Abstract
Morphological study of 1,795 spiders from sites across Pakistan placed these specimens in 27 families and 202 putative species. COI sequences >400 bp recovered from 1,782 specimens were analyzed using neighbor-joining trees, Bayesian inference, barcode gap, and Barcode Index Numbers (BINs). Specimens of 109 morphological species were assigned to 123 BINs with ten species showing BIN splits, while 93 interim species included representatives of 98 BINs. Maximum conspecific divergences ranged from 0–5.3% while congeneric distances varied from 2.8–23.2%. Excepting one species pair (Oxyopes azhari–Oxyopes oryzae), the maximum intraspecific distance was always less than the nearest-neighbor (NN) distance. Intraspecific divergence values were not significantly correlated with geographic distance. Most (75%) BINs detected in this study were new to science, while those shared with other nations mainly derived from India. The discovery of many new, potentially endemic species and the low level of BIN overlap with other nations highlight the importance of constructing regional DNA barcode reference libraries.
Introduction
With nearly 48,000 known species in 117 families [1], spiders are a major component of terrestrial ecosystems with important practical applications as biocontrol agents [2] and as bio-indicators [3,4]. Prior studies have documented 4,300 spider species in Europe [5] and a similar number (3,800) in the Nearctic [6]. By contrast, just 2,300 species have been reported from South Asia [7], suggesting that many species await detection in this region. Although studies on the spider fauna of Pakistan began nearly a century ago [8], work has recently intensified, but most of these studies have produced regional checklists (S1 Table). Unfortunately, these publications often employ invalid or incorrect species names or only identify specimens to a family [9], compromising their value [10–12]. It is likely that many species reported as new discoveries from Pakistan [13] await description. For example, in her dissertation research on spiders of Punjab, Parveen [13] reported the discovery of 33 new species but only one has been formally described [9]. Examination of prior taxonomic work (S1 Table) indicates that just 400 species of spiders have been documented from Pakistan. Considering the country’s diverse ecosystems [14], this count must seriously underestimate the true diversity of its fauna given the much higher numbers reported for India (1686) [15] and Iran (528) [16]. The limited knowledge of the spider fauna of Pakistan is a particular example of the barrier to our general understanding of spider biodiversity in a global context, a factor compromising both scientific progress and conservation efforts [17].
The poor documentation of spider diversity of Pakistan reflects, in part, the paucity of taxonomic specialists working on the group [18]. Moreover, spiders pose a challenge for morphological approaches because cryptic species are common [19], and sexual dimorphism is often striking [20]. DNA barcoding [21] provides an alternate approach to identifications. It employs sequence diversity in a standard gene region (COI-5′) to discriminate both morphologically cryptic species and all life stages, even for species with sexual dimorphism [22,23]. Although concerns about the use of single marker [24,25] or discordance between the barcode and other gene regions [26] have been voiced [27], the advantages of employing a single standard gene region for DNA barcoding is now very well established [28]. Fifteen years after its introduction, this approach has demonstrated its effectiveness in discriminating species in diverse groups, including spiders [29–34].
The use of DNA barcoding for specimen identification and species discovery is greatly facilitated by BOLD, the Barcode of Life Data System (http://www.boldsystems.org). This informatics platform assembles specimen metadata and sequences and provides tools to facilitate data analysis and publication [35]. It also enables species discrimination by assigning each COI sequence cluster to a Barcode Index Number (BIN) [36], which is an analogue of Operational Taxonomic Unit (OTU). Because BINs have high congruence with species recognized through morphological analysis [37–40], they are now routinely used as a species proxy [41,42]. Consequently, they have gained wide adoption [41,43] for cryptic species recognition [40,43], species discovery [44], taxonomic revisions [45], and faunal assessments [46,47]. The DNA barcode reference libraries available for diverse animal groups [48–54] are helping to identify newly collected specimens [45,54] and to speed taxonomic progress [33]. By assigning sequences from unidentified specimens to a species proxy [44], the BIN system has greatly augmented the application of barcode data in groups where taxonomic knowledge is poor. These barcode libraries are, in effect, forming the foundation for a global “DNA library of life” [55].
At present, BOLD holds 6.8 million records derived from specimens representing 587,000 BINs (accessed 13 April, 2019). This total includes 117,000 records from spiders that have been assigned to more than 10,000 BINs. Past work on spiders has had varied motivations [39,56–60], but just two prior studies have aimed to construct a comprehensive DNA barcode library for a national fauna–Canada [61] and Germany [62]. The need for similar work in other regions is evident, particularly in south Asia. For example, barcode records are only available for 73 species of spiders from India [35,63] and for 41 species from Pakistan [64–66]. The current study aimed to develop a barcode library for the spider fauna of Pakistan and investigate the spider diversity overlap with other regions using BINs. The study addresses the gap for reference data in the country by expanding DNA barcode coverage for Pakistan to 202 species.
Materials and methods
Ethics statement
No specific permissions were required for this study. The study did not involve endangered or protected species.
Spider collection
From 2010 to 2016, 1,795 spiders were collected at 225 sites in Pakistan (Fig 1). Each spider was provisionally identified by collectors in Pakistan before it was sequenced for the barcode region of the mitochondrial COI gene [21]. GB subsequently validated and refined identifications by examining (including genitalic dissections) representative specimens from each barcode cluster. Generic and species assignments generally followed taxonomic publications on Asian spiders (S1 Table), but nomenclature was updated as required to follow the World Spider Catalog [1]. Collection data, a photograph, and a taxonomic assignment for each specimen are available in the public dataset, "DS-MASPD DNA barcoding spiders of Pakistan" (dx.doi.org/10.5883/DS-MASPD) on BOLD. The 1,795 specimens are held in four repositories: Centre for Biodiversity Genomics, University of Guelph, Guelph, Canada (585); National Institute for Biotechnology and Genetic Engineering, Faisalabad, Pakistan (1126); University of Sargodha, Sargodha, Pakistan (84). The location of any particular specimen is reported in the dataset.
Fig 1. Map showing collection localities for the 1,795 spiders examined in this study.
The map was developed using www.simplemappr.net. The author of SimpleMapper has waived all copyrights and no permission is needed to use. GPS coordinates (Latitude, Longitude) for the collection localities were: 24.45, 70.8; 25.488, 67.821; 25.681, 67.781; 25.756, 67.739; 25.757, 67.732; 25.759, 67.737; 25.76, 67.732; 25.801, 67.733; 25.812, 67.739; 25.9, 69.85; 28.083, 70.283; 28.261, 70.647; 28.293, 70.115; 28.304, 70.134; 28.306, 70.128; 28.308, 70.132; 28.308, 70.134; 28.309, 70.13; 28.309, 70.131; 28.309, 70.133; 29.083, 69.083; 29.103, 70.324; 29.104, 70.324; 29.105, 70.328; 29.24, 71.415; 29.242, 71.413; 29.39, 71.68; 29.393, 71.688; 29.393, 71.684; 29.394, 71.682; 29.396, 71.683; 29.401, 71.627; 29.429, 71.548; 29.454, 71.161; 29.518, 71.645; 29.584, 71.439; 29.868, 71.291; 29.9167, 69.9667; 30, 70.6; 30.026, 71.381; 30.053, 71.385; 30.065, 71.363; 30.105, 71.417; 30.189, 71.455; 30.189, 71.458; 30.189, 71.457; 30.191, 71.457; 30.516, 72.583; 30.518, 72.624; 30.519, 72.606; 30.52, 72.624; 30.522, 72.635; 30.523, 72.629; 30.525, 72.624; 30.529, 72.63; 30.531, 72.655; 30.531, 72.632; 30.533, 72.63; 30.534, 72.633; 30.534, 72.606; 30.537, 72.638; 30.538, 72.641; 30.54, 72.608; 30.585, 72.993; 30.6, 73.0667; 30.65, 73.1; 30.66, 73.1; 30.6612, 73.1086; 30.791, 72.594; 30.8, 72.05; 30.832, 72.512; 30.85, 72.083; 30.85, 72.544; 30.854, 72.538; 30.855, 72.54; 30.855, 72.539; 30.856, 72.572; 30.857, 72.542; 30.859, 72.566; 30.862, 72.56; 30.862, 72.554; 30.866, 72.555; 30.875, 72.557; 30.959, 73.984; 31.024, 74.531; 31.033, 73; 31.0833, 73.95; 31.2167, 73.8667; 31.3333, 73.4167; 31.3833, 73.0167; 31.3833, 73; 31.393, 73.027; 31.394, 73.026; 31.4167, 73.05; 31.4167, 73.0667; 31.45, 73.7; 31.45, 73.6833; 31.45, 73.1333; 31.463, 74.436; 31.4667, 73.2; 31.496, 74.294; 31.5, 73.2667; 31.532, 73.063; 31.5333, 74.3333; 31.56, 72.54; 31.6167, 73.8667; 31.64, 74.13; 31.825, 72.541; 31.8424, 70.8952; 31.86, 73.276; 31.924, 72.863; 31.965, 72.867; 31.976, 72.328; 31.986, 72.832; 32.027, 72.653; 32.034, 72.703; 32.05, 73; 32.055, 72.946; 32.059, 73.011; 32.063, 73.042; 32.0667, 72.6667; 32.0667, 72.6833; 32.067, 73.05; 32.074, 72.684; 32.077, 72.671; 32.077, 72.67; 32.078, 72.672; 32.08, 72.9; 32.081, 72.667; 32.082, 72.675; 32.083, 73.067; 32.0837, 72.6719; 32.084, 72.68; 32.088, 72.673; 32.093, 72.684; 32.1, 73.067; 32.102, 72.957; 32.109, 72.846; 32.11, 72.655; 32.119, 72.679; 32.122, 72.681; 32.125, 72.693; 32.1333, 74.1833; 32.15, 74.1833; 32.17, 72.26; 32.19, 73.025; 32.267, 72.476; 32.275, 72.904; 32.287, 72.43; 32.3054, 72.3482; 32.5333, 69.85; 32.56, 72.02; 32.59, 72.999; 32.59, 72.008; 32.59, 73.049; 32.59, 73.999; 32.591, 73.008; 32.591, 72.999; 32.5916, 72.3446; 32.592, 73.011; 32.592, 72.999; 32.593, 72.999; 32.594, 73.02; 32.594, 72.999; 32.595, 72.999; 32.5964, 72.217; 32.597, 73.041; 32.601, 73.369; 32.601, 73.038; 32.603, 73.042; 32.624, 73; 32.629, 73.009; 32.63, 73.005; 32.632, 73.013; 32.637, 73.008; 32.637, 72.008; 32.652, 73; 32.656, 73.005; 32.657, 73.004; 32.658, 73.003; 32.6581, 73.0034; 32.659, 73.008; 32.6592, 72.2433; 32.755, 72.677; 33.686, 73.076; 33.714, 73.132; 33.714, 73.133; 33.714, 73.13; 33.715, 73.132; 33.716, 73.129; 33.7167, 73.0333; 33.7167, 73.05; 33.7667, 73.8833; 33.8, 72.9167; 33.8167, 73.8167; 33.9, 73.3833; 33.9167, 73.3833; 34.333, 73.204; 34.334, 73.201; 34.38, 73.52; 34.38, 73.54; 34.385, 73.544; 34.386, 73.546; 34.386, 73.545; 34.541, 73.348; 34.543, 73.348; 34.546, 73.349; 34.638, 73.461; 34.639, 73.461; 34.639, 73.462; 34.7333, 72.35; 34.7667, 72.35; 34.776, 73.527; 34.777, 73.526; 34.778, 73.528; 34.78, 73.53; 34.78, 73.531; 34.8167, 72.3333; 35.426, 74.098; 35.461, 72.588; 35.465, 72.584; 35.4667, 72.5833; 35.478, 72.588; 35.918, 74.29; 35.918, 74.289.
Molecular analysis
DNA extraction, PCR, and Sanger sequencing were performed at the Canadian Centre for DNA Barcoding (CCDB) (http://ccdb.ca/resources/) using standard protocols. A single leg was removed from each specimen with a sterile forceps and transferred into a well in a 96-well microplate pre-filled with 30 μl of 95% EtOH. DNA was subsequently extracted by tissue lysis at 56°C overnight followed by a column-based protocol [67]. PCR amplification of the COI-5′ barcode region employed the primer pair C_LepFolF and C_LepFolR (http://ccdb.ca/site/wp-content/uploads/2016/09/CCDB_PrimerSets.pdf). This primer cocktail includes equal volume of LepF1 [68] /LCO1490 [69] and LepR1 [68] /HCO2198 [69], respectively. The target COI region was amplified using 2 μL of DNA template in a 12.5 μL reaction containing standard PCR ingredients [30] employing the following PCR regime: 94°C (1 min), 5 cycles of 94°C (40 s), 45°C (40 s), 72°C (1 min); 35 cycles of 94°C (40 s), 51°C (40 s), 72°C (1 min) and final extension of 72°C (5 min). Amplicons were analyzed on a 2% agarose E-gel 96 system (Invitrogen Inc.) and were sequenced bidirectionally using the BigDye Terminator Cycle Sequencing Kit (v3.1) on an Applied Biosystems 3730XL DNA Analyzer. Sequences were assembled, aligned, and edited using CodonCode Aligner (CodonCode Corporation, USA) and validated in MEGA5 [70] to ensure they lacked a stop codon.
Data analysis
All sequences were submitted to BOLD (DS-MASPD) where those meeting required quality criteria (>507 bp, <1% Ns, no stop codon or contamination flag) were assigned to a BIN [36]. An accumulation curve, BIN discordance, genetic distance analysis, barcode gap analysis (BGA), and geo-distance correlation were determined using analytical tools on BOLD. The Accumulation Curve plots the rise in the number of BINs with increased sampling effort making it possible to ascertain if asymptotic diversity has been reached. The BGA determines if the maximum sequence divergence within members of a species or BIN is less than the distance to its Nearest-Neighbor (NN) species or BIN, a condition required for unambiguous identification [71,72]. The geo-distance correlation ascertains the correlation between geographic distance and genetic distance in each species or BIN employing two methods. The Mantel Test [73] examines the relationship between the geographic distance (km) and genetic divergence (K2P) matrices. The second approach compares the spread of the minimum spanning tree of collection sites and maximum intra-specific divergence [61]. The relationship between geographic and intraspecific distances was analyzed for each species with at least one individual from three or more sites. The analysis included all the conspecific records public on BOLD.
A neighbor-joining (NJ) tree was generated in MEGA5 using the Kimura-2-Parameter (K2P) [74] distance model along with pairwise deletion of missing sites. Nodal support on the NJ tree was estimated by 1000 bootstrap replicates. Bayesian inference (BI) was calculated by MrBayes v3.2.0 [75] using representative sequences of the 221 BINs and employing Phalangium opilio (Arachnida: Opiliones) and Galeodes sp. (Arachnida: Solifugae) as outgroups. The data was partitioned in two ways; i) a single partition with parameters estimated across all codon positions, ii) a codon-partition in which each codon position was allowed different parameter estimates. Sequence evolution was modelled by the GTR+Γ model independently for the two partitions using the ‘‘unlink” command in MrBayes. Analyses were run for 10 million generations using four chains with sampling every 1000 generations and the BI trees were obtained using the Markov Chain Monte Carlo (MCMC) technique. Posterior probabilities were calculated from the sample points once the MCMC algorithm converged. Convergence was determined when the standard deviation of split frequencies was less than 0.022 and the PSRF (potential scale reduction factor) approached 1, and both runs converged to a stationary distribution after the burn-in stage (the first 25% of samples were discarded by default). The resultant trees were visualized in FigTree v1.4.0. The NJ and Bayesian analyses were employed to assess support for the BINs detected in this study, not to reconstruct the phylogeny of Araneae.
Results
Coupling of the DNA sequence results with detailed morphological analysis made it possible to assign 1,574 of the 1,795 barcoded specimens to one of 109 species, but the other 221 specimens could only be placed into one of 93 interim species. Collectively, these specimens included representatives of 27 families, 113 genera, and 202 species (Table 1). Most species were only represented by a single sex, usually females. Two-thirds (1,256) of the specimens were immatures that lacked the diagnostic characters required for species assignment. However, their DNA barcodes allowed them to be linked to adults whose identification was established through morphology. Four families (Amaurobiidae, Atypidae, Ctenidae, Segestriidae), 43 genera, and 74 species identified here represent first records for Pakistan (Tables 1 and S1). As adults from 12 of the 93 interim species possessed clear morphological differences from any known species in their genus, they are likely new to science (Table 1).
Table 1. Species, maximum barcode divergence (K2P), nearest neighbor distance (NN), and BIN assignment of 1,795 spiders collected in Pakistan.
| No. | Taxa | N | K2P | NN | BINs | |
|---|---|---|---|---|---|---|
| Agelenidae C. L. Koch,1837 | ||||||
| 1 | Draconarius sp. 1GAB_PAK | 2 | 0 | 8.8 | BOLD:AAO2052 | |
| 2 | Draconarius sp. 2GAB_PAK | 2 | 0 | 8.8 | BOLD:AAO2053 | |
| *NP | 3 | Tegenaria domestica (Clerck, 1757) | 1 | N/A | 19 | BOLD:AAF1312 |
| NP | Amaurobiidae Thorell,1870 | |||||
| *NS | 4 | Himalmartensus cf. martensi Wang & Zhu, 2008 | 1 | N/A | 14 | BOLD:ACB2928 |
| Araneidae Clerck, 1757 | ||||||
| NP | 5 | Araneus affinis Zhu, Tu & Hu, 1988 | 2 | 0.6 | 12 | BOLD:AAV7611 |
| 6 | Araneus mitificus (Simon, 1886) | 20 | 1.7 | 13 | BOLD:AAV1598 | |
| 7 | Araniella sp. 1GAB_PAK | 4 | 0.8 | 10 | BOLD:AAV1625 | |
| 8 | Argiope aemula (Walckenaer, 1841) | 8 | 1.4 | 10 | BOLD:ACG0732 | |
| 9 | Argiope anasuja Thorell, 1887 | 1 | N/A | 9.5 | BOLD:ACB2926 | |
| NP | 10 | Argiope lobata (Pallas, 1772) | 2 | 0.5 | 12 | BOLD:ACI8559 |
| NP | 11 | Argiope pulchella Thorell, 1881 | 6 | 0.8 | 9.5 | BOLD:ACG0576 |
| 12 | Argiope trifasciata (Forsskål, 1775) | 15 | 1.1 | 10 | BOLD:AAQ2634 | |
| NP | 13 | Chorizopes wulingensis Yin, Wang & Xie, 1994 | 2 | 0.5 | 12 | BOLD:ABX7347 |
| U | 14 | Cyclosa confraga (Thorell, 1892) | 8 | 0.8 | 18 | BOLD:ADF2726 |
| 15 | Cyclosa hexatuberculata Tikader, 1982 | 4 | 0 | 11 | BOLD:ADD8756 | |
| NP | 16 | Cyclosa moonduensis Tikader, 1963 | 9 | 1.1 | 11 | BOLD:ACZ2455 |
| 17 | Cyrtophora citricola (Forsskål, 1775) | 66 | 1.6 | 13 | BOLD:AAO2032 | |
| 18 | Eriovixia excelsa (Simon, 1889) | 40 | 1.1 | 16 | BOLD:AAQ0105 | |
| 19 | Gea subarmata Thorell, 1890 | 1 | N/A | 10 | BOLD:ACG0733 | |
| *NS | 20 | Hypsosinga cf. alboria Yin, Wang, Xie & Peng, 1990 | 4 | 0 | 10 | BOLD:ABX7344 |
| *NP | 21 | Hypsosinga wanica Song, Qian & Gao, 1996 | 17 | 1.6 | 10 | BOLD:AAQ0134 |
| 22 | Larinia phthisica (L. Koch, 1871) | 5 | 0.9 | 10 | BOLD:AAO2160 | |
| 23 | Larinia sp. 1GAB_PAK | 1 | N/A | 11 | BOLD:ABX7407 | |
| * | 24 | Leviellus sp. 1GAB_PAK | 2 | 0 | 14 | BOLD:AAV1590 |
| NP | 25 | Neoscona polyspinipes Yin, Wang, Xie & Peng, 1990 | 20 | 0.8 | 4.9 | BOLD:AAO1983 |
| NP | 26a | Neoscona scylla (Karsch, 1879) | 13 | 1.9 | 7.8 | BOLD:ACI8762 |
| 26b | Neoscona scylla (Karsch, 1879) | 16 | BOLD:AAO1997 | |||
| 27 | Neoscona sp. 1BAG_PAK | 1 | N/A | 7.6 | BOLD:ACI2573 | |
| 28 | Neoscona sp. 2BAG_PAK | 1 | N/A | 9 | BOLD:ADD4537 | |
| NP | 29 | Neoscona subfusca (C. L. Koch, 1837) | 1 | N/A | 8.6 | BOLD:AAV3851 |
| 30 | Neoscona theisi (Walckenaer, 1841) | 160 | 2.5 | 7.6 | BOLD:ACM3489 | |
| 31 | Neoscona vigilans (Blackwall, 1865) | 38 | 1.5 | 4.9 | BOLD:AAO2202 | |
| 32 | Plebs himalayaensis (Tikader, 1975) | 2 | 0 | 17 | BOLD:ACI8675 | |
| NP | Atypidae Thorell, 1870 | |||||
| *NS | 33 | Calommata sp. 1GAB_PAK | 1 | N/A | 21 | BOLD:ACP9624 |
| Cheiracanthiidae Wagner, 1887 | ||||||
| NP | 34 | Cheiracanthium inornatum O. Pickard-Cambridge, 1874 | 5 | 2.3 | 7.4 | BOLD:ACC4872 |
| NP | 35 | Cheiracanthium insulanum (Thorell, 1878) | 20 | 3.3 | 6.9 | BOLD:AAQ0110 |
| 36 | Cheiracanthium sp. 1GAB_PAK | 2 | 0.2 | 11 | BOLD:ACA7676 | |
| 37 | Cheiracanthium sp. 2GAB_PAK | 2 | 1.1 | 4.9 | BOLD:ABW2880 | |
| 38 | Cheiracanthium sp. 3GAB_PAK | 2 | 0.2 | 4.9 | BOLD:AAU6055 | |
| Clubionidae Wagner, 1887 | ||||||
| 39 | Clubiona drassodes O. Pickard-Cambridge, 1874 | 28 | 0.9 | 13 | BOLD:AAV1620 | |
| 40 | Clubiona filicata O. Pickard-Cambridge, 1874 | 18 | 0.9 | 13 | BOLD:AAV1603 | |
| 41 | Clubiona sp. 1GAB_PAK | 1 | N/A | 8.8 | BOLD:AAV1602 | |
| 42 | Clubiona sp. 2GAB_PAK | 1 | N/A | 8.8 | BOLD:AAO2055 | |
| Corinnidae Karsch, 1880 | ||||||
| 43 | Castianeira sp. 1GAB_PAK | 1 | N/A | 16 | BOLD:ACP7698 | |
| NP | Ctenidae Keyserling, 1877 | |||||
| * | 44 | Anahita sp. 1GAB_PAK | 1 | N/A | 12 | BOLD:ADF5307 |
| * | 45 | Ctenus sp. 1GAB_PAK | 1 | N/A | 9 | BOLD:AAV1591 |
| * | 46 | Ctenus sp. 2GAB_PAK | 1 | N/A | 9 | BOLD:ABW2888 |
| Filistatidae Ausserer, 1867 | ||||||
| 47 | Kukulcania sp. 1GAB_PAK | 1 | N/A | 22 | BOLD:ABX7408 | |
| Gnaphosidae Pocock, 1898 | ||||||
| 48 | Berlandina afghana Denis, 1958 | 1 | N/A | 14 | BOLD:AAV1613 | |
| 49 | Drassodes sp. 1GAB_PAK | 1 | N/A | 12 | BOLD:AAV1404 | |
| *NP | 50 | Drassyllus coreanus Paik, 1986 | 2 | 0 | 14 | BOLD:AAV0899 |
| 51 | Gnaphosa jodhpurensis Tikader & Gajbe, 1977 | 2 | 1.2 | 15 | BOLD:ACR0656 | |
| *NP | 52 | Haplodrassus signifer (C. L. Koch, 1839) | 1 | N/A | 13 | BOLD:ACB2432 |
| 53 | Micaria sp. 1GAB_PAK | 1 | N/A | 13 | BOLD:ACP3811 | |
| * | 54 | Phaeocedus sp. 1GAB_PAK | 2 | 1.2 | 14 | BOLD:AAV1605 |
| *NP | 55 | Scopoides maitraiae (Tikader & Gajbe, 1977) | 2 | 0 | 16 | BOLD:ACZ1655 |
| *NP | 56 | Trachyzelotes kulczynskii (Bösenberg, 1902) | 1 | N/A | 13 | BOLD:AAQ2633 |
| NS | 57 | Zelotes cf. puritanus Chamberlin, 1922 | 2 | 0.8 | 12 | BOLD:AAQ0137 |
| NP | 58 | Zelotes shantae Tikader, 1982 | 1 | N/A | 12 | BOLD:ADD7482 |
| 59 | Zelotes sp. 1GAB_PAK | 1 | N/A | 12 | BOLD:ACZ4032 | |
| *NP | 60 | Zimiris diffusa Platnick & Penney, 2004 | 1 | N/A | 14 | BOLD:AAV1616 |
| Hersiliidae Thorell, 1870 | ||||||
| 61 | Hersilia savignyi Lucas, 1836 | 16 | 1.1 | 17 | BOLD:AAP4789 | |
| Linyphiidae Blackwall, 1859 | ||||||
| 62 | Gnathonarium dentatum (Wider, 1834) | 5 | 0 | 14 | BOLD:AAQ0150 | |
| * | 63 | Mermessus sp. 1GAB_PAK | 1 | N/A | 14 | BOLD:ACP3810 |
| *NP | 64 | Neriene emphana (Walckenaer, 1841) | 3 | 0.8 | 14 | BOLD:ACI8558 |
| Lycosidae Sundevall, 1833 | ||||||
| * | 65 | Alopecosa sp. 1GAB_PAK | 1 | N/A | 9.2 | BOLD:AAV1615 |
| NS | 66 | Arctosa cf. serrulata Mao & Song, 1985 | 1 | N/A | 9.4 | BOLD:ACB2931 |
| 67 | Arctosa sp. 1GAB_PAK | 1 | N/A | 13 | BOLD:AAV1608 | |
| 68 | Draposa oakleyi (Gravely, 1924) | 19 | 1.6 | 5.8 | BOLD:ABX7398 | |
| NS | 69 | Evippa sp. 1GAB_PAK | 5 | 1.4 | 8.3 | BOLD:ABX7397 |
| 70 | Evippa sp. 2GAB_PAK | 1 | N/A | 8.3 | BOLD:ABW2890 | |
| 71a | Hippasa pisaurina Pocock, 1900 | 16 | 4.1 | 5.8 | BOLD:AAO2058 | |
| 71b | Hippasa pisaurina Pocock, 1900 | 1 | BOLD:ADF3448 | |||
| 72 | Hippasa sp. 1GAB_PAK | 1 | N/A | 5.8 | BOLD:ADE8277 | |
| * | 73 | Hogna sp. 1GAB_PAK | 5 | 0.6 | 10 | BOLD:AAQ0158 |
| * | 74 | Hogna sp. 2GAB_PAK | 1 | N/A | 11 | BOLD:ADF5080 |
| 75 | Lycosa poonaensis Tikader & Malhotra, 1980 | 5 | 0.6 | 10 | BOLD:ABW2889 | |
| 76 | Lycosa sp. 1GAB_PAK | 1 | N/A | 10 | BOLD:AAO2168 | |
| E | 77 | Lycosa terrestris Butt, Anwar & Tahir, 2006 | 45 | 0.9 | 4.3 | BOLD:AAO2150 |
| NP | 78 | Pardosa mionebulosa Yin, Peng, Xie, Bao & Wang, 1997 | 3 | 1.6 | 5.3 | BOLD:ACZ3882 |
| 79 | Pardosa pseudoannulata (Bösenberg & Strand, 1906) | 5 | 0.6 | 5.9 | BOLD:AAO2149 | |
| 80 | Pardosa sp. 1GAB_PAK | 3 | 0.2 | 5.9 | BOLD:AAO2146 | |
| 81 | Pardosa sp. 2GAB_PAK | 1 | N/A | 4.9 | BOLD:AAO2148 | |
| 82 | Pardosa sp. 3GAB_PAK | 1 | N/A | 5.2 | BOLD:AAV1588 | |
| 83 | Pardosa sp. 4GAB_PAK | 13 | 2.4 | 4.6 | BOLD:AAO2147 | |
| 84 | Pardosa sp. 5GAB_PAK | 4 | 0.8 | 5.2 | BOLD:AAV1589 | |
| NP | 85 | Pardosa sutherlandi (Gravely, 1924) | 7 | 0.2 | 4.6 | BOLD:ABX7411 |
| NP | 86 | Trochosa aquatica Tanaka, 1985 | 17 | 0.6 | 5.9 | BOLD:AAV3200 |
| 87 | Trochosa sp. 1GAB_PAK | 3 | 0.3 | 5.9 | BOLD:ADF4175 | |
| *NP | 88 | Wadicosa fidelis (O. Pickard-Cambridge, 1872) | 75 | 1.9 | 7.2 | BOLD:AAG7456 |
| Oecobiidae Blackwall, 1862 | ||||||
| 89 | Oecobius putus O. Pickard-Cambridge, 1876 | 10 | 0.4 | 14 | BOLD:AAV1624 | |
| Oxyopidae Thorell, 1870 | ||||||
| E | 90 | Oxyopes azhari Butt & Beg, 2001 | 112 | 3.6 | 3.6 | BOLD:AAO1991 |
| E | 91 | Oxyopes chenabensis Mukhtar, 2017 | 5 | 0.9 | 6.4 | BOLD:ABX7410 |
| NP | 92 | Oxyopes heterophthalmus (Latreille, 1804) | 8 | 0.3 | 4.9 | BOLD:AAD0599 |
| 93 | Oxyopes hindostanicus Pocock, 1901 | 123 | 3 | 5.6 | BOLD:AAO1990 | |
| NP | 94 | Oxyopes macilentus L. Koch, 1878 | 8 | 1.5 | 1.3 | BOLD:AAF9665 |
| NP | 95a | Oxyopes matiensis Barrion & Litsinger, 1995 | 3 | 2.1 | 1.3 | BOLD:ACX5149 |
| 95b | Oxyopes matiensis Barrion & Litsinger, 1995 | 5 | BOLD:ABX7414 | |||
| E | 96 | Oxyopes oryzae Mushtaq & Qadar, 1999 | 52 | 1.9 | 3.6 | BOLD:AAO1989 |
| 97 | Oxyopes sp. 1GAB_PAK | 1 | N/A | 6.7 | BOLD:ACZ2323 | |
| NS | 98 | Oxyopes sp. 2GAB_PAK | 3 | 1.2 | 5.7 | BOLD:ACZ4097 |
| 99 | Oxyopes sp. 3GAB_PAK | 1 | N/A | 11 | BOLD:ACP4193 | |
| NP | 100 | Peucetia ranganathani Biswas & Roy, 2005 | 14 | 0.8 | 11 | BOLD:ACB4190 |
| 101 | Peucetia sp. 1GAB_PAK | 1 | N/A | 13 | BOLD:ACB4188 | |
| Philodromidae Thorell, 1870 | ||||||
| 102 | Philodromus sp. 1GAB_PAK | 1 | N/A | 12 | BOLD:ADD8987 | |
| 103 | Philodromus sp. 2GAB_PAK | 3 | 2 | 13 | BOLD:ABX7412 | |
| *NP | 104 | Pulchellodromus mainlingensis (Hu & Li, 1987) | 2 | 0 | 12 | BOLD:ACB4189 |
| NS | 105 | Rhysodromus cf. xinjiangensis (Tang & Song, 1987) | 4 | 0 | 13 | BOLD:AAO2159 |
| NP | 106 | Thanatus vulgaris Simon, 1870 | 2 | 0.3 | 15 | BOLD:AAQ0111 |
| Pholcidae C. L. Koch, 1850 | ||||||
| 107 | Artema sp. 1GAB_PAK | 1 | N/A | 19 | BOLD:ABW2886 | |
| NP | 108 | Artema transcaspica Spassky, 1934 | 2 | 1 | 19 | - |
| 109 | Crossopriza lyoni (Blackwall, 1867) | 2 | 0.3 | 16 | BOLD:AAG2795 | |
| 110a | Crossopriza maculipes (Spassky, 1934) | 4 | 5.3 | 16 | BOLD:ACN4846 | |
| 110b | Crossopriza maculipes (Spassky, 1934) | 7 | BOLD:AAU5412 | |||
| 110c | Crossopriza maculipes (Spassky, 1934) | 1 | BOLD:ACB2929 | |||
| Pisauridae Simon, 1890 | ||||||
| *NP | 111 | Pisaura mirabilis (Clerck, 1757) | 4 | 0.5 | 10 | BOLD:AAE4245 |
| 112 | Pisaura sp. 1GAB_PAK | 1 | N/A | 12 | BOLD:AAO2059 | |
| Salticidae Blackwall, 1841 | ||||||
| NP | 113 | Bianor albobimaculatus (Lucas, 1846) | 21 | 0.7 | 12 | BOLD:AAP4728 |
| 114 | Bianor sp. 1GAB_PAK | 1 | N/A | 13 | BOLD:ACI8750 | |
| NP | 115 | Epocilla sirohi Caleb, Chatterjee, Tyagi, Kundu, Kumar, 2018 | 7 | 1.9 | 11 | BOLD:ADD4346 |
| 116 | Euophrys sp. 1GAB_PAK | 1 | N/A | 13 | BOLD:ADD1307 | |
| *NS | 117 | Evarcha sp. 1GAB_PAK | 3 | 0.8 | 9 | BOLD:AAV1614 |
| 118 | Hasarius adansoni (Audouin, 1826) | 2 | 0 | 13 | BOLD:AAW0165 | |
| NP | 119 | Hyllus dotatus (Peckham & Peckham, 1903) | 3 | 0.7 | 11 | BOLD:AAV1597 |
| NP | 120 | Menemerus brevibulbis (Thorell, 1887) | 3 | 1.4 | 7.7 | BOLD:AAO2155 |
| 121 | Menemerus marginatus (Kroneberg, 1875) | 1 | N/A | 11 | BOLD:AAV1611 | |
| 122 | Menemerus nigli Wesolowska & Freudenschuss, 2012 | 12 | 1.1 | 7.7 | BOLD:AAQ0156 | |
| *NP | 123 | Modunda staintoni (O. Pickard-Cambridge, 1872) | 3 | 0.8 | 14 | BOLD:AAV0387 |
| NP | 124 | Mogrus cognatus Wesolowska & van Harten, 1994 | 12 | 1.4 | 8.1 | BOLD:AAV1599 |
| 125 | Mogrus sp. 1GAB_PAK | 1 | N/A | 8.1 | BOLD:ACZ1977 | |
| 126 | Mogrus sp. 2GAB_PAK | 6 | 0.8 | 10 | BOLD:AAQ2635 | |
| 127 | Myrmarachne melanocephala MacLeay, 1839 | 1 | N/A | 6.7 | BOLD:AAV1609 | |
| 128 | Myrmarachne robusta (Peckham & Peckham, 1892) | 5 | 1.4 | 6.7 | BOLD:ACS0377 | |
| *NP | 129 | Philaeus chrysops (Poda, 1761) | 1 | N/A | 9.8 | BOLD:ACE4347 |
| * | 130 | Philaeus sp. 1GAB_PAK | 1 | N/A | 9.8 | BOLD:AAV0574 |
| 131 | Phintella vittata (C. L. Koch, 1846) | 11 | 0 | 9.9 | BOLD:ACR1776 | |
| 132a | Plexippus paykulli (Audouin, 1826) | 34 | 5 | 8.8 | BOLD:AAO2152 | |
| 132b | Plexippus paykulli (Audouin, 1826) | 4 | BOLD:AAO2151 | |||
| 132c | Plexippus paykulli (Audouin, 1826) | 1 | BOLD:ACU8433 | |||
| 132d | Plexippus paykulli (Audouin, 1826) | 1 | BOLD:ABX7409 | |||
| 132e | Plexippus paykulli (Audouin, 1826) | 1 | BOLD:ACZ4027 | |||
| 133 | Plexippus sp. 1GAB_PAK | 2 | 0.2 | 8.8 | BOLD:AAV1604 | |
| 134a | Pseudicius admirandus Logunov, 2007 | 8 | 1.4 | 9.4 | BOLD:AAQ0115 | |
| 134b | Pseudicius admirandus Logunov, 2007 | 2 | BOLD:ADD4534 | |||
| NP | 135 | Rhene albigera (C. L. Koch, 1846) | 1 | N/A | 5.9 | BOLD:AAV5815 |
| NP | 136 | Rhene flavigera (C. L. Koch, 1846) | 4 | 0 | 5.4 | BOLD:ADD7823 |
| 137 | Rhene sp. 1GAB_PAK | 1 | N/A | 5.4 | BOLD:ACU6737 | |
| *NS | 138 | Sonoita cf. lightfooti Peckham & Peckham, 1903 | 1 | N/A | 13 | BOLD:ADD9560 |
| 139 | Stenaelurillus arambagensis (Biswas & Biswas, 1992) | 3 | 0.3 | 11 | BOLD:ABX7343 | |
| * | 140 | Talavera sp. 1GAB_PAK | 1 | N/A | 12 | BOLD:ACZ2472 |
| 141 | Telamonia dimidiata (Simon, 1899) | 17 | 1.4 | 10 | BOLD:ACG1123 | |
| 142 | Thyene imperialis (Rossi, 1846) | 56 | 3.5 | 9 | BOLD:AAO2153 | |
| 143 | Thyene sp. 1GAB_PAK | 1 | N/A | 11 | BOLD:AAV1607 | |
| *NS | 144 | Trite sp. 1GAB_PAK | 13 | 0.5 | 11 | BOLD:AAO2154 |
| NP | Segestriidae Simon, 1893 | |||||
| * | 145 | Ariadna sp. 1GAB_PAK | 1 | N/A | 20 | BOLD:AAO2054 |
| Sparassidae Bertkau, 1872 | ||||||
| NP | 146 | Heteropoda maxima Jäger, 2001 | 20 | 0.3 | 4.3 | BOLD:ACB5077 |
| 147a | Heteropoda sp. 3GAB_PAK | 1 | 2.3 | 5.4 | BOLD:ABW2881 | |
| 147b | Heteropoda sp. 3GAB_PAK | 1 | BOLD:AAO2057 | |||
| 148 | Heteropoda sp. 4GAB_PAK | 1 | N/A | 4.3 | BOLD:ACB5549 | |
| 149 | Olios sp. 1GAB_PAK | 1 | N/A | 3.9 | BOLD:ADD6859 | |
| 150 | Olios sp. 2GAB_PAK | 10 | 0.5 | 3.9 | BOLD:ADD7417 | |
| 151 | Olios sp. 3GAB_PAK | 4 | 0.3 | 4.1 | BOLD:ACB4191 | |
| 152 | Olios sp. 4GAB_PAK | 4 | 1.1 | 7.2 | BOLD:AAQ0159 | |
| 153 | Olios sp. 5GAB_PAK | 15 | 2.2 | 7.2 | BOLD:AAQ0157 | |
| 154a | Olios tener (Thorell, 1891) | 4 | 1.9 | 11 | BOLD:AAQ0107 | |
| 154b | Olios tener (Thorell, 1891) | 1 | BOLD:ADK3497 | |||
| 154c | Olios tener (Thorell, 1891) | 1 | BOLD:ADJ7965 | |||
| 155 | Pseudopoda prompta (O. Pickard-Cambridge, 1885) | 4 | 0.9 | 13 | BOLD:AAO2056 | |
| 156a | Spariolenus tigris Simon, 1880 | 1 | 4.1 | 12 | BOLD:ADF5077 | |
| 156b | Spariolenus tigris Simon, 1880 | 1 | BOLD:ABW2878 | |||
| Tetragnathidae Menge, 1866 | ||||||
| *NP | 157 | Glenognatha tangi (Zhu, Song & Zhang, 2003) | 3 | 1.2 | 18 | BOLD:AAQ0147 |
| 158 | Guizygiella indica (Tikader & Bal, 1980) | 8 | 1.1 | 14 | BOLD:ABX7345 | |
| 159 | Leucauge celebesiana (Walckenaer, 1841) | 7 | 0.2 | 11 | BOLD:AAO2068 | |
| 160 | Leucauge decorata (Blackwall, 1864) | 30 | 0.5 | 11 | BOLD:AAG8516 | |
| * | 161 | Metleucauge sp. 1GAB_PAK | 1 | N/A | 19 | BOLD:AAV1600 |
| NP | 162 | Tetragnatha boydi O. Pickard-Cambridge, 1898 | 3 | 0 | 15 | BOLD:ACB2930 |
| NP | 163 | Tetragnatha cavaleriei Schenkel, 1963 | 2 | 0.5 | 16 | BOLD:AAT8904 |
| 164 | Tetragnatha javana (Thorell, 1890) | 43 | 2.8 | 17 | BOLD:AAO2174 | |
| 165 | Tetragnatha mandibulata Walckenaer, 1841 | 1 | N/A | 15 | BOLD:AAK2567 | |
| NP | 166 | Tetragnatha maxillosa Thorell, 1895 | 4 | 0.3 | 15 | BOLD:AAK2560 |
| NP | 167 | Tetragnatha nitens (Audouin, 1826) | 6 | 0.8 | 15 | BOLD:AAD3790 |
| 168 | Tetragnatha sp. 1GAB_PAK | 1 | N/A | 16 | BOLD:ABW2885 | |
| Theraphosidae Thorell, 1869 | ||||||
| * | 169 | Chilobrachys sp. 1GAB_PAK | 1 | N/A | 4.3 | BOLD:ADD5278 |
| * | 170 | Chilobrachys sp. 2GAB_PAK | 1 | N/A | 4.3 | BOLD:AAQ0160 |
| Theridiidae Sundevall, 1833 | ||||||
| *NP | 171 | Emertonella taczanowskii (Keyserling, 1886) | 1 | N/A | 12 | BOLD:AAV1610 |
| 172 | Enoplognatha sp. 1GAB_PAK | 1 | N/A | 12 | BOLD:ACI8909 | |
| 173 | Enoplognatha sp. 2GAB_PAK | 1 | N/A | 15 | BOLD:ACP4208 | |
| * | 174 | Euryopis sp. 1GAB_PAK | 1 | N/A | 12 | BOLD:AAQ0155 |
| 175 | Latrodectus sp. 1GAB_PAK | 1 | N/A | 19 | BOLD:AAV1732 | |
| 176 | Latrodectus sp. 2GAB_PAK | 1 | N/A | 16 | BOLD:AAO3347 | |
| * | 177 | Meotipa sp. 1GAB_PAK | 2 | 2 | 12 | BOLD:AAQ0152 |
| 178 | Phylloneta sp. 1GAB_PAK | 11 | 0.3 | 11 | BOLD:AAV3043 | |
| *NP | 179 | Steatoda cingulata (Thorell, 1890) | 2 | 0 | 14 | BOLD:ABW2877 |
| NP | 180 | Theridion melanostictum O. Pickard-Cambridge, 1876 | 1 | N/A | 11 | BOLD:AAV1617 |
| 181 | Theridion sp. 1GAB_PAK | 1 | N/A | 11 | BOLD:ACB2932 | |
| 182 | Theridion sp. 3GAB_PAK | 1 | N/A | 12 | BOLD:AAV1623 | |
| Thomisidae Sundevall, 1833 | ||||||
| *NP | 183 | Coriarachne melancholica Simon, 1880 | 1 | N/A | 7.9 | BOLD:ACI8639 |
| *NP | 184 | Ebelingia kumadai (Ono, 1985) | 3 | 0 | 12 | BOLD:AAV1619 |
| 185 | Henriksenia hilaris (Thorell, 1877) | 1 | N/A | 11 | BOLD:AAV1618 | |
| NP | 186 | Lysiteles kunmingensis Song & Zhao, 1994 | 3 | 0 | 9.9 | BOLD:ACI8899 |
| * | 187 | Misumenoides sp. 1GAB_PAK | 1 | N/A | 11 | BOLD:AAV1594 |
| * | 188 | Misumenops sp. 1GAB_PAK | 2 | 1.9 | 11 | BOLD:AAV1596 |
| * | 189 | Ozyptila sp. 1GAB_PAK | 1 | N/A | 11 | BOLD:ADF5201 |
| 190a | Runcinia insecta (L. Koch, 1875) | 40 | 4.9 | 11 | BOLD:AAI0997 | |
| 190b | Runcinia insecta (L. Koch, 1875) | 2 | BOLD:AAQ0108 | |||
| *NP | 191 | Tharpyna indica Tikader & Biswas, 1979 | 1 | N/A | 12 | BOLD:AAV1606 |
| NP | 192 | Thomisus onustus Walckenaer, 1805 | 1 | N/A | 8.6 | BOLD:AAD7031 |
| E | 193a | Thomisus zaheeri Parveen, Khan, Mushtaq, Ahmad & Rana, 2008 | 30 | 4.3 | 11 | BOLD:AAP4819 |
| 193b | Thomisus zaheeri Parveen, Khan, Mushtaq, Ahmad & Rana, 2008 | 1 | BOLD:AAQ0153 | |||
| 194 | Tmarus dostinikus Barrion & Litsinger, 1995 | 13 | 0.2 | 11 | BOLD:ABX7413 | |
| NS | 195a | Tmarus sp. 1GAB_PAK | 3 | 2.9 | 11 | BOLD:ABX7346 |
| 195b | Tmarus sp. 1GAB_PAK | 5 | BOLD:ADJ6297 | |||
| 195c | Tmarus sp. 1GAB_PAK | 4 | BOLD:ADK4624 | |||
| 195d | Tmarus sp. 1GAB_PAK | 1 | BOLD:ADK4625 | |||
| NP | 196 | Xysticus joyantius Tikader, 1966 | 1 | N/A | 13 | BOLD:ADF4849 |
| 197 | Xysticus sp. 1GAB_PAK | 3 | 0.6 | 7.9 | BOLD:ACI8898 | |
| 198 | Xysticus sp. 2GAB_PAK | 1 | N/A | 12 | BOLD:ADF4647 | |
| Uloboridae Thorell, 1869 | ||||||
| * | 199 | Hyptiotes sp. 1GAB_PAK | 1 | N/A | 15 | BOLD:AAQ2632 |
| 200a | Uloborus sp. 1GAB_PAK | 4 | 4 | 14 | BOLD:AAW8359 | |
| 200b | Uloborus sp. 1GAB_PAK | 1 | BOLD:ABW2879 | |||
| Zodariidae Thorell, 1881 | ||||||
| * | 201 | Zodarion sp. 1GAB_PAK | 1 | N/A | 14 | BOLD:AAV1621 |
| * | 202 | Zodarion sp. 2GAB_PAK | 1 | N/A | 14 | BOLD:ACG0983 |
| Total | 1795 | 221 |
N = number of individuals; K2P = maximum Kimura 2-parameter distance; NN = distance to Nearest Neighbor species; BIN = Barcode Index Number; NP = new species or family to Pakistan; * = new genus to Pakistan; E = endemic species to Pakistan; U = undescribed opposite sex; NS = putative new species to science.
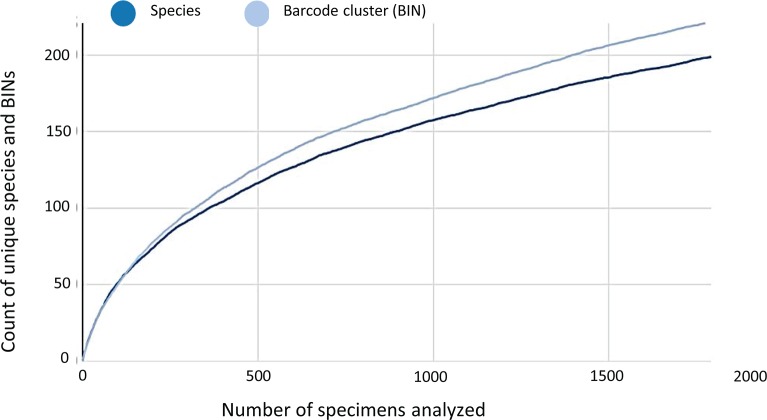
As the accumulation curve failed to approach an asymptote (Fig 2), it is certain that more species await detection. Although one species (Artema transcaspica) failed to qualify for a BIN assignment because its only sequence was too short, the other 108 morphological species were assigned to 123 BINs with 10 species showing a split to two or more BINs (Table 1 and Fig 3). The 93 interim species were allocated to 98 BINs with three showing BIN splits (Table 1), making the total BIN count 221 –with 94 of them singletons. NJ clustering (Fig 3) and Bayesian inference (Fig 4), supported the monophyly of all 221 BINs. Barcode distances (K2P) varied for differing taxonomic ranks with conspecific values ranging from 0.0–5.3% (mean = 0.8%), congenerics from 2.8–23.2% (mean = 8.8%), and confamilials from 4.3–26.7% (mean = 15.1%) (Table 2). Excepting 14 species, maximum intraspecific divergences did not exceed 2% in the 90 species that were represented by two or more specimens (Table 1). The barcode gap analysis showed that maximum intraspecific distance for all but one of the 90 species with two or more records was less than its NN distance (Oxyopes azhari was the exception, overlapping with Oxyopes oryzae) (Fig 5). The Mantel test was non-significant (P>0.01) for 60 of the 69 species and the regression line for all species showed a weak positive relationship (R2 = 0.08; y = 0.0003x + 2.62) (Fig 6).
Fig 2. Accumulation curve for morphological species and barcode index numbers (BINs) for 1,795 spiders from Pakistan.
Fig 3. NJ analysis of spider species based on the analysis of 1,782 COI sequences.
Bootstrap values (50% or higher; 1000 replicates) are shown above the branches. The scale bar shows K2P distances. The node for each species with multiple specimens is collapsed to a vertical line or triangle, with the horizontal depth indicating the level of intraspecific divergence. Species assigned to multiple BINs are indicated in bold. The tree is presented in two parts.
Fig 4. Bayesian phylogenetic analysis of spiders from Pakistan based on COI sequences.
Posterior probabilities are indicated at the nodes. Taxa are followed by the BINs. Phalangium opilio (Arachnida: Opiliones) and Galeodes sp. (Arachnida: Solifugae) were employed as outgroups. Due to its large size, the tree is presented in two parts.
Table 2. Sequence divergences (K2P) for differing levels of taxonomic affinity for the COI-5′ gene region for the spiders from Pakistan.
Analysis was restricted to sequences >400 bp.
| Distance class | n | Taxa | Comparisons | Min (%) | Mean (%) | Max (%) |
|---|---|---|---|---|---|---|
| Intraspecific | 1702 | 122 | 44347 | 0 | 0.8 | 5.3 |
| Congeners | 1338 | 44 | 56792 | 2.8 | 8.8 | 23.2 |
| Confamilial | 1662 | 15 | 137164 | 4.3 | 15.1 | 26.7 |
Fig 5. Barcode gap analysis for spider species represented by three or more records.

Points that fall above the 1:1 line (blue) indicate the presence of a local barcode gap. NN = Nearest-Neighbor species.
Fig 6. Intraspecific sequence divergence (K2P) for the COI gene (blue dots) versus geographic distance (km) for spider species from Pakistan with data from other regions.
The relationship between genetic and geographic distances is indicated by a regression line. P-values for the Mantel Test are indicated by red vertical lines.
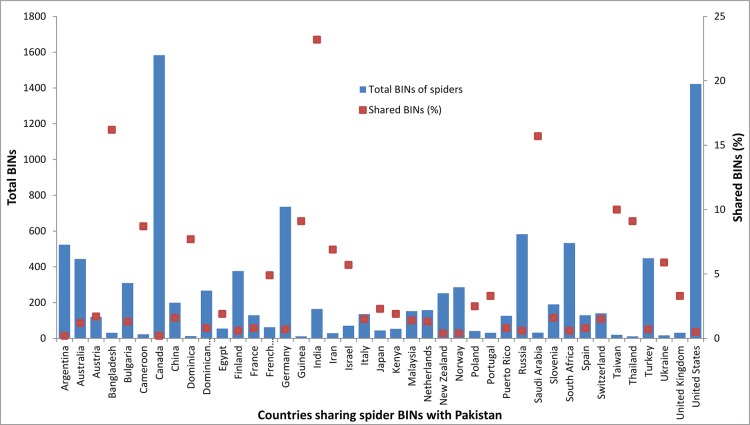
The similarity between the spider fauna in Pakistan and that of other nations was calculated by examining BIN overlap. Less than a quarter (52/221) of the BINs from Pakistan were represented among the 10,229 spider BINs reported in prior studies. As expected, the highest overlap (23%) was with India, but the proportion of shared BINs was far lower for the other 43 countries (Fig 7).
Fig 7. Percentage of spider BINs shared between Pakistan and 41 other nations.
Discussion
Most prior work on the spider fauna of Pakistan has had a regional focus and only employed morphological approaches. For example, 157 species were reported from the province of Punjab [9], 56 from the district of Sargodha [76], 23 from Peshawar [11], and 13 from Buner [77]. A recent checklist for the spiders of Pakistan [10] included records for 239 species, but the present study has substantially increased this total by adding first records for 84 described species and another 93 that could not be assigned to a known taxon. Most importantly, this study generated a DNA barcode reference library for 202 species, facilitating their future identification.
Because the spider fauna of Pakistan has seen such limited study, the discovery of new species was not unexpected, and follows a pattern seen for spiders in other regions. For example, the analysis of 80 species of Salticidae from Papua New Guinea revealed 34 species and five genera new to the country [78]. Likewise, 6% of the 136 spider species recovered from the Northern Cape Province, South Africa were new [79]. This study employed a mix of methods for spider collection, including beating, sweeping, and pitfalls. The choice of sampling method impacts species detection [80] and extensive sampling is critical to generate comprehensive species coverage [81]. Although the present study involved collections at 225 sites, the resultant species accumulation curve did not reach an asymptote, indicating that many more species await detection.
The present study revealed a close correspondence (93%) between BINs and morphospecies as 188 of the 202 species were assigned to a unique BIN, reinforcing a pattern seen in other groups [37,38,40]. For example, the concordance between BINs and species was 78% in a study that examined 30,000 Canadian spiders representing 1,018 species [61] with most discordances reflecting BIN splits suggestive of overlooked species. Stronger species-BIN correspondence has been reported in several insect groups; 96% for Erebidae (Lepidoptera) from the Iberian Peninsula [38], 94% for tiger moths from Brazil [82] and 92% for beetles from central Europe [40]. However, some arthropod groups have shown relatively low level of species-BIN concordance; for example, orthopterans in Central Europe (76%) [83], waterstriders in Germany (82%) [84] and katydids in China (75%) [85]. Thirteen (6%) species in this study were assigned to two or more BINs (BIN splits), and one species (Plexippus paykulli) was assigned to five. BIN splits often indicate the presence of a species complex [43]. For example, 13% of 1,018 species of Canadian spiders [61], 13% of 1,541 Canadian Noctuoidea [86], 5.7% of 1,872 Finnish beetles [87], and 20% of 62 global mealybugs [88] possessed BIN splits. Although in most cases the subsequent morphological investigation has revealed overlooked species [89], other factors can cause BIN splits/mergers, such as hybridization [90], incomplete lineage sorting [83], or rapid speciation [91].
K2P divergences >2% were found in 14 of the 202 spider species from Pakistan with a maximum value of 5.3%. There was, however, no significant relationship between intraspecific divergence and the number of specimens analyzed. For example, 12 specimens of Crossopriza maculipes (3 BINs) showed 5.3% divergence and were assigned to three BINs while 160 specimens of Neoscona theisi possessed a maximum divergence of 2.5%. High COI divergence is not uncommon in spiders. For example, the maximum intraspecific divergence in 561 spider species from Germany was 10.1%, but it was below 2.5% in 95% of the cases with an arithmetic mean of 0.7% [62]. The divergence could depend on several factors such as the number of specimens analyzed, the number of localities, the geographic distance between them and the dispersal capabilities of the particular species [92,93]. With the exception of a single species (Oxyopes azhari), high conspecific distances did not impede the capacity of DNA barcodes to discriminate the species encountered in our study. However, species with BIN splits and high divergences are likely to represent a cryptic species complex. Preliminary morphological analyses including genitalic dissections of specimens from taxa with BIN splits in this study reinforced this conclusion.
Correlation analysis revealed only a weak relationship between the geographic range of the species examined in this study and their intraspecific divergence value. The Mantel test was significant for a few (13%) species, but species identification was not impeded as maximum intraspecific distances were nearly always less than NN distances. Similar results have been reported for Lepidoptera from Europe [94], Pakistan [32] and Central Asia [95]. Although a study that examined a single tribe, Agabini, of aquatic beetles in Europe [96] argued that regional divergences were so great as to obscure species assignments, this result is clearly not the rule [72].
Because BINs are generally an effective species proxy [41], we used them to assess faunal overlap. This work revealed that most (76%) BINs detected in this study were first records. Just 52 BINs have records from other nations and 13 of these were shared only with India. The BIN overlap with other nations was considerably lower for the spiders (24%) of Pakistan than for its Lepidoptera (42%) [42], but this difference almost certainly reflects the intensive barcode studies on the latter group. Although DNA barcoding has been used to assess regional biodiversity [41,47] and to ascertain species connections [42], the limited data availability complicates interpretation. Although further sampling will add new BINs, it is also likely to raise BIN overlap with other regions, improving our understanding of faunal overlap. Such efforts to better document local biodiversity are also certain to reveal new species as evidenced by the discovery of 93 taxa in this study that could not be assigned to a known species.
Supporting information
(DOCX)
Data Availability
Collection data, a photograph, a taxonomic assignment, and DNA barcode (COI-5p) sequence for each specimen are available in the public dataset, "DS-MASPD DNA barcoding spiders of Pakistan" on the Barcode of Life Data System (BOLD) (www.boldsystems.org). (dx.doi.org/10.5883/DS-MASPD).
Funding Statement
This study was enabled by grant 106106-001 “Engaging Developing Nations in iBOL” from the International Development Research Centre in Canada and by grant HEC No. 20-1403/R& D/09 “Sequencing DNA Barcodes of Economically Important Insect Species from Pakistan” from the Higher Education Commission of Pakistan awarded to MA. Sequence analysis was made possible by a grant from the Government of Canada through Genome Canada and Ontario Genomics in support of the International Barcode of Life (iBOL) project awarded to PDNH. This is a contribution to the Food From Thought project supported by the Canada First Research Excellence Fund awarded to PDNH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.World Spider Catalog. World Spider Catalog. Version 20.0. Natural History Museum Bern, online at http://wsc.nmbe.ch, accessed on 7 January 2019. doi: 10.24436/2.
- 2.Riechert SE, Lockley T. Spiders as biological control agents. Annu Rev Entomol. 1984;29: 299–320. [Google Scholar]
- 3.Maelfait J-P, Hendrickx F. Spiders as bio-indicators of anthropogenic stress in natural and semi-natural habitats in Flanders (Belgium): some recent developments. In: Selden PA (ed.), Proceedings of the 17th European Colloquium of Arachnology, Edinburgh; 1998. pp. 293–300.
- 4.Rybak J. Accumulation of major and trace elements in spider webs. Water Air Soil Pollut. 2015;226: 105 10.1007/s11270-015-2369-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Helsdingen PJ. The merits of a European checklist of spiders (Arachnida: Araneae). Logunov DV, Penney D, (Eds). Eur Arachnology. 2003;1: 101–106. [Google Scholar]
- 6.Ubick D, Paquin P, Cushing P, Roth V. Spiders of North America: An Identification Manual, Second Edition 2nd Edition. American Arachnological Society; 2017: 425 pp. [Google Scholar]
- 7.Siliwal M, Molur S. Checklist of spiders (Arachnida: Araneae) of South Asia including the 2006 update of Indian spider checklist. Zoos' Print J. 2007; 22: 2551–2597. [Google Scholar]
- 8.Dyal S. Fauna of Lahore. 4. -Spiders of Lahore. Bulletin of the Department of Zoology of the Panjab University. 1935; 1, i–ii, 119–252. [Google Scholar]
- 9.Parveen R, Khan AA, Mushtaq S, Rana SA. A checklist of the spiders of the Punjab. Pak J Agric Sci. 2007;44: 625–626. [Google Scholar]
- 10.Ghazanfar M, Hussain M, Hashim M, Fahid AM. Checklist of spider (Araneae) fauna of Pakistan: A review. J Entomol Zool Studies. 2016;4: 245–256. [Google Scholar]
- 11.Perveen F, Jamal A. Checklist of spider fauna of FR Peshawar, FATA, Pakistan. Arthropods. 2012;1: 35−39. [Google Scholar]
- 12.Sial N, Ruby T, Malik S, Mushtaq S. A checklist of the spiders of Cholistan and neighbouring areas. Pak J Agric Sci. 2012;49: 301–304. [Google Scholar]
- 13.Parveen R. Taxonomic study on some spiders of Punjab, Pakistan. PhD Thesis. Department of Zoology and Fisheries, Agriculture University Faisalabad, Pakistan. 2003: 356 pp. http://prr.hec.gov.pk/jspui/handle/123456789//4123
- 14.Baig MB, Al-Subaiee FS. Biodiversity in Pakistan: Key issues. Biodiversity 2009;10: 20–29. [Google Scholar]
- 15.Keswani S, Hadole P, Rajoria A. Checklist of spiders (Arachnida: Araneae) from India-2012. Indian J Arachnol. 2012;1: 1–129. [Google Scholar]
- 16.Mirshamsi O, Marusik YM, Zamani A, Moradmand M, Kashefi R. Annotated checklist of the spiders of Iran (Arachnida: Araneae). Iranian J Biosyst Fauna Iranica. 2015;1: 1–108. [Google Scholar]
- 17.Borges PAV, Wunderlich J. Spider biodiversity patterns and their conservation in the Azorean archipelago, with descriptions of new species In: Systematics and Biodiversity; 2008. pp. 249–282. [Google Scholar]
- 18.Drew LW. Are we losing the science of taxonomy? BioScience. 2011;61: 942–946. [Google Scholar]
- 19.Zhang Y, Li S. A spider species complex revealed high cryptic diversity in South China caves. Mol Phylogenet Evol. 2014;79: 353–358. 10.1016/j.ympev.2014.05.017 [DOI] [PubMed] [Google Scholar]
- 20.Hamilton CA, Formanowicz DR, Bond JE. Species delimitation and phylogeography of Aphonopelma hentzi (Araneae, Mygalomorphae, Theraphosidae): cryptic diversity in North American tarantulas. PLOS ONE. 2011;6: e26207 10.1371/journal.pone.0026207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hebert PDN, Cywinska A, Ball SL, deWaard JR. Biological identifications through DNA barcodes. Proc Biol Sci. 2003;270: 313–321. 10.1098/rspb.2002.2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dhananjeyan KJ, Paramasivan R, Tewari SC, Rajendran R, Thenmozhi V, Leo SV, et al. Molecular identification of mosquito vectors using genomic DNA isolated from eggshells, larval and pupal exuvium. Trop Biomed. 2010;27: 47–53. [PubMed] [Google Scholar]
- 23.Iftikhar R, Ashfaq M, Rasool A, Hebert PDN. DNA barcode analysis of thrips (Thysanoptera) diversity in Pakistan reveals cryptic species complexes. PLOS ONE. 2016;11: e0146014 10.1371/journal.pone.0146014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dupuis JR, Roe AD, Sperling FAH. Multi-locus species delimitation in closely related animals and fungi: one marker is not enough. Mol Ecol. 2012;21: 4422–36. 10.1111/j.1365-294X.2012.05642.x [DOI] [PubMed] [Google Scholar]
- 25.Abe TA, Spence JR, Sperling FAH. Mitochondrial introgression is restricted relative to nuclear markers in a water strider (Hemiptera: Gerridae) hybrid zone. Can J Zool. 2005;83:432–444. [Google Scholar]
- 26.Pazhenkova EA, Lukhtanov VA. Nuclear genes (but not mitochondrial DNA barcodes) reveal real species: Evidence from the Brenthis fritillary butterflies (Lepidoptera, Nymphalidae). J Zool Syst Evol Res. 2018; 10.1111/jzs.12252 [DOI] [Google Scholar]
- 27.Hickerson MJ, Meyer CP, Moritz C. DNA Barcoding will often fail to discover new animal species over broad parameter space. Syst Biol. 2006;55: 729–739. 10.1080/10635150600969898 [DOI] [PubMed] [Google Scholar]
- 28.Stein ED, White BP, Mazor RD, Jackson JK, Battle JM, Miller PE, et al. Does DNA barcoding improve performance of traditional stream bioassessment metrics? Freshw Sci. 2014;33: 302–311. [Google Scholar]
- 29.Yang J, Zhang X, Zhang W, Sun J, Xie Y, Zhang Y, et al. Indigenous species barcode database improves the identification of zooplankton. PLOS ONE. 2017;12: e0185697 10.1371/journal.pone.0185697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hebert PDN, DeWaard JR, Zakharov EV, Prosser SW, Sones JE, McKeown JT, et al. A DNA 'barcode blitz': rapid digitization and sequencing of a natural history collection. PLOS ONE. 2013;8: e68535 10.1371/journal.pone.0068535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Slowik J, Blagoev GA. A survey of spiders (Arachnida: Araneae) of Prince of Wales Island, Alaska; combining morphological and DNA barcode identification techniques. Insecta Mundi. 2012;251: 1–12. [Google Scholar]
- 32.Blagoev GA, Nikolova NI, Sobel CN, Hebert PDN, Adamowicz SJ. Spiders (Araneae) of Churchill, Manitoba: DNA barcodes and morphology reveal high species diversity and new Canadian records. BMC Ecol. 2013;13: 44 10.1186/1472-6785-13-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cao XW, Liu J, Chen J, Zheng G, Kuntner M, Agnarsson I. Rapid dissemination of taxonomic discoveries based on DNA barcoding and morphology. Sci Rep. 2016;6: 13 10.1038/s41598-016-0006-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nadolny AA, Omelko MM, Marusik YM, Blagoev GA. A new species of spider belonging to the Pardosa lugubris-group (Araneae: Lycosidae) from Far East Asia. Zootaxa. 2016;4072: 263–281. 10.11646/zootaxa.4072.2.8 [DOI] [PubMed] [Google Scholar]
- 35.Ratnasingham S, Hebert PDN. BOLD: The Barcode of Life Data System (http://www.barcodinglife.org). Mol Ecol Notes. 2007;7: 355–364. 10.1111/j.1471-8286.2007.01678.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ratnasingham S, Hebert PDN. A DNA-based registry for all animal species: the Barcode Index Number (BIN) system. PLOS ONE. 2013;8: e66213 10.1371/journal.pone.0066213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ashfaq M, Akhtar S, Khan AM, Adamowicz SJ, Hebert PDN. DNA barcode analysis of butterfly species from Pakistan points towards regional endemism. Mol Ecol Resour. 2013;13: 832–43. 10.1111/1755-0998.12131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ortiz AS, Rubio RM, Guerrero JJ, Garre MJ, Serrano J, Hebert PDN, et al. Close congruence between Barcode Index Numbers (BINs) and species boundaries in the Erebidae (Lepidoptera: Noctuoidea) of the Iberian Peninsula. Biodivers Data J. 2017;5: e19840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wirta H, Varkonyi G, Rasmussen C, Kaartinen R, Schmidt NM, Hebert PDN, et al. Establishing a community-wide DNA barcode library as a new tool for arctic research. Mol Ecol Resour. 2016;16: 809–822. 10.1111/1755-0998.12489 [DOI] [PubMed] [Google Scholar]
- 40.Hendrich L, Morinière J, Haszprunar G, Hebert PDN, Hausmann A, Köhler F, et al. A comprehensive DNA barcode database for Central European beetles with a focus on Germany: adding more than 3500 identified species to BOLD. Mol Ecol Resour. 2015;15: 795–818. 10.1111/1755-0998.12354 [DOI] [PubMed] [Google Scholar]
- 41.Hebert PDN, Ratnasingham S, Zakharov EV, Telfer AC, Levesque-Beaudin V, Milton MA, et al. Counting animal species with DNA barcodes: Canadian insects. Phil Trans R Soc B. 2016;371: 20150333 10.1098/rstb.2015.0333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ashfaq M, Akhtar S, Rafi MA, Mansoor S, Hebert PD. Mapping global biodiversity connections with DNA barcodes: Lepidoptera of Pakistan. PLOS ONE. 2017;12: e0174749 10.1371/journal.pone.0174749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ashfaq M, Hebert PDN. DNA barcodes for bio-surveillance: Regulated and economically important arthropod plant pests. Genome. 2016;59: 933–945. 10.1139/gen-2016-0024 [DOI] [PubMed] [Google Scholar]
- 44.Mutanen M, Kekkonen M, Prosser SWJ, Hebert PDN, Kaila L. One species in eight: DNA barcodes from type specimens resolve a taxonomic quagmire. Mol Ecol Resour. 2015;15: 967–984. 10.1111/1755-0998.12361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller SE, Hausmann A, Hallwachs W, Janzen DH. Advancing taxonomy and bioinventories with DNA barcodes. Philos Trans Royal Soc B. 2016;371: 20150339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Telfer AC, Young MR, Quinn J, Perez K, Sobel CN, Sones JE, et al. Biodiversity inventories in high gear: DNA barcoding facilitates a rapid biotic survey of a temperate nature reserve. Biodivers Data J. 2015;30: 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ashfaq M, Sabir JSM, El-Ansary HO, Perez K, Levesque-Beaudin V, Khan AM, et al. Insect diversity in the Saharo-Arabian region: Revealing a little-studied fauna by DNA barcoding. PLOS ONE. 2018;13: e0199965 10.1371/journal.pone.0199965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kerr KC, Stoeckle MY, Dove CJ, Weigt LA, Francis CM, Hebert PDN. Comprehensive DNA barcode coverage of North American birds. Mol Ecol Notes. 2007;7: 535–543. 10.1111/j.1471-8286.2007.01670.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hawlitschek O, Moriniere J, Dunz A, Franzen M, Rodder D, Glaw F, et al. Comprehensive DNA barcoding of the herpetofauna of Germany. Mol Ecol Resour. 2016;16: 242–253. 10.1111/1755-0998.12416 [DOI] [PubMed] [Google Scholar]
- 50.Dinca V, Zakharov EV, Hebert PDN, Vila R. Complete DNA barcode reference library for a country's butterfly fauna reveals high performance for temperate Europe. Proc Biol Sci. 2010;278: 347–355. 10.1098/rspb.2010.1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raupach MJ, Hendrich L, Kuchler SM, Deister F, Moriniere J, Gossner MM. Building-up of a DNA barcode library for true bugs (Insecta: Hemiptera: Heteroptera) of Germany reveals taxonomic uncertainties and surprises. PLOS ONE. 2014;9: e106940 10.1371/journal.pone.0106940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moriniere J, Hendrich L, Balke M, Beermann AJ, Konig T, Hess M, et al. A DNA barcode library for Germany’s mayflies, stoneflies and caddisflies (Ephemeroptera, Plecoptera and Trichoptera). Mol Ecol Resour. 2017;17: 1293–1307. 10.1111/1755-0998.12683 [DOI] [PubMed] [Google Scholar]
- 53.Porco D, Chang CH, Dupont L, James S, Richard B, Decaens T. A reference library of DNA barcodes for the earthworms from Upper Normandy: Biodiversity assessment, new records, potential cases of cryptic diversity and ongoing speciation. Appl Soil Ecol. 2018;124: 362–371. [Google Scholar]
- 54.Garcia-Robledo C, Kuprewicz EK, Staines CL, Kress WJ, Erwin TL. Using a comprehensive DNA barcode library to detect novel egg and larval host plant associations in a Cephaloleia rolled-leaf beetle (Coleoptera: Chrysomelidae). Biol J Linnean Soc. 2013;110: 189–198. [Google Scholar]
- 55.Gall LLE, Delsuc F, Hourdez S, Lecointre G, Rasplus JY. Toward the DNA library of life. Eur J Taxon. 2017;266: 1–9. [Google Scholar]
- 56.Robinson EA, Blagoev GA, Hebert PDN, Adamowicz SJ. Prospects for using DNA barcoding to identify spiders in species-rich genera. ZooKeys. 2009;16: 27–46. [Google Scholar]
- 57.Blagoev GA, Dondale CD. A new species of Alopecosa (Araneae: Lycosidae) from Canada: a morphological description supported by DNA barcoding of 19 congeners. Zootaxa. 2014;3894: 152–160. 10.11646/zootaxa.3894.1.12 [DOI] [PubMed] [Google Scholar]
- 58.Magalhaes ILF, Martins PH, Nogueira AA, Santos AJ. Finding hot singles: matching males to females in dimorphic spiders (Araneidae: Micrathena) using phylogenetic placement and DNA barcoding. Invertebr Syst. 2017;31: 8–36. [Google Scholar]
- 59.Piacentini LN, Scioscia CL, Carbajal MN, Ott R, Brescovit AD, Ramirez MJ. A revision of the wolf spider genus Diapontia Keyserling, and the relationships of the subfamily Sosippinae (Araneae: Lycosidae). Arthropod Syst Phylo. 2017;75: 387–415. [Google Scholar]
- 60.Ivanov V, Lee KM, Mutanen M. Mitonuclear discordance in wolf spiders: Genomic evidence for species integrity and introgression. Mol Ecol. 2018;27: 1681–1695. 10.1111/mec.14564 [DOI] [PubMed] [Google Scholar]
- 61.Blagoev GA, deWaard JR, Ratnasingham S, deWaard SL, Lu L, Robertson J, et al. Untangling taxonomy: a DNA barcode reference library for Canadian spiders. Mol Ecol Resour. 2016;16: 325–341. 10.1111/1755-0998.12444 [DOI] [PubMed] [Google Scholar]
- 62.Astrin JJ, Hofer H, Spelda J, Holstein J, Bayer S, Hendrich L, et al. Towards a DNA barcode reference database for spiders and harvestmen of Germany. PLOS ONE. 2016;11: e0162624 10.1371/journal.pone.0162624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gaikwad S, Warudkar A, Shouche Y. Efficacy of DNA barcoding for the species identification of spiders from Western Ghats of India. Mitochondrial DNA Part A. 2017;28: 638–644. [DOI] [PubMed] [Google Scholar]
- 64.Tahir HM, Naseem S, Akhtar S, Ashfaq M, Butt A, Mukhtar MK. DNA barcode record of some common spiders from Punjab, Pakistan. Pak J Zool. 2016;48(1): 159–164. [Google Scholar]
- 65.Naseem S, Tahir HM. Use of mitochondrial COI gene for the identification of family Salticidae and Lycosidae of spiders. Mitochondrial DNA. 2018;29: 96–101. 10.1080/24701394.2016.1248428 [DOI] [PubMed] [Google Scholar]
- 66.Majeed N, Butt A, Krammer H-J, Astrin JJ. DNA barcoding in jumping spider communities of Pakistan reveals a new species (Araneae: Salticidae). ZooKeys. (In Press). [Google Scholar]
- 67.Ivanova NV, deWaard JR, Hebert PDN. An inexpensive, automation-friendly protocol for recovering high quality DNA. Mol Ecol Notes. 2006;6: 998–1002. [Google Scholar]
- 68.Hebert PDN, Penton EH, Burns JM, Janzen DH, Hallwachs W. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc Natl Acad Sci USA. 2004;101: 14812–14817. 10.1073/pnas.0406166101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3: 294–299. [PubMed] [Google Scholar]
- 70.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28: 2731‒2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Meyer CP, Paulay G. DNA barcoding: Error rates based on comprehensive sampling. PLOS Biol. 2005;3: e422 10.1371/journal.pbio.0030422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Candek K, Kuntner M. DNA barcoding gap: reliable species identification over morphological and geographical scales. Mol Ecol Resour. 2015;15: 268–277. 10.1111/1755-0998.12304 [DOI] [PubMed] [Google Scholar]
- 73.Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27: 209–220. [PubMed] [Google Scholar]
- 74.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16: 111–120. [DOI] [PubMed] [Google Scholar]
- 75.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61: 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mukhtar MK, Khan SY, Jabeen S, Tahir HM, Qadir A, Ahmad KR, et al. A preliminary checklist of the spider fauna of Sargodha (Punjab), Pakistan. Pak J Zool. 2012;44: 1245–1254. [Google Scholar]
- 77.Ahmad S, Akhtar N, Saeed K. Some observations on spider fauna of district Buner, Khyber Pakhtunkhwa, Pakistan. J Entomol Zool Studies. 2015;3: 47–52. [Google Scholar]
- 78.Zhang J-X, Maddison WP. New euophryine jumping spiders from Papua New Guinea (Araneae: Salticidae: Euophryinae). Zootaxa. 2012;3491: 1–74. [Google Scholar]
- 79.Dippenaar-Schoeman AS, Haddad CR, Lyle R, Lotz LN, Foord SH, Jocque R, et al. South African National Survey of Arachnida: A checklist of the spiders (Arachnida, Araneae) of the Tswalu Kalahari Reserve in the Northern Cape province, South Africa. KOEDOE. 2018;60: a1486. [Google Scholar]
- 80.King JR, Porter SD. Evaluation of sampling methods and species richness estimators for ants in upland ecosystems in Florida. Environ Entomol. 2005;34: 1566–1578. [Google Scholar]
- 81.Skalak SL, Sherwin RE, Brigham RM. Sampling period, size and duration influence measures of bat species richness from acoustic surveys. Methods Ecol Evol. 2012;3: 490–502. [Google Scholar]
- 82.Zenker MM, Rougerie R, Teston JA, Laguerre M, Pie MR, Freitas AVL. Fast census of moth diversity in the neotropics: A comparison of field-assigned morphospecies and DNA barcoding in tiger moths. PLOS ONE. 2016;11: e0148423 10.1371/journal.pone.0148423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hawlitschek O, Moriniere J, Lehmann GUC, Lehmann AW, Kropf M, Dunz A, et al. DNA barcoding of crickets, katydids and grasshoppers (Orthoptera) from Central Europe with focus on Austria, Germany and Switzerland. Mol Ecol Resour. 2017;17(5):1037–1053. 10.1111/1755-0998.12638 [DOI] [PubMed] [Google Scholar]
- 84.Havemann N, Gossner MM, Hendrich L, Moriniere J, Niedringhaus R, Schafer P, et al. From water striders to water bugs: the molecular diversity of aquatic Heteroptera (Gerromorpha, Nepomorpha) of Germany based on DNA barcodes. PeerJ. 2018;6: e4577 10.7717/peerj.4577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhou Z, Guo H, Han L, Chai J, Che X, Shi F. Singleton molecular species delimitation based on COI-5P barcode sequences revealed high cryptic/undescribed diversity for Chinese katydids (Orthoptera: Tettigoniidae). BMC Evol Biol. 2019;19: 79 10.1186/s12862-019-1404-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zahiri R, Lafontaine JD, Schmidt BC, DeWaard JR, Zakharov EV, Hebert PDN. A transcontinental challenge–a test of DNA barcode performance for 1,541 species of Canadian Noctuoidea (Lepidoptera). PLOS ONE. 2014;9: e92797 10.1371/journal.pone.0092797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pentinsaari M, Hebert PDN, Mutanen M. Barcoding beetles: a regional survey of 1872 species reveals high identification success and unusually deep interspecific divergences. PLOS ONE. 2014;9: e108651 10.1371/journal.pone.0108651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ren J-M, Ashfaq M, Hu X-N, Ma J, Liang F, Hebert PDN, et al. Barcode index numbers expedite quarantine inspections and aid the interception of nonindigenous mealybugs (Pseudococcidae). Biol Invasions. 2018;20: 449–460. [Google Scholar]
- 89.Landry J-F, Hebert PDN. Plutella australiana (Lepidoptera, Plutellidae), an overlooked diamondback moth revealed by DNA barcodes. Zookeys. 2013;327: 43–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gottsberger B, Mayer F. Behavioral sterility of hybrid males in acoustically communicating grasshoppers (Acrididae, Gomphocerinae). J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2007;193: 703–714. 10.1007/s00359-007-0225-y [DOI] [PubMed] [Google Scholar]
- 91.Hawlitschek O, Hendrich L, Espeland M, Toussaint EFA, Genner MJ, Balke M. Pleistocene climate change promoted rapid diversification of aquatic invertebrates in Southeast Australia. BMC Evolutionary Biology. 2012;12:142 10.1186/1471-2148-12-142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Papadopoulou A, Bergsten J, Fujisawa T, Monaghan MT, Barraclough TG, Vogler AP. Speciation and DNA barcodes: testing the effects of dispersal on the formation of discrete sequence clusters. Philos Trans R Soc Lond B Biol Sci. 2008;363: 2987–2996. 10.1098/rstb.2008.0066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Huemer P, Hebert PDN, Mutanen M, Wieser C, Wiesmair B, Hausmann A, et al. Large geographic distance versus small DNA barcode divergence: Insights from a comparison of European to South Siberian Lepidoptera. PLOS ONE. 2018;13: e0206668 10.1371/journal.pone.0206668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Huemer P, Mutanen M, Sefc KM, Hebert PDN. Testing DNA barcode performance in 1000 species of European Lepidoptera: large geographic distances have small genetic impacts. PLOS ONE. 2014;9: e115774 10.1371/journal.pone.0115774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lukhtanov VA, Sourakov A, Zakharov EV, Hebert PDN. DNA barcoding Central Asian butterflies: increasing geographical dimension does not significantly reduce the success of species identification. Mol Ecol Resour. 2009;9: 1302–1310. 10.1111/j.1755-0998.2009.02577.x [DOI] [PubMed] [Google Scholar]
- 96.Bergsten J, Bilton DT, Fujisawa T, Elliott M, Monaghan MT, Balke M, et al. The effect of geographical scale of sampling on DNA barcoding. Syst Biol. 2012;61: 851–869. 10.1093/sysbio/sys037 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Data Availability Statement
Collection data, a photograph, a taxonomic assignment, and DNA barcode (COI-5p) sequence for each specimen are available in the public dataset, "DS-MASPD DNA barcoding spiders of Pakistan" on the Barcode of Life Data System (BOLD) (www.boldsystems.org). (dx.doi.org/10.5883/DS-MASPD).