This colorectal tumor-on-chip model is an original and reliable platform for high-throughput drug screening and genetic expression.
Abstract
Awareness that traditional two-dimensional (2D) in vitro and nonrepresentative animal models may not completely emulate the 3D hierarchical complexity of tissues and organs is on the rise. Therefore, posterior translation into successful clinical application is compromised. To address this dearth, on-chip biomimetic microenvironments powered by microfluidic technologies are being developed to better capture the complexity of in vivo pathophysiology. Here, we describe a “tumor-on-a-chip” model for assessment of precision nanomedicine delivery on which we validate the efficacy of drug-loaded nanoparticles in a gradient fashion. The model validation was performed by viability studies integrated with live imaging to confirm the dose-response effect of cells exposed to the CMCht/PAMAM nanoparticle gradient. This platform also enables the analysis at the gene expression level, where a down-regulation of all the studied genes (MMP-1, Caspase-3, and Ki-67) was observed. This tumor-on-chip model represents an important development in the use of precision nanomedicine toward personalized treatment.
INTRODUCTION
Colorectal cancer (CRC) is now the third leading cause of cancer-related death in women and the second leading cause in men, mainly as a result of high incidence of cancer metastasis and chemotherapeutic treatment inefficiency (1). Concerning cancer research, currently available two-dimensional (2D) culture systems and animal models used to study the efficacy of chemotherapy do not reproduce the tumor physiology in the human body. As a deleterious consequence, more than 80% of drug candidates of all cancer drugs fail during phase 2 and phase 3 clinical trials (2).
The need for a more reliable and predictable screening approach to better assess drug responses in preclinical studies in vitro has led to the development of novel methodologies for cell culture, using, e.g., microfluidics (2, 3). Over the past decade, the expansion in the field of microfluidics has helped to establish a new set of standards in the study of basic biological phenomena, especially helpful in the study of pathogenesis and drug development (4). Microfluidics is by definition the science and technology of systems that process and manipulate small (10−9 to 10−18 liters) amounts of fluids by using channels with dimensions from tens to hundreds of micrometers (5). Microfluidic technology offers new opportunities for cell-based sensors and multifunctional platforms for biochemical and biomedical functions under physiologically relevant conditions (6, 7).
Microfluidic technologies enable precise control over small fluid/liquid volumes and compartmentalization and combinatorial factors/materials in a single device, allowing cells to coexist in a 3D complex microenvironment. This kind of platform has proven useful in unraveling key cellular behavioral aspects of cancer metastases and enabling precise and representative drug screening (6, 8). In addition, microfluidics has also been successful in the generation of vasculature-like structures, with special emphasis on the vital association between vasculature/angiogenesis and cancer (3, 9–11). These are useful not only for revealing the mechanisms in the development of vasculatures but also as experimental platforms for vascular disease models and drug screening. Another particular feature of microfluidics, which will be used in this study, is the ability to generate well-controlled gradients of molecules in solution. Gradients of molecules can be generated by advection, being the mass transport by the fluids bulk motion (flow-based gradient generation), or by diffusion, being the spreading of unequally distributed molecules by biased random walk induced by Brownian motion (diffusion-based gradient generation) (12). The importance of gradient generation lies in the fact that many cell processes work by recognizing the directional information present in gradients, including processes such as chemotaxis and angiogenesis and diseases such as cancer (13). In the in vivo scenario, chemical microenvironment includes nutrients and oxygen, delivered to cells and tissues through a network of vasculature that creates important biomolecular gradients within the tissue. The successful development of 3D microengineered gradients set the stage for physiologically relevant in vitro models, comprising, for instance, oxygen, chemokines, and drug gradients (6, 12, 14). Another important aspect of our model is the microvasculature. It defines the biological and physical characteristics of the microenvironment within tissues and plays a role in the initiation and progression of many pathologies, including cancer (15). Microfluidics has also been used in the generation of vasculature-like structures, with special emphasis on the association between vasculature/angiogenesis and cancer (3, 9–11). In the case of CRC, endothelial cells lining the microvasculature are known to play a critical “gatekeeper” role, which is why we developed our microvasculature model using human colonic microvascular endothelial cells (HCoMECs) (16).
Regarding CRC treatment, gemcitabine (2′,2′-difluoro-2′-deoxycytidine) (from now on designated as GEM) is now being tested as treatment for patients with advanced CRC (17, 18). However, because of its low molecular weight and high solubility in water, the clinical benefits of GEM are hampered by its short plasma half-life and relatively low concentration around tumor sites (18, 19). Therefore, a search for novel therapeutic strategies is imperative, such as the use of nanoparticles (19).
Our goal was to develop a complex 3D microfluidic chip-based in vitro model that emulates the human colorectal tumor microenvironment. This model enables the reconstitution of physiological functions of microvascular tissue, which is instrumental to assess the efficiency of delivery of anticancer drug–loaded nanoparticles through a dynamic controllable gradient in a core compartment of the microfluidic chip. This gradient is generated and maintained by perfused microvasculature-mimicking side channels. Together, these results demonstrate that this 3D platform is suitable for efficacy/toxicity screening in a more physiological environment than previously possible.
RESULTS
Microfluidic chip fabrication and characterization
The aim of this work was to develop an in-house customized 3D microfluidic model that emulates the human colorectal tumor microenvironment and enables the reconstitution of physiological functions of microvascular tissue. The model was designed to comprise features that permitted the generation of soluble compounds’ gradient for the assessment of the delivery efficiency of anticancer drug–loaded nanoparticles. Moreover, ease of use possibility of realtime imaging, and performance of immunocytochemistry and cell retrieval after the experiments for polymerase chain reaction (PCR) analysis were requirements of this platform. In addition to having to meet certain conditions in terms of operation and design, the choice of material for fabrication, the geometry, dimensions of the culture chamber, and fluid flow control are some important considerations when designing the device. The developed microfluidic chip is made using poly(dimethylsiloxane) (PDMS). The 3D map of the final design is created by laser-reading the wafer surface at 50× magnification, as can be observed in Fig. 1A. Our microfluidic device has three compartments: a circular central chamber for the extracellular matrix (ECM)–like hydrogel (Matrigel), 5 mm in diameter and 126 μm in depth, with a separate inlet and outlet. The central chamber is laterally sided by a pair of perfused channels, 100 μm in width and 126 μm in depth, that are not interconnected between them (Fig. 1B).
Fig. 1. Design and characterization of the microfluidic chip.
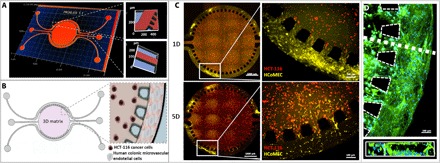
(A) Profilometer characterization: 3D map of the microfluidic chip. Representative image of quality control and characterization of the microfluidic chip features. The central chamber is 5 mm in diameter and 126 μm in depth, with a separate inlet and outlet for hydrogel injection. The two lateral channels, each 100 μm in width and 126 μm in depth, are not interconnected, opening up the possibility of perfusing two distinct solutions. (B) Schematic of chip design and zoom-in image of the concept of colorectal tumor-on-a-chip model: Round microfluidic central chamber in which HCT-116 cancer cells are embedded in Matrigel; HCoMECs are seeded in the side channels to form a 3D vessel-like assembly. (C) Establishment of a microvascular 3D microenvironment of colorectal tumor-on-a-chip: HCT-116 CRC cells embedded in Matrigel (in central chamber, stained in red) and HCoMECs (in lateral microchannels, stained in yellow) at day 1 (1D) and day 5 (5D) of culture. Endothelial cells start to invade the central chamber filled with Matrigel in response to VEGF presence. (D) Fluorescence microscopy image of microfluidic lateral channel mimicking prevascularization with HCoMECs after 5 days of culture [4′,6-diamidino-2-phenylindole (DAPI) blue, nuclei; phalloidin green, F-actin filaments]. Representative fluorescence microscopy close-up image of microchannel cross section showing endothelial cells aligning and creating an endothelialized lumen within the microchannel.
To evaluate the maintenance of the coculture over the course of 5 days, we labeled Matrigel-supported CRC cells with CellTrace Red (red) and HCoMECs with CellTrace Yellow (yellow), as shown in Fig. 1C. The endothelial cells were cultured in a 3D vessel-like fashion, attaching to the walls of the lateral channels of the microfluidic chip. As expected, these cells respond to a signal of vascular endothelial growth factor (VEGF) that has been loaded in the Matrigel, by migrating from the lateral channels toward the hydrogel through the interspacings between the pillars that separate the two compartments. The “microvasculature” component was characterized by obtaining a 3D representation (through actin/nuclei fluorescent stain of HCoMECs) to observe the morphology of the colonic endothelial cells lining the lateral channel structure after 5 days under perfusion of media (Fig. 1D).
Endothelial invasion
We examined the 3D formation of endothelial sprouts in the microfluidic device and the changes in cellular organization during early stages of invasion. This invasion is oriented directly toward the source of VEGF in the Matrigel despite the fact that cell migration from the endothelium is not physically constrained in any direction by the system design (Fig. 2A). Sprouting quantification shows that HCoMECs could progress up to 500 μm after 5 days. The leading tip cell is replete with filopodia-like protrusions, morphologically recapitulating in vivo endothelial cell sprouting. As the sprouts continued to invade and extend throughout the 3D matrix, they become longer, contain progressively more cells, and begin to form interconnected branches, as it occurs in early stages of tumor development (Fig. 2B, zoom in at day 3) (20). Although we are not mimicking the angiogenesis phenomenon per se, in this model, we observe that endothelial cells invade the hydrogel core but do not form anastomosis.
Fig. 2. Endothelial cell invasion.
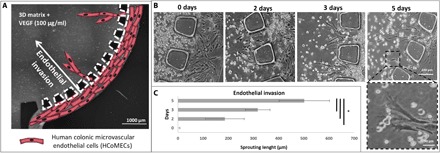
(A) Schematic representation of HCoMECs in the lateral channels invading the central chamber in response to the presence of VEGF mixed in the Matrigel. (B) Formation of endothelial sprouts in the microfluidic device: Representative bright-field images of the chip taken at determined time points were analyzed with ImageJ. (C) Quantification of endothelial invasion. Data are presented as means ± SD (n = 9). The asterisk (*) denotes statistical differences (P < 0.01).
Validation of the integrated microfluidic gradient chip device
Ultimately, in this work, we aim to establish a 3D cancer microfluidic cell culture for studying onco-nanomedicine efficacy. Through its architecture, the platform offers the possibility to observe the effect of several concentrations of GEM-loaded CMCht/PAMAM dendrimer nanoparticles as they penetrate the HCT-116–laden hydrogel in a gradient fashion. As a result, from one experiment, a real-time response of the cancer cells in a gradient-like fashion is obtained, as occurs in a physiological concentric tumor microenvironment. For this purpose, the formation of the gradient was validated using fluorescently labeled CMCht/PAMAM dendrimer nanoparticles before the GEM release. Note that these nanoparticles present remarkable physicochemical and biological properties such as solubility in water, biocompatibility, and ability to be easily surface modified. Therefore, they have great potential to be used in fundamental cell biology studies and for intracellular drug delivery (21).
The CMCht/PAMAM dendrimer nanoparticles were synthetized and characterized previously, showing an average size of 50 nm (22, 23) (Fig. 3A). As depicted in Fig. 3B, after 60 min, a gradient of fluorescence was established in the cell culture chamber and maintained for at least 12 hours of continuous lateral perfusion, exhibiting a quasi-linear fluorescent profile (from 100% at the source to 0% at the sink) (Fig. 3C). A 3D projection of fluorescein isothiocyanate (FITC)–labeled nanoparticles’ gradient according to surface fluorescence intensity (Fig. 3D) also confirms these findings.
Fig. 3. Schematic representation of fluorescent nanoparticle gradient formation.
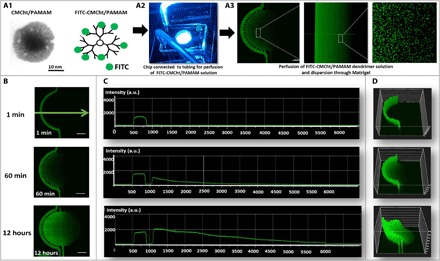
(A) Transmission electron microscopy image of individual CMCht/PAMAM dendrimer nanoparticle, representative image of chip connected to tubing perfusing the nanoparticle solution, and zoom-in confocal microscopy image confirming FITC-labeled CMCht/PAMAM dendrimer nanoparticles’ dispersion in Matrigel. (B) Panel of fluorescence microscopy images of FITC-labeled nanoparticle gradient up to 12 hours. (C) Quantification of fluorescence: Intensity [in arbitrary units (a.u.)] versus distance (in micrometers) across the chip up to 12 hours [measurements according to the direction of the green arrow indicated in (B)]. (D) 3D projection of FITC-labeled nanoparticle gradient according to fluorescence intensity.
Drug screening validation
We aimed to establish an image-based method to investigate the concentration-dependent response of cells inside the microfluidic device by creating a gradient of concentrations of GEM. To investigate the concentration-dependent toxicity on cancer cells inside the microfluidic chip, we examined the survival response of cells of the CRC cell line HCT-116 embedded within Matrigel. The cells were allowed to adhere and develop for 12 hours before drug exposure. The exposure of the cells to the delivery-induced gradient of GEM was generated from a maximum concentration of dendrimer nanoparticles (0.5 mg/ml) sourced from one microvasculature lumen perfused with them and diffused through the hydrogel core toward the opposite side (Fig 3).
GEM-loaded nanoparticles were studied in terms of encapsulation efficiency and drug-loading efficiency. The encapsulation efficiency of GEM-CMCht/PAMAM was 52.99 ± 3.5%, and the drug-loading efficiency was 9.42 ± 3.5%. We recorded the in vitro release profiles of GEM-loaded CMCht/PAMAM in a cumulative release curve during 5 days. As shown in Fig. 4, after 6 hours, around 40% of the drug was released, which shows that GEM-loaded CMCht/PAMAM can retain their stability in solution. Thereafter, the nanoparticles showed an accelerated release, where more than half of the GEM was released by 24 hours (Fig. 4A).
Fig. 4. GEM drug effect on colorectal tumor model.
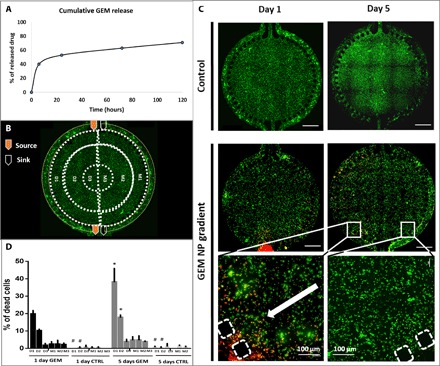
(A) Release profile of GEM: Cumulative release (in hours) of GEM at pH 7.4 in phosphate-buffered saline (PBS), at 37°C and stirred at 60 rpm, determined by ultraviolet (UV) spectrophotometer set to 275 nm. The results are expressed as means ± SD (n = 6). (B) Schematic representation of experimental setup [perfusion of culture medium through one inlet and one outlet and of culture medium supplemented with GEM-loaded dendrimer nanoparticles (0.5 mg/ml) through the other inlet and outlet]; the areas defined for cell death quantification are delimited by dashed lines, and each ring is assigned D1 to D3 and M1 to M3. (C) Representative images of fluorescence microscopy images of live/dead assay performed on microfluidic chip at days 1 and 5. (D) Quantification of cell death, represented as percentage, based on the microscopy data. Data are presented as means ± SD (n = 3). The asterisk (*) denotes statistical differences (P < 0.05). Scale bars, 1000 μm. NP, nanoparticle; CTRL, control.
In addition, an important event to notice is the immediate internalization of the nanoparticles after contact with cells, as shown in our previous study (22), decreasing the chance of drug degradation. This intracellular release mechanism represents a key factor of the importance of GEM-loaded nanoparticles as an antitumoral agent (24).
Live experiments were similarly performed using the same type and concentration profiles for the sake of reproducibility and final measurement correlation. To quantify cell response to GEM, we segmented the circular area of the chip (core) into six concentric areas, as represented in Fig. 4B. Thereby, the following nomenclature was used: We assigned M1, M2, and M3 to the areas of the chip subjected to culture medium (Fig. 4B, left side) and D1, D2, and D3 to the areas of the chip subjected to drug (Fig. 4B, right side).
After 1 and 5 days of exposure to GEM, we evaluated the viability of CRC cells in the 3D matrix under perfusion at side channels. The controls, at both time points (Fig. 4, C and D), indicate insignificant levels of cell death, demonstrating the ability of the microfluidic model support a stable and viable coculture of HCoMECs and HCT-116 CRC cells in contiguous compartments under perfusion. Cell viability in controls shows that oxygen can freely permeate to the cells either through PDMS or the hydrogel matrix while being fed for nutrients dynamically by perfusion of side channels (Fig. 4B). On the other hand, in the presence of GEM-loaded CMCht/PAMAM dendrimer nanoparticles, cell death occurs in a gradient manner, with a stepwise decay in toxicity, i.e., less linear than the concentration release profile of GEM previously quantified. This indicates that GEM could be efficiently released from the nanoparticles and taken up by the cancer cells while remaining an active anticancer drug, in its soluble form (Fig. 4C). The cell death induced by the anticancer drug was more efficient within the first 150 μm of the cell culture, which might be due to an eventual threshold of drug efficacy on cells. This concentration effect has been consistent over time, with the cell death values increasing from day 1 to day 5 (Fig 4C).
Gene expression analysis and immunocytochemistry on a chip
To the best of our knowledge, very few organ-on-chip models allow for gene expression analysis of the cells cultured therein because of the small mRNA content accessible from the chip’s culture chambers. On a subset of corresponding experiments, exposure to GEM-loaded CMCht/PAMAM dendrimer nanoparticles was tested as single-concentration exposure per device (GEM-loaded CMCht/PAMAM perfused through both inlets), not as a gradient, to demonstrate the potential of the platform for evaluating gene expression as function of dose response. To this end, we tested two representative conditions of previous experiments, i.e., 0 and 0.5 mg/ml (on continuously perfused media), as proof of concept for the suitability of the model for reverse transcription PCR (RT-PCR) and gene studying. Our control group consists of 0 mg/ml. At each time point (days 1 and 5), we extracted cells from the device and RNA from cells, after hydrogel removal (Fig. 5A), for analysis of mRNA expression of Caspase-3 (Casp-3), Ki-67, and MMP-1 (matrix metalloproteinase 1) by real-time PCR. These markers were chosen to allow the evaluation of the effect of cytotoxic compounds: MMP-1 because of its relationship with the metastatic phenotype (25), Casp-3 because of its involvement in apoptosis, and Ki-67 as a universal marker for proliferation.
Fig. 5. Gene expression analysis and immunocytochemistry on a chip.
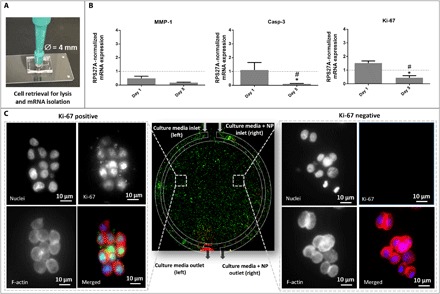
(A) Representation of hydrogel retrieval containing HCT-116 cells from the chip for gene expression analysis. (B) Gene expression of Ki-67, Casp-3, and MMP-1, in the absence (control) and presence of GEM, at days 1 and 5. The values for controls are represented by the dashed line. The asterisk (*) indicates significant differences when comparing the same condition at two different time points. The hash key (#) indicates significant differences when comparing to controls at the same time point. Data represent mean values of three independent experiments ± SD. RPS27A, ribosomal protein S27A. (C) Immunohistochemistry: In the center, a representative image of the entire chip is depicted. The lateral images represent the zoom in at the cell level showing the immunocytochemistry Ki-67 staining after 5 days in culture [panels showing DAPI nuclei staining (blue), Ki-67 staining (green), F-actin staining (red), and merged image in colors].
In general, we observed a down-regulation for all genes when comparing day 1 to day 5, as shown in Fig. 5. Compared to the controls, MMP-1 expression is down-regulated 0.5- and 0.2-fold for days 1 and 5, respectively. Similarly, for Casp-3, at day 1, no statistical differences between the control and GEM-treated cells were observed. For Ki-67 expression, no significant effect of the exposure to GEM was observed at day 1, which can be due to the limited time for the compound to act on the cells and affect their proliferation. However, at day 5, we observed a statistically significant effect of GEM, with a 0.4-fold decrease in the expression of Ki-67 compared to the control. These results demonstrate the suitability of our platform for assessing gene expression.
Concerning Ki-67 immunofluorescence on the microfluidic chip shown in Fig. 5C, the marker is expressed only when the nanoparticles are absent, which confirms the PCR data. These qualitative results translate the presence of Ki-67–positive cells, corresponding to proliferating tumor cells in the area of the chip more subjected to drug-free media, in contrast to Ki-67–negative cells present in the matrix regions close to the perfusion source of GEM-loaded CMCht/PAMAM dendrimer nanoparticle media.
DISCUSSION
The design of our new microfluidic chip resembles the previously shown ECM-containing devices, where endothelial monolayers are cultured onto the sidewall of an ECM material that separates two microfluidic channels (26). In our model, the “tumor” core defined by the hydrogel is supported by an adjacent “microvascular” network fed by external syringe pumps, supporting physiological events specific of each cell type. The lumen-like channels deliver nutrients to surrounding tissues sufficient to support robust growth. This opens up the possibility of perfusing two distinct solutions in each side of the central chamber containing Matrigel and the cancer cells encapsulated therein, and therefore, the ability to produce and maintain stable gradients. Around the inner part of the chamber, we designed micropillars to help retain Matrigel when injected into the chamber, creating enough superficial tension and avoiding leakage toward the lateral channels (Fig. 1C). Furthermore, the liquid continuity between the side channels and the culture chamber was ensured by these small pillars, which act as resistors, preventing the shear stress to be transmitted to the cell culture chamber (27). The chip is made using PDMS because of its several unique properties, which make it a suitable choice for microfluidic application: PDMS is inexpensive, easy to handle, gas permeable (assuring the supply of oxygen to the cells), and transparent—properties needed for imaging (28). Corroborating this, our results show the CRC cells in red and endothelial cells in yellow proliferating, elongating in the direction of the flow, and covering the lateral channels, consequently forming a vessel-like structure at day 5. We observed an endothelial 3D macrostructure invading the central chamber through the pillars, in response to the VEGF, defining an early stage of sprouting (Fig. 1, C and D).
When characterizing the microfluidic coculture, the sprouting phenomena caught our attention. Angiogenesis is a complex morphogenetic process where endothelial cells from preexisting vessels invade the matrix as multicellular sprouts to form new vessels (29). For capillary sprouting and blood vessel formation, both proliferation and migration are important, but how these two events are coordinated during the extension process is not fully clear (30). Proangiogenic signals, such as VEGF, promote processes such as proliferation, endothelial motility, and filopodia extension (31). Evidence from preclinical and clinical studies indicates that VEGF is the predominant angiogenic factor in human colon cancer and is associated with formation of metastases and poor prognosis (32). So far, innumerous models tried to recreate these phenomena (30, 33, 34). For instance, the tube formation assay involves the reorganization of endothelial cells seeded on Matrigel surface to form cords similar to vascular networks. Still, they lack important features observed in native angiogenesis, including directional invasion of cells into a 3D ECM, proper polarization of the luminal, lumen formation, and support of fluid flow (35). Although it has been stated before that we are not trying to recreate an anastomosis in the central chamber, the fact that we cannot observe it can be explained by several factors, for example, by the fact that the central chamber of the device is too large to observe these events. Nevertheless, from a technical perspective, this feature allows higher volumetric hosting of HCT-116 cells, which enables quantitative analysis of gene expression (i.e., enough cells for PCR analysis). Another factor is probably the lack of support cells, such as pericytes. This cell type would help to stabilize vessel formation and therefore permit the bridging of sprouts over longer distances of cell culture regions (35). As our goal was uniquely to use the side channels to create a perfusable lateral channel with a lumen, we seeded the endothelial cells in the lateral channel and not in the central chamber. However, it is interesting to see the natural behavior of the endothelial cells invading the Matrigel-filled chamber with the cancer cells and further characterize our model. As conclusion, this biomimetic model is able to reconstitute the beginning of the angiogenic sprouting process.
To generate a stable gradient, one of the main goals of this project, we chose the CMCht/PAMAM dendrimer nanoparticles. Their proven ability to act as drug release vehicles inside the cells, as well as cell trackers [when surface modified with a fluorescence molecule such as FITC (22, 23)], made them excellent candidates. One of the initial concerns was the ability of these nanoparticles to perfuse through Matrigel’s fiber network. As observed in Fig. 3A, the nanoparticles disperse widely when perfused in the lateral channels in a gradient fashion. The real-time observation and establishment of a fluorescent gradient generated by the perfusion of the FITC-labeled nanoparticles were essential to move on to drug screening assays, using the same flow and the same nanoparticles, this time releasing the drug over time.
Regarding the GEM release from the dendrimer nanoparticles in the microfluidic model, we chose a concentration of 0.5 mg/ml with the rationale to (i) avoid the cytotoxic effect of the nanoparticles and (ii) enable continuous and sustained intracellular drug delivery by the nanoparticles and therewith a high efficacy, as was demonstrated by the viability results (Fig. 4) and the quantitative PCR data (Fig. 5). In addition, nanoparticles are highly advantageous when comparing to the gold standard clinical delivery of one bulk delivery (every 2 weeks), more prone to result in secondary undesirable effects.
Given that the gradient we observed was more or less concentrically generated from the periphery toward the center of the chamber, the areas defined for quantification followed this geometry. Apparently, lower amounts of nanoparticles leading to drug release in the areas further away from the source are not sufficient to induce cell death. Higher drug efficacy at D1 and D2 regions seems to define a cutoff area on the action range of the nanomedicines in the matrix delineated by the Matrigel cell culture. Another important aspect to consider when analyzing the data is that, although we were not quantifying cell death among endothelial cells, it is visible that both the cancer cells and the vasculature are sensitive to standard-of-care chemotherapeutic GEM. Targeting of the nanoparticles toward CRC cells and minimizing deleterious effects on endothelial cells would be very useful and constitute the next step of this work. We observed no significant differences between the controls at both days or between the controls and the areas subjected to culture medium M1, M2, and M3. Even after 5 days of exposure to the maximum concentration of drug possible in this model (area D1), only about 40% of cell death was observed. This low cell death level is in agreement with previous studies, corroborating the fact that 3D cultures usually show a reduced sensitivity to anticancer drugs (36). This can be due to various factors, including matrix stiffness, which was reported to be closely related to tumor chemoresistance (6), decreased penetration of anticancer drugs, increased prosurvival signaling, and/or up-regulation of genes conferring drug resistance (7).
Moreover, cells could be in the process of dying at the analysis time but not yet dead and therefore not quantified in this screening, since the ethidium homodimer only stains dead cells. This microfluidic chip also allows the use of other stainings.
Moreover, the need to use hydrogels such as Matrigel to replicate the intricate 3D microenvironment in which cells live makes it harder to retrieve them for genetic analysis under good conditions. This analysis works as a proof of concept to demonstrate the versatility of this platform to both work as a drug screening platform and allow for genetic analysis. One of the main challenges of this task was to effectively retrieve HCT-116 cells from the Matrigel inside the 5-mm central chamber of the microfluidic device. The strategy we developed involves punching out the central chamber with a sterile surgical 5-mm punch and dissolving the Matrigel with encapsulated cancer cells in recovery solution. Results show a down-regulation of all genes tested. MMP-1 is a collagenase that degrades ECM, specifically targeting types I, II, and III collagens, the major components of the interstitial stroma (37). It has been suggested to be associated with an advanced stage of colon cancer with poor prognosis and has emerged as a new oncogenesis and metastasis target for treatment of diverse cancers (38). However, molecular factors that contribute directly to tumor cell vascular penetration and the extent of MMP-1 activity in the presence of chemotherapeutic drugs have not been fully identified (39, 40). For example, Khanna et al. (41) used small interfering RNA against proto-oncoprotein Ets-1 in GEM-resistant cells and demonstrated a reversal in GEM chemosensitivity, translated in a marked reduction in the expression of MMP-1 (41). Given this evidence, it makes sense that exposing HCT-116 cells to an anticancer drug would lead to the down-regulation of the expression of MMP-1, as observed, however, with no statistical differences. Regarding Casp-3 results, the down-regulation obtained over the two time points corroborates the previous live/dead assay results, where at day 1, the extent of cell death due to GEM exposure is less pronounced than that at day 5 (although with no statistical differences when compared to the respective control). Nevertheless, at the later time point, the expression of Casp-3 is down-regulated, although we see a clear increase in cell death in the live/dead experiments performed before. Recently, caspase activity identified in the absence of cell death has sparked strong interest in caspase functions in nonapoptotic cellular responses, suggesting that caspases can be activated without inducing or even before apoptosis occurs (42). Several pathways can be related to GEM-induced apoptosis. On one hand, literature evidence exists for caspase-dependent apoptosis (43), while other studies describe GEM-induced apoptosis via reactive oxygen species generation, without an increase in Casp-3 (44). In addition, GEM-induced apoptosis is also related to ERK (extracellular signal–regulated kinase), Akt, Bcl-2, and p38 MAPK (mitogen-activated protein kinase) pathways and not only to Casp-3 (45).
When looking at Ki-67 expression, a significant difference in cells exposed to the drug from day 1 to day 5 reveals that the drug decreased cell proliferation. Its increased expression in human cancer specimens generally denotes an aggressive phenotype, although new studies suggest that this process is more complex than initially thought (46). However, the fact that many aggressive cancers have high expression of Ki-67 relative to normal and quiescent cells suggests that Ki-67 might be an attractive target for cancer therapy if its role as a driver of carcinogenesis were validated.
The developed tumor-on-chip platform comprising a human CRC-like core and surrounding vascularized microtissue is a promising tool to be used in onco-medicine, either for high-content image-based screenings or gene expression analysis to study drug-dose responses. On the basis of a robust dynamic microfluidic system with a viable coculture of HCT-116 CRC cells and HCoMECS, this platform was validated as a tumor-on-chip in vitro platform suitable for assessing an anticancer nanoparticle-based deliverable.
Although very useful as a high-throughput system for drug testing, our model does not comprise an oxygen gradient, common to the in vivo cancer microenvironment. Generating such a gradient would require inclusion of another complex feature in this already complex model and will be considered as a part of future work. In this study, we used the in-house–synthetized dendrimer nanoparticles as a model for nanomaterial-based anticancer therapeutics. The fluorescently modified nanoparticles successfully acted as a tracking tool that allowed the real-time imaging of the nanoparticle location, whereas drug-loaded particles functioned as an effective delivery vehicle of GEM in a gradient fashion. The resulting datasets enabled the definition of precise ranges of action of these anticancer nanoparticles, taking into account the complexity of the tumor microenvironment, including the critical microvasculature component. Highlighting the importance of the high range of concentrations in a single experiment in this model, which has an impact on its robustness as a validation tool, this tumor-on-chip device enabled both the performance of RT-PCR and immunocytochemistry stainings as the drug was being delivered to the tumor core. Technically, we proved that cells can easily be extracted from the chip and studied in terms of gene expression, providing valuable information on the mechanisms underlying the drug effect. Through this 3D microfluidic cell culture, the study of phenomena such as vascularization and oncogenesis under dynamic conditions may provide a powerful insight on the cancer stage prognosis and eventually on depicting a treatment option. These capabilities are of great value, especially if using patient cells, and thus, for precision therapy screenings on personalized medicine.
Most of all, the greatest advantages of our platform compared to others of the same type (central chamber for 3D matrix laterally sided by perfusable channels) are (i) facile molecular cell analysis; (ii) physiologically inspired design of radial drug penetration into solid tumors; (iii) hybrid coculture system to show efficiency of drug penetration through a microvascular network into a cancer-mimicking tissue, composed of a supporting relevant matrix with CRC cells; (iv) real-time imaging of drug delivery and its effect on cell viability; (v) design compatible with assessing physiologically relevant cell migration and thus metastatic events (not the scope of the current manuscript); and (vi) system easily combined with organoid seeding of the central chamber.
This platform has the potential to further study the EPR effect by controlling the permeability of the vascular network, e.g., by readjusting the distance between pillars. Moreover, tumor cell stress can be studied by, e.g., varying the flow rates and, thus, exposing the cells to shear stresses. In addition, while tumor cell signaling was not studied here, this could be done by analyzing the medium collected in the outlet for, e.g., soluble factor profiling. The potential for further application is vast, from testing various types and sizes of particles and patient-derived cells to bringing this tool closer to a clinical and pharmaceutical translatable setting in further research. Together, this platform can potentially lead to breakthrough discoveries not only in basic research but also related to clinical applications, namely in the CRC field.
MATERIALS AND METHODS
Microfluidic chip design and fabrication
The microfluidic device composed of three main compartments was designed using CAD (computer-assisted design) on a photolithography mask, which consisted of a central chamber for an ECM-like hydrogel (diameter, 5 mm) and a pair of perfusable channels flanking the center chamber (connected to the chamber with a range of pillars that allow the generation of stable gradients by diffusion).
The microfluidic device in PDMS (Dow Corning) elastomer was fabricated by replica molding from an SU-8 (MicroChem)/silicon master based on a ratio of 10:1 to curing agent. The master mold was produced by ultraviolet (UV) lithography. After pouring PDMS on a mold, the mixture was degassed in a vacuum chamber for air bubble removal and cured at 80°C for 1 hour in the oven. The devices were then cut out by a razor blade; the fluidic connection ports were punched, and bonding to a glass slide was done after oxygen plasma of both surfaces and conformal contact (Harrick Plasma cleaner/sterilizer, 4 mbar, 120 s). Assembly quality control and characterization were done by optical 3D profilometry (VK-X250, KEYENCE, Germany).
Characterization of the microfluidic device
This microfluidic device functions as a multiphase microbioreactor, which was built on the basis of three major compartments: a central chamber seeded with cells embedded in a hydrogel matrix (Matrigel used as model gel), being sided by a pair of channels perfused with culture medium. The central chamber and the lateral channels are separated by small pillars to allow for hydrogel loading of the chamber retained before the channels. Quality check was done by optical microscopy.
Cell culture
HCT-116 cells (human colon cancer cell line) were originally obtained from the American Collection of Cell Cultures, USA. Cells were cultured in Dulbecco’s modified eagle medium (Gibco, Invitrogen) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin and streptomycin (pen/strep) solution under standard conditions (37°C in a humidified atmosphere, 5% CO2). HCoMECs were obtained from Innoprot and cultured in endothelial cell medium supplemented with 5% FBS, 1% of endothelial cell growth supplement, and 1% of pen/strep solution. The medium was replaced every 3 days. Subculture of cells was performed before 90% of confluence using 0.25% trypsin/EDTA (Gibco) for 5 min at 37°C.
Cell seeding on the chip
HCT-116 cancer cells were resuspended in Matrigel supplemented with VEGF (100 ng/ml; R&D Systems) in a density of 10 × 106 cells/ml, injected in the central chamber of the microfluidic device using a syringe, and let to incubate for 15 min at 37°C for cross-linking. After this, the lateral channels were coated using a 10% Matrigel solution and incubated for 30 min at 37°C. HCoMECs in the same density (10 × 106 cells/ml) were seeded on the lateral channels using a pipette and left to adhere for 6 hours. Then, the cells were exposed to a perfusion flow regime at a flow rate of 8 μl/hour for the duration of the experiment. At days 1 and 5, the cell culture was evaluated using fluorescence microscopy.
Validation of the coculture by fluorescence imaging
Before seeding in the microfluidic device, HCT-116 cells and HCoMECs were made fluorescent by means of CellTrace Far Red and Yellow (Life Technologies), respectively. These dyes were designed to be easily internalized, passing through the cell membrane into the cytoplasm. The desired concentration of cells were trypsinized, centrifuged, and resuspended in dye solution/phosphate-buffered saline (PBS) (at a concentration of 10 μM). The suspension was incubated for 20 min at 37°C (in the dark) and then diluted in five times the original volume of medium, let to incubate for 5 min, and centrifuged. Last, the pellet was resuspended in fresh medium into a final concentration, and the cell suspension was seeded. Cells in microfluidic culture were observed under fluorescence microscope at days 1 and 5. In addition, afterward, all microfluidic cultures were fixed for further analysis of the microfluidic coculture model. Endothelial cells were seeded on the lateral channels; cells were stained with 4′,6-diamidino-2-phenylindole (DAPI)/phalloidin. Cells were fixed with 4% paraformaldehyde in PBS for 30 min at room temperature and washed again with PBS. Permeabilization of cells was done with 0.1% Triton X-100 (Sigma-Aldrich) for 15 min at room temperature, followed by washing with PBS. High-affinity filamentous actin probe Alexa Fluor 594 phalloidin (1:80; Invitrogen) was injected through the channels, let to incubate for 30 min at room temperature, washed with PBS, and then incubated with DAPI (1:100; Invitrogen) diluted in PBS for 10 min at room temperature.
Endothelial invasion into microfluidic core and quantification
During cell culture, bright-field images were taken with an inverted microscope to analyze and quantify to which extent endothelial cells remain on the lateral microchannels or respond to VEGF present in the Matrigel in the central chamber. Three images of each time point (days 0, 2, 3, and 5) were analyzed. The length was measured using ImageJ (Fiji), from the outer limit of the microchannels/hydrogel interface to the furthest point of the elongated cells.
CMCht/PAMAM dendrimer nanoparticles
Chemical synthesis of CMCht/PAMAM dendrimer nanoparticles
Dendrimer nanoparticles were surface-modified as previously described by Oliveira et al. (23). CMCht with a degree of 80% deacetylation and 47% substitution was synthesized by a chemical modification route of chitin (Sigma-Aldrich, Germany), as described by Chen and Park (47).
Starburst PAMAM carboxylic acid–terminated dendrimers, hereafter designated as PAMAM-CT [generation 1.5, 20% (w/v) methanolic solution], with an ethylenediamine core were purchased from Sigma-Aldrich. CMCht/PAMAM dendrimer nanoparticles were prepared in a stepwise manner as follows: increasing the generation of the PAMAM-CT (G 1.5), obtaining a PAMAM methyl ester–terminated dendrimer, reaction between PAMAM and CMCht (the reaction occurs by a condensation reaction between the methyl ester and amine groups), and converting methyl ester groups that do not react into carboxylic groups in the CMCht/PAMAM dendrimer, followed by precipitation.
Labeling of CMCht/PAMAM dendrimer nanoparticles with FITC
The conjugates of CMCht/PAMAM-FITC were obtained by covalently bonding the amine group of CMCht and the isothyocianate group of FITC (10 mg ml−1; FITC, Sigma-Aldrich, Germany) in anhydrous dimethyl sulfoxide (DMSO; Norconcessus), creating a thiourea bond. First, a CMCht/PAMAM dendrimer nanoparticle solution (10 mg ml−1) was prepared in a carbonate-bicarbonate coupled buffer of pH 9.2. Then, a solution of FITC/DMSO was added under agitation and kept in the dark at 4°C for 8 hours. The FITC-labeled CMCht/PAMAM dendrimer nanoparticle solution was then dialyzed against ultrapure water. The final product was obtained after freeze-drying.
GEM incorporation into CMCht/PAMAM dendrimer nanoparticles
GEM was incorporated in the nanoparticles by mixing an aqueous solution of CMCht/PAMAM dendrimer nanoparticles with an aqueous GEM solution with a final concentration of 0.5 mM. Saturated sodium carbonate solution (Na2CO3; Sigma-Aldrich, Germany) and acetone (Pronalab, Portugal) were then added to the mixture. The resulting precipitates were dialyzed against ultrapure water. GEM-loaded CMCht/ PAMAM dendrimer nanoparticles were obtained by freezing the final solution at 80°C and freeze-drying (Telstar CRYODOS-80, Spain) the samples for 7 days.
Encapsulation efficiency, drug-loading efficiency, and release profile studies
For assessing encapsulation efficiency, CMCht/PAMAM (50 mg) was dissolved in 50 ml of deionized water. GEM (7 mg) in 2 ml of deionized water was added dropwise to the dendrimer solution and stirred for 30 min. Precipitation of the dendrimers and consequent encapsulation of the drug were induced, as described before. Samples were taken before and after precipitation, centrifuged, analyzed by a UV-visible (UV-vis) spectrophotometer at 275 nm in a Hellma quartz plate, and compared to a calibration curve.
For evaluating drug-loading efficiency, preweighted amount of GEM-loaded CMCht/PAMAM (50 mg) was added to distilled water and sonicated to extract GEM. The extracted GEM concentration was analyzed by a UV-Vis spectrophotometer at 270 nm and compared to a calibration curve. The absorbance was used to calculate the percentage of GEM encapsulated. The following formulas were used:
| (1) |
| (2) |
GEM release from nanoparticles was measured by UV spectrophotometry (Shimadzu) at 275 nm, in a Hellma quartz plate, after dissolution of 50 mg of GEM-loaded CMCht/PAMAM dendrimer nanoparticles in 5 ml of PBS solution with 0.01% sodium azide (Sigma-Aldrich). The in vitro release was performed at 37°C under stirring at 60 rpm between 0-hour and 5-day time points.
Characterization of the CMCht/PAMAM dendrimer nanoparticles by transmission electron microscope
The nanoparticle morphology was investigated by transmission electron microscopy (Philips CM-12; FEI Company, The Netherlands; equipped with a MegaView 2 DOCU camera and an image software analyzer SIS NT DOCU) after staining with 2% of phosphotungstic acid and fixation on copper grids.
Gradients generation and characterization inside the microfluidic chip
For the generation of a stable gradient through the Matrigel core compartment, a source-sink mechanism was created, with a solution of FITC-labeled nanoparticles (0.5 mg/ml) in PBS perfused through one side channel, and PBS perfused through other using a Nexus 3000 syringe pump (Chemyx). Three-milliliter syringes were used to introduce PBS with or without the nanoparticles at a rate of 8 μl/hour. The experiment was performed under a Nikon Eclipse Ti-E fluorescence microscope (Nikon Instruments, Japan) for time-lapse imaging of the gradient formation.
Profile and stability of the dynamic gradient were analyzed using NIS-Elements AR (Nikon) software. The chips were analyzed at different time points between 0 and 12 hours with continuous perfusion. Built-in “plot profile” module was used for assessing the gradient intensity and stability over time. The intensity profile corresponds to distance versus intensity. A 3D projection was also reconstructed using software.
GEM-loaded CMCht/PAMAM dendrimer nanoparticle gradient
After the establishment of the coculture model, a gradient of GEM release from dendrimer nanoparticles was generated by perfusing GEM-loaded nanoparticles (0.5 mg/ml) in one inlet and endothelial cell culture medium in the other inlet using exactly the same experimental source-sink mechanism used for the establishment and characterization of the nanoparticle gradient. This was achieved by connecting the chip to the tubings into lateral inlets fed by glass syringes (SGE Analytical Science), mounted on a Nexus 3000 syringe pump.
Live/dead assay
The viability of cells cultured in the microfluidic chips in the presence or absence of GEM-loaded dendrimer nanoparticles was assessed using the live/dead assay. For this, at endpoints, the chip side channels were washed with PBS (Sigma-Aldrich), incubated for 40 min with calcein AM (1 μM; live cells in green) and ethidium homodimer 1 (6 μM; dead cells in red), and then washed again. Microfluidic chips were then imaged with fluorescence microscope using automated acquisition for Z-stack and multicolor channel.
Image processing
Acquired multiframe multicolor images were combined using either Nikon AR or Fiji software (open-source software ImageJ focused on biological image analysis) for operations of processing such as rotation, crop, color balance, and related assemblies, considering initial scales and aspect ratios. High-resolution Z-stack images were processed for 3D reconstruction through deconvolution and 3D projections made by NIS-Elements AR (Nikon) software.
RT-PCR analysis
After each culture time, Matrigel from the central chamber was retrieved by punching with a 5-mm sterile biopsy punch (World Precision Instruments) and placed in 24-well plates. Samples were washed with PBS solution, and Matrigel was depolymerized by incubation with 1 ml of MatriSperse Cell Recovery Solution (BD Biosciences) for 30 min at 4°C. After complete release from the Matrigel, cells were transferred to an Eppendorf tube and centrifuged to a pellet by centrifugation at 300g for 5 min. Cells were washed with cold PBS and centrifuged again. Total RNA was extracted from cells recovered from the 3D cultures using Phasemaker Tubes (Invitrogen), following the manufacturer’s instructions. The pellet was then immersed in 500 ml of TRIzol Reagent and stored at 80°C until further use. RNA quantification and purity were assessed using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies). Complementary DNA (cDNA) was synthetized according to the protocol from the iScript cDNA Synthesis Kit (Bio-Rad). The obtained cDNA was used as template for the amplification of the target genes using the iQ SYBR Green Supermix (Bio-Rad), according to the manufacturer’s instructions. The primer sequence, specific for each gene, namely, ribosomal protein S27A (RPS27A), Caspase-3 (Casp-3), Ki-67, and MMP-1, is described in Table 1. The transcript expression data were normalized to the endogenous housekeeping gene RPS27A, and the relative quantification was calculated, normalizing the obtained results for each target gene at each time point in basal culture conditions (controls), according to the Livak (2ΔΔCT) method (48). Three samples of each group were tested at each time point in three independent experiments.
Table 1. Primer list for the studied genes.
| Oligo name | Sequence 5′ to 3′ |
| Casp-3 (human) F | GAAATTGTGGAATTGATGCGTGA |
| Casp-3 (human) R | CTACAACGATCCCCTCTGAAAAA |
| MMP-1 (human) F | GGGGCTTTGATGTACCCTAGC |
| MMP-1 (human) R | TGTCACACGCTTTTGGGGTTT |
| Ki-67 (human) F | GCTGGCTCCTGTTCACGTA |
| Ki-67 (human) R | CTGGGCTACACTGAGCACC |
| RPS27A (human) F | GCTTGCCAGCAAAGATCAGT |
| RPS27A (human) R | GAGGTTGAACCCTCGGATAC |
Immunohistochemistry
The microfluidic chip was processed for immunohistochemistry by perfusion of CORRESPONDING reagents, through both side inlets. After cell culture, the cells in the chip were washed with PBS, and the cells were fixed with 4% paraformaldehyde in PBS for 30 min at room temperature and washed again with PBS. Permeabilization of cells was done with 0.1% Triton X-100 (Sigma-Aldrich) for 15 min at room temperature, followed by washing and incubation with blocking solution CAS-Block (Thermo Fisher Scientific) for 30 min at 37°C. Primary antibody for Ki-67 diluted in CAS-Block (1:100; rabbit, ab833, R&D Systems) was subsequently incubated overnight at 4°C. Samples were washed with PBS and incubated with the secondary antibody Alexa Fluor 488 conjugated (1:500; anti-rabbit, R&D Systems) together with high-affinity filamentous actin probe phalloidin conjugated with Alexa Fluor 594 (1:80; Invitrogen) in CAS-Block for 3 hours at room temperature. The samples were then washed with PBS, incubated with DAPI (1:100 in PBS; Invitrogen) for 10 min at room temperature, and lastly washed twice with PBS. The samples were kept in PBS in the dark at 4°C until image analysis. High-magnification images were acquired with Nikon Eclipse Ti-E using automated acquisition for Z-stack and multicolor channel.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 5.0a software. The nonparametric Mann-Whitney test was used to compare two groups. A value of P < 0.05 was considered statistically significant. For multiple comparisons, the parametric test one-way analysis of variance (ANOVA) was used, with P < 0.05 considered statistically significant.
Acknowledgments
Funding: The project FronThera (NORTE-01-0145-FEDER-000023) was supported by the Norte Portugal Regional Operational Programme (NORTE 2020) under the Portugal 2020 Partnership Agreement through the European Regional Development Fund (ERDF). M.R.C. acknowledges her PhD scholarship NORTE-08-5369-FSE- 000044, funded by Programa Operacional Regional do Norte, Fundo Social Europeu, Norte2020 TERM&SC, and EMBO Short-Term Fellowship 7232. J.M.O. thanks FCT for his distinction attributed under the FCT Investigator Program (IF/00423/2012 and IF/01285/2015). This work was partially supported by the IET A. F. Harvey Engineering Research Award 2018 (ENG ThE CANCER) and by the Dutch Province of Limburg. S.G., R.T., and P.H. acknowledge financial support by the Province Limburg [program Limburg INvesteert in haar Kenniseconomie (LINK)]. Author contributions: M.R.C. is the main author of the manuscript and was involved in every aspect of the manuscript preparation, from its conception to laboratory work and writing. D.B. was largely involved in the conception of the work and responsible for training and supervision in microfluidic chip design and fabrication, as well as microscopy analysis, and contributed to writing and correcting the manuscript. L.M.T. assisted in the conception of the work and was involved in almost every aspect of the laboratory work and responsible for training and supervision in microfluidic chip design, as well as all biological assays. S.G. assisted in the conception of the work. R.L.R. is the director of 3Bs Research Group, Guimarães, Portugal, and the work developed at the 3B’s involving biomaterials development and characterization was done under his supervision and in his institute. J.M.O. is one of the main contributors to the conception of the manuscript and contributed to writing and correcting the manuscript. R.T. is an associate professor in the Complex Tissue Regeneration (CTR) Department [meanwhile, changed over to Instructive Biomaterials Engineering (IBE) Department] at the MERLN Institute for Technology-Inspired Regenerative Medicine. Part of the work was developed under his supervision in CTR Department. P.H. is the head of the IBE Department at the MERLN Institute (in the meantime also the director of MERLN). Part of the work was developed under her supervision in IBE Department. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper. Additional data related to this paper may be requested from the authors.
REFERENCES AND NOTES
- 1.Vatandoust S., Price T. J., Karapetis C. S., Colorectal cancer: Metastases to a single organ. World J. Gastroenterol. 21, 11767–11776 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carvalho M. R., Lima D., Reis R. L., Oliveira J. M., Correlo V. M., Anti-cancer drug validation: The contribution of tissue engineered models. Stem Cell Rev. 13, 347–363 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Phan D. T. T., Wang X., Craver B. M., Sobrino A., Zhao D., Chen J. C., Lee L. Y. N., George S. C., Lee A. P., Hughes C. C. W., A vascularized and perfused organ-on-a-chip platform for large-scale drug screening applications. Lab Chip 17, 511–520 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barata D., van Blitterswijk C., Habibovic P., High-throughput screening approaches and combinatorial development of biomaterials using microfluidics. Acta Biomater. 34, 1–20 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Foudeh A. M., Fatanat Didar T., Veres T., Tabrizian M., Microfluidic designs and techniques using lab-on-a-chip devices for pathogen detection for point-of-care diagnostics. Lab Chip 12, 3249–3266 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Orcheston-Findlay L., Hashemi A., Garrill A., Nock V., A microfluidic gradient generator to simulate the oxygen microenvironment in cancer cell culture. Microelectron. Eng. 195, 107–113 (2018). [Google Scholar]
- 7.Bott K., Upton Z., Schrobback K., Ehrbar M., Hubbell J. A., Lutolf M. P., Rizzi S. C., The effect of matrix characteristics on fibroblast proliferation in 3D gels. Biomaterials 31, 8454–8464 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Ma Y.-H. V., Middleton K., You L., Sun Y., A review of microfluidic approaches for investigating cancer extravasation during metastasis. Microsyst. Nanoeng. 4, 17104 (2018). [Google Scholar]
- 9.Wan L., Skoko J., Yu J., LeDuc P. R., Neumann C. A., Mimicking embedded vasculature structure for 3D cancer on a chip approaches through micromilling. Sci. Rep. 7, 16724 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen M. B., Whisler J. A., Fröse J., Yu C., Shin Y., Kamm R. D., On-chip human microvasculature assay for visualization and quantification of tumor cell extravasation dynamics. Nat. Protoc. 12, 865–880 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zervantonakis I. K., Hughes-Alford S. K., Charest J. L., Condeelis J. S., Gertler F. B., Kamm R. D., Three-dimensional microfluidic model for tumor cell intravasation and endothelial barrier function. Proc. Natl. Acad. Sci. U.S.A. 109, 13515–13520 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berthier E., Beebe D. J., Gradient generation platforms: New directions for an established microfluidic technology. Lab Chip 14, 3241–3247 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keenan T. M., Folch A., Biomolecular gradients in cell culture systems. Lab Chip 8, 34–57 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwarz J., Bierbaum V., Merrin J., Frank T., Hauschild R., Bollenbach T., Tay S., Sixt M., Mehling M., A microfluidic device for measuring cell migration towards substrate-bound and soluble chemokine gradients. Sci. Rep. 6, 36440 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fouad Y. A., Aanei C., Revisiting the hallmarks of cancer. Am. J. Cancer Res. 7, 1016–1036 (2017). [PMC free article] [PubMed] [Google Scholar]
- 16.Shangguan W., Fan C., Chen X., Lu R., Liu Y., Li Y., Shang Y., Yin D., Zhang S., Huang Q., Li X., Meng W., Xu H., Zhou Z., Hu J., Mo X., Endothelium originated from colorectal cancer stem cells constitute cancer blood vessels. Cancer Sci. 108, 1357–1367 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiménez-Fonseca P., Solis M. P., Garrido M., Faez L., Rodriguez D., Ruiz A. L., Sanchez Lorenzo M. L., Uriol E., Menendez M. D., Viéitez J. M., Gemcitabine plus capecitabine (Gem–Cape) biweekly in chemorefractory metastatic colorectal cancer. Clin. Transl. Oncol. 17, 384–392 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saif M. W., Kaley K., Penney R., Hotchkiss S., Syrigos K. N., Strimpakos A. S., The efficacy of gemcitabine as salvage treatment in patients with refractory advanced colorectal cancer (CRC): A single institution experience. Anticancer Res. 31, 2971–2974 (2011). [PubMed] [Google Scholar]
- 19.Yu X., Di Y., Xie C., Song Y., He H., Li H., Pu X., Lu W., Fu D., Jin C., An in vitro and in vivo study of gemcitabine-loaded albumin nanoparticles in a pancreatic cancer cell line. Int. J. Nanomedicine 10, 6825–6834 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacquemet G., Hamidi H., Ivaska J., Filopodia in cell adhesion, 3D migration and cancer cell invasion. Curr. Opin. Cell Biol. 36, 23–31 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Oliveira J. M., Sousa R. A., Kotobuki N., Tadokoro M., Hirose M., Mano J. F., Reis R. L., Ohgushi H., The osteogenic differentiation of rat bone marrow stromal cells cultured with dexamethasone-loaded carboxymethylchitosan/poly(amidoamine) dendrimer nanoparticles. Biomaterials 30, 804–813 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Carvalho M. R., Maia F. R., Silva-Correia J., Costa B. M., Reis R. L., Oliveira J. M., A semiautomated microfluidic platform for real-time investigation of nanoparticles' cellular uptake and cancer cells’ tracking. Nanomedicine (Lond.) 12, 581–596 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Oliveira J. M., Kotobuki N., Marques A. P., Pirraco R. P., Benesch J., Hirose M., Costa S. A., Mano J. F., Ohgushi H., Reis R. L., Surface engineered carboxymethylchitosan/poly(amidoamine) dendrimer nanoparticles for intracellular targeting. Adv. Funct. Mater. 18, 1840–1853 (2008). [Google Scholar]
- 24.Han H., Wang J., Chen T., Yin L., Jin Q., Ji J., Enzyme-sensitive gemcitabine conjugated albumin nanoparticles as a versatile theranostic nanoplatform for pancreatic cancer treatment. J. Colloid Interface Sci. 507, 217–224 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Bendardaf R., Buhmeida A., Ristamäki R., Syrjanen K., Pyrhönen S., MMP-1 (collagenase-1) expression in primary colorectal cancer and its metastases. Scand. J. Gastroenterol. 42, 1473–1478 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Bogorad M. I., DeStefano J., Karlsson J., Wong A. D., Gerecht S., Searson P. C., Review: In vitro microvessel models. Lab Chip 15, 4242–4255 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barata D., Spennati G., Correia C., Ribeiro N., Harink B., van Blitterswijk C., Habibovic P., van Rijt S., Development of a shear stress-free microfluidic gradient generator capable of quantitatively analyzing single-cell morphology. Biomed. Microdevices 19, 81 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Markov D. A., Lillie E. M., Garbett S. P., McCawley L. J., Variation in diffusion of gases through PDMS due to plasma surface treatment and storage conditions. Biomed. Microdevices 16, 91–96 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishida N., Yano H., Nishida T., Kamura T., Kojiro M., Angiogenesis in cancer. Vasc. Health Risk Manag. 2, 213–219 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yeon J. H., Ryu H. R., Chung M., Hu Q. P., Jeon N. L., In vitro formation and characterization of a perfusable three-dimensional tubular capillary network in microfluidic devices. Lab Chip 12, 2815–2822 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Weis S., Cui J., Barnes L., Cheresh D., Endothelial barrier disruption by VEGF-mediated Src activity potentiates tumor cell extravasation and metastasis. J. Cell Biol. 167, 223–229 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bendardaf R., El-Serafi A., Syrjänen K., Collan Y., Pyrhönen S., The effect of vascular endothelial growth factor-1 expression on survival of advanced colorectal cancer patients. Libyan J. Med. 12, 1290741 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tahergorabi Z., Khazaei M., A review on angiogenesis and its assays. Iran. J. Basic Med. Sci. 15, 1110–1126 (2012). [PMC free article] [PubMed] [Google Scholar]
- 34.Song W., Yan D., Wei T., Liu Q., Zhou X., Liu J., Tumor-derived extracellular vesicles in angiogenesis. Biomed. Pharmacother. 102, 1203–1208 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Nguyen D.-H., Stapleton S. C., Yang M. T., Cha S. S., Choi C. K., Galie P. A., Chen C. S., Biomimetic model to reconstitute angiogenic sprouting morphogenesis in vitro. Proc. Natl. Acad. Sci. U.S.A. 110, 6712–6717 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wen Z., Liao Q., Hu Y., You L., Zhou L., Zhao Y., A spheroid-based 3-D culture model for pancreatic cancer drug testing, using the acid phosphatase assay. Braz. J. Med. Biol. Res. 46, 634–642 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shen C.-J., Kuo Y.-L., Chen C.-C., Chen M.-J., Cheng Y.-M., MMP1 expression is activated by Slug and enhances multi-drug resistance (MDR) in breast cancer. PLOS ONE 12, e0174487 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Foley C. J., Luo C., O'Callaghan K., Hinds P. W., Covic L., Kuliopulos A., Matrix metalloprotease-1a promotes tumorigenesis and metastasis. J. Biol. Chem. 287, 24330–24338 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benbow U., Maitra R., Hamilton J. W., Brinckerhoff C. E., Selective modulation of collagenase 1 gene expression by the chemotherapeutic agent doxorubicin. Clin. Cancer Res. 5, 203–208 (1999). [PubMed] [Google Scholar]
- 40.Said A. H., Raufman J.-P., Xie G., The role of matrix metalloproteinases in colorectal cancer. Cancer 6, 366–375 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khanna A., Mahalingam K., Chakrabarti D., Periyasamy G., Ets-1 expression and gemcitabine chemoresistance in pancreatic cancer cells. Cell. Mol. Biol. Lett. 16, 101–113 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuranaga E., Beyond apoptosis: Caspase regulatory mechanisms and functions in vivo. Genes Cells 17, 83–97 (2012). [DOI] [PubMed] [Google Scholar]
- 43.Chandler N. M., Canete J. J., Callery M. P., Caspase-3 drives apoptosis in pancreatic cancer cells after treatment with gemcitabine. J. Gastrointest. Surg. 8, 1072–1078 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Guo Y., Ziesch A., Hocke S., Kampmann E., Ochs S., De Toni E. N., Göke B., Gallmeier E., Overexpression of heat shock protein 27 (HSP27) increases gemcitabine sensitivity in pancreatic cancer cells through S-phase arrest and apoptosis. J. Cell. Mol. Med. 19, 340–350 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kao Y.-T., Hsu W.-C., Hu H.-T., Hsu S.-H., Lin C.-S., Chiu C.-C., Lu C.-Y., Hour T.-C., Pu Y.-S., Huang A.-M., Involvement of p38 mitogen-activated protein kinase in acquired gemcitabine-resistant human urothelial carcinoma sublines. Kaohsiung J. Med. Sci. 30, 323–330 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Melling N., Kowitz C. M., Simon R., Bokemeyer C., Terracciano L., Sauter G., Izbicki J. R., Marx A. H., High Ki67 expression is an independent good prognostic marker in colorectal cancer. J. Clin. Pathol. 69, 209–214 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Chen X.-G., Park H.-J., Chemical characteristics of O-carboxymethyl chitosans related to the preparation conditions. Carbohydr. Polym. 53, 355–359 (2003). [Google Scholar]
- 48.Livak K. J., Schmittgen T. D., Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]


