Abstract
Light-driven sodium pumps (NaRs) are microbial rhodopsins that utilize light energy to actively transport sodium ions out of the cell. Here, we used targeted mutagenesis and electrophysiological methods in living cells to demonstrate that NaRs can be converted into light-activated cation channels by molecular engineering. Specifically, introduction of the R109Q mutation into the sodium ion pump of Dokdonia eikasta (KR2) results in passive ion conductance, with a high preference for potassium over sodium ions. However, in this mutant, residual active outward pumping of sodium ions competes with passive inward transport of potassium. Channel-like behavior could also be achieved by introduction of other mutations into the KR2 counterion complex, and further, these modifications were transferrable to other NaRs. Combining the R109Q replacement with modifications at position S70 removed the residual sodium pumping and greatly enhanced the channel-like activity. However, passive photocurrents were only observed in leak mutants if the KR2 counterions, D116 and D251, were deprotonated, which was only observed under alkaline conditions. Overall, our results reveal that interactions between R109 and the nearby residues, L75, S70, D116, and D251, prevent passive backflow during ion transport in NaRs.
Introduction
Microbial rhodopsins are light-sensitive proteins, containing seven-transmembrane domains, that are found in archaea, bacteria, and lower eukaryotes. These function as ion transporters, sensors, or enzymes, using covalently bound retinal as a light-sensitive chromophore (1). Light-gated ion channels (channelrhodopsins) and light-driven ion pumps are widely used in neuroscience to noninvasively investigate and manipulate neuronal activity with light (2, 3, 4). In addition to their optogenetic applicability, microbial rhodopsins offer an excellent opportunity to investigate the structural differences between channels and pumps and to elucidate how these differences affect the function of ion transporters (5). The high similarity between channels and pumps of the microbial rhodopsin family was initially noted with the discovery of the first channelrhodopsins (6, 7, 8), but only recently have active proton pumps been converted into passive proton channels by the introduction of specific mutations or synthetic chromophores (9, 10, 11). In 2013, the family of ion-pumping rhodopsins was further expanded by the discovery of light-driven Na+ pumps (NaRs) (12, 13, 14). These cation pumps further raised the question as to whether the structural mechanisms existing in proton pumps to prevent passive proton leakage are also preserved in sodium pumps. This question is challenging because both proton and sodium pumps differ in several respects. Proton pumps contain water molecules and residues, which are stepwise protonated and deprotonated, and the released proton is already bound in the dark state (1). In contrast, in sodium pumps, the same sodium ion has to enter and leave the protein during the photocycle (14, 15, 16).
Here, we assessed the potential for converting NaRs into channel-like cation transporters using mutations that are known to induce passive currents in proton pumps (Fig. 1 A). Introduction of these leak mutations into the sodium ion pump of Dokdonia eikasta (KR2) resulted in inward-directed photocurrents at physiological electrochemical loads, as observed by two-electrode voltage-clamp (TEVC) measurements in Xenopus laevis oocytes. These photocurrents were identified as light-controlled passive cation conductance. Notably, we found that the mutant KR2-R109Q promotes inward photocurrents and is highly selective for potassium over sodium. This observation is particularly intriguing because of the fact that light-gated potassium-selective channels are a valuable optogenetic tool for the inhibition of neuronal activity.
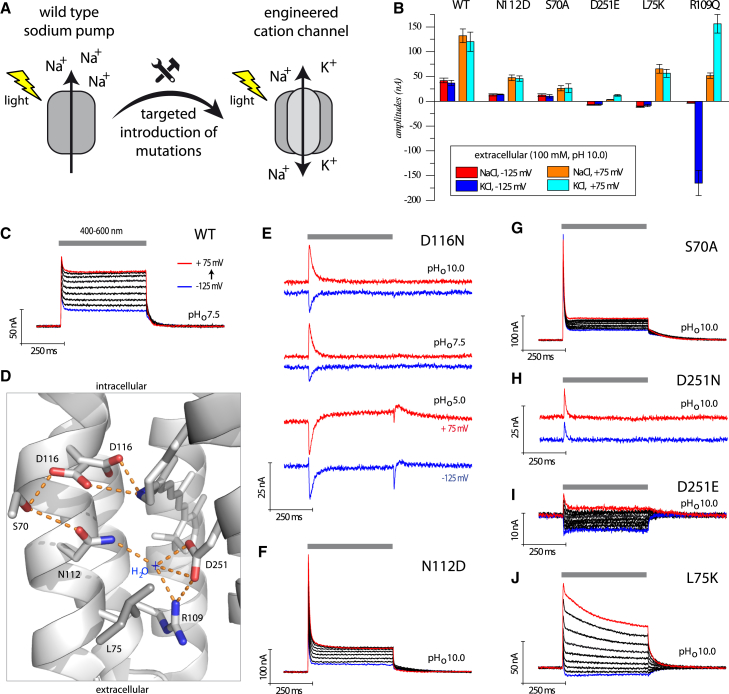
Figure 1.
Electrophysiological analysis of KR2βHK in X. laevis oocytes. (A) A sketch about the intention of this scientific work is shown. (B) Quantitative comparison of the stationary photocurrent amplitudes (see Fig. S6A for details). (C) Representative photocurrent traces of wild-type (WT) KR2 at pHo = 7.5 and different holding potentials (in 25 mV steps) are shown. (D) The KR2 crystal structure (PDB: 3X3C) at neutral pH is shown, illustrating the mutated positions of the counterion complex. (E) Substitution of the proposed primary proton acceptor (D116N) causes pHo-dependent transient photocurrents. Traces at −125 mV (blue) and +75 mV (red) for each pHo value are displayed. (F–J) Photocurrents of additional mutants are shown at pHo = 10. All traces were measured in extracellular buffer containing 100 mM NaCl. Oocytes were illuminated with 500 ms light pulses, using a xenon lamp and a 400–600 nm broadband filter. To see this figure in color, go online.
Materials and Methods
Molecular biology
We utilized the pGEMHE vector for expression in oocytes (T7-promotor, polyadenylation site), the pmKate2-N1 plasmid for expression in ND7/23 cells (CMV promotor), and the pET21a + vector for expression in Escherichia coli. Site-directed mutagenesis was performed using the QuikChange Kit (Agilent Technologies, Santa Clara, CA). NaRs containing specific targeting sequences were codon-optimized and synthesized by GenScript (Piscataway Township, NJ), using human-optimized codons for oocytes and ND7/23 cells and E. coli-optimized codons for pH assays and protein purification. Protein IDs for all tested NaRs are shown in Fig. S1 A, and a multiple sequence alignment is shown in Fig. S2 (prepared with Clustal Omega, European Molecular Biology Laboratory). The detailed sequences for all KR2 constructs are shown in Fig. S3.
TEVC measurements in X. laevis oocytes
Protocols for animal maintenance and oocyte harvesting were approved by the German administration for animal testing (Berlin, Germany). Female frogs were maintained in tap water at 18°C with a 12:12 h light/dark cycle. Frogs were anesthetized with 2 g/L tricaine (Sigma Aldrich, St. Louis, MO) for harvest of ovary lobes. Oocytes were then washed and stored in oocyte incubation buffer at 4°C (96 mM NaCl, 5 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 5 mM 3-(N-morpholino)propanesulfonic acid, 50 U penicillin/streptomycin (pH 7.5)). For injection, oocytes were first defolliculated by hand, after partial digestion with 2 mg/L collagenase solution (dissolved in antibiotic- and Ca2+-free oocyte incubation buffer), for 1.5 h at 18°C. Oocytes were then injected with 30 ng capped messenger RNA using the Nanoliter 2000 microinjection system (World Precision Instruments, Sarasota, FL) and incubated at 18°C for 4 days. This capped messenger RNA was synthesized in vitro from NheI-linearized pGEMHE plasmid, using the mMESSAGE mMACHINE T7 Kit (Life Technologies, Carlsbad, CA). After injection, the incubation buffer was supplemented with 5 μM all-trans retinal (Sigma Aldrich).
TEVC experiments were performed using a Turbo TEC-10X amplifier without transient compensation (npi electronic, Tamm, Germany). Buffer composition is shown in Fig. S4. Continuous light was provided by an XBO75/2 75 W xenon lamp (Carl Zeiss, Oberkochen, Germany) and was controlled by an LS3 shutter (Vincent Associates UNIBLITZ Shutter Systems, Rochester, NY). Light was passed through a 400–600 nm broadband filter (Optics Balzers, Liechtenstein, Germany), with an intensity of 10 mW/mm2. Microelectrodes were fabricated from borosilicate glass capillaries (1.50 mm outer diameter and 1.17 mm inner diameter; Harvard Apparatus, Holliston, MA), using a P-97 micropipette puller (Sutter Instrument, Novato, CA), and microelectrode resistance was maintained between 0.6 and 2.0 MΩ. Data acquisition and light triggering were controlled using the Axon pCLAMP 9.0 software via the Digidata 1322A interface (Molecular Devices, San Jose, CA). Currents were recorded at 10 kHz and filtered to 1 kHz using built-in circuits. The photocurrent traces were baseline corrected and filtered to 300 Hz (Gaussian low-pass cutoff filter, using pCLAMP). Reversal potentials (Erev) were estimated for each single buffer condition by linear regression. For current-voltage plots, measurements from each single oocyte (and ND7/23 cells) were normalized to a reference condition (indicated with small black boxes in the figures and in the legends). All normalized values from several cells are summarized and are shown as mean ± standard error.
Patch-clamp recordings with ND7/23 cells
ND7/23 cells were cultured in Dulbecco’s modified Eagle’s medium, supplemented with 5% (v/v) fetal bovine serum and 1 μg/mL penicillin/streptomycin at 37°C under a 5% CO2 atmosphere. For all experiments, 5 × 104 cells/mL were seeded in dishes on poly-D-lysine-coated glass coverslips in media supplemented with 1 μM all-trans retinal (Sigma Aldrich). The following day, cells were transiently transfected using FuGENE HD Transfection Reagent (Promega, Madison, WI). Electrophysiological recordings were conducted 24–30 h after transfection. Patch pipettes (2–3.5 MΩ) were prepared from standard wall (inner diameter 0.86 nm, outer diameter 1.50 nm) borosilicate capillaries with filament (Warner Instruments, Harnden, CT), using a micropipette puller (P1000; Sutter Instrument). The recordings were performed in the whole-cell configuration at room temperature. Light was provided by a CoolLED system (model pE-4000), coupled to the epifluorescence port of an Axiovert 100 TV inverted microscope (Carl Zeiss). Cells were illuminated with a 525 nm light-emitting diode (full width at half-maximum 31 nm), with a power of 23 mW/mm2 in the focal plane. The currents were filtered at 5 kHz with an ELC-03XS amplifier (npi electronic) and digitized at a sampling rate of 20 kHz (Digidata 1440 A; Molecular Devices). Buffer composition is listed in Fig. S4. For stability reasons, the patch and following measurements were always initiated in the extracellular high-sodium buffer (NaCl 9.0). Electrophysiology data were analyzed using Clampfit 10.4 (Molecular Devices).
Monitoring light-induced pH changes in E. coli suspension (pH assay)
The expression plasmid (pET21a+) containing KR2-wild type (WT) or KR2-R109Q was transformed into E. coli C41 (DE3) cells. Protein expression was induced in cells at an optical density at 600 nm (OD600) of 1.0 with 0.5 mM isopropyl β-D-1-thiogalactopyranocide (IPTG; Carl Roth, Karlsruhe, Germany), in lysogeny broth medium supplemented with 5 μM all-trans retinal (Sigma Aldrich). Induced cells were grown for 2.5 h at 37°C (OD600 ≈ 3.0), followed by centrifugation for 10 min at 3000 × g. The pellets were then resuspended in salt solution (either 100 mM NaCl or 100 mM KCl, no further ingredients) for 1.5 h at room temperature. These cells were centrifuged again, and the pellets were resuspended in fresh salt solution (concentrated to OD600 = 10) and stored in the dark at room temperature until measurement.
For analysis of light-induced pH changes, the E. coli suspension (8 mL) was stirred in a temperature-controlled glass chamber (25°C). The pH was measured every 5 s (WTW model inoLab pH 730; Weilheim, Germany), and the cells were illuminated with white light from a slide projector (model Noris Trumpf Halogen 150; Ernst Plank KG, Germany). The first measurement was obtained without any ionophore, followed by a second measurement after addition of the protonophore, carbonyl cyanide m-chlorophenyl hydrazone (CCCP; Sigma Aldrich), at a final concentration of 50 μM.
Purification and spectroscopy of KR2
Expression plasmid transformation, protein expression, and protein purification were performed as previously reported (15, 17). KR2 expression was induced in cells at an OD600 of 0.6 with IPTG (Carl Roth) in lysogeny broth medium supplemented with 5 μm all-trans retinal (Sigma Aldrich). Induced cells were grown for 3 h at 37°C and then harvested and disrupted using an EmulsiFlex-C3 Homogenizer (AVESTIN, Ottawa, ON, Canada). The membrane fraction was collected by ultracentrifugation at 45,000 rotations per minute for 1 h at 4°C, using a Type 45 Ti rotor (Beckman Coulter, Brea, CA). The membrane pellet was then resuspended in buffer containing 50 mM Tris-HCl (pH 8.0), 300 mM NaCl, 0.1 mM phenylmethylsulfonyl fluoride (PMSF), 1.5% n-dodecyl-D-maltoside (DDM) (GLYCON Biochemicals, Luckenwalde, Germany), and 0.3% cholesteryl hemisuccinate (CHS; Sigma Aldrich) and stirred overnight for solubilization. The insoluble fraction was removed by ultracentrifugation at 200,000 × g for 1 h at 4°C, and recombinant KR2 protein was purified by Ni-NTA affinity using the ÄKTAxpress protein purification system (GE Healthcare Life Sciences, Marlborough, MA), configured with a HisTrap HP Ni-NTA column. The collected protein fractions were concentrated in buffer containing 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 0.1 mM PMSF, 0.02% DDM, and 0.004% CHS.
To analyze the spectroscopic characteristics of purified KR2-WT and the R109Q mutant in variable ionic and pH conditions, different buffers were utilized. Steady-state absorption was measured at pH 8.0 (50 mM Tris-C6H8O7), pH 9.0 (50 mM glycine-N-methyl-D-glucamine [NMG]), and pH 10.0 (50 mM glycine-NMG). The salt conditions were 150 mM NaCl, LiCl, or KCl. In the case of steady-state absorption measurements at pH 9.0, a salt concentration of 110 mM was used. Flash photolysis measurements were performed with 150 mM NaCl at pH 8.0 (50 mM Tris-HCl) and with 150 mM KCl at pH 10.0 (50 mM glycine-NMG). All buffer exchange was performed using PD-10 Desalting Columns, with Sephadex G-25 resin (GE Healthcare Life Sciences).
Ultraviolet-visible spectroscopy
Steady-state ultraviolet-visible absorption spectra were recorded using a Cary 300 spectrophotometer (Varian, Palo Alto, CA) at a spectral resolution of 1 nm and at room temperature (22°C). The LKS.60 Laser Flash Photolysis Spectrometer (Applied Photophysics, Leatherhead, UK), with modifications, was used to measure microsecond-to-second changes in multiwavelength data sets at a resolution of 0.4 nm. To excite the sample, the laser pulse was tuned to 530 nm using the MagicPrism Inline Tunable Optical Parametric Oscillator (Opotek, Carlsbad, CA), which was pumped via the third harmonic of a BrilliantB Nd:YAG laser (Quantel, Newbury, UK). The laser energy was adjusted to 5 mJ/shot and a pulse duration of 10 ns, and changes in absorption were measured with a 150 W Xenon lamp (Osram, Munich, Germany). The transient spectra were recorded using an Andor iStar ICCD camera (DH734; Andor Technology, Belfast, UK), at 36 different time points between 1 μs and 1 s (five points per decade, isologarithmically). To ensure complete recovery of the dark state before the following recording, the sample was held in the dark for 30 s, and the resulting data sets were averaged over at least 10 cycles.
Primary data analysis was performed using MATLAB R2016b (The MathWorks, Natick, MA) to calculate difference spectra and reconstruct the three-dimensional spectra. Global analysis of the spectral data sets was performed with Glotaran 1.5.1 (Vrije Universiteit Amsterdam, Netherlands) (18). Singular value decomposition analysis of the data was used to determine the number of significant components needed to reconstruct the signal from the data sets, enabling noise reduction.
Results
WT photocurrents in oocytes
We initially measured the photocurrents of KR2-WT in Xenopus oocytes and found that these were barely detectable. However, addition of the H+/K+-ATPase β-subunit fragment (“βHK”) to the N-terminus improved the photocurrents substantially (Fig. S1, B and C), a strategy first established for bacteriorhodopsin (BR) (19). Therefore, unless otherwise stated, all measurements in Xenopus oocytes were performed with this KR2βHK construct (named KR2 in the following). KR2 photocurrents reached values up to 150 nA depending on the voltage, the extracellular pH value (pHo), and the ionic conditions (Fig. 1 B; Fig. S5 A). The characteristic initial transient peak after light on is followed by stationary photocurrents (Fig. 1 C), and overall, these results for KR2-WT are consistent with earlier reports (13, 15, 17). We further found that photocurrents and pHo dependence are unaffected by the replacement of extracellular NaCl with KCl (Fig. S5 A). In contrast, stationary photocurrent amplitudes are slightly reduced by replacement of extracellular sodium or potassium ions with NMG-Cl, LiCl, MgCl2, or CaCl2 (Figs. S5 A and S6 C). However, under all conditions, photocurrents were found to be positive, confirming the outward-directed pumping activity of KR2. Photocurrents of other sodium pumps, including those from Nonlabens dokdonensis (NdR2), N. marinus (NMR2), Nonlabens sp. YIK11, Gillisia limnaea, Indibacter alkaliphilus, and Truepera radiovictrix (TrNaR1 and TrNaR2) (12, 20, 21, 22, 23, 24, 25, 26, 27), were also measured with and without βHK-targeting, but in all cases, the currents remained poor compared to KR2βHK (Fig. S1, A, B, and D; alignment in Fig. S2).
Introduction of leak mutations into KR2
The counterion complex (also called the “active site”) is composed of residues interacting directly or indirectly with the protonated Schiff base and the extracellular half channel. It is a critical region in light-driven proton pumps that enables active pumping at substantial electrochemical loads and prevents passive proton backflow during illumination (9). Therefore, we mutated corresponding positions of the counterion complex in KR2, as illustrated in Fig. 1 D, using the published KR2 crystal structure (Protein Data Bank [PDB]: 3X3C) solved at neutral pH (15).
In KR2, position D116 is predicted to act as the primary proton acceptor of the protonated Schiff base. However, D116 is protonated at low pH (12, 15, 28), consistent with the low activity of KR2 observed at pHo 5.0 (Fig. S5 A). Indeed, the KR2-D116N mutant, which mimics the protonated state, showed no distinct stationary photocurrents (Fig. 1 E). Nevertheless, some diverse and highly pHo-dependent transient photocurrents—reflecting charge movements within the protein—remained and reversed to inward at low pHo. In addition, positions S70 and N112 are located in close proximity to D116 (Fig. 1 D), and both single mutants, S70A and N112D, showed reduced outward photocurrents with pronounced initial peak currents (Fig. 1, B, F, and G; Fig. S5, B and C).
Mutation of the second predicted counterion, D251 to N, resulted in only small transient photocurrents and a total loss of stationary photocurrents (Fig. 1 H). This outcome was unexpected because of the fact that the homologous mutation in the CsR proton pump (CsR-D211N) induces passive proton conductance (9). However, the conservative mutation, KR2-D251E, showed stationary inward photocurrents, indicating pump leakiness (Fig. 1, B and I), but with amplitudes smaller than 10 nA. Further, replacement of extracellular Na+ with either K+ or Li+ led to changes in the reversal potential, indicating nonselective passive cation conductance in KR2-D251E (Fig. S5 D). Interestingly, KR2-L75K also displayed inward stationary photocurrents at negative membrane voltage (Fig. 1, B and J). This mutation was also encouraged by our earlier experiments in which the homologous mutation (≙ CsR-Y57K, BR-Y57K) converted the proton pumps CsR and BR into outward rectifying proton channels (9). The strong pHo dependency was conserved in KR2-L75K, but more negative reversal potentials were observed for all ionic conditions, indicating that ion leakiness is poor compared to D251E (Fig. S5 E).
Potassium selectivity in KR2-R109Q
Nearly all microbial rhodopsins contain a highly conserved arginine (e.g., KR2-R109, BR-R82, CsR-R83, ChR2-R120) in the extracellular half channel, near the counterion complex (Fig. 1 D; alignment in Fig. S2), with the exception of channelrhodopsins from the alga Guillardia theta (29, 30) and inward H+-pumping xenorhodopsins (31). Notably, previous reports have indicated that mutation of this conserved arginine in proton pumps also results in proton leakage at moderate electrochemical load with residual pump activity (9). Here, we mutated the conserved arginine at position 109 in KR2 to generate KR2-R109Q. Our results suggest that this mutant is a passive potassium channel with unexpected properties, as described in the following.
Fig. 2 A shows representative photocurrent traces of KR2-R109Q at different extracellular cation conditions (all pHo 10, with 100 mM NaCl, 100 mM KCl, or 100 mM LiCl). The cytoplasm of X. laevis oocytes contains ∼5–10 mM Na+ and 80–140 mM K+ (32). With extracellular 100 mM NaCl, there exists an inward-directed chemical gradient for Na+ and an outward-directed gradient for K+. Therefore, the observed outward currents in Fig. 2 A (left) can be explained by either cation pump activity or passive transport of K+ ions. Further, pronounced bidirectional stationary photocurrents were only observed in presence of extracellular KCl (100 mM in Fig. 2 A, middle). In this case, inward currents can only be explained by passive K+ influx because the proton gradient was outward-directed (high pHo). Additionally, passive proton transport is excluded because the reversal potentials are not affected by the pH gradient (Erev in Fig. 2 B). Nevertheless, the current-voltage relationship for KR2-R109Q (Fig. 2 B) demonstrates that photocurrents are strongly promoted by alkaline pHo, even more pronounced than in KR2-WT (illustrated in Fig. S6 B as the ratio pHo 10/7.5, measured at 100 mM KCl). Lithium ions are also passively transported but with reduced efficiency because these are associated with smaller inward currents, despite the higher Li+ gradient (Fig. 2 A, right).
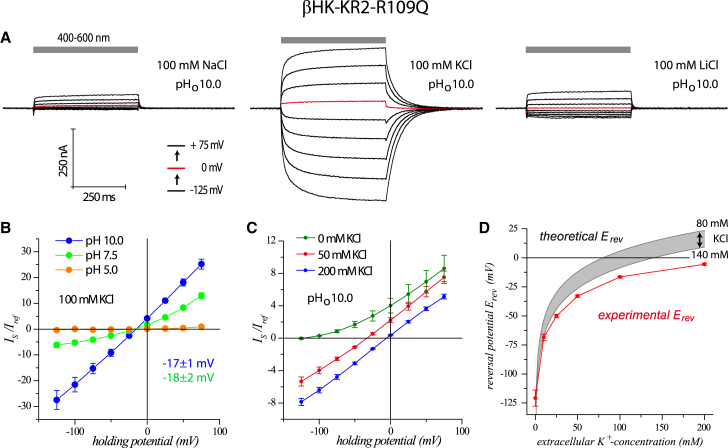
Figure 2.
Analysis of the K+-selective mutant, KR2βHK-R109Q, in X. laevis oocytes. (A) Photocurrent traces from a single oocyte at different extracellular ion conditions are shown: 100 mM NaCl (left), KCl (middle), or LiCl (right), and holding potentials (in 25 mV steps; red trace indicates 0 mV). (B) A current-voltage plot at different extracellular pH values (all 100 mM KCl, normalized to buffer with 100 mM NaCl (pHo 7.5) and 0 mV) is shown; pHo 10/7.5/5.0 (n = 16/8/3). (C) A current-voltage plot of extracellular K+ ion titration demonstrates that inward-directed photocurrents are carried by passive transport of K+ ions (normalized to buffer with 100 mM KCl, pHo 10.0, and 0 mV); 0/50/100 mM KCl (n = 4/4/5). (D) Comparison of the experimentally calculated reversal potentials with the theoretically expected reversal potentials for pure passive K+ conductance is shown (calculated with intracellular K+ concentrations of 80 and 140 mM); 0/10/25/50/100/200 mM KCl (n = 4/2/3/4/16/5). To see this figure in color, go online.
We next performed a stepwise reduction of extracellular K+ and observed changes of the reversal potential as expected for a passive K+ conductance (Fig. 2, C and D). The theoretically expected reversal potentials for an ideal passive potassium channel are shown in Fig. 2 D. However, the experimentally recorded reversal potentials (Fig. 2 D) are more positive than the calculated values (Δ15 mV for Ko = 200 mM) indicating either small residual pump activity or a small passive conductance for Na+. As support of small passive Na+ conductance, tiny inward currents were observed in the presence of extracellular Na+ already at pHo 7.5, which disappeared after replacement of NaCl with NMG-Cl, MgCl2, or CaCl2 (Fig. S6, D and E). Contribution of chloride is unlikely because current-voltage relationships are unchanged after replacement of 100 mM NaCl with 100 mM Na-gluconate (Fig. S6 E). Interestingly, we observed that currents were reduced by more than half in a mixture of “50 mM NaCl and 50 mM KCl” compared with “50 mM KCl,” suggesting that Na+ inhibits K+ conductance. Na+ itself is not passively conducted in this mixture of Na+/K+ ions because the reversal potentials remained unaffected (Fig. S6 F).
In addition, replacement of R109 with either alanine or asparagine (KR2-R109A and KR2-R109N, respectively) also resulted in pump leakiness but with reduced photocurrent amplitudes (Figs. S1 C and S6 A). For KR2-R109N in particular, the reversal potentials shifted to more positive values, and an extended passive conductance for Na+ was observed (Fig. S5, F–H). These results indicate that position R109 is not only involved in preventing leakiness but also plays a role in ion selectivity itself.
We then investigated the importance of position KR2-R109 in other NaRs. In particular, both NdR2 and NMR2 displayed small, but sufficient, photocurrents of up to 50 nA in our initial screen (Figs. S1 C and S7, A and B). Notably, the mutated pumps (NdR2-R109Q and NMR2-R108Q) also showed cation leakiness and strong pHo dependency but with ion selectivity that was distinct from KR2-R109Q. Unexpectedly, for the rhodopsin TrNaR2, bidirectional photocurrents were recorded with the WT protein (Fig. S7 C), and inward photocurrents disappeared after removal of extracellular sodium ions (Document S1. Figs. S1–S10, Document S2. Article plus Supporting Material). This indicates that TrNaR2 might have a natural leakiness for cations or is still a natural cation channel; however, further analysis of TrNaR2 was not possible because of the weak photocurrent amplitudes observed with this protein.
Voltage-clamp experiments in ND7/23-cells
We next performed voltage-clamp whole-cell analyses in ND7/23-cells, which allowed the control of intracellular buffer conditions. Because membrane targeting of KR2βHK is poor in ND7/23-cells, for these experiments, we used the well-targeted eKR2 construct instead (17). To ensure construct design had no effect on electrical properties, we compared KR2βHK and eKR2 in oocytes. Current-voltage relationships were similar with the exception that the K+ conductance was reduced in eKR2-R109Q, with Erev shifted from −17 to −31 mV (Fig. S8, A–E). In addition, inhibition of endogenous potassium channels with triethylamine (TEA, 20 mM), BaCl2 (5 mM), and CsCl (5 mM) in the buffers (Fig. S4) was critical for observing photocurrents of eKR2-R109Q at high intra- and extracellular K+ concentrations in ND7/23-cells.
The experiments in ND7/23-cells confirmed that the mutant eKR2-R109Q passively conducts potassium and lithium because inward currents were observed at low intracellular cation concentrations (1 mM NaCl, 1 mM KCl) and high extracellular K+ or Li+ concentrations (110 mM KCl or LiCl) (Fig. 3 A). Inward currents were reduced after increasing the intracellular K+ concentration from 1 to 110 mM (see Fig. 3 B).
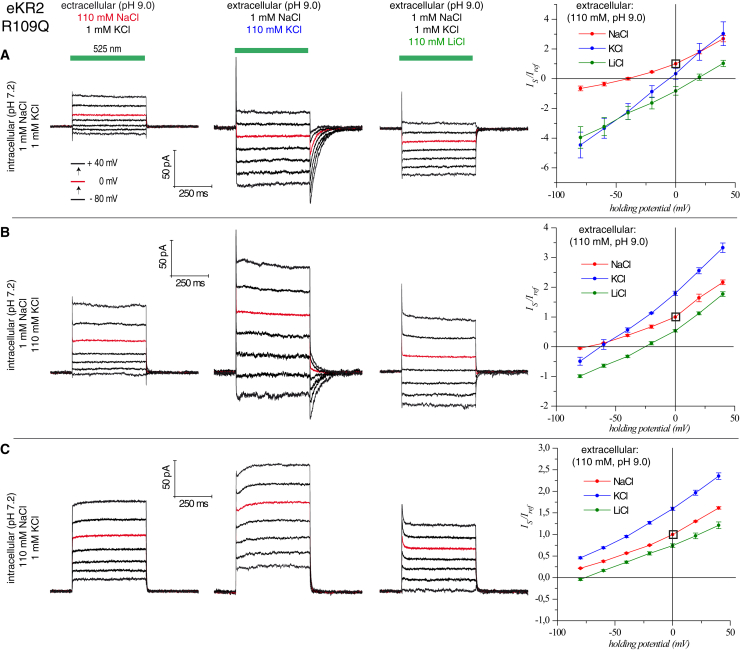
Figure 3.
Electrophysiological analysis of eKR2-R109Q with controlled intracellular ion conditions in ND7/23-cells and, therefore, with defined cation gradients. The extracellular buffers were varied, whereas the intracellular conditions remained constant during the patch. For all three intracellular conditions tested (A–C), the current curves of a single cell (holding potentials in 20 mV steps; red curve shows 0 mV) and the corresponding current-voltage diagrams from several measurements are shown (normalized to 110 mM NaCl (pHo 9.0) and 0 mV). (A) Intracellular: 1 mM Na+ and 110 mM K+. Extracellular: 110 mM NaCl, KCl, or LiCl (n = 4/4/4). (B) Intracellular: 1 mM Na+ and 110 mM K+. Extracellular: 110 mM NaCl, KCl, or LiCl (n = 6/5/5). (C) Intracellular: 110 mM Na+ and 1 mM K+. Extracellular: 110 mM NaCl, KCl, or LiCl (n = 6/5/5). To see this figure in color, go online.
In addition, we conclude that eKR2-R109Q was still capable of pumping Na+, and passive inward currents were only apparent if the intracellular Na+ concentration was low. This conclusion is based on the following measurement (Fig. 3 C): only outward photocurrents were observed at each voltage and extracellular ion state at high intracellular Na+ concentrations (110 mM NaCl, 1 mM KCl). Such residual pump activity is in line with the observed differences between the measured and calculated reversal potentials in oocytes (Fig. 2 D).
KR2-S70 is critical for sodium pump activity in R109Q
Within the hydrogen bonding network of the counterion complex, we identified KR2-S70 as a residue that may be critical for the residual Na+ pumping activity still observed in KR2-R109Q. The single mutant, KR2-S70A, remained an outward-directed pump without any leakiness in oocytes (Fig. 1 G; Fig. S6 A). Combination of KR2-R109Q with the S70A substitution, however, led to strong passive transport for all tested cations, with the exception of protons (Fig. 4, A–C). Inward-directed photocurrent amplitudes of this double mutant were also tenfold larger than for KR2-WT, highlighting its pronounced channel-like behavior (Fig. S6 A), although pHo dependence was further increased (Fig. 4 C; Fig. S6 B).
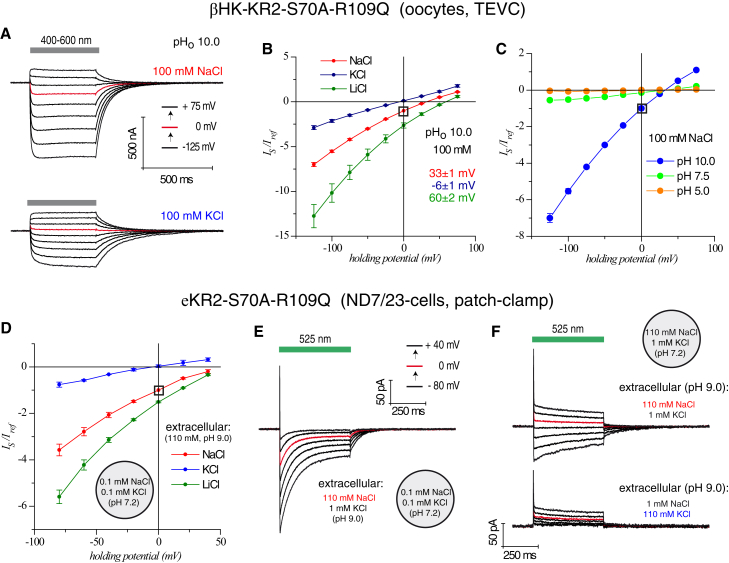
Figure 4.
Electrophysiological investigation of the Na+-conducting channel-like double mutant, KR2-S70A-R109Q, in oocytes (βHK) and ND7/23-cells (eKR2). (A) Photocurrent traces from a single oocyte at 100 mM NaCl or 100 mM KCl (both extracellular) and holding potentials (in 25 mV steps; red trace indicates 0 mV) are shown. (B and C) Current-voltage plots of the double mutant, KR2-S70A-R109Q, at different extracellular ion and pH conditions, measured in oocytes, are shown (normalized to 100 mM NaCl (pHo 10.0) and 0 mV); 110 mM NaCl/KCl/LiCl (n = 15/10/7), pHo 10/7.5/5.0 (n = 15/6/4). (D) Current-voltage plot of stationary photocurrents from patch-clamp measurements with strong inward-directed chemical gradients for Na+, K+, or Li+ are shown (normalized to 110 mM NaCl (pHo 9.0) and 0 mV); 110 mM NaCl/KCl/LiCl (n = 2/2/2). (E) Photocurrent traces from a single ND7/23-cell with a high inward-directed Na+ gradient are shown (in 20 mV steps; red trace indicates 0 mV). (F) Photocurrents from a single ND7/23-cell with a no-chemical gradient (upper traces) or with competition between an inward-directed Na+ and an outward-directed K+ gradient (bottom) are shown. To see this figure in color, go online.
We also tested the double mutant, eKR2-S70A-R109Q, in ND7/23-cells and observed negative photocurrents at inward-directed electrochemical gradients (Fig. 4, D and E). Here, the possibility of a small residual pump activity cannot be excluded because of the fact that small outward currents were observed at a high inward-directed K+ gradient, as shown in Fig. 4 D (extracellular: 110 mM KCl and 1 mM NaCl, intracellular: 0.1 mM NaCl and 0.1 mM KCl). Without a chemical gradient for Na+, bidirectional photocurrents were observed, depending on the electrical gradient (Fig. 4 F, top). Further, in this double mutant, Na+ transport outperformed K+ transport, as indicated by asymmetric Na+ and K+ distributions (Fig. 4 F, bottom). This can be explained by the residual pumping or higher passive conductance of Na+.
Reduction of the pHo dependence
Previous results (15) combined with our own observations indicate a strong pHo dependence of KR2 photocurrents at low internal Na+. Conversely, intracellular pH appears to exert only a minor influence on KR2 photocurrents (17). Interestingly, we found that pHo dependence is further increased for KR2 constructs containing the leak mutations, particularly in the double mutant KR2-S70A-R109Q (Fig. 4 B; Fig. S6 B), which limits their potential for use in optogenetic applications in neurons. These observations suggest that one or more charged residues are accessible from the extracellular side and reversibly protonated. Kato et al. reported (15) that at neutral pH, the KR2 structure contains the deprotonated D116 oriented toward the Schiff base and without a water molecule between D251 and R109 (as shown in Fig. 1 D). In the acidic structure, D116 is protonated and reoriented toward S70. Clearly, this protonated state is nonfunctional and exhibits no stationary photocurrents. However, beyond the deprotonation of D116, other residues also seem to be important for the pH dependence of KR2 function because the transient photocurrents of KR2-D116N (Fig. 1 E) were found to be pHo dependent (note that the KR2-R109Q-D116N/A mutants were nonfunctional).
Based on these considerations, various double mutant combinations with R109Q were tested to determine whether they can reduce pHo dependence (see Fig. S10). Of these, only the double mutant KR2-R109Q-D251N exhibited reduced pHo dependence between pHo 10 and pHo 7.5 (Fig. S6, A and B), suggesting that D251 is involved. However, this mutant also showed a restoration of ion-pumping activity, as indicated by the negative shift of Erev (Fig. S5 I). These data seem to be inconsistent with results from the single mutant, D251N (Fig. 1 H), which showed neither active nor passive photocurrents and only a transient charge movement upon light on, whereas the combination with R109Q rescued functionality.
Functional expression in E. coli and characterization of purified KR2-R109Q
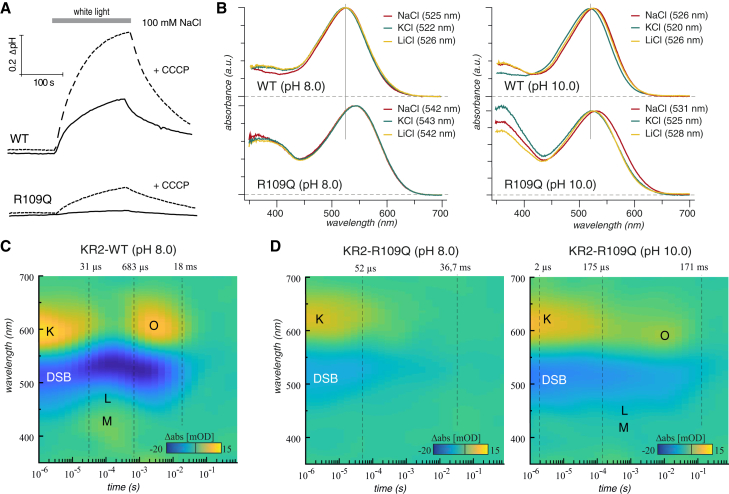
Lastly, we expressed KR2-R109Q in E. coli and found that illumination of cells expressing this protein did not promote substantial pH changes in the cell suspension but did lead to alkalization after addition of the protonophore, CCCP, in 100 mM NaCl (Fig. 5 A). This can be interpreted as residual outward-directed Na+ pumping at low membrane voltage (Fig. S9 B). We did not observe any light-induced pH changes for cell suspensions of KR2-R109A, R109N, or S70A-R109Q (Fig. S9).
Figure 5.
Expression and spectroscopic investigation of KR2-WT and KR2-R109Q in E. coli. (A) Light-induced pH changes in suspensions of E. coli cells expressing KR2-WT or KR2-R109Q (in 100 mM NaCl) are shown. (B) Absorption spectra of purified KR2 (upper panel) and KR2-R109Q (lower panel) recorded at pH 8.0 (left) and pH 10.0 (right) and variable ionic conditions are shown: 150 mM NaCl (red), 150 mM KCl (green), or 150 mM LiCl (yellow). (C and D) Reconstructed absorption difference surface plots are shown from flash photolysis measurements on purified recombinant (C) KR2 (150 mM NaCl) and (D) KR2-R109Q (150 mM NaCl (pH 8) and 150 mM KCl (pH 10)). The spectral data were globally fitted, and the resulting photointermediate decay τ values are indicated in the surface plots. To see this figure in color, go online.
We further found that the absorption maximum of recombinant KR2-R109Q was 16–21 nm red-shifted at pH 8.0 compared to KR2-WT, but this shift is reduced at pH 10 (Fig. 5 B; Fig. S8, F–H). These data suggest that the R109 substitution increases the pKa of D251 or D116, causing protonation at neutral pH and deprotonation only at high pH. In addition, time-resolved flash photolysis experiments revealed that KR2-R109Q shows a prolonged K-state decay, without an obvious L- and M-intermediate (Fig. 5, C and D). The O-intermediate was also less prominent because of protonation of D251 and was only detectable at alkaline pH when D251 deprotonates. Overall, the spectral data revealed that pH has a strong impact on the KR2-R109Q photocycle and its kinetics, which is consistent with our electrical recordings.
Discussion
In this study, we used mutagenesis, electrophysiology analysis, and spectroscopy to investigate the potential for conversion of NaRs, such as KR2, into passive cation channels. Our results reveal that cation leakiness can be introduced into these NaRs, and moreover, even discrimination between Na+ and K+ ions is possible. We further analyzed the KR2-R109Q mutant, which shows passive and selective conductance for potassium ions, in more detail in oocytes and ND7/23-cells. Despite its passive K+ conductance, KR2-R109Q still functions as an active outward-directed Na+ pump if intracellular Na+ ions are available and the counterion D116 is deprotonated in the dark state.
Residual pumping in KR2-R109Q is supported by the following observations: first, outward-directed photocurrents were detected against an inward-directed electrochemical gradient for Na+ in TEVC experiments in oocytes (Fig. 2 A, left). Second, only outward photocurrents were observed in whole-cell voltage-clamp measurements with high intracellular Na+ concentrations, as well as in those with a symmetrical chemical gradient for Na+ (Fig. 3 C). Finally, a slight alkalization of the extracellular medium (passive H+ influx facilitated by CCCP) was detected upon illumination of E. coli suspensions expressing KR2-R109Q in 100 mM NaCl (Fig. 5 A). In all cases, when the intracellular Na+ concentration is lower than ∼5–10 mM and extracellular K+ is high (110 mM), the KR2-R109Q mutant shows passive inward leakage for cations (preferentially K+) in both oocytes and ND7/23-cells (Figs. 2 and 3).
Moreover, as previously noted for KR2-WT in ND7/23-cells and suggested by the pH assay in E. coli, our measurements indicate a strong dependency of photocurrent amplitudes on extracellular pHo for all tested NaRs and their mutants under physiological, i.e., low, intracellular sodium concentrations (16, 17). Critically, an enhanced understanding of this characteristic feature of NaRs will be important for enabling the use of passive conducting mutants in optogenetic applications. Here, it was possible to reduce the pHo dependency of KR2-R109Q in the double mutant, KR2-R109Q-D251N, but only with a concomitant loss of passive conductance (Figs. S5 I and S6, A and B).
Based on our results, we conclude that leak mutations R109Q and L75K increase the pKa value of the aspartate D251 and that therefore alkaline pH values are necessary for maintenance of the deprotonated states. This is supported by the observation that the absorption spectrum maximum of KR2-R109Q is red-shifted at pH 8 (Fig. 5 B). For the proton pump BR, it is well known that residue R82 influences the protonation states of the counterion complex, as well as for other residues in the extracellular half channel (33, 34). We propose a similar importance of R109 in stabilization of pKa values in NaRs, and we further suggest that this residue serves a gate function for central restriction, which separates the cavity of the Schiff base from the ion-release cavity (16). It is likely that this restriction prevents the passive backflow of cations during the photocycle. Lastly, we hypothesize that pHo dependency arises because of the combination of the protonation states of D116 and D251 and their pH-dependent interactions with other residues of the counterion complex in both dark state and during the photocycle. Thus, this obstacle is not easy to overcome for development of a light-gated K+ channel that is fully functional at neutral pHo.
Conclusions
There continues to be a demand for the development of light-gated selective K+ channels, particularly for neuronal silencing, using light of moderate-to-low intensities. A number of attempts have been made to generate such channels (35, 36, 37, 38, 39, 40, 41), but all available constructs developed to date have specific drawbacks, such as weak expression, poor membrane targeting, slow kinetics, need for incubation with chemical compounds, and significant dark activity. Consistent with these limitations, all attempts to introduce high K+ selectivity directly in channelrhodopsins have also failed (42, 43). Here, our results show that ion selectivity for K+ over Na+ for passive transport can be introduced into a microbial rhodopsin starting from the Na+-pump KR2, but more work will be necessary to generate a channel that functions at neutral pH.
Author Contributions
A.V. and P.H. designed the study. A.V. designed and measured NaR constructs and their mutants in oocytes and ND7/23-cells and performed pH measurements with E. coli cells. A.S. cloned and purified the protein from E. coli and performed spectroscopic experiments. C.G. provided the targeting variant for mammalian cells, made early recordings in ND7/23 cells, and supported the E. coli measurements. F.H. and M.A.M. supported measurements in oocytes. A.V. wrote the manuscript, with support from P.H., C.G., and A.S. All authors approved the final version of the article.
Acknowledgments
We thank Altina Klein, Maila Reh, and Tharsana Thamalingam for technical assistance. We also thank Roman Fudim for helpful discussions.
This work was supported by the Deutsche Forschungsgemeinschaft (SPP 1926, SFB 1078, UniCat), the Gottfried Wilhelm Leibniz-Preis, and the European Union’s Horizon 2020 (Stardust) program. P.H. is senior professor for neuroscience, supported by the Hertie Foundation.
Editor: Ian Forster.
Footnotes
Christiane Grimm’s present address is NeuroTechnology Center, Department of Biological Sciences, Columbia University, New York, New York 10027.
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2019.04.001.
Supporting Material
References
- 1.Ernst O.P., Lodowski D.T., Kandori H. Microbial and animal rhodopsins: structures, functions, and molecular mechanisms. Chem. Rev. 2014;114:126–163. doi: 10.1021/cr4003769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hochbaum D.R., Zhao Y., Cohen A.E. All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins. Nat. Methods. 2014;11:825–833. doi: 10.1038/nmeth.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wiegert J.S., Mahn M., Yizhar O. Silencing neurons: tools, applications, and experimental constraints. Neuron. 2017;95:504–529. doi: 10.1016/j.neuron.2017.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rost B.R., Schneider-Warme F., Hegemann P. Optogenetic tools for subcellular applications in neuroscience. Neuron. 2017;96:572–603. doi: 10.1016/j.neuron.2017.09.047. [DOI] [PubMed] [Google Scholar]
- 5.Gadsby D.C. Ion channels versus ion pumps: the principal difference, in principle. Nat. Rev. Mol. Cell Biol. 2009;10:344–352. doi: 10.1038/nrm2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hegemann P., Fuhrmann M., Kateriya S. Algal sensory photoreceptors. J. Phycol. 2001;37:668–676. [Google Scholar]
- 7.Nagel G., Ollig D., Hegemann P. Channelrhodopsin-1: a light-gated proton channel in green algae. Science. 2002;296:2395–2398. doi: 10.1126/science.1072068. [DOI] [PubMed] [Google Scholar]
- 8.Nagel G., Szellas T., Bamberg E. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc. Natl. Acad. Sci. USA. 2003;100:13940–13945. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vogt A., Guo Y., Hegemann P. Conversion of a light-driven proton pump into a light-gated ion channel. Sci. Rep. 2015;5:16450. doi: 10.1038/srep16450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inoue K., Tsukamoto T., Sudo Y. Converting a light-driven proton pump into a light-gated proton channel. J. Am. Chem. Soc. 2015;137:3291–3299. doi: 10.1021/ja511788f. [DOI] [PubMed] [Google Scholar]
- 11.Takayama R., Kaneko A., Sudo Y. Production of a light-gated proton channel by replacing the retinal chromophore with its synthetic vinylene derivative. J. Phys. Chem. Lett. 2018;9:2857–2862. doi: 10.1021/acs.jpclett.8b00879. [DOI] [PubMed] [Google Scholar]
- 12.Inoue K., Ono H., Kandori H. A light-driven sodium ion pump in marine bacteria. Nat. Commun. 2013;4:1678. doi: 10.1038/ncomms2689. [DOI] [PubMed] [Google Scholar]
- 13.Tsunoda S.P., Prigge M., Kandori H. Functional characterization of sodium-pumping rhodopsins with different pumping properties. PLoS One. 2017;12:e0179232. doi: 10.1371/journal.pone.0179232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kandori H., Inoue K., Tsunoda S.P. Light-driven sodium-pumping rhodopsin: a new concept of active transport. Chem. Rev. 2018;118:10646–10658. doi: 10.1021/acs.chemrev.7b00548. [DOI] [PubMed] [Google Scholar]
- 15.Kato H.E., Inoue K., Nureki O. Structural basis for Na(+) transport mechanism by a light-driven Na(+) pump. Nature. 2015;521:48–53. doi: 10.1038/nature14322. [DOI] [PubMed] [Google Scholar]
- 16.Gushchin I., Shevchenko V., Gordeliy V. Crystal structure of a light-driven sodium pump. Nat. Struct. Mol. Biol. 2015;22:390–395. doi: 10.1038/nsmb.3002. [DOI] [PubMed] [Google Scholar]
- 17.Grimm C., Silapetere A., Hegemann P. Electrical properties, substrate specificity and optogenetic potential of the engineered light-driven sodium pump eKR2. Sci. Rep. 2018;8:9316. doi: 10.1038/s41598-018-27690-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Snellenburg J.J., Laptenok S., van Stokkum I.H.M. Glotaran: a Java-based graphical user interface for the R package TIMP. J. Stat. Softw. 2012;49:22. [Google Scholar]
- 19.Geibel S., Friedrich T., Bamberg E. The voltage-dependent proton pumping in bacteriorhodopsin is characterized by optoelectric behavior. Biophys. J. 2001;81:2059–2068. doi: 10.1016/S0006-3495(01)75855-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ivanova N., Rohde C., Lapidus A. Complete genome sequence of Truepera radiovictrix type strain (RQ-24) Stand. Genomic Sci. 2011;4:91–99. doi: 10.4056/sigs.1563919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H., Sineshchekov O.A., Spudich J.L. In vitro demonstration of dual light-driven Na+/H+ pumping by a microbial rhodopsin. Biophys. J. 2015;109:1446–1453. doi: 10.1016/j.bpj.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balashov S.P., Imasheva E.S., Lanyi J.K. Light-driven Na(+) pump from Gillisia limnaea: a high-affinity Na(+) binding site is formed transiently in the photocycle. Biochemistry. 2014;53:7549–7561. doi: 10.1021/bi501064n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwon Y.M., Kim S.Y., Kim S.J. Diversity and functional analysis of light-driven pumping rhodopsins in marine Flavobacteria. MicrobiologyOpen. 2016;5:212–223. doi: 10.1002/mbo3.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshizawa S., Kumagai Y., Kogure K. Functional characterization of flavobacteria rhodopsins reveals a unique class of light-driven chloride pump in bacteria. Proc. Natl. Acad. Sci. USA. 2014;111:6732–6737. doi: 10.1073/pnas.1403051111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwon S.K., Kim B.K., Kim J.F. Genomic makeup of the marine flavobacterium Nonlabens (Donghaeana) dokdonensis and identification of a novel class of rhodopsins. Genome Biol. Evol. 2013;5:187–199. doi: 10.1093/gbe/evs134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yi H., Chun J. Unification of the genera Nonlabens, Persicivirga, Sandarakinotalea and Stenothermobacter into a single emended genus, Nonlabens, and description of Nonlabens agnitus sp. nov. Syst. Appl. Microbiol. 2012;35:150–155. doi: 10.1016/j.syapm.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 27.Yoon J.H., Kang S.J., Oh T.K. Reclassification of the three species of the genus Krokinobacter into the genus Dokdonia as Dokdonia genika comb. nov., Dokdonia diaphoros comb. nov. and Dokdonia eikasta comb. nov., and emended description of the genus Dokdonia Yoon et al. 2005. Int. J. Syst. Evol. Microbiol. 2012;62:1896–1901. doi: 10.1099/ijs.0.035253-0. [DOI] [PubMed] [Google Scholar]
- 28.Shigeta A., Ito S., Kawamura I. Solid-state nuclear magnetic resonance structural study of the retinal-binding pocket in sodium ion pump rhodopsin. Biochemistry. 2017;56:543–550. doi: 10.1021/acs.biochem.6b00999. [DOI] [PubMed] [Google Scholar]
- 29.Govorunova E.G., Sineshchekov O.A., Spudich J.L. Structurally distinct cation channelrhodopsins from cryptophyte algae. Biophys. J. 2016;110:2302–2304. doi: 10.1016/j.bpj.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamauchi Y., Konno M., Kandori H. Molecular properties of a DTD channelrhodopsin from Guillardia theta. Biophys. Physicobiol. 2017;14:57–66. doi: 10.2142/biophysico.14.0_57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shevchenko V., Mager T., Gordeliy V. Inward H+ pump xenorhodopsin: mechanism and alternative optogenetic approach. Sci. Adv. 2017;3:e1603187. doi: 10.1126/sciadv.1603187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sobczak K., Bangel-Ruland N., Weber W.M. Endogenous transport systems in the Xenopus laevis oocyte plasma membrane. Methods. 2010;51:183–189. doi: 10.1016/j.ymeth.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 33.Otto H., Marti T., Heyn M.P. Substitution of amino acids Asp-85, Asp-212, and Arg-82 in bacteriorhodopsin affects the proton release phase of the pump and the pK of the Schiff base. Proc. Natl. Acad. Sci. USA. 1990;87:1018–1022. doi: 10.1073/pnas.87.3.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Subramaniam S., Marti T., Khorana H.G. Protonation state of Asp (Glu)-85 regulates the purple-to-blue transition in bacteriorhodopsin mutants Arg-82----Ala and Asp-85----Glu: the blue form is inactive in proton translocation. Proc. Natl. Acad. Sci. USA. 1990;87:1013–1017. doi: 10.1073/pnas.87.3.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Banghart M., Borges K., Kramer R.H. Light-activated ion channels for remote control of neuronal firing. Nat. Neurosci. 2004;7:1381–1386. doi: 10.1038/nn1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Janovjak H., Szobota S., Isacoff E.Y. A light-gated, potassium-selective glutamate receptor for the optical inhibition of neuronal firing. Nat. Neurosci. 2010;13:1027–1032. doi: 10.1038/nn.2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fortin D.L., Dunn T.W., Kramer R.H. Optogenetic photochemical control of designer K+ channels in mammalian neurons. J. Neurophysiol. 2011;106:488–496. doi: 10.1152/jn.00251.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caro L.N., Moreau C.J., Vivaudou M. Engineering of an artificial light-modulated potassium channel. PLoS One. 2012;7:e43766. doi: 10.1371/journal.pone.0043766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kang J.Y., Kawaguchi D., Wang L. In vivo expression of a light-activatable potassium channel using unnatural amino acids. Neuron. 2013;80:358–370. doi: 10.1016/j.neuron.2013.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cosentino C., Alberio L., Moroni A. Optogenetics. Engineering of a light-gated potassium channel. Science. 2015;348:707–710. doi: 10.1126/science.aaa2787. [DOI] [PubMed] [Google Scholar]
- 41.Bernal Sierra Y.A., Rost B.R., Schmitz D. Potassium channel-based optogenetic silencing. Nat. Commun. 2018;9:4611. doi: 10.1038/s41467-018-07038-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Plazzo A.P., De Franceschi N., Mongillo M. Bioinformatic and mutational analysis of channelrhodopsin-2 protein cation-conducting pathway. J. Biol. Chem. 2012;287:4818–4825. doi: 10.1074/jbc.M111.326207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kato H.E., Zhang F., Nureki O. Crystal structure of the channelrhodopsin light-gated cation channel. Nature. 2012;482:369–374. doi: 10.1038/nature10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.