Abstract
The present study aims to investigate if overexpressing the mitochondrial transcription factor A (TFAM) gene in a transgenic mouse model diminishes soleus and gastrocnemius atrophy occurring during hindlimb suspension (HLS). Additionally, we aim to observe if combining exercise training in TFAM transgenic mice prior to HLS has a synergistic effect in preventing skeletal muscle atrophy. Male C57BL/6J-based transgenic mice (12–14 weeks old) overexpressing TFAM were assigned to a control (T-Control), 7-day HLS (T-HLS), and 2-week exercise training prior to 7-day HLS (T-Ex+HLS) groups. These groups were compared to male C57BL/6J wild-type (WT) mice (12–14 weeks old) assigned to Control, 7-day HLS (HLS), 2week exercise training prior to 7-day HLS (Ex+HLS), and 2-week exercise training (Ex). Overexpressing TFAM results in a decrease of 8.3% in soleus and 2.6% in gastrocnemius muscle weight to bodyweight ratio after only HLS compared to wild-type mice incurring a loss of 27.1% in soleus and 21.5% in gastrocnemius muscle after HLS. Our data indicates TFAM may play a critical role in protecting skeletal muscle from disuse atrophy and is correlated with increased expression of antioxidants (SOD-2) and potential redox balance. TFAM may be an attractive molecule of interest for potential, future therapeutic development.
Keywords: TFAM, Exercise, Skeletal Muscle Atrophy, Mitochondria
Introduction
Skeletal muscle functions not only in propulsion for locomotion but also as a glucose and amino acid reservoir (31). It is the largest metabolically active organ in the human body and also functions as an endocrine secretory tissue (5, 28). Notably, skeletal muscle strength is a key predictor of life span (24) and vitality. Being that this tissue functions in a variety of important processes, pathological losses of skeletal muscle can greatly affect health and quality of life.
Skeletal muscle atrophy is the loss of muscle tissue and is characterized by a decline in muscle mass leading to reductions in force production, decreased cross-sectional area of the muscle fibers, and diminished oxidative ability (36). As this tissue atrophies, the ability to perform physical tasks is greatly reduced and can significantly decrease the physical independence of a patient as well as increase factors of morbidity and mortality (30). Furthermore, when patients enter a scenario in which atrophy occurs, resources must be devoted to the mitigation and recovery process, such as pharmacological interventions or the use of rehab specialists. Preventing the condition of skeletal muscle atrophy is an important research endeavor to improve the patient condition, both physically and fiscally.
While many scenarios may cause skeletal muscle atrophy (disease, starvation, and aging), the mechanical unloading and disuse of muscle results in specific atrophic changes. Skeletal muscle is abnormally disused or unloaded during circumstances such as bedrest, limb immobilization, spinal cord injury, microgravity, and a sedentary lifestyle (42). During all of these scenarios, significant reductions in fiber contraction and mechanical stress on the muscle tissue occur, leading to potentially abrupt and reductive structural and molecular changes (16).
Disuse-induced atrophy results in the loss of muscle fiber size, weight, and function (4). When this occurs, decreases in molecular signaling pathways leading to protein synthesis and increases in molecular pathways leading to protein degradation and apoptosis are observed resulting in a net loss of muscle protein over time (16, 37). These molecular mechanisms are also correlated with mitochondrial dysfunction and redox imbalance (26, 33). Dysfunctional mitochondria and excessive reactive oxygen species (ROS) accumulating during muscle atrophy reduces protein synthesis and activates protein catabolic mechanisms (7, 17, 25). ROS, in particular, can accumulate in the mitochondria and damage mtDNA, leading directly to dysfunction through the mutation of mtDNA and the transcription and translation of dysfunctional mitochondrial proteins (39).
Exercise powerfully stimulates mitochondrial function through small molecular changes that occur with physical activity that converge on the upregulation of the mitochondrial biogenesis master-regulator protein, peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α) (32). This master regulator co-activates with nuclear-respiratory factors 1 and 2, increasing the transcription of mitochondrial transcription factor A (TFAM). Translated TFAM is chaperoned and imported into the mitochondria by a heat-shock protein (HSP) complex, HSP 70 and HSP 60. It is then translocated into the inner-mitochondrial matrix and re-folded into its mature form where it can interact with mtDNA leading to increased mtDNA transcription. TFAM also abundantly binds mtDNA forming histone-like nucleoid structures to store, maintain, and protect mtDNA from binding and degradation via ROS (20).
Substantial evidence in prior research implicates the strong link of TFAM and mitochondrial health. Reviewing animal studies of TFAM, we see a germ line disruption of the mouse TFAM (TFAM −/−) gene leading to extreme reductions in mtDNA and embryonic lethality (29). A 2004 study using a combination of TFAM overexpression and knockout mice reveals mtDNA copy numbers are directly proportional to TFAM levels (8). Balliet et al. showed a lack of TFAM resulted in mitochondrial dysfunction due to the loss of OXPHOS proteins (38). In contrast, an in vivo treatment with a recombinant form of TFAM resulted in increased motor endurance and complex-I respiration in mice (15).
There is evidence in a limited number of in vivo and in vitro TFAM overexpression studies that reveal decreased left-ventricular remodeling after myocardial infarction and an overall protective role of TFAM in cardiomyocytes (11, 14), establishing the important role of TFAM in a muscle tissue. In skeletal muscle, a mouse model with a skeletal-muscle specific disruption of TFAM led to mitochondrial myopathy, ragged-red fibers, reduced muscle force, abnormal mitochondrial shape, and decreased respiratory chain activity (40) specifically indicating its vital role in skeletal muscle. During skeletal muscle differentiation, increases in TFAM mRNA levels corresponded with increases in mitochondrial content, while the authors concluded TFAM protein levels are regulated by the availability of TFAM mRNA (6). Furthermore, we see overexpressing PGC-1α in a mouse model prevents disuse-induced skeletal muscle atrophy of the soleus (3). With TFAM being transcribed downstream of PGC-1α and tightly associated with markers of mitochondrial function, a TFAM mechanism of skeletal muscle protection may exist. There are no known studies, to the best of our knowledge, analyzing TFAM overexpression in a disuse-induced skeletal muscle atrophy mouse model. Understanding the specific role of TFAM in skeletal muscle health may lead to atrophy treatments and increase patient quality of life in the future.
Therefore, we hypothesize overexpressing TFAM in mouse skeletal muscle will prevent or diminish unloading and disuse-induced atrophy. To test this, we use a 7-day hindlimb suspension (HLS) protocol to induce muscle atrophy in the hindlimbs of mice to be compared with wildtype mice entering the same HLS protocol. While we have separately shown short-term, concurrent exercise training prior to HLS diminishes atrophy in wild-type mice (35), here we further aim to observe if exercising TFAM overexpression transgenic mice prior to HLS results in a combination treatment effect to prevent muscle atrophy. Our results indicate overexpressing TFAM results in significantly diminished soleus and gastrocnemius atrophy prompting potential future research in genetic and molecularly directed mitochondrial therapies.
Methods
Animals and Ethical Approval
Male C57BL/6J mice (WT) and male C57-based transgenic mice overexpressing the TFAM gene (Cyagen Bioscience, Inc.) were used for analysis. Only male mice were used in the present study as the response to exercise between genders can be drastically different, producing varying results across a treatment group. TFAM-positive mice were purchased from Cyagen Biosciences, Inc., who created the mouse line using a plasmid-based transgenic vector. Genotyping confirmed the positive transgene in each mouse used in the present study. The TFAM transgenic groups consisted of a control (T-Control), hindlimb suspension (T-HLS), and exercise training prior to hindlimb suspension (T-Ex+HLS). Four groups of wild-type mice (WT) were used as comparison to transgenic groups including a Control, hindlimb suspension (HLS), exercise before hindlimb suspension (Ex+HLS), and exercise only (Ex) group. All data from WT mice are derived from our recent work (35). WT data will be used solely as comparative data for the objectives of this study. Mice that exercised prior to suspension were placed in HLS the same day after their last exercise session and all mice were sacrificed immediately after HLS. All mice were 13±1 weeks of age at the time of sacrifice. Wild-type mice were purchased from Jackson Laboratory (Bar Harbor, ME) and transgenic mice were purchased from Cyagen Biosciences, Inc. (Santa Clara, CA). All standard procedures and experiments involving animals were approved by the Institutional Animal Care and Use Committee of the University of Louisville and conformed with the National Institutes of Health guidelines.
Hindlimb Suspension
Custom built cages were used to unload the hindlimb musculature and induce atrophy for 7 continuous days. Cage construction followed similar guidelines as previously shown (10). Briefly, mice were first placed under continuous isoflurane anesthesia while a tail harness was attached. The tail was cleaned then taped cross-sectionally. A 27-gauge needle cap was cut down to ~2 centimeters in length with openings on each end. A small hole was created in the sidewall of the needle cap and a nylon string was used to tie a loop through this hole. This cap was placed over the tail and taped into place roughly 1/3 of the tail length from the base. The nylon loop and cap, now taped to the tail, could then be attached to the roof of the cage, suspending the animal’s hindlimbs. This also allowed the forelimbs to bear weight and the animal to move around the cage. Mice could also access food and water ad libitum in this manner. Bodyweights were recorded before and after suspension.
Exercise Protocol
Exercise consisted of 14 sessions over 18 days of treadmill running in a concurrent exercise program (i.e. combining different exercise styles into the same program) used in our previous work (35). Mice were acclimated to the treadmill during the first four sessions at low intensity (week 1). The subsequent 10 sessions (weeks 2 and 3) consisted of 6 endurance training style exercise sessions and 4 high-intensity interval style sprint exercise sessions. Sprint exercise consisted of alternating between high-intensity sprints and low-intensity walking speeds throughout a single session. Complete rest occurred every 100m during endurance training and after every 5 sprints during high-intensity training. All exercise program details (Table 1) were matched between exercising groups.
Table 1.
Exercise Protocol.
| WEEK 1 | Monday | Tuesday | Wednesday | Thursday | Friday |
|---|---|---|---|---|---|
| Warmup | 7m/min. for 50m | 7m/min. for 50m | 7m/min. for 50m | 7m/min. for 50m | |
| Work Rate | OFF | 7m/min. | 8m/min. | 9m/min | 10m/min. |
| Rest | 3min. off/100m | 3min. off/100m | 3min. off/100m | 3min. off/100m | |
| Distance | 300m | 300m | 300m | 300m | |
| WEEK 2 | Monday | Tuesday-Sprints | Wednesday | Thursday-Sprints | Friday |
| Warmup | 7m/min. for 50m | 7m/min. for 50m | 7m/min. for 50m | 7m/min. for 50m | 7m/min. for 50m |
| Work Rate | 11m/min. | 15m/min. for 10m | 12m/min. | 17m/min for 15m | 13m/min. |
| Walk Rate | - | 7m/min. for 10m | - | 7m/min. for 15m | - |
| # of Sprints | - | 8 | - | 10 | - |
| Rest | 3min. off/100m | 5mins after 5 sprints | 3min. off/100m | 5mins after 5 sprints | 3min. off/100m |
| Distance | 350m | 160m | 375m | 300m | 400m |
| WEEK 3 | Monday | Tuesday-Sprints | Wednesday | Thursday-Sprints | Friday |
| Warmup | 7m/min. for 50m | 7m/min. for 50m | 7m/min. for 50m | 7m/min. for 50m | 7m/min. for 50m |
| Work Rate | 13.5m/min. | 18m/min. for 15m | 14m/min. | 20m/min. for 20m | 14.5m/min. |
| Walk Rate | - | 7m/min. for 15m | - | 5x-7m/20m; 6x-7m/10m | - |
| # of Sprints | - | 11 | - | 11 | - |
| Rest | 3min. off/100m | 5mins after 5 sprints | 3min. off/100m | 5mins after 5 sprints | 3min. off/100m |
| Distance | 450m | 330m | 500m | 380m | 550m |
Table 1. Mouse concurrent treadmill exercise protocol consisting of a “Week 1” acclimation, followed by progressive training “Week 2” and “Week 3”. Three days per week (M,W,F) consisted of endurance-style exercise while two days per week (T,TH) consisted of higher-intensity interval sprint style of exercise.
Laser Doppler Imaging
Laser Doppler imaging (Moor FLPI, Wilmington, DE) of lower, hindlimb blood flow was done for each group and used to assess flux of the hindlimb after treatments. Mice were first administered anesthesia (tribromoethanol) standardized to bodyweight (mg/g). The musculature of the left, lower hindlimb was then exposed to reveal the skeletal muscle. Each mouse was placed in a prone position and the laser height was positioned roughly 15 cm from the posterior tibial vessel, a prominent vessel superficial to the gastrocnemius. Flux measurements were recorded for 2 mins and quantified.
Tissue Extraction and Muscle Weight
Both the soleus and gastrocnemius tissues were excised and separated from each leg for all experimental groups. Each muscle was washed with 50 mmol/L phosphate buffer saline (PBS) pH of 7.4, weighed individually, snap frozen in liquid nitrogen, and stored at −80°C until use. All tissue extraction methods and timings were equated across all groups.
Immunofluorescence
One of each muscle was cut cross-sectionally at the muscle midbelly and placed in tissue-freezing medium (Triangle Biomedical Sciences, Durham, NC, USA) in disposable plastic tissue-embedding molds (Polysciences Inc., Warrington, PA, USA). These tissue blocks were immediately frozen and stored at −80°C until use. Cryosections of each muscle, 7–10 µm thick, were obtained using a Cryocut System (CM 1850; Leica Microsystems, Buffalo Grove, IL, USA). These tissue sections were placed on Poly-L-Lysine coated microscope slides (Polysciences, Inc., Warrington, PA, USA) prior to use. Slides containing tissue sections were fixated in acetone for 20 mins, washed in 1X PBS-T solution, and incubated with a permeabilization solution (0.2 g Bovine Serum Albumin, 3 ul Triton X-100 in 10ml 1X PBS) for 1 hour at RT. This was followed by multiple washing steps with 1X PBS-T. The sections were incubated with a primary antibody (anti-Laminin; Abcam, Cambridge, MA, USA) solution with a dilution of 1:200 overnight at 4°C. After multiple washing steps with 1X PBS-T, the slides were incubated with a fluorescently labeled secondary antibody (goat anti-mouse Alexa flour 594; Invitrogen, Waltham, MA, USA) with a 1:300 dilution for 1 hour at RT. This was followed by another washing step, then slides were mounted with mounting medium and glass cover slips placed over the sections and air dried overnight in dark container. These samples could then be visualized using a laser scanning confocal microscope (Olympus Fluo View 1000; Center Valley, PA, USA). Cross-sectional area (CSA) measurements were acquired using these laminin images by hand tracing single, stained muscle fibers in ImageJ software.
Dihydroethidium Staining for Oxidative Stress
After tissue excision, each muscle was cross sectioned in half, immediately placed in tissuefreezing medium (Triangle Biomedical Sciences, Durham, NC, USA), snap frozen in liquid nitrogen and stored at −80°C until use. Samples were sectioned on a cryostat, placed on slides, and in situ superoxide generation was evaluated in each cryosection with the oxidative fluorescent dye dihydroethidium (DHE). Cryosections (10 µm) were incubated with DHE (2 µmol/l) in a PBS solution. After multiple washing steps, slides were mounted with mounting medium and a cover slip and allowed to dry. These samples were then visualized using a laser scanning confocal microscope (Olympus Fluo View 1000; Center Valley, PA, USA).
GSH/GSSG Ratio for Redox Status
Reduced glutathione (GSH) and oxidized glutathione (GSSG) was measured to assess oxidate stress and redox status of each group. A GSH/GSSG Ratio Detection Assay Kit (Abcam, ab138881) was used as previously described (34) and per the kit instructions. Briefly, protein lysates were first deproteinized by using trichloroacetic acid, incubated on ice (15 mins) and centrifuged (5 mins). All supernatant was removed and neutralized using the stock neutralizing solution (Abcam). The assay was performed against reduced glutathione (GSH) and oxidized glutathione (GSSG) standards. Samples were incubated and monitored between 10–60 mins and analyzed on 96-well plates at 490/520 nm wavelengths in a Spectra Max M2 plate reader (Molecular Devices Corporation, Sunnyvale, CA). All samples were loaded in duplicate.
Protein Extraction and Protein Estimation
Both muscle tissue samples from all groups were bead-homogenized in ice‐cold RIPA (1 mmol/L) buffer with PMSF and protease inhibitor cocktails (1 µL/mL of lysis buffer, Sigma Aldrich, St. Louis, MO). Centrifugation followed with these samples at 12,000 g for 20 min at 4°C. All supernatant was extracted, placed in Eppendorf tubes, and immediately stored at −80°C until use. Protein estimation of each sample was measured by the Bradford‐dye (Bio‐Rad, CA) method in a 96‐well microliter plate compared against a Bovine Serum Albumin (BSA) standards. This plate was analyzed at 594 nm in a Spectra Max M2 plate reader (Molecular Devices Corporation, Sunnyvale, CA).
Western Blot Analysis
Protein lysates (40 µg) were prepared and heated at 95°C for 5 min. These samples were then loaded into an SDS polyacrylamide gel submerged in running buffer and run at a constant current (75 Volts). Proteins were then transferred to a PVDF membrane overnight at 120 mA at 4°C. After the transfer, membranes were blocked in 5% nonfat milk for 1 hour at room temperature and then incubated overnight with the primary antibody (Anti-SOD2, Anti-HSP-60, Anti-MTCO1; Anti-GAPDH; Abcam, Cambridge, MA, USA.) at 4°C with slow agitation. Washing steps occurred the next day with a TBS-T buffer, then membranes were incubated with secondary antibody (horse radish peroxidase-conjugated goat anti-rabbit IgG, goat anti-mouse; Santa Cruz Biotechnology, Dallas, TX, USA) for 1 hour at RT with a 1:5000 dilution followed by another washing step. The membranes were developed using an ECL Western blotting detection system (GE Healthcare, Piscataway, NJ, USA) and all images were recorded in the gel documentation system ChemiDoc XRS (Bio-Rad, Richmond, CA, USA). Membranes, if being re-used, were stripped with stripping buffer (Boston BioProducts, Ashland, MA, USA) and blocked with 5% milk for 1 hour at RT under agitation. After washing, membranes were incubated with the anti-Gapdh antibody (Millipore, Billerica, MA, USA) as a loading control protein. The data were analyzed by Bio-Rad Image Lab densitometry software and normalized against anti-Gapdh bands.
Quantitative PCR
In the assessment of mRNA transcripts of specific genes in skeletal muscle, RNA was isolated with TRIzol® reagent (Life Technologies, Carlsbad, CA, USA) according to the instructions of the manufacturer. The RNA quantification and purity was assessed by nanodrop-1000 (Thermo Scientific, Walthan, MA, USA). Aliquots (2µg) of total RNA were reverse-transcribed into cDNA using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA) according to the protocol of the manufacturer. Quantitative PCR (q-PCR) was performed for different genes (TFAM, Myh4, SOD-1, SOD-2), in a final reaction volume of 20 µl containing 10 µl PerfeCTa SYBR Green SuperMix, Low ROX (Quanta Biosciences, Gaithersburg, MD), 6 µl nuclease free water, 2µl cDNA, 40 picomoles of forward, and reverse primers. Table 1 presents all sequence-specific oligonucleotide primers (Invitrogen, Carlsbad, CA, USA). The data was represented in fold expression. This was calculated as the cycle threshold difference between control and sample normalized with the housekeeping genes beta actin and 18s.
Statistical Analysis
All data were expressed as means ± SE. One-way ANOVA analyses were conducted on each data set with Tukey’s post-hoc analysis used between groups within the same mouse line and the student t-test used between mouse line groups of relevance. Significance was determined as a p-value<.05.
Results
TFAM overexpression reduces atrophic muscle changes
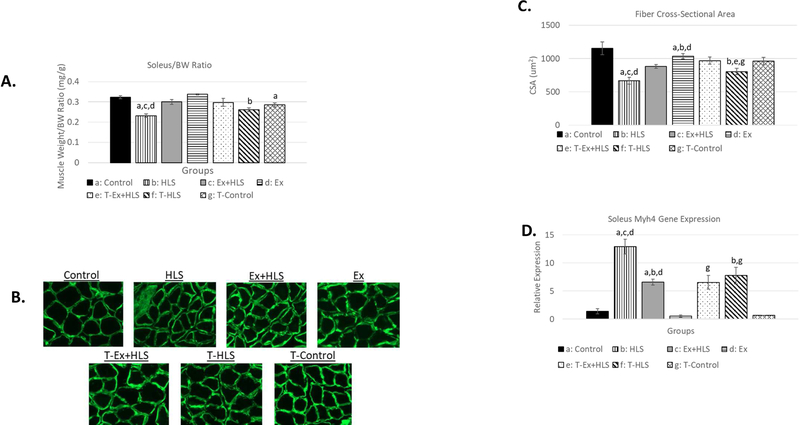
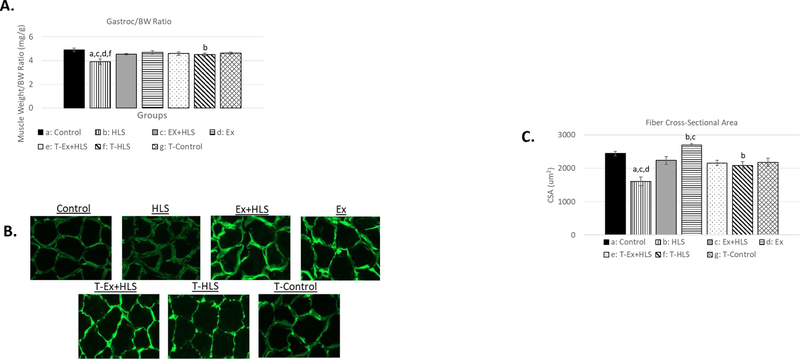
Wet muscle weights were recorded immediately after tissue excision. A muscle weight to bodyweight (mg/g) standardization ratio was used to account for individual size variation. HLS resulted in non-significant decreases of 8.3% in soleus (Fig. 1) and 2.6% in gastrocnemius (Fig. 2) muscle weight to bodyweight ratio in mice overexpressing TFAM (T-HLS), compared to TFAM Control, while HLS in wild-type mice revealed a 27.1% significant decrease in the soleus and a 21.5% significant decrease in the gastrocnemius in this ratio compared to wild-type Control (p<.05). TFAM Ex+HLS led to a non-significant increase of 4.5% in the soleus and decrease of 0.7% in this ratio in T-HLS, compared to TFAM Control (Fig. 1 and 2). Cross-sectional area measurements followed a similar trend with no significant difference between THLS and T-Control groups. Furthermore, T-HLS CSA was significantly greater (Fig. 3 and 4) than wild-type HLS CSA in both soleus [805um vs. 668um, p<.05] and gastrocnemius [2085um vs. 1605um, p<.01].
Figure 1.
A. Soleus muscle weight to bodyweight ratio (mg/g) of wild-type and TFAM transgenic groups (n=6). B. Immunofluorescence images of anti-laminin staining to visualize muscle fiber size and shape. All pictures are cropped to 200µm in width. C. Mean cross-sectional area measurement comparison between groups of anti-laminin staining images calculated using ImageJ software (n=6). D. Myosin Heavy chain 4 (Myh4) gene expression analysis of the soleus muscle (n=6). Statistical significance (p<.05) indicated by the corresponding group letter (means ± SE).
Figure 2.
A. Gastrocnemius muscle weight to bodyweight ratio (mg/g) of wild-type and TFAM transgenic groups (n=6). B. Immunofluorescence images of laminin staining to visualize muscle fiber size and shape. All pictures are cropped to 200µm in width. C. Mean cross-sectional area measurement comparison between groups of anti-laminin staining images calculated using ImageJ software (n=6).
Figure 3.
A. Soleus TFAM gene expression (n=6). B. Gastrocnemius TFAM gene expression. Statistical significance (p<.05) indicated by the corresponding group letter (means ± SE).
Figure 4.
A. Laser Doppler blood flow measurement of a prominent vessel in the dorsal hindlimb. B. Blood flux measurement comparison between groups (n=6).
Moreover, myosin heavy chain 4 (Myh4) was measured via qPCR to assess fiber-type transitioning of the soleus muscle. Here, similar transcript levels of Myh4 were observed in both wild-type Ex+HLS groups compared to T-Ex+HLS while T-HLS resulted in significantly lower levels of Myh4 compared to wild-type HLS (12.89 vs. 7.75, p<.01).
Greater TFAM expression in the gastrocnemius
To assess genetic expression of the TFAM transgenic model, we completed qPCR analysis of the TFAM transcript across both muscles (Fig. 3). Soleus TFAM expression in T-HLS mice was greater than wild-type HLS (1.75 vs. .82, p<.01) while T-Control was greater than wild-type Control (2.35 vs. 1.02, p<.01). In the gastrocnemius, T-HLS was significantly greater than wildtype HLS (4.23 vs. .49, p<.01) and T-Control was ~5-fold greater than wild-type Control (5.11 vs. 1.09, p<.01).
Hindlimb blood flow does not change
Laser Doppler imaging of the left hindlimb assessed blood flow through a prominent posterior limb artery superior to the gastrocnemius (Fig. 4). Flux is a function of red blood cell (RBC) content and its velocity through the vessel of interest. Flux between groups was not a factor, as there were no significant differences between all groups (p=.53).
TFAM overexpression improves redox status
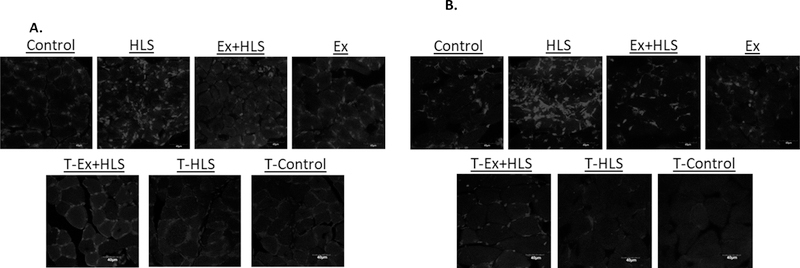
To observe ROS accumulation between groups, a DHE staining protocol was used. DHE staining binds ROS and has been noted to easily permeate the cell membrane and bind the superoxide anion with specificity (27). Here we observe clear differences between TFAM transgenic and wild-type groups. Specifically, T-HLS generally had low positive markers after staining while the wild-type HLS group typically had high levels of positive markers (Figure 5). T-Ex+HLS appeared to have similar positive markers compared to wild-type Ex+HLS.
Figure 5.
A. Observational soleus dihydroethidium staining for ROS. Figures are black and white. Bright spots are assumed to be positive staining for ROS (n=6, scale=40µm). B. Observational gastrocnemius dihydroethidium staining for ROS. Figures are black and white. Bright spots are assumed to be positive staining for superoxide anion (n=6, scale=40µm).
While DHE staining of tissues was purely observational, we followed up this assessment with a quantifiable measurement of oxidative stress using a GSH/GSSG ratio assay in the gastrocnemius. Total GSH and GSH/GSSG ratio was significantly greater in T-HLS compared to wild-type HLS (p<.05) in the gastrocnemius. Total GSH was also significantly greater in T-Control compared to wild-type Control (p<.01), however GSH/GSSG ratio between these groups was not significantly different (Fig. 8).
Figure 8.
A. Gastrocnemius oxidative stress measurement of Total GSH and GSH/GSSG ratio (n=6). B. Gastrocnemius protein expression analysis via Western blot for mitochondrial complex 1 (MTCO1) and heat shock protein 60 (HSP60).
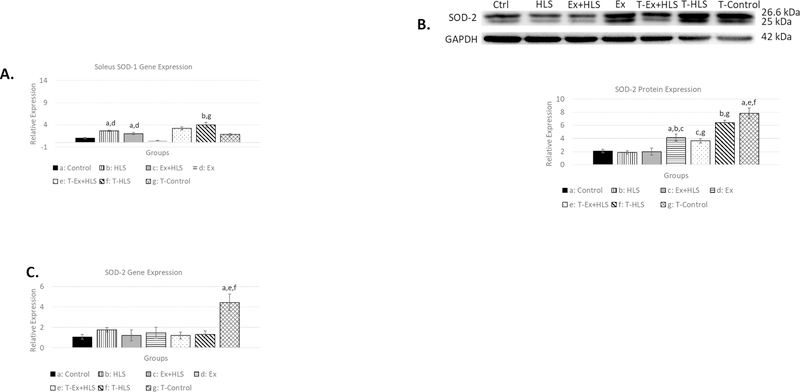
TFAM overexpression increases SOD-1 and SOD-2
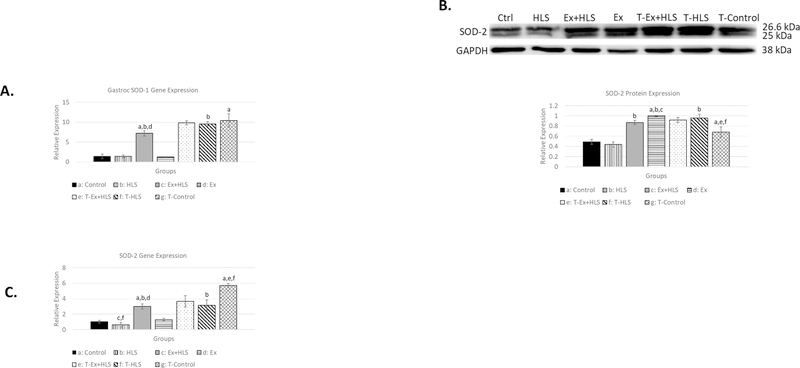
Skeletal muscle degradation and apoptotic pathways are induced by reactive oxygen species (7, 25). Superoxide dismutases (SODs) are antioxidant enzymes that catalyze the conversion of superoxide anions to hydrogen peroxide (12). Therefore, we measured SOD-1 (cytosolic) and SOD-2 (mitochondrial) transcripts via qPCR and SOD-2 protein expression via Western blot to assess inter-group antioxidant differences. SOD-1 mRNA analysis in the soleus (Fig. 6) revealed significantly greater expression of this gene in T-HLS mice compared to wild-type HLS (P<.05). In the gastrocnemius (Fig. 7), T-Control and T-HLS SOD-1 transcripts were both roughly 10fold greater than wild-type Control and HLS (p<.01).
Figure 6.
A. Soleus SOD-1 transcript expression (n=6). B. Soleus protein expression via Western blot analysis for antioxidant enzyme SOD-2 (n=6). C. Soleus SOD-2 gene transcript expression. Statistical significance (p<.05) indicated by the corresponding group letter (means ± SE).
Figure 7.
A. Gastrocnemius SOD-1 transcript expression (n=6). B. Gastrocnemius protein expression via Western blot analysis for antioxidant enzyme SOD-2 (n=6). C. Gastrocnemius SOD-2 gene transcript expression (n=6). Statistical significance (p<.05) indicated by the corresponding group letter (means ± SE).
Transcript analysis of SOD-2 in the soleus (Fig. 6) reveals T-Control being ~4.5x greater than wild-type Control (p<.01). SOD-2 soleus protein expression was greater in T-HLS compared to wild-type HLS (p<.01) as well as T-Control compared to wild-type Control (p<.01). We observed gastrocnemius SOD-2 gene expression (Fig. 7) resulting in the T-Control group having a nearly 5.5-fold increase compared to Control (p<.01) and T-HLS nearly 6-fold greater than HLS (p<.01). Analysis of protein expression of this antioxidant in the gastrocnemius (Fig. 7) similarly revealed significant increases in both T-HLS and T-Control compared to HLS and Control, respectively (p<.05).
TFAM overexpression increases downstream gastrocnemius mitochondrial markers
To further assess mitochondrial changes in the gastrocnemius, we first evaluated protein expression of HSP60. Here we see HSP60 expression is significantly greater in T-HLS compared to HLS (p<.05) while T-Control was roughly 2-fold greater than wild-type Control (p<.01). THLS was also significantly greater than wild-type Control (p<.05). MTCO1 protein expression was measured next in the gastrocnemius to further evaluate mitochondrial changes (Fig. 8). T-Control was significantly greater than wild-type Control (p<.01) while T-HLS was significantly greater than HLS (p<.05).
Discussion
Atrophy of skeletal muscle arises when protein degradation and apoptosis exceed protein synthesis and can occur when the musculature is unloaded or disused (36). Previously, we have shown evidence of atrophy in both the soleus and gastrocnemius skeletal muscle of wild-type mice following a hindlimb-unloading protocol (35). This skeletal muscle atrophy is associated with diminished markers of mitochondrial biogenesis and function, a reduced antioxidant state, and increased oxidative stress leading to redox imbalance. Feng et al. also showed reduced PGC-1α content after mouse hindlimb unloading, noting mitochondrial function may have a causal role in disuse-atrophy (9) while Lee et al. similarly note HLS in mice reduced mitochondrial signaling markers (22). In our work, we successfully reduced atrophy by exercising the musculature prior to hindlimb suspension (Table 3), which was associated with improvements in many of these mitochondrial and redox markers. Specifically, we observed diminished TFAM expression after HLS. Therefore, in the present study we attempt to build upon our previous work to more specifically analyze the role of TFAM in muscle atrophy using a C57BL/6J-based TFAM overexpression transgenic mouse model in HLS, while also observing the effects of combining exercise with this transgenic model.
Table 3.
Bodyweight and Muscle Weight
| Group | Pre-HLS BW (g) | BW (g) | Sol MW (mg) | Gas MW (mg) | Sol MW/BW (mg/g) | Gas MW/BW (mg/g) |
|---|---|---|---|---|---|---|
| Control | - | 24.5 ± .55 | 7.8 ± .17 | 121.1 ± 6.10 | .32 ± .01 | 4.9 ± .16 |
| HLS | 23.3 ± .87 | 22.0 ± .79 | 5.1 ± .15a,c,d | 85.4 ± 5.5a,d | .23 ± .01a,c,f | 3.9 ± .24a,c,f |
| Ex+HLS | 24.3 ± .78 | 22.4 ± .67 | 6.9 ± .33a,b,d | 103.8 ± 4.31 | .30 ± .01d | 4.5 ± .07 |
| Ex | - | 23.5 ± .22 | 7.9 ± .12 | 110.7 ± 4.30 | .34 ± .003 | 4.7 ± .15 |
| T-Ex+HLS | 23.8 ± .47 | 22.9 ± .55 | 6.8 ± .44 | 105.2 ± 1.77 | .30 ± .03 | 4.59 ± .14 |
| T-HLS | 26.4 ± .63 | 24.6 ± .59 | 6.3 ± .27b,g | 105.9 ± 2.60b | .261 ± .01b | 4.51 ± .12b |
| T-Control | - | 26.9 ± .27 | 7.7 ± .32f | 124.5 ± 3.42 | .29 ± .01a | 4.63 ± .09 |
Table 3. Bodyweight (BW) pre-HLS (g) and BW at time of sacrifice. Muscle weight of both the soleus and gastrocnemius. All data presented as means ± SE. Statistical significance (p<.05) between groups indicated by the superscript letter associated with each particular group
Control
HLS
Ex+HLS
Ex
Previous research using TFAM transgenic animal models is scarce. A 2005 study reported the use of a TFAM transgenic mouse model in a myocardial infarction study and results indicate TFAM overexpression ameliorated left-ventricular remodeling, providing a cardiomyocyte protective effect (14). Other in vitro studies similarly reveal a cardiomyocyte protective role via TFAM overexpression (11, 13). Moreover, mitochondrial dysfunction in skeletal muscle atrophy is highly associated with excessive ROS and oxidative stress. With TFAM protecting mtDNA from ROS as well as increasing mtDNA transcription, mtDNA copy number, and mitochondrial biogenesis, we chose to further investigate its role in skeletal muscle.
After HLS in transgenic mice we observe muscle wet weight, muscle weight/bodyweight ratio, and cross-sectional area are all significantly greater in both the soleus and gastrocnemius of the T-HLS group compared to the wild-type HLS group, indicating an initial, potential protective role of TFAM overexpression (Fig. 9, Table 3). TFAM abundantly binds and protects mtDNA from mitochondrial ROS and is correlated with mitochondrial function(19). As greater TFAM comes in contact with mtDNA, greater protection is expected (19). TFAM expression is also positively correlated with mtDNA copy number (8). Therefore, results from the current study suggest supra-physiologic gene expression of TFAM may lead to, speculatively, increased mtDNA protection during an atrophic setting with increased ROS, preserving muscle weight and cross-sectional area.
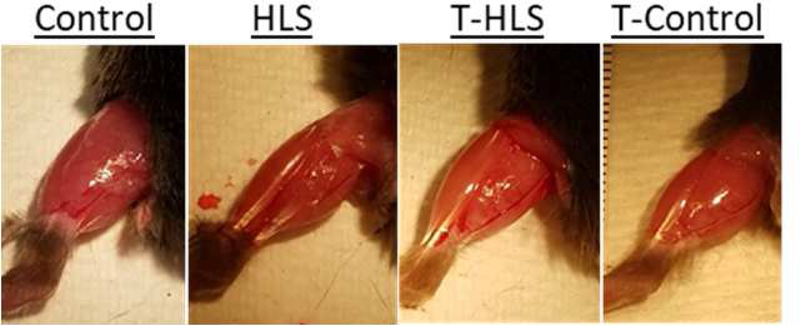
Figure 9.
Dorsal view of the exposed lower left hindlimb with mice, under anesthesia, placed in a prone position. Observational differences between groups.
In the present study, the transgenic gastrocnemius muscle seemed to have a more pronounced protective effect from atrophy with minimal reductions in muscle weight, size, and shape compared to the greater changes observed in the transgenic soleus. Our recent research indicates mitochondrial biogenesis markers, particularly TFAM gene and protein expression, in the wildtype gastrocnemius is more affected by 7 days of HLS (35). Yajid et al. also report muscle group mitochondrial differences where they conclude mitochondrial respiration is most affected in the gastrocnemius after HLS in rats. These researchers analyzed mitochondrial respiration in 4 separate muscles, including the soleus, and noted a 59% decrease in gastrocnemius state 3 respiration with disuse atrophy (41). The implication is muscle groups with very different fibertype makeups may respond to atrophy differently, and thus should be considered separately for analysis as well as therapeutic generation.
Nonetheless, while it appears the gastrocnemius may be more susceptible to mitochondrial changes during disuse and potentially able to respond better to mitochondrial treatment via the overexpression of the TFAM gene, we must also emphasize muscle fiber-type differences between muscle groups. In the analysis of C57BL/6J mouse muscle fiber-type composition, the soleus muscle reportedly consisted of much higher oxidative fibers with 37% type-I fibers compared to ~6% type-I fibers in the gastrocnemius. The soleus also consisted of ~38% type-IIA fibers while the gastrocnemius again consisted of much lower oxidative fibers with only ~6% type-IIA but over 54% type-IIB fibers (1). Oxidative fibers tend to have greater mitochondrial content and density while fast-glycolytic fibers tend to have less mitochondrial content (23). In speculation, due to the lower volume of mitochondria in the gastrocnemius, there may be a greater sensitivity to even minor mitochondrial changes, such as the decreases in TFAM we observed in HLS. By overexpressing this highly important gene in mitochondrial health and function that directly protects mtDNA copy number and activates mtDNA transcription, it may blunt the gastrocnemius sensitivity to mitochondrial fluctuations. While the gastrocnemius in wild-type mice revealed a significant decline in TFAM gene and protein expression after HLS, there was no significant difference in soleus TFAM gene and protein expression in wild-type mice after HLS. The soleus, with greater mitochondrial densities and no significant decreases in TFAM, did not benefit from increased TFAM transcript expression to the same extent while the gastrocnemius may have had the physiologic capacity for greater benefit from overexpression. We also observed differences in gene expression levels between muscles in the transgenic mice. To investigate this further, measurements of TFAM mRNA expression in transgenic mice against wild-type mice revealed a nearly 2.5-fold increase in transgenic soleus TFAM mRNA compared to ~5-fold increase in transgenic gastrocnemius TFAM mRNA (Fig. 3). This difference was similar across all transgenic treatment groups indicating a potential difference in fiber-type response to the random integration of the TFAM transgenic construct. Interestingly, in a study using a PGC-1α overexpression mouse model, the authors note the potential for certain promoters to be more highly expressed in fast-fibers compared to slow-fibers (3). Although purely speculation, this may play a role in the current study to account for expression differences between muscle types.
We previously observed increased superoxide anion after HLS in wild-type mice, which was similar to DHE staining results in skeletal muscle after 7-day HLS by Cannavino et al. (3). In the present study, we observed much lower expression of ROS across both muscles of all transgenic groups compared to wild-type. We also observed greater glutathione and GSH/GSSG ratios in the TFAM groups of the gastrocnemius (Fig. 8). These results combined indicate a connection between TFAM and redox balance with atrophy. Furthermore, there was an increase in SOD-1 gene expression and SOD-2 protein expression of both muscles, with the transgenic gastrocnemius revealing more pronounced levels of gene expression of both antioxidants compared to the transgenic soleus, possibly because of the greater TFAM expression in the gastrocnemius. The SOD findings are unexpected and links TFAM gene overexpression with increased antioxidant status. Interestingly, a 2009 study isolating mtDNA nucleoids revealed the presence of SOD-2 within each nucleoid, potentially acting as a front-line antioxidant defense barrier for mtDNA (18). Speculatively, it is possible overexpressing TFAM results in increased mtDNA binding and greater nucleoid formation, leading to a higher presence of SOD-2 expression and antioxidant defense overall. The exact mechanism between these two molecules appears to be unknown, to our knowledge, and worthy of future investigation.
With catabolic mechanisms, particularly degradation and apoptotic mechanisms, being activated by ROS and increased oxidative stress, a superior antioxidant status and redox balance within the TFAM overexpressed skeletal muscle may have prevented the physical reduction of the tissue resulting in diminished atrophy. While markers of degradation (Murf-1 and Atrogin-1) were measured in the current study, no reportable results were confirmed due to high variation. Future research should also elucidate not only the degradation pathway, but also protein synthesis markers (Akt/mTOR pathway) in this transgenic mouse model.
Moreover, investigation into mitochondrial markers in the gastrocnemius occurred due to the more robust response to gene overexpression. Our analysis of TFAM protein expression was inconclusive in the transgenic mice due to, potentially, the protein being GFP-tagged resulting in poor antibody adhesion. However, genotyping and increased TFAM mRNA expression indicated a viable transgenic mouse line. Increased TFAM mRNA transcripts would have to be chaperoned and imported into the mitochondria after protein translation for TFAM to be able to exert its effect. Indeed, here we observed HSP60, a mitochondrial chaperone and import protein of TFAM, to be significantly increased in TFAM transgenic mice (Fig. 8). Barone et al. found increased mtDNA copy numbers in mouse skeletal muscle after an HSP60 plasmid transfection while also finding increases in HSP60 after endurance exercise training (2). This increase in copy number may have been due to the increased ability to import TFAM into the mitochondria where it could interact with mtDNA and has also been shown to be correlated with the number of mtDNA copies (8). Furthermore, mtDNA specifically encodes for mitochondrial complex I subunits, the largest and most complicated structure of the electron transport chain (21). TFAM not only protects mtDNA from ROS, as discussed above, but also activates the transcription of mtDNA proteins. In the present study we not only found increases in HSP60 but also increases in mitochondrial complex I protein expression in TFAM transgenic mice, indicating an enhanced mitochondrial effect from TFAM overexpression. With these proteins being molecularly associated with mitochondrial function, we see a mechanism in which TFAM overexpression could diminish skeletal muscle atrophy.
Conclusion
Skeletal muscle atrophy is highly correlated with reduced mitochondrial biogenesis and function along with excessive ROS being previously shown to lead to the activation of protein degradation and apoptotic pathways. Here we follow up on our recent research that correlated diminished mitochondrial markers with skeletal muscle atrophy and attempt to display a protective role of TFAM in disuse-induced skeletal muscle atrophy. Our results indicate the overexpression of the TFAM gene is associated with increased antioxidant expression, redox balance, as well as increased TFAM-specific mitochondrial associated markers to potentially prevent mitochondrial dysfunction as a possible mechanism to prevent atrophy in skeletal muscle disuse. Mitochondrial and TFAM-directed pharmaceuticals or therapies to manipulate this molecule is an attractive area of research in preventing skeletal muscle atrophy. Future research into this molecule could also focus on other tissues and pathologic areas in which decreased mitochondrial biogenesis and increased dysfunction plays a major role.
Table 2.
Nucleotide Sequences.
| Gene | Forward Nucleotide Sequence | Reverse Nucleotide Sequence |
|---|---|---|
| Myh4 | 5’- TTGTGGTGGATGCTAAGGAGTC | 5’- GTACTTGGGAGGGTTCATGGAG |
| SOD-1 | 5’- GAACCAGTTGTGTTGTCAGGAC | 5’- GCCTTGTGTATTGTCCCCATAC |
| SOD-2 | 5’- GGTCACAGTTTCACAGTACACC | 5’- TCACAGCCTTGAGTTACAGAGT |
| TFAM | 5’ - GAGAGCTACACTGGGAAACCACA | 5’ - CATCAAGGACATCTGAGGAAAA |
| Rn18S | 5’- CACGGACAGGATTGACAGATTG | 5’- GACAAATCGCTCCACCAACTAA |
| ß-actin | 5’- CCCTGAAGTACCCCATTGAACA | 5’- CACACGCAGCTCATTGTAGAAG |
Table 2. Nucleotide sequences of all examined genes.
New and Noteworthy.
To the best of our knowledge, this is the first time a TFAM overexpression transgenic mouse model is being used in the analysis of disuse-induced skeletal muscle atrophy. Here we provide evidence of a potential role for TFAM in diminishing skeletal muscle atrophy.
Acknowledgements
Grants
The study was supported by NIH Grant HL74185 to SCT.
Footnotes
This manuscript is part of a dissertation thesis in the Department of Physiology, University of Louisville, KY, USA, and portions of this manuscript were presented at the Experimental Biology Conference- Chicago, IL, 2017 as well as at the Orthopaedic Research Society Musculoskeletal Workshop- Sun Valley, ID, 2017.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
No conflicts of interest, financial or otherwise, are declared by the authors.
References
- 1.Augusto VP, Carlos Roberto ; Campos, Gerson Eduardo Rocha. Skeletal muscle types in C57BL6J mice. Braz j morphol sci 21: 89–94, 2004. [Google Scholar]
- 2.Barone R, Macaluso F, Sangiorgi C, Campanella C, Marino Gammazza A, Moresi V, Coletti D, Conway de Macario E, Macario AJ, Cappello F, Adamo S, Farina F, Zummo G, and Di Felice V. Skeletal muscle Heat shock protein 60 increases after endurance training and induces peroxisome proliferator-activated receptor gamma coactivator 1 alpha1 expression. Sci Rep 6: 19781, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cannavino J, Brocca L, Sandri M, Bottinelli R, and Pellegrino MA. PGC1-alpha over-expression prevents metabolic alterations and soleus muscle atrophy in hindlimb unloaded mice. The Journal of physiology 592: 4575–4589, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chopard A, Hillock S, and Jasmin BJ. Molecular events and signalling pathways involved in skeletal muscle disuse-induced atrophy and the impact of countermeasures. Journal of cellular and molecular medicine 13: 3032–3050, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coleman SK, Rebalka IA, D’Souza DM, and Hawke TJ. Skeletal muscle as a therapeutic target for delaying type 1 diabetic complications. World journal of diabetes 6: 1323–1336, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collu-Marchese M, Shuen M, Pauly M, Saleem A, and Hood DA. The regulation of mitochondrial transcription factor A (Tfam) expression during skeletal muscle cell differentiation. Bioscience reports 35: 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dam AD, Mitchell AS, Rush JW, and Quadrilatero J. Elevated skeletal muscle apoptotic signaling following glutathione depletion. Apoptosis : an international journal on programmed cell death 17: 4860, 2012. [DOI] [PubMed] [Google Scholar]
- 8.Ekstrand MI, Falkenberg M, Rantanen A, Park CB, Gaspari M, Hultenby K, Rustin P, Gustafsson CM, and Larsson NG. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Human molecular genetics 13: 935–944, 2004. [DOI] [PubMed] [Google Scholar]
- 9.Feng HZ, Chen X, Malek MH, and Jin JP. Slow recovery of the impaired fatigue resistance in postunloading mouse soleus muscle corresponding to decreased mitochondrial function and a compensatory increase in type I slow fibers. American journal of physiology Cell physiology 310: C27–40, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferreira JA, Crissey JM, and Brown M. An Alternant Method to the Traditional NASA Hindlimb Unloading Model in Mice e2467, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujino T, Ide T, Yoshida M, Onitsuka K, Tanaka A, Hata Y, Nishida M, Takehara T, Kanemaru T, Kitajima N, Takazaki S, Kurose H, Kang D, and Sunagawa K. Recombinant mitochondrial transcription factor A protein inhibits nuclear factor of activated T cells signaling and attenuates pathological hypertrophy of cardiac myocytes. Mitochondrion 12: 449–458, 2012. [DOI] [PubMed] [Google Scholar]
- 12.Fukai T, and Ushio-Fukai M. Superoxide dismutases: role in redox signaling, vascular function, and diseases. Antioxidants & redox signaling 15: 1583–1606, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ikeda M, Ide T, Fujino T, Arai S, Saku K, Kakino T, Tyynismaa H, Yamasaki T, Yamada K, Kang D, Suomalainen A, and Sunagawa K. Overexpression of TFAM or twinkle increases mtDNA copy number and facilitates cardioprotection associated with limited mitochondrial oxidative stress. PLoS One 10: e0119687, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ikeuchi M, Matsusaka H, Kang D, Matsushima S, Ide T, Kubota T, Fujiwara T, Hamasaki N, Takeshita A, Sunagawa K, and Tsutsui H. Overexpression of mitochondrial transcription factor a ameliorates mitochondrial deficiencies and cardiac failure after myocardial infarction. Circulation 112: 683–690, 2005. [DOI] [PubMed] [Google Scholar]
- 15.Iyer S, Thomas RR, Portell FR, Dunham LD, Quigley CK, and Bennett JP Jr. Recombinant mitochondrial transcription factor A with N-terminal mitochondrial transduction domain increases respiration and mitochondrial gene expression. Mitochondrion 9: 196–203, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackman RW, and Kandarian SC. The molecular basis of skeletal muscle atrophy. American journal of physiology Cell physiology 287: C834–843, 2004. [DOI] [PubMed] [Google Scholar]
- 17.Kanazashi M, Tanaka M, Murakami S, Kondo H, Nagatomo F, Ishihara A, Roy RR, and Fujino H. Amelioration of capillary regression and atrophy of the soleus muscle in hindlimb-unloaded rats by astaxanthin supplementation and intermittent loading. Experimental physiology 99: 1065–1077, 2014. [DOI] [PubMed] [Google Scholar]
- 18.Kienhofer J, Haussler DJ, Ruckelshausen F, Muessig E, Weber K, Pimentel D, Ullrich V, Burkle A, and Bachschmid MM. Association of mitochondrial antioxidant enzymes with mitochondrial DNA as integral nucleoid constituents. Faseb j 23: 2034–2044, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kukat C, Davies KM, Wurm CA, Spåhr H, Bonekamp NA, Kühl I, Joos F, Polosa PL, Park CB, Posse V, Falkenberg M, Jakobs S, Kühlbrandt W, and Larsson N- G. Cross-strand binding of TFAM to a single mtDNA molecule forms the mitochondrial nucleoid. Proceedings of the National Academy of Sciences 112: 11288–11293, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kunkel GH, Chaturvedi P, and Tyagi SC. Mitochondrial pathways to cardiac recovery: TFAM. Heart failure reviews 21: 499–517, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lazarou M, McKenzie M, Ohtake A, Thorburn DR, and Ryan MT. Analysis of the assembly profiles for mitochondrial- and nuclear-DNA-encoded subunits into complex I. Molecular and cellular biology 27: 4228–4237, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee I, Huttemann M, and Malek MH. (−)-Epicatechin Attenuates Degradation of Mouse Oxidative Muscle Following Hindlimb Suspension. Journal of strength and conditioning research 30: 1–10, 2016. [DOI] [PubMed] [Google Scholar]
- 23.Lynch CJ, Xu Y, Hajnal A, Salzberg AC, and Kawasawa YI. RNA sequencing reveals a slow to fast muscle fiber type transition after olanzapine infusion in rats. PLoS One 10: e0123966, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McLeod M, Breen L, Hamilton DL, and Philp A. Live strong and prosper: the importance of skeletal muscle strength for healthy ageing. Biogerontology 17: 497–510, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muller FL, Song W, Jang YC, Liu Y, Sabia M, Richardson A, and Van Remmen H. Denervation-induced skeletal muscle atrophy is associated with increased mitochondrial ROS production. American journal of physiology Regulatory, integrative and comparative physiology 293: R1159–1168, 2007. [DOI] [PubMed] [Google Scholar]
- 26.Nuoc TN, Kim S, Ahn SH, Lee JS, Park BJ, and Lee TH. The analysis of antioxidant expression during muscle atrophy induced by hindlimb suspension in mice. The journal of physiological sciences : JPS 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Owusu-Ansah E, Yavari A, and Banerjee U. A protocol for _in vivo_ detection of reactive oxygen species 2008. [Google Scholar]
- 28.Pedersen BK. Muscle as a secretory organ. Comprehensive Physiology 3: 1337–1362, 2013. [DOI] [PubMed] [Google Scholar]
- 29.Peralta S, Wang X, and Moraes CT. Mitochondrial transcription: lessons from mouse models. Biochimica et biophysica acta 1819: 961–969, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Powers SK, Lynch GS, Murphy KT, Reid MB, and Zijdewind I. Disease-Induced Skeletal Muscle Atrophy and Fatigue. Medicine and science in sports and exercise 48: 2307–2319, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rudrappa SS, Wilkinson DJ, Greenhaff PL, Smith K, Idris I, and Atherton PJ. Human Skeletal Muscle Disuse Atrophy: Effects on Muscle Protein Synthesis, Breakdown, and Insulin Resistance-A Qualitative Review. Frontiers in physiology 7: 361, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Russell AP, Foletta VC, Snow RJ, and Wadley GD. Skeletal muscle mitochondria: a major player in exercise, health and disease. Biochimica et biophysica acta 1840: 1276–1284, 2014. [DOI] [PubMed] [Google Scholar]
- 33.Short KR, Bigelow ML, Kahl J, Singh R, Coenen-Schimke J, Raghavakaimal S, and Nair KS. Decline in skeletal muscle mitochondrial function with aging in humans. Proceedings of the National Academy of Sciences of the United States of America 102: 5618–5623, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smiles WJ, Parr EB, Coffey VG, Lacham-Kaplan O, Hawley JA, and Camera DM. Protein coingestion with alcohol following strenuous exercise attenuates alcohol-induced intramyocellular apoptosis and inhibition of autophagy. American journal of physiology Endocrinology and metabolism 311: E836–e849, 2016. [DOI] [PubMed] [Google Scholar]
- 35.Theilen NT, Jeremic N, Weber GJ, and Tyagi SC. Exercise preconditioning diminishes skeletal muscle atrophy after hindlimb suspension in mice. Journal of applied physiology (Bethesda, Md : 1985) 125: 999–1010, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Theilen NT, Kunkel GH, and Tyagi SC. The Role of Exercise and TFAM in Preventing Skeletal Muscle Atrophy. J Cell Physiol 232: 2348–2358, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tisdale MJ. The ubiquitin-proteasome pathway as a therapeutic target for muscle wasting. The journal of supportive oncology 3: 209–217, 2005. [PubMed] [Google Scholar]
- 38.van Gisbergen MW, Voets AM, Starmans MHW, de Coo IFM, Yadak R, Hoffmann RF, Boutros PC, Smeets HJM, Dubois L, and Lambin P. How do changes in the mtDNA and mitochondrial dysfunction influence cancer and cancer therapy? Challenges, opportunities and models. Mutation Research/Reviews in Mutation Research 764: 16–30, 2015. [DOI] [PubMed] [Google Scholar]
- 39.Wei YH. Oxidative stress and mitochondrial DNA mutations in human aging. Proceedings of the Society for Experimental Biology and Medicine Society for Experimental Biology and Medicine (New York, NY) 217: 53–63, 1998. [DOI] [PubMed] [Google Scholar]
- 40.Wredenberg A, Wibom R, Wilhelmsson H, Graff C, Wiener HH, Burden SJ, Oldfors A, Westerblad H, and Larsson NG. Increased mitochondrial mass in mitochondrial myopathy mice. Proceedings of the National Academy of Sciences of the United States of America 99: 15066–15071, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yajid F, Mercier JG, Mercier BM, Dubouchaud H, and Prefaut C. Effects of 4 wk of hindlimb suspension on skeletal muscle mitochondrial respiration in rats. Journal of applied physiology (Bethesda, Md : 1985) 84: 479–485, 1998. [DOI] [PubMed] [Google Scholar]
- 42.Zhang P, Chen X, and Fan M. Signaling mechanisms involved in disuse muscle atrophy. Medical hypotheses 69: 310–321, 2007. [DOI] [PubMed] [Google Scholar]