 To push back the growing tide of antibacterial resistance the discovery and development of new antibiotics is a must.
To push back the growing tide of antibacterial resistance the discovery and development of new antibiotics is a must.
Abstract
To push back the growing tide of antibacterial resistance the discovery and development of new antibiotics is a must. In recent years the calcium-dependent lipopeptide antibiotics (CDAs) have emerged as a potential source of new antibacterial agents rich in structural and mechanistic diversity. All CDAs share a common lipidated cyclic peptide motif containing amino acid side chains that specifically chelate calcium. It is only in the calcium bound state that the CDAs achieve their potent antibacterial activities. Interestingly, despite their common structural features, the mechanisms by which different CDAs target bacteria can vary dramatically. This review provides both a historic context for the CDAs while also addressing the state of the art with regards to their discovery, optimization, and antibacterial mechanisms.
1. Introduction
The rapid emergence and onset of drug-resistant bacteria is now considered one of the most urgent global threats to human health.1 In 2017 the Center for Disease Control and Prevention (CDC) revealed that more than 2 million people acquire an antibiotic-resistant infection per year which leads to at least 23 000 deaths in the US alone.2 The pernicious increase in the incidence of multi-drug-resistant (MDR) Gram-negative bacteria that are resistant to carbapenems3,4 and colistin5,6 in Enterobacteriaceae is considered a top priority by the CDC. The same holds true for infections caused by Gram-positive pathogens as exemplified by the infamous hospital bacteria methicillin-resistant Staphylococcus aureus (MRSA)7,8 and vancomycin-resistant enterococci (VRE).9 These developments have seen a steady decline in the availability of clinically approved antibiotics that are effective at combating MDR pathogens, leading some to speculate that the antibiotic era is coming to an end.10,11 During the so-called golden age of antibiotic discovery (1940s–1960s) a myriad of powerful and structurally unique antibiotics were discovered and brought to the clinic, including: sulfonamides, β-lactams, aminoglycosides, glycopeptides, macrolides, tetracyclines, chloramphenicol, lincosamides, quinolones and streptogramins.12 However, the decades that followed (1970s–2000s) are now known as the “discovery void” with only two new classes of structurally and mechanistically distinct antibiotics being discovered in this period as the pipeline of readily available antibiotics from nature dried up.13 Notably, the calcium-dependent lipopeptide antibiotic daptomycin is one of a rare number of first in class antibiotics to have entered the market since the year 2000.14,15
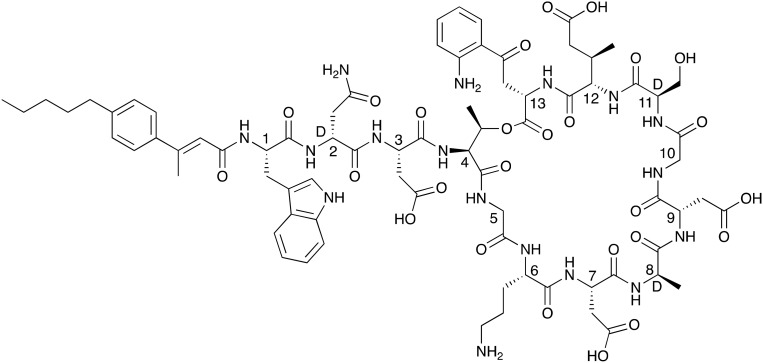
In this review we present a comprehensive summary of the calcium-dependent lipopeptide class of antibiotics (CDAs), with a primary focus on their structural features, antibacterial mechanisms and clinical potential. As the only currently clinically approved CDA, daptomycin (Fig. 1) is a special and representative case. First discovered by researchers at Eli Lily in 1983, daptomycin was later shepherded through clinical development by Cubist pharmaceuticals, gaining approval as cubicin in 2003 for the treatment of skin infections caused by Gram-positive bacteria and later in 2006 for bloodstream (bacteremia) infections caused by MRSA.15,16 Industrially, daptomycin is produced by the fermentation of Streptomyces roseosporus supplemented with decanoic acid to favor production of the compound containing the desired C10-fatty acid.16,17 In the years 2006–2014 annual sales of daptomycin reached 1 billion dollars making it a rare example of a blockbuster antibiotic drug. Based on these strong sales figures, Merck acquired Cubist in 2014 for a sum of 9.5 billion USD. As of 2016 daptomycin is off patent and various generic versions are now available.
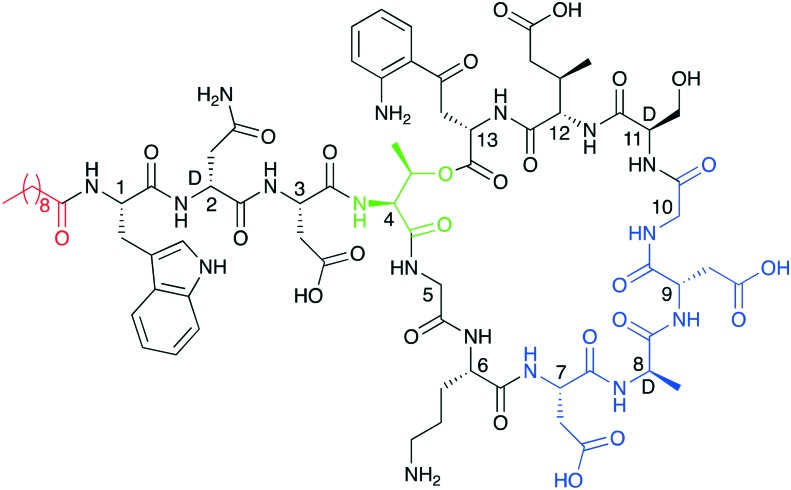
Fig. 1. Structure of daptomycin indicating N-terminal lipid in red and in blue the calcium binding motifs. The peptide macrocycles are formed biosynthetically via cyclization of the C-terminal residue with the side chain of Thr4 (linkages shown in green).
While daptomycin is the most renowned CDA, there are several other cyclic lipo(depsi)peptides of the same family that display potent antibacterial activity. These include a number of depsipeptides produced by Streptomyces coelicolor A(3)2 (ref. 18) and A 54145 produced by Streptomyces fradiaei.19 Other CDAs include the amphomycin,20 friulimicin,21 and laspartomycin/glycinocin22,23 classes which are also produced by various other streptomycetes.24 As illustrated by the structure of daptomycin (Fig. 1) nearly all CDAs share the same positioning of constituent d-amino or achiral amino acids in addition to a highly conserved Asp-X-Asp-Gly motif.25 This Asp-X-Asp-Gly sequence is imperative for calcium binding, which is needed for achieving full antibacterial activity as in the absence of calcium, the activity of the CDAs is significantly reduced.25 The structures of all CDAs consist of two main components: a cyclic peptide macrocycle and a long-chain fatty acid tail which is linked to the peptide core. In nature CDAs are produced by nonribosomal peptide synthetases (NRPSes). A number of excellent previous reviews provide in-depth information regarding the biosynthetic pathways and the specific enzymes involved in CDA production.24,25 In this review we will therefore focus more on the structure and mechanistic aspects of the CDAs as well as recent medicinal chemistry approaches to generating new analogues of these compounds and advanced screening techniques to identify novel family members.
2. Clinically used CDAs and compounds in advanced development
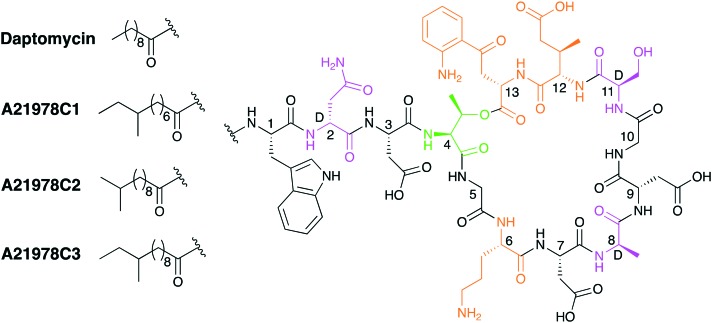
Daptomycin is the only clinically approved CDA and is primarily prescribed for the treatment of serious Gram-positive infections. The dosing of daptomycin is based on individual body weight and shows a concentration-dependent mode of action.15 Produced industrially by fermentation of the actinomycete Streptomyces roseosporus, daptomycin is actually formed as member of the so-called A21978C complex of structurally related CDAs (Fig. 2). All members of the A21978C complex consists of the same 13 amino acid peptide and are exemplified by their ten-membered ring which is achieved by macrolactonization between Thr4 and Kyn13.16 Other unique d- and non-proteinogenic amino acids present include: d-Asn2, d-Ala4, ornithine (Orn6), d-Ser11 and 3-methylglutamic acid (MeGlu12).25 The CDAs of the A21978C complex differ only in their aliphatic lipid tails and Fig. 2 illustrates this structural variation. Interestingly, daptomycin was initially identified as a minor component of the A21978C complex but demonstrated superior biological activity when compared with the other members of the complex.26 This led to an optimization of the production process whereby addition of exogenous decanoic acid to fermentation of S. roseosporus results in an increased production of daptomycin.27
Fig. 2. Structures of the A21978C family; the macrolactonization site is shaded in green, d-amino acids in purple and non-proteinogenic amino acids in orange.
Despite its clinical importance, the precise working mechanism by which daptomycin kills bacteria has yet to be fully elucidated. Over the past decade numerous studies have attempted to pinpoint its mechanism of action resulting in various, and in some cases conflicting models. These models range from calcium-dependent oligomerization and phosphatidylglycerol-mediated membrane pore formation, to alteration of membrane fluidity and inhibition of cell wall synthesis.28–30 Despite the continuing debate over its specific mechanism of action, daptomycin has been used for more than a decade in the clinic and reports are now emerging of resistance in strains of both E. faecium31 and MRSA.32 Resistance to daptomycin stems from mutation in a number of bacterial genes. These genes generally activate the pathogen's defenses to cell wall damage and stress as the bacterial cell attempts to prevent daptomycin from reaching the cytoplasmic membrane. Mutations in such genes can in turn result in increased MIC.33 Notably, clinical isolates from patients failing to respond to daptomycin therapy have shown frequent accumulation of mprF point mutations in S. aureus. The encoded protein MprF, is a transmembrane protein that caps phosphatidylglycerol in the cell membrane with lysine, thus reducing the net negative charge of the cell surface.34
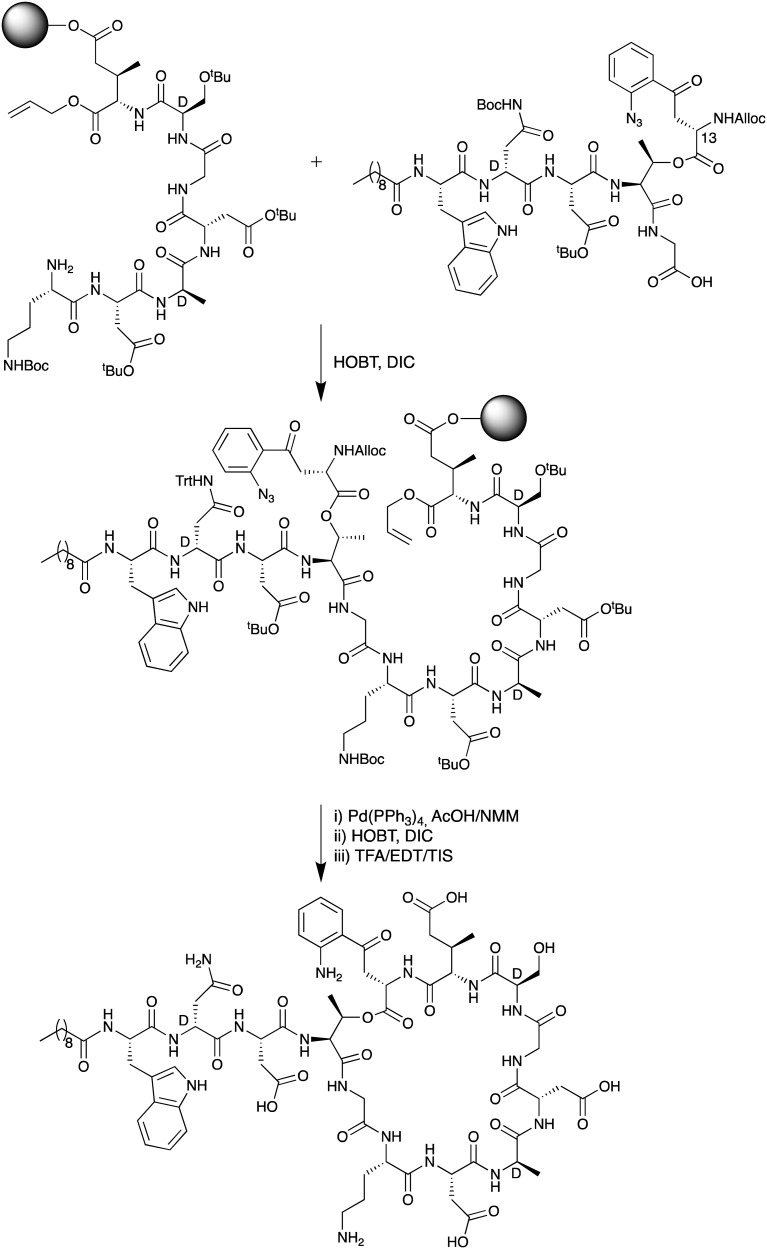
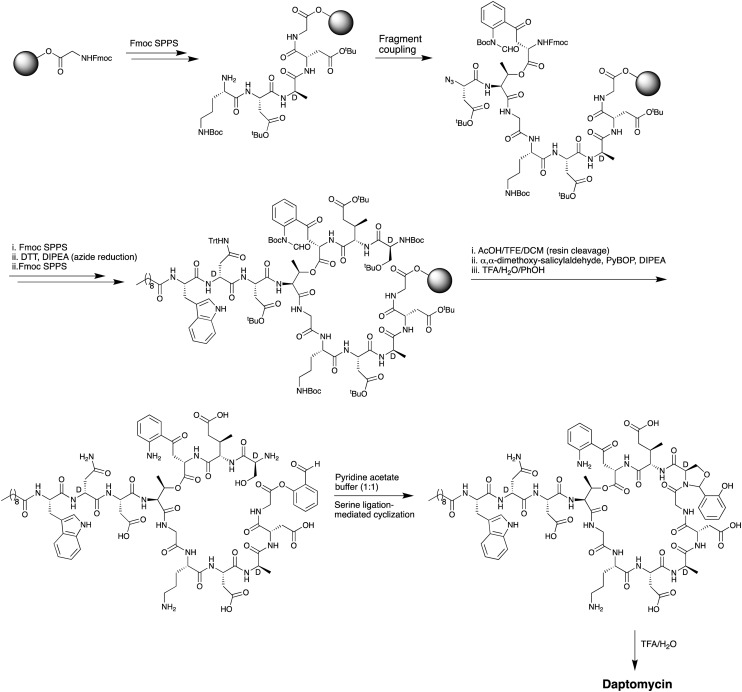
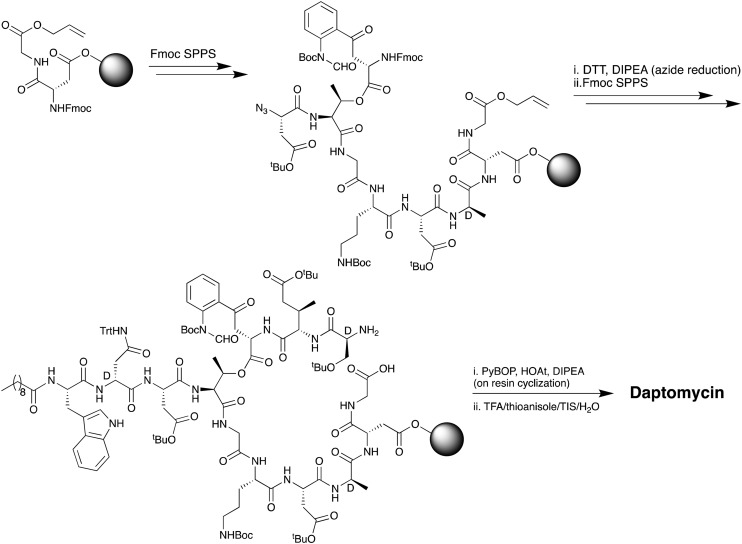
The first total chemical synthesis of daptomycin was achieved by researchers at Cubist Pharmaceuticals in 2006 and appears only in the patent literature.35 The Cubist team employed a convergent approach (Scheme 1) wherein one peptide fragment containing the synthetically challenging ester linkage between Thr4 and Kyn13 was coupled to a second resin bound peptide fragment attached to a solid support via the side chain of MeGlu12. Prudent use of alloc and allyl ester protecting groups and temporary azide masking of the aniline nitrogen of the kynurenine unit in the coupled fragment enabled the subsequent simultaneous deprotection/reduction of the resin bound intermediate using palladium catalysis. Following on-resin cyclization and cleavage/deprotection daptomycin was isolated in moderate yield. A second total synthesis of daptomycin was reported by the group of Li in 2013 who employed a similar convergent route combining solid- and solution-phase approaches with a chemoselective serine ligation providing the key cyclization step of the macrocycle (Scheme 2).36,37 Also notable in the Li synthesis was the practical introduction of an appropriately protected kynurenine by ozonolysis of the corresponding N-Boc-protected Trp. Judicious use of azide chemistry also provided the requisite orthogonality for elaboration of the resin bound peptide. More recently, Taylor and coworkers described an concise approach to synthesizing daptomycin entirely on solid support enabling the rapid synthesis of other daptomycin analogues (Scheme 3).38,39 To enable the total SPPS approach Taylor's group employed an elegant protecting group strategy, including a number of azide masked amino acid building blocks, as well as optimized Fmoc deprotection conditions to protect the sensitive ester linkage.
Scheme 1. SPPS approach used by Cubist team to achieve the first total synthesis of daptomycin.
Scheme 2. Serine ligation route used by Li and coworkers in synthesizing daptomycin.
Scheme 3. Entirely SPPS approach used by the Taylor group in the synthesis of daptomycin.
Semisynthetic approaches have also been used to produce daptomycin analogues with improved MIC profiles compared to that of the parent compound.40 Structure–activity relationship studies on the fatty acid tail revealed several critical factors for antimicrobial activity: lipophilicity, chain length and the location of key aromatic functionalities.40,41 The acyl tail is crucial for binding to the bacterial cell membrane and calcium-dependent insertion.42 One such semisynthetic analogue thus developed is surotomycin (Fig. 3) which differs from daptomycin only in the structure of its lipid tail. In surotomycin the lipid tail contains a conjugated substituted alkene and an aromatic moiety that was found to give enhanced activity against C. difficile. To date, two registered phase III clinical studies have been completed with surotomycin.43,44 Both were randomized, double blind, multi-center, global studies in adult patients with C. difficile-associated diarrhea. In these studies, the efficacy of surotomycin was compared to that of vancomycin for 10 day treatment regimens. At their conclusion these trials revealed that while surotomycin was generally well tolerated, it did not meet the primary efficacy endpoint of noninferiority compared to that of vancomycin. It also failed to meet the secondary endpoints, i.e. superiority over vancomycin and sustained clinical response at the end of the trial.
Fig. 3. Structure of surotomycin. This semisynthetic analogue of daptomycin bears an unnatural lipid tail imparting potent activity against C. difficile.
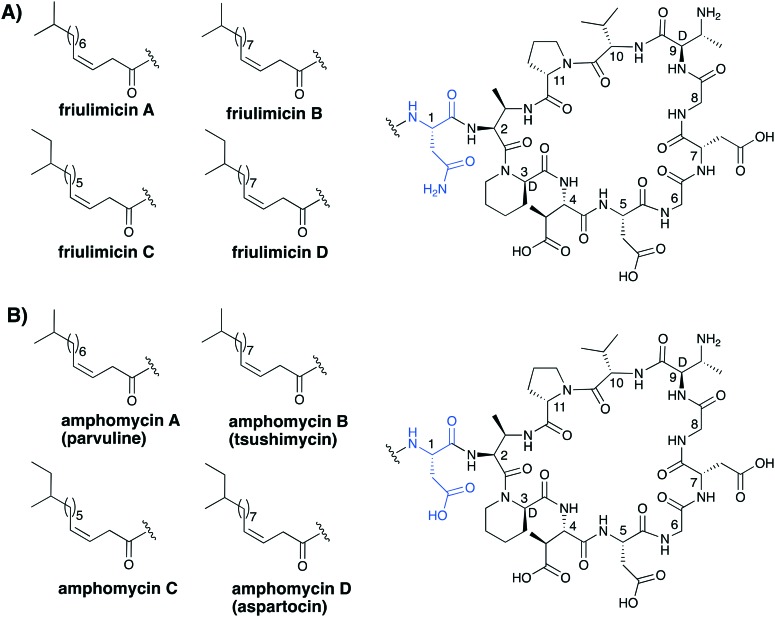
The lipodepsipeptides daptomycin and surotomycin are not the only CDAs to have been taken to clinical trials. Two others, amphomycin and friulimicin (Fig. 4), both belonging to a structurally distinct lipopeptide sub class of CDAs, have been evaluated as human therapeutics. Discovered in the 1950s, amphomycin was the first CDA reported in the literature.20 In the years following, a myriad of similar or identical compounds have been reported under different names, which has led to significant confusion in the nomenclature associated with the class.24,25,45,46 The reader is referred to a comprehensive 2005 review by Baltz and coworkers that serves to clarify much of the ambiguity associated with this group of CDAs.21,24 Typically these lipopeptides are subdivided into three subclasses, the amphomycins, the friulimicins and the laspartomycins. As indicated in Fig. 4, there is a logical convention used in the naming of the various friulimicins as A, B, C, or D depending on the lipid tail. We respectively propose to apply the same convention in the nomenclature used for describing the structurally similar amphomycin class of CDA (see Fig. 4 for proposed nomenclature). The amphomycins and the friulimicins are identical apart from the exocyclic Asp residue found in the amphomycin class, which in the friulimicins is instead an Asn. Variation in the lipid chains is responsible for additional diversity. The structure of the laspartomycin core peptide differs more significantly and is discussed in further detail in section 4.
Fig. 4. Structures of the (A) friulimicin and (B) amphomycin families of CDA. Indicated in brackets are the carious other names that have historically been assigned to these structures.
The amphomycins and friulimicins both share the same macrocyclic peptide consisting of 10 amino acids with cyclization between the diaminobutyric acid (Dab) at position 2 and Pro11. Several d- and nonproteinogenic amino acids, such as l-threo and d-allo-2,3-Dab, d-pipecolic acid (d-Pip) as well as l-thero-3-methyl-aspartate (MeAsp), are found at positions 2, 9, 3 and 4 in the peptide core respectively. Four main fatty acid tails are found throughout these compounds, consisting of either 13, 14 or 15 carbon atoms in total. The lipids commonly bear iso or anteiso branching groups at their terminal ends and all contain an unsaturated double bond between carbon three and four.
As is common to nearly all the CDAs, the amphomycin, friulimicin and laspartomycin lipopeptides contain the same Asp-X-Asp-Gly calcium binding motif as daptomycin. Despite this structural similarity, several investigations have revealed that the amphomycins, friulimicins, and laspartomycins operate via a mechanism different than that of daptomycin. Early studies with amphomycin indicated that it interferes with the first membrane associated step of bacterial cell wall biosynthesis by disrupting the transfer of the soluble UDP cell wall precursor to the phospholipid membrane anchor undecaprenyl phosphate (C55-P).47 More recent studies have revealed that members of this class of CDA form tight complexes with C55-P as a major driver of their antibacterial mechanisms.48,49
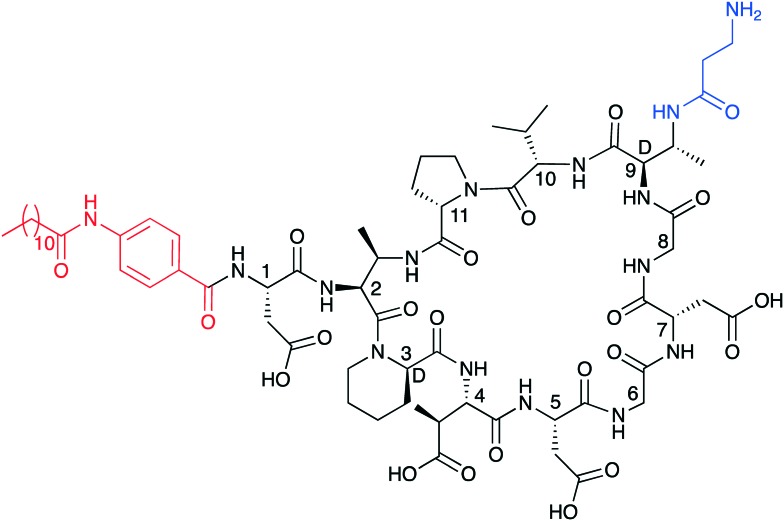
As stated above, both amphomycin and friulimicin have been investigated as possible leads for antibiotic drug development. To this end a semisynthetic analogue of amphomycin, MX-2401 (Fig. 5), was recently brought to clinical trials by BioWest Therapeutics Inc. for the treatment of serious Gram-positive infections.50,51 Compared to amphomycin, MX-2401 is rapidly bactericidal and was shown to be effective in several infections models with potent activity against VRSA, VRE and MRSA.48,49 Recent NMR studies demonstrated that S. aureus cells treated with MX-2401 exhibit a thinning of the cell wall, a decrease in d-alanine linked teichoic acids, and a reduction in peptidoglycan cross-linking.52 Although MX-2401 reached late stage preclinical development it has never been tested in human patients in a clinical setting. By comparison, in 2007 MerLion Pharmaceuticals initiated phase I clinical trial studies with friulimicin B. However, the trial was terminated soon thereafter due to unfavorable pharmacokinetics.
Fig. 5. Structure of the semisynthetic amphomycin analogue MX-2401. Highlighted in red is the 4-(dodecanamido)benzoic acid lipid tail and in blue the modified Dab9.
3. The A54145 and CDA classes
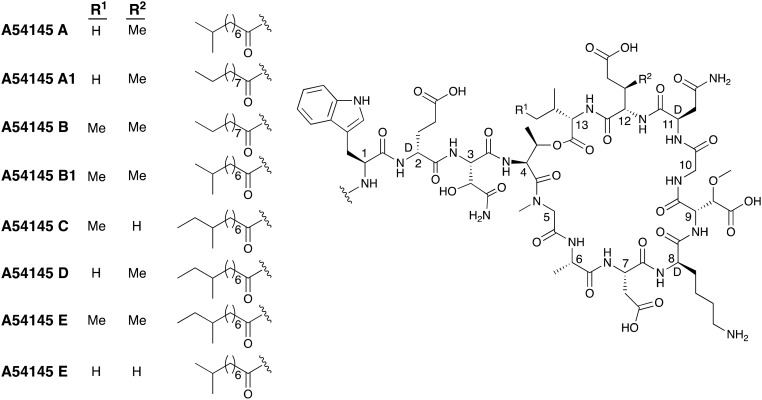
A54145 is a lipodepsipeptide like daptomycin and the A21978 family but is much more structurally complex (Fig. 6). The A54145 group is produced by Streptomyces fradiae and shows antibacterial activity against various strains of Staphylococcus aureus, Staphylococcus epidermidis, Clostridium, Streptococcus, and enterococci.19,53 The lipid tails of the A54145 family are fully saturated and often terminate with iso or anteiso branching patterns.19 The structural complexity of the A54145 group is found in their peptide sequences. Seven of the thirteen component amino acids are nonproteinogenic including three d-amino acids. Six of the seven unique amino acids (d-Glu2, 3-hydroxy-l-aspragine (hAsn3), sarcosine (Sar5), d-Lys8, 3-methoxyAsp (MeO-Asp9) and d-Asn11) are found in all members of the A54145 family. In addition, a MeGlu residue is found at position 12 in four of the known A54145 peptides and position 13 is either an Ile or a Val giving rise to additional structural variation.53,54 A54145 is homologous to daptomycin and permeabilizes the cell membrane of Gram-positive bacteria in a similar fashion. As for daptomycin, membrane permeabilization depends on the presence of calcium and phosphatidylglycerol and leads to the formation of oligomeric membrane pores that consist of 6–8 individual subunits.55,56
Fig. 6. Structures of A54145 class of CDAs.
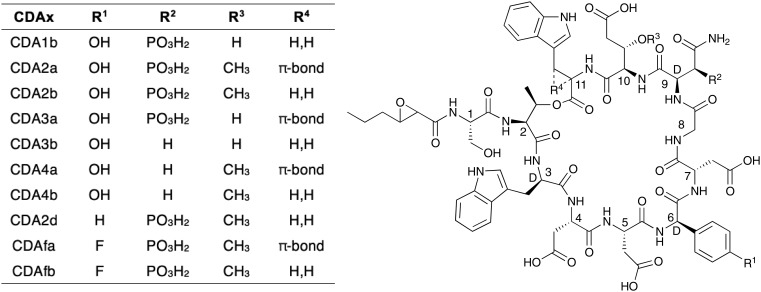
In 1983 a new class of calcium-dependent antibiotics produced by Streptomyces coelicolor was discovered and given the general family name CDA which subsequently led to some confusion in the field.18 These CDAs from S. coelicolor were later structurally characterized and found to belong to the same class of lipodepsipeptides as daptomycin and the A54145 family (Fig. 7).57 However, unlike daptomycin and the A54145s, in the so named CDA group the exocyclic motif consist of a single amino acid (Ser) and the fatty acid acyl tail is exclusively found to be 2,3-epoxy-hexanoyl.57,58 A high degree of structural variation is found within the macrocycles of the CDA peptides. Most notable is the d-hydroxy asparagine at position 9, in which the beta-hydroxyl is phosphorylated in CDA1 and CDA2. Two additional d-amino acids, d-Trp and d-4-hydroxyphenylglycine, are found at positions 3 and 6 respectively. Position 10 is either a Glu or 3-methyl-Glu. Another unique site of structural variation is found in the side chain of Trp-11: in CDA2a, CDA3a, ACD4a and CDAfa, Trp-11 is dehydrogenated to Z-2,3-dehydrotryptophan (ΔTrp).58
Fig. 7. Structural diversity among the CDA class.
4. Emerging insights and recent developments
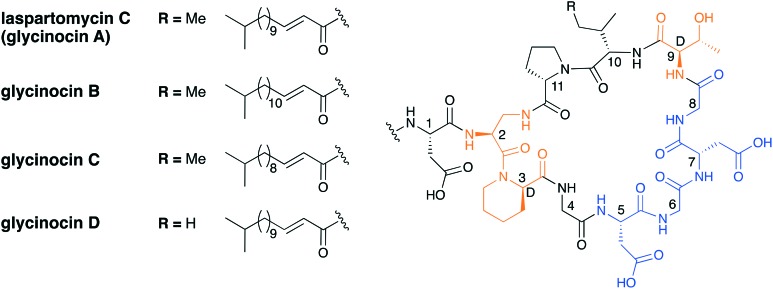
Recent studies conducted both in the Payne group59 as well as our own60 have focused on the synthesis and mechanism of action of the laspartomycin family of the CDAs. The laspartomycins belong to the same sub-family as the amphomycins/friulimicins and the first laspartomycin to be reported was isolated from Streptomyces viridochromogenes in 1968.22 Due to the analytical constraints of the time, a full chemical characterization was not reported until 2007 when advanced purification techniques and spectroscopic methods led to the structure elucidation of the major component, laspartomycin C.23 Somewhat parallel to this work, investigations aimed at identifying the active antibiotic components in the fermentation broth of an unidentified terrestrial actinomycetes species led to the discovery of the so-called glycinocin family of the CDAs.61 The glycinocins were subsequently shown to be largely identical to the laspartomycins with the dominant glycinocin A sharing the exact same structure as laspartomycin C (Fig. 8). Among the other glycinocins characterized, glycinocin B and C have the same peptide core as laspartomycin C and only differ in the length of their fatty acid, with the lipids both consisting of iso branching at the terminal end. Glycinocin D has the same lipid tail as laspartomycin C however, in place of a Ile at position 10, it contains Val (as for the amphomycins and friulimicins).61 Unlike the amphomycins and friulimicins, in both the laspartomycin and glycinocin classes, a diaminopropionic acid residue (Dap2) facilitates macrolactamization rather than 2S,3R-diaminobutyric acid (Dab2). Other differences include d-allo-Thr9 in place of 2R,3R-d-Dab9, and a simple Gly at position 4 instead of non-proteinogenic MeAsp4. Furthermore, the fatty acid of the laspartomycin/glycinocin family is 2,3-unstaurated and has E geometry versus the 3,4-unsaturation with Z geometry observed in the amphomycin and friulimicin families. Laspartomycin C shows antibacterial activity against a variety of Gram-positive pathogens including MRSA, VRE and VRSA.22,62 To further examine its potential as an antibiotic lead compound, a number of semisynthetic laspartomycin variants have been reported by incorporating a number of different lipid tails.62 These studies revealed that both the length and saturation of the lipid tail is essential for full activity.
Fig. 8. Laspartomycin C and the glycinocin A–D family. Highlighted in blue is the conserved Asp-X-Asp-Gly Ca2+ binding motif and highlighted in orange are nonproteinogenic amino acids.
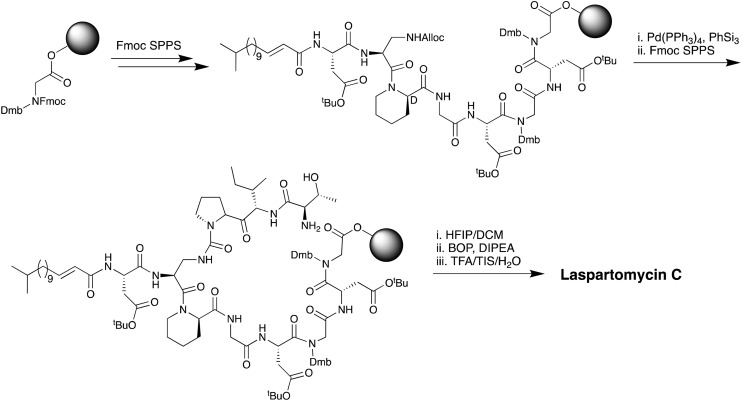
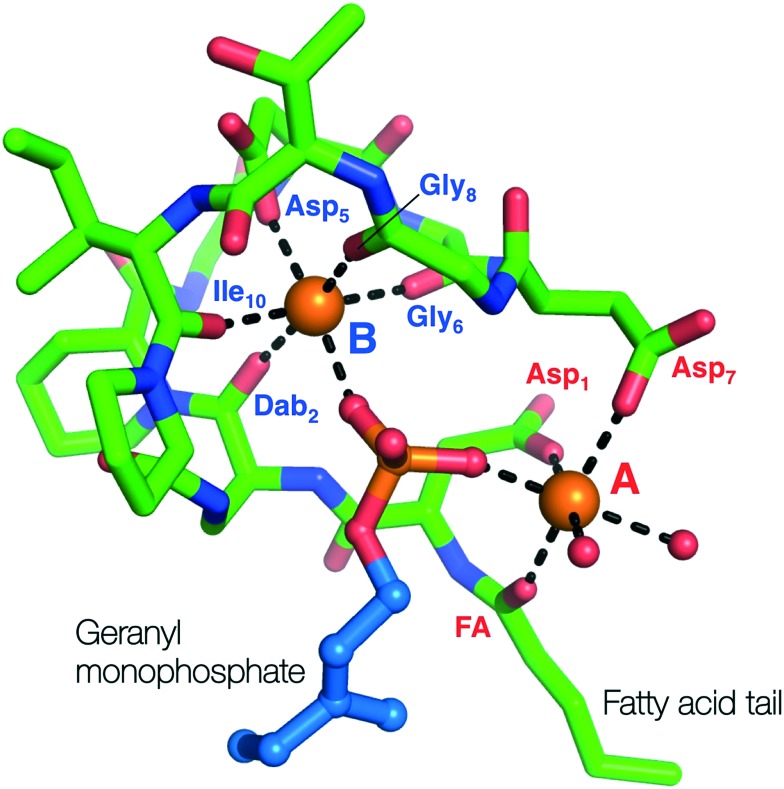
While laspartomycin C shares many structural similarities with the amphomycins and friulimicins the mechanistic details of its antibacterial mode of action remained unknown until recently elucidated by our group in 2016.60 In doing so we also developed a convenient combined solid- and solution-phase approach for the synthesis of laspartomycin C and structural analogues (Scheme 4). Notably, our synthesis of laspartomycin C represents only the second CDA to be prepared via total synthesis. Soon thereafter Payne and coworkers reported a similar approach to the synthesis of glycinocins A–C and variants thereof.63 Using our synthetic laspartomycin we demonstrated that it specifically and tightly binds undecaprenyl phosphate (C55-P) a bacterial phospholipid essential for cell wall synthesis. Using isothermal titration calorimetry, we quantified C55-P binding laspartomycin with a remarkably low nanomolar Kd value. Building upon this work, in a collaboration with the Janssen group, we recently solved the crystal structure of laspartomycin C bound to a more soluble truncated analogue of C55-P, geranyl phosphate (C10-P).64 The structure reveals a basic unit comprised of a 1 : 2 : 1 laspartomycin C/Ca2+/C55-P stoichiometry (Fig. 9). The structure also suggests that a dimeric species may be biologically relevant wherein two laspartomycin C fatty acid side chains and two C10-P tails orientated perpendicular to a hydrophobic plane. The phosphate head groups are then sequestered within the core of laspartomycin C dimer which lies slightly embedded in the bacterial membrane. While a previous crystal structure of tsushimycin (amphomycin B according to the nomenclature proposed in Fig. 4) provided insight into calcium binding,65 our laspartomycin C structure represents the first structure of a CDA bound to both calcium and its biomolecular target.
Scheme 4. Combined solution- and solid-phase approach used by our group in the first total synthesis of laspartomycin C.
Fig. 9. Crystal structure of laspartomycin C (green stick representation) bound to two bound Ca2+ ions (orange spheres) and the geranyl monophosphate ligand coordinated by the Ca2+ ions. The two Ca2+ ions are labelled A (red) and B (blue) with the interacting parts of laspartomycin C responsible for the chelation of the Ca2+ ions also indicated.
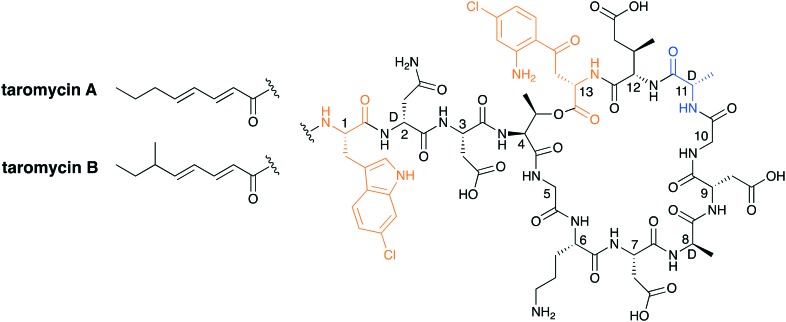
Advancements in the field of next-generation sequencing technologies have shed new light on microbial genomes as a potentially rich source for novel antibiotics. In a recent report from the Moore group, a transformation-associated recombination cloning technique was used to clone, refactor, and express a silent biosynthetic pathway to yield two CDAs named taromycin A and taromycin B (Fig. 10).66,67 While taromycin is structurally similar to daptomycin it also contains two chlorinated amino acids, namely a 6-chloro-tryptophan at position 1 and a 4-chloro-kynuenine at position 13. While these chlorinated amino acids are not found in any other CDAs, they do not seem to offer a significant advantage as the activity of taromycin does not match or surpass that of daptomycin.
Fig. 10. Taromycin A and B. Highlighted in orange are the chlorinated amino acids 6-chloro-tryptophan and 4-chloro-kynurenine. The taromycin family has a d-alanine in place of the d-serine found in daptomycin (highlighted in blue).
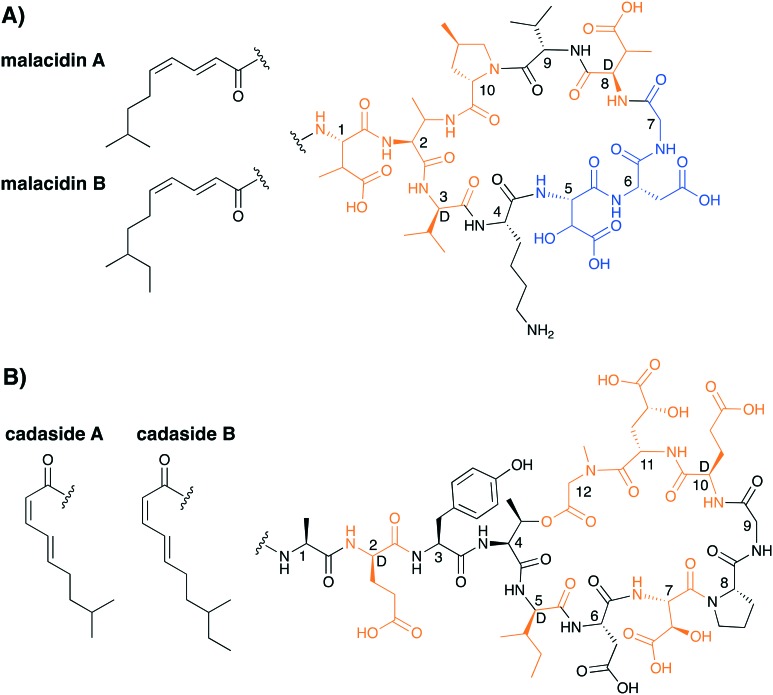
In another demonstration of the power of microbial genome mining, Brady and co-workers recently reported an entirely new class of CDAs, termed the malacidins (Fig. 11A).68 The malacidins are unique among the CDAs in that they lack the canonical Asp-X-Asp-Gly calcium binding motif which is instead replaced by the tri-peptide β-hydroxy-Asp-Asp-Gly. This change results in the malacidins having a nine amino acid peptide macrocycle rather than the ten amino acid macrocycle found in all other CDAs. Despite this difference, the malacidins demonstrate a strong calcium dependence for their antibacterial activity. Also, of note is the unique mechanism of action of the malacidins. Unlike the other CDAs, that act by membrane disruption (daptomycin, A54145, CDA) or by binding C55-P to inhibit cell-wall biosynthesis (amphomycins, friulimicins, laspartomycins), the malacidins inhibit bacterial cell-wall biosynthesis by binding to lipid II.68 The activity of the malacidins are on par with that of daptomycin, with MICs of 0.2–0.8 μg mL–1 against MRSA and 0.8–2.0 μg mL–1 against VRE. Malacidin's in vivo activity was also assessed in a rat cutaneous wound model where it successfully cleared MRSA infection.
Fig. 11. Structures of A) the malacidins and B) the cadasides. Highlighted in orange are the nonproteinogenic amino acids and highlighted in blue is the presumed Ca2+ binding motif for the malacidins.
Very recently, the Brady group reported another new member of the CDA family which they also identified using genome mining approaches and named the cadasides (Fig. 11B).69 Similar to the malacidins, the cadasides contain a nine amino macrocycle however, unlike the malacidins, the cycle is closed via an ester linkage to yield a depsipeptide. Also of note, while the malacidin lipid tail bears an E,Z unsaturation pattern, that of the cadasides contains the reversed Z,E geometry. The cadasides also lack the canonical calcium binding Asp-X-Asp-Gly motif found in other CDAs but do contain a β-hydroxy-Asp and a γ-hydroxy-Glu within their macrocycle. It is tempting to speculate that one or both of these hydroxylated amino acids may play a role in calcium binding. However, this hypothesis remains to be investigated experimentally. While metagenomic based antibiotic discovery is still in its infancy it may prove to be a useful tool in discovering new CDAs that are hidden in so called cryptic gene clusters.
5. Conclusions
The spread of multi-drug-resistant bacteria remains a concern and presents a serious threat to modern healthcare. While the first CDA was reported in the early 1950s, it was the approval of daptomycin in 2003 that brought this unique class of antibiotics into the spotlight. Enabled by advances in screening and synthetic chemistry, the past decade has witnessed a burst of activity in the field. With now over 40 unique CDAs reported, their structural diversity continues to provide opportunity for discovery. Of particular note is the growing appreciation that the different members of the CDA superfamily utilize a diverse range of antibacterial mechanisms. A more complete understanding of these different mechanisms, supported by structural insights, will be key to establishing a more complete structure–activity relationships for the various CDAs. Such information is in turn expected to inform the design of optimized CDAs accessible via synthetic means. While daptomycin remains the only clinically used CDA, the many advances and increasing interest in this compound class suggest there is good reason to expect additional family members to enter the clinic in the years to come.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgments
Financial support was provided by The Netherlands Organization for Scientific Research (NWO graduate school PhD fellowship to T. M. W.) and The European Research Council (ERC consolidator grant to N. I. M., grant agreement No. 725523).
References
- O'Neill J., Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations, 2014, http://www.jpiamr.eu/wp-content/uploads/2014/12/AMR-Review-Paper-Tackling-a-crisis-for-the-health-and-wealth-of-nations_1-2.pdf.
- CDCD, US Dep. Heal. Hum. Serv.
- Waterman P. E., McGann P., Snesrud E., Clifford R. J., Kwak Y. I., Munoz-Urbizo I. P., Tabora-Castellanos J., Milillo M., Preston L., Aviles R., Sutter D. E., Lesho E. P. Antimicrob. Agents Chemother. 2013;57:4584–4586. doi: 10.1128/AAC.00275-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tehrani K. H. M. E., Martin N. I. Med. Chem. Commun. 2018;9:1439–1456. doi: 10.1039/c8md00342d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang R., Li J., Wu Z., Yin W., Schwarz S., Tyrrell J. M., Zheng Y., Wang S., Shen Z., Liu Z., Liu J., Lei L., Li M., Zhang Q., Wu C., Zhang Q., Wu Y., Walsh T. R., Shen J. Nat. Microbiol. 2017;2:1–7. doi: 10.1038/nmicrobiol.2016.260. [DOI] [PubMed] [Google Scholar]
- Zhang X., Zhang B., Guo Y., Wang J., Zhao P., Liu J., He K. Int. J. Food Microbiol. 2019;291:87–90. doi: 10.1016/j.ijfoodmicro.2018.11.013. [DOI] [PubMed] [Google Scholar]
- Chambers H. F., DeLeo F. R. Nat. Rev. Microbiol. 2009;7:629–641. doi: 10.1038/nrmicro2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourramezan N., Ohadian Moghadam S., Pourmand M. R. New Microbes New Infect. 2019;27:29–35. doi: 10.1016/j.nmni.2018.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacconelli E., Cataldo M. A. Int. J. Antimicrob. Agents. 2008;31:99–106. doi: 10.1016/j.ijantimicag.2007.08.026. [DOI] [PubMed] [Google Scholar]
- Davies J. Can. J. Infect. Dis. Med. Microbiol. 2006;17:287–290. doi: 10.1155/2006/707296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventola C. L. Pharm. Ther. 2015;40:277–283. [PMC free article] [PubMed] [Google Scholar]
- Aminov R. Biochem. Pharmacol. 2017;133:4–19. doi: 10.1016/j.bcp.2016.10.001. [DOI] [PubMed] [Google Scholar]
- Gwynn M. N., Portnoy A., Rittenhouse S. F., Payne D. J. Ann. N. Y. Acad. Sci. 2010;1213:5–19. doi: 10.1111/j.1749-6632.2010.05828.x. [DOI] [PubMed] [Google Scholar]
- Muller A. E. and Gyssens I. C., Kucers Use Antibiot. A Clin. Rev. Antibacterial, Antifung. Antiparasit. Antivir. Drugs, 7th edn, 2017, vol. 2, pp. 866–907. [Google Scholar]
- Sauermann R., Rothenburger M., Graninger W., Joukhadar C. Pharmacology. 2008;81:79–91. doi: 10.1159/000109868. [DOI] [PubMed] [Google Scholar]
- Debono M., Barnhart M., Carrell C., Occolowitz J. L., Abbott B., Fukuda D., Hamil L., Hoffmann J. J. Antibiot. 1986;6:761–777. doi: 10.7164/antibiotics.40.761. [DOI] [PubMed] [Google Scholar]
- Mchenney M. A., Hosted T. J., Dehoff B. S., Rosteck P. R., Baltz R. H. J. Bacteriol. 1998;180:143–151. doi: 10.1128/jb.180.1.143-151.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakey J. H., Lea E. J., Rudd B. A., Wright H. M., Hopwood D. A. J. Gen. Microbiol. 1983;129:3565–3573. doi: 10.1099/00221287-129-12-3565. [DOI] [PubMed] [Google Scholar]
- Mertz F. P., Berry D. M., Boeck L. D., Papiska H. R., Wetzel R. W., Mynderse J. S., Fukuda D. S. J. Antibiot. 1989;XLIII:587–593. doi: 10.7164/antibiotics.43.587. [DOI] [PubMed] [Google Scholar]
- Heinemann I. R., Kaplan B., Muir M. A., Hooper R. D. Antibiot. Chemother. 1953;3:1239–1242. [PubMed] [Google Scholar]
- Aretz W., Meiwes J., Seibert G., Vobis G., Wink J. J. Antibiot. 2000;53:807–815. doi: 10.7164/antibiotics.53.807. [DOI] [PubMed] [Google Scholar]
- Naganawa H., Hamada M., Maeda K., Okami Y., Takeuchi T., Umezawa H. J. Antibiot. 1968;XXI:55–62. doi: 10.7164/antibiotics.21.55. [DOI] [PubMed] [Google Scholar]
- Borders D. B., Leese R. A., Jarolmen H., Francis N. D., Fantini A. A., Falla T., Fiddes J. C., Aumelas A. J. Nat. Prod. 2007;70:443–446. doi: 10.1021/np068056f. [DOI] [PubMed] [Google Scholar]
- Baltz R. H., Miao V., Wrigley S. K., Miao V. Nat. Prod. Rep. 2005:717–741. doi: 10.1039/b416648p. [DOI] [PubMed] [Google Scholar]
- Strieker M., Marahiel M. A. ChemBioChem. 2009;10:607–616. doi: 10.1002/cbic.200800546. [DOI] [PubMed] [Google Scholar]
- Debono M., Abbott B. J., Molloy R. M., Fukuda D. S., Hunt A. H., Daupert V. M., Counter F. T., Ott J. L., Carrell C. B., Howard L. C., Boeck L. D., Hamill R. L. J. Antibiot. 1988;XLI:1093–1105. doi: 10.7164/antibiotics.41.1093. [DOI] [PubMed] [Google Scholar]
- Huber F. M., Pieper R. L., Tietz A. J., Division A. D., Lilly E. J. Biotechnol. 1988;7:283–292. [Google Scholar]
- Zhang J., Scott W. R. P., Gabel F., Wu M., Desmond R., Bae J., Zaccai G., Algar W. R., Straus S. K. Biochim. Biophys. Acta, Proteins Proteomics. 2017;1865:1490–1499. doi: 10.1016/j.bbapap.2017.07.020. [DOI] [PubMed] [Google Scholar]
- Beriashvili D., Taylor R., Kralt B., Mazen N. A., Taylor S. D., Palmer M. Chem. Phys. Lipids. 2018;216:73–79. doi: 10.1016/j.chemphyslip.2018.09.015. [DOI] [PubMed] [Google Scholar]
- Müller A., Wenzel M., Strahl H., Grein F., Saaki T. N. V., Kohl B., Siersma T., Bandow J. E., Sahl H.-G., Schneider T., Hamoen L. W. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E7077–E7086. doi: 10.1073/pnas.1611173113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn J. P., Lolans K., Weinstein R. A., Hayden M. K. Antimicrob. Agents Chemother. 2005;49:1664–1665. [Google Scholar]
- Marty F. M., Yeh W. W., Wennersten C. B., Venkataraman L., Albano E., Alyea E. P., Gold H. S., Baden L. R., Pillai S. K. Antimicrob. Agents Chemother. 2006;44:595–597. doi: 10.1128/JCM.44.2.595-597.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster T. J. FEMS Microbiol. Rev. 2017:430–449. doi: 10.1093/femsre/fux007. [DOI] [PubMed] [Google Scholar]
- Bayer A. S., Mishra N. N., Chen L., Kreiswirth B. N., Rubio A., Yang S. Antimicrob. Agents Chemother. 2015;59:4930–4937. doi: 10.1128/AAC.00970-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopher Y. Z. A. D., Baltz R. H., Paul B., Marie-Franciose C. G., Sascha D., Xiaowei H. E., Kulkarni V., Leitheiser C., Miao V. P. W., Nguyen K. T., Parr I. B. and Ritz D., WO Pat., 2006110185A2, 2006.
- Lam H. Y., Zhang Y., Liu H., Xu J., Wong C. T. T., Xu C., Li X. J. Am. Chem. Soc. 2013;135:6272–6279. doi: 10.1021/ja4012468. [DOI] [PubMed] [Google Scholar]
- Lee C. L., Lam H. Y., Li X. Nat. Prod. Rep. 2015:1274–1279. doi: 10.1039/c5np00001g. [DOI] [PubMed] [Google Scholar]
- Lohani C. R., Taylor R., Palmer M., Taylor S. D. Org. Lett. 2015;17:748–751. doi: 10.1021/acs.orglett.5b00043. [DOI] [PubMed] [Google Scholar]
- Barnawi G., Noden M., Taylor R., Lohani C., Beriashvili D., Palmer M., Taylor S. D. Pept. Sci. 2019;111:e23094. doi: 10.1002/bip.23094. [DOI] [PubMed] [Google Scholar]
- Yin N., Li J., He Y., Herradura P., Pearson A., Mesleh M. F., Mascio C. T., Howland K., Steenbergen J., Thorne G. M., Citron D., Van Praagh A. D. G., Mortin L. I., Keith D., Silverman J., Metcalf C. J. Med. Chem. 2015;58:5137–5142. doi: 10.1021/acs.jmedchem.5b00366. [DOI] [PubMed] [Google Scholar]
- Knight V., Carmela C., Laurent M., Silverman J. J. Ind. Microbiol. Biotechnol. 2016;43:195–204. doi: 10.1007/s10295-015-1714-6. [DOI] [PubMed] [Google Scholar]
- Mukhtar T. A., Patel T., Koteva K., Waglechner N., Hughes D. W., Wright G. D., De Pascale G. Antimicrob. Agents Chemother. 2012:757–764. doi: 10.1128/AAC.05441-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boix V., Fedorak R. N., Mullane K. M., Pesant Y., Stoutenburgh U., Jin M., Adedoyin A., Chesnel L., Guris D., Larson K. B., Murata Y. Open Forum Infect. Dis. 2017;4:1–8. doi: 10.1093/ofid/ofw275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley P., Louie T., Lutz J. E., Khanna S., Stoutenburgh U., Jin M., Adedoyin A., Chesnel L., Guris D., Larson K. B., Murata Y. J. Antimicrob. Chemother. 2017;72:3462–3470. doi: 10.1093/jac/dkx299. [DOI] [PubMed] [Google Scholar]
- Jun-Ichi Shoji H. O., Kozuka S., Okamoto S., Sakazaki R. J. Antibiot. 1968;XXI:439–443. [PubMed] [Google Scholar]
- Tyurin A. P., Alferova V. A., Paramonov A. S., Shuvalov M. V., Malanicheva I. A., Grammatikova N. E., Solyev P. N., Liu S., Sun C., Prokhorenko I. A., Efimenko T. A., Terekhova L. P., Efremenkova O. V., Shenkarev Z. O., Korshun V. A. MedChemComm. 2018;9:667–675. doi: 10.1039/c8md00002f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H., Oiwa R., Matsukura S., Omura S. Biochem. Biophys. Res. Commun. 1979;86:902–908. doi: 10.1016/0006-291x(79)91797-2. [DOI] [PubMed] [Google Scholar]
- Rubinchik E., Schneider T., Elliott M., Scott W. R. P., Pan J., Anklin C., Yang H., Dugourd D., Müller A., Gries K., Straus S. K., Sahl H. G., Hancock R. E. W. Antimicrob. Agents Chemother. 2011;55:2743–2754. doi: 10.1128/AAC.00170-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider T., Gries K., Josten M., Wiedemann I., Pelzer S., Labischinski H., Sahl H. G. Antimicrob. Agents Chemother. 2009;53:1610–1618. doi: 10.1128/AAC.01040-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugourd D., Yang H., Elliott M., Siu R., Clement J. J., Straus S. K., Hancock R. E. W., Rubinchik E. Antimicrob. Agents Chemother. 2011;55:3720–3728. doi: 10.1128/AAC.00322-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig W. A., Andes D. R., Stamstad T. Antimicrob. Agents Chemother. 2010;54:5092–5098. doi: 10.1128/AAC.00238-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M., Chang J., Coffman L., Kim S. J. Sci. Rep. doi: 10.1038/srep31757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao V., Brost Æ. R., Chapple Æ. J., Marie-franc K. S. Æ. J. Ind. Microbiol. Biotechnol. 2006:129–140. doi: 10.1007/s10295-005-0028-5. [DOI] [PubMed] [Google Scholar]
- Counter F. T., Allen N. E., Fukuda D. S., Hobbs J. N., Ott J., Ensminger P. W., Mynderse J. S., Preston D. A., Wu C. Y. E., Lilly T., Lilly E. J. Antibiot. 1990:616–622. doi: 10.7164/antibiotics.43.616. [DOI] [PubMed] [Google Scholar]
- Taylor R., Butt K., Scott B., Zhang T., Muraih J. K., Mintzer E., Taylor S., Palmer M. Biochim. Biophys. Acta, Biomembr. 2016;1858:1999–2005. doi: 10.1016/j.bbamem.2016.05.020. [DOI] [PubMed] [Google Scholar]
- Zhang T., Taylor S. D., Palmer M., Duhamel J. Biophys. J. 2016;111:1267–1277. doi: 10.1016/j.bpj.2016.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempter C., Kaiser D., Haag S., Nicholson G., Gnau V., Walk T., Gierling K. H., Decker H., Zahner H., Jung G., Metzger J. W. Angew. Chem., Int. Ed. Engl. 1997;42:498–501. [Google Scholar]
- Hojati Z., Milne C., Harvey B., Gordon L., Borg M., Flett F., Wilkinson B., Sidebottom P. J., Rudd B. A. M., Hayes M. A., Smith C. P., Micklefield J. Chem. Biol. 2002;9:1175–1187. doi: 10.1016/s1074-5521(02)00252-1. [DOI] [PubMed] [Google Scholar]
- Corcilius L., Elias N. T., Ochoa J. L., Linington R. G., Payne R. J. J. Org. Chem. 2017;82:12778–12785. doi: 10.1021/acs.joc.7b01959. [DOI] [PubMed] [Google Scholar]
- Kleijn L. H. J., Oppedijk S. F., Hart P. T., Van Harten R. M., Martin-Visscher L. A., Kemmink J., Breukink E., Martin N. I. J. Med. Chem. 2016;59:3569–3574. doi: 10.1021/acs.jmedchem.6b00219. [DOI] [PubMed] [Google Scholar]
- Kong G. T. C. F. J. Antibiot. 2003;56:557–564. doi: 10.7164/antibiotics.56.557. [DOI] [PubMed] [Google Scholar]
- Curran W. V., Leese R. A., Jarolmen H., Borders D. B., Dugourd D., Chen Y., Cameron D. R. J. Nat. Prod. 2007;70:447–450. doi: 10.1021/np068062b. [DOI] [PubMed] [Google Scholar]
- Corcilius L., Liu D. Y., Ochoa J. L., Linington R. G., Payne R. J. Org. Biomol. Chem. 2018;16:5310–5320. doi: 10.1039/c8ob01268g. [DOI] [PubMed] [Google Scholar]
- Kleijn L. H. J., Vlieg H. C., Wood T. M., Sastre Toraño J., Janssen B. J. C., Martin N. I. Angew. Chem., Int. Ed. 2017;56:16546–16549. doi: 10.1002/anie.201709240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunkóczi G., Vértesy L., Sheldrick G. M. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2005;61:1160–1164. doi: 10.1107/S0907444905017270. [DOI] [PubMed] [Google Scholar]
- Yamanaka K., Reynolds K. A., Kersten R. D., Ryan K. S., Gonzalez D. J., Nizet B. S., Dorrestein V., Moore P. C. Proc. Natl. Acad. Sci. U. S. A. 2014;111:1957–1962. doi: 10.1073/pnas.1319584111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds K. A., Luhavaya H., Li J., Dahesh S., Nizet V., Yamanaka K., Moore B. S. J. Antibiot. 2018;71:333–338. doi: 10.1038/ja.2017.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hover B. M., Kim S. H., Katz M., Charlop-Powers Z., Owen J. G., Ternei M. A., Maniko J., Estrela A. B., Molina H., Park S., Perlin D. S., Brady S. F. Nat. Microbiol. 2018;3:415–422. doi: 10.1038/s41564-018-0110-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Shang Z., Lemetre C., Ternei M. A., Brady S. F., Wu C., Shang Z., Lemetre C., Ternei M. A., Brady S. F. J. Am. Chem. Soc. 2019;141:3910–3919. doi: 10.1021/jacs.8b12087. [DOI] [PMC free article] [PubMed] [Google Scholar]