Abstract
The interplay of microbiota and the human host is physiologically crucial in health and diseases. The beneficial effects of lactic acid bacteria (LAB), permanently colonizing the human intestine or transiently obtained from food, have been extensively reported. However, the molecular understanding of how LAB modulate human physiology is still limited. G protein-coupled receptors for hydroxycarboxylic acids (HCAR) are regulators of immune functions and energy homeostasis under changing metabolic and dietary conditions. Most mammals have two HCAR (HCA1, HCA2) but humans and other hominids contain a third member (HCA3) in their genomes. A plausible hypothesis why HCA3 function was advantageous in hominid evolution was lacking. Here, we used a combination of evolutionary, analytical and functional methods to unravel the role of HCA3 in vitro and in vivo. The functional studies included different pharmacological assays, analyses of human monocytes and pharmacokinetic measurements in human. We report the discovery of the interaction of D-phenyllactic acid (D-PLA) and the human host through highly potent activation of HCA3. D-PLA is an anti-bacterial metabolite found in high concentrations in LAB-fermented food such as Sauerkraut. We demonstrate that D-PLA from such alimentary sources is well absorbed from the human gut leading to high plasma and urine levels and triggers pertussis toxin-sensitive migration of primary human monocytes in an HCA3-dependent manner. We provide evolutionary, analytical and functional evidence supporting the hypothesis that HCA3 was consolidated in hominids as a new signaling system for LAB-derived metabolites.
Author summary
Although it has been known for 15 years that HCA3 is present in humans and other hominids but absent in all other mammals, no study so far aimed to understand why HCA3 was functionally preserved during evolution. Here, we take advantage of evolutionary analyses which we combine with functional assays of hominid HCA3 orthologs. In search for a reasonable scenario explaining the accumulated amino acid changes in HCA3 of hominids we discovered D-phenyllactic acid (D-PLA), a metabolite produced by lactic acid bacteria (LAB), as the so far most potent agonist specifically activating HCA3. Further, oral ingestion of Sauerkraut, known to contain high levels of D-PLA, caused subsequent plasma concentrations sufficient to activate HCA3. Our data interpreted in an evolutionary context suggests that the availability of a new food repertoire under changed ecological conditions triggered the fixation of HCA3 which took over new functions in hominids.
These findings are particularly important because they unveiled HCA3, which is not only expressed in various immune cells but also adipocytes, lung and skin, as a player that transfers signals of LAB-derived metabolites into a physiological response in humans. This opens up new directions towards the understanding of the versatile beneficial effects of LAB and their metabolites for humans.
Introduction
The interplay of microbiota and the human host is physiologically crucial in health and diseases. Lactic acid bacteria (LAB) are microorganisms present in many foods and the intestine of most mammals. There are extensive reports about the beneficial role of LAB on the immune system [1]. Short-chain fatty acids (SCFAs) and lactate are known metabolites of LAB that have been shown to play an important role in the maintenance of the gut barrier function [2]. SCFAs can induce effects in the host through activation of specific G protein-coupled receptors (GPCRs) expressed in intestinal epithelial cells and immune cells that are located in the intestinal mucosa [3].
The present study focuses on GPCRs belonging to the family of hydroxycarboxylic acid receptors (HCAR) which are regulators of immune functions and energy homeostasis under changing metabolic and dietary conditions. At least two HCAR subtypes are present in mammalian genomes.
HCA1 (formerly GPR81) is activated by lactate and HCA2 (formerly GPR109a, HM74A, PUMA-G) is activated by the ketone body D-3-hydroxybutyrate (D-3HB) but also by the SCFA butyrate. Both receptors mediate anti-lipolytic effects in adipocytes through Gi-protein coupling [4]. Further, HCA2 is known to be expressed in enterocytes, colonocytes and several types of immune cells including neutrophils and macrophages mediating anti-inflammatory effects [3].
A third HCAR subtype, HCA3 (formerly GPR109b, HM74), was recently identified in the human genome but is absent in the mouse genome [5]. The amino acid sequence of the human HCA3 differs from HCA2 in 16 positions and an extended C terminus (Fig 1A). These differences are sufficient to change agonist specificity of HCA3 towards being activated by the fatty acid β-oxidation intermediate 3-hydroxyoctanoate (3HO), but not by D-3HB, likely also mediating anti-lipolytic effects under fasting conditions [6]. Further, aromatic D-amino acids were found to activate HCA3 and elicit chemotactic responses in human neutrophils [7]. However, a plausible hypothesis why HCA3 function is of advantage in humans is currently missing.
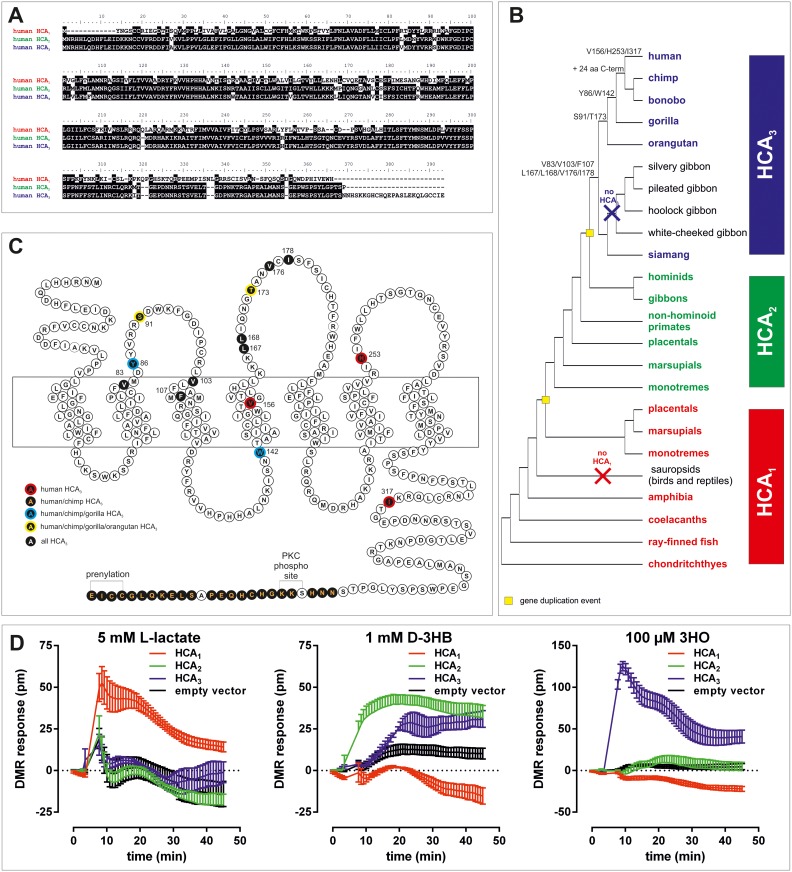
Fig 1. Phylogeny and evolutionary history of HCAR and agonist specificity of human HCA1, HCA2 and HCA3.
(A) Human HCA1 vs HCA2 and human HCA2 vs HCA3 show 50% and 95% amino acid identity, respectively. (B) Schematic HCAR phylogeny in vertebrates indicating the appearance of HCA3-specific amino acid positions and structural features (detailed tree in S1 Fig). (C) Snake-plot of human HCA3 with positions highlighted that are characteristic for HCA3 in comparison to HCA2 orthologs (detailed alignment in S3 Fig). (D) CHO-K1 cells were transiently transfected with human HCA1, HCA2, HCA3, and empty vector (control) and seeded in fibronectin-coated Epic plates. DMR responses were recorded upon stimulation with 5 mM lactate, 1 mM D-3-hydroxybutyrate (D-3HB) and 100 μM of 3 hydroxyoctanoate (3HO). Shown is the agonist-induced shift in pm as mean ± SEM of four independent experiments carried out in triplicates.
Here, we reconstructed the evolutionary history of HCARs and experimentally show that HCA3 is functionally present in humans and all other great apes. After gene duplication and distinct structural changes HCA3 gained the ability to recognize 3HO as an agonist but lost D-3HB specificity. Moreover, we identified D-phenyllactic acid (D-PLA), a metabolite produced by LAB as the so far most potent naturally occurring agonist acting at HCA3. High levels of D-PLA can be found in LAB-fermented products, such as Sauerkraut (S1 Table). LAB fermentation is an ancient process that happened even before humans took advantage of it. It is known that global transitions affected the last common ancestor of early hominoids causing changes in its diet, rendering ingested fruits and leaves more likely to be fermented before ingestion [8]. We provide functional and phylogenetic evidence supporting the hypothesis that increased intake of LAB-fermented food likely posed a positive selective pressure maintaining HCA3 function in hominids. We further hypothesize that HCA3 presence was advantageous in the interplay between ingested and gastrointestinal microbiome and the hominid host by taking over functions in the immune system.
Results
HCA3 arose from a duplication of HCA2 before the split of great apes and gibbons
Feedback regulation of the energy metabolism is vital for organisms exposed to variable dietary supply. Receptors for intermediates of the energy metabolism, such as hydroxycarboxylic acids, already appeared in early vertebrate evolution [9]. Mining of public sequence databases revealed the presence of at least one HCA1 ortholog in the genome of cartilaginous, lobe- and ray-finned fishes, amphibians, and mammals but not in any sauropsidian species (birds, reptiles) (Fig 1B, S1 Fig, S2 Table). HCA2 is present in all mammals and arose from an HCA1 gene duplication in early mammalian evolution (Fig 1B, S2 Fig, S2 Table).
We found that HCA3, the evolutionarily youngest HCAR, is present in the genomes of all great apes and siamang, but absent in all other gibbon genomes investigated so far [10]. Because automated genome assembly can cause problems in assigning highly homologous sequences, we manually reanalyzed the genomic sequence traces for the presence of HCA2 and HCA3 and verified the findings by amplifying, cloning and sequencing HCAR from great apes, siamang, and white-cheeked gibbon. Our analysis showed that HCA3 arose from a duplication of HCA2 before the split of great apes and gibbons (Fig 1B) but underwent pseudogenization in most gibbon species.
Subtypes resulting from gene duplication can have several fates, which include but are not limited to pseudogenization or gain of new function. In the latter case one copy may accumulate mutations and acquire unique functionality without risking the fitness of the organism, which is ensured by the remaining homolog. To test whether the persistence of HCA3 in great apes and some gibbons caused changes in evolutionary constraints of HCAR subtypes we performed Phylogenetic Analysis by Maximum Likelihood (PAML) [11]. Ape HCA1 evolved with an evolutionary rate (ω = 0.189) significantly (p = 0.0258) different compared to all other mammalian HCA1 (ω0 = 0.105) whereas no significant difference in evolutionary constraint of ape HCA2 compared to other mammalian HCA2 orthologs was detected (S3 Table). We further tested whether HCA3 evolved with a different evolutionary rate than HCA1 and HCA2 in species carrying all three HCAR subtypes. We found that HCA3 evolved with a significantly (p = 0.0064) higher evolutionary rate (ω = 0.257) than HCA1 and HCA2 (ω = 0.103), but significantly different from 1 (S3 Table).
Our data indicates that HCA3 is not drifting into neutrality (pseudogenization) in great apes but rather gained new functionality, a hypothesis we further addressed using functional analyses.
Functional analyses of hominoid HCAR reveal evolutionary conservation of known endogenous agonists
To study whether structural differences contribute to the distinct functional properties of the three HCAR we heterologously expressed the human HCA1, HCA2 and HCA3 in CHO-K1 cells and performed functional assays using the dynamic mass redistribution technology (DMR; Corning Epic System). Compared to classical second messenger assays, DMR assays provide the advantage that cellular responses are recorded time-resolved and independently of the activated signaling cascades. Receptor activation is monitored kinetically, thus revealing potential differences in activation kinetics mediated by different agonists.
As reported, lactate only activates HCA1 [12, 13], D-3HB activates HCA2 and with a delayed signal onset also HCA3 [14] and 3HO activates HCA3 but not HCA2 (Fig 1D) [6]. All HCAR are Gi protein-coupled receptors, i.e. activation of the receptor by an agonist leads to inhibition of adenylyl cyclases, thus resulting in a decrease in intracellular cAMP levels. Concentration-response curves of different agonists can be determined using cAMP inhibition assays. This extends the functional characterization since it reveals information about constitutive HCAR activity (basal cAMP) and allows the quantification of the agonistic activity at the respective HCAR by determination of EC50 and Emax values. EC50 values reflect the potency of an agonist, i.e. the concentration of an agonist required to produce 50% of its maximal effect; the lower the EC50, the higher the potency of an agonist. Emax (efficacy) is the maximum effect induced by the agonist, i.e. when Emax is reached increasing the agonist concentration will not produce a greater magnitude of the effect.
We performed cAMP inhibition assays on all hominoid HCARs as well as mouse HCA1 and HCA2 and analyzed them with the already established endogenous agonists lactate, D-3HB and 3HO (Table 1, Fig 2). Considering the observed differences in expression levels (Table 2), we detected no significant differences in Emax and EC50 values upon stimulation with the respective endogenous agonists when comparing within HCA1, HCA2, and HCA3 orthologs (Table 1). However, the fact that D-3HB stimulates HCA3 orthologs (Figs 1D, 2A and 2C) to some extent supports its evolutionary origination from HCA2. In contrast, 3HO had no activity on HCA2 orthologs, indicating a gain of functionality in great ape HCA3 after gene duplication (Figs 1D, 2D and 2E).
Table 1. Functional characterization of HCA1, HCA2, and HCA3 orthologs.
CHO-K1 cells were transfected with receptor constructs and cAMP accumulation was determined. The basal cAMP level of mock-transfected CHO-K1 stimulated with 2 μM forskolin (control) was 41.1 ± 1.9 nmol/well in DMEM and 13.8 ± 1.3 nmol/well in HBSS and set 100%. Analyses of HCA1 were carried out in DMEM. HCA2 and HCA3 were assayed in HBSS. Emax values are shown as % of mock-transfected CHO-K1 stimulated with 2 μM forskolin in absence of agonist. Emax and EC50 values were determined from concentration-response curves using GraphPad Prism. A general explanation of EC50 and Emax values is provided in the results section Functional hominoid HCAR analyses with known endogenous agonists. Data is given as mean ± SEM (basal and Emax) and geometric mean with 95% confidence interval (EC50) of at least three independent experiments each performed in triplicates.
| HCA1 | HCA2 | HCA3 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Basal cAMP (% of mock-transfected) |
L-lactate | Basal cAMP (% of mock-transfected) |
D-3HB | Basal cAMP (% of mock-transfected) |
3HO | ||||
| Emax (% of mock-transfected w/o agonist) |
EC50 (mM) (95% CI) |
Emax (% of mock-transfected w/o agonist) |
EC50 (mM) (95% CI) |
Emax (% of mock-transfected w/o agonist) |
EC50 (μM) (95% CI) |
||||
| human | 66.2 ± 5.2 | 31.1 ± 2.8 | 2.4 (1.1–5.1) | 260.5 ± 11.8 | 48.5 ± 6.9 | 0.53 (0.38–0.74) | 56.3 ±1.5 | 22.2 ± 2.6 | 1.6 (0.91–2.95) |
| chimpanzee | 73.8 ± 4.5 | 40.1 ± 2.0 | 4.1 (3.7–4.6) | 239.9 ± 13.9 | 61.3 ± 3.7 | 0.77 (0.55–1.1) | 51.5 ± 1.6 | 28.2 ± 1.0 | 3.7 (2.2–6.1) |
| gorilla | 87.1 ± 5.6 | 36.7 ± 5.7 | 2.3 (1.6–3.3) | 304.3 ± 18.5 | 69.5 ± 2.6 | 0.51 (0.37–0.70) | 53.4 ± 1.8 | 19.7 ± 1.0 | 2.4 (2.0–3.0) |
| orangutan | 61.2 ± 1.8 | 35.6 ± 3.7 | 4.6 (3.7–5.8) | 200.7 ± 3.9 | 65.7 ± 8.8 | 0.68 (0.60–0.78) | 58.0 ± 1.6 | 31.2 ± 3.6 | 7.4 (6.9–7.9) |
| siamang | 78.2 ± 1.0 | 32.1 ± 2.3 | 5.1 (4.9–5.3) | 138.1 ± 6.0 | 57.3 ± 4.9 | 1.02 (0.74–1.41) | 55.2 ± 1.6 | 24.2 ± 1.5 | 2.4 (1.2–4.6) |
| white-cheeked gibbon | 101.9 ± 4.7 | 28.2 ± 2.6 | 2.5 (2.0–3.1) | 200.1 ± 14.3 | 50.0 ± 7.8 | 0.45 (0.31–0.65) | - | - | - |
| mouse | 97.5 ± 0.6 | 37.1 ± 3.9 | 2.9 (2.2–3.8) | 206.7 ± 4.5 | 57.3 ± 9.2 | 1.03 (0.69–1.54) | - | - | - |
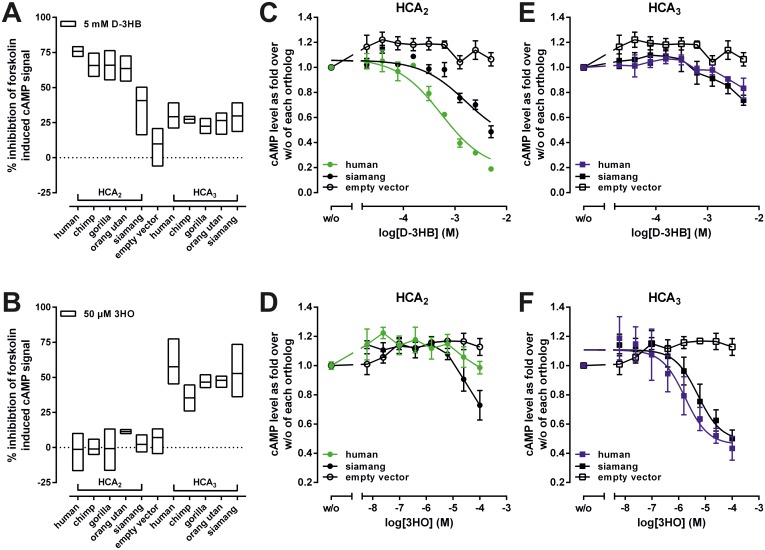
Fig 2. Functional characterization of ape HCA2 and HCA3 orthologs.
CHO-K1 cells were transiently transfected with receptor constructs and agonist-induced inhibition of forskolin-induced cAMP accumulation was determined. HCA2 and HCA3 orthologs were stimulated with (A) 5 mM D-3-hydroxybutyrate (D-3HB) and (B) 50 μM 3-hydroxyoctanoate (3HO). (A, B) Data is shown as percent inhibition of the forskolin-induced cAMP signal (min to max bars with line at mean). Concentration-response curves upon stimulation of human and siamang HCA2 (C, D) and HCA3 (E, F) with D-3HB (C, E), and 3HO (D, F) are shown as fold over forskolin-stimulated cAMP level in the absence of agonist of each ortholog. Human HCA2 is specificall activated by D-3HB (C), but not the HCA3 agonist 3HO (D). In contrast, siamang HCA2 is activated by D-3HB with lower potency when compared to the human ortholog (C), but also by 3HO to some extent (D). Data is given as mean ± SEM of at least three independent experiments each carried out in triplicates.
Table 2. Total and cell surface expression levels of HCA1, HCA2 and HCA3 orthologs as determined by ELISA.
Cell surface expression levels of mammalian HCAR orthologs were measured by a cell surface ELISA. Specific optical density (OD) readings are given as percentage of the respective HA-tagged human HCAR. The non-specific OD value (empty vector) was 0.014 ± 0.002 (set 0%). The OD value of the HA-tagged human HCA1 (0.091 ± 0.012), of the HA-tagged human HCA2 (0.755 ± 0.007) and of the HA-tagged human HCA3 (0.424 ± 0.038), each of which was set 100% to compare expression between orthologs. Total expression levels of mammalian HCAR orthologs were measured by a sandwich ELISA. Specific optical density (OD) readings are given as a percentage of HA-tagged respective human HCAR. The non-specific OD value (empty vector) was 0.011 ± 0.001 (set 0%). The OD value of the HA-tagged human HCA1 (0.057 ± 0.012), of the HA-tagged human HCA2 (0.591 ± 0.114) and of the HA-tagged human HCA3 (0.092 ± 0.006), each of which was set 100%. Data is given as mean ± SEM of three independent experiments carried out in triplicates.
| HCA1 | HCA2 | HCA3 | ||||
|---|---|---|---|---|---|---|
| cell surface expression (% of human HCA1) |
total expression (% of human HCA1) |
cell surface expression (% of human HCA2) |
total expression (% of human HCA2) |
cell surface expression (% of human HCA3) |
total expression (% of human HCA3) |
|
| human | 100 | 100 | 100 | 100 | 100 | 100 |
| chimp | 69 ± 3 | 87 ± 9 | 83 ± 4 | 57 ± 4 | 68 ± 5 | 68 ± 9 |
| gorilla | 59 ± 4 | 76 ± 3 | 81 ± 2 | 56 ± 3 | 138 ± 11 | 138 ± 12 |
| orang utan | 98 ± 10 | 41 ± 10 | 60 ± 9 | 22 ± 4 | 141 ± 15 | 126 ± 6 |
| siamang | 110 ± 3 | 91 ± 9 | 26 ± 2 | 20 ± 3 | 95 ± 6 | 106 ± 15 |
| white-cheeked gibbon | 225 ± 34 | 113 ± 11 | 60 ± 6 | 29 ± 2 | - | - |
| mouse | 185 ± 19 | 139 ± 21 | 48 ± 1 | 19 ± 1 | - | - |
D-3HB activates siamang HCA2 with about 3-5-fold lower potency when compared to the human ortholog (Fig 2B) and siamang HCA2 is activated by 3HO at high concentrations (Fig 2E). These findings and the fact that HCA3 is only present in siamang but not in any other gibbon species initiated further PAML analyses.
Evolutionary analyses suggest a loss of constraint on siamang HCA2 and HCA3
We found that HCA2 and HCA3 of siamang evolve with an ω that is not significantly different from 1 (S3 Table). This indicates a loss of constraint on siamang HCA2 and HCA3. Further, when compared within all other available gibbon HCA1 and HCA2 orthologs, we found that siamang HCA1 evolves under purifying selection, but indeed siamang HCA2 exhibits an ω (ω = 0.643), not significantly different from 1. Thus, we found that, in contrast to all other HCAR orthologs, the agonist profiles of siamang HCA2 and HCA3 are less distinguishable and therefore less conserved.
In sum, our combined evolutionary and functional analyses support a loss of evolutionary constraint on the siamang HCA2 and HCA3 orthologs (Fig 2, S3 Table).
Aromatic D-amino acids activate human HCA3 with highest potency
Sequence alignment of HCA3 orthologs revealed that from all amino acid positions which differ from HCA2 seven amino acid positions are conserved in all HCA3 orthologs (Fig 1B, 1C and S3 Fig). Further, we found that Ser91 and Thr173 (referring to positions in human HCA3 NP_006009.2) are great ape-specific HCA3 positions, while Tyr86 and Trp142 are only conserved in human, chimpanzee, bonobo, and gorilla HCA3 (Fig 1B, 1C and S3 Fig).
Since 3HO activated all hominoid HCA3 orthologs with comparable potency, we next analyzed whether potencies of the HCA3 agonists D-phenylalanine (D-Phe) and D-tryptophan (D-Trp) [7], vary between orthologs. We confirmed HCA3-specificity of both agonists (S4 Fig) and found that both, D-Phe and D-Trp, activate human HCA3 with highest potency when compared to all other ape orthologs (Table 3, S4 Fig). Although Irukayama-Tomobe et. al showed both aromatic D-amino acids to be HCA3-specific agonists, they did not identify relevant sources of D-Phe and D-Trp to put their findings in a physiological context [7]. By comprehensive research of literature, we here draw the link to fermented foods and beverages which are a likely source for D-Phe and D-Trp in concentrations sufficiently high to activate HCA3. Several classes of bacteria produce and secrete aromatic D-amino acids (e.g. Acetobacter, Bifidobacterium, Brevibacterium, Lactobacillus, Micrococcus, Propionibacterium, Streptococcus) [15–17] and for all of them residence in the human gastrointestinal tract has been demonstrated [18]. Moreover, D-amino acids were found to be present in body fluids and certain tissues in the μM range [19–22]. Connecting this previously established knowledge led us to further investigate whether other microbial metabolites activate HCA3.
Table 3. Functional characterization of HCA3 orthologs stimulated with aromatic D-amino acids and their metabolites.
CHO-K1 cells were transfected with receptor constructs and cAMP accumulation was determined. The basal cAMP level of mock-transfected CHO-K1 stimulated with 2 μM forskolin (control) in HBSS was 13.8 ± 1.3 nmol/well and set 100%. Emax values are referred to % of mock-transfected CHO-K1 stimulated with 2 μM forskolin in absence of agonist. Emax and EC50 values were determined from concentration-response curves of agonists using GraphPad Prism. A general explanation of EC50 and Emax values is provided in the results section Functional hominoid HCAR analyses with known endogenous agonists. Data is given as mean ± SEM (Emax) and geometric mean with 95% confidence interval (EC50) of at least three independent experiments each performed in triplicates.
| Emax (% of mock-transfected w/o agonist) |
EC50 (μM) (95% CI) |
Emax (% of mock-transfected w/o agonist) |
EC50 (μM) (95% CI) |
Emax (% of mock-transfected w/o agonist) |
EC50 (μM) (95% CI) |
|
|---|---|---|---|---|---|---|
| D-Phe | D-Trp | 3HDec | ||||
| human HCA3 | 21.0 ± 2.3 | 36.1 (17.7–73.5) | 14.7 ± 1.0 | 20.5 (11.7–36.2) | 18.7 ± 2.3 | 31.9 (22.3–45.8) |
| chimpanzee HCA3 | 23.4 ± 2.4 | 37.4 (23.2–60.2) | 22.2 ± 2.2 | 62.1 (47.6–81.0) | 25.7 ± 1.2 | 141(97.5–205) |
| gorilla HCA3 | 17.9 ± 2.3 | 45.7 (27.1–77.1) | 17.8 ±1.5 | 24.4 (23.2–25.8) | 25.8 ± 2.7 | 37.3 (23.0–60.3) |
| orangutan HCA3 | 24.6 ± 2.4 | 129 (103–161) | 24.7 ± 1.5 | 50.7 (30.8–83.4) | 28.4 ± 3.1 | 62.7 (39.8–98.7) |
| siamang HCA3 | 22.9 ± 5.2 | 202 (101–406) | 24.3 ± 1.2 | 56.7 (38.8–82.5) | 35.6 ± 1.0 | 102 (59.9–175) |
| D-PLA | L-PLA | ILA | ||||
| human HCA3 | 20.5 ± 2.8 | 0.15 (0.05–0.42) | 33.4 ± 2.9 | 5.2 (3.0–9.1) | 32.9 ± 2.9 | 0.18 (0.08–0.39) |
| chimpanzee HCA3 | 23.7 ± 1.8 | 0.13 (0.08–0. 22) | 25.4 ± 1.1 | 4.0 (2.4–6.5) | 32.6 ± 4.9 | 0.23 (0.16–0.33) |
| gorilla HCA3 | 19.9 ± 1.4 | 0.085 (0.032–0.21) | 36.5 ±3.0 | 6.2 (4.3–8.9) | 36.3 ±1.7 | 0.073 (0.053–0.10) |
| orangutan HCA3 | 29.3 ± 3.3 | 1.2 (0.60–2.2) | 39.3 ± 3.2 | 6.5 (3.1–13.7) | 42.0± 1.0 | 0.068 (0.030–0.15) |
| siamang HCA3 | 20.4 ± 1.7 | 0.28 (0.14–0.56) | 36.3 ± 3.5 | 17.2 (8.9–33.6) | 33.6 ± 1.2 | 0.17 (0.11–0.28) |
Lactic acid bacteria derived metabolites are highly potent agonists at HCA3
LAB are known to produce metabolites structurally related to 3HO and D-Phe, such as 3-hydroxydecanoate (3HDec) and D-PLA, respectively [23, 24]. PLA has been detected in μM concentrations in fermented food such as Sauerkraut for which remarkably stable microbial associations over time and region have been shown (Literature summarized in S1 Table) [25]. Moreover, LAB represent 0.01–1.8% of the total bacterial community found in the human intestine where some of them are colonizers and others are passengers [26]. Additional support for the link between LAB and D-PLA is provided by analyses of patients with short bowel syndrome (SBS) with their microbiota known to be imbalanced to Lactobacillus (L. mucosae, L. acidophilus, L. fermentum) [27, 28] as the major resident bacteria (increased from ≤ 1% up to 60% of the fecal flora). These patients exhibit highly increased urinary levels of D-PLA [29–33].
To our knowledge, the present study is the first in which D-PLA, L-phenyllactic acid (L-PLA) and indole 3-lactic acid (ILA), a metabolite derived from Trp metabolism, were functionally tested for their agonistic activity at the human HCA3. Using the DMR technology we found that all three metabolites activate the human HCA3, but not HCA1 and HCA2 (Fig 3A). D-PLA and ILA (each 1 μM) induced a response comparable to that of 100 μM 3HO, the known endogenous agonist indicating an about 100-fold higher potency of the LAB-derived compounds (Figs 1D and 3A).
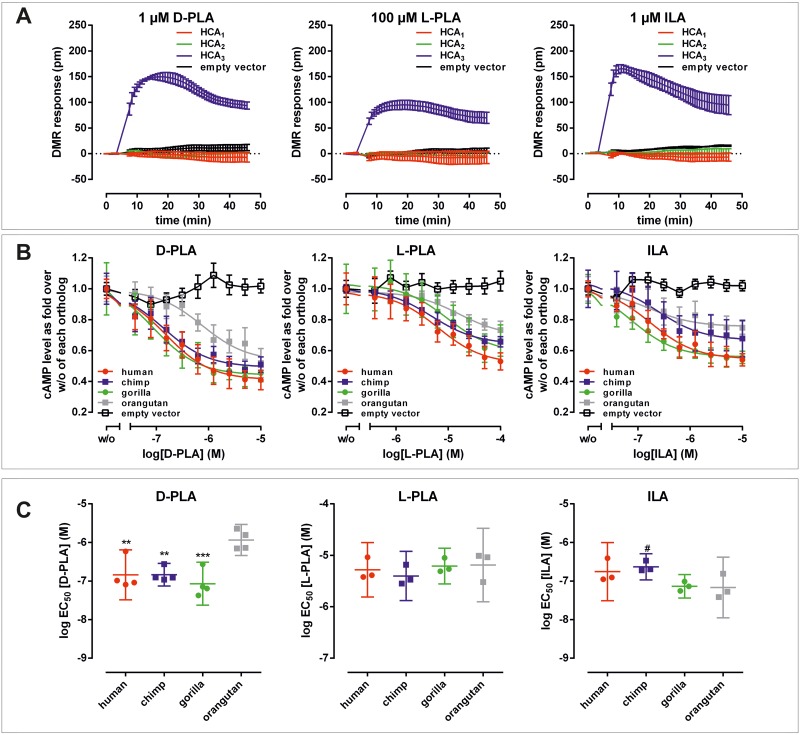
Fig 3. D-Phenyllactic acid is a potent agonist acting at HCA3.
(A) CHO-K1 cells were transiently transfected with human HCA1, HCA2, HCA3, and empty vector (control), seeded in fibronectin-coated Epic plates and DMR responses were recorded upon stimulation with 1 μM D-phenyllactic acid (D-PLA), 100 μM L-phenyllactic acid (L-PLA) and 1 μM indole lactic acid (ILA) to reveal potential differences in receptor activation kinetics. Shown is the agonist-induced shift in pm of three independent experiments carried out in triplicates. (B) The agonist-induced inhibition of forskolin-induced cAMP accumulation was determined in CHO-K1 cells transiently transfected with HCA3 orthologs and empty vector. Concentration-response curves upon stimulation with D-PLA, L-PLA and ILA are depicted as fold over over forskolin-stimulated cAMP level in the absence of agonist of each ortholog. Data is given as mean ± SEM of at least three independent experiments each carried out in triplicates. A general explanation of EC50 values is provided in the results section Functional hominoid HCAR analyses with known endogenous agonists. (C) log EC50 values (corresponding to EC50 values shown in Table 3) are plotted as mean ± 95% confidence interval. Statistical analyses were performed using an ordinary One-Way ANOVA (Dunnett’s multiple comparisons test) testing against orang utan HCA3. # ≤ 0.1, * P ≤ 0.05; ** P ≤ 0.01; *** P ≤ 0.001.
Next, we functionally analyzed all HCA3 orthologs with the newly identified agonists using cAMP inhibition assays to determine their potencies (Table 3). Unfortunately, the individual potencies of D- and L-ILA could not be determined since only the DL isomeric mixture was commercially available. L-PLA activates HCA3 orthologs in the μM range. However, D-PLA activates HCA3 with an EC50 value of 150 nM, thus being an about 35-fold more potent agonist than the L-enantiomer and about 10-fold and 240-fold more potent than the known agonists 3HO and D-Phe, respectively (Tables 1 and 3).
Further, both D-PLA and D-Phe exhibit higher potencies at the HCA3 of human, chimpanzee and gorilla when compared to the orangutan ortholog (Fig 3B, 3C and S4 Fig). In contrast, the more hydrophobic agonists 3HDec, D-Trp and ILA are less potent at the chimpanzee HCA3 ortholog when compared to the other great ape HCA3 orthologs (Fig 3B, 3C and S4 Fig). The Trp metabolite ILA acted at human HCA3 with a potency comparable to that of D-PLA. Thus, D-PLA is the most potent agonist at human HCA3 for which sufficient presence in LAB-fermented food has been previously described (S1 Table).
D-lactate dehydrogenase is abundant in the human gut microbiome
Accumulating evidence suggests that only a small number of LAB species are true inhabitants of the human intestinal tract. The majority of LAB is derived from fermented food, the oral cavity or more proximal parts of the gastrointestinal tract, like the esophagus [34]. However, we asked whether the gene for D-lactate dehydrogenase (EC 1.1.1.28), the enzyme that has been shown to produce D-PLA [35–37], is present in the human microbiome.
We found that genes encoding D-lactate dehydrogenase are very common among bacteria and are also found in some of the very common and abundant gut bacteria (S5 Fig, S4 Table). However, the question remains, if these D-lactate dehydrogenases are capable of converting the aromatic amino acids to the respective lactic acid in the human intestine. Even if this ability is restricted to LAB possessing this gene, they are still detectable to some degree in the human intestinal microbiome [38].
Oral uptake of pure or Sauerkraut-derived D-PLA results in increased plasma and urine levels
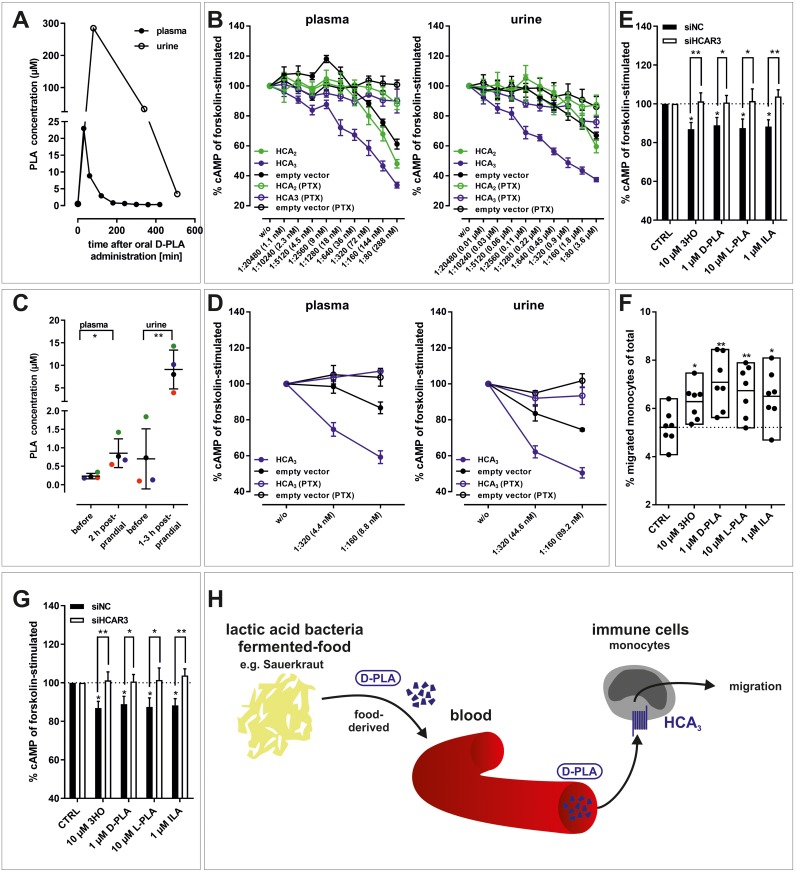
To further substantiate D-PLA resorption from gut and its urinary elimination, a pharmacokinetic study was performed determining PLA levels using Liquid Chromatography Mass Spectrometry (LC-MS) in human plasma and urine samples. As shown in Fig 4A, PLA plasma levels are increased already 30 minutes after oral application of 100 mg D-PLA and subsequently decline rapidly due to renal excretion as determined in concomitantly collected urine samples. PLA plasma levels reached concentrations (>20 μM) capable to maximally activate the human HCA3 (Fig 3B). To verify this, we tested whether PLA-containing plasma and urine samples elicit an HCA3-specific, pertussis toxin (PTX) sensitive decline in intracellular cAMP levels of transiently transfected CHO-K1 cells (Fig 4B, S5 Table). Thus, plasma and urine samples were diluted and assayed by addition to transfected CHO-K1 cells. Indeed, we found that PLA containing urine and to a lesser extend plasma activated specifically and concentration-dependent HCA3 transfected cells (Fig 4B, S5 Table).
Fig 4. D-PLA is absorbed from the human gut, reaches μM plasma concentrations and activates HCA3 in human monocytes.
(A) Upon oral ingestion of 100 mg D-PLA (one individual) plasma and urine PLA levels were measured using LC-MS. Details about experimental setup are stated in Materials and Methods, section Determination of PLA in human plasma and urine. (B) PLA containing plasma (23 μM, corresponding to the 30 min time point from (A)) and urine (285 μM, corresponding to the 80 min time point from (A)) were stepwise 1:2-diluted and tested in cAMP inhibition assays. (C) Upon oral uptake of 5–6 g Sauerkraut per kg body weight (n = 4 individuals, each individual labeled with a different color), plasma and urine PLA levels were measured using LC-MS. Details about experimental setup are stated in Materials and Methods, section Determination of PLA in human plasma and urine. (D) PLA containing plasma (1.4 μM, 2 h postprandial) and urine (14.3 μM, 3 h postprandial) were stepwise 1:2-diluted and tested in cAMP inhibition assays. (B, D) Data is shown as fold over unstimulated cAMP level, given as mean ± SEM (n = 3) and is summarized in S5 Table. (E) 3HO, D-PLA, L-PLA, and ILA induced a pertussis-toxin (PTX) sensitive reduction in cAMP levels and (F) migratory responses in human monocytes. (E) Data is given as percent of cAMP level in monocytes stimulated with 10 μM forskolin without agonist. The mean ± SEM (n = 3 different donors) is shown. (F) Data is shown as percent migrated monocytes of total monocytes (min to max bars with line at mean, n = 7 different donors). (G) siRNA-mediated knock-down of HCA3 in human monocytes diminished the agonist-induced reduction of cAMP levels still present in negative control siRNA (siNC) transfected monocytes. Data is shown as mean ± SEM (n = 7 different donors). (H) D-PLA is absorbed from ingested LAB-fermented food (Sauerkraut) and induces HCA3-dependent migration in human monocytes. This opens up new perspectives to study the role of HCA3 activation by LAB-derived metabolites in both immune function and energy homeostasis. (E, G) Paired two-tailed t-tests were performed to analyze the effect of PTX and siRNA transfection. (E, F, G) Statistical analyses were performed using an ordinary One-Way ANOVA (Dunnett’s multiple comparisons test) testing against control (CTRL, vehicle EtOH). # ≤ 0.1, * P ≤ 0.05; ** P ≤ 0.01; *** P ≤ 0.001.
Interestingly, we also found basal plasma levels of PLA of about 0.4 μM before oral D-PLA administration (Fig 4A, S6 Fig). Our established LC-MS measurement method does not discriminate between L-PLA and D-PLA. Blood levels of L-PLA in the μM range are only found in patients with phenylketonuria [39]. L-PLA is 35-fold less potent than D-PLA at HCA3 (Table 3) and plasma samples from basal conditions (0.4 μM) were sufficient to inhibit forskolin-induced cAMP formation via HCA3 (S5 Table), potentially due to presence of D-PLA derived from alimentary microbiota sources.
Sauerkraut is known to contain high levels of LAB as well as D-PLA. A recent study showed that Sauerkraut exhibits remarkably stable microbial associations over time and regions [40]. Therefore, we asked whether ingestion of Sauerkraut (5–6 g per kg body weight) can cause an increase in plasma and urinary D-PLA levels. Using LC-MS we found that levels of PLA in plasma (before = 0.2 μM; after = 0.9 μM) and urine (before = 0.7 μM; after = 9.1 μM) are increased 2 h postprandial in four individuals (Fig 4C, S6A Fig). Further, we found that plasma and urine containing increased concentrations of PLA after Sauerkraut ingestion induced an HCA3-specific, PTX-sensitive inhibition of intracellular cAMP levels (Fig 4D, S6B Fig, S5 Table). This clearly indicates that fermented food, such as Sauerkraut, can be a source of D-PLA leading to functionally relevant concentrations of this bacterial metabolite in humans.
mRNA expression of human HCA3 is highest in immune cells
Until recently, mRNA expression datasets based on microarrays constituted the main source for expression data. However, due to their high nucleotide sequence similarity (97%) these datasets did not enable the distinction between human HCA2 and HCA3. High quality, paired-end, RNA-Sequencing datasets are in principle suitable to distinguish between those highly similar sequences. Indeed, public resources provide quantitative expression data (FPKM: fragments per kilobase million, TPM: transcripts per kilobase million) for both HCA2 and HCA3. Quantification of transcripts in RNA-Sequencing projects is based on counting small RNA fragments mapped to a gene (read lengths 70–100 bp). Because long nucleotide sequence parts of HCA2 and HCA3 transcripts are identical it was not clear whether the used bioinformatic tools are precise enough to properly distinguish and quantify human HCA2 and HCA3 transcripts. Therefore, we manually reanalyzed a publicly available dataset (Bioproject: PRJNA326727) (S6 Table, S7 Fig). We mapped all relevant reads to HCA2 or HCA3 and excluded those matching to both receptors. We found no significant differences between the provided FPKM values and our manually curated read counts (S7 Fig).
Mining of publicly available RNA-Sequencing data reanalyzed and provided as TPM values revealed highest expression of HCA3 in immune cells such as neutrophils and monocytes and a pattern distinct from that of HCA2 (S7 Fig, S7 Table). Expression of HCA3 mRNA, especially in adipose tissue, skin and lung, is considerably lower compared to HCA2 (S7 Fig). However, both receptors are up-regulated in ulcerative colitis and inflammatory bowel disease patients compared to controls (S6 Table, S7 Fig).
LAB-derived metabolites induce a chemotactic response in human monocytes through activation of HCA3
As described above, HCA3 is expressed in a variety of human immune cells including macrophages, neutrophils, and monocytes (S7 Table). First, we used freshly isolated human peripheral blood mononuclear cells (PBMCs), which include lymphocytes (T cells, B cells, and NK cells), monocytes, and dendritic cells. We observed a significant reduction in cAMP levels for all HCA3 agonists except of 10 μM L-PLA in PBMCs (S6C Fig). However, PTX sensitivity of this reduction could only be observed for 3HO and D-PLA, but not the other HCA3 agonists (S6C Fig). We assume that this is likely due to inter-individual variation in cell counts of the different cell types in PBMCs. Thus, we isolated human monocytes and performed cAMP inhibition assays. We found that 10 μM 3HO as well as 1 μM D-PLA and ILA induced a decrease of cAMP in human monocytes that can be blocked by PTX (Fig 4E). Using a transwell migration assay we showed that D-PLA can trigger chemotactic responses with high potency in isolated monocytes (Fig 4F). Both, a PTX-sensitive reduction in cAMP levels and migratory responses were also observed for D-Phe and D-Trp with freshly isolated human monocytes, however, 1000-fold higher concentrations were required (S6D Fig).
Finally, we performed siRNA-mediated knock-down of HCA3 in freshly isolated human monocytes (S6E Fig) and determined cAMP levels upon stimulation with 3HO and D-PLA in comparison to scrambled negative control siRNA (siNC)-transfected cells (Fig 4G). In contrast to control (siNC)-transfected monocytes the reduction in intracellular cAMP levels was abolished in siHCAR3-transfected monocytes demonstrating HCA3 specificity of the D-PLA induced effect (Fig 4G).
Discussion
A recent paleogenetic study provides a comprehensive summary of existing evidence for large-scale ecological transitions occurring during hominid evolution that caused changes of the habitat accompanied by changes in diet and the microbial environment [8]. The exposure to an altered microbial environment most likely posed a selective pressure and required host adaptation but may also have provided the advantage to access new ecological niches.
In the present study, we provide evidence for such a unique example, where genetic events potentially improved the availability of a new food repertoire under changed ecological conditions that possibly triggered the fixation of a duplicated gene with new functions in hominids.
Through analyses of the evolutionary history of the HCA receptor family, we now show that HCA3, being absent in non-hominoid primates and all other vertebrates, resulted from a gene duplication that occurred before the split of gibbons from great apes (Fig 1B). In apes, we found gradual fixation of amino acid positions in HCA3 (Fig 1C). We discovered new LAB-derived agonists acting at all ape HCA3 orthologs. Both, D-PLA and ILA, activate human HCA3 with an about 10-fold higher potency when compared to the endogenous agonist 3HO (Tables 1 and 3). Further, our functional analyses of great ape HCA3 orthologs revealed, that potencies of D-PLA are significantly higher in human, chimpanzee and gorilla compared to orangutan (Fig 4C). This led us to do an extensive literature search to identify potential global environmental changes that conceivably coincided with dietary changes and are archaeologically accepted to have occurred when the last common ancestor of human, chimpanzee and gorilla lived on earth. As mentioned at the beginning of the discussion, a recent study showed that about 10 million years, ago a mutation in the class IV alcohol dehydrogenase appeared in the common ancestor that humans share with gorillas and chimpanzees, which allowed an increased tolerance of alcohol [8]. In this study, a scenario was projected in which the large-scale ecological transitions caused the common ancestor to move out of the trees and lead a more terrestrial life [8]. Carrigan et al. conclude that this new life style rendered fruit picking directly from the trees to be less likely but increased the probability that food that had started to ferment was collected from the ground [8]. This is suggested to have posed the selective pressure that increased the tolerance to dietary alcohol before human-directed fermentation [8]. In this context, not only the increased activity of ethanol-metabolizing enzymes (e.g. ADH4) might have provided a selective advantage but also an immune system that can sense high levels of D-PLA ingested with lacto-fermented food (including plants, fish, meats, milk) [35]. Our functional data combined with the described archaeologically documented evolutionary context gives rise to the hypothesis that in the last common ancestor of human, chimpanzee and gorilla, increased ingestion of fermented food was likely associated with increased ingestion of LAB accompanied by increased levels of D-PLA. D-PLA is a metabolite, produced in high concentrations by LAB in the process of lactic fermentation and has a broad antimicrobial activity against bacteria and fungi [35].
We demonstrate, using LC-MS that D-PLA is quickly absorbed from the gastrointestinal tract upon ingestion of LAB-fermented Sauerkraut, can reach physiologically relevant plasma concentrations and is renally eliminated in humans (Fig 4A and 4C). These diet-induced PLA plasma concentrations are sufficient to activate HCA3 in human monocytes and potentially modulate human immune functions (Fig 4E–4G).
The beneficial effects of LAB-fermented food on human health are numerous, ranging from protection of the intestinal barrier, limitation of inflammatory cytokine production to improved fasting insulin levels and glucose turnover rates among many others [41]. HCA3 is expressed in many different immune cells but also adipose tissue (S7 Table). We hypothesize that the LAB-mediated, HCA3-dependent physiological impact likely extends to influences on the human host energy storage. This would also be plausible in an evolutionary context, since it is known that the nutritional and functional properties of food are enhanced by LAB-fermentation due to the transformation of substrates and the formation of bioactive or bioavailable end-products [25].
In summary, our work provides multifaceted evidence supporting the hypothesis that HCA3 evolved as a GPCR activated by LAB metabolites. Our data suggests that this might have improved tolerance to the increased ingestion of LAB in the last common ancestor of human, chimpanzee and gorilla. As LAB-derived D-PLA is specifically recognized by HCA3 expressed in monocytes (Fig 4H) and numerous studies describe increased hypo-responsiveness of the immune system through anti-inflammatory processes for D-PLA and LAB (S1 Table), we speculate that HCA3 plays a role in mediating at least some of those effects (Fig 4H). In the present study, we identified a new set of microbial-derived metabolites acting as classical signaling molecules in the human host through recognition by a specific receptor. The question remains how D-PLA affects monocyte functions. Future studies shall address whether HCA3 activation by D-PLA impacts phagocytic capacity of monocytes or influences differentiation of monocytes to macrophages. Does D-PLA, through activation of HCA3, prime monocytes to either increase of pro-inflammatory host response to concomitantly ingested pathogenic bacteria or reduction of pro-inflammatory response to LAB? At last, our study opens-up the interesting question of how D-PLA ingested with LAB-fermented food can influence energy storage in HCA3-expressing adipocytes.
Materials and methods
Ethics statement
The studies on humans and with human materials were conducted in accordance with the Declaration of Helsinki and with the recommendations of “Ethik-Kommission an der Medizinischen Fakultät der Universität Leipzig” with written informed consent from all blood donors. The protocol was approved by the aforementioned committee (313/14-ek).
HCAR ortholog identification
Mining of NCBI trace archives
HCAR sequences of various mammalian species were obtained from extensive database mining followed by assembly, analysis and manual proof-reading. Trace identifier, NCBI accession or SRA accession numbers are listed in S2 Table. All newly obtained HCAR sequences have been deposited in the GenBank database: accession no. KU285431-KU285452, as listed in S2 Table.
Amplification, sequencing and cloning of HCAR orthologs from apes and mouse
To analyze the sequence of HCAR orthologs, genomic DNA samples were prepared from tissue of various species (sources are given in S8 Table). Tissue samples were digested in lysis buffer (50 mM Tris/HCl, pH 7.5, 100 mM EDTA, 100 mM NaCl, 1% SDS, 0.5 mg/ml proteinase K) and incubated at 55°C for 18 h. DNA was purified by phenol/chloroform extraction and ethanol precipitation. Degenerated primer pairs (S9 Table) were used to amplify HCAR-specific sequences. Primer pairs, positioned in the 5’- and 3’- UTR, were designed to simultaneously amplify HCA2 and HCA3. PCR reactions were performed with Taq and Pfu polymerase under variable annealing and elongation conditions. A standard PCR reaction (50 μl) contained genomic DNA (100 ng) with primers (1.5 μM each), ThermoPol reaction buffer (1x), dNTP (250 μM, each) as well as Taq and Pfu polymerase (1 U, NEB, Frankfurt am Main, Germany). The reactions were initiated with denaturation at 95 °C for 1 min, followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at 55 °C for 30 s and elongation at 72 °C for 1 min. A final extension step was performed at 72 °C for 10 min. Specific PCR products were subcloned into the pCR2.1-TOPO vector (Invitrogen, Paisley, UK) and sent to Seqlab for sequencing. All newly obtained HCAR sequences have been deposited in the GenBank database (accession no. KU285431-KU285452, S2 Table). Protein sequence alignments are depicted in S3 Fig.
The full-length HCAR were inserted into the mammalian expression vector pcDps [42] and epitope-tagged with an N-terminal hemagglutinin (HA) epitope and a C-terminal FLAG-tag by a PCR-based overlapping fragment approach. Identity of all constructs and correctness of all PCR-derived sequences were confirmed by restriction analysis and sequencing.
Sequence alignments and PAML analyses
HCA1, HCA2 and HCA3 of 54 selected vertebrate species were aligned to infer the phylogenetic model of HCAR evolution as shown in Fig 1 (S1 Fig). Mammalian HCA1 and HCA2 were aligned to infer a phylogenetic tree of 88 mammalian species. All nucleotide alignments were generated with the ClustalW algorithm (Bioedit Sequence Alignment Editor 7.0.9 [43]) followed by manual trimming where gaps were deleted. Phylogenetic evolutionary history was inferred by the Maximum Likelihood method based on the General Time Reversible model using MEGA6 [44]. The resulting tree (S2 Fig) with a 75% cut-off value was used for PAML analyses. Tests of selection (ω = dN/dS) were accomplished by maximum likelihood using a codon-based substitution model implemented in PAML version 4.2 [11]. Branch models [45] that allow ω to vary among branches in the phylogeny were applied to determine ω ratios along particular lineages. Likelihood ratio tests (LRT) were performed to test nested competing hypotheses (S3 Table).
Cell culture and functional assays
CHO-K1 cells were grown in Dulbecco’s Modified Eagle Medium: Nutrient Mixture F-12 (DMEM/F12) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin and 100 μg/ml streptomycin. Cells were maintained at 37 °C in a humidified 5% CO2 incubator. For transient transfection Lipofectamine 2000 (Life Technologies, Darmstadt, Germany) was used. Cells were split into 25 cm2-cell culture flasks (0.9 x 106 cells/flask) and transfected with a total amount of 3 μg of plasmid the following day.
ALPHAScreen cAMP assay
cAMP content of cell extracts was determined by a non-radioactive assay based on the ALPHAScreen technology according to the manufacturers’ protocol (Perkin Elmer LAS, Rodgau-Jügesheim, Germany) [46]. One day after transfection cells were split into 96-well plates (2 x 104 cells/well). Stimulation with various agonist concentrations (all compounds from Sigma-Aldrich, Seelze, Germany) or diluted human urine and plasma samples was performed 48 h after transfection. Reactions were stopped by aspiration of media and cells were lysed in 20 μl of lysis buffer containing 1 mM 3-isobutyl-1-methylxanthine (IBMX). From each well 5 μl of lysate were transferred to a 384-well plate. Acceptor beads and donor beads were added according to the manufacturers’ protocol. Cyclic AMP accumulation data were analyzed using GraphPad Prism version 6.03 (GraphPad Software, San Diego California USA).
Dynamic mass redistribution assay
To measure label-free receptor activation, a dynamic mass redistribution (DMR) assay (Corning Epic Biosensor Measurements) with CHO-K1 cells transiently expressing HCAR orthologs was performed. One day after transfection, cells were detached using Versene solution and transferred into a fibronectin-coated 384-well Epic microplate (1.2 x 104cells per well) and cultured for 24 h to reach confluent monolayers. After 2 h of equilibration in HBSS buffer with 20 mM HEPES (pH 7.4), incubation with various compound concentrations was performed and DMR was recorded for 60 min.
ELISA
Cell surface expression of N-terminal HA-tagged receptor constructs was determined using an indirect cellular ELISA and total receptor expression of full-length HA/FLAG double-tagged HCA constructs was assessed using a “sandwich-ELISA” both described in [46].
Manual reanalysis and comparison to FPKM values of a publicly available RNA-Sequencing dataset
All 134 RNA-sequencing samples belonging to Bioproject: PRJNA326727, (GEO Accession: GSE83687) [47] were manually reanalyzed and compared to available FPKM values as stated in S6 Table and S7 Fig.
Mining of human microbiome data for genes of enzymes with ability to produce D PLA
Two large collections of metagenomic data from the human gut microbiome were searched for D-lactate dehydrogenase (EC 1.1.1.28), the enzyme that catalyzes the conversion of D-lactate to pyruvate, but has also been shown for some LAB to convert phenylpyruvate to D-PLA. Metagenomic data from 1,885 human stool samples from the curated Metagenomic Dataset (https://github.com/waldronlab/ curatedMetagenomicData) [48] were searched for 1,443 EC 1.1.1.28-related UniRef100 entries. In addition, genes with the K03778 annotation were searched in the Integrated Gene Catalogue (IGC) [49] reference data set for the human gut microbiome, which comprises data from just over 1,000 individuals. Detailed information, a representative tree of the detected genes and a list of all entries are summarized in the Supplementary Information (S5 Fig, S4 Table).
Peripheral blood mononuclear cells (PBMCs) and monocytes
Monocyte isolation
PBMCs were obtained by Ficoll-Paque (GE Healthcare) density gradient centrifugation. After repeated washing in PBS containing 0.3 mM EDTA, untouched monocytes were isolated by negative magnetic depletion using the human Monocyte Isolation Kit II (Miltenyi Biotec) according to the manufacturer’s protocol. The cell preparations were >95% monocytes as determined by morphology and flow cytometry.
Monocyte migration
For migration analysis, ThinCert cell culture inserts (Greiner) with 8 μM pores were placed in a 24-well cell culture plate. To each well of the cell culture plate 1,000 μl of HBSS buffer supplemented with 20 mM HEPES (pH 7.4) and 1% FBS with or without chemoattractant were added. Pelleted freshly isolated monocytes in PBS with 0.3 mM EDTA and 1% FBS were labeled for 30 min at 4°C with 2 μM calcein AM (Thermo Scientific). Subsequently monocytes were washed and resuspended in HBSS buffer with 20 mM HEPES (pH 7.4) and 1% FBS to a final concentration of 5 × 105 monocytes per ml. 200 μl of the monocyte suspension were added to each insert or for control purposes directly to the bottom well and incubated at 37°C and 5% CO2. After 1 h the inserts were discarded, plates spun for 5 min at 700 rpm and imaged as well as analyzed using the Celigo S Imaging Cytometer (Nexcelom).
Monocyte siRNA transfection
Freshly isolated human monocytes were transfected with siRNA specifically targeting HCAR3 (OriGene), as shown in Stäubert et al. (2015) [50], using Viromer Green (Lipocalyx) according to the manufacturer’s protocol. 10 μL of siRNA-Viromer mix were preplated into a 96-well plate before the addition of freshly isolated monocytes resuspended in RPMI supplemented with 10% FBS (1.5 x 105 monocytes/well) which were then incubated 14 h at 37 °C in a humidified 5% CO2 incubator. cAMP assays were performed as described below. To monitor the transfection efficiency a red fluorescent siRNA (OriGene) and Hoechst 33342 (Sigma-Aldrich) were used. The viability of transfected monocytes was determined by staining with propidium iodide (PI) (Thermo Fisher Scientific) and Hoechst 33342. Transfection efficiency and viability were analyzed using the Celigo S Imaging Cytometer (Nexcelom). The transfection efficiency was found to be ≥ 80% with less than 50% of monocytes being PI-positive (S6 Fig).
Monocyte cAMP assay
Freshly isolated monocytes or PBMCs were resuspended in HBSS buffer with 20 mM HEPES (pH 7.4) and 1 mM IBMX, seeded in 96-well plates (1 x 105 monocytes/well, 2 x 105 PBMCs/well) with or without PTX (100 ng/ml) and incubated for 1 h at 37 °C. Stimulation with or without agonists was performed in presence of 10 μM forskolin for 1 h at 37 °C. Measurement of accumulated cAMP was performed as described above.
Determination of PLA in human plasma and urine
100 mg of D-PLA were dissolved in 30 ml H2O and ingested by a healthy male subject (78 kg, 1.80 m). 2.5 ml of EDTA blood samples were taken right before and 30 min, 60 min, and then every hour up to 7 h after ingestion. In a subsequent, chronically separate experiment the same healthy male subject adapted to a lacto-vegetarian diet, including uptake of 500 g per day of fresh (non-pasteurized) Sauerkraut (Spreewaldhof, Germany) for three days. 2.5 mL of EDTA blood samples were taken before diet, 2 h postprandial over three days and 5 days after having stopped the above described diet. Further, urine samples were taken over the time course. Similarly, three healthy female subjects ingested 5–6 g of fresh (non-pasteurized) Sauerkraut per kg body weight for one day. 2.5 mL of EDTA blood samples and urine samples were taken before ingestion and 2 h postprandial. Fresh whole blood samples were centrifuged for 10 min at 4 °C and 800xg to isolate plasma, which was stored in 500 μl aliquots at -80 °C until further analyses. Urine was collected and the total volume was determined. 10 μl of human plasma and urine were treated with 90 μl precipitating agent (acetonitrile including the internal standard phenyl-d5-lactate at 10 ng/ml), thoroughly mixed and centrifuged for 5 min at 13,000xg. Supernatants were transferred to autosampler vials and 10 μl were injected into the Liquid Chromatography Mass Spectrometry (LC-MS) system. It consisted of a Prominence UFLC system from Shimadzu (Duisburg, Germany) and a QTRAP 6500 from SCIEX (Framingham, MA, USA). Chromatographic separation took place on a Luna HILIC column (3 μm, 50 x 2 mm) from Phenomenex (Darmstadt, Germany) via gradient elution at a flow rate of 0.4 ml/min. The mobile phase was 15 mmol/l ammonium acetate buffer (pH 6) in acetonitrile and water. Electrospray ionization was applied in negative mode. Mass transitions were m/z 165>103 and m/z 170>108 for phenyl-lactate and phenyl-d5-lactate, respectively. Quantitation was carried out using a semi-quantitative approach following Eq 1 with c and A as the concentration and the peak area of the analyte (An) and the internal standard (IS). The response factor (RF) was determined from signal responses in standard solutions and equaled 1.
| (1) |
Supporting information
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Acknowledgments
We would like to thank the contributors of species samples (S8 Table) and Johannes Krause for suggestions and critical reading of the manuscript.
Data Availability
Newly generated sequences are available from GenBank (NCBI) (accession numbers KU285431-KU285452). All other relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was supported by the German Research Foundation (www.dfg.de/): C.S. (STA 1265/3-1), T.S. SFB 1052 (B6), by iDiv (FZT 118) to A.H.B and by the European Social Fonds (https://www.esf.de/) (A.P.) and research funding of the Medical Faculty, University Leipzig (https://www.uniklinikum-leipzig.de/wissenschaft-forschung/forschungs-administration/forschungsförderung) (C.S.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Perdigon G, Fuller R, Raya R. Lactic acid bacteria and their effect on the immune system. Current issues in intestinal microbiology. 2001;2(1):27–42. Epub 2001/11/17. . [PubMed] [Google Scholar]
- 2.Rios-Covian D, Ruas-Madiedo P, Margolles A, Gueimonde M, de Los Reyes-Gavilan CG, Salazar N. Intestinal Short Chain Fatty Acids and their Link with Diet and Human Health. Frontiers in microbiology. 2016;7:185 Epub 2016/03/01. 10.3389/fmicb.2016.00185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Husted AS, Trauelsen M, Rudenko O, Hjorth SA, Schwartz TW. GPCR-Mediated Signaling of Metabolites. Cell metabolism. 2017;25(4):777–96. Epub 2017/04/06. 10.1016/j.cmet.2017.03.008 . [DOI] [PubMed] [Google Scholar]
- 4.Offermanns S. Hydroxy-Carboxylic Acid Receptor Actions in Metabolism. Trends in endocrinology and metabolism: TEM. 2017;28(3):227–36. Epub 2017/01/15. 10.1016/j.tem.2016.11.007 . [DOI] [PubMed] [Google Scholar]
- 5.Zellner C, Pullinger CR, Aouizerat BE, Frost PH, Kwok PY, Malloy MJ, et al. Variations in human HM74 (GPR109B) and HM74A (GPR109A) niacin receptors. Hum Mutat. 2005;25(1):18–21. Epub 2004/12/08. 10.1002/humu.20121 . [DOI] [PubMed] [Google Scholar]
- 6.Ahmed K, Tunaru S, Langhans CD, Hanson J, Michalski CW, Kolker S, et al. Deorphanization of GPR109B as a receptor for the beta-oxidation intermediate 3-OH-octanoic acid and its role in the regulation of lipolysis. J Biol Chem. 2009;284(33):21928–33. Epub 2009/06/30. 10.1074/jbc.M109.019455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Irukayama-Tomobe Y, Tanaka H, Yokomizo T, Hashidate-Yoshida T, Yanagisawa M, Sakurai T. Aromatic D-amino acids act as chemoattractant factors for human leukocytes through a G protein-coupled receptor, GPR109B. Proc Natl Acad Sci U S A. 2009;106(10):3930–4. Epub 2009/02/25. 10.1073/pnas.0811844106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carrigan MA, Uryasev O, Frye CB, Eckman BL, Myers CR, Hurley TD, et al. Hominids adapted to metabolize ethanol long before human-directed fermentation. Proc Natl Acad Sci U S A. 2015;112(2):458–63. Epub 2014/12/03. 10.1073/pnas.1404167111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuei C, Yu J, Zhu J, Wu J, Zhang L, Shih A, et al. Study of GPR81, the lactate receptor, from distant species identifies residues and motifs critical for GPR81 functions. Mol Pharmacol. 2011;80(5):848–58. Epub 2011/08/25. 10.1124/mol.111.074500 . [DOI] [PubMed] [Google Scholar]
- 10.Carbone L, Harris RA, Gnerre S, Veeramah KR, Lorente-Galdos B, Huddleston J, et al. Gibbon genome and the fast karyotype evolution of small apes. Nature. 2014;513(7517):195–201. Epub 2014/09/12. 10.1038/nature13679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–91. 10.1093/molbev/msm088 . [DOI] [PubMed] [Google Scholar]
- 12.Ahmed K, Tunaru S, Tang C, Muller M, Gille A, Sassmann A, et al. An autocrine lactate loop mediates insulin-dependent inhibition of lipolysis through GPR81. Cell metabolism. 2010;11(4):311–9. Epub 2010/04/09. 10.1016/j.cmet.2010.02.012 . [DOI] [PubMed] [Google Scholar]
- 13.Liu C, Wu J, Zhu J, Kuei C, Yu J, Shelton J, et al. Lactate inhibits lipolysis in fat cells through activation of an orphan G-protein-coupled receptor, GPR81. J Biol Chem. 2009;284(5):2811–22. Epub 2008/12/03. 10.1074/jbc.M806409200 . [DOI] [PubMed] [Google Scholar]
- 14.Taggart AK, Kero J, Gan X, Cai TQ, Cheng K, Ippolito M, et al. (D)-beta-Hydroxybutyrate inhibits adipocyte lipolysis via the nicotinic acid receptor PUMA-G. J Biol Chem. 2005;280(29):26649–52. Epub 2005/06/03. 10.1074/jbc.C500213200 . [DOI] [PubMed] [Google Scholar]
- 15.Bruckner H, Becker D, Lupke M. Chirality of Amino-Acids of Microorganisms Used in Food Biotechnology. Chirality. 1993;5(5):385–92. 10.1002/chir.530050521 [DOI] [PubMed] [Google Scholar]
- 16.Mutaguchi Y, Ohmori T, Akano H, Doi K, Ohshima T. Distribution of D-amino acids in vinegars and involvement of lactic acid bacteria in the production of D-amino acids. SpringerPlus. 2013;2:691 Epub 2014/01/15. 10.1186/2193-1801-2-691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tamang JP, Watanabe K, Holzapfel WH. Review: Diversity of Microorganisms in Global Fermented Foods and Beverages. Frontiers in microbiology. 2016;7:377 Epub 2016/04/06. 10.3389/fmicb.2016.00377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rajilic-Stojanovic M, de Vos WM. The first 1000 cultured species of the human gastrointestinal microbiota. FEMS microbiology reviews. 2014;38(5):996–1047. Epub 2014/05/28. 10.1111/1574-6976.12075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bruckner H, Haasmann S, Friedrich A. Quantification of D-Amino Acids in Human Urine Using Gc-Ms and Hplc. Amino Acids. 1994;6(2):205–11. 10.1007/BF00805848 [DOI] [PubMed] [Google Scholar]
- 20.Lorenzo MP, Dudzik D, Varas E, Gibellini M, Skotnicki M, Zorawski M, et al. Optimization and validation of a chiral GC-MS method for the determination of free D-amino acids ratio in human urine: Application to a Gestational Diabetes Mellitus study. J Pharmaceut Biomed. 2015;107:480–7. 10.1016/j.jpba.2015.01.015 [DOI] [PubMed] [Google Scholar]
- 21.Bruckner H, Schieber A. Determination of free D-amino acids in mammalia by chiral gas chromatography-mass spectrometry. Hrc-J High Res Chrom. 2000;23(10):576–82. [Google Scholar]
- 22.Martinez-Rodriguez S, Martinez-Gomez AI, Rodriguez-Vico F, Clemente-Jimenez JM, Heras-Vazquez FJL. Natural Occurrence and Industrial Applications of D-Amino Acids: An Overview. Chemistry & biodiversity. 2010;7(6):1531–48. [DOI] [PubMed] [Google Scholar]
- 23.Broberg A, Jacobsson K, Strom K, Schnurer J. Metabolite profiles of lactic acid bacteria in grass silage. Applied and environmental microbiology. 2007;73(17):5547–52. Epub 2007/07/10. 10.1128/AEM.02939-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sjogren J, Magnusson J, Broberg A, Schnurer J, Kenne L. Antifungal 3-hydroxy fatty acids from Lactobacillus plantarum MiLAB 14. Applied and environmental microbiology. 2003;69(12):7554–7. Epub 2003/12/09. 10.1128/AEM.69.12.7554-7557.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marco ML, Heeney D, Binda S, Cifelli CJ, Cotter PD, Foligne B, et al. Health benefits of fermented foods: microbiota and beyond. Current opinion in biotechnology. 2017;44:94–102. Epub 2016/12/22. 10.1016/j.copbio.2016.11.010 . [DOI] [PubMed] [Google Scholar]
- 26.Pessione E. Lactic acid bacteria contribution to gut microbiota complexity: lights and shadows. Frontiers in cellular and infection microbiology. 2012;2:86 Epub 2012/08/25. 10.3389/fcimb.2012.00086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bongaerts GP, Tolboom JJ, Naber AH, Sperl WJ, Severijnen RS, Bakkeren JA, et al. Role of bacteria in the pathogenesis of short bowel syndrome-associated D-lactic acidemia. Microbial pathogenesis. 1997;22(5):285–93. Epub 1997/05/01. 10.1006/mpat.1996.0122 . [DOI] [PubMed] [Google Scholar]
- 28.Joly F, Mayeur C, Bruneau A, Noordine ML, Meylheuc T, Langella P, et al. Drastic changes in fecal and mucosa-associated microbiota in adult patients with short bowel syndrome. Biochimie. 2010;92(7):753–61. Epub 2010/02/23. 10.1016/j.biochi.2010.02.015 . [DOI] [PubMed] [Google Scholar]
- 29.Spaapen LJ, Ketting D, Wadman SK, Bruinvis L, Duran M. Urinary D-4-hydroxyphenyllactate, D-phenyllactate and D-2-hydroxyisocaproate, abnormalities of bacterial origin. Journal of inherited metabolic disease. 1987;10(4):383–90. Epub 1987/01/01. . [DOI] [PubMed] [Google Scholar]
- 30.Haan E, Brown G, Bankier A, Mitchell D, Hunt S, Blakey J, et al. Severe illness caused by the products of bacterial metabolism in a child with a short gut. European journal of pediatrics. 1985;144(1):63–5. Epub 1985/05/01. . [DOI] [PubMed] [Google Scholar]
- 31.Heil M, Podebrad F, Beck T, Mosandl A, Sewell AC, Bohles H. Enantioselective multidimensional gas chromatography-mass spectrometry in the analysis of urinary organic acids. Journal of chromatography B, Biomedical sciences and applications. 1998;714(2):119–26. Epub 1998/10/10. . [DOI] [PubMed] [Google Scholar]
- 32.Mccabe ERB, Goodman SI, Fennessey PV, Miles BS, Wall M, Silverman A. Glutaric, 3-Hydroxypropionic, and Lactic Aciduria with Metabolic Acidemia, Following Extensive Small Bowel Resection. Biochem Med Metab B. 1982;28(2):229–36. 10.1016/0006-2944(82)90074-6 [DOI] [PubMed] [Google Scholar]
- 33.Schoorel EP, Giesberts MA, Blom W, van Gelderen HH. D-Lactic acidosis in a boy with short bowel syndrome. Archives of disease in childhood. 1980;55(10):810–2. Epub 1980/10/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walter J. Ecological role of lactobacilli in the gastrointestinal tract: implications for fundamental and biomedical research. Applied and environmental microbiology. 2008;74(16):4985–96. Epub 2008/06/10. 10.1128/AEM.00753-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mu W, Yu S, Zhu L, Zhang T, Jiang B. Recent research on 3-phenyllactic acid, a broad-spectrum antimicrobial compound. Appl Microbiol Biotechnol. 2012;95(5):1155–63. Epub 2012/07/12. 10.1007/s00253-012-4269-8 . [DOI] [PubMed] [Google Scholar]
- 36.Xu GC, Zhang LL, Ni Y. Enzymatic preparation of D-phenyllactic acid at high space-time yield with a novel phenylpyruvate reductase identified from Lactobacillus sp CGMCC 9967. Journal of biotechnology. 2016;222:29–37. 10.1016/j.jbiotec.2015.12.011 [DOI] [PubMed] [Google Scholar]
- 37.Li L, Shin SY, Lee KW, Han NS. Production of natural antimicrobial compound D-phenyllactic acid using Leuconostoc mesenteroides ATCC 8293 whole cells involving highly active D-lactate dehydrogenase. Letters in Applied Microbiology. 2014;59(4):404–11. 10.1111/lam.12293 [DOI] [PubMed] [Google Scholar]
- 38.Douillard FP, de Vos WM. Lactic Acid Bacteria and the Human Intestinal Microbiome Biotechnology of Lactic Acid Bacteria: Novel Applications, 2nd Edition 2016:120–33. [Google Scholar]
- 39.Clemens PC, Schunemann MH, Hoffmann GF, Kohlschutter A. Plasma concentrations of phenyllactic acid in phenylketonuria. Journal of inherited metabolic disease. 1990;13(2):227–8. Epub 1990/01/01. . [DOI] [PubMed] [Google Scholar]
- 40.Zabat MA, Sano WH, Wurster JI, Cabral DJ, Belenky P. Microbial Community Analysis of Sauerkraut Fermentation Reveals a Stable and Rapidly Established Community. Foods. 2018;7(5). Epub 2018/05/15. 10.3390/foods7050077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Selhub EM, Logan AC, Bested AC. Fermented foods, microbiota, and mental health: ancient practice meets nutritional psychiatry. Journal of physiological anthropology. 2014;33:2 Epub 2014/01/16. 10.1186/1880-6805-33-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okayama H, Berg P. A cDNA cloning vector that permits expression of cDNA inserts in mammalian cells. Mol Cell Biol. 1983;3(2):280–9. Epub 1983/02/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–80. 10.1093/nar/22.22.4673 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–9. Epub 2013/10/18. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang Z. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol. 1998;15(5):568–73. 10.1093/oxfordjournals.molbev.a025957 . [DOI] [PubMed] [Google Scholar]
- 46.Staubert C, Bohnekamp J, Schoneberg T. Determinants involved in subtype-specific functions of rat trace amine-associated receptors 1 and 4. Br J Pharmacol. 2013;168(5):1266–78. Epub 2012/10/18. 10.1111/bph.12020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peters LA, Perrigoue J, Mortha A, Iuga A, Song WM, Neiman EM, et al. A functional genomics predictive network model identifies regulators of inflammatory bowel disease. Nature genetics. 2017;49(10):1437–49. Epub 2017/09/12. 10.1038/ng.3947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pasolli E, Schiffer L, Manghi P, Renson A, Obenchain V, Truong DT, et al. Accessible, curated metagenomic data through ExperimentHub. Nature methods. 2017;14(11):1023–4. Epub 2017/11/01. 10.1038/nmeth.4468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li J, Jia H, Cai X, Zhong H, Feng Q, Sunagawa S, et al. An integrated catalog of reference genes in the human gut microbiome. Nature biotechnology. 2014;32(8):834–41. Epub 2014/07/07. 10.1038/nbt.2942 . [DOI] [PubMed] [Google Scholar]
- 50.Staubert C, Broom OJ, Nordstrom A. Hydroxycarboxylic acid receptors are essential for breast cancer cells to control their lipid/fatty acid metabolism. Oncotarget. 2015. Epub 2015/04/04. 10.18632/oncotarget.3565 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability Statement
Newly generated sequences are available from GenBank (NCBI) (accession numbers KU285431-KU285452). All other relevant data are within the manuscript and its Supporting Information files.