). This provides the so-called glass curve. We assume a simple model of two springs in series between the cantilever and the virus particle. This type of AFM analysis has been previously performed for other viral systems18,53. We then obtain the spring constant for the viral capsid from the following equation: kv = kc keff (kc − keff)−1. For each case study, 40–60 force distance curves were collected for 15–20 viral particles. The spring constants were calculated from the slope of the force-distance curves providing a distribution of the spring constant values. The Gaussian fitting yields the s.d., which is converted to the s.e., shown as error bars in the plots.
SAXS.
SAXS measurements were conducted at the 12ID-B station at the Advanced Photon Source (APS), Argonne National Laboratory. A 14-KeV X-ray beam was used as the photon source with a sample-to-detector distance of about 2 m, covering the q (scattering momentum transfer, q = 4π sinθ/λ, where 2θ is the scattering angle and λ is the X-ray wavelength.) range of 0.006–0.54 Å−1. To reduce radiation damage, a flow-cell equipped with a quartz capillary of 1.5 mm in diameter was used for running samples. The flow rate was 10 μl/s, and the typical sample volume was ~120 μl. Samples were measured in the temperature range of 10–40 °C, and the sample temperature was controlled with a Peltier device with ± 0.5 °C accuracy. At each temperature, the equilibrium time was 15 min to ensure the solution reaches the desired temperature. 40 two-dimensional images were collected for each sample and buffer with X-ray exposure time of 1 s. The two-dimensional scattering images were converted into one-dimensional SAXS data (i.e., intensity versus q) through azimuthal averaging using the software package at the beamline. The one-dimensional SAXS data curves were grouped by sample and averaged, followed by buffer background subtraction. The peaks in the SAXS data below q of 0.1 Å−1, both HSV-1 A-capsid and C-capsid, mainly arise from the global shape of the particles, whereas the peak around 0.2 Å−1 is the diffraction peak of DNA array. As the DNA diffraction peak was distorted by the scattering background from the viral particle shape, it was fitted with the summation of a Gaussian curve and a linear background in the q range of 0.16 Å−1 to 0.3 Å−1, and this Gaussian function gave the accurate peak position and area. DNA-DNA interaxial spacing d can be calculated from the peak position qk, i.e., d = 4π/sqrt(3)qk, assuming DNA arrays adopt a hexagonal close packing.
Supplementary Material
Acknowledgments
We acknowledge T. Liu and I. Shefer for their substantial help with the manuscript preparation. We also thank B. Jönsson for discussions that have been inspiring for this work. We are grateful to G. Berry, M. Widom, P. LeDuc, M. Deserno and L. Walker for providing critically important feedback on data analysis. We acknowledge J. Shaw, B. Pittenger and M. Thompson from Bruker Nano Surfaces Division for outstanding support with AFM measurements. We thank A. Templeton for help with proofreading. The SAXS experiments were performed at beamline 12ID-B of the Advanced Photon Source at Argonne National Laboratory. We acknowledge the Advanced Photon Source, which is an Office of Science User Facility operated by Argonne National Laboratory for the US Department of Energy under contract no. DE-AC02–06CH11357. This work was supported by the Swedish Research Council, VR grant 622-2008-726 (A.E.) and US National Science Foundation grant CHE-1152770 (A.E.). Support was also provided by the Public Health Service Grant AI060836 from the US National Institutes of Health (NIH) (F.L.H.) and by the McWilliams Fellowship in the Mellon College of Science (to U.S.). This work was partially supported by the Intramural Research Program of the National Institutes of Child Health and Human Development-NIH (to D.R.).
Footnotes
Competing financial interests
The authors declare no competing financial interests.
Supplementary information is available in the online version of the paper. Reprints and permissions information is available online at http://www.nature.com/reprints/index.html. Correspondence and requests for materials should be addressed to A.E.
References
- 1.Roizmann B, Knipe DM & Whitley RJ in Fields Virology, Vol. 2 (eds. Knipe DM & Howley PM) 2501–2601 (Lippencott-Raven, New York, Y.Y., 2007). [Google Scholar]
- 2.Whitley RJ & Roizman B Herpes simplex virus infections. Lancet 357, 1513–1518 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Coen DM in Alpha Herpesviruses: Molecular and Cellular Biology (ed. Sandri-Goldin RM) 361–381 (Caister Academic Press, Norfolk, 2006). [Google Scholar]
- 4.Gilbert C, Bestman-Smith J & Boivin G Resistance of herpesviruses to antiviral drugs: clinical impacts and molecular mechanisms. Drug Resist. Updat 5, 88–114 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Flint SJ Principles of virology: molecular biology, pathogenesis, and control of animal viruses (ASM Press, Washington, D.C.; 2004). [Google Scholar]
- 6.Evilevitch A, Lavelle L, Knobler CM, Raspaud E & Gelbart WM Osmotic pressure inhibition of DNA ejection from phage. Proc. Natl. Acad. Sci. USA 100, 9292–9295 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.São-José C, de Frutos M, Raspaud E, Santos MA & Tavares P Pressure built by DNA packing inside virions: enough to drive DNA ejection in vitro, largely insufficient for delivery into the bacterial cytoplasm. J. Mol. Biol 374, 346–355 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Leforestier A et al. Bacteriophage T5 DNA ejection under pressure. J. Mol. Biol 384, 730–739 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Smith DE et al. The bacteriophage φ29 portal motor can package DNA against a large internal force. Nature 413, 748–752 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Booy FP et al. Liquid-crystalline, phage-like packing of encapsidated DNA in herpes simplex virus. Cell 64, 1007–1015 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bauer DW, Huffman JB, Homa FL & Evilevitch A Herpes virus genome, the pressure is on. J. Am. Chem. Soc 135, 11216–11221 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tzlil S, Kindt JT, Gelbart WM & Ben-Shaul A Forces and pressures in DNA packaging and release from viral capsids. Biophys. J 84, 1616–1627 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou ZH, Chen DH, Jakana J, Rixon FJ & Chiu W Visualization of tegument-capsid interactions and DNA in intact herpes simplex virus type 1 virions. J. Virol 73, 3210–3218 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berndsen ZT, Keller N, Grimes S, Jardine PJ & Smith DE Nonequilibrium dynamics and ultraslow relaxation of confined DNA during viral packaging. Proc. Natl. Acad. Sci. USA 111, 8345–8350 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riemer SC & Bloomfield VA Packaging of DNA in bacteriophage heads: some considerations on energetics. Biopolymers 17, 785–794 (1978). [DOI] [PubMed] [Google Scholar]
- 16.Earnshaw WC & Harrison SC DNA arrangement in isometric phage heads. Nature 268, 598–602 (1977). [DOI] [PubMed] [Google Scholar]
- 17.Homa FL & Brown JC Capsid assembly and DNA packaging in herpes simplex virus. Rev. Med. Virol 7, 107–122 (1997). [DOI] [PubMed] [Google Scholar]
- 18.Ivanovska I, Wuite G, Jonsson B & Evilevitch A Internal DNA pressure modifies stability of WT phage. Proc. Natl. Acad. Sci. USA 104, 9603–9608 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Purohit PK, Kondev J & Phillips R Mechanics of DNA packaging in viruses. Proc. Natl. Acad. Sci. USA 100, 3173–3178 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Odijk T Statics and dynamics of condensed DNA within phages and globules. Philos. Trans. A Math Phys. Eng. Sci 362, 1497–1517 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Gabashvili IS & Grosberg A Dynamics of double stranded DNA reptation from bacteriophage. J. Biomol. Struct. Dyn 9, 911–920 (1992). [DOI] [PubMed] [Google Scholar]
- 22.Evilevitch A et al. Effects of salts on internal DNA pressure and mechanical properties of phage capsids. J. Mol. Biol 405, 18–23 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Qiu X et al. Salt-dependent DNA-DNA spacings in intact bacteriophage lambda reflect relative importance of DNA self-repulsion and bending energies. Phys. Rev. Lett 106, 028102 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Evilevitch A et al. Effects of salt concentrations and bending energy on the extent of ejection of phage genomes. Biophys. J 94, 1110–1120 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geggier S, Kotlyar A & Vologodskii A Temperature dependence of DNA persistence length. Nucleic Acids Res 39, 1419–1426 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Purves D Neuroscience (Sinauer Associates, Sunderland, Massachusetts, 2012). [Google Scholar]
- 27.Cameron IL, Pool TB & Smith NK An X-ray microanalysis survey of the concentration of elements in the cytoplasm of different mammalian cell types. J. Cell. Physiol 101, 493–501 (1979). [DOI] [PubMed] [Google Scholar]
- 28.Cameron IL, Smith NK, Pool TB & Sparks RL Intracellular concentration of sodium and other elements as related to mitogenesis and oncogenesis in vivo. Cancer Res 40, 1493–1500 (1980). [PubMed] [Google Scholar]
- 29.Nagy IZ, Lustyik G, Nagy VZ, Zarandi B & Bertoni-Freddari C Intracellular Na+:K+ ratios in human cancer cells as revealed by energy dispersive X-ray microanalysis. J. Cell Biol 90, 769–777 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roos WH et al. Scaffold expulsion and genome packaging trigger stabilization of herpes simplex virus capsids. Proc. Natl. Acad. Sci. USA 106, 9673–9678 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grayson P, Han L, Winther T & Phillips R Real-time observations of single bacteriophage lambda DNA ejections in vitro. Proc. Natl. Acad. Sci. USA 104, 14652–14657 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Frutos M, Letellier L & Raspaud E DNA ejection from bacteriophage T5: analysis of the kinetics and energetics. Biophys. J 88, 1364–1370 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hanhijärvi KJ, Ziedaite G, Pietila MK, Haeggstrom E & Bamford DH DNA ejection from an archaeal virus—a single-molecule approach. Biophys. J 104, 2264–2272 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kindt J, Tzlil S, Ben-Shaul A & Gelbart WM DNA packaging and ejection forces in bacteriophage. Proc. Natl. Acad. Sci. USA 98, 13671–13674 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lander GC et al. DNA bending-induced phase transition of encapsidated genome in phage lambda. Nucleic Acids Res 41, 4518–4524 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parsegian VA, Rand RP, Fuller NL & Rau DC Osmotic stress for the direct measurement of intermolecular forces. Methods Enzymol 127, 400–416 (1986). [DOI] [PubMed] [Google Scholar]
- 37.Rau DC & Parsegian VA Direct measurement of temperature-dependent solvation forces between DNA double helices. Biophys. J 61, 260–271 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Livolant F & Leforestier A Condensed phases of DNA: structures and phase transitions. Prog. Polym. Sci 21, 1115–1164 (1996). [Google Scholar]
- 39.Livolant F Ordered phases of DNA in vivo and in vitro. Physica A 176, 117–137 (1991). [Google Scholar]
- 40.Pelta J Jr., Durand D, Doucet J & Livolant F DNA mesophases induced by spermidine: structural properties and biological implications. Biophys. J 71, 48–63 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Todd BA & Rau DC Interplay of ion binding and attraction in DNA condensed by multivalent cations. Nucleic Acids Res 36, 501–510 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gibson W & Roizman B Compartmentalization of spermine and spermidine in the herpes simplex virion. Proc. Natl. Acad. Sci. USA 68, 2818–2821 (1971). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Igarashi K & Kashiwagi K Polyamines: mysterious modulators of cellular functions. Biochem. Biophys. Res. Commun 271, 559–564 (2000). [DOI] [PubMed] [Google Scholar]
- 44.Davis RH, Morris DR & Coffino P Sequestered end products and enzyme regulation: the case of ornithine decarboxylase. Microbiol. Rev 56, 280–290 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hendrix RW Lambda II (Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory, 1983). [Google Scholar]
- 46.Ojala PM, Sodeik B, Ebersold MW, Kutay U & Helenius A Herpes simplex virus type 1 entry into host cells: reconstitution of capsid binding and uncoating at the nuclear pore complex in vitro. Mol. Cell. Biol 20, 4922–4931 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang X et al. Dengue structure differs at the temperatures of its human and mosquito hosts. Proc. Natl. Acad. Sci. USA 110, 6795–6799 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kol N et al. A stiffness switch in human immunodeficiency virus. Biophys. J 92, 1777–1783 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kornyshev AA, Lee DJ, Leikin S & Wynveen A Structure and interactions of biological helices. Rev. Mod. Phys 79, 943–996 (2007). [Google Scholar]
- 50.Zhang CX et al. Neuronal activity regulates viral replication of herpes simplex virus type 1 in the nervous system. J. Neurovirol 11, 256–264 (2005). [DOI] [PubMed] [Google Scholar]
- 51.Doceul V, Hollinshead M, van der Linden L & Smith GL Repulsion of superinfecting virions: a mechanism for rapid virus spread. Science 327, 873–876 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Elliott G & O’Hare P Live-cell analysis of a green fluorescent protein-tagged herpes simplex virus infection. J. Virol 73, 4110–4119 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 53.Ivanovska IL et al. Bacteriophage capsids: tough nanoshells with complex elastic properties. Proc. Natl. Acad. Sci. USA 101, 7600–7605 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hutter JL & Bechhoefer J Calibration of atomic-force microscope tips. Rev. Sci. Instrum 64, 1868–1873 (1993). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.