Abstract
Tomato spotted wilt tospovirus (TSWV), one of the most important plant viruses, causes yield losses to many crops including tomato. The current disease management for TSWV is based mainly on breeding tomato cultivars containing the Sw-5 locus. Unfortunately, several Sw-5 resistance-breaking strains of TSWV have been identified. Sw-7 is an alternative locus conferring resistance to a broad range of TSWV strains. In an effort to uncover gene networks that are associated with the Sw-7 resistance, we performed a comparative transcriptome profiling and gene expression analysis between a nearly-isogenic Sw-7 line and its susceptible recurrent parent (Fla. 8059) upon infection by TSWV. A total of 1,244 differentially expressed genes were identified throughout a disease progression process involving networks of host resistance genes, RNA silencing/antiviral defense genes, and crucial transcriptional and translational regulators. Notable induced genes in Sw-7 include those involved in callose accumulation, lignin deposition, proteolysis process, transcriptional activation/repression, and phosphorylation. Finally, we investigated potential involvement of PR-5 in the Sw-7 resistance. Interestingly, PR-5 overexpressed plants conferred enhanced resistance, resulting in delay in virus accumulation and symptom expression. These findings will facilitate breeding and genetic engineering efforts to incorporate this new source of resistance in tomato for protection against TSWV.
Subject terms: Molecular engineering in plants, Biotic
Introduction
Tomato spotted wilt tospovirus (TSWV), a member of the genus Tospovirus in the family Peribunyaviridae and the order Bunyavirales (https://talk.ictvonline.org/taxonomy/p/taxonomy-history?taxnode_id=20162190), is one of the most important viruses that infects tomato (Solanum lycopersicum), worldwide1. The TSWV genome consists of three RNA segments designated as large (L), medium (M), and small (S)2. This virus has a broad host range, infecting ~1,090 plant species3. Under field conditions, TSWV spreads from plant to plant by multiple species of thrips, primarily the Western flower thrips (Frankliniella occidentalis)4. TSWV causes plant stunting and chlorotic or necrotic spots on leaves and fruits, resulting in yield losses that can exceed $1 billion annually in the U.S.5.
Host resistance is the most effective and economical means of managing any disease, including TSWV. Conventional tomato breeding often begins by screening germplasm resources, typically wild tomato relatives, to identify sources of resistance. Once identified, a resistant accession is backcrossed to cultivated tomato to introgress the resistance allele. The first resistance source to TSWV was found in S. pimpinellifolium6. Over the years, seven TSWV resistance loci have been identified, designated as the dominant and allelic Sw-1a and Sw-1b; three recessive genes: sw-2, sw-3, and sw-4; and three dominant genes: Sw-5, Sw-6, and Sw-77–10. Sw-5, originally introgressed in the cultivar ‘Stevens’, is currently the primary source of TSWV resistance in commercial tomato varieties worldwide11. In addition to conferring a broad spectrum resistance to TSWV isolates, Sw-5 also confers resistance to closely related tospoviruses, including Tomato chlorotic spot tospovirus (TCSV) and Groundnut ringspot tospovirus (GRSV)12. Unfortunately, several Sw-5 resistance-breaking strains of TSWV have been identified in various regions around the world13, including the U.S. mainland14. Sequence comparison among TSWV isolates revealed that the ability of the virus to overcome Sw-5 is associated with C to Y amino acid substitutions at position 118 (C118Y) and T to N substitutions at position 120 (T120N) in the TSWV movement protein (NSm). The NSm protein is responsible for cell-to-cell movement, tubule formation, symptomology, host-range determination and interactions with the TSWV N protein14,15. There is therefore an urgent need to utilize other TSWV resistance loci in place of, or along with, Sw-5. The Sw-6 resistance locus confers only partial resistance under thrips inoculation and is effective against an even narrower range of TSWV isolates than Sw-516. Alternatively, Sw-7, is reported to exhibit field resistance against various isolates of TSWV, including those that overcome Sw-517. Sw-7 was introgressed from S. chilense accession LA 1938 and is generally mapped onto chromosome 129,18, but the molecular mechanism underlying this locus remains unknown.
In an effort to uncover the gene networks that are associated with Sw-7 resistance, we performed comprehensive comparative analysis of global gene expression profiles in response to TSWV infection between a TSWV-susceptible parental line (Fla. 8059) and a Sw-7 near isogenic line (with isogenicity estimated at 97.125% identity to the parental line Fla. 8059). From this analysis, 1,244 DEGs were identified between the two lines at five time points during disease progression from inoculation to symptom expression. Our findings provide a fundamental understanding of the virus-host interactions and identification of important candidate gene(s) for elucidation of the underlying mechanisms of Sw-7 resistance against TSWV, which may have broad implications for characterization of the mechanism of resistance in other plant-virus systems.
Results
Summary of RNA-Seq datasets and differentially expressed genes between Sw-7 and S-line
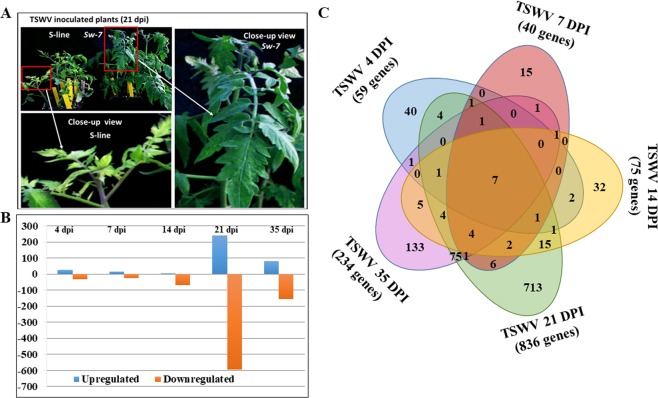
To provide a global view on differential gene expression between a near-isogenic line containing the Sw-7 resistance locus (hereafter referred to as Sw-7 line) and its susceptible recurrent parental line (Fla. 8059, hereafter referred to as S-line), comparative transcriptome profiling analysis was conducted using leaf samples collected throughout the virus infection process from inoculation to symptom expression. From these two lines, three biological replicate samples were taken at each of the five time points, 4, 7, 14, 21, and 35 days post inoculation (dpi). Typical disease symptoms, including chlorosis, mosaic, and necrotic lesions, were observed on the susceptible S-line plants at approximately 14–21 dpi. During the same period, symptoms were very mild to non-visible on TSWV-inoculated Sw-7 line plants (Fig. 1A). Real-time RT-PCR confirmed the presence of TSWV in the inoculated leaves as early as 4 dpi in both Sw-7 line (mean Ct: 27.02) and S-line plants (mean 27.43) (Supplementary Table S1), indicating virus infection had occurred and TSWV was replicating in the inoculated leaves. At 7 dpi, virus concentration continued to increase in the S-line (mean Ct: 22.46), but TSWV was nearly undetectable in systemic leaves in the Sw-7 line (mean Ct: 35.01). At later time points of 14, 21, and 35 dpi, these trends continued, with high levels of virus accumulation in three S-line plants (mean Ct: 22.54, 16.88, and 22.48, respectively), and much lower virus concentrations in the Sw-7 line plants (mean Ct: 33.04, 31.51, and 32.41, respectively) (Supplementary Table S1). These results indicated that although TSWV was initially capable of replicating in the inoculated leaves of the Sw-7 plants, virus movement or replication was restricted and did not become systemic. Over time, disease expression in the Sw-7 plants ranged from asymptomatic to mild disease symptoms with lower virus titer. On the other hand, the inoculated S-line plants exhibited severe disease symptoms with much higher virus titers in the systemic (upper uninoculated) leaves (Fig. 1 and Supplementary Table S1).
Figure 1.
Differential gene expression between resistant Sw-7 line and susceptible S-line after TSWV infection. (A) Plants of S-line (left), and the Sw-7 at 21 days post inoculation (dpi) with TSWV (inset close view of a single leaf). (B) Numbers of differentially expressed genes in Sw-7 line compared to S-line at 4, 7, 14, 21 and 35 dpi with TSWV. (C) Venn diagram showing the numbers of common, intersecting and specific DEGs at 4, 7, 14, 21 and 35 dpi with TSWV.
To achieve a comprehensive understanding on genes and pathways of tomato in response to TSWV infection, a total of 30 RNA-Seq libraries were constructed and sequenced. Differential gene expression was evaluated through an extensive comparative transcriptome analysis between the Sw-7 line and the S-line plants. We used fastqc (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) to assess the quality of both raw and final cleaned RNA-Seq reads, and ensured that the cleaned reads were of high quality. Overall, an average of 15.3 million raw reads per library were obtained. After adapter trimming and removal of low quality reads and rRNA sequences, an average of 9.9 million high quality cleaned reads were obtained, with 85% of those reads mapped to the tomato genome (version SL2.5) (Supplementary Dataset S1). Values of Pearson’s correlation coefficients for all biological replicates were high, suggesting highly reproducible data generated by RNA-Seq (Supplementary Dataset S2).
Comparative analysis of gene expression levels revealed that out of the 34,727 genes predicted in the tomato genome, a total of 1,244 (3.58%) differentially expressed genes (DEGs) were identified between TSWV-infected Sw-7 line and S-line plants during disease progression at 4, 7, 14, 21 and 35 dpi. Volcano plots were generated to illustrate distribution patterns of DEGs at each time point (Supplementary Fig. S1). Fewer genes were affected in the pre-symptom expression phase, as demonstrated at 4 and 7 dpi. Of 59 DEGs identified at 4 dpi (Supplementary Dataset S3), 27 were upregulated and 32 were down-regulated in the Sw-7 line compared to the S-line. Similarly, of 40 DEGs identified at 7 dpi (Supplementary Dataset S3), 16 were upregulated and 24 were down-regulated (Fig. 1B). However, as days after inoculation elapsed, the number of DEGs increased, peaking at 21 dpi, the time when symptoms began to appear in the plants of the S-line (Fig. 1). Most of the DEGs affected by TSWV infection were not constant from one time point to another, and only seven DEGs intersected all 5-time points (Fig. 1C). Among these were a mannan-endo-1,4-beta-mannosidase and a chloroplastic group IIA intron splicing facilitator CRS1 which were both down-regulated, while the other five genes had unknown functions (Supplementary Table S2).
Functional characterization of DEGs between the Sw-7 line and the S-line in response to TSWV infection
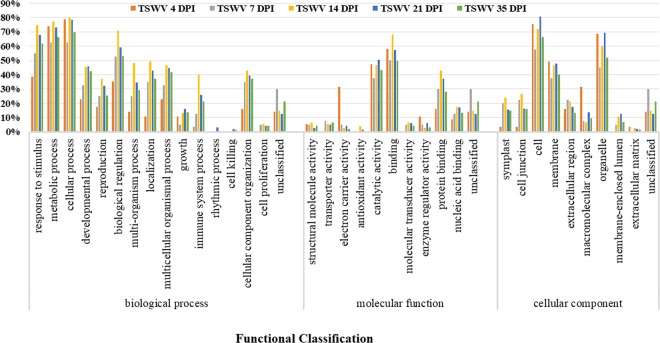
DEGs were functionally classified using the broad gene ontology (GO) categories. In the biological process category, a large number of DEGs were related to response to stimulus, metabolic process, cellular process, and biological regulation (Fig. 2). In the molecular function category, the majority of DEGs were responsible for catalytic activity and binding. In the cellular component category, a high proportion of DEGs were related to cell, membrane and organelles (Fig. 2). GO enrichment analysis was also performed on DEGs at each time point (Supplementary Fig. S2). In genes upregulated in the Sw-7 line at 4 dpi, GO terms including photosynthesis, electron carrier activity and chlorophyll binding were significantly enriched. However, in down-regulated genes at 4 dpi, only GO terms within the biological process category were enriched, including aldehyde and methylglyoxal metabolism. Although no enriched GO terms were identified from DEGs at 7 dpi, two genes encoding receptor-like proteins were upregulated in the Sw-7 line. At 14 dpi, in five upregulated genes identified in the Sw-7 line, no enriched GO terms were detected. At 21 dpi, in the stage of symptom expression, the majority of DEGs were induced in the S-line plants (Fig. 1). GO enrichment analysis revealed that these genes were related to plant defense and response to stimulus (Supplementary Fig. S2). A similar trend was observed in the post symptom stage at 35 dpi, when TSWV infection had resulted in severe symptom expression in the S-line plants. At this time point, genes in the photosynthesis, auxin homeostasis, and cellular carbohydrate metabolism pathways were upregulated in the Sw-7 line plants, and the susceptible S-line plants showed enrichment of genes related to plant wound response. S-line plants were also enriched for genes involved in xyloglucan transferase activity; these genes play a role in the organization of cellulose-xyloglucan matrix, which control the strength and extensibility of the plant primary cell wall.
Figure 2.
Gene Ontology (GO) functional classification of differentially expressed genes (DEGs). The percentage of genes assigned to each category were calculated at 4, 7, 14, 21 and 35 dpi, respectively.
To gain a better understanding of the mechanism of resistance and of possible candidate genes that are involved in the Sw-7 resistance response, several categories of genes, including those encoding nucleotide-binding site leucine-rich repeat (NBS-LRR) proteins, defense-related proteins, transcription factors, protein kinases, as well as those related to phytohormone signaling, cell wall, photosynthesis, gene silencing, and microRNA target genes, were further analyzed as follows.
Host defense-related genes
GO term enrichment analyses of DEGs between Sw-7 and S- line plants in response to TSWV infection revealed a total of 68 genes related to immunity, defense response, and disease resistance signaling molecules (Table 1). Among them, only two NBS-LRR genes were differentially expressed, and both were induced in the S-line plants at 21 dpi (Table 1).
Table 1.
Selected differentially expressed defense-related, RNA silencing pathway and microRNA target genes in Sw-7 compared to the S-line after TSWV inoculation.
| S. lycopersicum accession | Annotation | 4 dpi | 7 dpi | 14 dpi | 21 dpi | 35 dpi |
|---|---|---|---|---|---|---|
| DEFENSE-RELATED GENES | ||||||
| NBS-LRR Genes | ||||||
| Solyc08g006970 | LRR, resistance protein | — | — | — | −2.396 | — |
| Solyc01g014840 | TIR-NBS-LRR, resistance protein | — | — | — | −3.474 | — |
| MLO-like Protein | ||||||
| Solyc03g095650 | MLO-like protein | — | — | — | −3.322 | — |
| PATHOGENESIS-RELATED PROTEIN GENES | ||||||
| Solyc07g006710 | Pathogenesis-related-1: PR-1 | — | — | — | 1.546 | — |
| Solyc12g014310 | PR-like protein | — | — | −2.322 | — | — |
| Defensin | ||||||
| Solyc07g007760 | Defensin protein | — | — | — | −4.322 | −2.184 |
| Solyc07g006380 | Defensin-like protein | — | — | — | −2.144 | — |
| Nodulin | ||||||
| Solyc11g012930 | Nodulin family protein | — | 2.336 | — | — | — |
| Solyc05g005870 | Nodulin MtN21 family | — | — | — | 2.124 | — |
| Chitinase | ||||||
| Solyc07g005100 | Chitinase-like protein | — | — | — | −3.474 | — |
| Solyc05g050130 | Acidic chitinase | — | — | — | −4.644 | −2.737 |
| Solyc02g082920 | Endochitinase (Chitinase) | — | — | −3.184 | −5.059 | — |
| Protease | ||||||
| Solyc10g086600 | Subtilisin-like serine protease | — | — | — | — | 4.563 |
| Peroxidase | ||||||
| Solyc06g050440 | Peroxidase | — | — | — | −4.059 | — |
| Solyc01g006300 | Peroxidase | — | — | — | −2.120 | — |
| Osmotin-like protein, PR5 | ||||||
| Solyc08g080670 | Osmotin-like protein | — | — | — | 3.065 | — |
| Protease Inhibitor | ||||||
| Solyc00g071180 | Cysteine proteinase inhibitor | — | — | — | −6.644 | — |
| Solyc03g097270 | Cysteine proteinase inhibitor | — | — | — | −2.120 | — |
| Solyc03g098740 | Kunitz trypsin inhibitor | — | — | — | −3.837 | — |
| Solyc03g098790 | Kunitz-type | −1.889 | — | — | −6.644 | — |
| Solyc03g098780 | Kunitz-type | — | — | — | −2.644 | — |
| Solyc03g098760 | Kunitz-type like protein | — | — | — | −5.644 | — |
| Solyc09g089530 | Proteinase inhibitor I | — | — | — | −4.322 | — |
| Solyc09g089540 | Proteinase inhibitor I | — | — | — | −5.648 | — |
| Solyc09g089520 | Proteinase inhibitor I | −4.644 | — | — | −5.059 | — |
| Solyc09g089510 | Proteinase inhibitor I | — | — | — | −2.556 | — |
| Solyc09g084470 | Proteinase inhibitor I | — | — | — | −4.059 | — |
| Solyc09g084480 | Proteinase inhibitor I | — | — | — | −2.943 | — |
| Solyc09g083440 | Proteinase inhibitor I | — | — | — | −3.474 | — |
| Solyc09g084490 | Proteinase inhibitor I | — | — | — | −4.322 | — |
| OTHER DEFENSE GENES | ||||||
| Glycine-rich protein (Inducer of PR-1) | ||||||
| Solyc06g061200 | Glycine-rich protein TomR2 | — | 1.880 | — | — | — |
| Major latex-like protein | ||||||
| Solyc09g014530 | Major latex-like protein | — | — | — | −4.322 | — |
| Solyc08g023660 | Major latex-like protein | — | −1.889 | — | — | — |
| Solyc05g046150 | Major latex-like protein | −2.737 | — | — | — | — |
| Solyc04g005700 | Major latex-like protein | −1.556 | — | — | — | — |
| Universal stress protein | ||||||
| Solyc09g011660 | Universal stress protein | — | — | — | −2.396 | — |
| Solyc04g014600 | Universal stress protein | — | — | — | −3.059 | — |
| Solyc01g057000 | Universal stress protein | — | — | — | −2.943 | −5.059 |
| TMV response | ||||||
| Solyc04g082960 | TMV response-related | — | — | −3.059 | −2.837 | — |
| Disease resistance | ||||||
| Solyc01g021600 | Disease resistance response | — | — | — | −5.644 | — |
| Calmodulin | ||||||
| Solyc01g010020 | Calmodulin | — | — | — | — | −3.184 |
| Solyc02g091500 | Calmodulin | — | — | — | −2.252 | — |
| Solyc07g040710 | Calmodulin-binding protein | — | — | — | — | — |
| Solyc03g113940 | Calmodulin-binding protein | — | — | — | −5.644 | — |
| Solyc03g119250 | Calmodulin-binding protein | — | — | −2.474 | — | — |
| Solyc02g088090 | Calmodulin-like protein | — | — | −3.322 | — | — |
| Heat Shock Protein | ||||||
| Solyc06g076520 | class I heat shock protein | — | — | — | −2.322 | — |
| Solyc06g076570 | class I heat shock protein | — | — | — | −3.644 | — |
| Solyc06g076560 | class I heat shock protein | — | — | — | −3.059 | — |
| Solyc02g093600 | class I heat shock protein | — | — | — | −5.644 | — |
| Solyc04g014480 | class I heat shock protein 3 | — | — | — | — | −3.059 |
| Solyc08g062450 | class II heat shock protein | — | — | −3.837 | — | — |
| Solyc03g113930 | class IV heat shock protein | — | — | — | — | −3.644 |
| Solyc11g020330 | class IV heat shock protein | — | — | — | −4.644 | −3.644 |
| F-Box Protein | ||||||
| Solyc11g006740 | F-box protein | — | — | — | −4.322 | −3.644 |
| GENE SILENCING PATHWAY GENES | ||||||
| Solyc02g069260 | ARGONAUTE 1 | — | — | — | −3.059 | — |
| Solyc11g008540 | SlDCL2b | — | — | — | −2.737 | — |
| Solyc11g008530 | SlDCL2d | — | — | — | −2.252 | — |
| MICRORNA-TARGET GENES | ||||||
| Solyc03g115850 | miR164-NAC domain | — | — | — | −3.837 | — |
| Solyc06g069710 | miR164-NAC domain | — | — | — | −6.644 | — |
| Solyc06g075510 | miR172-AP2-like ERF | — | — | — | −2.252 | — |
| Solyc10g006710 | miR172-kinase receptor | — | — | — | −4.322 | — |
| Solyc06g007320 | miR396-Ubiquitin-activating enzyme E1 | — | — | — | −2.120 | — |
| Solyc01g005780 | miR6022-LRR -RLP kinase | — | — | — | −5.644 | — |
| Solyc01g006550 | miR6022-LRR- RLP kinase | — | — | — | −4.644 | — |
| Solyc01g009690 | miR6022-LRR- RLP kinase | — | — | −3.837 | −4.059 | — |
| Solyc01g009700 | miR6022-LRR- RLP kinase | — | — | — | −4.322 | — |
| Solyc01g009930a | N/A -LRR- RLP kinase | — | — | — | −3.251 | — |
aN/A referred to be a non-annotated microRNA (It is not registered at the miRBase registry; it can be considered as a new microRNA).
Pathogenesis-related (PR) proteins often accumulate in plants upon pathogen attack and are typically induced as a defense response through systemic acquired resistance. Interestingly, a total of 27 PR protein family genes with differential expression between the Sw-7 line and S-line plants were identified (Table 1). Among these, the pathogenesis related-1 (Solyc07g006710) gene was 1.5-fold higher expressed in the Sw-7 line at 21 dpi, and two nodulin family genes were ~2-fold higher expressed in the Sw-7 plants at 7 and 21 dpi. Two defensin (PR-12) genes were induced approximately 2- to 4-fold higher in the S-line at 21 dpi and 35 dpi. A total of 14 protease inhibitor genes, which belong to a pathogenesis-related protein subfamily (PR-6), were altered in our datasets. All of these were down-regulated (ranging from 2.1- to 6.6-fold) in the Sw-7 line, with the majority (14) being induced in the S-line at 21 dpi. A subtilisin-like serine protease gene (Solyc10g086600) was upregulated 4.6 fold in the Sw-7 line at 35 dpi. One gene encoding a member of glycine-rich protein (Solyc06g061200) was induced (~2-fold) in the Sw-7 line at 7 dpi. Interestingly, a gene encoding an osmotin-like protein (OLP) was induced approximately 3-fold at 21 dpi in the Sw-7 line (Table 1).
Three RNA silencing pathway genes, including one Argonaute 1 (Ago1) and two Dicer-like 2 (DCL 2), were down-regulated in the Sw-7 line (Table 1). In addition, 10 microRNA target genes were identified, including miR164 (2 genes), miR172 (2 genes), miR396, miR6022 (4 genes), and an un-annotated or new miRNA. Most of these were induced in the S-line at 21 dpi, ranging from 2.1 to 6.6-fold in differential expression relative to that of the Sw-7 line (Table 1).
Transcription factors (TFs)
In this study, a large number of TF genes (78 genes) exhibited differential expression between the Sw-7 line and the S-line plants (Table 2). Overall, only nine TF genes were up-regulated while 69 were down-regulated in the Sw-7 line (Table 2). The nine induced TFs include three in the basic helix-loop-helix (bHLH) family, one in the bZIP family, one in the MADS-box family, two Myb transcription factors, one nuclear transcription factor Y subunit B-3, and one Cycloidea transcription factor. There were nine AP2-like ethylene-responsive transcription factors that were induced in the S-line (2–4 fold) at 21 dpi. Five bHLH transcription factors were altered, with three being induced in the Sw-7 line at 21 or 35 dpi, and two being induced in the S-line. Interestingly, one of the bHLH genes was induced at a very high level (6.8-fold) in the Sw-7 line (Table 2).
Table 2.
List of differentially expressed transcription factors (TFs) in Sw-7 compared to the S-line after TSWV inoculation.
| S. lycopersicum accession | Annotationa | 4 dpi | 7 dpi | 14 dpi | 21 dpi | 35 dpi |
|---|---|---|---|---|---|---|
| AP2/ERF-AP2 Family | ||||||
| Solyc05g051380 | AP2-like ethylene-responsive | — | — | — | −4.644 | — |
| Solyc06g075510 | AP2-like ethylene-responsive | — | — | — | −2.252 | — |
| Solyc02g077370 | Ethylene responsive TF-2 | — | — | −5.059 | — | — |
| Solyc03g093540 | Ethylene responsive TF-1a | — | — | — | −2.396 | — |
| Solyc03g093560 | Ethylene responsive TF-2 | — | — | — | −2.556 | — |
| Solyc03g093610 | Ethylene responsive TF-1b | — | — | — | — | −2.837 |
| Solyc04g071770 | Ethylene responsive TF-2a | — | — | — | −3.184 | — |
| Solyc05g052410 | Ethylene responsive TF-1 | — | — | — | −5.059 | — |
| Solyc11g012980 | Ethylene responsive TF-9 | — | — | — | −4.322 | — |
| BHLH and BZIP Family | ||||||
| Solyc02g087860 | Transcription factor style2.1 | — | — | — | 1.840 | — |
| Solyc03g121240 | bHLH TF-like | — | — | — | — | 2.384 |
| Solyc04g007300 | bHLH TF | — | — | — | 6.759 | — |
| Solyc07g005400 | bHLH TF | — | — | — | −2.396 | — |
| Solyc12g036470 | bHLH TF | — | — | — | −4.644 | — |
| Solyc01g109880 | bZIP TF | — | — | — | −2.837 | — |
| Solyc04g082890 | bZIP TF | — | — | — | −1.889 | — |
| Solyc07g062710 | bZIP TF-family protein | — | — | — | 1.807 | — |
| Solyc10g078290 | bZIP TF-family protein | — | — | — | −2.000 | — |
| Zinc Finger Protein Family | ||||||
| Solyc05g009310 | ZF-CONSTANS-LIKE 16 | — | — | — | −6.644 | — |
| Solyc05g024010 | ZF-CONSTANS-LIKE | — | — | — | −3.184 | — |
| Solyc09g074560 | ZF-CONSTANS-LIKE | — | — | — | −2.474 | — |
| Solyc01g090840 | Cys2/His2 ZF | — | — | — | −3.837 | — |
| Solyc01g107170 | Zinc finger protein | — | — | — | −3.059 | — |
| Solyc06g075780 | Cys2/His2 ZF | — | — | — | −4.059 | −3.184 |
| Solyc11g073060 | ZF-family protein | — | — | — | −4.644 | −5.059 |
| Solyc06g071860 | ZF-CCCH-67 | — | — | — | −2.644 | — |
| Solyc01g102980 | Zinc finger-HD | −2.474 | — | — | — | — |
| Solyc04g080490 | Zinc finger-HD | — | — | — | 2.395 | — |
| Solyc09g005560 | Zinc finger and SCAN | — | — | — | −6.644 | — |
| GARP, MYB, GRAS and Scarecrow Family | ||||||
| Solyc06g061030 | GARP-ARR-B | — | — | — | −2.059 | — |
| Solyc10g076460 | Myb-like DNA-binding domain | — | — | — | −2.223 | — |
| Solyc05g053090 | GRAS family | — | — | — | −2.556 | — |
| Solyc05g054170 | Scarecrow-like 1 | — | — | — | −2.644 | — |
| HB-HD-ZIP Family | ||||||
| Solyc02g063520 | Homeobox leucine zipper | — | — | — | — | −2.252 |
| Solyc03g082550 | Homeobox leucine zipper | — | — | — | −3.322 | — |
| Solyc05g051460 | Homeobox-leucine zipper | — | — | — | −2.474 | — |
| Solyc06g053220 | Homeobox leucine zipper | — | — | — | −2.059 | — |
| Solyc08g083130 | Homeobox leucine zipper | — | — | — | −3.322 | — |
| Solyc03g034150 | Homeobox leucine zipper | — | — | — | — | −4.322 |
| HS-TF, LOB and MADS Family | ||||||
| Solyc02g090820 | Heat stress TF- | — | — | — | −3.474 | — |
| Solyc06g053960 | Heat stress TF-A3 | — | — | — | −2.474 | — |
| Solyc03g119530 | LOB domain protein 42 | — | — | — | — | −2.943 |
| Solyc04g077990 | LOB domain protein 38 | — | — | −2.396 | — | −5.059 |
| Solyc11g072470 | LOB domain protein 1 | — | — | — | −3.644 | — |
| Solyc03g114840 | MADS box | — | — | — | 1.915 | — |
| MYB Family | ||||||
| Solyc03g093890 | Myb-related TF | — | — | — | — | −2.737 |
| Solyc05g048830 | MYB TF | — | — | — | −2.737 | - |
| Solyc06g053610 | Myb-related TF | — | — | — | −2.644 | - |
| Solyc07g008010 | Myb TF | — | — | — | 1.891 | - |
| Solyc08g008480 | Myb TF | — | — | — | 2.118 | - |
| Solyc06g005310 | Myb TF | — | — | — | −2.737 | - |
| NAC Family | ||||||
| Solyc02g077610 | NAC domain TF | — | — | — | −3.184 | −3.322 |
| Solyc02g093420 | NAC domain TF | — | — | — | −2.837 | −2.737 |
| Solyc03g115850 | NAC domain TF | — | — | — | −3.837 | — |
| Solyc05g021090 | NAC domain TF | — | — | — | −6.644 | — |
| Solyc06g069710 | NAC domain TF | — | — | — | −6.644 | — |
| Solyc07g045030 | NAC domain TF | — | — | — | −4.322 | — |
| Solyc08g077110 | NAC domain TF | — | — | — | −4.644 | — |
| Solyc10g055760 | NAC domain TF | — | — | −2.837 | — | — |
| Solyc11g068620 | NAC domain TF | — | — | −4.322 | — | −2.644 |
| Solyc12g013620 | NAC domain TF | — | — | — | −4.644 | −2.837 |
| WRKY Family | ||||||
| Solyc02g080890 | WRKY TF-16 | — | — | — | −3.474 | — |
| Solyc04g051690 | WRKY TF-16 | — | — | −2.943 | — | — |
| Solyc06g008610 | WRKY TF-25 | — | — | — | −2.120 | — |
| Solyc07g051840 | WRKY TF | — | — | — | −2.396 | — |
| Solyc07g056280 | WRKY TF-16 | — | — | — | — | −2.737 |
| Solyc08g062490 | WRKY TF-16 | — | — | — | −3.837 | — |
| Solyc08g067340 | WRKY TF | — | — | — | — | −3.474 |
| Solyc08g067360 | WRKY TF-9 | — | — | — | −6.644 | — |
| Solyc09g014990 | WRKY-like TF | — | — | −2.252 | — | — |
| Solyc09g066010 | WRKY TF-25 | — | — | — | −2.252 | — |
| Solyc10g009550 | WRKY TF | — | — | −3.184 | −4.644 | — |
| Other Family members | ||||||
| Solyc05g056040 | Auxin response factor 14 | — | — | — | −2.184 | - |
| Solyc07g007220 | S/T phosphatase | — | — | — | −2.474 | — |
| Solyc09g074760 | Nuclear TF- Y subunit B-3 | — | — | — | 1.618 | — |
| Solyc10g076180 | Plant-specific domain | — | — | — | −5.059 | — |
| Solyc12g014140 | TF CYCLOIDEA | −3.644 | — | 3.474 | — | — |
| Solyc09g005560 | Zinc finger and SCAN | — | — | — | −6.644 | — |
aTF-Transcription Factor; S/T-Serine threonine; ZF-Zinc Finger.
On the other hand, four bZIP TFs were differentially expressed between Sw-7 and S-line plants. In addition, eight zinc finger proteins (C2C2-CO, C2H2 and C3H family) were differentially expressed. All eight of these were induced in the S-line at 21 dpi, two of which were also induced at 35 dpi. Furthermore, six homeobox leucine zipper proteins (HB-HD-ZIP) were induced in the S-line, mostly at 21 dpi after symptoms had already appeared on the infected susceptible plants. Among the six differentially expressed MYB transcription factor genes, two were induced and four were suppressed in the Sw-7 line. One nuclear transcription factor (NF-YB) and two Zinc finger-homeodomain protein 1 (zf-HD) genes were upregulated in the Sw-7 line (Table 2).
Protein kinases (PKs)
In the present study, only two of 42 protein kinase genes with differential expression were induced in the Sw-7 line. These were a Pto-like serine/threonine kinase from the RLK-Pelle_CrRLK1L-1 family and a RLK receptor-like kinase in the family of RLK-Pelle_LRR-XII-1 (Table 3).
Table 3.
List of differentially expressed protein kinases (PKs) in Sw-7 compared to the S-line after TSWV inoculation.
| S. lycopersicum accession | Annotationa | 4 dpi | 7 dpi | 14 dpi | 21 dpi | 35 dpi |
|---|---|---|---|---|---|---|
| CAMK_CAMKL, CAMK_CDPK Family | ||||||
| Solyc09g056430 | Kinase family protein | — | — | — | −3.059 | — |
| Solyc03g005330 | CBL-interacting PK-20 | — | — | — | −4.322 | — |
| Solyc09g018280 | Calcium/calmodulin-dependent | — | — | — | −2.474 | — |
| Solyc03g113390 | Calcium-dependent PK-1 | — | — | — | −2.474 | — |
| Solyc04g081910 | Calcium-dependent PK | — | — | — | −2.120 | — |
| Pto, S/T, LRR and RLK family | ||||||
| Solyc01g109950 | Pto-like, S/T PK, resistance protein | — | — | — | — | 3.123 |
| Solyc01g007990 | RLK, Receptor like protein | — | — | — | −2.943 | — |
| Solyc02g071800 | Receptor like kinase, RLK | — | — | — | −2.000 | — |
| Solyc02g080040 | Receptor-like PK | — | — | — | −3.474 | — |
| Solyc03g120110 | S/T kinase receptor | — | — | — | −3.322 | — |
| Solyc10g006710 | S/T kinase receptor | — | — | — | −4.322 | — |
| Solyc05g009040 | Receptor-like PK | — | — | — | −4.059 | — |
| Solyc07g006480 | LRR receptor S/T PK | — | — | −3.184 | — | — |
| Solyc03g123860 | Receptor like kinase, RLK | — | — | — | −2.943 | — |
| Solyc04g074000 | Receptor like kinase, RLK | — | — | −3.322 | — | — |
| Solyc04g074030 | Receptor like kinase, RLK | — | — | −3.474 | −3.059 | — |
| Solyc11g017270 | Receptor like kinase, RLK | — | — | −2.474 | −2.737 | — |
| Solyc03g111800 | Receptor like kinase, RLK | — | — | — | −3.059 | — |
| Solyc02g070890 | Receptor like kinase, RLK | — | — | — | 1.922 | — |
| Solyc04g009640 | Receptor like kinase, RLK | — | — | — | −3.474 | — |
| Solyc04g014900 | Receptor like kinase, RLK | — | — | — | −2.556 | — |
| Solyc06g006020 | Receptor like kinase, RLK | — | — | — | −4.322 | — |
| Solyc06g048740 | Receptor like kinase, RLK | — | — | — | −5.059 | — |
| Solyc12g089020 | Receptor-like protein kinase | — | — | — | −6.644 | — |
| Solyc10g075040 | Receptor-like protein kinase | — | — | — | −2.396 | — |
| Solyc04g057930 | Receptor-like kinase | — | — | — | −2.184 | — |
| Solyc11g072660 | Receptor PK-like protein | — | — | — | −2.396 | — |
| Solyc01g028830 | ATP binding; S/T kinase | — | — | — | −2.059 | — |
| Solyc01g112220 | Serine/threonine PK | — | — | — | −2.396 | — |
| Solyc03g032150 | Serine/threonine K-like protein | — | — | — | −2.059 | — |
| Solyc04g011520 | Serine/threonine K-like protein | — | — | — | −2.252 | — |
| Solyc04g082500 | ATP binding- S/T kinase | — | — | −4.059 | −3.837 | — |
| Solyc05g053930 | ATP binding-S/T kinase | — | — | - | −2.252 | — |
| Solyc03g078360 | Receptor-like PK | — | — | — | −3.837 | — |
| Solyc03g078370 | Receptor-like PK | — | — | — | −2.943 | — |
| Solyc07g055400 | Receptor-like kinase | — | — | — | −3.322 | — |
| Solyc05g008960 | Receptor-like PK | — | — | −3.644 | −4.322 | — |
| Solyc05g009010 | Serine/threonine PK | — | — | −2.252 | −2.184 | — |
| Solyc05g010530 | Serine/threonine PK | — | — | — | −2.644 | — |
| Solyc12g036330 | Receptor-like PK | — | — | — | −3.474 | — |
| STE_STE11, TKL-Pl-4 Family | ||||||
| Solyc07g051870 | Protein S/T K | — | — | — | — | −3.644 |
| Solyc03g006400 | Protein kinase | — | — | — | −3.644 | — |
aPK, protein kinase.
Phytohormone signaling
In the present study, a total of 33 phytohormone-related genes were identified as differentially expressed (Table 4). Among them, one geranylgeranyl prophosphate synthase pathway gene was induced in both 21 dpi and 35 dpi time points. Several DEGs in the Gibberellin and IAA Pathways were either up-regulated or down-regulated. However, a group of auxin pathway genes were highly induced in the Sw-7 line. In total, seven out of eight auxin pathway genes were induced in Sw-7 ranging from 1.6 to 8.4 folds.
Table 4.
Selected differentially expressed phytohormone-related genes in Sw-7 compared to the S-line after TSWV inoculation.
| S. lycopersicum Accession | Annotationa | 4 dpi | 7 dpi | 14 dpi | 21 dpi | 35 dpi |
|---|---|---|---|---|---|---|
| Cytokinin Pathway | ||||||
| Solyc04g016200 | CK-O-glucoside biosynthesis | — | — | — | −2.556 | — |
| Solyc10g079930 | CK-O-glucoside biosynthesis | — | — | −2.837 | — | — |
| Ethylene biosynthesis Pathway | ||||||
| Solyc00g095860 | ET biosynthesis | — | — | — | −3.322 | — |
| Solyc01g095080 | ET biosynthesis | — | — | — | −6.644 | — |
| Solyc08g081550 | ET biosynthesis | — | — | — | −3.184 | — |
| Solyc09g010000 | ET biosynthesis | — | — | — | −3.474 | — |
| Solyc09g089580 | ET biosynthesis | — | — | — | −2.474 | — |
| Geranylgeranyl Pyrophosphate Synthase Pathway | ||||||
| Solyc04g079960 | geranylgeranyldiphosphate biosynthesis | — | — | — | 1.541 | 1.727 |
| Gibberellin, IAA Pathways | ||||||
| Solyc03g006880 | gibberellin biosynthesis | — | — | — | 1.832 | — |
| Solyc11g072310 | gibberellin biosynthesis) | — | — | — | −1.888 | — |
| Solyc11g011210 | Gibberellin regulated protein | — | — | — | −4.059 | — |
| Solyc06g007890 | Gibberellin regulated protein | — | — | — | 2.384 | — |
| Solyc12g042520 | Gibberellin-regulated family protein | — | — | — | −2.737 | — |
| Solyc06g008870 | GID1-like gibberellin receptor | — | — | — | −2.837 | — |
| Solyc08g068480 | IAA-amido synthetase GH3.8 | — | — | — | 1.787 | — |
| Solyc01g107400 | IAA-amido synthetase GH3.8 | — | — | — | — | — |
| Solyc08g068490 | IAA-amido synthetase GH3.8 | — | — | — | 1.761 | −4.644 |
| Solyc07g043590 | IAA biosynthesis I | — | — | — | −1.889 | — |
| Solyc06g073060 | IAA biosynthesis II | — | — | — | −2.059 | — |
| Lipoxygenase Pathway | ||||||
| Solyc01g099160 | jasmonic acid biosynthesis | — | — | — | −4.059 | −2.644 |
| Solyc01g099170 | jasmonic acid biosynthesis | — | — | — | −4.059 | −2.737 |
| Solyc03g122340 | jasmonic acid biosynthesis | — | — | — | −2.644 | — |
| Solyc09g075860 | jasmonic acid biosynthesis | — | — | — | −1.943 | — |
| Solyc12g011040 | jasmonic acid biosynthesis | — | — | — | −6.644 | — |
| Phenylpropanoid Pathway | ||||||
| Solyc02g079490 | phenylpropanoid biosynthesis | — | — | — | — | 1.687 |
| Solyc02g093270 | phenylpropanoid biosynthesis | — | — | −2.556 | — | — |
| Solyc04g063210 | phenylpropanoid biosynthesis | — | — | — | −3.644 | — |
| Solyc06g074710 | phenylpropanoid biosynthesis | — | — | — | — | 1.727 |
| Solyc09g082660 | phenylpropanoid biosynthesis | — | — | — | — | −3.837 |
| Solyc11g071470 | phenylpropanoid biosynthesis | — | — | — | — | −4.644 |
| Solyc11g071480 | phenylpropanoid biosynthesis | — | — | — | — | −4.322 |
| Beta-carotenehydroxylase Pathway | ||||||
| Solyc03g007960 | lutein biosynthesis | — | — | — | — | −2.737 |
| Solyc03g007960 | zeaxanthin biosynthesis | — | — | — | — | −2.737 |
| Other Auxin Pathway Genes | ||||||
| Solyc01g110770 | Auxin-induced SAUR-like protein | — | — | — | 1.795 | — |
| Solyc11g011700 | Auxin-induced SAUR-like protein | — | — | — | 1.868 | — |
| Solyc11g011680 | Auxin-induced SAUR-like protein | — | — | — | 2.101 | — |
| Solyc04g052970 | Auxin-induced SAUR-like protein | — | — | — | 8.395 | — |
| Solyc12g005310 | Auxin-responsive GH3-like | — | — | — | 1.604 | — |
| Solyc03g082530 | Auxin-responsive family protein | — | — | — | −6.644 | — |
| Solyc11g011710 | Auxin-responsive protein | — | — | — | 2.928 | — |
aCK-Cytokinin, ET-stands for ethylene.
Cell wall-related genes
A total of 18 cell wall modification genes were altered in our datasets (Table 5). Interestingly, the majority of these genes were down-regulated in the Sw-7 line, which means they were highly induced in the S line. On the other hand, three in five pectinesterase genes were induced in the Sw-7 line.
Table 5.
List of differentially expressed cell wall pathway genes in Sw-7 compared to the S-line after TSWV inoculation.
| S. lycopersicum accession | Annotation | 4 dpi | 7 dpi | 14 dpi | 21 dpi | 35 dpi |
|---|---|---|---|---|---|---|
| Cellulose biosynthesis | ||||||
| Solyc04g016470 | cellulose biosynthesis | — | — | — | −5.644 | — |
| Solyc12g014430 | cellulose biosynthesis | — | — | — | −4.644 | — |
| Cuticular wax biosynthesis | ||||||
| Solyc01g088430 | cuticular wax biosynthesis | −1.599 | — | — | — | — |
| Solyc07g053890 | cuticular wax biosynthesis | — | — | — | −4.059 | — |
| Suberin biosynthesis | ||||||
| Solyc02g093270 | suberin biosynthesis | — | — | −2.556 | — | — |
| Solyc04g063210 | suberin biosynthesis | — | — | — | −3.644 | — |
| Solyc09g082660 | suberin biosynthesis | — | — | — | — | −3.837 |
| Wax esters biosynthesis | ||||||
| Solyc07g053890 | wax esters biosynthesis I | — | — | — | −4.059 | — |
| Solyc09g005940 | wax esters biosynthesis I | — | −1.786 | — | — | — |
| 3-oxoacyl-cyl-carrier-p-synthase | ||||||
| Solyc03g122120 | 3-oxoacyl-cyl-carrier-p-synthase | — | — | — | −2.474 | — |
| Expansin | ||||||
| Solyc06g051800 | Expansin | — | — | — | −6.644 | — |
| Pectate lyase | ||||||
| Solyc05g014000 | Pectate lyase | — | — | — | 2.057 | — |
| Solyc02g093580 | Pectate lyase | — | — | — | −5.644 | — |
| Pectinesterase | ||||||
| Solyc02g080210 | Pectinesterase | — | — | — | 1.674 | — |
| Solyc02g080200 | Pectinesterase | — | — | — | — | 1.546 |
| Solyc06g009190 | Pectinesterase | — | — | — | −3.837 | — |
| Solyc03g083770 | Pectinesterase | — | — | — | −4.644 | — |
| Solyc01g079180 | Pectinesterase | — | — | — | 1.669 | — |
Functional characterization of the osmotin-like protein (OLP) PR-5
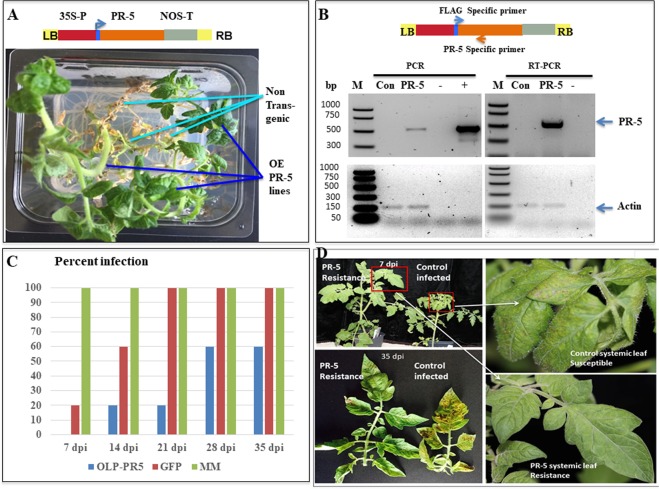
Based on the transcriptome analysis in the present study, several defense-related genes were induced in the Sw-7 line, including the pathogenesis-related family osmotin-like protein (OLP, a PR-5 protein). To gain a better understanding of the role of this gene in the Sw-7 resistance response, the OLP gene (PR-5) was chosen for evaluation through over-expression in the susceptible tomato cultivar ‘Moneymaker’. We initiated an Agrobacterium transformation with ~1,602 explants (leaf-disc) of ‘Moneymaker’, which resulted in producing ~50 plantlets in the selection media, from which we recovered 27 rooted plants. Among them, one T0 line was selected for multiplication by rooted cuttings and used for virus inoculation.
Using T0 transgenic plants expressing OLP-PR5 and those with GFP as a negative control, we compared levels of resistance to mechanical inoculation of TSWV in a greenhouse (Fig. 3). The typical disease symptoms of TSWV infection were observed as early as 7–14 dpi on the non-transformed ‘Moneymaker’ (MM) plants or transgenic plants with GFP, but transgenic OLP-PR5 plants showed no visible symptoms (Fig. 3). At 7 dpi, real-time RT-PCR detected the presence of TSWV on 100% of the control MM plants and 20% of GFP plants. In the case of OLP-PR5 plants, virus infection was delayed for at least one week and detected in only 20% of test plants at 14 dpi. At 21 dpi, 100% of control plants were infected, but only 20% of OLP-PR5 plants tested positive for TSWV. Likewise, when the bioassay concluded at 35 dpi, still only 60% of OLP-PR5 test plants were infected (Fig. 3).
Figure 3.
Functional validation of a candidate resistance gene PR-5 (OLP). (A) PR-5 gene was inserted between 35S and NOS terminator with an N-terminus FLAG tag. Transgenic plant lines regenerated on media containing phosphinothricin, but non-transformed plants could not survive (Top left). (B) PCR confirmation showed the presence of transgene (PR-5) and RT-PCR revealing the expression of transgene (PR-5) with positive (+), negative (−) controls and control (GFP) plants. Actin serves as the internal control in both cases for PCR reactions. (C) Percent of test plants infected as evaluated weekly over five weeks post inoculation on transgenic plants expressing OLP-PR5 gene, transgenic plants expressing GFP, and non-transformed ‘Moneymaker’ plants. (D) Evaluation of transgenic lines with resistance to TSWV: PR-5 over-expressing line (OLP-PR5) showed resistance to TSWV with no visible symptoms, whereas control plants (non-transformed Moneymaker) displayed chlorosis and necrotic spots upon TSWV inoculation.
Discussion
Using a near isogenic line generated through five backcrosses to the recurrent parental line, Fla. 8059, we developed a TSWV-resistant Sw-7 line, which has a highly similar genetic background (97.125%) to the susceptible parental line (Hutton et al., unpublished). Therefore, differential gene expression profiles identified between Sw-7 and S-line would be most likely to have resulted from their differential responses of the Sw-7 locus to the TSWV infection. Although TSWV is a thrips-transmitted virus, it can also infect tomato via mechanical inoculation19. In the current study, we have followed the standard mechanical inoculation protocol established for TSWV in tomato. Our experiments revealed the effectiveness of the mechanical inoculation, since parallel inoculations on both S-line and Sw-7 line plants showed similar virus load on the inoculated leaves at 4 dpi, but this load increased over time in the S-line plants relative to Sw-7 plants (Supplementary Table S1).
By analyzing differential gene expression profiles between the Sw-7 line and the S-line, we achieved a broad dynamic view of global gene expression and identified a number of induced defense-related genes in the Sw-7 line. Interestingly, relatively fewer genes exhibited differential expression during the early virus infection stages (59 and 40 DEGs at 4 and 7 dpi, respectively). Many more genes with differential expression were observed at 21 dpi (836 DEGs) and 35 dpi (234 DEGs), at which time points visible disease symptoms had appeared on the susceptible S-line plants, but little to no symptoms were observed on the Sw-7 line plants. Therefore, it is not surprising that the majority of DEGs at 21 dpi and 35 dpi were down-regulated in the Sw-7 line. It is likely that at the peak of a virus infection, greater engagement of virus and host plant interactions had occurred in the susceptible S-line plants. For the resistant Sw-7 plants, a greater proportion of genes with stronger expression at later time points were related to photosynthesis (13/14 genes) (Supplementary Table S3). In contrast, the inoculated susceptible S-line plants experienced significant changes in gene expression related to a variety of general immune system and defense response pathways, including phytohormone synthesis. Therefore, based on the various gene expression patterns, we were able to group and classify certain genes or gene families which were induced in the Sw-7 line and associated with positive regulation of virus resistance. On the other hand, another group of genes or gene families were induced in the S-line and were likely involved in symptom induction as well as in defending plants for survival from the virus infection. Finally, a third group of genes and gene families had split roles, some members were induced in the Sw-7 line and others in the S-line. Therefore, these genes might play dual roles in both positive and negative influence on virus resistance (Supplementary Fig. S3).
Transcription factors are proteins that control the rate of transcription, once bound to a specific DNA sequence. In the current study, some transcription factors were up-regulated, whereas many others were under down-regulated in the Sw-7 line. These results were in a general agreement with the transcriptome profiling of bean common mosaic virus (BCMV) infection in common bean20. The expression of certain genes could be dynamic or zig-zag, as shown here in the expression of the TF Cycloidea which is profiled as down-regulated at 4 dpi and upregulated at 14 dpi. At this stage, we are unsure of its function in responding to the virus infection, which would need to be further characterized. Zinc finger proteins have been shown to play a key role in disease resistance, particularly in virus resistance21. Interestingly, a large number of zinc finger proteins were induced in the S-line, at the time when plants began to show disease symptoms. This indicates a likely stronger antiviral response in the infected plants, which could have been defeated by viral-pathogenicity factors leading to enhanced symptoms.
Phytohormones are signal molecules produced within the plant cells that regulate plant growth and development22. Plant hormones can vary as a response to pathogen infection. During virus infection, many plant hormone-signaling molecules are either suppressed or induced, which, in turn, affect normal plant growth, resulting in disease-like appearance, such as developmental defects and/or plant stunting23. Auxins have key roles in determining patterns of plant development and growth, which has four families: glutathione S-transferases, auxin homeostasis proteins like GH3, SAUR genes, and Aux/IAA. In Arabidopsis, auxin (Aux/IAA) mutant constitutively represses auxin response that leads to suppression of plant growth24. Upon TSWV infection, the S-line plants showed a reduction in plant growth, which corresponded with the reduction in auxin pathway gene expression. In studying the mechanism of resistance to TYLCV, Yang and colleagues25 identified a MADS-box transcription factor as one of the candidate genes for Ty-2 resistance in tomato. Interestingly, in the present study, MADS-box genes were induced in the Sw-7 resistance to TSWV (Supplementary Fig. S3), implicating a possible involvement of a MADS-box gene in Sw-7 resistance.
Protein kinases (PKs) play a major role in disease resistance through phosphorylation of the interacting proteins to trigger active or functional processes. Although there are only a small number of PKs induced in the Sw-7 plants, previous studies26,27 have shown that a Pto-like serine/threonine kinase protein26 was induced in tomato with resistance to a bacterial disease. A large number of PK genes induced in the S-line during the virus infection may play a function by activating currently unknown susceptible host factors or downstream signaling components to promote symptom expression. A higher number of PK genes induced in the S-line plants pointed to a likely stronger antiviral response in the infected plants as they attempt to fight back for survival from the virus infection.
Our analysis revealed the induction of 10 microRNAs in the S-line. Non-coding RNAs (ncRNAs), including microRNAs and long ncRNA, play key roles in regulating gene expression in plants and animals28. Such induction may result from a physical binding (sequester) of the TSWV-encoded suppressor protein (NSs) to microRNAs, leading to an elevated expression of the target genes and as a consequence enhanced virus accumulation in the S-line.
NBS-LRRs are a major category of disease resistance genes (R-genes) in plants, which have been classified into two sub-families: TIR-NBS-LRR and CC-NBS-LRR29. Typically, there are hundreds of diverse NBS-LRR genes in a plant genome30. However, in the current study, only two NBS-LRR genes showed differential expression, both down-regulated in the Sw-7 line at 21 dpi, suggesting NBS-LRR genes are not likely the candidate resistance gene for Sw-7 against TSWV infection.
Induction of PR genes leads to local and systemic defense gene activation, which could restrict virus movement. Previous study revealed PR proteins could be involved in resistance against fungi, bacteria and viruses31. A recent report demonstrated that TSWV-derived siRNAs can effectively silence the host transcripts, such as ERF, NBS-LRR class R-genes and heat stress transcription factors32. In addition, viroid infection in tomato revealed viroid triggered immune responses, in particular, induction of host calcium-dependent protein kinases (CDPKs) PR1 and WRKY33. Defensins are small cysteine-rich basic proteins found in animals and plants that function as host defense peptides against pathogens (including fungi, bacteria and viruses) and are considered part of the innate immune response34. Nodulins are also among the PR protein family of genes and are important for transport of nutrients, amino acids and hormones for plant development, as well as for pathogen fitness in host colonization35. Nodulins are also considered to be resistance marker proteins induced by plants in response to pathogenic bacterial infection36. In the current study, PR-1 was induced in the Sw-7 line, which might play an important role in resistance against TSWV infection. This discovery is important, as PR-1 is a marker gene for disease resistance and it utilizes callose induction and deposition in the cell wall to restrict virus cell-to-cell movement37. Tomato and tobacco PR-1 proteins are also shown to have an antifungal activity against Phytophthora infestans38. The glycine/proline rich proteins have crucial roles in pathogen resistance by inducing PR proteins. Previous studies have demonstrated that glycine-rich proteins play a role in lignin biosynthesis and/or deposition39, and more importantly in enhancing callose deposition in the cell wall to block virus spread40. It is possible that the mechanism of resistance by Sw-7 is through blocking or slowing down virus cell-to-cell or systemic movement. This hypothesis is also supported by our quantitative measurement of virus titers in the inoculated leaves of both Sw-7 and S-line plants, which showed similar levels of virus titer in early infection stage at 4 dpi, but a gradual reduction in titer in systemic leaves of the Sw-7 line plants relative to S-line plants (Supplementary Table S1).
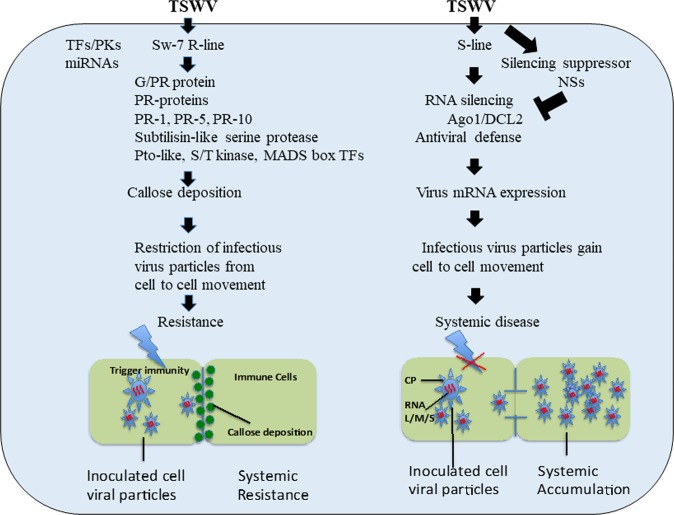
Interestingly, another member of PR proteins, OLP from PR-5, was also induced. Induction of an OLP in plants in response to a viroid pathogen41 has been demonstrated, and PR-5 also plays a defense role against the fungus P. infestans42. Previous studies have also demonstrated that transgenic plants overexpressing OLP are tolerant to other stress factors such as salt, drought, cold, and bacterial and fungal pathogens42–48. Although PR-5 is involved in multiple bacterial/fungal resistance, its involvement in virus resistance has not been characterized. Given the high induction in gene expression at 21 dpi when the susceptible plants were at the peak of showing disease symptoms, we suspected that it may also be involved in Sw-7 resistance to TSWV. To investigate its functional role in the defense response of Sw-7 against TSWV, we overexpressed OLP into a susceptible tomato line. Interestingly, resistance evaluation demonstrated that the over-expression transgenic plants showed moderate resistance to TSWV infection in comparison to the control plants. We propose a Sw-7 resistance model which involves OLP-PR5 to restrict virus movement from cell to cell through induction of callose deposition in the cell wall, resulting in virus resistance to TSWV (Fig. 4). Although it is quite clear that PR-5 was involved in the Sw-7 resistance to TSWV, it is not likely the actual resistance gene since over-expression transgenic plants did not offer the same level of resistance as its natural parent. Therefore, the Sw-7 gene (genes) remained to be identified. The discovery in association of a PR-5 gene for Sw-7 resistance against TSWV would offer an opportunity in future studies to determine whether PR-5 and Sw-7 have a direct or indirect interaction. Our future studies will also involve characterization of other identified PR-related candidate genes as well.
Figure 4.
A schematic model illustrating the predicted mechanisms of virus resistance to TSWV in Sw-7 tomato plants or of symptom expression in the susceptible (S) plants. The Sw-7 resistance requires defense-related signaling molecules, including pathogenesis-related 1 (PR-1) protein, pathogenesis-related 5 (PR-5) (osmotic-like protein), glycine/proline rich protein (GRP), nodulin (PR-10), Pto-like R-gene (bacterial resistance), MADS box transcription factors (candidate Ty-2 gene), and subtilisin serine protease, all of which showed elevated expression in the Sw-7 line relative to S-line. The potential functional roles of the above stated genes, and signaling pathways including GRP-triggered PR proteins, are to actively communicate to neighboring cells, resulting in callose, lignin, and suberin deposition to the cell wall and leading to restricted cell-to-cell movement of TSWV. This in turn leads to the resistance phenotype in the Sw-7 plants. For the susceptible response in the S-line, we speculate that the virus-encoded molecular factors would suppress the host immune pathways, leading to TSWV replication, transcription and translation. Abundance of viral RNA accumulation in the cells would trigger the expression of RNA silencing pathway genes in the S-line, including Argonaute 1 (Ago1) and Dicer-like 2 (DCL 2), resulting in antiviral defense. In the meantime, TSWV-encoded silencing suppressor protein (NSs) would suppress (sequester) the host antiviral defense pathway, leading to over-accumulation of viral particles. This in turn results in the opening of the cell wall/plasmodesmata to virus cell-to-cell and systemic movement, producing the disease phenotype.
Materials and Methods
Plant materials and generation of a near-isogenic line of Sw-7 with resistance to TSWV
Plant materials used for transcriptome experiments included the TSWV-susceptible inbred line, Fla. 805949, and a Sw-7 near-isogenic line (Sw-7 NIL) in the Fla. 8059 background. The NIL was developed by crossing Fla. 8059 with the Sw-7 donor line, Fla. 8516, followed by five backcrosses to the recurrent parent, Fla. 8059. Selection for the Sw-7 introgression was accomplished using a linked SCAR marker designed from the CT99 RFLP50. CT99 is located within the interval on chromosome 12 to which the Sw-7 locus is mapped51, and this location has been confirmed by recent fine-mapping (Hutton, unpublished data). Primer sequences (5′ to 3′) for the SCAR marker are F: GAAGGTGCCGACGGTGTA, R: AGGAATCAAGGTAAACCACCA. Amplicon sizes are 285 bp for the Solanum chilense allele and 241 bp for the S. lycopersicum allele. Seeds of the Sw-7 NIL were produced by self-pollinating plants from the BC5F1, and then saved from BC5F2 plants that were homozygous for the Sw-7 introgression.
TSWV culture collection and maintenance
TSWV culture was collected from local infected tomato plants in Charleston, SC, and maintained on tomato ‘Moneymaker’ plants in an insect-proof bug-dorm in an environment-controlled greenhouse maintained at 26 °C with 14 h daylight and 10 h dark. Systemic infected leaf tissues were collected and tested for the presence of TSWV using enzyme-linked immunosorbent assay (ELISA) following the manufacturer’s instructions (Agdia, Elkhart, IN).
Mechanical inoculation of TSWV on tomato
Seeds of the Sw-7 NIL and its recurrent parent Fla. 8059 were germinated in separate plastic pots (10 cm) filled with Profession Growing Mix (Sungro, Agawam, MA) in an environment-controlled greenhouse. Mechanical inoculation was conducted on tomato seedlings at the 2–3 leaf stage19. Virus inoculum was prepared by grinding a small piece of TSWV-infected leaf tissue in a plastic bag with addition of tissue extraction buffer [0.01 M sodium phosphate buffer (pH 7.0) with 0.4% β-mercaptoethanol] in a final dilution 1:10 (w/v). Mechanical inoculation was performed by using a cotton swab (Q-tips) soaked with the inoculum and gently rubbed on the surface of tomato leaves that had been dusted with Carborundum powder (600 mesh). The inoculated plants were maintained in a greenhouse to monitor for symptom expression.
Sample collection and real time quantification of TSWV
Virus-inoculated plants were maintained and monitored for symptom expression up to 35 dpi. TSWV-inoculated leaf tissues were collected at 4, 7, 14, 21 and 35 dpi, respectively. For sample collection, a small piece of leaf tissue (500 mg) from inoculated leaf (Sw-7 and S-line) was collected at 4 dpi and subsequent collections were performed on systemic young leaf at other time points (7, 14, 21 and 35 dpi). Inoculated plants were tested to confirm for the presence and concentration of TSWV using real-time RT-qPCR52. RT-qPCR was performed using the TaqMan probe 5′HEX-AAATCTAAGATTGCTTCCCACCCTTTGATTCAA-BHQ, with forward primer 5′GCTTGTTGAGGAAACTGGGAATT and reverse primer 5′AGCCTCACAGACTTTGCATCATC52 located in the N gene of TSWV and Takara One Step PrimeScript RT-PCR Kit (Clontech, USA) following manufacturer’s instructions. The One-step RT-qPCR reaction was carried out on a Stratagene MX3000P Real-Time PCR machine (Agilent, USA), under the following conditions: 50 °C for 30 min, denaturation at 95 °C for 2 min, followed by 40 cycles of 95 °C for 1 min and 55 °C for 30 sec.
Generation of plant transformation constructs
To functionally characterize some defense-related genes that are potentially contributing to TSWV resistance in the Sw-7 line, tomato OLP (PR5) gene was selected for evaluation. A synthetic gene (OLP or GFP) was designed (IDT, Coralville, IA) and inserted into pENTR-D TOPO vector and transformed into Top 10 Chemically competent cells (Invitrogen, USA). Plasmid DNA with inserts from selected colonies were confirmed through Sanger sequencing. Construct was recombined with Gateway vector PEG101 using clonase (Invitrogen, USA) between the Cauliflower mosaic virus (CaMV) 35S promoter and nopaline synthase (NOS) terminator. The sequence confirmed OLP and GFP inserted binary vectors were mobilized into Agrobactrium tumefaciens strain LBA4404 by electroporation and selected on YM agar containing kanamycin for PEG101 selection and Streptomycin for Agrobacterium.
Agrobacterium-mediated transformation and confirmation of transgenic plants
We followed an efficient protocol of tomato transformation and selection of transgenic plants53 with some modifications. Briefly, seeds of tomato “Moneymaker” after sterilization were germinated on a MS medium agar plate. Leaves and cotyledons of 2- to 3-week-old ‘Moneymaker’ seedlings were cut under sterile conditions to make small explants of about 2–3 mm. These explants were incubated for 10–15 min with the Agrobacterium suspension culture (infection solution). After incubation, explants were quickly wiped on a sterile filter paper and then transferred to a co-cultivation medium. After two days (48 h) of co-cultivation in the dark at 24 C, the explants were transferred to Petri dishes containing microshoot induction (MI) medium. After 3 weeks, the callus-forming explants that produced microshoots were cut and transferred to the shoot elongation medium. Shoots of 1/1.5 cm long after approximately three weeks of growth were cut and transferred into the rooting medium. Rooted plantlets were transferred into soil and maintained at 25 °C in a growth chamber. Confirmation of transgenic events in the regenerated plants was tested after 3–4 weeks of growth. The transgenic plants were self-pollinated. The T1 seeds were germinated on a MS basal medium containing 1 mg/L Phosphinotricin for selection. Surviving germinated seedlings were transferred to pots containing sterile soil and maintained in a glasshouse at 28–29 °C and 80–90% relative humidity. Transgene was confirmed by gene specific PCR and gene expression confirmed by RT-PCR using FLAG specific 5′- forward primers (KL17-151 FLAG-F:GACTACAAAGACGATGACGACA) and OLP specific reverse primers (KL14-397 OLP-1R:GCAACACATTGAATTGGATGACATT). For the internal control, an actin primer pair (forward primer KL17-071 03g078400F: TTGCTGGTCGTGACCTTACT and reverse primer KL17-072 03g078400R: TGCTCCTAGCGGTTTCAAGT) were used.
Evaluation of OLP-PR5 transgenic tomato plants for resistance to TSWV
To evaluate transgenic plants over-expressing the OLP-PR5 gene for their resistance against TSWV, five rooted plants (in 4–5 leaf stage) regenerated from PCR-confirmed OLP-PR5 transgenic T0 lines, along with similarly developed transgenic plants expressing GFP or non-transgenic ‘Moneymaker’ plants were mechanically inoculated with TSWV using the same method as described above. In addition to observe symptom expression on the inoculated plants, virus titers accumulated on systemic leaves in each of the test plants were also measured quantitatively using real-time RT-qPCR as described above52 using leaf tissue samples collected on 7, 14, 21, 28 and 35 dpi (Supplementary Table S4).
RNA extraction
Total RNA was extracted using TRIzol reagents (ThermoFisher Scientific, USA) from 500 mg of freshly collected leaf tissue in aplastic sample extraction bag and homogenized using Homex-6 homogenizer (BioReba, Swizerland) following manufacturer’s instructions. The DNase I-treated RNA was resuspended in NANOpure® water and its concentration measured with a NanoDrop spectrophotometer (ThermoFisher Scientific, USA). The cleaned DNA-free high quality RNA was also confirmed in a 1X bleach gel54.
RNA-Seq library preparation, Illumina sequencing and data analysis
Strand-specific RNA-Seq libraries were constructed using the protocol described55 and sequenced on an Illumina HiSeq 2500 system using the single-end 100-bp mode. Raw RNA-Seq reads were processed to remove adaptor and low quality sequences using Trimmomatic56. RNA-Seq reads were then aligned to the ribosomal RNA database57 using Bowtie58 and the mapped reads were discarded. The remaining high-quality cleaned reads were aligned to the tomato Heinz genome (The Tomato Genome Consortium, 2012) using HISAT59. Following alignments, raw counts for each tomato gene were derived and normalized to reads per kilobase of exon model per million mapped reads (RPKM). Raw counts were fed to the DESeq package60 to identify genes differentially expressed between the Sw-7 line and the S-line at each time point. Genes with adjusted p values less than 0.05 and fold changes greater than or equal to 1.5 were identified as DEGs.
The identified DEGs were uploaded into the Plant MetGenMAP system61 to identify enriched gene ontology terms, functional classifications, and biochemical pathways. Overlapping analysis of DEGs was performed with an online tool (http://bioinformatics.psb.ugent.be/webtools/Venn/). Summary plots of GO enrichment were created using Revigo62. The Tomato Functional Genomics Database63 and the iTAK database64 were used to identify plant transcription factors, receptor-like kinases, and miRNA targets. Identification of other genes of interest was performed using standalone BLAST65 by comparing homologs to genes of interest from Arabidopsis and S. lycopersicum in conjunction with utilizing annotated GO terms of tomato genes66 and manual annotation.
Supplementary information
Acknowledgements
We thank Andrea Gilliard, Deanna Dong, Louis William and Tyler Devaney for their excellent technical assistance. This work was supported in part by the USDA, National Institute of Food and Agriculture SCRI 2012-01507-229756 to K.S.L., Z.F. and J.W.S.
Author Contributions
K.S.L. and Z.F. conceived the idea and revised the manuscript; C.P. performed experiments, conducted data analysis and drafted the manuscript. Q.M. and K.S.S. performed RNA-Seq data and bioinformatics analysis. R.S., S.F.H. and J.W.S. developed near-isogenic line and tomato breeding lines used in this study.
Data Availability
RNA-Seq datasets were submitted to SRA database with the accession No. SRP119544.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zhangjun Fei, Email: zf25@cornell.edu.
Kai-Shu Ling, Email: kai.ling@ars.usda.gov.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-44100-x.
References
- 1.German TL, Ullman DE, Moyer WJ. Tospoviruses: diagnosis, molecular biology, phylogeny, and vector relationships. Annu. Rev. Phytopathol. 1992;30(315):348. doi: 10.1146/annurev.py.30.090192.001531. [DOI] [PubMed] [Google Scholar]
- 2.De Haan P, Wagemakers L, Peters D, Goldbach R. The S RNA segment of tomato spotted wilt virus has an ambisense character. J. Gen. Virol. 1990;71:1001–1007. doi: 10.1099/0022-1317-71-5-1001. [DOI] [PubMed] [Google Scholar]
- 3.Goldbach R, Peters D. Possible causes of the emergence of tospovirus diseases. Sem. Virol. 1994;5:113–120. doi: 10.1006/smvy.1994.1012. [DOI] [Google Scholar]
- 4.Rotenberg D, Jacobson AL, Schneweis DJ, Whitfield AE. Thrips transmission of tospoviruses. Curr. Opin. Virol. 2015;15:80–89. doi: 10.1016/j.coviro.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 5.Pappu HR, Jones RAC, Jain RK. Global status of Tospovirus epidemics in diverse cropping systems: Successes gained and challenges that lie ahead. Virus Res. 2009;141:219–236. doi: 10.1016/j.virusres.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 6.Samuel, G., Bald, J. G. & Pitman, H. A. Spotted wilt of tomatoes. Aust. Con. Sci. Ind. Res. Bull. 44 (1930).
- 7.Finlay KW. Inheritance of spotted wilt resistance in the tomato. II. Five genes controlling spotted wilt resistance in four tomato types. Aust. J. Sci. Res. 1953;6:153–163. [PubMed] [Google Scholar]
- 8.Paterson RG, Scott SJ, Gergerich RC. Resistance in two Lycopersicon species to an Arkansas isolate of tomato spotted wilt virus. Euphytica. 1989;43:173–178. doi: 10.1007/BF00037910. [DOI] [Google Scholar]
- 9.Price DL, Memmott FD, Scott JW, Olson SM, Stevens MR. Identification of molecular markers linked to a new tomato spotted wilt virus resistance source in tomato. Tomato Genet Coop. 2007;57:35–36. [Google Scholar]
- 10.Saidi M, Warade SD. Tomato breeding for resistance to tomato spotted wilt virus (TSWV): an overview of conventional and molecular approaches. Czech J. Genet. Plant Breed. 2008;44:83–92. doi: 10.17221/47/2008-CJGPB. [DOI] [Google Scholar]
- 11.Stevens MR, Scott SJ, Gergerich RC. Inheritance of a gene for resistance to tomato spotted wilt virus (TSWV) from Lycopersicon peruvianum Mill. Euphytica. 1992;59:9–17. [Google Scholar]
- 12.Soler S, Cebolla-Cornejo J, Nuez F. Control of diseases induced by tospoviruses in tomato: an update of the genetic approach. Phytopathol Mediterr. 2003;42:207–219. [Google Scholar]
- 13.Ciuffo M, Finetti-Sialer MM, Gallitelli D, Turina M. First report in Italy of a resistance-breaking strain of tomato spotted wilt virus infecting tomato cultivars carrying the Sw-5 resistance gene. Plant Pathol. 2005;54:564. doi: 10.1111/j.1365-3059.2005.01203.x. [DOI] [Google Scholar]
- 14.Batuman O, et al. First report of a resistance-breaking strain of tomato spotted wilt virus infecting tomatoes with the Sw-5 tospovirus-resistance gene in California. Plant disease. 2017;101:637. doi: 10.1094/PDIS-09-16-1371-PDN. [DOI] [PubMed] [Google Scholar]
- 15.Lopez C, et al. Evolutionary analysis of tomato Sw-5 resistance-breaking isolates of tomato spotted wilt virus. J Gen Virol. 2011;92:210–5. doi: 10.1099/vir.0.026708-0. [DOI] [PubMed] [Google Scholar]
- 16.Rosello S, Ricarte B, Diez MJ, Nuez F. Resistance to tomato spotted wilt virus introgressed from Lycopersicon peruvianum in line UPV 1 may be allelic to Sw-5 and can be used to enhance the resistance of hybrids cultivars. Euphytica. 2001;119:357–367. doi: 10.1023/A:1017506213974. [DOI] [Google Scholar]
- 17.Canady MA, Stevens MR, Barineau MS, Scott JW. Tomato spotted wilt virus (TSWV) resistance in tomato derived from Lycopersicon chilense Dun. LA 1938. Euphytica. 2001;117:19–25. doi: 10.1023/A:1004089504051. [DOI] [Google Scholar]
- 18.Scott, J. W., Hutton, S. F., Olson, S. M. & Stevens, M. R. Spotty results in our Sw-7 tomato spotted wilt virus research. 2011 Tomato Disease Workshop meeting abstract, http://vegetablemdonline.ppath.cornell.edu/TDW/Presentations/11%20Scott_TDW_2011.pdf (2011).
- 19.Mitter N, Koundal V, Williams S, Pappu H. Differential expression of tomato spotted wilt virus-derived viral small RNAs in infected commercial and experimental host plants. PLoS One. 2013;8:e76276. doi: 10.1371/journal.pone.0076276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin K, Singh J, Hill JH, Whitham SA, Cannon SB. Dynamic transcriptome profiling of bean common mosaic virus (BCMV) infection in common bean (Phaseolus vulgaris L.) BMC Genomics. 2016;17:613. doi: 10.1186/s12864-016-2976-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koshino-Kimura Y, et al. Construction of plants resistant to TYLCV by using artificial zinc-finger proteins. Nucleic Acids Symp Ser (Oxf). 2009;53:281–282. doi: 10.1093/nass/nrp141. [DOI] [PubMed] [Google Scholar]
- 22.Hanssen IM, et al. Differential tomato transcriptomic responses induced by pepino mosaic virus isolates with differential aggressiveness. Plant Physiol. 2011;156:301–318. doi: 10.1104/pp.111.173906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whitham SA, Yang C, Goodin MM. Global impact: elucidating plant responses to viral infection. Mol. Plant Microbe Interact. 2006;19:1207–1215. doi: 10.1094/MPMI-19-1207. [DOI] [PubMed] [Google Scholar]
- 24.Gray WM. Hormonal regulation of plant growth and development. PLoS Biol. 2014;2:e311. doi: 10.1371/journal.pbio.0020311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang X, et al. Fine mapping of the tomato yellow leaf curl virus resistance gene Ty-2 on chromosome 11 of tomato. Mol Breed. 2014;34:749–760. doi: 10.1007/s11032-014-0072-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin GB, et al. Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science. 1993;262:1432–1436. doi: 10.1126/science.7902614. [DOI] [PubMed] [Google Scholar]
- 27.Tang X, et al. Initiation of plant disease resistance by physical interaction of AvrPto and Pto kinase. Science. 1996;274:2060–2063. doi: 10.1126/science.274.5295.2060. [DOI] [PubMed] [Google Scholar]
- 28.Gomes AQ, Nolasco S, Soares H. Non-coding RNAs: Multi-tasking molecules in the cell. Int. J. Mol. Sci. 2013;14:16010–16039. doi: 10.3390/ijms140816010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McHale L, Tan X, Koehl P, Michelmore RW. Plant NBS-LRR proteins: adaptable guards. Genome Biology. 2006;7:212. doi: 10.1186/gb-2006-7-4-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andolfo. G, et al. Defining the full tomato NB-LRR resistance gene repertoire using genomic and cDNA RenSeq. BMC Plant Biology. 2014;14:120. doi: 10.1186/1471-2229-14-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elvira MI, Galdeano MM, Gilardi P, García-Luque I, Serra MT. Proteomic analysis of pathogenesis-related proteins (PRs) induced by compatible and incompatible interactions of pepper mild mottle virus (PMMoV) in Capsicum chinense L3 plants. J Exp Bot. 2008;59:1253–1265. doi: 10.1093/jxb/ern032. [DOI] [PubMed] [Google Scholar]
- 32.Ramesh SV, Williams S, Kappagantu M, Mitter N, Pappu HR. Transcriptome-wide identification of host genes targeted by tomato spotted wilt virus-derived small interfering RNAs. Virus Res. 2017;238:13–23. doi: 10.1016/j.virusres.2017.05.014. [DOI] [PubMed] [Google Scholar]
- 33.Zheng Y, Ding B, Fei Z, Wang Y. Comprehensive transcriptome analyses reveal tomato plant responses to tobacco rattle virus-based gene silencing vectors. Sci Rep. 2017;7:9771. doi: 10.1038/s41598-017-10143-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomma BPHJ, Cammue BPA, Thevissen K. Plant Defensins. Planta. 2002;216:193–202. doi: 10.1007/s00425-002-0902-6. [DOI] [PubMed] [Google Scholar]
- 35.Denance N, Szurek B, Noel LD. Emerging functions of nodulin-like proteins in non-nodulating plant species. Plant Cell Physiol. 2014;55:469–74. doi: 10.1093/pcp/pct198. [DOI] [PubMed] [Google Scholar]
- 36.Gamas P, de Billy F, Truchet G. Symbiosis-specific expression of two Medicago truncatula nodulin genes, MtN1 and MtN13, encoding products homologous to plant defense proteins. Mol Plant Microbe Interact. 1998;11:393–403. doi: 10.1094/MPMI.1998.11.5.393. [DOI] [PubMed] [Google Scholar]
- 37.Van Loon LC, Van Strien EA. The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol. Mol. Plant Pathol. 1999;55:85–97. doi: 10.1006/pmpp.1999.0213. [DOI] [Google Scholar]
- 38.Niderman T, et al. Pathogenesis-related PR-1 proteins are antifungal, isolation and characterization of three 14-kilodalton proteins of tomato and of a basic PR-1 of tobacco with inhibitory activity against Phytophthora infestans. Plant Physiol. 1995;108:17–27. doi: 10.1104/pp.108.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen AP, et al. Root and vascular tissue-specific expression of glycine-rich protein AtGRP9 and its interaction with AtCAD5, a cinnamyl alcohol dehydrogenase, in Arabidopsis thaliana. J Plant Res. 2007;120:337–343. doi: 10.1007/s10265-006-0058-8. [DOI] [PubMed] [Google Scholar]
- 40.Ueki S, Citovsky V. The systemic movement of a tobamovirus is inhibited by a cadmium-ion-induced glycine-rich protein. Nat. Cell Biol. 2002;4:478–486. doi: 10.1038/ncb806. [DOI] [PubMed] [Google Scholar]
- 41.Ruiz-Medrano R, Jimenez-Moraila B, Herrera-Estrella L, Rivera-Bustamante RF. Nucleotide sequence of an osmotin-like cDNA induced in tomato during viroid infection. Plant Mol. Biol. 1992;20:1199–1202. doi: 10.1007/BF00028909. [DOI] [PubMed] [Google Scholar]
- 42.Zhu B, Chen TH, Li PH. Expression of three osmotin-like protein genes in response to osmotic stress and fungal infection in potato. Plant Mol Biol. 1995;8:17–26. doi: 10.1007/BF00042034. [DOI] [PubMed] [Google Scholar]
- 43.Evers D, Overney S, Simon P, Greppin H, Hausman JF. Salt tolerance of Solanum tuberosum L. overexpressing an heterologous osmotin-like protein. Biol Plant. 1999;42:105–112. doi: 10.1023/A:1002131812340. [DOI] [Google Scholar]
- 44.Vasavirama K, Kirti PB. Increased resistance to late leaf spot disease in transgenic peanut using a combination of PR genes. Funct. Integr. Genomics. 2012;12:625–634. doi: 10.1007/s10142-012-0298-8. [DOI] [PubMed] [Google Scholar]
- 45.Choi DS, Hong JK, Hwang BK. Pepper Osmotin-like protein 1 (CaOSM1) is an essential component for defense response, cell death, and oxidative burst in plants. Planta. 2013;238:1113–1124. doi: 10.1007/s00425-013-1956-3. [DOI] [PubMed] [Google Scholar]
- 46.Weber RLM, et al. Expression of an osmotin-like protein from Solanum nigrum confers drought tolerance in transgenic soybean. BMC Plant Biol. 2014;14:343. doi: 10.1186/s12870-014-0343-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chowdhury S, Basu A, Kundu S. Cloning, characterization and bacterial over-expression of an osmotin-Like protein gene from Solanum nigrum L. with antifungal activity against three necrotrophic fungi. Mol. Biotechnol. 2015;57:371–381. doi: 10.1007/s12033-014-9831-4. [DOI] [PubMed] [Google Scholar]
- 48.Kumar SA, et al. Beyond just being foot soldiers – osmotin like protein (OLP) and chitinase (Chi11) genes act as sentinels to confront salt, drought, and fungal stress tolerance in tomato. Environ. Exp. Bot. 2016;132:53–65. doi: 10.1016/j.envexpbot.2016.08.007. [DOI] [Google Scholar]
- 49.Scott JW, et al. Fla.8153 Hybrid Tomato; Fla.8059 and Fla. 7907 Breeding lines. HortScience. 2008;43:2228–2230. doi: 10.21273/HORTSCI.43.7.2228. [DOI] [Google Scholar]
- 50.Tanksley SD, et al. High density molecular linkage maps of the tomato and potato genomes. Genetics. 1992;132:1141–60. doi: 10.1093/genetics/132.4.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stevens, M. R. Localization and mapping of Sw-7, a tomato spotted wild virus resistance gene. Proc. 42nd Tomato Breeders Roundtable, Sacramento, CA, http://tgc.ifas.ufl.edu/2009/Stevens%20SW7%20mapping.pdf (2009).
- 52.Roberts CA, Dietzgen RG, Heelan LA, Maclean DJ. Real-time RT-PCR fluorescent detection of tomato spotted wilt virus. J. Virol. Methods. 2000;88:1–8. doi: 10.1016/S0166-0934(00)00156-7. [DOI] [PubMed] [Google Scholar]
- 53.Khuong TTH, Crete P, Robaglia C, Caffarri S. Optimization of tomato Micro-tom regeneration and selection on glufosinate/Basta and dependency of gene silencing on transgenic copy number. Plant Cell Rep. 2013;32:1441–1454. doi: 10.1007/s00299-013-1456-8. [DOI] [PubMed] [Google Scholar]
- 54.Aranda PS, LaJoie DM, Jorcyk CL. Bleach gel: a simple agarose gel for analyzing RNA quality. Electrophoresis. 2012;33:366–369. doi: 10.1002/elps.201100335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhong S, et al. High-throughput illumina strand-specific RNA sequencing library preparation. Cold Spring Harb. Protoc. 2011;2011:940–949. doi: 10.1101/pdb.prot5652. [DOI] [PubMed] [Google Scholar]
- 56.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Quast C, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucl. Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Joung JG, et al. Plant MetGenMAP: an integrative analysis system for plant systems biology. Plant Physiol. 2009;151:1758–68. doi: 10.1104/pp.109.145169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Supek F, Bosnjak M, Skunca N, Smuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fei Z, et al. Tomato Functional Genomics Database: a comprehensive resource and analysis package for tomato functional genomics. Nucleic Acids Res. 2011;39:D1156–1163. doi: 10.1093/nar/gkq991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zheng Y, et al. iTAK: a program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Molecular Plant. 2016;9:1667–1670. doi: 10.1016/j.molp.2016.09.014. [DOI] [PubMed] [Google Scholar]
- 65.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 66.Fernandez-Pozo N, et al. The Sol Genomics Network (SGN) from genotype to phenotype to breeding. Nucleic Acids Res. 2015;43:D1036–D1041. doi: 10.1093/nar/gku1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-Seq datasets were submitted to SRA database with the accession No. SRP119544.