SUMMARY PARAGRAPH
Lack of reproducibility is a prominent problem in biomedical research. An important source of variation in animal experiments is the microbiome, but little is known about specific changes in the microbiota composition that cause phenotypic differences. Here we show that genetically similar laboratory mice obtained from four different commercial vendors exhibited marked phenotypic variation in their susceptibility to Salmonella infection. Fecal microbiota transplantation into germ-free mice replicated donor susceptibility, revealing that variability was due to changes in the gut microbiota composition. Co-housing of mice only partially transferred protection against Salmonella infection, suggesting that minority species within the gut microbiota might confer this trait. Consistent with this idea, we identified endogenous Enterobacteriaceae, a low abundance taxon, as keystone species responsible for variation in the susceptibility to Salmonella infection. Protection conferred by endogenous Enterobacteriaceae could be modeled by inoculating mice with probiotic Escherichia coli, which conferred resistance by using its aerobic metabolism to compete with Salmonella for resources. We conclude that a mechanistic understanding of phenotypic variation can accelerate development of strategies for enhancing the reproducibility of animal experiments.
A recent survey suggests that a majority of researchers have tried and failed to reproduce their own experiments or experiments from other scientists1. In animal experimentation this lack of reproducibility can be due to an unappreciated variation in the microbiota2,3. It has been suggested that a lack of reproducibility in animal experimentation could be addressed by co-housing animals or by comparing the gut microbiota of animals exhibiting differing phenotypes3. However, neither of these approaches is effective in addressing phenotypic variation caused by keystone species present in low abundance within the gut microbiota2. Instead, a return to a reductionist approach seems necessary to pinpoint how specific changes in the microbiota composition contribute to phenotypic variation4. Specific pathogens or pathobionts, including Helicobacter hepaticus, segmented filamentous bacteria (SFB), or enteric viruses, can contribute to phenotypic variation by inducing immune responses through microbe-host interactions3. For example, colonization with SFB induces differentiation of interleukin-22-producing T cells to confer resistance to Citrobacter rodentium infection5. Infection of laboratory mice with Salmonella enterica serovar (S.) Typhimurium is a commonly used model in bacterial pathogenesis research6. Induction of innate immune responses by viruses or the gut microbiota can increase resistance of mice to S. Typhimurium infection7,8. A disruption of the gut microbiota by antibiotic treatment increases susceptibility to S. Typhimurium infection9, suggesting that the resident microbial community is required for protection. However, it is not known whether more subtle changes in the microbiota composition, such as those observed between mice obtained from different vendors, could alter susceptibility to infection.
RESULTS
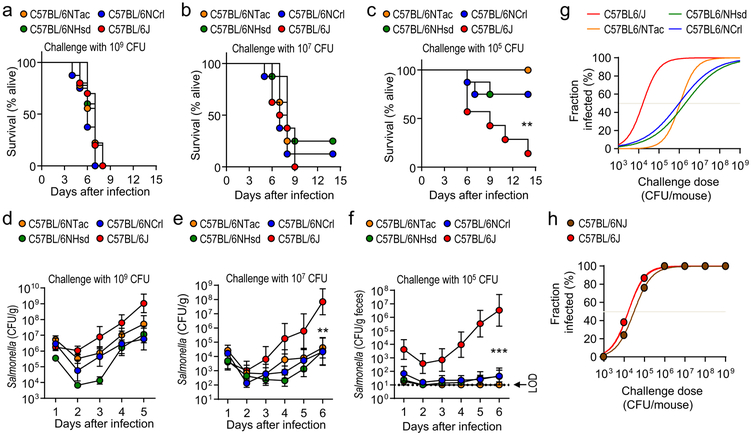
To determine if genetically similar strains of mice obtained from different commercial sources varied in their susceptibility to S. Typhimurium infection, C57BL/6 mice were obtained from Charles River Laboratories (C57BL/6NCrl), Harlan (C57BL/6NHsd), Taconic Farms (C57BL/6NTac), and Jackson Laboratories (C57BL/6J). While all mice inoculated intragastrically with high-doses of S. Typhimurium developed lethal morbidity (Fig. 1a and 1b), a low-dose challenge (105 colony-forming units [CFU]/animal) caused greater lethal morbidity in C57BL/6J mice than mice from other vendors (Fig. 1c). Susceptibility to lethal S. Typhimurium infection correlated with intestinal carriage, which developed in all mice challenged with high-doses of S. Typhimurium (Fig. 1d and 1e), but after a low-dose challenge (105 CFU/animal) most C57BL/6J mice became colonized whereas most mice obtained elsewhere did not (Fig. 1f). To investigate whether these differences in susceptibility varied with time, we repeated the experiment one year later, at which point Harlan had been integrated into Envigo. Differences in susceptibility to lethal S. Typhimurium infection and development of intestinal carriage were similar to those observed one year earlier (Supplementary Fig. 1 and 2), suggesting that variability was linked to differences between vendors.
Figure 1: Phenotypic variation in the susceptibility to Salmonella infection is observed in C57BL/6 mice from different vendors.
(a-g) Mice from Charles River Laboratories (C57BL/6NCrl), Harlan (C57BL/6NHsd), Taconic Farms (C57BL/6NTac) or Jackson Laboratories (C57BL/6J) were challenged with S. Typhimurium (n = ≥7). Lethal morbidity (a-c) and S. Typhimurium shedding with the feces (d-f) were monitored at the indicated time points after challenge with 109 CFU (a and d), 107 CFU (b and e) or 105 CFU (c and f) per animal. (g) Fraction of animals developing intestinal carriage at different S. Typhimurium challenge doses. (d-f) Dots represent geometric means ± standard error of the mean. (h) Two C57BL/6 substrains (C57BL/6J and C57BL/6NJ) from Jackson Laboratories were challenged with different S. Typhimurium doses and the fraction of animals developing intestinal carriage determined (n = 3–8 per dose). LOD, limit of detection; **, P ≤ 0.01; ***, P ≤ 0.001.
Combining data from both experiments suggested that the dose at which intestinal carriage with S. Typhimurium developed in 50% of animals (ID50) was approximately 100-fold higher in mice belonging to the C57BL/6N substrain than in C57BL/6J mice (Fig. 1g). Therefore, we hypothesized that genetic differences between substrains of C57BL/6 mice were responsible for phenotypic variation. The C57BL/6J substrain diverged in 1951 from the C57BL/6N substrain at the National Institutes of Health (NIH), which was subsequently passed to Charles River Laboratories in 1974 (C57BL/6NCrl), to Harlan in 1983 (C57BL/6NHsd) and to Taconic Farms in 1991 (C57BL/6NTac)10,11. Genetic drift produced a range of phenotypic differences between the C57BL/6J and C57BL/6N substrains12. To test our hypothesis, we obtained a C57BL/6N substrain that was established at Jackson laboratories in 1997 from frozen embryos provided by the NIH (C57BL/6NJ)12. Notably, C57BL/6NJ and C57BL/6J mice were equally susceptible to S. Typhimurium infection (Fig. 1h and Suppl Fig. 3), which did not support the idea that genetic differences between substrains of C57BL/6 mice were responsible for variation in the S. Typhimurium disease resistance phenotype.
Differences in S. Typhimurium susceptibility are caused by gut microbiota variation
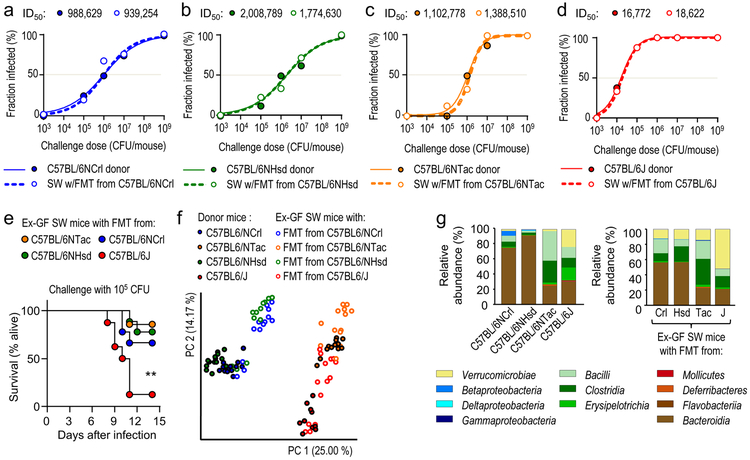
To determine whether the gut microbiota was an environmental factor causing phenotypic variation, fecal microbiota from C57BL/6NCrl mice, C57BL/6NHsd mice, C57BL/6NTac mice or C57BL/6J mice was transferred to germ-free Swiss Webster mice. After allowing the Swiss Webster recipients to adjust to the fecal microbiota transplant, mice were challenged orally with different doses of S. Typhimurium. Swiss Webster recipients of fecal microbiota transplants phenocopied the respective C57BL/6 donor mice in their susceptibility to intestinal S. Typhimurium carriage (Fig. 2a-d and Suppl Fig. 4), suggesting that vendor-linked variability in the gut microbiota was a dominant source of phenotypic variation. Furthermore, a low-dose challenge (105 CFU/animal) caused greater lethal morbidity in Swiss Webster mice with a fecal microbiota transplant from C57BL/6J mice than Swiss Webster mice with fecal microbiota transplants from other vendors (Fig. 2e).
Figure 2: The gut microbiota is a driver of phenotypic variation.
(a-d) Solid lines and closed circles: Mice from Charles River Laboratories (C57BL/6NCrl) (a), Harlan (C57BL/6NHsd) (b), Taconic Farms (C57BL/6NTac) (c) or Jackson Laboratories (C57BL/6J) (d) were challenged with S. Typhimurium (n = 3–11 per dose). Closed circles indicate the fraction of animals developing intestinal carriage at different S. Typhimurium challenge doses. Dashed lines and open circles: Germ-free Swiss Webster (SW) mice received a fecal microbiota transplant from Charles River mice (C57BL/6NCrl) (a), Harlan mice (C57BL/6NHsd) (b), Taconic mice (C57BL/6NTac) (c) or Jackson mice and were subsequently challenged with the indicated S. Typhimurium challenge doses (n = 3–11 per dose). Open circles indicate the fraction of animals developing intestinal carriage at different S. Typhimurium challenge doses. The S. Typhimurium ID50 for each group of donor mice (C57BL/6NCrl, C57BL/6NHsd, C57BL/6NTac, or C57BL/6J) and each corresponding group of ex-germ-free Swiss Webster recipient mice is indicated above each graph. (e) Germ-free Swiss Webster (SW) mice received fecal microbiota transplants from the indicated donor mice (ex-germ-free SW mice) and were subsequently challenged with S. Typhimurium (n is shown in panel f). Lethal morbidity was monitored at the indicated time points after challenging ex-germ-free SW mice with 105 CFU per animal. (f) Principle coordinate analysis of fecal bacterial communities from the indicated animal groups. Each dot represents data from one animal (n = 9–15 per group). (g) 16S profiling of bacterial communities from the indicated animal groups at the class level. Each bar represents the average relative abundance of animals shown in Figure S5. LOD, limit of detection; **, P ≤ 0.01.
Clostridia are a dominant taxon within the gut microbiota and the obligate anaerobic bacteria forming this class are important for colonization resistance against S. Typhimurium 13,14. To test the hypothesis that differences in the abundance of Clostridia might explain phenotypic variation, we performed 16S ribosomal RNA gene sequencing (microbiota profiling). Principal coordinate analysis revealed differences in the microbiota composition of mice obtained from different vendors, whereas fecal microbial communities from Swiss Webster mice receiving a fecal microbiota transplant resembled those of the respective C57BL/6 donor mice (Fig. 2f). However, microbiota profiling did not support the idea that increased resistance to S. Typhimurium infection was associated with an increased abundance of Clostridia (Fig. 1g and Suppli.Fig. 5 Neither did linear discriminant analysis identify any biomarkers of protection that had a consistently elevated relative abundance in resistant mice (C57BL/6NCrl, C57BL/6NHsd or C57BL/6NTac) compared to susceptible mice (C57BL/6J) (Suppl.Fig. 6).
Commensal Enterobacteriaceae are biomarkers for protection against Salmonella
To test whether bacterial taxa conferring protection in C57BL/6NCrl mice could be transferred to susceptible C57BL/6J mice, we co-housed these mice for 14 days. Principle coordinate analysis of bacterial communities suggested that co-housing changed the fecal microbiota composition of C57BL/6J mice (Fig. 3a), but increased resistance to S. Typhimurium challenge in only a fraction of C57BL/6J mice (Fig. 3b). A possible explanation for this observation was that conventional mice might impose a bottleneck on transfer of minority species within the fecal microbiota that might be responsible for phenotypic variation. To identify potential biomarkers associated with phenotypic variation after co-housing, we performed linear discriminant analysis of the microbiota composition from C57BL/6J mice prior to co-housing and the same animals after 14 days of co-housing. This analysis suggested that increased resistance to S. Typhimurium infection was associated with an increased abundance of Deferribacteraceae (phylum Deferribacteres), candidate phylum Saccharibacteria and Enterobacteriaceae (phylum Proteobacteria) (Fig. 3c). The latter taxon was of particular interest, because inoculation with probiotic Escherichia coli (family Enterobacteriaceae) strains can reduce the severity of S. Typhimurium infection in animal models15,16.
Figure 3: Enterobacteriaceae are biomarkers of phenotypic variation.
(a) Mice from Jackson Laboratories (C57BL/6J) were housed normally or cohoused with mice from Charles River (C57BL/6NCrl) and fecal bacterial communities analyzed by principle coordinate analysis (n = 7–8). (b) Mice from Jackson Laboratories (C57BL/6J) were housed normally (cohoused: no) or cohoused with mice from Charles River (C57BL/6NCrl) (cohoused: yes) and subsequently challenged with S. Typhimurium. S. Typhimurium shedding with the feces was quantified 5 days after challenge. Whiskers represent minimum to maximum points and the box extends from the 25th to 75th percentile with median plotted by a line. (c) The cladogram shows differences in taxa composition between Jackson mice (C57BL/6J) prior to co-housing and after 14 days of co-housing with Charles River mice (C57BL/6NCrl). (d) The presence of Enterobacteriaceae in mice from the cohousing experiment shown in panel a was determined by spreading dilutions of fecal homogenates on MacConkey agar plates prior to S. Typhimurium challenge. S. Typhimurium shedding with the feces was quantified 5 days after challenge. (e-f) Mice from Jackson Laboratories (C57BL/6J) were mock-treated or received Enterobacteriaceae from the indicated donor mice. (e) The presence of Enterobacteriaceae in feces of mice prior to S. Typhimurium challenge was determined by spreading dilutions on MacConkey agar plates (n = ≥11). The graph shows variation between animals (dots) and the geometric mean (line). (f) Mice were challenged with S. Typhimurium. Lethal morbidity was monitored over time after challenge with 105 CFU per animal. (a, b, d, and e) Each dot represents data from one animal, thus indicating the number (n) of repeats. LOD, limit of detection; ns, P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
We thus attempted to culture Enterobacteriaceae from the co-housing experiment on MacConkey agar, which revealed that feces of C57BL/6NCrl mice contained culturable bacteria, but these minority species were absent from C57BL/6J mice. Curiously, culturable bacteria had not been transferred during cohousing experiments to those C57BL/6J mice that remained susceptible to S. Typhimurium challenge (Fig. 3d). In contrast, C57BL/6J mice that acquired S. Typhimurium resistance during cohousing had also acquired bacteria that formed lactose positive and lactose negative colonies (Fig. 3d and Suppl. Fig. 7a), which persisted after separating mice again (Suppl. Fig. 7b). We recovered between 105 to 107 CFU/g feces on MacConkey agar from the feces of C57BL/6NCrl, C57BL/6NTac or C57BL/6NHsd mice, which was below the limit of detection by conventional microbiota profiling (Suppl. Fig. 8a). In contrast, we detected no increase in the number of bacteria culturable on Lactobacillus agar (MRS agar) in C57BL/6NCrl, C57BL/6NTac or C57BL/6NHsd mice compared to C57BL/6J mice (Suppl. Fig. 7c). Importantly, bacteria culturable on MacConkey agar had been efficiently transferred to germ-free Swiss Webster mice through fecal microbiota transplantation (Suppl. Fig. 8b). Collectively, these data suggested that the presence of bacteria culturable on MacConkey agar was associated with protection against S. Typhimurium infection.
Commensal Enterobacteriaceae are keystone species responsible for phenotypic variation
To determine causality, C57BL/6J mice were inoculated with mixtures of bacteria culturable on MacConkey agar, selected based on differences in colony morphology from resistant mice. C57BL/6J recipient mice shed bacteria culturable on MacConkey agar (Fig. 3e) at numbers similar to those observed in the respective donor mice (Suppl. Fig. 8a). Remarkably, transfer of bacteria rendered C57BL/6J recipient mice resistance to subsequent S. Typhimurium challenge, as indicated by reduced lethal morbidity (Fig. 3f), reduced pathogen burden in the feces (Suppl. Fig. 7d-g and 8c) and a reduced ID50 (Suppl. Fig. 8d). Next, we determined the identity of bacteria culturable on MacConkey agar by sequencing their 16S ribosomal RNA genes. This analysis revealed that all bacteria isolated on MacConkey agar were members of the family Enterobacteriaceae. Lactose-negative enteric bacteria included Proteus vulgaris and P. mirabilis, whereas lactose-positive colonies represented Klebsiella oxytoca, Enterobacter cloacae and E. coli isolates (Suppl. Fig. 8e). Next, C57BL/6J mice were colonized with individual Enterobacteriaceae isolates. With the exception of K. oxytoca isolate Tac148, all isolates colonized C57BL/6J recipient mice well (Suppl. Fig. 8f) and trended to confer protection against subsequent S. Typhimurium challenge, although this only reached statistical significance for E. coli Crl141 (Suppl. Fig. 8g). Collectively, these data supported the idea that commensal Enterobacteriaceae are keystone species responsible for phenotypic variation.
E. coli uses its aerobic metabolism to confer niche protection against Salmonella
To investigate the mechanism by which endogenous Enterobacteriaceae conferred protection, homogenized fecal pellets were inoculated with S. Typhimurium and incubated aerobically, which revealed that pathogen growth was slowed in fecal homogenates of resistant mice (C57BL/6NCrl, C57BL/6NHsd or C57BL/6NTac) compared to susceptible mice (C57BL/6J) (Suppl. Fig. 9a). S. Typhimurium growth inhibition by fecal homogenates from C57BL/6NCrl and C57BL/6NTac mice was not observed under anaerobic conditions (Suppl. Fig. 9a). Growth inhibition was not due to the presence of inhibitory metabolites, because S. Typhimurium grew equally well in sterile-filtered fecal homogenates from resistant or susceptible mice (Suppl. Fig. 9c). Homogenized fecal pellets from susceptible C57BL/6J mice spiked with P. vulgaris, P. mirabilis, E. cloacae, K. oxytoca or E. coli isolates from resistant mice suppressed S. Typhimurium growth (Suppl. Fig. 9d), suggesting that pathogen growth inhibition was due to the presence of Enterobacteriaceae. Interestingly, inhibition of pathogen growth by Enterobacteriaceae was no longer observed for most isolates when the experiment was repeated under anaerobic conditions (Suppl. Fig. 9e).
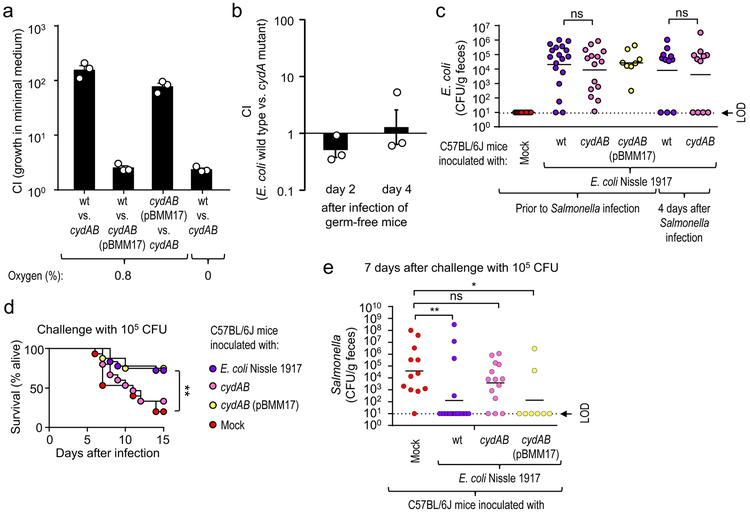
The large intestine is considered a largely anaerobic environment, but the bioavailability of oxygen is increased during conditions of intestinal inflammation17, which is associated with an expansion of facultative anaerobic Enterobacteriaceae. For instance, E. coli can use aerobic respiration to expand in the large intestine of mice with chemically-induced colitis18. Interestingly, oxygen is also a critical resource for S. Typhimurium, which uses its virulence factors to trigger intestinal inflammation to drive a pathogen expansion in the lumen of the large intestine using aerobic respiration13. We thus hypothesized that Enterobacteriaceae confer protection against S. Typhimurium by using their aerobic metabolism to compete with the pathogen for critical resources in vivo. To test this idea, we modeled colonization resistance conferred by Enterobacteriaceae using E. coli Nissle 1917. Inoculation of C57BL/6J mice with E. coli Nissle 1917 conferred protection against S. Typhimurium challenge (Suppl. Fig. 8g). We then generated E. coli Nissle 1917 mutants deficient for aerobic respiration under microaerophilic conditions by inactivating cytochrome bd oxidase encoded by the cydAB genes. The resulting cydAB mutant was deficient for growth under microaerophilic conditions, which could be complemented by introducing the cloned cydAB genes on a plasmid (pBMM17) (Fig. 4a). When germ-free mice were inoculated with a 1:1 mixture of the E. coli Nissle 1917 wild type and a cydAB mutant, similar numbers of both strains were recovered from the feces, suggesting that genetic ablation of cytochrome bd oxidase did not cause a general fitness defect in vivo (Fig. 4b). Compared to the cydAB mutant, the E. coli E. coli Nissle 1917 wild type competed more successfully with S. Typhimurium during in vitro growth under microaerophilic conditions (0.8% oxygen) in sterile-filtered fecal homogenates from susceptible mice (C57BL/6J) (Suppl. Fig. 8f). E. coli Nissle 1917 wild type or a cydAB mutant colonized C57BL/6J mice at similar levels (Fig. 4c). Challenge with S. Typhimurium revealed that genetic ablation of the ability to respire oxygen lowered the ability of E. coli Nissle 1917 to protect against lethal morbidity (Fig. 4d) and intestinal colonization by the pathogen (Fig. 4e), without reducing the numbers of E. coli during challenge (Fig. 4c). Collectively, these results supported the idea that commensal Enterobacteriaceae use their aerobic metabolism to compete with S. Typhimurium for resources critical for pathogen expansion in the intestinal lumen.
Figure 4: E. coli requires an aerobic metabolism to confer colonization resistance.
(a) Minimal medium containing 0.1% glucose was inoculated with a 1:1 ratio of the indicated bacterial strains and the competitive index (CI) determined after 24 hour incubation at the indicated oxygen concentrations. (b) Germ-free mice (n = 3) were inoculated with a 1:1 mixture of E. coli Nissle 1917 (wt) and an isogenic cydAB mutant and the CI determined in the feces at the indicated time points. (a-b) Bars represent geometric means ± standard error of the mean. (c-e) Mice from Jackson Laboratories (C57BL/6J) were mock-treated or were inoculated with 109 CFU/animal of E. coli Nissle 1917, a cydAB mutant or a cydAB mutant complemented with a plasmid carrying the cloned cydAB genes (pBMM17). Mice were challenged with S. Typhimurium and E. coli colonization (c), lethal morbidity (d) and S. Typhimurium shedding with the feces (c) were determined. (c) Prior to challenge (corresponding to 5 days after E. coli inoculation) or four days after Salmonella challenge the presence of E. coli in feces was determined. (c and e) Lines represent the geometric mean. Each dot represents data from one animal, thus indicating the number (n) of repeats. (d) The number of animals (n) is provided in panel e. LOD, limit of detection; ns, not statistically significantly different; wt, E. coli Nissle 1917 wild type; cydAB, E. coli Nissle 1917 cydAB mutant; cydAB (pBMM17), E. coli Nissle 1917 cydAB mutant complemented with plasmid pBMM17; *, P ≤ 0.05; **, P ≤ 0.01.
DISCUSSION
During homeostasis, physiologic hypoxia of the colonic epithelium limits the amount of oxygen emanating from the mucosal surface, thereby restricting growth of facultative anaerobic bacteria19. S. Typhimurium uses its virulence factors to induce intestinal inflammation accompanied by migration of neutrophils into the intestinal lumen, which deplete butyrate-producing Clostridia from the gut microbiota, thereby elevating epithelial oxygenation13. Oxygen emanating from the inflamed mucosa drives an expansion of S. Typhimurium through consumption of non-fermentable microbiota-derived carbon sources20,21. Whereas microbiota-derived short-chain fatty acids can inhibit growth of Enterobacteriaceae22, S. Typhimurium-induced inflammation triggers a luminal release of lactate by host cells23, which neutralizes growth inhibition by short-chain fatty acids24.
Commensal Enterobacteriaceae, such as E. coli, have been implicated in contributing to resistance against S. Typhimurium infection15,16,25. Metagenomic analysis of a defined microbial community that failed to confer colonization resistance against S. Typhimurium revealed an underrepresentation of pathways involved in microbial respiration compared to microbiota of conventional mice25. The addition of facultative anaerobic bacterial species that encode respiratory pathways, including E. coli, Streptococcus danieliae and Staphylococcus xylosus, correlated with a marked increase in colonization resistance the defined bacterial community conferred against S. Typhimurium25. Here we used bacterial genetics to establish a causal connection between aerobic respiration and the ability of E. coli to confer colonization resistance against S. Typhimurium. Our results suggest competition arises, because commensal Enterobacteriaceae have an aerobic metabolism that enables them to consume non-fermentable carbon sources. Genetic ablation of cytochrome bd oxidase synthesis prevents E. coli from consuming any of these non-fermentable carbon sources by aerobic respiration under microaerobic conditions and demonstrated aerobic respiration is required for E. coli to confer colonization resistance. The scarcity of respiratory electron acceptors is an important factor limiting the abundance of Enterobacteriaceae during gut homeostasis17, which explains why members of this taxon were difficult to detect by microbiota profiling in treatment naïve mice (Suppl. Fig. 7). However, a disruption of gut homeostasis by intestinal inflammation increases oxygen availability in the large intestine21 and other resources, such as iron, can become limiting during the consequent Enterobacteriaceae expansion26. As a result, E. coli Nissle 1917 also competes with S. Typhimurium for siderophore-bound iron during gut inflammation27. The emerging picture is that commensal Enterobacteriaceae are important keystone species that contribute to colonization resistance against S. Typhimurium by competing for critical resources, such as non-fermentable carbon sources and iron. Importantly, our work shows that these keystone species are a significant source of phenotypic variation in laboratory mice.
METHODS
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Andreas Bäumler (ajbaumler@ucdavis.edu).
Experimental Model and Subject Details
Mouse lines.
Female 5–7 week old C57BL/6NCrl, C57BL/6NHsd, and C57BL/6NTac (microbiological status equivalent; SPF) were purchased from The Charles River Laboratories (Wilmington, MA), Envigo (formerly Harlan; Indianapolis, IN), and Taconic Farms (Hudson, NY), respectively. C57BL/6J (stock no. 000664) and C57BL/6NJ (stock no. 005034) were purchased from The Jackson Laboratories (Bar Harbor, ME). Gnotobiotic Swiss Webster mice (Tac:SW) were initially purchased from Taconic Farms and bred at the UC Davis Genome and Biomedical Sciences Facility, maintained by investigators. All animal experiments were approved by the Institution of Animal Care and Use Committee at University of California, Davis. The exact number of mice used in each group is indicated on each graph, in the figure legend, or in Supplementary Table 1.
Bacterial strains.
All strains used in this study are listed in Supplementary Table 2. S. Typhimurium and E. coli Nissle 1917 were cultured in LB broth (10 g/L tryptone, 5 g/L1 yeast extract, 10 g/L NaCl) or LB plates (LB broth, 15g/L agar) and incubated at 37 °C. When needed, nalidixic acid (Nal), carbenicillin (Carb), kanamycin (Kan), and chloramphenicol (Cm) were added to LB broth and LB agar plates at a concentration of 50 mg/mL, 100 mg/mL, 100 mg/mL, and 15 mg/mL, respectively. Mouse commensal Proteus, Klebsiella, Enterobacter, and Escherichia were grown aerobically in LB broth or on MacConkey agar plates (10 g/L pancreatic digest of gelatin, 3 g/L peptone, 10 g/L lactose, 1.5 g/L bile salts, 5 g/L sodium chloride, 13.5 g/L agar, 30 mg/L neutral red, 1 mg/L crystal violet) at 37°C. All plasmids and primers used in this study are listed in Supplementary Table 2.
Method Details
Specific-pathogen free mouse husbandry.
Prior to transport, mice at Charles River, Envigo, Taconic and Jackson were fed (according to online rodent model information sheets) Purina 5L79 rodent chow, Teklad Global Rodent Diet 2018S, NIH #31M Rodent Diet, and LabDiet 5K52 formulation (6% fat), respectively. Upon arrival, mice from each cohort were randomly assigned into individually ventilated cages on one rack at a housing density of 3 to 4 animals per cage and allowed to acclimate in our vivarium for at least a week undisturbed. Feed was switched to irradiated TEKLAD GLOBAL 18% protein rodent diet 2918 (Envigo) and no breeding was performed. 70% ethanol was used to disinfect surfaces and gloves between groups. Clean (but not sterile) paper towels were utilized for fecal sample collections. None of the experiments performed in this study involved treatment or pretreatment of mice with antibiotics.
Preparation of fecal transplants.
Freshly voided pooled feces (approximately 200mg per cage) were collected from C57BL/6NCrl, C57BL/6NHsd, C57BL/6NTac, and C57BL/6J mice. Samples were maintained on ice and processed within 1 hour. An anaerobic chamber was not used. Feces were diluted at 1:50 (approximately 10mL) in sterile Phosphate Buffered Saline (PBS; pH = 7.4) and homogenized via vortex for 5 minutes at room temperature. Tubes were briefly centrifuged (500 x g for 1 minute) to pellet large particles. 500μl aliquots were mixed with 500μl glycerol (60%w/v) and immediately stored in −80°C.
S. Tm infection in C57BL/6 mice.
Mice were infected upon reaching eight to twelve-week of age. To generate contamination-free S. Typhimurium culture, LB broth supplemented with appropriate antibiotics in sterile flasks was aseptically inoculated from single colonies and incubated with shaking (200 rpm at 37°C) for 16 to 20 hours. Bacteria were then harvested by centrifugation (15 min, 4000 × g, 4°C), and adjusted to a density of 1 × 1010 CFU/mL in sterile LB. To generate lower density inocula, cultures were serially diluted ten-fold. Sterile LB broth (0.1 mL) containing S. Typhimurium at the indicated densities was inoculated by oral gavage (between 6am and noon). Primary experimental outcome assessed were: animal activity, weight loss, lethal morbidity and pathogen burdens. Weights were recorded daily and feces were collected at the indicated time points (between 1 and 7 days after infection). Mice were euthanized at the indicated time points or when they became moribund. Mice that were euthanized early due to health concerns were excluded from analysis, except for experiments determining lethal morbidity after challenge. Intestinal contents (approximately 15mg/animal) were homogenized in 1mL PBS. Samples were then serially diluted in PBS and plated on selective agar in order to distinguish pathogen CFU/g. Challenged mice were considered infected when a positive stool culture was obtained at >3 days after infection.
Germ-free mouse husbandry.
Animals were housed in flexible front glove-box isolators (Park Bioservices, LLC). Food (irradiated 2920X Teklad diet by Envigo) and autoclaved water access was ad libitum. Bedding changes were provided weekly. The room was maintained on a 12-h light/dark cycle. Principles of the established ‘out-of-the-isolator’ gnotobiotic husbandry system 28 were utilized and validated in our facility to preserve gnotobiotic status for up to 2 weeks by standard quality control (negative aerobic and anaerobic culture in brain-heart infusion broth (BHI) and 16S rRNA quantitative real-time PCR comparable to a no template control). Littermates of both sexes were removed from the isolator at 6 weeks of age and ascetically transferred to a biosafety cabinet and placed into autoclaved individually ventilated cages. Groups of 2 to 4 males or females were randomly assigned to experimental groups and housed together to facilitate social interactions and establishment of their fecal microbiota transplants. Animals were maintained in a biosafety cabinet for at least 5 days undisturbed after receiving fecal transplants and infections were carried out in the biosafety cabinet.
Fecal transplant into Germ-free Swiss Webster mice.
Previously frozen pooled fecal samples from cages of each C57BL/6 substrain were thawed on ice and delivered via oral gavage (200μl) once. Colonization was allowed to proceed for a minimum of 5 days before sample collection and infection with S. Typhimurium. Bedding changes were performed before infection, carried out under strictly aseptic conditions, and samples were collected from individual mice using autoclaved beakers before infection.
S. Typhimurium infections in Germ-free Swiss Webster mice.
Concentrated overnight cultures were prepared as described above. Animals were colonized with fecal microbiota transplant for 5 days. Each ex-germ-free mouse received 0.1 mL of a suspension containing the indicated CFU of S. Typhimurium via oral gavage. Fresh fecal pellets were collected aseptically using autoclaved beakers and plated on agar plates containing the appropriate antibiotics.
16S rDNA amplicon sequencing.
Fecal samples were collected from individual mice prior to infection for analysis and frozen at −20 C. DNA was extracted using the MoBio PowerSoil Kit according to the manufacturer instructions, with the following two recommended modifications. (1) Samples were heated at 70°C for 10 minutes following addition of Buffer 1 to enhance disruption of gram-positive bacteria. (2) Samples were homogenized using a mini bead beater for 60 seconds to achieve greater mechanical lysis of bacterial cells. Paired end library construction was performed as previously described 13. Primers 515F and 806R (Supplementary Table 2) were used to amplify the V4 domain of the 16S rRNA. Both forward and reverse primers contained a unique 8 nt barcode (N), a primer pad (underlined), a linker sequence (italicized), and the Illumina adaptor sequences (bold). Each sample was barcoded with a unique forward and reverse barcode combination. PCR contained 1 Unit Kapa2G Robust Hot Start Polymerase (Kapa Biosystems), 1.5mM MgCl2, 10 μmol of each primer, 10mM dNTP’s, and 1ul of DNA. PCR conditions were: an initial incubation at 95°C for 2 min, followed by 30 cycles of 95°C for 20 s, 50°C for 20 s, 72°C for 20 s and a final extension of 72°C for 3 min. The final product was quantified on the Qubit instrument using the Qubit High Sensitivity DNA kit and individual amplicon libraries were pooled, cleaned by Ampure XP beads (Beckman Coulter), and sequenced using a 250 bp paired-end method on an Illumina MiSeq instrument in the Genome Center DNA Technologies Core, University of California, Davis. Raw paired end-sequence data were de-multiplexed and trimmed, followed by filtering for quality. Samples containing less than 1000 quality reads were removed from dataset. Quantitative Insights into Microbial Ecology (QIIME) open source software package (version 1.9) was initially used to perform sequence alignment and closed-reference operational taxonomic units (OTUs) picking against the Greengenes reference collection (version 13_8) at 97% identity. Data were later re-analyzed when QIIME 2 became available (core 2018.2 distribution). Clustering, permutational multivariate analysis of variance (PERMANOVA), beta diversity measures (principle coordinate analysis of unweighted UniFrac distances) and phylogenetic profiling were similar between QIIME 1.9 and QIIME 2. Additionally, taxonomy was also assigned with the Ribsosomal Database Project (RDP release 11) classifier using the DADA2 R package version 1.6 pipeline (https://benjjneb.github.io/dada2/tutorial.html) to resolve more sequences down to the species level.
Cohousing.
Mice were received at 5–7 weeks of age. After allowing one week to acclimate in our vivarium, mice from Charles River Laboratories and The Jackson Laboratories were placed in clean cages (with new food and water) at a ratio of 1:1 and cohoused for 14 days. Ear punching facilitated long-term identification. In one experiment, cohoused mice were subsequently separated after two weeks. Fecal samples were flash frozen at the start of cohousing and every week thereafter. For challenge experiments, animals remained cohoused after receiving 1 × 108 CFU Salmonella. All mice were sacrificed at 5 days after infection.
Enteric strain isolation and identification.
Ten-fold serial dilutions of previously frozen fecal transplants in glycerol or flash frozen feces were plated on MacConkey agar and incubated aerobically at 37°C overnight. Single colonies with discernable unique morphologies were picked and streaked for isolation. Colonies were initially typed based on colony appearance into lactose-fermenting red colonies (Lac+) and lactose non-fermenting white colonies (Lac-). Approximately 100 colonies (30–40 colonies from each of the three vendors harboring Enterobacteriaceae) were isolated and identified biochemically using the EnteroPluri Test (BD) per the manufacturer’s directions. Isolates used for animal experiments were subjected to full-length 16S rRNA gene Sanger sequencing.
Quantification of Lactobacilli by culturing.
Fecal pellets from individual C57BL/6NCrl, C57BL/6NHsd, C57BL/6NTac, and C57BL/6J were plated on Difco Lactobacilli MRS Agar (10 g/L Protease Peptone No. 3, 10 g/L Beef Extract, 5 g/L Yeast Extract, 20 g/L Dextrose, 1 g/L Polysorbate 80, 2 g/L Ammonium Citrate, 5 g/L Sodium Acetate, 0.1 g/L Magnesium Sulfate, 0.05 g/L Manganese Sulfate, 2 g/L Dipotassium Phosphate, 15g/L Agar; pH 5.5). Feces were serially diluted in PBS and plates were pre-reduced for 2 days. Incubation was allowed for 3 days at 37°C in an anaerobic chamber (Shel Lab Bactron II; 5% Hydrogen, 5% Carbon Dioxide, 90% Nitrogen).
16S rDNA Sanger sequencing.
Bacterial strains were grown in LB broth and pelleted. After resuspension in distilled water, tubes were boiled for 1 minute and immediately cooled on ice. 1μl of the lysed bacterial suspension containing genomic DNA was used in a PCR reaction to amplify the 16S rRNA gene using primers 63F-1387R. PCR conditions: 1μl template, 1μl each forward and reverse primers, 17μl PCR Supermix high-fidelity. Thermocycler (MJ Research PTC-200 Peltier Thermal Cycler) conditions: thirty cycles of denaturation at 95°C for 1 minute, annealing at 48°C for 1 minute, elongation at 72C for 2 minutes, followed an additional 15 minutes at 72°C. Sizes (1.5 kb) were estimated using agarose gel electrophoresis. Following the QIAquick PCR purification kit, 1μl of DNA was ligated into pCR2.1-TOPO vector. Chemically competent TOP10 E. coli cells were transformed with ligation products by heat shock (30 s at 42°C). Recombinant cells were selected on LB plates containing kanamycin and X-Gal (5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside). DNA preparations for sequencing were made with a QIAprep mini as specified by the manufacturer. Plasmids were eluted with 50 μl of water, and the products were stored at −20°C. Sizes (5.4 kb) of plasmids containing inserts were checked by agarose gel electrophoresis and concentrations (200 ng/ul) were determined on a NanoDrop 1000 spectrophotometer (Thermo Scientific). Plasmid inserts were sequenced by the College of Biological Sciences UCDNA Sequencing Facility using M13 primers. Two contigs were aligned for each strain using Snapgene. Evolutionary analyses were conducted in MEGA7 29. The evolutionary history was inferred by using the Maximum Likelihood method based on the Tamura-Nei model 30. The analysis involved 10 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 1357 positions in the final dataset.
Colonization of C57BL/6J mice with Enterobacteriaceae.
Isolates from frozen fecal transplants of C57BL/6NCrl, C57BL/6NHsd, C57BL/6NTac, and C57BL/6J on MacConkey agar were maintained individually. Cultures were grown in LB broth overnight and 1 × 1010 CFU/ml preparations were made. In some experiments, strains were mixed in equal volumes to prepare CR-mix (E. coli Crl141, E. coli Crl142, P. mirabilis Crl143), Har-mix (E. coli Hsd145, E. cloacae Hsd146), and Tac-mix (K. oxytoca Tac148, P. vulgaris Tac149). A single dose of 100 μl was delivered via gavage to C57BL/6J mice. Colonization levels were determined by plating feces on MacConkey agar. E. coli Nissle 1917 and the E. coli Nissle 1917 cydAB mutant were transformed with plasmids (pCAL61 or pCAL62) prior to inoculation of mice to facilitate recovery from the feces.
In vitro growth assays.
For supplementary figure panels 9a-c, an overnight liquid culture of S. Typhimurium was diluted in PBS to a concentration of 1 × 103 CFU/mL and 100 μl aliquots were added to 900 μl of mouse fecal homogenate in glass tubes with loose-fitting caps. Feces was processed as follows: Approximately 100 to 300 mg of feces were collected from one cage of mice pooled. Samples were diluted 1:100 in PBS and homogenized by vortex for 10 minutes at room temperature. Large particles were pelleted by centrifugation at 500g for 1 minute. Half of the sample was filtered through a 0.45 μm low-protein binding membrane. Fecal homogenates were stored at 4°C for up to 1 hour prior to adding S. Typhimurium. Tubes were then incubated for 24 hours at 37°C while shaking under atmospheric oxygen. For supplementary figure panels 9d-e, pooled feces from C57BL/6J mice was diluted 1:100 in PBS, homogenized, briefly centrifuged, stored at 4°C for up to 1 hour prior to adding S. Typhimurium to achieve a final concentration of 1 × 103 CFU/mL fecal homogenate. 900 μl aliquots of Jackson feces containing S. Typhimurium were distributed to glass tubes with loose-fitting caps. Overnight cultures of each commensal Enterobacteriaceae strain were diluted in PBS to a concentration of 1 × 104 CFU/mL and 100 μl was added to tubes containing 900 μl of S. Typhimurium in fecal homogenate. For anaerobic conditions, a duplicate set of tubes was prepared and moved into the anaerobic chamber (Shel Lab Bactron II; 5% Hydrogen, 5% Carbon Dioxide, 90% Nitrogen) immediately and incubated without shaking at 37°C. CFU/ml were determined by selective plating of serial dilutions.
Construction of mutants by allelic exchange.
The E. coli Nissle cydAB mutant was constructed as described previously 19. Complementation was achieved by introducing cydAB on a plasmid under expression of the native promoter as follows. Primer pair “EcNcomp Fwd” and “EcNcomp Rev” (Supplementary Table 2) was used to amplify the coding sequence of E. coli Nissle 1917 cydAB operon plus 500bp upstream and downstream. The resulting PCR product was visualized via agarose gel electrophoresis and purified using Zymoclean Gel DNA Recovery kit (Zymo Research). The fragment was then cloned into the EcoRV restriction site of the low copy number plasmid pWSK129 31 to generate the plasmid pBMM17 via Gibson Assembly Master Mix (NEB).
Quantification and Statistical Analyses
The investigators were not blinded to allocation during experiments and outcome assessment. Sample size estimation based on published literature. Mice that were euthanized early due to health concerns were excluded from analysis, except for experiments determining lethal morbidity after challenge. Data analysis was performed in Microsoft Excel and GraphPad Prism v7.0. Precision measures are described in figure legends and details regarding the statistics of each experiment are reported in Supplementary Table 1. For survival analysis, the Log-rank (Mantel-Cox) test was used. The limit of detection via plating was 10 CFU/g. CFU/g feces were LN-transformed to normalize data. When samples were collected overtime and matched to individual mice, then a Two-way repeated measures ANOVA was used followed by Tukey’s multiple comparison test. When samples were not collected for all mice across time or were not matched to individual animals, then an Ordinary one-way ANOVA was used if variances were equal between groups. Tukey’s multiple comparison tests were used when all pairwise comparison were of interest or Dunnet’s multiple comparison test when comparisons to a reference were of interest. Otherwise, a Kruskal-Wallis Test was used when unequal variance was observed across groups (determined by Browne-Forsythe test and demonstrated by box and whisker plots) followed by Dunn’s multiple comparison test. All family-wise significance was set to 0.05 (95% confidence interval). For calculations of ID50s, a constrained nonlinear regression model was used. For analysis of community 16S rRNA amplicon sequences (microbiome), PERMANOVA was used to evaluate beta-diversity. For association testing, linear regression was used. For biomarker discovery, LEfSe was used with the following parameters: alpha = 0.05 for factorial Kurskal-Wallis test among classes, threshold = 2.0 on the logarithmic LDA score for discrimative features, and all-against-all multi-class analysis. For analysis of strain-specific full-length 16S rRNA Sanger sequences, MEGA was used to align sequences by MUSCLE with the following parameters: gap open penalty = −400, clustering method = UPGMC. A maximum likelihood tree was constructed under the Tamura-Nei model, assuming uniform variation rates among sites, gap deletion, and a bootstrap method (1000 replications) to test phylogeny. Differences with P ≤0.05 were considered to be significant.
Data and Software Availability
The following open-source software was used: QIIME (version 1.91; http://qiime.org/ and version 2; https://qiime2.org/), NIH nBLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi), MEGA (version 7; http://www.megasoftware.net/), R (http://cran.us.r-project.org/), DADA2 plugin for R (version 1.6; https://benjjneb.github.io/dada2/index.html), Greengenes version 13_5 (http://greengenes.secondgenome.com/), RDP Project (https://rdp.cme.msu.edu/) and LEfSE (http://huttenhower.sph.harvard.edu/galaxy). The following commercial software was used: Microsoft Excel (version 2011 for Mac and 2010 for Windows; www.microsoft.com/Buy/Excel), Graphpad Prism version 7 for Mac (https://www.graphpad.com/scientific-software/prism/), and Snapgene version 4.1 (http://www.snapgene.com/).
DATA AVAILABILITY
Illumina sequences obtained in the present study were deposited in the Sequence Read Archives (SRA) NCBI database under accession number SRP148888 (Available here). Sanger sequences were deposited in Genbank under accession numbers MH759762-MH759768 (Available here).
Supplementary Material
Acknowledgements.
This work was supported by PHS grants AI044170, AI112445, AI096528 and AI112949 to A.J.B. E.M.V. was supported by AI060555, OD010931, and OD010956. Y.L. was supported by Vaadia-BARD Postdoctoral Fellowship FI-505–2014. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the NIH.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nmicrobiol.
Competing Interests statement
The authors declare no competing financial interests.
REFERENCES
- 1.Baker M 1,500 scientists lift the lid on reproducibility. Nature 533, 452–454, doi: 10.1038/533452a (2016). [DOI] [PubMed] [Google Scholar]
- 2.Stappenbeck TS & Virgin HW Accounting for reciprocal host-microbiome interactions in experimental science. Nature 534, 191–199, doi: 10.1038/nature18285 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Franklin CL & Ericsson AC Microbiota and reproducibility of rodent models. Lab Anim (NY) 46, 114–122, doi: 10.1038/laban.1222 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanage WP Microbiology: Microbiome science needs a healthy dose of scepticism. Nature 512, 247–248, doi: 10.1038/512247a (2014). [DOI] [PubMed] [Google Scholar]
- 5.Ivanov II et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498, doi: 10.1016/j.cell.2009.09.033 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsolis RM, Xavier MN, Santos RL & Baumler AJ How to become a top model: impact of animal experimentation on human Salmonella disease research. Infect Immun 79, 1806–1814, doi: 10.1128/IAI.01369-10 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thiemann S et al. Enhancement of IFNgamma Production by Distinct Commensals Ameliorates Salmonella-Induced Disease. Cell Host Microbe 21, 682–694 e685, doi: 10.1016/j.chom.2017.05.005 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Fallon MT, Benjamin WH Jr., Schoeb TR & Briles DE Mouse hepatitis virus strain UAB infection enhances resistance to Salmonella typhimurium in mice by inducing suppression of bacterial growth. Infect Immun 59, 852–856 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bohnhoff M, Drake BL & Miller CP Effect of streptomycin on susceptibility of intestinal tract to experimental Salmonella infection. Proc Soc Exp Biol Med 86, 132–137 (1954). [DOI] [PubMed] [Google Scholar]
- 10.Mekada K et al. Genetic differences among C57BL/6 substrains. Exp Anim 58, 141–149 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Zurita E et al. Genetic polymorphisms among C57BL/6 mouse inbred strains. Transgenic Res 20, 481–489, doi: 10.1007/s11248-010-9403-8 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Simon MM et al. A comparative phenotypic and genomic analysis of C57BL/6J and C57BL/6N mouse strains. Genome Biol 14, R82, doi: 10.1186/gb-2013-14-7-r82 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rivera-Chavez F et al. Depletion of Butyrate-Producing Clostridia from the Gut Microbiota Drives an Aerobic Luminal Expansion of Salmonella. Cell Host Microbe 19, 443–454, doi: 10.1016/j.chom.2016.03.004 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim YG et al. Neonatal acquisition of Clostridia species protects against colonization by bacterial pathogens. Science 356, 315–319, doi: 10.1126/science.aag2029 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Splichalova A et al. Interference of Bifidobacterium choerinum or Escherichia coli Nissle 1917 with Salmonella Typhimurium in gnotobiotic piglets correlates with cytokine patterns in blood and intestine. Clin Exp Immunol 163, 242–249, doi: 10.1111/j.1365-2249.2010.04283.x (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lima-Filho JV, Vieira LQ, Arantes RM & Nicoli JR Effect of the Escherichia coli EMO strain on experimental infection by Salmonella enterica serovar Typhimurium in gnotobiotic mice. Braz J Med Biol Res 37, 1005–1013, doi:/S0100-879X2004000700009 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Rivera-Chavez F, Lopez CA & Baumler AJ Oxygen as a driver of gut dysbiosis. Free Radic Biol Med, doi: 10.1016/j.freeradbiomed.2016.09.022 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Hughes ER et al. Microbial Respiration and Formate Oxidation as Metabolic Signatures of Inflammation-Associated Dysbiosis. Cell Host Microbe 21, 208–219, doi: 10.1016/j.chom.2017.01.005 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Byndloss MX et al. Microbiota-activated PPAR-gamma signaling inhibits dysbiotic Enterobacteriaceae expansion. Science 357, 570–575, doi: 10.1126/science.aam9949 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faber F et al. Respiration of Microbiota-Derived 1,2-propanediol Drives Salmonella Expansion during Colitis. PLoS Pathog 13, e1006129, doi: 10.1371/journal.ppat.1006129 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spiga L et al. An Oxidative Central Metabolism Enables Salmonella to Utilize Microbiota-Derived Succinate. Cell Host Microbe 22, 291–301 e296, doi: 10.1016/j.chom.2017.07.018 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meynell GG Antibacterial mechanisms of the mouse gut. II. The role of Eh and volatile fatty acids in the normal gut. Br J Exp Pathol 44, 209–219 (1963). [PMC free article] [PubMed] [Google Scholar]
- 23.Gillis CC et al. Dysbiosis-Associated Change in Host Metabolism Generates Lactate to Support Salmonella Growth. Cell Host Microbe 23, 54–64 e56, doi: 10.1016/j.chom.2017.11.006 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bohnhoff M, Miller CP & Martin WR Resistance of the Mouse’s Intestinal Tract to Experimental Salmonella Infection. Ii. Factors Responsible for Its Loss Following Streptomycin Treatment. J Exp Med 120, 817–828 (1964). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brugiroux S et al. Genome-guided design of a defined mouse microbiota that confers colonization resistance against Salmonella enterica serovar Typhimurium. Nat Microbiol 2, 16215, doi: 10.1038/nmicrobiol.2016.215 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Nedialkova LP et al. Inflammation fuels colicin Ib-dependent competition of Salmonella serovar Typhimurium and E. coli in enterobacterial blooms. PLoS Pathog 10, e1003844, doi: 10.1371/journal.ppat.1003844 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deriu E et al. Probiotic bacteria reduce salmonella typhimurium intestinal colonization by competing for iron. Cell Host Microbe 14, 26–37, doi: 10.1016/j.chom.2013.06.007 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Faith JJ, Ahern PP, Ridaura VK, Cheng J & Gordon JI Identifying gut microbe-host phenotype relationships using combinatorial communities in gnotobiotic mice. Sci Transl Med 6, 220ra211, doi: 10.1126/scitranslmed.3008051 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar S, Stecher G & Tamura K MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol 33, 1870–1874, doi: 10.1093/molbev/msw054 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tamura K & Nei M Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10, 512–526, doi: 10.1093/oxfordjournals.molbev.a040023 (1993). [DOI] [PubMed] [Google Scholar]
- 31.Wang RF & Kushner SR Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100, 195–199 (1991). [PubMed] [Google Scholar]
- 32.Stojiljkovic I, Baumler AJ & Heffron F Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J Bacteriol 177, 1357–1366 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Winter SE et al. The TviA auxiliary protein renders the Salmonella enterica serotype Typhi RcsB regulon responsive to changes in osmolarity. Molecular microbiology 74, 175–193, doi: 10.1111/j.1365-2958.2009.06859.x (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grozdanov L et al. Analysis of the genome structure of the nonpathogenic probiotic Escherichia coli strain Nissle 1917. J Bacteriol 186, 5432–5441, doi: 10.1128/JB.186.16.5432-5441.2004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simon R, Priefer U & Puhler A A Broad Host Range Mobilization System for Invivo Genetic-Engineering - Transposon Mutagenesis in Gram-Negative Bacteria. Bio-Technology 1, 784–791, doi:Doi 10.1038/Nbt1183-784 (1983). [DOI] [Google Scholar]
- 36.Caporaso JG et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 108 Suppl 1, 4516–4522, doi: 10.1073/pnas.1000080107 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marchesi JR et al. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl Environ Microbiol 64, 795–799 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spees AM et al. Streptomycin-induced inflammation enhances Escherichia coli gut colonization through nitrate respiration. MBio 4, doi: 10.1128/mBio.00430-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Callahan BJ et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 13, 581–583, doi: 10.1038/nmeth.3869 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caporaso JG et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7, 335–336, doi: 10.1038/nmeth.f.303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McDonald D et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6, 610–618, doi: 10.1038/ismej.2011.139 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cole JR et al. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res 33, D294–296, doi: 10.1093/nar/gki038 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Segata N et al. Metagenomic biomarker discovery and explanation. Genome Biol 12, R60, doi: 10.1186/gb-2011-12-6-r60 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The following open-source software was used: QIIME (version 1.91; http://qiime.org/ and version 2; https://qiime2.org/), NIH nBLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi), MEGA (version 7; http://www.megasoftware.net/), R (http://cran.us.r-project.org/), DADA2 plugin for R (version 1.6; https://benjjneb.github.io/dada2/index.html), Greengenes version 13_5 (http://greengenes.secondgenome.com/), RDP Project (https://rdp.cme.msu.edu/) and LEfSE (http://huttenhower.sph.harvard.edu/galaxy). The following commercial software was used: Microsoft Excel (version 2011 for Mac and 2010 for Windows; www.microsoft.com/Buy/Excel), Graphpad Prism version 7 for Mac (https://www.graphpad.com/scientific-software/prism/), and Snapgene version 4.1 (http://www.snapgene.com/).
Illumina sequences obtained in the present study were deposited in the Sequence Read Archives (SRA) NCBI database under accession number SRP148888 (Available here). Sanger sequences were deposited in Genbank under accession numbers MH759762-MH759768 (Available here).