Abstract
Hepatocellular carcinoma (HCC) is the third most-common cause of cancer-related death worldwide. Most cases of HCC develop in patients that already have liver cirrhosis and have been recommended for surveillance for an early onset of HCC. Cirrhosis is the final common pathway for several etiologies of liver disease, including hepatitis B and C, alcohol, and increasingly non-alcoholic fatty liver disease. Only 20–30% of patients with HCC are eligible for curative therapy due primarily to inadequate early-detection strategies. Reliable, accurate biomarkers for HCC early detection provide the highest likelihood of curative therapy and survival; however, current early-detection methods that use abdominal ultrasound and serum alpha fetoprotein are inadequate due to poor adherence and limited sensitivity and specificity. There is an urgent need for convenient and highly accurate validated biomarkers for HCC early detection. The theme of this review is the development of new methods to discover glycoprotein-based markers for detection of HCC with mass spectrometry approaches. We outline the non-mass spectrometry based methods that have been used to discover HCC markers including immunoassays, capillary electrophoresis, 2-D gel electrophoresis, and lectin-FLISA assays. We describe the development and results of mass spectrometry-based assays for glycan screening based on either MALDI-MS or ESI analysis. These analyses might be based on the glycan content of serum or on glycan screening for target molecules from serum. We describe some of the specific markers that have been developed as a result, including for proteins such as Haptoglobin, Hemopexin, Kininogen, and others. We discuss the potential role for other technologies, including PGC chromatography and ion mobility, to separate isoforms of glycan markers. Analyses of glycopep-tides based on new technologies and innovative softwares are described and also their potential role in discovery of markers of HCC. These technologies include new fragmentation methods such as EThcD and stepped HCD, which can identify large numbers of glycopeptide structures from serum. The key role of lectin extraction in various assays for intact glycopeptides or their truncated versions is also described, where various core-fucosylated and hyperfucosylated glycopeptides have been identified as potential markers of HCC. Finally, we describe the role of LC-MRMs or lectin-FLISA MRMs as a means to validate these glycoprotein markers from patient samples. These technological advancements in mass spectrometry have the potential to lead to novel biomarkers to improve the early detection of HCC.
Keywords: cancer, hepatocellular carcinoma, biomarkers, early detection, lectins, glycans, glycopeptides, fucosylation, MALDI-MS, ESI-MS
I. INTRODUCTION
Hepatocellular carcinoma (HCC) is the third most-common cause of cancer-related death worldwide (Perz et al., 2006; Kanwal et al., 2011; El-Serag, 2012; White et al., 2017). The primary risk factors for development of cirrhosis are chronic hepatitis B (HBV) infection and presence of hepatic cirrhosis from any cause (e.g., hepatitis C [HCV], alcohol [ALD], non-alcoholic fatty liver disease [NAFLD]) (Table 1). Because patients with cirrhosis have a 2–8% annual risk to develop HCC, they are recommended to undergo surveillance for HCC with abdominal ultrasounds every 6 months (Kanwal et al., 2011; El-Serag, 2012). In the United States, Europe, and Japan, <30% of the patients are diagnosed with HCC at an early stage which would result in a 5-year survival rate near 70%, whereas patients diagnosed at later stages have a 2-year survival rate <16% (White et al., 2017). HCC is increasing in incidence in many countries worldwide and is responsible for over 700,000 deaths per year globally. Thus, early detection of HCC and the ability to distinguish early HCC from patients being monitored for cirrhosis is critical for patient survival and this is best achieved through reliable surveillance testing in patients with cirrhosis.
TABLE 1.
Risk factors for cirrhosis and subsequent hepatocellular carcinoma.
| Risk factors | Annual risk for HCC with cirrhosis (Bruix et al., 2011;Ioannou et al., 2018;Kanwal et al., 2018) |
|---|---|
| Hepatitis C (HCV) | 3–5% |
| Hepatitis B (HBV) | 3–8% |
| Alcoholic liver disease (ALD) | 0.8%-1.0% |
| Non-alcoholic fatty liver disease (NAFLD) | 0.9%-1.1% |
| Other Etiologies Cirrhosis | 1.0% |
The risk of HCC development depends on several different factors that vary depending on geography and demographic factors. HBV infection is the most-common etiologic factor to develop HCC and is associated with >80% of the HCC cases worldwide. There are more than 350 million people that are chronically infected with HBV globally, with the largest number in East Asia. Chronic hepatitis C infection is the most-common etiologic agent that leads to cirrhosis and HCC in many Western countries and the United States. The prevalence of cirrhosis has increased over the past 20 years, largely due to the prevalence of hepatitis C in the baby boomer population and the emergence of NAFLD-related liver disease which has resulted in steady increases in the incidence of HCC.
Only about 20–30% of patients with HCC are eligible for curative therapy with surgical resection, liver transplantation, or local ablative procedures (Bruix et al., 2005). Liver transplantation can cure HCC and the underlying liver disease; however, it is limited by recipient selection and organ availability. Surgical resection and local ablative therapies, although potentially curative, can only be applied to a minority of patients. The goal to development of new biomarkers for HCC detection is to increase the proportion of patients diagnosed at an early stage that would be eligible for such curative therapies.
The most-common non-invasive detection of HCC involves imaging, including ultrasound, computer tomography, and magnetic resonance imaging (Trevisani et al., 2001). Ultrasound is often used to detect small masses at an early stage in the liver but requires at least a 2 cm mass and often is not effective for early-stage detection. However, the American Association for the study of Liver Diseases guidelines recommends that surveil-lance for HCC be performed with ultrasonography at 6- to 12-month intervals. MRI is an effective method of detection, but the cost is excessive for routine screening. Serum biomarkers are often used together with these imaging techniques to improve the sensitivity for early detection of HCC (Marrero & Henley, 2011).
Serum biomarkers are used as a complementary method for early detection in patients with cirrhosis (Marrero & Henley, 2011; Singal et al., 2012). Serum alpha-fetoprotein (AFP) is the most widely used as a clinical HCC diagnostic marker; however, detection of AFP suffers from low sensitivity. The receiver operating characteristic curve (ROC), a plot of the data pairs for sensitivity and 1–specificity, and the area under the curve (AUC) (Hanley & Mcneil, 1982) have been widely used for the assessment of diagnostic ability of biomarkers. One study has shown that AFP has a specificity of 90.6% and a sensitivity of 60.0% at the cut-off value of 20 ng/mL (Flores & Marrero, 2014). AFP accuracy in HCC early detection varies by etiology of disease. The performance of AFP to differentiate cirrhosis versus HCC in patients with HCV is only an AUC of 0.64 whereas the AUC to differentiate HBV-related HCC from HBV-related cirrhosis is 0.9.
Des-gamma carboxy prothrombin (DCP) has been widely used as an alternative marker for AFP in Japan. However, the diagnostic value of DCP varies depending on the underlying characteristics of the patients (Marrero et al., 2009). AFP-L3 has also been used as a marker, but has not been shown to add significantly to the sensitivity for early detection of HCC (Marrero et al., 2009). All these markers still have relatively low sensitivity for early HCC detection, and current guidelines do not recommend their routine use in HCC surveillance. In any case, serum markers are urgently needed to enhance the sensitivity of HCC early detection over the currently available tests in order to increase the number of patients eligible for curative therapies.
Glycosylation has been associated with a majority of cancer serum biomarkers. Most prior work though has involved monitoring changes in the protein level rather than any distinctive changes in the actual glycan structure. However, new developments in mass spectrometry based technology and informatics have allowed the detailed study of glycan structure and site specificity. These developments have been applied to studies of glycoproteins in patient serum where it has been shown that changes in the structures of glycans on specific sites in a protein may provide potential markers for monitoring changes in disease state and as markers for early detection of cancer. Such work has been demonstrated in several different cancers but particular progress in this area has been made in the case of HCC where glycan and glycopeptide markers have been identified which can provide early detection for the development of HCC from patients being monitored for cirrhosis (Zhu et al., 2014; Yin et al., 2015).
In the current review, we discuss developments in state of the art methods to find new glycoprotein markers for early detection of HCC and the ability to distinguish HCC from cirrhosis based on changes in glycan structures identified via novel mass spectrometry techniques. These markers would be used for early detection of HCC, which includes screening and surveillance of potential patients at risk with cirrhosis or other liver diseases or as a diagnostic tool to differentiate patients with cirrhosis from those who have developed HCC. However, there are other uses for many of these markers, which include markers to stratify at-risk populations, stratify patients for clinical trials, and to predict treatment response to therapies. Some of the markers that might prove ineffective for early detection might prove valuable for these other purposes.
This article will cover various methodologies that have been used for biomarker glycoprotein analysis in the context of early detection of HCC, and will include methods for glycanand glycopeptide-based assays. Methods to be covered include mass spectrometric analysis of glycans and glycopeptides using both targeted and untargeted techniques, strategies to enrich glycopeptides and glycoproteins from biological fluids, micro-array assays, and gel-based methods of analysis and separation methods that can separate glycans and glycopeptides including isomeric forms of these units. The applications of these methods to important biomarker discoveries for liquid biopsies in serum for HCC will be discussed including efforts to validate some of these markers.
II. GLYCOSYLATION
Glycosylation is the most-frequent and important post-translational modification (PTM) of proteins, which has been demonstrated to participate in many key pathological steps during tumor development and progression (Fuster & Esko, 2005; Miyoshi et al., 2008; Pinho & Reis, 2015). Aberrant glycosylation is highly associated with the development of HCC, where increased fucosylation, sialylation, and branching structures have been determined in target serum proteins or total serum glycan analysis between HCC and liver cirrhosis patients (Mehta et al., 2015). These cancer-related alterations in glycosylation are, therefore, valuable sources of biomarkers for HCC (Miyoshi et al., 2012). We provide a brief overview of the rationale for glycosylation as a marker of cancer, different types of glycosylation, and methods to isolate/purify glycoproteins and their glycans/glycopeptides applied in glyco-marker studies for HCC.
A. Rationale for Glycosylation as a Marker of Cancer
Accumulating evidence has documented that alterations in the glycosylation patterns of cell surface and secreted glycoproteins are directly associated with malignant transformation and cancer progression (Dube & Bertozzi, 2005; Meany & Chan, 2011; Pinho & Reis, 2015). A recent review by Munkley and Elliott (2016) has elucidated in detail that aberrant glycosylation is not only itself a hallmark of cancer but also enables acquisition of all other recognized hallmarks of cancer. Aberrant glycosylation is a hallmark of many types of cancer, which indicates its clinical significance for cancer diagnosis, monitoring, and prognosis (Dube & Bertozzi, 2005; Meany & Chan, 2011; Kailemia et al., 2017). In the case of HCC, fucosylation levels in normal liver are relatively low, but distinctly increase during carcinogenesis (Miyoshi et al., 2008). The first notable glycosylation change identified as a more-specific marker for HCC was the a1–6 fucosylated (core-fucosylated) structure in serum AFP (Li et al., 2001). Subsequently, elevated levels of fucosylation in other serum proteins, such as alpha-1 anti-trypsin, alpha-1 acid glycoprotein, haptoglobin, fetuin, and transferrin were found in HCC patients. Increased fucosylated proteins in sera of patients with HCC originate from changes in fucosylation states in the liver and present a promising marker for cancer diagnosis (Mehta & Block, 2008; Miyoshi et al., 2012).
B. Different Types of Glycosylation
With glycans (also known as oligosaccharides) covalently attached to proteins at specific amino acid residues, glycosylation can be classified into two main categories: (1) N-glycosylation, linked to the amide group of asparagine (Asn) residues in the consensus sequence of N-X-S/T, where X can be any amino acid except proline and (2) O-glycosylation, linked to the hydroxyl group of serine (Ser) or threonine (Thr) residues. In humans, glycans are assembled from ten monosaccharides: mannose (Man), galactose (Gal), glucose (Glc), N-acetylglucosamine (GlcNAc), N-acetyl-galactosamine (GalNAc), sialic acid (Neu5Ac), fucose (Fuc), glucuronic acid (GlcA), iduronic acid (IdoA), and xylose (Xyl) (Stowell et al., 2015). According to the Swiss-Prot database, the majority of serum proteins are glycosylated, whereas about 90% of glycoproteins carry either N-linked glycans alone or N- and O-linked ones, and 10% carry O-linked glycans (Apweiler et al., 1999). N-linked glycans contain a conserved GlcNAc2Man3 core structure and several branches, whereas O-linked glycans are usually short, and contain one to four oligosaccharides. Due to the immense complexity and diversity of glycan structures, such as composition heterogeneity, branching, differences in linkages (1–3 vs. 1–4, etc.), and the different glycosylation sites within a protein, the characterization of glycome/glycoproteome is far more challenging than that of the proteome.
III. CURRENT MARKERS
There has been a marked advance in the discovery of HCC biomarkers over the last 10 years. Alpha-fetoprotein (Marrero et al., 2009), which can typically be monitored through a validated immunoassay, has been the standard biomarker for HCC and is the only FDA-approved marker for diagnosis. DCP (Marrero et al., 2009), also known as PIVKA-II, is also a popular alternative in East Asia for HCC detection (Yamamoto et al., 2010). Although these pre-approved clinical markers remain under research to improve the sensitivity and specificity of their use, several proteins have been studied as potential candidates to improve early detection, among which haptoglobin and GP73 (GOLPH2) are the closest to clinical advancement. In Table 2 are shown the currently used markers for early HCC detection with their performance in terms of the AUC and the sensitivity and specificity as determined by Marrero et al. (2005, 2009). Several other proteins such as A1AT (Comunale et al., 2010), Apo-J (Comunale et al., 2011), kininogen (Wang et al., 2009), HGF (Liu et al., 2010), and hemopexin (Benicky et al., 2014) have shown differential expression between HCC and cirrhosis. Further research on these markers is needed where some, such as hemopexin, have been demonstrated to confer only marginally better results than AFP (Kobayashi et al., 2012). More recently, fucosylated kininogen has been shown to have great potential as a marker in combination with other clinical variables (Wang et al., 2017). Only GP73 has undergone a multi-center epidemiological study to determine real-world sensitivity and specificity values (Mao et al., 2010).
TABLE 2.
Diagnostic performance of current markers for early HCC detection.
| Symbol | Name | AUC | Early Detection | Reference | |
|---|---|---|---|---|---|
| Sensitivity(%) | Specificity(%) | ||||
| AFP | Alpha-fetoprotein | 0.8 | 53 | 90 | (Marrero et al., 2009) |
| DCP | Des-gamma carboxy prothrombin | 0.72 | 61 | 70 | (Marrero et al., 2009) |
| AFP-L3 | Alpha-fetoprotein (binds to LCA) | 0.66 | 28 | 97 | (Marrero et al., 2009) |
| GP73 | Golgi protein 73 | 0.77 | 62 | 88 | (Marrero et al., 2005) |
| AFP + DCP | 0.83 | 78 | 62 | (Marrero et al., 2009) | |
Laboratory methods for protein biomarker discovery have involved quantitation through traditional methods such as ELISA, Western blots, microarrays, and immunoblotting. Cancer-related glyco-markers have also been extensively explored because aberrant glycosylation is recognized as a hallmark in oncogenic transformation (Fuster & Esko, 2005). More recently, mass spectrometry has proven critical in biomarker identification as well as glycoproteomic profiling. Research in the last 10 years on HCC biomarkers in blood is summarized in Table 3.
TABLE 3.
Research on serum biomarkers for HCC in recent 10 years.
| Biomarker | Type | Methodology | Ref. |
|---|---|---|---|
| Alpha-1-antitrypsin (A1AT) | core α-1,6 fucosylation | 2DE+MALDI-TOF MS; Lectin-FLISA | (Comunale et al., 2010) |
| fucosylated protein level | AAL affinity chromatography+LC-MRM-MS | (Ahn et al., 2013) | |
| AFP-L3 | core α-1,6 fucosylation | MALDI-TOF MS; LCA-affinity electrophoresis | (Nakagawa et al., 2008) |
| AFP-L3% | Liquid-phase binding assay | (Leerapun et al., 2007) | |
| hs-AFP-L3% | Microchip capillary electrophoresis; liquid-phase binding assay | (Oda et al., 2011) | |
| AFP | Fc glycopeptides | LC-MS/MS-PRM | (Kim et al., 2018) |
| AGP | multifucose index | AAL affinity chromatography+LC-TOF-MS | (Tanabe et al., 2016) |
| Apo-J | β-1,4 triantennary W-glycans | 2DE+LC-MS/MS; Lectin-FLISA | (Comunale et al., 2011) |
| β2-microgloblin (B2M) | protein level | SELDI-TOF-MS | (Saito et al., 2010) |
| Carboxylesterase 1 (hCE1) | protein level | LC-MS/MS; Antibody-based assay | (Na et al., 2009;Na et al., 2013) |
| Ceruloplasmin | core Fc ratio | Endo F3+LC-MS/MS/MS | (Yin et al., 2014) |
| Complement C3a | C3a fragment | ProteinChip arrays+SELDI-MS | (Kanmura et al., 2010) |
| Complement factor H (CFH) | site-specific core Fc | LC-MS/MS; LC-MS-MRM | (Benicky et al., 2014) |
| Des-gamma carboxyprothrombin (DCP) | protein level | ELISA | (Durazo et al., 2008;Marrero et al., 2009) |
| Fibronectin (FN) | core Fc peptides | iTRAQ+LCA affinity chromatography+LC-MS/MS | (Yin et al., 2015) |
| GP73 | protein level | 2DE; immunoblotting | (Block et al., 2005) |
| protein level | immunoblotting | (Marrero et al., 2005;Mao et al., 2010) | |
| protein level | ELISA | (Riener et al., 2009;Yamamoto et al., 2010) | |
| protein level | western blotting | (Hu et al., 2010) | |
| GP73, hemopexin, PIVKA-II | protein level | ELISA; lectin-ELISA; immunoassays | (Morota et al., 2011) |
| Haptoglobin (Hp) | bifucosylated N-glycans | MALDI-QIT-TOF MS | (Zhu et al., 2014) |
| fucosylated N-glycans | ESI-LC-MS | (Zhang et al., 2015b) | |
| Isomeric N-glycans | PGC-LC-MS/MS | (Huang et al., 2017) | |
| Fc-Hp level | Lectin-ELISA; CLEIA | (Asazawa et al., 2015) | |
| Multiply fucosylated glycopeptides | LC-MS/MS; exoglycosidase/MALDI-MS/MS | (Pompach et al., 2013) | |
| Multiply fucosylated glycopeptides | LC-MS-MRM | (Sanda et al., 2013) | |
| fucosylated N-glycans | MALDI-QIT-TOF MS | (Zhang et al., 2011) | |
| Fc-Hp/Hp ratio | magnetic beads-based lectin ELISA | (Shang et al., 2017) | |
| Hemopexin (Hpx) | Fc-Hpx | Lectin-FLISA; LC-MS/MS | (Comunale et al., 2009) |
| Fc-Hpx | ELISA; lectin-ELISA | (Kobayashi et al., 2012) | |
| N-glycans | DSA-FACE | (Debruyne et al., 2010) | |
| site-specific core Fc | LC-MS/MS; LC-MS-MRM | (Benicky et al., 2014) | |
| Hp, Hpx, Kng-1, CFH | site-specific core Fc | LC-MS/MS; MALDI-TOF MS | (Pompach et al., 2014) |
| HSP90 | protein level | 2-DE; MALDI-TOF MS | (Sun et al., 2010b) |
| IgGs | glycoforms | LC-MS-MRM | (Yuan et al., 2015) |
| Kininogen (Kng) | fucosylation level | Lectin-FLISA | (Wang et al., 2009;Wang et al., 2017) |
| Osteopontin (OPN) | protein level | LC-MS/MS; ELISA | (Shang et al., 2012) |
| protein level | ELISA | (El-Din Bessa et al., 2010;Abu El Makarem et al., 2011) | |
| Paraoxonase-1 (PON1) | fucosylation | Lectin-ELISA | (Zhang et al., 2015a) |
| PIVKA-II | protein level | LC-MRM-MS | (Sohn et al., 2017) |
| Vimentin (VIM) | protein level | MALDI-TOF/TOF MS; ELISA | (Sun et al., 2010a) |
| Vitronectin (VTN) | N-glycopeptides | iTRAQ+LC-MS/MS | (Lee et al., 2010) |
| N-glycopeptide ratios | TMT+LC-MS/MS | (Lee et al., 2014) | |
| WFA+−M2BP | GalNAc residues | lectin-antibody sandwich immunoassay | (Yamasaki et al., 2014) |
| global serum profiling | core Fc glycopeptides | LC-MS-MRM | (Ma et al., 2018) |
| core Fc glycopeptides | LC-MS/MS/MS; LC-MRM-MS | (Zhao et al., 2011) | |
| S90K, IGFBP-3, and TSP-1 | LC-MS/MS; ELISA | (Chen et al., 2011) | |
| C3, CE, HRG, CD14, and HGF | lectin affinity chromatography + LC-MS/MS; lectin-Ab arrays | (Liu et al., 2010) | |
| CP, ACT, and MMRN1 | 2D LC-MALDI-MS | (Ishihara et al., 2011) | |
| A1AG1, AACT, A1AT, and CERU | AAL affinity chromatography + LC-MRM-MS | (Ahn et al., 2012) | |
| N-glycans | LC-ESI-MS | (Tsai et al., 2014) | |
| G2890 and G3560 W-glycans | MALDI-TOF/TOF | (Kamiyama et al., 2013) | |
| glycoproteins/ N-glycans | LC-ESI-MS/MS; MALDI-TOF/TOF-MS | (Yang et al., 2013) | |
| PHA-L reactive glycoproteins | PHA-L affinity chromatography+LC-MS/MS | (Liu et al., 2017b) | |
| branch α(1,3)-fucosylated N-glycans | DSA-FACE | (Liu et al., 2007) | |
| N-glycans | MALDI-TOF-MS | (Tang et al., 2010) | |
| N-glycans | HPLC; Lectin-FLISA | (Comunale et al., 2013) | |
| N-glycans | MALDI-TOF/TOF MS | (Goldman et al., 2009) | |
| isomeric N-glycans | IMS-MS | (Isailovic et al., 2008;Isailovic et al., 2012) | |
| outer arm fucosylated N-glycans | LC-MS | (Tanabe et al., 2008) | |
| peptide and glycan panel | MALDI-TOF/TOF MS | (Ressom et al., 2008) | |
| peptides | MALDI-TOF/TOF MS | (Goldman et al., 2007) | |
| Glycoprotein proflie | lectin coupled IGOT-LC-MS/MS | (Kaji et al., 2013) | |
| protein profile | 2DE+LC-MS/MS | (Yang et al., 2007) | |
| protein profile | SELDI-TOF-MS | (Cui et al., 2007) | |
| protein profile | 2DE+MALDI-TOF-MS | (Wu et al., 2012) |
AAL, Aleuria aurantia lectin; CLEIA, chemiluminescent enzyme immunoassay; Core Fc, core fucosylation; DSA-FACE, DNA sequencer-assisted fluorophore-assisted carbohydrate electrophoresis; ELISA, enzyme linked immunosorbent assay; Fc-Hp, fucosylated haptoglobin; Fc-Hpx, fucosylated hemopexin; IGOT, isotope-coded glycosylation site-specific tagging; IMS-MS, ion mobility spectrometry-mass spectrometry; iTRAQ, isobaric tags for relative and absolute quantitation; LCA, Lens culinaris agglutinin; lectin-FLISA, Lectin fluoropore-linked immunosorbent assay; MRM, multiple reaction monitoring; PHA-L, Phaseolus vulgaris Leucoagglutinin; SELDI-TOF-MS, surface-enhanced laser desorption-ionization time-of-flight mass spectrometry; TMT, tandem mass tags; WFA+-M2BP, Wisteria floribunda agglutinin-positive human Mac-2 binding protein.
IV. NON-MASS SPECTROMETRY BASED METHODS
A. Immunoassays
HCC biomarker studies that do not involve mass spectrometry have predominantly centered on the use of ELISA kits or other related immunoassays. In these ELISAs, sample preprocessing is minimal and relatively simple absorbance measurements can be performed. These tests are ideal in a clinical setting where they can be performed with a simple set-up by a technician. The use of a clinical ELISA does require the use of a well-validated antibody for that assay. If such an antibody is available then it is difficult to surpass the performance of an ELISA in terms of analytical sensitivity and specificity. Additionally, diagnostic sensitivity, specificity, and AUC values can be quickly derived from these simple tests. Of course the results from these assays depend on the sample sets being tested where for different sets there may be a mix of different etiologies, early versus late stage samples, a mix of genders and the presence of confounding diseases. Nevertheless, ELISA assays have been generally used for large scale validation of HCC markers for early detection. The markers being tested for HCC are generally glycoproteins but these ELISAs measure the level of protein and do not take advantage of the glycan structure as a potential marker for early detection.
There have been several validation studies on the currently used markers for HCC including AFP, DCP, and AFP-L3. Among these is a study conducted by Marrero et al. (2009) under the auspices of the National Cancer Institute (NCI) Early Detection Research Network (EDRN). In this study there were a total of 836 patients of which 417 were cirrhosis controls and 419 were HCC cases of which 208 had early stage HCC. The results of these studies are shown in Table 2 where AFP had the best AUC value at 0.8, followed by DCP at 0.72 and AFP-L3 at 0.66. The optimal AFP cutoff value was 10.9 ng/mL, leading to a sensitivity of 66%. When only the samples with very early HCC were evaluated the AUC value was 0.78. Thus, according to this study AFP was more sensitive than DCP and AFP-L3 for the diagnosis of early stage HCC.
Another alternative marker for HCC is GP73 that has been analyzed as a biomarker with immunoassays. Although GP73 is a known glycoprotein with fucosylation suggested to play a discriminatory role in HCC diagnosis (Hu et al., 2010), recent studies have focused instead on relative abundance in serum protein level with primarily ELISA kits (Riener et al., 2009; Hu et al., 2010; Mao et al., 2010; Morota et al., 2011; Zhang et al., 2016), with AUC values that range from 0.89 via a western blot study (Hu et al., 2010) to 0.94 in a full epidemiologic study with over 4000 participants (Mao et al., 2010). In another study, immunoassays were used to compare GP73 with hemopexin, fucosylated hemopexin, PIVKA-II, and AFP (Morota et al., 2011). Interestingly, PIVKA-II was found to have the highest total discriminatory power (AUROC = 0.90) among several types of chronic liver diseases of different etiologies; however, GP73 was found to have higher discriminatory power (AUC = 0.90) for HCC and cirrhosis versus hepatitis and normal samples (Morota et al., 2011). The most recent study found the sensitivity and specificity of GP73 to be 74.6% and 97.4%, respectively, with a cutoff of 8.5 relative units (Mao et al., 2010), which is higher than any reported specificities for haptoglobin, but lower in sensitivity (Shang et al., 2017). In other recent work, GP73 and AFP-L3 were studied for their capabilities for diagnosing AFP-negative HCC (Zhao et al., 2018). It was found that, by using these two markers together, one could improve the diagnostic accuracy and sensitivity for detection of AFP-negative HCC. Other immunoassays have suggested beta-catenin as a biomarker for HCC with sensitivity and specificity of 96.9% and 92.6%, respectively (Zekri et al., 2011). However, because these results have not been pursued further, their use is limited. Nevertheless, immunoassay techniques have been essential for current clinical assays for HCC early detection.
B. CE-Based Assays for HCC Markers
Capillary electrophoresis (CE) has proven to be a valuable tool to separate glycans. This method is dependent on the differential migration of analytes in an applied electric field. It has been used to study the N-Glycan profile of the protein Hemopexin in patient serum in HCC versus cirrhosis to detect HCC (Debruyne et al., 2010). In this study, Hemopexin was purified from patient serum with heme agarose beads; IgG was depleted with protein A agarose, and Hemopexin glycans were removed and labeled for detection. The glycans were analyzed with CE and detected based on a fluorescent probe. Branching alpha-1,3-fucosylated multi-antennary glycans on Hemopexin were increased in the HCC group compared with cirrhosis, fibrosis, and healthy volunteers as compared to non-modified biantennary glycans, which decreased progressively across patients from fibrosis to cirrhosis to HCC. This Hemopexin glycan marker differentiated patients with HCC and cirrhosis from healthy volunteers and patients with cirrhosis or fibrosis with a sensitivity and specificity of 79% and 93%, respectively.
C. Lectin Fluorophore-Linked Immunoabsorbent Assays
1. Method
Methods for antibody and lectin arrays for glycoprotein detection have been reviewed elsewhere (Patwa et al., 2010). The lectin-FLISA method has advantages as a simple method to detect the response of various lectins to target proteins in biofluids (see Fig. 1). Often, the changes in glycan structures are more informative than the absolute changes in protein levels as determined by ELISAs. The lectin-FLISA uses an antibody that has had its glycans oxidized so they will not interfere with the analysis. The antibody selectively extracts the target protein from the serum, and a fluorescently labeled lectin is used to detect glycan structures with the appropriate structural moiety. The list of lectins and their corresponding recognized glycan structures has been summarized in the literature (Clark & Mao, 2012). Much of the published work has targeted fucosylated glycans (Wang et al., 2009; Liu et al., 2010) but other lectins that detect other structures could also be used. The method has the advantage that it can be used in an ELISA format or on a microarray-based format for high-throughput assays. A disadvantage of the method is that the assay is only as good as the antibody used, and there might be cross reactivity from other components in complex fluids such as serum. Also, the lectins can detect total amounts of the target carbohydrate, but cannot distinguish subtle differences in the number of sugars present, such as the case for mono-, di, or tri fucosylation, where individual minor structures might serve as the optimal biomarker but might be masked by the total sugar detection (Zhu et al., 2014).
FIGURE 1.
General lectin FLISA methodology for the measurement of glycoproteins with specific glycan motif. The method involves an antibody to selectively capture the target glycoprotein from biological samples and then a fluorophore-conjugated lectin is used to detect the glycan motif with the appropriate structural moiety. The antibody’s glycans are oxidized so they will not interfere with the analysis.
2. Lectin-FLISA Targeting Fucosylation
Taniguchi and coworkers (Kinoshita et al., 1991) have developed an antibody-lectin enzyme immunoassay to detect fucosylated alpha-fetoprotein in liver cancer. The use of fucosylated AFP has served as an alternative marker to AFP to detect HCC versus cirrhosis. The assay developed was similar to that described above where AAL was used as the lectin to detect AFP captured by an antibody on a microtiter plate. Taniguchi and coworkers also developed a method for enrichment of AFP to be used with this platform. The methodology measured highly fucosylated AFP diluted to 5–80 ng/mL in human serum.
Mehta and coworkers (Comunale et al., 2009, 2013) have also developed a lectin-FLISA that targeted fucosylation for early detection of HCC. They point out that AFP and core-fucosylated AFP can be produced under other circumstances than HCC, including other liver diseases, and is not present in all patients with HCC. Also, it is difficult to discriminate AFP levels in early-stage HCC from cirrhosis. They therefore used a lectin-FLISA to explore the levels of fuc-kininogen and fuc-A1AT in patient serum individually and in combination with the level of AFP and also GP73 to distinguish between a diagnosis of cirrhosis and HCC. They found that the levels of fuc-Kin and fuc-A1AT were significantly higher in patients with HCC compared to cirrhosis. The optimal performance was obtained with a combination of fuc-Kin, AFP, and GP73 to result in a sensitivity of 95% and a specificity of 70%, and an AUC of 0.94. Thus they concluded that the fucosylated proteins can act as markers to detect HCC by themselves or could improve detection in combination with other markers at the protein level.
In other work, Lubman and coworkers (Liu et al., 2010) used a lectin antibody array version of the lectin-FLISA to test 26 potential markers discovered with a mass spectrometry-based technique to distinguish early HCC from cirrhosis. In this method, the antibodies were printed on glass slides and incubated with AAL lectin to detect fucosylation differences. The AAL was tagged with a fluorescent probe, and each spot was quantitatively detected with a microarray scanner. C3, CE, HRG, CD14, and HGF were found to be potential biomarker candidates to distinguish early HCC from cirrhosis with a sensitivity of 72% and a specificity of 79%.
It should be noted that a mass spectrometry version of the lectin-FLISA was recently developed by Yoo and coworkers (Ahn et al., 2012) to detect HCC. This method will be discussed below.
D. 2-D Gels and Lectin Analysis
Lectin analysis has also been used with 2-D gel electrophoresis to search for HCC markers. Block and coworkers (Block et al., 2005) performed glycoproteomic analysis with 2-D gels to identify serum glycoproteins that correlate with liver cancer in woodchucks and humans. Their strategy used several lectins to extract fucosylation from serum and then analyzed the bound and unbound fractions with 2-D gels. This analysis showed there were clearly proteins that were fucosylated that were upregulated in the bound versus the unbound fraction. An HPLC analysis of the glycans from the bound fraction showed the presence of fucosylated glycans as compared to the unbound fraction. They performed the experiment for the bound fractions for HCC versus non-HCC (healthy), and found that there were proteins that were upregulated in the HCC sample; with further analysis these proteins were found to be highly fucosylated. One such protein of interest was fucosylated GP73, which was also tested in human samples and was found to be upregulated significantly compared to other control groups.
The summary of the four non-mass spectrometry methods is presented in Table 4.
TABLE 4.
Non-mass spectrometry methods for glycoprotein marker discovery.
| Immunoassays (ELISA) | Very sensitive and specific, but measures protein level but not sensitive to glycan structure |
| CE-based assays | Specific for separation and profiling of glycans |
| Lectin-FLISA | Specific for target glycoproteins and can detect changes in glycan structures |
| 2-D gels/lectin analysis | Can monitor global changes in the proteome for large number of proteins |
V. DISCOVERY—MASS SPECTROMETRY-BASED ASSAYS
A. Mass Spectrometry Versus Non-Mass Spectrometry Methods
In this section, we will review the advances in mass spectrometry for the analysis of glycans and glycopeptides related to early detection of HCC. There are several key advantages for mass spec based techniques (see Table 5). One of these advantages is that MS assays do not require antibodies, although antibodies are sometimes used to enhance MS based assays in the case of studies of target glycoproteins. The performance and availability of antibodies may vary and affect an assay whereas MS is a more general technique and can be performed on any protein. With no need of prior knowledge of the protein identity, MS can perform large scale screening of glyco-marker candidates in complex biological samples that underwent glycosylation changes, which can be efficiently conducted for in-depth quantitation of glyco-markers with the support of sophisticated MS softwares. An additional advantage of MS assays such as MRM for example is that the assays can be multiplexed for many different proteins simultaneously. Also, the advent of tagging methods such as iTRAQ and TMT labels has allowed quantitative multiplexing of samples not readily possible by current non-MS methods. This feature is essential in biomarker studies comparing multiple samples. Probably the most signifi-cant advantage of MS based techniques is the ability to obtain detailed structural analysis of glycan structures as well as to pinpoint the glycosylation sites that underwent glycan changes in complex biological samples, which might have direct impact on biomarker studies. Although CE/lectins and gel/lectins provide some information on glycan structure, assays using these methods cannot provide the level of detail that modern MS-based techniques can achieve, especially in the in-depth structure information (structural isomers, linkage information, etc.). The main advantage of these tools compared to MS is that they do provide a means of visually monitoring changes in glycan composition. Also, these non-MS based techniques do not require the sophisticated instrumentation or expense of modern mass spectrometers. Nevertheless, as discussed in the following sections, mass spectrometry can provide a level of detail in glycan analysis not available to any other method.
TABLE 5.
Advantages of mass spectrometry for glycan analysis and related assays.
| 1. | No antibodies or lectins required |
| 2. | Multiplex capabilities with quantitative analysis |
| 3. | Ability to obtain detailed glycan structures for biomarker studies |
B. Methods to Enrich Glycoproteins and Their Glycans/Glycopeptides
Due to low abundance of glycoproteins and the inherent structural complexity of protein glycosylation, isolation/purification of glycoproteins and their glycoforms such as glycans and glycopeptides is an essential element of successful characterization of the glycoproteome, glycome, and glycopeptidome. Advanced analytical technologies have been developed to selectively enrich glycoproteins and their glycans/glycopeptides (Table 6), including lectin-based affinity chromatography, hydrazide chemistry, hydrophilic interaction chromatography (HILIC), porous graphitized carbon (PGC) chromatography, and cotton wool SPE tips. Among these separation methods, lectin affinity chromatography and hydrazide chemistry can be performed either to capture glycoproteins at the protein level (Xu et al., 2007; Zhang, 2007; Wang et al., 2008; Liu et al., 2013) or capture glycopeptides at the peptide level (Chen et al., 2013; Yin et al., 2015; Tanabe et al., 2016). Whereas lectins have well-characterized sugar specificities to capture specific carbohydrate residues in glycoproteins/glycopeptides, hydra-zide chemistry can be used in the unbiased enrichment of glycoproteins/glycopeptides. PGC columns or tips are the most commonly used strategy to enrich glycans from protein mixtures (Zhu et al., 2014). HILIC is a well-recognized technique that can effectively enrich glycans/glycopeptides (Yang et al., 2017), whereas cotton SPE tips have been recently developed to purify glycopeptides and glycans (Selman et al., 2011). These strategies of glycoprotein/glycan/glycopeptide enrichment can be further coupled with advanced mass spectrometry analysis (An et al., 2009; Kailemia et al., 2014) with LC-MS/MS, QIT-TOF, MALDI-TOF/TOF, and MRM to discover more clinically relevant markers with greater sensitivity and specificity (Dai et al., 2009). Table 6 summarizes the methods for the enrichment of glycans/glycopeptides/glycoproteins.
TABLE 6.
Methods for enrichment of glycans/glycopeptides/glycoproteins.
| Method | Advantages |
|---|---|
| Lectins | Selective method for enrichment of specific glycan structures on glycoproteins and glycopeptides |
| Hydrazide | A general unbiased method for glycopeptide enrichment but with loss of some glycan information |
| Graphitized carbon Chromatography (PGC) | PGC tips can be used to enrich glycans from small amount of sample |
| HILIC chromatography | A general method that can effectively enrich glycans/glycopeptides based on hydrophilicity |
| MW cutoff filter | Can effectively separate large glycans/glycopeptides from smaller peptides |
| Size Exclusion Chromatography | Rapid method to enhance detection of N-linked glycosylation sites |
C. MALDI-MS Profiling of Glycans
1. Method
MALDI-MS has been used as a convenient method to profile glycans. The MALDI processed applications have been reviewed in prior publications (Michael et al., 1992; Zhao et al., 2006; Fukuyama et al., 2008). Because there is no intrinsic pre-separation in the MALDI process, the method is best for simple mixtures. In most glycan biomarker studies, the glycans from a single isolated protein or a limited number of proteins is usually studied. Also, generally the glycans must be derivatized, usually with permethylation, to enhance the volatility and to improve the limit of detection. It is also possible to perform an LCMALDI experiment (Young & Li, 2006; Chen et al., 2017b) for more-complex mixtures, where the glycans can be separated and placed on different spots on the MALDI plate to provide a simpler spectrum for interpretation. It should be noted that a major advantage of this method is that rapid screening of a large number of samples can be accomplished with a high-capacity MALDI plate and modern high repetition rate lasers.
2. Targeted MALDI Profiling for Haptoglobin (Hp) and Alpha-1-Acid Glycoprotein (AGP)
With a quantitative MALDI-QIT-MS/MS approach with only 10 μL of serum in individual patients, Lin and coworkers identified that fucosylated N-glycans in Hp were significantly elevated in pancreatic cancer compared to chronic pancreatitis (Lin et al., 2011). In this study, Hp was isolated with a monoclonal antibody, and the glycans removed for analysis with MALDI-MS where the glycans were also permethylated. Eight desialylated N-glycan structures of haptoglobin were identified, where a bifucosylated triantennary structure was reported for the first time in pancreatic cancer samples. Core and antennary fucosylation were both elevated in pancreatic cancer samples compared to samples from benign conditions. Structural analysis could be provided with the QIT-MS/MS capabilities (Nishikaze, 2017). Fucosylation degree indices were calculated, and showed a significant difference between pancreatic cancer patients of all stages and the benign conditions. This study showed the feasibility of MALDI-MS as an assay for cancer from serum samples.
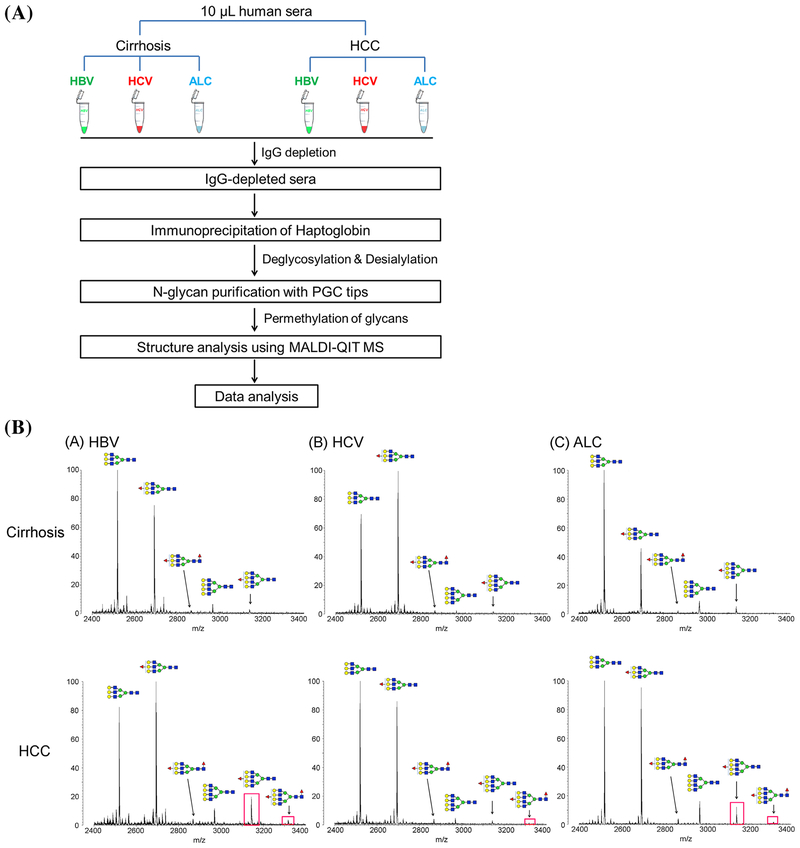
The MALDI-MS profiling method was applied by the same group to study the glycans expressed in the different HCC etiologies (see Fig. 2) (Zhu et al., 2014). A unique pattern of bifucosylated tetra-antennary glycan, with core and antennary fucosylation, was identified in HCC patients. Quantitative analysis indicated that the increased fucosylation degree was highly associated with HBV and ALD-related HCC patients compared to that of the corresponding cirrhosis patients. Notably, the bifucosylation degree was distinctly increased in HCC patients versus that in cirrhosis of all etiologies. The elevated bifucosylation degree of haptoglobin could discriminate early-stage HCC patients from cirrhosis in each etiologic category. This unique pattern of bifucosylated tetra-antennary N-glycan could outperform the clinically used AFP to discriminate early-stage HCC from cirrhosis (AUC = 0.834, P < 0.0001) (Zhu et al., 2014). More recently, Zhu et al. have developed an antibody-extraction column, which improves markedly over immunoprecipitation to isolate Hp for this experiment. This antibody column can extract Hp from 20 μL of serum in 40 min with >90% recovery (Zhu et al., 2015).
FIGURE 2.
(a) Workflow of N-glycan profiling of haptoglobin and fucosylation changes between HCC and liver cirrhosis of the three most common etiologies, infection with HBV or HCV, and heavy alcohol consumption (ALD). (b) MALDI-QIT-MS spectra showing the difference of fucosylation in tri- and tetra-antennary N-glycans of haptoglobin between HCC and cirrhosis in relation to the etiology, HBV (A), HCV (B), and ALD (C), respectively. The bifucosylated tetra-antennary (m/z 3316.69) glycan was predominantly present in HCC samples but not in liver cirrhosis. The tetra-antennary glycans were highly elevated in HBV- and ALD-related HCC compared with the corresponding levels in cirrhosis; however, no significant difference in tetra-antennary glycans was observed between HCV-related HCC and cirrhosis. The elevated presence of fucosylated tetra-antennary glycans in HCC samples compared to that in cirrhosis of each etiology is highlighted with a red rectangle. Reprinted with permission from Ref [J Proteome Res 2014, 13, 2986–2997] Copyright 2014 American Chemical Society.
In other work by Zhang et al. (2011), N-linked changes in serum Hp beta chain were studied with MALDI-QIT-MS. The Hp was isolated from serum using a Hp-antibody column and the glycans removed for analysis. In this work they specifically studied patients with HBV etiology which is predominant in China. Their studies included 20 each of HBV patients, cirrhosis patients, and HCC patients and also normal controls. They found that two fucosylated glycans, whose structures were identified with MS/MS in the QIT, were clearly elevated in the cirrhosis and HCC patients relative to HBV and normal. This result was also confirmed with lectin blot using AAL lectin.
In related work, Liang et al. applied MALDI-MS glycan profiling to AGP from serum where AGP was isolated with a chemical precipitation method. The AGP glycan profile was more complex than Hp, and yielded nine peaks in the MALDI MS spectrum. It was shown that a trifucosylated tetra-antennary glycan could distinguish HCC from cirrhosis samples with a performance for NASH-based samples that was comparable to AFP. Wang et al. studied glycans from IgGs isolated from serum and analyzed them with MALDI-MS. They found that several glycans from the IgGs improved detection of HCC versus cirrhosis relative to AFP. The ratio of galactose was particularly promising.
3. Global MALDI Screening to Profile HCC Markers in Serum
Goldman et al. (2009) evaluated the use of total glycan profiling with MALDI-MS to identify markers for HCC. In several studies they used 10 mL of serum; they removed the N-glycans with PNGaseF and solid-phase extraction, and permethylation was used to process the glycans for MALDI TOF-TOF MS. They studied HCC samples and controls with chronic liver disease and without liver disease. They used novel computational methods to analyze the complex set of glycan patterns obtained, and they found three selected N-glycans that could classify HCC with 90% sensitivity and 89% specificity in an independent set of patients with chronic liver disease. Because these samples were obtained from a hospital in Egypt, the marker performance might prove different for other populations. Although these glycans could identify HCC from cirrhosis and fibrosis, the proteins associated with the glycans are not known and are probably from high-abundance proteins. The latter makes it difficult to develop an antibody based version or ELISA to detect these markers.
4. MALDI-Imaging of Tissues in HCC
Although this review is focused on analysis from serum, it should be noted that there has been significant recent activity in the area of MALDI imaging which can evaluate changes in tissues during the progression of cancer. Recent work in this area related to HCC is that of Powers and coworkers, where they used MALDI-imaging to study changes in glycan structure between normal and HCC tissue samples (Powers et al., 2015). The method can be used in fresh tissues or in FFPE, where PNGaseF is sprayed on the sample to release N-glycans directly on the tissues mounted on glass slides before adding the MALDI matrix. They evaluated FFPE samples of HCC tissues using this method and detected over 30 N-glycans. Distinct differences were observed between the HCC versus normal samples. They also were able to study the distribution of singly fucosylated N-glycans detected in these tissues and could compare them to the staining pattern obtained using a core fucose binding lectin. In more recent work by this group, they analyzed 138 HCC tissues and compared the N-linked glycans in cancer tissue to tissue with liver cirrhosis (West et al., 2018). Ten glycans were found to be significantly elevated in HCC tissues compared to cirrhosis. These glycans were found to be with increased levels of fucosylation and/or with increased levels of branching. They also found that increased levels of fucosylated glycoforms were associated with a reduction in survival time (West et al., 2018).
D. ESI-MS Profiling of Glycans
1. Method
Electrospray ionization has been used to profile glycans from various biofluids in an attempt to find markers for HCC and other cancers. A major advantage of ESI is that it can readily be coupled to HPLC to pre-separate complex glycan mixtures. The method allows total glycan analysis from serum or other biofluids or of glycans from target proteins. The glycans can be analyzed without prior derivatization or with derivatization to increase detection sensitivity. The ESI source can be coupled to any type of mass spectrometer, and has been used with Orbitraps for structural analysis of the glycans using CID and HCD for MS/MS analysis. It can also be interfaced to ion mobility or PGC columns to separate glycan isoforms. The glycan profile often provides a distinct signature of a cancer versus control; however, the glycans cannot be associated with a specific protein so that an antibody-based assay cannot be readily developed.
2. Global Profiling of Glycans With LC-ESI-MS for Biomarkers of HCC
LC-ESI analysis has been used to profile glycans for a number of cancers, including ovarian (Leiserowitz et al., 2008; Kim et al., 2014), pancreatic (Zhao et al., 2007), HCC (Chandler et al., 2013; Tsai et al., 2014; Wang et al., 2017), and prostate cancer (Hua et al., 2011), among others. There have been a number of studies on global screening of glycans from serum for HCC biomarker studies. Ressom and coworkers (Tsai et al., 2014) used LC-ESI-MS to analyze N-glycans in sera from 183 patients to distinguish HCC from cirrhotic controls. N-glycans were released from serum proteins with PNGaseF and the glycans were permethylated with a solid-phase permethylation procedure. The glycans were profiled with LC-ESI interfaced to an Orbitrap Velos MS. Several glycans that were found to be significantly up- or down-regulated in their cohorts were identified for further analysis. These glycans were then analyzed with targeted MRM analysis, and 11 N-glycans were identified as being statistically significant between HCC cases and cirrhotic controls. This study demonstrated the use of an integrated approach for profiling and targeted assays for biomarker analysis of glycans.
3. Profiling of Glycans for Specific Proteins with LC-ESI-MS for Biomarkers of HCC
The alternative strategy to global profiling of glycans is to isolate a targeted protein from serum for analysis of its glycan content. This strategy has been performed for HCC for a number of different target proteins. One important target has been Haptoglobin, as shown in prior work on the MALDI-MS analysis of its glycan content. In the work of Lubman and coworkers (Zhang et al., 2015b), a workflow was developed that isolated Hp with an HPLC-based affinity column followed by glycan removal, extraction and desialylation. The fucosylated glycans from Hp were derivatized with Meladrazine which is a reagent developed in China (Tie & Zhang, 2012) that can significantly enhance the sensitivity for ESI-MS detection. The separation of the glycans with a HILIC column resulted in detection and quantitation of eight glycans with less than 1 mL of serum. The ratio of the various fucosylated peaks to their corresponding non-fucosylated forms showed that the fucosylated glycans are upregulated in the case of early HCC samples versus cirrhosis. In particular, a relatively low abundance bifucosylated tetra-antennary form might serve as a marker of HCC as shown in their prior MALDI-MS work.
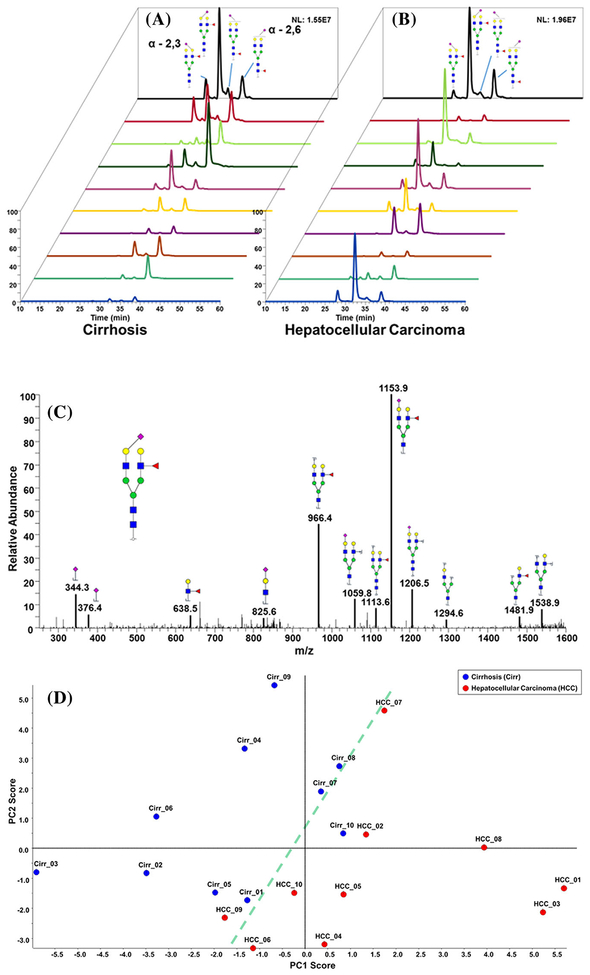
A different strategy developed by Mechref and coworkers (Huang et al., 2017) involved the use of isomeric profiling to identify glycan markers for cancer. Isomeric forms of glycans were separated and identified based upon separations with elevated temperatures in a PGC column followed by ESI-MS in an Orbitrap Velos MS (see Fig. 3). This method separated isoforms of sialic acid, including the isoforms of sialic acid with alpha-2,3 and alpha-2,6 linkages. A comparison of non-isomeric and isomeric permethylated glycan forms from Hp released from patient serum was achieved with C18 and porous graphitic carbon (PGC) columns. This method was used to distinguish early-stage HCC from cirrhosis, whereby 8 out of 34 glycans identified by PGC-LC-MS/MS were found to be significant due to the isomeric distributions of a particular glycan. This work represents the first example of the use of isoforms of glycans as potential cancer biomarkers. Other attempts have been made to distinguish glycan isoforms from serum with ion mobility spectrometry, although at this point with limited success (Gaye et al., 2012).
FIGURE 3.
EIC of biantennary monosialylated branch-fucosylated glycan linkage isomers derived from (a) cirrhotic and (b) HCC patients. (c) MS/MS interpretation of biantennary monosialylated branch-fucosylated glycan. (d) Unsupervised PCA plot of the glycans that were quantitatively determined by C18-LC-MS/MS analysis. Reprinted with permission from Ref [Electrophoresis 2017, 38, 2160–2167].
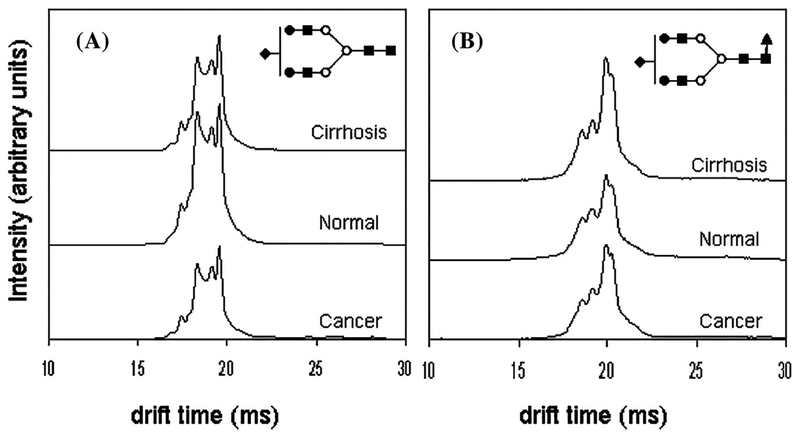
An alternative strategy developed by Clemmer and coworkers (Isailovic et al., 2012) used ion mobility spectrometry-mass spectrometry (IMS-MS) to profile serum N-linked glycans for HCC (see Fig. 4). In the ion mobility strategy, ions separate according to their mobility in a drift gas under the influence of an electric field. The drift time depends on the collisional cross section of the ion, which depends on its shape. The result is that isomers can often be separated that could not be detected in mass spectrometry alone. In this work, they profiled the glycans from HCC, cirrhosis, and normal with the IMS-MS technique, and they found that the ion mobility profiles for as many as ten different mass-to-charge ratios for glycans together with supervised PCA analysis distinguished the different disease states. The resolution of ion mobility is limited, but they nevertheless speculate that they were able to distinguish some isomer forms based on differences in the ion mobility. The method has distinct advantages in terms of speed of analysis compared to other separation methods. In other work, Li and coworkers developed a relative quantification platform for quantitative analysis of N-glycans released from human serum protein digests by combining aminoxy TMT labeling with CE-ESI-MS/MS, which has been demonstrated to be efficient in resolving glycan isomers and improving the relative quantification accuracy (Zhong et al., 2015). A review that details advances in the use of IMS to separate isomeric glycans has recently been published (Chen et al., 2017a).
FIGURE 4.
IMS profiles of glycan ions (A) [S1H5N4 + 3Na]3+ and (B) [S1F1H5N4 + 3Na]3+ glycans showing both conformational and intensity differences with respect to disease state. Note that in the case of S1H5N4, the disease states exhibit lower overall intensities than the healthy state, while S1F1H5N4 shows higher overall drift time intensities in the case of diseased states than in the healthy state. This might be due to increased fucosylation of glycans with cancer and cirrhosis. Reprinted with permission from Ref [J Proteome Res 2012, 11, 576–585] Copyright 2012 American Chemical Society.
E. ESI-MS of Glycoproteins/Glycopeptides to Profile Proteins Related to HCC
1. Method
The description for ESI-MS of glycoproteins and glycopep-tides is similar to that described above; however, there are several experimental issues that must be considered. In the case of glycopeptides, there can be other interfering peptides that must be removed or separated from the non-glycosylated peptides. The peptides tend to have a much higher ionization efficiency than the glycopeptides, and will suppress the signal from the glycopeptides. Also, an advantage to study glycopeptides is that one can identify site specificity for the glycans, which might be an important marker in itself for disease. An issue to study glycopeptides with MS/MS is that various methods are required for analysis of the peptide backbone and glycan structure. Some of these methods that have been used in HCC biomarker studies will be discussed in this section. Alternatively, one can study the glycoproteins based on the selective isolation of the glycoproteins with certain structures with lectin methods, and study of the peptide changes. There is some loss of structural information in this strategy, but it can differentiate protein levels according to the expression of specific glycan structures.
2. Lectin Extraction of Proteins
In one strategy used by several groups (Yang et al., 2006) for biomarker analysis in serum, lectin columns were used to quantitatively extract glycoproteins. This method has been used in studies of HCC versus cirrhosis in the work of Lubman and coworkers (Liu et al., 2010) to study differential changes in fucosylation. In this work, a lectin array was first used to establish that AAL and LCA provided the largest changes in response between HCC and cirrhosis in serum. The proteins in the sera samples were labeled with Exactag labels for quantitative analysis and passed through a lectin AAL or LCA column for further analysis. The extracted glycoproteins were digested with trypsin and analyzed with nanoLC-MS. The changes in the fucosylation level were monitored through the tagged peptides. They found five proteins, including complement C3, ceruloplasmin, histidine-rich glycoprotein, CD14 and hepatocyte growth factor, that were potential markers to detect early-stage HCC versus cirrhosis. The combination of the five proteins had an AUC of 0.81. This work showed that there was a significant increase in the level of fucosylation in serum proteins in early HCC versus cirrhosis as a potential means for early detection.
3. Lectin Extraction of Glycopeptides
Integrated analysis of lectin-extracted glycopeptides.
In a related but alternate strategy to that discussed in the above section, Narimatsu and coworkers (Kaji et al., 2013) developed an integrated strategy with lectins to extract glycopeptides from serum to analyze biomarker candidates for HCV/HBV Infection-associated liver fibrosis and HCC. The strategy focused on candidate glycoproteins that are expressed in the original tissues of the cancer and that carry glycan structures associated with carcinogenesis, in this case HCC. They analyzed the glycan profiles of culture media of HCC cell lines with lectin arrays, and found that AAL and DSA signals were significant. They used lectin affinity chromatography with different combinations of these lectins to extract glycopeptides from digested proteins from the culture media and patient serum, and analyzed the glycopep-tide profile with IGOT (isotope-coded glycosylation-site-specific tagging), where they remove the glycan and label the site with 18O and analyzed with nano-LC-MS on a QTOF MS. The glycoproteome profile was compared, where they found 744 first-step candidates from AAL extraction. They selected 21 of these candidates in patient serum based on liver expression and the availability of an effective antibody. A verification of the glycan alteration was conducted using pooled sera with lectin arrays to verify enhancement of fucosylation associated with HCC and detection of HCC-associated enhancement of fucosylation on the candidate glycoproteins with AAL-fractionation followed by Western blotting. However, they did not look for markers of early HCC in this study but rather were more focused on fibrosis.
In other work by Tanabe et al. (2016), a novel strategy was used to perform global screening of glycopeptides in serum to discover HCC markers. The proteins in patient serum were digested with trypsin, and enriched first with ultrafiltration to eliminate peptides and small glycopeptides, and further enriched with AAL lectin-based affinity chromatography. The glycopep-tides were analyzed with LC-ESI-QTOF MS, where custom software was used to screen thousands of AAL-enriched glycopeptide peaks over large numbers of samples. Glycopep-tide candidates were further isolated and identified with LC-MS/MS after removal of glycans with PNGaseF. The glycan structure was proposed based on MS/MS analysis. They identified a glycopeptide from AGP with multi-fucosylated tetraanten-nary N-glycans that was significantly elevated in HCC patients. The ROC curves for HCC versus cirrhosis provided values of an AUC = 0.86 for HCC HCV versus cirrhosis and an AUC = 0.93 for HCC HBV versus cirrhosis. However, because the stage of the patients was not provided, it was not clear if these were mainly early or late stage HCC patients.
Lectin extraction with truncation of glycan structure.
One of the issues to study glycopeptides has been to unravel the often complex structures of N-linked glycans. One method that has been used to study the structure of glycans involves initial removal of selected carbohydrate units with enzymes followed by analysis of the remaining glycan with CE (Varadi et al., 2013) or LC (Wu et al., 2010) or mass spectrometry. In one study by Yin and others, a method for mass-selected site specific core-fucosylation of serum proteins in HCC was developed (see Fig. 5) (Yin et al., 2015). This method involved initial depletion of high-abundance proteins, trypsin digestion of the remaining proteins, iTRAQ labeling of the peptides, LCA enrichment of core-fucosylated peptides, followed by endoglycosidase F3 digestion before mass spectrometry analysis. The endoglycosidase digestion removed most of the glycan structure and left the core-fucosylation structure intact for mass spectrometry analysis. This strategy simplified the detection of these core-fucosylated structures and increased detection sensitivity compared to the intact glycan. In this study, they detected 1,300 CF peptides from 613 CF proteins from patient sera, where 20 CF peptides were differentially expressed in ALD-related (alcohol) HCC samples compared with ALD-related cirrhosis samples and 26 CF peptides changed in hepatitis C virus (HCV)-related HCC samples compared with HCV-related cirrhosis samples. Among these there were three CF peptides from fibronectin upregulated in ALD-related HCC samples compared to cirrhosis with an AUC value of 0.89 at site 1007 for detection of early HCC. When combined with the AFP value, the AUC reached to 0.92; therefore, CF peptides of fibronectin might serve as potential biomarkers for early-stage HCC screening in ALD-related cirrhosis patients.
FIGURE 5.
Workflow for screening of changes in site-specific core-fucosylation (CF) of serum proteins in early stage HCC with different etiologies. The methods involve depletion of high abundance proteins, trypsin digestion of medium-to-low abundance proteins into peptides, iTRAQ labeling, and Lens culinaris Agglutinin (LCA) enrichment of CF peptides, followed by endoglycosidase F3 digestion before mass spectrometry analysis. 1300 CF peptides from 613 CF proteins were identified from patient sera, where 20 and 26 CF peptides were differentially expressed in alcohol (ALD)-related HCC samples compared with ALD-related cirrhosis samples and HCV-related HCC compared with HCV-related cirrhosis samples. Reprinted with permission from Ref [J Proteome Res 2015, 14, 4876–4884] Copyright 2015 American Chemical Society.
F. Analysis of Target Glycoproteins From Patient Serum With an Intact Glycopeptide Approach
1. Method
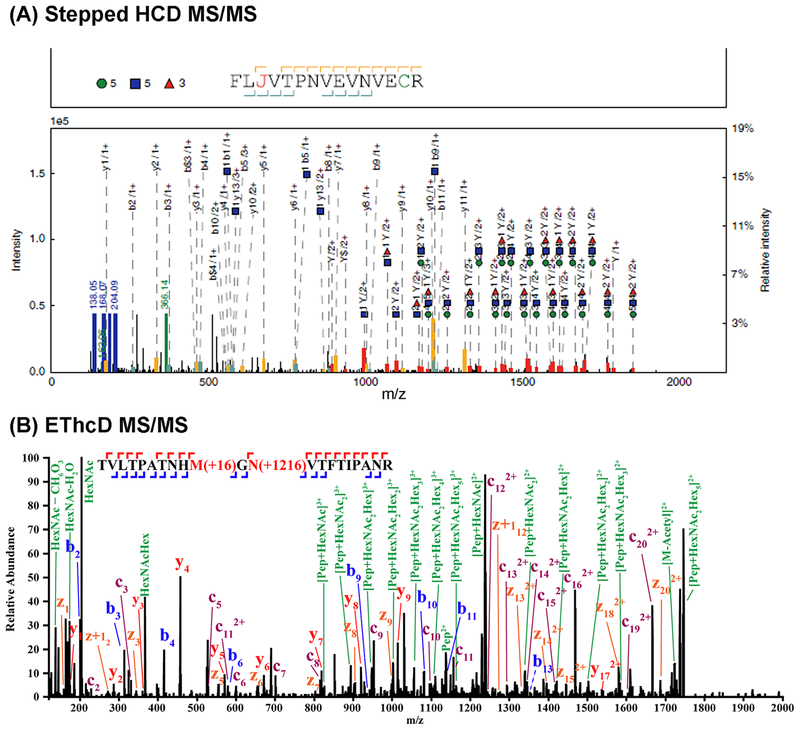
The ability to analyze intact glycopeptide analysis with mass spectrometry offers great advantages for biomarker discovery based on the ability to define glycan sites and structures. However, the analysis of intact glycopeptides has remained difficult due to the complexity of the glycopeptide structure, which contains a carbohydrate and a peptide structure, the low ionization efficiency and low abundance of glycopeptides compared to peptides in protein digests, and difficulties in data interpretation. The glycopeptides can be enriched from the peptides by a number of methods, including lectin or HILIC. However, the ability to analyze the glycopeptides has remained a challenge. In recent work, there have been a number of techniques developed to deal with this problem (Table 7), including collision-induced dissociation (CID)/electron-transfer dissociation (ETD) (Alley et al., 2009), CID/higher-energy collisional dissociation (HCD) (Segu & Mechref, 2010; Lee et al., 2016), stepped HCD (Liu et al., 2017a; Yin et al., 2018), and electron-transfer/higher-energy collision dissociation (EThcD) (Yu et al., 2017; Chen et al., 2018; Glover et al., 2018), for example. The use of CID/ETD MS to analyze glycopeptides has been recently reviewed by Mechref (Mechref, 2012). CID can provide information related to the composition of the glycan unit attached to the peptide, whereas ETD can sequence the peptide because it causes only peptide backbone fragmentation and keeps the glycans intact. Stepped collision energy (SCE) HCD-MS/MS uses different collision energies in HCD-MS/MS to produce complementary fragments of the glycan and peptide. It has been found that SCE-HCD-MS/MS under 20-30-40% energies generated highly informative fragment ions for peptide and glycan of a glycopeptide for structural analysis (see Fig. 6A) (Liu et al., 2017a; Yin et al., 2018). In other work by Heck and coworkers, ETD and HCD were combined to develop a hybrid technique called EThcD (Frese et al., 2012). In this method, a supplemental energy is applied to all ions formed by ETD to generate spectra with enhanced capabilities in glycopeptide studies (see Fig. 6B). There has been extensive work on some of these techniques towards analysis of glycoproteins/glycopep-tides for standard proteins, but more limited work for clinical samples as in serum.
TABLE 7.
MS/MS methods for MS analysis of intact glycopeptides.
| CID/ETD | CID provides information related to glycan structure/ETD provides sequence information of peptides |
| CID/HCD | Both CID and HCD result in glycan fragmentation/HCD also provides peptide backbone b-y ion fragmentation |
| HCD/ETD | HCD provides diagnostic glyco-oxonium ions which can be used to trigger the succeeding ETD dissociation and additional b and y ions of peptide backbones |
| Stepped HCD | Can generate highly informative fragmentations for peptide and glycan of a glycopeptide |
| EThcD | Combines ETD and HCD where a supplemental energy is applied to ions formed by ETD to generate both peptide and glycan structural information |
FIGURE 6.
Examples of (A) the stepped-energy HCD MS/MS collision for an intact glycopeptide, reprinted with permission from Ref [Nature Commun 2017, 8, 438] and (B) the EThcD MS/MS collision of N-glycopeptide TVLTPATNHMGNVTFTIPANR, reprinted with permission from Ref [J Am Soc Mass Spectrom 2017, 28, 1751–1764].
2. EThcD Analysis of Glycopeptides
A recent strategy to analyze intact glycopeptides with mass spectrometry developed a quantitative EThcD-MS/MS method to determine changes in intact N-glycopeptides between early HCC and liver cirrhosis for Hp in patient serum (Zhu et al., 2018). In this work, Hp was immunopurified from 20 μL of serum followed by digestion with trypsin and GluC, glycopep-tide enrichment with HILIC TopTips, and LC-EThcD-MS/MS analysis on an Orbitrap Fusion Lumos Tribrid mass spectrometer. The development of the Orbitrap mass spectrometer is an important advance for glycopeptide analysis. The EThcD method was developed by Li and coworkers (Yu et al., 2017), where HCD and ETD fragments could both be collected in a single spectrum to markedly improve intact glycopeptide characterization. Site-specific identification and quantitation of N-glycopeptides were achieved with novel software developed by Protein Metrics Inc.; that is, Byonic and Byologic softwares (Bern et al., 2012). Byonic provides a means to interpret the structure of the glycopeptides, whereas Byologic provides a means to quantitate the structures. This software is also an important advance because manual analysis would be difficult and time-consuming. The analysis of patient serum resulted in 279 N-glycopeptide spectral matches that corresponded to 98 site-specific N-glycopeptides (Zhu et al., 2018). In addition, several key structures were found to be quantitatively different between early stage HCC and cirrhosis, including a bifucosylated tetra-antennary form reported in earlier work (Zhu et al., 2014). The combination of LC-EThcD-MS/MS and Byonic/Byologic software represent a potential major breakthrough for the analysis of intact N-glycopeptides.
The group of An (Lee et al., 2018) has also identified and quantified the site-specific glycopeptides of serum haptoglobin between gastric cancer and healthy controls with Q-TOF LCMS/MS. Ninety-six glycopeptides of serum Hp were characterized across all cancer and control samples, where three glycopeptides exhibited exceptionally high fold-changes in gastric cancer (Lee et al., 2018).
VI. VERIFICATION LC-MRM ANALYSIS OF GLYCOSYLATION
A. Method
LC-MRM serves as an alternative method for targeted protein quantification. The method has been used in complex mixtures, including patient serum where the proteins are digested into peptides, which are monitored with mass spectrometry. The peptides monitored for each target protein are selected according to criteria to optimize sensitivity and at the same time provide a peptide that is unique to that protein. In order to quantitate the peptide, an isotopically labeled reference peptide with the same sequence is used. The target peptides are selected in the mass spectrometer and fragmented; where several transitions are monitored. The method has great advantages in that one can multiplex large numbers of target peptides in modern mass spectrometers, it does not require an antibody, the assay once developed can be automated, and the data are highly reproducible compared to standard ELISAs. In the case of glycoproteins, the assay becomes more difficult because glyco-peptide standards are generally not available. There have been several strategies developed for LC-MRMs of glycoproteins (Ahn et al., 2009; Song et al., 2012; Ruhaak & Lebrilla, 2015). Herein, we will just present some of those studies that directly pertain to detection of HCC.
B. LC-MRM of IgGs in Plasma
In recent work, Goldman and coworkers (Yuan et al., 2015) developed a method for quantitative analysis of immunoglobulin subclasses and subclass specific glycosylation with LC-MSMRM in liver disease. Immunoglobulins were first isolated from human plasma with protein A and G columns. The IgGs were digested with trypsin and subjected to LC-MS/MS. Goldman and coworkers developed a novel method whereby glycan oxonium ions and peptide-GlcNAc fragment ions were used to quantify glycoforms of IgG purified with affinity chromatography with normalization to a unique peptide associated with each IgG subclass. They found that HCC patients have increased circulating IgG1, IgG3, IgA1, and IgM compared to healthy controls. A comparison of HCC and Cirrhosis patients showed that HCC patients have a significantly higher concentration of IgG1 and IgM but lower IgG2 concentration. An increase in galactose-deficient core fucosylated glycoforms was observed in cirrhosis and HCC patients. They also found that specific glycoforms such as FA2G0 and FA2B0 increase in all IgG subclasses, whereas the FA2G2 form decreases. This work in effect developed a method whereby quantities and glycoforms of immunoglobulins both change significantly in liver disease progression to HCC.
Goldman and coworkers (Sanda et al., 2013) also developed a quantitative LC-MS-MRM method for site-specific glyco-forms of Hp in liver disease. In this work, they first isolated Hp from plasma samples, where the Hp was added to an internal standard and digested with trypsin. The digest was treated with alpha-neuraminidase to remove the sialic acids to enhance the sensitivity for detection. A further digestion of the glycan with beta (1–4)-galactosidase further simplified the glycan structure. The glycopeptides were subjected to LC-MS-MRM, where the oxonium ions and peptide-GlcNAc fragments were monitored as MRM transitions. The T3 glycopeptide of Hp was chosen for particular attention because of the large number of potential diagnostic isoforms. The combination of LC-MS-MRM with exoglycosidase digestion resolved isobaric glycoforms of the Hp T3 glycopeptide for quantification of multiply fucosylated glycoforms. They found that 14 multiply fucosylated glycoforms increased significantly in the liver disease group compared to healthy controls. They also found that the tri- and tetra-antennary singly fucosylated glycoforms are associated with MELD score and low platelet counts. The group recently carried out LC-MS-MRM quantification of core fucosylated N-glycopeptides of serum proteins and found increased core fucosylation of five glycopeptides at the stage of liver fibrosis (i.e., N630 of serotransferrin, N107 of alpha-1-antitrypsin, N253 of plasma protease C1 inhibitor, N397 of ceruloplasmin, and N86 of vitronectin), increase of additional six glycopeptides at the stage of cirrhosis (i.e., N138 and N762 of ceruloplasmin, N354 of clusterin, N187 of hemopexin, N71 of immunoglobulin J chain, and N127 of lumican) (Ma et al., 2018).
C. LC-PRM of AFP in Serum
Most recently, An and coworkers (Kim et al., 2018) developed a quantitative LC-MS/MS-PRM to monitor fucosylated glycopep-tides in serum AFP to distinguish between HCC and cirrhosis patients. Because AFP is normally present at low levels in serum, even in patients with liver disease, a more sensitive approach is required. To overcome the sensitivity issues, AFP was immunoprecipitated from serum, followed by trypsin digestion, neuraminidase treatment to remove sialic acids, and LC-MS/MS-based PRM analysis. With a combination of these approaches, the MS detection limit was significantly improved (LOD < 2 ng/mL). The result showed that the relative percentage of fucosylated AFP (AFP-fuc%) had a better performance than serum AFP levels to differentiate early-HCC and cirrhosis (AUC = 0.962) with a sensitivity of 92.3% (Kim et al., 2018).
D. Lectin-FLISA Based MRM
A mass spectrometry version of the lectin-FLISA was recently developed by Yoo and coworkers (Ahn et al., 2012, 2013) to detect HCC. In this work (Ahn et al., 2012), fucosylated proteins from plasma were captured onto AAL lectin immobilized on beads (see Fig. 7). The proteins were digested with trypsin and the samples were spiked with stable isotope-coded internal standards of the target peptides to be quantified with MRM mass spectrometry in a triple-quadrupole mass spectrometer. They studied HCC of HBV etiology and used controls from cirrhosis, HBV patients, and healthy people. They found that AGP, AACT, A1AT, and Ceruloplasmin were potential markers. The AUC from these markers ranged from 0.73 to 0.92, and combinations of these candidates yielded an AUC of >0.95. The mass spectrometry version of the lectin-FLISA has distinct advantages in that it eliminates the use of antibodies and associated problems such as cross reactivity. The MRM can allow marker multiplexing where large numbers of peptides can be analyzed simultaneously. The assay can also be performed on a very standard triple quadrupole mass spectrometer. Also, MRM assays can be automated to analyze large numbers of samples and tend to be more stable than conventional ELISAs.
FIGURE 7.
A scheme which shows the process for identifying aberrantly glycosylated biomarkers using the lectin-coupled MRM-based approach. Reprinted with permission from Ref [J Proteomics 2012, 75, 5507–5515].
E. Other Lectin-Based MRM Assays for HCC
In work by Qian and coworkers (Zhao et al., 2011), a method for site-specific quantification of core fucosylated glycoproteins with MRM-MS was developed. In this assay, a serum sample with bovine thyroglobulin added was first enriched at the protein level with LCH lectin. The CF proteins underwent trypsin digestion, and a normalized internal standard, which is a pool of CF peptides from normals with CF peptides from bovine thyroglobulin, were labeled with 18O was added to the sample and was further purified using ultrafiltration. The CF peptides underwent partial deglycosylation with Endo F3 to remove most of the glycan structure to thus increase ionization efficiency. The CF peptides underwent MRM-MS with a triple quadrupole MS; in the MS2 spectrum, product ions PGn+, yG+, and y+ appeared regularly with high abundance. These ions were used for MRM quantification, and were applied to HCC and normal serum samples with the peptide markers; see Comunale et al. (2006). Their studies did not show a significant difference between the healthy controls and HCC groups for the CF levels of seven peptides; however, these were preliminary results.
In more recent work by Lubman and coworkers, a similar assay was designed to validate markers of core-fucosylated proteins from previous work (Yin et al., 2015). In this method, serum samples from HCC, cirrhosis, and a pool of normal standards was first depleted of albumin and the IgGs with a spin column. At this point, standard proteins, which were targets of analysis, were added to the serum to enhance the ability to detect the peptides with MS/MS. The remaining proteins were digested with trypsin, and the CF-glycopeptides were extracted with LCA lectin. These CF-peptides were labeled with iTRAQ tags, and the samples were combined. The CF peptides were treated with endoF3 to remove most of the glycan structure and leave the truncated CF stem. The CF peptides were subjected to MRM analysis on an Orbitrap Q-Exactive. This method yielded a relative type of MRM where one can compare the changes in HCC CF-peptides to that of cirrhosis, and can use the pool of normal controls to standardize the signals. This work is currently underway to validate CF-fibronectin as a marker of early stage ALD-based HCC.
VII. CONCLUSION
There has been great progress in the development of methods to analyze glycoproteins from patient samples, including plasma and serum. These methods will be essential to discover and validate markers for detection of early-stage HCC where treatment can be effective. They might also be used for diagnosis, prognosis, and therapeutic treatment, where often the glycan structures might change during the course of cancer progression. These methods have included mass spectrometry methods to analyze glycans that have been removed from the protein or intact glycopeptides, which retain the glycan structure and site specificity. These methods can be applied to the global glycoprotein content of serum, or more often they can be applied to target proteins such as Haptoglobin, Hemopexin, or Kininogen, for example, which have been proven to have diagnostic utility in cancer.
Glycans have been analyzed with methods such as MALDI generally for target proteins and also with electrospray, where the glycans have been analyzed for either target proteins or large numbers of glycoproteins from serum. Other methods such as ion mobility have also been employed to separate isomeric structures. Glycan analysis has often proven diagnostic of HCC versus cirrhosis, but the diagnostic glycans might not be unique to HCC and are often found in other cancers. An antibody-based method to screen a large number of samples cannot be readily developed. Alternatively, there have been significant developments to analyze glycopeptides, where various methods, including stepped HCD and EThcD, have been developed to obtain the sequence of the peptide backbone and the glycans. In addition, the development of new software that can analyze the many glycopeptide isoforms and also quantitate these isoforms have made possible the identification of specific isoforms that might be potential markers of HCC. The use of PGC columns has recently shown the ability to separate these isoforms for analysis.
Although a number of potential biomarker candidates has been identified, validation of these markers in large Phase-3 biomarker validation sample sets such as the Hepatocellular Carcinoma Early Detection Strategy study (HEDS) set sponsored by the EDRN will be necessary before these markers can be further considered for clinical use. There have been various mass spectrometry-based techniques developed for such validations of glycopeptides such as a glyco-MRM and a lectin-FLISA based MRM. With the development of new technologies, we expect even more advances towards analysis of glycoproteins from patient samples. Nevertheless, the future of this field appears very exciting with many possibilities for clinical applications.
The success of future work will depend on the new instrumental developments described herein, but will also depend heavily on the quality, type, and number of clinical samples available. These issues will present important challenges in this work. The quality of the samples is essential and the NCI-EDRN has developed standard operating procedures (SOPs) for collection, processing, and storage of samples to eliminate these issues from affecting the biomarker discovery and verification process. Also the types of samples collected in regard to having available different etiologies of diseases available for study will be essential where each of these may have their own unique glycan structure specific markers. It is also important to eliminate biases in analysis by having a sufficient number of samples from different genders and ethnic groups and also sufficient clinical information available to evaluate biomarker performance. It will also be essential to have a sufficient number of samples to run properly powered discovery and validation sets to assess the clinical utility of these markers.
ACKNOWLEDGMENTS
This work was funded by the National Cancer Institute under grant nos. 1R01 CA160254 (D.M.L.), 1R01 CA154455 (D.M.L.), U01 CA225753 (D.M.L.), and R50 CA221808 (J.Z.) and received partial support from the National Institutes of Health through grant no. R01 GM 49500 (D.M.L.).
Contract grant sponsor: National Cancer Institute; Contract grant numbers: 1R01 CA160254, 1R01 CA154455, U01 CA225753, R50 CA221808; Contract grant sponsor: National Institutes of Health; Contract grant number: R01 GM 49500.
ABBREVIATIONS
- A1AT
alpha-1-antitrypsin
- AAL
Aleuria aurantia lectin
- AGP
alpha-1-acid-glycoprotein
- ALD-disease
alcohol-related disease
- AUC
area under the curve
- CID
collision induced dissociation
- CE
capillary electrophoresis
- CLEIA
chemiluminescent enzyme immunoassay
- DSA
Datura stramonium agglutinin
- DSA-FACE
DNA sequencer-assisted fluorophore-assisted carbohydrate electrophoresis
- EDRN
Early Detection Research Network
- ELISA
enzyme linked immunosorbent assay
- ESI
electrospray ionization
- ETD
electron transfer dissociation
- EThcD
Electron-Transfer/Higher-Energy Collision Dissociation
- FLISA
fluorescence-linked immunosorbent assay
- HCC
Hepatocellular Carcinoma
- HCD
higher-energy collisional dissociation
- HEDS
Hepatocellular Carcinoma Early Detection Strategy study
- HILIC
hydrophilic interaction chromatography
- Hp
Haptoglobin
- IGOT
isotope-coded glycosylation-site-specific tagging
- IMS
ion mobility spectrometry
- IMS-MS
ion mobility spectrometry-mass spectrometry
- iTRAQ
isobaric tags for relative and absolute quantitation
- LCA
lens culinaris agglutinin
- MALDI
matrix-assisted laser desorption ionization
- MRM
multiple reaction monitoring
- MS2
tandem mass spectrometry
- NAFLD
non-alcoholic fatty liver disease
- NCI
the National Cancer Institute
- PGC
porous graphitic carbon
- PHA-L
Phaseolus vulgaris leucoagglutinin
- PRM
parallel reaction monitoring
- PTM
post-translational modification
- QIT-TOF
quadrupole ion trap-Time of Flight
- SELDI-TOF-MS
surface-enhanced laser desorption-ionization time-of-flight mass spectrometry
- TMT
tandem mass tags
Biographies

Jianhui Zhu, PhD, is a Lead Research Scientist in the Laboratory of Cancer Proteomics in the Department of Surgery at the University of Michigan. Dr. Zhu received her PhD degree in Chemistry in 2009 under the direction of Professor Zijian Guo at the Nanjing University, China. She has worked with Professor David M. Lubman at the University of Michigan since 2010 initally as a postdoc. Her research has focused on the development of mass spectrometry-based methods to identify clinically relevant protein biomarkers from patient samples (tissue, serum) for early cancer detection. She has published 28 research articles in this field and has developed novel technologies and assays for biomarker discovery and validation phases for a variety of cancers, including pancreatic, liver, and ovarian cancers. She received the NCI Research Specialist Award in 2018. Her current NCI funded project aims at the discovery and validation of new glyco-markers based on the presence of characteristic glycans/glycopeptides in patient serum using mass spectrometry for early detection of hepatocellular carcinoma.

Elisa Warner is a PhD student in Computational Medicine and Bioinformatics at the University of Michigan, Ann Arbor, where she also earned her undergraduate degree and Master of Public Health. Her interests lie in the wide-ranging realm of disease detection, from wet lab to computational work. Her current work involves the development of computational models from EHR data to predict onset of specific diseases. Elisa is the recipient of numerous awards, including an NIH Training Grant in Bioinformatics. She has also been awarded the Hunein F. Massaab Award for Excellence in Molecular Epidemiology, and been a recipient of the DAAD scholarship from the German government to work at Forschungszentrum Jülich. She hopes her work will affect change for individuals with early signs of disease. Elisa has formerly trained under Dr. David M. Lubman, where she realized her passions in disease detection and academia. Although she works in the lab most days, she also enjoys playing violin and eating cookies.

Neehar Parikh, MD, MS, is an Assistant Professor in the Division of Gastroenterology and Hepatology at the University of Michigan. Dr. Parikh graduated from the University of Maryland with a degree in Microbiology. He obtained his medical degree from the University of Virginia and completed his internal medicine residency and fellowship training in gastroenterology and transplant hepatology at Northwestern University. He is currently medical director of the Liver Tumor program and the Living Donor Liver Transplantation program at the University of Michigan. Dr. Parikh’s main clinical interests include hepatocellular carcinoma treatment and chronic liver disease management and liver transplantation. Dr. Parikh has several NIH grants aimed at the early diagnosis of and personalized screening for hepatocellular carcinoma.

David M. Lubman, PhD, is currently the Maude T. Lane Professor of Surgery at the University of Michigan Medical Center and also a professor of Pathology. He is also an associate member of the Comprehensive Cancer Center and an associate member of the BioInformatics Program. He received his AB from Cornell University in 1975 and his PhD in 1979 under the direction of Professor Richard N. Zare at Stanford University in Physical Chemistry. He was a fellow at the Weizmann Institute in 1981 in the Department of Chemical Physics. He has been at the University of Michigan since 1983 initially in the Department of Chemistry where he developed a program in the areas of laser chemistry and mass spectrometry. In the last 15 years, he has undergone a career switch and joined the Department of Surgery to develop a program in new technologies aimed at discovering markers of early cancer and prognosis of disease. Much of this work is focused in on glycoproteomic changes in proteins during cancer progression and newer work on mapping pathways in disease using a mass spec-based approach. Other recent work has focused on the development of liquid biopsies via the use of exosomes and single amino acid variants as markers of cancer. The group has filed numerous patents resulting in the development of new companies and products involved in liquid mapping of proteins, protein microarrays, and new mass spec-based technology and hardware. His lab has published over 300 articles and graduated 57 PhDs with nearly 40 postdoctorals and visiting scientists in his laboratory many of whom have gone on to hold leadership positions in the pharmaceutical and biotech industries. Dr. Lubman is a fellow of the American Association for the Advancement of Science, an associate member of the Early Detection Research Network of the NCI and a member of the American Association of Cancer Research and American Chemical Society.
REFERENCES
- Abu El Makarem MA, Abdel-Aleem A, Ali A, Saber R, Shatat M, Rahem DA, Sayed D. Diagnostic significance of plasma osteopontin in hepatitis C virus-related hepatocellular carcinoma. Ann Hepatol 2011;10:296–305. [PubMed] [Google Scholar]
- Ahn YH, Lee JY, Lee JY, Kim YS, Ko JH, Yoo JS. Quantitative analysis of an aberrant glycoform of TIMP1 from colon cancer serum by L-PHA-enrichment and SISCAPA with MRM mass spectrometry. J Proteome Res 2009;8:4216–4224. [DOI] [PubMed] [Google Scholar]
- Ahn YH, Shin PM, Kim YS, Oh NR, Ji ES, Kim KH, Lee YJ, Kim SH, Yoo JS. Quantitative analysis of aberrant protein glycosylation in liver cancer plasma by AAL-enrichment and MRM mass spectrometry. Analyst 2013;138:6454–6462. [DOI] [PubMed] [Google Scholar]
- Ahn YH, Shin PM, Oh NR, Park GW, Kim H, Yoo JS. A lectin-coupled, targeted proteomic mass spectrometry (MRM MS) platform for identification of multiple liver cancer biomarkers in human plasma. J Proteomics 2012;75:5507–5515. [DOI] [PubMed] [Google Scholar]
- Alley WR Jr., Mechref Y, Novotny MV. Characterization of glycopeptides by combining collision-induced dissociation and electron-transfer dissociation mass spectrometry data. Rapid Commun Mass Spectrom 2009;23:161–170. [DOI] [PubMed] [Google Scholar]
- An HJ, Froehlich JW, Lebrilla CB. Determination of glycosylation sites and site-specific heterogeneity in glycoproteins. Curr Opin Chem Biol 2009;13:421–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apweiler R, Hermjakob H, Sharon N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim Biophys Acta 1999;1473:4–8. [DOI] [PubMed] [Google Scholar]
- Asazawa H, Kamada Y, Takeda Y, Takamatsu S, Shinzaki S, Kim Y, Nezu R, Kuzushita N, Mita E, Kato M, Miyoshi E. Serum fucosylated haptoglobin in chronic liver diseases as a potential biomarker of hepatocellular carcinoma development. Clin Chem Lab Med 2015; 53:95–102. [DOI] [PubMed] [Google Scholar]
- Benicky J, Sanda M, Pompach P, Wu J, Goldman R. Quantification of fucosylated hemopexin and complement factor H in plasma of patients with liver disease. Anal Chem 2014;86:10716–10723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bern M, Kil YJ, Becker C. Byonic: advanced peptide and protein identification software. Curr Protoc Bioinformatics Chapter 13 Unit 13.20 2012;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block TM, Comunale MA, Lowman M, Steel LF, Romano PR, Fimmel C, Tennant BC, London WT, Evans AA, Blumberg BS, Dwek RA, Mattu TS, Mehta AS. Use of targeted glycoproteomics to identify serum glycoproteins that correlate with liver cancer in woodchucks and humans. Proc Natl Acad Sci U S A 2005;102:779–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruix J, Sherman M, American Association for the Study of Liver Diseases. Management of hepatocellular carcinoma: an update. Hepatology 2011;53:1020–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruix J, Sherman M, Practice Guidelines Committee, American Association for the Study of Liver Diseases. Management of hepatocellular carcinoma. Hepatology 2005;42: 1208–1236. [DOI] [PubMed] [Google Scholar]
- Chandler KB, Pompach P, Goldman R, Edwards N. Exploring site-specific N-glycosylation microheterogeneity of haptoglobin using glycopeptide CID tandem mass spectra and glycan database search. J Proteome Res 2013;12:3652–3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Shah P, Zhang H. Solid phase extraction of N-linked glycopeptides using hydrazide tip. Anal Chem 2013;85:10670–10674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Tan Y, Wang M, Wang F, Yao Z, Dong L, Ye M, Wang H, Zou H. Development of glycoprotein capture-based label-free method for the high-throughput screening of differential glycoproteins in hepatocellular carcinoma. Mol Cell Proteomics 2011;10:M110 006445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Glover MS, Li L. Recent advances in ion mobility-mass spectrometry for improved structural characterization of glycans and glycoconju-gates. Curr Opin Chem Biol 2017a;42:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Yu Q, Hao L, Liu F, Johnson J, Tian Z, Kao WJ, Xu W, Li L. Site-specific characterization and quantitation of N-glycopeptides in PKM2 knockout breast cancer cells using DiLeu isobaric tags enabled by electron-transfer/higher-energy collision dissociation (EThcD). Analyst 2018;143:2508–2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Zhong X, Tie C, Chen B, Zhang X, Li L. Development of a hydrophilic interaction liquid chromatography coupled with matrix-assisted laser desorption/ionization-mass spectrometric imaging platform for N-glycan relative quantitation using stable-isotope labeled hydrazide reagents. Anal Bioanal Chem 2017b;409: 4437–4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark D, Mao L. Cancer biomarker discovery: lectin-based strategies targeting glycoproteins. Dis Markers 2012;33:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comunale MA, Lowman M, Long RE, Krakover J, Philip R, Seeholzer S, Evans AA, Hann HW, Block TM, Mehta AS. Proteomic analysis of serum associated fucosylated glycoproteins in the development of primary hepatocellular carcinoma. J Proteome Res 2006;5:308–315. [DOI] [PubMed] [Google Scholar]
- Comunale MA, Rodemich-Betesh L, Hafner J, Wang M, Norton P, Di Bisceglie AM, Block T, Mehta A. Linkage specific fucosylation of alpha-1-antitrypsin in liver cirrhosis and cancer patients: implications for a biomarker of hepatocellular carcinoma. PLoS ONE 2010;5: e12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comunale MA, Wang M, Anbarasan N, Betesh L, Karabudak A, Moritz E, Devarajan K, Marrero J, Block TM, Mehta A. Total serum glycan analysis is superior to lectin-FLISA for the early detection of hepatocellular carcinoma. Proteomics Clin Appl 2013;7: 690–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comunale MA, Wang M, Hafner J, Krakover J, Rodemich L, Kopenhaver B, Long RE, Junaidi O, Bisceglie AM, Block TM, Mehta AS. Identification and development of fucosylated glyco-proteins as biomarkers of primary hepatocellular carcinoma. J Proteome Res 2009;8:595–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comunale MA, Wang M, Rodemich-Betesh L, Hafner J, Lamontagne A, Klein A, Marrero J, Di Bisceglie AM, Gish R, Block T, Mehta A. Novel changes in glycosylation of serum Apo-J in patients with hepatocellular carcinoma. Cancer Epidemiol Biomarkers Prev 2011;20: 1222–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Kang X, Dai Z, Huang C, Zhou H, Guo K, Li Y, Zhang Y, Sun R, Chen J, Li Y, Tang Z, Uemura T, Liu Y. Prediction of chronic hepatitis B, liver cirrhosis and hepatocellular carcinoma by SELDI-based serum decision tree classification. J Cancer Res Clin Oncol 2007;133: 825–834. [DOI] [PubMed] [Google Scholar]
- Dai Z, Zhou J, Qiu SJ, Liu YK, Fan J. Lectin-based glycoproteomics to explore and analyze hepatocellular carcinoma-related glycoprotein markers. Electrophoresis 2009;30:2957–2966. [DOI] [PubMed] [Google Scholar]
- Debruyne EN, Vanderschaeghe D, Van Vlierberghe H, Vanhecke A, Callewaert N, Delanghe JR. Diagnostic value of the hemopexin N-glycan profile in hepatocellular carcinoma patients. Clin Chem 2010;56:823–831. [DOI] [PubMed] [Google Scholar]
- Dube DH, Bertozzi CR. Glycans in cancer and inflammation-potential for therapeutics and diagnostics. Nat Rev Drug Discov 2005;4:477–488. [DOI] [PubMed] [Google Scholar]
- Durazo FA, Blatt LM, Corey WG, Lin JH, Han S, Saab S, Busuttil RW, Tong MJ. Des-gamma-carboxyprothrombin, alpha-fetoprotein and AFP-L3 in patients with chronic hepatitis, cirrhosis and hepatocellular carcinoma. J Gastroenterol Hepatol 2008;23:1541–1548. [DOI] [PubMed] [Google Scholar]
- El-Din Bessa SS, Elwan NM, Suliman GA, El-Shourbagy SH. Clinical significance of plasma osteopontin level in Egyptian patients with hepatitis C virus-related hepatocellular carcinoma. Arch Med Res 2010;41:541–547. [DOI] [PubMed] [Google Scholar]
- El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology 2012;142:1264–1273 e1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores A, Marrero JA. Emerging trends in hepatocellular carcinoma: focus on diagnosis and therapeutics. Clin Med Insights Oncol 2014;8:71–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frese CK, Altelaar AF, Van Den Toorn H, Nolting D, Griep-Raming J, Heck AJ, Mohammed S. Toward full peptide sequence coverage by dual fragmentation combining electron-transfer and higher-energy collision dissociation tandem mass spectrometry. Anal Chem 2012;84: 9668–9673. [DOI] [PubMed] [Google Scholar]
- Fukuyama Y, Nakaya S, Yamazaki Y, Tanaka K. Ionic liquid matrixes optimized for MALDI-MS of sulfated/sialylated/neutral oligosaccharides and glycopeptides. Anal Chem 2008;80:2171–2179. [DOI] [PubMed] [Google Scholar]
- Fuster MM, Esko JD. The sweet and sour of cancer: glycans as novel therapeutic targets. Nat Rev Cancer 2005;5:526–542. [DOI] [PubMed] [Google Scholar]
- Gaye MM, Valentine SJ, Hu Y, Mirjankar N, Hammoud ZT, Mechref Y, Lavine BK, Clemmer DE. Ion mobility-mass spectrometry analysis of serum N-linked glycans from esophageal adenocarcinoma phenotypes. J Proteome Res 2012;11:6102–6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover MS, Yu Q, Chen Z, Shi X, Kent KC, Li L. Characterization of intact sialylated glycopeptides and phosphorylated glycopeptides from IMAC enriched samples by EThcD fragmentation: Toward combining phosphoproteomics and glycoproteomics. Int J Mass Spectrometry 2018;427:35–42. [Google Scholar]
- Goldman R, Ressom HW, Abdel-Hamid M, Goldman L, Wang A, Varghese RS, An Y, Loffredo CA, Drake SK, Eissa SA, Gouda I, Ezzat S, Moiseiwitsch FS. Candidate markers for the detection of hepatocellular carcinoma in low-molecular weight fraction of serum. Carcinogenesis 2007;28:2149–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman R, Ressom HW, Varghese RS, Goldman L, Bascug G, Loffredo CA, Abdel-Hamid M, Gouda I, Ezzat S, Kyselova Z, Mechref Y, Novotny MV. Detection of hepatocellular carcinoma using glycomic analysis. Clin Cancer Res 2009;15:1808–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley JA, Mcneil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 1982;143:29–36. [DOI] [PubMed] [Google Scholar]
- Hu JS, Wu DW, Liang S, Miao XY. GP73, a resident Golgi glycoprotein, is sensibility and specificity for hepatocellular carcinoma of diagnosis in a hepatitis B-endemic Asian population. Med Oncol 2010;27:339–345. [DOI] [PubMed] [Google Scholar]
- Hua S, An HJ, Ozcan S, Ro GS, Soares S, Devere-White R, Lebrilla CB. Comprehensive native glycan profiling with isomer separation and quantitation for the discovery of cancer biomarkers. Analyst 2011;136:3663–3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Zhou S, Zhu J, Lubman DM, Mechref Y. LC-MS/MS isomeric profiling of permethylated N-glycans derived from serum haptoglobin of hepatocellular carcinoma (HCC) and cirrhotic patients. Electrophoresis 2017;38:2160–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannou GN, Green P, Lowy E, Mun EJ, Berry K. Differences in hepatocellular carcinoma risk, predictors and trends over time according to etiology of cirrhosis. PLoS ONE 2018;13:e0204412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isailovic D, Kurulugama RT, Plasencia MD, Stokes ST, Kyselova Z, Goldman R, Mechref Y, Novotny MV, Clemmer DE. Profiling of human serum glycans associated with liver cancer and cirrhosis by IMS-MS. J Proteome Res 2008;7:1109–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isailovic D, Plasencia MD, Gaye MM, Stokes ST, Kurulugama RT, Pungpapong V, Zhang M, Kyselova Z, Goldman R, Mechref Y, Novotny MV, Clemmer DE. Delineating diseases by IMS-MS profiling of serum N-linked glycans. J Proteome Res 2012;11:576–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara T, Fukuda I, Morita A, Takinami Y, Okamoto H, Nishimura S, Numata Y. Development of quantitative plasma N-glycoproteomics using label-free 2-D LC-MALDI MS and its applicability for biomarker discovery in hepatocellular carcinoma. J Proteomics 2011; 74:2159–2168. [DOI] [PubMed] [Google Scholar]
- Kailemia MJ, Park D, Lebrilla CB. Glycans and glycoproteins as specific biomarkers for cancer. Anal Bioanal Chem 2017;409:395–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kailemia MJ, Ruhaak LR, Lebrilla CB, Amster IJ. Oligosaccharide analysis by mass spectrometry: a review of recent developments. Anal Chem 2014;86:196–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji H, Ocho M, Togayachi A, Kuno A, Sogabe M, Ohkura T, Nozaki H, Angata T, Chiba Y, Ozaki H, Hirabayashi J, Tanaka Y, Mizokami M, Ikehara Y, Narimatsu H. Glycoproteomic discovery of serological biomarker candidates for HCV/HBV infection-associated liver fibrosis and hepatocellular carcinoma. J Proteome Res 2013;12:2630–2640. [DOI] [PubMed] [Google Scholar]
- Kamiyama T, Yokoo H, Furukawa J, Kurogochi M, Togashi T, Miura N, Nakanishi K, Kamachi H, Kakisaka T, Tsuruga Y, Fujiyoshi M, Taketomi A, Nishimura S, Todo S. Identification of novel serum biomarkers of hepatocellular carcinoma using glycomic analysis. Hepatology 2013;57:2314–2325. [DOI] [PubMed] [Google Scholar]
- Kanmura S, Uto H, Sato Y, Kumagai K, Sasaki F, Moriuchi A, Oketani M, Ido A, Nagata K, Hayashi K, Stuver SO, Tsubouchi H. The complement component C3a fragment is a potential biomarker for hepatitis C virus-related hepatocellular carcinoma. J Gastroenterol 2010;45:459–467. [DOI] [PubMed] [Google Scholar]
- Kanwal F, Hoang T, Kramer JR, Asch SM, Goetz MB, Zeringue A, Richardson P, El-Serag HB. Increasing prevalence of HCC and cirrhosis in patients with chronic hepatitis C virus infection. Gastroenterology 2011;140:1182–1188 e1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanwal F, Kramer JR, Mapakshi S, Natarajan Y, Chayanupatkul M, Richardson PA, Li L, Desiderio R, Thrift AP, Asch SM, Chu J, El-Serag HB. Risk of hepatocellular cancer in patients with non-alcoholic fatty liver disease. Gastroenterology 2018. 10.1053/j.gastro.2018.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Ruhaak LR, Nguyen UT, Taylor SL, Dimapasoc L, Williams C, Stroble C, Ozcan S, Miyamoto S, Lebrilla CB, Leiserowitz GS. Evaluation of glycomic profiling as a diagnostic biomarker for epithelial ovarian cancer. Cancer Epidemiol Biomarkers Prev 2014; 23:611–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KH, Lee SY, Hwang H, Lee JY, Ji ES, An HJ, Kim JY, Yoo JS. Direct monitoring of fucosylated glycopeptides of alpha-fetoprotein in human serum for early hepatocellular carcinoma by liquid chromatography-tandem mass spectrometry with immunoprecipitation. Proteomics Clin Appl 2018;e1800062. [DOI] [PubMed] [Google Scholar]
- Kinoshita N, Ohno M, Nishiura T, Fujii S, Nishikawa A, Kawakami Y, Uozumi N, Taniguchi N. Glycosylation at the Fab portion of myeloma immunoglobulin G and increased fucosylated biantennary sugar chains: structural analysis by high-performance liquid chromatography and antibody-lectin enzyme immunoassay using Lens culinaris agglutinin. Cancer Res 1991;51:5888–5892. [PubMed] [Google Scholar]
- Kobayashi S, Nouso K, Kinugasa H, Takeuchi Y, Tomoda T, Miyahara K, Hagihara H, Kuwaki K, Onishi H, Nakamura S, Ikeda F, Miyake Y, Shiraha H, Takaki A, Yamamoto K. Clinical utility of serum fucosylated hemopexin in Japanese patients with hepatocellular carcinoma. Hepatol Res 2012;42:1187–1195. [DOI] [PubMed] [Google Scholar]
- Lee HJ, Cha HJ, Lim JS, Lee SH, Song SY, Kim H, Hancock WS, Yoo JS, Paik YK. Abundance-ratio-based semiquantitative analysis of site-specific N-linked glycopeptides present in the plasma of hepato-cellular carcinoma patients. J Proteome Res 2014;13:2328–2338. [DOI] [PubMed] [Google Scholar]
- Lee HJ, Na K, Choi EY, Kim KS, Kim H, Paik YK. Simple method for quantitative analysis of N-linked glycoproteins in hepatocellular carcinoma specimens. J Proteome Res 2010;9:308–318. [DOI] [PubMed] [Google Scholar]
- Lee J, Hua S, Lee SH, Oh MJ, Yun J, Kim JY, Kim JH, Kim JH, An HJ. Designation of fingerprint glycopeptides for targeted glycoproteomic analysis of serum haptoglobin: insights into gastric cancer biomarker discovery. Anal Bioanal Chem 2018;410:1617–1629. [DOI] [PubMed] [Google Scholar]
- Lee JY, Lee HK, Park GW, Hwang H, Jeong HK, Yun KN, Ji ES, Kim KH, Kim JS, Kim JW, Yun SH, Choi CW, Kim SI, Lim JS, Jeong SK, Paik YK, Lee SY, Park J, Kim SY, Choi YJ, Kim YI, Seo J, Cho JY, Oh MJ, Seo N, An HJ, Kim JY, Yoo JS. Characterization of site-specific N-glycopeptide isoforms of alpha-1-acid glycoprotein from an interlaboratory study using LC-MS/MS. J Proteome Res 2016;15:4146–4164. [DOI] [PubMed] [Google Scholar]
- Leerapun A, Suravarapu SV, Bida JP, Clark RJ, Sanders EL, Mettler TA, Stadheim LM, Aderca I, Moser CD, Nagorney DM, Larusso NF, De Groen PC, Menon KV, Lazaridis KN, Gores GJ, Charlton MR, Roberts RO, Therneau TM, Katzmann JA, Roberts LR. The utility of Lens culinaris agglutinin-reactive alpha-fetoprotein in the diagnosis of hepatocellular carcinoma: evaluation in a United States referral population. Clin Gastroenterol Hepatol 2007;5:394–402; quiz 267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiserowitz GS, Lebrilla C, Miyamoto S, An HJ, Duong H, Kirmiz C, Li B, Liu H, Lam KS. Glycomics analysis of serum: A potential new biomarker for ovarian cancer? Int J Gynecol Cancer 2008;18:470–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Mallory T, Satomura S. AFP-L3: A new generation of tumor marker for hepatocellular carcinoma. Clin Chim Acta 2001;313:15–19. [DOI] [PubMed] [Google Scholar]
- Liang J, Zhu J, Wang M, Zhang J, Parikh N, Liu S, Lubman DM. Evaluation of AGP fucosylation as a marker for hepatocellular carcinoma. (manuscript in preparation). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Simeone DM, Anderson MA, Brand RE, Xie X, Shedden KA, Ruffin MT, Lubman DM. Mass spectrometric assay for analysis of haptoglobin fucosylation in pancreatic cancer. J Proteome Res 2011;10: 2602–2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MQ, Zeng WF, Fang P, Cao WQ, Liu C, Yan GQ, Zhang Y, Peng C, Wu JQ, Zhang XJ, Tu HJ, Chi H, Sun RX, Cao Y, Dong MQ, Jiang BY, Huang JM, Shen HL, Wong CCL, He SM, Yang PY. PGlyco 2.0 enables precision N-glycoproteomics with comprehensive quality control and one-step mass spectrometry for intact glycopeptide identification. Nat Commun 2017a;8:438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Shang S, Li W, Qin X, Sun L, Zhang S, Liu Y. Assessment of hepatocellular carcinoma metastasis glycobiomarkers using advanced quantitative N-glycoproteome analysis. Front Physiol 2017b;8:472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XE, Desmyter L, Gao CF, Laroy W, Dewaele S, Vanhooren V, Wang L, Zhuang H, Callewaert N, Libert C, Contreras R, Chen C. N-glycomic changes in hepatocellular carcinoma patients with liver cirrhosis induced by hepatitis B virus. Hepatology 2007;46:1426–1435. [DOI] [PubMed] [Google Scholar]
- Liu Y, He J, Li C, Benitez R, Fu S, Marrero J, Lubman DM. Identification and confirmation of biomarkers using an integrated platform for quantitative analysis of glycoproteins and their glycosylations. J Proteome Res 2010;9:798–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, He J, Lubman DM. Characterization of membrane-associated glycoproteins using lectin affinity chromatography and mass spectrometry. Methods Mol Biol 2013;951:69–77. [DOI] [PubMed] [Google Scholar]
- Ma J, Sanda M, Wei R, Zhang L, Goldman R. Quantitative analysis of core fucosylation of serum proteins in liver diseases by LC-MS-MRM. J Proteomics 2018;DOI: 10.1016/j.jprot.2018.1002.1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Yang H, Xu H, Lu X, Sang X, Du S, Zhao H, Chen W, Xu Y, Chi T, Yang Z, Cai J, Li H, Chen J, Zhong S, Mohanti SR, Lopez-Soler R, Millis JM, Huang J, Zhang H. Golgi protein 73 (GOLPH2) is a valuable serum marker for hepatocellular carcinoma. Gut 2010;59:1687–1693. [DOI] [PubMed] [Google Scholar]
- Marrero JA, Feng Z, Wang Y, Nguyen MH, Befeler AS, Roberts LR, Reddy KR, Harnois D, Llovet JM, Normolle D, Dalhgren J, Chia D, Lok AS, Wagner PD, Srivastava S, Schwartz M. Alpha-fetoprotein, des-gamma carboxyprothrombin, and lectin-bound alpha-fetoprotein in early hepatocellular carcinoma. Gastroenterology 2009;137:110–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrero JA, Henley KS. The role of serum biomarkers in hepatocellular carcinoma surveillance. Gastroenterol Hepatol (N Y) 2011;7:821–823. [PMC free article] [PubMed] [Google Scholar]
- Marrero JA, Romano PR, Nikolaeva O, Steel L, Mehta A, Fimmel CJ, Comunale MA, D’amelio A, Lok AS, Block TM. GP73, a resident Golgi glycoprotein, is a novel serum marker for hepatocellular carcinoma. J Hepatol 2005;43:1007–1012. [DOI] [PubMed] [Google Scholar]
- Meany DL, Chan DW. Aberrant glycosylation associated with enzymes as cancer biomarkers. Clin Proteomics 2011;8:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechref Y. Use of CID/ETD mass spectrometry to analyze glycopeptides. Curr Protoc Protein Sci Chapter 12 Unit 12.11 2012;11–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta A, Block TM. Fucosylated glycoproteins as markers of liver disease. Dis Markers 2008;25:259–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta A, Herrera H, Block T. Glycosylation and liver cancer. Adv Cancer Res 2015;126:257–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael SM, Chien M, Lubman DM. An ion trap storage time-of-flight mass-spectrometer. Rev Sci Instrum 1992;63:4277–4284. [Google Scholar]
- Miyoshi E, Moriwaki K, Nakagawa T. Biological function of fucosylation in cancer biology. J Biochem 2008;143:725–729. [DOI] [PubMed] [Google Scholar]
- Miyoshi E, Moriwaki K, Terao N, Tan CC, Terao M, Nakagawa T, Matsumoto H, Shinzaki S, Kamada Y. Fucosylation is a promising target for cancer diagnosis and therapy. Biomolecules 2012;2:34–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morota K, Nakagawa M, Sekiya R, Hemken PM, Sokoll LJ, Elliott D, Chan DW, Dowell BL. A comparative evaluation of Golgi protein-73, fucosylated hemopexin, alpha-fetoprotein, and PIVKA-II in the serum of patients with chronic hepatitis, cirrhosis, and hepatocellular carcinoma. Clin Chem Lab Med 2011;49:711–718. [DOI] [PubMed] [Google Scholar]
- Munkley J, Elliott DJ. Hallmarks of glycosylation in cancer. Oncotarget 2016;7:35478–35489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na K, Jeong SK, Lee MJ, Cho SY, Kim SA, Lee MJ, Song SY, Kim H, Kim KS, Lee HW, Paik YK. Human liver carboxylesterase 1 outperforms alpha-fetoprotein as biomarker to discriminate hepatocellular carcinoma from other liver diseases in Korean patients. Int J Cancer 2013; 133:408–415. [DOI] [PubMed] [Google Scholar]
- Na K, Lee EY, Lee HJ, Kim KY, Lee H, Jeong SK, Jeong AS, Cho SY, Kim SA, Song SY, Kim KS, Cho SW, Kim H, Paik YK. Human plasma carboxylesterase 1, a novel serologic biomarker candidate for hepato-cellular carcinoma. Proteomics 2009;9:3989–3999. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Miyoshi E, Yakushijin T, Hiramatsu N, Igura T, Hayashi N, Taniguchi N, Kondo A. Glycomic analysis of alpha-fetoprotein L3 in hepatoma cell lines and hepatocellular carcinoma patients. J Proteome Res 2008;7:2222–2233. [DOI] [PubMed] [Google Scholar]
- Nishikaze T. Sensitive and structure-informative N-glycosylation analysis by MALDI-MS; ionization, fragmentation, and derivatization. Mass Spectrom (Tokyo) 2017;6:A0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda K, Ido A, Tamai T, Matsushita M, Kumagai K, Mawatari S, Saishoji A, Kure T, Ohno K, Toyokura E, Imanaka D, Moriuchi A, Uto H, Oketani M, Hashiguchi T, Tsubouchi H. Highly sensitive lens culinaris agglutinin-reactive alpha-fetoprotein is useful for early detection of hepatocellular carcinoma in patients with chronic liver disease. Oncol Rep 2011;26:1227–1233. [DOI] [PubMed] [Google Scholar]
- Patwa T, Li C, Simeone DM, Lubman DM. Glycoprotein analysis using protein microarrays and mass spectrometry. Mass Spectrom Rev 2010;29:830–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perz JF, Armstrong GL, Farrington LA, Hutin YJ, Bell BP. The contributions of hepatitis B virus and hepatitis C virus infections to cirrhosis and primary liver cancer worldwide. J Hepatol 2006;45:529–538. [DOI] [PubMed] [Google Scholar]
- Pinho SS, Reis CA. Glycosylation in cancer: mechanisms and clinical implications. Nat Rev Cancer 2015;15:540–555. [DOI] [PubMed] [Google Scholar]
- Pompach P, Ashline DJ, Brnakova Z, Benicky J, Sanda M, Goldman R. Protein and site specificity of fucosylation in liver-secreted glycoproteins. J Proteome Res 2014;13:5561–5569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pompach P, Brnakova Z, Sanda M, Wu J, Edwards N, Goldman R. Site-specific glycoforms of haptoglobin in liver cirrhosis and hepatocellular carcinoma. Mol Cell Proteomics 2013;12:1281–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers TW, Holst S, Wuhrer M, Mehta AS, Drake RR. Two-dimensional N-glycan distribution mapping of hepatocellular carcinoma tissues by MALDI-Imaging mass spectrometry. Biomolecules 2015;5: 2554–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ressom HW, Varghese RS, Goldman L, An Y, Loffredo CA, Abdel-Hamid M, Kyselova Z, Mechref Y, Novotny M, Drake SK, Goldman R. Analysis of MALDI-TOF mass spectrometry data for discovery of peptide and glycan biomarkers of hepatocellular carcinoma. J Proteome Res 2008;7:603–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riener MO, Stenner F, Liewen H, Soll C, Breitenstein S, Pestalozzi BC, Samaras P, Probst-Hensch N, Hellerbrand C, Mullhaupt B, Clavien PA, Bahra M, Neuhaus P, Wild P, Fritzsche F, Moch H, Jochum W, Kristiansen G. Golgi phosphoprotein 2 (GOLPH2) expression in liver tumors and its value as a serum marker in hepatocellular carcinomas. Hepatology 2009;49:1602–1609. [DOI] [PubMed] [Google Scholar]
- Ruhaak LR, Lebrilla CB. Applications of multiple reaction monitoring to clinical glycomics. Chromatographia 2015;78:335–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y, Oba N, Nishinakagawa S, Mizuguchi Y, Kojima T, Nomura K, Nakatsura T. Identification of beta2-microgloblin as a candidate for early diagnosis of imaging-invisible hepatocellular carcinoma in patient with liver cirrhosis. Oncol Rep 2010;23: 1325–1330. [DOI] [PubMed] [Google Scholar]
- Sanda M, Pompach P, Brnakova Z, Wu J, Makambi K, Goldman R. Quantitative liquid chromatography-mass spectrometry-multiple reaction monitoring (LC-MS-MRM) analysis of site-specific glyco-forms of haptoglobin in liver disease. Mol Cell Proteomics 2013;12:1294–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segu ZM, Mechref Y. Characterizing protein glycosylation sites through higher-energy C-trap dissociation. Rapid Commun Mass Spectrom 2010;24:1217–1225. [DOI] [PubMed] [Google Scholar]
- Selman MH, Hemayatkar M, Deelder AM, Wuhrer M. Cotton HILIC SPE microtips for microscale purification and enrichment of glycans and glycopeptides. Anal Chem 2011;83:2492–2499. [DOI] [PubMed] [Google Scholar]
- Shang S, Li W, Qin X, Zhang S, Liu Y. Aided diagnosis of hepatocellular carcinoma using serum fucosylated haptoglobin ratios. J Cancer 2017;8:887–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang S, Plymoth A, Ge S, Feng Z, Rosen HR, Sangrajrang S, Hainaut P, Marrero JA, Beretta L. Identification of osteopontin as a novel marker for early hepatocellular carcinoma. Hepatology 2012;55: 483–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singal AG, Conjeevaram HS, Volk ML, Fu S, Fontana RJ, Askari F, Su GL, Lok AS, Marrero JA. Effectiveness of hepatocellular carcinoma surveillance in patients with cirrhosis. Cancer Epidemiol Biomarkers Prev 2012;21:793–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn A, Kim H, Yu SJ, Yoon JH, Kim Y. A quantitative analytical method for PIVKA-II using multiple reaction monitoring-mass spectrometry for early diagnosis of hepatocellular carcinoma. Anal Bioanal Chem 2017;409:2829–2838. [DOI] [PubMed] [Google Scholar]
- Song E, Pyreddy S, Mechref Y. Quantification of glycopeptides by multiple reaction monitoring liquid chromatography/tandem mass spectrometry. Rapid Commun Mass Spectrom 2012;26:1941–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowell SR, Ju T, Cummings RD. Protein glycosylation in cancer. Annu Rev Pathol 2015;10:473–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Poon RT, Lee NP, Yeung C, Chan KL, Ng IO, Day PJ, Luk JM. Proteomics of hepatocellular carcinoma: serum vimentin as a surrogate marker for small tumors (<or¼2cm). J Proteome Res 2010a;9: 1923–1930. [DOI] [PubMed] [Google Scholar]
- Sun Y, Zang Z, Xu X, Zhang Z, Zhong L, Zan W, Zhao Y, Sun L. Differential proteomics identification of HSP90 as potential serum biomarker in hepatocellular carcinoma by two-dimensional electrophoresis and mass spectrometry. Int J Mol Sci 2010b;11:1423–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe K, Deguchi A, Higashi M, Usuki H, Suzuki Y, Uchimura Y, Kuriyama S, Ikenaka K. Outer arm fucosylation of N-glycans increases in sera of hepatocellular carcinoma patients. Biochem Biophys Res Commun 2008;374:219–225. [DOI] [PubMed] [Google Scholar]
- Tanabe K, Kitagawa K, Kojima N, Iijima S. Multifucosylated alpha-1-acid glycoprotein as a novel marker for hepatocellular carcinoma. J Proteome Res 2016;15:2935–2944. [DOI] [PubMed] [Google Scholar]
- Tang Z, Varghese RS, Bekesova S, Loffredo CA, Hamid MA, Kyselova Z, Mechref Y, Novotny MV, Goldman R, Ressom HW. Identification of N-glycan serum markers associated with hepatocellular carcinoma from mass spectrometry data. J Proteome Res 2010;9: 104–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tie C, Zhang XX. A new labelling reagent for glycans analysis by capillary electrophoresis-mass spectrometry. Anal Methods 2012;4: 357–359. [Google Scholar]
- Trevisani F, D’intino PE, Morselli-Labate AM, Mazzella G, Accogli E, Caraceni P, Domenicali M, De Notariis S, Roda E, Bernardi M. Serum alpha-fetoprotein for diagnosis of hepatocellular carcinoma in patients with chronic liver disease: influence of HBsAg and anti-HCV status. J Hepatol 2001;34:570–575. [DOI] [PubMed] [Google Scholar]
- Tsai TH, Wang M, Di Poto C, Hu Y, Zhou S, Zhao Y, Varghese RS, Luo Y, Tadesse MG, Ziada DH, Desai CS, Shetty K, Mechref Y, Ressom HW. LC-MS profiling of N-Glycans derived from human serum samples for biomarker discovery in hepatocellular carcinoma. J Proteome Res 2014;13:4859–4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadi C, Mittermayr S, Szekrenyes A, Kadas J, Takacs L, Kurucz I, Guttman A. Analysis of haptoglobin N-glycome alterations in inflammatory and malignant lung diseases by capillary electrophoresis. Electrophoresis 2013;34:2287–2294. [DOI] [PubMed] [Google Scholar]
- Wang M, Long RE, Comunale MA, Junaidi O, Marrero J, Di Bisceglie AM, Block TM, Mehta AS. Novel fucosylated biomarkers for the early detection of hepatocellular carcinoma. Cancer Epidemiol Biomarkers Prev 2009;18:1914–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Sanda M, Comunale MA, Herrera H, Swindell C, Kono Y, Singal AG, Marrero J, Block T, Goldman R, Mehta A. Changes in the glycosylation of kininogen and the Development of a Kininogen-Based Algorithm for the Early Detection of HCC. Cancer Epidemiol Biomarkers Prev 2017;26:795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Zhu J, Liang J, Gao C, Lubman DM. N-glycomic changes in serum immunoglobulins between hepatocellular carcinoma and liver cirrhosis. (manuscript in preparation). [Google Scholar]
- Wang Y, Ao X, Vuong H, Konanur M, Miller FR, Goodison S, Lubman DM. Membrane glycoproteins associated with breast tumor cell progression identified by a lectin affinity approach. J Proteome Res 2008;7:4313–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West CA, Wang M, Herrera H, Liang H, Black A, Angel PM, Drake RR, Mehta AS. N-linked glycan branching and fucosylation are increased directly in HCC tissue As determined through in situ glycan imaging. J Proteome Res 2018;17:3454–3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White DL, Thrift AP, Kanwal F, Davila J, El-Serag HB. Incidence of hepatocellular carcinoma in all 50 United States, from 2000 through 2012. Gastroenterology 2017;152:812–820 e815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Li J, Liu Y, Zhang C, Meng X, Zhou Z. Comparative proteomic studies of serum from patients with hepatocellular carcinoma. J Invest Surg 2012;25:37–42. [DOI] [PubMed] [Google Scholar]
- Wu Y, Mechref Y, Klouckova I, Mayampurath A, Novotny MV, Tang H. Mapping site-specific protein N-glycosylations through liquid chromatography/mass spectrometry and targeted tandem mass spectrometry. Rapid Commun Mass Spectrom 2010;24:965–972. [DOI] [PubMed] [Google Scholar]
- Xu Z, Zhou X, Lu H, Wu N, Zhao H, Zhang L, Zhang W, Liang YL, Wang L, Liu Y, Yang P, Zha X. Comparative glycoproteomics based on lectins affinity capture of N-linked glycoproteins from human Chang liver cells and MHCC97-H cells. Proteomics 2007; 7:2358–2370. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Imamura H, Matsuyama Y, Kume Y, Ikeda H, Norman GL, Shums Z, Aoki T, Hasegawa K, Beck Y, Sugawara Y, Kokudo N. AFP, AFP-L3, DCP, and GP73 as markers for monitoring treatment response and recurrence and as surrogate markers of clinicopathological variables of HCC. J Gastroenterol 2010;45: 1272–1282. [DOI] [PubMed] [Google Scholar]
- Yamasaki K, Tateyama M, Abiru S, Komori A, Nagaoka S, Saeki A, Hashimoto S, Sasaki R, Bekki S, Kugiyama Y, Miyazoe Y, Kuno A, Korenaga M, Togayachi A, Ocho M, Mizokami M, Narimatsu H, Yatsuhashi H. Elevated serum levels of Wisteria floribunda agglutinin-positive human Mac-2 binding protein predict the development of hepatocellular carcinoma in hepatitis C patients. Hepatology 2014;60:1563–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Cui T, Wang Y, Sun S, Ma T, Wang T, Chen Q, Li Z. Selective isolation and analysis of glycoprotein fractions and their glycomes from hepatocellular carcinoma sera. Proteomics 2013;13: 1481–1498. [DOI] [PubMed] [Google Scholar]
- Yang MH, Tyan YC, Jong SB, Huang YF, Liao PC, Wang MC. Identification of human hepatocellular carcinoma-related proteins by proteomic approaches. Anal Bioanal Chem 2007;388:637–643. [DOI] [PubMed] [Google Scholar]
- Yang W, Shah P, Hu Y, Toghi Eshghi S, Sun S, Liu Y, Zhang H. Comparison of enrichment methods for intact N- and O-linked glycopeptides using strong anion exchange and hydrophilic interaction liquid chromatography. Anal Chem 2017;89:11193–11197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Harris LE, Palmer-Toy DE, Hancock WS. Multilectin affinity chromatography for characterization of multiple glycoprotein bio-marker candidates in serum from breast cancer patients. Clin Chem 2006;52:1897–1905. [DOI] [PubMed] [Google Scholar]
- Yin H, An M, So PK, Wong MY, Lubman DM, Yao Z. The analysis of alpha-1-antitrypsin glycosylation with direct LC-MS/MS. Electrophoresis 2018;39:2351–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Lin Z, Nie S, Wu J, Tan Z, Zhu J, Dai J, Feng Z, Marrero J, Lubman DM. Mass-selected site-specific core-fucosylation of ceruloplasmin in alcohol-related hepatocellular carcinoma. J Proteome Res 2014;13: 2887–2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Tan Z, Wu J, Zhu J, Shedden KA, Marrero J, Lubman DM. Mass-selected site-specific core-fucosylation of serum proteins in hepatocellular carcinoma. J Proteome Res 2015;14:4876–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JB, Li L. An impulse-driven liquid-droplet deposition interface for combining LC with MALDI MS and MS/MS. J Am Soc Mass Spectrom 2006;17:325–334. [DOI] [PubMed] [Google Scholar]
- Yu Q, Wang B, Chen Z, Urabe G, Glover MS, Shi X, Guo LW, Kent KC, Li L. Electron-transfer/higher-energy collision dissociation (EThcD)-enabled intact glycopeptide/glycoproteome characterization. J Am Soc Mass Spectrom 2017;28:1751–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Sanda M, Wu J, Koomen J, Goldman R. Quantitative analysis of immunoglobulin subclasses and subclass specific glycosylation by LCMS-MRM in liver disease. J Proteomics 2015;116:24–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zekri AR, Bahnassy AA, Alam El-Din HM, Morsy HM, Shaarawy S, Moharram NZ, Daoud SS. Serum levels of beta-catenin as a potential marker for genotype 4/hepatitis C-associated hepatocellular carcinoma. Oncol Rep 2011;26:825–831. [DOI] [PubMed] [Google Scholar]
- Zhang H. Glycoproteomics using chemical immobilization. Curr Protoc Protein Sci Chapter 24 Unit 24.3 2007. [DOI] [PubMed] [Google Scholar]
- Zhang S, Jiang K, Zhang Q, Guo K, Liu Y. Serum fucosylated paraoxonase 1 as a potential glycobiomarker for clinical diagnosis of early hepatocellular carcinoma using ELISA Index. Glycoconj J 2015a;32:119–125. [DOI] [PubMed] [Google Scholar]
- Zhang S, Shu H, Luo K, Kang X, Zhang Y, Lu H, Liu Y. N-linked glycan changes of serum haptoglobin beta chain in liver disease patients. Mol Biosyst 2011;7:1621–1628. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhu J, Yin H, Marrero J, Zhang XX, Lubman DM. ESI-LC-MS method for haptoglobin fucosylation analysis in hepatocellular carcinoma and liver cirrhosis. J Proteome Res 2015b;14:5388–5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhang Y, Wang Y, Xu L, Xu W. Alpha-fetoprotein-L3 and Golgi protein 73 may serve as candidate biomarkers for diagnosing alpha-fetoprotein-negative hepatocellular carcinoma. Onco Targets Ther 2016;9:123–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Qiu W, Simeone DM, Lubman DM. N-linked glycosylation profiling of pancreatic cancer serum using capillary liquid phase separation coupled with mass spectrometric analysis. J Proteome Res 2007; 6:1126–1138. [DOI] [PubMed] [Google Scholar]
- Zhao J, Simeone DM, Heidt D, Anderson MA, Lubman DM. Comparative serum glycoproteomics using lectin selected sialic acid glycoproteins with mass spectrometric analysis: Application to pancreatic cancer serum. J Proteome Res 2006;5:1792–1802. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Jia W, Wang J, Ying W, Zhang Y, Qian X. Fragmentation and site-specific quantification of core fucosylated glycoprotein by multiple reaction monitoring-mass spectrometry. Anal Chem 2011; 83: 8802–8809. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Zhang L, Huo L, Pei L, Li Q, Li H, Jin L. Clinical significance of fucosylated GP73 in the differential diagnosis of hepatocellular carcinoma. Int J Biol Markers 2018. 10.1177/1724600818796646 [DOI] [PubMed] [Google Scholar]
- Zhong X, Chen Z, Snovida S, Liu Y, Rogers JC, Li L. Capillary electrophoresis-electrospray ionization-mass spectrometry for quantitative analysis of glycans labeled with multiplex carbonyl-reactive tandem mass tags. Anal Chem 2015;87: 6527–6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Chen Z, Zhang J, An M, Wu J, Yu Q, Skilton SJ, Bern M, Sen KI, Li L, Lubman DM. Differential quantitative determination of site-specific intact N-glycopeptides in serum haptoglobin between hepatocellular carcinoma and cirrhosis using LC-EThcD-MS/MS. J Proteome Res 2018. 10.1021/acs.jproteome.8b00654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Lin Z, Wu J, Yin H, Dai J, Feng Z, Marrero J, Lubman DM. Analysis of serum haptoglobin fucosylation in hepatocellular carcinoma and liver cirrhosis of different etiologies. J Proteome Res 2014;13: 2986–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Wu J, Yin H, Marrero J, Lubman DM. Mass spectrometric N-glycan analysis of haptoglobin from patient serum samples using a 96-well plate format. J Proteome Res 2015;14:4932–4939. [DOI] [PMC free article] [PubMed] [Google Scholar]