Abstract
The microbiome is receiving significant attention given its influence on a host of human diseases including cancer. Its role in response to cancer treatment is becoming increasingly apparent, with evidence suggesting that modulating the gut microbiome may impact responses to numerous forms of cancer therapy. A working knowledge of the microbiome is vital as we move forward in this age of precision medicine, and an understanding of the microbiome’s influence on immune responses and cancer is key. It is also important to understand factors influencing the gut microbiome and strategies to manipulate the microbiome to augment therapeutic responses.
Introduction
There is mounting evidence supporting the role of the microbiome in response to cancer therapy, with several recent studies demonstrating the influence of the gut microbiome specifically on the response to immune checkpoint blockade across cancer types (Chaput et al., 2017, Frankel et al., 2017, Gopalakrishnan et al., 2018, Matson et al., 2018, Routy et al., 2018). How and why this occurs necessitates an understanding of the intricate web of cancer, immunosurveillance, and the factors that influence both host and anti-tumor immunity – including the gut microbiome. Each of these factors as well as the impact of these factors on one another and on responses to immunotherapy will be discussed herein. Finally, strategies to modulate the gut microbiome and ongoing trials to do so will also be described.
Cancer immunosurveillance and response to cancer therapy
The immune system has long been recognized as a dominant force in cancer control, with defects in immunity contributing not only to carcinogenesis and cancer progression but also to poor responses to cancer therapy. Recent evidence substantiates the need for preserved overall systemic immunity in mediating responses to immunotherapy specifically (Chen and Mellman, 2013, Chen and Mellman, 2017, Spitzer et al., 2017).
Tremendous progress has been made in identifying specific factors contributing to response to cancer therapy - and have largely focused on tumor-centric markers including “foreignness” (including mutational load) (Snyder et al., 2014), aspects of tumor metabolism (such as glucose metabolism), and factors affecting tumor sensitivity to immune effectors (including HLA and interferon gamma gene expression)(Blank et al., 2016). However, markers of response to cancer therapy have continued to evolve and now include factors well beyond established tumor-centric markers – providing a more holistic paradigm that incorporates the multitude of factors that impact therapeutic response. These newer models have been extended to recognize components that contribute to overall immune status (Blank et al., 2016) and the tumor microenvironment including infiltrating immune cells that can either stimulate (such as CD8+ T cells) or inhibit an immune response (such as myeloid derived suppressor cells)(Sharma et al., 2017, Blank et al., 2016). Additionally, recent insights importantly highlight the impact of microbiota (particularly the gut microbiota) on responses across several cancer therapies (Kroemer and Zitvogel, 2018).
The microbiome and immunity
The microbiome is defined as the collective genomes of microbes within a community, whereas the term microbiota refers to the microbes themselves in aggregate. Within a human organism, there are trillions of microbes as numerous as human cells, that interact with the host constantly at numerous sites (including the skin and mucosal surfaces such as the gastrointestinal tract) throughout development. Therefore, it is not surprising that they play such a large role in numerous host functions including immunity (Sender et al., 2016, Morgan and Huttenhower, 2012)
The crosstalk between microbiota and the immune system at the level of the gut is extensive and critical, and not only allows for the tolerance of commensal bacteria and oral food antigens, but also enables the immune system to recognize and attack opportunistic bacteria thereby preventing bacterial invasion and infection. In addition to influencing localized immune responses, these microbiota also have broader effects contributing to innate and adaptive immunity at multiple levels. This concept is supported in preclinical models, as germ-free (GF) mice that lack intestinal microbiota are noted to have severe defects in immunity, with an absent mucous layer, altered IgA secretion and reduced size and functionality of Peyer’s patches and draining mesenteric lymph nodes (Johansson et al., 2015, Spiljar et al., 2017). Overall, there is compelling evidence that the microbiota helps to shape the immune system as a whole (Honda and Littman, 2016).
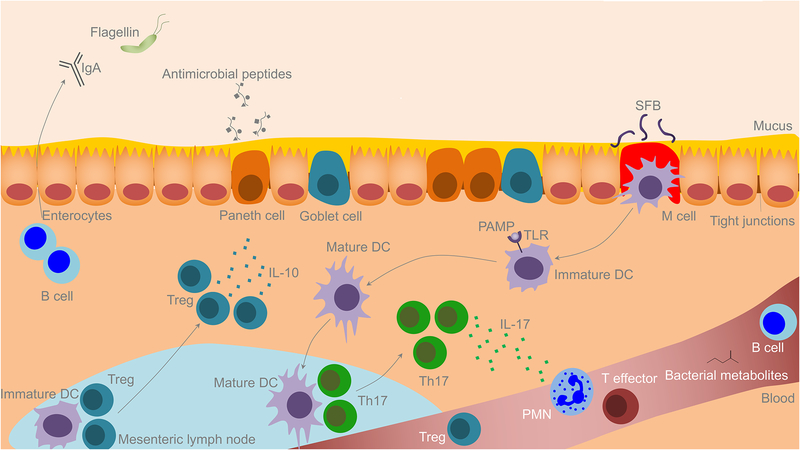
The structure of the gut, which contains a mucosa consisting of a single epithelial cell layer made up of intestinal epithelial cells (IECs) and intraepithelial lymphocytes (IELS), facilitates this interaction with the immune system. The IEC’s contain Paneth cells that secrete antimicrobial peptides and goblet cells that secrete mucus, which in turn coats the epithelial layer. Beneath the mucosal layer lies the lamina propria, a connective tissue layer containing Peyer’s patches and a host of other immune cells including antigen presenting cells (APCs) and innate lymphoid cells (ILCs) as well as CD4+ and CD8+ T cells and B cells. This gut-associated lymphoid tissue (GALT) represents the largest component of the immune system in the body, and influences immune responses both locally and systemically (Figure 1).
Figure 1. The Microbiome and Immunity.
Commensal organisms within the lumen of the gut have profound influences on the immune system at the local level within the gut mucosa, in draining mesenteric lymph nodes, and systemically. The immune system likewise can alter the gut microbiota. Goblet cells create a thick mucous protective layer covering the mucosa; this mucousal layer is largely deficient in germ-free animals. Plasma cells in the lamina propria secrete IgA into the lumen of the gut. Paneth cells secrete a number of anti-microbial peptides; their activity is amplified in response to signaling from local immune cells in response to the microbiota. Bacterial metabolites or bacteria themselves can activate local DCs which migrate to the draining lymph nodes to activate naïve T cells to effector T cells, Tregs or Th17 cells which can migrate back to the gut mucosa or enter systemic circulation. Specific metabolites or bacterial byproducts can alter the dendritic cell in a fashion that allows them to skew towards a Treg versus Th17 phenotype. Tregs function in secreting IL10, creating a local anti-inflammatory cytokine milieu. Th17 cells, meanwhile, produce IL17 which can increase Paneth cell production of antimicrobial peptides and can function in recruiting PMNs from the bloodstream. Some bacterial metabolites can enter the bloodstream directly further altering the systemic immune system.
Local immunity is promoted via recognition of pathogen-associated-molecular patterns (PAMPs) (such as lipopolysaccharide and flagellin) by pattern recognition receptors (PRRs) such as toll like receptors (TLRs) present on IECs as well as innate immune effectors within the gut. Metabolites produced by bacteria may also impact local immunity via production of short chain fatty acids (SCFAs), which, among a number of key activities, have been shown to augment immunity via IgA production by plasma cells (Pabst, 2012). IgA acts primarily by blocking bacterial adherence to epithelial cells; agglutination, entrapment and clearance; and also has direct effects on bacterial virulence (Mantis et al., 2011)
Draining lymph nodes for the gut lie within the mesentery (mLNs) of the small bowel and colon, where adaptive immune responses are further shaped by the gut commensals, impacting the differentiation of naïve T cells within the mLNs. PAMPs act to induce maturation of antigen-presenting cells such as dendritic cells (DCs) as they sample antigen from the lumen either directly via interdigitation of dendrites through the mucosal layer or indirectly after processing and transphagocytosis by specialized IECs called M cells. Once activated, DCs travel to mLNs where they interact with and stimulate naïve T cells to form CD4+ T cells (Lathrop et al., 2011), specifically CD4+ T regulatory cells (Tregs), and Th17 cells both of which have a tropism for the gut. DCs may also directly stimulate CD8+ T cells.
After education in the mLNs, T cells can influence immunity at a number of different sites. They play a critical role in gut homeostasis, highlighted by mucosal tolerance induced by Tregs, and via production of immunosuppressive cytokines such as interleukin 10 (IL-10). Importantly, there is ongoing crosstalk between gut commensals and mucosal T cells (such as Tregs), as maintenance of these cells at the level of the gut is facilitated by bacterial metabolites such as SCFAs; SCFAs here function in a manner dependent on their ability to inhibit histone deacetylase activity suggesting epigenetic regulation (Rooks and Garrett, 2016). Additionally, some bacterial species have been shown to drive Treg development via alternative pathways dependent on polysaccharide A and TLR signaling to dendritic cells (Round and Mazmanian, 2010, Shen et al., 2012).
A specific subset of CD4+ cells proven to be important in gut microbiota interactions are Th17 cells. These cells are prominent within the lamina propria of the small and large intestine and are critical in protecting against bacterial and fungal infections. Th17 cells also function in mucosal immunity as cytokine secretion from Th17 cells stimulates IECs to form tight junctions and to secrete anti-microbial proteins (Weaver et al., 2013). Th17 cells are markedly depleted in GF animals and can be induced by specific bacterial subsets such as segmented filamentous bacteria (SFB) (Ivanov et al., 2009, Ivanov et al., 2008). Interleukin 17 can cause further release of additional inflammatory cytokines and recruit neutrophils from the circulation to the gut microenvironment. In addition to influencing local immunity within the gut mucosa, microbiota can also shape systemic immune responses via immune cell priming. DCs primed by commensals typically do not pass into the circulation or travel to distant lymph nodes but can do so in certain settings. Toll-like receptor signaling from microbial peptides to DCs and other innate immune effectors generates cytokines and interferons that act in both a paracrine and endocrine manner at distant sites; it is thought that this signaling in the gut creates immune system “tone”. That is, the systemic immune system is primed (potentially at epigenetic or transcriptional level) to enact a robust response in the setting of pathogens, and in the absence of threat, to maintain a non-inflammatory state (Abt et al., 2012) Furthermore, B and T cells, including Tregs and Th17 cells can, upon being primed by DCs presenting antigen derived from commensal organisms in draining mLNs of the gut, circulate systemically to facilitate immune responses at distant sites against the same organism or against other antigens in cross-reaction to similar epitopes (Stary et al., 2015). Interestingly, Th17 cells that emigrate can have significant plasticity in function, changing their cytokine output based on the existing local inflammatory or non-inflammatory state (Hirota et al., 2011).
Disruption of the delicate balance of commensal bacteria is seen in the setting of dysbiosis, which is characterized by a less diverse and less stable microbiota, with potential enrichment of opportunistic pathogenic bacteria (Frosali et al., 2015). Dysbiosis can lead to impaired local, locoregional, and systemic immune responses with breakdown of mucosal barriers, translocation of gut bacteria to the mLNs and into the peripheral circulation, alteration of the cytokine milieu within the gut mucosa and draining mLNs towards an inflammatory phenotype with activation of Th17 cells and effector T cells causing an influx of neutrophils and inciting a profound inflammatory state both locally and systemically (Levy et al., 2017).
Exemplification of the importance of eubiosis in preserving immunity is seen in the case of response to vaccination. A highly diverse microbiota has been associated with improved adaptive immune responses to a variety of vaccines in infants (Huda et al., 2014). Specific components of the microbiota can prime the immune system by activating TLR signaling pathways serving as a natural vaccine adjuvant (Oh et al., 2014). Thus, it is increasingly clear that the gut microbiota may impact not only local immunity but also systemic immune responses.
The gut microbiota in response and toxicity to immunotherapy
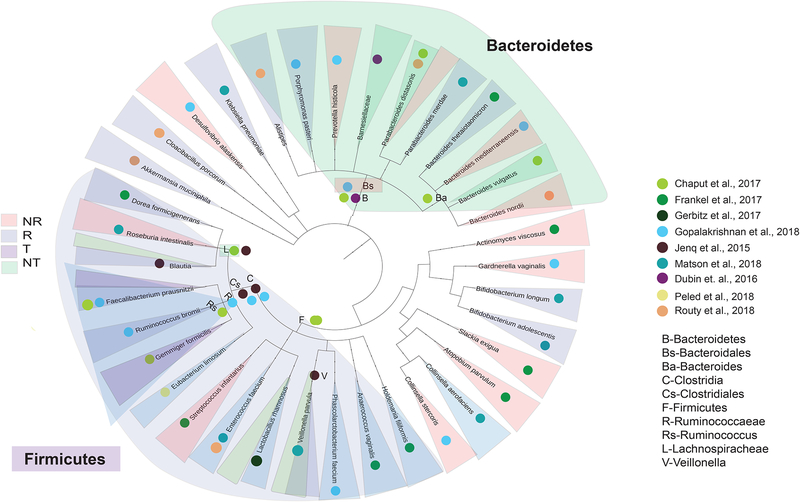
Given the role of the gut microbiota in modulating host immunity, it is fairly intuitive that it could significantly influence response and toxicity to various forms of cancer therapy. Though early studies primarily used murine models to assess these interactions, there is now mounting data from human cohorts suggesting that the gut microbiota is a dominant force in mediating both response and toxicity to these therapeutic strategies. Bacterial taxa implicated in response as well as toxicity to immunotherapy in human studies (Figure 2) and in murine models will be discussed herein.
Figure 2. Phylogenetic tree summarizing previously established links between the gut microbiome, treatment outcomes and toxicities in cancer patient populations.
A phylogenetic tree was constructed using evolutionary distances with the phyloT software (Letunic and Bork, 2016), to depict phylogenetic similarity (or lack thereof), of all bacterial taxa reported to be associated with response or toxicity to immune checkpoint blockade in human studies, moving from broader (kingdom) to more specific (species) taxonomies from inside-out. Bacterial taxa are labeled according to publication (colored dots) of origin and shaded (R=treatment response, light blue; NR=treatment non-response, light red; T=toxicity, light green; NT=non-toxicity, light purple) according to the phenotype(s) of association in the various studies included
The gut microbiota and stem cell transplantation
Perhaps one of the earliest demonstrations of the role of the gut microbiota in response and toxicity to cancer therapy was in the setting of allogeneic stem cell transplant (allo-HSCT) for hematologic malignancies. Dysbiosis and impaired systemic immunity is fairly common in these patients, as they are often treated with concurrent therapies that significantly alter immunity and composition of the gut microbiota - including immunosuppressants, broad-spectrum antibiotics and even total body irradiation (Routy et al., 2017, Shono et al., 2016). Several investigators hypothesized that dysbiosis could be associated with altered responses and potentially with toxicity to therapy. Analyses of longitudinal fecal samples demonstrated a disruption of the existing state of equilibrium of the gut microbiota post-HSCT with a relative dysbiosis and a loss of bacterial diversity and stability and dominance of Enterococcus, Streptococcus and various Proteobacteria (Holler et al., 2014, Taur et al., 2012). Importantly, health-promoting bacteria such as Faecalibacterium and Ruminococcus were reduced (Biagi et al., 2015).
Dysbiosis in the setting of HSCT has also been associated with differences in long-term survival, with patients having a lower diversity of microbiota in their gut at the time of HSCT having shortened overall survival and higher mortality rates (specifically transplant-related mortality) compared to those with a high diversity of gut microbiota (Taur et al., 2014). Further confirmation of this finding was reported when low levels of 3-indoxyl sulphate in the urine, a by-product of L-tryptophan metabolism by commensal microbiota and a marker for bacterial dysbiosis, was found to be associated with worse overall survival following allo- HSCT (Weber et al., 2015). In addition to diversity, compositional differences in the gut microbiota have also been studied in response and survival after HSCT, with a higher abundance of bacteria within the genus Blautia associated with improved overall survival (Jenq et al., 2015), and a higher abundance of Eubacterium limosum associated with a reduced risk of relapse (Peled et al., 2017).
In addition to the relationship to response and survival, the influence of the gut microbiota has also been studied in the context of toxicity to HSCT therapies – particularly with regard to graft versus host disease (GVHD). GVHD is characterized by the vigorous activation of immune-competent donor immune cells (mostly T cells) and causes significant damage to a variety of organs including the skin, liver, gut, as well as sites of hematopoiesis (Jacobsohn and Vogelsang, 2007). GVHD susceptibility varies with the type and extent of conditioning regimen, the degree of human leukocyte antigen (HLA) mismatch, and the activation status of donor cells. Severe acute GVHD has a long-term survival probability of less than 5% (Cahn et al., 2005) and chronic GVHD is also associated with significant morbidity and mortality.
The onset of acute-GVHD is associated with significant shifts in the composition of the microbiota with a loss of overall diversity and reduction of health-promoting obligate anaerobes such as Faecalibacterium, Ruminococcus, Lactobacillus, and Blautia, and an enrichment of Enterococcus and Clostridiales (Jenq et al., 2012, Heimesaat et al., 2010, Gerbitz et al., 2004, Biagi et al., 2015). A high abundance of the genus Blautia of the Clostridia class was found to be associated with reduced GVHD lethality in two independent cohorts of patients undergoing treatment with allo-HSCT (Jenq et al., 2015). The gut microbiota may also be impacted by antibiotic treatment for infectious complications during HSCT, with differences in GVHD-associated mortality with different antibiotic regimens (Shono et al., 2016).
Modulation of the gut microbiota to abrogate toxicity has been studied in pre-clinical models with mixed results. The administration of the probiotic Lactobacillus rhamnosus GG alone or in combination with Ciprofloxacin before and during transplantation in mice was associated with reduced rates of GVHD and improved overall survival (Gerbitz et al., 2004). In this work, the authors hypothesized that probiotic supplementation contributed to the preservation of gut bacterial integrity, as surveying the mesenteric lymph nodes revealed an absence of enteric pathogens (Gerbitz et al., 2004). However results in human studies have been more heterogeneous, with early studies demonstrating reduced rates of GVHD in patients treated with broad-spectrum antibiotics prior to HSCT (van Bekkum et al., 1974), and more recent studies demonstrating detrimental effects of antibiotic use with higher rates of GVHD (Routy et al., 2017, Shono et al., 2016). More refined strategies to modulate the gut microbiota to reduce the risk of GVHD are now being tested in clinical trials including dietary modifications and fecal microbiome transplant (FMT) (NCT03359980, NCT03148743, NCT03214289, NCT02763033). These trials are based on the central hypothesis that re-shaping the intestinal microbiota to its pre-treatment eubiotic state would lessen the risk of subsequent GVHD development, and are primarily exploratory in nature, seeking to assess the safety and feasibility of such modalities. Nevertheless, these trials will serve as a foundation and additional trials will be implemented based on insights gained.
The gut microbiota and immunotherapy
Though stem cell transplant may be considered one of the earliest effective forms of cancer immunotherapy, there are now a host of novel immunotherapeutic approaches, and, not unsurprisingly, these are similarly impacted by the gut microbiota. Important initial insights came from murine models (Sivan et al., 2015, Vetizou et al., 2015), and many of these findings have now been validated in several patient cohorts treated with immunotherapy, specifically immune checkpoint blockade.
Over a decade ago, investigators from the National Cancer Institute demonstrated that administration of antibiotics significantly abrogated anti-tumor activity in a murine model of adoptive cell therapy for melanoma. The authors posed that the proliferation of transferred T-cells in the tumor was augmented by the translocation of the gut microbiota to the mesenteric lymph nodes associated with total body irradiation, which was used as a preparative regimen. They surmised that the translocation of gut bacteria helped to prime an immune response via TLR4-signaling (Paulos et al., 2007). This notion is supported by studies demonstrating impaired responses to intra-tumoral injection of TLR agonists in GF or antibiotic-treated mice. In this setting, tumor-associated myeloid cells are primed by commensal gut bacteria (via TLR4 signaling) for the production of TNF and other inflammatory cytokines that mediate the anti-tumor effect of these agents (Iida et al., 2013).
Though not traditionally considered immunotherapy, effective treatment with conventional chemotherapy is also dependent on intact immune responses thus substantiating the notion that the gut microbiota could shape responses to these forms of therapy as well. This has certainly been demonstrated in platinum-based chemotherapies and cyclophosphamide therapy. During treatment with cyclophosphamide, translocation of commensal bacteria (specifically Gram-positive organisms such as Lactobacillus johnsonii and Enterococcus hirae) into mesenteric lymph nodes can potentially facilitate robust Th17 responses in the spleen and the induction of memory Th1 responses. Immune responses to cyclophosphamide have also been shown to be dependent on MyD88 and TLR signaling - suggesting that commensal microorganisms may play a role. Indeed, the effects of cyclophosphamide and other chemotherapy regimens were abrogated in GF or antibiotic- treated mice and were differentially impacted by the presence of particular bacterial species (Viaud et al., 2013, Iida et al., 2013).
Importantly, the impact of the gut microbiota has also been studied in the setting of treatment with immune checkpoint inhibitors, which target immunomodulatory molecules on the surface of T cells (or their ligands) to enhance anti-tumor immune responses. Despite the enthusiasm for treatment with these agents, a significant proportion of patients do not experience objective responses, and when responses do occur, may not be durable. Tremendous efforts have focused on identifying predictors of the response to immune checkpoint blockade as well as strategies to overcome therapeutic resistance (Cogdill et al., 2017, Sharma et al., 2017), and emerging evidence suggests that the gut microbiota may play a significant role in modulating responses to these therapies.
The impact of the gut microbiota on response to immune checkpoint blockade was first studied in mouse models, with landmark publications in Science in 2015 demonstrating that the composition of the gut microbiota could influence the response to immune checkpoint inhibitors targeting the cytotoxic T lymphocyte antigen-4 (CTLA-4) and the programmed death receptor-1 (PD-1) (Vetizou et al., 2015, Sivan et al., 2015). In the case of CTLA-4 blockade, notable changes in the abundance of gut microbiota in mice were seen following anti- CTLA-4 therapy, with a relative increase in Bacteroidales and Burkholderiales, and a decrease in Clostridiales. The efficacy of anti-CTLA-4 therapy was markedly reduced in GF mice and SPF mice with broad-spectrum antibiotics. Furthermore, oral feeding with Bacteroides fragilis in combination with either Bacteroides thetaiotaomicron or Burkholderia cepacia augmented the action of anti-CTLA-4 therapy by eliciting a Th1 response in the lymph nodes and facilitating the maturation of intra-tumoral dendritic cells. The translational impact of these findings was evidenced when FMT from patients having dominant Bacteroides species in their gut resulted in improved tumor control compared to FMT from patients with distinct Bacteroides or Prevotella species (Vetizou et al., 2015, Viaud et al., 2013). These findings were complemented by parallel studies in the context of treatment with PD-1 blockade, demonstrating significant differences in tumor outgrowth in genetically similar mice with differing gut microbiomes purchased from two separate vendors. Therapeutic responses also differed in these mice, and beneficial effects from mice with a more “favorable” microbiota could be transplanted to other mice using FMT or co-housing. Profiling of the gut microbiome revealed an over- representation of Bifidobacterium species in mice with delayed tumor outgrowth and favorable responses to PD- 1 based therapy. Furthermore, supplementation with an oral probiotic containing Bifidobacterium restored anti- tumor efficacy of PDL1-blockade in mice with an “unfavorable” gut microbiota, which primarily occurred through enhanced dendritic cell maturation resulting in increased tumor-specific CD8+ T-cell activity (Sivan et al., 2015).
These studies were further supplemented by multiple studies published in the past several months demonstrating a role for the gut microbiota in patients on immune checkpoint blockade (Chaput et al., 2017, Frankel et al., 2017, Gopalakrishnan et al., 2018, Matson et al., 2018, Routy et al., 2018). Several provocative findings were reported substantiating the role of the gut microbiota in shaping responses to therapy. First, the impact of antibiotic use on response to immune checkpoint blockade was shown in a large cohort of patients with non-small cell lung cancer, renal cell carcinoma or urothelial cancer. Patients treated with antibiotics for routine indications shortly before, during, or shortly after treatment with anti-PD1/PD-L1 mAB had significantly lower progression-free survival (PFS) and overall survival (OS) rates compared to patients who had not received antibiotics. This suggests that disrupting the gut microbiota (via antibiotic use) could potentially impair anti-tumor immune responses as well as response to immune checkpoint blockade (Routy et al., 2018). The group also studied the gut microbiota directly by performing whole metagenomic sequencing in fecal samples from these patients, demonstrating that responders to PD-1 blockade had differential composition of gut bacteria, including specific genera highlighted by the group as being enriched in responding patients (Akkermansia and Alistipes). In this study, FMT was performed in GF and SPF mice using stool from either responder (R) or non-responder (NR) patients prior to treatment with PD1 blockade, demonstrating enhanced responses in the setting of R-FMT. In these studies, the efficacy of anti-PD-1 in GF mice receiving NR-FMT could be restored by administration of Akkermansia muciniphila alone or in combination with Enterococcus hirae, and administration of A. muciniphilia in these mice was associated with increased intra-tumoral immune infiltrates mediated by the recruitment of CCR9+CXCR3+CD4+ T cells into the tumor bed and increased ratio of CD4+ T-cells to CD4+FoxP3+ T-cells (Tregs) in response to PD1 blockade (Routy et al., 2018).
These findings were corroborated in two additional papers published in the same issue of Science describing the impact of gut microbiota on responses to anti-PD-1 therapy in patients with metastatic melanoma (Gopalakrishnan et al., 2018, Matson et al., 2018). The study by Gopalakrishan et al. revealed that patients who responded to anti-PD-1 therapy had a significantly higher diversity of bacteria in their gut microbiota as well as a higher relative abundance of Clostridiales, Ruminococcaceae, and Faecalibacterium. In contrast, NR had significantly lower diversity of gut bacteria and higher abundance of Bacteroidales. Importantly, comparing the composition of bacteria in the gut with immune profiling in the tumor microenvironment revealed that patients with a “favorable” gut microbiota had increased expression of cytolytic T cell markers and antigen processing and presentation compared to patients with “unfavorable” gut microbiota. Mechanistic studies were performed in GF mice with FMT from R versus NR, recapitulating findings in parallel published studies that mice receiving FMT from R had significantly delayed tumor outgrowth and enhanced response to treatment with immune checkpoint blockade (Gopalakrishnan et al., 2018). Another cohort of patients with metastatic melanoma was studied by Matson et al and also demonstrated significant differences in response to treatment with immune checkpoint blockade based on profiles within the gut microbiota (Matson et al., 2018). Specifically, the group found that patients who responded to anti-PD-1 therapy had enrichment of Bifidobacterium longum, Collinsella aerofaciens and Enterococcus faecium in baseline fecal samples. Transfer of stool from patients to germ-free mice in this study also successfully recapitulated the phenotype, with mice that received responder-stool growing tumors at slower rate and having markedly improved efficacy to anti-PD- L1 immunotherapy when compared to mice that received non-responder stool. These effects were mediated by increased densities of CD8+ T-cells and reduced FoxP3+CD4+ Tregs in the tumor microenvironment (Matson et al., 2018).
While immune checkpoint blockade agents have been a qualified success in the treatment of various malignancies resulting in sustained responses, a significant proportion of patients continue to experience treatment-limiting toxicity with anti-PD-1 (16%), anti-CTLA-4 (27%), and combination therapies (65%) (Larkin et al., 2015). Approximately one-third of all patients undergoing anti-CTLA-4 therapy develop intestinal inflammation due to mucosal immune dysregulation (Berman et al., 2010, Weber et al., 2013). Efforts to characterize gut microbiota that contribute to toxicity to immune checkpoint blockade are underway. Pre- clinical models have demonstrated an improvement in toxicity scores in anti-CTLA-4-treated mice with oral gavage of Bacteroides fragilis and Burkholderia cepacia (Vetizou et al., 2015). The influence of the gut microbiota on toxicity has also been studied in human cohorts (Chaput et al., 2017, Dubin et al., 2016, Frankel et al., 2017) (Figure 2). Taxonomical and functional differences have been reported in anti-CTLA-4 treated melanoma patients who were colitis-free (with enrichment of Bacteroidetes and abundance of genetic pathways involved in polyamine transport and B vitamin synthesis) as opposed to those who developed colitis (Dubin et al., 2016). This may be related to the known influence of these factors in Treg differentiation (Round and Mazmanian, 2010, Faith et al., 2014). Additional cohorts have also been studied, showing that patients with a higher abundance of Faecalibacterium prausnitzii and other related Firmicutes and low abundance of Bacteroidetes had a higher risk of colitis on anti-CTLA-4 therapy (Chaput et al., 2017, Frankel et al., 2017). The group also reported that patients with colitis had increased expression of ICOS on the surface of effector CD4+ T-cells and low levels of Tregs and systemic inflammatory proteins such as IL-6, IL-8 and sCD25 in the blood at baseline, which may be related to the compositional differences in the microbiome (Chaput et al., 2017).
Based on the available literature, there are clearly bacterial taxa that are associated with response and toxicity – with some overlap in the bacterial signatures across the studies (Figure 2). Bacterial taxa within the Ruminococaceae family of the Firmicutes phylum (such as F. prausnitzii) have been associated with both response and toxicity to immune checkpoint blockade across studies (Chaput et al., 2017, Frankel et al., 2017, Gopalakrishnan et al., 2018) (Figure 2). Conversely, bacterial taxa within the Bacteroidales order of the Bacteroidetes phylum have been associated with a lack of response to immune checkpoint blockade (Bacteroidales), while a higher abundance of these taxa within the gut are also generally associated with a lower incidence of toxicity (Chaput et al., 2017, Dubin et al., 2016, Frankel et al., 2017, Gopalakrishnan et al., 2018). However, at lower levels of taxonomy within these phyla these generalizations do not apply, as some taxa within Firmicutes have been associated with a lack of response (Roseburia, Streptococcus) (Frankel et al., 2017, Matson et al., 2018), and some taxa within Bacteroidetes have been associated with response (Alistipes, Porphyromonas pasteri, and Colinsella aerofaciens) (Gopalakrishnan et al., 2018, Matson et al., 2018, Routy et al., 2018). Importantly, taxa outside of Firmicutes / Bacteroidetes have also been associated with response (such as Akkermansia muciniphila, Bifidobacterium longum and Bifidobacterium adolescentis, and Colinsela aerofaciens) and non-response (such as Actinomyces viscosus, Garnderella vaginalis) (Frankel et al., 2017, Gopalakrishnan et al., 2018, Matson et al., 2018, Routy et al., 2018). Overall there is not a great deal of overlap between specific bacterial taxa associated with response across these published studies, though several taxa that are implicated with either response or toxicity are phylogenetically related (such as members of the Ruminococcaceae and Lachnospiraceae families and the Bacteroidales order) (Figure 2). Importantly, differences may be related to several different factors – including differences in techniques used to analyze samples and reference databases used for analysis – which varied widely across the studies (Chaput et al., 2017, Dubin et al., 2016, Frankel et al., 2017, Gopalakrishnan et al., 2018, Jenq et al., 2015, Matson et al., 2018, Peled et al., 2017, Routy et al., 2018) -suggesting the importance of developing standardized approaches for microbiome analysis. Geographical influences also may exist, as these studies were performed in centers at different locations around the world. In line with this, dietary and lifestyle factors may also account for some of the differences in bacterial taxa observed across the studies. Nevertheless, the impact of the gut microbiota on therapeutic response is uncontested, and these data provide strong evidence that the gut microbiota can modulate anti-tumor immune responses and responses to immune checkpoint blockade.
Manipulating the gut microbiota to improve responses to therapy
Given this growing body of literature, it is becoming increasingly clear that modulation of the gut microbiota may represent a novel and important adjunct to current anti-cancer therapeutic modalities. While studies to further dissect the molecular interactions underlying the effects of the microbiota on cancer development and anti-tumor immune responses are underway, several ongoing and planned clinical trials will investigate the therapeutic potential of manipulation of the gut microbiota directly in cancer patients (Table 1).
Table 1.
Manipulation of the gut microbiome to enhance responses to cancer immunotherapy.
| Trial Number | Patient population | Intervention | Outcome(s) | Status |
|---|---|---|---|---|
| NCT02843425 | All cancer patients treated at MDACC | Addition of ½ cup beans per day to regular diet in a cross-over design |
Primary: Change in fecal microbiome profile from baseline (via 16S profiling) |
Open and recruiting (MDACC) |
| NCT02079662 | Stages II and III breast cancer patients treated at MDACC ages 18+ | Randomized intensive lifestyle change (Diet, exercise, psychosocial) |
Primary: Disease-free survival (DFS) Secondary: Change in fecal and oral microbiome (via 16S profiling) |
Open and recruiting (MDACC) |
| NCT01895530 | CRC patients ages 18+ undergoing elective CRC resection | Randomized probiotic (S. Boulardii) administration |
Primary: Cytokine expression in colonic mucosa (via qPCR) Secondary: Postoperative complications |
Completed (Consoli et al., 2016) |
| NCT03072641 | CRC patients ages 18+ | Randomized probiotic (ProBion Clinica B. lactis Bl-04, L.acidophilus NCFM + Inulin) administration |
Primary: Change in fecal and tumor microbiota from baseline Secondary: Changes in epigenetic patterns of tumor tissue from baseline |
Completed(Hibberd et al., 2017) |
| NCT03358511 | Postmenopausal breast cancer patients stages I–III | Single arm probiotic (Primal Defense Ultra multi-strain probiotic formula) administration |
Primary: Change in mean number of CD8+ cells from baseline |
Open and recruiting (Mayo Clinic) |
| NCT02928523 | Acute myeloid leukemia patients ages 18–65 treated with intensive chemo and antibiotics | Single arm autologous FMT (frozen inoculum) |
Primary: Diversity of the gut microbiome, multi-drug resistant bacteria eradication Secondary: Signature of dysbiosis of gut microbiome |
Ongoing, closed to recruiting (France) |
| NCT03353402 | Metastatic melanoma patients ages 18+ who previously failed standard therapies | Single arm FMT (colonoscopy or gastroscopy) from patient donors who responded to immunotherapy |
Primary: Safety (AEs associated with FMT), engraftment of FMT Secondary: Changes in immune cell populations and activity, objective response rate |
Open and recruiting (Israel) |
Diet
A major function of the gut microbiota is to aid in host food digestion, harvesting key nutrients that the host is incapable of metabolizing without the help of microbes (Backhed et al., 2005). However, dietary intake can also promote differential composition of the microbiome, with evidence that profound and intensive changes in dietary regimens can significantly alter the gut microbiota in a relatively short amount of time (David et al., 2014). The current body of literature in this area has mapped the responsiveness of specific 15 bacterial groups and their downstream metabolites to a variety of nutrients and immune parameters and provides preliminary insight into how dietary modulation could be used as a strategy to enrich the gut microbiome and immune health (Ma et al., 2018, Shortt et al., 2018), with parallels that can be drawn in the context of cancer treatment. Dietary fiber is one component that has been shown to have a profound influence on the composition of the gut microbiome, with a decrease in the abundance of immune-promoting F. prausnitzii (Benus et al., 2010) and the SCFA butyrate in stool samples after reducing dietary fiber in healthy human subjects. Other studies have focused on supplementation of the diet with plant polysaccharide inulin prebiotics, demonstrating significant increases of both Faecalibacterium and Bifidobacterium species with this dietary intervention (Ramirez-Farias et al., 2009). Conversely, elimination of animal fats in the human diet was associated with a decrease in detrimental Bacteroidales bacteria (Turnbaugh et al., 2007). Given the importance of these bacteria in implications for cancer therapy, it’s possible that diet could serve as a possible strategy to improve outcomes through modulation of the microbiome.
The favorable safety profile, cost and accessibility of dietary interventions could provide a simple and safe opportunity for assessing the implications of microbiota and downstream immune manipulation in cancer patient populations. Indeed, some groups have already begun to explore the dietary impact on the gut microbiota in cancer patients (Table 1). The “BE GONE” trial (NCT02843425) is designed to investigate fiber supplementation in cancer patients, through supplementation of a half cup of beans per day into the normal diets of cancer patients to measure shifts in bacterial populations. Meanwhile, “The Role of Lifestyle Factors in Breast Cancer-Related Outcomes” (NCT02079662) trial utilizes a comprehensive lifestyle overhaul, providing dietary counseling and meal delivery along with exercise and psychosocial services with a randomized design in patients with stage III breast cancer initiating radiation therapy. This study hypothesizes that women in the lifestyle intervention arm will have improved outcomes compared to those in the control arm. Though this trial is primarily powered to detect difference in recurrence rates, longitudinal gut and oral microbiome samples, along with a battery of questionnaires, are listed as secondary outcomes in order to better gauge how the microbiome changes in relation to behavior patterns in cancer patients. Blood samples are also being collected at the same time points to gain insight into mechanistic changes associated with lifestyle changes, microbial shifts over the course of the intervention and therapy. While both studies are in their infancy, they will provide valuable information on how lifestyle factors modulate the gut microbiome, disease markers and patient outcomes.
Administration of bacterial consortia or “designer probiotics”
While dietary interventions may seem relatively simple to design and implement, the effects on the microbiota can be modest, and patient compliance is difficult to enforce and monitor. Administration of bacterial consortia or “designer probiotics” could provide a more feasible method of microbial manipulation in the clinical setting. Several trials using probiotics in cancer patients have been initiated with some completed, and most studies have focused on safety and biomarker-related endpoints, with a minority including cancer- related outcomes (such as disease-free survival) as a primary endpoint (NCT02079662).
Some of the first studies initiated involved treatment with probiotics in patients with colorectal cancer (CRC) (Table 1). This includes a trial where patients with CRC were treated with probiotics containing strains of L. acidophilus and B. lactis, and were shown to have an increased abundance of butyrate-producing bacteria (particularly Faecalibacterium and other Clostridiales) within the tumor, and its associated non-tumor colonic mucosa and stool (NCT03072641) (Hibberd et al., 2017). Another study assessed preoperative probiotic therapy on mucosal immunity in CRC patients, demonstrating altered cytokine profiles within the colonic mucosa assessed at the time of colon resection, with lower IL-1β, IL-10, and IL-23A mRNA levels in the patients treated with probiotics compared to controls who received no probiotics (NCT01895530) (Consoli et al., 2016). These studies demonstrate mixed changes in colonic mucosa – with a decreased production of both pro-inflammatory and anti-inflammatory cytokines within healthy colonic mucosa; thus, it is difficult to interpret what these findings might imply in regards to colorectal cancer development and progression or response to therapy. Nonetheless it is proof of principle that probiotic therapy can alter immunity locally.
In addition to these studies in colorectal cancer, there are ongoing trials assessing the impact of administration of probiotics on other cancer types, including a trial investigating the effects of probiotics on CD8+ T cell infiltrate in patients with stage I-III breast cancer (NCT03358511). This is a single arm study and all patients will receive the same probiotic (Primal Defense Ultra multi-strain probiotic formula). Additional studies are currently in development, and it is important to highlight that assessment of changes to the microbiome as well as anti-tumor immunity in these studies is paramount so that we may best understand these interventions. Certainly, there is wide variability in probiotic formulations available with regard to their composition, stability, and authenticity (Huys et al., 2013) – thus significant caution should be taken in advocating their use to cancer patients until these can be carefully tested. Efforts to identify “ideal” bacterial consortia to be administered to cancer patients to enhance responses to cancer therapy are underway but have yet to be defined.
Fecal Microbiota Transplant (FMT)
FMT represents the most direct means to manipulate the microbiota, and FMT preparations can be administered to patients via oral administration of lyophilized or frozen pills or via direct delivery by colonoscopy or gastroscopy. FMT has already been employed in other patient populations, showing significant success in curing C. difficile infection resistant to conventional therapies (Borody et al., 2004).
Clinical trials utilizing FMT in cancer patients are in their infancy, but, based on results from preclinical studies discussed above, they have generated much excitement. Autologous FMT in Acute Myeloid Leukemia (AML) is being trialed in patients undergoing intensive treatment in an attempt to prevent dysbiosis and to increase diversity of the gut microbiota during the course of treatment (NCT02928523). Furthermore, FMT is being considered in patients undergoing immunotherapy for solid tumor malignancies, specifically those treated with immune checkpoint inhibitors (Table 1). A Phase 1 single-center trial for metastatic melanoma patients who failed prior immunotherapy opened recently (NCT03353402) wherein FMT from patients with a good response to immunotherapy is administered to refractory patients. Primary outcomes include safety and time to microbiota engraftment while secondary outcomes include immune cell shifts, alterations in immune cell activity and objective response. Design of additional trials is currently underway to test the hypothesis that modulation of the gut microbiota will improve response to treatment with immune checkpoint blockade (Gopalakrishnan et al., 2018).
Conclusions and future directions
The age of the microbiome is upon us, and seminal reports incorporating preclinical and clinical studies on the role of microbiota in cancer have brought this topic to light as a potentially dominant mediator in response to cancer therapy. We have gained insights into the influence of the microbiome on immunity and cancer – however, there is still a great deal to learn with regard to the mechanisms behind this, as well as optimal strategies to modulate the gut microbiome to enhance responses to cancer immunotherapy.
Provocative clinical questions are also raised from these studies, and call into question the potential need and utility of microbiome profiling in patients on cancer therapy – however complexities exist with regard to optimal methods for profiling (16S rRNA sequencing versus metagenomic shotgun sequencing and choice of reference databases). In addition to this, significant additional questions remain regarding how other factors impact the gut microbiome such as diet, medications (including probiotics, antibiotics, and other medications), mental health or other environmental factors and how they impact cancer therapy – and also call to question the potential need to monitor these factors during cancer therapy.
Furthermore, additional complexities exist as we move forward with efforts to modulate the gut microbiome to enhance therapeutic responses. It is not yet clear what composition of the gut microbiome is optimal to facilitate anti-tumor immune responses – and a diverse range of therapeutic options exist to change the microbiome that need to be tested carefully in the context of clinical trials. The use of preparative regimens prior to modulation of the gut microbiome (e.g. with antibiotics) and methods to sustain changes (via dietary and prebiotic supplementation) is also of important consideration. It is only through a comprehensive understanding of these interactions (in pre-clinical models and in the context of these trials) that we will learn how to optimally modulate the gut microbiota to enhance anti-tumor immunity and immunity as a whole, with the potential to enhance immune-surveillance and cancer therapies.
Acknowledgements:
JAW has honoraria from speakers’ bureau of Dava Oncology, Bristol-Myers Squibb, and Illumina and is an advisory board member for GlaxoSmithKline, Novartis and Roche/Genentech J.A.W. is supported by the Binational Science Foundation, the Melanoma Research Alliance, Stand Up To Cancer, an MD Anderson Cancer Center Multidisciplinary Research Program Grant, and MD Anderson Cancer Center’s Melanoma Moon Shots Program. J.A.W. is a member of the Parker Institute for Cancer Immunotherapy at MD Anderson Cancer Center. A.R. is supported by the Kimberley Clarke Foundation Award for Scientific Achievement provided by the Odyssey Fellowship program at The University of Texas MD Anderson Cancer Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests:
J.A.W. and V.G. are inventors on a U.S. patent application (PCT/US17/53717) submitted by The University of Texas MD Anderson Cancer Center that covers methods to enhance checkpoint blockade therapy by the microbiome. J.A.W is a clinical and scientific advisor at Microbiome DX. V.G. is a consultant at Microbiome DX. CS, AR and BH report no relevant conflicts of interest or financial disclosures.
References:
- ABT MC, OSBORNE LC, MONTICELLI LA, DOERING TA, ALENGHAT T, SONNENBERG GF, PALEY MA, ANTENUS M, WILLIAMS KL & ERIKSON J 2012. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity, 37, 158–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BACKHED F, LEY RE, SONNENBURG JL, PETERSON DA & GORDON JI 2005. Host-bacterial mutualism in the human intestine. Science, 307, 1915–20. [DOI] [PubMed] [Google Scholar]
- BENUS RF, VAN DER WERF TS, WELLING GW, JUDD PA, TAYLOR MA, HARMSEN HJ & WHELAN K 2010. Association between Faecalibacterium prausnitzii and dietary fibre in colonic fermentation in healthy human subjects. Br J Nutr, 104, 693–700. [DOI] [PubMed] [Google Scholar]
- BERMAN D, PARKER SM, SIEGEL J, CHASALOW SD, WEBER J, GALBRAITH S, TARGAN SR & WANG HL 2010. Blockade of cytotoxic T-lymphocyte antigen-4 by ipilimumab results in dysregulation of gastrointestinal immunity in patients with advanced melanoma. Cancer Immun, 10, 11. [PMC free article] [PubMed] [Google Scholar]
- BIAGI E, ZAMA D, NASTASI C, CONSOLANDI C, FIORI J, RAMPELLI S, TURRONI S, CENTANNI M, SEVERGNINI M, PEANO C, DE BELLIS G, BASAGLIA G, GOTTI R, MASETTI R, PESSION A, BRIGIDI P & CANDELA M 2015. Gut microbiota trajectory in pediatric patients undergoing hematopoietic SCT. Bone Marrow Transplant, 50, 992–8. [DOI] [PubMed] [Google Scholar]
- BLANK CU, HAANEN JB, RIBAS A & SCHUMACHER TN 2016. CANCER IMMUNOLOGY. The “cancer immunogram”. Science, 352, 658–60. [DOI] [PubMed] [Google Scholar]
- BORODY TJ, WARREN EF, LEIS SM, SURACE R, ASHMAN O & SIARAKAS S 2004. Bacteriotherapy using fecal flora: toying with human motions. J Clin Gastroenterol, 38, 475–83. [DOI] [PubMed] [Google Scholar]
- CAHN JY, KLEIN JP, LEE SJ, MILPIED N, BLAISE D, ANTIN JH, LEBLOND V, IFRAH N, JOUET JP, LOBERIZA F, RINGDEN O, BARRETT AJ, HOROWITZ MM, SOCIE G, SOCIETE FRANCAISE DE GREFFE DE MOELLE ET THERAPIE, C., DANA FARBER CANCER, I. & INTERNATIONAL BONE MARROW TRANSPLANT, R. 2005. Prospective evaluation of 2 acute graft-versus-host (GVHD) grading systems: a joint Societe Francaise de Greffe de Moelle et Therapie Cellulaire (SFGM-TC), Dana Farber Cancer Institute (DFCI), and International Bone Marrow Transplant Registry (IBMTR) prospective study. Blood, 106, 1495–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHAPUT N, LEPAGE P, COUTZAC C, SOULARUE E, LE ROUX K, MONOT C, BOSELLI L, ROUTIER E, CASSARD L, COLLINS M, VAYSSE T, MARTHEY L, EGGERMONT A, ASVATOURIAN V, LANOY E, MATEUS C, ROBERT C & CARBONNEL F 2017. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann Oncol, 28, 1368–1379. [DOI] [PubMed] [Google Scholar]
- CHEN DS & MELLMAN I 2013. Oncology meets immunology: the cancer-immunity cycle. Immunity, 39, 1–10. [DOI] [PubMed] [Google Scholar]
- CHEN DS & MELLMAN I 2017. Elements of cancer immunity and the cancer–immune set point. Nature, 541, 321. [DOI] [PubMed] [Google Scholar]
- COGDILL AP, ANDREWS MC & WARGO JA 2017. Hallmarks of response to immune checkpoint blockade. Br J Cancer, 117, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAVID LA, MAURICE CF, CARMODY RN, GOOTENBERG DB, BUTTON JE, WOLFE BE, LING AV, DEVLIN AS, VARMA Y, FISCHBACH MA, BIDDINGER SB, DUTTON RJ & TURNBAUGH PJ 2014. Diet rapidly and reproducibly alters the human gut microbiome. Nature, 505, 559–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DUBIN K, CALLAHAN MK, REN B, KHANIN R, VIALE A, LING L, NO D, GOBOURNE A, LITTMANN E, HUTTENHOWER C, PAMER EG & WOLCHOK JD 2016. Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat Commun, 7, 10391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAITH JJ, AHERN PP, RIDAURA VK, CHENG J & GORDON JI 2014. Identifying gut microbe-host phenotype relationships using combinatorial communities in gnotobiotic mice. Sci Transl Med, 6, 220ra11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FRANKEL AE, COUGHLIN LA, KIM J, FROEHLICH TW, XIE Y, FRENKEL EP & KOH AY 2017. Metagenomic Shotgun Sequencing and Unbiased Metabolomic Profiling Identify Specific Human Gut Microbiota and Metabolites Associated with Immune Checkpoint Therapy Efficacy in Melanoma Patients(). Neoplasia (New York, N.Y.), 19, 848–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FROSALI S, PAGLIARI D, GAMBASSI G, LANDOLFI R, PANDOLFI F & CIANCI R 2015. How the Intricate Interaction among Toll-Like Receptors, Microbiota, and Intestinal Immunity Can Influence Gastrointestinal Pathology. J Immunol Res, 2015, 489821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GERBITZ A, SCHULTZ M, WILKE A, LINDE HJ, SCHOLMERICH J, ANDREESEN R & HOLLER E 2004. Probiotic effects on experimental graft-versus-host disease: let them eat yogurt. Blood, 103, 4365–7. [DOI] [PubMed] [Google Scholar]
- GOPALAKRISHNAN V, SPENCER CN, NEZI L, REUBEN A, ANDREWS MC, KARPINETS TV, PRIETO PA, VICENTE D, HOFFMAN K, WEI SC, COGDILL AP, ZHAO L, HUDGENS CW, HUTCHINSON DS, MANZO T, PETACCIA DE MACEDO M, COTECHINI T, KUMAR T, CHEN WS, REDDY SM, SZCZEPANIAK SLOANE R, GALLOWAY-PENA J, JIANG H, CHEN PL, SHPALL EJ, REZVANI K, ALOUSI AM, CHEMALY RF, SHELBURNE S, VENCE LM, OKHUYSEN PC, JENSEN VB, SWENNES AG, MCALLISTER F, MARCELO RIQUELME SANCHEZ E, ZHANG Y, LE CHATELIER E, ZITVOGEL L, PONS N, AUSTIN-BRENEMAN JL, HAYDU LE, BURTON EM, GARDNER JM, SIRMANS E, HU J, LAZAR AJ, TSUJIKAWA T, DIAB A, TAWBI H, GLITZA IC, HWU WJ, PATEL SP, WOODMAN SE, AMARIA RN, DAVIES MA, GERSHENWALD JE, HWU P, LEE JE, ZHANG J, COUSSENS LM, COOPER ZA, FUTREAL PA, DANIEL CR, AJAMI NJ, PETROSINO JF, TETZLAFF MT, SHARMA P, ALLISON JP, JENQ RR & WARGO JA 2018. Gut microbiome modulates response to anti–PD-1 immunotherapy in melanoma patients. Science, 359, 97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HEIMESAAT MM, NOGAI A, BERESWILL S, PLICKERT R, FISCHER A, LODDENKEMPER C, STEINHOFF U, TCHAPTCHET S, THIEL E, FREUDENBERG MA, GOBEL UB & UHAREK L 2010. MyD88/TLR9 mediated immunopathology and gut microbiota dynamics in a novel murine model of intestinal graft-versus-host disease. Gut, 59, 1079–87. [DOI] [PubMed] [Google Scholar]
- HIROTA K, DUARTE JH, VELDHOEN M, HORNSBY E, LI Y, CUA DJ, AHLFORS H, WILHELM C, TOLAINI M, MENZEL U, GAREFALAKI A, POTOCNIK AJ & STOCKINGER B 2011. Fate mapping of IL-17-producing T cells in inflammatory responses. Nat Immunol, 12, 255–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOLLER E, BUTZHAMMER P, SCHMID K, HUNDSRUCKER C, KOESTLER J, PETER K, ZHU W, SPORRER D, HEHLGANS T, KREUTZ M, HOLLER B, WOLFF D, EDINGER M, ANDREESEN R, LEVINE JE, FERRARA JL, GESSNER A, SPANG R & OEFNER PJ 2014. Metagenomic analysis of the stool microbiome in patients receiving allogeneic stem cell transplantation: loss of diversity is associated with use of systemic antibiotics and more pronounced in gastrointestinal graft-versus-host disease. Biol Blood Marrow Transplant, 20, 640–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HONDA K & LITTMAN DR 2016. The microbiota in adaptive immune homeostasis and disease. Nature, 535, 75–84. [DOI] [PubMed] [Google Scholar]
- HUDA MN, LEWIS Z, KALANETRA KM, RASHID M, AHMAD SM, RAQIB R, QADRI F, UNDERWOOD MA, MILLS DA & STEPHENSEN CB 2014. Stool Microbiota and Vaccine Responses of Infants. Pediatrics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUYS G, BOTTELDOORN N, DELVIGNE F, DE VUYST L, HEYNDRICKX M, POT B, DUBOIS JJ & DAUBE G 2013. Microbial characterization of probiotics--advisory report of the Working Group “8651 Probiotics” of the Belgian Superior Health Council (SHC). Mol Nutr Food Res, 57, 1479–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IIDA N, DZUTSEV A, STEWART CA, SMITH L, BOULADOUX N, WEINGARTEN RA, MOLINA DA, SALCEDO R, BACK T, CRAMER S, DAI RM, KIU H, CARDONE M, NAIK S, PATRI AK, WANG E, MARINCOLA FM, FRANK KM, BELKAID Y, TRINCHIERI G & GOLDSZMID RS 2013. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science, 342, 967–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IVANOV II, ATARASHI K, MANEL N, BRODIE EL, SHIMA T, KARAOZ U, WEI D, GOLDFARB KC, SANTEE CA, LYNCH SV, TANOUE T, IMAOKA A, ITOH K, TAKEDA K, UMESAKI Y, HONDA K & LITTMAN DR 2009. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell, 139, 485–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IVANOV II, FRUTOS RDE L, MANEL N, YOSHINAGA K, RIFKIN DB, SARTOR RB, FINLAY BB & LITTMAN DR 2008. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe, 4, 337–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JACOBSOHN DA & VOGELSANG GB 2007. Acute graft versus host disease. Orphanet J Rare Dis, 2, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JENQ RR, TAUR Y, DEVLIN SM, PONCE DM, GOLDBERG JD, AHR KF, LITTMANN ER, LING L, GOBOURNE AC, MILLER LC, DOCAMPO MD, PELED JU, ARPAIA N, CROSS JR, PEETS TK, LUMISH MA, SHONO Y, DUDAKOV JA, POECK H, HANASH AM, BARKER JN, PERALES MA, GIRALT SA, PAMER EG & VAN DEN BRINK MR 2015. Intestinal Blautia Is Associated with Reduced Death from Graft-versus-Host Disease. Biol Blood Marrow Transplant, 21, 1373–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JENQ RR, UBEDA C, TAUR Y, MENEZES CC, KHANIN R, DUDAKOV JA, LIU C, WEST ML, SINGER NV, EQUINDA MJ, GOBOURNE A, LIPUMA L, YOUNG LF, SMITH OM, GHOSH A, HANASH AM, GOLDBERG JD, AOYAMA K, BLAZAR BR, PAMER EG & VAN DEN BRINK MR 2012. Regulation of intestinal inflammation by microbiota following allogeneic bone marrow transplantation. J Exp Med, 209, 903–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JOHANSSON ME, JAKOBSSON HE, HOLMEN-LARSSON J, SCHUTTE A, ERMUND A, RODRIGUEZ-PINEIRO AM, ARIKE L, WISING C, SVENSSON F, BACKHED F & HANSSON GC 2015. Normalization of Host Intestinal Mucus Layers Requires Long-Term Microbial Colonization. Cell Host Microbe, 18, 582–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KROEMER G & ZITVOGEL L 2018. Cancer immunotherapy in 2017: The breakthrough of the microbiota. Nat Rev Immunol, 18, 87–88. [DOI] [PubMed] [Google Scholar]
- LARKIN J, HODI FS & WOLCHOK JD 2015. Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. N Engl J Med, 373, 1270–1. [DOI] [PubMed] [Google Scholar]
- LATHROP SK, BLOOM SM, RAO SM, NUTSCH K, LIO CW, SANTACRUZ N, PETERSON DA, STAPPENBECK TS & HSIEH CS 2011. Peripheral education of the immune system by colonic commensal microbiota. Nature, 478, 250–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LETUNIC I & BORK P 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res, 44, W242–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEVY M, KOLODZIEJCZYK AA, THAISS CA & ELINAV E 2017. Dysbiosis and the immune system. Nat Rev Immunol, 17, 219–232. [DOI] [PubMed] [Google Scholar]
- MA N, GUO P, ZHANG J, HE T, KIM SW, ZHANG G & MA X 2018. Nutrients Mediate Intestinal Bacteria-Mucosal Immune Crosstalk. Front Immunol, 9, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MANTIS NJ, ROL N & CORTHÉSY B 2011. Secretory IgA’s Complex Roles in Immunity and Mucosal Homeostasis in the Gut. Mucosal immunology, 4, 603–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MATSON V, FESSLER J, BAO R, CHONGSUWAT T, ZHA Y, ALEGRE M-L, LUKE JJ & GAJEWSKI TF 2018. The commensal microbiome is associated with anti–PD-1 efficacy in metastatic melanoma patients. Science, 359, 104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MORGAN XC & HUTTENHOWER C 2012. Chapter 12: Human microbiome analysis. PLoS Comput Biol, 8, e1002808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OH JZ, RAVINDRAN R, CHASSAING B, CARVALHO FA, MADDUR MS, BOWER M, HAKIMPOUR P, GILL KP, NAKAYA HI, YAROVINSKY F, SARTOR RB, GEWIRTZ AT & PULENDRAN B 2014. TLR5-mediated sensing of gut microbiota is necessary for antibody responses to seasonal influenza vaccination. Immunity, 41, 478–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PABST O 2012. New concepts in the generation and functions of IgA. Nat Rev Immunol, 12, 821–32. [DOI] [PubMed] [Google Scholar]
- PAULOS CM, WRZESINSKI C, KAISER A, HINRICHS CS, CHIEPPA M, CASSARD L, PALMER DC, BONI A, MURANSKI P, YU Z, GATTINONI L, ANTONY PA, ROSENBERG SA & RESTIFO NP 2007. Microbial translocation augments the function of adoptively transferred self/tumor-specific CD8+ T cells via TLR4 signaling. J Clin Invest, 117, 2197–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PELED JU, DEVLIN SM, STAFFAS A, LUMISH M, KHANIN R, LITTMANN ER, LING L, KOSURI S, MALOY M, SLINGERLAND JB, AHR KF, POROSNICU RODRIGUEZ KA, SHONO Y, SLINGERLAND AE, DOCAMPO MD, SUNG AD, WEBER D, ALOUSI AM, GYURKOCZA B, PONCE DM, BARKER JN, PERALES MA, GIRALT SA, TAUR Y, PAMER EG, JENQ RR & VAN DEN BRINK MR 2017. Intestinal Microbiota and Relapse After Hematopoietic-Cell Transplantation. J Clin Oncol, 1650–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RAMIREZ-FARIAS C, SLEZAK K, FULLER Z, DUNCAN A, HOLTROP G & LOUIS P 2009. Effect of inulin on the human gut microbiota: stimulation of Bifidobacterium adolescentis and Faecalibacterium prausnitzii. Br J Nutr, 101, 541–50. [DOI] [PubMed] [Google Scholar]
- ROOKS MG & GARRETT WS 2016. Gut microbiota, metabolites and host immunity. Nat Rev Immunol, 16, 341–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROUND JL & MAZMANIAN SK 2010. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A, 107, 12204–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROUTY B, LE CHATELIER E, DEROSA L, DUONG CPM, ALOU MT, DAILLÈRE R, FLUCKIGER A, MESSAOUDENE M, RAUBER C, ROBERTI MP, FIDELLE M, FLAMENT C, POIRIER-COLAME V, OPOLON P, KLEIN C, IRIBARREN K, MONDRAGÓN L, JACQUELOT N, QU B, FERRERE G, CLÉMENSON C, MEZQUITA L, MASIP JR, NALTET C, BROSSEAU S, KADERBHAI C, RICHARD C, RIZVI H, LEVENEZ F, GALLERON N, QUINQUIS B, PONS N, RYFFEL B, MINARD-COLIN V, GONIN P, SORIA J-C, DEUTSCH E, LORIOT Y, GHIRINGHELLI F, ZALCMAN G, GOLDWASSER F, ESCUDIER B, HELLMANN MD, EGGERMONT A, RAOULT D, ALBIGES L, KROEMER G & ZITVOGEL L 2018. Gut microbiome influences efficacy of PD-1–based immunotherapy against epithelial tumors. Science, 359, 91–97. [DOI] [PubMed] [Google Scholar]
- ROUTY B, LETENDRE C, ENOT D, CHÉNARD-POIRIER M, MEHRAJ V, SÉGUIN NC, GUENDA K, GAGNON K, WOERTHER P-L, GHEZ D & LACHANCE S 2017. The influence of gut-decontamination prophylactic antibiotics on acute graft-versus-host disease and survival following allogeneic hematopoietic stem cell transplantation. Oncoimmunology, 6, e1258506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SENDER R, FUCHS S & MILO R 2016. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLOS Biology, 14, e1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHARMA P, HU-LIESKOVAN S, WARGO JA & RIBAS A 2017. Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell, 168, 707–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHEN Y, GIARDINO TORCHIA ML, LAWSON GW, KARP CL, ASHWELL JD & MAZMANIAN SK 2012. Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell Host Microbe, 12, 509–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHONO Y, DOCAMPO MD, PELED JU, PEROBELLI SM, VELARDI E, TSAI JJ, SLINGERLAND AE, SMITH OM, YOUNG LF, GUPTA J, LIEBERMAN SR, JAY HV, AHR KF, POROSNICU RODRIGUEZ KA, XU K, CALARFIORE M, POECK H, CABALLERO S, DEVLIN SM, RAPAPORT F, DUDAKOV JA, HANASH AM, GYURKOCZA B, MURPHY GF, GOMES C, LIU C, MOSS EL, FALCONER SB, BHATT AS, TAUR Y, PAMER EG, VAN DEN BRINK MRM & JENQ RR 2016. Increased GVHD-related mortality with broad-spectrum antibiotic use after allogeneic hematopoietic stem cell transplantation in human patients and mice. Science Translational Medicine, 8, 339ra71–339ra71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHORTT C, HASSELWANDER O, MEYNIER A, NAUTA A, FERNANDEZ EN, PUTZ P, ROWLAND I, SWANN J, TURK J, VERMEIREN J & ANTOINE JM 2018. Systematic review of the effects of the intestinal microbiota on selected nutrients and non-nutrients. Eur J Nutr, 57, 25–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SIVAN A, CORRALES L, HUBERT N, WILLIAMS JB, AQUINO-MICHAELS K, EARLEY ZM, BENYAMIN FW, LEI YM, JABRI B, ALEGRE ML, CHANG EB & GAJEWSKI TF 2015. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science, 350, 1084–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SNYDER A, MAKAROV V, MERGHOUB T, YUAN J, ZARETSKY JM, DESRICHARD A, WALSH LA, POSTOW MA, WONG P, HO TS, HOLLMANN TJ, BRUGGEMAN C, KANNAN K, LI Y, ELIPENAHLI C, LIU C, HARBISON CT, WANG L, RIBAS A, WOLCHOK JD & CHAN TA 2014. Genetic Basis for Clinical Response to CTLA-4 Blockade in Melanoma. N Engl J Med, 371, 2189–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SPILJAR M, MERKLER D & TRAJKOVSKI M 2017. The Immune System Bridges the Gut Microbiota with Systemic Energy Homeostasis: Focus on TLRs, Mucosal Barrier, and SCFAs. Front Immunol, 8, 1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SPITZER MH, CARMI Y, RETICKER-FLYNN NE, KWEK SS, MADHIREDDY D, MARTINS MM, GHERARDINI PF, PRESTWOOD TR, CHABON J, BENDALL SC, FONG L, NOLAN GP & ENGLEMAN EG 2017. Systemic Immunity Is Required for Effective Cancer Immunotherapy. Cell, 168, 487–502 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STARY G, OLIVE A, RADOVIC-MORENO AF, GONDEK D, ALVAREZ D, BASTO PA, PERRO M, VRBANAC VD, TAGER AM, SHI J, YETHON JA, FAROKHZAD OC, LANGER R, STARNBACH MN & VON ANDRIAN UH 2015. VACCINES. A mucosal vaccine against Chlamydia trachomatis generates two waves of protective memory T cells. Science, 348, aaa8205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAUR Y, JENQ RR, PERALES MA, LITTMANN ER, MORJARIA S, LING L, NO D, GOBOURNE A, VIALE A, DAHI PB, PONCE DM, BARKER JN, GIRALT S, VAN DEN BRINK M & PAMER EG 2014. The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood, 124, 1174–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAUR Y, XAVIER JB, LIPUMA L, UBEDA C, GOLDBERG J, GOBOURNE A, LEE YJ, DUBIN KA, SOCCI ND, VIALE A, PERALES MA, JENQ RR, VAN DEN BRINK MR & PAMER EG 2012. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin Infect Dis, 55, 905–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TURNBAUGH PJ, LEY RE, HAMADY M, FRASER-LIGGETT CM, KNIGHT R & GORDON JI 2007. The human microbiome project. Nature, 449, 804–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VAN BEKKUM DW, ROODENBURG J, HEIDT PJ & VAN DER WAAIJ D 1974. Mitigation of secondary disease of allogeneic mouse radiation chimeras by modification of the intestinal microflora. J Natl Cancer Inst, 52, 401–4. [DOI] [PubMed] [Google Scholar]
- VETIZOU M, PITT JM, DAILLERE R, LEPAGE P, WALDSCHMITT N, FLAMENT C, RUSAKIEWICZ S, ROUTY B, ROBERTI MP, DUONG CP, POIRIER-COLAME V, ROUX A, BECHAREF S, FORMENTI S, GOLDEN E, CORDING S, EBERL G, SCHLITZER A, GINHOUX F, MANI S, YAMAZAKI T, JACQUELOT N, ENOT DP, BERARD M, NIGOU J, OPOLON P, EGGERMONT A, WOERTHER PL, CHACHATY E, CHAPUT N, ROBERT C, MATEUS C, KROEMER G, RAOULT D, BONECA IG, CARBONNEL F, CHAMAILLARD M & ZITVOGEL L 2015. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science, 350, 1079–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VIAUD S, SACCHERI F, MIGNOT G, YAMAZAKI T, DAILLERE R, HANNANI D, ENOT DP, PFIRSCHKE C, ENGBLOM C, PITTET MJ, SCHLITZER A, GINHOUX F, APETOH L, CHACHATY E, WOERTHER PL, EBERL G, BERARD M, ECOBICHON C, CLERMONT D, BIZET C, GABORIAU-ROUTHIAU V, CERF-BENSUSSAN N, OPOLON P, YESSAAD N, VIVIER E, RYFFEL B, ELSON CO, DORE J, KROEMER G, LEPAGE P, BONECA IG, GHIRINGHELLI F & ZITVOGEL L 2013. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science, 342, 971–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WEAVER CT, ELSON CO, FOUSER LA & KOLLS JK 2013. The Th17 pathway and inflammatory diseases of the intestines, lungs, and skin. Annu Rev Pathol, 8, 477–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WEBER D, OEFNER PJ, HIERGEIST A, KOESTLER J, GESSNER A, WEBER M, HAHN J, WOLFF D, STAMMLER F, SPANG R, HERR W, DETTMER K & HOLLER E 2015. Low urinary indoxyl sulfate levels early after transplantation reflect a disrupted microbiome and are associated with poor outcome. Blood, 126, 1723–8. [DOI] [PubMed] [Google Scholar]
- WEBER JS, KUDCHADKAR RR, YU B, GALLENSTEIN D, HORAK CE, INZUNZA HD, ZHAO X, MARTINEZ AJ, WANG W, GIBNEY G, KROEGER J, EYSMANS C, SARNAIK AA & CHEN YA 2013. Safety, efficacy, and biomarkers of nivolumab with vaccine in ipilimumab-refractory or -naive melanoma. J Clin Oncol, 31, 4311–8. [DOI] [PMC free article] [PubMed] [Google Scholar]