Abstract
Mechanotransduction, or the process by which mechanical forces regulate cellular functions, is increasingly studied in a variety of different physiological and pathological contexts. Although these forces are most often studied at cell-matrix and cell-cell adhesions, recent work has shown that the nuclear LINC complex is also subject to mechanical forces. Here we describe how to use a FRET-based biosensor, known as TSmod, in the LINC complex protein nesprin-2G. This approach allows for measurement of LINC complex forces in living cells with spatial-temporal resolution.
Keywords: Mechanobiology, FRET tension biosensors, Nuclear LINC complex
1. Introduction
Evidence for mechanical forces at cell-matrix adhesions has existed for over 30 years [1]. More recently mechanical forces have been directly measured across cell-cell adhesions [2, 3]. The model of cellular tensegrity predicts that cell-matrix and cell-cell forces are readily transferred across the cytoskeleton and applied to intracellular structures such as the nucleus [4]. Over 20 years ago, Ingber and coworkers showed that changes in actomyosin forces altered nuclear shape [5]. Subsequent experiments showed that externally applied forces to the perimeter of the cell also altered nuclear shape [6–8], suggesting that the cytoskeleton is “hardwired” into the nuclear membrane [9]. More recent work in the field of cell biology has identified a group of nuclear membrane-associated proteins, known as the LINC (linker of nucleoskeleton and cytoskeleton) complex, that mediate the connection of the cytosolic cytoskeleton to the nuclear membrane [9]. The LINC complex is formed by nesprin and SUN proteins that cross both the inner and outer nuclear membranes to mechanically tether the cytosolic cytoskeleton to the nuclear lamina (Fig. 1). The LINC complex is evolutionarily conserved across virtually all eukaryotes [9], suggesting that the mechanical linkage of the cytoskeleton to the nucleus may be essential to cell function and homeostasis. One potential function of the LINC complex is the transfer of forces from the cytoplasmic cytoskeleton onto the nucleus.
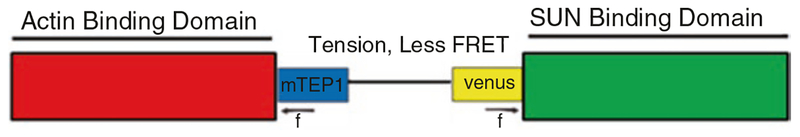
Fig. 1.
Diagram of nesprin-TS. TSmod, the FRET-based force sensing module is flanked by actin and sun binding domains to capture actin-based tensile forces exerted on sun proteins embedded in the nuclear membrane
The nucleus itself has been proposed to be a mechanosensor, where nuclear forces could regulate cellular functions, including nuclear transport, DNA structure, and gene expression [10]. However, studies to examine the role of nuclear forces have been hindered by a lack of techniques capable of directly measuring nuclear forces in living cells. Current knowledge of nuclear mechanical forces is based on experimental measurements of nuclear geometry and positioning, knockdown of LINC complex proteins, and overexpression of dominant negative proteins that disrupt the LINC complex. Disruption of the LINC complex perturbs cell migration, nuclear shape, and position [11, 12]. However, these experiments do not establish if forces directly regulate these functions—the LINC complex may also serve to regulate bio-chemical signaling pathways correlated with force [9].
Recently, a genetically encoded, calibrated Förster resonance energy transfer (FRET)-based tension biosensor (known as TSmod) was developed [13]. This sensor consists of a pair of fluorescent proteins capable of FRET separated by an elastic peptide (flagelliform). Application of force to the sensor results in increased elongation of the elastic peptide (strain), resulting in decreased FRET. The FRET-force relationship of the sensor was previously calibrated and found to have a dynamic range of 1–5 pN [13]. This sensor module was recently shown to respond to compressive loading, with higher FRET than in the unloaded state [14]. We and others have inserted the sensor into a number of proteins at cell-cell [15–18] and cell-matrix [13, 19] adhesions, indicating that the dynamic range of the sensor is well suited to study forces on proteins. In addition, the length of the flagelliform elastic linker in the TSmod sensor can be adjusted to shift the FRET-force dynamic range [20], which may be important for proteins subjected to higher or lower levels of force. Recently a new elastic peptide, known as HP35 (consisting of the villin headpiece), has been used to measure higher forces (7–10 pN) [19].
To extend this force measurement technique to the nuclear LINC complex, we recently developed a nesprin force sensor (known as nesprin-TS) in which TSmod was inserted into mininesprin-2G [21]. This sensor captures the mechanical forces exerted between the actin cytoskeleton and the nuclear envelope where nesprin-2G binds (see Notes 12–14 for additional considerations regarding how force is applied to the nucleus and limitations of the mini-nesprin-2G sensor). This sensor behaved similarly to nesprin-2G, in that it was able to rescue nuclear positioning in nesprin-2G-depleted cells [21]. Furthermore, the sensor exhibited FRET changes in response to changes in actomyosin contractility [21]. Nesprin-2G was found to be under constitutive mechanical tension in adherent fibroblasts. Mechanical forces on nesprin-2G were found to be spatially different (apical versus basal sides of the nucleus) and were increased in elongated cells [21]. Thus, TSmod-based biosensors are well suited to study LINC complex forces in living cells.
In this protocol, we provide a detailed methodology for using the nesprin-2G force sensor, which details how to express the nesprin tension sensor (nesprin-TS) in mammalian cells and how to acquire and analyze FRET images of these cells expressing nesprin-TS. Although this protocol discusses expressing nesprin-TS in fibroblasts, it is easily adaptable to other mammalian cells, including both cell lines and primary cells. In our hands nesprin-2G is under tensile force in a variety of different cells (fibroblasts, epithelial, endothelial), but these forces vary depending on the cell type (Arsenovic, unpublished) (see Note 10 concerning alternate FRET force senseors). Furthermore, this protocol as it relates to the use of the nesprin-TS can be readily adapted to other FRET-based force biosensors that have been developed for other proteins.
2. Materials
2.1. Growth and Amplification of Biosensor Constructs
DNA plasmids for nesprin-TS and the force-insensitive control nesprin-HL are available from Addgene (plasmids 68127 and 68128). Plasmids encoding mTFP1 (54613), venus (27793), and TSmod (26021) are can also be obtained from Addgene.
Standard lysogeny broth (LB).
LB plates and medium with ampicillin or kanamycin as appropriate.
1 L bacterial flask.
Temperature-controlled orbital shaker.
Midi-prep DNA isolation kit.
2.2. Cell Culture and Transfection
NIH3T3 fibroblasts (ATCC CRL-1658).
DMEM (Thermo Fisher 11995).
Bovine calf serum (Thermo Fisher 16170).
Lipofectamine 2000 (Thermo Fisher 11668).
#1.5 coverglass bottom dishes (Cellvis D35-20-1.5-N).
Bovine fibronectin (Alfa Aesar J65696).
Dulbecco’s phosphate buffered saline (Thermo 14190–144).
2.3. Imaging
Live Cell Imaging Solution (Thermo Fisher A14291DJ).
Temperature-controlled inverted confocal with 458 and 514 nm laser lines (Zeiss LSM 710 with spectral META detector).
3. Methods
3.1. Plasmid Amplification
The use of DNA-based biosensors requires E. coli amplification and DNA isolation. DNA obtained directly from Addgene arrives as a live bacterial stab and can be directly streaked onto an LB plate (with correct antibiotic). DNA obtained in water or Tris–HCl must first be transformed into a standard competent E. coli strains such as DH5-α.
After overnight growth at 37 °C, a single colony is selected from a plate and grown in 2 mL LB medium (with antibiotic) for 8 h with orbital shaking at 37 °C. After 8 h 1–2 mL is added to a large culture flask containing 150 mL LB medium (with antibiotic) and then grown overnight with orbital shaking at 37 °C. This larger flask is sometimes referred to as a midi-prep.
Spin down bacteria (5000 × g for 30 min), and isolate the DNA using a standard midi-prep DNA isolation kit, following manufacturer instructions.
3.2. Cell Transfection
Carry out all cell culture experiments in a BSL-2 equipped tissue culture facility, and follow all institutional guidelines for working with recombinant DNA.
NIH3T3 fibroblasts are cultured in DMEM with 10% bovine serum, using standard cell culture growth and passaging methods.
Nesprin-TS, nesprin-HL, and fluorescent protein control plasmids can readily be transfected into NIH3T3 fibroblasts using Lipofectamine 2000 per manufacturer instructions.
For all imaging experiments, cells are grown on #1.5 coverglass bottom dishes (Cellvis D35-20-1.5-N). Dishes are first coated with a layer of fibronectin at a concentration of 20 μg/mL (in PBS) for 20 min at room temperature, which is then removed immediately before cell seeding.
After 4–6 h of incubation with transfection reagents, cells are trypsinized, centrifuged, and resuspended on glass bottom dishes in fresh medium.
Cells are allowed to attach overnight and can be imaged the next day.
Viral methods of transfection (see Note 8) stable cell lines (see Note 9) can also be used.
3.3. Imaging
Prior to imaging phenol red containing culture medium is replaced with HEPES-buffered clear Live Cell Imaging Solution with 10% calf serum. The Live Cell Imaging Solution has improved clarity and signal to noise ratio and provides for proper pH at ambient atmosphere. Additionally, cells should be maintained in a 37 °C enclosure on the microscope, if possible. Maintaining cells at a constant physiological temperature prevents temperature-dependent focal drift and improves cell viability in longer experiments.
Image cells using a laser scanning confocal (Zeiss LSM 710 with spectral META detector). However, confocal imaging is not required for the sensors. Standard epifluorescence can be used with filters to separately excite mTFP1 (458 nm) and yellow (515 nm) with appropriate band pass filters to separately resolve blue and yellow emission; however this protocol will detail the use of spectral imaging-based FRET (see Note 1), not epifluorescence standard filter set-based FRET.
Collect spectral images (see Note 1) at 458 nm excitation using a 40× water or 63× oil objective. The 40× objective allows for acquisition of a larger group of cells, whereas the 63× objective (with additional optical zoom) can provide additional spatial information about the distribution of FRET across a single nucleus.
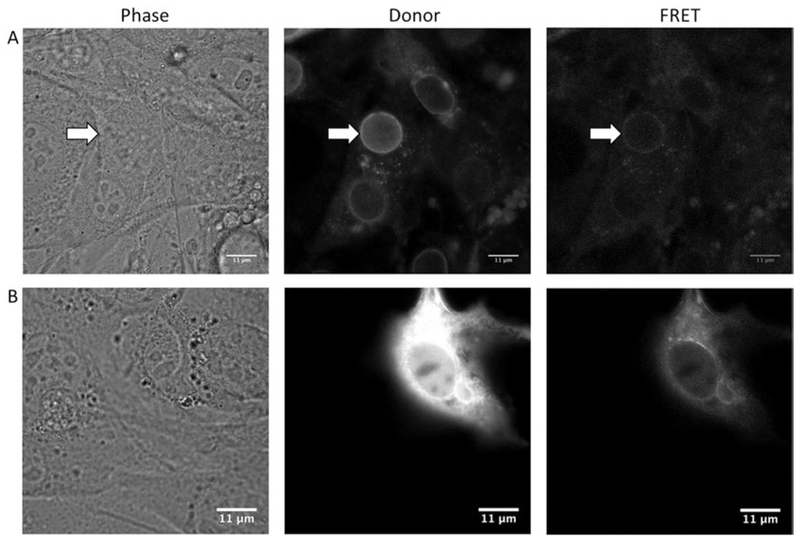
Select cells for imaging that are expressing the nesprin-TS at a high enough level to be readily visualized and imaged, but avoid imaging cells in which expression levels are too high (example images of appropriate and inappropriate cells are shown in Fig. 2A, B; also see Note 2). Additionally, cells that looked stressed or abnormal should be excluded from analysis.
To account for cell-cell variations in FRET, image at least 10–20 cells per condition (see Note 3).
Include a known high or low force control to confirm that the sensor and analysis of FRET is working. Examples include the force-insensitive nesprin-HL (headless) sensor, as well as myosin agonists and antagonists (see Notes 4–6).
Fig. 2.
Phase-contrast and fluorescent images of nesprin-TS expressing NIH3T3 fibroblasts. Row A: Phase-contrast, donor, and FRET channel fluorescent images of an optimally expressing cell. The best nucleus for analysis is indicated by the white arrow. Note that while other nuclei are discernable in the donor channel image, the intensity is too low in the FRET channel for accurate analysis. Row B: Phase-contrast, donor, and FRET fluorescent channel images of a cell with high expression. The high level of expression of nesprin-TS in both the ER and nucleoplasm makes it difficult to discern the boundaries of the nuclear envelope. These cells are excluded from analysis
3.4. Image Analysis
Use open-source ImageJ software (http://fiji.sc/) or similar software to open and further process FRET images.
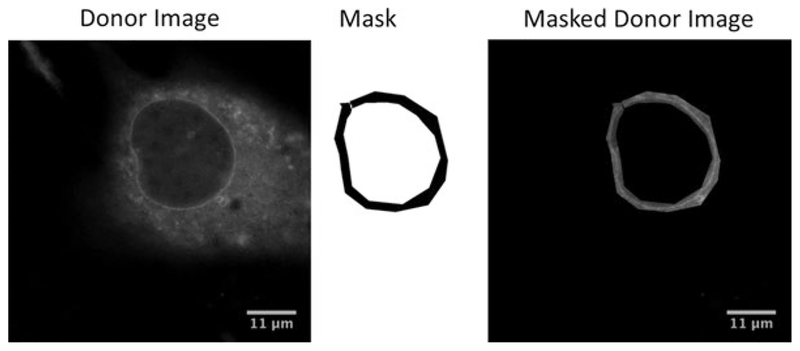
Manually mask images such that the only FRET signal is from the nuclear envelope (Fig. 2). Without masking, FRET signal from other regions of the cell may represent a large fraction of pixels and significantly affect the mean FRET for the image. Delineating the nuclear envelope is easiest for cells with lower expression of nesprin-TS (see Note 2).
Further process images by performing a background correction (also known as background subtraction) by subtracting the average pixel value of nonfluorescent cells. Untransfected cells imaged under the same power and gain are used to calculate the average background pixel value.
The masked and background FRET images need to be further processed to allow FRET images to be compared between conditions. Ratiometric FRET offers the most simple and straightforward method to measure relative FRET differences between conditions. To calculate the FRET ratio, background-corrected pixel values from the FRET channel are normalized to the background-corrected donor channel by dividing the unmixed acceptor image (FRET channel) by the unmixed donor image. Normalization to the acceptor-only signal (excited by the 514 nm acceptor laser) is also possible, but this will be less sensitive to changes in FRET with the benefit of reduced noise in the ratio image. Ratiometric FRET does not provide an absolute measure of FRET, but provides a relative measure of FRET changes between conditions. Because it can be influenced by background subtraction values, we do not recommend comparing FRET ratios obtained from experiments collected from different microscope sessions (see Note 7). Furthermore, it is essential to hold all laser power and detector gain settings constant across all samples. Repeated imaging of the same region may result in photobleaching effects (see Note 11).
4. Notes
FRET signal (acceptor emission obtained with donor excitation) can be contaminated with both donor and acceptor signals unrelated to FRET when using standard band pass filters. The FRET signal must be corrected to remove this bleed-through, and these corrections are beyond the scope of this protocol. Alternatively, spectral unmixing allows for real-time separation of donor and acceptor signal, eliminating the additional step of removing donor bleed-through. If a spectral detector is used, a single excitation wavelength (458 nm) may be selected to illuminate the sample and capture the entire fluorescent emission (420–720 nm) simultaneously. We have published a detailed methods paper in Journal of Visualized Experiments (JOVE) which details the use of the Zeiss 710 for spectral unmixing of the FRET signal from nesprin-TS [22].
In high-expressing cells, nesprin-TS frequently localizes inside the nucleus, in addition to the nuclear envelope. In these cells it is difficult to discern the fluorescence from the nuclear envelope from that inside of the nucleus, and therefore we exclude these cells from FRET analysis. Cells with lower levels of expression often give better contrast where it is easier to identify and mask the nuclear envelope. However this can be a trade-off as images acquired from lower intensity result in FRET with a lower signal to noise ratio.
Many times FRET differences are minimal between conditions. Furthermore, we have observed large cell-to-cell variations in FRET within the same condition. We therefore recommend the acquisition of many images (10–20 per condition) to more accurately determine if two conditions are significantly different in nesprin-2G forces. Paired comparisons of the same cell before and after treatment can be extremely useful for minimizing biological variability in baseline nesprin-2G force.
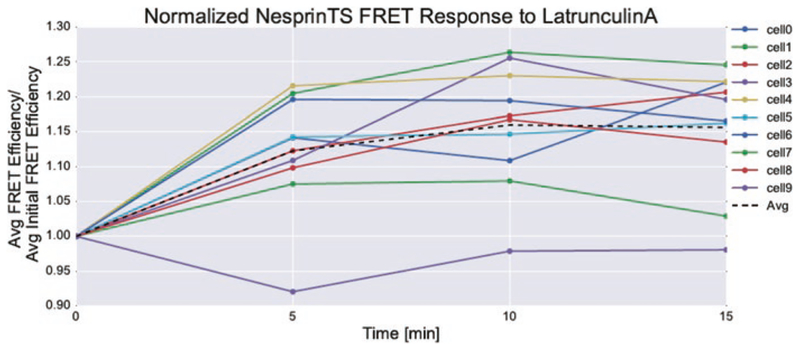
In addition to nesprin-HL, we frequently employ the use of ROCK and MLCK inhibitors (Y-27632 and ML7, respectively) to inhibit myosin. We have also used actin-destabilizing agents, such as latrunculin A, to reduce force (Fig. 3). Blebbistatin has yellow fluorescence and may exhibit phototoxicity at the wavelengths used to acquire nesprin-TS [23]. In our experience the yellow color of blebbistatin prevents its use with any mTFP1-venus TSmod sensor. The use of chemical inhibitors can be a quick way to determine whether measurable nesprin-2G tensile forces exist in your cell of interest. The use of these inhibitors has the added advantage of enabling before and after treatment imaging of the same cell, allowing for paired comparisons to be made (reducing cell-to-cell variability; see Fig. 3).
The largest variations in nesprin-2G forces we have observed were between cells grown on micropatterned 20 μm lines as compared to cells grown on non-patterned surfaces [24]. This may also be a useful positive control to confirm the responsiveness of the nesprin-TS to measure differences in force on the nucleus.
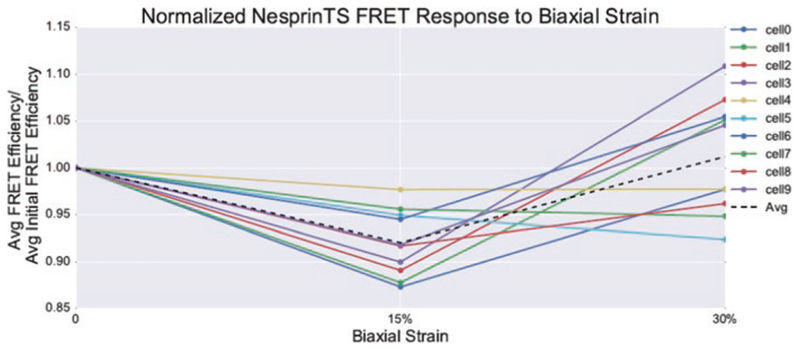
Although we have shown calyculin A and other activators of myosin can increase nesprin-2G force [24], these compounds, when used for too long or at too high of a dose, can cause cell detachment and rounding, eventually leading to a reduction in nuclear forces at longer time points. Recent work by our lab has shown that biaxial stretch can also be used to increase nesprin-2G force (Fig. 4). However too large of a deformation can also reduce nesprin-2G force in some cells, presumably due to dissociation or rupture of nesprin-2G linkage to cytoskeletal proteins (Fig. 5).
-
Alternate FRET methods which calculate FRET efficiency can be used [25] to provide more absolute quantities of FRET, which in turn can be compared across experiments. Additionally, with FRET efficiency, it is possible to estimate average force per molecule (in piconewtons) [13].
We caution against the use of ratiometric imaging if the cell of interest has very high measured tension on nesprin-2G. Systematic errors in the ratio images occur when the acceptor signal (venus) becomes very dim. We suggest using either FLIM or a quantitative FRET efficiency unmixing algorithm if you suspect the sensor is under high loads [25].
We have developed an adenovirus for nesprin-TS, which enables more uniform and longer-term expression of the sensor in a variety of cells, including primary cells. Lentiviral versions (particularly tetracycline inducible versions which could be used to control expression levels) may also provide a robust way to express sensors.
We have developed a MDCK stable cell line expressing nesprin-TS, as well as other force biosensors. Stable cell lines offer the advantage of a relatively uniform expressing population of cells, making imaging and analysis of images more straightfor-ward compared to transient transfections. Additionally this method allows for observation of forces across a group of cells.
Under high force conditions, we have observed better performance of nesprin-TS with a 25 amino acid linker replacing the 40 amino acid linker used in TSmod (nesprin TSmod with the 25 amino acid linker is located at the same insert site as the TSmod 40 nesprin shown in Fig. 1) (Arsenovic, unpublished). TSmod with the 25 amino acid linker has a higher dynamic range of FRET, but is less responsive at lower forces [20]. Although we have not used the newer HP35 elastic peptide [19] as the tension responsive element in nesprin tension sensors, it is possible that it may be more responsive under conditions when nesprin-2G forces are high.
When making repeated measurements of the same cell, it is important to control for photobleaching effects that may influence FRET. This can be done by imaging a control sample expressing nesprin-TS without treatment and comparing the change in FRET after repeated measurements.
Our sensor is based on the design of mini-nesprin-2G, a much smaller version of the giant form of nesprin-2G. This was chosen for ease of cloning and expression. However, this sensor lacks a number of important binding domains, such as FHOD1 [26], and does not contain binding sites for the microtubule motor proteins kinesin and dynein [27–29]. Thus, the sensor should be viewed only as a tool to study actin-based forces on the LINC complex, using a minimal form of nesprin-2G. Although we showed that this truncated form of nesprin retains the ability to rescue reward nuclear movement [24], it may be missing binding sites for key regulatory molecules that could influence forces. In addition, microtubule and intermediate-based connections to the LINC complex (through nesprin-2 and other nesprin isoforms) may also contribute tensile and compressive forces to the nucleus. It will be interesting to determine forces across other nesprin isoforms with similar biosensors for these proteins.
The mechanical model for how forces are transmitted into the nucleus remains unclear. While we hypothesize that forces applied to nesprin are transmitted across SUN1 and SUN2 and into the nucleus, this cannot be concluded from our observation that nesprin-2G is under tensile load. Additional force sensors for SUN1/2 or proteins inside the nucleus are needed to better understand force transmission.
Forces applied to the nucleus may not be exclusively through the LINC complex. Wirtz and colleagues have suggested that an actin cap may apply compressive forces onto the nucleus [30], which may or may not be LINC complex dependent. In addition Lele and colleagues have suggested that LINC-independent compressive forces from the plasma membrane may also be applied to the nucleus during cell spreading [31]. Results obtained from LINC complex biosensors may not capture all forces applied onto the nucleus. This raises the possibility that experiments which affect cell spread area (e.g., hydrogels of varying elastic moduli, experiments in which cell confluence is varied) may be altering both LINC-dependent and LINC-independent forces on cell nuclei.
Fig. 3.
Demonstration of image masking. Images may be manually masked using the polygon selection tool in ImageJ or a paintbrush tool of fixed width. A masked or binary image (values of 0 or 1) is created from the user selection. Donor image and FRET image (not shown) are each multiplied by the mask to create masked donor and FRET images of the nuclear envelope
Fig. 4.
Response of nesprin-TS to disruption of actin filaments. Exposure of NIH3T3 fibroblasts to 10 μM latrun-culin A results in a sustained increase in nesprin-TS FRET, indicating decreased force. The same cells were tracked before and during latrunculin treatment, which allowed for normalization to the initial FRET efficiency
Fig. 5.
Response of nesprin-TS to biaxial strain. Exposure of NIH3T3 fibroblasts to 15% biaxial strain results in decreased FRET for nesprin-TS, indicating increased force. However higher levels of stretch result in a varied response with some cells increasing and decreasing in force. The same cells were tracked before and during stretch
Acknowledgments
This work was supported by NIH grant R35GM119617 and NSF CAREER CMMI1653299.
References
- 1.Harris AK, Wild P, Stopak D (1980) Silicone rubber substrata: a new wrinkle in the study of cell locomotion. Science 208:177–179 [DOI] [PubMed] [Google Scholar]
- 2.Liu Z, Sniadecki NJ, Chen CS (2010) Mechanical forces in endothelial cells during firm adhesion and early transmigration of human monocytes. Cell Mol Bioeng 3:50–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maruthamuthu V, Sabass B, Schwarz US, Gardel ML (2011) Cell-ECM traction force modulates endogenous tension at cell-cell contacts. Proc Natl Acad Sci U S A 108:4708–4713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ingber DE (2003) Tensegrity I. Cell structure and hierarchical systems biology. J Cell Sci 116:1157–1173 [DOI] [PubMed] [Google Scholar]
- 5.Sims JR, Karp S, Ingber DE (1992) Altering the cellular mechanical force balance results in integrated changes in cell, cytoskeletal and nuclear shape. J Cell Sci 103(Pt 4):1215–1222 [DOI] [PubMed] [Google Scholar]
- 6.Maniotis AJ, Chen CS, Ingber DE (1997) Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc Natl Acad Sci U S A 94:849–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu S, Eberhard L, Chen J, Love JC, Butler JP, Fredberg JJ, Whitesides GM, Wang N (2004) Mechanical anisotropy of adherent cells probed by a three-dimensional magnetic twisting device. Am J Physiol Cell Physiol 287:C1184–C1191 [DOI] [PubMed] [Google Scholar]
- 8.Hu S, Chen J, Fabry B, Numaguchi Y, Gouldstone A, Ingber DE, Fredberg JJ, Butler JP, Wang N (2003) Intracellular stress tomography reveals stress focusing and structural anisotropy in cytoskeleton of living cells. Am J Physiol Cell Physiol 285:C1082–C1090 [DOI] [PubMed] [Google Scholar]
- 9.Alam S, Lovett DB, Dickinson RB, Roux KJ, Lele TP (2014) Nuclear forces and cell mechanosensing. Prog Mol Biol Transl Sci 126:205–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang N, Tytell JD, Ingber DE (2009) Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus. Nat Rev Mol Cell Biol 10:75–82 [DOI] [PubMed] [Google Scholar]
- 11.Lombardi ML, Jaalouk DE, Shanahan CM, Burke B, Roux KJ, Lammerding J (2011) The interaction between nesprins and sun proteins at the nuclear envelope is critical for force transmission between the nucleus and cytoskeleton. J Biol Chem 286:26743–26753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luxton GWG, Gomes ER, Folker ES, Vintinner E, Gundersen GG (2010) Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science 329:956–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grashoff C, Hoffman BD, Brenner MD, Zhou R, Parsons M, Yang MT, McLean MA, Sligar SG, Chen CS, Ha T, Schwartz MA (2010) Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature 466:263–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rothenberg KE, Neibart SS, LaCroix AS, Hoffman BD (2015) Controlling cell geometry affects the spatial distribution of load across Vinculin. Cell Mol Bioeng 8:364–382 [Google Scholar]
- 15.Conway DE, Breckenridge MT, Hinde E, Gratton E, Chen CS, Schwartz MA (2013) Fluid shear stress on endothelial cells modulates mechanical tension across VE-cadherin and PECAM-1. Curr Biol 23:1024–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuriyama S, Theveneau E, Benedetto A, Parsons M, Tanaka M, Charras G, Kabla A, Mayor R (2014) In vivo collective cell migration requires an LPAR2-dependent increase in tissue fluidity. J Cell Biol 206:113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borghi N, Sorokina M, Shcherbakova OG, Weis WI, Pruitt BL, Nelson WJ, Dunn AR (2012) E-cadherin is under constitutive actomyosin-generated tension that is increased at cell-cell contacts upon externally applied stretch. Proc Natl Acad Sci U S A 109:12568–12573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cai D, Chen S-C, Prasad M, He L, Wang X, Choesmel-Cadamuro V, Sawyer JK, Danuser G, Montell DJ (2014) Mechanical feedback through E-cadherin promotes direction sensing during collective cell migration. Cell 157:1146–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Austen K, Ringer P, Mehlich A, Chrostek-Grashoff A, Kluger C, Klingner C, Sabass B, Zent R, Rief M, Grashoff C (2015) Extracellular rigidity sensing by talin isoform-specific mechanical linkages. Nat Cell Biol 17:1597–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brenner MD, Zhou R, Conway DE, Lanzano L, Gratton E, Schwartz MA, Ha T (2016) Spider silk peptide is a compact, linear nano-spring ideal for intracellular tension sensing. Nano Lett 16(3):2096–2102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ostlund C, Folker ES, Choi JC, Gomes ER, Gundersen GG, Worman HJ (2009) Dynamics and molecular interactions of linker of nucleo-skeleton and cytoskeleton (LINC) complex proteins. J Cell Sci 122:4099–4108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arsenovic PT, Bathula K, Conway DE (2017) A protocol for using Förster resonance energy transfer (FRET)-force biosensors to measure mechanical forces across the nuclear LINC complex. J Vis Exp. 10.3791/54902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolega J (2004) Phototoxicity and photoinactivation of blebbistatin in UV and visible light. Biochem Biophys Res Commun 320:1020–1025 [DOI] [PubMed] [Google Scholar]
- 24.Arsenovic PT, Ramachandran I, Bathula K, Zhu R, Narang JD, Noll NA, Lemmon CA, Gundersen GG, Conway DE (2016) Nesprin-2G, a component of the nuclear LINC complex, is subject to myosin-dependent tension. Biophys J 110:34–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arsenovic PT, Mayer CR, Conway DE (2017) SensorFRET: a standardless approach to measuring pixel-based spectral bleed-through and FRET efficiency using spectral imaging. Sci Rep 7:15609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kutscheidt S, Zhu R, Antoku S, Luxton GWG, Stagljar I, Fackler OT, Gundersen GG (2014) FHOD1 interaction with nesprin-2G mediates TAN line formation and nuclear movement. Nat Cell Biol 16:708–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang W, Worman HJ, Gundersen GG (2015) Accessorizing and anchoring the LINC complex for multifunctionality. J Cell Biol 208:11–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson MH, Holzbaur ELF (2015) Nesprins anchor kinesin-1 motors to the nucleus to drive nuclear distribution in muscle cells. Development 142:218–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu R, Antoku S, Gundersen GG (2017) Centrifugal displacement of nuclei reveals multiple LINC complex mechanisms for homeostatic nuclear positioning. Curr Biol 27:3097–3110.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khatau SB, Hale CM, Stewart-Hutchinson PJ, Patel MS, Stewart CL, Searson PC, Hodzic D, Wirtz D (2009) A perinuclear actin cap regulates nuclear shape. Proc Natl Acad Sci U S A 106:19017–19022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y, Lovett D, Zhang Q, Neelam S, Kuchibhotla RA, Zhu R, Gundersen GG, Lele TP, Dickinson RB (2015) Moving cell boundaries drive nuclear shaping during cell spreading. Biophys J 109:670–686 [DOI] [PMC free article] [PubMed] [Google Scholar]