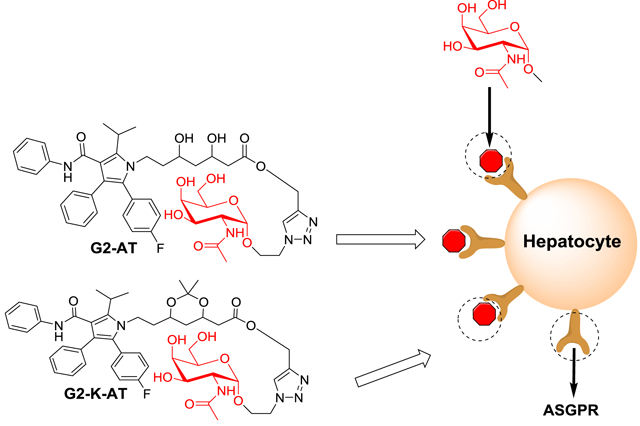
Abstract
Targeted drug delivery platforms can increase the concentration of drugs in specific cell populations, reduce adverse effects, and hence improve the therapeutic effect of drugs. Herein, we designed two conjugates by installing the targeting ligand GalNAc (N-acetylgalactosamine) onto atorvastatin (AT). Compared to the parent drug, these two conjugates, termed G2-AT and G2-K-AT, showed increased hepatic cellular uptake. Moreover, both conjugates were able to release atorvastatin, and consequently showed dramatic inhibition of β-hydroxy-β-methylglutaryl-CoA (HMG-CoA) reductase and increased LDL receptors on cell surface.
Keywords: atorvastatin, N-acetylgalactosamine, ligand conjugates, asialoglycoprotein receptor, targeted delivery
Graphical Abstract
1. Introduction.
Atorvastatin, a member of the statin drugs, is commonly used for the treatment of hypercholesterolemia patients, who suffer from cardiovascular disease and are more likely to develop atherosclerosis.1–3 Atorvastatin inhibits HMG-CoA reductase in order to increase the metabolism of LDL-cholesterol in hepatocytes. However, due to its low solubility4 and pre-systemic clearance in the gastrointestinal tract and liver,5, 6 the therapeutic effect of atorvastatin is impaired.7 Meanwhile, atorvastatin uptake by cells other than hepatocytes leads to various dose-related side effects such as myopathy and muscle pain.8, 9 In order to improve the effective delivery of atorvastatin to target organs and reduce its side effects, a new approach may be needed such as introducing targeting molecules, which can improve the concentration of atorvastatin in specific cell populations.
With advances in molecular biology, multiple receptors and transporters have been identified.10 They were found to play an important role in transmembrane transportation of drugs11and were utilized to improve pharmacological properties of active ingredients.12, 13 Based on these findings, a number of small molecule ligands have been exploited for modifying the properties of drugs such as water solubility, stability, and membrane permeability.14–18 Among those ligands, oligopeptide,19 folate acid,20 glucosamine,21 galactosamine,22 and bile acid23 were extensively used for targeting specific cells and tissues, which exhibited potential for therapeutic applications. In particular, N-acetylgalactosamine (GalNAc), a galactose derivative, was a well-characterized hepatocytes-targeting moiety due to its specific binding with asialoglycoprotein receptor (ASGPR) on the surface of hepatocytes.24 For example, GalNAc modified nanoparticle-siRNA system improved the gene silencing effects in the hepatocytes compared to free siRNA and naked nanoparticle.25 Moreover, the conjugation of antisense oligonucleotide and siRNA to GalNAc can dramatically enhance the entry of antisense oligonucleotide and siRNA into hepatocytes.26, 27 Building upon previous studies, GalNAc may serve as a promising ligand for targeted delivery of atorvastatin. Therefore, we designed and synthesized two conjugates of atorvastatin, termed G2-AT and G2-K-AT (Scheme 1), by covalently attaching GalNAc to atorvastatin using a click chemistry reaction.28 These two conjugates did not display inhibition of HMG-CoA reductase until release of the parent drug. G2-AT can be degraded by esterase in physiological condition (pH ~ 7.40), while G2-K-AT can be degraded in a more acidic pH (pH ~ 4.5). Importantly, cellular uptake of both conjugates was significantly improved compared to the parent drug atorvastatin in a cell model. Moreover, both conjugates enhanced the expression of LDL receptors in comparison to the parent drug.
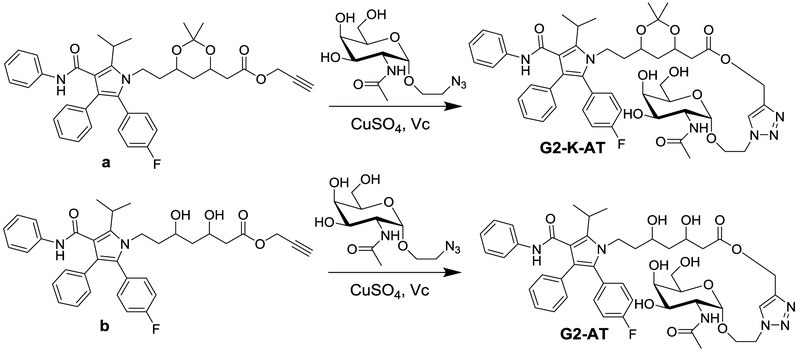
Scheme 1.
Synthetic routes to G2-AT and G2-K-AT.
2. Results and discussion
2.1. Design and synthesis of G2-AT and G2-K-AT
G2-AT and G2-K-AT were synthesized as shown in scheme 1. Firstly, two intermediates a and b were synthesized according to the reported methods.29 Then GalNAc was introduced through a ‘click reaction’ to yield G2-AT and G2-K-AT, respectively. Both conjugates were confirmed by 1H NMR and mass spectrometry (Supporting Information).
2.2. Hydrolysis assays
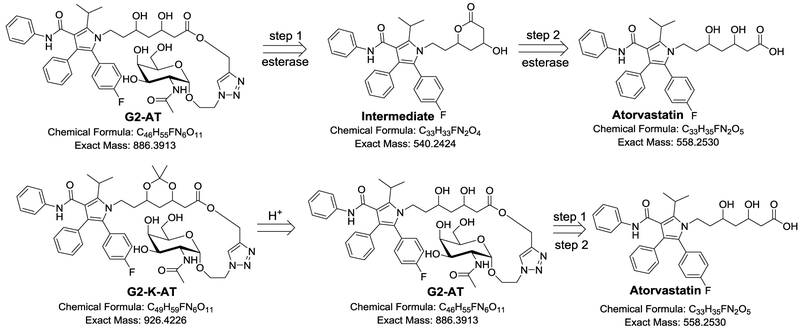
We further investigated the metabolism of G2-AT and G2-K-AT in the presence of esterase. Upon incubating with esterase in PBS (pH 7.40) at 37 °C, G2-AT underwent a two-step hydrolysis to release atorvastatin (Scheme 2), which had been proved in our previous publication.29 As shown in the mass spectra, an intermolecular ester formed while GalNAc was cleaved (Figure S1). The intermolecular ester was further transformed into atorvastatin (Figure S1). For G2-K-AT, an acidic (pH 4.5) environment was necessary to initiate the ketal deprotection, and then G2-K-AT was hydrolyzed by the esterase to release atorvastatin through the same route as G2-AT (Figure S2). These results showed that G2-K-AT requires an acidic condition as well as a process of ester hydrolysis in contrast to G2-AT to release atorvastatin. Atorvastatin was not detected upon incubating G2-K-AT in neutral PBS (pH 7.40, Figure S3), while another intermediate with a ketal group was found in the mass spectra (Figure S3).
Scheme 2.
Proposed metabolism pathways of G2-AT and G2-K-AT.
2.3. Inhibition of the activity of HMG-CoA Reductase
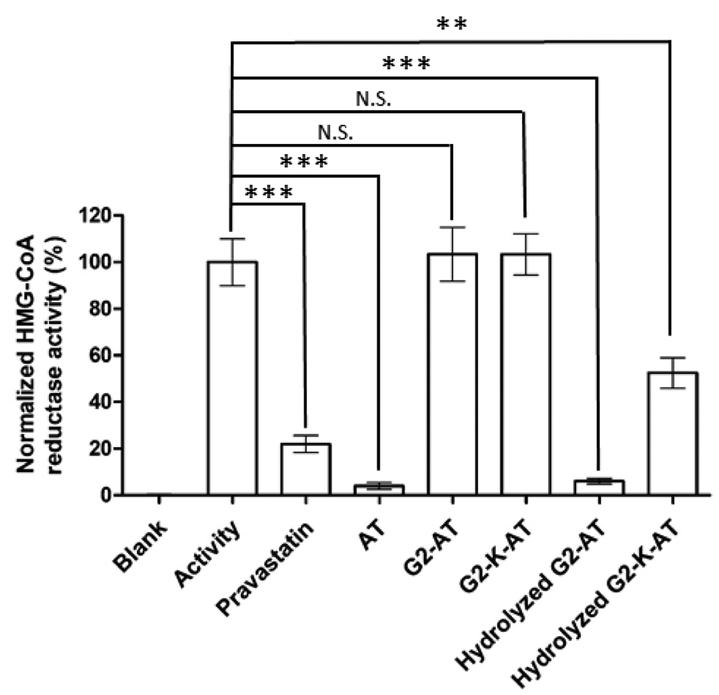
Next, we measured drug inhibition of the activity of HMG-CoA reductase using a well-established HMG-CoA reductase assay30. Both conjugates, G2-AT and G2-K-AT exhibited no inhibition of HMG-CoA reductase, while their hydrolyzed products displayed dramatic inhibition against HMG-CoA reductase (Figure 1). The results showed that both intact conjugates cannot inhibit the HMG-CoA reductase until the release of atorvastatin. After hydrolyzed by esterase, G2-AT exhibited comparable activity to free atorvastatin. After a two-step hydrolysis, G2-K-AT also inhibited HMG-CoA reductase, but to a less extent than atorvastatin, which might be due to a slower release of the parent drug. The inhibition activity was increased if the incubation time was extended (Figure S4).
Figure 1.
Inhibition of HMG-CoA reductase activity by atorvastatin, G2-AT, G2-K-AT and their hydrolysis products. (Triplicate; **, P <0.01; ***, P < 0.001; N.S., no significance; t test, double-tailed.)
2.4. Cellular Uptake of Atorvastatin, G2-AT, and G2-K-AT
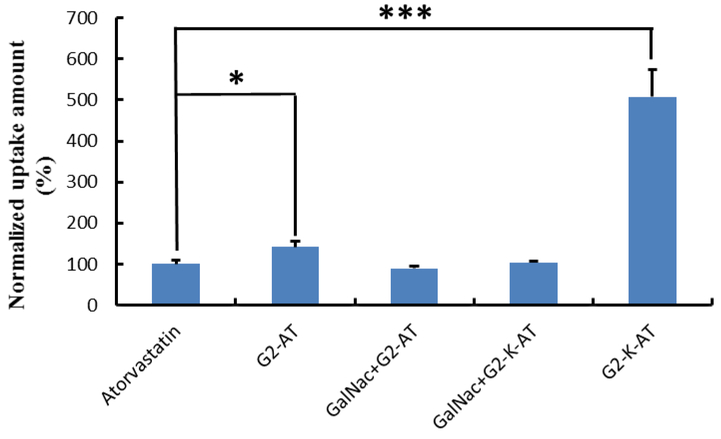
The cellular uptake of two conjugates were then quantified using a UV-spectra. Hep3B cells were treated with atorvastatin, G2-AT, G2-K-AT, G2-AT (in the presence of GalNac) and G2-K-AT (in the presence of GalNac), respectively. After one-day incubation, cells were washed and collected for UV-spectra analysis. The uptake of G2-AT and G2-K-AT in Hep3B cells were almost 1.4- and 5.1-fold of atorvastatin, respectively (Figure 2). Meanwhile, cellular uptake of G2-K-AT was much higher than that of the N-acetylglucosamine (GlcNAc, a glucose derivative) conjugates of atorvastatin, which was previously reported by our group.29 The higher cellular uptake of G2-K-AT compared to that of G2-AT may be due to its more stable chemical property. Meanwhile, the uptake of G2-AT (in the presence of GalNac) and G2-K-AT (in the presence of GalNac) significantly decreased than G2-AT and G2-K-AT group, which indicated that G2-AT and G2-K-AT were uptaken by ASGPR.
Figure 2.
Cellular uptake of atorvastatin, G2-AT, GalNac+G2-AT, GalNac+G2-K-AT or G2-K-AT. The data were normalized to the amount of atorvastatin. (Triplicate; *, P <0.05; ***, P < 0.001; t test, double-tailed).
2.5. Quantification Analysis of LDL Receptors
Subsequently, we investigated the effect of these two conjugates on regulating expression of LDL receptors using an immunofluorescence assay. According to the reportes31, after uptake by hepatocytes, atorvastatin can inhibit the activity of HMG-CoA, and hence enhance the expression of LDL receptors. In our study, the expression of LDL receptors on the Hep3B cell surface were quantified. It was found that LDL receptors expression were dramatically elevated after treated with atorvastatin, G2-AT, and G2-K-AT in comparison to the untreated group (Fig 3). These results combing with hydrolysis data confirmed that both conjugates can release the parent drug and hence up-regulate LDL receptors.
Figure 3.
Effects of atorvastatin, G2-AT, and G2-K-AT on the expression of LDL receptors in Hep3B cells. The expression of LDL receptors were quantified by an immunofluorescence assay. (Triplicate; *, P <0.05; ***, P < 0.001; t test, double-tailed).
3. Conclusion
In summary, we synthesized two atorvastatin conjugates, G2-AT, and G2-K-AT, by conjugating atorvastatin with the targeting ligand GalNAc. Both conjugates showed specific targeting capability. Moreover, two conjugates can be hydrolyzed and release the parent drug atorvastatin at different pH conditions, which can be further utilized to modify their pharmaceutical profiles. Atorvastatin and the hydrolyzed products of the two conjugates exhibited significant inhibition of HMG-CoA reductase, while no inhibition was observed with the intact conjugates.
Additionally, after being treated with the two conjugates, Hep3B cells showed increased amount of LDL receptors on cell surface. Combining the results mentioned above, we proved the concept of that GalNAc can be used as a ligand for cell specific delivery of atorvastatin.
4. Experimental procedures
4.1. Materials.
The Hep3B cell line was obtained from American Type Culture Collection (Manassas, VA). Eagle’s Minimum Essential Medium (EMEM) was purchased from Corning Incorporated (NY, USA). HMG-CoA Reductase Assay Kit, esterase from porcine liver and other chemicals were obtained from Alfa-Aesar or Sigma-Aldrich. Anti-LDL receptor clone 2H7.1 (mouse monoclonal) and Alexa Fluor® 647 labeled-Goat Anti-Mouse IgG H&L were purchased from EMD Millipore Corporation and Abcam respectively. Compounds a and b were obtained following the previous methods23.
4.2. Design of G2-AT
2-Azidoethyl-2-Acetamido-2-deoxy-β-D-galactopyranoside (0.104 mmole) was added into 6 mL ethanol solution of compound a (0.051 mmole) and stirred at room temperature under Argon (Ar) atmosphere overnight. Then 0.75mL water solution of CuSO4 (0.095 mmole) and sodium ascorbate (0.15 mmole) were separately added to the above reacting mixture with stirring. After 12h, the solution was washed with water and DCM, the organic layer was concentrated in vacuum. The residue was purified using a Combiflash Rf system with DCM/MeOH, (85/15 by volume) (yield 59.8%). 1H NMR (400 MHz, CD3OD) δ = 8.03 (1H, s), 7.32–7.22 (6H, m), 7.15–7.05 (7H, m), 5.28–5.20 (2H, m), 4.61–4.59 (2H, d, J = 8), 4.39–4.37 (1H, m), 4.27–4.22 (1H, m), 4.08–4.05 (2H, m), 3.98–3.84 (3H, m), 3.78–3.37 (7H, m), 2.54–2.42 (2H, m), 1.94 (3H, s), 1.72–1.67 (2H, m), 1.51 (6H, d, J = 4). ESI-MS m/z 909.3838 (M + Na)+.
4.3. Design of G2-K-AT
2-Azidoethyl-2-Acetamido-2-deoxy-β-D-galactopyranoside (0.104 mmole) was added into 6 mL ethanol solution of compound b (0.047 mmole) and stirred at room temperature under Argon (Ar) atmosphere overnight. Then 0.75mL water solution of CuSO4 (0.095 mmole) and sodium ascorbate (0.15 mmole) were separately added to the above reacting mixture above with stirring. After 12h, the solution was washed with water and dichloromethane (DCM), the organic layer was concentrated in vacuum. The residue was purified using a Combiflash Rf system with DCM/MeOH, (90/10 by volume) (yield 65.0%). 1H NMR (400 MHz, CD3Cl) δ = 8.01 (1H, s), 7.20–67.19 (6H, m), 7.15–7.05 (8H, m), 5.20 (2H, m), 4.53–4.44 (6H, m), 4.22–4.20 (2H, m), 4.10–4.05 (1H, m), 3.95 (1H, s), 3.84–3.82 (3H, m), 3.68 (2H, s), 3.59–3.56 (2H, m), 3.32 (1H, s), 2.52–2.46 (1H, m), 2.38–2.28 (1H, m), 2.07 (3H, s), 1.96 (3H, s), 1.67–1.64 (2H, m), 1.54–1.53 (6H, d, J = 4), 1.34 (6H, s). ESI-MS m/z 949.4150 (M + Na)+.
4.4. Hydrolysis assays
Stock solution of G2-AT and G2-K-AT (3.5 mM) were both prepared in DMSO. 30 μL of stock solution of G2-AT was mixed with 970 μL of PBS (pH 7.40, containing 0.5 mg/mL esterase) and incubated at 37 °C. Samples were collected from the mixture at three different time points post-mixing (0h, 3 h, 24 h) and 1 mL of DCM was added into the samples to terminate the hydrolysis reaction. The DCM layer of samples was used for mass spectrometry test. 30 μL of stock solution of G2-K-AT was mixed with 470 μL of PBS (pH 4.50) and incubated at 37 °C. After 24h, 1 M NaOH aqueous solution was added into the above solution to adjust the pH to 7.40. Then, 500 μL PBS (pH 7.40, containing 1.0 mg/mL esterase) was added into the solution. The mixture was incubated at 37 °C for one day. The hydrolysis reaction was stopped by adding 1 mL DCM. The DCM layer was pipetted for mass spectrometry analysis.
4.5. Inhibition of the activity of HMG-CoA Reductase
Inhibition of atorvastatin, G2-AT, and G2-K-AT on HMG-CoA Reductase was determined using a commercial HMG-CoA Reductase Assay Kit. The hydrolyzed G2-AT and G2-K-AT was obtained through the same hydrolysis procedure mentioned in 4.4 Hydrolysis assays section. After adding all the regents following the manufacturer’s procedure, the kinetic absorbance at wavelength of 340 nm was immediately measured for 10 minutes. The activity of HMG-CoA Reductase was obtained according to the following formula:
where 12.44 is εmM deducted the extinction coefficient for NADPH (6.22 mM−1 cm−1, 340 nm), 12.44 represents the 2 NADPH consumed in the reaction, V is the volume of enzyme used in the experiment in mL (12 × 10−3 mL), TV is the total volume of the reaction system and is given in mL (0.2 mL is used in this experiment), 0.6 is the enzyme concentration and given in mg/mL, and LP is light path in cm (0.55 cm for 96-well plates).
4.6. Cell culture
Human hepatocellular carcinoma Hep3B cells were cultured in EMEM supplemented with 10% FBS and maintained under a humidified atmosphere containing 5% CO2 at 37 °C.
4.7. Cellular Uptake of Atorvastatin, G2-AT, and G2-K-AT
Hep3B cells were seeded into 6-well plates at a density of 1×104 cells/cm2. Twenty-four hours after seeding, 10 μL 18 mM stock solutions of atorvastatin, G2-AT, G2-K-AT, G2-AT (in the presence of 90 mM GalNac) and G2-K-AT (in the presence of 90 mM GalNac) were added into each well respectively, the final concentration was 90 μM. After 3h treatment, cells were rinsed, trypsinized and collected by centrifugation (800rpm, 5min). Atorvastatin, G2-AT, and G2-K-AT were extracted from cells with a mixture of methanol (0.5mL) and DCM (0.5mL). After centrifugation, the supernatant was used for UV spectrum and mass spectrum test. The amount of atorvastatin, G2-AT, and G2-K-AT in Hep3B cells were then calculated using standard curves.
4.8. Quantification Analysis of LDL Receptors
Hep3B cells were seeded into 6-well plates at a density of 2×105 cells/cm2. Twenty-four hours after seeding, 5 μL stock solution of atorvastatin, G2-AT, and G2-K-AT (10 mM) were added into each well respectively, the final concentration was 25 μM. After 48 h incubation, cells were rinsed, trypsinized and collected by centrifugation (800rpm, 5min). After washed with cold PBS twice again, cells were resuspended and incubated with 100 μL anti-LDL receptor antibody (1/100 dilute, the primary antibody) solution for 30 min at RT. Cells were then rinsed twice with 1mL cold PBS. The cell pellets were collected by centrifugation (800rpm, 5min) and incubated with 100 μL Goat Anti-Mouse IgG H&L antibody (1/2000 dilute, the secondary antibody) for 30 min at RT in the dark environment. Finally, cells were rinsed twice with 1mL cold PBS, and then resuspended in 0.5 mL cold PBS for fluorescence detection immediately using a BD LSR II flow cytometer (BD Biosciences, San Jose, CA).
Supplementary Material
Acknowledgments
This work was supported by Maximizing Investigators’ Research Award R35GM119679 from the National Institute of General Medical Sciences as well as the start-up fund from the College of Pharmacy at The Ohio State University.
Abbreviations:
- GalNAc
N-acetylgalactosamine
- HMG-CoA
β-hydroxy-β-methylglutaryl-CoA
- AT
atorvastatin
- ASGPR
asialoglycoprotein receptor
- EMEM
eagle’s minimum essential medium
- DCM
dichloromethane
- MeOH
methonal
- DMSO
dimethyl sulfoxide
- Ar
Argon
- RT
room temperature
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of interest
The authors declare no competing financial interest.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- (1).Buhaescu I; Izzedine H Clin. Biochem 2007, 40(9–10), 575–584. [DOI] [PubMed] [Google Scholar]
- (2).Istvan ES Am. Heart J 2002, 144 (6 Suppl), S27–S32. [DOI] [PubMed] [Google Scholar]
- (3).Zhang L; Zhang S; Jiang H; Sun A; Zou Y; Ge J J Clin Cardiol. 2011, 34(2), 117–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Kim JS; Kim MS; Park HJ; Jin SJ; Lee S; Hwang SJ Int. J. Pharmaceut 2008, 359(1–2), 211–219. [DOI] [PubMed] [Google Scholar]
- (5).Ahmed IS; El-Hosary R; Shalaby S; Abd-Rabo MM; Elkhateeb DG; Nour S Int. J. Pharmaceut 2016, 504, 70–79. [DOI] [PubMed] [Google Scholar]
- (6).Choi DH; Shin WG; Choi JS Eur. J. Clin. Pharmacol 2008, 64(5), 445–449. [DOI] [PubMed] [Google Scholar]
- (7).Corsini A; Bellosta S; Baetta R; Fumagalli R; Paoletti R; Bernini F Pharmacol. Ther 1999, 84(3), 413–428. [DOI] [PubMed] [Google Scholar]
- (8).Wierzbicki AS; Poston R; Ferro A Pharmacol. Ther 2003, 99(1), 95–112. [DOI] [PubMed] [Google Scholar]
- (9).Sirtori CR Pharmacol. Res 2014, 88, 3–11. [DOI] [PubMed] [Google Scholar]
- (10).Majumdar S; Duvvuri S; Mitra AK Adv. Drug Deliver. Rev 2004, 56(10), 1437–1452. [DOI] [PubMed] [Google Scholar]
- (11).Zhang YX; Sun J; Sun YB; Wang YJ; He ZG Curr. Drug Metab 2013, 14, 675–687. [DOI] [PubMed] [Google Scholar]
- (12).Majumdar S; Duvvuri S; Mitra AK Adv Drug Deliv Rev. 2004, 56(10), 1437–1452. [DOI] [PubMed] [Google Scholar]
- (13).Puris E; Gynther M; Huttunen J; Petsalo A; Huttunen KM J. Control Release. 2017, 261(10), 93–104. [DOI] [PubMed] [Google Scholar]
- (14).Bhushan KR; Tanaka E; Frangioni JV Angew. Chem. Int. Ed 2007, 46, 7969–7971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Chen X; Bi Y; Wang T; Li P; Yan X; Hou S; Bammert CE; Ju J; Gibson KM; Pavan WJ; Bi L Sci. Rep 2015, 5, 9004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Low PS; Henne WA; Doorneweerd DD Acc. Chem. Res 2008, 41(1), 120–129. [DOI] [PubMed] [Google Scholar]
- (17).Weber PC; Ohlendorf DH; Wendoloski JJ; Salemme FR Science. 1989, 243 (4887), 85–88. [DOI] [PubMed] [Google Scholar]
- (18).Yin Q; Tang L; Cai K; Tong R; Sternberg R; Yang X; Dobrucki LW; Borst LB; Kamstock D; Song Z; Helferich WG; Cheng J; Fan TM Proc. Natl. Acad. Sci. U. S. A 2016, 113(32), E4601–E4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Zhang YX,; Sun J; Gao YK; Jin L; Xu YJ; Lian H; Sun YB; Sun YH; Liu JY; Fan R; Zhang TH; He ZG Mol. Pharmaceutics 2013, 10 (8), 3195–3202. [DOI] [PubMed] [Google Scholar]
- (20).Li H; Liu Y; Chen L; Liu Q; Qi S; Cheng X; Lee YB; Ahn CH; Kim DJ; Lee RJ J. Drug Target 2018, 26(5–6), 466–473. [DOI] [PubMed] [Google Scholar]
- (21).Chen X; Bi Y; Wang T; Li P; Yan X; Hou S; Bammert CE; Ju J; Gibson KM; Pavan WJ; Bi L Sci. Rep 2015, 5, 9004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Singh Y; Palombo M; Sinko PJ Curr. Med. Chem 2008, 15(18), 1802–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Sievänen E Molecules. 2007, 12(8), 1859–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Wall DA; Hubbard AL J Cell Biol. 1981, 90(3), 687–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Chen S; Tam YYC; Lin PJC; Leung AKK; Tam YK; Cullis PR J. Control. Release 2014, 196, 106–112. [DOI] [PubMed] [Google Scholar]
- (26).Huang YY. Mol Ther-Nucl Acids 2017, 6, 116–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Nair JK; Willoughby JL; Chan A; Charisse K; Alam MR; Wang Q; Hoekstra Menno.; Kandasamy P; Kel’in AV; Milstein S; Taneja N; O’Shea J; Shaikh S; Zhang LG; van der Sluis RJ; Jung ME; Akinc A; Hutabarat Renta.; Kuchimanchi S; Fitzgerald K; Zimmermann Tracy.; van Berkel TJ; Maier MA; Rajeev KG; Manoharan M J. Am. Chem. Soc 2014, 136 (49), 16958–16961. [DOI] [PubMed] [Google Scholar]
- (28).Kolb HC; Finn MG; Sharpless KB Angew. Chem. Int. Ed 2001, 40(11), 2004–2021. [DOI] [PubMed] [Google Scholar]
- (29).Zhang XF; Chen XF; Zhao WY; Zeng CX; Luo X; Li WQ; Li B; Jiang J; Dong YZ Bioconjugate Chem. 2017, 28(8), 2109–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Kleemann R; Kooistra T Curr. Drug Target Cardiovas. Haemat. Disord, 2005, 5, 441–453. [DOI] [PubMed] [Google Scholar]
- (31).Moon JH; Kang SB; Park JS; Lee BW; Kang ES; Ahn CW; Lee HC; Cha BS Metab. Clin. Exp 2011, 60(7), 930–940. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.