Abstract
The liver contains a mixture of hepatocytes with diploid or polyploid (tetraploid, octaploid, etc.) nuclear content. Polyploid hepatocytes are commonly found in adult mammals, representing ~90% of the entire hepatic pool in rodents. The cellular and molecular mechanisms that regulate polyploidization have been well-characterized; however, it is unclear if diploid and polyploid hepatocytes function similarly in multiple contexts. Answering this question has been challenging because proliferating hepatocytes can increase or decrease ploidy, and animal models with healthy diploid-only livers have not been available. Mice lacking E2f7 and E2f8 in the liver (LKO) were recently reported to have a polyploidization defect but were otherwise healthy. Herein, livers from LKO mice were rigorously characterized, demonstrating a 20-fold increase in diploid hepatocytes and maintenance of the diploid state even after extensive proliferation. Livers from LKO mice maintained normal function but became highly tumorigenic when challenged with tumor-promoting stimuli, suggesting that tumors in LKO mice were driven, at least in part, by diploid hepatocytes capable of rapid proliferation. Indeed, hepatocytes from LKO mice proliferate faster and out-compete control hepatocytes, especially in competitive repopulation studies. In addition, diploid or polyploid hepatocytes from wild-type mice were examined to eliminate potentially confounding effects associated with E2f7/E2f8 deficiency. The wild-type diploid cells also showed a proliferative advantage, entering and progressing through the cell cycle faster than polyploid cells, both in vitro and during liver regeneration. Diploid and polyploid hepatocytes responded similarly to hepatic mitogens, indicating that proliferation kinetics are unrelated to differential response to growth stimuli.
Conclusion:
Diploid hepatocytes proliferate faster than polyploids, suggesting that the polyploid state functions as a growth suppressor to restrict proliferation by the majority of hepatocytes.
Keywords: Ploidy, Diploid, Liver cancer, Cell cycle, E2f7/E2f8
Introduction
Most mammalian somatic cells are diploid and contain pairs of each chromosome, but there are also polyploid cells such as skeletal muscle, megakaryocytes, trophoblast giant cells, cardiac myocytes and hepatocytes that contain additional sets of chromosomes (1–3). Polyploid hepatocytes are among the best described, and in adult humans and mice, polyploids comprise more than 50% and 90%, respectively, of the hepatocyte population (4, 5). Polyploidization begins at day 14 in mice when proliferating hepatocytes fail to complete cytokinesis and produce binucleate tetraploid daughter cells containing two diploid nuclei (6–8). In turn, these binucleate tetraploid hepatocytes begin cycling, and, upon successful cytokinesis, generate a pair of mononucleate tetraploid daughters. This process continues, generating octaploid and even higher ploidy hepatocytes. The polyploid state is also reversible. Proliferating polyploid hepatocytes can undergo multipolar cell division to generate diploid or near-diploid daughters, a process termed the ploidy conveyor (5). Thus, the adult liver contains a heterogeneous mixture of diploid and polyploid hepatocytes, but it is unknown whether these different hepatic subsets perform specialized functions in normal homeostasis or liver regeneration.
The E2F family of transcription factors regulates progression through the cell cycle (10). In the liver, E2F1–6 are expressed at low levels throughout life, whereas E2F7 and E2F8 are elevated between weeks 1 and 7 of postnatal development, a period correlating with hepatocyte polyploidization (11). The role of E2F7 and E2F8 was studied using liver-specific E2f7/E2f8 double knockout mice (11, 12). Livers in these mice were reported to function normally through several months of age, but they were significantly depleted of polyploid hepatocytes. Similar, albeit less pronounced, phenotypes were seen in E2f7 and E2f8 single knockout mice. E2F7 and E2F8 regulate hepatic ploidy in two primary ways (11). First, E2F7 and E2F8 antagonize E2F1 activity, and, consistent with this relationship, transgenic mice overexpressing E2F1 were enriched for diploid hepatocytes, which is similar to the phenotype of E2f7/E2f8 double knockout mice (13). Secondly, E2F7 and E2F8 negatively regulate genes involved in cytokinesis. Since cytokinesis failure is the primary method hepatocytes become polyploid (7), loss of E2F7 and E2F8 promotes successful cytokinesis of proliferating mononucleate diploid cells.
Here, we utilized liver-specific E2f7/E2f8 double knockout (LKO) mice to investigate functional differences between diploid and polyploid hepatocytes. Similar to published reports, LKOs contained ~20 times more diploids and 3 times fewer polyploids. The reduced-polyploidy phenotype was stable at 2.5 and 5 months of age and following extensive hepatocyte proliferation. When adults were treated with a tumor induction/promotion model, LKO mice developed abundant liver tumors whereas control mice were resistant to tumorigenesis. We speculated that tumors in LKO mice were driven, at least in part, by diploid hepatocytes capable of rapid proliferation. Using in vivo and in vitro approaches, we showed that hepatocytes from LKO livers proliferated faster and could out-compete hepatocytes from control livers, especially in competitive repopulation. We also compared diploid and polyploid hepatocytes from WT mice. Cell culture and liver regeneration assays showed that diploids entered and completed the cell cycle earlier than polyploids. Diploid and polyploid hepatocytes responded to hepatic mitogens similarly, indicating that proliferation kinetics were unrelated to differential response to growth stimuli. Together, the data show for the first time that diploid hepatocytes proliferate faster than polyploids, suggesting that the polyploid state functions as a growth suppressor to restrict proliferation by the majority of hepatocytes.
Experimental Procedures
Mouse strains.
The Institutional Care and Use Committee of the University of Pittsburgh approved all mouse experiments. Mice were maintained in a standard 12-hour light/dark system. They were housed in Optimice cages (AnimalCare Systems, Centennial, CO) with Sani-Chip Coarse bedding (P. J. Murphy, Montville, NJ) and provided ad libitum access to water and standard mouse chow (LabDiet, St. Louis, MO, Purina ISO Pro Rodent 3000). Mice were provided huts and running wheels for enrichment. All animals were sacrificed between 9 am and noon daily. E2f7/E2f8 liver-specific knockout mice (LKO) were a kind gift from Alain deBruin and Gustavo Leone (11, 12). LKO mice, which were a mixed background but predominantly FVB, contained floxed E2f7 and E2f8 alleles, a Rosa26-lacZ reporter (R26R-lacZ) and Cre recombinase driven by the albumin promoter (Alb-Cre): E2f7loxP/loxP E2f8 loxP/loxP R26RlacZ/lacZ Alb-CreTg/0. Control mice were negative for Alb-Cre: E2f7loxP/loxP E2f8 loxP/loxP R26RlacZ/lacZ. FRGN mice (Fah−/− Rag2−/− Interleukin 2 common Gamma chain−/− Nod background) were used for liver repopulation studies (Yecuris, Inc., Tualatin, OR) (14, 15). FRGN mice were maintained on 8 mg/L 2-(2-nitro-4-trifluoro- methylbenzoyl)-1,3-cyclo-hexanedione (NTBC) in their drinking water (Ark Pharm, Libertyville, IL). Wild-type (WT) C57BL/6 mice were from Jackson Labs (Bar Harbor, ME). Unless noted, male mice were used for experiments.
Hepatocyte isolation.
Primary hepatocytes were isolated using a 2-step collagenase perfusion (16). Briefly, following general anesthesia induction, a catheter was inserted into the portal vein or inferior vena cava and 0.3 mg/ml collagenase II (Worthington, Lakewood, NJ) was circulated through the liver. Digested livers were placed in DMEM-F12 with 15 mM HEPES (Corning, Corning, NY) + 5% FBS (Sigma-Aldrich, St. Louis, MO), passed through a 70 μm cell strainer and washed twice using low-speed centrifugation (55g for 2 min) to remove non-parenchymal cells. Hepatocyte viability, determined by trypan blue staining, was typically >80%. Hepatocytes from FRGN transplantation recipients were isolated as described above, except the collagenase II concentration was increased to 1 mg/mL.
For additional methods, please refer to the Supporting Information.
Results
Liver-specific deletion of E2f7 and E2f8 blocks hepatic polyploidy.
To validate the effects of E2f7 and E2f8 deficiency in the liver, we obtained mice with floxed E2f7 and E2f8 alleles and a Rosa26-lacZ reporter (R26R-lacZ) that were crossed with Cre recombinase driven by the Albumin promoter (Alb-Cre) to specifically delete these genes in the liver (11, 12). E2f7/E2f8 liver-specific knockout (LKO) and control mice were compared. Taking advantage of R26R-lacZ, control hepatocytes lacking Alb-Cre did not express β-galactosidase (β-gal), as expected, whereas hepatocytes from LKO mice were β-gal+ (Fig. 1A). Consistent with the literature, liver weight to body weight (LW/BW) ratio (Fig. 1B), circulating liver enzymes (Supporting Fig. S1A) and hepatic zonation (Supporting Fig. S1B) were comparable (11, 12). Additionally, gene signatures were equivalent between LKO and control mice at 3 and 16 weeks of age (e.g., gene ontology categories, cell cycle, metabolism, proliferation and differentiation) and in livers depleted of E2f8 alone (e.g., differentiation genes such as Albumin, Foxa3 and Hnf4a; Cytochrome P450 family members; and cell cycle genes) (11, 17). To assess liver function following chronic injury, we maintained mice on high fat diet (HFD) for 8 weeks. LKO and control mice had equivalent ALT levels, proportional increases in body weight and LW/BW (Supporting Fig. S2A–C). Gross liver morphology and accumulation of neutral lipids was the same (Supporting Fig. S2D,E). Thus, together with the observation by Pandit et al. that LKO and control mice respond equivalently to DDC diet (11), E2f7/E2f8 deficient mice appeared to be functionally normal.
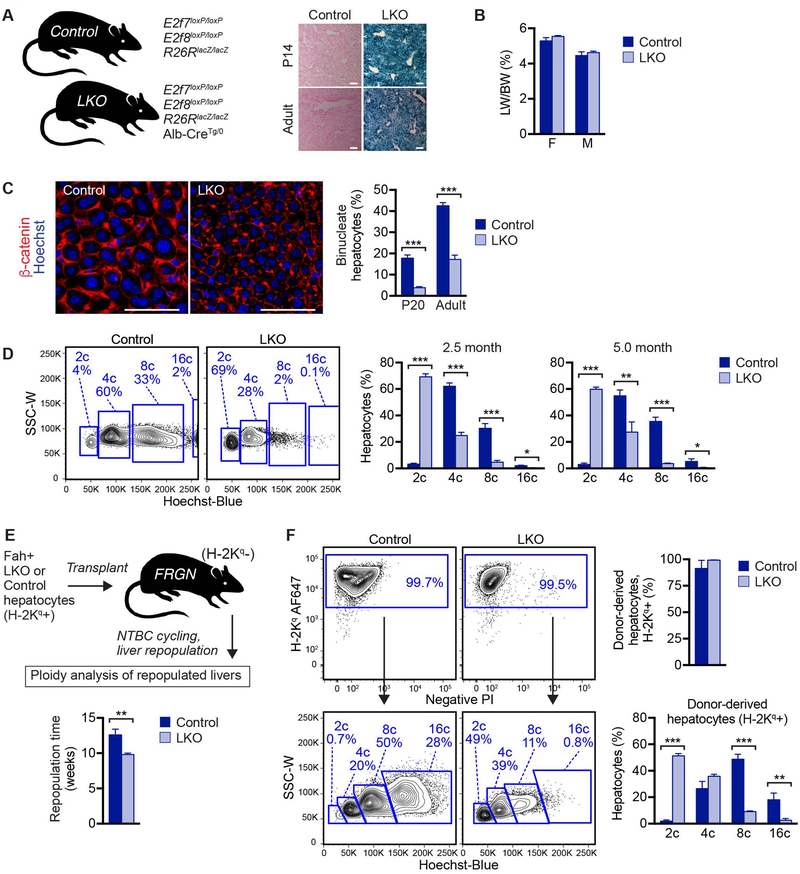
Fig. 1.
Liver-specific deletion of E2f7/E2f8 as a hepatic polyploidy knockout model. (A) Experimental mice contain floxed E2f7 and E2f8 alleles as well as R26R-lacZ. LKO mice are hemizygous for Alb-Cre and controls are negative. Livers isolated from 14-day-old (P14) and 2.5-month-old (adult) LKO and control mice were stained with X-gal to visualize β-gal (blue) and nuclear fast red to visualize nuclei (pink) (n = 3–4/genotype/age). β-gal activity confirms Cre activity in LKO livers. (B) LKO and control mice at 2.5 months have similar LW/BW ratios. (C) Livers from 20-day-old (P20) and 2.5-month-old (adult) LKO and control mice were stained for β-catenin to mark cell membranes (red) and Hoechst dye (blue) to mark nuclei. The percentage of binucleate hepatocytes was quantified (n = 3–5 mice/genotype; mixed gender), and representative images are shown at 2.5 months. (D) Freshly isolated hepatocytes from 2.5- or 5.0-month-old LKO and control mice were loaded with Hoechst to determine hepatocyte ploidy by FACS analysis. Representative FACS plots are shown (at 2.5 months) and the percentage of diploid, tetraploid, octaploid and hexadecaploid hepatocytes (2c, 4c, 8c, 16c, respectively) are shown for each age (n = 3–6/genotype/age; males). Here, ploidy populations are marked with chromatid number “c” since they contain a mixture of cycling and quiescent cells (although >99% are quiescent). The full gating strategy is shown in Supporting Fig. S3A. (E,F) LKO or control hepatocytes from 2-month-old male mice were transplanted into female FRGN mice that were subjected to NTBC cycling and harvested upon complete repopulation (E). The time to repopulation, as defined by maintenance of 100% body weight in the absence of NTBC, is indicated (n = 6–7/genotype). Representative FACS plots and compiled data are shown (n = 4–5/genotype) (F). Hepatocytes from repopulated recipients were ≥99.5% positive for the donor marker H-2Kq, indicating that LKO and control hepatocytes repopulated equivalently (F, top). Ploidy analysis revealed that LKO-repopulated livers were highly enriched for diploid hepatocytes, whereas livers repopulated with control hepatocytes were predominantly polyploid (F, bottom). See Supporting Fig. S3B for the full gating strategy. * P < 0.05, ** P < 0.01, *** P < 0.0004. Graphs show mean ± sem. Scale bars are 100 μm in (A) and 50 μm in (B).
The most striking effect associated with loss of E2f7/E2f8 is altered hepatic ploidy. Binucleate hepatocytes were reduced 3–4-fold in LKO livers as early as day 20 and into adulthood (Fig. 1C), and they were distributed evenly between all zones in the liver, similar to controls (Supporting Fig. S1B–D). We determined cellular ploidy by quantifying nuclear content using flow cytometry. In control adult livers, diploid cells comprised only 3–4% of hepatocytes and polyploids comprised the rest (Fig. 1D and Supporting Fig. S3A). The ploidy spectrum was dramatically altered in LKO livers, with diploids representing 60–70% of hepatocytes. This observed 20-fold enrichment of diploid hepatocytes in LKO livers was seen in 2.5- and 5-month-old livers.
We previously showed that diploid hepatocytes can polyploidize during therapeutic liver repopulation (5). We therefore wondered whether the LKO ploidy phenotype was stable during extensive proliferation. To test this, we utilized the fumarylacetoacetate hydrolase (Fah) liver repopulation model (Fig. 1E) (16). Mice lacking Fah develop liver failure but are kept alive and healthy by supplementing 2-(2-nitro-4-trifluoro- methylbenzoyl)-1,3-cyclo-hexanedione (NTBC) in the drinking water. Fah+ donor hepatocytes are transplanted into Fah−/− mice and NTBC is removed. In this environment, donor hepatocytes have a proliferative advantage, and cells proliferate 500–1000-fold to replace host hepatocytes. To prevent immune rejection of transplanted hepatocytes, which were a mixed background, we used immune deficient Fah−/− mice (termed “FRGN” for Fah−/− Rag2−/− Interleukin 2 common Gamma chain−/− Nod background) (14, 15). FRGN mice transplanted with adult LKO or control hepatocytes were completely repopulated (>99%), as seen by expression of the donor marker H-2Kq (Fig. 1F and Supporting Fig. S3B). Moreover, we noticed that LKO-transplanted recipients repopulated 25% faster than control-transplanted mice (Fig. 1E). After complete liver repopulation, control hepatocytes were >99% polyploid, and LKO hepatocytes remained predominantly diploid, indicating that the LKO ploidy phenotype is stable even after extensive in vivo proliferation (Fig. 1F). Together, the data show that LKO mice serve as a stable “hepatic polyploidy knockout” model.
DEN-induced liver tumors expand rapidly in LKO mice.
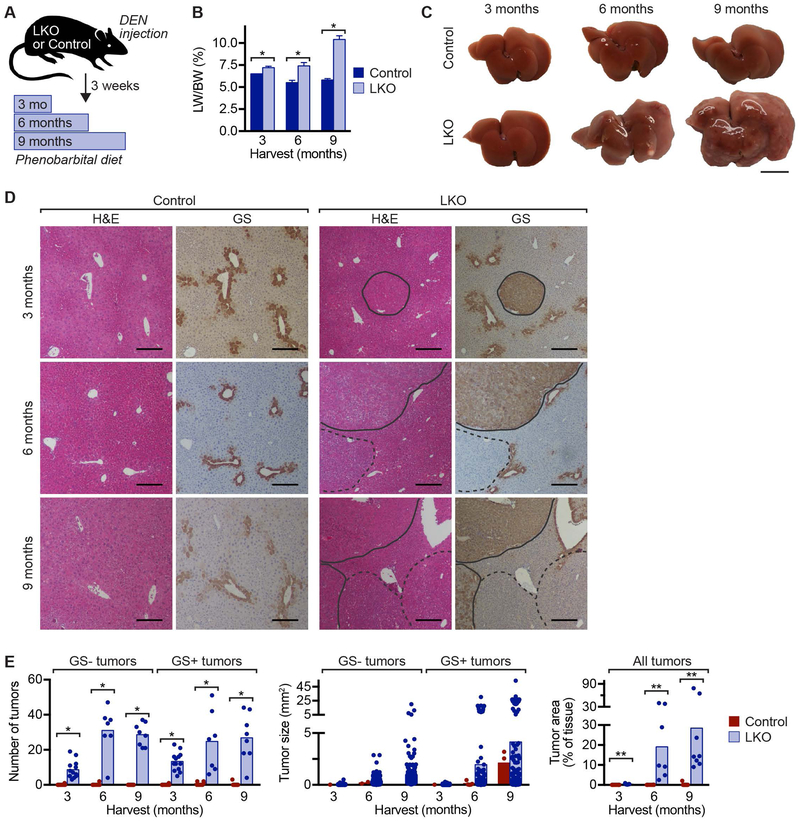
Polyploidy is a hallmark of cancer that has been shown to promote genomic instability and dysregulated gene expression, frequently leading to cellular transformation and tumorigenesis (18, 19). Intriguingly, in the liver, polyploidy has been suggested to protect against tumorigenesis (17, 20). Since LKO mice function normally and are predominantly diploid, these mice provided an opportunity to assess the role of polyploidy in hepatocellular carcinogenesis. Six-week-old LKO or control mice were injected with a single dose of diethylnitrosamine (DEN) to initiate tumor formation and 3 weeks later mice were placed on a phenobarbital (PB) diet to promote tumor expansion (Fig. 2A) (21–24). Genes involved in DEN metabolism (Cyp2b10) and PB-induced proliferation (Car and Cyp2e1) were expressed at similar levels, indicating that response to DEN/PB is not attributed to E2f7/E2f8 deficiency (Supporting Fig. S4A,B) (25–27). ALT, AST and bilirubin levels were similar in both groups, although ALT was increased 2–3-fold in LKO mice after 9 months (Supporting Fig. S4C). LW/BW was consistently higher in LKO mice (Fig. 2B). Macroscopically, tumors were not seen in control livers after 3, 6 or 9 months (Fig. 2C). In LKO livers, tumors were undetected after 3 months, discrete tumors were visible after 6 months, and by 9 months, livers were dramatically enlarged and filled with tumors. Liver tumors in the DEN/PB model typically contain activating β-catenin mutations, so we stained liver sections with H&E for general histology and glutamine synthetase (GS), a β-catenin target that serves as a surrogate marker for activated β-catenin (Fig. 2D) (22, 28, 29). LKO mice were highly enriched with GS- and GS+ liver tumors. Tumor number, size and area increased with time in the LKO mice (Fig. 2E). Together, these data are consistent with other reports (17, 30) and show that LKO mice are highly susceptible to DEN/PB-mediated tumorigenesis, suggesting that the hepatic polyploid state protects the liver from tumor formation.
Fig 2.
Liver tumors induced by the DEN model expand rapidly in LKO mice. (A) Experimental design is shown. Six-week-old LKO and control mice were injected with DEN (90 mg/kg) to induce liver tumors, and after 3 weeks, they were maintained on chow supplemented with 0.05% phenobarbital for 3, 6 or 9 months to promote tumor expansion (n = 7–9 mice/genotype/time point; males). (B) LW/BW was elevated in LKO mice at all time points. Graph shows mean ± sem. (C) Control livers contained no macroscopic tumors after 3–9 months. In LKO livers, there were few macroscopic tumors at 3 months, and they increased in size and number over time. Gross morphology is shown for representative livers. (D) Serial liver sections were stained with H&E and GS (brown) to identify liver tumors. GS- tumors (dashed line) and GS+ tumors (solid line) are indicated. (E) Compared to controls, tumors in LKO mice were more numerous (left), larger in size (middle) and occupied more liver tissue (left). Graphs show mean and dots represent individual mice (for tumor number and tumor area) or individual tumors (for tumor size). * P < 0.002, ** P < 0.0008. Scale bars are 1 cm (in C) or 200 μm (in E).
Adult LKO hepatocytes transit through the cell cycle rapidly in vitro.
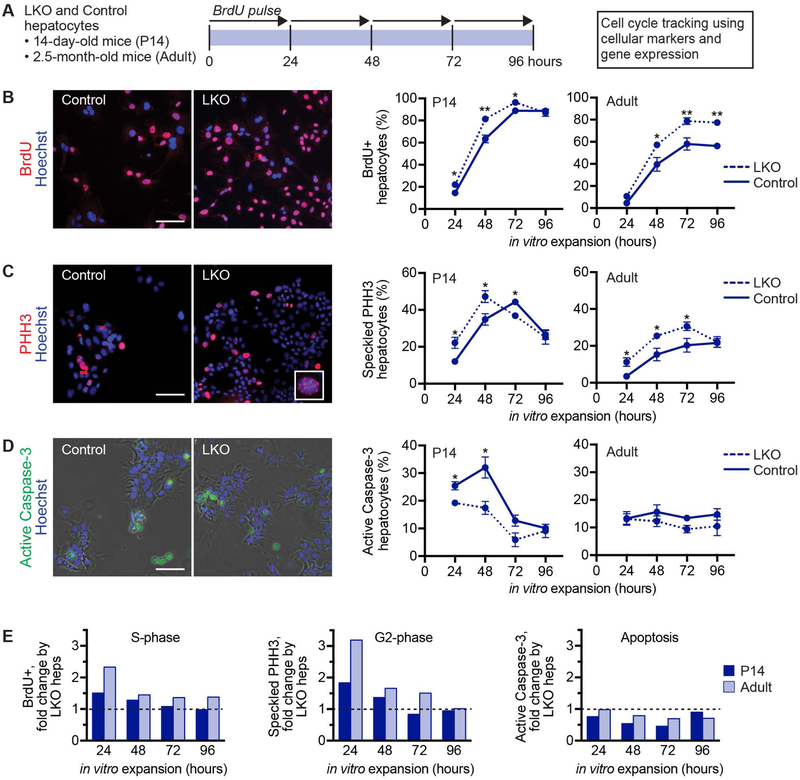
We observed that transplanted LKO hepatocytes repopulated FRGN livers 25% faster than control hepatocytes (Fig. 1E). Therefore, since LKO hepatocytes are predominantly diploid, we hypothesized that diploid hepatocytes are more proliferative than polyploids, which drives tumor formation in DEN/PB-treated LKO mice. To test this idea, we compared proliferation of LKO and control hepatocytes in vitro. Freshly isolated hepatocytes were seeded in cell culture dishes, and cells and nuclei in S-phase (BrdU incorporation), G2-phase (speckled phospho-histone H3 staining, PHH3) and undergoing apoptosis were tracked (Fig. 3A). First, we measured cell cycle progression using hepatocytes from 14-day-old mice where LKO and controls are equivalent in terms of ploidy (96% diploid) and cell cycle status (83% quiescent) (Supporting Fig. S5). Therefore, any variations in proliferation in vitro are unrelated to ploidy states or basal proliferation rates. Compared to controls, the percentage of LKO hepatocytes in S-phase was increased at all time points (Fig. 3B and Supporting Fig. S6A). LKO hepatocytes in G2-phase were modestly elevated at 24–48 hours, reduced at 72 hours and equivalent at 96 hours (Fig. 3C and Supporting Fig. S6B). There were slightly fewer LKO apoptotic hepatocytes throughout the culture period (Fig. 3D). Next, we tracked proliferation in 2.5-month-old adult hepatocytes. The percentage of LKO hepatocytes in S-phase (48–96 hours) and G2-phase (24–72 hours) was elevated throughout the culture period, while cell death was nearly equivalent (Fig. 3BD and Supporting Fig. S6A,B). Consistent with the cellular markers of proliferation, cyclin genes including Ccnd1, Ccne1, Ccna2 and Ccnb1, which are enriched at defined cell cycle stages, were also upregulated by proliferating LKO hepatocytes (Supporting Fig. S6C). To compare LKO proliferation kinetics between 14-day-old and adult hepatocytes, we normalized LKO values in each phase of the cell cycle to control values (Fig. 3E). The data indicate that cell cycling is modestly increased in 14-day-old LKO hepatocytes, whereas hepatocytes in S- and G2-phases in LKO adults are elevated consistently and to a greater extent. Since 14-day-old hepatocytes are inherently more proliferative than adult hepatocytes we cannot exclude the possibility that genotype-specific differences are masked at young ages. Nonetheless, the data suggest that non-ploidy differences contribute minimally to altered cell cycle kinetics by LKO hepatocytes and hepatic ploidy differences play a dominant role.
Fig. 3.
LKO hepatocytes that are predominantly diploid cycle faster than control hepatocytes. (A) Experimental design is shown. LKO and control hepatocytes isolated from 14-day-old (n = 3–5/genotype/time point) or 2.5-month-old mice (n = 4–6/genotype/time point; female) were seeded in cell culture dishes and harvested after 24, 48, 72 or 96 hours of proliferation to determine S-phase via BrdU incorporation, G2-phase via speckled PHH3 staining and apoptosis via active caspase-3 staining. For BrdU incorporation, cells were pulsed with BrdU for 24 hours prior to harvest. (B-D) Representative images are shown for cells after 72 hours in culture. Cells were stained for BrdU (red) (B), PHH3 (red, inset shows speckled staining pattern) (C) and active caspase-3 (green, overlaid with phase contrast) (D). Nuclei were stained with Hoechst dye (blue). Graphs show the percentage of positive cells at each time point; see Supporting Fig. S6 for the percentage of positive nuclei. (E) LKO proliferation kinetics by hepatocytes isolated from 14-day-old mice were compared to 2.5-month-old mice by calculating the fold-change of LKO hepatocytes in each phase of the cell cycle versus control values. The dashed lines indicate a normalized value of 1. * P < 0.04, ** P < 0.008. Graphs show mean ± sem (B-D) or mean only (E). Scale bars are 100 μm.
LKO hepatocytes proliferate faster than controls in vivo.
Since LKO hepatocytes progressed through the cell cycle faster than controls in vitro, we next characterized hepatocyte proliferation in vivo. First, we compared proliferation during postnatal liver development. LKO and control livers were harvested at 15–75 days of age and stained for Ki-67, a marker of G1/S/G2/M phases of the cell cycle. The percentage of Ki-67+ hepatocytes was equivalent at all ages (Fig. 4A). Next, we compared proliferation during liver regeneration in response to 2/3 partial hepatectomy (PH). In this classic procedure, 2/3 of the liver mass is surgically removed and the remaining liver tissue undergoes a wave of proliferation to restore the liver mass (31–33). LW/BW recovery and the percentage of Ki-67+ (G1/S/G2/M), PCNA+ (S-phase) and bright PHH3+ (M-phase) hepatocytes was nearly equivalent at all time points (Fig. 4B,C). While we cannot exclude the possibility that LKO hepatocytes entered the cell cycle earlier than controls between 0 and 48 hours, there was a trend toward increased Ki-67+ and PCNA+ hepatocytes in LKO livers at later time points, particularly at 72 hours (Fig. 4C).
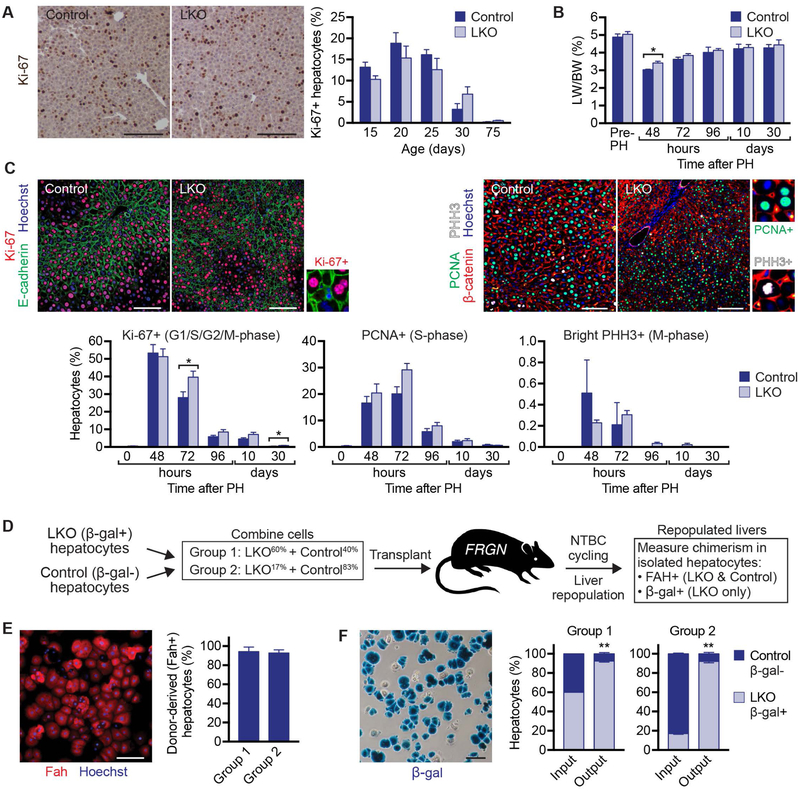
Fig. 4.
LKO hepatocytes proliferate faster than controls during liver regeneration and repopulation. (A) LKO and control livers harvested during postnatal development (15, 20, 25 and 30 days) and in adults (75 days), were stained for Ki-67 (brown) as a marker for proliferation (n = 4–5 mice/genotype/age; mixed gender). Representative micrographs are shown for 20-day-old mice (left), and percentage of Ki-67+ hepatocytes is shown for all ages (right). (B-C) Two-thirds PH was performed on 2.5-month-old LKO and control mice to induce liver regeneration. Livers were harvested before PH and at the indicated time points after PH (n=3–8 mice/genotype/age; mixed gender). LW/BW is shown (B). On the left, liver sections were stained with Ki-67 (red), E-cadherin (green, to mark cell membranes) and Hoechst (blue, to mark nuclei). On the right, liver sections were stained with β-catenin (red, to mark cell membranes), PCNA (green), PHH3 (white) and Hoechst (blue, to mark nuclei). Representative images are shown 48 hours after PH, and magnified insets depict Ki-67+ (G1/S/G2/M-phase), PCNA+ (S-phase) and bright PHH3+ (M-phase) nuclei. The percentage of hepatocytes in each phase of the cell cycle for all time points is indicated (bottom) (C). (D-F) Liver repopulation was compared using competitive transplantation. Hepatocytes from 2.5-month-old male LKO and control mice were mixed in defined ratios and transplanted into FRGN recipients. Group 1 contained 60% LKO and 40% control (n = 5 recipients); Group 2 contained 17% LKO and 83% control (n = 7 recipients). Mice were cycled off/on NTBC for ~3 months to promote expansion of donor cells. Following completed liver repopulation, hepatocytes were isolated and chimerism was determined. Cartoon shows the experimental design (D). Isolated hepatocytes were heterogeneous, containing Fah+ donor cells (LKO and control) and Fah- recipient cells and the degree of repopulation was determined by Fah staining. Representative staining is shown for a repopulated recipient from Group 1 (Fah, red and Hoechst, blue), and the graph shows the percentage of Fah+ cells in each group (E). Hepatocytes were stained with X-gal to visualize β-gal (blue). Representative image is shown for a repopulated recipient from Group 1. The graphs summarize the percentages of β-gal- and β-gal+ hepatocytes at the time of transplant (marked as “input”) and after liver repopulation (marked as “output”) (F). See Supporting Fig. S7 for data from individual recipients. * P < 0.03, ** P < 0.001. Graphs show mean ± sem. Scale bars are 100 μm.
In contrast to relatively mild proliferative programs during postnatal liver development and PH (where each hepatocyte divides 1–2 times to restore the liver mass), we compared hepatocyte proliferation using the Fah liver repopulation model where transplanted donor hepatocytes undergo 9–10 population doublings. We mixed LKO and control hepatocytes in ~equal ratios (Group 1: LKO60% and control40%) and transplanted mixtures of donor cells into FRGN recipients. Upon completed repopulation, we isolated hepatocytes and determined the degree of chimerism (Fig. 4D). More than 90% of hepatocytes were Fah+, indicating high-level repopulation (Fig. 4E). Since LKO hepatocytes express the β-gal reporter, we then quantified β-gal+ (LKO) and β-gal- (control) cells. The majority of cells were β-gal+ (93%), representing 1.5-fold expansion by LKO hepatocytes during liver repopulation (Fig. 4F and Supporting Fig. S7A). We also performed competitive transplants with the donor mixture skewed toward control hepatocytes (Group 2: LKO17% and control83%) (Fig. 4D). After repopulation, Fah+ donor hepatocytes comprised >90% of the livers (Fig. 4E) and 92% of hepatocytes were β-gal+, representing 5.4-fold expansion by LKO hepatocytes during liver repopulation (Fig. 4F and Supporting Fig. S7B).
To reconcile the observed cell proliferation differences between LKO and control hepatocytes during liver regeneration and competitive repopulation assays, we performed a modeling analysis. We compared the impact of cell size and proliferation rate on liver volume production, considering rapid proliferation by relatively small LKO hepatocytes and slower proliferation by relatively large control hepatocytes. As described in Supporting Fig. S8, slower-cycling, large control hepatocytes contributed greater cell volume after 1 or 2 cell divisions; but after 3 cell divisions, faster-cycling, small LKO cells generated increased cumulative volume with each successive division. Taken together, the data indicate that LKO hepatocytes are more proliferative than control hepatocytes, especially when induced to divide extensively.
WT diploid hepatocytes enter the cell cycle earlier than polyploids during liver regeneration.
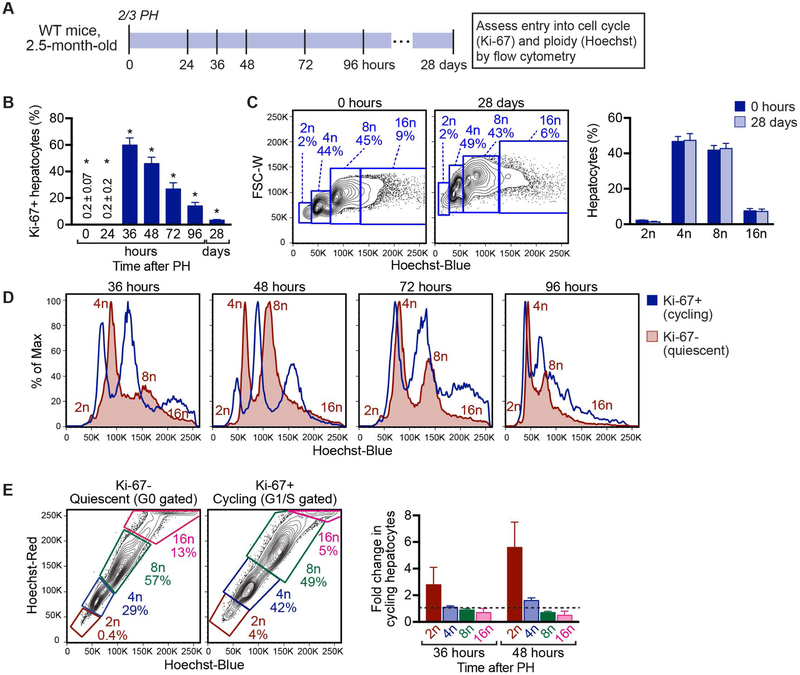
We next hypothesized that the polyploid state reduces proliferative potential, which enables diploid hepatocytes to proliferate quicker than polyploids. To test this idea, we compared proliferation by WT diploid and polyploid hepatocytes. Specifically, we performed 2/3 PH on 2.5-month-old C57BL/6J WT mice, harvested hepatocytes during liver regeneration and assessed proliferation and ploidy by flow cytometry (Fig. 5A). As expected, hepatocytes were nearly completely quiescent 0–24 hours post PH, but by 36 hours ~60% of hepatocytes were Ki-67+, indicating cell cycle entry (Fig. 5B and Supporting Fig. S9A). The number of cycling hepatocytes remained high 48 hours after PH (~50%) and decreased by 72–96 hours. Twenty-eight days after PH most hepatocytes returned to quiescence. The overall distribution of hepatic ploidy subsets was equivalent before (0 hours) and after (28 days) liver regeneration (Fig. 5C).
Fig. 5.
Diploid hepatocytes enter the cell cycle faster than polyploids during liver regeneration. (A) Experimental design for comparing regeneration by diploid and polyploid hepatocytes. Livers from 2.5-month-old male WT C57BL/6 mice were induced to regenerate by 2/3 PH and hepatocytes isolated during liver regeneration (24–96 hours) and after complete regeneration (28 days). Live hepatocytes were stained for Ki-67 and ploidy. (B) The percentage of Ki-67+ cells is indicated (n = 3–6/time point). Primary FACS plots are shown in Supporting Fig. S9A. (C) The ploidy spectrum is equivalent before (0 hours) and after (28 day) liver regeneration. Representative FACS plots are shown for Ki-67− hepatocytes (left) and the percentage of each ploidy population quantified (right, n = 4–5). (D) Representative histograms show ploidy overlays of Ki-67+ cycling and Ki-67− quiescent hepatocytes 36–96 hours after PH (n = 3–5). Individual ploidy populations are indicated for quiescent hepatocytes. Additional details are in Supporting Fig. S9B. (E) The percentage of each ploidy population was determined for quiescent and cycling hepatocytes during early-stage regeneration at 36 and 48 hours. Gates in the Ki-67− quiescent fraction represent cells in G0; gates in the Ki-67+ cycling fraction represent cells in G1/S. Representative FACS plots are shown at 48 hours (right) and the fold-change in cycling hepatocytes calculated (fold-change = Ki-67+/Ki-67−) for each ploidy population (left, n = 3–4). Dashed line represents fold-change of 1. * P < 0.05 in Fig. 5B for comparisons of Ki-67+ hepatocytes between each time point except 36 versus 48 hours and 36 versus 72 hours, which are not significant. Graphs show mean ± sem.
To determine the contribution of each ploidy subset during regeneration, we compared ploidy in quiescent and cycling cells. During early-stage regeneration (36–48 hours), ploidy subsets of cycling hepatocytes (Ki-67+) were right-shifted relative to quiescent cells (Ki-67−) in terms of Hoechst intensity (Fig. 5D and Supporting Fig. S9B). This increase in nuclear content by each cycling ploidy population indicates transition to S-phase and active DNA replication. In contrast, by late-stage regeneration (72–96 hours), ploidy profiles of cycling hepatocytes were more diffuse, consistent with cells in G1/S/G2/M phases, making it challenging to identify cycling populations with the same DNA content (e.g., cells with 4c DNA content could be diploids in G2 or tetraploids in G0/G1). We focused on early-stage regeneration where we could accurately identify all the ploidy populations. Qualitatively, at 36–48 hours the quiescent subpopulation contained few diploids and was enriched with tetraploid and octaploid hepatocytes. Ploidy distribution shifted markedly in the cycling subpopulation where there were more cycling diploids than quiescent diploids and fewer cycling octaploids than quiescent octaploids (Fig. 5D). We also quantified the percentage of hepatocytes within the quiescent and cycling subpopulations (Fig. 5E). At 36 hours, cycling diploid hepatocytes were increased 3-fold whereas cycling tetraploids, octaploids and hexadecaploids were unchanged or reduced. The trend was even more pronounced at 48 hours, with a 6-fold increase in cycling diploids and ploidy-dependent reduction in cycling cells. Although all hepatocytes proliferate in response to 2/3 PH (34), the data show that entry into the cell cycle is inversely proportional to hepatocyte ploidy. Thus, in response to regenerative stimuli, diploid hepatocytes enter the cell cycle quicker than polyploid hepatocytes.
WT diploid hepatocytes proliferate faster than polyploids in vitro, and ploidy subsets respond to mitogenic signals equivalently.
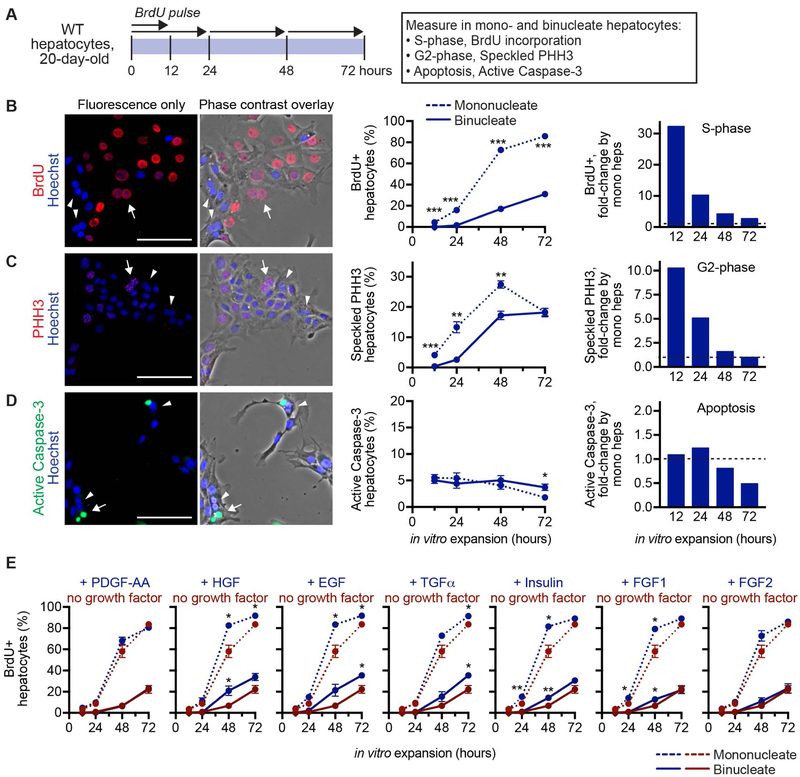
Liver regeneration is regulated by numerous signals that affect initiation, progression and termination of hepatocyte proliferation (35). To confirm our previous findings that diploid hepatocytes enter the cell cycle earlier than polyploids during liver regeneration, we compared proliferation by diploid and polyploid hepatocytes using a simplified culture model. We used hepatocytes from 20-day-old WT mice because, in contrast to adults, most mononucleate hepatocytes at this age are diploid, and nearly all binucleate hepatocytes are tetraploid (Supporting Fig. S10A–C). Thus, we quantified mono- and binucleate hepatocytes as a readout for diploid and tetraploid hepatocytes, respectively. Freshly isolated hepatocytes were cultured for 12–72 hours, and cells in S-phase, G2-phase and undergoing apoptosis were tracked (Fig. 6A and Supporting Fig. S10D). Binucleate hepatocytes started incorporating BrdU at 24–48 hours, and the percentage of BrdU+ cells slowly increased through 72 hours (Fig. 6B). In contrast, mononucleate hepatocytes entered S-phase earlier. Mononucleates incorporated BrdU by 12 hours, and the percentage of BrdU+ cells increased sharply at 24–48 hours and plateaued around 72 hours. In addition to entering S-phase earlier, the number of BrdU+ cells was consistently higher in the mononucleate subset, although the magnitude decreased with time. A similar trend was seen with cells in G2-phase, and the degree of apoptosis was similar for mono- and binucleate cells (Fig. 6C,D). Together with the in vivo findings, the data indicate that while all ploidy subsets are proliferative, diploid hepatocytes enter and transit the cell cycle faster than polyploids.
Fig. 6.
Diploid hepatocytes cycle faster than polyploids in vitro. (A) Experimental design for comparing in vitro proliferation by mono- and binucleate hepatocytes. Hepatocytes isolated from 20-day-old WT C57BL/6 mice were cultured in growth media containing 0.5% FBS + 1 μg/ml insulin and harvested after 12, 24, 48 or 72 hours to determine S-phase via BrdU incorporation, G2-phase via speckled PHH3 staining and apoptosis via active caspase-3 staining (n = 2–4 males and 2 females/time point). For BrdU incorporation, cells were pulsed with BrdU for 12 or 24 hours prior to harvest, as indicated. Note that the proliferative index is independent of gender (Supporting Fig. S10E). (B-D) Cells were stained for BrdU (red) (B), PHH3 (red) (C) or active caspase-3 (green) (D), and nuclei were stained with Hoechst dye (blue). Representative images after 48 hours in culture show fluorescence only or fluorescence overlaid with phase contrast, which is helpful for identifying mono- and binucleate cells (left panels). Binucleate cells are indicated with arrows (positive for BrdU, speckled PHH3 or active caspase-3) or arrow heads (negative for BrdU, speckled PHH3 or active caspase-3). Proliferation kinetics for mono- and binucleate hepatocytes are shown (middle panels). Additionally, graphs show the fold-change in positive mononucleate hepatocytes compared to binucleates (right panels). Normalized values for binucleates are indicated with a dashed line. (E) Hepatocytes isolated from 20-day-old WT C57BL/6 mice were cultured in serum-free media alone or with 40 ng/ml PDGF-AA, HGF, EGF, TGFα, FGF1, FGF2 or 1 μg/mL insulin for 12–72 hours (n = 2 males and 1 female/time point). Wells were harvested after BrdU pulse, as described in (A), and stained for BrdU incorporation. Each graph shows the percentage of BrdU+ mononucleate (dashed lines) and binucleate (solid lines) hepatocytes stimulated with (blue) and without (red) growth factor. * P < 0.05, ** P < 0.002, *** P < 0.00002. P values in (E) refer to comparisons between each population ± growth factor. Graphs show mean ± sem (B-middle, E) or mean only (B-right). Scale bars are 100 μm.
Finally, we wanted to understand the mechanism by which diploid hepatocytes enter the cell cycle and proliferate faster than polyploid hepatocytes. Recently, Kreutz et al. showed that diploid hepatocytes bound insulin with greater affinity than polyploids, suggesting that hepatic ploidy subsets could respond to insulin stimulation in different ways (36). Therefore, we hypothesized that differential growth factor binding by diploid and polyploid hepatocytes could regulate hepatocyte proliferation in a ploidy-specific manner. To test this idea, we stimulated primary hepatocytes with growth factors in serum-free media and compared BrdU incorporation by mononucleate and binucleate hepatocytes (Fig. 6A). We focused on growth factors that have been implicated in liver regeneration, including primary mitogens (HGF, EGF and TGFα) and auxiliary mitogens (insulin, FGF1 and FGF2) (35, 37). In the absence of exogenous growth factors, hepatocytes from 20-day-old WT mice were highly proliferative. Mononucleate hepatocytes entered S-phase 12–24 hours earlier than binucleates, and the percentage of BrdU+ cells was consistently higher (Fig. 6E). As a negative control, we stimulated with PDGF, which is produced by hepatocytes during liver regeneration but acts on stellate cells, and found that S-phase kinetics for mono- and binucleates were equivalent to controls (35). When we treated hepatocytes with mitogens, the number of BrdU+ cells trended upward throughout the culture period (e.g., compare BrdU incorporation for control and growth factor treatment). However, growth factor stimulation induced proliferation by mono- and binucleate hepatocytes to a similar degree, indicating that neither mononucleate nor binucleate hepatocytes responded preferentially (although we cannot exclude the possibility of subtle ploidy-specific effects). Thus, the data indicate the rapid proliferation by diploid hepatocytes is not caused by differential sensitivity to the growth stimuli tested here.
Discussion
The ability to study diploid and polyploid hepatocyte function in vivo is challenging. One reason is that the ploidy state of a hepatocyte can change during proliferation (5). Another reason is that, until recently, there have not been any loss-of-polyploidy genetic models that exhibit normal liver function. In 2012, Pandit et al. and Chen et al. described mice with liver-specific deletion of E2f7 and E2f8 and found the mice were depleted of polyploid hepatocytes (11, 12). We used liver-specific E2f7/E2f8 knockouts (LKO) to further study the effects of polyploidy-deficiency. Consistent with previous findings, LKO mice were functionally equivalent to controls. Most strikingly, diploid hepatocytes were increased 20-fold and LKO hepatocytes remained predominantly diploid even after 500–1000-fold expansion. Thus, LKO mice represent a stable “polyploidy knockout” model for testing the consequences of loss-of-polyploidy in vivo.
Herein we compared proliferation by diploid and polyploid hepatocytes. In response to mild proliferative cues during liver postnatal development, LKO and control livers had equivalent numbers of proliferating cells. But, in response to stronger proliferative cues, diploid hepatocytes proliferated faster. This was first seen during liver regeneration induced by 2/3 PH where there was a modest increase in proliferating LKO hepatocytes 3 days post PH and upon completion of liver regeneration. The effect was most pronounced during competitive repopulation when LKO hepatocytes outperformed controls. We also compared proliferation by diploid and polyploid hepatocytes from WT mice in vitro and in vivo. In the early stages of liver regeneration induced by 2/3 PH, diploid hepatocytes started proliferating sooner than polyploids. Additionally, among polyploid hepatocytes, tetraploids entered the cell cycle faster than octaploids, and octaploids began cycling earlier than hexadecaploids, suggesting that cell cycle entry is inversely proportional to ploidy. Collectively, the data demonstrate that diploid hepatocytes enter the cell cycle earlier and progress through the cell cycle faster than polyploids. Considering that diploid hepatocytes are the minority, representing ≤10% of all hepatocytes in mice (5), it remains to be determined which ploidy population contributes the most during a regenerative response.
At first glance, our current findings appear to conflict with earlier work where we compared repopulation by diploid and octaploid hepatocytes using competitive repopulation (5). In our earlier work, genetically marked WT diploid and octaploid hepatocytes were isolated by FACS and co-transplanted in defined ratios into Fah−/− mice. After repopulation, the ratio of diploid-derived and octaploid-derived hepatocytes was equivalent to the ratio that was originally transplanted, indicating equivalent repopulation potential by each group. However, these results were difficult to interpret because the ploidy status of donor cells changed during liver repopulation — diploid hepatocytes became polyploid and octaploid hepatocytes became tetraploid and diploid. We believe the current work provides a more accurate description of ploidy-dependent hepatocyte proliferation by focusing specifically on diploids and polyploids. Here we show that in response to proliferative cues, diploid hepatocytes enter the cell cycle earlier and progress through the cell cycle faster than polyploids. Taken together, we suggest that, at the level of individual cells (or even during clonal expansion), diploid hepatocytes cycle faster than polyploids. However, during extensive proliferation where diploid and polyploid hepatocytes generate mixed-ploidy daughters, these heterogeneous populations have equivalent proliferative capacity.
The molecular mechanisms by which diploid hepatocytes proliferate faster than polyploids are poorly understood. In 2007, Lu and colleagues compared gene expression between quiescent diploid and tetraploid hepatocytes from mice (38). Surprisingly, very few genes were differentially expressed, and the magnitude of change was small, suggesting that hepatic ploidy subsets are similar at the RNA level. Recently, it was reported that diploid hepatocytes bind insulin more effectively than polyploids, so we tested whether diploid and polyploid hepatocytes could have different responses to hepatic mitogens (36). We stimulated primary WT hepatocytes in vitro with growth factors associated with liver regeneration. Each growth factor induced hepatic proliferation, but the response by diploid and polyploid hepatocytes was equivalent, indicating that diploid hepatocytes are not hypersensitive to mitogenic stimulation. New work is necessary to determine how diploid hepatocytes proliferate faster than polyploids. We speculate that cell cycle regulation may be subtly different in diploids than polyploids (e.g., earlier replication licensing, shorter S-phase), which dramatically changes proliferation potential, especially over successive rounds of cell division. Additional mechanistic insights will come from understanding ploidy-specific proliferation in response to different forms of liver injury and by characterizing the liver microenvironment in this process.
In addition to comparing proliferation between diploid and polyploid hepatocytes in cell culture and during liver regeneration and repopulation, we also examined oncogenic proliferation. LKO livers developed numerous tumors of various sizes, whereas control mice were relatively tumor-free. These results are consistent with a report by Kent, Leone and colleagues where LKO mice spontaneously developed liver tumors during aging and developed more tumors than controls in the DEN tumor induction model (30). A number of mechanisms could explain the sensitivity of LKO mice to liver tumorigenesis. First, the E2F family of transcription factors regulate cell cycle progression, and dysregulated expression could promote oncogenic expansion. E2F8 (deleted in LKO) has been suggested to function as a tumor suppressor, and E2F1 (upregulated in LKO) has been implicated as both a tumor suppressor and oncogene (30, 39). Secondly, the increased tumorigenesis in LKO mice is consistent with our finding that diploid hepatocytes proliferate faster than polyploids. LKO mice are enriched with the most proliferative cells (diploids), and upon tumor initiation, the large number of rapidly cycling diploid hepatocytes in LKO mice drive tumorigenesis. A third mechanism was recently described by Zhang, Zhu and colleagues where they showed that polyploid hepatocytes protect from liver cancer (17). In a series of elegant experiments, polyploid hepatocytes were found to “buffer” against loss of tumor suppressors. Inactivation of one copy of a tumor suppressor in a diploid cell leads to loss-of-heterozygosity and increased potential for transformation. However, in the case of polyploid hepatocytes, the additional sets of chromosomes effectively provide “backup” copies of the tumor suppressor, which reduce the likelihood of transformation. We suggest that the large number of liver tumors in the LKO models arise through a combination of the aforementioned mechanisms. Notably, hepatocellular carcinomas (HCC) in humans and rodents are comprised predominantly of diploid hepatocytes, which strongly supports the notion that HCC formation is driven preferentially by diploid hepatocytes (40–42).
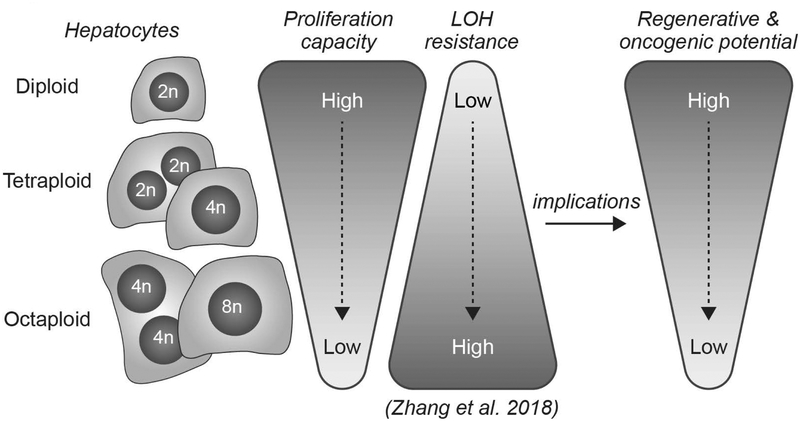
In summary, we identified a functional difference between ploidy subsets within the liver (Fig. 7). Diploid hepatocytes enter the cell cycle earlier, progress through the cell cycle faster and proliferate more extensively than polyploids. There are two major implications of this work. First, diploid hepatocytes are prone to tumorigenesis. Whereas polyploid hepatocytes have multiple copies of tumor suppressors that buffer against inactivating mutations (17), diploid hepatocytes with only 2 copies of each tumor suppressor are much more susceptible to mutational events leading to loss-of-heterozygosity. We speculate that the increased proliferative capacity associated with the diploid state promotes rapid expansion by these cells. Secondly, normal aging and liver diseases such as NAFLD, are associated with increased hepatocyte polyploidy and impaired liver regeneration (43, 44). Although polyploid hepatocytes can proliferate, we suggest that their reduced proliferative capacity depresses liver regeneration. Moreover, in the context of cell therapy, we hypothesize that transplantation of hepatocytes that are predominantly diploid rather than a mixture of all ploidy populations could facilitate rapid liver repopulation. Overall, we are just beginning to understand the functional roles played by diploid and polyploid hepatocytes, and a major goal for future work is to exploit these differences to develop new approaches for liver therapy.
Fig. 7.
Model illustrating functional differences between diploid and polyploid hepatocytes, including implications for liver regeneration and cancer. Diploid hepatocytes enter the cell cycle earlier and proliferate faster than polyploids, providing diploids with increased capacity for liver regeneration. In terms of liver cancer, diploids are uniquely sensitive to transformation due to tumor suppressor loss-of-heterozygosity (LOH) (17) and, when transformed, they are poised for rapid proliferation.
Supplementary Material
Acknowledgements
We thank Gustavo Leone (The Ohio State University) and Alain deBruin (Utrecht University) for sharing liver-specific E2f7/E2f8 knockout mice. Thanks also to Lynda Guzik (McGowan Institute Flow Cytometry Core at University of Pittsburgh) for flow cytometry assistance.
Financial Support
This work was supported by grants to AWD from the NIH (R01 DK103645) and the Commonwealth of Pennsylvania.
Abbreviations
- 2n, 4c, 8c
chromatid number for diploid, tetraploid, octaploid hepatocytes, respectively
- 2n, 4n, 8n
chromosome number for diploid, tetraploid, octaploid hepatocytes, respectively
- ALP
alkaline phosphatase
- ALT
alanine aminotransferase
- AST
aspartate aminotransferase
- β-gal
β-galactosidase
- BrdU
5-bromo-2’-deoxyuridine
- DEN
Diethylnitrosamine
- EGF
epidermal growth factor
- FACS
fluorescence-activated cell sorting
- Fah
fumarylacetoacetate hydrolase
- FBS
fetal bovine serum
- FGF
fibroblast growth factor
- FRGN
Fah−/− Rag2−/− Interleukin 2 common Gamma chain−/− Nod background
- GS
glutamine synthetase
- H&E
hematoxylin and eosin
- HFD
high fat diet
- HGF
hepatocyte growth factor
- LKO
liver-specific E2f7/E2f8 knockout
- LW/BW
Liver Weight/Body Weight
- NTBC
2-(2-nitro-4-trifluoro- methylbenzoyl)-1,3-cyclo-hexanedione
- PB
phenobarbital
- PCNA
proliferating cell nuclear antigen
- PDGF
platelet-derived growth factor
- PH
partial hepatectomy
- PHH3
phospho-histone H3
- qRT-PCR
quantitative reverse transcriptase polymerase chain reaction
- SEM
standard error of mean
- TGFα
transforming growth factor alpha
- WT
wild-type
References
- 1.Gentric G, Desdouets C. Polyploidization in liver tissue. Am J Pathol 2014;184:322–331. [DOI] [PubMed] [Google Scholar]
- 2.Duncan AW. Aneuploidy, polyploidy and ploidy reversal in the liver. Semin Cell Dev Biol 2013;24:347–356. [DOI] [PubMed] [Google Scholar]
- 3.Pandit SK, Westendorp B, de Bruin A. Physiological significance of polyploidization in mammalian cells. Trends Cell Biol 2013;23:556–566. [DOI] [PubMed] [Google Scholar]
- 4.Duncan AW, Hanlon Newell AE, Smith L, Wilson EM, Olson SB, Thayer MJ, Strom SC, et al. Frequent aneuploidy among normal human hepatocytes. Gastroenterology 2012;142:25–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duncan AW, Taylor MH, Hickey RD, Hanlon Newell AE, Lenzi ML, Olson SB, Finegold MJ, et al. The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature 2010;467:707–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guidotti JE, Bregerie O, Robert A, Debey P, Brechot C, Desdouets C. Liver cell polyploidization: a pivotal role for binuclear hepatocytes. J Biol Chem 2003;278:19095–19101. [DOI] [PubMed] [Google Scholar]
- 7.Margall-Ducos G, Celton-Morizur S, Couton D, Bregerie O, Desdouets C. Liver tetraploidization is controlled by a new process of incomplete cytokinesis. J Cell Sci 2007;120:3633–3639. [DOI] [PubMed] [Google Scholar]
- 8.Hsu SH, Delgado ER, Otero PA, Teng KY, Kutay H, Meehan KM, Moroney JB, et al. MicroRNA-122 regulates polyploidization in the murine liver. Hepatology 2016;64:599–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knouse KA, Wu J, Whittaker CA, Amon A. Single cell sequencing reveals low levels of aneuploidy across mammalian tissues. Proc Natl Acad Sci U S A 2014;111:13409–13414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xanthoulis A, Tiniakos DG. E2F transcription factors and digestive system malignancies: how much do we know? World J Gastroenterol 2013;19:3189–3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pandit SK, Westendorp B, Nantasanti S, van Liere E, Tooten PC, Cornelissen PW, Toussaint MJ, et al. E2F8 is essential for polyploidization in mammalian cells. Nat Cell Biol 2012;14:1181–1191. [DOI] [PubMed] [Google Scholar]
- 12.Chen HZ, Ouseph MM, Li J, Pecot T, Chokshi V, Kent L, Bae S, et al. Canonical and atypical E2Fs regulate the mammalian endocycle. Nat Cell Biol 2012;14:1192–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conner EA, Lemmer ER, Sanchez A, Factor VM, Thorgeirsson SS. E2F1 blocks and c-Myc accelerates hepatic ploidy in transgenic mouse models. Biochem Biophys Res Commun 2003;302:114–120. [DOI] [PubMed] [Google Scholar]
- 14.Azuma H, Paulk N, Ranade A, Dorrell C, Al-Dhalimy M, Ellis E, Strom S, et al. Robust expansion of human hepatocytes in Fah−/−/Rag2−/−/Il2rg−/− mice. Nat Biotechnol 2007;25:903–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilson EM, Bial J, Tarlow B, Bial G, Jensen B, Greiner DL, Brehm MA, et al. Extensive double humanization of both liver and hematopoiesis in FRGN mice. Stem Cell Res 2014;13:404–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Overturf K, Al-Dhalimy M, Tanguay R, Brantly M, Ou CN, Finegold M, Grompe M. Hepatocytes corrected by gene therapy are selected in vivo in a murine model of hereditary tyrosinaemia type I. Nat Genet 1996;12:266–273. [DOI] [PubMed] [Google Scholar]
- 17.Zhang S, Zhou K, Luo X, Li L, Tu HC, Sehgal A, Nguyen LH, et al. The Polyploid State Plays a Tumor-Suppressive Role in the Liver. Dev Cell 2018;44:447–459 e445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ganem NJ, Storchova Z, Pellman D. Tetraploidy, aneuploidy and cancer. Curr Opin Genet Dev 2007;17:157–162. [DOI] [PubMed] [Google Scholar]
- 19.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646–674. [DOI] [PubMed] [Google Scholar]
- 20.Zhang S, Li L, Kendrick SL, Gerard RD, Zhu H. TALEN-mediated somatic mutagenesis in murine models of cancer. Cancer Res 2014;74:5311–5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aydinlik H, Nguyen TD, Moennikes O, Buchmann A, Schwarz M. Selective pressure during tumor promotion by phenobarbital leads to clonal outgrowth of beta-catenin-mutated mouse liver tumors. Oncogene 2001;20:7812–7816. [DOI] [PubMed] [Google Scholar]
- 22.Loeppen S, Schneider D, Gaunitz F, Gebhardt R, Kurek R, Buchmann A, Schwarz M. Overexpression of glutamine synthetase is associated with beta-catenin-mutations in mouse liver tumors during promotion of hepatocarcinogenesis by phenobarbital. Cancer Res 2002;62:5685–5688. [PubMed] [Google Scholar]
- 23.Moennikes O, Buchmann A, Romualdi A, Ott T, Werringloer J, Willecke K, Schwarz M. Lack of phenobarbital-mediated promotion of hepatocarcinogenesis in connexin32-null mice. Cancer Res 2000;60:5087–5091. [PubMed] [Google Scholar]
- 24.Schmid A, Rignall B, Pichler BJ, Schwarz M. Quantitative analysis of the growth kinetics of chemically induced mouse liver tumors by magnetic resonance imaging. Toxicol Sci 2012;126:52–59. [DOI] [PubMed] [Google Scholar]
- 25.Honkakoski P, Zelko I, Sueyoshi T, Negishi M. The nuclear orphan receptor CAR-retinoid X receptor heterodimer activates the phenobarbital-responsive enhancer module of the CYP2B gene. Mol Cell Biol 1998;18:5652–5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li L, Bao X, Zhang QY, Negishi M, Ding X. Role of CYP2B in Phenobarbital-Induced Hepatocyte Proliferation in Mice. Drug Metab Dispos 2017;45:977–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shirakami Y, Gottesman ME, Blaner WS. Diethylnitrosamine-induced hepatocarcinogenesis is suppressed in lecithin:retinol acyltransferase-deficient mice primarily through retinoid actions immediately after carcinogen administration. Carcinogenesis 2012;33:268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bakiri L, Wagner EF. Mouse models for liver cancer. Mol Oncol 2013;7:206–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cieply B, Zeng G, Proverbs-Singh T, Geller DA, Monga SP. Unique phenotype of hepatocellular cancers with exon-3 mutations in beta-catenin gene. Hepatology 2009;49:821–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kent LN, Rakijas JB, Pandit SK, Westendorp B, Chen HZ, Huntington JT, Tang X, et al. E2f8 mediates tumor suppression in postnatal liver development. J Clin Invest 2016;126:2955–2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Michalopoulos GK. Liver regeneration. J Cell Physiol 2007;213:286–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michalopoulos GK, DeFrances MC. Liver regeneration. Science 1997;276:60–66. [DOI] [PubMed] [Google Scholar]
- 33.Higgins G, Anderson GM. Experimental pathology of the liver. Restoration of the liver of the white rat following partial surgical removal. Arch Pathol 1931;12:186–202. [Google Scholar]
- 34.Miyaoka Y, Ebato K, Kato H, Arakawa S, Shimizu S, Miyajima A. Hypertrophy and unconventional cell division of hepatocytes underlie liver regeneration. Curr Biol 2012;22:1166–1175. [DOI] [PubMed] [Google Scholar]
- 35.Michalopoulos GK. Liver regeneration after partial hepatectomy: critical analysis of mechanistic dilemmas. Am J Pathol 2010;176:2–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kreutz C, MacNelly S, Follo M, Waldin A, Binninger-Lacour P, Timmer J, Bartolome-Rodriguez MM. Hepatocyte Ploidy Is a Diversity Factor for Liver Homeostasis. Front Physiol 2017;8:862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bowen WC, Michalopoulos AW, Orr A, Ding MQ, Stolz DB, Michalopoulos GK. Development of a chemically defined medium and discovery of new mitogenic growth factors for mouse hepatocytes: mitogenic effects of FGF1/2 and PDGF. PLoS One 2014;9:e95487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu P, Prost S, Caldwell H, Tugwood JD, Betton GR, Harrison DJ. Microarray analysis of gene expression of mouse hepatocytes of different ploidy. Mamm Genome 2007;18:617–626. [DOI] [PubMed] [Google Scholar]
- 39.Johnson DG. The paradox of E2F1: oncogene and tumor suppressor gene. Mol Carcinog 2000;27:151–157. [DOI] [PubMed] [Google Scholar]
- 40.Anti M, Marra G, Rapaccini GL, Rumi C, Bussa S, Fadda G, Vecchio FM, et al. DNA ploidy pattern in human chronic liver diseases and hepatic nodular lesions. Flow cytometric analysis on echo-guided needle liver biopsy. Cancer 1994;73:281–288. [DOI] [PubMed] [Google Scholar]
- 41.Rua S, Comino A, Fruttero A, Torchio P, Bouzari H, Taraglio S, Torchio B, et al. Flow cytometric DNA analysis of cirrhotic liver cells in patients with hepatocellular carcinoma can provide a new prognostic factor. Cancer 1996;78:1195–1202. [DOI] [PubMed] [Google Scholar]
- 42.Saeter G, Schwarze PE, Nesland JM, Seglen PO. Diploid nature of hepatocellular tumours developing from transplanted preneoplastic liver cells. Br J Cancer 1989;59:198–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gentric G, Maillet V, Paradis V, Couton D, L’Hermitte A, Panasyuk G, Fromenty B, et al. Oxidative stress promotes pathologic polyploidization in nonalcoholic fatty liver disease. J Clin Invest 2015;125:981–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sigal SH, Rajvanshi P, Gorla GR, Sokhi RP, Saxena R, Gebhard DR Jr., Reid M, et al. Partial hepatectomy-induced polyploidy attenuates hepatocyte replication and activates cell aging events. Am J Physiol 1999;276:G1260–1272. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.