Most recently an epidemic of Zika virus in the Americas, affecting well over a million people, has caused substantial morbidity and mortality, including Guillain-Barre syndrome, microcephaly and other fetal developmental defects1,2. Preventive and therapeutic measures to specifically target the virus are not readily available. Transmission of Zika virus is predominantly mosquito-borne and Aedes aegypti mosquitoes serve as a key vector for Zika virus3. Here, to identify salivary factors that modulate mosquito-borne Zika virus infection, we focused on antigenic proteins in mice that were repeatedly bitten by mosquitoes and developed antibodies against salivary proteins. Using a yeast surface display screen, we identified five antigenic A. aegypti salivary proteins in mice. Antiserum against one of these five proteins - A. aegypti bacteria-responsive protein 1 (AgBR1) - suppressed early inflammatory responses in the skin of mice bitten by Zika virus-infected mosquitoes. AgBR1 antiserum also partially protected mice against lethal mosquito-borne - but not needle injected - Zika virus infection. These data suggest that AgBR1 is a target for the prevention of mosquito-transmitted Zika virus infection.
Mosquitoes inject numerous salivary proteins into the skin of a host during blood feeding4, and these molecules are capable of modulating various host responses5,6. Indeed, mosquito saliva enhances transmission and pathogenicity of specific arboviruses7,8. Although mosquito saliva can increase arboviral infectivity, only a limited number of specific salivary proteins have been characterized that influence these processes. The biogenic amine-binding D7 protein partially inhibits dengue infection, while saliva serine protease CLIPA3 enhances dissemination of dengue virus into the mammalian host9,10. In addition, salivary factor LTRIN from A. aegypti facilitates the transmission of Zika virus by inhibiting NFκB signaling during infection11. Despite these efforts, much remains to be discovered about how specific salivary factors facilitate mosquito-borne virus infection, and whether targeting these proteins can prevent or delay infection.
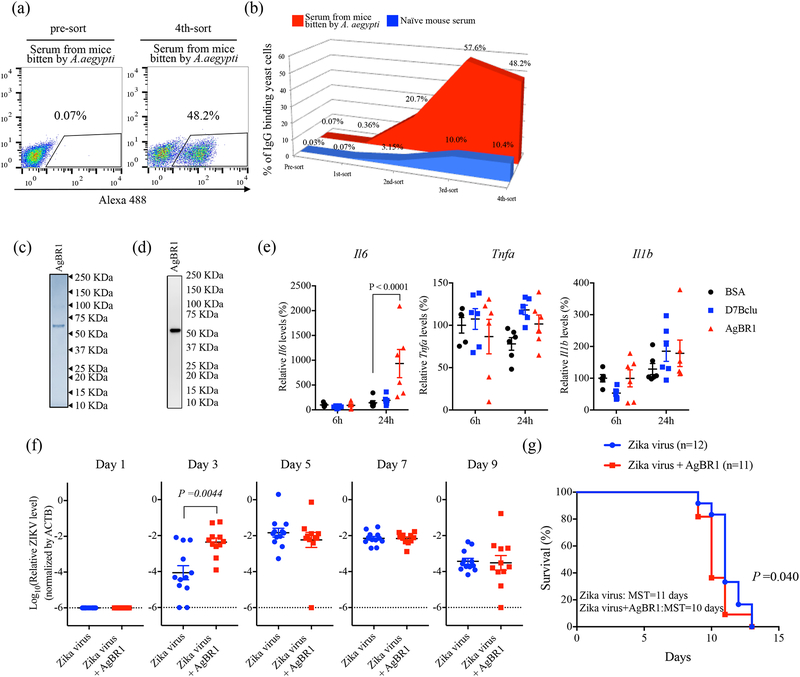
To identify salivary factors that modulate mosquito-borne Zika virus infection, we focused on antigenic proteins in a vertebrate host repeatedly bitten by A. aegypti mosquitoes. Although previous studies identified several antigenic proteins using SDS-PAGE and proteomics12, it is difficult for these methods to detect proteins of low-abundance and low-antigenicity13. Therefore, here, we employed a yeast surface display screening, which can identify uncommon proteins by iterative rounds of magnetic-activated cell sorting14. An A. aegypti salivary gland yeast surface display library was generated and probed with IgG from mice repeatedly bitten by A. aegypti (Supplementary Fig. 1). Individual yeast cell clones expressing salivary proteins identified using these sera (Fig. 1a and b) were enriched and isolated, and the recombinant plasmids were recovered and sequenced. Five unique mosquito genes were found, including previously identified mosquito proteins and some with unknown function (Supplementary Table 1). Among the five identified proteins, A. aegypti bacteria-responsive protein 1 (AgBR1), which we confirmed using immunoblot, was recognized by serum from mice bitten by mosquitoes (Supplementary Fig. 2), and had substantial homology (identities = 27%, positives = 43%) with murine chitinase 3 like-1 protein (Supplementary Fig. 3), a protein with putative functions in host defense, inflammation and repair15. AgBR1 is also known to be up-regulated in the salivary glands of mosquitoes after blood feeding16. The function of AgBR1 in the vertebrate host, however, remains unknown. Therefore, we examined whether AgBR1 stimulates inflammatory responses in vitro. Murine splenocytes stimulated with recombinant AgBR1 (Fig. 1c–d) produced in S2 cells (5 μg/ml) demonstrated significantly higher levels of Il6 expression compared with controls (Fig. 1e). As increased vascular permeability contributes to flavivirus pathogenicity17 and IL-6 is associated with these processes18, we next examined whether AgBR1 influences Zika virus infection in vivo. Given previous studies demonstrating that approximately half of the protein in the salivary glands is discharged during a blood meal19,20, the concentration of AgBR1 in mosquito saliva can be estimated to be between 1.6 – 8.2 μM (Supplementary Fig. 4, Supplementary Table 2). Therefore, we injected AG129 mice with Zika virus and AgBR1 (5.1 μM, 10 μg of AgBR1 in total volume 40 μl). At day 3 post infection, significantly higher viremia levels were observed in the group of mice inoculated with Zika virus in conjunction with AgBR1, compared to those of mice challenged with Zika virus alone (Fig. 1f). In addition, AgBR1 protein significantly impaired the survival of Zika virus-infected mice (Fig. 1g). These results demonstrate that AgBR1 can exacerbate Zika virus infection and disease in vivo.
Figure 1. AgBR1 is identified as an antigenic protein in mice and modulates host responses in vitro and in vivo.
(a-b) Yeast surface display (YSD) approach to identify mosquito proteins eliciting responses in mice bitten by Aedes aegypti. Flow cytometry (FACS) analysis of yeast cells using IgG from mice bitten repeatedly by mosquitoes (red) and IgG derived from naïve mouse serum (blue) of transformed yeast cells (left panel:pre-sort, right panel; 4th-sort). The percentages of IgG-binding yeast cells are shown in the right panel of Fig. 1A. Data are representative of two independent experiments with similar results. (c) Recombinant AgBR1 (0.25 μg protein) was run on SDS-PAGE and stained with Coomassie Brilliant Blue. (d) AgBR1 protein was detected using an anti-His antibody. (c-d) Data are representative of two independent experiments. (e) The expression levels of Il6, Tnfa and Il1b after BSA, D7Bclu or AgBR1 treatment. Data were analyzed by two-way ANOVA. n=5 or 6 biologically independent samples pooled from two separate experiments. Data are presented as mean ± s.e.m. (f) Zika virus level in blood after co-inoculation of Zika virus with AgBR1 protein (5.1 μM, 10 μg in 40 μl). Data are presented as mean ± s.e.m. Each data point represents one mouse. Normalized viral RNA levels were analyzed using the two-sided Wilcoxon–Mann–Whitney test. (Zika virus: n=12, Zika virus + AgBR1: n=11 pooled from two separate experiments) (g) Survival and median survival time (MST) were assessed using the Gehan-Wilcoxon test. (Zika virus: n=12, Zika virus + AgBR1: n=11 pooled from in two separate experiments)
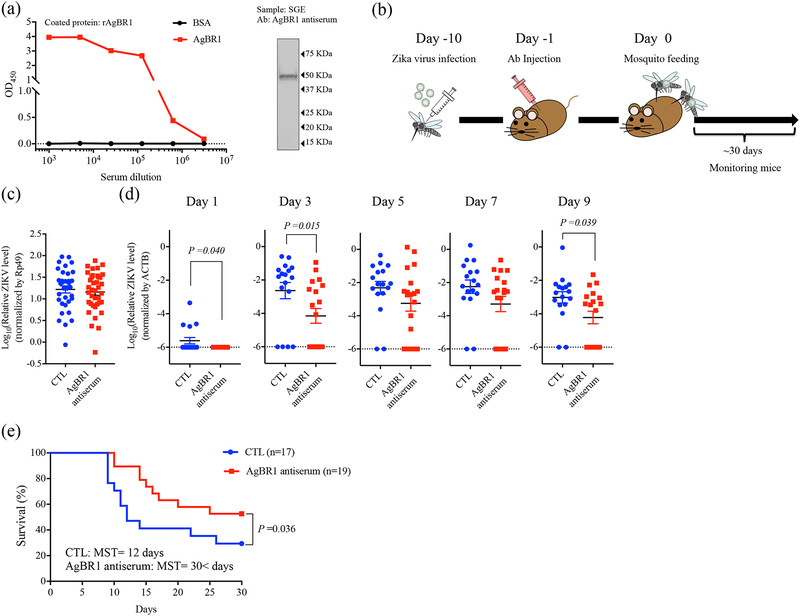
Next, we examined whether blocking AgBR1 in vivo affects mosquito-borne Zika virus infection. Rabbit antiserum against recombinant AgBR1 reacted strongly, and specifically, with the recombinant protein and recognized native AgBR1 in mosquito salivary gland extracts (Fig. 2a). We treated mice with AgBR1 antiserum to determine whether inhibiting this protein modulates pathogenesis during A. aegypti-borne Zika virus infection. AG129 mice were administered AgBR1- or control antiserum and 24 hours later were bitten by two Zika virus-infected A. aegypti mosquitoes (Fig. 2b). Zika virus levels in the salivary glands of all mosquitoes were similar, suggesting that mice were exposed to comparable levels of virus (Fig. 2c). We then determined whether the AgBR1 antiserum altered Zika virus infection in mice. AgBR1 antiserum significantly reduced Zika virus levels in mice over the course of viral infection (Fig. 2d) and provided partial protection against Zika virus-induced pathogenesis and death (Fig. 2e). We also found that the partial protective effect of AgBR1 antibodies was specific for mosquito-borne - and not needle-injected - Zika virus infection in mice (Supplementary Fig. 5).
Figure 2. AgBR1 antiserum protects mice from mosquito-borne Zika virus infection.
(a) AgBR1 antiserum recognized recombinant AgBR1 protein as confirmed by ELISA (left panel) and naïve AgBR1 in salivary gland extract (SGE) as confirmed by immunoblot (right panel). Data are representative of three independent experiments with similar results. (b) Workflow of passive immunization and mosquito-borne Zika virus infection. (c)Zika virus RNA levels in the salivary glands at 10 days after intrathoracic injection. n=34 (Control) or 38 (AgBR1 antiserum) biologically independent samples pooled from five separate experiments. Data represent mean ± s.e.m. (d) Zika virus RNA levels in blood in mice. Data represent mean ± s.e.m. Each data point represents one mouse. Normalized viral RNA levels were analyzed using two-sided Wilcoxon–Mann–Whitney test. n=17 (Control) or 19 (AgBR1 antiserum) biologically independent samples pooled from five separate experiments. (e) Survival and median survival time (MST) were assessed using the Gehan-Wilcoxon test. n=17 (Control) or 19 (AgBR1 antiserum) biologically independent samples pooled from five separate experiments.
We also assessed whether active immunization with AgBR1 could be effective at protection. AG129 mice were immunized three times with 10 μg of AgBR1 in complete or incomplete Freund’s adjuvant. After the final boost, high titers of AgBR1 antibodies were detected (Supplementary Fig. 6a). Actively immunized mice displayed significantly reduced viremia at day 5, and showed significantly better survival than control mice against mosquito-borne Zika virus infection (Supplementary Fig. 6b and c), suggesting that active immunization with AgBR1 delays the appearance of clinical signs and death of the animals as well as passive immunization. Here, we used AG129 mice that lack both Type I and II IFN receptors but can elicit B-cell and T-cell responses21,22. As type I interferon signaling can contribute to optimal antibody responses23, it is possible that further efforts using alternative adjuvants, protein concentration or different animal models could enhance the effect of active immunization. Overall, these results indicated that immunization with AgBR1 partially influenced mosquito-transmitted Zika virus infection.
To determine whether the effects observed with AgBR1 extended to other proteins identified in our screen, we chose two additional proteins, D7Bclu and SP, whose expression has been identified as upregulated salivary gland proteins during flavivirus infection10. We generated D7Bclu and SP antisera in a similar fashion to the AgBR1 antiserum and performed passive immunization experiments. Neither the D7Bclu nor SP antisera altered the viremia or protected mice from lethal mosquito-borne Zika virus infection (Supplementary Fig. 7).
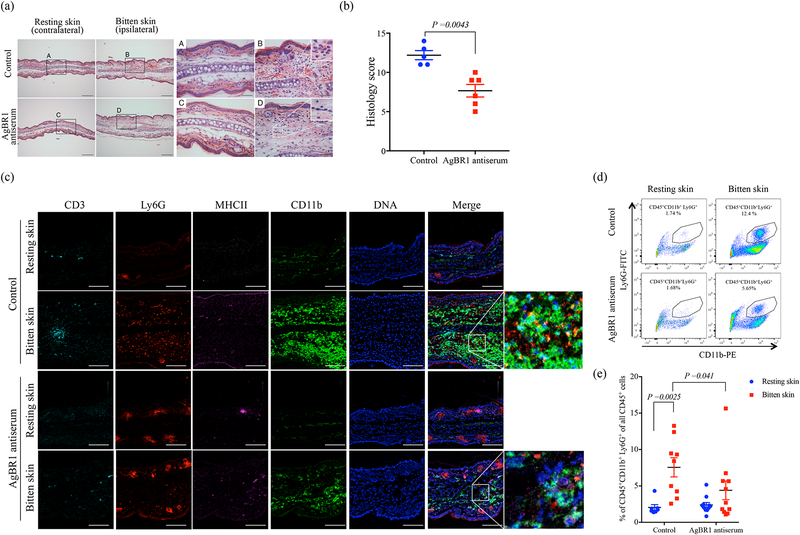
To more fully understand the underlying mechanism of protection of immunization with AgBR1, we examined whether AgBR1 antiserum influenced the early innate immune response at the bite site after exposure of mice to Zika virus-infected mosquitoes. Histological analysis of the bite site 24 h post-feeding by Zika virus-infected mosquitoes showed prominent inflammatory cell infiltration mainly composed of neutrophils in the dermis of mice administered naïve serum (control), which was less apparent in mice administered AgBR1 antiserum (Fig. 3a and Supplementary Table 3). Consistent with these findings, histology scores were significantly lower in mice administered AgBR1 antiserum compared with mice administered naïve serum (Fig. 3b). Furthermore, imaging mass cytometry showed that infiltrating cells at the bite site of Zika virus-infected mosquito bites were mainly Ly6G+ and CD11b+cells, supporting the observation made with hematoxylin and eosin staining that the infiltrating cells are predominantly composed of neutrophils, monocytes, and macrophages, with some minor populations of T cells or other immune cells (Fig. 3c). In addition, the infiltration of Ly6G+ cells and CD11b+ cells is reduced in mice administered AgBR1 antiserum, in contrast to control animals (Fig. 3c). The alteration of infiltrating cell populations in the skin of bitten mice administered AgBR1 antiserum indicates that the AgBR1 antiserum influenced the number of CD45+CD11b+Ly6G+ neutrophils at the bite site (Fig. 3d and e). These results suggest that AgBR1 antiserum suppressed acute inflammation, and particularly the neutrophilic response, at the mosquito bite site.
Figure 3. AgBR1 antiserum suppresses neutrophil infiltration at the mosquito bite site.
(a) Hematoxylin and eosin staining of the ears of mice treated with AgBR1 antiserum or control serum at 24 h post-feeding. Scale bar, 200 μm (left panels) and 50 μm (right panels). Data are representative of two independent experiments with similar results. (b) The total histology scores of the bite sites were compared between the AgBR1 antiserum and control group. Data are presented as mean ± s.e.m.. Statistical analysis was performed using two-sided Wilcoxon–Mann–Whitney test. n=5 (Control) or 6 (AgBR1 antiserum) biologically independent samples pooled from two separate experiments. (c) Imaging Mass Cytometry (IMC) labeling of ears of mice 24 h post Zika virus-infected mosquito feeding. Scale bar, 100 μm. Data are representative of two independent experiments with similar results. (d) The population of CD45+CD11b+Ly6G+ (neutrophils) was analyzed using flow cytometry. Data are representative of two independent experiments with similar results. (e) The percent of CD45+CD11b+Ly6G+ (neutrophils) cells in CD45+ leukocyte cells at 24h after Zika virus-infected mosquito feeding. Each dot represents one mouse. Significance was calculated using a two-way ANOVA test for multiple comparisons. Data are presented as mean ± s.e.m.. (Control-resting skin: n=7, Control-bitten skin: n=9, AgBR1 antiserum-resting skin: n=11, AgBR1 antiserum-bitten skin: n=11 biologically independent samples pooled from two separate experiments.)
To examine the direct effect of AgBR1 in the skin, we examined whether CD45+CD11b+Ly6G+ cells are recruited into the intradermally-injected skin site. More CD45+CD11b+Ly6G+ cells infiltrated into the AgBR1-injected skin compared with the resting skin, suggesting that AgBR1 can recruit CD45+CD11b+Ly6G+ cells (Supplementary Fig. 8). Since a mosquito bite represents a more natural introduction of AgBR1 into the skin, we investigated whether suppression of AgBR1 gene and AgBR1 protein expression in the salivary glands alters the levels of CD45+CD11b+Ly6G+ cells infiltration after mosquito bites (Supplementary Fig. 9a and b). Zika virus levels in salivary glands were similar in both dsRNA-treated groups of mosquitoes (Supplementary Fig. 9c), and we found that the levels of CD45+CD11b+Ly6G+ cells were significantly increased in mice bitten by control mosquitoes, but not in mice bitten by AgBR1 dsRNA-treated mosquitoes (Supplementary Fig. 9d). These results further demonstrate that AgBR1 plays a role in recruiting CD45+CD11b+Ly6G+ cells to the Zika virus infected-mosquito bite site.
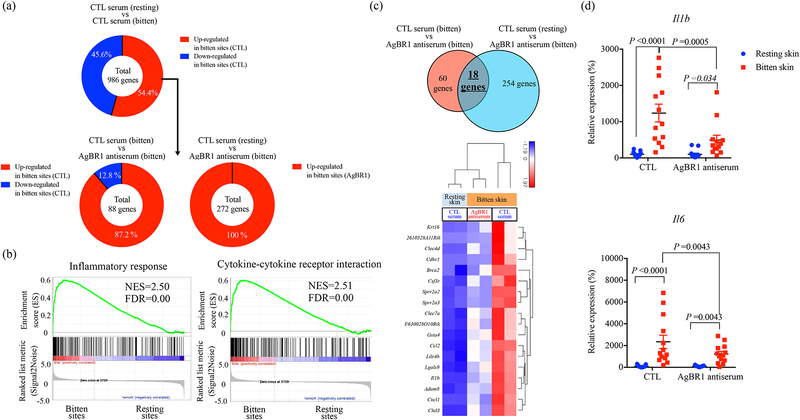
To further understand how AgBR1 may influence mosquito-borne Zika infection, we performed RNA sequencing on tissue collected at the bite site of mice 24 h after Zika virus-infected mosquito feeding. We found 536 upregulated genes out of 986 differentially expressed genes between the bite site and resting site in control mice following Zika virus-infected mosquito feedings (Fig. 4a, Supplementary Table. 4). A variety of cytokine and chemokine genes, including neutrophil-attracting chemokines Cxcl1, proinflammatory cytokine Il1b, monocytic chemoattractive chemokines Ccl2 and Ccl6, were significantly upregulated at the bite site compared to the resting site. This result was consistent with a previous report describing a detrimental role for inflammatory neutrophils that express IL-1β in the induction of cutaneous inflammatory responses at the bite site7,24. GSEA analysis also revealed that inflammatory responses and cytokine signaling, which are mediated by host immune cells, were highly enriched in bitten skin, supporting our histological findings (Fig. 4b, Supplementary Table 5).
Figure 4. AgBR1 antiserum modulates host responses at the mosquito bite site.
(a) (Top panel) 536 genes (54.4 %) within 986 differentially expressed genes (P < 0.05) were upregulated at the bitten sites of mice administered control serum. (Bottom left panel) Among these 536 genes, 78 genes were significantly upregulated at the bitten site of mice administered control serum compared with mice injected with AgBR1 antiserum. (Bottom right panel) Among the 536 genes, 272 genes were differentially upregulated in bitten sites of mice administered AgBR1 antiserum compared with the resting sites of mice inoculated with control serum. (b) GSEA of inflammatory responses (Hallmark) and cytokine-cytokine receptor interaction (KEGG) pathway enriched at bite sites of mice compared with resting sites in control mice. NES, normalized enrichment score. (c) (Top panel) Venn diagram depicting the overlap of genes differentially expressed across the conditions. (Bottom panel) Heat map of hierarchical clustering performed on 18 upregulated genes across the conditions (Fold change >1.5, P < 0.05). (a-c) Control-resting skin: n=2, Control-bitten skin: n=2, AgBR1 antiserum-bitten skin: n=2 biologically independent samples. Normalized read counts were statistically modeled using Partek Flow’s Gene Specific Analysis (GSA) approach. (d) QRT-PCR based analysis of Il1b and Il6 expression, which is normalized to mouse β actin RNA levels. Each dot represents one bitten or control site. Data are presented as mean ± s.e.m.. Significance was determined by two-way ANOVA test. (Control-resting skin: n=13, Control-bitten skin: n=13, AgBR1 antiserum-resting skin: n=13, AgBR1 antiserum-bitten skin: n=13 biologically independent samples pooled from two separate experiments.)
Next, we sought to evaluate the impact of AgBR1 antiserum on inflammatory responses induced by Zika virus-infected mosquito bites. To this end, we focused on genes that were upregulated in the bite site and identified 18 genes, including Il1b, Cxcl1 and Ccl2, which were attenuated in mice inoculated with AgBR1 antiserum (Fig. 4a and c). The reduction of Il1b was also confirmed by qPCR (Fig. 4d). In addition, we examined Il6 expression levels in each group because AgBR1 stimulates Il6 expression in vitro. Il6 was initially not included in the differential gene expression analysis due to the low expression in control skin. Il6 expression levels were nonetheless significantly suppressed in AgBR1 antiserum-treated mice compared with control mice, consistent with the in vitro data (Fig. 4d). We also found that the direct inoculation of AgBR1 into the skin significantly induces Il1b and Il6 expression (Supplementary Fig. 10).
Arboviral infection triggers the recruitment of peripheral neutrophils and monocytes to the site of infection7,25 and, previous studies showed that neutrophils are important targets of flaviviruses in vivo and that infiltration of neutrophils contribute to the initial flavivirus infection and dissemination7,26,27. Here, we show that AgBR1 induces neutrophil recruitment at the bite site and blocking this effect suppresses the early host response (Fig. 3 and 4). These data suggest that targeting AgBR1 blocks the early host responses caused by the bite of Zika virus-infected mosquitoes, leading to the suppression of viral dissemination and protection against lethal Zika virus infection.
In order to mimic mosquito salivation, we performed intradermal injections of AgBR1. A recent study demonstrated that mosquito-borne Zika virus infection alters virus tissue tropism and replication kinetics compared with needle-inoculation28. In addition, the results using needle injection of recombinant AgBR1 protein are modest, compared with those using AgBR1 antiserum or in the context of mosquito bites (Figs. 1–2, Supplementary Figs. 6, 8, 9 and 10). These results implied the possibility that AgBR1 may enhance its functions in cooperation with other molecules in saliva, since mosquito saliva contains many components. Therefore, further elucidation of the synergistic effect of AgBR1 with other salivary factors or the role that AgBR1 plays during natural mosquito feedings will pave the way for understandings how AgBR1 influences the vertebrate host at the local site and for the development of novel vaccines against Zika virus that target antigens produced by the arthropod vector.
The development of a vaccine against Zika virus is strongly needed. Conventional human vaccines against infectious diseases are based on components of specific pathogens21, which often lead to the emergence of escape mutants29. On the other hand, since the approach of targeting an arthropod protein does not exert a direct effect on the vectors themselves, the emergence of resistant mosquitoes is unlikely to occur. In addition, an arthropod-based target may enhance the efficacy of future Zika virus-specific vaccines currently in development. This approach also offers a functional paradigm for developing vaccines against other flaviviruses and arthropod-borne pathogens of medical importance. Moreover, it is likely that there are functional redundancies in facilitating infection with multiple pathogens, and this approach could lead to the generation of vaccines that are effective against diverse viruses transmitted by A. aegypti.
Materials and Methods
Ethics statement
All experiments were performed in accordance with guidelines from the Guide for the Care and Use of Laboratory Animals of the NIH. The animal experimental protocols were approved by the Institutional Animal Care and Use Committee (IACUC) at the Yale University School of Medicine (Assurance number A3230–01). All infection experiments were performed in a biosafety level 2 animal facility, according to Yale University regulations. Every effort was made to minimize murine pain and distress. Mice were anesthetized with ketamine/xylazine for mosquito infection experiments and euthanized as suggested by the Yale IACUC.
Viruses and cell lines
Vero cells (ATCC) were maintained in DMEM containing 10% FBS and antibiotics at 37°C with 5% CO2. Aedes albopictus C6/36 cells were grown in DMEM supplemented with 10% FBS, 1% tryptose phosphate, and antibiotics at 30°C with 5% CO2. Drosophila S2 cells (ATCC) were passaged in Schneider’s Drosophila media with 10 % FBS at 28 °C. A Mexican strain of Zika virus (Accession number KX446950), MEX2–81, was propagated in C6/36 insect cells.
Mosquitoes and animals
A. aegypti (Ho Chi Minh strain, obtained from the J. Powell laboratory at Yale) mosquitoes were maintained on 10% sucrose feeders inside a 12” × 12” × 12” metal mesh cage (BioQuip #1450B) at 28°C and ~80% humidity. Egg masses were generated via blood meal feeding on naïve mice. All mosquitoes were housed in a warm chamber in a space approved for BSL2 and ACL3 research. Four to six-week old gender mixed Ifnαr1−/−Ifnγr−/− mice (AG129 – SV129 background) were used in the Zika virus infection studies30. Mice were randomly chosen for experimental groups. For isolating splenocytes, 5-week old male C57BL/6 mice were purchased from the Jackson Laboratory. All mice were kept in a pathogen-free facility at Yale University.
Yeast display screening
To prepare RNA for library construction, salivary glands from about 300 A. aegypti mosquitoes, which had previously fed on mice once, were harvested. RNA was purified with the RNeasy Mini Kit (QIAGEN) and the purity of collected RNA was validated by gel electrophoresis confirm the presence of 18S and 28S rRNA. The cDNAs were synthesized by a modified SMART™ cDNA synthesis kit according to protocols by Bio S&T Inc. (Quebec, Canada). After generating double strand cDNAs by primer extension and cDNA normalization, cDNAs were directionally cloned into the yeast expression vector pYD1 (Invitrogen, CA) to generate a salivary gland expression library (Invitrogen, CA). Digestion of plasmids purified from 10 clones of the pYD1-salivary gland library, showed an average insert size of 1.2 kb and 100% of the clones contained inserts. The total number of primary clones was over 1 million. Plasmid DNA was purified from the library using the QIAGEN Plasmid Midi Kit (QIAGEN, CA, USA). Growth of transformed yeast cells and induction of recombinant protein production was done as previously described14,31. Briefly, fresh Saccharomyces cerevisiae EBY100 cells (Invitrogen, CA) with 2 μg of plasmid DNA were electroporated and subsequently grown in SDCAA medium (2% dextrose, 0.67% yeast nitrogen base, 0.5% bacto amino acids, 30 mM NaHPO4, 62 mM NaH2PO4) overnight at 30°C with shaking at 200 rpm.
The induction of surface protein expression was performed as described previously14,31. In brief, transformed yeast cells were grown for 24 hours at 30°C in SGCAA medium (2% galactose, 0.67% yeast nitrogen base, 0.5% bacto amino acids, 30 mM NaHPO4, 62 mM NaH2PO4). After induction with galactose, selection was performed by MACS separation (Miltenyi Biotec, Auburn, CA). Induced yeast cells were incubated with purified IgG derived from mice repeatedly bitten by A. aegypti. For MACS separation, an LS column (Mitenyi Biotec Cat# 130-042-401) was placed onto the magnet and stand assembly. After washing the column, induced yeast cells were applied to the column. After passing through the column, bound yeast cells were eluted by removing the column from the magnet and adding SDCAA medium to the column. Then, eluted yeast cells were propagated for additional rounds of sorting. After 4 rounds of magnetic sorting, plasmids were recovered using a Zymoprep™ II Yeast Plasmid Miniprep kit (Zymo Research), transformed into E. coli DH5α-competent cells (Invitrogen, CA), and sequenced.
Purification of recombinant proteins and antiserum preparation
AgBR1 (AAEL001965), SP (AAEL003600) and D7Bclu (AAEL006417) were cloned in-frame into the pMT-Bip-V5-His tag vector (Invitrogen, CA) and recombinant proteins expressed and purified using the Drosophila Expression System (Invitrogen, CA) as described earlier14. AgBR1, SP and D7Bclu were purified from the supernatant by TALON Metal Affinity Resin (Clontech, CA) and eluted with 150 mM imidazole. The eluted samples were filtered through a 0.22-μm filter and concentrated with a 10-kDa concentrator (Sigma-Aldrich, MO) by centrifugation at 4°C, washed and dialyzed against PBS. Recombinant protein purities were assessed by SDS-PAGE and quantified using the BCA protein estimation kit (Thermo scientific, IL). The PCR primer sequences for cloning are listed in Supplementary Table 6.
To generate rabbit sera against recombinant proteins, rabbits were immunized subcutaneously with 80–150 μg of recombinant proteins in complete Freund’s adjuvant and boosted twice at every 2 weeks with 80–150 μg of recombinant proteins in incomplete Freund’s adjuvant. Rabbits were euthanized and sera were obtained by cardiac puncture 2 weeks after the final boost. Reactivity to recombinant proteins was examined by immunoblot and ELISA.
Enzyme-linked immunosorbent assay (ELISA)
Recombinant AgBR1, SP, D7Bclu, OVA, or salivary gland extracts in PBS (0.1 μg/50 μl/well) were coated on 96 well plates overnight at 4 °C. After being blocked with 2% non-fat milk for 1 h at room temperature, the plates were then incubated with serum samples serially diluted in PBS for 1 h at room temperature. After being washed with PBS plus 0.05% Tween-20 (PBS-T) (Sigma) three times, the plates were incubated with HRP-conjugated secondary antibodies. Enzyme activity was detected by incubation with 100 μl of 3,3’,5,5’-Tetramethylbenzidine solution (KPL, USA) for 15 min at room temperature in the dark. The reaction was stopped by the addition of 1M H2SO4. The optical density (OD) at 450 nm was measured with a microplate reader.
Immunoblot
Recombinant proteins, BSA or salivary gland extracts were separated by SDS-PAGE using 4–20% Mini-Protean TGX gels (Bio-Rad) at 200 V for 25 min. Proteins were transferred onto a PVDF membrane for 60 min at 4 V. Blots were blocked in 1% non-fat milk in water for 60 min. Primary antibodies were diluted in 0.05% PBS-T and incubated with the blots for 1 h at room temperature or 4 °C overnight. HRP-conjugated secondary antibodies were diluted in PBS-T and incubated for 1 h at room temperature. After washing by PBS-T, the immunoblots were imaged with a LI-COR Odyssey imaging system.
Splenocyte stimulation with recombinant AgBR1
Splenocytes were isolated from C57BL/6 mice. Briefly, the spleens were minced in RPMI 1640 (Sigma-Aldrich) and forced gently through a 70 μm cell-strainer nylon mesh using a sterile syringe plunger and centrifuged at 400 g for 5 min. After washing once using cold PBS, spleen cells were incubated in 2 ml 0.83% NH4Cl for 5 min, then placed in 20 ml PBS, centrifuged at 400 g for 5 min and then resuspended in RPMI 1640. The total number of cells was calculated using a hemocytometer. Isolated splenocytes were stimulated with 5 μg/ml recombinant AgBR1 or BSA and cultured with serum-free RPMI medium for 6 h and 24 h. Total RNA was extracted by the RNeasy Mini Kit (QIAGEN) according to the instructions. The cDNA was generated with iScript cDNA Synthesis Kit (Bio-rad) according to manufacturer’s protocol. Gene expression was examined by quantitative RT-PCR (qRT-PCR) using IQ™ SYBR Green Supermix. Target gene mRNA levels were normalized to mouse β actin RNA levels according to the 2−ΔΔCt calculations. The qRT-PCR primer sequences are listed in Supplementary Table 6.
Co-inoculation of Zika virus with AgBR1
AG129 mice were inoculated via subcutaneous (footpad) with 3 PFU of Zika virus along with 10 μg AgBR1 (total volume; 40 μl). Survivals were monitored everyday post-infection. Mice exhibiting neurologic disease such as paralysis or weight loss of over 20% of body weight were euthanized. The weight loss is very rapid and usually begins three days before death, coinciding with neurological symptoms. Total RNA from murine blood was extracted in TRIzol Reagent and qRT-PCR was performed to examine Zika virus levels as previously described32.
Passive or active immunization studies
Zika virus injection was performed as previously described30. Briefly, Zika virus-filled needles were carefully inserted into the thorax of each mosquito and 69 nl of virus (100 PFU) was injected using a Nanoject II auto-nanoliter injector (Drummond). Infected mosquitoes were placed back in paper cups with mesh lids and maintained in triple containment for 10 days in a warm chamber. Mosquitoes were knocked-down on ice and salivary glands were dissected to examine the virus levels after mosquito feeding. RNA from salivary glands was purified with the RNeasy Mini Kit (QIAGEN), and cDNA was generated with iScript cDNA Synthesis Kit (Bio-rad) according to manufacturer’s protocol. Gene expression was examined by quantitative RT-PCR (qRT-PCR) using IQ™ SYBR Green Supermix. Viral RNA levels were normalized to mosquito Rp49 RNA levels according to the 2−ΔΔCt calculations.
For passive rabbit antiserum transfer experiments, mice were injected intraperitoneally with 150 μl per animal of antiserum against specific mosquito proteins or naive rabbit serum one day before challenge. On the same day, two infected mosquitoes were randomly aliquoted into individual cups with mesh covers. On the following day, mice were anesthetized with ketamine-xylazine and fed on by two Zika virus-infected mosquitoes. For active immunization, mice were immunized subcutaneously with 10 μg of AgBR1 or ovalbumin in complete Freund’s adjuvant and boosted twice every 2 weeks with the same amount of AgBR1 or ovalbumin in incomplete Freund’s adjuvant. Two weeks after the final immunization, mice were anesthetized with ketamine-xylazine and fed on by two Zika virus infected mosquitoes. The blood of fed mice was collected at 1, 3, 5, 7 and 9 days post infection. Survivals were monitored every day. Mice exhibiting weight loss of > 20% of initial body weight or neurologic disease were euthanized. The weight loss is very rapid and usually begins three days before death, coinciding with neurological symptoms. Viremia levels were examined at 1, 3, 5, 7, 9 days post infection (dpi) as described above.
Needle inoculation of Zika virus after passive immunization
Mice were injected intraperitoneally with control serum or AgBR1 antiserum one day before challenge. On the following day, mice were inoculated via subcutaneous footpad injection with 0.3 PFU of Zika virus. Survivals were monitored everyday post-infection. Mice exhibiting weight loss of > 20% of initial body weight or neurologic disease were euthanized. The weight loss is very rapid and usually begins three days before death, coinciding with neurological symptoms. Viremia levels were examined at 1, 3, 5, 7, 9 dpi as described above.
Gene silencing
RNA interference of genes expressed in the mosquito SGs was performed as previously described. Double stranded (ds) RNA targeting either a 400 bp region of the AgBR1 gene or an irrelevant green fluorescent protein (GFP) gene were transcribed using gene-specific primers designed with a T7 promoter and the MEGAScript RNAi kit (Thermo Fisher Scientific, Ambion). The primers for generating dsRNA are listed in Supplementary Table 6. For silencing the AgBR1 gene, adult female A. aegypti mosquitoes were kept on ice and then transferred to a cold tray to receive a dsRNA injection. Two hundred ng of dsRNA in PBS were microinjected into the thorax of each mosquito using a Nanoject II Auto-Nanoliter Injector (Drummond). At day 3 post dsRNA injection, mosquitoes were knocked-down on ice and injected with Zika virus described above. At day 10 post virus injection, salivary glands were dissected to examine the AgBR1 expression levels by qRT-PCR and AgBR1 protein production by immunoblot.
Analysis of local immune responses after bites of Zika virus infected mosquitoes or intradermal injection
For the analysis of local immune responses after bites of Zika virus infected mosquitoes, AG129 mice were allowed to be fed on the left ear by Zika virus-infected A. aegypti mosquitoes.
For the analysis of local immune responses after intradermal injection, AgBR1 was intradermally injected into the left ear. Briefly, the ear of an individual mouse was gently immobilized over a14 ml falcon tube covered with double stick tape. Five hundred nanoliters containing 0.5 μg/μl were injected intradermally into the dorsal ear using glass micropipettes with a 80 μm diameter beveled opening made as described elsewhere33 and a Nanoject II Auto-Nanoliter Injector (Drummond).
One day later, mice were euthanized and the locations bitten by mosquitoes or intradermally-injected locations and naïve skins were punched using a Disposable Biopsy Punch. Total RNA was extracted by the RNeasy Fibrous Tissue Mini Kit (QIAGEN) according to the manufacturer’s instructions.
For qRT-PCR, the cDNA generation and analysis of gene expression was conducted as described above. Gene expression was queried using IQ™ SYBR Green Supermix. Target gene mRNA levels were normalized to mouse β actin RNA levels according to the 2−ΔΔCt calculations. The qRT-PCR primer sequences are listed in Supplementary Table 6.
For RNA-seq library preparation and sequencing, barcoded libraries were generated by standard Truseq mRNA library protocol (Illumina) and sequenced with a 2 × 75 bp paired-end protocol on the HiSeq 4000 Sequencing System (Illumina).
All the analysis of RNA-seq data was performed using Partek flow (v7.0). RNA-seq data were trimmed and mapped to a mm10 genome reference using STAR (2.5.0e). The aligned reads were quantified to ENSEMBL transcripts release 91 using the Partek’ E/M algorithm and the subsequent steps were performed on gene-level annotation followed by total count normalization. The gene-level data were normalized by dividing the gene counts by total number of reads followed by addition of a small offset (0.001). Differential expression was assessed by fitting the Partek’s log-normal model with shrinkage (comparable in performance to limma-trend). Genes having geometric mean below value of 1.0 were filtered out from the analysis. Hierarchal clustering was performed on the genes, which were differentially expressed across the conditions (P value <0.05, fold change > 1.5 for each comparison). Gene set enrichment analysis (GSEA) was performed on normalized gene expression counts of RNA-seq data as described previously34. Gene sets with an estimated false discovery rate (FDR) of <0.05 were considered significant according to the GSEA guidelines.
Histopathology
Ear skins of the bite site and non-bite site on the contralateral ear were harvested by punch biopsy, fixed in 4% paraformaldehyde/PBS, paraffin embedded, and processed for hematoxylin and eosin staining. The histological findings were scored for the severity and character of the inflammatory response using a blinded grading scale that was previously described35, with minor modifications. Responses were graded as follows: 0, no response; 1, minimal response; 2, mild response; 3, moderate response; and 4, marked response. The responses were evaluated and graded on the histological sites with the most prominent responses in each specimen. The total histology score was calculated as the sum of scores, including inflammation, neutrophils, mononuclear cells and edema. The slides were blinded, randomized, and reread to determine the histology score by the same dermatopathologist throughout all studies.
Imaging Mass Cytometry (IMC)
IMC was performed on slides dewaxed in xylene for 20 min according to previously described studies36. After hydration in sequential concentrations of ethanol (100%, 95%, 80%, 70%) for 5 min, slides were incubated with antigen retrieval solution at 90–95°C for 20 min. Slides were then cooled to room temperature and washed with ddH2O and PBS (lacking Ca++ or Mg++) for 5 min. After blocking with 3% BSA in PBS for 45 min, slides were labeled with metal-conjugated antibodies against CD3 (170Er - Polyclonal, C-Terminal), CD11b (149Sm - EPR1344), MHCII (174Yb - M5/114.15.2) and Ly6G (141Pr – 1A8) diluted in PBS with 0.5% BSA at 4 °C overnight. After being washed with 0.1% Triton-X in PBS and then PBS, slides were labeled with intercalator-Ir (1:2,000 dilution) in PBS (lacking Ca++ or Mg++) for 30 min at room temperature. After being washed again with ddH2O for 5 min, the slides were dried. Tissues were laser ablated using a 200 Hz Hyperion™ Imaging System (Fluidigm Corp., South San Francisco, CA and the aerosol containing the ion cloud was directly transported to a Helios Mass Cytometer (Fluidigm). Images of labeled slides were obtained using the MCD viewer 1.0 (https://www.fluidigm.com/software).
Analysis of immune cells in mice following Zika virus-infected mosquito bites or intradermal injection.
For the analysis of immune cells following Zika virus-infected mosquito bites, AG129 mice were allowed to be fed by Zika virus-infected A. aegypti mosquitoes on the ear.
For the analysis of immune cells following AgBR1 injection, AG129 mice were intradermally injected with AgBR1 in the ear as described above.
After 24 hours, mice were sacrificed and both the bitten or injected and naive ears were cut off at the base and split into dorsal and ventral halves. Ears were incubated for 1.5 hours in 2 mg/ml of Dispase II (Sigma) in DMEM media with 10% FBS, and then cut into small pieces. Small pieces were then digested for 1.5 hours in 5 mg/ml of collagenase (Gibco) in media. Digested samples were then individually passed through 70 μM filters to obtain single-cell suspensions.
After washing once with PBS containing 2% FBS (FACS buffer), cells were stained using the LIVE/DEAD™ Fixable Violet Dead Cell Stain Kit (Thermo Fisher) and then incubated with fluorochrome-conjugated monoclonal antibodies against CD45 (PerCP - BD Pharmingen; Clone 30-F11), MHCII (APC-Cy7 – Biolegend; Clone M4/114.15.2), CD11b (PE – Biolegend; Clone M1/70), CD11c (PE-Cy7 – BD Pharmingen; Clone HL3), and Ly6G (FITC – Tonbo; Clone RB6–8C5) for 30 min at room temperature, washed twice with FACS buffer. Samples were run on a BD LSRII flow cytometer and analyzed using FlowJo software.
Statistical analysis
GraphPad Prism software was used to analyze all the data. Animals were randomly allocated into different groups. No statistical methods were used to predetermine sample size. Rp49 and mouse β actin normalized viral RNA levels were analyzed using the two-sided Wilcoxon–Mann–Whitney test. Host responses in vitro and in vivo was performed using a two-way ANOVA for multiple comparisons or using the two-sided Wilcoxon–Mann–Whitney or the two-sided Wilcoxon matched-pairs signed rank test for two sample comparisons, as indicated in the figure legends. Survival was assessed by a Gehan-Wilcoxon test. A p value of <0.05 was considered statistically significant.
Supplementary Material
Acknowledgements
We thank Dr. Sujan Shresta at the La Jolla Institute for Allergy and Immunology for originally providing us with the AG129 mouse strain. We also thank Shelly Ren, Yu-Min Chuang, Sarah Householder, Sydney Stanley, Hannah Sproch and Brent Vander Wyk for supporting experiments and analyzing data. The imaging mass cytometry was conducted at the Yale CyTOF facility, and the RNA-sequencing service was conducted at the Yale Stem Cell Center Genomics Core facility which was supported by the Connecticut Regenerative Medicine Research Fund and the Li Ka Shing Foundation. This work was supported by NIH grants AI089992 and AI127865, and the Japan Society for the Promotion of Science Overseas Research Fellowships. E. F. and A.I. are investigators with the Howard Hughes Medical Institute.
Footnotes
Competing Interests
The authors declare no competing interests.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Data availability
The data that support the findings of this study are available from the corresponding authors upon request. The RNA sequencing data have been deposited in the Gene Expression Omnibus under accession numbers GSE125194.
References
- 1.Ventura CV, Maia M, Bravo-Filho V, Gois AL & Belfort R Jr. Zika virus in Brazil and macular atrophy in a child with microcephaly. Lancet 387, 228 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Olsen B & Lundkvist A [Zika virus - ancient virus gets new life in a new ecosystem. Microcephaly and Guillain-Barre syndrome are possible consequences when there is no background herd immunity in the population]. Lakartidningen 113(2016). [PubMed] [Google Scholar]
- 3.Musso D & Gubler DJ Zika Virus. Clin Microbiol Rev 29, 487–524 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coutinho-Abreu IV, Guimaraes-Costa AB & Valenzuela JG Impact of Insect Salivary Proteins in Blood Feeding, Host Immunity, Disease, and in the Development of Biomarkers for Vector Exposure. Curr Opin Insect Sci 10, 98–103 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fontaine A, et al. Implication of haematophagous arthropod salivary proteins in host-vector interactions. Parasit Vectors 4, 187 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hayashi H, et al. Anopheline anti-platelet protein from a malaria vector mosquito has anti-thrombotic effects in vivo without compromising hemostasis. Thromb Res 129, 169–175 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Pingen M, et al. Host Inflammatory Response to Mosquito Bites Enhances the Severity of Arbovirus Infection. Immunity 44, 1455–1469 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruckert C & Ebel GD How Do Virus-Mosquito Interactions Lead to Viral Emergence? Trends Parasitol 34, 310–321 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conway MJ, et al. Mosquito saliva serine protease enhances dissemination of dengue virus into the mammalian host. J Virol 88, 164–175 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conway MJ, et al. Aedes aegypti D7 Saliva Protein Inhibits Dengue Virus Infection. PLoS Negl Trop Dis 10, e0004941 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin L, et al. Salivary factor LTRIN from Aedes aegypti facilitates the transmission of Zika virus by interfering with the lymphotoxin-beta receptor. Nat Immunol (2018). [DOI] [PubMed] [Google Scholar]
- 12.Wasinpiyamongkol L, et al. Blood-feeding and immunogenic Aedes aegypti saliva proteins. Proteomics 10, 1906–1916 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Rabilloud T, Chevallet M, Luche S & Lelong C Two-dimensional gel electrophoresis in proteomics: Past, present and future. J Proteomics 73, 2064–2077 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Schuijt TJ, et al. Identification and characterization of Ixodes scapularis antigens that elicit tick immunity using yeast surface display. PLoS One 6, e15926 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee CG, et al. Role of Chitin and Chitinase/Chitinase-Like Proteins in Inflammation, Tissue Remodeling, and Injury. Annual Review of Physiology, Vol 73 73, 479–501 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ribeiro JM, et al. An annotated catalogue of salivary gland transcripts in the adult female mosquito, Aedes aegypti. BMC Genomics 8, 6 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beatty PR, et al. Dengue virus NS1 triggers endothelial permeability and vascular leak that is prevented by NS1 vaccination. Science Translational Medicine 7(2015). [DOI] [PubMed] [Google Scholar]
- 18.Tanaka T & Kishimoto T The Biology and Medical Implications of Interleukin-6. Cancer Immunol Res 2, 288–294 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Calvo E, Mans BJ, Andersen JF & Ribeiro JM Function and evolution of a mosquito salivary protein family. J Biol Chem 281, 1935–1942 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Rathore APS & St John AL Immune responses to dengue virus in the skin. Open Biol 8(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kieny MP, Excler JL & Girard M Research and development of new vaccines against infectious diseases. Am J Public Health 94, 1931–1935 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sarathy VV, Milligan GN, Bourne N & Barrett AD Mouse models of dengue virus infection for vaccine testing. Vaccine 33, 7051–7060 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu J, Huang X & Yang Y Type I IFN signaling on both B and CD4 T cells is required for protective antibody response to adenovirus. J Immunol 178, 3505–3510 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Fong SW, Kini RM & Ng LFP Mosquito Saliva Reshapes Alphavirus Infection and Immunopathogenesis. J Virol 92(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi C & Pamer EG Monocyte recruitment during infection and inflammation. Nat Rev Immunol 11, 762–774 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Osuna CE, et al. Zika viral dynamics and shedding in rhesus and cynomolgus macaques. Nat Med 22, 1448–1455 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bai F, et al. A paradoxical role for neutrophils in the pathogenesis of West Nile virus. J Infect Dis 202, 1804–1812 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dudley DM, et al. Infection via mosquito bite alters Zika virus tissue tropism and replication kinetics in rhesus macaques. Nat Commun 8, 2096 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carman WF, et al. Vaccine-induced escape mutant of hepatitis B virus. Lancet 336, 325–329 (1990). [DOI] [PubMed] [Google Scholar]
- 30.Uraki R, Hastings AK, Gloria-Soria A, Powell JR & Fikrig E Altered vector competence in an experimental mosquito-mouse transmission model of Zika infection. PLoS Negl Trop Dis 12, e0006350 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chao G, et al. Isolating and engineering human antibodies using yeast surface display. Nat Protoc 1, 755–768 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Uraki R, et al. Zika virus causes testicular atrophy. Sci Adv 3, e1602899 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Balaban AE, Neuman K, Sinnis P & Balaban RS Robust fluorescent labelling of micropipettes for use in fluorescence microscopy: application to the observation of a mosquito borne parasite infection. J Microsc 269, 78–84 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakajima S, et al. Prostaglandin I2-IP signaling promotes Th1 differentiation in a mouse model of contact hypersensitivity. J Immunol 184, 5595–5603 (2010). [DOI] [PubMed] [Google Scholar]
- 36.Chang Q, Ornatsky O & Hedley D Staining of Frozen and Formalin-Fixed, Paraffin-Embedded Tissues with Metal-Labeled Antibodies for Imaging Mass Cytometry Analysis. Curr Protoc Cytom 82, 12 47 11–12 47 18 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.