Abstract
High content, phenotypic screens offer a powerful approach to systems biology at the cellular level. The approach employs cells carrying fluorescently-labeled molecules or organelles in 384 or 1536 well microplates, and an automated confocal screening microscope for capturing images from each well. Although some specifics vary according to the assay type, each will apply some degree of image processing and feature extraction followed by a data analysis pipeline to identify the perturbations (small molecules, etc.) of interest. We describe and discuss the advantages and limitations of high content assays and screens using the specific example of assaying mitochondrial dynamics in primary neurons. We provide a detailed description of our culturing methods, imaging and data analysis techniques and provide an open source, ready to use CellProfiler pipeline for high throughput image segmentation and quantification tool for mitochondrial parameters.
Keywords: high content phenotypic screen, automated image analysis, mitochondrial dynamics, mitochondrial morphology, primary neurons, CellProfiler, pintool, hit selection
1. Introduction
High content, phenotypic assays and screens offer powerful platforms for identifying and characterizing small molecules, genes, or other agents that modulate cellular processes (Singh, Carpenter, & Genovesio, 2014; Taylor, Haskins, & Giuliano, 2007; Zanella, Lorens, & Link, 2010). There are many advantages offered from high content, or image-based assays. Scientific data comes in many different forms but no type seems more powerful than a robust image for providing confidence in the conclusions. Image-based assays also can provide both temporal and spatial information at the subcellular level about the activities of organelles or other targets being monitored. Moreover, the investigator can design such assays to be multiplexed, revealing relationships between several cellular processes or targets being monitored. The latter advantage has been driven by the discovery and generation over the last two decades of an extraordinary number of new fluorescent proteins and dyes that provide the flexibility to label most cellular targets with fluorescent reporters with different spectral properties. Furthermore, imaging technology, including both hardware and software tools, have improved dramatically allowing the investigator to rapidly capture thousands of images from cells arrayed in either two- or three-dimensions. Phenotypic screens, which monitor cellular processes rather than a specific molecular target, cast a larger net and can offer increased value in the search for agents that modulate a cellular process or interest (Swinney & Anthony, 2011).
Nevertheless, high content, phenotypic assays do offer challenges compared to target-based assays. These assays require advanced imaging instrumentation that call for sophisticated knowledge of imaging techniques and software algorithms for processing the images. The advanced imaging instrumentation can increase costs and equipment breakdowns that create delays in obtaining experimental results. Moreover, the depth and rate of image capture demands external storage devices for terabytes of digital information. Although phenotypic screens cast a broader net in searches for agents of interest, they do not reveal the mechanism of action of such agents and this requires further studies compared to target-based screens based on a known mechanism of action.
Although such high content, phenotypic assays and screens are extraordinarily flexible and can be designed to query a myriad of cellular processes, providing essentially a systems-level snapshot of cellular processes, we focus here on techniques we have employed to monitor mitochondrial dynamics in primary neuronal cultures. Nevertheless, the principles employed in our studies are applicable to many other high content, phenotypic assays and screens that have or could be designed and implemented. Our focus on this particular organelle and its dynamics in neurons is driven by two fundamental points. First, mitochondrial loss and dysfunction are common hallmarks of neurodegenerative diseases such as Alzheimer’s disease, Parkinson’s disease, Huntington’s disease or Amyotrophic Lateral Sclerosis, which are characterized by a gradual, progressive and selective loss of certain neuronal systems in each disease (Nunnari & Suomalainen, 2012). The lack of biologically relevant screening platforms, validated targets and the high failure rate of clinical trials present an urgent need for novel screening technologies, new drugs and druggable targets (Cummings, 2018; Hawkes, 2018). Since the pathophysiology of neurodegenerative diseases is complex and not fully understood, the implementation of phenotypic screens for successful drug development is preferable over target-based approaches. Second, given the unique physiology of neurons, screening directly in the neuronal environment holds more promise towards the goal to identify novel CNS active compounds, than screening in non-neuronal cell types.
Neurons are highly dependent on energy, supplied predominantly by the mitochondrial system. Neuronal mitochondria are highly dynamic, continuously being born through biogenesis and turned over through mitophagy, being transported from the soma to distal cellular compartments in axons and dendrites, and fusing with each other and dividing (fission) in order to adapt to the local energy requirements. These mechanisms provide homeostatic control over intracellular calcium levels, and help with quality control over the mitochondrial system (Chen & Chan, 2009; Misgeld & Schwarz, 2017). Mixing mitochondrial content by fusion and isolating damaged parts by fission maintains healthy mitochondria and the effectiveness of the mitochondrial system. Excessive fission, which produces small, round mitochondria and can lead to cell death, has been reported in several neurodegenerative diseases (Johri & Beal, 2012). In contrast, elongation of mitochondria either by excessive fusion or the inhibition of fission increases efficiency and is protective under stressed conditions (Wang et al., 2012; Westermann, 2012). Mitochondrial mass has also been reported to decrease in neurodegenerative diseases (Sheng et al., 2012), either by the decrease of mitochondrial biogenesis and/or increased elimination by mitophagy. To capture the complexity of neuronal mitochondrial dynamics and screen for effective neuronal mitotherapeutics, we have employed high-content, phenotypic assays and screens (Daub, Sharma, & Finkbeiner, 2009).
2.0. HCS for Compounds Increasing Mitochondrial Content, Elongation and Improving Health
2.1. Labeling of mitochondria in primary neurons
High content assays and screening are centered on fluorescence microscopy, where a wide variety of new generation fluorescent probes, markers and dyes are available for visualizing whole cells, subcellular compartments, organelles or proteins. Live-cell fluorescence imaging enables the examination of the anatomy of cellular networks, subcellular structures, their dynamics, localization and even the mechanism of action of proteins. The cell type to be used, the specific phenotypic target, and the fluorescent reporter is dictated by the goal of assay development or screen.
2.1.1. Fluorescent dyes
Several cell-permeable mitochondrion-selective fluorescent dyes exist that allow live cell imaging among which many are also retained in mitochondria after fixation and therefore compatible with antibody-based imaging. The mitochondrial accumulation of lipophilic cationic dyes is dependent on the inner mitochondrial membrane potential and have been used widely to stain healthy mitochondria (DiOC1(3) (Korchak, Rich, Wilkenfeld, Rutherford, & Weissmann, 1982), Rhodamine 123 (Emaus, Grunwald, & Lemasters, 1986), TMRM (Ehrenberg, Montana, Wei, Wuskell, & Loew, 1988), TMRE (Ehrenberg et al., 1988), DASPMI (Bereiter-Hahn, 1976), DASPEI (Rafael, 1980)). Actively respiring mitochondria with high membrane potential will accumulate more of the fluorescent dye, therefore changes in mitochondrial membrane potential – the driving force for ATP synthase – allows for following alterations in mitochondrial function. Besides the classic cationic dyes, a newer generation of probes are also available that exhibit potential-dependent accumulation in mitochondria. For example, the cationic JC-1 (Smiley et al., 1991) carbocyanine dye exists as a monomer at low concentrations emitting green fluorescence in depolarized mitochondria, while when it is accumulated at high concentrations in hyperpolarized mitochondria, it aggregates and emits orange fluorescence. This provides the possibility of determining the ratio of polarized/depolarized mitochondria within a cell. A wide variety of MitoTracker dyes (Chazotte, 2011) with different fluorophores exist whose accumulation is dependent on or independent of the inner mitochondrial membrane potential, some of which are retained after fixation as well as after permeabilization of the membranes, giving researchers a wide range of dyes to choose from.
Staining of mitochondria is usually rapid (within an hour) compared to probes that require new protein expression, although dye concentration and incubation time should be optimized by the user for each cell type. The use of fluorescent dyes in HCS may be limited by increased variance occurring from differences in staining efficiency from experiment to experiment. The efficiency of the staining itself may be influenced by the state of the cell cultures as well and other factors. When applying dyes, their high cell permeability, specificity and non-toxicity to cells is crucial for the development of successful and robust assays. However, the main drawback of dyes in terms of high content assays and screens for neuronal mitochondria stems from the high density of mitochondria in neurons. In such cultures where all mitochondria are stained the high number of overlapping mitochondria hinders their individual identification for further analysis. On the other hand, primary neurons fail to thrive when plated at lower densities.
2.1.2. Expression of fluorescent reporters
Transient or stable expression of fluorescent reporter proteins in primary neurons usually provides better signal to noise ratio than dyes, persistent expression and the flexibility of performing cell-type or region-specific assays. Plasmid transfection of primary neurons has been a challenge in the past because of toxicity, low and variable efficiency across experiments. However, efficient viral transduction systems for neurons are available now with boosted uptake and expression such as modified baculoviral systems (Li, Yang, & Wang, 2005; Mansouri et al., 2016) or adeno-associated viral vectors (AAV) which can be used in vitro and in vivo as well. The different serotypes of AAV vectors offer a cell type specific infection of mixed cultures, and additional specificity can be obtained by using cell-type specific promoters to express the fluorescent reporter protein. Neurons can be infected with different AAV serotypes including AAV1, AAV2, AAV5, AAV6, AAV8 or AAV9 (Aschauer, Kreuz, & Rumpel, 2013; Hammond, Leek, Richman, & Tjalkens, 2017; Michelfelder & Trepel, 2009; Murlidharan, Samulski, & Asokan, 2014; Wu, Asokan, & Samulski, 2006). The different glycan architectures of the AAV capsids contribute to the variations of efficiency of the gene transfer, as well as how they manage endosomal escape, genome release or transcription (Murlidharan et al., 2014).
We have used a mouse line expressing a Cre-dependent dual reporter of mitochondrial tagGFP2 and nuclear tagBFP to visualize mitochondria and nuclei (Figure 1). The mitochondria targeted GFP2 (Rizzuto, Brini, Pizzo, Murgia, & Pozzan, 1995) is an improved mutant variant of GFP that matures faster, is more pH stable than EGFP, shows improved performance in protein fusions and is optimized for expression at 37°C. To induce the expression of the reporters specifically in neurons obtained from P0 mouse forebrains, we used a replication deficient AAV9 packaged with a plasmid containing a CaMKII neuronal-specific promoter upstream of the coding sequences for a mammalian codon optimized (improved) Cre-recombinase (iCRE).
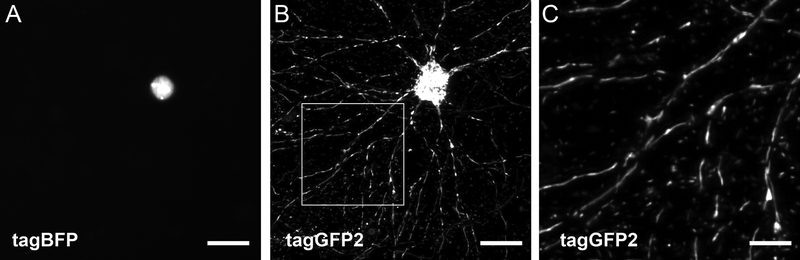
Figure 1. Neuron expressing a mitochondrial-localized tagGFP2 and a nuclear-localized tagBFP.
Independent labeling of the nucleus by tagBFP (A) and a representative display of several hundred neuritic mitochondria and a large, essentially contiguous mitochondrion in the soma labeled by tagGFP2 (B). The inset bordered with a solid line shows a magnified image of the neuritic mitochondria (C). Scalebars are 20 μm (A, B) and 8 μm (C).
2.2. Culturing of primary neurons
Primary neuronal cultures are powerful and relatively simple model systems to perform various screening projects towards the goal of finding effective compounds for the treatment of brain disorders. Homogenous cultures of neurons and/or glia can be maintained in 384- or 1536-well plates, in a highly controllable environment suitable for HTS and HCS.
Primary neuronal cultures are obtained by dissecting the brains of mice or rats - the two most commonly used mammals in research -, obtained from adults, newborn pups or embryos. Using newborn pups or embryos is preferable, since their brain is immature and less susceptible to damage. Different brain regions can be isolated and cultured, the cerebellum, cortex or hippocampus as examples, as well as the components of the peripheral system such as the dorsal root ganglia. Dissection should occur in a semi-sterile environment, with the freshly isolated tissue placed immediately in ice-cold buffered saline. Dissociation of the tissue into individual cells may involve both proteolytic (papain, trypsin) as well as mechanic procedures (mincing, trituration). Plating and maintenance include special supplements that support neuronal growth including vitamins, amino acids or glucose (insulin, transferrin, putrescine, progesterone, etc…) and protective supplements (anti-oxidants such as catalase, glutathione or superoxide dismutase, and l-carnitine for oxygen-glucose deprivation). Chemicals that prevent the growth of non-neuronal cells can also be added to the culture (e.g. 5-Fluoro-2′-deoxyuridine), or if astrocyte cultures are needed, special media and supplementation can be used. Neurons can be plated onto different dishes, plates or cover glasses placed into dishes, but surfaces need to be pre-treated with some extracellular matrix protein (poly-D-lysine, poly-amine, laminin) for proper attachment and to support growth. Neuronal processes of cells begin to extend a few hours after plating, but fully mature neurons are only developed after culturing at least 10–11 days in vitro. Cultures can be maintained for approximately a month, with a gradual decrease of the health of the neurons until the majority of cells die (Lesuisse & Martin, 2002).
Selecting the proper plate (material, coating, vendor) is an important decision for a successful HCS project. Primary neuronal cultures do not grow well on glass, so polystyrene or cycloolefin plates are used; the latter has better optical properties (Niles & Coassin, 2008). Poly-D-lysine or poly-amine both work well with primary forebrain neurons. If cells clump together after a few days of plating, or do not attach to the surface of the plate within the first 4 hours, the coating may be at fault. Figure 2 shows the same batch of primary neurons plated on the same cycloolefin plates but coated with different substrates. Unfortunately, the quality of plate coating from the same vendor can vary across different batches of plates.
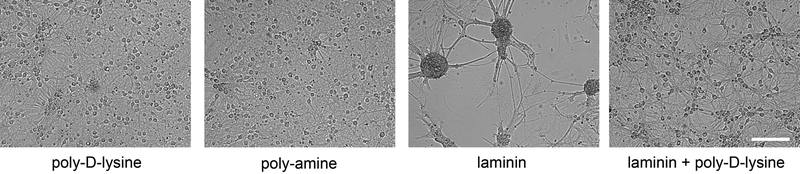
Figure 2. Culturing of neurons on surfaces with different coatings.
Bright-field images of DIV 14 primary forebrain neurons in 384-well plates plated from the same P0 pups at a seeding density of 15000 cells/well. Poly-D-lysine and poly-amine treated surfaces create a homogeneous layer of neurites with evenly scattered cell bodies, showing that neurons are healthy and not stressed. The stressed neurons plated on laminin form clumps of cell bodies with neurites that bundle together to form bridges between cell body clumps. Adding poly-D-lysine to laminin resolves the clumping, though these cell cultures are still less homogeneous than treating surfaces with solely poly-D-lysine or poly-amine. Scale bar is 200 μm.
In the following, we describe the maintenance of neuronal forebrain cultures isolated from newborn pups (P0). The expression of mitochondrial GFP2 was induced using optimized cell and viral particle numbers (Figure 3, see below).
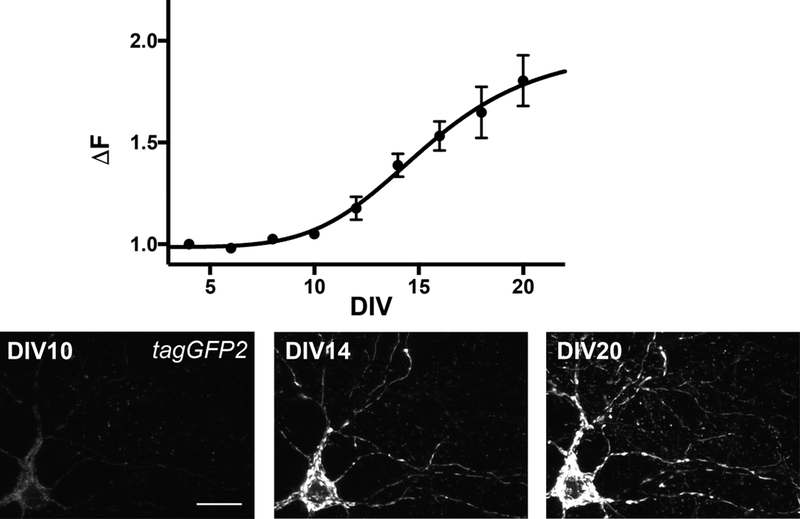
Figure 3. Expression of mitochondrial tagGFP2.
Expression of mitochondrial tagGFP2 in neurites over time shows that cultured neurons on DIV14 have an average 1.33 more intense signal than at DIV2, which signal plateaus at 1.97 (black curve, sigmoid fit). At DIV14 the signal is sufficiently strong to be analyzed and neurons are mature but still in good health. By DIV20, mitochondria show fragmentation and swelling, signs of mitochondrial degradation. Data are mean ± SD from 3 independent measurements. Scalebar is 20 μm.
2.2.1. Equipment and tools
Incubator (37°C, 5% CO2), microscope for dissection (Zeiss Stereoscope Discovery V8 or equivalent)
Forceps and scissors
Centrifuge, cell counter, 40 μm mesh-sized cell strainer
Biomek FXP liquid handling workstation or equivalent
45 μl Beckman Biomek XL filter tips (BA-0051–3FC)
Poly-D-lysine pre-coated Aurora 384-well plates with evaporation barrier (ABE1–01201-B), Aurora 1536-well plates ((ABI2–01000-B) or equivalent
2.2.2. Buffers and reagents
Dissection buffer (Ca2+ and Mg2+ free HBSS supplemented with 2 μg/ml gentamicin, 25 mM D-glucose, 1 mM pyruvate and 20 mM HEPES)
Plating media (Neurobasal supplemented with 0.75% FBS, 4 mM GlutaMAX and 2 μg/ml gentamicin, 2% B27, 16 μM 5-Fluoro-2′-deoxyuridine)
Feeding media (Neurobasal-A supplemented with 4 mM GlutaMAX, 2 μg/ml gentamicin, 2% B27)
Papain
2.2.3. Procedure
Collect forebrains of P0 pups in ice-cold dissection buffer and remove olfactory bulbs and meninges. Put 2–3 forebrains in 4 ml of ice-cold dissection buffer.
Digest tissue with 0.6 mg/ml pre-warmed papain at 37°C for 20 minutes, gently swirling the tube every 5 minutes.
Wash the brains extensively but carefully with pre-warmed plating media 3× while keeping them intact, leaving 2 ml of plating media on the tissue.
Triturate tissue carefully through a 1 ml regular plastic pipette tip several times (homogenize tissue), let the dissociated tissue sit for 60 seconds, then transfer 1 ml of the supernatant to a new tube and add 1 ml fresh plating media to the remaining tissue. Repeat this 3–5 times.
Dilute the cells collected in the supernatant to 10 ml with plating media and run it through a 40 μm mesh-sized cell strainer to filter out clumped tissue or small pieces of the meninges.
Centrifuge the cells at 420×g for 4 minutes.
Aspirate the supernatant and resuspend the cells carefully in ~5 ml warm plating media.
Check the live/dead cell ratio by using trypan blue (0.4%) with an automated cell counter or hematocytometer. A healthy cell suspension should have >90% live cells.
Dilute cells with warm plating media to achieve a final seeding density.. 5-Fluoro-2′-deoxyuridine in the plating media prevents glial cells or any other dividing cell types (e.g., cells from blood vessels) from overgrowing the neuronal culture. FBS is needed for the survival of ~10% astroglia that will support neuronal cell survival at plating and attachment, and it is not needed thereafter. B27 supports neuronal survival, especially during the first 4 days of culture. Initial seeding numbers are influenced by neuronal cell type, the type of plates used and the coating employed for attachment and survival. Each pup brain yields ~4–6×106 cells.
Add AVV9 containing the iCRE recombinase to the diluted cells at a final concentration of 1.5×108 viral particles/ml, mix gently and incubate for a few minutes, and then distribute the homogenous cell culture to the desired plates in the desired volume/wells. Using a robot for plating such as the Biomek FXP will increase consistency of plating across the plates. We add 80 μl/15000 cells/well (0.1875×106 cells/ml) on poly-D-lysine, pre-coated Aurora 384-well plates or 14 μl/7000 cells/well on pre-coated 1536-well Aurora plates. These give a nice confluency of monolayer cells which is needed for a healthy neuronal cell culture.
After 8 days, perform a 50% media change with feeding media and repeat it every 4 days thereafter.
2.2.4. Notes
Isolation of the forebrains (step 1) should not exceed 20–25 minutes since it will affect the viability of the cells.
Make sure to wash out papain well.
Trituration should be gentle, but the brain tissue should be totally homogenized at the end of the trituration cycles.
Make sure to use warm plating media throughout the whole process.
Cell number/well and the type of coating used for plating will substantially influence the growth and health of the neurons and should be optimized as an initial step for screens.
The number of viral particles added per well determines the number of neurons expressing the fluorophores and should be optimized for each batch of packaged virus and each screen.
2.3. Compound Transfer
Automated and standardized compound transfer is essential for a reproducible high throughput screen. Reproducibility not only depends on the method of delivery of compounds to the assay plates, but also on the type of library source plates, and how the compounds were prepared and how they were stored.
Compounds are usually dissolved at 10 mM concentration in DMSO:H2O=9:1, where the goal is to maximize compound dissolution and stability, and minimize the amount of solvent delivered onto the assay plates containing the cells. In case of primary neuronal cultures, cells will not tolerate more than 0.5% DMSO in the media, especially for 24–48-hour long treatments (C. Zhang et al., 2017). Compound transfer is also influenced by the material of the source plate that is used and its storage (room temperature, 4°C, −20°C or −80°C). Polypropylene and cycloolefin plates are preferable over polystyrene plates because polar molecules bind them less, they are more resistant to solvents and have high thermostability. Cycloolefin plates seem to perform even better than polypropylene plates when compounds were frozen previously (Smith et al., 2013). Compound potency seems to be the highest when plates have never been frozen, which is usually not feasible since source plates are usually kept frozen for the long term. If the compound plate is used frequently, it is advisable to store it well-sealed at room temperature then to subject it to multiple freeze-thaw cycles. Compounds can be stored at room temperature in DMSO:H2O=9:1 up to 6 months without significant loss of potency.
The delivery of compounds can be accomplished by pintooling them from a source plate into a destination assay plate containing the cell culture (Figure 4). In this contact-based method, each pin containing a slot or groove submerges into the liquid of a well and delivers a pre-set volume (few nanoliters to couple of hundreds of nanoliters) via capillary action and surface tension. The pins themselves are in a floating head, preventing pin damage and allowing the soaking of all the tips into the solution. Transfer depends very much on the properties of the solvent used for the source plates (Dunn & Feygin, 2000), the surface properties of the pins and the source plate, the amount of liquid it submerges into and the method of the cleaning of the tips (Cleveland & Koutz, 2005). As long as these variables are kept constant, pintooling is considered to be highly accurate and reproducible (Smith et al., 2013). Nevertheless, one should check precise transfer volumes using standardized procedures before starting a screen using pintooling (Cleveland & Koutz, 2005). One of its main drawbacks is its transfer of fixed volumes, which means the source concentrations may need to be adjusted to fit desired final concentrations. However, adjusting source concentrations is usually not applicable with preset concentrations and volumes of drug libraries available commercially.
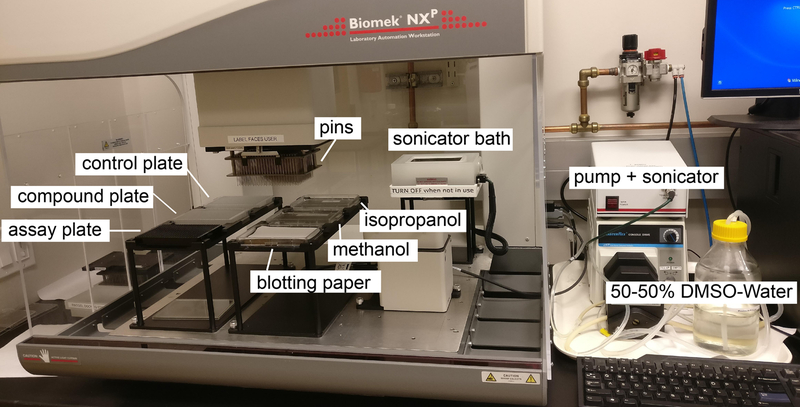
Figure 4. Deck layout of compound transfer via pintooling.
The pump is continuously circulating the DMSO:water mixture in the sonicator bath, where the pins are washed between each transfer, then soaked in isopropanol and methanol and dried on blotting paper. Pins pick up compounds first from either the control or compound plates and transfer them into the wells of the assay plate containing neurons. Usually pintooling is performed under non-sterile conditions, so minimizing the time the cells spend without a cover is important.
The non-contact acoustic droplet ejection (ADE) method offers the possibility of creating assay ready plates (ARPs), offering the advantage of custom designed layouts of the compounds from a fixed source plate. It uses acoustic energy to propel droplets (picoliters to nanoliters of volumes) with very high accuracy from a source plate into a destination plate that lies over the source plate in an inverted position. The source plate is stationary, while the transducer and the destination plate move to different locations, allowing the transfer of compounds from any well of the source plate to any other well of the destination plate. Although ARPs can be frozen and kept for a long periods of time, some evaporation of the droplets may occur during storage depending on temperature and humidity. Drying of the compounds onto the assay plates may lead to poor recovery and increased assay variability. Side-by-side comparisons of delivering compounds by pintooling versus using ARPs has produced the conclusion that ARPS provide lower potencies, reduced hit rates and higher false negative hit rates (Smith et al., 2013).
We have used pintooling from predefined library source plates for screens, and an automated 8-channel head liquid handling workstation for subsequent cherry picking compounds for re-screening or setting up new experimental layouts. Compounds are added to neurons on DIV13 at a final concentration of 12.5 μM and then incubated for 24–48 hours to measure compounds-induced changes in neuronal mitochondrial dynamics.
2.3.1. Equipment and tools
Liquid handling workstation with 100 nl pintool head (Biomek FXP or equivalent)
384-well polypropylene compound plates (Greiner Bio-One 784201 or equivalent)
384-well assay plates
Blotting paper
2.3.2. Buffers and reagents
Methanol, isopropanol, DMSO:H2O=1:1
2.3.3. Procedure
Soak pins in a bath sonicator containing DMSO:H2O=1:1 for 2 minutes.
Submerge pins into methanol then isopropanol and dry pins on blotter paper.
Submerge pins into the wells of the compound plate containing 8 μl of 10 mM compounds dissolved in DMSO, so that pins touch the bottom and are pushed up in their floating head, thereby making sure all pins come in contact with the compound irrespective of the volume in each well.
One can use additional compound plates containing control compounds occupying different wells than the experimental compounds, making sure the pins are submerged into the control liquid without the pins touching the bottom of the plate.
Wash the pins into the destination assay plates containing DIV13 cultured neurons, making sure pins do not touch the bottom of the wells. Incubate for 24–48 hours.
2.3.4. Notes
Ideally, the sonicator bath is attached to a peristaltic pump, which continuously circulates the DMSO:H2O=1:1 between the bath and a sealable reservoir bottle. By doing this, the volume of the washing mixture is increased considerably, eliminating the possibility of cross contaminations. If the equipment is not in use, store the DMSO:H2O mixture in the reservoir bottle.
DMSO:H2O can be reused multiple times. Methanol and isopropanol should be changed at least daily, depending on the pintooling cycles
Pins should be washed before and after pintooling as well.
The procedure can be performed similarly with 1536-well assay plates using a smaller volume 384-well pintool head (e.g. 10 nl) and pintooling from the four quadrants of the control plate to the assay plate.
2.4. Imaging
The resolution requirements for a HCS screen depend on the size of the objects of interest. Resolution in x and y is dependent on the emission wavelength (λEm) of the light source and the numerical aperture (NA) of the objective (Rayleigh resolutionx-y=(0.61*λEm)/NA). Resolution in the z direction if collecting stacks of z-plane images is also influenced by the refractive index (Ri) of the mounting media (Rayleigh resolutionz=(2*λEm*Ri)/NA2). The other parameter one must consider is the pixel size within the images, which is usually smaller than the resolution and depends on the pixel size of the camera and the magnification of the objective (pixel sizeimage=pixel sizecamera/magnification). When imaging small objects, like mitochondria (axonal mitochondria ~0.5–2 μm long (Kiryu-Seo, Ohno, Kidd, Komuro, & Trapp, 2010)), it is critical to ensure that each object is made up of a numerous of pixels. Otherwise identification and measurement of the object will differ significantly from its real size. Using high magnification objectives to increase resolution decreases the size of the field captured compared to lower magnification objectives (field size=(pixel sizeimage*number of pixelsimage)2). The net effect of this is that more fields within each well need to be imaged when using high magnification objectives to capture a sufficient number of objects to detect statistically significant differences between control and experimental wells.
A confocal fluorescent and automated screening microscope equipped with a 60× magnification (NA=0.95) air objective offers sufficient resolution needed to detect fluorescently labelled objects as small as mitochondria and neurites in living neurons. Environmental control that includes adjustable temperature, CO2 and humidity levels help to keep neurons alive and healthy during imaging. The highest signal-to-noise ratio possible is preferred to aid in the identification of objects of interest, and this can be elevated by increasing exposure time and laser intensity or imaging several planes in the z plane. However, these steps all contribute to phototoxicity, photobleaching and quenching of the fluorophores. Additionally, increasing exposure times, imaging several z-planes and several fields within each well increases imaging time significantly. These are all parameters that have to be optimized in pilot experiments when developing the assay for a screen. Binning pixels (e.g. 2×2) increases sensitivity four-fold, but decreases resolution two-fold. Brightness or fluorescence intensity of a given fluorophore should also be considered besides excitation and detection wavelengths when developing an assay, which is proportional to the product of its molar extinction coefficient (ε, efficiency of absorbing excitation light at a given wavelength) and quantum yield (ϕ, ratio of number of photons emitted to absorbed). Brightness of the tagGFP2 protein we used for targeting mitochondria is 33.9 ((56 500 M−1cm−1 × 0.6)/1000), which is not much brighter (5%) than EGFP, but it is much more photostable (Subach et al., 2008).
Using an air objective illuminating the samples from the bottom is preferred over oil objectives. Reliable autofocusing of the imaging system is a must in HCS, and also contributes to the speed of imaging. Autofocus performance may vary with objective and plate type and should be validated empirically before starting the screen. Finding the plane of interest automatically across a plate type prone to non-uniformity in the elevation of bottom height or bottom thickness can be a challenge for the hardware and the software of the microscope. Hardware or laser autofocus identifies the change in the refractive index of the air-to-plate bottom interface, and the bottom surface inside each well between the plate floor and medium where the cells are attached. The optimal plane of imaging can be fine-tuned by a given offset value. Software or image based autofocus on the other hand uses algorithms to find the highest contrast/strongest signal along the z-axis. This is not ideal if the objects of interest vary in brightness and reside at multiple z-elevations relative to the plate bottom, such as the dim mitochondria in flattened axons relative to bright mitochondria localized in the elevated soma. Software autofocus also adds a considerable time to image acquisition, which depends on the magnitude of the search range it uses to find the sharpest plane, and also on whether it uses the previous in-focus plane to start the search. Additionally, software autofocus repeatedly exposes cells to large doses of energy as it captures fluorescent images to determine the optimal focal plane, causing increased phototoxicity and photobleaching. In contrast, hardware autofocus uses low-energy infrared light for detecting differences in refractive index.
The time for collecting images after compound treatment, which depends on the time-course of the drug-induced phenotype, is another important parameter that needs to be optimized. If one begins a screen with an excellent positive control this can be used to establish the optimal incubation time of compounds in pilot experiments. However, after obtaining a set of positives from a screen, it is often necessary to collect images at multiple times after compound treatment in order to establish the maximum signal for each compound. For mitochondria in primary neurons, we have found that collecting images 24–48 hours after compound addition offers reliable signals (Figure 5).
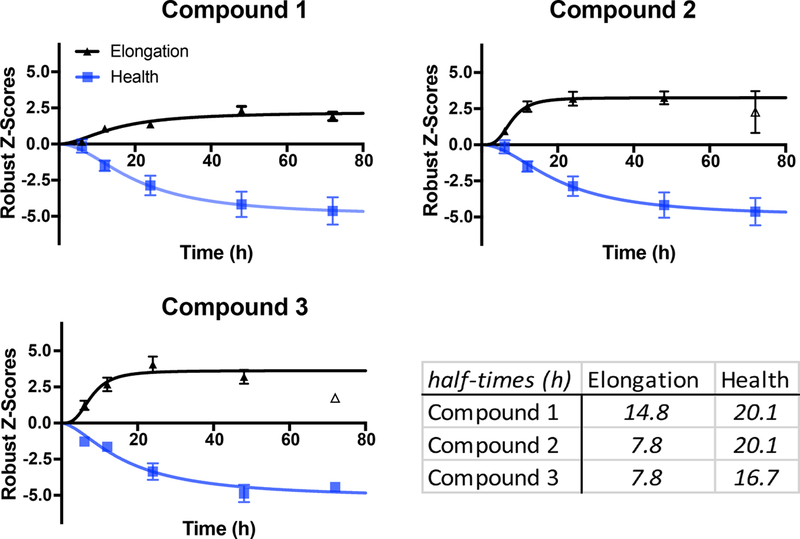
Figure 5. Time-courses of compound modulation of mitochondrial elongation and health.
Neurons were treated with 12.5 μM of the compounds and imaged 6, 12, 24, 48 and 72h later; data were normalized to DMSO treated control neurons using a robust Z-scoring method. Data are mean ± s.e.m. that were fit with a variable slope sigmoidal curve, yielding half-time values for each parameter. Open datapoints were not included in the fit. Half-time values for the two parameters were determined from the fits for each compound and for each parameter. The optimal time for image collection is a function of both the parameter of interest and the compound itself.
2.4.1. Equipment
InCell 6000 (automated confocal screening microscope) with environmental control (or equivalent)
2.4.2. Specifications and Settings
Camera: sCMOS 5.5 Mp, 6.5 μm pixel size
Objectives: For general use include 4X, 10X, 20X, 60×. For objects in the size range of axonal mitochondria: 60× air objective, 0.95 NA (Rayleigh Resolutionx-y: 0.33 μm, Rayleigh Resolutionz: 1.13 μm, pixel size: 0.11 μm, image size: 2048 × 2048 pixels, field size: 0.22 mm × 0.22 mm)
Imaging: λexc=488 nm, λem=515–535 nm, exposure time: 0.4 s, power: 50% (12.5 mW), aperture: 1.0 Au, 3D mode: 3 slices, Δz=0.7 μm, 4 fields/well, hardware autofocus, offset: 2.5
2.4.3. Notes
With these settings, a 384-well plate can be imaged in ~ 40 minutes.
The parameters of the plate used in the screen need to be precisely defined for the automated screening microscope precisely.
2.5. Automated Image Analysis
To extract biologically meaningful data from the captured images and quantify the various compound induced phenotypes, a robust and automated image analysis pipeline is essential in any HCS project. The main steps of such pipeline usually include: 1) image processing, 2) object segmentation and 3) feature extraction. In image processing, images are often corrected for noise (e.g. background noise or experimental artifacts) and uneven illumination. This is usually achieved by applying linear or nonlinear filters. Next, regions of interest (ROIs) are defined (e.g., cells or subcellular structures) in the images (fields) through the process of segmentation. This is most commonly performed by pixel binarization or using a thresholding algorithm, such as Otsu’s Method (Otsu, 1979), which calculates a threshold value separating the foreground and background pixels by minimizing the variance within the two groups. Once the objects are identified, their features (e.g. shape, intensity or texture) can be extracted by quantitative measurements.
Image analysis pipelines are constructed by employing one or several image processing algorithms. Different ways of implementing the same algorithms are readily available in image processing libraries such as the Matlab Image Processing Toolbox or the open source Python Scikit-image. The use of these packages requires programming expertise. Commercially-available HCS imaging systems are usually packaged with their own image analysis software that can also be used for constructing image analysis pipelines. In addition, there are free and open source image analysis software available. Two of the most widely used are ImageJ and CellProfiler. ImageJ can be used for HCS, but its design is more applicable for the analysis of individual images or image stacks rather than the hundreds or thousands of serial images obtained from HCS. Although ImageJ offers a programmable macro mode that supports many scripting languages, many of the macro functions activate GUI elements and cannot run in headless mode (van der Walt et al., 2014). CellProfiler is a free, open-source software for quantitative analysis of biological images designed for biologists without programming expertise in constructing automated image analysis pipelines (Carpenter et al., 2006). It is a collection of advanced image analysis algorithms in forms of modules, which modules can be placed in a sequential order to create an image analysis pipeline. For automated analysis of mitochondria in primary neurons, we constructed an image analysis pipeline using CellProfiler.
Neuronal mitochondria can be divided into 3 classes that include somatic, axonal and dendritic mitochondria. The morphology of these classes is different: somatic mitochondria form a highly interconnected, irregularly shaped network in the soma while axonal and dendritic mitochondria are individual tubular structures located in neurites. Axonal mitochondria are short (usually<1.4 μm) and dendritic mitochondria are larger (usually >2.4 μm) (Kiryu-Seo et al., 2010; Lewis et al., 2018). We separated the mitochondria labeled with the same fluorophore into those 3 classes based on morphology, and focused on the mitochondria found in neurites.
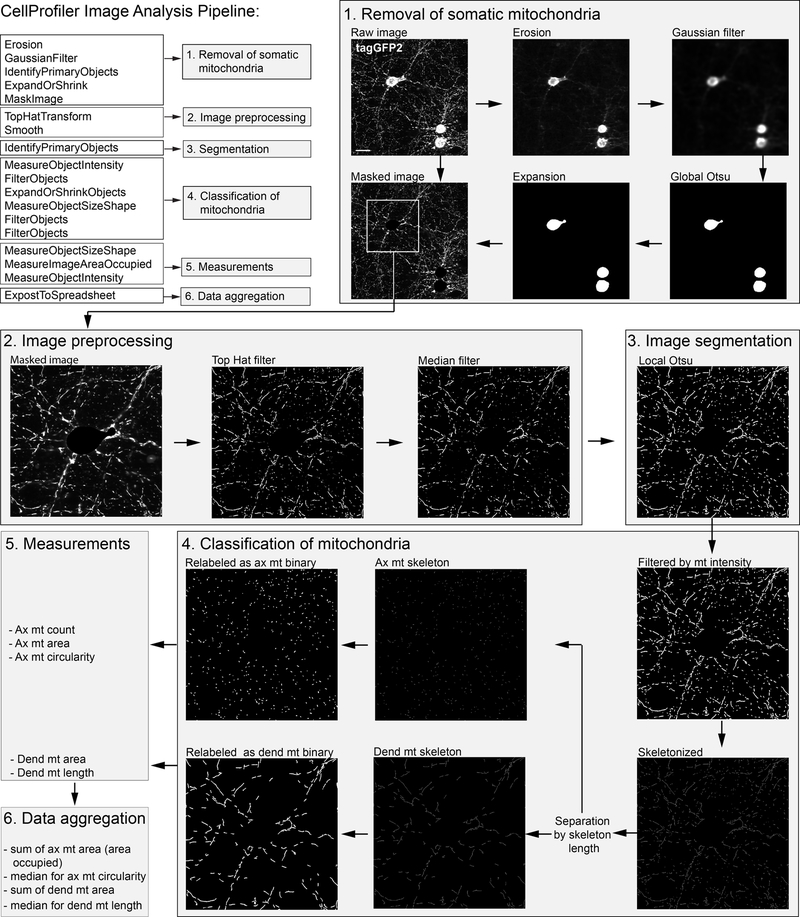
In the first part of our image analysis pipeline, densely packed somatic mitochondria are identified and removed from the original image (Figure 6). This is achieved by 5 sequential steps: 1) erosion, 2) Gaussian filtering to erode tubular structures (axonal and dendritic mitochondria), 3) global Otsu thresholding for the segmentation of somatic mitochondria, 4) expansion of the somatic ROI and 5) removal of the segmented somatic mitochondria from the original image. Then, top-hat filtering is applied for background subtraction and the resulting images are median filtered to reduce noise. Segmentation of GFP labeled objects is achieved by adaptive Otsu thresholding, while the resulting objects are intensity filtered to remove faint objects. Axonal and dendritic mitochondria are then separated based on their length. Length measurement is achieved by first skeletonizing the objects and then measuring their area. The width of a skeleton is 1 pixel, so the area in this case is equal to the length. Then, the segmented GFP-labeled objects are relabeled using the separated axonal and dendritic skeletons, resulting in the axonal and dendritic mitochondrial binaries. Besides the already performed length measurements of the skeletons, additional measurements like the area occupied by the axonal and the dendritic binaries, which is the sum of the individual mitochondrial areas, as well as the circularity of the axonal binaries are also measured. Then, the individual length and circularity values are aggregated using their median, and the data is exported..
Figure 6. Image analysis pipeline.
Raw images of mitochondrial tagGFP2 expressed in primary neurons were processed by a CellProfiler image analysis pipeline. ax=axonal; dend=dendritic; mt=mitochondria. Scalebar is 25 μm.
2.5.1. Step-by-step procedure for CellProfiler
The names of input and output images are arbitrary. Algorithm and parameter names are consistent with the naming convention used in CellProfiler.
Erosion: structuring element=disk, size=11 (input=raw, output=soma_erosion)
GaussianFilter: sigma=20 (input=soma_erosion, output=soma_gaussian)
IdentifyPrimaryObjects: threshold strategy=global, thresholding method=Otsu, three classes, assign pixels in the middle intensity class to the background, threshold smoothing scale=1.3488, threshold correction factor=1, lower and upper bounds on threshold= 0.0001 and 1, method to distinguished clumped objects=None, maximum number of objects=20 (input=soma_gaussian, output=soma_binary)
ExpandOrShrinkObjects: expand objects by a specific number of pixels, number of pixels by which to expand=5 (input=soma_binary, output=soma_binary_expanded)
MaskImage: select the input image=raw, name the output image=raw_masked, use objects or an image as objects, select object for mask=soma_binary_expanded, invert the mask=yes
TopHatTransform: white top-hat transform, structuring element=disk, size=3 (input=raw_masked, output=tophat)
Smooth: median filter, typical artifact diameter=3 (input=tophat, output=median)
IdentifyPrimaryObjects: threshold strategy=adaptive, thresholding method=Otsu, three classes, assign pixels in the middle intensity class to the background, threshold smoothing scale=0, threshold correction factor=0.6, lower and upper bounds on threshold=0.0003 and 1, size of adaptive window=25, method to distinguished clumped objects=None (input=median, output=mito_binary)
MeasureObjectIntensity: select an image=raw, select objects to measure=mito_binary
FilterObjects: objects to filter= mito_binary, output objects=mito_binary_skeleton, filtering method=limits, category=intensity, measurement=MeanIntensity, minimum value=0.004 (input=mito_binary, output=mito_binary_int_thresh)
ExpandOrShrinkObjects: skeletonize each object (input=mito_binary_int_thresh, output=mito_binary_skeleton)
MeasureObjectSizeShape (input=mito_binary_skeleton)
FilterObjects: objects to filter=mito_binary_skeleton, output objects=axonal_mito_skeleton, filtering mode=measurements, filtering method=limits, category=AreaShape, measurement=area, minimum value=4, maximum value=13, select additional objects to relabel=mito_binary_int_thresh, name the relabeled objects=axonal_mito_binary
FilterObjects: objects to filter=mito_binary_skeleton, output objects=dend_mito_skeleton, filtering mode=measurements, filtering method=limits, category=AreaShape, measurement=area, minimum value=22, select additional objects to relabel=mito_binary_int_thresh, name the relabeled objects=dend_mito_binary
MeasureObjectSizeShape: select objects=mito_binary_skeleton, dend_mito_binary, dend_mito_skeleton)
MeasureImageAreaOccupied: measure the area occupied in objects, objects to measure=mito_binary_int_thresh, axonal_mito_binary, dend_mito_binary
2.5.2. Notes:
Parameters of the different modules should be optimized for each assay.
If the intensity values of the images vary from field to filed and/or well to well, intensity normalization can be applied.
2.6. Analyzing the Data
After obtaining raw data generated from the image analysis pipeline, it is processed further in multiple steps. HCS data is somewhat different from HTS data, in that it also provides cellular level data (individual measurements of the segmented objects) besides well-level values (aggregated individual values). So, data analysis is a two-part process, first cellular measurements are converted to well-level aggregates, then well-level data is assessed further similar to other screening technologies (normalization, assay, statistics…).
2.6.1. Individual object measurements of cell-level data
Cell-level analysis, examining the individual measurements of each object as a population, involves examining hundreds or thousands of data points for each image. Thus, the number of objects can be extremely high even when comparing 2 single images, making statistical comparisons strong and valid. In addition, even if the mean values of a population fail to change with compound treatment, there may be changes in the distribution pattern of the population. So, the first task with the large data of individual object measurements is to aggregate it to a few concise, informative summary statistics without losing the effect a compound might have on the population. If the data generated is influenced by independent random factors with additive effects on variability, a symmetric, bell shaped normal distribution of population will emerge, where the mean, median and mode values are the same. However, biological processes often deviate from normal distributions and produce skewed population distributions.
The most popular summary statistics are the mean and standard deviation (SD) of the population and the coefficient of variation (CV). Since CV is normalized to the sample mean, it is unitless and allows comparisons of the dispersion of the data. The mean, SD and CV are all sensitive to outliers and are appropriate aggregate values for describing normal distributions. In contrast, the median, median absolute deviation (MAD) and interquartile range (IQR) are derived from rank ordering the elements of the population. These measures are less influenced by outliers and thus appropriate as aggregated values for skewed distributions. The shape of the distribution of the population can be described by skewness and kurtosis. Skewness measures the degree of asymmetry in a distribution, positive values refer to right-skewed distributions while negative values indicate left-skewed data. A symmetrical distribution has a skewness of 0. Kurtosis measures how heavy the tails are in a distribution. A normal distribution has a kurtosis of 3; values greater than this indicate heavy tails and a narrow peak, while lower values indicate a wide peak and thinner tails.
Transformation of non-normally distributed data provides an option for generating normal distribution (log: for positively skewed data, square root: for Poisson distribution, power: negatively skewed data, reciprocal: extremely right skewed data) if the transformation makes biological sense. It would enable the use of parametric tests which are more sensitive, more powerful and easier to interpret than non-parametric statistics. On the other hand, the distribution of the population may change due to experimental artefacts (autofocus or image analysis errors, edge effects) that increase the number of outliers or due to compound treatment. Therefore, careful consideration needs to be taken of whether data transformation can be applied across the entire dataset. Changes in population distribution will also change the variance of the samples, prohibiting the use of many statistical tests for comparisons that assume a constant variance across samples.
Our individual measures of elongation (dendritic mitochondrial length) and health (axonal mitochondrial circularity) generate non-normally distributed data, whose transformation would not make biological sense. Thus, median values are used to generate summary statistics for well-level data. The sum of individual axonal and dendritic mitochondrial area (area occupied by mitochondria) is used as well-level data to assess mitochondrial content in axons and dendrites (Figure 7 and Figure 8).
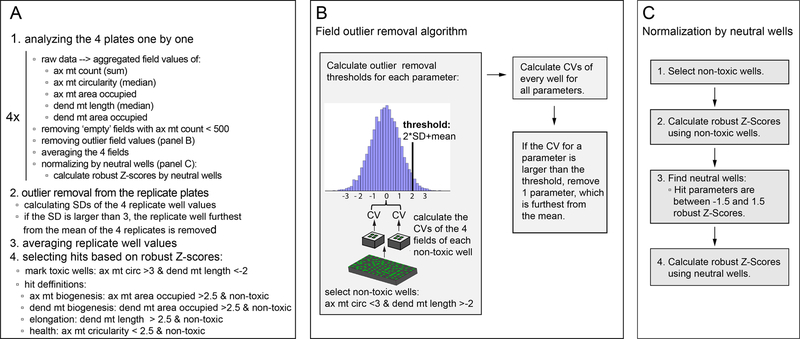
Figure 7. Data analysis pipeline.
Summary of the data analysis (A), and detailed description and illustration of the field outlier removal (B) and normalization (C) performed within step 1 of the data analysis pipeline. ax=axonal; dend=dendritic; mt=mitochondria; circ=circularity
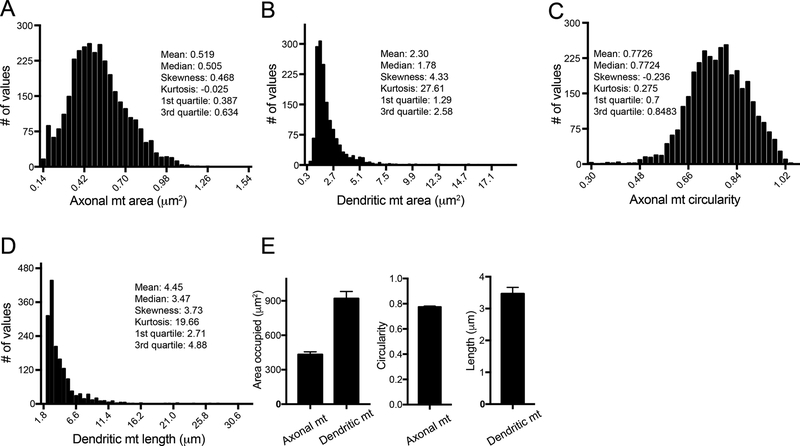
Figure 8. Cell-level, well-level and normalized data of mitochondrial features in primary neurons.
A-D) Cell-level measurements of individual mitochondria of vehicle-treated (DMSO) DIV15 neurons from 4 imaged fields show that distributions of axonal mitochondrial area and circularity are the closest to a Gaussian distribution (R2Gaussian, ax mt area=0.97, R2Gaussian, ax mt circ=0.98), while the two dendritic features are highly right-skewed. E) Well-level data was generated from the cell-level data as the average of the sum of the areas of individual mitochondria across each of the four imaged fields and the average of the field medians of axonal mitochondrial circularity and dendritic mitochondrial length. Well-level data are presented as the mean ± SD of the 4 aggregated field values.
2.6.2. Well-level data
Once the aggregated values from individual cell-level measurements are generated, they can be used to calculate summary statistics. Trends and artefacts can be seen when plotting data for each well (numbers of outliers, positional effects across the plate, etc.). Individual object measures are not always provided automatically by HCS software, only summary statistics. Examining how far the means and medians are from each other can be an implication of distribution and skewness of the cell-level data. By examining the 1st and 3rd quartiles in the summary statistics one can assess whether the majority of the signal comes from a minority of the population. In case it comes from the minority, images and the image analysis process should be re-checked for possible artifacts and cell-level data should be examined.
Normalization of the raw data is necessary to compare screening data across plates and across multiple experiments. Normalization can be performed to within-plate controls, to all wells on a plate (provided the majority of compounds are inactive) or to a subset of within-plate compounds (“neutral” compounds). The use of neutral compounds in a plate offers a convenient approach, since they are scattered across the plate which will attenuate edge or column/row effects. Often, the layout of the libraries for a primary screen do not allow the experimenter to distribute controls scattered across the plate, relegating them primarily to positions along the outer columns. Even alternating positive, vehicle-treated and negative controls along these columns can produce serious issues when used to normalize experimental wells (Bray & Carpenter, 2004; Malo, Hanley, Cerquozzi, Pelletier, & Nadon, 2006). When normalizing values to within plate compounds, the use of robust descriptive statistics is preferred because of its relative insensitivity to outliers.
Several methods exist for normalizing screening data (Goktug, Chai, & Chen, 2013), among the most used are percent of control, Z-score, robust Z-score and B score (Table 1). Percent of control is simple, but it does not take into consideration the variability of controls. Z-score (used for normally distributed data) accounts for the variability of controls, as it tells how many standard deviations a compound is away from the mean of the controls. In robust Z-score calculations (for non-normally distributed data), mean and SD is replaced by median and MAD, respectively; it is more resistant to outliers yielding more reliable results. All of these methods correct for plate-to-plate variability but assume a random distribution of error within each plate. Besides normalization, the B score method uses a two-way median polish algorithm to minimize within-plate column and row effects (Brideau, Gunter, Pikounis, & Liaw, 2003). Other noise reduction methods based on Tukey median polish algorithm or using polynomial least squares fit have also been developed to correct for systematic within plate effects (Makarenkov et al., 2007; Mpindi et al., 2015). The Loess local polynomial fit is based on the least squares polynomial approximation, and provides more reliable data and less false positives on plates with higher hit rates than 20% than the B score method, which could be attributed to B score’s dependency on the median polish algorithm (Mpindi et al., 2015). Combining a polynomial least squares fit method with normalization of data to scattered controls may provide the best option for screens with high hit rates and also for generating accurate dose-response curves.
Table 1. Methods for normalizing screening data.
Si=activity of the ith sample; C=control; Ci=ith control; SD=standard deviation; MAD=median absolute deviation; rijp= residual in ith row, jth column and pth plate; Sijp= sample value at ith row, jth column and pth plate; μp= plate average; rowi= median of ith row; colj= median of jth column
| Percent of control | (Si/Cmean)×100 |
| Z-score | (Si-Cmean)/CSD |
| Robust Z-score | (Si-Cmedian)/CMAD ; CMAD= 1.4826 × median(|Ci- Cmedian|) |
| B score | rijp/MADp; rijp=Sijp-μp-rowi-colj (median polish); MADp=1.4826 × median(|(rijp)all-median((rijp)all)|) |
We have normalized our screening data by using robust Z-scores and within plate neutral compounds scattered randomly across each plate (~100 compounds/ 384-well plate).
2.6.3. Assay performance and hit selection
The robustness of a screen highly depends on the nature of selected positive and negative controls. Positive controls with huge effect sizes but without biological relevance may provide false expectations and few or no hits at all. Therefore, strong positive controls with more modest effect sizes are preferred in a HCS (Davies & Shamu, 2014).
Assay performance may be quantified based on signal to background (S/B) or signal to noise ratio (S/N), where S/B is the ratio of the mean of the effect of the positive controls to the negative controls and S/N is the difference between the mean of the positive and negative controls divided by the variance of the negative controls. The most widely used HTS assay performance measure is the Z-factor (J. H. Zhang, Chung, & Oldenburg, 1999). It involves the means (μ) and standard deviations (SD) of both positive (p) and negative (n) controls (1-(3 (SDp-SDn)/ |μp-μn|)). Generally, assays with Z-factor values above 0.5 are considered reliable and robust, most screening facilities use this value as an acceptance criterion to run an assay as a HTS. This parameter plays a less important role for HCS data, since assays performed on living cells are usually noisier than data generated from biochemical or other assays employed in the more classical HTS.
Several hit selection methods have been described in the literature (Bhinder & Djaballah, 2012; Birmingham et al., 2009; Malo et al., 2006; X. D. Zhang, 2011), many having been developed originally for RNAi screens. The simplest method to select hits is to identify the upper 1% of the normalized and rank ordered data for further re-screening. Other methods use a pre-defined threshold to select hits, based on Z-score, robust Z-score or quartiles (X. D. Zhang et al., 2006). Z-Score or robust Z-score methods define the threshold for hits usually to be at least 3 SD or 3 MAD away from the population mean or median, respectively, although one can be more or less stringent depending on the assay. The quartile-based method usually used in RNAi screens seems to perform better with extremely skewed data and selects more true hits than Z-score based methods. Calculating strictly standardized mean difference (SSMD) values for each well, also developed originally for RNAi screens, takes into account the variability of both a hit compound and the control given that both groups have several replicates. This can lead to fewer false negatives or false positives (X. D. Zhang et al., 2007).
Hit detection also depends on compound concentration, which influences its potency and variability. Variables that include solvent evaporation, compound solubility and transfer efficiency will all affect the measured potency of a compound. Replicate plate screening provides a way of normalizing for this variability. For our screens for mitochondrial dynamics, we utilized 4 replicate plates with each compound plate represented in 1 well per plate. Once compounds are confirmed in a re-screen, dose-response curves are generated to yield EC50 values (half maximal effect concentration). A dose-response relationship assumes that there exists a low dose where the compound has no effect, as well as concentration producing a maximum response beyond which increasing compound concentration is not accompanied by further increase of the effect.
Here we present our data analysis pipeline with outlier removal optimized for 4 replicate fields/well and 4 replicate plates with 1 well/plate (Figure 7).
2.6.4. Step-by-step data analysis procedure using 4 fields/well and 4 replicate plates
Remove fields which do not contain a sufficient number of objects (e.g., >500 axonal mitochondrial/field for the mitochondrial dynamics assay).
- Remove outlier field values (Figure 7B):
- Select non-toxic wells:
- In our example, a well is considered toxic if there exists dendritic mitochondrial fragmentation and increased axonal mitochondrial circularity. To identify such wells, field values are averaged (without outlier removal), and robust Z-scores are calculated using the whole plate population. A well is classified as non-toxic if axonal mitochondrial circularity <3 and dendritic mitochondrial length >−2 robust Z-scores.
- Calculate outlier field removal thresholds for each feature:
- CVs of the field values are calculated for the non-toxic wells.
- Mean and standard deviations of the CVs are calculated.
- Outlier removal thresholds are calculated by taking the mean+2SD of the CVs for the given parameter.
- Remove outliers:
- Recalculate the CVs for every well.
- If the CV for a given feature is larger for a well than the outlier removal threshold for that feature, 1 field value is removed from the four, which is furthest from the mean.
Average field values into well values.
- Normalize data by neutral wells (Figure 7C):
- Select non-toxic wells as described above.
- Select neutral wells from non-toxic wells:
- A well is considered to be neutral if its robust Z-scores are between −1.5 and 1.5 for the hit parameters.
- Calculate robust Z-scores using neutral wells.
Repeat the same data processing for all the 4 plates.
- Remove outlier well values for each feature across the replicate plates:
- Calculate Z-scores for the replicate well values using the mean and standard deviation of the 4 replicate wells.
- Identify those replicates, whose Z-score>3 for a given feature.
- Remove one value from the four, which is furthest from the mean.
Average the replicate wells for each feature.
- Select the hits:
- Health: axonal mitochondrial circularity <−2.5 robust Z-score
- Elongation: dendritic mitochondrial length >2.5 robust Z-score
- Axonal mitochondrial content: axonal mitochondrial area occupied >2.5 robust Z-score
- Dendritic mitochondrial content: dendritic mitochondrial area occupied > robust Z-score
2.6.5. Notes:
The Pandas library of Python is a great tool to implement data analysis pipelines.
3.0. Summary and Conclusions
High content screens offer a powerful platform for probing biological processes at a “cell systems” level of analysis. This can be accomplished using many types of cells including primary neurons. The approach does require a high level of sophisticated image analysis and data processing knowledge, along with hardware for liquid handling robotics and a confocal screening microscope for the collection of images from microtiter plates. However, the approach offers a reward in a wealth of cellular data useful as basic science knowledge on the effects of small molecules, libraries of RNAi vectors, etc., on processes of interest to the investigator.
References
- Aschauer DF, Kreuz S, & Rumpel S (2013). Analysis of transduction efficiency, tropism and axonal transport of AAV serotypes 1, 2, 5, 6, 8 and 9 in the mouse brain. PLoS One, 8(9), e76310. doi: 10.1371/journal.pone.0076310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bereiter-Hahn J (1976). Dimethylaminostyrylmethylpyridiniumiodine (daspmi) as a fluorescent probe for mitochondria in situ. Biochim Biophys Acta, 423(1), 1–14. [DOI] [PubMed] [Google Scholar]
- Bhinder B, & Djaballah H (2012). A simple method for analyzing actives in random RNAi screens: introducing the “H Score” for hit nomination & gene prioritization. Comb Chem High Throughput Screen, 15(9), 686–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birmingham A, Selfors LM, Forster T, Wrobel D, Kennedy CJ, Shanks E, … Kelleher D (2009). Statistical methods for analysis of high-throughput RNA interference screens. Nature methods, 6(8), 569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray MA, & Carpenter A (2004). Advanced Assay Development Guidelines for Image-Based High Content Screening and Analysis In Sittampalam GS, Coussens NP, Brimacombe K, Grossman A, Arkin M, Auld D, Austin C, Baell J, Bejcek B, Chung TDY, Dahlin JL, Devanaryan V, Foley TL, Glicksman M, Hall MD, Hass JV, Inglese J, Iversen PW, Kahl SD, Kales SC, Lal-Nag M, Li Z, McGee J, McManus O, Riss T, Trask OJ Jr., Weidner JR, Xia M, & Xu X (Eds.), Assay Guidance Manual. Bethesda (MD). [PubMed] [Google Scholar]
- Brideau C, Gunter B, Pikounis B, & Liaw A (2003). Improved statistical methods for hit selection in high-throughput screening. J Biomol Screen, 8(6), 634–647. doi: 10.1177/1087057103258285 [DOI] [PubMed] [Google Scholar]
- Cagalinec M, Safiulina D, Liiv M, Liiv J, Choubey V, Wareski P, … Kaasik A (2013). Principles of the mitochondrial fusion and fission cycle in neurons. J Cell Sci, 126(Pt 10), 2187–2197. doi: 10.1242/jcs.118844 [DOI] [PubMed] [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, … Sabatini DM (2006). CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol, 7(10), R100. doi: 10.1186/gb-2006-7-10-r100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chazotte B (2011). Labeling mitochondria with MitoTracker dyes. Cold Spring Harb Protoc, 2011(8), 990–992. doi: 10.1101/pdb.prot5648 [DOI] [PubMed] [Google Scholar]
- Chen H, & Chan DC (2009). Mitochondrial dynamics--fusion, fission, movement, and mitophagy--in neurodegenerative diseases. Hum Mol Genet, 18(R2), R169–176. doi: 10.1093/hmg/ddp326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland PH, & Koutz PJ (2005). Nanoliter dispensing for uHTS using pin tools. Assay Drug Dev Technol, 3(2), 213–225. doi: 10.1089/adt.2005.3.213 [DOI] [PubMed] [Google Scholar]
- Cummings J (2018). Lessons Learned from Alzheimer Disease: Clinical Trials with Negative Outcomes. Clin Transl Sci, 11(2), 147–152. doi: 10.1111/cts.12491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daub A, Sharma P, & Finkbeiner S (2009). High-content screening of primary neurons: ready for prime time. Curr Opin Neurobiol, 19(5), 537–543. doi: 10.1016/j.conb.2009.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies A, & Shamu C (2014). An introduction to high content screening: imaging technology, assay development, and data analysis in biology and drug discovery: John Wiley & Sons. [Google Scholar]
- Dunn DA, & Feygin I (2000). Challenges and solutions to ultra-high-throughput screening assay miniaturization: submicroliter fluid handling. Drug Discov Today, 5(12 Suppl 1), 84–91. [DOI] [PubMed] [Google Scholar]
- Ebrahimi-Fakhari D, Saffari A, Wahlster L, Di Nardo A, Turner D, Lewis TL Jr., … Sahin M (2016). Impaired Mitochondrial Dynamics and Mitophagy in Neuronal Models of Tuberous Sclerosis Complex. Cell Rep, 17(4), 1053–1070. doi: 10.1016/j.celrep.2016.09.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenberg B, Montana V, Wei MD, Wuskell JP, & Loew LM (1988). Membrane potential can be determined in individual cells from the nernstian distribution of cationic dyes. Biophys J, 53(5), 785–794. doi: 10.1016/S0006-3495(88)83158-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emaus RK, Grunwald R, & Lemasters JJ (1986). Rhodamine 123 as a probe of transmembrane potential in isolated rat-liver mitochondria: spectral and metabolic properties. Biochim Biophys Acta, 850(3), 436–448. [DOI] [PubMed] [Google Scholar]
- Goktug AN, Chai SC, & Chen T (2013). Data analysis approaches in high throughput screening Drug Discovery: Intech.
- Hammond SL, Leek AN, Richman EH, & Tjalkens RB (2017). Cellular selectivity of AAV serotypes for gene delivery in neurons and astrocytes by neonatal intracerebroventricular injection. PLoS One, 12(12), e0188830. doi: 10.1371/journal.pone.0188830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkes N (2018). Pfizer abandons research into Alzheimer’s and Parkinson’s diseases. BMJ, 360, k122. doi: 10.1136/bmj.k122 [DOI] [PubMed] [Google Scholar]
- Johri A, & Beal MF (2012). Mitochondrial dysfunction in neurodegenerative diseases. J Pharmacol Exp Ther, 342(3), 619–630. doi: 10.1124/jpet.112.192138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiryu-Seo S, Ohno N, Kidd GJ, Komuro H, & Trapp BD (2010). Demyelination increases axonal stationary mitochondrial size and the speed of axonal mitochondrial transport. J Neurosci, 30(19), 6658–6666. doi: 10.1523/JNEUROSCI.5265-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korchak HM, Rich AM, Wilkenfeld C, Rutherford LE, & Weissmann G (1982). A carbocyanine dye, DiOC6(3), acts as a mitochondrial probe in human neutrophils. Biochem Biophys Res Commun, 108(4), 1495–1501. [DOI] [PubMed] [Google Scholar]
- Lesuisse C, & Martin LJ (2002). Long-term culture of mouse cortical neurons as a model for neuronal development, aging, and death. J Neurobiol, 51(1), 9–23. [DOI] [PubMed] [Google Scholar]
- Lewis TL, Kwon S-K, Lee A, Shaw R, & Polleux F (2018). MFF-dependent mitochondrial fission regulates presynaptic release and axon branching by limiting axonal mitochondria size. bioRxiv, 276691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Yang Y, & Wang S (2005). Neuronal gene transfer by baculovirus-derived vectors accommodating a neurone-specific promoter. Exp Physiol, 90(1), 39–44. doi: 10.1113/expphysiol.2004.028217 [DOI] [PubMed] [Google Scholar]
- Makarenkov V, Zentilli P, Kevorkov D, Gagarin A, Malo N, & Nadon R (2007). An efficient method for the detection and elimination of systematic error in high-throughput screening. Bioinformatics, 23(13), 1648–1657. doi: 10.1093/bioinformatics/btm145 [DOI] [PubMed] [Google Scholar]
- Malo N, Hanley JA, Cerquozzi S, Pelletier J, & Nadon R (2006). Statistical practice in high-throughput screening data analysis. Nat Biotechnol, 24(2), 167–175. doi: 10.1038/nbt1186 [DOI] [PubMed] [Google Scholar]
- Mansouri M, Bellon-Echeverria I, Rizk A, Ehsaei Z, Cianciolo Cosentino C, Silva CS, … Berger P (2016). Highly efficient baculovirus-mediated multigene delivery in primary cells. Nat Commun, 7, 11529. doi: 10.1038/ncomms11529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelfelder S, & Trepel M (2009). Adeno-associated viral vectors and their redirection to cell-type specific receptors. Adv Genet, 67, 29–60. doi: 10.1016/S0065-2660(09)67002-4 [DOI] [PubMed] [Google Scholar]
- Misgeld T, & Schwarz TL (2017). Mitostasis in Neurons: Maintaining Mitochondria in an Extended Cellular Architecture. Neuron, 96(3), 651–666. doi: 10.1016/j.neuron.2017.09.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mpindi JP, Swapnil P, Dmitrii B, Jani S, Saeed K, Wennerberg K, … Kallioniemi O (2015). Impact of normalization methods on high-throughput screening data with high hit rates and drug testing with dose-response data. Bioinformatics, 31(23), 3815–3821. doi: 10.1093/bioinformatics/btv455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murlidharan G, Samulski RJ, & Asokan A (2014). Biology of adeno-associated viral vectors in the central nervous system. Front Mol Neurosci, 7, 76. doi: 10.3389/fnmol.2014.00076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niles WD, & Coassin PJ (2008). Cyclic olefin polymers: innovative materials for high-density multiwell plates. Assay Drug Dev Technol, 6(4), 577–590. doi: 10.1089/adt.2008.134 [DOI] [PubMed] [Google Scholar]
- Nunnari J, & Suomalainen A (2012). Mitochondria: in sickness and in health. Cell, 148(6), 1145–1159. doi: 10.1016/j.cell.2012.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsu N (1979). A threshold selection method from gray-level histograms. IEEE transactions on systems, man, and cybernetics, 9(1), 62–66. [Google Scholar]
- Rafael J (1980). 2-(Dimethylaminostyryl)-1-ethylpyridinium iodide: a fluorescent probe of energetic conditions in brown-adipose-tissue mitochondria. Hoppe Seylers Z Physiol Chem, 361(3), 437–444. [DOI] [PubMed] [Google Scholar]
- Rizzuto R, Brini M, Pizzo P, Murgia M, & Pozzan T (1995). Chimeric green fluorescent protein as a tool for visualizing subcellular organelles in living cells. Curr Biol, 5(6), 635–642. [DOI] [PubMed] [Google Scholar]
- Sheng B, Wang X, Su B, Lee HG, Casadesus G, Perry G, & Zhu X (2012). Impaired mitochondrial biogenesis contributes to mitochondrial dysfunction in Alzheimer’s disease. J Neurochem, 120(3), 419–429. doi: 10.1111/j.1471-4159.2011.07581.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Carpenter AE, & Genovesio A (2014). Increasing the Content of High-Content Screening: An Overview. J Biomol Screen, 19(5), 640–650. doi: 10.1177/1087057114528537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smiley ST, Reers M, Mottola-Hartshorn C, Lin M, Chen A, Smith TW, … Chen LB (1991). Intracellular heterogeneity in mitochondrial membrane potentials revealed by a J-aggregate-forming lipophilic cation JC-1. Proc Natl Acad Sci U S A, 88(9), 3671–3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T, Ho PI, Yue K, Itkin Z, MacDougall D, Paolucci M, … Auld DS (2013). Comparison of compound administration methods in biochemical assays: effects on apparent compound potency using either assay-ready compound plates or pin tool-delivered compounds. J Biomol Screen, 18(1), 14–25. doi: 10.1177/1087057112455434 [DOI] [PubMed] [Google Scholar]
- Subach OM, Gundorov IS, Yoshimura M, Subach FV, Zhang J, Gruenwald D, … Verkhusha VV (2008). Conversion of red fluorescent protein into a bright blue probe. Chem Biol, 15(10), 1116–1124. doi: 10.1016/j.chembiol.2008.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinney DC, & Anthony J (2011). How were new medicines discovered? Nat Rev Drug Discov, 10(7), 507–519. doi: 10.1038/nrd3480 [DOI] [PubMed] [Google Scholar]
- Taylor H, Haskins J, & Giuliano K (2007). High content screening: A powerful approach to systems cell biology and drug discovery totowa. New Jersery: Humana Press Inc. [Google Scholar]
- van der Walt S, Schonberger JL, Nunez-Iglesias J, Boulogne F, Warner JD, Yager N, … scikit-image, c. (2014). scikit-image: image processing in Python. PeerJ, 2, e453. doi: 10.7717/peerj.453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Wang J, Bonamy GM, Meeusen S, Brusch RG, Turk C, … Schultz PG (2012). A small molecule promotes mitochondrial fusion in mammalian cells. Angew Chem Int Ed Engl, 51(37), 9302–9305. doi: 10.1002/anie.201204589 [DOI] [PubMed] [Google Scholar]
- Westermann B (2012). Bioenergetic role of mitochondrial fusion and fission. Biochim Biophys Acta, 1817(10), 1833–1838. doi: 10.1016/j.bbabio.2012.02.033 [DOI] [PubMed] [Google Scholar]
- Wu Z, Asokan A, & Samulski RJ (2006). Adeno-associated virus serotypes: vector toolkit for human gene therapy. Mol Ther, 14(3), 316–327. doi: 10.1016/j.ymthe.2006.05.009 [DOI] [PubMed] [Google Scholar]
- Zanella F, Lorens JB, & Link W (2010). High content screening: seeing is believing. Trends Biotechnol, 28(5), 237–245. doi: 10.1016/j.tibtech.2010.02.005 [DOI] [PubMed] [Google Scholar]
- Zhang C, Deng Y, Dai H, Zhou W, Tian J, Bing G, & Zhao L (2017). Effects of dimethyl sulfoxide on the morphology and viability of primary cultured neurons and astrocytes. Brain Res Bull, 128, 34–39. doi: 10.1016/j.brainresbull.2016.11.004 [DOI] [PubMed] [Google Scholar]
- Zhang JH, Chung TD, & Oldenburg KR (1999). A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen, 4(2), 67–73. doi: 10.1177/108705719900400206 [DOI] [PubMed] [Google Scholar]
- Zhang XD (2011). Illustration of SSMD, z score, SSMD*, z* score, and t statistic for hit selection in RNAi high-throughput screens. J Biomol Screen, 16(7), 775–785. doi: 10.1177/1087057111405851 [DOI] [PubMed] [Google Scholar]
- Zhang XD, Ferrer M, Espeseth AS, Marine SD, Stec EM, Crackower MA, … Strulovici B (2007). The use of strictly standardized mean difference for hit selection in primary RNA interference high-throughput screening experiments. J Biomol Screen, 12(4), 497–509. doi: 10.1177/1087057107300646 [DOI] [PubMed] [Google Scholar]
- Zhang XD, Yang XC, Chung N, Gates A, Stec E, Kunapuli P, … Espeseth AS (2006). Robust statistical methods for hit selection in RNA interference high-throughput screening experiments. Pharmacogenomics, 7(3), 299–309. doi: 10.2217/14622416.7.3.299 [DOI] [PubMed] [Google Scholar]