Abstract
Extracellular vesicles (EVs) have garnered significant interest in the biotechnology field due to their intrinsic therapeutic properties as well as their ability to serve as vehicles for bioactive cargo. However, the lack of an established biomanufacturing platform and limited potency of EVs in vivo remain critical bottlenecks for clinical translation. In this study, we utilized a 3D-printed scaffold-perfusion bioreactor system to assess the response of dynamic culture on extracellular vesicle production from endothelial cells (ECs). We also investigated whether ethanol conditioning, which was previously shown to enhance vascularization bioactivity of EC-derived EVs produced in standard 2D culture conditions, could be employed successfully for the same purpose in a 3D production system. Our results indicate that dynamic culture in a perfusion bioreactor significantly enhances EV production from human ECs. Moreover, the use of ethanol conditioning in conjunction with dynamic culture induces pro-vascularization bioactivity of EC-derived EVs that is correlated with increased EV levels of pro-angiogenic lncRNAs HOTAIR and MALAT1. Thus, this study represents one of the first reports of rationally-designed EV potency enhancement that is conserved between static 2D and dynamic 3D EV production systems, increasing the potential for scalable biomanufacturing of therapeutic EC-derived EVs for a variety of applications.
Keywords: extracellular vesicles, exosomes, bioreactor, scaffold, vascularization, alcohol
Graphical Abstract
1. Introduction
Extracellular vesicles (EVs) are crucial players in intercellular communication that are also being explored as therapeutic vectors for numerous applications [1,2]. The recent surge of interest in EVs as therapeutics has been fueled by reports of their status as purely biological vehicles with putative low immunogenicity, [3] high stability in circulation [3], amenability to broad cargo loading approaches [4], and ability to cross biological barriers [5,6]. Yet, as with all therapeutic platforms, EVs have shortcomings. These include potentially low potency due to limited amounts of bioactive cargo [7] as well as the lack of an established production scheme for large-scale EV generation [8].
Addressing scalable EV biomanufacturing, bioreactor systems have been successfully used to enhance EV production from multiple cell lines [9–11]. Notably, culturing HEK293T cells in a hollow-fiber bioreactor system enhanced EV yield by 40-fold compared to conventional cell culture [11]. This system was later integrated into a cGMP-compatible purification procedure for generation of engineered EVs with immunostimulatory properties [8]. While these and other reports of scalable EV production represent critical advances in the field, additional development leading to discovery of rational design parameters may enable further critical progress towards clinical translation of therapeutic EVs.
Here we present the use of a tubular perfusion bioreactor culture system with 3D-printed scaffolds to enhance EV production. 3D printing technology provides unique design opportunities to define scaffold geometries with intended therapeutic responses [12]. Moreover, computational fluid dynamic (CFD) studies can be used to simulate in vivo conditions and gain further insight into flow and shear characteristics of scaffolds with specific architectures [13]. Tuning the fluid characteristics of the bioreactor can be achieved by altering geometry (e.g. porosity, roughness, spacing, etc.) and flow characteristics [14]. Tubular perfusion systems have previously been used for large-scale cell-based therapeutic production [15,16] and are broadly amenable to controlled, reproducible examination of design parameters. In this study, we investigated the potential for increasing production of EVs from human dermal microvascular endothelial cells (HDMECs), which may be therapeutically useful in promoting vascularization for wound repair, critical limb ischemia, and other injuries or diseases [17]. Further, our group previously discovered that ethanol conditioning can improve vascularization bioactivity of EC-derived EVs via upregulation of angiogenic lncRNAs HOTAIR and MALAT1 [18]. Thus, we additionally examined whether this phenomenon – which was demonstrated in a conventional 2D cell culture system for EV generation – was compatible with enhanced EV production within a 3D platform. Our data show that perfusion bioreactor culture significantly increases the amount of HDMEC EVs generated per cell and that, despite this increase, ethanol conditioning-mediated pro-angiogenic effects on these EVs can be retained, likely via conserved upregulation of lncRNAs HOTAIR and MALAT1. Overall, this study addresses two of the most crucial limitations to EV-based therapeutics, production scalability and low potency, and describes a platform for enhancing both to promote translation into the clinic.
2. Methods
2.1. Cell culture
Human dermal microvascular endothelial cells (HDMECs) were obtained commercially from Promocell (C-12212). Cell passage was designated as P1 upon arrival from the manufacturer. HDMECs were cultured in tissue culture polystyrene flasks coated with 0.1% gelatin (30 min at 37°C) with Endothelial Growth Medium-2 (EGM2; Lonza CC-3162 or Promocell C-22111) for expansion and in EV-depleted EGM2 for experiments. EGM2 was depleted of EVs as previously described [19]. Briefly, fetal calf serum (FCS) or fetal bovine serum (FBS) provided in the EGM2 kits were centrifuged at 100,000 x g for 16 h, sterile filtered with a 0.2 μm syringe filter, and combined with other components of the kit to make EV-depleted EGM2.
2.2. Bioreactor scaffold fabrication
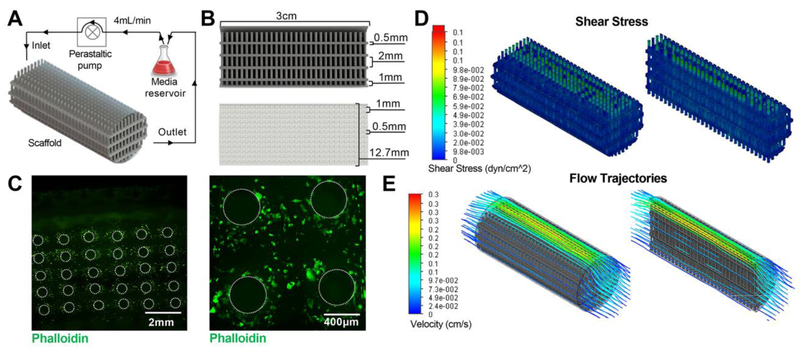
Scaffold computational models were created using SolidWorks (Dassault Systèmes, Velizy-Villacoublay, France) as shown in Figure 1A. The scaffold consists of small pillars that are 0.5 mm in size spaced 1 mm apart in a multi-layered shelf geometry. The structure has a cell growth surface area of 72 cm2 within a 0.6 cm3 volume construct. This design was chosen to provide a culture surface with slight curvature in a highly ordered pillared array. The architecture allows for flow to perfuse the circuit around and between the pillars and across the levels, facilitating active transport of nutrients and gases from gas permeable tubing, and provides a mechanism to control fluid characteristics predictably throughout. Computational fluid modeling was accomplished using the SolidWorks Flow Simulation add-in. Fluid flow was analyzed at flow rates of 2-6 mL/min by modulating the inlet volume rate with surface shear stress, flow profiles, and fluid velocity (computed and recorded).
Figure 1. Bioreactor setup and scaffold characterization.
(A) Schematic representation of perfusion bioreactor setup. Scaffold seeded with HDMECs is connected to a peristaltic pump and media reservoir using tubing. Inlet and outlet of the media circulating at 4 mL/min flow rate are noted. (B) Cross-sectioned image of the scaffold generated in Solid Works showing dimensions of the scaffold as well as individual pillar height, diameter, and spacing. (C) Confocal images of the Phalloidin-stained scaffold seeded with HDMECs after 1 day of dynamic culture. Visual representation of varying levels of (D) shear stress and (E) flow trajectories at different areas within the scaffold as evaluated by computational flow simulation are shown.
A commercially available stereolithography apparatus was used (EnvisionTEC Perfactory 4 Mini Multilens; Gladbeck Germany) to 3D-print scaffolds. Scaffold designs were exported into stereolithography (.stl) files following computational design and scaffold models were oriented, fixed, and supported using Magics 18 (Materialise, Leuven, Belgium). Solid objects were fabricated on a Perfactory 4 DSP out of E-Shell 300, a clear, biocompatible acrylate-based material (EnvisionTEC, Inc., Dearborn, MI). Excess resin was removed by submerging printed objects in 99% isopropanol (Pharmco-Aaper, Shelbyville, KY) for 5 min, followed by flowing 99% isopropanol through the scaffolds, and blowing the interior dry with air. The process was repeated until all excess material was removed from the interior of the object. Complete resin curing was achieved with 2000 flashes of broad spectrum light (Otoflash, EnvisionTEC, Inc.). Scaffolds were cleaned in 100% ethanol (Pharmco-Aaper, Shelbyville, KY) for >30 min to leach any remaining soluble contaminants. Scaffolds were disinfected under fresh 100% ethanol and UV light for 20 min. Scaffolds were rehydrated in a serial dilution of ethanol to pH 7.4 sterile PBS (principal chemicals sourced from Sigma-Aldrich St. Louis, MO) in 4 steps (75:25, 50:50, 25:75, and 0:100 ethanol:PBS), with each intermediary step allowed to soak for 5 min. Scaffolds were stored submerged in sterile PBS at 4°C until ready to use. Scaffolds were reused up to 5 times, disinfecting and rehydrating before each use.
2.3. Scaffold coating and cell seeding within bioreactor
Scaffolds were enclosed in ½” platinum cured tubing with 1-1.5 mL (sufficient for complete coverage) of 0.1% gelatin. Open ends of the tubing were closed off using binder clips to prevent leakage and scaffolds were subsequently incubated at 37°C for 30 min. After draining the scaffolds of the coating solution, both open ends were connected with ¼” tubing using ½” to ¼” connectors (Cole-Parmer, Vernon Hills. IL). To seed HDMECs on scaffolds, 1 mL of EV-depleted EGM2 containing HDMECs at passage 4 (P4) was transferred into each tubing holding the scaffolds. The ends of the tubing were clamped with a binder clip to retain media containing cells. Seeding density of experiments was 4,000 cells/cm2 for all experiments with EtOH treatment and 15,000 cells/cm2 per scaffold for all other conditions. Cells were allowed to adhere for 4 h at 37°C before fully connecting the scaffold and holder tubing to the pump; cells cultured in scaffolds under flow from the pump constituted the “Bioreactor” configuration referred to in the rest of the manuscript. Alternatively, some scaffolds were removed from the tubing after cell seeding and transferred to individual T-75 flasks (one scaffold per flask) with 50 mL of EV-depleted EGM2 for 3D scaffold static control; this constituted the “Scaffold” configuration referred to in the rest of the manuscript. In parallel, cells were also seeded in 75 cm2 tissue culture flasks at the same density as in scaffolds for each condition in EV depleted endothelial cell growth medium (EGM2; Lonza CC-3162 or Promocell C-22111) as a 2D static control (constituting the “Flask” configuration referred to in the rest of the manuscript).
2.4. Bioreactor assembly
After initial cell seeding, the scaffolds were connected to the bioreactor system as previously described [15]. Briefly, binder clips were removed and replaced with bioreactor lines as shown in Figure 1E. Both open ends of the tubing holding the scaffold were connected to 14-gauge pump head tubing (Cole-Parmer) using gas permeable 1/8” tubes in between. The 14-gauge tubing was inserted through the pump head (Cole-Parmer) attached to a Masterflex L/S Digital Drive (Cole-Parmer) to circulate medium at 4 mL/min. Open ends of the two 1/8” tubing on either end of the scaffold were fed through a 2-hole rubber stopper and affixed to a 125 mL Erlenmeyer flask containing 50 mL of EV-depleted EGM2. The inlet tube was submerged in the media reservoir and the outlet tube was situated just above the media line to avoid excessive bubble formation. The pump head allowed for up to 4 lines to be connected at a time. The assembly was then placed within a cell culture incubator at 37°C with 5% CO2, with the scaffold chamber secured on the incubator sidewall with flow anti-parallel to gravity. The complementary Flask and Scaffold controls were also incubated at 37°C with 5% CO2. Scaffolds were situated vertically to ensure complete submergence in medium.
2.5. Cell staining and imaging
After 24 h culture in the bioreactor, HDMECs were fixed in the scaffolds with 4% paraformaldehyde and 1% sucrose for 15 min and washed 3 times with PBS. Prior to staining, cells were permeabilized for 5 min with a 300 μM sucrose, 100 μM sodium chloride, 6 μM magnesium chloride, 20 μM HEPES, and 0.5% Triton-X-100 solution. Cellular actin was stained in a 1:100 dilution of AlexaFluor 488 Phalloidin in PBS for 20 min and visualized on a Nikon Eclipse Ti2 microscope.
2.6. EV collection
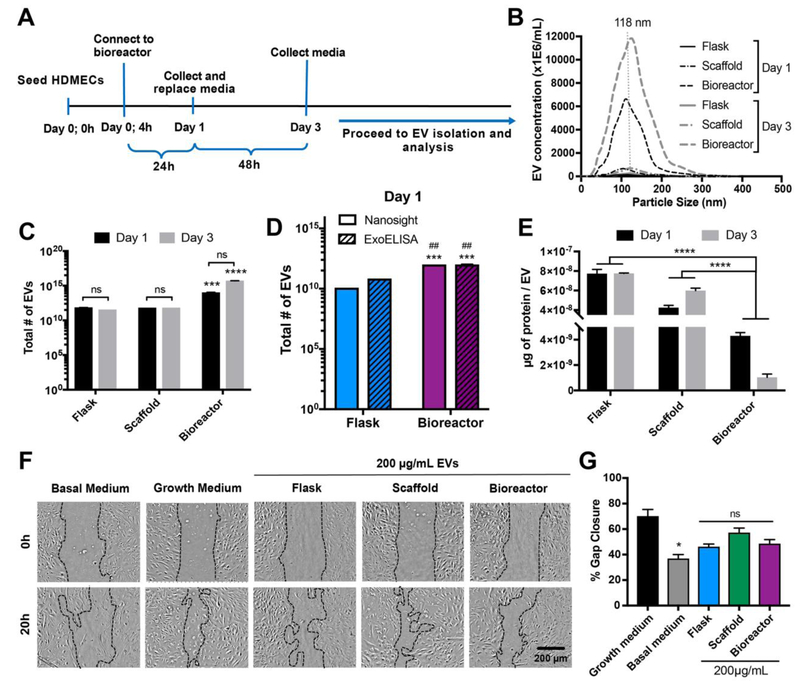
For experiments reported in Figure 2, conditioned medium containing EVs was collected from HDMECs initially seeded at 15,000 cells/cm2 after 24 h (Day 1) and after 48 h (Day 3) of the Day 1 collection as shown in the timeline (Figure 2A) for Bioreactor culture. Conditioned medium was collected from Scaffold and Flask culture groups at the same time points. For the ExoELISA in Figure 2D, HDMECs were seeded at 15,000 cell/cm2 and conditioned media containing EVs was collected after 24 h (Day 1) of culture. Experiments were carried out in duplicate with 3 biological replicates per experiment.
Figure 2. Bioreactor culture enhances EV production from HDMECs.
(A) Schematic of bioreactor experimental procedure. (B) Concentration and size distribution of HDMEC-derived EVs produced in Flask, Scaffold, and Bioreactor cultures from Day 1 and Day 3 as assessed by NTA. (C) Total numbers of EVs produced under each condition analyzed by NTA and (D) CD63 ExoELISA (Day 1) and (E) the ratio of total surface protein amount (μg) to total number of EVs are shown. No significant difference in EV production and protein content was calculated between Day 1 and Day 3 (p>0.05). Significant increase in EV production was observed in Bioreactor culture compared to both Scaffold and Flask cultures regardless of collection time (p<0.001). Significant increase in EV production was also observed as analyzed by CD63 ExoELISA from Bioreactor culture compared to Flask culture (D; * - compared to Nanosight Flask, # - compared to ExoELISA Flask). (F) HDMECs were treated with growth media (EGM2; positive control), basal media (EBM2; negative control), or 200 μg/mL EVs isolated from HDMECs in Flask, Scaffold, and Bioreactor culture conditions. Representative images for both Day 1 and Day 3 EV samples captured at 0 and 20 h are shown. (G) Cell gap area after 20 h as a percentage of gap area at 0 h (illustrated with dotted black lines in (F)) were determined and calculated using ImageJ. Images and data representative of three independent experiments (n=3). No significant differences in bioactivity of EVs between the three different culture conditions were calculated. Significant difference between growth and basal media was observed (p<0.05). Statistical significance was calculated using two-way ANOVA with Bonferroni’s multiple comparison test (ns – p>0.05, * p<0.05, *** p<0.001, **** p<0.0001). Scale bar = 200 μm.
For ethanol (EtOH) treatment, HDMECs seeded at a lower seeding density of 4,000 cells/cm2 in the Bioreactor, Scaffold, and Flask culture groups were expanded for 5 days or until 80% confluent to allow conditioning to their respective culture environment prior to ethanol treatment. Growth culture medium was replaced every other day. On Day 5, culture medium was replaced with EV-depleted EGM2 with or without 100 mM EtOH (ethanol conditioning). Conditioned medium was collected after 24 h for EV isolation and downstream characterization. Experiments were carried out in duplicate with 3 biological replicates per experiment.
2.7. EV isolation
EVs were isolated from conditioned media by differential centrifugation, with the last centrifugation step of 100,000 x g for 2 h, as previously described [20]. After resuspending the pelleted EVs in 1X PBS, they were transferred to a Nanosep 300kDa MWCO spin column (Pall) and centrifuged at 8,000 x g until all PBS was removed (~8-12 min). EVs collected at the top of the column were again resuspended in 1X PBS to a desired volume, sterile filtered using 0.2μm syringe filters (Pall) and stored at −20°C for up to one month with less than 2 freeze/thaw cycles. The total surface protein concentration of the EVs was measured by BCA assay.
2.8. EV quantification by NTA
EVs were diluted to a concentration of 1-10 μg of surface protein/mL and manually subjected to analysis using a NanoSight LM10 (Malvern Instruments; Malvern, UK). To capture and analyze the data, NanoTracking Analysis (NTA) analytical software version 2.3 was used. Each sample was measured three times with camera level set at 14 and acquisition time of 30 s. Approximately, 20-100 objects per frame with more than 200 completed tracks were analyzed for each video. The detection threshold was set at the beginning of each sample and kept constant for each repeat. Data were collected as EVs/mL and total EVs were calculated based on initial volume and dilution factor.
2.9. EV quantification by ExoELISA
EV concentration was also assessed via quantification of the amount of total immunoreactive CD63 present in a previously separated EV sample using the ExoELISA-ULTRA Complete Kit (CD63 Detection; System Biosciences, Mountain View, CA). Briefly, EVs were isolated from Day 1 conditioned media and quantified by NTA and BCA as described above. 50 μl of an EV suspension (in PBS) were added to each well of 96-well microtiter plates and incubated for 1 h at 37°C (binding step). After washing the plates three times for 5 min using wash buffer provided in the kit, plates were incubated with CD63 specific primary antibody (1:100) at room temperature (RT) for 1 h under agitation. Plates were subsequently washed 3 × 5 min with wash buffer and incubated with secondary antibody (1:5000) at RT for 1 h. The plates were washed and incubated with Supersensitive TMB ELISA Substrate at RT for 5 minutes under agitation. The reaction was terminated using Stop Buffer Solution. Absorbance was measured at 450 nm using a plate reader. The number of EVs/mL was obtained using an exosomal CD63 standard curve calibrated against NTA data (number of exosomes) (Figure S2) and the total number of EVs was calculated.
2.10. Long non-coding RNA (IncRNA) extraction and analysis
For RNA isolation, pelleted EVs were lysed using RNeasy Mini Kit (Qiagen; 74134) according to the manufacturer’s protocol immediately following 100,000 x g centrifugation. Subsequently, total RNA was isolated and 0.5 μg of total RNA from each sample was converted to complementary DNA (cDNA) using an iScript cDNA Synthesis Kit (Biorad; 1708891) following the manufacturer’s protocol. The cDNA was stored at −20°C for up to 1 month and used within 2 freeze/thaw cycles. Up to 10 ng of RNA transcribed to cDNA was used for each qPCR reaction. SsoAdvanced Universal SYBR Green Supermix PCR (Biorad; 172-5270) reaction mix was set up according to the provided protocol for a 10μL reaction. A RealTime PCR System (Applied Biosystems 7900HT) was used to detect lncRNA levels specific for HOTAIR and MALAT1 and β-actin was used as a control. The reaction cycle was 95°C for 30 s, followed by 40 cycles of 90°C for 15 s and 60°C for 30 s. Primer sequences were as follows:
HOTAIR:
Forward: 5’-GCA GTA GAA AAA TAG ACA TAG GAG A-3’
Reverse: 5’-ATA GCA GGA GGA AGT TCA GGC ATT G-3’
MALAT1:
Forward: 5’-GAA TTG CGT CAT TTA AAG CCT AG-3’
Reverse: 5’-GTT TCA TCC TAC ACA TCC CAA TT-3’
β-actin:
Forward: 5’-ACT TAG TTG CGT TAC ACC CTT-3’
Reverse: 5’-GTC ACC TTC ACC GTT CCA-3’
2.11. Immunoblotting
Approximately 20 μg of EVs and 2.5 μg of HDMEC protein lysate as determined by protein content (BCA assay) were used for immunoblots. Levels of exosome markers CD63 [21], CD9 [21], Alix [22], and TSG101 [23], intracellular makers HSP-90α/β, Calnexin, and Argonaut 2 (Ago2), as well as β-actin and GAPDH as controls were quantified by immunoblot analysis as described previously [20]. Primary antibodies for CD63 (ABM; Y402575), CD9 (Abcam; 92726), Alix (Abcam; 186429), TSG101 (Abcam 125011), Calnexin (Cell Signaling; C5C9), and Ago2 (Cell Signaling; C34C6) at 1:1000, HSP-90α/β (Santa Cruz; sc-13119) at 1:500, and β-actin (Cell Signaling; 13E5) and GAPDH (Cell Signaling, 14C10) at 1:2000 were used. Goat anti-rabbit IRDye 800CW (925-32210; LICOR) and goat anti-mouse secondary antibody were used at a dilution of 1:10,000. Bands were detected with a LI-COR Odyssey CLX Imager. Median intensity was measured for each band using Image Studio™ Software (LI-COR) and subtracted from that of the PBS (negative control) lane.
2.12. Gap closure assay
P4 or P5 HDMECs were seeded in gelatin-coated 48-well plates at 40,000 cells/well in EGM2 and allowed to grow until a uniform monolayer was formed (24 h). The cell monolayer was denuded using a pipette tip to create a linear gap through the middle of each well. Medium was replaced with endothelial cell basal medium (EBM2; Lonza, CC-3156) to serum starve the cells for 2 h. After serum starvation, medium was replaced with fresh EBM2 with or without the addition of EVs at 100 μg/mL based on BCA quantification of EV-associated protein. EBM2 or EV-depleted EGM2-treated cells were used as negative or positive controls, respectively. The closure of the cell gap was imaged at 0 h and 20 h. Overall gap closure was determined as a percentage of area covered by HDMECs versus the gap area after 20 h using ImageJ as previously described [19].
2.13. Statistics
Data are presented as mean ± SEM. Two-way ANOVA with Bonferroni’s multiple comparison test was used for grouped data sets to determine statistical significance (p<0.05) in Prism 7 (GraphPad Software, La Jolla, CA). Notation for significance in figures are as follows: ns – P>0.05, # or * - P<0.05; ## or ** - P<0.01; *** - P<0.001; **** - P<0.0001.
3. Results
3.1. Culture of HDMECs in 3D-printed scaffold perfusion bioreactor
Endothelial cells are physiologically subjected to hemodynamic forces that modulate cell phenotype [24] and potentially influence secretion, uptake, and cargo content of EVs. We designed and 3D-printed a scaffold with total surface area of 72 cm2 consisting of a pillared array that permitted medium and atmospheric oxygen flow from gas permeable tubing through the interior (Figure 1A,B). Computational fluid modeling was carried out for flow rates of 2-6 mL/min to evaluate theoretical shear stresses and flow trajectories throughout the scaffold (Figure S1, Figure 1D-E). Heat maps show the distribution of theoretical shear stresses across the scaffold growth surfaces, with the highest values found in the top shelf of the scaffold near the outer wall. Although shear stress has been shown to significantly impact cell behavior and genetic composition, we sought only to culture cells under a dynamic environment with minimal shear stress. Based on CFD analysis (Figures 1D-E, S1), we cultured cells under a flow rate of 4 mL/min for varying times; average and maximum theoretical shear stress values at this flow rate were calculated to be 1.5×10−2 dyn/cm2 and 1.3 dyn/cm2, respectively. Future work analyzing the impact of different flow rates is required to completely understand the impact of shear stress on EV production. For cell viability analysis, HDMECs were fixed and stained with Phalloidin after exposure to dynamic culture at 4 mL/min. Images confirmed the presence of live cells throughout the scaffold base and along the side walls of the pillars (Figure 1C), which is consistent with our previous findings [25].
3.2. HDMEC EV production is impacted by dynamic culture
The effects of 3D scaffold culturing (static and dynamic) on EV production were initially assessed via NTA. EVs were isolated from HDMECs cultured in each of the Scaffold, Flask, and Bioreactor systems for a total of 3 days. Conditioned medium was collected 24 hours after initial seeding (Day 1) and 48 hours after first collection (Day 3) (Figure 2A). The size distributions and peak sizes of EV samples from Flask, Scaffold, and Bioreactor for both collection times were similar and more than 90% of the total EV populations were within the typical exosomal diameter range (40-200 nm) (Figure 2B) [26]. NTA results showed approximately 100-fold (Day 1; p<0.0001) and 10,000-fold (Day 3; p<0.0001) increase in EV production when cultured in the Bioreactor compared to static Scaffold and Flask controls (Figure 2C). CD63 ExoELISA analysis confirmed increase in EV production from Bioreactor culture compared to Flask culture, however the observed increase was only ~14-fold (Figure 2D). As measured by protein mass using a BCA assay, total EVs produced in the Bioreactor (−1000 μg) far exceeded those produced in the static Flask (−150 μg) or Scaffold (−400 μg) culture for both collection time points. However, protein content per EV was inversely proportional to EV production rates. BCA analysis of Day 1 and Day 3 EV samples normalized per EV revealed more than 15-fold (p<0.001) and 70-fold (p<0.001) decreases in total protein content, respectively (Figure 2E). HDMEC EV bioactivity was subsequently assessed using an in vitro gap closure assay. No significant difference in gap closure of HDMECs treated with basal media alone or with 200 μg/mL EVs from Flask, Scaffold, or Bioreactor culture groups was observed (Basal: 40 ± 5%; Flask: 46 ± 4%; Scaffold: 57 ± 6%; Bioreactor; 48 ± 6%; p>0.05) (Figure 2F-G), matching expectations based on previous results [18].
3.3. Ethanol conditioning enhances vascularization bioactivity of EVs produced in scaffold culture systems (static and dynamic)
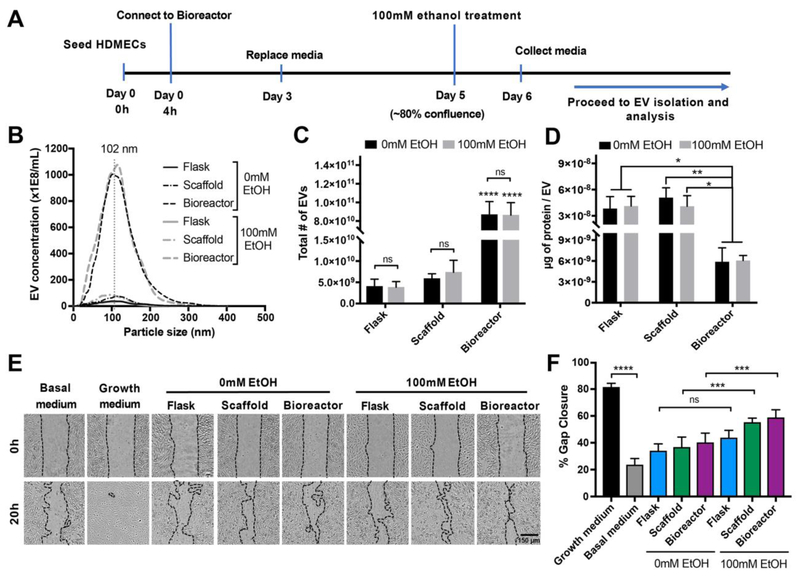
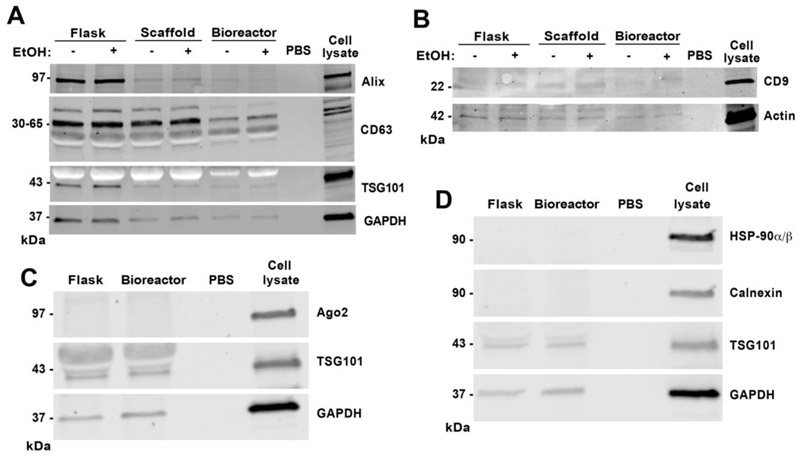
We previously demonstrated that ethanol conditioning of parent ECs in a 2D tissue culture environment (Flask) can enhance EV vascularization bioactivity [18]. To investigate whether this enhanced bioactivity effect could be retained in 3D culture systems, HDMECs were cultured in Flask, Scaffold, and Bioreactor conditions for up to 5 days or until they reached −80% confluence; at which point culture medium was replaced with medium containing 100 mM ethanol. After 24 hours of ethanol exposure, conditioned medium was collected and EVs were isolated for characterization (Figure 3A). NTA revealed no significant difference in size distribution and production between ethanol-treated and untreated EC-derived EVs for each of the three culture systems respectively (Figure 3B-C). Peak size values for EVs from Flask, Scaffold, and Bioreactor cultures with ethanol treatment were 90 ± 27 nm, 82 ± 22 nm, and 97 ± 21 nm and without ethanol treatment were 104 ± 20 nm, 100 ± 23 nm, and 89 ± 23 nm, respectively. Total EV counts for the Flask, Scaffold, and Bioreactor groups were similar with ethanol treatment (3.8E9 ± 2.3E9 EVs, 7.4E9 ± 4.9E9 EVs, and 8.6E10 ± 2.3E10 EVs, respectively) and without ethanol treatment (4.0E9 ± 2.9E9 EVs, 5.9E9 ± 1.9E9 EVs, and 8.7E10 ± 2.4E10 EVs, respectively) (Figure 3C), and EV production levels overall were consistent between experiments. Protein content per EV was also similar between ethanol and no ethanol groups (Figure 3D). Immunoblot analyses of protein markers associated with exosomes confirmed the presence of Alix, TSG101, CD63, and CD9 in EVs isolated from Flask, Scaffold, and Bioreactor cultures (Figure 4), consistent with prior observations in a hollow fiber bioreactor system [11]. Moreover, EV samples from Flask and Bioreactor culture had negligible levels of intracellular markers including endoplasmic reticulum (ER) markers HSP-90α/β and calnexin as well as Ago2, a RNA-induced silencing complex (RISC)-associated protein that is localized to the nucleus or cytoplasm depending on cell type (Figure 4).
Figure 3. Ethanol conditioning improves EV bioactivity in static and dynamic scaffold cultures.
(A) Schematic of experimental system for integrating ethanol conditioning with perfusion bioreactor culture. (B) Concentration and size distribution, (C) total number of EVs produced, and (D) surface protein content (μg) normalized to total number of EVs derived from HDMECs in Flask, Scaffold, and Bioreactor culture conditions with or without 100 mM ethanol (EtOH) treatment are shown. No statistical differences were observed between ethanol treated and untreated (0 mM EtOH) groups for EV production and protein amount per EV. Statistical significance between total EV production and protein content was detected between Bioreactor and Flask or Scaffold cultures (p<0.0001). (E) HDMECs were stimulated with growth media (EGM2; positive control), basal media (EBM2; negative control), or 200 μg/mL EVs isolated from the three indicated culture conditions with or without 100 mM ethanol conditioning. Representative images captured at 0 and 20 h are shown. (F) ImageJ analysis of cell gap area at 20 h relative to the gap area at 0 h (gap area depicted with black dotted lines). Data and images represent three independent experiments in triplicates each (n=3). A significant increase in EV bioactivity was observed in 100 mM EtOH treated EV groups from Scaffold and Bioreactor (p<0.001), but not from Flask culture (p>0.05). Statistical difference was calculated using two-way ANOVA with Bonferroni’s multiple comparison test (ns-p>0.05, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001). Scale bar= 150 μm.
Figure 4. Impact of ethanol and different culture conditions on exosome-associated protein markers and intracellular markers in the EV population.
(A) Immunoblot analyses of exosomal markers CD63, TSG101, and Alix and control protein marker GAPDH; (B) exosomal marker CD9 and control B-actin; (C) exosomal marker TSG101, intracellular marker Ago2, and GAPDH control; and (D) exosomal marker TSG101, endoplasmic reticulum (ER) markers HSP-90α/β and calnexin, and GAPDH control were conducted for EVs from each of the indicated culture conditions with 20 μg of EVs per lane (based on bicinchonic acid (BCA) analysis of EV protein content). A total of 2.5 μg of cell lysate (total cellular protein; cell marker control) and PBS (negative) were used as controls.
To analyze the impact of ethanol on EV vascularization bioactivity, EC gap closure was again assessed. In Flask culture, a trend towards increased gap closure that did not reach significance (0 mM EtOH: 34 ± 13%; 100 mM EtOH: 44 ± 13%; P>0.05)) was observed, however significant increases in gap closure were seen for Bioreactor and Scaffold EVs produced with 100 mM EtOH treatment (Scaffold: 55 ± 8%; Bioreactor: 59 ± 14%) compared to 0 mM EtOH (Scaffold: 37 ± 18%; Bioreactor: 40 ± 17%) (P<0.001). Gap closure values for 100 mM EtOH-treated Scaffold and Bioreactor EV groups were also significantly higher than no EV controls (basal medium: 24 ± 11%; p<0.001) (not indicated on the graph) (Figure 3E-F).
3.4. Ethanol-induced HDMEC EV lncRNA cargo changes are retained in perfusion bioreactor culture
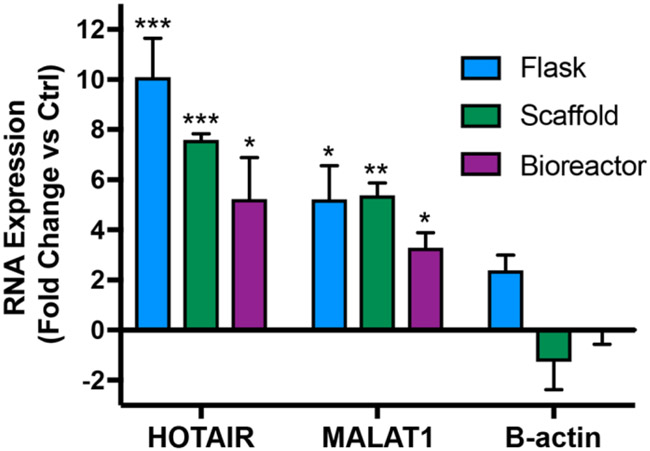
Our group previously showed that, in conventional tissue culture (Flask), ECs conditioned with ethanol (100 mM EtOH) produce EVs with increased vascularization bioactivity in part via a long non-coding RNA (lncRNA)-mediated mechanism. Specifically, ethanol conditioning induced higher levels of lncRNAs HOTAIR and MALAT1 – which are positively associated with angiogenesis – and these levels directly impacted EV-mediated vascularization [18]. Thus, we sought to determine whether increases in vascularization bioactivity of EVs from ethanol-conditioned ECs in Scaffold and Bioreactor culture were correlated with changes in expression of HOTAIR and MALAT1. LncRNA levels in EVs produced in the presence or absence of 100 mM EtOH were assessed by qPCR. Ethanol-conditioned HDMEC-derived EVs from Flask, Scaffold, and Bioreactor groups had significantly greater levels of both lncRNAs compared to respective controls as determined by the ΔΔCt method (Figure 5B). β-actin was used a negative control that was not expected to change with ethanol treatment.
Figure 5. Ethanol conditioning enhances angiogenic lncRNA cargo in HDMEC-derived EVs.
Expression levels of lncRNAs HOTAIR and MALAT1 were assessed by qPCR in EVs derived from HDMECs cultured with or without 100 mM ethanol for 24 h in each of the culture conditions. B-actin levels were assessed as a control. Data are shown as fold change of expression levels in ethanol treated group vs. expression levels in untreated group. No statistical significance was observed between the treated and untreated groups for B-actin. Significant variance was calculated for HOTAIR and MALAT1 for all three indicated culture conditions between ethanol treated and untreated groups. Data represent three independent experiments, each performed in triplicate (n=3). Statistical significance was calculated using two-way ANOVA with Bonferroni’s multiple comparison test (*p<0.05, **p<0.01, ***p<0.001).
4. Discussion
Perfusion bioreactor systems have been widely used for culturing cells to overcome limited mass transport in 2D static cultures [15,16]. Culture of ECs in a perfusion bioreactor to develop functional arteries in vitro has been especially successful due to the ability to recapitulate physiological hemodynamic forces, namely circumferential stretch and shear stress [27–29], as ECs in the vasculature experience a wide range of shear stresses [30,31]. Here, we utilized the perfusion bioreactor system with 3D-printed scaffolds to attempt to enhance HDMEC EV production and vascularization bioactivity. Thus, the design of the scaffold used throughout this study was motivated primarily by the need for large culture surface area amenable to medium perfusion and mass transport. Further, we used flow rates that theoretically produced shear stresses (1.5×10−2 dyn/cm2) near the lower end of reported physiological values for ECs to demonstrate proof-of-concept for a tubular perfusion bioreactor to improve EV production relative to current approaches. Based on total EV counts, our results showed greater than 100-fold increase in HDMEC EV production under dynamic culture vs. 2D flask culture (Figure 2C). However, we observed only a 14-fold increase in production of CD63-positive EVs as determined by ExoELISA. Given the status of CD63 as an essential component of exosome secretion and a validated marker of exosomes [21], these data suggest that other, non-exosome EV subtypes such as microvesicles may be preferentially produced in Bioreactor culture conditions.
The data also indicate that increased EV production in the Bioreactor culture came at a cost of total protein content per EV (Figure 2D). This result is consistent with the findings of Watson and colleagues in the hollow-fiber bioreactor system for HEK293 EVs, where the ratio of EVs to total protein increased substantially under bioreactor culture [11]. Both the overall reduction in protein:EV ratio as well as the relatively lower percentage of CD63-positive EVs of the total Bioreactor EV population may be explained by flow-induced cell membrane shedding. Cells are exposed to low shear stress due to media flow in the bioreactor (Figure 1C) that could induce increased shedding of the HDMEC membrane, resembling the process of microvesicle budding (as opposed to the multivesicular body (MVB)-mediated release of exosomes) [1]. Importantly, immunoblot data indicated a lack of calnexin, HSP-90α/β, and Ago2 in Flask and Bioreactor EVs; thereby suggesting that detected vesicles were unlikely to be apoptotic bodies or otherwise the result of mechanical disruption of cells during dynamic culture. Still, in-depth analysis of cellular changes and resulting EV cargo composition is required to elucidate any potential mechanism of shear stress-mediated increases in EV production. Future studies aiming to characterize the impact of scaffold geometry and topography are also necessary to delineate their role in EV production from that of fluid flow.
EC-derived EVs are known regulators of vascular remodeling via intercellular communication. They have been shown to promote neovascularization [32,33] as well as provide atheroprotective [34] and anti-inflammatory [35] effects, thereby alluding to the potential of EC-derived EVs as therapeutic agents in vascular diseases. However, several researchers have also observed adverse effects of EC-derived EVs, including destabilization of the vasculature [36–38]. Notably, the basal potency of HDMEC-derived EVs in our gap closure assay was negligible (Figure 2F-G). Thus, to improve therapeutic efficacy, we employed an ethanol conditioning approach, which we previously described, that induced increased levels of lncRNAs HOTAIR and MALAT1 in HUVEC EVs, resulting in enhanced vascularization bioactivity [18]. The current data show significant increases in HOTAIR and MALAT1 content in HDMEC EVs produced in Flask culture after ethanol conditioning, however no significant increase in bioactivity was observed (Figure 3E-F). This could be due to other bioactive cargo components in EVs. For example, our group previously identified an important role for ethanol-induced downregulation of anti-angiogenic miRNAs in EC EV bioactivity [18]. Similar lncRNA increases were observed in Scaffold and Bioreactor cultures, however, crucially, ethanol conditioning enhanced vascularization bioactivity of these EVs (Figure 3E-F). Thus, these results support the conclusion that enhancement of HDMEC EV potency via ethanol-induced regulation of lncRNAs MALAT1 and HOTAIR may be accomplished in a bioreactor. This represents, to our knowledge, the first demonstration of transference of an EV potency-enhancing mechanism from conventional 2D tissue culture to a scalable 3D culture system.
4.1. Conclusions
Overall, the use of perfusion bioreactor systems may be broadly applicable for production of therapeutic EVs. Specifically, this study demonstrates that ethanol conditioning is a scalable method for enhancing EC EV bioactivity. More generally, the data show that increased EV production in a bioreactor system need not come at the cost of reduced therapeutic potential. However, further investigation of potential adverse effects of dynamic culturing on parent cells and their EV competence is warranted. The present study also establishes the utility of 3D-printed scaffolds for EV generation. The precise control endowed by 3D printing will enable future systematic investigation of scaffold features on EV production and bioactivity. Ultimately, the combination of advances presented here may bring rational design of therapeutic EV biomanufacturing closer to realization.
Supplementary Material
Figure S1. Computational fluid modeling results of theoretical shear stresses throughout the scaffold (average and maximum) at varying flow rates shown in (A) the graph and (B) the table.
Figure S2. Calibration curves for the number of exosome particles versus OD 450 nm reading of exosomal CD63 standard for data shown in Figure 2D. Equation and R2 for the line of best fit were determined using linear regression analysis.
Statement of Significance.
Extracellular vesicles (EVs) have substantial therapeutic potential in a variety of applications. However, translation of EV-based therapies may be hindered by biomanufacturing challenges. EV production to date has predominantly involved the use of tissue culture flasks. Here, we report, for the first time, the use of a tubular perfusion bioreactor system with an integrated 3D-printed biomaterial scaffold for EV production from human endothelial cells. This system increases EV yield by over 100-fold compared to conventional tissue culture systems. Further, we show that an ethanol-conditioning approach that our group previously developed in 2D culture for enhancing EV potency is compatible with this new system. Thus, potency enhancement of EVs for vascularization applications is possible even with significantly increased production rate.
Acknowledgements:
Any opinions, findings, conclusions, or recommendations expressed in the material are those of the authors and do not necessarily reflect the views of NIST.
Funding:
This work was supported by the American Heart Association (16PRE30770016 to DBP), the National Institute of Standards and Technology through the 2014-NIST-MSE-01 fellowship program, (to MJL) the National Institutes of Health (HL141611 to SMJ), the National Science Foundation (1750542 to SMJ), the National Institute of Biomedical Imaging and Bioengineering / National Institutes of Health (NIBIB/NIH) Center for Engineering Complex Tissues (P41 EB023833 to JPF), and the University of Maryland.
Footnotes
Conflicts of Interest:
JPF is a founding member of 3DBioWorks, which is seeking to license technology related to 3D bioprinting and perfusion bioreactors.
Data Availability:
All data will be made available upon request.
Part of the Cell and Tissue Biofabrication Special Issue, edited by Professors Guohao Dai and Kaiming Ye.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- [1].El Andaloussi S, Mäger I, Breakefield XO, a Wood MJ, Extracellular vesicles: biology and emerging therapeutic opportunities., Nat. Rev. Drug Discov 12 (2013) 347–57. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- [2].Lamichhane TN, Sokic S, Schardt JS, Raiker RS, Lin JW, Jay SM, Emerging roles for extracellular vesicles in tissue engineering and regenerative medicine, Tissue Eng Part B Rev. 21 (2015)45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Zhu X, Badawi M, Pomeroy S, Sutaria DS, Xie Z, Baek A, Jiang J, Elgamal OA, Mo X, La Perle K, Chalmers J, Schmittgen TD, Phelps MA, Comprehensive toxicity and immunogenicity studies reveal minimal effects in mice following sustained dosing of extracellular vesicles derived from HEK293T cells, J. Extracell. Vesicles 6 (2017) 1324730. doi: 10.1080/20013078.2017.1324730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Sutaria DS, Badawi M, Phelps MA, Schmittgen TD, Achieving the Promise of Therapeutic Extracellular Vesicles: The Devil is in Details of Therapeutic Loading, Pharm. Res 34 (2017) 1053–1066. doi: 10.1007/s11095-017-2123-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ha D, Yang N, Nadithe V, Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: current perspectives and future challenges, Acta Pharm. Sin. B 6 (2016) 287–296. doi: 10.1016/j.apsb.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Yang T, Martin P, Fogarty B, Brown A, Schurman K, Phipps R, Yin VP, Lockman P, Bai S, Exosome Delivered Anticancer Drugs Across the Blood-Brain Barrier for Brain Cancer Therapy in Danio Rerio , Pharm. Res 32 (2015) 2003–2014. doi: 10.1007/s11095-014-1593-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chevillet JR, Kang Q, Ruf IK, Briggs HA, Vojtech LN, Hughes SM, Cheng HH, Arroyo JD, Meredith EK, Gallichotte EN, Pogosova-Agadjanyan EL, Morrissey C, Stirewalt DL, Hladik F, Yu EY, Higano CS, Tewari M, Quantitative and stoichiometric analysis of the microRNA content of exosomes, Proc. Natl. Acad. Sci. U. S. A 111 (2014) 14888–14893. doi: 10.1073/pnas.1408301111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Watson DC, Yung BC, Bergamaschi C, Chowdhury B, Bear J, Stellas D, Morales-Kastresana A, Jones JC, Felber BK, Chen X, Pavlakis GN, Scalable, cGMP-compatible purification of extracellular vesicles carrying bioactive human heterodimeric IL-15/lactadherin complexes, J. Extracell. Vesicles 7 (2018) 1442088. doi: 10.1080/20013078.2018.1442088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Jarmalavičiūtė A, Tunaitis V, Pivoraite U, Venalis A, Pivoriūnas A, Exosomes from dental pulp stem cells rescue human dopaminergic neurons from 6-hydroxy-dopamine–induced apoptosis, Cytotherapy. 17 (2015) 932–939. doi: 10.1016/j.jcyt.2014.07.013. [DOI] [PubMed] [Google Scholar]
- [10].Mitchell JP, Court J, Mason MD, Tabi Z, Clayton A, Increased exosome production from tumour cell cultures using the Integra CELLine Culture System, J. Immunol. Methods 335 (2008) 98–105. doi: 10.1016/j.jim.2008.03.001. [DOI] [PubMed] [Google Scholar]
- [11].Watson DC, Bayik D, Srivatsan A, Bergamaschi C, Valentin A, Niu G, Bear J, Monninger M, Sun M, Morales-Kastresana A, Jones JC, Felber BK, Chen X, Gursel I, Pavlakis GN, Efficient production and enhanced tumor delivery of engineered extracellular vesicles, Biomaterials. 105 (2016) 195–205. doi: 10.1016/j.biomaterials.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].An J, Teoh JEM, Suntomnond R, Chua CK, Design and 3D Printing of Scaffolds and Tissues, Engineering. 1 (2015)261–268. doi: 10.15302/J-ENG-2015061. [DOI] [Google Scholar]
- [13].Zhao F, Vaughan TJ, McNamara LM, Quantification of fluid shear stress in bone tissue engineering scaffolds with spherical and cubical pore architectures, Biomech. Model. Mechanobiol 15 (2016) 561–577. doi: 10.1007/s10237-015-0710-0. [DOI] [PubMed] [Google Scholar]
- [14].Giannitelli SM, Accoto D, Trombetta M, Rainer A, Current trends in the design of scaffolds for computer-aided tissue engineering, Acta Biomater. 10 (2014) 580–594. doi: 10.1016/j.actbio.2013.10.024. [DOI] [PubMed] [Google Scholar]
- [15].Yeatts AB, Fisher JP, Tubular perfusion system for the long-term dynamic culture of human mesenchymal stem cells., Tissue Eng. Part C. Methods 17 (2011) 337–348. doi: 10.1089/ten.tec.2010.0172. [DOI] [PubMed] [Google Scholar]
- [16].Yu L, Ferlin KM, Nguyen BB, Fisher JP, Tubular perfusion system for chondrocyte culture and superficial zone protein expression, (2014) 1–11. doi: 10.1002/jbm.a.35321. [DOI] [PubMed] [Google Scholar]
- [17].Jansen F, Li Q, Pfeifer A, Werner N, Endothelial- and Immune Cell-Derived Extracellular Vesicles in the Regulation of Cardiovascular Health and Disease, JACC Basic to Transl. Sci 2 (2017) 790–807. doi: 10.1016/j.jacbts.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lamichhane TN, Leung CA, Douti LY, Jay SM, Ethanol Induces Enhanced Vascularization Bioactivity of Endothelial Cell-Derived Extracellular Vesicles via Regulation of MicroRNAs and Long Non-Coding RNAs, Sci. Rep 7 (2017) 1–10. doi: 10.1038/s41598-017-14356-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Patel DB, Gray KM, Santharam Y, Lamichhane TN, Stroka KM, Jay SM, Impact of Cell Culture Parameters on Production and Vascularization Bioactivity of Mesenchymal Stem Cell-Derived Extracellular Vesicles, Bioeng. Transl. Med (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lamichhane TN, Jeyaram A, Patel DB, Parajuli B, Livingston NK, Arumugasaamy N, Schardt JS, Jay SM, Oncogene Knockdown via Active Loading of Small RNAs into Extracellular Vesicles by Sonication., Cell. Mol. Bioeng 9 (2016) 315–324. doi: 10.1007/s12195-016-0457-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Andreu Z, Yáñez-Mó M, Tetraspanins in Extracellular Vesicle Formation and Function, Front. Immunol 5 (2014) 442. doi: 10.3389/fimmu.2014.00442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Baietti MF, Zhang Z, Mortier E, Melchior A, Degeest G, Geeraerts A, Ivarsson Y, Depoortere F, Coomans C, Vermeiren E, Zimmermann P, David G, Syndecan–syntenin–ALIX regulates the biogenesis of exosomes, Nat. Cell Biol 14 (2012) 677–685. doi: 10.1038/ncb2502. [DOI] [PubMed] [Google Scholar]
- [23].Buschow SI, Liefhebber JMP, Wubbolts R, Stoorvogel W, Exosomes contain ubiquitinated proteins, Blood Cells, Mol. Dis 35 (2005) 398–403. doi: 10.1016/j.bcmd.2005.08.005. [DOI] [PubMed] [Google Scholar]
- [24].Punchard MA, Stenson-Cox C, O’Cearbhaill ED, Lyons E, Gundy S, Murphy L, Pandit A, McHugh PE, Barron V, Endothelial cell response to biomechanical forces under simulated vascular loading conditions, J. Biomech 40 (2007) 3146–3154. doi: 10.1016/j.jbiomech.2007.03.029. [DOI] [PubMed] [Google Scholar]
- [25].Lembong J, Lerman MJ, Kingsbury TJ, Civin CI, Fisher JP, A Fluidic Culture Platform for Spatially Patterned Cell Growth, Differentiation, and Cocultures, Tissue Eng. Part A (2018). doi: 10.1089/ten.tea.2018.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].El Andaloussi S, Mäger I, Breakefield XO, a Wood MJ, EL Andaloussi S, Mäger I, Breakefield XO, a Wood MJ, Extracellular vesicles: biology and emerging therapeutic opportunities., Nat. Rev. Drug Discov 12 (2013) 347–57. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- [27].Lovett M, Lee K, Edwards A, Kaplan DL, Vascularization Strategies for Tissue Engineering, Tissue Eng. Part B. Rev 15 (2009) 353–370. doi: 10.1089/ten.teb.2009.0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Williams C, Wick TM, Endothelial cell-smooth muscle cell co-culture in a perfusion bioreactor system, Ann. Biomed. Eng 33 (2005) 920–928. doi: 10.1007/s10439-005-3238-0. [DOI] [PubMed] [Google Scholar]
- [29].Williams C, Wick TM, Perfusion bioreactor for small diameter tissue-engineered arteries., Tissue Eng. 10 (2004) 930–41. doi: 10.1089/1076327041348536. [DOI] [PubMed] [Google Scholar]
- [30].Whitmore RL, The flow behaviour of blood in the circulation, Nature. 215 (1967) 123–126. file:///C:/Users/tzhou3/AppData/Local/MendeleyLtd./MendeleyDesktop/Downloaded/DeYoung,Axford-1967-©1967NaturePublishingGroup.pdf. [DOI] [PubMed] [Google Scholar]
- [31].Lipowsky HH, Kovalcheck S, Zweifach BW, The distribution of blood rheological parameters in the microvasculature of cat mesentery, Circ. Res 43 (1978) 738–749. doi: 10.1161/01.RES.43.5.738. [DOI] [PubMed] [Google Scholar]
- [32].Sheldon H, Heikamp E, Turley H, Dragovic R, Thomas P, Oon CE, Leek R, Edelmann M, Kessler B, Sainson RCA, Sargent I, Li J-L, Harris AL, New mechanism for Notch signaling to endothelium at a distance by Delta-like 4 incorporation into exosomes, Blood. 116 (2010) 2385 LP–2394. http://www.bloodjournal.org/content/116/13/2385.abstract. [DOI] [PubMed] [Google Scholar]
- [33].van Balkom BWM, de Jong OG, Smits M, Brummelman J, den Ouden K, de Bree PM, van Eijndhoven MAJ, Pegtel DM, Stoorvogel W, Würdinger T, Verhaar MC, Endothelial cells require miR-214 to secrete exosomes that suppress senescence and induce angiogenesis in human and mouse endothelial cells, Blood. 121 (2013) 3997 LP–4006. http://www.bloodjournal.org/content/121/19/3997.abstract. [DOI] [PubMed] [Google Scholar]
- [34].Hergenreider E, Heydt S, Treguer K, Boettger T, Horrevoets AJG, Zeiher AM, Scheffer MP, Frangakis AS, Yin X, Mayr M, Braun T, Urbich C, Boon RA, Dimmeler S, Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs, Nat Cell Biol. 14 (2012) 249–256. 10.1038/ncb2441. [DOI] [PubMed] [Google Scholar]
- [35].Jansen F, Yang X, Baumann K, Przybilla D, Schmitz T, Flender A, Paul K, Alhusseiny A, Nickenig G, Werner N, Endothelial microparticles reduce ICAM-1 expression in a microRNA-222-dependent mechanism, J. Cell. Mol. Med 19 (2015) 2202–2214. doi: 10.1111/jcmm.12607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ju R, Zhuang ZW, Zhang J, Lanahan AA, Kyriakides T, Sessa WC, Simons M, Angiopoietin-2 Secretion by Endothelial Cell Exosomes: REGULATION BY THE PHOSPHATIDYLINOSITOL 3-KINASE (PI3K)/Akt/ENDOTHELIAL NITRIC OXIDE SYNTHASE (eNOS) AND SYNDECAN-4/SYNTENIN PATHWAYS, J. Biol. Chem 289 (2014) 510–519. doi: 10.1074/jbc.M113.506899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Shan Z, Qin S, Li W, Wu W, Yang J, Chu M, Li X, Huo Y, Schaer GL, Wang S, Zhang C, An Endocrine Genetic Signal Between Blood Cells and Vascular Smooth Muscle Cells: Role of MicroRNA-223 in Smooth Muscle Function and Atherogenesis, J. Am. Coll. Cardiol 65 (2015) 2526–2537. doi: 10.1016/j.jacc.2015.03.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Jansen F, Yang X, Franklin BS, Hoelscher M, Schmitz T, Bedorf J, Nickenig G, Werner N, High glucose condition increases NADPH oxidase activity in endothelial microparticles that promote vascular inflammation, Cardiovasc. Res 98 (2013) 94–106. http://dx.doi.Org/10.1093/cvr/cvt013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Computational fluid modeling results of theoretical shear stresses throughout the scaffold (average and maximum) at varying flow rates shown in (A) the graph and (B) the table.
Figure S2. Calibration curves for the number of exosome particles versus OD 450 nm reading of exosomal CD63 standard for data shown in Figure 2D. Equation and R2 for the line of best fit were determined using linear regression analysis.