Abstract
RDH1 is one of the several enzymes that catalyze the first of the two reactions to convert retinol into all-trans-retinoic acid (atRA). Here, we show that Rdh1-null mice fed a low-fat diet gain more weight as adiposity (17% males, 13% females) than wild-type mice by 20 weeks old, despite neither consuming more calories nor decreasing activity. Glucose intolerance and insulin resistance develop following increased adiposity. Despite the increase in white fat pads, epididymal white adipose does not express Rdh1, nor does muscle. Brown adipose tissue (BAT) and liver express Rdh1 at relatively high levels compared to other tissues. Rdh1 ablation lowered body temperatures during ambient conditions. Given the decreased body temperature, we focused on BAT. A lack of differences in BAT adipogenic gene expression between Rdh1-null mice and wild-type mice, including Pparg, Prdm16, Zfp516 and Zfp521, indicated that the phenotype was not driven by brown adipose hyperplasia. Rather, Rdh1 ablation eliminated the increase in BAT atRA that occurs after re-feeding. This disruption of atRA homeostasis increased fatty acid uptake, but attenuated lipolysis in primary brown adipocytes, resulting in increased lipid content and larger lipid droplets. Rdh1 ablation also decreased mitochondrial proteins, including CYCS and UCP1, the mitochondria oxygen consumption rate, and disrupted the mitochondria membrane potential, further reflecting impaired BAT function, resulting in both BAT and white adipose hypertrophy. RNAseq revealed dysregulation of 424 BAT genes in null mice, which segregated predominantly into differences after fasting vs after re-feeding. Exceptions were Rbp4 and Gbp2b, which increased during both dietary conditions. Rbp4 encodes the serum retinol-binding protein—an insulin desensitizer. Gbp2b encodes a GTPase. Because Gbp2b increased several hundred-fold, we overexpressed it in brown adipocytes. This caused a shift to larger lipid droplets, suggesting that GBP2b affects signaling downstream of the β-adrenergic receptor during basal thermogenesis. Thus, Rdh1-generated atRA in BAT regulates multiple genes that promote BAT adaptation to whole-body energy status, such as fasting and re-feeding. These gene expression changes promote optimum mitochondria function and thermogenesis, limiting adiposity. Attenuation of adiposity and insulin resistance suggests that RDH1 mitigates metabolic syndrome.
Electronic supplementary material
The online version of this article (10.1007/s00018-019-03046-z) contains supplementary material, which is available to authorized users.
Keywords: Adipose, Lipid metabolism, Mitochondria membrane potential, Retinol dehydrogenase, Retinoic acid, Retinol, Thermogenesis
Introduction
The retinol (vitamin A) metabolite all-trans-retinoic acid (atRA) supports multiple processes in vertebrates starting with conception and embryonic development, and continuing throughout postnatal life [1]. Postnatal functions of atRA include regulating energy balance [2, 3], inhibiting differentiation of white preadipocytes, controlling mature adipocyte function [3], and inducing Ucp1 expression in brown adipose tissue (BAT) [4, 5]. Chronic supplementation with larger amounts of retinol, or with pharmacological doses of atRA, reduces obesity in rodent models [6–8]. Retinol and atRA, however, exhibit hormesis, i.e., their effects depend on the amount dosed. Inverted U or J-shaped dose–response curves relative to beneficial effects denote hormesis: as concentrations increase beneficial effects subside and atypical or toxic effects develop [9]. Thus, dosing high amounts of retinol or atRA confounds toxicology with physiology, especially in animals fed chow diets, which contain copious amounts of vitamin A. Chow diets also contain pre-vitamin A carotenoids, which raise atRA levels [10]. The phenomenon of hormesis has impaired understanding physiological effects of atRA in the adult, particularly in adipose.
As low as 1 nM atRA arrests conversion of pre-adipocytes into mature white adipocytes in primary cultures and cell lines [11–13]. atRA treatment of mature white adipocytes increases lipolysis, decreases triacylglycerol (TAG) content and increases fatty acid oxidation [14, 15]. Much less is known about the impact of atRA on BAT. Most studies have focused on transcriptional induction of the thermogenic gene Ucp1. However, the effect of atRA on Ucp1 mRNA expression differs among species. atRA alone induces Ucp1 mRNA in mouse brown adipocytes, but requires T3 and/or or a β-adrenergic agonist in rat brown adipocytes [16, 17]. Systematic actions of atRA and their mechanisms on brown adipogenesis and function remain partially understood.
Two consecutive reactions activate retinol into atRA [18, 19]. The first reaction, catalyzed by retinol dehydrogenases (RDH) of the short-chain dehydrogenase/reductase gene family, converts retinol into retinal. Retinal dehydrogenases (RALDH), of the aldehyde dehydrogenase gene family, convert retinal into atRA. Carotenoid cleavage and retinal reductases also contribute to steady-state atRA concentrations by regulating the concentration of retinal [20, 21]. The catabolic CYP26 isozymes, induced by atRA, also support atRA homeostasis by catabolizing atRA [22, 23]. Dissimilar but overlapping spatial, temporal and intracellular expression patterns of the multiple isoforms of each enzyme suggest distinct contributions to atRA functions [24, 25]. Differences in the phenotypes of ablated retinoid metabolon genes provide further support for this hypothesis. For example, heterozygous Rdh10-null mice fed a high-fat diet (HFD) gain additional adiposity, and males develop more severe liver steatosis than wild-type mice (WT), but do not undergo these changes when fed a low-fat diet (LFD) [26]. Homozygous Rdh1-null mice (KO), in contrast, gain additional adiposity relative to WT when fed a LFD, and do not develop liver steatosis [27].
In our initial characterization, KO were fed low-fat vitamin A-depleted diets [27]. These mice were born in Mendelian frequency with no obvious developmental defects. KO, however, gained more weight than WT, such that at 20 weeks old, they become 19% heavier. The increased weight reflected an increased adiposity index from increases in multiple visceral and subcutaneous white fat pads. In contrast to a diet restricted in vitamin A, a diet with 30 IU vitamin A/g (i.e., copious vitamin A) eliminated weight differences between KO and WT.
Here, we extend our original findings by reporting the mechanisms underlying Rdh1 actions. Surprisingly, WAT and muscle lack detectable Rdh1 expression, eliminating a direct effect within these tissues. In contrast, liver and BAT both express Rdh1 relatively intensely compared to most other tissues surveyed. BAT consumes energy disproportionately to its mass to defend body temperature—the process of non-shivering thermogenesis. As such, BAT mass and activity relate inversely to adiposity [28, 29]. We find that Rdh1-ablation does not affect brown adipocyte differentiation (hyperplasia), but disrupts retinoid homeostasis in differentiated brown adipocytes, despite partial compensation by other enzymes in the retinoid metabolon. KO have altered regulation of > 400 genes in BAT, attenuated mobilization of stored TAG, and impaired mitochondria function. These defects result in a cooler body temperature under ambient conditions. Increased adiposity and up-regulation of Rbp4 promote glucose intolerance and hepatic insulin resistance. These data indicate that RDH1 regulates genes controling BAT adaptation to whole-body energy status, such as fasting and re-feeding; its loss contributes to aspects of metabolic syndrome.
Materials and methods
Mice and diets
For initial studies, KO mice were backcrossed 6 times into a C57BL/6 J background. KO progeny were interbred and compared to age- and diet-matched WT controls. Mice were housed 2–5 per cage under standard conditions: 21 °C, 12-h light–dark cycle (07:00–19:00) in ventilated cages with an air circulation system (Techniplast TouchSLIM Line), unless housed in metabolic cages. Mice were provided extra enrichment materials in their home cages, such as a piece of shredded paper and a cotton ball for nest building. Mice were fed a purified AIN93G diet (i.e., a low-fat diet, LFD) available from Dyets, Inc. (#110700) at least 2 generations before use, unless noted otherwise. The AIN93G diet contains sufficient (4 IU vitamin A/g), as recommended by the National Research Council for rodents [30]. The AIN93G diet avoids the copious vitamin A found in chow diets that exceed recommendations by four to eightfold, not including contributions from pre-vitamin A carotenoids. For the HFD study, 7-week-old mice were fed an AIN93G diet identical with the standard AIN93G diet, with the exception of 50% fat-derived calories (Dyets #180614), with a decrease in dextrose to maintain a diet isocaloric with the LFD [3, 12]. To facilitate the switch, animals were fasted 16 h starting near the end of the light period. “Re-fed/re-feeding” indicates that mice were fasted 16 h and then provided food for 6 h, unless stated otherwise. “Ad lib” refers to experiments that did not control food intake. Samples were collected during the light period. Animal experiments were approved by The University of California Berkeley Animal Care and Use Committee.
Genotyping
Primers used for genotyping by PCR were: forward 5′-GGT TTA CAC AGC TGC TTT CAG GAC A; reverse 5′-GAG TCA TTA GGC TTA GAC GAT CTC to generate a ~ 300 bp WT Rdh1 amplicon. Other primers: forward 5′-TGT ATG CTA TAC GAA GTT ATG AAT TC; reverse 5′-CCT CTG GGT TCT AAG TCC A generated a ~ 1.7 kbp KO amplicon.
Mouse evaluation
Mice were weighed weekly to the nearest 0.1 g. Long-term food intake was monitored by weighing food once or twice weekly and normalizing to mouse number and average mouse weight. Total body fat was measured in live mice using an EchoMRI Whole Body Composition Analyzer. Tissues harvested for retinoid and gene expression were flash frozen in liquid nitrogen. Serum was prepared from blood collected via post-mortem cardiac puncture, kept on ice 30 min, and centrifuged 10 min at 10,000×g. Serum samples and tissues were stored at − 80 °C until analysis. To measure glucose, blood was obtained from a nick in the tail vein. Measurements were done with an Accu-chek glucometer and Accu-chek Comfort Curve test strips (Roche). For glucose tolerance tests (GTT), mice were fasted 16 h. A sterile 20% d-glucose solution was delivered intraperitoneally (2 g glucose/kg body weight, 10 µL/g). For insulin tolerance test (ITT), mice were fasted 5 h beginning ~ 08:00 AM. Shortly before use, an insulin solution was prepared from Humulin (Eli Lilly) [31]. Insulin (0.5 U/kg body weight, 3.6 µL/g) was administered intraperitoneally. For the pyruvate tolerance test (PTT), mice were fed ad lib. Sodium pyruvate (2 g/kg body weight, 10 µL/g) was dosed intraperitoneally. During all three tests, mice had access to water.
Retinoid quantification
Tissue collection and extraction for retinoid quantification were done under yellow light. atRA was quantified either as reported by LC/MS/MS [32] or with an alternative LC procedure. The alternative LC consisted of a Suplex pkb-100 column (Supelco, 2.1 × 250 mm, 5-µm particles) with 3 min of 80% acetonitrile/20% water/0.1% formic acid, followed by a linear gradient to 95% acetonitrile/5% water/0.1% formic acid over 9 min, held for 4 min, returned by linear gradient to 80/20/0.1 over 1 min, and held 8 min, all at 0.4 mL/min.
Histological analysis and tissue lipid droplet size determination
These analyses were done by standard techniques [3, 33, 34]. Briefly, pieces (5 mm × 5 mm) of freshly dissected BAT were fixed for 7 days in 10% formalin at 4 °C. Samples were cryoprotected in 20% sucrose/PBS, frozen in isopentane on dry ice, cut into 9-µm sections with a cryostat (Leica), stained with oil red-O (Sigma) and mounted in water-based mounting medium (Sure Mount, #LC-W, General Data Healthcare). A second set of tissue sections was stained with H&E (Sigma) to examine tissue integrity. A third set was stained with Lillie’s modified Masson’s Trichrome Stain for connective tissue to examine fibrosis. To measure lipid droplet (LD) sizes, 3 non-overlapping sections per mouse at least 100 µm apart in tissue depth were digitally imaged using a Zeiss M2 microscope with a 20x lens. An analyst blinded to experimental conditions used ImageJ software to quantify LD areas in at least 200 adipocytes per mouse, for a total of 5700 LD per genotype [34, 35]. For macrophage infiltration analysis, BAT sections were incubated in 0.3% H2O2 to block endogenous peroxidases, followed by rabbit anti-F4/80 antibody (Immunocruz® ABC Staining System) and were counterstained using H&E [3].
Serum lipids and hormones
Serum-free fatty acids (FFA) and glycerol levels were determined using EnzyChrom ™ FFA Assay Kit (BioAssay Systems, #EFFA-100) and Free Glycerol Reagent (Sigma, #F6428) [36], according to the manufacturer protocol. Serum TAG were determined using the Cayman Chemical Company kit (#10010303) [37]. Serum insulin, TNFα, monocyte chemoattractive protein 1 (MCP1) and IL6 levels were determined using the Milliplex Map Mouse Serum Adipokine Panel (Millipore, #MADPK-71 K) [38, 39]. Additional insulin measurements were made using an ultra-sensitive enzyme-linked immunosorbent assay (Crystal Chem, Cat. #90080) [40].
Metabolic analyses
Mice were housed individually in metabolic cages (Comprehensive Lab Animal Monitoring System, Columbus Instruments) with airflow of 0.5 L/min. Respiration rates (oxygen consumption, carbon dioxide production) were recorded using Oxymax software under standard housing temperature and light/dark cycle conditions. For energy balance data, mice were housed 24–25 h. A 1-h acclimation was allowed before data collection. Movement was quantified based on total breaks of infrared beams in the x-axis (XM), breaks of two or more beams in the x-axis (AM), and breaks in the z-axis (ZM). For β3-adrenergic studies, mice were allowed to acclimate to the metabolic cages ~ 16 h and were fasted beginning 09:00. At 12:00, mice were injected i.p. with 1 mg/kg CL316,243 (CL) in sterile saline at 10 μL/g body weight. Metabolic data were collected during 3 h post-injection. Dilute solutions of CL were made fresh on the day of use from frozen stock. The procedure was repeated using saline-only injections. β3-adrenergic induction was calculated by averaging metabolic data during low activity periods (XM < 100 per 10 min) for each mouse after CL injection relative to the average of similar periods after saline injection. Heat was calculated by Oxymax software. In β3-adrenergic studies, respiration was normalized to an effective body mass (mass0.75).
Stable isotope use
Six- to eight-week-old mice were fasted 16 h and then re-fed 5–6 h. Upon re-feeding, mice were injected with 100% deuterium-labeled water at 35 µL/g pre-fasted body weight and then provided 8% deuterium-labeled drinking water ad lib. Analyses were done by mass isotopic dilution analysis as described [41, 42]. Body water enrichment was determined empirically to facilitate accurate enrichment measures.
Temperature studies
Temperatures were taken 3 times during the light period (~ 10:00, 13:00 and 16:00) rectally using a probe (Physitemp RET-3) attached to a thermometer (Physitemp BAT-12) [43]. Mean temperatures for individual mice were then averaged by genotype. To test effects of cold, mice were housed individually at 4 °C for 24 h under standard light/dark conditions [44].
Lipolysis in tissue explants
BAT and epididymal WAT (eWAT) from 5-week-old mice were removed quickly from euthanized animals and placed into a room temperature lipolysis buffer (121 mM NaCl, 4.9 mM KCl, 1.2 mM MgSO4, 0.33 mM CaCl2, 12 mM HEPES, 0.1% glucose and 2% fatty acid-free BSA, pH 7.3). Approximately, 20-mg BAT or 50-mg eWAT (wet weights) was placed into cell culture wells containing 2-mL lipolysis buffer plus 10-µM isoproterenol [45]. Aliquots of medium (0.15 mL) were removed after 1-, 2- and 4-h incubation at 37 °C. Glycerol was measured by adding 20-µL sample or Triolein standard to 80-µL Free Glycerol Reagent and incubating 15 min at 37 °C [46]. Absorbance was read at 540 nm. Total glycerol release was plotted for each explant/well at each time point. Lines of best fit were determined. Rates were normalized to explant protein (BCA assay, Pierce) [47].
Isolation and culture of mouse primary brown adipocytes
BAT stromal vascular fraction (SVF) cells were harvested from mice backcrossed 12 times into the C57BL/6 J background. Six- to seven-week-old mice were euthanized by isoflurane followed by cervical dislocation. Interscapular BAT depots were dissected and minced finely in sterile Ringer balanced salt solution, as described [48]. BAT from 3 mice was pooled per replicate. Minced tissue was incubated in 0.1% collagenase II (Promega) solution with 5% fatty acid-free bovine serum albumin (Sigma #A8806) for 60 min at 37 °C with regular mixing. Tissue homogenates were filtered through a sterile 100-μm nylon cell strainer (Fisher Scientific). Mature adipocytes were separated from SVF cells by 5-min centrifugation at 100×g. The cell pellet was suspended in erythrocyte lysis buffer and incubated 10 min at room temperature. Cells were re-centrifuged as above, then suspended in DMEM (high glucose, sodium pyruvate and glutamate) containing 20% bovine calf serum (BCS) and 1% penicillin/streptomycin, and plated on plastic 12-well cell culture plates. Primary pre-adipocytes were kept in growth medium until confluent (7 days). Adipogenesis was induced at confluence (differentiation day 0, dd0) with 1 μM insulin, 10 nM triiodothyronine (T3), 1 μM dexamethasone, and 500 μM 3-isobutyl-1-methylxanthine in DMEM containing 7.5% BCS and 1% penicillin/streptomycin. After 2 days, the medium was changed to 1 μM insulin, 10 nM T3, and 20 nM rosiglitazone in DMEM containing 7.5% BCS, and 1% penicillin/streptomycin.
Western blots
Primary brown adipocytes were cultured in 12-well cluster plates. Mature adipocytes (dd8) were then conditioned to low glucose (5.6 mM) DMEM without serum for 18 h. For lipolysis, adipocytes were incubated 4 h in 1 x HBBS supplemented with 5% fatty acid-free BSA (Sigma #A8806) without serum. For induced lipolysis, cells were incubated 4 h in the same medium with 10 µM isoproterenol [45, 49]. For oleate treatment, cells were incubated 4 or 24 h in the same medium with 400 µM oleate [50]. Cells were lysed in RIPA buffer (Thermo Fisher Scientific, Cat. #89900). Fifteen µg protein lysates per lane was loaded onto 4–20% gradient Mini-PROTEAN® TGX Stain-Free™ Precast Gels (BioRad). Equal loading of each lane was verified in gel using UV-induced fluorescent signals from trihalo compounds covalently bound to tryptophan residues in cellular proteins, and imaged with ChemiDoc XRS+ gel imager (BioRad). Proteins were transferred onto a nitrocellulose membrane. Primary antibodies used for protein analyses were: rabbit anti-UCP1 (Sigma cat. U6382), Lipolysis Activation Antibody Sampler Kit (Cell Signaling Technology cat. #8334), rat monoclonal anti-CD36/SR-B3 (R&D Systems cat. MAB2519), mouse monoclonal anti-β-tubulin and rabbit mAb anti-ATGL (Cell Signaling Technology Cat. #2146, #2439), mouse monoclonal anti-p-perilipin 1-serine 522 (Vala Sciences Cat. #4856), and mouse monoclonal anti-cytochrome C (Abcam Cat. #37BA11) [51]. Fluorescently labeled secondary antibodies used were: IRDye® 800CW Goat anti-Mouse IgG (Cat. #926-68050), IRDye® 680RD Goat anti-Rabbit IgG (Cat. #925-32211), and IRDye® 680LT Goat anti-Rat IgG (Cat. #926-68029) (Li-COR). Fluorescent signals were quantified using an Odyssey Li-COR infrared fluorescent imager (Li-COR). Signal intensity analyses were done in Image Studio™ Lite (Li-COR). The study was repeated 4 times, with separate groups of mice.
FFA uptake by brown adipocytes
Primary brown SVF cells from male mice were differentiated until dd8 for gene expression analyses in 12-well plastic culture plates, and until dd10 for lipid uptake studies on 8-well Nunc® Lab-Tek® II chambered cover glass [52]. One day before assay, the medium was switched to FA uptake medium (HBSS with 5 mM glucose, 5% BSA, no serum) for 18 h to allow cells to adapt to serum-free medium. On the day of assay, chambers cells were fitted onto an inverted fluorescent microscope Aixovert (Zeiss) equipped with a 40x oil immersion objective for time-lapse image acquisition. During data collection, the microscope stage temperature was kept at 37 °C, with 5% CO2 in the imaging chamber. The first 10 frames were collected as background. At the assay start, the medium was switched to FA uptake media containing 2 µM BODIPY (C1-BODIPYC12: 4,4-difluoro-5-methyl-4-bora-3a,4a-diaza-s-indacene-3-dodecanoic acid, Molecular Probes), 5% BSA and 0.1% Tripan Blue. This medium quenches the fluorescent signal of BODIPY outside of cells, allowing fluorescent only of intracellular BODIPY [52]. Imaging data were collected every 10 s for 20 min. At the end of the assay, cells were fixed in 4% PFA and stained with DAPI. Time-lapse image analyses were performed using ImageJ. A region of interest in each frame was set manually over 20–30 LD from single cells. Background corrected fluorescent signals were measured from each region of interest for a minimum of 10 adipocytes for each genotype. This study was repeated three times with separate brown adipocyte primary cultures from different groups of mice.
Lipid droplet (LD) quantification in adipocytes
Primary brown adipocytes differentiated from SVF cells in 12-well cluster plates were fixed with 4% PFA at dd8 and stained with oil red O. Cells were imaged using the Zeiss Axiovert inverted microscope with a 20 × objective. Oil red O was extracted three times with isopropanol and absorbance was read at 510 nm. The amount of dye was calculated from a standard curve and normalized to the number of adipocytes in each well [3, 49].
Induced lipolysis was quantified by assaying glycerol in the medium of primary brown adipocytes (dd8) after 24-h treatment with 10 µM isoproterenol [45, 49]. Aliquots of the medium (60 µL) were removed after 0, 2, 4, 6, 8, 12, and 24 h. Glycerol levels were measured as above. Protein was measured with the BCA Protein Assay Kit, as above. To account for variability in differentiation, data were normalized to the fraction of cells containing LD in each well. Lines of best fit were plotted for 0-, 4-, and 6-h data and used to calculate the initial rate of glycerol release.
For LD quantification, primary adipocytes (dd8) cultured in 8-well Nunc® Lab-Tek® II chambered cover glass (Sigma, Cat. #Z734853-96EA) were switched to HBSS with 10 µM isoproterenol, as above for live cell imaging. A separate set of cells was supplemented 24 h with 400 μM oleate. Adipocytes were imaged at 0, 2, 4, 6, 8, 12 and 24 h of isoproterenol-induced lipolysis, or after 24 h of oleate supplement using a Zeiss Axiovert inverted microscope with a 20 × objective [49]. At least 12 images were taken for each genotype and analyzed for LD areas with ImageJ, by an investigator blind to treatment conditions. Every LD was traced manually using the manual region of interest tool. Each area and perimeter were measured. For each time point, an equal number of adipocytes per genotype were analyzed (minimum 250), allowing calculation of an average LD size. This study was repeated with primary adipocytes from an independent group of mice using LD fluorescently labeled with BODIPY™ 493/503 (Cat. #D3922, Molecular Probes), after 24-h induced lipolysis or 24-h oleate treatment. Cells were fixed after treatment in 4% PFA and labeled with BODIPY and DAPI [49]. A series of z-stacks (20–30 slices 2 µm thickness with 0.6–0.8 µm between each slice) was collected using a Zeiss 710 confocal microscope with 20 × and 63 × immersion objectives at 488 nm excitation laser. The 3D volumetric data sets were reconstructed using the Bitplane IMARIS program for surface rendering of LD. Analysis of LD number per adipocyte, surface area and volume was computer assisted based on a 3D surface model from calibrated images. The XY diameter was constant for all experimental conditions to optimize automatic detection of LD < 10 µm. The region and global threshold values were optimized for each data set to maximize detection of the smallest LD. With set parameters, the “SPOTS” custom written program automatically reconstructed all LD labeled with BOPIPY and calculated volume for each object.
Mitochondria function of brown adipocytes
Mitochondria respiratory rates were measured with a Agilent Seahorse XF24 extracellular flux analyzer [53]. Primary brown adipocytes (dd12) were differentiated in V7 microplates (Agilent, Cat. #101037-004) and cultured overnight without serum and with 5.6 mM glucose to condition use of oleate as substrate. For assay, the medium glucose concentration was decreased to 2.5 mM. Two % BSA was added to sequester FFA to decrease intracellular concentrations. The OCR (oxygen consumption rate) was measured from 0 to 30 min during these conditions of basal lipolysis. Seventy µM oleate complexed to 15 µM BSA (0.1%) was added at 30 min to measure the oleate-driven OCR. Lipolysis and FFA use were stimulated with 10 µM isoproterenol (90–170 min). The ATP-synthase inhibitor oligomycin (oligo) was added at 170 min to assess ATP synthesis. Treatment with the chemical uncoupler carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP) at 205 min was used to assess maximum respiration. Antimycin A (AA) and rotenone were added at 230 min to inhibit complex I and III, i.e., all respiration. This study was repeated five times with cells from independent groups of mice. For the rescue experiment, cells were pretreated 18 h with 100 nM TTNPB in serum-free medium.
For mitochondria membrane potential analysis, primary brown adipocytes were cultured on 8-well Nunc® Lab-Tek® II chambered cover glass (Sigma, Cat. #Z734853-96EA) [54, 55]. Adipocytes (dd8) were first conditioned 18 h to serum-free, low glucose DMEM, and then labeled 20 min in the dark at room temperature with MitoTracker™ Red FM (Life Technologies cat. M22425), and Hoechst 33342 nuclear dye for live cell imaging (Thermo Scientific, Cat. #62249). Cells were then incubated for 4 h in either basal lipolysis media or stimulated 4 h with 10 µM isoproterenol, and imaged using the Zeiss Axiovert inverted microscope with a 40x immersion objective. At least ten non-overlapping images were collected from each genotype for each condition. The microscope stage temperature was set to 37 °C under 5% CO2. Images (12-bit) were captured with a Retiga CCD camera. Signal intensity was quantified using ImageJ.
qPCR
Total RNA was isolated either via phenol–chloroform extraction using Trizol Reagent (Invitrogen, Cat. #15596026) or a spin kit (Aurum Total RNA Fatty and Fibrous Tissue Pack, Bio-Rad, Cat. #7-6870; RNeasy Mini Kit, Qiagen, Cat. #74104; RNeasy Micro Kit, Qiagen, Cat. #74004; RNeasy Lipid Tissue Mini Kit, Qiagen, Cat. #74804) according to manufacturer’s instructions. RNA was either DNAse treated on-column (Aurum kit) or following isolation (Turbo DNA-free, Ambion, #AM1907). For some qPCR, reverse transcriptase (Superscript II, Invitrogen) in the presence of RNAse Inhibitors (RNAse Out, Invitrogen) and poly-thymine primer, were used to prepare cDNA. Reverse transcription was followed by RNAse treatment (RNAse H, Invitrogen). For additional qPCR, a reverse transcriptase with intact RNAse activity (iScript, Bio-Rad) was used with random hexamer and poly-thymine primers. Similar results were obtained regardless of the RTase used.
qPCR reactions were run in 96-well plates on an ABI7000HT (Applied Biosystems) or CFX Connect (Bio-Rad) thermocyclers. Reactions were done in 20 μL with 1 μL cDNA solution, 1 μL TaqMan primer/probe mix (Applied Biosystems/ThermoFisher, Cat. #4369016), 10 μL master mix (TaqMan Gene Expression Mix, Applied Biosystems/ThermoFisher) and 8 μL RNase- and DNAse-free water. Primer/probe sets were purchased pre-designed from Applied Biosystems/ThermoFisher. Design variables such as %CG content, Tm, and amplicon length were optimized such that the TaqMan Gene Expression Assays in this study have amplification efficiencies of 100% or approaching 100% [56]. Samples were run as technical duplicates or triplicates. In tissues, mean Ct for β-actin (Actb) was subtracted from mean Ct for genes of interest. In SVF experiments, expression was normalized to the geometric mean of β-glucuronidase (Gusb) and TATA-box binding protein (Tbp) [57]. The linear fold change was calculated using the formula 2ΔCt. Linear values for control groups were averaged. Individual values were normalized to the control group mean.
RNAseq
BAT from 13 KO (8 fasted and 5 refed) and 7 WT (4 fasted and 3 refed) was collected and immediately frozen in liquid N2. Total RNA from each individual was isolated with an Aurum Total RNA Fatty and Fibrous Tissue Kit, and DNAse was treated on-column (Bio-Rad, Cat. #7-6870). Pooled RNA libraries were generated using the Illumina TruSeq RNA sample preparation kit (Cat. #RS-122-2001). RNA libraries were submitted to the Vincent J. Coates Genomic Sequencing Laboratory at UC Berkeley. Single-read, 100 bp experiments were run on a HiSeq 2000 instrument, with multiplexing (2 libraries per lane). Fastq sequencing files were filtered for common contaminants and mapped to the mm10 mouse reference genome using Tophat (v2.0.6). Cufflinks (2.0.2) was used to process the resulting BAM read alignments. We determined differentially expressed genes using Cuffmerge and Cuffdiff with a false discovery rate cutoff of 0.05. All spice variants were sequenced and included in the analysis. Data presented here will be deposited in NCBI’s Gene Expression Omnibus, and will be accessible through GEO (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?). Gene ontogeny (GO) analysis was performed using the DAVID database (https://david.ncifcrf.gov) and the Gene Ontology Consortium database (http://www.geneontology.org). Over-representation of GO terms was identified using a hypergeometric test with a Benjamini–Yekutieli-corrected p value < 0.05.
Gbp2b over expression in brown adipocytes
We obtained immortalized brown preadipocytes from Shingo Kajimura, UCSF [58]. We constructed plasmids pLJM-HygroB-EGFP and pLJM-HygroB-Gbp2b from the pLJM1-EGFP plasmid (Addgene #19319). Cells were transduced with either overexpression vector using Lentivirus particles (http://www.addgene.org/tools/protocols/plko/). Hygromycin-resistant, stably transduced cells were selected and propagated in culture. Gbp2b and Egfp overexpression were confirmed by qPCR. Adipogenesis was induced at confluence (dd0) with 5 µg/mL insulin, 1 nM triiodothyronine (T3), 5.1 µM dexamethasone, 500 μM 3-isobutyl-1-methylxanthine and 125 µM indomethacin in DMEM containing 10% FBS, and 1% pen/strep. After 2 days, the medium was changed to 5 µg/mL insulin and 1 nM T3 in DMEM containing 10% FBS, and 1% pen/strep. Cells were cultured for an additional 3 days before use (dd5). Cells were mixed in 4% PFA, washed and incubated in BODIPY493/503 (Cat. #D-3922) and DAPI (Life Technologies). Images were captured using a Zeiss Axiovert microscope with Retiga CCD camera (300 ms exposure, 25% fluorescent lamp intensity). Quantification of LD area was done using ImageJ.
Statistics
Data were evaluated with Graph Pad Prism 7 using parametric two-tailed unpaired student’s t tests with Welch’s correction, or linear regression analyses, or two-way ANOVA. Values are means ± SEM. A Grubb’s test was used to identify outliers at α = 0.05.
Results
Increased adiposity in Rdh1-null mice (KO)
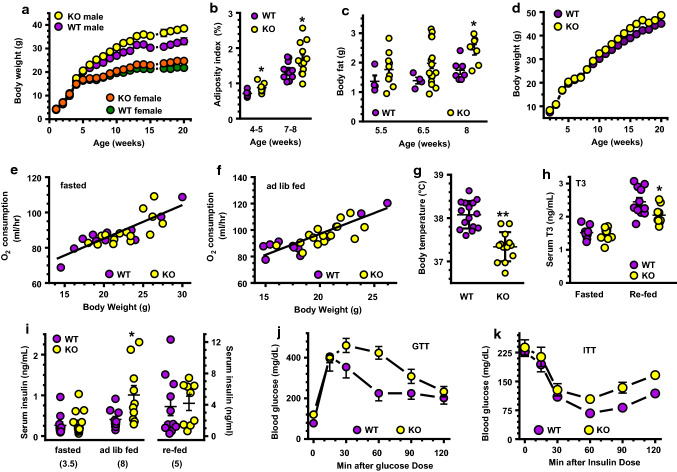
We tested feeding a vitamin A-sufficient AIN93G diet with 4 IU vitamin A/g, i.e., a low-fat diet (LFD), as recommended by the National Research Council for rodents [59]. Male KO fed this LFD gained ~ 10% more weight than WT by 7 weeks old and ~ 17% more weight by 20 weeks old: 33.1 vs 38.5 g (Fig. 1a). Female KO gained ~ 13% more weight than WT by 20 weeks old: 21.8 vs 24.7 g. The male KO adiposity index increased ~ 23% by 4–5 weeks old; an increase sustained at 7–9 weeks old (Fig. 1b). Male KO had ~ 40% more body fat at 6.5 and 8 weeks old compared to WT (Fig. 1c). We continued study with males because of an earlier onset of a more severe phenotype.
Fig. 1.
The KO phenotype. Each experiment was done at least twice with independent groups of mice. a Weight increases with age in mice fed a LFD (male WT, n = 7–8; male KO, n = 9–12; female WT, n = 7–8; female KO, n = 11): Two-way ANOVA p < 0.001 genotypic and age differences for both sexes. b Male adiposity indices (sum of one interstitial and one epididymal fat depot as percentage body weight) at age 4–5 weeks (n = 6–7) and 7–8 weeks (n = 11–12); *p < 0.05. c EchoMRI quantification of fat in males (WT, n = 4–7; KO, n = 7–17): *p < 0.009. d Weights of males fed a HFD: WT, (n = 7–10); KO (n = 7–8) switched from a LFD to a HFD at 7 weeks old; Two-way ANOVA p < 0.0001 genotype and age. e, f Oxygen consumption rates during fasting e or ad lib feeding f (n = 12 WT, 15 KO). g Average daytime temperatures of 6-week-old mice (n = 13–15). Data represent three independent experiments: **p < 0.0001 vs. WT. h Serum T3 in 5- to 7-week-old mice, n = 9–11 mice each group; *p < 0.03. i Serum insulin in 3.5-week-old mice fasted 16 h, 5-week-old mice 2–3.5 h after re-feeding, and 8-week-old ad lib fed mice (n = 10–11 WT, 9–14 KO); *p < 0.03 vs. WT. j Blood glucose of 24- to 25-week-old mice during a GTT (n = 7 WT, 9 KO): Two-way ANOVA genotype (p < 0.02), time (p < 0.0001). k Blood glucose of 25- to 26-week-old mice during an ITT (n = 7 WT, 9 KO): genotype p = 0.0009, time p < 0.0001
Expanded adiposity in genetically modified mice usually requires feeding a high-fat diet (HFD) to stimulate adipogenesis. To determine whether HFD feeding exacerbated the KO phenotype, we fed males a AIN93G rodent diet modified with 50% fat-derived calories beginning at 7 weeks old (Fig. 1d). Both KO and WT gained more weight than mice fed the LFD. By 20 weeks old, WT and KO weighed 45.2 and 48.7 g, respectively. Thus, the HDF maintained a weight difference between KO and WT at 20 weeks old of 3.5 g, compared to 5.4 g when mice were fed the LFD.
A HFD causes both hyperplasia and hypertrophy of adipose tissue in mice [60]. As expected, introduction of the HFD caused both KO and WT to increase adiposity, consistent with increased hyperplasia and hypertrophy. Yet a weight difference remained between the two genotypes as occurred during LFD feeding. The increased weight of KO during LFD feeding and its maintenance during HFD feeding indicate a fundamental difference in energy use and/or storage compared to WT.
Similar activity, food consumption and use of carbohydrates vs fat
Whole-body oxygen consumption during ad lib feeding and fasting in two independent groups of males at 4 or 8 weeks old varied relative to mouse weight, but not by genotype, indicated by genotypic slopes that did not differ significantly (Fig. 1e, f) [61]. LFD-fed KO consumed amounts of calories comparable to WT (Supplemental Fig. 1a). Activity levels did not differ between KO and WT during fasting or ad lib feeding (Supplemental Fig. 1b). The respiratory exchange ratio (CO2/O2) did not differ between WT and KO (Supplemental Fig. 1c). These data indicate that neither activity levels nor use of carbohydrate vs fat as fuel differed significantly between KO and WT. Thus, these variables do not make major contributions to the mechanisms that cause the phenotype.
Decreased core body temperature in KO
Despite the insulating value of additional adipose, male KO fed a LFD have an average body temperature ~ 0.8 °C lower than WT under normal vivarium conditions (Fig. 1g). KO also had a serum T3 level decreased by 16% relative to WT after re-feeding (Fig. 1h). The decrease in T3 would reflect a decrease in DIO2, the enzyme that converts T4 into T3, an activator of BAT metabolism. The decrease in body temperature reflects a decrease in BAT activity under ambient (basal) conditions, whereas the decrease in T3 after re-feeding would contribute to a decrease in diet-induced thermogenesis.
KO develop glucose intolerance and insulin resistance
Increased adiposity often accompanies other risk factors of metabolic syndrome, such as glucose intolerance, insulin resistance, dyslipidemia, and adipose inflammation. As assessed by tolerance tests, young (age 3.5–5 weeks old) KO have normal glucose (Supplemental Fig. 2a), insulin (Supplemental Fig. 2b), and pyruvate tolerances (measure of gluconeogenesis, Supplemental Fig. 2c). Young KO also have normal fasting and re-fed insulin levels (Fig. 1i, left and right). Beginning at 8 weeks old, KO insulin levels increased 2.5-fold (Fig. 1i, middle) and by ~ 4 months old, KO develop glucose intolerance (Fig. 1j). Hepatic insulin resistance also occurs, as shown by increased blood glucose 60 min after an insulin dose (Fig. 1k).
Serum androgens and fatty acids
RDH1 recognizes the steroid 5α-androstane-3α,17β-diol as substrate in vitro—a precursor to dihydrotestosterone, in addition to retinol [62]. Therefore, we measured serum dihydrotestosterone and testosterone in adult mice, but observed no differences between genotypes (data not shown), eliminating a major change in androgens as contributing to the phenotype. To determine whether RDH1 affects circulating lipid levels, we measured FFA, glycerol, and TAG in 7- to 8.5-week-old and 29- to 31-week-old mice during fasting, ad lib feeding, and/or re-feeding (Supplemental Table 1). Neither younger nor older mice had statistically significant changes in these three metabolites, suggesting that WAT hypertrophy maintained normal blood levels of lipids.
Inflammation
We also tested systemic and BAT inflammation in 7- to 8.5-week-old mice. Serum TNFα, monocyte chemoattractant protein-1 (MCP-1), and interleukin 6 (IL-6) did not increase significantly in KO (Supplemental Table 2). Imaging of the macrophage marker F4/80 revealed no significant increase in KO BAT inflammation relative to WT (Supplemental Fig. 3a, b). Further, mRNA of macrophage expression markers Intg, Itgam (Mac-1), Arg1, Soc2 and Nos2 did not increase significantly in KO relative to WT (Supplemental Fig. 3c). These data indicate that inflammation does not make a major contribution to the phenotype.
Tissue expression levels of Rdh1
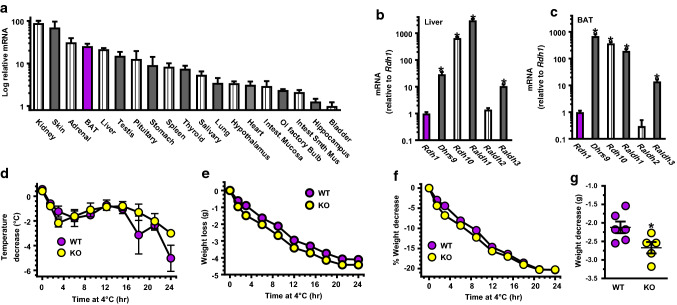
Rdh1 mRNA levels ranged through two orders of magnitude in the 24 tissues examined, with kidney and skin having the highest expression (Fig. 2a). Notably, BAT and liver were among the tissues that express Rdh1 mRNA more intensely than most other tissues examined. qPCR did not amplify Rdh1 mRNA in eWAT, pancreas, skeletal muscle, cerebellum, or cortex. These data draw attention to BAT as contributing to the KO phenotype.
Fig. 2.
Rdh1 mRNA expression and its effects on cold tolerance. a Relative Rdh1 mRNA expression levels in tissues of 4-week-old WT mice (n = 3 mice). Rdh1 mRNA was not detected in pancreas, eWAT, skeletal muscle, cerebellum, or brain cortex by qPCR. b mRNA levels relative to Rdh1 in liver (n = 10). c mRNA levels relative to Rdh1 in BAT (n = 7). Liver and BAT data were obtained from 4- to 5-week-old WT mice: *p ≤ 0.0005 from Rdh1. d Decreases in temperature with time at 4 °C in 5- to 7-week-old fasted mice (n = 6 WT, 5 KO). e Decreases in body weight: two-way ANOVA genotype p = 0.0001, time p = 0.0001. f % Body weight change: time from 0 to 12 h, Two-way ANOVA p = 0.0009 (genotype). g Degree of body weight decreases after 9-h exposure to 4 °C: *p < 0.04
We compared Rdh1 mRNA expression relative to other Rdh and Raldh in liver (Fig. 2b) and BAT (Fig. 2c). Liver expresses Dhrs9, Rdh10, Raldh1 (Aldh1a1) and Raldh3 (Aldh1a3) much more intensely than Rdh1 (Supplemental Table 3). BAT also expresses Dhrs9, Rdh10, Raldh1, and Raldh3 more intensely than Rdh1. Liver and BAT express Rdh1 and Raldh2 (Aldh1a2) at similar levels. These data reveal that Rdh1 mRNA levels are similar to the mRNA levels of another enzyme crucial to atRA biosynthesis (RALDH2) [63], and indicate that mRNA levels do not reflect significance of a retinoid metabolon enzyme to atRA biogenesis or function.
KO defend body temperature under stress by burning more fat than WT
Decreased body temperature, relatively high Rdh1 mRNA in BAT, and undetectable Rdh1 mRNA in WAT and muscle, drew our attention to BAT. KO seemed as capable as WT to defend body temperature when exposed to 4 °C for 24 h (Fig. 2d). KO, however, did so at the expense of losing more weight than WT (Fig. 2e). Thereafter, the rate of weight loss relative to initial body weight did not exceed that of WT (Fig. 2f). Overall, in the first 9 h of cold exposure, KO lost ~ 0.55 g more weight than WT (Fig. 2g).
Abnormal atRA homeostasis in KO BAT
Fasting and re-feeding cycles regulate Rdh1 and Rdh10 mRNA and atRA concentrations in liver [64]. Therefore, we quantified retinoid concentrations in BAT as a function of energy status and genotype. Neither retinol nor retinyl esters differed as a function of fasting vs re-feeding or with genotype (Supplemental Fig. 4). Two h after re-feeding, however, atRA begins to increase in WT and reached ~ twofold higher than fasted levels by 4 h (Fig. 3a). By 6 h after re-feeding, atRA returned to fasted levels. In contrast to WT, atRA did not increase in KO BAT upon re-feeding, but remained at fasted levels (Fig. 3b). This demonstrates a crucial contribution of RDH1 specifically to the increase in atRA after re-feeding.
Fig. 3.
Retinoid metabolism in KO BAT. Data were obtained after a 16-h fast (time 0) or at the times indicated after re-feeding. a atRA concentrations in 9- to 10-week-old WT mice (n = 6–8): 0 vs 4 h, *p < 0.008. b Relative atRA levels in 4- to 10-week-old mice. Data were pooled from two independent experiments (16–18 mice/point): WT 0 vs 4 h, **p < 0.0001; KO vs WT at 4 h re-fed, #p < 0.002. Boxes indicate 25–75th percentiles. Whiskers extend from minimum to maximum values with lines at medians. c–f mRNAs in 4- to 8-week-old mice (0 h, 9–27 mice; 4 h, 4–5 mice; 6 h, 11–26 mice). c WT Rdh1 mRNA vs. time, 0 h, *p < 0.0004, **p < 0.0001. dRdh10 mRNA. eDhrs9 mRNA: WT vs KO at 0 h, #p < 0.04; KO 0 h vs 6 h, **p < 0.02. fRaldh1 mRNA: WT vs KO at 0 h, #p < 0.03; both WT and KO 0 h vs 4 h, **p < 0.0004; both WT and KO 0 h vs 6 h, ***p ≤ 0.0001. gRaldh2 mRNA. hRaldh3 mRNA: WT vs KO 0 vs 6 h, #p < 0.0001. iCyp26b1 mRNA: WT vs KO at 0 h, #p = 0.0006; WT 0 vs 4 h, **p < 0.09; KO 0 vs 4 h, *p < 0.02; KO 0 vs 6 h, ***p < 0.0001. j mRNA expression of retinoid-binding proteins after 6-h re-feeding, relative to expression after a 16-h fast. Rbp1 (encodes Crbp1); Rbp2 (encodes Crbp2); Rbp7 (encodes Crbp3): no significant differences between WT vs KO, but all three showed feeding effects by two-way ANOVA. Boxes indicate 25–75th percentiles. Whiskers extend from minimum to maximum values with lines at medians
Fasting and re-feeding exerted gene-specific effects on the mRNA of enzymes that catalyze retinoid metabolism in WT BAT (summarized in Supplemental Table 4). Compared to fasting, Rdh1 mRNA declined ~ 80% 4–6 h after re-feeding (Fig. 3c). Neither Rdh10 nor Dhrs9 changes significantly in the fasting/re-feeding cycle in WT BAT (Fig. 3d, e). Raldh1 mRNA decreased ~ 66% after re-feeding (Fig. 3f). Raldh2 mRNA does not vary with fasting and re-feeding (Fig. 3g). Raldh3 mRNA decreased ~ 53% after re-feeding (Fig. 3h). The mRNA encoding the atRA catabolic enzyme CYP26b1 deviated from the biosynthetic enzymes by increasing 76% 4 h after re-feeding, but returned to fasting levels 6 h after re-feeding. Neither Cyp26a1 nor Cyp26c1 mRNA was detected by qPCR in BAT during fasting or 6 h after re-feeding.
Rdh1-ablation resulted in gene-specific effects on retinoid metabolon enzymes. Rdh10, Raldh2 and Raldh3 mRNA were not affected by Rdh1 ablation (Fig. 3d, g, h). Dhrs9 mRNA in KO after 16-h fasting increased 45% relative to WT, but 6 h after re-feeding decreased 33% from its peak at 0 h and did not differ significantly from WT (Fig. 3e). Raldh1 mRNA increased 31% in KO after fasting, but did not differ from WT 6 h after re-feeding (Fig. 3f). Rdh1-ablation caused a 46% decrease in Cyp26b1 after fasting relative to WT. Cyp26b1 mRNA in KO increased two to threefold 6 h after re-feeding, such that it did not differ from WT (Fig. 3i).
The totality of these data reveal that RDH1 contributes to the fasting pool of atRA, as indicated by compensatory increases in the synthetic Dhrs9 and Raldh1, and a decrease in the catabolic Cyp26b1. RDH1, however, seems to be solely responsible for the increase in atRA after re-feeding.
Ablation of Rdh1 did not change mRNA expression of the retinol-binding proteins CRBP1, CRBP2 and CRBP3, encoded by Rbp1, Rbp2 and Rbp7, respectively (Fig. 3j).
RDH1 regulates lipid metabolism in BAT
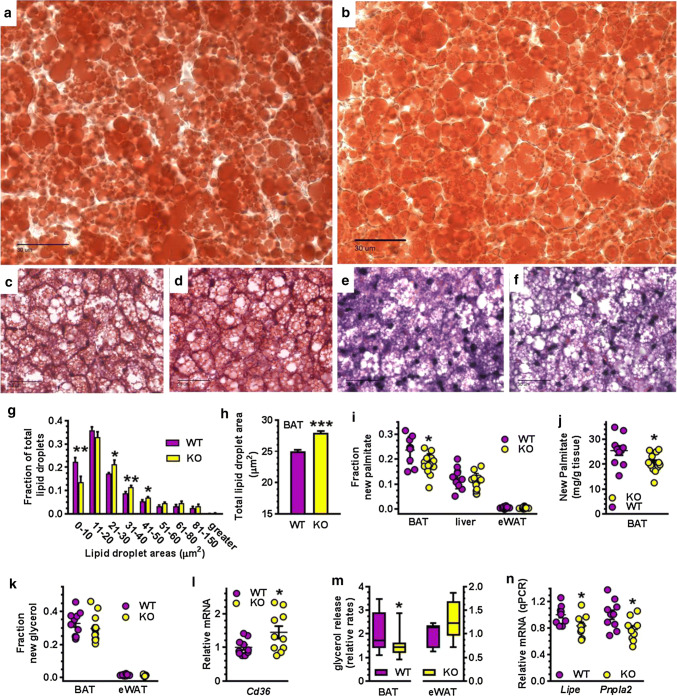
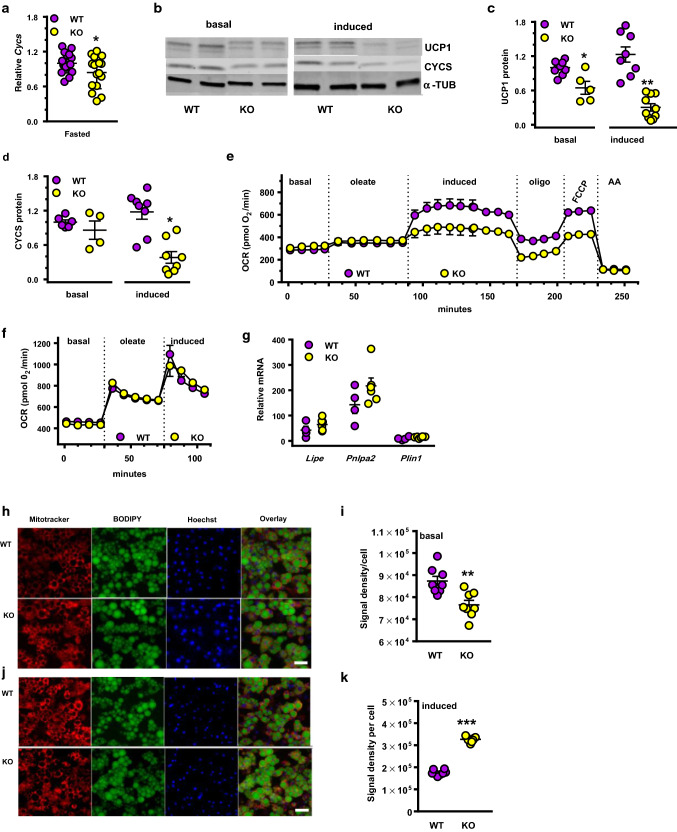
Lipid droplet (LD) sizes in BAT from fasted KO shifted away from smaller (0–10 µm2) toward medium (21–50 µm2) LD (Fig. 4a–g) compared to WT. This shift increased the total LD area in KO BAT ~ 12% (Fig. 4h), with no change in the number of LD, signifying an increase in lipids relative to WT. Increased lipid storage can result from increased fatty acid uptake, increased lipogenesis, and/or decreased lipolysis. We used mass isotopic dilution analysis to measure tissue fatty acid synthesis and TAG turnover rates [41]. During 5- to 6-h re-feeding, KO BAT made 20–25% less newly synthesized palmitate, either as a fraction of the total pool (Fig. 4i) or as mg palmitate/g tissue (Fig. 4j). Newly synthesized palmitate was unchanged in liver and eWAT. The fraction of labeled glycerol hydrolyzed in situ and extracted from the lipid pool did not differ between WT and KO, or between BAT and eWAT, suggesting similar rates of newly synthesized TAG (Fig. 4k). Therefore, increased fatty acids stored as TAG in KO BAT derive from increased fatty acid uptake and/or decreased mobilization of in situ stores. Accordingly, mRNA expression of the lipid uptake protein CD36 in KO BAT was 1.8-fold higher than WT during re-feeding, suggesting an increased rate of fatty acid uptake (Fig. 4i). Isoproterenol-induced lipolysis in BAT explants from fasted KO was ~ 26% lower than WT (Fig. 4m). In contrast, lipolysis in KO eWAT was not significantly different from WT; which agrees with the serum lipid measurements (Supplemental Table 1). Consistent with the decrease in BAT lipolysis, mRNAs of hormone sensitive lipase (Lipe) and ATGL/Desnutrin (Pnpla2) were 25–30% lower in KO than WT during fasting (Fig. 4n). These data are consistent with an increased rate of fatty acid uptake and a decreased rate of lipolysis contributing to increased intracellular lipids in KO BAT.
Fig. 4.
Increased lipid storage and decreased lipolysis in KO BAT. Representative images of BAT from fasted males. a WT oil red O. b KO oil red O. c WT H&E. d KO H&E. e WT trichrome. f KO trichrome. Scale bars in all = 30 µm. g Relative frequency distribution of BAT LD sizes. h Total LD areas in 6-week-old fasted male mice (n = 5 WT, 6 KO, 6–8 sections per mouse, > 5700 droplets per genotype). Representative of two independent experiments. i Fraction of newly synthesized palmitate relative to total palmitate after 5–6-h re-feeding in tissues of 6- to 8-week-old male WT (n = 10) and KO (n = 10–13). j Newly synthesized palmitate in BAT of mice described in i. k Fraction of newly synthesized glycerol after 5- to 6-h re-feeding in BAT and eWAT (n = 10 WT, 13 KO). l Expression of Cd36 in BAT from re-fed 4- to 8-week-old mice (n = 10). m Isoproterenol-stimulated lipolysis rates of BAT (n = 16 WT, 19 KO) and eWAT (n = 6 WT, 5 KO) in explants from 5-week-old fasted mice. n Relative expression of Lipe (HSL) and Pnpla2 (ATGL/Desnutrin) in BAT from fasted 4- to 8-week-old mice (n = 9 to 10). mRNA values were normalized to β-actin and to WT. All graphs: *p < 0.05, **p < 0.001, ***p < 0.0001 vs. WT
RDH1 does not affect stromal vascular fraction (SVF) cell differentiation
SVF cells isolated from KO BAT differentiated into brown adipocytes to a similar extent as WT (Supplemental Fig. 5). mRNA levels of genes that direct brown adipocyte differentiation remained the same in WT and KO throughout differentiation, including Pparg, Prdm16, Zfp516 and Zfp521. Expression levels of Ucp1 and Cidea also did not change.
RDH1 regulates lipid homeostasis in mature brown adipocytes
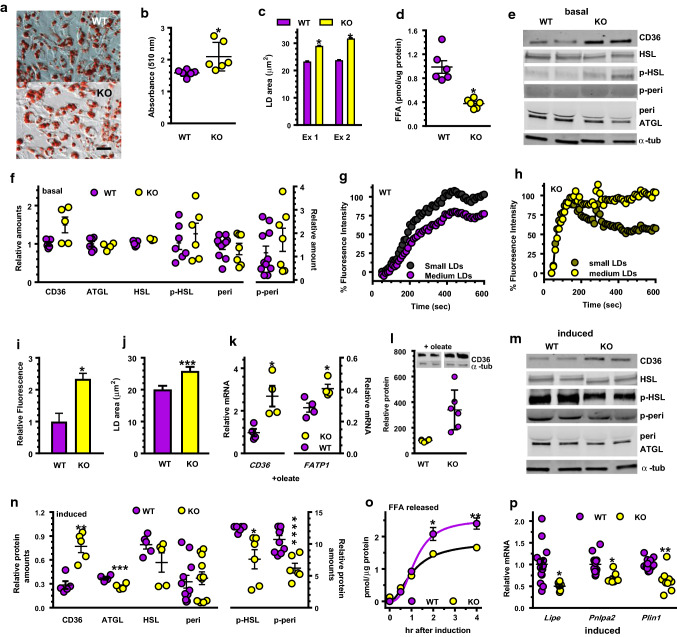
Lipid accumulation in primary brown adipocytes differentiated from KO SVF cells increased 20% relative to WT, measured by quantification of oil red O staining (Fig. 5a, b). LD in these adipocytes was 25–35% larger than that in WT (Fig. 5c). KO adipocytes during basal lipolysis (serum free, no isoproterenol or oleate in the medium) released only 38% of the FFA released by WT cells (Fig. 5d). Cell uptake contributed to the decreased medium FFA, indicated by a ~ 50% increase in CD36 (mRNA and protein) in KO brown adipocytes compared to WT (Fig. 5e, f). In differentiated cells, p-HSL, perilipin, p-perilipin, and ATGL did not differ between KO and WT, under basal conditions.
Fig. 5.
Lipid homeostasis in primary brown adipocytes differentiated from SVF cells. a Oil red O staining on dd8 done on cells in medium without added oleate: scale bar = 50 µm. b Quantitation of oil red O staining in a; n = 6 wells/genotype: *p < 0.05 c Average LD sizes in cells cultured in medium without added oleate. Data represent two independent experiments (Ex): *p < 0.0001. d FFA released into the medium from cells cultured 48 h: *p < 0.002 (n = 6). e Representative western blots during basal lipolysis (no serum, no isoproterenol). f Quantitation of western blots in d: n = 5–11 wells/genotype. g Rate of BODIPY uptake normalized to maximum WT uptake: n = 13–20. h Rate of BODIPY uptake normalized to maximum KO uptake: n = 17–20. i Relative BODIPY uptake in KO vs WT, n = 8: *p < 0.0001. j LD sizes in adipocytes treated 24 h with 400 µM oleate: n = 2000 LD, ***p < 0.0007. kCD36 and Fatp1 mRNA expression (n = 4 wells/genotype) after 24-h incubation with 400 µM oleate: *p < 0.04. l Representative western blots and quantitation of CD36 after 24-h incubation with 400 µM oleate: n = 6 wells/genotype, *p < 0.02. m Representative western blots after 4 h isoproterenol induction. n Quantification of western blots in m: 5–10 wells/genotype; *p < 0.03; **p < 0.002; ***p < 0.007; ****p = 0.0008. o Time-course of isoproterenol-induced lipolysis, measured by FFA in the medium (n = 6): *p < 0.03; **p < 0.004. p Relative mRNA expression during isoproterenol induction (mean ± SE of two experiments, n = 6–16 total): *p < 0.002; **p < 0.0001
We used the fatty acid analog BODIPY to assess rates of fatty acid uptake. Primary WT brown adipocytes incorporated BODIPY in two phases, which differed depending on LD size (Fig. 5g). For small LD, the rate in the first 200 s was 0.42 units/s. The rate from 200 to 400 s was 0.19 units/s. For medium-size LD, the rate in the first 200 s was 0.28 units/s and from 200 to 400 s was 0.17 units/s. Primary brown adipocytes from KO had one relatively rapid phase with an uptake rate of 1 unit/s for small LD and 0.9 unit/s for medium-size LD, which lasted until 140 s (Fig. 5h). Thus, brown adipocytes from KO have an initial saturation rate for fatty acid uptake ~ threefold faster than WT (140 vs 400 s). In addition, total uptake in KO exceeded that of WT by 2.3-fold (Fig. 5i). LD in brown adipocytes from KO was 25% larger than WT 24 h after treatment with oleate (Fig. 5j). This seems the result of a ~ 2.8-fold increase in CD36 mRNA and a 42% increase in Fatp1 mRNA in KO (Fig. 5k) and a fourfold increase in CD36 protein after 24-h incubation with oleate (Fig. 5l). KO also had a 28% decrease in the isoproterenol-induced amount of HSL, a 28% decrease in ATGL, a 39% decrease in p-HSL and a 42% decrease in p-perilipin 1 (Fig. 5m, n). Initial rates of isoproterenol-stimulated FFA release were similar, but after 2 h, KO medium had ~ 26% lower FFA concentration than WT (Fig. 5o). The decreased rate of lipolysis corresponded to decreases in the mRNAs of Lipe (encodes HSL), Pnlpa2 (encodes ATGL), and Plin1 (encodes perilipin) (Fig. 5p). Larger LD in KO, therefore, result from an increased rate of FFA uptake and a decreased rate of isoproterenol-induced lipolysis, without major differences in transcriptional regulation of differentiation.
Mitochondrial function in primary brown adipocytes
Lipolysis in brown adipocytes provides fuel for thermoregulation via UCP1 in mitochondria. BAT from fasted KO mice had 16% lower mRNA expression of the essential mitochondria gene cytochrome C (Cycs) compared to WT, indicating impaired mitochondria function (Fig. 6a). In primary cultures differentiated from BAT SVF cells, Ucp1 mRNA did not differ between WT and KO (Supplemental Fig. 5). UCP1 levels in KO cells, however, were 35% lower than WT under basal conditions and 76% lower after isoproterenol induction (Fig. 6b, c). CYCS protein was ~ 68% lower in KO cells after 4-h isoproterenol induction compared to WT (Fig. 6d).
Fig. 6.
Mitochondrial respiration and induced lipolysis. aCycs mRNA in BAT of fasted mice: n = 18 WT and 16 KO: *p = 0.056 relative to WT. b Representative western blots of UCP1 and CYCS from primary brown adipocytes during basal conditions or isoproterenol induction. c Quantitation of UCP1 western blots (n = 5–7 wells): *p < 0.04, **p < 0.0001 relative to WT. d Quantification of CYSC western blots (4–8 wells): *p < 0.0001. e Mitochondrial respiratory rates in primary adipocytes: basal (rate with no added oleate); oleate (70 µM oleate added); induced (isoproterenol added); oligo (inhibition of ATP-synthase by oligomycin); FCCP (treated with FCCP to measure uncoupler-induced respiration); AA (total inhibition of respiration by antimycin A inhibiting complex III). Two-way ANOVA from 94 through 225 min: p < 0.0001 genotype. f Primary brown adipocytes were pretreated with 100 nM of the RAR pan-agonist TTNPB 18 h prior to assay. g mRNA expression of lipolysis genes Lipe, Pnlpa2 and Plin1 after TTNPB treatment described in f (5–6 wells). h–k Semi-quantitative analysis of mitochondrial membrane potential in primary adipocytes (dd10) during basal and induced lipolysis: MitoTracker™ Red FM (red), C1-BODIPY-C12 (green), and Hoechest 33342 (blue): scale bar = 25 µm. h Representative images of MitoTracker™ labeling after 4 h of basal lipolysis (no serum in medium). i Quantification of MitoTracker™ fluorescent signal presented in h (8–9 images per genotype, 1000 adipocytes total): **p < 0.003. j Representative images of MitoTracker™ labeling after 4-h isoproterenol induction (-serum, +isoproterenol added). k quantification of MitoTracker™ fluorescent signal presented in j (8 images per genotype, 1000 adipocytes total): ***p < 0.0001
We analyzed mitochondrial respiration rates using oleate as the main substrate (Fig. 6e). No differences emerged between KO and WT during basal and oleate-supported metabolism. KO cells had a 28% lower isoproterenol-induced respiration rate and 37% lower UCP1-directed uncoupled respiration in the presence of the ATP-synthase inhibitor oligomycin. KO adipocytes also had 33% lower maximally stimulated respiration (FCCP). Pre-treatment 18 h with the RAR pan-agonist TTNPB restored mitochondria respiration in KO adipocytes to the same levels as WT (Fig. 6f); and also restored Lipe, Pnlpa2 and Plin1 mRNA to WT levels (Fig. 6g). We used MitoTracker™ Red FM, a fluorescent mitochondria dye sensitive to the inner mitochondrial membrane potential, to image mitochondria in live primary brown adipocytes (Fig. 6h–k). During basal lipolysis (serum-free medium with 5.56 mM glucose and 5% BSA to promote efflux of endogenous FA) the MitoTracker™ Red FM fluorescent signal was 13% lower in KO cells than WT, reflecting decreased mitochondrial membrane potential. Four h of isoproterenol stimulation increased the MitoTracker fluorescent signal by 46% in KO compared to WT cells. Decreases in UCP1 and CYCS, and alterations in mitochondrial membrane potential, therefore, underlie the impaired phenotype of KO.
RNAseq analyses
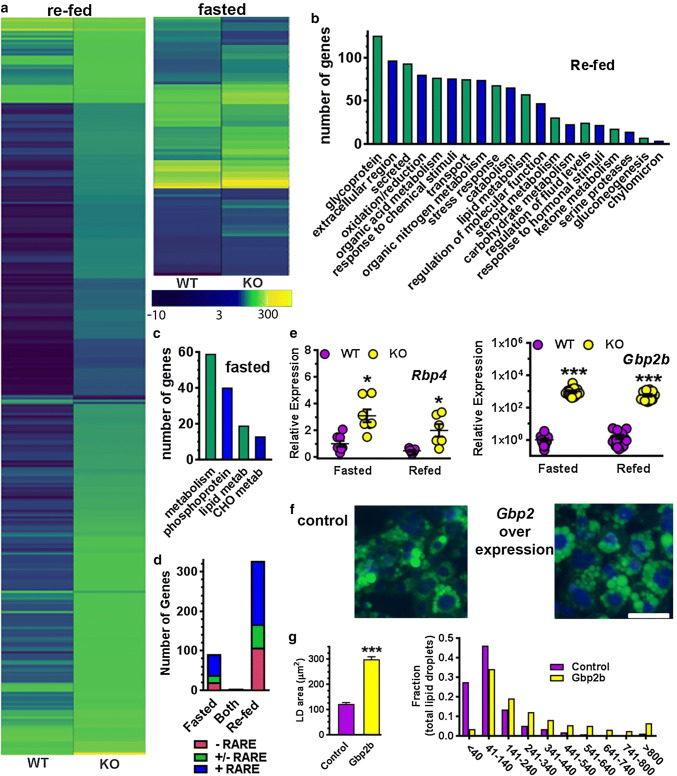
Rdh1-loss exerted a remarkable impact on the BAT transcriptome, which mostly segregated into changes after re-feeding vs changes during fasting (Fig. 7a). Only 4 genes in KO differed from WT during both fasting and re-feeding (Supplemental Table 5). Three hundred and twenty-eight genes differed in expression after re-feeding (Supplemental Table 6), whereas 92 genes differed during fasting (Supplemental Table 7). Rdh1-loss altered expression of the transcription factors Hnf3 g (Fox3a), Nr1h4 (FXR), HNF4α and Creb313 after re-feeding, and Fos, Atf3, Nr4a1 (Nurr77), Nr4a3 (Nor1), Mixipl (ChREBP) and Egr during fasting. Genes that encode proteins of the extracellular region and other secreted proteins differed from WT extensively (Fig. 7b, c). These included Ithi1-4 (heavy chains of the inter α-trypsin inhibitors), Vtn (vitronectin), and Fga, Fgb, and Fgg (fibrinogens). Extensive differences also occurred in genes of intermediary metabolism, including Gyk (glycerol kinase), Gcgr (glucagon receptor), Gck (glucokinase), Pklr (pyruvate kinase), Slc2a2 (Glut2), Acly (ATP citrate lyase) and Dio2 (deiodinase 2). Lep (leptin) expression decreased, despite increased adiposity. Rdh1-loss also triggered changes in genes associated with obesity, including the oxidoreductase Cyp2e1 and the neutral carboxylesterases Ces1b,c,g, Ces2a,e, Ces3a,b, and Acot11 (acyl-CoA thioesterase 11).
Fig. 7.
RNAseq analysis of differentially expressed genes in BAT. a Heat map diagrams of clustered genes with significant differences in expression from mice fasted 16 h and re-fed 6 h (328 genes) or fasted 16 h (92 genes). The majority of differentially expressed genes were upregulated in KO mice (101 after re-feeding and 63 during fasting). b GO analysis of differentially expressed genes during re-feeding confirms KO effects on metabolic processes such as oxidation/reduction, lipid, carbohydrate and steroid metabolism, and reveals new targets in extracellular matrix remodeling, BAT secretory pathways, and chylomicron particle uptake. c GO analysis of differentially expressed genes fasting confirms KO effects on phosphorylation signaling and metabolism. d Distribution of differentially expressed BAT genes according to the presence of RARE. e Verification of Rbp4 expression by qPCR (2 experiments): *p < 0.03. Up-regulation of guanylate binding protein 2b (Gbp2b) in KO BAT regardless of feeding status (4 experiments): ***p < 0.0001. f Representative images of control (left) and Gbp2b overexpressing (right) immortalized cultured brown adipocytes stained with BODIPY 493/503 and DAPI: scale bar = 50 µm. g Average LD areas (left) and relative frequency distribution of LD sizes (right) in the GFP control (n = 959 cells) vs. Gbp2b overexpressing brown adipocytes (n = 912 cells): ***p < 0.0001
Sixty % of genes that differed during fasting had confirmed RARE, and another 20% had putative RARE (Fig. 7d) [65–68]. Forty-nine % of genes that differed after re-feeding contained RARE, and 18% had putative RARE. We confirmed RNAseq results for Rbp4 and Gbp2b by qPCR, in part because these two deviated from WT during both re-feeding and fasting (Fig. 7e).
GBP2B function in a brown adipocyte cell line
Expression of the GTPase Gbp2b differed the most of any gene in KO and differed during both fasting and re-feeding (200- to 400-fold increase by RNAseq and 580- to 960-fold increase by qPCR). Therefore, we overexpressed Gbp2b in immortalized mouse brown adipocytes (Fig. 7f, g). Gbp2b-expressing cells accumulated LD with an overall shift to larger sizes and an average area ~ 2.4 greater than control cells. Overexpression of Gbp2b did not affect differentiation, indicated by unchanged mRNA expression of Pparg, Cebp/α and Prdm16, which drive brown adipocyte differentiation, and Ap2, which reflects adipocyte differentiation.
Discussion
Here, we show that adaptations in endogenous atRA concentrations during fasting and re-feeding in BAT, generated through RDH1 activity, regulate > 400 genes, extensively divided between either fasting or feeding. These gene changes affect BAT function by attenuating lipid uptake, supporting efficient lipolysis, and enabling optimal mitochondrial performance. These effects defend body temperature and restrain whole-body adiposity and its metabolic sequelae. The significant impact of RDH1 occurs despite its low expression level, relative to the other enzymes that contribute to the retinoid metabolon, except for RALDH2. Compensation by the increases in Dhrs9 and Raldh1 and the decrease in Cyp26b1 in KO during fasting mitigates loss of Rdh1. These compensations do not occur after re-feeding in KO, resulting in a lower than normal atRA concentration. The compensation shows that RDH1 contributes to atRA concentrations during fasting and seems solely responsible for the increases in atRA after feeding. These data also indicate that an increase in atRA after feeding is essential to optimum BAT thermogenesis.
Mouse models of obesity usually use diets copious in vitamin A (chow diets) consisting of high fat to induce adiposity [6–8]. The plentiful amount of vitamin A in a chow diet suppresses adipogenesis, partially reducing the effects of a HFD, which stimulate adipogenesis. Rescue of the KO phenotype by a chow diet illustrates normalization of weight gain by copious dietary vitamin A [27].
BAT functions as an exceptionally metabolically active thermogenic tissue in humans and other mammals. In humans, BAT metabolism limits adiposity [69–71]. The 40–60 g of BAT in a lean human may prevent up to a 9-pound increase in body weight per year, and if maximally stimulated, as many as 44 lb per year [72, 73]. Studies of human BAT ex vivo have shown similarities with constitutive rodent BAT [74]. The adrenergically responsive UCP1 operates as one, but not the only, thermogenic uncoupler in BAT [75]. UCP1 performance relies on the fatty acid concentration and fully functional mitochondria. Fatty acids activate UCP1 allosterically, and provide fuel to drive the proton electromotive gradient. Mitochondria oxidize the fatty acids to form the proton gradient dissipated by UCP1 and other uncouplers to generate heat. Primary BAT adipocytes lacking Rdh1 had a greater than 50% decrease in UCP1 protein during isoproterenol stimulation. This would reduce the OCR in KO to about half of WT cells, reflecting a large decrease in fatty acid oxidation. In addition, decreased CYCS and changes in membrane potential revealed dysfunctional mitochondria. The decrease in membrane potential under basal conditions would reflect impaired proton extrusion, whereas increased membrane potential during adrenergic stimulation would reflect decreased UCP1 activity. Thus, RDH1 generates atRA as a component vital to normal thermogenesis.
Decreased fatty acid use to safeguard body temperature by KO evidently diverts fatty acids from BAT to expand WAT, despite increased fatty acid uptake by BAT. WAT expansion leads to glucose intolerance, insulin insensitivity, and hyperinsulinemia—aspects of metabolic syndrome. Cold challenge overrides Rdh1 loss. KO mice resist a decrease in body heat during severe cold at the expense of greater weight loss than WT, i.e., they overcome the predisposition to store fatty acids in WAT that BAT normally would use. Lack of detectable differences in daily global energy expenditure by KO probably reflects alterations that escape quantification in the short term, but are significant cumulatively.
The atRA increase during 0- to 4-h re-feeding would induce Cyp26b1 mRNA and decrease the mRNAs of the biosynthetic enzymes, which by the time the atRA concentration reaches its peak, would have completed the enzymological adaptation to reduce atRA biosynthesis. This decrease in atRA as a function of feeding would amplify the effect of the increase in insulin, as atRA and insulin have opposing effects on gluconeogenesis and de novo lipogenesis [76, 77].
Serum retinol and atRA concentrations do not represent tissue retinoid levels, limiting studies of physiological retinoid effects on obesity [10, 78, 79]. The conclusions that vitamin A and atRA reduce adiposity have been supported by dosing vitamin A and atRA in higher amounts to animals that already have copious levels of vitamin A [6, 8, 80]. Although this approach assesses a potential clinical use (or toxic effect) of retinoids, it does not convey insight into physiological atRA functions. This ensues because of hormesis. Even though vertebrates require atRA to live, modest decreases or increases in tissue concentrations result in impaired function or toxicity, respectively [81–84]. As a result, insight has been limited into the postnatal effects of endogenous atRA and its physiological impact on adiposity. The modest but significant decreases in atRA, provided by knockouts of retinoid metabolon proteins, such as Rdh1 and Rdh10− allow investigation of physiological atRA contributions to postnatal health. The present data also complement a report that a vitamin A-deficient diet increases the adiposity index in mice fed a HFD [80].
RNAseq data confirmed the extensive influence of RDH1 and its production of atRA on BAT cell regulation. Genes that encode enzymes of intermediary metabolism were affected, as well as transcription factors that regulate metabolism. KO also had changes in “obesity” genes, which also may contribute directly to the phenotype. Cyp2e1, which increased twofold after re-feeding, encodes an enzyme that oxidizes fatty acids: its knockout prevents diet-induced obesity [85]. Ces isozyme mRNA, which increased 9- to 43-fold during re-feeding, contribute to lipid homeostasis [86]. Dosing a CES1 and CES3 inhibitor to obese diabetic mice enhanced glucose tolerance, improved dyslipidemia and attenuated obesity [87]. Ablation of Acot11, which increased ~ 30-fold during re-feeding, protects against diet-induced obesity [88]. The 45% decrease in Dio2 expression also would contribute directly to the phenotype, as activation of T4 into T3 stimulates thermogenesis. Ablation of Dio2 results in diet-induced obesity [89]. These effects on genes associated with glucose and lipid metabolism suggest extensive RDH1 function in normalizing BAT metabolism and consequently the propensity to adiposity.
Elevated serum RBP4 acting through STRA6 provokes insulin resistance, but its function in BAT has not been reported [90]. The increase in BAT Rbp4 expression with Rdh1 loss during both the fasted and fed states implies a contribution of RDH1 to counteracting insulin resistance. This shows that physiological atRA contributes to insulin sensitivity and extends the observation that pharmacological dosing with atRA suppresses Rbp4 expression in adipose [91].
Gbp2b overexpression in brown adipocytes increased lipid content, consistent with a direct contribution to the phenotype. Gbp2b belongs to a family of GTPases that affords protection against infection by regulating cell–cell interaction of extra-cellular matrix proteins, including integrins and fibrinogen [92, 93]. The impact of GBP2B on extracellular matrix proteins seems notable, because adipocyte shape and microenvironment affect function [94, 95]. Some of the largest gene expression changes in KO included extra-cellular matrix proteins mRNAs, such as Fga, Fgg and Fgg (fibrinogens) (~ 20-fold each), Itih1,2,3,4 (10–24-fold) and Vtn (vitronectin) (~ fivefold). Itih interacts with and stabilizes the cell surface polysaccharide hyaluronan. Hyaluronan binds with cell surface receptors to activate signaling pathways [96]. Integrins target vitronectin. It is reasonable to propose that GBP2B links β-adrenergic receptor stimulation and adipocyte function through effects on extracellular matrix proteins.
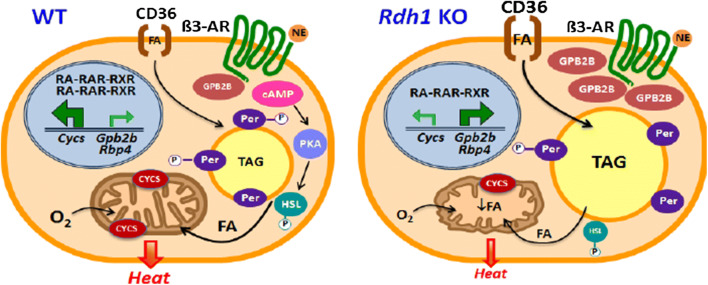
The results are summarized in the model of Fig. 8. RDH1 regulates genes controlling BAT adaptation to whole-body energy status, such as fasting and re-feeding. RDH1 contributes to the fasting atRA pools in BAT and has a unique function in increasing the atRA concentration after re-feeding. Other members of the retinoid metabolon in BAT do not substitute for the crucial increase in atRA after re-feeding. Loss of RDH1 impairs thermogenesis by decreasing lipolysis and mitochondria function. Lower body temperature and increased adiposity ensues. BAT Rbp4 expression increases in the absence of Rdh1, contributing to insulin resistance. Finally, intense expression of Gpb2b in KO and its association with increased LD offer insight into GBP2B function outside of the immune system, and suggest that RDH1 may contribute to combatting infection and/or regulating composition of the extra-cellular matrix. These data suggest that variations in Rdh1 expression or function contribute to disparities in adiposity and health.
Fig. 8.
Rdh1 supports lipid homeostasis and mitochondria function in brown adipocytes. In brown adipocytes, RDH1 generates atRA that regulates expression levels of numerous genes, which promote lipid homeostasis, mitochondria fitness, and thermogenesis. Rdh1 deletion results in increased fatty acid uptake, decreased lipolysis of FA from lipid droplets, decreased FA oxidation from impaired mitochondria function, and decreased thermogenesis. Cycs, Gpb2b and Rbp4 illustrate three genes and/or their proteins of the more than 400 affected by Rdh1 deletion. Brown adipocyte functional impairment shunts lipids to white adipose, increasing obesity, insulin resistance and glucose intolerance—contributors to metabolic syndrome. β3-AR β-androgenic receptor, Cycs cytochrome C, FA fatty acid, Gbp2b GTPase, HSL hormone sensitive lipase, Per perilipin, PKA phosphokinase A, Rbp4 serum retinol binding-protein, TAG triacylglycerol
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 2 (DOCX 52620 kb)
Acknowledgements
This work was supported by grants from the National Institutes of Health DK102014 and DK112754 (JLN), UC Berkeley College of Natural Resources Sponsored Projects for Undergraduate Research (MW), and a UC Berkeley NIH Bridges to Baccalaureate Grant R25GM095401 (CEG). CRK and KMO were supported by NIH Predoctoral Training Grant DK061918. The authors would like to thank Milena Tintcheva for KO mouse colony management and technical help with cell culture studies. Di Yang preformed immunolabeling for F4/80 in BAT. We thank Valerie Hynh, Jared Williams, and AgroSUP Dijon interns Julie Contini, Jeremy Rolland, Fabrice Orellana and Paul Snocks for assisting with microscopy, data collection and LD morphology analysis, and Julie Brayer for assistance with western blot experiments. Special thanks to Steve Ruzin and Denise Schichnes from the UC Berkeley CNR Biological Imaging Facility (BIF) for support of this project. Research reported in this publication was supported in part by the National Institutes of Health S10 program under award number 1S10RR026866-01 to UC Berkeley BIF. Maureen Kane and Jinshan Wang contributed to preliminary retinoid measurements. Matthew Bruss and Simply FlorCruz performed preliminary MIDA experiments. Mariarita Perri assisted with serum lipid measurements. Lilyana Chandra, Audrey Kim, Jane Kim, William Whang, Nicholas Bonniot, and Delphine Foucault assisted with animal studies.
Author contributions
CRK, MGV, and JLN contributed to designing the study, analyzing the data and writing the manuscript. CRK, MGV, PH, CG, CZ, CSL, KMO, JHM, CBH, MRW and ACT performed experimental work and reviewed the manuscript. SPG analyzed RNAseq data. IDS analyzed RARE data. MKH reviewed mass isotopic dilution analysis studies.
Compliance with ethical standards
Conflict of interests
The authors declare no competing interests.
Availability of data and materials
Data generated during this project are reported in this article and the supplementary information. Reagents specific to this project are available upon request from the corresponding author.
Declaration
These experiments comply with the laws of the USA.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.McLaren DS, Kraemer K. Vitamin A in health. World Rev Nutr Diet. 2012;103:33–51. doi: 10.1159/000170954. [DOI] [PubMed] [Google Scholar]
- 2.Li Y, Wong K, Walsh K, et al. Retinoic acid receptor β stimulates hepatic induction of fibroblast growth factor 21 to promote fatty acid oxidation and control whole-body energy homeostasis in mice. J Biol Chem. 2013;288:10490–10504. doi: 10.1074/jbc.M112.429852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang D, Vuckovic MG, Smullin CP, et al. Modest decreases in endogenous all-trans-retinoic acid produced by a mouse Rdh10 heterozygote provoke major abnormalities in adipogenesis and lipid metabolism. Diabetes. 2018;67:662–673. doi: 10.2337/db17-0946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alvarez R, de Andrés J, Yubero P, et al. A novel regulatory pathway of brown fat thermogenesis. Retinoic acid is a transcriptional activator of the mitochondrial uncoupling protein gene. J Biol Chem. 1995;270:5666–5673. doi: 10.1074/jbc.270.10.5666. [DOI] [PubMed] [Google Scholar]
- 5.Mercader J, Palou A, Bonet ML. Induction of uncoupling protein-1 in mouse embryonic fibroblast-derived adipocytes by retinoic acid. Obes Silver Spring Md. 2010;18:655–662. doi: 10.1038/oby.2009.330. [DOI] [PubMed] [Google Scholar]
- 6.Berry DC, DeSantis D, Soltanian H, et al. Retinoic acid upregulates preadipocyte genes to block adipogenesis and suppress diet-induced obesity. Diabetes. 2012;61:1112–1121. doi: 10.2337/db11-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeyakumar SM, Vajreswari A, Giridharan NV. Chronic dietary vitamin A supplementation regulates obesity in an obese mutant WNIN/Ob rat model. Obes Silver Spring Md. 2006;14:52–59. doi: 10.1038/oby.2006.7. [DOI] [PubMed] [Google Scholar]
- 8.Mercader J, Ribot J, Murano I, et al. Remodeling of white adipose tissue after retinoic acid administration in mice. Endocrinology. 2006;147:5325–5332. doi: 10.1210/en.2006-0760. [DOI] [PubMed] [Google Scholar]
- 9.Hayes DP. Nutritional hormesis. Eur J Clin Nutr. 2007;61:147–159. doi: 10.1038/sj.ejcn.1602507. [DOI] [PubMed] [Google Scholar]
- 10.Obrochta KM, Kane MA, Napoli JL. Effects of diet and strain on mouse serum and tissue retinoid concentrations. PLoS One. 2014;9:e99435. doi: 10.1371/journal.pone.0099435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stone RL, Bernlohr DA. The molecular basis for inhibition of adipose conversion of murine 3T3-L1 cells by retinoic acid. Differ Res Biol Divers. 1990;45:119–127. doi: 10.1111/j.1432-0436.1990.tb00465.x. [DOI] [PubMed] [Google Scholar]
- 12.Muenzner M, Tuvia N, Deutschmann C, et al. Retinol-binding protein 4 and its membrane receptor STRA6 control adipogenesis by regulating cellular retinoid homeostasis and retinoic acid receptor α activity. Mol Cell Biol. 2013;33:4068–4082. doi: 10.1128/MCB.00221-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang D, Krois CR, Huang P, et al. Raldh1 promotes adiposity during adolescence independently of retinal signaling. PLoS One. 2017;12:e0187669. doi: 10.1371/journal.pone.0187669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berry DC, Noy N. All-trans-retinoic acid represses obesity and insulin resistance by activating both peroxisome proliferation-activated receptor beta/delta and retinoic acid receptor. Mol Cell Biol. 2009;29:3286–3296. doi: 10.1128/MCB.01742-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mercader J, Madsen L, Felipe F, et al. All-trans retinoic acid increases oxidative metabolism in mature adipocytes. Cell Physiol Biochem. 2007;20:1061–1072. doi: 10.1159/000110717. [DOI] [PubMed] [Google Scholar]
- 16.Hernandez A, de Mena RM, Martin E, Obregon M-J. Differences in the response of UCP1 mRNA to hormonal stimulation between rat and mouse primary cultures of brown adipocytes. Cell Physiol Biochem. 2011;28:969–980. doi: 10.1159/000335810. [DOI] [PubMed] [Google Scholar]
- 17.Murholm M, Isidor MS, Basse AL, et al. Retinoic acid has different effects on UCP1 expression in mouse and human adipocytes. BMC Cell Biol. 2013;14:41. doi: 10.1186/1471-2121-14-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kedishvili NY. Retinoic acid synthesis and degradation. Subcell Biochem. 2016;81:127–161. doi: 10.1007/978-94-024-0945-1_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Napoli JL. Cellular retinoid binding-proteins, CRBP, CRABP, FABP5: effects on retinoid metabolism, function and related diseases. Pharmacol Ther. 2017;173:19–33. doi: 10.1016/j.pharmthera.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harrison EH. Mechanisms involved in the intestinal absorption of dietary vitamin A and provitamin A carotenoids. Biochim Biophys Acta. 2012;1821:70–77. doi: 10.1016/j.bbalip.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang S, Yu J, Jones JW, et al. Retinoic acid signaling promotes the cytoskeletal rearrangement of embryonic epicardial cells. FASEB J. 2018 doi: 10.1096/fj.201701038R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ross AC, Zolfaghari R. Cytochrome P450 s in the regulation of cellular retinoic acid metabolism. Annu Rev Nutr. 2011;31:65–87. doi: 10.1146/annurev-nutr-072610-145127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nelson CH, Buttrick BR, Isoherranen N. Therapeutic potential of the inhibition of the retinoic acid hydroxylases CYP26A1 and CYP26B1 by xenobiotics. Curr Top Med Chem. 2013;13:1402–1428. doi: 10.2174/1568026611313120004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhai Y, Sperkova Z, Napoli JL. Cellular expression of retinal dehydrogenase types 1 and 2: effects of vitamin A status on testis mRNA. J Cell Physiol. 2001;186:220–232. doi: 10.1002/1097-4652(200102)186:2<220::AID-JCP1018>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 25.Jiang W, Napoli JL. The retinol dehydrogenase Rdh10 localizes to lipid droplets during acyl ester biosynthesis. J Biol Chem. 2013;288:589–597. doi: 10.1074/jbc.M112.402883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang D, Vuckovic MG, Smullin CP, et al. Modest decreases in endogenous all-trans-retinoic acid produced by a MouseRdh10Heterozygote provoke major abnormalities in adipogenesis and lipid metabolism. Diabetes. 2018;67:662–673. doi: 10.2337/db17-0946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang M, Hu P, Krois CR, et al. Altered vitamin A homeostasis and increased size and adiposity in the rdh1-null mouse. FASEB J. 2007;21:2886–2896. doi: 10.1096/fj.06-7964com. [DOI] [PubMed] [Google Scholar]
- 28.Pradhan RN, Zachara M, Deplancke B. A systems perspective on brown adipogenesis and metabolic activation. Obes Rev. 2017;18(Suppl 1):65–81. doi: 10.1111/obr.12512. [DOI] [PubMed] [Google Scholar]
- 29.Trayhurn P. Origins and early development of the concept that brown adipose tissue thermogenesis is linked to energy balance and obesity. Biochimie. 2017;134:62–70. doi: 10.1016/j.biochi.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 30.Reeves PG, Nielsen FH, Fahey GC., Jr AIN-93 purified diets for laboratory rodents: final report of the American Institute of Nutrition ad hoc writing committee on the reformulation of the AIN-76A rodent diet. J Nutr. 1993;123:1939–1951. doi: 10.1093/jn/123.11.1939. [DOI] [PubMed] [Google Scholar]
- 31.Heikkinen S, Argmann CA, Champy M-F, Auwerx J (2001) Evaluation of glucose homeostasis. In: Current protocols in molecular biology. Wiley [DOI] [PubMed]
- 32.Kane MA, Napoli JL. Quantification of endogenous retinoids. Methods Mol Biol. 2010;652:1–54. doi: 10.1007/978-1-60327-325-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Dam AD, Nahon KJ, Kooijman S, et al. Salsalate activates brown adipose tissue in mice. Diabetes. 2015;64:1544–1554. doi: 10.2337/db14-1125. [DOI] [PubMed] [Google Scholar]
- 34.Parlee SD, Lentz SI, Mori H, MacDougald OA. Quantifying size and number of adipocytes in adipose tissue. Methods Enzymol. 2014;537:93–122. doi: 10.1016/B978-0-12-411619-1.00006-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Dam AD, Nahon KJ, Kooijman S, et al. Salsalate activates brown adipose tissue in mice. Diabetes. 2015;64:1544–1554. doi: 10.2337/db14-1125. [DOI] [PubMed] [Google Scholar]
- 36.Rowan S, Jiang S, Korem T, et al. Involvement of a gut-retina axis in protection against dietary glycemia-induced age-related macular degeneration. Proc Natl Acad Sci USA. 2017;114:E4472–E4481. doi: 10.1073/pnas.1702302114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khalil A, Cevik SE, Hung S, et al. Developmental exposure to 2,2′,4,4′-tetrabromodiphenyl ether permanently alters blood-liver balance of lipids in male mice. Front Endocrinol. 2018;9:548. doi: 10.3389/fendo.2018.00548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mackay H, Patterson ZR, Khazall R, et al. Organizational effects of perinatal exposure to bisphenol-A and diethylstilbestrol on arcuate nucleus circuitry controlling food intake and energy expenditure in male and female CD-1 mice. Endocrinology. 2013;154:1465–1475. doi: 10.1210/en.2012-2044. [DOI] [PubMed] [Google Scholar]
- 39.Wahlang B, Falkner KC, Gregory B, et al. Polychlorinated biphenyl 153 is a diet-dependent obesogen that worsens nonalcoholic fatty liver disease in male C57BL6/J mice. J Nutr Biochem. 2013;24:1587–1595. doi: 10.1016/j.jnutbio.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davidson SM, Papagiannakopoulos T, Olenchock BA, et al. environment impacts the metabolic dependencies of ras-driven non-small cell lung cancer. Cell Metab. 2016;23:517–528. doi: 10.1016/j.cmet.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turner SM, Murphy EJ, Neese RA, et al. Measurement of TG synthesis and turnover in vivo by 2H2O incorporation into the glycerol moiety and application of MIDA. Am J Physiol Endocrinol Metab. 2003;285:E790–E803. doi: 10.1152/ajpendo.00402.2002. [DOI] [PubMed] [Google Scholar]
- 42.Lee WN, Bassilian S, Guo Z, et al. Measurement of fractional lipid synthesis using deuterated water (2H2O) and mass isotopomer analysis. Am J Physiol. 1994;266:E372–E383. doi: 10.1152/ajpcell.1994.266.2.C319. [DOI] [PubMed] [Google Scholar]
- 43.Meyer CW, Ootsuka Y, Romanovsky AA. Body temperature measurements for metabolic phenotyping in mice. Front Physiol. 2017;8:520. doi: 10.3389/fphys.2017.00520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Christoffolete MA, Linardi CCG, de Jesus L, et al. Mice with targeted disruption of the Dio2 gene have cold-induced overexpression of the uncoupling protein 1 gene but fail to increase brown adipose tissue lipogenesis and adaptive thermogenesis. Diabetes. 2004;53:577–584. doi: 10.2337/diabetes.53.3.577. [DOI] [PubMed] [Google Scholar]
- 45.Schweiger M, Eichmann TO, Taschler U, et al. Measurement of lipolysis. Methods Enzymol. 2014;538:171–193. doi: 10.1016/B978-0-12-800280-3.00010-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rotondo F, Ho-Palma AC, Remesar X, et al. Glycerol is synthesized and secreted by adipocytes to dispose of excess glucose, via glycerogenesis and increased acyl-glycerol turnover. Sci Rep. 2017;7:8983. doi: 10.1038/s41598-017-09450-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith PK, Krohn RI, Hermanson GT, et al. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 48.Aune UL, Ruiz L, Kajimura S. Isolation and differentiation of stromal vascular cells to beige/brite cells. J Vis Exp. 2013 doi: 10.3791/50191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Briand N, Prado C, Mabilleau G, et al. Caveolin-1 expression and cavin stability regulate caveolae dynamics in adipocyte lipid store fluctuation. Diabetes. 2014;63:4032–4044. doi: 10.2337/db13-1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Listenberger LL, Brown DA. Fluorescent detection of lipid droplets and associated proteins. Curr Protoc Cell Biol Chapter. 2007;24:Unit 24.2. doi: 10.1002/0471143030.cb2402s35. [DOI] [PubMed] [Google Scholar]
- 51.Harjes U, Bridges E, Gharpure KM, et al. Antiangiogenic and tumour inhibitory effects of downregulating tumour endothelial FABP4. Oncogene. 2017;36:912–921. doi: 10.1038/onc.2016.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liao J, Sportsman R, Harris J, Stahl A. Real-time quantification of fatty acid uptake using a novel fluorescence assay. J Lipid Res. 2005;46:597–602. doi: 10.1194/jlr.D400023-JLR200. [DOI] [PubMed] [Google Scholar]
- 53.Tharp KM, Kang MS, Timblin GA, et al. Actomyosin-mediated tension orchestrates uncoupled respiration in adipose tissues. Cell Metab. 2018;27:602.e4–615.e4. doi: 10.1016/j.cmet.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nguyen TB, Louie SM, Daniele JR, et al. DGAT1-dependent lipid droplet biogenesis protects mitochondrial function during starvation-induced autophagy. Dev Cell. 2017;42:9.e5–21.e5. doi: 10.1016/j.devcel.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jayaraman S. Flow cytometric determination of mitochondrial membrane potential changes during apoptosis of T lymphocytic and pancreatic beta cell lines: comparison of tetramethylrhodamineethylester (TMRE), chloromethyl-X-rosamine (H2-CMX-Ros) and MitoTracker Red 580 (MTR580) J Immunol Methods. 2005;306:68–79. doi: 10.1016/j.jim.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 56.Nolan T, Hands RE, Bustin SA. Quantification of mRNA using real-time RT-PCR. Nat Protoc. 2006;1:1559–1582. doi: 10.1038/nprot.2006.236. [DOI] [PubMed] [Google Scholar]
- 57.Fink T, Lund P, Pilgaard L, et al. Instability of standard PCR reference genes in adipose-derived stem cells during propagation, differentiation and hypoxic exposure. BMC Mol Biol. 2008;9:98. doi: 10.1186/1471-2199-9-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shinoda K, Luijten IHN, Hasegawa Y, et al. Genetic and functional characterization of clonally derived adult human brown adipocytes. Nat Med. 2015;21:389–394. doi: 10.1038/nm.3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reeves PG, Nielsen FH, Fahey GC., Jr AIN-93 purified diets for laboratory rodents: final report of the American Institute of Nutrition ad hoc writing committee on the reformulation of the AIN-76A rodent diet. J Nutr. 1993;123:1939–1951. doi: 10.1093/jn/123.11.1939. [DOI] [PubMed] [Google Scholar]
- 60.Jo J, Gavrilova O, Pack S, et al. Hypertrophy and/or hyperplasia: dynamics of adipose tissue growth. PLoS Comput Biol. 2009;5:e1000324. doi: 10.1371/journal.pcbi.1000324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tschop MH, Speakman JR, Arch JRS, et al. A guide to analysis of mouse energy metabolism. Nat Methods. 2012;9:57–63. doi: 10.1038/nmeth.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang M, Chen W, Smith SM, Napoli JL. Molecular characterization of a mouse short chain dehydrogenase/reductase active with all-trans-retinol in intact cells, mRDH1. J Biol Chem. 2001;276:44083–44090. doi: 10.1074/jbc.M105748200. [DOI] [PubMed] [Google Scholar]
- 63.Niederreither K, Vermot J, Messaddeq N, et al. Embryonic retinoic acid synthesis is essential for heart morphogenesis in the mouse. Dev Camb Engl. 2001;128:1019–1031. doi: 10.1242/dev.128.7.1019. [DOI] [PubMed] [Google Scholar]
- 64.Obrochta KM, Krois CR, Campos B, Napoli JL. Insulin regulates retinol dehydrogenase expression and all-trans-retinoic acid biosynthesis through FoxO1. J Biol Chem. 2015;290:7259–7268. doi: 10.1074/jbc.M114.609313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moutier E, Ye T, Choukrallah M-A, et al. Retinoic acid receptors recognize the mouse genome through binding elements with diverse spacing and topology. J Biol Chem. 2012;287:26328–26341. doi: 10.1074/jbc.M112.361790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mazzoni EO, Mahony S, Peljto M, et al. Saltatory remodeling of Hox chromatin in response to rostrocaudal patterning signals. Nat Neurosci. 2013;16:1191–1198. doi: 10.1038/nn.3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.He Y, Tsuei J, Wan Y-JY. Biological functional annotation of retinoic acid alpha and beta in mouse liver based on genome-wide binding. Am J Physiol Gastrointest Liver Physiol. 2014;307:G205–G218. doi: 10.1152/ajpgi.00105.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chatagnon A, Veber P, Morin V, et al. RAR/RXR binding dynamics distinguish pluripotency from differentiation associated cis-regulatory elements. Nucleic Acids Res. 2015;43:4833–4854. doi: 10.1093/nar/gkv370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lowell BB, S-Susulic V, Hamann A, et al. Development of obesity in transgenic mice after genetic ablation of brown adipose tissue. Nature. 1993;366:740–742. doi: 10.1038/366740a0. [DOI] [PubMed] [Google Scholar]
- 70.Enerbäck S. Human brown adipose tissue. Cell Metab. 2010;11:248–252. doi: 10.1016/j.cmet.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 71.Kajimura S, Saito M. A new era in brown adipose tissue biology: molecular control of brown fat development and energy homeostasis. Annu Rev Physiol. 2014;76:225–249. doi: 10.1146/annurev-physiol-021113-170252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rothwell NJ, Stock MJ. Regulation of energy balance. Annu Rev Nutr. 1981;1:235–256. doi: 10.1146/annurev.nu.01.070181.001315. [DOI] [PubMed] [Google Scholar]
- 73.Virtanen KA, Lidell ME, Orava J, et al. Functional brown adipose tissue in healthy adults. N Engl J Med. 2009;360:1518–1525. doi: 10.1056/NEJMoa0808949. [DOI] [PubMed] [Google Scholar]
- 74.Cypess AM, White AP, Vernochet C, et al. Anatomical localization, gene expression profiling and functional characterization of adult human neck brown fat. Nat Med. 2013;19:635–639. doi: 10.1038/nm.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Long JZ, Svensson KJ, Bateman LA, et al. The secreted enzyme PM20D1 regulates lipidated amino acid uncouplers of mitochondria. Cell. 2016;166:424–435. doi: 10.1016/j.cell.2016.05.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang XL, Herzog B, Waltner-Law M, et al. The synergistic effect of dexamethasone and all-trans-retinoic acid on hepatic phosphoenolpyruvate carboxykinase gene expression involves the coactivator p300. J Biol Chem. 2004;279:34191–34200. doi: 10.1074/jbc.M403455200. [DOI] [PubMed] [Google Scholar]
- 77.He Y, Gong L, Fang Y, et al. The role of retinoic acid in hepatic lipid homeostasis defined by genomic binding and transcriptome profiling. BMC Genomics. 2013;14:575. doi: 10.1186/1471-2164-14-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Palmer AC, West KP, Dalmiya N, Schultink W. The use and interpretation of serum retinol distributions in evaluating the public health impact of vitamin A programmes. Public Health Nutr. 2012;15:1201–1215. doi: 10.1017/S1368980012000560. [DOI] [PubMed] [Google Scholar]
- 79.Tanumihardjo SA, Russell RM, Stephensen CB, et al. Biomarkers of nutrition for development (BOND)-vitamin A review. J Nutr. 2016;146:1816S–1848S. doi: 10.3945/jn.115.229708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ribot J, Felipe F, Bonet ML, Palou A. Changes of adiposity in response to vitamin A status correlate with changes of PPAR gamma 2 expression. Obes Res. 2001;9:500–509. doi: 10.1038/oby.2001.65. [DOI] [PubMed] [Google Scholar]
- 81.Kamm JJ. Toxicology, carcinogenicity, and teratogenicity of some orally administered retinoids. J Am Acad Dermatol. 1982;6:652–659. doi: 10.1016/S0190-9622(82)70054-4. [DOI] [PubMed] [Google Scholar]
- 82.Park SH, Gray WC, Hernandez I, et al. Phase I trial of all-trans retinoic acid in patients with treated head and neck squamous carcinoma. Clin Cancer. 2000;6:847–854. [PubMed] [Google Scholar]
- 83.Cheruvattath R, Orrego M, Gautam M, et al. Vitamin A toxicity: when one a day doesn’t keep the doctor away. Liver Transpl. 2006;12:1888–1891. doi: 10.1002/lt.21007. [DOI] [PubMed] [Google Scholar]
- 84.Benn CS, Aaby P, Arts RJW, et al. An enigma: why vitamin A supplementation does not always reduce mortality even though vitamin A deficiency is associated with increased mortality. Int J Epidemiol. 2015;44:906–918. doi: 10.1093/ije/dyv117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zong H, Armoni M, Harel C, et al. Cytochrome P-450 CYP2E1 knockout mice are protected against high-fat diet-induced obesity and insulin resistance. Am J Physiol Endocrinol Metab. 2012;302:E532–E539. doi: 10.1152/ajpendo.00258.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Quiroga AD, Lehner R. Pharmacological intervention of liver triacylglycerol lipolysis: the good, the bad and the ugly. Biochem Pharmacol. 2018;155:233–241. doi: 10.1016/j.bcp.2018.07.005. [DOI] [PubMed] [Google Scholar]
- 87.Delacroix L, Moutier E, Altobelli G, et al. Cell-specific interaction of retinoic acid receptors with target genes in mouse embryonic fibroblasts and embryonic stem cells. Mol Cell Biol. 2010;30:231–244. doi: 10.1128/MCB.00756-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang Y, Li Y, Niepel MW, et al. Targeted deletion of thioesterase superfamily member 1 promotes energy expenditure and protects against obesity and insulin resistance. Proc Natl Acad Sci USA. 2012;109:5417–5422. doi: 10.1073/pnas.1116011109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Marsili A, Aguayo-Mazzucato C, Chen T, et al. Mice with a targeted deletion of the type 2 deiodinase are insulin resistant and susceptible to diet induced obesity. PLoS One. 2011;6:e20832. doi: 10.1371/journal.pone.0020832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Berry DC, Jin H, Majumdar A, Noy N. Signaling by vitamin A and retinol-binding protein regulates gene expression to inhibit insulin responses. Proc Natl Acad Sci USA. 2011;108:4340–4345. doi: 10.1073/pnas.1011115108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mercader J, Granados N, Bonet ML, Palou A. All-trans retinoic acid decreases murine adipose retinol binding protein 4 production. Cell Physiol Biochem. 2008;22:363–372. doi: 10.1159/000149815. [DOI] [PubMed] [Google Scholar]
- 92.Kim B-H, Shenoy AR, Kumar P, et al. A family of IFN-γ-inducible 65-kD GTPases protects against bacterial infection. Science. 2011;332:717–721. doi: 10.1126/science.1201711. [DOI] [PubMed] [Google Scholar]
- 93.Laudanna C, Campbell JJ, Butcher EC. Role of Rho in chemoattractant-activated leukocyte adhesion through integrins. Science. 1996;271:981–983. doi: 10.1126/science.271.5251.981. [DOI] [PubMed] [Google Scholar]
- 94.McBeath R, Pirone DM, Nelson CM, et al. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell. 2004;6:483–495. doi: 10.1016/S1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 95.Pope BD, Warren CR, Parker KK, Cowan CA. Microenvironmental control of adipocyte fate and function. Trends Cell Biol. 2016;26:745–755. doi: 10.1016/j.tcb.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dicker KT, Gurski LA, Pradhan-Bhatt S, et al. Hyaluronan: a simple polysaccharide with diverse biological functions. Acta Biomater. 2014;10:1558–1570. doi: 10.1016/j.actbio.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 2 (DOCX 52620 kb)
Data Availability Statement
Data generated during this project are reported in this article and the supplementary information. Reagents specific to this project are available upon request from the corresponding author.