ABSTRACT
The Bacillus cereus group includes several Bacillus species with closely related phylogeny. The most well-studied members of the group, B. anthracis, B. cereus, and B. thuringiensis, are known for their pathogenic potential. Here, we present the historical rationale for speciation and discuss shared and unique features of these bacteria. Aspects of cell morphology and physiology, and genome sequence similarity and gene synteny support close evolutionary relationships for these three species. For many strains, distinct differences in virulence factor synthesis provide facile means for species assignment. B. anthracis is the causative agent of anthrax. Some B. cereus strains are commonly recognized as food poisoning agents, but strains can also cause localized wound and eye infections as well as systemic disease. Certain B. thuringiensis strains are entomopathogens and have been commercialized for use as biopesticides, while some strains have been reported to cause infection in immunocompromised individuals. In this article we compare and contrast B. anthracis, B. cereus, and B. thuringiensis, including ecology, cell structure and development, virulence attributes, gene regulation and genetic exchange systems, and experimental models of disease.
TAXONOMY: HISTORICAL AND SOCIOECONOMIC ASPECTS
The microorganisms constituting the Bacillus cereus group are Gram-positive low-GC-content bacteria belonging to the phylum Firmicutes. The group of spore-forming, aerobic, facultative anaerobic, rod-shaped bacteria comprises at least eight closely related species: B. anthracis, B. cereus, B. thuringiensis, B. mycoides, B. pseudomycoides, B. weihenstephanensis, B. cytotoxicus, and B. toyonensis (1). With the exception of B. cytotoxicus, which is the most divergent of the group, with a chromosome of 4.085 Mb (2), the genomes of the B. cereus group species are highly conserved, with sizes of 5.2- to 5.9-Mb and very similar 16S rRNA gene sequences.
The definition of these species and the distribution of the strains within them were based on phenotypes and their clinical and economic significance and mainly associated with plasmid content (3, 4). B. anthracis harbors two plasmids, pXO1 (182 kb) and pXO2 (95 kb), carrying the structural genes for the toxin proteins and the biosynthetic genes for capsule formation, respectively (5). The emetic B. cereus strains harbor plasmid pCER270, encoding enzymatic components required for the nonribosomal biosynthesis of the toxin cereulide (6, 7). B. thuringiensis harbors several plasmids encoding a large variety of the insecticidal toxins Cry and Cyt, which form the parasporal crystal characteristic of the species (8–10). The other five species are discriminated by morphological or physiological traits. B. mycoides and B. pseudomycoides have typical rhizoid growth (11, 12). B. weihenstephanensis strains are psychrotolerant. Like the mostly mesophilic members of the B. cereus group, the optimal growth temperature for B. weihenstephanensis is between 25 and 35°C, but the species is distinguished by its ability to grow at temperatures as low as 4°C (13). On the other hand, B. cytotoxicus strains are thermotolerant and can grow in up to 50°C (14). Finally, B. toyonensis was characterized for probiotic properties and is used as such in animal feed (15).
Speciation based on plasmid content and on morphologic or certain physiological features is confusing and can result in subjective conclusions. A recent genomic analysis of 224 strains shows that the B. cereus group can be divided into 5 major clades which do not match the 8 species described above (see Fig. 1). This lack of congruence between genomic-based phylogenetic analysis and specific phenotypes leads to the conclusion that plasmids carrying toxin genes such as the pXO plasmids of B. anthracis and the cry plasmids of B. thuringiensis cannot serve as signatures for species identification (1). As proposed by Helgason and colleagues (16) and by Rasko and coworkers (4), it is more useful to consider the B. cereus group a unique species comprising extremely diverse strains whose properties differ due to plasmid content or gene expression associated with key regulatory genes. For example, the global transcription regulator PlcR is active in B. cereus and B. thuringiensis but inactive in B. anthracis, abolishing the expression of a major regulon in that species (17, 18).
FIGURE 1.
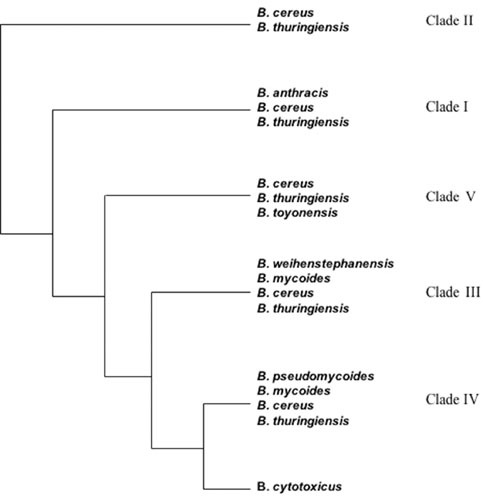
The five major phylogenetic clades of the B. cereus group. Clade I is known as the B. anthracis clade but also includes emetic B. cereus and several clinical B. thuringiensis isolates. Clade II is known as the B. cereus/B. thuringiensis clade and includes most of the commercially used B thurigiensis as well as the B. cereus and B. thuringiensis type strains. Clade III is known as the B. weihenstephanensis clade and comprises most of the psychrotolerant isolates of the B. cereus group. Clade IV includes strains belonging to various B. cereus group species. These clades are based on multilocus sequence typing, amplified fragment length polymorphism, and whole-genome sequence data (429–431). The thermotolerant strains belonging to the species B. cytotoxicus cluster together in a separate group.
While it is arguably appropriate to refer to members of this highly related group of bacteria as B. cereus subsp. cereus, B. cereus subsps. anthracis, B. cereus subsp. thuringiensis, etc., in keeping with current practice, we will refer to the individual subspecies by their traditional names. We focus here on B. anthracis, B. cereus, and B. thuringiensis because of the abundance of research related to their clinical and socioeconomic importance. We first present some unique and overlapping traits of these bacteria.
B. anthracis
B. anthracis, a major scourge of humanity, was first characterized by Koch and Pasteur as the etiologic agent of anthrax. Their investigations of this bacterium in the late 19th century represented early breakthroughs in microbiology and vaccination. Koch’s postulates, criteria designed to establish specific causal relationships between microbes and diseases, were based on studies of B. anthracis. In addition, an attenuated strain of B. anthracis served as the first live bacterial vaccine (19, 20).
Historically, strains harboring pXO1 and/or pXO2 were designated B. anthracis. However, recent reports of closely related species harboring similar plasmids indicate that speciation can be complex. B. cereus strains that harbor pXO1- and pXO2-like plasmids, named B. cereus bv. anthracis, have been isolated as the causative agents of anthrax-like infections in primates in Cameroon and Côte d’Ivoire (21, 22). Similar B. cereus strains that produce the anthrax toxins were identified as the etiological agents of an anthrax-like cutaneous infection and anthrax-like respiratory infections in metalworkers (23–27). The well-studied B. cereus strain G9241, isolated from a welder with a severe anthrax-like respiratory illness, contains two virulence plasmids, pBCXO1 and pBC210 (also known as pBC218), as well as pBClin29, a linear plasmid that harbors cryptic prophage genes. The plasmid pBCXO1 has high similarity to pXO1 and contains the toxin genes pagA, lef, and cya, encoding toxin proteins with amino acid sequences of 96% or greater identity to their counterparts in B. anthracis (24). Plasmid pBC210 encodes a toxin protein paralogue, PA2, and the novel ADP-ribosyltransferase certhrax (24, 28–30). Each plasmid contains genes required for synthesis of a unique polysaccharide capsule; the enzymes needed for hyaluronic acid capsule production are encoded by the hasACB operon on pBCXO1, and the proteins required for elaboration of a putative tetrasaccharide capsule are encoded by the bps locus on pBC210 (24, 31). Interestingly, although B. cereus G9241 and B. anthracis Ames both produce capsule(s) and the anthrax toxin proteins, the virulence of G9241 in rabbits is more similar to that of B. anthracis Sterne strain 34F2 (pXO2–), which produces the toxins but not capsule (32).
B. cereus
B. cereus was first isolated from air in a cow shed and was characterized as a highly motile bacterium, generally appearing as single cells but occasionally forming longer threads and causing rapid liquefaction of gelatin (33). In the second half of the 20th century, B. cereus was recognized as a common food contaminant responsible for two types of poisonings. More recently, B. cereus has been increasingly recognized as an etiological agent of localized wound and eye infections as well as systemic infections (34, 35).
In the 1950s, Hauge showed that consumption of B. cereus-contaminated vanilla sauce could cause diarrhea (36). Since Hauges’ remarkable discovery, two protein toxin complexes (Nhe and Hbl) and a single-protein toxin (CytK) have been described, which are generally thought to contribute to the diarrheal type of food poisoning (37). More recently, several exoproteins with enzymatic activities, such as proteases or phospholipases, have been discussed as putative virulence factors contributing to the diarrheal syndrome (38, 39). Notably, these diarrhea-associated known and putative virulence factors are encoded by chromosome genes. In contrast, genes encoding the biosynthetic machinery for production of the depsipeptide toxin cereulide, which is the causative agent of the emetic type of B. cereus-mediated food poisoning, are on the 270-kb megaplasmid pCER270 (35). In contrast to the enterotoxin genes, which are broadly distributed among the members of the B. cereus group and show a high degree of molecular diversity (40), the cereulide synthetase gene is restricted to genetically closely related strains and shows rather low molecular diversity (41, 42). The ecological and evolutionary mechanisms driving the diversification processes of B. cereus virulence were hitherto poorly understood.
B. thuringiensis
B. thuringiensis was first isolated in 1901 in Japan from infected silkworm larvae (Bombyx mori). The bacterium was identified as the causal agent of the sotto disease (sudden-collapse disease), a lethal infection of this economically important insect (43). Subsequently, Berliner isolated the bacterium from the cadavers of flour moth larvae (Ephestia kuehniella) collected in a mill in Thuringia (44) and named it Bacillus thuringiensis, after the German province.
Based on its entomopathogenic properties, B. thuringiensis was rapidly commercialized and used as a biopesticide for pest control. The insecticidal activity spectrum of the bacterium was initially thought to be restricted to a few lepidopteran species. However, in 1977 discovery of the israelensis strain of B. thuringiensis, which is active against mosquitoes (45), accelerated a search for additional strains active against several species of insects and other invertebrates.
Today, a large number of strains have been shown to have activity against coleopteran insects or nematodes (46, 47). B. thuringiensis-based biopesticides account for approximately 75% of the global bioinsecticide market, representing about 4% of global insecticides (48). Several thousand B. thuringiensis strains have been shown to form crystalline Cry protein inclusions during sporulation, which in many cases are key to the insecticidal or nematicidal activity of the bacteria. However, not all B. thuringiensis isolates that produce Cry proteins have been found to be toxic. It is possible that some Cry proteins are inactive or have unidentified target hosts.
All the insecticidal toxin genes (cry, cyt, and vip) of B. thuringiensis are located on large plasmids (9, 49, 50). Generally, these plasmids are conjugative (51), and the cry genes are frequently found in the vicinity of various mobile genetic elements, such as the transposon Tn4430 and the insertion sequences IS231 and IS232 (8, 52, 53). The localization of the insecticidal toxin genes on elements associated with horizontal gene transfer is likely responsible for the great diversity of these genes and their multiplicity within the B. thuringiensis strains.
ECOLOGY: ENVIRONMENTS AND PATHOGENICITY
The B. cereus group species are endemic soil bacteria that occupy diverse ecological habitats as depicted in Fig. 2 (41, 54). Due to the formation of heat-, UV-, acid-, and desiccation-resistant endospores, the bacteria can persist in a dormant state, making it difficult to elucidate their primary ecological niches. In addition to soil, the species have been isolated from fresh and stored foods, invertebrates, and plants (55–60). Some reports suggest that the B. cereus group species do not germinate and grow in nonsterile soils unless a nutrient source, such as decomposing plants, insects, or animals is present (61–64). Reports of the isolation of B. cereus associated with plants may even point toward an endophytic lifestyle as an alternative niche for B. cereus (65, 66). Due to their ubiquitous presence, these organisms are hard to control and can enter food production and processing at various points (Fig. 2). Certain B. cereus strains have been reported to promote plant growth and to suppress plant diseases (67, 68). Recently, it was reported that the B. cereus AR156 strain induces systemic resistance to Pseudomonas syringae pv. tomato in Arabidopsis thaliana (69), suggesting that plant rhizospheres represent a natural niche for B. cereus.
FIGURE 2.
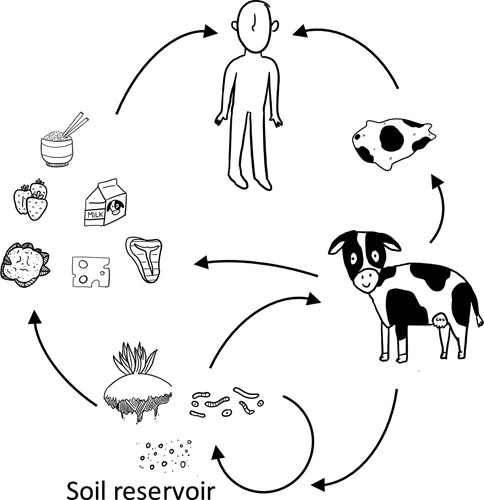
Transmission of the B. cereus group species from the soil reservoir to humans via food and textile production. Soil and soil-associated organisms including plants, insects, nematodes, and amoebae, serve as the major reservoirs for acquisition of spores. The bacteria are transferred to humans through agricultural products including food and animal-associated textiles, entering humans and other mammals through ingestion, inhalation, and breaks in the skin. Illustration credit: Olive E. Morrison.
Genome and physiology studies revealed that unlike true soil microorganisms, bacteria of the B. cereus group have few genes involved in the degradation of carbohydrate polymers. Conversely, these species contain a wide arsenal of genes encoding extracellular factors such as degradative enzymes (phospholipases, proteases, and chitinases), cytotoxic proteins (hemolysins, enterotoxins, and cytotoxins), and cell surface proteins (70, 71). These factors are potentially involved in the virulence and propagation of the bacteria in an animal host, thus conferring an obvious pathogenic lifestyle to the species (72). Outside of their hosts, the B. cereus group species are generally considered to reside as spores. Nevertheless, it is likely that the bacteria undergo some cycling of saprophytic growth, sporulation and germination in the soil. Epidemiological studies of clinical and nonclinical isolates suggest loss and horizontal movement of plasmids (22, 73–75).
The ecology of B. thuringiensis remains unclear despite multiple investigations (54, 76), and B. cereus ecology is even less explored (66). Studies of B. anthracis ecology have been fueled by the severity and worldwide occurrence of anthrax disease. B. anthracis can infect all mammals, some birds, and possibly even reptiles. The most severely affected parts of the world are central Asia and western areas of Africa. An “anthrax belt” exists from Turkey to Pakistan. In North America, anthrax is endemic in northern Alberta and the Northwest Territories. In the United States, sporadic outbreaks of anthrax occur in northwest Mississippi/southeast Arkansas and western Texas. The disease is also endemic throughout Mexico, Central America, and many South American countries. “Anthrax seasons” are characterized by hot dry weather which stresses animals and reduces their innate resistance to infection. Data from studies of central Asia support seasonal outbreaks, with the largest number of cases occurring in late summer (77, 78).
The prevalence of B. anthracis spores in the soil is affected by mean annual temperature, annual precipitation, elevation, amount of vegetation, soil moisture content, and soil pH (77, 78). Multiplication of B. anthracis outside of the host has been controversial. Concentrations of spores following disease outbreaks are thought to result from rains that pool spores in low-lying areas, as opposed to bacterial proliferation. Decomposition of infected animals at anthrax carcass sites leads to increased numbers of vegetative cells, but ultimately the numbers decrease as cells sporulate or die (79). Some studies suggest that B. anthracis competes poorly against indigenous bacteria; however, germination and amplification of anthrax spores by soil-dwelling amoebas and in the context of the plant rhizosphere have been reported (63, 80).
Humans and scavenging animals acquire gastrointestinal (GI) anthrax by ingesting spores in meat from infected animals, while wild and domestic herbivores ingest or inhale B. anthracis spores from the soil while grazing (81). Acquisition of spores from water appears to be less common (82). The infectious dose and severity of disease vary widely in different hosts. Domestic livestock, including sheep and cattle, and wild herbivores, such as bison, elephants, hippopotami, and kudu, are particularly susceptible to infection (83).
Inhalation anthrax in humans, also known as wool-sorter’s disease, was once common among textile workers who inhaled spores in dust generated from contaminated wool and animal hides. Today, most naturally acquired human cases of anthrax are cutaneous infections resulting from contact with dead infected animals or their products. Intentional dissemination of B. anthracis spores occurred in the United States in 2001. Eleven inhalation and eleven cutaneous cases of anthrax in humans were traced to four envelopes containing B. anthracis spores in powder form. The envelopes were sent to different locations via the U.S. Postal Service, and most of the patients were either mail-handlers or were exposed at worksites where contaminated letters were processed or received. Others are thought to have been exposed to cross-contaminated mail (84). This tragic event demonstrated the apparent ease with which the organism can be dispersed and solidified the placement of B. anthracis on the list of biothreat agents.
B. anthracis: the Anthrax Pathogen
Systemic anthrax, generally resulting from inhalation or ingestion of B. anthracis spores, has a high fatality rate. When B. anthracis spores are inhaled, alveolar macrophages and dendritic cells transport the organism to the proximal lymph nodes. Following spore germination, vegetative cells produce the antiphagocytic capsule and the anthrax toxin proteins, virulence factors which are critical for initiation of systemic disease (85, 86). In humans, bacteremia begins after an asymptomatic incubation period of 1 to 6 days. Following the onset of bacteremia, patients can experience flu-like symptoms for 1 to 5 days prior to an acute disease stage that lasts 1 to 2 days. Ultimately, death results from respiratory failure, sepsis, and shock (84, 87).
B. anthracis virulence is attributed primarily to production of anthrax toxin and capsule. The toxin structural genes and capsule biosynthesis genes are encoded by plasmids pXO1 and pXO2, respectively, and both plasmids are required for full virulence in immunocompetent hosts. The tripartite anthrax toxin, which comprises three secreted proteins—protective antigen (PA), edema factor (EF), and lethal factor (LF)—has been the focus of investigations of anthrax disease. PA facilitates entry of EF and LF into host cells. EF is a calmodulin-dependent adenyl cyclase, and LF is a zinc protease. Together, the toxins disable the host’s innate and adaptive immune systems to induce lethal disease (88, 89).
The poly-γ-d-glutamic acid (PGA) capsule of B. anthracis, provides protection of the bacterium from phagocytosis, as is true for many bacterial capsules which are more commonly made of polysaccharide. High concentrations of capsule released from B. anthracis cells during infection are important also for host-pathogen interactions (90–93). The PGA capsule has been shown to augment lethal toxin-mediated cytotoxicity and induce production of inflammatory cytokines and nitric oxide, which likely contribute to systemic inflammation and sepsis at the terminal stage of infection (94–97).
In addition to toxin and capsule, other secreted and nonsecreted factors have been reported to affect host-pathogen interactions and affect host pathology (98, 99). Among these are extracellular proteases that interfere with the immune response by cleaving antimicrobial peptides (100). The proteases NprB/Npr599 (also designated NprA in B. thuringiensis and B. cereus) and InhA1 can degrade host tissues, resulting in increasing barrier permeability (101–103). Crossing of the brain barrier is considered to be key to the extensive brain pathology that has been reported for human disease (104).
B. cereus and Gastrointestinal Disease
B. cereus pathogenicity is based on a panoply of virulence factors, which are far from understood, contributing to a range of human disease. B. cereus can cause two types of food poisoning, which are primarily characterized by diarrhea and emesis (37, 105, 106). The diarrheal syndrome has been linked to different enterotoxins thought to be produced after outgrowth of spores, taken up with contaminated foods, and active in the small intestine. The emetic syndrome is the result of an intoxication caused by the single depsipeptide toxin cereulide, which is preformed in food. The genes encoding the cereulide synthetase toxin genes are primarily found in a distinct subgroup of B. cereus, while the enterotoxin genes are broadly distributed among the members of the B. cereus group (41, 107). Usually, these GI-related diseases are self-limiting, but more severe forms require hospitalization, and occasionally fatalities have been reported (58, 108, 109). Intoxication related to cereulide has been linked to severe clinical manifestation, such as acute liver failures and acute encephalopathy (110). Due to the high biological activity of the cereulide toxin, occasionally liver transplantation is necessary as a life saving measure (111, 112). Generally, the pathogenic potential of B. cereus strains is highly variable and ranges from strains that show no cytotoxic activity in cell culture assays to strains that are highly cytotoxic (113, 114). In addition to intrinsic strain-associated differences in pathogenic potential, virulence is affected by environmental factors (58, 115, 116).
Apart from its food poisoning potential, B. cereus is increasingly recognized as an opportunistic pathogen that can cause local as well as systemic infections. Immunocompromised people and neonates are at special risk for hospital-acquired B. cereus infections, such as infections of the central nervous system, endocarditis, respiratory and urinary tract infections, wound infections, and septicemia (34). Outside of hospital-acquired infections, B. cereus has been linked to eye infections, such as endophthalmitis and keratitis (117). Furthermore, B. cereus can cause severe mammary gland infections in ruminants (118, 119). The virulence factors contributing to infections in ruminants and to most non-GI-related human infections are unknown.
B. thuringiensis Infection of Invertebrates
In addition to virulence genes common to the B. cereus group and to specific insecticidal toxin genes, the B. thuringiensis genome contains several genes encoding chitinases, suggesting a close relationship between this bacterium and the insects. Many well-studied B. thuringiensis strains were isolated from insects (44–46), and large screens of natural environments have shown that insects were more fruitful sources of the bacterium than soil samples (120). About 40% of the invertebrates tested were found to harbor B. thuringiensis spores. While insects appear to be the preferred target of B. thuringiensis, the bacterium can propagate in other invertebrates (121, 122). Nematode-B. thuringiensis association is highly probable in soil and other natural environments, and several B. thuringiensis strains harbor cry genes that are specifically active against nematodes (123). When nutrients are available, B. thuringiensis might be a multihost pathogen (124) with two pathogenic lifestyles: one resulting from its specific toxins (Cry or Vip) encoded by plasmid genes and targeting insects, nematodes, and other invertebrates (49, 125) and one nonspecific, depending on various chromosomal factors providing opportunistic properties. These nonspecific factors may allow the bacteria to propagate and eventually to kill various organisms if they are weakened by any cause, for example, by Cry toxins in the case of insects (126). This second pathogenic lifestyle may be shared with B. cereus, which possesses the same chromosome-encoded virulence factors (70). In this point of view, the Cry toxins of B. thuringiensis are “public goods” which can act cooperatively and thus profit cheater bacteria such as B. cereus (127).
The genome of B. thuringiensis contains a 41-gene regulon devoted to its saprophytic or necrotrophic lifestyle (128). This regulon, which includes genes encoding various degradative enzymes, is activated by the quorum sensing regulator NprR and allows B. thuringiensis to survive in the insect cadaver. The NprR regulon is also transcribed in all other B. cereus group species, suggesting that the bacteria of this group have similar saprophytic properties (129). Altogether, these pathogenic and saprophytic functions are highly efficient adaptive traits that allow the species to grow and survive in various natural conditions and niches.
CELL STRUCTURE AND DEVELOPMENT
Vegetative cell and spore structures do not vary greatly among the B. cereus group species. Generally, vegetative cells appear in chains, with the square ends of the cells fitting closely together. Variations in chain length occur as a normal aspect of growth in different media and environments (34, 130). Interestingly, B. anthracis vegetative cell chains in infected tissues are generally shorter than those formed during growth in vitro. Chaining of B. anthracis cells impedes invasion of host cells and is associated with escape from phagocytic clearance (131). Spores of the three species also have similar morphology. Spore size and morphology can vary between strains of the same species, but most strains produce spores with diameters of approximately 1 μm that are surrounded by a loose-fitting outer layer termed the exosporium (Fig. 3). Fine features of spores can be altered in different growth and sporulation conditions.
FIGURE 3.
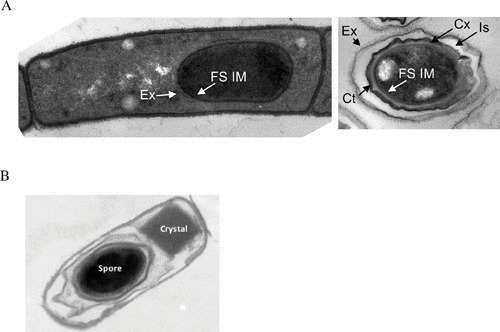
(A) Thin-section transmission electron micrograph of B. anthracis cell after 5 hours of sporulation (left image) and a mature spore (right image). The forespore inner membrane (FS IM) and nascent exosporium (Ex) (left image), and forespore inner membrane (FS IM), mature exosporium (Ex), coat (Ct), interspace, (Is), and cortex (Cx) (right image) are labeled. (B) B. thuringiensis sporulating cell with parasporal crystal. Cells were sporulated, prepared, and imaged as described in reference 432. Image credit: (A) Tyler Boone and Adam Driks. (B) Fuping Song.
Vegetative Cell Envelope
The surface of B. anthracis, including secondary cell wall polysaccharides (SCWPs), the surface layer (S-layer), and capsule, has been the most well studied of the B. cereus group species. The B. anthracis SCWP comprises trisaccharide repeats [→4(-β-ManNAc-(1→4)-β-GlcNAc-(1→6)-α-GlcNAc-)1→] with α-Gal and β-Gal substitutions tethered to the peptidoglycan by acid-labile phosphodiester bonds. SCWP is essential for cell growth, shape, and division. Although the enzymes for SCWP synthesis have not been fully characterized, two proteins, WpaA and WpaB, have been proposed as glycosyltransferases that act to assemble the SCWP polymers prior to attachment to the peptidoglycan. The B. anthracis SCWP is critical for maintenance of cell shape and for full virulence. Depletion of the glycosyltransferases results in irregularly shaped, nonviable cells (132). In addition, a mutant lacking a UDP-glucose 4-epimerase required for SCWP galactosylation exhibited reduced encapsulation and decreased virulence in a murine model of anthrax (133). Recent investigations of B. cereus isolates causing anthrax-like disease indicate SCWP with a structure similar to that of B. anthracis but with an additional α-Gal substitution at O3 of N-acetylmannosamine (134).
In B. anthracis and some pathogenic B. cereus strains, S-layer proteins have been shown to attach to the cell wall via noncovalent interactions with SCWPs (135). The S-layers are paracrystalline arrays of protein that serve as protective barriers and as scaffolds for housekeeping enzymes and virulence factors (136). Major S-layer proteins of B. anthracis are Sap (for surface associated protein) and EA1 (for extractable antigen 1). Fully assembled and acetylated SCWP is important for assembly of some S-layer proteins. Mutants deleted for putative acetyltransferases are unable to deposit the S-layer-associated murein hydrolase BslO at cell division septa and have altered chain lengths (137).
Finally, a key distinguishing feature of B. anthracis is its capsule. The B. anthracis capsule comprises PGA. As is true for typical polysaccharide capsules, the B. anthracis capsule protects cells from phagocytosis during infection. In addition, the B. anthracis capsule renders the bacterium somewhat resistant to mammalian proteases and is a poor immunogen (138, 139). An immunoassay for PGA using monoclonal antibodies has revealed that PGA is shed into the blood during infection of mice and nonhuman primates (91, 140, 141), likely due to the activity of a capsule depolymerizing enzyme, CapD, that is coexpressed with the capsule biosynthesis operon. The capsule degradation products are thought to inhibit the innate immune response through targeting of cytokine pathways (142).
The PGA capsule is also produced by some but not all B. cereus isolates that cause anthrax-like disease. These strains harbor plasmids that allow strain-specific differences in synthesis of the PGA capsule, a hyaluronic acid capsule, and a tetrasaccharide capsule. The results of virulence studies to assess the role(s) of the B. cereus capsules are somewhat contradictory and incomplete (22, 24, 31, 75). Further investigations are needed to determine the specific functions of these capsules.
Spore Architecture
The architecture of the B. cereus group species spore is comparable to the spores of other Bacillus species. The innermost compartment of the spore contains the DNA. The DNA is housed with cations and pyridine-2,6-dicarboxylic acid in a 1:1 chelate with divalent calcium ions, which serve to maintain the spore’s dormant state and resistance to DNA-damaging agents. Germinant receptors in the inner membrane initiate the signal transduction pathway that leads to outgrowth of the spore and the vegetative cell state (143). Germinants of the B. cereus group species can be a subset of sugars, amino acids, and ribonucleosides, as well as other small molecules, depending on the species and strain (144). A shell of peptidoglycan containing two sublayers, the cortex and the germ cell wall, surrounds the inner membrane. The cortex constricts the core’s volume to retain the relatively dry state of the core. At outgrowth of the spore, the germ cell wall becomes the nascent cell wall of the vegetative cell. Surrounding the cortex is the spore coat, which contains approximately 70 proteins arranged in layers to form a somewhat flexible structure (143).
The exosporium, an additional layer surrounding the spore coat of some Bacillus species that has been most well studied in B. cereus and B. anthracis (145), can respond to changes in spore hydration. The exosporium comprises a crystalline basal layer overlaid by a “hairy nap.” The composition and assembly process of the exosporium are poorly understood. BclA and related proteins form the hairy nap. Among the approximately 20 proteins of the basal layer is ExsY, which can self-assemble into structures resembling the intact basal layer and thus appears to be a key player in exosporium formation (146). The exosporium excludes large molecules. It also provides resistance to specific chemicals in vitro and to nitric oxide in the context of the macrophage (147–149). Removal of the exosporium from B. anthracis spores does not impede virulence in several animal models (150–152). However, the exosporium protein BclA binds macrophage receptors to mediate interactions with epithelial cells and extracellular matrix proteins (153, 154), suggesting a role for the exosporium in pathogenesis (145).
Germination Dynamics and Signals
The overall process of germination, in which spores transition to metabolically active vegetative cells, is highly conserved in the Bacillus genus, but the specific signals and associated receptors vary among species. B. anthracis germinates in response to alanine or purine ribonucleotides, with one or more additional amino acids acting as cogerminants (155–157). Many germinant receptors have been described in the inner membrane of Bacillus species. Notably, a unique set of germination genes, the gerX operon, is located on pXO1 of B. anthracis. Mutants missing the operon are altered for germination in vivo (158, 159). Once Bacillus spores germinate, spore cortex lytic enzymes degrade the cortex peptidoglycan, allowing core rehydration and cell outgrowth (160). B. anthracis differs from other species examined in that it contains an additional spore coat lytic enzyme which appears to work synergistically with the common enzymes (161).
Germination in the Host
Multiple routes of infection with B. anthracis spores lead to disease, suggesting that germination can be triggered by signals in different host niches. Studies of B. anthracis germination in vivo have yielded intriguing results. Inhalation anthrax models have shown that phagocytes and dendritic cells phagocytose spores and transport the bacteria to the lymph nodes (162, 163). Spores and vegetative cells appear in phagocytic cells, but it is unclear whether spores germinate before or after phagocytosis. Moreover, some strains germinate quickly in the lung, while others remain ungerminated for days (164, 165), suggesting that germinants plus additional unknown factors influence development in vivo. Finally, germination occurs in cell-free macrophage-conditioned media, indicating that phagocytes are not required for B. anthracis spore germination (157).
Studies of B. thuringiensis germination include experiments simulating conditions encountered in the insect midgut. The pH of the lepidopteran midgut is 10, and pretreatment of B. thuringiensis spores with larval midgut fluid or with an alkaline buffer stimulates germination (166, 167). This alkaline activation of spores requires plasmid-encoded germination genes (168). Cry proteins associated with the spore surface allow B. thuringiensis spores to bind brush border membrane vesicles prepared from lepidopteran insects, and the binding has been proposed to be responsible for stimulation of alkali-activated germination (169). It should be noted however, that the spore surface-Cry protein association may simply result from concomitant synthesis rather than from a specific interaction. Finally, germination of B. thuringiensis spores in the intestines of nematodes has also been reported (122).
Sporulation
The sporulation signal cascade has been well studied in Bacillus subtilis, and the pathways and regulators of sporulation in the B. cereus group species appear to be similar to those of B. subtilis. Nevertheless, variations among species have evolved with specific niches. B. anthracis does not sporulate during infection. In the wild, spores (which are the infectious form of the bacterium) only form when the blood of an infected carcass is exposed to the environment (78). The lack of sporulation in the mammalian host appears to result from a specific configuration of the phosphorylation cascade in this species (170–172). In contrast, it has been shown that part of the B. thuringiensis population sporulates during infection in insect larvae, and this process is under the control of the bifunctional regulator NprR (173, 174). The Rap phosphatases negatively control sporulation by stimulating a component of the phosphorelay involved in the phosphorylation of Spo0A, the master regulator of sporulation initiation (175). Rap activity is controlled by the Phr signaling peptides through a quorum sensing system. In B. anthracis and B. thuringiensis, plasmids harbor rap-phr genes involved in sporulation (176). These plasmid-borne Rap-Phr systems may provide advantages for adaptation of these pathogenic bacteria to their respective ecological niches.
TOXINS
Toxins, the first bacterial virulence factors to be described (177), represent the major drivers of pathogenicity within the B. cereus group. A variety of chromosome- and plasmid-encoded toxins, some shared among the species and some species specific, have been described. Table 1 shows an overview on the most important B. cereus group exotoxins. The species-specific toxins, namely, the B. anthracis tripartite anthrax toxin complex and the B. thuringiensis insecticidal Cry toxins, as well as the depsipeptide toxin cereulide of the emetic type of B. cereus, are encoded on plasmids, while many other B. cereus group toxins are encoded by the chromosome. Some B. anthracis and B. cereus isolates harbor duplicated and/or closely related toxin genes (35, 178), and B. thuringiensis strains can harbor several cry genes.
TABLE 1.
Exotoxins, the major virulence factors of the B. cereus group
| Toxin | Toxin type | B. anthracis | B. cereus | B. thuringiensis |
|---|---|---|---|---|
| Anthrax toxin (PA, LF, EF) | AB toxin | + | +a | – |
| Emetic toxin (cereulide) | Heat-stable depsipeptide toxin | – | + | – |
| Enterotoxins (Nhe, Hbl, CytK) | Pore-forming toxins | –b | + | + |
| Insecticidal toxins (Cry, Cyt, Vip, Sip) | Pore-forming toxins | – | – | + |
Some rare strains.
nhe genes are present but inactive.
B. anthracis Toxins
Anthrax toxin has been a major focus of anthrax research since the initial discovery of the toxigenic potential of B. anthracis culture supernates. B. anthracis secretes three proteins, PA, LF, and EF, known collectively as anthrax toxin. Purified toxin proteins in binary combinations have distinct effects when injected into experimental animals. Lethal toxin (LeTx), a mixture of LF and PA, causes death, while edema toxin (EdTx), a mixture of EF and PA, produces edema. PA (83 kD) mediates binding and translocation of the enzymatic proteins LF (90 kD) and EF (89 kD) into host cells.
PA is a nonenzymatic but critical component of LeTx and EdTX because it mediates toxin binding to host cells and facilitates translocation of LF and EF into cells. PA binds to one of two related proteins, capillary morphogenesis protein 2 (CMG2 or ANTRX2) and tumor endothelial marker 8 (TEM8 or ANTRX1), which are found on cells of many different tissues. CMG-2 is the most important for pathogenesis, while TEM-8 has a minor role (179, 180). In the model of anthrax toxin entry, furin and possibly additional host-associated proteases cleave PA into two fragments, PA63 (63 kDa) and PA20 (20 kDa) (89). The receptor-bound carboxyterminal 63-kDa fragment (PA63) oligomerizes to form a ring-shaped structure comprising seven or eight PA63 molecules. One to three molecules of LF and/or EF bind to the PA ring, and the complex is endocytosed by clathrin-coated vesicles. Endosome acidification induces a conformational change in the ring such that it converts from a “prepore” to a pore (channel) in the membrane, facilitating translocation of EF and LF across the membrane (181).
LF, a zinc-dependent metalloproteinase, is the central effector of shock and death from systemic anthrax. Initial investigations of LF activity revealed that the protein cleaves mitogen-activated protein kinase kinases, resulting in disruption of multiple signaling pathways and causing broad downstream effects on cell physiology (182). More recent reports show that low levels of LeTx inhibit the glucocorticoid receptor, an important component of host defense against inflammation. LF targets the inflammasome sensor NLRP1, causing activation of the caspase-1-dependent cell death signaling pathway (183). Finally, murine susceptibility to lethal toxin has been associated with genetic polymorphisms in the Ltx gene on mouse chromosome 11. The Ltx gene encodes a kinesin-like motor protein, Kif1C, but the relationship between lethal toxin and Kif1C is not clear (184).
EF is a calmodulin-dependent, Ca+2-dependent adenylyl cyclase (185). As is true for other bacterial toxins that elevate cAMP levels, disruption of normal second-messenger signaling has many effects on host cell physiology. EdTx weakens antibacterial responses of phagocytes, impairing apoptosis, motility, and release of inflammatory cytokines. Human neutrophils treated with the toxin have reduced phagocytic and oxidative burst abilities (186).
A comprehensive model of the molecular basis of anthrax toxin effects on mammalian hosts is still evolving, but a wealth of current information reveals that together, the toxin components severely impinge on the immune functions of susceptible hosts, which are key to the outcome of B. anthracis infection (89, 187, 188).
Anthrolysin O is a cholesterol-dependent cytolysin with properties similar to the pore-forming toxins of other Gram-positive pathogens (189). Initially discovered as a hemolysin (190), ALO has lytic activity against phagocytes (191), decreases the barrier function of human polarized epithelial cells (192), and appears to play a role in gut epithelial disruption during B. anthracis infection (193). Expression of the alo gene is highly responsive to in vitro growth conditions (190). During infection, alo transcripts are present in newly germinated bacteria after phagocytosis of the spores by mouse macrophages (194, 195). Concurrent deletion of alo and phospholipase genes attenuates virulence and reduces growth and survival in phagocytes, suggesting a role for the toxin in destabilization of the phagocytic membrane (196).
B. cereus Toxins
Various B. cereus exotoxins have been associated with pathogenicity, but the actual contribution and importance of these toxins for GI and non-GI related diseases is largely unknown. So far, four B. cereus exotoxins have been clearly linked to food poisoning. Three pore-forming enterotoxins (Nhe, a three-component nonhemolytic enterototoxin, Hbl, a three-component hemolytic enterotoxin, and the single protein cytotoxin CytK) are thought to provoke diarrhea after toxicoinfections with B. cereus spore-contaminated food (37). Hbl enterotoxic activity has been demonstrated in vivo on rabbit intestine (197), but direct in vivo proof for Nhe and CytK activity is lacking. Nhe was identified from a strain isolated from a foodborne outbreak in Norway (198), and CytK was originally isolated from a B. cereus strain that was responsible for a severe foodborne outbreak in which several people developed bloody diarrhea and three elderly people died (199). Enterotoxins may act synergistically with each other and with other virulence factors, making it difficult to predict the overall enterotoxic potential of a given strain (39, 106, 200–204). Nhe, Hbl, and CytK show cytotoxic activity on Vero and Caco2 cells as well as on other human cells. Different cell lines show distinct susceptibilities to the different toxins (205), suggesting different modes of toxin action.
Typically, symptoms occur after 6 to 12 hours and are self-limiting. However, in rare cases the severity of symptoms requires hospitalization, and fatalities among neonates and immunocompromised patients have been reported (108, 109, 199).
The fourth toxin, the so-called cereulide, is linked to the emetic type of B. cereus food poisoning (206). Cereulide is a depsipeptide toxin, composed of alternating α-amino and α-hydroxy acids (d-O-Leu–d-Ala–l-O-Val–l-Val)3 (Fig. 4), that is structurally related to the potassium ionophore valinomycin (206). It is produced by the cereulide peptide synthetase Ces (6, 42, 207, 208), which represents a novel type of nonribosomal peptide synthetase (6, 207–209). Cereulide is usually preformed in food, and the onset of symptoms occurs between 15 minutes and 6 hours after uptake of contaminated food (105). In the past, quantification of cereulide has been hampered by the lack of appropriate methods (58). The recently developed ISO method (EN ISO 18465), based on a stable isotope mass spectrometry-based dilution assay (210), will allow accurate cereulide quantitation in the future. Recently, 18 cereulide isoforms were described, with cytotoxic potential ranging from nontoxic to 10-fold more toxic than the classic cereulide (211). Cereulide is not inactivated during stomach passage in the host because it is resistant to cleavage by pepsin and trypsin (105, 212). Due to its hepatotoxic activity, cereulide can cause rhabdomyolysis, liver damage, and severe multiorgan failure (110, 112). Recently, cereulide was linked to the induction of diabetes by causing beta cell dysfunctions (213).
FIGURE 4.
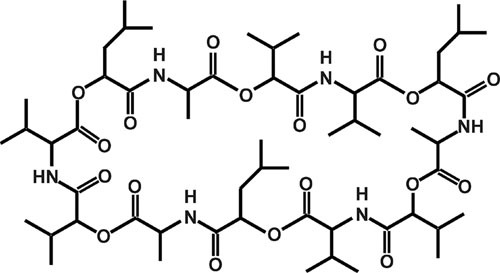
Cereulide, the emetic toxin of B. cereus.
B. thuringiensis Toxins
Except for the cereulide toxin, B. thuringiensis strains generally produce the same toxins as B. cereus (214). The phenotype distinguishing B. thuringiensis from the other species of the B. cereus group is the production of a crystal inclusion during sporulation (Fig. 3). This crystal generally appears beside the spore in the mother cell compartment of the bacteria (215), but other situations have been reported in a few strains, including a crystal between the exosporium and the spore coat (216), and production of spores and crystals in two distinct subpopulations of bacterial cells (217). The crystal inclusion can account for up to 25% of the dry weight of B. thuringiensis cells, which means that a massive production of proteins should occur during sporulation or stationary phase. This massive protein production results from several mechanisms, including efficient transcription regulation, an exceptionally high stability of cry gene mRNA, and the crystallization of the proteins (218). In most of the B. thuringiensis strains the expression of the cry genes depends on sporulation-specific sigma factors, Sigma E and Sigma K, which are functional in the mother cell compartment during a long period of the sporulation process. Some exceptions have been observed. For example, the cry3 genes encoding coleopteran-active toxins are expressed during stationary phase independently of the sporulation sigma factors (218).
The crystal inclusions are composed mainly of two types of proteins, the Cry and the Cyt proteins, also called δ-endotoxins (49, 219). Various combinations of toxins can be found on the crystal, depending on the B. thuringiensis strain. The commercial strain kurstaki HD1 Dipel® contains five cry genes (cry1Aa, cry1Ab, cry1Ac, cry2Aa, and cry2Ab), all encoding proteins active against lepidopteran larvae (220). The strain israelensis contains four cry genes (cry4Aa, cry4Ba, cry10Aa, cry11Aa) and two cyt genes (cyt1Aa and cyt2Ba), all encoding proteins toxic for the mosquito larvae (9). The Cry proteins are distributed into 75 distinct classes, with less than 45% identity between them (http://www.lifesci.sussex.ac.uk/home/Neil_Crickmore/Bt/). They are pore-forming proteins whose activity requires the binding to specific receptors on the surface of midgut cells (194). The Cyt toxins are cytotoxins specifically active against dipteran larvae (199); they also have a synergistic action by serving as receptors for dipteran-active Cry proteins (221).
In addition to the Cry proteins forming parasporal inclusions, some B. thuringiensis strains secrete insecticidal proteins: the Sip (secreted insecticidal protein) and Vip (vegetative insecticidal protein) proteins. Sip1A is active against coleopteran insects and was isolated from a B. thuringiensis strain harboring a coleopteran-specific cry3 gene (207). The toxin Vip3A is specifically toxic against lepidopteran insects and is generally produced by strains that also produce Cry1A lepidopteran-active toxins (209). The insecticidal activity of the exported proteins Vip and Sip and their association with Cry proteins showing related activity spectra suggest that the larvicidal activity of the Cry proteins is reinforced by the secretion of Sip and Vip proteins, thus increasing the entomopathogenicity of the B. thuringiensis cells multiplying in the infected insect larvae.
Common Secreted Enzymes of the B. cereus Group Species
Phospholipases, sphingomyelinases, hemolysins, proteinases, and peptidases likely represent additional virulence factors of the B. cereus group species (39, 106, 202–204). In a murine model of anthrax, protease inhibitors enhance the positive effect of antibiotics, suggesting that one or more proteases contribute to anthrax disease (222).
Immune inhibitor A (InhA1) is a particularly well-studied protease that has homologues in all three species. B. anthracis, B. cereus, and B. thuringiensis each carry multiple apparent inhA genes, with transcription of each gene being subject to different regulation (223, 224). Autolytically processed active InhA1 of B. anthracis cleaves multiple host proteins and other proteins in the B. anthracis secretome (225, 226). A B. anthracis inhA1-null mutant is attenuated in a murine model of anthrax (227). The reduced virulence of the mutant likely results from its broad activity on multiple targets, ultimately facilitating bacterial dissemination. InhA1 disturbs the host coagulation machinery by cleaving von Willebrand factor and its regulator ADAMTS13 (102). The enzyme also cleaves zonula occluden-1, resulting in disruption of the blood-brain barrier (103). Degradation of hemoglobin by InhA in the bloodstream has been proposed to allow B. anthracis access to essential amino acids required for proliferation in the blood (227). In B. cereus and B. thuringiensis, deletion of the three inhA genes strongly affects virulence (202). InhA1 is required for efficient escape from macrophages (228).
MODELLING HOST INTERACTIONS
Animal Models of Anthrax
Multiple animal models differing in host species, bacterial strain, and delivery route are used to investigate B. anthracis-host interactions. The significant variability among laboratory animals in susceptibility to B. anthracis infection makes modeling human anthrax particularly challenging. Nonhuman primates are considered superior models for studies of potential therapeutics and vaccines, but the high cost and ethical considerations limit their use. Rabbits serve as good models because the disease pathology is similar to that observed in nonhuman primate and clinical human cases of anthrax. Mice and guinea pig models are used frequently because of experimental ease and moderate cost. For most animal studies, the spore form of the bacterium is used as the infectious agent, modeling natural infection. Methods for pulmonary delivery of spores include intranasal inoculation, aerosol exposure, and tracheal injection. Spores can also be introduced subcutaneously. To model late-stage disease, vegetative cells are inoculated directly into the bloodstream, bypassing the requirement for in vivo germination of spores (229).
Mice are the most common experimental animal for investigations of B. anthracis. Inbred mice infected via the parenteral and pulmonary routes develop acute disease, with vegetative cells in multiple tissues. Immunodeficient (C5–) mouse strains such as A/J and DBA/2J are susceptible to relatively low doses (103 to 104 CFU) of capsule-deficient (pXO1+pXO2–) Sterne-type strains and are used often for studies of the roles of anthrax toxin in vivo and for testing the efficacy of toxin-based therapeutics and vaccines. Exposure of C5– mice to aerosolized Sterne spores results in pathology similar to that observed in rabbit and nonhuman primate models, with lethal toxin required for dissemination and development of protective immunity (230).
Studies of pathogenesis and host responses related to nontoxin factors have been commonly assessed in infections of immunocompetent mice with nontoxigenic (pXO1–pXO2+) strains. Interestingly, guinea pigs, rabbits, and nonhuman primates are not susceptible to infection by pXO1–pXO2+ strains (231). It is also notable that mice are not protected from fully virulent (pXO1+ pXO2+) strains by the PA-based vaccine AVA, whereas AVA is fully protective for rabbits and nonhuman primates but only partially protective for guinea pigs (232, 233).
Mouse models have revealed interesting aspects of germination and dissemination of B. anthracis during infection (229). Carr and coworkers (234) demonstrated functional redundancy among the five known germinant receptors, and McKevitt and coworkers (159) showed that the germination inhibitor d-alanine reduced the infectious dose of spores, suggesting that timing of germination is important for disease. Experiments conducted by Glomski and colleagues (235, 236) using luminescent recombinant strains showed that germination and dissemination from pulmonary, skin, and GI entry points does not require transport to lymph nodes, but spore dose and method of delivery can affect the timing and location of germination. Their data also revealed a role for capsule in dissemination.
Inbred mice have also been used extensively to explore the host response to B. anthracis infections. Investigations of host systems important for recognition of bacterial factors and activation of immunity have revealed a model in which B. anthracis activates Toll-like receptors and the nucleotide-binding oligomerization domain-like receptor (NLR) inflammasome, resulting in tumor necrosis factor alpha production by a mitogen-activated protein kinase signaling pathway. LeTx suppresses this response by cleaving mitogen-activated protein kinase kinases. EdTx-mediated increases in cAMP also disrupt cell signaling pathways. In the battle between host and pathogen, the immunosuppressive effects of LeTx are countered by Nlrp1b, the product of a “lethal toxin sensitivity” locus on chromosome 11. In response to lethal toxin, the lethal toxin-sensitive Nlrp1b allele activates caspase-1, inducing proinflammatory cytokines that recruit neutrophils and mononuclear cells. Thus, in mice, LeTx-mediated Nlrp1b-dependent activation of caspase-1 is a major factor in the control of B. anthracis infection (229).
Guinea pigs are keenly susceptible to B. anthracis spore infection but not very sensitive to the anthrax toxins. While other animal models show some inconsistencies in terminal loads of bacteria, intramuscular challenge of guinea pigs with a fully virulent (pXO1+ pXO2+) strain leads to specially and temporally consistent infections. Aerosol infection of guinea pigs results in edema and hemorrhage in the spleen, lungs, and lymph nodes, facilitating studies of pathology. Guinea pigs have been used in competitive infection studies employing different strains and mutants. These experiments have revealed roles for biosynthetic genes and surface proteins in disease (229).
Rabbits have been useful to determine virulence differences in B. anthracis strains and mutants and to examine pathology, which is similar to that observed in humans and nonhuman primates. Multiple investigations of vaccines and therapeutics have employed rabbit models. Rabbits exhibit a strong immune response to PA and have played a key role in the development of correlates of immunity. A quantitative anti-PA IgG enzyme-linked immunosorbent assay and a toxin neutralization assay developed using sera from vaccinated rabbits are used to evaluate the human immune response to PA-based vaccines (237–240).
Nonhuman primates provide a valuable model for anthrax disease because infection with aerosolized spores results in a disease course that most closely matches human infection. Nonhuman primates, including rhesus macaques, cynomolgus macaques, African green monkeys, and marmosets, represent the only animal models of inhalational anthrax in which anthrax meningitis is consistently observed. Infected nonhuman primates, like humans, also develop bacteremia, lymphadenitis, splenitis, mediastinitis, pneumonia, pleural effusions, hepatitis, adenitis, and vasculitis. Tissues of diseased animals and humans also show hemorrhage, necrosis, and edema. Sera from infected nonhuman primates have been used to develop biomarkers for early diagnosis of infection. Capsule and toxin, detected using enzyme-linked immunosorbent assay, chemiluminescence, and mass spectrometry, serve as reliable markers.
Because nonhuman primate responses to infection most closely resemble human physiological and immune responses, these animals are best suited for immunogenicity, toxicity, and safety testing of vaccines and therapeutics for anthrax. Nonhuman primates have been used extensively for studies of antibiotic efficiency. Investigations of the dose, pharmacokinetics, and time course of antibiotic treatment have shown that antibiotics are only effective against germinating spores and vegetative cells. The long-term persistence of spores and extended time periods to germination mean that complete protection from high doses of aerosolized spores requires prolonged antibiotic treatments over more than 30 days.
Rats are somewhat resistant to challenge with B. anthracis spores, but they are exquisitely sensitive to injection of LeTx. Thus, the Fisher 344 strain has been used as a model system for toxin activity. The rat toxin model was key to determining the role of the NLR sensor Nlrp1 in animal susceptibility to LeTx (241, 242). The model also provides a sensitive means to test the efficacy of small-molecule toxin inhibitors, monoclonal antibodies, dominant-negative toxin components, and other potential antitoxin countermeasures (229).
Animal Models of B. thuringiensis and B. cereus Infection
The opportunistic properties of B. thuringiensis and B. cereus have been investigated using nasal instillation in mice (243, 244) and intraocular infection in mice and rabbits (245). In all cases, the results highlight the importance of the PlcR regulon in the pathogenic properties of these bacteria. However, established mammalian models of GI infections caused by B. cereus are lacking. It has been hypothesized that rodents are not suitable models of GI-related B. cereus infection because their gut physiology does not support germination of spores and subsequent enterotoxin production (246). In a more recent study, Rolny et al. (247) used intragastric gavage to test the effect of vegetative B. cereus cells on the host immune system. They found some modifications in immune cells in the spleen, Peyer’s patches, and mesenteric lymph nodes but did not report any enterotoxin production. Thus, an appropriate mammal model for enterotoxigenic B. cereus awaits development.
The emetic activity of the B. cereus toxin cereulide was first shown in a rhesus monkey model (248, 249). Results from a study in Suncus murinus suggest that cereulide causes emesis through the 5-HT receptor (206). However, it is still unclear if the S. murinus model reflects the physiology of the human gut. Thus, further studies will be needed using model organisms, such as pig, more closely resembling the human gut physiology.
Insect Models of B. cereus and B. thuringiensis Infection
In insects susceptible to B. thuringiensis infection, the first stage of the disease results from gut paralysis due to the crystal inclusion (250, 251). Cry toxins are responsible for this initial step. The toxins damage the peritrophic membrane and cause lysis of the intestinal cells by producing osmotic shock (252–254). Depending on the amount of Cry toxins and the susceptibility of the insect larvae to a given Cry toxin, the toxemia resulting from toxin ingestion may result in insect death. This toxemia and the paralysis of the gut allow spore germination. Several studies have shown that the presence of B. thuringiensis cells synergizes the toxic effect of the crystals (243, 255–259).
As natural targets of B. thuringiensis, insect larvae are a valuable model for studying the virulence of these bacteria. The utility of this infection model is increased by using insects that are poorly susceptible to the ingestion of crystals alone. A commonly used insect is the lepidopteran species Galleria mellonella (the greater wax moth) because the larvae are killed by the ingestion of B. thuringiensis spore-crystal mixtures but are only weakly affected by the ingestion of crystals alone (243, 258, 259). This model host has also been used to assess the virulence of B. cereus. Many other human pathogens are unable to kill G. mellonella in this infection model, reflecting the entomopathogenic specificity of these two species (260).
The G. mellonella host can be used in two infection methods. In association with Cry toxins to impair the insect defenses, the pathogenicity of B. thuringiensis and B. cereus can be assayed via the oral route. At the last instar, G. mellonella larvae are large enough (2 cm long and 250 mg) to be force-fed, thus allowing the ingestion of defined doses of bacteria (243). Larvae can be raised at various temperatures to mimic different natural conditions, including the mammal environment. This infection model allows determination of a 50% lethal concentration, measurement of bacterial survival, and assessment of the infectious process. The behavior of the bacteria in the intestinal environment, including bacterial factors interacting with the intestinal barrier, can be investigated (261). G. mellonella was used to demonstrate the major role of the PlcR regulon in the virulence of B. thuringiensis and B. cereus (243). The regulon consists of 45 genes encoding a large number of extracellular virulence factors such as degradative enzymes, hemolysins, and cytotoxins (17, 70). The PlcR-regulated metalloproteases InhA2, Bel, and ColB were demonstrated to be involved in B. thuringiensis virulence (38, 262, 263). In addition to G. mellonella, a nematode oral infection model in association with Cry proteins has also revealed virulence factors of B. thuringiensis (264). The ColB protease and another metalloprotease (Bmp1) were found to be involved in the pathogenicity of B. thuringiensis against Caenorhabditis elegans (262, 265).
A second mode of insect infection is direct injection of the bacteria into the hemocoelic cavity of the larva. This model bypasses the intestinal barrier and therefore does not require the crystal proteins. In addition to G. mellonella, the silk worm, B. mori, and several other insect species can be used. Bacteria are injected through the intersegmental membrane between the fourth and fifth abdominal legs of fourth-instar larvae (38). Most insect larvae are highly susceptible to the direct injection of vegetative cells or spores of B. thuringiensis and B. cereus into the hemocoel. This mode of infection led to the identification of multiple virulence factors, including genes associated with (i) iron metabolism and acquisition (266–268), (ii) resistance to the host defenses, including antimicrobial peptides, phagocytic cells, or nitric stress (202, 269–271), (iii) motility (272), and (iv) key transcription factors for B. cereus pathogenicity (273). The major role of the sphingomyelinase in the pathogenicity of B. cereus was also revealed by this method (39), thus confirming the observation that clinical strains of B. cereus overproducing sphingomyelinase were more virulent than environmental strains (203).
Data from these experimental infections clearly demonstrate that B. thuringiensis is a true entomopathogenic bacterium (Fig. 5), producing specific toxins that weaken or kill invertebrates and other virulence factors, shared with other B. cereus group isolates, that allow bacteria to provoke septicemia and death of the host (72). Moreover, it has been shown that these bacteria harbor a regulon conferring the ability to survive in the insect cadaver through a pathway independent of the sporulation and corresponding to a necrotrophic lifestyle (128, 174). This regulon is activated during the stationary phase of the bacteria by the NprR-NprX quorum sensing system and consists of 41 genes encoding several degradative enzymes, including proteases and chitinases. Although the role of each component is not determined, altogether they should contribute to maximizing the consumption of the nutriments found in the host cadaver. Furthermore, the insect model was successfully employed to demonstrate antibiotic-induced small colony variants, a novel persister phenotype of B. cereus (274).
FIGURE 5.
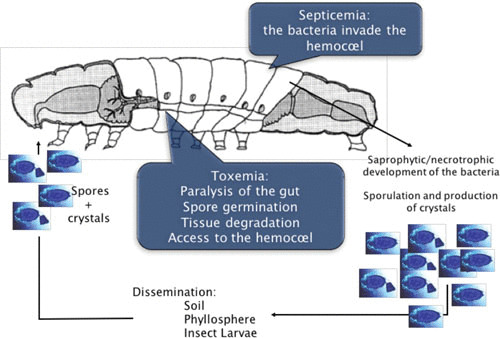
The infectious cycle of B. thuringiensis in a susceptible insect larva. Ingestion of spores and crystals is followed by the dissolution of the crystal in the alkaline environment of the midgut. Insect proteases activate Cry proteins. Spores germinate in the paralyzed insect gut. Cry toxins degrade the peritrophic membrane. Bacteria multiply and express PlcR-regulated genes. The intestinal barrier is disrupted and bacteria gain access to the hemocoel. The bacteria resist host defenses and cause a fatal septicemia. The NprR regulon is activated, and the bacteria survive in the host cadaver. Finally, spores and vegetative cells are disseminated outside of the host.
Cell Culture
Immortalized cell lines are employed for testing the cytotoxic potential of B. cereus strains. Hep2 cells are used to check strains for the emetic toxin cereulide, while Vero cells are commonly used to check strains for their enterotoxic potential (35). However, more recently, it has been reported that different human cell lines show distinct susceptibility for specific enterotoxins, which must be taken into consideration when functional studies on enterotoxins are carried out (205). These results are in line with a previous study from Doll et al. (39) showing that in polarized colon epithelial cells B. cereus sphingomyelinase is a strong inducer of epithelial cell death, by synergistically interacting with Nhe, while most of the cytotoxic effect on primary endothelial cells can be explained by the cytotoxic activity of Nhe (205). Studies carried out on pancreatic beta cells revealed a detrimental effect of cereulide toxin on these cells, even in low concentrations, rendering it possible that long-term uptake of cereulide could have some implications in diabetes (213). Since beta cell dysfunction and cell death play key roles in the pathophysiology of diabetes, it will be important to study the long-term effects of subemetic doses of cereulide in more detail in appropriate eukaryotic cell models.
Furthermore, boar sperm has been used to show the mitochondrial activity of the B. cereus emetic toxin cereulide (275), and the enterotoxic activity of Hbl has been demonstrated in a rabbit ligated ileal loop test (197).
In vitro Simulation of GI Transit
Several systems that mimic gastric passage of B. cereus have been described (276–278). B. cereus spores, and to a certain extent vegetative cells, survive in these models, but enterotoxin production has not been detected, making these models suboptimal for studies of B. cereus enteropathogenicity. Such systems mimicking the physicochemical conditions from mouth to ileum have been successfully used to study probiotic bacteria (279) but lack the influence of host factors, which have been shown to be important for enterotoxin production in B. cereus (116).
VIRULENCE GENE EXPRESSION
Virulence gene expression within the B. cereus group is controlled by complex regulatory circuits. In addition to global transcription regulators, such as CodY and AbrB, that play key roles in the integration of virulence and bacterial metabolism (273, 280, 281), B. cereus group-specific regulators exert control of virulence gene expression on the transcriptional and posttranscriptional levels. The global transcriptional regulator AtxA, encoded by pXO1 of B. anthracis, plays a key role in the pathogenicity of B. anthracis, while the chromosome-encoded pleiotropic transcriptional regulator PlcR is the key regulator in B. cereus virulence (70, 282). Most of the virulence factors in B. cereus described so far are at least partially under the control of PlcR, except cereulide toxin synthesis, which belongs to the Spo0A-AbrB regulon (283). In addition, some reports highlight the importance of posttranscriptional regulation of virulence factor expression, which is largely unexplored (114, 284, 285).
Global Transcription Regulators
CodY, a global transcriptional regulator integrating virulence and metabolism, plays an essential role in regulation and timing of virulence gene expression in the B. cereus group. CodY, which is highly conserved in low-G+C-content Gram-positive bacteria, senses the nutritional and energetic status of a cell by binding to branched chain amino acids and GTP (286, 287). It often exerts control of virulence factors indirectly by acting on downstream regulators (288, 289). Although the majority of CodY-regulated genes are repressed during rapid growth, CodY is a positive regulator of some virulence genes (286, 290). In B. cereus, CodY governs the expression of virulence factors by impacting key regulatory circuits (66). CodY directly represses cereulide toxin synthesis in emetic B. cereus during early growth stages but acts indirectly on virulence genes belonging to the PlcR regulon by controlling import of the signaling peptide PapR, which is required for PlcR activity (273, 281, 433). CodY was also reported to be essential for full virulence of B. anthracis. Data suggest that CodY exerts indirect positive control on posttranslational regulation of AtxA accumulation (284). By acting, either directly or indirectly, on other virulence regulators, CodY adds an additional level of complexity to the regulatory circuits governing metabolic activities and virulence within the B. cereus group.
AbrB is a global transition state regulator that links virulence and cell development. It has been extensively characterized in B. subtilis but also highly conserved in other Gram-positive bacteria (291, 292). As is true for B. subtilis, AbrB is negatively regulated in the B. cereus group by the response regulator Spo0A at the onset of the transition phase. AbrB has been shown to control the expression of the anthrax toxin genes pagA, lef, and cya in B. anthracis as well as the cereulide (ces) synthetase genes in emetic B. cereus. In B. thuringiensis, abrB represses expression of inhA1 during vegetative growth (293). During early exponential growth phase, abrB-null mutants synthesize elevated levels of anthrax toxin in B. anthracis and cereulide levels in emetic B. cereus (283, 294). Thus, it can be assumed that AbrB plays a pivotal role in the correct timing of toxin expression in B. anthracis as well as in emetic B. cereus.
Alternative sigma factors have also been implicated in transcription of virulence factors of the B. cereus group. The sporulation gene regulator SigH represents a target of the AbrB repressor in B. subtilis (295), and it has been postulated that AbrB acts on anthrax toxin gene expression via control of sigH (296). However, the B. subtilis consensus sequence for recognition by SigH has not been found upstream of the anthrax toxin genes nor upstream of the ces genes in emetic B. cereus, and no direct SigH-dependent toxin promotor activity could be shown (283, 297). Therefore, it is tempting to speculate that SigH is not directly linked to toxin gene regulation in B. anthracis or emetic B. cereus. SigB, another alternative sigma factor, may be a minor regulator of virulence in B. anthracis. Although there is no evidence that SigB affects toxin or capsule gene expression, a sigB mutant exhibits reduced virulence in a mouse model of anthrax (298).
RNPP Quorum Sensors
A major trait of the B. cereus group is the unique presence of two transcriptional regulators, PlcR and NprR, forming two distinct quorum sensing systems in association with their cognate signaling peptides, PapR and NprX, respectively (129, 299). In addition to these specific regulators, the bacteria of the B. cereus group produce proteins homologous to the Rap phosphatases of B. subtilis (175, 300). These quorum sensors define the RNPP family according to the name of the prototypical members Rap, NprR, PlcR, and PrgX (301). Quorum sensing systems connect gene expression to cell density, thus coordinating the behavior of the whole bacterial community able to respond to the same signaling peptide. In insects, it was shown that the sequential activity of RNPP regulators (PlcR, NprR, and Rap) controls the physiological stages of B. thuringiensis throughout the infectious cycle, from virulence to sporulation (302).
PlcR positively regulates 45 genes, most of which encode exported virulence factors such as the enterotoxins Hbl and Nhe, the cytotoxin CytK, the cholesterol-dependent hemolysins, and several degradative enzymes (70). As discussed above, PlcR plays a key role in the virulence of B. thuringiensis and B. cereus in their invertebrate or mammal hosts (70, 72, 245). The plcR gene is present in all B. cereus group species but does not always produce a functional protein (303). Notably, in B. anthracis a nonsense mutation inactivates the PlcR function (17, 18). PlcR is a 34-kDa helix-turn-helix-type transcription factor activated upon binding of PapR. This binding induces a drastic conformational change of the dimeric form of PlcR, allowing an appropriate positioning of the helix-turn-helix domains and their binding to the PlcR-boxes located upstream from the −10 region of the PlcR-regulated promoters (299, 301, 304). Binding of PapR to PlcR is highly specific, and polymorphisms exist within the B. cereus group. Four specificity groups, or pherotypes, can be distinguished (305, 306).
NprR is also found in all B. cereus group species and acts as a transcriptional regulator at the onset of sporulation (128). As indicated above, NprR is an RNPP regulator whose transcriptional activity depends on a signaling peptide (NprX) that switches NprR from a dimeric apo form to a tetrameric form (307). Similar to PlcR-PapR, the NprR-NprX system is strain specific, and seven pherotypes were defined within the B. cereus group (129). NprR-NprX activates the transcription of a regulon including genes encoding degradative enzymes (proteases, lipases, and chitinases) and a lipopeptide (kurstakin) involved in biofilm formation (128, 308). Using insect larvae as an infection model, it was shown that the NprR regulon allows bacteria to enter in a necrotrophic lifestyle essential for the survival of the bacilli in the cadavers of infected insects. In the absence of its cognate peptide, the apo form of NprR functions as a phosphatase inhibiting the sporulation process (309). It has been shown that sporulation occurs only in bacteria expressing the NprR regulon and following a necrotrophic lifestyle in the insect cadaver (174). These results are in agreement with the dual function of NprR and indicate that its phosphatase activity should be turned off by NprX to allow the sporulation of the bacteria.
AtxA, the Key Regulator in B. anthracis Virulence
AtxA, named for its control of toxin gene expression (310) and encoded by a pXO1 gene, is the master virulence regulator of B. anthracis. AtxA positively controls transcript levels of the structural genes for anthrax toxin, pagA, lef, and cya, located on pXO1; the capsule biosynthetic operon capBCADE located on pXO2; and multiple other genes on the plasmids and chromosome (5, 282, 311–318). An atxA-null B. anthracis strain is highly attenuated in a murine model of anthrax disease (318, 319). The atxA gene has also been associated with control of B. anthracis development. AtxA activates expression of a pXO2-encoded sporulation kinase inhibitor to repress sporulation in conditions in which AtxA is highly expressed and active (320). This activity may be related to the lack of B. anthracis sporulation during infection.
Paralogous regulators, AcpA and AcpB encoded by pXO2 genes, were first identified as regulators of capBCADE (321, 322). AcpA and AcpB do not appear to affect toxin gene expression but may affect expression of noncapsule genes (282, 323). Data from experiments employing null mutants reveal that expression of the atxA, acpA, and acpB genes is interdependent and suggest that AcpA and AcpB have similar functions. The primary means for AtxA control of capBCADE is via positive regulation of acpA transcription. The acpA gene is located upstream of the cap locus and in the opposite orientation. Two apparent transcriptional start sites, one expressed at a low constitutive level and one controlled positively by AtxA, drive expression of acpA. AcpA can positively affect transcription of capBCADE in the absence of AtxA and AcpB. The acpB gene, located downstream of capBCADE and in the same orientation, is expressed as a monocistronic transcript initiated from a weak constitutive promoter. AcpB, like AcpA, can positively affect capBCADE expression in the absence of the other regulators. A weak transcription terminator located between capE and acpB results in cotranscription of acpB, with capBCADE in roughly 10% of transcripts, thus forming a positive feedback loop (324).
AtxA expression is also subject to complex regulation. Transcription of atxA is affected by temperature, carbohydrate availability, redox potential, metabolic state, and growth phase (325–328). In addition, CodY controls AtxA protein levels posttranscriptionally by an unknown mechanism (284), and AbrB represses atxA expression by binding to the atxA promoter region upstream of the P1 transcriptional start site (294, 329). Finally, an unknown additional repressor binds to the atxA promoter to negatively regulate expression (319).
AtxA, AcpA, and AcpB belong to an emerging class of regulators termed PCVRs (phosphoenolpyruvate-dependent phosphotransferase system regulation domain [PRD]-containing virulence regulators) (330, 331). Generally, PCVRs contain nucleic acid-binding domains and PRDs subject to phosphorylation by the phosphotransferase system (332). AtxA crystalizes as a dimer, and distinct associated folds predicted that DNA-binding domains and PRDs are apparent in each chain. The model for PRD control of AtxA function is that phosphorylation events at two specific histidines, one in each PRD, have opposing effects on AtxA function.
Specific DNA-binding activity of AtxA has not been reported, but phosphorylation of H199 of PRD1 is predicted to affect the interaction of AtxA with DNA, while phosphorylation of H379 in PRD2 disrupts AtxA dimerization and abolishes activity (333). Phosphorylation of AcpA and AcpB has not been demonstrated. Although comparisons of the AcpA and AcpB amino acid sequences and structure predictions are highly suggestive of the presence of PRDs, the placement of histidines within the PRDs differ between the three regulators, suggesting large differences in the potential phosphorylation sites (330). If indeed the AcpA and AcpB PRDs control PCVR function, the relationships between phosphorylation and protein structure may differ between the PCVRs, affecting their target specificity and relative activity.
GENETIC TOOLS
Natural Genetic Exchange: Intra- and Interspecies
Historically, facile identification of the B. cereus group species has relied on their plasmid content: plasmids carrying the crystal insecticidal genes of B. thuringiensis, the anthrax toxin and capsule genes of B. anthracis and more recently B. cereus bv. anthracis, and the biosynthetic gene cluster for cereulide synthesis of B. cereus (6, 16, 49, 334). However, genetic experiments have revealed that plasmids can move among the strains of the B. cereus group by conjugation and transduction (335–338), thus complicating the speciation of strains belonging to this bacterial group. Horizontal genetic transfer of plasmid and chromosome DNA has likely contributed to the remarkable diversity of the strains composing the B. cereus group.
Transducing phage have been characterized for all three B. cereus species. The bacteriophage CP-51, isolated from B. cereus, was the first generalized transducing phage discovered for the group and has been the most utilized for genetic manipulation (339). CP-51 and CP-54, a phage with a broader host range than CP-51, also mediate generalized transduction in B. thuringiensis (340, 341). They can transduce various plasmids between the B. cereus group species and are also useful for intraspecies transduction of chromosomal markers (338, 342). CP-51-mediated transduction of pXO2 from B. anthracis to B. cereus was instrumental in demonstrating that the plasmid was associated with capsule synthesis (343), while the first genome maps of B. thuringiensis were established using this phage (340, 341, 344).
Plasmid conjugation in bacteria of the B. cereus group was first demonstrated by showing the transfer of plasmid-borne cry genes from B. thuringiensis strains to B. cereus (51). These observations were then extended to other plasmids and species, including B. anthracis (343, 345). Conjugative plasmids native to B. thuringiensis have been shown to transfer to B. anthracis and B. cereus and in some cases to mobilize other non-self-transmissible plasmids. Among these are B. thuringiensis plasmids harboring insecticidal toxin genes, such as pXO12 (348) and pHT73 (347). The conjugation experiments performed between B. anthracis and B. thuringiensis revealed transfer of plasmids by a conduction process involving the formation of cointegrate structures between a conjugative plasmid (i.e., pXO12) and the nonconjugative plasmid pXO1. These experiments showed that the transposon Tn4430, located in various conjugative plasmids of B. thuringiensis (8, 346), mediates the transfer of nonconjugative plasmids by a conduction process (348). These findings indicate that transposable elements associated with conjugative plasmids may have important adaptive functions by mediating the horizontal transfer of genetic material, including toxin genes, within the B. cereus group.
Plasmid transfer among the B. cereus group has been reported to occur in various niches, including the insect gut (349–351), gnotobiotic rats (352), the plant rhizosphere (63), soil (61), water (353), and dairy products (354, 355), and a growing database of DNA sequences from clinical and environmental isolates reveals evidence for extensive genetic exchange among the B. cereus group. Multiple isolates from soil carry pXO1- and pXO2-like plasmids (24, 355–358). In some cases, clinical isolates from humans and animals have been reported with newly discovered combinations of potential plasmid-borne virulence genes. Interestingly, pAW63, from a B. thuringiensis kurstaki strain, shares a common backbone sequence with pXO2 of B. anthracis and with pBT9727 from the pathogenic B. thuringiensis subspecies konkukian (359, 360). B. cereus strain G9241, isolated from a patient with life-threatening pneumonia, carries a specific PA gene (pagA) on plasmid pBCXO1 and a gene on plasmid pBC218 that encodes a polysaccharide capsule (24).
Some reports have also described genetic exchange between B. subtilis and members of the B. cereus group. Plasmid pLS20 from B. subtilis subspecies natto is self-transmissible and facilitates transfer of other plasmids from B. subtilis to the B. cereus group species (361). Although integrative and conjugative elements (ICEs) have not been reported in the B. cereus group, B. anthracis has been shown to be an effective recipient of ICEBs1 from B. subtilis (362). The presence of cry genes encoding insecticidal toxins in Bacillus popilliae (363) and Clostridium bifermentans (364) strongly suggests a horizontal transfer of B. thuringiensis plasmids to other spore-forming Gram-positive bacteria.
Generation of Recombinant Strains and Genetic Analysis
B. cereus group members are not naturally transformable, and the commercialization of the BioRad electroporation system in the 1980s was a crucial milestone in the development of genetic approaches for these bacteria. In 1989, several electroporation procedures were published for the transformation of B. thuringiensis and B. cereus (365–367, 434) and were followed by procedures functional in B. anthracis (311) or in the emetic B. cereus strains (42, 370).
Interestingly, it was shown that electrotransformation is significantly improved when plasmids are prepared in DNA methyltransferase-deficient (dam) E. coli strains or from B. subtilis 168, a strain that is not known to possess adenine-methylating activities (371, 372). Plasmids can also be transferred from Escherichia coli to the bacteria of the B. cereus group by heterogramic conjugation (368, 369, 373). A second important milestone for the development of genetic studies in the B. cereus group was the construction of cloning and expression vectors. This was notably achieved thanks to the use of the pHT1030 replicon, which allowed the construction of a series of highly stable vectors (374, 435). Due to the relatively low transformation frequency of the B. cereus strains, site-directed mutagenesis and direct allelic exchange is not possible using linear DNA and requires the use of shuttle plasmids such as pMAD or pRN5101, whose replication in Gram-positive bacteria is thermosensitive (375, 436). More recent systems draw upon methods first developed for other bacteria. They include group II intron targeting (376) and systems using tyrosine site-specific recombinases that catalyze cutting and joining reactions between short specific DNA sequences. The systems employing tyrosine site-specific recombinases and their cognate target sequences have been derived from E. coli bacteriophage P1 Cre-loxP, Saccharomyces cerevisiae Flp-FRT, and a newly discovered IntXO-PSL system from B. anthracis plasmid pXO1 (377). A method for conducting I-SceI-mediated allelic exchange in which recombinational loss of a chromosomally integrated allelic exchange vector is stimulated by creation of a double-stranded break within the vector by the homing endonuclease I-SceI has also been described (378, 379). This system has been used to make in-frame deletions in genetic elements of B. anthracis (152, 161, 380), B. cereus (381, 382), and B. thuringiensis (383).
For random mutagenesis, Tn10- and mariner-based mini-transposons have proved useful for the detection of virulence genes or for the study of genetic systems (384–388). Insertion mutant libraries have also been generated using Enterococcus faecalis transposons Tn916 and Tn917. However, insertions of these transposons can be unstable and are sometimes accompanied by local deletions and DNA rearrangements (389, 390).
An in vivo expression technology was developed for the identification of B. cereus genes specifically expressed during infection in an insect host (266). This genetic tool allows identification of transient promoter activation by using site-specific recombination. The activity of a promoter cloned upstream from a recombinase gene results in the loss of a resistance marker and the activation of a new resistance gene. The in vivo screening of a B. cereus genomic library led to the characterization of genes expressed during infection in the insect G. mellonella. One of these genes was found specifically expressed in the hemocoel and was required for iron acquisition (267), while a five-gene cluster, specifically expressed in the larval gut, was essential for glucose-6-phosphate uptake (391).
Measurements of Gene Expression
Whole-transcriptome sequences of B. anthracis and B. cereus cultured in different growth conditions have been described (392–394). Not surprisingly, the data predict gene functions and reveal multiple transcripts not associated with previous genome annotations. As is true for other bacteria, the advent of RNA whole transcriptome shotgun sequencing (RNASeq) and quantitative reverse transcriptase PCR (qRT-PCR) has lessened the need for recombinant methods to measure gene expression. Nevertheless, for some studies of the bacteria of the B. cereus group, expression of specific genes can be readily monitored using transcriptional fusions employing promoterless genes for β-galactosidase and chloramphenicol acetyltransferase (369, 396, 397). Investigations of transcript and protein function are also aided by the use of recombinant constructs harboring promoters inducible by isopropyl-β-Dthiogalactopyranoside or xylose (223, 273, 299, 398) or by use of recombinant constructs harboring promoters inducible by cold (283).
Bioluminescent reporter systems, based on luxABCDE promoter fusions, for real-time monitoring of gene transcription have been described for B. anthracis and for B. cereus. A sspBp::lux plasmid was used for in vivo spore germination of B. anthracis in mice (164), and a cesP1::lux plasmid was successfully employed for monitoring cereulide synthetase (ces) promoter activity in complex environments, such as foods (58, 370). Various fluorescent reporters were used to record gene expression at the single-cell level and to follow the physiological stages of B. thuringiensis in different growth conditions, including biofilm and infection in insect larvae (174, 399). Furthermore, the E. coli/B. cereus shuttle vector pAD123 with a promoterless red-shifted green fluorescent protein gene (gfpmut3a) was shown to be an effective tool for promoter studies in B. cereus (370, 400).
THERAPEUTICS AND VACCINES
Antibiotic therapy is the primary treatment for anthrax and for severe infections caused by B. cereus and B. thuringiensis. The B. cereus group species have similar antibiotic susceptibility profiles and are susceptible to the majority of frontline antibiotics. Historically, penicillin was the antibiotic of choice for the treatment of known or suspected anthrax. However, intrinsic or inducible resistance to β-lactam antibiotics has been reported in clinical isolates (401–403).
Because B. cereus and B. thuringiensis infections are relatively rare, are generally treatable with antibiotics, and in the case of B. cereus food-poisoning, are self-limiting, vaccines have not been developed for these infections. Anthrax vaccines for humans have been available for decades, but the nature of the vaccine, as described below, provides the impetus for continued research in anthrax vaccine development.
Anthrax Prophylaxis and Treatment
The Centers for Disease Control and Prevention recommends fluoroquinolones, such as ciprofloxacin or doxycycline, as first-line drugs for postexposure prophylaxis and treatment of B. anthracis infections. The bactericidal activity and exceptional central nervous system penetration make ciprofloxacin recommended over doxycycline. For treatment of severe systemic and life-threatening disease and for cases with fulminant bacteremia, ciprofloxacin with additional antibiotics, especially clindamycin, is recommended. Clindamycin inhibits protein synthesis, potentially reducing toxin production (404, 405). Treatment with additional antibiotics is also recommended because of reports of ciprofloxacin-resistance generated following serial passage in the presence of increasing concentrations of the antibiotic. Ciprofloxacin resistance is associated with a point mutation in the gyrA gene encoding gyrase A (406, 407).
Long courses of antibiotics are recommended for anthrax prophylaxis and treatment because B. anthracis spores have been reported to remain viable for up to 60 days in nonhuman primates. Recently, the U.S. Food and Drug Administration approved the anthrax vaccine AVA for postexposure prophylaxis in conjunction with antimicrobials. The efficacy of antibiotic therapy for anthrax decreases as the time between exposure and treatment increases (408–411), likely due to increasing toxin levels, indicating a need for antitoxin therapeutics. Toxin inhibitors in development include a variety of peptides and antibodies (412, 413).
Anthrax Vaccines
Current human vaccines for anthrax are the U.S.-licensed AVA (anthrax vaccine adsorbed, BioThrax) and the United Kingdom-licensed AVP (anthrax vaccine precipitated). The anthrax toxin protein PA is the major immunogenic component of both vaccines. PA elicits toxin-neutralizing antibodies that correlate with protection against anthrax disease in humans and many animal models (414–418). AVA is an aluminum hydroxide-adsorbed, PA-containing supernate from a culture of a toxin-producing (pXO1+) but noncapsulated (pXO2–) strain of B. anthracis. The standard AVA vaccination protocol requires five doses administered intramuscularly over 18 months, followed by annual booster injections (419), while a more recently approved regimen requires three doses over 6 months. AVP is precipitated with aluminum potassium sulfate (alum) from culture supernates of a different toxinogenic noncapsulated strain. The AVP vaccine is administered as four doses over 32 weeks with annual boosters. Unlike AVA, AVP contains low levels of LF and EF, and some reports indicate that humoral responses to LF and EF generated by AVP may contribute to toxin neutralization (420, 421).
The prolonged immunization regimen with reports of reactogenicity, cold-storage requirement, and 4-year half-life make the anthrax vaccines nonoptimal for pre-exposure prophylaxis and postexposure therapy. Thus, various expression systems, immunogens, adjuvants, and delivery vehicles are in development to achieve a higher-performing and more user-friendly vaccine.
Efforts include investigations of purified recombinant PA preparations, Lactobacillus- and Salmonella-producing PA for oral delivery, DNA vaccines, and non-PA-based vaccines with alternative targets such as the B. anthracis capsule, cell wall S-layer proteins, spore components, and the toxin proteins EF and LF (422–427). The veterinary anthrax vaccine is a live spore vaccine developed in 1937 by Max Sterne (428). The strain, 34F2 or “Sterne,” was first isolated as a stable noncapsulated isolate and was later shown to carry pXO1+ but not pXO2–. Animals injected with the Sterne vaccine develop humoral immunity within 4 weeks, but the immunity is not long-lived, and annual revaccination is recommended. Although the vaccine can be effective in livestock populations, in many parts of the world, underreporting, logistics, and limited resources make implementing vaccination campaigns difficult.
FUTURE DIRECTIONS
Substantial effort has gone into determining biomarkers and distinguishing features of the closely related members of the B. cereus group. However, distinctions between the species and between virulent and nonvirulent strains of the species are not always clear. A key issue for B. cereus (and for B. thuringiensis) is the detection of pathogenic strains responsible for food poisoning resulting in diarrheal syndromes. These syndromes are suspected to be caused by the various cytotoxins and degradative enzymes produced by bacteria under the control of the virulence regulator PlcR. However, most strains produce these factors and are not generally pathogenic, as is likely the case with B. thuringiensis strains that are used as biopesticides.
B. thuringiensis is defined by the ability to produce a parasporal inclusion made of Cry proteins. Numerous Cry proteins are insecticidal or nematicidal toxins, but some have no known targets. The search for the presumed targets, whether in arthropods or other organisms, is of scientific and economical interest and will facilitate increased understanding of the expanded ecology of these bacteria. Efforts to identify biomarkers specific for B. anthracis are a significant part of biodefense research. While such studies have affirmed some novel features of the species, the recent reports of B. cereus isolates causing anthrax-like disease suggest that strains of other species may also present a bioweapon threat. Moreover, increased environmental surveillance and expanded genome sequencing are revealing newly recognized B. cereus group isolates with unique physiology, genetics, and virulence capabilities.
The emergence of clinical isolates with novel combinations of plasmids and recombinant forms of well-characterized plasmids suggests an evolving virulence repertoire. It will be interesting to explore the mechanisms by which these changes occur and the frequencies of apparent horizontal gene transfer in nonhost environments. What relationships, if any, exist between plasmid content and specific chromosomes? What happens to the signaling networks controlling gene expression in these atypical isolates? For some B. cereus isolates, plasmids can represent up to 20% of the total DNA. With the exception of genes involved in pathogenicity, the adaptive functions of these plasmids are still largely unknown.
Much has been learned about host-pathogen interactions associated with virulent strains of the B. cereus group species. What are the core genomic factors of the B. cereus group species that are required to cause distinct diseases? Can such factors actually be defined? Do the “virulence” plasmids play roles in the nonhost environments of the B. cereus group species? To answer these questions, more advances must be made in the study of the ecology of these organisms, their lifestyles outside of their hosts, and their interactions with each other.
REFERENCES
- 1.Liu Y, Lai Q, Göker M, Meier-Kolthoff JP, Wang M, Sun Y, Wang L, Shao Z. 2015. Genomic insights into the taxonomic status of the Bacillus cereus group. Sci Rep 5:14082 10.1038/srep14082. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lapidus A, Goltsman E, Auger S, Galleron N, Ségurens B, Dossat C, Land ML, Broussolle V, Brillard J, Guinebretiere MH, Sanchis V, Nguen-The C, Lereclus D, Richardson P, Wincker P, Weissenbach J, Ehrlich SD, Sorokin A. 2008. Extending the Bacillus cereus group genomics to putative food-borne pathogens of different toxicity. Chem Biol Interact 171:236–249 10.1016/j.cbi.2007.03.003. [PubMed] [DOI] [PubMed] [Google Scholar]
- 3.Kolstø AB, Lereclus D, Mock M. 2002. Genome structure and evolution of the Bacillus cereus group. Curr Top Microbiol Immunol 264:95–108. [PubMed] [Google Scholar]
- 4.Rasko DA, Altherr MR, Han CS, Ravel J. 2005. Genomics of the Bacillus cereus group of organisms. FEMS Microbiol Rev 29:303–329. [PubMed] [DOI] [PubMed] [Google Scholar]
- 5.Okinaka R, Cloud K, Hampton O, Hoffmaster A, Hill K, Keim P, Koehler T, Lamke G, Kumano S, Manter D, Martinez Y, Ricke D, Svensson R, Jackson P. 1999. Sequence, assembly and analysis of pX01 and pX02. J Appl Microbiol 87:261–262 10.1046/j.1365-2672.1999.00883.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 6.Ehling-Schulz M, Fricker M, Grallert H, Rieck P, Wagner M, Scherer S. 2006. Cereulide synthetase gene cluster from emetic Bacillus cereus: structure and location on a mega virulence plasmid related to Bacillus anthracis toxin plasmid pXO1. BMC Microbiol 6:20 10.1186/1471-2180-6-20. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rasko DA, Rosovitz MJ, Økstad OA, Fouts DE, Jiang L, Cer RZ, Kolstø AB, Gill SR, Ravel J. 2007. Complete sequence analysis of novel plasmids from emetic and periodontal Bacillus cereus isolates reveals a common evolutionary history among the B. cereus-group plasmids, including Bacillus anthracis pXO1. J Bacteriol 189:52–64 10.1128/JB.01313-06. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lereclus D, Ribier J, Klier A, Menou G, Lecadet M-M. 1984. A transposon-like structure related to the delta-endotoxin gene of Bacillus thuringiensis. EMBO J 3:2561–2567 10.1002/j.1460-2075.1984.tb02174.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berry C, O’Neil S, Ben-Dov E, Jones AF, Murphy L, Quail MA, Holden MT, Harris D, Zaritsky A, Parkhill J. 2002. Complete sequence and organization of pBtoxis, the toxin-coding plasmid of Bacillus thuringiensis subsp. israelensis. Appl Environ Microbiol 68:5082–5095 10.1128/AEM.68.10.5082-5095.2002. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kronstad JW, Whiteley HR. 1984. Inverted repeat sequences flank a Bacillus thuringiensis crystal protein gene. J Bacteriol 160:95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Franco C, Beccari E, Santini T, Pisaneschi G, Tecce G. 2002. Colony shape as a genetic trait in the pattern-forming Bacillus mycoides. BMC Microbiol 2:33 10.1186/1471-2180-2-33. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakamura LK. 1998. Bacillus pseudomycoides sp. nov. Int J Syst Bacteriol 48:1031–1035 10.1099/00207713-48-3-1031. [PubMed] [DOI] [PubMed] [Google Scholar]
- 13.Lechner S, Mayr R, Francis KP, Prüss BM, Kaplan T, Wiessner-Gunkel E, Stewart GS, Scherer S. 1998. Bacillus weihenstephanensis sp. nov. is a new psychrotolerant species of the Bacillus cereus group. Int J Syst Bacteriol 48:1373–1382 10.1099/00207713-48-4-1373. [PubMed] [DOI] [PubMed] [Google Scholar]
- 14.Guinebretière MH, Auger S, Galleron N, Contzen M, De Sarrau B, De Buyser ML, Lamberet G, Fagerlund A, Granum PE, Lereclus D, De Vos P, Nguyen-The C, Sorokin A. 2013. Bacillus cytotoxicus sp. nov. is a novel thermotolerant species of the Bacillus cereus group occasionally associated with food poisoning. Int J Syst Evol Microbiol 63:31–40 10.1099/ijs.0.030627-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 15.Jiménez G, Urdiain M, Cifuentes A, López-López A, Blanch AR, Tamames J, Kämpfer P, Kolstø AB, Ramón D, Martínez JF, Codoñer FM, Rosselló-Móra R. 2013. Description of Bacillus toyonensis sp. nov., a novel species of the Bacillus cereus group, and pairwise genome comparisons of the species of the group by means of ANI calculations. Syst Appl Microbiol 36:383–391 10.1016/j.syapm.2013.04.008. [PubMed] [DOI] [PubMed] [Google Scholar]
- 16.Helgason E, Økstad OA, Caugant DA, Johansen HA, Fouet A, Mock M, Hegna I, Kolstø AB. 2000. Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis: one species on the basis of genetic evidence. Appl Environ Microbiol 66:2627–2630 10.1128/AEM.66.6.2627-2630.2000. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Agaisse H, Gominet M, Økstad OA, Kolstø AB, Lereclus D. 1999. PlcR is a pleiotropic regulator of extracellular virulence factor gene expression in Bacillus thuringiensis. Mol Microbiol 32:1043–1053 10.1046/j.1365-2958.1999.01419.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 18.Mignot T, Mock M, Robichon D, Landier A, Lereclus D, Fouet A. 2001. The incompatibility between the PlcR- and AtxA-controlled regulons may have selected a nonsense mutation in Bacillus anthracis. Mol Microbiol 42:1189–1198 10.1046/j.1365-2958.2001.02692.x. [DOI] [PubMed] [Google Scholar]
- 19.Koch R. 1876. Die Ätiologie der Milzbrand-Krankheit, begründet auf die Entwicklungsgeschichte des Bacillus anthracis. Beitr Biol Pflanz 2:277–310. [Google Scholar]
- 20.Pasteur L. 1881. Le vaccin du charbon. C R Acad Sci 92:666–668. [Google Scholar]
- 21.Klee SR, Ozel M, Appel B, Boesch C, Ellerbrok H, Jacob D, Holland G, Leendertz FH, Pauli G, Grunow R, Nattermann H. 2006. Characterization of Bacillus anthracis-like bacteria isolated from wild great apes from Cote d’Ivoire and Cameroon. J Bacteriol 188:5333–5344 10.1128/JB.00303-06. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brézillon C, Haustant M, Dupke S, Corre JP, Lander A, Franz T, Monot M, Couture-Tosi E, Jouvion G, Leendertz FH, Grunow R, Mock ME, Klee SR, Goossens PL. 2015. Capsules, toxins and AtxA as virulence factors of emerging Bacillus cereus biovar anthracis. PLoS Negl Trop Dis 9:e0003455 10.1371/journal.pntd.0003455. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller JM, Hair JG, Hebert M, Hebert L, Roberts FJ Jr, Weyant RS. 1997. Fulminating bacteremia and pneumonia due to Bacillus cereus. J Clin Microbiol 35:504–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoffmaster AR, Ravel J, Rasko DA, Chapman GD, Chute MD, Marston CK, De BK, Sacchi CT, Fitzgerald C, Mayer LW, Maiden MC, Priest FG, Barker M, Jiang L, Cer RZ, Rilstone J, Peterson SN, Weyant RS, Galloway DR, Read TD, Popovic T, Fraser CM. 2004. Identification of anthrax toxin genes in a Bacillus cereus associated with an illness resembling inhalation anthrax. Proc Natl Acad Sci U S A 101:8449–8454 10.1073/pnas.0402414101. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Avashia SB, Riggins WS, Lindley C, Hoffmaster A, Drumgoole R, Nekomoto T, Jackson PJ, Hill KK, Williams K, Lehman L, Libal MC, Wilkins PP, Alexander J, Tvaryanas A, Betz T. 2007. Fatal pneumonia among metalworkers due to inhalation exposure to Bacillus cereus containing Bacillus anthracis toxin genes. Clin Infect Dis 44:414–416 10.1086/510429. [PubMed] [DOI] [PubMed] [Google Scholar]
- 26.Wright AM, Beres SB, Consamus EN, Long SW, Flores AR, Barrios R, Richter GS, Oh SY, Garufi G, Maier H, Drews AL, Stockbauer KE, Cernoch P, Schneewind O, Olsen RJ, Musser JM. 2011. Rapidly progressive, fatal, inhalation anthrax-like infection in a human: case report, pathogen genome sequencing, pathology, and coordinated response. Arch Pathol Lab Med 135:1447–1459 10.5858/2011-0362-SAIR.1. [PubMed] [DOI] [PubMed] [Google Scholar]
- 27.Gee JE, Marston CK, Sammons SA, Burroughs MA, Hoffmaster AR. 2014. Draft genome sequence of Bacillus cereus strain BcFL2013, a clinical isolate similar to G9241. Genome Announc 2:e00469-14 10.1128/genomeA.00469-14. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Visschedyk D, Rochon A, Tempel W, Dimov S, Park HW, Merrill AR. 2012. Certhrax toxin, an anthrax-related ADP-ribosyltransferase from Bacillus cereus. J Biol Chem 287:41089–41102 10.1074/jbc.M112.412809. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simon NC, Vergis JM, Ebrahimi AV, Ventura CL, O’Brien AD, Barbieri JT. 2013. Host cell cytotoxicity and cytoskeleton disruption by CerADPr, an ADP-ribosyltransferase of Bacillus cereus G9241. Biochemistry 52:2309–2318 10.1021/bi300692g. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simon NC, Barbieri JT. 2014. Bacillus cereus Certhrax ADP-ribosylates vinculin to disrupt focal adhesion complexes and cell adhesion. J Biol Chem 289:10650–10659 10.1074/jbc.M113.500710. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oh SY, Budzik JM, Garufi G, Schneewind O. 2011. Two capsular polysaccharides enable Bacillus cereus G9241 to cause anthrax-like disease. Mol Microbiol 80:455–470 10.1111/j.1365-2958.2011.07582.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson MK, Vergis JM, Alem F, Palmer JR, Keane-Myers AM, Brahmbhatt TN, Ventura CL, O’Brien AD. 2011. Bacillus cereus G9241 makes anthrax toxin and capsule like highly virulent B. anthracis Ames but behaves like attenuated toxigenic nonencapsulated B. anthracis Sterne in rabbits and mice. Infect Immun 79:3012–3019 10.1128/IAI.00205-11. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frankland GC, Frankland PF. 1887. Studies on some new micro-organisms obtained from air. R Soc Lond Philos Trans B 178:257–287 10.1098/rstb.1887.0011. [DOI] [Google Scholar]
- 34.Bottone EJ. 2010. Bacillus cereus, a volatile human pathogen. Clin Microbiol Rev 23:382–398 10.1128/CMR.00073-09. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ehling-Schulz M, Knutsson R, Scherer S. 2011. Bacillus cereus, p 147–164. In Kathariou S, Fratamico P, Lui Y (ed), Genomes of Food- and Water-Borne Pathogens. ASM Press, Washington DC. [Google Scholar]
- 36.Hauge S. 1955. Food poisoning caused by aerobic spore forming bacilli. J Appl Bacteriol 18:591–595 10.1111/j.1365-2672.1955.tb02116.x. [DOI] [Google Scholar]
- 37.Stenfors Arnesen LP, Fagerlund A, Granum PE. 2008. From soil to gut: Bacillus cereus and its food poisoning toxins. FEMS Microbiol Rev 32:579–606 10.1111/j.1574-6976.2008.00112.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 38.Fedhila S, Nel P, Lereclus D. 2002. The InhA2 metalloprotease of Bacillus thuringiensis strain 407 is required for pathogenicity in insects infected via the oral route. J Bacteriol 184:3296–3304 10.1128/JB.184.12.3296-3304.2002. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Doll VM, Ehling-Schulz M, Vogelmann R. 2013. Concerted action of sphingomyelinase and non-hemolytic enterotoxin in pathogenic Bacillus cereus. PLoS One 8:e61404 10.1371/journal.pone.0061404. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Böhm ME, Huptas C, Krey VM, Scherer S. 2015. Massive horizontal gene transfer, strictly vertical inheritance and ancient duplications differentially shape the evolution of Bacillus cereus enterotoxin operons hbl, cytK and nhe. BMC Evol Biol 15:246 10.1186/s12862-015-0529-4. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ehling-Schulz M, Svensson B, Guinebretiere MH, Lindbäck T, Andersson M, Schulz A, Fricker M, Christiansson A, Granum PE, Märtlbauer E, Nguyen-The C, Salkinoja-Salonen M, Scherer S. 2005. Emetic toxin formation of Bacillus cereus is restricted to a single evolutionary lineage of closely related strains. Microbiology 151:183–197 10.1099/mic.0.27607-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 42.Ehling-Schulz M, Vukov N, Schulz A, Shaheen R, Andersson M, Märtlbauer E, Scherer S. 2005. Identification and partial characterization of the nonribosomal peptide synthetase gene responsible for cereulide production in emetic Bacillus cereus. Appl Environ Microbiol 71:105–113 10.1128/AEM.71.1.105-113.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ishiwata I. 1901. On a kind of severe flacherie (sotto diseases). Dainihon Sanshi Kaiho 114:1–5. [Google Scholar]
- 44.Berliner E. 1915. Uber die Schlaffsucht der Mehlmottenraupe (Ephestia kuhniella, Zell.) ihren Erreger, Bacillus thuringiensisn. sp. Z Angew Entomol 2:29–56 10.1111/j.1439-0418.1915.tb00334.x. [DOI] [Google Scholar]
- 45.Goldberg LH, Margalit J. 1977. A bacterial spore demonstrating rapid larvicidal activity against Anopheles sergentii, Uranotaenia unguiculata, Culex univitatus, Aedes aegypti and Culex pipiens. Mosq News 37:355–358. [Google Scholar]
- 46.Krieg A, Huger AM, Langenbruch GA, Schnetter W. 1983. Bacillus thuringiensis var. tenebrionis: ein neuer gegenuber arven von Coleopteren wirksamer athotyp. Z Ang Ent 96:500–508 10.1111/j.1439-0418.1983.tb03704.x. [DOI] [Google Scholar]
- 47.Wei JZ, Hale K, Carta L, Platzer E, Wong C, Fang SC, Aroian RV. 2003. Bacillus thuringiensis crystal proteins that target nematodes. Proc Natl Acad Sci U S A 100:2760–2765 10.1073/pnas.0538072100. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanchis V. 2011. From microbial sprays to insect-resistant transgenic plants: history of the biospesticide Bacillus thuringiensis. A review. Agron Sustain Dev 31:217–231 10.1051/agro/2010027. [DOI] [Google Scholar]
- 49.Schnepf E, Crickmore N, Van Rie J, Lereclus D, Baum J, Feitelson J, Zeigler DR, Dean DH. 1998. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol Mol Biol Rev 62:775–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Espinasse S, Chaufaux J, Buisson C, Perchat S, Gohar M, Bourguet D, Sanchis V. 2003. Occurrence and linkage between secreted insecticidal toxins in natural isolates of Bacillus thuringiensis. Curr Microbiol 47:501–507 10.1007/s00284-003-4097-2. [PubMed] [DOI] [PubMed] [Google Scholar]
- 51.González JMJ Jr, Brown BJ, Carlton BC. 1982. Transfer of Bacillus thuringiensis plasmids coding for delta-endotoxin among strains of B. thuringiensis and B. cereus. Proc Natl Acad Sci U S A 79:6951–6955 10.1073/pnas.79.22.6951. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mahillon J, Rezsöhazy R, Hallet B, Delcour J. 1994. IS231 and other Bacillus thuringiensis transposable elements: a review. Genetica 93:13–26 10.1007/BF01435236. [PubMed] [DOI] [PubMed] [Google Scholar]
- 53.Menou G, Mahillon J, Lecadet MM, Lereclus D. 1990. Structural and genetic organization of IS232, a new insertion sequence of Bacillus thuringiensis. J Bacteriol 172:6689–6696 10.1128/jb.172.12.6689-6696.1990. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jensen GB, Hansen BM, Eilenberg J, Mahillon J. 2003. The hidden lifestyles of Bacillus cereus and relatives. Environ Microbiol 5:631–640 10.1046/j.1462-2920.2003.00461.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 55.Martin PAW, Travers RS. 1989. Worldwide abundance and distribution of Bacillus thuringiensis isolates. Appl Environ Microbiol 55:2437–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meadows MP, Ellis DJ, Butt J, Jarrett P, Burges HD. 1992. Distribution, frequency, and diversity of Bacillus thuringiensis in an animal feed mill. Appl Environ Microbiol 58:1344–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smith RA, Couche GA. 1991. The phyllophane as a source of Bacillus thuringiensis variants. Appl Environ Microbiol 57:311–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Messelhäusser U, Frenzel E, Blöchinger C, Zucker R, Kämpf P, Ehling-Schulz M. 2014. Emetic Bacillus cereus are more volatile than thought: recent foodborne outbreaks and prevalence studies in Bavaria (2007-2013). BioMed Res Int 2014:465603 10.1155/2014/465603. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hoton FM, Fornelos N, N’guessan E, Hu X, Swiecicka I, Dierick K, Jääskeläinen E, Salkinoja-Salonen M, Mahillon J. 2009. Family portrait of Bacillus cereus and Bacillus weihenstephanensis cereulide-producing strains. Environ Microbiol Rep 1:177–183 10.1111/j.1758-2229.2009.00028.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 60.Monnerat RG, Soares CM, Capdeville G, Jones G, Martins ES, Praça L, Cordeiro BA, Braz SV, dos Santos RC, Berry C. 2009. Translocation and insecticidal activity of Bacillus thuringiensis living inside of plants. Microb Biotechnol 2:512–520 10.1111/j.1751-7915.2009.00116.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vilas-Bôas LA, Vilas-Bôas GF, Saridakis HO, Lemos MV, Lereclus D, Arantes OM. 2000. Survival and conjugation of Bacillus thuringiensis in a soil microcosm. FEMS Microbiol Ecol 31:255–259 10.1016/S0168-6496(00)00002-7. [DOI] [PubMed] [Google Scholar]
- 62.West AW, Burges HD, Dixon TJ, Wyborn CH. 1985. Survival of Bacillus thuringiensis and Bacillus cereus spore inocula in soil: effects of pH, moisture, nutrient availability and indigenous microorganisms. Soil Biol Biochem 17:657–665 10.1016/0038-0717(85)90043-4. [DOI] [Google Scholar]
- 63.Saile E, Koehler TM. 2006. Bacillus anthracis multiplication, persistence, and genetic exchange in the rhizosphere of grass plants. Appl Environ Microbiol 72:3168–3174 10.1128/AEM.72.5.3168-3174.2006. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fricker M, Ågren J, Segerman B, Knutsson R, Ehling-Schulz M. 2011. Evaluation of Bacillus strains as model systems for the work on Bacillus anthracis spores. Int J Food Microbiol 145(Suppl 1):S129–S136 10.1016/j.ijfoodmicro.2010.07.036. [PubMed] [DOI] [PubMed] [Google Scholar]
- 65.Hoornstra D, Andersson MA, Teplova VV, Mikkola R, Uotila LM, Andersson LC, Roivainen M, Gahmberg CG, Salkinoja-Salonen MS. 2013. Potato crop as a source of emetic Bacillus cereus and cereulide-induced mammalian cell toxicity. Appl Environ Microbiol 79:3534–3543 10.1128/AEM.00201-13. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ehling-Schulz M, Frenzel E, Gohar M. 2015. Food-bacteria interplay: pathometabolism of emetic Bacillus cereus. Front Microbiol 6:704 10.3389/fmicb.2015.00704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Halverson LJ, Clayton MK, Handelsman J. 1993. Variable stability of antibiotic-resistance markers in Bacillus cereus UW85 in the soybean rhizosphere in the field. Mol Ecol 2:65–78 10.1111/j.1365-294X.1993.tb00001.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 68.Dutta S, Rani TS, Podile AR. 2013. Root exudate-induced alterations in Bacillus cereus cell wall contribute to root colonization and plant growth promotion. PLoS One 8:e78369 10.1371/journal.pone.0078369. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang S, Zheng Y, Gu C, He C, Yang M, Zhang X, Guo J, Zhao H, Niu D. 2017. Bacillus cereus AR156 activates defense responses to Pseudomonas syringae pv. tomato in Arabidopsis thaliana similarly to flg22. Mol Plant Microbe Interact 31:311–322. [PubMed] [DOI] [PubMed] [Google Scholar]
- 70.Gohar M, Faegri K, Perchat S, Ravnum S, Økstad OA, Gominet M, Kolstø AB, Lereclus D. 2008. The PlcR virulence regulon of Bacillus cereus. PLoS One 3:e2793 10.1371/journal.pone.0002793. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ivanova N, Sorokin A, Anderson I, Galleron N, Candelon B, Kapatral V, Bhattacharyya A, Reznik G, Mikhailova N, Lapidus A, Chu L, Mazur M, Goltsman E, Larsen N, D’Souza M, Walunas T, Grechkin Y, Pusch G, Haselkorn R, Fonstein M, Ehrlich SD, Overbeek R, Kyrpides N. 2003. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 423:87–91 10.1038/nature01582. [PubMed] [DOI] [PubMed] [Google Scholar]
- 72.Raymond B, Johnston PR, Nielsen-LeRoux C, Lereclus D, Crickmore N. 2010. Bacillus thuringiensis: an impotent pathogen? Trends Microbiol 18:189–194 10.1016/j.tim.2010.02.006. [PubMed] [DOI] [PubMed] [Google Scholar]
- 73.Braun P, Grass G, Aceti A, Serrecchia L, Affuso A, Marino L, Grimaldi S, Pagano S, Hanczaruk M, Georgi E, Northoff B, Schöler A, Schloter M, Antwerpen M, Fasanella A. 2015. Microevolution of anthrax from a young ancestor (M.A.Y.A.) suggests a soil-borne life cycle of Bacillus anthracis. PLoS One 10:e0135346 10.1371/journal.pone.0135346. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Antonation KS, Grützmacher K, Dupke S, Mabon P, Zimmermann F, Lankester F, Peller T, Feistner A, Todd A, Herbinger I, de Nys HM, Muyembe-Tamfun JJ, Karhemere S, Wittig RM, Couacy-Hymann E, Grunow R, Calvignac-Spencer S, Corbett CR, Klee SR, Leendertz FH. 2016. Bacillus cereus biovar anthracis causing anthrax in sub-Saharan Africa: chromosomal monophyly and broad geographic distribution. PLoS Negl Trop Dis 10:e0004923 10.1371/journal.pntd.0004923. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Scarff JM, Raynor MJ, Seldina YI, Ventura CL, Koehler TM, O’Brien AD. 2016. The roles of AtxA orthologs in virulence of anthrax-like Bacillus cereus G9241. Mol Microbiol 102:545–561 10.1111/mmi.13478. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Argôlo-Filho RC, Loguercio LL. 2013. Bacillus thuringiensis is an environmental pathogen and host-specificity has developed as an adaptation to human-generated ecological niches. Insects 5:62–91 10.3390/insects5010062. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Blackburn JK, Matakarimov S, Kozhokeeva S, Tagaeva Z, Bell LK, Kracalik IT, Zhunushov A. 2017. Modeling the ecological niche of Bacillus anthracis to map anthrax risk in Kyrgyzstan. Am J Trop Med Hyg 96:550–556. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hugh-Jones M, Blackburn J. 2009. The ecology of Bacillus anthracis. Mol Aspects Med 30:356–367 10.1016/j.mam.2009.08.003. [PubMed] [DOI] [PubMed] [Google Scholar]
- 79.Valseth K, Nesbø CL, Easterday WR, Turner WC, Olsen JS, Stenseth NC, Haverkamp THA. 2017. Temporal dynamics in microbial soil communities at anthrax carcass sites. BMC Microbiol 17:206 10.1186/s12866-017-1111-6. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dey R, Hoffman PS, Glomski IJ. 2012. Germination and amplification of anthrax spores by soil-dwelling amoebas. Appl Environ Microbiol 78:8075–8081 10.1128/AEM.02034-12. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Barandongo ZR, Mfune JKE, Turner WC. 2017. Dust-bathing behaviors of African herbivores and the potential risk of inhalational anthrax. J Wildl Dis 54:34–44. [PubMed] [DOI] [PubMed] [Google Scholar]
- 82.Turner WC, Kausrud KL, Beyer W, Easterday WR, Barandongo ZR, Blaschke E, Cloete CC, Lazak J, Van Ert MN, Ganz HH, Turnbull PC, Stenseth NC, Getz WM. 2016. Lethal exposure: an integrated approach to pathogen transmission via environmental reservoirs. Sci Rep 6:27311 10.1038/srep27311. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Turnbull PC. 2002. Introduction: anthrax history, disease and ecology. Curr Top Microbiol Immunol 271:1–19 10.1007/978-3-662-05767-4_1. [PubMed] [DOI] [PubMed] [Google Scholar]
- 84.Jernigan DB, Raghunathan PL, Bell BP, Brechner R, Bresnitz EA, Butler JC, Cetron M, Cohen M, Doyle T, Fischer M, Greene C, Griffith KS, Guarner J, Hadler JL, Hayslett JA, Meyer R, Petersen LR, Phillips M, Pinner R, Popovic T, Quinn CP, Reefhuis J, Reissman D, Rosenstein N, Schuchat A, Shieh WJ, Siegal L, Swerdlow DL, Tenover FC, Traeger M, Ward JW, Weisfuse I, Wiersma S, Yeskey K, Zaki S, Ashford DA, Perkins BA, Ostroff S, Hughes J, Fleming D, Koplan JP, Gerberding JL, National Anthrax Epidemiologic Investigation Team. 2002. Investigation of bioterrorism-related anthrax, United States, 2001: epidemiologic findings. Emerg Infect Dis 8:1019–1028 10.3201/eid0810.020353. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fouet A. 2009. The surface of Bacillus anthracis. Mol Aspects Med 30:374–385 10.1016/j.mam.2009.07.001. [PubMed] [DOI] [PubMed] [Google Scholar]
- 86.Liu S, Moayeri M, Leppla SH. 2014. Anthrax lethal and edema toxins in anthrax pathogenesis. Trends Microbiol 22:317–325 10.1016/j.tim.2014.02.012. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Meselson M, Guillemin J, Hugh-Jones M, Langmuir A, Popova I, Shelokov A, Yampolskaya O. 1994. The Sverdlovsk anthrax outbreak of 1979. Science 266:1202–1208 10.1126/science.7973702. [PubMed] [DOI] [PubMed] [Google Scholar]
- 88.Agrawal A, Lingappa J, Leppla SH, Agrawal S, Jabbar A, Quinn C, Pulendran B. 2003. Impairment of dendritic cells and adaptive immunity by anthrax lethal toxin. Nature 424:329–334 10.1038/nature01794. [PubMed] [DOI] [PubMed] [Google Scholar]
- 89.Moayeri M, Leppla SH, Vrentas C, Pomerantsev AP, Liu S. 2015. Anthrax pathogenesis. Annu Rev Microbiol 69:185–208 10.1146/annurev-micro-091014-104523. [PubMed] [DOI] [PubMed] [Google Scholar]
- 90.Makino S, Watarai M, Cheun HI, Shirahata T, Uchida I. 2002. Effect of the lower molecular capsule released from the cell surface of Bacillus anthracis on the pathogenesis of anthrax. J Infect Dis 186:227–233 10.1086/341299. [PubMed] [DOI] [PubMed] [Google Scholar]
- 91.Kozel TR, Murphy WJ, Brandt S, Blazar BR, Lovchik JA, Thorkildson P, Percival A, Lyons CR. 2004. mAbs to Bacillus anthracis capsular antigen for immunoprotection in anthrax and detection of antigenemia. Proc Natl Acad Sci U S A 101:5042–5047 10.1073/pnas.0401351101. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sutherland MD, Kozel TR. 2009. Macrophage uptake, intracellular localization, and degradation of poly-gamma-d-glutamic acid, the capsular antigen of Bacillus anthracis. Infect Immun 77:532–538 10.1128/IAI.01009-08. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ezzell JW, Abshire TG, Panchal R, Chabot D, Bavari S, Leffel EK, Purcell B, Friedlander AM, Ribot WJ. 2009. Association of Bacillus anthracis capsule with lethal toxin during experimental infection. Infect Immun 77:749–755 10.1128/IAI.00764-08. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jang J, Cho M, Chun JH, Cho MH, Park J, Oh HB, Yoo CK, Rhie GE. 2011. The poly-γ-d-glutamic acid capsule of Bacillus anthracis enhances lethal toxin activity. Infect Immun 79:3846–3854 10.1128/IAI.01145-10. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lee HR, Jeon JH, Park OK, Chun JH, Park J, Rhie GE. 2015. The poly-γ-d-glutamic acid capsule surrogate of the Bacillus anthracis capsule induces nitric oxide production via the platelet activating factor receptor signaling pathway. Mol Immunol 68(2 Pt A):244–252 10.1016/j.molimm.2015.08.015. [PubMed] [DOI] [PubMed] [Google Scholar]
- 96.Lee HR, Jeon JH, Rhie GE. 2017. The poly-γ-D-glutamic acid capsule of Bacillus licheniformis: a surrogate of Bacillus anthracis capsule induces interferon-gamma production in NK cells through interactions with macrophages. J Microbiol Biotechnol 27:1032–1037 10.4014/jmb.1612.12043. [PubMed] [DOI] [PubMed] [Google Scholar]
- 97.Cho MH, Ahn HJ, Ha HJ, Park J, Chun JH, Kim BS, Oh HB, Rhie GE. 2010. Bacillus anthracis capsule activates caspase-1 and induces interleukin-1beta release from differentiated THP-1 and human monocyte-derived dendritic cells. Infect Immun 78:387–392 10.1128/IAI.00956-09. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chitlaru T, Gat O, Gozlan Y, Ariel N, Shafferman A. 2006. Differential proteomic analysis of the Bacillus anthracis secretome: distinct plasmid and chromosome CO2-dependent cross talk mechanisms modulate extracellular proteolytic activities. J Bacteriol 188:3551–3571 10.1128/JB.188.10.3551-3571.2006. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chitlaru T, Gat O, Grosfeld H, Inbar I, Gozlan Y, Shafferman A. 2007. Identification of in vivo-expressed immunogenic proteins by serological proteome analysis of the Bacillus anthracis secretome. Infect Immun 75:2841–2852 10.1128/IAI.02029-06. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Thwaite JE, Hibbs S, Titball RW, Atkins TP. 2006. Proteolytic degradation of human antimicrobial peptide LL-37 by Bacillus anthracis may contribute to virulence. Antimicrob Agents Chemother 50:2316–2322 10.1128/AAC.01488-05. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chung MC, Popova TG, Millis BA, Mukherjee DV, Zhou W, Liotta LA, Petricoin EF, Chandhoke V, Bailey C, Popov SG. 2006. Secreted neutral metalloproteases of Bacillus anthracis as candidate pathogenic factors. J Biol Chem 281:31408–31418 10.1074/jbc.M605526200. [PubMed] [DOI] [PubMed] [Google Scholar]
- 102.Chung MC, Popova TG, Jorgensen SC, Dong L, Chandhoke V, Bailey CL, Popov SG. 2008. Degradation of circulating von Willebrand factor and its regulator ADAMTS13 implicates secreted Bacillus anthracis metalloproteases in anthrax consumptive coagulopathy. J Biol Chem 283:9531–9542 10.1074/jbc.M705871200. [PubMed] [DOI] [PubMed] [Google Scholar]
- 103.Mukherjee DV, Tonry JH, Kim KS, Ramarao N, Popova TG, Bailey C, Popov S, Chung MC. 2011. Bacillus anthracis protease InhA increases blood-brain barrier permeability and contributes to cerebral hemorrhages. PLoS One 6:e17921 10.1371/journal.pone.0017921. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Grinberg LM, Abramova FA, Yampolskaya OV, Walker DH, Smith JH. 2001. Quantitative pathology of inhalational anthrax I: quantitative microscopic findings. Mod Pathol 14:482–495 10.1038/modpathol.3880337. [PubMed] [DOI] [PubMed] [Google Scholar]
- 105.Ehling-Schulz M, Fricker M, Scherer S. 2004. Bacillus cereus, the causative agent of an emetic type of food-borne illness. Mol Nutr Food Res 48:479–487 10.1002/mnfr.200400055. [PubMed] [DOI] [PubMed] [Google Scholar]
- 106.Schoeni JL, Wong AC. 2005. Bacillus cereus food poisoning and its toxins. J Food Prot 68:636–648 10.4315/0362-028X-68.3.636. [PubMed] [DOI] [PubMed] [Google Scholar]
- 107.Prüss BM, Dietrich R, Nibler B, Märtlbauer E, Scherer S. 1999. The hemolytic enterotoxin HBL is broadly distributed among species of the Bacillus cereus group. Appl Environ Microbiol 65:5436–5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ehling-Schulz M, Messelhäusser U. 2012. One pathogen but two different types of food borne outbreaks; Bacillus cereus in catering facilities in Germany , p 63–70. In Hoorfar J (ed), Case Studies in Food Safety and Quality Management: Lessons from Real-Life Situations. Woodhead Publishing, Cambridge, United Kingdom. [Google Scholar]
- 109.Lotte R, Hérissé AL, Berrouane Y, Lotte L, Casagrande F, Landraud L, Herbin S, Ramarao N, Boyer L, Ruimy R. 2017. Virulence analysis of Bacillus cereus isolated after death of preterm neonates, Nice, France, 2013. Emerg Infect Dis 23:845–848 10.3201/eid2305.161788. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dierick K, Van Coillie E, Swiecicka I, Meyfroidt G, Devlieger H, Meulemans A, Hoedemaekers G, Fourie L, Heyndrickx M, Mahillon J. 2005. Fatal family outbreak of Bacillus cereus-associated food poisoning. J Clin Microbiol 43:4277–4279 10.1128/JCM.43.8.4277-4279.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pósfay-Barbe KM, Schrenzel J, Frey J, Studer R, Korff C, Belli DC, Parvex P, Rimensberger PC, Schäppi MG. 2008. Food poisoning as a cause of acute liver failure. Pediatr Infect Dis J 27:846–847 10.1097/INF.0b013e318170f2ae. [PubMed] [DOI] [PubMed] [Google Scholar]
- 112.Tschiedel E, Rath PM, Steinmann J, Becker H, Dietrich R, Paul A, Felderhoff-Müser U, Dohna-Schwake C. 2015. Lifesaving liver transplantation for multi-organ failure caused by Bacillus cereus food poisoning. Pediatr Transplant 19:E11–E14 10.1111/petr.12378. [PubMed] [DOI] [PubMed] [Google Scholar]
- 113.Stark T, Marxen S, Rütschle A, Lücking G, Scherer S, Ehling-Schulz M, Hofmann T. 2013. Mass spectrometric profiling of Bacillus cereus strains and quantitation of the emetic toxin cereulide by means of stable isotope dilution analysis and HEp-2 bioassay. Anal Bioanal Chem 405:191–201 10.1007/s00216-012-6485-6. [PubMed] [DOI] [PubMed] [Google Scholar]
- 114.Jeßberger N, Krey VM, Rademacher C, Böhm ME, Mohr AK, Ehling-Schulz M, Scherer S, Märtlbauer E. 2015. From genome to toxicity: a combinatory approach highlights the complexity of enterotoxin production in Bacillus cereus. Front Microbiol 6:560. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kranzler M, Stollewerk K, Rouzeau-Szynalski K, Blayo L, Sulyok M, Ehling-Schulz M. 2016. Temperature exerts control of Bacillus cereus emetic toxin production on post-transcriptional levels. Front Microbiol 7:1640 10.3389/fmicb.2016.01640. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jeßberger N, Rademacher C, Krey VM, Dietrich R, Mohr AK, Böhm ME, Scherer S, Ehling-Schulz M, Märtlbauer E. 2017. Simulating intestinal growth conditions enhances toxin production of enteropathogenic Bacillus cereus. Front Microbiol 8:627 10.3389/fmicb.2017.00627. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pinna A, Sechi LA, Zanetti S, Usai D, Delogu G, Cappuccinelli P, Carta F. 2001. Bacillus cereus keratitis associated with contact lens wear. Ophthalmology 108:1830–1834 10.1016/S0161-6420(01)00723-0. [DOI] [PubMed] [Google Scholar]
- 118.Jones TO, Turnbull PC. 1981. Bovine mastitis caused by Bacillus cereus. Vet Rec 108:271–274 10.1136/vr.108.13.271. [PubMed] [DOI] [PubMed] [Google Scholar]
- 119.Mavangira V, Angelos JA, Samitz EM, Rowe JD, Byrne BA. 2013. Gangrenous mastitis caused by Bacillus species in six goats. J Am Vet Med Assoc 242:836–843 10.2460/javma.242.6.836. [PubMed] [DOI] [PubMed] [Google Scholar]
- 120.Chaufaux J, Marchal M, Gilois N, Jehanno I, Buisson C. 1997. Investigation of natural strains of Bacillus thuringiensis in different biotopes through the world. Can J Microbiol 43:337–343 10.1139/m97-047. [DOI] [Google Scholar]
- 121.Ruan L, Crickmore N, Peng D, Sun M. 2015. Are nematodes a missing link in the confounded ecology of the entomopathogen Bacillus thuringiensis? Trends Microbiol 23:341–346 10.1016/j.tim.2015.02.011. [PubMed] [DOI] [PubMed] [Google Scholar]
- 122.Borgonie G, Van Driessche R, Leyns F, Arnaut G, De Waele D, Coomans A. 1995. Germination of Bacillus thuringiensis spores in bacteriophagous nematodes (Nematoda: rhabditida). J Invertebr Pathol 65:61–67 10.1006/jipa.1995.1008. [PubMed] [DOI] [PubMed] [Google Scholar]
- 123.van Frankenhuyzen K. 2009. Insecticidal activity of Bacillus thuringiensis crystal proteins. J Invertebr Pathol 101:1–16 10.1016/j.jip.2009.02.009. [PubMed] [DOI] [PubMed] [Google Scholar]
- 124.Loguercio LL, Argôlo-Filho RC. 2015. Anthropogenic action shapes the evolutionary ecology of Bacillus thuringiensis: response to Ruan et al. Trends Microbiol 23:519–520 10.1016/j.tim.2015.06.002. [PubMed] [DOI] [PubMed] [Google Scholar]
- 125.Hendriksen NB. 2016. The two lives of Bacillus thuringiensis: response to Ruan et al. and Loguercio and Argôlo-Filho. Trends Microbiol 24:1–2 10.1016/j.tim.2015.10.010. [PubMed] [DOI] [PubMed] [Google Scholar]
- 126.Lereclus D, Agaisse H, Grandvalet C, Salamitou S, Gominet M. 2000. Regulation of toxin and virulence gene transcription in Bacillus thuringiensis. Int J Med Microbiol 290:295–299 10.1016/S1438-4221(00)80024-7. [DOI] [PubMed] [Google Scholar]
- 127.Raymond B, West SA, Griffin AS, Bonsall MB. 2012. The dynamics of cooperative bacterial virulence in the field. Science 337:85–88 10.1126/science.1218196. [PubMed] [DOI] [PubMed] [Google Scholar]
- 128.Dubois T, Faegri K, Gélis-Jeanvoine S, Perchat S, Lemy C, Buisson C, Nielsen-LeRoux C, Gohar M, Jacques P, Ramarao N, Slamti L, Kolstø AB, Lereclus D. 2016. Correction: necrotrophism is a quorum-sensing-regulated lifestyle in Bacillus thuringiensis. PLoS Pathog 12:e1006049 10.1371/journal.ppat.1006049. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Perchat S, Dubois T, Zouhir S, Gominet M, Poncet S, Lemy C, Aumont-Nicaise M, Deutscher J, Gohar M, Nessler S, Lereclus D. 2011. A cell-cell communication system regulates protease production during sporulation in bacteria of the Bacillus cereus group. Mol Microbiol 82:619–633 10.1111/j.1365-2958.2011.07839.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 130.Vilain S, Luo Y, Hildreth MB, Brözel VS. 2006. Analysis of the life cycle of the soil saprophyte Bacillus cereus in liquid soil extract and in soil. Appl Environ Microbiol 72:4970–4977 10.1128/AEM.03076-05. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ruthel G, Ribot WJ, Bavari S, Hoover TA. 2004. Time-lapse confocal imaging of development of Bacillus anthracis in macrophages. J Infect Dis 189:1313–1316 10.1086/382656. [PubMed] [DOI] [PubMed] [Google Scholar]
- 132.Oh SY, Lunderberg JM, Chateau A, Schneewind O, Missiakas D. 2016. Genes required for Bacillus anthracis secondary cell wall polysaccharide synthesis. J Bacteriol 199:e00613-16. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chateau A, Lunderberg JM, Oh SY, Abshire T, Friedlander A, Quinn CP, Missiakas DM, Schneewind O. 2018. Galactosylation of the secondary cell wall polysaccharide of Bacillus anthracis and its contribution to anthrax pathogenesis. J Bacteriol 200:e00562-17. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Forsberg LS, Choudhury B, Leoff C, Marston CK, Hoffmaster AR, Saile E, Quinn CP, Kannenberg EL, Carlson RW. 2011. Secondary cell wall polysaccharides from Bacillus cereus strains G9241, 03BB87 and 03BB102 causing fatal pneumonia share similar glycosyl structures with the polysaccharides from Bacillus anthracis. Glycobiology 21:934–948 10.1093/glycob/cwr026. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sychantha D, Chapman RN, Bamford NC, Boons GJ, Howell PL, Clarke AJ. 2018. Molecular basis for the attachment of S-layer proteins to the cell wall of Bacillus anthracis. Biochemistry 57:1949–1953 10.1021/acs.biochem.8b00060. [PubMed] [DOI] [PubMed] [Google Scholar]
- 136.Missiakas D, Schneewind O. 2017. Assembly and function of the Bacillus anthracis S-layer. Annu Rev Microbiol 71:79–98 10.1146/annurev-micro-090816-093512. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Anderson VJ, Kern JW, McCool JW, Schneewind O, Missiakas D. 2011. The SLH-domain protein BslO is a determinant of Bacillus anthracis chain length. Mol Microbiol 81:192–205 10.1111/j.1365-2958.2011.07688.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Goodman JW, Nitecki DE. 1967. Studies on the relation of a prior immune response to immunogenicity. Immunology 13:577–583. [PMC free article] [PubMed] [Google Scholar]
- 139.Scorpio A, Chabot DJ, Day WA, O’Brien DK, Vietri NJ, Itoh Y, Mohamadzadeh M, Friedlander AM. 2007. Poly-gamma-glutamate capsule-degrading enzyme treatment enhances phagocytosis and killing of encapsulated Bacillus anthracis. Antimicrob Agents Chemother 51:215–222 10.1128/AAC.00706-06. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.AuCoin DP, Sutherland MD, Percival AL, Lyons CR, Lovchik JA, Kozel TR. 2009. Rapid detection of the poly-gamma-d-glutamic acid capsular antigen of Bacillus anthracis by latex agglutination. Diagn Microbiol Infect Dis 64:229–232 10.1016/j.diagmicrobio.2009.02.001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Boyer AE, Quinn CP, Hoffmaster AR, Kozel TR, Saile E, Marston CK, Percival A, Plikaytis BD, Woolfitt AR, Gallegos M, Sabourin P, McWilliams LG, Pirkle JL, Barr JR. 2009. Kinetics of lethal factor and poly-d-glutamic acid antigenemia during inhalation anthrax in rhesus macaques. Infect Immun 77:3432–3441 10.1128/IAI.00346-09. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Jelacic TM, Chabot DJ, Bozue JA, Tobery SA, West MW, Moody K, Yang D, Oppenheim JJ, Friedlander AM. 2014. Exposure to Bacillus anthracis capsule results in suppression of human monocyte-derived dendritic cells. Infect Immun 82:3405–3416 10.1128/IAI.01857-14. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Swick MC, Koehler TM, Driks A. 2016. Surviving between hosts: sporulation and transmission. Microbiol Spectr 4:10.1128/microbiolspec.VMBF-0029-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Moir A. 2006. How do spores germinate? J Appl Microbiol 101:526–530 10.1111/j.1365-2672.2006.02885.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 145.Bozue JA, Welkos S, Cote CK. 2015. The Bacillus anthracis exosporium: what’s the big “hairy” deal? Microbiol Spectr 3:3 10.1128/microbiolspec.TBS-0021-2015. [DOI] [PubMed] [Google Scholar]
- 146.Terry C, Jiang S, Radford DS, Wan Q, Tzokov S, Moir A, Bullough PA. 2017. Molecular tiling on the surface of a bacterial spore: the exosporium of the Bacillus anthracis/cereus/thuringiensis group. Mol Microbiol 104:539–552 10.1111/mmi.13650. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Weaver J, Kang TJ, Raines KW, Cao GL, Hibbs S, Tsai P, Baillie L, Rosen GM, Cross AS. 2007. Protective role of Bacillus anthracis exosporium in macrophage-mediated killing by nitric oxide. Infect Immun 75:3894–3901 10.1128/IAI.00283-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Johnson MJ, Todd SJ, Ball DA, Shepherd AM, Sylvestre P, Moir A. 2006. ExsY and CotY are required for the correct assembly of the exosporium and spore coat of Bacillus cereus. J Bacteriol 188:7905–7913 10.1128/JB.00997-06. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ball DA, Taylor R, Todd SJ, Redmond C, Couture-Tosi E, Sylvestre P, Moir A, Bullough PA. 2008. Structure of the exosporium and sublayers of spores of the Bacillus cereus family revealed by electron crystallography. Mol Microbiol 68:947–958 10.1111/j.1365-2958.2008.06206.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 150.Giorno R, Mallozzi M, Bozue J, Moody KS, Slack A, Qiu D, Wang R, Friedlander A, Welkos S, Driks A. 2009. Localization and assembly of proteins comprising the outer structures of the Bacillus anthracis spore. Microbiology 155:1133–1145 10.1099/mic.0.023333-0. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Giorno R, Bozue J, Cote C, Wenzel T, Moody KS, Mallozzi M, Ryan M, Wang R, Zielke R, Maddock JR, Friedlander A, Welkos S, Driks A. 2007. Morphogenesis of the Bacillus anthracis spore. J Bacteriol 189:691–705 10.1128/JB.00921-06. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Brahmbhatt TN, Janes BK, Stibitz ES, Darnell SC, Sanz P, Rasmussen SB, O’Brien AD. 2007. Bacillus anthracis exosporium protein BclA affects spore germination, interaction with extracellular matrix proteins, and hydrophobicity. Infect Immun 75:5233–5239 10.1128/IAI.00660-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Oliva C, Turnbough CL Jr, Kearney JF. 2009. CD14-Mac-1 interactions in Bacillus anthracis spore internalization by macrophages. Proc Natl Acad Sci U S A 106:13957–13962 10.1073/pnas.0902392106. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Oliva CR, Swiecki MK, Griguer CE, Lisanby MW, Bullard DC, Turnbough CL Jr, Kearney JF. 2008. The integrin Mac-1 (CR3) mediates internalization and directs Bacillus anthracis spores into professional phagocytes. Proc Natl Acad Sci U S A 105:1261–1266 10.1073/pnas.0709321105. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Fisher N, Hanna P. 2005. Characterization of Bacillus anthracis germinant receptors in vitro. J Bacteriol 187:8055–8062 10.1128/JB.187.23.8055-8062.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ireland JA, Hanna PC. 2002. Amino acid- and purine ribonucleoside-induced germination of Bacillus anthracis DeltaSterne endospores: gerS mediates responses to aromatic ring structures. J Bacteriol 184:1296–1303 10.1128/JB.184.5.1296-1303.2002. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Weiner MA, Read TD, Hanna PC. 2003. Identification and characterization of the gerH operon of Bacillus anthracis endospores: a differential role for purine nucleosides in germination. J Bacteriol 185:1462–1464 10.1128/JB.185.4.1462-1464.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Guidi-Rontani C, Pereira Y, Ruffie S, Sirard JC, Weber-Levy M, Mock M. 1999. Identification and characterization of a germination operon on the virulence plasmid pXO1 of Bacillus anthracis. Mol Microbiol 33:407–414 10.1046/j.1365-2958.1999.01485.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 159.McKevitt MT, Bryant KM, Shakir SM, Larabee JL, Blanke SR, Lovchik J, Lyons CR, Ballard JD. 2007. Effects of endogenous d-alanine synthesis and autoinhibition of Bacillus anthracis germination on in vitro and in vivo infections. Infect Immun 75:5726–5734 10.1128/IAI.00727-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Setlow P. 2014. Spore resistance properties. Microbiol Spectr 2:10.1128/microbiolspec.TBS-0003-2012 10.1128/microbiolspec.TBS-0003-2012. [PubMed] [DOI] [PubMed] [Google Scholar]
- 161.Giebel JD, Carr KA, Anderson EC, Hanna PC. 2009. The germination-specific lytic enzymes SleB, CwlJ1, and CwlJ2 each contribute to Bacillus anthracis spore germination and virulence. J Bacteriol 191:5569–5576 10.1128/JB.00408-09. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ross JM. 1955. On the histopathology of experimental anthrax in the guinea-pig. Br J Exp Pathol 36:336–339. [PMC free article] [PubMed] [Google Scholar]
- 163.Cleret A, Quesnel-Hellmann A, Vallon-Eberhard A, Verrier B, Jung S, Vidal D, Mathieu J, Tournier JN. 2007. Lung dendritic cells rapidly mediate anthrax spore entry through the pulmonary route. J Immunol 178:7994–8001 10.4049/jimmunol.178.12.7994. [PubMed] [DOI] [PubMed] [Google Scholar]
- 164.Sanz P, Teel LD, Alem F, Carvalho HM, Darnell SC, O’Brien AD. 2008. Detection of Bacillus anthracis spore germination in vivo by bioluminescence imaging. Infect Immun 76:1036–1047 10.1128/IAI.00985-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Cote CK, Van Rooijen N, Welkos SL. 2006. Roles of macrophages and neutrophils in the early host response to Bacillus anthracis spores in a mouse model of infection. Infect Immun 74:469–480 10.1128/IAI.74.1.469-480.2006. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wilson GR, Benoit TG. 1990. Activation and germination of Bacillus thuringiensis spores in Manduca sexta larval gut fluid. J Invertebr Pathol 56:233–236 10.1016/0022-2011(90)90105-F. [DOI] [PubMed] [Google Scholar]
- 167.Wilson GR, Benoit TG. 1993. Alkaline pH activates Bacillus thuringiensis spores. J Invertebr Pathol 62:87–89 10.1006/jipa.1993.1079. [DOI] [Google Scholar]
- 168.Abdoarrahem MM, Gammon K, Dancer BN, Berry C. 2009. Genetic basis for alkaline activation of germination in Bacillus thuringiensis subsp. israelensis. Appl Environ Microbiol 75:6410–6413 10.1128/AEM.00962-09. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Du C, Nickerson KW. 1996. Bacillus thuringiensis HD-73 spores have surface-localized Cry1Ac toxin: physiological and pathogenic consequences. Appl Environ Microbiol 62:3722–3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Bongiorni C, Stoessel R, Shoemaker D, Perego M. 2006. Rap phosphatase of virulence plasmid pXO1 inhibits Bacillus anthracis sporulation. J Bacteriol 188:487–498 10.1128/JB.188.2.487-498.2006. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Brunsing RL, La Clair C, Tang S, Chiang C, Hancock LE, Perego M, Hoch JA. 2005. Characterization of sporulation histidine kinases of Bacillus anthracis. J Bacteriol 187:6972–6981 10.1128/JB.187.20.6972-6981.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.White AK, Hoch JA, Grynberg M, Godzik A, Perego M. 2006. Sensor domains encoded in Bacillus anthracis virulence plasmids prevent sporulation by hijacking a sporulation sensor histidine kinase. J Bacteriol 188:6354–6360 10.1128/JB.00656-06. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Perchat S, Talagas A, Zouhir S, Poncet S, Bouillaut L, Nessler S, Lereclus D. 2016. NprR, a moonlighting quorum sensor shifting from a phosphatase activity to a transcriptional activator. Microb Cell 3:573–575 10.15698/mic2016.11.542. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Verplaetse E, Slamti L, Gohar M, Lereclus D. 2015. Cell differentiation in a Bacillus thuringiensis population during planktonic growth, biofilm formation, and host infection. MBio 6:e00138-15 10.1128/mBio.00138-15. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Perego M. 2013. Forty years in the making: understanding the molecular mechanism of peptide regulation in bacterial development. PLoS Biol 11:e1001516 10.1371/journal.pbio.1001516. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Fazion F, Perchat S, Buisson C, Vilas-Boas G, Lereclus D. 2017. A plasmid-borne Rap-Phr system regulates sporulation of Bacillus thuringiensis in insect larvae. Environ Microbiol 20:145–155. [PubMed] [DOI] [PubMed] [Google Scholar]
- 177.Roux E, Yersin A. 1888. Contribution a à l’étude de la diphthérie. Ann Inst Pasteur 2:629–661. [Google Scholar]
- 178.Böhm ME, Krey VM, Jeßberger N, Frenzel E, Scherer S. 2016. Comparative bioinformatics and experimental analysis of the intergenic regulatory regions of Bacillus cereushbl and nhe enterotoxin operons and the impact of CodY on virulence heterogeneity. Front Microbiol 7:768 10.3389/fmicb.2016.00768. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Bradley KA, Mogridge J, Mourez M, Collier RJ, Young JA. 2001. Identification of the cellular receptor for anthrax toxin. Nature 414:225–229 10.1038/n35101999. [PubMed] [DOI] [PubMed] [Google Scholar]
- 180.Scobie HM, Rainey GJ, Bradley KA, Young JA. 2003. Human capillary morphogenesis protein 2 functions as an anthrax toxin receptor. Proc Natl Acad Sci U S A 100:5170–5174 10.1073/pnas.0431098100. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Feld GK, Brown MJ, Krantz BA. 2012. Ratcheting up protein translocation with anthrax toxin. Protein Sci 21:606–624 10.1002/pro.2052. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Park JM, Greten FR, Li ZW, Karin M. 2002. Macrophage apoptosis by anthrax lethal factor through p38 MAP kinase inhibition. Science 297:2048–2051 10.1126/science.1073163. [PubMed] [DOI] [PubMed] [Google Scholar]
- 183.Greaney AJ, Leppla SH, Moayeri M. 2015. Bacterial exotoxins and the inflammasome. Front Immunol 6:570 10.3389/fimmu.2015.00570. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Nakajima K, Tanaka Y. 2010. Exclusion of Kif1c as a candidate gene for anthrax toxin susceptibility. Microb Pathog 48:188–190 10.1016/j.micpath.2010.02.001. [PubMed] [DOI] [PubMed] [Google Scholar]
- 185.Leppla SH. 1982. Anthrax toxin edema factor: a bacterial adenylate cyclase that increases cyclic AMP concentrations of eukaryotic cells. Proc Natl Acad Sci U S A 79:3162–3166 10.1073/pnas.79.10.3162. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.O’Brien J, Friedlander A, Dreier T, Ezzell J, Leppla S. 1985. Effects of anthrax toxin components on human neutrophils. Infect Immun 47:306–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Tournier JN, Rossi Paccani S, Quesnel-Hellmann A, Baldari CT. 2009. Anthrax toxins: a weapon to systematically dismantle the host immune defenses. Mol Aspects Med 30:456–466 10.1016/j.mam.2009.06.002. [PubMed] [DOI] [PubMed] [Google Scholar]
- 188.Moayeri M, Crown D, Dorward DW, Gardner D, Ward JM, Li Y, Cui X, Eichacker P, Leppla SH. 2009. The heart is an early target of anthrax lethal toxin in mice: a protective role for neuronal nitric oxide synthase (nNOS). PLoS Pathog 5:e1000456 10.1371/journal.ppat.1000456. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Tweten RK. 2005. Cholesterol-dependent cytolysins, a family of versatile pore-forming toxins. Infect Immun 73:6199–6209 10.1128/IAI.73.10.6199-6209.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Shannon JG, Ross CL, Koehler TM, Rest RF. 2003. Characterization of anthrolysin O, the Bacillus anthracis cholesterol-dependent cytolysin. Infect Immun 71:3183–3189 10.1128/IAI.71.6.3183-3189.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Mosser EM, Rest RF. 2006. The Bacillus anthracis cholesterol-dependent cytolysin, anthrolysin O, kills human neutrophils, monocytes and macrophages. BMC Microbiol 6:56 10.1186/1471-2180-6-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Bourdeau RW, Malito E, Chenal A, Bishop BL, Musch MW, Villereal ML, Chang EB, Mosser EM, Rest RF, Tang WJ. 2009. Cellular functions and X-ray structure of anthrolysin O, a cholesterol-dependent cytolysin secreted by Bacillus anthracis. J Biol Chem 284:14645–14656 10.1074/jbc.M807631200. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Bishop BL, Lodolce JP, Kolodziej LE, Boone DL, Tang WJ. 2010. The role of anthrolysin O in gut epithelial barrier disruption during Bacillus anthracis infection. Biochem Biophys Res Commun 394:254–259 10.1016/j.bbrc.2010.02.091. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Bergman NH, Anderson EC, Swenson EE, Janes BK, Fisher N, Niemeyer MM, Miyoshi AD, Hanna PC. 2007. Transcriptional profiling of Bacillus anthracis during infection of host macrophages. Infect Immun 75:3434–3444 10.1128/IAI.01345-06. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Klichko VI, Miller J, Wu A, Popov SG, Alibek K. 2003. Anaerobic induction of Bacillus anthracis hemolytic activity. Biochem Biophys Res Commun 303:855–862 10.1016/S0006-291X(03)00440-6. [DOI] [PubMed] [Google Scholar]
- 196.Heffernan BJ, Thomason B, Herring-Palmer A, Hanna P. 2007. Bacillus anthracis anthrolysin O and three phospholipases C are functionally redundant in a murine model of inhalation anthrax. FEMS Microbiol Lett 271:98–105 10.1111/j.1574-6968.2007.00713.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 197.Beecher DJ, Schoeni JL, Wong AC. 1995. Enterotoxic activity of hemolysin BL from Bacillus cereus. Infect Immun 63:4423–4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Lund T, Granum PE. 1996. Characterisation of a non-haemolytic enterotoxin complex from Bacillus cereus isolated after a foodborne outbreak. FEMS Microbiol Lett 141:151–156 10.1111/j.1574-6968.1996.tb08377.x. [DOI] [PubMed] [Google Scholar]
- 199.Lund T, De Buyser ML, Granum PE. 2000. A new cytotoxin from Bacillus cereus that may cause necrotic enteritis. Mol Microbiol 38:254–261 10.1046/j.1365-2958.2000.02147.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 200.Beecher DJ, Wong AC. 2000. Cooperative, synergistic and antagonistic haemolytic interactions between haemolysin BL, phosphatidylcholine phospholipase C and sphingomyelinase from Bacillus cereus. Microbiology 146:3033–3039 10.1099/00221287-146-12-3033. [PubMed] [DOI] [PubMed] [Google Scholar]
- 201.Callegan MC, Jett BD, Hancock LE, Gilmore MS. 1999. Role of hemolysin BL in the pathogenesis of extraintestinal Bacillus cereus infection assessed in an endophthalmitis model. Infect Immun 67:3357–3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Guillemet E, Cadot C, Tran SL, Guinebretière MH, Lereclus D, Ramarao N. 2010. The InhA metalloproteases of Bacillus cereus contribute concomitantly to virulence. J Bacteriol 192:286–294 10.1128/JB.00264-09. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Oda M, Hashimoto M, Takahashi M, Ohmae Y, Seike S, Kato R, Fujita A, Tsuge H, Nagahama M, Ochi S, Sasahara T, Hayashi S, Hirai Y,Sakurai J. 2012. Role of sphingomyelinase in infectious diseases caused by Bacillus cereus. PLoS One 7:e38054 10.1371/journal.pone.0038054. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Ramarao N, Sanchis V. 2013. The pore-forming haemolysins of bacillus cereus: a review. Toxins (Basel) 5:1119–1139 10.3390/toxins5061119. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Jeßberger N, Dietrich R, Bock S, Didier A, Märtlbauer E. 2014. Bacillus cereus enterotoxins act as major virulence factors and exhibit distinct cytotoxicity to different human cell lines. Toxicon 77:49–57 10.1016/j.toxicon.2013.10.028. [PubMed] [DOI] [PubMed] [Google Scholar]
- 206.Agata N, Ohta M, Mori M, Isobe M. 1995. A novel dodecadepsipeptide, cereulide, is an emetic toxin of Bacillus cereus. FEMS Microbiol Lett 129:17–20. [PubMed] [DOI] [PubMed] [Google Scholar]
- 207.Magarvey NA, Ehling-Schulz M, Walsh CT. 2006. Characterization of the cereulide NRPS alpha-hydroxy acid specifying modules: activation of alpha-keto acids and chiral reduction on the assembly line. J Am Chem Soc 128:10698–10699 10.1021/ja0640187. [PubMed] [DOI] [PubMed] [Google Scholar]
- 208.Marxen S, Stark TD, Rütschle A, Lücking G, Frenzel E, Scherer S, Ehling-Schulz M, Hofmann T. 2015. Depsipeptide intermediates interrogate proposed biosynthesis of cereulide, the emetic toxin of Bacillus cereus. Sci Rep 5:10637 10.1038/srep10637. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Medema MH, Kottmann R, Yilmaz P, Cummings M, Biggins JB, Blin K, de Bruijn I, Chooi YH, Claesen J, Coates RC, Cruz-Morales P, Duddela S, Düsterhus S, Edwards DJ, Fewer DP, Garg N, Geiger C, Gomez-Escribano JP, Greule A, Hadjithomas M, Haines AS, Helfrich EJN, Hillwig ML, Ishida K, Jones AC, Jones CS, Jungmann K, Kegler C, Kim HU, Kötter P, Krug D, Masschelein J, Melnik AV, Mantovani SM, Monroe EA, Moore M, Moss N, Nützmann H-W, Pan G, Pati A, Petras D, Reen FJ, Rosconi F, Rui Z, Tian Z, Tobias NJ, Tsunematsu Y, Wiemann P, Wyckoff E, Yan X, Yim G, Yu F, et al. 2015. Minimum information about a biosynthetic gene cluster. Nat Chem Biol 11:625–631 10.1038/nchembio.1890. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Bauer T, Stark T, Hofmann T, Ehling-Schulz M. 2010. Development of a stable isotope dilution analysis for the quantification of the Bacillus cereus toxin cereulide in foods. J Agric Food Chem 58:1420–1428 10.1021/jf9033046. [PubMed] [DOI] [PubMed] [Google Scholar]
- 211.Marxen S, Stark TD, Frenzel E, Rütschle A, Lücking G, Pürstinger G, Pohl EE, Scherer S, Ehling-Schulz M, Hofmann T. 2015. Chemodiversity of cereulide, the emetic toxin of Bacillus cereus. Anal Bioanal Chem 407:2439–2453 10.1007/s00216-015-8511-y. [PubMed] [DOI] [PubMed] [Google Scholar]
- 212.Rajkovic A, Uyttendaele M, Vermeulen A, Andjelkovic M, Fitz-James I, in ’t Veld P, Denon Q, Verhe R, Debevere J. 2008. Heat resistance of Bacillus cereus emetic toxin, cereulide. Lett Appl Microbiol 46:536–541. [PubMed] [DOI] [PubMed] [Google Scholar]
- 213.Vangoitsenhoven R, Rondas D, Crèvecoeur I, D’Hertog W, Baatsen P, Masini M, Andjelkovic M, Van Loco J, Matthys C, Mathieu C, Overbergh L, Van der Schueren B. 2014. Foodborne cereulide causes beta-cell dysfunction and apoptosis. PLoS One 9:e104866 10.1371/journal.pone.0104866. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Johler S, Kalbhenn EM, Heini N, Brodmann P, Gautsch S, Bagcioglu M, Contzen M, Stephan R, Ehling-Schulz M. 2018. Enterotoxin production of Bacillus thuringiensis isolates from biopesticides, foods, and outbreaks. Front Microbiol 9:1915 10.3389/fmicb.2018.01915. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Deng C, Peng Q, Song F, Lereclus D. 2014. Regulation of cry gene expression in Bacillus thuringiensis. Toxins (Basel) 6:2194–2209 10.3390/toxins6072194. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Zhu Y, Ji F, Shang H, Zhu Q, Wang P, Xu C, Deng Y, Peng D, Ruan L, Sun M. 2011. Gene clusters located on two large plasmids determine spore crystal association (SCA) in Bacillus thuringiensis subsp. finitimus strain YBT-020. PLoS One 6:e27164 10.1371/journal.pone.0027164. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Deng C, Slamti L, Raymond B, Liu G, Lemy C, Gominet M, Yang J, Wang H, Peng Q, Zhang J, Lereclus D, Song F. 2015. Division of labour and terminal differentiation in a novel Bacillus thuringiensis strain. ISME J 9:286–296 10.1038/ismej.2014.122. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Agaisse H, Lereclus D. 1995. How does Bacillus thuringiensis produce so much insecticidal crystal protein? J Bacteriol 177:6027–6032 10.1128/jb.177.21.6027-6032.1995. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Crickmore N, Zeigler DR, Feitelson J, Schnepf E, Van Rie J, Lereclus D, Baum J, Dean DH. 1998. Revision of the nomenclature for the Bacillus thuringiensis pesticidal crystal proteins. Microbiol Mol Biol Rev 62:807–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Ibrahim MA, Griko N, Junker M, Bulla LA. 2010. Bacillus thuringiensis: a genomics and proteomics perspective. Bioeng Bugs 1:31–50 10.4161/bbug.1.1.10519. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Pérez C, Fernandez LE, Sun J, Folch JL, Gill SS, Soberón M, Bravo A. 2005. Bacillus thuringiensis subsp. israelensis Cyt1Aa synergizes Cry11Aa toxin by functioning as a membrane-bound receptor. Proc Natl Acad Sci U S A 102:18303–18308 10.1073/pnas.0505494102. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Popov SG, Popova TG, Hopkins S, Weinstein RS, MacAfee R, Fryxell KJ, Chandhoke V, Bailey C, Alibek K. 2005. Effective antiprotease-antibiotic treatment of experimental anthrax. BMC Infect Dis 5:25 10.1186/1471-2334-5-25. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Pflughoeft KJ, Sumby P, Koehler TM. 2011. Bacillus anthracis sin locus and regulation of secreted proteases. J Bacteriol 193:631–639 10.1128/JB.01083-10. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Dubois T, Faegri K, Perchat S, Lemy C, Buisson C, Nielsen-LeRoux C, Gohar M, Jacques P, Ramarao N, Kolstø AB, Lereclus D. 2012. Necrotrophism is a quorum-sensing-regulated lifestyle in Bacillus thuringiensis. PLoS Pathog 8:e1002629 10.1371/journal.ppat.1002629. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Pflughoeft KJ, Swick MC, Engler DA, Yeo HJ, Koehler TM. 2014. Modulation of the Bacillus anthracis secretome by the immune inhibitor A1 protease. J Bacteriol 196:424–435 10.1128/JB.00690-13. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Gomis-Rüth FX. 2013. A different look for AB5 toxins. Structure 21:1909–1910 10.1016/j.str.2013.10.004. [PubMed] [DOI] [PubMed] [Google Scholar]
- 227.Terwilliger A, Swick MC, Pflughoeft KJ, Pomerantsev A, Lyons CR, Koehler TM, Maresso A. 2015. Bacillus anthracis overcomes an amino acid auxotrophy by cleaving host serum proteins. J Bacteriol 197:2400–2411 10.1128/JB.00073-15. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Ramarao N, Lereclus D. 2005. The InhA1 metalloprotease allows spores of the B. cereus group to escape macrophages. Cell Microbiol 7:1357–1364 10.1111/j.1462-5822.2005.00562.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 229.Welkos S, Bozue J, Twenhafel N, Cote C. 2015. Animal models for the pathogenesis, treatment, and prevention of infection by Bacillus anthracis. Microbiol Spectr 3:TBS-0001–TBS-2012. [DOI] [PubMed] [Google Scholar]
- 230.Loving CL, Khurana T, Osorio M, Lee GM, Kelly VK, Stibitz S, Merkel TJ. 2009. Role of anthrax toxins in dissemination, disease progression, and induction of protective adaptive immunity in the mouse aerosol challenge model. Infect Immun 77:255–265 10.1128/IAI.00633-08. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Friedlander AM, Welkos SL, Ivins BE. 2002. Anthrax vaccines. Curr Top Microbiol Immunol 271:33–60 10.1007/978-3-662-05767-4_3. [PubMed] [DOI] [PubMed] [Google Scholar]
- 232.Ivins BE, Welkos SL. 1986. Cloning and expression of the Bacillus anthracis protective antigen gene in Bacillus subtilis. Infect Immun 54:537–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Fellows PF, Linscott MK, Ivins BE, Pitt ML, Rossi CA, Gibbs PH, Friedlander AM. 2001. Efficacy of a human anthrax vaccine in guinea pigs, rabbits, and rhesus macaques against challenge by Bacillus anthracis isolates of diverse geographical origin. Vaccine 19:3241–3247 10.1016/S0264-410X(01)00021-4. [DOI] [PubMed] [Google Scholar]
- 234.Carr KA, Lybarger SR, Anderson EC, Janes BK, Hanna PC. 2010. The role of Bacillus anthracis germinant receptors in germination and virulence. Mol Microbiol 75:365–375 10.1111/j.1365-2958.2009.06972.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 235.Glomski IJ, Corre JP, Mock M, Goossens PL. 2007. Noncapsulated toxinogenic Bacillus anthracis presents a specific growth and dissemination pattern in naive and protective antigen-immune mice. Infect Immun 75:4754–4761 10.1128/IAI.00575-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Glomski IJ, Dumetz F, Jouvion G, Huerre MR, Mock M, Goossens PL. 2008. Inhaled non-capsulated Bacillus anthracis in A/J mice: nasopharynx and alveolar space as dual portals of entry, delayed dissemination, and specific organ targeting. Microbes Infect 10:1398–1404 10.1016/j.micinf.2008.07.042. [PubMed] [DOI] [PubMed] [Google Scholar]
- 237.Little SF, Webster WM, Ivins BE, Fellows PF, Norris SL, Andrews GP. 2004. Development of an in vitro-based potency assay for anthrax vaccine. Vaccine 22:2843–2852 10.1016/j.vaccine.2003.12.027. [PubMed] [DOI] [PubMed] [Google Scholar]
- 238.Pittman PR, Gibbs PH, Cannon TL, Friedlander AM. 2001. Anthrax vaccine: short-term safety experience in humans. Vaccine 20:972–978 10.1016/S0264-410X(01)00387-5. [DOI] [PubMed] [Google Scholar]
- 239.Pittman PR, Norris SL, Barrera Oro JG, Bedwell D, Cannon TL, McKee KT Jr. 2006. Patterns of antibody response in humans to the anthrax vaccine adsorbed (AVA) primary (six-dose) series. Vaccine 24:3654–3660 10.1016/j.vaccine.2006.01.054. [PubMed] [DOI] [PubMed] [Google Scholar]
- 240.Williamson ED, Hodgson I, Walker NJ, Topping AW, Duchars MG, Mott JM, Estep J, Lebutt C, Flick-Smith HC, Jones HE, Li H, Quinn CP. 2005. Immunogenicity of recombinant protective antigen and efficacy against aerosol challenge with anthrax. Infect Immun 73:5978–5987 10.1128/IAI.73.9.5978-5987.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Nye SH, Wittenburg AL, Evans DL, O’Connor JA, Roman RJ, Jacob HJ. 2008. Rat survival to anthrax lethal toxin is likely controlled by a single gene. Pharmacogenomics J 8:16–22 10.1038/sj.tpj.6500448. [PubMed] [DOI] [PubMed] [Google Scholar]
- 242.Newman ZL, Crown D, Leppla SH, Moayeri M. 2010. Anthrax lethal toxin activates the inflammasome in sensitive rat macrophages. Biochem Biophys Res Commun 398:785–789 10.1016/j.bbrc.2010.07.039. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Salamitou S, Ramisse F, Brehélin M, Bourguet D, Gilois N, Gominet M, Hernandez E, Lereclus D. 2000. The plcR regulon is involved in the opportunistic properties of Bacillus thuringiensis and Bacillus cereus in mice and insects. Microbiology 146:2825–2832 10.1099/00221287-146-11-2825. [PubMed] [DOI] [PubMed] [Google Scholar]
- 244.Hernandez E, Ramisse F, Gros P, Cavallo J. 2000. Super-infection by Bacillus thuringiensis H34 or 3a3b can lead to death in mice infected with the influenza A virus. FEMS Immunol Med Microbiol 29:177–181 10.1111/j.1574-695X.2000.tb01520.x. [DOI] [PubMed] [Google Scholar]
- 245.Callegan MC, Kane ST, Cochran DC, Gilmore MS, Gominet M, Lereclus D. 2003. Relationship of plcR-regulated factors to Bacillus endophthalmitis virulence. Infect Immun 71:3116–3124 10.1128/IAI.71.6.3116-3124.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Wilcks A, Hansen BM, Hendriksen NB, Licht TR. 2006. Fate and effect of ingested Bacillus cereus spores and vegetative cells in the intestinal tract of human-flora-associated rats. FEMS Immunol Med Microbiol 46:70–77. [PubMed] [DOI] [PubMed] [Google Scholar]
- 247.Rolny IS, Minnaard J, Racedo SM, Pérez PF. 2014. Murine model of Bacillus cereus gastrointestinal infection. J Med Microbiol 63:1741–1749. [PubMed] [DOI] [PubMed] [Google Scholar]
- 248.Shinagawa K, Konuma H, Sekita H, Sugii S. 1995. Emesis of rhesus monkeys induced by intragastric administration with the HEp-2 vacuolation factor (cereulide) produced by Bacillus cereus. FEMS Microbiol Lett 130:87–90. [PubMed] [DOI] [PubMed] [Google Scholar]
- 249.Melling J, Capel BJ, Turnbull PC, Gilbert RJ. 1976. Identification of a novel enterotoxigenic activity associated with Bacillus cereus. J Clin Pathol 29:938–940 10.1136/jcp.29.10.938. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Angus TA. 1954. A bacterial toxin paralysing silkworm larvae. Nature 173:545–546 10.1038/173545a0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 251.Hannay CL. 1953. Crystalline inclusions in aerobic spore-forming bacteria. Nature 172:1004 10.1038/1721004a0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 252.Bravo A, Likitvivatanavong S, Gill SS, Soberón M. 2011. Bacillus thuringiensis: a story of a successful bioinsecticide. Insect Biochem Mol Biol 41:423–431 10.1016/j.ibmb.2011.02.006. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Rees JS, Jarrett P, Ellar DJ. 2009. Peritrophic membrane contribution to Bt Cry delta-endotoxin susceptibility in Lepidoptera and the effect of calcofluor. J Invertebr Pathol 100:139–146 10.1016/j.jip.2009.01.002. [PubMed] [DOI] [PubMed] [Google Scholar]
- 254.Caccia S, Di Lelio I, La Storia A, Marinelli A, Varricchio P, Franzetti E, Banyuls N, Tettamanti G, Casartelli M, Giordana B, Ferré J, Gigliotti S, Ercolini D, Pennacchio F. 2016. Midgut microbiota and host immunocompetence underlie Bacillus thuringiensis killing mechanism. Proc Natl Acad Sci U S A 113:9486–9491 10.1073/pnas.1521741113. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Johnson DE, McGaughey WH. 1996. Contribution of Bacillus thuringiensis spores to toxicity of purified Cry proteins towards Indianmeal moth larvae. Curr Microbiol 33:54–59 10.1007/s002849900074. [PubMed] [DOI] [PubMed] [Google Scholar]
- 256.Dubois NR, Dean DH. 1995. Synergism between Cry1A insecticidal crystal proteins and spores of Bacillus thuringiensis, other bacterial spores and vegetative cells against Lymantria dispar (Lepidoptera: Lymantriidae) larvae. Environ Entomol 24:1741–1747 10.1093/ee/24.6.1741. [DOI] [Google Scholar]
- 257.Miyasono M, Inagaki S, Yamamoto M, Ohba K, Ishiguro T, Takeda R, Hayashi Y. 1994. Enhancement of δ-endotoxin activity by toxin-free spore of Bacillus thuringiensis against the diamondback moth, Plutella xylostella. J Invertebr Pathol 63:111–112 10.1006/jipa.1994.1021. [DOI] [Google Scholar]
- 258.Burges HD, Thomson EM, Latchford RA. 1976. Importance of spores and d-endotoxin protein crystals of Bacillus thuringiensis in Galleria mellonella. J Invertebr Pathol 27:87–94 10.1016/0022-2011(76)90032-X. [DOI] [Google Scholar]
- 259.Li RS, Jarrett P, Burges HD. 1987. Importance of spores, crystals, and δ-endotoxins in the pathogenicity of different varieties of Bacillus thuringiensis in Galleria mellonela and Pieris brassicae. J Invertebr Pathol 50:277–284 10.1016/0022-2011(87)90093-0. [DOI] [Google Scholar]
- 260.Fedhila S, Buisson C, Dussurget O, Serror P, Glomski IJ, Liehl P, Lereclus D, Nielsen-LeRoux C. 2010. Comparative analysis of the virulence of invertebrate and mammalian pathogenic bacteria in the oral insect infection model Galleria mellonella. J Invertebr Pathol 103:24–29 10.1016/j.jip.2009.09.005. [PubMed] [DOI] [PubMed] [Google Scholar]
- 261.Nielsen-LeRoux C, Gaudriault S, Ramarao N, Lereclus D, Givaudan A. 2012. How the insect pathogen bacteria Bacillus thuringiensis and Xenorhabdus/Photorhabdus occupy their hosts. Curr Opin Microbiol 15:220–231 10.1016/j.mib.2012.04.006. [PubMed] [DOI] [PubMed] [Google Scholar]
- 262.Peng D, Lin J, Huang Q, Zheng W, Liu G, Zheng J, Zhu L, Sun M. 2016. A novel metalloproteinase virulence factor is involved in Bacillus thuringiensis pathogenesis in nematodes and insects. Environ Microbiol 18:846–862 10.1111/1462-2920.13069. [PubMed] [DOI] [PubMed] [Google Scholar]
- 263.Fang S, Wang L, Guo W, Zhang X, Peng D, Luo C, Yu Z, Sun M. 2009. Bacillus thuringiensis bel protein enhances the toxicity of Cry1Ac protein to Helicoverpa armigera larvae by degrading insect intestinal mucin. Appl Environ Microbiol 75:5237–5243 10.1128/AEM.00532-09. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Ruan L, Crickmore N, Sun M. 2015. Is there sufficient evidence to consider Bacillus thuringiensis a multihost pathogen? Response to Loguercio and Argôlo-Filho. Trends Microbiol 23:587 10.1016/j.tim.2015.08.004. [PubMed] [DOI] [PubMed] [Google Scholar]
- 265.Luo X, Chen L, Huang Q, Zheng J, Zhou W, Peng D, Ruan L, Sun M. 2013. Bacillus thuringiensis metalloproteinase Bmp1 functions as a nematicidal virulence factor. Appl Environ Microbiol 79:460–468 10.1128/AEM.02551-12. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Fedhila S, Daou N, Lereclus D, Nielsen-LeRoux C. 2006. Identification of Bacillus cereus internalin and other candidate virulence genes specifically induced during oral infection in insects. Mol Microbiol 62:339–355 10.1111/j.1365-2958.2006.05362.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 267.Daou N, Buisson C, Gohar M, Vidic J, Bierne H, Kallassy M, Lereclus D, Nielsen-LeRoux C. 2009. IlsA, a unique surface protein of Bacillus cereus required for iron acquisition from heme, hemoglobin and ferritin. PLoS Pathog 5:e1000675 10.1371/journal.ppat.1000675. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Harvie DR, Vílchez S, Steggles JR, Ellar DJ. 2005. Bacillus cereus Fur regulates iron metabolism and is required for full virulence. Microbiology 151:569–577 10.1099/mic.0.27744-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 269.Kamar R, Réjasse A, Jéhanno I, Attieh Z, Courtin P, Chapot-Chartier MP, Nielsen-Leroux C, Lereclus D, El Chamy L, Kallassy M, Sanchis-Borja V. 2017. DltX of Bacillus thuringiensis is essential for d-alanylation of teichoic acids and resistance to antimicrobial response in insects. Front Microbiol 8:1437 10.3389/fmicb.2017.01437. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Guillemet E, Leréec A, Tran SL, Royer C, Barbosa I, Sansonetti P, Lereclus D, Ramarao N. 2016. The bacterial DNA repair protein Mfd confers resistance to the host nitrogen immune response. Sci Rep 6:29349 10.1038/srep29349. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Tran SL, Guillemet E, Gohar M, Lereclus D, Ramarao N. 2010. CwpFM (EntFM) is a Bacillus cereus potential cell wall peptidase implicated in adhesion, biofilm formation, and virulence. J Bacteriol 192:2638–2642 10.1128/JB.01315-09. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Mazzantini D, Celandroni F, Salvetti S, Gueye SA, Lupetti A, Senesi S, Ghelardi E. 2016. FlhF is required for swarming motility and full pathogenicity of Bacillus cereus. Front Microbiol 7:1644 10.3389/fmicb.2016.01644. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Frenzel E, Doll V, Pauthner M, Lücking G, Scherer S, Ehling-Schulz M. 2012. CodY orchestrates the expression of virulence determinants in emetic Bacillus cereus by impacting key regulatory circuits. Mol Microbiol 85:67–88 10.1111/j.1365-2958.2012.08090.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 274.Frenzel E, Kranzler M, Stark TD, Hofmann T, Ehling-Schulz M. 2015. The endospore-forming pathogen Bacillus cereus exploits a small colony variant-based diversification strategy in response to aminoglycoside exposure. MBio 6:e01172-15 10.1128/mBio.01172-15. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Andersson MA, Mikkola R, Helin J, Andersson MC, Salkinoja-Salonen M. 1998. A novel sensitive bioassay for detection of Bacillus cereus emetic toxin and related depsipeptide ionophores. Appl Environ Microbiol 64:1338–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Wijnands LM, Dufrenne JB, Zwietering MH, van Leusden FM. 2006. Spores from mesophilic Bacillus cereus strains germinate better and grow faster in simulated gastro-intestinal conditions than spores from psychrotrophic strains. Int J Food Microbiol 112:120–128 10.1016/j.ijfoodmicro.2006.06.015. [PubMed] [DOI] [PubMed] [Google Scholar]
- 277.Ceuppens S, Van de Wiele T, Rajkovic A, Ferrer-Cabaceran T, Heyndrickx M, Boon N, Uyttendaele M. 2012. Impact of intestinal microbiota and gastrointestinal conditions on the in vitro survival and growth of Bacillus cereus. Int J Food Microbiol 155:241–246 10.1016/j.ijfoodmicro.2012.02.013. [PubMed] [DOI] [PubMed] [Google Scholar]
- 278.Tsilia V, Kerckhof FM, Rajkovic A, Heyndrickx M, Van de Wiele T. 2015. Bacillus cereus NVH 0500/00 can adhere to mucin but cannot produce enterotoxins during gastrointestinal simulation. Appl Environ Microbiol 82:289–296 10.1128/AEM.02940-15. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Possemiers S, Marzorati M, Verstraete W, Van de Wiele T. 2010. Bacteria and chocolate: a successful combination for probiotic delivery. Int J Food Microbiol 141:97–103 10.1016/j.ijfoodmicro.2010.03.008. [PubMed] [DOI] [PubMed] [Google Scholar]
- 280.Sonenshein AL. 2007. Control of key metabolic intersections in Bacillus subtilis. Nat Rev Microbiol 5:917–927 10.1038/nrmicro1772. [PubMed] [DOI] [PubMed] [Google Scholar]
- 281.Lindbäck T, Mols M, Basset C, Granum PE, Kuipers OP, Kovács AT. 2012. CodY, a pleiotropic regulator, influences multicellular behaviour and efficient production of virulence factors in Bacillus cereus. Environ Microbiol 14:2233–2246 10.1111/j.1462-2920.2012.02766.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 282.Bourgogne A, Drysdale M, Hilsenbeck SG, Peterson SN, Koehler TM. 2003. Global effects of virulence gene regulators in a Bacillus anthracis strain with both virulence plasmids. Infect Immun 71:2736–2743 10.1128/IAI.71.5.2736-2743.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Lücking G, Dommel MK, Scherer S, Fouet A, Ehling-Schulz M. 2009. Cereulide synthesis in emetic Bacillus cereus is controlled by the transition state regulator AbrB, but not by the virulence regulator PlcR. Microbiology 155:922–931 10.1099/mic.0.024125-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 284.van Schaik W, Château A, Dillies MA, Coppée JY, Sonenshein AL, Fouet A. 2009. The global regulator CodY regulates toxin gene expression in Bacillus anthracis and is required for full virulence. Infect Immun 77:4437–4445 10.1128/IAI.00716-09. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Lücking G, Frenzel E, Rütschle A, Marxen S, Stark TD, Hofmann T, Scherer S, Ehling-Schulz M. 2015. Ces locus embedded proteins control the non-ribosomal synthesis of the cereulide toxin in emetic Bacillus cereus on multiple levels. Front Microbiol 6:1101 10.3389/fmicb.2015.01101. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Shivers RP, Sonenshein AL. 2004. Activation of the Bacillus subtilis global regulator CodY by direct interaction with branched-chain amino acids. Mol Microbiol 53:599–611 10.1111/j.1365-2958.2004.04135.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 287.Handke LD, Shivers RP, Sonenshein AL. 2008. Interaction of Bacillus subtilis CodY with GTP. J Bacteriol 190:798–806 10.1128/JB.01115-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Molle V, Nakaura Y, Shivers RP, Yamaguchi H, Losick R, Fujita Y, Sonenshein AL. 2003. Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis. J Bacteriol 185:1911–1922 10.1128/JB.185.6.1911-1922.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Dineen SS, Villapakkam AC, Nordman JT, Sonenshein AL. 2007. Repression of Clostridium difficile toxin gene expression by CodY. Mol Microbiol 66:206–219 10.1111/j.1365-2958.2007.05906.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 290.Shivers RP, Dineen SS, Sonenshein AL. 2006. Positive regulation of Bacillus subtilis ackA by CodY and CcpA: establishing a potential hierarchy in carbon flow. Mol Microbiol 62:811–822 10.1111/j.1365-2958.2006.05410.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 291.Strauch MA, Hoch JA. 1993. Transition-state regulators: sentinels of Bacillus subtilis post-exponential gene expression. Mol Microbiol 7:337–342 10.1111/j.1365-2958.1993.tb01125.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 292.Bobay BG, Benson L, Naylor S, Feeney B, Clark AC, Goshe MB, Strauch MA, Thompson R, Cavanagh J. 2004. Evaluation of the DNA binding tendencies of the transition state regulator AbrB. Biochemistry 43:16106–16118 10.1021/bi048399h. [PubMed] [DOI] [PubMed] [Google Scholar]
- 293.Grandvalet C, Gominet M, Lereclus D. 2001. Identification of genes involved in the activation of the Bacillus thuringiensis inhA metalloprotease gene at the onset of sporulation. Microbiology 147:1805–1813 10.1099/00221287-147-7-1805. [PubMed] [DOI] [PubMed] [Google Scholar]
- 294.Saile E, Koehler TM. 2002. Control of anthrax toxin gene expression by the transition state regulator abrB. J Bacteriol 184:370–380 10.1128/JB.184.2.370-380.2002. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Britton RA, Eichenberger P, Gonzalez-Pastor JE, Fawcett P, Monson R, Losick R, Grossman AD. 2002. Genome-wide analysis of the stationary-phase sigma factor (sigma-H) regulon of Bacillus subtilis. J Bacteriol 184:4881–4890 10.1128/JB.184.17.4881-4890.2002. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Hadjifrangiskou M, Chen Y, Koehler TM. 2007. The alternative sigma factor sigmaH is required for toxin gene expression by Bacillus anthracis. J Bacteriol 189:1874–1883 10.1128/JB.01333-06. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Bongiorni C, Fukushima T, Wilson AC, Chiang C, Mansilla MC, Hoch JA, Perego M. 2008. Dual promoters control expression of the Bacillus anthracis virulence factor AtxA. J Bacteriol 190:6483–6492 10.1128/JB.00766-08. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Fouet A, Namy O, Lambert G. 2000. Characterization of the operon encoding the alternative sigma(B) factor from Bacillus anthracis and its role in virulence. J Bacteriol 182:5036–5045 10.1128/JB.182.18.5036-5045.2000. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Slamti L, Lereclus D. 2002. A cell-cell signaling peptide activates the PlcR virulence regulon in bacteria of the Bacillus cereus group. EMBO J 21:4550–4559 10.1093/emboj/cdf450. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Pottathil M, Lazazzera BA. 2003. The extracellular Phr peptide-Rap phosphatase signaling circuit of Bacillus subtilis. Front Biosci 8:d32–d45 10.2741/913. [DOI] [PubMed] [Google Scholar]
- 301.Declerck N, Bouillaut L, Chaix D, Rugani N, Slamti L, Hoh F, Lereclus D, Arold ST. 2007. Structure of PlcR: insights into virulence regulation and evolution of quorum sensing in Gram-positive bacteria. Proc Natl Acad Sci U S A 104:18490–18495 10.1073/pnas.0704501104. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Slamti L, Perchat S, Huillet E, Lereclus D. 2014. Quorum sensing in Bacillus thuringiensis is required for completion of a full infectious cycle in the insect. Toxins (Basel) 6:2239–2255 10.3390/toxins6082239. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Slamti L, Perchat S, Gominet M, Vilas-Bôas G, Fouet A, Mock M, Sanchis V, Chaufaux J, Gohar M, Lereclus D. 2004. Distinct mutations in PlcR explain why some strains of the Bacillus cereus group are nonhemolytic. J Bacteriol 186:3531–3538 10.1128/JB.186.11.3531-3538.2004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Grenha R, Slamti L, Nicaise M, Refes Y, Lereclus D, Nessler S. 2013. Structural basis for the activation mechanism of the PlcR virulence regulator by the quorum-sensing signal peptide PapR. Proc Natl Acad Sci U S A 110:1047–1052 10.1073/pnas.1213770110. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Bouillaut L, Perchat S, Arold S, Zorrilla S, Slamti L, Henry C, Gohar M, Declerck N, Lereclus D. 2008. Molecular basis for group-specific activation of the virulence regulator PlcR by PapR heptapeptides. Nucleic Acids Res 36:3791–3801 10.1093/nar/gkn149. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Slamti L, Lereclus D. 2005. Specificity and polymorphism of the PlcR-PapR quorum-sensing system in the Bacillus cereus group. J Bacteriol 187:1182–1187 10.1128/JB.187.3.1182-1187.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Zouhir S, Perchat S, Nicaise M, Perez J, Guimaraes B, Lereclus D, Nessler S. 2013. Peptide-binding dependent conformational changes regulate the transcriptional activity of the quorum-sensor NprR. Nucleic Acids Res 41:7920–7933 10.1093/nar/gkt546. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Gélis-Jeanvoine S, Canette A, Gohar M, Caradec T, Lemy C, Gominet M, Jacques P, Lereclus D, Slamti L. 2017. Genetic and functional analyses of krs, a locus encoding kurstakin, a lipopeptide produced by Bacillus thuringiensis. Res Microbiol 168:356–368 10.1016/j.resmic.2016.06.002. [PubMed] [DOI] [PubMed] [Google Scholar]
- 309.Perchat S, Talagas A, Poncet S, Lazar N, Li de la Sierra-Gallay I, Gohar M, Lereclus D, Nessler S. 2016. How quorum sensing connects sporulation to necrotrophism in Bacillus thuringiensis. PLoS Pathog 12:e1005779 10.1371/journal.ppat.1005779. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Uchida I, Hornung JM, Thorne CB, Klimpel KR, Leppla SH. 1993. Cloning and characterization of a gene whose product is a trans-activator of anthrax toxin synthesis. J Bacteriol 175:5329–5338 10.1128/jb.175.17.5329-5338.1993. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Koehler TM, Dai Z, Kaufman-Yarbray M. 1994. Regulation of the Bacillus anthracis protective antigen gene: CO2 and a trans-acting element activate transcription from one of two promoters. J Bacteriol 176:586–595 10.1128/jb.176.3.586-595.1994. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Sirard JC, Guidi-Rontani C, Fouet A, Mock M. 2000. Characterization of a plasmid region involved in Bacillus anthracis toxin production and pathogenesis. Int J Med Microbiol 290:313–316 10.1016/S1438-4221(00)80030-2. [DOI] [PubMed] [Google Scholar]
- 313.Okinaka RT, Cloud K, Hampton O, Hoffmaster AR, Hill KK, Keim P, Koehler TM, Lamke G, Kumano S, Mahillon J, Manter D, Martinez Y, Ricke D, Svensson R, Jackson PJ. 1999. Sequence and organization of pXO1, the large Bacillus anthracis plasmid harboring the anthrax toxin genes. J Bacteriol 181:6509–6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Mignot T, Mock M, Fouet A. 2003. A plasmid-encoded regulator couples the synthesis of toxins and surface structures in Bacillus anthracis. Mol Microbiol 47:917–927 10.1046/j.1365-2958.2003.03345.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 315.Makino S, Uchida I, Terakado N, Sasakawa C, Yoshikawa M. 1989. Molecular characterization and protein analysis of the cap region, which is essential for encapsulation in Bacillus anthracis. J Bacteriol 171:722–730 10.1128/jb.171.2.722-730.1989. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Guignot J, Mock M, Fouet A. 1997. AtxA activates the transcription of genes harbored by both Bacillus anthracis virulence plasmids. FEMS Microbiol Lett 147:203–207 10.1111/j.1574-6968.1997.tb10242.x. [DOI] [PubMed] [Google Scholar]
- 317.Candela T, Mock M, Fouet A. 2005. CapE, a 47-amino-acid peptide, is necessary for Bacillus anthracis polyglutamate capsule synthesis. J Bacteriol 187:7765–7772 10.1128/JB.187.22.7765-7772.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Dai Z, Sirard JC, Mock M, Koehler TM. 1995. The atxA gene product activates transcription of the anthrax toxin genes and is essential for virulence. Mol Microbiol 16:1171–1181 10.1111/j.1365-2958.1995.tb02340.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 319.Dale JL, Raynor MJ, Dwivedi P, Koehler TM. 2012. cis-Acting elements that control expression of the master virulence regulatory gene atxA in Bacillus anthracis. J Bacteriol 194:4069–4079 10.1128/JB.00776-12. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Dale JL, Raynor MJ, Ty MC, Hadjifrangiskou M, Koehler TM. 2018. A dual role for the Bacillus anthracis master virulence regulator AtxA: control of sporulation and anthrax toxin production. Front Microbiol 9:482 10.3389/fmicb.2018.00482. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Drysdale M, Bourgogne A, Hilsenbeck SG, Koehler TM. 2004. atxA controls Bacillus anthracis capsule synthesis via acpA and a newly discovered regulator, acpB. J Bacteriol 186:307–315 10.1128/JB.186.2.307-315.2004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Vietri NJ, Marrero R, Hoover TA, Welkos SL. 1995. Identification and characterization of a trans-activator involved in the regulation of encapsulation by Bacillus anthracis. Gene 152:1–9 10.1016/0378-1119(94)00662-C. [DOI] [PubMed] [Google Scholar]
- 323.Drysdale M, Bourgogne A, Koehler TM. 2005. Transcriptional analysis of the Bacillus anthracis capsule regulators. J Bacteriol 187:5108–5114 10.1128/JB.187.15.5108-5114.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Drysdale M, Heninger S, Hutt J, Chen Y, Lyons CR, Koehler TM. 2005. Capsule synthesis by Bacillus anthracis is required for dissemination in murine inhalation anthrax. EMBO J 24:221–227 10.1038/sj.emboj.7600495. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Chiang C, Bongiorni C, Perego M. 2011. Glucose-dependent activation of Bacillus anthracis toxin gene expression and virulence requires the carbon catabolite protein CcpA. J Bacteriol 193:52–62 10.1128/JB.01656-09. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Wilson AC, Hoch JA, Perego M. 2009. Two small c-type cytochromes affect virulence gene expression in Bacillus anthracis. Mol Microbiol 72:109–123 10.1111/j.1365-2958.2009.06627.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Dai Z, Koehler TM. 1997. Regulation of anthrax toxin activator gene (atxA) expression in Bacillus anthracis: temperature, not CO2/bicarbonate, affects AtxA synthesis. Infect Immun 65:2576–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.van Schaik W, Prigent J, Fouet A. 2007. The stringent response of Bacillus anthracis contributes to sporulation but not to virulence. Microbiology 153:4234–4239 10.1099/mic.0.2007/010355-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 329.Strauch MA, Ballar P, Rowshan AJ, Zoller KL. 2005. The DNA-binding specificity of the Bacillus anthracis AbrB protein. Microbiology 151:1751–1759 10.1099/mic.0.27803-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 330.Tsvetanova B, Wilson AC, Bongiorni C, Chiang C, Hoch JA, Perego M. 2007. Opposing effects of histidine phosphorylation regulate the AtxA virulence transcription factor in Bacillus anthracis. Mol Microbiol 63:644–655 10.1111/j.1365-2958.2006.05543.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 331.Hammerstrom TG, Roh JH, Nikonowicz EP, Koehler TM. 2011. Bacillus anthracis virulence regulator AtxA: oligomeric state, function and CO(2)-signalling. Mol Microbiol 82:634–647 10.1111/j.1365-2958.2011.07843.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Hondorp ER, Hou SC, Hause LL, Gera K, Lee CE, McIver KS. 2013. PTS phosphorylation of Mga modulates regulon expression and virulence in the group A streptococcus. Mol Microbiol 88:1176–1193 10.1111/mmi.12250. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Hammerstrom TG, Horton LB, Swick MC, Joachimiak A, Osipiuk J, Koehler TM. 2015. Crystal structure of Bacillus anthracis virulence regulator AtxA and effects of phosphorylated histidines on multimerization and activity. Mol Microbiol 95:426–441 10.1111/mmi.12867. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Kolstø AB, Tourasse NJ, Økstad OA. 2009. What sets Bacillus anthracis apart from other Bacillus species? Annu Rev Microbiol 63:451–476 10.1146/annurev.micro.091208.073255. [PubMed] [DOI] [PubMed] [Google Scholar]
- 335.Timmery S, Modrie P, Minet O, Mahillon J. 2009. Plasmid capture by the Bacillus thuringiensis conjugative plasmid pXO16. J Bacteriol 191:2197–2205 10.1128/JB.01700-08. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Andrup L, Jørgensen O, Wilcks A, Smidt L, Jensen GB. 1996. Mobilization of “nonmobilizable” plasmids by the aggregation-mediated conjugation system of Bacillus thuringiensis. Plasmid 36:75–85 10.1006/plas.1996.0035. [PubMed] [DOI] [PubMed] [Google Scholar]
- 337.Wilcks A, Jayaswal N, Lereclus D, Andrup L. 1998. Characterization of plasmid pAW63, a second self-transmissible plasmid in Bacillus thuringiensis subsp. kurstaki HD73. Microbiology 144:1263–1270 10.1099/00221287-144-5-1263. [PubMed] [DOI] [PubMed] [Google Scholar]
- 338.Thorne CB. 1993. Bacillus anthracis, p 113–124. In Sonenshein AL, Hoch JA, Losick R (ed), Bacillus subtilis and Other Gram-Positive Bacteria: Biochemistry, Physiology, and Molecular Genetics. American Society for Microbiology, Washington, DC. [Google Scholar]
- 339.Thorne CB. 1968. Transducing bacteriophage for Bacillus cereus. J Virol 2:657–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Lecadet MM, Blondel MO, Ribier J. 1980. Generalized transduction in Bacillus thuringiensis var. berliner 1715 using bacteriophage CP-54Ber. J Gen Microbiol 121:203–212. [PubMed] [DOI] [PubMed] [Google Scholar]
- 341.Thorne CB. 1978. Transduction in Bacillus thuringiensis. Appl Environ Microbiol 35:1109–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Lecadet MM, Chaufaux J, Ribier J, Lereclus D. 1992. Construction of novel Bacillus thuringiensis strains with different insecticidal activities by transduction and transformation. Appl Environ Microbiol 58:840–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Battisti L, Green BD, Thorne CB. 1985. Mating system for transfer of plasmids among Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis. J Bacteriol 162:543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Barsomian GD, Robillard NJ, Thorne CB. 1984. Chromosomal mapping of Bacillus thuringiensis by transduction. J Bacteriol 157:746–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Lereclus D, Menou G, Lecadet M-M. 1983. Isolation of a DNA sequence related to several plasmids from Bacillus thuringiensis after a mating involving the Streptococcus faecalis plasmid pAM beta 1. Mol Gen Genet 191:307–313 10.1007/BF00334831. [DOI] [PubMed] [Google Scholar]
- 346.Lereclus D, Mahillon J, Menou G, Lecadet M-M. 1986. Identification of Tn4430, a transposon of Bacillus thuringiensis functional in Escherichia coli. Mol Gen Genet 204:52–57 10.1007/BF00330186. [DOI] [PubMed] [Google Scholar]
- 347.Yuan Y, Zheng D, Hu X, Cai Q, Yuan Z. 2010. Conjugative transfer of insecticidal plasmid pHT73 from Bacillus thuringiensis to B. anthracis and compatibility of this plasmid with pXO1 and pXO2. Appl Environ Microbiol 76:468–473 10.1128/AEM.01984-09. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Reddy A, Battisti L, Thorne CB. 1987. Identification of self-transmissible plasmids in four Bacillus thuringiensis subspecies. J Bacteriol 169:5263–5270 10.1128/jb.169.11.5263-5270.1987. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Jarrett P, Stephenson M. 1990. Plasmid transfer between strains of Bacillus thuringiensis infecting Galleria mellonella and Spodoptera littoralis. Appl Environ Microbiol 56:1608–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Vilas-Bôas G, Vilas-Bôas LA, Lereclus D, Arantes O. 1998. Bacillus thuringiensis conjugation under environmental conditions. FEMS Microbiol Ecol 25:369–374 10.1016/S0168-6496(98)00005-1. [DOI] [PubMed] [Google Scholar]
- 351.Thomas DJ, Morgan JA, Whipps JM, Saunders JR. 2000. Plasmid transfer between the Bacillus thuringiensis subspecies kurstaki and tenebrionis in laboratory culture and soil and in lepidopteran and coleopteran larvae. Appl Environ Microbiol 66:118–124 10.1128/AEM.66.1.118-124.2000. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Wilcks A, Smidt L, Bahl MI, Hansen BM, Andrup L, Hendriksen NB, Licht TR. 2008. Germination and conjugation of Bacillus thuringiensis subsp. israelensis in the intestine of gnotobiotic rats. J Appl Microbiol 104:1252–1259 10.1111/j.1365-2672.2007.03657.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 353.Thomas DJ, Morgan JA, Whipps JM, Saunders JR. 2001. Plasmid transfer between Bacillus thuringiensis subsp. israelensis strains in laboratory culture, river water, and dipteran larvae. Appl Environ Microbiol 67:330–338 10.1128/AEM.67.1.330-338.2001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Modrie P, Beuls E, Mahillon J. 2010. Differential transfer dynamics of pAW63 plasmid among members of the Bacillus cereus group in food microcosms. J Appl Microbiol 108:888–897 10.1111/j.1365-2672.2009.04488.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 355.Van der Auwera GA, Timmery S, Hoton F, Mahillon J. 2007. Plasmid exchanges among members of the Bacillus cereus group in foodstuffs. Int J Food Microbiol 113:164–172 10.1016/j.ijfoodmicro.2006.06.030. [PubMed] [DOI] [PubMed] [Google Scholar]
- 356.Hu X, Van der Auwera G, Timmery S, Zhu L, Mahillon J. 2009. Distribution, diversity, and potential mobility of extrachromosomal elements related to the Bacillus anthracis pXO1 and pXO2 virulence plasmids. Appl Environ Microbiol 75:3016–3028 10.1128/AEM.02709-08. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Pannucci J, Okinaka RT, Sabin R, Kuske CR. 2002. Bacillus anthracis pXO1 plasmid sequence conservation among closely related bacterial species. J Bacteriol 184:134–141 10.1128/JB.184.1.134-141.2002. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Hoffmaster AR, Hill KK, Gee JE, Marston CK, De BK, Popovic T, Sue D, Wilkins PP, Avashia SB, Drumgoole R, Helma CH, Ticknor LO, Okinaka RT, Jackson PJ. 2006. Characterization of Bacillus cereus isolates associated with fatal pneumonias: strains are closely related to Bacillus anthracis and harbor B. anthracis virulence genes. J Clin Microbiol 44:3352–3360 10.1128/JCM.00561-06. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Hernandez E, Ramisse F, Ducoureau JP, Cruel T, Cavallo JD. 1998. Bacillus thuringiensis subsp. konkukian (serotype H34) superinfection: case report and experimental evidence of pathogenicity in immunosuppressed mice. J Clin Microbiol 36:2138–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Van der Auwera GA, Timmery S, Mahillon J. 2008. Self-transfer and mobilisation capabilities of the pXO2-like plasmid pBT9727 from Bacillus thuringiensis subsp. konkukian 97-27. Plasmid 59:134–138 10.1016/j.plasmid.2007.11.007. [PubMed] [DOI] [PubMed] [Google Scholar]
- 361.Koehler TM, Thorne CB. 1987. Bacillus subtilis (natto) plasmid pLS20 mediates interspecies plasmid transfer. J Bacteriol 169:5271–5278 10.1128/jb.169.11.5271-5278.1987. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.DeWitt T, Grossman AD. 2014. The bifunctional cell wall hydrolase CwlT is needed for conjugation of the integrative and conjugative element ICEBs1 in Bacillus subtilis and B. anthracis. J Bacteriol 196:1588–1596 10.1128/JB.00012-14. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Zhang J, Hodgman TC, Krieger L, Schnetter W, Schairer HU. 1997. Cloning and analysis of the first cry gene from Bacillus popilliae. J Bacteriol 179:4336–4341 10.1128/jb.179.13.4336-4341.1997. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Qureshi N, Chawla S, Likitvivatanavong S, Lee HL, Gill SS. 2014. The cry toxin operon of Clostridium bifermentans subsp. malaysia is highly toxic to Aedes larval mosquitoes. Appl Environ Microbiol 80:5689–5697 10.1128/AEM.01139-14. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Bone EJ, Ellar DJ. 1989. Transformation of Bacillus thuringiensis by electroporation. FEMS Microbiol Lett 49:171–177 10.1111/j.1574-6968.1989.tb03039.x. [DOI] [PubMed] [Google Scholar]
- 366.Lereclus D, Arantes O, Chaufaux J, Lecadet M. 1989. Transformation and expression of a cloned delta-endotoxin gene in Bacillus thuringiensis. FEMS Microbiol Lett 51:211–217. [DOI] [PubMed] [Google Scholar]
- 367.Schurter W, Geiser M, Mathé D. 1989. Efficient transformation of Bacillus thuringiensis and B. cereus via electroporation: transformation of acrystalliferous strains with a cloned delta-endotoxin gene. Mol Gen Genet 218:177–181 10.1007/BF00330581. [DOI] [PubMed] [Google Scholar]
- 368.Pezard C, Berche P, Mock M. 1991. Contribution of individual toxin components to virulence of Bacillus anthracis. Infect Immun 59:3472–3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Sirard JC, Mock M, Fouet A. 1994. The three Bacillus anthracis toxin genes are coordinately regulated by bicarbonate and temperature. J Bacteriol 176:5188–5192 10.1128/jb.176.16.5188-5192.1994. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Dommel MK, Frenzel E, Strasser B, Blöchinger C, Scherer S, Ehling-Schulz M. 2010. Identification of the main promoter directing cereulide biosynthesis in emetic Bacillus cereus and its application for real-time monitoring of ces gene expression in foods. Appl Environ Microbiol 76:1232–1240 10.1128/AEM.02317-09. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Macaluso A, Mettus AM. 1991. Efficient transformation of Bacillus thuringiensis requires nonmethylated plasmid DNA. J Bacteriol 173:1353–1356 10.1128/jb.173.3.1353-1356.1991. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Marrero R, Welkos SL. 1995. The transformation frequency of plasmids into Bacillus anthracis is affected by adenine methylation. Gene 152:75–78 10.1016/0378-1119(94)00647-B. [DOI] [PubMed] [Google Scholar]
- 373.Trieu-Cuot P, Carlier C, Martin P, Courvalin P. 1987. Plasmid transfer by conjugation from Escherichia coli to Gram-positive bacteria. FEMS Microbiol Lett 48:289–294 10.1111/j.1574-6968.1987.tb02558.x. [DOI] [Google Scholar]
- 374.Arantes O, Lereclus D. 1991. Construction of cloning vectors for Bacillus thuringiensis. Gene 108:115–119 10.1016/0378-1119(91)90495-W. [DOI] [PubMed] [Google Scholar]
- 375.Arnaud M, Chastanet A, Débarbouillé M. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, Gram-positive bacteria. Appl Environ Microbiol 70:6887–6891 10.1128/AEM.70.11.6887-6891.2004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Saldanha RJ, Pemberton A, Shiflett P, Perutka J, Whitt JT, Ellington A, Lambowitz AM, Kramer R, Taylor D, Lamkin TJ. 2013. Rapid targeted gene disruption in Bacillus anthracis. BMC Biotechnol 13:72 10.1186/1472-6750-13-72. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Pomerantsev AP, McCall RM, Chahoud M, Hepler NK, Fattah R, Leppla SH. 2017. Genome engineering in Bacillus anthracis using tyrosine site-specific recombinases. PLoS One 12:e0183346 10.1371/journal.pone.0183346. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Janes BK, Stibitz S. 2006. Routine markerless gene replacement in Bacillus anthracis. Infect Immun 74:1949–1953 10.1128/IAI.74.3.1949-1953.2006. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Plaut RD, Stibitz S. 2015. Improvements to a markerless allelic exchange system for Bacillus anthracis. PLoS One 10:e0142758 10.1371/journal.pone.0142758. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Lee JY, Janes BK, Passalacqua KD, Pfleger BF, Bergman NH, Liu H, Håkansson K, Somu RV, Aldrich CC, Cendrowski S, Hanna PC, Sherman DH. 2007. Biosynthetic analysis of the petrobactin siderophore pathway from Bacillus anthracis. J Bacteriol 189:1698–1710 10.1128/JB.01526-06. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Vörös A, Simm R, Slamti L, McKay MJ, Hegna IK, Nielsen-LeRoux C, Hassan KA, Paulsen IT, Lereclus D, Økstad OA, Molloy MP, Kolstø AB. 2014. SecDF as part of the Sec-translocase facilitates efficient secretion of Bacillus cereus toxins and cell wall-associated proteins. PLoS One 9:e103326 10.1371/journal.pone.0103326. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Simm R, Vörös A, Ekman JV, Sødring M, Nes I, Kroeger JK, Saidijam M, Bettaney KE, Henderson PJ, Salkinoja-Salonen M, Kolstø AB. 2012. BC4707 is a major facilitator superfamily multidrug resistance transport protein from Bacillus cereus implicated in fluoroquinolone tolerance. PLoS One 7:e36720 10.1371/journal.pone.0036720. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Klimowicz AK, Benson TA, Handelsman J. 2010. A quadruple-enterotoxin-deficient mutant of Bacillus thuringiensis remains insecticidal. Microbiology 156:3575–3583 10.1099/mic.0.039925-0. [PubMed] [DOI] [PubMed] [Google Scholar]
- 384.Gominet M, Slamti L, Gilois N, Rose M, Lereclus D. 2001. Oligopeptide permease is required for expression of the Bacillus thuringiensis plcR regulon and for virulence. Mol Microbiol 40:963–975 10.1046/j.1365-2958.2001.02440.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 385.Espinasse S, Gohar M, Lereclus D, Sanchis V. 2002. An ABC transporter from Bacillus thuringiensis is essential for beta-exotoxin I production. J Bacteriol 184:5848–5854 10.1128/JB.184.21.5848-5854.2002. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Fedhila S, Guillemet E, Nel P, Lereclus D. 2004. Characterization of two Bacillus thuringiensis genes identified by in vivo screening of virulence factors. Appl Environ Microbiol 70:4784–4791 10.1128/AEM.70.8.4784-4791.2004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Tam C, Glass EM, Anderson DM, Missiakas D. 2006. Transposon mutagenesis of Bacillus anthracis strain Sterne using Bursa aurealis. Plasmid 56:74–77 10.1016/j.plasmid.2006.01.002. [PubMed] [DOI] [PubMed] [Google Scholar]
- 388.McGillivray SM, Ebrahimi CM, Fisher N, Sabet M, Zhang DX, Chen Y, Haste NM, Aroian RV, Gallo RL, Guiney DG, Friedlander AM, Koehler TM, Nizet V. 2009. ClpX contributes to innate defense peptide resistance and virulence phenotypes of Bacillus anthracis. J Innate Immun 1:494–506 10.1159/000225955. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Ivins BE, Welkos SL, Knudson GB, Leblanc DJ. 1988. Transposon Tn916 mutagenesis in Bacillus anthracis. Infect Immun 56:176–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Hoffmaster AR, Koehler TM. 1997. The anthrax toxin activator gene atxA is associated with CO2-enhanced non-toxin gene expression in Bacillus anthracis. Infect Immun 65:3091–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Song F, Peng Q, Brillard J, Buisson C, de Been M, Abee T, Broussolle V, Huang D, Zhang J, Lereclus D, Nielsen-LeRoux C. 2012. A multicomponent sugar phosphate sensor system specifically induced in Bacillus cereus during infection of the insect gut. FASEB J 26:3336–3350 10.1096/fj.11-197681. [PubMed] [DOI] [PubMed] [Google Scholar]
- 392.Passalacqua KD, Bergman NH, Lee JY, Sherman DH, Hanna PC. 2007. The global transcriptional responses of Bacillus anthracis Sterne (34F2) and a Delta sodA1 mutant to paraquat reveal metal ion homeostasis imbalances during endogenous superoxide stress. J Bacteriol 189:3996–4013 10.1128/JB.00185-07. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Passalacqua KD, Varadarajan A, Byrd B, Bergman NH. 2009. Comparative transcriptional profiling of Bacillus cereus sensu lato strains during growth in CO2-bicarbonate and aerobic atmospheres. PLoS One 4:e4904 10.1371/journal.pone.0004904. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Martin J, Zhu W, Passalacqua KD, Bergman N, Borodovsky M. 2010. Bacillus anthracis genome organization in light of whole transcriptome sequencing. BMC Bioinformatics 11(Suppl 3):S10 10.1186/1471-2105-11-S3-S10. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Mols M, Mastwijk H, Nierop Groot M, Abee T. 2013. Physiological and transcriptional response of Bacillus cereus treated with low-temperature nitrogen gas plasma. J Appl Microbiol 115:689–702 10.1111/jam.12278. [PubMed] [DOI] [PubMed] [Google Scholar]
- 396.Agaisse H, Lereclus D. 1994. Structural and functional analysis of the promoter region involved in full expression of the cryIIIA toxin gene of Bacillus thuringiensis. Mol Microbiol 13:97–107 10.1111/j.1365-2958.1994.tb00405.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 397.Bartkus JM, Leppla SH. 1989. Transcriptional regulation of the protective antigen gene of Bacillus anthracis. Infect Immun 57:2295–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Han H, Wilson AC. 2013. The two CcdA proteins of Bacillus anthracis differentially affect virulence gene expression and sporulation. J Bacteriol 195:5242–5249 10.1128/JB.00917-13. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Ben Rejeb S, Lereclus D, Slamti L. 2017. Analysis of abrB expression during the infectious cycle of Bacillus thuringiensis reveals population heterogeneity. Front Microbiol 8:2471 10.3389/fmicb.2017.02471. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Dunn AK, Handelsman J. 1999. A vector for promoter trapping in Bacillus cereus. Gene 226:297–305 10.1016/S0378-1119(98)00544-7. [DOI] [PubMed] [Google Scholar]
- 401.Turnbull PC, Sirianni NM, LeBron CI, Samaan MN, Sutton FN, Reyes AE, Peruski LF Jr. 2004. MICs of selected antibiotics for Bacillus anthracis, Bacillus cereus, Bacillus thuringiensis, and Bacillus mycoides from a range of clinical and environmental sources as determined by the Etest. J Clin Microbiol 42:3626–3634 10.1128/JCM.42.8.3626-3634.2004. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Weber DJ, Rutala WA. 1988. Bacillus species. Infect Control Hosp Epidemiol 9:368–373 10.2307/30145465. [PubMed] [DOI] [PubMed] [Google Scholar]
- 403.Chen Y, Succi J, Tenover FC, Koehler TM. 2003. Beta-lactamase genes of the penicillin-susceptible Bacillus anthracis Sterne strain. J Bacteriol 185:823–830 10.1128/JB.185.3.823-830.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Inglesby TV, O’Toole T, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, Friedlander AM, Gerberding J, Hauer J, Hughes J, McDade J, Osterholm MT, Parker G, Perl TM, Russell PK, Tonat K, Working Group on Civilian Biodefense. 2002. Anthrax as a biological weapon, 2002: updated recommendations for management. JAMA 287:2236–2252 10.1001/jama.287.17.2236. [PubMed] [DOI] [PubMed] [Google Scholar]
- 405.Hendricks KA, Wright ME, Shadomy SV, Bradley JS, Morrow MG, Pavia AT, Rubinstein E, Holty JE, Messonnier NE, Smith TL, Pesik N, Treadwell TA, Bower WA, Workgroup on Anthrax Clinical Guidelines. 2014. Centers for disease control and prevention expert panel meetings on prevention and treatment of anthrax in adults. Emerg Infect Dis 20:20 10.3201/eid2002.130687. [PubMed] [DOI] [Google Scholar]
- 406.Bast DJ, Athamna A, Duncan CL, de Azavedo JC, Low DE, Rahav G, Farrell D, Rubinstein E. 2004. Type II topoisomerase mutations in Bacillus anthracis associated with high-level fluoroquinolone resistance. J Antimicrob Chemother 54:90–94 10.1093/jac/dkh294. [PubMed] [DOI] [PubMed] [Google Scholar]
- 407.Price LB, Vogler A, Pearson T, Busch JD, Schupp JM, Keim P. 2003. In vitro selection and characterization of Bacillus anthracis mutants with high-level resistance to ciprofloxacin. Antimicrob Agents Chemother 47:2362–2365 10.1128/AAC.47.7.2362-2365.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Kalns J, Morris J, Eggers J, Kiel J. 2002. Delayed treatment with doxycycline has limited effect on anthrax infection in BLK57/B6 mice. Biochem Biophys Res Commun 297:506–509 10.1016/S0006-291X(02)02226-X. [DOI] [PubMed] [Google Scholar]
- 409.Peterson JW, Moen ST, Healy D, Pawlik JE, Taormina J, Hardcastle J, Thomas JM, Lawrence WS, Ponce C, Chatuev BM, Gnade BT, Foltz SM, Agar SL, Sha J, Klimpel GR, Kirtley ML, Eaves-Pyles T, Chopra AK. 2010. Protection afforded by fluoroquinolones in animal models of respiratory infections with Bacillus anthracis, Yersinia pestis, and Francisella tularensis. Open Microbiol J 4:34–46 10.2174/1874285801004010034. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Holty JE, Kim RY, Bravata DM. 2006. Anthrax: a systematic review of atypical presentations. Ann Emerg Med 48:200–211 10.1016/j.annemergmed.2005.11.035. [PubMed] [DOI] [PubMed] [Google Scholar]
- 411.Jernigan JA, Stephens DS, Ashford DA, Omenaca C, Topiel MS, Galbraith M, Tapper M, Fisk TL, Zaki S, Popovic T, Meyer RF, Quinn CP, Harper SA, Fridkin SK, Sejvar JJ, Shepard CW, McConnell M, Guarner J, Shieh WJ, Malecki JM, Gerberding JL, Hughes JM, Perkins BA, Anthrax Bioterrorism Investigation Team. 2001. Bioterrorism-related inhalational anthrax: the first 10 cases reported in the United States. Emerg Infect Dis 7:933–944 10.3201/eid0706.010604. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Mourez M, Kane RS, Mogridge J, Metallo S, Deschatelets P, Sellman BR, Whitesides GM, Collier RJ. 2001. Designing a polyvalent inhibitor of anthrax toxin. Nat Biotechnol 19:958–961 10.1038/nbt1001-958. [PubMed] [DOI] [PubMed] [Google Scholar]
- 413.Hull AK, Criscuolo CJ, Mett V, Groen H, Steeman W, Westra H, Chapman G, Legutki B, Baillie L, Yusibov V. 2005. Human-derived, plant-produced monoclonal antibody for the treatment of anthrax. Vaccine 23:2082–2086 10.1016/j.vaccine.2005.01.013. [PubMed] [DOI] [PubMed] [Google Scholar]
- 414.Ionin B, Hopkins RJ, Pleune B, Sivko GS, Reid FM, Clement KH, Rudge TL Jr, Stark GV, Innes A, Sari S, Guina T, Howard C, Smith J, Swoboda ML, Vert-Wong E, Johnson V, Nabors GS, Skiadopoulos MH. 2013. Evaluation of immunogenicity and efficacy of anthrax vaccine adsorbed for postexposure prophylaxis. Clin Vaccine Immunol 20:1016–1026 10.1128/CVI.00099-13. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Chen L, Schiffer JM, Dalton S, Sabourin CL, Niemuth NA, Plikaytis BD, Quinn CP. 2014. Comprehensive analysis and selection of anthrax vaccine adsorbed immune correlates of protection in rhesus macaques. Clin Vaccine Immunol 21:1512–1520 10.1128/CVI.00469-14. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Little SF, Ivins BE, Webster WM, Fellows PF, Pitt ML, Norris SL, Andrews GP. 2006. Duration of protection of rabbits after vaccination with Bacillus anthracis recombinant protective antigen vaccine. Vaccine 24:2530–2536 10.1016/j.vaccine.2005.12.028. [PubMed] [DOI] [PubMed] [Google Scholar]
- 417.Larkin M. 2002. Anthrax vaccine is safe and effective-but needs improvement, says IOM. Lancet 359:951 10.1016/S0140-6736(02)08051-0. [DOI] [PubMed] [Google Scholar]
- 418.Fay MP, Follmann DA, Lynn F, Schiffer JM, Stark GV, Kohberger R, Quinn CP, Nuzum EO. 2012. Anthrax vaccine-induced antibodies provide cross-species prediction of survival to aerosol challenge. Sci Transl Med 4:151ra126 10.1126/scitranslmed.3004073. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Sivko GS, Stark GV, Tordoff KP, Taylor KL, Glaze E, VanRaden M, Schiffer JM, Hewitt JA, Quinn CP, Nuzum EO. 2016. Evaluation of early immune response-survival relationship in cynomolgus macaques after Anthrax Vaccine Adsorbed vaccination and Bacillus anthracis spore challenge. Vaccine 34:6518–6528 10.1016/j.vaccine.2016.04.048. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Dumas EK, Gross T, Larabee J, Pate L, Cuthbertson H, Charlton S, Hallis B, Engler RJM, Collins LC Jr, Spooner CE, Chen H, Ballard J, James JA, Farris AD. 2017. Anthrax vaccine precipitated induces edema toxin-neutralizing, edema factor-specific antibodies in human recipients. Clin Vaccine Immunol 24:24 10.1128/CVI.00165-17. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Dumas EK, Garman L, Cuthbertson H, Charlton S, Hallis B, Engler RJM, Choudhari S, Picking WD, James JA, Farris AD. 2017. Lethal factor antibodies contribute to lethal toxin neutralization in recipients of anthrax vaccine precipitated. Vaccine 35:3416–3422 10.1016/j.vaccine.2017.05.006. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Zegers ND, Kluter E, van Der Stap H, van Dura E, van Dalen P, Shaw M, Baillie L. 1999. Expression of the protective antigen of Bacillus anthracis by Lactobacillus casei: towards the development of an oral vaccine against anthrax. J Appl Microbiol 87:309–314 10.1046/j.1365-2672.1999.00900.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 423.Garmory HS, Titball RW, Griffin KF, Hahn U, Böhm R, Beyer W. 2003. Salmonella enterica serovar Typhimurium expressing a chromosomally integrated copy of the Bacillus anthracis protective antigen gene protects mice against an anthrax spore challenge. Infect Immun 71:3831–3836 10.1128/IAI.71.7.3831-3836.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Galloway DR, Baillie L. 2004. DNA vaccines against anthrax. Expert Opin Biol Ther 4:1661–1667 10.1517/14712598.4.10.1661. [PubMed] [DOI] [PubMed] [Google Scholar]
- 425.Price BM, Liner AL, Park S, Leppla SH, Mateczun A, Galloway DR. 2001. Protection against anthrax lethal toxin challenge by genetic immunization with a plasmid encoding the lethal factor protein. Infect Immun 69:4509–4515 10.1128/IAI.69.7.4509-4515.2001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Schneerson R, Kubler-Kielb J, Liu TY, Dai ZD, Leppla SH, Yergey A, Backlund P, Shiloach J, Majadly F, Robbins JB. 2003. Poly(gamma-d-glutamic acid) protein conjugates induce IgG antibodies in mice to the capsule of Bacillus anthracis: a potential addition to the anthrax vaccine. Proc Natl Acad Sci U S A 100:8945–8950 10.1073/pnas.1633512100. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Jones RM, Burke M, Dubose D, Chichester JA, Manceva S, Horsey A, Streatfield SJ, Breit J, Yusibov V. 2017. Stability and pre-formulation development of a plant-produced anthrax vaccine candidate. Vaccine 35:5463–5470 10.1016/j.vaccine.2016.12.009. [PubMed] [DOI] [PubMed] [Google Scholar]
- 428.Sterne M. 1946. Avirulent anthrax vaccine. Onderstepoort J Vet Sci Anim Ind 21:41–43. [PubMed] [Google Scholar]
- 429.EFSA Panel on Biological Hazards. 2016. Risks for public health related to the presence of Bacillus cereus and other Bacillus spp. including Bacillus thuringiensis in foodstuffs. EFSA J. 10.2903/j.efsa.2016.4524. [DOI] [Google Scholar]
- 430.Guinebretière MH, Thompson FL, Sorokin A, Normand P, Dawyndt P, Ehling-Schulz M, Svensson B, Sanchis V, Nguyen-The C, Heyndrickx M, De Vos P. 2008. Ecological diversification in the Bacillus cereus group. Environ Microbiol 10:851–865 10.1111/j.1462-2920.2007.01495.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 431.Méric G, Mageiros L, Pascoe B, Woodcock DJ, Mourkas E, Lamble S, Bowden R, Jolley KA, Raymond B, Sheppard SK. 2018. Lineage-specific plasmid acquisition and the evolution of specialized pathogens in Bacillus thuringiensis and the Bacillus cereus group. Mol Ecol 27:1524–1540 10.1111/mec.14546. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Margolis PS, Driks A, Losick R. 1993. Sporulation gene spoIIB from Bacillus subtilis. J Bacteriol 175:528–540 10.1128/jb.175.2.528. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Slamti L, Lemy C, Henry C, Guillot A, Huillet E, Lereclus D 2016. CodY Regulates the Activity of the Virulence Quorum Sensor PlcR by Controlling the Import of the Signaling Peptide PapR in Bacillus thuringiensis. Front Microbiol 6:1501 10.3389/fmicb.2015.01501. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Mahillon J, Chungjatupornchai W, Decock J, Dierickx S, Michiels F, Peferoen M, Joos H. 1989. Transformation of Bacillus thuringiensis by electroporation. FEMS Microbiol Lett 60:205–210 10.1111/j.1574-6968.1989.tb03447.x. [DOI] [Google Scholar]
- 435.Lereclus D, Arantes O. 1992. spbA locus ensures the segregational stability of pHT1030, a novel type of Gram-positive replicon. Mol. Microbiol 7:35–36 10.1111/j.1365-2958.1992.tb00835.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 436.Lereclus D, Vallade M, Chaufaux J, Arantes O, Rambaud S. 1992. Expansion of insecticidal host range of Bacillus thuringiensis by in vivo genetic recombination. Biotechnology (N Y)10:418–21 10.1038/nbt0492-418. [PubMed] [DOI] [PubMed] [Google Scholar]


